Lmx1b Influences Correct Post-mitotic Coding of Mesodiencephalic Dopaminergic Neurons
- Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
The Lim Homeobox transcription factor 1 beta (LMX1b) has been identified as one of the transcription factors important for the development of mesodiencephalic dopaminergic (mdDA) neurons. During early development, Lmx1b is essential for induction and maintenance of the Isthmic Organizer (IsO), and genetic ablation results in the disruption of inductive activity from the IsO and loss of properly differentiated mdDA neurons. To study the downstream targets of Lmx1b without affecting the IsO, we generated a conditional model in which Lmx1b was selectively deleted in Pitx3-expressing cells from embryonic day (E)13 onward. Supporting previous data, no significant changes could be observed in general dopamine (DA) marks, like Th, Pitx3 and Vmat2 at E14.5. However, in depth analysis by means of RNA-sequencing revealed that Lmx1b is important for the mRNA expression level of survival factors En1 and En2 and for the repression of mdDA subset mark Ahd2 during (late) development. Interestingly, the regulation of Ahd2 by Lmx1b was found to be Pitx3 independent, since Pitx3 mRNA levels were not altered in Lmx1b conditional knock-outs (cKOs) and Ahd2 expression was also up-regulated in Lmx1b/Pitx3 double mutants compared to Pitx3 mutants. Further analysis of Lmx1b cKOs showed that post-mitotic deletion of Lmx1b additional leads to a loss of TH+ cells at 3 months age both in the ventral tegmental area (VTA) and substantia nigra pars compacta (SNc). Remarkably, different cell types were affected in the SNc and the VTA. While TH+AHD2+ cells were lost the SNc, TH+AHD2- neurons were affected in the VTA, reflected by a loss of Cck expression, indicating that Lmx1b is important for the survival of a sub-group of mdDA neurons.
Introduction
Dopamine (DA) is one of the catecholaminergic neurotransmitters found in the central nervous system. Although cell bodies of DA neurons can be found in several positions within the mammalian brain, the largest population of DA neurons is located in the ventral mesencephalon (Björklund and Dunnett, 2007; Smidt and Burbach, 2007; Roeper, 2013). The mesodiencephalic dopaminergic (mdDA) neuronal population consists of specific subsets with distinct functions (Smits et al., 2006), including the substantia nigra pars compacta (SNc) and the ventral tegmental area (VTA). The SNc innervates the dorsolateral striatum and caudate putamen forming the nigrostriatal pathway. The nigrostriatal pathway is integrated in a complex network that controls voluntary movement and body posture, and the degeneration of this pathway is the characteristic of Parkinson’s disease (Braak et al., 2003). The VTA has projections to the ventral striatum, the amygdala and the prefrontal cortex. These pathways are involved in regulating emotion-related behavior and are linked to addiction, depression and schizophrenia (Prakash and Wurst, 2006; Smidt and Burbach, 2007). The different subsets of mdDA neurons can already be distinguished during embryonic development based on their molecular profile and anatomical position (Smits et al., 2013; La Manno et al., 2016; Tiklová et al., 2019). It has been shown that the rostrolateral population will eventually form the largest part of the SNc, while the mediocaudal population is destined to become the VTA (Hökfelt et al., 1980; Veenvliet et al., 2013). Recent studies have revealed that each subset is dependent on a different transcriptional program for their development and survival (Smits et al., 2006; Jacobs et al., 2011; Veenvliet et al., 2013; Panman et al., 2014; Veenvliet and Smidt, 2014; Kouwenhoven et al., 2017). The rostrolateral fate is determined by a complex interplay between Pitx3 and En1. En1 induces the general DA phenotype and influences Pitx3 expression, while Pitx3 is required for the suppression of the caudomedial phenotype by down-regulating En1 mRNA levels ((Veenvliet et al., 2013). After the formation of the rostrolateral population the remaining DA population will obtain a caudomedial fate under the control of En1 (Bye et al., 2012; Veenvliet et al., 2013). Another factor that has been shown to be crucial for mdDA development and neuronal survival is LMX1B (Adams et al., 2000; Smidt et al., 2000; Deng et al., 2011; Laguna et al., 2015; Doucet-Beaupré et al., 2016). Analysis of the Lmx1b complete knock out mouse revealed that LMX1B is an essential component of a positive feedback loop required to maintain genes associated with the formation and functioning of the Isthmic Organizer (IsO), including Wnt1, En1, En2, Pax2 and Fgf8 (Adams et al., 2000; Guo et al., 2007). In the absence of Lmx1b the initiation, maintenance and inductive activity of the IsO were found to be severely impaired, affecting the development of the midbrain in general (Guo et al., 2007). Further analysis of Lmx1b−/− embryonic midbrains demonstrated a reduction in TH-expressing cells in the ventral mesencephalon (Smidt et al., 2000; Deng et al., 2011). Although the loss of Lmx1b initially seemed to primarily affect the lateral group of the DA progenitor domain (Deng et al., 2011), the medially located TH-expressing cells failed to induce Pitx3 and were lost during further development (Smidt et al., 2000). In contrast to the Lmx1b null mutant, the conditional deletion of Lmx1b in mdDA progenitors, but not in the IsO, resulted in normal development of mdDA neurons (Yan et al., 2011). It was suggested that Lmx1a compensates for the loss of Lmx1b, since the deletion of both led to a severe reduction in both DA progenitors and mature mdDA neurons (Yan et al., 2011). Further studies into the functional redundancy between Lmx1a and Lmx1b suggest that the function of Lmx1b in mdDA development and postnatal neuronal survival can be mostly compensated by Lmx1a (Nakatani et al., 2010; Deng et al., 2011; Yan et al., 2011; Doucet-Beaupré et al., 2016). However, a recent study Laguna et al. (2015) identified a critical role for Lmx1b in the functioning of mdDA neurons. They showed that DatCre driven deletion of Lmx1b reduces the protein levels of DAT and TH in the nerve terminals of mdDA neurons in the dorsal and ventral Striatum of 18-month-old mutant mice. In the present study we aimed to further elucidate the role of Lmx1b in postmitotic development and neuronal survival of mdDA neurons. We generated a mouse model that deletes Lmx1b in postmitotic mdDA neurons, by crossing a floxed Lmx1b mutant (Suleiman et al., 2007) with a Pitx3 driven iCre (Smidt et al., 2012). The loss of Lmx1b did not affect the development of the general DA phenotype, however a loss of TH+ cells was observed in 3-month-old animals, suggesting that Lmx1b plays a role in the survival and/or maintenance of mdDA neurons. During (late) development we found reduced mRNA levels of En1 and En2, which relates to the function of Lmx1b during early development (Guo et al., 2007). In addition, the rostrolateral subset mark, Ahd2, was increased and ectopically expressed at embryonic day (E)14.5, however, the caudomedial mark Cck was unaffected. To verify whether mdDA subsets were also affected during adult stages, the amount of TH+AHD2+ cells were analyzed in both the SNc and the VTA. Interestingly the amount of TH+AHD2+ cells was less in the SNc of the mutant compared to controls, but not in the VTA. In the VTA, TH+AHD2- cells were affected, which was reflected in lower mRNA levels of Cck. Taken together, our data shows that Lmx1b plays a role during the late development and survival of mdDA neurons. During development Lmx1b is important for maintaining proper En1 and En2 mRNA levels and the repression of the subset mark Ahd2, while during adult stages Lmx1b is important for the survival of a sub-group of mdDA neurons.
Materials and Methods
Animals
All lines were maintained on a C57BL/6J background (Charles River). Lmx1b-floxed animals were generated by R.L. Johnson and obtained from R.Witzgall (University of Regensburg, Germany) and have been previously described (Suleiman et al., 2007). The Pitx3Cre has been previously generated in our lab (Smidt et al., 2012). Pitx3CreCre; Lmx1b L/+ animals were crossed with Lmx1b L/+ or Pitx3CreCre; Lmx1b L/+ animals to generate Pitx3Cre/+; Lmx1b +/+, Pitx3Cre/+; Lmx1b L/L and Pitx3CreCre; Lmx1b +/+, Pitx3CreCre; Lmx1b L/+ littermates. Embryos were isolated at E14.5, considering the morning of plug formation as E0.5. Pregnant and adult mice were euthanized by CO2 asphyxiation and embryos and brain were collected in 1× PBS and immediately frozen on dry-ice (fresh frozen) or fixed by immersion in 4% paraformaldehyde (PFA) for 4–8 h at 4°C. After PFA incubation, samples were cryoprotected O/N in 30% sucrose at 4°C. Embryos and brains were frozen on dry-ice and stored at −80°C. Cryosections were slices at 16 μm, mounted at Superfrost plus slides, air-dried and stored at −80°C until further use.
Genotyping
Genotyping of the Lmx1b-Lox animals was done by PCR using primers: forward 5′-AGGCTCCATCCATTCTTCTC, and reverse: 5′-CCACAATAAGCAAGAGGCAC, resulting in a wild-type product of 243 bp, or a LoxP-inserted product of 277 bp. Pitx3-Cre genotyping was done by two different PCR’s. To discriminate between wild-type and Pitx3-Cre: forward 5′-GCATGATTTCAGGGATGGAC and reverse 5′-ATGCTCCTGTCTGTGTGCAG, resulting in a product of 750 bp for a mutant allele, and no product in wild-type animals. To additionally discriminate between heterozygous and homozygous Pitx3-Cre animals, primers were designed around Pitx3 exon 2 and exon 3: forward 5′-CAAGGGGCAGGAGCACA and reverse 5′-GTGAGGTTGGTCCACACCG, resulting in a product of 390 bp for the wildtype allele and no product for the mutant allele.
In situ Hybridization
In situ hybridization with digoxigenin (DIG)-labeled RNA probes was performed as described (Smits et al., 2003; Smidt et al., 2004). DIG-Labeled RNA probes for Th, Vmat2, Dat, Aadc, Pitx3, En1, Ahd2 and Cck have been previously described (Grima et al., 1985; Smits et al., 2003; Smidt et al., 2004; Jacobs et al., 2007; Hoekstra et al., 2013). The used Lmx1b probe is a 310 bp fragment containing exon 4–6 of the Lmx1b coding sequence.
Fluorescence Immunohistochemistry
Cryosections were blocked with 4% HIFCS in 1× THZT (50 mM Tris-HCL, pH 7.6; 0.5M NaCl; 0.5% Triton X-100) and incubated with a primary antibody [Rb-TH (Pelfreeze 1:1,000), Sh-TH (Millipore AB1542, 1:750), Rb-AHD2 (Abcam AB24343, 1:500)] in 1× THZT overnight at room temperature. The following day the slides were washed and incubated for 2 h at room temperature with secondary Alexafluor antibody [anti-rabbit, anti-sheep (Invitrogen, 1:1,000)] in 1× TBS. After immunostaining nuclei were staining with DAPI (1:3,000) and embedded in Fluorsave (Calbiogen) and analyzed with the use of a fluorescent microscope (Leica). All washes were done in TBS and double stainings were performed sequential, with immunostaining for TH being done first, followed by the staining for AHD2. The antibody against AHD2 requires antigen retrieval, which was executed as follows; slides were incubates 10 min in PFA after which they were submersed in 0.1 M citrate buffer pH 6.0 for 3 min at 1,000 Watts followed by 9 min at 200 Watts. Slides were left to cool down, after which protocol was followed as described above.
Quantitative Analysis
Quantification of TH-expressing neurons, TH+AHD2+ cells and TH+AHD- cells in 3-month-old and 12-month-old midbrain was performed in ImageJ as follows. First, TH-positive cells were counted independently of whether or not they expressed AHD2. Cells were counted in 10–12 matching coronal sections covering the whole midbrain dopaminergic neuronal pool [3 months old, n = 3 wildtype, n = 4 conditional Lmx1b knock-out (cKO), n = 3 for Pitx3 KOs, n = 3 for double Pitx3/Lmx1b KOs; 12 months old, n = 4 wildtype, n = 4 Lmx1b cKO]. Cells were counted as TH+neurons when TH staining co-localized with nuclear DAPI staining. For the double stained sections, a color overlay was made and the double stained cells were counted as TH+AHD2+ and the green cells were considered as TH+AHD2- cells. Similar to the single TH quantifications, cells were counted as neurons when the staining co-localized with nuclear DAPI staining. The separation of the SNc and VTA were made based on anatomical landmarks. Everything rostral of the supramammillary decussation was considered as SNc and distinction between the SNc and the VTA was made based on the tracts medial lemniscus, positioned between the SNc and VTA. Cell numbers from all sections were pooled and statistical analysis was performed via a student’s T-test. Graphs represent the mean of counted cells divided by wildtype ± the SEM.
RNA-Sequencing
RNA was isolated from dissected E14.5 midbrains of Pitx3Cre/+; Lmx1b +/+ and Lmx1b cKO embryos. RNA was isolated with Trizol (ThermoFisher) according to manufacturer’s protocol. After isolation RNA clean-up was performed with an RNA mini kit from Qiagen according to manufacturer’s protocol. E14.5 embryos were obtained from four different litters and RNA isolated of six wildtype or six mutant embryos from different nests was pooled to eventually form an n = 3 per group. Pair-end RNA-sequencing (minimal 2 * 10∧7 reads per sample), mapping on the mouse genome and DESeq2 statistical analysis on read counts was performed by Service XS (Leiden, Netherlands). The mapping was done with the mouse ensemble GRCm38.p4 data set.
Quantitative PCR (qPCR)
RNA was isolated from dissected E14.5 midbrains of Pitx3Cre/+; Lmx1b +/+ and Lmx1b cKO embryos and of E14.5 Pitx3CreCre; Lmx1b +/+ and Pitx3CreCre; Lmx1b L/L. RNA was isolated with Trizol (ThermoFisher) according to manufacturer’s protocol. For the Lmx1b cKO three midbrains were pooled per samples (n = 4 wildtype, n = 4 cKO) and further purified on a column, according to the manufacturer’s protocol (Qiagen, RNeasy mini kit). For the Pitx3/Lmx1b mutants, a single midbrain was used per sample (n = 3 Pitx3CreCre; Lmx1b+/+, n = 3 Pitx3CreCre; Lmx1b L/L) and no RNA clean-up was performed. Relative mRNA levels were determined by using the QuantiTect SYBR green PCR lightCycler kit (Qiagen) according to the manufacturer’s instructions. For each reaction 10 ng (dissected midbrain) of total RNA was used as input. Primers used for Th, Ahd2, En1 and Cck were previously published (Jacobs et al., 2011), primers for Lmx1b forward 5′-GAGCAAAGATGAAGAAGCTGGC and reverse 5′-CTCCATGCGGCTTGACAGAA (product size 98 bp). Quantitative PCR (qPCR) data was analyzed using the delta-delta Ct method and statistical analysis was performed via a student’s T-test Graphs represent the fold change divided by wildtype ± the SEM.
MN9D Cell Culture and Transfection
MN9D cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 4.5 g/L glucose, supplemented with 10% (v/v) HIFCS, 100 units/mL penicillin, 100 units/ml streptomycin and 2 mM L-glutamine. Cells were grown on 10 cm-diameter plates manually coated with poly-D-Lysine, in a humidified atmosphere with 5% CO2 at 37°C. Transfection of MN9D cells was performed using Lipofectamine 2,000 reagent (Invitrogen) on 10 cm-diameter plates-cultured MN9D cells on 70%–90% confluency. Lipofectamine 2000 Reagent was diluted 1:50 (v/v) in high glucose DMEM containing 2.5 μg of DNA vector, in a total volume of 1.2 mL of DMEM. After 20 min of incubation at room temperature, the DNA-lipid complexes mix was diluted in medium without antibiotics (DMEM, 4.5 g/L glucose, 10% HIFCS) to a final volume of 10 mL and MN9D cells were incubated with this medium for 6 h in a humidified atmosphere with 5% CO2 at 37°C. Cells were harvested for RNA isolation with Trizol (Thermofisher) according to the manufacturer’s protocol.
Results
Specific Deletion of Lmx1b in Developing Pitx3 Positive Neurons
Studies on Lmx1b null mutants have shown that Lmx1b is essential for the development of mdDA neurons. In the absence of Lmx1b TH+ cells fail to induce Pitx3 and are lost during development (Smidt et al., 2000). In addition to an effect on the mdDA neuron population, Lmx1b was also found to be essential for the expression of several genes associated with IsO functioning, including Fgf8, Wnt1, En1, En2, Pax2 and Gbx2, and that the loss of Lmx1b severely affects the initiation, maintenance and the inductive activity of the IsO during midbrain development (Guo et al., 2007). To study the role of Lmx1b in mdDA development, without affecting the IsO, we generated a conditional Lmx1b knock-out. Lmx1b-floxed animals (Suleiman et al., 2007) were crossed with Pitx3Cre animals (Smidt et al., 2012), to generate mice that lack Lmx1b exclusively in Pitx3 positive neurons. Pitx3 expression is initiated around E11.5 (Smidt et al., 1997) and it has been shown that the onset of CRE activity of Pitx3-Locus driven Cre expression is around E13.5 (Smidt et al., 2012), so to guarantee a complete loss of Lmx1b transcript we decided to study E14.5 embryos. To verify that these animals lack Lmx1b only in the Pitx3 positive domain, we assessed the expression of Lmx1b in Pitx3Cre/+; Lmx1b L/L compared to Pitx3Cre/+; Lmx1b +/+ littermates by means of in situ hybridization (Figure 1A). A clear loss of Lmx1b expression could be observed in the mdDA area (arrowheads) in Lmx1b L/L embryos, while expression in other regions, like the diencephalon and the hindbrain, remains unaffected (arrows). To examine whether the loss of a Pitx3 allele and the complete loss of Lmx1b influenced Pitx3 expression we analyzed its expression in adjacent slides (Figure 1B). When comparing wild-type to mutant embryos no clear difference in Pitx3 expression was observed. Taken together, our analysis shows that crossing Lmx1b-floxed mice with Pitx3Cre animals, generates a Lmx1b cKO, in which the deletion is confined to the mdDA neuronal field, leaving the rostral and caudal expression domains of Lmx1b intact.
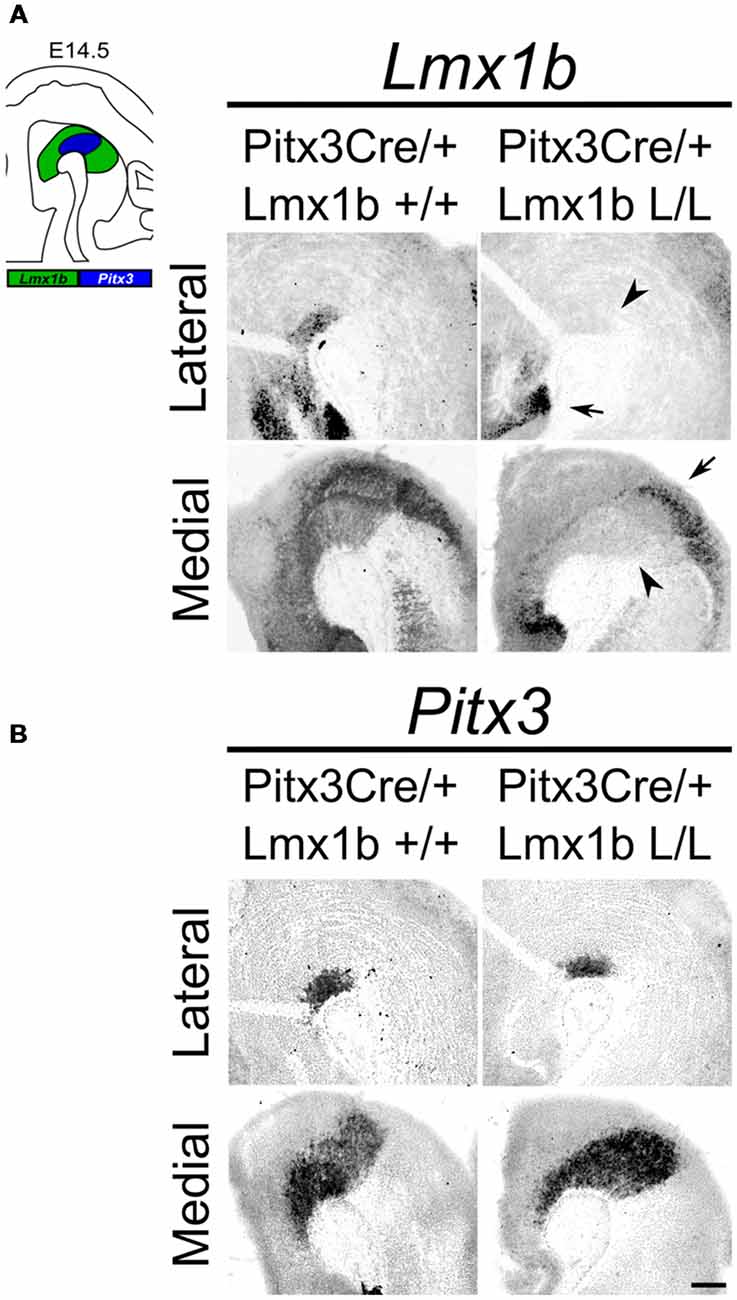
Figure 1. Lmx1b expression is specifically lost in the Pitx3 positive domain of Lmx1b conditional knock-outs (cKOs) embryos. Analysis of Lmx1b and Pitx3 expression via in situ hybridization in embryonic day (E)14.5 midbrain sagittal sections. (A) When comparing the Pitx3Cre/+; Lmx1b L/L to the Pitx3Cre/+; Lmx1b+/+ a clear loss of Lmx1b is observed in the Pitx3 positive area (arrowheads). However Lmx1b expression is maintained in regions that are negative for Pitx3 (arrows). (B) Pitx3 expression seems unaffected. Scale bar = 300 μM.
Lmx1b Is Not Essential for Early Post-mitotic Development of mdDA Neurons
As described above, the onset of Pitx3 driven CRE activity is around E13.5 and by E14.5 most genes that define a mature mdDA neuron, like Th, Vmat2 and Dat, are expressed (Iversen, 2010; Arenas et al., 2015). The expression of these genes is dependent on the combined functioning of Nurr1 and Pitx3 (Saucedo-Cardenas et al., 1998; Hwang et al., 2003; Nunes et al., 2003; Smits et al., 2003; van den Munckhof et al., 2003; Smidt et al., 2004; Jacobs et al., 2009). Loss of function studies showed that Nurr1 mutants fail to induce Th, Vmat2 and Dat, leading to a developmental arrest of mdDA progenitors and cell death (Zetterström et al., 1997; Saucedo-Cardenas et al., 1998; Smits et al., 2003). While Pitx3 was found to be crucial for the correct specification of mdDA neurons by acting as an activator of the NURR1 transcriptional complex (Jacobs et al., 2009). Since we removed Lmx1b in post-mitotic cells, we hypothesized that any effect on the differentiation of mdDA neurons would become apparent in the spatial expression of Th, Vmat2 and Dat at E14.5. In addition to these three factors, we also examined Aadc, which is also required for the proper DA neurotransmitter phenotype and is affected in both Nurr1 and the Pitx3 mutants, similar to the other marks we examined (Saucedo-Cardenas et al., 1998; Smidt et al., 2004). However, this gene is induced in an early state at E10.5 in a Nurr1-independent manner (Smits et al., 2003). When analyzing the spatial expression of Th via in situ hybridization no clear differences could be observed in the lateral or medial sections (Figure 2A). In addition, the expression of Vmat2, Aadc and Dat also seem unaffected by the post-mitotic loss of Lmx1b (Figures 2B–D). Together these results suggest that post-mitotic deletion of Lmx1b does not influence the expression of general DA marks Th, Vmat2, Aadc and Dat.
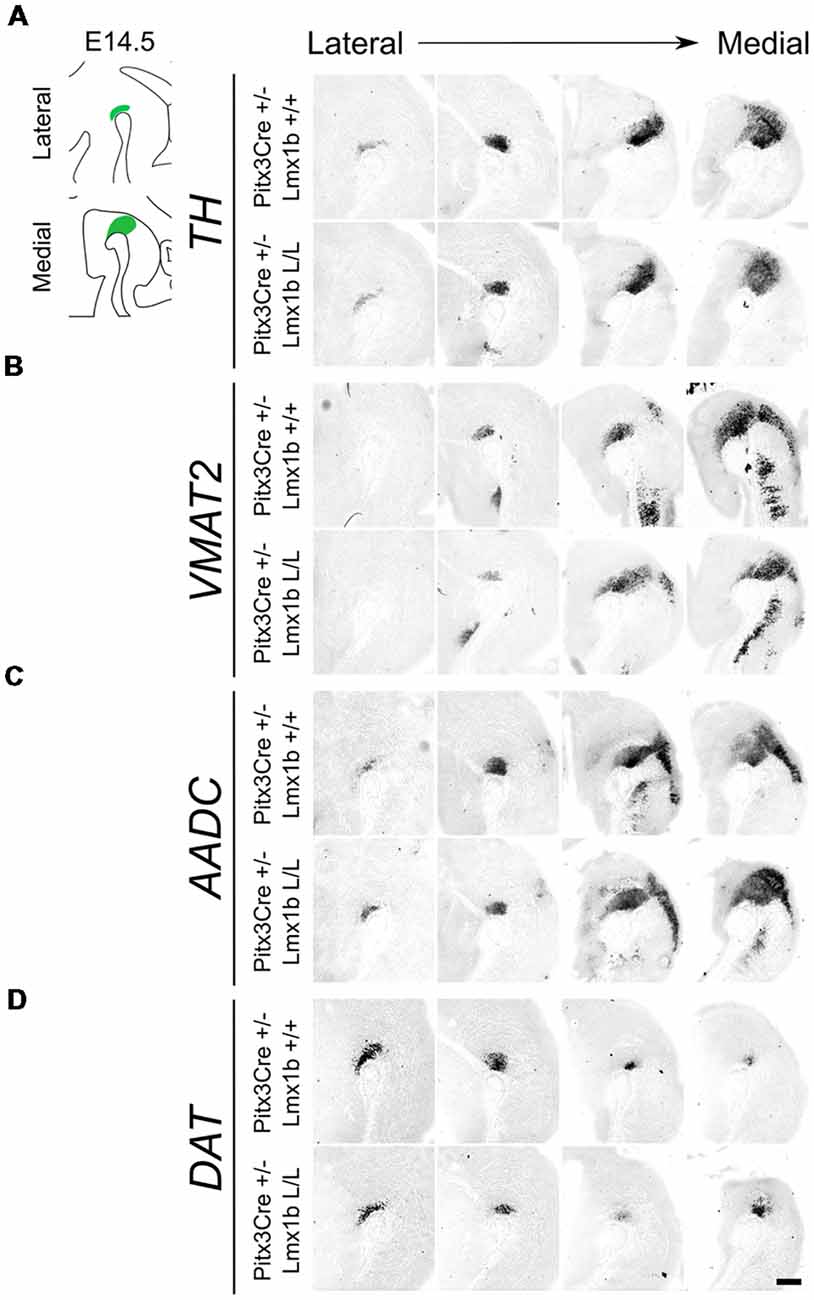
Figure 2. Several genes important for the dopamine (DA) neurotransmitter phenotype are not clearly affected by the Pitx3Cre driven loss of Lmx1b at E14.5. (A–D) In situ hybridization of Th, Vmat2, Aadc and Dat in E14.5 midbrain sagittal sections. (A–D) In both lateral and medial sections, expression patterns of Th, Vmat2, Aadc and Dat do not seem to be affected in Pitx3Cre/+; Lmx1b L/L embryos. Scale bar = 200 μM.
Conditional Removal of Lmx1b Results in the Loss of TH+ Cells in the Adult Midbrain
Previous studies have already shown that Lmx1b expression is continued throughout life (Smidt et al., 2000; Dai et al., 2008; Laguna et al., 2015), suggesting that Lmx1b might have a role in neuronal identity, maintenance and survival. A study performed by Doucet-Beaupré et al. (2016) demonstrated that the combined conditional genetic ablation of Lmx1a and Lmx1b under the control of the Dat promoter causes a degeneration of TH+ cells in 2-month-old mice in both the SNc and the VTA. To verify whether Pitx3 driven deletion of Lmx1b also influenced neuronal identity and/or maintenance, we examined the expression of Th, Vmat2, Aadc, and Dat in 3-month-old midbrains (Figure 3). When comparing Pitx3Cre; Lmx1b +/+ animals to Pitx3Cre; Lmx1b L/L animals no obvious alterations in the distribution of Th and Vmat2 can be observed (Figures 3A,B), however when analyzing the expression of Aadc, alterations can be observed (Figure 3C, arrowheads) and when comparing Dat expression between the wildtype and the Lmx1b cKOs an overall reduction can be seen (Figure 3D, arrow heads). To further establish whether changes in expression are caused by the influence of Lmx1b on specific gene expression or on neuronal survival in general, we aimed to quantify the total amount of TH+ cells in the mdDA population. We performed immunohistochemistry for TH and counted the cells in both the SNc and the VTA (Figure 4A; Supplementary Table S1). The total amount of TH+ cells is reduced with ~15% (n = 3, **P < 0.01, one-tailed) in the cKO compared to wildtype littermates (Figure 4B). The loss of TH+ cells is largest in the VTA (~ 20% loss, **P < 0.01, one-tailed), while ~13% of the cells in the SNc are lost (Figure 4B). Since Laguna et al. (2015) found a progressive loss of cells between 2 months old cKO mice and 18-month-old DatCre; Lmx1a L/L; Lmx1b L/L animals, we aimed to investigate whether the reduction in TH+ cells in our model would also further progress. We quantified the amount of TH+ cells in the SNc and the VTA in 12-month-old midbrains (Figure 5A; Supplementary Table S1). A reduction of ~10% of the total TH+ population is observed in Pitx3Cre; Lmx1b L/L midbrains (n = 4, P < 0.05, one-tailed; Figure 5B). In contrast to 3-month-old animals, the loss of TH+ cells is only observed in the SNc, where the amount of TH+ cells is reduced with 15% (n = 4, P < 0.05, one-tailed) in the cKO midbrains, whereas the amount of TH+ cells in the VTA are not significantly altered (n = 4; Figure 5B). Together these results demonstrate that ablation of Lmx1b leads to the loss of TH+ neurons. In the VTA, an accelerated loss of cells is observed in the Lmx1b cKO, leading to reduced numbers of TH+ cells at 3 months of age. However, over time the number of cells between the wildtype and Pitx3Cre/+; Lmx1b L/L animals are equalized, suggesting that during aging Pitx3Cre/+; Lmx1b +/+ animals also lose cells in the VTA. Interestingly, the defect observed in the SNc at 3 months of age, resides with time and is still present at 12 months of age.
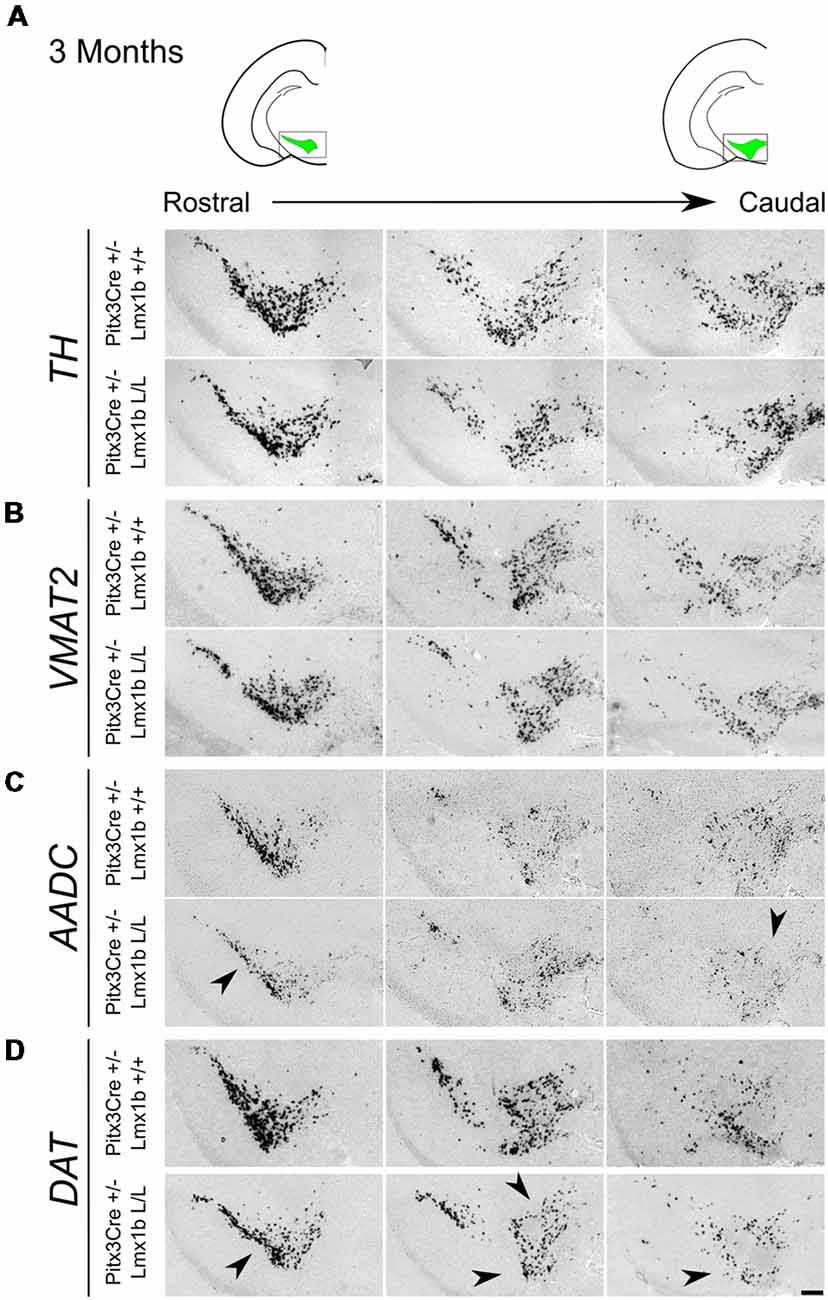
Figure 3. Pitx3 driven deletion of Lmx1b results in a change in Aadc and Dat expression in the adult midbrain. Analysis of Th, Vmat2, Aadc and Dat in coronal adult section in the Pitx3Cre/+; Lmx1b L/L mutant via in situ hybridization. (A,B) Expression of Th and Vmat2 seem unaffected in Pitx3Cre/+; Lmx1b L/L animals, but the Pitx3 driven deletion of Lmx1b results in alteration in the expression of Aadc (C, arrowheads) and an overall reduction in Dat expression (D) in both rostral and caudal sections (arrowheads). Scale bar = 150 μM.
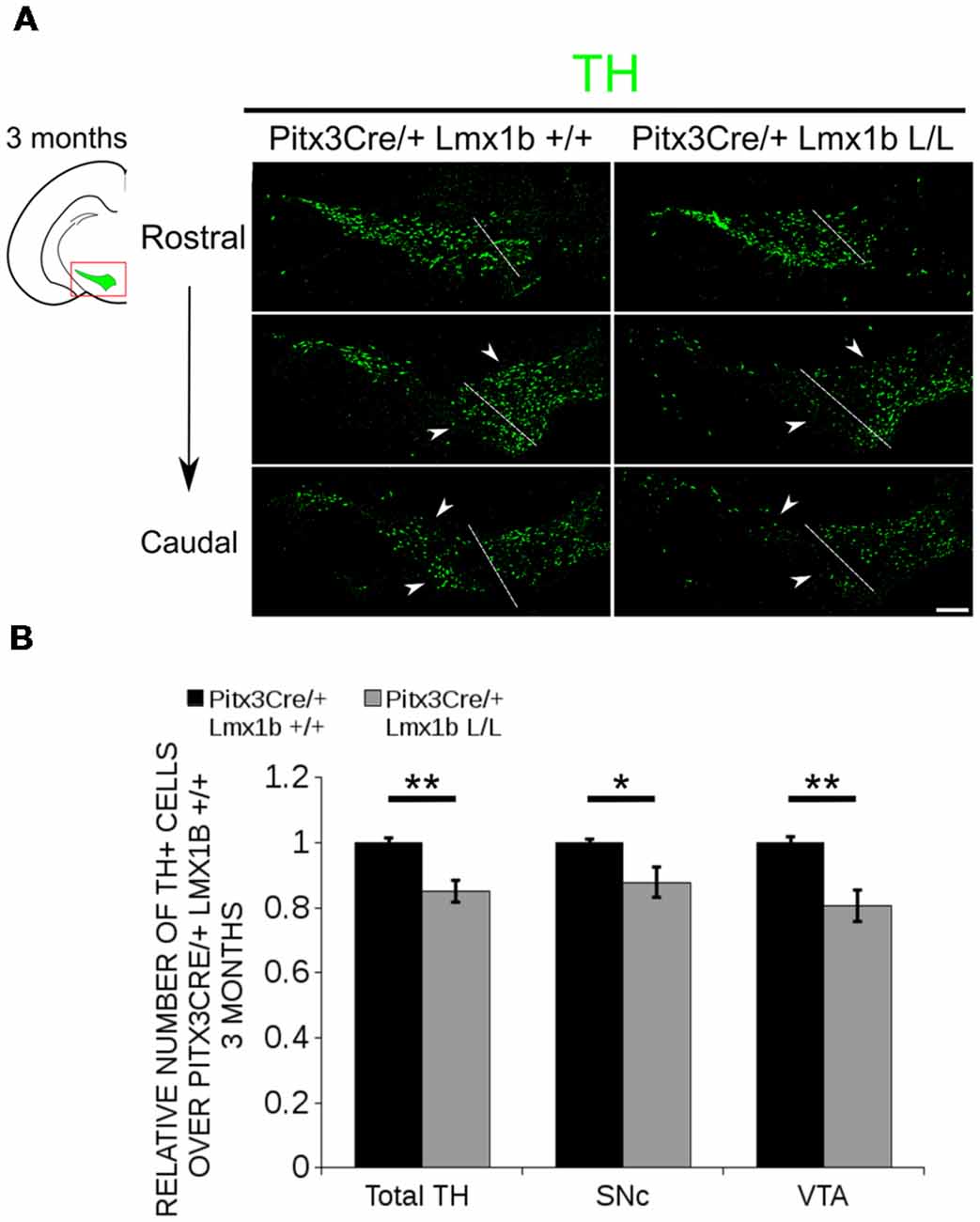
Figure 4. The number of TH+ cells is decreased in Pitx3Cre/+; Lmx1b L/L 3-month-old midbrains. (A) Protein expression of TH (green) was evaluated via immunohistochemistry in the adult midbrain of 3-month-old Lmx1b cKO animals. A loss of TH signal was observed in both the ventral tegmental area (VTA) and the substantia nigra pars compacta (SNc; white arrowheads). The white dotted line represents the border between what is considered SNc and VTA. (B) Quantification of TH+ cells in the adult midbrain of Pitx3Cre/+; Lmx1b L/L (n = 3, gray bars) and Pitx3Cre/+; Lmx1b +/+ controls (n = 3, black bars) shows that the total amount of TH+ neurons is significantly lower (~ 15% loss, **P < 0.05, one-tailed) and that neurons are lost both in the SNc (~ 13% loss, *P < 0.05) and the VTA (~ 20% loss, **P < 0.01, one-tailed). Pitx3Cre/+ Lmx1b+/+ animals were set at 1. Scale bar = 100 μM.
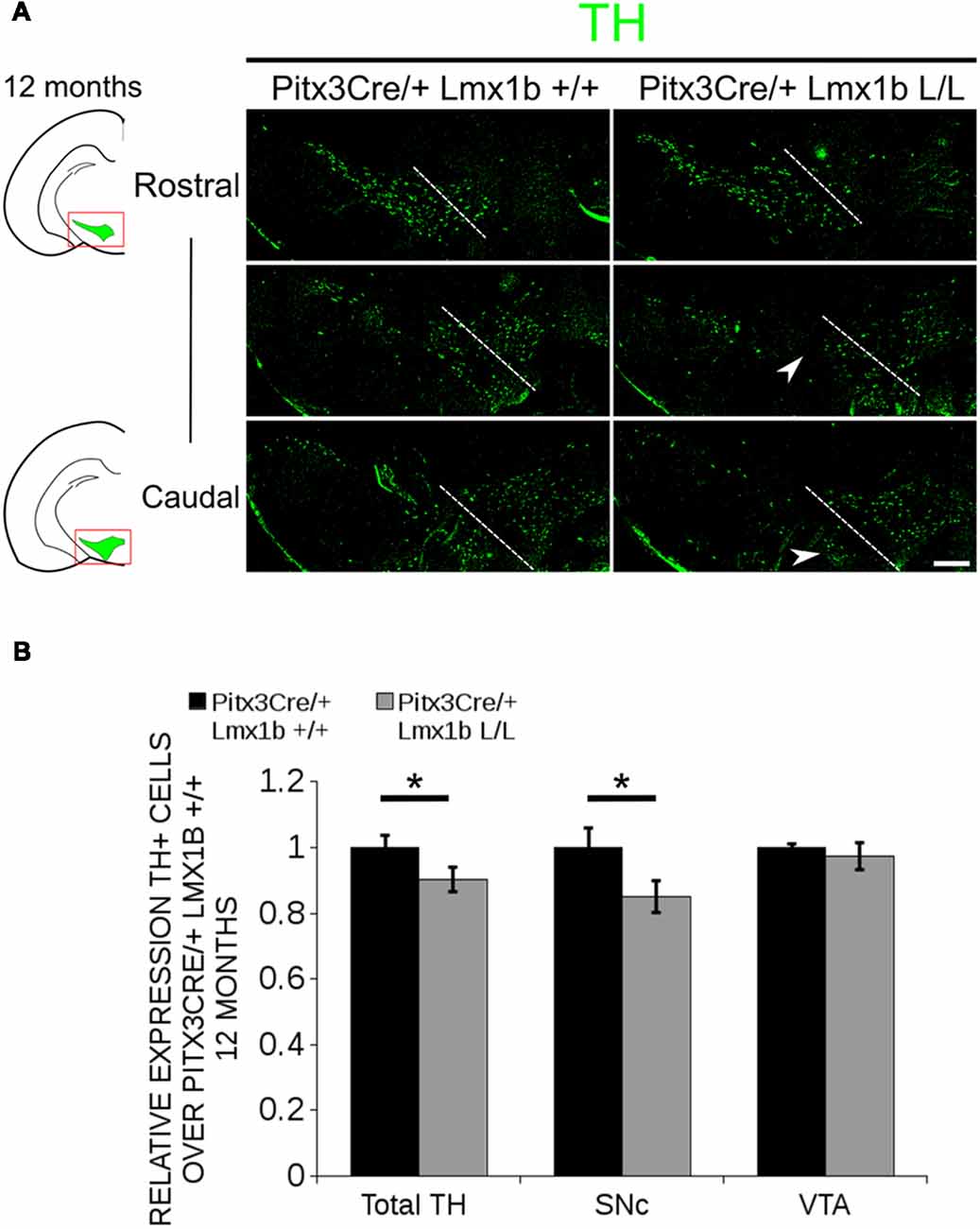
Figure 5. In 12-month-old midbrains, a reduction in TH+ neurons is observed in Pitx3Cre driven Lmx1b cKO animals. (A) Protein expression of TH (green) was evaluated by immunohistochemistry in adult midbrains of 12-month-old mutants and wildtypes. A loss of TH signal was observed in the SNc (white arrowheads). The white dotted line represents the border between what is considered SNc and VTA. (B) Quantification of TH+ cells in the adult midbrains of Pitx3Cre/+; Lmx1b L/L (n = 4, gray bars) and Pitx3Cre/+; Lmx1b +/+ controls (n = 4, black bars) shows that the total amount of TH+ neurons is significantly lower (~10% loss, **P < 0.05, one-tailed) and that neurons are lost in the SNc (~15% loss, *P < 0.05), but no difference was observed in the VTA (n = 4, one-tailed). Pitx3Cre/+ Lmx1b +/+ animals were set at 1. Scale bar = 100 μM.
Lmx1b Regulates Genes Involved in Neuronal Development and Acts as a Repressor of Ahd2
In order to obtain a better insight into which molecular mechanisms are affected during development after the conditional removal of Lmx1b, we performed next-generation RNA-sequencing on dissected E14.5 midbrains of Pitx3Cre/+; Lmx1b +/+ and Pitx3Cre/ Lmx1b L/L embryos (n = 3; two pooled embryos per biological replicate; Figure 6A). When using a P-value cut-off of P < 0.05 285 genes were identified, of which 105 were up-regulated and 180 are down-regulated. A PANTHER over-representation tests showed that these genes are mainly associated with head and nervous system development at E14.5 (Supplementary Figure S1). Among the 20 most regulated genes we found Aldh1a1 (Ahd2), En1 and En2 (Figure 6B). En1 and En2 have been implicated in mdDA neuronal survival before Albéri et al. (2004) and Simon et al. (2001). It was shown that within animals heterozygous for En1 lose DA neurons specifically in the SNc (Sonnier et al., 2007). In addition to a role in neuronal survival En1 has been implicated to play a role in the specification of the mdDA subpopulations. En1-deficient embryos demonstrate a down-regulation of Th, Dat, Vmat2 and D2R in rostrolateral sub-population of mdDA neurons, which are destined to become the SNc (Veenvliet et al., 2013). Interestingly, this rostrolateral sub-population is marked by the expression of Ahd2 (Jacobs et al., 2007; Veenvliet et al., 2013), which shows an upregulation of approximately two-fold (Figure 6B), suggesting that the loss of Lmx1b might specifically affect the rostrolateral subset. To identify other genes associated with mdDA neuronal development and the rostrolateral subset, we cross-referenced the list of 285 possible target genes to a list of genes known to be involved in mdDA development (Figure 6C). In addition, we performed an overlay of transcripts of Lmx1b target genes with the MAANOVA-FDR analysis of genes regulated by either En1 (Veenvliet et al., 2013) or Pitx3 (Jacobs et al., 2011), two genes that have been shown to be essential for the formation of rostrolateral mdDA neurons (Veenvliet et al., 2013). We found nine genes that were previously associated with mdDA development of which En1 and Ahd2 were considered for further study (Figure 6D, red arrows). Furthermore, genes regulated by both Lmx1b and En1 were found to be regulated in the same direction (Figure 6E). This in contrast to genes that are regulated by both Lmx1b and Pitx3, which are mostly regulated reciprocal (Figure 6F), similar to the Pitx3-En1 interplay (Veenvliet et al., 2013). En1, En2 and CD9 were found to be regulated by all three genes, Lmx1b, En1 and Pitx3 (Figure 6G), and while they are up-regulated in the Pitx3 mutant (Figure 6F), they are down-regulated in both En1 mutants and Pitx3Cre/+; Lmx1b L/L embryos (Figure 6E). Based on the results from the genome-wide expression analysis and the association of En1 and Ahd2 with the SNc (Jacobs et al., 2007; Sonnier et al., 2007; Veenvliet et al., 2013), we decided to examine the spatial expression of these two genes. In independent sets, the mRNA levels were confirmed using qPCR and the spatial expression patterns were studied using in situ hybridization. The levels of En1 were 30% reduced (n = 4, *P < 0.05, one-tailed) in Lmx1b mutant embryos compared to wildtype (Figure 7A, right panel), validating our RNA-sequencing results. Interestingly, the spatial expression of En1 is not altered in the Pitx3Cre/+; Lmx1b L/L embryos compared to Pitx3Cre/+; Lmx1b +/+ embryos at E14.5 (Figure 7A, left panel), suggesting that Lmx1b is important for the mRNA expression level of En1 in all mdDA neurons. When examining the expression pattern of Ahd2, an increase in expression can be found in the entire Ahd2 positive area (Figure 7B) and clear ectopic Adh2 expression is found in the more medial sections (Figure 7B, left panel, arrowheads). Interestingly, the Ahd2 positive domain seems to exceed that of Th in the lateral sections of E14.5 Lmx1b cKO midbrains, while in the medial sections it remains restricted to the Th positive domain (Figure 7C, arrowhead). Quantification of the mRNA expression level showed a two-fold increase in Adh2 (n = 4, **p < 0.01, one-tailed) in the mutant compared to wildtype (Figure 7B, right panel). Interestingly, initial observation in Lmx1b overexpression studies in MN9D cells pointed already to this regulatory role of Lmx1b, since we found a 48% reduction in Ahd2 levels after Lmx1b overexpression (n = 4, **P < 0.01, two-tailed; Supplementary Figure S2). In summary, we show that Lmx1b is important for the regulation of genes associated with mdDA neuronal development, including En1, and that Lmx1b acts as a repressor of Ahd2.
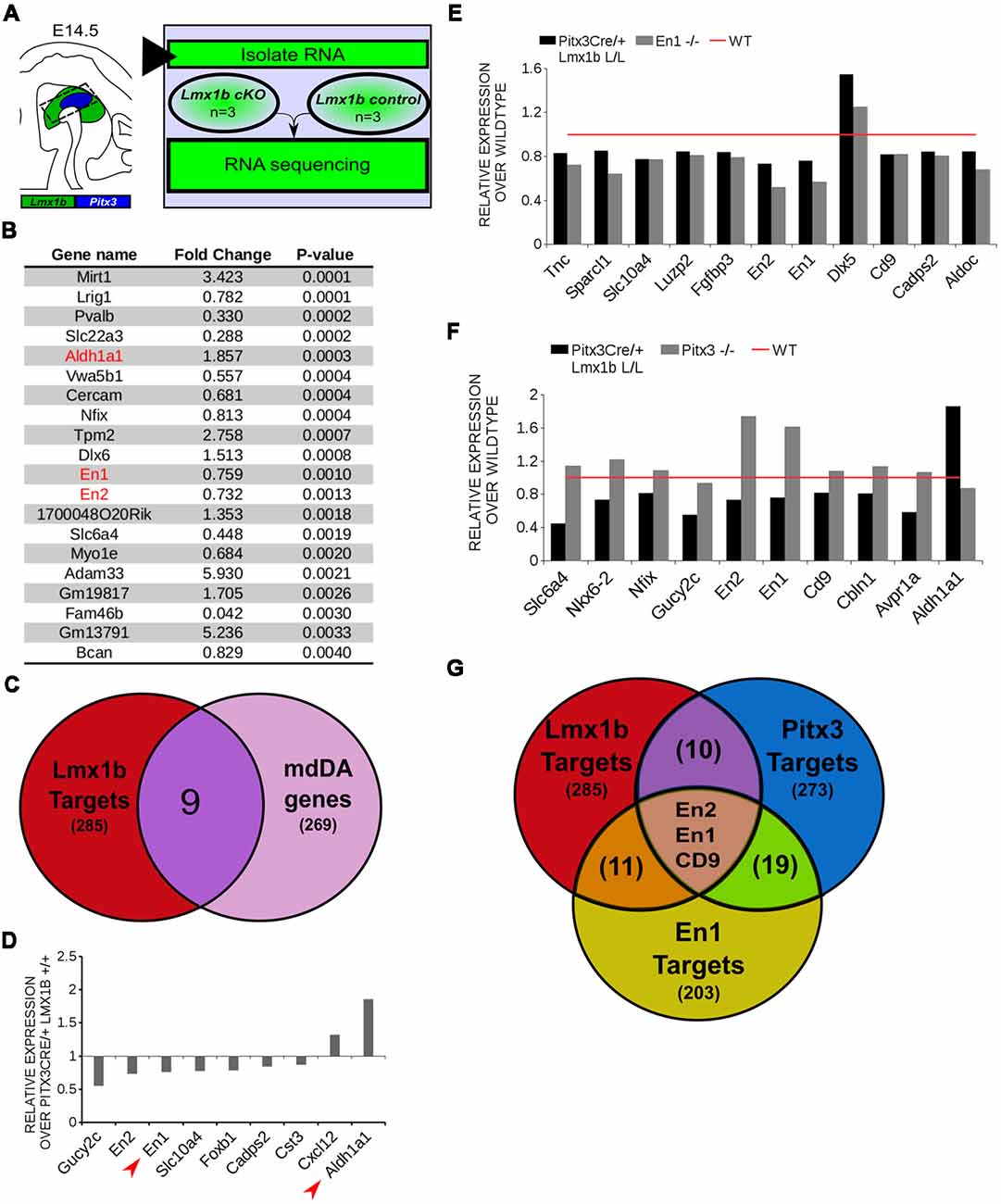
Figure 6. In vivo genome-wide expression analysis reveals that Lmx1b regulates genes involved in neuronal development, including En1 and En2 and acts as a repressor of Ahd2 expression. (A) Schematic representation of the experimental setup for sample preparation and sample comparison. (B) Among the 20 most regulated genes are Ahd2 (Aldh1a1, red), which shows an 85% increase, and En1 (red) and En2 (red) which are both down-regulated (En1, ~25% loss, En2, ~27% loss). (C) Venn diagram illustrating that nine genes regulated by Lmx1b are mdDA enriched. (D) Relative expression of nine genes that were mdDA enriched. Among the 9 genes En1 and Ahd2 were considered most interesting (red arrows). (E) Relative expression of genes regulated by both En1 and Lmx1b demonstrate that genes that are regulated by the loss of En1 (gray bars) are affected in the same manner in the Pitx3Cre/+ Lmx1b L/L E14.5 embryos (black bars). Wildtype was set at 1. (F) When looking at the relative expression of genes regulated by Pitx3 and Lmx1b the opposite effect is observed as with the En1 mutant. Genes up-regulated in the Pitx3 mutant are down-regulated in the Lmx1b depleted embryos and genes up-regulated in the Pitx3Cre driven Lmx1b mutant are down-regulated in embryos missing Pitx3. (G) Venn diagram showing that Lmx1b, En1 and Pitx3 are all involved in regulating En1, En2 and Cd9.
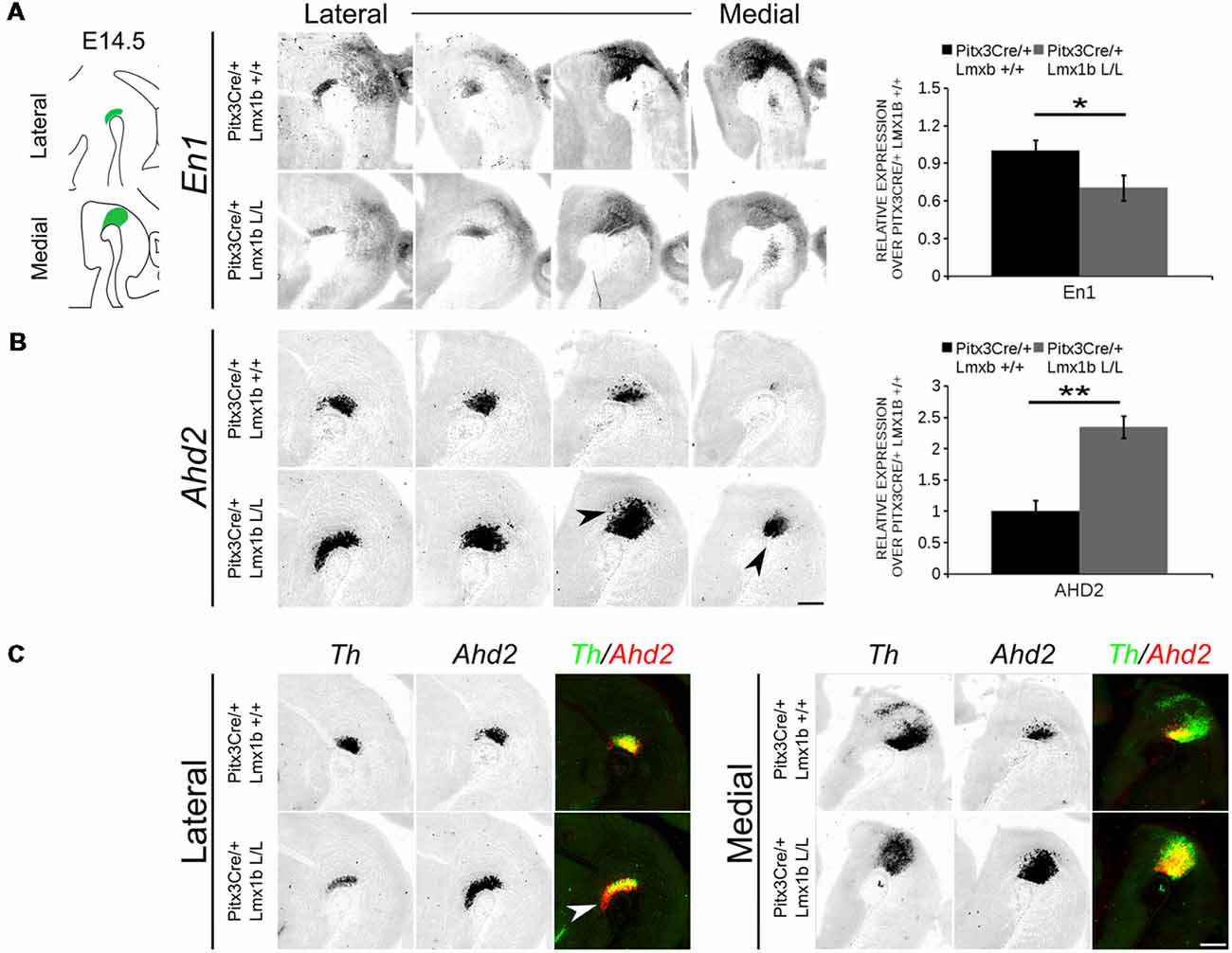
Figure 7. Ahd2 is up-regulated and ectopically expressed in Pitx3Cre driven Lmx1b mutants, together with a generic reduction in En1 expression in the midbrain of E14.5 mutant embryos. The expression patterns of En1 and Ahd2 in E14.5 midbrains of Lmx1b cKO and wildtype were analyzed via in situ hybridization, followed by quantitative PCR (qPCR) to quantify the mRNA levels. (A) The mRNA expression level of En1 is significantly reduced (~30% loss, *p < 0.05, one-tailed) in the Pitx3Cre/+ Lmx1b L/L E14.5 midbrain (n = 4, gray bars) compared to wildtype littermate controls (n = 4, black bars). Wildtype mRNA levels were set at 1. When visualizing the expression pattern an overall reduction in En1 expression can be seen in both lateral and medial sections. (B) Ahd2 expression can be found ectopically in medial section (arrowheads) and an up-regulation can be observed in more lateral sections. Quantification of the mRNA levels via qPCR shows an increase of ~130% (n = 4, **P < 0.01, one-tailed) in the mutant (gray bar) compared to the wildtype (black bar). Wildtype was set at 1. (C) Pseudo-overlay of the lateral and medial expression pattern of Ahd2 (red) and Th (green). In the lateral sections the Ahd2 expression domain exceeds the Th positive domain (white arrowhead), while the ectopic expression of Ahd2 in the medial sections is restricted to the spatial expression of Th. Scale bar = 300 μM.
TH+ Cells Are Lost in the SNc and the VTA of Pitx3Cre/+ Lmx1b L/L Adult Mice
During embryonic development different subsets of mdDA neurons can already be distinguished based on their molecular profile and anatomical position (Smits et al., 2013; Panman et al., 2014; La Manno et al., 2016; Tiklová et al., 2019). As described above, the rostrolateral subset, that will later form most of the SNc, is marked by the expression of Ahd2 (Jacobs et al., 2011; Veenvliet et al., 2013), while the caudomedial population, destined to become the VTA, expresses Cck (Hökfelt et al., 1980; Veenvliet et al., 2013). It was shown that these subsets are dependent on different transcriptional programs for their proper specification and survival (Smits et al., 2006; Jacobs et al., 2011; Veenvliet et al., 2013; Panman et al., 2014; Kouwenhoven et al., 2017). The rostrolateral population is dependent on a complex interplay between Pitx3 and En1, in which En1 is required for the induction of DA phenotype and influences Pitx3 expression and Pitx3 is required for antagonizing the caudomedial phenotype by regulating EN1 functioning (Veenvliet et al., 2013). When the rostrolateral population is formed, the remaining DA population will obtain a caudomedial phenotype under the influence of En1 (Bye et al., 2012; Veenvliet et al., 2013). Our results show lower mRNA levels of En1 and an expansion of the rostrolateral mark Ahd2 (Figure 7), suggesting that the specification of mdDA subsets is affected. In order to determine whether the loss of Lmx1b influences the caudomedial population at E14.5 we examined the expression of Cck (Figure 8A). No differences in spatial expression could be observed between the wildtype and Pitx3Cre/+; Lmx1b L/L embryos when analyzing Cck using in situ hybridization (Figure 8A, left panel). In addition, the mRNA levels of Cck are not significantly different between the wildtype and the cKO (n = 4; Figure 8A, right panel). Next to analyzing the expression at E14.5, we also decided to examine the spatial expression in 3-month-old adult midbrains (Figure 8B). Interestingly, a clear loss of Cck expression can be observed in the more lateral positions of the VTA (arrowheads; n = 2 observation), which seems to match the position where we observed the initial loss in TH+ cells (Figure 4B, arrowheads). However, Th expression can still be found in corresponding areas in adjacent slides (Figure 8C), suggesting that Cck is down-regulated in these areas. To further substantiate the effect of the deletion of Lmx1b on the adult mdDA subsets, we also examined the expression of Adh2 in the adult midbrain and quantified the amount of TH+AHD2+ cells in the SNc and the VTA (Figure 9A). The ectopic Ahd2 expression observed in the embryo could not be detected in the adult stages (Supplementary Figure S3) and a ~18% loss of TH+AHD2+ cells (n = 3, *P < 0.05, one-tailed) is even observed in the SNc (Figure 9B; Supplementary Table S1). While the TH+AHD2+ are affected in the SNc, the AHD2 negative population is significantly reduced in the VTA, where ~21% of the TH+AHD2- cells are lost (n = 3, **P < 0.01, one-tailed) in the mutant compared to Pitx3Cre/+; Lmx1b +/+ midbrains (Figure 9C; Supplementary Table S1). Together, our data shows that the DA-subsets are differential affected by the loss of Lmx1b in both the embryonic and adult stage.
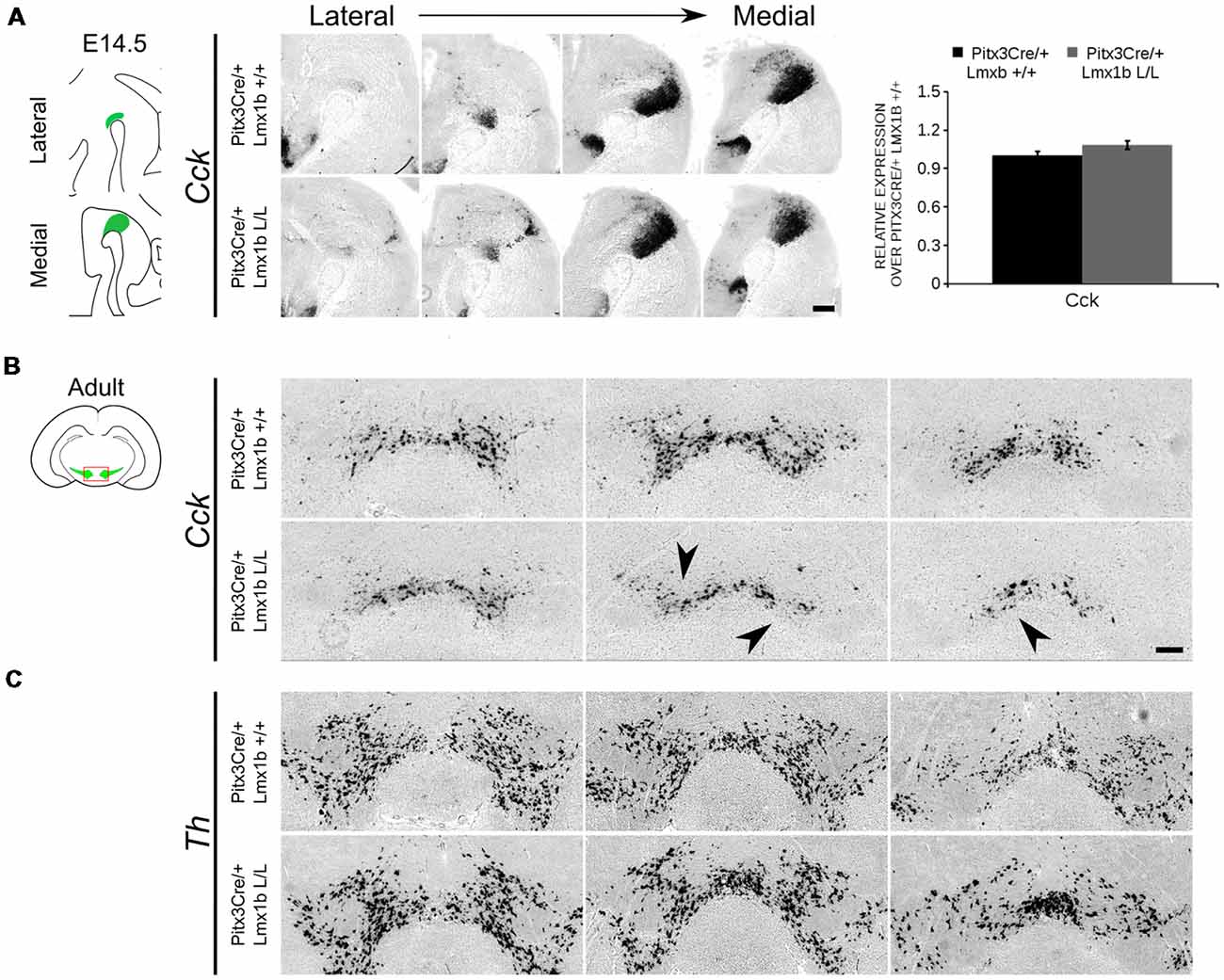
Figure 8. Cck expression is not affected in the midbrain of E14.5 Pitx3cre/+ Lmx1b L/L embryos, but shows a clear loss in the adult mutant midbrain. (A) Expression of Cck was examined using in situ hybridization and qPCR. No significant difference in mRNA level was found between Lmx1b conditional mutant E14.5 midbrain (n = 4, gray bar) and wildtype littermate midbrains (n = 4, black bar). Wildtype mRNA levels were set at 1. In addition, the expression pattern of Cck is comparable between Pitx3Cre/+; Lmx1b L/L E14.5 midbrain sections and Pitx3Cre/+ Lmx1b +/+ E14.5 midbrain sections. (B) In situ hybridization on the adult midbrain demonstrates a loss of Cck expression in the lateral parts of the VTA (black arrowheads, n = 4 observation). (C) The expression pattern of Th was visualized using in situ hybridization on adjacent sections. Scale bars are 200 μM.
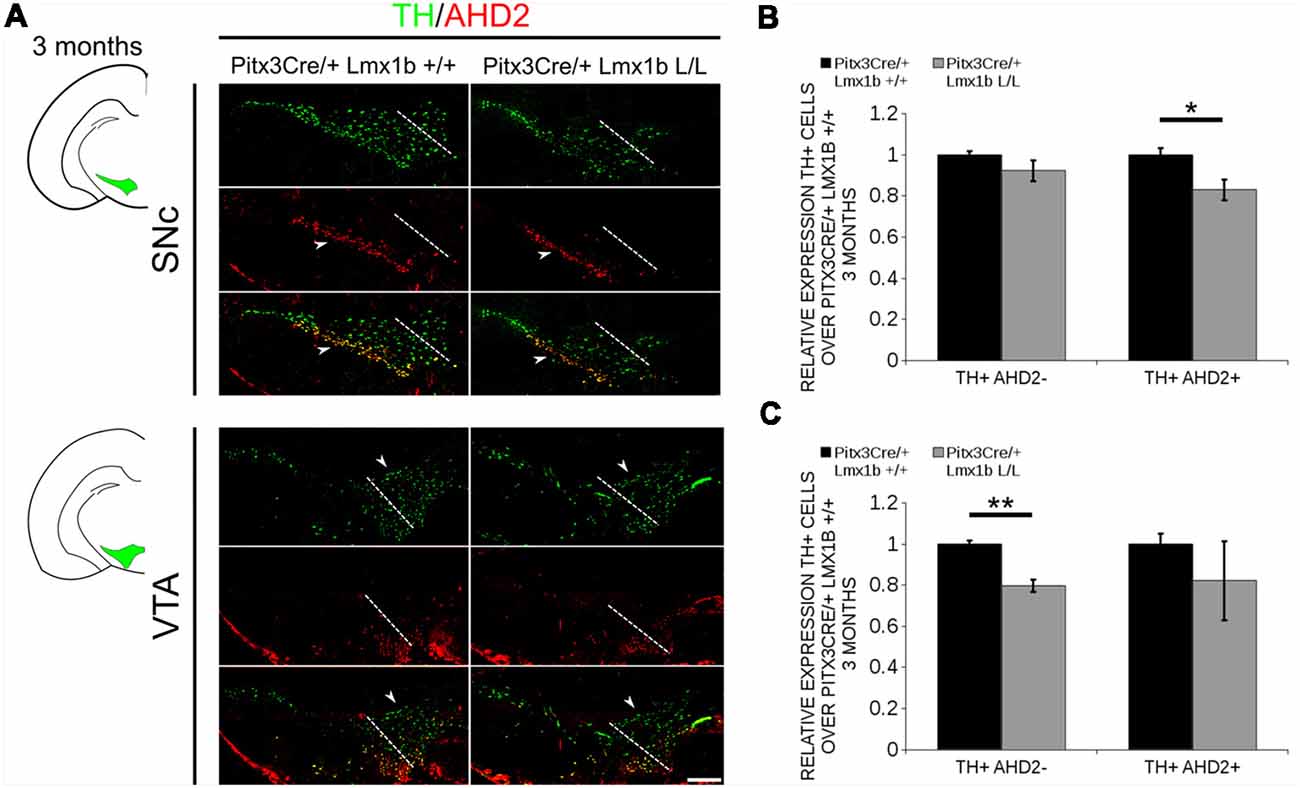
Figure 9. The TH+ cells that are lost in the SNc of 3-month-old mice conditionally depleted of Lmx1b are also AHD2+, while the TH+AHD2- population is affected in the VTA. Immunohistochemistry of TH (green) and AHD2 (red) in the adult midbrain of 3-month-old animals. (A) Assessment of the TH+AHD2+ (yellow) and the TH+AHD2- (green) population in both the SNc and VTA of Pitx3Cre/+ Lmx1b L/L animals shows that in the SNc (upper panel) the TH+ AHD2+ neurons are affected, while TH+ neurons affected in the VTA (lower panel) are AHD2- (white arrowheads). The white dotted line represents the border between what is considered SNc and VTA. (B) Quantification of the amount of cells in the SNc shows that the number of TH+AHD2+ neurons are reduced (~18%, n = 3, *p < 0.05, one-tailed), while the number TH+AHD2- cells are similar to the Pitx3Cre/+ Lmx1b +/+ (n = 3, black bar). (C) This in contrast to the VTA, where TH+AHD2- cells are lost (~ 21% loss, n = 3, **P < 0.01, one-tailed) in the Lmx1b cKO (gray bar) and TH+AHD2+ cells are not significantly reduced. The number of cells of in the midbrain of Pitx3Cre/+; Lmx1b +/+ animals were set at 1. Scale bar = 100 μM.
Lmx1b Represses Ahd2 Expression Independent of Pitx3
The transcriptional activation of Ahd2 in the rostrolateral population has been demonstrated to be dependent on the expression of Pitx3 (Jacobs et al., 2007). Pitx3 can interact with a region close to the transcriptional start site of Ahd2 and loss of expression is observed in Pitx3−/− embryos and adult midbrains (Jacobs et al., 2007). As mentioned above, the Pitx3 driven deletion of Lmx1b causes an up-regulation of Adh2 and to get a better insight into the mechanisms via which Lmx1b influences Ahd2 expression we generated a double mutant for Pitx3 and Lmx1b (Pitx3CreCre; Lmx1b L/L). We analyzed the expression of Ahd2, En1 and Th in Pitx3CreCre; Lmx1b L/L and Pitx3CreCre; Lmx1b +/+ embryos with in situ hybridization (Figures 10A–C). When analyzing the expression of Ahd2 we observed a clear increase in signal in several sections (arrows) in embryos depleted of both Pitx3 and Lmx1b (Figure 10A). In more lateral sections this increase in Ahd2 was less pronounced (Figure 10A). Importantly, The expression of En1 and Th seem unaffected (Figures 10B,C), suggesting that the ablation of Lmx1b only leads to an increase in Ahd2 expression in a Pitx3 mutant background. To quantify the mRNA levels, we isolated RNA from E14.5 dissected midbrains of Pitx3CreCre: Lmx1b L/L and Pitx3CreCre: Lmx1b +/+ embryos (Figure 10D). As expected, the Lmx1b mRNA levels were severely reduced as a consequence of the conditional ablation (n = 3, *P < 0.05, one-tailed). In addition to the loss of Lmx1b, Ahd2 was up-regulated to ~160% (n = 3, *p < 0.05, one-tailed), indicating that repression of Ahd2 by Lmx1b does not require Pitx3.
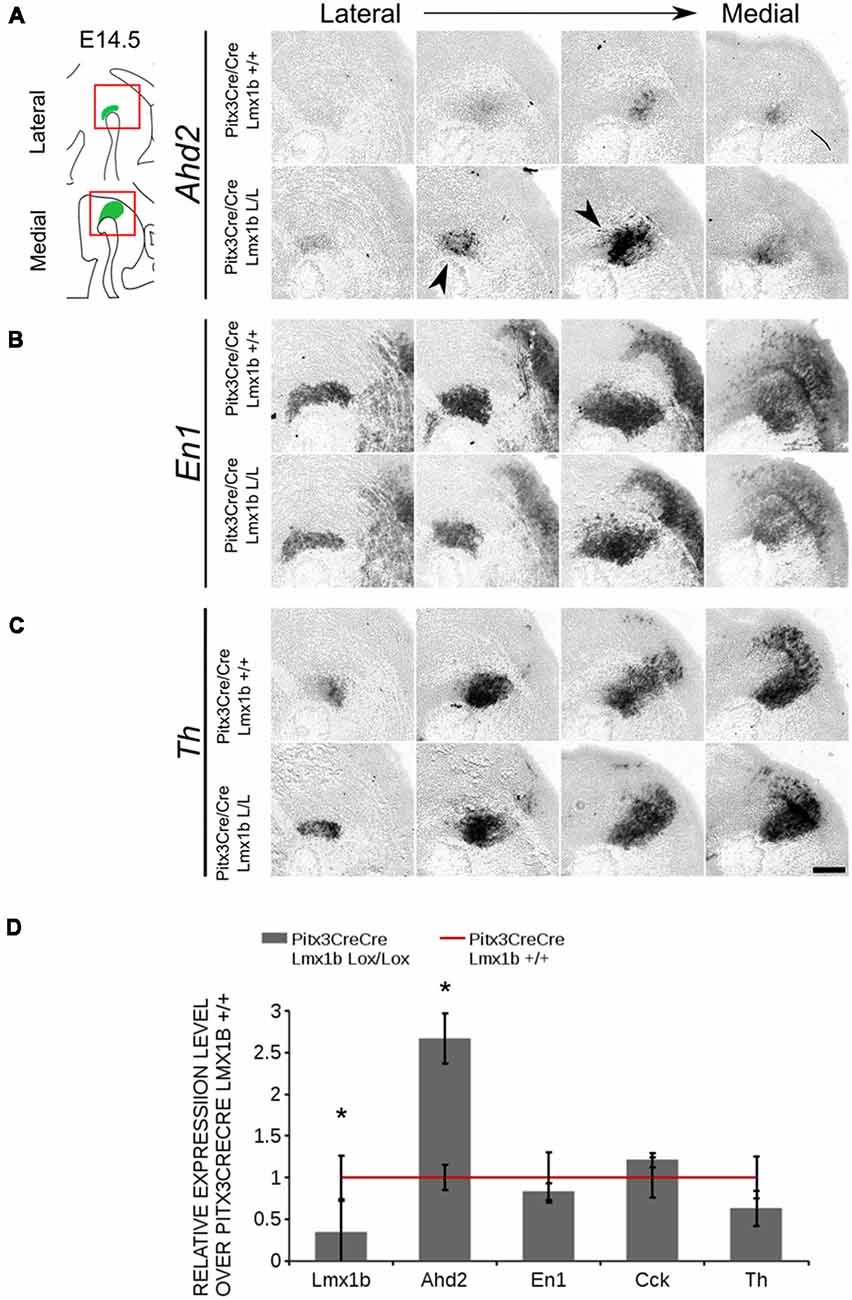
Figure 10. Lmx1b represses Ahd2 independent of Pitx3. The expression patterns of Adh2, En1 and Th were examined using in situ hybridization and the mRNA levels for Lmx1b, Ahd2, En1, Cck and Th were verified using qPCR. (A) Pitx3Cre driven loss of Lmx1b in Pitx3CreCre animals caused an increase in Ahd2 expression in the medial sections of the E14.5 midbrain (black arrowheads). (B,C) The expression pattern of En1 did not show clear differences between Pitx3CreCre; Lmx1b +/+ and the Pitx3CreCre Lmx1b mutant and neither did Th. (D) The mRNA expression levels of Lmx1b were down-regulated with ~76% in conditional mutant (gray bar, n = 3, *p < 0.05, one-tailed), while Ahd2 shows an increase of ~160% in the Pitx3CreCre; Lmx1b L/L (gray bar, n = 3, *p < 0.05, one-tailed) compared to Pitx3CreCre; Lmx1b +/+ littermates (red line, n = 4). The mRNA expression levels of En1, Cck and Th did not show significant changes. Wildtype was set at 1. Scale bar = 100 μM.
AHD2 is an aldehyde dehydrogenase and generates RA and in previous studies it has been shown that a partial recovery of TH+ neurons in the rostrolateral population can be accomplished by maternal supplementation with RA in Pitx3 ablated animals (Jacobs et al., 2007). In addition to the generation of RA, Ahd2 has also been found to have a protective role for SNc neurons by detoxifying aldehydes (Wey et al., 2012). With partial rescue of Adh2 expression in the Pitx3/Lmx1b double mutant we hypothesized that this might also rescue part of the TH+ cells that are lost in full Pitx3 mutants in adults. We therefore analyzed the amount of TH+ and TH+AHD2+ cells in the adult midbrain of Pitx3 mutants and Pitx3CreCre/Lmx1b L/L animals by performing immunohistochemistry for TH (green) and AHD2 (red; Figure 11A). More TH+ cells could be observed in the lateral tier of the SNc (Figure 11A, arrows) and in the VTA (Figure 11A, arrowheads). Quantification showed an ~16% increase in TH+ cells (n = 3, **p < 0.01, one-tailed) and an upward trend for the amount of TH+AHD2+ cells (n = 3; p = 0.12, one-tailed) in Pitx3/Lmx1b double mutant midbrains compared to Pitx3 mutants (Figure 11B; Supplementary Table S1). To summarize our data, by conditionally removing Lmx1b in a Pitx3 mutant background a recovery of Ahd2 expression is observed at E14.5 and an increase in TH+ cells can be detected in the adult midbrain of Pitx3CreCre; Lmx1b L/L animals compared to Pitx3 mutants alone.
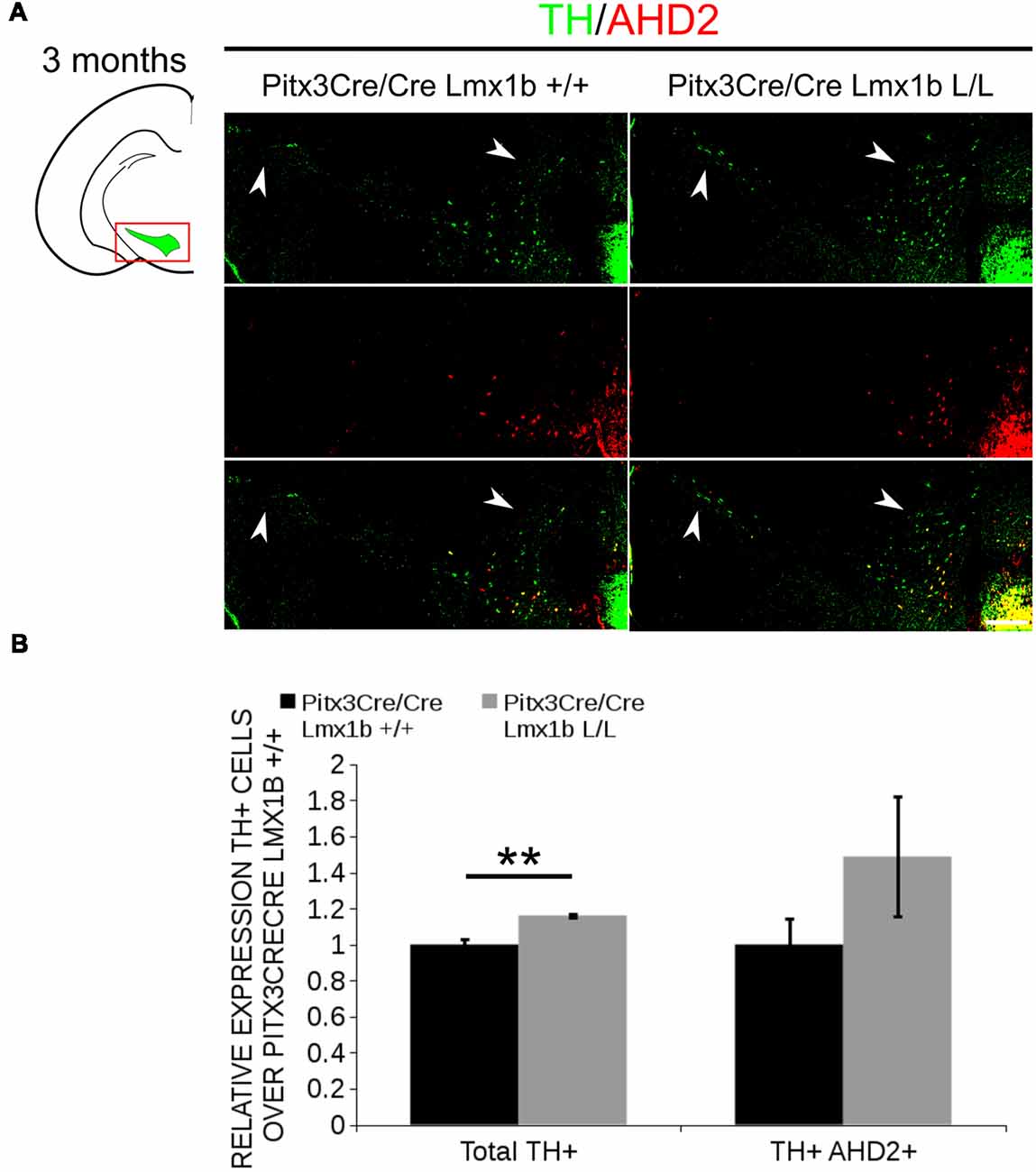
Figure 11. Additional Lmx1b deletion partially rescues the cell loss observed in Pitx3 mutants alone. TH (green) and AHD2 (red) expression was assessed using immunohistochemistry in coronal midbrain sections of 3-month-old animals. (A) More TH+ cells were observed in lateral tier of the SNc (white arrows) and in the VTA (white arrowhead). The white dotted line represents the border between what is considered SNc and VTA. (B) Quantification of the total amount of TH+ cells shows an increase in TH+ cells of ~16% (n = 3, **P < 0.01, one-tailed). The number of TH+AHD2+ cells are not significant increased, but shows an upward trend (n = 3, P = 0.12, one-tailed). Pitx3CreCre; Lmx1b +/+ was set at 1. Scale bar = 100 μM.
Discussion
Lmx1b has been associated with several developmental processes (Chen et al., 1998; Adams et al., 2000; Smidt et al., 2000; Guo et al., 2007; Deng et al., 2011; Yan et al., 2011). During early development Lmx1b is a critical component of the positive feedback loop that maintains genes associated with IsO functioning. When studying Lmx1b null mutants, it was demonstrated that Wnt1, Fgf8, En1 and En2 all require Lmx1b to maintain their expression at the IsO and in mdDA progenitors (Adams et al., 2000; Guo et al., 2007; Anderegg et al., 2013; Sherf et al., 2015). The loss of Lmx1b initially seems to predominantly affect the lateral positioned mdDA progenitor pool, however the medial located progenitors are arrested in their development as they fail to co-express Th and Pitx3 and are lost during later stages (Smidt et al., 2000; Deng et al., 2011). Next to a developmental role, a recent study also linked Lmx1b to cellular homeostasis of mdDA neurons in adult mice (Laguna et al., 2015). By using a Dat driven Cre, Lmx1b was removed in postmitotic mdDA neurons, which led to dysregulation of the autophagic-lysosal pathway, abnormal synaptic responses and reduced protein levels of striatal TH and DAT expression, all processes that contribute to a Parkinson’s disease pathology (Laguna et al., 2015). Here, we showed that Lmx1b has a variety of functions during postmitotic development, including subset specification and neuronal survival. In line with its function in early development, we found that a loss of Lmx1b in postmitotic neurons leads to reduced mRNA levels of En1 and En2 at E14.5. Both of these factors have be associated with the survival of mdDA neurons (Simon et al., 2001; Albéri et al., 2004) and En1 ± animals show a progressive loss of TH+ cells in the ventral midbrain from 3 weeks of age onward (Sonnier et al., 2007). The lower mRNA levels of En1, if equally represented by a lower protein level, might contribute to the observed reduction in the amount of TH+ cells in 3-month-old Pitx3Cre/+; Lmx1b L/L animals, however in heterozygous En1 mice the SNc is mostly affected, while in our model initially the VTA demonstrates the largest percentile reduction in TH+ cells. Notably, after 1 year the defect resides in the SNc suggesting a change in timing of neuronal survival which culminates in loss of SNc neurons after 12 months, which is in line with observations done in En1 mutant animals. In addition to a role in neuronal survival we also found a role for Lmx1b in subset specification. Pitx3 driven deletion of Lmx1b led to a significant increase in the rostrolateral mark Ahd2 in vivo and in vitro. In E14.5 embryos, an extension of the Ahd2 positive domain was observed, as Ahd2 expression was found ectopically in medial sections of the ventral mesencephalon, overlapping with the Cck positive domain, and in lateral sections exceeding the Th positive domain. These findings corroborated initial observations that Lmx1b overexpression in a dopaminergic cell line (MN9D) led to the repression of Ahd2 mRNA expression. Interestingly, the shift in Ahd2 towards the more medial DA domain did not affect the expression of Cck during development. The actual increased level of Ahd2 mRNA could be attributed to a higher level per cell in addition to the ectopic presence. The transcriptional activation of Ahd2 has been demonstrated to be dependent on the expression of Pitx3 and En1 (Jacobs et al., 2007; Veenvliet et al., 2013). Pitx3 can interact with a region close to the transcriptional start site of Ahd2 and loss of expression is observed in Pitx3−/− embryos and adult midbrains (Jacobs et al., 2007). The ablation of Lmx1b did not influence the mRNA expression levels of Pitx3 itself (not shown) and also the spatial expression of Pitx3 was not affected. In addition, spatial expression of Ahd2 was partially recovered in Pitx3/Lmx1b double mutants compared to Pitx3 mutants, suggesting that the regulation of Ahd2 by Lmx1b is a PITX3-independent process. Interestingly, the Ahd2 promoter contains several conserved FLAT-elements, to which LMX1B can bind directly (Rascle et al., 2009), suggesting that LMX1B might directly influence Ahd2 expression. AHD2 is involved in the generation of retinoic acid (RA) from retinol and Jacobs et al. (2007) demonstrated that administration of RA to the maternal diet of Pitx3−/− animals led to an increase in TH+ cells during development and an increased innervation of the dorsal Striatum. Although we only observed a partial rescue of Ahd2 expression, we did observe an increase in TH+ cells in the midbrain of adult Pitx3CreCre; Lmx1b L/L animals.
Overall, our data shows that Lmx1b is important for the mRNA expression level of survival factors En1 and En2 during development and for the survival of a specific group of mdDA neurons postnatally. Next to a role in survival we found that Lmx1b functions as a transcriptional repressor of Ahd2 independent of Pitx3. This underscores the presence of developmental programs that lead to mdDA subsets and that Lmx1b is part of the complex network of interacting transcription factors that specify these subsets.
Data Availability
The datasets generated for this study can be found in GEO, GSE121120.
Ethics Statement
All animal studies were performed in accordance with local animal welfare regulations, as this project has been approved by the animal experimental committee (Dier ethische commissie, Universiteit van Amsterdam; DEC-UvA), and international guidelines.
Author Contributions
IW performed the experiments, analyzed the data and wrote the mansuscript. PL-B and EH performed the experiments. MS initiated the study, analyzed the data, wrote the manuscript, funded the study.
Funding
This work was sponsored by the NWO-ALW (Nederlandse Organisatie voor Wetenschappelijk Onderzoek-Aard en Levenswetenschappen) VICI grant (865.09.002) awarded to Prof. Dr. MS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Ralph Witzgall of the University of Regensburg (Germany) for the kind gift of the Lmx1b-floxed mouse.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2019.00062/full#supplementary-material
FIGURE S1 | Lmx1b mostly regulates genes associated with head and nervous system development. PANTHER over-representation tests show that genetic ablation of Lmx1b mainly affects genes associated with head and nervous system development at E14.5.
FIGURE S2 | Over-expression of Lmx1b reduces Ahd2 expression in MN9D cells. Ahd2 and Th mRNA levels were examined in MN9D cells over-expressing Lmx1b. A loss of 48% in Ahd2 mRNA levels was observed in cells over-expressing Lmx1b (n = 4. **P < 0.01, two-tailed), while mRNA levels of Th were not significantly altered (N = 4).
FIGURE S3 | No differences could be detected in the expression pattern of Ahd2 between Pitx3Cre/+; Lmx1b+/+ 3-month-old animals and Pitx3Cre/+; Lmx1b L/L animals. Analysis of Ahd2 in coronal adult section in the Pitx3Cre/+; Lmx1b L/L mutant via in situ hybridization. Expression of Ahd2 seems similar in Pitx3Cre/+; Lmx1b L/L animals compared to wildtype. Scale bar = 200 μM.
TABLE S1 | Absolute numbers of cells counted. (A) The actual amount of TH+ cells counted in total and the SNc and VTA in 3-month-old wildtype and Lmx1b cKO used for Figure 4. (B) The total amount of TH+cells counted in 12-month-old Pitx3Cre/+; Lmx1b +/+ and Pitx3Cre/+; Lmx1b L/L and subdivided in the SNc and VTA, used for Figure 5. (C) Actual cell numbers used for the graphs in Figure 9 in 3-month-old Lmx1b cKO and wildtypes. (D) Actual amount of cells counted in the Pitx3Cre/Cre; Lmx1b +.+ and the Pitx3Cre/Cre; Lmx1b L/L at 3 months of age and used for Figure 11.
References
Adams, K. A., Maida, J. M., Golden, J. A., and Riddle, R. D. (2000). The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development 127, 1857–1867.
Albéri, L., Sgadò, P., and Simon, H. H. (2004). Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development 131, 3229–3236. doi: 10.1242/dev.01128
Anderegg, A., Lin, H.-P., Chen, J.-A., Caronia-Brown, G., Cherepanova, N., Yun, B., et al. (2013). An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 9:e1003973. doi: 10.1371/journal.pgen.1003973
Arenas, E., Denham, M., and Villaescusa, J. C. (2015). How to make a midbrain dopaminergic neuron. Development 142, 1918–1936. doi: 10.1242/dev.097394
Björklund, A., and Dunnett, S. B. (2007). Fifty years of dopamine research. Trends Neurosci. 30, 185–187. doi: 10.1016/j.tins.2007.03.004
Braak, H., Del Tredici, K., Rüb, U., de Vos, R. A., Jansen Steur, E. N., and Braak, E. (2003). Staging of brain pathology related to sporadic Parkinson’s disease. Neurobiol. Aging 24, 197–211. doi: 10.1016/S0197-4580(02)00065-9
Bye, C. R., Thompson, L. H., and Parish, C. L. (2012). Birth dating of midbrain dopamine neurons identifies A9 enriched tissue for transplantation into Parkinsonian mice. Exp. Neurol. 236, 58–68. doi: 10.1016/j.expneurol.2012.04.002
Chen, H., Lun, Y., Ovchinnikov, D., Kokubo, H., Oberg, K. C., Pepicelli, C. V., et al. (1998). Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat. Genet. 19, 51–55. doi: 10.1038/ng0598-51
Dai, J.-X., Hu, Z.-L., Shi, M., Guo, C., and Ding, Y.-Q. (2008). Postnatal ontogeny of the transcription factor Lmx1b in the mouse central nervous system. J. Comp. Neurol. 509, 341–355. doi: 10.1002/cne.21759
Deng, Q., Andersson, E., Hedlund, E., Alekseenko, Z., Coppola, E., Panman, L., et al. (2011). Specific and integrated roles of Lmx1a, Lmx1b and Phox2a in ventral midbrain development. Development 138, 3399–3408. doi: 10.1242/dev.065482
Doucet-Beaupré, H., Gilbert, C., Profes, M. S., Chabrat, A., Pacelli, C., Giguère, N., et al. (2016). Lmx1a and Lmx1b regulate mitochondrial functions and survival of adult midbrain dopaminergic neurons. Proc. Natl. Acad. Sci. U S A 113, E4387–E4396. doi: 10.1073/pnas.1520387113
Grima, B., Lamouroux, A., Blanot, F., Biguet, N. F., and Mallet, J. (1985). Complete coding sequence of rat tyrosine hydroxylase mRNA. Proc. Natl. Acad. Sci. U S A 82, 617–621. doi: 10.1073/pnas.82.2.617
Guo, C., Qiu, H.-Y., Huang, Y., Chen, H., Yang, R.-Q., Chen, S.-D., et al. (2007). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development 134, 317–325. doi: 10.1242/dev.02745
Hökfelt, T., Skirboll, L., Rehfeld, J. F., Goldstein, M., Markey, K., and Dann, O. (1980). A subpopulation of mesencephalic dopamine neurons projecting to limbic areas contains a cholecystokinin-like peptide: evidence from immunohistochemistry combined with retrograde tracing. Neuroscience 5, 2093–2124. doi: 10.1016/0306-4522(80)90127-x
Hoekstra, E. J., von Oerthel, L., van der Heide, L. P., Kouwenhoven, W. M., Veenvliet, J. V., Wever, I., et al. (2013). Lmx1a encodes a rostral set of mesodiencephalic dopaminergic neurons marked by the Wnt/B-catenin signaling activator R-spondin 2. PLoS One 8:e74049. doi: 10.1371/journal.pone.0074049
Hwang, D.-Y., Ardayfio, P., Kang, U. J., Semina, E. V., and Kim, K.-S. (2003). Selective loss of dopaminergic neurons in the substantia nigra of Pitx3-deficient aphakia mice. Mol. Brain Res. 114, 123–131. doi: 10.1016/s0169-328x(03)00162-1
Jacobs, F. M. J., Smits, S. M., Noorlander, C. W., von Oerthel, L., van der Linden, A. J. A., Burbach, J. P. H., et al. (2007). Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development 134, 2673–2684. doi: 10.1242/dev.02865
Jacobs, F. M. J., van Erp, S., van der Linden, A. J. A., von Oerthel, L., Burbach, J. P. H., and Smidt, M. P. (2009). Pitx3 potentiates Nurr1 in dopamine neuron terminal differentiation through release of SMRT-mediated repression. Development 136, 531–540. doi: 10.1242/dev.029769
Jacobs, F. M. J., Veenvliet, J. V., Almirza, W. H., Hoekstra, E. J., von Oerthel, L., van der Linden, A. J. A., et al. (2011). Retinoic acid-dependent and -independent gene-regulatory pathways of Pitx3 in meso-diencephalic dopaminergic neurons. Development 138, 5213–5222. doi: 10.1242/dev.071704
Kouwenhoven, W. M., von Oerthel, L., and Smidt, M. P. (2017). Pitx3 and En1 determine the size and molecular programming of the dopaminergic neuronal pool. PLoS One 12:e0182421. doi: 10.1371/journal.pone.0182421
La Manno, G., Gyllborg, D., Codeluppi, S., Nishimura, K., Salto, C., Zeisel, A., et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580. doi: 10.1016/j.cell.2016.09.027
Laguna, A., Schintu, N., Nobre, A., Alvarsson, A., Volakakis, N., Jacobsen, J. K., et al. (2015). Dopaminergic control of autophagic-lysosomal function implicates Lmx1b in Parkinson’s disease. Nat. Neurosci. 18, 826–835. doi: 10.1038/nn.4004
Nakatani, T., Kumai, M., Mizuhara, E., Minaki, Y., and Ono, Y. (2010). Lmx1a and Lmx1b cooperate with Foxa2 to coordinate the specification of dopaminergic neurons and control of floor plate cell differentiation in the developing mesencephalon. Dev. Biol. 339, 101–113. doi: 10.1016/j.ydbio.2009.12.017
Nunes, I., Tovmasian, L. T., Silva, R. M., Burke, R. E., and Goff, S. P. (2003). Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc. Natl. Acad. Sci. U S A 100, 4245–4250. doi: 10.1073/pnas.0230529100
Panman, L., Papathanou, M., Laguna, A., Oosterveen, T., Volakakis, N., Acampora, D., et al. (2014). Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep. 8, 1018–1025. doi: 10.1016/j.celrep.2014.07.016
Prakash, N., and Wurst, W. (2006). Development of dopaminergic neurons in the mammalian brain. Cell. Mol. Life Sci. 63, 187–206. doi: 10.1007/s00018-005-5387-6
Rascle, A., Neumann, T., Raschta, A.-S., Neumann, A., Heining, E., Kastner, J., et al. (2009). The LIM-homeodomain transcription factor LMX1B regulates expression of NF-kappa B target genes. Exp. Cell Res. 315, 76–96. doi: 10.1016/j.yexcr.2008.10.012
Roeper, J. (2013). Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci. 36, 336–342. doi: 10.1016/j.tins.2013.03.003
Saucedo-Cardenas, O., Quintana-Hau, J. D., Le, W.-D., Smidt, M. P., Cox, J. J., Mayo, F. D., et al. (1998). Nurr1 is essential for the induction of the dopaminergic phenotype and the survival of ventral mesencephalic late dopaminergic precursor neurons. Proc. Natl. Acad. Sci. U S A 95, 4013–4018. doi: 10.1073/pnas.95.7.4013
Sherf, O., Nashelsky Zolotov, L., Liser, K., Tilleman, H., Jovanovic, V. M., Zega, K., et al. (2015). Otx2 requires Lmx1b to control the development of mesodiencephalic dopaminergic neurons. PLoS One 10:e0139697. doi: 10.1371/journal.pone.0139697
Simon, H. H., Saueressig, H., Wurst, W., Goulding, M. D., and O’Leary, D. D. M. (2001). Fate of midbrain dopaminergic neurons controlled by the engrailed genes. J. Neurosci. 21, 3126–3134. doi: 10.1523/JNEUROSCI.21-09-03126.2001
Smidt, M. P., Asbreuk, C. H. J., Cox, J. J., Chen, H., Johnson, R. L., and Burbach, J. P. H. (2000). A second independent pathway for development of mesencephalic dopaminergic neurons requires Lmx1b. Nat. Neurosci. 3, 337–341. doi: 10.1038/73902
Smidt, M. P., and Burbach, J. P. H. (2007). How to make a mesodiencephalic dopaminergic neuron. Nat. Rev. Neurosci. 8, 21–32. doi: 10.1038/nrn2039
Smidt, M. P., Smits, S. M., Bouwmeester, H., Hamers, F. P. T., van der Linden, A. J. A., Hellemons, A. J. C. G. M., et al. (2004). Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development 131, 1145–1155. doi: 10.1242/dev.01022
Smidt, M. P., van Schaick, H. S. A., Lanctôt, C., Tremblay, J. J., Cox, J. J., van der Kleij, A. A. M., et al. (1997). A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc. Natl. Acad. Sci. U S A 94, 13305–13310. doi: 10.1073/pnas.94.24.13305
Smidt, M. P., von Oerthel, L., Hoekstra, E. J., Schellevis, R. D., and Hoekman, M. F. M. (2012). Spatial and temporal lineage analysis of a Pitx3-driven cre-recombinase knock-in mouse model. PLoS One 7:e42641. doi: 10.1371/journal.pone.0042641
Smits, S. M., Burbach, J. P. H., and Smidt, M. P. (2006). Developmental origin and fate of meso-diencephalic dopamine neurons. Prog. Neurobiol. 78, 1–16. doi: 10.1016/j.pneurobio.2005.12.003
Smits, S. M., Ponnio, T., Conneely, O. M., Burbach, J. P. H., and Smidt, M. P. (2003). Involvement of Nurr1 in specifying the neurotransmitter identity of ventral midbrain dopaminergic neurons. Eur. J. Neurosci. 18, 1731–1738. doi: 10.1046/j.1460-9568.2003.02885.x
Smits, S. M., von Oerthel, L., Hoekstra, E. J., Burbach, J. P. H., and Smidt, M. P. (2013). Molecular marker differences relate to developmental position and subsets of mesodiencephalic dopaminergic neurons. PLoS One 8:e76037. doi: 10.1371/journal.pone.0076037
Sonnier, L., Le Pen, G., Hartmann, A., Bizot, J.-C., Trovero, F., Krebs, M.-O., et al. (2007). Progressive loss of dopaminergic neurons in the ventral midbrain of adult mice heterozygote for engrailed1. J. Neurosci. 27, 1063–1071. doi: 10.1523/JNEUROSCI.4583-06.2007
Suleiman, H., Heudobler, D., Raschta, A.-S., Zhao, Y., Zhao, Q., Hertting, I., et al. (2007). The podocyte-specific inactivation of Lmx1b, Ldb1 and E2a yields new insight into a transcriptional network in podocytes. Dev. Biol. 304, 701–712. doi: 10.1016/j.ydbio.2007.01.020
Tiklová, K., Björklund, Å. K., Lahti, L., Fiorenzano, A., Nolbrant, S., Gillberg, L., et al. (2019). Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat. Commun. 10:581. doi: 10.1038/s41467-019-08453-1
van den Munckhof, P., Luk, K. C., Ste-Marie, L., Montgomery, J., Blanchet, P. J., Sadikot, A. F., et al. (2003). Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development 130, 2535–2542. doi: 10.1242/dev.00464
Veenvliet, J. V., and Smidt, M. P. (2014). Molecular mechanisms of dopaminergic subset specification: fundamental aspects and clinical perspectives. Cell. Mol. Life Sci. 71, 4703–4727. doi: 10.1007/s00018-014-1681-5
Veenvliet, J. V., Dos Santos, M. T. M. A., Kouwenhoven, W. M., von Oerthel, L., Lim, J. L., van der Linden, A. J. A., et al. (2013). Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 140, 4116–4116. doi: 10.1242/dev.094565
Wey, M. C.-Y., Fernandez, E., Martinez, P. A., Sullivan, P., Goldstein, D. S., and Strong, R. (2012). Neurodegeneration and motor dysfunction in mice lacking cytosolic and mitochondrial aldehyde dehydrogenases: implications for Parkinson’s disease. PLoS One 7:e31522. doi: 10.1371/journal.pone.0031522
Yan, C. H., Levesque, M., Claxton, S., Johnson, R. L., and Ang, S.-L. (2011). Lmx1a and Lmx1b function cooperatively to regulate proliferation, specification, and differentiation of midbrain dopaminergic progenitors. J. Neurosci. 31, 12413–12425. doi: 10.1523/JNEUROSCI.1077-11.2011
Keywords: dopamine, development, transcription, LMX1B, substantia nigra
Citation: Wever I, Largo-Barrientos P, Hoekstra EJ and Smidt MP (2019) Lmx1b Influences Correct Post-mitotic Coding of Mesodiencephalic Dopaminergic Neurons. Front. Mol. Neurosci. 12:62. doi: 10.3389/fnmol.2019.00062
Received: 04 December 2018; Accepted: 25 February 2019;
Published: 14 March 2019.
Edited by:
Ildikó Rácz, Universitätsklinikum Bonn, GermanyReviewed by:
Sandra Blaess, University of Bonn, GermanyJerome Mertens, Salk Institute for Biological Studies, United States
Larissa Traxler, University of Innsbruck, Innsbruck, Austria, in collaboration with reviewer JM
Copyright © 2019 Wever, Largo-Barrientos, Hoekstra and Smidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marten P. Smidt, m.p.smidt@uva.nl
 Iris Wever
Iris Wever Pablo Largo-Barrientos
Pablo Largo-Barrientos Elisa J. Hoekstra
Elisa J. Hoekstra  Marten P. Smidt
Marten P. Smidt