Genetic Modification of Mucor circinelloides for Canthaxanthin Production by Heterologous Expression of β-carotene Ketolase Gene
- 1Colin Ratledge Center for Microbial Lipids, School of Agricultural Engineering and Food Science, Shandong University of Technology, Zibo, China
- 2Department of Food Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, Bangi, Malaysia
- 3Department of Botany and Microbiology, Faculty of Science, Al-Azhar University, Assiut, Egypt
- 4University Institute of Diet and Nutritional Sciences, Faculty of Allied Health Sciences, The University of Lahore, Lahore, Pakistan
- 5Departamento de Genética y Microbiología (Unidad asociada al Instituto de Química Física Rocasolano-Consejo Superior de Investigaciones Científicas (IQFR-CSIC)), Facultad de Biología, Universidad de Murcia, Murcia, Spain
Canthaxanthin is a reddish-orange xanthophyll with strong antioxidant activity and higher bioavailability than carotenes, primarily used in food, cosmetics, aquaculture, and pharmaceutical industries. The spiking market for natural canthaxanthin promoted researchers toward genetic engineering of heterologous hosts for canthaxanthin production. Mucor circinelloides is a dimorphic fungus that produces β-carotene as the major carotenoid and is considered as a model organism for carotenogenic studies. In this study, canthaxanthin-producing M. circinelloides strain was developed by integrating the codon-optimized β-carotene ketolase gene (bkt) of the Haematococcus pluvialis into the genome of the fungus under the control of strong promoter zrt1. First, a basic plasmid was constructed to disrupt crgA gene, a negative regulator of carotene biosynthesis resulted in substantial β-carotene production, which served as the building block for canthaxanthin by further enzymatic reaction of the ketolase enzyme. The genetically engineered strain produced a significant amount (576 ± 28 μg/g) of canthaxanthin, which is the highest amount reported in Mucor to date. Moreover, the cell dry weight of the recombinant strain was also determined, producing up to more than 9.0 g/L, after 96 h. The mRNA expression level of bkt in the overexpressing strain was analyzed by RT-qPCR, which increased by 5.3-, 4.1-, and 3-folds at 24, 48, and 72 h, respectively, compared with the control strain. The canthaxanthin-producing M. circinelloides strain obtained in this study provided a basis for further improving the biotechnological production of canthaxanthin and suggested a useful approach for the construction of more valuable carotenoids, such as astaxanthin.
Introduction
Canthaxanthin is a valuable ketocarotenoid, which is naturally present in algae, bacteria, and some fungi (1). Canthaxanthin is also commonly present in wild bird tissue, egg yolk, crustaceans, and fish at low levels. It was first isolated as a major coloring pigment from the edible mushroom Cantharellus cinnabarinus, from where it was named canthaxanthin (2). It is formed as an intermediate compound in β-carotene metabolisms to astaxanthin. It has more effective antioxidant potential than β-carotene (3) due to which it is regarded as one among the important xanthophylls of commercial significance and has extensive utilization in industries (4). In the poultry industry, it is used as a feed additive to obtain red color in egg yolks and skins (1), while in the cosmetics industry, it is used as a pigmenting agent for human skin applications.
In the past few decades, the demand for canthaxanthin has increased owing to its beneficial properties on human health such as anticancer, anti-inflammatory, anti-dermatosis, and coloring agent (5). It has been documented as an effective protective agent against skin cancers in the laboratory for the treatment of polymorphous light eruptions and idiopathic photodermatosis (6). It can effectively stimulate the immune defensive system as compared to other carotenoid species (7) and owns excellent medicinal properties for treating rashes and itching. It is anticipated that the natural canthaxanthin market will grow from 75 million to 85 million from 2018 to 2024 with a compound annual growth rate (CAGR) of 3.5% due to change in consumer preferences toward fermentation products instead of synthetic compounds (https://www.gminsights.com/industry-analysis/canthaxanthin-market).
Canthaxanthin is produced by plants, bacteria, microalgae, and halophilic archaeon (Haloferax alexandrines) in relatively low concentrations, hence these organisms are unable to compete economically with synthetic canthaxanthin (8). Synthetic canthaxanthin is produced from petrochemicals and is widely preferred due to its low cost. The applicability of plant-based canthaxanthin is also hampered due to geographical, and seasonal variation and marketing. However, microbial production of canthaxanthin by biotechnological approaches is a major development in comparison to chemical synthesis due to safety concerns connected with chemical-based canthaxanthin.
Microbial production of carotenoid using non-native platforms are appealing due to many drawbacks associated with natural hosts such as low abundance and more sophisticated growth requirements, etc. Mucor circinelloides is a well-known model organism to study the molecular background of carotene biosynthesis in zygomycetes due to the availability of the whole-genome sequence. M. circinelloides has been reported as a natural competent organism compared to other related species and also acts as a good host for heterologous gene expression (9, 10). These attributes make Mucor an amenable candidate for genetic modification for desired and high-value products. Although β-carotene is the principal carotenoid of M. circinelloides, it has also shown weak β-carotene hydroxylase activity, because it has shown the accumulation of zeaxanthin and β-cryptoxanthin in minute amounts (11). It is possible to produce new carotenoids such as xanthophylls in Mucor by exogenous expression of genes involved in carotenogenesis. For example, Papp et al. introduced canthaxanthin-producing genes crtW from Paracoccus sp. N81106 into M. circinelloides (12). The subsequent transformants produced canthaxanthin and astaxanthin but in low amounts, which was attributed to the low copy number of autonomously replicating plasmids. To solve this problem they introduced crtW gene into the genome of Mucor to get stable expression of the ketolase gene and obtained a higher amount of canthaxanthin with canthaxanthin as the principal carotenoid (13).
The present study attempted to construct a canthaxanthin-producing cell factory by integrating the β-carotene ketolase–encoded gene (bkt) from Haematococcus pluvialis inside the genome of M. circinelloides CBS 277.49. Since precursor accumulation is considered as a prerequisite for xanthophyll production so in the present study higher β-carotene availability as a substrate for β-carotene ketose enzyme was ensured by disrupting the crgA locus, which is a negative regulator of the carotene biosynthesis pathway. Then codon-optimized gene of bkt from H. pluvialis was overexpressed under the control of strong promoter zrt1. Integration can be forced by the transformation with linear fragments that have extensive homologous regions at their ends to direct homologous recombination and gene replacement (13). Thus, this study attempted to construct a mutant of M. circinelloides CBS 277.49 strain harboring the algal bkt gene for canthaxanthin production.
Materials and Methods
Strains, Media, and Growth Conditions
The double auxotroph (leucine and uracil) strain MU402 (14), derived from the wild-type M. circinelloides CBS277.49 was used as a recipient strain in all the transformation procedures. MU402 was grown on solid YPG media (3 g/L yeast extract, 10 g/L peptone, and 20 g/L glucose) at 28°C for 5–7 days to obtain spores for transformation. Transformants were grown on MMC media (10 g/L casamino acids, 0.5 g/L yeast nitrogen base (w/o amino acids), and 20 g/L glucose). Media were supplemented with uridine (200 μg m/L) when required. The pH was adjusted to 3 and 4.5 for colonial and mycelial growth, respectively.
For carotenoid extraction, all strains were initially grown by inoculating 100 μl of spore suspension (~107 spores/mL) in 500 ml baffled flasks holding 150 ml Kendrick and Ratledge (K&R) medium for 24 h with 150 rpm at 28°C (15). Then these seed cultures were inoculated at 10% v/v into 2 L fermenters (BioFlo/CelliGen 115, New Brunswick Scientific, Edison, NJ, United States) containing 1.5 L modified K&R medium supplemented with 0.6 g/L of leucine to compensate leucine auxotrophy of transformants. Fermenters were maintained at 28°C with stirring at 700 rpm and aeration of 1 v/v per min and the pH was maintained at 6.0 by automatic addition of 2 M NaOH. All the transformants were cultivated for 4 days under continuous light. Escherichia coli strain DH5α was used for maintenance and amplification of recombinant plasmids and grown in lysogeny broth (LB) containing 100 μg/ml ampicillin at 37°C and 220 rpm (16).
Construction of Recombinant Plasmids
According to the genomic data of H. pluvialis available at NCBI, the complete coding sequence of the β-carotene ketolase (GenBank accession No: AF534876.1) was synthesized by GeneWiz (GeneWiz, Suzhou, China). Codon optimization of the bkt genes was done according to the genome of M. circinelloides CBS 277.49 with the given GenBank accession number MZ020513 to increase the chances of successful expression. The codon-optimized sequence of bkt is given in Supplementary File 1. The gene sequence was provided in the modified vector, named pMAT1552+bkt. To disrupt crgA locus, a new basic plasmid was constructed and named pCRC53, which contained pyrG gene of M. circinelloides as a selection marker surrounded by 1 kb up- and down-stream of crgA sequences. The pyrG gene encodes orotidine 5′-phosphate decarboxylase and produces uridine to compensate uridine auxotrophy in Mucor and allows targeted genomic integration of the whole construct by homologous recombination. pCRC53 was used as a basic plasmid used for the generation of bkt overexpressing vector pCRC55. Plasmids used in the present study are listed in Supplementary Table S1.
For the construction of pCRC53, crgA gene (JGI accession number: 39344) with 1 kb up- and down-stream region was amplified from the genome of M. circinelloides CBS 277.49 by PCR using the primer pair F1/R1 (Supplementary Table S2). The crgA fragment was digested with SphI and SnaBI and the pUC18 vector was digested with SmaI and SphI restriction enzymes followed by ligation of these two linear fragments by T4 DNA ligase. From this ligated circular vector, crgA coding sequence was deleted by performing reverse PCR (RPCR) using primers F2/R2, resulting in a linear fragment of pUC18 that contained crgA up-and down-stream sequences. This linear fragment was then digested with SpeI and SnaBI restriction endonuclease. The pyrG fragment was amplified from the plasmid pMAT1552+bkt using primers F3/R3 and digested with SmaI and SpeI. Then both the digested fragments were ligated by T4 DNA ligase to make the basic plasmid pCRC53, which was used for the construction of recombinant plasmid pCRC55 carrying the bkt gene. For this, the joined PzrtI and bkt fragment was isolated from modified plasmid pMAT1552+bkt using primers F4/R4 as mentioned in Supplementary Table S2. Then pCRC53 was digested with SpeI and XhoI. The PCR amplified product containing joined Pzrt1 and bkt was also digested with SpeI and Xhol and then ligated by T4 DNA ligase to obtain the final recombinant plasmid pCRC55. The chemically competent E. coli DH5α was used for maintaining and propagating recombinant plasmids during gene cloning experiments. The presence of recombinant plasmids in these E. coli cells was confirmed by DNA sequencing (Sangon Biotech, Shanghai Co., Ltd, China). The steps of the gene cloning strategy with the complete map of pCRC53 and pCRC55 are presented in Figure 1.
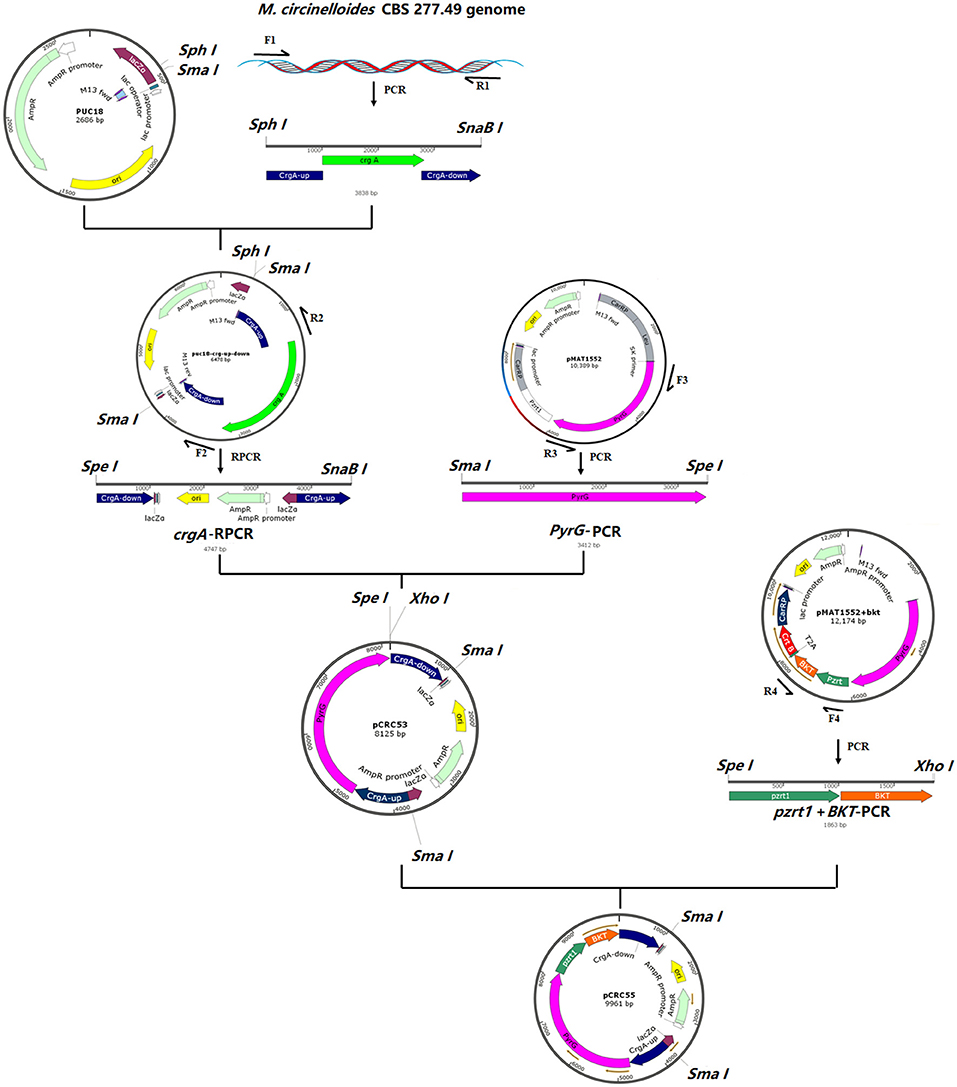
Figure 1. The structure of plasmids pCRC53 and pCRC55. The gene bkt was isolated by PCR amplification with specific primers. The PCR fragments were ligated by T4 DNA ligation to generate these plasmids.
Both pCRC53 and pCRC55 were digested with SmaI and AatII to release 5.45 and 7.29 kb fragments, respectively, containing overexpression cassettes, which were used to transform MU402 double auxotrophic strain as described previously (17). In all the transformation experiments, transformants were selected based on auxotrophy complementation and change in color under dark conditions (18).
Transformation Experiment
For electroporation-mediated transformation, the previously described procedure was followed with some minor modifications (17). Fresh spores of double auxotrophic strain MU402, grown on YPG media for 5–7 days were harvested and nearly 12.5 × 107 spores were inoculated into 25 mL YPG media supplemented with uridine and leucine and placed at 4°C overnight without shaking and then incubated at 28°C at 200 rpm to get germinated spores (2–4 h). Germinated spores were recovered at room temperature by centrifugation at 1,100 rpm for 5 min and then washed with ice-cold phosphate buffer. For protoplast generation, germinated spores were resuspended in 5 ml of PS buffer treated with 0.3 μL chitosanase RD (C0794, Sigma–Aldrich) and 1 mg/mL lysing enzymes (L-1412; Sigma–Aldrich). Removal of the cell wall was assured by using a phase-contrast microscope to monitor the loss of cell wall-associated refringence in the protoplast. Then the prepared protoplasts were suspended in 800 μl of 0.5 M sorbitol, which could be used for electroporation of eight different transformations.
For electroporation, 4–10 μg of linear DNA fragment in a volume of 10–20 μL was added to 100 μL of protoplasts and electric pulse was applied by setting the following condition of electroporator: field strength of 0.8 kV, constant resistance of 400 Ω, and capacitance of 25 μF. After electroporation, 1 ml of ice-cold YPG was added immediately and the solution mixture was incubated at 26°C and 100 rpm for 1 h. Then centrifugation was done to recover protoplasts, followed by resuspension in 500 μl of YNB plus 0.5 M sorbitol and spreading on selective medium (MMC plus 0.5 M sorbitol). These plates were covered with aluminum foil and placed in an incubator at 28°C for 3–4 days.
Molecular Techniques
Extraction and purification of plasmid DNA were performed with the plasmid mini kit and cycle pure kit (Omega-Biotek, Norcross, United States). PCR products were purified using an Exbio PCR Product purification kit according to the instructions of the manufacturer. For the preparation of genomic DNA, mycelium was disrupted with a pestle and mortar in liquid nitrogen, and DNA was extracted using DNA quick Plant system kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the instructions of the manufacturer. DNA fragments were purified from agarose gel using the Gel Extraction Kit (Omega Biotek, Norcross, United States).
Determination of CDW, Glucose, and Nitrogen Concentration
Control and overexpressing strains were grown in a 2 L fermenter containing a 1.5 L modified K&R medium. Mycelia of both strains were collected at 3, 6, 9, 12, 24, 48, 72, and 96 h from the fermenters for analysis. Biomass was harvested on a dried and pre-weighed filter paper by filtration through a Buchner funnel under reduced pressure and washed three times with distilled water, frozen overnight at −80°C, and then freeze dried. The weight of the biomass was determined gravimetrically. A glucose oxidase Perid-test kit was used to determine the concentration of glucose in the culture media according to the instructions of the manufacturer (Shanghai Rongsheng Biotech Co., Ltd.). For ammonium concentration, the indophenol method was used (19).
Carotenoid Extraction and Quantification
Carotenoid was extracted as described in our previous study (20). High-performance liquid chromatography (HPLC) was performed for carotenoid quantification in samples. Samples in a volume of 10 μL were loaded on an infinity Lab Proshell 120 EC-C18 column (4.6 × 150, ODS 4 μm). Two solvents A (96% methanol) and B (100% methyl-terc-butyl ether) were used as mobile phase in the following gradient to analyze carotenoid: min/solvent A%/solvent B% was (0/99/1; 8/60/40; 13/46/54; 15/0/100; 18/0/100; 21/99/1; 25/99/1) at a flow rate of 1 ml/min. Column thermostat temperature was set as 35°C and detection wavelength was set as 450 nm, using a diode-array detector (Agilent Technologies, Santa Clara, CA, United States). The following standards were used to identify the carotenoids in transformants: β-carotene, canthaxanthin, astaxanthin, echinenone (Sigma–Aldrich). The total carotenoids were quantified by a spectrophotometer at 450 nm and measured using an extinction coefficient of 2,500 (A1% = 2,500) as previously described (21).
RNA Extraction and Analysis of Genes Expression
For RNA extraction, Mc-55 was grown in a 2 L fermenter with 1.5 L K&R media, and biomass were collected at 3, 24, 48, and 72 h. Total RNA was extracted by using TRIzol after disruption of biomass in pestle and mortar, using liquid nitrogen as described previously (22). RNA was reverse transcribed using the Prime ScriptRT reagent kit (Takara Biotechnology, Dalian Co., Ltd, Dalian, China) according to the instructions of the manufacturer. To investigate the expression levels of the canthaxanthin biosynthesis gene bkt, real-time quantitative PCR (RT-qPCR) was conducted using specifically designed primers (Supplementary Table S2) on Light Cycler 96 Instrument (Roche Diagnostics GmbH, Switzerland) with FastStart Universal SYBR Green Master (ROX) Supermix (Roche) according to the instructions of the manufacturer. The mRNA expression level was normalized to levels of actin gene and the results were determined as relative expression levels. The data were quantified by the method of 2–ΔΔCt.
Statistical Analysis
The mean values were calculated from the data obtained from three independent experiments. Statistical analysis was conducted by one-/two-way ANOVA with multiple comparison tests wheresoever applicable. These calculations were done by the GraphPad Prism software for Windows (San Diego, CA, United States) and p < 0.05 was considered as statistically significant.
Results
Generation of bkt Overexpressing Strains of M. circinelloides
To overexpress the bkt gene in M. circinelloides CBS 277.49, an expression vector pCRC55 was constructed to allow targeted integration of bkt in the crgA locus. The strong promoter zrt1 was used for the overexpression of bkt gene. Basic plasmid pCRC53 was constructed to disrupt crgA locus and act as a control plasmid to generate control strain Mc-53. Both basic pCRC53 and recombinant plasmids pCRC55 were digested with SmaI and AatII to obtain linear fragments. These linear fragments were then used to transform the recipient strain MU402 generating a control strain named Mc-53 and recombinant strain Mc-55, respectively. Two independent bkt overexpressing transformants, named Mc-55, Mc-55-1 were selected.
Due to the syncytial nature of Mucor hyphae, and the multinucleate nature of protoplasts (23), and spores, heterokaryotic transformants were obtained initially. Therefore, transformants were grown on selective media for several consecutive vegetative cycles to obtain stable pyrG+ transformants. After more than 10 vegetative cycles, homokaryotic transformants were obtained for both the linear fragments of plasmids pCRC53 and pCRC55. The successful expression of bkt gene in M. circinelloides was confirmed by the accumulation of ketocarotenoids such as echinenone, canthaxanthin, and astaxanthin in small amounts. Gene replacement in the crgA locus resulted in deep yellow colonies that are recognizable among the pale-yellow colonies of wild-type M. circinelloides in dark. This color change was an indication of a higher amount of β-carotene accumulation due to disruption of crgA gene locus, resulting in deep yellow mycelia for control strain Mc-53 and reddish-orange in case of overexpressing strain Mc-55 in dark (Figure 2). Lack of crgA function is associated with enhanced production of carotene under both light and dark conditions (24).
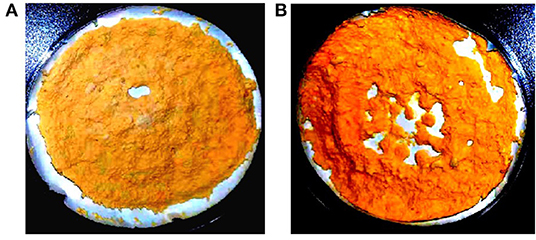
Figure 2. Lyophilized samples of Mucor circinelloides: (A) control strain Mc-53 and (B) bkt overexpressing strain Mc-55 grown on K&R media.
The integration of the exogenous gene in all transformants was confirmed by PCR amplification using primers (Ch-F1/R1 and Ch-F2/R2) listed in Supplementary Table S2. Genomic DNA was used as a template. The first primer pair (Ch-F1/R1) was designed in such a way that it could amplify few base pairs (bp) beyond crgA 1 kb upstream (inside genome) to some portion of pyrG (Figure 3A). This primer pair amplified the expected size, 1,513 bp sequence from up-streams of crgA verifying the presence of marker PyrG inside the genome. The same band size was also obtained for Mc-53 as our control strain also contained pyrG selection marker (Figure 3B). The other primer pair (Ch-F2/R2) was designed to amplify a region of 54 bp beyond crgA 1 kb downstream to almost half of the segment of the exogenous bkt gene. Amplification reaction produced an expected 1,533 bp fragment as shown in Figure 3C, which confirmed the presence of bkt gene inside the overexpressing strain Mc-55 while no fragment was amplified for control strain Mc-53 (Figure 3C). Thus, PCR amplification results confirmed the integration of the target gene into the genome of the transformants. The presence of a single band indicated the homokaryotic nature of the transformants.
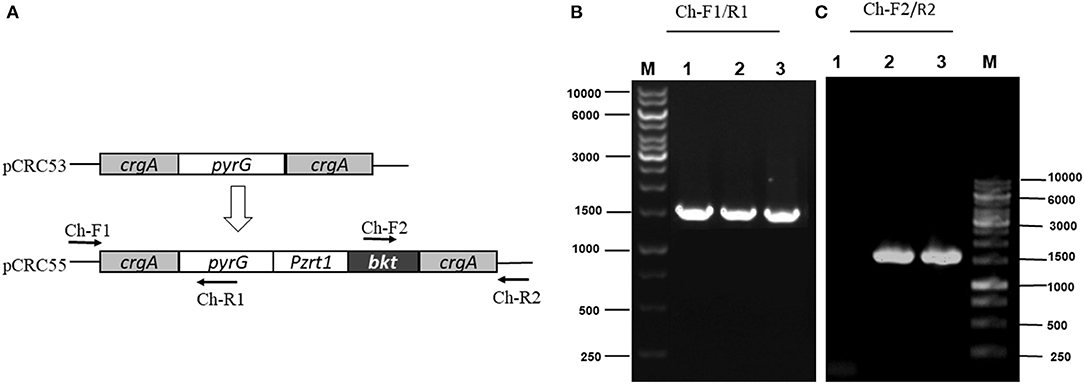
Figure 3. PCR amplification of genomic DNA of control (Mc-53) and recombinant strains (Mc-55) with the indicated primers. Lane 1 representing the control strain and Lane 2, 3 showing the presence of bkt in recombinant strains Mc-55 and Mc-55-1, respectively. (A) Plasmid structure of pCRC53 and pCRC55 for bkt overexpression. (B) Verification of the presence of marker pyrG inside transformants. Lane M: Marker, 1: control strain, 2-3: Recombinant strains (Ch-F1/R1primers). (C) Verification of the presence of bkt inside transformants. Lane M: Marker, 1: control strain, 2-3: Recombinant strains (Ch-F2/R2 primers).
Two overexpression strains Mc-55, Mc-55-1, and one control strain Mc-53 were grown in 150 mL K&R medium for 3–4 days in 500 ml baffled flasks to perform additional screening. Since CDW and canthaxanthin of these two strains were comparable (data not shown), only one strain Mc-55 was selected for further experiments.
Growth of bkt Overexpressing Strain
CDW and consumption of ammonium and glucose by Mc-55 were analyzed and compared with control strain Mc-53 as shown in Figure 4. In general, both the control and the recombinant strain showed a similar and typical growth profile. Ammonium was completely exhausted in 12–24 h in Mc-55 but it was consumed more rapidly by the control strain Mc-53, while glucose remained in sufficient amount during the entire fermentation time (Figures 4A,B). CDW was increased rapidly until 24 h and slowed down (Figure 4C). However, cell growth was slightly affected in both strains as compared to wild-type CBS 277.49, which might be attributed to crgA disruption in these strains.
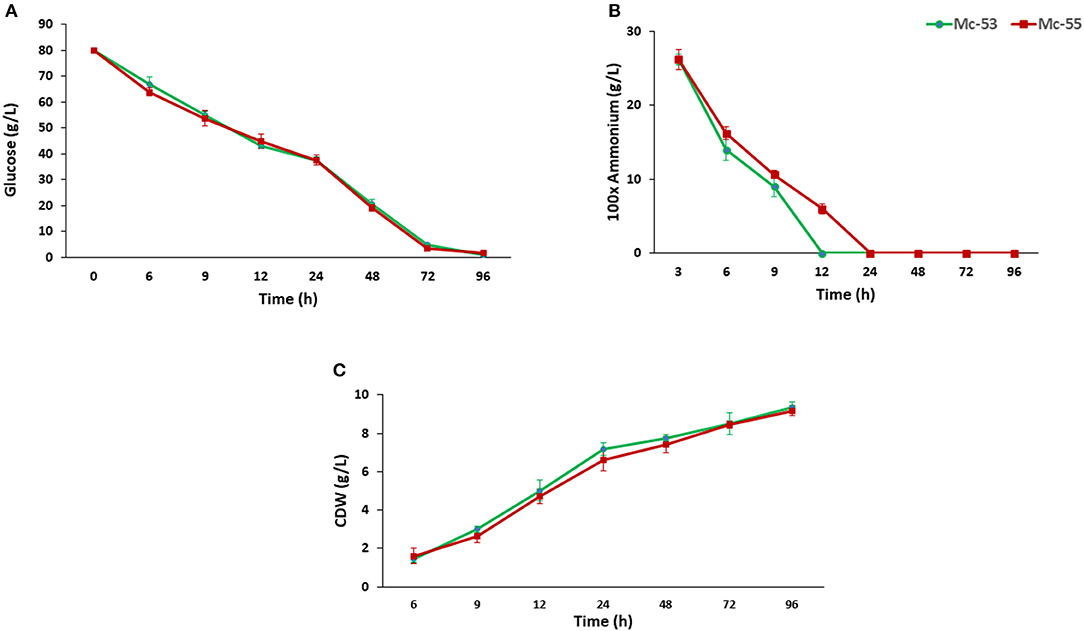
Figure 4. Determination of glucose and nitrogen consumption and cell CDW in recombinant strain Mc-55 and control strain Mc-53 at indicated time intervals. (A) Glucose concentration, (B) ammonium concentration, and (C) CDW. Values were the mean of three independent experiments. Error bars represent the standard deviation of the data.
Carotenoid Accumulation in bkt Overexpressing Strain
Detailed carotenoid profile of recombinant strain is presented in Table 1. The mycelia were collected from fermenters at the specified intervals and disrupted using pestle and mortar into fine powder for carotenoid extraction. HPLC analysis showed β-carotene, canthaxanthin, echinenone as major carotenoids in Mc-55. While in the control strain only β-carotene was observed as a major carotenoid, verifying that bkt gene was successfully expressed in Mc-55. Nevertheless, the control strain Mc-53 produced an elevated level of β-carotene (2211 ± 47 μg/g of CDW), which was 8-folds higher as compared to recipient strain MU402, that could only produce 275 ± 13 μg/g of β-carotene under the same cultivation condition (data not shown). This substantial increase in β-carotene production could be possible due to crgA disruption in Mc-53 since disruption of crgA caused over-accumulation of β-carotene even in dark by increasing the mRNA levels of carB and carRP (24).
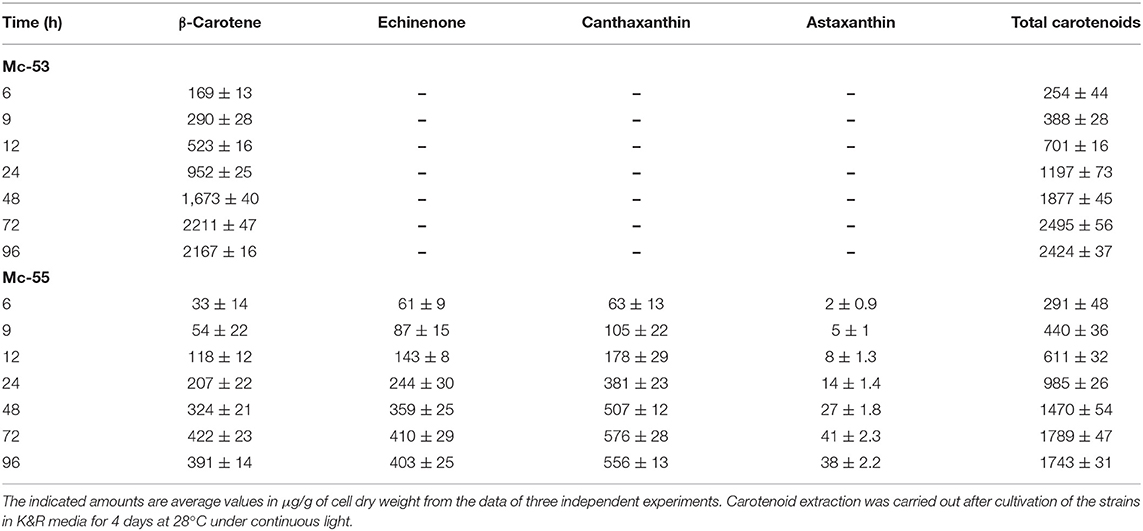
Table 1. Total carotenoid content and major carotenoid composition of control Mc-53 and recombinant Mc-55 strains.
Overexpression of the algal bkt in the Mc-55 gene led to the production of ketoderivatives (Figure 5). The accumulation of canthaxanthin started at 6 h and substantially increased with the progress of cultivation time. Mc-55 produced the maximum amount of canthaxanthin at 72 h. Other measured carotenoids and total carotenoids also showed the same trend. The highest amount of canthaxanthin (576 ± 28 μg/g) and echinenone (410 ± 29 μg/g) and a comparatively small amount of astaxanthin (41 ± 2.3 μg/g) were produced, which corresponded to 32, 23, and 3% of total carotenoid in Mc-55 at 72 h, respectively. This significant amount of canthaxanthin production might be possible in the current study due to the availability of a higher amount of β-carotene substrate as it is the closest intermediate to canthaxanthin in the introduced pathway (Figure 5).
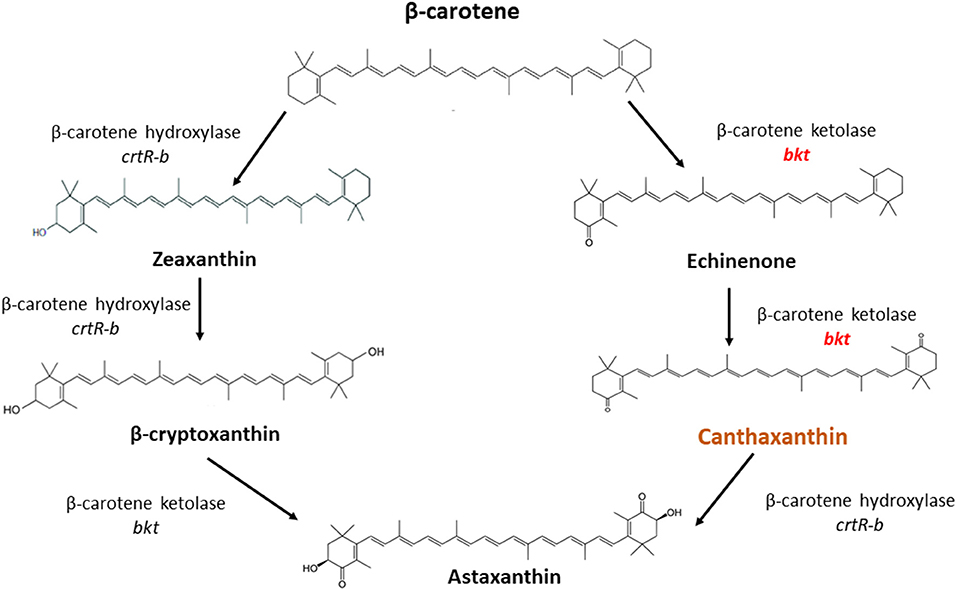
Figure 5. The possible pathway for conversion of β-carotene to canthaxanthin and astaxanthin in M. circinelloides as proposed by Misawa et al. (25).
The Expression Level of bkt Gene in Overexpressing Strain
The mRNA expression level of bkt in overexpressing strain Mc-55 was analyzed by RT-qPCR at specified intervals. In control strain Mc-53, the mRNA level of bkt was undetectable throughout the culture time. The mRNA expression level of Mc-55 was considered as 1 at 3 h, and by comparing with this value the expression level was determined at other time intervals. The expression level of Mc-55 was quickly increased at the start of fermentation till 24 h but showed a decreasing trend afterward. It was observed that the expression level of bkt was increased by 5.3-, 4.1-, and 3-folds at 24, 48, and 72 h, respectively, and maintained at elevated levels throughout the cultivation time, suggesting that it was overexpressed successfully in Mc-55 (Figure 6).
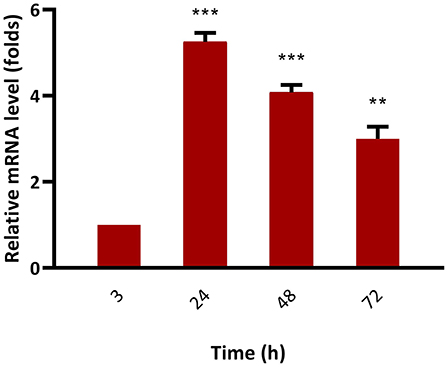
Figure 6. Determination of expression levels of bkt genes by RT-qPCR in the overexpressing strains Mc-55. Values were the mean of three independent experiments. Error bars represent the standard deviation of the data. Asterisks indicate that the difference is significant.***p < 0.001 in comparison between 3, 24, and 48 h, and **p < 0.01 in comparison between 24, 48, and 72 h of growth.
Discussion
In M. circinelloides β-carotene is produced as a major carotenoid via the mevalonate pathway along with a lower amount of zeaxanthin and β-cryptoxanthin due to weak hydroxylase activity. However, no astaxanthin could be produced due to the absence of ketolase enzymes (11). β-carotene can be further converted into canthaxanthin by β-carotene ketolase encoded by gene bkt in algae and crtW in bacteria, which add keto group in the ring of β-carotene at 4,4-position. The efficient conversion of β-carotene into canthaxanthin depends on the ketolase enzyme with higher catalytic activity. Haematococcus pluvialis is a marine, astaxanthin producing algae, in which the conversion of β-carotene to astaxanthin is carried out by the enzymes β-carotene ketolase and hydroxylase with canthaxanthin as an intermediate compound (Figure 5). Haematococcus pluvialis, being a natural source of valuable xanthophylls, is grown for ketocarotenoid production, but commercial-scale cultivation of this algae is embedded with few drawbacks. For example, high light requirement, slow growth at room temperature, as well as vulnerable to contamination by other algae (26). Over the past two decades, M. circinelloides has been successfully engineered for various useful metabolites by either overexpressing indigenous genes or by introducing the exogenous biosynthetic genes (11, 27–30).
In the present study, bkt gene from H. pluvialis was successfully overexpressed in M. circinelloides under the control of strong promoter zrt1 to construct the canthaxanthin biosynthesis pathway. The successful integration of bkt was indicated by the accumulation of ketocarotenoid in Mc-55, producing ~576 ± 28 μg/g canthaxanthin, which is the highest titer of canthaxanthin ever reported in M. circinelloides. This amount was almost 3-folds more than the canthaxanthin content of the wild-type strain of Gordonia jacobaea (200 μg/g) (31), but almost comparable with 600 μg/g of canthaxanthin in Brevibacterium KY-4313 (32), both of them are considered as a natural source of canthaxanthin. Similarly, the bacterium Dietza natronolimnaea HS-1 was reported to produce a maximum of 8,923 ± 18 μg/L canthaxanthin in a fed-batch process (33). Previously, Papp et al. had successfully engineered crtW gene in M. circinelloides from marine bacterium but the amount of canthaxanthin produced was very low, i.e., 6–13 μg/g (12). The transformants obtained in our study accumulated a significantly higher amount of canthaxanthin than that obtained by a previous study in which a maximum of 443 ± 71 μg/g canthaxanthin was produced by Mucor after co-expression of crtW and crtZ and cultivation of the mutant strain on the combination of glucose and dihydroxyacetone (34). Similarly, in another report overexpression of crtW gene from Paracoccus into M. circinelloides produced a maximum amount of canthaxanthin in the range of 100–240 μg/g after genetic modification and medium optimization (13). Csernetics et al. also introduced two genes crtR and crtS from the yeast Xanthophyllomyces dendrorhous in M. circinelloides, which are responsible for the conversion of β-carotene to astaxanthin. After laborious genetic modification, a maximum of 190 μg/g of canthaxanthin was produced in a mutant strain of M. circinelloides (29). So, the higher production of canthaxanthin could be attributed to the higher catalytic efficiency of bkt from an algal source in comparison to its bacterial and yeast counterpart.
However, control strain Mc-53 in the present study showed superior production of β-carotene up to 2211 ± 47 μg/g in the fermenter at 72 h with increased total carotenoid as compared to bkt overexpressing strain Mc-55 (Table 1). In overexpressing strain Mc-55, the concentration of β-carotene was drastically reduced to 422 ± 23 at 72 h which might be attributed to the conversion of β-carotene into ketocarotenoids such as echinenone and canthaxanthin. This could also be explained as a possible feedback effect of β-carotene and its derivatives produced during the β-carotene biosynthesis. So, it might be possible that β-carotene production has decreased in Mc-55 due to its conversion into canthaxanthin and affected the functioning of genes involved in carotenogenesis (13).
The successful overexpression of the bkt gene from an algal source was also verified by RT-qPCR. The result showed that mRNA levels of the bkt gene remained elevated throughout the fermentation time. The expression level of bkt showed maximum expression reaching up to 5.3-folds at 24 h after which a decrease in the expression level was observed. However, the maximum production of canthaxanthin was achieved in our study at 72 h, which might be explained as bkt gene regulated the canthaxanthin accumulation by post-transcriptional or post-translational regulatory processes (35).
In our study, the higher canthaxanthin production might be possible because of two reasons (1): The expression of codon-optimized bkt gene under the control of strong promoter zrt1; (2): Availability of a high amount of β-carotene precursor, achieved by the disruption of crgA, a well-known repressor of carotenogenesis (24). Codon optimization of the algal bkt gene was done with the codon preference of M. circinelloides. Zhou et al. also used codon-optimized genes for astaxanthin synthesis by Saccharomyces cerevisiae and found increased expression levels of bkt and crtZ (36) as compared to the non-optimized aforementioned gene sequence. Moreover, it is well-known from the literature that initial astaxanthin synthesis utilizes the existing β-carotene as a precursor, so the production of the bulk astaxanthin esters depends on de novo β-carotene synthesis (37). CrgA is identified as a repressor of carotenoid biosynthesis in M. circinelloides because disruption of crgA caused over-accumulation of β-carotene even in dark by increasing the mRNA levels of structural carotenogenic genes, that is, carB and carRP (24). The crgA gene suppresses carotenoid biosynthesis in Mucor by proteolysis-independent mono-ubiquitilation and di-ubiquitilation of white collar wc-1b, which triggers the transcription of carB and carRP genes when non-ubiquitilated (38). A previous study has also shown that the deletion of crgA resulted in higher lycopene accumulation in mutant M. circinelloides strain (39). Similarly, Zhang et al. found that disruption of crgA caused over-accumulation of β-carotene in mutant strains of M. circinelloides (18).
The canthaxanthin-producing strain of M. circinelloides obtained in our study can be a strong putative candidate for industrial production because of two main reasons; first, the fermentation process using M. circinelloides would be much simpler than the complicated growth requirement of H. pluvialis. Second, dimorphic nature favors its biotechnological application. The highest canthaxanthin production of 576 ± 28 μg/g was achieved in Mucor, which is the maximum reported canthaxanthin in Mucor to date. This study can be regarded as a good extension of previous genetic modifications done in M. circinelloides for canthaxanthin production. In conclusion, canthaxanthin production in our recombinant strain could be further augmented by improving cultivation conditions or by employing a fed-batch process to enhance the biomass productivity for the volumetric production of canthaxanthin. Genetic engineering of early steps of the mevalonate pathway to increase the supply of rate-limiting precursor, suppressing the competitive fatty acid and sterol biosynthesis pathway, and improving the fermentation process could be attractive strategies for future improvement of canthaxanthin production in the obtained recombinant strains of M. circinelloides.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary Material.
Author Contributions
TN and JY performed the experiments and manuscript writing. SN worked in experimental design. CS reviewed the initial draft and helped in editing. YN and HM were involved in the experimental design and data analysis. AF and SL helped with the HPLC analysis and quantification. SU helped in statistical analysis. WY helped in drawing images. VG involved in results interpretations. YS proposed the project and was involved in data analysis, and review of the final draft. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31670064 and 31972851), the TaiShan Industrial Experts Program tscy 20160101, and the Shandong provincial key technology R&D plan (2018GNC110039, 2018GSF121013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Faiza Asghar, Kiren Mustafa, Hafiy Halim, and Pranesha for their assistance during fermentation and RNA extraction and Atif Taj for careful reading of the manuscript and discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2021.756218/full#supplementary-material
References
1. Esatbeyoglu T, Rimbach G. Canthaxanthin: from molecule to function. Mol Nutr Food Res. (2017) 61:1600469. doi: 10.1002/mnfr.201600469
2. Haxo F. Carotenoids of the mushroom Cantharellus cinnabarinus. Bot Gaz. (1950) 112:228–32. doi: 10.1086/335653
3. Naguib Y. Pioneering astaxanthin. Nutr Sci News. (2001) 1–6. Available online at: https://chiro.org/nutrition/FULL/Pioneering_Astaxanthin.shtm
4. Perera CO, Yen GM. Functional properties of carotenoids in human health. Int J Food Prop. (2007) 10:201–30. doi: 10.1080/10942910601045271
5. Chan KC, Mong MC, Yin MC. Antioxidative and anti-inflammatory neuroprotective effects of astaxanthin and canthaxanthin in nerve growth factor differentiated PC12 cells. J Food Sci. (2009) 74:H225–31. doi: 10.1111/j.1750-3841.2009.01274.x
6. Santamaria L, Bianchi A, Arnaboldi A, Ravetto C, Bianchi L, Pizzala R, et al. Chemoprevention of indirect and direct chemical carcinogenesis by carotenoids as oxygen radical quenchers. Ann NY Acad Sci. (1988) 534:584–96. doi: 10.1111/j.1749-6632.1988.tb30149.x
7. Jyonouchi H, Sun S, Mizokami M, Gross MD. Effects of various carotenoids on cloned, effector-stage T-helper cell activity. Nutr Cancer. (1996) 26:313–24. doi: 10.1080/01635589609514487
8. Sanchez S, Ruiz B, Rodríguez-Sanoja R, Flores-Cotera LB. Microbial production of carotenoids. In: McNeil B, Archer D, Giavasis I, Harvey L, editors. Microbial Production of Food Ingredients, Enzymes and Nutraceuticals. Mexico: Elsevier; Woodhead Publishing (2013). p. 194–233.
9. Iturriaga EA, Díaz-Mínguez JM, Benito EP, Alvarez MI, Eslava AP. Heterologous transformation of Mucor circinelloides with the Phycomyces blakesleeanus leul gene. Curr Genet. (1992) 21:215–23. doi: 10.1007/BF00336844
10. Garre V, Barredo JL, Iturriaga EA. Transformation of Mucor circinelloides f. lusitanicus Protoplasts. In: Genetic Transformation Systems in Fungi. Cham: Springer (2015). p. 49–59.
11. Csernetics Á, Nagy G, Iturriaga EA, Szekeres A, Eslava AP, Vágvölgyi C, et al. Expression of three isoprenoid biosynthesis genes and their effects on the carotenoid production of the zygomycete Mucor circinelloides. Fungal Genet Biol. (2011) 48:696–703. doi: 10.1016/j.fgb.2011.03.006
12. Papp T, Velayos A, Bartók T, Eslava AP, Vágvölgyi C, Iturriaga EA. Heterologous expression of astaxanthin biosynthesis genes in Mucor circinelloides. Appl Microbiol Biotechnol. (2006) 69:526–31. doi: 10.1007/s00253-005-0026-6
13. Papp T, Csernetics Á, Nagy G, Bencsik O, Iturriaga EA, Eslava AP, et al. Canthaxanthin production with modified Mucor circinelloides strains. Appl Microbiol Biotechnol. (2013) 97:4937–50. doi: 10.1007/s00253-012-4610-2
14. Benito EP, Díaz-Mínguez JM, Iturriaga EA, Campuzano V, Eslava AP. Cloning and sequence analysis of the Mucor circinelloides pyrG gene encoding orotidine-5′-monophosphate decarboxylase: use of pyrG for homologous transformation. Gene. (1992) 116:59–67. doi: 10.1016/0378-1119(92)90629-4
15. Kendrick A, Ratledge C. Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur J Biochem. (1992) 209:667–73. doi: 10.1111/j.1432-1033.1992.tb17334.x
16. Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. (1983) 166:557–80. doi: 10.1016/S0022-2836(83)80284-8
17. Gutiérrez A, López-García S, Garre V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J Microbiol Methods. (2011) 84:442–6. doi: 10.1016/j.mimet.2011.01.002
18. Zhang Y, Navarro E, Cánovas-Márquez JT, Almagro L, Chen H, Chen YQ, et al. A new regulatory mechanism controlling carotenogenesis in the fungus Mucor circinelloides as a target to generate β-carotene over-producing strains by genetic engineering. Microb Cell Fact. (2016) 15:1–14. doi: 10.1186/s12934-016-0493-8
19. Chaney AL, Marbach EP. Modified reagents for determination of urea and ammonia. Clin Chem. (1962) 8:130–2.
20. Naz T, Nazir Y, Nosheen S, Ullah S, Halim H, Fazili ABA, et al. Redirecting metabolic flux towards the mevalonate pathway for enhanced β-carotene production in M. circinelloides CBS 277.49. Biomed Res Int. (2020) 2020:8890269. doi: 10.1155/2020/8890269
21. Scott KJ. Detection and measurement of carotenoids by UV/VIS spectrophotometry. Curr Protoc food Anal Chem. (2001), (1): F2–2.
22. Nosheen S, Yang J, Naz T, Nazir Y, Ahmad MI, Fazili ABA, Li S, et al. Annotation of AMP-activated protein kinase genes and its comparative transcriptional analysis between high and low lipid producing strains of Mucor circinelloides. Biotechnol Lett. (2020) 43:1–10.
23. Binder U, Navarro-Mendoza MI, Naschberger V, Bauer I, Nicolas FE, Pallua JD, Lass-Flörl C, Garre V. Generation of a Mucor circinelloides reporter strain—A promising new tool to study antifungal drug efficacy and mucormycosis. Genes (Basel). (2018), 9(12): 613. doi: 10.3390/genes9120613
24. Navarro E, Lorca-Pascual J, Quiles-Rosillo M, Nicolás F, Garre V, Torres-Martínez S, et al. negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol Genet Genomics. (2001) 266:463–70. doi: 10.1007/s004380100558
25. Misawa N, Satomi Y, Kondo K, Yokoyama A, Kajiwara S, Saito T, et al. Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol. (1995) 177:6575–84. doi: 10.1128/jb.177.22.6575-6584.1995
26. Masojídek J, Torzillo G. “Mass Cultivation of Freshwater Microalgae,” in Encyclopedia of Ecology, Five-Volume Set doi: 10.1016/B978-008045405-4.00830-2
27. Khan MAK, Yang J, Hussain SA, Zhang H, Liang L, Garre V, Song Y. Construction of DGLA producing cell factory by genetic modification of Mucor circinelloides. Microb Cell Fact. (2019), 18(1): 1–8. doi: 10.1186/s12934-019-1110-4
28. Yang J, Khan MAK, López-García S, Nosheen S, Nazir Y, Zhang H, et al. Improved SDA production in high lipid accumulating strain of Mucor circinelloides WJ11 by genetic modification. Am J Biochem Biotechnol. (2020) 16:138–47. doi: 10.3844/ajbbsp.2020.138.147
29. Csernetics Á, Tóth E, Farkas A, Nagy G, Bencsik O, Vágvölgyi C, et al. Expression of Xanthophyllomyces dendrorhous cytochrome-P450 hydroxylase and reductase in Mucor circinelloides. World J Microbiol Biotechnol. (2015) 31:321–36. doi: 10.1007/s11274-014-1784-z
30. Hussain SA, Hameed A, Khan MAK, Zhang Y, Zhang H, Garre V, et al. Engineering of fatty acid synthases (FASs) to boost the production of medium-chain fatty acids (MCFAs) in Mucor circinelloides. Int J Mol Sci. (2019) 20:786. doi: 10.3390/ijms20030786
31. Veiga-Crespo P, Blasco L, Do Santos FR, Poza M, Villa TG. Influence of culture conditions of Gordonia jacobaea MV-26 on canthaxanthin production. Int Microbiol. (2005) 8:55–8. doi: 10.2436/im.v8i1.9498
32. Nelis HJ, De Leenheer AP. Reinvestigation of Brevibacterium sp. strain KY-4313 as a source of canthaxanthin. Appl Environ Microbiol. (1989) 55:2505–10. doi: 10.1128/aem.55.10.2505-2510.1989
33. Nasrabadi MRN, Razavi SH. Enhancement of canthaxanthin production from dietzia natronolimnaea HS-1 in a fed-batch process using trace elements and statistical methods. Brazilian J Chem Eng. (2010) 27:517–29. doi: 10.1590/s0104-66322010000400003
34. Csernetics Á, Tóth E, Farkas A, Nagy G, Bencsik O, Manikandan P, et al. Expression of a bacterial ß-carotene hydroxylase in canthaxanthin producing mutant Mucor circinelloides strains. Acta Biol Szeged. (2014) 58:139–46. Available online at: http://abs.bibl.u-szeged.hu/index.php/abs/articleview/2830
35. Zhao Y, Yue C, Geng S, Ning D, Ma T, Yu X. Role of media composition in biomass and astaxanthin production of Haematococcus pluvialis under two-stage cultivation. Bioprocess Biosyst Eng. (2019) 42:593–602. doi: 10.1007/s00449-018-02064-8
36. Zhou P, Ye L, Xie W, Lv X, Yu H. Highly efficient biosynthesis of astaxanthin in Saccharomyces cerevisiae by integration and tuning of algal crtZ and bkt. Appl Microbiol Biotechnol. (2015) 99:8419–28. doi: 10.1007/s00253-015-6791-y
37. Han D, Li Y, Hu Q. Astaxanthin in microalgae: Pathways, functions and biotechnological implications. Algae. (2013) 28:131–47. doi: 10.4490/algae.2013.28.2.131
38. Silva F, Navarro E, Peñaranda A, Murcia-Flores L, Torres-Martínez S, Garre V, et al. RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a White Collar-1-like activator. Mol Microbiol. (2008) 70:1026–36. doi: 10.1111/j.1365-2958.2008.06470.x
Keywords: Mucor circinelloides, crgA, canthaxanthin, β-carotene ketolase, overexpression
Citation: Naz T, Yang J, Nosheen S, Sun C, Nazir Y, Mohamed H, Fazili ABA, Ullah S, Li S, Yang W, Garre V and Song Y (2021) Genetic Modification of Mucor circinelloides for Canthaxanthin Production by Heterologous Expression of β-carotene Ketolase Gene. Front. Nutr. 8:756218. doi: 10.3389/fnut.2021.756218
Received: 10 August 2021; Accepted: 06 September 2021;
Published: 13 October 2021.
Edited by:
Liguang Xu, Jiangnan University, ChinaReviewed by:
Enrique A. Iturriaga, University of Salamanca, SpainYonghong Meng, Shaanxi Normal University, China
Copyright © 2021 Naz, Yang, Nosheen, Sun, Nazir, Mohamed, Fazili, Ullah, Li, Yang, Garre and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanda Song, ysong@sdut.edu.cn
†These authors have contributed equally to this work
 Tahira Naz
Tahira Naz Junhuan Yang
Junhuan Yang Shaista Nosheen
Shaista Nosheen Caili Sun
Caili Sun Yusuf Nazir
Yusuf Nazir Hassan Mohamed
Hassan Mohamed Abu Bakr Ahmad Fazili
Abu Bakr Ahmad Fazili Samee Ullah
Samee Ullah Shaoqi Li
Shaoqi Li Wu Yang1
Wu Yang1  Victoriano Garre
Victoriano Garre Yuanda Song
Yuanda Song