PhbZIP2 regulates photosynthesis-related genes in an intertidal macroalgae, Pyropia haitanensis, under stress
- 1Fisheries College, Jimei University, Xiamen, China
- 2Fujian Engineering Research Center of Aquatic Breeding and Healthy Aquaculture, Xiamen, China
- 3Key Laboratory of Healthy Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, Xiamen, China
- 4Freshwater Fisheries Research Institute of Fujian, Fuzhou, China
Intertidal macroalgae are important research subjects in stress biology. Basic region-leucine zipper transcription factors (bZIPs) play an important regulatory role in the expression of target genes under abiotic stress. We herein identified a bZIP2 gene PhbZIP2 to regulate abiotic stress tolerance in Pyropia haitanensis, a representative intertidal macroalgal species. Cloning and sequencing of the cDNA characterized a BRLZ structure and an α coiled-coil structure between amino acids and Expression of PhbZIP2 was detected to upregulate under both high temperature and salt stresses. A DAP-seq analysis revealed the PhbZIP2-binding motifs of (T/C)TCCA(C/G) and A (A/G)AAA (G/A), which differed from the conserved motifs in plants. Overexpression of PhbZIP2 was indicative of a high temperature and salt stress tolerances in transgenic Chlamydomonas reinhardtii. It was suggested that PhbZIP2 was probably involved in regulating expression of the photosynthetic-related genes and the response to the abiotic stresses in P. haitanensis, which provide new insights for elucidating efficient adaptation strategies of intertidal macroalgae.
1 Introduction
The intertidal zone experiences some of the most drastic environmental changes of any of the world’s ecosystems, but a variety of benthic macroalgae are widely distributed within it. At low tide, these intertidal macroalgae are completely exposed to the air and face multiple abiotic stresses, such as water loss, high temperature, hypersalinity, and high irradiance; at high tide, however, their thalli are completely submerged in seawater, where they quickly emerge from the ‘dormant’ state and return to normal growth and development (Blouin et al., 2011; Bartlett et al., 2017; Wang et al., 2020; Chen et al., 2021). Intertidal macroalgae are therefore an important subject of research in stress biology, and analysis of the adaptive strategies of intertidal macroalgae can provide new ideas for the study of plant adaptive evolution.
Bangiomorpha pubescens, possibly extant 1,198–24 million years ago according to fossil evidence, is presumed to have had the strong ability to live in a harsh intertidal habitat during this ancient era (Nicholas, 2000). Pyropia/Porphyra, a representative genus of intertidal Bangiales, is shaped by stress (Blouin et al., 2011). Many studies have been devoted to the molecular mechanism behind the stress tolerance of this economic seaweed. Under external stress, a potential self-amplifying signaling loop involving Ca2+, phosphatidylinositol, and reactive oxygen species (ROS) is generated in this alga (Zheng et al., 2019; Suda and Mikami, 2020; Wang et al., 2020; Guan et al., 2022), and the rapid, unique Ca2+ signal is then decoded by calmodulin (CaM) to activate various CaM-dependent kinases (Brawley et al., 2017; Wang et al., 2022). Potential downstream pathways, such as the antioxidant system, are then activated to maintain redox homeostasis (Wang et al., 2018; Wang et al., 2019a; Wang et al., 2020a; Wang et al., 2020b), while heat shock proteins (HSPs), the ubiquitin–proteasome system, and protein processing systems are induced to maintain intracellular protein synthesis, folding, and degradation homeostasis (Wang et al., 2019b; Wen et al., 2020; Niu et al., 2022; Zheng et al., 2022). This ‘sensing-transduction-activation’ model is crucial to the ability of Pyropia/Porphyra thalli to acclimate and adapt to various stresses. A number of important candidate genes involved in the stress tolerance of Pyropia/Porphyra have also been identified by omics technology and molecular biology methods; examples include the superoxide dismutase (SOD) gene PhSODs (Ji et al., 2016), PhHSPs (Uji et al., 2019; Chang et al., 2021), the RING-type ubiquitin ligase PhCUL4 (Wang et al., 2021), and the D-group mitogen-activated protein kinase PyMAPK2 (Kong et al., 2021). The manner in which these candidate genes are activated and regulated is nonetheless still unclear.
When plants respond to abiotic stress, transcription factors can control the expressions of downstream target genes at a specific intensity at a specific time and space to regulate this process. Basic region-leucine zipper transcription factors (bZIPs), which constitute one of the largest, most conserved transcription factor families in plants, play an important role in the regulation of target gene expression in plants under abiotic stress (Jakoby et al., 2002; Droge-Laser et al., 2018). bZIPs contain two domains: a basic region in the N-terminus that recognizes and binds conserved sequences on gene promoters, and a leucine zipper region in the C-terminus that drives activation and repression functions (Hurst, 1995). bZIPs can bind to a downstream gene promoter containing the ACGT core motif in the N-terminus and then regulate the transcriptional expression of downstream stress-related genes to improve plant stress resistance. For example, glutathione activates the promoters of HSP90.1 via bZIP10 in Arabidopsis (Kumar and Chattopadhyay, 2018). Similarly, overexpression of ZmbZIP4 can increase the expression of abscisic acid synthesis-related genes, including NCED, ABA1, AAO3, and LOS5, and further improve the abiotic stress tolerance of maize (Ma et al., 2018). The role of bZIPs in regulating the response of intertidal macroalgae to abiotic stress, however, is not clear.
The comprehensive identification of transcription factor binding sites helps to characterize regulatory elements and transcription factor functions. Chromatin immunoprecipitation sequencing (ChIP-seq) is currently the leading technology for capturing transcription factor binding site information. This method is highly dependent on gene-specific antibodies, however, and has disadvantages, such as difficult operation and high cost, that limit its popularization and use (Pouya and Manols, 2014). DNA affinity purification sequencing (DAP-seq) is a novel technique for analyzing information on the binding of transcription factors. This approach relies on the construction and expression of transcription-factor proteins in vitro, which are then bound to target genome fragments and sequenced. The motif sequences obtained by Dap-seq are compared with the known motif database, and the detected motifs are annotated by the known motifs. DAP-seq is suitable for analyzing species for which transcription factor antibodies cannot be prepared and for investigating less-studied transcription factors. In a comparison of DAP-seq and ChIP-seq data obtained for multiple transcription factors, the overlap rate of DAP-seq and ChIP-seq peaks reached 81%, and the overlap rate with high-scoring motif data was as high as 97% (Brawley et al., 2017). DAP-seq data can therefore be used for peak-calling and motif analysis to obtain potential target genes of transcription factors.
In this study, we identified the key bZIP transcription factor, PhbZIP2, from whole genome data of P. haitanensis and analyzed its gene structure and expression pattern. After cloning, we obtained information on the binding site and target proteins of PhbZIP2 by combining ChIP-seq and DAP-seq technology. Finally, we confirmed the function of PhbZIP2 in abiotic stress response via heterologous expression. Our results can serve as a theoretical foundation for a comprehensive understanding of the abiotic stress tolerance of intertidal macroalgae.
2 Materials and methods
2.1 Treatment of experimental materials
The experimental strain, Pyropia haitanensis Z-61, was selected from the Laboratory of Germplasm Improvements and Applications of P. haitanensis at Jimei University (Chen et al., 2019). The control group was cultured at 21°C ± 0.5°C under a light intensity of 50–60 μmol photons m−2 s−1 and a 12-h light/12-h dark photoperiod. The culture medium, fresh Provasoli’s enrichment solution, was replaced every 2 days. When the cultured algae reached a length of 15 ± 2 cm, undamaged, non-distorted, smooth-planar thalli exhibiting good growth were selected for subsequent experiments. For the high temperature stress treatment, thalli were maintained in a constant temperature incubator at 32°C for 0, 5, 10, 15, 30, 60, 180, or 360 min. According to our previous study, 4 h under 100‰ and 110‰ were selected as hypersaline stress conditions (Chen et al., 2019), and 0‰ and 5‰ were selected as hyposaline stress conditions (Wang et al., 2020a).
The Chlamydomonas reinhardtii strain used in this study was selected from the cell wall deletion strain ‘CC-400 CW15 MT+’ cultured in our laboratory. The control group was cultured in a 150-mL conical flask with shaking at 100 rpm min−1 and a temperature of 25°C ± 0.5°C under a light intensity of 50 μmol photons m−2 s−1 and a 14-h light/10-h dark photoperiod. High temperature treatment conditions were the same as those used for P. haitanensis. As a hypersaline treatment, the NaCl salinity of the C. reinhardtii medium was adjusted to 150 mM, with all other conditions unchanged. Variation in the biomass of treated samples over time (0, 24, 48, and 72 h) was determined by observing sample phenotypes and measuring OD values at 750 nm.
2.2 Isolation and purification of total RNA and synthesis of cDNA
Total RNA was extracted from P. haitanensis samples using an EZNA plant RNA extraction kit (Omega, USA). The purity and quantity of the extracted RNA were initially estimated from OD260 and OD280 values measured with a Cary50 UV spectrophotometer (Varian, USA), and RNA integrity and quality subsequently confirmed by 1% agarose gel electrophoresis. The extracted RNA was then reverse transcribed into cDNA using a PrimeScript RT Reagent kit (Takara).
2.3 Cloning and expression pattern analysis of PhbZIP2 from P. haitanensis
A bZIP family-related gene, designated as phaG00003535, was screened from the previously generated database of P. haitanensis genome and transcriptome (Cao et al., 2020). The identified sequence was cloned by PCR using specific primers. Next, forward and reverse primers for real-time fluorescence quantitative PCR (qRT-PCR) were designed from the cloned sequence, and their quality was assessed using an amplification efficiency of 95%–105% as a criterion. PhUBC and β-tubulin were used as reference genes with P. haitanensis and C. reinhardtii, respectively (Supplementary Table S1). A qRT-PCR amplifications were performed using 10× gradient-diluted cDNA as a template to produce standard gene curves on a Step One Plus fluorescence quantitative PCR instrument (ABI, USA). The qRT-PCR cycling protocol for P. haitanensis and C. reinhardtii was as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 58.1°C for 30 s.
2.4 Bioinformatics analysis of PhbZIP2
The online tool Open Reading Frame Finder (ORF Finder) at NCBI (https://www.ncbi.nlm.nih.gov) was used to analyze PhbZIP2 nucleotide and amino acid sequences, predict and analyze the open reading frame position, and obtain start and stop codons. Conserved function domains were analyzed with the SMART online tool (http://smart.emblheidelberg.de/), and the primary structure of the protein encoded by the gene was investigated using ExPASy (http://web.expasy.org/protparam/). A phylogenetic tree of bZIP2 protein sequences was constructed by the neighbor-joining method in MEGA6.
2.5 Expression of an PhbZIP2 fusion protein and DNA affinity purification sequencing
A DAP genomic DNA library was prepared and DAP reactions were performed as previously described (Brawley et al., 2017). Briefly, the homology-arm base sequences of a HaloTag expression vector were added to both ends of the start and stop codons of the PhbZIP2 gene to construct a recombinant vector containing a HaloTag and PhbZIP2 (Supplementary Table S1). The TnT SP6 high-yield wheat germ protein expression system was used to express the PhbZIP2–HaloTag fusion protein, which was purified with HaloTag magnetic beads, washed, and filtered to remove non-specific or unconnected protein. Next, 30–100 ng DNA was added to the fusion protein for co-incubation and further recovery of binding fragments. The DNA library was detected, filtered, and sequenced. Finally, a peak analysis of the read data was performed on the whole P. haitanensis genome to identify PhbZIP2 binding sites. Peaks were scanned with MACS2 analysis software at a threshold of q < 0.05, and the information on peak locations was used to screen out peak-related genes. Peak-related genes were annotated with ChIPseeker R package. In addition, MEME (http://meme-suite.org/tools/meme) and DREME (https://meme-suite.org/meme/tools/dreme) were used to detect the sequence motif.
2.6 Construction and transformation of the PhbZIP2 expression vector in C. reinhardtii
Base sequences of KpnI and PstI restriction sites were added to the target-gene start and stop codons, respectively. The target gene and the C. reinhardtii expression vector pChlamy_3 was then digested with KpnI and Pst I endonucleases, respectively. After double-enzyme digestion, the target gene and the expression vector were ligated overnight at 16°C, and the ligation product was transformed into E. coli DH5α receptive cells. Positive monoclonal colonies with ampicillin resistance were selected for PCR verification and expansion culture. The plasmid was extracted with an EndoFree Mini Plasmid kit II to obtain a pChlamy_3 expression vector containing the PhbZIP2 gene. The circular plasmid vector was linearized by single-enzyme digestion to optimize gene integration into the C. reinhardtii genome. The target gene was then transferred into C. reinhardtii by the glass bead transformation method (Kindle, 1990), and monoclonal C. reinhardtii with hygromycin resistance was screened for propagation and culture for use in subsequent experiments.
2.7 Transcriptome analysis
Total RNA from C. reinhardtii was sent to Gene Denovo Biotechnology Co., Ltd. (Guangzhou, China) for cDNA synthesis, and construct cDNA library for sequencing. Removing the low quality raw data reads, all remaining high quality clean sequencing reads were mapped onto C. reinhardtii genome reference to reconstruct transcripts using Stringtie. After calculating the expression levels of all single genes, the single gene expression levels of two samples were compared. The false discovery rate (FDR) was used to determine the threshold p-value for multiple tests. The following criteria were used to identify differentially expressed genes (DEGs): FDR <0.05 and a log2|fold-change| ≥ 1. Mapping all differentially expressed genes to each pathway of the KEGG database (https://www.kegg.jp/) and counting the number of genes in each pathway to identify the pathways that are significantly enriched in differentially expressed genes. Tbtools (v1.098769) was used to map and present the analysis of DEGs.
2.8 Statistical analysis
Each experimental treatment was performed with three replicates. SPSS 23.0 and Excel were used for statistical analysis of the experimental results. One-way ANOVA was used to compare differences between groups, with p < 0.05 and p < 0.01 considered to be significant and extremely significant differences, respectively.
3 Results
3.1 Cloning and expression pattern analysis of PhbZIP2
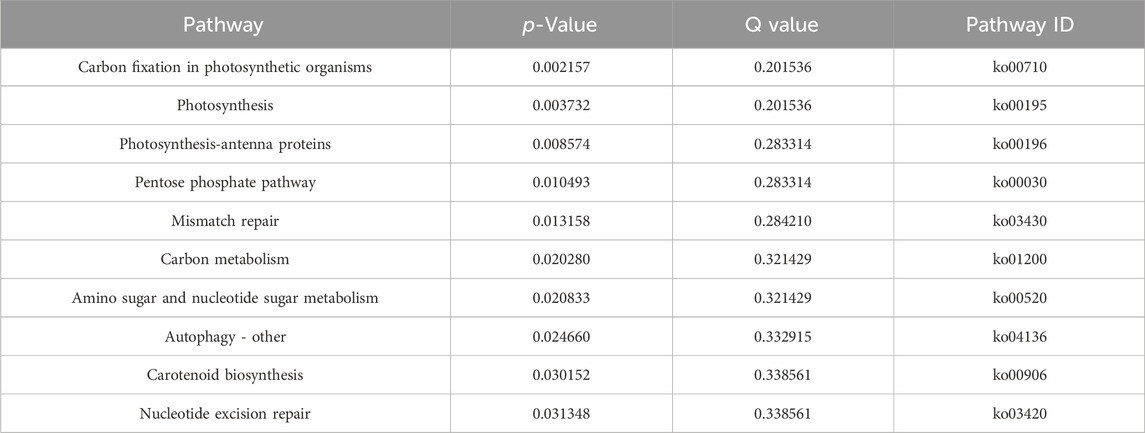
The cDNA clone of PhbZIP2 was verified with a PCR-based sequence. The PhbZIP2 open reading frame, comprised 1,314 bp encoding 437 amino acids. The predicted protein had a molecular weight of 44,761.56 Da and a theoretical isoelectric point of 8.92. Conserved domain analysis revealed that the gene contained one BRLZ (337–418 aa) domain (Figure 1A). Phylogenetic analysis placed PhbZIP2 in a sister relationship with a bZIP2 from Porphyra umbilicalis (Figure 1B). The detected expression of PhbZIP2 was significantly induced by high temperature and hypersaline and hyposaline stresses (Figures 1C, D).
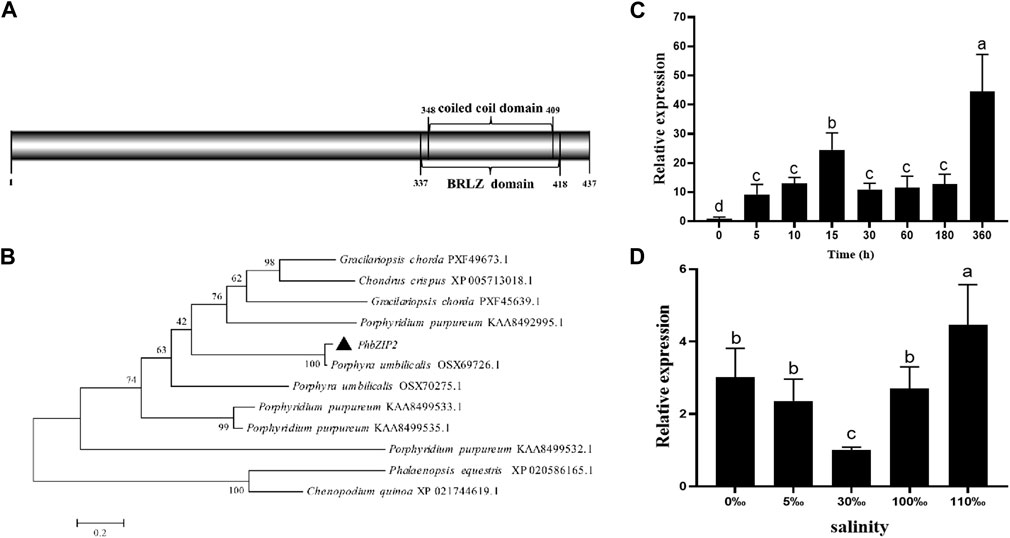
Figure 1. Sequence and expression pattern of PhbZIP2. (A) Protein domain analysis of PhbZIP2; (B) Neighbor-joining phylogenetic tree of PhbZIP2 amino acid sequences, The accession number of each sequence are PXF49673.1 Gracilariopsis chorda, XP_005713018.1 Chondrus crispus, PXF45639.1 Gracilariopsis chorda, KAA8492995.1 Porphyridium purpureum, OSX69726.1 Porphyra umbilicalis, OSX70275.1 Porphyra umbilicalis, KAA8499533.1 Porphyridium purpureum, KAA8499535.1 Porphyridium purpureum, KAA8499532.1 Porphyridium purpureum, XP_020586165.1 Phalaenopsis equestris, XP_021744619.1 Chenopodium quinoa; (C) Changes in transcript levels of PhbZIP2 under high temperature (32°C) stress; (D) Relative transcript levels of PhbZIP2 following 4 h' different salinity treatments. Salinity means the number of grams of NaCl per liter of water. Data are presented as means ± standard error (n = 3). Different superscript letters indicate significance compared to the control (p < 0.05).
3.2 Global analysis of PhbZIP2 binding sites in gene functional elements
The DAP-seq detected a total of 2,981 peaks with a total length of 1,027,663 bp, an average length of 344 bp, a total stacking depth of 47,973, and an average stacking depth of 16 bp. The peak intervals accounted for 2.00% of the genome. The observed distribution of the peaks clustered around 400 bp upstream of transcription start sites (TSSs) (Figure 2A). The overall distribution of peaks was 34.12% in promoters, 0.03% in 5′UTRs, 18.35% in exons, 6.24% in introns, 9.46% in downstream regions, and 31.8% in intergenic regions (Figure 2B). PhbZIP2 can bind to conserved motifs, such as (T/C)TCCA(C/G), A (A/G)AAA (G/A) and A (T/C)A (G/T)AC (A/G)T (Figure 2C). The motif-enriched regions detected by the DAP-seq were mainly located on Chromosome1, two and 3 (Figure 2D). The significantly enriched KEGG pathways (p < 0.01) of the target genes of PhbZIP2 were involved in Carbon fixation in photosynthetic organisms, Photosynthesis, and Photosynthesis-antenna proteins (Table 1).
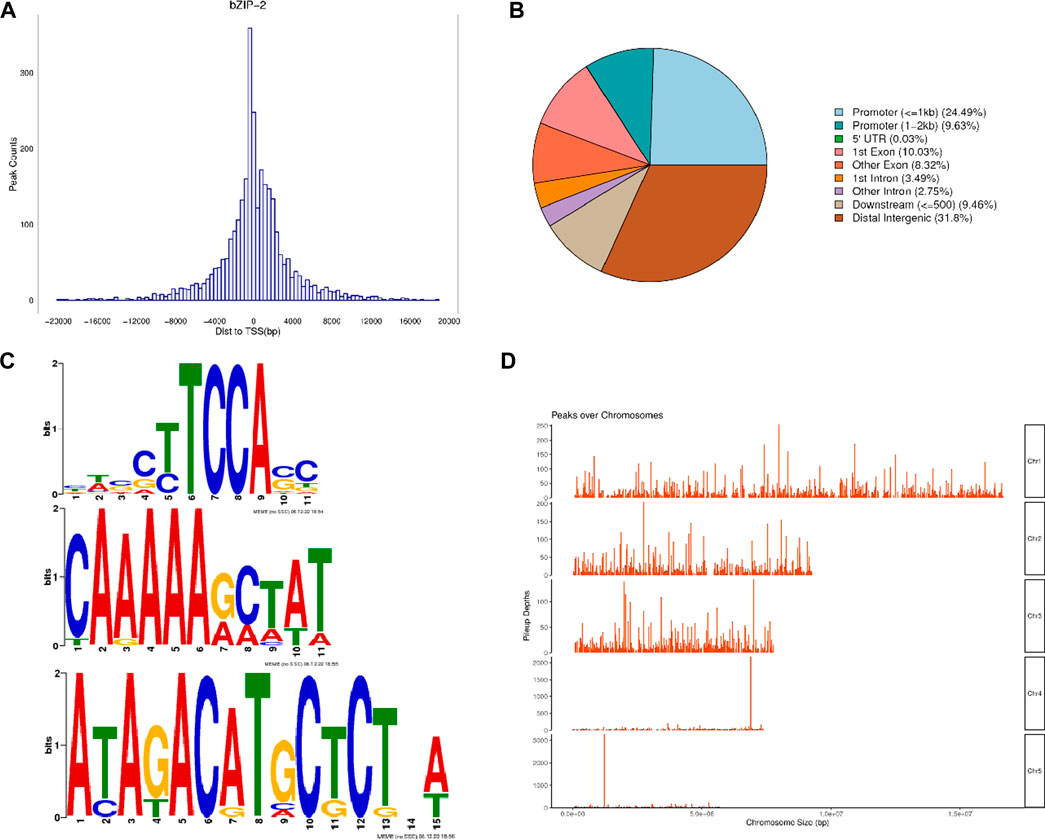
Figure 2. Analysis of PhbZIP2 binding motifs. (A) Number of peaks vs. distance from transcription start sites (TSSs); (B) Proportion of peaks in different gene functional elements; (C) PhbZIP2 gene binding-motif sequences; (D) Distribution of motif-enriched regions (Peaks) on chromosomes.
3.3 Identification of stress tolerance in PhbZIP2-expressed C. reinhardtii
As confirmed by agarose gel electrophoresis, PhbZIP2 target fragments were amplified from C. reinhardtii, whereas no PCR products were detected from wild type C. reinhardtii (Supplementary Figure S1). In transgenic individuals maintained at 32°C, the transcription level of PhbZIP2 was significantly increased at 5 min and gradually increased as the high temperature stress was prolonged up to 360 min (Figure 3A). In transgenic C. reinhardtii subjected to hypersaline stress, the expression level of PhbZIP2 was relatively stable for the first 30 min. After 60 min of treatment, however, the expression level was significantly increased and then remained at a high level (Figure 3B). It was observed that the survival rate of transgenic C. reinhardtii after 48 h of high temperature treatment was always higher than that of the wild type, and the biomass difference between the two lines became increasingly significant with lengthening treatment duration (Figures 3C, D). After treatment with 150 mM NaCl for 72 h, most of the wild type individuals died, whereas transgenic C. reinhardtii was still alive (Figures 3E, F).
Figure 3. Stress tolerance of wild type (WT) and PhbZIP2-expressed transgenic (T2) C. reinhardtii. (A,B): Changes in transcript levels of PhbZIP2 in transformed C. reinhardtii at 32°C (A) and hypersaline (B) stresses; (C,D): Changes in the phenotype (C) and biomass (D) of transgenic and wild type C. reinhardtii under high temperature (32°C) stress. (E,F): Changes in the phenotype (E) and biomass (F) of transgenic and wild type C. reinhardtii under hypersalinity stress. Data are presented as means ± standard error (n = 3). In (A,B), different superscript letters indicate significant differences between groups (p < 0.05). In d and f, asterisk indicate significant differences between samples at the same time point (*, p < 0.05; **, p < 0.01).
3.4 KEGG enrichment of differentially expressed stress--related genes in transgenic C. reinhardtii with PhbZIP2
Analysis of the KEGG pathway enrichment displayed that the DEGs employed in response to 12 h salt stress were mainly involved in photosynthesis, photosynthesis, photosynthesis-antenna proteins and Ribosome biogenesis in eukaryotes (Figure 4A). The subsequent analysis identified most genes related to photosynthesis-antenna protein had downregulated expression trends in response to salt stress. In the wild type, 17 of 20 and 19 of 20 genes involved in photosynthesis and photosynthesis-antenna proteins, respectively, were downregulated under salt stress treatment. Compared with the wild type, the expression of photosynthesis-related genes (PSBS1, PSBS2, petF) and photosynthesis antenna protein-related genes (LHCB7, LHCSR1, LHCSR3.1, CAB8) were upregulated in transgenic lines under salt stress (Figures 4B, C).
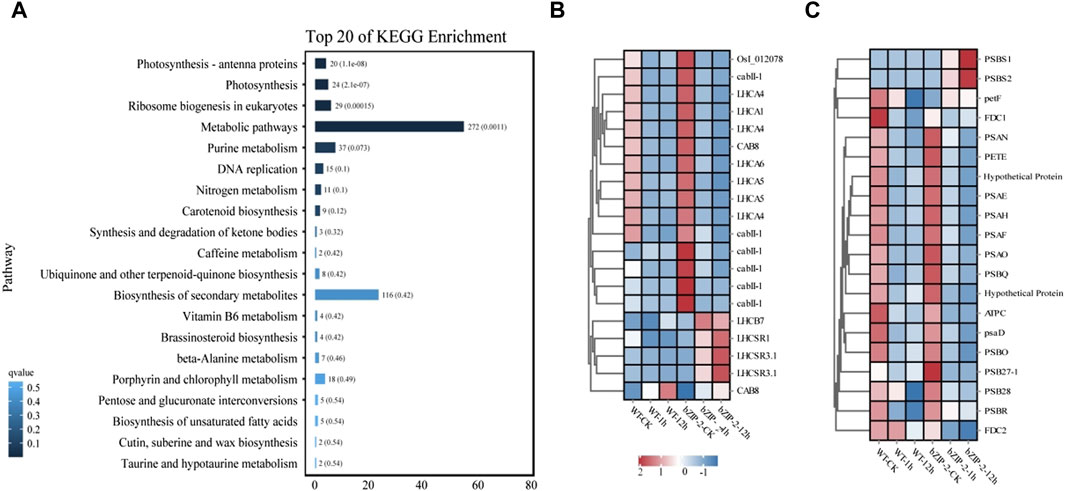
Figure 4. Comparative transcriptome analysis of the wild type (WT) strain and the transgenic strain of Chlamydomonas reinhardtii with PhbZIP2 under salt stress. (A) KEGG pathway enrichment analysis of genes under 12 h of salt stress treatment. (B,C) represents the effect of stress on the expression of genes annotated by photosynthesis–antenna proteins and photosynthesis, respectively. WT-CK, the sample of WT strain under normal condition; WT-1h, the sample of WT strain treated with salt stress for 1 h; WT-12h, the sample of WT strain treated with salt stress for 12 h; bZIP-2-CK, the sample of the transgenic C. reinhardtii strain with PhbZIP2 under normal condition; bZIP-2-1h, the sample of the transgenic C. reinhardtii strain with PhbZIP2 treated with salt stress for 1 h; bZIP-2-12h, the sample of the transgenic C. reinhardtii strain with PhbZIP2 treated with salt stress for 12 h; Abbreviated gene names are listed in Supplementary Table S2.
4 Discussion
In this study, we found that PhbZIP2 of P. haitanensis contains a BRLZ domain as well as an α coiled-coil structure between amino acids 337 and 418 (Figure 1A), which is typical of bZIP family members (Jakoby et al., 2002). According to the structural differences of bZIP transcription factors, they can be divided into 10 subfamilies. Different family members are widely involved in a variety of biological processes, among which H class is mainly involved in the regulation of photosynthesis (Blouin et al., 2011). The results of phylogenetic tree showed that PhbZIP2 was clustered with AtbZIP56 (H class) and other transcription factors, indicating that PhbZIP2 had high homology with Arabidopsis thaliana, and the protein structure type and function had certain similarity. PhbZIP2 expression was significantly upregulated in response to high temperature almost immediately (starting at 5 min of stress). This outcome is similar to results obtained in studies of wheat and rice, where significant increases in TabZIP60 and OsbZIP23 expressions were observed during early stages of high temperature and drought stresses, respectively (Geng et al., 2018; Zong et al., 2020). The upregulation of PhbZIP2 may therefore serve as a rapid response to abiotic stress to activate downstream stress-resistance genes. The expression of PhbZIP2 obviously decreased after 15 min of high temperature treatment, however, and then significantly increased at 360 min (Figure 1C). Wang et al. (2018) have reported that the response of P. haitanensis to high temperature stress can be divided into two stages. First, in the early stage of stress, the thallus had a stress response, which reduced photosynthesis to reduce the production of reactive oxygen species and unnecessary energy consumption. Afterwards, recombination was performed at the transcriptional level to further activate stress-resistant pathways such as photosynthesis, energy metabolism, and antioxidant systems to resist long-term high temperature stress. At 360 min of heat stress, in the present study, the upregulation of bZIP transcription factor may help activate stress-related genes such as photosynthesis to meet the energy and material needs of the thallus. (Wang et al., 2018). In addition, PhbZIP2 expression in transgenic C. reinhardtii was significantly increased at 5 min and 60 min of high temperature and hypersaline stress treatments, respectively (Figures 3A,B). The survival rate of PhbZIP2-expressing transgenic C. reinhardtii was always higher than that of the wild type under continuous high temperature or hypersaline stress (Figures 3C–F). These results indicate that PhbZIP2 is a key transcription factor regulating the transcriptional expression of downstream stress resistance-related genes in P. haitanensis.
Most PhbZIP2 peaks obtained from DAP-seq were concentrated near the TSS, with 34.12% and 31.8% distributed in gene promoters and intergenic regions, respectively (Figures 2A, B). This result suggests that PhbZIP2 regulates the transcription of downstream genes by binding to their promoters or to intergenic regions. Plant bZIP proteins preferentially bind palindromic or pseudo-palindromic cis-acting elements containing an ACGT core sequence, such as ABREs (CCACGTGG) and A-box (TACGTA), C-box (GACGTC), and G-box (CACGTG) elements (Dröge-Laser et al., 2018). These proteins can also bind to some motifs of non-palindromic structures (Sornaraj et al., 2016). For example, rice RF2a and tomato VSF-1 bZIP transcription factors can bind to the non-palindromic promoter sequences CCCACCTACCA and TCACCAACCGTTGGATGTGG, respectively (Yin et al., 1997; Ringli and Keller, 1998). Our analysis revealed that (T/C)TCCA(C/G) and A (A/G)AAA (G/A) motif sequences are abundant in PhbZIP2 (Figure 2C). We hypothesize that the motifs bound by PhbZIP2 differ from those with conserved binding regions in plants.
The significantly enriched (p < 0.01) KEGG pathways that were common to both the target genes of PhbZIP2 were Carbon fixation in photosynthetic organisms, Photosynthesis, and Photosynthesis-antenna proteins (Table 1). Photosynthesis of Pyropia/Porphyra is found to be highly sensitive to abiotic stresses (Wang et al., 2018; Terada et al., 2021; Shao et al., 2022). Increasing in light-harvesting efficiency and light-energy use, and suppressing accumulation of ROS are crucial for P. haitanensis responses to abiotic stress (Ji et al., 2023). bZIP transcription factor BLZ8 expression increased the photosynthetic linear electron transfer rate, decreasing the excitation pressure of the photosynthetic electron transport chain, and in turn inhibiting the accumulation of ROS in C. reinhardtii under oxidative stress (Choi et al., 2022). The Arabidopsis transgenics overexpressing a bZIP gene from Triticum aestivum, TabZIP, also exhibited higher photosynthetic efficiency and increased tolerance to salt stress (Agarwal et al., 2019). Similarly, the DEGs between the transgenics C. reinhardtii overexpressing PhbZIP2 and wild type under salt stress were also enriched (p < 0.01) by Photosynthesis and Photosynthesis-antenna proteins (Figure 4). Overall, the major of these DEGs showed downregulation in wild type and transgenics lines under salt stress, which was consistent with the results of reduction of biomass (Figure 3), indicating salt treatment impaired the photosynthesis of algae. But PhbZIP2 increased the expression levels of most DEGs involved in photosynthesis and photosynthesis-antenna proteins under normal condition. Additionally, in comparison with wild type, one PsbR genes, two PsbS genes, three LHCSR genes, and one LHCB genes exhibited obvious upregulation in transgenics lines under salt stress. This was beneficial for transgenics lines to protect against photo-oxidative damage to photosynthesis organelles during prolonged salt stress. Therefore, photosynthesis-related genes may be important target genes for transcription factor PhbZIP2 regulation in response to abiotic stress.
5 Conclusion
In this study, the bZIP family transcription factor PhbZIP2 in P. haitanensis was cloned and characterized for the first time. Transcription of PhbZIP2 were rapidly accumulated during the high temperature and saline stresses. DAP-seq and heterologous expression analyses suggested that PhbZIP2 was involved in the response of P. haitanensis to abiotic stress by regulating photosynthesis, which shed light on the stress resistance mechanisms of intertidal seaweed and provide important information for breeding new varieties with stronger stress resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA839642.
Author contributions
HZ: data curation, validation, writing–original draft. GZ: data curation, validation, writing–original draft. JX: writing–review and editing. YZ: supervision, methodology, writing–original draft. DJ: supervision, methodology, writing–original draft. YX: supervision, methodology, writing–original draft. CX: conceptualization, project administration, writing–review and editing. WW: conceptualization, writing–original draft, writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant numbers: 42176117 and U21A20265), and the China Agriculture Research System of MOF and MARA (grant number: CARS-50), and The Young Elite Scientists Sponsorship Program by the China Association of Science and Technology (grant number: 2021QNRC001).
Acknowledgments
We are grateful to Guangzhou Genedenovo Biotechnology Co., Ltd for assisting in Dap sequencing. We thank Liwen Bianji (Edanz) (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2024.1345585/full#supplementary-material
References
Agarwal, P., Baranwal, V. K., and Khurana, P. (2019). Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 9, 4608. doi:10.1038/s41598-019-40659-7
Bartlett, A., O'Malley, R. C., Huang, S., Galli, M., Nery, J. R., Gallavotti, A., et al. (2017). Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12 (8), 1659–1672. doi:10.1038/nprot.2017.055
Blouin, N. A., Brodie, J. A., Grossman, A. C., Xu, P., and Brawley, S. H. (2011). Porphyra: a marine crop shaped by stress. Trends Plant Sci. 16 (1), 29–37. doi:10.1016/j.tplants.2010.10.004
Brawley, S. H., Blouin, N. A., Ficko-Blean, E., Wheeler, G. L., Lohr, M., Goodson, H. V., et al. (2017). Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. U. S. A. 114, E6361–E6370. doi:10.1073/pnas.1703088114
Cao, M., Xu, K. P., Yu, X. Z., Bi, G. Q., Liu, Y., Kong, F. N., et al. (2020). A chromosome-level genome assembly of Pyropia haitanensis (Bangiales, Rhodophyta). Mol. Ecol. Resour. 20, 216–227. doi:10.1111/1755-0998.13102
Chang, J., Shi, J. Z., Lin, J. Z., Ji, D. H., Xu, Y., Chen, C. S., et al. (2021). Molecular mechanism underlying Pyropia haitanensis PhHsp22-mediated increase in the high-temperature tolerance of Chlamydomonas reinhardtii. J. Appl. Phycol. 33, 1137–1148. doi:10.1007/s10811-020-02351-6
Chen, H., Shih-Chieh, C. J., Chen, J., Luo, Q., Wang, H., Lu, R., et al. (2021). Insights into the ancient adaptation to intertidal environments by red algae based on a genomic and multiomics investigation of Neoporphyra haitanensis. Mol. Biol. Evol. 39, b315. doi:10.1093/molbev/msab315
Chen, T. X., Wang, W. L., Xu, K., Xu, Y., Ji, D. H., Chen, C. S., et al. (2019). K+ and Na+ transport contribute to K+/Na+ homeostasis in Pyropia haitanensis under hypersaline stress. Algal Res. 40, 101526. doi:10.1016/j.algal.2019.101526
Choi, B. Y., Kim, H., Shim, D., Jang, S., Yamaoka, Y., Shin, S., et al. (2022). The Chlamydomonas bZIP transcription factor BLZ8 confers oxidative stress tolerance by inducing the carbon-concentrating mechanism. Plant Cell 34 (2), 910–926. doi:10.1093/plcell/koab293
Dröge-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP transcription factor family—an update. Curr. Opin. Plant Biol. 45, 36–49. doi:10.1016/j.pbi.2018.05.001
Geng, X., Zang, X., Li, H., Liu, Z., Zhao, A., Liu, J., et al. (2018). Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 274, 252–260. doi:10.1016/j.plantsci.2018.05.029
Guan, X., Mao, Y., Stiller, J. W., Shu, S., Pang, Y., Qu, W., et al. (2022). Comparative gene expression and physiological analyses reveal molecular mechanisms in wound-induced spore formation in the edible seaweed nori. Front. Plant Sci. 13, 840439. doi:10.3389/fpls.2022.840439
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends plant Sci. 7 (3), 106–111. doi:10.1016/S1360-1385(01)02223-3
Ji, D. H., Xu, Y., Xiao, H. D., Chen, C. S., Xu, K., and Xie, C. T. (2016). Superoxide dismutase genes in Pyropia haitanensis: molecular cloning, characterization and mRNA expression. Acta Oceanol. Sin. 35, 101–111. doi:10.1007/s13131-016-0873-2
Ji, D. H., Zhang, Y. C., Zhang, B., Xu, Y., Xu, K., Chen, C. S., et al. (2023). Investigating the mechanisms underlying the low irradiance-tolerance of the economically important seaweed species Pyropia haitanensis. Life 13 (2), 481. doi:10.3390/life13020481
Kindle, K. L. (1990). High frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 87, 1228–1232. doi:10.1073/pnas.87.3.1228
Kong, F., Dong, D., Li, N., Sun, B., and Sun, M. (2021). Characterization of PyMAPK2, a D group mitogen-activated protein kinase gene from Pyropia yezoensis responding to various abiotic stress. Algal Res. 59, 102445. doi:10.1016/j.algal.2021.102445
Kumar, D., and Chattopadhyay, S. (2018). Glutathione modulates the expression of heat shock proteins via the transcription factors BZIP10 and MYB21 in Arabidopsis. J. Exp. Bot. 69, 3729–3743. doi:10.1093/jxb/ery166
Ma, H. Z., Liu, C., Li, Z. X., Ran, Q. J., Xie, G. N., Wang, B. M., et al. (2018). ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant physiol. 178, 753–770. doi:10.1104/pp.18.00436
Nicholas, J. (2000). Bangiomorpha pubescensn. gen., n. sp.: implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 26, 386–404. doi:10.1666/0094-8373(2000)026<0386:bpngns>2.0.co;2
Niu, J. F., Yang, J. L., Feng J, H., Feng, Z. Z., Wang, X., Yu, B., et al. (2022). Ubiquitin-proteasome pathway plays an essential regulatory role during spermatangium formation in Neopyropia yezoensis. Algal Res. 62, 102623. doi:10.1016/j.algal.2021.102623
Pouya, K., and Manolis, K. (2014). Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 42 (5), 2976–2987. doi:10.1093/nar/gkt1249
Ringli, C., and Keller, B. (1998). Specific interaction of the tomato bZIP transcription factor VSF-1 with a non-palindromic DNA sequence that controls vascular gene expression. Plant Mol. Biol. 37, 977–988. doi:10.1023/A:1006030007333
Shao, Z., Xie, X., Liu, X., Zheng, Z., Huan, L., Zhang, B., et al. (2022). Overexpression of mitochondrial γCAL1 reveals a unique photoprotection mechanism in intertidal resurrection red algae through decreasing photorespiration. Algal Res. 66, 102766. doi:10.1016/j.algal.2022.102766
Sornaraj, P., Luang, S., Lopato, S., and Hrmova, M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochimica Biophysica Acta 1860, 46–56. doi:10.1016/j.bbagen.2015.10.014
Suda, M., and Mikami, K. (2020). Reproductive responses to wounding and heat stress in gametophytic thalli of the red alga Pyropia yezoensis. Front. Mar. Sci. 7, 394. doi:10.3389/fmars.2020.00394
Terada, R., Nishihara, G. N., Arimura, K., Watanabe, Y., Mine, T., and Morikawa, T. (2021). Photosynthetic response of a cultivated red alga, Neopyropia yezoensis f. narawaensis (= Pyropia yezoensis f. narawaensis; Bangiales, Rhodophyta) to dehydration stress differs with between two heteromorphic life-history stages. Algal Res. 55, 102262. doi:10.1016/j.algal.2021.102262
Uji, T., Gondaira, Y., Fukuda, S., Mizuta, H., and Saga, N. (2019). Characterization and expression profiles of small heat shock proteins in the marine red alga Pyropia yezoensis. Cell Stress Chaperones 24, 223–233. doi:10.1007/s12192-018-00959-9
Wang, D. M., Yu, X. Z., Xu, K. P., Bi, G. Q., Cao, M., Zelzion, E., et al. (2020). Pyropia yezoensis genome reveals diverse mechanisms of carbon acquisition in the intertidal environment. Nat. Commun. 11, 4028. doi:10.1038/s41467-020-17689-1
Wang, W. L., Chang, J., Zheng, H. Y., Ji, D. H., Xu, Y., Chen, C. S., et al. (2019a). Full-length transcriptome sequences obtained by a combination of sequencing platforms ap-plied to heat shock proteins and polyunsaturated fatty acids biosynthesis in Pyropia haitanensis. J. Appl. Phycol. 31, 1483–1492. doi:10.1007/s10811-018-1624-x
Wang, W. L., Chen, T. X., Xu, Y., Xu, K., Xu, Y., Ji, D. H., et al. (2020a). Investigating the mechanisms underlying the hyposaline tolerance of intertidal seaweed, Pyropia haitanensis. Algal Res. 47, 101886. doi:10.1016/j.algal.2020.101886
Wang, W. L., Lin, J. Z., Chang, J., Ji, D. H., Xu, Y., Chen, C. S., et al. (2021). A RING type ubiquitin ligase PhCUL4 is involved in thermotolerance of Pyropia haitanensis. Algal Res. 59, 102448. doi:10.1016/j.algal.2021.102448
Wang, W. L., Teng, F., Lin, Y. H., Ji, D. H., Xu, Y., Che, C. S., et al. (2018). Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PLoS One 13, e0195842. doi:10.1371/journal.pone.0195842
Wang, W. L., Xing, L., Xu, K., Ji, D. H., Xu, Y., Chen, C. S., et al. (2020b). Salt stress-induced H2O2 and Ca2+ mediate K+/Na+ homeostasis in Pyropia haitanensis. J. Appl. Phycol. 32, 4199–4210. doi:10.1007/s10811-020-02284-0
Wang, W. L., Xu, Y., Chen, T. X., Xing, L., Xu, K., Xu, Y., et al. (2019b). Regulatory mechanisms underlying the maintenance of homeostasis in Pyropia haitanensis under hypersaline stress conditions. Sci. Total Environ. 662, 168–179. doi:10.1016/j.scitotenv.2019.01.214
Wang, W. L., Zheng, H. Y., Xu, K., Ji, D. H., Xu, Y., Chen, C. S., et al. (2022). Early signaling events in the heat stress response of Pyropia haitanensis revealed by phosphoproteomic and lipidomic analyses. Algal Res. 67, 102837. doi:10.1016/j.algal.2022.102837
Wen, J., Wang, W. L., Xu, K., Ji, D. H., Xu, Y., Chen, C. S., et al. (2020). Comparative Analysis of proteins involved in energy metabolism and protein processing in Pyropia haitanensis at different salinity levels. Front. Mar. Sci. 7, 415. doi:10.3389/fmars.2020.00415
Yin, Y. H., Chen, L. L., and Beachy, R. (1997). Promoter elements required for phloem-specific gene expression from the RTBV promoter in rice. Plant J. 12, 1179–1188. doi:10.1046/j.1365-313X.1997.12051179.x
Zheng, H. Y., Wang, W. L., Xu, K., Xu, Y., Ji, D. H., Chen, C. S., et al. (2019). Ca2+ influences heat shock signal transduction in Pyropia haitanensis. Aquaculture 516, 734618. doi:10.1016/j.aquaculture.2019.734618
Zheng, H. Y., Xu, Y., Ji, D. H., Xu, K., Chen, C. S., Wang, W. L., et al. (2022). Calcium-calmodulin-involved heat shock response of Neoporphyra haitanensis. Front. Mar. Sci. 9, 875308. doi:10.3389/fmars.2022.875308
Keywords: intertidal macroalgae, abiotic stress tolerance, bZIP transcription factor, DAP-seq, heterologous expression
Citation: Zhang H, Zeng G, Xie J, Zhang Y, Ji D, Xu Y, Xie C and Wang W (2024) PhbZIP2 regulates photosynthesis-related genes in an intertidal macroalgae, Pyropia haitanensis, under stress. Front. Mol. Biosci. 11:1345585. doi: 10.3389/fmolb.2024.1345585
Received: 28 November 2023; Accepted: 11 March 2024;
Published: 15 April 2024.
Edited by:
Ying Lu, Shanghai Ocean University, ChinaReviewed by:
Bahman Panahi, Agricultural Biotechnology Research Institute of Iran, IranNianjun Xu, Ningbo University, China
Binbin Chen, Wenzhou University, China
Copyright © 2024 Zhang, Zeng, Xie, Zhang, Ji, Xu, Xie and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaotian Xie, ctxie@jmu.edu.cn; Wenlei Wang, wlwang@jmu.edu.cn
†These authors have contributed equally to this work
 Han Zhang1,2,3†
Han Zhang1,2,3†  Jiajia Xie
Jiajia Xie Dehua Ji
Dehua Ji Chaotian Xie
Chaotian Xie Wenlei Wang
Wenlei Wang