- 1Institute of Molecular Infection Biology, University of Würzburg, Würzburg, Germany
- 2Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo, Egypt
Old yellow enzymes (OYEs) are widely found in the bacterial, fungal, and plant kingdoms but absent in humans and have been used as biocatalysts for decades. However, OYEs’ physiological function in bacterial stress response and infection situations remained enigmatic. As a pathogen, the Gram-positive bacterium Staphylococcus aureus adapts to numerous stress conditions during pathogenesis. Here, we show that in S. aureus genome, two paralogous genes (ofrA and ofrB) encode for two OYEs. We conducted a bioinformatic analysis and found that ofrA is conserved among all publicly available representative staphylococcal genomes and some Firmicutes. Expression of ofrA is induced by electrophilic, oxidative, and hypochlorite stress in S. aureus. Furthermore, ofrA contributes to S. aureus survival against reactive electrophilic, oxygen, and chlorine species (RES, ROS, and RCS) via thiol-dependent redox homeostasis. At the host–pathogen interface, S. aureusΔofrA has defective survival in macrophages and whole human blood and decreased staphyloxanthin production. Overall, our results shed the light onto a novel stress response strategy in the important human pathogen S. aureus.
Introduction
Staphylococcus aureus colonization is linked with an increased risk of infection (Krismer et al., 2017). S. aureus can cause minor (skin and soft tissue) and life-threatening infections (pneumonia, osteomyelitis, and bacteremia) (Tong et al., 2015). S. aureus is an ESKAPE pathogen, being increasingly resistant to the commonly prescribed antibiotics (Renner et al., 2017). Methicillin-resistant S. aureus increased prevalence leads to treatment failure due to multiple drug resistance (Fischbach and Walsh, 2009). Hence, we need a better understanding of the microbial factors impacting the host–pathogen interplay.
In cellular respiration, energy is produced via redox reactions in which electrons migrate through biomolecules to oxygen as final acceptor (Imlay, 2019). Reactive oxygen species (ROS) are generated either as an inevitable cost of oxidative respiration, a result of antibiotic exposure, or a consequence from the host’s immune response (Van Acker and Coenye, 2017; Imlay, 2019). Myeloperoxidase in activated macrophages and neutrophils generates reactive chlorine species (RCS). In particular, ROS and RCS are the main bacterial killing mechanisms in the phagolysosome (Klebanoff et al., 2013). S. aureus must cope with endogenous reactive electrophilic species (RES: menaquinones, siderophores, and methylglyoxal), reactive electrophilic species generated secondarily to ROS, and from host interaction (formaldehyde) (Groitl and Jakob, 2014; Chen et al., 2016). Therefore, S. aureus maintains defense systems against reactive oxygen, chlorine, and electrophilic species to quench their toxicities and repair the damaged biomolecules (Guerra et al., 2017; Reichmann et al., 2018; Linzner et al., 2020).
Electrophilic species have electron-deficient carbon centers such as α,β-unsaturated carbonyl compounds, quinones, and N-ethylmaleimide (NEM) (Farmer and Davoine, 2007). In Escherichia coli, NemA, a member of the old yellow enzyme (OYE) family, reduces NEM in vitro (Miura et al., 1997; Gray et al., 2013; Ozyamak et al., 2013). OYEs reduce activated C = C bonds in α,β-unsaturated carbonyl compounds via bound flavin mononucleotide cofactor and have broad substrate specificity (Williams and Bruce, 2002; Shi et al., 2020). OYEs are phylogenetically classified into the following: Class-I (from plants and bacteria), Class-II (from fungi), and Class-III (from bacteria) (Scholtissek et al., 2017). YqiG and YqjM are the two OYEs isolated from Bacillus subtilis and are orthologs to SAUSA300_0859 and SAUSA300_0322 in S. aureus USA300_FPR3757, respectively (Kitzing et al., 2005; Sheng et al., 2016). Recently, our group proved that SAUSA300_0859 exhibits a type-I nitroreductase activity against the DNA-binding antibacterial agent MT02 (El-Hossary et al., 2018). Despite the fact that many OYEs are useful biocatalysts, the physiological role of bacterial OYEs, to our knowledge, is still mysterious (Toogood et al., 2010).
Here, we show that OfrA has a role in preventing intoxication by RES, RCS, and ROS conditions and contributes to S. aureus survival in human blood and RAW 264.7 macrophage cell line. Furthermore, OfrA is associated with the carotenoid pigment (staphyloxanthin) production via upper mevalonate pathway.
Materials and Methods
Bacterial Strains, Growth Conditions, and Materials
A summary of the bacterial strains and oligos used in this study is listed in Supplementary Tables 1, 2, respectively.
B-medium is a modified LB medium suitable for staphylococci cultivation by adding 1 g/L potassium phosphate (Brückner, 2006). In cultivation steps, the ratio of the bacterial suspension to the total volume of the flask was less than or equal to 1:3 to ensure sufficient aeration.
RPMI medium (catalog number 72400021) was purchased from LIFE Technologies. Formaldehyde (FA), diamide, and NaOCl was bought from Fisher Scientific, MP Biomedicals, and Alfa Aesar, respectively. 4-Methylumbelliferyl-β-D-glucuronide hydrate (MUG), methylhydroquinone (MHQ), methylglyoxal (MG), H2O2, cumene hydroperoxide (CHP), and mevalonate were obtained from Sigma-Aldrich. Thiourea, N-acetylcysteine (NAC), and catalase was purchased from Carl Roth, Hölzel Diagnostika, and MP Biomedicals, respectively. Stressors were dissolved in sterilized Milli-Q water for β-galactosidase and survival assays.
Bioinformatic and Phylogenetic Analyses
Completely assembled chromosomal sequences of S. aureus strains were retrieved from the NCBI website1 in May 2021. TBLASTN program from the standalone BLAST ncbi-blast-2.11.0+ used WP_000838037.1 as a query to search for possible proteins. Then, we used BLASTP program to identify the homology between the proteins retrieved via TBLASTN to WP_000838037.1. We consider 35% amino acids identity and protein length 375 ± 38 amino acids (10% deviation from WP_000838037.1) as a cutoff to limit OfrA-like proteins (Shi et al., 2020). Identical proteins were filtered out using SDDC program (Ibrahim et al., 2017).
Multiple sequence alignments were done using Clustal Omega2 with the default parameters. Phylogenetic trees were constructed using RAxML 8.0.0 software with the following setup (−f a −# autoMRE −m PROTGAMMAAUTO) (Stamatakis, 2014). Tree visualization and annotation were done using ggtree v2.0.1 (Yu, 2020).
Chromosomal Manipulation of Staphylococcus aureus JE2
To construct EI011, we exchanged Phla in pKO10 with a 1-kb fragment upstream of ofrA (Ohlsen et al., 1997). The reporter plasmid was transformed into E. coli IM08B and then electroporated into S. aureus JE2 strain (Monk et al., 2015). We confirmed the single crossover event by sequencing of the amplified fragment using primer in the plasmid and another one upstream of the 1-kb fragment. To construct EI046 (JE2ΔofrA), the allelic exchange in S. aureus JE2 strain was mediated by cloning the upstream and downstream fragments into pBASE6 shuttle vector (Geiger et al., 2012). After the double crossover with counter-selection, polymerase chain reaction (PCR) and sequencing were used to identify the mutant (Geiger et al., 2012).
Minimum Inhibitory Concentration Assay
Overnight cultures in RPMI were diluted into final OD600 = 0.05 and incubated with serial dilutions of each compound. Minimum inhibitory concentration (MIC) is the minimum concentration that results in not more than (OD600 = 0.1) after 24-h incubation at 37°C with shaking at 200 rpm. OD600 were measured using Synergy H1 plate reader.
β-Galactosidase Assay
The reporter strain was conditioned in RPMI for 24 h. We diluted the overnight culture 1:100 into fresh medium. In the overnight and the diluted culture, 10 mg/ml chloramphenicol was added as a final concentration. The resulting culture was grown in 37°C until transition from exponential phase to stationary phase (OD600 = 1.25 ± 0.05). 500 μl of the bacterial culture was supplemented with the stressor at the specified concentrations. After 2 h in 37°C with shaking at 200 rpm, samples were taken for analysis as indicated in the study of Vidal-Aroca et al. (2006).
RNA Isolation and Reverse Transcription Quantitative Polymerase Chain Reaction
Overnight cultures grown in RPMI were diluted 1:100 to reach OD600 = 0.5. Samples were taken to represent the control before stress. Substances were added to the indicated concentrations and incubated for 15 min. After the incubation, the cultures were immediately put on ice to transfer to −80°C. RNA was isolated using RNAeasy Mini Kit following the manufacturer’s instructions. DNase I treatment was done using RapidOut DNA removal kit followed by cDNA synthesis via SuperScript IV Reverse Transcriptase utilizing random hexamer primers and ofrA-specific primer with non-staphylococcal tag (Supplementary Table 2). Quantitative PCR was performed with Biozym Blue S’Green qPCR Kit and rho and rpoB as the internal controls (Sihto et al., 2014).
Bacterial Survival Assay
We diluted overnight cultures 1:100 in fresh RPMI followed by incubation at 37°C with shaking at 200 rpm until mid-logarithmic phase. OD600 were adjusted to be 0.4 after collecting the bacteria by centrifugation for 10 min at 4°C and 4,000 rpm. After adding the indicated concentration and incubation at 37°C for the indicated time interval, serial dilutions of the bacterial suspension were made followed by plating of 80 μl on LB agar using single plate-serial dilution spotting (SP-SDS) method (Thomas et al., 2015).
The exposure time to 1.5 mM NaOCl was 30 min, while we challenged the bacteria against 40 or 30 mM H2O2 for 1 h. MHQ and methylglyoxal were exposed for 3 h.
In H2O2 survival assay and after 1-h exposure, samples were centrifuged at 4,000 rpm for 10 min and then resuspended in sterile PBS supplemented with 10 mg/ml catalase. In NaOCl survival assay, the serial dilutions of the bacteria were made in sterile LB to quench the remaining NaOCl.
Genomic DNA Isolation, Whole Genome Sequencing, and Variant Calling
Overnight cultures of S. aureus JE2 and EI046 (JE2ΔofrA) strains were grown at 37°C with shaking at 200 rpm in B-medium. Genomic DNA was extracted using DNeasy Blood and Tissue Kit (from Qiagen) according to the manufacturer’s protocol modification for Gram-positive bacteria. Whole genome sequencing and variant calling were done by MicrobesNG. The raw sequenced reads are deposited in SRA database (BioProject ID: PRJNA812552).
Macrophage Survival Assay
Similar protocol was applied as done in the study of Flannagan et al. (2018). RAW 264.1 macrophage cell line was prepared by passaging in RPMI medium supplemented with 10% FCS and Pen/Strep. Passages 12–15 were used in the bacterial survival assays. Bacteria grown in BHI to logarithmic phase were washed in sterile PBS, resuspended in RPMI, and added (∼5 × 107 CFU) to ∼5 × 106 RAW 264.1 cells in 24-well plates (MOI = 1:10).
Extracellular bacteria were killed by treatment with 150 μg/ml gentamicin in 1 h. We considered zero-time by adding fresh RPMI + 10% FCS medium. Samples (n = 5) were taken at time = 4, 24, and 48 h while the 4-h samples were set as the normalization factor as indicated (Fritsch et al., 2019). Bacterial counts were achieved as indicated above using SP-SDS method on LB agar.
Whole Blood Killing
Venous blood specimens were collected from four healthy human blood donors (age: 21–32 years, gender: two women and two men) in tubes supplemented with anti-coagulant (1.6 mg/ml EDTA). Blood was kept at room temperature (RT) until use. We used a similar protocol to van der Maten et al. (2017) with some modifications.
Bacteria grown in BHI to logarithmic phase were washed two times with sterile PBS. Afterward, 30 μl of the bacterial suspension (2.2 × 107 CFU/ml) was mixed with 100 μl of human blood (final concentration = 5 × 106 CFU/ml). Saponin (final concentration = 1%), immediately or after 60-min incubation at 37°C with shaking, was added to blood–bacteria mixture for cell lysis. After incubation for 20 min at 4°C, viable bacterial cells were determined using SP-SDS method on LB agar in two technical replicates.
RNA Isolation for RNA-Seq Experiment and Bioinformatic Analysis
Dilutions (1:100) of three independent overnight cultures of S. aureus JE2 and EI046 (JE2ΔofrA) strains were grown at 37°C with shaking at 200 rpm in RPMI medium until OD600 = 0.5. We extracted total RNA using RNeasy Mini Kit (from Qiagen) as in the manufacturer’s manual. DNA digestion was done using RapidOut DNA Removal Kit. Negative amplification in PCR using 16S rDNA primers was taken as an evidence of successful DNase treatment. Evaluation of RNA quality, rRNA depletion, cDNA library generation, and sequencing were done by the Core Unit Systems Medicine Facility at the University Hospital Würzburg. Adaptor trimming was done using Cutadapt software. Trimmed reads were aligned to the reference genome (NC_007793). We used READemption pipeline for reads mapping, coverage calculations, gene quantification, and differential gene expression analysis (Förstner et al., 2014). We developed scripts for gene set enrichment analysis (GSEA) using clusterProfiler (Wu et al., 2021). Regulon analysis was done using self-written R scripts. RNA-seq data are available in NCBI’s Gene Expression Omnibus (GSE196683).
Staphyloxanthin Assay
We diluted overnight cultures 1:100 in the respective medium (with supplementation if necessary) and allowed the bacterial growth in 37°C and shaking at 200 rpm for 16 h (stationary phase) (Sullivan and Rice, 2021). 2 ml of the bacteria was centrifuged at 16,000 rpm for 2 min and then washed in sterilized water. OD600 were recorded for normalization. 400 μl of methanol was added to the washed bacterial pellets and incubated at 55°C for 3 min. After centrifugation at 16,000 rpm for 2 min, 300 μl of the methanolic extract was added to 700 μl methanol. 200 μl of the solution was measured in three technical replicates at A465 with infinite 200Pro machine and methanol as a blank.
Growth Inhibition Assay
Overnight cultures in RPMI were 1:100 diluted into fresh RPMI medium and incubated in 37°C with shaking at 200 rpm until OD600 = 0.5. Then, we diluted the bacteria down to ∼5 × 105 cells and mixed with different concentration of streptonigrin (0–2 μg/ml). OD600 were measured using Synergy H1 plate reader.
Statistical Analysis
Statistical analysis was done under R version 3.6.1 using rstatix R package version 0.6.0 and ggpubr R package version 0.4.0. Statistical tests were indicated in the corresponding figure legends. We considered statistical significance if p < 0.05.
Results
OfrA Is an Old Yellow Enzyme Flavin Oxidoreductase
Utilizing TBLASTN of WP_000838037.1 (SAUSA300_0859 gene product) against S. aureus USA300_FPR3757 genome, we found that OYEs are encoded from two paralogous genes (SAUSA300_0859 and SAUSA300_0322). We propose to name SAUSA300_0859 as old yellow enzyme flavin oxidoreductase A (ofrA) and SAUSA300_0322 as ofrB. Upon NCBI’s CDD search, OfrA and OfrB contain “OYE_like_4_FMN” domain. OfrA and OfrB orthologs are conserved in B. subtilis as YqiG and YqjM, respectively (Figure 1A). Multiple sequence alignment shows that E. coli_NemA and P. fluorescens_XenB do not belong to the same class of Gram-positive OYEs (Figure 1A); rather, NemA and XenB belong to Class-I of classical OYEs whereas YqjM belongs to Class-III OYEs (Scholtissek et al., 2017). OfrA does not belong to any of the studied OYEs classes and represent a novel class of OYEs (Scholtissek et al., 2017). Therefore, we hypothesized that OfrA could play different roles in S. aureus than NemA in E. coli. Since S. aureus has a wide spectrum of genomic lineages, we were interested to study ofrA conservation in S. aureus strains and Firmicutes.

Figure 1. OfrA conservation in Firmicutes and staphylococci. (A) Phylogenetic analysis of OYE examples in Gram-positive S. aureus (OfrA, OfrB) and B. subtilis (YqiG, YqjM) compared to the Gram-negative E. coli (NemA) and Pseudomonas fluorescens (XenB) with distinctive multiple sequence alignment generated by Clustal Omega. (B) Maximum likelihood tree showing the evolutionary relationship of OfrA in different Firmicutes chromosomes. Multiple sequence alignment was utilized to build the phylogenetic tree using RAxML software and visualized with ggtree. (C) Bar chart shows the presence or absence of OfrA in seven Firmicutes genera (Bacillus, Paenibacillus, Streptococcus, Clostridium, Staphylococcus, Lactobacillus, and Enterococcus). Filtration criteria were based on 35% amino acid identity cutoff and protein length = 375 ± 38 amino acids, refer to section “Materials and Methods”. (D) OfrA conservation across the different staphylococci compared to the conservation of OfrB, MvaA (mevalonate pathway), and CrtM (staphyloxanthin biosynthesis) in the same genomes. Filled and unfilled circles indicate gene presence and absence, respectively. CrtM, squalene desaturase; MvaA, hydroxymethylglutaryl-CoA reductase; OfrA, old yellow enzyme flavin oxidoreductase A; OfrB, old yellow enzyme flavin oxidoreductase B.
OfrA Is Conserved in Staphylococci and Some Firmicutes
OfrA is encoded in all publicly available 749 chromosomes of S. aureus strains with 98–100% amino acid identities (Supplementary Table 3 and Supplementary Figure 1A). On genus level, OfrA is encoded from the 28 staphylococcal representative chromosomes. Staphylococcal OfrA orthologs cluster in three distinct clades (Figure 1B, Supplementary Figure 1B, and Supplementary Table 4). The phylogenetic tree illustrates that OYEs are not subjected to horizontal transfer; rather, they evolved within the encoding organism to adapt to certain function.
Although different OYEs are encoded in a number of representative Firmicutes’ chromosomes, OfrA-like orthologs are limited to only a few species (Figure 1C and Supplementary Figure 2). However, OfrB is not conserved across the different staphylococci (Figure 1D). In fact, BLASTP retrieved OfrA orthologs from the ofrB-minus genomes (such as S. saprophyticus, S. hominis, and S. epidermidis). From our analysis, we learned that some of the ofrB-minus genomes encode variants of the OYEs, other than OfrB, with varying lengths and/or other fused protein domains. Apparently, an earlier speciation event in a common ancestor resulted in OfrA and OfrB differences. Since OfrA is associated with MT02 resistance and is found conserved in staphylococci, we intended to understand the function of ofrA in the human pathogen S. aureus as an example of OYEs.
Electrophilic Stress Conditions Induce ofrA
Previously, we showed that the bisquaternary bisnaphthalimide MT02 induces ofrA (El-Hossary et al., 2018). Thus, we hypothesized that compounds with an electron-deficient center (such as electrophilic stress generators) could similarly induce ofrA. To avoid the quenching activity of standard laboratory media (TSB and LB), we used RPMI as a well-defined medium, which, in addition, mimics the host environment (Meerwein et al., 2020).
To screen for important induction conditions, we constructed a reporter strain (EI011) that harbors a chromosomally encoded β-galactosidase from a promoter-less lacZ gene under the control of PofrA (Supplementary Figure 3). Since β-galactosidase assay is protein-based, we chose 2-h exposure time to report for ofrA induction. We tested a range of RES conditions such as diamide, fosfomycin (Fosfo), formaldehyde (FA), methylglyoxal (MG), and MHQ at the MIC against EI011 to avoid false negative results from stressors’ toxicities in higher concentrations. The MIC concentration did not significantly affect the bacterial growth in the 2-h experimental time (Supplementary Figure 4).
β-Galactosidase assays suggest that diamide, formaldehyde, methylglyoxal, and MHQ induce ofrA (Figure 2A). MHQ results in the highest upregulation (21-folds), while formaldehyde, methylglyoxal, and diamide result in approximately fourfold upregulation. However, there is no upregulation upon exposure to fosfomycin (Figure 2A). Furthermore, the β-galactosidase assays show a dose-dependent induction by diamide, formaldehyde, methylglyoxal, and MHQ (Figure 2B).
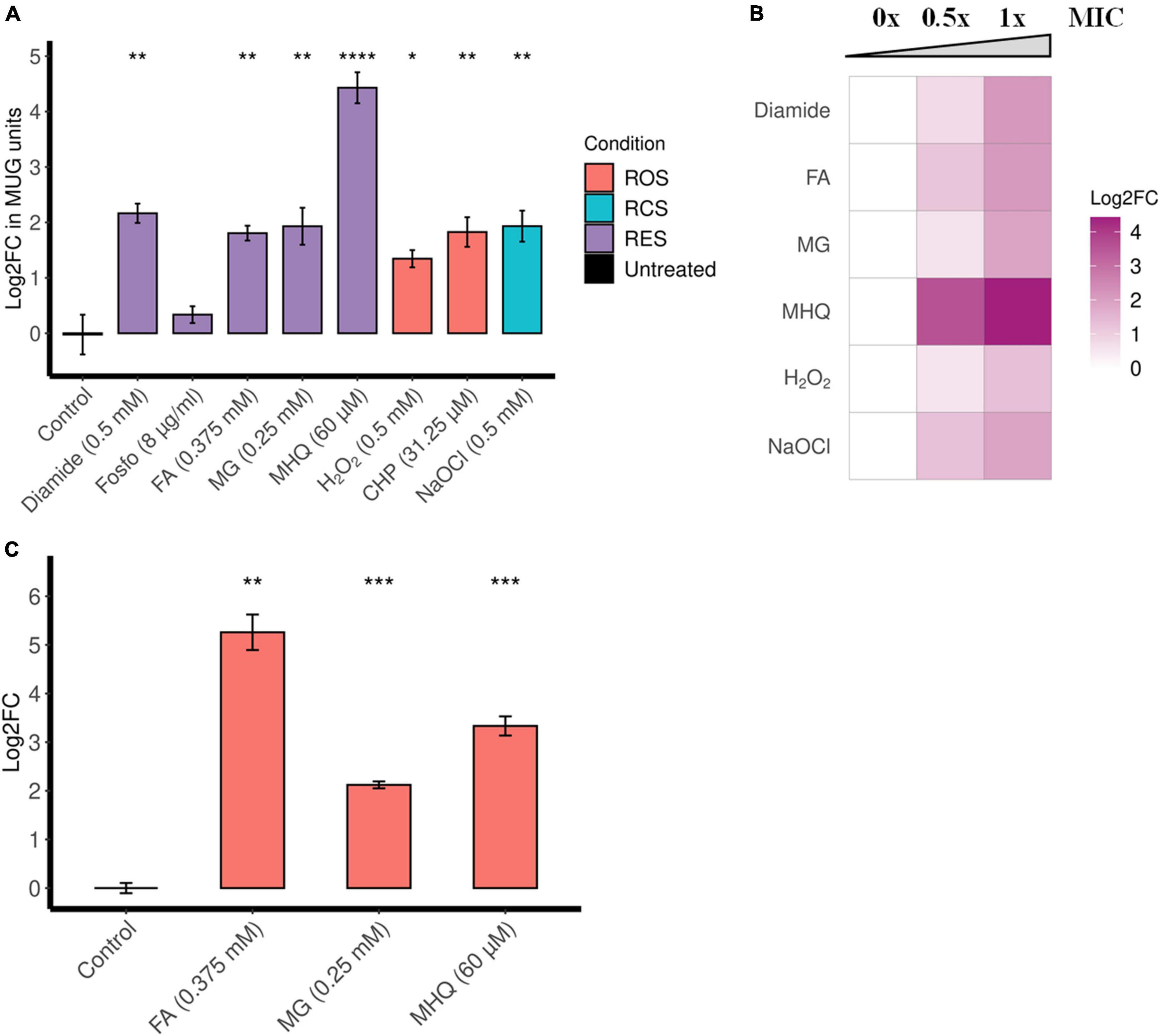
Figure 2. ofrA is induced in RES, ROS, and RCS in a dose-dependent manner. (A) ofrA induction conditions using the reporter strain EI011, which harbors a chromosomally encoded lacZ under PofrA (Supplementary Figure 3). After 2-h incubation with shaking at 37°C, β-galactosidase assay was used to report ofrA transcriptional level. The corresponding concentrations were indicated in the graph. A total of four biological replicates were compared to untreated controls using unpaired two-tailed Student’s t-test. Error bars represent the standard error of the means. (B) Dose dependency of ofrA induction using β-galactosidase assays. The highest concentration is the minimum inhibitory concentration (1 × MIC), the intermediate concentration is 0.5 × MIC, and compared to control (no compounds were added = 0 × MIC). Log2FC was calculated as average from three biological replicates. (C) RT-qPCR shows ofrA induction in S. aureus JE2 background in agreement with the reporter system. JE2 strain was cultivated in RPMI until mid-logarithmic phase (OD600 = 0.5). Samples were taken before adding the compounds as a control. After adding the compounds, the bacterial pellets were collected after 15 min of incubation at 37°C with shaking. A total of three biological replicates were compared to untreated controls via unpaired two-tailed Student’s t-test. Error bars represent the standard error of the means. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. CHP, cumene hydroperoxide; FA, formaldehyde; Fosfo, fosfomycin; MG, methylglyoxal; MHQ, methylhydroquinone; MIC, minimum inhibitory concentration; RCS, reactive chlorine species; RES, reactive electrophilic species; ROS, reactive oxygen species.
Since diamide is a non-specific disulfide-stress inducer, we were interested in the induction with toxic aldehydes (formaldehyde and methylglyoxal) and quinone-stress (MHQ). Reverse transcription quantitative polymerase chain reaction (RT-qPCR), comparing ofrA mRNA levels after 15-min exposure in S. aureus JE2 strain background, confirms the results obtained by the reporter strain (Figure 2C). After formaldehyde, methylglyoxal, and MHQ exposure for 15 min, there were 38-, 4-, and 10-fold upregulation in ofrA, respectively.
OfrA Protects Staphylococcus aureus in Quinone Stress and Against Toxic Aldehydes
Staphylococcus aureus faces electrophilic stress in many natural niches including host–pathogen interface. Therefore, we were interested in elucidating the role of OfrA in S. aureus survival in electrophilic stress conditions in more detail. To address this, we used a marker-less deletion mutant in S. aureus JE2 (EI046 = ΔofrA) as well as a complemented strain (EI047 = pofrA) which harbors a plasmid-based expression of ofrA from its natural promoter. To assure the absence of secondary mutations that could affect any phenotype, we sequenced the whole genome of JE2 and ΔofrA. The results show that there are no discriminating mutations in ΔofrA compared to JE2 except for ofrA mutation.
In bacterial survival assays, we compared the survival of ΔofrA vs. JE2 strain after 3-h exposure of 0.5 mM MHQ and 2 mM methylglyoxal compared to an untreated control. In quinone stress, JE2 strain survived 90%; however, ΔofrA survived only 61%. pofrA with restored ofrA expression complemented the survival defect phenotype (Figure 3A).
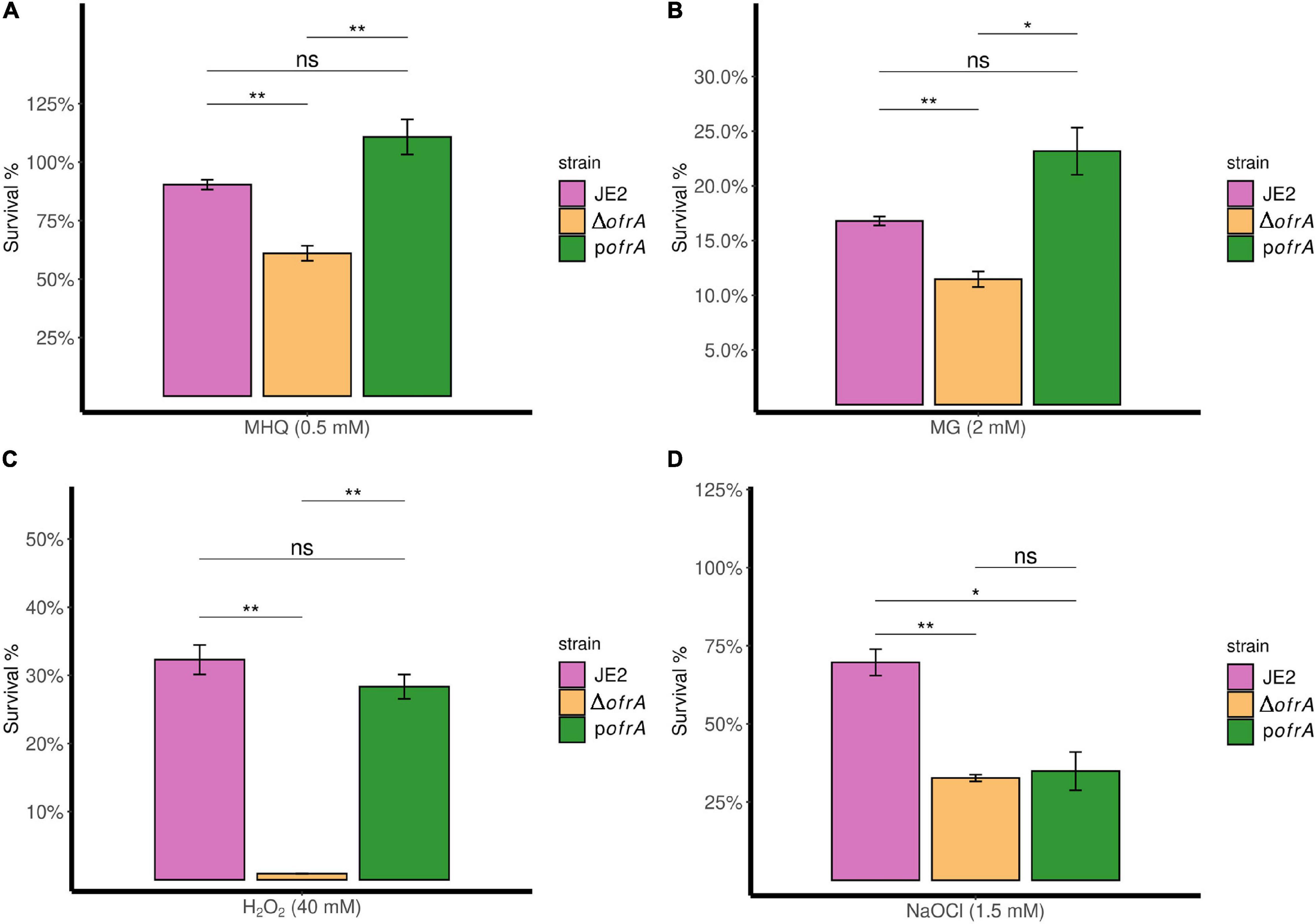
Figure 3. OfrA provides S. aureus resistance against quinone stress, toxic aldehydes, oxidative, and hypochlorite stresses. Bacterial survival assays in RES (MHQ and MG), ROS (H2O2), and RCS (NaOCl). The three strains; JE2, ΔofrA, and pofrA were allowed to grow until the logarithmic phase (OD600 = 0.4–0.6). Bacterial pellets were washed with sterile 1 × PBS, and the OD600 were adjusted to 0.4 in fresh RPMI. We added: (A) 0.5 mM MHQ for 3 h, (B) 2 mM MG for 3 h, (C) 40 mM H2O2 for 1 h, or (D) 1.5 mM NaOCl for 30 min. Samples were taken from the untreated control (for normalization) or with the stress conditions after the indicated time points for CFU determination using SP-SDS method on LB agar. Data represent four–five biological replicates. Error bars represent the standard error of the means. Statistical analysis was carried out using one-way ANOVA and pairwise t-test with Bonferroni p-value adjustment; ns, not significant; *p < 0.05; **p < 0.01. MG, methylglyoxal; MHQ, methylhydroquinone; RCS, reactive chlorine species; RES, reactive electrophilic species; ROS, reactive oxygen species.
ΔofrA exhibits a similar survival defect in methylglyoxal compared to the parental strain which we could restore in the complemented strain pofrA (Figure 3B). We concluded that OfrA is important to mediate quinone-stress and toxic aldehydes and that OfrA is an important factor in S. aureus defense against electrophilic stress conditions.
OfrA Affects the Survival of Staphylococcus aureus in Oxidative and Hypochlorite Stress
Since NemA was reported to be important in hypochlorite stress in E. coli, we also analyzed the role of OfrA in ROS and hypochlorite stress conditions. β-Galactosidase assays suggest that oxidative stress [H2O2 and cumene hydroperoxide (CHP)] and hypochlorite (NaOCl) stress induce ofrA (Figure 2A). Moreover, ofrA induction follows a dose response at different concentrations of H2O2 and NaOCl (Figure 2B).
Next, we analyzed the survival of the deletion mutant ΔofrA vs. JE2 wild-type (WT) after 1-h exposure of 40 mM H2O2 compared to untreated control. The ofrA mutant strain had decreased survival in 40 mM H2O2 compared to WT (Figure 3C). Moreover, the complementation in pofrA restored the WT phenotype indicating that OfrA enhances S. aureus survival in oxidative stress.
In 1.5 mM NaOCl, JE2 survived (70%) after 30 min compared to 33% survival of ΔofrA denoting OfrA importance in S. aureus survival against NaOCl (Figure 3D). However, pofrA failed to complement the mutant phenotype. We hypothesized that complementation with a high-copy number plasmid could result in overconsumption of cellular resources and therefore a decreased resistance against NaOCl taking in consideration the devastating non-specific effects of HOCl (da Cruz Nizer et al., 2020). From the whole genome sequencing results, there are no secondary mutations that could affect NaOCl survival phenotype. In addition, NaOCl results in ofrA induction (Figure 2A). Hence, we concluded that OfrA is also an important factor in S. aureus defense against ROS and hypochlorite stress conditions. To assure that there are no effects of strain growth behavior on the survival phenotypes, we observed the growth kinetics of the logarithmic phase cells of the three strains in RPMI medium (Supplementary Figure 5). Similar growth kinetics suggested that the growth behavior did not contribute to any of the measured survival phenotypes.
Staphylococcus aureus USA300 JE2ΔofrA Shows Decreased Fitness at the Host–Pathogen Interface by Survival Defect in Murine Macrophages RAW 264.7 Cell Line and Whole Human Blood
Macrophages produce reactive oxygen, chlorine, and electrophilic species as the killing factors against internalized S. aureus (Moldovan and Fraunholz, 2019). Since OfrA is important in survival in these stress conditions, we wondered whether ofrA mutation results in defective macrophage survival.
After 24 h, ΔofrA survival was reduced in RAW 264.7 macrophages compared to JE2 but this was not statistically significant. However, the difference became significant after 48 h. After 48 h, ΔofrA survived significantly (∼50%) less than JE2 in RAW 264.7 cell line (Figure 4A). In the complemented strain, the difference between pofrA and ΔofrA was statistically significant even after 24 h. The bigger difference could be explained by ofrA dosage effect from the high-copy number plasmid utilized in the complementation. We concluded that OfrA affects the bacterial fitness by enhancing S. aureus JE2 survival in macrophages.
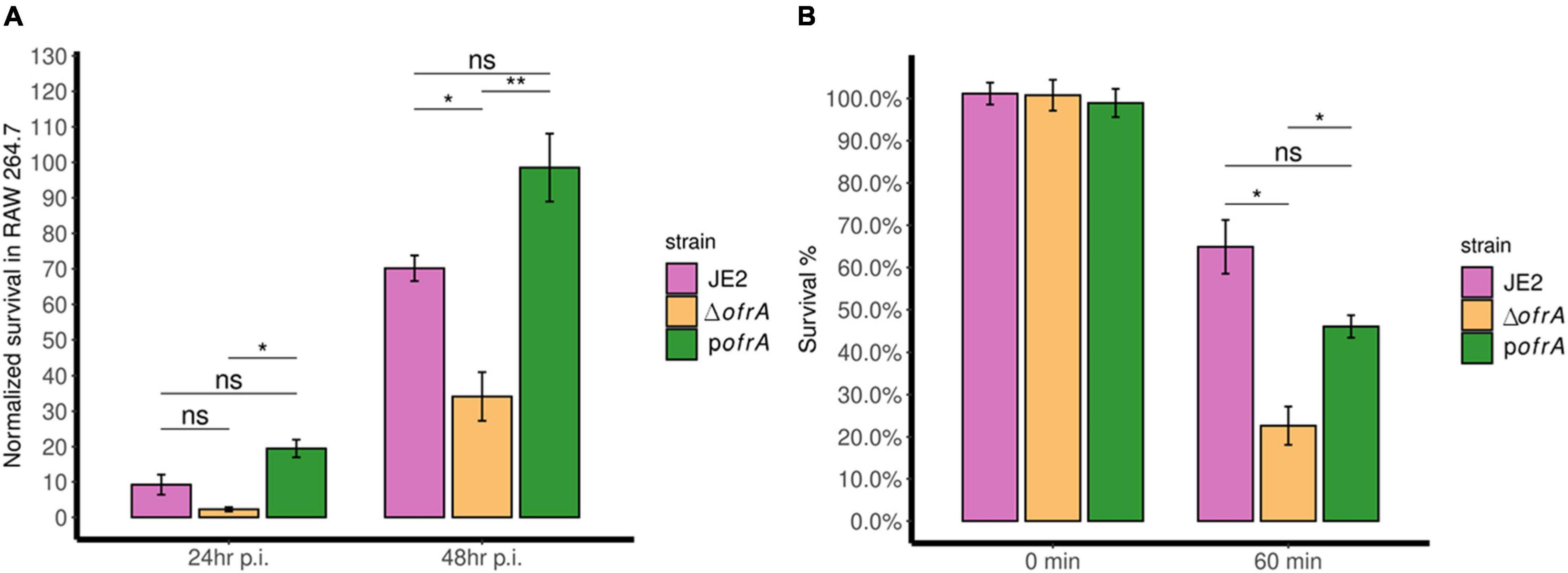
Figure 4. OfrA promotes S. aureus fitness at the host-pathogen interface by enhancing survival in RAW 264.7 macrophages and whole human blood. (A) Macrophage survival assay. JE2, ΔofrA, and pofrA were added to RAW 264.7 macrophage cell lines in 1:10 MOI. After 1 h of infection, gentamicin (150 μg/ml) was used to kill extracellular bacteria for 1 h. Fresh RPMI + 10% FCS was added (t = 0). At (t = 4 h), viable intracellular bacteria were determined and the CFU/ml was used as a normalization factor. Samples were taken at (t = 24 and 48 h). The assay was repeated for three independent experiments. Data represent five biological replicates from one of the three experiments. (B) Whole human blood killing assay. A total of 5 × 106 CFU/ml of each strain were incubated in whole human blood for 60 min at 37°C with continuous shaking. The number of viable bacteria was enumerated after serial dilutions in sterile 1 × PBS using SP-SDS method on LB agar and normalized to the viable cells without incubation. The experiment was repeated in blood taken from four different blood donors. Data represent four biological replicates from one of the four experiments. Error bars represent the standard error of the means. Statistical analysis was carried out using one-way ANOVA and pairwise t-test with Bonferroni correction p-value adjustment; ns, not significant; *p < 0.05; **p < 0.01.
The bacteria–immune response interaction in human blood determines the fatality of S. aureus-mediated bacteremia. We wondered whether OfrA contributes to S. aureus JE2 virulence via promoting survival in human blood. After 1 h of incubation with whole human blood, ΔofrA survives (∼23%) compared to the WT (∼65%) (Figure 4B). The complementation in pofrA restores the survivability of the mutant back to ∼46% (Figure 4B). In conclusion, ofrA contributes to S. aureus survival in whole human blood.
ofrA Deletion Promotes Transcriptional Changes in Some Redox and Stress-Related Genes
To understand ofrA function in S. aureus, we compared the transcriptome of ΔofrA vs. JE2 in mid-logarithmic phase in RPMI. Through RNA-seq experiment, we found that the ofrA mutant had decreased RNA abundances corresponding to 93 genes and increased RNA abundances corresponding to 95 genes (Supplementary Table 5). Several redox-related (SAUSA300_0339, SAUSA300_0340, SAUSA300_0212, SAUSA300_0213, ypdA, and cymR) and stress-related genes (csbD, clpB, sigB, and rsbW) are deregulated. Using regulon analysis and GSEA, we observed the following: (1) one-carbon metabolism is inhibited in ΔofrA indicating an unbalanced redox status (Shetty and Varshney, 2021), and (2) the carotenoid biosynthesis (crtOPQMN) is suppressed in the mutant (Supplementary Table 6).
To validate the results of RNA-seq analysis, we performed RT-qPCR to quantify the mRNA abundances of crtM, acuA, and rocD genes. RT-qPCR confirmed the results obtained by the RNA-seq analysis. In RT-qPCR, log2 (fold change) of crtM expression is −0.7 ± 0.1 in ΔofrA compared to JE2 (Supplementary Figure 6). Moreover, log2 (fold change) of acuA and rocD expression is −0.6 ± 0.2 and 2.9 ± 0.2, respectively (Supplementary Figure 6).
Suppressed Staphyloxanthin Production in ΔofrA Is Glucose-Independent But Mevalonate-Dependent
The carotenoid pigment (staphyloxanthin) production is mediated via the crtOPQMN operon (Götz, 2005). Staphyloxanthin (STX) is a virulence factor that affects the survival of S. aureus against oxidative stress and human neutrophils, so we were interested in quantifying STX levels in the ofrA mutant (Clauditz et al., 2006). Indeed, STX is decreased in ΔofrA compared to JE2 and pofrA (Figure 5A).
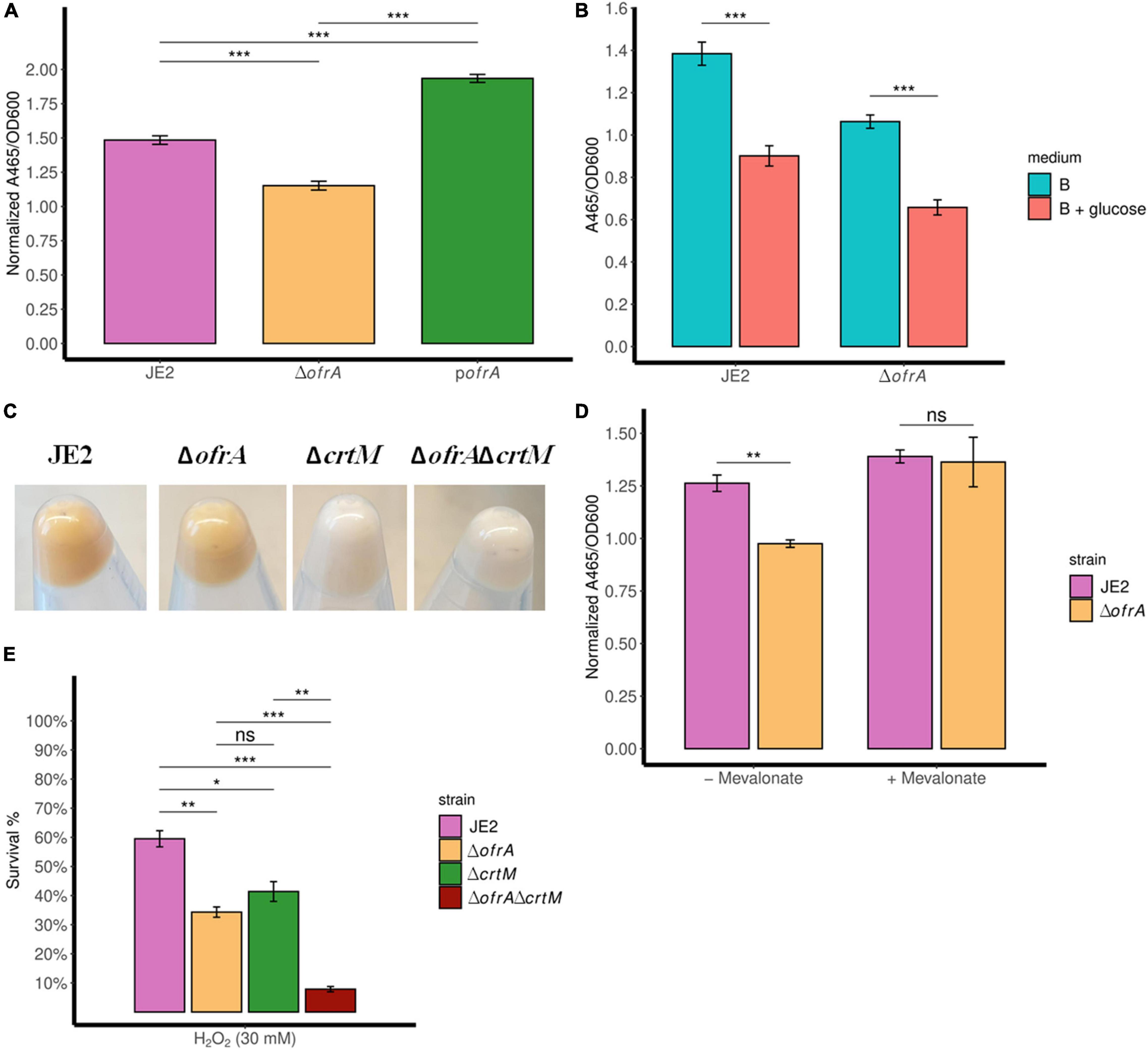
Figure 5. ofrA mutation decreases STX production via the upper mevalonate pathway but cannot solely explain ROS hypersensitivity. Staphyloxanthin assay showing STX levels in TSB medium (A,C), B-medium (B), and RPMI (D). The strains were grown in overnight culture in the respective medium without any supplementation. Then, we diluted the overnight cultures 1:100 in fresh medium without or with supplementation; 0.5% glucose (B) or 1 mM mevalonate (D). After 24 h, the bacteria were collected and washed with sterile water. OD600 were recorded for normalization. STX was extracted using methanol (refer to section “Materials and Methods”). A465 were used for measuring the extracted STX. Error bars represent the standard error of the means (A,D) and standard deviation (B) of four biological replicates. Statistical analysis was carried out using unpaired two-tailed Student’s t-test (B,D) or one-way ANOVA and pairwise t-test with Bonferroni p-value adjustment (A); ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001. (E) Bacterial survival assays showing crtM mutation additive effect to ofrA mutation in ROS hypersensitivity. The strains were grown in overnight culture in RPMI medium. We diluted the overnight cultures 1:100 in fresh RPMI until mid-logarithmic phase. Cells were harvested by centrifugation and washed with sterile PBS. OD600 were adjusted to 0.4. Bacteria were challenged with 30 mM H2O2. After 1 h of exposure to 30 mM H2O2, viable cells were diluted in PBS after catalase treatment for residual H2O2. Samples were taken from the untreated control (for normalization) or with the stress condition after 1 h for CFU determination using SP-SDS method on LB agar. STX, staphyloxanthin.
Acetyl-CoA is the key input of mevalonate pathway to produce farnesyl pyrophosphate (FPP), which enters the crtOPQMN pathway (Pelz et al., 2005). STX was previously shown to be decreased with glucose due to intracellular acetyl-CoA loss (Tiwari et al., 2018). To test the acetyl-CoA-dependency of STX phenotype in the ofrA mutant, we measured STX level in B-medium (contains no glycolytic substrates) ± 0.5% glucose.
As expected, glucose decreased STX levels in WT (Figure 5B). However, the intracellular acetyl-CoA loss did not affect the ratio between ΔofrA and JE2 in STX production (Figure 5B). This result suggests that ΔofrA does not have decreased STX production via change in acetyl-CoA concentration. Conversely, the loss of crtM in JE2 and ΔofrA, transduced from strain Newman (Clauditz et al., 2006), resulted in the disappearance of the ofrA-dependent phenotype, and both strains become white (Figure 5C; Reichert et al., 2018). Therefore, we concluded that ofrA mutation could affect the mevalonate pathway.
The mevalonate pathway is classified into upper (mvaS, mvaA) and lower (mvaK1, mvaK2, and mvaD) mevalonate pathways (Reichert et al., 2018). The output of the upper mevalonate pathway is the mevalonate itself. So, we were interested to understand the dependency of ofrA-mediated STX phenotype on the presence of mevalonate.
We compared the STX production ±1 mM mevalonate in RPMI medium. The presence of mevalonate results in the disappearance of ofrA-mediated phenotype in ΔofrA compared to JE2 (Figure 5D). Therefore, we deduced that the ofrA mutant has decreased STX production via the upper mevalonate pathway in S. aureus.
Lower Staphyloxanthin Generation Cannot Solely Explain Reactive Oxygen Species Hypersensitivity in ΔofrA
To understand whether ROS hypersensitivity is linked to decreased STX, we challenged JE2ΔcrtM and JE2ΔcrtMΔofrA strains against H2O2 in the survival assay. As expected, crtM and ofrA mutations in JE2 resulted in decreased survival in ROS (Figure 5E). If low STX production is responsible for ROS-mediated killing, the double deletion mutants shall behave as ΔcrtM and ΔofrA. Contrary to this hypothesis, the double mutation in both genes, JE2ΔcrtMΔofrA, causes H2O2 hypersensitivity and more killing in 30 mM H2O2 (Figure 5E). Thus, crtM and ofrA are important in ROS survival but independent of each other.
OfrA Contributes to Reactive Oxygen Species Tolerance Through Affecting Thiol-Dependent Redox Homeostasis
From RNA-seq analysis, we know that ofrA mutation does not result in upregulation of sodA, sodM, katA, peroxidases, and hmp, which indicates that intracellular levels of O2– and H2O2 are within the WT levels (Supplementary Table 5). Therefore, ofrA-dependent ROS hypersensitivity is downstream to H2O2 production.
The only plausible explanation of ROS hypersensitivity we had is that ofrA contributes to the repair mechanism of thiol-oxidation caused by H2O2. This notion is supported by the fact that ofrA is generally induced with electrophilic, hypochlorite, and oxidative stress. Thiourea scavenges the hydroxyl radical that should decrease the H2O2-mediated killing (Wasil et al., 1987). As expected, the survival of ΔofrA was lower than JE2 strain in H2O2 (Figure 6A). Addition of 120 mM thiourea resulted in increased S. aureus JE2 survival and ΔofrA up to a similar level (Figure 6A). Therefore, we concluded that ofrA contributes to oxidative stress tolerance via a repair mechanism downstream to H2O2 but upstream to hydroxyl radical-mediated lethality.
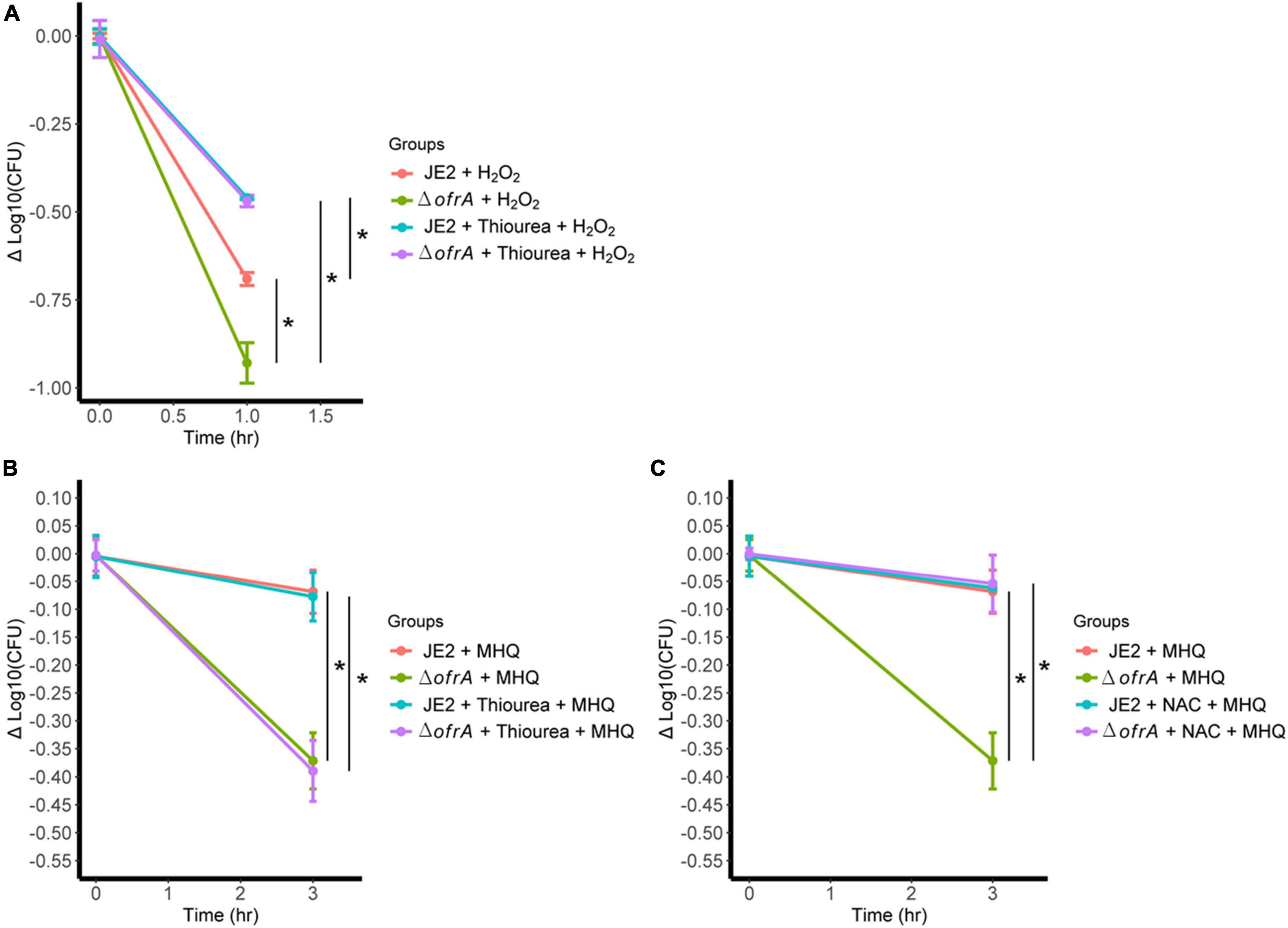
Figure 6. ofrA mutation increases ROS-mediated killing via disturbing thiol-dependent redox homeostasis. (A) Bacterial survival assay in 40 mM H2O2 with or without 120 mM thiourea. The strains were grown in overnight culture in RPMI medium. We diluted the overnight cultures 1:100 in fresh RPMI until mid-logarithmic phase. Cells were harvested by centrifugation and washed with sterile PBS. OD600 were adjusted to 0.4. Bacteria were challenged with H2O2 with or without 120 mM thiourea. Bacterial survival assay in 0.5 mM MHQ with or without 120 mM thiourea (B) or 1.25 mM NAC (C). Data represent four biological replicates. Error bars represent the standard error of the means. Statistical analysis was carried out using one-way ANOVA and pairwise t-test with Bonferroni p-value adjustment; *p < 0.05. MHQ, methylhydroquinone; NAC, N-acetyl cysteine.
Since MHQ is the highest induction condition (Figure 2A) and the hydroxyl radical is the main killing mechanism after H2O2 challenge (Figure 6A), we hypothesized that the survival defect of the mutant in ROS is secondary to disruption of thiol-dependent homeostasis upon ROS challenging.
In S. aureus, MHQ imposes oxidative and electrophilic stress (Fritsch et al., 2019). To test our hypothesis, we conducted MHQ survival assay ± N-acetyl cysteine (NAC). NAC supports the thiol-dependent redox homeostasis that acts as both reactive oxygen and electrophilic species scavenger (Pedre et al., 2021), and thiourea as ROS scavenger via thiol-independent mechanism. If our hypothesis was correct, thiourea would not be able to quench the electrophilic stress. 120 mM thiourea does not abolish MHQ toxicity in ofrA mutation; however, 1.25 mM NAC does (Figures 6B,C). We, therefore, concluded that ofrA plays a role in the thiol-dependent redox homeostasis, which affects the survival in oxidative, electrophilic, and hypochlorite stress, and that is an essential function during infection inside macrophages, and in human blood.
Discussion
Old yellow enzyme family proteins are widely distributed in the bacterial kingdom with yet-to-be explored functions. In this study, we aimed at identifying the physiological role of the staphylococcal conserved OYE OfrA in S. aureus. We learnt that OfrA is an important resistance factor against reactive species (RES, RCS, and ROS). Moreover, the virulence of S. aureus is decreased by compromised survival in murine RAW 264.7 macrophages and whole human blood after ofrA deletion.
We noticed that ofrA mRNA levels are stable in different media and growth phases and were only slightly (approximately two to fourfolds) upregulated under all tested stress conditions except for MHQ induction. One reason for that behavior could be the promiscuity known to OYEs so that higher protein levels could cause cellular toxicity from the low-substrate specificities (Lee et al., 2013). Noteworthy, in the complementation analysis, we tried to use a complemented strain in which ofrA transcription is initiated via xylose-dependent promoter. No growth could be noticed using 0.5% xylose for overexpression possibly because of the aforementioned cellular-mediated toxicities from inducing a high gene dosage of ofrA or overconsuming the reducing equivalents NAD(P)H.
In agreement to our β-galactosidase reporter system, and RT-qPCR validation, ofrA (SACOL0959 in S. aureus COL and SA0817 in S. aureus N315) upregulation could be observed in previous transcriptome studies in the presence of MHQ (Fritsch et al., 2019), NaOCl (Loi et al., 2018), and H2O2 (Chang et al., 2006). Moreover, reactive sulfur species (RSS) result in ofrA induction in S. aureus (Peng et al., 2017; Loi et al., 2019). Since ofrA induction conditions include RES, ROS, RCS, and RSS, we believe that ofrA transcriptional regulation responds to a wide variety of conditions that disrupts the redox homeostasis.
The OYE NemA of E. coli was reported to be important in hypochlorite stress (Gray et al., 2013; Lee et al., 2013; Ozyamak et al., 2013). Remarkedly, in S. aureus, we observed that OfrA is important in protecting against intoxication by ROS and toxic aldehydes in addition to hypochlorite stress. Therefore, we conclude that the compromised survival phenotype of ofrA mutant after oxidative, electrophilic, and hypochlorite stress could be due to a defect in a common redox-balancing mechanism important in the three conditions. Most likely, this involves thiol-disulfide homeostasis of so far unknown proteins as shown by our quenching experiments using NAC and thiourea. In relation, methylglyoxal is detoxified via both thiol-dependent and -independent pathways. In agreement to our latter conclusion, the thiol-dependent mechanism is the essential pathway for S. aureus survival against methylglyoxal (Imber et al., 2018).
In the classical mevalonate pathway, HMG-CoA reductase is the rate-limiting step for the mevalonate production and essential for S. aureus growth in the absence of mevalonate supplementation (Wilding et al., 2000; Matsumoto et al., 2016). HMG-CoA reductase uses NAD(P)H as a reducing equivalent for the mevalonate production. Therefore, the availability of NAD(P)H could be the critical factor to explain the decreased mevalonate production and hence the staphyloxanthin production. S. aureus JE2ΔofrA has a decreased levels of staphyloxanthin compared to its parental strain (Figures 5A–D). Since OYEs use the reducing equivalents NAD(P)H to regenerate their prosthetic FMN group as an integral part of their activity (Toogood et al., 2010), we believe that ofrA mutation affects the NAD(P)H/NAD(P) ratio in S. aureus and staphyloxanthin production.
S. aureusΔofrA shows a quick survival defect in whole human blood. Neutrophils in the human blood represent 60% of the leukocyte population and kill the invading bacteria via ROS. We assume that ofrA mutation-dependent killing mechanism in whole blood is due to the ROS generated by neutrophils. One possibility could be that higher levels of intracellular iron could indirectly enhance the production of HO⋅ via Fenton reaction and result in higher toxicities from the same dose of H2O2 (Wang and Zhao, 2009). However, we could exclude these mechanisms as growth inhibition experiments using streptonigrin, which requires intracellular iron for its antimicrobial activities (White and Yeowell, 1982; Duggan et al., 2020), have shown a similar growth of WT and mutant strains indicating that both strains contain comparable amounts of intracellular iron (Supplementary Figure 7). Since also the survival rate of the mutant in macrophages was reduced, we conclude that OfrA is an important factor to resist killing of S. aureus by redox-based molecules produced within phagocytes. Interestingly, a knockout of NTR2 gene, which encodes for an OYE orthologous to OfrA, in the parasite Leishmania results in reduced replication within macrophages (Wyllie et al., 2016). Therefore, OYEs could function as anti-stress mechanism included in different eukaryotic and bacterial backgrounds with chromosomal evolution for better fitting the special niche of the organism.
In our attempt to understand the role of OfrA in S. aureus, we investigated a transcriptomic approach. ofrA mutation leads to slight transcriptomic changes at standard growth conditions in RPMI. Although no specific pattern of deregulated genes could be found, a number of genes involved in redox and stress-related mechanisms were affected in the mutant which reflects the proposed broad substrate specificity of OYEs. The transcriptome data are in line with our hypothesis that OfrA is a member of redox buffering systems that regularly functions under stress and is linked to energy metabolism. Since OfrA has a proposed function in thiol-dependent redox homeostasis, we believe that a targeted thiol redox proteomic approach will be a promising approach in studying the effect of ofrA mutation.
We present our current understanding of OfrA functions in S. aureus based on our results in Figure 7. Our findings suggest that OfrA participates in oxidative, hypochlorite, and electrophilic stress mediation. This has relevance at the bacteria–host interface as OfrA supports intra-macrophage replication and survival. Moreover, OfrA protects S. aureus against killing in whole human blood. In addition, STX production is inhibited in the ofrA mutant via the upper mevalonate pathway, which is, however, not the main mechanism of OfrA-mediated protection against ROS. Overall, we provide evidence that OfrA protects S. aureus against numerous stress types through thiol-dependent redox homeostasis.
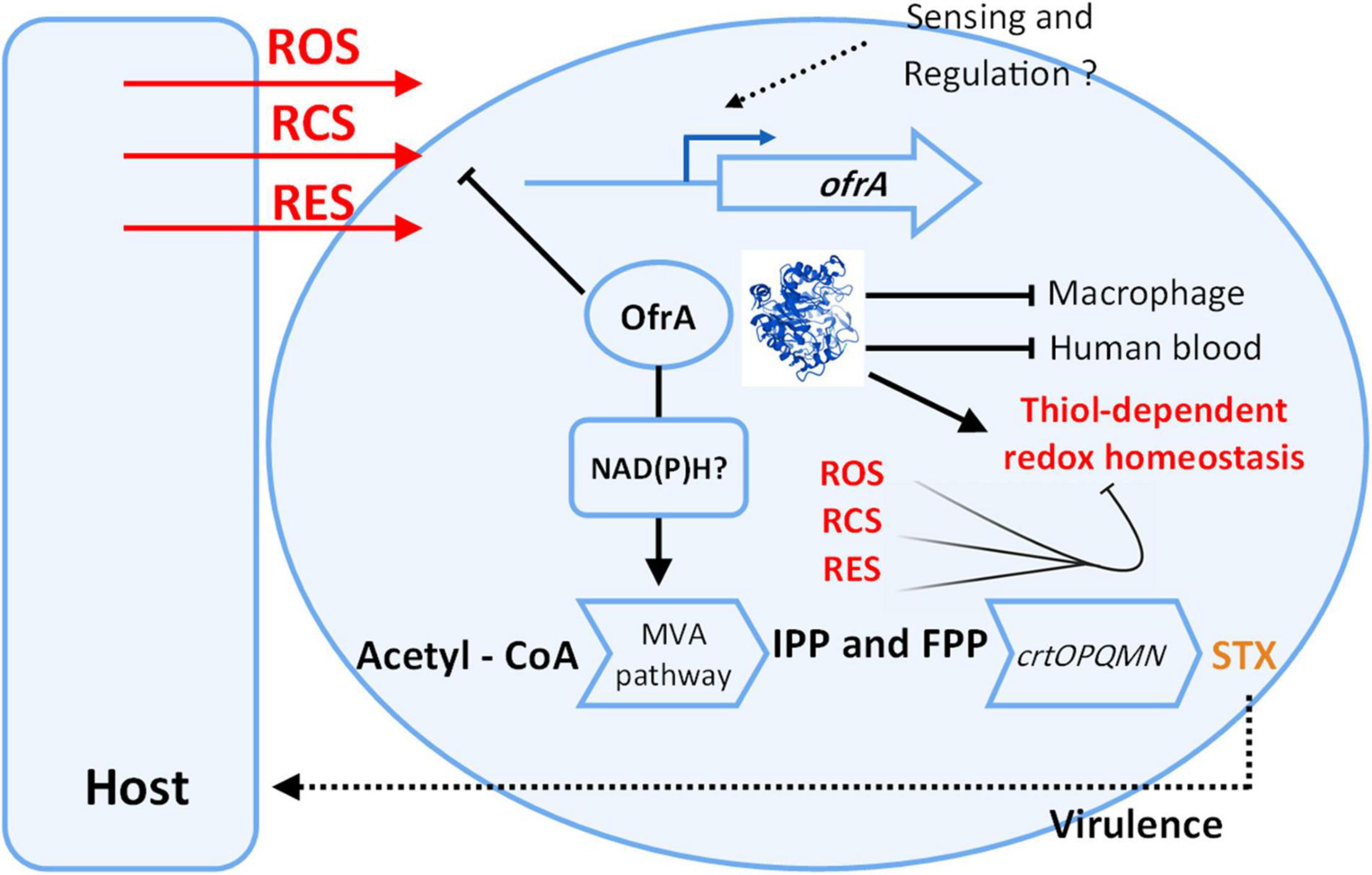
Figure 7. Cartoon representation shows our current understanding of ofrA function in S. aureus. OfrA protein 3D structure was predicted via AlphaFold. For 3D visualization, refer to alphafold.ebi.ac.uk/entry/Q2FZU7. ofrA is induced in ROS, RES, and RCS conditions which are available at the host-S. aureus interface. We showed that ofrA is an important factor in S. aureus resistance to the aforementioned stress conditions. ofrA contributes to S. aureus virulence via human blood and macrophage survival. ofrA mutation is involved in decreased STX production via MVA pathway. Both STX and ofrA protects S. aureus against oxidative stress via different mechanisms. ofrA supports the thiol-dependent redox homeostasis. FPP, farnesyl pyrophosphate; IPP, isopentenyl pyrophosphate; MVA, mevalonate; STX, staphyloxanthin.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The RNA-seq datasets for this study can be found in the NCBI’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196683). The raw sequenced reads of the whole-genome sequencing are deposited in the SRA database (BioProject ID: PRJNA812552).
Author Contributions
EI performed the experimental work, designed the experiments, analyzed the data, and wrote the manuscript. KO supervised the project, obtained the funding, discussed the data, and revised the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
EI was supported by a grant of the German Excellence Initiative to the Graduate School of Life Sciences, University of Würzburg. Furthermore, this work was supported by a Seed Grant of the Helmholtz-Institut für RNA-basierte Infektionsforschung (HIRI), and DFG project OH97/8-1. This publication was supported by the Open Access Publication Fund of the University of Würzburg.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge Friedrich Götz for crtM mutant. We thank Wilma Ziebuhr, Martin Fraunholz, and Tobias Hertlein for critical thoughts and discussions. We would also like to thank Jessica Brock for technical assistance in the whole human blood killing assay.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.888140/full#supplementary-material
Footnotes
References
Brückner, R. (2006). Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151, 1–8. doi: 10.1111/j.1574-6968.1997.tb10387.x
Chang, W., Small, D. A., Toghrol, F., and Bentley, W. E. (2006). Global Transcriptome Analysis of Staphylococcus aureus Response to Hydrogen Peroxide. J. Bacteriol. 188, 1648–1659. doi: 10.1128/jb.188.4.1648-1659.2006
Chen, N. H., Djoko, K. Y., Veyrier, F. J., and McEwan, A. G. (2016). Formaldehyde Stress Responses in Bacterial Pathogens. Front. Microbiol. 7:257. doi: 10.3389/fmicb.2016.00257
Clauditz, A., Resch, A., Wieland, K.-P., Peschel, A., and Götz, F. (2006). Staphyloxanthin Plays a Role in the Fitness of Staphylococcus aureus and Its Ability To Cope with Oxidative Stress. Infect. Immun. 74, 4950–4953. doi: 10.1128/IAI.00204-06
da Cruz Nizer, W. S., Inkovskiy, V., and Overhage, J. (2020). Surviving Reactive Chlorine Stress: responses of Gram-Negative Bacteria to Hypochlorous Acid. Microorganisms 8:1220. doi: 10.3390/microorganisms8081220
Duggan, S., Laabei, M., Alnahari, A. A., O’Brien, E. C., Lacey, K. A., Bacon, L., et al. (2020). A small membrane stabilizing protein critical to the pathogenicity of staphylococcus aureus. Infect. Immun. 88, e162–e120. doi: 10.1128/IAI.00162-20
El-Hossary, E. M., Förstner, K. U., François, P., Baud, D., Streker, K., Schrenzel, J., et al. (2018). A Novel Mechanism of Inactivating Antibacterial Nitro Compounds in the Human Pathogen Staphylococcus aureus by Overexpression of a NADH-Dependent Flavin Nitroreductase. Antimicrob. Agents Chemother. 62, e1510–e1517. doi: 10.1128/AAC.01510-17
Farmer, E. E., and Davoine, C. (2007). Reactive electrophile species. Curr. Opin. Plant Biol. 10, 380–386. doi: 10.1016/J.PBI.2007.04.019
Fischbach, M. A., and Walsh, C. T. (2009). Antibiotics for Emerging Pathogens. Science 325, 1089–1093. doi: 10.1126/science.1176667
Flannagan, R. S., Kuiack, R. C., McGavin, M. J., and Heinrichs, D. E. (2018). Staphylococcus aureus uses the GraXRS regulatory system to sense and adapt to the acidified phagolysosome in macrophages. MBio 9, e1143–e1118. doi: 10.1128/mBio.01143-18
Förstner, K. U., Vogel, J., and Sharma, C. M. (2014). READemption—a tool for the computational analysis of deep-sequencing–based transcriptome data. Bioinformatics 30, 3421–3423. doi: 10.1093/BIOINFORMATICS/BTU533
Fritsch, V. N., Van Loi, V., Busche, T., Sommer, A., Tedin, K., et al. (2019). The MarR-Type Repressor MhqR Confers Quinone and Antimicrobial Resistance in Staphylococcus aureus. Antioxid. Redox Signal. 31, 1235–1252. doi: 10.1089/ars.2019.7750
Geiger, T., Francois, P., Liebeke, M., Fraunholz, M., Goerke, C., Krismer, B., et al. (2012). The Stringent Response of Staphylococcus aureus and Its Impact on Survival after Phagocytosis through the Induction of Intracellular PSMs Expression. PLoS Pathog. 8:e1003016. doi: 10.1371/journal.ppat.1003016
Götz, F. (2005). “Genetic and Biochemical Analysis of the Biosynthesis of the Orange Carotenoid Staphyloxanthin of Staphylococcus aureus,” in Microbial Fundamentals of Biotechnology, eds V. Braun and F. Götz (Hoboken: Wiley), 284–294. doi: 10.1002/3527602720.CH17
Gray, M. J., Wholey, W. Y., Parker, B. W., Kim, M., and Jakob, U. (2013). NemR is a bleach-sensing transcription factor. J. Biol. Chem. 288, 13789–13798. doi: 10.1074/jbc.M113.454421
Groitl, B., and Jakob, U. (2014). Thiol-based redox switches. Biochim. Biophys. Acta Proteins Proteom. 1844, 1335–1343. doi: 10.1016/j.bbapap.2014.03.007
Guerra, F. E., Borgogna, T. R., Patel, D. M., Sward, E. W., and Voyich, J. M. (2017). Epic Immune Battles of History: neutrophils vs. Staphylococcus aureus. Front. Cell. Infect. Microbiol. 7:286. doi: 10.3389/fcimb.2017.00286
Ibrahim, E. S., Kashef, M. T., Essam, T. M., and Ramadan, M. A. (2017). A Degradome-Based Polymerase Chain Reaction to Resolve the Potential of Environmental Samples for 2,4-Dichlorophenol Biodegradation. Curr. Microbiol. 74, 1365–1372. doi: 10.1007/s00284-017-1327-6
Imber, M., Van Loi, V., Reznikov, S., Fritsch, V. N., Pietrzyk-Brzezinska, A. J., et al. (2018). The aldehyde dehydrogenase AldA contributes to the hypochlorite defense and is redox-controlled by protein S-bacillithiolation in Staphylococcus aureus. Redox Biol. 15, 557–568. doi: 10.1016/j.redox.2018.02.001
Imlay, J. A. (2019). Where in the world do bacteria experience oxidative stress? Environ. Microbiol. 21, 521–530. doi: 10.1111/1462-2920.14445
Kitzing, K., Fitzpatrick, T. B., Wilken, C., Sawa, J., Bourenkov, G. P., Macheroux, P., et al. (2005). The 1.3 Å crystal structure of the flavoprotein YqjM reveals a novel class of old yellow enzymes. J. Biol. Chem. 280, 27904–27913. doi: 10.1074/jbc.M502587200
Klebanoff, S. J., Kettle, A. J., Rosen, H., Winterbourn, C. C., and Nauseef, W. M. (2013). Myeloperoxidase: a front-line defender against phagocytosed microorganisms. J. Leukoc. Biol. 93:185. doi: 10.1189/JLB.0712349
Krismer, B., Weidenmaier, C., Zipperer, A., and Peschel, A. (2017). The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat. Rev. Microbiol. 15, 675–687. doi: 10.1038/nrmicro.2017.104
Lee, C., Shin, J., and Park, C. (2013). Novel regulatory system nemRA-gloA for electrophile reduction in Escherichia coliK-12. Mol. Microbiol. 88, 395–412. doi: 10.1111/mmi.12192
Linzner, N., Van Loi, V., Fritsch, V. N., and Antelmann, H. (2020). Thiol-based redox switches in the major pathogen Staphylococcus aureus. Biol. Chem. 0, 333–361. doi: 10.1515/hsz-2020-0272
Van Loi, V., Busche, T., Tedin, K., Bernhardt, J., Wollenhaupt, J., et al. (2018). Redox-Sensing Under Hypochlorite Stress and Infection Conditions by the Rrf2-Family Repressor HypR in Staphylococcus aureus. Antioxid. Redox Signal. 29, 615–636. doi: 10.1089/ars.2017.7354
Van Loi, V., Huyen, N. T. T., Busche, T., Tung, Q. N., Gruhlke, M. C. H., et al. (2019). Staphylococcus aureus responds to allicin by global S-thioallylation – Role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress. Free Radic. Biol. Med. 139, 55–69. doi: 10.1016/j.freeradbiomed.2019.05.018
Matsumoto, Y., Yasukawa, J., Ishii, M., Hayashi, Y., Miyazaki, S., and Sekimizu, K. (2016). A critical role of mevalonate for peptidoglycan synthesis in Staphylococcus aureus. Sci. Rep. 6:22894. doi: 10.1038/srep22894
Meerwein, M., Tarnutzer, A., Böni, M., Van Bambeke, F., Hombach, M., and Zinkernagel, A. S. (2020). Increased Azithromycin Susceptibility of Multidrug-Resistant Gram-Negative Bacteria on RPMI-1640 Agar Assessed by Disk Diffusion Testing. Antibiotics 9:218. doi: 10.3390/antibiotics9050218
Miura, K., Tomioka, Y., Suzuki, H., Yonezawa, M., Hishinuma, T., and Mizugaki, M. (1997). Molecular cloning of the nemA gene encoding N-ethylmaleimide reductase from Escherichia coli. Biol. Pharm. Bull. 20, 110–112. doi: 10.1248/BPB.20.110
Moldovan, A., and Fraunholz, M. J. (2019). In or out: phagosomal escape of Staphylococcus aureus. Cell. Microbiol. 21:e12997. doi: 10.1111/cmi.12997
Monk, I. R., Tree, J. J., Howden, B. P., Stinear, T. P., and Foster, T. J. (2015). Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6:e00308-15. doi: 10.1128/mBio.00308-15
Ohlsen, K., Koller, K. P., and Hacker, J. (1997). Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect. Immun. 65, 3606–3614. doi: 10.1128/iai.65.9.3606-3614.1997
Ozyamak, E., Almeida, C., Moura, A. P. S., Miller, S., and Booth, I. R. (2013). Integrated stress response of Escherichia coli to methylglyoxal: transcriptional readthrough from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol. Microbiol. 88, 936–950. doi: 10.1111/mmi.12234
Pedre, B., Barayeu, U., Ezeri na, D., and Dick, T. P. (2021). The mechanism of action of N-acetylcysteine (NAC): the emerging role of H2S and sulfane sulfur species. Pharmacol. Ther. 228:107916. doi: 10.1016/J.PHARMTHERA.2021.107916
Pelz, A., Wieland, K.-P., Putzbach, K., Hentschel, P., Albert, K., and Götz, F. (2005). Structure and Biosynthesis of Staphyloxanthin from Staphylococcus aureus. J. Biol. Chem. 280, 32493–32498. doi: 10.1074/jbc.M505070200
Peng, H., Shen, J., Edmonds, K. A., Luebke, J. L., Hickey, A. K., Palmer, L. D., et al. (2017). Sulfide Homeostasis and Nitroxyl Intersect via Formation of Reactive Sulfur Species in Staphylococcus aureus. mSphere 2, e82–e17. doi: 10.1128/mSphere.00082-17
Reichert, S., Ebner, P., Bonetti, E.-J., Luqman, A., Nega, M., Schrenzel, J., et al. (2018). Genetic Adaptation of a Mevalonate Pathway Deficient Mutant in Staphylococcus aureus. Front. Microbiol. 9:1539. doi: 10.3389/FMICB.2018.01539
Reichmann, D., Voth, W., and Jakob, U. (2018). Maintaining a Healthy Proteome during Oxidative Stress. Mol. Cell 69, 203–213. doi: 10.1016/j.molcel.2017.12.021
Renner, L. D., Zan, J., Hu, L. I., Martinez, M., Resto, P. J., Siegel, A. C., et al. (2017). Detection of ESKAPE Bacterial Pathogens at the Point of Care Using Isothermal DNA-Based Assays in a Portable Degas-Actuated Microfluidic Diagnostic Assay Platform. Appl. Environ. Microbiol. 83, e2449–e2416. doi: 10.1128/AEM.02449-16
Scholtissek, A., Tischler, D., Westphal, A., van Berkel, W., and Paul, C. (2017). Old Yellow Enzyme-Catalysed Asymmetric Hydrogenation: linking Family Roots with Improved Catalysis. Catalysts 7:130. doi: 10.3390/catal7050130
Sheng, X., Yan, M., Xu, L., and Wei, M. (2016). Identification and characterization of a novel Old Yellow Enzyme from Bacillus subtilis str.168. J. Mol. Catal. B Enzym. 130, 18–24. doi: 10.1016/j.molcatb.2016.04.011
Shetty, S., and Varshney, U. (2021). Regulation of translation by one-carbon metabolism in bacteria and eukaryotic organelles. J. Biol. Chem. 296:100088. doi: 10.1074/JBC.REV120.011985
Shi, Q., Wang, H., Liu, J., Li, S., Guo, J., Li, H., et al. (2020). Old yellow enzymes: structures and structure-guided engineering for stereocomplementary bioreduction. Appl. Microbiol. Biotechnol. 104, 8155–8170. doi: 10.1007/s00253-020-10845-z
Sihto, H.-M., Tasara, T., Stephan, R., and Johler, S. (2014). Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol. Lett. 356, 134–140. doi: 10.1111/1574-6968.12491
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sullivan, L. E., and Rice, K. C. (2021). ““Measurement of Staphylococcus aureus Pigmentation by Methanol Extraction,” in Staphylococcus aureus. Methods in Molecular Biology, Vol. 2341, ed. K. C. Rice (New York, NY: Humana), doi: 10.1007/978-1-0716-1550-8_1
Thomas, P., Sekhar, A. C., Upreti, R., Mujawar, M. M., and Pasha, S. S. (2015). Optimization of single plate-serial dilution spotting (SP-SDS) with sample anchoring as an assured method for bacterial and yeast cfu enumeration and single colony isolation from diverse samples. Biotechnol. Rep. 8, 45–55. doi: 10.1016/j.btre.2015.08.003
Tiwari, K., Gatto, C., and Wilkinson, B. (2018). Interrelationships between Fatty Acid Composition, Staphyloxanthin Content, Fluidity, and Carbon Flow in the Staphylococcus aureus Membrane. Molecules 23:1201. doi: 10.3390/molecules23051201
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus Infections: epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 28, 603–661. doi: 10.1128/CMR.00134-14
Toogood, H. S., Gardiner, J. M., and Scrutton, N. S. (2010). Biocatalytic Reductions and Chemical Versatility of the Old Yellow Enzyme Family of Flavoprotein Oxidoreductases. ChemCatChem 2, 892–914. doi: 10.1002/CCTC.201000094
Van Acker, H., and Coenye, T. (2017). The Role of Reactive Oxygen Species in Antibiotic-Mediated Killing of Bacteria. Trends Microbiol. 25, 456–466. doi: 10.1016/j.tim.2016.12.008
van der Maten, E., de Jonge, M. I., de Groot, R., van der Flier, M., and Langereis, J. D. (2017). A versatile assay to determine bacterial and host factors contributing to opsonophagocytotic killing in hirudin-anticoagulated whole blood. Sci. Rep. 7, 1–10. doi: 10.1038/srep42137
Vidal-Aroca, F., Giannattasio, M., Brunelli, E., Vezzoli, A., Plevani, P., Muzi-Falconi, M., et al. (2006). One-step high-throughput assay for quantitative detection of β-galactosidase activity in intact Gram-negative bacteria, yeast, and mammalian cells. Biotechniques 40, 433–440. doi: 10.2144/000112145
Wang, X., and Zhao, X. (2009). Contribution of Oxidative Damage to Antimicrobial Lethality. Antimicrob. Agents Chemother. 53, 1395–1402. doi: 10.1128/AAC.01087-08
Wasil, M., Halliwell, B., Grootveld, M., Moorhouse, C. P., Hutchison, D. C., and Baum, H. (1987). The specificity of thiourea, dimethylthiourea and dimethyl sulphoxide as scavengers of hydroxyl radicals. Their protection of alpha 1-antiproteinase against inactivation by hypochlorous acid. Biochem. J. 243:867. doi: 10.1042/BJ2430867
White, J. R., and Yeowell, H. N. (1982). Iron enhances the bactericidal action of streptonigrin. Biochem. Biophys. Res. Commun. 106, 407–411. doi: 10.1016/0006-291X(82)91125-1
Wilding, E. I., Kim, D. Y., Bryant, A. P., Gwynn, M. N., Lunsford, R. D., McDevitt, D., et al. (2000). Essentiality, expression, and characterization of the class II 3-hydroxy-3-methylglutaryl coenzyme a reductase of Staphylococcus aureus. J. Bacteriol. 182, 5147–5152. doi: 10.1128/JB.182.18.5147-5152.2000
Williams, R. E., and Bruce, N. C. (2002). ‘New uses for an Old Enzyme’ – the Old Yellow Enzyme family of flavoenzymes. Microbiology 148, 1607–1614. doi: 10.1099/00221287-148-6-1607
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov 2:100141. doi: 10.1016/j.xinn.2021.100141
Wyllie, S., Roberts, A. J., Norval, S., Patterson, S., Foth, B. J., Berriman, M., et al. (2016). Activation of Bicyclic Nitro-drugs by a Novel Nitroreductase (NTR2) in Leishmania. PLoS Pathog. 12:e1005971. doi: 10.1371/JOURNAL.PPAT.1005971
Keywords: MRSA, blood, phagocytes, quinone, ROS, stress response, electrophilic stress
Citation: Ibrahim ES and Ohlsen K (2022) The Old Yellow Enzyme OfrA Fosters Staphylococcus aureus Survival via Affecting Thiol-Dependent Redox Homeostasis. Front. Microbiol. 13:888140. doi: 10.3389/fmicb.2022.888140
Received: 02 March 2022; Accepted: 31 March 2022;
Published: 17 May 2022.
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Dorte Frees, University of Copenhagen, DenmarkHwan Kim, Stony Brook University, United States
Jeffrey Michael Boyd, Rutgers, The State University of New Jersey, United States
Copyright © 2022 Ibrahim and Ohlsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Knut Ohlsen, knut.ohlsen@uni-wuerzburg.de
 Eslam S. Ibrahim
Eslam S. Ibrahim Knut Ohlsen
Knut Ohlsen