Inactivation of Fluorescent Lipid Bilayers by Irradiation With 300 keV Electrons Using Liquid Cell Transmission Electron Microscopy
- 1Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, Richland, WA, United States
- 2School of Biological Sciences, Washington State University, Pullman, WA, United States
Liquid cell transmission electron microscopy allows for imaging of samples in a fully hydrated state at high resolution and has the potential for visualizing static or dynamic biological structures. However, the ionizing nature of the electron beam makes it difficult to discern real physiological dynamics from radiation induced artifacts within liquid cell samples. Electron flux thresholds for achieving high resolution structures from biological samples frozen in ice have been described extensively by the cryo-electron microscopy field, while electron flux thresholds which do not result in a functional change for biological samples within the hydrated environment of a transmission electron microscope liquid cell is less clear. Establishing these functional thresholds for biologically relevant samples is important for accurate interpretation of results from liquid cell experiments. Here we demonstrate the electron damage threshold of fluorescently tagged lipid bilayers by quantifying the change in fluorescence before and after electron exposure. We observe the reduction of fluorescent signal in bilayers by 25% after only 0.0005 e−/Å2 and a reduction of over 90% after 0.01 e−/Å2. These results indicate that the loss of function occurs at irradiation thresholds far below a typical single high resolution (scanning) transmission electron microscopy image and orders of magnitude below fluxes used for preserving structural features with cryo-electron microscopy.
Introduction
Visualizing biological dynamics at high spatial resolutions in their native, hydrated environment is an exciting prospect made possible with liquid cell transmission electron microscopy (LC-TEM). Significant work within the LC-TEM field in recent years has showcased the ability the technique has for probing dynamics of biological and soft materials systems (Woehl et al., 2020; Wu et al., 2020; Gibson and Patterson 2021). Yet imaging biological or organic samples in an aqueous environment using electrons raises concerns over the transfer of energy from the probe to the sample, manifesting as knock on damage (Baker and Rubinstein 2010), radiolysis (Glaeser 2016), charging (Jiang 2017), and heating (Egerton et al., 2004). For the application of imaging biological dynamics, understanding the impacts of electron irradiation damage on the sample is an important step in distinguishing representative physiological behavior from damage driven artifacts. A necessary distinction for the discussion of radiation damage is the difference between dose, the amount of energy transferred to the sample expressed in Grays (Gy), and number of irradiating particles per unit area which for electron microscopy is typically expressed as e−/Å2 and referred to as electron flux or density in literature. The relationship between flux and dose is primarily dependent on the energy of the electrons in the probe, the atomic composition, density, and thickness of the sample. For 300 keV electrons, a flux of 1 e−/Å2 on a thin layer of water is equal to approximately 4 × 106 Gy (Schneider et al., 2014), where the absorbed dose decreases as the electron accelerating energy increases (Baker and Rubinstein 2010).
In hydrated systems the primary damage mechanism of ionizing radiation is radiolysis of water which results in the production of the primary radical species H+, OH•, eh− (a.k.a eaq− or aqueous/hydrated/solvated electron), H•, OH, H2O2, H2, and HO2• (Kempner 2011; Schneider et al., 2014). Understanding how the radical species produced as a result of radiation exposure affect physiological function has been characterized extensively for applications in radiotherapy for electron (Sowa et al., 2005; Acharya et al., 2011), neutron (Kraft et al., 2010; Doria et al., 2012), and photon (gamma, x-ray, and ultraviolet) (Tokalov and Iagunov 2011; Acosta-Elias et al., 2017; Eom et al., 2017) sources. Ionizing radiation effects from gamma, and x-rays on cellular growth and biomolecule functionality has been characterized suggesting physiological impacts after doses below 50 Gy (Tokalov and Iagunov 2011; Reisz et al., 2014). While often the effects of irradiation on a cell or tissue is characterized as a physiological response, the impact of gamma irradiation on cell morphology and structure has also been shown where erythrocytes irradiated by gamma rays at doses from 15 to 50 Gy resulted in increased osmotic fragility, and alteration of cell membrane roughness (Acosta-Elias et al., 2017). The biological response of cells under electron irradiation has also been studied, where micronuclei formation suggests chromosomal dose sensitivity in similar ranges to those found for gamma and x-rays (Acharya et al., 2011). The ability for living cells to resist the effects of cellular damage caused by ionizing radiation by activation of stress response pathways makes establishing a specific lethal dose range difficult. General activation of measurable cellular response has been documented within the 2–50 Gy range which is significantly lower than what is currently used in LC-TEM experiments. However, most of the work done on the effects of ionizing radiation on biological samples has not been performed in an environment comparable to the conditions experienced by samples which are imaged using LC-TEM.
Extensive characterization on the loss of structural information with increasing electron exposure has been described for biological samples using cryogenic electron microscopy (cryo-EM) (Baumeister, Fringeli et al., 1976; Baumeister, Hahn et al., 1976; Baker et al., 2010; Glaeser et al., 2011), and more recently in liquid environments (Mirsaidov et al., 2012; Keskin and de Jonge 2018), although the effect of structure loss on a molecules function is not clear. Electron dose thresholds for functional activity in liquids is not well described, although dehydrated catalase monolayers have been demonstrated to lose functional activity from electron fluxes as low as 0.05 e−/Å2 (Hahn et al., 1976). Post mortem MALDI imaging mass spectrometry on LC-TEM systems has demonstrated the radiation induced fragmentation and damage of short peptides and poly-ethylene glycol, indicating that radiolysis induced damage results in structural and functional changes for other soft, organic based materials (Touve et al., 2019; Korpanty et al., 2021). Some concern has been raised about the impact of the electron beam on biological activity in LC-TEM (Glaeser 2012; de Jonge and Peckys 2016), although electron flux thresholds for biomolecules in the thin liquid environments used in LC-TEM has not been determined experimentally. Here we explore the functional inactivation of fluorescent lipid bilayers by electron irradiation within the constrained environment of a TEM liquid cell.
Methods
Bilayer Generation
Free standing silicon nitride membranes for use with liquid cell TEM holders were fabricated as described previously (Moser et al., 2018). Each silicon device contained five membranes separated by 100 µm edge to edge. Membranes were approximately 10 nm in thickness, 30 µm in width, and 600 µm in length. Devices were plasma cleaned under an argon/oxygen plasma for 3 minutes to remove surface contaminants and render the surface hydrophilic. Lipids were purchased from Avanti Polar lipids and spread using Langmuir-Blodgett (LB) deposition using an LB trough from KSV Nima. In short, a mixture of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-carboxyfluorescein (DOPE-CF) at a ratio of 19:1 and a concentration of 1 mM in chloroform was made. Hydrophilic silicon nitride devices were fixed to a hydrophilic silicon oxide substrate and immersed in the subphase (18.5 MΩ⋅cm deionized water) of the LB-trough, and 50 µL of the lipid solution was deposited onto the surface of the subphase. The chloroform solvent was allowed to evaporate fully for 30 min, and the resulting lipid film was compressed to a pressure of 20 mN/m. The substrate was then withdrawn and resubmerged in the subphase vertically at a speed of 10 cm/min resulting in a bilayer of the lipid mixture depositing on the surface of the silicon nitride membranes.
Fluorescence and Electron Imaging
Fluorescence imaging was performed on a Leica Confocal DMi8 microscope with a 5x objective, a 488 nm laser at 1.5%, a zoom factor of 3, and a 1024 × 1024 pixel scan area with a dwell time of 600 ms. Electron irradiation was performed on a 300 keV Thermo Fischer probe corrected Titan equipped with a monochromator and Gatan Imaging Filter (GIF) and calibrated with a Faraday cup holder. Fluorescent lipid bilayers were held in a hydrated environment using a Hummingbird Liquid Stage (Hummingbird). The liquid cell was imaged first with confocal fluorescence to confirm the presence of lipid bilayers, followed by loading into the electron microscope for irradiation. The electron beam diameter was 450 μm, the beam current was 2 nA for an electron flux of 0.0005 e−/Å2·s. The sample was then irradiated at predetermined stage locations such that there was not overlap of the electron beam between adjacent regions, and the cumulative electron flux was varied by changing the irradiation duration. Thickness of the liquid cell was then estimated using electron energy loss spectroscopy, where spectra were acquired in the corners of each window and the inelastic mean free path (IMFP) was estimated using the log ratio method (Malis et al., 1988; Jungjohann et al., 2012).
Data Analysis
For each image set, an optical bright field and fluorescent image was collected before and after irradiation with the electron beam. Fluorescent and bright field optical images were opened with and pixel signal count integration was performed with ImageJ. The bright field optical image was used to determine the dimensions in pixels of the window area where the same region was used to integrate signal in pixel counts in the corresponding fluorescent image. For the image collected after electron irradiation, the same procedure was used to integrate fluorescent signal in the corresponding window region using an integration region identical in pixel dimensions used to determine the fluorescent signal in the image prior to electron irradiation. The resulting difference in integrated fluorescent pixel intensity was used to measure the loss in fluorescent activity. Pixel intensity data was plotted and fit using R.
Results
Inactivation of Fluorescence/Dose Threshold
An illustration of our custom LC-TEM tip is shown in Figure 1A, where the inset shows a cross sectional view of the assembled liquid cell with a deposited lipid bilayer on the upper silicon nitride membrane in Figure 1B. Figure 1C shows the structures of the lipids in the bilayer, DOPC and DOPE-CF, which were deposited in a 19:1 ratio. Liquid cells assembled without a deposited lipid bilayer and filled with deionized water did not show significant fluorescence as shown in Figure 2A (left), Imaging of a liquid cell where the upper window had a deposited lipid bilayer resulted in a significant increase in fluorescent signal shown in Figure 2A (right). The integrated fluorescent signal is plotted in Figure 2B where a significant increase in fluorescent signal is observed when lipid bilayers are present. Imaging conditions and detector gain were identical for all imaging experiments and while there was some background autofluorescence for the water filled liquid cells, the signal was significantly higher for liquid cells with deposited lipid bilayers.
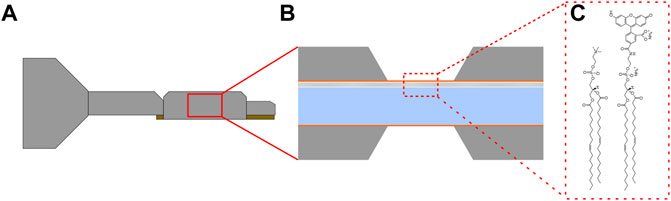
FIGURE 1. (A) Illustration of a liquid cell transmission electron microscope holder which contains an assembled set of silicon device filled with the sample. (B) Cross section illustration of a liquid cell, with a lipid bilayer on the upper silicon nitride membrane. (C) The lipids used for monolayer formation, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-carboxyfluorescein (DOPE-CF) (left and right respectively).
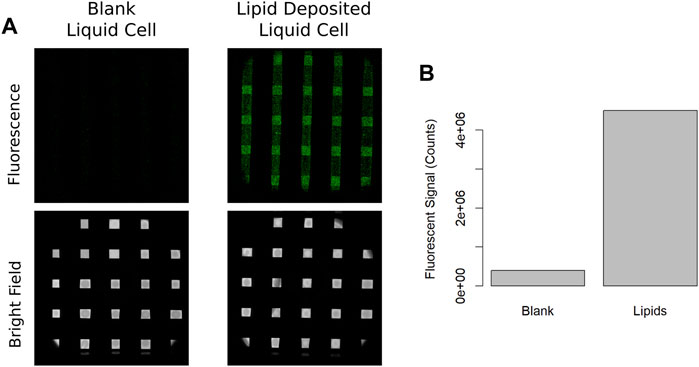
FIGURE 2. (A) Fluorescence image of a set of blank silicon nitride windows assembled only with distilled water and fluorescence image of a liquid cell membranes where the top membrane has been coated with a fluorescent lipid bilayer containing carboxyfluorescein. (B) Total fluorescent signal for blank and lipid deposited images.
Serial confocal imaging of the lipid bilayers was also performed to determine the sensitivity of the lipid bilayers to photobleaching. Assembled liquid cells with bilayers were imaged with confocal fluorescence microscopy, and videos of fluorescence were acquired for 2 h of sustained imaging. The integrated fluorescent signal from each frame was measured and plotted (shown in Supplementary Figure S1) where no significant change in fluorescence was observed over the first 60 min (about 1,000 frames) of imaging, followed by a decrease in fluorescent signal over the final 60 min due to photobleaching. For the following described electron irradiation experiments less than 50 cumulative frames were used to find focus and acquire images used for data processing suggesting that any observed changes in fluorescence signal during electron irradiation experiments were not due to photobleaching. Bilayers were also cycled in and out of the vacuum of the microscope without electron irradiation to ensure that fluorescence signal was not lost due to silicon nitride membrane flexing under pressure differentials of the microscope (Supplementary Figure S2). The above described control experiments indicate that any change in fluorescence signal during electron irradiation was not the product of photobleaching or mechanical strain the samples experienced under typical experiment conditions.
Subsequently, irradiation experiments consisted of fluorescence images acquired after device assembly (and before electron irradiation) and were acquired with the optical objective located below the holder as illustrated in Figure 3A. Irradiation of the sample with 300 keV electrons (at a constant flux) was then performed as depicted in Figure 3B, followed by fluorescent imaging to observe functional changes induced by the electron beam (Figure 3C). Initial irradiation experiments were performed by first collecting a fluorescent and bright field image, shown in Figure 4A, and then loading the sample into the microscope and left in a neutral stage position (0 μm, 0 μm x and y coordinates). The sample was then exposed with an electron beam with a diameter of 450 µm and a cumulative flux of 0.1 e−/Å2 as depicted in Figure 4B. After electron irradiation the sample was taken back to the confocal microscope and fluorescence and bright field images were again acquired shown in Figure 4C. Figure 4 illustrates that fluorescence signal can be used as a functional reporter for lipid bilayer function where the position of the electron beam can be seen by the region of lost fluorescence when compared to the non-irradiated region.

FIGURE 3. (A) Illustration of the fluorescent imaging strategy, where the liquid cell was imaged from below the liquid cell tip with a 488 nm laser generating a bright field and fluorescent image (B) Illustration of electron irradiation, where the orientation of the liquid holder was always such that the lipid bilayer was always on the top of the liquid cell. (C) Illustration of the fluorescent imaging strategy after electron irradiation, where bright field and fluorescent images were collected of the liquid cell immediately post irradiation.
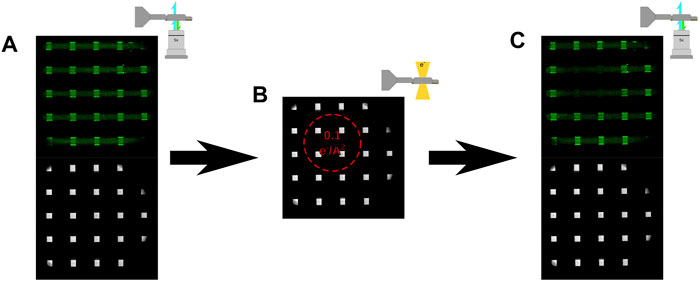
FIGURE 4. (A) Fluorescent and bright field optical image of a liquid cell assembled with deionized water and fluorescent bilayers prior to irradiating with 300 keV electrons. (B) Bright field optical image showing the approximate location of the electron beam with a flux of (0.1 e/Å2). (C) Fluorescent and bright field optical images of the same liquid cell shown in (A) after irradiation with 300 keV electrons. The region of exposure can be seen by the loss of fluorescent signal corresponding to the noted area in (B).
To improve throughput of subsequent experiments multiple electron irradiation experiments were performed within a single assembled liquid cell. Figure 5A shows a fluorescence image of a liquid cell with a deposited lipid bilayer prior to electron irradiation. The sample was then loaded into the microscope and irradiation was performed by moving the stage to locations notated by Figure 5B and irradiating with differing electron fluxes at each position. The stage x and y positions and beam diameters (approximately depicted by the dotted circles in Figure 5B) were such that there was no overlap of electron irradiation in any of the positions depicted. The beam diameter was maintained consistent for each location so that several windows would be irradiated for each region and the electron flux was varied by changing the exposure time. Figure 5A shows the fluorescence image for one of these experiments prior to electron irradiation and Figure 5C shows the fluorescence image after exposing each region successively, where a gradient of decreased signal relative to the incident electron flux can be seen. These experiments were repeated for different electron flux conditions and sample thicknesses was measured with EELS after the final fluorescence image was taken to verify the presence of liquid. The loss of fluorescence signal for each window region irradiated, across multiple samples, was quantified and plotted as shown in Figure 5D for cumulative electron fluxes of 0.0001, 0.0005, 0.001, 0.005, and 0.01 e−/Å2. The number of observations for each electron flux was 10, 6, 14, 13, and 11 respectively, and the red curve in Figure 5D is an exponential fit to the measured data.
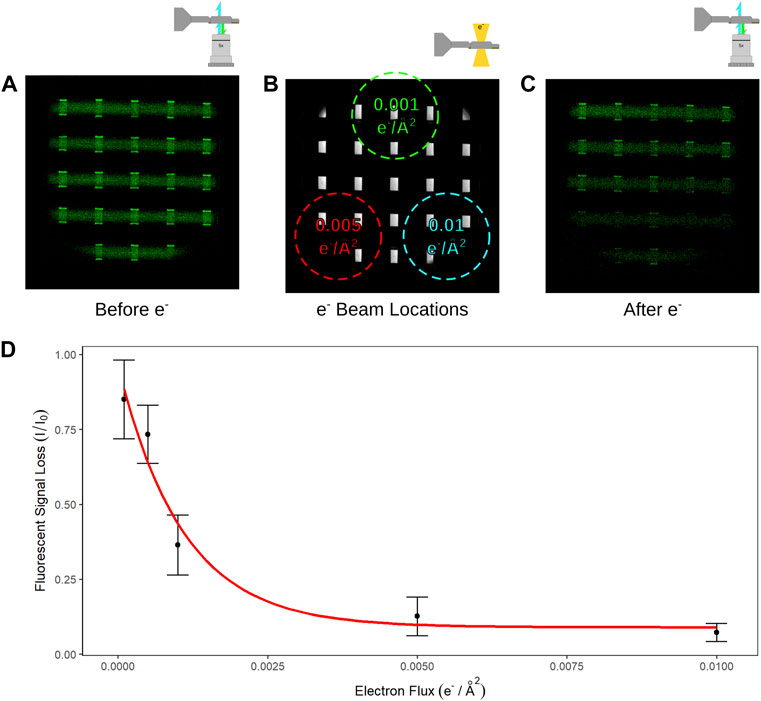
FIGURE 5. (A) Fluorescence image of the liquid cell prior to electron irradiation. (B) Bright field optical image of an assembled liquid cell were the size, location, and fluxes of 3 different electron exposures of the sample are noted. (C) Fluorescence image of the same liquid cell from (A) after electron irradiation in the locations and fluxes as noted in (B). (D) Measured signal loss from window regions which have been irradiated with electrons at fluxes of 0.01, 0.005, 0.001, 0.0005, and 0.0001 e−/Å2.
Discussion
The results above describe the loss of fluorescence signal due to electron irradiation and suggest the disruption of the fluorescent carboxyfluorescein tag or supporting lipid bilayer by primary or secondary damage. Observed fluorescence signal was decreased by 25% after just 0.0005 e−/Å2 and decreased by over 90% after 0.01 e−/Å2. Notably, the approximate absorbed dose by a thin liquid sample from a flux of 0.0001 e−/Å2 is ∼4 × 102 Gy, while the dose from a flux of 0.01 e−/Å2 is ∼4 × 104 Gy. These doses are still considerably higher than irradiation dose values which have been described as impacting the physiology of living systems. These results suggest that biological function in a liquid cell is affected by a relatively low electron flux compared to what is traditionally used in LC-TEM imaging. So, why the discrepancy with other electron microscopy techniques such as cryo-EM which still uses single image doses 4,000x higher (10–40 e−/Å2) for structural annotation of 3D structure at atomic resolution?
Thresholds Showing Functional Loss are Much Lower Than Thresholds for Atomic Imaging
A central concept of biology is that structure and function are directly related, where the function of a protein is dictated by its sequence and 3D structure. Any alteration of amino acid composition or the structural folding of a protein can have significant impact on its functional activity. In many cases, functional activity of a protein can be altered by reducing a disulfide bond critical to the active site. As mentioned earlier, the thresholds determined for cryo-EM have been based on the loss of structural resolution and not the loss of function of the molecule. For example, Peet et al. found that the 4 Å reflection from the diffraction pattern of paraffin crystals at 80 K faded by 50% after ∼15–20 e−/Å2 of a 300 keV beam, and the 4 Å reflection from the diffraction pattern of a bacteriorhodopsin protein crystal faded by 50% after ∼5 e−/Å2 (Peet et al., 2019). Similar results have shown fading of reflections from catalase crystals at similar electron flux values at 200 keV and 80oK (Baker et al., 2010). For most single particle cryo-EM studies, better than 5 Å structural resolution can be determined from electron fluxes below 50 e−/Å2 (Grant and Grigorieff 2015). Indeed, the highest resolution single particle 3D reconstruction to date using cryo-EM (Yip et al., 2020) achieved ∼1.2 Å and was solved with data acquired at 50 e−/Å2. The discrepancy in published electron flux thresholds for cryo-EM structural imaging and the LC-TEM results described here can be explained by the advantage of using cryogenic temperatures and embedding the sample within a frozen matrix of vitreous ice, where the effects of radiation damage can be reduced by decreasing the temperature of the sample (Stark et al., 1996). Cryo-EM samples also benefit from being trapped in a solid matrix of ice, where the radical species produced by hydrolysis of the surrounding water molecules are not free to diffuse and react with adjacent water molecules or the sample as they are in a liquid system (Schneider et al., 2014). Finally, it may be that there are damage events which are beyond the resolution limit of cryo-EM (such as single atom knock on damage) that would still result in a chemical and functional change in the molecule. For example, it is well described that water undergoes hydrolysis during electron irradiation and forms a number of complex radical species. While these hydrolysis events cause a substantial change in the chemistry and resulting behavior of the water (Park et al., 2015; Abellan et al., 2017), they are not detectable through imaging alone for LC-TEM or cryo-EM with current detectors and microscopes. Therefore, the damage events which result in the inactivation of the fluorescent bilayers might result from structural changes which occur at spatial frequencies that are below what is achievable using cryo-EM. These differences indicate that LC-TEM experiment design cannot rely solely on the previously established cumulative electron fluxes outlined by the cryo-EM field when attempting to observe biological dynamics in liquid.
Electron Beam Induced Functional Loss of Fluorescence Could Manifest Multiple Ways
While our results demonstrate loss of fluorescence due to damage to the molecules of the fluorescent lipid bilayer, it is not immediately apparent where the damage is taking place or the chemical changes that are occurring due to the inability to resolve individual lipid molecules. In the work above, the DOPC lipids consisted of two 18 carbon fatty acid chains, a phosphocholine head group, and in the case of the fluorescent lipid (DOPE), a covalently linked carboxyfluorescein dye which consists of several aromatic rings. While this is a relatively simple biological molecule compared to larger proteins or DNA oligomers, there are at least 5 different ways that primary and secondary damage from electron irradiation could result in a loss of fluorescence function and signal. Both primary and secondary damage to any of the aromatic rings of the carboxyfluorescein dye would affect its conjugation and therefore its ability to fluoresce, resulting in signal loss. The slightly increased electron density of the aromatic rings of this molecule may result in an increased scattering cross section and resulting increase in likelihood of inelastic collisions. Indeed, recent cryo-EM reports have demonstrated site-selective damage in proteins occurring at other electron dense regions such as disulfide bridges, charged amino acids, and metal binding sites (Hattne et al., 2018). The reliance of biological molecules on critical bonds or interactions for function is well known and hints at the difficulty for biological imaging with LC-TEM where the loss of a single bond due to electron irradiation could cause the loss of function but not structure.
Besides disrupting the pi-conjugation of the aromatic rings of carboxyfluorescein, other forms of damage to the carboxyfluorescein-DOPE or the DOPC molecules could also cause the loss of fluorescent signal. For example, primary or secondary damage disruption (knock on or chemical cleavage) of the linker that connects the lipid head group to the carboxyfluorescein moiety would cause a release of the fluorophore into bulk solution while the lipid bilayer stays attached to the window and cause the loss of fluorescent signal. Additionally, damage to the alkane fatty acid chain or the polar phosphate head group of the lipid molecules could also disrupt the hydrophobic/hydrophilic forces which hold the bilayer together on the silicon nitride membrane causing regions to delaminate and thereby lose fluorescence. Altering the chemistry of the lipid molecules which affect its amphiphilic nature would affect its ability to stay anchored over the window and could explain the loss of fluorescence over the irradiated regions. Secondary damage resulting from a reaction of the lipid bilayer with the radical species produced from hydrolysis could also react with and change the chemistry of the lipid fatty acid chain, phosphate head group, or carboxyfluorescein dye. Indeed, the chemistry of lipid oxidation from ROS has been described extensively in the literature (Frankel 1984; Frankel 1987). Unsaturated lipids in particular are susceptible to oxidation by radicals, where the carbon-carbon double bond is a site for hydrogen abstraction by OH• radicals, forming a lipid radical (L•) and H2O (Girotti 1985). Once generated, lipid radical formation can be propagated where lipid radicals are oxidized by O2, which can then abstract a hydrogen from a neighboring lipid to form LOOH and a new lipid radical. This cycle can continue and form new lipid molecules until the L• radical concentration climbs high enough for lipid radicals to react with each other resulting in cross linking (Gardner 1989). In addition to oxidation and crosslinking, other structural changes can include fragmentation and cyclization (Gardner 1989). Functionally, the impact of lipid oxidation in membranes and biological systems have been demonstrated to manifest as damage to integral membrane proteins (Stark 2005) and increased membrane permeability (Nakazawa and Nagatsuka 1980; Wong-Ekkabut et al., 2007). It is worthwhile to note that radical scavengers, such as an alcohol to scavenge OH• radicals, have been used to mitigate the effects of radiolysis induced damage in LC-TEM experiments (Sutter et al., 2014; Korpanty et al., 2021). In this case however, it is important to also consider how the sample is impacted by the presence of the scavenger or any intermediary radical formed during the scavenging reaction. While the damage observed is likely the product of some combination of primary and secondary damage events it is not possible with our current experimental process to determine which, if either, is the dominant mechanism.
Prospects for Biological Imaging With LC-TEM
Determining the irradiation thresholds for inactivation of lipid bilayers is relevant to LC-TEM work with biological systems as they are key components of cellular membranes where they play important roles in regulating intra/extracellular gradients, anchoring integral and peripheral membrane proteins, and packaging or compartmentalizing cellular components. Most all these functions are tied to the amphiphilic properties of lipids which allow them to assemble and anchor membrane associated proteins. The loss of fluorescence signal from the bilayers irradiated in the experiments described here could possibly be due to the disruption of the hydrophobic or hydrophilic regions of the lipid molecules through electron irradiation, as they anchor to the silicon nitride membranes through these intramolecular forces. Previous results have demonstrated the shrinking of bacteria imaged in LC-TEM under electron irradiation, suggesting a loss of cellular membrane integrity (Woehl et al., 2014; Moser et al., 2018; Moser et al., 2019), and as noted above similar observations have been made for cell membranes exposed to gamma radiation (Acosta-Elias et al., 2017). As such, the thresholds determined for inactivation of lipid bilayers are a relative starting point for dose thresholds for imaging dynamics in whole cells using LC-TEM. An important note is that the presence of damage does not necessarily preclude the possibility of performing LC-TEM experiments on biological samples. Rather, reliable interpretation of LC-TEM data must account for all the types of damage to different biomolecules at various electron irradiation thresholds to accurately conclude whether any observed motion or dynamics was truly linked to functional biomolecules. The work here establishes a starting point for working with electron fluxes which may allow for functional activity within the environment of a liquid cell, although it is by no means comprehensive of all classes of physiological molecules at this point. While the work here focuses on lipids, a number of other biomolecules such as proteins and DNA must also be investigated to determine their functional damage thresholds. The wide range of biological interactions and environmental states however suggests that characterization of electron beam impacts on all biological systems will not be a trivial task. For example, some proteins function in their native environment to reduce or oxidize substrates and the inherent reducing environment caused by electron beam irradiation may make it difficult to observe native dynamics for one or both of these protein types. Similarly, some proteins rely on transition metal centers for electron transfer while others simply create a channel for movement of ions across a membrane. The lack or presence of metal atoms within the protein of interest may significantly impact the functional damage threshold and its ability to be studied by LC-TEM. As a result, electron thresholds for functional activity should be determined in an experimentally specific manner and relative to the system under investigation. While the diversity of biological samples may be overwhelming for complete characterization of sample-beam interactions, it also presents exciting options for exploring the real boundaries of LC-TEM and ensuring physiological interpretation of movies. The results reported here are a first step for identifying electron fluxes required to maintain the functional activity of biomolecules under electron irradiation in a liquid cell, an important step towards realizing the visualization of biological dynamics with LC-TEM.
Conclusion
We describe a method for experimentally measuring the functional activity of fluorescent lipid bilayers anchored to silicon nitride membranes in a liquid cell before and after electron irradiation, where the change in lipid functional activity is inferred from the absolute change in activity from an optical reporter. Functional activity, via our fluorescent reporter, is observed to decrease by decreased by 25% after just 0.0005 e−/Å2 and decreased by over 90% after 0.01 e−/Å2. These thresholds are substantially lower than those typically used in transmission electron microscopy imaging for liquid, cryogenic, or material samples. Damage resulting in functional change or inactivity may range in molecular location and mechanism, and are likely the product of several (or all) of the above proposed damage mechanisms in some combination. Additional LC-TEM experiments will be needed to determine both functional electron dose thresholds as well as the types of functional changes which occur across a wide range of biologically relevant molecules in order to accurately interpret the results of future work on biological samples with LC-TEM.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
TM and JE conceived the research, performed all data analysis and wrote the manuscript. TM acquired all light and electron microscopy data.
Funding
This work was supported in part by DOE-BER projects FWP 66382 and FWP 76295. A portion of this research was performed at EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2022.772469/full#supplementary-material
References
Abellan, P., Moser, T. H., Lucas, I. T., Grate, J. W., Evans, J. E., and Browning, N. D. (2017). The Formation of Cerium(iii) Hydroxide Nanoparticles by a Radiation Mediated Increase in Local pH. RSC Adv. 7 (7), 3831–3837. doi:10.1039/c6ra27066b
Acharya, S., Bhat, N. N., Joseph, P., Sanjeev, G., Sreedevi, B., and Narayana, Y. (2011). Dose Rate Effect on Micronuclei Induction in Human Blood Lymphocytes Exposed to Single Pulse and Multiple Pulses of Electrons. Radiat. Environ. Biophys. 50 (2), 253–263. doi:10.1007/s00411-011-0353-1
Acosta-Elías, M. A., Burgara-Estrella, A. J., Sarabia-Sainz, J. A.-i., Silva-Campa, E., Angulo-Molina, A., Santacruz-Gómez, K. J., et al. (2017). Nano Alterations of Membrane Structure on Both γ-irradiated and Stored Human Erythrocytes. Int. J. Radiat. Biol. 93 (12), 1306–1311. doi:10.1080/09553002.2017.1393581
Baker, L. A., and Rubinstein, J. L. (2010). Radiation Damage in Electron Cryomicroscopy. Methods Enzymol. 481, 371–388. doi:10.1016/s0076-6879(10)81015-8
Baker, L. A., Smith, E. A., Bueler, S. A., and Rubinstein, J. L. (2010). The Resolution Dependence of Optimal Exposures in Liquid Nitrogen Temperature Electron Cryomicroscopy of Catalase Crystals. J. Struct. Biol. 169 (3), 431–437. doi:10.1016/j.jsb.2009.11.014
Baumeister, W., Hahn, M., Seredynski, J., and Herbertz, L. M. (1976). Radiation Damage of Proteins in the Solid State: Changes of Amino Acid Composition in Catalase. Ultramicroscopy 1 (3-4), 377–382. doi:10.1016/0304-3991(76)90052-8
Baumeister, W., Fringeli, U. P., Hahn, M., and Seredynski, J. (1976). Radiation Damage of Proteins in the Solid State: Changes of β-lactoglobulin Secondary Structure. Biochim. Biophys. Acta (Bba) - Protein Struct. 453 (1), 289–292. doi:10.1016/0005-2795(76)90276-2
de Jonge, N., and Peckys, D. B. (2016). Live Cell Electron Microscopy Is Probably Impossible. ACS Nano 10 (10), 9061–9063. doi:10.1021/acsnano.6b02809
Doria, D., Kakolee, K. F., Kar, S., Litt, S. K., Fiorini, F., Ahmed, H., et al. (2012). Biological Effectiveness on Live Cells of Laser Driven Protons at Dose Rates Exceeding 109Gy/s. AIP Adv. 2 (1), 011209-1–011209-6. doi:10.1063/1.3699063
Egerton, R. F., Li, P., and Malac, M. (2004). Radiation Damage in the TEM and SEM. Micron 35 (6), 399–409. doi:10.1016/j.micron.2004.02.003
Eom, H. S., Park, H. S., You, G. E., Kim, J. Y., and Nam, S. Y. (2017). Identification of Cellular Responses to Low-Dose Radiation by the Profiling of Phosphorylated Proteins in Human B-Lymphoblast IM-9 Cells. Int. J. Radiat. Biol. 93 (11), 1207–1216. doi:10.1080/09553002.2017.1377362
Frankel, E. N. (1987). Secondary Products of Lipid Oxidation. Chem. Phys. Lipids 44 (2-4), 73–85. doi:10.1016/0009-3084(87)90045-4
Frankel, E. N. (1984). Chemistry of Free Radical and Singlet Oxidation of Lipids. Prog. Lipid Res. 23 (4), 197–221. doi:10.1016/0163-7827(84)90011-0
Gardner, H. W. (1989). Oxygen Radical Chemistry of Polyunsaturated Fatty Acids. Free Radic. Biol. Med. 7 (1), 65–86. doi:10.1016/0891-5849(89)90102-0
Gibson, W., and Patterson, J. P. (2021). Liquid Phase Electron Microscopy Provides Opportunities in Polymer Synthesis and Manufacturing. Macromolecules 54 (11), 4986–4996. doi:10.1021/acs.macromol.0c02710
Girotti, A. W. (1985). Mechanisms of Lipid Peroxidation. J. Free Radicals Biol. Med. 1 (2), 87–95. doi:10.1016/0748-5514(85)90011-x
Glaeser, R. M. (2012). Electron Microscopy of Biological Specimens in Liquid Water. Biophysical J. 103 (1), 163–164. doi:10.1016/j.bpj.2012.05.042
Glaeser, R. M., McMullan, G., Faruqi, A. R., and Henderson, R. (2011). Images of Paraffin Monolayer Crystals with Perfect Contrast: Minimization of Beam-Induced Specimen Motion. Ultramicroscopy 111 (2), 90–100. doi:10.1016/j.ultramic.2010.10.010
Glaeser, R. M. (2016). Specimen Behavior in the Electron Beam. Methods Enzymol. 579, 19–50. doi:10.1016/bs.mie.2016.04.010
Grant, T., and Grigorieff, N. (2015). Measuring the Optimal Exposure for Single Particle Cryo-EM Using a 2.6 Å Reconstruction of Rotavirus VP6. Elife 4, e06980. doi:10.7554/eLife.06980
Hahn, M., Seredynski, J., and Baumeister, W. (1976). Inactivation of Catalase Monolayers by Irradiation with 100 keV Electrons. Proc. Natl. Acad. Sci. U.S.A. 73 (3), 823–827. doi:10.1073/pnas.73.3.823
Hattne, J., Shi, D., Glynn, C., Zee, C.-T., Gallagher-Jones, M., Martynowycz, M. W., et al. (2018). Analysis of Global and Site-specific Radiation Damage in Cryo-EM. Structure 26 (5), 759–766. doi:10.1016/j.str.2018.03.021
Jiang, N. (2017). Note on In Situ (Scanning) Transmission Electron Microscopy Study of Liquid Samples. Ultramicroscopy 179, 81–83. doi:10.1016/j.ultramic.2017.04.012
Jungjohann, K. L., Evans, J. E., Aguiar, J. A., Arslan, I., and Browning, N. D. (2012). Atomic-scale Imaging and Spectroscopy for In Situ Liquid Scanning Transmission Electron Microscopy. Microsc. Microanal 18 (3), 621–627. doi:10.1017/s1431927612000104
Kempner, E. S. (2011). Direct Effects of Ionizing Radiation on Macromolecules. J. Polym. Sci. B Polym. Phys. 49 (12), 827–831. doi:10.1002/polb.22250
Keskin, S., and de Jonge, N. (2018). Reduced Radiation Damage in Transmission Electron Microscopy of Proteins in Graphene Liquid Cells. Nano Lett. 18, 7435–7440. doi:10.1021/acs.nanolett.8b02490
Korpanty, J., Parent, L. R., and Gianneschi, N. C. (2021). Enhancing and Mitigating Radiolytic Damage to Soft Matter in Aqueous Phase Liquid-Cell Transmission Electron Microscopy in the Presence of Gold Nanoparticle Sensitizers or Isopropanol Scavengers. Nano Lett. 21 (2), 1141–1149. doi:10.1021/acs.nanolett.0c04636
Kraft, S. D., Richter, C., Zeil, K., Baumann, M., Beyreuther, E., Bock, S., et al. (2010). Dose-dependent Biological Damage of Tumour Cells by Laser-Accelerated Proton Beams. New J. Phys. 12 (8), 1–12. doi:10.1088/1367-2630/12/8/085003
Malis, T., Cheng, S. C., and Egerton, R. F. (1988). EELS Log-Ratio Technique for Specimen-Thickness Measurement in the TEM. J. Elec. Microsc. Tech. 8 (2), 193–200. doi:10.1002/jemt.1060080206
Mirsaidov, U. M., Zheng, H., Casana, Y., and Matsudaira, P. (2012). Imaging Protein Structure in Water at 2.7 Nm Resolution by Transmission Electron Microscopy. Biophysical J. 102 (4), L15–L17. doi:10.1016/j.bpj.2012.01.009
Moser, T. H., Mehta, H., Park, C., Kelly, R. T., Shokuhfar, T., and Evans, J. E. (2018). The Role of Electron Irradiation History in Liquid Cell Transmission Electron Microscopy. Sci. Adv. 4 (4), eaaq1202. doi:10.1126/sciadv.aaq1202
Moser, T. H., Shokuhfar, T., and Evans, J. E. (2019). Considerations for Imaging Thick, Low Contrast, and Beam Sensitive Samples with Liquid Cell Transmission Electron Microscopy. Micron 117, 8–15. doi:10.1016/j.micron.2018.10.007
Nakazawa, T., and Nagatsuka, S. (1980). Radiation-induced Lipid Peroxidation and Membrane Permeability in Liposomes. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 38 (5), 537–544. doi:10.1080/09553008014551351
Park, J. H., Schneider, N. M., Grogan, J. M., Reuter, M. C., Bau, H. H., Kodambaka, S., et al. (2015). Control of Electron Beam-Induced Au Nanocrystal Growth Kinetics through Solution Chemistry. Nano Lett. 15 (8), 5314–5320. doi:10.1021/acs.nanolett.5b01677
Peet, M. J., Henderson, R., and Russo, C. J. (2019). The Energy Dependence of Contrast and Damage in Electron Cryomicroscopy of Biological Molecules. Ultramicroscopy 203, 125–131. doi:10.1016/j.ultramic.2019.02.007
Reisz, J. A., Bansal, N., Qian, J., Zhao, W., and Furdui, C. M. (2014). Effects of Ionizing Radiation on Biological Molecules-Mechanisms of Damage and Emerging Methods of Detection. Antioxid. Redox Signaling 21 (2), 260–292. doi:10.1089/ars.2013.5489
Schneider, N. M., Norton, M. M., Mendel, B. J., Grogan, J. M., Ross, F. M., and Bau, H. H. (2014). Electron-Water Interactions and Implications for Liquid Cell Electron Microscopy. J. Phys. Chem. C 118 (38), 22373–22382. doi:10.1021/jp507400n
Sowa, M. B., Kathmann, L. E., Holben, B. A., Thrall, B. D., and Kimmel, G. A. (2005). Low-LET Microbeam Investigation of the Track-End Dependence of Electron-Induced Damage in Normal Human Diploid Fibroblasts. Radiat. Res. 164 (5), 677–679. doi:10.1667/rr3464.1
Stark, G. (2005). Functional Consequences of Oxidative Membrane Damage. J. Membr. Biol 205 (1), 1–16. doi:10.1007/s00232-005-0753-8
Stark, H., Zemlin, F., and Boettcher, C. (1996). Electron Radiation Damage to Protein Crystals of Bacteriorhodopsin at Different Temperatures. Ultramicroscopy 63 (2), 75–79. doi:10.1016/0304-3991(96)00045-9
Sutter, E., Jungjohann, K., Bliznakov, S., Courty, A., Maisonhaute, E., Tenney, S., et al. (2014). In Situ liquid-cell Electron Microscopy of Silver-Palladium Galvanic Replacement Reactions on Silver Nanoparticles. Nat. Commun. 5, 4946. doi:10.1038/ncomms5946
Tokalov, S. V., and Iagunov, A. S. (2011). Radiation-induced Cell Cycle Arrests in Ehrlich Ascites Carcinoma Cells In Vivo. Radiat. Environ. Biophys. 50 (2), 265–270. doi:10.1007/s00411-011-0354-0
Touve, M. A., Carlini, A. S., and Gianneschi, N. C. (2019). Self-assembling Peptides Imaged by Correlated Liquid Cell Transmission Electron Microscopy and MALDI-Imaging Mass Spectrometry. Nat. Commun. 10 (1), 4837. doi:10.1038/s41467-019-12660-1
Woehl, T. J., Kashyap, S., Firlar, E., Perez-Gonzalez, T., Faivre, D., Trubitsyn, D., et al. (2014). Correlative Electron and Fluorescence Microscopy of Magnetotactic Bacteria in Liquid: toward In Vivo Imaging. Sci. Rep. 4, 6854. doi:10.1038/srep06854
Woehl, T. J., Moser, T., Evans, J. E., and Ross, F. M. (2020). Electron-beam-driven Chemical Processes during Liquid Phase Transmission Electron Microscopy. MRS Bull. 45 (9), 746–753. doi:10.1557/mrs.2020.227
Wong-Ekkabut, J., Xu, Z., Triampo, W., Tang, I.-M., Peter Tieleman, D., and Monticelli, L. (2007). Effect of Lipid Peroxidation on the Properties of Lipid Bilayers: a Molecular Dynamics Study. Biophysical J. 93 (12), 4225–4236. doi:10.1529/biophysj.107.112565
Wu, H., Friedrich, H., Patterson, J. P., Sommerdijk, N. A. J. M., and de Jonge, N. (2020). Liquid-Phase Electron Microscopy for Soft Matter Science and Biology. Adv. Mater. 32 (25), e2001582. doi:10.1002/adma.202001582
Keywords: LC-TEM, liquid cell transmission electron microscopy, in situ TEM, radiation chemistry, fluorescence inactivation
Citation: Moser T and Evans JE (2022) Inactivation of Fluorescent Lipid Bilayers by Irradiation With 300 keV Electrons Using Liquid Cell Transmission Electron Microscopy. Front. Nanotechnol. 4:772469. doi: 10.3389/fnano.2022.772469
Received: 08 September 2021; Accepted: 10 March 2022;
Published: 29 March 2022.
Edited by:
Ajeet Kaushik, Florida Polytechnic University, United StatesReviewed by:
Suresh K. Verma, Uppsala University, SwedenRahul Kumar Dhaka, Chaudhary Charan Singh Haryana Agricultural University, India
Copyright © 2022 Moser and Evans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James E. Evans, James.Evans@PNNL.Gov
 Trevor Moser
Trevor Moser James E. Evans
James E. Evans