An efficient computer vision-based approach for acute lymphoblastic leukemia prediction
- 1Department of Computer Engineering and Networks, College of Computer and Information Sciences, Jouf University, Sakaka, Saudi Arabia
- 2Department of Management Science, Beaconhouse National University, Lahore, Pakistan
- 3Computer Science Department, Faculty of Computers & Information Technology, University of Tabuk, Tabuk, Saudi Arabia
- 4Department of Computer Science and Software Engineering, International Islamic University, Islamabad, Pakistan
- 5Department of Management Information Systems, Prince Sattam Bin Abdulaziz University, Al-Kharj, Saudi Arabia
- 6Department of Computer Engineering and Networks, College of Computer and Information Sciences, Jouf University, Sakaka, Saudi Arabia
Leukemia (blood cancer) diseases arise when the number of White blood cells (WBCs) is imbalanced in the human body. When the bone marrow produces many immature WBCs that kill healthy cells, acute lymphocytic leukemia (ALL) impacts people of all ages. Thus, timely predicting this disease can increase the chance of survival, and the patient can get his therapy early. Manual prediction is very expensive and time-consuming. Therefore, automated prediction techniques are essential. In this research, we propose an ensemble automated prediction approach that uses four machine learning algorithms K-Nearest Neighbor (KNN), Support Vector Machine (SVM), Random Forest (RF), and Naive Bayes (NB). The C-NMC leukemia dataset is used from the Kaggle repository to predict leukemia. Dataset is divided into two classes cancer and healthy cells. We perform data preprocessing steps, such as the first images being cropped using minimum and maximum points. Feature extraction is performed to extract the feature using pre-trained Convolutional Neural Network-based Deep Neural Network (DNN) architectures (VGG19, ResNet50, or ResNet101). Data scaling is performed by using the MinMaxScaler normalization technique. Analysis of Variance (ANOVA), Recursive Feature Elimination (RFE), and Random Forest (RF) as feature Selection techniques. Classification machine learning algorithms and ensemble voting are applied to selected features. Results reveal that SVM with 90.0% accuracy outperforms compared to other algorithms.
1. Introduction
The whole body receives crucial nutrients from the blood. Blood cells are divided into three primary types in the human body such as erythrocytes (red blood cells), leukocytes (white blood cells), and thrombocytes (platelets). Red blood cells (RBCs) ensure oxygen gets to all the body's cells. Platelets contribute to blood coagulation in the event of an accident. White blood cells (WBCs) defend against viruses and stop diseases in people. Only 1% of blood is made up of White blood cells because white blood cells are essential to the human immune system. Even small changes significantly impact (Sharif et al., 2020; Winter et al., 2021; Abir et al., 2022).
Diseases are indicated by an increase or decrease in the quantity of White Blood cells (WBCs) in blood plasma. There are five types of White Blood cells: lymphocyte, monocyte, neutrophil, eosinophil, and basophil. Any variation in the white blood cell count is cause for concern. Leukemia is the most frequently associated with a low White Blood cell (WBCs) count (Ghanem et al., 2019; Abir et al., 2022). It can be harmful and a factor in disease when our bodies have an unusually high quantity of WBC (Hegde et al., 2019).
Leukemia is a potentially deadly disease and is a type of cancer (Alfayez et al., 2021). It affects the blood and bone marrow due to the rapid growth of aberrant white blood cells. These aberrant WBCs cannot resist disease and affect the bone marrow's ability to make RBCs and platelets. There are two types of leukemia: acute and chronic. Acute leukemia grows faster and has more severe symptoms than chronic leukemia. The types of leukemia include lymphocytic and myelogenous. A type of white blood cell known as a lymphocyte contributes to the immune system. Lymphocytic leukemia describes excessive tumor growth in the cells in the bone marrow that become those lymphocytes. The hematopoietic cells that develop into RBCs, WBCs, and platelets grow abnormally in myelogenous leukemia. Four types of diseases classify leukemia, namely, Acute lymphocytic leukemia (ALL), Acute myelogenous leukemia (AML), Chronic lymphocytic leukemia (CLL), and Chronic myelogenous leukemia (CML).1
The most frequent type of childhood cancer is acute lymphoblastic leukemia (ALL) (Pan et al., 2017). The percentage of ALL cases involve healthy individuals, and only a small number of patients have genetic factors, including familial vulnerability or environmental factors. Chromosome abnormalities and genetic modifications related to lymphoid are its defining features (Godoy et al., 2020; Fujita et al., 2021). While ALL accounts for 80% of children's leukemia, it only accounts for 20% of adult leukemia. ALL is diagnosed using time-consuming and sophisticated procedures. The survival rate in developed nations has grown above 90% due to modern risk-adapted medicines and supportive care (Pan et al., 2017).
1.1. Motivation
In increasing healthcare problems, automated systems are essential (Kumar et al., 2022b). These systems can predict and classify acute lymphoblastic leukemia and are required to offer patients appropriate care and reduce its risks. The therapy and recovery of the patient depend heavily on the timely and precise detection of this type of cancer. Machine learning (ML) is a well-known scientific area based on artificial intelligence concepts that predict healthcare problems (Alsalem et al., 2018; Lalotra et al., 2021; Kumar et al., 2022a). Many researchers proposed techniques based on machine learning and deep learning algorithms for the early detection of acute lymphoblastic leukemia. Still, they are limited in providing better performance (Daqqa et al., 2017; Nazari et al., 2020) and did not consider feature extraction and selection techniques (Hossain et al., 2021). Considering these limitations, this research proposed an approach in which features are extracted using pre-trained CNN-based deep network architectures. The feature selection techniques are used to choose the relevant features from the large-size feature set acquired, and finally, classification is accomplished through machine learning techniques.
The main contributions performed for this proposed approach are given below:
• The pre-trained Convolutional Neural Network (CNN) based architectures VGG19, ResNet50, or ResNet101 are applied to enhance images by reducing noise and extracting the critical features.
• The feature selection techniques ANOVA, Recursive Feature Elimination (RFE), and Random Forest (RF) is applied to select the essential features that help to improve accuracy.
• The classification machine learning algorithms RF, SVM, KNN, NB, and Ensemble voting model, are applied to selected dataset features.
• The proposed model outperforms the classification of acute lymphocytic leukemia diseases and is also helpful for medical experts and patients by diagnosing acute lymphocytic leukemia early on time.
The rest of the paper is organized as follows. Section 2 presents the relevant literature. Section 3 provides the proposed approach. Section 4 presents the results and discussion. Finally, Section 5 concludes the research. Table 1 provides the list of abbreviations.
2. Literature review
The techniques based on machine and deep learning algorithms used for the prediction of leukemia are explained in this section.
2.1. Machine learning techniques
Machine learning (ML) is gaining fast attention in various areas of cancer research (Abbas et al., 2021; Sacca et al., 2022; Safdar et al., 2022). ML is a promising tool for managing hematologic malignancies in the future due to its ability to interpret data from many diagnostic modalities, estimate mortality, and recommend treatment plans. ML-based techniques deal with different medical data and diseases. These ML approaches can be incorporated into numerous applications to guarantee rapid and accurate diagnosis, risk classification, and effective treatment (Eckardt et al., 2020). This article predicts leukemia using data mining techniques by examining the associations between blood characteristics and leukemia, gender, age, and patient health. Support Vector Machine (SVM), k-nearest neighbor (KNN), and decision tree (DT) are used for the prediction. 4,000 patient records are gathered from Gaza hospital. The experiment is performed, and accuracy and f1-measure evaluation metrics are used. Decision trees from all classifiers performed well with 77.30% accuracy (Daqqa et al., 2017).
The author proposed a supervised machine-learning approach for the early prediction of leukemia (Hossain et al., 2021). They primarily concentrate on typical symptoms and the possibility that a patient would eventually acquire leukemia. Typically, information about the features is provided at routine checkups. The used dataset is split into training and testing. Naive Bayes (NB), K-Nearest Neighbor (KNN), Random Forest (RF), Linear Regression (LR), Adaboost, etc., are used as machine learning algorithms to check out the performance of the model. Determined 17 parameters with a discussion with the doctor. Data is gathered from hospitals on various leukemia and non-leukemia patients. The highest result is achieved with a Decision Tree (DT) of 98%. In this article (Alsuwaidi et al., 2021), the author investigates novel genes demonstrating optimistic clinical and molecular signatures and uses a lymphoid cancer dataset. Lymphoblastic and lymphoid leukemia research comprising 1706 individuals and 2144 data are found in three different research. Only samples of B-lymphoblastic leukemia are chosen for additional examination. Using the cBioPortal tool, chromosomal alterations are evaluated to identify novel genomic loci to analyze clinical and molecular characteristics for the leukemia of lymphoid genesis. Homozygous deletions of the ADAM6 gene are found in 59.60% of the individuals analyzed and are linked to poor 10-year survival rates.
The author proposed an approach of supervised machine learning algorithms for the prediction of leukemia in the early stage by using symptoms. Dataset is gathered from the hospitals in Bangladesh. A survey on leukemia and non-leukemia patients is conducted with the help of a specialized doctor to gather 16 features for the datasets. Decision tree (DT) algorithms are supervised machine learning models. The Apriori algorithm is used to produce understandable principles for leukemia prediction. For comparison, many classifiers are applied to check the model's generalizability. Two feature selection and analysis steps are executed on the dataset to increase the model performance. The proposed model outperforms with 97.45% accuracy, 0.63 Mathew's Correlation Coefficient (MCC), and 0.783 of the area under the Receiver Operating Characteristic (ROC) curve (Hossain et al., 2022).
2.2. Deep learning techniques
The author of this research describes a model for detecting acute lymphocytic leukemia with separate depthwise convolutions (Clinton et al., 2020; Chatila et al., 2021). Diagnosing these tumor tissues as soon as possible is necessary to reduce the physical strain on the patient and the treatment difficulties that the disease brings (Sleiman et al., 2019; Aoun et al., 2020; Ali et al., 2022; Rizwan et al., 2022). Convolutional neural network (CNN) is tested using the transfer learning method. Xception model performance is evaluated on an extended microscopic blood smear image dataset. The model achieved 99% for the training dataset and 91% for the testing dataset. The 22283 gene leukemia microarray training dataset is taken from the Gene Expression Omnibus source. After designing and applying a deep neural network model to the data, classifiers cross-check the findings. Two preprocessing techniques are used for the data: normalization and principal component analysis (PCA). The findings demonstrate the PCA gene's capacity for independently segregating cancerous and healthy cells. The accuracy of the proposed model with three hidden layers and single-layer neural networks is 63.33 and 96.67, correspondingly (Nazari et al., 2020). Author in Genovese et al. (2021) proposed a methodology based on machine learning that uses an adaptive un-sharpening technique to improve images of blood samples. The approach normalizes the cell's radius using image processing algorithms, and deep learning estimates the focus reliability, adaptively enhances the sharpness of the images, and then does the classification. We tested the technique using an ALL publically available images dataset, taking into account several cutting-edge CNNs to carry out the classifying. The conclusions demonstrate the reliability of the proposed approach.
Author in de Oliveira and Dantas (2021) presents straightforward modifications to conventional neural network topologies to attain excellent performance in categorizing malignant leukocyte problems. Emulation, spinning, fading, shearing, and inserting salt-and-pepper noise are some of the changes used. The testing model used for the testing is VGG16, VGG19, and Xception. The model uses data augmentation to counteract the training and validation datasets. The model outperforms the evaluation metric f1-score 92.60%. The author proposed an Internet of Medical Things- (IoMT-) based approach to improve and nourish a fast and secure prediction of leukemia. The proposed system facilitates real-time leukemia screening, diagnosis, and treatment scenarios that save time and effort for both patients and medical experts and also address patients' issues in pandemics like COVID-19. Two convolutional neural networks (CNN) architectures, ResNet-34 and DenseNet-121, are used as a proposed model. The proposed model performed well-compared to other traditional algorithms. An experiment is performed on ASH and ALL-IDB images dataset accessible to the general public (Bibi et al., 2020).
In conclusion, many techniques are proposed based on machine learning and deep learning algorithms for predicting acute lymphoblastic leukemia (ALL). Still, they are limited in giving better performance and did not consider feature selection and extraction techniques. Considering these issues, this research proposed an efficient approach based on machine learning algorithms and ensemble algorithms to classify leukemia disease. Timely prediction of ALL diseases can reduce the mortality rate.
3. Proposed methodology
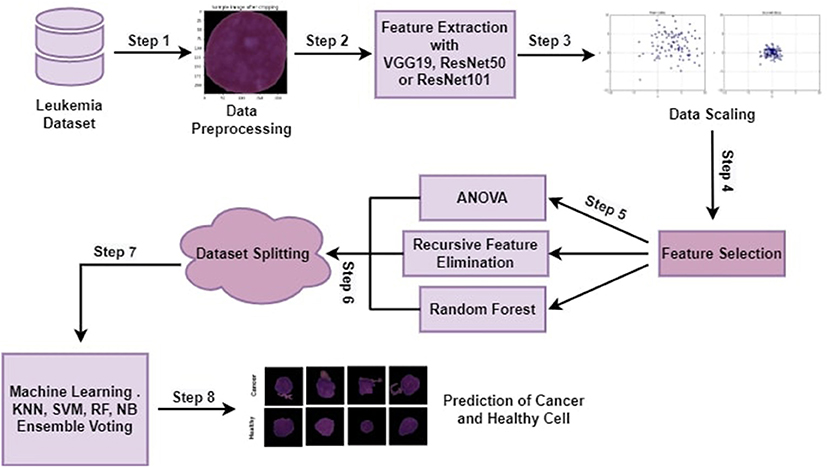
The proposed methodology with all steps is explained in this section in detail. Machine learning algorithms and ensemble algorithms are used to classify leukemia disease. Evaluation measurements such as accuracy, precision, recall, and f1-score are used to evaluate the proposed approach. This research experimented using a jupyter notebook on Anaconda. Python programming language is used for this experiment. This testing environment shows how programmers may create and evaluate machine learning (ML) models on a well-structured platform. Dynamic semantics are a feature of the high-level programming and interpreting language Python. The proposed approach has the following main steps cropping all the images by using minimum and maximum points, feature extraction through pre-trained CNN architectures (VGG16, ResNet50, or ResNet 101), Data scaling by applying MinMax normalization technique, feature selection through ANOVA, RFE, and RF. Selected features are utilized by machine learning algorithms and ensemble voting algorithms to classify leukemia. Figure 1 depicts the proposed approach steps.
3.1. Dataset
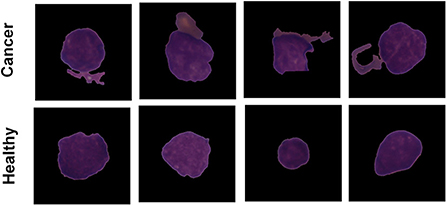
The Proposed model performance is evaluated on the C-NMC leukemia dataset for leukemia cancer prediction. Dataset is available publicly and is collected from the Kaggle repository. The user can search and submit various datasets on the Kaggle repository, collaborate with other machine and deep learning publishers and data scientists, and develop and explore various models in various data science environments (Dahiwade et al., 2019). The leukemia dataset has 10,661 total images in the form of data. The dataset contains three leading folder names fold_0, fold_1, and fold_2. Every folder has two sub-folder with the name “all” and “hem.” The folder “all” has 0:7272 data equal to 1 data label, and “hem” has 7272:10661 data equal to 0 data labels. Figure 2 shows classes of the leukemia dataset.
3.2. Dataset preprocessing
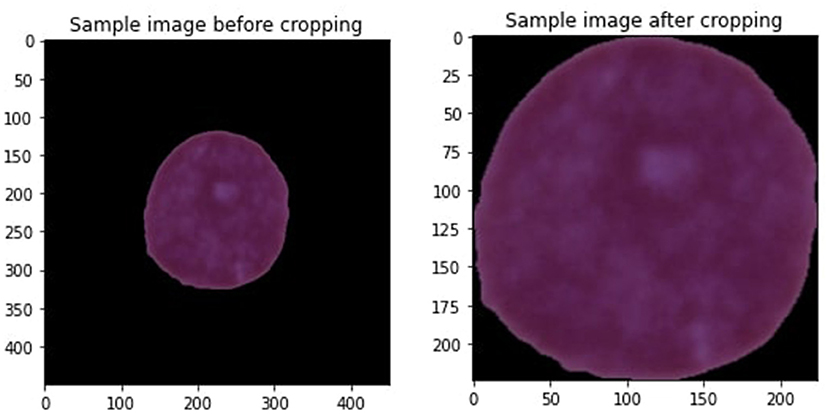
Data preprocessing is the preliminary step when designing any machine learning (ML) model for predicting diseases (Khan et al., 2019). Data preprocessing is executed to terminate the noisy data that are the reason for decreasing model performance. Every image of a leukemia dataset contains undesirable areas and empty spaces. Hence, cropping the images is essential to eliminate unnecessary space and use only vital information. We applied the extreme point calculation methodology for cropping images used in this study (Dahiwade et al., 2019). Figure 3 is the sample image we are cropping.
Some steps involve cropping the image, such as loading the leukemia image, transforming it into a grayscale, and then applying a threshold to the image to convert it to a binary image. Then, compute four separate minimum and maximum points (x,y) using the four most significant contours determined from the threshold images. At last, crop the image using the data gathered from minimum and maximum points and contours. All the images in the leukemia dataset have various sizes, heights, and widths, so it is essential to keep them in the same size with equal height and weight to improve the model's performance. After resizing all the images, the crop image size is 224 * 224. Range from 0 to 255 are used for all the images. Since there is greater noise around the edge, cropping the images is the best choice. Figure 3 is the sample image after cropping the image. Some more steps are involved in conducting data preprocessing that are explained below.
3.2.1. Feature extraction
After cropping all images of the leukemia dataset, the next step is feature extraction. Feature extraction is the numerical representation of the used dataset. All the raw data is converted into useful information for applying machine learning (ML) models on the dataset. Model performance increased by applying this step compared to raw data for the ML model. This research used pre-trained Convolutional Neural network-based deep neural network architectures (VGG19, ResNet50, or ResNet101) to extract features from images. From all three CNN deep neural networks, we applied the ResNet50 architecture with a reshaped size (−1, 224, 224, 3). Several features are in 2,048 before applying feature selection techniques.
3.2.2. Data scaling
It is essential to place dataset features into a scale equivalent when they are available on various scales. The standardization process involves rescaling the features to give them the properties of a typical normal distribution (Dahiwade et al., 2019). This research applied a MinMaxScaler normalization technique for scaling the leukemia dataset feature. Range defines for normalizing the features are 0–1.
3.2.3. Feature selection method
Feature selection plays a significant role in the preprocessing stage of data analysis. Features must offer sufficient characteristics to categorize the data into classes or groups for further implication and decision-making in intelligent systems to achieve high accuracy. If only a few components are used, the results will not be sufficient. Performance becomes a problem if there are many features chosen. More essential features can be added to increase accuracy. In this research, we applied three feature selection techniques: ANOVA, Recursive Feature Elimination (RFE), and Random Forest (RF).
3.2.3.1. ANOVA
Analysis of Variance (ANOVA) F-test statistics is a feature selection technique that removes insignificant features from the dataset (Olaolu et al., 2018). The parametric statistical hypothesis test determines whether the means from two or more data samples are from the same range. ANOVA is applied in this research as the first feature selection technique, and the k-value equals 500 (number of features).
3.2.3.2. Recursive feature elimination
Recursive feature elimination (RFE), a feature selection technique, eliminates the weakest feature from a model until the required number of features is attained. RFE is applied in this research as the second feature selection technique. RFE aims to choose the best feature subset considering the learned model and classification accuracy (Jeon and Oh, 2020).
3.2.3.3. Random forest
All the features in the dataset have different functions to classify. So some features that have less influence on the model's performance can be removed by applying the feature selection technique. Random forest algorithm has many benefits for selecting the features because it can quickly process the completion of high-dimensional data. Random feature selection can enhance classification performance (Li et al., 2020). RF is applied in this research as the third feature selection technique, and the value of the n-estimator is 200. After applying all techniques of feature selection, the number of features selected is 584. The leukemia dataset after preprocessing is split into 80% training and 20% testing for experimenting.
3.3. Machine learning algorithms
Machine learning algorithms such as K-nearest neighbors, Random forest, Support vector machine, Naive Bayes, and voting ensemble algorithm are applied as a proposed approach.
3.3.1. K-nearest neighbors
K Nearest Neighbor (KNN) algorithm is a supervised machine learning algorithm applied to classification and regression problems. The knn algorithm has demonstrated impressive performance on data with many examples, such as near infinity, where its error rate roughly reaches the Bayes optimum under relatively moderate conditions. The performance of the KNN algorithm increases by choosing an optimal value of k (Zhang et al., 2017).
3.3.2. Support vector machine
Support Vector Machine (SVM) algorithm is a supervised machine learning classifier that applies kernel trick procedure to determine the non-linear divisible problem by augmenting the data across a multi-resolution area. To find the best hyper-plane among the classes in the dataset, SVM classifiers extend the distance between the points closest to each class. The maximal hyper-plane gap widens the separation between the two classes (Pisner and Schnyer, 2020). This research used the SVM machine learning algorithm, and parameter values such as C = 100, gamma = 0.01, and kernel = “rbf” are used.
3.3.3. Random forest
The random forest approach is based on developing many decision trees, each serving as a classifier. In random forests, a sub-dataset is created by sampling every tree in the forest from the given dataset. Each decision tree receives the sub-datasets, and each decision tree produces a conclusion. All decision trees vote to establish the final decision outcome (Li et al., 2020).
3.3.4. Naive bayes
Naive Bayes is an essential learning algorithm that uses the Bayes rule and the fundamental presumption that, given the class, the attributes are conditionally independent (Webb et al., 2010). This independence assumption is frequently broken in practice, but naive Bayes produces competitive classification accuracy. It is also called a probabilistic classifier.
3.3.5. Voting classifier
A voting algorithm is a machine learning classifier that develops several base models or classifiers. It makes predictions by combining their results. Voting for each classifier result can be integrated with the aggregating criteria. This research used the following base classifiers, RF, SVM, and NB, for making a voting classifier.
In Algorithm 1, this proposed approach work of all steps is described in detail. In this proposed approach, the input is the Leukemia dataset Lds, and Mp model performance is the output. Dataset is preprocessed by applying some steps such as minimum and maximum cropping techniques (Ci) for cropping, an important feature is extracted using feature extraction (fextract), and CNN architectures VGG16, ResNet50, or ResNet101 are used. When the architecture of CNN is selected feature then reshapes (ImgReshape) the data. When the architecture of CNN is selected feature then reshapes (ImgReshape) the data. The next step is data scaling (Dscaling) using the MinMax Normalization technique. The last step of preprocessing is feature selection fs. An important feature is selected by applying three techniques ANOVA, RFE, and RF. The leukemia dataset is split with 80% training data and 20% testing data. Simple machine learning classifier ClassifiersML is applied to the training dataset (KNN, SVM, NB, RF). Then an ensemble voting (soft) model is applied, which contains three algorithms (RF, KNN, and SVM) with their hyper-parameters. The proposed model evaluated accuracy, precision, recall, and f1-score measurements.
4. Result and discussion
This section explains the result of the proposed approach and used evaluation measurements in this research. The effectiveness of the proposed model for the prediction of ALL is discussed.
4.1. Evaluation measurements
Many evaluation measurements are utilized to identify the proposed model's classification results. This research used accuracy, precision, recall, and f1-score as evaluation measurements because many researchers used these measurements to evaluate the proposed methodology. Accuracy defines as calculating the ratio of true positive, false positive, true negative, and false negative. To find precision, compute the ratio of all positive values in the data. The recall is known as sensitivity, defined as computing the true positive by the true negative and false negative. F1-measure is calculated by taking the average of precision and recall measurements.
4.2. Experimental result
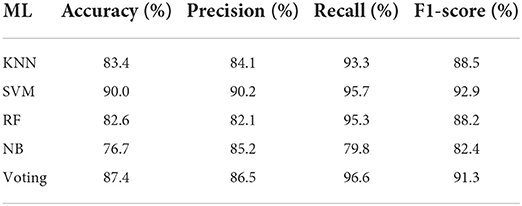
Table 2 demonstrates the overall performance of machine learning (ML) algorithms on the leukemia dataset. K-nearest Neighbor (KNN) algorithm achieved an accuracy of 83.4%, a precision of 84.1%, a recall of 93.3%, and an f1-score of 88.5%. The support Vector Machine (SVM) algorithm achieved an accuracy of 90.0%, a precision of 90.2%, a recall of 95.7%, and 92.9%. Random Forest (RF) achieved an accuracy of 82.6%, a precision of 82.1%, a recall of 95.3%, and an f1-score of 88.2%. Naive Bayes (NB) achieved an accuracy of 76.7%, a precision of 85.2% recall of 79.8%, and an f1-score of 82.4%.
The voting classifier achieved an accuracy of 87.4%, a precision of 86.5%, a recall of 96.6%, and an f1-score of 91.3%. SVM performed better in accuracy, precision, and f1-score than other machine learning algorithms. The voting classifier performed well in terms of recall as compared to other machine learning algorithms.
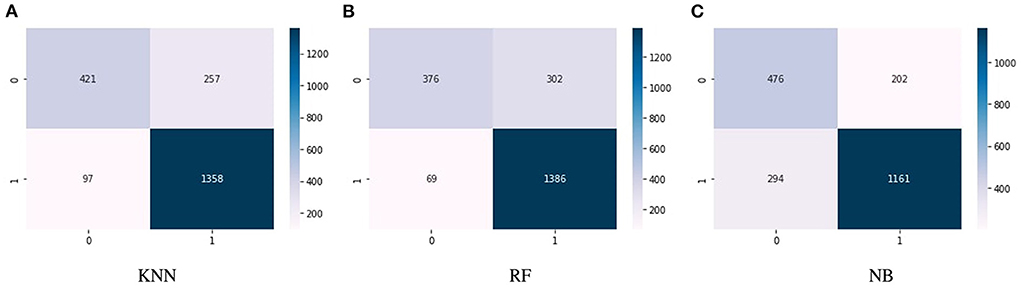
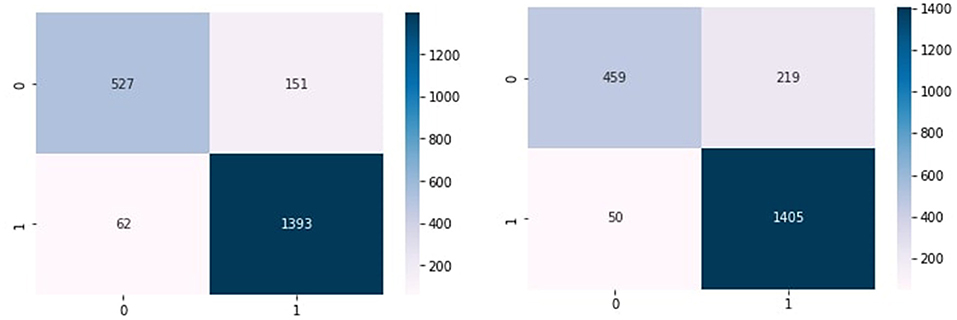
Figures 4A–C show the confusion matrix of KNN, RF, and NB algorithms. These graphs express machine learning working. The proposed model performance increases if true positive and negative values are higher than false positive and negative values. Figure 5 presents the confusion matrix of the SVM (left side) and voting classifier (right side).
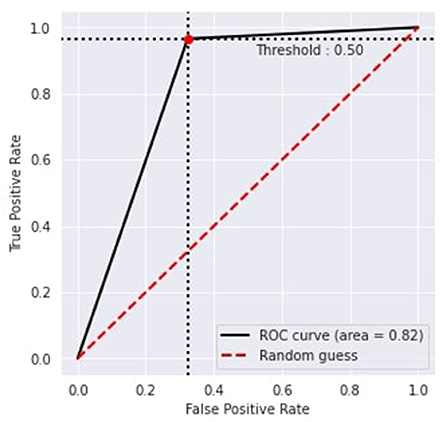
Figure 6 represents the proposed approach's recursive operating characteristics (ROC). The area of the ROC curve is 0.82% with a threshold value of 0.50, which makes the model better than the traditional model.
5. Conclusion
This research proposed an approach for the prediction of Acute Leukemia (ALL) based on machine learning algorithms (RF, SVM, KNN, NB) and ensemble voting classifier with a pre-defined CNN architecture (VGG16, ResNet50, or ResNet101) based feature extraction techniques. SVM performed well-compared to other algorithms with 90.0% accuracy for predicting ALL diseases. The voting classifier has 87.4% accuracy, which is less than the SVM algorithm but better than other algorithms. The result demonstrates that the proposed approach delivered more precise results. The C-NMC leukemia dataset is used from a kaggle repository. Following data preprocessing steps are performed, such as, every image is cropped by using minimum and maximum points techniques. The feature is extracted by applying ResNet50 architecture, and MinMax Normalization is used for data scaling. Three techniques: ANOVA, Recursive Feature Elimination (RFE), and Random Forest (RF), are applied for feature selection. Then preprocessed data are utilized as input for the ML algorithms to predict ALL. Thus, the proposed approach provides practitioners with a reasonable means of determining whether or not a patient has ALL. In the future, model generalizability will be checked by applying multiple datasets.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was funded by the Deanship of Scientific Research at Jouf University under Grant No. (DSR-2021-02-0354).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abbas, S., Jalil, Z., Javed, A. R., Batool, I., Khan, M. Z., Noorwali, A., et al. (2021). BCD-WERT: a novel approach for breast cancer detection using whale optimization based efficient features and extremely randomized tree algorithm. PeerJ Comput. Sci. 7, e390. doi: 10.7717/peerj-cs.390
Abir, W. H., Uddin, M., Khanam, F. R., Tazin, T., Khan, M. M., Masud, M., et al. (2022). Explainable AI in diagnosing and anticipating leukemia using transfer learning method. Comput. Intell. Neurosci. 2022, 5140148. doi: 10.1155/2022/5140148
Alfayez, M., Issa, G. C., Patel, K. P., Wang, F., Wang, X., Short, N. J., et al. (2021). The clinical impact of PTPN11 mutations in adults with acute myeloid leukemia. Leukemia 35, 691–700. doi: 10.1038/s41375-020-0920-z
Ali, T., Nawaz, A., Ur Rehman, A., Ahmad, R., Javed, A., Gadekallu, T., et al. (2022). A sequential machine learning-cum-attention mechanism for effective segmentation of brain tumor. Front. Oncol. 12, 873268. doi: 10.3389/fonc.2022.873268
Alsalem, M., Zaidan, A., Zaidan, B., Hashim, M., Albahri, O. S., Albahri, A. S., et al. (2018). Systematic review of an automated multiclass detection and classification system for acute leukaemia in terms of evaluation and benchmarking, open challenges, issues and methodological aspects. J Med. Syst. 42, 1–36. doi: 10.1007/s10916-018-1064-9
Alsuwaidi, L., Hachim, M., and Senok, A. (2021). Novel markers in pediatric acute lymphoid leukemia: the role of ADAM6 in B cell leukemia. Front. Cell Dev. Biol. 9, 1680. doi: 10.3389/fcell.2021.706129
Aoun, R., Kaul, M., and Sahni, A. (2020). Excessive daytime sleepiness due to brain tumor. J. Clin. Sleep Med. 16, 2117–2119. doi: 10.5664/jcsm.8788
Bibi, N., Sikandar, M., Ud Din, I., Almogren, A., and Ali, S. (2020). Iomt-based automated detection and classification of leukemia using deep learning. J. Healthcare Eng. 2020, 6648574. doi: 10.1155/2020/6648574
Chatila, R., Mansour, J., Mugharbil, A., Nsouli, G., O'Son, L., Sayad, E., et al. (2021). Epidemiology and survival of colorectal cancer in lebanon: a sub-national retrospective analysis. Cancer Control 28, 10732748211041221. doi: 10.1177/10732748211041221
Clinton, L. P. Jr., Somes, K. M., Chu, Y., and Javed, F. (2020). Acute lymphoblastic leukemia detection using depthwise separable convolutional neural networks. SMU Data Sci. Rev. 3, 4.
Dahiwade, D., Patle, G., and Meshram, E. (2019). “Designing disease prediction model using machine learning approach,” in 2019 3rd International Conference on Computing Methodologies and Communication (ICCMC) (IEEE), 1211–1215. doi: 10.1109/ICCMC.2019.8819782
Daqqa, K. A. A., Maghari, A. Y., and Al Sarraj, W. F. (2017). “Prediction and diagnosis of leukemia using classification algorithms,” in 2017 8th International Conference on Information Technology (ICIT) (IEEE), 638–643. doi: 10.1109/ICITECH.2017.8079919
de Oliveira, J. E. M., and Dantas, D. O. (2021). “Classification of normal versus leukemic cells with data augmentation and convolutional neural networks,” in VISIGRAPP (4: VISAPP), 685–692.
Eckardt, J.-N., Bornhäuser, M., Wendt, K., and Middeke, J. M. (2020). Application of machine learning in the management of acute myeloid leukemia: current practice and future prospects. Blood Adv. 4, 6077–6085. doi: 10.1182/bloodadvances.2020002997
Fujita, T. C., Sousa-Pereira, N., Amarante, M. K., and Watanabe, M. A. E. (2021). Acute lymphoid leukemia etiopathogenesis. Mol. Biol. Rep. 48, 817–822. doi: 10.1007/s11033-020-06073-3
Genovese, A., Hosseini, M. S., Piuri, V., Plataniotis, K. N., and Scotti, F. (2021). “Acute lymphoblastic leukemia detection based on adaptive unsharpening and deep learning,” in ICASSP 2021-2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (IEEE), 1205–1209. doi: 10.1109/ICASSP39728.2021.9414362
Ghanem, P., Zouein, A., Mohamad, M., Hodroj, M. H., Haykal, T., Abou Najem, S., et al. (2019). The vitamin e derivative gamma tocotrienol promotes anti-tumor effects in acute myeloid leukemia cell lines. Nutrients. 11, 2808. doi: 10.3390/nu11112808
Godoy, P. B. G., Simionato, N. M., de Mello, C. B., and Suchecki, D. (2020). Assessment of executive functions after treatment of childhood acute lymphoid leukemia: a systematic review. Neuropsychol. Rev. 30, 386–406. doi: 10.1007/s11065-020-09446-4
Hegde, R. B., Prasad, K., Hebbar, H., and Singh, B. M. K. (2019). Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybernet. Biomed. Eng. 39, 382–392. doi: 10.1016/j.bbe.2019.01.005
Hossain, M. A., Islam, A. M., Islam, S., Shatabda, S., and Ahmed, A. (2022). Symptom based explainable artificial intelligence model for leukemia detection. IEEE Access. doi: 10.1109/ACCESS.2022.3176274
Hossain, M. A., Sabik, M. I., Rahman, M., Sakiba, S. N., Muzahidul Islam, A., Shatabda, S., et al. (2021). “An effective leukemia prediction technique using supervised machine learning classification algorithm,” in Proceedings of International Conference on Trends in Computational and Cognitive Engineering (Springer), 219–229. doi: 10.1007/978-981-33-4673-4_19
Jeon, H., and Oh, S. (2020). Hybrid-recursive feature elimination for efficient feature selection. Appl. Sci. 10, 3211. doi: 10.3390/app10093211
Khan, A., Zubair, S., and Al Sabri, M. (2019). “An improved pre-processing machine learning approach for cross-sectional mr imaging of demented older adults,” in 2019 First International Conference of Intelligent Computing and Engineering (ICOICE) (IEEE), 1–7. doi: 10.1109/ICOICE48418.2019.9035164
Kumar, V., Lalotra, G. S., and Kumar, R. K. (2022a). Improving performance of classifiers for diagnosis of critical diseases to prevent covid risk. Comput. Electric. Eng. 102, 108236. doi: 10.1016/j.compeleceng.2022.108236
Kumar, V., Lalotra, G. S., Sasikala, P., Rajput, D. S., Kaluri, R., Lakshmanna, K., et al. (2022b). “Addressing binary classification over class imbalanced clinical datasets using computationally intelligent techniques,” in Healthcare, Vol. 10 (MDPI), 1293. doi: 10.3390/healthcare10071293
Lalotra, G. S., Kumar, V., and Rajput, D. S. (2021). “Predictive performance analysis of ensemble learners on bcd dataset,” in 2021 IEEE International Conference on Technology, Research, and Innovation for Betterment of Society (TRIBES) (IEEE), 1–6. doi: 10.1109/TRIBES52498.2021.9751648
Li, X., Chen, W., Zhang, Q., and Wu, L. (2020). Building auto-encoder intrusion detection system based on random forest feature selection. Comput. Sec. 95, 101851. doi: 10.1016/j.cose.2020.101851
Nazari, E., Farzin, A. H., Aghemiri, M., Avan, A., Tara, M., and Tabesh, H. (2020). Deep learning for acute myeloid leukemia diagnosis. J. Med. Life 13, 382.
Olaolu, A. M., Abdulsalam, S. O., Mope, I. R., and Kazeem, G. A. (2018). A comparative analysis of feature selection and feature extraction models for classifying microarray dataset. Comput. Inf. Syst. J. 29, 1.
Pan, L., Liu, G., Lin, F., Zhong, S., Xia, H., Sun, X., et al. (2017). Machine learning applications for prediction of relapse in childhood acute lymphoblastic leukemia. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-07408-0
Pisner, D. A., and Schnyer, D. M. (2020). “Support vector machine,” in Machine Learning (Elsevier), 101–121. doi: 10.1016/B978-0-12-815739-8.00006-7
Rizwan, M., Shabbir, A., Javed, A. R., Shabbir, M., Baker, T., and Obe, D. A.-J. (2022). Brain tumor and glioma grade classification using gaussian convolutional neural network. IEEE Access 10, 29731–29740. doi: 10.1109/ACCESS.2022.3153108
Sacca, L., Maroun, V., and Khoury, M. (2022). Predictors of high trust and the role of confidence levels in seeking cancer-related information. Inform. Health Soc. Care 47, 53–61. doi: 10.1080/17538157.2021.1925676
Safdar, S., Rizwan, M., Gadekallu, T. R., Javed, A. R., Rahmani, M. K. I., Jawad, K., et al. (2022). Bio-imaging-based machine learning algorithm for breast cancer detection. Diagnostics. 12, 1134. doi: 10.3390/diagnostics12051134
Sharif, M., Amin, J., Siddiqa, A., Khan, H. U., Malik, M. S. A., Anjum, M. A., et al. (2020). Recognition of different types of leukocytes using yolov2 and optimized bag-of-features. IEEE Access. 8, 167448–167459. doi: 10.1109/ACCESS.2020.3021660
Sleiman, Z., Hussein, S., Mohsen, A., Khazzaka, A., Tropea, A., and Biondi, A. (2019). Laparoscopic management of uncommon benign uterine tumors: a systematic review. Updates Surg. 71, 637–643. doi: 10.1007/s13304-019-00651-2
Webb, G. I., Keogh, E., and Miikkulainen, R. (2010). Naïve bayes. Encycloped. Mach. Learn. 15, 713–714. doi: 10.1007/978-0-387-30164-8_576
Winter, G., Kirschner-Schwabe, R., Groeneveld-Krentz, S., Escherich, G., Möricke, A., von Stackelberg, A., et al. (2021). Clinical and genetic characteristics of children with acute lymphoblastic leukemia and li-fraumeni syndrome. Leukemia 35, 1475–1479. doi: 10.1038/s41375-021-01163-y
Keywords: leukemia prediction, acute lymphocytic leukemia, feature extraction, feature selection, machine learning, voting algorithm
Citation: Almadhor A, Sattar U, Al Hejaili A, Ghulam Mohammad U, Tariq U and Ben Chikha H (2022) An efficient computer vision-based approach for acute lymphoblastic leukemia prediction. Front. Comput. Neurosci. 16:1083649. doi: 10.3389/fncom.2022.1083649
Received: 29 October 2022; Accepted: 14 November 2022;
Published: 24 November 2022.
Edited by:
Abdul Mueed Hafiz, University of Kashmir, IndiaReviewed by:
Vinod Kumar, K L University, IndiaSaurabh Singh, Dongguk University Seoul, South Korea
Copyright © 2022 Almadhor, Sattar, Al Hejaili, Ghulam Mohammad, Tariq and Ben Chikha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmad Almadhor, aaalmadhor@ju.edu.sa; Usman Sattar, usman.sattar@bnu.edu.pk
 Ahmad Almadhor
Ahmad Almadhor Usman Sattar
Usman Sattar Abdullah Al Hejaili3
Abdullah Al Hejaili3  Uzma Ghulam Mohammad
Uzma Ghulam Mohammad Usman Tariq
Usman Tariq Haithem Ben Chikha
Haithem Ben Chikha