Bacterial extracellular vesicles: towards realistic models for bacterial membranes in molecular interaction studies by surface plasmon resonance
- 1Drug Transport and Delivery Research Group, Department of Pharmacy, Faculty of Health Sciences, UiT the Arctic University of Norway, Tromsø, Norway
- 2Chemical Synthesis and Analysis Research Group, Department of Chemistry, Faculty of Natural Sciences and Technology, UiT the Arctic University of Norway, Tromsø, Norway
- 3Natural Products and Medicinal Chemistry Research Group, Department of Pharmacy, Faculty of Health Sciences, UiT the Arctic University of Norway, Tromsø, Norway
- 4Pediatric Research Group, Department of Clinical Medicine, Faculty of Health Sciences, UiT the Arctic University of Norway, Tromsø, Norway
- 5Research Group for Host Microbe Interactions, Department of Medical Biology, Faculty of Health Sciences, UiT the Arctic University of Norway, Tromsø, Norway
One way to mitigate the ongoing antimicrobial resistance crisis is to discover and develop new classes of antibiotics. As all antibiotics at some point need to either cross or just interact with the bacterial membrane, there is a need for representative models of bacterial membranes and efficient methods to characterize the interactions with novel molecules -both to generate new knowledge and to screen compound libraries. Since the bacterial cell envelope is a complex assembly of lipids, lipopolysaccharides, membrane proteins and other components, constructing relevant synthetic liposome-based models of the membrane is both difficult and expensive. We here propose to let the bacteria do the hard work for us. Bacterial extracellular vesicles (bEVs) are naturally secreted by Gram-negative and Gram-positive bacteria, playing a role in communication between bacteria, as virulence factors, molecular transport or being a part of the antimicrobial resistance mechanism. bEVs consist of the bacterial outer membrane and thus inherit many components and properties of the native outer cell envelope. In this work, we have isolated and characterized bEVs from one Escherichia coli mutant and three clinical strains of the ESKAPE pathogens Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. The bEVs were shown to be representative models for the bacterial membrane in terms of lipid composition with speciesstrain specific variations. The bEVs were further used to probe the interactions between bEV and antimicrobial peptides (AMPs) as model compounds by Surface Plasmon Resonance (SPR) and provide proof-of-principle that bEVs can be used as an easily accessible and highly realistic model for the bacterial surface in interaction studies. This further enables direct monitoring of the effect induced by antibiotics, or the response to host-pathogen interactions.
Introduction
The development of antimicrobial resistance (AMR) is a serious threat to public health and the main obstacle to successful treatment of infectious diseases. AMR caused 4.95 million deaths globally already in 2019 and the numbers are growing (WHO, 2014; Antimicrobial Resistance Collaborators, 2022). The antimicrobial resistant bacteria responsible for more than 15% of hospital acquired infections are classified as high priority pathogens by the World Health Organization (WHO). The Gram-negative ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) strains K. pneumoniae, A. baumannii, and P. aeruginosa are on top of the list and are thus representing the strains of critical priority (WHO, 2017; Mulani et al., 2019).
To fight the ongoing AMR crisis effort is put into the discovery and optimization of new antimicrobial compounds. As most aspiring antibiotics at some point need to either cross or interact with the bacterial membrane, there is a need for cost-effective native-like models and efficient methods to characterize such interactions (Delcour, 2009).
The cell envelope of Gram-negative bacteria is a complex structure consisting of inner and outer membranes. The inner membrane is mainly composed of phospholipids, while the outer membrane is asymmetric, mainly containing phospholipids (PLs) at the inner leaflet and lipopolysaccharides (LPS) at the outer leaflet. Lipid A is the major structural unit of LPS resulting in a negative net charge. During infection, or in response to antibiotic treatment, bacteria can modify lipid A to alter the surface charge which could lead to reduced binding of cationic antimicrobials such as colistin (Olaitan et al., 2014; Llobet et al., 2015; Jeannot et al., 2017; Jasim et al., 2018; Hobby et al., 2019). The periplasmic space between the inner and outer membrane is enriched in peptidoglycans. The cell envelope is also the site for various proteins such as ion pumps and channels, receptors and membrane bound enzymes (Beveridge, 1999; Silhavy et al., 2010; Roier et al., 2016). Simple synthetic lipids in the form of liposomes, supported lipid bilayers, or nanodiscs are often utilized to investigate new antimicrobial compounds. However, they lack the intricate complexity observed in the native cell envelope (Denisov et al., 2004; Sezgin and Schwille, 2012; Akbarzadeh et al., 2013; Scheidelaar et al., 2015; Denisov et al., 2019; Jackman and Cho, 2020; Bailey-Hytholt et al., 2021). Incorporation of all native outer membrane components into synthetic model liposomes is a possible but laborious and expensive process (Prenner et al., 1999; Pirc and Ulrih, 2015; Elderdfi and Sikorski, 2018; Paulowski et al., 2020; Jakubec et al., 2021). There is thus a need for affordable but relevant model systems that can accurately mimic the bacterial membrane.
Bacterial extracellular vesicles (bEVs) are formed mainly from outer membranes and maintain similar composition containing PLs, LPS and OM-proteins, and thus, can represent naturally assembled nano-models of the outer membranes (Haurat et al., 2015; Roier et al., 2016). For bacteria, the bEVs play an extensive role in physiological and pathogenic processes in, for example, molecular transport, biofilm formation, antibiotic resistance and cell-cell communication. As a part of the bacterial defense mechanism, bEVs serve as a decoy, absorbing and neutralizing membrane active agents, including bacteriophages. bEVs can also be a central part of a disposal mechanism by which bacteria can get rid of damaged/affected membranes (Manning and Kuehn, 2011; Balhuizen et al., 2021). EVs are ideal candidates to serve as the outer membrane model for the bacterial strain it is produced from.
The aim of this work was to explore the applicability of native bEVs as bacterial membrane models in interaction studies. To achieve this, bEVs secreted by overproducing E. coli ΔtolA mutant (Reimer et al., 2021), LPS deficient E. coli NR698 strain (Ruiz et al., 2005; Krüger et al., 2019) and three clinical multi-resistant strains of critical importance A. baumannii K47-42, P. aeruginosa K34-7 and K. pneumoniae K47-25 (Paulsen et al., 2021) were isolated and characterized. The clinical strains were previously shown to be beta-lactam resistant caused by beta-lactamase production (Paulsen et al., 2021). The K. pneumoniae and A. baumannii strains are also colistin resistant, being able to modify lipid A in the outer membrane (Paulsen et al., 2021). The kinetic rates of interaction between the bEVs and membrane active compounds were measured by surface plasmon resonance (SPR). Antimicrobial peptides (AMPs) are receiving increasing attention in the fight against AMR, as their different mode of actions lead to destabilization of bacterial membranes and bacterial death (Czaplewski et al., 2016; Mahlapuu et al., 2016; Ageitos et al., 2017; Kumar et al., 2018). A test panel of five cyclic AMPs with various distributions of charged and hydrophobic amino acids, known to interact with bacterial membranes was used (Junkes et al., 2008; Bagheri, 2010; Gopal et al., 2012; Finger et al., 2015; Rainsford et al., 2022; Jakubec et al., 2023). This set of peptides was originally designed to study how the peptide charge distribution affects membrane interactions and was used by our lab to explore kinetics of binding towards synthetic liposomes, nanodiscs and to a series of E. coli strains (Rainsford et al., 2022; Jakubec et al., 2023).
Results and discussions
In this work we aimed to apply bEVs as a relevant model of the native bacterial membrane for interaction studies by SPR. Antimicrobial peptides were used as test compounds to establish proof of principle. An E. coli strain with a deletion of the tolA gene, which results in higher bEV yields (Reimer et al., 2021), was used as a starting point to establish the production- and isolation protocols. The LPS deficient E. coli strain NR698 (Ruiz et al., 2005; Krüger et al., 2019) as well as clinical multidrug resistant strains of A. baumannii, P. aeruginosa, and K. pneumoniae were further selected. The latter belong to the group of critical priority ESKAPE pathogens listed by WHO (WHO, 2017). First, bEVs were characterized in terms of morphology and lipid composition and compared to bacterial lipid isolates from the same strains. Then bEVs were directly applied to SPR measurements of interactions with the selected test compounds. In addition to the bEVs, liposomes were prepared from lipids isolated from E. coli ΔtolA and used as control vesicles in this study.
Characterization of native bEVs
Morphology and zeta potential of isolated bEVs
All strains, except the LPS deficient E. coli NR698, produced bEVs in sufficient amounts and quality. Isolated bEVs were visualized by transmission electron microscopy (TEM) to assess morphology and sample quality. Figure 1A–D shows micrographs of representative samples of bEVs from all strains, except E. coli NR698. The samples appear to be free from contamination with only minor traces of cell debris, confirming the successful isolation of the bEVs. All vesicles observed were spherical with a visible double layer. The bEVs from P. aeruginosa strain seemed aggregated (Figure 1D), a feature commonly observed during the study of this strain. For the LPS deficient E. coli NR698 strain, although some bEVs were present, the sample was dominated by deformed bEVs (Supplementary Figure S1). The strain has mutations causing deficiency of LPS in the outer membrane, possibly resulting in a higher content of phospholipids (Ruiz et al., 2005; Krüger et al., 2019). This sample represents one limitation of bEVs application and experimental design regarding genetic alterations affecting the properties of the outer membrane. Due to the inadequate quality of the isolated bEVs, this strain was not further pursued.
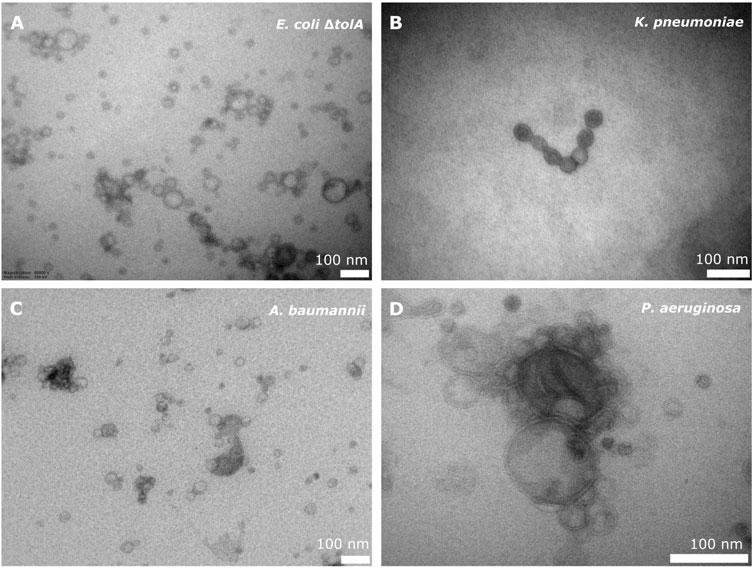
FIGURE 1. TEM micrograph of bEVs secreted by E. coli ΔtolA (A), K. pneumoniae (B), A. baumannii (C) (50,000 x magnification), and P. aeruginosa (D) (80,000 x magnification). 100 nm scalebar is presented at the bottom right corner of each micrograph.
The average vesicle diameter was determined to be in the range from 194.8 ± 1.0 nm for K. pneumoniae, 175.0 ± 1.3 nm for A. baumannii and 143.3 ± 1.1 nm for P. aeruginosa, to 82.7 ± 1.3 nm for E. coli ΔtolA (Figure 2A). When comparing the zeta potentials of bEVs and bacterial cells (Figure 2B; Supplementary Table S1), the values are found to be significantly lower for bEVs in all the cases except P. aeruginosa, where no significant difference was observed (t-test, p > 0.05). The lowest zeta potential of −14.4 ± 0.6 mV was measured for bEVs from A. baumannii. bEVs from K. pneumoniae and E. coli ΔtolA showed similar values for zeta potentials of −9.9 ± 0.3 mV and −9.4 ± 0.8 mV, respectively. The zeta potential of bEVs from P. aeruginosa was measured to −3.90 ± 0.6 mV, which suggests a less stable dispersion due to less repulsion between the vesicles, and could, thus, explain the aggregation observed (Figure 1D). Liposomes prepared from E. coli ΔtolA lipid isolate were used as a control in our studies and were also characterized according to size distribution and zeta potential. They showed an average size of 139.9 ± 1.6 nm and had a significantly lower zeta potential, −17.8 ± 0.5 mV, compared to both bEVs produced by the same bacteria, and the whole bacteria itself. This is an indication that the bEVs better represent the bacterial surface than liposomes prepared from lipid isolates. The size of the bEVs isolated from E. coli ΔtolA was reanalyzed after 1 month of storage at 4°C. No significant change in size was observed, suggesting that the bEVs were stable for at least 1 month. All samples were analyzed in triplicates.
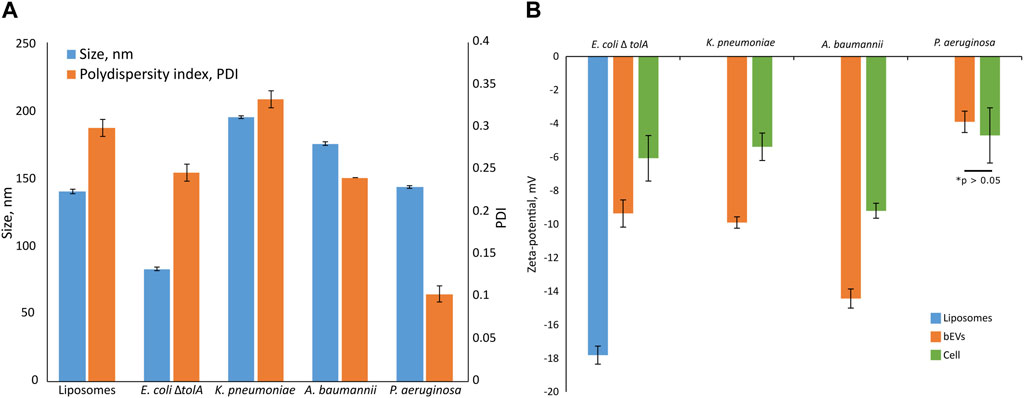
FIGURE 2. Summary of size and polydispersity index (PDI) (A) and zeta-potential measurements (B) for isolated bEVs in comparison to zeta-potential of the bacterial cells. The sample of liposomes prepared from E. coli ΔtolA lipid isolate is also included. Samples were held in 10 mM TRIS buffer (100 mM NaCl and 5 mM KCl, pH 8.0) at 25°C. All samples were analyzed in triplicates.
Lipid composition of native bacterial membranes
The compositions of the bacteria lipid isolates were quantified by 31P NMR, and the qualitative pattern was acquired by MS. The corresponding pattern was subsequently acquired for lipid isolates of the corresponding secreted bEVs from the same strains.
For gram negative bacteria in general, the bacterial membrane is expected to be dominated by Phosphatidylethanolamine (PE) supplemented by the negative phospholipids Phosphatidylglycerol (PG) and cardiolipin (CL). The results from the 31P NMR studies on the whole bacterial lipid isolates are shown in Figure 3A and confirmed the expected general pattern that PE was the dominant phospholipid, ranging from 50%–52% (for A. baumanii and K. pneumoniae) up to 64%–68% (for P. aeruginosa and E. coli ΔtolA) of the total phospholipid content. The CL content was found to be 7% for K. pneumoniae, 10%–11% for P. aeruginosa and E. coli ΔtolA and up to 21% for A. baumannii. Lyso-phosphatidylethanolamine (LPE) was determined to be at 24%, 17% and 13% levels for K. pneumoniae, A. baumannii and E. coli ΔtolA respectively, while no LPE was detected in P. aeruginosa. Phosphatidylcholine (PC) contributed to 3% and 9% in A. baumannii and P. aeruginosa respectively, while it was not detected in E. coli ΔtolA and K. pneumoniae. PG contributed to 9% of the total phospholipids in E. coli ΔtolA and A. baumannii, and up to 17% for K. pneumoniae and P. aeruginosa. In conclusion, the expected pattern for gram negative bacteria was confirmed, where PE lipid was the major lipid of the studied bacterial membranes, together with a handful of other phospholipids in smaller quantities, in agreement with previous reports (Wassef, 1976; Sohlenkamp and Geiger, 2016; Deschamps et al., 2021; Tao et al., 2021).
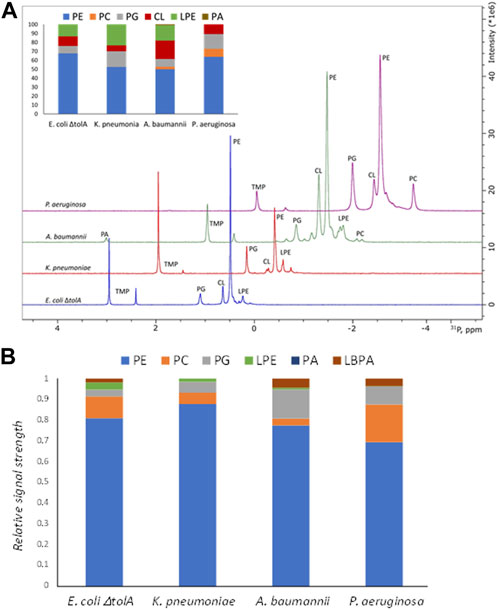
FIGURE 3. Summary of lipidomics analysis of bacterial lipid isolates by 31P NMR (A) and Mass Spectrometry (B). Intersection in the panel A shows quantified composition of the lipids in %. Traces of the NMR spectra were shifted up and right to ease the display. Here PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; LPE, lyso-phosphatidylethanolamine; CL, cardiolipin; PA, phosphatidic acid; LBPA, lysobisphosphatidic acid.
The MS analysis (Figure 3B) confirmed the presence of the same major lipid classes as in the NMR. Although the available MS approach only allowed for non-quantitative comparison of the relative signal within each sample, the results are in qualitative agreement with the 31P NMR in that PE was the dominant lipid class in all samples (Figure 4). The relative abundance of PC is the highest for bEVs from P. aeruginosa and A. baumannii, and the observation was consistent with 31P NMR. While PC is commonly found in P. aeruginosa, this phospholipid is normally not present in the bacterial membrane of E. coli strains (Sohlenkamp and Geiger, 2016; Deschamps et al., 2021). What is presented is however the relative signal, and a high signal of PC does not only reflect the concentration of the lipid but also the ability to ionize. Further, only PC with 18°C-atoms in the fatty acid chains were detected, which is not what is expected from a naturally occurring class of lipids.
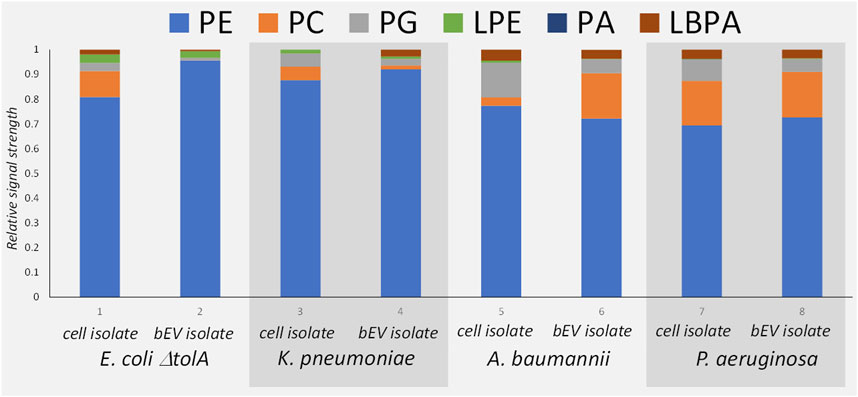
FIGURE 4. Mass Spectrometry patterns of the phospholipid compositions of the bEVs in comparison to the lipid isolates of corresponding strains: E. coli ΔtolA, K. pneumoniae, A. baumannii and P. aeruginosa. Here PE, phosphatidylethanolamine; PC, phosphatidylcholine; PG, phosphatidylglycerol; LPE, lyso-phosphatidylethanolamine; PA, phosphatidic acid; LBPA, lysobisphosphatidic acid.
Bacteria are typically missing the biosynthetic pathways for atypical lipids, but they can sometimes utilize fatty acids, phospholipids or their precursors available in the environment to adapt and, for example, evade the host immune defense system (Heung et al., 2006; Hobby et al., 2019). The strains in this work were cultured in rich lysogeny broth (LB) medium, which is based on yeast extracts (Alvarez and Steinbuchel, 2002; Montefusco et al., 2014). Phospholipid analysis of LB media by 31P NMR revealed only trace amounts of PE and PA lipids (Supplementary Figure S2), thus it is possible that LB media is a source of minor PA or LBPA contaminations. We therefore concluded that the growth media did not significantly influence the phospholipid composition of the bacteria or the vesicles.
In general, quantitative comparison between NMR (quantitative relative abundance) and MS (non-quantitative relative abundance) were interpreted conservatively. The higher sensitivity of MS allowed us to get data also from the lipid isolates of the bEVs, and to compare the pattern to that of the total lipid isolates of the same bacteria.
The comparison between the lipid isolates from the whole cells and the corresponding bEVs (Figure 4) provided a qualitatively good match between the lipid patterns of the bEVs and the parent bacteria. Since the lipid isolate contains lipids from the whole bacteria not just the membrane, the lipid isolate from the bEVs are not expected to be identical to the mother bacteria but contain the same pattern of the most dominant lipid classes as the bacteria. The results again confirmed that PE is the dominating lipid, and over all the same minor lipids are identified. The main deviation was observed for A. baumannii, which showed different distributions of PC between the bacterial extracellular vesicles and the whole cell isolate, having more PC in the outer membrane (Figure 4). The MS approach also allowed fatty acid composition analysis of the detected lipid species, but was limited to the PE phospholipid as it is the most dominant species in all samples. Out of the detected fatty acids, the hits with a relative abundance higher than 1% were selected for analysis (Supplementary Table S2). Mono-unsaturated fatty acids dominated in all samples. The top hits with relative abundance above 10% are mono-unsaturated or saturated fatty acids with a chain length of 16 or 18 carbon atoms in different combinations. The results also suggested higher preferences of K. pneumoniae for shorter fatty acid chains with 14 carbons, or chains with odd number of carbons (17 or 19) instead of 18 carbons. For the E. coli ΔtolA sample, a fatty acid with 20 carbons and 4 double bonds was detected. When comparing the bEVs’ fatty acid composition to the lipids isolated from the whole bacteria it was observed that E. coli ΔtolA had similar lipid compositions in both the bEVs and the native cell, while for the ESKAPE strains some differences could be observed. In particular this was evident for K. pneumoniae where the bEVs to a higher extent contained even numbered fatty acid chains (e.g., PE 16:0_16:1) compared to the whole bacteria where the PE 16:0_17:1 and PE 16:0_19:1 were the most prevalent. This could affect its lipid phase and physical properties like permeability, flexibility and fluidity. The dynamic nature of membrane lipid metabolism is another factor that can allow bacteria to survive antimicrobials by modifying lipids, fatty acids distribution or their whole lipid composition (Staubitz et al., 2004; Shireen et al., 2012; Schenk et al., 2015; Han et al., 2018; Bisignano et al., 2019; Tao et al., 2021).
In conclusion, the bEVs are shown to broadly inherit the lipid composition of the parent bacterial membrane, including species specific variations. Thus, they show potential to be an elegant model system for targeted studies, carrying authentic bacterial outer membrane in a highly stable and reasonably sized nano-construct. As such they can be seen as a much more relevant mimic of the bacterial cell surface compared to synthetic liposome membranes.
Interaction studies by SPR
The potential to use bEVs as models resembling the bacterial membrane in interaction studies was explored. As proof-of-principle, the interactions between bEVs and AMPs were measured using SPR.
For the SPR analysis, samples of either bEVs or liposomes were immobilized at the surface of L1 chips, and the dilution series of the antimicrobial peptides were run over the layers, measuring the response. A set of synthetic cyclic AMPs with alternating- or clumped distribution of charged (Lysine or Arginine) and hydrophobic (Tryptophan) amino acids was used: c-WKWKWK, c-WRWRWR, c-WWWKKK and c-WWWRRR (Figure 5). The set was selected as a useful chemical model to study the effects of charge distribution on the interactions. The peptide c-LWwNKr (Figure 5) was included in the study as a control that is chemically similar, but without antimicrobial activity (MIC > 256 μg/mL) (Junkes et al., 2008; Bagheri, 2010; Gopal et al., 2012; Finger et al., 2015; Rainsford et al., 2022; Jakubec et al., 2023).
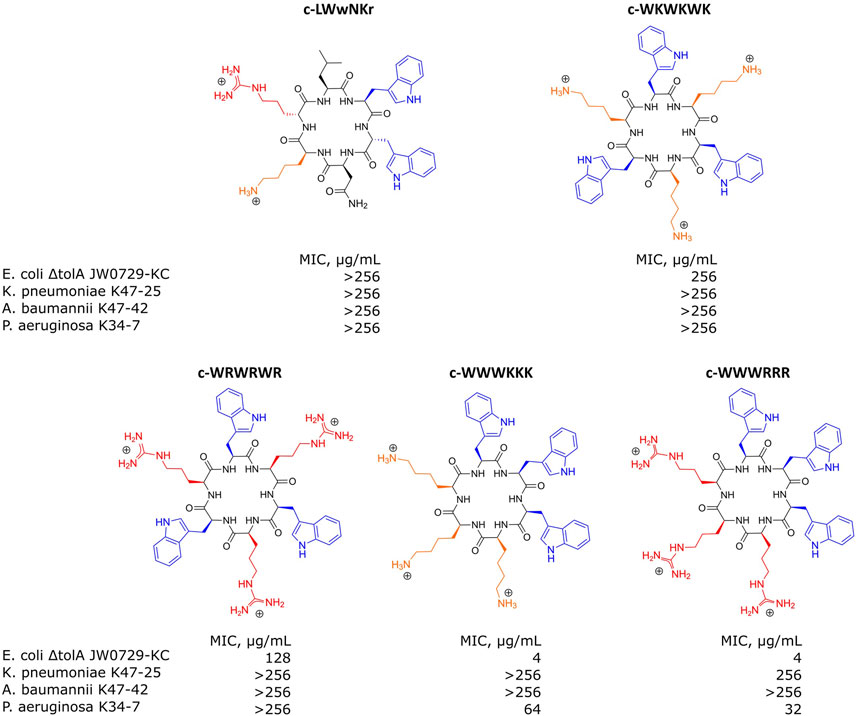
FIGURE 5. Structures of antimicrobial peptides used in this study and their minimal inhibitory concentrations (MIC) values determined for E. coli ΔtolA JW0729-KC, K. pneumoniae K47-25, A. baumannii K47-42 and P. aeruginosa K34-7. Hydrophobic Tryptophans highlighted in blue, positively charged Arginines and Lysines are highlighted in red and orange correspondingly.
Interactions were analyzed using the model described by (Figueira et al., 2017). By this, the dissociation constant KD and the disassociation rate koff were obtained. These constants allowed us to calculate kon rate, using Eq. 4. Figures 6A–D present examples of the raw response curves obtained during a run and the analytical processing steps of data fitting and analysis. The results for all samples are summarized in Figures 6E–G; Supplementary Tables S3–S5.
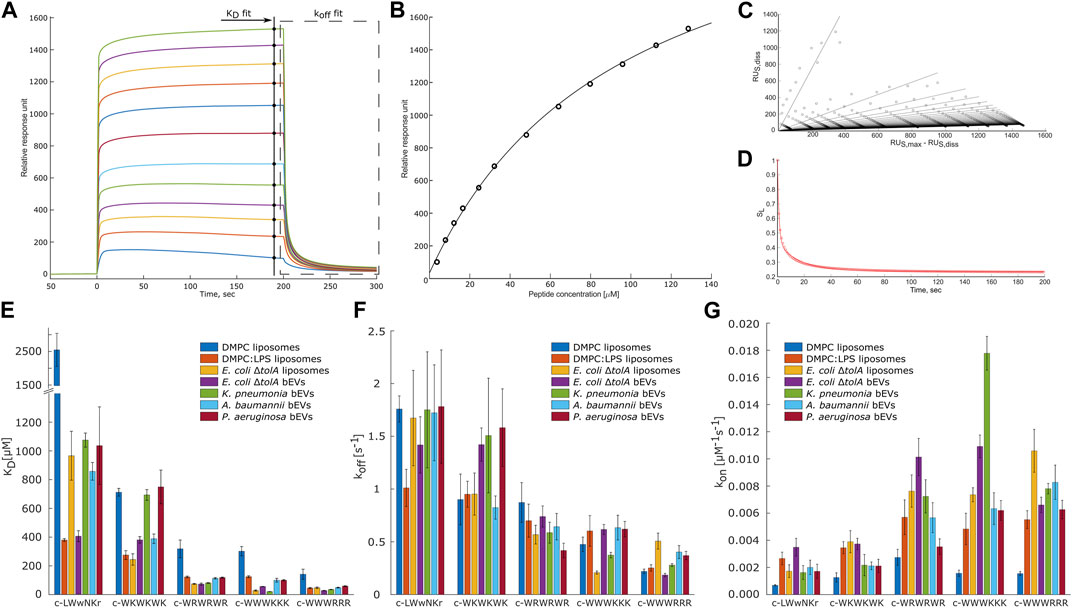
FIGURE 6. Overview panel of the SPR method and results. (A) – example of typical raw response signals collected during a run. Marked data points used for fitting to models to obtain KD (B) and koff values. To obtain koff values the raw data of the marked region first was linearized (C) and then a model was fitted into in-time changing linear coefficients (D). (B–D) represent typical data fit plots for our work. (E,F) summarize measured KD and koff values. (G) shows calculated kon values. Three parallels were performed for each combination. Values are available in Supplementary Tables S3–S5 in Supplementary Material. T-test statistical analysis of the data is summarized in Supplementary Table S6 in Supplementary Material. Results for DMPC and DMPC:LPS (90:10) are reproduced from Jakubec et al. (2023).
MIC determinations for each of the peptides against the bacterial strains used in this study are shown in Figure 5. The non-resistant E. coli ΔtolA displayed strong binding (low KD) and high antimicrobial activity (MIC = 4 μg/mL for both c-WWWKKK and c-WWWRRR). However, for the clinical strains none of the peptides showed any activity (MIC ≥ 256 μg/mL), except c-WWWKKK and c-WWWRRR that were inhibiting growth of P. aeruginosa at ≥64 μg/mL and ≥32 μg/mL, respectively. This does not directly reflect the fact that 3 out of the 4 antimicrobially active peptides bind as efficiently to the surface of the bEVs from resistant strains as to the bEVs from susceptible strains. The c-WKWKWK displayed a weaker interaction with the bEVs from K. pneumoniae and P. aeruginosa while the control peptide c-LWwNKr showed no antimicrobial activity as expected.
Quantification of the non-lipid components’ contribution
Any detailed analysis of the non-lipid contents of the bEVs have been out of the scope of this work. However, comparing the observed interactions of the peptides with E. coli ΔtolA bEVs and liposomes assembled from the lipid isolate of the same strain gives an indication of how the kinetic barrier imposed by LPS together with membrane proteins and other native components affects the interaction. For lysine-containing peptides, both the koff and kon rates were positively affected for the interactions with the bEVs, but the effect was more pronounced for koff, resulting in an overall increase in KD. A corresponding increase in the kon and koff rates for the lysine-containing peptides was also observed in the recently published comparison between homogeneous DMPC liposomes and DMPC liposomes containing 10% LPS (Jakubec et al., 2023) (results included in Figures 6E–G). In this case the balance was however different, resulting in slightly improved KD and Kp.
The opposite picture is observed for the c-WWWRRR peptide, where LPS and other components decreased both the koff and kon rates, shifting the balance towards decreased dissociation. In the recent study with dimyristoylphosphatidylcholine (DMPC) liposomes, LPS similarly improved KD, but in that case, it was driven by a significant increase of the kon rate (Jakubec et al., 2023). This peptide demonstrated the strongest binding of all tested peptides, and had the lowest MIC for E. coli ΔtolA. For the control peptide, the native bEV mostly contributed to an increase in association rate, kon, and a slight decrease in dissociation rate. The resulting change in the KD for this sample was the most prominent among the studied peptides. This observation was fully consistent with the trends in the synthetic DMPC/DMPC + LPS system, suggesting that LPS is likely responsible for the stronger binding of c-WWWRRR also in the bEV system.
Interactions of bEVs produced by ESKAPE strains
The overarching focus was to isolate and characterize bEVs from resistant clinical strains of ESKAPE pathogens, and to explore their applicability in interaction studies with antimicrobial model compounds. Comparison of the results from the interaction studies with bEVs to those with liposomes made from E. coli lipid isolate showed that the trend is consistent, with the clumped peptides being the strongest binders and the control peptide and c-WKWKWK being the weakest, reflected by both KD and koff (Figures 6E–G). There are, however, notable peculiarities in the results. The c-WWWKKK peptide was the peptide in this study that had the highest affinity for bEVs from K. pneumoniae and was even binding stronger to the BEVs than to the E. coli lipid isolate liposomes. The bEVs from K. pneumoniae had a prominent positive effect on the kon rate and only a slight increase in the koff rate, resulting in an approximately 5-fold lower KD compared to the KD for other clinical strains. The outer membrane of A. baumannii and P. aeruginosa had on the other hand a negative effect on the koff rate and KD for this peptide.
The results for the control peptide, c-LWwNKr, displayed a strong preference for the outer membrane of E. coli ΔtolA over the lipid isolate liposomes. A similar increase was observed in DMPC + LPS liposomes compared to pure DMPC liposomes, and in both cases this increase is driven by an increase in kon. Even though this peptide is inactive, it exhibits this apparent affinity for LPS. This increase was not observed in the resistant strains. However, similar trends in interaction of resistant strains and the c-WKWKWK peptide could be found. Here the kon rate (Figure 6E) was reduced similarly for bEVs from the resistant strains compared to the E. coli lipid isolate liposomes, while for the bEVs from the non-resistant E. coli ΔtolA there was no statistically significant change in kon. The koff rate (Figure 6F) was slightly decreased for A. baumannii while significantly increasing for the other strains, including the susceptible E. coli ΔtolA. Thus, as a result the KD values (Figure 6G) of bEVs from A. baumannii is similar with bEVs from E. coli ΔtolA, while for bEVs from K. pneumoniae and P. aeruginosa a twofold increase in KD was displayed.
For the interactions with the c-WRWRWR peptide, the bEVs from the resistant strains displayed a decrease in kon rates compared to the E. coli lipid isolate liposomes, especially for A. baumannii and P. aeruginosa. In contrast, for the susceptible E. coli ΔtolA, the kon rate instead increased for this peptide compared to the lipid isolate liposomes. The c-WWWRRR peptide showed a consistent behavior for all samples, both kon and koff rates being slower compared to the lipid isolate, resulting in only minor changes in KD.
The overall trend when comparing the peptides to each other is, however, that the peptides that have a strong affinity for the E. coli lipid isolate and the bEVs of the susceptible strain, also show a strong affinity for the bEVs from the resistant strains. The only exception is the c-WKWKWK peptide with a moderate affinity, which displayed a weaker affinity for two out of the three resistant strains.
bEVs from ESKAPE pathogens further appear to have inherited the strain specific properties of the outer membranes, reflecting the native surface of the bacteria. Their different properties resulted in corresponding differences in their interactions with AMPs measured by SPR. Further, comparison to liposomes made from whole bacteria lipid isolates allowed us to distinguish between the contributions from the lipids and the other outer membrane components, presumably dominated by LPS. The results demonstrated that the affinity of the AMPs to ESKAPE strains did not directly reflect their antimicrobial activity. The strongest binder was inactive towards the resistant strains, while being active in non-resistant strain with comparable binding parameters. Some potential explanations why there are no close correlations between the binding affinity and the antimicrobial effect could be that the resistant bacteria either maintain the membrane integrity upon association, or the bacteria have a mechanism to efficiently deactivate the AMPs (Manning and Kuehn, 2011; Balhuizen et al., 2021). Another possible explanation why the peptides would bind but not cause bacterial death is that the peptide did not manage to penetrate the kinetic conditions set by LPS and its modifications (Olaitan et al., 2014; Llobet et al., 2015; Jeannot et al., 2017).
Conclusion
The traditionally used synthetic membrane models for the bacterial membrane cannot effectively reconstruct the full complexity of native bacterial membranes. Herein, we have for the first time shown that the bacteria can do the hard work for us and produce sufficiently large quantities of native bEVs that enable interaction studies by SPR between compounds of interest and authentic bacterial outer membranes. The isolated bEVs were found to broadly inherit the lipid composition of the parent bacterial membrane, including species specific variations of the outer membrane composition, which in turn dictated the kinetic rates of the interaction processes. bEVs can thus be seen as a much more relevant mimic of the bacterial cell surface compared to synthetic liposome membranes.
Thus, we have provided proof of principle that native bEVs produced by clinical strains of Gram-negative ESKAPE strains, carrying specific antimicrobial resistance genes, can be used as tools to study processes on the bacterial surface. This sets the conditions to enable direct quantification of, for example, the effect of induction by antibiotics, or the response to host-pathogen interactions.
Materials and methods
Materials
All common chemicals and solvents were purchased from Sigma Aldrich (Merk KGaA, Darmstadt, Germany). Escherichia coli ΔtolA strain was obtained from National BioResource Project (NIG, Japan): E. coli (Keio Collection JW0729-KC). Strains of clinical isolates of multi-drug resistant K. pneumoniae K47-25, A. baumannii K47-42 and P. aeruginosa K34-7 strains were provided by The Norwegian National Advisory Unit on Detection of Antimicrobial Resistance (K-res), University Hospital of Northern-Norway (UNN). The peptides c-WKWKWK, c-WRWRWR, c-WWWKKK, c-WWWRRR and c-LWwNKr were synthesized as previously reported in (Rainsford et al., 2022; Jakubec et al., 2023). PhosSTOP tablets were purchased from Roche via Sigma Aldrich. Lipids were purchased from Avanti Lipids via Sigma Aldrich.
Minimum inhibitory concentration (MIC) determination
MIC for all test peptides was determined following EUCAST clinical standard (Hasselmann, 2003). Briefly, 3–4 colonies were picked by a cotton swab from freshly plated cultures on agar and resuspended in sterile saline solution. Turbidity of inoculum was adjusted to 0.5 McFarland standard units prior to ×100 dilution in Mueller-Hinton broth. Antimicrobial peptides were dissolved in the broth at the stock concentration of 512 μg/mL. MIC was determined in series of dilutions of antimicrobial peptides at concentrations ranging from 256 to 2 μg/mL in microdilution plates, keeping concentration of bacterial cells at constant. Positive control of bacterial culture diluted with broth and negative control of blank broth were included for each bacterial strain. The MICs were determined after incubation of the plates at 37°C for 18 h and red as the concentration of antimicrobial peptide at the next to the last well where bacterial growth was detected.
Bacterial extracellular vesicles isolation
Bacterial cultures were grown in lysogeny broth (LB) media at 37°C at 120 rpm until stationary growth phase for 16 h and harvested by centrifugation at 10,000 × g for 20 min. For further lipidomic analysis the bacterial pellet was lysed by immediate resuspension in 5 mL of TRIS buffer (10 mM TRIS, 100 mM NaCl and 5 mM KCl, pH 8.0) supplemented with 1 tablet of PhosSTOP, 2 mg/mL of 2-butoxyphenylboronic acid (BPBA), and 500 µL of mixture of mutanolysin (50 µg), lysozyme (10 mg), RNAase (200 µg) per 1 mL of glycerol/PBS 1:1 (the stock stored at −80°C). The mixture was left overnight at room temperature on a soft shaker and then frozen at −80°C before being freeze dried.
For isolation of BEVs the collected supernatant was filtered through 0.45 µm PES bottle-top vacuum filters and further upconcentrated to approx. 50–100 mL using Sartorius VivaFlow 200 30,000 PES membrane according to the instruction manual. Concentrate was filtered through 0.22 µm filters using a syringe, and then was ultracentrifuged (Beckman Coulter, Optima XPN-100) at 30,000 rpm for 2 h. The supernatant was discarded, and the pellet was resuspended in TRIS buffer, following another run of centrifugation at 30,000 rpm for 2 h. The remaining pellet, which contains the outer membrane vesicles, was finally resuspended in 300 µL TRIS buffer and inspected by negative-stain electron microscopy to assess the quality of the samples and analyzed on Zetasizer (see below). Protein concentration was used as a reference for the samples and was measured by Bradford protein assay from Bio-Rad.
Lipids isolation
To isolate lipids a modified Bligh and Dyer method was used (Bligh and Dyer, 1959; Furse et al., 2017). Briefly, lysed bacterial pellet was freeze dried for 48 h after being treated with the enzymatic mixture as described above. The freeze-dried material was mixed with 10–15 mL of dichloromethane/methanol mixture (2:1), properly vortexed and centrifuged (6,000 × g for 10 min). The organic solution was decanted to a separation funnel and the remaining pellet was mixed with another 10–15 mL of the same dichloromethane/methanol mixture. After vortexing the dispersion was transferred to the same separation funnel. The same volume of 5% NaCl solution in water was added to the funnel. The content of the funnel was gently mixed, and the mixture was left for the phase separation. After separation, the bottom organic phase was collected, and the remaining water phase was washed by addition of the same volume of dichloromethane/methanol mixture. Again, the mixture was allowed to form the phases and the bottom organic one was collected and combined with the previously collected fraction. The collected pooled organic mixture was filtered through filter paper and evaporated on a rotary evaporator. The resulting lipid film of E. coli ΔtolA was used to prepare liposomes for SPR analysis (by hydration with TRIS buffer, see below). Otherwise, the dried lipid films were redissolved in smaller volume of isopropanol and transferred to a smaller pre-weighted glass container through a cotton filter. After drying the lipid isolates under a nitrogen stream the resulting lipids were weighed. These isolates were used to prepare samples for 31P NMR analysis by dissolving in CUBO solvent or for MS analysis by dissolving in isopropanol (see below).
Lipid isolation from bEVs was performed in a similar fashion but with a simpler procedure. Freeze dried bEVs were resuspended in 3–5 mL of 2:1 mixture of dichloromethane and methanol, sonicated for 2 min of 2 s on/8 s off cycles on ice using probe sonicator (Ultrasonic processor 500 W, Sigma Aldrich, United States). 3–5 mL of 5% NaCl was added to the samples and after thorough vortexing the phases were separated by centrifugation at 3,000 × g for 5 min. The organic solution was transferred to a clean glass vial with glass pipet. The remaining aqueous phase was mixed with another 3–5 mL of the same dichloromethane/methanol mixture, vortexed and centrifuged. After separation, the organic phases were pooled together into the glass vial and evaporated under nitrogen. Dried isolates were redissolved in a smaller volume of isopropanol and transferred into a smaller pre-weighted glass container through a cotton filter. After drying under nitrogen, the lipid isolates were weighed. The samples were dissolved in isopropanol for mass spectrometry analysis.
Mass spectrometry
All lipid samples were dissolved in isopropanol at 1 mg/mL concentration. Identification and relative quantification of lipids were done on a Thermo IdX Tribrid Orbitrap MS with a Thermo Vanquish UPLC. Separation was done on a Waters Acquity Premier BEH C18 1.7 um 2.1 × 100 mm column with gradient elution. Mobile phase A consisted of 50% ACN in MilliQ-water and mobile phase B of 47.5% ACN, 47.5% isopropanol and 5% MilliQ-water. Both mobile phases were buffered with 1 mM ammonium formate and added formic acid to pH 3. The gradient started with 30% B, followed by a linear increase to 75% B at 20 min, again followed by a linear increase to 95% B at 25 min, which were held for 5 min, giving a total analysis time of 30 min. The injection volume was 2 μL, the column temperature 60°C, the flow rate 0.6 mL/min, and the sample compartment was held at room temperature to avoid precipitation of lipids. Identification of lipids were done by MS2 and MS3 experiments with the orbitrap mass analyzer. The analysis was done with positive ionization, the orbitrap full scan analysis was run with a resolution of 120,000, while MS2 and MS3 experiments were run at 15,000. Identification was obtained by first running a full scan in the mass range m/z 250–1,500, followed by data dependent MS2 (ddMS2) where the most intense ions were selected for HCD fragmentation with stepped collision energy (25, 30, 35%). Based on the results from the first fragmentation the ions could be sent for a second MS2 analysis if the result showed that the molecule was a phosphocholine (based on m/z 184 corresponding to the head group) for full identification by CID fragmentation. If the first ddMS2 showed that the molecule was a triacylglycerol (by neutral loss of a fatty acid) the ions were sent for MS3 analysis by CID fragmentation for full identification. All other lipids were identified after the first ddMS2 fragmentation. To identify as many lipids as possible in the samples 5 consecutive injections of a pooled sample were analysed with MSn fragmentation. In the first injection the most intense signals are fragmented for identification, thereafter, put on an exclusion list for injection 2 so that signals with lower intensity are fragmented and identified in the second injection. The process is repeated for all 5 injections to ensure identification of all lipids with significant signal intensity. The samples were analyzed in full scan with a resolution of 120,000, and after analysis the results were aligned with the results from the MS2 and MS3 experiment for identification. Lipid identification and relative quantification were done with the software LipidSearch from Thermo. LipidSearch data were filtered to only include Grade A and B identifications of phospholipids, in addition a manual inspection of identifications was performed to have as high degree of identification certainty as possible. All details for the LC-MS method can be found in the Supplementary Material.
Liposomes preparation
The lipid film from E. coli ΔtolA culture lipid isolate was prepared by removing the solvent under vacuo at 45°C on a rotavapor with final step of holding at vacuum of 55 hPa at room temperature for 3 h. The lipid film was then hydrated with TRIS buffer. Before extrusion through 100 nm polycarbonate membrane filters (Millipore) the liposome dispersions were subjected to 5 min of sonication cycles (5 s impulse followed by 10 s hold) at room temperature using probe sonicator. Size and zeta-potential of resulting liposomes were analyzed by zetasizer (see below).
Size and zeta potential measurements
Samples of isolated bEVs and freshly extruded liposomes prepared from lipid isolate dispersed in TRIS buffer were subjected to size and zeta potentials measurements using a Malvern Zetasizer Nano ZS (Malvern, Oxford, United Kingdom) instrument. In addition to size measurements, the polydispersity index (PDI) was estimated to assess population homogeneity. For comparison zeta-potential of the bacterial cells of the presented strains were determined by growing a 10 mL mini-prep of bacterial culture in LB media for 6 h and measuring the zeta-potential by diluting the samples in TRIS buffer. All zeta-potential measurements were performed using the disposable folded capillary cells (DTS1070). All samples were measured in triplicates at 25°C.
Electron microscopy
For negative stain electron microscopy freshly carbon-coated 400 mesh grids were used. The grids were absorbed on the droplets of isolated bEVs for 5 min, washed with double distilled water and stained with 1:9 mixture of 3% uranyl acetate and 2% methylcellulose for 2 min. The grids with the sample were picked up with loops, excess uranyl acetate was removed with filter paper, and the loops with the grids were dried on the loop holder for 10 min prior the imaging. The micrographs were obtained with HT7800 Series transmission electron microscope (Hitachi High-Tech Corp., Tokyo, Japan) operating at accelerated voltage of 100 kV coupled with Morada camera.
SPR assays
bEVs isolates resuspended in TRIS buffer at 0.5 mg/mL protein concentration were used as is without additional treatment. It was not possible to keep track on the final concentration of liposomes prepared from lipid isolate. For this sample optimal dilution was empirically determined by monitoring coverage of the chip. The SPR experiments were performed as was described in (Rainsford et al., 2022; Jakubec et al., 2023). Briefly, the analysis was carried out with a T200 Biacore instrument (GE Healthcare, Chicago, Ill, United States), at 25°C. Samples of bEVs or liposomes were immobilized on the surface of L1 chip (Cytiva, Marlborough, MA, United States) at 2 μL/min flow rate for 2,400 s. Chip coverage was assessed by injection of 0.5 mg/mL of BSA at 30 μL/min flow rate for 60 s. Change in response units (RU) of less than 400 was considered as sufficient coverage of the chip. The peptides were tested at increasing range of concentrations (from 4 to 128 µM for c-WRWRWR, c-WWWKKK and c-WWWRRR or 24–768 µM for c-WKWKWK and c-LWwNKr) in 10 mM HEPES buffer (with 150 mM NaCl). The peptides were injected over immobilized samples at 15 μL/min flow rate for 200 s followed by 400 s of dissociation phase. After each injection, the surface of immobilized vesicles was stabilized by three subsequent injections of 10 mM NaOH at 30 μL/min flow rate for 30 s each. Each run was concluded with cleaning of the chip surface by subsequently injecting 10 mM CHAPS, 40 mM octyl-β-D-glucopyranoside and 30% ethanol for 1 min at 30 μL/min flow rate. The control channel of the chip was treated the same way, except only HEPES buffer was injected instead of peptide solutions. The resulting data sets were processed and analyzed by in-house developed MATLAB scripts (MATLAB R2020a; available at: https://github.com/MarJakubec). KD was obtained using steady-state fit (Eq. 1) and koff rate constant was determined using formalism suggested by Figueira et al., 2017 (Eqs 2, 3). The association kon rate was calculated from obtained KD and koff values using Eq. 4.
where RU is measured SPR response; RUMAX is RU response at saturation; [P] is peptide concentration, and off is the offset of the response; SL is the linearised ratio of responses of the solute, which is plotted against the time of dissociation; α and β are individual populations, and SL,r is the retained solute fraction.
31P NMR spectroscopy
NMR spectra were acquired using a Bruker 400 MHz Avance III HD spectrometer equipped with an RT SmartProbe. Lipid isolates from bacterial membranes were dissolved in 500 µL of CUBO solvent (800 mM guanidine chloride dissolved in 3:1 mixture of dimethylformamide (DMF) and trimethylamine). For lipid quantification trimethyl phosphate (TMP) standard was added to the samples at the concentration of 0.7 µM. All experiments were acquired at 298 K in 5 mm NMR tubes. Spectra were processed in TopSpin 4.1.4 (Bruker, Billerica, MA, United States).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
MB’k: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing–original draft. VS: Data curation, Investigation, Writing–review and editing. MJ: Methodology, Writing–review and editing. TV: Data curation, Methodology, Writing–review and editing. TK: Methodology, Writing–review and editing. JC: Funding acquisition, Resources, Supervision, Writing–review and editing. JE: Funding acquisition, Resources, Writing–review and editing. JI: Funding acquisition, Resources, Supervision, Writing–review and editing. GF: Funding acquisition, Project administration, Resources, Supervision, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article. The authors acknowledge research funding support from CANS Centre for New Antibacterial Strategies (TFS Grant No. 18_CANS_AS) at UIT The Arctic University of Norway, the Digital Life project DigiBiotics (ID 269425) granted by the Research Council of Norway as well as funding from NordForsk for the Nordic University Hub project #85352 (Nordic POP, Patient Oriented Products). The APC was covered by the open access publishing fund, UiT.
Acknowledgments
Prof. Hanne Cecilie Winther-Larsen and Verena Mertes at Department of Pharmacy, University of Oslo, Norway, are greatly acknowledge for hosting a mobility action, scientific discussions and knowledge transfer on bEVs production. We also gratefully acknowledge K-res, University Hospital Northern Norway, for providing clinical isolates of resistant bacterial strains. TEM-imaging was done with help from The Advanced Microscopy Core Facility, Department of Medical Biology, UiT The Arctic University of Norway and the peptides acquired from the Digibiotics pipeline.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1277963/full#supplementary-material
Abbreviations
AMPs, antimicrobial peptides; AMR, antimicrobial resistance; bEVs, bacterial extracellular vesicles; DMPC, dimyristoylphosphatidylcholine; ESKAPE, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.; LPS, lipopolysaccharides; MIC, minimum inhibitory concentration; MS, mass spectrometry; NMR, nuclear magnetic resonance spectroscopy; PLs, phospholipids; SPR, surface plasmon resonance.
References
Ageitos, J. M., Sánchez-Pérez, A., Calo-Mata, P., and Villa, T. G. (2017). Antimicrobial peptides (AMPs): ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 133, 117–138. doi:10.1016/j.bcp.2016.09.018
Akbarzadeh, A., Rezaei-Sadabady, R., Davaran, S., Joo, S. W., Zarghami, N., Hanifehpour, Y., et al. (2013). Liposome: classification, preparation, and applications. Nanoscale Res. Lett. 8 (1), 102. doi:10.1186/1556-276X-8-102
Alvarez, H. M., and Steinbuchel, A. (2002). Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biot. 60 (4), 367–376. doi:10.1007/s00253-002-1135-0
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399 (10325), 629–655. doi:10.1016/S0140-6736(21)02724-0
Bagheri, M. (2010). Synthesis and thermodynamic characterization of small cyclic antimicrobial arginine and tryptophan-rich peptides with selectivity for Gram-negative bacteria. Methods Mol. Biol. 618, 87–109. doi:10.1007/978-1-60761-594-1_7
Bailey-Hytholt, C. M., LaMastro, V., and Shukla, A. (2021). Assembly of cell mimicking supported and suspended lipid bilayer models for the study of molecular interactions. J. Vis. Exp. 2021 (174). doi:10.3791/62599-v
Balhuizen, M. D., van Dijk, A., Jansen, J. W. A., van de Lest, C. H. A., Veldhuizen, E. J. A., and Haagsman, H. P. (2021). Outer membrane vesicles protect gram-negative bacteria against host defense peptides. mSphere 6 (4), e0052321. doi:10.1128/mSphere.00523-21
Beveridge, T. J. (1999). Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181 (16), 4725–4733. doi:10.1128/JB.181.16.4725-4733.1999
Bisignano, C., Ginestra, G., Smeriglio, A., La Camera, E., Crisafi, G., Franchina, F. A., et al. (2019). Study of the lipid profile of ATCC and clinical strains of Staphylococcus aureus in relation to their antibiotic resistance. Molecules 24 (7), 1276. doi:10.3390/molecules24071276
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Phys. 37 (8), 911–917. doi:10.1139/o59-099
Czaplewski, L., Bax, R., Clokie, M., Dawson, M., Fairhead, H., Fischetti, V. A., et al. (2016). Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16 (2), 239–251. doi:10.1016/S1473-3099(15)00466-1
Delcour, A. H. (2009). Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 1794 (5), 808–816. doi:10.1016/j.bbapap.2008.11.005
Denisov, I. G., Grinkova, Y. V., Lazarides, A. A., and Sligar, S. G. (2004). Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 126 (11), 3477–3487. doi:10.1021/ja0393574
Denisov, I. G., Schuler, M. A., and Sligar, S. G. (2019). Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol. Biol. 2003, 645–671. doi:10.1007/978-1-4939-9512-7_25
Deschamps, E., Schaumann, A., Schmitz-Afonso, I., Afonso, C., Dé, E., Loutelier-Bourhis, C., et al. (2021). Membrane phospholipid composition of Pseudomonas aeruginosa grown in a cystic fibrosis mucus-mimicking medium. Biochim. Biophys. Acta Biomembr. 1863 (1), 183482. doi:10.1016/j.bbamem.2020.183482
Elderdfi, M., and Sikorski, A. F. (2018). Interaction of membrane palmitoylated protein-1 with model lipid membranes. Gen. Physiol. Biophys. 37 (6), 603–617. doi:10.4149/gpb_2018029
Figueira, T. N., Freire, J. M., Cunha-Santos, C., Heras, M., Gonçalves, J., Moscona, A., et al. (2017). Quantitative analysis of molecular partition towards lipid membranes using surface plasmon resonance. Sci. Rep. 7, 45647. doi:10.1038/srep45647
Finger, S., Kerth, A., Dathe, M., and Blume, A. (2015). The efficacy of trivalent cyclic hexapeptides to induce lipid clustering in PG/PE membranes correlates with their antimicrobial activity. Biochim. Biophys. Acta 1848 (11), 2998–3006. doi:10.1016/j.bbamem.2015.09.012
Furse, S., Jakubec, M., Rise, F., Williams, H. E., Rees, C. E. D., and Halskau, O. (2017). Evidence that Listeria innocua modulates its membrane's stored curvature elastic stress, but not fluidity, through the cell cycle. Sci. Rep-Uk 7, 8012. doi:10.1038/s41598-017-06855-z
Gopal, R., Na, H., Seo, C. H., and Park, Y. (2012). Antifungal activity of (KW)n or (RW)n peptide against Fusarium solani and Fusarium oxysporum. Int. J. Mol. Sci. 13 (11), 15042–15053. doi:10.3390/ijms131115042
Han, M. L., Zhu, Y., Creek, D. J., Lin, Y. W., Anderson, D., Shen, H. H., et al. (2018). Alterations of metabolic and lipid profiles in polymyxin-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62 (6), e02656-17. doi:10.1128/AAC.02656-17
Hasselmann, C. ESCM (2003). Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 9 (8). doi:10.1046/j.1469-0691.2003.00790.x
Haurat, M. F., Elhenawy, W., and Feldman, M. F. (2015). Prokaryotic membrane vesicles: new insights on biogenesis and biological roles. Biol. Chem. 396 (2), 95–109. doi:10.1515/hsz-2014-0183
Heung, L. J., Luberto, C., and Del Poeta, M. (2006). Role of sphingolipids in microbial pathogenesis. Infect. Immun. 74 (1), 28–39. doi:10.1128/IAI.74.1.28-39.2006
Hobby, C. R., Herndon, J. L., Morrow, C. A., Peters, R. E., Symes, S. J. K., and Giles, D. K. (2019). Exogenous fatty acids alter phospholipid composition, membrane permeability, capacity for biofilm formation, and antimicrobial peptide susceptibility in Klebsiella pneumoniae. Microbiologyopen 8 (2), e00635. doi:10.1002/mbo3.635
Jackman, J. A., and Cho, N. J. (2020). Supported lipid bilayer formation: beyond vesicle fusion. Langmuir 36 (6), 1387–1400. doi:10.1021/acs.langmuir.9b03706
Jakubec, M., Bariås, E., Furse, S., Govasli, M. L., George, V., Turcu, D., et al. (2021). Cholesterol-containing lipid nanodiscs promote an α-synuclein binding mode that accelerates oligomerization. FEBS J. 288 (6), 1887–1905. doi:10.1111/febs.15551
Jakubec, M., Rylandsholm, F. G., Rainsford, P., Silk, M., Bril'kov, M., Kristoffersen, T., et al. (2023). Goldilocks dilemma: LPS works both as the initial target and a barrier for the antimicrobial action of cationic AMPs on E. coli. Biomolecules 13 (7), 1155. doi:10.3390/biom13071155
Jasim, R., Han, M. L., Zhu, Y., Hu, X., Hussein, M. H., Lin, Y. W., et al. (2018). Lipidomic analysis of the outer membrane vesicles from paired polymyxin-susceptible and -resistant Klebsiella pneumoniae clinical isolates. Int. J. Mol. Sci. 19 (8), 2356. doi:10.3390/ijms19082356
Jeannot, K., Bolard, A., and Plésiat, P. (2017). Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 49 (5), 526–535. doi:10.1016/j.ijantimicag.2016.11.029
Junkes, C., Wessolowski, A., Farnaud, S., Evans, R. W., Good, L., Bienert, M., et al. (2008). The interaction of arginine- and tryptophan-rich cyclic hexapeptides with Escherichia coli membranes. J. Pept. Sci. 14 (4), 535–543. doi:10.1002/psc.940
Krüger, M., Richter, P., Strauch, S. M., Nasir, A., Burkovski, A., Antunes, C. A., et al. (2019). What an Escherichia coli mutant can teach us about the antibacterial effect of chlorophyllin. Microorganisms 7 (2), 59. doi:10.3390/microorganisms7020059
Kumar, P., Kizhakkedathu, J. N., and Straus, S. K. (2018). Antimicrobial peptides: diversity, mechanism of action and Strategies to improve the activity and biocompatibility in vivo. Biomolecules 8 (1), 4. doi:10.3390/biom8010004
Llobet, E., Martínez-Moliner, V., Moranta, D., Dahlström, K. M., Regueiro, V., Tomás, A., et al. (2015). Deciphering tissue-induced Klebsiella pneumoniae lipid A structure. Proc. Natl. Acad. Sci. U. S. A. 112 (46), E6369–E6378. doi:10.1073/pnas.1508820112
Mahlapuu, M., Håkansson, J., Ringstad, L., and Björn, C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell Infect. Microbiol. 6, 194. doi:10.3389/fcimb.2016.00194
Manning, A. J., and Kuehn, M. J. (2011). Contribution of bacterial outer membrane vesicles to innate bacterial defense. BMC Microbiol. 11, 258. doi:10.1186/1471-2180-11-258
Montefusco, D. J., Matmati, N., and Hannun, Y. A. (2014). The yeast sphingolipid signaling landscape. Chem. Phys. Lipids 177, 26–40. doi:10.1016/j.chemphyslip.2013.10.006
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging Strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 10, 539. doi:10.3389/fmicb.2019.00539
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643. doi:10.3389/fmicb.2014.00643
Paulowski, L., Donoghue, A., Nehls, C., Groth, S., Koistinen, M., Hagge, S. O., et al. (2020). The beauty of asymmetric membranes: reconstitution of the outer membrane of gram-negative bacteria. Front. Cell Dev. Biol. 8, 586. doi:10.3389/fcell.2020.00586
Paulsen, M. H., Engqvist, M., Ausbacher, D., Anderssen, T., Langer, M. K., Haug, T., et al. (2021). Amphipathic barbiturates as mimics of antimicrobial peptides and the marine natural products eusynstyelamides with activity against multi-resistant clinical isolates. J. Med. Chem. 64 (15), 11395–11417. doi:10.1021/acs.jmedchem.1c00734
Pirc, K., and Ulrih, N. P. (2015). α-Synuclein interactions with phospholipid model membranes: key roles for electrostatic interactions and lipid-bilayer structure. Biochim. Biophys. Acta 1848 (10), 2002–2012. doi:10.1016/j.bbamem.2015.06.021
Prenner, E. J., Lewis, R. N., and McElhaney, R. N. (1999). The interaction of the antimicrobial peptide gramicidin S with lipid bilayer model and biological membranes. Biochim. Biophys. Acta 1462 (1-2), 201–221. doi:10.1016/s0005-2736(99)00207-2
Rainsford, P., Rylandsholm, F., Jakubec, M., Juskewitz, E., Silk, M., Engh, R. A., et al. (2022). Label-free measurement of antimicrobial peptide interactions with lipid vesicles and nanodiscs using microscale thermophoresis. Sci. Rep. 13 (1), 12619. doi:10.1038/s41598-023-39785-0
Reimer, S. L., Beniac, D. R., Hiebert, S. L., Booth, T. F., Chong, P. M., Westmacott, G. R., et al. (2021). Comparative analysis of outer membrane vesicle isolation methods with an Escherichia coli tolA mutant reveals a hypervesiculating phenotype with outer-inner membrane vesicle content. Front. Microbiol. 12, 628801. doi:10.3389/fmicb.2021.628801
Roier, S., Zingl, F. G., Cakar, F., Durakovic, S., Kohl, P., Eichmann, T. O., et al. (2016). A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat. Commun. 7, 10515. doi:10.1038/ncomms10515
Ruiz, N., Falcone, B., Kahne, D., and Silhavy, T. J. (2005). Chemical conditionality: a genetic strategy to probe organelle assembly. Cell 121 (2), 307–317. doi:10.1016/j.cell.2005.02.014
Scheidelaar, S., Koorengevel, M. C., Pardo, J. D., Meeldijk, J. D., Breukink, E., and Killian, J. A. (2015). Molecular model for the solubilization of membranes into nanodisks by styrene maleic Acid copolymers. Biophys. J. 108 (2), 279–290. doi:10.1016/j.bpj.2014.11.3464
Schenk, E. R., Nau, F., Thompson, C. J., Tse-Dinh, Y. C., and Fernandez-Lima, F. (2015). Changes in lipid distribution in E. coli strains in response to norfloxacin. J. Mass Spectrom. 50 (1), 88–94. doi:10.1002/jms.3500
Sezgin, E., and Schwille, P. (2012). Model membrane platforms to study protein-membrane interactions. Mol. Membr. Biol. 29 (5), 144–154. doi:10.3109/09687688.2012.700490
Shireen, T., Singh, M., Dhawan, B., and Mukhopadhyay, K. (2012). Characterization of cell membrane parameters of clinical isolates of Staphylococcus aureus with varied susceptibility to alpha-melanocyte stimulating hormone. Peptides 37 (2), 334–339. doi:10.1016/j.peptides.2012.05.025
Silhavy, T. J., Kahne, D., and Walker, S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2 (5), a000414. doi:10.1101/cshperspect.a000414
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40 (1), 133–159. doi:10.1093/femsre/fuv008
Staubitz, P., Neumann, H., Schneider, T., Wiedemann, I., and Peschel, A. (2004). MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 231 (1), 67–71. doi:10.1016/S0378-1097(03)00921-2
Tao, Y., Acket, S., Beaumont, E., Galez, H., Duma, L., and Rossez, Y. (2021). Colistin treatment affects lipid composition of acinetobacter baumannii. Antibiot. (Basel) 10 (5), 528. doi:10.3390/antibiotics10050528
Wassef, M. K. (1976). Lipids of Klebsiella pneumoniae: the presence of phosphatidyl choline in succinate-grown cells. Lipids 11 (5), 364–369. doi:10.1007/BF02532842
WHO (2014). Antimicrobial resistance: global report on surveillance. World Health Organization. Geneva, Switzerland.
WHO (2017). WHO publishes list of bacteria for which new antibiotics are urgently needed 2017. Available at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
Keywords: antimicrobial resistance, outer membrane vesicles, protein-lipid interactions, membrane interactions, native barriers, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa
Citation: Bril’kov MS, Stenbakk V, Jakubec M, Vasskog T, Kristoffersen T, Cavanagh JP, Ericson JU, Isaksson J and Flaten GE (2023) Bacterial extracellular vesicles: towards realistic models for bacterial membranes in molecular interaction studies by surface plasmon resonance. Front. Mol. Biosci. 10:1277963. doi: 10.3389/fmolb.2023.1277963
Received: 15 August 2023; Accepted: 01 December 2023;
Published: 13 December 2023.
Edited by:
Martin Van Der Laan, Saarland University, GermanyReviewed by:
Anton Le Brun, Australian Nuclear Science and Technology Organisation, AustraliaTapani Viitala, University of Helsinki, Finland
Copyright © 2023 Bril’kov, Stenbakk, Jakubec, Vasskog, Kristoffersen, Cavanagh, Ericson, Isaksson and Flaten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gøril Eide Flaten, goril.flaten@uit.no
†These authors share last authorship
 Maxim S. Bril’kov
Maxim S. Bril’kov Victoria Stenbakk1
Victoria Stenbakk1  Martin Jakubec
Martin Jakubec Jorunn Pauline Cavanagh
Jorunn Pauline Cavanagh Johanna U. Ericson
Johanna U. Ericson Johan Isaksson
Johan Isaksson Gøril Eide Flaten
Gøril Eide Flaten