- 1Guangdong Provincial Key Laboratory of Food Quality and Safety, Guangdong Provincial Key Laboratory of Nutraceuticals and Functional Foods, College of Food Science, South China Agricultural University, Guangzhou, China
- 2Guangdong Moyanghua Grains and Oils Co., Ltd., Yangjiang, China
Microbial degradation is considered as an attractive method to eliminate exposure to aflatoxin B1 (AFB1), the most toxic mycotoxin that causes great economic losses and brings a serious threat to human and animal health, in food and feed. In this study, Bacillus amyloliquefaciens WF2020, isolated from naturally fermented pickles, could effectively degrade AFB1 ranging from 1 to 8 μg/ml, and the optimum temperature and pH value were 37–45°C and 8.0, respectively. Moreover, B. amyloliquefaciens WF2020 was considered to be a potential probiotic due to the synthesis of active compounds, absence of virulence genes, susceptibility to various antibiotics, and enhanced lifespan of Caenorhabditis elegans. Extracellular enzymes or proteins played a major role in AFB1 degradation mediated by B. amyloliquefaciens WF2020 into metabolites with low or no mutagenicity and toxicity to C. elegans. AFB1 degradation by the cell-free supernatant was stable up to 70°C, with an optimal pH of 8.0, and the cell-free supernatant could still degrade AFB1 by 37.16% after boiling for 20 min. Furthermore, B. amyloliquefaciens WF2020 caused a slight defect in fungal growth and completely inhibited AFB1 production when co-incubated with Aspergillus flavus. Additionally, B. amyloliquefaciens WF2020 suppressed the expression of 10 aflatoxin pathway genes and 2 transcription factors (alfR and alfS), suggesting that B. amyloliquefaciens WF2020 might inhibit AFB1 synthesis in A. flavus. These results indicate that B. amyloliquefaciens WF2020 and/or its extracellular enzymes or proteins have a promising potential to be applied in protecting food and feed from AFB1 contamination.
Introduction
Aflatoxins, a group of mycotoxins produced mainly by Aspergillus flavus and Aspergillus parasiticus, contaminate about 60–80% of food and feed around the world (Lee and Ryu, 2017; Eskola et al., 2019), and are also perceived as a severe threat to human health due to their hepatotoxicity, nephrotoxicity, immunotoxicity, etc. (Silvia et al., 2018). Among the 20 types of aflatoxins identified, aflatoxin B1 (AFB1) is the most toxic, mutagenic, and carcinogenic to both humans and livestock and is classified as a group-1 carcinogen by the International Agency for Research on Cancer (IARC, 1993). AFB1 contamination in crops has become a widespread problem, and considerable investigations have been directed at finding methods, such as physical, chemical, and biological methods, to prevent its toxicity.
Microbial degradation was considered as an attractive method due to its specificity, efficiency, environmental friendliness, protection of the quality and flavor of food, and feasibility of the processes when applied in industries (Mishra and Das, 2003; Wu et al., 2009). In the last decade, beneficial microorganisms substantially were found to be capable of reducing AFB1 in contaminated media, including Actinobacteria (e.g., Brachybacterium sp., Rhodococcus, Streptomyces, Nocardia, and Mycobacterium), Bacillus (e.g., Bacillus, Lysinibacillus, Streptococcus, and Staphylococcus), -Proteobacteria (e.g., Enterobacter sp., Klebsiella, Pseudomonas, and Brevundimonas), Ascomycota (e.g., Aspergillus, Alternaria, Neurospora, and Trichoderma), Basidiomycota (e.g., Pleurotus), Zygomycota (e.g., Mucor, Rhizopus, and Absidia), etc. (Verheecke et al., 2016). However, bacteria have more applications for AFB1 remediation due to some advantages such as more elimination within a shorter time and producing no pigments (Laciakova et al., 2008), and among them, Bacillus becomes an attractive candidate because of its high tolerance to various environmental stresses and application as a kind of potential probiotics (Yan et al., 2017). For instance, AFB1 was reduced by 92.1% by Bacillus shackletonii L7 for 72 h (Liang et al., 2017), 85.61% by Bacillus subtilis UTBSP1 for 96 h (Farzaneh et al., 2012), 91.5% by Bacillus velezensis DY3108 for 96 h (Shu et al., 2018), 94.70% by Bacillus licheniformis CFR1 for 72 h (Rao et al., 2016), and 100% by Bacillus TUBF1 for 72 h (El-Deeb et al., 2013). Although more and more Bacillus were reported to degrade AFB1, few studies have performed the safety assessment of selected strains. In addition, the narrow working temperature range and unsuitability for the processing environment in AFB1-degrading bacteria reported previously also become challenges in commercial applications. Therefore, it is still worth exploring safe bacteria, including Bacillus, which are suitable for food and feed processing and detoxify AFB1 into less toxic metabolites with excellent degradation efficiency and wide temperature ranges in the future.
Bacillus amyloliquefaciens, which was ubiquitously found in various environments, including food, plants, animals, soil, and aquatic environments, was reported as a potential probiotic due to its strong antimicrobial activity, the synthesis of bioactive compounds, including peptides and exopolysaccharides, its survival in gastrointestinal conditions, etc. (WoldemariamYohannes et al., 2020; Ngalimat et al., 2021). Moreover, B. amyloliquefaciens could be a multifunctional microbe and potentially applied in the animal food and feed industry and in functional food processing due to the improvement in the functional, sensory, and shelf life of end products and the production of several enzymes, including -glutamyl transpeptidase pectinase, xylanase, β-glucosidase, and amylase, which can hydrolyze complex compounds, including insoluble proteins, carbohydrates, fibers, hemicellulose, and lignans, and then increase the digestion and absorption of nutrients from food and feed and form novel functional and bioactive compounds (WoldemariamYohannes et al., 2020; Chen et al., 2021). In addition, some strains of B. amyloliquefaciens, such as B. amyloliquefaciens UTB2, UNRC52, and UNRCLR, could suppress AFB1 synthesis (Bluma and Etcheverry, 2006; Siahmoshteh et al., 2018). However, except for B. amyloliquefaciens S8C, Y1-B1, SWUN-TP23, SG-16, and HSP-5 (Xu et al., 2015; Guo et al., 2017; Wang J. et al., 2018; Ali et al., 2021; Zhang et al., 2021), little was known about the AFB1 degradation potential of B. amyloliquefaciens as well as the molecular mechanism of the loss in AFB1 production. Here, the AFB1-degrading bacterium in naturally fermented pickles was isolated and identified as B. amyloliquefaciens (WF2020), and the toxicities of the strain and its AFB1 degradation products were also assessed based on sequenced genome information, antibiotic susceptibility, the changes in the lifespan of Caenorhabditis elegans, and Ames mutagenicity. Moreover, the effects of cultivation conditions on AFB1 degradation mediated by B. amyloliquefaciens WF2020 and its active components were investigated by measuring the concentrations of residue AFB1 in media with different temperatures, pH values, and metal ions. Lastly, the effects of B. amyloliquefaciens WF2020 on the fungal growth and synthesis of AFB1 were investigated when B. amyloliquefaciens WF2020 was co-incubated with A. flavus, a producer of aflatoxins. The results indicated that B. amyloliquefaciens WF2020 is a potential probiotic applied in the protection of food and feed from AFB1 contamination.
Materials and Methods
Isolation of Potential Aflatoxin B1-Degrading Bacteria From Fermented Foods
About 1 g of fermented food was mixed with 10 ml of sterile saline and then diluted to 10–3, 10–4, 10–5, 10–6, and 10–7 levels. All dilutions were spread evenly on coumarin medium (CM: 1% coumarin, 0.025% KH2PO4, 0.1% NH4NO3, 0.1% CaCl2, 0.025% MgSO4⋅7H2O, 0.0001% FeSO4, and 1.5% agar) and cultured at 37°C for 4 days. Single colonies were isolated and transferred to fresh CM plates three times. Colonies growing on CM plates were selected and tested for AFB1 degradation.
Aflatoxin B1 Degradation in Liquid Culture
Overnight cultured bacterial cells were diluted to an optical density at 600 nm (OD600) of 0.01 with fresh Luria-Bertani (LB) medium, and then AFB1 purchased from J&K Scientific (Beijing, China) was added into 1 ml of dilution for a final concentration of 2 μg/ml. Sterile LB medium with AFB1 was used as the control. After 3-day incubation at 37°C by shaking at 180 rpm, the supernatant was extracted using chloroform according to previous reports (Guan et al., 2008), and the chloroform fractions were evaporated and dissolved using dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA). The redissolved solution was filtered using the 0.22-μm pore filter (Merck-Millipore, Darmstadt, Germany) and stored at −20°C for high-performance liquid chromatography (HPLC) detection. About 94–96% of AFB1 could be recovered from the liquid culture using chloroform extraction.
Quantification of Aflatoxin B1 by High-Performance Liquid Chromatography
Aflatoxin B1 was analyzed by HPLC according to the procedure reported by Fang et al. (2020). The percentage of AFB1 degradation was calculated using the following formula: the percentage of AFB1 degradation = (1 − Ca/Cb) × 100%, where Ca and Cb are the concentration of remaining AFB1 in the sample and total AFB1 in the control sample, respectively.
Analysis of Aflatoxin B1 Metabolites by HPLC-Quadrupole-Time-of-Flight-Mass Spectrometry
Aflatoxin B1 metabolites were extracted with chloroform after a 72-h incubation of AFB1 degrading bacterium in LB medium with 2 μg/ml of AFB1 and analyzed by HPLC-Q-TOF-MS according to the procedure described by Fang et al. (2020). Extractions from the AFB1-degrading bacterium in LB and sterile LB media with AFB1 were used as controls.
Genome Sequencing and Analysis
Genomic DNA was extracted using Wizard® Genomic DNA Purification kit (Promega, Beijing, China) according to the manufacturer’s protocol and sequenced using a combination of PacBio RS II Single Molecule Real Time (SMRT, Pacific Biosciences, MenloPark, CA, United States) and Illumina sequencing platforms (Hiseq X Ten; Illumina, San Diego, CA, United States). The PacBio reads and Illumina reads were used to assemble the complete genome sequence into a contig using the hierarchical genome assembly process (HGAP) and CANU (Version 1.71). The last circular step was manually checked and finished, generating a complete genome with seamless chromosomes and plasmids. Finally, error correction of the PacBio assembly results was performed with Illumina reads using Pilon. Sequence data were deposited at the US National Center for Biotechnology Information (NCBI) under accession number CP092778.
The coding sequences (CDSs) were predicted with Glimmer (Version 3.022) and annotated from the databases of Non-Redundant (NR Protein Sequence Database), Swiss-Prot, Pfam, Gene Ontology (GO), Clusters of Orthologous Group (COG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) using sequence alignment tools such as Basic Local Alignment Search Tool (BLAST, Version 2.3.03), Diamond (Version 0.8.34), and HMMER (Version 3.1b25), and annotations were obtained from the best-matched subjects (E-value < 10–5) for gene annotation. All data were analyzed on the free online Majorbio Cloud Platform6.
Antibiotic Susceptibility
Disk diffusion susceptibility tests were conducted according to the procedure reported by the National Committee for Clinical Laboratory Standards. Briefly, a bacterial dilution (OD600 = 0.01) was spread on the Mueller-Hinton agar (MHA: 0.2% beef dehydrated infusion, 1.75% casein hydrolyzate, 0.15% starch, and 2% agar) plates and the disks with 2 μg lincomycin, 5 μg ciprofloxacin or rifampin, 10 μg gentamicin, streptomycin, ampicillin penicillin, imipenem, or norfloxacin, 15 μg erythromycin, or 30 μg tetracycline, cefalexin, kanamycin, chloramphenicol, or vancomycin were put on the plates. After 12 h of incubation at 37°C, the diameters of the inhibition zones were recorded.
Assay for the Lifespan of Caenorhabditis elegans
Lifespans were monitored as described previously (Donato et al., 2017). Briefly, L4 worms of C. elegans N2 were grown on a nematode growth medium (NGM) agar plate seeded with Escherichia coli OP50 at 20°C and treated with alkaline hypochlorite to collect embryos. Embryos were cultivated to obtain a synchronized population. Synchronized L4 worms were transferred to fresh NGM plates seeded with the tested bacterium or E. coli OP50 or E. coli OP50 plus AFB1 or degradation metabolites every 2 days. Worms were considered dead when they stopped pharyngeal pumping and did not respond to prodding with a platinum wire. The number of dead/live worms was recorded every day.
Ames Mutagenicity Assay
To evaluate the mutagenicity of the degradation metabolites, the Salmonella (Ames) test was conducted with the S9 Enzyme Activation kit (Iphase Pharma Service, Beijing, China) according to the manufacturer’s instructions and the procedure described by Fang et al. (2020). Briefly, the degradation metabolites extracted from a 96-h culture co-incubated with the AFB1-degrading bacterium and AFB1 were incubated with Salmonella typhimurium TA98 or TA100 at 37°C for 48 h. The number of S. typhimurium colonies was recorded, and the data were given as the number of reversed colony-forming units (CFUs). Samples extracted from LB medium with AFB1 were used as positive controls, and extracts from LB medium were used as negative controls.
Aflatoxin B1 Degradation by Extracellular Extracts, Intracellular Extracts, and Dead Cells
The dilution of bacterial cells (OD600 = 0.02, the same below unless specified) was cultured in LB medium with shaking for 48 h at 37°C, and the supernatant and cells were collected, respectively, after centrifugation at 12,000 rpm for 5 min at 4°C. The supernatant filtered with a 0.22-μm pore filter served as the extracellular extracts for AFB1 degradation. After washing with 10 mM phosphate buffer (pH 8.0) three times, the cells were broken by ultrasonication (25 kHz, ultrasound for 4 s interval 1 s, 15 min) in the ice bath, and centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant filtered with 0.22-μm pore filter served as the intracellular extracts for AFB1 degradation. Meanwhile, cells washed with phosphate buffer were boiled for 20 min, resuspended in an equal volume of 10 mM phosphate buffer (pH 8.0), and served as dead cells for AFB1 degradation. Extracellular extracts, intracellular extracts, and dead cells were co-incubated with 2 μg/ml of AFB1 at 37°C with shaking at 180 rpm for 72 h, respectively. Cultures of LB medium or phosphate buffer supplemented with 2 μg/ml AFB1 were used as the control, and all variables of control groups were similar to those of the corresponding extracts and dead cells. Residual AFB1 was tested as described above.
Effect of Proteinase K, Sodium Dodecyl Sulfate, and Heat Treatment on Aflatoxin B1 Degradation
Extracellular extracts were divided into four fractions to investigate the influence of proteinase K, SDS, and heat on AFB1 degradation. One fraction was boiled for 20 min, and other fractions were treated with proteinase K (1 mg/ml), SDS (1%), or SDS plus proteinase K for 6 h, respectively. Subsequently, each fraction was incubated with 2 μg/ml AFB1 at 37°C with shaking at 180 rpm, and phosphate buffer with 2 μg/ml AFB1 was used as the control. After 24 h, residual AFB1 was monitored as described above.
Effects of Aflatoxin B1 Concentrations, Temperature, pH Values, and Metal Ions on Aflatoxin B1 Degradation by the Aflatoxin B1-Degrading Bacterium and Its Cell-Free Supernatant
To investigate the effects of AFB1 concentrations on AFB1 degradation mediated by the AFB1-degrading bacterium, bacterial cells were incubated with 1, 2, 5, and 8 μg/ml AFB1, respectively, at 37°C for 96 h by shaking at 180 rpm. LB medium with the corresponding concentration of AFB1 was used as the control. The effects of temperature, pH, and metal ions were determined by setting the cultivation temperature at 25, 30, 37, 40, 45, or 50°C, adjusting the initial pH values to 5.0, 6.0, 7.0, 8.0, or 9.0, and adding MgSO4 (0.5 mg/ml), ZnSO4⋅7H2O (0.5 mg/ml), CuSO4⋅5H2O (0.5 mg/ml), MnSO4⋅H2O (0.5 mg/ml), FeSO4⋅7H2O (0.5 mg/ml), or CaCl2 (0.5 mg/ml). Bacterial cells were incubated in LB medium with 2μg/ml AFB1 at 37°C for 24, 48, or 72 h by shaking at 180 rpm. Correspondingly, LB medium with 2 μg/ml AFB1 in each incubation was used as the control. Residual AFB1 was detected by the HPLC described as above. In addition, bacterial growth was also investigated by measuring the OD600 value.
To investigate the effects of initial pH values, temperature, and metal ions on AFB1 degradation by the cell-free supernatant of AFB1-degrading bacterial culture, the cell-free supernatant was collected as described and exposed to 2 μg/ml of AFB1, and the mixture was incubated at 37°C by shaking at 180 rpm. The effects of initial pH values were analyzed by adjusting the mixture to 5.0, 6.0, 7.0, 8.0, or 9.0. In the temperature test, the mixture was incubated at 20, 30, 37, 40, 50, 60, or 70°C, respectively. In terms of metal ions, the reaction mixture was supplemented with 0.5 mg/ml of MgSO4, ZnSO4⋅7H2O, CuSO4⋅5H2O, MnSO4⋅H2O, FeSO4⋅7H2O, and CaCl2, respectively. Correspondingly, LB medium with 2 μg/ml of AFB1 in each incubation was used as the control. After a 48-h incubation, residual AFB1 was analyzed by HPLC as described above.
Assays for the Fungal Growth and the Production of Aflatoxin B1 in Aspergillus flavus
To investigate an effect of the AFB1-degrading bacterium on the fungal growth of A. flavus, the antagonistic effect and dry weight were tested after the AFB1-degrading bacterium was co-incubated with A. flavus for 2 days in potato dextrose agar (PDA) and potato dextrose broth (PDB), respectively. About 1 μl of the bacterial cell dilution was spotted in the upper part of a PDA plate and 1 μl of a conidial suspension (1.0 × 106 conidia/ml) of A. flavus was spotted in the lower part of the plate. The plates were incubated at 30°C for 2 days, and fungal growth was observed. Meanwhile, 100 μl of a conidial suspension (1.0 × 109 conidia/ml) of A. flavus was added into 100 ml of PDB supplemented with bacterial cells (OD600 = 0.02) and incubated at 30°C by shaking at 180 rpm. PDB with A. flavus conidia was set as the control. After 2 days, fungal mycelia and the supernatant were collected by centrifugation. The collected mycelia were dried at 60°C and weighted. The supernatant was filtered by the 0.22 μm pore filter and analyzed by HPLC to detect the content of AFB1.
Transcriptional Profiling of Genes Related to Aflatoxin B1 Synthesis
About 100 μl of a conidial suspension (1.0 × 109 conidia/ml) of A. flavus was added to 100 ml of PDB supplemented with bacterial cells (OD600 = 0.02) and incubated at 30°C by shaking at 180 rpm. PDB with A. flavus conidia was set as the control. After 2 days, fungal mycelia were harvested and used to extract total RNA using the RNAiso™ Plus reagent (TaKaRa, Dalian, China). Total RNA was reversely transcribed to cDNA using the PrimeScript® RT reagent kit (TaKaRa). Transcripts of targeted genes were quantified via real-time quantitative polymerase chain reaction (qRT-PCR) with paired primers (Supplementary Table 1) under the action of SYBR® Premix Ex TaqTM (TaKaRa). The transcript of the fungal β-tublin gene was used as an internal standard. The relative transcript level of each gene was calculated as the ratio of its transcript in the group of A. flavus plus bacterial cells to the control group, using the threshold-cycle (2–ΔΔCt) method.
Statistical Analysis
All the above experiments were conducted three times. The results of three replicates were expressed as mean ± standard deviation (SD), and statistical analysis was subjected to one-factor analysis of variance (ANOVA) performed with SPSS software. It is considered statistically significant when p < 0.05 in all the experiments.
Results
Isolation and Identification of Aflatoxin B1-Degrading Bacteria
After primary screening using coumarin as the sole carbon source and secondary screening by addition of 2 μg/ml AFB1 in LB medium, four isolates showed the ability to degrade AFB1 after a 48-h incubation (Supplementary Figure 1A). Among the four isolates, WF2020, which was isolated from naturally fermented pickles, displayed the maximum degradation ability up to 70.22% (Supplementary Figure 1A). When AFB1 concentration was not more than 5 μg/ml, except those in the first 24 h, the percentages of degrading AFB1 for WF2020 during the 96-h incubation were nearly similar among the treatments at the same cultivation time and nearly reached the maximum at 72 h where the percentage of AFB1 degradation was more than 84% (Figure 1A). Moreover, when AFB1 concentration was up to 8 μg/ml, WF2020 could degrade AFB1 in a time-dependent manner, and a reduction of more than 75% was observed at 96 h (Figure 1A).
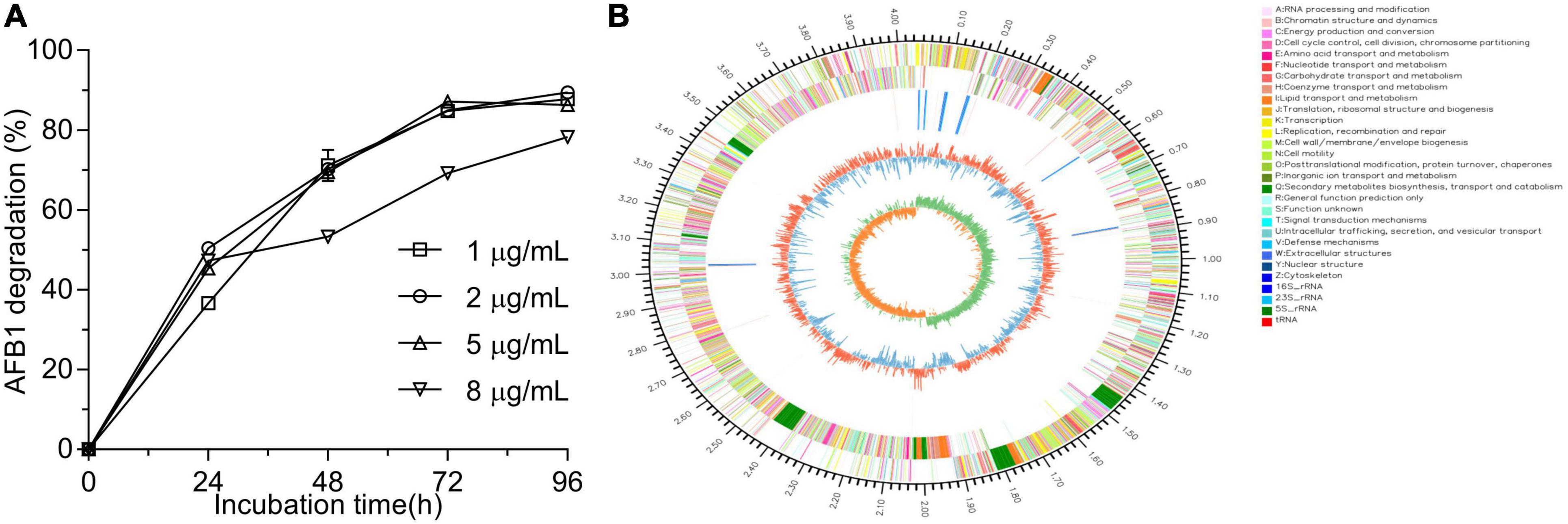
Figure 1. Aflatoxin B1 (AFB1) degradation mediated by Bacillus amyloliquefaciens WF2020 at different concentrations of AFB1 (A) and circular representation of the complete genome of B. amyloliquefaciens WF2020 (B). From outermost to innermost circle: circle 1, genome size; circle 2, genes on forward strand; circle 3, genes on reverse strand; circle 4, rRNA and tRNA; circle 5, GC content; and circle 6, GC skew.
WF2020 is a Gram-positive bacterium with the typical colony characteristics of Bacillus sp. (Supplementary Figure 1B). According to genome sequences obtained using Illumina Hiseq and a PacBio system, the complete genome sequence of WF2020 comprises a 4,043,726 bp circular chromosome, consisting of 4,133 predicted genes, 27 rRNA genes, and 86 tRNA genes (Figure 1B). In addition, no plasmid was observed in the genome. Based on the sequence analysis of 16s rRNA and other 31 housekeeping genes, including dnaG, frr, infC, nusA, pgk, pyrG, rplA, rplB, rplC, rplD, rplE, rplF, rplK, rplL, rplM, rplN, rplP, rplS, rplT, rpmA, rpoB, rpsB, rpsC, rpsE, rpsI, rpsJ, rpsK, rpsM, rpsS, smpB, and tsf, the closest relative of WF2020 was B. amyloliquefaciens strain (Supplementary Figures 1C,D). Therefore, this isolate was termed B. amyloliquefaciens WF2020.
The Active Component to Degrade Aflatoxin B1 in Bacillus amyloliquefaciens WF2020 and Its Characteristics
Adsorption and degradation are the two main approaches in the removal of mycotoxins by microbes (Hathout and Aly, 2014). Here, cell-free supernatant (i.e., extracellular extracts) of B. amyloliquefaciens WF2020 was more effective than dead cells and intracellular extracts in reducing AFB1 during a 72-h incubation (Figure 2A). The percentage reduction of AFB1 for cell-free supernatant, intracellular extracts, and dead cells is 60.67, 14.11, and 26.95% at 24 h, 71.01, 19.80, and 27.87% at 48 h, and 71.01, 20.95, and 27.85% at 72 h (Figure 2A), respectively. Additionally, bacterial cells harvested from the cultivation of 48-h incubation in LB medium removed 12.25% of AFB1 on average after incubated with 2 μg/ml AFB1 for 1 h at 37°C by shaking at 180 rpm. These findings suggested that the removal of AFB1 mediated by B. amyloliquefaciens WF2020 was mainly dependent on the degradation and the cell-free supernatant was the main active ingredient during AFB1 degradation. Moreover, AFB1 degradation capacity of the cell-free supernatant decreased by 20.50, 93.40, and 100% after pretreatment with proteinase K, SDS, and SDS plus proteinase K (Figure 2B), respectively. Furthermore, cell-free supernatant still could degrade AFB1 by 37.16% after boiling for 20 min (Figure 2B). These results indicated that thermostable extracellular proteins or enzymes secreted by B. amyloliquefaciens WF2020 were involved in AFB1 degradation.
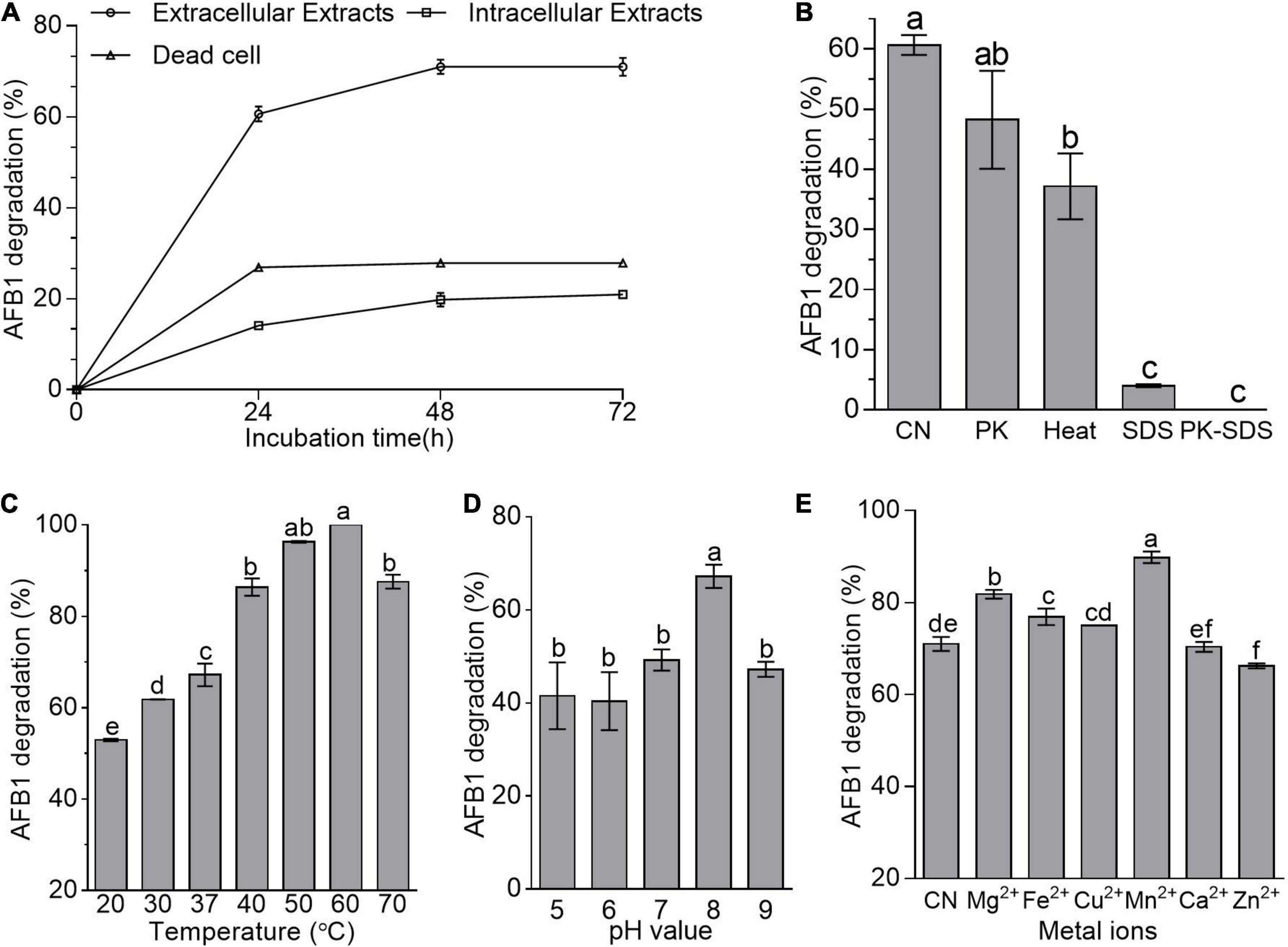
Figure 2. AFB1 degradation among diverse cell components of B. amyloliquefaciens WF2020. (A) AFB1 degradation by extracellular extracts, intracellular extracts, and dead cells during 72-h incubation with 2 μg/ml AFB1 at 37°C. (B) Effects of heat, proteinase K (PK), SDS, and proteinase K plus SDS on AFB1 degradation mediated by the cell-free supernatant after co-incubation for 24 h. (C–E) Effects of different temperatures (C), pH values (D), and metal ions (E) on AFB1 degradation mediated by the cell-free supernatant after co-incubation for 48 h. Different lowercase letters in the bars of each group indicate significant differences between treatments (Tukey’s test, p < 0.05).
pH, temperature, and metal ions affected the AFB1 degradation ability of the cell-free supernatant from bacteria and fungi (Zhang et al., 2014; Rao et al., 2016; Wang et al., 2017; Xu et al., 2017; Shu et al., 2018). Here, AFB1 degradation studies with different incubation temperatures after 48-h incubation showed that the cell-free supernatant of B. amyloliquefaciens WF2020 could degrade AFB1 at temperatures ranging from 20°C to 70°C and the percentage of AFB1 degradation at 70°C remained more than 70% (Figure 2C), implying that the active constituents or components of the cell-free supernatant were thermostable and could work well within a wide range of working temperature. Moreover, the percentage of AFB1 degradation increased with the increase of temperature up to 60°C which was the optimum temperature for 100% AFB1 degradation (Figure 2C). In addition, the cell-free supernatant of B. amyloliquefaciens WF2020 could degrade AFB1 over a broad pH from 5.0 to 9.0 and the maximum percentage displayed at pH 8 (Figure 2D). Lastly, the effects of metal ions on the AFB1 degradation ability of the cell-free supernatant were evaluated (Figure 2E). Mn2+, Mg2+, Fe2+, and Cu2+ stimulated AFB1 degradation by 26.52, 15.19, 8.29, and 5.69%, respectively, whereas Ca2+ had no significant effect, but Zn2+ inhibited the degradation by 6.73% (Figure 2E), inferring that Mn2+, Mg2+, Fe2+, and Cu2+ may act as enzyme activators, membrane stabilizers, and help to maintain the structural integrity of proteins.
Safety and Toxicity of Bacillus amyloliquefaciens WF2020 and Its Aflatoxin B1 Degradation Products
Based on the genomic sequence analysis, there are 12 secondary metabolic gene clusters via an antiSMASH analysis, but only six gene clusters harbored 100% similarity to those of known secondary metabolites (Table 1). The metabolites of the six gene clusters were macrolactin, bacillaene, fengycin, difficidin, bacillibactin, and bacilysin, respectively (Table 1), which are active substances with antibacterial, antifungal, anticancer, antiviral, anti-biofilm activities, biocontrol activity, etc. (Chen et al., 2008; Ryohei et al., 2015; Wu et al., 2015; Cochrane and Vederas, 2016; Zhou et al., 2018, 2021; Catherine et al., 2020; Erega et al., 2021; Kaushik et al., 2021). Moreover, a total of 35 genes with up to 50% similarity were found after blasting in the database of virulence factors, but they were not virulence genes but rather regulatory genes that played important roles in regulating biological processes, including virulence in other bacteria (Table 2). Additionally, a total of 19 genes with up to 50% similarity were found after blasting in the Comprehensive Antibiotic Resistance Database, and there is only one gene, imrB important for the resistance to lincosamide antibiotics, with up to 85% similarity (Table 3). Susceptibility to the corresponding antibiotics showed that, except for lincomycin belonging to a member of lincosamide antibiotics, B. amyloliquefaciens WF2020 was sensitive to other 14 antibiotics, including tetracycline, penicillin, cefalexin, ampicillin, streptomycin, kanamycin, gentamicin, ciprofloxacin, chloramphenicol, vancomycin, imipenem, rifampin, erythromycin, and norfloxacin (Figure 3A), suggesting their lower likelihood of being antibiotic-resistant bacterium.

Table 1. Secondary metabolites predicted by the antiSMASH analysis of Bacillus amyloliquefaciens WF2020.
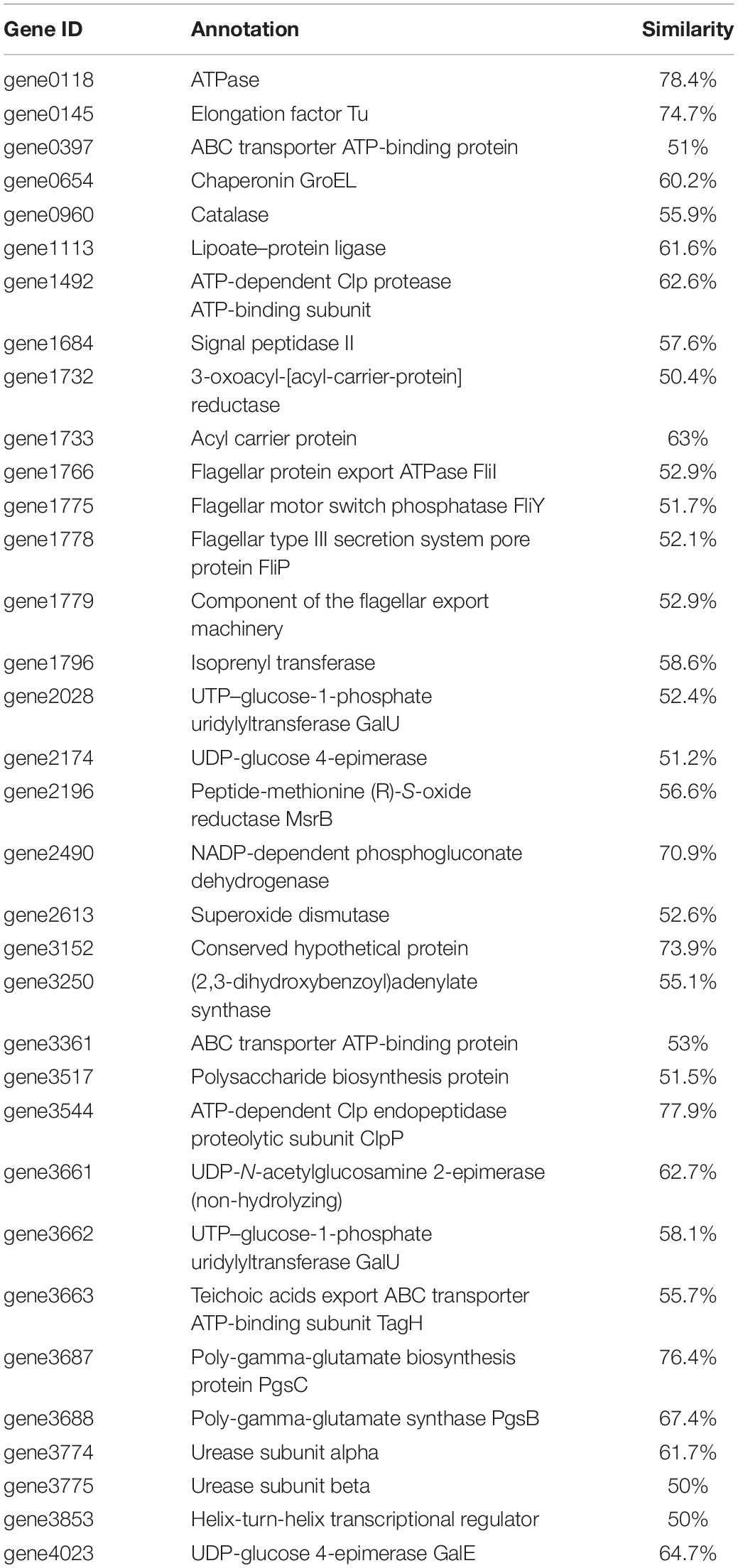
Table 2. Genes with up to 50% similarity found in B. amyloliquefaciens WF2020 genome according to the database of virulence factors.
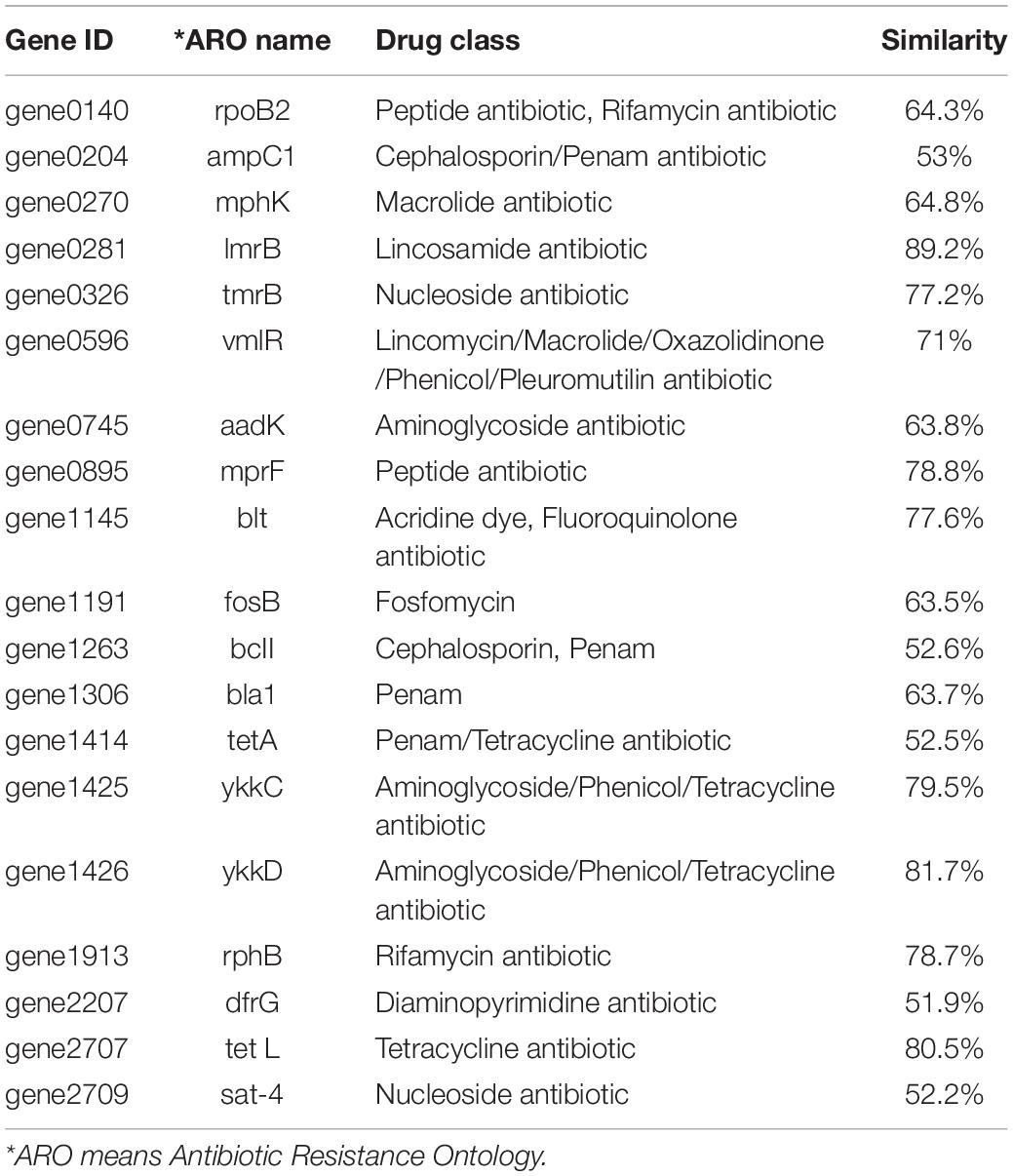
Table 3. Genes with up to 50% similarity found in B. amyloliquefaciens WF2020 genome according to the Comprehensive Antibiotic Resistance Database.
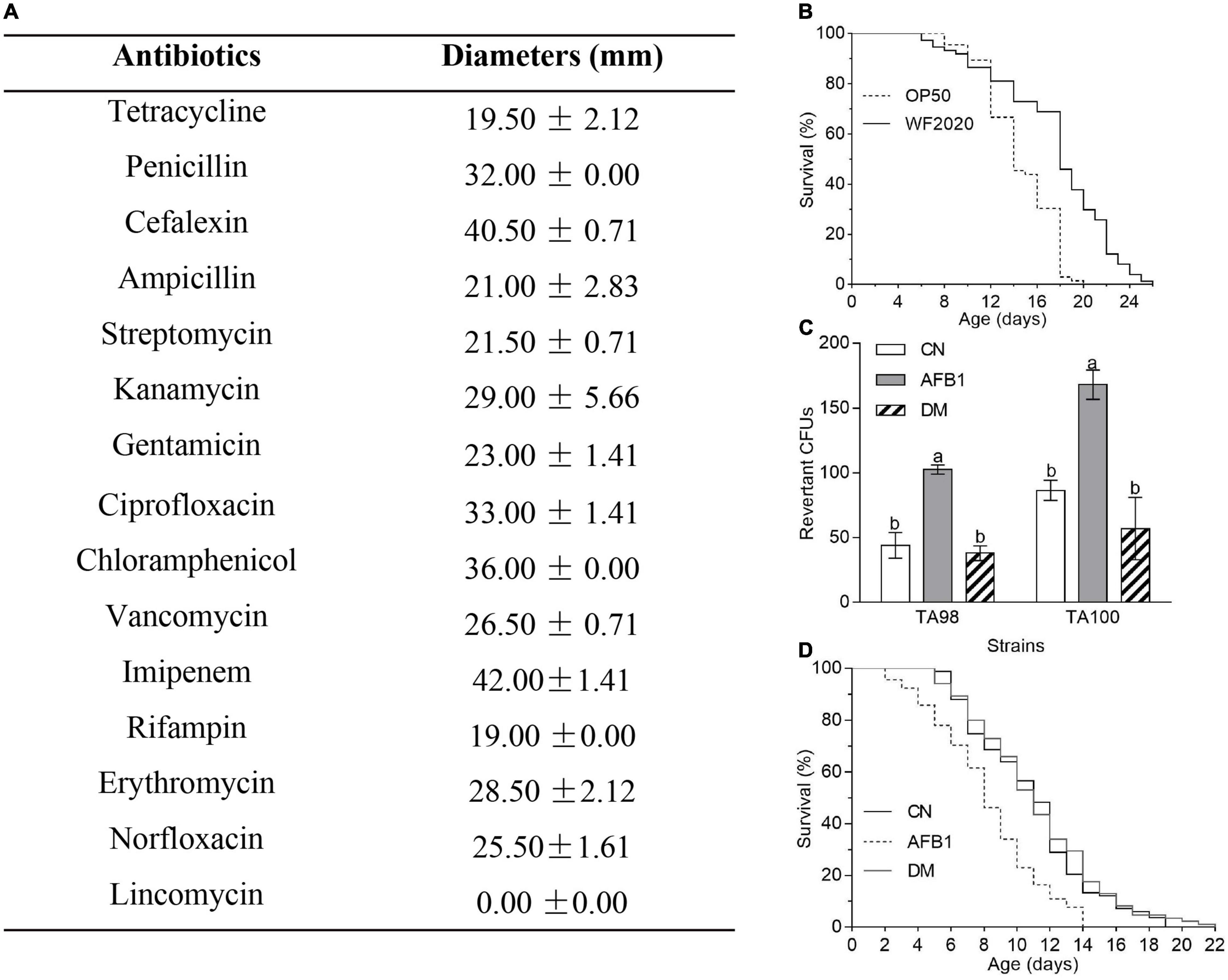
Figure 3. The safety of B. amyloliquefaciens WF2020 and its AFB1 degradation products. (A) Susceptibility of B. amyloliquefaciens WF2020 to different antibiotics by the disk diffusion test. (B) Changes in the lifespan of C. elegans N2 caused by B. amyloliquefaciens WF2020. (C,D) Reduction of AFB1 mutagenic effects (C) and the toxicity to C. elegans N2 (D) caused by B. amyloliquefaciens WF2020. The AFB1 group means extracts from the media supplemented with 20 μg AFB1. The DM group refers to the culture extracts from the supernatant of the 96 h co-incubation of 20 μg AFB1 and B. amyloliquefaciens WF2020. The CN group means the control group. Different lowercase letters in the bars of each group indicate significant differences between treatments (Tukey’s test, p < 0.05).
Moreover, C. elegans has emerged as an invertebrate model to study host–pathogen interactions since its first documentation by Sydney Brenner (Brenner, 1974; Kumar et al., 2020). In the current study, C. elegans was used to evaluate the toxicity of B. amyloliquefaciens WF2020 to animals. C. elegans fed on B. amyloliquefaciens WF2020 cells showed significantly increased longevity compared with the effect of the laboratory-feeding bacterium E. coli OP50 cells when used as a food source (Figure 3B). The survival of worms fed on WF2020 cells increased by an average of 20.78% (average survival: 14.58 days, 95% confidence interval (CI): 13.83–15.32) compared with the strain OP50 (average survival: 17.61 days, 95% CI: 16.47–18.75). Maximum lifespans of worms fed on WF2020 were prolonged by 6 days compared with the strain OP50.
Except for the safety of the AFB1-degrading bacterium, the toxicity of AFB1 degradation products should not be neglected as some degradable products might be toxic like AFB1. In this case, B. amyloliquefaciens WF2020 might degrade AFB1 into C15H11O (m/z 207.08), C15H15O2 (m/z 227.11), and C15H19O4 (m/z 263.13), according to the HPLC-Q-TOF-MS analysis of the 72-h co-incubation culture of AFB1 and B. amyloliquefaciens WF2020 (Supplementary Figure 2), compared with those of AFB1 solution and the fermentation culture of B. amyloliquefaciens WF2020. Firstly, the Ames test was used to assess the mutagenicity of AFB1 degradation products by B. amyloliquefaciens WF2020. Compared with the control group, an approximately twofold increase in the number of revertant CFUs from S. typhimurium TA98 and TA100 was observed in the AFB1 group, but there was no significant difference in revertant CFUs of the degradation products and the control group (Figure 3C), indicating that B. amyloliquefaciens WF2020 converted AFB1 to the metabolites with a loss of mutagenicity. Except for mutagenicity, AFB1 decreased the lifespan and increased the mortality rate of C. elegans (Yang et al., 2015). Therefore, the effect of AFB1 and its degradation products on the lifespan of C. elegans was performed to further evaluate the toxicity of AFB1 degradation products mediated by B. amyloliquefaciens WF2020. The mean lifespan exposed to AFB1 significantly decreased by 25.14% compared with the control, but there was no significant difference in the survival rates of C. elegans exposed to degradation products and the control (Figure 3D), implying that AFB1 degradation products mediated by B. amyloliquefaciens WF2020 were not toxic to the lifespan of C. elegans. These findings demonstrated that B. amyloliquefaciens WF2020 degraded AFB1 into metabolites, which exhibited no mutagenicity or toxicity to the lifespan of C. elegans.
These collective results demonstrated that B. amyloliquefaciens WF2020 might be used as a potential probiotic to degrade AFB1 in food and feed.
Effect of Fermentation Conditions on Aflatoxin B1 Degradation by Bacillus amyloliquefaciens WF2020
To evaluate the effects of fermentation conditions on AFB1 degradation mediated by B. amyloliquefaciens WF2020, incubation temperature, the initial pH of the culture, and metal ions were chosen as the tested fermentation conditions. In this study, AFB1 was degraded by B. amyloliquefaciens WF2020 at all incubation temperatures after 72-h incubation. The percentage of AFB1 degradation was 31.20, 46.99, 86.53, 85.16, 89.24, and 48.79% on average at 25, 30, 37, 40, 45, and 50°C, respectively (Figure 4A). However, the degradation rate showed no significant difference in the range of 37–45°C (Figure 4A). The growth of B. amyloliquefaciens WF2020 at 25, 30, 45, and 50°C decreased by 32.94, 19.47, 22.44, and 32.23%, respectively, compared with that at 37°C, and bacterial growth at 37 and 40°C showed no significant difference (Figure 4B). Combined with the effects of different temperatures on the AFB1 degradation ability of the active component of B. amyloliquefaciens WF2020, we speculated the lower degradation of AFB1 mediated by B. amyloliquefaciens WF2020 at 25 and 30°C might be due to the lower bacterial growth and lower activities of the active components at 25 and 30°C, and the lower degradation of AFB1 at 50°C might be attributed to the lower bacterial growth of B. amyloliquefaciens WF2020.
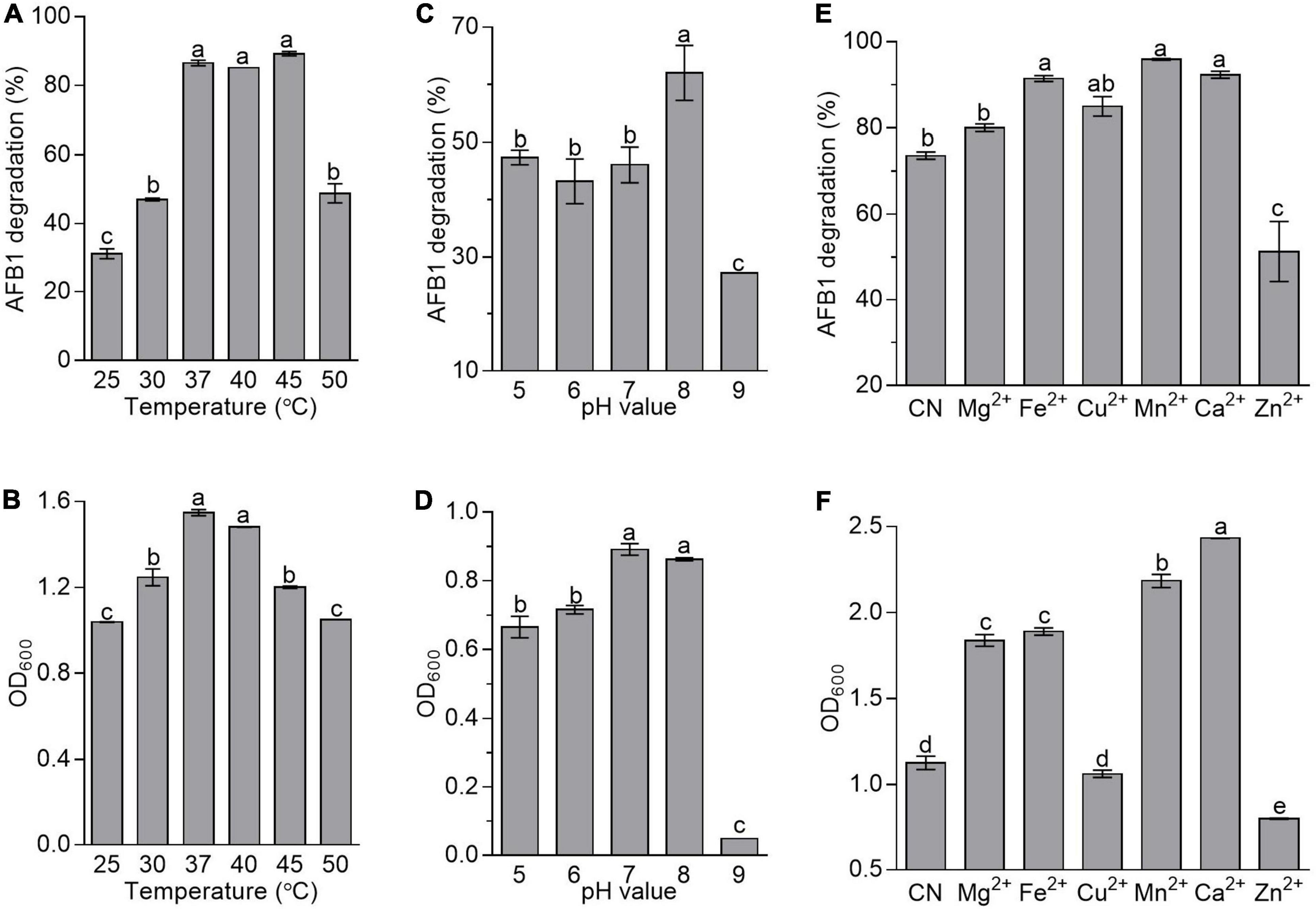
Figure 4. Effects of temperature (A,B), initial pH value (C,D), and metal ions (E,F) on the AFB1 degradation (A,C,E) and bacterial growth (B,D,F) in B. amyloliquefaciens WF2020. In terms of temperature, the residual AFB1 was analyzed after B. amyloliquefaciens WF2020 was co-incubated with 2 μg/ml AFB1 for 72 h. With respect to the effects of initial pH value and metal ions, the residual AFB1 was analyzed after B. amyloliquefaciens WF2020 was co-incubated with 2 μg/ml AFB1 at 37°C for 24 and 48 h, respectively. Different lowercase letters on the bars of each group indicate significant differences between the treatments (Tukey’s test, p < 0.05).
In B. velezensis DY3108, AFB1 degradation capability decreased in parallel with a decrease in initial pH (Shu et al., 2018), suggesting that the initial pH of the medium might be a critical factor in AFB1 degradation mediated by Bacillus. Here, AFB1 degradation mediated by B. amyloliquefaciens WF2020 was also sensitive to the initial pH of the medium. The percentage of AFB1 degradation was 47.36, 43.20, 46.08, 62.02, and 27.01% on average at an initial pH of 5, 6, 7, 8, and 9, respectively (Figure 4C), indicating that an initial pH value at 8.0 favored degradation when AFB1 was co-incubated with B. amyloliquefaciens WF2020. Moreover, the growth of B. amyloliquefaciens WF2020 at an initial pH of 5, 6, and 9 was inhibited by 25.36, 19.70, and 94.50%, respectively, compared with that at an initial pH of 7, and bacterial growth at an initial pH of 7 and 8 showed no significant difference (Figure 4D). Thus, the significant decrease in the percentage of AFB1 degradation mediated by B. amyloliquefaciens WF2020 at pH 9.0 might be due to the severe bacterial growth defects at pH 9.0.
With respect to the effect of metal ions on AFB1 degradation mediated by B. amyloliquefaciens WF2020, it was observed that, compared with the control, Mn2+, Ca2+, Fe2+, and Cu2+ stimulated degradation by 30.24, 25.35, 24.14, and 15.36%, respectively, and Mg2+ showed no significant difference though the percentage of AFB1 degradation increased by 8.61%, whereas Zn2+ inhibited degradation by 30.39% (Figure 4E). Moreover, compared with the control group, the growth of B. amyloliquefaciens WF2020 treated with Mg2+, Fe2+, Mn2+, and Ca2+ increased by 63.22, 67.84, 94.00, and 116.13%, respectively, but Zn2+ inhibited bacterial growth by 29.05% and Cu2+ had no significant effect on bacterial growth (Figure 4F). Combined with the effects of different metal ions on the AFB1 degradation ability of the active component of B. amyloliquefaciens WF2020, we speculated that changes in AFB1 degradation by B. amyloliquefaciens WF2020 caused by Fe2+, Mn2+, and Zn2+ might be due to the effects of corresponding metal ions on bacterial growth and active component capacities, and the increase in AFB1 degradation by B. amyloliquefaciens WF2020 caused by Ca2+ and Cu2+ might be attributed to the increase in bacterial growth and active component capacities caused by the corresponding metal ions, respectively.
Effects of Bacillus amyloliquefaciens WF2020 on the Fungal Growth and Production of Aflatoxin B1 in Aspergillus flavus
Except for AFB1 degradation, B. amyloliquefaciens WF2020 could inhibit the fungal growth of A. flavus and reduce AFB1 production. Pairwise interaction on agar plates proved that B. amyloliquefaciens WF2020 inhibited the fungal growth of A. flavus (Figure 5A). Moreover, the dry weight of the co-incubation culture of B. amyloliquefaciens WF2020 and A. flavus was reduced by 6.55% compared with that of A. flavus culture (Figure 5B). Additionally, B. amyloliquefaciens WF2020 completely inhibited AFB1 production when co-incubated with A. flavus in PDB for 2 days (Figure 5C). Accompanied by a reduction of AFB1 production, B. amyloliquefaciens WF2020 suppressed the transcriptional expression of 10 aflatoxin pathway genes (aflA, aflB, aflE, aflG, aflH, aflJ, aflK, aflL, aflO, and aflQ) and 2 gene encoding transcription factor aflR and aflS by 22.44–100% but increased the expression of aflM, an aflatoxin pathway gene, by 146.98% (Figure 5D). The downregulated expression of 10 aflatoxin pathway genes and 2 transcription factors suggested that AFB1 synthesis might be inhibited by B. amyloliquefaciens WF2020, which might result in reduced AFB1 production caused by B. amyloliquefaciens WF2020.
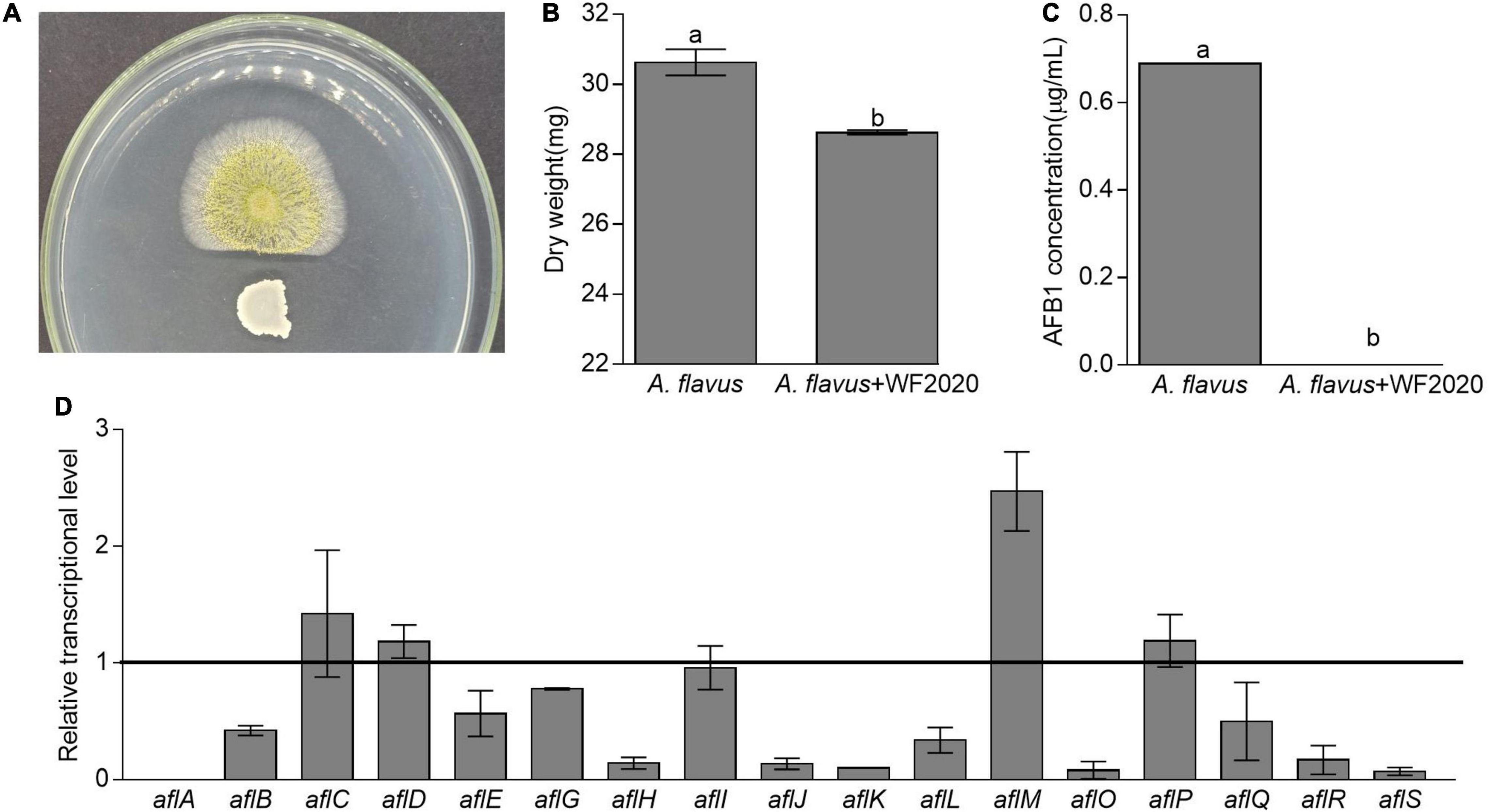
Figure 5. Changes in fungal growth, AFB1 production, and transcriptional expression of genes involved in AFB1 synthesis in Aspergillus flavus. (A) The antagonistic effect of B. amyloliquefaciens WF2020 on fungal growth after the bacterium was co-incubated with A. flavus for 2 days on a plate of potato dextrose agar (PDA). (B,C) The reduction of dry weight (B) and AFB1 production (C) caused by B. amyloliquefaciens WF2020 after the bacterium was co-incubated with A. flavus for 2 days in the potato dextrose broth (PDB). (D) Quantitation of relative transcriptional levels of selected genes associated with AFB1 synthesis in A. flavus via real-time quantitative polymerase chain reaction (qRT-PCR) after B. amyloliquefaciens WF2020 was co-incubated with A. flavus for 2 days in PDB. The line represents the transcriptional levels of genes in control experiments, which were defined as 1. Different lowercase letters in the bars of each group indicate significant differences between treatments (Tukey’s test, p < 0.05).
Discussion
Generally, B. amyloliquefaciens was considered as a safe and non-toxic producing microbe and could be used for food and pharmaceutical purposes (WoldemariamYohannes et al., 2020). It was also reported that some strains of B. amyloliquefaciens, such as B. amyloliquefaciens UTB2, UNRC52, UNRCLR, S8C, Y1-B1, SWUN-TP23, SG-16, and HSP-5, could inhibit AFB1 synthesis or accelerate AFB1 degradation (Bluma and Etcheverry, 2006; Xu et al., 2015; Guo et al., 2017; Siahmoshteh et al., 2018; Wang J. et al., 2018; Ali et al., 2021; Zhang et al., 2021). Here, our results indicated that B. amyloliquefaciens WF2020 derived from naturally fermented pickles could act as a potential probiotic to efficiently detoxify AFB1 in a time dependent manner in ranges of 1–8 μg/ml and inhibit the fungal growth of A. flavus and AFB1 production, as discussed below.
Firstly, B. amyloliquefaciens WF2020 can degrade AFB1 ranging from 1 to 5 μg/ml by more than 80% after a 72-h incubation, which was similar to the 85.50% reduction of AFB1 at the concentration of 0.5 μg/ml reported in B. amyloliquefaciens SG16 (Wang J. et al., 2018) and was significantly higher than the 42.13 and 58.77% reduction reported in B. amyloliquefaciens SWUN-TP23 and HSP-5, respectively (Xu et al., 2015; Guo et al., 2017) and the 40 and 73.2% reduction of AFB1 at the concentration of 0.5 μg/ml reported in B. amyloliquefaciens S8C and Y1-B1, respectively (Ali et al., 2021; Zhang et al., 2021). Compared with the degradation abilities of AFB1 in the reported Bacillus species, the degradation ability in B. amyloliquefaciens WF2020 is similar to that in B. licheniformis BL010 (Wang Y. et al., 2018), B. velezensis DY3108 (Shu et al., 2018), and B. subtilis UTBSP1 (Farzaneh et al., 2012), higher than that in B. subtilis JSW-1 (Xia et al., 2017) but slightly lower than that in Bacillus sp. TUBF1 (El-Deeb et al., 2013) and B. licheniformis CFR1 (Rao et al., 2016), suggesting that there were great differences in degradation efficiency from one strain to other. Moreover, AFB1 degradation by B. amyloliquefaciens WF2020 was affected by fermentation temperatures, initial pH values, and metal ions. The temperature and initial pH value at the maximum degradation of AFB1 were 45°C and pH 8.0, respectively. Mn2+, Ca2+, Fe2+, and Cu2+ stimulated AFB1 degradation, and Mg2+ had no effect but Zn2+ inhibited the degradation. Compared with the reported Bacillus strains, the temperature was higher than the estimates of 30°C observed in B. velezensis DY3108 and 37°C observed in B. cereus CaG6 (Abdel-Shafi et al., 2018; Shu et al., 2018), and the pH value was the same to that observed in B. velezensis DY3108 (Shu et al., 2018). The stimulation induced by Ca2+ and the inhibition induced by Zn2+ were in agreement with the results in Myroides odoratimimus 3J2MO, but the stimulation induced by Fe2+, and Cu2+ and unchanged degradation caused by Mg2+ were opposite to the findings in M. odoratimimus 3J2MO (Mwakinyli et al., 2019). Meanwhile, the stimulation induced by Mn2+ was also opposite to that in M. odoratimimus 3J2MO (Mwakinyli et al., 2019) but was in well agreement with that in B. cereus CaG6 (Abdel-Shafi et al., 2018).
Secondly, the removal of mycotoxins by microbes was mainly attributed to adsorption and degradation (Hathout and Aly, 2014). In B. amyloliquefaciens WF2020, the removal of AFB1 was mainly dependent on degradation, and extracellular proteins or enzymes were the main active ingredient, which was similar to previous studies on AFB1 degradation mediated by Bacillus, such as B. amyloliquefaciens SG16 (Wang J. et al., 2018), B. licheniformis CFR1 (Rao et al., 2016), B. subtilis UTBSP1 and JSW-1 (Farzaneh et al., 2012; Xia et al., 2017), B. velezensis DY3108 (Shu et al., 2018), and B. shackletonii L7 (Xu et al., 2017). Moreover, the AFB1 degradation ability of extracellular proteins or enzymes was affected by temperature, the pH value, and metal ions. Increased temperatures may have promoted the bioavailability of organic compounds and facilitated biodegradation (Müller et al., 1998). Here, the percentage of AFB1 degradation mediated by the cell-free supernatant increased with the increase of temperature up to 60°C where 100% of AFB1 was removed, and the percentage of AFB1 degradation at 70°C remained more than 70%. Compared with the reported Bacillus, the thermostability of the cell-free supernatant of B. amyloliquefaciens WF2020 was similar to that from B. shackletonii L7 (Xu et al., 2017) and higher than that from B. amyloliquefaciens SG16 (Wang J. et al., 2018), B. licheniformis CFR1 (Rao et al., 2016), and B. subtilis UTBSP1 (Farzaneh et al., 2012), but slightly lower than that of B. velezensis DY3108 (Shu et al., 2018). Additionally, the cell-free supernatant of B. amyloliquefaciens WF2020 could still degrade AFB1 by 37.16% after boiling for 20 min, which was lower than that of B. amyloliquefaciens Y1-B1 (Zhang et al., 2021). These results demonstrated that extracellular proteins or enzymes were thermostable and could work well within a wide range of working temperature, which was helpful for application in food and feed processing and industry for AFB1 degradation. With respect to pH values, the optimal pH value of the cell-free supernatant from B. amyloliquefaciens WF2020 was 8.0, which was the same to that of extracellular enzymes from E. coli CG1061 (Wang et al., 2019), Stenotrophomonas maltophilia 35-3 (Guan et al., 2008), B. shackletonii L7 (Xu et al., 2017), and B. velezensis DY3108 (Shu et al., 2018) and was slightly higher than 7.5 reported in B. amyloliquefaciens SG16 (Wang J. et al., 2018). In addition, the AFB1 degradation ability of the cell-free supernatant from B. amyloliquefaciens WF2020 was increased by Mn2+, Mg2+, Fe2+, and Cu2+ and inhibited by Zn2+ but was not affected by Ca2+, inferring that Mn2+, Mg2+, Fe2+, and Cu2+ may act as enzyme activators, membrane stabilizers, and help to maintain the structural integrity of proteins. The enhancement of AFB1 degradation ability induced by Cu2+ and the inhibition of AFB1 degradation caused by Zn2+ were in agreement with the findings of extracellular enzymes or culture supernatant in B. shackletonii L7 (Xu et al., 2017), B. licheniformis CFR1 (Rao et al., 2016), and B. velezensis DY3108 (Shu et al., 2018). Cu2+ may take part in the redox reaction in electron transport, transferring an oxygen atom to the AFB1 substrate, and the oxidized AFB1 would then be hydrolyzed into non-toxic products (Xu et al., 2017). It has been reported that the inhibition of AFB1 degradation by Zn2+ might be due to the change in enzyme conformation caused by Zn2+, which resulted in decreased affinity of AFB1 (D’souza and Brackett, 1998). The stimulation of AFB1 degradation caused by Mg2+ was similar to that of B. licheniformis CFR1 (Rao et al., 2016), but opposite to that of B. amyloliquefaciens SG16 (Wang J. et al., 2018) and B. shackletonii L7 (Xu et al., 2017). The increase in AFB1 degradation induced by Mn2+ was opposite to that in B. amyloliquefaciens SG16 (Wang J. et al., 2018), B. shackletonii L7 (Xu et al., 2017), and B. velezensis DY3108 (Shu et al., 2018) and different from no obvious changes in B. licheniformis CFR1 (Rao et al., 2016). The increase of AFB1 degradation induced by Fe2+ was opposite to that in B. amyloliquefaciens SG16 (Wang J. et al., 2018) and B. licheniformis CFR1 (Rao et al., 2016).
Thirdly, the application of B. amyloliquefaciens WF2020 in AFB1 degradation was safe. On one hand, B. amyloliquefaciens WF2020 could act as a safe and non-toxic producing microbe. Based on the genomic sequencing analysis, B. amyloliquefaciens WF2020 produces several active compounds such as macrolactin, bacillaene, fengycin, difficidin, bacillibactin, and bacilysin and does not contain virulence genes and any plasmid. Additionally, B. amyloliquefaciens WF2020 is not an antibiotic-resistant bacterium due to susceptibility to various antibiotics, including tetracycline, penicillin, cefalexin, ampicillin, streptomycin, kanamycin, gentamicin, ciprofloxacin, chloramphenicol, vancomycin, imipenem, rifampin, erythromycin, and norfloxacin. Moreover, B. amyloliquefaciens WF2020 significantly enhanced the lifespan of C. elegans by an average of 20.78%, which was slightly lower than that of B. amyloliquefaciens EnB-alf1 isolated from alfalfa (Medicago sativa L.) seeds (Zhang et al., 2019). On the other hand, B. amyloliquefaciens WF2020 converted AFB1 into metabolites with a loss of mutagenicity and non-toxicity to the lifespan of C. elegans. The loss of mutagenicity was also observed in Aspergillus oryzae MAO103 and MAO104, Aspergillus niger RAF106, B. licheniformis CFR1, and Rhodococcus erythropolis (Alberts et al., 2006; Rao et al., 2016; Lee et al., 2017; Fang et al., 2020). The detoxification of AFB1 was mainly focused on the damage of the AFB1 toxic group of coumarin, which is a carcinogenic group, and bifuran nucleus, which are basic toxic structures (Xie et al., 2019). The loss of mutagenicity and the mortality rate of C. elegans suggested that B. amyloliquefaciens WF2020 might detoxify AFB1 into non-toxic compounds with the damage of coumarin and/or bifuran nucleus. These findings demonstrated that B. amyloliquefaciens WF2020 could act as a probiotic used to degrade AFB1 in food and feed.
Lastly, B. amyloliquefaciens WF2020 could slightly inhibit the fungal growth of A. flavus, completely reduce AFB1 production, and significantly suppress the expression of some important genes involved in the synthesis of aflatoxins, such as aflA, aflB, alfE, alfG, alfH, alfJ, alfK, alfL, alfO, alfQ, alfR, and alfS. The inhibition of the fungal growth of A. flavus was lower than that in B. amyloliquefaciens UNRC52, UNRCLR, and HSP-5, Bacillus safensis RF69, Bacillus. sp. RP103, and Bacillus sp. RP242 (Bluma and Etcheverry, 2006; Xu et al., 2015; Einloft et al., 2021). The reduction in AFB1 production was similar to that in B. amyloliquefaciens UTB2, B. amyloliquefaciens UNRC52, and B. amyloliquefaciens UNRCLR, but greater than that in B. safensis RF69, Bacillus. sp. RP103, and Bacillus sp. RP242 (Bluma and Etcheverry, 2006; Siahmoshteh et al., 2018; Einloft et al., 2021). aflA, aflB, alfE, alfG, alfH, alfJ, alfK, alfL, alfO, and alfQ are important aflatoxin pathway genes, which encode two fatty acid synthases, a norsolorinic acid ketoreductase, a P450 monooxygenase, an alcohol dehydrogenase, an esterase, versicolorin B synthase, a cytochrome P450 monooxygenase, O-methyltransferase B, and a P450 monooxygenase, respectively (Yu, 2012). alfR, encoding the positive-acting transcription factor, is required for the transcriptional activation of most, if not all, structural genes in the aflatoxin gene cluster, such as aflB, alfE, alfG, alfH, alfJ, alfK, alfL, alfO, and alfQ (Price et al., 2006; Yu, 2012). alfS, bidirectionally transcribed from aflR, is necessary for aflatoxin formation by regulating several aflatoxin pathway genes, such as alfA and aflB (Yu, 2012). Therefore, it was speculated that B. amyloliquefaciens WF2020 might inhibit AFB1 synthesis by downregulating the expression of aflR, aflS, and several important aflatoxin pathway genes. The reduction in AFB1 production might be attributed to the inhibition of fungal growth and AFB1 synthesis and AFB1 degradation caused by B. amyloliquefaciens WF2020.
Conclusion
Bacillus amyloliquefaciens WF2020 could act as a potential probiotic with susceptibility to various antibiotics, the synthesis of several active substances, and beneficial effects on the lifespan of C. elegans to degrade AFB1 into non-toxic products over a wide pH range from 5 to 9 and the temperature from 25 to 50°C. Bacterial growth and AFB1 degradation ability of B. amyloliquefaciens WF2020 were also affected by metal ions, including Mg2+, Fe2+, Cu2+, Mn2+, Ca2+, and Zn2+. This degradation was mainly attributed to extracellular proteins or enzymes possessing a wide reaction temperature ranging from 20 to 70°C and pH ranging from 5 to 9, which will be helpful for their application in the harsh conditions during food and feed processing. Moreover, B. amyloliquefaciens WF2020 also could inhibit fungal growth, reduce AFB1 production, and downregulate the expression of several aflatoxin pathway genes and two transcription factors (aflR and aflS) in A. flavus. Therefore, B. amyloliquefaciens WF2020 and/or its enzymes or proteins in the supernatant are new promising agents to protect food and feed from AFB1 contamination. However, the structure of degradation products and the purification of enzymes or proteins merit further investigation to elucidate the mechanisms of AFB1 degradation mediated by B. amyloliquefaciens WF2020, which will be helpful to exploit the probable agents used in food and feed processing to reduce AFB1 contamination.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, CP092778.
Author Contributions
GC and QF designed and performed the experiments, analyzed the data, and prepared this manuscript. ZheL performed the experiments and revised this manuscript. CX and ZhiL analyzed the data. QZ and LW designed the experiments. XF contributed to the revision of this manuscript. JW contributed to the revision of this manuscript and overall support of this study. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Guangdong Province Science and Technology Innovation Strategy Special Fund (2020B020226008), Guangzhou Science and Technology Research Projects (201904010274), Yangjiang Science and Technology Research Projects (SDZX2020030), and the opening foundation from State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology (SKLAM003-2018).
Conflict of Interest
CX and ZhiL are employed by Guangdong Moyanghua Grains and Oils Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.891091/full#supplementary-material
Footnotes
- ^ http://canu.readthedocs.io/en/latest/
- ^ http://ccb.jhu.edu/software/glimmer/index.shtml
- ^ ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.3.0/
- ^ https://github.com/bbuchfink/diamond
- ^ http://www.hmmer.org/
- ^ www.majorbio.com
References
Abdel-Shafi, S., Shehata, S., Shindia, A., Ei-Meligy, K., and Khidr, A. (2018). Biodegradation of aflatoxins by bacteria. Egyptian J. Microbiol. 53, 241–254. doi: 10.21608/ejm.2018.5752.1078
Alberts, J. F., Engelbrecht, Y., Steyn, P. S., Holzapfel, W. H., and van Zyl, W. H. (2006). Biological degradation of Aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 109, 121–126. doi: 10.1016/j.ijfoodmicro.2006.01.019
Ali, S., Hassan, M., Essam, T., Ibrahim, M. A., and Al-Amry, K. (2021). Biodegradation of aflatoxin by bacterial species isolated from poultry farms. Toxicon 195, 7–16. doi: 10.1016/j.toxicon.2021.02.005
Bluma, R. V., and Etcheverry, M. G. (2006). Influence of Bacillus spp. isolated from maize agroecosystem on growth and aflatoxin B1 production by Aspergillus section Flavi. Pest Manag. Sci. 62, 242–252. doi: 10.1002/ps.1154
Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi: 10.1093/genetics/77.1.71
Catherine, N., Quynh, V. H., Annika, G., Simon, C., Thi, N. T. T., and Jacques, M. (2020). Bacilysin within the Bacillus subtilis group: gene prevalence versus antagonistic activity against gram-negative foodborne pathogens. J. Biotechnol. 327, 28–35. doi: 10.1016/j.jbiotec.2020.12.017.327
Chen, N., Liu, Y., Qin, P., Li, Y., Ma, Y., Li, J., et al. (2021). Antibacterial activities of Bacillus amyloliquefaciens DQB-1 isolated from the cecum of Dezhou donkeys. J. Equine Vet. Sci. 102:103616. doi: 10.1016/j.jevs.2021.103616
Chen, X., Scholz, R., Borriss, M., Junge, H., Mögel, G., Kunz, S., et al. (2008). Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 140, 38–44. doi: 10.1016/j.jbiotec.2008.10.015
Cochrane, S. A., and Vederas, J. C. (2016). Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. 36, 4–31. doi: 10.1002/med
D’souza, D. H., and Brackett, R. E. (1998). The role of trace metal ions in aflatoxin B1 degradation by Flavobacterium aurantiacum. J. Food Prot. 61, 1666–1669. doi: 10.1089/cmb.1998.5.747
Donato, V., Ayala, F. R., Cogliati, S., Bauman, C., Costa, J. G., Le Ini, C., et al. (2017). Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signalling pathway. Nat. Commun. 8:14332. doi: 10.1038/ncomms14332
Einloft, T. C., Oliveria, P. B. D., Radünz, L. L., and Dionello, R. G. (2021). Biocontrol capabilities of three Bacillus isolates towards aflatoxin B1 producer A. flavus in vitro and on maize grains. Food Control. 125:107978. doi: 10.1016/j.foodcont.2021.107978
El-Deeb, B., Altalhi, A., Khiralla, G., Hassan, S., and Gherbawy, Y. (2013). Isolation and characterization of endophytic bacilli bacterium from maize grains able to detoxify Aflatoxin B1. Food Biotechnol. 27, 199–212. doi: 10.1080/08905436.2013.811083
Erega, A., Stefanic, P., Dogsa, I., Danevcic, T., Simunovic, K., Klancnik, A., et al. (2021). Bacillaene mediates the inhibitory effect of Bacillus subtilis on Campylobacter jejuni biofilm. Appl. Environ. Microbiol. 87:e0295520. doi: 10.1128/AEM.02955-2920
Eskola, M., Kos, G., Elliott, C. T., Hajšlová, J., Mayar, S., and Krska, R. (2019). Worldwide contamination of food-crops with mycotoxins: validity of the widely cited ‘FAO estimate’ of 25%. Critical Rev. Food Sci. Nutrition 60, 2773–2789. doi: 10.1080/10408398.2019.1658570
Fang, Q., Du, M., Chen, J., Liu, T., Zheng, Y., Liao, Z., et al. (2020). Degradation and detoxification of aflatoxin B1 by tea-derived Aspergillus niger RAF106. Toxins 12:777. doi: 10.3390/toxins12120777
Farzaneh, M., Shi, Z., Ghassempour, A., Sedaghat, N., Ahmadzadeh, M., Mirabolfathy, M., et al. (2012). Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control. 23, 100–106. doi: 10.1016/j.foodcont.2011.06.018
Guan, S., Cheng, J., Ting, Z., Junxia, L., Qiugang, M., and Tiangui, N. (2008). Aflatoxin B1 degradation by Stenotrophomonas Maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 9, 1489–1503. doi: 10.3390/ijms9081489
Guo, Z., Zhou, B., and Yang, H. (2017). Study on probiotic function, safety and application of Bacillus amyloliquefaciens from Tibetan pigs. Jiangsu Agricultural Sci. 45, 137–140. doi: 10.15889/j.issn.1002-1302.2017.14.038
Hathout, A. S., and Aly, S. E. (2014). Biological detoxification of mycotoxins: a review. Annals Microbiol. 64, 905–919. doi: 10.1007/s13213-014-0899-897
IARC (1993). Monographs on the evaluation of carcinogenic risks to humans: some naturally occurring substances, food items and constituents. Heterocyclic Aromatic Amines Mycotoxins 56, 498–521.
Kaushik, K. B., Tanmay, S., Arabinda, G., Debabrat, B., Bijuli, R., Manasa, K. P., et al. (2021). Macrolactin a as a novel inhibitory agent for SARS-CoV-2 Mpro: bioinformatics approach. Appl. Biochem. Biotechnol. 193, 1–24. doi: 10.1007/s12010-021-03608-3607
Kumar, A., Baruah, A., Tomioka, M., Lino, Y., Kalita, M., and Khan, M. (2020). Caenorhabditis elegans: a model to understand host-microbe interactions. Cell. Mol. Life Sci. 77, 1229–1249. doi: 10.1007/s00018-019-03319-3317
Laciakova, A., Cicooova, P., Mate, D. M., and Laciak, V. (2008). Aflatoxins and possibilities for their biological detoxification. Medycyna Weterynaryjna 64, 276–279.
Lee, H. J., and Ryu, D. (2017). Worldwide occurrence of mycotoxins in cereals and cereal-derived food products: public health perspectives of their co-occurrence. J. Agric. Food Chem. 65, 7034–7051. doi: 10.1021/acs.jafc.6b04847
Lee, K. R., Yang, S. M., Cho, S. M., Kim, M., Hong, S. Y., and Chung, S. H. (2017). Aflatoxin B1 detoxification by Aspergillus oryzae from Meju, a traditional Korean fermented soybean starter. J. Microbiol. Biotechnol. 27, 57–66. doi: 10.4014/jmb.1607.07064
Liang, X., Mohamed, F. E. A., Lancine, S., Yueju, Z., Jonathan, N. S., Fuguo, X., et al. (2017). Novel aflatoxin-degrading enzyme from Bacillus shackletonii L7. Toxins 9:36. doi: 10.3390/toxins9010036
Mishra, H. N., and Das, C. (2003). A review on biological control and metabolism of aflatoxin. Crit. Rev. Food Sci. Nutr. 43, 245–264. doi: 10.1080/10408690390826518
Müller, R., Antranikian, G., Maloney, S., and Sharp, S. (1998). Thermophilic degradation of environmental pollutants. Biotechnol. Extremophiles 61, 155–169. doi: 10.1007/BFb0102292
Mwakinyli, S. E., Ming, Z., Xie, H., Zhang, Q., and Li, P. (2019). Investigation and characterization of Myroides odoratimimus strain 3J2MO aflatoxin B1 degradation. J. Agric. Food Chem. 67, 4595–4602. doi: 10.1021/acs.jafc.8b06810
Ngalimat, M. S., Yahaya, R. S. R., Baharudin, M. M. A., Yaminudin, S. M., Karim, M., Ahmad, S. A., et al. (2021). A review on the biotechnological applications of the operational group Bacillus amyloliquefaciens. Microorganisms 9:614. doi: 10.3390/microorganisms9030614
Price, M. S., Yu, J., Nierman, W. C., Kim, H. S., Pritchard, B., Jacobus, C. A., et al. (2006). The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol. Lett. 255, 275–279. doi: 10.1111/j.1574-6968.2005.00084.x
Rao, R. K., Vipin, A. V., Hariprasad, P., Appaiah, K. A. A., and Venkateswaran, G. (2016). Biological detoxification of aflatoxin B1 by Bacillus licheniformis CFR1. Food Control. 71, 234–241. doi: 10.1016/j.foodcont.2016.06.040
Ryohei, T., Prof, D. F. K., Mario, N. P. D., and Tadashi, E. (2015). Epimerization at C-3” in butirosin biosynthesis by an NAD(+)-dependent dehydrogenase BtrE and an NADPH-dependent reductase BtrF. ChemBioChem 16, 487–495. doi: 10.1002/cbic.201402612
Shu, X., Wang, Y., Zhou, Q., Li, M., Hu, H., Ma, Y., et al. (2018). Biological degradation of Aflatoxin B1 by cell-free extracts of Bacillus velezensis DY3108 with broad PH stability and excellent thermostability. Toxins 10:330. doi: 10.3390/toxins10080330
Siahmoshteh, F., Hamidi-Esfahani, Z., Spadaro, D., Shams-Ghahfarokhi, M., and Razzaghi-Abyaneh, M. (2018). Unraveling the mode of antifungal action of Bacillus subtilis and Bacillus amyloliquefaciens as potential biocontrol agents against aflatoxigenic Aspergillus parasiticus. Food Control. 89, 300–307. doi: 10.1016/j.foodcont.2017.11.010
Silvia, M., Andrea, P., Andrea, A., Salvatore, V., Susan, C., and Lorella, S. (2018). Aflatoxin B1 and M1: biological properties and their involvement in cancer development. Toxins 10:214. doi: 10.3390/toxins10060214
Verheecke, C., Liboz, T., and Mathieu, F. (2016). Microbial degradation of aflatoxin B1: current status and future advances. Int. J. Food Microbiol. 237, 1–9. doi: 10.1016/j.ijfoodmicro.2016.07.028
Wang, C., Li, Z., Wang, H., Qiu, H., Zhang, M., Li, S., et al. (2017). Rapid biodegradation of Aflatoxin B1 by metabolites of Fusarium sp. WCQ3361 with broad working temperature range and excellent thermostability. J. Sci. Food Agriculture 97, 1342–1348. doi: 10.1002/jsfa.7872
Wang, J., Song, G., Yang, R., Han, L., and Zhao, Z. (2018). Isolation and analysis of aflatoxin degrading bacterial strain from compost soil. China Feed 23, 18–23. doi: 10.15906/j.cnki.cn11-2975/s.20182304
Wang, L., Wu, J., Liu, Z., Shi, Y., Liu, J., Xu, X., et al. (2019). Aflatoxin B1 degradation and detoxification by Escherichia coli CG1061 isolated from Chicken Cecum. Front. Pharmacol. 9:1548. doi: 10.3389/fphar.2018.01548
Wang, Y., Zhang, H., Yan, H., Yin, C., Liu, Y., Xu, Q., et al. (2018). Effective biodegradation of aflatoxin B1 using the Bacillus licheniformis (BL010) Strain. Toxins 10:497. doi: 10.3390/toxins10120497
WoldemariamYohannes, K., Wan, Z., Yu, Q., Li, H., Wei, X., Liu, Y., et al. (2020). Prebiotic, probiotic, antimicrobial, and functional food applications of Bacillus amyloliquefaciens. J. Agric. Food Chem. 2020, 14709–147327. doi: 10.1021/acs.jafc.0c06396
Wu, L., Wu, H., Chen, L., Yu, X., Borriss, R., and Gao, X. (2015). Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 5:12975. doi: 10.1038/srep12975
Wu, Q., Jezkova, A., Yuan, Z., Pavlikova, L., Dohnal, V., and Kuca, K. (2009). Biological degradation of aflatoxins. Drug Metab. Rev. 41, 1–7. doi: 10.1080/03602530802563850
Xia, X., Zhang, Y., Li, M., Garba, B., Zhang, Q., Wang, Y., et al. (2017). Isolation and characterization of a Bacillus subtilis strain with aflatoxin B-1 biodegradation capability. Food Control. 75, 92–98. doi: 10.1016/j.foodcont.2016.12.036
Xie, Y., Wang, W., and Zhang, S. (2019). Purification and identification of an aflatoxin B1 degradation enzyme from Pantoea sp. T6. Toxicon 157, 35–42. doi: 10.1016/j.toxicon.2018.11.290
Xu, L., Ahmed, M. F. E., Sangare, L., Zhao, Y., Selvaraj, J. N. S., Xing, Y., et al. (2017). Novel aflatoxin-degrading enzyme from Bacillus shackletonii L7. Toxins 9:36.
Xu, M., Cai, G., Zhu, D., Wang, L., and Lu, J. (2015). Isolation and identification of strain degrading aflatoxin B1 and its application in peanut meal. China Oils Fats 40, 20–24.
Yan, Z., Yousef, H., Dion, L., Shao, S., and Zhou, T. (2017). Strategies and methodologies for developing microbial detoxification systems to mitigate mycotoxins. Toxins 9:130. doi: 10.3390/toxins9040130
Yang, Z., Xue, K. S., Sun, X., Tang, L., and Wang, J. (2015). Multi-toxic endpoints of the foodborne mycotoxins in nematode Caenorhabditis elegans. Toxins 7, 5224–5235. doi: 10.3390/toxins7124876
Yu, J. (2012). Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins 4, 1024–1057. doi: 10.3390/toxins4111024
Zhang, H. L., Jia, F., Li, M., Yu, F., Zhou, B., Hao, Q. H., et al. (2019). Endophytic Bacillus strains isolated from alfalfa (Medicago sativa L.) seeds: enhancing the lifespan of Caenorhabditis elegans. Lett. Appl. Microbiol. 68, 226–233. doi: 10.1111/lam.13102
Zhang, W., Xue, B., Li, M., Mu, Y., Chen, Z., Li, J., et al. (2014). Screening a strain of Aspergillus niger and optimization of fermentation conditions for degradation of aflatoxin B1. Toxins 6, 2072–6651. doi: 10.3390/toxins6113157
Zhang, X., Zhang, Y., Zhang, X., Qiao, H., and Bian, C. (2021). Isolation and identification of aflatoxin B1 degrading Bacillus amyloliquefaciens and its in vitro detoxification effect. Acta Vet. Zootechnica Sinica 52, 2291–2301.
Zhou, M., Liu, F., Yang, X., Jin, J., Dong, X., Zeng, K., et al. (2018). Bacillibactin and bacillomycin analogues with cytotoxicities against human cancer cell lines from marine Bacillus sp. PKU-MA00093 and PKU-MA00092. Mar. Drugs 16:22. doi: 10.3390/md16010022
Keywords: aflatoxin B1, Bacillus amyloliquefaciens, Aspergillus flavus, genome sequence, Ames test, Caenorhabditis elegans
Citation: Chen G, Fang Q, Liao Z, Xu C, Liang Z, Liu T, Zhong Q, Wang L, Fang X and Wang J (2022) Detoxification of Aflatoxin B1 by a Potential Probiotic Bacillus amyloliquefaciens WF2020. Front. Microbiol. 13:891091. doi: 10.3389/fmicb.2022.891091
Received: 07 March 2022; Accepted: 28 March 2022;
Published: 10 May 2022.
Edited by:
Alicia Rodríguez, University of Extremadura, SpainReviewed by:
Fuguo Xing, Institute of Food Science and Technology (CAAS), ChinaZhendong Cai, Ningbo University, China
Copyright © 2022 Chen, Fang, Liao, Xu, Liang, Liu, Zhong, Wang, Fang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Wang, wangjielangjing@126.com; Xiang Fang, fxiang@scau.edu.cn
†These authors have contributed equally to this work
 Guojun Chen1†
Guojun Chen1† Qian’an Fang
Qian’an Fang Zhenlin Liao
Zhenlin Liao Qingping Zhong
Qingping Zhong Li Wang
Li Wang Xiang Fang
Xiang Fang