Transfer RNA and Origins of RNA Interference
- Department of Biology, Center for Computational and Integrative Biology, Rutgers University, Camden, NY, Uinted States
Introduction
Almost 30 years ago the first microRNA (miRNA) was detected (Lee et al., 1993), later put in mechanistical context by the discovery of the RNA interference (RNAi) and RNA-induced silencing complex (RISC). miRNA is the variable RISC element (guide) that recognizes messenger RNA (mRNA) targets in the RNAi process facilitated by Argonaute (Ago) proteins, the fixed key players in RISC. Alas, poetically sounding Argonautes were named not after the adventurers from Greek mythology but octopus-shaped leaves of an Arabidopsis thaliana mutant (Bohmert et al., 1998). Production of miRNAs is performed by a sophisticated orchestra of players with even groovier names, like Drosha, Pasha or Dicer. While roles of miRNA in post-transcriptional regulation have been established, its evolution has been typically described as expansion of new miRNA genes from the existing ones, not their first appearance (Berezikov, 2011). The emergence of RNAi has been attributed to its defense against pathogens, which involves other participants, e.g., double-stranded RNA binding proteins (dsRBPs). But details of such emergence are lacking.
This is a short opinion piece, not a full review, so I apologetically skip some of these players and hundreds of relevant miRNA/RNAi citations (and those for subsections below). The complexity of the RISC is bewildering. Intricate steps of miRNA production in the nucleus are followed by unequal strand loading to Ago, and finally resulting in cytoplasmic events, sometimes involving “slashing” of mRNA. All that (for simplicity, ignoring nuclear events upon Ago transport from cytoplasm) is based on weak and imperfect binding of short “seeds” (5′ 6-8 nucleotides in length in animals) to targets and on weak outcomes of slight tuning of target translation. How can such weak binding be a driving force for emergence and evolution of this complicated system and its parts, acting in concert in different cellular compartments?
Plant RNAi requires full-length small RNA hybridization, significantly limiting the number of target genes, yet the system appears even more complex (four Dicers, seven dsRBPs versus a single Dicer/couple of dsRBPs in humans). Further, the miRNA generation machineries are not entirely homologous, mature miRNAs are produced and modified in the plant nucleus, but these processes are divided between the nucleus and cytoplasm in animals. How would these different processes evolve in parallel with creating the elaborate and divergent Ago functionalities (four different Agos in humans, ten in A. thaliana, 26 in Caenorhabditis elegans), able to utilize these precisely cut miRNAs for mRNA regulation? This scenario would require a small RNA already acting together with some Ago prototypic protein. Compatible with this view, the last common ancestor of eukaryotes likely appeared after the RNAi functional principles (and some components, except for miRNA) had been invented (Cerutti and Casas-Mollano, 2006). And many prokaryotes do not have miRNAs but possess Ago protein homologs.
Transfer RNAs and Their Fragments
Enter transfer RNAs (tRNAs), fundamental elements in mRNA translation. Each tRNA is a link of informational (anticodon) and corresponding chemical (aminoacid) units, which jointly provide the basis of the central dogma. These are ancient molecules, potentially capable of performing primitive replication (Kühnlein et al., 2021) or aiding in it (Maizels and Weiner, 1994). While the genetic code requires <64 anticodons for translation, tRNA genes are very numerous, with several hundred copies in human genome. From a regulation standpoint, these numbers far exceed potential codon adaptation mechanisms. Perhaps fitting to this view, many tRNA genes have been described as inactive, raising questions about their actual role (Torres, 2019).
The early sequence/structure determination and clearly defined function of tRNAs established their place in molecular biology textbooks, hardly revisited until the arrival of tRNA fragments (tRFs). Hypotheses of further roles were occasionally entertained, based on additional function observed, e.g., in viral replication, etc. (see also a review (Avcilar-Kucukgoze and Kashina, 2020) in this article collection for more details on these roles). However, an avalanche of data from small RNA sequencing experiments have challenged such perceptions, revealing numerous and ubiquitous tRFs in a multitude of sequenced samples. These fragments, detected in datasets produced to study miRNA, have been mostly dismissed as noise. It took >10 years to get acceptance even after detailed studies (Cole et al., 2009; Lee et al., 2009; Haussecker et al., 2010; Ivanov et al., 2011; Gebetsberger et al., 2012), even though tRNA breakage products were detected in urine of cancer patients much earlier (Speer et al., 1979). The finding of Ago proteins loaded with tRFs, in addition to their cargo of miRNAs, have prompted speculations about similar functionality of tRFs and search for their targets (Table 1).
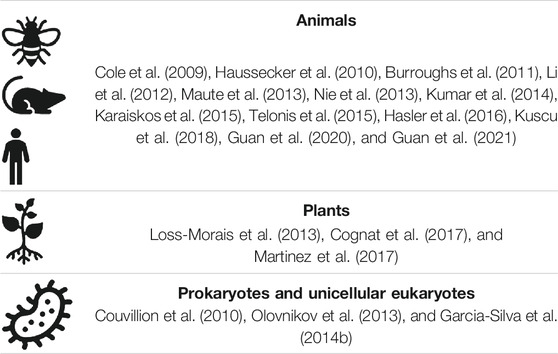
TABLE 1. Representative studies on detection and analysis of Argonaute-loaded tRFs in different organisms.
By now, tRFs have been observed across all domains of life. There are multiple databases, containing different tRFs ever identified. For example, >28,000 tRFs are listed in MINTbase (Pliatsika et al., 2016), although the exact roles of most tRFs remain largely unknown, and only a dozen have been assigned a somewhat specific function. Yet, during the short period since their acceptance as bona fide cellular products and not sequencing noise, tRFs have provided several evolutionary surprises, exemplifying their regulatory effects not only within an organism but across time and space barriers.
Time: tRFs have been described as leading to metabolic disorders in the progeny of male mice on high protein or high fat diet (Chen et al., 2016; Sharma et al., 2016). This is an example of a non-Darwinian short-term trait propagation (to avoid the term “inheritance”) via incorporating tRFs into sperm. A bit like sending into the nearest future an information packet with father’s current environmental conditions. Did such signaling originate to get progeny better prepared for abundant food?
Space: a Pseudomonas aeruginosa vesicle-delivered tRF has been reported to affect the host immune response (Koeppen et al., 2016), while Trypanosoma cruzi tRFs—as contributing to susceptibility to infection of mammalian cells (Garcia-Silva et al., 2014a). Crossing species and kingdom barriers expands the idea of small RNA signals, e.g., the loading of tRFs into extracellular vesicles for likely cell-to-cell communication, such as T-cell de/activation (Chiou et al., 2018).
If regulation based on the weak binding of short RNAs to their targets is an ancient mechanism, we should see many more examples of intra-and interorganismal signals, exploiting this common principle. Notably, similar effects were reported in the classical RNAi papers on C. elegans. Progeny produced from the eggs of nematodes injected with dsRNAs showed interference phenotypes (Fire et al., 1998), while long dsRNA expressed in bacteria induced repression of genes with complementary sequences in worms fed with such bacteria (Timmons and Fire, 1998). Could these roles originate from tRFs (which apparently continue performing them, too)? If the tRF-driven regulation had led to the emergence of Ago functionality followed by the invention of RNAi, then one could imagine how divergent parts of plant and animal miRNA production arose based on this early mechanism.
Turning a Weak Interaction Into a Strong Selective Force
The classical role of tRNAs co-exists with that of tRFs. What factors determine the fate of a given tRNA? And how does this relate to the origin of miRNA/RNAi?
A look at the evolutionary distant Archaea reveals some unusual but frequent tRNAs, potentially very relevant for dissecting the tRF function. These carry one or more introns, with the genomic order of tRNA parts occasionally permuted (Soma et al., 2007). Strikingly, units corresponding to tRNA exons sometimes also form 2 to 3 separate genes, as in the hyperthermophilic Archaea Nanoarchaeum equitans (Randau et al., 2005) or Caldivirga maquilingensis (Fujishima et al., 2009).
In protein-coding genes, exon borders are correlated with domain borders (Liu and Grigoriev, 2004), indicating how introns separate functional and evolutionarily mobile protein parts (Liu et al., 2005). This principle may also exist in tRNAs, although the introns were hypothesized to be mobile there (Fujishima et al., 2010). But what about the multi-gene tRNAs above? These unusual tRNAs may indicate separate functional roles of their parts, in addition to being involved in forming a whole tRNA from pieces. The split genes have been discussed in the context of tRNA origin (Di Giulio, 2008; Kanai, 2015). However, they also may represent traces/features of early tRFs. It would be interesting to check whether such exon units correspond to functional tRFs in these archaeal species. Notably, a map of intron insertion sites (Sugahara et al., 2009) shows they occur nearly everywhere between positions 12 and 60 on the canonical cloverleaf structure (encompassing the D- and T-loops). The majority of tRFs stored in the databases have ends in these positions—does this reflect sequencing noise, diversity of tRFs or potential ancient links with tRF production?
Such arrangements of tRNA exons quite possibly indicate additional functions of these exons, especially when they separate so much as to form individual sub-tRNA genes. And not just of the exons but of broken/digested tRNA pieces. Or, at least, this may be how all this had started, with random fragments present in the right place at the right time.
Role of tRNAs in translation makes their fragments ideal candidates for regulating this fundamental process. Proximity (essentially a macro-term for elevated concentration) is a very significant factor in evolution. For example, several ribosomes translating the same mRNA in bacteria produce identical proteins. These proteins are always formed close to each other, hence any mutation that carried even a small fitness advantage from enhancing their homotypic interactions would likely be selected for (compared to proteins separated in space and time). This had possibly led to the formation of homodimers and higher-order homo-multimers seen in a very large number of modern proteins (Grigoriev, 2001).
Similarly, a constant presence of fragments of tRNA right next to translated mRNAs could turn any slight fitness advantage from interactions between these molecules into a strong evolutionary force, despite interactions themselves being relatively weak. A similar force could then further select RNA-binding proteins for Ago-like functionality of enhancing the tRF-mRNA recognition to form a first prototypic RISC. The theoretical multitude of possible primordial tRF-target interactions suggests that many of them probably had not followed the rules that would later involve Ago. And many still do not: LeuCAG tRF seems to unwind the helices of a target mRNA to enable its translation (Kim et al., 2017) in Ago-less manner. Is this process catalyzed by another protein? Possibly. The very existence of these interactions could give rise to positive selection of other proteins for such catalysts (as with Ago, enabling more targeted tRF binding).
After a rudimentary RISC with this Ago-like functionality on tRFs was invented, a simple change would allow it to accept other small RNAs. After all, current Agos are very promiscuous RNA binders, “inviting” other short RNAs. While tRNA have constraints imposed on the sequence by their structure and essential function, candidate miRNAs may have seemingly any sequence, as long as it produces a recognizable hairpin for Drosha, Dicer & Co.
Ubiquitous small hairpins in the transcribed parts of a genome (from existing introns or appearing de novo), appropriately cut to provide a short RNA, could greatly enrich the repertoire of regulatory guides (and targets they regulate). That would drive the birth to thousands of miRNAs (and notably, some of miRNAs have recently been recognized as tRFs). Given the reports on likely roles of tRFs in regulating some of the more recent inventions in organism capabilities, such as neuronal or immunological, tRFs may be outnumbered by relatively unconstrained miRNAs but unlikely to be replaced.
Finally, I briefly revisit the emergence of RNAi for anti-pathogen defense. Although an attractive explanation for the RNAi existence, such defense seems unlikely to appear on its own, without a slashing agent, like Ago. How would a short RNA matching an arbitrary pathogen sequence “defend” against it without this protein? Perhaps by interfering with pathogen’s translation? This would again suggest that tRNA breakage products, abundant near translating ribosomes, might have been potential drivers of such interference. Then the defensive functionality of RNAi and its ability to regulate own mRNAs could have emerged almost simultaneously, following the same principles, and there is no need to develop defense as a predecessor of regulation.
In summary, while divergent modes of action seem to exist for tRFs, the outlined evolutionary path for an emergence of Argonaute-mediated gene regulation appears plausible.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
The work in AG’s lab is supported by NIH R15CA220059 and NSF MCB-2027611 to AG.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
I thank Lingyu Guan for discussions of these topics.
References
Avcilar-Kucukgoze, I., and Kashina, A. (2020). Hijacking tRNAs from Translation: Regulatory Functions of tRNAs in Mammalian Cell Physiology. Front. Mol. Biosci. 7 (388). doi:10.3389/fmolb.2020.610617
Berezikov, E. (2011). Evolution of microRNA Diversity and Regulation in Animals. Nat. Rev. Genet. 12 (12), 846–860. doi:10.1038/nrg3079
Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 Defines a Novel Locus of Arabidopsis Controlling Leaf Development. EMBO J. 17 (1), 170–180. doi:10.1093/emboj/17.1.170
Burroughs, A. M., Ando, Y., de Hoon, M. L., Tomaru, Y., Suzuki, H., Hayashizaki, Y., et al. (2011). Deep-sequencing of Human Argonaute-Associated Small RNAs Provides Insight into miRNA Sorting and Reveals Argonaute Association with RNA Fragments of Diverse Origin. RNA Biol. 8 (1), 158–177. doi:10.4161/rna.8.1.14300
Cerutti, H., and Casas-Mollano, J. A. (2006). On the Origin and Functions of RNA-Mediated Silencing: from Protists to Man. Curr. Genet. 50 (2), 81–99. doi:10.1007/s00294-006-0078-x
Chen, Q., Yan, M., Cao, Z., Li, X., Zhang, Y., Shi, J., et al. (2016). Sperm tsRNAs Contribute to Intergenerational Inheritance of an Acquired Metabolic Disorder. Science 351 (6271), 397–400. doi:10.1126/science.aad7977
Chiou, N.-T., Kageyama, R., and Ansel, K. M. (2018). Selective export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cel Rep. 25 (12), 3356–3370. doi:10.1016/j.celrep.2018.11.073
Cognat, V., Morelle, G., Megel, C., Lalande, S., Molinier, J., Vincent, T., et al. (2017). The Nuclear and Organellar tRNA-Derived RNA Fragment Population in Arabidopsis thaliana Is Highly Dynamic. Nucleic Acids Res. 45 (6), 3460–3472. doi:10.1093/nar/gkw1122
Cole, C., Sobala, A., Lu, C., Thatcher, S. R., Bowman, A., Brown, J. W. S., et al. (2009). Filtering of Deep Sequencing Data Reveals the Existence of Abundant Dicer-dependent Small RNAs Derived from tRNAs. RNA 15 (12), 2147–2160. doi:10.1261/rna.1738409
Couvillion, M. T., Sachidanandam, R., and Collins, K. (2010). A Growth-Essential Tetrahymena Piwi Protein Carries tRNA Fragment Cargo. Genes Develop. 24 (24), 2742–2747. doi:10.1101/gad.1996210
Di Giulio, M. (2008). Permuted tRNA Genes of Cyanidioschyzon Merolae, the Origin of the tRNA Molecule and the Root of the Eukarya Domain. J. Theor. Biol. 253 (3), 587–592. doi:10.1016/j.jtbi.2008.04.022
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis elegans. nature 391 (6669), 806–811. doi:10.1038/35888
Fujishima, K., Sugahara, J., Kikuta, K., Hirano, R., Sato, A., Tomita, M., et al. (2009). Tri-split tRNA Is a Transfer RNA Made from 3 Transcripts that Provides Insight into the Evolution of Fragmented tRNAs in Archaea. Pnas 106 (8), 2683–2687. doi:10.1073/pnas.0808246106
Fujishima, K., Sugahara, J., Tomita, M., and Kanai, A. (2010). Large-scale tRNA Intron Transposition in the Archaeal Order Thermoproteales Represents a Novel Mechanism of Intron Gain. Mol. Biol. Evol. 27 (10), 2233–2243. doi:10.1093/molbev/msq111
Garcia-Silva, M. R., Cura das Neves, R. F., Cabrera-Cabrera, F., Sanguinetti, J., Medeiros, L. C., Robello, C., et al. (2014a). Extracellular Vesicles Shed by Trypanosoma Cruzi Are Linked to Small RNA Pathways, Life Cycle Regulation, and Susceptibility to Infection of Mammalian Cells. Parasitol. Res. 113 (1), 285–304. doi:10.1007/s00436-013-3655-1
Garcia-Silva, M. R., Sanguinetti, J., Cabrera-Cabrera, F., Franzén, O., and Cayota, A. (2014b). A Particular Set of Small Non-coding RNAs Is Bound to the Distinctive Argonaute Protein of Trypanosoma Cruzi: Insights from RNA-Interference Deficient Organisms. Gene 538 (2), 379–384. doi:10.1016/j.gene.2014.01.023
Gebetsberger, J., Zywicki, M., Kunzi, A., and Polacek, N. (2012). tRNA-Derived Fragments Target the Ribosome and Function as Regulatory Non-coding RNA in Haloferax Volcanii. Archaea-an Int. Microbiol. J. 2012, 260909. doi:10.1155/2012/260909
Grigoriev, A. (2001). A Relationship Between Gene Expression and Protein Interactions on the Proteome Scale: Analysis of the Bacteriophage T7 and the Yeast Saccharomyces cerevisiae. Nucleic Acids Res. 29 (17), 3513–3519. doi:10.1093/nar/29.17.3513
Guan, L., Karaiskos, S., and Grigoriev, A. (2020). Inferring Targeting Modes of Argonaute-Loaded tRNA Fragments. RNA Biol. 17 (8), 1070–1080. doi:10.1080/15476286.2019.1676633
Guan, L., Lam, V., and Grigoriev, A. (2021). Large-Scale Computational Discovery of Binding Motifs in tRNA Fragments. Front. Mol. Biosci. 8, 499.
Hasler, D., Lehmann, G., Murakawa, Y., Klironomos, F., Jakob, L., Grässer, F. A., et al. (2016). The Lupus Autoantigen La Prevents Mis-Channeling of tRNA Fragments into the Human MicroRNA Pathway. Mol. Cel 63 (1), 110–124. doi:10.1016/j.molcel.2016.05.026
Haussecker, D., Huang, Y., Lau, A., Parameswaran, P., Fire, A. Z., and Kay, M. A. (2010). Human tRNA-Derived Small RNAs in the Global Regulation of RNA Silencing. RNA 16 (4), 673–695. doi:10.1261/rna.2000810
Ivanov, P., Villen, J., Gygi, S. P., Anderson, P., and Anderson, P. (2011). Angiogenin-induced tRNA Fragments Inhibit Translation Initiation. Mol. Cel 43 (4), 613–623. doi:10.1016/j.molcel.2011.06.022
Kanai, A. (2015). Disrupted tRNA Genes and tRNA Fragments: A Perspective on tRNA Gene Evolution. Life 5 (1), 321–331. doi:10.3390/life5010321
Karaiskos, S., Naqvi, A. S., Swanson, K. E., and Grigoriev, A. (2015). Age-driven Modulation of tRNA-Derived Fragments in Drosophila and Their Potential Targets. Biol. Direct 10 (1), 1. doi:10.1186/s13062-015-0081-6
Kim, H. K., Fuchs, G., Wang, S., Wei, W., Zhang, Y., Park, H., et al. (2017). A Transfer-RNA-Derived Small RNA Regulates Ribosome Biogenesis. Nature 552 (7683), 57–62. doi:10.1038/nature25005
Koeppen, K., Hampton, T. H., Jarek, M., Scharfe, M., Gerber, S. A., Mielcarz, D. W., et al. (2016). A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 12 (6), e1005672. doi:10.1371/journal.ppat.1005672
Kühnlein, A., Lanzmich, S. A., and Braun, D. (2021). tRNA Sequences Can Assemble into a Replicator. Elife 10, e63431. doi:10.7554/elife.63431
Kumar, P., Anaya, J., Mudunuri, S. B., and Dutta, A. (2014). Meta-analysis of tRNA Derived RNA Fragments Reveals that They Are Evolutionarily Conserved and Associate with AGO Proteins to Recognize Specific RNA Targets. Bmc Biol. 12 (1), 78. doi:10.1186/s12915-014-0078-0
Kuscu, C., Kumar, P., Kiran, M., Su, Z., Malik, A., and Dutta, A. (2018). tRNA Fragments (tRFs) Guide Ago to Regulate Gene Expression post-transcriptionally in a Dicer-independent Manner. RNA 24 (8), 1093–1105. doi:10.1261/rna.066126.118
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 75 (5), 843–854. doi:10.1016/0092-8674(93)90529-y
Lee, Y. S., Shibata, Y., Malhotra, A., and Dutta, A. (2009). A Novel Class of Small RNAs: tRNA-Derived RNA Fragments (tRFs). Genes Dev. 23 (22), 2639–2649. doi:10.1101/gad.1837609
Li, Z., Ender, C., Meister, G., Moore, P. S., Chang, Y., and John, B. (2012). Extensive Terminal and Asymmetric Processing of Small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40 (14), 6787–6799. doi:10.1093/nar/gks307
Liu, M., and Grigoriev, A. (2004). Protein Domains Correlate Strongly with Exons in Multiple Eukaryotic Genomes - Evidence of Exon Shuffling?. Trends Genet. 20 (9), 399–403. doi:10.1016/j.tig.2004.06.013
Liu, M., Walch, H., Wu, S., and Grigoriev, A. (2005). Significant Expansion of Exon-Bordering Protein Domains during Animal Proteome Evolution. Nucleic Acids Res. 33 (1), 95–105. doi:10.1093/nar/gki152
Loss-Morais, G., Waterhouse, P. M., and Margis, R. (2013). Description of Plant tRNA-Derived RNA Fragments (tRFs) Associated with Argonaute and Identification of Their Putative Targets. Biol. direct 8 (1), 1–5. doi:10.1186/1745-6150-8-6
Maizels, N., and Weiner, A. M. (1994). Phylogeny from Function: Evidence from the Molecular Fossil Record that tRNA Originated in Replication, Not Translation. Proc. Natl. Acad. Sci. 91 (15), 6729–6734. doi:10.1073/pnas.91.15.6729
Martinez, G., Choudury, S. G., and Slotkin, R. K. (2017). tRNA-derived Small RNAs Target Transposable Element Transcripts. Nucleic Acids Res. 45 (9), 5142–5152. doi:10.1093/nar/gkx103
Maute, R. L., Schneider, C., Sumazin, P., Holmes, A., Califano, A., Basso, K., et al. (2013). tRNA-derived microRNA Modulates Proliferation and the DNA Damage Response and Is Down-Regulated in B Cell Lymphoma. Proc. Natl. Acad. Sci. USA 110 (4), 1404–1409. doi:10.1073/pnas.1206761110
Nie, Z., Zhou, F., Li, D., Lv, Z., Chen, J., Liu, Y., et al. (2013). RIP-seq of BmAgo2-Associated Small RNAs Reveal Various Types of Small Non-coding RNAs in the Silkworm, Bombyx mori. BMC genomics 14 (1), 1–15. doi:10.1186/1471-2164-14-661
Olovnikov, I., Chan, K., Sachidanandam, R., Newman, D. K., and Aravin, A. A. (2013). Bacterial Argonaute Samples the Transcriptome to Identify Foreign DNA. Mol. Cel 51 (5), 594–605. doi:10.1016/j.molcel.2013.08.014
Pliatsika, V., Loher, P., Telonis, A. G., and Rigoutsos, I. (2016). MINTbase: a Framework for the Interactive Exploration of Mitochondrial and Nuclear tRNA Fragments. Bioinformatics 32 (16), 2481–2489. doi:10.1093/bioinformatics/btw194
Randau, L., Münch, R., Hohn, M. J., Jahn, D., and Söll, D. (2005). Nanoarchaeum Equitans Creates Functional tRNAs from Separate Genes for Their 5′- and 3′-halves. Nature 433 (7025), 537–541. doi:10.1038/nature03233
Sharma, U., Conine, C. C., Shea, J. M., Boskovic, A., Derr, A. G., Bing, X. Y., et al. (2016). Biogenesis and Function of tRNA Fragments during Sperm Maturation and Fertilization in Mammals. Science 351 (6271), 391–396. doi:10.1126/science.aad6780
Soma, A., Onodera, A., Sugahara, J., Kanai, A., Yachie, N., Tomita, M., et al. (2007). Permuted tRNA Genes Expressed via a Circular RNA Intermediate in Cyanidioschyzon Merolae. Science 318 (5849), 450–453. doi:10.1126/science.1145718
Speer, J., Gehrke, C. W., Kuo, K. C., Waalkes, T. P., and Borek, E. (1979). tRNA Breakdown Products as Markers for Cancer. Cancer 44 (6), 2120–2123. doi:10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6
Sugahara, J., Fujishima, K., Morita, K., Tomita, M., and Kanai, A. (2009). Disrupted tRNA Gene Diversity and Possible Evolutionary Scenarios. J. Mol. Evol. 69 (5), 497–504. doi:10.1007/s00239-009-9294-6
Telonis, A. G., Loher, P., Honda, S., Jing, Y., Palazzo, J., Kirino, Y., et al. (2015). Dissecting tRNA-Derived Fragment Complexities Using Personalized Transcriptomes Reveals Novel Fragment Classes and Unexpected Dependencies. Oncotarget 6 (28), 24797–24822. doi:10.18632/oncotarget.4695
Timmons, L., and Fire, A. (1998). Specific Interference by Ingested dsRNA. Nature 395 (6705), 854. doi:10.1038/27579
Keywords: tRNA, tRNA fragments, argonaute, microRNA, RISC, RNAi
Citation: Grigoriev A (2021) Transfer RNA and Origins of RNA Interference. Front. Mol. Biosci. 8:708984. doi: 10.3389/fmolb.2021.708984
Received: 13 May 2021; Accepted: 08 July 2021;
Published: 23 July 2021.
Edited by:
Lluís Ribas De Pouplana, Institute for Research in Biomedicine, SpainReviewed by:
Clement Carre, Institut de biologie Paris Seine, FranceCopyright © 2021 Grigoriev. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrey Grigoriev, andrey.grigoriev@rutgers.edu
 Andrey Grigoriev
Andrey Grigoriev