- 1National Engineering Research Center of Edible Fungi, Institute of Edible Fungi, Shanghai Academy of Agricultural Sciences, Shanghai, China
- 2College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 3Shandong Provincial Key Laboratory of Applied Mycology, School of Life Sciences, Qingdao Agricultural University, Qingdao, China
Most of the sequenced wood-rotting edible mushroom produce fruiting body at relatively low temperatures. Little information has been known about the high-temperature wood-rotting mushroom. Here, we performed de novo sequencing and assembly of the genome of a high-temperature edible mushroom Pleurotus giganteus from a monokaryotic strain zhudugu2 using the Illumina and Pac-Bio CLR sequencing technologies. P. giganteus, also known as Zhudugu in China, is a well-known culinary edible mushroom that has been widely distributed and cultivated in China, Southeast Asia, and South Asia. The genome consists of 40.00 Mb in 27 contigs with a contig N50 of 4.384 Mb. Phylogenetic analysis reveals that P. giganteus and other strains in Pleurotus clustered in one clade. Phylogenetic analysis and average nucleotide identity analysis indicated that the P. giganteus genome showed a closer relationship with other Pleurotus species. Chromosome collinearity analysis revealed a high level of collinearity between P. ostreatus and P. giganteus. There are 12,628 protein-coding genes annotated in this monoploid genome. A total of 481 enzymes accounting for 514 carbohydrate-active enzymes (CAZymes) terms were identified in the P. giganteus genome, including 15 laccases and 10 class II peroxidases predicted in the genome, which revealed the robustness of lignocellulose degradation capacity of P. giganteus. The mating-A type locus of P. giganteus consisted of a pair of homeodomain mating-type genes HD1 and HD2. The mating-B type locus of P. giganteus consisted of at least four pheromone receptor genes and three pheromone genes. The genome is not only beneficial for the genome-assisted breeding of this mushroom but also helps us to understand the high-temperature tolerance of the edible mushroom.
Introduction
Pleurotus giganteus is a culinary edible mushroom that has been recorded in tropical and subtropical regions, such as China, Malaysia, Sri Lanka, Indonesia, Vietnam, Laos, and Thailand, and has been commercially cultivated in recent years (Karunarathna et al., 2012; Phan et al., 2019). P. giganteus is known as “Zhudugu” in China due to the organoleptic properties that resemble pork stomach, and it is also named “Dabeisangu,” “Dabeixungu,” “Dalougougu,” “Dabeixianggu,” and “Sungu” in China because that the mature fruiting body of P. giganteus is like a goblet and funnel (Phan et al., 2019). P. giganteus is a wood-rotting fungus that uses sawdust and cottonseed hull as the main growth substrates. Soil casing methods are the major method for the fruiting production of this mushroom, either buried on the ground or the shelf in the factory. P. giganteus was also regarded as a multi-functional food supplement due to the high levels of nutrients and bioactive compounds in this species, such as polysaccharide, uridine, lipids, and feruloyl esterase, with neurite outgrowth, antioxidant, anti-candida, antitumor, hepatoprotective, and amylase inhibitory activities (Phan et al., 2012, 2013, 2014, 2015; Wang et al., 2014; Tian et al., 2016; Baskaran et al., 2017; Debnath et al., 2019).
By studying the mating system, Dong et al. (2010) Reported that P. giganteus has a typical tetrapolar heterothallic mating system. The distribution of the four mating types among spore monokaryons was ∼1:1:1:1 (Dong et al., 2010; Yu et al., 2021). Intraspesies et al. (2020) selected new hydrides of P. giganteus with high biological efficiency using intraspecific mating (Intraspesies et al., 2020; Li et al., 2021). Bioactive compounds and the mating system of this mushroom have been studied for many years; however, molecular and genetic studies on P. giganteus are rare due to the lack of genomic information. Owing to the advent of single-molecule real-time sequencing technologies, the continuity of mushroom genome assemblies [such as Lentinus edodes (Zhang et al., 2021; Yu et al., 2022), Agrocybe cylindracea (Liang et al., 2020), Auricularia heimuer (Yuan et al., 2019; Fang et al., 2020), Stropharia rugosoannulata (Li et al., 2022), Russula griseocarnosa (Yu et al., 2020), Hericium erinaceus (Gong et al., 2020), Phellinus gilvus (Huo et al., 2020), etc.] reached several orders of magnitude higher when compared with Illumina assemblies.
Here, we report a high-quality de novo genome assembly of P. giganteus through a combination of PacBio CLR and Illumina sequencing. Comparative analysis was conducted between the genome of P. giganteus and the other 23 published fungi genomes. Repeat sequences, carbohydrate-active enzymes (CAZymes), lignocellulose degradation enzymes, mating related genes were also analyzed. The P. giganteus genome sequence will help understand the molecular mechanisms and evolution of this important edible mushroom.
Materials and methods
Strains and culture condition
The P. giganteus strain “Shen Xun 1 Hao” was selected for de novo genome sequencing and maintained in the Improved and Standardized Spawn Breeding Center (ISSBC), Shanghai Academy of Agricultural Sciences, China. The strain was domesticated from the wild strain collected in Fujian province in China and identified as a new mushroom variety by Shanghai Agricultural Technology Promotion Center. The P. giganteus strains “Shen Xun 1 Hao” were cultivated and maintained on potato dextrose agar (PDA) plates. For fruiting body production, strain Shen Xun 1 Hao was inoculated into the solid media [40% (w/w) sawdust, 40% (w/w) cottonseed hull, 18% (w/w) wheat bran, and 2% (w/w) gypsum powder] in the polypropylene bag. Vegetative growth of P. giganteus mycelia was carried out at 25°C with a humidity of 70-80%. After the mycelia occupied the full culture bag, the polypropylene bag was open and soil was covered on the top of the media. The fruiting body formed under the stimulation of temperature, water, and light. The monokaryotic strain zhudugu2 was obtained by selecting from the protoplasm of the strain “Shen Xun 1 Hao.”
Genome sequencing
One monokaryon (zhudugu2) of the dikaryon strain “Shen Xun 1 Hao” was originally isolated by protoplast monokaryotization (Zhao and Chang, 1993). The obtained monokaryon zhudugu2 was cultured on 20 Potato Dextrose Agar (PDA) plates covered with cellophane at 25°C in darkness for 16 days. These mycelia were then collected, frozen in liquid nitrogen, and used for genome sequencing and chromosome-level genome construction. The genome of strain zhudugu2 was extracted using the NucleoBond HMW DNA kit (Macherey-Nagel, Düren, Germany). The concentration and quality of the DNA were analyzed using Thermal Nanodrop One (United States, CA) and Agilent 2100 Bioanalyzer System (United States, CA). The P. giganteus genome was sequenced using Illumina NovaSeq 6000 (paired-end, 2 × 150 bp, insert size, 400 bp) and PacBio Sequel sequencing platforms (CLR mode) by Personalbio Technology (Shanghai, China).
Genome assembling and gene prediction
The PacBio reads were de novo assembled using Falcon and CANU software (Koren et al., 2017). Illumina sequencing reads were filtered using FastQC software. The assembled contigs were corrected using pilon v1.24 software Illumina short reads (Walker et al., 2014). The assembly completeness was evaluated with QUAST v5.1.0rc1 software (Gurevich et al., 2013) with the Illumina reads. The ab initio gene prediction was performed using Augustus v 3.03, glimmerHMM v 3.0.1, and GeneMark-ES v 4.35 software (Majoros et al., 2004; Stanke et al., 2006; Ter-Hovhannisyan et al., 2008). The predicted genes were integrated using EVidenceModeler v r2012-06-25 software (Haas et al., 2008). The completeness of the assembled genome was also evaluated using BUSCO v5.1.2 software with comparison to lineage dataset fungi_odb10 (creation date: 2020-09-10, number of BUSCO markers: 758) (Manni et al., 2021). Repeat sequence was analyzed using RepeatModeler and RepeatMasker software (Chen, 2004; Flynn et al., 2020). RepBase database was used to predict sequences similar to known repeat sequences. Ab initio structure prediction was performed using RepeatModeler software. RepeatMasker was used to make the prediction using the constructed repeat sequence library.
Functional annotation
Functional annotations of the predicted protein-coding sequences (CDSs) were obtained using eggNGOmapper software (Cantalapiedra et al., 2021). Pfam and SwissProt function annotation was performed by sequence alignment against Pfam-A database (database version: Pfam35.0) and SwissProt database (2022-04-30) by Hmmer 3.3.2 and diamond 0.9.21, respectively.
Comparative genomic analysis
The pairwise average nucleotide identity (ANI) values between genomes were analyzed using FastANI software (Jain et al., 2018). Collinearity analysis was performed by MCScanX-jcvi software based on the protein sequence from the GFF3 files of P. giganteus, P. ostreatus, and Agaricus bisporus (Wang et al., 2012). Gene families and single-copy orthologous genes were analyzed using OrthoFinder v2.5.4 software (Emms and Kelly, 2019). The species tree was constructed using concatenate single-copy orthologous genes and visualized using FastTree software (Price et al., 2009).
Identification of CAZymes
Annotation of CAZymes for the genome of P. giganteus was performed using dbcan version v3.0.2 software (Cantarel et al., 2009; Zhang et al., 2018). The database was downloaded from the dbCAN meta server1 (version of the database is V10). Hmmer software was used for the annotation of proteins with default parameters (HMMER E-Value < 1e-15, HMMER Coverage > 0.35).
Identification of the mating locus
The mating-type locus of P. giganteus was analyzed using sequence alignment with diamond software (Buchfink et al., 2015). Mating type genes in other Pleurotus strains as the reference sequences. The genome of P. eryngii ATCC90797 was downloaded from the MycoCosm portal of the Joint Genome Institution (JGI). The A mating type locus of strain ATCC90797 is in scaffold1 between 196670 and 871073, and the B mating type locus is in scaffold49 between 196670 and 254852. The genome of P. ostreatus strain PC9 was also downloaded from the JGI database. The A mating type locus of strain PC9 is in scaffold4 between 2032971 and 2039163. Gene cluster structure was visualized using integrative genomics viewer software (Thorvaldsdóttir et al., 2013).
Results and discussion
Cultivation of Pleurotus giganteus and monokaryotic strain isolation
Figure 1 showed the cultivation situation and the main process of fruiting body development of P. giganteus (Shanghai, China, by our lab). At the S1 primordium stage, mycelium kinked to form fruiting body. The S2 growing stage showed the continued growth period after primordium formation. S3 phase is the elongation stage and the fruiting bodies continue to grow until the funnel is formed at this stage. S4 is the harvest stage when the fruiting body grew to 70–80% maturity and the cap is funnel-shaped. At mature stage S5, the fruiting body was fully mature and the cap was in the shape of a large funnel. The monokaryotic zhudugu2 strain was isolated from the protoplasm of the “Shen Xun 1 Hao” strain (Figure 1C). Figures 1C,D show the mycelia of the dikaryotic and monokaryotic mycelia of P. giganteus, respectively, and no clamp connection was observed on monokaryotic mycelia (Figure 1D).

Figure 1. Life cycle of P. giganteus. (A) Industrially cultivated P. giganteus. (B) Fruiting body of P. giganteus at different stage of life cycle. (C) Dikaryotic mycelia of the P. giganteus Shen Xun 1 Hao strain. Clamp connections were indicated by arrows. (D) Monokaryotic mycelia of the P. giganteus zhudugu2 strain.
Genome assembly and annotation of Pleurotus giganteus
The genome of the P. giganteus zhudugu2 strain was sequenced using PacBio and Illumina sequencing platforms. A total of 36,251,924 clean reads (∼130 ×) were obtained from Illumina sequencing which was used for k-mer analysis and genome polish. GenomeScope was used to generate a histogram of the depth distribution of the sequencing (k = 19) (Supplementary Figure 1). A single k-mer coverage peak was observed and the heterozygous rate was 0.02%. The results confirmed that strain zhudugu2 was monokaryotic. The approximately 877,248 million clean Nanopore reads (∼200 ×) were de novo assembled into 27 contigs with an N50 of 2.61 Mbp and an N90 of 1.36 Kbp. The total sequence length was 40,035,591 bp (Table 1) and the length of the largest contig is 4.38 Mbp (Figure 2). The GC content of P. giganteus genome was 50.5%. The integrity of the genome was evaluated using QUAST v5.1.0 software and determined to be 98.2%. The size of the P. giganteus genome is similar to those of other species in the Pleurotus genus. It a little larger than those of Pleurotus tuber-regium, Pleurotus ostreatus and Pleurotus citrinopileatus, but less than the genome size of Pleurotus eryngii and Pleurotus tuoliensis. The GC content of P. giganteus is similar to most of the Pleurotus species but larger than those of the P. tuber-regium strains.
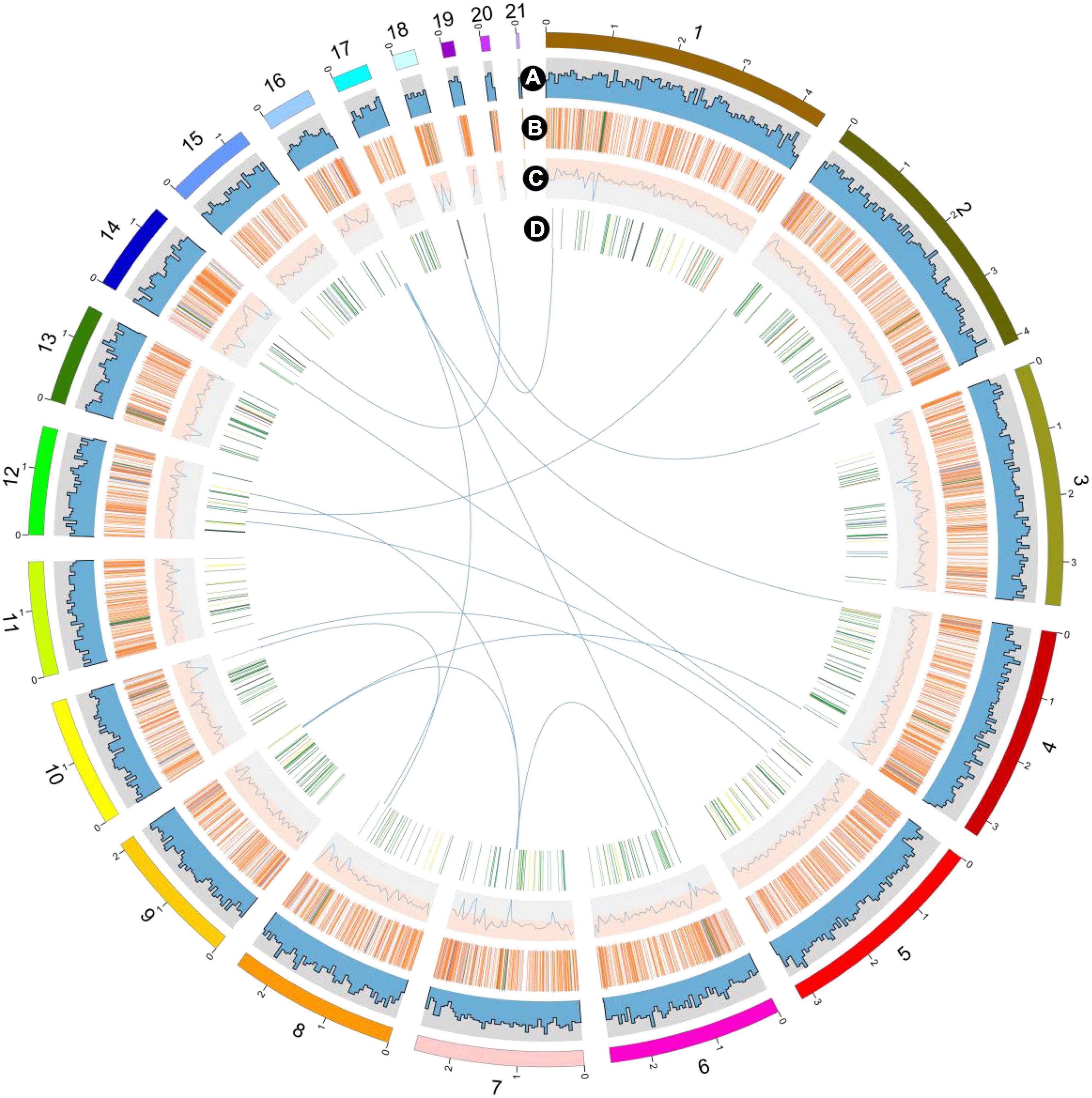
Figure 2. Overview of the P. giganteus genome assembly. The outermost layer of colored blocks is a circular representation of the 21 contigs with coding sequences, with a scale mark labeling each 1 Mb. (A) Gene density per window. (B) Repeat sequence density per window. (C) GC content per window. (D) carbohydrate-active enzymes (CAZymes). Links within and between chromosomes represent collinear blocks generated from MCScanX and JCVI software. For tracks (A–C), the window size is 50 kb. The plot was visualized using Circos software.
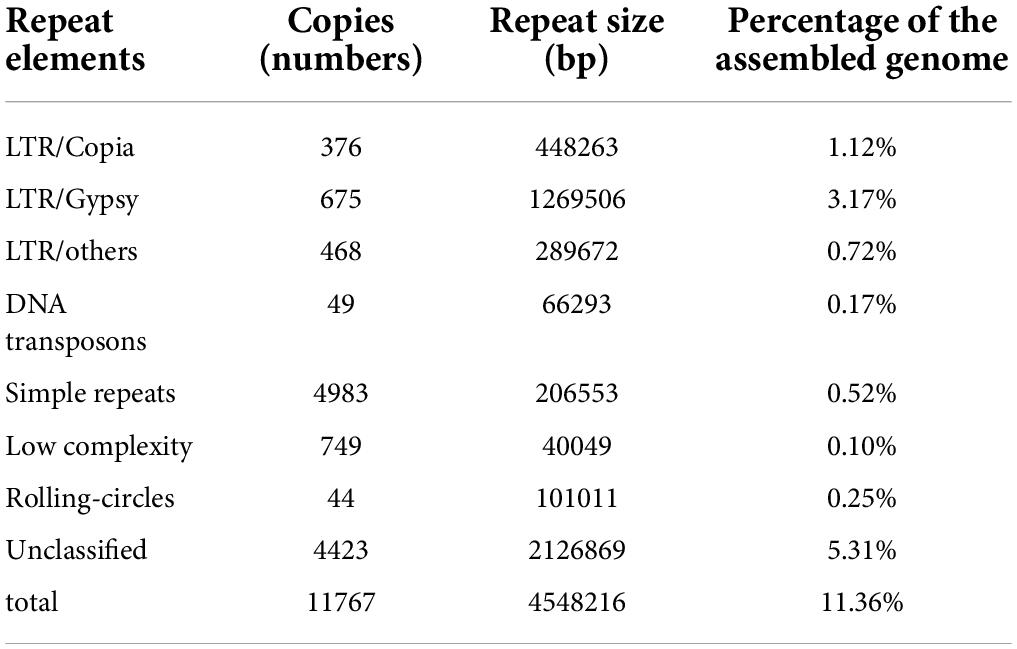
Repeat sequence was identified using RepeatMasker based on homology alignment and ab initio prediction and accounted for 11.36% of the P. giganteus genome (Figure 2B). The majority of repetitive sequences were LTR retrotransposons (5.01%), where 0.17% and 0.52% of the repeat element types were DNA transposons and simple repeats (Table 2). However, no LINEs and Satellites were predicted in the P. giganteus genome.
A total of 12,628 gene models were predicted from the genome of P. giganteus with an average sequence length of 1761 bp. The concatenated length of CDSs was 2.22 Mbp, which accounted for 55.5% of the total genome (Supplementary Tables 1, 2). Of the identified genes, 9801 (77.6%) and 5494 (43.5%) genes were annotated by the EggNOG database and SwissProt database, respectively. Based on the similarity of protein domains, 7802 (61.8%) genes were annotated by the Pfam database. The completeness of P. giganteus genome assembly and gene prediction was also evaluated using the BUSCO software with fungi_odb10. The completeness of the zhudugu2 genome was 94.2% (Supplementary Figure 2). These results and the assembly parameters indicated that we generated a high-quality genome of P. giganteus.
Comparative genomic analysis
Pleurotus giganteus was originally described as Lentinus giganteus and Panus giganteus due to its different fruiting body shapes at different stages in the life cycle (Samantha et al., 2016; Phan et al., 2019). Karunarathna et al. (2012) have corrected the classification of this strain to the Pleurotus genus based on ITS sequences (Samantha et al., 2016). To confirm the evolutionary relationship of P. giganteus, a comparative analysis of the P. giganteus genome and 23 fully sequenced fungi genomes (21 Basidiomycetes and 2 Ascomycetes) was performed. A phylogenetic tree constructed based on conserved single-copy orthologous gene alignment showed that P. giganteus had a close evolutionary relationship with other Pleurotus species (Figure 3). The strains in Pleurotus species were clustered into two main clades. P. giganteus, P. tuber-regium, P. citrinopileatus, and P. salmoneostramineus form one clade, while the other seven species formed the other clear clade. P. giganteus showed a closer evolutionary relationship with P. tuber-regium and P. citrinopileatus. Indeed, the fruiting bodies of the three species shared similar characteristics. They all have a complete circle cap with a dent in the middle, which look like a funnel. However, the fruiting body of the species on the other clade has a round cap on one side and a notch on the other side.

Figure 3. Comparison of genomes between P. giganteus with other 23 fungal species (21 Basidiomycetes and 2 Ascomycetes). The evolutionary relationship analysis was constructed based on 312 single-copy orthologous gene. Tree scale = 0.2.
Average Nucleotide Identity (ANI) analysis is a high-resolution taxonomic analysis method. To further confirm the evolutionary relationship, ANI analysis was performed to estimate genomic differences and relatedness between Pleurotus strains’ genomes. As a result in Figure 4, Pleurotus species in Clade B in the species tree showed lower genomic similarities (74 to 75%) with each other and Pleurotus species in Clade A. Whereas species in Clade A showed high genomic similarities between each other (85 to 100%). In a word, the results above confirmed that P. giganteus belong to the genus of Pleurotus according to the current classification.
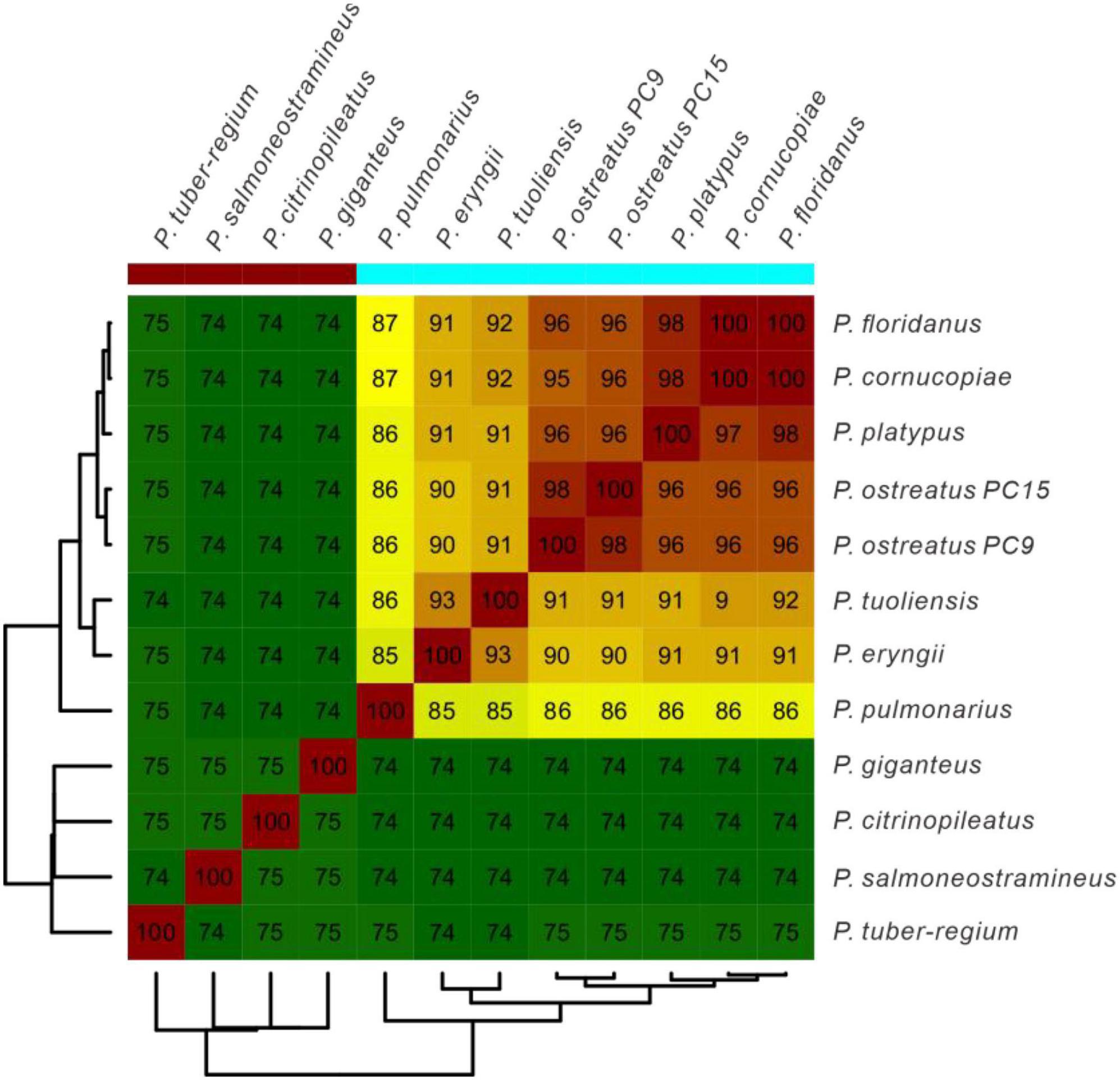
Figure 4. The average nucleotide identity (ANI) values based on the fastANI algorithm generated matrix for Pleurotus genomes. The clustering was constructed using Euclidean distance matrix.
To explore the genetic factor for the fruiting body shape and thermo-adaptation of P. giganteus, orthogroups of five commercially produced Pleurotus species were analyzed (Supplementary Table 4). As shown in Supplementary Figure 3 P. giganteus and P. tuber-regium shared 142 orthogroups that were missed in the other three species. Both P. giganteus and P. tuber-regium have a cup-shaped fruiting body and are distributed in the tropical zone. Therefore, these 142 orthogroups might related with fruiting body shape and thermo-adaptation. The 142 orthogroups contain 353 genes in P. giganteus genome, and 164 were annotated by the Pfam database (Supplementary Table 5). According to the Pfam annotation, most of these 164 genes were related to regulation, transposon, and protein digestion. For example, 43 genes contained F-box-like domain, which was first characterized as components of SCF ubiquitin-ligase complexes (Kipreos and Pagano, 2000). Ubiquitin was related with many important biological processes including thermotolerance (Panek et al., 2020; Zhang et al., 2022). The process of ubiquitination plays a key role in plants’ thermotolerance by eliminating denatured proteins (Zhang et al., 2022). In our previous study, proteins related to the ubiquitin-dependent protein catabolic process were enriched in heat-shock-induced proteins (Xu et al., 2021), which also indicates that ubiquitination played a key role in thermotolerance in mushroom. Therefore, regulation of gene expression, protein expression, and protein degradation might one of the important reasons for the characteristics of fruiting body shape and thermo-adaptation of P. giganteus. A detailed molecular mechanism needs to be investigated through differential expression analysis of genes or proteins in these species.
Chromosome collinearity analysis of P. giganteus and other two edible mushrooms, which have chromosome levels of the genome, was performed using JCVI and MCScanX software (Figure 5). The results revealed high levels of collinearity between P. ostreatus and P. giganteus. According to the collinearity results, contig 5 and contig 13 may belong to the same chromosome, contig 9, 14, and 15 may belong to the same chromosome, and contig 12, 16, 17, and 20 may belong to the same chromosome. The connection of these contigs could be further confirmed using PCR or Hi-C technology (Teh et al., 2017). Rupture and fusion events were identified in contig 5, contig 8, contig 15, contig 9, contig 3 of P. giganteus compared to chromosomes from A. bisporus. More rupture and fusion events occurred between P. giganteus and A. bisporus than those between P. ostreatus and P. giganteus, which is in accordance with the phylogenetic analysis results.
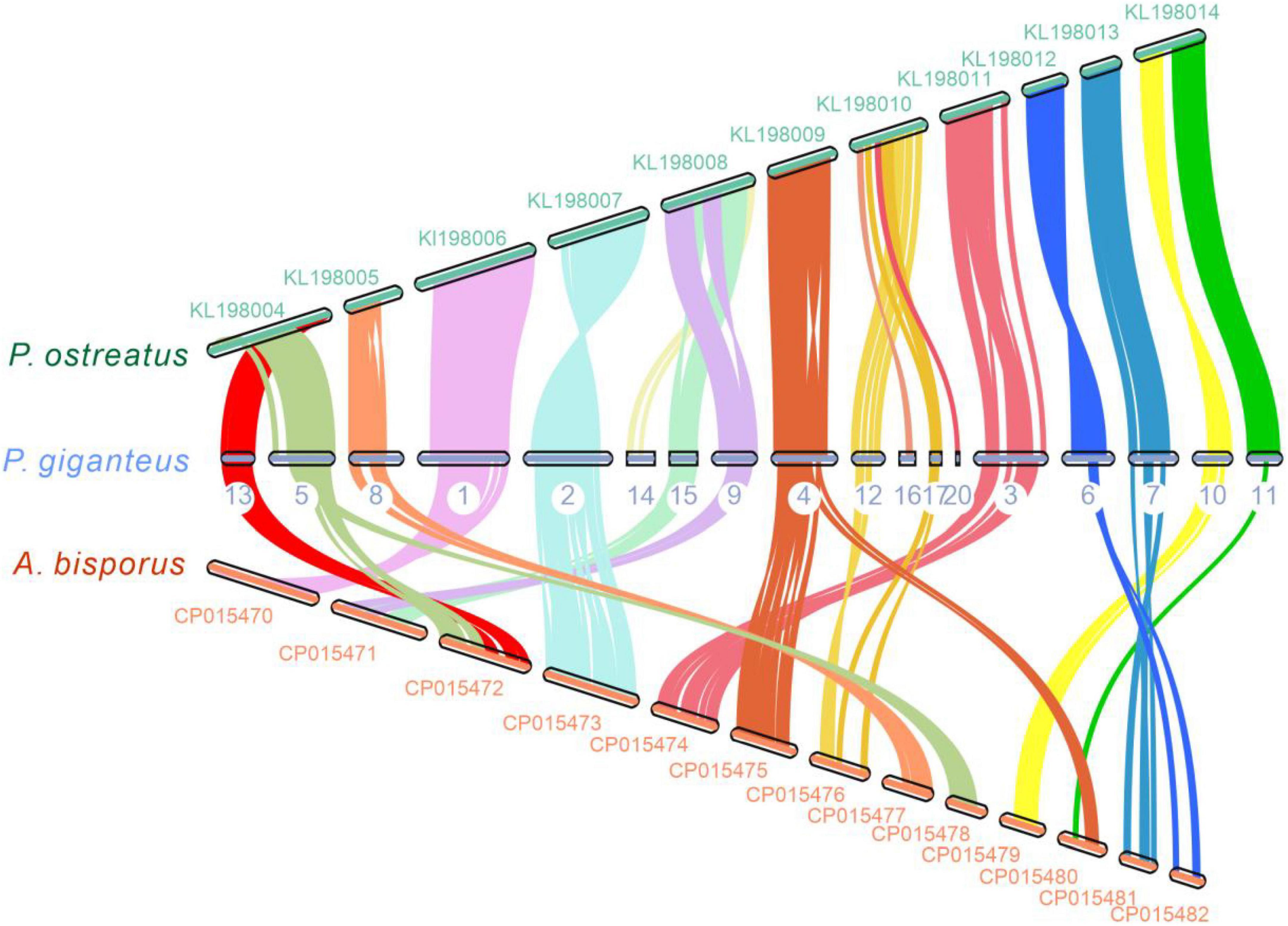
Figure 5. The genome collinearity among P. giganteus, P. ostreatus, and Agaricus bisporus. Each line connects a pair of collinearity blocks between two genomes. Different colors represent different scaffolds of P. giganteus. Scaffold with no collinearity was not shown.
CAZymes in Pleurotus giganteus genome
CAZymes are one of the most important gene families in the fungal genome, which are responsible for lignocellulose degradation and many other biological processes, such as development and stress response (Garron and Henrissat, 2019; Pallister et al., 2020). A total of 481 enzymes accounting for 514 CAZymes terms were identified in the P. giganteus genome with Hmmer software, including 232 GHs, 72 GTs, 23 PLs, 27 CEs, 139 AAs, and 21 CBMs (Figure 6A and Supplementary Table 3). The CAZymes of other 23 fungal species were analyzed using the same parameters with dbCAN2 database. As shown in Figure 6B, the number of CAZymes in P. giganteus is a little less than those in other Pleurotus species, except for P. eryngii and P. tuoliensis. The different CAZymes number might be caused by gene prediction methods, structural variation of genome, and alignment parameters. It can be sure that P. giganteus also has a powerful lignocellulose degradation capacity just like other members of Pleurotus genus. The results are consistent with the production processed, that we can use different lignocellulosic biomass to produce P. giganteus fruiting body.
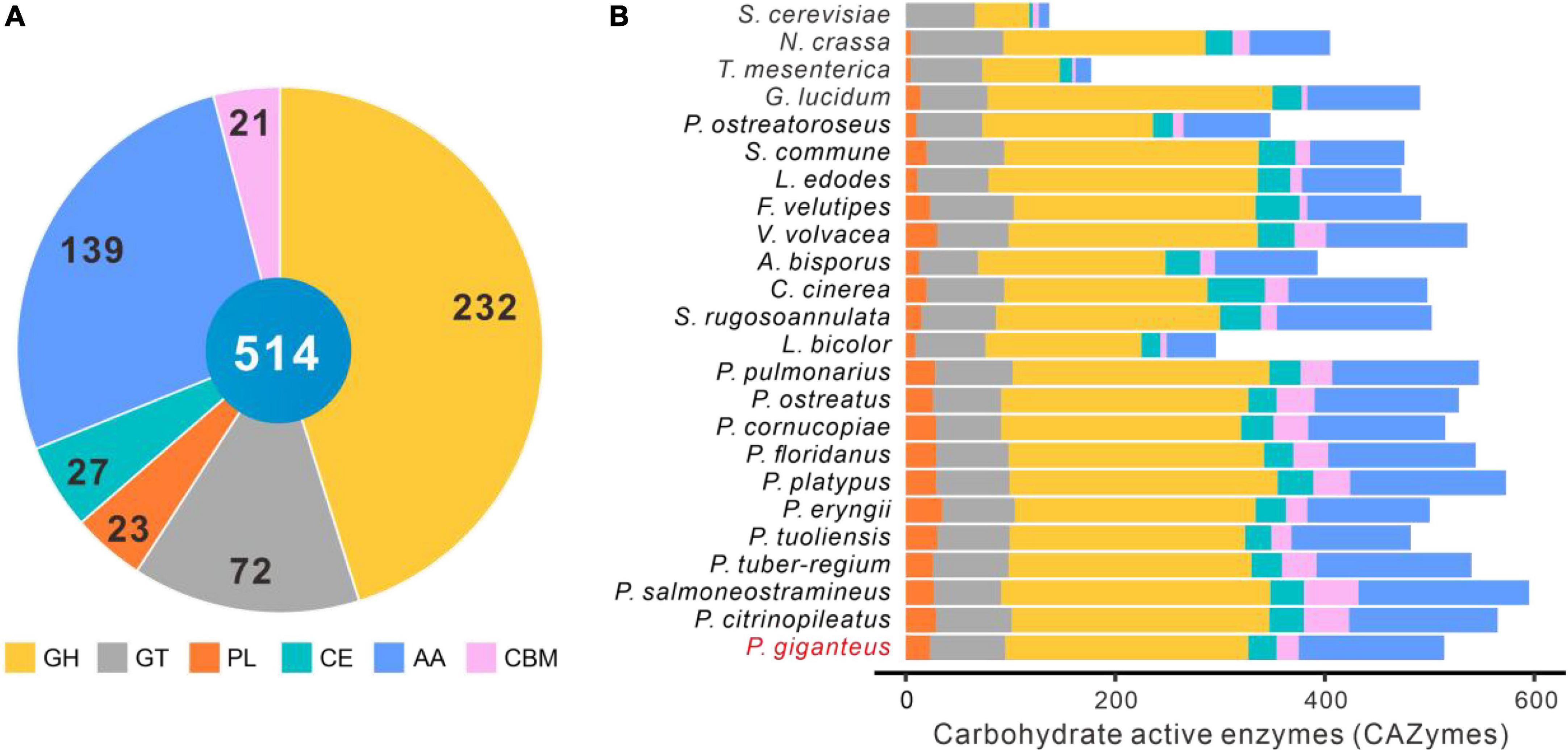
Figure 6. Carbohydrate-active enzymes (CAZymes) in P. giganteus and other 23 fungi. (A) The distribution of CAZymes categories in P. giganteus. (B) The distribution of CAZymes in other 23 fungi. The strain names of these fungi are the same as those in Figure 3. GH, glycoside hydrolase; GT, glycosyltransferase; PL, polysaccharide lyase; CE, carbohydrate esterase; CBM, carbohydrate-binding module; AA, auxiliary activity.
Pleurotus giganteus had 139 AAs. Proteins in the AA category were mainly distributed in AA3, AA9, AA7, AA5, AA1, and AA2 families (Supplementary Table 3). Proteins in AA3 and AA9 categories were related to the degradation of cellumose and hemicellulose. The lignin-degrading enzymes, such as laccases and manganese peroxidases, were in the AA1 and AA2 categories. A total of 15 laccases and 10 class II peroxidases were annotated in the genome of P. giganteus with SwissProt database. These enzymes play key roles in the degradation of plant cell wall components (Manavalan et al., 2015; Suryadi et al., 2022). Therefore, the results indicated a robustness lignocellulose degradation of this strain, which is in consistent with the woody materials used in the fruiting body production.
Identification of the mating locus
Mating-type genes from P. ostreatus PC9 and P. eryngii ATCC 90797 were used as query sequences for the alignment to identify mating type locus in strain zhudugu2. The A mating type locus of P. giganteus was on scaffold 1. The total length of the locus was over 4 kb, consisting of two homeodomain genes (HD) of similar sizes. The two HD genes were transcribed in opposite directions as in P. ostreatus (Figure 7A) (Ju et al., 2020). Different from P. ostreatus PC9 and P. giganteus zhudugu2, P. eryngii ATCC 90797 has one HD1 gene and one HD2 gene transcribed in opposite directions and one extra HD2 gene is located on the other side of HD1. The amino acid sequence showed 52.7 and 51.1% sequence similarity with the HD1 in P. ostreatus PC9 and P. eryngii ATCC 90797, respectively. The HD2 showed 57.4 and 50.0% (49.5%) sequence similarity with those in strain PC9 and ATCC 90797, respectively.
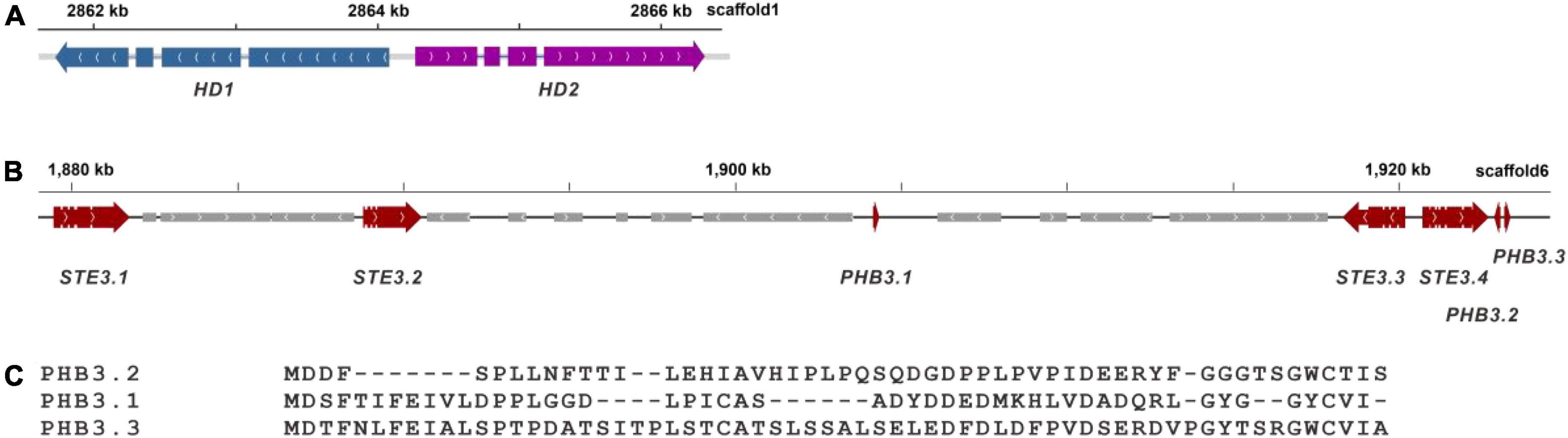
Figure 7. Gene structure of the mating type locus of P. giganteus. (A) Structure of A mating type locus in P. giganteus. (B) Structure of B mating type locus in P. giganteus zhudugu2. (C) Comparison of the mating pheromones predicted in P. giganteus.
Four pheromone receptor genes (STE3.1-STE3.4) and 3 pheromone genes (PHB3.1-PHB3.3) were predicted and clustered in a ∼45 kb gene cluster (Figures 7B,C). TMHMM prediction showed that all four pheromone receptors contained 7 trans-membrane domains. The gene organization of the B mating locus is different from that in P. eryngii (Ju et al., 2020) and Pleurotus djamor (James et al., 2004), which indicated the B mating type locus has evolved through multiple events of genome rearrangement like in other mushroom such as in Flammulina velutipes (Wang et al., 2016) and Lentinula edodes (Ha et al., 2019). The information provided here is important for the development of molecular markers for crossbreeding of P. giganteus.
Conclusion
In summary, this is the first report of the whole genome of P. giganteus. Integrity, completeness, and collinearity analysis revealed the high quality of genome assembly. Comparative genome analysis confirmed that P. giganteus should be classified as Pleurotus genus instead of Lentinus and Panus. Identification of CAZymes revealed that P. giganteus had robustness lignocellulose degradation capacity. Repeat sequence analysis and mating locus could be used for the development of molecular markers in strain identification and breeding. The P. giganteus genome provided insights for basic research on the cultivation, nutrition, and medicinal utility of this mushroom.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/gsa, CRA006857.
Author contributions
HLY, LZ, and HY conceived and designed the project and wrote the manuscript. HLY, MZ, YS, QL, JL, and CS performed the experiments. QT and HLY contributed reagents and materials. HLY, MZ, XS, and HY analyzed the data. All authors have read and approved the manuscript.
Funding
This research was funded by China Agriculture Research System (CARS20), SAAS Program for Excellent Research Team([2022]001), the Shanghai Science and Technology Commission The belt and road project (20310750500), and Shandong provincial key research and development plan (2021ZDSYS28).
Acknowledgments
We thank Yuan Bin from Zhangzhou Institution of Agricultural Sciences for providing the wild strain of P. giganteus.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.941889/full#supplementary-material
Footnotes
References
Baskaran, A., Chua, K. H., Sabaratnam, V., Ravishankar Ram, M., and Kuppusamy, U. R. (2017). Pleurotus giganteus (Berk. Karun & Hyde), the giant oyster mushroom inhibits NO production in LPS/H2O2 stimulated RAW 264.7 cells via STAT 3 and COX-2 pathways. BMC Complem. Altern. Med. 17:40. doi: 10.1186/s12906-016-1546-6
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60.
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). EggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chen, N. (2004). Using repeat masker to identify repetitive elements in genomic sequences. Curr. Protocol. Bioinformatics 5, 4–10.
Debnath, G., Das, P., and Saha, A. K. (2019). Green synthesis of silver nanoparticles using mushroom extract of Pleurotus giganteus: characterization, antimicrobial, and α-amylase inhibitory activity. Bionanoscience 9, 611–619.
Dong, H. X., Cai, D. H., and Li, Y. (2010). Analysis of mating types of basidiospores in Panus giganteus. Microbiol. China 37, 1617–1620.
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Fang, M., Wang, X., Chen, Y., Wang, P., Lu, L., Lu, J., et al. (2020). Genome sequence analysis of Auricularia heimuer combined with genetic linkage map. J. Fungi 6:37. doi: 10.3390/jof6010037
Flynn, J. M., Hubley, R., Goubert, C., Rosen, J., Clark, A. G., Feschotte, C., et al. (2020). RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. U.S.A. 117, 9451–9457. doi: 10.1073/pnas.1921046117
Garron, M. L., and Henrissat, B. (2019). The continuing expansion of CAZymes and their families. Curr. Opin. Chem. Biol. 53, 82–87.
Gong, W., Wang, Y., Xie, C., Zhou, Y., Zhu, Z., and Peng, Y. (2020). Whole genome sequence of an edible and medicinal mushroom, Hericium erinaceus (Basidiomycota, Fungi). Genomics 112, 2393–2399. doi: 10.1016/j.ygeno.2020.01.011
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075.
Ha, B., Moon, Y. J., Song, Y., Kim, S., Kim, M., Yoon, C. W., et al. (2019). Molecular analysis of B mating type diversity in Lentinula edodes. Sci. Hortic. 243, 55–63.
Haas, B. J., Salzberg, S. L., Zhu, W., Pertea, M., Allen, J. E., Orvis, J., et al. (2008). Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, 1–22. doi: 10.1186/gb-2008-9-1-r7
Huo, J., Zhong, S., Du, X., Cao, Y., Wang, W., Sun, Y., et al. (2020). Whole-genome sequence of Phellinus gilvus (mulberry Sanghuang) reveals its unique medicinal values. J. Adv. Res. 24, 325–335. doi: 10.1016/j.jare.2020.04.011
Intraspesies, K., Yun, H. K., Boon, C. S., Shin, T., and Sabaratnam, V. (2020). Breeding and evaluation of Pleurotus giganteus (Berk.) Karunarathna & KD Hyde Hybrids via Intraspecific Mating. Sains Malays. 49, 1223–1236.
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 1–8.
James, T. Y., Liou, S. R., and Vilgalys, R. (2004). The genetic structure and diversity of the A and B mating-type genes from the tropical oyster mushroom, Pleurotus djamor. Fungal Genet. Biol. 41, 813–825. doi: 10.1016/j.fgb.2004.04.005
Ju, Y., Kim, S., Kim, M., San Ryu, J., and Ro, H. S. (2020). Structure analysis of A and B mating type loci in a representative commercial strain of Pleurotus eryngii. Sci. Hortic. 274:109686.
Karunarathna, S. C., Yang, Z. L., Raspé, O., Ko Ko, T. W., Vellinga, E. C., Zhao, R. L., et al. (2012). Lentinus giganteus revisited: new collections from Sri Lanka and Thailand. Mycotaxon 118, 57–71.
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Li, Q. Z., Shen, X. F., Zhang, M. Y., Shang, X. D., Zhang, D., and Yu, H. L. (2021). Effects of different cultivation modes on agronomic characters and quality of Panus giganteus fruiting body. Acta Agric. Shanghai 37, 23–28.
Li, S., Zhao, S., Hu, C., Mao, C., Guo, L., Yu, H., et al. (2022). Whole genome dequence of an edible mushroom Stropharia rugosoannulata (Daqiugaigu). J. Fungi 8:99. doi: 10.3390/jof8020099
Liang, Y., Lu, D., Wang, S., Zhao, Y., Gao, S., Han, R., et al. (2020). Genome assembly and pathway analysis of edible mushroom Agrocybe cylindracea. Genom. Proteom. Bioinformatics 18, 341–351. doi: 10.1016/j.gpb.2018.10.009
Majoros, W. H., Pertea, M., and Salzberg, S. L. (2004). TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics 20, 2878–2879. doi: 10.1093/bioinformatics/bth315
Manavalan, T., Manavalan, A., and Heese, K. (2015). Characterization of lignocellulolytic enzymes from white-rot fungi. Curr. Microbiol. 70, 485–498.
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Pallister, E., Gray, C. J., and Flitsch, S. L. (2020). Enzyme promiscuity of carbohydrate active enzymes and their applications in biocatalysis. Curr. Opin. Struc. Biol. 65, 184–192.
Panek, J., Gang, S. S., Reddy, K. C., Luallen, R. J., Fulzele, A., Bennett, E. J., et al. (2020). A cullin-RING ubiquitin ligase promotes thermotolerance as part of the intracellular pathogen response in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 117, 7950–7960. doi: 10.1073/pnas.1918417117
Phan, C. W., David, P., Tan, Y. S., Naidu, M., Wong, K. H., Kuppusamy, U. R., et al. (2014). Intrastrain comparison of the chemical composition and antioxidant activity of an edible mushroom, Pleurotus giganteus, and its potent neuritogenic properties. Sci. World J. 2014:378651. doi: 10.1155/2014/378651
Phan, C. W., David, P., Wong, K. H., Naidu, M., and Sabaratnam, V. (2015). Uridine from Pleurotus giganteus and its neurite outgrowth stimulatory effects with underlying mechanism. PLoS One 10:e0143004. doi: 10.1371/journal.pone.0143004
Phan, C. W., Lee, G. S., Macreadie, I. G., Malek, S. N. A., Pamela, D., and Sabaratnam, V. (2013). Lipid constituents of the edible mushroom, Pleurotus giganteus demonstrate anti-Candida activity. Nat. Prod. Commun. 8:1763–1765.
Phan, C. W., Wang, J. K., Tan, E. Y. Y., Tan, Y. S., Sathiya Seelan, J. S., Cheah, S. C., et al. (2019). Giant oyster mushroom, Pleurotus giganteus (Agaricomycetes): current status of the cultivation methods, chemical composition, biological, and health-promoting properties. Food Rev. Int. 35, 324–341.
Phan, C. W., Wong, W. L., David, P., Naidu, M., and Sabaratnam, V. (2012). Pleurotus giganteus (Berk.) Karunarathna & KD Hyde: nutritional value and in vitro neurite outgrowth activity in rat pheochromocytoma cells. BMC Complem. Altern. Med. 12:102. doi: 10.1186/1472-6882-12-102
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Samantha, C., Peter, E., Jie, C., Guojie Li, M.Q., and Zhao, R. (2016). Correct names of two cultivated mushrooms from the genus Pleurotus in China. Phytotaxa 260, 036–046.
Stanke, M., Keller, O., Gunduz, I., Hayes, A., Waack, S., and Morgenstern, B. (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439. doi: 10.1093/nar/gkl200
Suryadi, H., Judono, J. J., Putri, M. R., Eclessia, A. D., Ulhaq, J. M., Agustina, D. N., et al. (2022). Biodelignification of lignocellulose using ligninolytic enzymes from white-rot fungi. Heliyon 8:e08865.
Teh, B. T., Lim, K., Yong, C. H., Ng, C. C. Y., Rao, S. R., Rajasegaran, V., et al. (2017). The draft genome of tropical fruit durian (Durio zibethinus). Nat. Genet. 49, 1633–1641. doi: 10.1038/ng.3972
Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y. O., and Borodovsky, M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18, 1979–1990.
Thorvaldsdóttir, H., Robinson, J. T., and Mesirov, J. P. (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 14, 178–192. doi: 10.1093/bib/bbs017
Tian, Y., Zhao, Y., Zeng, H., Zhang, Y., and Zheng, B. (2016). Structural characterization of a novel neutral polysaccharide from Lentinus giganteus and its antitumor activity through inducing apoptosis. Carbohyd. Polym. 154, 231–240. doi: 10.1016/j.carbpol.2016.08.059
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Wang, L., Ma, Z., Du, F., Wang, H., and Ng, T. B. (2014). Feruloyl esterase from the edible mushroom Panus giganteus: a potential dietary supplement. J. Agr. Food Chem. 62, 7822–7827. doi: 10.1021/jf405654u
Wang, W., Lian, L., Xu, P., Chou, T., Mukhtar, I., Osakina, A., et al. (2016). Advances in understanding mating type gene organization in the mushroom-forming fungus Flammulina velutipes. G3 6, 3635–3645. doi: 10.1534/g3.116.034637
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Xu, L., Guo, L., and Yu, H. (2021). Label-free comparative proteomics analysis revealed heat stress responsive mechanism in Hypsizygus marmoreus. Front. Microbiol. 11:541967. doi: 10.3389/fmicb.2020.541967
Yu, F., Song, J., Liang, J., Wang, S., and Lu, J. (2020). Whole genome sequencing and genome annotation of the wild edible mushroom, Russula griseocarnosa. Genomics 112, 603–614. doi: 10.1016/j.ygeno.2019.04.012
Yu, H. L., Zhai, D. D., Shen, X. F., Zhang, M. Y., Wang, Y. X., Shang, X. D., et al. (2021). Variation and probability classification of fruiting body quantitative characteristics in Pleurotus giganteus germplasm resources. Acta Edulis Fungi 28, 42–44.
Yu, H., Zhang, L., Shang, X., Peng, B., Li, Y., Xiao, S., et al. (2022). Chromosomal genome and population genetic analyses to reveal genetic architecture, breeding history and genes related to cadmium accumulation in Lentinula edodes. BMC Genomics 23:120. doi: 10.1186/s12864-022-08325-x
Yuan, Y., Wu, F., Si, J., Zhao, Y. F., and Dai, Y. C. (2019). Whole genome sequence of Auricularia heimuer (Basidiomycota. Fungi), the third most important cultivated mushroom worldwide. Genomics 111, 50–58. doi: 10.1016/j.ygeno.2017.12.013
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101.
Zhang, H., Zhou, J. F., Kan, Y., Shan, J. X., Ye, W. W., Dong, N. Q., et al. (2022). A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376, 1293–1300. doi: 10.1126/science.abo5721
Zhang, J., Shen, N., Li, C., Xiang, X., Liu, G., Gui, Y., et al. (2021). Population genomics provides insights into the genetic basis of adaptive evolution in the mushroom-forming fungus Lentinula edodes. J. Adv. Res. 38, 91–106. doi: 10.1016/j.jare.2021.09.008
Keywords: Pleurotus giganteus, genome, edible mushroom, white-rot fungi, mating locus, CAZymes
Citation: Yu H, Zhang M, Sun Y, Li Q, Liu J, Song C, Shang X, Tan Q, Zhang L and Yu H (2022) Whole-genome sequence of a high-temperature edible mushroom Pleurotus giganteus (zhudugu). Front. Microbiol. 13:941889. doi: 10.3389/fmicb.2022.941889
Received: 11 May 2022; Accepted: 29 July 2022;
Published: 16 August 2022.
Edited by:
Yao-Cheng Lin, Academia Sinica, TaiwanReviewed by:
Chao-Li Huang, National Cheng Kung University, TaiwanWankuan Shen, South China Agricultural University, China
Copyright © 2022 Yu, Zhang, Sun, Li, Liu, Song, Shang, Tan, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lujun Zhang, zhanglujun@saas.sh.cn; Hao Yu, yuhao@qau.edu.cn
†These authors have contributed equally to this work
 Hailong Yu
Hailong Yu Meiyan Zhang1†
Meiyan Zhang1† Qiaozhen Li
Qiaozhen Li Qi Tan
Qi Tan Lujun Zhang
Lujun Zhang Hao Yu
Hao Yu