- 1University of Cincinnati College of Medicine, Cincinnati, OH, United States
- 2Division of Asthma Research, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, United States
MYC is a transcription factor crucial for a host of cellular functions from proliferation to metabolism, and MYC dysregulation contributes to disease pathogenesis. A growing body of evidence suggests that MYC signaling is regulated by the caspase activation and recruitment domain-coiled-coil (CARD-CC) proteins: a family of immunological signaling mediators that canonically drive NF-κB signaling across nearly all tissues. MYC regulation coordinated by the CARD-CC proteins occurs by multiple mechanisms, including transcription, physical binding, and subcellular localization. Herein, we highlight the hallmark studies that collectively broaden the sphere of influence of CBM complexes beyond NF-κB to include MYC, which has functional impact on cells within and likely beyond the immune system. The studies reviewed herein provide rationale for future studies that examine non-canonical CBM-MYC signaling, its relationship with canonical NF-κB signaling, and its contribution to human health and disease.
Introduction
MYC, or c-Myc, is a critical transcription factor that, when appropriately regulated, regulates physiological processes ranging from proliferation and metabolism to differentiation and apoptosis in virtually all tissues (Jha et al., 2023). Precise regulation of MYC is crucial, however, as its dysregulation mediates a wide range of diseases (Jha et al., 2023). Separately, the caspase activation and recruitment domain-coiled-coil (CARD-CC) family of proteins are important signaling mediators within the innate and adaptive immune systems in particular (DeVore and Hershey, 2022). These proteins form signalosomes with B-cell lymphoma/leukemia 10 (BCL10) and mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1) known as “CBM complexes,” which canonically drive pro-inflammatory NF-κB signaling. To date, the literature on CBM complexes has largely focused on this NF-κB signaling; however, there is emerging evidence that these signalosomes are novel regulators of MYC within and beyond the immune system, which we will discuss in the following sections. After introducing MYC and the CARD-CC family, we will review the published literature supporting the regulatory roles of CARD11 and CARD14 in MYC signaling and how this influences tissue homeostasis and disease. Though no studies have yet directly confirmed CARD9-MYC and CARD10-MYC signaling, we will also speculate on these potential pathways based on available studies. Further, we will highlight future research directions that may impact how the scientific community approaches the study and treatment of CBM- or MYC-dependent diseases.
MYC in cellular function and disease
Mammalian cells rely on the complex coordination of numerous signaling pathways to regulate cellular function, and the ubiquitous transcription factor MYC, encoded by MYC, is central to many of them. Though MYC can canonically drive target gene expression by directly binding DNA, it is also considered to be a global amplifier of all active genes because of its influence on both transcriptional activators and transcriptional machinery (Jha et al., 2023). As such, MYC exerts significant control over cellular processes crucial for tissue homeostasis, including proliferation, differentiation, and cellular survival.
Within the immune system, MYC directs the balance between self-renewal and differentiation in hematopoietic stem cells, mediates the development of B- and T-cells, and sustains myeloid populations (Ahmadi et al., 2021; Delgado and León, 2010). MYC is also crucial for maintaining the epithelial barriers that serve as the body’s first line of defense against environmental threats (e.g., allergens, microbes) by regulating self-renewal of basal keratinocytes and initiating terminal differentiation of suprabasal keratinocytes into the stratum corneum (DeVore et al., 2024).
The fundamental role of MYC is highlighted by the disease states resulting from its dysregulation (Zacarías-Fluck et al., 2024). Unchecked MYC signaling is oncogenic by orchestrating uncontrolled proliferation and cellular metabolism, impeding tumor-suppressive mechanisms and promoting genomic instability (Dhanasekaran et al., 2022). Several hematological malignancies are driven by or associated with MYC dysregulation (Delgado and León, 2010), including neoplasms of B-cells, T-cells, plasma cells, and myeloid lineages (Delgado and León, 2010). MYC dysregulation is also associated with epithelial malignancies, including cutaneous squamous cell carcinomas and their precursor actinic keratoses (Hameetman et al., 2013; Kim et al., 2022; Toll et al., 2009) as well as carcinomas of the head and neck, lung, esophagus, and colon (Kalkat et al., 2017).
Since even modest changes in MYC protein levels can alter cellular responses or drive oncogenesis (Dhanasekaran et al., 2022; Liu et al., 2023), MYC is subject to multiple levels of regulation which are comprehensively reviewed elsewhere (Jha et al., 2023; Lee et al., 2023). Briefly, MYC gene transcription is controlled by multiple promoters and initiation sites and influenced cis-regulatory elements, DNA conformational changes, and other transcription factors (e.g., SP1, TGFβ, p53). MYC transcript stability and translation are modulated by post-transcriptional modification, RNA-binding proteins and micro-RNAs. At the protein level, subcellular localization, binding partners, and post-translational modifications (PTMs; e.g., phosphorylation, acetylation, and ubiquitination) influence MYC stability and transcriptional activity (Lee et al., 2023). Finally, MYC is canonically downregulated by proteasomal degradation (Jha et al., 2023). However, the number of recognized MYC regulatory mechanisms continues to grow. As discussed below, our group recently demonstrated that MYC can be degraded by selective autophagy in keratinocytes (DeVore et al., 2024)—a mechanism since noted in colorectal cancer (Ye et al., 2024). Further, MYC in myeloid cells is subjected to K63-linked polyubiquitination by TRAF6, which counters an activating acetylation PTM to prevent leukemogenesis (Jha et al., 2023; Muto et al., 2022). The elucidation and study of MYC regulatory mechanisms are key to future interventions targeting MYC signaling for the prevention or treatment of MYC-associated diseases.
The CARD-CC-BCL10-MALT1 (CBM) complex
Upon detection of an environmental threat, the innate and adaptive immune systems synergistically mount a protective immune response. This outcome is dependent on multiple families of pro-inflammatory signaling complexes that are activated in response to upstream triggers, such as pattern recognition receptors ligated by pathogen-associated molecular patterns. Upon activation, specific proteins nucleate the assembly of large supramolecular complexes that amplify and modulate signal transduction, which is necessary for immune effector functions. One notable signalosome is the Myddosome: in response to upstream toll-like receptors (TLRs), the Myddosome is nucleated by myeloid differentiation factor 88 (MyD88) and signals through TRAF6 to induce NF-κB and other pathways that induce cytokine and chemokine expression (Balka and De Nardo, 2019). Numerous other signalosomes, including inflammasomes (Broz and Dixit, 2016), have been described and are reviewed elsewhere (Xia et al., 2021).
CARD-CC-BCL10-MALT1 (CBM) signalosomes are one family of immunological signaling complexes that have gained significant attention because of their roles in T-cell receptor (TCR) and B-cell receptor (BCR) signal transduction. CBM complexes are nucleated by the four paralogs of the CARD-CC family: CARD9, CARD10, CARD11, and CARD14. Though their expression is particularly prominent within the immune system, the CARD-CC paralogs are collectively expressed in nearly all tissues. Broadly, CARD9 is expressed in myeloid cells, CARD10 in endothelia and solid organs, CARD11 in lymphoid cells, and CARD14 in epidermal and mucosal epithelial cells (Figure 1) (DeVore and Hershey, 2022).
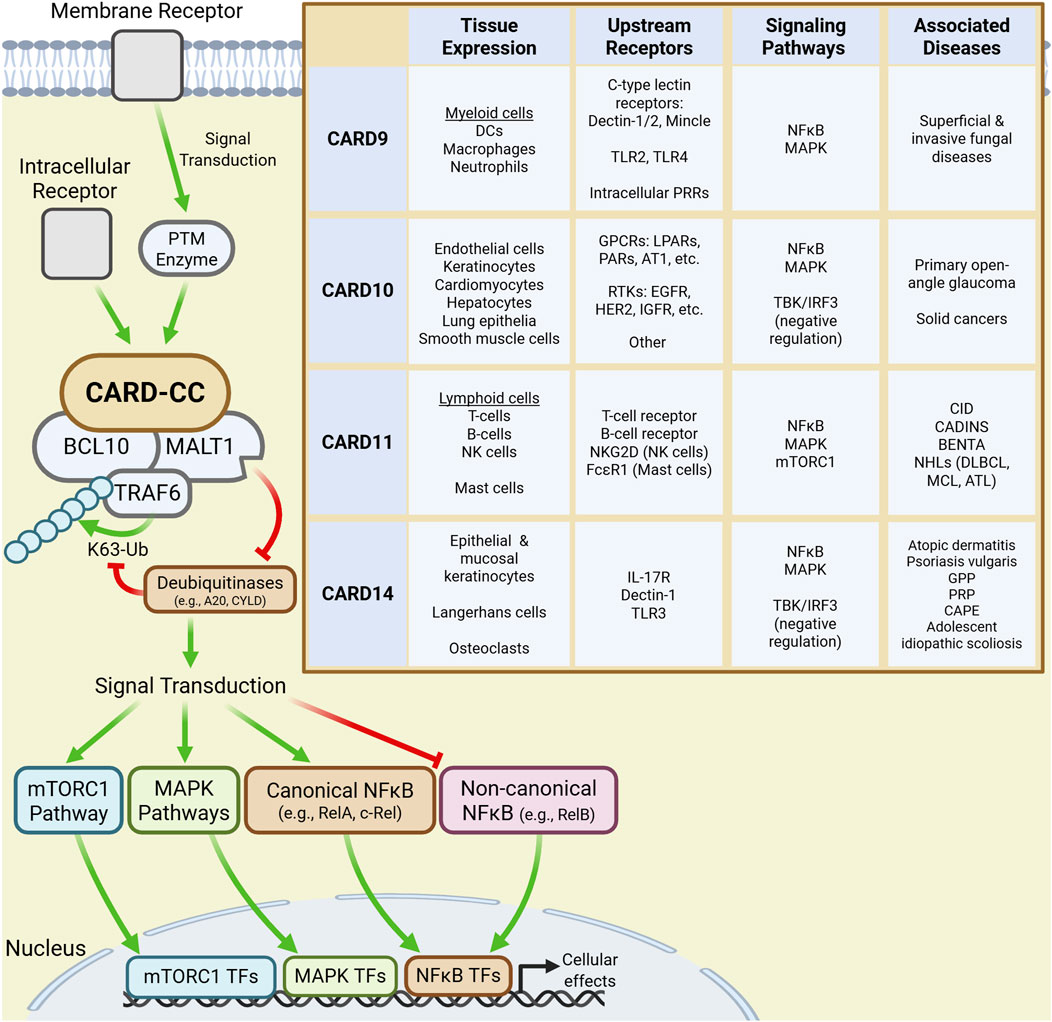
Figure 1. CARD-CC signaling and characteristics. Signal transduction downstream of membrane and intracellular receptors activate CARD-CC paralogs leading to the recruitment of BCL10 and MALT1 into a CBM complex. Recruited TRAF6 then K63-polyubiquitinates CBM components which scaffold modulators of downstream signaling pathways leading to changes in cellular function. The inset table summarizes the tissue expression profile, upstream receptors, signaling pathways and major disease associations for each CARD-CC paralog.
Inactive CARD-CC paralogs are maintained in an autoinhibited conformation by an inhibitor domain. Upstream ligand-receptor pairs, which differ by paralog and cellular context, activate CARD-CC proteins by mediating PTMs (e.g., phosphorylation) of inhibitory domain residues (Bedsaul et al., 2018). Notably, many disease-associated gain-of-function (GoF) mutations affect residues in autoinhibitory regions, thereby bypassing PTM-dependent activation (Bedsaul et al., 2018; Mellett, 2020). Canonically, each activated paralog recruits BCL10 and MALT1 into a supramolecular CBM complex which, like the Myddosome, uses TRAF6-mediated K63-linked polyubiquitination to activate NF-κB signaling. The protease activity of MALT1 potentiates this signaling by cleaving deubiquitinases that would otherwise disassemble the polyubiquitin chains (DeVore and Hershey, 2022; Bedsaul et al., 2018). Ultimately, CBM signaling drives the expression of pro-inflammatory cytokines, chemokines, and even antimicrobial peptides (AMPs) that offer primary protection and modulate downstream immune responses (DeVore and Hershey, 2022).
However, CBM signalosomes also influence immunological pathways other than the canonical NF-κB pathway (DeVore and Hershey, 2022; Mellett, 2020; Mei et al., 2016), such as the non-canonical NF-κB and antiviral TBK/IRF3 pathways. CBMs also signal through pathways that influence cellular growth, proliferation, differentiation and metabolism, including the mitogen-activated protein kinase (MAPK) and mTORC1 pathways (DeVore and Hershey, 2022). Further, MALT1 paracaspase activity modulates cellular activity by cleaving proteins that affect NF-κB and MAPK signaling transduction, RNA transcript stability (regnase-1, roquin-1/2), cytoskeletal regulation and cell growth (LIM domain and actin-binding protein 1) and even components of the CBM itself (Ruland and Hartjes, 2019).
Only recently has it become evident that CBM signalosomes can regulate MYC, which expands the sphere of influence of the CBM to include MYC-driven processes central to the immune system. The evidence supporting CBM-MYC signaling downstream of each paralog is discussed in the following sections.
CARD11-MYC signaling
CARD11 (CARMA1) is expressed in lymphoid cells including T-cells, B-cells, natural killer cells; and in mast cells. CARD11 is the most well-understood paralog given its crucial roles in BCR and TCR signal transduction: it is essential for the activation, proliferation, and survival of lymphocytes and for the differentiation of specific effector T-cell (Teff) subsets including regulatory T-cells (Treg), T follicular helper cells, and T-helper 17 (TH17) cells (Lee et al., 2017; Brüstle et al., 2017; Molinero et al., 2009; Molinero et al., 2012; Egawa et al., 2003; Barnes et al., 2009). Germline absence of CARD11 causes combined immunodeficiency (CID), whereas germline hypomorphic and GoF mutations drive the immune dysregulation syndromes CARD11-associated atopy with dominant interference of NF-κB signaling (CADINS) and B-cell expansion with NF-κB and T-cell anergy (BENTA), respectively (DeVore and Hershey, 2022). The significant influence of CARD11 signaling in lymphocytes is also demonstrated by its role in hematologic malignancies: GoF mutations and activating gene fusions (i.e., CARD11-PIK3R3) are associated with hematologic malignancies including B-cell and T-cell non-Hodgkin lymphomas (Ruland and Hartjes, 2019; Garcia et al., 2024).
MYC regulation by the CBM signalosome was first noted in B-cell lymphoma, where CARD11 can drive MYC activity to promote cellular proliferation (Figure 2). In mantle cell lymphoma (MCL) BCR-CARD11-MALT1 signaling supports MYC activity by reducing its proteasomal degradation. Further, proliferation and survival of MCL cell lines are MYC-dependent, demonstrating that CARD11-MYC is a crucial pathway for sustaining MCL (Dai et al., 2017). CARD11-MYC signaling also occurs in untransformed B-cells: MYC levels are reduced in splenocytes with catalytically-inactive MALT1 (Dai et al., 2017). However, in a separate study on diffuse large B-cell lymphoma, GoF CARD11 mutants behaved as a “second-hit” to constitutively-expressed MYC in lymphomagenesis, suggesting CARD11 and MYC participate in independent but complementary pathways (Reimann et al., 2021). However, CARD11-MYC signaling was not directly tested.
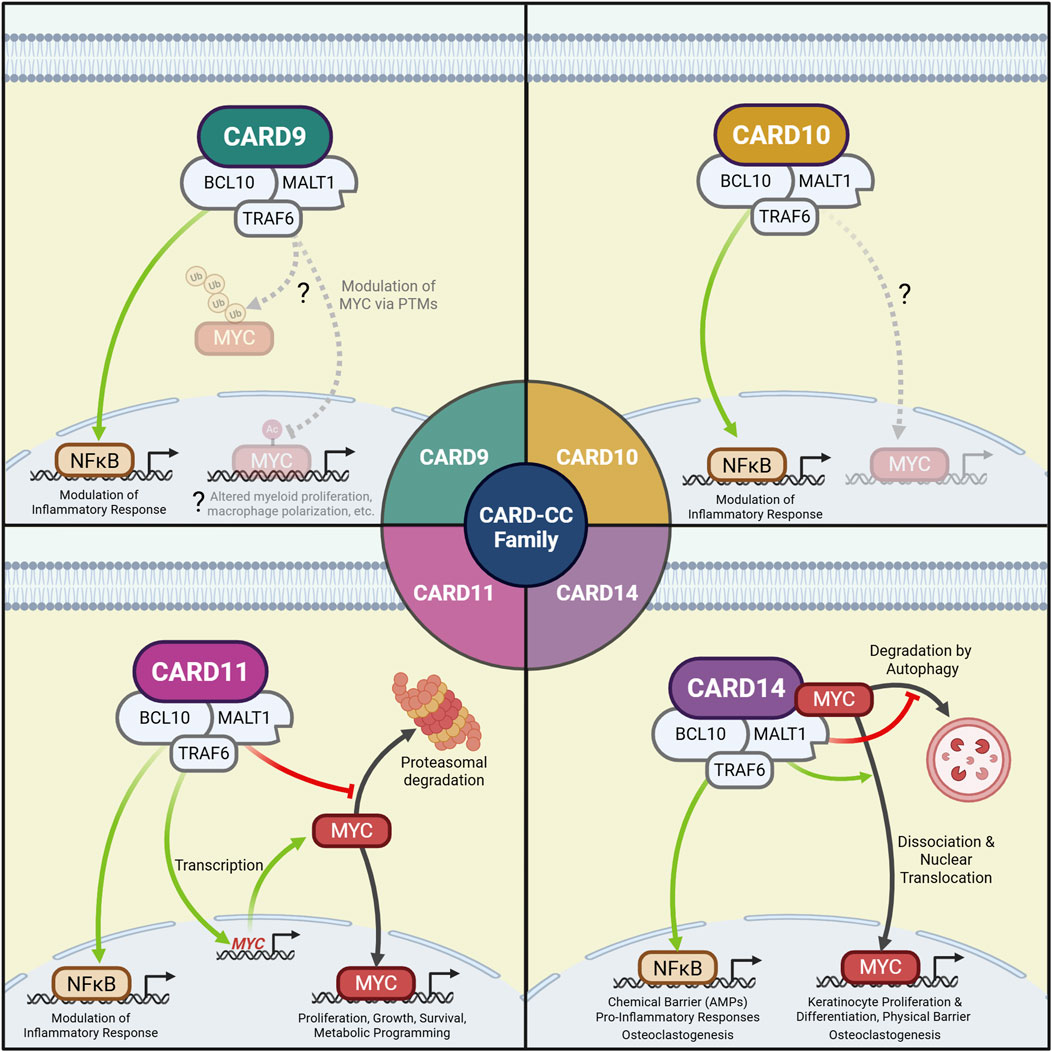
Figure 2. CBM complexes may regulate MYC at multiple levels to coordinate cellular function. CARD9: CARD9-MYC signaling has yet to be confirmed but may include altered post-translational modification of MYC. CARD10: CARD10-MYC signaling has yet to be directly observed. CARD11: The CARD11 CBM promotes MYC gene transcription in Treg cells and prevents MYC proteasomal degradation in B-cells, resulting in altered proliferation, growth, survival and metabolic programming. CARD14: The CARD14 CBM directly binds MYC, which dissociates upon CARD14 activation to mediate transcriptional activity in the nucleus. CARD14 and MALT1 signaling also prevent MYC autophagic degradation. These events lead to keratinocyte proliferation, differentiation, and physical barrier formation, which complement the immunomodulatory effects of CARD14-NF-κB. In bone marrow-derived macrophages, CARD14-MYC and CARD14-NF-κB signaling together drive osteoclastogenesis.
The CARD11 CBM is important for thymic Treg development (Molinero et al., 2009; Barnes et al., 2009). While human versus mice phenotypes differ, deficient or impaired CARD11 or MALT1 signaling can cause systemic inflammatory disease possibly due to greater impairment of Treg cells than Teff cells (Carter and Pomerantz, 2022; Dorjbal et al., 2019; Gewies et al., 2014; Bornancin et al., 2015; Charbit-Henrion et al., 2017; Ma et al., 2017; Jaworski et al., 2014). NF-κB is important in Treg development (Oh et al., 2017); however, Treg defects associated with CARD11 deficiency are not fully complemented by NF-κB rescue (Lee et al., 2010), suggesting involvement of other downstream pathways. Notably, CARD11-MYC signaling has recently been demonstrated in Tregs. Rosenbaum et al. demonstrated that CARD11-MYC signaling induces Myc mRNA expression in Treg cells (Rosenbaum et al., 2022). Notably, this mechanism enhances Treg proliferation and the mitochondrial metabolism that is preferentially utilized by Treg cells (Rosenbaum et al., 2022; Newton et al., 2016). Unlike B-cells, however, CBM signaling does not affect MYC proteasomal degradation (Rosenbaum et al., 2022). MALT1-MYC signaling also protects against fatal autoimmune disease (Rosenbaum et al., 2022). Interestingly, mice with protease-dead MALT1 are phenotypically similar to mice with Treg-specific deficiency in MYC (Saravia et al., 2020). These data support that CARD11-MYC signaling may indeed play a separate but cooperative role to CARD11-NF-κB signaling in Treg biology.
Like Treg cells, Teff cells are also dependent on both MYC and CARD11: MYC is required for Teff proliferation, growth and metabolic reprogramming upon activation (Wang et al., 2011), while CARD11 is required for Teff cytokine expression and proliferation. Teff proliferation is even sensitive to disease-associated hypomorphic or GoF CARD11 mutations (Egawa et al., 2003; Dorjbal et al., 2019; Snow et al., 2012). However, in the same study by Rosenbaum et al., MALT1 paracaspase activity did not appear to influence MYC mRNA levels or mitochondrial metabolism in conventional CD4+ T-cells (Rosenbaum et al., 2022). Beyond this, no other studies have directly investigated CARD11-MYC signaling in Teff cells. Future studies should consider other possible mechanisms by which CARD11 could regulate MYC to influence Teff function, including by altering its proteasomal degradation (as in B-cells), or influencing its physical binding or autophagic degradation (as CARD14 does in keratinocytes; see next section). Such studies would also help determine whether CARD11-MYC signaling could contribute to T-cell malignancies such as adult T-cell leukemia/lymphoma, which harbors CARD11 GoF mutations in about 25% of cases (Kataoka et al., 2015) and can demonstrate increased MYC activity (Kameda et al., 2022; Yamagishi et al., 2021).
CARD14-MYC signaling
CARD14 (CARMA2) is expressed in epidermal keratinocytes, mucosal epithelia and osteoclasts (DeVore et al., 2024; Luo et al., 2025). It is associated with inflammatory skin diseases, including atopic dermatitis (AD), psoriasis vulgaris (PsV), and rarer psoriatic entities like generalized pustular psoriasis (GPP), pityriasis rubra pilaris (PRP), and CARD14-associated papulosquamous eruption (CAPE) (DeVore et al., 2024). AD is associated with hypomorphic CARD14 variants, while the psoriatic diseases are associated with CARD14 GoF variants (DeVore et al., 2024; Mellett, 2020; DeVore et al., 2021; Peled et al., 2019; Craiglow et al., 2018). Functionally, CARD14 stimulates the expression of cytokines, chemokines, AMPs and crucial barrier genes like FLG (encoding filaggrin) (DeVore and Hershey, 2022) that are collectively important for a protective skin barrier.
CARD14-MYC signaling in keratinocytes was recently identified as a determinant of skin barrier integrity (Figure 2). After finding an association between the CARD14 R820W (rs11652075) variant and reduced epidermal FLG expression in children with AD (DeVore et al., 2021), our group noted prominently decreased MYC transcriptional signatures in keratinocytes with the variant despite only subtle decreases in NF-κB signaling (DeVore et al., 2024). MYC was similarly attenuated by MALT1 inhibition. Though CARD14 did not affect MYC expression or proteasomal degradation, we found CARD14 physically interacted with MYC in a manner dependent on CARD14 variants and activation status. Wild-type CARD14 activation promoted MYC dissociation, nuclear localization and transcriptional activity; however, the R820W variant and MALT1 inhibition both attenuated dissociation from the CBM and promoted MYC autophagic degradation (DeVore et al., 2024). Additional studies are required to explore the underlying mechanisms, such as testing whether MYC is purposefully targeted to the autophagosome (i.e., the CBM triggers its ubiquitination by TRAF6) (Ye et al., 2024; Muto et al., 2022) or if the undissociated MYC is collaterally degraded in complex with the CBM (which can by downregulated by autophagy) (DeVore and Hershey, 2022; Paul et al., 2012; Yang et al., 2012; Moreno-García et al., 2013). Functionally, dysregulated CARD14-MYC signaling impaired keratinocyte proliferation, keratinocyte differentiation (e.g., reduced FLG), and barrier function (DeVore and Hershey, 2022; DeVore et al., 2024). Elevated MYC signaling is also implicated in psoriasis (Elder et al., 1990; Leng et al., 2024), and skin expressing psoriasis-associated GoF CARD14 mutants have both elevated MYC signatures and psoriatic features (e.g., epidermal hyperproliferation, parakeratosis) (DeVore et al., 2024). These findings indicate that CARD14 promotes physical barrier immunity through MYC that complements chemical barrier immunity through NF-κB. Interestingly, some CARD14 mutations impact CARD14-NF-κB signaling independent of CARD14-MYC signaling (DeVore et al., 2024), which may explain the pleiotropic presentations of CARD14-associated skin disease.
Most recently, CARD14 has been shown to promote differentiation of bone marrow-derived macrophages into osteoclasts in a MYC-dependent fashion (Luo et al., 2025). Like keratinocytes, CARD14 orchestrates this by directly binding MYC, enhancing its stability, and promoting its nuclear translation. CARD14 concomitantly promotes NF-κB and MAPK signaling which are also crucial for osteoclastogenesis (Luo et al., 2025), again demonstrating cooperation between multiple CARD14-dependent pathways.
CARD9-MYC signaling
CARD9 is predominantly expressed in myeloid cells and is crucial for antifungal immunity. In antigen presenting cells, receptors sensing fungal carbohydrates stimulate CARD9 which mediates the secretion of cytokines (e.g., IL-12, IL-23, IL-1β) that polarize T-cells towards antifungal TH1/TH17 immunophenotypes (Liu et al., 2022). Indeed, patients with deleterious CARD9 mutations often develop fungal infections and allergic inflammation attributed to abrogated Th17 and Th1 polarization, respectively (DeVore and Hershey, 2022; Ruland, 2008). Though no studies have yet definitively linked CARD9 to MYC regulation, there are data suggesting that CARD9 not only regulates MYC but that, in contrast to CARD11 and CARD14, it uniquely suppresses MYC signaling.
CARD9 is crucial for polarization of resting M0 macrophages to pro-inflammatory M1 macrophages and mediating their effector function downstream of inflammatory stimuli (Campuzano et al., 2020). MYC, however, is suppressed in M1 macrophages (Liu et al., 2016; Pello, 2016; Pello et al., 2012). In contrast, CARD9-deficiency promotes polarization towards anti-inflammatory M2 macrophages (Campuzano et al., 2020), which require MYC for activation and for which MYC is a cellular marker (Pello et al., 2012). These observations suggest antagonistic regulation of CARD9 on MYC in macrophages: increased CARD9 signaling may suppress MYC to drive M1 polarization, while reduced CARD9 signaling may disinhibit MYC to permit M2 polarization. However, specific studies are needed to explore this hypothesis.
Dysregulated MYC in myeloid progenitors can mediate progression to AML (Huang et al., 2006; Laurenti et al., 2008). In mice, myeloid TLR2 signaling through MyD88 and TRAF6 suppresses MYC and subsequent progression to AML (Muto et al., 2022). Notably, TLR2-MyD88 signaling can also activate the CARD9 CBM complex (Wang et al., 2020; Hara et al., 2007; Dong et al., 2006), which also employs TRAF6 (Gorjestani et al., 2012). It would be interesting to test if CARD9 is either contributory or necessary for TRAF6-dependent MYC downregulation.
CARD10-MYC signaling
The fourth CARD-CC paralog is CARD10 (CARMA3), whose expression is more variable and includes endothelial cells, smooth muscle cells, epidermal and respiratory epithelia, and the mesenchyme of several solid organs. CARD10 mutations are associated with primary open-angle glaucoma and are implicated in various neoplasms (e.g., bladder and breast) (Zhou et al., 2016; Ekambaram et al., 2018; Man et al., 2019). CARD10 CBM signaling is primarily pro-inflammatory, and is the only paralog known to be activated by receptor tyrosine kinase and G-protein coupled receptors (DeVore and Hershey, 2022; Ruland and Hartjes, 2019).
No focused studies have directly investigated a CARD10-MYC signaling pathway. However, many cancer-associated CARD10 mutations do not influence NF-κB signaling (Staal et al., 2024), suggesting other pathways—possibly including MYC—may be affected instead. In one study, CARD10 knockdown reduced β-catenin-mediated MYC expression and attenuated the invasion and proliferation of bladder carcinoma cells. β-catenin overexpression, however, rescued MYC and cellular invasion and proliferation, suggesting there may be a CARD10-β-catenin-MYC signaling axis (Man et al., 2019). Further, CARD10 is activated by the GPCRs lysophosphatidic acid receptor 2 and angiotensin II type 1 receptor which can also activate MYC (Ekambaram et al., 2018; Taghavi et al., 2008; Sun, 2010)—two other contexts in which CARD10-MYC signaling may be possible. However, additional studies are required to robustly test for CARD10-MYC signaling in CARD10-expressing tissues and how this may contribute to CARD10-associated disease.
Conclusion and outlook
The elucidation of novel signaling pathways is crucial for not only understanding homeostatic function of cellular systems, but also for identifying previously unrecognized disease mechanisms that could be targeted therapeutically. Given both the widespread expression of CBM components and the significant cellular influence and pathogenic potential of MYC, the emerging evidence supporting CBM-MYC regulation is worth highlighting.
CBM complexes have been noted to regulate MYC by not only affecting its transcription but also by influencing its subcellular localization and protein stability (Figure 2). We believe it is likely that CBM complexes affect MYC by other mechanisms as well, including transcript stability, translation, or even post-translational modification (such as by TRAF6-mediated ubiquitination). Further, these regulatory mechanisms appear to be cell-type and/or paralog-specific, exemplified by the effect of CARD11 on MYC proteasomal degradation in B-cells but not Treg cells, and the influence of CARD11 (but not CARD14) on MYC transcription. Future studies centered on the CBM should thus not only consider CBM-dependent MYC signaling but also consider the various levels of MYC regulation it may influence.
While CARD11 and CARD14 promote MYC signaling, early data suggest CARD9 may downregulate MYC. This could be explained by lack of the protein-binding membrane-associated guanylate kinase (MAGUK) domain in CARD9 (DeVore and Hershey, 2022), which is predicted in silico to bind MYC (DeVore et al., 2024). However, future studies are required to investigate whether the MAGUK domain is needed to promote MYC signaling and if its absence could permit MYC suppression by CARD9 or even the MAGUK-deficient splice-variant of CARD14 (CARD14sh).
Finally, CBM-dependent MYC signaling appears to be a functionally complementary to canonical CBM-NF-κB signaling, which may explain the pleiotropy of CBM-associated disease: the development or proliferation of B-cells and Treg cells is partially dependent on both CARD11-MYC and CARD11-NF-κB signaling; the physical skin barrier induced by CARD14-MYC signaling complements the chemical barrier (e.g., AMP and cytokine expression) driven by CARD14-NF-κB; and both CARD14-MYC and CARD14-NF-κB signaling promote osteoclastogenesis. Additional studies are needed to define the roles of CBM signaling through MYC versus NF-κB (or other pathways) and how they interact. This work would contribute to a broader understanding of how the CBM controls cellular function and may elucidate novel approaches to treating associated pathologies.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SD: Conceptualization, Writing – original draft, Writing – review and editing. GK: Conceptualization, Funding acquisition, Supervision, Writing – review and editing.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This work was supported by NIH grant U19AI070235 (GK) and T32GM063483 (SD).
Acknowledgements
The figures were created using BioRender.com. Figure 1: https://BioRender.com/rtpwgeo; Figure 2: https://BioRender.com/dihdc6l.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
AD, Atopic dermatitis; AML, Acute myeloid leukemia; AMP, Antimicrobial peptide; BCL10, B-cell lymphoma/leukemia 10; BCR, B-cell receptor; BENTA, B-cell expansion with NF-κB and T-cell anergy; CADINS, CARD11-associated atopy with dominant interference of NF-κB signaling; CAPE, CARD14-associated papulosquamous eruption; CARD-CC, Caspase activation and recruitment domain coiled-coil; CBM complex, CARD-CC-BCL10-MALT1 complex; CID, Combined immunodeficiency; FLG, Filaggrin; GPP, Generalized pustular psoriasis; MAGUK, Membrane-associated guanylate kinase; MALT, Mucosa-associated lymphoid tissue lymphoma translocation protein 1; MAPK, Mitogen-activated protein kinase; MCL, Mantle cell lymphoma; PAR, Protease-activated receptor; PTM, Post-translational modification; TCR, T-cell receptor; TGFβ, Tumor growth factor β; Teff cell, Effector T-cell; TH cell, T-helper cell; Treg cell, Regulatory T-cell; TLR, Toll-like receptor; TNFα, Tumor necrosis factor α.
References
Ahmadi, S. E., Rahimi, S., Zarandi, B., Chegeni, R., and Safa, M. (2021). MYC: a multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J. Hematol. and Oncol. 14, 121. doi:10.1186/s13045-021-01111-4
Balka, K. R., and De Nardo, D. (2019). Understanding early TLR signaling through the myddosome. J. Leukoc. Biol. 105, 339–351. doi:10.1002/JLB.MR0318-096R
Barnes, M. J., Krebs, P., Harris, N., Eidenschenk, C., Gonzalez-Quintial, R., Arnold, C. N., et al. (2009). Commitment to the regulatory T cell lineage requires CARMA1 in the thymus but not in the periphery. PLoS Biol. 7, e51. doi:10.1371/journal.pbio.1000051
Bedsaul, J. R., Carter, N. M., Deibel, K. E., Hutcherson, S. M., Jones, T. A., Wang, Z., et al. (2018). Mechanisms of regulated and dysregulated CARD11 signaling in adaptive immunity and disease. Front. Immunol. 9, 2105. doi:10.3389/fimmu.2018.02105
Bornancin, F., Renner, F., Touil, R., Sic, H., Kolb, Y., Touil-Allaoui, I., et al. (2015). Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J. Immunol. 194, 3723–3734. doi:10.4049/jimmunol.1402254
Broz, P., and Dixit, V. M. (2016). Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420. doi:10.1038/nri.2016.58
Brüstle, A., Brenner, D., Knobbe-Thomsen, C. B., Cox, M., Lang, P. A., Lang, K. S., et al. (2017). MALT1 is an intrinsic regulator of regulatory T cells. Cell Death Differ. 24, 1214–1223. doi:10.1038/cdd.2015.104
Campuzano, A., Castro-Lopez, N., Martinez, A. J., Olszewski, M. A., Ganguly, A., Leopold Wager, C., et al. (2020). CARD9 is required for classical macrophage activation and the induction of protective immunity against pulmonary cryptococcosis. mBio 11, e03005-19. doi:10.1128/mBio.03005-19
Carter, N. M., and Pomerantz, J. L. (2022). CARD11 signaling in regulatory T cell development and function. Adv. Biol. Regul. 84, 100890. doi:10.1016/j.jbior.2022.100890
Charbit-Henrion, F., Jeverica, A. K., Bègue, B., Markelj, G., Parlato, M., Avcin, S. L., et al. (2017). Deficiency in mucosa-associated lymphoid tissue lymphoma translocation 1: a novel cause of IPEX-like syndrome. J. Pediatr. Gastroenterology Nutr. 64, 378–384. doi:10.1097/MPG.0000000000001262
Craiglow, B. G., Boyden, L. M., Hu, R., Virtanen, M., Su, J., Rodriguez, G., et al. (2018). CARD14-associated papulosquamous eruption: a spectrum including features of psoriasis and pityriasis rubra pilaris. J. Am. Acad. Dermatol 79, 487–494. doi:10.1016/j.jaad.2018.02.034
Dai, B., Grau, M., Juilland, M., Klener, P., Höring, E., Molinsky, J., et al. (2017). B-cell receptor-driven MALT1 activity regulates MYC signaling in mantle cell lymphoma. Blood 129, 333–346. doi:10.1182/blood-2016-05-718775
Delgado, M. D., and León, J. (2010). Myc roles in hematopoiesis and leukemia. Genes and Cancer 1, 605–616. doi:10.1177/1947601910377495
DeVore, S. B., and Hershey, G. K. K. (2022). The role of the CBM complex in allergic inflammation and disease. J. Allergy Clin. Immunol. 150, 1011–1030. doi:10.1016/j.jaci.2022.06.023
DeVore, S. B., Stevens, M. L., He, H., Biagini, J. M., Kroner, J. W., Martin, L. J., et al. (2021). Novel role for caspase recruitment domain family member 14 and its genetic variant rs11652075 in skin filaggrin homeostasis. J. Allergy Clin. Immunol. S0091-6749 (21), 708–717. doi:10.1016/j.jaci.2021.07.003
DeVore, S. B., Schuetz, M., Alvey, L., Lujan, H., Ochayon, D. E., Williams, L., et al. (2024). Regulation of MYC by CARD14 in human epithelium is a determinant of epidermal homeostasis and disease. Cell Rep. 43, 114589. doi:10.1016/j.celrep.2024.114589
Dhanasekaran, R., Deutzmann, A., Mahauad-Fernandez, W. D., Hansen, A. S., Gouw, A. M., and Felsher, D. W. (2022). The MYC oncogene — the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 19, 23–36. doi:10.1038/s41571-021-00549-2
Dong, W., Liu, Y., Peng, J., Chen, L., Zou, T., Xiao, H., et al. (2006). The IRAK-1-BCL10-MALT1-TRAF6-TAK1 cascade mediates signaling to NF-kappaB from toll-like receptor 4. J. Biol. Chem. 281, 26029–26040. doi:10.1074/jbc.M513057200
Dorjbal, B., Stinson, J. R., Ma, C. A., Weinreich, M. A., Miraghazadeh, B., Hartberger, J. M., et al. (2019). Hypomorphic caspase activation and recruitment domain 11 (CARD11) mutations associated with diverse immunologic phenotypes with or without atopic disease. J. Allergy Clin. Immunol. 143, 1482–1495. doi:10.1016/j.jaci.2018.08.013
Egawa, T., Albrecht, B., Favier, B., Sunshine, M.-J., Mirchandani, K., O’Brien, W., et al. (2003). Requirement for CARMA1 in antigen receptor-induced NF-κB activation and lymphocyte proliferation. Curr. Biol. 13, 1252–1258. doi:10.1016/S0960-9822(03)00491-3
Ekambaram, P., Lee, J.-Y., Hubel, N. E., Hu, D., Yerneni, S., Campbell, P. G., et al. (2018). The CARMA3-Bcl10-MALT1 signalosome drives NF-κB activation and promotes aggressiveness in angiotensin II receptor-positive breast cancer. Cancer Res. 78, 1225–1240. doi:10.1158/0008-5472.CAN-17-1089
Elder, J. T., Tavakkol, A., Klein, S. B., Zeigler, M. E., Wicha, M., and Voorhees, J. J. (1990). Protooncogene expression in normal and psoriatic skin. J. Investigative Dermatology 94, 19–25. doi:10.1111/1523-1747.ep12873313
Garcia, J., Daniels, J., Lee, Y., Zhu, I., Cheng, K., Liu, Q., et al. (2024). Naturally occurring T cell mutations enhance engineered T cell therapies. Nature 626, 626–634. doi:10.1038/s41586-024-07018-7
Gewies, A., Gorka, O., Bergmann, H., Pechloff, K., Petermann, F., Jeltsch, K. M., et al. (2014). Uncoupling Malt1 threshold function from paracaspase activity results in destructive autoimmune inflammation. Cell Rep. 9, 1292–1305. doi:10.1016/j.celrep.2014.10.044
Gorjestani, S., Darnay, B. G., and Lin, X. (2012). Tumor necrosis factor receptor-associated factor 6 (TRAF6) and TGFβ-activated kinase 1 (TAK1) play essential roles in the C-type lectin receptor signaling in response to Candida albicans infection. J. Biol. Chem. 287, 44143–44150. doi:10.1074/jbc.M112.414276
Hameetman, L., Commandeur, S., Bavinck, J. N. B., Wisgerhof, H. C., de Gruijl, F. R., Willemze, R., et al. (2013). Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 13, 58. doi:10.1186/1471-2407-13-58
Hara, H., Ishihara, C., Takeuchi, A., Imanishi, T., Xue, L., Morris, S. W., et al. (2007). The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and toll-like receptors. Nat. Immunol. 8, 619–629. doi:10.1038/ni1466
Huang, M.-J., Cheng, Y., Liu, C.-R., Lin, S., and Liu, H. E. (2006). A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 34, 1480–1489. doi:10.1016/j.exphem.2006.06.019
Jaworski, M., Marsland, B. J., Gehrig, J., Held, W., Favre, S., Luther, S. A., et al. (2014). Malt1 protease inactivation efficiently dampens immune responses but causes spontaneous autoimmunity. EMBO J. 33, 2765–2781. doi:10.15252/embj.201488987
Jha, R. K., Kouzine, F., and Levens, D. (2023). MYC function and regulation in physiological perspective. Front. Cell Dev. Biol. 11, 1268275. doi:10.3389/fcell.2023.1268275
Kalkat, M., De Melo, J., Hickman, K. A., Lourenco, C., Redel, C., Resetca, D., et al. (2017). MYC deregulation in primary human cancers. Genes (Basel) 8, 151. doi:10.3390/genes8060151
Kameda, T., Shide, K., Kamiunten, A., Kogure, Y., Morishita, D., Koya, J., et al. (2022). CARD11 mutation and HBZ expression induce lymphoproliferative disease and adult T-cell leukemia/lymphoma. Commun. Biol. 5, 1309. doi:10.1038/s42003-022-04284-x
Kataoka, K., Nagata, Y., Kitanaka, A., Shiraishi, Y., Shimamura, T., Yasunaga, J.-I., et al. (2015). Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 47, 1304–1315. doi:10.1038/ng.3415
Kim, Y.-S., Shin, S., Jung, S.-H., Park, Y. M., Park, G. S., Lee, S. H., et al. (2022). Genomic progression of precancerous actinic keratosis to squamous cell carcinoma. J. Invest Dermatol 142, 528–538.e8. doi:10.1016/j.jid.2021.07.172
Laurenti, E., Varnum-Finney, B., Wilson, A., Ferrero, I., Blanco-Bose, W. E., Ehninger, A., et al. (2008). Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611–624. doi:10.1016/j.stem.2008.09.005
Lee, A. J., Wu, X., Cheng, H., Zhou, X., Cheng, X., and Sun, S.-C. (2010). CARMA1 regulation of regulatory T cell development involves modulation of Interleukin-2 receptor signaling. J. Biol. Chem. 285, 15696–15703. doi:10.1074/jbc.M109.095190
Lee, P., Zhu, Z., Hachmann, J., Nojima, T., Kitamura, D., Salvesen, G., et al. (2017). Differing requirements for MALT1 function in peripheral B cell survival and differentiation. J. Immunol. 198, 1066–1080. doi:10.4049/jimmunol.1502518
Lee, J. M., Hammarén, H. M., Savitski, M. M., and Baek, S. H. (2023). Control of protein stability by post-translational modifications. Nat. Commun. 14, 201. doi:10.1038/s41467-023-35795-8
Leng, X., Wang, S., Zhuang, D., Feng, T., Jiang, X., Xu, S., et al. (2024). Topical application of phenformin ameliorates the psoriasis-like inflammatory response via the inhibition of c-Myc expression in keratinocytes. Biochem. Biophysical Res. Commun. 736, 150503. doi:10.1016/j.bbrc.2024.150503
Liu, L., Lu, Y., Martinez, J., Bi, Y., Lian, G., Wang, T., et al. (2016). Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc. Natl. Acad. Sci. 113, 1564–1569. doi:10.1073/pnas.1518000113
Liu, X., Jiang, B., Hao, H., and Liu, Z. (2022). CARD9 signaling, inflammation, and diseases. Front. Immunol. 13, 880879. doi:10.3389/fimmu.2022.880879
Liu, C., Kudo, T., Ye, X., and Gascoigne, K. (2023). Cell-to-cell variability in Myc dynamics drives transcriptional heterogeneity in cancer cells. Cell Rep. 42, 112401. doi:10.1016/j.celrep.2023.112401
Luo, H., Lin, S., Xiong, J., Tan, W., Lv, H., Liu, Z., et al. (2025). CARD14-Mediated MYC interaction promotes osteoclastogenesis and bone density reduction in adolescent idiopathic scoliosis. J. Bone Min. Res., zjaf127. doi:10.1093/jbmr/zjaf127
Ma, C. A., Stinson, J. R., Zhang, Y., Abbott, J. K., Weinreich, M. A., Hauk, P. J., et al. (2017). Germline hypomorphic CARD11 mutations in severe atopic disease. Nat. Genet. 49, 1192–1201. doi:10.1038/ng.3898
Man, X., Liu, T., Jiang, Y., Zhang, Z., Zhu, Y., Li, Z., et al. (2019). Silencing of CARMA3 inhibits bladder cancer cell migration and invasion via deactivating β-catenin signaling pathway. Onco Targets Ther. 12, 6309–6322. doi:10.2147/OTT.S191502
Meininger, I., and Krappmann, D. (2016). Lymphocyte signaling and activation by the CARMA1-BCL10-MALT1 signalosome. Biol. Chem. 397, 1315–1333. doi:10.1515/hsz-2016-0216
Mellett, M. (2020). Regulation and dysregulation of CARD14 signalling and its physiological consequences in inflammatory skin disease. Cell. Immunol. 354, 104147. doi:10.1016/j.cellimm.2020.104147
Molinero, L. L., Yang, J., Gajewski, T., Abraham, C., Farrar, M. A., and Alegre, M.-L. (2009). CARMA1 controls an early checkpoint in the thymic development of FoxP3+ regulatory T cells. J. Immunol. 182, 6736–6743. doi:10.4049/jimmunol.0900498
Molinero, L. L., Cubre, A., Mora-Solano, C., Wang, Y., and Alegre, M.-L. (2012). T cell receptor/CARMA1/NF-κB signaling controls T-helper (Th) 17 differentiation. Proc. Natl. Acad. Sci. U. S. A. 109, 18529–18534. doi:10.1073/pnas.1204557109
Moreno-García, M. E., Sommer, K., Rincon-Arano, H., Brault, M., Ninomiya-Tsuji, J., Matesic, L. E., et al. (2013). Kinase-independent feedback of the TAK1/TAB1 complex on BCL10 turnover and NF-κB activation. Mol. Cell Biol. 33, 1149–1163. doi:10.1128/MCB.06407-11
Muto, T., Guillamot, M., Yeung, J., Fang, J., Bennett, J., Nadorp, B., et al. (2022). TRAF6 functions as a tumor suppressor in myeloid malignancies by directly targeting MYC oncogenic activity. Cell Stem Cell 29, 298–314.e9. doi:10.1016/j.stem.2021.12.007
Newton, R., Priyadharshini, B., and Turka, L. A. (2016). Immunometabolism of regulatory T cells. Nat. Immunol. 17, 618–625. doi:10.1038/ni.3466
Oh, H., Grinberg-Bleyer, Y., Liao, W., Maloney, D., Wang, P., Wu, Z., et al. (2017). An NF-κB transcription-factor-dependent lineage-specific transcriptional program promotes regulatory T cell identity and function. Immunity 47, 450–465.e5. doi:10.1016/j.immuni.2017.08.010
Paul, S., Kashyap, A. K., Jia, W., He, Y.-W., and Schaefer, B. C. (2012). Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity 36, 947–958. doi:10.1016/j.immuni.2012.04.008
Peled, A., Sarig, O., Sun, G., Samuelov, L., Ma, C. A., Zhang, Y., et al. (2019). Loss-of-function mutations in caspase recruitment domain-containing protein 14 (CARD14) are associated with a severe variant of atopic dermatitis. J. Allergy Clin. Immunol. 143, 173–181.e10. doi:10.1016/j.jaci.2018.09.002
Pello, O. M. (2016). Macrophages and c-Myc cross paths. Oncoimmunology 5, e1151991. doi:10.1080/2162402X.2016.1151991
Pello, O. M., De Pizzol, M., Mirolo, M., Soucek, L., Zammataro, L., Amabile, A., et al. (2012). Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 119, 411–421. doi:10.1182/blood-2011-02-339911
Reimann, M., Schrezenmeier, J., Richter-Pechanska, P., Dolnik, A., Hick, T. P., Schleich, K., et al. (2021). Adaptive T-cell immunity controls senescence-prone MyD88- or CARD11-mutant B-cell lymphomas. Blood 137, 2785–2799. doi:10.1182/blood.2020005244
Rosenbaum, M., Schnalzger, T., Engleitner, T., Weiß, C., Mishra, R., Mibus, C., et al. (2022). MALT1 protease function in regulatory T cells induces MYC activity to promote mitochondrial function and cellular expansion. Eur. J. Immunol. 52, 85–95. doi:10.1002/eji.202149355
Ruland, J. (2008). CARD9 signaling in the innate immune response. Ann. N. Y. Acad. Sci. 1143, 35–44. doi:10.1196/annals.1443.024
Ruland, J., and Hartjes, L. (2019). CARD–BCL-10–MALT1 signalling in protective and pathological immunity. Nat. Rev. Immunol. 19, 118–134. doi:10.1038/s41577-018-0087-2
Saravia, J., Zeng, H., Dhungana, Y., Bastardo Blanco, D., Nguyen, T.-L. M., Chapman, N. M., et al. (2020). Homeostasis and transitional activation of regulatory T cells require c-Myc. Sci. Adv. 6, eaaw6443. doi:10.1126/sciadv.aaw6443
Snow, A. L., Xiao, W., Stinson, J. R., Lu, W., Chaigne-Delalande, B., Zheng, L., et al. (2012). Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J. Exp. Med. 209, 2247–2261. doi:10.1084/jem.20120831
Staal, J., Driege, Y., Van Gaever, F., Steels, J., and Beyaert, R. (2024). Chimeric and mutant CARD9 constructs enable analyses of conserved and diverged autoinhibition mechanisms in the CARD-CC protein family. FEBS J. 291, 1220–1245. doi:10.1111/febs.17035
Sun, J. (2010). CARMA3: a novel scaffold protein in regulation of NF-κB activation and diseases. World J. Biol. Chem. 1, 353–361. doi:10.4331/wjbc.v1.i12.353
Taghavi, P., Verhoeven, E., Jacobs, J. J. L., Lambooij, J. P., Stortelers, C., Tanger, E., et al. (2008). In vitro genetic screen identifies a cooperative role for LPA signaling and c-Myc in cell transformation. Oncogene 27, 6806–6816. doi:10.1038/onc.2008.294
Toll, A., Salgado, R., Yébenes, M., Martín-Ezquerra, G., Gilaberte, M., Baró, T., et al. (2009). MYC gene numerical aberrations in actinic keratosis and cutaneous squamous cell carcinoma. Br. J. Dermatol 161, 1112–1118. doi:10.1111/j.1365-2133.2009.09351.x
Wang, R., Dillon, C. P., Shi, L. Z., Milasta, S., Carter, R., Finkelstein, D., et al. (2011). The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882. doi:10.1016/j.immuni.2011.09.021
Wang, Y., Zhang, D., Hou, Y., Shen, S., and Wang, T. (2020). The adaptor protein CARD9, from fungal immunity to tumorigenesis. Am. J. Cancer Res. 10, 2203–2225.
Xia, S., Chen, Z., Shen, C., and Fu, T.-M. (2021). Higher-order assemblies in immune signaling: supramolecular complexes and phase separation. Protein and Cell 12, 680–694. doi:10.1007/s13238-021-00839-6
Yamagishi, M., Kubokawa, M., Kuze, Y., Suzuki, A., Yokomizo, A., Kobayashi, S., et al. (2021). Chronological genome and single-cell transcriptome integration characterizes the evolutionary process of adult T cell leukemia-lymphoma. Nat. Commun. 12, 4821. doi:10.1038/s41467-021-25101-9
Yang, C.-S., Rodgers, M., Min, C.-K., Lee, J.-S., Kingeter, L., Lee, J.-Y., et al. (2012). The autophagy regulator rubicon is a feedback inhibitor of CARD9-Mediated host innate immunity. Cell Host Microbe 11, 277–289. doi:10.1016/j.chom.2012.01.019
Ye, W.-L., Huang, L., Yang, X.-Q., Wan, S., Gan, W.-J., Yang, Y., et al. (2024). TRIM21 induces selective autophagic degradation of c-Myc and sensitizes regorafenib therapy in colorectal cancer. Proc. Natl. Acad. Sci. 121, e2406936121. doi:10.1073/pnas.2406936121
Zacarías-Fluck, M. F., Soucek, L., and Whitfield, J. R. (2024). MYC: there is more to it than cancer. Front. Cell Dev. Biol. 12, 1342872. doi:10.3389/fcell.2024.1342872
Keywords: Myc (c-Myc), CARD11 (CARMA1), CARD14 (CARMA2), CARD9, MALT1, CBM complex, NF-kappaB (NF-κB), CARD10 (CARMA3)
Citation: DeVore SB and Khurana Hershey GK (2025) MYC is in the CARDs: CBM complexes coordinate immune and MYC-dependent cellular function. Front. Mol. Med. 5:1731823. doi: 10.3389/fmmed.2025.1731823
Received: 24 October 2025; Accepted: 13 November 2025;
Published: 19 November 2025.
Edited by:
Haseeb Ahsan, Jamia Millia Islamia, IndiaReviewed by:
Jens Staal, Ghent University, BelgiumTakuro Kameda, Miyazaki Daigaku - Kiyotake Campus, Japan
Copyright © 2025 DeVore and Khurana Hershey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gurjit K. Khurana Hershey, Z3Vyaml0LmhlcnNoZXlAY2NobWMub3Jn
 Stanley B. DeVore
Stanley B. DeVore Gurjit K. Khurana Hershey1,2*
Gurjit K. Khurana Hershey1,2*