Do We Need a Human post mortem Whole-Brain Anatomical Ground Truth in in vivo Magnetic Resonance Imaging?
- Integrative Model-Based Cognitive Neuroscience Research Unit, University of Amsterdam, Amsterdam, Netherlands
Non-invasive in vivo neuroimaging techniques provide a wide array of possibilities to study human brain function. A number of approaches are available that improve our understanding of the anatomical location of brain activation patterns, including the development of probabilistic conversion tools to register individual in vivo data to population based neuroanatomical templates. Two elegant examples were published by Horn et al. (2017) in which a method was described to warp DBS electrode coordinates, and histological data to MNI-space (Ewert et al., 2017). The conversion of individual brain scans to a standard space is done assuming that individual anatomical scans provide a reliable image of the underlying neuroanatomy. It is unclear to what extent spatial distortions related to tissue properties, or MRI artifacts exist in these scans. Therefore, the question rises whether the anatomical information from the individual scans can be considered a real ground truth. To accommodate the knowledge-gap as a result of limited anatomical information, generative brain models have been developed circumventing these challenges through the application of assumption sets without recourse to any ground truth. We would like to argue that, although these efforts are valuable, the definition of an anatomical ground truth is preferred. Its definition requires a system in which non-invasive approaches can be validated using invasive methods of investigation. We argue that the application of post mortem MRI studies in combination with microscopy analyses brings an anatomical ground truth for the human brain within reach, which is of importance for all research within the human in vivo neuroimaging field.
Introduction
In recent years, within the cognitive neurosciences, functional magnetic resonance imaging (fMRI), measuring blood oxygen level dependent (BOLD) signal has become the tool of choice (Bandettini, 2009). The field of non-invasive neuroimaging techniques continues to progress, providing new opportunities with increased spatial and temporal resolution (Bandettini, 2009; De Martino et al., 2017). Registration to average brain templates such as the Montreal Neurological Institute (MNI-) template provide a platform for the creation of an average human brain, as well as an attractive framework in which data from separate studies can be combined, reused and reanalyzed, providing an important contribution to resolving of the reproducibility crisis (Ioannidis, 2005).
In parallel to the ongoing development of non-invasive imaging techniques, the number of clinical autopsies performed is decreasing world-wide (Burton and Underwood, 2007; Shojania and Burton, 2008; Gaensbacher et al., 2012; Blokker et al., 2016). Although these autopsies do not necessarily include the brain, it raises the question whether more classic neuroimaging techniques, in particular post mortem microscopy studies can, and should be replaced by modern non-invasive alternatives. Post mortem whole-brain atlases based on microscopic dissections, such as the atlases of Schaltenbrand and Wahren (1977), Talairach and Tournoux (1988), Mai et al. (2015), and BigBrain (Amunts et al., 2013) represent laborious investigations of small numbers or even single brains. Combined, these atlases provide information on fewer than 10 whole brains, a number that does not even start to compare to the number of MRI scans that are produced of individual human brains every single day. It is important to acknowledge efforts of several groups world-wide that perform comparisons between MRI with or without the combination with histological data on smaller tissue blocks (e.g., Bürgel et al., 1999; Castellanos et al., 2008; Makris et al., 2013; Adler et al., 2014; Annese et al., 2014; Augustinack et al., 2014; Plantinga et al., 2016). These studies are of great value for understanding of tissue contrast, but are not incorporated in atlasing efforts. Therefore, we conclude that efforts that could potentially provide a post mortem ground truth are not funneled back into in vivo neuroimaging studies. Confounding factors are an inherent part of human post mortem brain research, and include a bias toward old age, potential effects of pharmacological treatment, nutritional status, ante mortem disease state, cause of death, and post mortem factors such as the interval between death and fixation, as well as the known effects of the fixative on the shape and MR characteristics of the tissue (Chu et al., 2005; Schmierer et al., 2008; van Duijn et al., 2011; Stüber et al., 2014). Given these limitations and challenges, we would like to discuss the potential value for human post mortem brain research in the validation of state-of-the-art non-invasive functional neuroimaging techniques. Such studies could entail efforts to help to determine to what extent structural MRI scans reliably reflect the underlying neuroanatomy, and to what extent structure fits structure?
We would like to initiate the discussion on the value of the definition of a post mortem ground truth for in vivo functional neuroimaging studies bringing forward four arguments against, as well as four (related) arguments in favor of these efforts. The first two arguments and their counterarguments relate to inherent limitations of working with post mortem tissues. Argument 3 and 4 relate to unresolved issues that require additional experimenting and technical developments.
Argument 1
Post mortem studies by definition provide no information on brain function. Anatomical training represents a cornerstone of medical curriculum, and is historically performed on human cadavers. Today’s development in training includes a shift toward using 3D models of anatomical structures (Dirnhofer et al., 2006). Both digital and 3D printed models are of great value, and could gradually replace cadaver training. It can be argued that the lack of insight in the interindividual variability, and function when studying the post mortem brain are in sharp contrast with the large number of in vivo MRI images that are freely accessible through various data sharing platforms (e.g., NITRC and DataDryad). Studies on BOLD signal have the important advantage that they provide direct information on the activation of specific parts of the brain during specific tasks, allowing to study human brain function in a reliable and detailed fashion (Bandettini, 2009; De Martino et al., 2017).
Counter Argument 1
A multimodal approach is needed to understand brain function. BOLD MRI signal provides information on blood oxygenation building on the basic assumption that increased neuronal activity results in increased blood flow, and thereby increasing oxygenated hemoglobin, which can be detected using MRI. Like all hemodynamic-based technologies, BOLD MRI measures a surrogate signal, with limited spatial and temporal specificity due to inherent physical and biological limitations (Logothetis, 2008). Interestingly, information on the geometric and topographic characteristics of the vasculature that allow the size estimation of the area from which BOLD signals originates are now available (Turner, 2002). Linking localization of BOLD signal to detailed post mortem investigations of the anatomy of the vasculature allows further pinpointing of the anatomical location of the neuronal activity, thereby providing additional anatomical specificity.
Other techniques used to assess brain activity, including invasive micro-electrical recordings (MER), as well as computed tomography (CT) and positron emission tomography (PET) scanning all have their individual benefits, and face their own challenges and limitations. The combined value of individual techniques is recognized within the scientific community, and our as well as other research groups have argued for pipelines to integrate valuable information from complementing research approaches (Logothetis, 2008; Ewert et al., 2017; Forstmann et al., 2017; Horn et al., 2017).
Argument 2
Not all neuropathological alterations can be detected using post mortem investigations. The (potential) value of non-invasive neuroimaging methods for diagnostics is enormous. Dissection of specific brain areas using invasive autopsy procedures inevitably causes damage to brain structures located on the cutting plane. Thereby not allowing complete analyses of the brain. Additionally, it is feasible that fixation procedures can obscure the presence of neuropathological alterations.
Counter Argument 2
Not all Neuropathological Alterations can be detected using non-invasive neuroimaging. Both qualitative and quantitative diagnostics can greatly benefit from MRI techniques. Probable Alzheimer disease and Parkinson’s disease can be diagnosed in the clinic, and non-invasive imaging techniques play an important role (Jack et al., 2011; Heim et al., 2017). It is evident that we can visualize neuropathology, but at the same time it is generally accepted that not all abnormalities are visible on non-invasive scans, and misdiagnosis of neuropathological conditions using MRI scans can occur (Kloppel et al., 2008). A probable diagnosis of these neurodegenerative diseases, which fits the clinical definition, but can only be confirmed after death using histological approaches. Through the histological analyses, it also becomes clear which smaller pathological changes cannot be detected. This is of grave importance for diagnostic and research purposes in early stage disease, and for the study of disease progression.
Argument 3
Individual anatomical scans can provide an anatomical ground truth. Structural MRI scans acquired for fMRI studies are undergoing continuous development and signal to noise ratios have been shown to improve proportional to the applied field strength (Chandran et al., 2016). Additionally, development of novel contrasts tailored to the specific tissue properties of individual brain structures, such as their iron content, contribute to improved visualization of small subcortical nuclei for, e.g., the subthalamic nucleus (Ogg et al., 1999; Rauscher et al., 2005; Manova et al., 2009; Schafer et al., 2009; Vertinsky et al., 2009; Alkemade et al., 2017). At the same time progress is being made to improve white matter imaging and tractography (Maier-Hein et al., 2017). The impressive and constantly improving quality of in vivo MRI scans has the potential to increasingly replace post mortem investigations.
Counter Argument 3
The ground truth provided by individual anatomical scans requires testing. At present, we cannot exclude that MRI techniques may suffer from undefined spatial distortions in the living brain. Therefore, the reliability of the assumed ground truth provided by individual MRI images requires testing. It is unclear to what extent spatial distortions exist in anatomical scans, due to tissue or physiological properties, or MR artifacts. Therefore, the information provided by the individual scans can be considered a surrogate ground truth at best. Horn and other researchers have therefore developed methods to accommodate the knowledge-gap, and created generative brain models circumventing these challenges through the application of assumption sets without recourse to any ground truth or individual scan (Horn et al., 2017). Interestingly, this technique also allows the warping of histological data into MNI-space (Ewert et al., 2017). Even though these efforts are valuable, in our opinion closing of the knowledge-gap, and the definition of an anatomical ground truth without potential distortions is preferred. Definition of such a ground truth requires a system in which non-invasive approaches can be validated using invasive methods of investigation. This requires the application of post mortem MRI studies followed by histological validation of the images in the same tissue specimens, through which potential distortions can be assessed. Valuable progress is being made in this field, including the application of polarized light imaging, which allows detailed imaging of white matter and its directionality in microscopy analyses (Palm et al., 2010). Additionally, techniques are being developed for registration of results obtained in dissected tissue to MRI space (Zemmoura et al., 2014, 2016).
Argument 4
Post mortem MRI studies provide unchallenged spatial resolution and contrast. State of the art in vivo MRI studies provide a high level of anatomical detail, which will most likely continue to improve in the future. Today’s high quality images allow the parcellation of a number of subcortical brain nuclei, including the subthalamic nucleus (STN), which is a target for deep brain stimulation (DBS) surgery (Benabid et al., 2009). Using post mortem tissue specimens, we have been able to obtain a 60 μm isotropic resolution image of the STN using ultra-high field MRI scanning, in which all borders of the STN are clearly discernible, providing a higher level of anatomical detail than can be obtained in vivo. (Figure 1; Weiss et al., 2015). It can be argued that in vivo MRI scanning techniques will improve further, and motion correction will allow to compensate for movement artifacts, allowing for improved in vivo scan resolutions which will potentially approach the level of detail that can be obtained with post mortem MRI scans for subcortical nuclei as well as smaller fiber bundles.
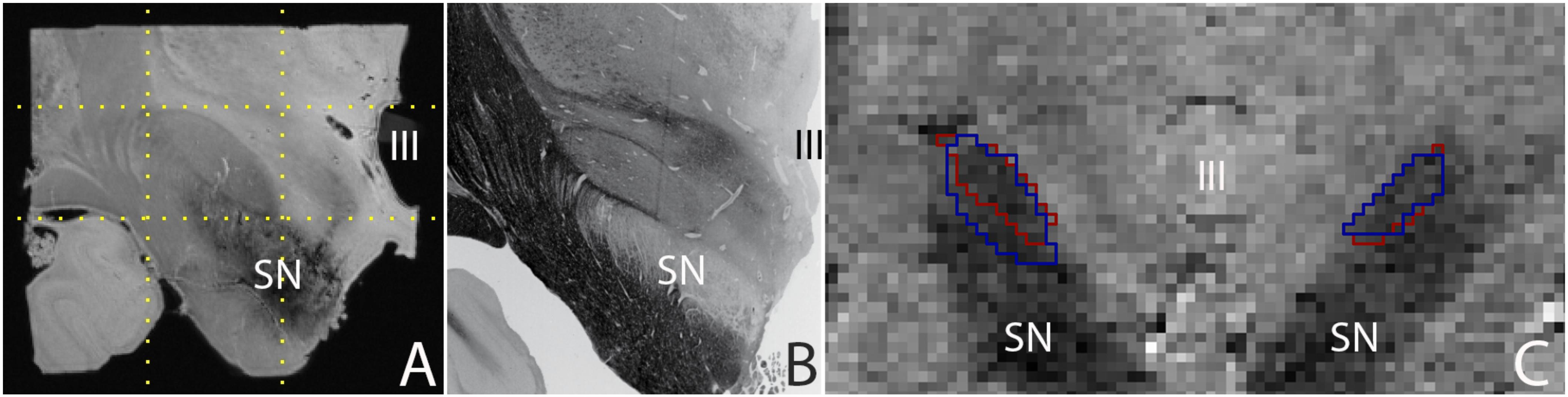
FIGURE 1. (A) 0.06 mm isotropic MRI image of a human post mortem subthalamic nucleus (one hemisphere). The STN is located within the square indicated by the dotted lines (adapted from Weiss et al., 2015), (B) Histological preparation of the STN (adapted from Mai et al., 2015), and (C) 0.8 mm isotropic MRI image of an in vivo subthalamic nucleus (left and right hemisphere). Outlines indicate the location of the structure as assessed by two independent raters (adapted from Alkemade et al., 2017). SN, substantia nigra; III, third ventricle.
Counter Argument 4
A majority of subcortical nuclei, as well as smaller fiber bundles still cannot be visualized using (post mortem) MRI. Indistinguishable brain structures include a subset of individual thalamic, and hypothalamic nuclei, among which a number of potential DBS targets. As a result, any (functional) MRI signal ascribed to these structures can only be based on anatomical orientation using landmarks. To what extent orientation based on anatomical landmarks is reliable, requires further confirmation using post mortem microscopy sectioning approaches, and cannot be achieved using a ground truth based on post mortem MRI. Additionally, given the high spatial resolution that can be obtained post mortem (see Figure 1), such an approach also has the potential to provide more detailed information on the interindividual variation by more accurate delineations of the borders of individual brain structures. It is important to note that post mortem scans registered to MNI-space have been subject to the same registration limitations as present in vivo. Additionally, individual specimens do not provide probabilistic information on the location of individual structures. A combined effort of research groups performing post mortem MRI scanning and delineating individual brain structures has the potential to create a probabilistic brain atlas that can be registered to MNI-space.
For any neuroscientist using fMRI techniques, as well as for surgeons performing DBS surgery it is important to know the answer to two important questions. The first question is: To what extent are anatomical brain scans representative of the underlying anatomy? Here, we argue that it is possible to answer this question by performing post mortem MRI scans, followed by histological validation at the level of the whole brain. The second question is: To what extent are the observations representative for the human brain? Answering this question requires laborious studies on a substantial number of post mortem human brain specimens, but is not impossible. Quantitative comparisons to in vivo data can subsequently be performed. Quantitative comparisons across modalities could include volume estimates and calculations of differences in the location of individual brain nuclei (Keuken et al., 2014). Challenges to execute the required post mortem MRI and histological validation studies are largely logistic. We would like to conclude that post mortem MRI and microscopy studies of the human brain will provide an important contribution to the field.
Author Contributions
All authors were involved in formulating the arguments and in writing the manuscript.
Funding
This work was financially supported by the European Research Council (313481) (BF), Stichting Internationaal Parkinson Fonds (AA and BF), and The Netherlands Organisation for Research and Development, Applied and Engineering Sciences Domain (STW14017) (AA and BF).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adler, D. H., Pluta, J., Kadivar, S., Craige, C., Gee, J. C., Avants, B. B., et al. (2014). Histology-derived volumetric annotation of the human hippocampal subfields in postmortem MRI. Neuroimage 84, 505–523. doi: 10.1016/J.NEUROIMAGE.2013.08.067
Alkemade, A., de Hollander, G., Keuken, M. C., Schäfer, A., Ott, D. V. M., Schwarz, J., et al. (2017). Comparison of T2∗-weighted and QSM contrasts in Parkinson’s disease to visualize the STN with MRI. PLoS One 12:e0176130. doi: 10.1371/journal.pone.0176130
Amunts, K., Lepage, C., Borgeat, L., Mohlberg, H., Dickscheid, T., Rousseau, M. E., et al. (2013). BigBrain: an ultrahigh-resolution 3D human brain model. Science 340, 1472–1475. doi: 10.1126/science.1235381
Annese, J., Schenker-Ahmed, N. M., Bartsch, H., Maechler, P., Sheh, C., Thomas, N., et al. (2014). Postmortem examination of patient H.M.’s brain based on histological sectioning and digital 3D reconstruction. Nat. Commun. 5, 3122. doi: 10.1038/ncomms4122
Augustinack, J. C., Magnain, C., Reuter, M., van der Kouwe, A. J. W., Boas, D., and Fischl, B. (2014). MRI parcellation of ex vivo medial temporal lobe. Neuroimage 93, 252–259. doi: 10.1016/J.NEUROIMAGE.2013.05.053
Bandettini, P. A. (2009). What’s new in neuroimaging methods? Ann. N. Y. Acad. Sci. 1156, 260–293. doi: 10.1111/j.1749-6632.2009.04420.x
Benabid, A. L., Chabardes, S., Mitrofanis, J., and Pollak, P. (2009). Deep brain stimulation of the subthalamic nucleus for the treatment of Parkinson’s disease. Lancet Neurol. 8, 67–81. doi: 10.1016/S1474-4422(08)70291-6
Blokker, B. M., Wagensveld, I. M., Weustink, A. C., Oosterhuis, J. W., and Hunink, M. G. M. (2016). Non-invasive or minimally invasive autopsy compared to conventional autopsy of suspected natural deaths in adults: a systematic review. Eur. Radiol. 26, 1159–1179. doi: 10.1007/s00330-015-3908-8
Bürgel, U., Schormann, T., Schleicher, A., and Zilles, K. (1999). Mapping of histologically identified long fiber tracts in human cerebral hemispheres to the mri volume of a reference brain: position and spatial variability of the optic radiation. Neuroimage 10, 489–499. doi: 10.1006/NIMG.1999.0497
Burton, J. L., and Underwood, J. (2007). Clinical, educational, and epidemiological value of autopsy. Lancet 369, 1471–1480. doi: 10.1016/S0140-6736(07)60376-6
Castellanos, F. X., Margulies, D. S., Kelly, C., Uddin, L. Q., Ghaffari, M., Kirsch, A., et al. (2008). Cingulate-precuneus interactions: a new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biol. Psychiatry 63, 332–337. doi: 10.1016/j.biopsych.2007.06.025
Chandran, A. S., Bynevelt, M., and Lind, C. R. P. (2016). Magnetic resonance imaging of the subthalamic nucleus for deep brain stimulation. J. Neurosurg. 124, 96–105. doi: 10.3171/2015.1.JNS142066
Chu, W.-S., Furusato, B., Wong, K., Sesterhenn, I. A., Mostofi, F. K., Wei, M. Q., et al. (2005). Ultrasound-accelerated formalin fixation of tissue improves morphology, antigen and mRNA preservation. Mod. Pathol. 18, 850–863. doi: 10.1038/modpathol.3800354
De Martino, F., Yacoub, E., Kemper, V., Moerel, M., Uludag, K., De Weerd, P., et al. (2017). The impact of ultra-high field MRI on cognitive and computational neuroimaging. Neuroimage 168, 366–382. doi: 10.1016/j.neuroimage.2017.03.060
Dirnhofer, R., Jackowski, C., Vock, P., Potter, K., and Thali, M. J. (2006). Virtopsy: minimally invasive, imaging-guided virtual autopsy. Radiographics 26, 1305–1333. doi: 10.1148/rg.265065001
Ewert, S., Plettig, P., Li, N., Chakravarty, M. M., Collins, L., Herrington, T. M., et al. (2017). Toward defining deep brain stimulation targets in MNI space: a subcortical atlas based on multimodal MRI, histology and structural connectivity. Neuroimage 170, 271–282. doi: 10.1016/j.neuroimage.2017.05.015
Forstmann, B. U., de Hollander, G., van Maanen, L., Alkemade, A., and Keuken, M. C. (2017). Towards a mechanistic understanding of the human subcortex. Nat. Rev. Neurosci. 18, 57–65. doi: 10.1038/nrn.2016.163
Gaensbacher, S., Waldhoer, T., and Berzlanovich, A. (2012). The slow death of autopsies: a retrospective analysis of the autopsy prevalence rate in Austria from 1990 to 2009. Eur. J. Epidemiol. 27, 577–580. doi: 10.1007/s10654-012-9709-3
Heim, B., Krismer, F., De Marzi, R., and Seppi, K. (2017). Magnetic resonance imaging for the diagnosis of Parkinson’s disease. J. Neural Transm. 124, 915–964. doi: 10.1007/s00702-017-1717-8
Horn, A., Kühn, A. A., Merkl, A., Shih, L., Alterman, R., and Fox, M. (2017). Probabilistic conversion of neurosurgical DBS electrode coordinates into MNI space. Neuroimage 150, 395–404. doi: 10.1016/j.neuroimage.2017.02.004
Ioannidis, J. P. A. (2005). Why most published research findings are false. PLoS Med. 2:e124. doi: 10.1371/journal.pmed.0020124
Jack, C. R., Albert, M. S., Knopman, D. S., McKhann, G. M., Sperling, R. A., Carrillo, M. C., et al. (2011). Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7, 257–262. doi: 10.1016/j.jalz.2011.03.004
Keuken, M. C., Bazin, P. L., Crown, L., Hootsmans, J., Laufer, A., Muller-Axt, C., et al. (2014). Quantifying inter-individual anatomical variability in the subcortex using 7T structural MRI. Neuroimage 94, 40–46. doi: 10.1016/j.neuroimage.2014.03.032
Kloppel, S., Stonnington, C. M., Chu, C., Draganski, B., Scahill, R. I., Rohrer, J. D., et al. (2008). Automatic classification of MR scans in Alzheimer’s disease. Brain 131, 681–689. doi: 10.1093/brain/awm319
Logothetis, N. K. (2008). What we can do and what we cannot do with fMRI. Nature 453, 869–878. doi: 10.1038/nature06976
Mai, J. K., Majtanik, M., and Paxinos, G. (2015). Atlas of the Human Brain, 4th Edn. Cambridge, MA: Academic Press.
Maier-Hein, K. H., Neher, P. F., Houde, J.-C., Côté, M.-A., Garyfallidis, E., Zhong, J., et al. (2017). The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 8:1349. doi: 10.1038/s41467-017-01285-x
Makris, N., Swaab, D. F., van der Kouwe, A., Abbs, B., Boriel, D., Handa, R. J., et al. (2013). Volumetric parcellation methodology of the human hypothalamus in neuroimaging: normative data and sex differences. Neuroimage 69, 1–10. doi: 10.1016/j.neuroimage.2012.12.008
Manova, E. S., Habib, C. A., Boikov, A. S., Ayaz, M., Khan, A., Kirsch, W. M., et al. (2009). Characterizing the mesencephalon using susceptibility-weighted imaging. Am. J. Neuroradiol. 30, 569–574. doi: 10.3174/ajnr.A1401
Ogg, R. J., Langston, J. W., Haacke, E. M., Steen, R. G., and Taylor, J. S. (1999). The correlation between phase shifts in gradient-echo MR images and regional brain iron concentration. Magn. Reson. Imaging 17, 1141–1148.
Palm, C., Axer, M., Gräßel, D., Dammers, J., Lindemeyer, J., Zilles, K., et al. (2010). Towards ultra-high resolution fibre tract mapping of the human brain - registration of polarised light images and reorientation of fibre vectors. Front. Hum. Neurosci. 4:9. doi: 10.3389/neuro.09.009.2010
Plantinga, B. R., Roebroeck, A., Kemper, V. G., Uludağ, K., Melse, M., Mai, J., et al. (2016). Ultra-high field mri post mortem structural connectivity of the human subthalamic nucleus, substantia nigra, and globus pallidus. Front. Neuroanat. 10:66. doi: 10.3389/fnana.2016.00066
Rauscher, A., Sedlacik, J., Barth, M., Mentzel, H. J., and Reichenbach, J. R. (2005). Magnetic susceptibility-weighted MR phase imaging of the human brain. Am. J. Neuroradiol. 26, 736–742.
Schafer, A., Wharton, S., Gowland, P., and Bowtell, R. (2009). Using magnetic field simulation to study susceptibility-related phase contrast in gradient echo MRI. Neuroimage 48, 126–137. doi: 10.1016/j.neuroimage.2009.05.093
Schaltenbrand and Wahren (1977). Neurosurgery Atlas for Stereotaxy of the Human Brain. Thieme. Available at: http://www.thieme.com/books-main/neurosurgery/product/168-atlas-for-stereotaxy-of-the-human-brain
Schmierer, K., Wheeler-Kingshott, C. A. M., Tozer, D. J., Boulby, P. A., Parkes, H. G., Yousry, T. A., et al. (2008). Quantitative magnetic resonance of postmortem multiple sclerosis brain before and after fixation. Magn. Reson. Med. 59, 268–277. doi: 10.1002/mrm.21487
Shojania, K. G., and Burton, E. C. (2008). The Vanishing nonforensic autopsy. N. Engl. J. Med. 358, 873–875. doi: 10.1056/NEJMp0707996
Stüber, C., Morawski, M., Schäfer, A., Labadie, C., Wähnert, M., Leuze, C., et al. (2014). Myelin and iron concentration in the human brain: a quantitative study of MRI contrast. Neuroimage 93, 95–106. doi: 10.1016/j.neuroimage.2014.02.026
Talairach, J. J., and Tournoux, P. (1988). Co-Planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to cerebral imaging. Stuttgart: Thieme.
Turner, R. (2002). How much cortex can a vein drain? downstream dilution of activation-related cerebral blood oxygenation changes. Neuroimage 16, 1062–1067. doi: 10.1006/nimg.2002.1082
van Duijn, S., Nabuurs, R. J., van Rooden, S., Maat-Schieman, M. L., van Duinen, S. G., van Buchem, M. A., et al. (2011). MRI artifacts in human brain tissue after prolonged formalin storage. Magn. Reson. Med. 65, 1750–1758. doi: 10.1002/mrm.22758
Vertinsky, A. T., Coenen, V. A., Lang, D. J., Kolind, S., Honey, C. R., Li, D., et al. (2009). Localization of the subthalamic nucleus: optimization with susceptibility-weighted phase MR imaging. Am. J. Neuroradiol. 30, 1717–1724. doi: 10.3174/ajnr.A1669
Weiss, M., Alkemade, A., Keuken, M. C., Müller-Axt, C., Geyer, S., Turner, R., et al. (2015). Spatial normalization of ultrahigh resolution 7 T magnetic resonance imaging data of the postmortem human subthalamic nucleus: a multistage approach. Brain Struct. Funct. 220, 1695–1703. doi: 10.1007/s00429-014-0754-4
Zemmoura, I., Blanchard, E., Raynal, P.-I., Rousselot-Denis, C., Destrieux, C., and Velut, S. (2016). How Klingler’s dissection permits exploration of brain structural connectivity? An electron microscopy study of human white matter. Brain Struct. Funct. 221, 2477–2486. doi: 10.1007/s00429-015-1050-7
Keywords: post mortem neuroanatomy, non-invasive neuroimaging, spatial distortions, 7 Tesla MRI, subcortex
Citation: Alkemade A, Groot JM and Forstmann BU (2018) Do We Need a Human post mortem Whole-Brain Anatomical Ground Truth in in vivo Magnetic Resonance Imaging? Front. Neuroanat. 12:110. doi: 10.3389/fnana.2018.00110
Received: 27 September 2018; Accepted: 23 November 2018;
Published: 05 December 2018.
Edited by:
Laurent Petit, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Ilyess Zemmoura, Université de Tours, FrancePiotr Majka, Nencki Institute of Experimental Biology (PAS), Poland
Copyright © 2018 Alkemade, Groot and Forstmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anneke Alkemade, jmalkemade@gmail.com
 Anneke Alkemade
Anneke Alkemade Josephine M. Groot
Josephine M. Groot Birte U. Forstmann
Birte U. Forstmann