Evolution and functional diversity of the Calcium Binding Proteins (CaBPs)
- The Physiological Laboratory, Department of Cellular and Molecular Physiology, Institute of Translational Medicine, University of Liverpool, Liverpool, UK
The mammalian central nervous system (CNS) exhibits a remarkable ability to process, store, and transfer information. Key to these activities is the use of highly regulated and unique patterns of calcium signals encoded by calcium channels and decoded by families of specific calcium-sensing proteins. The largest family of eukaryotic calcium sensors is those related to the small EF-hand containing protein calmodulin (CaM). In order to maximize the usefulness of calcium as a signaling species and to permit the evolution and fine tuning of the mammalian CNS, families of related proteins have arisen that exhibit characteristic calcium binding properties and tissue-, cellular-, and sub-cellular distribution profiles. The Calcium Binding Proteins (CaBPs) represent one such family of vertebrate specific CaM like proteins that have emerged in recent years as important regulators of essential neuronal target proteins. Bioinformatic analyses indicate that the CaBPs consist of two subfamilies and that the ancestral members of these are CaBP1 and CaBP8. The CaBPs have distinct intracellular localizations based on different targeting mechanisms including a novel type-II transmembrane domain in CaBPs 7 and 8 (otherwise known as calneuron II and calneuron I, respectively). Recent work has led to the identification of new target interactions and possible functions for the CaBPs suggesting that they have multiple physiological roles with relevance for the normal functioning of the CNS.
Introduction
Communication between neurons lies at the heart of abstract higher level cognitive processes including memory acquisition, learning, and complex reasoning. Fundamental to the mechanism by which all mammalian neurons communicate is intracellular calcium signaling (Berridge, 1998; Lohmann, 2009). Complex patterns of spatial and temporal calcium signals drive alterations in synaptic plasticity and neuronal gene expression which in turn affect neuronal architecture and connectivity to influence higher brain function (Catterall and Few, 2008; Greer and Greenberg, 2008). At the single neuron level, unique pre- and post-synaptic calcium signals generated by the opening of plasma membrane (PM) voltage sensitive- or ligand gated-ion channels are initially decoded by families of small calcium sensing proteins that exhibit distinct calcium binding characteristics in combination with specific patterns of cellular expression and sub-cellular localization (Haeseleer et al., 2000; Burgoyne and Weiss, 2001; Burgoyne et al., 2004; Burgoyne, 2007). Calcium binding typically elicits a conformational switch in the sensor (Haynes and Burgoyne, 2008; Ames and Lim, 2011) which in turn permits association with specific downstream effectors to modulate intracellular signaling cascades and ultimately neuronal activity and local synaptic structure.
In mammals, the largest class of calcium sensing proteins are those belonging to the calmodulin (CaM) superfamily that is defined by the EF-hand calcium binding motif (Kawasaki et al., 1998). CaM is expressed in all plants and animals and exerts essential functions in many aspects of normal cellular physiology (Klee et al., 1980). One CaM-related sub-family of calcium sensors, the Calcium Binding Proteins or CaBPs, has recently been shown to have co-evolved with vertebrate animals (McCue et al., 2010a). The CaBPs share a similar domain organization with CaM and have four EF-hand motifs (Haeseleer et al., 2000; Mikhaylova et al., 2006) however, they exhibit significant sequence divergence from their common ancestor and this is reflected in unique structural and functional properties (McCue et al., 2010b; Mikhaylova et al., 2011). The CaBPs are enriched in neuronal tissues where they have been shown to act as important regulators of key calcium influx channels. Coupled to their vertebrate specific expression profile the available evidence implicates CaBPs specifically as mediators of central nervous system (CNS) behavior in higher animals. Perhaps not unexpectedly members of the CaBP family regulate target effectors in common with CaM however, in every instance thus far examined there has been no detectable redundancy and CaBP target regulation appears distinct to that exerted by CaM (Lee et al., 2002; Haynes et al., 2004; Kinoshita-Kawada et al., 2005; Zhou et al., 2005; Tang et al., 2007; Li et al., 2009; Findeisen and Minor, 2010; Minor and Findeisen, 2010; Few et al., 2011; Oz et al., 2011). The importance and specificity of CaBP function is further highlighted by the distinct phenotypes exhibited by CaBP4 and CaBP5 knock-out mice and the visual impairment observed in human patients carrying mutations in the CaBP4 gene (Jeziorski et al., 2000; Williams, 2006; Zeitz et al., 2006; Rieke et al., 2008; Littink et al., 2009; Aldahmesh et al., 2010). It has also been discovered that the CaBPs exhibit specific target interactions independent of CaM and intriguingly some of these appear to have co-evolved with the CaBP family during the emergence of vertebrates (McCue et al., 2010a).
A picture is now emerging whereby the CaBP family can be viewed as providing an expansion in CaM functionality by exerting additional levels of target regulation for shared effectors in addition to executing novel functions through unique effector interactions. This increase in signaling complexity maximizes the range of physiological calcium signals that can be utilized and may have been instrumental in the evolution of complex vertebrate nervous systems that we observe today (Williams, 2006). This paper will focus on the evolution, mechanisms of targeting, and emerging roles of the CaBP protein family and discuss the importance of recent experimental findings in the context of mammalian CNS function.
Methods
Hela cells were cultured in 75 cm2 flasks at 37°C in a humidified atmosphere of 5% CO2/95% air. Cells for transfection were seeded onto coverslips in a 24-well tray at a density of ∼4 × 105 cells/well. 1 μg of mCherry-TM7 [residues 188–215 of human CaBP7 (transmembrane domain: residues 189–205)] or mCherry-CaBP7 plasmid was transfected per well using Genejuice transfection reagent (Novagen) according to manufacturer's instructions. Cells were maintained for 24 h post-transfection before fixation. Cells were washed three times in phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, 2 mM NaH2PO4, pH 7.4) then fixed in 4% formaldehyde in PBS at room temperature for 10 min. Cells were then washed a further three times in PBS then permeabilized using 0.2% triton X-100 in PBS for 6 min at room temperature. Cells were washed three times in PBS to remove detergent then twice in PBS containing 5% (w/v) BSA (PBSB). Cells were incubated with anti-TOM20 (1:1000, BD Biosciences) or anti-Calnexin (1:200, Sigma) primary antibodies for 1 h at room temperature in PBSB. Cells were washed three times in PBS and twice in PBSB prior to incubation with the relevant species specific fluorophore conjugated secondary antibody for 1 h at room temperature. Cells were washed again three times in PBS, rinsed with deionized H2O, dried, and mounted onto glass slides using ProLong Gold antifade reagent (Invitrogen). Imaging of transfected and immunostained cells was carried out using a Leica AOBS SP2 microscope (Leica microsystems, Heidelberg, Germany) using a 63x oil immersion objective with a 1.4 numerical aperture. In most cases the pinhole was set to Airy1 to give the optimum signal to noise ratio and hence minimum thickness confocal section for the excitation laser used.
CaBP Evolution
The CaBP family in humans comprises six proteins: Caldendrin/CaBP1, 2, 4, 5, 7, and 8 (Haeseleer et al., 2000; Wu et al., 2001; Mikhaylova et al., 2006). The CaBP3 that was originally identified is likely to be a pseudogene and no CaBP6 gene exists. CaBP7 and CaBP8 have been referred to by the alternative names calneuron II and calneuron I, respectively, in other studies (Wu et al., 2001; Mikhaylova et al., 2006, 2009; Hradsky et al., 2011) but will henceforth be referred to as CaBP7 and CaBP8 in this paper. We have taken this approach for self-consistency (our previously published work has used these protein identifiers) and also because bioinformatics analyses links CaBP 7 and 8 to CaBPs 1–5 more than to any other small EF-hand Ca2+-sensors (McCue et al., 2010a). Alternative splicing of the CaBP1 and 2 genes generates additional novel transcripts so that the final complement of CaBPs numbers nine distinct proteins in humans (McCue et al., 2010a). These proteins share a core domain comprised of four EF-hand motifs but differ in unique regions located at the extreme N- or C-termini (McCue et al., 2010b). The EF-hand core represents the region with greatest similarity to CaM however, the most closely related family member, CaBP2-Short, only shares 37.8% total sequence identity with its primordial ancestor suggesting that the unique N- and C-terminal domains of the CaBPs are highly divergent and likely confer specific cellular functions and target regulation distinct from that exerted by CaM (McCue et al., 2010a). The EF-hand core additionally exhibits divergence within the CaBP family and there are unique patterns of EF-hand inactivation coupled to sequence substitutions conferring binding specificity preferentially toward either Ca2+ or Mg2+ ions (McCue et al., 2010a). The unique cation binding properties of individual CaBP proteins will further influence their biological activity and increase the specificity of cellular Ca2+-signals to which they are able to respond.
How these proteins evolved from their CaM ancestor was unclear until a recent bioinformatic analyses examined available invertebrate and vertebrate genome sequence databases in an effort to locate CaBP-related transcripts (McCue et al., 2010a). From these analyses it was determined that the oldest species harboring CaBP-related genomic DNA sequences was the cartilaginous fish, the elephant shark (Callorhincus milli). No invertebrate genomes analyzed, including the widely used experimental model organisms Drosophila melanogaster and Caenorhabditis elegans, where genome coverage is >10X, contained CaBP like sequences (C.elegans, 1998; Adams et al., 2000; McCue et al., 2010a). The elephant shark genome sequencing project currently stands at 1.4X coverage however, sequences with homology to CaBPs 1, 2, 5, 7, and 8 are clearly present based on BLAST searches against the human coding sequences [(McCue et al., 2010a) and Table 1].
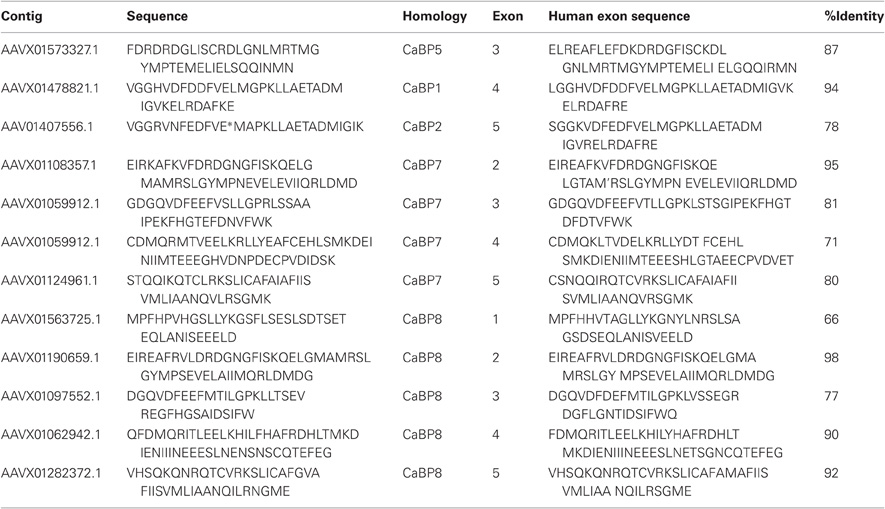
Table 1. Protein sequence fragments identified in BLAST searches against the elephant shark genome (left column) along with the sequence of the corresponding CaBP exon in the human genome (right column) and the percentage identity between the two sequences.
Further detailed analysis of protein alignment data showed that the elephant shark genome contains a CaBP1 like sequence having 46% sequence coverage of the human orthologue and 90% identity (Figure 1A). The coverage for elephant shark CaBP7 and CaBP8 sequences was found to be significantly greater and these again shared considerable identity with the corresponding human sequences (Figure 1B). In the study of McCue et al. (McCue et al., 2010a) no CaBP like sequences were discovered in the jawless fish, lamprey which is thought to represent the oldest living organism related to the human vertebrate ancestor. Coverage of the Lamprey genome has since improved and a new series of specific BLAT and BLAST searches performed here has uncovered two partial CaBP like sequences with highest homology to human CaBP1 and CaBP8 (Figures 1A,B). It would appear, therefore, that there were two ancestral members of the CaBP family, CaBP1 and CaBP8. This observation is consistent with the CaBP family forming two distinct sub-families based on unique sequence characteristics, the first comprising CaBPs1–5 and the second consisting of CaBP7 and CaBP8 (McCue et al., 2010a). Collectively, these results suggest that the CaBP protein family arose specifically with the emergence of vertebrate species and that whole genome duplication event, acting initially on CaBP1- and CaBP8-like genes present in the jawless fish, likely gave rise to the full complement of CaBP proteins found from the bony fish onwards through vertebrate evolution (McCue et al., 2010a).

Figure 1. Bioinformatic analysis of CaBP1, 7, and 8 proteins during vertebrate evolution. (A) Protein sequence alignment for human CaBP1-Short (Genbank accession: NM_004276) with related protein sequences identified from BLAT/BLAST searches of Lamprey and Elephant Shark (E. Shark) genomes. Identical residues between = 50% aligned sequences are highlighted in blue, similar residues are highlighted in green. Lamprey CaBP1 shares 80% coverage and 76% identity with human CaBP1-Short. Elephant Shark CaBP1 shares 46% coverage and 90% identity with human CaBP1-Short. (B) Protein sequence alignment for human CaBP7 (Genbank accession: NM_182527) and CaBP8 (Genbank accession: AY007302) with BLAT/BLAST identified related sequences from Lamprey (CaBP8) and Elephant Shark (CaBP7 and CaBP8). Identical residues between = 50% aligned sequences are highlighted in blue, similar residues are highlighted in green. Lamprey CaBP8 exhibited 61% and 60% coverage and 56% and 65% identity with human CaBP7 and human CaBP8, respectively. Elephant Shark CaBP7 exhibited 83% and 82% coverage and 84% and 66% identity with human CaBP7 and human CaBP8, respectively. Elephant Shark CaBP8 exhibited 102% and 100% coverage and 63% and 87% identity with human CaBP7 and human CaBP8, respectively.
In addition to examining the evolution of CaBP proteins by performing searches based on the full-length human coding sequences (McCue et al., 2010a), domains unique to individual CaBPs were also analyzed to examine if there was evolutionary conservation or divergence which might indicate correspondingly conserved or unique cellular functions. It was determined that the novel N-terminal domain of CaBP1-Long (residues 16–75) was relatively well conserved from amphibians to humans which may correlate with an important vertebrate specific activity in the inhibition of Ca2+-release from ER resident inositol 1, 4, 5-trisphosphate (IP3) receptors (IP3Rs) (Haynes et al., 2004; Kasri et al., 2004; Li et al., 2009; McCue et al., 2010a). In contrast, the variable N-terminal regions of CaBP2 and CaBP4 both exhibited a progressive increase in sequence similarity from the bony fish to humans. As both of these proteins are expressed in sensory neurons these observations may correspond to an increase in the complexity of the audio-visual sensory system during vertebrate evolution. Specific domains of CaBP7 and CaBP8 were not included in this analysis but recent evidence characterizing the unique functional properties of the hydrophobic C-termini of both of these family members (McCue et al., 2011) led us to evaluate the evolutionary conservation of these regions (Figure 1B). The hydrophobic C-terminal 38 residues of both CaBP7 (residues 177–215) and CaBP8 (residues 181–219) exhibit significant sequence conservation across vertebrate evolution. There is 92% sequence similarity between lamprey and human CaBP8 sequences in the C-terminal region and 94% similarity between elephant shark and human CaBP7 in the corresponding region. This high degree of domain conservation is indicative of a conserved function and is consistent with this region of CaBP7 and CaBP8 controlling the normal sub-cellular targeting of both proteins to the trans-Golgi network (TGN) and post-TGN vesicles (McCue et al., 2009; Mikhaylova et al., 2009; Hradsky et al., 2011).
The conservation of CaBP family members throughout vertebrate evolution raised a further interesting question regarding the potential co-evolution of specific interacting proteins (McCue et al., 2010a) which might shed light on vertebrate specific functions for this protein family. There are numerous documented interactions between CaBPs and various classes of cellular calcium channels including IP3Rs (Haynes et al., 2004; Kasri et al., 2004) and voltage gated calcium channels (VGCCs) (Lee et al., 2002; Haeseleer et al., 2004; Zhou et al., 2004, 2005; Cui et al., 2007; Lee et al., 2007b; Tippens and Lee, 2007; Few et al., 2011). These channels are also targets of CaM, however, in mammalian systems it would appear that the CaBPs and CaM have the capacity to differentially regulate common channel targets (Lee et al., 2002; Haynes et al., 2004; Kasri et al., 2006; Minor and Findeisen, 2010). Within the CaBP family further examples of regulatory diversity exist and both Caldendrin and CaBP1-Short modulate L-type VGCCs in distinct ways even though they share a common C-terminal EF-hand domain (Tippens and Lee, 2007). VGCCs and IP3Rs are present in invertebrates, however, the number of specific isoforms has increased with the appearance of vertebrates (Jeziorski et al., 2000; Iwasaki et al., 2002; Zhang et al., 2007; McCue et al., 2010a). It seems likely that the co-evolution of key calcium channels tailored to discrete functions within the mammalian CNS in conjunction with new types of calcium sensor proteins including the CaBPs has been instrumental in permitting vertebrates to generate and utilize an extended repertoire of spatio-temporal calcium signals. This in turn has likely influenced the evolution of increasingly complex modes of neuronal communication and enhanced CNS processing power.
CaBP Targeting
CaM is a cytosolic protein that lacks primary sequence information or post-translational modifications which would mediate its specific localization to subcellular organelles (Mikhaylova et al., 2006). This feature of CaM perhaps underlies its versatility as a Ca2+-sensor and helps explain its ability to interact with a wide spectrum of cellular effectors, only becoming recruited to specific cellular domains at specific time points through formation of multiple distinct complexes. The cellular activity of CaM consequently often relies on Ca2+ increases that are generated globally throughout the cytoplasm (Parekh, 2011). In order to maximize the usefulness of Ca2+ as a second messenger, mechanisms have evolved for the generation Ca2+-signals that are highly restricted in both space and time to ensure that only specific cell signaling pathways are activated or inhibited without perturbing other Ca2+-sensitive events within the cell. For Ca2+ to be used in this manner it follows that calcium sensing proteins dedicated to the detection of such specific signals must be similarly localized or harbor the ability to become localized in response to the signal. A variety of mechanisms are employed by the CaBPs to restrict their localization to particular cellular organelles and domains. Like CaM, a number of the CaBP proteins (Caldendrin, CaBP4 and CaBP5) contain no inherent primary sequence targeting information or consensus sequences for post translational modification that might influence their retention at specific subcellular locations (Haeseleer et al., 2000). The strategy employed in these instances relies on interactions with target effectors that in turn are specifically localized (Haeseleer et al., 2004; Rieke et al., 2008).
Caldendrin is enriched in post-synaptic density protein fractions (Seidenbecher et al., 1998) and specifically interacts with a light chain of the microtubule associated protein MAP1A/B (Seidenbecher et al., 2004). More recently a novel interaction between Caldendrin and a retinal specific small EF-hand containing calcium sensor, recoverin, was reported (Fries et al., 2010). Recoverin is a photoreceptor-enriched, myristoylated protein that exhibits a so called Ca2+-myristoyl switch mechanism (Tanaka et al., 1995; Ames et al., 1997; Ames and Ikura, 2002) whereby the acyl group is sequestered within the protein core in the absence of Ca2+ and, on Ca2+-binding and a conformational rearrangement (Yap et al., 1999), is extruded to permit dynamic association with cellular membranes. Caldendrin interacted with recoverin in a calcium dependent manner and this was reported to traffic the normally cytosolic Caldendrin protein to Golgi membranes (Fries et al., 2010). This mechanism of targeting is potentially interesting since many neuronal populations often express multiple different Ca2+-sensing proteins. Further investigations will be required to validate this targeting mechanism for the endogenous proteins, in particular it would be of interest to examine what effect depletion of recoverin has on the ability of Caldendrin to redistribute in response to elevations in cytoplasmic Ca2+ concentration. Since the C-terminal EF-hand containing domain of Caldendrin and not its variable N-terminal region was able to bind to recoverin (Fries et al., 2010) and because this domain is identical in the other CaBP1 splice isoforms, CaBP1-Long and CaBP1-Short, it would also be of interest to test the promiscuity of recoverin binding with respect to other CaBP1 interactions.
Shorter splice variants of the Caldendrin/CaBP1 gene, CaBP1-Long, and CaBP1-Short (Landwehr et al., 2003; Haynes et al., 2004; Kasri et al., 2004) incorporate a distinct exon, not present in the Caldendrin transcript, that encodes for an N-terminal myristoylation consensus site (Landwehr et al., 2003). N-myristoylation of both proteins has been proven essential for localization to the PM and membranes of the Golgi apparatus and for functional inhibition of IP3Rs (Haynes et al., 2004; Kasri et al., 2004).
The remaining two members of the CaBP family, CaBP7 and CaBP8, like CaM, Caldendrin, CaBP4 and CaBP5 contain no consensus motifs for post-translational modifications that are known to mediate association with cellular membranes (Wu et al., 2001; Mikhaylova et al., 2006). Surprisingly, when the localization of these proteins was examined in mammalian cell lines, they were found to specifically localize to membranes of the TGN and vesicular compartments of the constitutive secretory pathway (McCue et al., 2009; Mikhaylova et al., 2009). Closer inspection of the CaBP7 and CaBP8 primary sequences (McCue et al., 2009) using transmembrane domain prediction tools uncovered the presence of a highly hydrophobic 38 residue C-terminal extension not present in the other CaBPs that was strongly predicted to form a transmembrane domain. Cellular and biochemical characterization of these sequences determined that they do indeed operate as transmembrane domains which are essential for normal CaBP7 and CaBP8 localization (McCue et al., 2009). Furthermore it was demonstrated that chimeric constructs encoding normally cytosolic proteins fused to the CaBP7 and CaBP8 transmembrane domains were efficiently targeted to membranes of the secretory pathway (McCue et al., 2009).
The CaBP7 and CaBP8 transmembrane domains are located 10 residues from the C-terminus in each protein and this topology is consistent with that of the tail-anchor class of type-II transmembrane proteins that have cytosolically oriented N-terminal functional domains, a transmembrane domain and a short lumenal C-terminal tail (Borgese et al., 2003, 2007; Brambillasca et al., 2006; McCue et al., 2011). Tail-anchor proteins are defined by their post-translational insertion into biological membranes through a series of novel pathways, the molecular details of which remain to be fully resolved (Abell et al., 2004; Stefanovic and Hegde, 2007; Borgese and Fasana, 2011). CaBP7 and CaBP8 have recently been shown to adopt the expected tail-anchor protein topology (Hradsky et al., 2011; McCue et al., 2011) and to interact with the ATPase TRC40/Asna1 (Hradsky et al., 2011) (Figure 3) a protein implicated in the post-translational membrane insertion of other tail-anchored proteins (Stefanovic and Hegde, 2007; Rabu et al., 2008). Tail-anchor proteins are initially post-translationally inserted either into the endoplasmic reticulum, outer mitochondrial- or peroxisomal-membranes (Borgese et al., 2007; Borgese and Fasana, 2011). Peroxisomes and mitochondria represent terminal destinations, however, ER targeted tail-anchor proteins can subsequently traffic along the secretory pathway and access a number of possible cellular membranes. Analysis of the CaBP7 and CaBP8 transmembrane domains explains why these proteins are observed at the TGN and on transport vesicles (McCue et al., 2009, 2011; Hradsky et al., 2011). When fused to the normally cytosolic fluorescent tag protein mCherry, a C-terminal domain fragment of CaBP7 incorporating the predicted transmembrane domain [residues 188–215 (transmembrane domain: residues 189–205)] showed no colocalization with the outer mitochondrial membrane tail-anchored protein TOM20 (Figure 2A). In contrast, it exhibited a reticular distribution in a proportion of HeLa cells where it colocalized extensively with the ER marker calnexin (Figure 2B). This construct is also competent for correct traffic to the TGN in another proportion of HeLa cells (data not shown) as has been observed in the neuronal like N2A cell line (McCue et al., 2009). This observation may be consistent with differential expression levels between cells: in low to moderate expressing situations the CaBP7 transmembrane domain fusion remains ER-trapped, however, at higher expression levels ER-based retention mechanisms are likely saturated and the construct escapes to latter compartments of the secretory pathway. Collectively these data illustrate that the transmembrane domain of CaBP7 is of the class that directs initial insertion into the ER membrane. Additional intrinsic targeting information or specific protein-protein interactions must be required for the efficient direction of CaBP7 and CaBP8 for forward traffic through the secretory pathway to the TGN and beyond as can be observed for full-length CaBP7 protein which exhibits a punctate distribution that does not significantly overlap with either calnexin or TOM20 (Figure 2C,D).
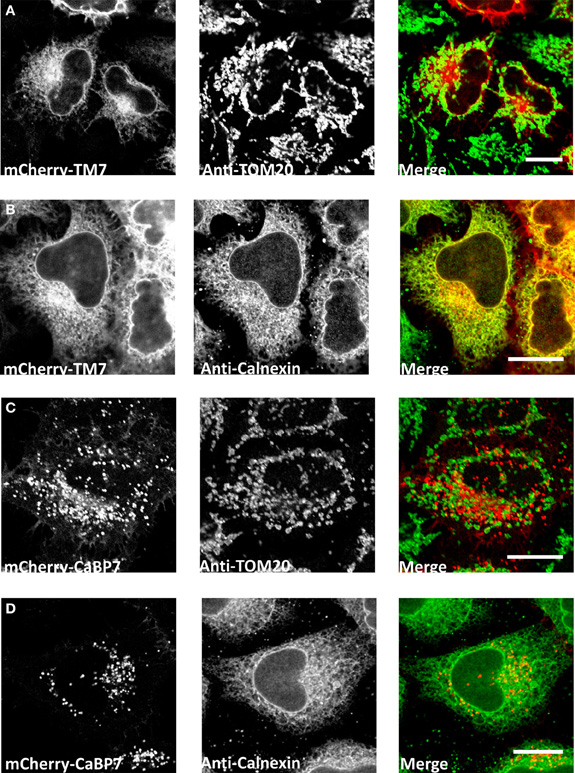
Figure 2. Analysis of localization of mCherry-CaBP7 or mCherry-CaBP7 transmembrane containing domain (residues 188–215) fusion (mCherry-TM7) compared with markers of the mitochondria (TOM20) and the ER (Calnexin). (A and B) mCherry-TM7 expression in HeLa cells (red) and co-staining with antibodies against endogenous TOM20 (A, green) or Calnexin (B, green). Cells with low to moderate levels of mCherry-TM7 were analyzed in this experiment, more highly expressing cells exhibit a localization pattern more consistent with the full-length CaBP7 protein (see text for further discussion). (C and D) mCherry-CaBP7 expression in HeLa cells (red) and co-staining with antibodies against endogenous TOM20 (C, green) or Calnexin (D, green). Regions of colocalization appear yellow in overlay images. Scale bars = 10 μm.
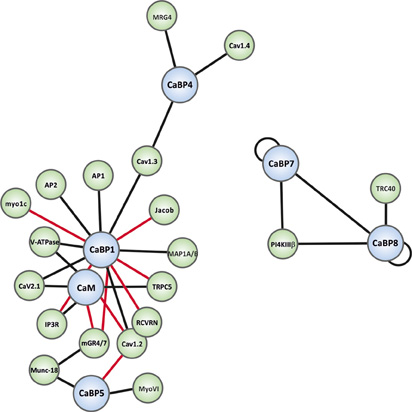
Figure 3. Interaction map of the known binding partners for the CaBP family. CaBP proteins are shown in blue spheres and specific effectors in green spheres. Black connecting lines indicate Ca2+-independent interactions and red connecting lines Ca2+-dependent interactions. Binding partner abbreviations are: MRG4 (Unc-119), Cav1.4 (L-type Ca2+-channel), Cav1.3 (L-type Ca2+-channel), Jacob, AP1 (Clathrin adaptor complex 1), AP2 (Clathrin adaptor complex 2), Myo1c (Myosin1c), V-ATPase (Vacuolar ATPase), Cav2.1 (P/Q-type Ca2+-channel), IP3R (Inositol 1,4,5-trisphosphate Receptor), Cav1.2 (L-type Ca2+-channel), RCVRN (Recoverin), TRPC5 (Transient receptor potential channel 5), MAP1A/B (Microtubule associated protein light chain 3 isoform A and B, LC3α/β), PI4KIIIβ (Phosphatidylinositol4-kinaseIIIβ), TRC40 (Transmembrane domain recognition complex-40), mGR4/7 (Group III metabotropic glutamate receptor 4/7), Munc-18 (nSec1), MyoVI (myosinVI).
CaBP7 and CaBP8 represent the only known CaM-related small EF-hand containing calcium sensors that have adopted the use of a tail-anchor targeting strategy. Tail-anchor proteins perform highly conserved and essential functions in mammalian cells and include SNARE proteins required for membrane fusion events throughout the secretory pathway (Borgese and Fasana, 2011). A number of issues remain to be resolved with regards to the precise targeting of CaBP7 and CaBP8. Although CaBP8 was shown to associate with TRC40 (Hradsky et al., 2011) it remains to be demonstrated that this chaperone is capable of post-translationally inserting recombinantly expressed or in vitro translated CaBP7 or CaBP8 across endoplasmic reticulum/microsomal membranes. It will also be of interest to understand the exact molecular determinants and protein-protein or protein-lipid interactions that control the final targeting of CaBP7 and CaBP8 to latter compartments of the secretory pathway. This information will further our understanding of the emerging functions of CaBP7 and CaBP8 as regulatory factors of protein trafficking from the TGN (Mikhaylova et al., 2009).
CaBP-Effector Interactions
As noted above, CaBPs along with CaM have been found to be able to regulate several types of VGCCs (Lee et al., 2002; Zhou et al., 2005; Cui et al., 2007; Findeisen and Minor, 2010; Minor and Findeisen, 2010). For a detailed overview of these and other well-characterized CaBP-effector interactions readers are directed to the recent comprehensive review articles (McCue et al., 2010b; Mikhaylova et al., 2011). The current article will focus on novel CaBP interactions that have been reported only in the past few years and which hint at further important physiological functions for the CaBP family in the mammalian CNS. A graphical overview of all presently characterized CaBP interactions is presented in network diagram form in Figure 3.
Caldendrin/CaBP1
Group III Metabotropic Glutamate Receptors (mGluRs)
Group III mGluRs function presynaptically to modulate synaptic vesicle exocytosis and neurotransmitter release (Pinheiro and Mulle, 2008). A recent study by Nakajima (Nakajima, 2011) reported an in vitro Ca2+-dependent interaction between rat CaBP1-Long and the membrane proximal portion of the cytoplasmic C-terminal tail of mGluR4 and mGluR7. This interaction was sensitive to phosphorylation of the mGluR fragment by protein kinase C mimicking the characterized binding of CaM to these receptors (Nakajima et al., 1999; O'Connor et al., 1999; Nakajima et al., 2009). There was competitive binding between CaM and CaBP1-Long suggesting that both proteins could regulate this receptor type through interaction with the same target motif. This is reminiscent of CaBP1/CaM dual regulation of VGCCs (Lee et al., 2002; Few et al., 2005, 2011) and it will be fascinating to examine whether, in the case of mGluR regulation, both Ca2+-sensors exert identical or differential modulation of these important neuronal receptors. CaM binding to mGluRs is able to displace the regulatory protein Munc-18 (Nakajima et al., 2009) and it will of interest to test if CaBP1 can elicit the same loss of Munc-18 binding.
The study of Nakajima (Nakajima, 2011) was conducted entirely using in vitro biochemical techniques and there is currently no information validating these findings in intact cells. The biochemical characteristics reported in this paper are, however, consistent with documented CaM/CaBP1 in vivo interactions (Lee et al., 2002; Few et al., 2005, 2011) and this work remains of great potential interest as Ca2+-influx downstream of mGluR activation is known to influence short term facilitation and higher level neuronal function (Cosgrove et al., 2011; Fioravante and Regehr, 2011).
Unconventional Myosin 1c
Class I unconventional myosins organize the cortical actin cytoskeleton and are involved in processes encompassing membrane trafficking and cell migration (Soldati, 2003). Myosin1c (myo1c), a protein essential for inner hair cell (IHC) adaptation in the inner ear (Gillespie and Cyr, 2004), has been shown to interact with CaBP1 through its IQ motif containing regulatory domain (Tang et al., 2007). CaBP1 binding was found to be competitive with that of CaM suggesting that myo1c may represent yet another example of a target protein regulated by multiple small EF-hand Ca2+-sensors. No functional data was presented in this study to confirm a physiological role for the CaBP1/myo1c interaction and further work is required to clarify this issue. Intriguingly, CaBP1 expression in rat and chicken auditory IHCs has been confirmed by both reverse transcription PCR and immunostaining (Yang et al., 2006; Lee et al., 2007b) and these may represent prime cellular models to study the functional implications of a putative CaBP1/myo1c interaction. IHCs have also been demonstrated to express CaBP4 (Yang et al., 2006; Lee et al., 2007b and CaBP7 Lee et al., 2007b) and, therefore, an analysis of myo1c binding by these family members might also further our understanding of potential mechanisms of cytoskeleton organization and regulation by multiple CaBPs in a key special sensory tissue.
Jacob
The interplay of calcium signaling through synaptic and extra-synaptic NMDA receptors influences complex aspects of neuronal function including synaptic connectivity and survival (Hardingham and Bading, 2003). These changes are brought about by alterations in neuronal gene expression (Greer and Greenberg, 2008; Lyons and West, 2011) and one pathway involved in such regulation in mammalian neurons is controlled by Caldendrin (Dieterich et al., 2008). This detailed study isolated and characterized a brain specific Caldendrin binding protein, Jacob, and demonstrated a role for this interaction in modulating activity of the CREB transcription factor to ultimately influence neuronal gene expression. Activation of synaptic NMDA receptors and subsequent Ca2+-binding by Caldendrin localized at the post-synaptic density was shown to drive its interaction with the Jacob nuclear localization signal thereby preventing import of Jacob into the nucleus. Conversely, activation of extra-synaptic NMDA receptors was shown to induce Jacob redistribution to the nucleus where it was able to induce CREB dephosphorylation and inactivation (Dieterich et al., 2008). Jacob mediated inhibition of CREB dependent gene expression elicited a loss of synaptic contacts and extensive simplification of dendritic architecture. This mechanism, therefore, couples synaptic activity to maintenance/loss of synaptic contacts through Ca2+-signaling via Caldendrin. Intriguingly, Jacob is a vertebrate specific protein (McCue et al., 2010a) that may have co-evolved specifically with the CaBP family to regulate this important aspect of synaptic plasticity.
CaBP4
Unc-119 (MRG4)
CaBP4 is the only CaBP member thus far characterized that has a proven role in human disease and mutations in the gene lead to rod and cone dysfunction and visual impairments of varying severity (Zeitz et al., 2006; Littink et al., 2009; Aldahmesh et al., 2010). CaBP4 regulates Cav1.4 channels in the retina (Haeseleer et al., 2004) and Cav1.3 channels in auditory IHCs (Yang et al., 2006; Cui et al., 2007; Lee et al., 2007b), however, non-channel binding partners are also emerging (Lee et al., 2007a).
Vertebrate orthologues of the C. elegans Unc-119 protein (also known as MRG4), like CaBP4, exhibit retinal specific expression profiles (Higashide et al., 1998; Higashide and Inana, 1999). Functional deficits due to mutations in MRG4 mirror those observed for CaB4 mutations and have been found to lead to rod/cone dystrophy in animal models and humans (Kobayashi et al., 2000). Unc-119 orthologues are expressed from invertebrates onwards (Maduro et al., 2000) and appear to have a nervous system specific function in C. elegans and D. melanogaster (Maduro and Pilgrim, 1995, 1996) which has apparently become restricted to the visual system in vertebrate animals.
MRG4 was found to interact with CaBP4 in a calcium independent manner consistent with binding to the variable N-terminal domain of CaBP4 (Haeseleer, 2008). This observation itself is intriguing as the vast majority of CaBP target effectors characterized to date have been shown to bind to the conserved C-terminal EF-hand containing core of the CaBP under investigation. An interaction with the novel N-terminus of CaBP4 seems to be important for stabilizing MRG4 protein (Haeseleer, 2008) and further hints at a co-evolution of tissue expression and cellular function between these proteins which has likely been important for the development and normal function of vertebrate visual systems. Further investigations into the molecular pathway linking CaBP4 and MRG4 to normal retinal activity are required to gain a complete understanding regarding the roles that both proteins play in human disease pathologies and to ascertain whether this retinal signaling system might provide a useful therapeutic target in the future treatment of some visual impairments.
CaBP5
Munc-18 (nSec-1) and Myosin VI
CaBP5 has been shown to be expressed in IHCs, however, the available data suggests that, in this particular cell type, it is CaBP1 and CaBP4 that functionally regulate Cav1.3 channels (Cui et al., 2007). CaBP5 is able to modulate the activity of both Cav1.2 and Cav1.3 channels when co-expressed in HEK293 cells (Cui et al., 2007; Rieke et al., 2008) and CaBP5 knock-out mice exhibit a reduction in light sensitivity of their retinal ganglion cells suggesting that CaBP5 may instead perform an important modulatory function in the visual system (Rieke et al., 2008).
Munc-18 is a highly conserved protein related to yeast Sec1 that is critical for neurotransmission throughout the mammalian CNS where it interacts with multiple effectors to control membrane fusion. Originally isolated as a syntaxin1 binding partner, Munc-18 has since been shown to interact with the synaptic SNARE complex in multiple unique “modes” to regulate synaptic vesicle fusion with the presynaptic PM (Burgoyne et al., 2009; Sudhof and Rothman, 2009). New, SNARE independent, Munc-18 binding proteins have been discovered suggesting that this protein itself is tightly regulated through multiple routes perhaps to further modulate SNARE complex activity and ultimately neurotransmission (Okamoto and Sudhof, 1997; Verhage et al., 1997; Graham et al., 2008; Hikita et al., 2009; Nakajima et al., 2009; Huang et al., 2011). Recently, a calcium independent interaction has been described between Munc-18 and CaBP5 in the retina establishing a potential new mechanism of linking calcium sensor function to SNARE activity in the visual system (Sokal and Haeseleer, 2011). CaBP5 was also shown to influence neurotransmitter release when overexpressed in a model neuroendocrine cell line, further implicating it as a regulator of secretion in selected regions of the nervous system (Sokal and Haeseleer, 2011).
In the same study, myosin VI was identified as a second specific binding partner for CaBP5 (Sokal and Haeseleer, 2011). Myosin VI has been implicated in endocytic processes. The related myosin V, a protein associated with synaptic vesicles and involved in their movement to the PM, was also detected in CaBP5 affinity chromatography pull-downs. For technical reasons a myosin V interaction was not characterized further in this study, however, these observations hint at a possible role for CaBP5 in synaptic vesicle recruitment to the presynaptic membrane of retinal neurons, a potentially important role which warrants further investigation. The CaBP5 knock-out mouse is available and it would be interesting to examine if there are defects in synaptic vesicle recruitment to the presynaptic active zone (myosin V pathway) or retrieval of exocytosed synaptic membrane components (myosin VI pathway) in retinal neurons from these animals. It would also be of interest to examine whether CaBP5 mediated stimulation of neurotransmitter release and neurite outgrowth (Sokal and Haeseleer, 2011) can be supported by a CaBP5 EF-hand mutant incapable of binding to Ca2+.
CaBP7 and CaBP8
Phosphatidylinositol 4-Kinaseiiiβ (PI4Kβ)
PI4Kβ is a lipid modifying enzyme that associates with the TGN to generate phosphatidylinositol 4-phosphate (PI4P) from phosphatidylinositol (Graham and Burd, 2011). PI4P is an essential lipid for budding of transport vesicles from the TGN and, therefore, regulation of PI4Kβ is a key control checkpoint of the secretory pathway. Known regulators of PI4Kβ activity include the small GTPase ARF1 and the small calcium sensing protein NCS-1 (Hendricks et al., 1999; Audhya et al., 2000; Haynes et al., 2005, 2007; Burgoyne and Haynes, 2010). NCS-1 modulation of PI4Kβ has been reported to be both Ca2+-independent (Zhao et al., 2001) and Ca2+-dependent (Haynes et al., 2005). It is known, however, that the Golgi complex stores luminal Ca2+ and uses this to generate local Ca2+-signals suggesting a link between Ca2+-release and membrane trafficking events (Dolman and Tepikin, 2006). A further level of regulation in this system was reported with the finding that both CaBP7 and CaBP8 could also interact with and inhibit the activity of PI4Kβ in mammalian cells to influence trafficking of specific secretory cargo to the PM (Mikhaylova et al., 2009).
The study of Mikhaylova et al. (Mikhaylova et al., 2009) uncovered an inhibitory interaction of CaBP7 and CaBP8 with PI4Kβ at resting (∼100 nM) cytosolic calcium concentrations [(Ca2+)i] that was competitively displaced by NCS-1 at increasing free [Ca2+]i to elicit stimulation of kinase activity. The interplay between NCS-1 and CaBP7 and CaBP8, therefore, permits dual control of TGN transport events over a range of physiological [Ca2+]i and importantly prevents activation of the enzyme in the absence of a threshold Ca2+-signal. The interaction of CaBP7 and CaBP8 with PI4Kβ was specific and was not observed with Caldendrin, however, it would be of interest to test PI4Kβ regulation with CaBP5, the family member exhibiting highest homology to CaBP7 and CaBP8 (McCue et al., 2010a).
Crosstalk between ARF1 and NCS-1/CaBP7 and CaBP8 was not examined in this study and since the NCS-1-PI4K-ARF1 network has been implicated in a developmental setting (Petko et al., 2009) it would be of interest to examine if CaBP7 and CaBP8 repression of PI4K activity can be linked to a similar physiological model.
VGCCs?
Until recently, the only documented regulatory role for CaBP7 and CaBP8 was that involving the interaction with PI4K discussed in the previous section. As small EF-hand Ca2+-sensing proteins often display promiscuity in their target interactions (Haynes et al., 2006; Burgoyne, 2007) it seems reasonable to expect further binding partners for CaBP7 and CaBP8 to appear in future studies. A new potential interaction between CaBP8 and various VGCC's has been reported which may provide the first clues regarding additional CaBP7 and CaBP8 cellular functions (Shih et al., 2009). This study examined the effect of CaBP8 over-expression in bovine adrenal chromaffin cells on PM Ca2+-channel activity and found that wild-type protein or a mutant deficient in Ca2+-binding both inhibited currents generated through N-, L-, and P/Q-type channels (Shih et al., 2009). A mutant CaBP8 lacking the hydrophobic C-terminal domain mislocalized in this cell type and did not support inhibition of channel currents reinforcing the idea that correct sub-cellular targeting on this Ca2+-sensor is critical for its normal cellular function.
The most profound inhibitory phenotype was observed on N-type currents and N-type channels are widely expressed in the mammalian CNS where they operate presynaptically to regulate neurotransmitter release (Delcour et al., 1993). No direct interaction between CaBP8 and the various Ca2+-channels analyzed was reported in the study of Shih et al. (Shih et al., 2009), however, this merits examination in future investigations in view of the importance of N-type channels, coupled to the widespread expression of CaBP7 and CaBP8, in the mammalian CNS.
Since CaBP7 and CaBP8 regulate PI4K, the possibility exists that these Ca2+-sensors are able to inhibit Ca2+-currents not by direct channel gating but rather by restricting the traffic of Ca2+-channels to the PM (Shih et al., 2009). Other small EF-hand Ca2+-sensors are known to modulate channel activity in this manner, the most well characterized example being the regulation of Kv4 potassium channel traffic to the PM by the KChIP family of Ca2+-sensors (Burgoyne, 2007; Flowerdew and Burgoyne, 2009). Since a mutant CaBP8 incapable of Ca2+-binding was able to inhibit N-type currents and since PI4K activity is inhibited at resting [Ca2+]i, these observations are consistent with this regulatory model. Distinguishing between channel traffic and direct channel modulation mechanisms should prove straightforward and will provide further insights into the role of CaBP7 and CaBP8 in the normal function of the mammalian CNS.
Discussion
Calcium sensing in the vertebrate CNS is of fundamental importance in establishing and maintaining normal neuronal activity. The complexity of Ca2+-signals generated in the mammalian brain is only just beginning to be understood, however, the identity of many of the protein factors involved in decoding them is already known. The CaM superfamily of small EF-hand containing Ca2+-sensors have emerged as key regulators of multiple important neuronal Ca2+-channels that influence all aspects of neuronal plasticity. Sub-groups of vertebrate specific CaM-related proteins including the CaBPs have seemingly evolved in parallel with increasing levels of CNS complexity in higher animals. These proteins exhibit diversity in patterns of expression, sub-cellular localization and Ca2+-sensing dynamics and are, therefore, well suited to functioning as transducers of the highly specific Ca2+-signals that underpin complex neuronal activity.
We are now aware that the CaBPs not only regulate neuronal specific Ca2+-channels but also targets involved in membrane trafficking, the organization of the cytoskeleton and proteins that influence neuronal gene expression and synaptic connectivity. The list of CaBP effector interactions is steadily increasing as is our appreciation of CaBP functionality. Testing the importance of CaBP activity in the mammalian CNS is largely restricted to studies utilizing cell lines and would benefit greatly from the generation of transgenic animal models. In particular, the application of Cre-Lox approaches (Sauer, 1998; Nickerson et al., 2011) to test loss of function of CaBPs in particular tissues and cell types during both development and in the adult animal will significantly enhance our understanding of the key physiological functions performed by these multifunctional signaling molecules.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a Wellcome Trust Prize Studentship to Hannah V. McCue.
References
Abell, B. M., Pool, M. R., Schlenker, O., Sinning, I., and High, S. (2004). Signal recognition particle mediates post-translational targeting in eukaryotes. EMBO J. 23, 2755–2764.
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., George, R. A., Lewis, S. E., Richards, S., Ashburner, M., Henderson, S. N., Sutton, G. G., Wortman, J. R., Yandell, M. D., Zhang, Q., Chen, L. X., Brandon, R. C., Rogers, Y. H., Blazej, R. G., Champe, M., Pfeiffer, B. D., Wan, K. H., Doyle, C., Baxter, E. G., Helt, G., Nelson, C. R., Gabor, G. L., Abril, J. F., Agbayani, A., An, H. J., Andrews-Pfannkoch, C., Baldwin, D., Ballew, R. M., Basu, A., Baxendale, J., Bayraktaroglu, L., Beasley, E. M., Beeson, K. Y., Benos, P. V., Berman, B. P., Bhandari, D., Bolshakov, S., Borkova, D., Botchan, M. R., Bouck, J., Brokstein, P., Brottier, P., Burtis, K. C., Busam, D. A., Butler, H., Cadieu, E., Center, A., Chandra, I., Cherry, J. M., Cawley, S., Dahlke, C., Davenport, L. B., Davies, P., de Pablos, B., Delcher, A., Deng, Z., Mays, A. D., Dew, I., Dietz, S. M., Dodson, K., Doup, L. E., Downes, M., Dugan-Rocha, S., Dunkov, B. C., Dunn, P., Durbin, K. J., Evangelista, C. C., Ferraz, C., Ferriera, S., Fleischmann, W., Fosler, C., Gabrielian, A. E., Garg, N. S., Gelbart, W. M., Glasser, K., Glodek, A., Gong, F., Gorrell, J. H., Gu, Z., Guan, P., Harris, M., Harris, N. L., Harvey, D., Heiman, T. J., Hernandez, J. R., Houck, J., Hostin, D., Houston, K. A., Howland, T. J., Wei, M. H., Ibegwam, C., Jalali, M., Kalush, F., Karpen, G. H., Ke, Z., Kennison, J. A., Ketchum, K. A., Kimmel, B. E., Kodira, C. D., Kraft, C., Kravitz, S., Kulp, D., Lai, Z., Lasko, P., Lei, Y., Levitsky, A. A., Li, J., Li, Z., Liang, Y., Lin, X., Liu, X., Mattei, B., McIntosh, T. C., McLeod, M. P., McPherson, D., Merkulov, G., Milshina, N. V., Mobarry, C., Morris, J., Moshrefi, A., Mount, S. M., Moy, M., Murphy, B., Murphy, L., Muzny, D. M., Nelson, D. L., Nelson, D. R., Nelson, K. A., Nixon, K., Nusskern, D. R., Pacleb, J. M., Palazzolo, M., Pittman, G. S., Pan, S., Pollard, J., Puri, V., Reese, M. G., Reinert, K., Remington, K., Saunders, R. D., Scheeler, F., Shen, H., Shue, B. C., Siden-Kiamos, I., Simpson, M., Skupski, M. P., Smith, T., Spier, E., Spradling, A. C., Stapleton, M., Strong, R., Sun, E., Svirskas, R., Tector, C., Turner, R., Venter, E., Wang, A. H., Wang, X., Wang, Z. Y., Wassarman, D. A., Weinstock, G. M., Weissenbach, J., Williams, S. M., Woodage, T., Worley, K. C., Wu, D., Yang, S., Yao, Q. A., Ye, J., Yeh, R. F., Zaveri, J. S., Zhan, M., Zhang, G., Zhao, Q., Zheng, L., Zheng, X. H., Zhong, F. N., Zhong, W., Zhou, X., Zhu, S., Zhu, X., Smith, H. O., Gibbs, R. A., Myers, E. W., Rubin, G. M., and Venter, J. C. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195.
Aldahmesh, M. A., Al-Owain, M., Alqahtani, F., Hazzaa, S., and Alkuraya, F. S. (2010). A null mutation in CABP4 causes Leber's congenital amaurosis-like phenotype. Mol. Vis. 16, 207–212.
Ames, J. B., and Ikura, M. (2002). Structure and membrane-targeting mechanism of retinal Ca2+-binding proteins, recoverin and GCAP-2. Adv. Exp. Med. Biol. 514, 333–348.
Ames, J. B., Ishima, R., Tanaka, T., Gordon, J. I., Stryer, L., and Ikura, M. (1997). Molecular mechanics of calcium-myristoyl switches. Nature 389, 198–202.
Ames, J. B., and Lim, S. (2011). Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta. [Epub ahead of print].
Audhya, A., Foti, M., and Emr, S. D. (2000). Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673–2689.
Borgese, N., Brambillasca, S., and Colombo, S. (2007). How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375.
Borgese, N., Brambillasca, S., Soffientini, P., Yabal, M., and Makarow, M. (2003). Biogenesis of tail-anchored proteins. Biochem. Soc. Trans. 31 (Pt 6), 1238–1242.
Borgese, N., and Fasana, E. (2011). Targeting pathways of C-tail-anchored proteins. Biochim. Biophys. Acta 1808, 937–946.
Brambillasca, S., Yabal, M., Makarow, M., and Borgese, N. (2006). Unassisted translocation of large polypeptide domains across phospholipid bilayers. J. Cell Biol. 175, 767–777.
Burgoyne, R. D. (2007). Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat. Rev. Neurosci. 8, 182–193.
Burgoyne, R. D., Barclay, J. W., Ciufo, L. F., Graham, M. E., Handley, M. T., and Morgan, A. (2009). The functions of Munc18-1 in regulated exocytosis. Ann. N.Y. Acad. Sci. 1152, 76–86.
Burgoyne, R. D., and Haynes, L. P. (2010). Neuronal calcium sensor proteins: emerging roles in membrane traffic and synaptic plasticity. F1000 Biol. Rep. 2, 5.
Burgoyne, R. D., O'Callaghan, D. W., Hasdemir, B., Haynes, L. P., and Tepikin, A. V. (2004). Neuronal Ca2+-sensor proteins: multitalented regulators of neuronal function. Trends Neurosci. 27, 203–209.
Burgoyne, R. D., and Weiss, J. L. (2001). The neuronal calcium sensor family of Ca2+-binding proteins. Biochem. J. 353(Pt 1), 1–12.
C.elegans, S. C. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018.
Catterall, W. A., and Few, A. P. (2008). Calcium channel regulation and presynaptic plasticity. Neuron 59, 882–901.
Cosgrove, K. E., Galvan, E. J., Barrionuevo, G., and Meriney, S. D. (2011). mGluRs modulate strength and timing of excitatory transmission in hippocampal area CA3. Mol. Neurobiol. 44, 93–101.
Cui, G., Meyer, A. C., Calin-Jageman, I., Neef, J., Haeseleer, F., Moser, T., and Lee, A. (2007). Ca2+-binding proteins tune Ca2+-feedback to Cav1.3 channels in mouse auditory hair cells. J. Physiol. 585(Pt 3), 791–803.
Delcour, A. H., Lipscombe, D., and Tsien, R. W. (1993). Multiple modes of N-type calcium channel activity distinguished by differences in gating kinetics. J. Neurosci. 13, 181–194.
Dieterich, D. C., Karpova, A., Mikhaylova, M., Zdobnova, I., Konig, I., Landwehr, M., Kreutz, M., Smalla, K. H., Richter, K., Landgraf, P., Reissner, C., Boeckers, T. M., Zuschratter, W., Spilker, C., Seidenbecher, C. I., Garner, C. C., Gundelfinger, E. D., and Kreutz, M. R. (2008). Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 6, e34. doi: 10.1371/journal.pbio.0060034
Dolman, N. J., and Tepikin, A. V. (2006). Calcium gradients and the Golgi. Cell Calcium 40, 505–512.
Few, A. P., Lautermilch, N. J., Westenbroek, R. E., Scheuer, T., and Catterall, W. A. (2005). Differential regulation of CaV2.1 channels by calcium-binding protein 1 and visinin-like protein-2 requires N-terminal myristoylation. J. Neurosci. 25, 7071–7080.
Few, A. P., Nanou, E., Scheuer, T., and Catterall, W. A. (2011). Molecular determinants of CaV2.1 channel regulation by calcium-binding protein-1. J. Biol. Chem. 286, 41917–41923.
Findeisen, F., and Minor, D. L. Jr. (2010). Structural basis for the differential effects of CaBP1 and calmodulin on Ca(V)1.2 calcium-dependent inactivation. Structure 18, 1617–1631.
Fioravante, D., and Regehr, W. G. (2011). Short-term forms of presynaptic plasticity. Curr. Opin. Neurobiol. 21, 269–274.
Flowerdew, S. E., and Burgoyne, R. D. (2009). A VAMP7/Vti1a SNARE complex distinguishes a non-conventional traffic route to the cell surface used by KChIP1 and Kv4 potassium channels. Biochem. J. 418, 529–540.
Fries, R., Reddy, P. P., Mikhaylova, M., Haverkamp, S., Wei, T., Muller, M., Kreutz, M. R., and Koch, K. W. (2010). Dynamic cellular translocation of caldendrin is facilitated by the Ca2+-myristoyl switch of recoverin. J. Neurochem. 113, 1150–1162.
Gillespie, P. G., and Cyr, J. L. (2004). Myosin-1c, the hair cell's adaptation motor. Annu. Rev. Physiol. 66, 521–545.
Graham, M. E., Handley, M. T., Barclay, J. W., Ciufo, L. F., Barrow, S. L., Morgan, A., and Burgoyne, R. D. (2008). A gain-of-function mutant of Munc18-1 stimulates secretory granule recruitment and exocytosis and reveals a direct interaction of Munc18-1 with Rab3. Biochem. J. 409, 407–416.
Graham, T. R., and Burd, C. G. (2011). Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 21, 113–121.
Greer, P. L., and Greenberg, M. E. (2008). From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860.
Haeseleer, F. (2008). Interaction and colocalization of CaBP4 and Unc119 (MRG4) in photoreceptors. Invest. Ophthalmol. Vis. Sci. 49, 2366–2375.
Haeseleer, F., Imanishi, Y., Maeda, T., Possin, D. E., Maeda, A., Lee, A., Rieke, F., and Palczewski, K. (2004). Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat. Neurosci. 7, 1079–1087.
Haeseleer, F., Sokal, I., Verlinde, C. L., Erdjument-Bromage, H., Tempst, P., Pronin, A. N., Benovic, J. L., Fariss, R. N., and Palczewski, K. (2000). Five members of a novel Ca(2+)-binding protein (CABP) subfamily with similarity to calmodulin. J. Biol. Chem. 275, 1247–1260.
Hardingham, G. E., and Bading, H. (2003). The Yin and Yang of NMDA receptor signalling. Trends Neurosci. 26, 81–89.
Haynes, L. P., and Burgoyne, R. D. (2008). Unexpected tails of a Ca2+ sensor. Nat. Chem. Biol. 4, 90–91.
Haynes, L. P., Fitzgerald, D. J., Wareing, B., O'Callaghan, D. W., Morgan, A., and Burgoyne, R. D. (2006). Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin delta. Proteomics 6, 1822–1832.
Haynes, L. P., Sherwood, M. W., Dolman, N. J., and Burgoyne, R. D. (2007). Specificity, promiscuity and localization of ARF protein interactions with NCS-1 and phosphatidylinositol-4 kinase-III beta. Traffic 8, 1080–1092.
Haynes, L. P., Tepikin, A. V., and Burgoyne, R. D. (2004). Calcium-binding protein 1 is an inhibitor of agonist-evoked, inositol 1,4,5-trisphosphate-mediated calcium signaling. J. Biol. Chem. 279, 547–555.
Haynes, L. P., Thomas, G. M., and Burgoyne, R. D. (2005). Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J. Biol. Chem. 280, 6047–6054.
Hendricks, K. B., Wang, B. Q., Schnieders, E. A., and Thorner, J. (1999). Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat. Cell Biol. 1, 234–241.
Higashide, T., and Inana, G. (1999). Characterization of the gene for HRG4 (UNC119), a novel photoreceptor synaptic protein homologous to unc-119. Genomics 57, 446–450.
Higashide, T., McLaren, M. J., and Inana, G. (1998). Localization of HRG4, a photoreceptor protein homologous to Unc-119, in ribbon synapse. Invest. Ophthalmol. Vis. Sci. 39, 690–698.
Hikita, T., Taya, S., Fujino, Y., Taneichi-Kuroda, S., Ohta, K., Tsuboi, D., Shinoda, T., Kuroda, K., Funahashi, Y., Uraguchi-Asaki, J., Hashimoto, R., and Kaibuchi, K. (2009). Proteomic analysis reveals novel binding partners of dysbindin, a schizophrenia-related protein. J. Neurochem. 110, 1567–1574.
Hradsky, J., Raghuram, V., Reddy, P. P., Navarro, G., Hupe, M., Casado, V., McCormick, P. J., Sharma, Y., Kreutz, M. R., and Mikhaylova, M. (2011). Post-translational membrane insertion of tail-anchored transmembrane EF-hand Ca2+ sensor calneurons requires the TRC40/Asna1 rrotein chaperone. J. Biol. Chem. 286, 36762–36776.
Huang, C. C., Yang, D. M., Lin, C. C., and Kao, L. S. (2011). Involvement of Rab3A in vesicle priming during exocytosis: interaction with Munc13-1 and Munc18-1. Traffic 12, 1356–1370.
Iwasaki, H., Chiba, K., Uchiyama, T., Yoshikawa, F., Suzuki, F., Ikeda, M., Furuichi, T., and Mikoshiba, K. (2002). Molecular characterization of the starfish inositol 1,4,5-trisphosphate receptor and its role during oocyte maturation and fertilization. J. Biol. Chem. 277, 2763–2772.
Jeziorski, M. C., Greenberg, R. M., and Anderson, P. A. (2000). The molecular biology of invertebrate voltage-gated Ca(2+) channels. J. Exp. Biol. 203(Pt 5), 841–856.
Kasri, N. N., Holmes, A. M., Bultynck, G., Parys, J. B., Bootman, M. D., Rietdorf, K., Missiaen, L., McDonald, F., De Smedt, H., Conway, S. J., Holmes, A. B., Berridge, M. J., and Roderick, H. L. (2004). Regulation of InsP3 receptor activity by neuronal Ca2+-binding proteins. EMBO J. 23, 312–321.
Kasri, N. N., Torok, K., Galione, A., Garnham, C., Callewaert, G., Missiaen, L., Parys, J. B., and De Smedt, H. (2006). Endogenously bound calmodulin is essential for the function of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 281, 8332–8338.
Kawasaki, H., Nakayama, S., and Kretsinger, R. H. (1998). Classification and evolution of EF-hand proteins. Biometals 11, 277–295.
Kinoshita-Kawada, M., Tang, J., Xiao, R., Kaneko, S., Foskett, J. K., and Zhu, M. X. (2005). Inhibition of TRPC5 channels by Ca2+-binding protein 1 in Xenopus oocytes. Pflugers Arch. 450, 345–354.
Kobayashi, A., Higashide, T., Hamasaki, D., Kubota, S., Sakuma, H., An, W., Fujimaki, T., McLaren, M. J., Weleber, R. G., and Inana, G. (2000). HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest. Ophthalmol. Vis. Sci. 41, 3268–3277.
Landwehr, M., Redecker, P., Dieterich, D. C., Richter, K., Bockers, T. M., Gundelfinger, E. D., and Kreutz, M. R. (2003). Association of Caldendrin splice isoforms with secretory vesicles in neurohypophyseal axons and the pituitary. FEBS Lett. 547, 189–192.
Lee, A., Jimenez, A., Cui, G., and Haeseleer, F. (2007a). Phosphorylation of the Ca2+-binding protein CaBP4 by protein kinase C zeta in photoreceptors. J. Neurosci. 27, 12743–12754.
Lee, A., Westenbroek, R. E., Haeseleer, F., Palczewski, K., Scheuer, T., and Catterall, W. A. (2002). Differential modulation of Ca(v)2.1 channels by calmodulin and Ca2+-binding protein 1. Nat. Neurosci. 5, 210–217.
Lee, S., Briklin, O., Hiel, H., and Fuchs, P. (2007b). Calcium-dependent inactivation of calcium channels in cochlear hair cells of the chicken. J. Physiol. 583(Pt 3), 909–922.
Li, C., Chan, J., Haeseleer, F., Mikoshiba, K., Palczewski, K., Ikura, M., and Ames, J. B. (2009). Structural insights into Ca2+-dependent regulation of inositol 1,4,5-trisphosphate receptors by CaBP1. J. Biol. Chem. 284, 2472–2481.
Littink, K. W., van Genderen, M. M., Collin, R. W., Roosing, S., de Brouwer, A. P., Riemslag, F. C., Venselaar, H., Thiadens, A. A., Hoyng, C. B., Rohrschneider, K., den Hollander, A. I., Cremers, F. P., and van den Born, L. I. (2009). A novel homozygous nonsense mutation in CABP4 causes congenital cone-rod synaptic disorder. Invest. Ophthalmol. Vis. Sci. 50, 2344–2350.
Lohmann, C. (2009). Calcium signaling and the development of specific neuronal connections. Prog. Brain Res. 175, 443–452.
Lyons, M. R., and West, A. E. (2011). Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog. Neurobiol. 94, 259–295.
Maduro, M., and Pilgrim, D. (1995). Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141, 977–988.
Maduro, M., and Pilgrim, D. (1996). Conservation of function and expression of unc-119 from two Caenorhabditis species despite divergence of non-coding DNA. Gene 183, 77–85.
Maduro, M. F., Gordon, M., Jacobs, R., and Pilgrim, D. B. (2000). The UNC-119 family of neural proteins is functionally conserved between humans, Drosophila and C. elegans. J. Neurogenet. 13, 191–212.
McCue, H. V., Burgoyne, R. D., and Haynes, L. P. (2009). Membrane targeting of the EF-hand containing calcium-sensing proteins CaBP7 and CaBP8. Biochem. Biophys. Res. Commun. 380, 825–831.
McCue, H. V., Burgoyne, R. D., and Haynes, L. P. (2011). Determination of the membrane topology of the small EF-Hand Ca2+-sensing proteins CaBP7 and CaBP8. PLoS One 6, e17853. doi: 10.1371/journal.pone.0017853
McCue, H. V., Haynes, L. P., and Burgoyne, R. D. (2010a). Bioinformatic analysis of CaBP/calneuron proteins reveals a family of highly conserved vertebrate Ca2+-binding proteins. BMC Res. Notes 3, 118.
McCue, H. V., Haynes, L. P., and Burgoyne, R. D. (2010b). The diversity of calcium sensor proteins in the regulation of neuronal function. Cold Spring Harb. Perspect. Biol. 2, a004085.
Mikhaylova, M., Hradsky, J., and Kreutz, M. R. (2011). Between promiscuity and specificity: novel roles of EF-hand calcium sensors in neuronal Ca2+ signalling. J. Neurochem. 118, 695–713.
Mikhaylova, M., Reddy, P. P., Munsch, T., Landgraf, P., Suman, S. K., Smalla, K. H., Gundelfinger, E. D., Sharma, Y., and Kreutz, M. R. (2009). Calneurons provide a calcium threshold for trans-Golgi network to plasma membrane trafficking. Proc. Natl. Acad. Sci. U.S.A. 106, 9093–9098.
Mikhaylova, M., Sharma, Y., Reissner, C., Nagel, F., Aravind, P., Rajini, B., Smalla, K. H., Gundelfinger, E. D., and Kreutz, M. R. (2006). Neuronal Ca2+ signaling via caldendrin and calneurons. Biochim. Biophys. Acta 1763, 1229–1237.
Minor, D. L. Jr., and Findeisen, F. (2010). Progress in the structural understanding of voltage-gated calcium channel (CaV) function and modulation. Channels (Austin) 4, 459–474.
Nakajima, Y. (2011). Ca2+-dependent binding of calcium-binding protein 1 to presynaptic group III metabotropic glutamate receptors and blockage by phosphorylation of the receptors. Biochem. Biophys. Res. Commun. 412, 602–605.
Nakajima, Y., Mochida, S., Okawa, K., and Nakanishi, S. (2009). Ca2+-dependent release of Munc18-1 from presynaptic mGluRs in short-term facilitation. Proc. Natl. Acad. Sci. U.S.A. 106, 18385–18389.
Nakajima, Y., Yamamoto, T., Nakayama, T., and Nakanishi, S. (1999). A relationship between protein kinase C phosphorylation and calmodulin binding to the metabotropic glutamate receptor subtype 7. J. Biol. Chem. 274, 27573–27577.
Nickerson, P. E., Ronellenfitch, K., McEwan, J., Kim, H., McInnes, R. R., and Chow, R. L. (2011). A transgenic mouse line expressing cre recombinase in undifferentiated postmitotic mouse retinal bipolar cell precursors. PLoS One 6, e27145. doi: 10.1371/journal.pone.0027145
O'Connor, V., El Far, O., Bofill-Cardona, E., Nanoff, C., Freissmuth, M., Karschin, A., Airas, J. M., Betz, H., and Boehm, S. (1999). Calmodulin dependence of presynaptic metabotropic glutamate receptor signaling. Science 286, 1180–1184.
Okamoto, M., and Sudhof, T. C. (1997). Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J. Biol. Chem. 272, 31459–31464.
Oz, S., Tsemakhovich, V., Christel, C. J., Lee, A., and Dascal, N. (2011). CaBP1 regulates voltage-dependent inactivation and activation of Ca(V)1.2 (L-type) calcium channels. J. Biol. Chem. 286, 13945–13953.
Petko, J. A., Kabbani, N., Frey, C., Woll, M., Hickey, K., Craig, M., Canfield, V. A., and Levenson, R. (2009). Proteomic and functional analysis of NCS-1 binding proteins reveals novel signaling pathways required for inner ear development in zebrafish. BMC Neurosci. 10, 27.
Pinheiro, P. S., and Mulle, C. (2008). Presynaptic glutamate receptors: physiological functions and mechanisms of action. Nat. Rev. Neurosci. 9, 423–436.
Rabu, C., Wipf, P., Brodsky, J. L., and High, S. (2008). A precursor-specific role for Hsp40/Hsc70 during tail-anchored protein integration at the endoplasmic reticulum. J. Biol. Chem. 283, 27504–27513.
Rieke, F., Lee, A., and Haeseleer, F. (2008). Characterization of Ca2+-binding protein 5 knockout mouse retina. Invest. Ophthalmol. Vis. Sci. 49, 5126–5135.
Seidenbecher, C. I., Landwehr, M., Smalla, K. H., Kreutz, M., Dieterich, D. C., Zuschratter, W., Reissner, C., Hammarback, J. A., Bockers, T. M., Gundelfinger, E. D., and Kreutz, M. R. (2004). Caldendrin but not calmodulin binds to light chain 3 of MAP1A/B: an association with the microtubule cytoskeleton highlighting exclusive binding partners for neuronal Ca(2+)-sensor proteins. J. Mol. Biol. 336, 957–970.
Seidenbecher, C. I., Langnaese, K., Sanmarti-Vila, L., Boeckers, T. M., Smalla, K. H., Sabel, B. A., Garner, C. C., Gundelfinger, E. D., and Kreutz, M. R. (1998). Caldendrin, a novel neuronal calcium-binding protein confined to the somato-dendritic compartment. J. Biol. Chem. 273, 21324–21331.
Shih, P. Y., Lin, C. L., Cheng, P. W., Liao, J. H., and Pan, C. Y. (2009). Calneuron I inhibits Ca(2+) channel activity in bovine chromaffin cells. Biochem. Biophys. Res. Commun. 388, 549–553.
Sokal, I., and Haeseleer, F. (2011). Insight into the role of Ca2+-binding protein 5 in vesicle exocytosis. Invest. Ophthalmol. Vis. Sci. 52, 9131–9141.
Soldati, T. (2003). Unconventional myosins, actin dynamics and endocytosis: a menage a trois? Traffic 4, 358–366.
Stefanovic, S., and Hegde, R. S. (2007). Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell 128, 1147–1159.
Sudhof, T. C., and Rothman, J. E. (2009). Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477.
Tanaka, T., Ames, J. B., Harvey, T. S., Stryer, L., and Ikura, M. (1995). Sequestration of the membrane-targeting myristoyl group of recoverin in the calcium-free state. Nature 376, 444–447.
Tang, N., Lin, T., Yang, J., Foskett, J. K., and Ostap, E. M. (2007). CIB1 and CaBP1 bind to the myo1c regulatory domain. J. Muscle Res. Cell Motil. 28, 285–291.
Tippens, A. L., and Lee, A. (2007). Caldendrin, a neuron-specific modulator of Cav/1.2 (L-type) Ca2+ channels. J. Biol. Chem. 282, 8464–8473.
Verhage, M., de Vries, K. J., Roshol, H., Burbach, J. P., Gispen, W. H., and Sudhof, T. C. (1997). DOC2 proteins in rat brain: complementary distribution and proposed function as vesicular adapter proteins in early stages of secretion. Neuron 18, 453–461.
Williams, R. J. (2006). The evolution of calcium biochemistry. Biochim. Biophys. Acta 1763, 1139–1146.
Wu, Y. Q., Lin, X., Liu, C. M., Jamrich, M., and Shaffer, L. G. (2001). Identification of a human brain-specific gene, calneuron 1, a new member of the calmodulin superfamily. Mol. Genet. Metab. 72, 343–350.
Yang, P. S., Alseikhan, B. A., Hiel, H., Grant, L., Mori, M. X., Yang, W., Fuchs, P. A., and Yue, D. T. (2006). Switching of Ca2+-dependent inactivation of Ca(v)1.3 channels by calcium binding proteins of auditory hair cells. J. Neurosci. 26, 10677–10689.
Yap, K. L., Ames, J. B., Swindells, M. B., and Ikura, M. (1999). Diversity of conformational states and changes within the EF-hand protein superfamily. Proteins 37, 499–507.
Zeitz, C., Kloeckener-Gruissem, B., Forster, U., Kohl, S., Magyar, I., Wissinger, B., Matyas, G., Borruat, F. X., Schorderet, D. F., Zrenner, E., Munier, F. L., and Berger, W. (2006). Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am. J. Hum. Genet. 79, 657–667.
Zhang, D., Boulware, M. J., Pendleton, M. R., Nogi, T., and Marchant, J. S. (2007). The inositol 1,4,5-trisphosphate receptor (Itpr) gene family in Xenopus: identification of type 2 and type 3 inositol 1,4,5-trisphosphate receptor subtypes. Biochem. J. 404, 383–391.
Zhao, X., Varnai, P., Tuymetova, G., Balla, A., Toth, Z. E., Oker-Blom, C., Roder, J., Jeromin, A., and Balla, T. (2001). Interaction of neuronal calcium sensor-1 (NCS-1) with phosphatidylinositol 4-kinase beta stimulates lipid kinase activity and affects membrane trafficking in COS-7 cells. J. Biol. Chem. 276, 40183–40189.
Zhou, H., Kim, S. A., Kirk, E. A., Tippens, A. L., Sun, H., Haeseleer, F., and Lee, A. (2004). Ca2+-binding protein-1 facilitates and forms a postsynaptic complex with Cav1.2 (L-type) Ca2+ channels. J. Neurosci. 24, 4698–4708.
Keywords: calcium, Calcium Binding Protein, bioinformatics, protein evolution, protein targeting, protein-protein interaction
Citation: Haynes LP, McCue HV and Burgoyne RD (2012) Evolution and functional diversity of the Calcium Binding Proteins (CaBPs). Front. Mol. Neurosci. 5:9. doi: 10.3389/fnmol.2012.00009
Received: 09 January 2012; Paper pending published: 23 January 2012;
Accepted: 25 January 2012; Published online: 21 February 2012.
Edited by:
Karl-Wilhelm Koch, Carl von Ossietzky University Oldenburg, GermanyReviewed by:
Michael R. Kreutz, Leibniz-Institute for Neurobiology, GermanyKarl H. Braunewell, Southern Research Institute, USA
Copyright: © 2012 Haynes, McCue and Burgoyne. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Lee P. Haynes, The Physiological Laboratory, Department of Cellular and Molecular Physiology, Institute of Translational Medicine, University of Liverpool, Crown Street, Liverpool L69 3BX, UK. e-mail: leeh@liv.ac.uk