Glial Derived TGF-β Instructs Axon Midline Stopping
- Department of Molecular Cell Biology, Weizmann Institute of Sciences, Rehovot, Israel
A fundamental question that underlies the proper wiring and function of the nervous system is how axon extension stops during development. However, our mechanistic understanding of axon stopping is currently poor. The stereotypic development of the Drosophila mushroom body (MB) provides a unique system in which three types of anatomically distinct neurons (γ, α’/β’, and α/β) develop and interact to form a complex neuronal structure. All three neuronal types innervate the ipsi-lateral side and do not cross the midline. Here we find that Plum, an immunoglobulin (Ig) superfamily protein that we have previously shown to function as a TGF-β accessory receptor, is required within MB α/β neurons for their midline stopping. Overexpression of Plum within MB neurons is sufficient to induce retraction of α/β axons. As expected, rescue experiments revealed that Plum likely functions in α/β neurons and mediates midline stopping via the downstream effector RhoGEF2. Finally, we have identified glial-derived Myoglianin (Myo) as the major TGF-β ligand that instructs midline stopping of MB neurons. Taken together, our study strongly suggests that TGF-β signals originating from the midline facilitate midline stopping of α/β neuron in a Plum dependent manner.
Introduction
During development of the nervous system, individual neurons send axons and dendrites to form intricate neuronal circuitry whose precise connectivity underlies the proper function of the brain. Roger Sperry, who pioneered the inception of the chemoaffinity hypothesis (Sperry, 1963), postulated that the proper development of neuronal connectivity is orchestrated by a complex system of molecular cues. This hypothesis has since been solidified as a wide variety of neuronal guidance molecules have been identified (Tessier-Lavigne and Goodman, 1996; Dickson, 2002; Kolodkin and Tessier-Lavigne, 2011). These include molecules that can act from a distance and those that act only in close contact. While most of the molecules, such as slits and netrins, were initially identified as either being attractive or repulsive, it is becoming clear today that most if not all molecules can have both attractive and repulsive roles, depending on the context and identity of the interacting receptors (Kolodkin and Tessier-Lavigne, 2011; Kaplan et al., 2014). However, while there is a growing understanding of the mechanisms that promote axon growth and guidance, much less is known about the mechanisms that induce axon stopping.
The Drosophila mushroom body (MB) is a bilaterally symmetrical central nervous system (CNS) neuropil structure that is mostly involved in associative learning and memory (reviewed in Heisenberg, 2003; Aso et al., 2014). It is comprised of three types of intrinsic neurons (γ, α’/β’, and α/β), sequentially born from four identical neuroblasts per hemisphere (Crittenden et al., 1998; Lee et al., 1999). Each neuron type exhibits a unique morphology, connectivity and function (Trannoy et al., 2011; Aso et al., 2014). Interestingly, all three MB neuronal types are confined to one hemisphere and do not cross the midline. Therefore, the stereotypic anatomical morphology of specific MB neurons and its relative simplicity offer a unique model to study axon guidance, branching and stopping.
Several studies have reported of mutations in which α/β neurons fail to stop at the midline and therefore cross to the contralateral hemisphere. This phenotype has been attributed to mutants in both intrinsic and extrinsic cues (Moreau-Fauvarque et al., 1998; Michel et al., 2004; Kobayashi et al., 2006; Grillenzoni et al., 2007; Kurusu and Zinn, 2008). Despite these findings, the cellular and molecular mechanisms responsible for axonal stopping remain mostly elusive. One of the best-documented examples in which α/β-axons fail to stop at the midline is in TGF-β type I receptor Baboon (babo) mutants. Ng (2008) has used the mosaic analysis with a repressible cell marker (MARCM) technique to show that MB neuroblast clones mutant for babo exhibit β-axon midline crossing. Additionally, he showed that in this context babo functions in a non-canonical fashion, signaling via LIMK rather than Smads. However, the identity of the TGF-β ligand, its source, and the precise cellular mechanisms are still not known.
In previous work we showed that Plum, an immunoglobulin super family (IgSF) trans-membrane protein, is required to promote pruning of MB γ-neurons, most likely functioning as a TGF-β accessory receptor (Yu et al., 2013). Here, we explore the role of Plum in the midline stopping of α/β neurons. Interestingly, we uncovered that glial-derived TGF-β ligands instruct midline stopping of MB α/β-axons.
Materials and Methods
Drosophila Strains
The plumΔ1 allele and the following Plum transgenes: UAS-Plum::Flag, UAS-PlumΔcyt::Flag, UAS-PlumΔFNIII(1-5)::Flag, UAS-PlumΔIG1-4::Flag, UAS-PlumΔIg1-3::Flag, UAS-PlumΔIg4::Flag, UAS-PlumECD::Flag and UAS-PlumECD::TM-CD8::Flag were previously described (Yu et al., 2013). G0050-Gal4 was kindly provided by A.S. Chiang. UAS-Myo RNAi (31200 and 31114), GMR71G10-Gal4, GMR44E04-Gal4 (originally made as part of the FlyLight project at the Janelia farm, Rubin lab), c305a-Gal4, c155-Gal4, Repo-Gal4 and UAS-Plum RNAi (TRiP.HMC05055, originally made by DRSC) were obtained from the Bloomington Drosophila Stock Center (BDSC).
Construction of plumV11X CRISPR Mutant Flies
Two guide RNA sequences were cloned into pCFD4 plasmid using Transfer-PCR as described previously (Port et al., 2014; Meltzer et al., 2019). The pCFD4Plum plasmid was injected into attP86Fb landing site using ϕC31 integration (BestGene). Injected flies were crossed with nanos-Cas9 flies (Bloomington stock #54591). After two generations, single males were crossed with balancers and checked for deletions/indels using specific primers. gRNA plum specific sequences (5′-3′):
Forward: CAATCAATTGAATCACAAAG.
Reverse: TCACCCATTGGGATCCACCT.
Genotypes Abbreviations
Full genotypes are detailed in the figure legends and are abbreviated as follows: hsFlp is y, w, hsFlp122; CD8 is UAS-mCD8::GFP; UAS-PlumFL is UAS-Plum::Flag; UAS-PlumΔcyt is UAS-PlumΔcyt::Flag; UAS-PlumΔFNIII(1-5) is UAS-PlumΔFNIII(1-5)::Flag; UAS-PlumΔIG1-4 is UAS-PlumΔIG1-4::Flag; UAS-PlumΔIg1-3 is UAS-PlumΔIg1-3::Flag; UAS-PlumΔIg4 is UAS-PlumΔIg4::Flag; UAS-PlumECD is UAS-PlumECD::Flag; UAS-PlumECD::TM-CD8 is UAS-PlumECD::TM-CD8::Flag; plumΔ1 is 82B, plumΔ1; plumV11X is 82B, plumV11X; 40A, and 82B are FRTs on 2L and 3R, respectively; 201Y is 201Y-Gal4; OK107 is OK107-Gal4; R71G10 is GMR71G10-Gal4; R44E04 is GMR44E04-Gal4; Repo is Repo-Gal4; c155 is c155-Gal4; TIFR is TIFR-Gal4; G0050 is G0050-Gal4; c305a is c305a-Gal4; Gal80 is TubP-Gal80; UAS-plumRNAi is TRiP.HMC05055 from DRSC/TRiP Functional Genomics Resources; UAS-MyoRNAi (31200 or 31114) are either TRiP.JF01717 or TRiP.JF01587, respectively, from DRSC/TRiP Functional Genomics Resources.
MARCM and Antibody Conditions
MB MARCM neuroblast clones were generated by heat shocking newly hatched larvae and examined as described previously (Lee et al., 1999).
Antibody staining conditions were as follows: mouse monoclonal anti-FasII (ID4; 1:25); anti-Trio (9.4A; 1:100), both from the Hybridoma bank (DSHB); chicken polyclonal anti GFP (1:500; Aves); rat polyclonal anti mCD8 (1:200; Invotrogen MCD0800). FITC (ENCO), Alexa488 or Alexa647 (Invitrogen) conjugated secondary antibodies were used at 1:300. Brains were mounted in Slowfade (Invitrogen) and imaged on LSM710 or LSM800 confocal microscopes (Zeiss).
Ranking and Statistical Analyses
A single scorer performed ranking, in a double-blind manner. The ranking was performed by reviewing the entire z-stack. Ranking range was set as follows: WT = 0, β-lobes almost touching each other but not crossing the midline = 1 and β-lobes crossing the midline with increasing severity = 2–5. In Figures 1–3, five statistical significance was calculated by performing non-parametric Kruskal Wallis analysis of variance (ANOVA) test followed by, if necessary, post hoc pairwise Wilcoxon test. In Figure 6, statistical significance was calculated by performing one-tailed Wilcox test, and the p-values were adjusted by FDR correction.
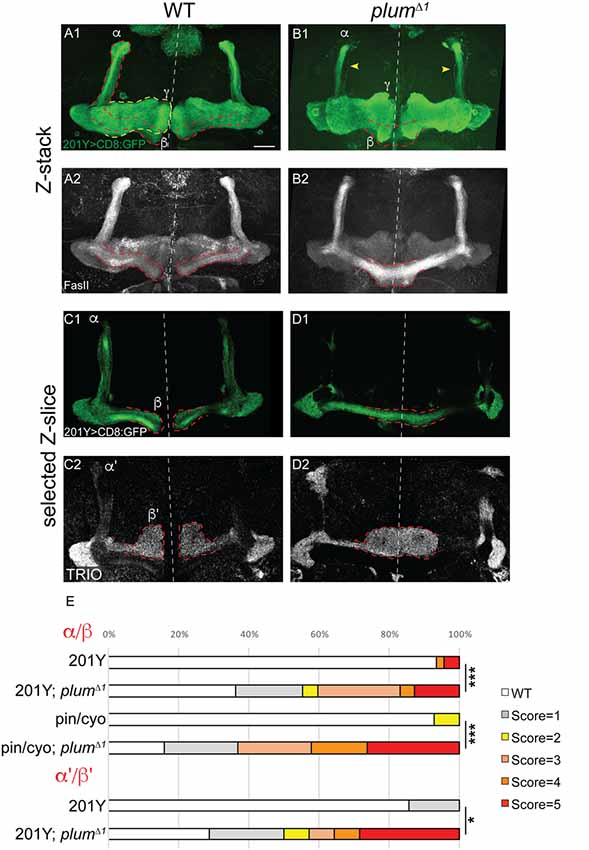
Figure 1. Mushroom body (MB) α/β and α’/β’ neurons over-extend beyond the midline in plumΔ1 homozygous brains. (A–D) Confocal Z-projections of WT (A,C) or plumΔ1 (B,D) brains expressing CD8::GFP driven by the 201Y-Gal4. mCD8::GFP (green, A1–D1); FasII (A2,B2, gray); TRIO (C2,D2, gray); Scale bars, 20 μm. (A,C) While in a WT brain the β-lobes (A) or β’-lobes (C) from both hemispheres terminate in a fair distance from each other and do not cross the midline, (B,D) the β- (B) or β’-axons (D) often cross the midline and sometime seem to fuse in plumΔ1 brains. Dashed lines demarcate the extent of the β or β’-lobes, yellow arrows in (B) indicate unpruned γ neurons. (E) Quantification of midline crossing phenotypes in (A–D; 5 being the most severe), as well as additional genotypes as mentioned below (for more information, see Supplementary Figure S1 and the “Materials and Methods” section). Genotypes: (A) 201Y, CD8/+ (n = 45). (B) 201Y, CD8/+; 82B, plumΔ1 (n = 42) p-value: p < 0.001. (C) 201Y, CD8/+; 82B, plumΔ1 (n = 14). (D) 201Y, CD8/+ (n = 7) p-value: p = 0.012. (Additionally, quantified in E): Pin/CyO (n = 14) and Pin/CyO; 82B, PlumΔ1 (n = 19), p-value: p < 0.001.
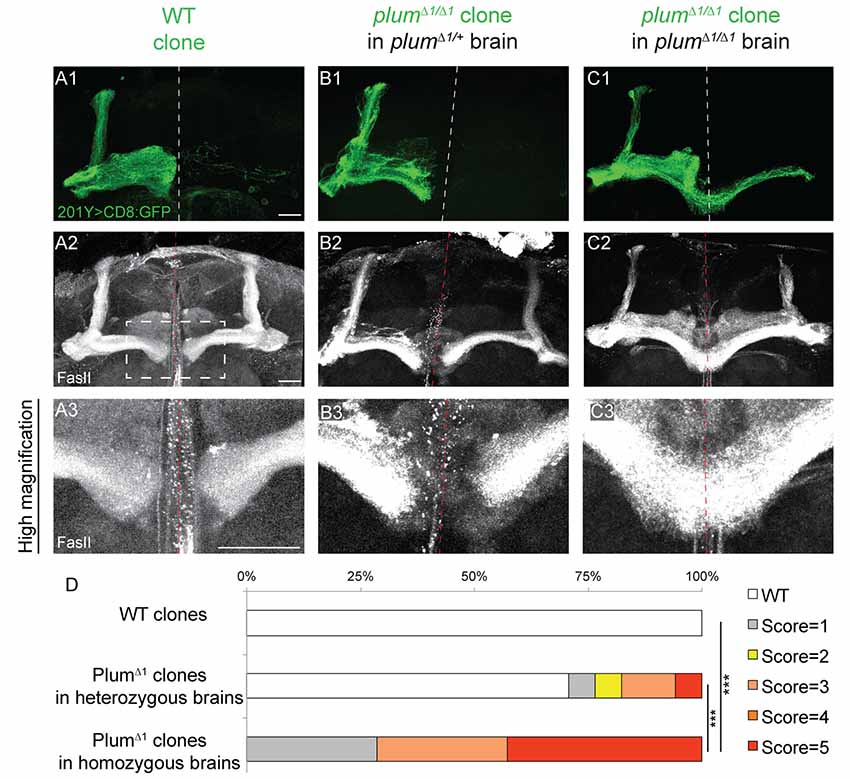
Figure 2. Plum is both autonomously and non-autonomously required for β-lobe midline stopping. (A–C) Confocal Z-projections of (A) WT mosaic analysis with a repressible cell marker (MARCM) neuroblast clone, (B) plumΔ1 MARCM clone in a plumΔ1/+ heterozygous brain or (C) plumΔ1 MARCM clone in a plumΔ1 homozygous mutant brains, labeled with 201Y-Gal4. CD8::GFP (green, A1–C1); FasII (gray, A2–C3). Scale bars, 20 μm. (D) Quantification of midline crossing phenotypes in (A–C). p-values compared to plumΔ1 clone in plumΔ1 brain—WT clones: ***p < 0.001. plumΔ1 clone in heterozygous brain: p = 0.0074. Genotypes: (A) hsFlp, CD8/Y or +; 201Y, CD8/+; 82B, Gal80/82B (n = 10). (B) hsFlp, CD8/Y or +; 201Y, CD8/+; 82B, Gal80/82B, plumΔ1 (n = 17). (C) hsFlp, CD8/Y or +; Gal80, 40A, 201Y, CD8/40A; 82B, plumΔ1 (n = 7).
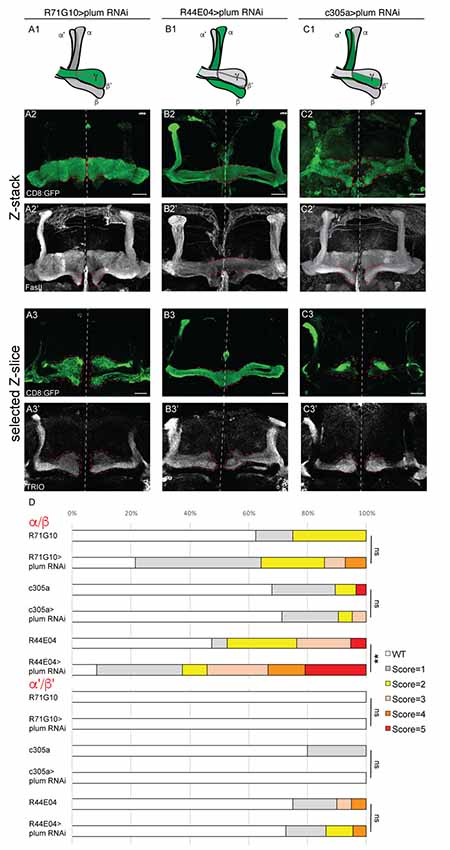
Figure 3. Plum is required in MB α/β neurons to initiate β-lobe midline stopping. (A–C) Confocal Z-projections of brains expressing PlumRNAi specifically in γ neurons using the R71G10 driver (A), in α/β neurons using the R44E04 driver (B) or in α’/β’ neurons using the c305a driver (C). Top panel in each column represents a scheme of MB-axons, where the expression of each specific driver is illustrated in green. (D) Quantifications of the midline crossing phenotypes in (A–C and Supplementary Figure S4). p-values compared to controls in Supplementary Figure S4: in the case of R44E04 driven RNAi: **p = 0.003; ns=not significant. CD8::GFP (green, A2–C3); FasII (A2’–C2’, gray); TRIO (A3’–C3’, gray); Scale bars, 20 μm. Genotypes: (A) CD8/UAS-plumRNAi; R71/+ (A2: n = 14, A3: n = 18). (B) CD8/UAS-plumRNAi; R44/+ (B2: n = 24, B3: n = 22). (C) c305/UAS-plumRNAi; CD8/+ (C2: n = 21, C3: n = 8). Additionally, quantified in (D): CD8; R71G10 (n = 16, 16, respectively). CD8; R44E04 (n = 38, 20, respectively). c305a; CD8 (n = 28, 20, respectively).
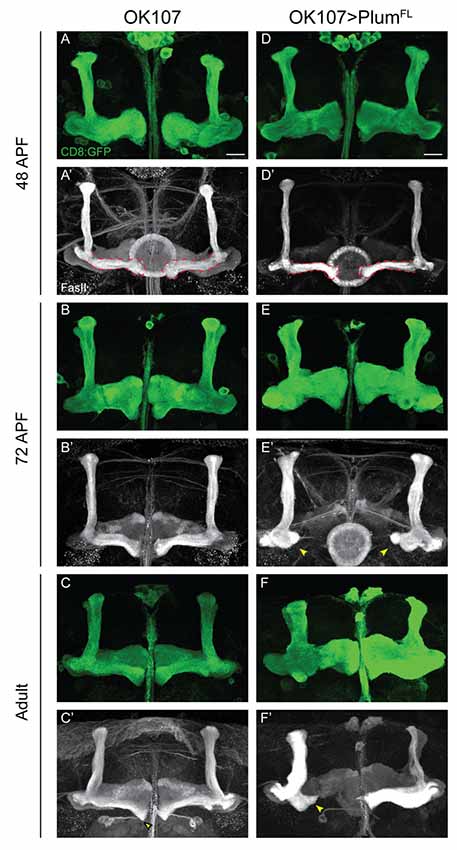
Figure 4. Plum over-expression within MB neurons is sufficient to induce β-lobe retraction. (A–F) Confocal Z-projections of (A–C) WT brains expressing CD8::GFP and (D–F) WT brains expressing CD8::GFP as well as PlumFL driven by OK107-Gal4. Arrowheads in (E,F) depict aberrant growth of β-lobes. CD8::GFP (green, A–F); FasII (gray, A’–F’). Scale bars, 20 μm. Genotypes: (A–C) CD8/+; OK107 (n = 5, 5, 18, respectively). (D–F) UAS-PlumFL/CD8;; OK107 (n = 7, 15, 22, respectively).
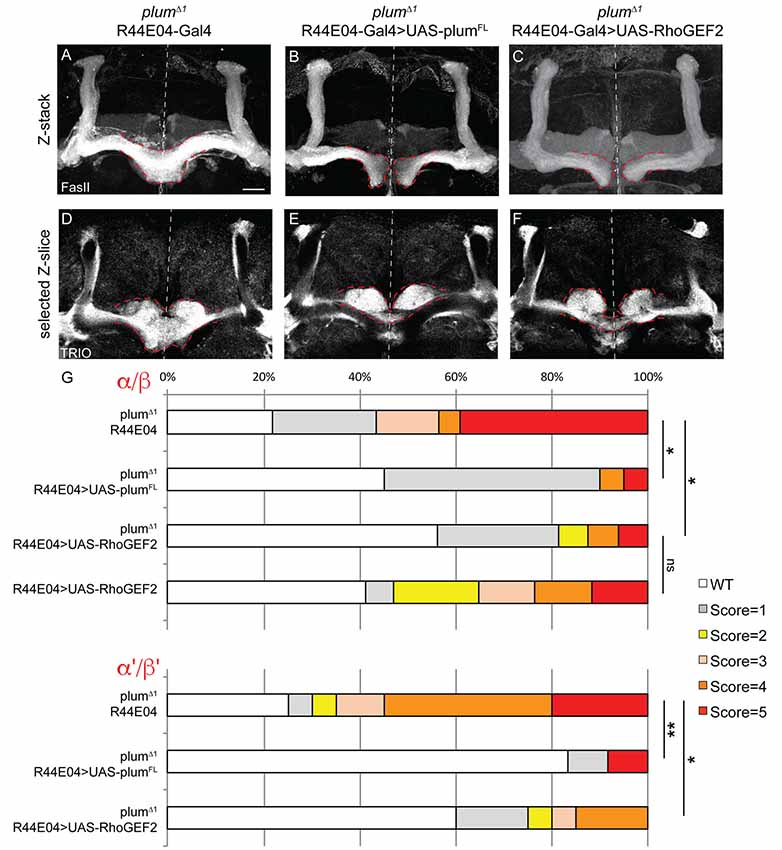
Figure 5. Plum acts through RhoGEF2 in α/β neurons to facilitate β-axon midline stopping. (A–F) Confocal Z-projections or selected Z-slices of adult (A,D) plumΔ1 brains expressing CD8::GFP driven by the R44-Gal4 driver, or brains additionally expressing (B,E) UAS-PlumFL or (C,F) UAS-RhoGEF2. (G) Quantification of β-lobe midline crossing phenotype in (A–F). p-values compared to plumΔ1 R44E04: plumΔ1 R44E04>PlumFL, *p = 0.038, **p=0.0098, for α/β and α’/β’, respectively. plumΔ1 R44E04>RhoGEF2, *p = 0.049, *p = 0.0123 for α/β and α’/β’, respectively. Dashed red lines demarcate the extent of the β-lobes. FasII (in A–C, gray); TRIO (in D–F, gray). Scale bars, 20 μm. Genotypes: (A,D) CD8::GFP/+; R44-Gal4, PlumΔ1/PlumΔ1 (n = 23, 39, respectively), (B,E) UAS-PlumFL/+; R44-Gal4, PlumΔ1/PlumΔ1(n = 20, 27, respectively), (C,F) UAS-RhoGEF2/+; R44-Gal4, PlumΔ1/PlumΔ1 (n = 16, 16, respectively).
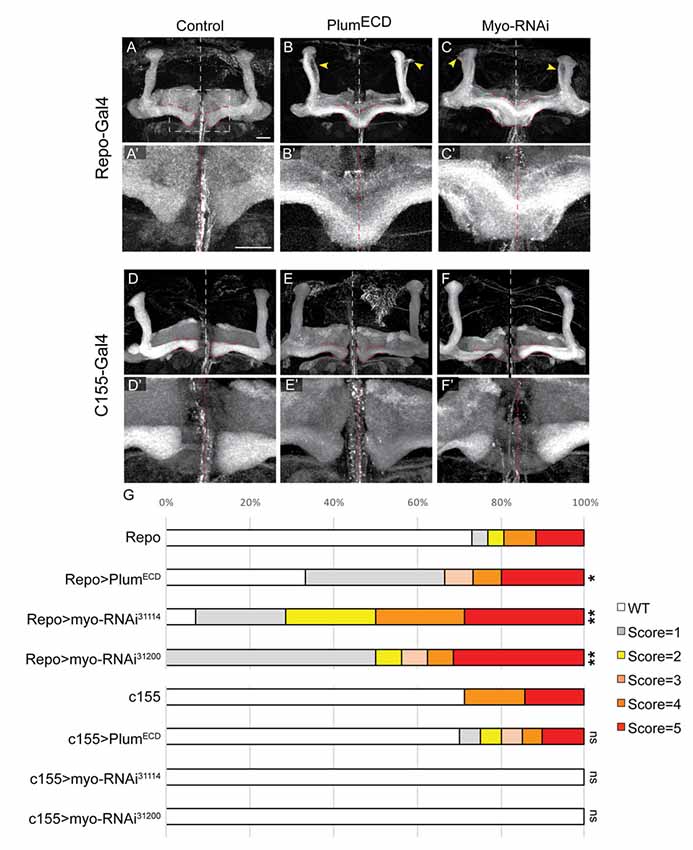
Figure 6. Glial-derived TGF-β ligand Myo is required for β-lobe midline stopping. (A–F) Confocal Z-projections of adult brains expressing (A) Repo-Gal4 as control or those expressing (B) PlumECD driven by Repo-Gal4 or (C) MyoRNAi-31114 driven by Repo-Gal4 or (D) those expressing c155-Gal4 as control or (E) PlumECD driven by c155-Gal4 or (C) MyoRNAi-31114 driven by c155-Gal4. Yellow arrows in (B,C) indicate unpruned γ neurons. Panels (A’–F’) are high magnification images of (A–F) FasII (gray). Scale bars, 20 μm. (G) Quantification of the β-lobes midline crossing in experiments. p-values compared to Repo: Repo-Gal4>PlumECD, *p = 0.037; Repo-Gal4>Myo RNAi31114, **p = 0.0018; Myo RNAi31200, **p = 0.0012; p-values compared to c155: ns = not significant. By lanes from top to bottom in (E): (1) Repo/+ (n = 26); (2) UAS-PlumECD/Repo (n = 15); (3) Repo/UAS-MyoRNAi-31114 (n = 11); (4) Repo/UAS-MyoRNAi-31200 (n = 18); (5) c155 (n = 7); (6) c155(y or +); UAS-PlumECD/+ (n = 20); (7) c155/(y or +); UAS-MyoRNAi-31114/+ (n = 10) and (8) C155/y or +; UAS-MyoRNAi-31200/+ (n = 9).
Results
Plum Is Required for Normal MB β-Lobe Development
We have previously focused on the role of plum during the development of MB γ neurons and have shown that it is cell-autonomously required for axon pruning (Yu et al., 2013). While analyzing plumΔ1 homozygous mutant brains, we noticed that another type of MB neurons, the late-born α/β neurons, were also defective. Specifically, when plum was homozygous mutant, we found that β-axons over-extended beyond the midline in 47% of the brains and often the β-lobes from both hemispheres seemed to fuse (17%; Figures 1A,B; see Supplementary Figure S1A and “Materials and Methods” section for ranking and statistics). This morphology was in contrast to WT brains where midline crossing was observed in only 7% of the brains while in the vast majority of the brains the β-lobe normally arrested at a considerable distance from the midline (compare Figure 1B with 1A, quantified in 1E). This low incidence of β-lobe midline crossing in WT brains is normal and consistent with previously described occasional wild-type β-lobe axon midline crossing (Moreau-Fauvarque et al., 1998; Michel et al., 2004).
The β-lobe midline crossing phenotype was even more pronounced when we looked at antibody staining for the FasII adhesion molecule (compare Figure 1B1 with 1B2). The difference in phenotype expressivity when looking at GFP vs. FasII expression might stem from the fact that FasII is highly expressed endogenously in most if not all β-axons while the expression of GFP driven by the 201Y-Gal4 driver is weaker and is limited to core β-lobe axons (Crittenden et al., 1998; Kurusu et al., 2002). Since plumΔ1 is a tailored deficiency that encompasses both plum as well as CG5455 (Yu et al., 2013), we wanted to validate that the β-lobe midline crossing phenotype we observed is indeed caused by plum loss-of-function. Therefore, we generated a CRISPR/Cas9 mediated indel, plumV11X, a deletion of 10 bp resulting in a premature stop codon after 10 amino acids (Supplementary Figure S1B). A MARCM analysis of plumV11X revealed that clones of MB γ neurons demonstrate a severe pruning defect (Supplementary Figures S1C,D), similar to plumΔ1 mutant clones (Yu et al., 2013). Indeed, in plumV11X homozygous mutant brains, β-axons cross the midline in similar fashion as in plumΔ1 (Supplementary Figures S1E,F, quantified in Supplementary Figure S1G).
Plum is therefore required for midline stopping of α/β but not γ MB neurons. We next questioned whether Plum is required for the midline stopping of α’/β’ neurons. To test that, we stained plumΔ1 homozygous brains with TRIO antibody, which labels both γ and α’/β’ neurons, and were surprised to find that α’/β’ neurons also cross the midline in plumΔ1 brains (Figures 1C,D, quantified in 1E). These results suggest that in addition to its role in γ neuron pruning, Plum is required for normal midline stopping of β and β’-axons.
Plum Is Required Within MB Neurons for β-Lobe Midline Stopping
We next questioned if Plum functions cell-autonomously, similar to its role during pruning. We, therefore, compared the morphology of plumΔ1 MB MARCM clones within plumΔ1 heterozygous or homozygous mutant brains (Figures 2B,C, respectively). Seventy percentage of β-axons in plumΔ1 mutant clones crossed the midline in homozygous brains, while only 20% crossed the midline in heterozygous brains and 0% control β-axons crossed the midline (Figures 2A,D for quantification).
The genotype of the mutant α/β MARCM clone in these experiments was identical with the only difference being the genotype of the surrounding cells. One possible interpretation of the difference between the phenotypes of plum mutant clone in heterozygous vs. homozygous brains (20% compared with 70%, respectively), could potentially be explained by protein perdurance. The clonal cells generated within plumΔ1 heterozygous brain, did express Plum before the FRT-mediated recombination occurred and therefore some plum mRNA and protein might still be present in low quantities. We think this interpretation is less likely due to the many cell divisions occurring in a neuroblast clone, resulting in constant dilution of Plum’s RNA and protein. Therefore, the most plausible interpretation suggests a non-autonomous role for Plum in the non-clonal cells, either in other α/β MB neurons, or in other MB or non-MB cells. Taken together, although Plum acts cell-autonomously to instruct pruning of MB-γ neurons (Yu et al., 2013), our results suggest that Plum midline stopping function also reflects a non-cell autonomous activity.
To investigate if the non-cell autonomous function of Plum is required in MB neurons, we knocked down plum expression specifically within MB neurons by expressing plum RNAi using a strong, pan-MB driver, OK107-Gal4 (Supplementary Figures S2A–D). Indeed, knocking down plum in all MB neurons resulted in 60% of the brains exhibiting severe β-lobe midline crossing (Supplementary Figures S2B,B’ compared to S2A,A’, quantified in S2E), and 45% of the brains exhibiting severe β-lobe midline crossing (Supplementary Figures S2D,D’ compared to S2C,C’, quantified in S2E), indicating that Plum expression is required within the MB for proper α/β or α’/β’ neuronal development.
Plum Expression in α/β Neurons Is Required for β Lobe Midline Stopping
To specifically determine in which type of MB neurons Plum is required to induce midline stopping, we specifically knocked down plum in γ, α/β or α’/β’ neurons (Figure 3, see developmental expression pattern of α’/β’ and α/β-specific drivers in Supplementary Figures S3A–F). Knocking down plum in γ neurons (using R71G10-Gal4) did not affect midline stopping of either γ, β or β’-axons (Figure 3A quantified in 3D, compare to control in Supplementary Figure S4A). In contrast, knocking down plum only in α/β neurons (using the R44E04-Gal4) resulted in severe β lobe midline crossing in 30% of the brains (Figure 3B, quantified in 3D, compare to control in Supplementary Figure S4B), while the β’ lobes were unaffected and stopped at the midline (Figure 3B3’). Although the R44E04-Gal4 driver is not thought to be expressed in γ neurons, we also observed what seems to be a pruning defect when plum was knocked down using this driver (Figures 3B2’,B3’), perhaps due to a very low expression level of R44E04 also in γ neurons. Because α’/β’ neurons also cross the midline in plumΔ1 animals (Figure 1D), we expected Plum to have a role in their midline stopping as well. However, to our surprise, knocking down plum specifically in α’/β’ neurons (using both c305a-Gal4; Figure 3C, Supplementary Figure S4C; and G0050-Gal4; data not shown) did not induce midline crossing of β or β’-axons (Figures 3C2’,3C3’, respectively, quantified in 3D), however we cannot rule out the possibility that the α’/β’ drivers are either too weak or expressed only in a subset of α’/β’ neurons. In summary, our results indicate that Plum is important in α/β for their own midline stopping and also contribute to α’/β’ crossing to some degree.
Plum Over-Expression in MB Neurons Is Sufficient to Induce β-Lobe Retraction
Our results show that Plum is required within MB neurons to induce midline stopping of β-axons. We, therefore, hypothesized that Plum over-expression within MB neurons could induce a premature stop signal and result in short α/β-lobes. Indeed, we found that over-expressing Plum by the strong, pan-MB driver OK107-Gal4, resulted in shorter β but not α or γ lobes (Figure 4F compare to 4C). We were intrigued to find, however, that β’ lobes length was not affected by over-expression of Plum, even in brains in which the β lobe has been retracted (Supplementary Figure S5). Because two branches of the same neuron form the α and β lobes, this result implies that the length of α and β branches of the α/β neurons is determined independently.
The manifestation of a short β-lobe in adult brains could be a result of at least two different scenarios. Either the growth of the β-lobe is impaired, leading to premature axon stopping as suggested by other studies focusing on β-lobe length (Ng, 2008; Abe et al., 2014), or the β-lobe could initially form normally but later undergo shortening either by axon retraction or fragmentation. To differentiate between these possibilities, we performed a developmental time course study of pupal brains over-expressing Plum driven by OK107-Gal4. To our surprise, we found that at 48 h after pupae formation (APF) the β-lobe seemed to develop normally and axons extend up to the midline (Figure 4D compare with 4A). However, as development proceeded, the β-lobe appeared progressively shorter at 60 h APF (data not shown) and even more so at 72 h APF (Figure 4E compare with 4B). As we did not observe axon fragments at any of the developmental time points, we conclude that β-lobe shortening by ectopic expression of Plum likely results by β-axon retraction.
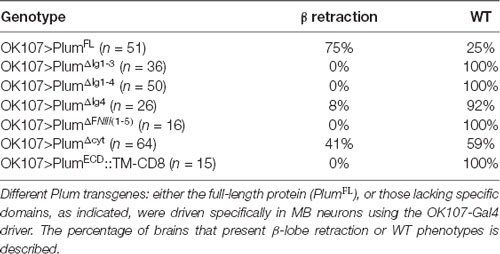
In our previous work, we performed a detailed structure-function analysis and found that Plum functions in the context of MB γ neuron pruning as an accessory receptor, which can bind a ligand but cannot transduce a signal by itself. This observation was based on our findings that while Plum’s extracellular domains were required for its function in axon pruning, its cytoplasmic domain was not. To test if Plum functions similarly in α/β neurons, we performed a structure-function analysis. As expected, we found that deletions of specific extracellular domains that are required for pruning also abrogated Plum’s ability to induce β lobe retraction (Table 1). To our surprise, over-expression of a Plum transgene encoding for its extracellular domain fused to a CD8 transmembrane domain did not induce retraction (UAS::plumECD::TM-CD8; n = 15; Table 1). Because over-expression of Plum lacking its cytoplasmic domain did induce axon retraction in 40% of the brains, these results suggest that in contrast to pruning, Plum transmembranal domain is required to induce retraction, likely via interactions with another signaling transmembrane receptor.
Plum Acts Through the Downstream Effector RhoGEF2 in α/β Neurons to Facilitate β-Axon Midline Stopping
To further unravel Plum’s mode of action, we asked whether expression of Plum in MB α/β neurons within homozygous mutant brains is sufficient to rescue β-lobe midline crossing. We expressed different Plum rescue transgenes in MB α/β neurons using the R44E04-Gal4 driver, which is expressed in α/β neurons but also in few non-MB cells (Supplementary Figures S3D–F). indeed, expressing Plum in α/β neurons was sufficient to significantly rescue β-lobe over-extension phenotype (Figures 5A,B, quantified in 5G). Interestingly, expressing plumΔcyt, plumΔIG1-4, plumΔIG-1-3 or plumECD-TM:CD8 could not rescue β-lobe crossing (Supplementary Figure S6), suggesting that unlike Plum’s domain requirement in γ axon pruning, where only the IG4 domain is important, in the context of β midline stopping, all of the tested extracellular, membranal and intracellular Plum domains are required. Interestingly, Babo, the TGF-β type I receptor, was shown to regulate axon midline stopping through a non-canonical Smad independent pathway, signaling via RhoGEF2 and LIMK (Ng, 2008). We, therefore, hypothesized that Plum might also act through this pathway in order to facilitate β-axon midline stopping. To test that, we expressed RhoGEF2 transgene in MB α/β neurons, in plumΔ1 homozygous brains. As expected, expressing RhoGEF2 specifically in α/β neurons was sufficient to significantly suppress the β-axon midline crossing phenotype (Figures 5A,C, Supplementary Figure S7, quantified in 5G). To our surprise, expressing either PlumFL or RhoGEF2 specifically in α/β neurons was also sufficient to suppress the crossing of the β’-axons (Figures 5D–F, quantified in 5G), suggesting that Plum can mediate a non-cell autonomous interaction between these two neuronal types to induce midline stopping.
The TGF-β Ligand Myoglianin Is Required for β-Axon Midline Stopping
Like plum, the TGF-β receptors—babo, wit and punt, were also previously shown to be required for normal β-lobe development (Ng, 2008). Loss of either one of these receptors results in β-lobe over-extension, while over-expression of a constitutively active version of Babo (BaboCA) results in short α and β-lobes. Furthermore, a recent study associated Myo as the activating ligand of Babo during pruning (Awasaki et al., 2011) and our own work has demonstrated genetic interaction between plum and baboon and between plum and myo (Yu et al., 2013). We were, therefore, interested to investigate whether TGF-β ligands, and perhaps Myo specifically, were involved in inducing a β-lobe stop signal.
We have previously shown that expressing a secreted form of Plum extra cellular domain (PlumECD) has a dominant negative effect on MB γ neuron pruning, presumably by sequestering its ligand, which in the context of pruning is Myo. To test if secreted ligands are also involved in β-lobe stop signal, we first over-expressed a PlumECD in glia by using the pan-glial driver, Repo-Gal4. Indeed, glial over-expression of PlumECD resulted in β-lobe midline crossing in 34% of the brains (Figure 6B compare with 6A, quantified in 6G), while neuronal expression of PlumECD had no effect on midline crossing (Figure 6E compared with 6D, quantified in 6G), suggesting that Plum might interact with a secreted ligand.
The Drosophila genome encodes for three TGF-β ligands: Myoglianin (Myo), Dawdle (Daw) and Activin (dAct). Due to its known interaction with Plum in the context of axon pruning, we decided to focus on Myo; we over-expressed two different Myo-RNAi constructs in glial cells using the pan-glial driver Repo-Gal4 and in neurons using the pan-neuronal driver c155-Gal4. While 40% of β-lobes crossed the midline when we knocked down Myo expression in glia (Figure 6C compared with 6A, quantified in 6G), 0% crossed the midline when Myo was knocked down in neurons (Figure 6F compared to 6D, quantified in 6G) indicating that glial-derived Myo is required for proper β-lobe formation. The fact that only 40% of the β-lobes cross the midline in brains expressing Myo RNAi could suggest one of three options: Myo RNAi knockdown could be incomplete; Myo could be expressed in and secreted by cells other than glia; or Myo might function cooperatively with one of the other two TGF- β ligands, in determining β-lobes length. These results together demonstrate that Myo and possibly also Daw are the ligands required for β-axon stop signal at the midline.
Discussion
In this study, we have uncovered a novel mechanism by which axon stopping is regulated. Using loss of function analyses combined with rescue and gain of function experiments, we found that Plum, an Ig superfamily plasma membrane receptor, is required for the midline stopping of α/β MB neurons, and acts through RhoGEF2. Additionally, we found that glial-derived TGF-β ligand Myo induces the midline stopping of α/β MB neurons.
Genetic evidence suggests that Plum is both required and sufficient in MB neurons to regulate midline stopping of β-axons (Figures 3, 4, respectively). However, the fact that plumΔ1 MARCM MB clones show a more severe β-lobe midline crossing in homozygous brains compared to heterozygous brains (Figure 2) suggests that Plum may also play an important non-cell autonomous role in β-lobe development. This is intriguing, especially in light of the fact that Plum is a cell surface receptor. Because in mosaic brains containing neuroblast MARCM clones, of the neurons are heterozygous while only of the neurons are homozygous to the mutation, one simple explanation would be that Plum functions cell-autonomously in α/β neurons but axon stopping might be a communal decision.
In addition to Plum being important in α/β neurons to instruct their midline stopping, we also observed midline crossing of α’/β’ neurons in plumΔ1 animals. While α’/β’ midline crossing has never been reported, many studies have identified conditions in which α/β neurons extend beyond the midline; yet the underlying cellular mechanisms remain unclear (Moreau-Fauvarque et al., 1998; Michel et al., 2004; Kobayashi et al., 2006; Grillenzoni et al., 2007; Kurusu and Zinn, 2008; Ng, 2008). To our surprise, although α’/β’ neurons cross the midline in plumΔ1 animals, when we attempted to knock down plum specifically in α’/β’ neurons (using two different drivers), there was no midline crossing in either α’/β’ or α/β neurons. This inconsistency may be due to technical reasons such as weak or late expression of the Gal4 driver or non-cell autonomous effects. Since expressing Plum or its downstream effector RhoGEF2 specifically in α/β neurons partially suppressed the midline crossing of both β and β’-lobes to a similar degree, this indeed hints for a non-cell autonomous role of Plum in the context of midline stopping or retraction. Since α’/β’ neurons are born during the late-larval stage, and already send their axons by the time the α/β neurons are born, this non-cell autonomous interaction needs to be further and carefully studied and evaluated. Therefore, we cannot rule out an additional role for Plum in α’/β’ neurons in β’ or β-axon midline stopping.
One potential strategy to restrict axon extension into the opposite hemisphere would be for the existence of a signal at the midline. Indeed, during embryonic development, the midline provides major instructive signals. These signals, such as slit, are mainly secreted by midline glia in the embryo (Sonnenfeld and Jacobs, 1994). How the developing adult brain continues to exhibit bilateral symmetry is much less studied. The transient interhemispheric fibrous ring (TIFR), a transient glia structure, was proposed as a source of signaling molecules required during pupal development. TIFR glial cells are required for olfactory receptor neurons (ORN) axons to project across the midline and establish the contralateral wiring in the antennal lobes (AL; Chen and Hing, 2008) and more importantly, were proposed to restrict β-lobes from crossing the midline, via an unknown molecular mechanism (Simon et al., 1998). We found that glial-derived Myo was required for normal stopping of α/β neurons. Therefore, our data suggest that Myo is at least one of these midline signals that instruct β-axon stopping. Unfortunately, the available tools prevented us from making a clear conclusion regarding the role of the TIFR-glia at this time but further studies are definitely warranted.
Our findings suggest that the Plum domains that are required for axon stopping (all tested extracellular, transmembranal and intracellular domains) are different from than those required for pruning (mainly the IG4). Nonetheless, our data suggest that Plum functions downstream to the TGF-β ligand myo and upstream to RhoGEF2, which was previously shown to mediate TGF-β type I receptor Babo (Ng, 2008). Therefore, our data is most consistent with Plum functioning as a TGF-β accessory receptor that facilitates TGF-β signaling. Whether the transmembranal domain of Plum is important to mediate its potential interaction with Babo or another unknown component of the receptor complex, and also the mechanistic differences in Plum function in axon pruning or stopping, remain to be further studied. Therefore, variations on TGF-β signaling between glia and neurons play diverse roles in neuronal development.
Significance Statement
How axons stop growing during development is not well understood. Here, we use a Drosophila neuropil structure called the MB to address this issue. All three MB neuronal type refrain from crossing the midline. We found that glial-derived TGF-β signals bind to MB α/β neurons to mediate their midline stopping. Thus, we have identified the source of the patterning signal and its identity.
Data Availability Statement
This manuscript contains previously unpublished data. Previously published stocks and their source are mentioned in the “Materials and Methods” section.
Author Contributions
NM-K and IG designed, performed and analyzed experiments and wrote the manuscript. NI-Z provided technical assistance and performed specific experiments. OS designed and analyzed experiments, wrote the manuscript and procured funding.
Funding
This work was supported by the Israeli Science Foundation (ISF), grant #683/11 (legacy heritage program) and #2573/18, the David and Fela Shapell Family Center for Genetic Disorders Research, and the Estate of Olga Klein Astrachan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Zeiss LSM710 microscope was purchased with the help of the Adelis foundation. OS is an incumbent of the Prof. Erwin Netter Professorial Chair of Cell Biology. We thank, Takeshi Awasaki, Ann-Shyn Chiang, Yuh-Nung Jan, Julian Ng, Michael O’Connor, the Vienna Drosophila Resource Center, the Bloomington Stock Center and the Kyoto Drosophila Genomic Resource Center for reagents; Ron Rotkopf for statistical analysis; A. Yaron and the members of the Schuldiner lab for discussions and critical readings of the manuscript; the FasII (1D4) and Trio (9.4A) monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2019.00232/full#supplementary-material
FIGURE S1 | Midline crossing ranking, plum locus and CRISPR plumV11X description, relates to Figure 1. (A) Examples of midline crossing severity ranking, for scores 1 (A1), 3 (A2) and 5 (A3). for more information, see “Materials and Methods” section. (B) Scheme of plum locus including available mutant and gRNAs used for generating plumV11X. (C,D) Confocal Z-projections of (C) WT or (D) plumV11X MARCM neuroblast clones labeled with 201Y-Gal4. Yellow arrowheads indicate severe pruning defects. CD8::GFP (green); FasII (magenta). (E,F) Confocal Z-projections of WT (E) or plumV11X (F) brains. Dashed red lines demarcate the extent of the β-lobes. FasII (gray); Scale bars, 20 μm. (G) Quantification of midline crossing phenotypes in (E,F). p-value: ***p < 0.001. Genotypes: (C) y, w, hsFlp/Y or +; CD8::GFP, 201Y/+; 82B, Gal80/82B (n = 6). (D) y, w, hsflp/Y or +; CD8::GFP, 201Y/+; 82B, Gal80/82B, plumV11X (n = 21). (E) 201Y, CD8:GFP/+ (n = 45). (F) 82B, plumV11X (n = 13). p-value: ***p < 0.001.
FIGURE S2 | plum RNAi in all MB neurons, relates to Figure 3. (A–D) Confocal Z-projections or selected slices, as depicted in the figure, of (A,C) brains expressing CD8::GFP or (B,D) those additionally expressing plumRNAi. Dashed red lines demarcate the extent of the β or β’-lobes. CD8::GFP (green); FasII (in A’,B’, gray); TRIO (in C’,D’, gray). Scale bars, 20 μm. (E) Quantification of the midline crossing phenotypes in (A,B). p-values: ***p < 0.001. Genotypes: (A,C) CD8/+; OK107/+ (n = 11, 20, respectively). (B,D) UAS- plumRNAi/CD8::GFP; OK107/+ (n = 19, 21, respectively).
FIGURE S3 | Expression pattern of α’/β’ and α/β-specific Gal4s, relates to Figure 3. (A–F) Confocal Z-projections of WT brains expressing CD8::GFP driven by either c305a-Gal4 (A–C) or R44E04-Gal4 (D–F) driver at third-instar (L3) larva (A,D), 0 h APF pupa (B), 30 h APF pupa (E) and adult brains (C,F). CD8::GFP (green); FasII (magenta). Scale bars, 20 μm. Genotypes: (A–C) c305a-Gal4/+; CD8::GFP/+. (D–F) CD8::GFP/+; R44-Gal4/+.
FIGURE S4 | No midline crossing in controls expressing cell type specific Gal4 drivers, relates to Figure 3. (A–C) Confocal Z-projections of WT brains expressing CD8::GFP with the γ-specific R71G10 driver (A), α/β-specific R44E04 driver (B), or α’/β’-specific c305a driver (C). Dashed red lines demarcate the extent of the β or β’-lobes. CD8::GFP (green in A1–C1 and A2–C2), FasII (in A1’–C1’, gray), TRIO (in A2’–C2’, gray). Scale bars, 20 μm. Genotypes: (A) CD8::GFP/+; R71/+ (n = 16, 16, respectively). (B) c305a/+; CD8::GFP/+ (n = 28, 20, respectively). (C) CD8::GFP/+; R44/+ (n = 38, 20, respectively).
FIGURE S5 | Plum over-expression within MB neurons does not affect β’-lobe length, relates to Figure 4. (A,B) Confocal Z-projections of (A) WT brains expressing CD8::GFP and (B) WT brains expressing CD8::GFP as well as PlumFL driven by OK107-Gal4. Arrowheads in (B1) depict aberrant growth of β-lobes. CD8::GFP (green, A1–B1, A3–B3); TRIO (gray, A2–B2, magenta, A3–B3). Scale bars, 20 μm. Genotypes: (A) CD8::GFP/+;; OK107/+ (n = 20). (B) CD8::GFP/PlumFL;; OK107/+ (n = 5).
FIGURE S6 | Plum’s intracellular, extracellular and transmembranal domains are required to induce midline stopping, relates to Figure 5. (A–D) Confocal Z-projections of adult plumΔ1 brains expressing CD8::GFP driven by the R44-Gal4 driver, and additionally expressing (A) UAS-PlumΔcyt, (B) UAS-PlumΔIG1-4, (C) UAS-PlumΔIG1-3, or (D) UAS-PlumΔECD-TM:CD8. (E) Quantification of β-lobe midline crossing phenotypes in (B–E). p-value: *p = 0.017. ns = not significant. Dashed red lines demarcate the extent of the β-lobes. FasII (gray). Scale bars, 20 μm. Genotypes: (A) UAS-PlumΔcyt/+; 82B, plumΔ1/R44E04-Gal4, 82B, plumΔ1 (n = 18). (B) UAS-PlumΔIG1-4/+; 82B, plumΔ1/R44E04-Gal4, 82B, plumΔ1 (n = 10). (C) UAS-PlumΔIG1-3/+; 82B, plumΔ1/R44E04-Gal4, 82B, plumΔ1 (n = 17). (D) UAS-PlumΔECD-TM:CD8/+; 82B, plumΔ1/R44E04-Gal4, 82B, plumΔ1 (n = 23). (Additionally, quantified in E): R44E04-Gal4, 82B, plumΔ1/82B, plumΔ1 (n = 39). UAS-PlumFL/+; 82B, plumΔ1/R44E04-Gal4, 82B, plumΔ1 (n = 20).
FIGURE S7 | RhoGEF2 does not cause retraction when expressed in α/β neurons, relates to Figure 5. (A) Confocal Z-projection of adult brain expressing UAS-RhoGEF2 with the R44E04-Gal4 driver. Genotype: (A) R44E04-Gal4/UAS-RhoGEF2 (n = 17).
References
Abe, T., Yamazaki, D., Murakami, S., Hiroi, M., Nitta, Y., Maeyama, Y., et al. (2014). The NAV2 homolog sickie regulates f-actin-mediated axonal growth in Drosophila mushroom body neurons via the non-canonical Rac-cofilin pathway. Development 141, 4716–4728. doi: 10.1242/dev.113308
Aso, Y., Hattori, D., Yu, Y., Johnston, R. M., Iyer, N. A., Ngo, T. T., et al. (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 3:e04577. doi: 10.7554/eLife.04577
Awasaki, T., Huang, Y., O’Connor, M. B., and Lee, T. (2011). Glia instruct developmental neuronal remodeling through TGF-β signaling. Nat. Neurosci. 14, 821–823. doi: 10.1038/nn.2833
Chen, W., and Hing, H. (2008). The L1-CAM, Neuroglian, functions in glial cells for Drosophila antennal lobe development. Dev. Neurobiol. 68, 1029–1045. doi: 10.1002/dneu.20644
Crittenden, J. R., Skoulakis, E. M., Han, K. A., Kalderon, D., and Davis, R. L. (1998). Tripartite mushroom body architecture revealed by antigenic markers. Learn. Mem. 5, 38–51.
Dickson, B. J. (2002). Molecular mechanisms of axon guidance. Science 298, 1959–1964. doi: 10.1126/science.1072165
Grillenzoni, N., Flandre, A., Lasbleiz, C., and Dura, J. M. (2007). Respective roles of the DRL receptor and its ligand WNT5 in Drosophila mushroom body development. Development 134, 3089–3097. doi: 10.1242/dev.02876
Heisenberg, M. (2003). Mushroom body memoir: from maps to models. Nat. Rev. Neurosci. 4, 266–275. doi: 10.1038/nrn1074
Kaplan, A., Kent, C. B., Charron, F., and Fournier, A. E. (2014). Switching responses: spatial and temporal regulators of axon guidance. Mol. Neurobiol. 49, 1077–1086. doi: 10.1007/s12035-013-8582-8
Kobayashi, M., Michaut, L., Ino, A., Honjo, K., Nakajima, T., Maruyama, Y., et al. (2006). Differential microarray analysis of Drosophila mushroom body transcripts using chemical ablation. Proc. Natl. Acad. Sci. U S A 103, 14417–14422. doi: 10.1073/pnas.0606571103
Kolodkin, A. L., and Tessier-Lavigne, M. (2011). Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb. Perspect. Biol. 3:a001727. doi: 10.1101/cshperspect.a001727
Kurusu, M., Awasaki, T., Masuda-Nakagawa, L. M., Kawauchi, H., Ito, K., and Furukubo-Tokunaga, K. (2002). Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development 129, 409–419.
Kurusu, M., and Zinn, K. (2008). Receptor tyrosine phosphatases regulate birth order-dependent axonal fasciculation and midline repulsion during development of the Drosophila mushroom body. Mol. Cell. Neurosci. 38, 53–65. doi: 10.1016/j.mcn.2008.01.015
Lee, T., Lee, A., and Luo, L. (1999). Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development 126, 4065–4076.
Meltzer, H., Marom, E., Alyagor, I., Mayseless, O., Berkun, V., Segal-Gilboa, N., et al. (2019). Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat. Commun. 10:2113. doi: 10.1038/s41467-019-10140-0
Michel, C. I., Kraft, R., and Restifo, L. L. (2004). Defective neuronal development in the mushroom bodies of Drosophila fragile X mental retardation 1 mutants. J. Neurosci. 24, 5798–5809. doi: 10.1523/jneurosci.1102-04.2004
Moreau-Fauvarque, C., Taillebourg, E., Boissoneau, E., Mesnard, J., and Dura, J. M. (1998). The receptor tyrosine kinase gene linotte is required for neuronal pathway selection in the Drosophila mushroom bodies. Mech. Dev. 78, 47–61. doi: 10.1016/s0925-4773(98)00147-6
Ng, J. (2008). TGF-β signals regulate axonal development through distinct Smad-independent mechanisms. Development 135, 4025–4035. doi: 10.1242/dev.028209
Port, F., Chen, H. M., Lee, T., and Bullock, S. L. (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U S A 111, E2967–E2976. doi: 10.1073/pnas.1405500111
Simon, A. F., Boquet, I., Synguelakis, M., and Preat, T. (1998). The Drosophila putative kinase linotte (derailed) prevents central brain axons from converging on a newly described interhemispheric ring. Mech. Dev. 76, 45–55. doi: 10.1016/s0925-4773(98)00104-x
Sonnenfeld, M. J., and Jacobs, J. R. (1994). Mesectodermal cell fate analysis in Drosophila midline mutants. Mech. Dev. 46, 3–13. doi: 10.1016/0925-4773(94)90033-7
Sperry, R. W. (1963). Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl. Acad. Sci. U S A 50, 703–710. doi: 10.1073/pnas.50.4.703
Tessier-Lavigne, M., and Goodman, C. S. (1996). The molecular biology of axon guidance. Science 274, 1123–1133. doi: 10.1126/science.274.5290.1123
Trannoy, S., Redt-Clouet, C., Dura, J. M., and Preat, T. (2011). Parallel processing of appetitive short- and long-term memories in Drosophila. Curr. Biol. 21, 1647–1653. doi: 10.1016/j.cub.2011.08.032
Keywords: Drosophila, axon, midline, glia, axon stopping, mushroom body, develepment, axon-glia interaction
Citation: Marmor-Kollet N, Gutman I, Issman-Zecharya N and Schuldiner O (2019) Glial Derived TGF-β Instructs Axon Midline Stopping. Front. Mol. Neurosci. 12:232. doi: 10.3389/fnmol.2019.00232
Received: 30 April 2019; Accepted: 11 September 2019;
Published: 27 September 2019.
Edited by:
Robert W. Burgess, Jackson Laboratory, United StatesReviewed by:
Kai Zinn, California Institute of Technology, United StatesGreg J. Bashaw, University of Pennsylvania, United States
Copyright © 2019 Marmor-Kollet, Gutman, Issman-Zecharya and Schuldiner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oren Schuldiner, oren.schuldiner@weizmann.ac.il
† These authors have contributed equally to this work
 Neta Marmor-Kollet
Neta Marmor-Kollet Itai Gutman†
Itai Gutman†  Oren Schuldiner
Oren Schuldiner