The need for unbiased genetic screens to dissect aggression in Drosophila melanogaster
- 1Department of Molecular & Human Genetics, Baylor College of Medicine, Houston, TX, United States
- 2Department of Neuroscience, Baylor College of Medicine, Houston, TX, United States
Aggression is an evolutionarily conserved behavior present in most animals and is necessary for survival when competing for limited resources and mating partners. Studies have shown that aggression is modulated both genetically and epigenetically, but details of how the molecular and cellular mechanisms interact to determine aggressive behavior remain to be elucidated. In recent decades, Drosophila melanogaster has emerged as a powerful model system to understand the mechanisms that regulate aggression. Surprisingly most of the findings discovered to date have not come from genetic screens despite the fly’s long and successful history of using screens to unravel its biology. Here, we highlight the tools and techniques used to successfully screen for aggression-linked behavioral elements in Drosophila and discuss the potential impact future screens have in advancing our knowledge of the underlying genetic and neural circuits governing aggression.
Introduction
For many decades mutant screens have been a powerful approach to investigate the molecular mechanisms underlying the basic processes of life (Beadle and Tatum, 1941; Tatum and Lederberg, 1947; Hartwell et al., 1970; Hartwell, 1971; Nurse, 1975; Kenyon and Walker, 1980; Schekman et al., 1983; Tsukada and Ohsumi, 1993). Genetic screens only became possible after the discovery that ionizing radiation is mutagenic (Gager and Blakeslee, 1927; Muller, 1927, 1928; Calabrese, 2018), which vastly increased the number of mutants that could be generated and analyzed in a screen. Beadle and Tatum used X-ray mutagenesis in a conceptually simple screen in Neurospora to isolate mutants responsible for specific metabolic functions essential for life by switching mutants from complete media to minimal media supplemented with specific metabolites (Beadle and Tatum, 1941). Their screen, in combination with the selection approach isolated strains that were deficient in three essential metabolic enzymes necessary for the synthesis of pyridoxine, thiazole, and para-amino benzoic acid respectively (Beadle and Tatum, 1941). The approach revolutionized the identification of essential genes and has even been credited as the origin of molecular biology (Horowitz et al., 2004; Strauss, 2016). Screens also revealed other basic and universal processes such as gene recombination in E. coli (Tatum and Lederberg, 1947), genetic control of cell division (Hartwell et al., 1970; Hartwell, 1971; Nurse, 1975), vesicle secretion (Schekman et al., 1983), and autophagy (Tsukada and Ohsumi, 1993) to name just a few.
The idea turned out to be equally powerful in multicellular organisms. In worms and flies, screens identified the molecular players that control development (Brenner, 1974; Riddle et al., 1981; Jürgens et al., 1984; Nüsslein-Volhard et al., 1984; Wieschaus et al., 1984), cell death (Ellis and Horvitz, 1986), aging (Kenyon et al., 1993), immunity (Lemaitre et al., 1995), and many other biological processes. Many of the genes identified in these screens turned out to be conserved across the animal kingdom (Wieschaus and Nüsslein-Volhard, 2016). One of the pathways uncovered by the screen for embryonic lethal fly mutants is the Toll pathway, which plays a role in dorso-ventral polarity in Drosophila embryos (Anderson et al., 1985a, b; Nüsslein-Volhard, 2022). This pathway was later discovered to regulate part of the innate immune responses in flies (Lemaitre, 2004) and eventually led to the discovery of Toll-like receptors (TLRs) as regulators of immunity in mammals (Medzhitov et al., 1997; Takeda and Akira, 2004). Not surprisingly, other genetic model systems, including plants and even vertebrates like zebrafish and mice, have successfully used mutagenesis to uncover basic mechanisms of development and identify unknown gene functions (Patton and Zon, 2001; Page and Grossniklaus, 2002; Kile and Hilton, 2005).
In the 60s, Benzer who had originally used mutant screens to dissect the nature of the gene in phage (Benzer, 1956; Benzer and Champe, 1961) became convinced this approach would allow him to dissect the genetic underpinnings of behavior. Just a few years earlier, Margaret Bastock had shown that a yellow pigmentation mutant affected courtship in flies (Bastock, 1956), which was the first demonstration that a single gene can affect behavior. This finding may have inspired Benzer (Cobb, 2007), and his group to embark on a series of screens premised on simple behavioral paradigms in Drosophila melanogaster (Greenspan, 1990). This effort again turned out to be very successful and led to the isolation of a series of behavior mutants, one of which was the first courtship mutant isolated from a screen (Gill, 1963). The mutant (fru1) affected the fruitless (fru) locus (Ito et al., 1996; Lee and Hall, 2001), which encodes a transcription factor necessary and sufficient for male courtship and mating (Villella and Hall, 2008; Sato and Yamamoto, 2020). In another screen, they isolated the first learning and memory mutant, dunce1 (dnc1) (Dudai et al., 1976). The dunce gene was later cloned and shown to encode cAMP phophodiesterase (Chen et al., 1986), further providing evidence to a growing body of knowledge that cAMP signaling plays an important role in memory formation across species (Alberini, 1999). Perhaps the most famous mutants isolated in those early days were the first circadian mutants (Konopka and Benzer, 1971). In a small screen of less than 2,000 mutants, they identified three alleles that mapped to the period locus: one arrhythmic (per0), one with a short period (perS), and the last one with a long period (perL) , one with a short period, and the last one with a long period (Konopka and Benzer, 1971). This simple screen changed the field of circadian biology although it took several more decades before the gene was cloned (Bargiello et al., 1984; Reddy et al., 1984) and the basic aspects of its molecular mechanisms became better understood (Hall, 2005). This process was further aided by additional screens that isolated other components of the pathway (Price, 2005) and by discoveries in mammalian systems, which helped further establish the transcriptional feedback loop that maintains the circadian clock (Honma, 2018).
Despite the potential of mutant screens to elucidate the underlying networks governing many facets of biology, they come with some drawbacks. Arguably the biggest hurdle to overcome in a saturation screen, in particular, is the sheer amount of labor involved. Depending on the number of genes that control the process of interest, performing a screen to saturation can take years (Wieschaus and Nüsslein-Volhard, 2016), and does not guarantee that most or all genes are identified. The workload is based on the genetic effort to make mutant strains (St Johnston, 2002) and on the time it takes to phenotype these mutants. For behavioral screens, this adds an additional layer of difficulty because the organism must be monitored for a sufficient amount of time to ensure that a statistically significantly measurable phenotype can be recorded. Once the phenotype in a specific mutant is confirmed, the causal locus has to be identified. This typically involves a mapping step that can itself be time-consuming (St Johnston, 2002). However, as whole-genome sequencing platforms have become cheaper, mapping can be skipped as long as the screen is deep enough that multiple alleles for each gene can be isolated in so-called complementation groups (Sarin et al., 2008, 2010; Hobert, 2010; Haelterman et al., 2014). When the mutants from the same complementation group are sequenced, only the causal gene will have a deleterious mutation in all the mutants. The assay that is used for phenotyping mutants is also very important for the success of a screen. It is critical that it is sensitive and specific enough to limit both false positives and false negatives. Despite these limitations, the expansive toolkit for genetic and molecular manipulation in fruit flies is one of the major factors enabling screens to be carried out efficiently in this organism even for behavior. This has led to many successful screens that have helped elucidate a range of behavioral phenotypes in flies such as olfaction (Helfand and Carlson, 1989; McKenna et al., 1989), hearing (Eberl et al., 1997), alcohol sensitivity (Singh and Heberlein, 2000), pain (Tracey et al., 2003), sleep (Cirelli et al., 2005; Stavropoulos and Young, 2011), and gravity sensing (Armstrong et al., 2006) to name only a few. Nevertheless, very few screens so far have been done to elucidate complex social behaviors such as aggression.
Here, we highlight the few small-scale unbiased screens (Hoopfer et al., 2015; Davis et al., 2018; Eddison, 2021) that have so far been performed to help figure out the underlying molecular and circuit mechanisms that control aggression in flies to illustrate the power of this approach. We also briefly discuss a recent behavioral pipeline (Chowdhury et al., 2021) that was optimized to perform high throughput screens for aggression in flies to capitalize on the strength of this approach to better understand this elusive phenotype.
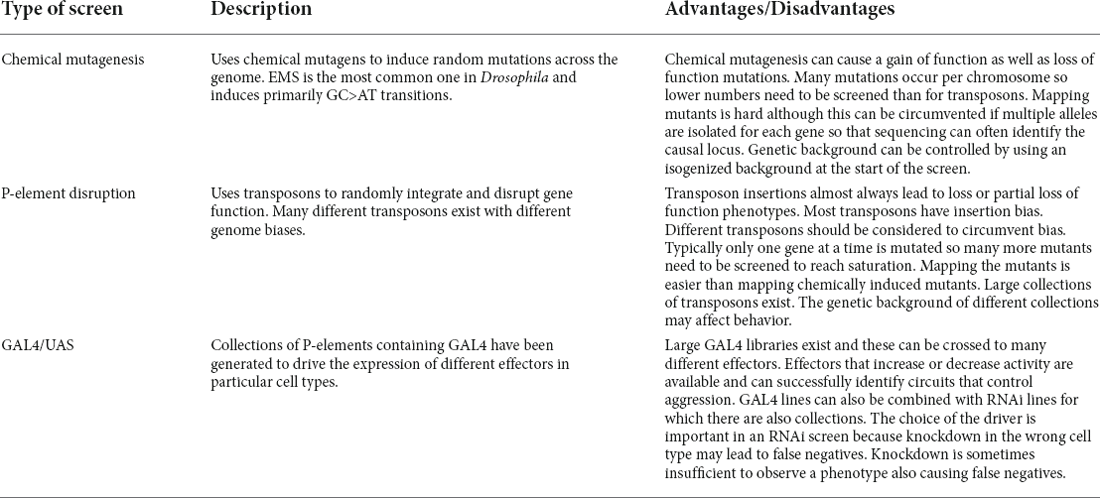
Chemical Mutagenesis Screen for Aggression
The original method to induce random mutations in the genome in most early screens including Drosophila was done with ionizing radiation, but most random mutant screens now use chemicals or transposons (see below, Table 1). The most commonly used chemical mutagen in flies is ethyl methanesulfonate (EMS) although other agents can and have also been used (St Johnston, 2002; Greenspan, 2004; Venken and Bellen, 2014). EMS is an alkylating agent that primarily induces random mutations via GC > AT transitions although other changes are also possible, including small deletions and insertions (Arrizabalaga and Lehmann, 1999). In the standard protocol male flies are fed a solution of 25 mM EMS in 1% sucrose overnight and crossed to untreated females to produce mutant offspring each unique in their repertoire of induced mutations (Lewis and Bacher, 1968). Lower concentrations have also been used as they lead to a lower number of mutations (Haelterman et al., 2014), which may be beneficial for screens where flies have to behave instead of merely survive. The use of EMS inevitably results in lower mutation rates in small genes creating a bias towards mutating larger genes. Thus, the chances of recovering novel alleles of small genes are very low in a small-scale mutagenesis screen, and often require significantly increasing the number of flies that are mutagenized. Despite this drawback, mutagenesis screens are still useful in identifying novel genes involved in a given process.
Genetically the easiest and quickest EMS-induced mutant screen is an F1 screen for viable mutants on the X-chromosome. To make this happen, mutagenized males are crossed to attached-X females (X^X/Y females) and pass on their mutagenized X’s patroclinously to their sons (Bridges, 1914; Morgan, 1922) because the attached-X chromosome passes to the next generation as two X’s, inevitably making a female. To produce fertile males in this cross, the attached-X females also carry a Y-chromosome that they pass on to their sons, making them mutagenized-X/Y and fertile (XO males are sterile). This cross scheme was used in the original circadian mutant screen that identified the three per alleles (Konopka and Benzer, 1971). Because aggression cannot be tested on a single male, an extra generation is required by simply crossing each unique F1 mutant male again to X^X/Y females to produce F2 males that all have the same mutant X as their F1 fathers. Rather than testing these F2 males against each other in an assay to measure aggression directly, Davis and colleagues used a secondary phenotype to perform the screen (Davis et al., 2018). They had found that hyper-aggressive males cause wing damage to each other’s wings when they are group-housed. Using this simple proxy phenotype, they screened approximately 1,400 viable fertile mutants and found five that also had an increased aggression phenotype. After performing whole genome sequencing on all the mutants, they found that each had on average about 30 altered coding variations that could be responsible for the mutant phenotype. To rapidly sort through these, they crossed each mutant to a defined set of duplications on the X-chromosome (Venken et al., 2010) that each covered one of the candidate mutations to test whether any of these rescued the mutant phenotype. This strategy led them to find a novel mutation in Shaker responsible for one of the mutant’s increased aggression phenotype (Davis et al., 2018). Shaker is the major voltage-gated potassium channel in Drosophila but how this mutation affects aggression remains unclear. This screen was relatively fast as the primary screen including mutagenesis and F2 screen took only 3 months while the secondary screen to specifically assess aggression took two more months. However, of the 40 mutant hits in the primary wing damage screen, only five also showed increased aggression. This suggests that there were both false positives and likely false negatives given the low overall number of positives. Clearly, a better primary assay would improve the hit rate on the screen. Below we will discuss a newer assay with high specificity, high sensitivity, and is amenable to high throughput analysis (Chowdhury et al., 2021).
P-Element Insertion Screens for Aggression
P-elements are transposable elements whose sequence encodes a transposase and terminally located inverted repeats that direct transposition (Engels, 1983). Random insertion of the P-element into a gene can disrupt its function, which has led to its use as a mutagen for screens (Cooley et al., 1988). However, because P-element insertions are biased toward certain regions of the genome (Venken and Bellen, 2014) the most common method of using P-elements in a screen is to screen through P-element insertion libraries. These are collections of flies where each strain contains a P-element insertion in a unique gene. For example, the library generated by the Berkeley Drosophila Genome Project comprises 1,045 strains with single P-element insertions disrupting more than 25% of essential genes (Spradling et al., 1999). Many of the insertion sites have been precisely mapped, greatly reducing the amount of time and labor required in traditional mutagenesis screens to pinpoint and characterize the disruptive mutation. Similarly, the Kyoto stock center has a collection of 6,900 PGS lines that were generated by the Drosophila Gene Search Project and that can be used for gain-of-function approaches (Toba et al., 1999).
Some P-element insertion libraries are not publicly available but are instead generated and housed by individual labs. Some of these libraries incorporate the GAL4/UAS system (Brand and Perrimon, 1993) to allow for a broader range of manipulations than simply disrupting gene function. A recent forward genetic screen used a P[GAL4] insertion library of 1,606 lines with random insertions of GAL4 throughout the fly genome to screen for mutants with altered behavioral susceptibility to social isolation, including aggression (Eddison, 2021). Forced social isolation in D. melanogaster is known to increase aggression levels (Hoffmann and Cacoyianni, 1990; Wang et al., 2008), locomotor activity (Panova et al., 2013), and resistance to ethanol sedation (Eddison et al., 2011), but most of the underlying genes remain unknown. The screen identified a mutant, sex pistols (sxp) that enhances the effect of social isolation on aggression and resistance to ethanol sedation, and also showed strong male-male courtship. The P-element in this strain is inserted between the first two non-coding exons of hu-li tai shao (hts, an ortholog of adducin) and is close to the start codon of CalpA (encoding a calcium-dependent protease). By capitalizing on the embedded GAL4 driver in this transposon insertion, the author was able to use RNAi lines against hts and CalpA to test the causality of the two genes in the phenotypes affected in this mutant to further unravel the phenotype (Eddison, 2021). Knockdown experiments and genomic rescue suggested that the increased aggression phenotype is due to the reduction of hts and that increased courtship is due to reduced levels of CalpA in sxp mutants.
The use of GAL4 insertion libraries in screens is not limited to just dissecting gene function to identify the genetic interactions that give rise to behaviors, but also to understand the underlying circuits. This is important because understanding the mechanisms that govern behavior not only requires mapping the underlying genetic networks, but also the neural circuitry that drives the behavior. For this reason, a collection of almost 7,000 GAL4 lines was created using site-specific integration to insert GAL4 driven by defined genomic DNA fragments from some 1,200 neuronal genes to serve as specific transcriptional enhancers (Jenett et al., 2012). All of these driver lines were analyzed for expression in the adult brain and ventral nerve cord by crossing them to a fluorescent reporter (Jenett et al., 2012). Many of these are likely expressed outside of the nervous system and most of these fragments only show limited resemblance to the expression pattern of the gene from which the fragment is derived, but the collection is nevertheless very useful to investigate circuit connectivity and function. Hoopfer and colleagues used a subset of this collection with the narrowest expression patterns to look for neurons that promote aggression (Hoopfer et al., 2015). They examined aggression in about 2,200 strains that were crossed to UAS-TrpA1. Expression of this thermosensitive ion channel increases the neuronal activity in the circuit acutely by raising the temperature of the flies (Hamada et al., 2008). Nineteen of the lines they tested showed a strong increase in aggression at the elevated temperature and three of them also showed increased male-male courtship, a phenotype associated with mutations in the fruitless (fru) locus, one of the mutants originally isolated in the Benzer lab (Gill, 1963). Two of these showed overlapping expression in a small set of neurons known as P1 neurons, which are part of the fruitless circuit in the brain known to play a key role in male-specific behaviors (Villella and Hall, 2008). Depending on the strength of activation of these neurons, the males either engaged in fighting or courtship (Hoopfer et al., 2015). P1 neurons are part of a larger network of so-called pC1 neurons, some of which also express doublesex, another sex determination regulator. A subset of these pC1 neurons that are fru-negative and dsx-positive strongly increase aggression when activated, while fru-positive, dsx-negative pC1 neurons drive courtship when activated, suggesting this circuit acts as a switch between these normally mutually exclusive behaviors (Koganezawa et al., 2016).
An in-depth analysis of a different line identified in the screen turned out to be important to regulate wing threat (Duistermars et al., 2018), a phenotype also long known to be part of the aggression repertoire in Drosophila males (Jacobs, 1960).
Divider Assay for High Throughput Aggression Analysis
One of the important bottlenecks in genetic screens is the workload to generate the unique mutants and phenotype them. One of the screens (Davis et al., 2018) we highlighted used a simple-to-score wing damage phenotype, which reduced phenotyping workload but also reduced the sensitivity and specificity of the screen. The other two screens (Hoopfer et al., 2015; Eddison, 2021) used existing genetic resources but directly measured aggression by using automated video analysis methods (Dankert et al., 2009; Kabra et al., 2013; Eyjolfsdottir et al., 2014). Despite the automation of behavioral analysis, these screens were still slow because setting up aggression experiments is also labor-intensive. Flies need to be collected, isolated, and loaded in multiple pairs to have enough statistical power to measure a significant phenotypic effect.
A recently developed Divider Assay (Figure 1; Chowdhury et al., 2021) solves the phenotyping bottleneck and should significantly improve the ability to perform high throughput screens. This method uses a cheap 3D printed chamber with 12 square arenas. Each arena can be divided in half by a removable opaque divider allowing each pair of males to be separated on either side of the divider. Flies can now be collected, loaded, and isolated in a single step rather than in separate steps, reducing the time it takes to set up an experiment to just a few minutes. Combined with automated behavioral video analysis that has high specificity and sensitivity, this assay makes phenotyping mutants fast and should be a useful advancement to perform high throughput genetic screens.
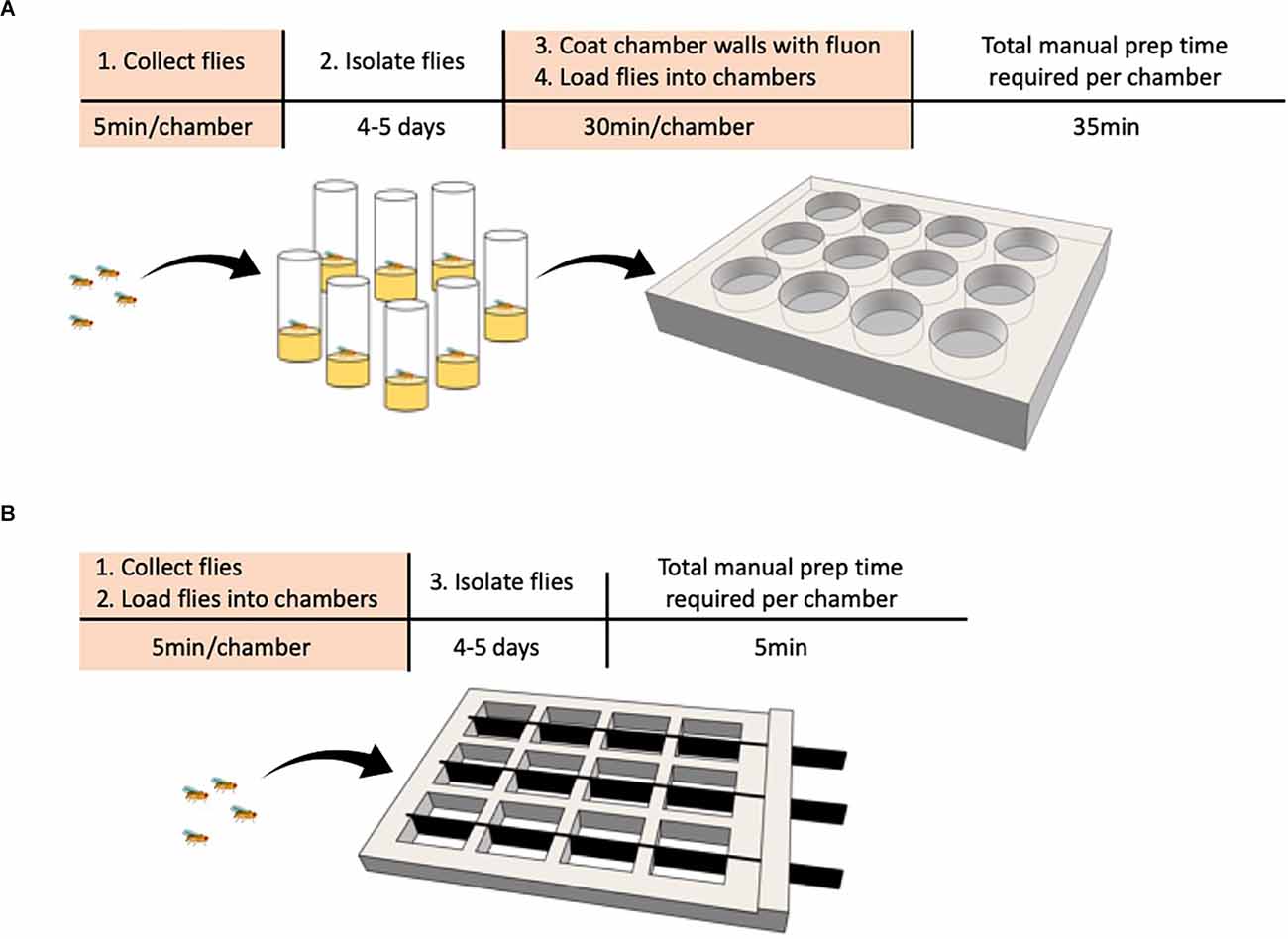
Figure 1. Comparison of behavioral chamber setup to streamline phenotyping in a screen. (A,B) Steps requiring manual labor are highlighted in orange. (A) Traditional method. The experimenter collects male flies and loads one male fly per vial. The flies are grown in isolation for 4–5 days, then transferred to the behavioral chamber with one pair per circular well before behavior is recorded. (B) Divider assay. The experimenter collects male flies and loads two flies per chamber, one on each side of the removable divider. The flies are grown in isolation for 4–5 days, the dividers removed, and behavior is recorded.
Discussion
Aggression is a complex behavioral phenotype that, despite decades of work, is still not very well understood. In the last two decades, Drosophila has emerged as a powerful model to study this behavior. Since its original detailed description (Jacobs, 1960) and repeated rediscovery (Dow and Schilcher, 1975; Hoffmann, 1987a, b, Hoffmann, 1989; Chen et al., 2002), we have learned much about the biology of aggression in this species. Aggression is heritable (Hoffmann, 1988) and selectable (Hoffmann and Cacoyianni, 1989; Dierick and Greenspan, 2006), modulated by biogenic amines and neuropeptides (Certel et al., 2007; Dierick and Greenspan, 2007; Hoyer et al., 2008; Zhou et al., 2008; Alekseyenko et al., 2010, 2014; Andrews et al., 2014; Asahina et al., 2014; Davis et al., 2014; Thomas et al., 2015; Wu et al., 2020), some of which are controlled by conserved transcriptional regulators (Davis et al., 2014; Thomas et al., 2015), influenced by pheromonal cues and receptors (Fernández et al., 2010; Wang and Anderson, 2010; Liu et al., 2011; Wang et al., 2011; Andrews et al., 2014), and controlled by sex determination pathways (Vrontou et al., 2006; Chan and Kravitz, 2007; Asahina et al., 2014; Hoopfer et al., 2015; Koganezawa et al., 2016; Wohl et al., 2020) to name only some of the recent mechanistic discoveries. Many of these findings were discovered by analyzing conserved pathways or by pursuing educated guesses rather than through unbiased screens. In addition, female aggression has also been described in Drosophila and found to be both qualitatively and quantitatively different from male aggression (Nilsen et al., 2004; Ueda and Wu, 2009; Schretter et al., 2020).
Despite the strong and long history of Drosophila as a genetic model system (Roberts, 2006) with tremendous impact on neuroscience (Bellen et al., 2010) and behavior (Vosshall, 2007), genetic screens have been sparsely used to uncover the mysteries of aggression (Hoopfer et al., 2015; Davis et al., 2018; Eddison, 2021) and so far only in males. This is likely in part due to the workload associated with genetic screens with the added difficulty of the need to carefully measure a complex behavioral phenotype. However, automated video analysis paradigms for aggression have been around for more than a decade (Hoyer et al., 2008; Dankert et al., 2009), and they have been further improved since then (Kabra et al., 2013; Eyjolfsdottir et al., 2014) and yet only three genetic screens have been done. Two of these even used existing strain collections combined with automated video analysis paradigms (Hoopfer et al., 2015; Eddison, 2021), and one of the screens used a simple proxy phenotype as a primary screen (Davis et al., 2018), suggesting phenotyping remains a bottleneck. A recent simplified experimental setup reduces the phenotyping workload further (Chowdhury et al., 2021) and it will be interesting to see if this reduces the roadblocks to future genetic screens.
What should be the focus going forward? History tells us that unbiased genetic screens are the most successful strategy to identify novel mechanisms for any phenotype. Circuit screens will identify the specific circuits that affect behavior and mutant screens will identify the genes that are important for aggression. Many genetic screens look for mutant phenotypes in both directions, but for aggression that is not advisable because increasing aggression in a mutant is much more likely to be specific than decreasing aggression. For example, a mutant that is sick due to a metabolic defect is likely to fight less, but the underlying cause may have nothing to do with regulatory mechanisms of aggression. In addition, the baseline level of aggression of wild-type flies is quite low (although it can be modulated using food and social isolation, and is also dependent on the assay being used) and in order to reliably observe a decrease in the phenotype, it would be necessary to start with a high aggression mutant. This may be a useful strategy but this type of suppressor screen should not be the first priority in screening. As long as we have no good idea how many genes are involved in aggression regulation, a forward genetic screen is the most straightforward strategy to find comprehensive answers. Once a mutant is identified, it has to first be confirmed. A mutant can be confirmed through rescue with a genomic rescue construct or by recreating it in a different strain through genome editing, for example with Crispr/Cas9 (Zirin et al., 2021). An added benefit of using this method is that a GAL4 can be knocked into the locus and expression of the gene can be evaluated. This can link gene function to circuit function. Structure-function analysis of the gene can further help elucidate the mechanism of action of the gene.
Large-scale screens are daunting because they can take a long time (Wieschaus and Nüsslein-Volhard, 2016). However, they also have significant benefits. First, when enough mutants are screened, multiple alleles will likely be identified. Mutants are placed in complementation groups by crossing them together (St Johnston, 2002). As most mutants are recessive, an outcross to wild-type will make the phenotype disappear. Crossing recessive mutants together will identify the ones that fail to complement. Most of these will be part of the same complementation group and represent different alleles of the same gene. This will significantly simplify finding the causal variant without a time-consuming mapping step through whole-genome sequencing (Sarin et al., 2008, 2010; Hobert, 2010; Haelterman et al., 2014). Sometimes mutants will fail to complement without being alleles of the same gene, known as non-allelic non-complementation or compound haploinsufficiency. These mutants almost certainly are interesting because they likely encode components of a complex and this will give mechanistic insight into pathways that regulate the phenotype. Identifying the expression pattern of such mutants may narrow down the circuit where these genes are important. Most genes are pleiotropic but components of a complex almost certainly work together in the same cells in their effect on the phenotype and finding the overlap in their expression will likely refine the possible circuit involved in the phenotype again linking gene function to circuit function.
While genetic screens are often viewed as fishing expeditions, many have generated discoveries that have transformed our understanding of biology. It is clear that much remains to be discovered in this exciting field and genetic screens will likely play an important role in that endeavor.
Author Contributions
GH and HD co-wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Institutes of Health (RO1GM109938 and RO1MH107474). GH is supported by Training Grant (T32) GM139534.
Acknowledgments
We thank Dr. Christophe Herman for valuable discussions and feedback on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alberini, C. M. (1999). Genes to remember. J. Exp. Biol. 202, 2887–2891. doi: 10.1242/jeb.202.21.2887
Alekseyenko, O. V., Chan, Y.-B., Fernandez, M. d. l. P., Bülow, T., Pankratz, M. J., and Kravitz, E. A. (2014). Single serotonergic neurons that modulate aggression in Drosophila. Curr. Biol. 24, 2700–2707. doi: 10.1016/j.cub.2014.09.051
Alekseyenko, O. V., Lee, C., and Kravitz, E. A. (2010). Targeted manipulation of serotonergic neurotransmission affects the escalation of aggression in adult male Drosophila melanogaster. PLoS One 5:e10806. doi: 10.1371/journal.pone.0010806
Anderson, K. V., Bokla, L., and Nüsslein-Volhard, C. (1985a). Establishment of dorsal-ventral polarity in the Drosophila embryo: the induction of polarity by the toll gene product. Cell 42, 791–798. doi: 10.1016/0092-8674(85)90275-2
Anderson, K. V., Jürgens, G., and Nüsslein-Volhard, C. (1985b). Establishment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell 42, 779–789. doi: 10.1016/0092-8674(85)90274-0
Andrews, J. C., Fernández, M. P., Yu, Q., Leary, G. P., Leung, A. K. W., Kavanaugh, M. P., et al. (2014). Octopamine neuromodulation regulates Gr32a-linked aggression and courtship pathways in Drosophila males. PLoS Genet. 10:e1004356. doi: 10.1371/journal.pgen.1004356
Armstrong, J. D., Texada, M. J., Munjaal, R., Baker, D. A., and Beckingham, K. M. (2006). Gravitaxis in Drosophila melanogaster: a forward genetic screen. Genes Brain Behav. 5, 222–239. doi: 10.1111/j.1601-183X.2005.00154.x
Arrizabalaga, G., and Lehmann, R. (1999). A selective screen reveals discrete functional domains in Drosophila nanos. Genetics 153, 1825–1838. doi: 10.1093/genetics/153.4.1825
Asahina, K., Watanabe, K., Duistermars, B. J., Hoopfer, E., González, C. R., Eyjólfsdóttir, E. A., et al. (2014). Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell 156, 221–235. doi: 10.1016/j.cell.2013.11.045
Bargiello, T. A., Jackson, F. R., and Young, M. W. (1984). Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 312, 752–754. doi: 10.1038/312752a0
Bastock, M. (1956). A gene mutation which changes a behavior pattern. Evolution 10, 421–439. doi: 10.1111/j.1558-5646.1956.tb02868.x
Beadle, G. W., and Tatum, E. L. (1941). Genetic control of biochemical reactions in neurospora. Proc. Natl. Acad. Sci. U S A 27, 499–506. doi: 10.1073/pnas.27.11.499
Bellen, H. J., Tong, C., and Tsuda, H. (2010). 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 11, 514–522. doi: 10.1038/nrn2839
Benzer, S. (1956). Genetic fine structure and its relation to the DNA molecule. Brookhaven Symp. Biol. 8, 3–5.
Benzer, S., and Champe, S. P. (1961). Ambivalent rII mutants of phage T4. Proc. Natl. Acad. Sci. U S A 47, 1025–1038. doi: 10.1073/pnas.47.7.1025
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. doi: 10.1242/dev.118.2.401
Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi: 10.1093/genetics/77.1.71
Bridges, C. B. (1914). Direct proof through non-disjunction that the sex-linked genes of Drosophila are borne by the X-chromosome. Science 40, 107–109. doi: 10.1126/science.40.1020.107
Calabrese, E. J. (2018). Was Muller’s 1946 Nobel Prize research for radiation-induced gene mutations peer-reviewed? Philos. Ethics Humanit. Med. 13:6. doi: 10.1186/s13010-018-0060-5
Certel, S. J., Savella, M. G., Schlegel, D. C. F., and Kravitz, E. A. (2007). Modulation of Drosophila male behavioral choice. Proc. Natl. Acad. Sci. U S A 104, 4706–4711. doi: 10.1073/pnas.0700328104
Chan, Y.-B., and Kravitz, E. A. (2007). Specific subgroups of FruM neurons control sexually dimorphic patterns of aggression in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 104, 19577–19582. doi: 10.1073/pnas.0709803104
Chen, C. N., Denome, S., and Davis, R. L. (1986). Molecular analysis of cDNA clones and the corresponding genomic coding sequences of the Drosophila dunce+ gene, the structural gene for cAMP phosphodiesterase. Proc. Natl. Acad. Sci. U S A 83, 9313–9317. doi: 10.1073/pnas.83.24.9313
Chen, S., Lee, A. Y., Bowens, N. M., Huber, R., and Kravitz, E. A. (2002). Fighting fruit flies: a model system for the study of aggression. Proc. Natl. Acad. Sci. U S A 99, 5664–5668. doi: 10.1073/pnas.082102599
Chowdhury, B., Wang, M., Gnerer, J. P., and Dierick, H. A. (2021). The divider assay is a high-throughput pipeline for aggression analysis in Drosophila. Commun. Biol. 4:85. doi: 10.1038/s42003-020-01617-6
Cirelli, C., Bushey, D., Hill, S., Huber, R., Kreber, R., Ganetzky, B., et al. (2005). Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092. doi: 10.1038/nature03486
Cobb, M. (2007). A gene mutation which changed animal behaviour: margaret Bastock and the yellow fly. Anim. Behav. 74, 163–169. doi: 10.1016/j.anbehav.2007.05.002
Cooley, L., Kelley, R., and Spradling, A. (1988). Insertional mutagenesis of the Drosophila genome with single P elements. Science 239, 1121–1128. doi: 10.1126/science.2830671
Dankert, H., Wang, L., Hoopfer, E. D., Anderson, D. J., and Perona, P. (2009). Automated monitoring and analysis of social behavior in Drosophila. Nat. Methods 6, 297–303. doi: 10.1038/nmeth.1310
Davis, S. M., Thomas, A. L., Liu, L., Campbell, I. M., and Dierick, H. A. (2018). Isolation of aggressive behavior mutants in Drosophila using a screen for wing damage. Genetics 208, 273–282. doi: 10.1534/genetics.117.300292
Davis, S. M., Thomas, A. L., Nomie, K. J., Huang, L., and Dierick, H. A. (2014). Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat. Commun. 5:3177. doi: 10.1038/ncomms4177
Dierick, H. A., and Greenspan, R. J. (2006). Molecular analysis of flies selected for aggressive behavior. Nat. Genet. 38, 1023–1031. doi: 10.1038/ng1864
Dierick, H. A., and Greenspan, R. J. (2007). Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat. Genet. 39, 678–682. doi: 10.1038/ng2029
Dow, M. A., and Schilcher, F. V. (1975). Aggression and mating success in Drosophila melanogaster. Nature 254, 511–512. doi: 10.1038/254511a0
Dudai, Y., Jan, Y. N., Byers, D., Quinn, W. G., and Benzer, S. (1976). dunce, a mutant of Drosophila deficient in learning. Proc. Natl. Acad. Sci. U S A 73, 1684–1688. doi: 10.1073/pnas.73.5.1684
Duistermars, B. J., Pfeiffer, B. D., Hoopfer, E. D., and Anderson, D. J. (2018). A brain module for scalable control of complex, multi-motor threat displays. Neuron 100, 1474–1490.e4. doi: 10.1016/j.neuron.2018.10.027
Eberl, D. F., Duyk, G. M., and Perrimon, N. (1997). A genetic screen for mutations that disrupt an auditory response in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 94, 14837–14842. doi: 10.1073/pnas.94.26.14837
Eddison, M. (2021). A genetic screen for Drosophila social isolation mutants and analysis of sex pistol. Sci. Rep. 11:17395. doi: 10.1038/s41598-021-96871-x
Eddison, M., Guarnieri, D. J., Cheng, L., Liu, C.-H., Moffat, K. G., Davis, G., et al. (2011). arouser reveals a role for synapse number in the regulation of ethanol sensitivity. Neuron 70, 979–990. doi: 10.1016/j.neuron.2011.03.030
Ellis, H., and Horvitz, H. (1986). Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829. doi: 10.1016/0092-8674(86)90004-8
Engels, W. R. (1983). The P family of transposable elements in Drosophila. Annu. Rev. Genet. 17, 315–344. doi: 10.1146/annurev.ge.17.120183.001531
Eyjolfsdottir, E., Branson, S., Burgos-Artizzu, X. P., Hoopfer, E. D., Schor, J., Anderson, D. J., et al. (2014). “Detecting social actions of fruit flies,” in Computer Vision - ECCV 2014 Lecture Notes in Computer Science (Vol. 8690), eds D. Fleet, T. Pajdla, B. Schiele, and T. Tuytelaars (Cham: Springer International Publishing), 772–787. doi: 10.1007/978-3-319-10605-2_50
Fernández, M. d. l. P., Chan, Y.-B., Yew, J. Y., Billeter, J.-C., Dreisewerd, K., Levine, J. D., et al. (2010). Pheromonal and behavioral cues trigger male-to-female aggression in Drosophila. PLoS Biol. 8:e1000541. doi: 10.1371/journal.pbio.1000541
Gager, C. S., and Blakeslee, A. F. (1927). Chromosome and gene mutations in datura following exposure to radium rays. Proc. Natl. Acad. Sci. U S A 13, 75–79. doi: 10.1073/pnas.13.2.75
Gill, K. (1963). A mutation causing abnormal mating behavior Drosophila information service. Am. Zool. 38:33.
Greenspan, R. J. (2004). Fly Pushing: The Theory And Practice Of Drosophila Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Haelterman, N. A., Jiang, L., Li, Y., Bayat, V., Sandoval, H., Ugur, B., et al. (2014). Large-scale identification of chemically induced mutations in Drosophila melanogaster. Genome Res. 24, 1707–1718. doi: 10.1101/gr.174615.114
Hall, J. C. (2005). Systems approaches to biological rhythms in Drosophila. Methods Enzymol. 393, 61–185. doi: 10.1016/S0076-6879(05)93004-8
Hamada, F. N., Rosenzweig, M., Kang, K., Pulver, S. R., Ghezzi, A., Jegla, T. J., et al. (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. doi: 10.1038/nature07001
Hartwell, L. H. (1971). Genetic control of the cell division cycle in yeast. J. Mol. Biol. 59, 183–194. doi: 10.1016/0022-2836(71)90420-7
Hartwell, L. H., Culotti, J., and Reid, B. (1970). Genetic control of the cell-division cycle in yeast, I. detection of mutants. Proc. Natl. Acad. Sci. U S A 66, 352–359. doi: 10.1073/pnas.66.2.352
Helfand, S. L., and Carlson, J. R. (1989). Isolation and characterization of an olfactory mutant in Drosophila with a chemically specific defect. Proc. Natl. Acad. Sci. U S A 86, 2908–2912. doi: 10.1073/pnas.86.8.2908
Hobert, O. (2010). The impact of whole genome sequencing on model system genetics: get ready for the ride. Genetics 184, 317–319. doi: 10.1534/genetics.109.112938
Hoffmann, A. A. (1987a). A laboratory study of male territoriality in the sibling species Drosophila melanogaster and D. simulans. Anim. Behav. 35, 807–818. doi: 10.1016/S0003-3472(87)80117-3
Hoffmann, A. A. (1987b). Territorial encounters between Drosophila males of different sizes. Anim. Behav. 35, 1899–1901. doi: 10.1016/S0003-3472(87)80085-4
Hoffmann, A. A. (1988). Heritable variation for territorial success in two Drosophila melanogaster populations. Anim. Behav. 36, 1180–1189. doi: 10.1016/S0003-3472(88)80077-0
Hoffmann, A. A. (1989). Geographic variation in the territorial success of Drosophila melanogaster males. Behav. Genet. 19, 241–255. doi: 10.1007/BF01065908
Hoffmann, A. A., and Cacoyianni, Z. (1989). Selection for territoriality in Drosophila melanogaster: correlated responses in mating success and other fitness components. Anim. Behav. 38, 23–34. doi: 10.1016/S0003-3472(89)80062-4
Hoffmann, A. A., and Cacoyianni, Z. (1990). Territoriality in Drosophila melanogaster as a conditional strategy. Anim. Behav. 40, 526–537. doi: 10.1016/S0003-3472(05)80533-0
Honma, S. (2018). The mammalian circadian system: a hierarchical multi-oscillator structure for generating circadian rhythm. J. Physiol. Sci. 68, 207–219. doi: 10.1007/s12576-018-0597-5
Hoopfer, E. D., Jung, Y., Inagaki, H. K., Rubin, G. M., and Anderson, D. J. (2015). P1 interneurons promote a persistent internal state that enhances inter-male aggression in Drosophila. eLife 4:e11346. doi: 10.7554/eLife.11346
Horowitz, N. H., Berg, P., Singer, M., Lederberg, J., Susman, M., Doebley, J., et al. (2004). A centennial: George W. Beadle, 1903–1989. Genetics 166, 1–10. doi: 10.1534/genetics.166.1.1
Hoyer, S. C., Eckart, A., Herrel, A., Zars, T., Fischer, S. A., Hardie, S. L., et al. (2008). Octopamine in male aggression of Drosophila. Curr. Biol. 18, 159–167. doi: 10.1016/j.cub.2007.12.052
Ito, H., Fujitani, K., Usui, K., Shimizu-Nishikawa, K., Tanaka, S., and Yamamoto, D. (1996). Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. U S A 93, 9687–9692. doi: 10.1073/pnas.93.18.9687
Jacobs, M. E. (1960). Influence of light on mating of Drosophila melanogaster. Ecology 41, 182–188. doi: 10.2307/1931952
Jenett, A., Rubin, G. M., Ngo, T.-T. B., Shepherd, D., Murphy, C., Dionne, H., et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. doi: 10.1016/j.celrep.2012.09.011
Jürgens, G., Wieschaus, E., Nüsslein-Volhard, C., and Kluding, H. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: II. Zygotic loci on the third chromosome. Wilhelm. Roux Archiv. Dev. Biol. 193, 283–295. doi: 10.1007/BF00848157
Kabra, M., Robie, A. A., Rivera-Alba, M., Branson, S., and Branson, K. (2013). JAABA: interactive machine learning for automatic annotation of animal behavior. Nat. Methods 10, 64–67. doi: 10.1038/nmeth.2281
Kenyon, C. J., and Walker, G. C. (1980). DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc. Natl. Acad. Sci. U S A 77, 2819–2823. doi: 10.1073/pnas.77.5.2819
Kenyon, C., Chang, J., Gensch, E., Rudner, A., and Tabtiang, R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. doi: 10.1038/366461a0
Kile, B. T., and Hilton, D. J. (2005). The art and design of genetic screens: mouse. Nat. Rev. Genet. 6, 557–567. doi: 10.1038/nrg1636
Koganezawa, M., Kimura, K., and Yamamoto, D. (2016). The neural circuitry that functions as a switch for courtship versus aggression in Drosophila males. Curr. Biol. 26, 1395–1403. doi: 10.1016/j.cub.2016.04.017
Konopka, R. J., and Benzer, S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 68, 2112–2116. doi: 10.1073/pnas.68.9.2112
Lee, G., and Hall, J. C. (2001). Abnormalities of male-specific FRU protein and serotonin expression in the CNS of fruitless mutants in Drosophila. J. Neurosci. 21, 513–526. doi: 10.1523/JNEUROSCI.21-02-00513.2001
Lemaitre, B., Kromer-Metzger, E., Michaut, L., Nicolas, E., Meister, M., Georgel, P., et al. (1995). A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. U S A 92, 9465–9469. doi: 10.1073/pnas.92.21.9465
Lewis, E., and Bacher, F. (1968). Method of feeding ethyl methane sulfonate (EMS) to Drosophila males. Drosophila Info. Serv. 43:193.
Liu, W., Liang, X., Gong, J., Yang, Z., Zhang, Y.-H., Zhang, J.-X., et al. (2011). Social regulation of aggression by pheromonal activation of Or65a olfactory neurons in Drosophila. Nat. Neurosci. 14, 896–902. doi: 10.1038/nn.2836
McKenna, M., Monte, P., Helfand, S. L., Woodard, C., and Carlson, J. (1989). A simple chemosensory response in Drosophila and the isolation of acj mutants in which it is affected. Proc. Natl. Acad. Sci. U S A 86, 8118–8122. doi: 10.1073/pnas.86.20.8118
Medzhitov, R., Preston-Hurlburt, P., and Janeway, C. A. (1997). A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397. doi: 10.1038/41131
Morgan, L. V. (1922). Non-criss-cross inheritance in Drosophila melanogaster. Biol. Bull. 42, 267–274. doi: 10.2307/1536473
Muller, H. (1927). Artificial transmutation of the gene. Science 66, 84–87. doi: 10.1126/science.66.1699.84
Muller, H. J. (1928). The production of mutations by X-rays. Proc. Natl. Acad. Sci. U S A 14, 714–726. doi: 10.1073/pnas.14.9.714
Nilsen, S. P., Chan, Y.-B., Huber, R., and Kravitz, E. A. (2004). Gender-selective patterns of aggressive behavior in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A 101, 12342–12347. doi: 10.1073/pnas.0404693101
Nurse, P. (1975). Genetic control of cell size at cell division in yeast. Nature 256, 547–551. doi: 10.1038/256547a0
Nüsslein-Volhard, C. (2022). The toll gene in Drosophila pattern formation. Trends Genet. 38, 231–245. doi: 10.1016/j.tig.2021.09.006
Nüsslein-Volhard, C., Wieschaus, E., and Kluding, H. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: I. Zygotic loci on the second chromosome. Wilhelm Roux Archiv. Dev. Biol. 193, 267–282. doi: 10.1007/BF00848156
Page, D. R., and Grossniklaus, U. (2002). The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet. 3, 124–136. doi: 10.1038/nrg730
Panova, A. A., Bragina, J. V., Danilenkova, L. V., Besedina, N. G., Kamysheva, E. A., Fedotov, S. A., et al. (2013). Group rearing leads to long-term changes in locomotor activity of Drosophila males. OJAS 03, 31–35. doi: 10.4236/ojas.2013.34a2004
Patton, E. E., and Zon, L. I. (2001). The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2, 956–966. doi: 10.1038/35103567
Price, J. L. (2005). Genetic screens for clock mutants in Drosophila. Methods Enzymol. 393, 35–60. doi: 10.1016/S0076-6879(05)93003-6
Reddy, P., Zehring, W. A., Wheeler, D. A., Pirrotta, V., Hadfield, C., Hall, J. C., et al. (1984). Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710. doi: 10.1016/0092-8674(84)90265-4
Riddle, D. L., Swanson, M. M., and Albert, P. S. (1981). Interacting genes in nematode dauer larva formation. Nature 290, 668–671. doi: 10.1038/290668a0
Roberts, D. B. (2006). Drosophila melanogaster: the model organism. Entomol. Exp. Appl. 121, 93–103. doi: 10.1111/j.1570-8703.2006.00474.x
Sarin, S., Bertrand, V., Bigelow, H., Boyanov, A., Doitsidou, M., Poole, R. J., et al. (2010). Analysis of multiple ethyl methanesulfonate-mutagenized caenorhabditis elegans strains by whole-genome sequencing. Genetics 185, 417–430. doi: 10.1534/genetics.110.116319
Sarin, S., Prabhu, S., O’Meara, M. M., Pe’er, I., and Hobert, O. (2008). Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat. Methods 5, 865–867. doi: 10.1038/nmeth.1249
Sato, K., and Yamamoto, D. (2020). The mode of action of fruitless: is it an easy matter to switch the sex? Genes Brain Behav. 19:e12606. doi: 10.1111/gbb.12606
Schekman, R., Esmon, B., Ferro-Novick, S., Field, C., and Novick, P. (1983). Yeast secretory mutants: isolation and characterization. Methods Enzymol. 96, 802–815. doi: 10.1016/s0076-6879(83)96068-8
Schretter, C. E., Aso, Y., Robie, A. A., Dreher, M., Dolan, M.-J., Chen, N., et al. (2020). Cell types and neuronal circuitry underlying female aggression in Drosophila. eLife 9:e58942. doi: 10.7554/eLife.58942
Singh, C. M., and Heberlein, U. (2000). Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol. Clin. Exp. Res. 24, 1127–1136. doi: 10.1111/j.1530-0277.2000.tb02075.x
Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., et al. (1999). The berkeley Drosophila genome project gene disruption project: single p-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177. doi: 10.1093/genetics/153.1.135
St Johnston, D. (2002). The art and design of genetic screens: Drosophila melanogaster. Nat. Rev. Genet. 3, 176–188. doi: 10.1038/nrg751
Stavropoulos, N., and Young, M. W. (2011). insomniac and cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72, 964–976. doi: 10.1016/j.neuron.2011.12.003
Strauss, B. S. (2016). Biochemical genetics and molecular biology: the contributions of George Beadle and Edward Tatum. Genetics 203, 13–20. doi: 10.1534/genetics.116.188995
Takeda, K., and Akira, S. (2004). TLR signaling pathways. Semin. Immunol. 16, 3–9. doi: 10.1016/j.smim.2003.10.003
Tatum, E. L., and Lederberg, J. (1947). Gene recombination in the bacterium Escherichia coli. J. Bacteriol. 53, 673–684. doi: 10.1128/jb.53.6.673-684.1947
Thomas, A. L., Davis, S. M., and Dierick, H. A. (2015). Of fighting flies, mice and men: are some of the molecular and neuronal mechanisms of aggression universal in the animal kingdom. PLoS Genet. 11:e1005416. doi: 10.1371/journal.pgen.1005416
Toba, G., Ohsako, T., Miyata, N., Ohtsuka, T., Seong, K. H., and Aigaki, T. (1999). The gene search system. a method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151, 725–737. doi: 10.1093/genetics/151.2.725
Tracey, W. D., Wilson, R. I., Laurent, G., and Benzer, S. (2003). Painless, a Drosophila gene essential for nociception. Cell 113, 261–273. doi: 10.1016/s0092-8674(03)00272-1
Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. doi: 10.1016/0014-5793(93)80398-e
Ueda, A., and Wu, C.-F. (2009). Effects of social isolation on neuromuscular excitability and aggressive behaviors in Drosophila: altered responses by Hk and gsts1, two mutations implicated in redox regulation. J. Neurogenet. 23, 378–394. doi: 10.3109/01677060903063026
Venken, K. J. T., and Bellen, H. J. (2014). Chemical mutagens, transposons and transgenes to interrogate gene function in Drosophila melanogaster. Methods 68, 15–28. doi: 10.1016/j.ymeth.2014.02.025
Venken, K. J. T., Popodi, E., Holtzman, S. L., Schulze, K. L., Park, S., Carlson, J. W., et al. (2010). A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics 186, 1111–1125. doi: 10.1534/genetics.110.121285
Villella, A., and Hall, J. C. (2008). Neurogenetics of courtship and mating in Drosophila. Adv. Genet. 62, 67–184. doi: 10.1016/S0065-2660(08)00603-2
Vrontou, E., Nilsen, S. P., Demir, E., Kravitz, E. A., and Dickson, B. J. (2006). fruitless regulates aggression and dominance in Drosophila. Nat. Neurosci. 9, 1469–1471. doi: 10.1038/nn1809
Wang, L., and Anderson, D. J. (2010). Identification of an aggression-promoting pheromone and its receptor neurons in Drosophila. Nature 463, 227–231. doi: 10.1038/nature08678
Wang, L., Dankert, H., Perona, P., and Anderson, D. J. (2008). A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc. Natl. Acad. Sci. U S A 105, 5657–5663. doi: 10.1073/pnas.0801327105
Wang, L., Han, X., Mehren, J., Hiroi, M., Billeter, J.-C., Miyamoto, T., et al. (2011). Hierarchical chemosensory5663 regulation of male-male social interactions in Drosophila. Nat. Neurosci. 14, 757–762. doi: 10.1038/nn.2800
Wieschaus, E., and Nüsslein-Volhard, C. (2016). The heidelberg screen for pattern mutants of Drosophila: a personal account. Annu. Rev. Cell Dev. Biol. 32, 1–46. doi: 10.1146/annurev-cellbio-113015-023138
Wieschaus, E., Nüsslein-Volhard, C., and Jürgens, G. (1984). Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: III. Zygotic loci on the X-chromosome and fourth chromosome. Wilhelm Roux Archiv. Dev. Biol. 193, 296–307. doi: 10.1007/BF00848158
Wohl, M., Ishii, K., and Asahina, K. (2020). Layered roles of fruitless isoforms in specification and function of male aggression-promoting neurons in Drosophila. eLife 9:e52702. doi: 10.7554/eLife.52702
Wu, F., Deng, B., Xiao, N., Wang, T., Li, Y., Wang, R., et al. (2020). A neuropeptide regulates fighting behavior in Drosophila melanogaster. eLife 9:e54229. doi: 10.7554/eLife.54229
Zhou, C., Rao, Y., and Rao, Y. (2008). A subset of octopaminergic neurons are important for Drosophila aggression. Nat. Neurosci. 11, 1059–1067. doi: 10.1038/nn.2164
Keywords: aggression, Drosophila melanogaster, genetic screen, chemical mutagenesis, P-element insertions
Citation: Huang G and Dierick HA (2022) The need for unbiased genetic screens to dissect aggression in Drosophila melanogaster. Front. Behav. Neurosci. 16:901453. 10.3389/fnbeh.2022.901453
Received: 19 May 2022; Accepted: 14 July 2022;
Published: 01 August 2022.
Edited by:
Maria de la Paz Fernandez, Columbia University, United StatesReviewed by:
Sarah J. Certel, University of Montana, United StatesSéverine Trannoy, UMR5169 Centre de Recherches sur la Cognition Animale (CRCA), France
Copyright © 2022 Huang and Dierick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Herman A. Dierick, Dierick@bcm.edu
 Gary Huang
Gary Huang Herman A. Dierick
Herman A. Dierick