Protein folding as a driving force for dual protein targeting in eukaryotes
- 1Department of Microbiology and Molecular Genetics, Faculty of Medicine, Institute for Medical Research Israel-Canada, Hebrew University of Jerusalem, Jerusalem, Israel
- 2CREATE-NUS-HUJ Cellular and Molecular Mechanisms of Inflammation Program, National University of Singapore, Singapore, Singapore
It is well documented that in eukaryotic cells molecules of one protein can be located in several subcellular locations, a phenomenon termed dual targeting, dual localization, or dual distribution. The differently localized identical or nearly identical proteins are termed “echoforms.” Our conventional definition of dual targeted proteins refers to situations in which one of the echoforms is translocated through/into a membrane. Thus, dual targeted proteins are recognized by at least one organelle's receptors and translocation machineries within the lipid bilayer. In this review we attempt to evaluate mechanisms and situations in which protein folding is the major determinant of dual targeting and of the relative distribution levels of echoforms in the subcellular compartments of the eukaryotic cell. We show that the decisive folding step can occur prior, during or after translocation through the bilayer of a biological membrane. This phenomenon involves folding catalysts in the cell such as chaperones, proteases and modification enzymes, and targeting processes such as signal recognition, translocation through membranes, trapping, retrotranslocation and reverse translocation.
Introduction
Dual localization of proteins can be achieved by a variety of molecular mechanisms all described in depth in reviews on this topic (Karniely and Pines, 2005; Regev-Rudzki and Pines, 2007; Avadhani, 2011; Yogev and Pines, 2011; Duchene and Giege, 2012; Carrie and Small, 2013; Carrie and Whelan, 2013). The dual localized, identical or nearly identical proteins, are termed “echoforms” indicating repetitious forms of the same protein distinctly placed in the cell (Yogev and Pines, 2011). Dual targeting mechanisms can be divided into two types, according to the number of translation products involved. Dual targeting by two translation products can occur due to the existence of multiple mRNAs that are derived from a single gene. This can be achieved either by alternative transcription initiation or mRNA splicing, in which the coding for a targeting sequence is removed. One mRNA can also give rise to several proteins by translation initiation from a downstream in frame start codon or stop codon read-through (see reviews above and Mitrpant et al., 2009; Freitag et al., 2012). In all these cases, two translation products (one containing and one lacking the targeting signal) are made and are targeted to different cellular locations. Thus, dual targeting is determined prior to synthesis of the protein(s) and these mechanisms do not necessitate a decision involving protein folding. Dual targeting of a single translation product on the other hand may or may not include protein folding as a driving force. In the next section we consider the participation of protein folding in the dual targeting mechanisms of single translation products. Figure 1 presents types of dual targeting mechanisms (Figures 1A–F) and other theoretically possible mechanisms (Figures 1G–I) that could lead to dual localization of echoforms in eukaryotic cells.
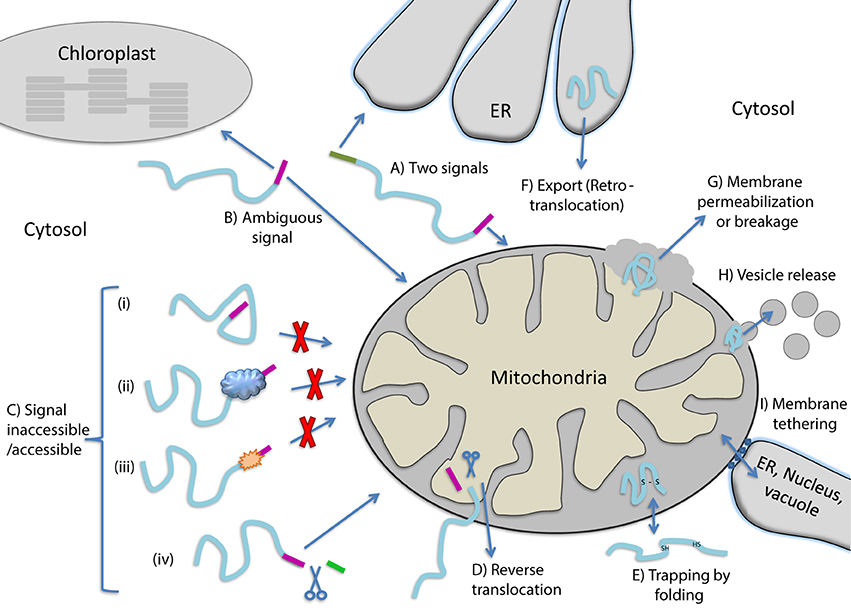
Figure 1. Mechanisms allowing dual targeting of a single translation product. (A) Competition between two signals for different organelles on the same polypeptide. (B) An ambiguous targeting signal is recognized by two organelles (C) Changes in the targeting signal accessibility caused by protein (i) folding, (ii) binding to cellular factors, (iii) modification or (iv) cleavage by a protease that exposes a targeting signal. (D) Reverse translocation, polypeptides move back to the cytosol during translocation into an organelle. (E) Trapping of proteins in an organelle by folding. (F) Export of proteins out of an organelle. (G) Release of proteins from organelles due to membrane permeablization or breakage. (H) Release of proteins from organelles via vesicles. (I) Release of proteins from organelles through tethering of membranes.
Considerations of Protein Folding and Dual Targeting
Dual localization of proteins may be affected by folding of proteins prior to their targeting to an organelle, during translocation through membranes or even after translocation into an organelle (Figure 2). In the first situation, dual targeting can be determined for instance by an ambiguous targeting sequence on a single polypeptide that can be recognized by more than one organelle (Figure 1B). Similarly, two (or more) targeting signals on a single polypeptide can provide a mechanism of dual targeting (Figure 1A). Here the balance of echoform amounts between the different organelles is determined by the affinity of each signal for its target. In these cases single translation products harboring one or more specific targeting signals can be dual targeted, when one of the signals is inaccessible under certain conditions or there is a change in the affinity of the signal for its receptor. We have chosen to present examples in which this change in accessibility or affinity is probably due to protein folding, or related processes which affect protein conformation such as modification or binding of another protein (Figure 1C).
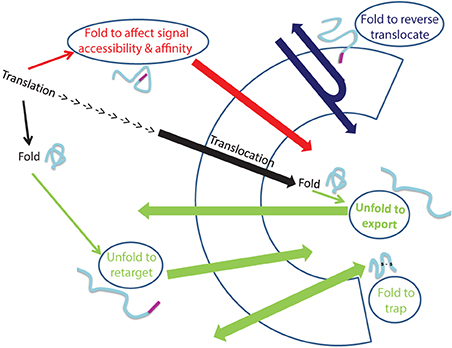
Figure 2. Folding and unfolding “decisions” that determine dual targeting. The normal process of protein translocation into an organelle (such as the ER and mitochondria) is depicted in black. Dual localization of proteins may be determined by folding of proteins, indicated by ellipses, prior to their targeting to an organelle (Red arrow), during translocation through membranes (Blue arrow) or after translocation into an organelle (Green arrows). Folding of proteins can affect signal accessibility and affinity (Red arrow; e.g., Adk1, Apn1, Gus1, CYP1A1, Fis1, b5R, CYP2E1), cause reverse translocation (Blue; e.g., Fum1, Aco1), trap proteins in an organelle (Green bottom arrow: e.g., Ccs1, COX19) or by unfolding, allow export of a protein out of an organelle (Green: top arrow, e.g., PrPc) or allow retargeting of a protein into an organelle (Green middle arrow, e.g., TERT, p53).
An intriguing example in which dual targeting is determined by protein folding during translocation is the mechanism termed “reverse translocation” (Figures 1D, 2). In this case all protein molecules are first targeted to an organelle, begin their translocation but then a sub-population of the molecules moves back to the cytosol. Protein folding and assembly appears to be the driving force for this dual targeting and we will review this matter employing yeast fumarase as a paradigm.
We can identify a number of situations in which dual targeting is determined after translocation of substrates into organelles. For instance, newly synthesized proteins that fail to achieve their native active conformation in the endoplasmic reticulum (ER) are recognized and degraded by the ubiquitin–proteasome machinery which functions as a quality control system. This process is termed ER−associated degradation (ERAD) and is characterized by incorrect protein folding. Given that ubiquitin, ubiquitination machinery and proteasomes are not in the ER lumen and are associated with the cytosol and nucleus, ERAD substrates must be exported from the ER lumen and/or extracted from the ER membrane. We will discuss whether this can be considered as a basic mechanism for dual targeting of proteins (Figure 1F).
Dual targeting can also potentially occur after translocation of proteins such as the small TIMs, and superoxide dismutase (SOD). These proteins are imported into the mitochondrial intermembrane space, there they are trapped by folding and intramolecular disulfide bond formation (Figure 1E). In the absence of disulfide bond formation some of these protein molecules can move back through the TOM complex into the cytosol. In these cases, if such a protein is dual localized to the IMS and cytosol, one may refer to a mechanism of dual targeting involving “trapping of proteins” in a subcellular compartment due to protein folding.
How certain mitochondrial matrix proteins, under specific circumstances, can also be located outside the organelle has not been completely understood. This is troubling since in contrast to ERAD, above, a defined export system of proteins out of mitochondria or chloroplasts has not been detected. For example mitochondrial matrix proteins found outside mitochondria include the molecular chaperones mtHsp60 and mtHsp70 in mammalian cells and WHIRLY in plants (Soltys et al., 1996; Chandra et al., 2007; Kaul et al., 2007; Iosefson and Azem, 2010; Krause et al., 2012). Mechanisms that may explain the above observations include (i) the breakdown of the outer membrane or both inner and outer membranes, for instance during apoptosis (Figure 1G) (reviewed in Kluck et al., 1997; Green and Kroemer, 2004; Brenner and Mak, 2009), (ii) release of proteins by vesicles (Figure 1H, e.g., McLelland et al., 2014; Sugiura et al., 2014), or (iii) transfer between organelles by tethering of membranes such as those of the ER, vacuole/lysosome and mitochondria (Figure 1I, e.g., Kornmann et al., 2009; Elbaz and Schuldiner, 2011; Elbaz-Alon et al., 2014). In these cases we cannot assess whether protein folding is involved let alone whether it is the driving force for protein distribution. Nevertheless, we should keep in mind that, these mechanisms can theoretically lead to dual targeting of proteins in eukaryotes.
Finally, targeting or localization of proteins to particular compartments in the cell is not an unchangeable process. Actually the change in the destination of a protein is one of the mechanisms that should allow response to changes within the cell or in its environment. In this sense, relocalization can result in dual targeting of proteins (Figure 2). We shall discuss examples in which relocalization is apparently determined by folding of the retargeted protein.
When discussing examples in most cases mechanisms as presented in Figure 1 may overlap. For instance a protein may have two signals (1A) in which one of the signals becomes inaccessible (1C). In such cases, for the sake of clarity, we will refer only to the mechanism which is defined by folding.
Accessibility of Targeting Signals
Single translation products can be dual targeted due accessibility or inaccessibility of a targeting signal. For some of the molecules of a specific protein the signal becomes inaccessible, thus inhibiting the contact with the specific receptor (Figures 1C, 2). Inaccessibility of the targeting sequence can be the consequence of (i) folding of the protein (ii) interaction of the protein with other proteins, or (iii) modification of the polypeptide chain. The yeast adenylate kinase 2 (Adk1) is distributed between the mitochondrial intermembrane space and the cytosol. Its rapid folding into a protease resistant structure hinders the accessibility of targeting signals (N terminal and internal sequences) to the mitochondrial receptor and some of these molecules are therefore retained in the cytosol (Bandlow et al., 1998; Strobel et al., 2002). In fact, x-ray crystallographic analysis (Egner et al., 1987), shows that both the N terminus and internal import relevant sequences are concealed in the folded protein structure of the native conformation, and the protein cannot be imported post translationally. In this case, competition between folding and targeting determines protein localization (Figures 1Ci, 2). A relevant study by Pfeiffer et al. (2013) shows that certain signal peptides promote efficient ER import when artificially fused to α-helical domains, but target unstructured polypeptides to mitochondria. Thus, targeting is affected not only by the targeting sequence but also by the structure of the nascent chain to which these sequences are attached.
The import efficiency may also affect accessibility of signals and substrates. ATFS-1, a key regulator of UPRmt (mitochondrial unfolded protein response), has both a nuclear localization sequence (NLS) and an MTS (Mitochondrial targeting sequence) that is essential for UPRmt repression. Normally, ATFS-1 is imported into mitochondria and degraded, however, upon stress, reduction in mitochondrial import efficiency causes a percentage of unprocessed ATFS-1 to accumulate in the cytosol and traffic to the nucleus (Nargund et al., 2012; Pellegrino et al., 2014) In this regard, the import efficiency could also be affected by modification of the translocation apparatus which may in turn affect signal accessibility (Schmidt et al., 2011; Harbauer et al., 2014).
The protein Apurinic/apyrimidinic endonuclease 1 (Apn1) (Vongsamphanh et al., 2001) is a case in which the targeting signal becomes inaccessible by binding of another protein (Figure 1Cii). Apn1 has two targeting signals: a nuclear localization sequence (NLS) and an MTS. Apn1 is bound by the protein Pir1 thereby concealing its NLS and allowing more pronounced mitochondrial targeting of the protein by the MTS. Apn1 (above) like Gus1 (below) belongs to a group of dual targeted proteins that are nucleic acid-transacting proteins (e.g., DNA damage response and RNA metabolism enzymes) that perform similar activities in mitochondria and outside the organelle. Glutamyl-tRNA synthetase (Gus1), in yeast, displays a similar mechanism to Apn1, by which it is localized to the mitochondria and cytosol. Import into mitochondria is driven by an MTS like sequence located following the first 190 residues. Binding of Arc1p to Gus1 inhibits mitochondrial import and causes the accumulation of Gus1 in the cytoplasm, while absence of Arc1 leads to exclusive mitochondrial localization. Arc1 levels are regulated by a metabolic change from fermentation to respiration, in which a reduction in Arc1 leads to an increase in mitochondrial Gus1 import and mitochondrial protein synthesis (Frechin et al., 2009). Apn1/Pir1 and Gus1/Arc1 are examples of signals whose accessibility is determined by the binding of another protein (Figure 1Cii).
For CYP1A1, the polypeptide modification makes one of the signals inaccessible, by cleaving off that signal (Figure 1Civ). In this sense proteases (and other protein modification enzymes) are reminiscent of molecular chaperones that bind the substrate protein, affect its conformation and ultimately its fate, which in this case is its final destination. CYP1A1 contains an ER signal peptide at its N-terminus followed by a cryptic MTS. The majority of the signal-peptide containing molecules are, as expected, translocated into the ER, yet a quarter of the polypeptides are cleaved by a cytosolic protease. These latter molecules escape ER membrane insertion and concomitantly expose an active cryptic mitochondrial targeting sequence (Addya et al., 1997; Avadhani et al., 2011). Thus, CYP1A1 dual distribution can be regulated by inducing the protease (e.g., with b-naphthoflavone).
Changes in Affinity of Targeting Signals
Human Fis1 is a tail-anchored membrane protein that regulates the membrane fission of both peroxisomes and mitochondria. The C-terminus of this protein is an ambiguous signal, which is affected by the binding of another protein (Figure 1Cii). The C-terminal 26 amino acids bind Pex19, a peroxisomal membrane protein import factor or alternatively function as a mitochondrial tail anchor (Delille and Schrader, 2008). Even though, both targeting events to mitochondria and peroxisomes are dependent on the same C-terminal sequence, they seem to be independent; down-regulation of Pex19 reduces peroxisome targeting but not targeting to mitochondria.
The affinity of an ambiguous targeting signal to the separate subcellular compartments can be modulated by protein modification (Figure 1Ciii). Modification of the mammalian NADH–cytochrome b(5)reductase (b5R) ambiguous targeting sequence, is a good example for this mechanism (Colombo et al., 2005). b5R is found both in the outer mitochondrial and ER membranes. Within the ER membrane, b5R is involved in lipid metabolism through its function as an electron acceptor. Within the mitochondrial outer membrane b5R mediates the regeneration of ascorbate from ascorbate free radical and is involved in transfer of electrons from cytosolic NADH to cytochrome C in the intermembrane space. b5R is translated as a single translation product from a single mRNA. This product contains an N-terminal targeting signal, required for targeting both to the ER and mitochondria. The targeting signal consists of a myristoylation consensus sequence followed by a 14 amino acids sequence which is moderately hydrophobic. The myristoylation consensus sequence is modified in about half of the b5R molecules. Nascent chains of b5R that are not myristoylated, remain bound to the signal recognition particle (SRP) and are consequently translocated into the ER. However, myristoylation of the N-terminal signal lowers its affinity for SRP, situating nascent chains on membrane free polysomes which are consequently imported into mitochondria (Colombo et al., 2005).
A change in the relative affinity of each targeting sequence for its target can also be affected by protein phosphorylation, which can change the relative amounts of the subcellular populations of the protein (Figure 1Ciii). A number of Cytochrome P450 monooxygenases (CYPs), mainly by the comprehensive studies of Avadahni and colleagues, have been shown to be dual targeted (e.g., see the CYP1A1 in the previous section). Phosphorylation plays a major role in the dual targeting of CYP2B1, CYP2E1, and CYP2D6 (Avadhani et al., 2011). CYP2E1, for example, which plays an important role in alcohol-induced toxicity and oxidative stress, is dual targeted to the ER and mitochondria. It contains an N-terminal 30 amino acids that constitute a bimodal signal for dual targeting. The model of dual targeting proposes that the low affinity of the CYP2E1 signal sequence for the SRP, causes half of the nascent chains to escape ER targeting and their translation as membrane free protein. Notably, CYP2E1 also harbors a cryptic signal located at the N-terminus of the protein (Robin et al., 2002; Avadhani et al., 2011). The twist in this case is that, cAMP-dependent phosphorylation of CYP2E1 on Ser129 by PKA results in the activation of the cryptic targeting signal, which increases the association of the protein with the cytoplasmic Hsp70 and Hsp90 chaperones and in turn binding to mitochondrial translocase subunits TOM70 and TOM40, thereby favoring its mitochondrial import (Anandatheerthavarada et al., 2009; Avadhani et al., 2011).
Reverse Translocation out of an Organelle (During Translocation)
An exceptional dual targeting mechanism is based on retrograde movement of a protein during its import (Figure 1D). The enzyme fumarase in the yeast S. cerevisiae is a paradigm of this mechanism. Fumarase is a TCA cycle enzyme in mitochondria and functions in the DNA damage response in the cytosol/nucleus (Yogev et al., 2010). In this case, all molecules are first targeted to mitochondria, begin their translocation and are processed by Mitochondrial Processing Peptidase (MPP). Nevertheless, a sub-population of the molecules moves back to the cytosol (Sass et al., 2001). According to the model, the driving force for this distribution is protein folding; if during import the nascent chain starts to fold in the mitochondrial matrix, it will complete its import and be localized in mitochondria. On the other hand, if the nascent chain starts its folding in the cytosol, thereby blocking its forward movement, the protein will withdraw from the import machinery, and will be localized in the cytosol (Sass et al., 2003). An indication of this mechanism are identical MPP processed echoforms found both inside and outside the organelle (Sass et al., 2001). The relative distribution of fumarase between the mitochondria and cytosol can be affected by the level of Hsp70 molecular chaperones in these respective compartments and by the rate of mitochondrial import (Karniely et al., 2006; Yogev et al., 2007; Regev-Rudzki et al., 2008). In addition, mutations or short deletions within the polypeptide sequence which disturb the fumarase structure (required for reverse translocation) cause full import of the protein (Sass et al., 2003). These data support the notion that folding is the major driving force for reverse translocation. Recently this model gained additional support; the bacterial fumarase homolog, fumC, was evolved by in vitro evolution into a dual targeted protein in yeast, suggesting that the natural folding of this protein was harnessed by evolution to distribute the protein in the cell (Burak et al., 2013). Worth pointing out is the fact, that upon expression of fumarase from the mitochondrial genome, no fumarase is detected outside the organelle, ruling out export or release of fumarase from mitochondria as the mechanism of dual targeting (Yogev et al., 2010). Dual targeting in S. cerevisiae of Nfs1 and Aco1 has been suggested to occur via the reverse translocation mechanism. Aconitase is targeted to the mitochondria and the cytosol, while Nfs is detected in the mitochondria and the nucleus (Regev-Rudzki et al., 2005; Naamati et al., 2009; Ben-Menachem et al., 2011a). Of importance is the finding that the MTS strength can determine the relative distribution of aconitase (and fumarase) between the two locations. The mechanism suggested is that the mitochondrial-targeting signal affects the translocation rate thereby determining the time (opportunity) required for the protein to fold or bind factors in the cytosol that block import (Regev-Rudzki et al., 2008). Consequently, slowing down translocation by reducing membrane potential or by using translocase mutant causes accumulation of cytosolic fumarase.
Trapping of Proteins in an Organelle by Protein Folding (Figure 1E)
Numerous proteins of the intermembrane space (IMS) are imported by the mitochondrial disulfide relay system. These polypeptides which lack MTSs are recognized and oxidized by the IMS located receptor Mia40. Reoxidation of Mia40 is facilitated by the sulfhydryl oxidase Erv1 and the respiratory chain. The majority of the substrates of the mitochondrial disulfide relay system are small proteins with simple helix-loop-helix folds in which the helices are connected by two disulfide bonds; twin CX3C and twin CX9C proteins (Chacinska et al., 2009; Herrmann and Riemer, 2012). Other IMS proteins containing disulfide bonds include the yeast superoxide dismutase Sod1 and its copper chaperone Ccs1, both which are dually localized in the IMS of mitochondria and the cytosol. Sod1 and Ccs1 form part of the anti-oxidative system that dismutates superoxide anions to hydrogen peroxide. Sod1 is a dimeric copper- and zinc-containing protein that contains one disulfide bond per subunit. The insertion of this disulfide bond and of the copper ion is facilitated by Ccs1 (Sturtz et al., 2001; Reddehase et al., 2009; Kloppel et al., 2010, 2011; Gross et al., 2011) whose mitochondrial form also contains a stable disulfide bond between cysteine residues C27 and C64. In the absence of these cysteines, the levels of Ccs1 and Sod1 in mitochondria are strongly reduced. Accordingly, enhanced Ccs1 levels lead to an increase in the levels of active Sod1. Thus, the Mia40/Erv1 disulfide relay system introduces a structural disulfide bond in Ccs1 between the cysteine residues C27 and C64, thereby trapping Ccs1 in the IMS of mitochondria and controlling its distribution between the IMS and the cytosol. The distribution of Ccs1, in turn, determines the distribution of Sod1 by determining its oxidation and copper binding.
The cytochrome oxidase assembly factor COX19 which contains a twin Cx9C motif partitions between mitochondria and the cytosol in human cells (Nobrega et al., 2002; Leary et al., 2013). The cytosol is relatively enriched for COX19 when intracellular copper concentrations are elevated, suggesting that trapping of COX19 in the IMS is affected by copper levels. Proper function of SCO1 and SCO2 within the IMS is essential for the COX19-mediated transduction of appropriate redox signals outside mitochondria to regulate cellular copper homeostasis. The full mechanism of how copper affects disulfide formation and trapping of COX19 remains to be determined.
Export of Proteins out of Organelles
Quality control of protein folding inside the ER includes chaperone-mediated assistance in folding and the selective targeting of terminally misfolded species to a pathway called ER-associated protein degradation, ERAD. Once selected for ERAD, substrates will be transported (back) into the cytosol, a step called retrotranslocation. Although still ill defined, retrotranslocation likely involves a protein conducting channel that is in part formed by specific membrane-embedded E3 ubiquitin ligases. A common mechanism of how individual misfolded proteins in the ER are first recognized is not known but it involves the exposure of hydrophobic domains on these proteins and the binding to chaperones. The selected substrates are then targeted to the membrane-embedded E3 ligase complexes where they undergo ubiquitination on their cytosolically exposed protein domains during or after retrotranslocation. Ubiquitin on substrates was originally thought to be a permanent modification that promotes the late steps of retrotranslocation. However, not all ERAD substrates are ubiquinated and degraded (Bernardi et al., 2010; Li et al., 2010). The enzymatic A1 chain of cholera toxin in the absence of ubiquitination retrotranslocates across the endoplasmic reticulum membrane into the cytosol, where it induces toxicity. The force that drives this retrotranslocation is not known. Viruses, like simian virus 40 (SV40), travel to the ER through the secretory pathway and then these viruses exploit ERAD components to reach the cytosol and nucleus (Tsai et al., 2001; Schelhaas et al., 2007; Bernardi et al., 2008).
PrPC is a GPI-anchored secretory pathway protein which is found on the cell surface, in endocytic vesicles and endosomes. Several functions have been proposed for PrPC, including roles in cell adhesion, neurite outgrowth, neuronal excitability and neuroprotection. Whereas the proposed functions of PrPC are a matter of controversy the fact that the protein can be detected in the endomembrane system and the cytosol is widely accepted (Biasini et al., 2012). The identification of interacting proteins with PrPC which are cell surface or secreted molecules but also cytoplasmic, supports this conclusion (Biasini et al., 2012). Studies have shown that treating cells with proteasome inhibitors causes ubiquitylated PrPC to accumulate, implying that PrPC is degraded by the proteasome in the cytosol (Ma and Lindquist, 2001; Yedidia et al., 2001). In addition the PrPC that was detected in the cytosol appeared to have undergone post-translational modifications in the ER (removal of N-terminal signal peptide and C-terminal GPI anchor signal sequence) strongly suggesting that PrPC was reaching the cytoplasm via retrotranslocation. Like other ERAD substrates one can assume that it is the conformation or folding of the PrPC molecules that determines retrotranslocation out of the ER (Figure 1F).
Retargeting of Echoforms
Dual or exclusive localization of a protein to specific compartments in the cell is a changeable process. In fact protein relocalization is one of the mechanisms that allows response to changes within the cell or in its environment. Recently, changes in the localization of hundreds of proteins have been shown to occur in response to different stress conditions in yeast (Breker et al., 2014). TERT, the enzyme telomerase reverse transcriptase is required to counteract shortening the ends of chromosomes in the cell nucleus. Nevertheless, there is evidence for a TERT function in mitochondria where it is proposed to reduce reactive oxygen species, protect mitochondrial DNA and reduce apoptosis. TERT harbors a bipartite NLS (nuclear localization sequence) which can be phosphorylated on a serine residue by protein kinase B/Akt (Chung et al., 2012). TERT also contains an NES (nuclear export signal) at its C-terminus, which can interact with the nuclear export receptor CRM1/exportin 1. Besides the NLS and NES, an N-terminal mitochondrial targeting sequence (MTS) has been identified on TERT (Santos et al., 2004). Mitochondrial TERT to a large extent is found in the matrix and IMS there it interacts with the mitochondrial translocases of the outer membrane Tom20 and Tom40 and Tim23 of the inner membrane and binds mitochondrial RNAs (Haendeler et al., 2009; Sharma et al., 2012). Upon oxidative stress, TERT is excluded from the nucleus and imported into mitochondria in which phosphorylation plays a regulatory role (Ale-Agha et al., 2014). Based on the decrease in nuclear TERT and concomitant mitochondrial targeting, the model is that the protein is exported from the nucleus and imported into the mitochondria (Figure 1Ciii).
A second possible example for protein relocalization is p53, which is a transcription factor that mediates apoptosis by transcription activation of pro-apoptotic genes or repression of anti-apoptotic genes. In response to stress, a fraction of p53 rapidly localizes to mitochondria prior to p53 nuclear accumulation, triggering mitochondrial outer membrane permeabilization and caspase activation (Mihara et al., 2003; Erster et al., 2004; Zhao et al., 2005). Whether mitochondrial p53 is derived from a cytosolic or nuclear pool of p53 molecules, remains to be determined. p53 can interact with mitochondrial proteins located in the mitochondrial matrix, such as the DNA polymerase γ, which is involved in the synthesis and repair of mtDNA (Bergeaud et al., 2013). In fact this interaction accounts for a higher mtDNA copy number in wild type vs. p53 knockout cells (de Souza-Pinto et al., 2004). Most of the intramitochondrial pool of p53 is present in two soluble compartments of mitochondria, the intermembrane space (IMS) and the matrix. Even though p53 is detected within mitochondria, the p53 polypeptide sequence lacks a mitochondrial-targeting signal, which is true for other nuclear factors that are detected in mitochondria such as CREB, NFκ B, or STAT3 (Cogswell et al., 2003; Lee et al., 2005; Wegrzyn et al., 2009). p53 can be targeted to the translocase of mitochondria by other proteins such as mtHsp70, Tid1, mitochondrial helicase RECQL4, or OKL38 (Marchenko et al., 2000; Ahn et al., 2010; De et al., 2012; Hu et al., 2012). p53 contains several reactive cysteines and may also be imported into mitochondria via the disulfide relay import system which was referred to in previous sections of this review (Herrmann and Riemer, 2012) Thus, although we do not know the precise molecular events by which p53 is retargeted to mitochondria this obviously involves protein binding to p53, unfolding and folding of p53 that are required for its import (Figure 2).
Concluding Remarks
It is now established that dual targeting of proteins in eukaryotic cells is a highly abundant phenomenon (Introduction and (Dinur-Mills et al., 2008; Ben-Menachem et al., 2011b; Yogev and Pines, 2011). Why are proteins dual localized and why is this phenomenon so abundant? We have discussed this question (Ben-Menachem et al., 2011b; Kisslov et al., 2014) and have concluded that dual targeting is driven by dual function. There are numerous examples of separate functions of dual localized proteins in the different compartments. Recently, dual targeted proteins were shown to be more evolutionarily conserved (by a number of criteria) which is consistent with a double selective pressure based on dual function (Kisslov et al., 2014). In addition, homologous proteins in different organisms can be dual targeted by different mechanisms (e.g., Yogev et al., 2011) indicating that what matters is the function in the different compartments and not the mechanism by which they are targeted. Current research indicates that dual localization is not a sloppy or leaky process but it is rather based on precise molecular mechanisms and serves distinct functional requirements. The regulation of dual targeting has profound influence on the way we comprehend the control of gene expression and function in eukaryotes.
In this review we have discussed how dual targeting can be affected and in some cases controlled by protein folding. We show that dual targeting can be affected via protein folding through the action of chaperones, proteases, kinases/phosphotases, oxidases, ubiquitin-ligases/ubiquitylases and other modification enzymes and binding proteins. The decisive folding step in each case can occur prior, during or after translocation through the bilayer of a biological membrane (Figure 2). The concept that we present is that the cell can regulate the distribution of many proteins, in concert, by modifying the level and activity of these folding catalysts and folding conditions. With all that said, it is quite clear that we do not have the full picture of protein dual targeting mechanisms in the cell and moreover we are only beginning to understand the impact of protein folding on this phenomenon. For example, a lot is yet to be learned regarding dual targeting involving membrane permeabilization/breakage, vesicle release or membrane tethering (Figures 1G–I).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the Israel Science Foundation (ISF), The USA-Israel Binational Science Foundation (BSF) and the CREATE project of the National Research Foundation of Singapore.
References
Addya, S., Anandatheerthavarada, H. K., Biswas, G., Bhagwat, S. V., Mullick, J., and Avadhani, N. G. (1997). Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J. Cell Biol. 139, 589–599. doi: 10.1083/jcb.139.3.589
Ahn, B. Y., Trinh, D. L., Zajchowski, L. D., Lee, B., Elwi, A. N., and Kim, S. W. (2010). Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene 29, 1155–1166. doi: 10.1038/onc.2009.413
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ale-Agha, N., Dyballa-Rukes, N., Jakob, S., Altschmied, J., and Haendeler, J. (2014). Cellular functions of the dual-targeted catalytic subunit of telomerase, telomerase reverse transcriptase–potential role in senescence and aging. Exp. Gerontol. 56, 189–193. doi: 10.1016/j.exger.2014.02.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Anandatheerthavarada, H. K., Sepuri, N. B., and Avadhani, N. G. (2009). Mitochondrial targeting of cytochrome P450 proteins containing NH2-terminal chimeric signals involves an unusual TOM20/TOM22 bypass mechanism. J. Biol. Chem. 284, 17352–17363. doi: 10.1074/jbc.M109.007492
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Avadhani, N. G. (2011). Targeting of the same proteins to multiple subcellular destinations: mechanisms and physiological implications. FEBS J. 278, 4217. doi: 10.1111/j.1742-4658.2011.08355.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Avadhani, N. G., Sangar, M. C., Bansal, S., and Bajpai, P. (2011). Bimodal targeting of cytochrome P450s to endoplasmic reticulum and mitochondria: the concept of chimeric signals. FEBS J. 278, 4218–4229. doi: 10.1111/j.1742-4658.2011.08356.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bandlow, W., Strobel, G., and Schricker, R. (1998). Influence of N-terminal sequence variation on the sorting of major adenylate kinase to the mitochondrial intermembrane space in yeast. Biochem. J. 329(Pt 2), 359–367.
Ben-Menachem, R., Regev-Rudzki, N., and Pines, O. (2011a). The aconitase C-terminal domain is an independent dual targeting element. J. Mol. Biol. 409, 113–123. doi: 10.1016/j.jmb.2011.03.045
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ben-Menachem, R., Tal, M., Shadur, T., and Pines, O. (2011b). A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics 11, 4468–4476. doi: 10.1002/pmic.201100199
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergeaud, M., Mathieu, L., Guillaume, A., Moll, U. M., Mignotte, B., Le Floch, N., et al. (2013). Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F(1)F0-ATP synthase. Cell Cycle 12, 2781–2793. doi: 10.4161/cc.25870
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bernardi, K. M., Forster, M. L., Lencer, W. I., and Tsai, B. (2008). Derlin-1 facilitates the retro-translocation of cholera toxin. Mol. Biol. Cell 19, 877–884. doi: 10.1091/mbc.E07-08-0755
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bernardi, K. M., Williams, J. M., Kikkert, M., van Voorden, S., Wiertz, E. J., Ye, Y., et al. (2010). The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol. Biol. Cell 21, 140–151. doi: 10.1091/mbc.E09-07-0586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Biasini, E., Turnbaugh, J. A., Unterberger, U., and Harris, D. A. (2012). Prion protein at the crossroads of physiology and disease. Trends Neurosci. 35, 92–103. doi: 10.1016/j.tins.2011.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Breker, M., Gymrek, M., Moldavski, O., and Schuldiner, M. (2014). LoQAtE–Localization and Quantitation ATlas of the yeast proteomE. A new tool for multiparametric dissection of single-protein behavior in response to biological perturbations in yeast. Nucleic Acids Res. 42, D726–D730. doi: 10.1093/nar/gkt933
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenner, D., and Mak, T. W. (2009). Mitochondrial cell death effectors. Curr. Opin. Cell Biol. 21, 871–877. doi: 10.1016/j.ceb.2009.09.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burak, E., Yogev, O., Sheffer, S., Schueler-Furman, O., and Pines, O. (2013). Evolving dual targeting of a prokaryotic protein in yeast. Mol. Biol. Evol. 30, 1563–1573. doi: 10.1093/molbev/mst039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carrie, C., and Small, I. (2013). A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim. Biophys. Acta 1833, 253–259. doi: 10.1016/j.bbamcr.2012.05.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carrie, C., and Whelan, J. (2013). Widespread dual targeting of proteins in land plants: when, where, how and why. Plant Signal. Behav. 8:e25034. doi: 10.4161/psb.25034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T., and Pfanner, N. (2009). Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644. doi: 10.1016/j.cell.2009.08.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chandra, D., Choy, G., and Tang, D. G. (2007). Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. J. Biol. Chem. 282, 31289–31301. doi: 10.1074/jbc.M702777200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chung, J., Khadka, P., and Chung, I. K. (2012). Nuclear import of hTERT requires a bipartite nuclear localization signal and Akt-mediated phosphorylation. J. Cell Sci. 125, 2684–2697. doi: 10.1242/jcs.099267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cogswell, P. C., Kashatus, D. F., Keifer, J. A., Guttridge, D. C., Reuther, J. Y., Bristow, C., et al. (2003). NF-kappa B and I kappa B alpha are found in the mitochondria. Evidence for regulation of mitochondrial gene expression by NF-kappa B. J. Biol. Chem. 278, 2963–2968. doi: 10.1074/jbc.M209995200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Colombo, S., Longhi, R., Alcaro, S., Ortuso, F., Sprocati, T., Flora, A., et al. (2005). N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 168, 735–745. doi: 10.1083/jcb.200407082
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Delille, H. K., and Schrader, M. (2008). Targeting of hFis1 to peroxisomes is mediated by Pex19p. J. Biol. Chem. 283, 31107–31115. doi: 10.1074/jbc.M803332200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De, S., Kumari, J., Mudgal, R., Modi, P., Gupta, S., Futami, K., et al. (2012). RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J. Cell Sci. 125, 2509–2522. doi: 10.1242/jcs.101501
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
de Souza-Pinto, N. C., Harris, C. C., and Bohr, V. A. (2004). p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene 23, 6559–6568. doi: 10.1038/sj.onc.1207874
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dinur-Mills, M., Tal, M., and Pines, O. (2008). Dual targeted mitochondrial proteins are characterized by lower MTS parameters and total net charge. PLoS ONE 3:e2161. doi: 10.1371/journal.pone.0002161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Duchene, A. M., and Giege, P. (2012). Dual localized mitochondrial and nuclear proteins as gene expression regulators in plants? Front. Plant Sci. 3:221. doi: 10.3389/fpls.2012.00221
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Egner, U., Tomasselli, A. G., and Schulz, G. E. (1987). Structure of the complex of yeast adenylate kinase with the inhibitor P1,P5-di(adenosine-5′-)pentaphosphate at 2.6 A resolution. J. Mol. Biol. 195, 649–658. doi: 10.1016/0022-2836(87)90188-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elbaz, Y., and Schuldiner, M. (2011). Staying in touch: the molecular era of organelle contact sites. Trends Biochem. Sci. 36, 616–623. doi: 10.1016/j.tibs.2011.08.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Elbaz-Alon, Y., Rosenfeld-Gur, E., Shinder, V., Futerman, A. H., Geiger, T., and Schuldiner, M. (2014). A dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 30, 95–102. doi: 10.1016/j.devcel.2014.06.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Erster, S., Mihara, M., Kim, R. H., Petrenko, O., and Moll, U. M. (2004). In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol. Cell Biol. 24, 6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frechin, M., Senger, B., Braye, M., Kern, D., Martin, R. P., and Becker, H. D. (2009). Yeast mitochondrial Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 23, 1119–1130. doi: 10.1101/gad.518109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Freitag, J., Ast, J., and Bolker, M. (2012). Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525. doi: 10.1038/nature11051
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Green, D. R., and Kroemer, G. (2004). The pathophysiology of mitochondrial cell death. Science 305, 626–629. doi: 10.1126/science.1099320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gross, D. P., Burgard, C. A., Reddehase, S., Leitch, J. M., Culotta, V. C., and Hell, K. (2011). Mitochondrial Ccs1 contains a structural disulfide bond crucial for the import of this unconventional substrate by the disulfide relay system. Mol. Biol. Cell 22, 3758–3767. doi: 10.1091/mbc.E11-04-0296
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haendeler, J., Drose, S., Buchner, N., Jakob, S., Altschmied, J., Goy, C., et al. (2009). Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler. Thromb. Vasc. Biol. 29, 929–935. doi: 10.1161/ATVBAHA.109.185546
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Harbauer, A. B., Zahedi, R. P., Sickmann, A., Pfanner, N., and Meisinger, C. (2014). The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab. 19, 357–372. doi: 10.1016/j.cmet.2014.01.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herrmann, J. M., and Riemer, J. (2012). Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J. Biol. Chem. 287, 4426–4433. doi: 10.1074/jbc.R111.270678
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, J., Yao, H., Gan, F., Tokarski, A., and Wang, Y. (2012). Interaction of OKL38 and p53 in regulating mitochondrial structure and function. PLoS ONE 7:e43362. doi: 10.1371/journal.pone.0043362
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iosefson, O., and Azem, A. (2010). Reconstitution of the mitochondrial Hsp70 (mortalin)-p53 interaction using purified proteins–identification of additional interacting regions. FEBS Lett. 584, 1080–1084. doi: 10.1016/j.febslet.2010.02.019
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karniely, S., and Pines, O. (2005). Single translation–dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 6, 420–425. doi: 10.1038/sj.embor.7400394
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karniely, S., Regev-Rudzki, N., and Pines, O. (2006). The presequence of fumarase is exposed to the cytosol during import into mitochondria. J. Mol. Biol. 358, 396–405. doi: 10.1016/j.jmb.2006.02.023
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaul, S. C., Deocaris, C. C., and Wadhwa, R. (2007). Three faces of mortalin: a housekeeper, guardian and killer. Exp. Gerontol. 42, 263–274. doi: 10.1016/j.exger.2006.10.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kisslov, I., Naamati, A., Shakarchy, N., and Pines, O. (2014). Dual-targeted proteins tend to be more evolutionarily conserved. Mol. Biol. Evol. 31, 2770–2779. doi: 10.1093/molbev/msu221
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloppel, C., Michels, C., Zimmer, J., Herrmann, J. M., and Riemer, J. (2010). In yeast redistribution of Sod1 to the mitochondrial intermembrane space provides protection against respiration derived oxidative stress. Biochem. Biophys. Res. Commun. 403, 114–119. doi: 10.1016/j.bbrc.2010.10.129
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloppel, C., Suzuki, Y., Kojer, K., Petrungaro, C., Longen, S., Fiedler, S., et al. (2011). Mia40-dependent oxidation of cysteines in domain I of Ccs1 controls its distribution between mitochondria and the cytosol. Mol. Biol. Cell 22, 3749–3757. doi: 10.1091/mbc.E11-04-0293
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kluck, R. M., Bossy-Wetzel, E., Green, D. R., and Newmeyer, D. D. (1997). The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science 275, 1132–1136. doi: 10.1126/science.275.5303.1132
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kornmann, B., Currie, E., Collins, S. R., Schuldiner, M., Nunnari, J., Weissman, J. S., et al. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477–481. doi: 10.1126/science.1175088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krause, K., Oetke, S., and Krupinska, K. (2012). Dual targeting and retrograde translocation: regulators of plant nuclear gene expression can be sequestered by plastids. Int. J. Mol. Sci. 13, 11085–11101. doi: 10.3390/ijms130911085
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Leary, S. C., Cobine, P. A., Nishimura, T., Verdijk, R. M., de Krijger, R., de Coo, R., et al. (2013). COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol. Biol. Cell 24, 683–691. doi: 10.1091/mbc.E12-09-0705
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, J., Kim, C. H., Simon, D. K., Aminova, L. R., Andreyev, A. Y., Kushnareva, Y. E., et al. (2005). Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J. Biol. Chem. 280, 40398–40401. doi: 10.1074/jbc.C500140200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, S., Spooner, R. A., Allen, S. C., Guise, C. P., Ladds, G., Schnoder, T., et al. (2010). Folding-competent and folding-defective forms of ricin A chain have different fates after retrotranslocation from the endoplasmic reticulum. Mol. Biol. Cell 21, 2543–2554. doi: 10.1091/mbc.E09-08-0743
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, J., and Lindquist, S. (2001). Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc. Natl. Acad. Sci. U.S.A. 98, 14955–14960. doi: 10.1073/pnas.011578098
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marchenko, N. D., Zaika, A., and Moll, U. M. (2000). Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275, 16202–16212. doi: 10.1074/jbc.275.21.16202
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McLelland, G. L., Soubannier, V., Chen, C. X., McBride, H. M., and Fon, E. A. (2014). Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33, 282–295. doi: 10.1002/embj.201385902
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mihara, M., Erster, S., Zaika, A., Petrenko, O., Chittenden, T., Pancoska, P., et al. (2003). p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11, 577–590. doi: 10.1016/S1097-2765(03)00050-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mitrpant, C., Fletcher, S., Iversen, P. L., and Wilton, S. D. (2009). By-passing the nonsense mutation in the 4 CV mouse model of muscular dystrophy by induced exon skipping. J. Gene Med. 11, 46–56. doi: 10.1002/jgm.1265
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Naamati, A., Regev-Rudzki, N., Galperin, S., Lill, R., and Pines, O. (2009). Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J. Biol. Chem. 284, 30200–30208. doi: 10.1074/jbc.M109.034694
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M., and Haynes, C. M. (2012). Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. doi: 10.1126/science.1223560
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nobrega, M. P., Bandeira, S. C., Beers, J., and Tzagoloff, A. (2002). Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J. Biol. Chem. 277, 40206–40211. doi: 10.1074/jbc.M207348200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pellegrino, M. W., Nargund, A. M., Kirienko, N. V., Gillis, R., Fiorese, C. J., and Haynes, C. M. (2014). Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. doi: 10.1038/nature13818. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pfeiffer, N. V., Dirndorfer, D., Lang, S., Resenberger, U. K., Restelli, L. M., Hemion, C., et al. (2013). Structural features within the nascent chain regulate alternative targeting of secretory proteins to mitochondria. EMBO J. 32, 1036–1051. doi: 10.1038/emboj.2013.46
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reddehase, S., Grumbt, B., Neupert, W., and Hell, K. (2009). The disulfide relay system of mitochondria is required for the biogenesis of mitochondrial Ccs1 and Sod1. J. Mol. Biol. 385, 331–338. doi: 10.1016/j.jmb.2008.10.088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Regev-Rudzki, N., Karniely, S., Ben-Haim, N. N., and Pines, O. (2005). Yeast aconitase in two locations and two metabolic pathways: seeing small amounts is believing. Mol. Biol. Cell 16, 4163–4171. doi: 10.1091/mbc.E04-11-1028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Regev-Rudzki, N., and Pines, O. (2007). Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. Bioessays 29, 772–782. doi: 10.1002/bies.20609
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Regev-Rudzki, N., Yogev, O., and Pines, O. (2008). The mitochondrial targeting sequence tilts the balance between mitochondrial and cytosolic dual localization. J. Cell Sci. 121, 2423–2431. doi: 10.1242/jcs.029207
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Robin, M. A., Anandatheerthavarada, H. K., Biswas, G., Sepuri, N. B., Gordon, D. M., Pain, D., et al. (2002). Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J. Biol. Chem. 277, 40583–40593. doi: 10.1074/jbc.M203292200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santos, J. H., Meyer, J. N., Skorvaga, M., Annab, L. A., and Van Houten, B. (2004). Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell 3, 399–411. doi: 10.1111/j.1474-9728.2004.00124.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sass, E., Blachinsky, E., Karniely, S., and Pines, O. (2001). Mitochondrial and cytosolic isoforms of yeast fumarase are derivatives of a single translation product and have identical amino termini. J. Biol. Chem. 276, 46111–46117. doi: 10.1074/jbc.M106061200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sass, E., Karniely, S., and Pines, O. (2003). Folding of fumarase during mitochondrial import determines its dual targeting in yeast. J. Biol. Chem. 278, 45109–45116. doi: 10.1074/jbc.M302344200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schelhaas, M., Malmstrom, J., Pelkmans, L., Haugstetter, J., Ellgaard, L., Grunewald, K., et al. (2007). Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131, 516–529. doi: 10.1016/j.cell.2007.09.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmidt, O., Harbauer, A. B., Rao, S., Eyrich, B., Zahedi, R. P., Stojanovski, D., et al. (2011). Regulation of mitochondrial protein import by cytosolic kinases. Cell 144, 227–239. doi: 10.1016/j.cell.2010.12.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sharma, N. K., Reyes, A., Green, P., Caron, M. J., Bonini, M. G., Gordon, D. M., et al. (2012). Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria. Nucleic Acids Res. 40, 712–725. doi: 10.1093/nar/gkr758
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Soltys, B. J., Falah, M., and Gupta, R. S. (1996). Identification of endoplasmic reticulum in the primitive eukaryote Giardia lamblia using cryoelectron microscopy and antibody to Bip. J. Cell Sci. 109(Pt 7), 1909–1917.
Strobel, G., Zollner, A., Angermayr, M., and Bandlow, W. (2002). Competition of spontaneous protein folding and mitochondrial import causes dual subcellular location of major adenylate kinase. Mol. Biol. Cell 13, 1439–1448. doi: 10.1091/mbc.01-08-0396
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sturtz, L. A., Diekert, K., Jensen, L. T., Lill, R., and Culotta, V. C. (2001). A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J. Biol. Chem. 276, 38084–38089. doi: 10.1074/jbc.M105296200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sugiura, A., McLelland, G. L., Fon, E. A., and McBride, H. M. (2014). A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33, 2142–2156. doi: 10.15252/embj.201488104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsai, B., Rodighiero, C., Lencer, W. I., and Rapoport, T. A. (2001). Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104, 937–948. doi: 10.1016/S0092-8674(01)00289-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vongsamphanh, R., Fortier, P. K., and Ramotar, D. (2001). Pir1p mediates translocation of the yeast Apn1p endonuclease into the mitochondria to maintain genomic stability. Mol. Cell Biol. 21, 1647–1655. doi: 10.1128/MCB.21.5.1647-1655.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wegrzyn, J., Potla, R., Chwae, Y. J., Sepuri, N. B., Zhang, Q., Koeck, T., et al. (2009). Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797. doi: 10.1126/science.1164551
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yedidia, Y., Horonchik, L., Tzaban, S., Yanai, A., and Taraboulos, A. (2001). Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. EMBO J. 20, 5383–5391. doi: 10.1093/emboj/20.19.5383
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yogev, O., Karniely, S., and Pines, O. (2007). Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J. Biol. Chem. 282, 29222–29229. doi: 10.1074/jbc.M704201200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yogev, O., Naamati, A., and Pines, O. (2011). Fumarase: a paradigm of dual targeting and dual localized functions. FEBS J. 278, 4230–4242. doi: 10.1111/j.1742-4658.2011.08359.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yogev, O., and Pines, O. (2011). Dual targeting of mitochondrial proteins: mechanism, regulation and function. Biochim. Biophys. Acta 1808, 1012–1020. doi: 10.1016/j.bbamem.2010.07.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yogev, O., Singer, E., Shaulian, E., Goldberg, M., Fox, T. D., and Pines, O. (2010). Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 8:e1000328. doi: 10.1371/journal.pbio.1000328
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhao, Y., Chaiswing, L., Velez, J. M., Batinic-Haberle, I., Colburn, N. H., Oberley, T. D., et al. (2005). p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer Res. 65, 3745–3750. doi: 10.1158/0008-5472.CAN-04-3835
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: echoforms, membranes, organelles, signal peptide, MTS (mitochondrial targeting sequence), chaperones, reverse translocation, retrotranslocation
Citation: Kalderon B and Pines O (2014) Protein folding as a driving force for dual protein targeting in eukaryotes. Front. Mol. Biosci. 1:23. doi: 10.3389/fmolb.2014.00023
Received: 30 September 2014; Accepted: 28 October 2014;
Published online: 25 November 2014.
Edited by:
Abdussalam Azem, Tel Aviv University, IsraelReviewed by:
Johannes Herrmann, University of Kaiserslautern, GermanyDoron Rapaport, University of Tuebingen, Germany
Copyright © 2014 Kalderon and Pines. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ophry Pines, Department of Microbiology and Molecular Genetics, Faculty of Medicine, Institute for Medical Research Israel-Canada, Hebrew University of Jerusalem, Jerusalem 91120, Israel e-mail: ophryp@ekmd.huji.ac.il
 Bella Kalderon
Bella Kalderon Ophry Pines
Ophry Pines