Emerging roles of long non-coding RNAs in osteosarcoma
- 1Frontiers Medical Center, Tianfu Jincheng Laboratory, Chengdu, Sichuan, China
- 2Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu, China
Osteosarcoma (OS) is a highly aggressive and lethal malignant bone tumor that primarily afflicts children, adolescents, and young adults. However, the molecular mechanisms underlying OS pathogenesis remain obscure. Mounting evidence implicates dysregulated long non-coding RNAs (lncRNAs) in tumorigenesis and progression. These lncRNAs play a pivotal role in modulating gene expression at diverse epigenetic, transcriptional, and post-transcriptional levels. Uncovering the roles of aberrant lncRNAs would provide new insights into OS pathogenesis and novel tools for its early diagnosis and treatment. In this review, we summarize the significance of lncRNAs in controlling signaling pathways implicated in OS development, including the Wnt/β-catenin, PI3K/AKT/mTOR, NF-κB, Notch, Hippo, and HIF-1α. Moreover, we discuss the multifaceted contributions of lncRNAs to drug resistance in OS, as well as their potential to serve as biomarkers and therapeutic targets. This review aims to encourage further research into lncRNA field and the development of more effective therapeutic strategies for patients with OS.
1 Introduction
Osteosarcoma (OS) is a highly prevalent and aggressive bone tumor that affects individuals across various age groups, including children, adolescents, and adults over 60 years of age (Mirabello et al., 2009). It is typically located in the metaphysis of the long bones, most commonly in the knee (50%), humerus (15%), or pelvic girdle (12%) (Bielack et al., 2002). Approximately 80% of patients initially present with a localized form of OS and no distant metastases (Bacci et al., 1997). Currently, the primary treatment modalities for OS include surgical resection, chemotherapy, and radiotherapy. Advances in surgical techniques and neoadjuvant chemotherapy have resulted in a 65%–70% cure rate when using cisplatin (DDP), doxorubicin (DOX) and methotrexate (MTX) as adjuvant chemotherapies (Whelan and Davis, 2018). However, the overall survival rate for individuals with distant metastases or multi-drug resistance (MDR) remains below 20% and has not shown significant improvement over an extended period of time (Bacci et al., 1997; Sakamoto and Iwamoto, 2008). The pathogenesis of OS is believed to originate from the malignant transformation of mesenchymal cells of an undefined stage of differentiation into osteoblasts (Klein and Siegal, 2006). Furthermore, numerous genetic alterations have been identified in OS, including dysregulation of p53 and Rb tumor suppressor genes, aneuploidies, and extensive disruption in the regulation of chromosomal arrangement, hinting at a possible primary defect in the DNA repair mechanisms. However, the detailed molecular mechanisms of OS remain unclear. Therefore, unraveling the mechanisms driving oncogenesis, progression and chemoresistance in OS, as well as identifying novel biomarkers for alternative therapies or improved efficacy of existing treatments, is of paramount importance in order to improve clinical outcomes.
Previous studies of OS primarily focused on protein-coding genes due to their importance in biological regulation, however, analysis of the human genome has revealed that less than 2% of its sequence is composed of protein-coding genes, while the remaining 98% is actively transcribed into non-coding RNAs (ncRNAs), previously considered as transcriptional “noise” (Rosenbloom et al., 2012). Recent evidence, however, suggests that these evolutionarily conserved ncRNAs have a significant role in a range of physiological and pathological processes (Huarte, 2015; Chi et al., 2019). lncRNAs are non-coding transcripts longer than 200 nucleotides and can be classified into sense, antisense, bidirectional, intronic, and intergenic transcripts (Ma et al., 2013; Mattick et al., 2023). To date, over 50,000 genes have been identified to generate lncRNAs, with tissue-, lineage-, and spatiotemporal-specific patterns (Iyer et al., 2015). Certain lncRNAs, such as HOTAIR, MALAT1 and H19, have been implicated in OS oncogenesis and progression by modulating signaling pathways (Wang B. et al., 2015; Li E. et al., 2017; Li H. et al., 2023). Furthermore, lncRNAs are associated with the response of OS patients to chemotherapy (Hahne and Valeri, 2018). Intriguingly, some lncRNAs are specific to OS and can be detected in body fluids, making them attractive biomarkers in liquid biopsy (Cai et al., 2017; Han et al., 2017; Yang Q. et al., 2019). For example, alkaline phosphatase (ALP) has displayed both good specificity and sensitivity when used for the diagnosis of OS(Levine and Rosenberg, 1979), however, serum HNF1A-AS1 is more effective than ALP in distinguishing OS from healthy individuals (Cai et al., 2017). Furthermore, pre-clinical studies have already demonstrated therapeutic targeting of lncRNAs, such as MALAT1 and HOXB-AS3 in cancer (Arun et al., 2016; Papaioannou et al., 2019), suggesting their potential clinical value in cancer diagnosis and treatment. In this review, we discuss the dysregulation and regulatory role of lncRNAs in the oncogenesis, progression, as well as chemoresistance of OS. Finally, we discuss the diagnostic and therapeutic potential of lncRNAs in OS.
2 Regulatory roles of lncRNAs on gene expression in cancers
LncRNAs have emerged as crucial regulators of gene expression in cancer, exerting their influence through diverse mechanisms. Depending on their subcellular localization, lncRNAs perform distinct functions within the nucleus and cytoplasm. In the nucleus, lncRNAs orchestrate epigenetic modifications by recruiting chromatin remodeling and modification complexes (Figure 1A, B). They also collaborate with transcription factors to act as transcriptional regulators, modulating the expression of neighboring messenger RNAs (Figure 1C). Moreover, lncRNAs can interact with splicing factors or pre-mRNA molecules themselves, thereby influencing the splicing process (Figure 1D). Furthermore, lncRNAs participate in post-transcriptional gene regulation, impacting mRNA stability, translation, protein stability, and competing endogenous RNA (ceRNA) networks (Figures 1E–G). Intriguingly, studies have revealed that certain lncRNAs can encode peptides (Figure 1H). Collectively, lncRNAs are implicated in gene expression regulation at multiple levels.
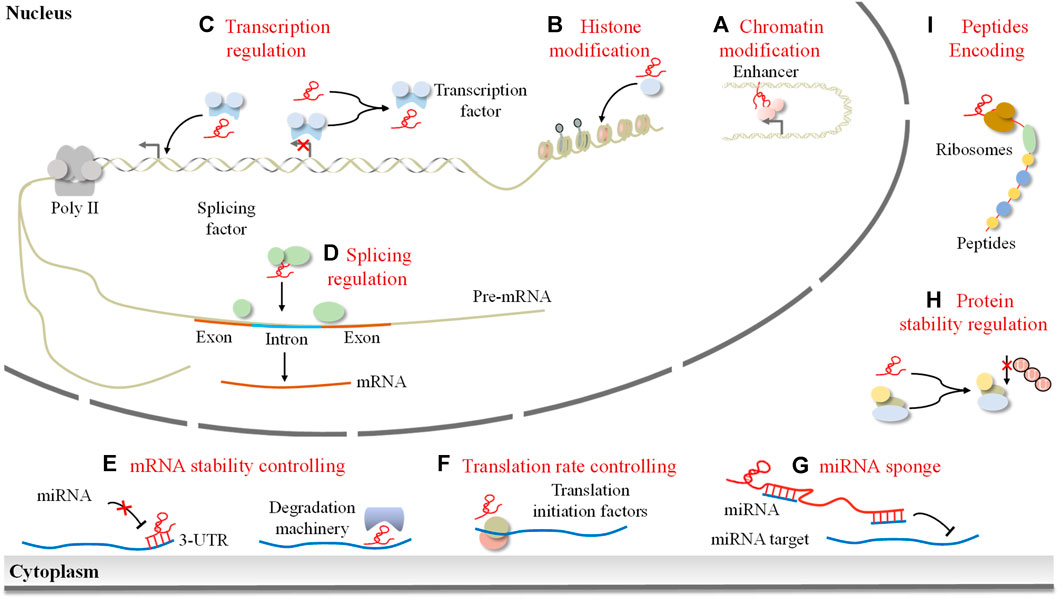
FIGURE 1. The mode of lncRNAs in the regulation of gene expression. LncRNAs exert their effects depending on their subcellular localization. In the nucleus, lncRNAs can control the epigenetic state of genes by (A) modifying chromatin architecture and (B) mediating chromatin/histone modifications, participating in the transcriptional process by (C) guiding transcription factors to induce or inhibit gene transcription and (D) regulating splicing of pre-mRNAs. Cytoplasmic lncRNAs can (E) regulate mRNA stability, (F) control translation rate, (G) act as a miRNA sponge, (H) mediate protein stability, and (I) encode peptides.
2.1 LncRNAs in epigenetic regulation
LncRNAs are involved in epigenetic regulation, which can influence gene expression in a heritable manner without changing the underlying DNA sequence (Xiao et al., 2018). These enigmatic molecules have been unveiled as influential regulators, recruiting esteemed chromatin-modifying enzymes such as histone acetyltransferases (HATs), histone deacetylases (HDACs), and histone methyltransferases (HMTs). Through their interactions, these enzymes shape the expression patterns of transcription factors and their coregulators, thereby exerting a profound impact on gene expression. For example, the lncRNA HOTAIR guides a chromatin modification complex towards target genes, altering chromatin state and modulating their transcription (Tsai et al., 2010). HOTTIP, in turn, interacts with WDR5 through chromatin looping, enabling the WDR5-MLL complex to target HOXA gene promoters, promoting histone H3K4 trimethylation (Luo et al., 2019). LncRNAs also regulate DNA methylation through modulation of DNMT expression. TARID binds GADD45α, inducing demethylation of 5-methylcytosine on the TCF21 promoter, activating transcription, and suppressing tumor formation (Arab et al., 2014).
2.2 LncRNAs in transcriptional regulation
LncRNAs have been shown to regulate gene transcription through both cis- and trans-regulation. In cis-regulation, lncRNAs can act as transcriptional regulators by binding to regulatory sequences in the promoter region of their target genes, thus modulating the transcription of those genes. For instance, lncRNAs can recruit SWI/SNF complexes to the promoter of target genes, such as TCF7, to modulate the expression of these genes and consequently activate the Wnt signaling pathway, thus promoting the self-renewal of liver cancer stem cells (CSCs) and tumor proliferation (Wang Y. et al., 2015). On the other hand, in trans-regulation, lncRNAs can act as scaffolds, recruiting transcription factors and chromatin modifiers to regulate gene expression. For example, lncRNA AGAP2-AS1 has been demonstrated to recruit the enhancers of zeste homolog 2 (EZH2) and LSD1 to the promoter regions of KLF2 and large tumor suppressor 2 (LATS2), thus suppressing the transcription of KLF2 and LATS2 and promoting the progression of non-small cell lung cancer (Li W. et al., 2016). Additionally, lncRNA GAS5 has been found to fold into a DNA-like structure that binds to the glucocorticoid receptor (GR) and inhibits its transcription activity (Mourtada-Maarabouni and Williams, 2013).
2.3 LncRNAs in post-transcriptional regulation
At the post-transcriptional level, lncRNAs interact with molecules to modulate mRNA splicing, stability, protein stability, and subcellular localization (Yoon et al., 2013). They can influence mRNA translation as stabilizers, destabilizers, ribosomal recruiters, or mRNA decoys. Antisense lncRNAs regulate pre-mRNA alternative splicing, alone or with splicing factors. For example, lnc-Spry1 binds U2 small nuclear ribonucleoprotein auxiliary factor 65 kD, controlling variable splicing of fibroblast growth factor receptor pre-mRNA during the epithelial-mesenchymal transition (EMT) (Rodríguez-Mateo et al., 2017). LncRNAs also affect mRNA stability through their 3′UTR. For instance, lncRNA PXN-AS1-L binds the 3′UTR of Paxillin (PXN) mRNA, preventing miRNA-24 binding and reducing its degradation (Yuan et al., 2017). Moreover, lncRNAs influence protein translation by recruiting translation initiation factors. LncRNA GAS5 binds eukaryotic translation initiation factor 4E (eIF4E), inhibiting c-Myc protein translation and downregulating its expression (Hu et al., 2014). It is worth noting that lncRNAs also play a role in regulating protein stability. Notably, lncRNAs regulate protein stability. For instance, lncRNA UPAT enhances UHRF1 stability by inhibiting β-transducin repeat-containing protein (TrCP)-mediated ubiquitination (Taniue et al., 2016).
Noncoding RNA molecules, such as lncRNAs (Salmena et al., 2011), circular RNAs (circRNAs) (Qin et al., 2023), or other RNA transcripts containing specific miRNA response elements (MREs), can serve as ceRNAs or miRNA sponges that bind to miRNAs and sequester them from their target mRNAs, thus modulating gene expression at post-transcriptional level. Many well-known lncRNAs have been demonstrated to exert important ceRNA roles in cancer. For example, lncRNA H19 promotes the angiogenesis of glioma by acting as a sponge for miR-29a and upregulating the angiogenesis factor vasohibin-2 (Jia et al., 2016). LncRNA CCAT1 can promote the proliferation and metastasis of esophageal squamous cell carcinoma (ESCC) by sponging miR-7 to enhance the expression of homeobox gene B1 (Zhang E. et al., 2017). Moreover, DANCR decoys two miRNAs, miR-335-5p and miR-1972, to facilitate ROCK1-mediated proliferation and metastasis of OS via a ceRNA network (Wang et al., 2018b).
LncRNAs were initially assumed to be ncRNAs that could not encode proteins. Nevertheless, recent evidence suggests that some lncRNAs contain open reading frames capable of encoding peptides, thereby allowing them to exert biological effects through the proteins they encode. For instance, the 53-amino acid peptide encoded by long non-coding RNA HOXB-AS3 has the capacity to suppress colon cancer (Huang et al., 2017). These transcripts are no longer considered bona fide ncRNAs, yet they are still categorized as a specific class of lncRNAs as they may play dual roles as both non-coding RNA or protein-coding transcripts in cancer.
3 Role of lncRNAs in OS oncogenesis and progression
LncRNAs are involved in the regulation of cell proliferation, survival, metastasis, and invasion in OS via modulation of multiple oncogenic pathways, such as the Wnt/β-catenin, PI3K/AKT/mTOR, NF-κB, Notch, Hippo and HIF-1α pathways (Figure 2).
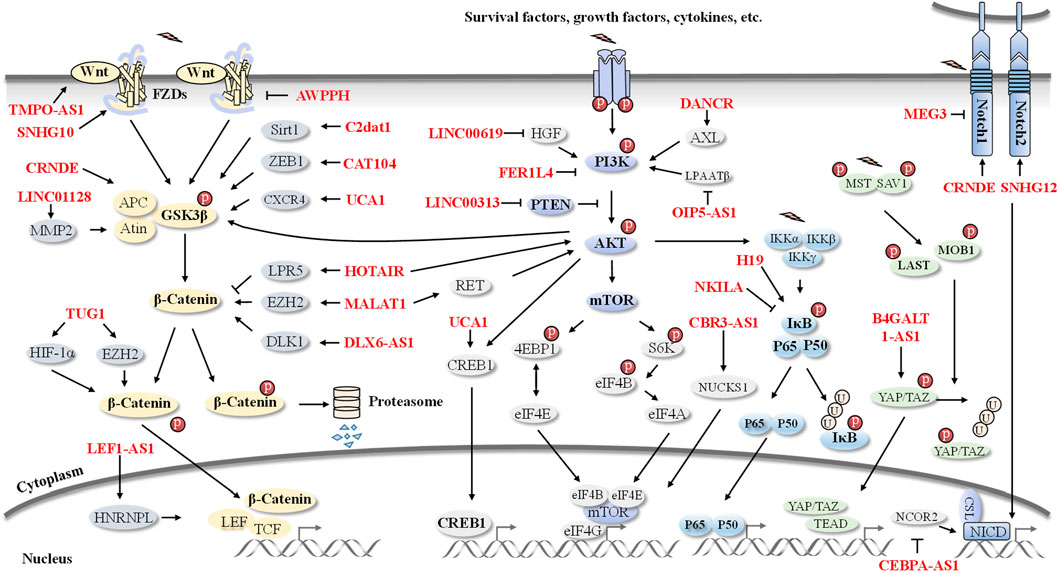
FIGURE 2. An overview of lncRNAs-involved signaling pathways in OS. Pathogenesis and progression of OS are regulated by several lncRNAs, including HOTAIR, MALAT1, UCA1, CRNDE, NKILA, CEBPA-AS1, H19, DANCR, and SNHG12 et al. These OS-related lncRNAs are involved in the Wnt/β-catenin, PI3K/AKT, NF-κB, Notch, and Hippo signaling pathways.
3.1 The Wnt/β-catenin pathway
The Wnt/β-catenin signaling pathway is a major player in the tumorgenesis of OS. This cascade is initiated by the binding of Wnt ligands to their receptors, Frizzled proteins, leading to the activation of Dishevelled (Dvl). Upon activation, Dvl triggers the phosphorylation of β-catenin, preventing its degradation by the proteasome and thus allowing its accumulation in the cytoplasm. Subsequently, β-catenin is translocated to the nucleus, where it binds to TCF/LEF transcription factors and induces the expression of target genes, including c-Myc and cyclin D1. This pathway can regulate processes such as proliferation, migration, apoptosis, and differentiation, while its dysregulation is associated with the development of various cancers, including OS (Nusse and Clevers, 2017).
3.1.1 Regulation of proliferation
Several lncRNAs have been shown to control cell proliferation of OS via the Wnt/β-catenin signaling pathway. For instance, upregulation of LINC01128 facilitates cell proliferation in OS and exhibits a negative correlation with overall survival. In vitro and in vivo studies supported its role in modulating MMP2, a protease responsible for cleaving and releasing β-catenin from its cytoplasmic complex, by acting as a sponge of miR-299-3p, ultimately activating the Wnt/β-catenin signaling pathway (Yao and Chen, 2020). Similarly, lncRNA SNHG10 promotes OS growth by acting as a miR-182-5p sponge, which subsequently leads to increased FZD3 expression, thereby sustaining the activation of the Wnt/β-catenin signaling pathway (Zhu S. et al., 2020). Moreover, NF1A-AS1, overexpressed in OS, promotes cell proliferation by activating the Wnt/β-catenin signaling pathway, however, the underlying mechanism requires further exploration (Zhao et al., 2016). lncRNA LEF1-AS1 has been investigated to bind with HNRNPL and enhance the mRNA stability of LEF1, the nuclear effector of β-catenin, thereby promoting OS cell proliferation (Lu et al., 2020).
3.1.2 Regulation of apoptosis
Wnt signaling has been proven to regulate apoptosis in OS, and recently, lncRNAs associated with Wnt signaling have also played an important role in regulating apoptosis (Brock et al., 2019). Depletion of TMPO-AS1 has been observed to restrain the Wnt/β-catenin pathway, thereby facilitating cell apoptosis (Cui and Zhao, 2020). CAT104 was highly expressed in OS cell lines and its knockdown significantly promoted OS-732 cell apoptosis. Mechanistically, CAT104 was found to regulate miR-381 to target ZEB1, as well as Wnt/β-catenin pathways, thus exerting its oncogenic effects on OS cells (Xia et al., 2018).
3.1.3 Regulation of metastasis and invasion
Aggressive metastasis at early stages has been suggested to drive the rapid progression and poor prognosis of OS. Increasing evidence has pointed to the crucial role of Wnt cascade-associated lncRNAs in regulating the metastasis and invasion of OS. For instance, the knockdown of lncRNA CAMK2D-associated transcript 1 (C2dat1) reduced OS cell invasion and migration by modulating the miR-34a-5p/Sirt1 network and Wnt signaling pathway (Jia et al., 2018). Additionally, urothelial carcinoma associated 1 (UCA1) was reported to enhance cell viability, migration, and invasion by activating the Wnt/β-catenin signaling pathway (Zhu et al., 2018). EMT is an important process in which epithelial cells are transitioned into mesenchymal phenotype to gain invasive and metastatic capacities, thus worsening cancer patients’ survival rate (Pastushenko and Blanpain, 2019). CRNDE has been associated with the development of OS tumors, owing to its ability to trigger glycogen synthase kinase-3 (GSK-3) phosphorylation and consequent activation of Wnt/β-catenin target genes c-Myc and CCND1 expression (Ding et al., 2020). EZH2 is a key component of this pathway, acting as a negative regulator of Wnt/β-catenin signaling. EZH2 is known to interact with several components of the Wnt/β-catenin signaling pathway, including TCF/LEF transcription factors, β-catenin, and the co-activator CBP/p300, thus playing a crucial role in modulating their activity. EZH2 also interacts with other proteins involved in the Wnt/β-catenin pathway, such as GSK-3, which is involved in the phosphorylation and degradation of β-catenin. It was found that many lncRNAs can affect the Wnt/β-catenin pathway by regulating EZH2. For instance, MALAT1 can inhibit the expression of E-cadherin and promote the expression of β-catenin due to MALAT1-induced EZH2 activation (Zhao et al., 2022). Moreover, it has also been reported that TUG1 is involved in OS development, via mediating miRNA-144-3p/EZH2/Wnt/β-catenin pathway (Cao et al., 2017).
3.1.4 Regulation of cell cycle
The dysregulation of the Wnt/β-catenin signaling pathway has been implicated in the disruption of cell cycle progression, which can lead to uncontrolled cellular proliferation (Ameratunga et al., 2019). Several studies have sought to uncover the association of lncRNAs with the Wnt signaling-driven cell growth in OS. For instance, BE503655 has been demonstrated to exert its effects on OS cell proliferation through regulation of the Wnt/β-catenin pathway. Silencing of BE503655 arrested OS cells in G0 and G1 phases and downregulated β-catenin and a range of Wnt cascades downstream targets, such as c-Myc, Cyclin D and MMP2(Huang et al., 2019). The lncRNA HOTTIP is the first lncRNA documented to regulate Wnt expression in OS. Its upregulation stimulates OS cell proliferation and cell cycle progression via activation of the Wnt/β-catenin pathway (Li Z. et al., 2016).
3.1.5 Regulation of CSCs
Studies are increasingly suggesting a crucial role of lncRNAs in maintaining the stemness of CSCs (Shen et al., 2021). For instance, lncRNAs distal-less homeobox 6 antisense 1 (DLX6-AS1) was found to be highly expressed in OS tissue and cell lines. Moreover, its expression was positively correlated with tumor progression and metastasis, as well as poor prognosis. Mechanistically, DLX6-AS1 might act as a competing endogenous RNA for miR-129-5p to upregulate DLK1, consequently activating the Wnt pathway and promoting the stemness of OS CSCs (Zhang RM. et al., 2018).
3.2 The PI3K/AKT/mTOR pathway
The PI3K/AKT/mTOR pathway is a signaling pathway involved in the regulation of a variety of cellular processes such as growth, survival, metabolism, and motility in normal and cancer cells. This pathway is regulated by the phosphoinositide 3-kinase (PI3K) enzyme and its downstream effectors, the serine/threonine kinase AKT (also known as PKB) and the mechanistic target of rapamycin (mTOR) (Fruman et al., 2017). Researchers discovered that lncRNAs may exert their roles in OS evolution by targeting different components of this pathway. For example, HOTAIR silencing was found to inhibit the proliferation of MG-63 cells by inducing G1 phase arrest, which was associated with the decreased phosphorylation of mTOR and its upstream kinase AKT (Li E. et al., 2017). On the other hand, MALAT1 was found to be upregulated in both OS tissues and cell lines compared to normal controls, and its overexpression was associated with the proliferation, migration, and upregulation of stemness markers such as CD133, CD90 and SOX2 (Cai et al., 2016; Gao and Lian, 2016). Mechanistically, MALAT1 was found to bind competitively to miRNA-129-5p, blocking miRNA-129-5p-mediated degradation of RET, and thus activating the PI3K/AKT signaling pathway (Chen et al., 2018). LINC00313 has been reported to suppress PTEN expression, leading to AKT phosphorylation and promoting cell proliferation and metastasis in vitro and in vivo (Xing et al., 2022). Additionally, DANCR, an oncogenic lncRNA, has been found to promote the expression of the receptor tyrosine kinase AXL, activating the PI3K/AKT signaling pathway (Jiang et al., 2017). Furthermore, FER1L4 knockdown increases PI3K levels and AKT phosphorylation, consequently facilitating cell apoptosis and inhibiting EMT (Ye et al., 2019). Conversely, LINC00619 has been shown to inhibit the proliferation, migration, and invasion of OS cells by inactivating the HGF-mediated PI3K/AKT signaling pathway (Zi et al., 2022). Other lncRNAs such as UCA1(Ma et al., 2019), LINC00968(Liu et al., 2018), LINC00628(He et al., 2018), HULC (Kong and Wang, 2018), CCAT2 (Liu J. et al., 2019), PVT1 (Sun et al., 2019), LOXL1-AS1 (Chen et al., 2019), CBR3-AS1 (Yao et al., 2022) and DBH-AS1 (Liu ZB. et al., 2019) have also been implicated in the pathogenesis and progression of OS through the regulation of the PI3K/AKT signaling pathway.
3.3 The NF-κB pathway
Nuclear factor-κB (NF-κB) is a pivotal transcription factor that regulates cell growth, metastasis, and inflammation in cancer (Liu et al., 2015; Taniguchi and Karin, 2018). IκB (inhibitor of NF-κB), which binds to NF-κB in the cytoplasm and serves as a major brake to restraining NF-κB in an inactive form, is subject to phosphorylation by IκB kinases (IKK) after being activated by multiple stimuli, leading to its breakdown and eventual NF-κB activation (Taniguchi and Karin, 2018). Recently, a study concerning OS revealed that NKILA, a tumor suppressor that interferes with the NF-κB pathway in various cancer types, has lower expression levels in OS tissues and is associated with metastasis, tumor size, and the Enneking stage. Further investigation demonstrated that increased expression of NKILA suppresses the migration and invasion of OS cells by directly impeding the phosphorylation of IκB, thus inhibiting IKK phosphorylation and NF-κB activation (Liu M. et al., 2019). In addition, patients with high H19 expression had a shorter survival rate than those with low expression. H19 knockdown has been shown to reduce OS cell migration and invasion by affecting the levels of p-PI3K, p-AKT, and IκBα with no alteration in PI3K and AKT, suggesting that H19 knockdown interdicts migration and invasion of OS cells through blocking the NF-κB pathway (Zhao and Ma, 2018). Moreover, lncRNA NORAD (Xie et al., 2020) and XIST (Lu et al., 2017) have been reported to control cellular biological processes in OS by stimulating the NF-κB signaling pathway.
3.4 The Notch pathway
Notch signaling is known to be indispensable for development and is implicated in neoplastic transformation in multiple cancers. The Notch ligand binds to the Notch receptor on the surface of a contiguous target cell, thereby initiating the Notch signaling cascade. This cascade is then transmitted through the Notch effectors, the Notch-responsive transcription factors, and downstream target genes to determine cell fate through the regulation of cell proliferation, differentiation, and apoptosis processes (Radtke and Raj, 2003; Zhang et al., 2008; Takebe et al., 2014). Studies have demonstrated that the Notch pathway contributes to the pathogenesis of OS(Engin et al., 2009; Tao et al., 2014). The oncogenic role of CRNDE in OS is suggested to be mediated by regulating Notch1 signaling and EMT (Li et al., 2018b). Furthermore, SNHG12 promotes OS oncogenesis and metastasis by sponging miR-195-5p and consequently activating Notch2 (Zhou S. et al., 2018). On the contrary, tumor suppressor lncRNA MEG3 suppresses the proliferative and metastatic capabilities of MG63 and U2OS cells by reducing protein expressions of Jagged1, Notch1 and NICD1(Chen et al., 2020). Additionally, tumor suppressor lncRNA CEBPA-AS1 overexpression impedes OS progression by upregulating NCOR2 and attenuating Notch signaling pathway activation (Xia et al., 2020).
3.5 The Hippo signaling pathway
The Hippo signaling pathway is firmly established as a crucial regulator of tissue homeostasis and is implicated in the development of multiple human cancers, including OS (Badouel and McNeill, 2011; Morice et al., 2020). Activation of the Hippo kinase cascade leads to the phosphorylation of transcriptional co-activators YAP and TAZ, causing the retention of these proteins in the cytoplasm and subsequent degradation (Ji et al., 2017). Conversely, when the Hippo pathway is inactive, dephosphorylated YAP/TAZ are translocated to the nucleus, where they bind to the TEAD family and other transcription factors, thus stimulating gene transcription and consequently promoting tumorigenesis through cell proliferation and inhibition of apoptosis (Gumbiner and Kim, 2014). Recently, lncRNAs have been demonstrated to be involved in critical cancer-related signaling pathways, such as the Hippo signaling pathway. For instance, SOX2 and MYC, two downstream effectors of the Hippo pathway, are transcriptionally activated by ZMIZ1-AS1, resulting in augmented OS growth in vivo due to the stabilization of ZMIZ1 (Zhou et al., 2022). Moreover, silencing B4GALT1-AS1 impedes OS cell proliferation, migration, stemness, and chemotherapeutic sensitivity. Mechanistically, B4GALT1-AS1 recruits HuR to prolong YAP mRNA stability and thus its transcriptional activity (Li et al., 2018c). Additionally, LINC01089 impedes the progression of OS by activating the Hippo pathway (Zhang et al., 2022).
3.6 The HIF-1α pathway
As the most influential modulator in the reaction to hypoxia, hypoxia-inducible factor-1alpha (HIF-1α) has been corroborated to control hypoxic gene expression through diverse cellular components, including lncRNAs. LincRNA for kinase activation (LINK-A), an oncogenic activator of HIF1α signaling in breast cancer (Lin et al., 2016), displays a greater expression of plasma circulating LINK-A in metastatic OS patients in comparison to non-metastatic OS patients, demonstrating its potential role in OS metastasis. Augmented expression of LINK-A is shown to inhibit the expression of HIF1α. This inhibition can be reversed by treatment with the HIF inhibitor LW6, resulting in improved migration and invasion of OS cells. Hence, these results suggest that LINK-A might serve as an upstream activator of HIF1α to trigger the metastasis of OS (Zhao B. et al., 2019). TUG1 acts as a ceRNA to annul the endogenous effect of miR-143-5p, which repressed HIF1α expression to regulate OS cell metastasis (Yu et al., 2019).
3.7 Other signaling pathways
In addition to the signaling pathways already elucidated, some pathways are also correlated with lncRNAs in OS. The Mitogen-activated protein kinase (MAPK) signaling pathway is a pivotal modulator of cellular processes. Zinc finger antisense 1 (ZFAS1) has been reported to upregulate OS cells and promote colony formation, migration, and invasion through the activation of the MAPK signaling pathway (Liu CW. et al., 2019). Moreover, lncRNA H19, a key regulator of OS pathogenesis, is regulated by Hedgehog signaling and Yap1 overexpression (Chan et al., 2014). Additionally, LINC01116 is identified to upregulate IL6R in OS via targeting miR-520a-3p and stimulating the JAK/STAT signaling pathway (Zhang B. et al., 2018). With ongoing research, we may gain a better understanding of the significance of lncRNAs in OS pathogenesis by exploring their correlation with various cellular signaling pathways.
4 LncRNAs in OS drug resistance
The management of OS usually involves neoadjuvant chemotherapy, surgical excision, and post-operative adjuvant chemotherapy. Nevertheless, the effectiveness of this traditional approach is often impeded by drug resistance, which encompasses both primary and acquired resistance (Holohan et al., 2013; Housman et al., 2014). Primary resistance is innate, whereas acquired resistance is a result of the adaptability of cancer cells, and is driven by various aspects such as drug efflux, aberrant drug metabolism, autophagy, apoptosis, and upregulation of MDR-associated genes (Holohan et al., 2013; Leary et al., 2018). Common chemotherapeutic drugs used include cisplatin, doxorubicin, methotrexate, and gemcitabine (GEM) (Isakoff et al., 2015; Zheng et al., 2018). Of these, the emergence of secondary resistance to DOX and DDP is particularly concerning. Recently, several ncRNAs have been identified with the potential to impact chemosensitivity or chemoresistance in OS (Figure 3).
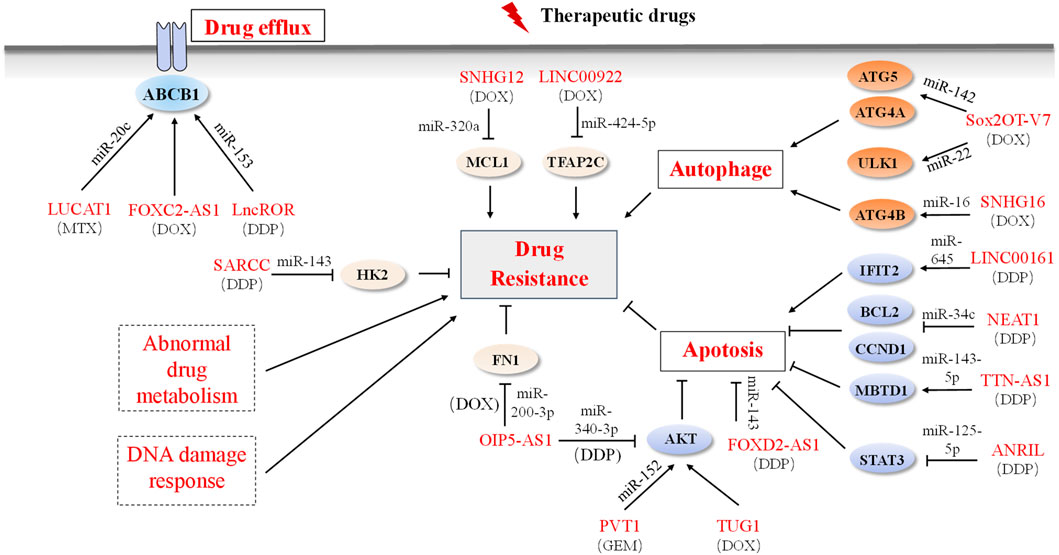
FIGURE 3. LncRNAs and mechanisms of drug resistance in OS. Dysregulated lncRNAs are involved in OS drug resistance by directly binding to related proteins or sponging miRNAs to alter the expression of downstream target genes involved in the drug efflux system, apoptosis, autophagy, and drug resistance-related signaling pathways. Solid boxes indicate the reported mechanism and dashed boxes indicate no OS-related lncRNAs participation mechanism.
4.1 Drug efflux
The ATP-binding cassette (ABC) transporter family of proteins, which is implicated in ATP-dependent drug efflux, is essential for the maintenance of drug concentrations and successful treatments (Kathawala et al., 2015; Serra et al., 2021). However, augmented outflow and diminished inflow of drugs can have an antithetical effect, resulting in treatment failure (Gottesman et al., 2002; Belisario et al., 2020). LncRNAs have been demonstrated to modulate the expression of ABCs, conferring resistance in tumor cells (Ma et al., 2016). For example, lncRNA LUCAT1 has also been shown to modulate the classical drug resistance-related ABC subfamily B, member 1 (ABCB1) via sponging of miR-200c, leading to methotrexate resistance in OS (Han and Shi, 2018). Additionally, FOXC2-AS1 and its antisense transcript FOXC2 are observed to be highly expressed in DOX-resistant OS cell lines and tissues, predicting a poor prognosis. FOXC2 promotes chemotherapy resistance by inducing ABCB1 (Zhang CL. et al., 2017). The lncRNA ROR (regulator of reprogramming), markedly elevated in MG63/DDP and U2OS/DDP cells, as well as in relapsed OS tissues, is implicated in DDP resistance in OS through the miR-153/ABCB1 axis (Cheng et al., 2019).
4.2 Apoptosis
Apoptosis is a process by which a cell undergoes programmed cell death. There are two pathways by which this occurs: an intrinsic pathway, which is regulated by proteins of the BCL-2 family, caspase-9, and AKT, and an extrinsic pathway, which is mediated by death receptors on the cell surface. Ultimately, the intrinsic and extrinsic pathways converge and activate caspase-3, resulting in apoptosis (Elmore, 2007). Apoptosis resistance is a key factor in cancer cells’ ability to evade the effects of chemotherapeutic treatment (Song et al., 2009). LncRNAs have been demonstrated to modulate drug resistance by regulating apoptosis. For example, LINC00161 is induced by DDP and upregulates IFIT2, reversing the DDP-resistant phenotype of OS cells. Mechanistic studies show that LINC00161 sponges endogenous miR-645, leading to an increase in IFIT2 expression and enhanced DDP-induced apoptosis (Wang et al., 2016). On the contrary, NEAT1 has been observed to reduce sensitivity to DDP and impede DDP-induced apoptosis and cell cycle arrest, possibly by targeting miR-34c and disabling anti-apoptotic signals of the BCL-2 pathway and cell cycle-related cyclin D1 (CCND1) signals (Hu Y. et al., 2018). Similarly, LncRNA ANRIL, upregulated in OS, has been found to confer DDP resistance to U2OS and Saos-2 cells by targeting miR-125a-5p/STAT3 (Li and Zhu, 2019), while ANRIL knockdown significantly enhances DDP-induced cell apoptosis (Lee et al., 2021). LncRNA TTN-AS1 has been reported to increase the expression of MBTD1 by targeting miR-134-5p, thereby regulating cell growth, apoptosis, and drug resistance in OS (Fu et al., 2019). Furthermore, knockdown of lncRNA FOXD2-AS1 attenuates drug resistance, stimulates apoptosis, and weakens the invasion and migration capabilities of OS cells, plausibly due to the inhibition of miR-143 expression in drug-resistant cell lines (Zhang et al., 2021). Moreover, overexpression of lncRNA TUG1, a natural anti-tumor agent, is discovered in DOX-resistant OS cells, while downregulation of TUG1 by polydatincan promotes DOX-induced apoptosis in vitro and inhibits tumor growth in the DOX-resistant model in vivo by suppressing the AKT signaling pathway (Zhou Q. et al., 2020). Additionally, OIP5-AS1 inhibits apoptosis and stimulates various types of drug resistance by sponging different miRNAs. For example, OIP5-AS1 induces DDP resistance in OS by activating the LPAATβ/PI3K/AKT/mTOR signaling pathway through sponging miR-340-5p (Song et al., 2019). Another study reveals that OIP5-AS1confers DOX resistance in OS by upregulating fibronectin 1 (FN1) expression via sponging miR-200b-3p. This effect is reversed by inhibition of FN1, enhancing the sensitivity of OS cells to DOX both in vitro and in vivo (Kun-Peng et al., 2019). Moreover, lncRNA PVT1 plays a contributory role in the GEM resistance of OS cells through c-MET/PI3K/AKT pathway activation, which is largely dependent on miR-152 (Sun et al., 2019).
4.3 Autophagy
Autophagy is a lysosome-mediated process of intracellular degradation and recycling. It has been linked to drug resistance in cancer cells as it degrades pro-apoptotic proteins that might otherwise induce apoptosis. Autophagy can also increase cancer cell survival in response to drug treatments by degrading potentially harmful substances (Li YJ. et al., 2017). LncRNAs regulate autophagy and play an important role in drug resistance. For example, lncRNA Sox2OT-V7 contributes to the chemoresistance of OS to DOX (Wang W. et al., 2018). The underlying mechanism is that Sox2OT-V7 directly targets tumor-suppressive miR142/miR-22 to inhibit their expression, leading to the inhibition of downstream core ATG genes and consequently promotes DOX-induced autophagy and chemoresistance in OS cells (Zhu K. et al., 2020). Similarly, SNHG16 confers resistance to DDP in OS Saos-2 cells by upregulating ATG4B via miR-16, which induces autophagy (Liu Y. et al., 2019).
4.4 Abnormal drug metabolism
Abnormal drug metabolism is another potential mechanism of drug resistance in OS; lncRNAs can affect the expression of key metabolic enzymes, such as the cytochrome P450 (CYP) system, glutathione-S-transferase (GST) superfamily, and uridine diphosphate glucuronosyltransferase (UGT) superfamily, which play a role in the activation and inactivation of drugs (Michael and Doherty, 2005). Notably, tumor cells can increase their resistance to drugs by reducing their activation, as many anticancer drugs require metabolic activation (Housman et al., 2014). However, research into chemoresistance in OS has largely focused on drug efflux and cell death induction; the interplay between lncRNAs and drug-metabolizing enzymes has yet to be elucidated.
4.5 DNA damage response
The DNA damage response (DDR) is activated when cellular DNA is exposed to physical, chemical or biological factors, thus ensuring chromosomal stability (Jackson and Bartek, 2009). Chemotherapeutic agents are capable of inducing DNA damage and cytotoxicity; when repairable, this damage may either enable tumor cells to survive or become more resilient to chemotherapeutic agents. To our knowledge, chemotherapeutic drugs generate either direct (DDP) or indirect (DOX) DNA damage. Previous studies have revealed an intricate interplay between DDR genes and lncRNAs in cancer (Liu et al., 2013; Sharma et al., 2015). However, the association between lncRNAs and DDR in OS remains unexplored.
4.6 Other signal pathways involved
Apart from mediating drug resistance in OS via the mechanisms mentioned above, some lncRNAs have been identified as involved in other genes associated with drug resistance. For instance, LncRNA NORAD has been shown to inhibit the proliferation of OS HOS/DDP cells by targeting miR-410-3p, increasing their sensitivity to DDP (Xie et al., 2020). Furthermore, studies in both in vivo and in vitro settings have confirmed the role of lncRNA SARCC in enhancing OS DDP sensitivity. It can inhibit glycolysis by regulating Hexokinase 2 (HK2), a direct target of miR-143 in OS(Wen et al., 2020). Additionally, LINC00922 and SNHG12 have been revealed to contribute to OS DOX resistance via the miR-424-5p/TFAP2C axis and miR-320a/MCL1 axis respectively (Zhou B. et al., 2018; Gu et al., 2020). Nonetheless, further research is required to elucidate the exact mechanisms by which these lncRNAs are involved in drug resistance in OS and to explore ways to target them therapeutically.
Aberrantly expressed lncRNAs in OS show promising ability to predict chemotherapy response. Consequently, dynamic monitoring of these lncRNA levels may be a useful strategy for accurately assessing chemotherapy response in OS patients. Simultaneously, altering the expression of pertinent lncRNAs may be a strategy for reversing chemoresistance. In summary, combinatorial targeting of lncRNAs and conventional chemotherapy offers a novel approach for overcoming chemoresistance in OS, allowing for the selection of the best regimens and increased the efficacy of chemotherapy.
4.7 Regulatory network of lncRNA-miRNA-mRNA in OS
Compelling evidence highlights the regulatory network of lncRNA-miRNA-mRNA in OS. This competition for miRNA binding can influence the expression of genes involved not only in drug resistance but also in OS progression and outcome. Fascinatingly, it is noteworthy that a singular lncRNA can manifest diverse roles through its interactions with distinct miRNAs. For instance, the lncRNA PVT1 augments the chemoresistance of OS to GEM via its interaction with miR-152 (Sun et al., 2019). Simultaneously, PVT1 engages in miR-183-5p/ERG axis to propel the proliferation and metastasis of OS (Zhao W. et al., 2019). Likewise, the lncRNA NEAT1 has been observed to attenuate the sensitivity of OS cells to DDP and hinder DDP-induced apoptosis by targeting miR-34c and its downstream BCL-2 and CCND1 signals (Hu Y. et al., 2018). Additionally, NEAT1 facilitates the EMT and metastasis of osteosarcoma cells by sequestering miR-483, thereby upregulating STAT3 expression (Chen et al., 2021). Moreover, DANCR is engaged in miR-33a-5p/AXL (Jiang et al., 2017), miR-335-5p and miR-1972/ROCK1 axis (Wang et al., 2018b), thereby modulating OS progression through different signaling pathways. Given the substantial length of lncRNAs, it is plausible that distinct lncRNAs may share interaction sites with a particular miRNA. For instance, both GAS5 (Wang et al., 2021) and PINT (Liu, 2019) have shown the capacity to interact with miR-21, thereby exerting their tumor-suppressive roles.
It is worth highlighting that the utilization of a dual luciferase reporter assay and a RIP assay furnishes tangible evidence that fortify the ceRNA hypothesis. Nevertheless, the precise mechanistic model of ceRNA remains a subject of contention, with varying conclusions due to the abundance and affinity of these intracellular RNAs in ceRNA crosstalk. Several experimental investigations have unveiled the interplay between some lncRNAs and their ability to either foster or restrain tumorigenesis through distinctive ceRNA regulatory axes and subsequent downstream pathways. Further investigations are much warranted to unveil the intricacies inherent in lncRNA-miRNA-mRNA and/or lncRNA-protein networks. These captivating regulatory axes orchestrated by lncRNAs hold immense promise as novel therapeutic targets for OS. Here, we briefly describe the expression of some representative lncRNAs and their roles in OS (Table 1).
5 Potential application of lncRNAs in OS
The current diagnosis of OS heavily relies on imaging tests, including X-rays, CT, MRI, and PET scans, which provide information on the tumor’s size, location, and spread within the body (Meyer et al., 2008; Strauss et al., 2021). However, despite the molecular features that identify patients at higher risk, such as RB1 loss, MYC amplification, and VEGFA amplification, none of these factors have been validated sufficiently to serve as a basis for risk stratification in clinical settings. Furthermore, the lack of reliable biomarkers and limited therapeutic options contributes to the high mortality rate of OS. Surgical resection remains the only possible curative approach for early OS, while patients with unresectable or widely metastatic OS are deemed incurable. Therefore, there is an urgent need to identify novel biomarkers and therapeutic targets for the improved diagnosis and treatment of OS.
5.1 LncRNAs as diagnostic biomarkers in OS
As previously discussed, many lncRNAs are aberrantly expressed in OS compared with normal tissue, which are useful to distinguish OS patients from healthy cohorts. Several studies reveal that the levels of OS-related lncRNAs can affect the activity, proliferation, and differentiation of osteoblasts, thereby directly reflecting the pathological staging and prognosis of patients. So far, Enneking staging has become an important basis for surgical staging of OS and is a better indicator of prognosis, aiding in the selection of appropriate treatment (Jawad and Scully, 2010). Various lncRNAs are associated with the clinical stage of Enneking, with higher malignancy correlating with higher stages and increased risk of distant metastasis. For instance, GAPLINC expression is increased in OS tissues and cell lines and is associated with poor histological grade, distant metastasis, and advanced Enneking stage. Increased GAPLINC expression is found to be an independent risk factor and inversely correlated with overall survival (Liao et al., 2018). Similarly, lncRNA GNAS-AS1 significantly increase in OS cells and tissues, which were positively correlated with the Enneking stage and distant metastasis (Mi et al., 2021). Moreover, lncRNA TUG1 (Yu et al., 2019), HOTAIR (Wang B. et al., 2015), DLX6-AS1 (Zhang N. et al., 2019) and HOTTIP (Li et al., 2015) are prominently increased in OS, which are associated with tumor size, distant metastases, TNM stage, overall survival and recurrence-free survival. These lncRNA biomarkers may provide a valuable method for the diagnosis and clinical staging of OS, offering a useful complement to existing biomarkers for more accurate clinical guidance.
Liquid biopsy is significantly less invasive than sampling solid tumor tissues; as a result, it makes an excellent diagnostic strategy. Intriguingly, several lncRNAs are identified as being cancer-specific and detected in body fluids (whole blood, plasma, urine, gastric juice, and saliva), which makes them promising biomarkers for cancer diagnosis and prognosis (Qi et al., 2016). The ability of lncRNAs to be released from cells and found encapsulated in exosomes in the extracellular space and bodily fluids is one of the confirmed methods. Recent research demonstrates that lncRNAs can be found in exosomes that have been extracted from the bodily fluids of cancer patients with a variety of malignancies (Li S. et al., 2018; Zhou R. et al., 2018; Li and Wang, 2021). As an alternative to traditional biomarkers, lncRNA-based tests in the circulatory system can be considered. Some plasma lncRNAs are linked to the growth and progression of tumors and may act as non-invasive circulating biomarkers for the detection of OS. Their classification, length, cellular localization, and mechanism of action have been discussed in another review (Moonmuang et al., 2021). For instance, UCA1 expression is much higher in OS tissues than in normal bone tissues, and it was also significantly higher in the serum of patients with OS than in the serum of healthy controls. Additionally, the tumor status, metastasis, and worse prognosis were all associated with its overexpression (Wen et al., 2017). Receiving operating characteristic (ROC) curve study shows that HNF1A-AS1 may distinguish patients with OS from healthy people. Upregulation of lncRNA HNF1A-S1 in OS serum is related to patients’ status (Cai et al., 2017). FAL1 may serve as a potential diagnostic marker for OS because it has recently been discovered that it is elevated in OS and that its serum level is related to the condition of the disease (Wang et al., 2018c). The low degree of invasiveness, affordability and timesaving procedures make a few circulating lncRNAs, whose expression coincides with the clinical stage of the disease, promising novel biomarkers to be added to current clinical practices for the management of OS.
5.2 LncRNAs as prognostic biomarkers in OS
Pulmonary metastasis and drug resistance are the two main factors that adversely affect overall survival prognosis. To improve existing treatment plans and establish the optimal timing for chemotherapy and surgery, the connection between lncRNAs biomarkers and OS prognosis must be explored. It has been found that lncRNAs have significant effects on cancer metastasis and medication sensitivity. Research shows that lncRNAs have a substantial impact on cancer metastasis and medication sensitivity. For instance, LncRNA BC050642 is found to be considerably more abundant in OS tissues and cell lines, and it was also proposed as an independent biomarker of OS prognosis (Yang Y. et al., 2019). The highly elevated LINC01614 in OS tissues predicts a poorer prognosis for patients. Knocking down LINC01614 inhibits OS cell proliferation, invasion, and metastasis. The miR-520a-3p/SNX3 regulatory axis, considered a potential clinical prognostic indicator for OS (Cai et al., 2021), is responsible for this effect. Additionally, TTN-AS1 may act as a carcinogenic agent by disrupting the miR-134-5p/MBTD1 gene and, given its high expression, can serve as a biomarker correlated with a poor prognosis (Fu et al., 2019). Other lncRNAs, such as FRFR-AS1 (Sun et al., 2016), BCAR4 (Wei et al., 2019), ZEB-AS1 (Liu and Lin, 2016), MALAT1 (Liu M. et al., 2019), DICER1-AS1 (Hu XH. et al., 2018), EWSAT1 (Zhang GY. et al., 2017), SOX2-OT (Wang et al., 2017) CCAL (Zhou et al., 2017), ATB (Han et al., 2017), and MEG3 (Shen et al., 2019), have been identified as potential prognostic biomarkers of OS. Although some lncRNAs have been pinpointed as prognostic markers of OS, their functions have yet to be validated and implemented in clinical practice. Thus, it is essential to identify lncRNA biomarkers for the prognosis of OS, particularly for patients with metastasis, to enable early diagnosis and potential therapeutic targets.
5.3 LncRNAs as promising OS therapeutic targets
Currently, the standard treatment for OS is surgery to remove the tumor followed by chemotherapy. Targeted drugs are being developed to treat OS, but none have been approved yet (Martin Broto et al., 2022). Chemoresistance resistance is frequently observed during OS treatment. Therefore, the prognosis of OS has not been improved for a long time, which is at least in part due to a lack of therapeutic target and chemoresistance. Thus, a novel target is required for improving the prognosis of OS patients. LncRNAs have emerged as attractive candidates due to their cell-, tissue-, and disease-specific expression patterns, and pre-clinical studies have demonstrated promising results in the treatment of disorders, including cancer (Arun et al., 2016; Papaioannou et al., 2019). Moreover, lncRNA-targeting approaches have advantages over protein-targeting methods, as the base-pairing principle is simpler than designing a specific protein-binding inhibitor. Targeting lncRNAs can be achieved using small interfering RNA (siRNA), antisense oligonucleotides (ASOs), ribozymes, aptamers, and miRNAs, which have all been evaluated for targeting cancer-associated genes (Parasramka et al., 2016; Pierce et al., 2022). For instance, Arun et al. administered subcutaneously injected mice with 16-mer gapmer targeting MALAT1 modified with alternative cEt-constrained nucleotides and phosphorothioate bonds before the spontaneous development of mammary tumors, resulting in a 50% reduction in metastatic burden, and a highly differentiated, cystic/ductular histological phenotype resembling MALAT1−/− genomic knockout mice (Arun et al., 2016). Similarly, patient-derived xenograft (PDX) mouse models demonstrated that the treatment with lipid nanoparticle (LNP)-formulated anti-HOXB-AS3 gapmers effectively downregulated HOXB-AS3 expression in human CD45-selected AML blasts isolated from the bone marrow of the treated mice. This treatment significantly prolonged the survival of the treated mice compared to the control group, with the median survival of the scramble-treated mice being only 63 days, whereas it was not reached after 100 days of treatment for the HOXB-AS3 KD group (Papaioannou et al., 2019).
Drug resistance-associated lncRNAs, such as LUCAT1(Han and Shi, 2018), ANRIL (Lee et al., 2021), Sox2OT-V7 (Zhu K. et al., 2020) and others, may be useful as therapeutic targets to reverse chemotherapy resistance in the future. In addition, lncRNAs, such as LINC00588 (Zhou FC. et al., 2020), GAPLINC (Liao et al., 2018), HOTAIR (Wang B. et al., 2015) and DNACR (Wang et al., 2018b), which are up- or downregulated in OS and regulate OS metastasis, could be targets for therapeutic intervention. For instance, lncRNA LINC00588 is expressed differently in cancer in bone and metastatic carcinoma in the lung and may act as a ceRNA to affect the expression of TP53. Thus, the LINC00588/miRNA-1972/TP53 axis may serve as a potential therapeutic target in OS patients (Zhou FC. et al., 2020). Overall, targeting these lncRNAs could be used to reverse drug resistance or prevent OS metastasis. However, further clinical trials and transformation research are needed to determine which lncRNAs are key targets and to elucidate the mechanism of lncRNAs in OS drug resistance.
6 Conclusion
LncRNAs, a prominent topic in RNA biology and oncology, are involved in multiple aspects of OS progression and development. Recent studies have revealed that alterations in transcription or processing can lead to alterations in the expression of both oncogenic and tumor-suppressive lncRNAs. These lncRNAs bind to DNA, RNA, or proteins, affecting the expression, localization, stability, and activity of their binding partners, thus driving cancerous phenotypes such as persistent proliferation, metastasis, increased stemness, and drug resistance, resulting in tumorigenesis and OS progression. As a result, lncRNAs present in bodily fluids can be utilized as biomarkers for OS, offering high sensitivity, or in combination with other molecules to enhance clinical efficacy and accuracy. Further research into lncRNA roles and functions in OS may lead to novel strategies to modulate lncRNA expression and benefit OS treatment and management.
Functional studies of lncRNAs can be challenging due to their overall lower sequence conservation compared with protein-coding genes (Necsulea et al., 2014). Recent research has revealed that lncRNAs tend to maintain evolutionarily conserved secondary structures in the presence of considerable sequence variation (Juan et al., 2000). It has been suggested that selective pressure may act on lncRNAs at the secondary structure level to constrain sequence variation, resulting in reduced mutations in structurally critical regions (Pegueroles and Gabaldón, 2016). This contrasts with protein-coding genes, which are generally more highly conserved since even a single nucleotide insertion or deletion can cause a frameshift and complete loss of function. This implies that the secondary structure of lncRNAs may not only be an evolutionary constraint but also the functional unit involved in biological processes. However, the secondary structure of lncRNAs has not been sufficiently studied, nor has the relationship between lncRNAs secondary structure and its biological functions been revealed. Another study also points out that lncRNAs exhibit common motifs and motif combinations, conserved secondary structures, and small sequence elements that are key determinants of their function (Ross et al., 2021). Despite the progress made in understanding lncRNA structure-function relationships, much more research needs to be done to comprehend the structure and functionality of the thousands of lncRNAs. Moreover, a multitude of investigations have revealed the pivotal role of ncRNAs, particularly circRNAs (Qin et al., 2023; Zeng et al., 2023) and lncRNAs (Wang et al., 2020), as ceRNA in the regulation of malignant biological properties, encompassing cell proliferation, invasion, metastasis, and chemotherapy resistance in OS (Table 1). However, the direct experimental evidence elucidating the involvement of ceRNA remains insufficient, necessitating further exploration into the intricate mechanisms governing their regulation.
Another concern when studying lncRNAs is the absence of effective tools to manipulate their expression or function. Traditional methods for studying gene function, such as gene knockout or overexpression, are often not possible for lncRNAs due to their large size and complex structure. Additionally, lncRNAs often lack conserved functional domains or motifs, making it difficult to predict their function based on sequence alone. Thus, CRISPR/Cas9-mediated knockout (Yang et al., 2018) is often not suitable for the study of lncRNAs because lncRNAs have biological functions that are less likely to be perturbed by small indels. Moreover, RNA interference (RNAi) (Heale et al., 2005), and antisense oligonucleotide (ASO) (Meng et al., 2015) and Cas13-based methods (Zhang et al., 2020) for knockdown have a significant risk of false positives caused by off-target effects, and the introduction of exogenous molecules may be toxic to organisms. Meanwhile, CRISPR interference (CRISPRi) works by using a deactivated Cas9 (dCas9) protein fused to a transcriptional repressor domain to selectively silence the expression of a particular lncRNA without affecting neighboring genes or ncRNAs. The advantages of CRISPRi over other methods for modulating lncRNA expression include its specificity, reversibility, and the potential to reveal the function of lncRNAs. However, more accurate mapping of lncRNA TSSs, optimizations to the design of sgRNA sequences, and potential off-target effects are still needed.
Obtaining reliable clinical biomarkers for early diagnosis and treatment of OS is a key issue. Circulating lncRNAs may be more reliable cancer biomarkers than CTCs, cfDNA (including circulating tumor DNA, ctDNA) and exosomes due to their high stability and relative abundance. They may be used as a biomarker for early diagnosis of OS; however, challenges remain in establishing a real-time and minimally invasive monitoring system using circulating lncRNA signatures. The main challenge at this stage is the difficulty in meeting the demand for both sensitivity and specificity. Circulating lncRNAs may struggle with sensitivity and specificity when targeting specific cancer species for indicated detection. Thus, a combination of several blood-based lncRNAs for signature offers better diagnostic performance than most individual circulating lncRNAs. For example, a signature composed of three lncRNAs (PTENP1, LSINCT-5, and CUDR) significantly outperforms CEA and CA19-9 in gastric cancer diagnosis studies (Dong et al., 2015). Additionally, new technologies, such as dual electrochemical genosensor (Sánchez-Salcedo et al., 2021), can help advance circulating lncRNAs as tumor markers for clinical applications. Moreover, to improve feasibility as a biomarker for OS, larger and more diverse cohorts are also needed to confirm its diagnostic potential.
Author contributions
XL: Writing–original draft, Writing–review and editing. RW: Writing–original draft. JXZ and KW: Writing–review and editing. JL: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Science and Technology Foundation of Sichuan Province, China (24NSFSC1838, 2022YSF0211, and 2022YFS0091), and the National Natural Science Foundation of China (81903082).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ameratunga, M., Kipps, E., Okines, A. F. C., and Lopez, J. S. (2019). To cycle or fight-CDK4/6 inhibitors at the crossroads of anticancer immunity. Clin. Cancer Res. 25 (1), 21–28. doi:10.1158/1078-0432.ccr-18-1999
Arab, K., Park, Y. J., Lindroth, A. M., Schafer, A., Oakes, C., Weichenhan, D., et al. (2014). Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell. 55 (4), 604–614. doi:10.1016/j.molcel.2014.06.031
Arun, G., Diermeier, S., Akerman, M., Chang, K. C., Wilkinson, J. E., Hearn, S., et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30 (1), 34–51. doi:10.1101/gad.270959.115
Bacci, G., Mercuri, M., Briccoli, A., Ferrari, S., Bertoni, F., Donati, D., et al. (1997). Osteogenic sarcoma of the extremity with detectable lung metastases at presentation. Cancer 79 (2), 245–254. doi:10.1002/(sici)1097-0142(19970115)79:2<245::aid-cncr7>3.3.co;2-m
Badouel, C., and McNeill, H. (2011). SnapShot: the hippo signaling pathway. Cell 145 (3), 484. doi:10.1016/j.cell.2011.04.009
Belisario, D. C., Akman, M., Godel, M., Campani, V., Patrizio, M. P., Scotti, L., et al. (2020). ABCA1/ABCB1 ratio determines chemo- and immune-sensitivity in human osteosarcoma. Cells 9 (3), 647. doi:10.3390/cells9030647
Bielack, S. S., Kempf-Bielack, B., Delling, G., Exner, G. U., Flege, S., Helmke, K., et al. (2002). Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 20 (3), 776–790. doi:10.1200/jco.2002.20.3.776
Brock, C. K., Wallin, S. T., Ruiz, O. E., Samms, K. M., Mandal, A., Sumner, E. A., et al. (2019). Stem cell proliferation is induced by apoptotic bodies from dying cells during epithelial tissue maintenance. Nat. Commun. 10 (1), 1044. doi:10.1038/s41467-019-09010-6
Cai, L., Lv, J., Zhang, Y., Li, J., Wang, Y., and Yang, H. (2017). The lncRNA HNF1A-AS1 is a negative prognostic factor and promotes tumorigenesis in osteosarcoma. J. Cell. Mol. Med. 21 (11), 2654–2662. doi:10.1111/jcmm.12944
Cai, Q., Zhao, X., Wang, Y., Li, S., Wang, J., Xin, Z., et al. (2021). LINC01614 promotes osteosarcoma progression via miR-520a-3p/SNX3 axis. Cell. Signal. 83, 109985. doi:10.1016/j.cellsig.2021.109985
Cai, X., Liu, Y., Yang, W., Xia, Y., Yang, C., Yang, S., et al. (2016). Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J. Orthop. Res. 34 (6), 932–941. doi:10.1002/jor.23105
Cao, J., Han, X., Qi, X., Jin, X., and Li, X. (2017). TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int. J. Oncol. 51 (4), 1115–1123. doi:10.3892/ijo.2017.4110
Chan, L. H., Wang, W., Yeung, W., Deng, Y., Yuan, P., and Mak, K. K. (2014). Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene 33 (40), 4857–4866. doi:10.1038/onc.2013.433
Chen, L., Wang, J., Li, J. W., Zhao, X. W., and Tian, L. F. (2020). LncRNA MEG3 inhibits proliferation and promotes apoptosis of osteosarcoma cells through regulating Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 24 (2), 581–590. doi:10.26355/eurrev_202001_20034
Chen, S., Li, W. G., and Guo, A. (2019). LOXL1-AS1 predicts poor prognosis and promotes cell proliferation, migration, and invasion in osteosarcoma. Biosci. Rep. 39, Unsp Bsr20190447. doi:10.1042/Bsr20190447
Chen, Y., Huang, W., Sun, W., Zheng, B., Wang, C., Luo, Z., et al. (2018). LncRNA MALAT1 promotes cancer metastasis in osteosarcoma via activation of the PI3K-akt signaling pathway. Cell. Physiol. biochem. 51 (3), 1313–1326. doi:10.1159/000495550
Chen, Y., Li, J., Xiao, J. K., Xiao, L., Xu, B. W., and Li, C. (2021). The lncRNA NEAT1 promotes the epithelial-mesenchymal transition and metastasis of osteosarcoma cells by sponging miR-483 to upregulate STAT3 expression. Cancer Cell Int. 21 (1), 90. doi:10.1186/s12935-021-01780-8
Cheng, F. H., Zhao, Z. S., and Liu, W. D. (2019). Long non-coding RNA ROR regulated ABCB1 to induce cisplatin resistance in osteosarcoma by sponging miR-153-3p. Eur. Rev. Med. Pharmacol. Sci. 23 (17), 7256–7265. doi:10.26355/eurrev_201909_18828
Chi, Y., Wang, D., Wang, J., Yu, W., and Yang, J. (2019). Long non-coding RNA in the pathogenesis of cancers. Cells 8 (9), 1015. doi:10.3390/cells8091015
Cui, H., and Zhao, J. (2020). LncRNA TMPO-AS1 serves as a ceRNA to promote osteosarcoma tumorigenesis by regulating miR-199a-5p/WNT7B axis. J. Cell. Biochem. 121 (3), 2284–2293. doi:10.1002/jcb.29451
Ding, Q., Mo, F., Cai, X., Zhang, W., Wang, J., Yang, S., et al. (2020). LncRNA CRNDE is activated by SP1 and promotes osteosarcoma proliferation, invasion, and epithelial-mesenchymal transition via Wnt/β-catenin signaling pathway. J. Cell. Biochem. 121 (5-6), 3358–3371. doi:10.1002/jcb.29607
Dong, L., Qi, P., Xu, M. D., Ni, S. J., Huang, D., Xu, Q. H., et al. (2015). Circulating CUDR, LSINCT-5 and PTENP1 long noncoding RNAs in sera distinguish patients with gastric cancer from healthy controls. Int. J. Cancer. 137 (5), 1128–1135. doi:10.1002/ijc.29484
Elmore, S. (2007). Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35 (4), 495–516. doi:10.1080/01926230701320337
Engin, F., Bertin, T., Ma, O., Jiang, M. M., Wang, L., Sutton, R. E., et al. (2009). Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum. Mol. Genet. 18 (8), 1464–1470. doi:10.1093/hmg/ddp057
Fruman, D. A., Chiu, H., Hopkins, B. D., Bagrodia, S., Cantley, L. C., and Abraham, R. T. (2017). The PI3K pathway in human disease. Cell 170 (4), 605–635. doi:10.1016/j.cell.2017.07.029
Fu, D., Lu, C., Qu, X., Li, P., Chen, K., Shan, L., et al. (2019). LncRNA TTN-AS1 regulates osteosarcoma cell apoptosis and drug resistance via the miR-134-5p/MBTD1 axis. Aging 11 (19), 8374–8385. doi:10.18632/aging.102325
Gao, K. T., and Lian, D. (2016). Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 20 (17), 3561–3565.
Gottesman, M. M., Fojo, T., and Bates, S. E. (2002). Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer. 2 (1), 48–58. doi:10.1038/nrc706
Gu, Z., Zhou, Y., Cao, C., Wang, X., Wu, L., and Ye, Z. (2020). TFAP2C-mediated LINC00922 signaling underpins doxorubicin-resistant osteosarcoma. Biomed. Pharmacother. 129, 110363. doi:10.1016/j.biopha.2020.110363
Gumbiner, B. M., and Kim, N. G. (2014). The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 127 (4), 709–717. doi:10.1242/jcs.140103
Hahne, J. C., and Valeri, N. (2018). Non-coding RNAs and resistance to anticancer drugs in gastrointestinal tumors. Front. Oncol. 8, 226. doi:10.3389/fonc.2018.00226
Han, F., Wang, C., Wang, Y., and Zhang, L. (2017). Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am. J. Cancer Res. 7 (4), 770–783.
Han, X., Yin, M., Gong, C., Zhang, C., Zhu, G., Hu, M., et al. (2023). A1BG-AS1 promotes the biological functions of osteosarcoma cells via regulating the microRNA-148a-3p/USP22 axis and stabilizing the expression of SIRT1 through deubiquitinase function. Expert Opin. Ther. Targets. 27 (10), 1017–1029. doi:10.1080/14728222.2023.2263908
Han, Z., and Shi, L. (2018). Long non-coding RNA LUCAT1 modulates methotrexate resistance in osteosarcoma via miR-200c/ABCB1 axis. Biochem. Biophys. Res. Commun. 495 (1), 947–953. doi:10.1016/j.bbrc.2017.11.121
He, R., Wu, J. X., Zhang, Y., Che, H., and Yang, L. (2018). LncRNA LINC00628 overexpression inhibits the growth and invasion through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 22 (18), 5857–5866. doi:10.26355/eurrev_201809_15915
Heale, B. S., Soifer, H. S., Bowers, C., and Rossi, J. J. (2005). siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res. 33 (3), e30. doi:10.1093/nar/gni026
Holohan, C., Van Schaeybroeck, S., Longley, D. B., and Johnston, P. G. (2013). Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 13 (10), 714–726. doi:10.1038/nrc3599
Housman, G., Byler, S., Heerboth, S., Lapinska, K., Longacre, M., Snyder, N., et al. (2014). Drug resistance in cancer: an overview. Cancers (Basel) 6 (3), 1769–1792. doi:10.3390/cancers6031769
Hu, G., Lou, Z., and Gupta, M. (2014). The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One 9 (9), e107016. doi:10.1371/journal.pone.0107016
Hu, X. H., Dai, J., Shang, H. L., Zhao, Z. X., and Hao, Y. D. (2018a). High levels of long non-coding RNA DICER1-AS1 are associated with poor clinical prognosis in patients with osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 22 (22), 7640–7645. doi:10.26355/eurrev_201811_16379
Hu, Y., Yang, Q., Wang, L., Wang, S., Sun, F., Xu, D., et al. (2018b). Knockdown of the oncogene lncRNA NEAT1 restores the availability of miR-34c and improves the sensitivity to cisplatin in osteosarcoma. Biosci. Rep. 38 (3). doi:10.1042/BSR20180375
Huang, J. Z., Chen, M., Chen, D., Gao, X. C., Zhu, S., Huang, H., et al. (2017). A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell 68 (1), 171–184. doi:10.1016/j.molcel.2017.09.015
Huang, Q., Shi, S. Y., Ji, H. B., and Xing, S. X. (2019). LncRNA BE503655 inhibits osteosarcoma cell proliferation, invasion/migration via Wnt/β-catenin pathway. Biosci. Rep. 39 (7). doi:10.1042/bsr20182200
Huang, X., Zhang, W., Pu, F., and Zhang, Z. (2023). LncRNA MEG3 promotes chemosensitivity of osteosarcoma by regulating antitumor immunity via miR-21-5p/p53 pathway and autophagy. Genes Dis. 10 (2), 531–541. doi:10.1016/j.gendis.2021.11.004
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21 (11), 1253–1261. doi:10.1038/nm.3981
Isakoff, M. S., Bielack, S. S., Meltzer, P., and Gorlick, R. (2015). Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 33 (27), 3029–3035. doi:10.1200/JCO.2014.59.4895
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47 (3), 199–208. doi:10.1038/ng.3192
Jackson, S. P., and Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature 461 (7267), 1071–1078. doi:10.1038/nature08467
Jawad, M. U., and Scully, S. P. (2010). In brief: classifications in brief: enneking classification: benign and malignant tumors of the musculoskeletal system. Clin. Orthop. Relat. Res. 468 (7), 2000–2002. doi:10.1007/s11999-010-1315-7
Ji, X. Y., Zhong, G., and Zhao, B. (2017). Molecular mechanisms of the mammalian Hippo signaling pathway. Yi Chuan 39 (7), 546–567. doi:10.16288/j.yczz.17-094
Jia, D., Niu, Y., Li, D., and Liu, Z. (2018). lncRNA C2dat1 promotes cell proliferation, migration, and invasion by targeting miR-34a-5p in osteosarcoma cells. Oncol. Res. 26 (5), 753–764. doi:10.3727/096504017x15024946480113
Jia, P., Cai, H., Liu, X., Chen, J., Ma, J., Wang, P., et al. (2016). Long non-coding RNA H19 regulates glioma angiogenesis and the biological behavior of glioma-associated endothelial cells by inhibiting microRNA-29a. Cancer Lett. 381 (2), 359–369. doi:10.1016/j.canlet.2016.08.009
Jiang, N., Wang, X., Xie, X., Liao, Y., Liu, N., Liu, J., et al. (2017). lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405, 46–55. doi:10.1016/j.canlet.2017.06.009
Ju, C., Zhou, R., Sun, J., Zhang, F., Tang, X., Chen, K. K., et al. (2018). LncRNA SNHG5 promotes the progression of osteosarcoma by sponging the miR-212-3p/SGK3 axis. Cancer Cell Int. 18, 141. doi:10.1186/s12935-018-0641-9
Juan, V., Crain, C., and Wilson, C. (2000). Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucleic Acids Res. 28 (5), 1221–1227. doi:10.1093/nar/28.5.1221
Kathawala, R. J., Gupta, P., Ashby, C. R., and Chen, Z. S. (2015). The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug resist. updat. 18, 1–17. doi:10.1016/j.drup.2014.11.002
Klein, M. J., and Siegal, G. P. (2006). Osteosarcoma: anatomic and histologic variants. Am. J. Clin. Pathol. 125 (4), 555–581. doi:10.1309/uc6k-qhld-9lv2-kenn
Kong, D., and Wang, Y. (2018). Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J. Cell. Biochem. 119 (1), 1050–1061. doi:10.1002/jcb.26273
Kun-Peng, Z., Chun-Lin, Z., Xiao-Long, M., and Lei, Z. (2019). Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J. Cell. Physiol. 234 (5), 6927–6939. doi:10.1002/jcp.27435
Leary, M., Heerboth, S., Lapinska, K., and Sarkar, S. (2018). Sensitization of drug resistant cancer cells: a matter of combination therapy. Cancers (Basel) 10 (12), 483. doi:10.3390/cancers10120483
Lee, A. M., Ferdjallah, A., Moore, E., Kim, D. C., Nath, A., Greengard, E., et al. (2021). Long non-coding RNA ANRIL as a potential biomarker of chemosensitivity and clinical outcomes in osteosarcoma. Int. J. Mol. Sci. 22 (20), 11168. doi:10.3390/ijms222011168
Levine, A. M., and Rosenberg, S. A. (1979). Alkaline phosphatase levels in osteosarcoma tissue are related to prognosis. Cancer 44 (6), 2291–2293. doi:10.1002/1097-0142(197912)44:6<2291::aid-cncr2820440643>3.0.co;2-s
Li, C., Wang, F., Wei, B., Wang, L., and Kong, D. (2019). LncRNA AWPPH promotes osteosarcoma progression via activation of Wnt/β-catenin pathway through modulating miR-93-3p/FZD7 axis. Biochem. Biophys. Res. Commun. 514 (3), 1017–1022. doi:10.1016/j.bbrc.2019.04.203
Li, E., Zhao, Z., Ma, B., and Zhang, J. (2017a). Long noncoding RNA HOTAIR promotes the proliferation and metastasis of osteosarcoma cells through the AKT/mTOR signaling pathway. Exp. Ther. Med. 14 (6), 5321–5328. doi:10.3892/etm.2017.5248
Li, F., Cao, L., Hang, D., Wang, F., and Wang, Q. (2015). Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int. J. Clin. Exp. pathology 8 (9), 11414–11420.
Li, G., and Zhu, Y. (2019). Effect of lncRNA ANRIL knockdown on proliferation and cisplatin chemoresistance of osteosarcoma cells in vitro. Pathol. Res. Pract. 215 (5), 931–938. doi:10.1016/j.prp.2019.01.042
Li, H., Fu, S., and Wang, W. (2023a). Long non-coding RNA HOTAIR promotes human osteosarcoma proliferation, migration through activation of the wnt/b-catenin signaling pathway. J. Oncol. 2023, 9667920. doi:10.1155/2023/9667920
Li, L., Zhang, Y., Gao, Y., Hu, Y., Wang, R., Wang, S., et al. (2023b). LncSNHG14 promotes nutlin3a resistance by inhibiting ferroptosis via the miR-206/SLC7A11 axis in osteosarcoma cells. Cancer Gene Ther. 30 (5), 704–715. doi:10.1038/s41417-022-00581-z
Li, S., Li, Y., Chen, B., Zhao, J., Yu, S., Tang, Y., et al. (2018a). exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 46 (1), D106–d112. doi:10.1093/nar/gkx891
Li, S., and Wang, X. (2021). The potential roles of exosomal noncoding RNAs in osteosarcoma. J. Cell. Physiol. 236 (5), 3354–3365. doi:10.1002/jcp.30101
Li, W., Sun, M., Zang, C., Ma, P., He, J., Zhang, M., et al. (2016a). Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 7 (5), e2225. doi:10.1038/cddis.2016.126
Li, Y. J., Lei, Y. H., Yao, N., Wang, C. R., Hu, N., Ye, W. C., et al. (2017b). Autophagy and multidrug resistance in cancer. Chin. J. Cancer. 36 (1), 52. doi:10.1186/s40880-017-0219-2
Li, Z., Tang, Y., Xing, W., Dong, W., and Wang, Z. (2018b). LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp. Mol. Pathol. 104 (1), 19–25. doi:10.1016/j.yexmp.2017.12.002
Li, Z., Wang, Y., Hu, R., Xu, R., and Xu, W. (2018c). LncRNA B4GALT1-AS1 recruits HuR to promote osteosarcoma cells stemness and migration via enhancing YAP transcriptional activity. Cell Prolif. 51 (6), e12504. doi:10.1111/cpr.12504
Li, Z., Zhao, L., and Wang, Q. (2016b). Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway. Am. J. Transl. Res. 8 (5), 2385–2393.
Lian, H., Xie, P., Yin, N., Zhang, J., Zhang, X., Li, J., et al. (2019). Linc00460 promotes osteosarcoma progression via miR-1224-5p/FADS1 axis. Life Sci. 233, 116757. doi:10.1016/j.lfs.2019.116757
Liao, S., Zhou, S., and Wang, C. (2018). GAPLINC is a predictor of poor prognosis and regulates cell migration and invasion in osteosarcoma. Biosci. Rep. 38 (5). doi:10.1042/BSR20181171
Lin, A., Li, C., Xing, Z., Hu, Q., Liang, K., Han, L., et al. (2016). The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 18 (2), 213–224. doi:10.1038/ncb3295
Liu, B., Sun, L., Liu, Q., Gong, C., Yao, Y., Lv, X., et al. (2015). A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27 (3), 370–381. doi:10.1016/j.ccell.2015.02.004
Liu, C., and Lin, J. (2016). Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am. J. Transl. Res. 8 (10), 4095–4105.
Liu, C. W., Liu, D., and Peng, D. (2019a). Long non-coding RNA ZFAS1 regulates NOB1 expression through interacting with miR-646 and promotes tumorigenesis in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 23 (8), 3206–3216. doi:10.26355/eurrev_201904_17679
Liu, G., Yuan, D., Sun, P., Liu, W., Wu, P. F., Liu, H., et al. (2018). LINC00968 functions as an oncogene in osteosarcoma by activating the PI3K/AKT/mTOR signaling. J. Cell. Physiol. 233 (11), 8639–8647. doi:10.1002/jcp.26624
Liu, J., Kong, D., Sun, D., and Li, J. (2019b). Long non-coding RNA CCAT2 acts as an oncogene in osteosarcoma through regulation of miR-200b/VEGF. Artif. Cells Nanomed Biotechnol. 47 (1), 2994–3003. doi:10.1080/21691401.2019.1640229
Liu, M., Yang, P., Mao, G., Deng, J., Peng, G., Ning, X., et al. (2019c). Long non-coding RNA MALAT1 as a valuable biomarker for prognosis in osteosarcoma: a systematic review and meta-analysis. Int. J. Surg. 72, 206–213. doi:10.1016/j.ijsu.2019.11.004
Liu, W. (2019). LncRNA LINC-PINT inhibits cancer cell proliferation, invasion, and migration in osteosarcoma by downregulating miRNA-21. Cancer biother. Radiopharm. 34 (4), 258–263. doi:10.1089/cbr.2018.2684
Liu, Y., Gu, S., Li, H., Wang, J., Wei, C., and Liu, Q. (2019d). SNHG16 promotes osteosarcoma progression and enhances cisplatin resistance by sponging miR-16 to upregulate ATG4B expression. Biochem. Biophys. Res. Commun. 518 (1), 127–133. doi:10.1016/j.bbrc.2019.08.019
Liu, Z., Sun, M., Lu, K., Liu, J., Zhang, M., Wu, W., et al. (2013). The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One 8 (10), e77293. doi:10.1371/journal.pone.0077293
Liu, Z. B., Wang, J. A., and Lv, R. Q. (2019e). Downregulation of long non-coding RNA DBH-AS1 inhibits osteosarcoma progression by PI3K-AKT signaling pathways and indicates good prognosis. Eur. Rev. Med. Pharmacol. Sci. 23 (4), 1418–1427. doi:10.26355/eurrev_201902_17098
Lu, X., Qiao, L., and Liu, Y. (2020). Long noncoding RNA LEF1-AS1 binds with HNRNPL to boost the proliferation, migration, and invasion in osteosarcoma by enhancing the mRNA stability of LEF1. J. Cell. Biochem. 121 (10), 4064–4073. doi:10.1002/jcb.29579
Lu, Z., Li, Y., Wang, J., Che, Y., Sun, S., Huang, J., et al. (2017). Long non-coding RNA NKILA inhibits migration and invasion of non-small cell lung cancer via NF-κB/Snail pathway. J. Exp. Clin. Cancer Res. 36 (1), 54. doi:10.1186/s13046-017-0518-0
Luo, H., Zhu, G., Xu, J., Lai, Q., Yan, B., Guo, Y., et al. (2019). HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell 36 (6), 645–659. doi:10.1016/j.ccell.2019.10.011
Ma, H., Su, R., Feng, H., Guo, Y., and Su, G. (2019). Long noncoding RNA UCA1 promotes osteosarcoma metastasis through CREB1-mediated epithelial-mesenchymal transition and activating PI3K/AKT/mTOR pathway. J. Bone Oncol. 16, 100228. doi:10.1016/j.jbo.2019.100228
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10 (6), 925–933. doi:10.4161/rna.24604
Ma, Y., Yang, Y., Wang, F., Moyer, M.-P., Wei, Q., Zhang, P., et al. (2016). Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut 65 (9), 1494–1504. doi:10.1136/gutjnl-2014-308392
Martin Broto, J., Martinez Garcia, J., Moura, D. S., Redondo, A., Gutierrez, A., Lopez-Pousa, A., et al. (2022). Phase II trial of palbociclib in advanced sarcoma overexpressing CDK4 gene excluding dedifferentiated liposarcoma (DD LPS): a study from the Spanish Group for Research on Sarcoma (GEIS). J. Clin. Oncol. 40 (16), 11511. doi:10.1200/JCO.2022.40.16_suppl.11511
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447. doi:10.1038/s41580-022-00566-8
Meng, L., Ward, A. J., Chun, S., Bennett, C. F., Beaudet, A. L., and Rigo, F. (2015). Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 518 (7539), 409–412. doi:10.1038/nature13975
Meyer, J. S., Nadel, H. R., Marina, N., Womer, R. B., Brown, K. L., Eary, J. F., et al. (2008). Imaging guidelines for children with ewing sarcoma and osteosarcoma: a report from the children's oncology group bone tumor committee. Pediatr. Blood Cancer. 51 (2), 163–170. doi:10.1002/pbc.21596
Mi, Z., Dong, Y., Wang, Z., and Ye, P. (2021). Biomarker potential of lncRNA GNAS-AS1 in osteosarcoma prognosis and effect on cellular function. J. Orthop. Surg. Res. 16 (1), 470. doi:10.1186/s13018-021-02611-2
Michael, M., and Doherty, M. M. (2005). Tumoral drug metabolism: overview and its implications for cancer therapy. J. Clin. Oncol. 23 (1), 205–229. doi:10.1200/JCO.2005.02.120
Mirabello, L., Troisi, R. J., and Savage, S. A. (2009). Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer 115 (7), 1531–1543. doi:10.1002/cncr.24121
Moonmuang, S., Chaiyawat, P., Jantrapirom, S., Pruksakorn, D., and Lo Piccolo, L. (2021). Circulating long non-coding RNAs as novel potential biomarkers for osteogenic sarcoma. Cancers (Basel) 13 (16), 4214. doi:10.3390/cancers13164214
Morice, S., Mullard, M., Brion, R., Dupuy, M., Renault, S., Tesfaye, R., et al. (2020). The YAP/TEAD Axis as a new therapeutic target in osteosarcoma: effect of verteporfin and CA3 on primary tumor growth. Cancers (Basel) 12 (12), 3847. doi:10.3390/cancers12123847
Mourtada-Maarabouni, M., and Williams, G. T. (2013). Growth arrest on inhibition of nonsense-mediated decay is mediated by noncoding RNA GAS5. Biomed. Res. Int. 2013, 358015. doi:10.1155/2013/358015
Necsulea, A., Soumillon, M., Warnefors, M., Liechti, A., Daish, T., Zeller, U., et al. (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505 (7485), 635–640. doi:10.1038/nature12943
Nusse, R., and Clevers, H. (2017). Wnt/β-Catenin signaling, disease, and emerging therapeutic modalities. Cell 169 (6), 985–999. doi:10.1016/j.cell.2017.05.016
Pan, X., Tan, J., Tao, T., Zhang, X., Weng, Y., Weng, X., et al. (2021). LINC01123 enhances osteosarcoma cell growth by activating the Hedgehog pathway via the miR-516b-5p/Gli1 axis. Cancer Sci. 112 (6), 2260–2271. doi:10.1111/cas.14913
Papaioannou, D., Petri, A., Dovey, O. M., Terreri, S., Wang, E., Collins, F. A., et al. (2019). The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat. Commun. 10 (1), 5351. doi:10.1038/s41467-019-13259-2
Parasramka, M. A., Maji, S., Matsuda, A., Yan, I. K., and Patel, T. (2016). Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol. Ther. 161, 67–78. doi:10.1016/j.pharmthera.2016.03.004
Pastushenko, I., and Blanpain, C. (2019). EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29 (3), 212–226. doi:10.1016/j.tcb.2018.12.001
Pegueroles, C., and Gabaldón, T. (2016). Secondary structure impacts patterns of selection in human lncRNAs. BMC Biol. 14, 60. doi:10.1186/s12915-016-0283-0
Pierce, J. B., Zhou, H., Simion, V., and Feinberg, M. W. (2022). Long noncoding RNAs as therapeutic targets. Adv. Exp. Med. Biol. 1363, 161–175. doi:10.1007/978-3-030-92034-0_9
Qi, P., Zhou, X. Y., and Du, X. (2016). Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol. Cancer. 15 (1), 39. doi:10.1186/s12943-016-0524-4
Qin, S., Wang, Y., Ma, C., and Lv, Q. (2023). Competitive endogenous network of circRNA, lncRNA, and miRNA in osteosarcoma chemoresistance. Eur. J. Med. Res. 28 (1), 354. doi:10.1186/s40001-023-01309-x
Radtke, F., and Raj, K. (2003). The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 3 (10), 756–767. doi:10.1038/nrc1186
Rodríguez-Mateo, C., Torres, B., Gutiérrez, G., and Pintor-Toro, J. A. (2017). Downregulation of Lnc-Spry1 mediates TGF-β-induced epithelial-mesenchymal transition by transcriptional and posttranscriptional regulatory mechanisms. Cell Death Differ. 24 (5), 785–797. doi:10.1038/cdd.2017.9
Rosenbloom, K. R., Dreszer, T. R., Long, J. C., Malladi, V. S., Sloan, C. A., Raney, B. J., et al. (2012). ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 40, D912–D917. doi:10.1093/nar/gkr1012
Ross, C. J., Rom, A., Spinrad, A., Gelbard-Solodkin, D., Degani, N., and Ulitsky, I. (2021). Uncovering deeply conserved motif combinations in rapidly evolving noncoding sequences. Genome Biol. 22 (1), 29. doi:10.1186/s13059-020-02247-1
Sakamoto, A., and Iwamoto, Y. (2008). Current status and perspectives regarding the treatment of osteo-sarcoma: chemotherapy. Rev. Recent Clin. Trials. 3 (3), 228–231. doi:10.2174/157488708785700267
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146 (3), 353–358. doi:10.1016/j.cell.2011.07.014
Sánchez-Salcedo, R., Miranda-Castro, R., de-Los-Santos-Álvarez, N., and Lobo-Castañón, M. J. (2021). Dual electrochemical genosensor for early diagnosis of prostate cancer through lncRNAs detection. Biosens. Bioelectron. 192, 113520. doi:10.1016/j.bios.2021.113520
Serra, M., Hattinger, C. M., Pasello, M., Casotti, C., Fantoni, L., Riganti, C., et al. (2021). Impact of ABC transporters in osteosarcoma and ewing's sarcoma: which are involved in chemoresistance and which are not? Cells 10 (9), 2461. doi:10.3390/cells10092461
Sharma, V., Khurana, S., Kubben, N., Abdelmohsen, K., Oberdoerffer, P., Gorospe, M., et al. (2015). A BRCA1-interacting lncRNA regulates homologous recombination. EMBO Rep. 16 (11), 1520–1534. doi:10.15252/embr.201540437
Shen, B., Zhou, N., Hu, T., Zhao, W., Wu, D., and Wang, S. (2019). LncRNA MEG3 negatively modified osteosarcoma development through regulation of miR-361-5p and FoxM1. J. Cell. Physiol. 234 (8), 13464–13480. doi:10.1002/jcp.28026
Shen, C., Yang, C., Xia, B., and You, M. (2021). Long non-coding RNAs: emerging regulators for chemo/immunotherapy resistance in cancer stem cells. Cancer Lett. 500, 244–252. doi:10.1016/j.canlet.2020.11.010
Song, B., Wang, Y., Xi, Y., Kudo, K., Bruheim, S., Botchkina, G. I., et al. (2009). Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28 (46), 4065–4074. doi:10.1038/onc.2009.274
Song, L., Zhou, Z., Gan, Y., Li, P., Xu, Y., Zhang, Z., et al. (2019). Long noncoding RNA OIP5-AS1 causes cisplatin resistance in osteosarcoma through inducing the LPAATβ/PI3K/AKT/mTOR signaling pathway by sponging the miR-340-5p. J. Cell. Biochem. 120 (6), 9656–9666. doi:10.1002/jcb.28244
Strauss, S. J., Frezza, A. M., Abecassis, N., Bajpai, J., Bauer, S., Biagini, R., et al. (2021). Bone sarcomas: ESMO-EURACAN-GENTURIS-ERN PaedCan clinical practice guideline for diagnosis, treatment and follow-up. Ann. Oncol. 32 (12), 1520–1536. doi:10.1016/j.annonc.2021.08.1995
Sun, J., Wang, X., Fu, C., Wang, X., Zou, J., Hua, H., et al. (2016). Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol. Biol. Rep. 43 (5), 427–436. doi:10.1007/s11033-016-3975-1
Sun, Z. Y., Jian, Y. K., Zhu, H. Y., and Li, B. (2019). lncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to gemcitabine through activating c-MET/PI3K/AKT pathway. Pathol. Res. Pract. 215 (3), 555–563. doi:10.1016/j.prp.2018.12.013
Takebe, N., Nguyen, D., and Yang, S. X. (2014). Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol. Ther. 141 (2), 140–149. doi:10.1016/j.pharmthera.2013.09.005
Taniguchi, K., and Karin, M. (2018). NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 18 (5), 309–324. doi:10.1038/nri.2017.142
Taniue, K., Kurimoto, A., Sugimasa, H., Nasu, E., Takeda, Y., Iwasaki, K., et al. (2016). Long noncoding RNA UPAT promotes colon tumorigenesis by inhibiting degradation of UHRF1. Proc. Natl. Acad. Sci. U. S. A. 113 (5), 1273–1278. doi:10.1073/pnas.1500992113
Tao, J., Jiang, M. M., Jiang, L., Salvo, J. S., Zeng, H. C., Dawson, B., et al. (2014). Notch activation as a driver of osteogenic sarcoma. Cancer Cell 26 (3), 390–401. doi:10.1016/j.ccr.2014.07.023
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329 (5992), 689–693. doi:10.1126/science.1192002
Vimalraj, S., Subramanian, R., and Dhanasekaran, A. (2021). LncRNA MALAT1 promotes tumor angiogenesis by regulating MicroRNA-150-5p/VEGFA signaling in osteosarcoma: in-vitro and in-vivo analyses. Front. Oncol. 11, 742789. doi:10.3389/fonc.2021.742789
Wang, B., Qu, X. L., Liu, J., Lu, J., and Zhou, Z. Y. (2019). HOTAIR promotes osteosarcoma development by sponging miR-217 and targeting ZEB1. J. Cell. Physiol. 234 (5), 6173–6181. doi:10.1002/jcp.27394
Wang, B., Su, Y., Yang, Q., Lv, D., Zhang, W., Tang, K., et al. (2015a). Overexpression of long non-coding RNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma. Mol. Cells. 38 (5), 432–440. doi:10.14348/molcells.2015.2327
Wang, J. Y., Yang, Y., Ma, Y., Wang, F., Xue, A., Zhu, J., et al. (2020). Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed. Pharmacother. 121, 109627. doi:10.1016/j.biopha.2019.109627
Wang, W., Chen, D., and Zhu, K. (2018a). SOX2OT variant 7 contributes to the synergistic interaction between EGCG and Doxorubicin to kill osteosarcoma via autophagy and stemness inhibition. J. Exp. Clin. Cancer Res. 37 (1), 37. doi:10.1186/s13046-018-0689-3
Wang, Y., He, L., Du, Y., Zhu, P., Huang, G., Luo, J., et al. (2015b). The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16 (4), 413–425. doi:10.1016/j.stem.2015.03.003
Wang, Y., Ren, X., Yuan, Y., and Yuan, B. S. (2021). Downregulated lncRNA GAS5 and upregulated miR-21 lead to epithelial-mesenchymal transition and lung metastasis of osteosarcomas. Front. Cell Dev. Biol. 9, 707693. doi:10.3389/fcell.2021.707693
Wang, Y., Zeng, X., Wang, N., Zhao, W., Zhang, X., Teng, S., et al. (2018b). Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol. Cancer. 17 (1), 89. doi:10.1186/s12943-018-0837-6
Wang, Y., Zhang, L., Zheng, X., Zhong, W., Tian, X., Yin, B., et al. (2016). Long non-coding RNA LINC00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the miR-645-IFIT2 axis. Cancer Lett. 382 (2), 137–146. doi:10.1016/j.canlet.2016.08.024
Wang, Y., Zhao, Z., Zhang, S., Li, Z., Li, D., Yang, S., et al. (2018c). LncRNA FAL1 is a negative prognostic biomarker and exhibits pro-oncogenic function in osteosarcoma. J. Cell. Biochem. 119 (10), 8481–8489. doi:10.1002/jcb.27074
Wang, Z., Tan, M., Chen, G., Li, Z., and Lu, X. (2017). LncRNA SOX2-OT is a novel prognostic biomarker for osteosarcoma patients and regulates osteosarcoma cells proliferation and motility through modulating SOX2. IUBMB Life 69 (11), 867–876. doi:10.1002/iub.1681
Wei, L., Yi, Z., Guo, K., and Long, X. (2019). Long noncoding RNA BCAR4 promotes glioma cell proliferation via EGFR/PI3K/AKT signaling pathway. J. Cell Physiol. 234 (12), 23608–23617. doi:10.1002/jcp.28929
Wen, J. F., Jiang, Y. Q., Li, C., Dai, X. K., Wu, T., and Yin, W. Z. (2020). LncRNA-SARCC sensitizes osteosarcoma to cisplatin through the miR-143-mediated glycolysis inhibition by targeting Hexokinase 2. Cancer Biomark. 28 (2), 231–246. doi:10.3233/cbm-191181
Wen, J. J., Ma, Y. D., Yang, G. S., and Wang, G. M. (2017). Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 21 (3), 498–503.
Whelan, J. S., and Davis, L. E. (2018). Osteosarcoma, chondrosarcoma, and chordoma. J. Clin. Oncol. 36 (2), 188–193. doi:10.1200/JCO.2017.75.1743
Xia, B., Wang, L., Feng, L., Tian, B., Tan, Y., and Du, B. (2018). Knockdown of long noncoding RNA CAT104 inhibits the proliferation, migration, and invasion of human osteosarcoma cells by regulating MicroRNA-381. Oncol. Res. 27 (1), 89–98. doi:10.3727/096504018x15199511344806
Xia, P., Gu, R., Zhang, W., and Sun, Y. F. (2020). lncRNA CEBPA-AS1 overexpression inhibits proliferation and migration and stimulates apoptosis of OS cells via notch signaling. Mol. Ther. Nucleic Acids 19, 1470–1481. doi:10.1016/j.omtn.2019.10.017
Xiao, Y., Xu, J., and Yin, W. (2018). Aberrant epigenetic modifications of non-coding RNAs in human disease. Adv. Exp. Med. Biol. 1094, 65–75. doi:10.1007/978-981-13-0719-5_7
Xie, X., Liu, W., Duan, Z., Li, X., Zhang, L., and Yang, G. (2020). LncRNA NORAD targets miR-410-3p to regulate drug resistance sensitivity of osteosarcoma. Cell. Mol. Biol. (Noisy-Le-Grand) 66 (3), 143–148. doi:10.14715/cmb/2020.66.3.22
Xing, C. Y., Zhang, Y. Z., Hu, W., and Zhao, L. Y. (2022). LINC00313 facilitates osteosarcoma carcinogenesis and metastasis through enhancing EZH2 mRNA stability and EZH2-mediated silence of PTEN expression. Cell. Mol. Life Sci. 79 (7), 382. doi:10.1007/s00018-022-04376-1
Yang, J., Meng, X., Pan, J., Jiang, N., Zhou, C., Wu, Z., et al. (2018). CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol. 15 (1), 35–43. doi:10.1080/15476286.2017.1391443
Yang, Q., Yu, H., Yin, Q., Hu, X., and Zhang, C. (2019a). lncRNA-NEF is downregulated in osteosarcoma and inhibits cancer cell migration and invasion by downregulating miRNA-21. Oncol. Lett. 17 (6), 5403–5408. doi:10.3892/ol.2019.10276
Yang, Y., Fei, M., Zhou, X., Li, Y., and Jin, D. (2019b). The potential value of lncRNA-BC050642 in osteosarcoma origination and outcomes. Artif. Cells Nanomed Biotechnol. 47 (1), 1859–1866. doi:10.1080/21691401.2019.1611593
Yao, Q., and Chen, T. (2020). LINC01128 regulates the development of osteosarcoma by sponging miR-299-3p to mediate MMP2 expression and activating Wnt/β-catenin signalling pathway. J. Cell. Mol. Med. 24 (24), 14293–14305. doi:10.1111/jcmm.16046
Yao, W., Hou, J., Liu, G., Wu, F., Yan, Q., Guo, L., et al. (2022). LncRNA CBR3-AS1 promotes osteosarcoma progression through the network of miR-140-5p/DDX54-NUCKS1-mTOR signaling pathway. Mol. Ther. Oncolytics 25, 189–200. doi:10.1016/j.omto.2022.03.001
Yao, X. Y., Liu, J. F., Luo, Y., Xu, X. Z., and Bu, J. (2021). LncRNA HOTTIP facilitates cell proliferation, invasion, and migration in osteosarcoma by interaction with PTBP1 to promote KHSRP level. Cell Cycle 20 (3), 283–297. doi:10.1080/15384101.2020.1870820
Ye, F., Tian, L., Zhou, Q., and Feng, D. (2019). LncRNA FER1L4 induces apoptosis and suppresses EMT and the activation of PI3K/AKT pathway in osteosarcoma cells via inhibiting miR-18a-5p to promote SOCS5. Gene 721, 144093. doi:10.1016/j.gene.2019.144093
Yoon, J. H., Abdelmohsen, K., and Gorospe, M. (2013). Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425 (19), 3723–3730. doi:10.1016/j.jmb.2012.11.024
Yu, X., Hu, L., Li, S., Shen, J., Wang, D., Xu, R., et al. (2019). Long non-coding RNA Taurine upregulated gene 1 promotes osteosarcoma cell metastasis by mediating HIF-1α via miR-143-5p. Cell Death Dis. 10 (4), 280. doi:10.1038/s41419-019-1509-1
Yuan, J. H., Liu, X. N., Wang, T. T., Pan, W., Tao, Q. F., Zhou, W. P., et al. (2017). The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 19 (7), 820–832. doi:10.1038/ncb3538
Zeng, L., Liu, L., Ni, W. J., Xie, F., and Leng, X. M. (2023). Circular RNAs in osteosarcoma: an update of recent studies (Review). Int. J. Oncol. 63 (5), 123. doi:10.3892/ijo.2023.5571
Zhang, B., Yu, L., Han, N., Hu, Z., Wang, S., Ding, L., et al. (2018a). LINC01116 targets miR-520a-3p and affects IL6R to promote the proliferation and migration of osteosarcoma cells through the Jak-stat signaling pathway. Biomed. Pharmacother. 107, 270–282. doi:10.1016/j.biopha.2018.07.119
Zhang, C., Xie, L., Liang, H., and Cui, Y. (2019a). LncRNA MIAT facilitates osteosarcoma progression by regulating mir-128-3p/VEGFC axis. IUBMB Life 71 (7), 845–853. doi:10.1002/iub.2001
Zhang, C. L., Zhu, K. P., and Ma, X. L. (2017a). Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 396, 66–75. doi:10.1016/j.canlet.2017.03.018
Zhang, E., Han, L., Yin, D., He, X., Hong, L., Si, X., et al. (2017b). H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 45 (6), 3086–3101. doi:10.1093/nar/gkw1247
Zhang, G. Y., Zhang, J. F., Hu, X. M., Luo, Z. P., and Ma, Y. Z. (2017c). Clinical significance of long non-coding RNA EWSAT1 as a novel prognostic biomarker in osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 21 (23), 5337–5341. doi:10.26355/eurrev_201712_13918
Zhang, H., Zhou, L., Hu, S., Gu, W., Li, Z., Sun, J., et al. (2022). The crosstalk between LINC01089 and hippo pathway inhibits osteosarcoma progression. J. Bone Min. Metab. 40 (6), 890–899. doi:10.1007/s00774-022-01377-9
Zhang, N., Meng, X., Mei, L., Zhao, C., and Chen, W. (2019b). LncRNA DLX6-AS1 promotes tumor proliferation and metastasis in osteosarcoma through modulating miR-641/HOXA9 signaling pathway. J. Cell. Biochem. 120, 11478–11489. doi:10.1002/jcb.28426
Zhang, P., Yang, Y., Zweidler-McKay, P. A., and Hughes, D. P. (2008). Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin. Cancer Res. 14 (10), 2962–2969. doi:10.1158/1078-0432.ccr-07-1992
Zhang, Q. Q., Xu, S. L., Ding, C., Ma, C. C., Yuan, T. S., Hua, C. C., et al. (2021). LncRNA FOXD2-AS1 knockdown inhibits the resistance of human osteosarcoma cells to cisplatin by inhibiting miR-143 expression. Eur. Rev. Med. Pharmacol. Sci. 25 (2), 678–686. doi:10.26355/eurrev_202101_24629
Zhang, R. M., Tang, T., Yu, H. M., and Yao, X. D. (2018b). LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem. Biophys. Res. Commun. 507 (1-4), 260–266. doi:10.1016/j.bbrc.2018.11.019
Zhang, Y., Dai, Q., Zeng, F., and Liu, H. (2018c). MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the Rac1/JNK pathway via targeting MiR-509. Oncol. Res. 2018, 26111. doi:10.3727/096504017x14957939026111
Zhang, Z., Chen, J., Zhu, Z., Zhu, Z., Liao, X., Wu, J., et al. (2020). CRISPR-Cas13-Mediated knockdown of lncRNA-GACAT3 inhibited cell proliferation and motility, and induced apoptosis by increasing p21, bax, and E-cadherin expression in bladder cancer. Front. Mol. Biosci. 7, 627774. doi:10.3389/fmolb.2020.627774
Zhao, B., Liu, K., and Cai, L. (2019a). LINK-A lncRNA functions in the metastasis of osteosarcoma by upregulating HIF1α. Oncol. Lett. 17 (6), 5005–5011. doi:10.3892/ol.2019.10177
Zhao, H., Hou, W., Tao, J., Zhao, Y., Wan, G., Ma, C., et al. (2016). Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am. J. Transl. Res. 8 (8), 3503–3512.
Zhao, H., Wang, Y., Hou, W., Ding, X., and Wang, W. (2022). Long non-coding RNA MALAT1 promotes cell proliferation, migration and invasion by targeting miR-590-3p in osteosarcoma. Exp. Ther. Med. 24 (5), 672. doi:10.3892/etm.2022.11608
Zhao, J., and Ma, S. T. (2018). Downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathway. Mol. Med. Rep. 17 (5), 7388–7394. doi:10.3892/mmr.2018.8746
Zhao, W., Qin, P., Zhang, D., Cui, X., Gao, J., Yu, Z., et al. (2019b). Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging (Albany N. Y.). 11 (21), 9581–9596. doi:10.18632/aging.102406
Zheng, C., Li, R., Zheng, S., Fang, H., Xu, M., and Zhong, L. (2023). The knockdown of lncRNA DLGAP1-AS2 suppresses osteosarcoma progression by inhibiting aerobic glycolysis via the miR-451a/HK2 axis. Cancer Sci. 114 (12), 4747–4762. doi:10.1111/cas.15989
Zheng, Y., Wang, G., Chen, R., Hua, Y., and Cai, Z. (2018). Mesenchymal stem cells in the osteosarcoma microenvironment: their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell. Res. Ther. 9 (1), 22. doi:10.1186/s13287-018-0780-x
Zhou, B., Li, L., Li, Y., Sun, H., and Zeng, C. (2018a). Long noncoding RNA SNHG12 mediates doxorubicin resistance of osteosarcoma via miR-320a/MCL1 axis. Biomed. Pharmacother. 106, 850–857. doi:10.1016/j.biopha.2018.07.003
Zhou, D. K., Yang, X. W., Li, H., Yang, Y., Zhu, Z. J., and Wu, N. (2017). Up-regulation of long noncoding RNA CCAL predicts poor patient prognosis and promotes tumor metastasis in osteosarcoma. Int. J. Biol. Markers 32 (1), e108–e112. doi:10.5301/jbm.5000240
Zhou, F. C., Zhang, Y. H., Liu, H. T., Song, J., and Shao, J. (2020a). LncRNA LINC00588 suppresses the progression of osteosarcoma by acting as a ceRNA for miRNA-1972. Front. Pharmacol. 11, 255. doi:10.3389/fphar.2020.00255
Zhou, Q., Hu, T., and Xu, Y. (2020b). Anticancer potential of TUG1 knockdown in cisplatin-resistant osteosarcoma through inhibition of MET/Akt signalling. J. Drug Target. 28 (2), 204–211. doi:10.1080/1061186x.2019.1644651
Zhou, R., Chen, K. K., Zhang, J., Xiao, B., Huang, Z., Ju, C., et al. (2018b). The decade of exosomal long RNA species: an emerging cancer antagonist. Mol. Cancer. 17 (1), 75. doi:10.1186/s12943-018-0823-z
Zhou, S., Yu, L., Xiong, M., and Dai, G. (2018c). LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem. Biophys. Res. Commun. 495 (2), 1822–1832. doi:10.1016/j.bbrc.2017.12.047
Zhou, Y., Jin, Q., Chang, J., Zhao, Z., and Sun, C. (2022). Long non-coding RNA ZMIZ1-AS1 promotes osteosarcoma progression by stabilization of ZMIZ1. Cell Biol. Toxicol. 38 (6), 1013–1026. doi:10.1007/s10565-021-09641-w
Zhu, G., Liu, X., Su, Y., Kong, F., Hong, X., and Lin, Z. (2018). Knockdown of urothelial carcinoma-associated 1 suppressed cell growth and migration through regulating miR-301a and CXCR4 in osteosarcoma MHCC97 cells. Oncol. Res. 27 (1), 55–64. doi:10.3727/096504018x15201143705855
Zhu, K., Yuan, Y., Wen, J., Chen, D., Zhu, W., Ouyang, Z., et al. (2020a). LncRNA Sox2OT-V7 promotes doxorubicin-induced autophagy and chemoresistance in osteosarcoma via tumor-suppressive miR-142/miR-22. Aging (Albany N. Y.). 12 (8), 6644–6666. doi:10.18632/aging.103004
Zhu, S., Liu, Y., Wang, X., Wang, J., and Xi, G. (2020b). lncRNA SNHG10 promotes the proliferation and invasion of osteosarcoma via wnt/β-catenin signaling. Mol. Ther. Nucleic Acids 22, 957–970. doi:10.1016/j.omtn.2020.10.010
Zi, X., Zhang, G., and Qiu, S. (2022). Up-regulation of LINC00619 promotes apoptosis and inhibits proliferation, migration and invasion while promoting apoptosis of osteosarcoma cells through inactivation of the HGF-mediated PI3K-Akt signalling pathway. Epigenetics 17 (2), 147–160. doi:10.1080/15592294.2021.1890873
Keywords: osteosarcoma, long non-coding RNAs, signaling pathways, drug resistance, biomarkers
Citation: Liao X, Wei R, Zhou J, Wu K and Li J (2024) Emerging roles of long non-coding RNAs in osteosarcoma. Front. Mol. Biosci. 11:1327459. doi: 10.3389/fmolb.2024.1327459
Received: 25 October 2023; Accepted: 12 February 2024;
Published: 07 March 2024.
Edited by:
Alessio Colantoni, Sapienza University of Rome, ItalyReviewed by:
Fabrizio Ferrè, University of Bologna, ItalyArijita Sarkar, University of Southern California, United States
Copyright © 2024 Liao, Wei, Zhou, Wu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiao Li, lijiaospace@163.com
 Xun Liao
Xun Liao Rong Wei2
Rong Wei2  Jiao Li
Jiao Li