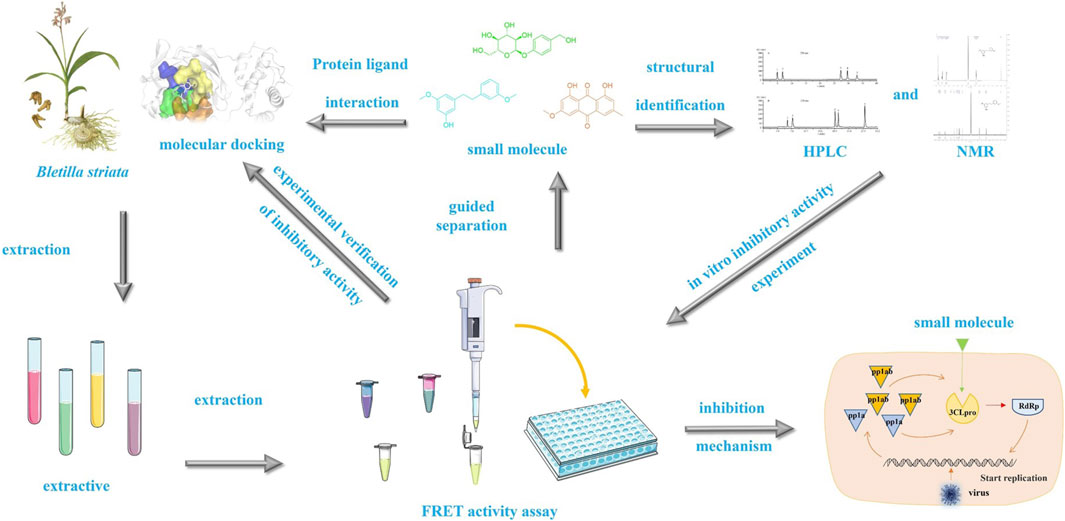
A study of the protease inhibitory activity component of SARS-CoV-2 3CL in Bletilla striata
- 1Hubei University of Science and Technology, Xianning, China
- 2Shenzhen University, Shenzhen, China
The protease Mpro (referred to as 3CLpro or 3CL protease) is a cysteine protease that is highly conserved in coronavirus and is indispensable for viral replication. Because there is no homologous protein for MPro in the human body, SARS-CoV-2 3CL protease is an ideal target against coronavirus. Bletilla striata (Reich. Bf.) is a well-known form of Traditional Chinese Medicine and can exert many pharmacological effects, including hemostasis, anti-microbial and anti-virus activities. Our preliminary screening showed that the n-butanol component of a methanol extract of B. striata exhibited potent inhibitory activity against SARS-CoV-2 3CL protease (58.82% at 200 μg/mL). In this study, we biologically evaluated ten isolated chemical compounds from B. striata and investigated the inhibitory activities of its constituents on SARS-CoV-2 3CL protease. Following bioactivity-guided fractionation, four bibenzyls (1, 4, 6 and 7), three phenols (5, 8 and 9), two anthraquinones (2 and 3) and one glucosyloxybenzyl 2-isobutylmalate (10) were isolated and evaluated for their ability to inhibit SARS-CoV-2 3CL protease by fluorescence resonance energy transfer (FRET) analysis. The binding mode between compounds and enzymes was investigated by molecular docking and Saturation Transfer Differences - nuclear magnetic resonance (STD-NMR). Moreover, Pleiobibenzynin B (7), Blestritin B, Gymconopin D (4), Physcion, 3′-O-methyl dioscin III (6), Gastrodin (8) and caffeic acid (5) all exhibited inhibitory activity against 3CL protease in vitro. The four bibenzyls (1, 4, 6 and 7) exhibited good inhibitory activity against SARS-CoV-2 3CL protease (7.37–39.86 μM). These findings identify potential new inhibitors against SARS-CoV-2 3CL protease, which offers promising lead compounds for the development of novel anti-SARS-CoV-2 drugs.
1 Introduction
Since 2002, there have been three serious global outbreaks caused by coronaviruses: Severe acute respiratory symdrom (SARS, 2002), Middle East Respiratory Syndrome (MERS, 2012) and Corona Virus Disease 2019 (COVID-19) (Liu et al., 2020; Shang et al., 2020). The COVID-19 epidemic, which began in December 2019, was a global public health event caused by a novel coronavirus (SARS-CoV-2). This epidemic caused significant social and economic losses across the entire world. SARS-CoV-2 is an enveloped, positive, and single-stranded RNA virus. The main protease in SARS-CoV-2 is Mpro (also known as 3C-like protease, 3CLpro, or 3CL protease), a cysteine protease that can hydrolyze viral polyprotein at more than 11 different sites. SARS-CoV-2 is a highly conserved form of coronavirus that infects cells expressing ACE2 and enters such cells by direct cell surface fusion or via an endosomal route. With regards to direct cell surface fusion, a host membrane protease (TMPRSS2) cleaves the virus spike protein to initiate the fusion of the virus and cell membranes in the host (Dai et al., 2020). Upon release into the cytoplasm, viral RNA will be translated into two polyproteins: PP1A and PP1AB. Once cleaved by two viral proteases, the main protease (Mpro) and the papain-like protease (PL pro), the released viral proteins can be assembled to form a viral polymerase RDRP (RNA dependent RNA polymerase, RdRP) complex to catalyze the replication of viral RNA (Dai et al., 2020; Hoffmann et al., 2020). SARS-CoV-2 3CL protease represents an ideal target against coronavirus because it has no homologous protein in the human body (Jin et al., 2020; Jo et al., 2020; Zhang et al., 2020).
On the 12th of February 2022, Paxlovid, a peptidoid SARS-CoV-2 3CL protease inhibitor developed by Pfizer Inc., was granted emergency authorization by the FDA (Chia et al., 2022; Najjar-Debbiny et al., 2023). At present, most patent applications for anti-coronavirus 3CL protease inhibitors in China are mainly submitted by scientific research institutes and universities. Only a limited number of applications are submitted from other enterprises. This demonstrates that this field of technology remains in the early phase of research and development. Moreover, the current technical application is predominantly biased towards extracting and separating appropriate compounds from plants (Chia et al., 2022). Historically, the extraction of active ingredients from plants has been an established practice for many decades. Monomer drugs extracted from plants have made indelible contributions to the lives and health of the global population and include artemisinin, morphine, ephedrine, and aspirin. These examples also remind us that medicinal plants represent a critical resource for novel drugs that have yet to be fully developed. However, the chemical compositions of most medicinal plants have yet to be fully characterized; consequently, their specific application in medicine remains unclear. Although a large number of chemical components have been extracted and isolated from plants, it is very difficult to identify new chemical structures from such plants. Therefore, the focus of research on medicinal plants has gradually shifted from identifying new chemical structures in traditional medicines to identifying new applications for substances with known chemical structures (Marzi et al., 2022; Jin and Huang, 2023). In the present study, we used FRET to search for a SARS-CoV-2 3CL protease inhibitor from natural sources and identified that the n-butanol fraction of a methanol extract of Bletilla striata exhibited inhibitory activity against 3CL proteases.
Bletilla striata is a dried tuber of the Orchidaceae and is predominantly distributed in Asia, North America, and Europe. Bletilla species are considered to be of significant medicinal importance in Asia, especially in China, Japan, and Korea. Bletillae Rhizoma (the tubers of Bletilla) have been widely used for the treatment of various hemorrhagic diseases, including gastrointestinal bleeding, cough and hemoptysis, gastric and duodenal ulcers, and traumatic diseases (He et al., 2009; Jiang et al., 2021). The aqueous and alcohol extracts of B. striata have also been shown to exert effects against influenza in vitro by inhibiting viral RNA synthesis and neuraminidase activity. However, the mechanisms of action associated with these molecules have yet to be investigated. Stilbenes (bibenzyls and phenanthrenes) and polysaccharides are considered as the main constituents of B. striata. Stilbenes compounds have previously been reported to exert antimicrobial and antitumor activities (Woo et al., 2014; Shi et al., 2017). In the present study, we evaluated ten chemical constituents identified in a 90% methanol extract of Bletilla tuber and performed biological evaluations to investigate whether these constituents exerted inhibitory activity of the SARS-CoV-2 3CL protease in vitro (Scheme 1).
2 Materials and methods
2.1 Separation of Bletilla striata extract
A dried powder of white B. striata (6 kg) was extracted with methyl alcohol (3 × 15 L) for a week at room temperature. The concentrated extract (456 g) was suspended in H2O (1 L) and the aqueous layer was sequentially partitioned with petroleum ether, ethyl acetate, and n-butanol until the extract became colorless. The obtained n-butanol layer (58 g) was separated by polyamide column chromatography and eluted with water, followed by 30%, 60% and 90% ethanol. Next, the extract was concentrated under reduced pressure and freeze-dried, and the 3CL protease inhibitory activity of different components present in the freeze-dried powder was determined by FRET.
The 30% ethanol elution component (18 g) was separated by silica gel column chromatography (inner diameter: 4 cm, 120 g), and eluted with chloroform:methanol (100:1–100:60). Five components (A-E) were obtained. Component A (3.2 g) was separated by silica gel column chromatography (inner diameter: 2.5 cm, 20 g) and cleaned with petroleum ether:ethyl acetate (30:1) to obtain 7 mg of 1(2-[(4-hydroxyphenyl)methyl]-5-methoxy-3-[2-(3-methoxyphenyl)ethyl]phenol, Gymconopin D). Component B (4.8 g) was separated by silica gel column chromatograph (inner diameter: 2.5 cm; 30 g) and cleaned by petroleum ether:ethyl acetate (16:1) to obtain four components. Component B3 (2.5 g) was repeatedly separated with petroleum ether and ethyl acetate (petroleum ether:ethyl acetate; 24:1) to obtain 2(2- methyl anthraquinone, 7 mg). Component C (5.6 g) was separated by silica gel column chromatography (petroleum ether:ethyl acetate; 12:1) to obtain five components. Component C3 (2.6 g) was repeatedly separated with petroleum ether and ethyl acetate (chloroform:methanol; 24:1) to obtain 3 (Physcion 8 mg) and 4(3-[2-(4-hydroxy-3-methoxyphenyl) ethyl]-2,4-bis[(4-hydroxyphenyl)methyl]-5-methoxyphenol, Blestritin B 7 mg). Component E (2.3 g) was separated by petroleum ether:ethyl acetate (8:1) and four components were obtained by silica gel column chromatography. Component E3 (0.9 g) was eluted with petroleum ether and ethyl acetate (petroleum ether:ethyl acetate; 12:1) to obtain 5 (caffeic acid, 5 mg).
The 60% ethanol elution component (21 g) was separated by silica gel column chromatography (inner diameter: 4 cm; 130 g), and eluted with chloroform:methanol (100:1–100:60). Five components (A-E) were obtained. Component A (3.2 g) was separated by silica gel column chromatography (inner diameter: 2.5 cm; 20 g) and cleaned by chloroform:methanol (100:60) to obtain 7 mg of 6(3′-O- methyl dioscin III). Component B (5.8 g) was separated by silica gel column chromatograph (inner diameter: 2.5 cm; 30 g) and cleaned by petroleum ether:ethyl acetate (24:1) to obtain four components. Component B3 (1.5 g) was repeatedly separated with petroleum ether and ethyl acetate (chloroform:methanol; 36:1) to obtain 7(3-[2-(3-hydroxyphenyl)ethyl]-2,4,6-tris[(4-hydroxyphenyl)methyl]-5-methoxyphenol (Pleiobibenzynin B; 8 mg). Component C (4.6 g) was separated by silica gel column chromatography (chloroform:methanol; 100:50) to obtain five components. Component C3 (1.8 g) was repeatedly separated with petroleum ether and ethyl acetate (chloroform:methanol; 100: 40) to obtain 8(Gastrodin 10 mg) and 9(Arbutin, 9 mg). Component E (2.3 g) was separated by chloroform:methanol (100:30) and four components were obtained by silica gel column chromatography. Component E3 (0.9 g) was eluted with petroleum ether and ethyl acetate (chloroform:methanol; 100: 60) to obtain 10 (Militarine 25 mg).
2.2 Conditions for 1H-NMR, 13C-NMR and ESI-MS
1H-NMR and 13C-NMR experiments were carried out on a 298 K Bruker 400 MHz instrument (AVANCE III, Bruker, Switzerland). Hydrogen spectrum scanning was performed 32 times, with 1,024 carbon spectrum scans; the excitation angles remained at 30 throughout. Experimental data were processed by Mest Renova software (https://mestrelab.com/download/mnova/) based on 1H-NMR and 13C-NMR data of known compounds that had been published previously and the structures of the compounds were preliminarily determined.
We used an electrospray ion source (ESI) with positive ion detection. The heating block temperature was set to 400°C and DL temperature was set to 250°C. The scanning time was 0.1 S and we applied multiple reaction monitoring (MRM). The electrospray voltage was 3.5 KV and N2 was used as the atomizing gas with a velocity of 3.0 L/min with a drier flow of 15.0 L/min. The collision gas was high-purity nitrogen and the collision gas pressure was 230 KPa.
2.3 Chromatographic methods and determination
2.3.1 HPLC analysis conditions for Arbutin, Gastrodin, Pleiobibenzynin B, Militarine, 3′-O-methyl dioscin III
HPLC analysis involved a Shimadzu-LC-16 liquid chromatography system (Japan), including an InertsilODS-3 C18 chromatographic column (250 mm × 4.6 mm, 5 µm) with a detection wavelength of 270 nm. The column was maintained at 35°C, with a flow rate of 0.8 mL/min, and a sample volume of 20 µL. With regards to the mobile phase, pump A included 0.1% phosphoric acid water while pump B included acetonitrile. The elution procedure was as follows: 0–10 min, 6%–15% B, 10–15 min, 15%–55% B, 15–20 min, 55%–65% B, 20–25 min, 65%–80% B, 25–30 min, and 80% B.
2.3.2 HPLC analysis conditions for Physcion, Blestritin B, 2-methyl anthraquinone, Gymconopin D, and Caffeic acid
HPLC analysis involved a Shimadzu-LC-16 liquid chromatograph system (Japan), including an InertsilODS-3 C18 chromatographic column (250 mm × 4.6 mm, 5 µm) with a detection wavelength of 254 nm. The column was maintained at 35°C, with a flow rate of 0.8 mL/min, and a sample volume of 20 µL. With regards to the mobile phase, pump A included 0.1% phosphoric acid water while pump B included acetonitrile. The elution procedure was as follows: 0–4 min, 35%–44% B, 4–6 min, 44%–40% B, 6–10 min, 40%–45% B, 10–15 min, 45%–65% B, 15–25 min, 65%–75% B, 25–30 min, 75%–85% B, 30–40 min, and 85%–95% B.
2.4 FRET-based enzymatic activity assays for the inhibition of SARS-CoV-2 3CLpro
Fluorescence resonance energy transfer (FRET) was used to determine the inhibitory activity of the compounds on SARS-CoV-2 3CL protease in vitro (Su et al., 2021). First, a 3CL protease fluorescent substrate peptide (Dabcyl-KTSAVLQSGFRKME-Edans) was synthesized by Nanjing Peptide Industry Biotechnology Co., Ltd.; a SARS-CoV-2 3CL protease was also prepared by Sigma (Sigma Aldrich Company, USA). The SARS-CoV-2 3CL protease (2.4 μg/mL at a final concentration) was mixed with serial dilutions of the sample compounds or herbal extracts (the final concentration for primary screening was 200 μg/mL) in 50 µL of assay buffer (50 mM Tris, pH 7.3, 1 mM EDTA) and incubated at 37°C for 10 min. The reaction was initiated by adding 200 µL of fluorogenic substrate at a final concentration of 10 μg/mL. Next, the fluorescence signal at 360 nm (excitation)/460 nm (emission) was measured every 45 s for 10 min with a Biotek synergy2 plate reader. The initial velocity of reactions with compounds was compared to a reaction with DMSO; these data were used to generate IC50 curves. For each compound, at least three independent experiments were performed for the determination of IC50 values and each experiment was carried out in triplicate. IC50 values are expressed as the mean from three independent experiments and were determined by non-linear regression analysis using GraphPad Prism software 9.5 (https://www.graphpad.com/).
2.5 STD-NMR experiments
All STD-NMR experiments were performed using a 400 MHz solution-state instrument comprising a Bruker Instruments superconducting magnet, Bruker Avance III console, and TCI probe head. Experiments were performed using TopSpin (Bruker). The Pulse program (stddiffesgp.3, Germany) was used for all STD-NMR experiments, with excitation sculpting for water suppression. For STD-NMR experiments, samples consisted of 100 μM of 3CL protease and 100 mM of each ligand compound formulated in NMR buffer supplement (10 mM Tris-d11 pH 7.8 and 100 mM NaCl) with 10% v/v D2O and deuterated dimethyl sulfoxide to a 20% v/v final DMSO-d6 concentration. All sample volumes were 600 µL (Kantsadi et al., 2021).
Saturation was achieved by applying a train of 50 ms Gaussian pulses (0.40 mW), resulting in a total saturation time of 3.5 s; the relaxation delay was set to 4 s, at 0.5 ppm (on-resonance experiments) and 36 ppm (off-resonance experiments). Protein signals were suppressed in STD-NMR by the application of a 30 ms spin-lock pulse. Data for each STD NMR experiment were acquired by a total of 1,056 scans. On- and off-resonance spectra were stored and processed separately, and the final STD-NMR spectra were obtained by subtracting the on-resonance from the off-resonance spectra. All STD effects were quantified using the following equation: ASTD = (I0 - Isat)/I0 = ISTD/I0. Thus, the signal intensities of the STD NMR spectrum (ISTD) were compared with the corresponding signal intensities of a reference spectrum (I0). The strongest STD signal in the spectrum was assigned a value of 100% and used as a reference to calculate relative STD effects accordingly (Guillon et al., 2014; Tamigney Kenfack et al., 2017; Kantsadi et al., 2021).
2.6 Molecular docking
Molecular docking was employed to investigate the interaction and types of interaction of each chemical compound with SARS-CoV-2 3CL protease. The binding modes of each compound with protease were studied by molecular docking and compared with the crystal structure of baicalein-3CL protease (PDB ID: 6m2n) (Su et al., 2021). We generated the structural formula of small molecules in Chem Draw 2D 20.0 software (https://revvitysignals.com/products/research/chemdraw), and determined the energy optimization of the small molecule structure by using MM2 force field in Chem Draw 3D 20.0 (https://revvitysignals.com/products/research/chemdraw). The crystal structure of SARS-CoV-2 3CL protease was downloaded from the protein database RCSB (www.rcsb.org) (PDB ID: 6m2n). PyMol (https://pymol.org/2/) was then used to remove water molecules and original eutectic ligands from the protein. Docking was completed by the software AutoDockTools-1.5.7 (https://autodocksuite.scripps.edu/adt/). Docking was completed in four steps. The first step was protein hydrogenation, charge calculation, and protein type assignment. The second step was the hydrogenation of small molecules to maintain their flexibility. The third step was to set the docking ranges X50, Y60, and Z50, with a grid spacing of 0.375, and the 3CL protein active center located at (−16.09, 19.895, 70, and 16) in the grid. The last step was to use the default parameters and Genetic Algorithm to dock molecules with the protein. Finally, we visualized docking in PyMol (https://pymol.org/2/).
3 Results and discussion
3.1 Isolation and identification
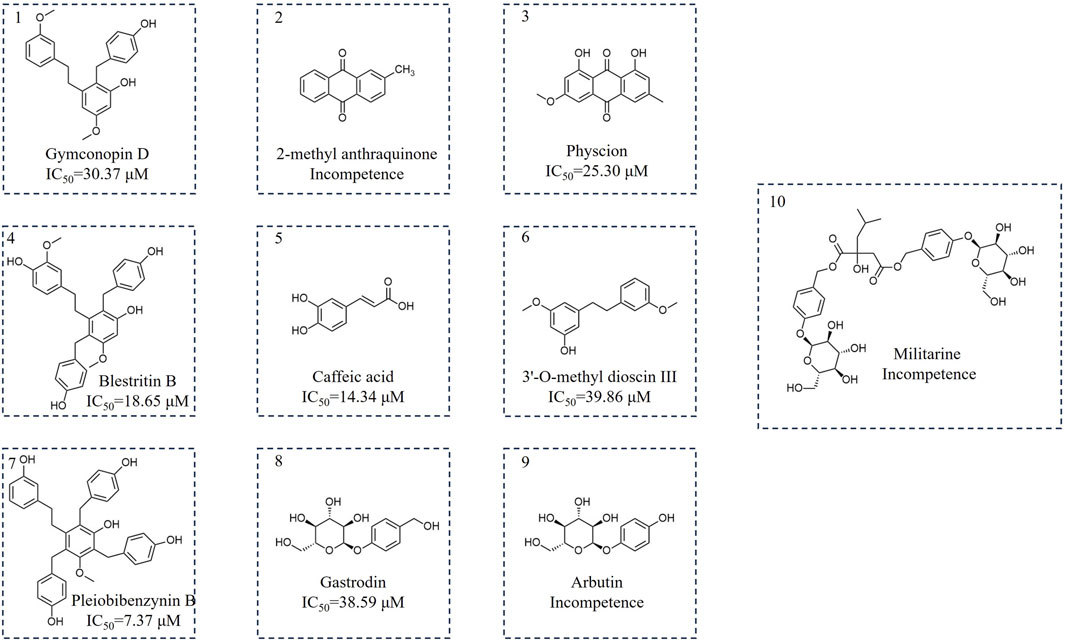
Following bioactivity-guided fractionation, we evaluated the inhibitory effects of the B. striata MeOH extract on SARS-CoV-2 3CL protease, along with petroleum ether, ethyl acetate and n-butanol fractions (the final concentration of the extract was 200 μg/mL). Analysis indicated that n-butanol (58.82% at 200 μg/mL) exhibited good inhibitory activity against 3CL protease. Then, we used a polyamide chromatographic column to isolate bioactive compounds from the fractions for further phytochemical investigation. The n-butanol extract was passed through a polyamide chromatographic column and eluted with water, followed by 30%, 60% and 90% ethanol. Analysis indicated that the 30% (63.06% at 200 μg/mL) and 60% (65.07% at 200 μg/mL) fractions had good inhibitory activity against 3CL protease. Then, we used other silica gel columns to isolate bioactive compounds from both fractions for further phytochemical investigation. The components were preliminarily identified by ESI-MS, 1H-NMR, and 13C-NMR, and the properties of the compounds were compared to previously published data. The compounds isolated from the 30% fraction (1–5) were identified as Gymconopin D, 2-methyl anthraquinone, Physcion, Blestritin B, and Caffeic acid; these were all known species. The compounds isolated from the 60% fraction (6–10) were identified as 3′-O-methyl dioscin III, Pleiobibenzynin B, Gastrodin, Arbutin, and Militarine; these were all known species (Figure 1).
Compound 1 was a white powder, 1H-NMR (400 MHz, CDCl3) δ 7.12 (t, J = 2.2 Hz, 1H), 6.93 (d, J = 2 Hz, 2H), 6.68 (br, 1H), 6.64 (br, 3H), 6.55 (tt, J = 2.2, 2.1 Hz, 1H), 6.45 (dt, J = 2.2, 2.1 Hz, 1H), 6.31 (d, J = 2.2 Hz, 1H), 3.87 (s, 2H), 3.73 (s, 3H), 3.66 (s, 3H), 2.72 (m, 2H), 2.57 (m, 2H). 13C-NMR (100 MHz, CDCl3) δ 160.14, 159.34, 156.74, 142.03, 141.57, 131.97, 130.24, 129.69, 121.83, 121.40, 119.70, 115.91, 114.24, 107.20, 98.88, 55.59, 55.18, 36.08, 35.20, 33.70. ESI-MS m/z 365.17 [M + H]+. This compound was tentatively identified as 2-[(4-hydroxyphenyl) methyl]-5-methoxy-3-[2-(3-methoxyphenyl) ethyl] phenol (Gymconopin D) (Matsuda et al., 2004).
Compound 2 was a yellowish amorphous powder. 1H-NMR (400 MHz, CDCl3) δ 8.29 (dd, J = 8 Hz, 2H), 8.19 (d, J = 8 Hz, 1H), 8.09 (d, 1H), 7.79 (m, J = 8 Hz 2 H), 7.58 (dd, J = 8 Hz, 1H), 2.53 (s, 3H). 13C-NMR (100 MHz, CDCl3) δ 183.56, 183.12, 145.42, 135.07, 134.17, 134.05, 133.74, 133.54, 131.43, 127.61 (d, J = 5.2 Hz), 127.28 (d, J = 2.2 Hz), 22.05. ESI-MS m/z 223.07 [M + H]+. Compound 2 was tentatively identified as 2-methyl anthraquinone (Sun et al., 2013).
Compound 3 was a yellow needle-like crystal. 1H-NMR (400 MHz, CDCl3) δ 12.30 (s, 1H), 12.10 (s, 1H), 7.61 (d, J = 4 Hz, 1H), 7.35 (d, J = 4 Hz, 1H), 7.07 (s, 1H), 6.68 (d, J = 4 Hz, 1H), 3.93 (s, 3H), 2.45 (s, 3H).13C-NMR (100 MHz, CDCl3) δ 190.94, 182.15, 166.70, 165.34, 162.65, 148.59, 135.40, 133.36, 124.65, 121.43, 113.82, 110.40, 108.36, 106.91, 56.23, 22.31. ESI-MS m/z 285.07 [M + H]+. Compound 3 was tentatively identified as Physcion (Guetchueng et al., 2023).
Compound 4 was a white amorphous powder. 1H-NMR (400 MHz, CDCl3) δ 6.94 (d, J = 8.4 Hz, 1H), 6.85 (d, J = 8.4 Hz, 1H), 6.65 (m, 3H), 6.59 (m, 2H), 6.48 (s, 1H), 6.44 (dd, J = 8.0, 1.8 Hz, 1H), 6.38 (d, J = 1.4 Hz, 1H), 3.90 (s, 4H), 3.79 (s, 3H), 3.77 (s, 3H), 2.66 (t, J = 8.2 Hz, 2H), 2.19 (m, 2H). 13C- NMR (100 MHz, CDCl3) δ 157.73, 156.74, 154.63, 147.85, 145.67, 141.74, 132.92, 131.88, 131.51, 130.22, 123.11, 121.94, 115.89, 115.55, 113.06, 96.59, 56.03, 35.56, 34.28, 33.89, 31.89. ESI-MS m/z 487.21 [M + H]+. Compound 4 was tentatively identified as 4(3-[2-(4-hydroxy-3-methoxyphenyl)eth-yl]-2,4-bis[(4-hydroxyphenyl)methyl]-5-methoxyphenol (Blestritin B) (Feng et al., 2008).
Compound 5 was a faint yellow crystalline powder. 1H-NMR (400 MHz, DMSO-d6) δ 12.05 (s, 1H), 9.69 (s, 2H), 7.43 (d, J = 16 Hz, 1H), 7.02 (d, 1H), 6.97 (dd, J = 12 Hz, 1H), 6.76 (d, J = 8 Hz, 1H), 6.18 (d, J = 8 Hz, 1H). 13C-NMR (100 MHz, DMSO-d6) δ 167.93, 148.15, 145.58, 144.60, 125.71, 121.18, 115.76, 115.14, 114.64. ESI-MS m/z 181.05 [M + H]+. Compound 5 was tentatively identified as caffeic acid (Hu et al., 2019).
Compound 6 was a white amorphous powder. 1H-NMR (400 MHz, CDCl3) δ 7.20 (t, J = 16 Hz, 1H), 6.73–6.80 (m, 3H), 6.33 (s, 1H), 6.26 (dd, J = 4 Hz, 2H), 3.77 (d, J = 16 Hz, 6H), 2.87–2.83 (m, 4H). 13C-NMR (100 MHz, CDCl3) δ 161.02, 159.74, 156.72, 144.64, 143.43, 129.46, 121.01, 114.35, 111.45, 108.09, 106.92, 99.21, 55.41, 55.31, 37.95, 37.73. ESI-MS m/z 259.13 [M + H]+. Compound 6 was tentatively identified as 3′-O-methyl dioscin III (Yamaki et al., 1989).
Compound 7 was a white amorphous powder. 1H-NMR (400 MHz, CDCl3) δ 7.09–7.04 (m, 1H), 7.03 (d, J = 8.1 Hz, 2H), 6.91 (d, J = 8.0 Hz, 2H), 6.84 (d, J = 8.0 Hz, 2H), 6.65 (d, J = 2.0 Hz, 2H), 6.64 (d, J = 8.0 Hz, 2H), 6.62 (d, J = 8.0 Hz, 2H), 6.48–6.43 (m, 3H), 4.03 (s, 3H), 3.94 (s, 2H), 3.46 (s, 3H), 2.67 (t, 2H), 2.41–2.29 (m, 2H).13C-NMR (100 MHz, CDCl3) δ 157.64, 156.73, 156.34, 154.74, 142.99, 138.77, 131.88, 131.67, 131.36, 130.23,130.02, 122.86, 120.84, 116.24, 115.90, 114.42, 61.35, 35.56, 34.31, 34.14, 31.85, 29.94. ESI-MS m/z 563.24 [M + H]+. Compound 7 was tentatively identified as 3-[2-(3-hydroxyphenyl)ethyl]-2,4,6-tris[(4-hydroxyphenyl)methyl]-5-methoxyphenyl (Pleiobibenzynin B) (Shiao et al., 2009).
Compound 8 was a yellowish amorphous powder. 1H-NMR (400 MHz, DMSO-d6) δ 7.22 (d, J = 8 Hz, 2H), 7.03–6.93 (m, J = 8 Hz, 2H), 5.29 (d, J = 4 Hz, 1H), 5.07 (t, J = 12 Hz, 1H), 5.01 (d, J = 4 Hz, 1H), 4.82 (d, J = 8 Hz, 1H), 4.55 (t, J = 12 Hz, 1H), 4.42 (d, J = 4 Hz, 1H), 3.68 (ddd, J = 4, 4, 4 Hz, 1H), 3.45 (dt, J = 24, 12 Hz, 1H), 3.31–3.12 (m, 2H). 13C-NMR (100 MHz, DMSO-d6) δ 156.32, 135.85, 127.71, 115.94, 100.55, 77.01, 76.65, 73.27, 69.75, 62.52, 60.72. ESI-MS m/z 287.11 [M + H]+. Compound 8 was tentatively identified as Gastrodin (Cai et al., 2007).
Compound 9 was a white crystalline powder. 1H-NMR (400 MHz, D2O) δ 7.06 (d, J = 8.0 Hz, 2H), 6.92–6.83 (d, J = 8.0 Hz 2H), 5.41 (d, J = 8 Hz, 1H), 4.06–3.92 (m, 2 H), 3.86–3.83 (m, 2H), 3.78–3.73 (m, 2H), 3.62–3.46 (m, 5H). 13C-NMR (100 MHz, D2O) δ 151.24, 150.40, 118.42, 116.21, 101.31, 76.04, 75.56, 72.96, 69.43, 60.54. ESI-MS m/z 273.09 [M + H]+. Compound 9 was tentatively identified as Arbutin (CHEN, 2019).
Compound 10 was a white crystalline powder. 1H-NMR (400 MHz, D2O) δ 7.33–7.23 (dd, J = 8, 8 Hz, 4 H), 7.11 (t, J = 16 Hz, 4H), 5.00–5.06 (m, 5H), 3.89 (t, 2H), 3.74 (dt, 2 H), 3.66–3.45 (m, 8 H), 3.08 (d, J = 16 Hz, 1H), 2.71 (d, J = 16 Hz, 1H), 1.73–1.56 (m, 1H), 0.87 (d, J = 8 Hz, 3H), 0.75 (d, J = 8 Hz, 3H). 13C-NMR (100 MHz, D2O) δ 175.71, 171.73, 156.78, 130.45 (d, J = 3.4 Hz), 129.93, 129.59, 116.70 (d, J = 5.1 Hz), 100.24 (d, J = 4.3 Hz), 76.05, 75.53, 72.87, 69.35, 67.23, 66.78, 60.49, 47.74, 45.09, 23.50, 22.51. ESI-MS m/z 727.28 [M + H]+. Compound 10 was tentatively identified as Militarine (CHEN, 2019).
3.2 Chromatographic methods
3.2.1 Screening of the mobile phase and chromatographic conditions
For the mobile phase, we used acetonitrile (A) and 0.1% formic acid water, acetonitrile (A) and 0.1% acetic acid water, acetonitrile (A) and 0.1% phosphoric acid water, methanol (A) and 0.1% acetic acid water and methanol(A) and 0.1% phosphoric acid water. We found that acetonitrile (A) and 0.1% phosphoric acid water had the best effect. We preliminarily determined the detection wavelengths of these compounds by Bypre-UV scanning. We found that Physcion, Blestritin B, 2-methyl anthraquinone, Gymconopin D, and Caffeic acid had the high absorption intensity at 254 nm. Therefore, these compounds were determined with acetonitrile (A) and 0.1% phosphoric acid water at a wavelength of 254 nm (Figure 2A). We found that Arbutin, Gastrodin, Pleiobibenzynin B, Militarine, and 3′-O-methyl dioscin III had the best detection effect at 270 nm. Consequently, these compounds were detected by acetonitrile (A) and 0.1% phosphoric acid aqueous solution at a wavelength of 270 nm (Figure 2C).
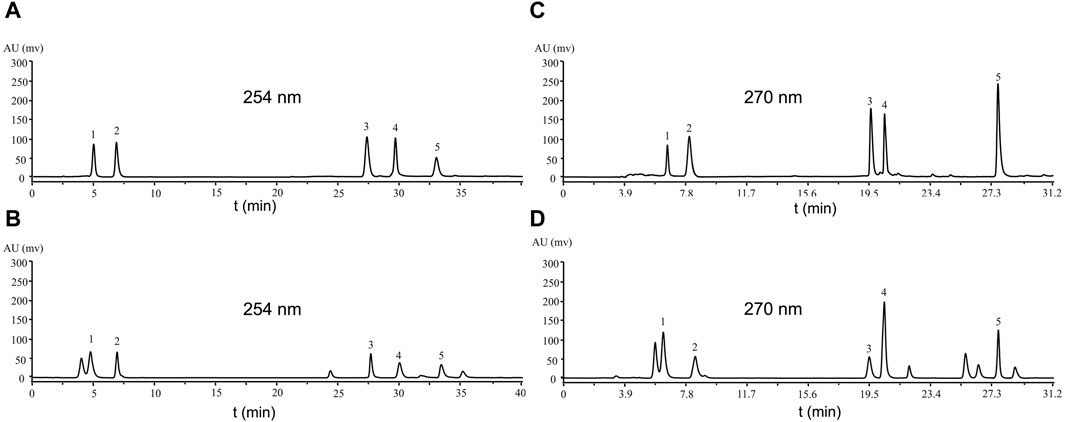
Figure 2. Analysis of the compound standard by HPLC. (A, C) standards; (B) eluted by 30% ethanol when n-butanol was partially passed through a polyamide chromatographic column; (D) eluted by 60% ethanol when n-butanol was partially passed through a polyamide chromatographic column. (A) Caffeic acid 5.029 s (1), Blestritin B 6.942 s (2), 2-methyl anthraquinone 27.854 s (3), Gymconopin D 30.158 s (4), Physcion 33.674 s (5), (C) Arbutin 6.533 s (1), Gastrodin 7.946 s (2), Pleiobibenzynin B 19.582 s (3), Militarine 20.461 s (4), 3′-O-methyl dioscin III 27.699 s (5).
3.2.2 HPLC analysis and identification
Next, the n-butanol extract was passed through a polyamide chromatographic column and eluted with water, followed by 30%, 60% and 90% ethanol, for HPLC analysis. Compared with the standard sample, the 30% ethanol eluate contained Physcion, Blestritin B, 2-methyl anthraquinone, Gymconopin D, and Caffeic acid (Figure 2B). The 60% ethanol eluate contained Arbutin, Gastrodin, Pleiobibenzynin B, Militarine and 3′-O-methyl dioscin III (Figure 2D). By HPLC, we calculated the content of these separated compounds in the n-butanol extract. The 30% and 60% ethanol eluate extract of B. striata contained 0.14% Caffeic acid, 0.19% Blestritin B, 0.01% 2-methyl anthraquinone, 0.15% 3′-O- methyl dioscin III, 0.18% Gymconopin D, 0.16% Physcion, 0.31% Arbutin, 0.58% Gastrodin, 0.18% Pleiobibenzynin B and 2.34% Militarine. The highest content was Militarine (2.34%), followed by Gastrodin (0.58%) and Arbutin (0.31%). Militarine is composed of one molecule of 2-isobutyl apple and two molecules of Gastrodin, a metabolite of Militarine. Furthermore, Militarine can be metabolized into Gastrodin in vivo (Yang et al., 2019). Gastrodin was first isolated from Gastrodia elata by Feng et al., in 1978 (Feng et al., 1979; Zhou et al., 1979). Among its various pharmacological properties, Gastrodin can exert particularly strong actions when used to treat CNS diseases. Considering its low toxicity and remarkable pharmacological performance, Gastrodin may therefore represent a potentially valuable therapeutic for preventing and treating certain CNS disorders (Chen, 2019; Lu et al., 2020). In mice, the oral administration of Gastrodin at a dosage of up to 5,000 mg/kg caused no mortality or apparent toxic effects (Han et al., 2001; Zeng et al., 2021; Zhang et al., 2023). Gastrodin is also known to have good neuroprotective effects. Studies have shown that infection with the COVID-19 virus can lead to nerve damage (Pezzini and Padovani, 2020; Wang et al., 2023). Therefore, Gastrodin may represent a potentially useful anti-COVID-19 drug, because it can inhibit the proliferation of the virus while exerting good protective effects on nerve injury.
3.2.3 Methodological investigation
Next, we established standard curves by taking the ratio of compound/internal standard peak area as the ordinate and the concentration of compound as the abscissa. Analysis showed that the correlation coefficient (R2) of the standard curve for the target compound was >0.99 in the effective concentration range, and that the linear relationship was good. Thus, the standard curve could be used effectively to calculate the concentration of the compound in unknown samples. The precision of the RSD values for each compound in the sample were all <1.5%, thus indicating that the precision of the instrument was good. The RSD of each compound within 48 h was <1.5%, thus indicating that the target compound remained basically stable within a 48 h period. The RSD of each compound in the samples prepared in parallel was <1.5%, thus showing that the sample treatment method had good repeatability. In addition, the average recovery rate for each component was 90.59%–115.26%, and the RSD value ranged from 2.5% to 3.8%, thus showing that the accuracy of the method was good. The results of the methodological investigation are shown in Supplementary Table S1 (the relevant content is provided in the Supplementary Material).
3.3 FRET-based enzymatic assays to determine inhibitory activity against SARS-CoV-2 3CLpro
Next, FRET was used to determine the inhibitory activity of the compounds on protease; Baicalein was selected as the positive control (Su et al., 2021). Figure 3A shows the relationship between the inhibition rate of baicalein at different concentrations. The petroleum ether (PE), ethyl acetate (EAC) and n-butanol (NBA) components of the 90% methanol extract of B. striata (all at a concentration of 200 μg/mL) were all found to exert inhibitory activity on protease (Figure 3B). The inhibitory rate of the n-butanol component on 3CL protease exceeded 50% at this concentration; therefore, we further separated the n-butanol component. After screening, we investigated the inhibitory potency of the 10 compounds (1–10) against SARS-CoV-2 3CL protease. All ten of the components separated from n-butanol, except 2-methyl anthraquinone, Arbutin, and Militarine (2, 9 and 10), exhibited SARS-CoV-2 3CL protease inhibitory activities in a dose-dependent manner. The average inhibitory rates of Pleiobibenzynin B, Caffeic acid, Blestritin B, Physcion, Gymconopin D, Gastrodin and 3′-O-methyl dioscin III were 98.11%, 74.30%, 63.30%, 36.77%, 35.16%, 20.78% and 16.79% respectively (at a final concentration of 20 μM, Figure 3C). We calculated the 50% inhibitory concentration of these compounds with baicalein as control. The 50% inhibitory concentration of baicalein is 0.98 µM, which is not much different from the literature 0.94 µM (Su et al., 2021). The 50% inhibitory concentrations of Pleiobibenzynin B, Caffeic acid, Blestritin B, Physcion, Gymconopin D, Gastrodin and 3′-O-methyl dioscin III were 7.37 µM, 14.34 µM, 18.65 µM, 25.30 µM, 30.27 µM, 38.59 µM and 39.86 µM respectively (Figure 4).
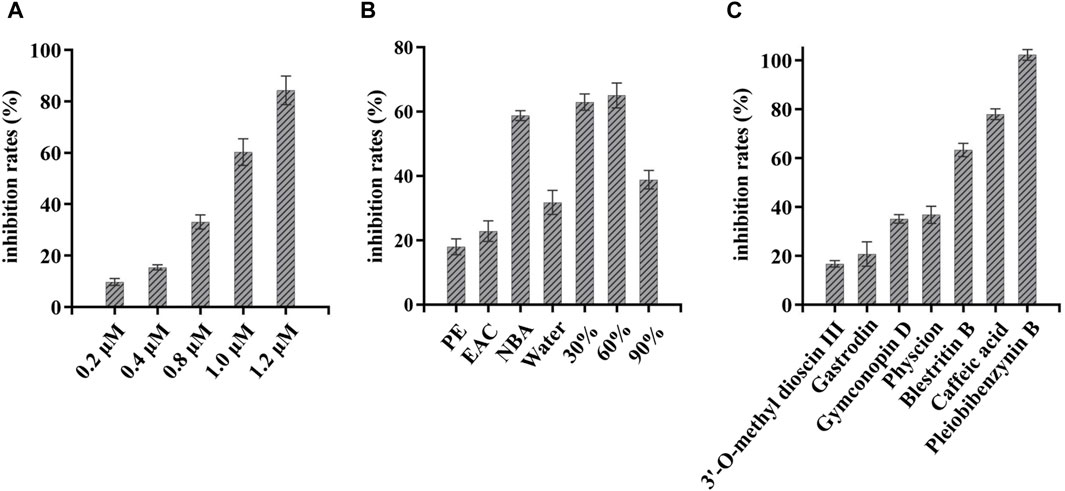
Figure 3. FRET-based enzymatic assay to determine inhibitory activity against SARS-CoV-2 3CLpro assay. (A) Inhibitory rate of different concentrations of baicalein on 3CL protease; (B) The 3CL protease inhibition rate of extracts from Bletilla striata prepared with petroleum ether (PE), ethyl acetate (EAC) and n-butanol (NBA). The inhibition rate of 3CL protease exhibited by elutions by water, 30%, 60% and 90% ethanol when n-butanol was partially passed through a polyamide chromatographic column (at a final concentration of 200 μg/mL); (C) Inhibition rate of 3CL protease in isolated components (at a final concentration of 20 µM).
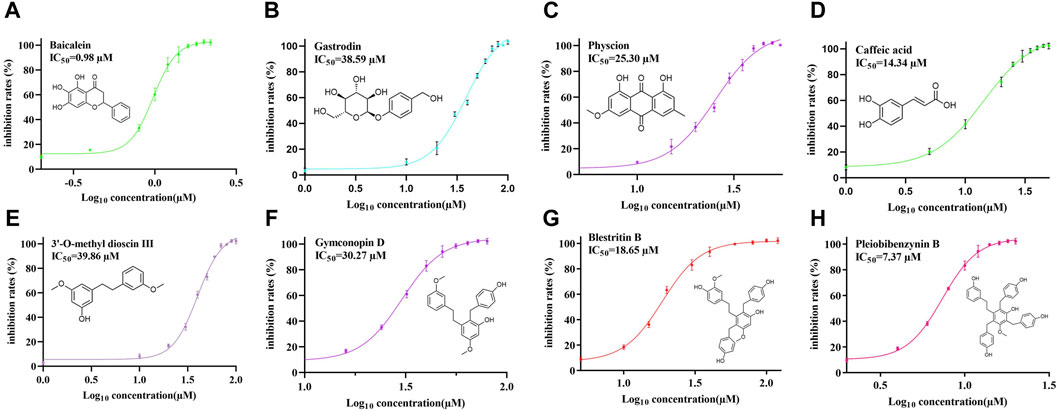
Figure 4. The 50% inhibitory concentrations of Baicalein (A), Gastrodin (B), Physcion (C), caffeic acid (D), 3′-O-methyl dioscin III (E), Gymconopin D (F), Blestritin B (G), and Pleiobibenzynin B (H).
Four types of bibenzyls, 3′-O-methyl dioscin III, Blestritin B, Gymcopind and Pleiobibenzynin B, exhibited good inhibitory activity against 3CL protease. Bletilla species are known to be rich in bibenzyls; these are the characteristic compounds of this genus. This category of compounds exhibits a variety of biological activities. It has been reported that bibenzyl exhibited moderate to good antibacterial activities on strains of Gram-positive bacteria (Jiang et al., 2019). In this study, we found that bibenzyl compounds may have better inhibitory activity on 3CL protease, although this requires further investigation. However, at this concentration (a final concentration of 20 µM), Arbutin, Militarine and 2-methyl anthraquinone showed no obvious inhibitory activity against 3CL protease inhibitory. When compared with the structure of Gastrodin, Arbutin differs only by one methylene group, although our FRET analysis showed that arbutin had no obvious protease inhibitory activity, indicating that the methylene group on the benzene ring is the key group responsible for Gastrodin activity. The inhibitory activity of caffeic acid on 3CL protease has been reported previously; the half inhibitory concentration of caffeic acid on 3CL protease was between 9.33 and 13.51 µM (Wang et al., 2024). Our research showed that the median inhibitory concentration of caffeic acid was 14.34 µM; this value was similar to that reported previously. However, the 3CL protease inhibitory activity of Gastrodin, Blestritin B, Gymcopind and Pleiobibenzynin B has not been reported previously. We found that the activity of these compounds ranged from 7.37 to 39.86 µM against 3CL protease. In this study, Pleiobibenzynin B had the strongest protease inhibitory activity of 7.37 µM. It is well established that Gastrodin has a good neuroprotective effect and that infection with SARS-CoV-2 can cause nerve damage (Meinhardt et al., 2024). Our research showed that Gastrodin may inhibit SARS-CoV-2 proliferation by inhibiting the activity of 3CL protease, and may therefore represent a promising anti-SARS-CoV-2 agent.
We studied the kinetic mechanism of the interaction of these inhibitors with SARS-CoV-2 3CL protease. The enzymatic activity was measured with a series of substrate concentrations and various inhibitor concentrations. The inhibition data were globally fit to all of the possible kinetic models. As shown in Supplementary Figure S23, the kinetic plots show that Pleiobibenzynin B (7), Blestritin B, Gymconopin D (4), Physcion, 3′-O-methyl dioscin III (6), Gastrodin (8) and caffeic acid (5) has competitive inhibition profiles because the Lineweaver–Burk plot (1/V vs. 1/[S]) is a series of straight lines with the same y-axis intercept for the SARS-CoV-2 3CL protease inhibitors. All of the isolated inhibitors exhibited the same mode of action for inhibition. The Ki values were calculated from the Dixon plots, which are useful for determining the Ki value of the inhibitor (see Supplementary Figure S24). The Ki values of Pleiobibenzynin B, Caffeic acid, Blestritin B, Physcion, Gymconopin D, Gastrodin and 3′-O-methyl dioscin III were 5.26 µM, 12.78 µM, 10.56 µM, 12.74 µM, 15.17 µM, 19.46 µM and 32.13 µM respectively.
3.4 STD-NMR analysis
STD-NMR analysis depends on magnetization exchange from the protein-bound state to the ligand-free state. This method includes selectively saturating the protein NMR signal by using a certain radio frequency radiation without interfering with the resonance signal of small molecules. After saturation, the signal passes through NOE and spin diffusion. Therefore, all protein resonances are saturated. When a molecule binds to a protein surface, the saturation of the protein proton is transferred to the molecule through the spin diffusion of intermolecular NOE, which leads to the proton saturation of the small molecule bound to the protein. As the protein and ligand are constantly bound and dissociated, the small molecule is constantly saturated, but the molecule that is not bound to the protein will not change. Thus, STD-NMR can be used to determine the conjugate in a mixture, and the binding epitope (the part that binds the protein most closely) can also be defined (Fejzo et al., 1999; Wan et al., 2020; Pomin and Rajarathnam, 2023).
STD NMR is a technique used to characterize ligand/protein binding interactions at an atomic level. In STD NMR analysis, the magnetization status is transferred from the target protein to the bound ligand; the building block of the ligand which has the stronger contact with the protein will obtain more magnetization from the irradiated target protein, thus resulting in more intense NMR signals. Based on this principle, STD NMR can acquire information about which of its structural features directly interact with the receptor proteins (Mayer and Meyer, 2001; Pereira et al., 2009).
Next, we performed group epitope mapping analysis (Figure 5). The differential STD effects within bound ligands can yield information relating to the proximity of the individual protons to the protein surface. In Gastrodin, hydroxyl protons (H-20) showed the closest proximity to the protein with the largest STD integral value, and was assigned to receive 100% saturation. Other protons were normalized against H-20. Aromatic protons H-1, H-3 and methylene protons H-7 received less saturation (between 60% and 40%). The pyropyranose methylene proton H-9 and H10 received the lowest saturation, thus indicating that these were located furthest away from the interacting residues of 3CL protease (Figure 5A).
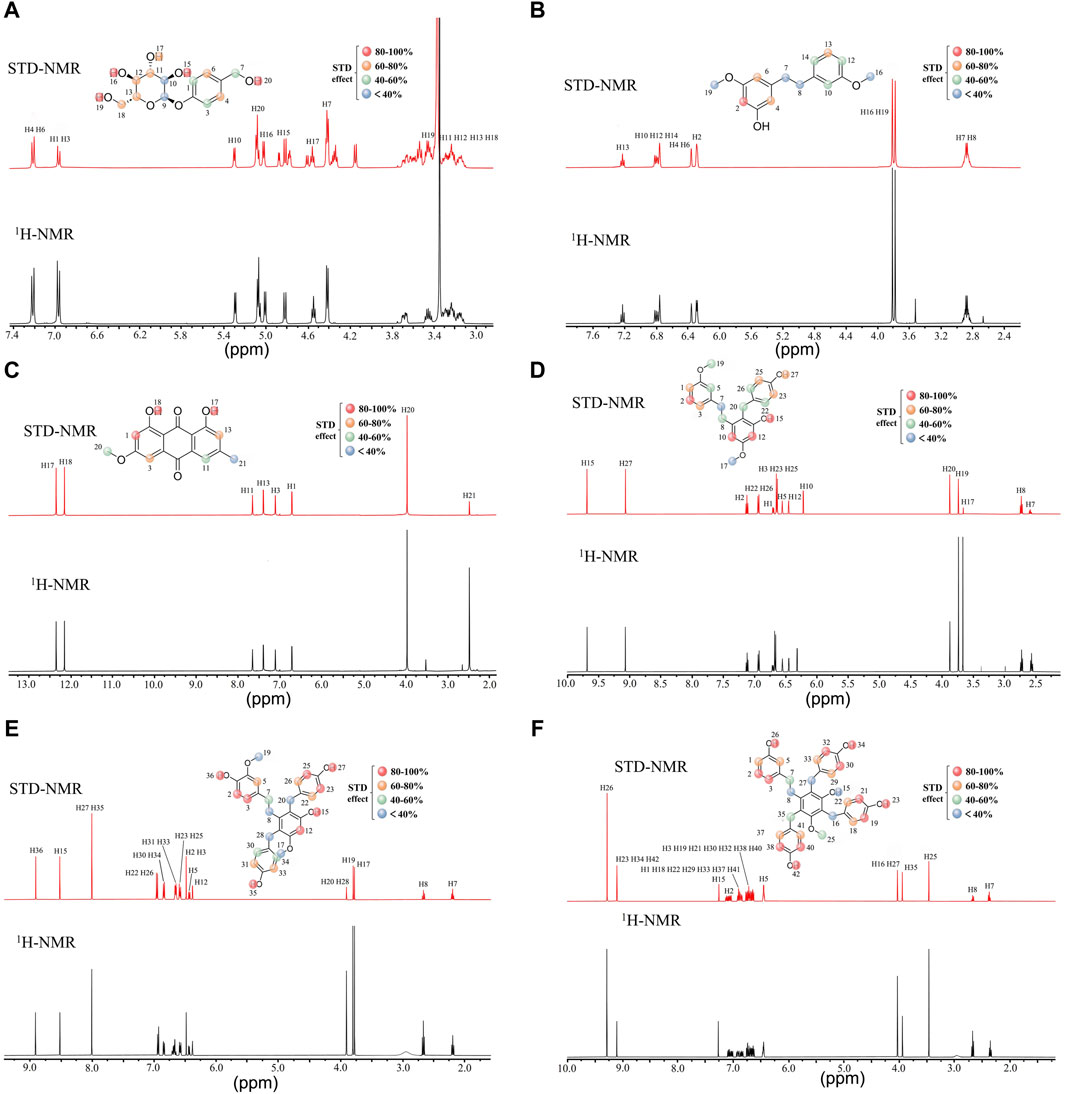
Figure 5. Characterization of group epitope mapping between compounds and 3CL protease using STD-NMR. STD (red) and reference 1H-NMR (black) NMR spectra acquired. Relative STD intensities are coded according to the color scale shown. The 100% STD effect corresponds to the resonance showing the highest intensity in each case and all other STD effects were calculated accordingly. Gastrodin and 3CL protease (A), 3′-O-methyl dioscin III and 3CL protease (B), Physcion and 3CL protease(C), Gymconopin D and 3CL protease(D), Blestritin B and 3CL protease (E), Pleiobibenzynin B and 3CL protease (F).
In 3′-O-methyl dioscin III, the aromatic protons H-2 could receive 100% saturation as it showed the highest integral value. Hydroxyl protons H-17 and H-18 received 100%–80% saturation, thus suggesting that these are located away from the residues interacting with 3CL protease. Aromatic protons H-3 and H-13 received 80%–60% saturation. Methyl and methylene protons H-7, H-8, H16, and H19 had the lowest saturation, thus suggesting that these were located furthest away from the interacting residues of 3CL protease (Figure 5B).
For Physcion, H-1 exhibited the highest STD integral value (100%). Hydroxyl protons H-17 and H-18 received saturation in between 100% and 80%. suggesting that these were located approach from (close) interacting residues of 3CL protease. Aromatic H3 and H-13 received 80%–60% saturation. The methoxy group and methyl group H-20 and H-21 received the lowest saturation, perhaps due to their longer distances from interacting residues of 3CL protease (Figure 5C).
For caffeic acid, Aromatic H-3 received 100% saturation as it exhibited the highest integral value. Styrene protons H-9 and H-10 received saturation in between 100% and 80%. Hydroxyl protons H-7, H8 and H-12 received saturation in between 80% and 60%. Aromatic H-3 received the lowest level of saturation, suggesting that this was located far away from the interacting residues in the 3CL protease (Supplementary Figure S21).
For Gymconopin D, H-12 showed the highest STD integral value (100%). Aromatic H2 and H-10 received 100%–80% saturation while H-7, H-8, H-19, H-20, H-22 and H-26 received 60%–40% saturation, thus suggesting a longer distance to the interacting protein residues. H-7 and H-17 received the lowest levels of saturation, thus suggesting that these residues were located away from the interacting residues in 3CL protease. (Figure 5D).
For Blestritin B, H-27 and H35 showed the highest STD integral value (100%). Aromatic H-2, H-3, H12, H-23 and H-25 received 100%–80% saturation, while H-5, H-22, H-26, H-31 and H33 received 80%–60% saturation. Methyl and methylene protons H-8, H17, H19, H20, and H28 received the lowest saturation, most likely due to their longer distances from interacting residues in the 3CL protease (Figure 5E).
For Pleiobibenzynin B, H-26 showed the highest STD integral value (100%). Aromatic H-2, H3, H-19, H21, H-26, H30, H-32, H-38 and H-40 received 80%–60% saturation, while H-1, H5, H18, H22, H29, H33, H37 and H41 received 60%–40%. Methylene and hydroxyl protons H–8, H15, H16 and H27 received the lowest saturation, thius indicating that these were located away from the interacting residues of 3CL protease (Figure 5F).
3.5 Correlation analysis between STD-NMR and molecular docking
To gain further insight into the mechanisms responsible for inhibition, we employed in silico docking simulation. Docking analysis revealed that the binding sites of Gymconopin D, Blestritin B, Pleiobibenzynin B, Physcion, 3′-O-methyl dioscin III, caffeic acid and Gastrodin with SARS-CoV-2 3CL protease were all within the active sites. The active sites of 3CL protease are represented by the blue areas in Figure 6. The eutectic structure of baicalein and protease showed that baicalein formed hydrogen bonds with amino acid residues Asn142 and Glu166 of protease, and that Cys145 was the key amino acid residue for 3CL protease to bind to the substrate. From the eutectic structure of baicalein and 3CL protease (PDB ID: 6m2n; visualized by Pymol), baicalein is located in front of Cys145, which effectively prevents the combination of substrate and protease (Figure 6A). Physcion forms hydrogen bonds with amino acid residues Asn142 and His164 of 3CL protease (Figure 6B) with a binding energy of −6.74 kcal/mol. 3′-O-methyl dioscin III forms hydrogen bonds with amino acid residues Thr26, Gly143 and Cys145 of 3CL protease (Figure 6C) with a binding energy of −6.47 kcal/mol. Caffeic acid forms hydrogen bonds with amino acid residues Cys145, Gly143, Ser144, Leu141 and Glu166 of 3CL protease (Figure 6E) with a binding energy of −5.36 kcal/mol. Gastrodin forms hydrogen bonds with amino acid residues Cys145, Ser144, His163, His164 and Glu166 of 3CL protease (Figure 6D), with a binding energy of −5.48 kcal/mol. The binding sites of Caffeic acid, 3′-O-methyl dioscin III, Gastrodin and caffeic acid with protease are all located in the active site of the enzyme, thus preventing the substrate peptide from approaching the key catalytic residue Cys145.
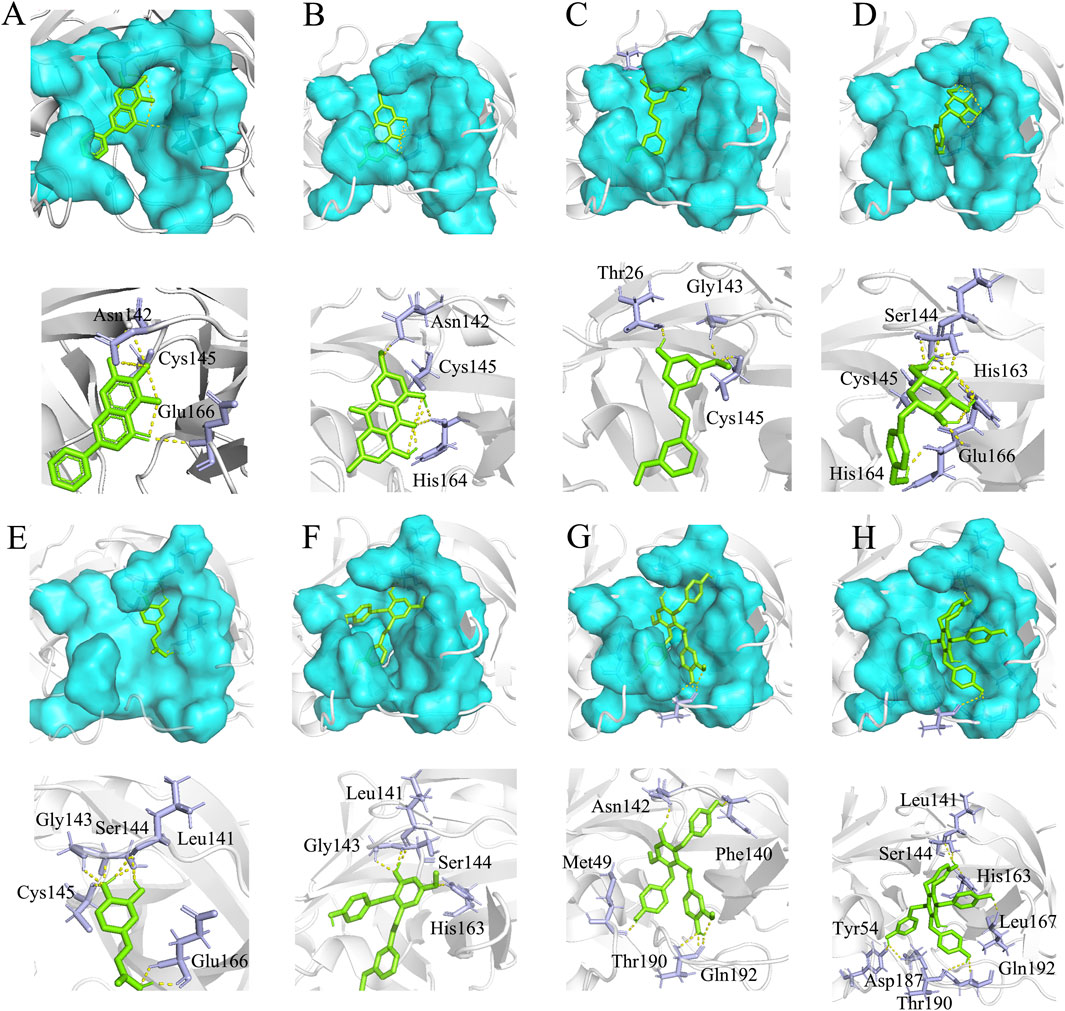
Figure 6. Molecular docking results. (A) The eutectic structure of 3CL protease and baicalein (PDB ID: 6m2n) (Su et al., 2021). (B) Physcion and 3CL protease binding energy: −6.74 kcal/mol, (C) 3′-O-methyl dioscin III and 3CL protease binding energy: −6.47 kcal/mol, (D) Gastrodin and 3CL protease binding energy: −5.48 kcal/mol, (E) caffeic acid and 3CL protease binding energy: −5.36 kcal/mol, (F) Gymconopin D and 3CL protease binding energy: −8.11 kcal/mol, (G) Blestritin B and 3CL protease binding energy: −8.75 kcal/mol, (H) Pleiobibenzynin B and 3CL protease binding energy: −10.08 kcal/mol. The blue region represents the critical amino acid residue site where the enzyme interacts with the substrate (the active site of 3CL protease), the green stick represents the small molecule fraction, and the purple stick represents the amino acid residue where the small molecule forms hydrogen bonds with the protease; Cys145 is the key amino acid residue where the protease binds to the substrate (Su et al., 2021).
The methoxy (H20) part of Physcion acid is located in the hydrophilic part of the active pocket in the 3CL protease. STD-NMR of this part revealed a low STD effect (Figure 5C), thus indicating that this part may need to be optimized into a more hydrophilic group. Furthermore, methyl (H21) is located in the hydrophobic pocket of 3CL protease (Figure 6B), although STD-NMR revealed a low effect (Figure 5C), thus indicating that the methyl part of Physcion is relatively weak when in contact with the enzyme; this part may need to be optimized to be a larger hydrophobic group to increase contact with the protease. One methoxy (H16) group of 3′-O-methyl dioscin III is located in the hydrophobic region and the other (H19) is located in the hydrophilic region of protease; STD-NMR of these two methoxy groups revealed weak STD effects (Figure 5B). It may be necessary to change the methoxy group located in the hydrophilic region to a more hydrophilic group to increase contact with the protease. The glucopyranoside part of Gastrodin is located in the hydrophilic part of the active pocket in protease, and the benzene ring part is located in the hydrophobic part of the protease (Figure 6D). However, STD-NMR showed that the protons of the benzene ring part (H1, H3) and hydroxyl protons on the benzene ring methylene (H7) showed weak STD effects (Figure 5A).
Compared with the structure of Gastrodin, Arbutin differs only by one methylene group, although our FRET analysis showed that Arbutin has no protease inhibitory activity, indicating that the methylene group on the benzene ring is the key group for Gastrodin activity; consequently, it is important to retain this region when optimizing the structure. However, the hydrogen on the benzene ring of Gastrodin may need to be replaced by hydrophobic groups to increase contact with protease. The benzene ring of caffeic acid is located in the hydrophilic region of protease (Figure 6E), but the STD effect of proton (H3) in the benzene ring is weak (Supplementary Figure S21); therefore, proton (H3) may need to be replaced by hydrophilic substituents (such as hydroxyl groups).
Three types of bibenzyls Blestritin B, Gymcopind and Pleiobibenzynin B, showed good levels of inhibitory activity against 3CL protease. The methylene and methoxy groups of these compounds all showed weak STD-NMR effects, thus indicating that the hydrogen atoms in these parts exhibit only weak contact with protease; therefore, it is necessary to increase the appropriate substituents to increase contact with protease (Figures 5D–F). The hydroxyl proton on the benzene ring of the parent nucleus (H15) of Pleiobibenzynin B showed weak STD-NMR effects (Figure 5F). Molecular docking showed that the hydroxyl proton of Pleiobibenzynin B did not form hydrogen bonds with 3CL protease (Figure 6H). Thus, this part needs to be modified to enhance the contact with protease.
4 Conclusion
Collectively, our findings show that the n-butanol extract of B. striata exerted inhibitory activity on 3CL protease in vitro. Ten components were separated from this extract and analyzed by in vitro activity tests. For the first time, we report that Gymconopin D, Blestritin B, Pleiobibenzynin B, and Gastrodin represent potential inhibitors of SARS-CoV-2 3CL protease. Moreover, we used molecular docking to investigate interactions between the compounds and the protease. Molecular docking revealed that the binding sites of compounds were all within the active site of the enzyme binding with substrate, and that the combination of enzyme and substrate could be effectively prevented. Furthermore, we used STD-NMR to investigate the epitopes responsible for binding of the compounds with protease. These investigations indicated that chemical compounds isolated from B. striata can inhibit SARS-CoV-2 3CL protease efficiently, thereby making them potential leads for further research towards the treatment of coronavirus disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materail, further inquiries can be directed to the corresponding author.
Author contributions
YY: Conceptualization, Data curation, Investigation, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing, Formal Analysis. FZ: Data curation, Formal Analysis, Visualization, Validation, Writing–original draft. SL: Investigation, Writing–original draft, Methodology, Project administration, Validation. WJ: Formal Analysis, Investigation, Project administration, Validation, Writing–original draft. QS: Formal Analysis, Investigation, Validation, Writing–original draft. CL: Investigation, Visualization, Writing–original draft, Project administration. BF: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing–review and editing, Visualization, Methodology.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Special Fund for Major Science and Technology in Hubei Province (2022BCE066).
Acknowledgments
The authors would like to express their gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fntpr.2024.1372623/full#supplementary-material
References
Cai, J.-Y., Zhao, L., and Zhang, D.-Z. (2007). Chemical constituents from Bletilla ochracea schltr. Chem. Res. Chin. Univ. 23, 705–707. doi:10.1016/s1005-9040(07)60153-6
Chen, C.-F. (2019). Glycoside constituents from Bletilla striata. Chin. Traditional Herb. Drugs, 4879–4883.
Chia, C. S. B., Xu, W., and Shuyi Ng, P. (2022). A patent review on SARS coronavirus main protease (3CLpro) inhibitors. ChemMedChem 17, e202100576. doi:10.1002/cmdc.202100576
Dai, W., Zhang, B., Jiang, X.-M., Su, H., Li, J., Zhao, Y., et al. (2020). Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368, 1331–1335. doi:10.1126/science.abb4489
Fejzo, J., Lepre, C. A., Peng, J. W., Bemis, G. W., Murcko, M. A., Moore, J. M., et al. (1999). The SHAPES strategy: an NMR-based approach for lead generation in drug discovery. Chem. Biol. 6, 755–769. doi:10.1016/s1074-5521(00)80022-8
Feng, J.-Q., Zhang, R.-J., and Zhao, W.-M. (2008). Novel bibenzyl derivatives from the tubers of Bletilla striata. Helvetica Chim. Acta 91, 520–525. doi:10.1002/hlca.200890056
Feng, X., Chen, Y., and Yang, J. (1979). Studies on the chemical constituents of Gastrodia elata. Chin. Pharm. J. 80.
Guetchueng, S. T., Djouonzo, P. T., Lame, Y., Kopa Kowa, T., Dotse, E., Tchokouaha, L. R. Y., et al. (2023). Antileishmanial anthraquinones from the rhyzomes of rumex abyssinicus jacq (polygonaceae). Nat. Prod. Res. 37, 2935–2939. doi:10.1080/14786419.2022.2137797
Guillon, P., Dirr, L., El-Deeb, I. M., Winger, M., Bailly, B., Haselhorst, T., et al. (2014). Structure-guided discovery of potent and dual-acting human parainfluenza virus haemagglutinin–neuraminidase inhibitors. Nat. Commun. 5, 5268. doi:10.1038/ncomms6268
Han, G., Wang, L., Zhang, W., Wang, M., Liu, H., and Xiao, J. (2001). Studies on chemical constituents of Bletilla striata Ⅱ. Academic Journal of Second Military Medical University.
He, X., Wang, A., Li, Y., Wang, Y., and Zhou, S. (2009). Determination of militarine in rhizoma bletillae by HPLC. Zhongguo Zhong yao za zhi 34, 2076–2078.
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. doi:10.1016/j.cell.2020.02.052
Hu, M., Jiang, M., Zhang, G., Liu, H., He, Y., and Wu, Z. (2019). Chemical composition of tubers of Bletilla striata. Chem. Nat. Compd. 55, 555–556. doi:10.1007/s10600-019-02741-4
Jiang, S., Wan, K., Lou, H.-Y., Yi, P., Zhang, N., Zhou, M., et al. (2019). Antibacterial bibenzyl derivatives from the tubers of Bletilla striata. Phytochemistry 162, 216–223. doi:10.1016/j.phytochem.2019.03.022
Jiang, S., Wang, M., Jiang, L., Xie, Q., Yuan, H., Yang, Y., et al. (2021). The medicinal uses of the genus Bletilla in traditional Chinese medicine: a phytochemical and pharmacological review. J. Ethnopharmacol. 280, 114263. doi:10.1016/j.jep.2021.114263
Jin, C., Huang, L., Du, H., Lu, Y., Yu, N., Nie, X., et al. (2023). Gardenia (Gardenia jasminoides Ellis) fruit: a critical review of its functional nutrients, processing methods, health-promoting effects, comprehensive application and future tendencies. Chin. J. New Drugs 32, 1–28. doi:10.1080/10408398.2023.2270530
Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293. doi:10.1038/s41586-020-2223-y
Jo, S., Kim, S., Kim, D. Y., Kim, M.-S., and Shin, D. H. (2020). Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhibition Med. Chem. 35, 1539–1544. doi:10.1080/14756366.2020.1801672
Kantsadi, A. L., Cattermole, E., Matsoukas, M.-T., Spyroulias, G. A., and Vakonakis, I. (2021). A COVID moonshot: assessment of ligand binding to the SARS-CoV-2 main protease by saturation transfer difference NMR spectroscopy. J. Biomol. NMR 75, 167–178. doi:10.1007/s10858-021-00365-x
Liu, C., Zhou, Q., Li, Y., Garner, L. V., Watkins, S. P., Carter, L. J., et al. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Central Sci. 6, 315–331. doi:10.1021/acscentsci.0c00272
Lu, P., Xiong, K., Huang, M., Zhang, B., and Kong, X. (2020). Research progress on pharmacology and adverse reactions of gastrodin. Asian Toxicol. Res. 2, 170–175. doi:10.53388/atr20201005
Marzi, M., Vakil, M. K., Bahmanyar, M., and Zarenezhad, E. (2022). Paxlovid: mechanism of action, synthesis, and in silico study. BioMed Res. Int. 2022, 1–16. doi:10.1155/2022/7341493
Matsuda, H., Morikawa, T., Xie, H., and Yoshikawa, M. (2004). Antiallergic phenanthrenes and stilbenes from the tubers of gymnadenia conopsea. Planta Med. 70, 847–855. doi:10.1055/s-2004-827234
Mayer, M., and Meyer, B. (2001). Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117. doi:10.1021/ja0100120
Meinhardt, J., Streit, S., Dittmayer, C., Manitius, R. V., Radbruch, H., and Heppner, F. L. (2024). The neurobiology of SARS-CoV-2 infection. Nat. Rev. Neurosci. 25, 30–42. doi:10.1038/s41583-023-00769-8
Najjar-Debbiny, R., Gronich, N., Weber, G., Khoury, J., Amar, M., Stein, N., et al. (2023). Effectiveness of Paxlovid in reducing Severe coronavirus disease 2019 and mortality in high-risk patients. Clin. Infect. Dis. 76, e342–e349. doi:10.1093/cid/ciac443
Pereira, A., Pfeifer, T. A., Grigliatti, T. A., and Andersen, R. J. (2009). Functional cell-based screening and saturation transfer double-difference NMR have identified haplosamate A as a cannabinoid receptor agonist. ACS Chem. Biol. 4, 139–144. doi:10.1021/cb800264k
Pezzini, A., and Padovani, A. (2020). Lifting the mask on neurological manifestations of COVID-19. Nat. Rev. Neurol. 16, 636–644. doi:10.1038/s41582-020-0398-3
Pomin, V. H., and Rajarathnam, K. (2023). “NMR methods for characterization of glycosaminoglycan–chemokine interactions,” in Chemokine-glycosaminoglycan interactions: methods and protocols. Editor A. R. Lucas (New York, NY: Springer US), 143–157.
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., et al. (2020). Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 117, 11727–11734. doi:10.1073/pnas.2003138117
Shi, Y., Zhang, B., Lu, Y., Qian, C., Feng, Y., Fang, L., et al. (2017). Antiviral activity of phenanthrenes from the medicinal plant Bletilla striata against influenza A virus. BMC Complementary Altern. Med. 17, 273. doi:10.1186/s12906-017-1780-6
Shiao, Y.-J., Chen, W.-P., and Lin, Y.-L. (2009). New polyphenols and triterpene from the pseudobulbs of pleione formosana. J. Chin. Chem. Soc. 56, 828–833. doi:10.1002/jccs.200900122
Su, H., Yao, S., Zhao, W., Zhang, Y., Liu, J., Shao, Q., et al. (2021). Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 12, 3623. doi:10.1038/s41467-021-23751-3
Sun, R.-R., Miao, F.-P., Zhang, J., Wang, G., Yin, X.-L., and Ji, N.-Y. (2013). Three new xanthone derivatives from an algicolous isolate of Aspergillus wentii. Magnetic Reson. Chem. 51, 65–68. doi:10.1002/mrc.3903
Tamigney Kenfack, M., Mazur, M., Nualnoi, T., Shaffer, T. L., Ngassimou, A., Blériot, Y., et al. (2017). Deciphering minimal antigenic epitopes associated with Burkholderia pseudomallei and Burkholderia mallei lipopolysaccharide O-antigens. Nat. Commun. 8, 115. doi:10.1038/s41467-017-00173-8
Wan, H., Tian, Y., Jiang, H., Zhang, X., and Ju, X. (2020). A NMR-based drug screening strategy for discovering active substances from herbal medicines: using Radix Polygoni Multiflori as example. J. Ethnopharmacol. 254, 112712. doi:10.1016/j.jep.2020.112712
Wang, P., Jin, L., Zhang, M., Wu, Y., Duan, Z., Guo, Y., et al. (2023). Blood–brain barrier injury and neuroinflammation induced by SARS-CoV-2 in a lung–brain microphysiological system. Nat. Biomed. Eng. doi:10.1038/s41551-023-01054-w
Wang, R., Chen, X., Li, H., Chen, X., Sun, D., Yu, D., et al. (2024). Danshensu inhibits SARS-CoV-2 by targeting its main protease as a specific covalent inhibitor and discovery of bifunctional compounds eliciting antiviral and anti-inflammatory activity. Int. J. Biol. Macromol. 257, 128623. doi:10.1016/j.ijbiomac.2023.128623
Woo, K. W., Park, J. E., Choi, S. U., Kim, K. H., and Lee, K. R. (2014). Phytochemical constituents of Bletilla striata and their cytotoxic activity. Nat. Product. Sci. 20, 91–94.
Yamaki, M., Bai, L., Inoue, K., and Takagi, S. (1989). Biphenanthrenes from Bletilla striata. Phytochemistry 28, 3503–3505. doi:10.1016/0031-9422(89)80373-5
Yang, C., Xia, T., Wang, C., Sun, H., Li, Y., Gong, Z., et al. (2019). Using the UPLC-ESI-Q-TOF-MSE method and intestinal bacteria for metabolite identification in the nonpolysaccharide fraction from Bletilla striata. Biomed. Chromatogr. 33, e4637. doi:10.1002/bmc.4637
Zeng, Y.-Q., Gu, J.-H., Chen, L., Zhang, T.-T., and Zhou, X.-F. (2021). Gastrodin as a multi-target protective compound reverses learning memory deficits and AD-like pathology in APP/PS1 transgenic mice. J. Funct. Foods 77, 104324. doi:10.1016/j.jff.2020.104324
Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., et al. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368, 409–412. doi:10.1126/science.abb3405
Zhang, Y., Jin, W., Chen, J., Wei, S., Cai, W., Zhong, Y., et al. (2023). Gastrodin alleviates rat chondrocyte senescence and mitochondrial dysfunction through Sirt3. Int. Immunopharmacol. 118, 110022. doi:10.1016/j.intimp.2023.110022
Keywords: Bletilla striata, SARS-CoV-2 3CL protease, FRET, molecular docking, STD-NMR
Citation: Yang Y, Zhang F, Liu S, Jin W, Shao Q, Li C and Fan B (2024) A study of the protease inhibitory activity component of SARS-CoV-2 3CL in Bletilla striata. Front. Nat. Produc. 3:1372623. doi: 10.3389/fntpr.2024.1372623
Received: 22 January 2024; Accepted: 18 March 2024;
Published: 28 March 2024.
Edited by:
Seoung Rak Lee, Pusan National University, Republic of KoreaReviewed by:
Kittikhun Wangkanont, Chulalongkorn University, ThailandNanjaraj Urs Ankanahalli Nanjaraje Urs, Washington University in St. Louis, United States
Copyright © 2024 Yang, Zhang, Liu, Jin, Shao, Li and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baolei Fan, fanbl_1980@163.com
 Yuyu Yang1
Yuyu Yang1  Baolei Fan
Baolei Fan