Emerging Alternative Proteinases in APP Metabolism and Alzheimer’s Disease Pathogenesis: A Focus on MT1-MMP and MT5-MMP
- Aix-Marseille Univ, CNRS, INP, Inst Neurophysiopathol, Marseille, France
Processing of amyloid beta precursor protein (APP) into amyloid-beta peptide (Aβ) by β-secretase and γ-secretase complex is at the heart of the pathogenesis of Alzheimer’s disease (AD). Targeting this proteolytic pathway effectively reduces/prevents pathology and cognitive decline in preclinical experimental models of the disease, but therapeutic strategies based on secretase activity modifying drugs have so far failed in clinical trials. Although this may raise some doubts on the relevance of β- and γ-secretases as targets, new APP-cleaving enzymes, including meprin-β, legumain (δ-secretase), rhomboid-like protein-4 (RHBDL4), caspases and membrane-type matrix metalloproteinases (MT-MMPs/η-secretases) have confirmed that APP processing remains a solid mechanism in AD pathophysiology. This review will discuss recent findings on the roles of all these proteinases in the nervous system, and in particular on the roles of MT-MMPs, which are at the crossroads of pathological events involving not only amyloidogenesis, but also inflammation and synaptic dysfunctions. Assessing the potential of these emerging proteinases in the Alzheimer’s field opens up new research prospects to improve our knowledge of fundamental mechanisms of the disease and help us establish new therapeutic strategies.
Alzheimer’s Disease, A Proteolytic Problem
Alzheimer’s disease (AD) is the most common type of neurodegenerative disorder for which only a few drugs have shown transient and moderate anti-symptomatic effects, but there is no treatment that slows down or prevents the progression of the disease. A minority of AD cases find their cause in deterministic genetic mutations in three genes: PSEN1, PSEN2 and APP, encoding, respectively, aspartyl proteinase presenilin 1 and 2 (PS1/PS2) and amyloid precursor protein (APP; Van Cauwenberghe et al., 2016). These mutations account for the so-called “familial forms” of the disease. The overwhelming majority of AD cases (∼95%) are sporadic forms of unknown etiology. Despite controversies over the causes of AD, it is still recognized that brain accumulation of the amyloid peptide-β (Aβ) plays a central role in the pathogenic process (Selkoe and Hardy, 2016). Aβ results from the proteolysis of APP, a type I transmembrane protein targeted first at the plasma membrane, then rapidly endocytosed to endosomes to be metabolized to Aβ or subsequently sent to the lysosomal compartment for degradation (Wang X. et al., 2017; Van Acker et al., 2019). Endosomes are thought to be the main locus of Aβ production, which is ensured by canonical β- and γ-secretases (Vassar et al., 1999).
β-site APP cleaving enzyme 1 (BACE-1) is the main β-secretase. This type I transmembrane protein of the aspartyl proteinase family needs an acidic environment (optimum pH 4.5) to be enzymatically active (Saric et al., 2013). BACE-1 cleaves numerous substrates, which confers this enzyme a wide spectrum of physiological and pathological activities (Kuhn et al., 2012; Zhou et al., 2012; Vassar et al., 2014), but it is indisputably its ability to process APP that has attracted much attention, especially in relation to AD. As illustrated in Figure 1, BACE-1 cleaves APP between Met671 and Asp672 to generate the soluble APP-β fragment (sAPPβ) and its complementary C-terminal counterpart of 99 amino acids termed β-CTF or C99. This cleavage signals the first emblematic proteolytic step to the production of Aβ, considered to be one of the main driving forces in AD pathogenesis. BACE-1 can also catalyze amyloidolytic processing by cleavage between Tyr10 and Glu11 of the Aβ sequence (Huse et al., 2002; Liu et al., 2002; Kimura et al., 2016), the so-called β’-cleavage site (Figure 1). Interestingly, β’-cleavage is favored in the neuroprotective Icelandic APP mutation while the canonic β-cleavage at Asp1 is reduced (Kimura et al., 2016). However, a causal effect between β’-cleavage and neuroprotection is not straight forward as Aβ11–40 is found in insoluble Aβ pools of post-mortem AD brains (Huse et al., 2002). More recently, it has been shown that BACE-1 can also cleave Aβ40 or Aβ42 to generate a C-terminal truncated Aβ34 form, which appears to be a new biomarker of Aβ clearance in AD as it is noticeably increased in mild cognitive impaired patients along with strong BACE-1 activity (Liebsch et al., 2019). Together, these data confirm early studies (Fluhrer et al., 2003; Shi et al., 2003), placing BACE-1 as a prominent Aβ-generating, but also as an occasional Aβ-“degrading” enzyme under some circumstances. Physiologically relevant Aβ degradation has been mainly attributed to 4 metalloproteinases: neprilysin, insulin degrading enzyme, endothelin converting enzyme and angiotensin converting enzyme, the regulation and functions of which have been extensively reviewed elsewhere (De Strooper, 2010; Nalivaeva et al., 2012). Matrix metalloproteinases (MMPs), including MMP-2, MMP-3, MMP-7 and MMP-9 also cleave within the Aβ sequence (Rivera et al., 2019) and these cleavages have been compared to those of membrane-type MMPs (MT-MMPs) in Figure 1.
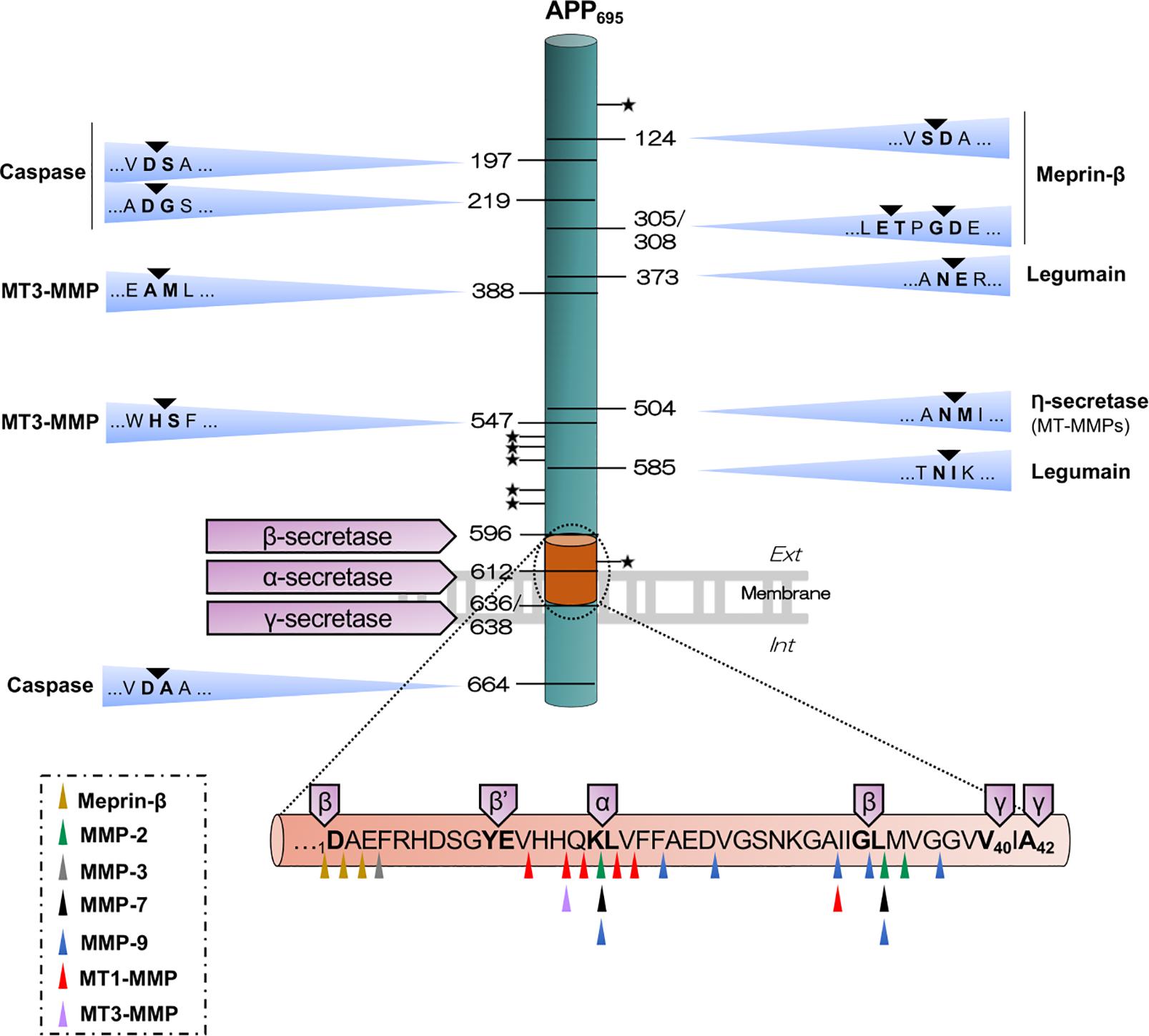
Figure 1. Illustrative outline of the different APP cleavage sites. The scheme shows the canonical (β-, α- and γ-secretases) and non-canonical APP-cleavages that result in the different APP fragments. Meprin-β cleaves APP in five sites, possibly generating soluble APP fragments of different size, sAPPβ or sAPPβ-like (sAPPβ with one or two additional amino acids) and its complementary C-terminal β-CTF or β-CTF-like (β-CTF with one or two amino acids less), which is further processed by γ-secretase, thus producing Aβ1–X, Aβ2–X or Aβ3–X. Legumain cleaves in two sites, giving rise to two soluble fragments and one C-terminal transmembrane fragment than can be further processed by the canonical enzymes. Caspase can cleave in the N-terminal domain, generating truncated APP-species, and within the intracytoplasmic domain, generating an APP-Ncas fragment and a C-terminal residual peptide of 31 amino acids. RHBDL4 cleavages within the ectodomain at several sites are not yet identified (represented with a star), generating different N- and C-terminal fragments. Cleavage by η-secretase generates a soluble fragment (sAPPη/sAPP95) and a paired transmembrane product η-CTF/CTF-30 that can be further processed by β- or α-secretase to release Aη-β and Aη-α, respectively. MT3-MMP can cleave APP in 4 sites, including the η-secretase site and also within the Aβ sequence. The scheme below represents the Aβ sequence with multiple cleavage sites for various soluble MMPs and MT1-MMP, for meprin-β and for canonical secretases. All the residues are termed using APP695 numbering.
The γ-secretase complex is formed by a PS1 or PS2 catalytic subunit and 3 partner proteins, Aph-1, pen-2 and nicastrin (Haass et al., 2012; Rajendran and Annaert, 2012; Masters et al., 2015; Selkoe and Hardy, 2016). Presenilins are acidic proteinases [optimum pH 6.3, (Campbell et al., 2003)] that belong to the family of seven transmembrane domain proteins. Like BACE-1, γ-secretase targets many substrates in addition to APP, with the consequent impact on a vast array of physiological and pathological processes (Haapasalo and Kovacs, 2011). The subcellular location and substrate specificity of γ-secretase may vary depending on the presence of PS1 or PS2 in the complex. While the complex containing PS1 is widely distributed in the cell, a single acidic-dileucine sorting motif present in PS2 directs the γ-secretase complex to late endosomes/lysosomes (Sannerud et al., 2016). γ-secretase performs regulated intramembrane proteolysis to process C99 and release Aβ and the remaining APP intracellular domain (AICD). The latter can be translocated in the nucleus and engages in transcriptional activities, which are to some extent still controversial in the AD field (reviewed in Pardossi-Piquard and Checler, 2012) (Table 1).
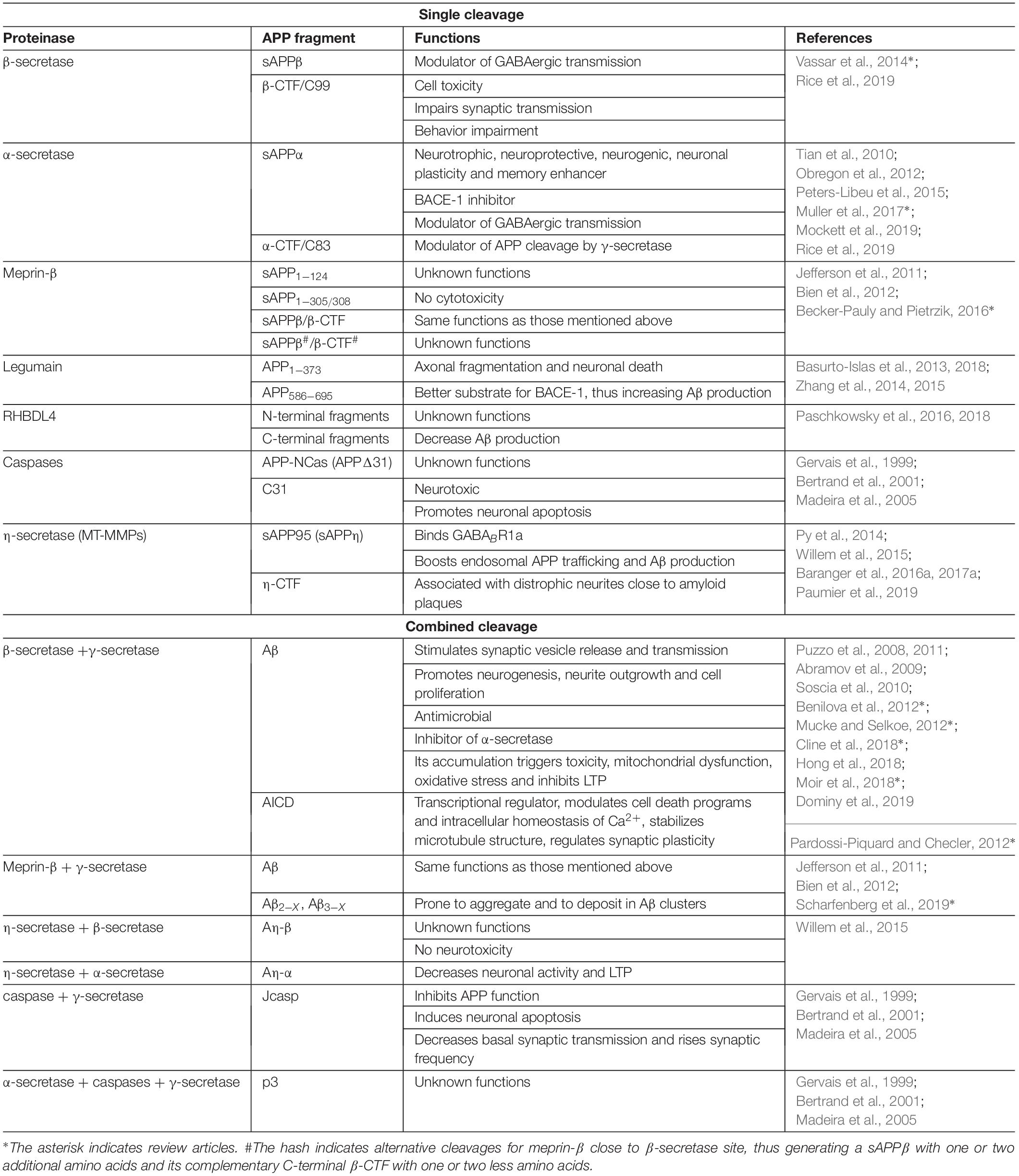
Table 1. Representative of single or combined cleavages of APP by proteinases and known functions of generated fragments.
Along the secretory pathway, APP can also undergo α-secretase cleavage when reaching the plasma membrane. This is performed by a disintegrin and metalloproteinases (ADAMs) of the metzincin superfamily of metalloproteinases (Rivera et al., 2010; Paschkowsky et al., 2019), in a constitutive (ADAM10) or regulated manner (ADAM17; also known as TACE for TNF-α converting enzyme) (Lammich et al., 1999; Jorissen et al., 2010; Kuhn et al., 2010). ADAMs cleave APP in the middle of the Aβ sequence, thereby preventing its generation and releasing at the same time a soluble APP fragment (sAPPα) of ∼110 kDa. In cultured neurons from rodents and humans, sAPPα has shown anti-excitotoxic properties through a mechanism involving the reduction of Ca2+ cytotoxic influx (Mattson et al., 1993). These data linking sAPPα to neuroprotection were later confirmed by studies showing that moderate neuronal overexpression of ADAM10 in transgenic AD mice favored an increase in sAPPα levels and a decrease in Aβ, concomitant with a reduction of deficits in long-term potentiation (LTP) and learning (Postina et al., 2004). Likewise, sAPPα could rescue LTP in acute hippocampal slices from mice lacking both APP and its family homolog APP-like protein 2 (APLP2) (Hick et al., 2015), and prevented memory deficits after lentiviral overexpression in the APPswe/PS1dE9 AD mouse model (Tan et al., 2018). While sAPPα release is usually considered as the result of physiological APP processing, Aβ oligomers may be responsible for sAPPα release by cultured neurons and also in vivo, which could be considered as a neuroprotective cellular response to the amyloid challenge (Rose et al., 2018). This idea is all the more interesting in the context of data indicating that sAPPα interacts with BACE-1 and inhibits Aβ production (Obregon et al., 2012), in clear contrast with sAPPβ, which fails to interact with BACE-1 because the truncation of 16 aminoacids at the carboxy end (sequence between the α- and β-cleavage sites) is sufficient to impose significant structural changes compared to sAPPα (Peters-Libeu et al., 2015). Recent studies have highlighted new mechanisms by which sAPPα could influence synaptic function through the modulation of the GABA and glutamate neurotransmitter systems. Thus, sAPPα has been shown to act as a ligand of GABABR1a -a metabotropic receptor for GABA neurotransmitter-, which results in modulation of hippocampal synaptic plasticity and neurotransmission in vivo by decreasing the release of synaptic vesicles (Rice et al., 2019). In addition, sAPPα stimulates trafficking of GluA2-lacking AMPA and NMDA receptors to the synapse, as well as de novo protein synthesis of GluA1 protein, thereby providing mechanistic ground for promotion of LTP and synaptic plasticity (Mockett et al., 2019). Consistent with the aforementioned anti-amyloidogenic properties of sAPPα, its complementary C-terminal fragment of 83 amino acids (C83) can also inhibit Aβ generation by interfering with C99 processing by γ-secretase in HEK cells (Tian et al., 2010). The idea that there is cross-regulation between amyloidogenic and non-amyloidogenic pathways is further supported by the fact that Aβ inhibits ADAM10 in mouse primary neuronal cultures, thus favoring endogenous APP processing by β-secretase (Spilman et al., 2016). Since activation and inhibition of ADAMs may be a determining factor for AD pathology, the regulatory potential of the tissue inhibitors of metalloproteinases (TIMPs) should be considered. TIMPs are mainly known as inhibitors of MMPs (see section below), but they also modulate ADAMs activity with low Ki values of 0.5 and 0.1 nM for TIMP-1 and TIMP-3, respectively (Rapti et al., 2008; Baranger et al., 2014). One of our studies shows that TIMP-1 expression is highly upregulated in the brain of transgenic 5xFAD mice at early stages of the pathology and remains durably high, likely matching the progression of neuroinflammation and gliosis (Py et al., 2014). In the diseased brain, reactive astrocytes are the main cellular source of TIMP-1 (Rivera et al., 1997, 2002; Pagenstecher et al., 1998), which in turn promotes astrocyte proliferation (Ogier et al., 2005; Hernandez-Guillamon et al., 2009), suggesting altogether that TIMP-1 could contribute to AD neuroinflammation and/or downregulate ADAM10 activity. As for TIMP-3, one study in neuroblastoma cells reported that this inhibitor not only reduces α-cleavage of APP but also the cell surface levels of ADAM10 and APP, which leads to increased endocytosis and β-secretase cleavage. The potential relevance of TIMP-3 in promoting AD pathogenesis is reinforced in this study by the elevated neuronal immunostaining found in 3xTg transgenic mouse and by increased levels in AD brain homogenates (Hoe et al., 2007). However, another study in 5xFAD mice, showed mild variations of TIMP-3 mRNA levels at different ages, compared to wild type (Py et al., 2014). Clearly, studies are lacking to confirm or refute the idea that a regulatory action of TIMPs on ADAMs could impact on the progression of the disease.
The precise neurotoxicity mechanisms of Aβ are still poorly understood, probably reflecting a combination of events triggered by its accumulation, including stimulation of Tau hyperphosphorylation and subsequent formation of neurofibrillary tangles (NFTs), promotion of inflammation and also synaptic dysfunctions (Benilova et al., 2012; Mucke and Selkoe, 2012; Selkoe and Hardy, 2016; Cline et al., 2018). Monomeric Aβ can be assembled into oligomers, protofibrils or fibrils, the latter being the main constituent of amyloid plaques. Although oligomers are increasingly considered to be the most toxic Aβ assemblage, recent data suggest that only a limited fraction of highly diffusible small oligomers have adverse effects, for instance on the disruption of synaptic activity (Hong et al., 2018). Missing relevant Aβ structures as possible therapeutic targets might be one of the reasons why anti-Aβ strategies developed to fight AD have not yet demonstrated significant clinical benefits (Panza et al., 2019). In the case of therapies based on β- and γ-secretase inhibitors, a possible cause of clinical failure could be the difficulty of inhibiting these proteinases without causing side effects resulting from non-targeted proteolytic inhibition on their many physiological substrates (Karran et al., 2011; Vassar et al., 2014; De Strooper and Chavez Gutierrez, 2015; Barao et al., 2016; Ohno, 2016; Yan, 2017). Besides Aβ, other potential neurotoxic APP metabolites generated with the contribution of BACE-1 activity were initially reported. This was the case of a 35 kDa sAPP NTF fragment (1-286) and sAPPβ whose binding to death receptor 6 (DR6) induced axonal pruning and neuronal apoptosis mediated by caspase activation (Nikolaev et al., 2009). The implication of this 35 kDa fragment was later refuted; instead sAPPβ was reported to induce axonal pruning but not neuronal death, and to interact with DR6 through the more C-terminal E2 domain placed above the β-cleavage site (Olsen et al., 2014). Furthermore, the involvement of DR6 in Alzheimer’s neurodegeneration could not be proved in two transgenic mouse models of AD deficient for DR6 (Kallop et al., 2014). The roles of C-terminal APP fragments resulting from the proteolytic activity of canonical secretases have also been investigated. Thus, many studies using genetic or pharmacological approaches to increase C99 levels in animal models consistently demonstrate the pathogenic properties of C99 linked with AD phenotypes (Neve et al., 1996; Oster-Granite et al., 1996; Song et al., 1998; Matsumoto et al., 2002; Lauritzen et al., 2012, 2016; Bourgeois et al., 2018). It is noteworthy that C99 accumulates in the brains of AD patients (Pera et al., 2013; Kim et al., 2016) and shows a better correlation than Aβ with the degree of vulnerability to neurodegeneration (Pulina et al., 2019). The other C-terminal APP fragment that has focused a great deal of research activity in recent years is the AICD. Despite controversial data has been often reported on its physiological and pathological functions, AICD has gained consensus as signaling molecule that can translocate to the nucleus and regulate the expression of manifold genes, some of them involved in AD pathogenic mechanisms (Kim et al., 2003; Chang et al., 2006), but also in cytoskeleton dynamics, cell cycle control or synaptic plasticity. A recent review has identified nearly 40 genes targeted by the AICD in relation with these and other processes (Bukhari et al., 2017). These genes will need to be tested for their possible contribution to signaling pathways that depend on APP and their consequent impact on brain pathology and physiology.
Other APP fragments generated by emerging APP processing enzymes are currently being investigated to determine their potential physiological or pathological actions and they will be discussed below.
Insufficiently Explored Territories
The idea that APP metabolites other than Aβ could actually impact on AD pathology is gaining momentum. However, the assessment of their physiological functions should also be carried out in parallel in order to have a more accurate picture of the pathophysiological scenario. The same holds true for Aβ, which remains the main driving force behind AD pathology, but whose physiological activities are still poorly understood and certainly insufficiently studied. Thus, for example, a few studies indicate that Aβ has antimicrobial activities, leading some authors to propose that it could act as an innate immunity protein in the brain (Soscia et al., 2010; Moir et al., 2018; Dominy et al., 2019). In addition, Aβ has been shown to be a positive modulator of synaptic vesicle release (Abramov et al., 2009) and synaptic transmission (Puzzo et al., 2008, 2011). In fact, picomolar concentrations of Aβ are sufficient to stimulate nicotinic receptors and subsequently LTP and learning and memory in healthy rodents (Puzzo et al., 2008, 2012; Morley et al., 2010). To what extent the failure of current anti-Aβ therapies could be due to interference with these (or other unknown) physiological roles of Aβ, remains an open question.
Despite the importance of APP proteolysis and the resulting fragments in the context of AD, APP role as a cell adhesion protein or receptor also deserves consideration insofar as the alteration of these functions may indeed contribute to the disease process. Accordingly, it is proposed that APP contributes to cell adhesion responsible for maintaining the structure of synapses and neural circuits through its interactions with extracellular matrix and adhesion molecules, but also through its own dimerization at the cell surface (see van der Kant and Goldstein, 2015; Montagna et al., 2017; Muller et al., 2017; Sosa et al., 2017), which is regulated, inter alia, by APP binding with Cu2+ and Zn2+ (Wild et al., 2017). Disruptions of physiological functions of APP could cause synaptic dysfunctions that occur in the early phases of AD, but it could also alter the subcellular location of APP and thus the degree of pathogenicity of proteolytic fragments depending on whether they are mainly generated in endosomes (e.g., C99, Aβ) or in the cell membrane (e.g., sAPPα). APP has also been described as a functional cellular receptor for Aβ to the point that Aβ oligomers isolated from AD brains require APP expression to induce synaptotoxic effects on mice hippocampi (Wang Z. et al., 2017). By the same time, it was shown that Aβ oligomers and also Tau could cause APP-dependent impairment of LTP and spatial learning in mice (Puzzo et al., 2017). Another work highlights the potential of full length APP as functional receptor for the regulation of cholesterol/lipoprotein metabolism and Aβ clearance (Fong et al., 2018). In this case, human astrocytes derived from induced pluripotent stem cells (iPS) deficient for APP exhibited reduced levels of intracellular cholesterol, as well as a reduced ability to endocyte apolipoprotein E (APOE) and Aβ, which are major processes disturbed in AD and possible cause of deficient Aβ clearance. Similar deficits have been observed in astrocytes expressing APP harboring the Swedish familial mutation, characterized by reduced canonical levels of APP due to exacerbated cleavage of APP by β-secretase. It is interesting to note that the inhibition of β-secretase reversed these effects, thereby highlighting the potential benefits of such inhibition specifically in astrocytes (Fong et al., 2018). These few examples illustrate the importance of canonical APP as a regulator of tissue homeostasis and the extent to which subtle alterations in its biology could trigger or accompany pathological processes. Further research will be needed to increase our knowledge of the biology of APP and thus determine whether maintaining APP levels could be a therapeutic strategy of interest.
The complexity of interactions between APP and canonical secretases results in multifunctional derivatives of APP (Chen et al., 2015; Liu et al., 2019). The emergence of proteinases with new APP transformation activities and new fragments does not precisely simplify this scenario, but may be useful to broaden the spectrum of mechanisms to be targeted in a therapeutic context. We will focus our attention in the following section on a subfamily of MMPs, the MT-MMPs, but will also discuss recent findings linking meprin-β, legumain, rhomboid-like protein-4 (RHBDL4) and caspases to APP/Aβ metabolism (Figure 1 and Table 1).
Some Emerging APP Processing Proteinases
Meprin-β
Meprin-β is a type I transmembrane protein of the astacin group of the metzincin superfamily of metalloproteinases (Villa et al., 2003). The enzyme is naturally inhibited by fetuin-A (or alpha2-Heremans-Schmid glycoprotein) and fetuin-B, but not by cystatin C, a closely related inhibitor (Hedrich et al., 2010; Karmilin et al., 2019). Meprin-β activity has been linked to the processing of a variety of substrates, including inflammatory cytokines, and cell adhesion and extracellular matrix molecules in different organs. Overall, this proteinase exerts control on inflammatory/immune and cell migration processes. In the nervous system, meprin-β has been described as a new alternative APP processing enzyme, which links its activity to AD (reviewed in Scharfenberg et al., 2019). In this context, several studies have reported increased mRNA and protein levels of meprin-β in AD patients compared to age-matched healthy individuals (Bien et al., 2012; Schlenzig et al., 2018). Produced by brain neurons, meprin-β exists in plasma membrane-bound form or in soluble form after shedding by ADAM10 or ADAM17. Using a synthetic peptide mimicking APP/Aβ sequence, it was shown in vitro that meprin-β was able to cleave APP at the β-cleavage site and after the first and second amino acids of the Aβ sequence, suggesting that meprin-β can behave as a β-secretase. This was further supported by the fact that meprin-β expression in BACE-1/2 knockout fibroblasts was sufficient to generate Aβ (Bien et al., 2012). In addition, meprin-β can cleave APP near the N-terminal end to generate truncated fragments of 11 and 20 kDa (N-APP), as demonstrated by terminal amine isotopic labeling of substrates. N-APP were detected in mice brains as well as in the brains of healthy individuals and patients with AD, but not in meprin-β knockout mice, validating the role of meprin-β in the physiological processing of APP (Jefferson et al., 2011). These APP fragments did not induce toxicity in neuronal cultures and therefore further work will be needed to elucidate their physiological functions. Unlike BACE-1, meprin-β cleaves APP on the plasma membrane before it undergoes endocytosis, and preferentially generates Aβ1–40 and a N-terminally truncated Aβ (Aβ2–X), which is more prone to aggregation than Aβ1–40. This is of interest, as truncated Aβ species may promote the formation of highly toxic Aβ oligomers. Also important, Aβ2–X can only be generated by membrane-bound meprin-β and not by the soluble form after ADAM10/17-mediated shedding (Bien et al., 2012). An additional regulatory interplay between meprin-β and α-secretase has been identified; meprin-β deletion in mice correlates with higher levels of sAPPα, thereby suggesting in vivo competition with α-secretase for membrane-bound APP (Schonherr et al., 2016). It has to be noted that APP mutations proximal to the β-cleavage site, like the Swedish (K670N-M671L) or the “protective” Icelandic (A673T), prevent the generation of Aβ2–X by meprin-β as opposed to the lack of effect of distal mutations (i.e., London (V717I), highlighting the importance for meprin-β of aminoacid composition around the β-cleavage site (Schonherr et al., 2016). Overall, meprin-β bears great interest as alternative APP processing enzyme with amyloidogenic features. It remains to be determined to which extent the relatively small amount of Aβ2–X generated in AD compared to Aβ42 provides meprin-β with a relevant pathogenic role in AD. Further investigations will also be necessary to assess the possible impact of meprin-β as a regulatory factor of neuroinflammatory processes operating in AD and other brain disorders.
Legumain
Legumain, also known as asparagine endopeptidase, is a soluble cysteine proteinase mainly found in endo-lysosomal compartments that provide the optimal functional pH of 6. Its proteolytic activity is inhibited by cystatin C and closely related cystatins E/M and F (Chen et al., 1997; Alvarez-Fernandez et al., 1999). The enzyme was later renamed δ-secretase (Zhang et al., 2015) to highlight its role in APP cleavage at Asn586 that generates the δ-CTF first identified in the nineties (Simons et al., 1996; Scharfenberg et al., 2019). As lysosomal dysfunction is a transversal pathogenic mechanism, legumain has been involved in numerous pathological settings, including atherosclerosis, osteoporosis, cancer, ischemic stroke, and neurodegenerative diseases (Lunde et al., 2019). Legumain has been described in relation with AD as a modulator of Tau phosphorylation (Basurto-Islas et al., 2013), and as a Tau- or APP-cleaving enzyme (Zhang et al., 2014, 2015). Under acidic conditions (e.g., brain ischemia, hypoxia, or AD), legumain can translocate from neuronal lysosomes into the cytoplasm, where it cleaves I2PP2A (also known as SET). This cleavage generates two fragments, I2NTF and I2CTF that inhibit protein phosphatase 2A (PP2A), a key phosphatase that limits Tau hyperphosphorylation both in vitro (Basurto-Islas et al., 2013) and in vivo (Basurto-Islas et al., 2018). In total, legumain activity promotes the hyperphosphorylation of Tau protein, which is a major hallmark of AD pathogenesis.
In addition to controlling Tau phosphorylation, legumain generates neurotoxic fragments after cleavage of Tau and APP. The brain of AD patients show increased levels of the Tau1–368 fragment compared to healthy individuals, in correlation with increased levels of legumain activity (Zhang et al., 2014). This study showed that Tau1–368 generated by legumain inhibits microtubule polymerization in vitro and promotes apoptosis in rat cultured neurons. Consistent with these in vitro findings, the deletion of legumain in the Tau P301S transgenic mouse model of AD prevents synaptic dysfunction and improves learning and memory. Furthermore, mice virally infected with uncleavable Tau mutant showed reduced pathological and behavioral defects as compared with mice infected with Tau P301S. Of note, antibodies specifically raised against the Tau1–368 neoepitope were found in AD brains and absent in legumain knockout mice, thus validating in vivo legumain-mediated Tau processing (Zhang et al., 2014). The same group also demonstrated that legumain can cleave APP at positions 373 and 585 (APP695 numbering), generating 2 fragments with distinctive cytotoxicity (Zhang et al., 2015). While APP1–373 triggers axonal fragmentation and neuronal death in primary cultured neurons, the APP586–695 fragment appears to be a better pro-amyloidogenic substrate for BACE-1 than full-length APP and the other legumain-derived C-terminal fragment (APP374–695). In support of these in vitro data, legumain deficiency in the 5xFAD transgenic mouse model of AD (Oakley et al., 2006) causes a drop of Aβ40, Aβ42 and amyloid plaque burden, while it increases spine density, and prevents deficits in LTP and learning and memory (Zhang et al., 2015). The first step for pharmacological validation of legumain in AD was obtained using a chemical inhibitor, which reduced the formation of neurotoxic Tau fragments and Aβ accumulation in P301S and 5xFAD mouse models, respectively. In addition, legumain inhibition reduced synaptic loss, improved LTP, prevented microglial activation and reduced the levels of inflammatory mediators TNF-α and IL-1β (Zhang et al., 2017).
Rhomboid-Like Protein-4
There are five active intramembrane proteinases Rhomboids in humans, RHBDL1-4 and PARL. RHBDL1-4 are located in the secretory pathway, while PARL is found in mitochondria. This family of serine proteinases is involved in many cellular processes, such as inflammatory signaling, cell migration, proliferation and mitochondria homeostasis (reviewed in Dusterhoft et al., 2017; Paschkowsky et al., 2019). Recently, Paschkowsky et al. (2016) showed in HEK cells expressing APP695 that RHBDL4 but not RHDBL1, 2 and 3, cleaves at multiple sites within the ectodomain of APP (Figure 1) and APLP1 and APLP2, generating a N-terminal fragment of ∼70 kDa and different C-terminal fragments whose functions are still unknown. The use of specific inhibitors for a wide spectrum of proteinases, including BACE-1, α- and γ-secretase, aspartyl and cysteine proteinases, and metalloproteinases did not modify the APP fragmentation profile generated by RHBDL4, suggesting a specific action of this proteinase. Moreover, it was shown that RHBDL4 activity decreases the levels of Aβ38, Aβ40 and Aβ42. RHBDL4 is mainly located in the endoplasmic reticulum, which contains low levels of membrane cholesterol. Interestingly, RHBDL4-mediated cleavage of APP is negatively regulated by cholesterol when it binds to two transmembrane domains of the proteinase, but not to APP. Thus, it is possible that lowering cholesterol could increase RHBDL4 activity on full-length APP, with the consequent generation of CTFs and the decrease in Aβ generation (Paschkowsky et al., 2018). Overall, these data underline the unexpected important role of RHBDL4, which is not a classical secretase, in the metabolism of APP/Aβ. Nevertheless, much remains to be done to validate most of the referred in vitro observations in in vivo models with relevance for AD pathogenesis.
Caspases
Caspases are pleiotropic cysteine proteinases that specifically cleave target proteins after an aspartic acid residue (Hyman and Yuan, 2012). Several studies have demonstrated in a variety of culture cell systems the ability of caspase-3, -6 and -8 to cleave APP, mainly between amino acids V664-D665 (APP695 numbering) within the intracytoplasmic domain (Gervais et al., 1999; LeBlanc et al., 1999; Pellegrini et al., 1999; Weidemann et al., 1999). Cleavage of APP by caspases generates a membrane bound N-terminal fragment, named APP-Ncas, and an intracellular C-terminal peptide of 31 amino acids, named APP-Ccas or C31. When γ-secretase processes C99 into Aβ, the remaining AICD is further cleaved by caspase to generate C31 and JCasp fragments, both of which promote neuronal apoptosis (Bertrand et al., 2001; Madeira et al., 2005). For this reason, it was suggested that the cytotoxicity of C99 might actually result from subsequent formation of C31 by caspases (Lu et al., 2000). It is also noteworthy that AD brains show elevated levels of caspases and that APP fragments resulting from caspase cleavage colocalize with amyloid plaques (Gervais et al., 1999; Lu et al., 2000). Eventually, caspase-mediated APP cleavage stimulates Aβ production in B103 and NT2 cells (Gervais et al., 1999). However, other authors have challenged these data in a study where caspase removed the internalization signal (YENPTY) located in the extremity of the AICD, thereby leading to impaired APP internalization and Aβ production in B103 cells (Soriano et al., 2001). These results were recently confirmed using CRISPR/Cas9 editing in human iPS to remove the last 36 amino acids of APP containing the internalization motif. The expression of this C-terminal truncated APP in human iPS-derived neurons prevented the production sAPPβ and Aβ, while stimulating α-secretase cleavage. In addition, it was shown that cultured hippocampal neurons with truncated APP gene accumulate APP on the plasma membrane and present reduced colocalization of BACE-1 and APP in endosomes, confirming a key role of the YENPTY sequence in APP internalization and Aβ production by endosomes (Sun et al., 2019).
Caspases have also been proposed as a possible molecular link between amyloid and Tau pathologies in AD, since Aβ simulation of neuronal apoptosis is accompanied by an increase of Tau cleavage by caspases at Asp421. This is consistent with the observation that C-terminal truncated forms of Tau show increased polymerization capacity in vitro and are associated with NFTs formation in the brain of AD patients (Gamblin et al., 2003; Rissman et al., 2004).
MT-MMPs
MT-MMPs are new players in the field of AD, mainly because of their ability to regulate APP metabolism and therefore amyloidogenesis. In the next sections, we will focus on two MT-MMPs, MT1-MMP and MT5-MMP, which have been associated with the pathophysiological mechanisms that support AD. More generally, the involvement of other MMPs in AD and other neurodegenerative disorders has been discussed elsewhere (Rivera et al., 2019).
Structure of MT1-MMP and MT5-MMP
MMPs constitute a multigenic family of 24 endopeptidases that belong to the metzincin superfamily of metalloproteinases. MMPs show pleiotropic regulatory actions in many processes in the nervous system, including axonal growth (Pastrana et al., 2006; Ould-yahoui et al., 2009; Trivedi et al., 2019), neurogenesis (Wojcik et al., 2009), learning and memory (Beroun et al., 2019), glial reactivity and inflammation (Chopra et al., 2019; Montaner et al., 2019; Muri et al., 2019), cell migration (Ould-Yahoui et al., 2013) or neuronal death (Gu et al., 2002; Jourquin et al., 2003). The many aspects of MMP functions and action mechanisms in the nervous system have been extensively discussed earlier (Rivera et al., 2010, 2019; Baranger et al., 2014).
MMPs are mostly secreted proteinases, but 6 transmembrane proteins form the so-called subfamily of MT-MMPs: MT1-MMP (MMP-14), MT2-MMP (MMP-15), MT3-MMP (MMP-16), MT4-MMP (MMP-17), MT5-MMP (MMP-24) and MT6-MMP (MMP-25). All MT-MMPs share closely related structural features, described in detail elsewhere (Itoh, 2015). These include a signal peptide that targets the MT-MMP to the endoplasmic reticulum, where it is proteolytically excised, and then a pro-domain bearing a conserved cysteine that interacts with the catalytic domain and maintains the enzyme as an inactive zymogen. The pro-peptide can be separated from the rest of the molecule by redox-mediated chemical reactions or after proteolytic cleavage by serine endoproteinase furin. This mechanism, called “cysteine switch,” implies the dissociation between the Cys residue of the pro-peptide and the Zn2+ of the catalytic domain, which leads to enzymatic activation. The catalytic domain is highly conserved domain across MT-MMPs (de facto, across MMPs) and is linked by a hinge region to the hemopexin domain, which has a more variable sequence, thus conferring certain specificity to the binding of substrates and endogenous TIMPs. MT-MMPs are linked to the plasma membrane either by a glycosylphosphatidyl inositol (GPI) bond (MT2-MMP and MT6-MMP) or by a transmembrane domain (MT1-, MT2-, MT3- and MT5-MMP), followed in this case by an intracytoplasmic domain that can control MT-MMP cell traffic and its proteolytic activity (Uekita et al., 2001; Wang et al., 2004; Sakamoto and Seiki, 2009). Analysis of the amino acid sequence reveals a variable percentage of sequence identity between human MT1- and MT5-MMP, ordered from N- to C-terminus as follows: pro-domain (44%), catalytic (72%), hinge (41%), hemopexin (66%), stem part (17%), transmembrane (37%) and intracytoplasmic (20%). Despite the high degree of sequence identity between MT1- and MT5-MMP, human and mouse MT5-MMP carry unique dibasic motifs in their stem regions, which are recognized by proteinases from the furin proprotein convertase family. Proteolytic cleavage by furin gives MT5-MMP singular biochemical properties, in particular an ephemeral presence on the plasma membrane since it is released into the medium in soluble form (Pei, 1999; Wang and Pei, 2001).
Physiological Functions of MT1- and MT5-MMP
MT1-MMP
MT1-MMP is ubiquitously distributed in the body. In the nervous system it is mainly expressed by microglia, astrocytes and neurons (Liao and Van Nostrand, 2010; Langenfurth et al., 2014; Py et al., 2014; Itoh, 2015). As most MMPs, MT1-MMP degrade extracellular matrix (ECM) proteins, in particular type I and III collagen, fibronectin, laminin, vitronectin and proteoglycans (Ohuchi et al., 1997). MT1-MMP catalyzes the conversion of pro-MMP-2 into the active MMP-2 form through a mechanism involving one of its endogenous inhibitors (see below) (Sato et al., 1996; Llano et al., 1999; Pei, 1999; Baranger et al., 2014). MT1-MMP is the most studied of the MT-MMPs and the progressive identification of new substrates beyond ECM has led to an expansion of the biological functions of the proteinase. MT1-MMP has an impact on inflammatory processes by modulating the action of cytokines, chemokines, proteinase inhibitors or receptors. For example, MT1-MMP can activate pro-TNF-α into active TNF-α, and inactivate CXCL12 (SDF1α), as well as complement protein C3, secretory leukocyte proteinase inhibitor (SLPI), IL-8 and other chemokines. Cellular receptors and membrane proteins such as DR6, the VEGF receptor neuropilin, the prion protein or a ligand of Notch receptor Delta-like I, are also processed by MT1-MMP (Overall et al., 2004; Tam et al., 2004; Jin et al., 2011; Starr et al., 2012; Kojima et al., 2014; Itoh, 2015). MT1-MMP knockout mice are not viable beyond one month of age, suggesting an important role of this proteinase in development (Holmbeck et al., 1999). In this context, it is now well documented that MT1-MMP contributes to cell migration including monocytes/macrophages, endothelial cells or neural stem cells (Galvez et al., 2001; Matias-Roman et al., 2005; Ould-Yahoui et al., 2013). It is noteworthy that in some cases the stimulation of migration or chemotaxis by MT1-MMP does not depend on its catalytic activity, but rather on the properties of its cytoplasmic tail. Thus, proteolytic inhibition of MT1-MMP does not affect migration of isolated macrophages on matrigel invasion tests. In addition, the expression of catalytically active and inactive forms of MT1-MMP in MT1-MMP knockout macrophages rescue chemotaxis properties, while a mutant form lacking the cytoplasmic tail does not (Sakamoto and Seiki, 2009). Recently, Aguirre and collaborators elegantly demonstrated that MT1-MMP is crucial in the inflammatory response upon intraperitoneal LPS injection in wild type and MT1-MMP knockout pups (5–8 days post-natal). Under these experimental conditions, they observed that the absence of MT1-MMP reduces life span, which is associated with lung enlargement and higher neutrophil recruitment (Aguirre et al., 2017). The effects of MT1-MMP appear to be related to MMP-2 activation, as lower concentrations of active MMP-2 are found in MT1-MMP knockout pups, associated with higher levels of MMP-2 substrate S100A9 (Aguirre et al., 2017). The latter is an inflammatory protein (Vogl et al., 2012) also implicated in AD pathogenesis (Chang et al., 2012). Taken together, these results clearly reflect the multifaceted mode of action of MT1-MMP, with functions that depend on proteolysis, but also on protein-protein interactions mediated by the C-terminal intracytoplasmic tail. It remains to be demonstrated whether this functional diversity is limited to certain cell types or organs, in particular with regard to the role of MT1-MMP in the nervous system.
MT5-MMP
To date, fewer substrates have been described for MT5-MMP, compared to MT1-MMP (reviewed in Itoh, 2015). MT5-MMP preferentially cleaves proteoglycans and to a lesser extent fibronectin, but not type I collagen or laminin (Wang et al., 1999). MT5-MMP expression is mainly confined to the nervous system, with levels in rodents reaching their maximum before birth and remaining high into adulthood in areas like the hippocampus or the cerebellum (Pei, 1999; Jaworski, 2000; Hayashita-Kinoh et al., 2001; Sekine-Aizawa et al., 2001; Warren et al., 2012). Such a distribution is consistent with the proposed role of MT5-MMP in brain development and more broadly in neural plasticity. Similarly, MT5-MMP has been reported to promote axonal growth (Hayashita-Kinoh et al., 2001) and to control the activation of adult neural stem cells under physiological and regenerative conditions by a mechanism that requires the cleavage of N-cadherin (Porlan et al., 2014), one of the non-ECM substrates of MT5-MMP. Despite the presumed developmental role of MT5-MMP, knockout mice are viable and have no apparent phenotypes, indicating that there is functional redundancy with other proteinases in physiological conditions. However, phenotypes linked to MT5-MMP deficiency become evident when mice are subjected to stressful conditions. This is for instance the case after sciatic nerve injury, where MT5-MMP deletion prevents aberrant sprouting of nociceptive Aβ-fibers and the resulting mechanical allodynia (Komori et al., 2004). The implication of MT5-MMP in post-lesion axonal regeneration has also been suggested in the context of concerted work with ADAM10 in reactive astrocytes to control post-lesion synaptic remodeling (Warren et al., 2012). We will see later that the combined action of MT5-MMP and ADAM10 has also been proposed in AD, with different functional implications. Deletion of MT5-MMP also revealed a reduction in hyperalgesia after intraplantar injections of IL-1β or TNF-α in a model of thermal pain, indicating that MT5-MMP plays role in the inflammatory pathways driven by these cytokines. The proposed mechanism associates the absence of MT5-MMP with deficient cleavage of its substrate N-cadherin, a cell adhesion molecule involved in synapse architecture. N-cadherin cleavage is required to ensure communication between mast cells and sensory neurons for the transmission of nociceptive stimuli (Folgueras et al., 2009). It is therefore concluded that MT5-MMP is capable of regulating the neuroinflammatory process (Baranger et al., 2016a).
The implication of MT5-MMP in synaptic processes has been suggested by the discovery that the proteinase interacts with the AMPA binding protein and the glutamate receptor-interacting protein. Both are postsynaptic density-95/Discs large/zona occludens-1 (PDZ) protein containing domains. The interactions between these PDZ proteins and MT5-MMP are mediated by the last three carboxyl terminus amino acids of the proteinase, thus facilitating its targeting to synapses and the cleavage of N-cadherin (Monea et al., 2006). In addition, it has been reported that MT5-MMP could act in synergy with γ-secretase to affect synaptic protein levels of GluA2 and PSD95, eventually resulting in a decrease in synaptic transmission (Restituito et al., 2011); however, functional evidence of such interaction is still lacking.
MT5-MMP processing and metabolism are tightly controlled; before reaching the plasma membrane, MT5-MMP can be cleaved by a furin-like activity in its stem part, leading to generation of a catalytically active soluble form of the enzyme that lacks the C-terminal part (Wang and Pei, 2001). Once on the membrane, MT5-MMP can be internalized in the endosomes, where it interacts with the PDZ Mint3 protein, allowing proteinase to be recycled into the membrane (Wang et al., 2004). Despite having the structure of a membrane protein, MT5-MMP can mostly be found in intracellular and extracellular compartments due to its shedding and cellular routing.
TIMPs
As with the other members of the MMP family, TIMPs control the catalytic activities of MT1- and MT5-MMP. Of the four TIMPs, TIMP-2, -3 and -4 inhibit MT1-MMP, while MT5-MMP is only targeted by TIMP-2. Both MT-MMPs can activate MMP-2 through a process that requires the formation of a tripartite molecular complex formed by two MT-MMPs, a TIMP-2 and a pro-MMP-2; the N-terminal domain of TIMP-2 interacts with MT1-MMP on the plasma membrane, while its C-terminal domain binds to the hemopexin domain of pro-MMP-2, then the pro-peptide of pro-MMP-2 is readily removed by an adjacent MT1-MMP eventually resulting in MMP-2 activation (Strongin et al., 1995). It should be noted that TIMP-1, which targets soluble MMPs and ADAM10, is a very poor inhibitor of MT-MMPs (reviewed in Baranger et al., 2014).
MT1-MMP and MT5-MMP Contribute to Alzheimer’s Pathogenesis
MT1-MMP
Higashi and Miyazaki were the first to show that MT1-MMP could cleave APP between residues Asn579 and Met580 (VLAN579-M580ISEPR) of APP770 after being activated by concanavalin A in the HT1080 fibrosarcoma cell line (Higashi and Miyazaki, 2003b). Three years later, Ahmad and collaborators showed that MT1-, MT3-, and MT5-MMP, but not MT2-, MT4- and MT6-MMP were able to cleave APP770 when co-expressed in HEK293 cells (Ahmad et al., 2006). Mass spectrometry analysis for MT3-MMP cleavage-sites revealed the same VLAN579-M580ISEPR site previously identified for MT1-MMP and 3 other cleavage sites at Ala463-Met464, His622-Ser623 and His685-Gln686 (Higashi and Miyazaki, 2003b), the latter being located in the Aβ sequence, just upstream of the α-cleavage site (Figure 1). Despite this, Aβ levels remained stable in HEK293 cells expressing MT3-MMP, probably indicating that MT3-MMP processes APP in cell compartments that do not influence amyloidogenesis. Although the APP cleavage sites for MT1-MMP and MT5-MMP were not specifically identified in the Ahmad study, the analysis of the APP degradation profile by western blot suggested that both MT-MMPs should cleave APP at the same location as MT3-MMP (Ahmad et al., 2006). Unlike MT3-MMP, the functional interaction between MT1-MMP and Aβ was shown when MT1-MMP overexpressed in COS cells degraded exogenous Aβ. In addition, the recombinant catalytic domain of MT1-MMP could degrade amyloid plaques when incubated ex vivo on brain slices of transgenic AD Tg2576 mice (Liao and Van Nostrand, 2010). In this way, MT1-MMP joins the list of metalloproteinases with Aβ-degrading, activity, such as MMP-2, MMP-3, MMP-7 and MMP-9 (Roher et al., 1994; Backstrom et al., 1996; Deb et al., 2003; White et al., 2006; Yan et al., 2006; Yin et al., 2006; Hernandez-Guillamon et al., 2010, 2015; Liao and Van Nostrand, 2010; Py et al., 2014; Brkic et al., 2015; Taniguchi et al., 2017; Rivera et al., 2019) (Figure 1). In complement of these results, we have recently reported that MT1-MMP can also prominently increase Aβ levels in the HEK293 cell model of amyloidogenesis, which stably expresses APP with the familial Swedish mutation (HEKswe) (Paumier et al., 2019). Transiently expressed MT1-MMP in HEKswe interacts with APP and induces the release of a soluble APP fragment of ∼95 kDa (sAPP95), distinct of sAPPα or sAPPβ generated by α- and β-secretase, respectively. The complementary transmembrane fragment of ∼30 kDa (CTF-30) and C99 are also dramatically increased and their levels highly correlated, suggesting that CTF-30 may be the precursor of C99 (Paumier et al., 2019). It is also noteworthy that MT1-MMP content is strongly upregulated in the cortex and hippocampus of 6-month old 5xFAD mice (Py et al., 2014). This mouse model mimics the symptoms and pathology of AD, including exacerbated amyloidosis, neuroinflammation and synaptotoxicity (Oakley et al., 2006), deficits in LTP (Crouzin et al., 2013) and cognition (Girard et al., 2013, 2014; Giannoni et al., 2016; Baranger et al., 2017a, b), and eventually neuronal death (Oakley et al., 2006; Jawhar et al., 2012; Eimer and Vassar, 2013). MT1-MMP upregulation in 5xFAD mice timely and spatially correlates with the increase in C99 levels (Py et al., 2014), thus providing indirect in vivo support for the pro-amyloidogenic features of this MMP described in HEKswe cells (Paumier et al., 2019).
Although functional interactions between MT1-MMP and APP/Aβ are not fully understood, it has been shown that the release of sAPP95 by MT1-MMP does not involve BACE-1, but it may involve MMP-2. It is estimated that about 50% of the sAPP95 released after MT1-MMP expression in HEKswe cells results from concerted action with MMP-2, MT1-MMP being in any case the limiting factor in this enzymatic tandem (Paumier et al., 2019). A plausible scenario would be that activation of MMP-2 by MT1-MMP (Sato et al., 1996) allows MMP-2 access to membrane substrates (i.e., APP), which would not otherwise be accessible for this soluble MMP. MMP-2, mainly considered as a soluble Aβ-degrading proteinase (Yin et al., 2006), could thus extend its range of action to the pericellular environment thanks to membrane docking provided by MT1-MMP. Another interesting feature linking MT1-MMP, MMP-2 and APP is that MT1-MMP processing of APP releases a soluble APP fragment (likely sAPP95) lacking a potent inhibitor of MMP-2 activity (Miyazaki et al., 1993). In this study, purified soluble forms of APP could efficiently inhibit MMP-2 degradation of gelatin as well as Aβ cleavage between K16-L17 (α-cleaving site) (Miyazaki et al., 1993). Years later, the authors identified a decapeptide sequence ISYGNDALMP, called APP-derived peptide inhibitor (APP-IP), located 6 amino acids downstream the cleavage site (N504-M505ISEPRISYGNDALMP) shared by MT1-MMP and MT3-MMP (and also MT5-MMP, see below). APP-IP shows low nanomolar range potency toward MMP-2 (IC50 value of 30 nM) and is a weaker inhibitor for MT1-MMP, MMP-3, -7 and -9, with IC50 values between 2 and 10 μM. Although sAPPα contains the decapeptide, it appears to be a weaker MMP-2 inhibitor than APP-IP (Higashi and Miyazaki, 2003a,b, 2008).
The mechanism by which MT1-MMP could promote amyloidogenesis is still elusive. Nevertheless, reminiscent of legumain, MT1-MMP cleavage upstream of the β-cleavage site could facilitate β-secretase cleavage of APP and thus the subsequent production of C99 and Aβ. Not exclusively, MT1-MMP could favor amyloidogenic β-secretase processing by promoting APP sorting into BACE-1-enriched early endosomes (Paumier et al., 2019). Most interestingly, it has been proposed that MT1-MMP could also function as a surrogate β-secretase when BACE-1 is inhibited; this is supported by experiments in HEKswe cells where MT1-MMP is able to restore the basal levels of Aβ, almost abolished in the presence of BACE-1 inhibitor IV (or C3) (Paumier et al., 2019). It should be noted that a β-cleavage site for MT1-MMP has not yet been identified. MT1-MMP replacing BACE-1 to maintain physiological levels of Aβ could be relevant in the event of therapeutic inhibition of BACE-1 if it is assumed that reducing Aβ levels below a physiological threshold could have adverse consequences.
Overall, these data highlight potential new mechanisms in the control of APP processing involving functional interactions of α- and β-secretases with MT1-MMP and MMP-2. It is now time to develop in vivo experimental models where genetic (i.e., conditional MT1-MMP knockout mice in transgenic AD mouse) and/or pharmacological (i.e., neutralizing antibodies) manipulation of MT1-MMP activity should further confirm the pathophysiological relevance of the aforementioned in vitro data. This includes a functional assessment of the apparent dual activity of MT1-MMP as an amyloidolytic and amyloidogenic enzyme, as well as the in vivo validation of APP cleavage and the resulting NTFs and CTFs.
MT5-MMP
MT5-MMP was detected in neurons and around senile plaques in the brains of AD patients (Sekine-Aizawa et al., 2001) and it was later suggested to cleave APP, yielding a fragmentation profile similar to that of MT1- and MT3-MMP (Ahmad et al., 2006). More recently, two independent studies highlighted in the same time the involvement of MT5-MMP on APP processing and the subsequent functional consequences (Willem et al., 2015; Baranger et al., 2016b). Willem and collaborators identified by mass spectrometry the VLAN504-M505ISEPR (APP695 numbering) as a cleavage site for MT5-MMP on APP, which was named η-cleavage site (Willem et al., 2015). This site was previously identified for MT1-MMP (Higashi and Miyazaki, 2003b) and MT3-MMP (Ahmad et al., 2006). Intriguingly, the Willem’s study did not identify MT1-MMP as cleaving at this VLAN504-M505ISEPR/η-site and it was concluded that only MT5-MMP could act as a η-secretase in vivo (Willem et al., 2015). η-secretase cleavage of APP generates sAPPη and a residual transmembrane C-terminal APP fragment-η (η-CTF), which accumulates in dystrophic neurites around amyloid plaques in APP/PS1 mice. Using specific antibodies against the neoepitopes of the α-, β- and η-sites, it was observed that BACE-1 inhibition promoted the release of a peptide Aη-α (for amyloid eta-alpha) resulting from the combined action of MT5-MMP and α-secretase. Recombinant Aη-α was able to impair LTP when incubated on primary rat neuronal cultures. In addition, the inhibition of ADAM10 promoted the concerted action of MT5-MMP and BACE-1 to release a shorter peptide, Aη-β, unexpectedly innocuous (Willem et al., 2015). Counter-intuitively, ADAM10 is presented in this work as conveying neurotoxic actions by contributing to the generation of Aη-α together with MT5-MMP. Moreover, the fact that Aη-α levels are exacerbated upon BACE-1 inhibition may raise concerns of possible side effects of therapeutic anti-BACE-1 strategies. Aη-γ peptides, potentially generated by a combined action of MT5-MMP and γ-secretase, were not detected, probably revealing a lack of functional interaction between η- and γ-secretase as it is the case with BACE-1, ADAM10 and γ-secretase (Chen et al., 2015; Liu et al., 2019). Indeed, a recent study has shown that MT5-MMP does not co-purify in the high molecular weight complex formed by BACE-1 and γ-secretase. Replacing the transmembrane domain (TMD) of BACE-1 by that of MT5-MMP did not prevent BACE-1 complex with γ-secretase and Aβ production, suggesting that motifs other than TMD are involved in the formation of the BACE-1/γ-secretase interactions (Liu et al., 2019). Aη-α and Aη-β peptides were both detected in human cerebrospinal fluid (CSF) of healthy and AD patients, but no differences in their levels were observed between groups (Willem et al., 2015). Another study identified a ∼25 kDa η-CTF in CSF, but in this case the peptide content was higher in AD patients with a PSEN1 familial mutation, in patients with sporadic AD and in Down Syndrome individuals with Alzheimer’s type dementia, compared to age-matched non-demented controls, raising the possibility that η-CTF could be a new disease biomarker (Garcia-Ayllon et al., 2017).
By the same time, our group demonstrated that MT5-MMP induced the production of a soluble APP fragment of sAPP95 in HEKswe, concomitant with elevated levels of C99 and Aβ40 (Baranger et al., 2016b). Although the exact nature of sAPP95 produced upon MT1-MMP and MT5-MMP expression needs to be established, it likely corresponds to what was defined as sAPPη by others (Willem et al., 2015). The function of sAPP95/sAPPη is not yet known, but a recent study showed that it binds GABABR1a, which also acts as a receptor for sAPPα and sAPPβ, resulting in a reduced probability of presynaptic release (Rice et al., 2019). These findings link MT5-MMP (as well as α- and β-secretases) with the regulation of a major inhibitory neurotransmitter system coupled to Ca2+ and K+ channels, and expands the range of possible mechanisms by which APP proteolysis may influence changes in synaptic activity occurring at early stages of the disease. Gaining insight into the functional interplay between proteinases and neurotransmission will require precise assessment of the functional specificities of 3 APP NTFs generated by 3 different enzymes sharing a common receptor.
Similarly to MT1-MMP, we found that MT5-MMP could interact with APP and promote its endosomal sorting, thereby providing mechanistic support for MT5-MMP pro-amyloidogenic activity (Baranger et al., 2017a). Moreover, to evaluate the genuine role of MT5-MMP in vivo, we generated a bigenic mouse model by crossing 5xFAD mice (Oakley et al., 2006) and MT5-MMP–/– (MT5–/–) mice (Komori et al., 2004). Compared to 5xFAD, 5xFAD/MT5–/– mice showed significantly lower levels of sAPP95, validating our in vitro observations and APP being an in vivo physiological substrate of MT5-MMP (Baranger et al., 2016b). Most important, 5xFAD/MT5–/– mice showed strikingly lower levels of Aβ (soluble Aβ40 and Aβ42, oligomers and plaques) and C99 at prodromal-like stages of the pathology (Baranger et al., 2016b, 2017a). Moreover, decreased APP processing and amyloidogenesis upon MT5-MMP deficiency were concomitant with fewer reactive microglial cells and astrocytes around amyloid plaques, as well as reduced levels of IL-1β and TNF-α (Baranger et al., 2016b, 2017a). From a functional standpoint, the absence of MT5-MMP prevented deficits in LTP and spatial and working memory observed in 5xFAD mice (Baranger et al., 2016b, 2017a). Most interestingly, some of these beneficial effects were also observed at advanced stages of the pathology in 16-month-old 5xFAD/MT5–/– mice, which presented a better preservation of neuronal networks and synaptic integrity compared to age-matched 5xFAD control mice (Baranger et al., 2016b).
Mechanistically, it is tempting to speculate that MT5-MMP could share with MT1-MMP and legumain proteolytic actions by which cleavage of APP upstream of the β-cleavage site would facilitate the processing of APP by β-secretase, especially in endosomes. Two sets of findings support such hypothesis: (1) bigenic 5xFAD/legumain–/– and 5xFAD/MT5–/– mice both show significant reductions in Aβ burden and glial reactivity, as well as prevention of LTP and learning and memory deficits (Zhang et al., 2015; Baranger et al., 2016b, 2017a), reminiscent of observations made in bigenic 5xFAD/BACE-1–/– or 5xFAD/BACE-1+/– mice (Ohno et al., 2007; Kimura et al., 2010) and (2) MT5-MMP can be internalized in endosomes (Wang et al., 2004; Baranger et al., 2017a) and thus promote endosomal APP sorting (Baranger et al., 2017a). In such hypothetical model, the first cleavage by MT5-MMP at the η-site could have place on the membrane and be followed by joint internalization with the APP transmembrane breakdown product (CTF-30/η-CTF). It is noteworthy that MT5-MMP trafficking involves interactions between the last three residues at carboxyl terminus and Mint3, a protein that contains two type III PDZ domains (Wang et al., 2004). Remarkably, Mint3 can also function as an adaptor protein to promote APP export from the Golgi complex (Caster and Kahn, 2013). These observations highlight Mint3 as a potential molecular bridge between MT5-MMP and APP, which could facilitate their interactions in the context of traffic-based mechanisms for the control C99/Aβ production. This is consistent with the increasing importance granted to traffic regulation in the amyloidogenic process, but also as means of exchanging relevant biofactors between cells. In this vein, η-CTF was detected in extracellular vesicles released by N2a neuroblastoma cells expressing APP Swedish. The vesicles were uptaken by neurons, suggesting the possibility that η-CTF could function as a vector for intercellular pathogenic propagation (Laulagnier et al., 2018).
Secretase Inhibitors/Modulators Can Be Linked to AD
An indirect way to assess the physiological impact of the new APP processing enzymes is through the study of their endogenous inhibitors, assuming that the control of proteolytic activities is an important component of the final proteolytic balance. Such an evaluation is not an easy task because the inhibitory selectivity is not always completely elucidated, as is often the case with metalloproteinases, which have a highly conserved catalytic site. In general, the available literature on proteinase inhibitors and AD remains essentially correlative and the link between both is to date inconclusive or, at best, speculative. In this context, the levels of fetuin-A, an inhibitor of meprin-β, were found to be reduced in CSF of AD patients compared to healthy individuals (Puchades et al., 2003). In addition, a polymorphism in the promotor sequence of the fetuin-A encoding gene has been associated with late onset AD (LOAD) (Geroldi et al., 2005), and lower plasma levels of fetuin-A correlated with severe to mild cognitive impairment (Smith et al., 2011). Likewise, a polymorphism in the cystatin C gene (CST3) was significantly associated with LOAD patients, while lower levels of this legumain inhibitor were found in their plasma (Chuo et al., 2007). Other studies have shown that overexpression of cystatin C in brains of APP-transgenic mice reduces amyloid plaque formation (Kaeser et al., 2007). A potential protective role of cystatin C in AD could be related to its ability to bind soluble Aβ and thus prevent Aβ deposition (Mi et al., 2007). A clear link between high cholesterol levels and the pathogenesis of AD has not yet been confirmed, but patients taking statins, a cholesterol-lowering drug, are still less likely to develop the disease (Eckert et al., 2005; Fonseca et al., 2010; Wood et al., 2014). In this context, it has been shown that high cholesterol levels inhibit RHBDL4 activity, which has been correlated with an increase in the formation of Aβ (Paschkowsky et al., 2018). For TIMP-2, the main inhibitor of MT-MMPs, clinical data are rather inconsistent. For example, one study showed no change in plasma TIMP-2 concentrations in patients with AD compared to healthy individuals (Lorenzl et al., 2003b), while in the CSF, two independent studies showed no difference (Mroczko et al., 2014) or increased (Lorenzl et al., 2003a) TIMP-2 levels in AD patients. More recently, Duits and collaborators reported decreases in TIMP-2 (and also TIMP-1) in the CSF of AD patients with microbleeds (Duits et al., 2015). Pre-clinical work has reported TIMP-2 as a possible anti-aging factor as it promotes neuronal plasticity and hippocampal-dependent cognition in aging mice (Castellano et al., 2017). In this study, the authors used parabiosis experiments to demonstrate that plasma from young donor mice (3 months) enriched with TIMP-2 reversed cognitive deficits in elderly mice (18 months). The confirmation that TIMP-2 was responsible for the observed effects was obtained in experiments where the plasma of young TIMP-2 knockout mice failed to rescue the aged phenotype. It is noteworthy that the levels of many immune and trophic factors were upregulated in plasma from TIMP-2 knockout mice, indicating a possible regulatory role of TIMP-2 on molecules that influence the functional cross talk between the CNS and peripheral tissues. It is therefore tempting to speculate on TIMP-2 as a pleiotropic “rejuvenating” factor with anti-MMP activities and, as shown earlier, with anti-mitotic and neural cell-differentiation promoting effects independent of MMP-inhibitory activity (Perez-Martinez and Jaworski, 2005). The idea of a “global” anti-aging factor is particularly appealing because aging is by far the main risk factor for AD and other degenerative disorders of nervous and non-nervous tissues.
Overall, the available partial data on proteinase inhibitors highlight the need for more functional studies. These should confirm their potential in modulating proteolytic balance or non-proteinase-dependent actions as a means of deciphering new AD pathophysiological pathways and developing new therapeutic strategies.
Concluding Remarks and Future Directions
The importance of APP proteolysis in AD mechanisms is not in doubt. The emergence of new APP cleaving enzymes significantly broadens the scope of study and makes evident the need for a comprehensive understanding of the complex molecular processes that encompasses APP proteolysis and the subsequent range of beneficial/pathological effects. Gaining insight in such processes will depend on our ability to determine the nature and functions of new APP fragments generated by these enzymes in a given cell type, subcellular compartment or stage of the disease, in particular at early stages where they could affect neurotransmitter release, synaptic function or incipient neuroinflammation. APP fragments can also be a new source of inspiration for developing therapeutic approaches based on blocking or promoting their formation. This may intrinsically require specific modulation of a given proteolytic cleavage, which turns out to be extremely difficult when the targeted proteinase has many substrates. This is the case for BACE-1 or γ-secretase, with dozens of substrates identified to date (Haapasalo and Kovacs, 2011; Kuhn et al., 2012; Zhou et al., 2012; Vassar et al., 2014). The list of substrates for most of the emerging proteinases discussed here is still relatively short and could therefore offer a therapeutic advantage in this respect. Time will tell whether the number of substrates will increase as new studies are conducted. Alternatively, APP proteolytic fragments may become full-fledged targets and their functions inhibited or potentiated, thus making it unnecessary to specifically inhibit the proteolytic processes that produce them. This option logically relies on a thorough knowledge of the biological functions of these fragments but also of the structural APP features. An additional alternative to modulating proteinase catalytic activity is to gain insight into the trafficking of APP and its metabolites. Mounting evidence indicates that the way these molecules circulate in the cell (or between cells) affects their functions and their availability for proteolytic processing. Similarly, understanding the cellular routing of proteinases may also be crucial to better target them. Beyond proteolytic activity, proteinases can also exert non-proteolytic actions through domains of interaction with other proteins; the question arises of the importance that these phenomena could have in the pathogenesis of AD and the new avenues they open to targeting domains other than the catalytic domain. This idea works as a mirror when it comes to the implications of proteinase inhibitors (e.g., TIMPs), which are known to control cell cycle, cell migration or cell fate also independent of anti-proteinase activities (Baranger et al., 2014). Since inhibitors are usually circulating proteins, they could favor communication between the CNS and peripheral systems (e.g., cardiovascular and immune systems), a process on which brain function and aging may depend.
In summary, this review highlights APP as a central molecule in the pathophysiology of AD and as a common substrate for MT-MMPs and other emerging proteinases. However, in the context of a more holistic approach to the disease that goes beyond APP/Aβ, we anticipate that in the coming years proteolytic and non-proteolytic interaction models will be established between proteinases, inhibitors and their partner proteins to continue to reveal the diversity of pathogenic pathways in AD and to learn how better control them.
Author Contributions
All the authors discussed, wrote and approved the manuscript. KB and SR supervised the writing of the manuscript.
Funding
This work was supported by funding from the CNRS and Aix-Marseille Université and by public grants to SR overseen by the French National Research Agency (ANR), MAD5 as part of the second “Investissements d’Avenir” program. The work was also supported by the DHUNE Centre of Excellence and grants from CoEN, “Fondation Plan Alzheimer,” France Alzheimer and Vaincre l’Alzheimer to SR. LG-G was granted by the ANR. DP was granted by the Excellence Initiative of Aix-Marseille Université – A∗MIDEX, a French “Investissements d’Avenir” program through the ICN Ph.D. program and NeuroSchool. KB was granted by the Excellence Initiative of Aix-Marseille Université – A∗MIDEX, a French “Investissements d’Avenir” program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
5xFAD, transgenic mouse model of AD bearing 3 familial mutations on human APP and 2 on PSEN1 genes; AD, Alzheimer’s disease; ADAM, a disintegrin and metalloproteinase; AICD, APP intracellular domain; AMPA, α-amino -3-hydroxy-5-methyl-4- isoxazolepropionic acid; APLP1/2, amyloid precursor like protein 1/2; APOE, apolipoprotein E; APP, amyloid-beta precursor protein; APP-IP, APP-derived inhibitor peptide; A β, amyloid-beta peptide; BACE-1, beta-site APP cleaving enzyme 1; C99/C83, APP-CTF of 99/83 amino acids; CSF, cerebrospinal fluid; CST3, cystatin C encoding gene; CXCL12, C-X -C motif chemokine ligand 12; DR6, death receptor 6; ECM, extracellular matrix; GABA, gamma-aminobutyric acid; GluA1/A2, glutamate A1/A2; GPI, glycosylphosphatidyl inositol; HEKswe, Human Embryonic Kidney cells 293 stably expressing APP with the familial Swedish mutation; IL-1 β, interleukin-1 beta; IL-8, interleukin-8; iPS, induced pluripotent stem cells; LOAD, late onset Alzheimer’s disease; LTP, long-term potentiation; mEPSCs, mini excitatory postsynaptic currents; mIPSCs, mini inhibitory postsynaptic currents; MMP, matrix metalloproteinase; MT-MMP, membrane-type matrix metalloproteinase; N/CTF, N-terminal or C-terminal APP fragments generated by APP-cleaving enzymes; NFT, neurofibrillary tangles; NMDA, N-methyl -D-aspartate; PARL, presenilin associated rhomboid like; PP2A, protein phosphatase 2; PS1/2, presenilin 1 and 2; PSD95, postsynaptic density protein 95; RHBDL4, rhomboid-like protein-4; sAPP α / β, soluble APP α / β; SDF1 α, stromal cell-derived factor 1; SLPI, secretory leukocyte proteinase inhibitor; TIMP, tissue inhibitor of MMPs; TMD, transmembrane domain; TNF- α, tumor necrosis factor alpha; VEGF, vascular endothelial growth factor.
References
Abramov, E., Dolev, I., Fogel, H., Ciccotosto, G. D., Ruff, E., and Slutsky, I. (2009). Amyloid-beta as a positive endogenous regulator of release probability at hippocampal synapses. Nat. Neurosci. 12, 1567–1576. doi: 10.1038/nn.2433
Aguirre, A., Blazquez-Prieto, J., Amado-Rodriguez, L., Lopez-Alonso, I., Batalla-Solis, E., Gonzalez-Lopez, A., et al. (2017). Matrix metalloproteinase-14 triggers an anti-inflammatory proteolytic cascade in endotoxemia. J. Mol. Med. 95, 487–497. doi: 10.1007/s00109-017-1510-z
Ahmad, M., Takino, T., Miyamori, H., Yoshizaki, T., Furukawa, M., and Sato, H. (2006). Cleavage of amyloid-beta precursor protein (APP) by membrane-type matrix metalloproteinases. J. Biochem. 139, 517–526. doi: 10.1093/jb/mvj054
Alvarez-Fernandez, M., Barrett, A. J., Gerhartz, B., Dando, P. M., Ni, J., and Abrahamson, M. (1999). Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J. Biol. Chem. 274, 19195–19203.
Backstrom, J. R., Lim, G. P., Cullen, M. J., and Tokes, Z. A. (1996). Matrix metalloproteinase-9 (MMP-9) is synthesized in neurons of the human hippocampus and is capable of degrading the amyloid-beta peptide (1-40). J. Neurosci. 16, 7910–7919.
Baranger, K., Bonnet, A. E., Girard, S. D., Paumier, J. M., Garcia-Gonzalez, L., Elmanaa, W., et al. (2017a). MT5-MMP promotes Alzheimer’s pathogenesis in the frontal cortex of 5xFAD mice and APP trafficking in vitro. Front. Mol. Neurosci. 9:163. doi: 10.3389/fnmol.2016.00163
Baranger, K., Giannoni, P., Girard, S. D., Girot, S., Gaven, F., Stephan, D., et al. (2017b). Chronic treatments with a 5-HT4 receptor agonist decrease amyloid pathology in the entorhinal cortex and learning and memory deficits in the 5xFAD mouse model of Alzheimer’s disease. Neuropharmacology 126, 128–141. doi: 10.1016/j.neuropharm.2017.08.031
Baranger, K., Khrestchatisky, M., and Rivera, S. (2016a). MT5-MMP, just a new APP processing proteinase in Alzheimer’s disease? J. Neuroinflammation 13:167. doi: 10.1186/s12974-016-0633-4
Baranger, K., Marchalant, Y., Bonnet, A. E., Crouzin, N., Carrete, A., Paumier, J. M., et al. (2016b). MT5-MMP is a new pro-amyloidogenic proteinase that promotes amyloid pathology and cognitive decline in a transgenic mouse model of Alzheimer’s disease. Cell Mol. Life Sci 73, 217–236. doi: 10.1007/s00018-015-1992-1
Baranger, K., Rivera, S., Liechti, F. D., Grandgirard, D., Bigas, J., Seco, J., et al. (2014). Endogenous and synthetic MMP inhibitors in CNS physiopathology. Prog. Brain Res. 214, 313–351. doi: 10.1016/B978-0-444-63486-3.00014-1
Barao, S., Moechars, D., Lichtenthaler, S. F., and De Strooper, B. (2016). BACE1 physiological functions may limit its use as therapeutic target for Alzheimer’s disease. Trends Neurosci. 39, 158–169. doi: 10.1016/j.tins.2016.01.003
Basurto-Islas, G., Grundke-Iqbal, I., Tung, Y. C., Liu, F., and Iqbal, K. (2013). Activation of asparaginyl endopeptidase leads to Tau hyperphosphorylation in Alzheimer disease. J. Biol. Chem. 288, 17495–17507. doi: 10.1074/jbc.M112.446070
Basurto-Islas, G., Gu, J. H., Tung, Y. C., Liu, F., and Iqbal, K. (2018). Mechanism of Tau hyperphosphorylation involving lysosomal enzyme asparagine endopeptidase in a mouse model of brain ischemia. J. Alzheimers Dis. 63, 821–833. doi: 10.3233/JAD-170715
Becker-Pauly, C., and Pietrzik, C. U. (2016). The metalloprotease meprin beta is an alternative beta-secretase of APP. Front. Mol. Neurosci. 9:159. doi: 10.3389/fnmol.2016.00159
Benilova, I., Karran, E., and De Strooper, B. (2012). The toxic Abeta oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat. Neurosci. 15, 349–357. doi: 10.1038/nn.3028
Beroun, A., Mitra, S., Michaluk, P., Pijet, B., Stefaniuk, M., and Kaczmarek, L. (2019). MMPs in learning and memory and neuropsychiatric disorders. Cell Mol. Life. Sci. 76, 3207–3228. doi: 10.1007/s00018-019-03180-8
Bertrand, E., Brouillet, E., Caille, I., Bouillot, C., Cole, G. M., Prochiantz, A., et al. (2001). A short cytoplasmic domain of the amyloid precursor protein induces apoptosis in vitro and in vivo. Mol. Cell. Neurosci. 18, 503–511. doi: 10.1006/mcne.2001.1030
Bien, J., Jefferson, T., Causevic, M., Jumpertz, T., Munter, L., Multhaup, G., et al. (2012). The metalloprotease meprin beta generates amino terminal-truncated amyloid beta peptide species. J. Biol. Chem. 287, 33304–33313. doi: 10.1074/jbc.M112.395608
Bourgeois, A., Lauritzen, I., Lorivel, T., Bauer, C., Checler, F., and Pardossi-Piquard, R. (2018). Intraneuronal accumulation of C99 contributes to synaptic alterations, apathy-like behavior, and spatial learning deficits in 3xTgAD and 2xTgAD mice. Neurobiol. Aging 71, 21–31. doi: 10.1016/j.neurobiolaging.2018.06.038
Brkic, M., Balusu, S., Van Wonterghem, E., Gorle, N., Benilova, I., Kremer, A., et al. (2015). Amyloid beta oligomers disrupt blood-CSF barrier integrity by activating matrix metalloproteinases. J. Neurosci. 35, 12766–12778. doi: 10.1523/JNEUROSCI.0006-15.2015
Bukhari, H., Glotzbach, A., Kolbe, K., Leonhardt, G., Loosse, C., and Muller, T. (2017). Small things matter: implications of APP intracellular domain AICD nuclear signaling in the progression and pathogenesis of Alzheimer’s disease. Prog. Neurobiol. 156, 189–213. doi: 10.1016/j.pneurobio.2017.05.005
Campbell, W. A., Reed, M. L., Strahle, J., Wolfe, M. S., and Xia, W. (2003). Presenilin endoproteolysis mediated by an aspartyl protease activity pharmacologically distinct from gamma-secretase. J. Neurochem. 85, 1563–1574. doi: 10.1046/j.1471-4159.2003.01799.x
Castellano, J. M., Mosher, K. I., Abbey, R. J., McBride, A. A., James, M. L., Berdnik, D., et al. (2017). Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492. doi: 10.1038/nature22067
Caster, A. H., and Kahn, R. A. (2013). Recruitment of the Mint3 adaptor is necessary for export of the amyloid precursor protein (APP) from the Golgi complex. J. Biol. Chem. 288, 28567–28580. doi: 10.1074/jbc.M113.481101
Chang, K. A., Kim, H. J., and Suh, Y. H. (2012). The role of S100a9 in the pathogenesis of Alzheimer’s disease: the therapeutic effects of S100a9 knockdown or knockout. Neurodegener. Dis. 10, 27–29. doi: 10.1159/000333781
Chang, K. A., Kim, H. S., Ha, T. Y., Ha, J. W., Shin, K. Y., Jeong, Y. H., et al. (2006). Phosphorylation of amyloid precursor protein (APP) at Thr668 regulates the nuclear translocation of the APP intracellular domain and induces neurodegeneration. Mol. Cell. Biol. 26, 4327–4338. doi: 10.1128/MCB.02393-05
Chen, A. C., Kim, S., Shepardson, N., Patel, S., Hong, S., and Selkoe, D. J. (2015). Physical and functional interaction between the alpha- and gamma-secretases: a new model of regulated intramembrane proteolysis. J. Cell Biol. 211, 1157–1176. doi: 10.1083/jcb.201502001
Chen, J. M., Dando, P. M., Rawlings, N. D., Brown, M. A., Young, N. E., Stevens, R. A., et al. (1997). Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J. Biol. Chem. 272, 8090–8098.
Chopra, S., Overall, C. M., and Dufour, A. (2019). Matrix metalloproteinases in the CNS: interferons get nervous. Cell Mol. Life Sci. 76, 3083–3095. doi: 10.1007/s00018-019-03171-9
Chuo, L. J., Sheu, W. H., Pai, M. C., and Kuo, Y. M. (2007). Genotype and plasma concentration of cystatin C in patients with late-onset Alzheimer disease. Dement. Geriatr. Cogn. Disord. 23, 251–257. doi: 10.1159/000100021
Cline, E. N., Bicca, M. A., Viola, K. L., and Klein, W. L. (2018). The amyloid-beta oligomer hypothesis: beginning of the third decade. J. Alzheimers Dis. 64, S567–S610. doi: 10.3233/JAD-179941
Crouzin, N., Baranger, K., Cavalier, M., Marchalant, Y., Cohen-Solal, C., Roman, F. S., et al. (2013). Area-specific alterations of synaptic plasticity in the 5XFAD mouse model of Alzheimer’s disease: dissociation between somatosensory cortex and hippocampus. PLoS One 8:e74667. doi: 10.1371/journal.pone.0074667
De Strooper, B. (2010). Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol. Rev. 90, 465–494. doi: 10.1152/physrev.00023.2009
De Strooper, B., and Chavez Gutierrez, L. (2015). Learning by failing: ideas and concepts to tackle gamma-secretases in Alzheimer’s disease and beyond. Annu. Rev. Pharmacol. Toxicol. 55, 419–437. doi: 10.1146/annurev-pharmtox-010814-124309
Deb, S., Wenjun Zhang, J., and Gottschall, P. E. (2003). Beta-amyloid induces the production of active, matrix-degrading proteases in cultured rat astrocytes. Brain Res. 970, 205–213.
Dominy, S. S., Lynch, C., Ermini, F., Benedyk, M., Marczyk, A., Konradi, A., et al. (2019). Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 5:eaau3333. doi: 10.1126/sciadv.aau3333
Duits, F. H., Hernandez-Guillamon, M., Montaner, J., Goos, J. D., Montanola, A., Wattjes, M. P., et al. (2015). Matrix metalloproteinases in Alzheimer’s disease and concurrent cerebral microbleeds. J. Alzheimers Dis. 48, 711–720. doi: 10.3233/JAD-143186
Dusterhoft, S., Kunzel, U., and Freeman, M. (2017). Rhomboid proteases in human disease: mechanisms and future prospects. Biochim. Biophys. Acta Mol. Cell Res. 1864(11 Pt B), 2200–2209. doi: 10.1016/j.bbamcr.2017.04.016
Eckert, G. P., Wood, W. G., and Muller, W. E. (2005). Statins: drugs for Alzheimer’s disease? J. Neural Transm. 112, 1057–1071. doi: 10.1007/s00702-004-0273-1
Eimer, W. A., and Vassar, R. (2013). Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Abeta42 accumulation and Caspase-3 activation. Mol. Neurodegener. 8:2. doi: 10.1186/1750-1326-8-2
Fluhrer, R., Multhaup, G., Schlicksupp, A., Okochi, M., Takeda, M., Lammich, S., et al. (2003). Identification of a beta-secretase activity, which truncates amyloid beta-peptide after its presenilin-dependent generation. J. Biol. Chem. 278, 5531–5538. doi: 10.1074/jbc.M211485200
Folgueras, A. R., Valdes-Sanchez, T., Llano, E., Menendez, L., Baamonde, A., Denlinger, B. L., et al. (2009). Metalloproteinase MT5-MMP is an essential modulator of neuro-immune interactions in thermal pain stimulation. Proc. Natl. Acad. Sci. U.S.A. 106, 16451–16456. doi: 10.1073/pnas.0908507106
Fong, L. K., Yang, M. M., Dos Santos Chaves, R., Reyna, S. M., Langness, V. F., Woodruff, G., et al. (2018). Full-length amyloid precursor protein regulates lipoprotein metabolism and amyloid-beta clearance in human astrocytes. J. Biol. Chem. 293, 11341–11357. doi: 10.1074/jbc.RA117.000441
Fonseca, A. C., Resende, R., Oliveira, C. R., and Pereira, C. M. (2010). Cholesterol and statins in Alzheimer’s disease: current controversies. Exp. Neurol. 223, 282–293. doi: 10.1016/j.expneurol.2009.09.013
Galvez, B. G., Matias-Roman, S., Albar, J. P., Sanchez-Madrid, F., and Arroyo, A. G. (2001). Membrane type 1-matrix metalloproteinase is activated during migration of human endothelial cells and modulates endothelial motility and matrix remodeling. J. Biol. Chem. 276, 37491–37500. doi: 10.1074/jbc.M104094200
Gamblin, T. C., Chen, F., Zambrano, A., Abraha, A., Lagalwar, S., Guillozet, A. L., et al. (2003). Caspase cleavage of tau: linking amyloid and neurofibrillary tangles in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 100, 10032–10037. doi: 10.1073/pnas.1630428100
Garcia-Ayllon, M. S., Lopez-Font, I., Boix, C. P., Fortea, J., Sanchez-Valle, R., Lleo, A., et al. (2017). C-terminal fragments of the amyloid precursor protein in cerebrospinal fluid as potential biomarkers for Alzheimer disease. Sci. Rep. 7:2477. doi: 10.1038/s41598-017-02841-7
Geroldi, D., Minoretti, P., Bianchi, M., Di Vito, C., Reino, M., Bertona, M., et al. (2005). Genetic association of alpha2-Heremans-Schmid glycoprotein polymorphism with late-onset Alzheimer’s disease in Italians. Neurosci. Lett. 386, 176–178. doi: 10.1016/j.neulet.2005.06.014
Gervais, F. G., Xu, D., Robertson, G. S., Vaillancourt, J. P., Zhu, Y., Huang, J., et al. (1999). Involvement of caspases in proteolytic cleavage of Alzheimer’s amyloid-beta precursor protein and amyloidogenic A beta peptide formation. Cell 97, 395–406.
Giannoni, P., Arango-Lievano, M., Neves, I. D., Rousset, M. C., Baranger, K., Rivera, S., et al. (2016). Cerebrovascular pathology during the progression of experimental Alzheimer’s disease. Neurobiol. Dis. 88, 107–117. doi: 10.1016/j.nbd.2016.01.001
Girard, S. D., Baranger, K., Gauthier, C., Jacquet, M., Bernard, A., Escoffier, G., et al. (2013). Evidence for early cognitive impairment related to frontal cortex in the 5XFAD mouse model of Alzheimer’s disease. J. Alzheimers Dis. 33, 781–796. doi: 10.3233/JAD-2012-120982
Girard, S. D., Jacquet, M., Baranger, K., Migliorati, M., Escoffier, G., Bernard, A., et al. (2014). Onset of hippocampus-dependent memory impairments in 5XFAD transgenic mouse model of Alzheimer’s disease. Hippocampus 24, 762–772. doi: 10.1002/hipo.22267
Gu, Z., Kaul, M., Yan, B., Kridel, S. J., Cui, J., Strongin, A., et al. (2002). S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science 297, 1186–1190. doi: 10.1126/science.1073634
Haapasalo, A., and Kovacs, D. M. (2011). The many substrates of presenilin/gamma-secretase. J. Alzheimers Dis. 25, 3–28. doi: 10.3233/JAD-2011-101065
Haass, C., Kaether, C., Thinakaran, G., and Sisodia, S. (2012). Trafficking and proteolytic processing of APP. Cold Spring Harb. Perspect. Med. 2:a006270. doi: 10.1101/cshperspect.a006270
Hayashita-Kinoh, H., Kinoh, H., Okada, A., Komori, K., Itoh, Y., Chiba, T., et al. (2001). Membrane-type 5 matrix metalloproteinase is expressed in differentiated neurons and regulates axonal growth. Cell Growth Differ. 12, 573–580.
Hedrich, J., Lottaz, D., Meyer, K., Yiallouros, I., Jahnen-Dechent, W., Stocker, W., et al. (2010). Fetuin-A and cystatin C are endogenous inhibitors of human meprin metalloproteases. Biochemistry 49, 8599–8607. doi: 10.1021/bi1004238
Hernandez-Guillamon, M., Delgado, P., Ortega, L., Pares, M., Rosell, A., Garcia-Bonilla, L., et al. (2009). Neuronal TIMP-1 release accompanies astrocytic MMP-9 secretion and enhances astrocyte proliferation induced by beta-amyloid 25-35 fragment. J. Neurosci. Res. 87, 2115–2125. doi: 10.1002/jnr.22034
Hernandez-Guillamon, M., Mawhirt, S., Blais, S., Montaner, J., Neubert, T. A., Rostagno, A., et al. (2015). Sequential amyloid-beta degradation by the matrix metalloproteases MMP-2 and MMP-9. J. Biol. Chem. 290, 15078–15091. doi: 10.1074/jbc.M114.610931
Hernandez-Guillamon, M., Mawhirt, S., Fossati, S., Blais, S., Pares, M., Penalba, A., et al. (2010). Matrix metalloproteinase 2 (MMP-2) degrades soluble vasculotropic amyloid-beta E22Q and L34V mutants, delaying their toxicity for human brain microvascular endothelial cells. J. Biol. Chem. 285, 27144–27158. doi: 10.1074/jbc.M110.135228
Hick, M., Herrmann, U., Weyer, S. W., Mallm, J. P., Tschape, J. A., Borgers, M., et al. (2015). Acute function of secreted amyloid precursor protein fragment APPsalpha in synaptic plasticity. Acta Neuropathol. 129, 21–37. doi: 10.1007/s00401-014-1368-x
Higashi, S., and Miyazaki, K. (2003a). Identification of a region of beta-amyloid precursor protein essential for its gelatinase A inhibitory activity. J. Biol. Chem. 278, 14020–14028. doi: 10.1074/jbc.M212264200
Higashi, S., and Miyazaki, K. (2003b). Novel processing of beta-amyloid precursor protein catalyzed by membrane type 1 matrix metalloproteinase releases a fragment lacking the inhibitor domain against gelatinase A. Biochemistry 42, 6514–6526. doi: 10.1021/bi020643m
Higashi, S., and Miyazaki, K. (2008). Identification of amino acid residues of the matrix metalloproteinase-2 essential for its selective inhibition by beta-amyloid precursor protein-derived inhibitor. J. Biol. Chem. 283, 10068–10078. doi: 10.1074/jbc.M709509200
Hoe, H. S., Cooper, M. J., Burns, M. P., Lewis, P. A., van der Brug, M., Chakraborty, G., et al. (2007). The metalloprotease inhibitor TIMP-3 regulates amyloid precursor protein and apolipoprotein E receptor proteolysis. J. Neurosci. 27, 10895–10905. doi: 10.1523/JNEUROSCI.3135-07.2007
Holmbeck, K., Bianco, P., Caterina, J., Yamada, S., Kromer, M., Kuznetsov, S. A., et al. (1999). MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 99, 81–92.
Hong, W., Wang, Z., Liu, W., O’Malley, T. T., Jin, M., Willem, M., et al. (2018). Diffusible, highly bioactive oligomers represent a critical minority of soluble Abeta in Alzheimer’s disease brain. Acta Neuropathol. 136, 19–40. doi: 10.1007/s00401-018-1846-7
Huse, J. T., Liu, K., Pijak, D. S., Carlin, D., Lee, V. M., and Doms, R. W. (2002). Beta-secretase processing in the trans-Golgi network preferentially generates truncated amyloid species that accumulate in Alzheimer’s disease brain. J. Biol. Chem. 277, 16278–16284. doi: 10.1074/jbc.M111141200
Hyman, B. T., and Yuan, J. (2012). Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat. Rev. Neurosci. 13, 395–406. doi: 10.1038/nrn3228
Itoh, Y. (2015). Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 44-46, 207–223. doi: 10.1016/j.matbio.2015.03.004
Jawhar, S., Trawicka, A., Jenneckens, C., Bayer, T. A., and Wirths, O. (2012). Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Abeta aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 33, 196.e29–196.e140. doi: 10.1016/j.neurobiolaging.2010.05.027
Jaworski, D. M. (2000). Developmental regulation of membrane type-5 matrix metalloproteinase (MT5-MMP) expression in the rat nervous system. Brain Res. 860, 174–177.
Jefferson, T., Causevic, M., auf dem Keller, U., Schilling, O., Isbert, S., Geyer, R., et al. (2011). Metalloprotease meprin beta generates nontoxic N-terminal amyloid precursor protein fragments in vivo. J. Biol. Chem. 286, 27741–27750. doi: 10.1074/jbc.M111.252718
Jin, G., Zhang, F., Chan, K. M., Xavier Wong, H. L., Liu, B., Cheah, K. S., et al. (2011). MT1-MMP cleaves Dll1 to negatively regulate Notch signalling to maintain normal B-cell development. EMBO J. 30, 2281–2293. doi: 10.1038/emboj.2011.136
Jorissen, E., Prox, J., Bernreuther, C., Weber, S., Schwanbeck, R., Serneels, L., et al. (2010). The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J. Neurosci. 30, 4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010
Jourquin, J., Tremblay, E., Decanis, N., Charton, G., Hanessian, S., Chollet, A. M., et al. (2003). Neuronal activity-dependent increase of net matrix metalloproteinase activity is associated with MMP-9 neurotoxicity after kainate. Eur. J. Neurosci. 18, 1507–1517.
Kaeser, S. A., Herzig, M. C., Coomaraswamy, J., Kilger, E., Selenica, M. L., Winkler, D. T., et al. (2007). Cystatin C modulates cerebral beta-amyloidosis. Nat. Genet. 39, 1437–1439. doi: 10.1038/ng.2007.23
Kallop, D. Y., Meilandt, W. J., Gogineni, A., Easley-Neal, C., Wu, T., Jubb, A. M., et al. (2014). A death receptor 6-amyloid precursor protein pathway regulates synapse density in the mature CNS but does not contribute to Alzheimer’s disease-related pathophysiology in murine models. J. Neurosci. 34, 6425–6437. doi: 10.1523/JNEUROSCI.4963-13.2014
Karmilin, K., Schmitz, C., Kuske, M., Korschgen, H., Olf, M., Meyer, K., et al. (2019). Mammalian plasma fetuin-B is a selective inhibitor of ovastacin and meprin metalloproteinases. Sci. Rep. 9:546. doi: 10.1038/s41598-018-37024-5
Karran, E., Mercken, M., and De Strooper, B. (2011). The amyloid cascade hypothesis for Alzheimer’s disease: an appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 10, 698–712. doi: 10.1038/nrd3505
Kim, H. S., Kim, E. M., Lee, J. P., Park, C. H., Kim, S., Seo, J. H., et al. (2003). C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 17, 1951–1953. doi: 10.1096/fj.03-0106fje
Kim, S., Sato, Y., Mohan, P. S., Peterhoff, C., Pensalfini, A., Rigoglioso, A., et al. (2016). Evidence that the rab5 effector APPL1 mediates APP-betaCTF-induced dysfunction of endosomes in Down syndrome and Alzheimer’s disease. Mol. Psychiatry 21, 707–716. doi: 10.1038/mp.2015.97
Kimura, A., Hata, S., and Suzuki, T. (2016). Alternative selection of beta-site APP-cleaving enzyme 1 (BACE1) cleavage sites in amyloid beta-protein precursor (APP) harboring protective and pathogenic mutations within the abeta sequence. J. Biol. Chem. 291, 24041–24053. doi: 10.1074/jbc.M116.744722
Kimura, R., Devi, L., and Ohno, M. (2010). Partial reduction of BACE1 improves synaptic plasticity, recent and remote memories in Alzheimer’s disease transgenic mice. J. Neurochem. 113, 248–261. doi: 10.1111/j.1471-4159.2010.06608.x
Kojima, A., Konishi, M., and Akizawa, T. (2014). Prion fragment peptides are digested with membrane type matrix metalloproteinases and acquire enzyme resistance through Cu(2)(+)-binding. Biomolecules 4, 510–526. doi: 10.3390/biom4020510
Komori, K., Nonaka, T., Okada, A., Kinoh, H., Hayashita-Kinoh, H., Yoshida, N., et al. (2004). Absence of mechanical allodynia and Abeta-fiber sprouting after sciatic nerve injury in mice lacking membrane-type 5 matrix metalloproteinase. FEBS Lett. 557, 125–128.
Kuhn, P. H., Koroniak, K., Hogl, S., Colombo, A., Zeitschel, U., Willem, M., et al. (2012). Secretome protein enrichment identifies physiological BACE1 protease substrates in neurons. EMBO J. 31, 3157–3168. doi: 10.1038/emboj.2012.173
Kuhn, P. H., Wang, H., Dislich, B., Colombo, A., Zeitschel, U., Ellwart, J. W., et al. (2010). ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 29, 3020–3032. doi: 10.1038/emboj.2010.167
Lammich, S., Kojro, E., Postina, R., Gilbert, S., Pfeiffer, R., Jasionowski, M., et al. (1999). Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc. Natl. Acad. Sci. U.S.A. 96, 3922–3927. doi: 10.1073/pnas.96.7.3922
Langenfurth, A., Rinnenthal, J. L., Vinnakota, K., Prinz, V., Carlo, A. S., Stadelmann, C., et al. (2014). Membrane-type 1 metalloproteinase is upregulated in microglia/brain macrophages in neurodegenerative and neuroinflammatory diseases. J. Neurosci. Res. 92, 275–286. doi: 10.1002/jnr.23288
Laulagnier, K., Javalet, C., Hemming, F. J., Chivet, M., Lachenal, G., Blot, B., et al. (2018). Amyloid precursor protein products concentrate in a subset of exosomes specifically endocytosed by neurons. Cell Mol. Life Sci. 75, 757–773. doi: 10.1007/s00018-017-2664-0
Lauritzen, I., Pardossi-Piquard, R., Bauer, C., Brigham, E., Abraham, J. D., Ranaldi, S., et al. (2012). The beta-secretase-derived C-terminal fragment of betaAPP, C99, but not Abeta, is a key contributor to early intraneuronal lesions in triple-transgenic mouse hippocampus. J. Neurosci. 32, 16243–11655a. doi: 10.1523/JNEUROSCI.2775-12.2012
Lauritzen, I., Pardossi-Piquard, R., Bourgeois, A., Pagnotta, S., Biferi, M. G., Barkats, M., et al. (2016). Intraneuronal aggregation of the beta-CTF fragment of APP (C99) induces Abeta-independent lysosomal-autophagic pathology. Acta Neuropathol. 132, 257–276. doi: 10.1007/s00401-016-1577-6
LeBlanc, A., Liu, H., Goodyer, C., Bergeron, C., and Hammond, J. (1999). Caspase-6 role in apoptosis of human neurons, amyloidogenesis, and Alzheimer’s disease. J. Biol. Chem. 274, 23426–23436. doi: 10.1074/jbc.274.33.23426
Liao, M. C., and Van Nostrand, W. E. (2010). Degradation of soluble and fibrillar amyloid beta-protein by matrix metalloproteinase (MT1-MMP) in vitro. Biochemistry 49, 1127–1136. doi: 10.1021/bi901994d
Liebsch, F., Kulic, L., Teunissen, C., Shobo, A., Ulku, I., Engelschalt, V., et al. (2019). Abeta34 is a BACE1-derived degradation intermediate associated with amyloid clearance and Alzheimer’s disease progression. Nat. Commun. 10:2240. doi: 10.1038/s41467-019-10152-w
Liu, K., Doms, R. W., and Lee, V. M. (2002). Glu11 site cleavage and N-terminally truncated A beta production upon BACE overexpression. Biochemistry 41, 3128–3136. doi: 10.1021/bi015800g
Liu, L., Ding, L., Rovere, M., Wolfe, M. S., and Selkoe, D. J. (2019). A cellular complex of BACE1 and gamma-secretase sequentially generates Abeta from its full-length precursor. J. Cell Biol. 218, 644–663. doi: 10.1083/jcb.201806205
Llano, E., Pendas, A. M., Freije, J. P., Nakano, A., Knauper, V., Murphy, G., et al. (1999). Identification and characterization of human MT5-MMP, a new membrane-bound activator of progelatinase a overexpressed in brain tumors. Cancer Res. 59, 2570–2576
Lorenzl, S., Albers, D. S., LeWitt, P. A., Chirichigno, J. W., Hilgenberg, S. L., Cudkowicz, M. E., et al. (2003a). Tissue inhibitors of matrix metalloproteinases are elevated in cerebrospinal fluid of neurodegenerative diseases. J. Neurol. Sci. 207, 71–76.
Lorenzl, S., Albers, D. S., Relkin, N., Ngyuen, T., Hilgenberg, S. L., Chirichigno, J., et al. (2003b). Increased plasma levels of matrix metalloproteinase-9 in patients with Alzheimer’s disease. Neurochem. Int. 43, 191–196.
Lu, D. C., Rabizadeh, S., Chandra, S., Shayya, R. F., Ellerby, L. M., Ye, X., et al. (2000). A second cytotoxic proteolytic peptide derived from amyloid beta-protein precursor. Nat. Med. 6, 397–404. doi: 10.1038/74656
Lunde, N. N., Bosnjak, T., Solberg, R., and Johansen, H. T. (2019). Mammalian legumain - A lysosomal cysteine protease with extracellular functions? Biochimie doi: 10.1016/j.biochi.2019.06.002 [Epub ahead of print].
Madeira, A., Pommet, J. M., Prochiantz, A., and Allinquant, B. (2005). SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. FASEB J. 19, 1905–1907. doi: 10.1096/fj.05-3839fje
Masters, C. L., Bateman, R., Blennow, K., Rowe, C. C., Sperling, R. A., and Cummings, J. L. (2015). Alzheimer’s disease. Nat. Rev. Dis. Primers 1:15056. doi: 10.1038/nrdp.2015.56
Matias-Roman, S., Galvez, B. G., Genis, L., Yanez-Mo, M., de la Rosa, G., Sanchez-Mateos, P., et al. (2005). Membrane type 1-matrix metalloproteinase is involved in migration of human monocytes and is regulated through their interaction with fibronectin or endothelium. Blood 105, 3956–3964. doi: 10.1182/blood-2004-06-2382
Matsumoto, Y., Watanabe, S., Suh, Y. H., and Yamamoto, T. (2002). Effects of intrahippocampal CT105, a carboxyl terminal fragment of beta-amyloid precursor protein, alone/with inflammatory cytokines on working memory in rats. J. Neurochem. 82, 234–239. doi: 10.1046/j.1471-4159.2002.00944.x
Mattson, M. P., Cheng, B., Culwell, A. R., Esch, F. S., Lieberburg, I., and Rydel, R. E. (1993). Evidence for excitoprotective and intraneuronal calcium-regulating roles for secreted forms of the beta-amyloid precursor protein. Neuron 10, 243–254.
Mi, W., Pawlik, M., Sastre, M., Jung, S. S., Radvinsky, D. S., Klein, A. M., et al. (2007). Cystatin C inhibits amyloid-beta deposition in Alzheimer’s disease mouse models. Nat. Genet. 39, 1440–1442. doi: 10.1038/ng.2007.29
Miyazaki, K., Hasegawa, M., Funahashi, K., and Umeda, M. (1993). A metalloproteinase inhibitor domain in Alzheimer amyloid protein precursor. Nature 362, 839–841. doi: 10.1038/362839a0
Mockett, B. G., Guevremont, D., Elder, M. K., Parfitt, K. D., Peppercorn, K., Morrissey, J., et al. (2019). Glutamate receptor trafficking and protein synthesis mediate the facilitation of ltp by secreted amyloid precursor protein-alpha. J Neurosci. 39, 3188–3203. doi: 10.1523/JNEUROSCI.1826-18.2019
Moir, R. D., Lathe, R., and Tanzi, R. E. (2018). The antimicrobial protection hypothesis of Alzheimer’s disease. Alzheimers Dement 14, 1602–1614. doi: 10.1016/j.jalz.2018.06.3040
Monea, S., Jordan, B. A., Srivastava, S., DeSouza, S., and Ziff, E. B. (2006). Membrane localization of membrane type 5 matrix metalloproteinase by AMPA receptor binding protein and cleavage of cadherins. J. Neurosci. 26, 2300–2312. doi: 10.1523/JNEUROSCI.3521-05.2006
Montagna, E., Dorostkar, M. M., and Herms, J. (2017). The role of APP in structural spine plasticity. Front. Mol. Neurosci. 10:136. doi: 10.3389/fnmol.2017.00136
Montaner, J., Ramiro, L., Simats, A., Hernandez-Guillamon, M., Delgado, P., Bustamante, A., et al. (2019). Matrix metalloproteinases and ADAMs in stroke. Cell Mol. Life Sci. 76, 3117–3140. doi: 10.1007/s00018-019-03175-5
Morley, J. E., Farr, S. A., Banks, W. A., Johnson, S. N., Yamada, K. A., and Xu, L. (2010). A physiological role for amyloid-beta protein:enhancement of learning and memory. J. Alzheimers Dis. 19, 441–449. doi: 10.3233/JAD-2009-1230
Mroczko, B., Groblewska, M., Zboch, M., Kulczynska, A., Koper, O. M., Szmitkowski, M., et al. (2014). Concentrations of matrix metalloproteinases and their tissue inhibitors in the cerebrospinal fluid of patients with Alzheimer’s disease. J. Alzheimers Dis. 40, 351–357. doi: 10.3233/JAD-131634
Mucke, L., and Selkoe, D. J. (2012). Neurotoxicity of amyloid beta-protein: synaptic and network dysfunction. Cold Spring Harb. Perspect. Med. 2:a006338. doi: 10.1101/cshperspect.a006338
Muller, U. C., Deller, T., and Korte, M. (2017). Not just amyloid: physiological functions of the amyloid precursor protein family. Nat. Rev. Neurosci. 18, 281–298. doi: 10.1038/nrn.2017.29
Muri, L., Leppert, D., Grandgirard, D., and Leib, S. L. (2019). MMPs and ADAMs in neurological infectious diseases and multiple sclerosis. Cell Mol. Life Sci. 76, 3097–3116. doi: 10.1007/s00018-019-03174-6
Nalivaeva, N. N., Beckett, C., Belyaev, N. D., and Turner, A. J. (2012). Are amyloid-degrading enzymes viable therapeutic targets in Alzheimer’s disease? J. Neurochem. 120 (Suppl. 1), 167–185. doi: 10.1111/j.1471-4159.2011.07510.x
Neve, R. L., Boyce, F. M., McPhie, D. L., Greenan, J., and Oster-Granite, M. L. (1996). Transgenic mice expressing APP-C100 in the brain. Neurobiol. Aging 17, 191–203.
Nikolaev, A., McLaughlin, T., O’Leary, D. D., and Tessier-Lavigne, M. (2009). APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457, 981–989. doi: 10.1038/nature07767
Oakley, H., Cole, S. L., Logan, S., Maus, E., Shao, P., Craft, J., et al. (2006). Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006
Obregon, D., Hou, H., Deng, J., Giunta, B., Tian, J., Darlington, D., et al. (2012). Soluble amyloid precursor protein-alpha modulates beta-secretase activity and amyloid-beta generation. Nat. Commun. 3:777. doi: 10.1038/ncomms1781
Ogier, C., Creidy, R., Boucraut, J., Soloway, P. D., Khrestchatisky, M., and Rivera, S. (2005). Astrocyte reactivity to Fas activation is attenuated in TIMP-1 deficient mice, an in vitro study. BMC Neurosci. 6:68. doi: 10.1186/1471-2202-6-68
Ohno, M. (2016). Alzheimer’s therapy targeting the beta-secretase enzyme BACE1: benefits and potential limitations from the perspective of animal model studies. Brain Res. Bull. 126(Pt 2), 183–198. doi: 10.1016/j.brainresbull.2016.04.007
Ohno, M., Cole, S. L., Yasvoina, M., Zhao, J., Citron, M., Berry, R., et al. (2007). BACE1 gene deletion prevents neuron loss and memory deficits in 5XFAD APP/PS1 transgenic mice. Neurobiol. Dis. 26, 134–145. doi: 10.1016/j.nbd.2006.12.008.
Ohuchi, E., Imai, K., Fujii, Y., Sato, H., Seiki, M., and Okada, Y. (1997). Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 272, 2446–2451. doi: 10.1074/jbc.272.4.2446
Olsen, O., Kallop, D. Y., McLaughlin, T., Huntwork-Rodriguez, S., Wu, Z., Duggan, C. D., et al. (2014). Genetic analysis reveals that amyloid precursor protein and death receptor 6 function in the same pathway to control axonal pruning independent of beta-secretase. J. Neurosci. 34, 6438–6447. doi: 10.1523/JNEUROSCI.3522-13.2014
Oster-Granite, M. L., McPhie, D. L., Greenan, J., and Neve, R. L. (1996). Age-dependent neuronal and synaptic degeneration in mice transgenic for the C terminus of the amyloid precursor protein. J. Neurosci. 16, 6732–6741.
Ould-Yahoui, A., Sbai, O., Baranger, K., Bernard, A., Gueye, Y., Charrat, E., et al. (2013). Role of matrix metalloproteinases in migration and neurotrophic properties of nasal olfactory stem and ensheathing cells. Cell Transplant. 22, 993–1010. doi: 10.3727/096368912X657468
Ould-yahoui, A., Tremblay, E., Sbai, O., Ferhat, L., Bernard, A., Charrat, E., et al. (2009). A new role for TIMP-1 in modulating neurite outgrowth and morphology of cortical neurons. PLoS One 4:e8289. doi: 10.1371/journal.pone.0008289
Overall, C. M., Tam, E. M., Kappelhoff, R., Connor, A., Ewart, T., Morrison, C. J., et al. (2004). Protease degradomics: mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol. Chem. 385, 493–504. doi: 10.1515/BC.2004.058
Pagenstecher, A., Stalder, A. K., Kincaid, C. L., Shapiro, S. D., and Campbell, I. L. (1998). Differential expression of matrix metalloproteinase and tissue inhibitor of matrix metalloproteinase genes in the mouse central nervous system in normal and inflammatory states. Am. J. Pathol. 152, 729–741.
Panza, F., Lozupone, M., Logroscino, G., and Imbimbo, B. P. (2019). A critical appraisal of amyloid-beta-targeting therapies for Alzheimer disease. Nat Rev Neurol 15, 73–88. doi: 10.1038/s41582-018-0116-6
Pardossi-Piquard, R., and Checler, F. (2012). The physiology of the beta-amyloid precursor protein intracellular domain AICD. J. Neurochem. 120 (Suppl. 1), 109–124. doi: 10.1111/j.1471-4159.2011.07475.x.
Paschkowsky, S., Hamze, M., Oestereich, F., and Munter, L. M. (2016). Alternative processing of the amyloid precursor protein family by rhomboid protease RHBDL4. J. Biol. Chem. 291, 21903–21912. doi: 10.1074/jbc.M116.753582
Paschkowsky, S., Hsiao, J. M., Young, J. C., and Munter, L. M. (2019). The discovery of proteases and intramembrane proteolysis (1). Biochem. Cell Biol. 97, 265–269. doi: 10.1139/bcb-2018-0186
Paschkowsky, S., Recinto, S. J., Young, J. C., Bondar, A. N., and Munter, L. M. (2018). Membrane cholesterol as regulator of human rhomboid protease RHBDL4. J. Biol. Chem. 293, 15556–15568. doi: 10.1074/jbc.RA118.002640
Pastrana, E., Moreno-Flores, M. T., Gurzov, E. N., Avila, J., Wandosell, F., and Diaz-Nido, J. (2006). Genes associated with adult axon regeneration promoted by olfactory ensheathing cells: a new role for matrix metalloproteinase 2. J. Neurosci. 26, 5347–5359. doi: 10.1523/JNEUROSCI.1111-06.2006
Paumier, J. M., Py, N. A., Garcia-Gonzalez, L., Bernard, A., Stephan, D., Louis, L., et al. (2019). Proamyloidogenic effects of membrane type 1 matrix metalloproteinase involve MMP-2 and BACE-1 activities, and the modulation of APP trafficking. FASEB J. 33, 2910–2927. doi: 10.1096/fj.201801076R
Pei, D. (1999). Identification and characterization of the fifth membrane-type matrix metalloproteinase MT5-MMP. J. Biol. Chem. 274, 8925–8932.
Pellegrini, L., Passer, B. J., Tabaton, M., Ganjei, J. K., and D’Adamio, L. (1999). Alternative, non-secretase processing of Alzheimer’s beta-amyloid precursor protein during apoptosis by caspase-6 and -8. J. Biol. Chem. 274, 21011–21016. doi: 10.1074/jbc.274.30.21011
Pera, M., Alcolea, D., Sanchez-Valle, R., Guardia-Laguarta, C., Colom-Cadena, M., Badiola, N., et al. (2013). Distinct patterns of APP processing in the CNS in autosomal-dominant and sporadic Alzheimer disease. Acta Neuropathol. 125, 201–213. doi: 10.1007/s00401-012-1062-9
Perez-Martinez, L., and Jaworski, D. M. (2005). Tissue inhibitor of metalloproteinase-2 promotes neuronal differentiation by acting as an anti-mitogenic signal. J. Neurosci. 25, 4917–4929. doi: 10.1523/JNEUROSCI.5066-04.2005
Peters-Libeu, C., Campagna, J., Mitsumori, M., Poksay, K. S., Spilman, P., Sabogal, A., et al. (2015). sAbetaPPalpha is a Potent Endogenous Inhibitor of BACE1. J. Alzheimers Dis. 47, 545–555. doi: 10.3233/JAD-150282
Porlan, E., Marti-Prado, B., Morante-Redolat, J. M., Consiglio, A., Delgado, A. C., Kypta, R., et al. (2014). MT5-MMP regulates adult neural stem cell functional quiescence through the cleavage of N-cadherin. Nat. Cell Biol. 16, 629–638. doi: 10.1038/ncb2993
Postina, R., Schroeder, A., Dewachter, I., Bohl, J., Schmitt, U., Kojro, E., et al. (2004). A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J. Clin. Invest. 113, 1456–1464. doi: 10.1172/JCI20864
Puchades, M., Hansson, S. F., Nilsson, C. L., Andreasen, N., Blennow, K., and Davidsson, P. (2003). Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer’s disease. Brain Res. Mol. Brain Res. 118, 140–146.
Pulina, M., Hopkins, M., Haroutunian, V., Greengard, P., and Bustos, V. (2019). C99, not beta-amyloid, is associated with selective death of vulnerable neurons in Alzheimer’s disease. bioRxiv doi: 10.1101/527572
Puzzo, D., Piacentini, R., Fa, M., Gulisano, W., Li Puma, D. D., Staniszewski, A., et al. (2017). LTP and memory impairment caused by extracellular Abeta and Tau oligomers is APP-dependent. eLife 6:e26991. doi: 10.7554/eLife.26991
Puzzo, D., Privitera, L., Fa, M., Staniszewski, A., Hashimoto, G., Aziz, F., et al. (2011). Endogenous amyloid-beta is necessary for hippocampal synaptic plasticity and memory. Ann. Neurol. 69, 819–830. doi: 10.1002/ana.22313
Puzzo, D., Privitera, L., Leznik, E., Fa, M., Staniszewski, A., Palmeri, A., et al. (2008). Picomolar amyloid-beta positively modulates synaptic plasticity and memory in hippocampus. J. Neurosci. 28, 14537–14545. doi: 10.1523/JNEUROSCI.2692-08.2008
Puzzo, D., Privitera, L., and Palmeri, A. (2012). Hormetic effect of amyloid-beta peptide in synaptic plasticity and memory. Neurobiol. Aging 33, 1484.e15–1484e24. doi: 10.1016/j.neurobiolaging.2011.12.020
Py, N. A., Bonnet, A. E., Bernard, A., Marchalant, Y., Charrat, E., Checler, F., et al. (2014). Differential spatio-temporal regulation of MMPs in the 5xFAD mouse model of Alzheimer’s disease: evidence for a pro-amyloidogenic role of MT1-MMP. Front. Aging Neurosci. 6:247. doi: 10.3389/fnagi.2014.00247
Rajendran, L., and Annaert, W. (2012). Membrane trafficking pathways in Alzheimer’s disease. Traffic 13, 759–770. doi: 10.1111/j.1600-0854.2012.01332.x
Rapti, M., Atkinson, S. J., Lee, M. H., Trim, A., Moss, M., and Murphy, G. (2008). The isolated N-terminal domains of TIMP-1 and TIMP-3 are insufficient for ADAM10 inhibition. Biochem. J. 411, 433–439. doi: 10.1042/BJ20071430
Restituito, S., Khatri, L., Ninan, I., Mathews, P. M., Liu, X., Weinberg, R. J., et al. (2011). Synaptic autoregulation by metalloproteases and gamma-secretase. J. Neurosci. 31, 12083–12093. doi: 10.1523/JNEUROSCI.2513-11.2011
Rice, H. C., de Malmazet, D., Schreurs, A., Frere, S., Van Molle, I., Volkov, A. N., et al. (2019). Secreted amyloid-beta precursor protein functions as a GABABR1a ligand to modulate synaptic transmission. Science 363:eaao4827. doi: 10.1126/science.aao4827
Rissman, R. A., Poon, W. W., Blurton-Jones, M., Oddo, S., Torp, R., Vitek, M. P., et al. (2004). Caspase-cleavage of tau is an early event in Alzheimer disease tangle pathology. J. Clin. Invest. 114, 121–130. doi: 10.1172/JCI20640
Rivera, S., García-González, L., Khrestchatisky, M., and Baranger, K. (2019). Metalloproteinases and their tissue inhibitors in Alzheimer’s disease and other neurodegenerative disorders. Cell Mol. Life Sci. 76, 3167–3191. doi: 10.1007/s00018-019-03178-2
Rivera, S., Khrestchatisky, M., Kaczmarek, L., Rosenberg, G. A., and Jaworski, D. M. (2010). Metzincin proteases and their inhibitors: foes or friends in nervous system physiology? J. Neurosci. 30, 15337–15357. doi: 10.1523/JNEUROSCI.3467-10.2010
Rivera, S., Ogier, C., Jourquin, J., Timsit, S., Szklarczyk, A. W., Miller, K., et al. (2002). Gelatinase B and TIMP-1 are regulated in a cell- and time-dependent manner in association with neuronal death and glial reactivity after global forebrain ischemia. Eur. J. Neurosci. 15, 19–32.
Rivera, S., Tremblay, E., Timsit, S., Canals, O., Ben-Ari, Y., and Khrestchatisky, M. (1997). Tissue inhibitor of metalloproteinases-1 (TIMP-1) is differentially induced in neurons and astrocytes after seizures: evidence for developmental, immediate early gene, and lesion response. J. Neurosci. 17, 4223–4235.
Roher, A. E., Kasunic, T. C., Woods, A. S., Cotter, R. J., Ball, M. J., and Fridman, R. (1994). Proteolysis of A beta peptide from Alzheimer disease brain by gelatinase A. Biochem. Biophys. Res. Commun. 205, 1755–1761. doi: 10.1006/bbrc.1994.2872
Rose, C., Dorard, E., Audrain, M., Gorisse-Hussonnois, L., Cartier, N., Braudeau, J., et al. (2018). Transient increase in sAPPalpha secretion in response to Abeta1-42 oligomers: an attempt of neuronal self-defense? Neurobiol. Aging 61, 23–35. doi: 10.1016/j.neurobiolaging.2017.09.008
Sakamoto, T., and Seiki, M. (2009). Cytoplasmic tail of MT1-MMP regulates macrophage motility independently from its protease activity. Genes Cells 14, 617–626. doi: 10.1111/j.1365-2443.2009.01293.x
Sannerud, R., Esselens, C., Ejsmont, P., Mattera, R., Rochin, L., Tharkeshwar, A. K., et al. (2016). Restricted location of PSEN2/gamma-secretase determines substrate specificity and generates an intracellular Abeta pool. Cell 166, 193–208. doi: 10.1016/j.cell.2016.05.020
Saric, A., Brugge, L., Muller-Pompalla, D., Rysiok, T., Ousson, S., Permanne, B., et al. (2013). Development and characterization of a novel membrane assay for full-length BACE-1 at pH 6.0. J. Biomol. Screen 18, 277–285. doi: 10.1177/1087057112462237
Sato, H., Takino, T., Kinoshita, T., Imai, K., Okada, Y., Stetler Stevenson, W. G., et al. (1996). Cell surface binding and activation of gelatinase A induced by expression of membrane-type-1-matrix metalloproteinase (MT1-MMP). FEBS Lett. 385, 238–240.
Scharfenberg, F., Armbrust, F., Marengo, L., Pietrzik, C., and Becker-Pauly, C. (2019). Regulation of the alternative beta-secretase meprin beta by ADAM-mediated shedding. Cell Mol. Life Sci. 76, 3193–3206. doi: 10.1007/s00018-019-03179-1
Schlenzig, D., Cynis, H., Hartlage-Rubsamen, M., Zeitschel, U., Menge, K., Fothe, A., et al. (2018). Dipeptidyl-peptidase activity of meprin beta links N-truncation of Abeta with Glutaminyl Cyclase-Catalyzed pGlu-Abeta formation. J. Alzheimers Dis. 66, 359–375. doi: 10.3233/JAD-171183.
Schonherr, C., Bien, J., Isbert, S., Wichert, R., Prox, J., Altmeppen, H., et al. (2016). Generation of aggregation prone N-terminally truncated amyloid beta peptides by meprin beta depends on the sequence specificity at the cleavage site. Mol. Neurodegener. 11:19. doi: 10.1186/s13024-016-0084-5
Sekine-Aizawa, Y., Hama, E., Watanabe, K., Tsubuki, S., Kanai-Azuma, M., Kanai, Y., et al. (2001). Matrix metalloproteinase (MMP) system in brain: identification and characterization of brain-specific MMP highly expressed in cerebellum. Eur. J. Neurosci. 13, 935–948.
Selkoe, D. J., and Hardy, J. (2016). The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608. doi: 10.15252/emmm.201606210.
Shi, X. P., Tugusheva, K., Bruce, J. E., Lucka, A., Wu, G. X., Chen-Dodson, E., et al. (2003). Beta-secretase cleavage at amino acid residue 34 in the amyloid beta peptide is dependent upon gamma-secretase activity. J. Biol. Chem. 278, 21286–21294. doi: 10.1074/jbc.M209859200
Simons, M., de Strooper, B., Multhaup, G., Tienari, P. J., Dotti, C. G., and Beyreuther, K. (1996). Amyloidogenic processing of the human amyloid precursor protein in primary cultures of rat hippocampal neurons. J. Neurosci. 16, 899–908.
Smith, E. R., Nilforooshan, R., Weaving, G., and Tabet, N. (2011). Plasma fetuin-A is associated with the severity of cognitive impairment in mild-to-moderate Alzheimer’s disease. J. Alzheimers Dis. 24, 327–333. doi: 10.3233/JAD-2011-101872
Song, D. K., Won, M. H., Jung, J. S., Lee, J. C., Kang, T. C., Suh, H. W., et al. (1998). Behavioral and neuropathologic changes induced by central injection of carboxyl-terminal fragment of beta-amyloid precursor protein in mice. J. Neurochem. 71, 875–878. doi: 10.1046/j.1471-4159.1998.71020875.x
Soriano, S., Lu, D. C., Chandra, S., Pietrzik, C. U., and Koo, E. H. (2001). The amyloidogenic pathway of amyloid precursor protein (APP) is independent of its cleavage by caspases. J. Biol. Chem. 276, 29045–29050. doi: 10.1074/jbc.M102456200
Sosa, L. J., Caceres, A., Dupraz, S., Oksdath, M., Quiroga, S., and Lorenzo, A. (2017). The physiological role of the amyloid precursor protein as an adhesion molecule in the developing nervous system. J. Neurochem. 143, 11–29. doi: 10.1111/jnc.14122.
Soscia, S. J., Kirby, J. E., Washicosky, K. J., Tucker, S. M., Ingelsson, M., Hyman, B., et al. (2010). The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One 5:e9505. doi: 10.1371/journal.pone.0009505
Spilman, P. R., Corset, V., Gorostiza, O., Poksay, K. S., Galvan, V., Zhang, J., et al. (2016). Netrin-1 interrupts amyloid-beta amplification, increases sabetappalpha in vitro and in vivo, and improves cognition in a mouse model of Alzheimer’s disease. J. Alzheimers Dis. 52, 223–242. doi: 10.3233/JAD-151046
Starr, A. E., Dufour, A., Maier, J., and Overall, C. M. (2012). Biochemical analysis of matrix metalloproteinase activation of chemokines CCL15 and CCL23 and increased glycosaminoglycan binding of CCL16. J. Biol. Chem. 287, 5848–5860. doi: 10.1074/jbc.M111.314609
Strongin, A. Y., Collier, I., Bannikov, G., Marmer, B. L., Grant, G. A., and Goldberg, G. I. (1995). Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 270, 5331–5338. doi: 10.1074/jbc.270.10.5331
Sun, J., Carlson-Stevermer, J., Das, U., Shen, M., Delenclos, M., Snead, A. M., et al. (2019). CRISPR/Cas9 editing of APP C-terminus attenuates beta-cleavage and promotes alpha-cleavage. Nat. Commun. 10:53. doi: 10.1038/s41467-018-07971-8
Tam, E. M., Morrison, C. J., Wu, Y. I., Stack, M. S., and Overall, C. M. (2004). Membrane protease proteomics: Isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. U.S.A. 101, 6917–6922. doi: 10.1073/pnas.0305862101
Tan, V. T. Y., Mockett, B. G., Ohline, S. M., Parfitt, K. D., Wicky, H. E., Peppercorn, K., et al. (2018). Lentivirus-mediated expression of human secreted amyloid precursor protein-alpha prevents development of memory and plasticity deficits in a mouse model of Alzheimer’s disease. Mol. Brain 11:7. doi: 10.1186/s13041-018-0348-9
Taniguchi, M., Matsuura, K., Nakamura, R., Kojima, A., Konishi, M., and Akizawa, T. (2017). MMP-7 cleaves amyloid beta fragment peptides and copper ion inhibits the degradation. Biometals 30, 797–807. doi: 10.1007/s10534-017-0048-4
Tian, Y., Crump, C. J., and Li, Y. M. (2010). Dual role of alpha-secretase cleavage in the regulation of gamma-secretase activity for amyloid production. J. Biol. Chem. 285, 32549–32556. doi: 10.1074/jbc.M110.128439
Trivedi, A., Noble-Haeusslein, L. J., Levine, J. M., Santucci, A. D., Reeves, T. M., and Phillips, L. L. (2019). Matrix metalloproteinase signals following neurotrauma are right on cue. Cell Mol. Life Sci. 76, 3141–3156. doi: 10.1007/s00018-019-03176-4
Uekita, T., Itoh, Y., Yana, I., Ohno, H., and Seiki, M. (2001). Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. J. Cell Biol. 155, 1345–1356. doi: 10.1083/jcb.200108112
Van Acker, Z. P., Bretou, M., and Annaert, W. (2019). Endo-lysosomal dysregulations and late-onset Alzheimer’s disease: impact of genetic risk factors. Mol. Neurodegener. 14:20. doi: 10.1186/s13024-019-0323-7
Van Cauwenberghe, C., Van Broeckhoven, C., and Sleegers, K. (2016). The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet. Med. 18, 421–430. doi: 10.1038/gim.2015.117
van der Kant, R., and Goldstein, L. S. (2015). Cellular functions of the amyloid precursor protein from development to dementia. Dev. Cell 32, 502–515. doi: 10.1016/j.devcel.2015.01.022
Vassar, R., Bennett, B. D., Babu-Khan, S., Kahn, S., Mendiaz, E. A., Denis, P., et al. (1999). Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286, 735–741.
Vassar, R., Kuhn, P. H., Haass, C., Kennedy, M. E., Rajendran, L., Wong, P. C., et al. (2014). Function, therapeutic potential and cell biology of BACE proteases: current status and future prospects. J. Neurochem. 130, 4–28. doi: 10.1111/jnc.12715
Villa, J. P., Bertenshaw, G. P., Bylander, J. E., and Bond, J. S. (2003). Meprin proteolytic complexes at the cell surface and in extracellular spaces. Biochem. Soc. Symp. 70, 53–63.
Vogl, T., Gharibyan, A. L., and Morozova-Roche, L. A. (2012). Pro-inflammatory S100A8 and S100A9 proteins: self-assembly into multifunctional native and amyloid complexes. Int. J. Mol. Sci. 13, 2893–2917. doi: 10.3390/ijms13032893
Wang, P., Wang, X., and Pei, D. (2004). Mint-3 regulates the retrieval of the internalized membrane-type matrix metalloproteinase, MT5-MMP, to the plasma membrane by binding to its carboxyl end motif EWV. J. Biol. Chem. 279, 20461–20470. doi: 10.1074/jbc.M400264200
Wang, X., and Pei, D. (2001). Shedding of membrane type matrix metalloproteinase 5 by a furin-type convertase: a potential mechanism for down-regulation. J. Biol. Chem. 276, 35953–35960. doi: 10.1074/jbc.M103680200
Wang, X., Yi, J., Lei, J., and Pei, D. (1999). Expression, purification and characterization of recombinant mouse MT5-MMP protein products. FEBS Lett. 462, 261–266.
Wang, X., Zhou, X., Li, G., Zhang, Y., Wu, Y., and Song, W. (2017). Modifications and trafficking of app in the pathogenesis of Alzheimer’s disease. Front. Mol. Neurosci. 10:294. doi: 10.3389/fnmol.2017.00294
Wang, Z., Jackson, R. J., Hong, W., Taylor, W. M., Corbett, G. T., Moreno, A., et al. (2017). Human brain-derived Abeta oligomers bind to synapses and disrupt synaptic activity in a manner that requires APP. J. Neurosci. 37, 11947–11966. doi: 10.1523/JNEUROSCI.2009-17.2017
Warren, K. M., Reeves, T. M., and Phillips, L. L. (2012). MT5-MMP, ADAM-10, and N-cadherin act in concert to facilitate synapse reorganization after traumatic brain injury. J. Neurotrauma 29, 1922–1940. doi: 10.1089/neu.2012.2383
Weidemann, A., Paliga, K., Durrwang, U., Reinhard, F. B., Schuckert, O., Evin, G., et al. (1999). Proteolytic processing of the Alzheimer’s disease amyloid precursor protein within its cytoplasmic domain by caspase-like proteases. J. Biol. Chem. 274, 5823–5829. doi: 10.1074/jbc.274.9.5823
White, A. R., Du, T., Laughton, K. M., Volitakis, I., Sharples, R. A., Xilinas, M. E., et al. (2006). Degradation of the Alzheimer disease amyloid beta-peptide by metal-dependent up-regulation of metalloprotease activity. J. Biol. Chem. 281, 17670–17680. doi: 10.1074/jbc.M602487200
Wild, K., August, A., Pietrzik, C. U., and Kins, S. (2017). Structure and synaptic function of metal binding to the amyloid precursor protein and its proteolytic fragments. Front. Mol. Neurosci. 10:21. doi: 10.3389/fnmol.2017.00021
Willem, M., Tahirovic, S., Busche, M. A., Ovsepian, S. V., Chafai, M., Kootar, S., et al. (2015). eta-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 526, 443–447. doi: 10.1038/nature14864
Wojcik, L., Sawicka, A., Rivera, S., and Zalewska, T. (2009). Neurogenesis in gerbil hippocampus following brain ischemia: focus on the involvement of metalloproteinases. Acta Neurobiol. Exp. 69, 52–61.
Wood, W. G., Li, L., Muller, W. E., and Eckert, G. P. (2014). Cholesterol as a causative factor in Alzheimer’s disease: a debatable hypothesis. J. Neurochem. 129, 559–572. doi: 10.1111/jnc.12637
Yan, P., Hu, X., Song, H., Yin, K., Bateman, R. J., Cirrito, J. R., et al. (2006). Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. J. Biol. Chem. 281, 24566–24574. doi: 10.1074/jbc.M602440200
Yan, R. (2017). Physiological functions of the beta-site amyloid precursor protein cleaving enzyme 1 and 2. Front. Mol. Neurosci. 10:97. doi: 10.3389/fnmol.2017.00097
Yin, K. J., Cirrito, J. R., Yan, P., Hu, X., Xiao, Q., Pan, X., et al. (2006). Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J. Neurosci. 26, 10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006
Zhang, Z., Obianyo, O., Dall, E., Du, Y., Fu, H., Liu, X., et al. (2017). Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer’s disease. Nat. Commun. 8:14740. doi: 10.1038/ncomms14740
Zhang, Z., Song, M., Liu, X., Kang, S. S., Kwon, I. S., Duong, D. M., et al. (2014). Cleavage of tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer’s disease. Nat. Med. 20, 1254–1262. doi: 10.1038/nm.3700
Zhang, Z., Song, M., Liu, X., Su Kang, S., Duong, D. M., Seyfried, N. T., et al. (2015). Delta-secretase cleaves amyloid precursor protein and regulates the pathogenesis in Alzheimer’s disease. Nat. Commun. 6:8762. doi: 10.1038/ncomms9762
Keywords: amyloid precursor protein, matrix metalloproteinases, eta-secretase, meprin-beta, legumain, rhomboid-like protein-4, caspase, neurodegenerative disease
Citation: García-González L, Pilat D, Baranger K and Rivera S (2019) Emerging Alternative Proteinases in APP Metabolism and Alzheimer’s Disease Pathogenesis: A Focus on MT1-MMP and MT5-MMP. Front. Aging Neurosci. 11:244. doi: 10.3389/fnagi.2019.00244
Received: 30 June 2019; Accepted: 20 August 2019;
Published: 24 September 2019.
Edited by:
Elena Marcello, University of Milan, ItalyReviewed by:
Michael Willem, Ludwig Maximilian University of Munich, GermanyMarcia Regina Cominetti, Federal University of São Carlos, Brazil
Copyright © 2019 García-González, Pilat, Baranger and Rivera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kévin Baranger, kevin.baranger@univ-amu.fr; Santiago Rivera, santiago.rivera@univ-amu.fr
 Laura García-González
Laura García-González Dominika Pilat
Dominika Pilat Kévin Baranger
Kévin Baranger Santiago Rivera
Santiago Rivera