- 1Department of Clinical Neurophysiology, BABA Center, Children’s Hospital, Helsinki University Hospital and University of Helsinki, Helsinki, Finland
- 2Department of Signal Processing and Acoustics, Aalto University, Espoo, Finland
- 3Department of Paediatrics, Children’s Hospital Helsinki University Hospital, Helsinki, Finland
- 4Neuroscience Center, Helsinki Institute of Life Science, University of Helsinki, Helsinki, Finland
- 5Brain Modeling Group, QIMR Berghofer Medical Research Institute, Brisbane, QLD, Australia
Objective: To develop a non-invasive and clinically practical method for a long-term monitoring of infant sleep cycling in the intensive care unit.
Methods: Forty three infant polysomnography recordings were performed at 1–18 weeks of age, including a piezo element bed mattress sensor to record respiratory and gross-body movements. The hypnogram scored from polysomnography signals was used as the ground truth in training sleep classifiers based on 20,022 epochs of movement and/or electrocardiography signals. Three classifier designs were evaluated in the detection of deep sleep (N3 state): support vector machine (SVM), Long Short-Term Memory neural network, and convolutional neural network (CNN).
Results: Deep sleep was accurately identified from other states with all classifier variants. The SVM classifier based on a combination of movement and electrocardiography features had the highest performance (AUC 97.6%). A SVM classifier based on only movement features had comparable accuracy (AUC 95.0%). The feature-independent CNN resulted in roughly comparable accuracy (AUC 93.3%).
Conclusion: Automated non-invasive tracking of sleep state cycling is technically feasible using measurements from a piezo element situated under a bed mattress.
Significance: An open source infant deep sleep detector of this kind allows quantitative, continuous bedside assessment of infant’s sleep cycling.
Introduction
Recent studies on sleep quality in the intensive care units have prompted interest in early sleep monitoring due to its association with general well-being and distress (van den Hoogen et al., 2017; Werth et al., 2017a). Compromised sleep in infancy is also considered to increase the risk of neurodevelopmental delay (Paruthi et al., 2016; van den Hoogen et al., 2017). Several studies have indicated that infant’s ability to fluctuate between sleep states, a.k.a. sleep-wake cycling (SWC) carries important prognostic information. Bedside tracking of SWC is still based on visual assessment of a compressed trend display of scalp-recorded electroencephalograph (Thoresen et al., 2010; Klebermass et al., 2011), which essentially identifies alternation between deep sleep (quiet sleep) and other vigilance states.
A wide range of methods have been used for the assessment of sleep in infants (Werth et al., 2017a,b). The gold standard in short-term infant sleep studies is polysomnography (PSG), a non-invasive technique that combines a large set of physiological signals, recorded overnight, to generate an assessment of sleep behavior (Grigg-Damberger et al., 2007). For long-term studies, infant sleep behavior is assessed with sleep diaries and questionnaires (Sadeh, 2004; Paavonen et al., 2019). Recent work has also used wrist- or ankle-worn actigraphy (Sadeh, 2011; Paavonen et al., 2019) to provide rough assessments of sleep-wake cycles. All of these methods have significant limitations. The use of PSG is hampered by its relative obtrusiveness and is labor-intensive in both recording and analysis, questionnaires have only limited accuracy and reliability, while actigraphy is challenged in infants due to many factors that confound interpretation (Sokoloff et al., 2020).
Several alternative solutions have been recently proposed for infant sleep studies, based on one or more physiological signals, such as cardiac, respiratory, or body movements. These works have shown clearly that wake and sleep states exhibit characteristic changes in respiration variability, body movements, and heart rate variability (Haddad et al., 1987; Harper et al., 1987). As a result, rules have been proposed for sleep scoring based on body movements and changes in the pattern of respiration, both of which can be reliably recorded with bed mattress sensors (BMS) (Thoman and Tynan, 1979; Erkinjuntti et al., 1990; Kirjavainen et al., 1996).
Recent developments in computational analyses have introduced several sleep state classifiers that are based on one or more signals in the PSG recording (Held et al., 2006; Gerla et al., 2009), such as electroencephalography (EEG) (Dereymaeker et al., 2017; Koolen et al., 2017; Ansari et al., 2020), electrocardiography (ECG) (Werth et al., 2017b) and respiratory inductive plethysmography (RIP) (Sazonova et al., 2006; Terrill et al., 2010; Terrill et al., 2012; Long et al., 2015; Isler et al., 2016). These classifiers have used heuristics (Haddad et al., 1987), computational thresholds (Terrill et al., 2012; Isler et al., 2016), discriminant analyses (Harper et al., 1987) and machine learning approaches (Sazonova et al., 2006; Koolen et al., 2017; Werth et al., 2017a; Ansari et al., 2020) to classify sleep states.
Measuring respiration with BMS may allow non-invasive long-term monitoring in the neonatal intensive care unit (NICU). In this context, the clinical need is focused on tracking cycling between deep sleep and wake—rather than an accurate detection of sleep states as in the clinical sleep medicine unit. While several BMS-based classifiers have been developed for adults in both research (Kortelainen et al., 2010; Mendez et al., 2010; Jansen and Shankar, 1993) and commercial consumer products (Tal et al., 2017; Ranta et al., 2019), there is a dearth of BMS based sleep classifiers for infants (Thoman and Glazier, 1987).
Here, we aimed to develop a BMS-based classifier for an automated assessment of deep sleep to allow observing infant’s sleep cycling. We also studied the effects of classification architecture and augmentation with the ECG on classification accuracy.
Methods
This study used a retrospective collection of PSG and BMS recordings. Study design is outlined in Figure 1 including examples of BMS data in different vigilance states, and a schematic diagram of classifier construction. All classifiers were trained using labels generated by the visual interpretation of the hypnogram. The signal pre-processing, feature extraction, and selection, SVM training and testing, in addition to final analysis were conducted in Matlab [MATLAB 2016. version 9.1.0 (R2016b), The MathWorks Inc., Natick, Massachusetts]. The neural networks were trained and tested in TensorFlow (Abadi et al., 2015). The classifiers are publicly available at GitHub (Ranta, 2020).
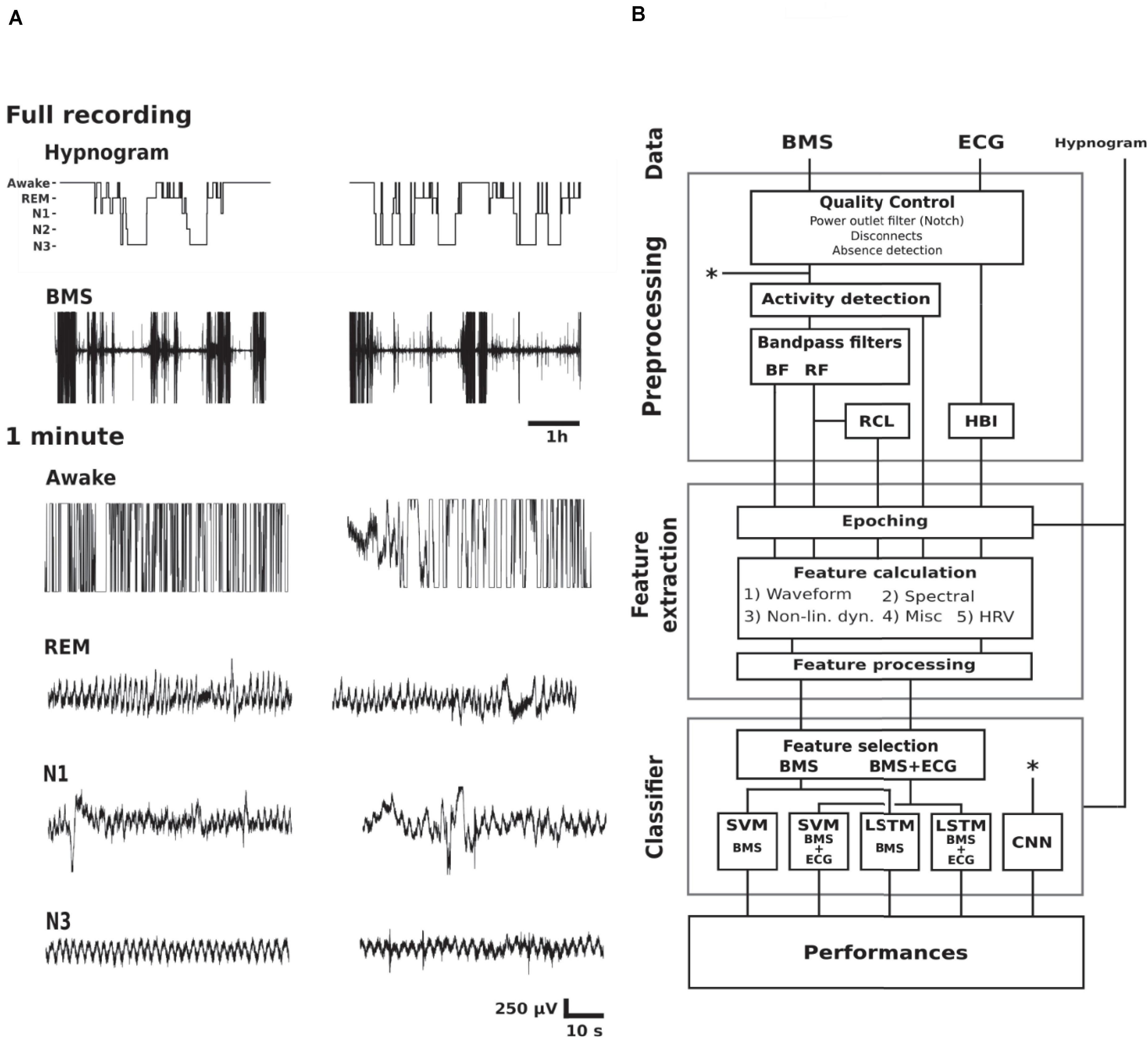
Figure 1. Study design. (A) Examples of hypnogram and BMS data of two subjects (columns) in different vigilance states. The BMS signals below the hypnograms show the full length of BMS signal where movement epochs are readily observed. The shorter 1 min epochs show examples of awake (lots of high amplitude artifacts), REM sleep (variable respiration frequency with relatively stable baseline), light sleep (N1; variable respiration frequency with baseline instability due to movements), as well as N3 (relatively stationary respiration, i.e., steady respiration frequency and amplitude). (B) A schematic diagram of classifier construction divided into functional blocks. Abbreviations: Rapid eye movement (REM), non-REM 1–3 (N1–3), bed mattress sensor (BMS), ballistocardiographic bandpass filtered BMS (BF), respiration frequency bandpass filtered BMS (RF), respiration cycle length series (RCL), heart beat interval series (HBI), non-linear dynamics feature category (Non-lin. dyn.), heart rate variability (HRV), support vector machine (SVM), Long Short-Term Memory neural network (LSTM), convolutional neural network (CNN). Asterisks refer to the shortcut of the CNN pipeline from the raw signal (upper asterisk) to the classifier training (lower asterisk).
Data Acquisition
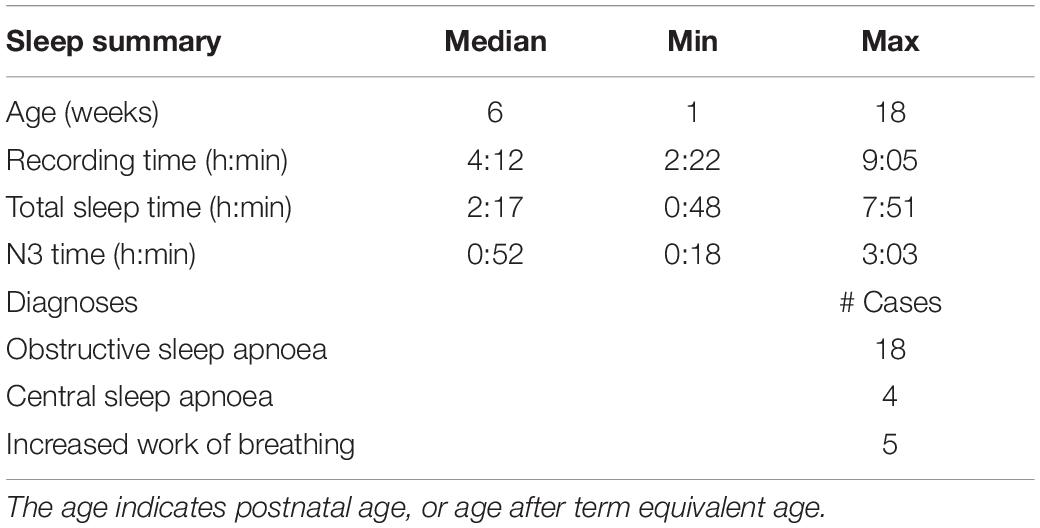
A data set of 51 infants was collected at the Department of Children’s Clinical Neurophysiology, Children’s Hospital, Helsinki University Hospital (see Table 1 and Supplementary Table S1), including all infants (< 6 months) that were assigned to a clinical sleep study in 2016. The PSG data was gathered using Embla N700 equipment and RemLogic 3.2.0 software (Natus, United States) as per routine clinical protocol. A thin 40 cm × 50 cm × 6.35 mm electromechanical ferroelectric sleep mattress sensor (model L-4060SLC, Emfit, Finland) was recorded as an additional channel in the analog input of the PSG recorder (pass band between 0.07 and 48 Hz). All signals were sampled at 200 Hz. The thin, cellular, quasi-piezoelectric film sensor generates a charge when the external pressure changes (Paajanen et al., 2000, 2001). The data set was visually reviewed to exclude recordings with poor signal quality, i.e., no visually detectable respiration, leaving 43 PSG recordings for classifier development. The study was approved by the Institutional Review Ethics Committee of Helsinki Hospital.
In addition, we tested the real-life feasibility of this method by recording two newborn infants during their stay in the NICU. These infants both represent the key target group where SWC monitoring is of interest during the early stay in the NICU (Thoresen et al., 2010; Klebermass et al., 2011): They were both born at full-term and recorded during days 2–3 after birth. One infant was admitted to NICU due to a placenta ablation during labor, leading to resuscitation. The other infant was taken to NICU due to severe birth asphyxia. No other brain pathology was known by the time of recording. The recordings included bed mattress as above, as well as a standard four electrode EEG to be inspected as the amplitude integrated trend (aEEG), the basis of current bedside detection of SWC (Thoresen et al., 2010; Klebermass et al., 2011). The SWC in the aEEG trend was visually compared to the output of BMS-based classification to verify the correspondence.
Clinical review of PSG studies included sleep scoring, i.e., generation of hypnogram, followed by analysis of respiratory events. The hypnogram was scored every 30 s according to the AASM guidelines by a clinical expert (T.K.) who was unaware of this study at the time of clinical sleep scoring (Grigg-Damberger et al., 2007).
We chose to focus on the distinction between deep sleep (N3) and other states for two reasons: First, the clinical need for online sleep monitoring is mainly to recognize sleep-wake cycles (SWC, Kidokoro et al., 2012; van den Hoogen et al., 2017), so identifying likelihood of N3 sleep would be sufficient to quantify cyclicity in sleep states (Stevenson et al., 2014). Second, our clinical experience as well as preliminary analyses showed that distinction of more superficial sleep states, including REM, from the respiratory signals may not be as accurate as needed for SWC quantification (Figure 1A). Therefore, we designed the study to classify N3 from the BMS signals alone, or by using additional input from the ECG signal available in PSG recordings. The full processing pipeline (Figure 1B) consisted of three main blocks; pre-processing, feature extraction, and sleep classifier construction. We trained and evaluated five different classifiers, four feature-based classifiers, and one classifier with representation learning capability.
Pre-processing
A notch filter was used to remove mains artifact (50 Hz). We applied a detection for infant’s presence/absence by using simple thresholding within 6–16 Hz band power, as infants were occasionally removed from the bed. Gross-body movements and disconnects were also identified using simple thresholds applied to the smoothed root-mean-square value.
Next, we separated respiratory and cardiac activity with Butterworth bandpass filters using 0.2–1 Hz band (12–60/min respiratory rate) for respiration frequency range (RF), and 6–16 Hz band was used for identifying ballistocardiography frequency (BF). An algorithm was developed to derive respiratory cycle length time series (RCL). Since the infant heart beats were found to be too weak for robust and reliable detection from BF, we decided to extract heart beat interval series (HBI) from the R-peaks of the ECG that was part of PSG. R-peaks were identified using the Pan Tompkins algorithm (Pan and Tompkins, 1985).
Feature Extraction
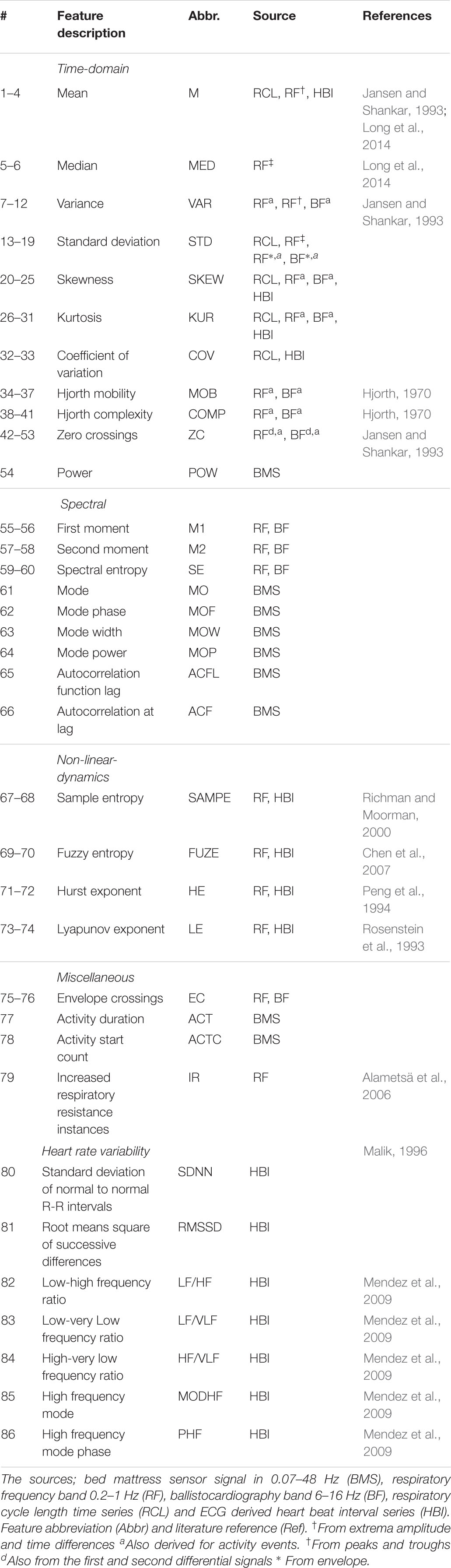
All features were computed for each 30 s epochs corresponding to the visually interpreted hypnograms. The feature set was designed to cover a wide range of physiologically reasoned features from four overall categories; waveform, spectral, non-linear dynamics, and miscellaneous. In addition, we also calculated common heart rate variability features from the ECG data (Malik, 1996). This resulted in a total of 71 BMS-derived features and 15 ECG-based features. See Table 2 for details of the feature set.
Epochs judged to contain excessive movement artifacts were labeled as missing, and replaced by linear interpolation of surrounding feature values. Features with a heavy tailed distribution were log-modulus transformed (John and Draper, 1980) and a subject specific Z-score normalization was applied to reduce interpatient variability.
Classifiers
We evaluated four feature-based classifiers using support vector machine (SVM) (Cortes and Vapnik, 1995) and Long Short-Term Memory recurrent neural network (LSTM) (Hochreiter and Schmidhuber, 1997), as well as a classifier using deep convolutional neural network (CNN) on BMS signal with a representation learning capability. For SVM and LSTM, we tested two feature sets, one with solely BMS-based features (SVM BMS, LSTM BMS) and extensions with additional ECG-based features (SVM BMS + ECG, LSTM BMS + ECG).
We deployed SVMs with radial basis kernels. In addition to treating epoch level features as discrete inputs, we also decided to build temporal context into the SVM classifier (Sazonova et al., 2006). This was reasoned by the presence of significant natural autocorrelation in hypnograms, as well as our attempt to emulate the clinical practice where sleep scores are assumed to inherit conditional inputs from the past epochs (Grigg-Damberger et al., 2007). We included feature values of 6 epochs into our classification building, thus, the classification of each 30 s epoch considers the past 3 min.
To investigate the impact of a longer temporal context we decided to use LSTM with 32 units, that takes preceding feature time series as an input. Temporally distributed layer with softmax activation function yields the class output. The network uses the same features as selected with the SVMs.
We also used an end-to-end CNN classifier, which is capable of feature representation learning from raw input data. The architecture of the utilized CNN is presented in Supplementary Table S2.
Training and Testing
The SVM classifier was trained with a feature selection stage to understand which features are important for classification and reduce the number of unhelpful features. We used a two-stage feature selection algorithm relying on minimal-redundancy maximal-relevance criterion (mRMR) followed by a forward selection procedure (Peng et al., 2005). The kernel scale and box constraint hyperparameters were tuned within each training iteration using three-fold cross-validation and Bayesian optimization (Snoek et al., 2012).
The CNN and LSTM were trained with backpropagation using stochastic gradient descent with the Adam algorithm (Kingma and Ba, 2014) (learning rate = 10–4, β1 = 0.9, β2 = 0.99, CNN batch size = 128 epochs, LSTM batch size = 6 sleep periods) minimizing the sigmoid cross-entropy loss. For CNN, five recordings of the training set were held out as evaluation data for the early stopping criterion.
Performance Evaluation
We used area under receiving operating characteristic curve (AUC) as the primary performance measure. The accuracy (ACC), sensitivity (Sens), specificity (Spes), positive predictive values (PPV) as well as confusion matrices were also calculated.
where TP, TN, FP, and FN denote the numbers of true positives, true negatives, false positives and false negatives, respectively. A true positive is when a N3 epoch is correctly identified as N3. Further methodological description is provided in the online repository (Ranta, 2020).
Results
Our final training dataset from the 43 infant PSG recordings included 21,913 epochs. Of these, 1,891 (8.6%) were excluded due to artifacts or absence periods. The remaining 20,022 epochs included 4,802 (24.0%) of N3 and 15,220 (76.0%) of other sleep stages (N1, N2, REM, and awake).
Feature Selection
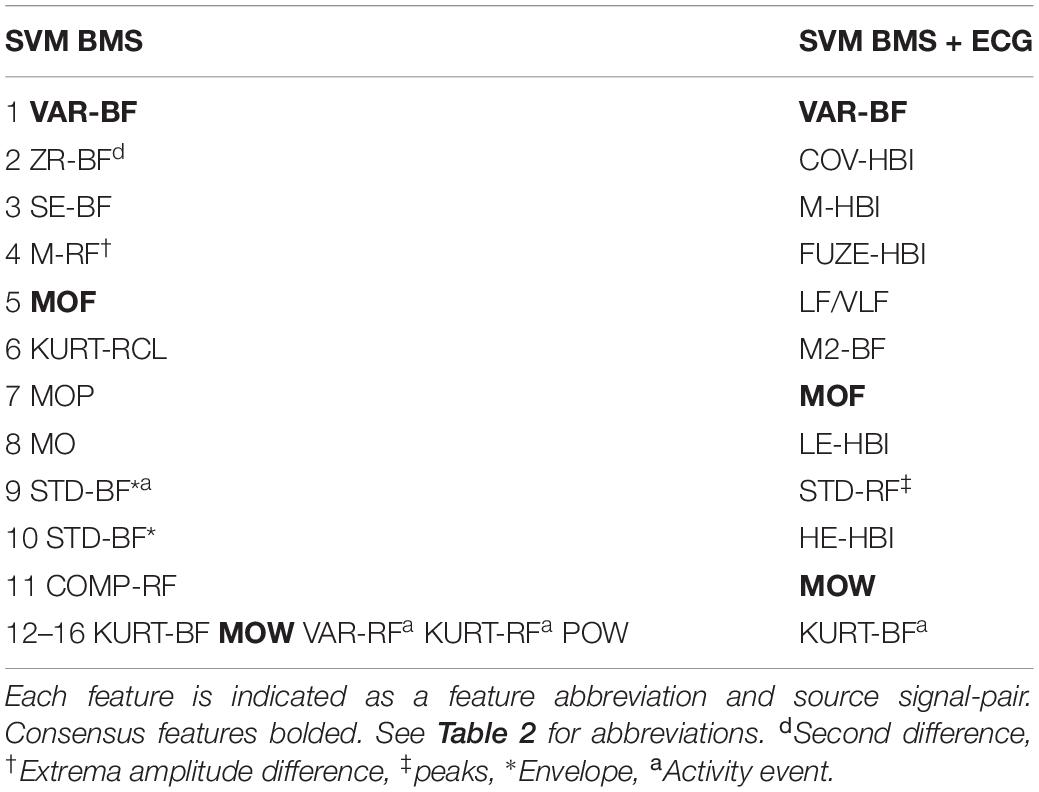
Individual features of the BMS and ECG signals are insufficient for the detection of N3 (see Figures 2C,D,G,H). Feature selection for the SVM BMS yielded 41 features in the first filter phase; the second wrapper phase resulted in 16 final features (Figures 2A,B). Feature selection for the SVM BMS + ECG yielded 31 features in the first phase and 12 features were selected in the second phase (Figures 2E,F). The selected features are listed in Table 3.
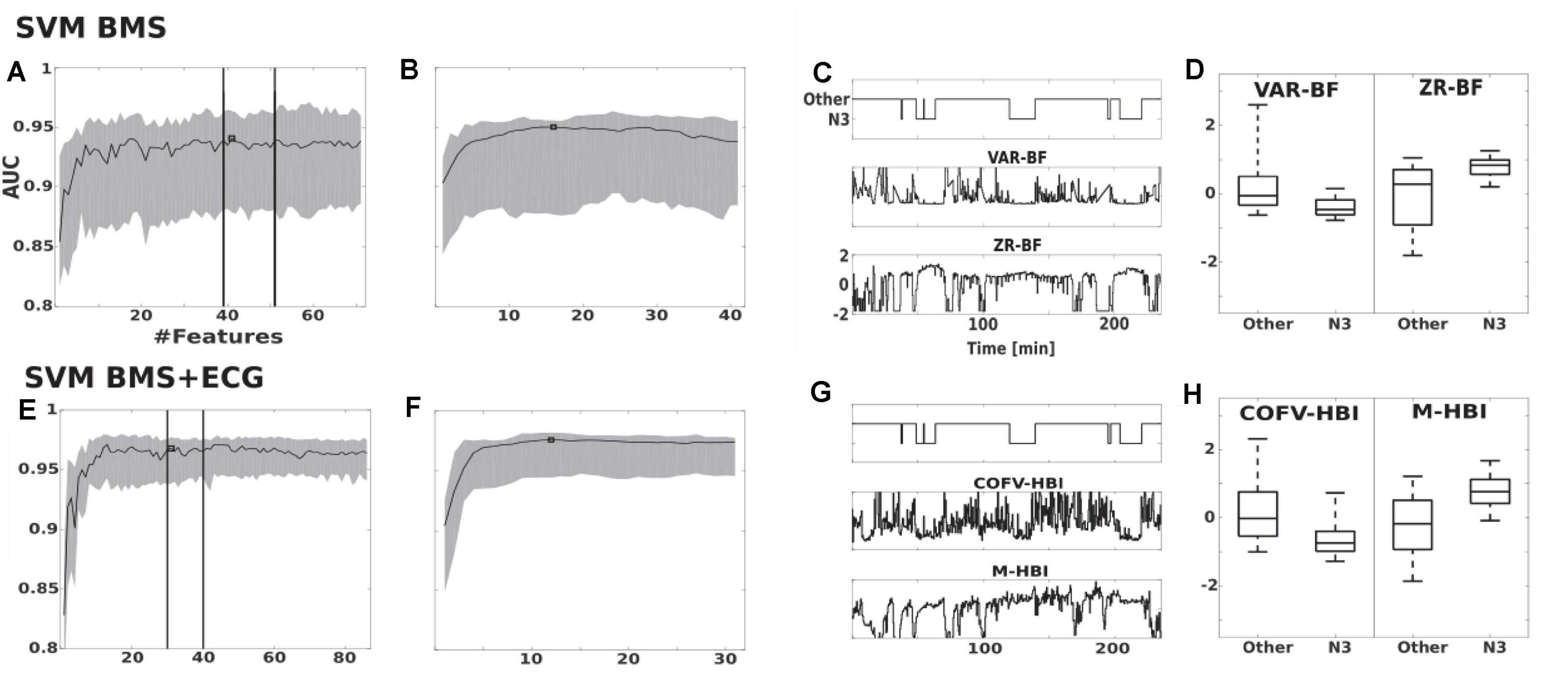
Figure 2. Feature selection of SVM BMS and SVM BMS + ECG classifiers. (A,B) The effect on AUC of adding features in the SVM BMS. The traces show the median and interquartile range of AUC in the two-step maximum-relevance-minimum-redundancy criteria feature selection (A) filter and (B) wrapper phase. Vertical lines indicate the consistent performance interval and square shows the selected feature set. (C,D) Examples of the two most important features, as a function of PSG study as well as comparison of all epochs. (E–H) Corresponding results for SVM BMS + ECG classifier. VAR-BF, variance in 6–16 Hz bed mattress signal band (BF), was the first selected feature for both SVM BMS and SVM BMS + ECG classifiers. The second selected feature was ZR-BF, the number of zero crossing of twice differentiated BF band, and COVF-HBI, the coefficient of variation of heart beat intervals (HBI), for SVM BMS and SVM BMS + ECG, respectively. The third selected feature was M-HBI, the mean of HBI for SVM BMS + ECG.
Classifier Performances
The leave-one-patient-out cross-validation performance of SVM BMS over all 43 patients yielded a median AUC of 95.0% (IQR 87.7–95.6%). The SVM BMS with ECG derived features, had slightly increased performance with a median AUC of 97.6% (94.4–98.2%). Using a LSTM resulted in reduced performance (BMS features: median AUCs 93.9%, IQR 89.2–96.4%; BMS + ECG, median AUC 96.4%, IQR 94.2–98.2%. The CNN applied to the BMS signal resulted in a median AUC of 93.3% (IQR: 90.5–96.1%). However, there were no statistically significant differences in AUC between BMS classifiers (Friedman test, p = 0.977). Similarly, the Wilcoxon signed-rank test did not indicate a significant difference between BMS + ECG classifiers (p = 0.633). Additional results are in Figure 3 and Supplementary Table S3.
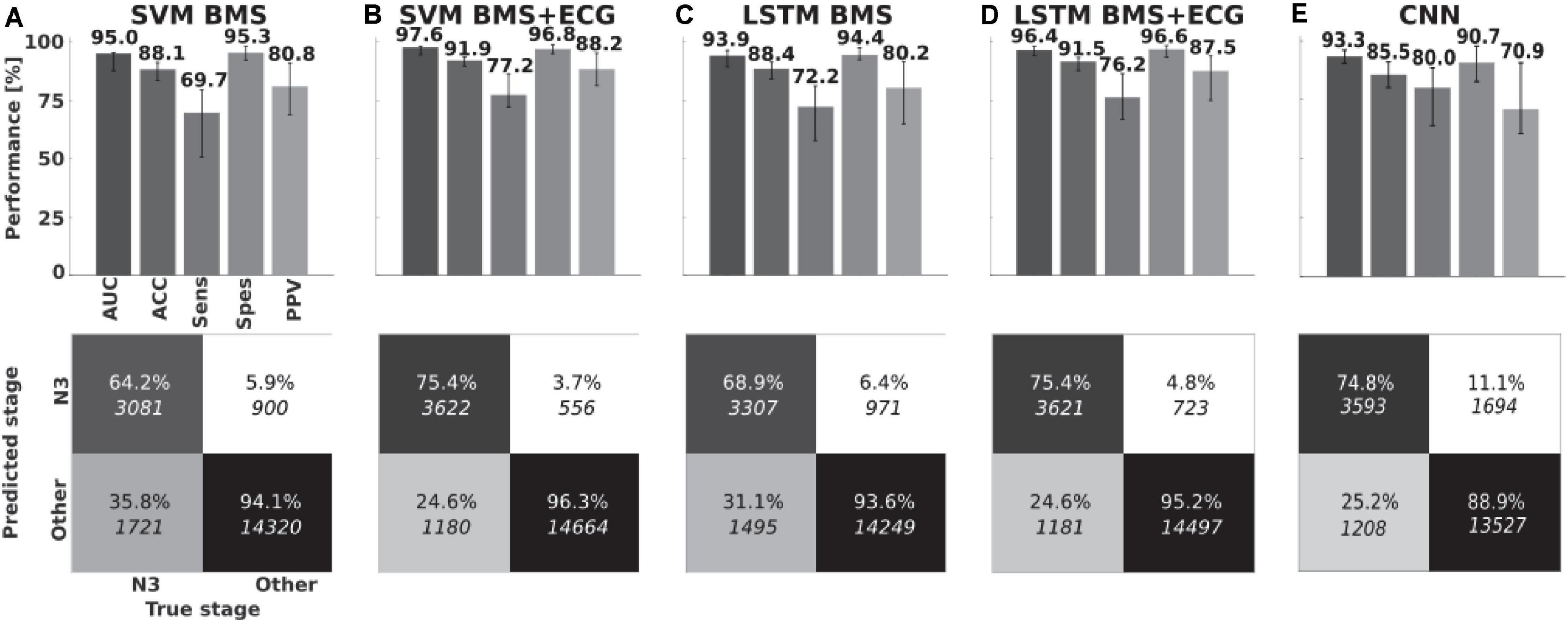
Figure 3. Classifier performances. Medians, interquartile ranges and pooled confusion matrices for (A) BMS feature-based Support Vector Machine classifier and (B) with ECG feature extension. (C,D) Long Short-Term Memory neural networks with same features. (E) Convolutional neural network on bed mattress signal. Area Under Receiving Operating Character Curve (AUC), accuracy (ACC), Sensitivity (Sens), Specificity (Spes), and positive predictive value (PPV).
We also assessed the effect of clinical context on classifier performance (Figure 4), in particular presence of respiratory diagnoses (yes/no, N = 26/17) or infant’s age (under/above 10 weeks, N = 31/12). The accuracy of BMS-based classifiers was slightly affected by the clinical context: The SVM classifier performed a bit worse in infants with respiratory diagnoses (Mann-Whitney U test, p = 0.03 median AUC 92.9 vs. 95.3%), while the LSTM classifier showed somewhat better performance in younger infants (Mann-Whitney U-Test, p = 0.03; median AUC 94.6 vs. 89.8%). There was, however, no significant correlation between AUC and age (Spearman’s rank correlation; rho = −0.23 p = 0.14).
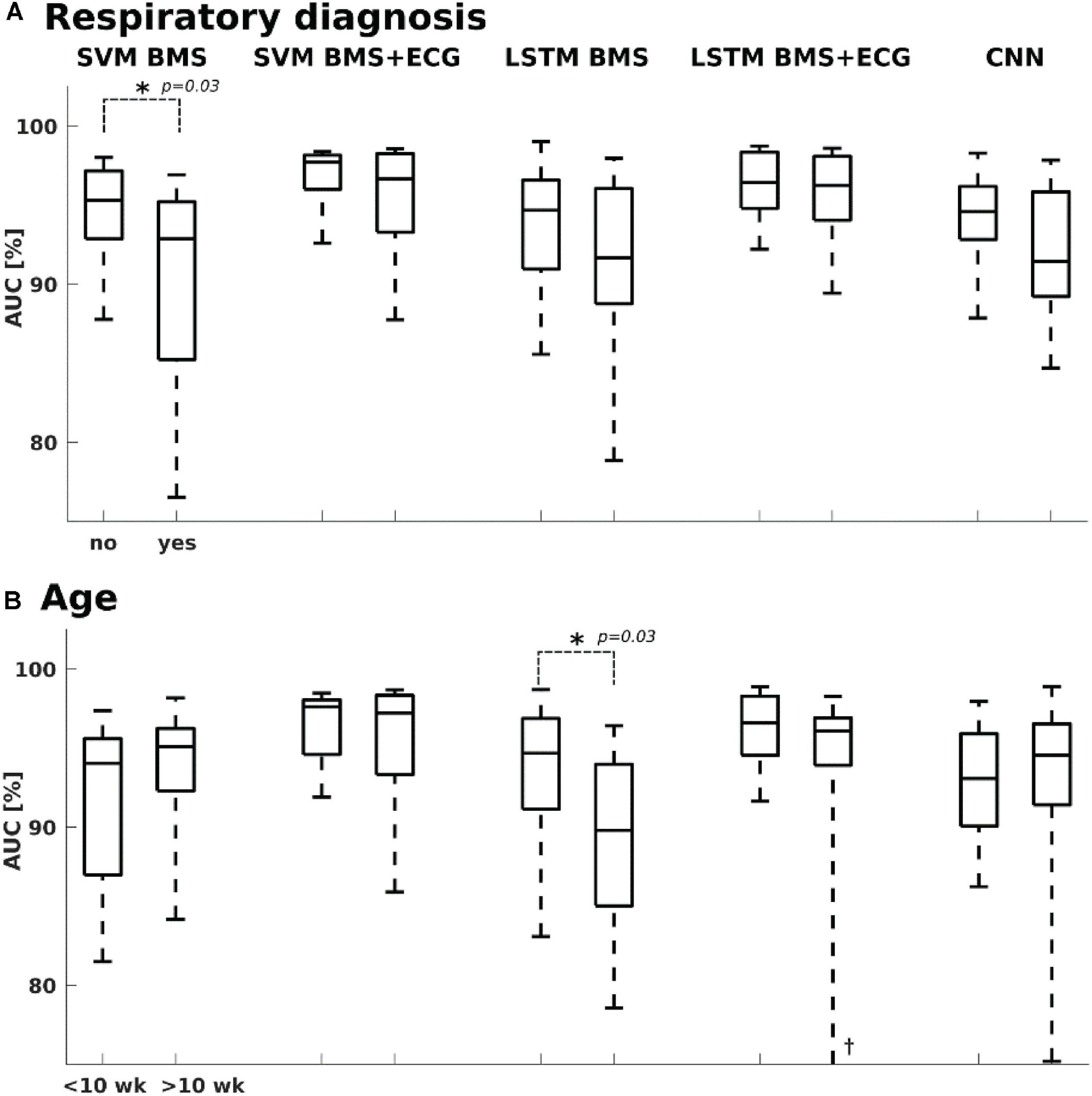
Figure 4. Classifier performance vs. clinical context. (A) SVM BMS classifier showed slightly higher performance with infants who did not have a diagnosis related to respiratory issues, while no significant difference was seen with other classifiers. (B) Only the performance of LSTM BMS was slightly lower with the older age group (> 10 weeks). †Lower whisker at 58%. The age refers to postnatal age, or age after term equivalent age.
Analysis of Features Selected for the Classifier
The most commonly selected features for BMS-based classifiers represent both cardiac and respiratory parameters. The variance of the ballistocardiogram pass-band (VAR-BF) reflects: (1) the reduced cardiac output which manifests the power of BF related to N3 sleep and (2) reduced gross-body movements during N3 which provides similar information to actigraphy. The number of zero crossings from the second difference of BF signal (ZR-BF) and BF spectral entropy (SE-BF) respond similarly. The mean difference of respiration band-pass peaks and troughs (M-RF†) measures the maximum momentary pressure changes in the BMS signal, and as such they are related to the depth of breathing movements. The respiration frequency is reduced in N3.
Feature selection yielded different feature sets on BMS + ECG classifiers, and only three features were in common with the list selected for BMS alone (Table 3). The ECG features selected focus on features describing heart rate variability (COV-HBI, FUZE-HBI, and LF/VLF) and the RR interval (M-HBI) (Figure 2F).
Pilot Test in the NICU
Finally, we performed a recording on two newborn infants in the NICU to see how the BMS-based classifier would compare with the state-of-the-art visual inspection of the aEEG trend that is now routinely available at bedside. As shown in the Figure 5, there was a high correlation between the time course of BMS classifier and the “thickening” epochs in the aEEG trend that are known to correspond to quiet sleep. This correspondence between the “aEEG thickening” and the N3-detection is the ultimate aim for the BMS classifier as it will allow detection of cycling, or SWC. The results from our other pilot recording are shown in the Supplementary Figure S1, which demonstrated how the inherent ambiguity between sleep states may be seen in the dichotomic classifier output before post-processing. Some of the comparable ambiguity is also seen in Figure 5 (near 5 and 7 h). Much of that could be readily removed by post-processing, or by replacing the dichotomic classification with a sleep state probability index as shown before for a comparable EEG-based classifier (Koolen et al., 2017).
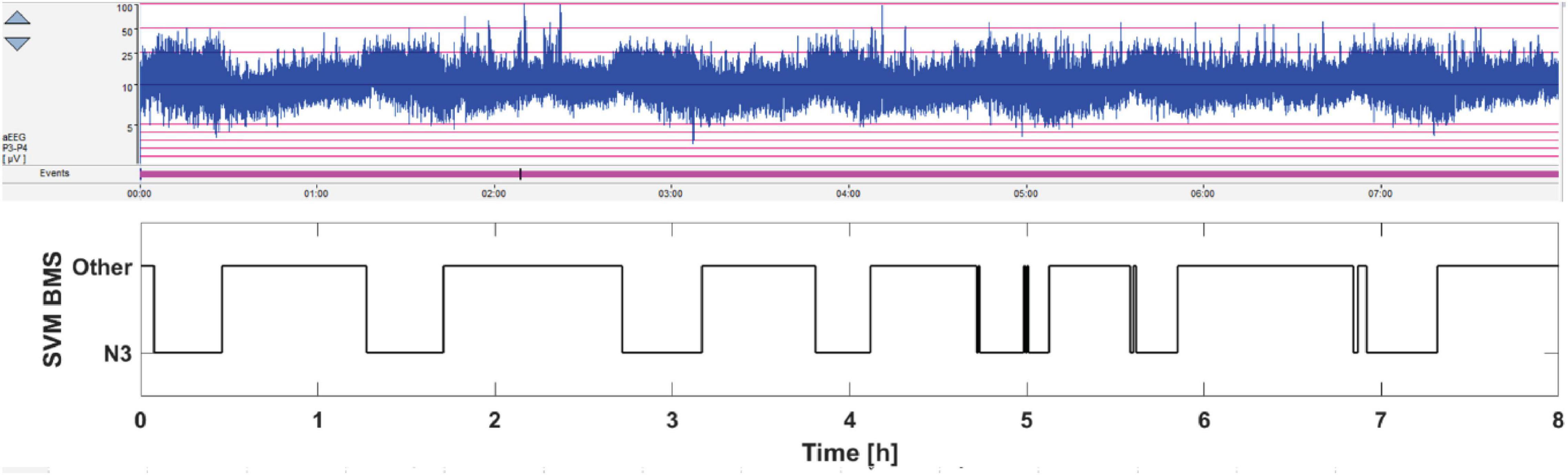
Figure 5. Pilot 8 h monitoring of sleep cycling in the NICU. The current standard practice is to observe SWC pattern in the aEEG trend (above), which refers to the alternation between thicker and thinner trends. The thicker aEEG trend comes from the intermittent, trace alternant/discontinue background pattern, which characterizes quiet (“deep”) sleep state. In contrast, the thinner aEEG trends come from the more continuous EEG pattern that characterizes the active sleep state. The BMS-based sleep monitoring is shown below. Note the faithful correspondence between thicker epochs in the aEEG trend and the deep sleep output of the BMS classifier.
Discussion
We show that an automated detection of deep sleep vs. other sleep states is feasible using fully non-invasive recording of BMS signal alone. We extend on previous work by providing an open source algorithm for infant sleep, which allows automated recognition of SWC. We show further with a pilot recording that such a method could be used as a real-time bedside trend of SWC in infants during intensive care (van den Hoogen et al., 2017) facilitating further development of clustered care and other procedures to minimize infants’ stress during care (van den Hoogen et al., 2017; Knauert et al., 2018). Measuring the amount of deep/quiet sleep (Shellhaas et al., 2017) or the cycling between sleep states (Thoresen et al., 2010; Klebermass et al., 2011) have prognostic value as they are associated with neurodevelopmental outcomes.
The feature selection protocol showed that a reasonable classification accuracy can be achieved with only a handful of features, while the majority of the examined 71 computational features are essentially unhelpful or redundant. This demonstrates that smaller feature sets could be sufficient if computational complexity needs to be optimized, e.g., for an online algorithm or embedded implementation. The addition of features derived from the ECG heart beat increased classifier performance; however, it comes at the cost of needing a merger of BMS and ECG sensor signals.
The two BMS classifier designs, one based on SVM and the other based on LSTM, demonstrated a comparable performance in identifying N3 sleep stage at the full cohort level. However, the SVM-based classifier showed more variability between subjects, and the LSTM-based classifier showed a better performance with the younger subjects. A respiratory diagnosis lowered performance of the classifier. This could be due to pathophysiological changes in breathing stability. Comparable performance between CNN and SVM classifiers indicates that the feature representation can be learned directly from the BMS signal.
Our findings are in agreement with findings from prior studies in infants (Thoman and Glazier, 1987; Sazonova et al., 2006; Terrill et al., 2012; Isler et al., 2016). Thoman and Glazier (1987) reported the total accuracy of 80.6% (active, quiet, transition, awake) for their BMS respiration and gross-body movement-based classifier. However, they used a small sample size (n = 10) and their in-house behavioral scoring criteria as a reference (Thoman, 1975). Deploying different source signals for respiration and gross-body movements has yielded variable though lower (53–70%) accuracies in distinguishing active and quiet sleep states from the RIP or actigraphy signals (N = 26) (Sazonova et al., 2006). Terrill et al. developed a novel computational measure based on recurrence quantification analysis of RIP signals, which allowed sleep classification at a significantly higher accuracy (80%) (Terrill et al., 2012). Likewise, the variance of instantaneous breathing rate was shown to yield over 80% accuracy (Isler et al., 2016). An exact comparison of classifier algorithms between our study and the earlier works is challenging because we used an updated sleep scoring reference (Grigg-Damberger et al., 2007), and there are poorly translatable differences between the physiological signals used in different studies. Our fully non-obtrusive BMS signal might be more sensitive to, e.g., movement artifacts than the body-attached RIP sensor used in prior studies (e.g., Terrill et al., 2012). However, our presently introduced classifier is in a general agreement with the prior works.
The presently introduced BMS-based detector has a limited focus: It aims to detect quiet sleep only. It is not aimed to provide a full multiclass classification of all sleep states, neither does it allow diagnostic measures of specific sleep–related adversities. Such in-depth sleep analyses would require more comprehensive measures of physiological parameters which are routinely available in the existing PSG paradigms. Moreover, it will be important to validate the use of our BMS-based classifier in different kinds of user scenarios in the NICU, including, e.g., infants born preterm, undergoing ventilatory support, or receiving sedative medication or hypothermia therapy; all these factors may have different effects on the relationship between respiration and sleep states. These prospective studies are on-going, and they will become possible in many independent centers with the open-access algorithm provided in this work.
The common challenge with sleep classifiers is in detecting the exact timing of sleep state transition, which lowers the nominal performance. This is also a challenge in clinicians’ visual scoring, and it may substantially lower the inter-rater agreement (Satomaa et al., 2016). Notably, visual scoring of PSG studies relies heavily on multiple physiological signals, including cortical activity from the EEG, while our presently developed classifier relies on respiratory and cardiac activity alone. Adding the other PSG signals to the classifier would undoubtedly improve classifier performance, however it would also directly affect the utility of the classifier as a non-invasive measure for longer term monitoring. It is crucial in this context to consider the aimed use of the automated classifier of this kind. The ultimate purpose is to estimate cycling/fluctuation of vigilance states rather than to estimate accurate transition times (SWC, Kidokoro et al., 2012; van den Hoogen et al., 2017). This implies that identifying any sleep state, such as N3 in this work, with reasonable accuracy would allow quantification of SWC (Stevenson et al., 2014), or its fragmentation. This deflates the importance of exact transition times as long as the overall pattern is detected with a reasonable accuracy.
Conclusion
We introduce an automated signal processing pipeline for infant deep sleep detection from the BMS signal with or without ECG signal. The proposed method allows long-term monitoring of sleep-wake cycling, a key bedside index in the NICU brain monitoring. We also provide pilot proof of concept evidence that this closely corresponds to the SWC observed in the currently available aEEG-based review, hence the method can be applied in the NICU to allow future development of treatments and nursing practices with a minimally disturbed sleep (van den Hoogen et al., 2017).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Ethics Committee of Helsinki Hospital, Helsinki, Finland. Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author Contributions
JR developed the support vector machine and Long Short-Term Memory neural network infant sleep classifiers, conducted sleep classifier evaluation, and comparison and contributed writing the article. MA developed the convolutional neural network classifier and contributed writing the article. TK conducted the infant polysomnygraphy and sleep mattress data collection in addition to sleep stage scoring. SV coordinated the project, contributed clinical, and physiological knowledge on designing the classifiers and took part in writing. NS coordinated the project, took part in designing the classifiers, and contributed writing the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by EU Marie Skłodowska Action (H2020-MCSA-IF-656131), Academy of Finland (335778, 314450, 332017, and 314573), Helsinki University Hospital, Lastentautien tutkimussaatio, Aivosaatio, and Sigrid Juselius Foundation.
Conflict of Interest
JR was a shareholder and a part time employee in sensor manufacturer Emfit Ltd. Emfit did not have any role in study design, data analysis or publication process. This work was mostly done before JR was employed by Emfit.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2020.602852/full#supplementary-material
References
Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z., and Citro, C. (2015). TensorFlow: Large-scale Machine Learning on Heterogeneous Systems. Software. Available online at: http://tensorflow.org/ (accessed December 23, 2020).
Alametsä, J., Rauhala, E., Huupponen, E., Saastamoinen, A., Värri, A., Joutsen, A., et al. (2006). Automatic detection of spiking events in EMFi sheet during sleep. Med. Eng. Phys. 28, 267–275. doi: 10.1016/j.medengphy.2005.07.008
Ansari, A. H., De Wel, O., Pillay, K., Dereymaeker, A., Jansen, K., Van Huffel, S., et al. (2020). A convolutional neural network outperforming state-of-the-art sleep staging algorithms for both preterm and term infants. J. Neural Eng. 17:016028. doi: 10.1088/1741-2552/ab5469
Chen, W., Wang, Z., Xie, H., and Yu, W. (2007). Characterization of surface EMG signal based on fuzzy entropy. IEEE Trans. Neural Syst. Rehabil. Eng. 15, 266–272. doi: 10.1109/tnsre.2007.897025
Dereymaeker, A., Pillay, K., Vervisch, J., Van Huffel, S., Naulaers, G., Jansen, K., et al. (2017). An automated quiet sleep detection approach in preterm infants as a gateway to assess brain maturation. Intl. J. Neural Syst. 27:1750023. doi: 10.1142/s012906571750023x
Erkinjuntti, M., Kero, P., Halonen, J.-P., Mikola, H., and Sainio, K. (1990). Scsb method compared to eeg-based polygraphy in sleep state scoring of newborn infants. Acta Pædiatrica 79, 274–279. doi: 10.1111/j.1651-2227.1990.tb11456.x
Gerla, V., Paul, K., Lhotska, L., and Krajca, V. (2009). Multivariate analysis of full-term neonatal polysomnographic data. IEEE Trans. Inform. Technol. Biomed. 13, 104–110. doi: 10.1109/titb.2008.2007193
Grigg-Damberger, M., Gozal, D., Marcus, C. L., Quan, S. F., Rosen, C. L., Chervin, R. D., et al. (2007). The visual scoring of sleep and arousal in infants and children. J. Clin. Sleep Med. 3, 201–240. doi: 10.5664/jcsm.26819
Haddad, G. G., Jeng, H. J., Lai, T. L., and Melling, R. B. (1987). Determination of sleep state in infants using respiratory variability. Pediatr. Res. 21, 556–562. doi: 10.1203/00006450-198706000-00010
Harper, R. M., Schechtman, V. L., and Kluge, K. A. (1987). Machine classification of infant sleep state using cardiorespiratory measures. Electroencephalogr. Clin. Neurophysiol. 67, 379–387. doi: 10.1016/0013-4694(87)90126-x
Held, C. M., Heiss, J. E., Estévez, P. A., Perez, C. A., Garrido, M., Algarín, C., et al. (2006). Extracting fuzzy rules from polysomnographic recordings for infant sleep classification. IEEE Trans. Biomed. Eng. 53, 1954–1962. doi: 10.1109/tbme.2006.881798
Hjorth, B. (1970). EEG analysis based on time domain properties. Electroencephalogr. Clin. Neurophysiol. 29, 306–310. doi: 10.1016/0013-4694(70)90143-4
Isler, J. R., Thai, T., Myers, M. M., and Fifer, W. P. (2016). An automated method for coding sleep states in human infants based on respiratory rate variability. Dev. Psychobiol. 58, 1108–1115. doi: 10.1002/dev.21482
Jansen, B. H., and Shankar, K. (1993). Sleep staging with movement-related signals. Intl. J. Bio-Med. Comput. 32, 289–297. doi: 10.1016/0020-7101(93)90021-w
John, J. A., and Draper, N. R. (1980). An alternative family of transformations. Appl. Stat. 29, 190–197. doi: 10.2307/2986305
Kingma, D. P., and Ba, J. (2014). Adam: a method for stochastic optimization. arXiv [Preprint]. Available online at: https://arxiv.org/abs/1412.6980 (accessed December 23, 2020).
Kidokoro, H., Inder, T., Okumura, A., and Watanabe, K. (2012). What does cyclicity on amplitude-integrated EEG mean? J. Perinatol. 32, 565–569. doi: 10.1038/jp.2012.25
Kirjavainen, T., Cooper, D., Polo, O., and Sullivan, C. (1996). Respiratory and body movements as indicators of sleep stage and wakefulness in infants and young children. J. Sleep Res. 5, 186–194. doi: 10.1046/j.1365-2869.1996.t01-1-00003.x
Klebermass, K., Olischar, M., Waldhoer, T., Fuiko, R., Pollak, A., and Weninger, M. (2011). Amplitude-integrated EEG pattern predicts further outcome in preterm infants. Pediatr. Res. 70, 102–108. doi: 10.1203/pdr.0b013e31821ba200
Knauert, M. P., Redeker, N. S., Yaggi, H. K., Bennick, M., and Pisani, M. A. (2018). Creating naptime: an overnight, nonpharmacologic intensive care unit sleep promotion protocol. J. Patient Exp. 5, 180–187. doi: 10.1177/2374373517747242
Koolen, N., Oberdorfer, L., Rona, Z., Giordano, V., Werther, T., Klebermass-Schrehof, K., et al. (2017). Automated classification of neonatal sleep states using EEG. Clin. Neurophysiol. 128, 1100–1108. doi: 10.1016/j.clinph.2017.02.025
Kortelainen, J. M., Mendez, M. O., Bianchi, A. M., Matteucci, M., and Cerutti, S. (2010). Sleep staging based on signals acquired through bed sensor. IEEE Trans. Inform. Technol. Biomed. 14, 776–785. doi: 10.1109/titb.2010.2044797
Long, X., Arends, J. B., Aarts, R. M., Haakma, R., Fonseca, P., and Rolink, J. (2015). Time delay between cardiac and brain activity during sleep transitions. Appl. Phys. Lett. 106:143702.
Long, X., Foussier, J., Fonseca, P., Haakma, R., and Aarts, R. M. (2014). Analyzing respiratory effort amplitude for automated sleep stage classification. Biomed. Signal Process. Control 14, 197–205. doi: 10.1016/j.bspc.2014.08.001
Malik, M. (1996). Heart rate variability: standards of measurement, physiological interpretation, and clinical use: task force of the european society of cardiology and the north american society for pacing and electrophysiology. Ann. Noninvasive Electrocardiol. 1, 151–181. doi: 10.1111/j.1542-474x.1996.tb00275.x
Mendez, M., Matteucci, M., Cerutti, S., Bianchi, A., and Kortelainen, J. M. (2009). Automatic detection of sleep macrostructure based on bed sensors. Ann. Intl. Conf. IEEE Eng. Med. Biol. Soc. 2009, 5555–5558.
Mendez, M. O., Migliorini, M., Kortelainen, J. M., Nistico, D., Arce-Santana, E., Cerutti, S., et al. (2010). Evaluation of the sleep quality based on bed sensor signals: time-variant analysis. Annu. Int. IEEE Eng. Med. Biol. Soc. 2010, 3994–3997.
Paajanen, M., Lekkala, J., and Kirjavainen, K. (2000). ElectroMechanical Film (EMFi) - a new multipurpose electret material. Sens. Actuators 84, 95–102. doi: 10.1016/s0924-4247(99)00269-1
Paajanen, M., Lekkala, J., and Valimaki, H. (2001). Electromechanical modeling and properties of the electret film EMFI. IEEE Trans. Dielectr. Electr. Insul. 8, 629–636. doi: 10.1109/94.946715
Paavonen, E. J., Morales-Muñoz, I., Pölkki, P., Paunio, T., Porkka-Heiskanen, T., Kylliäinen, A., et al. (2019). Development of sleep–wake rhythms during the first year of age. J. Sleep Res. 29:e12918.
Pan, J., and Tompkins, W. J. (1985). A real-time qrs detection algorithm. IEEE Trans. Biomed. Eng. 32, 230–236. doi: 10.1109/tbme.1985.325532
Paruthi, S., Brooks, L. J., D’Ambrosio, C., Hall, W. A., Kotagal, S., Lloyd, R. M., et al. (2016). Consensus statement of the american academy of sleep medicine on the recommended amount of sleep for healthy children: methodology and discussion. J. Clin. Sleep Med. 12, 1549–1561. doi: 10.5664/jcsm.6288
Peng, C.-K., Buldyrev, S. V., Havlin, S., Simons, M., Stanley, H. E., and Goldberger, A. L. (1994). Mosaic organization of dna nucleotides. Physical Rev. 49, 1685–1689. doi: 10.1103/physreve.49.1685
Peng, H., Long, F., and Ding, C. (2005). Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 8, 1226–1238. doi: 10.1109/tpami.2005.159
Ranta, J. R. (2020). Infant Bed Mattress Sensor Sleep Classifier. Available online at: https://github.com/yuccaRanta/BMSSC (accessed December 23, 2020).
Ranta, J. R., Aittokoski, T., Tenhunen, M., and Alasaukko-Oja, M. (2019). Emfit QS heart rate and respiration rate validation. Biomed. Phys. Eng. Exp. 5:025016. doi: 10.1088/2057-1976/aafbc8
Richman, J. S., and Moorman, J. R. (2000). Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 278, H2039–H2049.
Rosenstein, M. T., Collins, J. J., and De Luca, C. J. (1993). A practical method for calculating largest Lyapunov exponents from small data sets. Physica D: Nonlinear Phenomena 65, 117–134. doi: 10.1016/0167-2789(93)90009-p
Sadeh, A. (2004). A brief screening questionnaire for infant sleep problems: validation and findings for an internet sample. Pediatrics 113, e570–e577.
Sadeh, A. (2011). The role and validity of actigraphy in sleep medicine: an update. Sleep Med. Rev. 15, 259–267. doi: 10.1016/j.smrv.2010.10.001
Satomaa, A.-L., Saarenpää-Heikkilä, O., Paavonen, E. J., and Himanen, S.-L. (2016). The adapted American academy of sleep medicine sleep scoring criteria in one month old infants: a means to improve comparability? Clin. Neurophysiol. 127, 1410–1418. doi: 10.1016/j.clinph.2015.08.013
Sazonova, N. A., Sazonov, E. E., Tan, B., Schuckers, S. A., and Chime Study, and Group. (2006). Sleep state scoring in infants from respiratory and activity measurements. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 2462–2465.
Shellhaas, R. A., Burns, J. W., Hassan, F., Carlson, M. D., Barks, J. D., and Chervin, R. D. (2017). Neonatal sleep–wake analyses predict 18-month neurodevelopmental outcomes. Sleep 40:zsx144.
Snoek, J., Larochelle, H., and Adams, R. P. (2012). Practical Bayesian optimization of machine learning algorithms. Adv. Neural Inform. Process. Syst. 2, 2951–2959.
Sokoloff, G., Hickerson, M. M., Wen, R. Y., Tobias, M. E., McMurray, B., and Blumberg, M. S. (2020). Spatiotemporal organization of myoclonic twitching in sleeping human infants. Dev. Psychobiol. 62, 697–710. doi: 10.1002/dev.21954
Stevenson, N. J., Palmu, K., Wikström, S., Hellström-Westas, L., and Vanhatalo, S. (2014). Measuring brain activity cycling (BAC) in long term EEG monitoring of preterm babies. Physiol. Meas. 35, 1493–1508. doi: 10.1088/0967-3334/35/7/1493
Tal, A., Shinar, Z., Shaki, D., Codish, S., and Goldbart, A. (2017). Validation of contact-free sleep monitoring device with comparison to polysomnography. J. Clin. Sleep Med. 13, 517–522. doi: 10.5664/jcsm.6514
Terrill, P. I., Wilson, S. J., Suresh, S., Cooper, D. M., and Dakin, C. (2010). Attractor structure discriminates sleep states: recurrence plot analysis applied to infant breathing patterns. IEEE Trans. Biomed. Eng. 57, 1108–1116. doi: 10.1109/tbme.2009.2038362
Terrill, P. I., Wilson, S. J., Suresh, S., Cooper, D. M., and Dakin, C. (2012). Application of recurrence quantification analysis to automatically estimate infant sleep states using a single channel of respiratory data. Med. Biol. Eng. Comput. 50, 851–865. doi: 10.1007/s11517-012-0918-4
Thoman, E. B. (1975). Sleep and wake behaviors in neonates: consistencies and consequences. Merrill Palmer Q. Behav. Dev. 21, 295–314.
Thoman, E. B., and Glazier, R. C. (1987). Computer scoring of motility patterns for states of sleep and wakefulness: human infants. Sleep 10, 122–129. doi: 10.1093/sleep/10.2.122
Thoman, E. B., and Tynan, W. D. (1979). Sleep states and wakefulness in human infants: profiles from motility monitoring. Physiol. Behav. 23, 519–525. doi: 10.1016/0031-9384(79)90052-0
Thoresen, M., Hellström-Westas, L., Liu, X., and de Vries, L. S. (2010). Effect of hypothermia on amplitude-integrated electroencephalogram in infants with asphyxia. Pediatrics 126, e131–e139.
van den Hoogen, A., Teunis, C. J., Shellhaas, R. A., Pillen, S., Benders, M., and Dudink, J. (2017). How to improve sleep in a neonatal intensive care unit: a systematic review. Early. Hum. Dev. 113, 78–86. doi: 10.1016/j.earlhumdev.2017.07.002
van den Oord, A., Dieleman, S., Zen, H., Simonyan, K., Vinyals, O., Graves, A., et al. (2016). Wavenet: a generative model for raw audio. arXiv [Preprint]. Available online at: https://arxiv.org/abs/1609.03499 (accessed December 23, 2020).
Werth, J., Atallah, L., Andriessen, P., Long, X., Zwartkruis-Pelgrim, E., and Aarts, R. M. (2017a). Unobtrusive sleep state measurements in preterm infants–a review. Sleep Med. Rev. 32, 109–122. doi: 10.1016/j.smrv.2016.03.005
Werth, J., Long, X., Zwartkruis-Pelgrim, E., Niemarkt, H., Chen, W., Aarts, R. M., et al. (2017b). Unobtrusive assessment of neonatal sleep state based on heart rate variability retrieved from electrocardiography used for regular patient monitoring. Early Hum. Dev. 113, 104–113. doi: 10.1016/j.earlhumdev.2017.07.004
Keywords: infant sleep, non-invasive monitoring, intensive care monitoring, NICU, bed mattress sensor, sleep-wake cycling
Citation: Ranta J, Airaksinen M, Kirjavainen T, Vanhatalo S and Stevenson NJ (2021) An Open Source Classifier for Bed Mattress Signal in Infant Sleep Monitoring. Front. Neurosci. 14:602852. doi: 10.3389/fnins.2020.602852
Received: 14 September 2020; Accepted: 15 December 2020;
Published: 14 January 2021.
Edited by:
Alessandro Silvani, University of Bologna, ItalyReviewed by:
Flora Wong, Monash University, AustraliaErwan Stéphan-Blanchard, UPJV, University of Picardy Jules Verne, France
Copyright © 2021 Ranta, Airaksinen, Kirjavainen, Vanhatalo and Stevenson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jukka Ranta, jukka.r.ranta@helsinki.fi; Sampsa Vanhatalo, sampsa.vanhatalo@helsinki.fi
 Jukka Ranta
Jukka Ranta Manu Airaksinen
Manu Airaksinen Turkka Kirjavainen
Turkka Kirjavainen Sampsa Vanhatalo
Sampsa Vanhatalo Nathan J. Stevenson
Nathan J. Stevenson