Corrigendum: Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis
- 1National Research Council, Institute of Biomedical Technologies, Bari Unit, Bari, Italy
- 2Department of Basic Sciences, Neurosciences and Sense Organs, University of Bari, Bari, Italy
A Corrigendum on
Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis
by Liguori, M., Nuzziello, N., Introna, A., Consiglio, A., Licciulli, F., D'Errico, E., et al. (2018). Front. Mol. Neurosci. 11:288. doi: 10.3389/fnmol.2018.00288
In the original article, there was a mistake in Figure 1 as published. We used the Volcano Plot, which was automatically generated using the Expression Suite v1.0.3 software (Thermo Fisher Scientific). This software visualizes the p-values and fold change values after each qRT-PCR run. A log2 (Fold Change) greater than or equal to one, is the standard boundary field.
Therefore, targets with a fold less than the lower boundary were automatically colored in green. However, as stated in the article, according to published guidelines, we validated the miRNAs with p-values < 0.05 (dot over the blue line) independently from their fold change. We have therefore adjusted the representation in the figure to avoid confusion.
The corrected Figure 1 appears below. The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
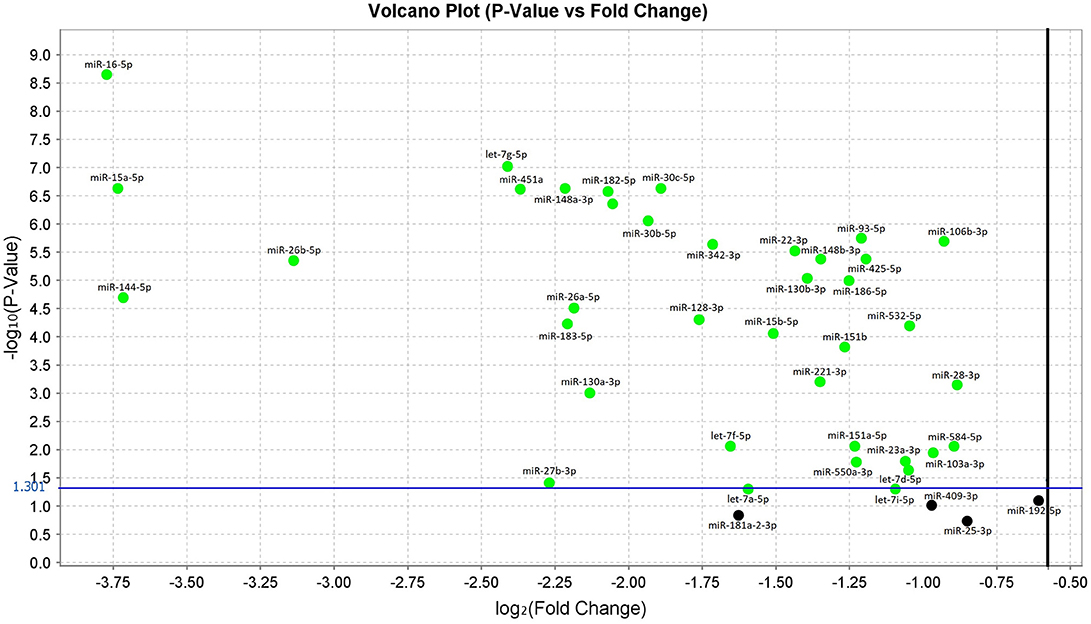
Figure 1. Volcano plot of validated miRNAs. Green dots represent the 38 differentially expressed miRNAs obtained from the comparison between sALS and HC subjects by qRT-PCR (p < 0.05). All black dots below the blue line did not discriminate sALS from HC. The Y-axis represents the log10 of the p-value and the X-axis represents log fold change of miRNA expression in the sALS versus HC.
Keywords: sporadic amyotrophic lateral sclerosis, microRNA, target genes, peripheral blood markers, high throughput next-generation sequencing (HT-NGS), clinical parameters, bioinformatics, pathway analysis
Citation: Liguori M, Nuzziello N, Introna A, Consiglio A, Licciulli F, D'Errico E, Scarafino A, Distaso E and Simone IL (2019) Corrigendum: Dysregulation of MicroRNAs and Target Genes Networks in Peripheral Blood of Patients With Sporadic Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 11:488. doi: 10.3389/fnmol.2018.00488
Received: 30 November 2018; Accepted: 14 December 2018;
Published: 10 January 2019.
Edited and reviewed by: Giuseppe Filomeni, Danish Cancer Society, Denmark
Copyright © 2019 Liguori, Nuzziello, Introna, Consiglio, Licciulli, D'Errico, Scarafino, Distaso and Simone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Liguori, maria.liguori@cnr.it;maria.liguori@ba.itb.cnr.it
†These authors have contributed equally to this work
 Maria Liguori
Maria Liguori Nicoletta Nuzziello
Nicoletta Nuzziello Alessandro Introna
Alessandro Introna Arianna Consiglio1
Arianna Consiglio1  Flavio Licciulli
Flavio Licciulli Eustachio D'Errico
Eustachio D'Errico Antonio Scarafino
Antonio Scarafino Eugenio Distaso
Eugenio Distaso Isabella L. Simone
Isabella L. Simone