- 1Istituto di Biomembrane, Bioenergetica e Biotecnologie Molecolari, Consiglio Nazionale delle Ricerche, Bari, Italy
- 2Dipartimento di Scienze del Suolo, della Pianta e degli Alimenti, Università degli Studi di Bari “Aldo Moro”, Bari, Italy
- 3Dipartimento di Bioscienze, Biotecnologie e Biofarmaceutica, Università degli Studi di Bari “Aldo Moro”, Bari, Italy
- 4Food Safety Lab s.r.l., Corato, BA, Italy
Microbial stability of fresh pasta depends on heat treatment, storage temperature, proper preservatives, and atmosphere packaging. This study aimed at improving the microbial quality, safety, and shelf life of fresh pasta using modified atmosphere composition and packaging with or without the addition of bioprotective cultures (Lactobacillus acidophilus, Lactobacillus casei, Bifidobacterium spp., and Bacillus coagulans) into semolina. Three fresh pasta variants were made using (i) the traditional protocol (control), MAP (20:80 CO2:N2), and barrier packaging, (ii) the experimental MAP (40:60 CO2:N2) and barrier packaging, and (iii) the experimental MAP, barrier packaging, and bioprotective cultures. Their effects on physicochemical properties (i.e., content on macro elements, water activity, headspace O2, CO2 concentrations, and mycotoxins), microbiological patterns, protein, and volatile organic compounds (VOC) were investigated at the beginning and the end of the actual or extended shelf-life through traditional and multi-omics approaches. We showed that the gas composition and properties of the packaging material tested in the experimental MAP system, with or without bioprotective cultures, positively affect features of fresh pasta avoiding changes in their main chemical properties, allowing for a storage longer than 120 days under refrigerated conditions. These results support that, although bioprotective cultures were not all able to grow in tested conditions, they can control the spoilage and the associated food-borne microbiota in fresh pasta during storage by their antimicrobials and/or fermentation products synergically. The VOC profiling, based on gas-chromatography mass-spectrometry (GC-MS), highlighted significant differences affected by the different manufacturing and packaging of samples. Therefore, the use of the proposed MAP system and the addition of bioprotective cultures can be considered an industrial helpful strategy to reduce the quality loss during refrigerated storage and to increase the shelf life of fresh pasta for additional 30 days by allowing the economic and environmental benefits spurring innovation in existing production models.
Introduction
Pasta is a cereal-based food of the traditional Mediterranean diet, popular worldwide due to its convenience, palatability, and nutritional quality, ideal for easy and quick meals (Antognelli, 1980; Arendt and Zannini, 2013; Carini et al., 2014; Zardetto et al., 2021). According to Italian law, “fresh pasta” is defined as the product obtained by extrusion or lamination of a dough made of durum wheat semolina or alternative flours and water, having moisture content between 24% and 30%, water activity (aw) between 0.92 and 0.97, and stored at 4 (±2)°C (Dpr, 2001; Costa et al., 2010; Carini et al., 2014). In this form, fresh pasta has on average 2–3 days-long shelf life even though this depends on the microbial cell density found at the end of the production process (Tabanelli, 2020). The heat treated (equivalent to pasteurization) industrial fresh pasta, stored at an appropriate temperature, has a shelf life of 30–90 days (Pagani et al., 2007; Angiolillo et al., 2017). However, its shelf life after packaging depends on microbial survival rates to thermal treatments, overcoming the hurdles determined by thermal treatment, aw, and storage temperature (Sanguinetti et al., 2011, 2016). The high moisture content and aw, as well as the nutrient content, lead to microbial metabolic activities (Guerzoni et al., 1994; Zardetto, 2005a; Del Nobile et al., 2009a) determining fresh pasta as a foodstuff easily perishable and compromising both safety and sensorial characteristics of the end product (Del Nobile et al., 2009a; Costa et al., 2010; Oliveira et al., 2014). For this reason, based on nationality, different laws allowed for using chemical preservatives and bacteriostatic compounds, such as potassium sorbate or sodium benzoate, to maintain the microbial safety and quality of fresh or filled pasta (Fda, 2006; Li et al., 2011; Shahmohammadi et al., 2016; Reg. UE 1129/2011).
Nowadays, the broad diffusion of fresh pasta and, particularly, the increased consumers’ demand for reducing the use of synthetic preservatives have provided the interest to extend the product shelf life (90–120 days) taking advantage of “clean-label” methods (Del Nobile et al., 2009b; Angiolillo et al., 2017; Schettino et al., 2019). Innovative approaches are those based on modified atmosphere packaging (MAP) and biopreservatives, both aimed to reduce the growth of microorganisms surviving heat thermal treatments while maintaining the traditional organoleptic properties and assuring the hygienic quality of fresh pasta (Del Nobile et al., 2009a).
Modified atmosphere packaging is a well-established technique for preserving fresh pasta quality by taking advantage of gas ratios surrounding the product that is different from the air (Zardetto, 2005b; Del Nobile et al., 2009a). The ratios 70:30 and 60:40 N2:CO2 are the most commonly used MAP gas mixtures to preserve the microbial quality of fresh pasta (Zardetto et al., 2022). These ratios limit microbial overgrowth and toxin production reducing the physicochemical deterioration of packaged foods (Zardetto, 2005b; Chaix et al., 2015). The un-/success of MAP depends on the structure, thickness, area, and permeability of the used film for packaging (e.g., thermoformed trays or flexible films containing specific barrier properties), gradient concentrations, differences in pressure across the film, and temperature (Gholizadeh et al., 2007). Furthermore, recent advances in MAP technologies were based on the application of environmentally friendly film materials in fresh food preservation (Qu et al., 2022; Zardetto et al., 2022).
Biopreservation is a biotechnological strategy based on the use of bioprotective cultures (BCs) or their antimicrobials and fermentation products, such as bacteriocins and organic acids, aimed at preserving foods and extending the shelf life in terms of spoilage and pathogen control (Ananou et al., 2007; Oliveira et al., 2018). Lactic acid bacteria (LAB) have antagonistic properties, proven antimicrobial properties, and safe history that make them ideal biopreservative candidates (Ghanbari et al., 2013; Cifuentes Bachmann and Leroy, 2015). In previous works, LAB were added during pasta-making to have a final product with a lower glycemic index, low gluten (Calasso et al., 2018), or higher content of B2 vitamin (Capozzi et al., 2011) than controls. Furthermore, LAB strains were recently used to preserve both the microbial and sensory quality of fresh or filled pasta (Angiolillo et al., 2017; Tabanelli, 2020). Also, spore-forming microorganisms (e.g., Bacillus spp.) have been used in research studies to produce heat-treated probiotic pasta (Fares et al., 2015; Konuray and Erginkaya, 2020).
Based on these considerations, this work explored two different fields. First, different MAP conditions (gas concentration and packaging materials) have been tested. As a second-step evaluation, the experimental MAP was combined with a multi-strain probiotic mixture acting as BCs during pasta production. Therefore, both were compared against control pasta to assess differences in chemical, microbiological, and metabolomic parameters considering also temporal dynamics (storage) based on a multi-omics approach. Hence, the microbiota was profiled through culturomics and rDNA gene-target metagenetics, while chemical, proteomic, and metabolomic profiles of the fresh pasta samples have been also studied.
Materials and methods
Pasta making
Fresh pasta samples were produced in a semi-industrial factory (Altamura, Bari, Italy), in duplicate on three consecutive days, using commercial durum wheat semolina (Triticum turgidum L. var. durum; moisture 14.6 ± 1.3 g/100 g, ash 0.63 ± 0.06 g/100 g, proteins 13.8 ± 0.6g/100 g, dry gluten 11.2 g/100 g, all on dry weight). All samples were prepared in a semi-industrial plant equipped with a bronze die. The chosen shape of pasta was a short, thin twisted pasta type, named “trofie.”
Three different sets of pasta were manufactured (Figure 1 and Appendix A): (i) control fresh pasta made following the conventional protocol and MAP (1MA); (ii) fresh pasta obtained by conventional protocol and packaged in experimental MAP (2MA); and (iii) fresh pasta made with the addition of bioprotective cultures (BCs) and packaged in experimental MAP (2MA-BC).
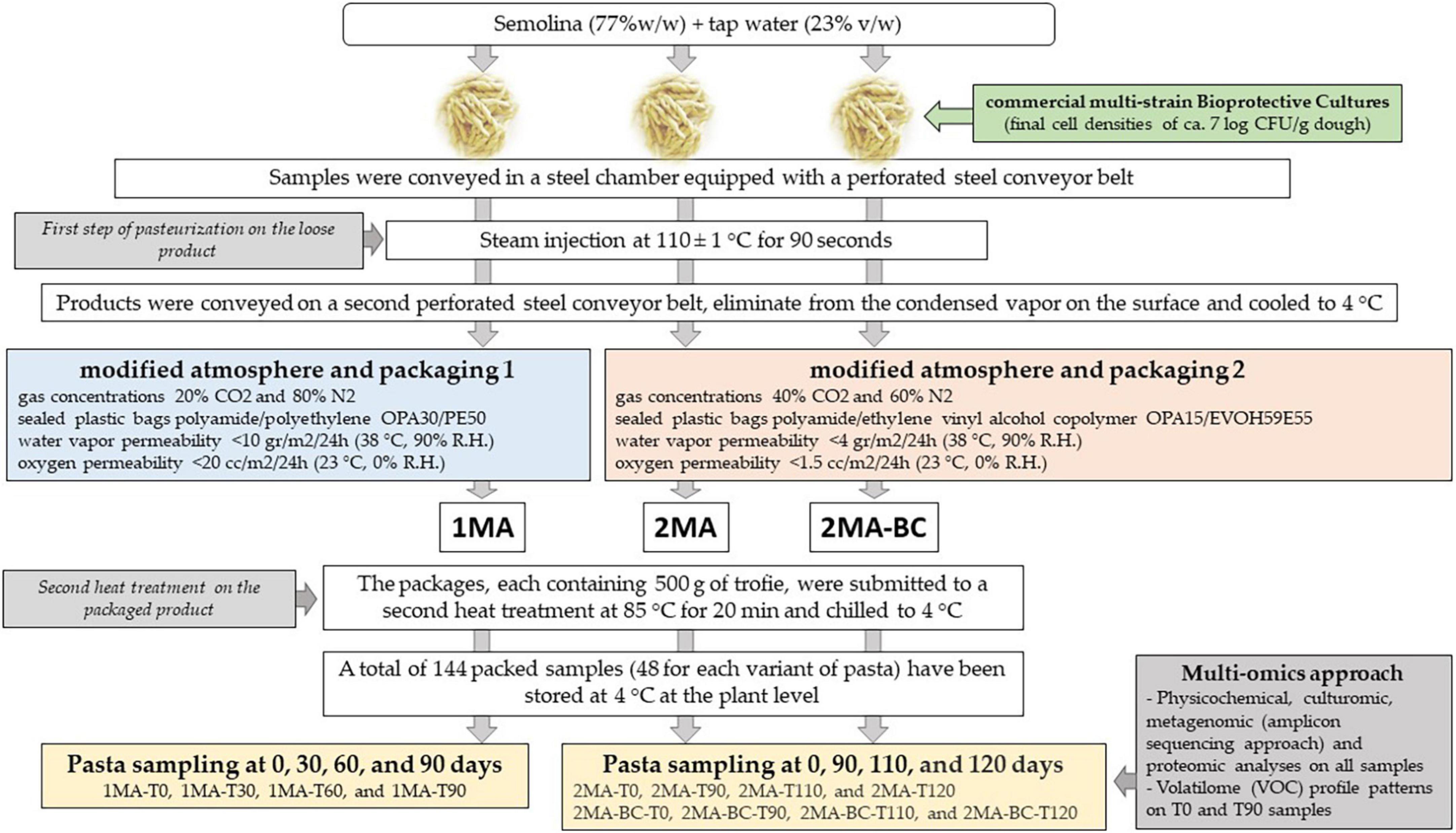
Figure 1. Schematic design showing the production of fresh pasta at the semi-industrial factory and the analyses performed.
In 2MA-BC, a commercial freeze-dried multi-strain probiotic mixture commercially available was used. The mixture contained Lactobacillus acidophilus, Bifidobacterium animalis, Lacticaseibacillus paracasei (basonym Lactobacillus paracasei), Lacticaseibacillus casei (basonym Lactobacillus casei), and Bacillus coagulans (Montefarmaco OTC SpA, Bollate, Milan, Italy). Freeze-dried BCs were added to semolina (final cell density for each strain ∼7 log CFU/g). BCs were prepared by dissolving lyophilized microorganisms in a liquid contained in the provided bottle immediately before using, as recommended by the manufacturer’s instructions, and added to semolina and water during pasta dough-making (final cell densities ∼7 log CFU/g dough).
For 1MA batches, the sampling was performed at 0, 30, 60, and 90 days (actual shelf-life) of storage at 4 ± 2°C. For both 2MA (2MA and 2MA-BC) batches, the sampling was performed at 0, 90, 110, and 120 days (expected shelf-life) of storage at 4 ± 2°C. All samples were shipped to the laboratory under refrigerated conditions (∼4°C) and immediately chemically and microbiologically analyzed. An aliquot of each sample was frozen (-80°C) until metagenomic and metabolomic analyses were performed.
Chemical characterization
Lipids, proteins (total nitrogen × 6.25) (Reg. CE 1169/2011), ash, and total dietary fibers were assessed according to AOAC methods 945.38F, 979.09, 923.03, and 991.43 (AOAC International, 1990, 2006), respectively. Moisture content was determined by an automatic moisture analyzer at 105°C (Mod. MAC 110/NP, Rodwang Wagi Elektroniczne, Poland). The carbohydrate content was determined by difference [100 − (moisture + proteins + lipids + ash)]. The energy was determined by multiplying the protein and carbohydrate contents by their calorific value (4 kcal/100 g), while the fat amount by its calorific value (9 kcal/100 g). Water activity in fresh pasta samples was determined according to ISO 18787:2017 method.
Headspace changes of O2 and CO2 concentrations were measured in packaged samples according to L-MI056 rev.0 Ed.2018 internal method. In detail, the LMI056 method was based on measurements with electrochemical cells of the packaging headspace in terms of gas (percentage) concentrations by a DANSENSOR instrument (Ametek MOCON, Inc., Brooklyn Park, MN, USA). Additionally, total aflatoxins (AFTs), deoxynivalenol (DON), ochratoxin A (OTA), and zearalenone (ZEA) were assessed according to the L-MI067 rev.0 2020 method, which was previously accredited by the Italian certification organization Accredia, based on measurements with SCIEX model 5500 + HPLC-MS/MS (AB Sciex LLC, Framingham, MA, USA) using an isotopically labeled internal standard.
Cultivable microbiota characterization
Microbiological analyses enumerated total aerobic mesophilic bacteria (UNI EN ISO 4833-1:2013 method), mesophilic lactic acid bacteria (LAB) (ISO 15214:1998), Bifidobacterium spp., spore-forming bacteria (SFB), presumptive B. coagulans, coliform bacteria (ISO 4832:2006 method), beta-glucuronidase-positive Escherichia coli (ISO 16649-2:2001 method), Enterobacteriaceae (ISO 21528-2:2017 method), Salmonella spp. (AFNOR BIO 12/32-10/11 method), coagulase-positive Staphylococcus (UNI EN ISO 6888-2:2004 method), Listeria monocytogenes (AFNOR BIO 12/27-02/10 method), Clostridium perfrigens (ISO 7937 method), yeasts, and molds (standard method ISO 21527-2:2008 method). The sample preparation was performed in accordance with UNI EN ISO 6887-1:2000 and UNI EN ISO 6887-4:2012. Aliquots (10 g) of semolina and fresh pasta were aseptically removed from each package and homogenized [90 ml of Buffered Peptone Water (BPW)] for total counts of aerobic mesophilic bacteria at 30°C, LAB, bifidobacteria, SFB and Bacillus, coliforms, β-glucuronidase-positive E. coli, coagulase-positive Staphylococcus, L. monocytogenes, sulfite reducing clostridia, yeasts, and molds. The determination of Enterobacteriaceae and Salmonella spp. was carried out in 25 g of sample homogenized in 225 mL BPW. For each sample, appropriate dilutions were performed. Bifidobacterium was enumerated according to Tharmaraj and Shah (2003), using MRS agar which is added 0.05% L-cysteine hydrochloride, 0.1 g/L neomycin sulfate, 0.15 g/L nalidixic acid, 3 g/L lithium chloride, and 0.2 g/L paromomycin sulfate, and then plates were anaerobically incubated (37°C for 72 h). To determine SFM densities, dilutions were heat-treated (90 °C for 10 min in a water bath) and spread-plated on nutrient agar plates, then incubated (at 37 °C for 48 h). For the enumeration of presumptive B. coagulans, dilutions were heat-treated (75 °C for 30 min), spread-plated on glucose yeast extract agar, and incubated (40 °C for 48–72 h) as described by Konuray and Erginkaya (2020). Except for bifidobacteria, SFB, and Bacillus, all used methods were validated and accredited by the Italian certification organization Accredia. All used culture media were previously controlled in accordance with the UNI EN ISO 11133:2014. For each of the following parameters, productivity, selectivity, and specificity scores were considered.
DNA extraction, sequencing, and bioinformatics analysis
Total genomic DNA was extracted from 1MA, 2MA, and 2MA-BC pasta samples at different time points. About 5 g of each pasta sample was homogenized with 45 ml of sterile saline solution (Minervini et al., 2010). Pellet was treated with FastPrep (BIO 101, C lsbad, Canada) and DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Illkrich, France), following the manufacturer’s instructions. Qualitative and quantitative analyses of the extracted DNA were performed using agarose gel (1%) electrophoresis and the Quant-iTTM PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA), respectively. The V5–V6 hypervariable regions of the 16S rDNA and the ITS1 (Internal Transcribed Spacer) region, within the ITS region of the gene locus for ribosomal RNA, were chosen as targets for prokaryotic and fungal characterization, respectively (Chakravorty et al., 2007; Bokulich and Mills, 2013). Amplicon libraries strategy agreed with Manzari et al. (2015). In the first PCR round, the overhang primer pairs, BV5 (Next For), and AV6 (Next Rev) (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG[ATTAG ATACCCYGGTAGTCC]-3′/5′-GTCTCGTGGGCTCGGAGAT GTGTATAAGAGACAG[ACGAGCTGACGACARCCATG]-3′) were used for the analysis of the V5–V6 regions (Manzari et al., 2015), while BITS (Next For) and B58S3 (Next Rev) (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG[ACCT GCGGARGGATCA]-3′/5′-GTCTCGTGGGCTCGGAGATGT GTATAAGAGACAG[GAGATCCRTTGYTRAAAGTT]-3′) were used for the ITS1 region (Bokulich and Mills, 2013). Finally, equimolar ratios of the purified amplicons were pooled and subjected to 2 × 250 bp paired-end sequencing on the Illumina MiSeq platform. To increase the genetic diversity, the phage PhiX genomic DNA library was added to the mix and co-sequenced (Kozich et al., 2013). The V5--V6 hypervariable regions and ITS1 raw sequencing data were initially quality checked by using FastQC1 and multiQC (Ewels et al., 2016). Illumina adapters and PCR primers were removed from raw reads by applying cutadapt (Martin, 2011). The 16S rRNA and ITS1 data were analyzed by using two different pipelines: QIIME2 (Bolyen et al., 2019) and BioMaS (Fosso et al., 2015). The first one relies on ASVs (Amplicon Sequence Variants) estimation and classification, and the latter one performs the taxonomic classification of Illumina data, especially for barcodes characterized by a wide variation in length (i.e., ITS) that may not properly be profiled by using ASV-based approaches. Trimmed V5-V6 data were analyzed by using the QIIME2 suite (version 2019.7). Fastq files were imported as QIIME2 artifacts by using the tools plugin and denoised into ASVs (Callahan et al., 2017) by applying DADA2 (Callahan et al., 2016). The obtained ASVs were taxonomically annotated by using the fit-classifier-sklearn (Pedregosa et al., 2011) plugin and the release 132 of the SILVA database (Pruesse et al., 2007) as the 16S rRNA reference collection and taxonomy. Trimmed ITS1 paired-end reads were merged into consensus sequences using PEAR (version 0.9.6) (Zhang et al., 2013) and then dereplicated through VSEARCH (version 2.15) (Rognes et al., 2016). The unmerged reads were trimmed of low-quality regions (Phred score cutoff of 25), and paired ends containing reads shorter than 50 nt were removed. Both the merged and the unmerged sequences were mapped against the ITSoneDB database (release 141.1) (Santamaria et al., 2012, 2018) by using Bowtie 2 (version 2.3.5.1). The mapping data were filtered according to query coverage (≥70%) and similarity percentage (≥97%) and taxonomically classified by applying TANGO (Alonso-Alemany et al., 2014; Fosso et al., 2018) on the NCBI taxonomy (Kim et al., 2013).
Protein characterization
Protein fractions (albumins and globulins, gliadins, and glutenins) were extracted from pasta following the method originally described by Osborne (1907) and further modified by Weiss et al. (1993) and Rizzello et al. (2007). All extracts were stored at -80°C until they were used. The protein concentration of the various fractions was determined by the Bradford method (Bradford, 1976).
Aliquots of ca. 15 μg of protein from extracted fractions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the Laemmli protocol (Laemmli, 1970).
Two-dimensional electrophoresis (2-DE) of ca. 30 μg of proteins was carried out with the Immobiline-polyacrylamide system as previously described (Bjellqvist et al., 1993). The second dimension was carried out in a Laemmli system (Laemmli, 1970). Gels were silver stained, and spot intensities were normalized (Bini et al., 1997). Three gels from each sampling time were analyzed.
Profiles of volatile organic compounds
To profile volatile organic compounds (VOC)by gas chromatography–mass spectrometry (GC-MS) analyses, 7.25 g of crushed fresh pasta was placed into a 20-ml vial with 5 μl of internal standard 4-methyl-2-pentanol (final concentration of 0.67 μg/g) (Cavallo et al., 2017). A PAL COMBI-xt autosampler (CTC combiPAL, CTC Analysis AG, Zwingen, Switzerland) was used to standardize the extraction procedure. To optimize the headspace solid phase microextraction (HS-SPME), a divinylbenzene/carboxen/polydimethylsiloxane (DVB/CARB/PDMS) (Supelco, Bellefonte, PA, USA) fiber was exposed to the sample headspace for 60 min at 75°C (Giannetti et al., 2021). Adsorbed molecules were desorbed in the injector with the analytical conditions reported by Montemurro et al. (2020), using a Clarus 680 (PerkinElmer, Beaconsfield, UK) gas chromatography equipped with an Rtx-WAX column (30 m × 0.25 mm i. d., 0.25 μm film thickness) (Restek Superchrom, Milano, Italy) and a single-quadrupole mass spectrometer Clarus SQ8MS (PerkinElmer) detector. Each chromatogram was analyzed for peak identification by comparing (i) the retention time (RT) of the detected compound with those of pure standard for HPLC (Sigma-Aldrich, St. Louis, MO, USA) and (ii) experimental mass spectra with those of the National Institute of Standards and Technology database (NIST/EPA/NIH Mass Spectral Library with Search Program, data version NIST 05, software version 2.0 d). Quantitative data for the identified compounds were obtained by the interpolation of the relative areas versus the internal standard area, expressed as μg/g of 2-methyl-4-pentanol.
Statistical analyses
Two-way ANOVA was applied on the means of data obtained from three biological replicates (analyzed in duplicate), using the statistical software Statistica 12.5 (TIBCO Software Inc., Palo Alto, CA, USA) for Windows. GraphPad Prism version 8.0.1 (GraphPad Software, San Diego, CA, USA) and Permut-MatrixEN software have been used to display bacterial and fungi abundances. Principal Components Analysis was performed through Xlstat 2014 (Addinsoft, New York, NY, USA).
Results
Chemical characterization
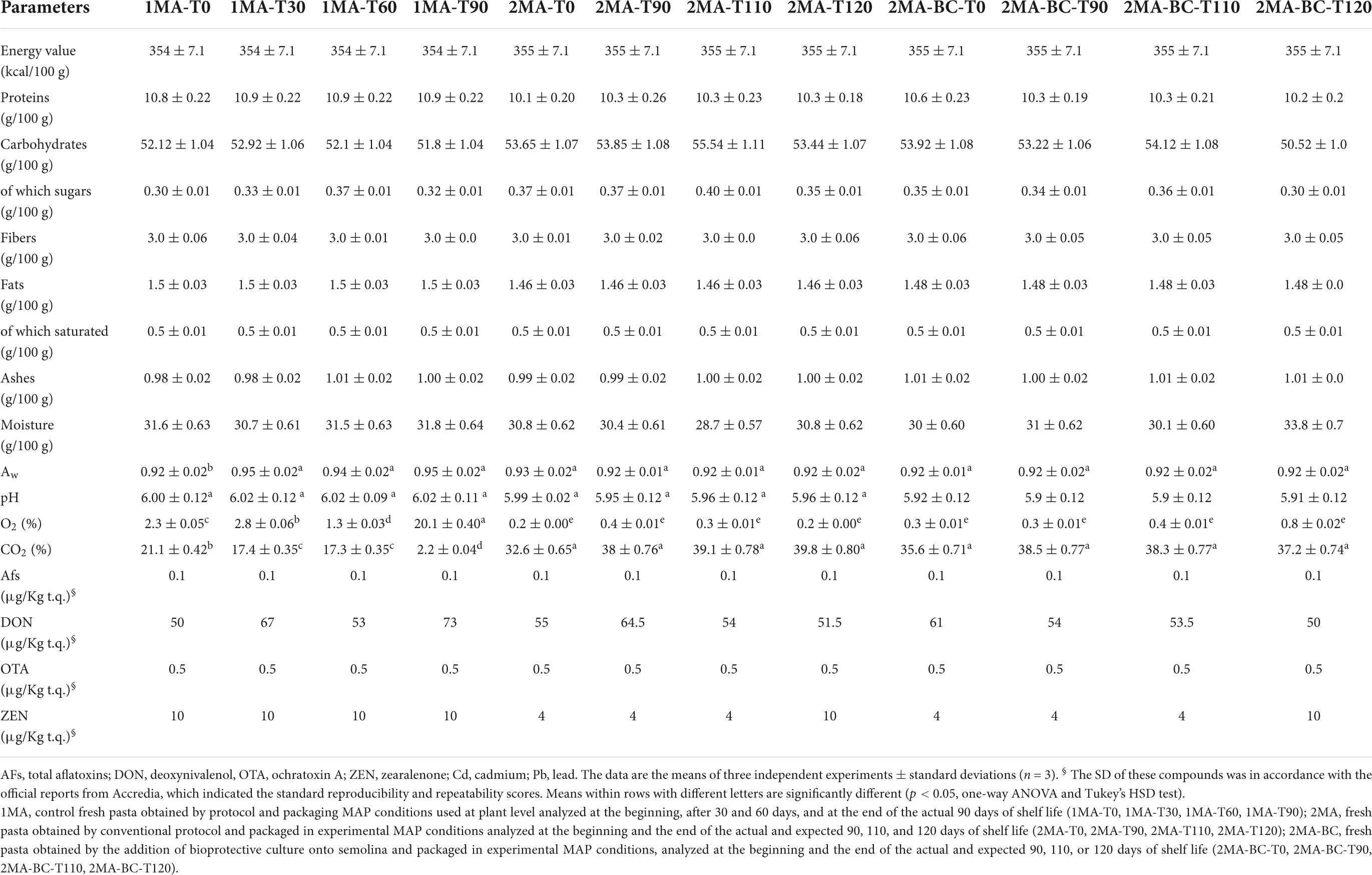
1MA pasta was produced using T. durum flour and tap water by conventional protocol, packaged in CO2:N2 = 20:80 PACT30 PE50 MAP system, and used as control. 2MA and 2MA-BC pasta samples were packaged in CO2:N2 = 40:60 PA15/PEEVOH5/PE60 MAP system. 2MA-BC pasta was produced using the commercial spray-dried probiotic bioprotective cultures (BCs) added to pasta dough.
Pasta packaged in experimental MAP with or without the addition of BCs were not different in terms of energy value, proteins, carbohydrates, fibers, fats, and ashes content than 1MA samples (Table 1).
Compared to the control 1MA, 2MA, and 2MA-BC pasta showed similar levels of moisture content at T0 (31.6, 30, and 30.8%, respectively).
The aw was similar (ca. 0.92) between 2MA and 2MA-BC samples and no differences were found during storage. In contrast, in 1MA, the aw was lowest at the beginning of storage (0.92) while it increased (p < 0.05) up to 0.95 at 90 days.
At T0, 1MA, 2MA, and 2MA-BC had pH values of 6.00 ± 0.14, 5.99 ± 0.02, and 5.92 ± 0.12, respectively. No statistical differences (p > 0.05) were found during storage.
The headspace gas analysis in 1MA samples showed some changes during storage. In particular, the O2 concentration increased from 2.3% (at T0) to 20.1% (at T90) while, concerning the same mentioned time-points of storage, the CO2 decreased from 21.1% to 2.2%. Compared to 1MA samples, both 2MA (with or without the addition of probiotic BCs) showed the same percentage (p < 0.05) of gas concentration during 120 days of storage, and non-significant differences (p > 0.05) were observed between samples.
In all pasta samples, values of mycotoxins (AFTs, DON, OTA, and ZEN) were below the safety limits according to the Regulation (CE) N. 401/2006, and non-significant differences (p < 0.05) were found between sample types as well as based on storage.
Cultivable microbiota of fresh pasta
The baseline cell densities of semolina flour used for pasta manufacturing were higher than final products, mainly due to total aerobic mesophilic microorganisms at 30°C (ca. 3.80 log CFU/g), coliforms (ca. 2.5 log CFU/g), and Enterobacteriaceae (ca. 2.76 log CFU/g). Moreover, in the used semolina flour LAB were 1.6 log CFU/g, whereas Salmonella and L. mononocytogenes were absent in 25 g of samples. Beta-glucuronidase-positive E. coli, sulfite-reducing clostridia, and coagulase-positive Staphylococcus densities were below the detection limit (<1 log CFU/g). Yeasts and molds were <1 and 2.08 log CFU/g, respectively.
In the experimental trial, the use of experimental MAP conditions with or without the addition of BCs (2MA-BC and 2MA, respectively) was carried out and compared against the 1MA product at the beginning and during 90 or 120 days of storage at 4°C.
Pathogens (Salmonella and L. monocytogenes) were absent in all samples. Beta-glucuronidase-positive E. coli, sulfite-reducing clostridia, and coagulase-positive Staphylococcus densities were below the detection limit (<1 log CFU/g) in all samples. Coliforms and Enterobacteriaceae densities were below 1 log CFU/g in all samples during the monitored 120 days.
In 1MA, total aerobic mesophilic microorganisms at 30°C grew from 3.00 to 4.34 log CFU/g after 90 days of storage (Table 2). When 2MA condition without BCs was used, aerobic mesophilic cell density was ca. 2.52 log CFU/g and remain almost constant until 120 days of storage. In 2MA-BCs, total aerobic microorganisms were <2.0 log CFU/g during storage.
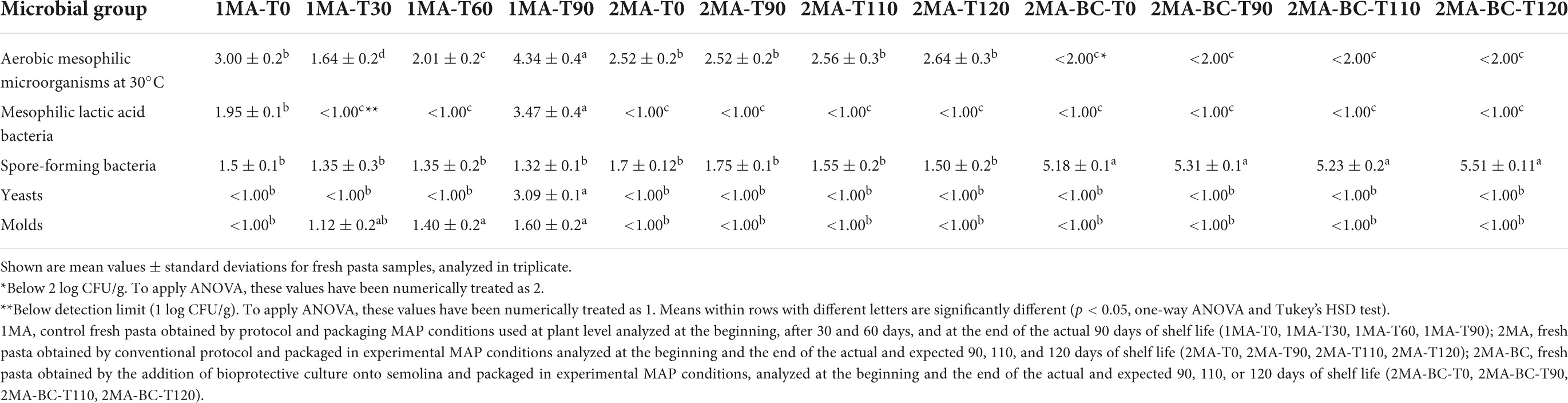
Table 2. Viable cell counts (log CFU/g) of different microbial groups during storage at 4°C of fresh pasta “trofie.”.
The LAB cell densities in 1MA at the beginning of refrigerated storage was ca. 1.95 ± 0.1 log CFU/g. In 1MA, LAB increased during 90 days of storage and reached 3.47 log CFU/g, while for samples stored with the innovative gas concentration and film (with or without the addition of BCs), it was <1 log CFU/g until the end of 120 days of storage, without significant differences (p < 0.05) between samples (Table 2). Bifidobacteria were not detected in all samples.
The range of counts for SFB and Bacillus spp. were similar in all samples analyzed (Table 2). 1MA and 2MA showed low counts of aerobic spore-forming bacteria (1.5 and 1.7 log CFU/g, respectively), without significant differences (p > 0.05) during storage, whereas 2MA-BC showed higher (p < 0.05) counts of spore-forming bacteria than both 1MA and 2MA.
Yeast and mold densities were below the detection limit (1 log CFU/g) in 2MA and 2MA-BC without significant differences (p > 0.05) driven by the addition of BCs. In the 1MA, yeasts and molds grew up to 3.09 and 1.6 log CFU/g, respectively, at 90 days of storage (Table 2 and Supplementary Figure 1). 2MA and 2MA-BC samples did not report either signs of spoilage or microbial alterations until 120 days of storage.
Proteomic characterization of fresh pasta
No differences (p < 0.05) were observed for the concentration of protein fractions in pasta samples, whose levels were stable during storage (data not shown). 1MA pasta protein fractions were separated and visualized in triplicate by SDS-PAGE (Supplementary Figure 2). The intensities of the albumins, globulins, and gliadins bands were similar between the sampling times (Supplementary Figure 2). Protein fractions were also separated and visualized by 2DE. The reproducibility of the 2DE gel performance was analyzed by comparing samples in triplicate (data not shown). Based on SDS-PAGE, no differences were found for the total number of spots and protein expression between samples of 1MA profiled at different times of sampling, indicating the absence of substantial differences or protein degradation on wheat protein fractions of pasta affected by storage. Therefore, no additional samples were investigated in terms of proteomic profiling.
Study of microbial community dynamics through amplicon sequencing and metagenomics
Profiling the prokaryotic microbiota, nine libraries of dual indexed amplicons of 420 bp related to the V5–V6 hypervariable regions of the 16S rRNA gene were successfully sequenced on the MiSeq platform, using a 2 × 250 bp paired-end (PE) sequencing strategy. No amplicon has been obtained and sequenced for 2MA-BC-T120 pasta samples. All sequenced samples generated reads of high quality with the expected length of 250 bp. Raw sequencing data are available in the SRA repository under the BioProject PRJNA782807. About 88.8% of PE reads were retained as ASVs (Amplicon Sequence Variants), following the denoising procedure, and were subjected to the taxonomic analysis.
The identified taxa at phylum and genus levels showing a relative abundance higher than 0.1% were reported as stacked bar plots for each analyzed sample (Figure 2). At the phylum level, Firmicutes were dominant in all the samples during storage. Compared to 1MA, 2MA samples with or without the addition of BCs showed a slight increase of gamma-Proteobacteria during storage. Among phyla, 2MA-BC exhibited the presence of Actinobacteria for a relevant proportion of the total microbiota (average abundance of 18% of ASVs from the beginning until the end of the storage). Taxa of the Bacteroidetes and Cyanobacteria were detected in all the samples even though the total relative abundance was lower than 1%.
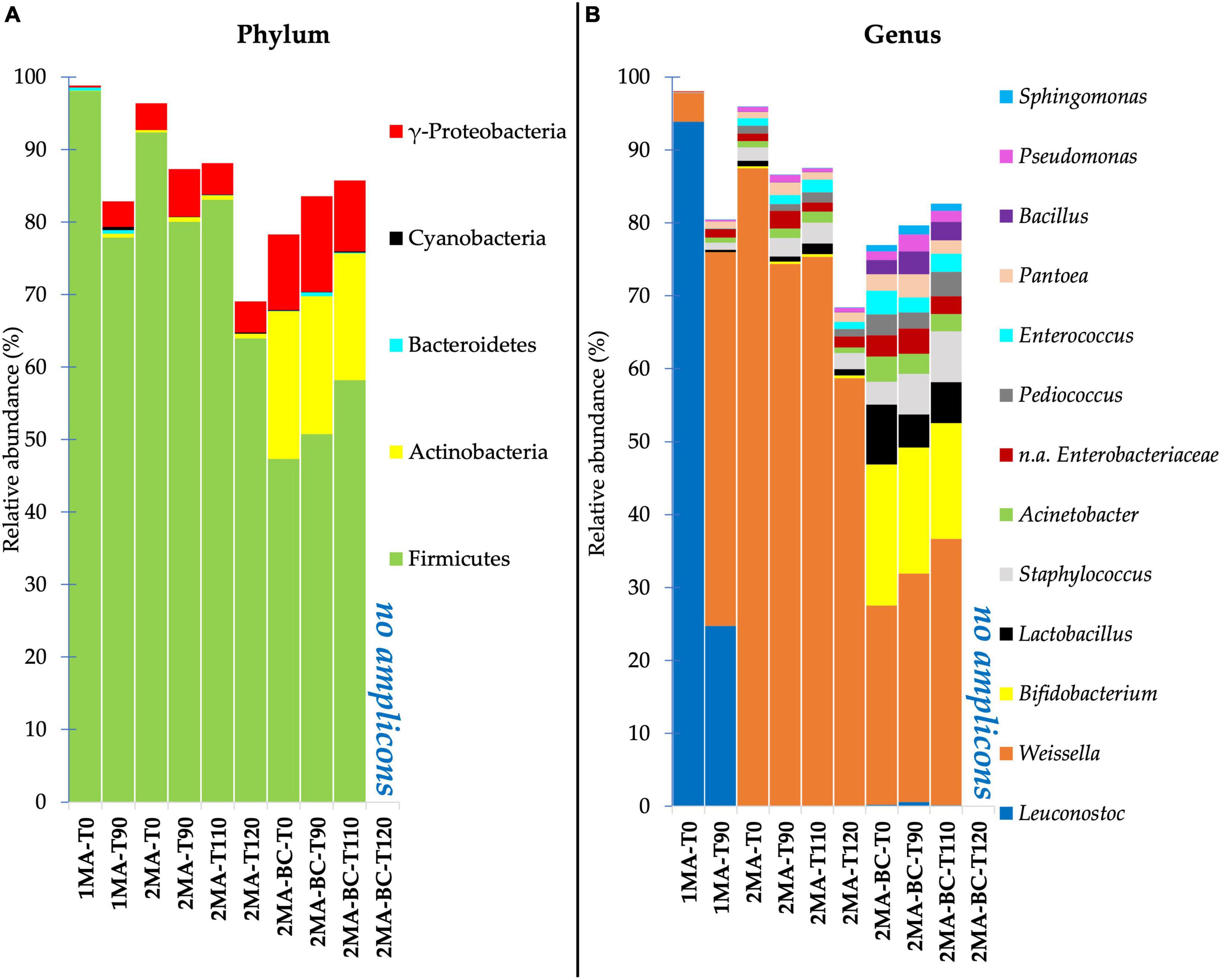
Figure 2. Bacterial community composition at phylum (A) and genus (B) level for fresh pasta samples by 16S rRNA gene V5–V6 region sequencing. 1MA, control fresh pasta obtained by protocol and packaging MAP conditions used at plant level analyzed at the beginning and the end of the actual 90 days of shelf life (1MA-T0, 1MA-T90); 2MA, fresh pasta obtained by conventional protocol and packaged in experimental MAP conditions analyzed at the beginning and the end of the actual and expected 90, 110, and 120 days of shelf life (2MA-T0, 2MA-T90, 2MA-T110, 2MA-T120); 2MA-BC, fresh pasta obtained by the addition of bioprotective culture onto semolina and packaged in experimental MAP conditions, analyzed at the beginning and the end of the actual and expected 90, 110, or 120 days of shelf life (2MA-BC-T0, 2MA-BC-T90, 2MA-BC-T110, 2MA-BC-T120).
Leuconostoc and Weissella were the dominant genera (93.87 and 3.96% of ASVs, respectively) in 1MA-T0 (Figure 2B). Leuconostoc was the most relevant genus in 1MA until 90 days of shelf life despite its relative abundance decreasing to 24.7% in 1MA-T90. Instead, Weissella increased up to 51.25% in 1MA-T90. Staphylococcus and Enterobacteriaceae (not assigned at genus level) were ca. 1% till 90 days of storage.
The use of the experimental gas concentration and barrier film combined with the addition of BCs changed the quantitative composition of the prokaryotic microbiota at the genus level. Weissella was the most relevant genus despite its relative abundance being ca. 87.5 and 27% of ASVs in 2MA-T0 and 2MA-BC-T0, respectively. The relative abundance of Weissella significantly decreased in 2MA-T120 (58.7% of the total ASVs) while increasing up to 37% in 2MA-BC-T110.
In 2MA-BC, the ASVs detection of Bifidobacterium, Lactobacillus, and Bacillus confirmed the presence of the BCs, and these on average were 17.5, 6.12, and 2.55% of the ASVs, respectively, without significant differences determined by storage.
During 120 days of shelf life of samples packaged in 2MA, Staphylococcus, Acinetobacter, Pediococcus, Enterococcus, Pseudomonas, and Enterobacteriaceae (not assigned at genus level) were found each at a lower abundance than 2.85% without significant differences determined by storage.
To profile the fungal microbiota, ITS1 sequencing was performed. No amplicons were retrieved from the total DNA of 2MA and 2MA-BC. Therefore, the ITS1 sequencing was performed only for 1MA samples at the beginning and 90 days of shelf life. Wickerhamomyces was the genus more present (94 and 91.7% of the ASVs in 1MA-T0 and 1MA-T90 samples, respectively) (Figure 3A). Besides Wickerhamomyces, Torulaspora was most represented (3.18 and 1.93% of ASVs in 1MA-T0 and 1MA-T90, respectively) followed by Saccharomyces (1.08 and 1.27% of ASVs in 1MA-T0 and 1MA-T90, respectively). Cyberlindnera, Malassezia, Alternaria, Theobroma, Fusarium, and Cladosporium genera had a relative abundance lower than 1% even though a slight increase in their relative abundance was found at T90. The heatmap with normalized values of fungal species’ relative abundance (Figure 3B), including those poor-representative species (0.01–0.1%), showed a more heterogeneous fungal microbiota at T90 than T0.
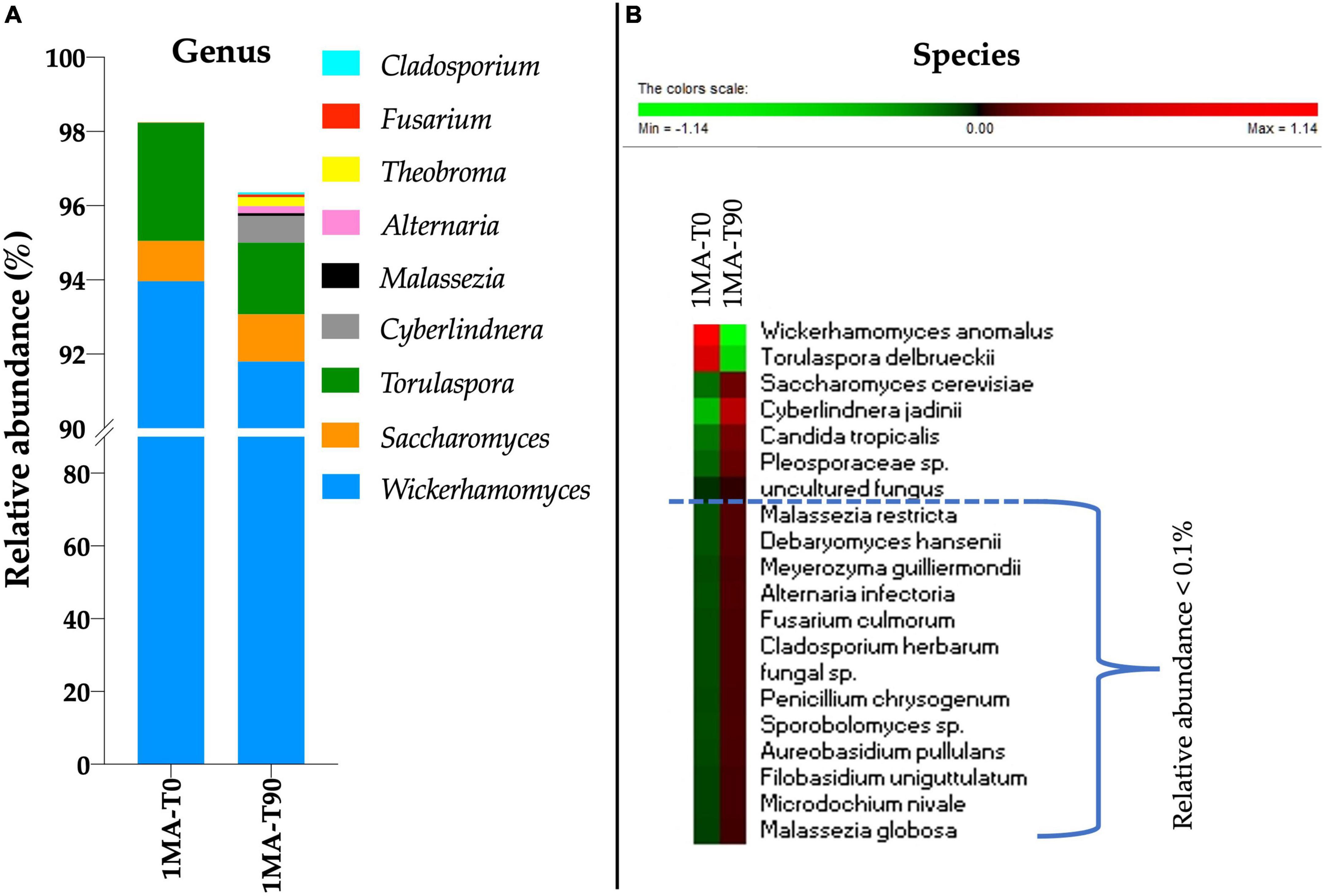
Figure 3. Fungi microbiota (yeasts and molds) composition in fresh pasta samples by ITS1 region sequencing. 1MA, control fresh pasta obtained by protocol and packaging MAP conditions used at plant level analyzed at the beginning and the end of the actual 90 days of shelf life (1MA-T0, 1MA-T90). (A) Fungal genera with a relative abundance >0.1% in at least one sample. To note, the scale values started from 90% due to the relative abundance of Wickerhamomyces that was >91%. (B) Heatmap based on relative abundance (normalized for row) of the top 20 abundant fungal species. Colors correspond to normalized mean data levels from low (green) to high (red).
Profiling the volatile organic compounds of fresh pasta
Volatile organic compounds of fresh pasta samples at the beginning and the end of 90 days of refrigerated storage have been investigated by HS-SPME/GC-MS. Forty-two VOCs were detected (Supplementary Table 1). Overall, after 90 days of storage, the total amount of fatty acids and aldehydes in 1MA-T90 was higher compared to 2MA and 2MA-BC pasta samples. The highest concentration of hexanal (p < 0.05) was found in 1MA-T90. The highest concentration of total alcohol was observed in 2MA-BC samples. Comparing the different fresh pasta samples, 24 out of 42 VOCs were statistically different. 1MA-T90 showed the highest (p < 0.05) amount of hexanal, 2-methylpentanoic anhydride, nonanoic acid, hexanoic acid, 2H-pyran-2,6(3H)-dione, and other hydrocarbons. Both 2MA samples reported a higher concentration (p < 0.05) of hexanol and octanoic acid compared to 1MA. Aromatic compounds, benzyl alcohol, and propanedioic acid-diethyl ester were found only in 2MA-BC (Supplementary Table 1). VOC profiles were plotted based on a principal component analysis (PCA) (Figure 4). The PCA plot of samples (score plot) and variables (loading plot) accounted for 68% of the total variance (PC1: 52.9% and PC2: 15.1%). According to PC1, a different VOC profile was observed between 1MA and both 2MA fresh pasta samples. Differently, the PC2 discriminates samples based on both the time of storage and the use of 2MA combined with BCs.
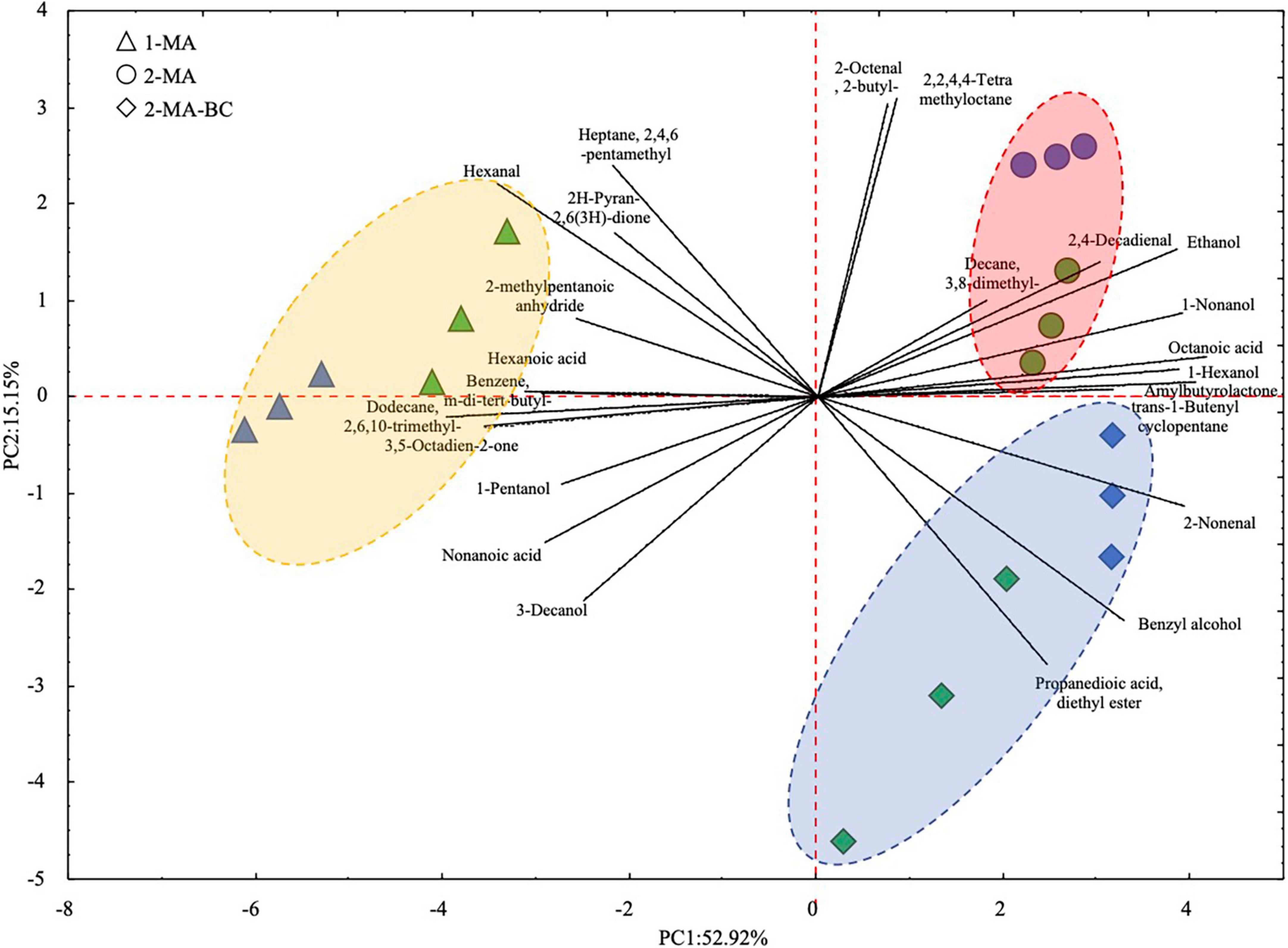
Figure 4. Principal component analysis (PCA) of 24 volatile organic compounds that significantly differed (p < 0.05; one-way ANOVA) between fresh pasta samples. ▲, 1MA, control fresh pasta obtained by conventional protocol and packaging MAP conditions used at the plant level; □, 2MA, fresh pasta obtained by conventional protocol and packaged in experimental MAP conditions; ⋄, 2MA-BC, fresh pasta obtained by the addition of bioprotective culture onto semolina and packaged in experimental MAP conditions. Three biological replicates were analyzed at the beginning (0 days, green color) and the end (blue color) of the actual 90 days of storage.
Discussion
Homemade, artisanal, or industrial fresh pasta products, for example, filled pasta or one heat-treated, are vulnerable under a microbiological point of view based on values of moisture and aw. When values of these parameters were too high, they favor the growth of pathogenic or potentially pathogenic microorganisms limiting, therefore, both the healthiness and shelf life of products (Ricci et al., 2017). To preserve fresh pasta and extend its shelf-life avoiding food loss and waste, double-time pasteurization, packaging, temperature of refrigeration, and the use of preservatives are the most studied approaches (Del Nobile et al., 2009a; Costa et al., 2010; Zardetto et al., 2021). With this purpose, the present study describes a “clean-label” approach that uses MAP combined with and without bioprotective cultures (BCs) to increase the shelf life of fresh pasta from 90 to 120 days of storage at 4°C. During storage, several chemical, microbiological, and metabolomic characteristics can be affected by the different technologies that have, in turn, an effect on pasta properties to extend/limit its shelf-life (Costa et al., 2010; Sanguinetti et al., 2011; Tabanelli, 2020; Zardetto et al., 2021). We discovered the suitability of a multi-omics approach to microbial community (bacterial and fungal) profiling through culturomics and metagenomics (amplicon sequencing-based), proteomics, and metabolomics techniques.
First, the influence of the innovative sealed bags and MAP conditions, used in this research, lead to a different O2 and CO2 concentration during storage being a strong deliverable of the present work, since previous evidence allowed for extended storage thanks to increased concentrations of CO2 (Zardetto et al., 2022). Visible molds were found in control fresh pasta packaged under routinary CO2:N2 (20:80) ratios in PACT30-PE50-bags, in which a decreasing CO2 was found after 90 days of storage. Hence, we hypothesized that a gas diffusion between the packaging headspace and the environment was affected by sealed bag characteristics (e.g., mechanical resistance), as suggested by previous works (Lucera et al., 2014; Chaix et al., 2015; Zardetto et al., 2022). In turn, the altered gas composition harbored mold growth on the pasta surface before the use-by date. On the contrary, the experimental CO2:N2 (40:60) concentration combined with PA15/PEEVOH5/PE60 films for packaging was more stable during storage. Values of WVTR and OTR (Appendix A) of the two films were considered and, as reported in technical sheets, results from permeation tests showed substantial differences. The OTR values particularly differed in terms of magnitude order; the 1MA film being more permeable to O2 than bags used for 2MA.
Culture-dependent and -independent approaches allowed for obtaining an integrated overview in terms of microbial dynamics during storage. Microbial cell densities increased during the storage of 1MA samples, which are products no longer marketable after 90 days. A microbial inhibition was found in 2MA based on MAP with 40% CO2 in line with previous results (Zardetto, 2005a; Kimbuathong et al., 2020). The CO2 permits prolonging of both the lag phase and the time for spoilage inhibiting pivotal enzymatic activities in Krebs’s cycle of microorganisms (Daniels et al., 1985). Instead, in control samples, culturomics seems to indicate a survival rate of several microbial groups despite the heat treatment. Hence, the effectiveness of pasteurization can not be absolute due to the intrinsic and specific characteristics of foods and, as recognized, the shelf life is affected by both the number of surviving microorganisms and textural/structural changes deriving from thermal treatments (Lopez et al., 1998; Tabanelli, 2020). Nonetheless, all used pasta variants were suitable under the hygienic profile, as emphasized by the absence of pathogenic microorganisms (e.g., Salmonella spp., L. monocytogenes). These outcomes, indeed, are related to the strictness in observing hygienic practices during the manufacturing, as well as to the use of raw materials with good quality and, overall, the effectiveness of the thermal treatment (Aureli et al., 1989; Alamprese et al., 2008; Ricci et al., 2017). No fungi growth and, indeed, no ITS amplicons were retrieved from 2MA pasta samples. As already reported for fresh-filled pasta, molds can be responsible for spoilage, but their multiplication can be inhibited using a concentration of CO2 higher than 15% in MAP packaging (Zardetto, 2005b; Tabanelli, 2020). Consistent with these results, AFTs, DON, OTA, and ZEA fungi mycotoxins did not exceed during storage, as they were below the maximum limits established by Commission regulation (No. 401/2006). Mycotoxins are natural food contaminants and the quality and safety of the final pasta depend on the flours, the technological process (thermal treatment and drying process), and the conditions used during preparation (de Nijs et al., 2016; Bouafifssa et al., 2018). The metagenomics explored mycotoxins-producers’ fungi such as Penicillium, Alternaria, and Fusarium being below the 0.1% of relative abundance. The present technological process did not avoid the presence of mycotoxigenic fungal taxa even though mycotoxin production was always under the safety limit confirming the inhibitory effect of MAP toward these important chronic dietary risk factors (Bouafifssa et al., 2018). Living microbes were macroscopically found in 1MA-T90; however, chemical and proteomic analyses did not emphasize a considerable metabolism of macronutrients and proteins for which microbes are well known (De Angelis et al., 2021). The proteomic profiles of all 1MA samples overlapped for both the presence and intensity of gel bands, as well as, albumins, gliadins, and glutenins spots. For this reason, 2MA samples were not profiled with respect to the proteomic field. According to the ITS1-seq, the time of storage determined an increase in alpha diversity in 1MA samples. On the contrary, samples of 1MA were characterized by lower alpha diversity scores of bacteria domains due to the main presence of Leuconostoc (at T0), while Weissella (at T90) in relative microbiota. Probably due to lower microbial competition, the presence of only these two bacterial genera was not sufficient to avoid mold overgrowth. The presence of ASVs belonging to Lactobacillus, Bacillus, and Bifidobacterium confirmed the addition of BCs (L. acidophilus, L. casei, Bifidobacterium spp., B. coagulans) and that these taxa were detectable till 110 days of storage. Therefore, this result confirms the previously stated evidence that probiotic multistrain cultures used for bioprotection are metabolically active preparations applied to inhibit undesired microbes (Spanu et al., 2017). Herein, culturomics highlighted the thermal treatment effectiveness in decreasing cell densities of lactobacilli and bifidobacteria, whereas the spore-forming B. coagulans preserved its viability during pasta production and storage as previously discussed (Konuray and Erginkaya, 2020). In the present study, the addition of BCs reduced the growth of autochthonous microbes allowing for improved stability of pasta during storage. This may be attributed to the release of heat-stable bacteriocins synthesized by BCs, which represent an extra hurdle against spoilage and/or pathogen microorganisms during the processing and/or storage and/or selling of the foodstuff (Baka et al., 2014). Antimicrobials could be present in 2MA-BC fresh pasta independently of the viability of the producing microorganisms while contributing to inhibiting the growth of spoilage and pathogenic bacteria and allowing for extending the storage of pasta up to 120 days.
Concerning VOCs, 42 compounds were overall detected in fresh pasta samples. As shown by the multivariate analyses of VOC profiles, samples were mainly differentiated based on the production process (Figure 4). The main differences were related to aldehydes, fatty acids, and alcohols derived from lipid oxidation. Nonanoic and hexanoic acids, both deriving from autoxidation (Giannetti et al., 2014), were significantly abundant in control pasta (1MA-T90) rather than in 2MA-T90 and 2MA-BC-T90 samples. Hence, the experimental MAP substantially extended the shelf-life, probably also delaying lipid oxidation as a result of a reduced O2 content (Kimbuathong et al., 2020). Lipid oxidation processes are one of the most important parameters influencing the shelf-life of products affecting their quality through deteriorative events (Calligaris et al., 2007; Difonzo et al., 2018). Microorganisms can synthesize lipase and phospholipase contributing to an increase in free fatty acids, molecules that are susceptible to oxidation, such as aldehydes (Nirmal and Benjakul, 2011). In a similar line, hexanal, which is known as a low-quality marker for artisanal and industrial pasta being responsible for fresh pasta off-flavors, decreased when the CO2:N2 = 40:60 MAP system and BCs were used (Giannetti et al., 2014; Hong et al., 2021). It is also well known that fungal growth leads to VOC metabolism (Sunesson et al., 1997). With this respect, 2-methylpentanoic anhydride, being a recognized fungal metabolite, was only found in 1MA samples that, in turn, were characterized by the greatest fungal spoilage (Ewen et al., 2004).
Conclusion
This research highlights the novel exploitation of MAP with or without the addition of BCs to produce fresh pasta. The results indicate that the MAP (40:60 CO2:N2), high barrier packaging, and BCs and their metabolites acted in a synergistic way to control the microbial spoilage of fresh pasta during refrigerated storage at 4°C and can be introduced at the industrial level giving an increase of 30 days in shelf life compared to low barrier conventional MAP packaging, with potential benefits on the economy and on the environment, spurring innovation in existing production models. The microbial assembly and function varied depending on gas concentration, affecting the microbial stability of fresh pasta. The multi-omics approach used in this study can be applied, combined with traditional protocols, to evaluate which of the studied factors is the most influential to drive the shelf-life of fresh pasta. Even if the added BCs were not all detected by traditional protocols during storage of fresh pasta, their antimicrobials and/or fermentation products leads to quantitative and qualitative effect on the bacterial and fungal microbiota associated with pasta. Data herein reported are encouraging additional tests aimed at setting up new protocols for the use of BCs also in other types of cereal-based products, considering the possible implication that viable microorganisms resting in foods can exert potential positive effects on the gut microbiota of consumers, with consequences that remain to explore.
Data availability statement
The data presented in this study are deposited in the SRA repository, accession number BioProject PRJNA782807 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA782807).
Author contributions
MM, GP, FD, and MDA: conceptualization. MM, MC, GP, FD, and MDA: methodology. MM and MC: validation. MM, MC, GC, GRC, BF, and MV: formal analysis. MM, MC, GRC, GC, BF, DD, MV, and EN: investigation and writing—original draft preparation. FD, GP, and MDA: resources. MM, MC, GRC, GC, BF, DD, and MV: data curation. MM, GRC, GC, BF, and MV: visualization. GP, FD, and MDA: supervision and funding acquisition. GP and FD: project administration. All authors: writing—review and editing.
Funding
This project has received funding from Regione Puglia Asse 1 - Azione 1.6 Bando INNONETWORK “sostegno alle attivita’ di r&s per lo sviluppo di nuove tecnologie sostenibili, di nuovi servizi e prodotti” under grant agreement No: 1JLZKD0 (OMICS4FOOD). This work was also supported by ELIXIR-IT, the research infrastructure for life-science data, under grant agreement No: 871075 (ELIXIR-CONVERGE) and by the Rete LAIFF-Rete di laboratori per l’innovazione nel campo degli alimenti funzionali.
Acknowledgments
We thank Tommaso Depalma and Pasta Apulia srl for the fresh pasta used in this study.
Conflict of interest
Author DD was employed by Food Safety Lab s.r.l.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1003437/full#supplementary-material
Footnotes
References
Alamprese, C., Casiraghi, E., and Rossi, M. (2008). Structural and cooking properties of fresh egg pasta as a function of pasteurization treatment intensity. J. Food Eng. 89, 1–7. doi: 10.1016/j.jfoodeng.2008.03.022
Alonso-Alemany, D., Barre, A., Beretta, S., Bonizzoni, P., Nikolski, M., and Valiente, G. (2014). Further steps in TANGO: Improved taxonomic assignment in metagenomics. Bioinformatics 30, 17–23. doi: 10.1093/bioinformatics/btt256
Ananou, S., Maqueda, M., Martínez-Bueno, M., and Valdivia, E. (2007). “Biopreservation, an ecological approach to improve the safety and shelf-life of foods,” in Communicating current research and educational topics and trends in applied microbiology, Vol. 1, ed. A. M. éndez Vilas (Guadalajara: FORMATEX), 475–487.
Angiolillo, L., Conte, A., and del Nobile, M. A. (2017). Biotechnological Approach to preserve fresh pasta quality. J. Food Prot. 80, 2006–2013. doi: 10.4315/0362-028X.JFP-16-382
Antognelli, C. (1980). The manufacture and applications of pasta as a food and as a food ingredient: A review. Int. J. Food Sci. Technol. 15, 125–145. doi: 10.1111/j.1365-2621.1980.tb00926.x
AOAC International (1990). Official methods of analysis of AOAC International, 15th Edn, Vol. I., Rockville: AOAC International.
AOAC International (2006). Official methods of analysis of AOAC International, 18. Edn. Rockville: AOAC International.
Arendt, E., and Zannini, E. (2013). Cereal grains for the food and beverage industries. Sawston: Woodhead Publishing.
Aureli, P., Papasso, A., Fenicia, L., Ferrini, A. M., and Gianfranceschi, M. (1989). ISTISAN reports: Metodiche analitiche per il controllo microbiologico delle paste alimentari - ISTISAN 89/9-ISSN-0391-1675.
Baka, M., Noriega, E., Mertens, L., Van Derlinden, E., and Van Impe, J. F. M. (2014). Protective role of indigenous Leuconostoc carnosum against Listeria monocytogenes on vacuum packed frankfurter sausages at suboptimal temperatures. Food Res. Int. 66, 197–206. doi: 10.1016/j.foodres.2014.08.011
Bini, L., Magi, B., Marzocchi, B., Arcuri, F., Tripodi, S., Cintorino, M., et al. (1997). Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis 18, 2832–2841. doi: 10.1002/elps.1150181519
Bjellqvist, B., Hughes, G. J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J.-C., et al. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031. doi: 10.1002/elps.11501401163
Bokulich, N. A., and Mills, D. A. (2013). Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 79:2519. doi: 10.1128/AEM.03870-12
Bolyen, E., Rideout, J. R., and Dillon, M. R. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bouafifssa, Y., Manyes, L., Rahouti, M., Mañes, J., Berrada, H., Zinedine, A., et al. (2018). Multi-Occurrence of twenty mycotoxinsin pasta and a risk assessment in the moroccan population. Toxins 10:432. doi: 10.3390/toxins10110432
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Calasso, M., Francavilla, R., Cristofori, F., De Angelis, M., and Gobbetti, M. (2018). New Protocol for production of reduced-gluten wheat bread and pasta and clinical effect in patients with irritable bowel syndrome: A randomised, double-blind, cross-over study. Nutrients 10:1873. doi: 10.3390/nu10121873
Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 11, 2639–2643. doi: 10.1038/ismej.2017.119
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Calligaris, S., Manzocco, L., Kravina, G., and Nicoli, M. C. (2007). Shelf-life modeling of bakery products by using oxidation indices. J. Agric. Food Chem. 55, 2004–2009. doi: 10.1021/jf063004h
Capozzi, V., Menga, V., Digesù, A. M., De Vita, P., van Sinderen, D., Cattivelli, L., et al. (2011). Biotechnological production of vitamin B2-enriched bread and pasta. J. Agric. Food Chem. 59, 8013–8020. doi: 10.1021/jf201519h
Carini, E., Curti, E., Minucciani, M., Antoniazzi, F., and Vittadini, E. (2014). “Chapter 10: Pasta,” in Contemporary food engineering, eds R. de Pinho Ferreira Guinè and P. M. dos Reis Correia (Boca Raton, FL: CRC Press), 21238. ISBN: 978-1-4398-8702-8.
Cavallo, N., Angelis, M. D., Calasso, M., Quinto, M., Mentana, A., Minervini, F., et al. (2017). Microbial cell-free extracts affect the biochemical characteristics and sensorial quality of sourdough bread. Food Chem. 237, 159–168. doi: 10.1016/j.foodchem.2017.05.089
Chaix, E., Couvert, O., Guillaume, C., Gontard, N., and Guillard, V. (2015). Predictive Microbiology coupled with gas (O 2/CO 2) transfer in food/packaging systems: How to develop an efficient decision support tool for food packaging dimensioning: A decision support tool for map. Compr. Rev. Food Sci. Food Saf. 14, 1–21. doi: 10.1111/1541-4337.12117
Chakravorty, S., Helb, D., Burday, M., Connell, N., and Alland, D. (2007). A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69, 330–339. doi: 10.1016/j.mimet.2007.02.005
Cifuentes Bachmann, D. E., and Leroy, F. (2015). Use of bioprotective cultures in fish products. Curr. Opin. Food Sci. 6, 19–23. doi: 10.1016/j.cofs.2015.11.009
Costa, C., Lucera, A., Mastromatteo, M., Conte, A., and Del Nobile, M. A. (2010). Shelf life extension of durum semolina-based fresh pasta: Shelf life extension of durum semolina-based fresh pasta. Int. J. Food Sci. Technol. 45, 1545–1551. doi: 10.1111/j.1365-2621.2010.02277.x
Daniels, J. A., Krishnamurthi, R., and Rizvi, S. S. H. (1985). A review of effects of carbon dioxide on microbial growth and food quality. J. Food Prot. 48, 532–537. doi: 10.4315/0362-028X-48.6.532
De Angelis, M., Siragusa, S., Vacca, M., Di Cagno, R., Cristofori, F., Schwarm, M., et al. (2021). Selection of gut-resistant bacteria and construction of microbial consortia for improving gluten digestion under simulated gastrointestinal conditions. Nutrients 13:992. doi: 10.3390/nu13030992
de Nijs, M., van den Top, H., de Stoppelaar, J., Lopez, P., and Mol, H. (2016). Fate of enniatins and deoxynivalenol during pasta cooking. Food Chem. 213, 763–767. doi: 10.1016/j.foodchem.2016.07.024
Del Nobile, M. A., Di Benedetto, N., Suriano, N., Conte, A., Corbo, M. R., and Sinigaglia, M. (2009a). Combined effects of chitosan and MAP to improve the microbial quality of amaranth homemade fresh pasta. Food Microb. 26, 587–591. doi: 10.1016/j.fm.2009.03.012
Del Nobile, M. A., Di Benedetto, N., Suriano, N., Conte, A., Lamacchia, C., Corbo, M. R., et al. (2009b). Use of natural compounds to improve the microbial stability of Amaranth-based homemade fresh pasta. Food Microbiol. 26, 151–156. doi: 10.1016/j.fm.2008.10.003
Difonzo, G., Pasqualone, A., Silletti, R., Cosmai, L., Summo, C., Paradiso, V. M., et al. (2018). Use of olive leaf extract to reduce lipid oxidation of baked snacks. Food Res. Int. 108, 48–56. doi: 10.1016/j.foodres.2018.03.034
DPR (2001). Decreto del Presidente della Repubblica 9 Febbraio 2001, n. 187. Regolamento per la revisione della normativa sulla produzione e commercializzazione di sfarinati e paste alimentari, a norma dell’articolo 50 della legge 22 febbraio 1994. 146., Available online at: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/2772 (accessed October, 2022).
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Ewen, R. J., Jones, P. R. H., Ratcliffe, N. M., and Spencer-Phillips, P. T. N. (2004). Identification by gas chromatography-mass spectrometry of the volatile organic compounds emitted from the wood-rotting fungi Serpula lacrymans and Coniophora puteana, and from Pinus sylvestris timber. Mycol. Res. 108, 806–814. doi: 10.1017/S095375620400022X
Fares, C., Menga, V., Martina, A., Pellegrini, N., Scazzina, F., and Torriani, S. (2015). Nutritional profile and cooking quality of a new functional pasta naturally enriched in phenolic acids, added with β-glucan and Bacillus coagulans GBI-30, 6086. J. Cereal Sci. 65, 260–266. doi: 10.1016/j.jcs.2015.07.017
FDA (2006). Food and drug administration department of health and human services. eCFR ID 21CFR582.3640. Available online at: https://www.ecfr.gov/current/title-21/chapter-I/subchapter-E/part-582/subpart-D/section-582.3640 (accessed October, 2022).
Fosso, B., Pesole, G., Rosselló, F., and Valiente, G. (2018). Unbiased taxonomic annotation of metagenomic samples. J. Comput. Biol. A J. Comput. Mol. Cell Biol. 25, 348–360. doi: 10.1089/cmb.2017.0144
Fosso, B., Santamaria, M., Marzano, M., Alonso-Alemany, D., Valiente, G., Donvito, G., et al. (2015). BioMaS: A modular pipeline for Bioinformatic analysis of Metagenomic AmpliconS. BMC Bioinform. 16:203. doi: 10.1186/s12859-015-0595-z
Ghanbari, M., Jami, M., Domig, K. J., and Kneifel, W. (2013). Seafood biopreservation by lactic acid bacteria – A review. LWT Food Sci. Technol. 54, 315–324. doi: 10.1016/j.lwt.2013.05.039
Gholizadeh, M., Razavi, J., and Mousavi, S. A. (2007). Gas permeability measurement in polyethylene and its copolymer films. Mater. Des. 28, 2528–2532. doi: 10.1016/j.matdes.2006.09.018
Giannetti, V., Boccacci Mariani, M., Mannino, P., and Testani, E. (2014). Furosine and flavour compounds in durum wheat pasta produced under different manufacturing conditions: Multivariate chemometric characterization. LWT Food Sci. Technol. 56, 15–20. doi: 10.1016/j.lwt.2013.10.045
Giannetti, V., Boccacci Mariani, M., Marini, F., and Biancolillo, A. (2021). Effects of thermal treatments on durum wheat pasta flavour during production process: A modelling approach to provide added-value to pasta dried at low temperatures. Talanta 225:121955. doi: 10.1016/j.talanta.2020.121955
Guerzoni, M. E., Sinigaglia, M., Gardini, F., and Lanciotti, R. (1994). “Microbiological aspects of processed pasta products,” in Pasta based meals: Qualitative, nutritional and technological aspects (AITA-ASTER), 115–128.
Hong, T., Zhao, Q., Xu, D., Yuan, Y., Ma, Y., Wu, F., et al. (2021). Effect of heat-treated flour on the quality and storage stability of fresh noodles. LWT 146:111463. doi: 10.1016/j.lwt.2021.111463
Kim, M., Lee, K.-H., Yoon, S.-W., Kim, B.-S., Chun, J., and Yi, H. (2013). Analytical tools and databases for metagenomics in the next-generation sequencing era. Genomics Inform. 11, 102–113. doi: 10.5808/GI.2013.11.3.102
Kimbuathong, N., Leelaphiwat, P., and Harnkarnsujarit, N. (2020). Inhibition of melanosis and microbial growth in Pacific white shrimp (Litopenaeus vannamei) using high CO2 modified atmosphere packaging. Food Chem. 312:126114. doi: 10.1016/j.foodchem.2019.126114
Konuray, G., and Erginkaya, Z. (2020). Quality evaluation of probiotic pasta produced with Bacillus coagulans GBI-30. Innov. Food Sci. Emerg. Technol. 66:102489. doi: 10.1016/j.ifset.2020.102489
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79:5112. doi: 10.1128/AEM.01043-13
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Li, M., Zhu, K., Guo, X., Peng, W., and Zhou, H. (2011). Effect of water activity (aw) and irradiation on the shelf-life of fresh noodles. Innov. Food Sci. Emerg. Technol. 12, 526–530. doi: 10.1016/j.ifset.2011.06.005
Lopez, C. C., Vannini, L., Lanciotti, R. A., and Guerzoni, M. E. (1998). Microbiological quality of filled pasta in relation to the nature of heat treatment. J. Food Prot. 61, 994–999. doi: 10.4315/0362-028X-61.8.994
Lucera, A., Mastromatteo, M., Conte, A., Zambrini, A. V., Faccia, M., and Del Nobile, M. A. (2014). Effect of active coating on microbiological and sensory properties of fresh mozzarella cheese. Food Packag. Shelf Life 1, 25–29. doi: 10.1016/j.fpsl.2013.10.002
Manzari, C., Fosso, B., Marzano, M., Annese, A., Caprioli, R., D’Erchia, A. M., et al. (2015). The influence of invasive jellyfish blooms on the aquatic microbiome in a coastal lagoon (Varano, SE Italy) detected by an Illumina-based deep sequencing strategy. Biol. Invasions 17, 923–940. doi: 10.1007/s10530-014-0810-2
Minervini, F., De Angelis, M., Di Cagno, R., Pinto, D., Siragusa, S., Rizzello, C. G., et al. (2010). Robustness of Lactobacillus plantarum starters during daily propagation of wheat flour sourdough type I. Food Microbiol. 27, 897–908. doi: 10.1016/j.fm.2010.05.021
Montemurro, M., Celano, G., De Angelis, M., Gobbetti, M., Rizzello, C. G., and Pontonio, E. (2020). Selection of non-Lactobacillus strains to be used as starters for sourdough fermentation. Food Microbiol. 90:103491. doi: 10.1016/j.fm.2020.103491
Nirmal, N. P., and Benjakul, S. (2011). Retardation of quality changes of Pacific white shrimp by green tea extract treatment and modified atmosphere packaging during refrigerated storage. Int. J. Food Microbiol. 149, 247–253. doi: 10.1016/j.ijfoodmicro.2011.07.002
Oliveira, M., Ferreira, V., Magalhães, R., and Teixeira, P. (2018). Biocontrol strategies for Mediterranean-style fermented sausages. Food Res. Int. 103, 438–449. doi: 10.1016/j.foodres.2017.10.048
Oliveira, P. M., Zannini, E., and Arendt, E. K. (2014). Cereal fungal infection, mycotoxins, and lactic acid bacteria mediated bioprotection: From crop farming to cereal products. Food Microbiol. 37, 78–95. doi: 10.1016/j.fm.2013.06.003
Osborne, T. B. (1907). The proteins of the wheat kernel. Wasington, DC: Carnegie Inst. doi: 10.5962/bhl.title.26152
Pagani, M. A., Lucisano, M., and Mariotti, M. (2007). “Traditional italian products from wheat and other starchy flours,” in Handbook of food products manufacturing, ed. Y. H. Hui (Hoboken, NJ: John Wiley & Sons, Inc), 327–388. doi: 10.1002/9780470113554.ch17
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2011). Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 12, 2825–2830.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Qu, P., Zhang, M., Fan, K., and Guo, Z. (2022). Microporous modified atmosphere packaging to extend shelf life of fresh foods: A review. Crit. Rev. Food Sci. Nutr. 62, 51–65. doi: 10.1080/10408398.2020.1811635
Ricci, V., Barone, F., and Petrella, L. (2017). Microbiological quality of industrial and artisanal pasta from italian market. J. Food Chem. Nanotechnol. 03, 44–49.
Rizzello, C. G., De Angelis, M., Di Cagno, R., Camarca, A., Silano, M., Losito, I., et al. (2007). Highly efficient gluten degradation by Lactobacilli and fungal proteases during food processing: New perspectives for celiac disease. Appl. Environ. Microbiol. 73, 4499–4507. doi: 10.1128/AEM.00260-07
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Sanguinetti, A. M., Del Caro, A., Mangia, N. P., Secchi, N., Catzeddu, P., and Piga, A. (2011). Quality changes of fresh filled pasta during storage: Influence of modified atmosphere packaging on microbial growth and sensory properties. Food Sci. Technol. Int. 17, 23–29. doi: 10.1177/1082013210368742
Sanguinetti, A. M., Del Caro, A., Scanu, A., Fadda, C., Milella, G., Catzeddu, P., et al. (2016). Extending the shelf life of gluten-free fresh filled pasta by modified atmosphere packaging. LWT Food Sci. Technol. 71, 96–101. doi: 10.1016/j.lwt.2016.03.010
Santamaria, M., Fosso, B., Consiglio, A., De Caro, G., Grillo, G., Licciulli, F., et al. (2012). Reference databases for taxonomic assignment in metagenomics. Brief. Bioinform. 13, 682–695. doi: 10.1093/bib/bbs036
Santamaria, M., Fosso, B., Licciulli, F., Balech, B., Larini, I., Grillo, G., et al. (2018). ITSoneDB: A comprehensive collection of eukaryotic ribosomal RNA Internal Transcribed Spacer 1 (ITS1) sequences. Nucleic Acids Res. 46, D127–D132. doi: 10.1093/nar/gkx855
Schettino, R., Pontonio, E., and Rizzello, C. G. (2019). Use of fermented hemp, chickpea and milling by-products to improve the nutritional value of semolina pasta. Foods 8:604. doi: 10.3390/foods8120604
Shahmohammadi, M., Javadi, M., and Nassiri-Asl, M. (2016). An overview on the effects of sodium benzoate as a preservative in food products. Biotechnol. Health Sci. 3, 7–11. doi: 10.17795/bhs-35084
Spanu, C., Scarano, C., Piras, F., Spanu, V., Pala, C., Casti, D., et al. (2017). Testing commercial biopreservative against spoilage microorganisms in MAP packed Ricotta fresca cheese. Food Microbiol. 66, 72–76. doi: 10.1016/j.fm.2017.04.008
Sunesson, A., Nilsson, C., Carlson, R., Blomquist, G., and Andersson, B. (1997). Production of volatile metabolites from Streptomyces albidoflavus cultivated on gypsum board and tryptone glucose extract agar-influence of temperature, oxygen and carbon dioxide levels. Ann. Occup. Hyg. 41, 393–413. doi: 10.1016/S0003-4878(96)00046-4
Tabanelli, G. (2020). Biogenic amines and food quality: Emerging challenges and public health concerns. Foods 9:859. doi: 10.3390/foods9070859
Tharmaraj, N., and Shah, N. P. (2003). Selective Enumeration of Lactobacillus delbrueckii ssp. Bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Bifidobacteria, Lactobacillus casei, Lactobacillus rhamnosus, and Propionibacteria. J. Dairy Sci. 86, 2288–2296. doi: 10.3168/jds.S0022-0302(03)73821-1
Weiss, W., Vogelmeier, C., and Görg, A. (1993). Electrophoretic characterization of wheat grain allergens from different cultivars involved in bakers’ asthma. Electrophoresis 14, 805–816. doi: 10.1002/elps.11501401126
Zardetto, S. (2005a). Potential applications of near infrared spectroscopy for evaluating thermal treatments of fresh egg pasta. Food Control 16, 249–256. doi: 10.1016/j.foodcont.2004.03.002
Zardetto, S. (2005b). Effect of modified atmosphere packaging at abuse temperature on the growth of Penicillium aurantiogriseum isolated from fresh filled pasta. Food Microbiol. 22, 367–371. doi: 10.1016/j.fm.2004.09.008
Zardetto, S., Fregonese, M., and Pasini, G. (2022). Effects of modified atmospheric packaging configuration on spoilage mould growth in damaged packages of fresh pasta. J. Food Eng. 314:110760.
Zardetto, S., Pasini, G., Romani, S., Rocculi, P., and Dalla Rosa, M. (2021). Evaluation of physico-chemical changes and FT-NIR spectra in fresh egg pasta packed in modified atmosphere during storage at different temperatures. Food Pack. Shelf Life 28:100648. doi: 10.1016/j.fpsl.2021.100648
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2013). PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Appendix A
After dough making, the 1MA, 2MA, and 2MA-BC fresh pasta samples were conveyed in a steel chamber equipped with a perforated steel conveyor belt. By steam injection at 110 ± 1°C for 90 s, the first step of pasteurization (SARP srl, Padua, Italy, model SST200/V) on the loose product was carried out. After pasteurization, the product was conveyed on a second perforated steel conveyor belt, eliminated from the condensed vapor on the surface, and then cooled to 4°C. Two modified atmospheric conditions and polymeric films with different characteristics as packaging bags were used. In the first, fresh pasta samples obtained by the conventional protocol were packed under MAP and film conventionally used in the semi-industrial factory (1MA). To realize the 1MA headspace conditions, the following gas concentrations were used: 20% CO2 and 80% N2 (CO2:N2 = 20:80), using sealed plastic bags polyamide/polyethylene (PACT30/PE50) (Carton Pack, R&D LAB, Rutigliano, Bari, Italy). The film was characterized by a water vapor permeability (WVTR) of 10 g/m2/24 h (38°C, 90% R.H.) and oxygen permeability (OTR) of 20 cc/m2/24 h (23°C, 0% R.H.). In the second MAP condition (experimental), fresh pasta samples obtained by the conventional protocol (2MA) or by the addition of the bioprotective mixture (2MA-BC) were packed in PA15/PEEVOH5/PE60 film (Carton Pack) under a modified atmosphere (CO2:N2 = 40:60). The film was characterized by a WVTR <5 g/m2/24 h (38°C, 90% R.H.) and an OTR <2 cc/m2/24 h (23°C, 0% R.H.).
Keywords: fresh pasta, modified atmosphere packaging, bioprotective cultures, multi-omics approach, metagenomics, shelf-life
Citation: Marzano M, Calasso M, Caponio GR, Celano G, Fosso B, De Palma D, Vacca M, Notario E, Pesole G, De Leo F and De Angelis M (2022) Extension of the shelf-life of fresh pasta using modified atmosphere packaging and bioprotective cultures. Front. Microbiol. 13:1003437. doi: 10.3389/fmicb.2022.1003437
Received: 26 July 2022; Accepted: 05 October 2022;
Published: 02 November 2022.
Edited by:
Fausto Gardini, University of Bologna, ItalyReviewed by:
Stefano Zardetto, Voltan Spa, ItalyPablo Fuciños, International Iberian Nanotechnology Laboratory (INL), Portugal
Copyright © 2022 Marzano, Calasso, Caponio, Celano, Fosso, De Palma, Vacca, Notario, Pesole, De Leo and De Angelis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca De Leo, f.deleo@ibiom.cnr.it; Mirco Vacca, mirco.vacca@uniba.it
 Marinella Marzano
Marinella Marzano Maria Calasso
Maria Calasso Giusy Rita Caponio
Giusy Rita Caponio Giuseppe Celano
Giuseppe Celano Bruno Fosso
Bruno Fosso Domenico De Palma
Domenico De Palma Mirco Vacca
Mirco Vacca Elisabetta Notario
Elisabetta Notario Graziano Pesole
Graziano Pesole Francesca De Leo
Francesca De Leo Maria De Angelis
Maria De Angelis