m6A methylation: Critical roles in aging and neurological diseases
- 1Department of Neurology, Xiangya Hospital, Central South University, Changsha, China
- 2National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, China
- 3Hunan Key Laboratory of Medical Epigenomics, Department of Dermatology, The Second Xiangya Hospital of Central South University, Changsha, China
N6-methyladenosine (m6A) is the most abundant internal RNA modification in eukaryotic cells, which participates in the functional regulation of various biological processes. It regulates the expression of targeted genes by affecting RNA translocation, alternative splicing, maturation, stability, and degradation. As recent evidence shows, of all organs, brain has the highest abundance of m6A methylation of RNAs, which indicates its regulating role in central nervous system (CNS) development and the remodeling of the cerebrovascular system. Recent studies have shown that altered m6A levels are crucial in the aging process and the onset and progression of age-related diseases. Considering that the incidence of cerebrovascular and degenerative neurologic diseases increase with aging, the importance of m6A in neurological manifestations cannot be ignored. In this manuscript, we focus on the role of m6A methylation in aging and neurological manifestations, hoping to provide a new direction for the molecular mechanism and novel therapeutic targets.
1. Introduction
Epigenetic modifications are crucial posttranscriptional regulations of gene expression, which play important regulatory roles in organogenesis, homeostasis and pathological process (Wang Y. et al., 2021). Since 2013, epigenetic alterations have been listed as one of the nine hallmarks of aging and important changes of cellular senescence (López-Otín et al., 2013; Hernandez-Segura et al., 2018). RNA modifications are important parts of epigenetic modifications. At present, over 160 types of chemical modifications have been identified in RNA, which participate in the regulation of the structural properties of RNA or changing the affinity of mRNA for ribosomes (Zhao et al., 2020). Among all the modifications, N-Methyl adenosine (mA) is the most prevalent internal one and has been found to be highly conserved and hard-coded in mammals and other eukaryotic species (Huang et al., 2020). Studies have shown that N6-methyladenosine (m6A) is abundant in the brain and is involved in the regulation of brain volume, memory formation and consolidation and mammalian postnatal cortical neurogenesis (Ma et al., 2018). Abnormal RNA m6A methylation level is associated with aging associated central nervous system (CNS) changes and the onset and prognosis of various neurological diseases, such as transient focal ischemia, ischemic stroke, Parkinson’s disease (PD), Alzheimer’s disease (AD), multiple sclerosis, depression, epilepsy, and gliomas, etc. (Chokkalla et al., 2019; Chang et al., 2022; Li et al., 2022; Zhang N. et al., 2022).
Aging is a natural process of organismal decay, which is characterized by the functional decline of tissues and organs and the increased risk of aging-associated disorders (Zhang et al., 2020). Brain aging is a complex process, which influences brain structure and functional connectivity (Damoiseaux, 2017). Morphologically, brain aging is characterized by volume loss, cortical thinning, white matter degradation, loss of gyrification, and ventricular enlargement. Pathologically, brain aging is associated with neuron cell shrinking, dendritic degeneration, demyelination, small vessel disease, metabolic slowing, microglial activation, and the formation of white matter lesions (Blinkouskaya et al., 2021). Mechanisms under these changes are not clear, resulting in the lack of effective therapeutic methods (Hou et al., 2019). Epigenetic alterations have been regarded as important hallmarks of aging and cellular senescence (López-Otín et al., 2013; Hernandez-Segura et al., 2018). Considering the abundance and aging related changes of RNA m6A methylation in CNS, it must play critical roles in aging and degenerative neurological diseases. Altered m6A methylation modifications and mutated RNA m6A methyltransferases are associated with diverse neurological pathological processes, which provide new aspects for brain aging research.
The CNS is a complex regulatory network that requires the homeostasis and functional connectivity between neurons and other constituents such as endothelial cells, astrocytes, pericytes, microglia, oligodendrocytes, basement membrane, as well as surrounding extracellular matrix (ECM; Edison, 2020; Schaeffer and Iadecola, 2021). To describe the situation in the brain more realistically, the concept of neurovascular unit (NVU) was proposed, which is composed of neurons, blood–brain barrier (BBB), microglia, pericyte, astrocyte and surrounding ECM, etc. (Seo et al., 2021; Figure 1). Intercellular communication and signaling within the NVU is fundamental to the CNS homeostasis and function (Zagrean et al., 2018). Functionally, the NVU is responsible for maintaining the integrity of the BBB, regulating the cerebral blood flow (CBF) and promoting the signal transmission of local neurons (Yu et al., 2020). Some internal and external factors, such as ischemic stroke or age-related degeneration, can disrupt the balance of NVU. If that happens, neuronal cell death, glial reaction, and immune cell infiltration would subsequently occur, resulting in various neurological diseases (Cai et al., 2017a; Wang L. et al., 2021). Therefore, functional recovery of NVU through remodeling these cellular networks have become an emerging therapeutic target for aging related neurological diseases and ischemic CNS diseases (Ozaki et al., 2019; Forró et al., 2021). Published papers have found that RNA m6A methylation participates in promoting angiogenesis and nervous system development, which are important parts of NVU remodeling. Therefore, m6A methylation could be the cross hub linking aging, NVU remodeling and neurological diseases. In this manuscript, we summarize recent findings in the field of RNA m6A methylation and NVU remodeling and discuss the potential application of m6A methylation in the treatment of different neurological diseases. We hope to provide novel therapeutic targets for future disease management.
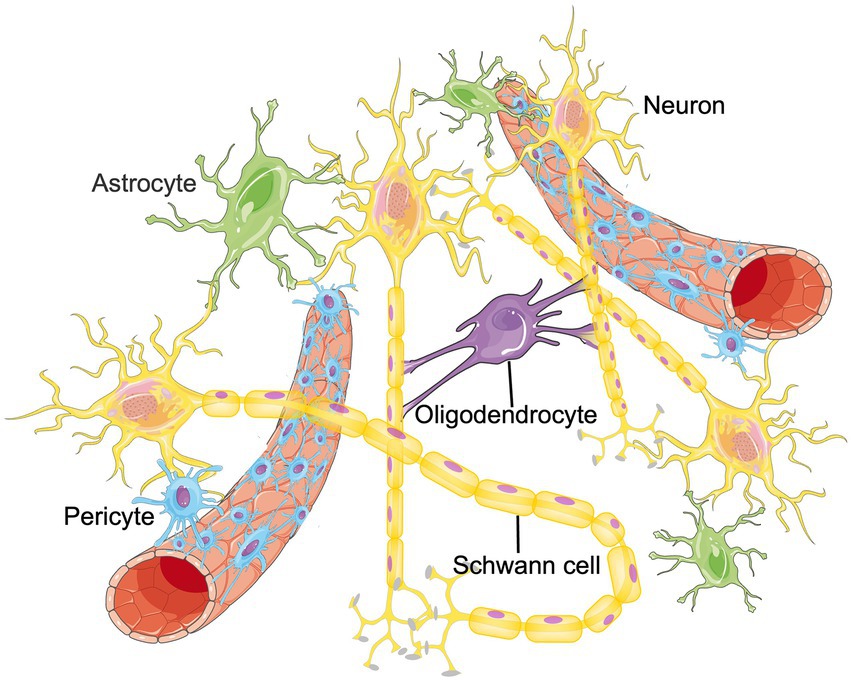
Figure 1. The structure of NVU. The neurovascular unit (NVU) is the minimal functional unit of the brain, consisting of astrocytes, pericytes, microglia, neurons, oligodendrocytes and endothelial cells. The crosstalk between these cellular networks and surrounding extracellular matrix (ECM) plays an essential role in the onset and progression of central nervous system diseases.
2. The CNS system: Remodeling during aging and diseases
The development of the human CNS requires the precise orchestration and coordination of myriad molecular and cellular processes across a staggering array of cell types and over a long period of time (Bohlen et al., 2019). The formation of the CNS begins early in development with the induction of the neural ectoderm on the dorsal surface of the embryo. Subsequently, the neural ectoderm plate changes its shape to form a neural groove and eventually, a neural tube. The wall of the neural tube is composed of germinal cells, collectively called the neuroepithelium, that produces neurons and glia throughout the CNS (Bayer, 1989). After that, neuron begin to migration, glial cells differentiate and mature, laminar organized and form regional patterning and lateralization of the human neocortex, which facilitates neural circuit assembly, maturation, and plasticity (Silbereis et al., 2016). To better understand the function and structure changes of CNS, the concept of NVU was proposed as a structural and functional unit of brain which is composed of neurons, BBB, microglia, pericyte, astrocyte and surrounding ECM, etc. NVU emphasizes the symbiotic relationship between the nervous system and the microenvironment, which is a dynamic interaction between multiple cells instead of a simple nutritional relationship. The dysfunction of NVU is an important pathological feature underlying neurological disorders and during aging process (Sato et al., 2022). When that happens, cells and other constituents in NVU interact with each other to maintain homeostasis and promote NVU recovery. These changes are called NVU remodeling, which has significant influence on the onset and prognosis of various neurological manifestations (Zagrean et al., 2018).
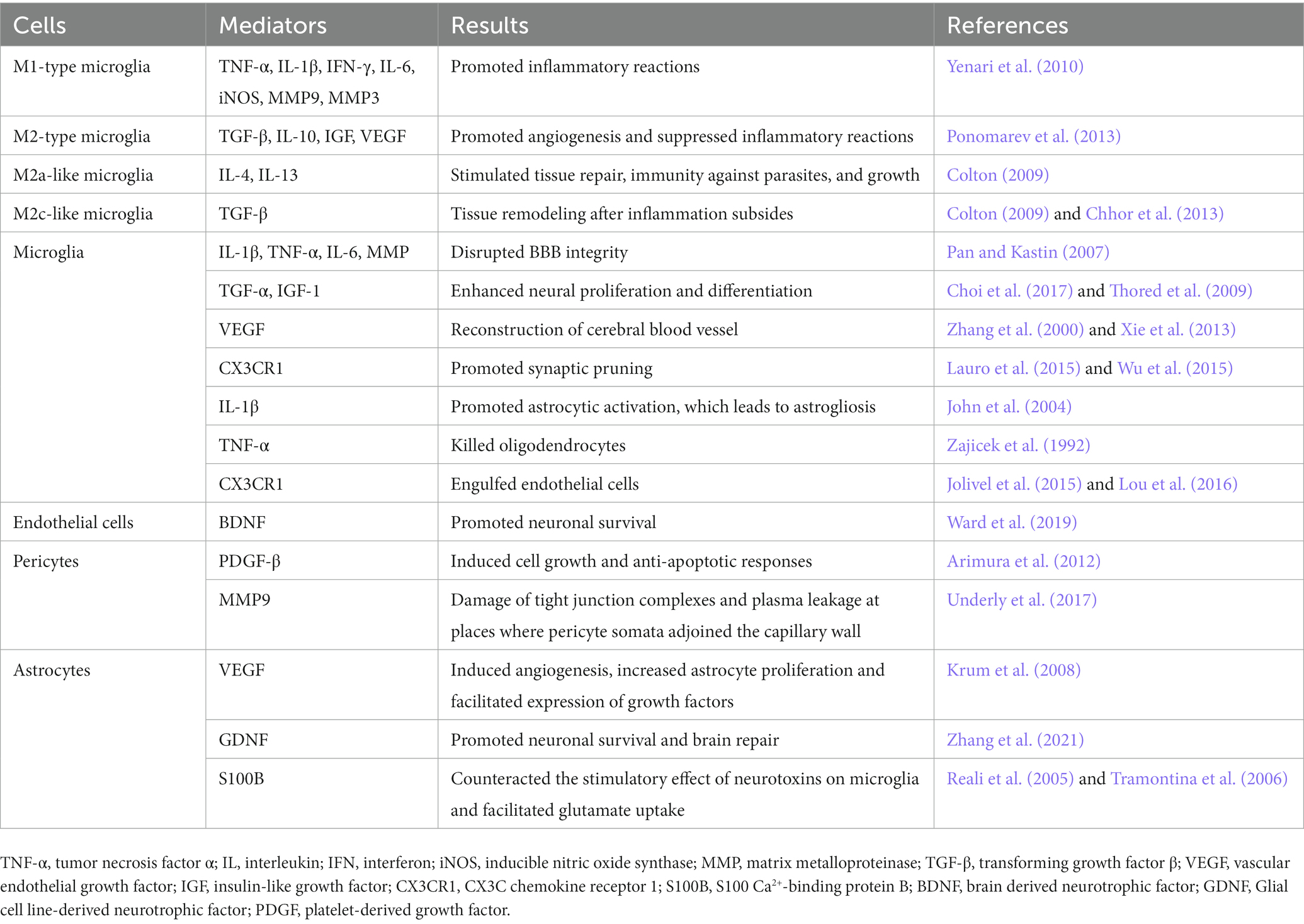
The process of NVU remodeling involves the interactions between neurons, glial and microvascular cells that create a microenvironment in which neurological recovery may ensue (Zagrean et al., 2018). Take NVU remodeling during ischemia/reperfusion as an example. When ischemia occurs, decreased cerebral perfusion leads to neuronal injury and death, which is the beginning of NVU remodeling. Diverse proinflammatory mediators released by damaged neurons leads to astrocyte end-foot swelling and increased endothelial vesicles (Haley and Lawrence, 2017). Impaired astrocytes secrete proinflammatory mediators that cause further NVU disruption and neuronal injury. Matrix metalloproteinases (MMPs) produced by endothelial cells and pericytes degrade the basement membrane and cause the breakdown of tight junctions (Underly et al., 2017). Microglia translocate to the penumbra and exacerbate BBB breakdown (Yenari et al., 2010). Leukocytes then transmigrate across the breached BBB and cause subsequent neuroinflammation (Zhang et al., 2019). After reperfusion, neuronal networks and brain capillaries start to develop, which involves the migration of neural progenitor cells (NPCs), the remodeling of functional axons and synapses and the formation of ECM (Andres et al., 2011). Angiogenesis is the key and the first step for NVU remodeling after cerebral ischemia, which includes proliferation of vessel composing cells, recruitment of pericytes, coverage of endothelial tube by pericytes, and maturation of neo-vessels (Hatakeyama et al., 2020). Pericytes function as vital modulators in angiogenesis and help remodel the BBB and support the neurogenesis (Cai et al., 2017b). Endogenous vascular endothelial growth factor (VEGF) produced by astrocytes promotes angiogenesis and the proliferation of astrocytes themselves (Krum et al., 2008). NPCs are attracted to the damaged area accompanied with the process of angiogenesis, which initiates the neurogenesis process (Hermann and Zechariah, 2009). Interaction between matrix and receptors on NVU cells regulates cell survival and focal bioavailability of growth factors, which is essential for NVU remodeling processes (Stamatovic et al., 2019). Restoring the function of neurons is the ultimate therapeutic target of ischemic diseases. Neuron function restoration requires the local formation of functional axons and synapses along pyramidal tract (Andres et al., 2011; Reitmeir et al., 2011) and within motor cortex (Clarkson et al., 2010; Hermann and ElAli, 2012). Meanwhile, neurons adjacent to the infarct induce axonal growth (Li et al., 2010; Overman et al., 2012; Li et al., 2015; Joy et al., 2019) and synapse formation (Luke et al., 2004) via regulating growth differentiation factor 10 (GDF10), ephrin-A5, and C-C chemokine receptor type 5 (CCR5) signals. Also, new axonal projections are formed, which can project into premotor, motor, sensory (Overman et al., 2012; Li et al., 2015; Joy et al., 2019) and retrosplenial cortices (Brown et al., 2009). The remodeling of ECM mainly depends on the glial cells. In vivo and in vitro studies have shown that, glial cell regeneration occurs before neuron regeneration. After 24 h of reperfusion, microglia fully enwrap small blood vessels in the peri-infarct region. The remodeling process is regulated by many cytokines, such as VEGF, angiogenin, Netrin-4, etc. (Table 1) They participate in mediating cerebral angiogenesis and restoring the function of neurological diseases (Lemons and Condic, 2006). Administration of exogenous endostatin, an angiogenesis antagonist, can not only inhibit angiogenesis, but also inhibit the migration and survival of newborn neurons (Guo and Lo, 2009; Xiong et al., 2010). In the process of neurons remodeling, VEGF secreted by the proliferating microvascular cells promotes the migration of neural precursor cells and the remodeling of neurons and glial cells (Hermann and Zechariah, 2009).
3. Basic science of RNA m6A methylation
The dynamic nature and increasing number of RNA modifications provide new possibilities for adapting to specific environments by changing gene expression rapidly. At present, over 160 types of chemical modifications have been identified in RNA (Zhao et al., 2020), among which RNA methylation is the most abundant modification. The most abundant and diverse epigenetic modification of mRNAs in eukaryotes is m6A methylation which mainly modified mRNA and lncRNA (Huang et al., 2020). Considering that RNA methylation play an important role in nervous system development (Widagdo and Anggono, 2018) and angiogenesis (Qin et al., 2020), it may play an important role in NVU remodeling.
M6A methylation, first reported in 1974, plays a conservative role in the evolution of meiosis and cell differentiation (Desrosiers et al., 1974; Yue et al., 2015). The abundance of m6A methylation varies in different organs, tissues and cell lines but peaks in the brain (Meyer et al., 2012; Chang et al., 2017). Molecularly, m6A mainly locates near the termination codon of the protein coding sequence (CDS) of the mRNA and the 3 ‘untranslated region (3′UTRs; Dominissini et al., 2012; Ke et al., 2015). The specific modification site of m6A methylation is mainly on the adenine of the RRACH sequence (R is guanine or adenine, A is adenine, C is cytosine, H is uracil, adenine or cytosine), which regulates the stability, location, transportation, splicing and translation of RNA at the post-transcription level (Deng et al., 2018). According to recent studies, m6A methylation plays an important role in multiple processes including mRNA splicing regulation (Haussmann et al., 2016; Lence et al., 2016), mRNA translatability and stability (Bodi et al., 2015; Liu et al., 2017), and alternative polyadenylation site selection (Ke et al., 2015), etc.
In the process of RNA methylation, three types of molecules are involved: writers, erasers, and readers. Writers refer to methyltransferase, which can add methylation modifications to RNA and mediate the process of RNA methylation modification (Zhou et al., 2021a; Satterwhite and Mansfield, 2022). Erasers are demethylase that erase the RNA methylation modification and mediate the process of RNA demethylation modification (Qu et al., 2022). Readers, known as m6A methylation recognition protein, can read the information of RNA methylation modification, guiding, and participating in the translation and degradation of the downstream sequence of the modified RNA (Wei et al., 2022). These three types of molecules are indispensable for RNA methylation regulation and are powerful tools in the studies of specific mechanism, physiological and pathological role of RNA methylation (Figure 2).
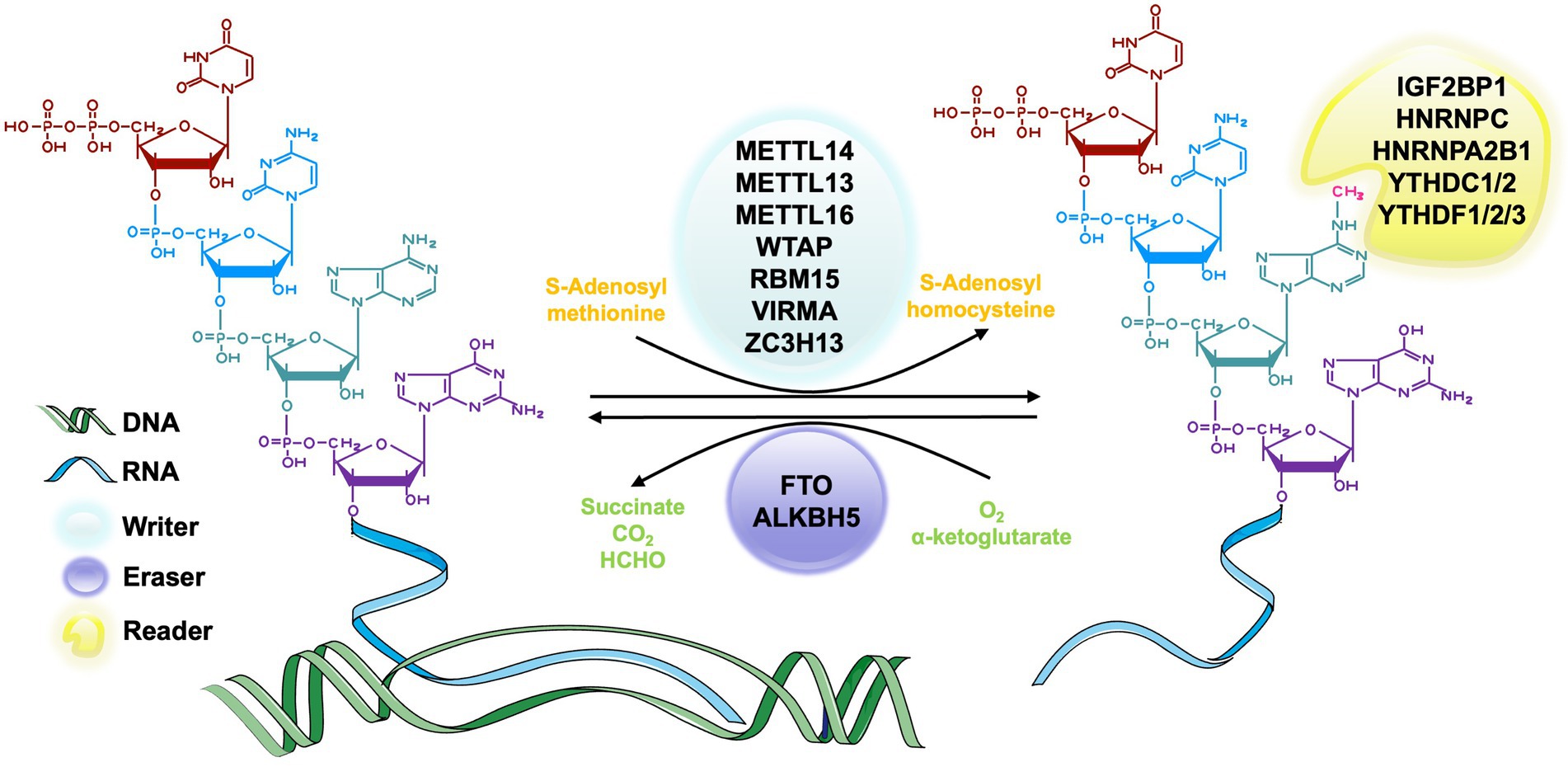
Figure 2. The process of m6A RNA methylation. The m6A methyltransferase, also called writers, catalyze the transfer of methyl groups from the donor S-adenosylmethionine (SAM) to the adenine nucleotides in the recipient RNA subunit. The m6A demethylase, also known as erasers, is responsible for removing the SAM on the adenine nucleotide of m6A-containing substrates. m6A methylation recognition proteins, which are known as readers, function in recognizing and binding to the m6A modified site and decoding the methylation code by recruiting or weakening the connection between the target RNAs and RNA binding-proteins (RBPs) of different functions.
The m6A methyltransferase complex, also known as writer, is responsible for catalyzing the transfer of methyl groups from the donor S-adenosylmethionine (SAM) to the adenine nucleotides in the recipient RNA subunit (Bokar et al., 1997). M6A methyltransferase complex consists of METTL3 (McGraw et al., 2007), METTL14 (Liu et al., 2014), Wilms’ tumor 1-associated protein (WTAP; Ping et al., 2014), vir-like m6A methyltransferase-associated protein (VIRMA, originally known as KIAA1429; Yue et al., 2018), and RNA binding motifs protein 15/15B (RBM15/15B; Patil et al., 2016; Chen X.-Y. et al., 2019). The most common molecular components of this complex are METTL3 and METTL14 (Liu et al., 2014). METTL3 is a highly conserved catalytic subunit and the core component of this complex, which has the ability to bind SAM (Lin et al., 2019). METTL14 is highly homologous to METTL3 and can combine with METTL3 to form a stable heterodimer and greatly enhance the catalytic activity (Liu et al., 2014). METTL3 and METTL4 maintain the main function of this complex together (Liu et al., 2016; Wang et al., 2017a). WTAP is also a core component in the m6A methyltransferase complex that can interact with METTL3-METTL14 complex in catalyzing methylation (Ping et al., 2014). The zebrafish embryos with WTAP knockdown can reduce the abundance of m6A methylation in cells significantly (Ping et al., 2014). It may result from the ability of WTAP to bind the alternative splicing pattern of mRNA (Ping et al., 2014). KIAA1429 is a homologous protein of the Virilizer protein that participates in regulating the catalytic activity of the methyltransferase complex by aggregating the core components (Yue et al., 2018). There are also some other relevant molecules of writers, such as METTL5 (van Tran et al., 2019), tRNA methyltransferase 11-2 (TRMT112; van Tran et al., 2019), Cbl proto-oncogene like 1 (CBLL1; Figueroa et al., 2009) and so on.
The m6A demethylase, also known as erasers, is responsible for removing the SAM on the adenine nucleotide of m6A-containing substrates. According to current research, two m6A demethylases, fat mass and obesity-associated (FTO; Jia et al., 2011) and a-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5; Zheng et al., 2013), have been identified in eukaryotes. These two molecules belong to the AlkB family and have similar catalytic centers, but differ in their substrates and positioning (Zou et al., 2016). FTO is the first discovered RNA deacetylase and mainly catalyzes RNA demethylation with its C-terminal structure in the nucleus (Bartosovic et al., 2017). Studies showed that FTO played an important role in neural differentiation (Li L. et al., 2017), lipogenesis (Zhang et al., 2015) and bone mesenchymal stem cells (BMSCs) ossification (Wang et al., 2017b). ALKBH5 is the second identified m6A erasers in mammalian. It has the similar catalytic ability to FTO (Zheng et al., 2013) but has different organ distribution. FTO has an abundance in the brain (Aas et al., 2017), but ALKBH5 is mainly found in the tests and has an important effect on spermatogenesis (Zheng et al., 2013).
m6A methylation recognition proteins, which are known as readers, functions in recognizing and binding to the m6A modified site and decoding the methylation code by recruiting or weakening the connection between the target RNAs and RNA binding-proteins (RBPs) of different functions (Roost et al., 2015; Spitale et al., 2015; Adhikari et al., 2016; Maity and Das, 2016; Wu et al., 2017). The most important reader in eukaryotes is a group of proteins that have YT521-B homology (YTH) domains. These domains include conserved C-terminal for RNA recognition and the variable N-terminal for RNA binding, which are also considered to be the most primitive m6A readers (Zhang et al., 2010), YTHDF family, including YTHDF1, YTHDF2, YTHDF3 in the cytoplasm, and YTHDC1, YTHDC2 in nucleus (Wang et al., 2014, 2015; Zhou et al., 2015; Li L.-J. et al., 2018) all belong to this group. YTHDF proteins regulate the metabolism of RNA cooperatively (Li A. et al., 2017; Shi et al., 2017). Other readers include but not limited to heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1; Alarcón et al., 2015), heterogeneous nuclear ribonucleoproteins C (HNRNPC; Liu et al., 2015), heterogeneous nuclear ribonucleoproteins G (HNRNPG; Liu et al., 2017), fragile X messenger ribonucleoprotein (FMRP; Edupuganti et al., 2017), insulin-like growth factor 2 mRNA-binding protein1-3 (IGF2BP1-3; Huang et al., 2018) and eukaryotic initiation factor 3 (eIF3; Meyer et al., 2015).
4. Effect of RNA m6A methylation
The m6A modification can regulate gene transcription, influencing the cellular location, stability and translation efficiency of targeted coding and non-coding RNAs (Zaccara et al., 2019). For example, m6A methylation participates in the pre-mRNA spicing (Berulava and Horsthemke, 2010; Zheng et al., 2013; Liu et al., 2014; Alarcón et al., 2015), mRNA stability (Batista et al., 2014) and the translation process (Li L.-J. et al., 2018) of coding RNA. As for noncoding RNA, such as rRNA, miRNAs and lncRNA, m6A methylation can increase the translation efficiency of rRNA (van Tran et al., 2019), influence the pre-rRNA processing of snoRNA (Sergeeva et al., 2020), re-miRNA and lncRNA (Fazi and Fatica, 2019). RNA methylation shows a significant effect on regulating gene expression efficiency. According to recent studies, m6A methylation and other related molecules such as FTO and NSun2 are abundant in brain (Blanco et al., 2011; Lence et al., 2016; Aas et al., 2017; Chang et al., 2017). These molecules play an important role in the differentiation and growth of nervous system and blood vessels. Following, we introduce the effect of RNA methylation in NVU remodeling from the aspect of blood vessel repair, neurons regeneration and other CNS cell function.
4.1. RNA m6A methylation in blood vessels repair and angiogenesis
Cell differentiation, especially mesenchymal stem cells (MSCs) and hematopoietic stem cells (HSCs), is a key process in blood vessels repair and angiogenesis, which is also a new therapeutic target of ischemic stroke (Hao et al., 2014). m6A methylation plays an important role in various developmental decisions including angiogenesis (Mathiyalagan et al., 2019). Studies using zebrafish showed that RNA m6A methylation determines the differentiation stage of cells during the endothelial-to-hematopoietic transition (EHT) process in the development of embryos. This is achieved via the continuous activation of Notch signal transduction mediated by the m6A methyltransferase METTL3 in arterial endothelial cells (Zhang et al., 2017; Lv et al., 2018). In malignant hematological diseases, such as acute myeloid leukemia (AML), the quantity of the mRNA modified by METTL3 in blood cells is significantly increased (Vu et al., 2017). Rong et al. found that the density of FTO protein in cells was negatively correlated with micro-vessel density (MVD; Rong et al., 2019). Zhu et al. showed that total Panax notoginseng saponin (TPNS) can modulate the WTAP/p16 signaling axis through m6A modification in vascular smooth muscle cell (VSMC). Increased m6A modification also inhibited vascular intimal hyperplasia, intravascular smooth muscle migration and hyperplasia potential (Zhu et al., 2020).
RNA methylation also regulates angiogenesis and blood vessel repair. In vitro studies shows that RNA methylation affects the endothelial cells viability, proliferation, migration, and tube formation (Goyal and Goyal, 2019). Studies showed that m6A methylation level was significantly upregulated in endothelial cells following hypoxic stress and had positive effects on blood vessel repair and angiogenesis (Wang L.-J. et al., 2020; Yao et al., 2020). For example, METTL3 promotes angiogenesis via up-regulating putative arterial endothelial marker, hairy and enhancer of split-related with YRPW motif 2 (HEY2), which plays an important role in the formation of capillary-like tubes and endothelial cell migration (Yao et al., 2020).
RNA methylation also promotes angiogenesis via affecting cytokine secretion such as angiogenic growth factor (Zaitseva et al., 2019). METTL3 can promote angiogenesis by catalyzing m6A methylation and improving the stability of hepatoma-derived growth factor (HDGF) mRNA (Wang Q. et al., 2020). IGF2BP3 can recognize and bound to the m6A methylation on the mRNA of HDGF and VEGF mRNA. This combination leads to increased expression and stability of HDGF and VEGF and subsequently promote the angiogenesis (Wang Q. et al., 2020; Yang et al., 2020). METTL14/ALKBH5 are also proven to be important molecules that affect angiogenesis. They constitute a positive feedback loop with the RNA stability factor HuR, and promotes epithelial-mesenchymal transition by activating the gene expression of transforming growth factor β (TGF β) signaling pathway (Bertero et al., 2018). Hypoxia is found to reduce the m6A methylation and angiogenesis effects of METTL14/ALKBH5 by affecting the activity of related molecules (Panneerdoss et al., 2018).
4.2. RNA m6A methylation in neuron regeneration
According to current studies, the effect of RNA methylation in nervous system regeneration and repair is mainly achieved via promoting neuronal development and repair. m6A modification played an important role in regulating gene expression and cell differentiation of NPCs (radial keratinocytes; Yoon et al., 2017) and adult neural stem cells (aNSCs; Li L. et al., 2017) in the process of neurogenesis in the mammalian brain. For example, m6A modification can prolong cell cycle and delay cellular differentiation by promoting the decay of key mRNAs. m6A methylation can also affect the self-renewal, differentiation and lineage determination of various stem cells (Yoon et al., 2017; Wang et al., 2018). m6A methylation is enriched in highly conserved motifs of aNSCs transcriptome. This partially explains its role in promoting the proliferation of aNSCs and the morphological maturation of newborn neurons in the adult brain (Chen J. et al., 2019). By knocking out METTL14 or FTO in mouse embryos, the occurrence of cortical nerves can be delayed, and the development of nervous system is defects to varying degrees (Li L. et al., 2017). A study in Drosophila has found that Nito (RBM14 in human) in the m6A methyltransferase complex control and regulate neuronal development activities, such as axon growth and branch, synapse formation by regulating m6A methylation activity in CCAP/bursicon neurons (Gu et al., 2017). Research by Li has shown that conditional exhaustion of the m6A reader protein Ythdf2 in mice can cause irreversible damage in embryonic neuronal differentiation and development, resulting in lethality in the later stages of embryonic development (Li M. et al., 2018).
Neuron injury was found to result in the increased m6A mRNA methylation modification and cellular level of METTL14 and YTHDF1 in adult mice dorsal root ganglia (DRG). Knocking down METTL14 and YTHDF1 genes results in significantly reduced regeneration of sensory axon (Weng et al., 2018). Methylated recognition proteins also play an important role in regulating neurodevelopment. For example, FMRP has the priority to bind RNA probes containing m6A chemical modifications (Edupuganti et al., 2017). FMRP is involved in nervous system development and synaptic plasticity (Hagerman and Polussa, 2015), which is mainly achieved by regulating alternative mRNA splicing, mRNA stability, mRNA dendritic transport and partial post-synaptic local protein synthesis of mRNA (Didiot et al., 2008; Bechara et al., 2009; Ascano et al., 2012; Guo et al., 2015). Some researchers have found abundant m6A methylation on the target mRNA of FMRP (Chang et al., 2017). Moreover, the abundance of FMRP target mRNA in the cytoplasm decreased in FMRP-KO mice, which indicates that FMRP may affect the nuclear export of m6A-modified RNA (Hsu et al., 2019). These studies suggest that methylation recognition protein plays an important role in promoting the differentiation and development of neurons.
4.3. RNA m6A methylation in other CNS cells
Glial cells are the most abundant cells and induce several changes in pathological conditions such as inflammation, demyelination and disruption of BBB (You et al., 2022). RNA m6A modification is observed to regulate microglia’s inflammatory processes (Zhang F. et al., 2022). Studies suggest that METTL3 promotes lipopolysaccharide (LPS)-induced microglial inflammation by activating the TNF receptor associated factor 6 (TRAF6)-NF-κB pathway (Wen et al., 2022) and improves neuronal apoptosis and microglial activation by inactivating MyD88/NF-κB pathway (Chen Y. et al., 2022). In addition, m6A reader Igf2bp1 is reported to regulate the inflammatory processes of microglia via enhancing the m6A methylation and stabilizing Gbp11 and Cp mRNAs (Ding et al., 2022). It is reported that microRNA-421–3p could prevent inflammatory response in cerebral ischemia/reperfusion injury through targeting m6A reader YTHDF1 to inhibit p65 mRNA translation, which may provide a target for ischemia treatment (Zheng et al., 2020). The m6A methylation also participates in the development of autogenic immune diseases (Zhou et al., 2021b) and the change of dopaminergic neuron function (Teng et al., 2021).
RNA m6A methylation plays an essential role in the development of glia cells and brain tumor (Wang J. et al., 2021). Research conducted by Chang G et al. shows that YTHDF3 promotes cancer cell interactions with brain endothelial cells and astrocytes, BBB extravasation and angiogenesis via enhancing the translation of m6A-enriched transcripts for ST6GALNAC5, GJA1 and epidermal growth factor receptor (EGFR), which are all associated with cancer brain metastasis (Chang et al., 2020). Another study suggests that glioma stem-like cells (GSCs) radio resistance is mediated by m6A modification (Visvanathan et al., 2018). Cytoplasmic polyadenylation element binding protein 2 (CPEB2) m6A methylation regulates BBB permeability via regulating splicing factor SRSF5 stability, which could serve as a target for improving glioma-specific chemotherapeutic effects (Zhang M. et al., 2022). Besides, m6A regulation is also associated with spinal cord injury (SCI) and may contribute to spinal cord regeneration (Xing et al., 2021). RNA m6A methylation is proven to be crucial for oligodendrocyte maturation and CNS myelination (Xu et al., 2020). Current study shows that Prrc2a plays an important role in oligodendrocyte specification through functioning as a novel m6A reader, suggesting a therapeutic strategy for hypomyelination-related neurological diseases (Wu R. et al., 2019).
Pericytes are also important parts of NVU. m6A RNA modification in pericyte can lead to pericyte dysfunction, which induces vascular complication. Study suggests that METTL3-mediated m6A methylation regulates diabetes-induced pericyte dysfunction, which could be a potential therapeutical target for diabetes-induced retinal vascular complication treatment (Suo et al., 2022). Moreover, the results of m6A high-throughput sequencing suggests that hypertension is potentially involved in the changes in m6A methylation level in microvascular pericytes (Wu Q. et al., 2019). Although there are no studies on nervous system diseases, RNA m6A methylation in pericyte is supposed to play an essential part in NVU remodeling.
5. RNA m6A methylation in CNS diseases
NVU homeostasis disturbance and function loss are observed in neurological diseases caused by ischemia (Cai et al., 2017a; Zagrean et al., 2018). The persistent NVU dysfunction is thought to underlie the development of post-traumatic brain injury (TBI) neurodegeneration and late-onset neurodegenerative diseases (Zhou et al., 2020). Therefore, the treatment and recovery also depend on neurovascular remodeling. Research by Chokkalla has shown that compared with the control group, m6A methylation level in the ischemic stroke group increased significantly, mainly through suppressing m6A demethylase (such as FTO; Chokkalla et al., 2019). According to the effect of RNA methylation in the generation and repair of nervous system and blood vessels, RNA methylation can be regarded as a crucial target in the prevention, diagnosis and treatment of related diseases (Wei et al., 2017). For NVU remodeling, nutrition supply is the foremost requirement, which can be achieved by the newborn blood vessels. Considering the promising effect of m6A RNA methylation in angiogenesis, it is possible to be applied to facilitate nervous system repair and regeneration. For example, TPNS can prevent the proliferation of vascular intima and smooth muscle by downregulating m6A methylation level, which can be applied to arterial restenosis (Zhu et al., 2020).
Researchers are making great efforts to seek breakthrough in neuropathy based on RNA methylation and NVU remodeling. Wang et al. revealed the mechanism of m6A modification in regulating angiogenesis and provided potential pharmacological targets to prevent the formation and progression of cerebral arteriovenous malformation (Wang L.-J. et al., 2020). Zhang et al.’s work suggests that the neuroprotective effects of 2-(2-benzofuranyl)-2-imidazoline (2-BFI) in acute ischemic brain damage are at least partly due to the drug’s ability to improve the functions of NVU (Zhang et al., 2018). Brooks et al. found that modulating endothelial barrier function of NVU may provide new therapeutic approaches to improving outcomes in cerebral malaria (Brooks and Hawkes, 2017). However, most studies describe m6A RNA methylation or NVU remodeling separately, but few investigate the combined influence of these two or the mechanisms and effects of m6A in NVU remodeling. More studies are needed in the future to find therapeutic approaches based on the m6A RNA methylation and NVU remodeling.
6. Outlook
NVU is a holistic concept that includes cellular components such as neurons, blood vessels, and local microenvironmental components such as ECM and regulatory factors. Previous studies have found newborn neurons in ischemic brain, which can gradually mature over time, replace old neurons in structure and function, and integrate into the neural network to restore brain function (Gu et al., 2000; Zhang et al., 2006). It has been widely accepted that neuroprotective approaches to prevent brain deficits or restore neurofunction should target NVU as a whole rather than neurons alone. Both newborn neurons and neural stem cells count on the special microenvironment created by the surrounding blood vessels and glial cells (Osipova et al., 2018). Existing research confirmed that NVU played an important role in brain development (Yoon et al., 2017), BBB formation and maintenance (Andreone et al., 2015), etc. Targeting NVU remodeling is a promising therapeutic approach for treating cerebral ischemic diseases (del Zoppo, 2010), neurodegenerative diseases (De Strooper and Karran, 2016) and vascular dementia (Iadecola, 2013). RNA m6A methylation is attracting more attention in current gene expression regulation research. At present, many studies focus on the role of RNA m6A methylation in the nervous system or blood vessels, but few on the combination of these two. Meanwhile, studies on RNA m6A methylation in NVU remodeling and brain function restoration is still limited. Finding the relationship and mechanism between NVU remodeling and RNA m6A methylation is of great significance for the prognosis of various nervous system diseases.
RNA m6A methylation related studies in other diseases may provide directions for future research in NVU remodeling and neurofunction recovery. For example, excessive METTL3-mediated m6A modification attenuated the RNA stability of autophagy-related 7 (ATR7) in osteoarthritic chondrocytes. Decreased ATR7 level prevented the formation of autophagosomes and promoted cellular senescence (Chen X. et al., 2022), which could be applied to studying neurodegenerative diseases, such as AD. However, attention also should be paid that reduced expression of methyltransferase complex and subsequent decreased m6A methylation is related to cell proliferation as well as tumorigenicity (Liu et al., 2018). Therefore, a precise spatiotemporal control of m6A methylation is crucial for future clinical application. Orchestrating the m6A RNA methylation level of different cells of NVU is another challenge. Besides, noncell components also influence NVU remodeling. Further studies are needed in this area to provide advanced findings before we can apply to clinical research.
Author contributions
MZ conceptualized the study, acquired funding, and administered the project. YF and XL wrote the original draft. ZC and YP reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Natural Science Foundations for Excellent Young Scholars of Hunan Province (no. 2021JJ20095), the Key Research and Development Program of Hunan Province (no. 2020SK2063), Research Project on Education and Teaching Innovation of Central South University (no. 2021jy145), the Natural Science Foundations of Hunan Province (no. 2020JJ4134), the National Natural Science Foundation of China (no. 81501025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aas, A., Isakson, P., Bindesbøll, C., Alemu, E. A., Klungland, A., and Simonsen, A. (2017). Nucleocytoplasmic shuttling of FTO does not affect starvation-induced autophagy. PLoS One 12:e0168182. doi: 10.1371/journal.pone.0168182
Adhikari, S., Xiao, W., Zhao, Y. L., and Yang, Y. G. (2016). M(6)a: signaling for mRNA splicing. RNA Biol. 13, 756–759. doi: 10.1080/15476286.2016.1201628
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cells 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Andreone, B. J., Lacoste, B., and Gu, C. (2015). Neuronal and vascular interactions. Annu. Rev. Neurosci. 38, 25–46. doi: 10.1146/annurev-neuro-071714-033835
Andres, R. H., Horie, N., Slikker, W., Keren-Gill, H., Zhan, K., Sun, G., et al. (2011). Human neural stem cells enhance structural plasticity and axonal transport in the ischaemic brain. Brain 134, 1777–1789. doi: 10.1093/brain/awr094
Arimura, K., Ago, T., Kamouchi, M., Nakamura, K., Ishitsuka, K., Kuroda, J., et al. (2012). PDGF receptor β Signaling in pericytes following ischemic brain injury. Curr. Neurovasc. Res. 9, 1–9. doi: 10.2174/156720212799297100
Ascano, M., Mukherjee, N., Bandaru, P., Miller, J. B., Nusbaum, J. D., Corcoran, D. L., et al. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386. doi: 10.1038/nature11737
Bartosovic, M., Molares, H. C., Gregorova, P., Hrossova, D., Kudla, G., and Vanacova, S. (2017). N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing. Nucleic Acids Res. 45, 11356–11370. doi: 10.1093/nar/gkx778
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi: 10.1016/j.stem.2014.09.019
Bechara, E. G., Didiot, M. C., Melko, M., Davidovic, L., Bensaid, M., Martin, P., et al. (2009). A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 7:e16. doi: 10.1371/journal.pbio.1000016
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 interactome reveals that TGFβ controls m6A mRNA methylation in pluripotency. Nature 555, 256–259. doi: 10.1038/nature25784
Berulava, T., and Horsthemke, B. (2010). The obesity-associated SNPs in intron 1 of the FTO gene affect primary transcript levels. Eur. J. Hum. Genet. 18, 1054–1056. doi: 10.1038/ejhg.2010.71
Blanco, S., Kurowski, A., Nichols, J., Watt, F. M., Benitah, S. A., and Frye, M. (2011). The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7:e1002403. doi: 10.1371/journal.pgen.1002403
Blinkouskaya, Y., Caçoilo, A., Gollamudi, T., Jalalian, S., and Weickenmeier, J. (2021). Brain aging mechanisms with mechanical manifestations. Mech. Ageing Dev. 200:111575. doi: 10.1016/j.mad.2021.111575
Bodi, Z., Bottley, A., Archer, N., May, S. T., and Fray, R. G. (2015). Yeast m6A methylated mRNAs are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PLoS One 10:e0132090. doi: 10.1371/journal.pone.0132090
Bohlen, C. J., Friedman, B. A., Dejanovic, B., and Sheng, M. (2019). Microglia in brain development, homeostasis, and neurodegeneration. Annu. Rev. Genet. 53, 263–288. doi: 10.1146/annurev-genet-112618-043515
Bokar, J. A., Shambaugh, M. E., Polayes, D., Matera, A. G., and Rottman, F. M. (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3(11, 1233–1247.
Brooks, H. M., and Hawkes, M. T. (2017). Repurposing pharmaceuticals as neuroprotective agents for cerebral malaria. Curr. Clin. Pharmacol. 12, 62–72. doi: 10.2174/1574884712666170704144042
Brown, C. E., Aminoltejari, K., Erb, H., Winship, I. R., and Murphy, T. H. (2009). In vivo voltage-sensitive dye imaging in adult mice reveals that somatosensory maps lost to stroke are replaced over weeks by new structural and functional circuits with prolonged modes of activation within both the peri-infarct zone and distant sites. J. Neurosci. 29, 1719–1734. doi: 10.1523/JNEUROSCI.4249-08.2009
Cai, W., Liu, H., Zhao, J., Chen, L. Y., Chen, J., Lu, Z., et al. (2017b). Pericytes in brain injury and repair after ischemic stroke. Transl. Stroke Res. 8, 107–121. doi: 10.1007/s12975-016-0504-4
Cai, W., Zhang, K., Li, P., Zhu, L., Xu, J., Yang, B., et al. (2017a). Dysfunction of the neurovascular unit in ischemic stroke and neurodegenerative diseases: an aging effect. Ageing Res. Rev. 34, 77–87. doi: 10.1016/j.arr.2016.09.006
Chang, M., Lv, H., Zhang, W., Ma, C., He, X., Zhao, S., et al. (2017). Region-specific RNA m6A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7:170166. doi: 10.1098/rsob.170166
Chang, G., Shi, L., Ye, Y., Shi, H., Zeng, L., Tiwary, S., et al. (2020). YTHDF3 induces the translation of mA-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell 38, 857–871.e7. doi: 10.1016/j.ccell.2020.10.004
Chang, H., Yang, J., Wang, Q., Zhao, J., and Zhu, R. (2022). Role of N6-methyladenosine modification in pathogenesis of ischemic stroke. Expert. Rev. Mol. Diagn. 22, 295–303. doi: 10.1080/14737159.2022.2049246
Chen, Y., Cao, P., Xiao, Z., and Ruan, Z. (2022). M(6)a methyltransferase METTL3 relieves cognitive impairment of hyperuricemia mice via inactivating MyD88/NF-κB pathway mediated NLRP3-ASC-Caspase1 inflammasome. Int. Immunopharmacol. 113:109375. doi: 10.1016/j.intimp.2022.109375
Chen, X., Gong, W., Shao, X., Shi, T., Zhang, L., Dong, J., et al. (2022). METTL3-mediated m6A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann. Rheum. Dis. 81, 85–97. doi: 10.1136/annrheumdis-2021-221091
Chen, J., Zhang, Y. C., Huang, C., Shen, H., Sun, B., Cheng, X., et al. (2019). mA regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinformatics 17, 154–168. doi: 10.1016/j.gpb.2018.12.007
Chen, X.-Y., Zhang, J., and Zhu, J.-S. (2019). The role of mA RNA methylation in human cancer. Mol. Cancer 18:103. doi: 10.1186/s12943-019-1033-z
Chhor, V., le Charpentier, T., Lebon, S., Oré, M. V., Celador, I. L., Josserand, J., et al. (2013). Characterization of phenotype markers and neuronotoxic potential of polarised primary microglia in vitro. Brain Behav. Immun. 32, 70–85. doi: 10.1016/j.bbi.2013.02.005
Choi, J. Y., Kim, J. Y., Kim, J. Y., Park, J., Lee, W. T., and Lee, J. E. (2017). M2 phenotype microglia-derived cytokine stimulates proliferation and neuronal differentiation of endogenous stem cells in ischemic brain. Exp. Neurobiol. 26, 33–41. doi: 10.5607/en.2017.26.1.33
Chokkalla, A. K., Mehta, S. L., Kim, T. H., Chelluboina, B., Kim, J., and Vemuganti, R. (2019). Transient focal ischemia significantly alters the m6A Epitranscriptomic tagging of RNAs in the brain. Stroke 50, 2912–2921. doi: 10.1161/STROKEAHA.119.026433
Clarkson, A. N., Huang, B. S., MacIsaac, S. E., Mody, I., and Carmichael, S. T. (2010). Reducing excessive GABA-mediated tonic inhibition promotes functional recovery after stroke. Nature 468, 305–309. doi: 10.1038/nature09511
Colton, C. A. (2009). Heterogeneity of microglial activation in the innate immune response in the brain. J. Neuroimmune Pharmacol. 4, 399–418. doi: 10.1007/s11481-009-9164-4
Damoiseaux, J. S. (2017). Effects of aging on functional and structural brain connectivity. NeuroImage 160, 32–40. doi: 10.1016/j.neuroimage.2017.01.077
De Strooper, B., and Karran, E. (2016). The cellular phase of Alzheimer's disease. Cells 164, 603–615. doi: 10.1016/j.cell.2015.12.056
del Zoppo, G. J. (2010). The neurovascular unit in the setting of stroke. J. Intern. Med. 267, 156–171. doi: 10.1111/j.1365-2796.2009.02199.x
Deng, X., Su, R., Weng, H., Huang, H., Li, Z., and Chen, J. (2018). RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28, 507–517. doi: 10.1038/s41422-018-0034-6
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Didiot, M.-C., Tian, Z., Schaeffer, C., Subramanian, M., Mandel, J. L., and Moine, H. (2008). The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 36, 4902–4912. doi: 10.1093/nar/gkn472
Ding, L., Wu, H., Wang, Y., Li, Y., Liang, Z., Xia, X., et al. (2022). m6A reader Igf2bp1 regulates the inflammatory responses of microglia by stabilizing and mRNAs. Front. Immunol. 13:872252. doi: 10.3389/fimmu.2022.872252
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Edison, P. (2020). Brain connectivity: structural and functional neuronal integrity and its relationship with pathological substrates. Brain Connect. 10, 106–107. doi: 10.1089/brain.2020.29008.ped
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N-methyladenosine (mA) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi: 10.1038/nsmb.3462
Fazi, F., and Fatica, A. (2019). Interplay between-Methyladenosine (mA) and non-coding RNAs in cell development and cancer. Front. Cell Dev. Biol. 7:116. doi: 10.3389/fcell.2019.00116
Figueroa, A., Kotani, H., Toda, Y., Mazan-Mamczarz, K., Mueller, E. C., Otto, A., et al. (2009). Novel roles of hakai in cell proliferation and oncogenesis. Mol. Biol. Cell 20, 3533–3542. doi: 10.1091/mbc.e08-08-0845
Forró, T., Bajkó, Z., Bălașa, A., and Bălașa, R. (2021). Dysfunction of the neurovascular unit in ischemic stroke: highlights on microRNAs and exosomes as potential biomarkers and therapy. Int. J. Mol. Sci. 22:5621. doi: 10.3390/ijms22115621
Goyal, D., and Goyal, R. (2019). Angiogenic transformation in human brain micro endothelial cells: whole genome DNA methylation and transcriptomic analysis. Front. Physiol. 10:1502. doi: 10.3389/fphys.2019.01502
Gu, W., Brännström, T., and Wester, P. (2000). Cortical neurogenesis in adult rats after reversible photothrombotic stroke. J. Cereb. Blood Flow Metab. 20, 1166–1173. doi: 10.1097/00004647-200008000-00002
Gu, T., Zhao, T., Kohli, U., and Hewes, R. S. (2017). The large and small SPEN family proteins stimulate axon outgrowth during neurosecretory cell remodeling in drosophila. Dev. Biol. 431, 226–238. doi: 10.1016/j.ydbio.2017.09.013
Guo, S., and Lo, E. H. (2009). Dysfunctional cell-cell signaling in the neurovascular unit as a paradigm for central nervous system disease. Stroke 40, S4–S7. doi: 10.1161/STROKEAHA.108.534388
Guo, W., Polich, E. D., Su, J., Gao, Y., Christopher, D. M., Allan, A. M., et al. (2015). Fragile X proteins FMRP and FXR2P control synaptic GluA1 expression and neuronal maturation via distinct mechanisms. Cell Rep. 11, 1651–1666. doi: 10.1016/j.celrep.2015.05.013
Hagerman, R. J., and Polussa, J. (2015). Treatment of the psychiatric problems associated with fragile X syndrome. Curr. Opin. Psychiatry 28, 107–112. doi: 10.1097/YCO.0000000000000131
Haley, M. J., and Lawrence, C. B. (2017). The blood-brain barrier after stroke: structural studies and the role of transcytotic vesicles. J. Cereb. Blood Flow Metab. 37, 456–470. doi: 10.1177/0271678X16629976
Hao, L., Zou, Z., Tian, H., Zhang, Y., Zhou, H., and Liu, L. (2014). Stem cell-based therapies for ischemic stroke. Biomed. Res. Int. 2014:468748. doi: 10.1155/2014/468748
Hatakeyama, M., Ninomiya, I., and Kanazawa, M. (2020). Angiogenesis and neuronal remodeling after ischemic stroke. Neural Regen. Res. 15, 16–19. doi: 10.4103/1673-5374.264442
Haussmann, I. U., Bodi, Z., Sanchez-Moran, E., Mongan, N. P., Archer, N., Fray, R. G., et al. (2016). m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540, 301–304. doi: 10.1038/nature20577
Hermann, D. M., and ElAli, A. (2012). The abluminal endothelial membrane in neurovascular remodeling in health and disease. Sci. Signal. 5:re4. doi: 10.1126/scisignal.2002886
Hermann, D. M., and Zechariah, A. (2009). Implications of vascular endothelial growth factor for postischemic neurovascular remodeling. J. Cereb. Blood Flow Metab. 29, 1620–1643. doi: 10.1038/jcbfm.2009.100
Hernandez-Segura, A., Nehme, J., and Demaria, M. (2018). Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453. doi: 10.1016/j.tcb.2018.02.001
Hou, Y., Dan, X., Babbar, M., Wei, Y., Hasselbalch, S. G., Croteau, D. L., et al. (2019). Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 15, 565–581. doi: 10.1038/s41582-019-0244-7
Hsu, P. J., Shi, H., Zhu, A. C., Lu, Z., Miller, N., Edens, B. M., et al. (2019). The RNA-binding protein FMRP facilitates the nuclear export of-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895. doi: 10.1074/jbc.AC119.010078
Huang, H., Weng, H., and Chen, J. (2020). The biogenesis and precise control of RNA m(6)a methylation. Trends Genet. 36, 44–52. doi: 10.1016/j.tig.2019.10.011
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Iadecola, C. (2013). The pathobiology of vascular dementia. Neuron 80, 844–866. doi: 10.1016/j.neuron.2013.10.008
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
John, G. R., Chen, L., Rivieccio, M. A., Melendez-Vasquez, C. V., Hartley, A., and Brosnan, C. F. (2004). Interleukin-1beta induces a reactive astroglial phenotype via deactivation of the rho GTPase-rock axis. J. Neurosci. 24, 2837–2845. doi: 10.1523/JNEUROSCI.4789-03.2004
Jolivel, V., Bicker, F., Binamé, F., Ploen, R., Keller, S., Gollan, R., et al. (2015). Perivascular microglia promote blood vessel disintegration in the ischemic penumbra. Acta Neuropathol. 129, 279–295. doi: 10.1007/s00401-014-1372-1
Joy, M. T., Ben Assayag, E., Shabashov-Stone, D., Liraz-Zaltsman, S., Mazzitelli, J., Arenas, M., et al. (2019). CCR5 is a therapeutic target for recovery after stroke and traumatic brain injury. Cells 176, 1143–1157.e13. doi: 10.1016/j.cell.2019.01.044
Ke, S., Alemu, E. A., Mertens, C., Gantman, E. C., Fak, J. J., Mele, A., et al. (2015). A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation. Genes Dev. 29, 2037–2053. doi: 10.1101/gad.269415.115
Krum, J. M., Mani, N., and Rosenstein, J. M. (2008). Roles of the endogenous VEGF receptors flt-1 and flk-1 in astroglial and vascular remodeling after brain injury. Exp. Neurol. 212, 108–117. doi: 10.1016/j.expneurol.2008.03.019
Lauro, C., Catalano, M., Trettel, F., and Limatola, C. (2015). Fractalkine in the nervous system: neuroprotective or neurotoxic molecule? Ann. N. Y. Acad. Sci. 1351, 141–148. doi: 10.1111/nyas.12805
Lemons, M. L., and Condic, M. L. (2006). Combined integrin activation and intracellular cAMP cause rho GTPase dependent growth cone collapse on laminin-1. Exp. Neurol. 202, 324–335. doi: 10.1016/j.expneurol.2006.06.008
Lence, T., Akhtar, J., Bayer, M., Schmid, K., Spindler, L., Ho, C. H., et al. (2016). m6A modulates neuronal functions and sex determination in drosophila. Nature 540, 242–247. doi: 10.1038/nature20568
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. doi: 10.1038/cr.2017.10
Li, L.-J., Fan, Y. G., Leng, R. X., Pan, H. F., and Ye, D. Q. (2018). Potential link between mA modification and systemic lupus erythematosus. Mol. Immunol. 93, 55–63. doi: 10.1016/j.molimm.2017.11.009
Li, Y., Li, H., Luo, Y., Li, X., Chen, Z., Zhang, W., et al. (2022). The alteration profiles of m6A-tagged circRNAs in the peri-infarct cortex after cerebral ischemia in mice. Front. Neurosci. 16:869081. doi: 10.3389/fnins.2022.869081
Li, S., Nie, E. H., Yin, Y., Benowitz, L. I., Tung, S., Vinters, H. V., et al. (2015). GDF10 is a signal for axonal sprouting and functional recovery after stroke. Nat. Neurosci. 18, 1737–1745. doi: 10.1038/nn.4146
Li, S., Overman, J. J., Katsman, D., Kozlov, S. V., Donnelly, C. J., Twiss, J. L., et al. (2010). An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat. Neurosci. 13, 1496–1504. doi: 10.1038/nn.2674
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411. doi: 10.1093/hmg/ddx128
Li, M., Zhao, X., Wang, W., Shi, H., Pan, Q., Lu, Z., et al. (2018). Ythdf2-mediated mA mRNA clearance modulates neural development in mice. Genome Biol. 19:69. doi: 10.1186/s13059-018-1436-y
Lin, S., Liu, J., Jiang, W., Wang, P., Sun, C., Wang, X., et al. (2019). METTL3 promotes the proliferation and mobility of gastric cancer cells. Open Med. 14, 25–31. doi: 10.1515/med-2019-0005
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Liu, K., Ding, Y., Ye, W., Liu, Y., Yang, J., Liu, J., et al. (2016). Structural and functional characterization of the proteins responsible for N6-Methyladenosine modification and recognition. Curr. Protein Pept. Sci. 17, 306–318. doi: 10.2174/1389203716666150901113553
Liu, J., Eckert, M. A., Harada, B. T., Liu, S. M., Lu, Z., Yu, K., et al. (2018). m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 20, 1074–1083. doi: 10.1038/s41556-018-0174-4
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, N., Zhou, K. I., Parisien, M., Dai, Q., Diatchenko, L., and Pan, T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063. doi: 10.1093/nar/gkx141
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M., and Kroemer, G. (2013). The hallmarks of aging. Cells 153, 1194–1217. doi: 10.1016/j.cell.2013.05.039
Lou, N., Takano, T., Pei, Y., Xavier, A. L., Goldman, S. A., and Nedergaard, M. (2016). Purinergic receptor P2RY12-dependent microglial closure of the injured blood-brain barrier. Proc. Natl. Acad. Sci. U. S. A. 113, 1074–1079. doi: 10.1073/pnas.1520398113
Luke, L. M., Allred, R. P., and Jones, T. A. (2004). Unilateral ischemic sensorimotor cortical damage induces contralesional synaptogenesis and enhances skilled reaching with the ipsilateral forelimb in adult male rats. Synapse 54(4, 187–199. doi: 10.1002/syn.20080
Lv, J., Zhang, Y., Gao, S., Zhang, C., Chen, Y., Li, W., et al. (2018). Endothelial-specific mA modulates mouse hematopoietic stem and progenitor cell development via notch signaling. Cell Res. 28, 249–252. doi: 10.1038/cr.2017.143
Ma, C., Chang, M., Lv, H., Zhang, Z. W., Zhang, W., He, X., et al. (2018). RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19:68. doi: 10.1186/s13059-018-1435-z
Maity, A., and Das, B. (2016). N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J. 283, 1607–1630. doi: 10.1111/febs.13614
Mathiyalagan, P., Adamiak, M., Mayourian, J., Sassi, Y., Liang, Y., Agarwal, N., et al. (2019). FTO-dependent N-Methyladenosine regulates cardiac function during Remodeling and repair. Circulation 139, 518–532. doi: 10.1161/CIRCULATIONAHA.118.033794
McGraw, S., Vigneault, C., and Sirard, M.-A. (2007). Temporal expression of factors involved in chromatin remodeling and in gene regulation during early bovine in vitro embryo development. Reproduction 133, 597–608. doi: 10.1530/REP-06-0251
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m6A promotes cap-independent translation. Cells 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cells 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Osipova, E. D., Semyachkina-Glushkovskaya, O. V., Morgun, A. V., Pisareva, N. V., Malinovskaya, N. A., Boitsova, E. B., et al. (2018). Gliotransmitters and cytokines in the control of blood-brain barrier permeability. Rev. Neurosci. 29, 567–591. doi: 10.1515/revneuro-2017-0092
Overman, J. J., Clarkson, A. N., Wanner, I. B., Overman, W. T., Eckstein, I., Maguire, J. L., et al. (2012). A role for ephrin-A5 in axonal sprouting, recovery, and activity-dependent plasticity after stroke. Proc. Natl. Acad. Sci. U. S. A. 109, E2230–E2239. doi: 10.1073/pnas.1204386109
Ozaki, T., Nakamura, H., and Kishima, H. (2019). Therapeutic strategy against ischemic stroke with the concept of neurovascular unit. Neurochem. Int. 126, 246–251. doi: 10.1016/j.neuint.2019.03.022
Pan, W., and Kastin, A. J. (2007). Tumor necrosis factor and stroke: role of the blood-brain barrier. Prog. Neurobiol. 83, 363–374. doi: 10.1016/j.pneurobio.2007.07.008
Panneerdoss, S., Eedunuri, V. K., Yadav, P., Timilsina, S., Rajamanickam, S., Viswanadhapalli, S., et al. (2018). Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Advances 4:eaar8263. doi: 10.1126/sciadv.aar8263
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Ping, X.-L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Ponomarev, E. D., Veremeyko, T., and Weiner, H. L. (2013). MicroRNAs are universal regulators of differentiation, activation, and polarization of microglia and macrophages in normal and diseased CNS. Glia 61, 91–103. doi: 10.1002/glia.22363
Qin, Y., Li, L., Luo, E., Hou, J., Yan, G., Wang, D., et al. (2020). Role of m6A RNA methylation in cardiovascular disease (review). Int. J. Mol. Med. 46, 1958–1972. doi: 10.3892/ijmm.2020.4746
Qu, J., Yan, H., Hou, Y., Cao, W., Liu, Y., Zhang, E., et al. (2022). RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential. J. Hematol. Oncol. 15:8. doi: 10.1186/s13045-022-01224-4
Reali, C., Scintu, F., Pillai, R., Donato, R., Michetti, F., and Sogos, V. (2005). S100b counteracts effects of the neurotoxicant trimethyltin on astrocytes and microglia. J. Neurosci. Res. 81, 677–686. doi: 10.1002/jnr.20584
Reitmeir, R., Kilic, E., Kilic, Ü., Bacigaluppi, M., ElAli, A., Salani, G., et al. (2011). Post-acute delivery of erythropoietin induces stroke recovery by promoting perilesional tissue remodelling and contralesional pyramidal tract plasticity. Brain 134, 84–99. doi: 10.1093/brain/awq344
Rong, Z.-X., Li, Z., He, J. J., Liu, L. Y., Ren, X. X., Gao, J., et al. (2019). Downregulation of fat mass and obesity associated (FTO) promotes the progression of intrahepatic cholangiocarcinoma. Front. Oncol. 9:369. doi: 10.3389/fonc.2019.00369
Roost, C., Lynch, S. R., Batista, P. J., Qu, K., Chang, H. Y., and Kool, E. T. (2015). Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 137, 2107–2115. doi: 10.1021/ja513080v
Sato, Y., Falcone-Juengert, J., Tominaga, T., Su, H., and Liu, J. (2022). Remodeling of the neurovascular unit following cerebral ischemia and Hemorrhage. Cells 11:2823. doi: 10.3390/cells11182823
Satterwhite, E. R., and Mansfield, K. D. (2022). RNA methyltransferase METTL16: targets and function. Wiley Interdiscip Rev. RNA 13:e1681. doi: 10.1002/wrna.1681
Schaeffer, S., and Iadecola, C. (2021). Revisiting the neurovascular unit. Nat. Neurosci. 24, 1198–1209. doi: 10.1038/s41593-021-00904-7
Seo, S., Choi, C. H., Yi, K. S., Kim, S. U., Lee, K., Choi, N., et al. (2021). An engineered neurovascular unit for modeling neuroinflammation. Biofabrication 13:035039. doi: 10.1088/1758-5090/abf741
Sergeeva, O., Sergeev, P., Melnikov, P., Prikazchikova, T., Dontsova, O., and Zatsepin, T. (2020). Modification of Adenosine196 by Mettl3 methyltransferase in the 5′-external transcribed spacer of 47S pre-rRNA affects rRNA maturation. Cells 9:1061. doi: 10.3390/cells9041061
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017.15
Silbereis, J. C., Pochareddy, S., Zhu, Y., Li, M., and Sestan, N. (2016). The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268. doi: 10.1016/j.neuron.2015.12.008
Spitale, R. C., Flynn, R. A., Zhang, Q. C., Crisalli, P., Lee, B., Jung, J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. doi: 10.1038/nature14263
Stamatovic, S. M., Phillips, C. M., Martinez-Revollar, G., Keep, R. F., and Andjelkovic, A. V. (2019). Involvement of epigenetic mechanisms and non-coding RNAs in blood-brain barrier and neurovascular unit injury and recovery after stroke. Front. Neurosci. 13:864. doi: 10.3389/fnins.2019.00864
Suo, L., Liu, C., Zhang, Q. Y., Yao, M. D., Ma, Y., Yao, J., et al. (2022). METTL3-mediated-methyladenosine modification governs pericyte dysfunction during diabetes-induced retinal vascular complication. Theranostics 12, 277–289. doi: 10.7150/thno.63441
Teng, Y., Liu, Z., Chen, X., Liu, Y., Geng, F., le, W., et al. (2021). Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J. Cell. Mol. Med. 25, 8567–8572. doi: 10.1111/jcmm.16740
Thored, P., Heldmann, U., Gomes-Leal, W., Gisler, R., Darsalia, V., Taneera, J., et al. (2009). Long-term accumulation of microglia with proneurogenic phenotype concomitant with persistent neurogenesis in adult subventricular zone after stroke. Glia 57, 835–849. doi: 10.1002/glia.20810
Tramontina, F., Tramontina, A. C., Souza, D. F., Leite, M. C., Gottfried, C., Souza, D. O., et al. (2006). Glutamate uptake is stimulated by extracellular S100B in hippocampal astrocytes. Cell. Mol. Neurobiol. 26, 81–86. doi: 10.1007/s10571-006-9099-8
Underly, R. G., Levy, M., Hartmann, D. A., Grant, R. I., Watson, A. N., and Shih, A. Y. (2017). Pericytes as inducers of rapid, matrix metalloproteinase-9-dependent capillary damage during ischemia. J. Neurosci. 37, 129–140. doi: 10.1523/JNEUROSCI.2891-16.2016
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733. doi: 10.1093/nar/gkz619
Visvanathan, A., Patil, V., Arora, A., Hegde, A. S., Arivazhagan, A., Santosh, V., et al. (2018). Essential role of METTL3-mediated mA modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37, 522–533. doi: 10.1038/onc.2017.351
Vu, L. P., Pickering, B. F., Cheng, Y., Zaccara, S., Nguyen, D., Minuesa, G., et al. (2017). The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376. doi: 10.1038/nm.4416
Wang, Q., Chen, C., Ding, Q., Zhao, Y., Wang, Z., Chen, J., et al. (2020). METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 69, 1193–1205. doi: 10.1136/gutjnl-2019-319639
Wang, X., Feng, J., Xue, Y., Guan, Z., Zhang, D., Liu, Z., et al. (2017a). Corrigendum: structural basis of N-adenosine methylation by the METTL3-METTL14 complex. Nature 542:260. doi: 10.1038/nature21073
Wang, X., Huang, N., Yang, M., Wei, D., Tai, H., Han, X., et al. (2017b). FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 8:e2702. doi: 10.1038/cddis.2017.122
Wang, Y., Li, Y., Yue, M., Wang, J., Kumar, S., Wechsler-Reya, R. J., et al. (2018). N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 21, 195–206. doi: 10.1038/s41593-017-0057-1
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Wang, J., Sha, Y., and Sun, T. (2021). mA modifications play crucial roles in glial cell development and brain tumorigenesis. Front. Oncol. 11:611660. doi: 10.3389/fonc.2021.611660
Wang, L., Xiong, X., Zhang, L., and Shen, J. (2021). Neurovascular unit: a critical role in ischemic stroke. CNS Neurosci. Ther. 27, 7–16. doi: 10.1111/cns.13561
Wang, L.-J., Xue, Y., Huo, R., Yan, Z., Xu, H., Li, H., et al. (2020). N6-methyladenosine methyltransferase METTL3 affects the phenotype of cerebral arteriovenous malformation via modulating notch signaling pathway. J. Biomed. Sci. 27:62. doi: 10.1186/s12929-020-00655-w
Wang, Y., Zhang, X., Liu, H., and Zhou, X. (2021). Chemical methods and advanced sequencing technologies for deciphering mRNA modifications. Chem. Soc. Rev. 50, 13481–13497. doi: 10.1039/D1CS00920F
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cells 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Ward, R., Li, W., Abdul, Y., Jackson, L. D., Dong, G., Jamil, S., et al. (2019). NLRP3 inflammasome inhibition with MCC950 improves diabetes-mediated cognitive impairment and vasoneuronal remodeling after ischemia. Pharmacol. Res. 142, 237–250. doi: 10.1016/j.phrs.2019.01.035
Wei, K., Gao, Y., Wang, B., and Qu, Y. X. (2022). Methylation recognition protein YTH N6-methyladenosine RNA binding protein 1 (YTHDF1) regulates the proliferation, migration and invasion of osteosarcoma by regulating m6A level of CCR4-NOT transcription complex subunit 7 (CNOT7). Bioengineered 13, 5236–5250. doi: 10.1080/21655979.2022.2037381
Wei, W., Ji, X., Guo, X., and Ji, S. (2017). Regulatory role of N6-methyladenosine (m6A) methylation in RNA processing and human diseases. J. Cell. Biochem. 118, 2534–2543. doi: 10.1002/jcb.25967
Wen, L., Sun, W., Xia, D., Wang, Y., Li, J., and Yang, S. (2022). The m6A methyltransferase METTL3 promotes LPS-induced microglia inflammation through TRAF6/NF-κB pathway. Neuroreport 33, 243–251. doi: 10.1097/WNR.0000000000001550
Weng, Y.-L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325.e6. doi: 10.1016/j.neuron.2017.12.036
Widagdo, J., and Anggono, V. (2018). The m6A-epitranscriptomic signature in neurobiology: from neurodevelopment to brain plasticity. J. Neurochem. 147, 137–152. doi: 10.1111/jnc.14481
Wu, Y., Dissing-Olesen, L., MacVicar, B. A., and Stevens, B. (2015). Microglia: dynamic mediators of synapse development and plasticity. Trends Immunol. 36, 605–613. doi: 10.1016/j.it.2015.08.008
Wu, B., Li, L., Huang, Y., Ma, J., and Min, J. (2017). Readers, writers and erasers of N6-methylated adenosine modification. Curr. Opin. Struct. Biol. 47, 67–76. doi: 10.1016/j.sbi.2017.05.011
Wu, R., Li, A., Sun, B., Sun, J. G., Zhang, J., Zhang, T., et al. (2019). A novel mA reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 29, 23–41. doi: 10.1038/s41422-018-0113-8
Wu, Q., Yuan, X., Han, R., Zhang, H., and Xiu, R. (2019). Epitranscriptomic mechanisms of N6-methyladenosine methylation regulating mammalian hypertension development by determined spontaneously hypertensive rats pericytes. Epigenomics 11, 1359–1370. doi: 10.2217/epi-2019-0148
Xie, L., Mao, X., Jin, K., and Greenberg, D. A. (2013). Vascular endothelial growth factor-B expression in postischemic rat brain. Vasc. Cell 5:8. doi: 10.1186/2045-824X-5-8
Xing, L., Cai, Y., Yang, T., Yu, W., Gao, M., Chai, R., et al. (2021). Epitranscriptomic m6A regulation following spinal cord injury. J. Neurosci. Res. 99, 843–857. doi: 10.1002/jnr.24763
Xiong, Y., Mahmood, A., and Chopp, M. (2010). Angiogenesis, neurogenesis and brain recovery of function following injury. Curr. Opin. Investig. Drugs. 11(3, 298–308.
Xu, H., Dzhashiashvili, Y., Shah, A., Kunjamma, R. B., Weng, Y. L., Elbaz, B., et al. (2020). mA mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron 105, 293–309.e5. doi: 10.1016/j.neuron.2019.12.013
Yang, Z., Wang, T., Wu, D., Min, Z., Tan, J., and Yu, B. (2020). RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J. Exp. Clin. Cancer Res. 39:203. doi: 10.1186/s13046-020-01714-8
Yao, M.-D., Jiang, Q., Ma, Y., Liu, C., Zhu, C. Y., Sun, Y. N., et al. (2020). Role of METTL3-dependent N-Methyladenosine mRNA modification in the promotion of angiogenesis. Mol. Ther. 28, 2191–2202. doi: 10.1016/j.ymthe.2020.07.022
Yenari, M. A., Kauppinen, T. M., and Swanson, R. A. (2010). Microglial activation in stroke: therapeutic targets. Neurotherapeutics 7, 378–391. doi: 10.1016/j.nurt.2010.07.005
Yoon, K.-J., Ringeling, F. R., Vissers, C., Jacob, F., Pokrass, M., Jimenez-Cyrus, D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m6A methylation. Cells 171, 877–889.e17. doi: 10.1016/j.cell.2017.09.003
You, S., Su, X., Ying, J., Li, S., Qu, Y., and Mu, D. (2022). Research Progress on the role of RNA m6A modification in glial cells in the regulation of neurological diseases. Biomol. Ther. 12:1158. doi: 10.3390/biom12081158
Yu, X., Ji, C., and Shao, A. (2020). Neurovascular unit dysfunction and neurodegenerative disorders. Front. Neurosci. 14:334. doi: 10.3389/fnins.2020.00334
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential mA mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10. doi: 10.1038/s41421-018-0019-0
Yue, Y., Liu, J., and He, C. (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. doi: 10.1101/gad.262766.115
Zaccara, S., Ries, R. J., and Jaffrey, S. R. (2019). Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624. doi: 10.1038/s41580-019-0168-5
Zagrean, A.-M., Hermann, D. M., Opris, I., Zagrean, L., and Popa-Wagner, A. (2018). Multicellular crosstalk between exosomes and the neurovascular unit after cerebral ischemia. Therapeutic implications. Front. Neurosci. 12:811. doi: 10.3389/fnins.2018.00811
Zaitseva, T. S., Alcazar, C., Zamani, M., Hou, L., Sawamura, S., Yakubov, E., et al. (2019). Aligned nanofibrillar scaffolds for controlled delivery of modified mRNA. Tissue Eng. Part A 25, 121–130. doi: 10.1089/ten.tea.2017.0494
Zajicek, J. P., Wing, M., Scolding, N. J., and Compston, D. A. S. (1992). Interactions between oligodendrocytes and microglia. Brain 115, 1611–1631. doi: 10.1093/brain/115.6.1611-a
Zhang, C., Chen, Y., Sun, B., Wang, L., Yang, Y., Ma, D., et al. (2017). m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276. doi: 10.1038/nature23883
Zhang, N., Ding, C., Zuo, Y., Peng, Y., and Zuo, L. (2022). N6-methyladenosine and neurological diseases. Mol. Neurobiol. 59, 1925–1937. doi: 10.1007/s12035-022-02739-0
Zhang, W., Qu, J., Liu, G. H., and Belmonte, J. C. I. (2020). The ageing epigenome and its rejuvenation. Nat. Rev. Mol. Cell Biol. 21, 137–150. doi: 10.1038/s41580-019-0204-5
Zhang, F., Ran, Y., Tahir, M., Li, Z., Wang, J., and Chen, X. (2022). Regulation of N6-methyladenosine (m6A) RNA methylation in microglia-mediated inflammation and ischemic stroke. Front. Cell. Neurosci. 16:955222. doi: 10.3389/fncel.2022.955222
Zhang, Z., Sun, G. Y., and Ding, S. (2021). Glial cell line-derived neurotrophic factor and focal ischemic stroke. Neurochem. Res. 46, 2638–2650. doi: 10.1007/s11064-021-03266-5
Zhang, Z., Theler, D., Kaminska, K. H., Hiller, M., de la Grange, P., Pudimat, R., et al. (2010). The YTH domain is a novel RNA binding domain. J. Biol. Chem. 285, 14701–14710. doi: 10.1074/jbc.M110.104711
Zhang, R., Xue, Y. Y., Lu, S. D., Wang, Y., Zhang, L. M., Huang, Y. L., et al. (2006). Bcl-2 enhances neurogenesis and inhibits apoptosis of newborn neurons in adult rat brain following a transient middle cerebral artery occlusion. Neurobiol. Dis. 24, 345–356. doi: 10.1016/j.nbd.2006.07.012
Zhang, M., Yang, C., Ruan, X., Liu, X., Wang, D., Liu, L., et al. (2022). CPEB2 m6A methylation regulates blood-tumor barrier permeability by regulating splicing factor SRSF5 stability. Commun. Biol. 5:908. doi: 10.1038/s42003-022-03878-9
Zhang, Z., Zhang, L., Chen, J., Cao, Y., Qu, M., Lin, X., et al. (2018). 2-(2-benzofuranyl)-2-imidazoline mediates neuroprotection by regulating the neurovascular unit integrity in a rat model of focal cerebral ischemia. J. Stroke Cerebrovasc. Dis. 27, 1481–1489. doi: 10.1016/j.jstrokecerebrovasdis.2017.12.041
Zhang, Z. G., Zhang, L., Jiang, Q., Zhang, R., Davies, K., Powers, C., et al. (2000). VEGF enhances angiogenesis and promotes blood-brain barrier leakage in the ischemic brain. J. Clin. Invest. 106, 829–838. doi: 10.1172/JCI9369
Zhang, M., Zhang, Y., Ma, J., Guo, F., Cao, Q., Zhang, Y., et al. (2015). The demethylase activity of FTO (fat mass and obesity associated protein) is required for preadipocyte differentiation. PLoS One 10:e0133788. doi: 10.1371/journal.pone.0133788
Zhang, W., Zhao, J., Wang, R., Jiang, M., Ye, Q., Smith, A. D., et al. (2019). Macrophages reprogram after ischemic stroke and promote efferocytosis and inflammation resolution in the mouse brain. CNS Neurosci. Ther. 25, 1329–1342. doi: 10.1111/cns.13256
Zhao, L. Y., Song, J., Liu, Y., Song, C. X., and Yi, C. (2020). Mapping the epigenetic modifications of DNA and RNA. Protein Cell 11, 792–808. doi: 10.1007/s13238-020-00733-7
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Zheng, L., Tang, X., Lu, M., Sun, S., Xie, S., Cai, J., et al. (2020). microRNA-421-3p prevents inflammatory response in cerebral ischemia/reperfusion injury through targeting m6A reader YTHDF1 to inhibit p65 mRNA translation. Int. Immunopharmacol. 88:106937. doi: 10.1016/j.intimp.2020.106937
Zhou, Y., Chen, Q., Wang, Y., Wu, H., Xu, W., Pan, Y., et al. (2020). Persistent neurovascular unit dysfunction: pathophysiological substrate and trigger for late-onset neurodegeneration after traumatic brain injury. Front. Neurosci. 14:581. doi: 10.3389/fnins.2020.00581
Zhou, J., Wan, J., Gao, X., Zhang, X., Jaffrey, S. R., and Qian, S. B. (2015). Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. doi: 10.1038/nature15377
Zhou, H., Xu, Z., Liao, X., Tang, S., Li, N., and Hou, S. (2021b). Low expression of YTH domain-containing 1 promotes microglial M1 polarization by reducing the stability of Sirtuin 1 mRNA. Front. Cell. Neurosci. 15:774305. doi: 10.3389/fncel.2021.774305
Zhou, H., Yin, K., Zhang, Y., Tian, J., and Wang, S. (2021a). The RNA m6A writer METTL14 in cancers: roles, structures, and applications. Biochim. Biophys. Acta Rev. Cancer 1876:188609. doi: 10.1016/j.bbcan.2021.188609
Zhu, B., Gong, Y., Shen, L., Li, J., Han, J., Song, B., et al. (2020). Total panax notoginseng saponin inhibits vascular smooth muscle cell proliferation and migration and intimal hyperplasia by regulating WTAP/p16 signals via m6A modulation. Biomed. Pharmacother. 124:109935:109935. doi: 10.1016/j.biopha.2020.109935
Zou, S., Toh, J. D. W., Wong, K. H. Q., Gao, Y. G., Hong, W., and Woon, E. C. Y. (2016). N6-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep. 6:25677. doi: 10.1038/srep25677
Glossary
Keywords: neurovascular unit, 6-methyladenosine, CNS diseases, RNA methylation, m6A
Citation: Fan Y, Lv X, Chen Z, Peng Y and Zhang M (2023) m6A methylation: Critical roles in aging and neurological diseases. Front. Mol. Neurosci. 16:1102147. doi: 10.3389/fnmol.2023.1102147
Edited by:
Andreas Vlachos, University of Freiburg, GermanyReviewed by:
Tao Tao Lu, Anhui University of Chinese Medicine, ChinaBen Meir Maoz, Tel Aviv University, Israel
Copyright © 2023 Fan, Lv, Chen, Peng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengqi Zhang, ✉ zhangmengqi8912@163.com
 Yishu Fan
Yishu Fan Xinyi Lv
Xinyi Lv Zhuohui Chen1,2
Zhuohui Chen1,2  Mengqi Zhang
Mengqi Zhang