Kinesin-7 CENP-E in tumorigenesis: Chromosome instability, spindle assembly checkpoint, and applications
- 1Department of Cell Biology and Genetics, The School of Basic Medical Sciences, Fujian Medical University, Fuzhou, China
- 2Key Laboratory of Stem Cell Engineering and Regenerative Medicine, Fujian Province University, Fuzhou, China
- 3Medical Research Center, Fujian Maternity and Child Health Hospital, Fuzhou, China
- 4College of Clinical Medicine for Obstetrics and Gynecology and Pediatrics, Fujian Medical University, Fuzhou, China
Kinesin motors are a large family of molecular motors that walk along microtubules to fulfill many roles in intracellular transport, microtubule organization, and chromosome alignment. Kinesin-7 CENP-E (Centromere protein E) is a chromosome scaffold-associated protein that is located in the corona layer of centromeres, which participates in kinetochore-microtubule attachment, chromosome alignment, and spindle assembly checkpoint. Over the past 3 decades, CENP-E has attracted great interest as a promising new mitotic target for cancer therapy and drug development. In this review, we describe expression patterns of CENP-E in multiple tumors and highlight the functions of CENP-E in cancer cell proliferation. We summarize recent advances in structural domains, roles, and functions of CENP-E in cell division. Notably, we describe the dual functions of CENP-E in inhibiting and promoting tumorigenesis. We summarize the mechanisms by which CENP-E affects tumorigenesis through chromosome instability and spindle assembly checkpoints. Finally, we overview and summarize the CENP-E-specific inhibitors, mechanisms of drug resistances and their applications.
1 Introduction
The human genome contains 45 different kinesins, which can be divided into 14 subfamilies according to the phylogenetic analysis and classification of the motor domain (Lawrence et al., 2004; Rath and Kozielski, 2012). Kinesin-7 CENP-E (Centromere protein E) was first discovered as a 312 kDa chromosome scaffold-associated protein, which is located at the centromere of chromosome at metaphase and then redistributed to the midbody at telophase (Yen et al., 1991; 1992). CENP-E is a plus-end-directed kinesin at the outer kinetochore plate and the fibrous corona of kinetochores (Thrower et al., 1996; Cooke et al., 1997; Kim et al., 2008). The 230-nm-long coiled-coil of CENP-E serves as a motile kinetochore tether for microtubule capture and chromosome alignment (Kim et al., 2008). CENP-E is required for chromosome congression, alignment, and metaphase-to-anaphase transition during cell division (McEwen et al., 2001; Putkey et al., 2002; Weaver et al., 2003).
The expression of the CENP-E gene varies in different types of cancer, and most of them are upregulated (Yuan et al., 2023). CENP-E acts both as an oncogene and as a tumor suppressor during tumorigenesis (Weaver et al., 2007). Low levels of chromosome instability can promote tumor initiation, while high levels of aneuploidy result in the suppression of tumor growth and eventually cell death (Weaver and Cleveland, 2007; Weaver et al., 2007; Silk et al., 2013). CENP-E participates in mitotic checkpoint and cell cycle control to prevent chromosome missegregation that leads to aneuploidy (Weaver et al., 2007).
Kinesin family motors are key regulators in cell division and have become potential targets for chemotherapeutic intervention and cancer treatment (Rath and Kozielski, 2012). Considering the relationship between CENP-E and tumorigenesis, CENP-E’s specific inhibitors (Henderson et al., 2009; Ding et al., 2010; Qian et al., 2010; Hirayama et al., 2013; Kung et al., 2014; Ohashi et al., 2015a; Yamane et al., 2019) have also been synthesized and verified. And only GSK923295 entered clinical phase I (Chung et al., 2012). In this review, we summarize molecular mechanisms of CENP-E and tumorigenesis from the perspectives of expression patterns, cell division, and aneuploidy. Furthermore, we highlight the applications of CENP-E inhibitors and drug resistance mechanisms in tumor research and treatment.
2 Structure and molecular kinetics of kinesin-7 CENP-E
CENP-E consists of an N-terminal motor domain, a central coiled-coil domain, and a C-terminal tail domain (Figure 1A). The N-terminal motor domain is highly conserved in diverse organisms (Craske et al., 2022) (Figure 1B). Unlike conventional kinesins, CENP-E has a 230-nm-long discontinuous coiled-coil, which forms different conformations in vitro and carries cargos in a compact configuration (Kim et al., 2008; Gudimchuk et al., 2018). The long coiled-coil domain mediates the motor functions and structural flexibility of CENP-E (Vitre et al., 2014; Taveras et al., 2019). The adjustable stalk configuration is required for physical interactions between CENP-E and spindle microtubules (Gudimchuk et al., 2018). The neck linker domain is responsible for the processivity of CENP-E motors (Hariharan and Hancock, 2009; Shastry and Hancock, 2011).
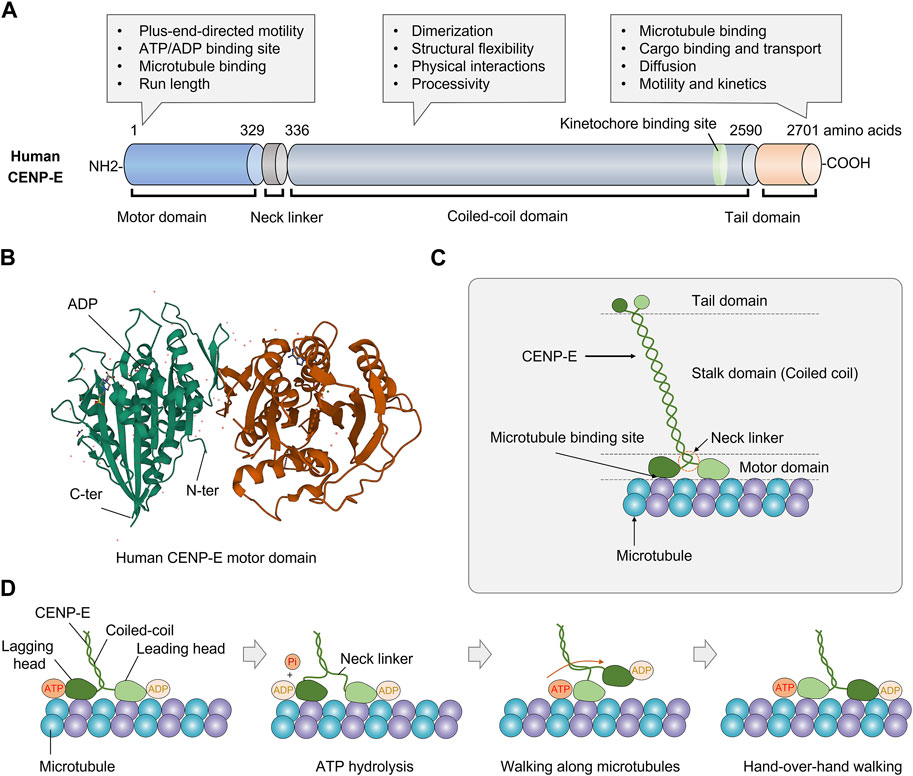
FIGURE 1. Structure and molecular kinetics of kinesin-7 CENP-E. (A) CENP-E is comprised of an N-terminal motor domain, a coiled-coil domain, and a C-terminal tail domain. The motor domain is required for plus-end-directed motility, ATP hydrolysis, microtubule binding, and run length. The coiled-coil domain is required for dimerization, structural flexibility, physical interactions with partner proteins, and processivity. The tail domain is essential for microtubule binding, cargo transport, diffusion along microtubules, motility, and kinetics. (B) Three-dimensional structure of human CENP-E motor domain (PDB database, No. 1T5C). The globular N-terminal motor domain contains the ATP/ADP binding site, which is essential for ATP hydrolysis and movement. (C) Schematic structure of kinesin-7 CENP-E. (D) The coordinated movement of CENP-E’s head domains along a microtubule. CENP-E is a processive motor that takes 8 nm steps along microtubules for each adenosine triphosphate hydrolyzed via a hand-over-hand mechanism.
Human full-length CENP-E is predominantly inactive and becomes processive after microtubule binding (Craske et al., 2022). Most full-length CENP-E motors move at a slow velocity of 46.4 ± 1.88 nm/s (Craske et al., 2022). CENP-E motors show a higher run length and residency time than truncated CENP-E motors, which may be due to the non-motor microtubule-binding site at the C-terminal tail domain (Craske et al., 2022). Full-length CENP-E can walk to the microtubule plus end and maintain at the microtubule end for 20 s (Gudimchuk et al., 2013). In the tail domain, there is a kinetochore-targeting region (2055-2,450 amino acids) (Legal et al., 2020), a centrosome-targeting domain (2,260-2,608 amino acids), and a second microtubule-binding site (Gudimchuk et al., 2013; Ciossani et al., 2018) (Figure 1C). The C-terminal region (2091-2,358 amino acids) is essential for the recruitment of BubR1 at the kinetochores (Legal et al., 2020). The C-terminal domain also recruits the ROD-Zwilch-ZW10 (RZZ) complex, Spindly and Mad1 to the kinetochores (Weber et al., 2024). The tail domain is intrinsically disordered, which is a common structural feature of kinesins (Seeger et al., 2012). The tail domain truncated protein can diffuse along the microtubules with an average binding time of 0.5 s, suggesting a weak microtubule-binding affinity (Gudimchuk et al., 2013). Interestingly, the tail domain of CENP-E can inhibit the motility of CENP-E through the motor-tail interaction (Espeut et al., 2008). In different species, CENP-E is not conserved in the coiled-coil and tail domains, which are required to interact with partner proteins in vivo. This also contributes to the divergence in the kinetics and functions of CENP-E.
The crystal structure of the CENP-E motor domain associated with MgADP (PDB entry 1t5c) was reported (Garcia-Saez et al., 2004). The release of ADP is a rate-limiting step in the ATPase cycle of CENP-E (Sardar and Gilbert, 2012; Shibuya et al., 2021). The α0 helix is conserved in kinesin Eg5, however, these residues are disordered (Shibuya et al., 2021). The regions of α0 helix and Loop L1 are flexible in the motor domain of CENP-E (Shibuya et al., 2021) (Figure 1D). A recent study has revealed the crystal structure of CENP-E motor domain in complex with AMPPNP. And the helix α4 is required for the slow binding of CENP-E to microtubules (Shibuya et al., 2023). In the future, crystal structures of CENP-E in complex with specific inhibitors will help to elucidate the mechanisms of kinesin motors and the development of anticancer drugs.
3 Expression patterns of CENP-E in cancers: the contradiction between protective factor and oncogene
The high-level expression of CENP-E is closely related to its important functions during cell division. However, it is still unclear which factors regulate the high-level expression of CENP-E in the G2/M phase, which are fascinating questions that remain to be uncovered in the future. The upregulation of the expression level of CENP-E was involved in the tumorigenesis of various cancers. CENP-E is upregulated in human neuroblastoma (Balamuth et al., 2010), retinoblastoma (Shi et al., 2021), melanoma (Uzdensky et al., 2014), esophageal cancer (Zhu et al., 2019), lung adenocarcinoma (Shan et al., 2019), gliomas (Rahane et al., 2019; Xu et al., 2020), non-small cell lung cancer (Hao and Qu, 2019; Ma et al., 2019), basal-like subtype among breast cancer (Kung et al., 2014), chemotherapy-resistant epithelial ovarian cancer (Ju et al., 2009), and castration-resistant PCa (Liang et al., 2017). In addition, evaluation based on TCGA, GEPIA, and Oncomine databases has also revealed that the upregulation of the CENP-E gene in multiple tumor types, including colorectal cancer, cervical cancer, gastric cancer, breast cancer, lung cancer, and sarcoma (Shi et al., 2021). Expression patterns and key functions of CENP-E in multiple cancers are shown in Table 1.
In breast cancer cells, the CENP-E gene is overexpressed and associated with poor prognosis (Agarwal et al., 2009). The elevated expression levels of the CENP-E gene enhance the sensitivity of breast cancers to a drug, (+)—JQ1 (Tian et al., 2021). Moreover, the high expression of the CENP-E gene is associated with poor overall survival in patients with esophageal cancer and adenocarcinoma (Zhu et al., 2019). In retinoblastoma cell lines, the elevated expression of CENP-E positively correlates with tumor cell invasiveness (Shi et al., 2021), which suggests its potential role as a biomarker and drug target. CENP-E expression correlates with survival analyses in primary and recurrent synovial sarcomas, which may serve as a biomarker to indicate prognostic significance between metastasis and recurrence (Yao et al., 2021). Overexpression of CENP-E correlates with poor prognosis in the low-grade gliomas (Qi et al., 2020). And the expression level of CENP-E can be used as an indicator to evaluate the prognosis of esophageal squamous cell carcinoma (Shi et al., 2020) and osteosarcoma (Wang et al., 2020). These findings suggest that high expression level of CENP-E is closely related to poor prognosis and overall survival. Accumulating evidence has revealed that CENP-E is a candidate biomarker in cancer diagnosis and treatment.
siRNA or GSK923295-mediated CENP-E inhibition can activate TP53 or TP73 and cell death signaling pathways, suggesting that CENP-E may be a potential therapeutic target for medulloblastoma (Iegiani et al., 2021). Furthermore, proliferation of the aneuploid cells induced by CENP-E partial deletion using RNAi interference is counteracted by the p14ARF tumor suppressor, indicating that p14ARF-p53 pathway is critical for preventing aneuploidy and chromosome instability in human cells (Veneziano et al., 2019). The interactions between CENP-E with kinesin-14 KIFC1 promote cell proliferation, migration, and epithelial-mesenchymal transition in ovarian cancers (Li et al., 2020). In castration-resistant PCa, genetic deletion or drug inhibition of CENP-E suppresses cell proliferation of prostate cancers (Liang et al., 2017). CENP-E is highly expressed in lung adenocarcinoma, and the downregulation of CENP-E is associated with the inhibition of the proliferation of lung cancer cells (Shan et al., 2019). In addition, the transcriptomic analysis revealed that CENP-E knockdown results in the downregulation of the pathways associated with G2/M checkpoint, mitotic spindle assembly checkpoint and the stress response in human primary fibroblasts (Cilluffo et al., 2021).
In contrast, the expression levels of CENP-E mRNAs and proteins are low in human hepatocellular carcinoma, and the low expression of CENP-E leads to aneuploidy in normal liver cell line LO2 cells (Liu et al., 2009). Furthermore, CENP-E functions as a tumor suppressor in human hepatocellular carcinoma (He et al., 2020). In addition, the CENP-E gene has also been shown to be associated with the prognosis of hepatocellular carcinoma cells and may be used as a biomarker or therapeutic target (Liang et al., 2021). But strangely, the CENP-E gene is highly expressed in LIHC (Yuan et al., 2023). It is still inconclusive as to why CENP-E is lowly expressed in hepatocellular carcinoma. Considering that only part of HCC shows consensus subtypes of chromosome instability (Lee et al., 2022), this difference may be caused by tumor heterogeneity. In contrast to most cancers, the low CENP-E expression in hepatocellular carcinoma is associated with increased cell proliferation, poor prognosis, and adverse clinical pathology (He et al., 2020). CENP-E is required for cell cycle control to prevent chromosome missegregation (Weaver et al., 2003). Reduction of CENP-E may produce aneuploidy (He et al., 2020), and chromosome instability, which is one of the subtypes of hepatocellular carcinoma (Lee et al., 2022). However, the specific roles of CENP-E in liver cancers as a tumor suppressor need to be further elucidated.
Furthermore, in acute lymphoblastic leukemia, there is a new alternative transcript of CENP-E (NAT-CENP-E) in patients, which is downregulated in 3/4 of the patients and upregulated in 1/4 of the patients (Jiménez-Ávila et al., 2018). In addition, XAB2 interacts with the promoter of CENP-E and transcriptionally activates the expression of CENP-E in HeLa cells (Hou et al., 2016). Taken together, these findings suggest that there are different factors or specific pathways for regulating the expression level of CENP-E in different types of tumors. However, the generality of the regulation of the CENP-E expression levels in different tumors needs to be further studied in the future.
4 Functions and mechanisms of kinesin-7 CENP-E in cell division
In mammalian cells, kinetochore fibers comprise 20-30 microtubules, which are essential for end-on attachment between the microtubule plus-ends and the kinetochore (McEwen et al., 1997). During mitosis, CENP-E proteins are enriched at unattached and misaligned kinetochores at prometaphase (Craske and Welburn, 2020), but detached from the aligned kinetochores at metaphase (Brown et al., 1994; Vitre et al., 2014). CENP-E proteins translocate from the kinetochores to the midbody at anaphase and telophase (Yen et al., 1991; Yao et al., 1997). Accumulating studies have revealed that BubR1 (Chan et al., 1999), Bub3 (Jiang et al., 2014; Li et al., 2016), Bub1 (Johnson et al., 2004), CENP-F, Mad1 (Akera et al., 2015), Astrin (Chung et al., 2016), SKAP (Huang et al., 2012) and small ubiquitin-related modifier (SUMO) proteins (Zhang et al., 2008; Wang and Dasso, 2009) are associated with kinetochore targeting of CENP-E proteins in mitosis. In turn, CENP-E recruits the kinetochore-associated proteins, including CLASP1 and CLASP2, to mediate microtubule turnover and poleward flux at the kinetochores (Maffini et al., 2009). CENP-E has also been shown to interact with CLASP through the C-terminal domain of CLASP (Gareil et al., 2023). Aurora A and B kinases phosphorylate CENP-E by releasing it from an autoinhibited state. At kinetochores, Aurora B phosphorylates CENP-E to inhibit its premature removal from kinetochores by dynein (Eibes et al., 2023).
The antibody injection, dominant negative constructs, and genetic deletion of CENP-E both results in chromosome misalignment, which indicates that CENP-E is essential for chromosome congression and alignment (Schaar et al., 1997; Wood et al., 1997; Yao et al., 2000; McEwen et al., 2001; Maiato et al., 2017). CENP-E inhibition/deletion results in metaphase arrest with several mono-oriented chromosomes (McEwen et al., 2001), a delayed mitotic progression (Tanudji et al., 2004), and a decreased number of microtubules at kinetochore fibers (McEwen et al., 2001; Putkey et al., 2002; Weaver et al., 2003). Chromosome misalignment induced by CENP-E depletion is accompanied by mitotic spindle assembly defects, mitotic catastrophe, and severe spindle positioning defects (Tame et al., 2016; Iegiani et al., 2021; Owa and Dynlacht, 2021). In addition, CENP-E is related to microtubule flux in early mitosis, which is required for the conversion from lateral to end-on attachment and chromosome congression (Shrestha and Draviam, 2013; Barisic and Rajendraprasad, 2021) (Figure 2).
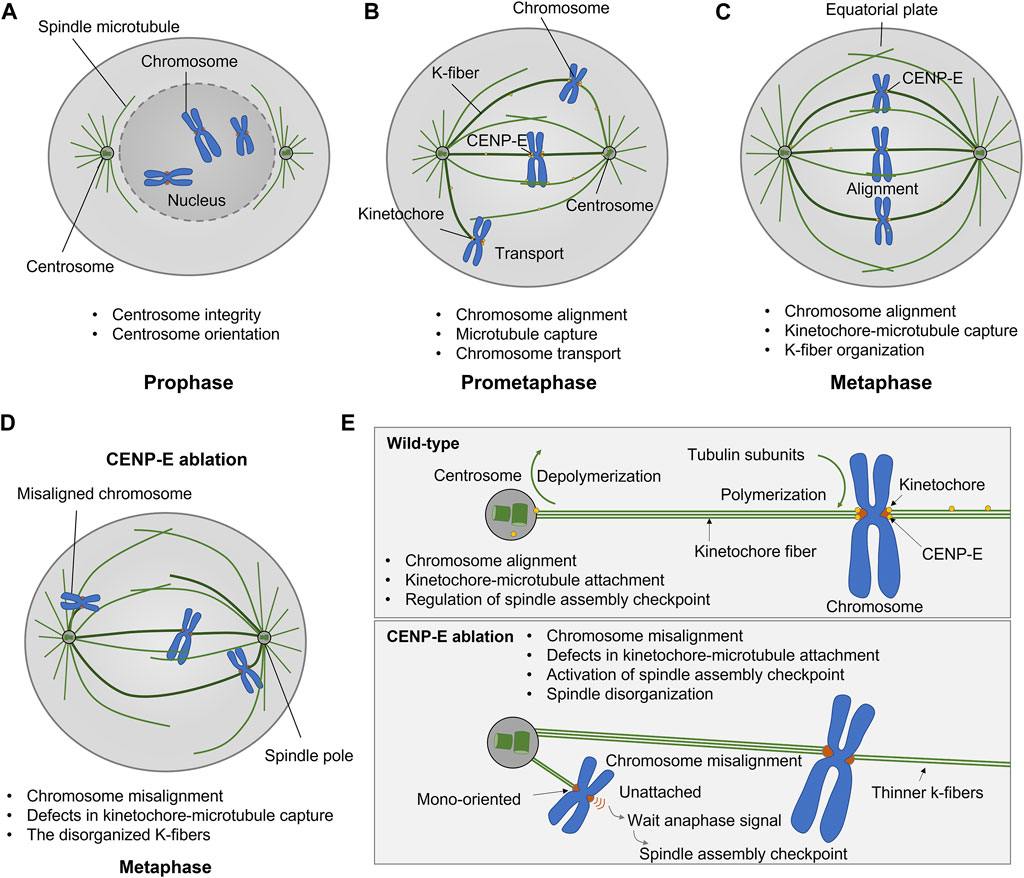
FIGURE 2. Kinesin-7 CENP-E is essential for kinetochore-microtubule attachment, chromosome alignment, and spindle assembly checkpoint in cell division. (A–C) CENP-E proteins are located at microtubules in prophase and accumulate at the kinetochores in prometaphase. CENP-E plays a key role in kinetochore-microtubule attachment and chromosome alignment during prometaphase and metaphase. (D) CENP-E ablation results in chromosome misalignment, spindle disorganization, and the activation of the spindle assembly checkpoint. (E) In wild-type cells, CENP-E proteins are essential for chromosome alignment, kinetochore-microtubule attachment, and the regulation of spindle assembly checkpoint. In the absence of CENP-E, the chromosomes are mono-oriented and misaligned, which further forms a wait anaphase signal and activates the spindle assembly checkpoint.
During prometaphase, CENP-E motors convert from a lateral mode to an end-on attachment mode in both the assembling and disassembling of microtubule plus-ends (Gudimchuk et al., 2013), which is required for chromosome movements and positioning. The combination of CENP-E and kinetochore protein Ndc80 supports lateral transport and microtubule wall-to-end transition at stabilized microtubules (Chakraborty et al., 2019). CENP-E binds to PRC1 through a conserved hydrophobic motif and promotes the antiparallel PRC1-crosslinked microtubules (Gluszek-Kustusz et al., 2023). During spindle assembly, PRC1-crosslinked microtubules undergo a network-to-bundles transition, and CENP-E promotes further microtubule bundling and kinetochore-mediated overlap formation (Matković et al., 2022).
CENP-E transports chromosomes to the spindle equator (Kapoor et al., 2006). CENP-E inhibition results in large chromosomes more vulnerable to defects in chromosome congression (Tovini and McClelland, 2019). CENP-E cooperates with chromokinesin KID and KIF4A to transport chromosomes toward the spindle equator along microtubules (Kapoor et al., 2006; Barisic and Rajendraprasad, 2021). The lateral kinetochore-microtubule attachment is mediated by CENP-E and dynein, which is required for chromosome congression (Maiato et al., 2017). During the initial poleward movement of peripheral chromosomes along astral microtubules, dynein is the dominant force counteracting the forces from CENP-E and chromokinesin in early mitosis (Barisic et al., 2014). During chromosome congression, CENP-E-mediated traction forces, in coordination with Kid-mediated forces on chromosome arms, are responsible for the loss of spindle pole integrity and multipolarity in CLASP1/2-depleted cells (Logarinho et al., 2012). Once the peripheral chromosomes reach the spindle pole, CENP-E becomes dominant over dynein and chromokinesin (Barisic and Maiato, 2016). There is a potential molecular switch between dynein and CENP-E activities on polar chromosomes (Barisic and Maiato, 2016) (Figure 2).
CENP-E is also essential for spindle assembly checkpoint in cell division (Chan et al., 1999; Abrieu et al., 2000; Guo et al., 2012). CENP-E interacts with multiple kinetochore proteins, including BubR1 (Legal et al., 2020), CENP-F (Chan et al., 1998), CLASP1 (Maffini et al., 2009), MAD1 (Akera et al., 2015). CENP-E is recruited by Bub1-Bub3 and BubR1-Bub3 complex at unattached kinetochores (Johnson et al., 2004). CENP-E is an activator of the BubR1 kinase, and CENP-E-dependent BubR1 autophosphorylation enhances chromosome alignment and the spindle assembly checkpoint (Mao et al., 2003; Guo et al., 2012). The basic C-terminal helix of BubR1 interacts with the minimal key acidic patch at the kinetochore-targeting domain of CENP-E to fulfill the recruitment of CENP-E to the kinetochores (Legal et al., 2020) (Figure 3). CENP-E is required for kinetochore recruitment of the corona’s building block consisting of ROD, Zwilch, ZW10, and the DD adaptor Spindly (RZZS). CENP-E proteins translocate to kinetochore through interactions with BubR1 and RZZS, and then mediate the kinetochore targeting of dynein-dynactin (Cmentowski et al., 2023). During fibrous corona formation, CENP-E interacts with Spindly and recruits RZZS to kinetochores through a farnesyl-dependent modification of its C-terminal kinetochore- and microtubule-binding domain (Wu et al., 2024).
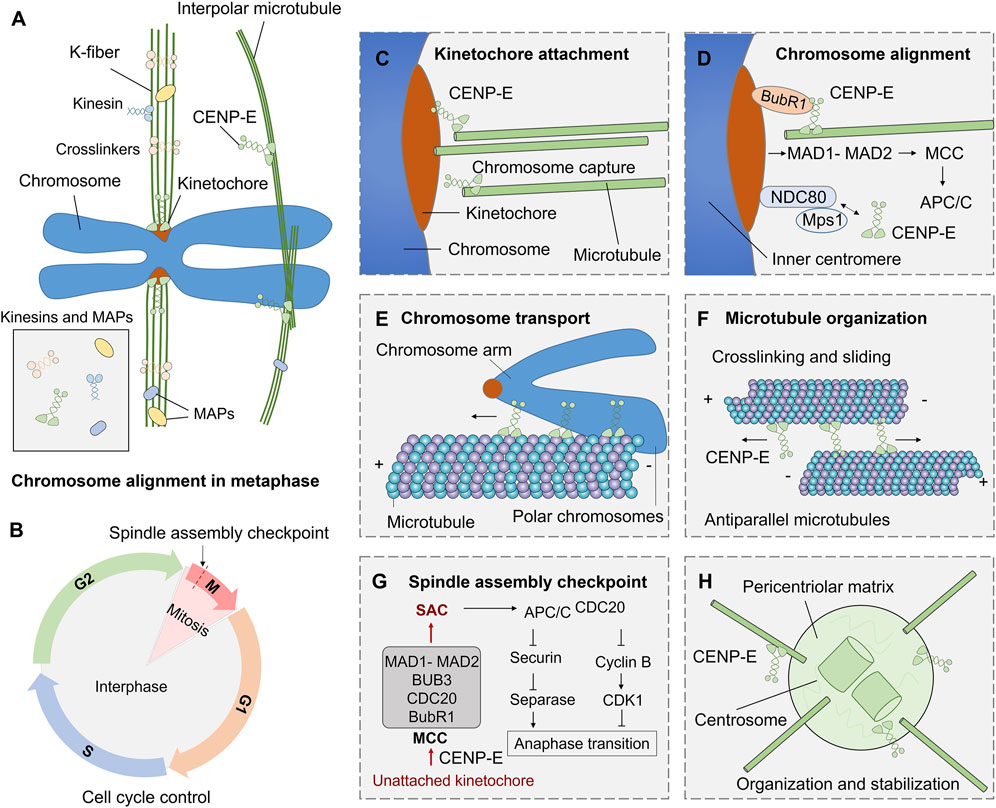
FIGURE 3. Functions and mechanisms of kinesin-7 CENP-E in cell division. (A) During mitosis, kinesins and microtubule-associated proteins (MAPs) are involved in microtubule crosslinking, kinetochore fiber assembly, and chromosome alignment. (B) The G1, S, G2, and M phases in the cell cycle are regulated by a complex cell cycle control system. (C) CENP-E associates with the plus ends of k-fibers and promotes kinetochore-microtubule attachment. (D) CENP-E interacts with BubR1, NDC80, Mps1, and kinetochore proteins to mediate chromosome alignment during metaphase. (E) CENP-E can transport polar chromosome arms along microtubules during prometaphase. (F) Both the motor and tail domains of CENP-E can bind to antiparallel microtubules and crosslink microtubules during spindle assembly. (G) The spindle assembly checkpoint pathway in mitosis. The unattached kinetochores on misaligned chromosomes can result in the formation of the mitotic checkpoint complex (MCC), including MAD1-MAD2, BUB3, CDC20, and BubR1 proteins, and then trigger the spindle assembly checkpoint. The checkpoint activates APC/CCDC20, inhibits Securin and separase, and then inhibits chromosome separation and regulates metaphase-to-anaphase transition. (H) CENP-E also mediates the organization of spindle poles and regulates centrosome organization and stabilization.
5 Dual roles of CENP-E: synthesis requirement, chromosomal instability, or spindle assembly checkpoint
Aneuploidy was recognized as a characteristic of human cancer cells (Zasadil et al., 2016), and is usually accompanied by chromosome instability (McGranahan et al., 2012; Zasadil et al., 2013). Both aneuploidy and chromosome instability are the markers of poor prognosis in many tumor types (McGranahan et al., 2012; Zasadil et al., 2013). In yeast and murine cells, aneuploidy is associated with growth defects under optimal conditions (Torres et al., 2007; Williams et al., 2008). However, specific aneuploidy karyotypes can confer a growth advantage in response to certain stresses (Zasadil et al., 2016). In yeast, preexisting aneuploidy leads to accelerated growth in response to environmental stresses. Specific aneuploidy can evolve to overcome functional insufficiencies or adapt to environmental challenges (Rancati et al., 2008; Pavelka et al., 2010; Millet et al., 2015).
Tumorigenesis is associated with a lack of genomic integrity and genomic instability in cells, while chromosomal instability (CIN) and microsatellite instability (MIN) are also thought to be different mechanisms of cancer development (Yuen et al., 2005; Abbas et al., 2013; Pussila et al., 2018). It has been found that CENP-E heterozygosity cells can quickly induce aneuploidy in vitro, while aneuploidy can inhibit and promote tumorigenesis (Weaver et al., 2007), suggesting a dual role of CENP-E in tumorigenesis. CENP-E heterozygous deletion results in a low rate of chromosome segregation in liver cells, and causes high chromosomal instability and tumor suppression in the Mad2+/− mice (Silk et al., 2013). Furthermore, CENP-E heterozygous deletion also induces an increased level of aneuploidy and then leads to an elevated level of spontaneous lymphomas and lung tumors in the aged mice (Weaver et al., 2007; Silk et al., 2013).
A lower level of aneuploidy provides a growth advantage for tumorigenesis and promotes tumorigenesis (Weaver and Cleveland, 2006), while a higher level of instability inhibits its growth (Weaver et al., 2007). But whether it is “promoting cancer” or “inhibiting cancer” depends on the cell type and whether there is additional genetic damage. The functional defects of CENP-E can induce CIN in different tissues. For example, in human soft tissue sarcoma, loss of NF-kB activating protein (NKAP) leads to CENP-E mislocalization, which in turn leads to chromosomal missegregation and aneuploidy dysregulation that ultimately promotes tumorigenesis (Li et al., 2016). Meanwhile, depletion of CENP-E in epithelial tissues unable to activate the apoptosis has also been observed to induce significant levels of aneuploidy and drive tumor-like growth (Clemente-Ruiz et al., 2014). The rate of chromosome missegregation based on CENP-E has also been found to have such a dual effect and is synchronized with the similar effect of aneuploidy (Silk et al., 2013). But when compared with the expression levels of CENP-E in tumors, CENP-E is more likely to promote tumor growth, and may only play a role as a tumor suppressor in liver cancer and acute lymphoblastic leukemia. The ability of CENP-E to inhibit and promote cancer in acute lymphoblastic leukemia may be the result of alternative splicing of CENP-E transcripts of mRNA (Jiménez-Ávila et al., 2018). This dual mechanism may mean that increasing the rate of chromosome missegregation can be used as a successful chemotherapy strategy (Funk et al., 2016).
Cancer cells usually harbor chromosome abnormalities and abnormal ploidy, which can result in specific constraints on the evolution of genetic changes (Gordon et al., 2012; Podgornaia and Laub, 2015; Bakhoum and Cantley, 2018). In uveal melanomas, CENP-E is a significantly mutated gene. CENP-E mutations are correlated with a higher percentage of chromosome copy number alterations (Johansson et al., 2020), but the underlying mechanisms are obscure. Furthermore, in follow-up studies, high levels of chromosomal instability based on CENP-E heterozygous have been shown to not inhibit tumor cell initiation, but inhibit subsequent cell growth (Zasadil et al., 2016).
CENP-E is a crucial regulator in mitotic checkpoint, and the absence of mitotic checkpoint will lead to tumorigenesis (Kops et al., 2005). Cells with a reduced level of CENP-E can enter the anaphase in the presence of misaligned chromosomes due to the weakened mitotic checkpoint. This results in a low rate of chromosome instability (Weaver et al., 2003; Weaver et al., 2007). In primary mouse embryonic fibroblasts with reduced levels of CENP-E, polar chromosomes are missegregated in 25% of divisions (Weaver et al., 2003). In head and neck cancers, polar chromosomes produced by decreased levels of CENP-E proteins lead to the occurrence of chromosomal instability, which may lead to tumorigenesis (Cosper et al., 2023). Resveratrol exhibits a biphasic effect on chromosomal instability, low doses of Resveratrol may reduce spontaneous chromosome instability, while high doses may induce chromosomal instability in human normal cells (Guo et al., 2018). Cells with a reduced level of CENP-E can enter the anaphase in the presence of misaligned chromosomes due to the weakened mitotic checkpoint, which further suggest that the dual effect occurs through the comprehensive regulation of the spindle assembly checkpoint pathway (Figure 4).
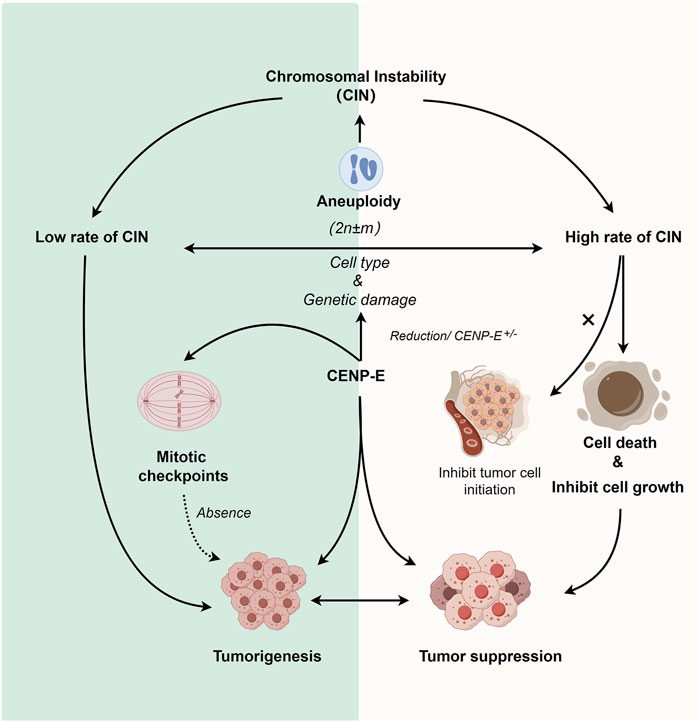
FIGURE 4. Dual roles of CENP-E in tumorigenesis. Reduction of CENP-E or CENP-E+/− can induce the occurrence of aneuploidy, and aneuploidy is highly related to chromosomal instability (CIN). CENP-E+/− can induce high or low rates of chromosomal instability, which depends on the cell type and genetic damage. A low rate of chromosomal instability can promote tumorigenesis, while a high rate of chromosomal instability will lead to cell death or tumor cell growth inhibition (but not tumor cell initiation). CENP-E is also involved in mitotic checkpoint, and the loss of mitotic checkpoint can also lead to tumorigenesis, suggesting another pathway for CENP-E-induced tumorigenesis.
6 Discovery and applications of CENP-E inhibitors in cancer treatment and therapy
Cancer cells are a population of cells with the ability to proliferate. The cytotoxic agents of cancers can be divided into four main kinds, including DNA alkylating agents, topoisomerases I and II inhibitors, antimetabolite agents, and microtubule targeting agents (Calligaris and Lafitte, 2011; Tcherniuk et al., 2011). Microtubule targeting agents can disrupt spindle assembly and microtubule dynamics, which are excellent cancer chemotherapeutic targets (Jordan and Wilson, 2004). Paclitaxel and the Vinca alkaloids are the most successful microtubule-target chemotherapeutic drugs that suppress microtubule dynamics and chromosome alignment, which results in mitotic arrest and apoptosis (Jordan and Wilson, 2004; Maiato et al., 2017). However, considering the neurotoxicity, neutropenia, and chemical resistance of microtubule-target agents, the discovery of novel anti-mitotic agents that do not disrupt microtubules is an emerging trend in cancer treatment (Jordan and Wilson, 2004). To date, seven kinds of CENP-E inhibitors have been found and synthesized (Table 2), which mainly inhibit chromosome alignment, and induce cell cycle arrest and eventually cell death. These CENP-E inhibitors might be novel anti-mitotic agents for cancer treatment (Figure 5).
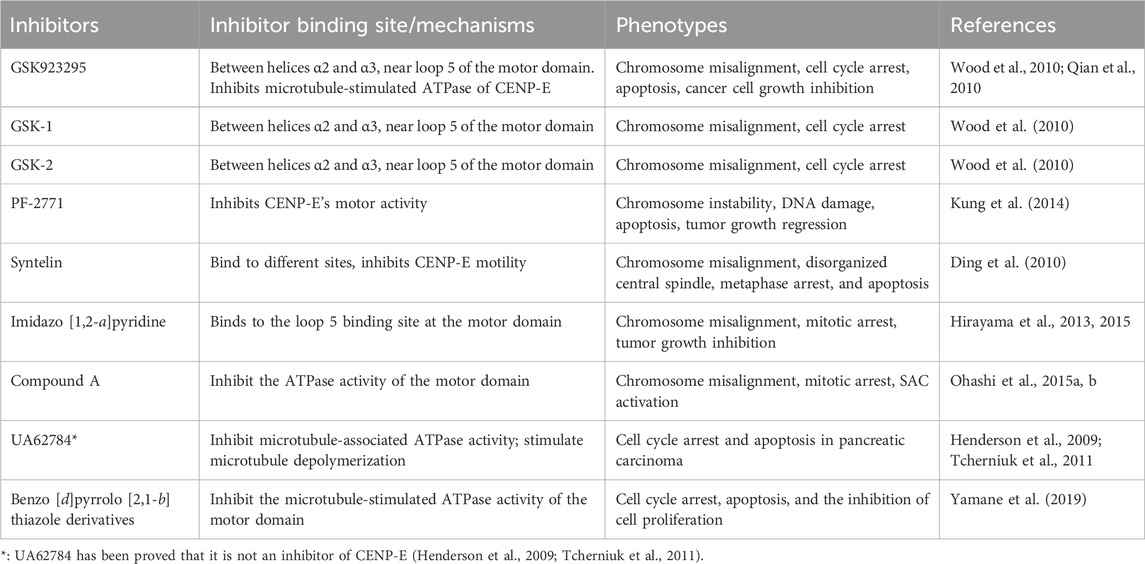
TABLE 2. Summaries and characterizations of the binding sites, mechanisms, and phenotypes of CENP-E inhibitors.
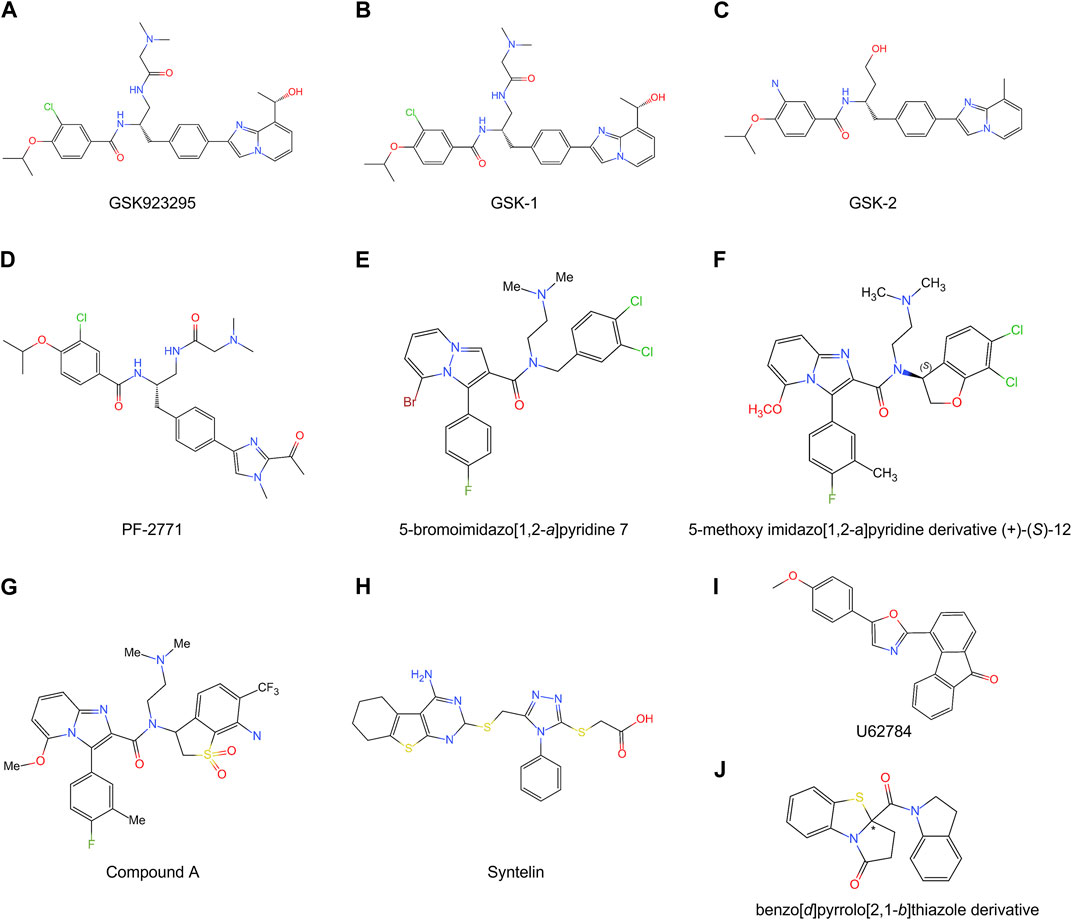
FIGURE 5. Chemical structures of multiple CENP-E inhibitors. (A) GSK923295; (B) GSK-1; (C) GSK-2; (D) PF-2771; (E) 5-bromoimidazo [1,2-a]pyridine 7; (F) 5-methoxy imidazo [1,2-a]pyridine derivative (+)-(S)-12; (G) Compound A; (H) Syntelin; (I) U62784; (J) benzo [d]pyrrolo [2,1-b]thiazole derivative.
6.1 GSK923295 and its derivatives
GSK923295 is an allosteric and uncompetitive CENP-E inhibitor of both ATP and microtubules (Qian et al., 2010), which specifically binds to the motor domain of CENP-E and inhibits CENP-E microtubule-stimulated ATPase activity with a Ki of 3.2 ± 0.2 nM (Wood et al., 2010) (Figure 5A). Site-directed mutagenesis reveals that GSK923295 interacts with Ile182 and Thr183, and interacts with CENP-E as sandwiched between helices α2 and α3 and near loop 5 (Wood et al., 2010). GSK923295 inhibits the release of inorganic phosphate and locks the motor domain of CENP-E at microtubules (Wood et al., 2010). In cultured cancer cells and mouse tumor xenografts, CENP-E inhibition by GSK923295 leads to chromosome misalignment, cell apoptosis, and tumor regression (Qian et al., 2010; Wood et al., 2010).
In addition, GSK-1 (Figure 5B) shows an ATP competitive behavior, which is different from the ATP uncompetitive behavior of GSK923295 (Wood et al., 2010). These differences are caused by chemical modifications of the carbon extension of a sidechain (Qian et al., 2010). Due to this small difference, GSK-1 may bind to the sites overlapping with the binding site of GSK923295. GSK-2 (Figure 5C) is a closely related inhibitor of GSK923295 (Wood et al., 2010). GSK923295 and GSK-2 result in cell cycle arrest in mitosis and tumor regression in vivo (Wood et al., 2010). The examination of the growth inhibitory activity of GSK923295 in 237 tumor cell lines shows that the GI50 values of 212 cell lines are less than 100 nM (Wood et al., 2010). Further studies have revealed that GSK923295 shows antitumor activity in neuroblastoma cells (Balamuth et al., 2010), Ewing sarcoma, rhabdoid, rhabdomyosarcoma xenografts (Lock et al., 2012), and hepatocellular carcinoma (Tang et al., 2019). As the only CENP-E-specific inhibitor entering clinical trials, the synthesis process, modification method, and target site of GSK923295 can provide a reference for the subsequent development of new CENP-E inhibitors.
The Phase I, first-in-human study has revealed that the maximum-tolerated dose of GSK923295 is 190 mg/m2 and examined the safety, dose-proportional pharmacokinetics, and preliminary clinical activity of GSK923295 (Chung et al., 2011). Among all 39 patients, 33% of patients had a response of stable disease, 54% had progressive disease, and most patients had mild adverse events, including fatigue, gastrointestinal toxicities of diarrhea, nausea, vomiting, and anemia (Chung et al., 2012). GSK923295 can inhibit CENP-E with high penetrance and at a low effective dose in medulloblastoma cells (Iegiani et al., 2021). The combination of GSK923295 and pharmacologic inhibitors of mitogen-activated ERK kinase (MEK1/2) shows a significant synergistic growth inhibition on neuroblastoma, lung, pancreatic, and colon carcinoma cell lines, which further results in mitotic arrest and apoptosis (Mayes et al., 2013). GSK923295 significantly inhibits the proliferation of tetraploid cells compared with diploids, suggesting superior generality of CENP-E-targeted tetraploidy inhibition (Yoshizawa et al., 2023). These findings indicate that in cancer treatment, exploring the combination of GSK923295 with other antitumor drugs might improve its clinical application.
Furthermore, the in-depth exploration of the off-target effects of GSK923295, the half-life of the drug in vivo, pharmacokinetics, and tumor targeting efficiency can improve its clinical effects. The binding site of GSK923295 to CENP-E can be further clarified by site-directed mutagenesis of CENP-E protein, CRISPR-Cas9 gene editing technology, and protein-drug crystal structure analysis in the future. By modifying the chemical moiety of GSK923295, its tissue and cell penetration ability, solubility and half-life can be further improved, which can enhance its effects and clinical applications.
6.2 PF-2771
PF-2771 is a non-competitive and selective inhibitor of CENP-E, which specifically suppresses cell growth of basal breast cancer cells, resulting in chromosome instability, increased phosphor-HH3-Ser10 levels, and tumor growth regression (Kung et al., 2014) (Figure 5D). PF-2771 inhibits the motor activity of CENP-E with an IC50 of 16.1 ± 1.2 nM (Kung et al., 2014). The treatment of PF-2771 results in elevated expression of BubR1, Aurora B, securin, and Cyclin B, increased DNA damage, and apoptosis (Kung et al., 2014). PF-2771, similar to GSK923295, induces a high effect on chromosome instability and loss of human artificial chromosomes (Kim et al., 2016). PF2771 and GSK923295, along with paclitaxel, olaparib, and talazoparib (Lee et al., 2016) can be candidates for cancer therapy when chromosome instability is a therapeutic target.
To date, researchers have claimed that a variety of CENP-E inhibitors induce “high chromosome instability” (Kim et al., 2016). However, few studies concerning the effectiveness and differences of CENP-E inhibitors in chromosome instability. In addition, the binding sites and inhibition modes of these CENP-E inhibitors, such as PF-2771, remain largely unknown. Site-directed mutagenesis and in vitro experiments, as well as the analyses of the structures of the drug-bound CENP-E proteins, would help to discover the specific binding sites and mechanisms of the inhibitors in the future. Furthermore, cross-detection of the responses of different drug-resistant cell lines to different CENP-E inhibitors can verify whether the binding sites of the inhibitors are consistent.
6.3 Imidazo [1,2-a]pyridine scaffold derivatives and compound A
The imidazo [1,2-a]pyridine scaffold derivatives are another inhibitors of CENP-E, including 5-bromoinidazo [1,2-a]pyridine 7 (Hirayama et al., 2013) (Figure 5E) and 5-methoxy imidazo [1,2-a]pyridine derivative (+)-(S)-12 (Hirayama et al., 2015) (Figure 5F). Based on a fused bicyclic compound, 4, 5-dihydrothieno [3, 4-c]pyridine-6-carboxamide 1a, researchers synthesized a new 5-bromoimidazo [1,2-a]pyridine 7, which shows the potent in CENP-E inhibition with an IC50 at 50 nM and binds to the loop 5 binding sites at the motor domain of CENP-E (Hirayama et al., 2013). By site-direct mutagenesis and electrostatic potential map analyses, the modification of imidazo [1,2-a]pyridine scaffold led to the discovery of 5-methoxy imidazo [1,2-a]pyridine derivative (+)-(S)-12, which inhibits CENP-E with an IC50 at 3.6 nM, suppresses cell growth of HeLa cells at GI50 at 130 nM and shows antitumor activities in a Colo205 xenograft model (Hirayama et al., 2015). The docking model suggests that the imidazo [1,2-a]pyridine inhibitors interact with Pro107, Ile182, and loop 5 at CENP-E (Hirayama et al., 2015).
Based on imidazo [1,2-a]pyridine scaffold derivatives, researchers further synthesized 6-cyano-7-trifluoromethyl-2,3-dihydro-1-benzothiophene 1,1-dioxide derivative (+)-5d (Compound A) (Hirayama et al., 2015) (Figure 5G). Compound A is a time-dependent CENP-E inhibitor with ATP competitive behavior, which effectively inhibits the motor activity of CENP-E (Ohashi et al., 2015a). Compound A induces chromosome misalignment, prolonged mitotic arrest, and antiproliferation in multiple cancer cell lines (Ohashi et al., 2015a). Furthermore, Compound A shows strong anti-tumor activity in the COLO205 xenograft nude mouse tumor model and induces the activation of spindle assembly checkpoint in a variety of tumor cell lines (Ohashi et al., 2015a). In addition, CENP-E inhibition by Compound A causes chromosome missegregation, the p53 gene-mediated post-mitotic apoptosis, which finally leads to proteotoxic stress and DNA damage in spindle assembly checkpoint-attenuated cells. However, polyploidy caused by Eg5 inhibition using Ispinesib under the same conditions does not result in proteotoxic stress and DNA damage (Ohashi et al., 2015b).
6.4 Syntelin
Syntelin is a novel class of CENP-E inhibitor, which inhibits the motility of CENP-E in a dose-dependent manner with an IC50 value of 160 nM (Ding et al., 2010) (Figure 5H). Compared with GSK923295, syntelin interacts with different regions outside the GSK923295s binding site and induces the inhibition of GSK923295-resistant cells (Ding et al., 2010). In HeLa cells, syntelin treatment results in misaligned chromosomes, reduced centromere stretches (Ding et al., 2010), and the disruption of the PRC1-organized central spindle (Liu et al., 2020a). The inhibition of CENP-E by syntelin causes metaphase arrest of HeLa cells and a syntelic attachment of spindle on chromosomes (Ding et al., 2010; Liu et al., 2020b). Syntelin treatment in triple-negative breast cancer, such as MDA-MB-231 cells, results in chromosome misalignment, the suppression of cell proliferation, and Bax-elicited apoptosis (Mullen et al., 2021). In a recent study, Syntelin also showed inhibition of proliferation and metastasis of triple-negative breast cancer and rarely led to cell necrosis (Mullen et al., 2021).
6.5 UA62784 and its derivatives
UA62784 is a novel fluorenone that specifically inhibits pancreatic cancer cell lines (Henderson et al., 2009) (Figure 5I). UA62784 inhibits microtubule-associated ATPase activity and leads to reversible cell cycle arrest and apoptosis in pancreatic carcinoma (Henderson et al., 2009). Previously, UA62784 was revealed as an inhibitor of CENP-E, and showed effective anti-tumor activity in the treatment of pancreatic cancer (Henderson et al., 2009). More than eighty UA62784 analogs have been synthesized and tested, however, there is no improvement in the selectivity pancreatic cancer of and kinesin-specific inhibitory patterns of the lead analog UA62784, excluding two analogs PC-046 and PC-053 (Shaw et al., 2009). However, Tcherniuk et al. (2011) have shown that UA62784 does not inhibit CENP-E ATPase activity but stimulates microtubule depolymerization through the interactions with microtubule near colchicine binding site using biophysical binding studies and in vivo imaging (Tcherniuk et al., 2011). The utilization of biophysical methods, molecular mass spectrometry imaging, live cell imaging, and optical tweezers would gain insight into the targets of small molecular compounds (Calligaris et al., 2010; Calligaris and Lafitte, 2011), which may reveal the truth and resolve disputes. The effect of UA62784 is superimposed with other microtubule targeting drugs currently used in the clinic, such as vinblastine, which makes it possible that UA62784 may be used in combination with vinblastine to avoid drug resistance of tumor cells. In addition, though UA62784 does not inhibit CENP-E, it also shows specific cytotoxicity to pancreatic cancer locus 4 (DPC4)-deficient cancer cells (Wang et al., 2009), and the mechanisms remain to be studied in the future.
6.6 Benzo [d]pyrrolo [2,1-b] thiazole derivatives
A new kind of CENP-E inhibitor, benzo [d]pyrrolo [2,1-b] thiazole derivatives (Figure 5J), was identified through the screening of a small-molecule chemical library (Yamane et al., 2019). This compound suppresses the microtubule-stimulated ATPase of CENPE’s motor domain with an IC50 of 17 μM in an ATP-competitive behavior (Yamane et al., 2019). Benzo [d]pyrrolo [2,1-b] thiazole derivatives induce cell cycle arrest, apoptosis, and the inhibition of cell proliferation in HeLa and HCT116 cells (Yamane et al., 2019).
CENP-E inhibition results in the aneuploidy-mediated p53-dependent post-mitotic apoptosis, which is different from Eg5 inhibition (Ohashi et al., 2015b). CENP-E inhibitors can suppress spindle assembly checkpoint-deficient cancers, which may expand the treatment window for the other chemotherapeutics. Previous studies have shown that radiotherapy combined with cell cycle inhibitors can enhance antitumor activity (Hauge et al., 2023). In-depth studies can focus on whether CENP-E inhibitors can be estimated as novel radiosensitizers for radiotherapy. Future studies on the in vitro antitumor activity of CENP-E inhibitors may also measure whether their effects on the cell cycle contribute to tumor radiation therapy. This may serve as adjuvant therapy in addition to chemotherapy. Considering that CENP-E is active in mitotic cells, the inhibitory effect of CENP-E inhibitors on undivided cells is limited, for example, CENP-E inhibitors are more likely to have low neurotoxicity (low levels of peripheral neuropathy), which has also been preliminarily confirmed in the results of clinical trials (Chung et al., 2012), and CENP-E remains important as a potential low-neurotoxicity antitumor target.
In summary, these CENP-E inhibitors provide useful backbones for future structural modifications and modeling studies. At present, there are various inhibitors of CENP-E, but the binding site of these inhibitors is very single. In the future, new inhibitors with other binding sites can be screened. In addition, animal experiments are necessary for existing inhibitors of CENP-E. Novel inhibitors may focus on improving their antitumor activity and minimizing adverse reactions in vivo.
7 Drug resistance mechanisms of CENP-E inhibitors: mutations, transporters, or expression alterations
Cancer cells usually contain chromosomal translocation, inversion, duplication, and aneuploidy, which lead to specific constraints on the evolution of genetic changes and chemotype-specific resistance (Bakhoum and Cantley, 2018). A recent study has indicated that different chromosome copy numbers in cancer cells result in distinct modes of GSK923295-specific resistance (Pisa et al., 2020) (Figure 6). The diploid HCT116 cells form the drug-specific resistance through the mutations at the GSK923295-binding site (M97V and R189M) near loop 5 of the CENP-E motor domain, which suggests that a single point mutation in CENP-E motor domain is sufficient to confer drug resistance through inhibiting GSK923295 recognition (Pisa et al., 2020). However, the near-haploid mammalian KBM7 cells show an approximately 300 kb deletion of genomic DNA, which results in the deletion of the CENP-E tail domain and GSK923295-specific resistance (Pisa et al., 2020). Together, these results suggest that distinct mechanisms of resistance can arise in cancer cells with different ploidies or karyotypes. However, how the deletion of the C-terminal domain of CENP-E increases the resistance of haploid cells to GSK923295 is unknown and remains to be revealed.
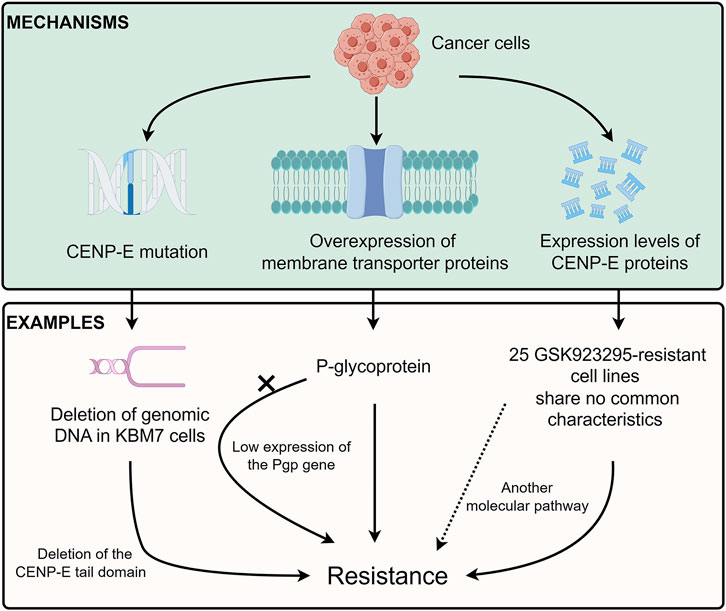
FIGURE 6. Drug resistance of CENP-E inhibitors. Tumor cells can develop resistance to CENP-E inhibitors through the CENP-E gene mutation, membrane transporter proteins overexpression, or their own CENP-E expression level. For example, the deletion of DNA in KBM7 cells will lead to the deletion of the CENP-E tail domain, which in turn leads to specific resistance to GSK923295. The overexpression of P-glycoprotein (Pgp) can also lead to resistance to GSK923295, but the low expression of the Pgp gene in GSK923295-resistant KBM7 cells suggests that cells may generate different mechanisms of drug resistance. No common characteristics of 25 GSK923295-resistant cell lines indicate that other molecular pathways lead to drug resistance.
Previous studies have suggested that multidrug resistance efflux transporter P-glycoprotein (referred to as Pgp or ABCB1) is responsible for the GSK923295 resistance (Tcherniuk and Oleinikov, 2015). However, there is low or no expression of the Pgp gene in both parental and GSK923295-resistant KBM7 cells (Pisa et al., 2020). These results indicate that there might be different mechanisms in diverse cancer cells to generate resistance to GSK923295. In addition, according to the hints of these studies, exploring the mutations of CENP-E in drug-resistant cells may help to discover several important sites for drug binding.
Unlike multiple cancer cell lines, the SW620, CAPAN-2, and MRC5 cancer cell lines are resistant to Compound A (Ohashi et al., 2015a). These results indicate that not only low expression of CENP-E but also another molecular pathway, may be involved in the sensitivity and resistance to Compound A. Among 237 tumor cell lines, there are 25 GSK923295-resistant cell lines with no common characteristics (Wood et al., 2010). Moreover, a group of basal subtype breast cancer cells is most sensitive to GSK923295, while nonmalignant cancer cells are more resistant to GSK923295 (Wood et al., 2010). The similarities and differences between GSK923295-sensitive and non-sensitive tumor cells deserve to be studied and will help to discover the possible mechanisms of CENP-E drug resistance.
There are many determinants of sensitivity and resistance to antimitotic drugs, including the overexpression of a class of membrane transporter proteins, ABC-transporters (ATP-dependent drug efflux pumps or ATP-binding cassettes), such as the P-glycoprotein (Dumontet and Sikic, 1999; Ambudkar et al., 2003; Chanel-Vos and Giannakakou, 2010; Pote and Gacche, 2023). Moreover, cancer cells also have microtubule-related mechanisms to confer chemical resistance and generate intrinsic insensitivity to antimitotic drugs, including the expression or binding of regulatory proteins, post-translational modifications of tubulin, and abnormal expression of tubulin isotypes (Dumontet and Sikic, 1999; Gonçalves et al., 2001; Jordan and Wilson, 2004). CENP-E is related to microtubule-resistance drugs (Chanel-Vos and Giannakakou, 2010). The interactions between CENP-E and BubR1 are diminished in epothilone B-resistant A549 cells (Yang et al., 2010). In addition, whether CENP-E inhibitor-resistant tumor cells lead to cells acquiring broad-spectrum resistance to other inhibitors of mitotic kinesins deserves further exploration. Moreover, it is still unknown whether the microtubule dynamics and properties of CENP-E inhibitor-resistant cells change compared with normal tumor cells, which can be further verified by the cold treatment, the response of microtubule inhibitors colchicine, or paclitaxel.
In summary, there are three main mechanisms of drug resistance mechanisms of CENP-E inhibitors, including the gene mutations in the CENP-E gene (Pisa et al., 2020), the expression of the P-glycoprotein (Tcherniuk and Oleinikov, 2015), and the different expression levels of CENP-E proteins in diverse cancer cells (Wood et al., 2010) (Figure 6). These results indicate that several unidentified factors, including the overexpression of functionally redundant genes, the silence of spindle assembly checkpoints, or the resistance to cell death after chromosome instability, may contribute to different inhibitory effects of CENP-E inhibitors and drug resistance.
8 Conclusions and future perspectives
To date, there is a key scientific question that remains largely obscure: how does kinesin −7 CENP-E achieve high expression levels in a wide range of tumor tissues and cancer cells? The transcriptional regulation and intracellular environment of the CENP-E gene might be a reason for the tissue-specific expression and upregulated expression of CENP-E. The expression of CENP-E has a stable pattern in the cell cycle, but the transcription factors or regulatory proteins that regulate CENP-E gene expression are less studied. Thus, the in-depth studies of the promoter region, transcription factor binding site, and enhancer elements of the CENP-E gene will help to explain the molecular basis of CENP-E periodic expression and its high expression in tumors. Tumors are characterized by rapid proliferation and stronger requirements for vigorous mitosis compared with normal tissue. Thus, tumors appear to have higher expression of cell cycle regulated genes such as CENP-E, which are strictly cell cycle regulated. In particular, the low expression of CENP-E in liver cancer cells is different from the high expression in most tumors, and the underlying reasons and specific mechanisms need to be further elucidated.
The specificity and effective concentrations of the drugs on the targets are important for cancer treatment. The question of whether GSK923295 specifically targets CENP-E and its effective concentrations in vivo. In the future, the construct of the CENP-E knockout cell line using CRISPR-Cas9 gene-editing technology, together with the observations of the phenotypes and responses of CENP-E knockout cancer cells in the presence of inhibitors, would help to explore the binding specificity, off-target effects, and side effects of CENP-E inhibitors. Moreover, the differences in the effects of GSK923295 in vivo and in vitro should also take into account the following factors, including the half-life of the drugs, the metabolic pathways in vivo, the methods of drug administration, the target sites of action, and the cumulative concentrations of the drugs at the tumors.
At present, the development of drug combinations, including different drugs on one single target, is a new strategy to overcome drug resistance (Wylie et al., 2017; Real et al., 2020). To date, most CENP-E inhibitors bind to the motor domain and inhibit the ATPase activity of CENP-E. Therefore, the search, development, and chemical modifications of novel inhibitors targeting CENP-E’s coiled-coil or tail domain are conducive to solving the issues of drug-induced mutations in the motor domain and related drug resistances. Furthermore, a combination therapy based on CENP-E inhibitors and microtubule-targeting agents would be of high clinical advantage in wide applicability, lower toxicity, and better antitumor activity. In addition, tumor-specific targeting should also be taken into account to reduce the effects on rapidly dividing, normal cells in vivo.
Author contributions
Z-YS: Writing–review and editing, Writing–original draft, Supervision, Project administration, Funding acquisition. Y-HY: Writing–review and editing, Writing–original draft. Y-LW: Writing–review and editing, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the following grants: National Natural Science Foundation of China (grant numbers 82001608 and 82101678), Natural Science Foundation of Fujian Province, China (grant number 2023J01306), Fujian Medical University high-level talents scientific research start-up funding project (grant number XRCZX2017025), and College Students’ innovation and entrepreneurship training program (C21015).
Acknowledgments
We are grateful to all members of the Cytoskeleton Laboratory at Fujian Medical University for the discussions. Figure 4 and Figure 6 were drawn using the Figdraw software (www.figdraw.com).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, T., Keaton, M. A., and Dutta, A. (2013). Genomic instability in cancer. Cold Spring Harb. Perspect. Biol. 5, a012914. doi:10.1101/cshperspect.a012914
Abrieu, A., Kahana, J. A., Wood, K. W., and Cleveland, D. W. (2000). CENP-E as an essential component of the mitotic checkpoint in vitro. Cell 102, 817–826. doi:10.1016/s0092-8674(00)00070-2
Agarwal, R., Gonzalez-Angulo, A. M., Myhre, S., Carey, M., Lee, J. S., Overgaard, J., et al. (2009). Integrative analysis of cyclin protein levels identifies cyclin b1 as a classifier and predictor of outcomes in breast cancer. Clin. Cancer Res. 15, 3654–3662. doi:10.1158/1078-0432.CCR-08-3293
Akera, T., Goto, Y., Sato, M., Yamamoto, M., and Watanabe, Y. (2015). Mad1 promotes chromosome congression by anchoring a kinesin motor to the kinetochore. Nat. Cell Biol. 17, 1124–1133. doi:10.1038/ncb3219
Ambudkar, S. V., Kimchi-Sarfaty, C., Sauna, Z. E., and Gottesman, M. M. (2003). P-glycoprotein: from genomics to mechanism. Oncogene 22, 7468–7485. doi:10.1038/sj.onc.1206948
Bakhoum, S. F., and Cantley, L. C. (2018). The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360. doi:10.1016/j.cell.2018.08.027
Balamuth, N. J., Wood, A., Wang, Q., Jagannathan, J., Mayes, P., Zhang, Z., et al. (2010). Serial transcriptome analysis and cross-species integration identifies centromere-associated protein E as a novel neuroblastoma target. Cancer Res. 70, 2749–2758. doi:10.1158/0008-5472.CAN-09-3844
Barisic, M., Aguiar, P., Geley, S., and Maiato, H. (2014). Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 16, 1249–1256. doi:10.1038/ncb3060
Barisic, M., and Maiato, H. (2016). The tubulin code: a navigation system for chromosomes during mitosis. Trends Cell Biol. 26, 766–775. doi:10.1016/j.tcb.2016.06.001
Barisic, M., and Rajendraprasad, G. (2021). Mitotic poleward flux: finding balance between microtubule dynamics and sliding. Bioessays 43, e2100079. doi:10.1002/bies.202100079
Brown, K. D., Coulson, R. M., Yen, T. J., and Cleveland, D. W. (1994). Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J. Cell Biol. 125, 1303–1312. doi:10.1083/jcb.125.6.1303
Calligaris, D., and Lafitte, D. (2011). Chemical inhibitors: the challenge of finding the right target. Chem. Biol. 18, 555–557. doi:10.1016/j.chembiol.2011.05.003
Calligaris, D., Verdier-Pinard, P., Devred, F., Villard, C., Braguer, D., and Lafitte, D. (2010). Microtubule targeting agents: from biophysics to proteomics. Cell. Mol. Life Sci. 67, 1089–1104. doi:10.1007/s00018-009-0245-6
Chakraborty, M., Tarasovetc, E. V., Zaytsev, A. V., Godzi, M., Figueiredo, A. C., Ataullakhanov, F. I., et al. (2019). Microtubule end conversion mediated by motors and diffusing proteins with no intrinsic microtubule end-binding activity. Nat. Commun. 10, 1673. doi:10.1038/s41467-019-09411-7
Chan, G. K., Jablonski, S. A., Sudakin, V., Hittle, J. C., and Yen, T. J. (1999). Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146, 941–954. doi:10.1083/jcb.146.5.941
Chan, G. K., Schaar, B. T., and Yen, T. J. (1998). Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143, 49–63. doi:10.1083/jcb.143.1.49
Chanel-Vos, C., and Giannakakou, P. (2010). CENP-E checks in microtubule-drug resistance. Cell Cycle 9, 1456. doi:10.4161/cc.9.8.11382
Chung, H. J., Park, J. E., Lee, N. S., Kim, H., and Jang, C. Y. (2016). Phosphorylation of Astrin regulates its kinetochore function. J. Biol. Chem. 291, 17579–17592. doi:10.1074/jbc.M115.712745
Chung, V., Heath, E. I., Schelman, W. R., Johnson, B. M., Kirby, L. C., Lynch, K. M., et al. (2012). First-time-in-human study of GSK923295, a novel antimitotic inhibitor of centromere-associated protein E (CENP-E), in patients with refractory cancer. Cancer Chemother. Pharmacol. 69, 733–741. doi:10.1007/s00280-011-1756-z
Cilluffo, D., Chiavetta, R. F., Bivona, S., Contino, F., Coronnello, C., Feo, S., et al. (2021). Transcriptomic changes following partial depletion of CENP-E in normal human fibroblasts. Genes (Basel) 12, 1322. doi:10.3390/genes12091322
Ciossani, G., Overlack, K., Petrovic, A., Huis, In ’t V. P. J., Koerner, C., Wohlgemuth, S., et al. (2018). The kinetochore proteins CENP-E and CENP-F directly and specifically interact with distinct BUB mitotic checkpoint Ser/Thr kinases. J. Biol. Chem. 293, 10084–10101. doi:10.1074/jbc.RA118.003154
Clemente-Ruiz, M., Muzzopappa, M., and Milán, M. (2014). Tumor suppressor roles of CENP-E and Nsl1 in Drosophila epithelial tissues. Cell Cycle 13, 1450–1455. doi:10.4161/cc.28417
Cmentowski, V., Ciossani, G., d’Amico, E., Wohlgemuth, S., Owa, M., Dynlacht, B., et al. (2023). RZZ-Spindly and CENP-E form an integrated platform to recruit dynein to the kinetochore corona. EMBO J. 42, e114838. doi:10.15252/embj.2023114838
Cooke, C. A., Schaar, B., Yen, T. J., and Earnshaw, W. C. (1997). Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106, 446–455. doi:10.1007/s004120050266
Cosper, P. F., Hrycyniak, L. C. F., Paracha, M., Lee, D. L., Wan, J., Jones, K., et al. (2023). HPV16 E6 induces chromosomal instability due to polar chromosomes caused by E6AP-dependent degradation of the mitotic kinesin CENP-E. Proc. Natl. Acad. Sci. U. S. A. 120, e2216700120. doi:10.1073/pnas.2216700120
Craske, B., Legal, T., and Welburn, J. P. I. (2022). Reconstitution of an active human CENP-E motor. Open Biol. 12, 210389. doi:10.1098/rsob.210389
Craske, B., and Welburn, J. P. I. (2020). Leaving no-one behind: how CENP-E facilitates chromosome alignment. Essays Biochem. 64, 313–324. doi:10.1042/EBC20190073
Ding, X., Yan, F., Yao, P., Yang, Z., Wan, W., Wang, X., et al. (2010). Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 20, 1386–1389. doi:10.1038/cr.2010.167
Dumontet, C., and Sikic, B. I. (1999). Mechanisms of action of and resistance to antitubulin agents: microtubule dynamics, drug transport, and cell death. J. Clin. Oncol. 17, 1061–1070. doi:10.1200/JCO.1999.17.3.1061
Eibes, S., Rajendraprasad, G., Guasch-Boldu, C., Kubat, M., Steblyanko, Y., and Barisic, M. (2023). CENP-E activation by Aurora A and B controls kinetochore fibrous corona disassembly. Nat. Commun. 14, 5317. doi:10.1038/s41467-023-41091-2
Espeut, J., Gaussen, A., Bieling, P., Morin, V., Prieto, S., Fesquet, D., et al. (2008). Phosphorylation relieves autoinhibition of the kinetochore motor Cenp-E. Cenp-E. Mol. Cell 29, 637–643. doi:10.1016/j.molcel.2008.01.004
Funk, L. C., Zasadil, L. M., and Weaver, B. A. (2016). Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev. Cell 39, 638–652. doi:10.1016/j.devcel.2016.10.023
Garcia-Saez, I., Yen, T., Wade, R. H., and Kozielski, F. (2004). Crystal structure of the motor domain of the human kinetochore protein CENP-E. J. Mol. Biol. 340, 1107–1116. doi:10.1016/j.jmb.2004.05.053
Gareil, N., Gervais, A., Macaisne, N., Chevreux, G., Canman, J. C., Andreani, J., et al. (2023). An unconventional TOG domain is required for CLASP localization. Curr. Biol. 33, 3522–3528.e7. doi:10.1016/j.cub.2023.07.009
Gluszek-Kustusz, A., Craske, B., Legal, T., McHugh, T., and Welburn, J. P. (2023). Phosphorylation controls spatial and temporal activities of motor-PRC1 complexes to complete mitosis. EMBO J. 42, e113647. doi:10.15252/embj.2023113647
Gonçalves, A., Braguer, D., Kamath, K., Martello, L., Briand, C., Horwitz, S., et al. (2001). Resistance to Taxol in lung cancer cells associated with increased microtubule dynamics. Proc. Natl. Acad. Sci. U. S. A. 98, 11737–11742. doi:10.1073/pnas.191388598
Gordon, D. J., Resio, B., and Pellman, D. (2012). Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 13, 189–203. doi:10.1038/nrg3123
Gudimchuk, N., Tarasovetc, E. V., Mustyatsa, V., Drobyshev, A. L., Vitre, B., Cleveland, D. W., et al. (2018). Probing mitotic CENP-E kinesin with the tethered cargo motion assay and laser tweezers. Biophys. J. 114, 2640–2652. doi:10.1016/j.bpj.2018.04.017
Gudimchuk, N., Vitre, B., Kim, Y., Kiyatkin, A., Cleveland, D. W., Ataullakhanov, F. I., et al. (2013). Kinetochore kinesin CENP-E is a processive bi-directional tracker of dynamic microtubule tips. Nat. Cell Biol. 15, 1079–1088. doi:10.1038/ncb2831
Guo, X., Ni, J., Dai, X., Zhou, T., Yang, G., Xue, J., et al. (2018). Biphasic regulation of spindle assembly checkpoint by low and high concentrations of resveratrol leads to the opposite effect on chromosomal instability. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 825, 19–30. doi:10.1016/j.mrgentox.2017.11.004
Guo, Y., Kim, C., Ahmad, S., Zhang, J., and Mao, Y. (2012). CENP-E--dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J. Cell Biol. 198, 205–217. doi:10.1083/jcb.201202152
Hao, X., and Qu, T. (2019). Expression of CENPE and its prognostic role in non-small cell lung cancer. Open Med. (Wars) 14, 497–502. doi:10.1515/med-2019-0053
Hariharan, V., and Hancock, W. O. (2009). Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell. Mol. Bioeng. 2, 177–189. doi:10.1007/s12195-009-0059-5
Hauge, S., Eek Mariampillai, A., Rødland, G. E., Bay, L. T. E., Landsverk, H. B., and Syljuåsen, R. G. (2023). Expanding roles of cell cycle checkpoint inhibitors in radiation oncology. Int. J. Radiat. Biol. 99, 941–950. doi:10.1080/09553002.2021.1913529
He, P., Hu, P., Yang, C., He, X., Shao, M., and Lin, Y. (2020). Reduced expression of CENP-E contributes to the development of hepatocellular carcinoma and is associated with adverse clinical features. Biomed. Pharmacother. 123, 109795. doi:10.1016/j.biopha.2019.109795
Henderson, M. C., Shaw, Y. J., Wang, H., Han, H., Hurley, L. H., Flynn, G., et al. (2009). UA62784, a novel inhibitor of centromere protein E kinesin-like protein. Mol. Cancer Ther. 8, 36–44. doi:10.1158/1535-7163.MCT-08-0789
Hirayama, T., Okaniwa, M., Banno, H., Kakei, H., Ohashi, A., Iwai, K., et al. (2015). Synthetic studies on Centromere-Associated Protein-E (CENP-E) inhibitors: 2. Application of electrostatic potential map (EPM) and structure-based modeling to Imidazo[1,2-a]pyridine derivatives as anti-tumor agents. J. Med. Chem. 58, 8036–8053. doi:10.1021/acs.jmedchem.5b00836
Hirayama, T., Okaniwa, M., Imada, T., Ohashi, A., Ohori, M., Iwai, K., et al. (2013). Synthetic studies of centromere-associated protein-E (CENP-E) inhibitors: 1.Exploration of fused bicyclic core scaffolds using electrostatic potential map. Bioorg. Med. Chem. 21, 5488–5502. doi:10.1016/j.bmc.2013.05.067
Hou, S., Li, N., Zhang, Q., Li, H., Wei, X., Hao, T., et al. (2016). XAB2 functions in mitotic cell cycle progression via transcriptional regulation of CENPE. Cell Death Dis. 7, e2409. doi:10.1038/cddis.2016.313
Huang, Y., Wang, W., Yao, P., Wang, X., Liu, X., Zhuang, X., et al. (2012). CENP-E kinesin interacts with SKAP protein to orchestrate accurate chromosome segregation in mitosis. J. Biol. Chem. 287, 1500–1509. doi:10.1074/jbc.M111.277194
Iegiani, G., Gai, M., Di Cunto, F., and Pallavicini, G. (2021). CENPE inhibition leads to mitotic catastrophe and DNA damage in medulloblastoma cells. Cancers (Basel) 13, 1028. doi:10.3390/cancers13051028
Jiang, H., He, X., Wang, S., Jia, J., Wan, Y., Wang, Y., et al. (2014). A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting. Dev. Cell 28, 268–281. doi:10.1016/j.devcel.2013.12.013
Jiménez-Ávila, C. E., Villegas-Ruíz, V., Zapata-Tarres, M., Rubio-Portillo, A. E., Pérez López, E. I., Zenteno, J. C., et al. (2018). Centromere-associated protein E expresses a novel mRNA isoform in acute lymphoblastic leukemia. Int. J. Mol. Epidemiol. Genet. 9, 43–54.
Johansson, P. A., Brooks, K., Newell, F., Palmer, J. M., Wilmott, J. S., Pritchard, A. L., et al. (2020). Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat. Commun. 11, 2408. doi:10.1038/s41467-020-16276-8
Johnson, V. L., Scott, M. I., Holt, S. V., Hussein, D., and Taylor, S. S. (2004). Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J. Cell Sci. 117, 1577–1589. doi:10.1242/jcs.01006
Jordan, M. A., and Wilson, L. (2004). Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265. doi:10.1038/nrc1317
Ju, W., Yoo, B. C., Kim, I. J., Kim, J. W., Kim, S. C., and Lee, H. P. (2009). Identification of genes with differential expression in chemoresistant epithelial ovarian cancer using high-density oligonucleotide microarrays. Oncol. Res. 18, 47–56. doi:10.3727/096504009789954672
Kapoor, T. M., Lampson, M. A., Hergert, P., Cameron, L., Cimini, D., Salmon, E. D., et al. (2006). Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388–391. doi:10.1126/science.1122142
Kim, J. H., Lee, H. S., Lee, N. C., Goncharov, N. V., Kumeiko, V., Masumoto, H., et al. (2016). Development of a novel HAC-based "gain of signal" quantitative assay for measuring chromosome instability (CIN) in cancer cells. Oncotarget 7, 14841–14856. doi:10.18632/oncotarget.7854
Kim, Y., Heuser, J. E., Waterman, C. M., Cleveland, D. W., et al. (2008). CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J. Cell Biol. 181, 411–419. doi:10.1083/jcb.200802189
Kops, G. J., Weaver, B. A., Cleveland, D. W., et al. (2005). On the road to cancer: aneuploidy and the mitotic checkpoint. Nat. Rev. Cancer 5, 773–785. doi:10.1038/nrc1714
Kung, P. P., Martinez, R., Zhu, Z., Zager, M., Blasina, A., Rymer, I., et al. (2014). Chemogenetic evaluation of the mitotic kinesin CENP-E reveals a critical role in triple-negative breast cancer. Mol. Cancer Ther. 13, 2104–2115. doi:10.1158/1535-7163.MCT-14-0083-T
Lawrence, C. J., Dawe, R. K., Christie, K. R., Cleveland, D. W., Dawson, S. C., Endow, S. A., et al. (2004). A standardized kinesin nomenclature. J. Cell Biol. 167, 19–22. doi:10.1083/jcb.200408113
Lee, H. S., Lee, N. C., Kouprina, N., Kim, J. H., Kagansky, A., Bates, S., et al. (2016). Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res. 76, 902–911. doi:10.1158/0008-5472.CAN-15-1617
Lee, S. H., Yim, S. Y., Jeong, Y. S., Li, Q. X., Kang, S. H., Sohn, B. H., et al. (2022). Consensus subtypes of hepatocellular carcinoma associated with clinical outcomes and genomic phenotypes. Hepatology 76, 1634–1648. doi:10.1002/hep.32490
Legal, T., Hayward, D., Gluszek-Kustusz, A., Blackburn, E. A., Spanos, C., Rappsilber, J., et al. (2020). The C-terminal helix of BubR1 is essential for CENP-E-dependent chromosome alignment. J. Cell Sci. 133, jcs246025. doi:10.1242/jcs.246025
Li, J., Diao, H., Guan, X., and Tian, X. (2020). Kinesin family member C1 (KIFC1) regulated by Centrosome Protein E (CENPE) promotes proliferation, migration, and epithelial-mesenchymal transition of ovarian cancer. Med. Sci. Monit. 26, e927869. doi:10.12659/MSM.927869
Li, T., Chen, L., Cheng, J., Dai, J., Huang, Y., Zhang, J., et al. (2016). SUMOylated NKAP is essential for chromosome alignment by anchoring CENP-E to kinetochores. Nat. Commun. 7, 12969. doi:10.1038/ncomms12969
Liang, Q., Tan, C., Xiao, F., Yin, F., Liu, M., Lei, L., et al. (2021). Integrated profiling identifies ITGB3BP as prognostic biomarker for hepatocellular carcinoma. Bosn. J. Basic Med. Sci. 21, 712–723. doi:10.17305/bjbms.2021.5690
Liang, Y., Ahmed, M., Guo, H., Soares, F., Hua, J. T., Gao, S., et al. (2017). LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res. 77, 5479–5490. doi:10.1158/0008-5472.CAN-17-0496
Liu, X., Liu, X., Wang, H., Dou, Z., Ruan, K., Hill, D. L., et al. (2020b). Phase separation drives decision making in cell division. J. Biol. Chem. 295, 13419–13431. doi:10.1074/jbc.REV120.011746
Liu, X., Xu, L., Li, J., Yao, P. Y., Wang, W., Ismail, H., et al. (2020a). Mitotic motor CENP-E cooperates with PRC1 in temporal control of central spindle assembly. J. Mol. Cell Biol. 12, 654–665. doi:10.1093/jmcb/mjz051
Liu, Z., Ling, K., Wu, X., Cao, J., Liu, B., Li, S., et al. (2009). Reduced expression of cenp-e in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 28, 156. doi:10.1186/1756-9966-28-156
Lock, R. B., Carol, H., Morton, C. L., Keir, S. T., Reynolds, C. P., Kang, M. H., et al. (2012). Initial testing of the CENP-E inhibitor GSK923295A by the pediatric preclinical testing program. Pediatr. Blood Cancer 58, 916–923. doi:10.1002/pbc.23176
Logarinho, E., Maffini, S., Barisic, M., Marques, A., Toso, A., Meraldi, P., et al. (2012). CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat. Cell Biol. 14, 295–303. doi:10.1038/ncb2423
Ma, Q., Xu, Y., Liao, H., Cai, Y., Xu, L., Xiao, D., et al. (2019). Identification and validation of key genes associated with non-small-cell lung cancer. J. Cell Physiol. 234, 22742–22752. doi:10.1002/jcp.28839
Maffini, S., Maia, A. R., Manning, A. L., Maliga, Z., Pereira, A. L., Junqueira, M., et al. (2009). Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr. Biol. 19, 1566–1572. doi:10.1016/j.cub.2009.07.059
Maiato, H., Gomes, A. M., Sousa, F., and Barisic, M. (2017). Mechanisms of chromosome congression during mitosis. Biol. (Basel) 6, 13. doi:10.3390/biology6010013
Mao, Y., Abrieu, A., and Cleveland, D. W. (2003). Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. BubR1. Cell 114, 87–98. doi:10.1016/s0092-8674(03)00475-6
Matković, J., Ghosh, S., Ćosić, M., Eibes, S., Barišić, M., Pavin, N., et al. (2022). Kinetochore- and chromosome-driven transition of microtubules into bundles promotes spindle assembly. Nat. Commun. 13, 7307. doi:10.1038/s41467-022-34957-4
Mayes, P. A., Degenhardt, Y. Y., Wood, A., Toporovskya, Y., Diskin, S. J., Haglund, E., et al. (2013). Mitogen-activated protein kinase (MEK/ERK) inhibition sensitizes cancer cells to centromere-associated protein E inhibition. Int. J. Cancer 132, E149–E157. doi:10.1002/ijc.27781
McEwen, B. F., Chan, G. K., Zubrowski, B., Savoian, M. S., Sauer, M. T., and Yen, T. J. (2001). CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776–2789. doi:10.1091/mbc.12.9.2776
McEwen, B. F., Heagle, A. B., Cassels, G. O., Buttle, K. F., Rieder, C. L., et al. (1997). Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 137, 1567–1580. doi:10.1083/jcb.137.7.1567
McGranahan, N., Burrell, R. A., Endesfelder, D., Novelli, M. R., and Swanton, C. (2012). Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 13, 528–538. doi:10.1038/embor.2012.61
Millet, C., Ausiannikava, D., Le Bihan, T., Granneman, S., and Makovets, S. (2015). Cell populations can use aneuploidy to survive telomerase insufficiency. Nat. Commun. 6, 8664. doi:10.1038/ncomms9664
Mullen, M., Yang, F., Cao, J., Cao, Y., Liu, X., Lee, G. Y., et al. (2021). Syntelin inhibits triple-negative breast cancer cell proliferation and metastasis. J. Mol. Cell Biol. 13, 834–837. doi:10.1093/jmcb/mjab054
Ohashi, A., Ohori, M., Iwai, K., Nakayama, Y., Nambu, T., Morishita, D., et al. (2015b). Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat. Commun. 6, 7668. doi:10.1038/ncomms8668
Ohashi, A., Ohori, M., Iwai, K., Nambu, T., Miyamoto, M., Kawamoto, T., et al. (2015a). A novel time-dependent CENP-E inhibitor with potent antitumor activity. PLoS One 10, e0144675. doi:10.1371/journal.pone.0144675
Owa, M., and Dynlacht, B. (2021). A non-canonical function for Centromere-associated protein-E controls centrosome integrity and orientation of cell division. Commun. Biol. 4, 358. doi:10.1038/s42003-021-01861-4
Pavelka, N., Rancati, G., Zhu, J., Bradford, W. D., Saraf, A., Florens, L., et al. (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325. doi:10.1038/nature09529
Pisa, R., Phua, D. Y. Z., and Kapoor, T. M. (2020). Distinct mechanisms of resistance to a CENP-E inhibitor emerge in near-haploid and diploid cancer cells. Cell Chem. Biol. 27, 850–857. doi:10.1016/j.chembiol.2020.05.003
Podgornaia, A. I., and Laub, M. T. (2015). Protein evolution. Pervasive degeneracy and epistasis in a protein-protein interface. Science 347, 673–677. doi:10.1126/science.1257360
Pote, M. S., and Gacche, R. N. (2023). ATP-binding cassette efflux transporters and MDR in cancer. Drug Discov. Today 28, 103537. doi:10.1016/j.drudis.2023.103537
Pussila, M., Törönen, P., Einarsdottir, E., Katayama, S., Krjutškov, K., Holm, L., et al. (2018). Mlh1 deficiency in normal mouse colon mucosa associates with chromosomally unstable colon cancer. Carcinogenesis 39, 788–797. doi:10.1093/carcin/bgy056
Putkey, F. R., Cramer, T., Morphew, M. K., Silk, A. D., Johnson, R. S., McIntosh, J. R., et al. (2002). Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev. Cell 3, 351–365. doi:10.1016/s1534-5807(02)00255-1
Qi, C., Lei, L., Hu, J., Wang, G., Liu, J., and Ou, S. (2020). Serine incorporator 2 (SERINC2) expression predicts an unfavorable prognosis of low-grade glioma (LGG): evidence from bioinformatics analysis. J. Mol. Neurosci. 70, 1521–1532. doi:10.1007/s12031-020-01620-w
Qian, X., McDonald, A., Zhou, H. J., Adams, N. D., Parrish, C. A., Duffy, K. J., et al. (2010). Discovery of the first potent and selective inhibitor of Centromere-Associated Protein E: GSK923295. ACS Med. Chem. Lett. 1, 30–34. doi:10.1021/ml900018m
Rahane, C. S., Kutzner, A., and Heese, K. (2019). A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature. J. Neurooncol. 141, 57–70. doi:10.1007/s11060-018-03029-3
Rancati, G., Pavelka, N., Fleharty, B., Noll, A., Trimble, R., Walton, K., et al. (2008). Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135, 879–893. doi:10.1016/j.cell.2008.09.039
Rath, O., and Kozielski, F. (2012). Kinesins and cancer. Nat. Rev. Cancer. 12, 527–539. doi:10.1038/nrc3310
Real, A. M., Marsiglia, W. M., and Dar, A. C. (2020). Ploidy leads a molecular motor to walk different paths to drug resistance. Cell Chem. Biol. 27, 770–772. doi:10.1016/j.chembiol.2020.06.019
Sardar, H. S., and Gilbert, S. P. (2012). Microtubule capture by mitotic kinesin centromere protein E (CENP-E). J. Biol. Chem. 287, 24894–24904. doi:10.1074/jbc.M112.376830
Schaar, B. T., Chan, G. K., Maddox, P., Salmon, E. D., and Yen, T. J. (1997). CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373–1382. doi:10.1083/jcb.139.6.1373
Seeger, M. A., Zhang, Y., and Rice, S. E. (2012). Kinesin tail domains are intrinsically disordered. Proteins 80, 2437–2446. doi:10.1002/prot.24128
Shan, L., Zhao, M., Lu, Y., Ning, H., Yang, S., Song, Y., et al. (2019). CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. Int. J. Oncol. 55, 257–266. doi:10.3892/ijo.2019.4805
Shastry, S., and Hancock, W. O. (2011). Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. U. S. A. 108, 16253–16258. doi:10.1073/pnas.1102628108
Shaw, A. Y., Henderson, M. C., Flynn, G., Samulitis, B., Han, H., Stratton, S. P., et al. (2009). Characterization of novel diaryl oxazole-based compounds as potential agents to treat pancreatic cancer. J. Pharmacol. Exp. Ther. 331, 636–647. doi:10.1124/jpet.109.156406
Shi, K., Zhu, X., Wu, J., Chen, Y., Zhang, J., and Sun, X. (2021). Centromere protein E as a novel biomarker and potential therapeutic target for retinoblastoma. Bioengineered 12, 5950–5970. doi:10.1080/21655979.2021.1972080
Shi, X., Li, Y., Sun, Y., Zhao, X., Sun, X., Gong, T., et al. (2020). Genome-wide analysis of lncRNAs, miRNAs, and mRNAs forming a prognostic scoring system in esophageal squamous cell carcinoma. PeerJ 8, e8368. doi:10.7717/peerj.8368
Shibuya, A., Ogo, N., Sawada, J. I., Asai, A., and Yokoyama, H. (2021). Structure and comparison of the motor domain of centromere-associated protein E. D. Struct. Biol. 77, 280–287. doi:10.1107/S2059798321000176
Shibuya, A., Suzuki, A., Ogo, N., Sawada, J. I., Asai, A., and Yokoyama, H. (2023). Crystal structure of the motor domain of centromere-associated protein E in complex with a non-hydrolysable ATP analogue. FEBS Lett. 597, 1138–1148. doi:10.1002/1873-3468.14602
Shrestha, R. L., and Draviam, V. M. (2013). Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 23, 1514–1526. doi:10.1016/j.cub.2013.06.040
Silk, A. D., Zasadil, L. M., Holland, A. J., Vitre, B., Cleveland, D. W., and Weaver, B. A. (2013). Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc. Natl. Acad. Sci. U. S. A. 110, E4134–E4141. doi:10.1073/pnas.1317042110
Steblyanko, Y., Rajendraprasad, G., Osswald, M., Eibes, S., Jacome, A., Geley, S., et al. (2020). Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J. 39, e105432. doi:10.15252/embj.2020105432
Tang, J. C., Wu, K., Zheng, X., Xu, M., Dai, Y., Wei, S. S., et al. (2019). GSK923295 as a potential antihepatocellular carcinoma agent causing delay on liver regeneration after partial hepatectomy. Chin. Med. J. Engl. 132, 311–318. doi:10.1097/CM9.0000000000000053
Tanudji, M., Shoemaker, J., L’Italien, L., Russell, L., Chin, G., and Schebye, X. M. (2004). Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell 15, 3771–3781. doi:10.1091/mbc.e03-07-0482
Taveras, C., Liu, C., and Mao, Y. (2019). A tension-independent mechanism reduces Aurora B-mediated phosphorylation upon microtubule capture by CENP-E at the kinetochore. Cell Cycle 18, 1349–1363. doi:10.1080/15384101.2019.1617615
Tcherniuk, S., Deshayes, S., Sarli, V., Divita, G., and Abrieu, A. (2011). UA62784 Is a cytotoxic inhibitor of microtubules, not CENP-E. E. Chem. Biol. 18, 631–641. doi:10.1016/j.chembiol.2011.03.006
Tcherniuk, S. O., and Oleinikov, A. V. (2015). Pgp efflux pump decreases the cytostatic effect of CENP-E inhibitor GSK923295. Cancer Lett. 361, 97–103. doi:10.1016/j.canlet.2015.02.040
Thrower, D. A., Jordan, M. A., and Wilson, L. (1996). Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskelet. 35, 121–133. doi:10.1002/(SICI)1097-0169(1996)35:2<121::AID-CM5>3.0.CO;2-D
Tian, S., Fu, L., Zhang, J., Xu, J., Yuan, L., Qin, J., et al. (2021). Identification of a DNA methylation-driven genes-based prognostic model and drug targets in breast cancer: in silico screening of therapeutic compounds and in vitro characterization. Front. Immunol. 12, 761326. doi:10.3389/fimmu.2021.761326
Torres, E. M., Sokolsky, T., Tucker, C. M., Chan, L. Y., Boselli, M., Dunham, M. J., et al. (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317, 916–924. doi:10.1126/science.1142210
Tovini, L., and McClelland, S. E. (2019). Impaired CENP-E function renders large chromosomes more vulnerable to congression failure. Biomolecules 9, 44. doi:10.3390/biom9020044
Uzdensky, A., Demyanenko, S., Bibov, M., Sharifulina, S., Kit, O., Przhedetski, Y., et al. (2014). Expression of proteins involved in epigenetic regulation in human cutaneous melanoma and peritumoral skin. Tumour Biol. 35, 8225–8233. doi:10.1007/s13277-014-2098-3
Veneziano, L., Barra, V., Cilluffo, D., and Di Leonardo, A. (2019). Proliferation of aneuploid cells induced by CENP-E depletion is counteracted by the p14ARF tumor suppressor. Mol. Genet. Genomics 294, 149–158. doi:10.1007/s00438-018-1495-5
Vitre, B., Gudimchuk, N., Borda, R., Kim, Y., Heuser, J. E., Cleveland, D. W., et al. (2014). Kinetochore-microtubule attachment throughout mitosis potentiated by the elongated stalk of the kinetochore kinesin CENP-E. Mol. Biol. Cell 25, 2272–2281. doi:10.1091/mbc.E14-01-0698
Wang, F., Zhao, Q., Liu, W., and Kong, D. (2020). CENPE, PRC1, TTK, and PLK4 may play crucial roles in the osteosarcoma progression. Technol. Cancer Res. Treat. 19, 1533033820973278. doi:10.1177/1533033820973278
Wang, H., Stephens, B., Von Hoff, D. D., and Han, H. (2009). Identification and characterization of a novel anticancer agent with selectivity against deleted in pancreatic cancer locus 4 (DPC4)-deficient pancreatic and colon cancer cells. Pancreas 38, 551–557. doi:10.1097/MPA.0b013e31819d7415
Wang, Y., and Dasso, M. (2009). SUMOylation and deSUMOylation at a glance. J. Cell Sci. 122, 4249–4252. doi:10.1242/jcs.050542
Weaver, B. A., Bonday, Z. Q., Putkey, F. R., Kops, G. J., Silk, A. D., and Cleveland, D. W. (2003). Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162, 551–563. doi:10.1083/jcb.200303167
Weaver, B. A., and Cleveland, D. W. (2006). Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 18, 658–667. doi:10.1016/j.ceb.2006.10.002
Weaver, B. A., and Cleveland, D. W. (2007). Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 67, 10103–10105. doi:10.1158/0008-5472.CAN-07-2266
Weaver, B. A., Silk, A. D., Montagna, C., Verdier-Pinard, P., and Cleveland, D. W. (2007). Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11, 25–36. doi:10.1016/j.ccr.2006.12.003
Weber, J., Legal, T., Lezcano, A. P., Gluszek-Kustusz, A., Paterson, C., Eibes, S., et al. (2024). A conserved CENP-E region mediates BubR1-independent recruitment to the outer corona at mitotic onset. Curr. Biol. S0960-9822 (24), 00076–00079. doi:10.1016/j.cub.2024.01.042
Williams, B. R., Prabhu, V. R., Hunter, K. E., Glazier, C. M., Whittaker, C. A., Housman, D. E., et al. (2008). Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322, 703–709. doi:10.1126/science.1160058
Wood, K. W., Lad, L., Luo, L., Qian, X., Knight, S. D., Nevins, N., et al. (2010). Antitumor activity of an allosteric inhibitor of centromere-associated protein-E. Proc. Natl. Acad. Sci. U. S. A. 107, 5839–5844. doi:10.1073/pnas.0915068107
Wood, K. W., Sakowicz, R., Goldstein, L. S., and Cleveland, D. W. (1997). CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell 91, 357–366. doi:10.1016/s0092-8674(00)80419-5
Wu, J., Raas, M. W. D., Alcaraz, P. S., Vos, H. R., Tromer, E. C., Snel, B., et al. (2024). A farnesyl-dependent structural role for CENP-E in expansion of the fibrous corona. J. Cell Biol. 223, e202303007. doi:10.1083/jcb.202303007
Wylie, A. A., Schoepfer, J., Jahnke, W., Cowan-Jacob, S. W., Loo, A., Furet, P., et al. (2017). The allosteric inhibitor ABL001 enables dual targeting of BCR-ABL1. Nature 543, 733–737. doi:10.1038/nature21702
Xu, C., Su, W., and Jiang, X. (2020). Identification of differentially expressed genes and signaling pathways in glioma by integrated bioinformatics analysis. J. Craniofac. Surg. 31, 2360–2363. doi:10.1097/SCS.0000000000006743
Yamane, M., Sawada, J. I., Ogo, N., Ohba, M., Ando, T., and Asai, A. (2019). Identification of benzo[d]pyrrolo[2,1-b]thiazole derivatives as CENP-E inhibitors. Biochem. Biophys. Res. Commun. 519, 505–511. doi:10.1016/j.bbrc.2019.09.028
Yang, C. P., Liu, L., Ikui, A. E., and Horwitz, S. B. (2010). The interaction between mitotic checkpoint proteins, CENP-E and BubR1, is diminished in epothilone B-resistant A549 cells. Cell Cycle 9, 1207–1213. doi:10.4161/cc.9.6.11122
Yao, Q., He, Y. L., Wang, N., Dong, S. S., Tu He Ta Mi Shi, M. E., Feng, X., et al. (2021). Identification of potential genomic alterations and the circRNA-miRNA-mRNA regulatory network in primary and recurrent synovial sarcomas. Front. Mol. Biosci. 8, 707151. doi:10.3389/fmolb.2021.707151
Yao, X., Abrieu, A., Zheng, Y., Sullivan, K. F., and Cleveland, D. W. (2000). CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2, 484–491. doi:10.1038/35019518
Yao, X., Anderson, K. L., and Cleveland, D. W. (1997). The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139, 435–447. doi:10.1083/jcb.139.2.435
Yen, T. J., Compton, D. A., Wise, D., Zinkowski, R. P., Brinkley, B. R., Earnshaw, W. C., et al. (1991). CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10, 1245–1254. doi:10.1002/j.1460-2075.1991.tb08066.x
Yen, T. J., Li, G., Schaar, B. T., Szilak, I., and Cleveland, D. W. (1992). CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature 359, 536–539. doi:10.1038/359536a0
Yoshizawa, K., Matsura, A., Shimada, M., Ishida-Ishihara, S., Sato, F., Yamamoto, T., et al. (2023). Tetraploidy-linked sensitization to CENP-E inhibition in human cells. Mol. Oncol. 17, 1148–1166. doi:10.1002/1878-0261.13379
Yuan, Y., Deng, X., Wang, S., and Han, S. (2023). Comprehensive pan-cancer analysis identifies centromere-associated protein E as a novel prognostic and immunological biomarker in human tumors. Biochim. Biophys. Acta Gen. Subj. 1867, 130346. doi:10.1016/j.bbagen.2023.130346
Yuen, K. W., Montpetit, B., and Hieter, P. (2005). The kinetochore and cancer: what’s the connection? Curr. Opin. Cell Biol. 17, 576–582. doi:10.1016/j.ceb.2005.09.012
Zasadil, L. M., Britigan, E. M., Ryan, S. D., Kaur, C., Guckenberger, D. J., Beebe, D. J., et al. (2016). High rates of chromosome missegregation suppress tumor progression but do not inhibit tumor initiation. Mol. Biol. Cell 27, 1981–1989. doi:10.1091/mbc.E15-10-0747
Zasadil, L. M., Britigan, E. M., and Weaver, B. A. (2013). 2n or not 2n: aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Semin. Cell Dev. Biol. 24, 370–379. doi:10.1016/j.semcdb.2013.02.001
Zhang, X. D., Goeres, J., Zhang, H., Yen, T. J., Porter, A. C., and Matunis, M. J. (2008). SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol. Cell 29, 729–741. doi:10.1016/j.molcel.2008.01.013
Keywords: kinesin, CENP-E, chromosome instability, cancer, aneuploidy, tumorigenesis
Citation: Yang Y-H, Wei Y-L and She Z-Y (2024) Kinesin-7 CENP-E in tumorigenesis: Chromosome instability, spindle assembly checkpoint, and applications. Front. Mol. Biosci. 11:1366113. doi: 10.3389/fmolb.2024.1366113
Received: 05 January 2024; Accepted: 04 March 2024;
Published: 15 March 2024.
Edited by:
Gloria C Ferreira, University of South Florida, United StatesReviewed by:
Viviana Barra, University of Palermo, ItalySergiy Borysov, Saint Leo University, United States
Copyright © 2024 Yang, Wei and She. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Yu She, zhenyushe@fjmu.edu.cn
 Yu-Hao Yang
Yu-Hao Yang Ya-Lan Wei
Ya-Lan Wei Zhen-Yu She
Zhen-Yu She