Modeling the Heterodimer Interfaces of Melatonin Receptors
- 1Division of Life Science and the Biotechnology Research Institute, Hong Kong University of Science and Technology, Hong Kong, SAR China
- 2State Key Laboratory of Molecular Neuroscience, Molecular Neuroscience Center, Hong Kong University of Science and Technology, Hong Kong, SAR China
- 3Hong Kong Center for Neurodegenerative Diseases, Hong Kong Science Park, Hong Kong, SAR China
Melatonin receptors are Class A G protein-coupled receptors (GPCRs) that regulate a plethora of physiological activities in response to the rhythmic secretion of melatonin from the pineal gland. Melatonin is a key regulator in the control of circadian rhythm and has multiple functional roles in retinal physiology, memory, immunomodulation and tumorigenesis. The two subtypes of human melatonin receptors, termed MT1 and MT2, utilize overlapping signaling pathways although biased signaling properties have been reported in some cellular systems. With the emerging concept of GPCR dimerization, melatonin receptor heterodimers have been proposed to participate in system-biased signaling. Here, we used computational approaches to map the dimerization interfaces of known heterodimers of melatonin receptors, including MT1/MT2, MT1/GPR50, MT2/GPR50, and MT2/5-HT2C. By homology modeling and membrane protein docking analyses, we have identified putative preferred interface interactions within the different pairs of melatonin receptor dimers and provided plausible structural explanations for some of the unique pharmacological features of specific heterodimers previously reported. A thorough understanding of the molecular basis of melatonin receptor heterodimers may enable the development of new therapeutic approaches against aliments involving these heterodimeric receptors.
Introduction
Melatonin (N-acetyl-5-methoxytryptamine) is a neuroendocrine hormone which regulates multiple physiological and neuroendocrine functions. In humans, the functions of melatonin are mainly mediated by two subtypes of melatonin receptors: MT1 and MT2. Melatonin receptors are expressed in various brain regions such as the hypothalamus, hippocampus, and pineal gland, as well as in the retina and a host of peripheral tissues and organs that range from arteries, liver, and skin to the immune system [reviewed in Liu et al. (2016) and Cecon et al. (2018)]. One of the major roles of melatonin receptors is to regulate the circadian rhythm by directly acting on the suprachiasmatic nucleus of the hypothalamus (Waly and Hallworth, 2015). Melatonin receptors also have regulatory actions on sleep, immune functions, retinal physiology, and blood glucose [reviewed in Pandi-Perumal et al. (2008)].
The melatonin MT1 and MT2 receptors belong to the Class A G protein-coupled receptor (GPCR) superfamily and they share a high sequence identity of 68%. Upon activation, both receptors regulate similar signaling pathways primarily via Gi/o proteins and β-arrestin1/2 (Witt-Enderby et al., 2003), although they can also signal through Gq/11 and G16 proteins depending on the cellular milieu (Brydon et al., 1999; Chan et al., 2002). Despite the high sequence similarity, differences in the intrinsic signaling capacity of MT1 and MT2 have been documented: MT2 has been shown to inhibit cGMP production (Petit et al., 1999) while MT1 can activate Gs (Chen et al., 2014) in some experimental models.
In the classical view, GPCR signaling is based on a ternary complex consisting of a receptor monomer, a ligand, and a G protein heterotrimer. However, a new paradigm involving receptor dimerization (and even oligomerization) has emerged as an important concept in GPCR signaling (Terrillon and Bouvier, 2004; Wang et al., 2018). The notion of GPCR dimerization/oligomerization has garnered substantial evidence from cross-linking assays, quantitative luminescence/fluorescence studies (e.g., FRET and BRET), X-ray resolved oligomeric structures, as well as computational analyses (e.g., molecular dynamics simulations). A large number of GPCR homo- and heterodimers have been documented (Prinster et al., 2005; Milligan et al., 2019). This change in the fundamental concept of GPCR signaling has brought along a huge impact on drug development. Hence, a thorough understanding on the specificity of GPCR connectivity is essential for the design of novel drugs that target receptor dimers.
Dimerization of melatonin MT1 and MT2 receptors with each other or with other GPCRs has been reported (Table 1). Co-immunoprecipitation and BRET assays in HEK 293 cells have confirmed that MT1 and MT2 can form constitutive homo- and heterodimers (Ayoub et al., 2002, 2004). A distinct signaling capacity of the MT1/MT2 heterodimer was further demonstrated in W661 cells that express both receptors endogenously (Sánchez-Bretaño et al., 2019), wherein the heterodimer mediates the melatonin-induced inhibition of adenylyl cyclase and phosphorylation of AKT/FoxO1. MT1/MT2 heterodimers are also reported to mediate the effect of melatonin on light sensitivity of mouse rod photoreceptor cells (Baba et al., 2013). In both studies, depletion of either protomer would disrupt signaling, indicating the requirement of a cooperative action of MT1 and MT2. GPR50, an orphan receptor which is structurally related to the melatonin receptors with no melatonin binding capacity, can also heterodimerize with either of the melatonin receptors in a constitutive manner (Levoye et al., 2006; Dufourny et al., 2008). The heterodimerization of GPR50 with MT1 abolishes the agonist binding and G protein-coupling capacity of the MT1 protomer, whereas it does not alter that of MT2 in the MT2/GPR50 heterodimer (Levoye et al., 2006). Another Class A GPCR, serotonin receptor 2C (5-HT2C) has been reported to form dimers with MT2, wherein the serotonin-induced Gq signaling pathway of 5-HT2C is amplified upon dimerization with MT2 (Kamal et al., 2015). Interestingly, a melatonin-mediated unidirectional transactivation of 5-HT2C protomer is associated with the MT2/5-HT2C heterodimer (Kamal et al., 2015).
The formation of different melatonin receptor heterodimers is likely to serve as a regulatory mechanism in specific cell types to exert distinct functions. However, despite the validation of heterodimerization of melatonin receptor subtypes by proximity-based assays and co-immunoprecipitation (Cecon et al., 2018), the extent of the cooperativity across the dimer interface remains largely unexplored. GPCR heterodimerization has been extensively studied by biophysical and biochemical methods (Angers et al., 2001; Vidi et al., 2010; Borroto-Escuela et al., 2016; Fuxe and Borroto-Escuela, 2016), yet the structural basis of their interactions remains poorly understood. For a GPCR such as MT2 that can form distinct heterodimers, a key question is whether it dimerizes with different partners via the same interface. Here, we used computational approaches to examine the preferred interface on the MT2 protomer across different MT2 heterodimers to further understand the structural basis of dimer formation.
Materials and Methods
Homology Modeling
The homology models of human CXCR4, MOR, DOR, MT1, MT2, GPR50, and 5-HT2C in an inactive conformation were constructed using the Modeler 9.24 (Webb and Sali, 2016). Protein sequences of the human CXCR4, MOR, DOR, MT1, MT2, GPR50, and 5-HT2C were obtained from the UniProt database.1 Mutations in the crystal structures of CXCR4 (PDB code 3ODU), MOR (PDB code 4DKL), DOR (PDB code 4N6H), MT1 (PDB code 6ME2), MT2 (PDB code 6ME6), and 5-HT2C (PDB code 6BQH) were rebuilt into wild-type structures. Intracellular loop 3 (IL3) of the receptors was omitted by chain break to avoid uncertainty during the loop prediction. The model of GPR50 was generated based on the crystal structure of its most closely related receptor, MT1 (51.1% identity, PDB code 6ME2). Selection of the final homology models was based on the DOPE scoring function (Shen and Sali, 2006) and visual inspection, followed by validation using PROCHECK2 (Laskowski et al., 1993; Rullmann, 1996).
Membrane Protein Docking
Rosetta 3.12 MPdock was used to study the potential interfaces of melatonin receptor heterodimers (Gray et al., 2003; Chaudhury et al., 2011; Alford et al., 2015). The homology models of CXCR4, MOR, DOR, MT1, MT2, GPR50, and 5-HT2C were firstly superposed on GPCR dimer structures representative of interfaces I, II, and III to obtain initial dimer structures of CXCR4/CXCR4, MOR/DOR, MT1/MT2, MT1/GPR50, MT2/GPR50, and MT2/5-HT2C with each pair in three distinct orientations. The dimer templates 6OFJ (Interface I), 5O9H (Interface II), and 4DKL (Interface III) were chosen based on the inactive conformation of receptors and the relative orientation between protomers that allow more flexible docking. The membrane spanning information of the initial dimers were obtained from PPM server (Lomize et al., 2012) and span files that contain the spanning topology of proteins were generated with Rosetta 3.12 (Alford et al., 2015). A pre-packing step was undertaken in membrane embedding constant, which optimizes side chains of protomers before docking. Ten models were generated for each heterodimer pair and each interface, and the model with the lowest Rosetta total energy was selected as the input for the membrane protein–protein docking using Rosetta MPdock (Alford et al., 2015). For each heterodimer pair, 1,000 models were generated for interfaces I, II, and III. The top 50% model output with lower Rosetta total score were sorted according to their Rosetta interface score, and the best 10 models for each interface were exported for further analysis.
Scoring, Analysis, and Mutagenesis of the Predicted Heterodimer Models
The top 10 models for each orientation of a dimer pair were then submitted to the PRODIGY web server3 to assess the free energy of binding and dissociation constant (at 37°C; Xue et al., 2016). The interface area of models was measured by UCSF Chimera using default parameters (Pettersen et al., 2004; Ray et al., 2005). Receptor structures visualization and in silico mutagenesis was performed using PyMOL Molecular Graphic System 2.1 (Schrodinger, LLC).
Results
Homology Models
The homology models of C–X–C chemokine receptor type 4 (CXCR4), μ-opioid receptor (MOR), δ-opioid receptor (DOR), MT1, MT2, GPR50, and 5-HT2C were built by Modeler 9.24 and assessed with the PROCHECK server with the Ramachandran plots shown (Supplementary Figure 1). According to the PROCHECK evaluation, 94.4, 97.8, 96.6, 96.3, 95.1, 91.1, and 90.3% of the residues are in the most favored regions for CXCR4, MOR, DOR, MT1, MT2, GPR50, and 5-HT2C, respectively. No residues are found in disallowed regions for all the models. All homology models share a high structural similarity with their corresponding crystal structures (i.e., CXCR4 vs. 3ODU, MOR vs. 4DKL, DOR vs. 4N6H, MT1 vs. 6ME2, MT2 vs. 6ME6, GPR50 vs. 6ME2, and 5-HT2C vs. 6BQH) with RMSD values of 0.287, 0.140, 0.141, 0.155, 0.129, 0.118, and 0.300 Å, respectively.
Heterodimer Interfaces
The assembly of GPCR dimers or oligomers can be achieved via multiple interfaces. Both symmetric [protomers interact with each other using the same set of transmembrane helices (TMs)] and asymmetric (protomers interact with each other via different sets of TMs) dimer conformations have been described, with most of the Class A GPCRs adopting the symmetric dimer interface. A list of known symmetric dimers with crystal poses is shown in Table 2. The available receptor dimer structures (Table 2) together with the previous computational studies (Kaczor et al., 2015) have unveiled three major interfaces for GPCR dimerization or oligomerization: (I) TM1, 2, 7, and/or helix 8 (H8); (II) TM3, 4, and/or 5; and (III) TM5 and 6 (Figure 1). Due to the architecture of GPCRs, it is unlikely and energy-unfavorable for dimerization to occur via the interfaces of TM2, 3 and TM6, 7 (Kaczor et al., 2015). Besides, although H8 is not in the transmembrane region, the interaction between residues on H8 can also affect the dimeric interactions and the relative orientation of protomers.
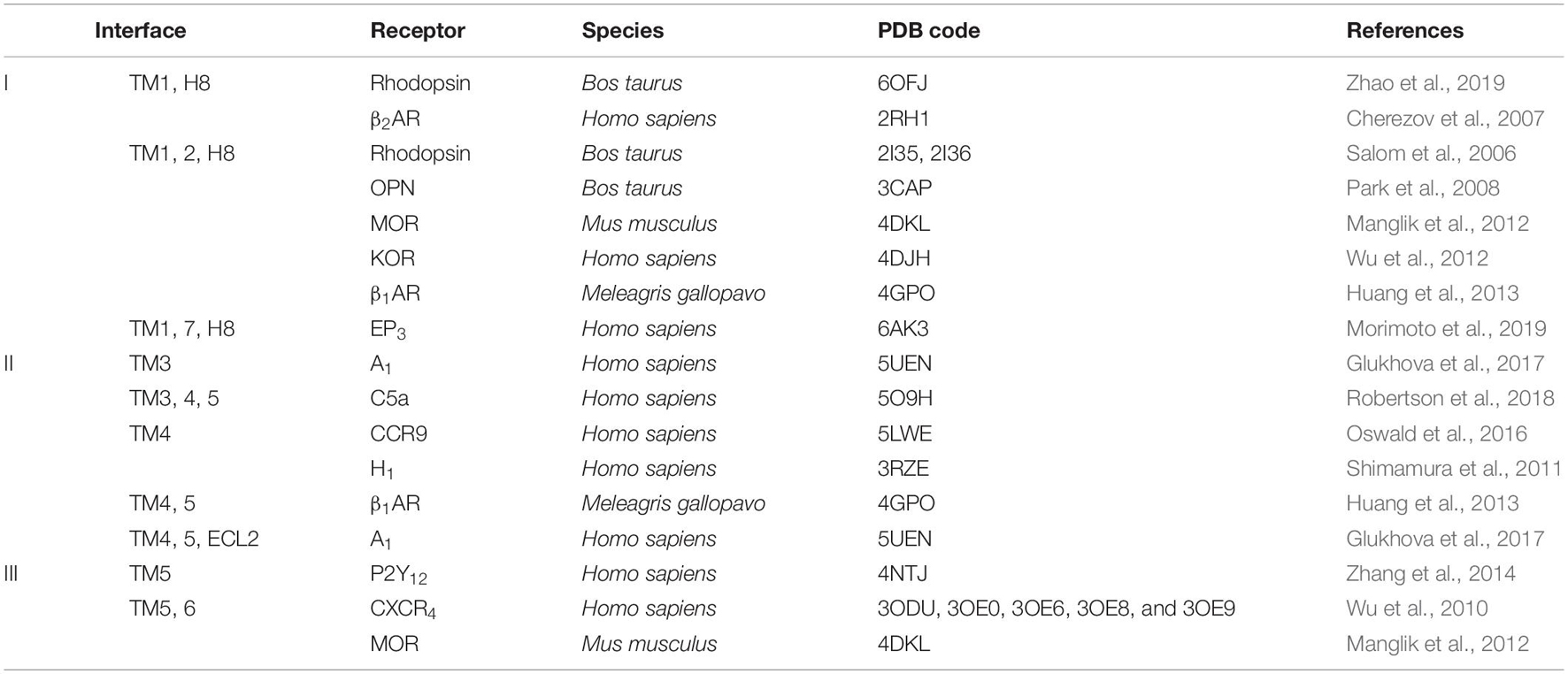
Table 2. Examples of Class A G protein-coupled receptor (GPCR) dimer structures in inactive conformation.
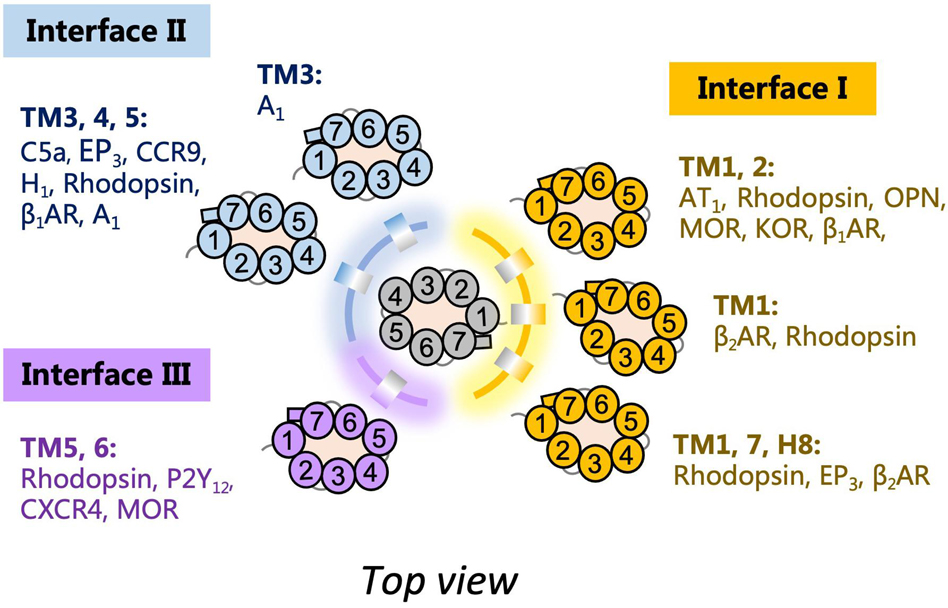
Figure 1. Schematic diagram of G protein-coupled receptor (GPCR) dimer interfaces I, II, and III as depicted from the top. Examples of receptor dimers bound at the different transmembrane helices (TM) regions are shown.
The heterodimers of melatonin receptors (i.e., MT1/MT2, MT1/GPR50, MT2/GPR50, and MT2/5-HT2C) were docked into the general interfaces I, II, and III as described in the methods. For all heterodimer pairs, the best ten models for each interface were exported for protein binding energy prediction and interface area measurement (Figure 2). An interface with the lowest free energy of binding and dissociation constant would be considered as the preferred interface. In order to validate the computational pipeline, a homodimer pair CXCR4/CXCR4 with a published crystal structure was included (Wu et al., 2010). The predicted dimer models of CXCR4/CXCR4 suggested a preference of dimerization interfaces I and III (Figure 2), which recapitulated known structural knowledge and computational simulations (Wu et al., 2010; Rodríguez and Gutiérrez-de-Terán, 2012; Gahbauer et al., 2018). Another heterodimer, MOR/DOR, which was predicted by molecular dynamic simulations (Liu et al., 2009; Provasi et al., 2015) to favor dimerization via TM1, 7 and TM 4, 5 (Interfaces I and II), was also tested by our computational pipeline. Indeed, the same preference of interfaces was observed (Figure 2). Since the interface area of the predicted CXCR4/CXCR4 as well as MOR/DOR models did not show a direct relationship with the preference of dimer interfaces (Figure 2), the interface area parameter is only taken as additional supportive structural information of dimer models.
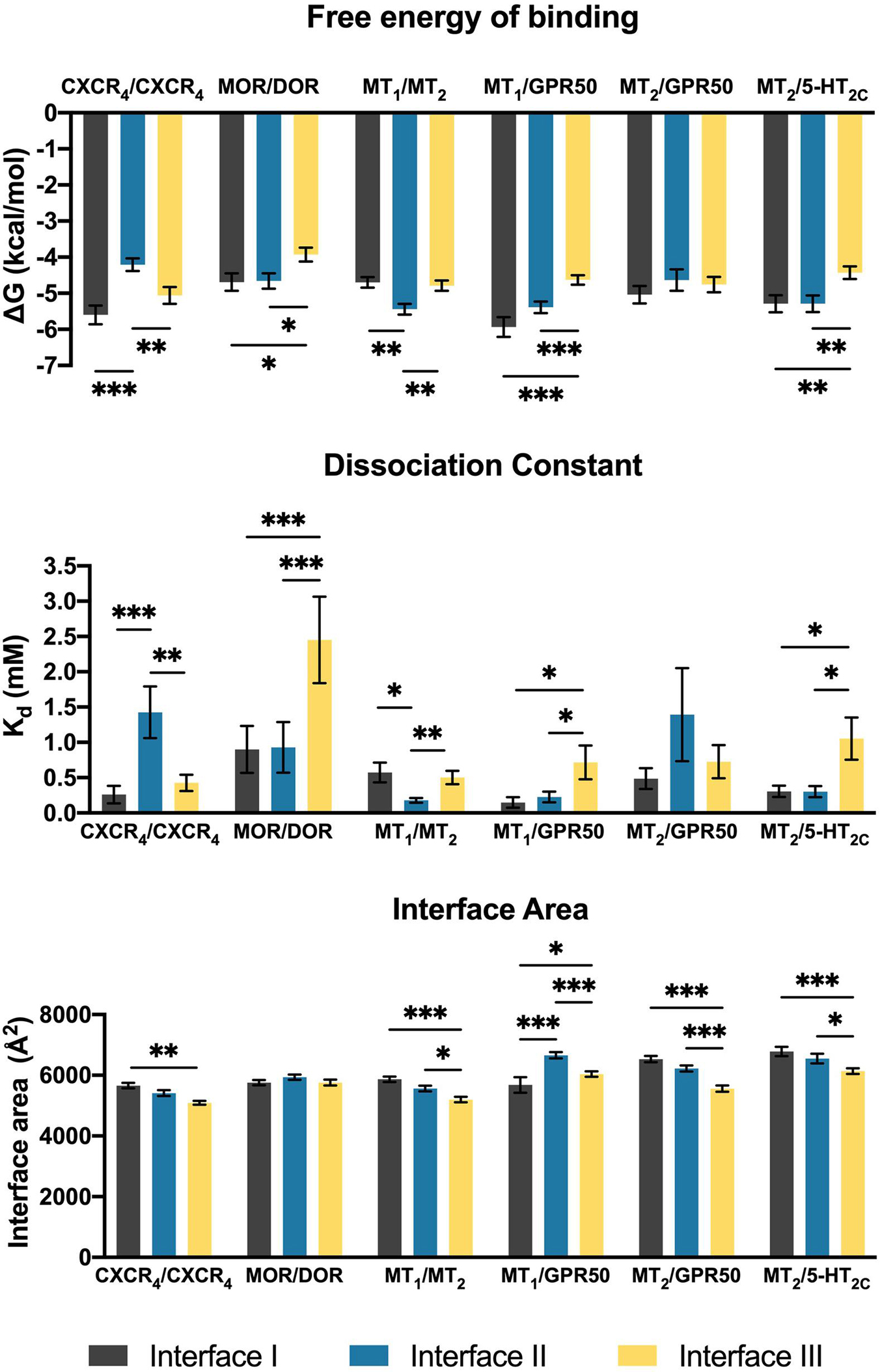
Figure 2. Scoring parameters calculated as average scores of top 10 models of each heterodimer with the given interface I, II, or III. Mean ± SEM is shown. Welch’s t-test is performed within each dimer pair; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
In contrast to the CXCR4/CXCR4 homodimer, interface III is generally less preferred by the melatonin receptor heterodimers (Figure 2). The MT1/MT2 heterodimer exhibited a clear preference for interface II in accordance with the parameters of free energy of binding and dissociation constant, while both interfaces I and II of MT1/GPR50 showed lower scores in these two parameters as compared to interface III (Figure 2). Different from MT1/GPR50, the MT2/GPR50 heterodimer did not show specific preference on any of the interfaces in terms of both parameters, while the area of interfaces I and II are significantly greater than that of interface III. As for MT2/5-HT2C, interfaces I and II were similarly preferred in all the scoring parameters (Figure 2). It should be noted that the dimerization between GPCRs can be constitutive or dynamic depending on the receptors. Rearrangement of the interfaces or dissociation of dimer may occur upon GPCR activation (Petersen et al., 2017; Xue et al., 2019). The dimeric models in this study are predicted as in their inactive ground states.
Interface II Is the Most Favored Heterodimer Interface for MT1/MT2
For the MT1/MT2 heterodimer, both the free energy of binding and the dissociation constant favor interface II over either interface I or III, while interface I provides the greatest interface area (Figure 2). The best model of MT1/MT2 heterodimer via interface II is depicted in Figure 3. The proposed lateral ligand entrances of MT1 and MT2 that lie between TM4 and 5 (Johansson et al., 2019; Stauch et al., 2019) and the residues forming the lateral ligand channel of both receptors [at position 4.56, 5.38, and 5.46 based on Ballesteros–Weinstein numbering scheme (Ballesteros and Weinstein, 1995)] are involved in the dimerization interface (Figures 3A,B). Therefore, dimerization via interface II is likely to hinder ligand binding. Yet, a gap was observed along L1604.58 and R1644.62 of MT1, and Q1995.37, A2035.41, and I2075.45 of MT2 with an estimated diameter of 2.08 Å, which is sufficiently large to enable melatonin to gain access to the lateral ligand entrances, while the other side of the melatonin receptor heterodimer is completely sealed off by A1865.37, A1905.41 of MT1 and L1664.51, V1704.55, and L1734.58 of MT2 (Figure 3C). The lateral ligand entrance of MT1 remains accessible in the lipid bilayer (Figure 3A), whereas an upward torsion of H2085.46 was observed in the MT2 protomer as compared with the reported crystal structure of MT2 (PDB code 6ME6), resulting in a complete seal of the lateral entrance by residues A1714.56, Y2005.38, H2085.46, and V2045.42, disallowing the ligand access (Figures 3A, 4A,B). On the other hand, an extra opening is present in MT2, which is formed by the open-lid conformation of ECL2 (Johansson et al., 2019) lined by residues T191ECL2, Q194ECL2, and Y2947.39, remains accessible for ligand entry in the MT2 protomer of the heterodimer. A disulfide bridge between C1133.25 and C190ECL2 is apparently important for holding the ECL2 in an open-lid conformation (Figure 4C). It has been shown that the ligand binding pockets of both protomers remain functional in the heterodimeric state (Ayoub et al., 2004). Our predicted model suggests that the ligand would access the binding pocket of MT1 via the lateral opening between protomers whilst ligand binding to MT2 would occur through the ECL entrance which is more accessible.
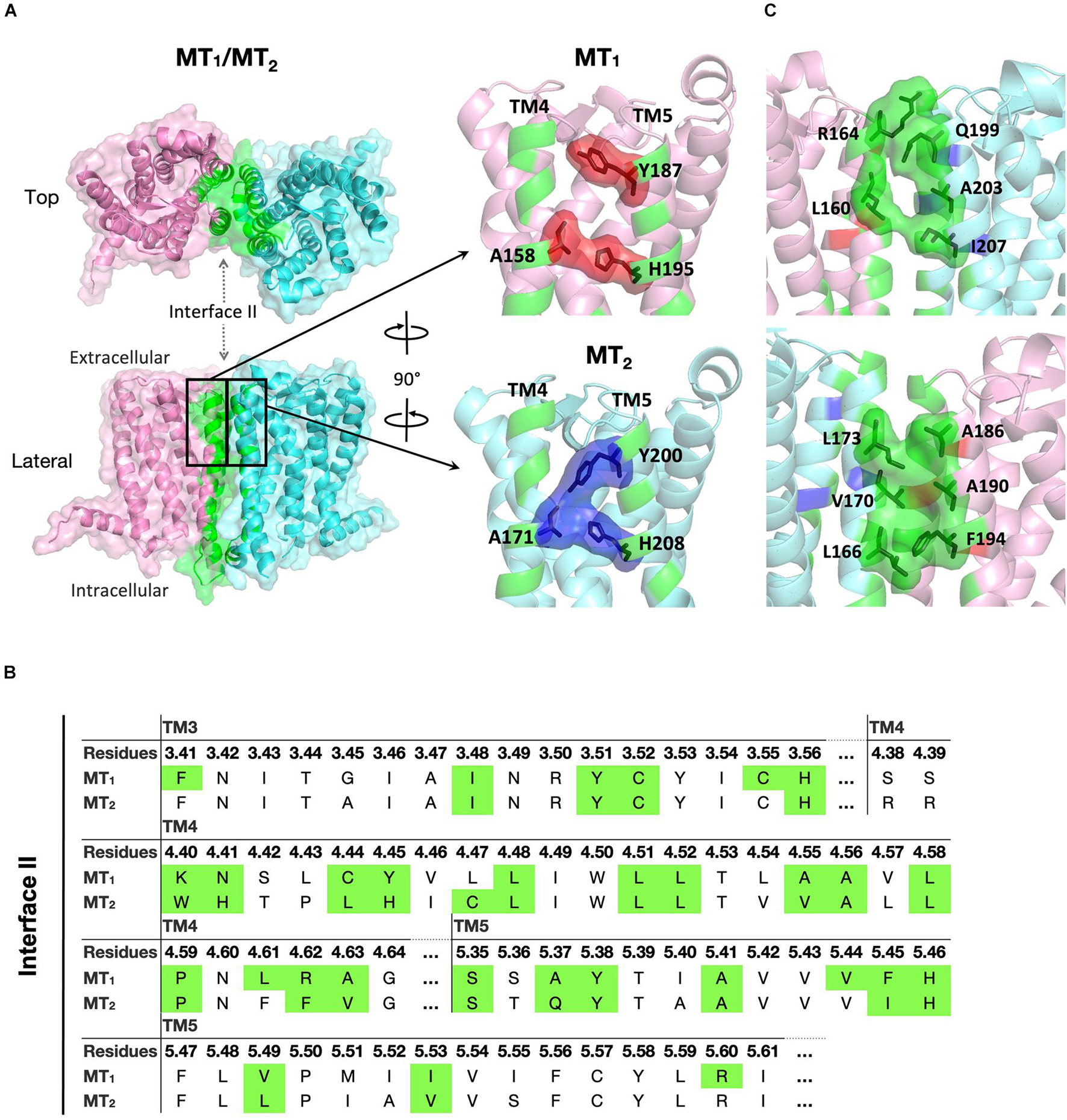
Figure 3. Predicted MT1/MT2 heterodimer. (A) MT1 (pink) and MT2 (cyan) dimerize with each other via interface II. Residues located in the interface with cutoff dASA = 1.0 are labeled in green. Residues forming the lateral ligand entrances of MT1 and MT2 are involved in the dimerization interface, and are shown in red and blue, respectively. (B) Predicted interface residues involved in the MT1/MT2 heterodimer. Residues involved in dimer interface II (as selected by cutoff dASA = 1.0) are labeled in green. Residues are shown in the Ballesteros–Weinstein numbering scheme (Ballesteros and Weinstein, 1995). (C) Side views of MT1/MT2 near the lateral ligand entrance with the orientation the same (top) or 180° rotated (bottom) as bracketed in panel (A).
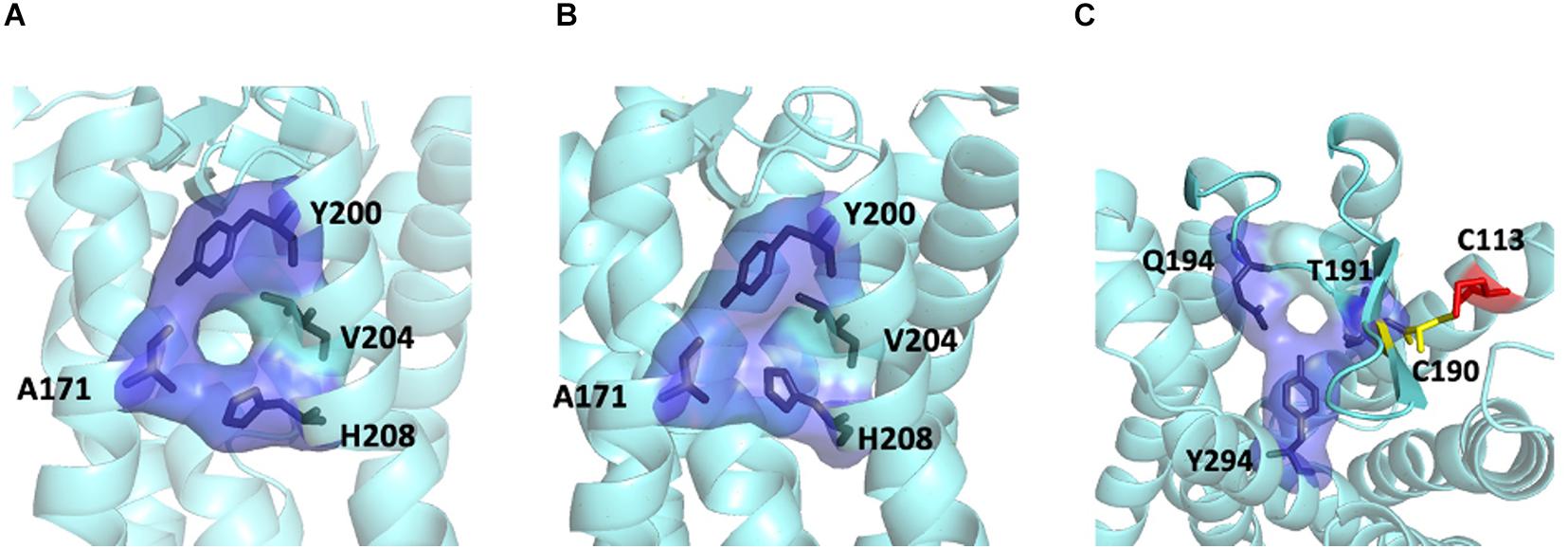
Figure 4. Ligand entrances of MT2. The lateral ligand channel of MT2 under the condition of crystallization (PDB code 6ME6) (A) or dimerization (B). (C) The ECL opening of MT2 protomer in MT1/MT2 heterodimer in a top-down view, with potential disulfide bridge between C1133.25 (red) and C190ECL2 (yellow).
Differential Interface Preference by MT1/GPR50 and MT2/GPR50
Although MT1 and MT2 share high structural similarity, our results suggested that MT1/GPR50 and MT2/GPR50 heterodimers have distinct preferences for dimer interfaces, which may explain the previous finding that GPR50 specifically decreases 125I-melatonin binding to MT1 but not MT2 (Levoye et al., 2006). The heterodimer MT1/GPR50 has lower free energy of binding and dissociation constant for interfaces I and II, while a larger interface area is formed at interface II (Figure 2). On the other hand, no specific preferential interface was found in the heterodimer MT2/GPR50 in terms of free energy and dissociation constant, while the greatest interface area formed by the MT2/GPR50 heterodimer is at interface I, and the smallest at interface III (Figure 2). Moreover, the dissociation constants of interface I and II of MT2/GPR50 are generally greater than that of MT1/GPR50 (Interface I: P-value of 0.0308 and interface II: P-value of 0.0042 by Welch’s t-test), revealing a potentially less stable condition for the MT2/GPR50 interhelical heterodimerization. The interface residues that lie in the transmembrane helical regions were further extracted from the top models of MT1/GPR50 and MT2/GPR50 in interfaces I and II (Figure 5). The models were selected based on the scoring parameters of free energy of binding and dissociation constant. Fewer residues are involved in the dimerization interfaces of MT2/GPR50 than that of MT1/GPR50: a total of 42 and 48 residues are involved in the dimerization of MT1/GPR50 heterodimer in interface I and II, respectively, as compared to 30 and 38 residues for that of the MT2/GPR50 heterodimer (Figure 5B).
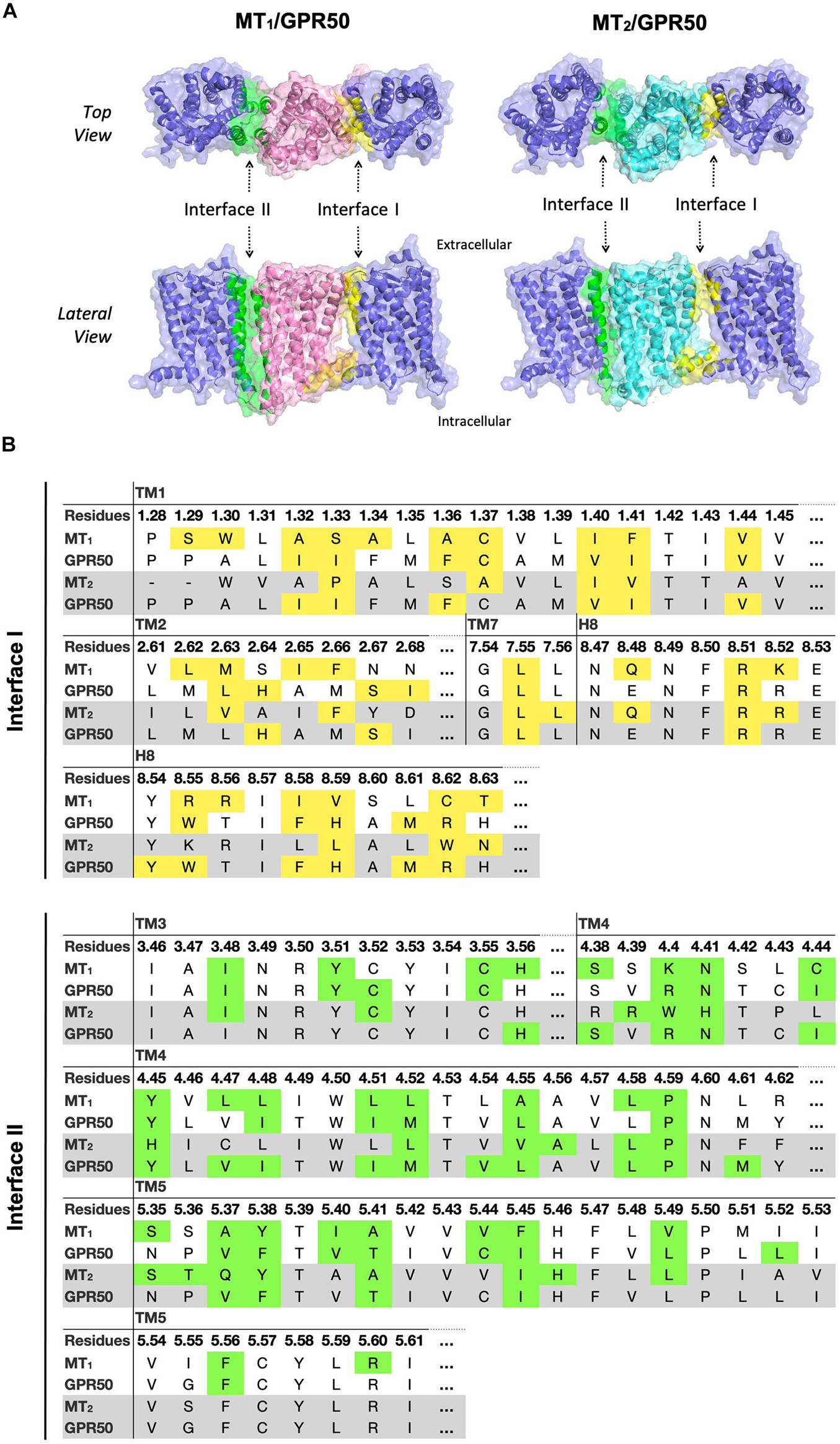
Figure 5. Predicted heterodimer models of MT1/GPR50 and MT2/GPR50. (A) MT1 (pink), MT2 (cyan), and GPR50 (slate blue) form heterodimers via interface I and II. Residues located in interface I and II with cutoff dASA = 1.0 are labeled in yellow and green, respectively. (B) Detailed interface residues of MT1/GPR50 (white background) and MT2/GPR50 (gray background) in dimer interface I and interface II are highlighted in yellow and green, respectively.
Two Potential Dimerization Interfaces for the MT2/5-HT2C Heterodimer
In the MT2/5-HT2C heterodimer, both interfaces I and II have higher scores in free energy of binding and dissociation constant than interface III, while interface I contributes the greatest surface area among all interfaces (Figure 2). The best predicted heterodimer model of MT2/5-HT2C which dimerizes through interface I or II is shown in Figure 6. A previous study using the cysteine crosslinking approach has reported that 5-HT2C can be assembled into homodimers via interface I or II (Mancia et al., 2008). Cysteine substitution of residues N541.32 and W551.33 successfully crosslinked the two 5-HT2C protomers via interface I, and I1924.63C and N2135.37C mutations linked up the receptors via interface II (Mancia et al., 2008). All these residues are similarly involved in the predicted interface between MT2 and 5-HT2C (Figure 6). Our results support two potential dimerization interfaces for the MT2/5-HT2C heterodimer.
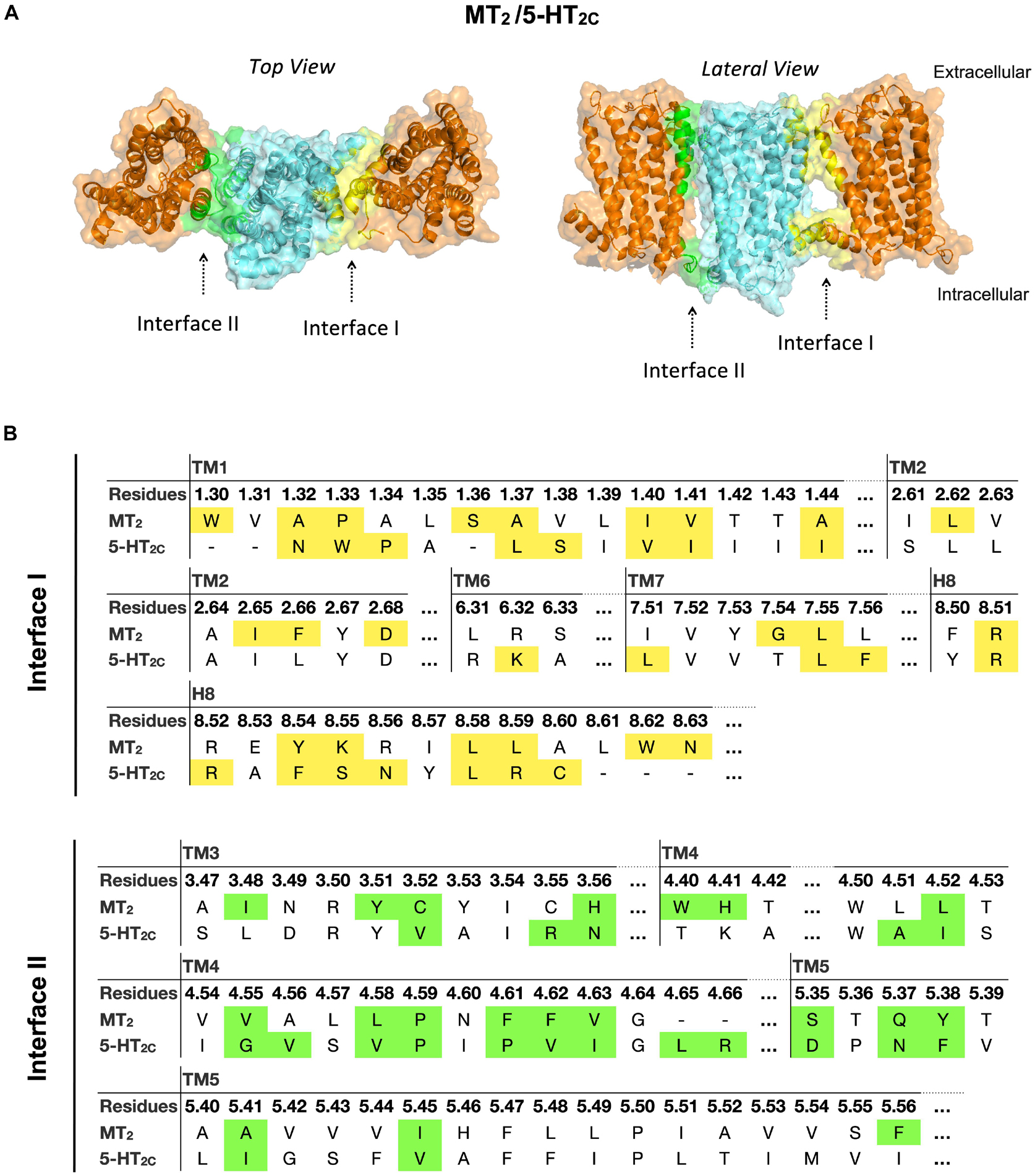
Figure 6. Models of MT2/5-HT2C heterodimers. (A) MT2 (cyan) and 5-HT2C (orange) are predicted to form heterodimers via interface I or II. The top view (left) and lateral view (right) of the MT2/5-HT2C models are shown. (B) Predicted interface residues involved in the MT2/5-HT2C heterodimer. Residues involved in dimer interface I and II (as selected by cutoff dASA = 1.0) are labeled in yellow and green, respectively.
Interface Residues in Melatonin Receptor Heterodimers
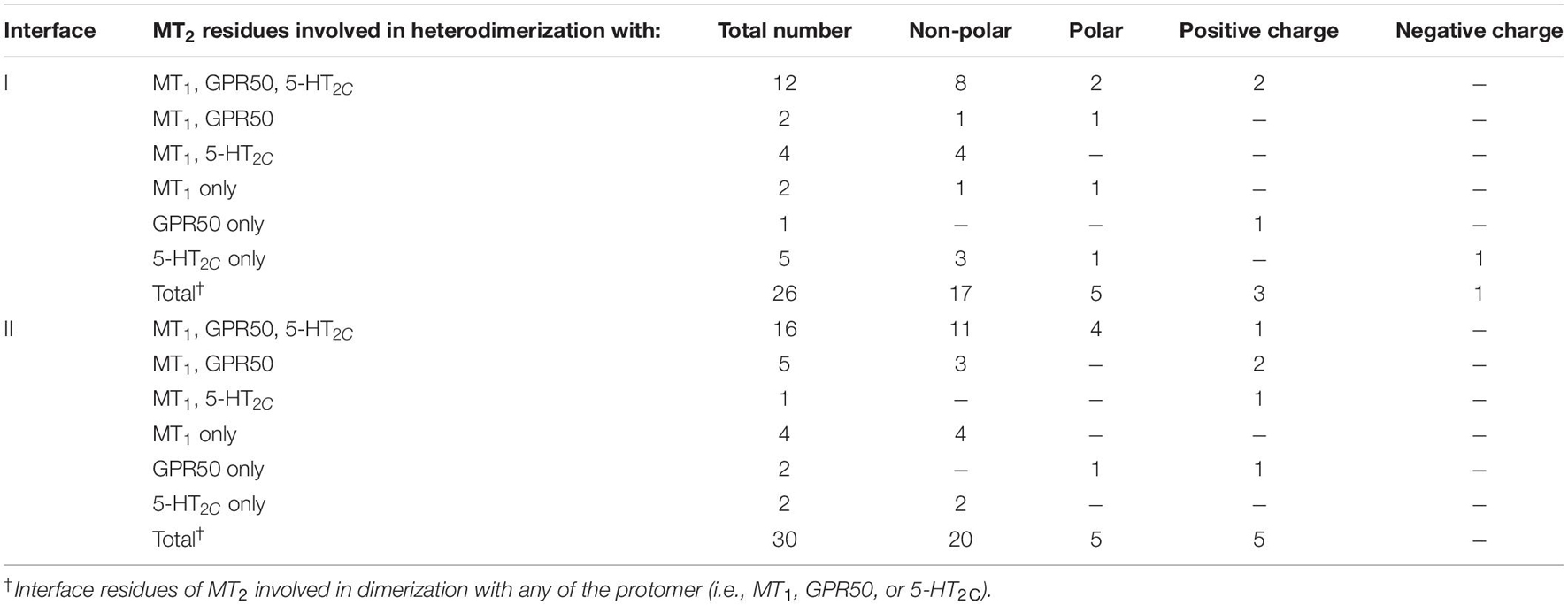
Since MT2 can dimerize with receptors not only from the melatonin receptor subfamily (i.e., MT1 and GPR50) but also with 5-HT2C, we further compared the interface residues used by MT2 protomer in the MT1/MT2, MT2/GPR50, and MT2/5-HT2C heterodimers in interfaces I and II (Figure 7 and Table 3). The highlighted interface residues in Figure 7 are generally separated into three types: (a) interface residues commonly involved in all the three described heterodimers of MT2; (b) interface residues involved in two pairs of heterodimers of MT2 (i.e., MT1/MT2 and MT2/GPR50; MT1/MT2 and MT2/5-HT2C); (c) interface residues solely involved in the specific heterodimer pairs.
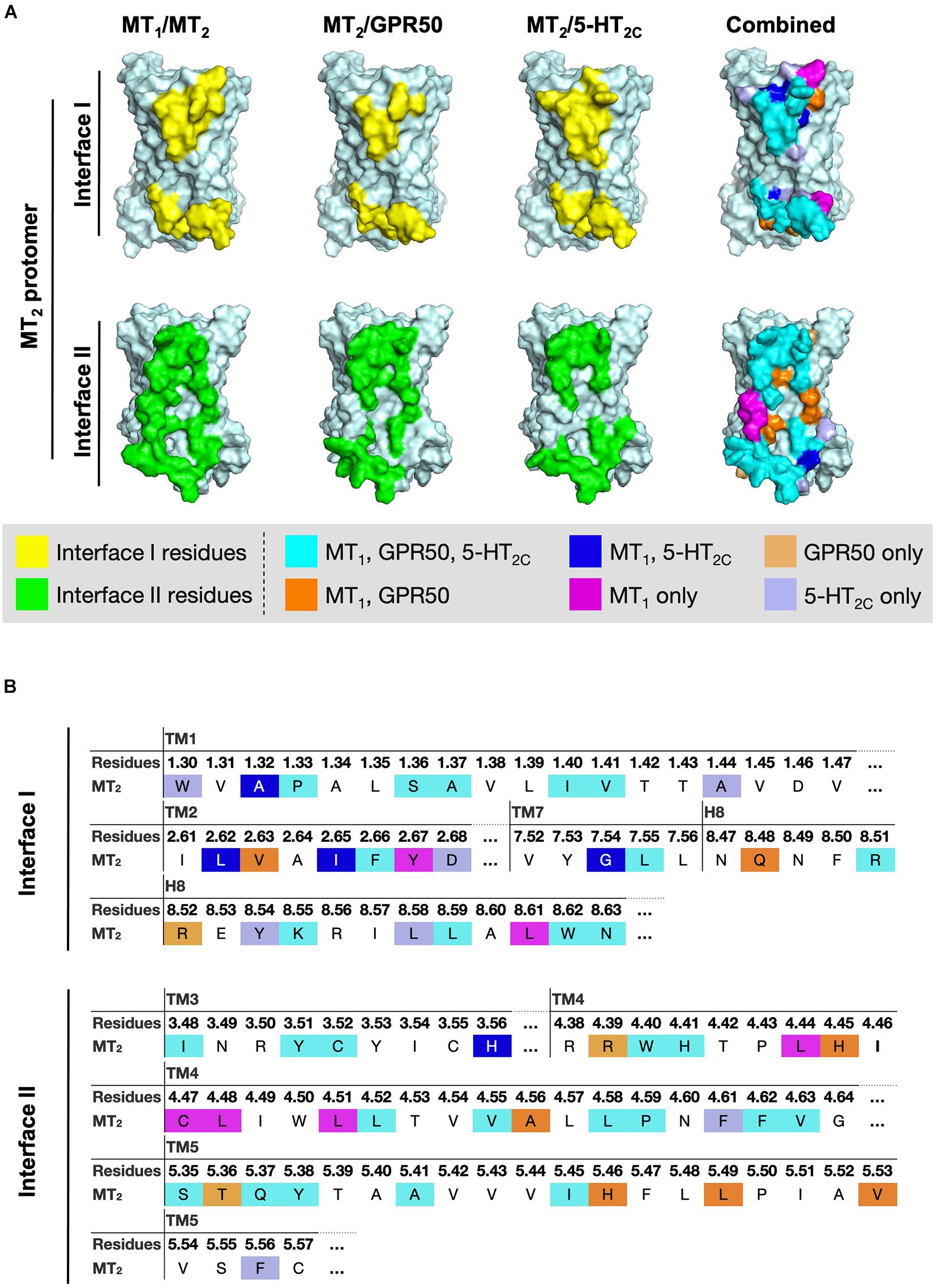
Figure 7. Residues of MT2 protomer in interface I and II of the MT2 heterodimer models. (A) The lateral interfaces I and II of the MT2 receptor model (pale cyan) are shown. Predicted interface residues on the MT2 protomer interface I or II involving in the heterodimer pairs (i.e., MT1/MT2, MT2/GPR50, or MT2/5-HT2C) with cutoff dASA = 1.0 are highlighted in yellow or green, respectively, and overlayed into one combined image with the indicated color codes. (B) The highlighted interface residues on the MT2 protomer in heterodimerization with MT1, GPR50, or 5-HT2C, with cutoff dASA = 1.0 are labeled with the color codes: cyan for residues involved in the interfaces of all the three heterodimers MT1/MT2, MT2/GPR50, and MT2/5-HT2C, orange for interface residues in both MT1/MT2 and MT2/GPR50, light orange for interface residues in MT2/GPR50 only, magenta for interface residues in MT1/MT2 only, blue for interface residues in both MT1/MT2 and MT2/5-HT2C, and light blue for interface residues in MT2/5-HT2C only.
Because of their geometrical location, some surface residues invariably contribute to the interfaces of all three MT2 heterodimers examined in this study. The common interface residues range from non-polar amino acids (Ala, Val, Cys, Pro, Leu, Ile, Trp, and Phe) to polar (Ser, Try, Asn, and Gln) and charged (Arg, His) ones, with the majority being non-polar residues; only one and two positively charged residues are involved in interfaces I and II, respectively (Figure 7B, residues in cyan; Table 3).
A smaller subset of interface residues is additionally involved when MT2 dimerizes with distinct protomers, which may act as anchors for establishing interaction with specific protomers (Figure 7 and Table 3). Residues including Ala, Val, Leu, Gln, and His on the MT2 protomer are implicated in both MT1/MT2 and MT2/GPR50 heterodimers (Figure 7B, residues in orange). Most of these residues are found in interface II while only two residues are in interface I of MT2 (Table 3). Specifically shared interface residues on MT2 are also observed in MT1/MT2 and MT2/5-HT2C heterodimers. Interface residues that are only involved in MT1/MT2 and MT2/5-HT2C heterodimers in the interface I are all hydrophobic (A401.32, L982.61, I1012.65, and G3097.54) and may help to stabilize the inter-helical interactions, while a polar His residue (H1443.56) is involved in the interface II of both MT1/MT2 and MT2/5-HT2C (Figure 7, residues in blue; Table 3). No overlapping interface residue is uniquely shared between MT2/GPR50 and MT2/5-HT2C heterodimers except the commonly involved interface residues (Figure 7).
As for the interface residues solely involved in specific heterodimer pairs, the MT2 protomer has the fewest residues for unique interaction with GPR50 at both interfaces I and II (Figure 7B, residues in light orange). Four non-polar residues were specifically involved in the MT1/MT2 heterodimer at interface II while only two residues (one non-polar and one polar) were located on interface I (Figure 7B, residues in magenta). Conversely, interface I of MT2 contains more residues than interface II for specific MT2/5-HT2C dimerization (Figure 7B, residues in light purple).
In silico Mutations of Interface Residues on MT2
Alanine mutagenesis is a common approach in computational studies of protein-protein interactions, as replacement of Ala residue at the interaction hot-spots can perturb the interaction and increase the free energy of binding (Massova and Kollman, 1999; Moreira et al., 2007). Each subset of the interface residues on MT2 (as defined in the previous section and highlighted in Figure 7) were mutated into Ala residues in silico, except those that are originally Ala. MPDock was performed using the wildtype (WT) or mutated MT2 with MT1, GPR50, or 5-HT2C WTs. The best ten models for interfaces I or II were exported and the free energy of binding and dissociation constant were estimated (Figure 8). Mutating the common interface residues was expected to introduce the greatest disturbance on dimerization, as 11 out of 26 interface residues and 15 out of 30 interface residues were mutated in interfaces I and II, respectively. Indeed, for the MT1/MT2 heterodimer, mutation of any subset of interface residues (i.e., common interface residues, residues involved in both MT1/MT2 and MT2/5-HT2C or MT2/GPR50 heterodimers, or those in MT1/MT2 only) significantly affected the scoring parameters in interface II but not interface I (Figure 8, top panels). Mutation of common interface residues in all three heterodimers (MT1/MT2, MT2/GPR50, and MT2/5-HT2C) led to the greatest perturbation in dimerization in interface II, with the free energy of binding increased from −5.59 to −3.7 kcal/mol and the dissociation constant increased from 0.276 to 2.99 mM (Figure 8, top panels). The docking result further supported that MT1/MT2 may heterodimerize via interface II. On the other hand, relatively high free energy of binding and dissociation constant were observed in the modeling of MT2/GPR50 heterodimer, alanine mutation of interface residues on MT2 did not further increase the two parameters except for the residue Arg8.52 (which is specifically involved in MT2/GPR50), which increased the free energy of binding in interface I but not the dissociation constant (Figure 8, middle panels). Interestingly, alanine mutagenesis on interface II even reduced the scoring parameters, indicating a more stable dimeric form (Figure 8, middle panels). The interaction between MT2 and GPR50 seems to be less dependent on the transmembrane domains. As for MT2/5-HT2C, the mutation of common interface residues and the interface residues shared by MT1/MT2 and MT2/5-HT2C increased the free energy of binding and dissociation constant, respectively, at the interface I of the heterodimer (Figure 8, lower panels); the scoring parameters of interface II were not affected. These results suggested that all subsets of interface residues are important for MT1/MT2 dimerization, in which the mutagenesis on any group of residues can perturb the interaction between protomers. Contrastingly, in silico mutation of a certain subset of interface residues is insufficient to disrupt the dimerization of MT2/5-HT2C in terms of free energy, the dimerization of protomers is dependent on multiple factors.
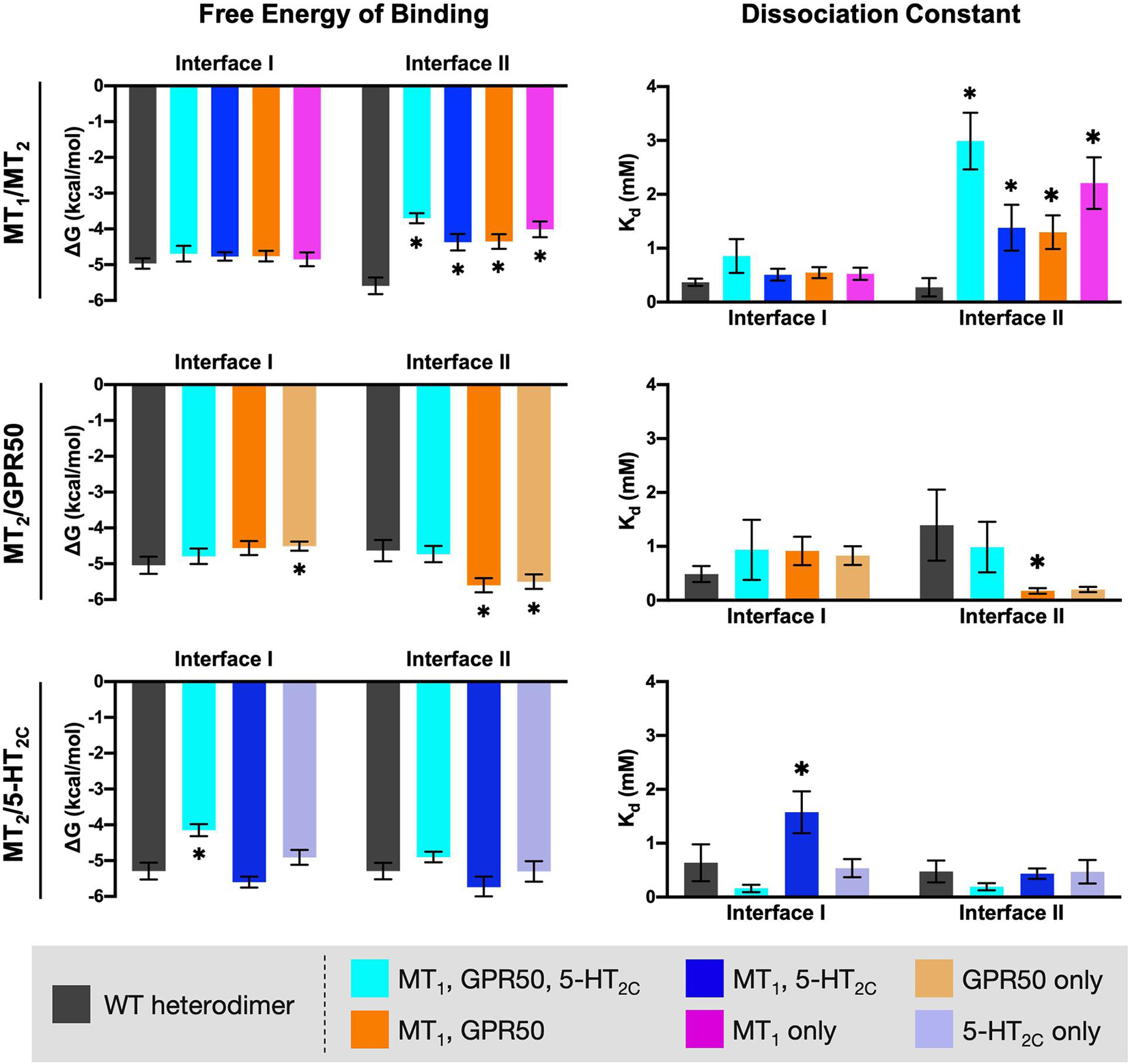
Figure 8. In silico alanine mutagenesis of MT2 residues putatively involved in heterodimerization. MT2 residues involved in the predicted interfaces of different heterodimers were mutated into Ala residues. MPDock was performed to predict the heterodimers formed by the corresponding WT protomer (MT1, GPR50, or 5-HT2C) with the WT MT2 (dark gray), or with the alanine mutants of MT2 (as depicted by color codes), at interfaces I and II. Free energy of binding (left) and dissociation constant (right) of each heterodimer were calculated as average scores of top 10 models of each heterodimer with the given interface I or II. Welch’s t-test is performed within each interface compared to the WT dimer pair; ∗p < 0.05.
Discussion
Melatonin, GPR50, and 5-HT2C receptors are expressed in multiple regions of the central nervous system including the hypothalamus, cortex, and hippocampus (Giorgetti and Tecott, 2004; Hamouda et al., 2007; Gupta et al., 2017), and heterodimerization between these receptors have been reported. With the recent revelation of melatonin receptors utilizing a unique lateral channel embedded in the lipid bilayer for ligand entry (Johansson et al., 2019; Stauch et al., 2019), it becomes pertinent to establish if melatoninergic ligand binding is obstructed or unaffected in GPCR dimers containing one or more melatonin receptor protomers, since a dimer interface involving TM4 and 5 may seal off the lateral channel. The molecular modeling and computational approaches used in this study suggest that the melatonin receptors may employ interface I or II to form heterodimers, with the latter perhaps being more prevalent. Although interface II encompasses TM4 and 5, molecular modeling reveals that the lateral ligand channel remains accessible in the MT1/MT2 heterodimer. Moreover, the predicted dimerization interfaces seemingly provide a structural basis for unique pharmacological features that have been reported for the melatonin receptor and GPR50 heterodimers. It has been noted that an outward movement of TM6 is a hallmark of GPCR activation (Zhou et al., 2019). Such GPCR activation was hypothesized to impede the conformational shift of dimeric/oligomeric GPCR-G protein structures upon activation when the receptors are interacting through TM5, 6 bundles (Cordomí et al., 2015). The GPCR activation may also affect the dimer interface. A study on class C metabotropic glutamate receptors has demonstrated that the dimer interface was changed from interface II (TM4, 5) to III (TM6) upon activation (Xue et al., 2015). In this study, we are focusing on the inactive state of the melatonin receptor heterodimers and therefore the potential alteration in the activation induced- structural changes of dimers is beyond the current scope and remains to be investigated by future work.
Our computational analysis suggested a more favorable dimer condition at interface II (Figure 3) for the MT1/MT2 heterodimer, where the lateral ligand entrances of MT1 and MT2 is located (Johansson et al., 2019; Stauch et al., 2019). The ligand binding sites of both MT1 and MT2 protomers are functional in the heterodimer as revealed by radioligand binding and BRET assays (Ayoub et al., 2004), which makes dimerizing via TM4 and 5 (interface II) seems unreasonable. However, a closer look at the entrance area of the model revealed a gap between the MT1 and MT2 protomers that allows ligand access to the binding pocket of MT1, while the lateral entrance of MT2 is completely sealed. Nonetheless, the ECL ligand entrance of MT2 remains an open conformation maintained by the disulfide linkage between a pair of residues C1133.25 or C190ECL2 (Figures 3, 4), which allows molecules to pass through. In agreement with our prediction, mutagenesis study has demonstrated that alanine substitution of either residues C1133.25 or C190ECL2 in MT2 resulted in the loss of 125I-melatonin binding without altering the receptor surface expression (Mseeh et al., 2002), implying the importance of ECL ligand path for MT2. Moreover, the C113A mutant of MT2 remains able to heterodimerize with MT1 and does not prohibit the binding of 125I-melatonin to the MT1 protomer (Levoye et al., 2006). Taken together, ligand binding is likely to take place via the lateral ligand channel of the MT1 protomer and the extracellular opening of the MT2 protomer in the MT1/MT2 heterodimer.
Albeit MT1 and MT2 share high sequence identity, their dimerization with GPR50 was shown to have different functional outcomes. The ligand binding capacity of MT1 but not MT2 is diminished upon interaction with GPR50 (Levoye et al., 2006). This effect is presumably brought on by the C-terminal tail of GPR50, which impedes Gi protein recruitment by MT1 and hence disfavors the conformation for ligand binding (Drew et al., 1997; Levoye et al., 2006). Given that the C-terminal truncated GPR50 was able to dimerize with both MT1 and MT2, it remains unresolved as to why GPR50 specifically hinders the ligand binding of MT1 but not MT2. Our study suggested that MT1/GPR50 is more energy favorable to form heterodimer via interfaces I and II, while all the potential interfaces of MT2/GPR50 are less stable as compared to other heterodimer pairs in terms of energy (Figure 2). A relatively more stable dimeric form of MT1/GPR50 may accentuate the inhibition brought upon by the C-terminal tail of GPR50. Moreover, the crystal structure of MT1 has revealed a membrane-buried lateral ligand entrance which lies between TM4 and 5 (interface II; Stauch et al., 2019), hindrance of ligand binding of MT1 by GPR50 could potentially be attributed to the blockade of ligand entrance through interface II or through the allosteric conformational change induced by dimerization at the interface I. In contrast, MT2 has an additional extracellular ligand entry through the ECL (Johansson et al., 2019), thus blocking the lateral ligand entrance would not completely abolish its ligand binding. The structural information and our predicted interface proclivity of MT1 or 2/GPR50 heterodimers supports the notion that GPR50 does not affect ligand binding of MT2 while it abolishes that of MT1.
In the MT2/5-HT2C heterodimer, dimerization via interfaces I and II is more favorable than via interface III. Indeed, the 5-HT2C homodimer was also shown to dimerize through interfaces I and II (Mancia et al., 2008). Dimer crosslinking by I1924.64C and P2125.37C mutations can only be observed in an activated state of 5-HT2C, indicating a conformational rearrangement at the TM5 of interface II upon receptor activation, whereas crosslinking through interface I (N541.32C and W551.33C mutations) was observed in both active and inactive receptor states (Mancia et al., 2008). Hence, interface II is speculated to be the site for allosteric communication between receptor protomers while the role of interface I-mediated dimerization remains unclear.
The interface residues on MT2 involved in its heterodimer partners were highlighted and separated into different groups (Figure 7 and Table 3). As suggested by a helix packing study, amino acids with small side chains (Gly, Ala, Ser, and Cys) enable tight helix-helix packing via van der Waals interactions, and the proximity between helices further facilitates inter-helical hydrogen bonding between polar residues (Liu et al., 2004). As the commonly involved interface residues among the heterodimers of MT2 cover both non-polar and polar residues, this group of residues potentially represent a conserved mechanism for MT2 to establish receptor-receptor interactions.
It is noted that His residues in interface II of the MT2 protomer (H1443.56, H1604.45, and H2085.46) frequently participate in the interactions with specific protomers while only one His residue (H1564.41) is commonly involved for all the MT2 heterodimer pairs. In general, His amino acids putatively serve as structural determinants in specific inter-helical interactions for heterodimerization (Figure 7B). Since GPCRs are membrane proteins that also interact with lipid molecules in the membrane bilayer, residues in the TM regions may also interact with cholesterol and sphingolipids. However, the identified cholesterol binding domains [i.e., (L/V)–X1–5–(Y)–X1–5–(K/R) from N- to C-terminal direction, or (K/R)–X1–5–(Y/F/W)–X1–5–(L/V) from N- to C-terminal direction, with X can be any residue] predominantly interact with positively charged residues (Lys or Arg) instead of His, while there is no positively charged residue involved in the identified sphingomyelin-binding motif (V–X2–TL–X2–IY; Fantini and Yahi, 2015). His residues are uncommon for protein-lipid interactions, and thus the His residues in the predicted interfaces are more likely to participate in protein-protein interactions. The H1443.56 residue is located close to the ICL2, one of the regions known to regulate G protein coupling/activation (Flock et al., 2017), and cooperation between two protomers is observed for the G protein signals of MT1/MT2 and MT2/5-HT2C (Baba et al., 2013; Kamal et al., 2015; Sánchez-Bretaño et al., 2019). Thus, H1443.56 may have the potential to affect G protein coupling properties upon dimerization.
Clinically relevant mutations of the GPCRs have been documented. The association between MT2 and T2D has been well established by experimental and genome-wide association studies (Bonnefond et al., 2012; Tuomi et al., 2016), and the identified MT2 variants related to T2D have been broadly categorized into three types based on their locations: at/near the ligand binding site; on the solvent-exposed intracellular side; or on the lipid-exposed intramembrane region (Bonnefond et al., 2012; Stauch et al., 2020). While the former two types of variants target the receptor functions by affecting ligand binding and downstream signaling of MT2, the latter type of variants may interfere with receptor dimerization or oligomerization (Stauch et al., 2020). In our predicted models, two of the T2D-associated mutants, A521.44T and L1664.51I, lie in the heterodimer interfaces of MT2 with 5-HT2C (interface I) and MT1 (interface II), respectively. The A521.44 residue is only involved in interface I of the MT2/5-HT2C heterodimer. As demonstrated by immunofluorescence staining, 5-HT2C receptor expressed alone was mainly intracellular whilst its cell surface expression was significantly increased in the presence of MT2 (Kamal et al., 2015), hence the dimerization between MT2 and 5-HT2C receptors may be crucial for the functioning of 5-HT2C. Besides, MT2/5-HT2C displays distinct heterodimer-specific signaling with melatonin acquiring the ability to transactivate Gq-coupled downstream responses (Kamal et al., 2015). Polymorphisms in the promoter region of 5-HT2C are also associated with T2D, in which a lower promoter activity is correlated with the predisposition of obesity and T2D (Yuan et al., 2000). Therefore, the T2D associated MT2 variant A521.44T may lead to a reduced 5-HT2C-mediated signaling or a functional impairment in the MT2/5-HT2C heterodimer in the pathogenesis of T2D. This speculation is further supported by evidence that the A521.44T mutation does not result in a significant functional change of MT2 receptor alone as compared to wild-type (including Gi1 and Gz protein activation, β-arrestin 2 recruitment and ERK activation; Karamitri et al., 2018). As for the L1664.51I variant, the mutation can cause a defect in the melatonin-induced β-arrestin 2 recruitment while increasing the spontaneous Gz activation by MT2 (Karamitri et al., 2018). However, the spontaneous β-arrestin 2 recruitment and Gi1 activation were similar to that of wild-type MT2 (Karamitri et al., 2018). One should not rule out the possibility that the L1664.51I mutation interferes MT1/MT2 heterodimerization at interface II. Although the association between MT1 and T2D has not been established, the implication of MT1 in T2D has been demonstrated in genetic knock-out mice, since MT1-deficient mice exhibit increased insulin resistance and impaired glucose metabolism (Contreras-Alcantara et al., 2010).
Apart from the T2D-associated MT2 variants, the A1574.55V variant of MT1 mutant with no obvious functional defect was apparently associated with non-24-h sleep-wake syndrome (Ebisawa et al., 1999; Chaste et al., 2010). The A1574.55 residue is located at the interface II of MT1/GPR50 and MT1/MT2 heterodimers. Given that both MT1 and MT2 are involved in mediating neuronal firing of the suprachiasmatic nucleus (known to control circadian rhythms) of the hypothalamus and regulate sleep (Liu et al., 1997; Gerdin et al., 2004; Gobbi and Comai, 2019), and GPR50 is also expressed in multiple regions of the hypothalamus (Regard et al., 2008; Sidibe et al., 2010), the A1574.55V mutation may affect the sleep-wake cycle by interrupting the formation or the function of heterodimers composed of MT1. A thorough understanding of the structural basis of melatonin receptor heterodimerization may bring new insights into disease pathogeneses and therapeutic designs. However, as computational study is speculative by nature, the predicted dimer structures remain to be tested experimentally. Mutagenesis studies that map the dimerization interface between protomers might further contribute to the structure-function relationship of melatonin receptor heterodimers and heterodimer-specific signaling mechanisms.
Concluding Remarks
Our study has assessed putative dimerization interfaces and established plausible dimerization models for MT1/MT2, MT1/GPR50, MT2/GPR50, and MT2/5-HT2C heterodimers. Computation models of these heterodimers have provided putative structural mechanisms/novel hypothesis on differential ligand binding properties between MT1/GPR50 and MT2/GPR50 heterodimers; distinct ligand access channels for protomers in the MT1/MT2 heterodimer (dimerize via interface II); two potential dimerization interfaces for MT2/5-HT2C with interface II potentially affects the G protein coupling properties. Besides, we have identified interface residues that potentially determine the specificity of receptor-receptor interactions. Since the mapping of the dimer interfaces and residues are based on computational predictions, validation studies should be performed. Our modeling approach may facilitate the design of experimental studies (such as mutagenesis) in understanding GPCR dimers and their structure and function. Furthermore, by mutagenesis or using small molecules that disrupt the heterodimer interface, one may shift the equilibrium of receptor monomer, dimer, or even oligomer, and alter the compositions of heterodimers in cells. Manipulating the population of heterodimers in cells could also be a novel approach to study the physiological relevance of GPCR heterodimer and therapeutic development, which without directly interfering with the function of receptor but the communication between different signaling routes.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
LT and YW contributed to the conception and design of the study and reviewed and edited the writing of the manuscript. Methodology and formal analysis were performed by LT with assistance from Ning Chen and Eunah Kim. LT prepared the original draft. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants from the University Grants Committee of Hong Kong (AoE/M-604/16), Research Grants Council, Hong Kong (16103015 and T13-605/18-W), the National Key Basic Research Program of China (2013CB530900), Innovation and Technology Commission of Hong Kong (ITCPD/17-9), and the Hong Kong Jockey Club.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ning Chen and Eunah Kim for assistance in data analysis and modeling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2021.725296/full#supplementary-material
Supplementary Figure 1 | Ramachandran plots for homology models of CXCR4, MOR, DOR, MT1, MT2, GPR50, and 5-HT2C. The red, brown, and yellow regions represent the favored, allowed, and “generously allowed” regions as defined by ProCheck.
Footnotes
- ^ www.uniprot.org/
- ^ https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/
- ^ https://wenmr.science.uu.nl/prodigy/
References
Alford, R. F., Koehler Leman, J., Weitzner, B. D., Duran, A. M., Tilley, D. C., Elazar, A., et al. (2015). An integrated framework advancing membrane protein modeling and design. PLoS Comput. Biol. 11:e1004398. doi: 10.1371/journal.pcbi.1004398
Angers, S., Salahpour, A., and Bouvier, M. (2001). Biochemical and biophysical demonstration of GPCR oligomerization in mammalian cells. Life Sci. 68, 2243–2250. doi: 10.1016/S0024-3205(01)01012-8
Ayoub, M. A., Couturier, C., Lucas-Meunier, E., Angers, S., Fossier, P., Bouvier, M., et al. (2002). Monitoring of ligand-independent dimerization and ligand-induced conformational changes of melatonin receptors in living cells by bioluminescence resonance energy transfer. J. Biol. Chem. 277, 21522–21528. doi: 10.1074/jbc.M200729200
Ayoub, M. A., Delagrange, P., and Jockers, R. (2004). Preferential formation of MT1/MT2 melatonin receptor heterodimers with distinct ligand interaction properties. Mol. Pharmacol. 66, 312–321. doi: 10.1124/mol.104.000398
Baba, K., Benleulmi-Chaachoua, A., Journé, A.-S., Kamal, M., Guillaume, J.-L., Dussaud, S., et al. (2013). Heteromeric MT1/MT2 melatonin receptors modulate photoreceptor function. Sci. Signal. 6:e4302. doi: 10.1126/scisignal.2004302
Ballesteros, J. A., and Weinstein, H. (1995). “Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors,” in Receptor Molecular Biology, ed. N. Sealfon (Cambridge, MA: Academic Press), 366–428. doi: 10.1016/s1043-9471(05)80049-7
Bonnefond, A., Clément, N., Fawcett, K., Yengo, L., Vaillant, E., Guillaume, J.-L., et al. (2012). Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat. Genet. 44:297. doi: 10.1038/ng.1053
Borroto-Escuela, D. O., Hagman, B., Woolfenden, M., Pinton, L., Jiménez-Beristain, A., Oflijan, J., et al. (2016). “In situ proximity ligation assay to study and understand the distribution and balance of GPCR homo- and heteroreceptor complexes in the brain,” in Receptor and Ion Channel Detection in the Brain: Methods and Protocols, eds R. Luján and F. Ciruela (New York: Springer), 109–124. doi: 10.1007/978-1-4939-3064-7_9
Brydon, L., Roka, F., Petit, L., de Coppet, P., Tissot, M., Barrett, P., et al. (1999). Dual signaling of human Mel1a melatonin receptors via Gi2, Gi3, and Gq/11 proteins. Mol. Endocrinol. 13, 2025–2038. doi: 10.1210/mend.13.12.0390
Cecon, E., Oishi, A., and Jockers, R. (2018). Melatonin receptors: molecular pharmacology and signaling in the context of system bias. Br. J. Pharmacol. 175, 3263–3280. doi: 10.1111/bph.13950
Chan, A. S. L., Lai, F. P. L., Lo, R. K. H., Voyno-Yasenetskaya, T. A., Stanbridge, E. J., and Wong, Y. H. (2002). Melatonin mt1 and MT2 receptors stimulate c-Jun N-terminal kinase via pertussis toxin-sensitive and -insensitive G proteins. Cell. Signal. 14, 249–257. doi: 10.1016/S0898-6568(01)00240-6
Chaste, P., Clement, N., Mercati, O., Guillaume, J.-L., Delorme, R., Botros, H. G., et al. (2010). Identification of pathway-biased and deleterious melatonin receptor mutants in autism spectrum disorders and in the general population. PLoS One 5:e11495. doi: 10.1371/journal.pone.0011495
Chaudhury, S., Berrondo, M., Weitzner, B. D., Muthu, P., Bergman, H., and Gray, J. J. (2011). Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One 6:e22477. doi: 10.1371/journal.pone.0022477
Chen, L., He, X., Zhang, Y., Chen, X., Lai, X., Shao, J., et al. (2014). Melatonin receptor type 1 signals to extracellular signal-regulated kinase 1 and 2 via Gi and Gs dually coupled pathways in HEK-293 cells. Biochemistry 53, 2827–2839. doi: 10.1021/bi500092e
Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G. F., Thian, F. S., Kobilka, T. S., et al. (2007). High-resolution crystal structure of an engineered human β2-adrenergic G protein–coupled receptor. Science 318, 1258–1265. doi: 10.1126/science.1150577
Contreras-Alcantara, S., Baba, K., and Tosini, G. (2010). Removal of melatonin receptor type 1 induces insulin resistance in the mouse. Obesity 18, 1861–1863. doi: 10.1038/oby.2010.24
Cordomí, A., Navarro, G., Aymerich, M. S., and Franco, R. (2015). Structures for G-protein-coupled receptor tetramers in complex with G proteins. Trends Biochem. Sci. 40, 548–551. doi: 10.1016/j.tibs.2015.07.007
Drew, J. E., Williams, L. M., Hannah, L. T., Barrett, P., Abramovich, D. R., and Morgan, P. J. (1997). Identification and characterisation of 2-(125I)iodomelatonin binding and Mel1a melatonin receptor expression in the human fetal leptomeninges. Brain Res. 761, 87–92. doi: 10.1016/S0006-8993(97)00335-1
Dufourny, L., Levasseur, A., Migaud, M., Callebaut, I., Pontarotti, P., Malpaux, B., et al. (2008). GPR50 is the mammalian ortholog of Mel1c: evidence of rapid evolution in mammals. BMC Evol. Biol. 8:105. doi: 10.1186/1471-2148-8-105
Ebisawa, T., Kajimura, N., Uchiyama, M., Katoh, M., Sekimoto, M., Watanabe, T., et al. (1999). Alleic variants of human melatonin 1a receptor: function and prevalence in subjects with circadian rhythm sleep disorders. Biochem. Biophys. Res. Commun. 262, 832–837. doi: 10.1006/bbrc.1999.1308
Fantini, J., and Yahi, N. (2015). Protein–Lipid Interactions in the Brain. San Diego: Academic Press, 135–162.
Flock, T., Hauser, A. S., Lund, N., Gloriam, D. E., Balaji, S., and Babu, M. M. (2017). Selectivity determinants of GPCR–G-protein binding. Nature 545, 317–322. doi: 10.1038/nature22070
Fuxe, K., and Borroto-Escuela, D. O. (2016). Heteroreceptor complexes and their allosteric receptor–receptor interactions as a novel biological principle for integration of communication in the CNS: targets for drug development. Neuropsychopharmacology 41, 380–382. doi: 10.1038/npp.2015.244
Gahbauer, S., Pluhackova, K., and Böckmann, R. A. (2018). Closely related, yet unique: distinct homo- and heterodimerization patterns of G protein coupled chemokine receptors and their fine-tuning by cholesterol. PLoS Comput. Biol. 14:e1006062. doi: 10.1371/journal.pcbi.1006062
Gerdin, M. J., Masana, M. I., Rivera-Bermúdez, M. A., Hudson, R. L., Earnest, D. J., Gillette, M. U., et al. (2004). Melatonin desensitizes endogenous MT2 melatonin receptors in the rat suprachiasmatic nucleus: relevance for defining the periods of sensitivity of the mammalian circadian clock to melatonin. FASEB J. 18, 1646–1656. doi: 10.1096/fj.03-1339com
Giorgetti, M., and Tecott, L. H. (2004). Contributions of 5-HT2C receptors to multiple actions of central serotonin systems. Eur. J. Pharmacol. 488, 1–9. doi: 10.1016/j.ejphar.2004.01.036
Glukhova, A., Thal, D. M., Nguyen, A. T., Vecchio, E. A., Jörg, M., Scammells, P. J., et al. (2017). Structure of the adenosine a1 receptor reveals the basis for subtype selectivity. Cell 168, 867.e13–877.e13. doi: 10.1016/j.cell.2017.01.042
Gobbi, G., and Comai, S. (2019). Differential function of melatonin MT1 and MT2 receptors in REM and NREM Sleep. Front. Endocrinol. 10:87. doi: 10.3389/fendo.2019.00087
Gray, J. J., Moughon, S., Wang, C., Schueler-Furman, O., Kuhlman, B., Rohl, C. A., et al. (2003). Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J. Mol. Biol. 331, 281–299. doi: 10.1016/S0022-2836(03)00670-3
Gupta, T., Sahni, D., Gupta, R., and Gupta, S. K. (2017). Expanding the horizons of melatonin use: an immunohistochemical neuroanatomic distribution of MT1 and MT2 receptors in human brain and retina. J. Anat. Soc. India 66, 58–66. doi: 10.1016/j.jasi.2017.05.007
Hamouda, H. O., Chen, P., Levoye, A., Sözer-Topçular, N., Daulat, A. M., Guillaume, J.-L., et al. (2007). Detection of the human GPR50 orphan seven transmembrane protein by polyclonal antibodies mapping different epitopes. J. Pineal Res. 43, 10–15. doi: 10.1111/j.1600-079X.2007.00437.x
Huang, J., Chen, S., Zhang, J. J., and Huang, X.-Y. (2013). Crystal structure of oligomeric β1-adrenergic G protein–coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 20, 419–425. doi: 10.1038/nsmb.2504
Johansson, L. C., Stauch, B., McCorvy, J. D., Han, G. W., Patel, N., Huang, X.-P., et al. (2019). XFEL structures of the human MT2 melatonin receptor reveal the basis of subtype selectivity. Nature 569, 289–292. doi: 10.1038/s41586-019-1144-0
Kaczor, A. A., Guixà-González, R., Carriõ, P., Poso, A., Dove, S., Pastor, M., et al. (2015). Multi-component protein-protein docking based protocol with external scoring for modeling dimers of G protein-coupled receptors. Mol. Inform. 34, 246–255. doi: 10.1002/minf.201400088
Kamal, M., Gbahou, F., Guillaume, J. L., Daulat, A. M., Benleulmi-Chaachoua, A., Luka, M., et al. (2015). Convergence of melatonin and serotonin (5-HT) signaling at MT2/5-HT2C receptor heteromers. J. Biol. Chem. 290, 11537–11546. doi: 10.1074/jbc.M114.559542
Karamitri, A., Plouffe, B., Bonnefond, A., Chen, M., Gallion, J., and Guillaume, J. L. (2018). Type 2 diabetes–associated variants of the MT2 melatonin receptor affect distinct modes of signaling. Sci. Signal. 11:eaan6622. doi: 10.1126/scisignal.aan6622
Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. doi: 10.1107/s0021889892009944
Levoye, A., Dam, J., Ayoub, M. A., Guillaume, J.-L., Couturier, C., Delagrange, P., et al. (2006). The orphan GPR50 receptor specifically inhibits MT1 melatonin receptor function through heterodimerization. EMBO J. 25, 3012–3023. doi: 10.1038/sj.emboj.7601193
Liu, C., Weaver, D. R., Jin, X., Shearman, L. P., Pieschl, R. L., Gribkoff, V. K., et al. (1997). Molecular dissection of two distinct actions of melatonin on the suprachiasmatic circadian clock. Neuron 19, 91–102. doi: 10.1016/S0896-6273(00)80350-5
Liu, J., Clough, S. J., Hutchinson, A. J., Adamah-Biassi, E. B., Popovska-Gorevski, M., and Dubocovich, M. L. (2016). MT1 and MT2 melatonin receptors: a therapeutic perspective. Annu. Rev. Pharmacol. Toxicol. 56, 361–383. doi: 10.1146/annurev-pharmtox-010814-124742
Liu, W., Eilers, M., Patel, A. B., and Smith, S. O. (2004). Helix packing moments reveal diversity and conservation in membrane protein structure. J. Mol. Biol. 337, 713–729. doi: 10.1016/j.jmb.2004.02.001
Liu, X., Kai, M., Jin, L., and Wang, R. (2009). Computational study of the heterodimerization between μ and δ receptors. J. Comput. Aided. Mol. Des. 23, 321–332. doi: 10.1007/s10822-009-9262-7
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I., and Lomize, A. L. (2012). OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376. doi: 10.1093/nar/gkr703
Mancia, F., Assur, Z., Herman, A. G., Siegel, R., and Hendrickson, W. A. (2008). Ligand sensitivity in dimeric associations of the serotonin 5-HT2C receptor. EMBO Rep. 9, 363–369. doi: 10.1038/embor.2008.27
Manglik, A., Kruse, A. C., Kobilka, T. S., Thian, F. S., Mathiesen, J. M., Sunahara, R. K., et al. (2012). Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature 485, 321–326. doi: 10.1038/nature10954
Massova, I., and Kollman, P. A. (1999). Computational alanine scanning to probe protein-protein interactions: a novel approach to evaluate binding free energies. J. Am. Chem. Soc. 121, 8133–8143. doi: 10.1021/ja990935j
Milligan, G., Ward, R. J., and Marsango, S. (2019). GPCR homo-oligomerization. Curr. Opin. Cell Biol. 57, 40–47. doi: 10.1016/j.ceb.2018.10.007
Moreira, I. S., Fernandes, P. A., and Ramos, M. J. (2007). Computational alanine scanning mutagenesis—an improved methodological approach. J. Comput. Chem. 28, 644–654. doi: 10.1002/jcc.20566
Morimoto, K., Suno, R., Hotta, Y., Yamashita, K., Hirata, K., Yamamoto, M., et al. (2019). Crystal structure of the endogenous agonist-bound prostanoid receptor EP3. Nat. Chem. Biol. 15, 8–10. doi: 10.1038/s41589-018-0171-8
Mseeh, F., Gerdin, M. J., and Dubocovich, M. L. (2002). Identification of cysteines involved in ligand binding to the human melatonin MT2 receptor. Eur. J. Pharmacol. 449, 29–38. doi: 10.1016/S0014-2999(02)01903-9
Oswald, C., Rappas, M., Kean, J., Doré, A. S., Errey, J. C., Bennett, K., et al. (2016). Intracellular allosteric antagonism of the CCR9 receptor. Nature 540, 462–465. doi: 10.1038/nature20606
Pandi-Perumal, S. R., Trakht, I., Srinivasan, V., Spence, D. W., Maestroni, G. J. M., Zisapel, N., et al. (2008). Physiological effects of melatonin: role of melatonin receptors and signal transduction pathways. Prog. Neurobiol. 85, 335–353. doi: 10.1016/j.pneurobio.2008.04.001
Park, J. H., Scheerer, P., Hofmann, K. P., Choe, H.-W., and Ernst, O. P. (2008). Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187. doi: 10.1038/nature07063
Petersen, J., Wright, S. C., Rodríguez, D., Matricon, P., Lahav, N., Vromen, A., et al. (2017). Agonist-induced dimer dissociation as a macromolecular step in G protein-coupled receptor signaling. Nat. Commun. 8:226. doi: 10.1038/s41467-017-00253-9
Petit, L., Lacroix, I., Coppet, P. De, Strosberg, A. D., and Jockers, R. (1999). Differential Signaling of human Mel1a and Mel1b melatonin receptors through the cyclic guanosine 3′-5′-monophosphate pathway. Biochem. Pharmacol. 58, 633–639. doi: 10.1016/S0006-2952(99)00134-3
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Prinster, S. C., Hague, C., and Hall, R. A. (2005). Heterodimerization of G protein-coupled receptors: specificity and functional significance. Pharmacol. Rev. 57, 289–298. doi: 10.1124/pr.57.3.1
Provasi, D., Boz, M. B., Johnston, J. M., and Filizola, M. (2015). Preferred supramolecular organization and dimer interfaces of opioid receptors from simulated self-association. PLoS Comput. Biol. 11:e1004148. doi: 10.1371/journal.pcbi.1004148
Ray, N., Cavin, X., Paul, J. C., and Maigret, B. (2005). Intersurf: dynamic interface between proteins. J. Mol. Graph. Model. 23, 347–354. doi: 10.1016/j.jmgm.2004.11.004
Regard, J. B., Sato, I. T., and Coughlin, S. R. (2008). Anatomical profiling of G protein-coupled receptor expression. Cell 135, 561–571. doi: 10.1016/j.cell.2008.08.040
Robertson, N., Rappas, M., Doré, A. S., Brown, J., Bottegoni, G., Koglin, M., et al. (2018). Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 553, 111–114. doi: 10.1038/nature25025
Rodríguez, D., and Gutiérrez-de-Terán, H. (2012). Characterization of the homodimerization interface and functional hotspots of the CXCR4 chemokine receptor. Proteins Struct. Funct. Bioinform. 80, 1919–1928. doi: 10.1002/prot.24099
Rullmann, J. A. C. (1996). AQUA, Computer Program, Department of NMR Spectroscopy. Netherlands: Bijvoet Center for Biomolecular Research, Utrecht University.
Salom, D., Lodowski, D. T., Stenkamp, R. E., Trong, I. L., Golczak, M., Jastrzebska, B., et al. (2006). Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 103, 16123–16128. doi: 10.1073/pnas.0608022103
Sánchez-Bretaño, A., Suen, T.-C., Baba, K., DeBruyne, J., and Tosini, G. (2019). Melatonin receptor heterodimerization in a photoreceptor-like cell line endogenously expressing melatonin receptors. Mol. Vis. 25, 791–799.
Shen, M., and Sali, A. (2006). Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524. doi: 10.1110/ps.062416606
Shimamura, T., Shiroishi, M., Weyand, S., Tsujimoto, H., Winter, G., Katritch, V., et al. (2011). Structure of the human histamine H1 receptor complex with doxepin. Nature 475, 65–70. doi: 10.1038/nature10236
Sidibe, A., Mullier, A., Chen, P., Baroncini, M., Boutin, J. A., Delagrange, P., et al. (2010). Expression of the orphan GPR50 protein in rodent and human dorsomedial hypothalamus, tanycytes and median eminence. J. Pineal Res. 48, 263–269. doi: 10.1111/j.1600-079X.2010.00750.x
Stauch, B., Johansson, L. C., and Cherezov, V. (2020). Structural insights into melatonin receptors. FEBS J. 287, 1496–1510. doi: 10.1111/febs.15128
Stauch, B., Johansson, L. C., McCorvy, J. D., Patel, N., Han, G. W., Huang, X. P., et al. (2019). Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 569, 284–288. doi: 10.1038/s41586-019-1141-3
Terrillon, S., and Bouvier, M. (2004). Roles of G-protein-coupled receptor dimerization. EMBO Rep. 5, 30–34. doi: 10.1038/sj.embor.7400052
Tuomi, T., Nagorny, C. L. F., Singh, P., Bennet, H., Yu, Q., Alenkvist, I., et al. (2016). Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 23, 1067–1077. doi: 10.1016/j.cmet.2016.04.009
Vidi, P.-A., Przybyla, J. A., Hu, C.-D., and Watts, V. J. (2010). Visualization of G protein-coupled receptor (GPCR) interactions in living cells using bimolecular fluorescence complementation (BiFC). Curr. Protoc. Neurosci. 51, 5.29.1–5.29.15. doi: 10.1002/0471142301.ns0529s51
Waly, N. E., and Hallworth, R. (2015). Circadian pattern of melatonin MT1 and MT2 receptor localization in the rat suprachiasmatic nucleus. J. Circ. Rhythms 13:1. doi: 10.5334/jcr.ab
Wang, W., Qiao, Y., and Li, Z. (2018). New insights into modes of GPCR activation. Trends Pharmacol. Sci. 39, 367–386. doi: 10.1016/j.tips.2018.01.001
Webb, B., and Sali, A. (2016). Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 54, 5.6.1–5.6.37. doi: 10.1002/cpbi.3
Witt-Enderby, P. A., Bennett, J., Jarzynka, M. J., Firestine, S., and Melan, M. A. (2003). Melatonin receptors and their regulation: biochemical and structural mechanisms. Life Sci. 72, 2183–2198. doi: 10.1016/S0024-3205(03)00098-5
Wu, B., Chien, E. Y. T., Mol, C. D., Fenalti, G., Liu, W., Katritch, V., et al. (2010). Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071. doi: 10.1126/science.1194396
Wu, H., Wacker, D., Mileni, M., Katritch, V., Han, G. W., Vardy, E., et al. (2012). Structure of the human κ-opioid receptor in complex with JDTic. Nature 485, 327–332. doi: 10.1038/nature10939
Xue, L., Rovira, X., Scholler, P., Zhao, H., Liu, J., Pin, J.-P., et al. (2015). Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol. 11, 134–140. doi: 10.1038/nchembio.1711
Xue, L., Sun, Q., Zhao, H., Rovira, X., Gai, S., He, Q., et al. (2019). Rearrangement of the transmembrane domain interfaces associated with the activation of a GPCR hetero-oligomer. Nat. Commun. 10:2765. doi: 10.1038/s41467-019-10834-5
Xue, L. C., Rodrigues, J. P., Kastritis, P. L., Bonvin, A. M., and Vangone, A. (2016). PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32, 3676–3678. doi: 10.1093/bioinformatics/btw514
Yuan, X., Yamada, K., Ishiyama-Shigemoto, S., Koyama, W., and Nonaka, K. (2000). Identification of polymorphic loci in the promoter region of the serotonin 5-HT2C receptor gene and their association with obesity and type II diabetes. Diabetologia 43, 373–376. doi: 10.1007/s001250050056
Zhang, K., Zhang, J., Gao, Z.-G., Zhang, D., Zhu, L., Han, G. W., et al. (2014). Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509, 115–118. doi: 10.1038/nature13083
Zhao, D. Y., Pöge, M., Morizumi, T., Gulati, S., Van Eps, N., Zhang, J., et al. (2019). Cryo-EM structure of the native rhodopsin dimer in nanodiscs. J. Biol. Chem. 294, 14215–14230. doi: 10.1074/jbc.RA119.010089
Keywords: G protein-coupled receptors, dimerization, melatonin receptor, modeling, interface
Citation: Tse LH and Wong YH (2021) Modeling the Heterodimer Interfaces of Melatonin Receptors. Front. Cell. Neurosci. 15:725296. doi: 10.3389/fncel.2021.725296
Received: 15 June 2021; Accepted: 16 August 2021;
Published: 07 October 2021.
Edited by:
Yamina Berchiche, Dr. GPCR, United StatesReviewed by:
Xavier Periole, The University of Auckland, New ZealandQing Fan, Columbia University, United States
Copyright © 2021 Tse and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yung Hou Wong, boyung@ust.hk
 Lap Hang Tse
Lap Hang Tse Yung Hou Wong
Yung Hou Wong