Personalized Dietary Recommendations Based on Lipid-Related Genetic Variants: A Systematic Review
- 1Laboratorio Integral de Investigación en Alimentos, Instituto Tecnológico de Tepic/Instituto Nacional de México, Tepic, Mexico
- 2Departamento de Reproducción Humana, Crecimiento y Desarrollo Infantil, Centro Universitario de Ciencias de la Salud, Universidad de Guadalajara, Guadalajara, Mexico
- 3Departamento de Ciencias Médicas y de la Vida, Centro Universitario de la Ciénega, Instituto de Investigación en Genética Molecular, Universidad de Guadalajara, Guadalajara, Mexico
- 4Grupo de Investigación en Nutrición y Ciencias de los Alimentos, Departamento de Psicología, Educación y Salud, ITESO, Universidad Jesuita de Guadalajara, Tlaquepaque, Mexico
- 5Departamento de Salud, Universidad Iberoamericana (IBERO), Mexico City, Mexico
- 6Department of Food Technology, Engineering, and Nutrition, Lund University, Lund, Sweden
Background: Obesity and dyslipidemias are risk factors for developing cardiovascular diseases, the leading causes of morbidity and mortality worldwide. The pathogenesis of these diseases involves environmental factors, such as nutrition, but other aspects like genetic polymorphisms confer susceptibility to developing obesity and dyslipidemias. In this sense, nutrigenetics is being used to study the influence of genetic variations on the circulating lipid responses promoted by certain nutrients or foods to provide specific dietary strategies considering the genetic factors in personalized nutrition interventions.
Objective: To identify throughout a systematic review the potential nutrigenetic recommendations that demonstrate a strong interaction between gene-diet and circulating lipid variations.
Methods: This systematic review used the PRISMA-Protocol for manuscript research and preparation using PubMed and ScienceDirect databases. Human studies published in English from January 2010 to December 2020 were included. The main results were outcomes related to gene-diet interactions and plasmatic lipids variation.
Results: About 1,110 articles were identified, but only 38 were considered to fulfill the inclusion criteria established based on the reported data. The acquired information was organized based on gene-diet interaction with nutrients and components of the diet and dietary recommendation generated by each interaction: gene-diet interaction with dietary fats, carbohydrates or dietary fiber, gene-diet interaction with nutraceutical or dietary supplementation, and gene-diet interaction with proteins.
Conclusion: Findings included in this systematic review indicated that a certain percentage of dietary macronutrients, the consumption of specific amounts of polyunsaturated or monounsaturated fatty acids, as well as the ingestion of nutraceuticals or dietary supplements could be considered as potential strategies for the development of a wide range of nutrigenetic interventions since they have a direct impact on the blood levels of lipids. In this way, specific recommendations were identified as potential tools in developing precision diets and highlighted the importance of personalized nutrition. These recommendations may serve as a possible strategy to implement as dietary tools for the preventive treatment and control alterations in lipid metabolism.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42021248816, identifier [CRD42021248816].
Introduction
Overweight and obesity contribute to the development of various comorbidities associated with metabolic syndrome (MS), such as type 2 diabetes (T2D), hypertension, dyslipidemias, and cardiovascular diseases (CVD), which are the first causes of morbidity and mortality in the world (1). The pathogenesis of obesity is complex, and it involves environmental, sociocultural, physiological, medical, behavioral, genetic, epigenetic, and many other factors.
Although the main characteristic of obesity is the excess of adipose tissue, certain adipose tissue spots are associated with a higher risk of chronic diseases, such as that leading to increased waist circumference. Deposition of visceral fat leads to increased plasma levels of free-fatty acids and cytokines which can contribute to chronic inflammation, insulin resistance, and consequently, to the development of T2D, non-alcoholic fatty liver disease, dyslipidemias, and high blood pressure (2). Dyslipidemias comprise abnormally high concentrations and altered proportions of the different blood lipids and prelude to diverse CVD (3). Dyslipidemias are related to multiple factors such as genetic/hereditary (primary dyslipidemias) or an inadequate lifestyle (secondary dyslipidemias), which includes sedentary lifestyle, unbalanced diet, and excessive alcohol intake (4).
In this regard, it is emphasized that lifestyle habits, especially a healthy diet, are essential modifiable determinants for overweight and obesity, dyslipidemias, and CVD. However, genetic factors also play an important role in developing these pathologies (5). Individual responses to lifestyle modification vary, partially due to the genetic factor (6, 7). Data based on genome-wide association studies (GWAS) suggest that genotypic variations in specific nucleotides [single nucleotide polymorphisms (SNPs)] associated with obesity and dyslipidemias are factors of great importance behind this variable susceptibility (8). This has opened a new field for nutritional genomics around nutrigenetics, especially precision nutrition. These sciences have provided innovative evidence of the inter-individual interindividual variability and its relation with SNPs and nutrient intake since the human genome was completed (7). The postulates of nutrigenomics as applied science state that food components can act on the human genome, directly or indirectly, to alter the expression of genes and gene products. Diet can potentially compensate or accentuate the effects of genetic polymorphisms and the consequences of a particular diet depending on the balance of health, disease states, and an individual's genetic background (9).
Today's population nutrition guidelines are stratified to general characteristics (sex, age, or particular conditions like pregnancy). These recommendations are good for what they are made for, wide-reaching guidelines. Nonetheless, even with stratification for specific characteristics and conditions, they are not tailored to meet the needs of all individuals, especially for treating health conditions or reducing the risk of a disease integrating the genetic variants that confer susceptibility (10). A more exhaustive and individualized evaluation is required to include a variety of disease indicators, and therefore, a better disease stratification can be achieved (11). Thus, this systematic review aims to identify potential nutrigenetic recommendations that consider established interactions between gene-diet and blood lipid variations to provide possible strategies for personalized gene-diet treatment for dyslipidemias. However, it is crucial to consider that these critical points and potential nutrigenetic recommendations are subjects to specific populations and the results are reproducible in populations with the characteristics specified in each study.
Methods
Search Strategy for Systematic Review
This systematic review was developed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Protocol (PRISMA-P) guidelines (12). The review protocol was registered at PROSPERO under the CRD42021248816 registration number.
Studies published between January 2010 and December 2020, were identified by performing a comprehensive search in PubMed, the database of the National Center for Biotechnology Information (NCBI) of the US National Library of Medicine (US NLM), and ScienceDirect. In some cases, dietary recommendations were extracted directly from the original publications. The search strategy was composed of several MeSH terms (i.e., “Dyslipidemia” AND “polymorphism” AND “Diet,” “lipid polymorphism” AND “nutrients,” “Gene dietary recommendation” OR “genetic dietary recommendation”), which were revised and approved by all co-authoring researchers. Additionally, filters such as language restriction to English and studies that only involved humans, were applied. The detailed search strategies are available as Supplementary Table S1.
Study Selection and Eligibility Criteria
Once titles and abstracts were filtered from the research databases, duplicate records were removed, the eligibility criteria were checked at three different stages, first applied only to titles, then after reading the abstracts, and once more by going through the full article. Inclusion criteria were: nutrigenetics, personalized diet, diet-gene interaction, mutation, genetics, genes, genomics, genotype, genetic variants, polymorphism, lipid alterations, lipid abnormalities, hyperlipidemia, dyslipidemia, cholesterol, hypercholesterolemia, triglycerides, hypertriglyceridemia, HDL-c, and LDL-c. This revision included studies conducted only in adults, randomized controlled trials, studies without randomization, crossover, cohort, case-control, and cross-sectional studies. Article abstracts were excluded if they included the following criteria: children or infants, maternal, pregnant women, elderly, reviews, meta-analysis, mental-related diseases, renal diseases, ocular diseases, respiratory diseases, familial dyslipidemias, hepatitis, dental diseases, inflammatory diseases, alcohol consumption, use of any drugs or medicine, or if the approached genes not directly related to lipid metabolism, did not measure lipid parameters, articles not focused on diet-gene interaction and lipid responses or if no statistically significant changes in the lipid profile were detected, either positive or negative changes.
Data Collection Process
Two or three reviewers thoroughly analyzed all articles included in this review to determine eligibility. The most relevant information was captured in an electronic spreadsheet using the Population, Intervention, Comparison, Outcome (PICO) system (13). The data considered in the sample section were: sample size, ethnicity, age range, sex, physiological alterations or diseases of relevance to the study, allele frequency reported in the studies was also included. Regarding Intervention criteria, the following were considered: the administration of diets or structured feeding schemes, consumption of nutraceuticals or dietary supplements, and the application of Food Frequency Questionnaires (FFQ) or some other validated dietary assessment instrument to capture an individual's regular food consumption. A placebo, the consumption of a different diet or feeding scheme, the presence or absence of some polymorphism, and the genotype were considered as comparison criteria between treatments. The main results were outcomes related to gene-diet interactions and plasmatic lipids variation. Supplementary Table S2 shows the eligibility criteria of original studies in this review according to PICOS format.
Quality Assessment and Risk Bias
The quality and the risk of bias for selected articles were assessed individually using either the JADAD Scale (14) for Randomized Control Trials, Crossover studies and interventional studies, or the Newcastle-Ottawa Scale (NOS) (15, 16) for cohort and cross-sectional studies. Both scales are procedures to assess the methodological quality of a clinical trial independently; JADAD evaluates a total of 11 quality items (maximum score: 13 points) and NOS 8 items (maximum score: 10 points). Criteria such as population selection, comparability between groups or treatments, exposure determination, clarity of methodology and results, and risk of bias are evaluated on both scales. The articles selected were those that obtained a quality assessment score greater or equal (≥) than 8 points on the Jadad scale (adequate quality) or 7 ≥ points according to NOS (good and excellent quality). In this manner, studies were categorized as excellent, good, and satisfactory quality, and records that failed the quality assessment were excluded. The risk of bias and quality assessment was performed by 2 reviewers and cross-checked to solve inconsistencies; in case of discrepancy, a third external reviewer evaluated the article. Besides, the level of evidence was categorized according to the US Preventive Service Task Force report (17).
Data Analyses
The acquired information was organized based on gene-diet interaction with nutrients and components of the diet, as well as dietary recommendation generated by each interaction: gene-diet interaction with dietary fats (Table 1), gene-diet interaction with carbohydrates or dietary fiber (Table 2), gene-diet interaction with nutraceutical or dietary supplementation (Table 3) and, gene-diet interaction with proteins (Table 4).
Results
Study Selection and Characteristics
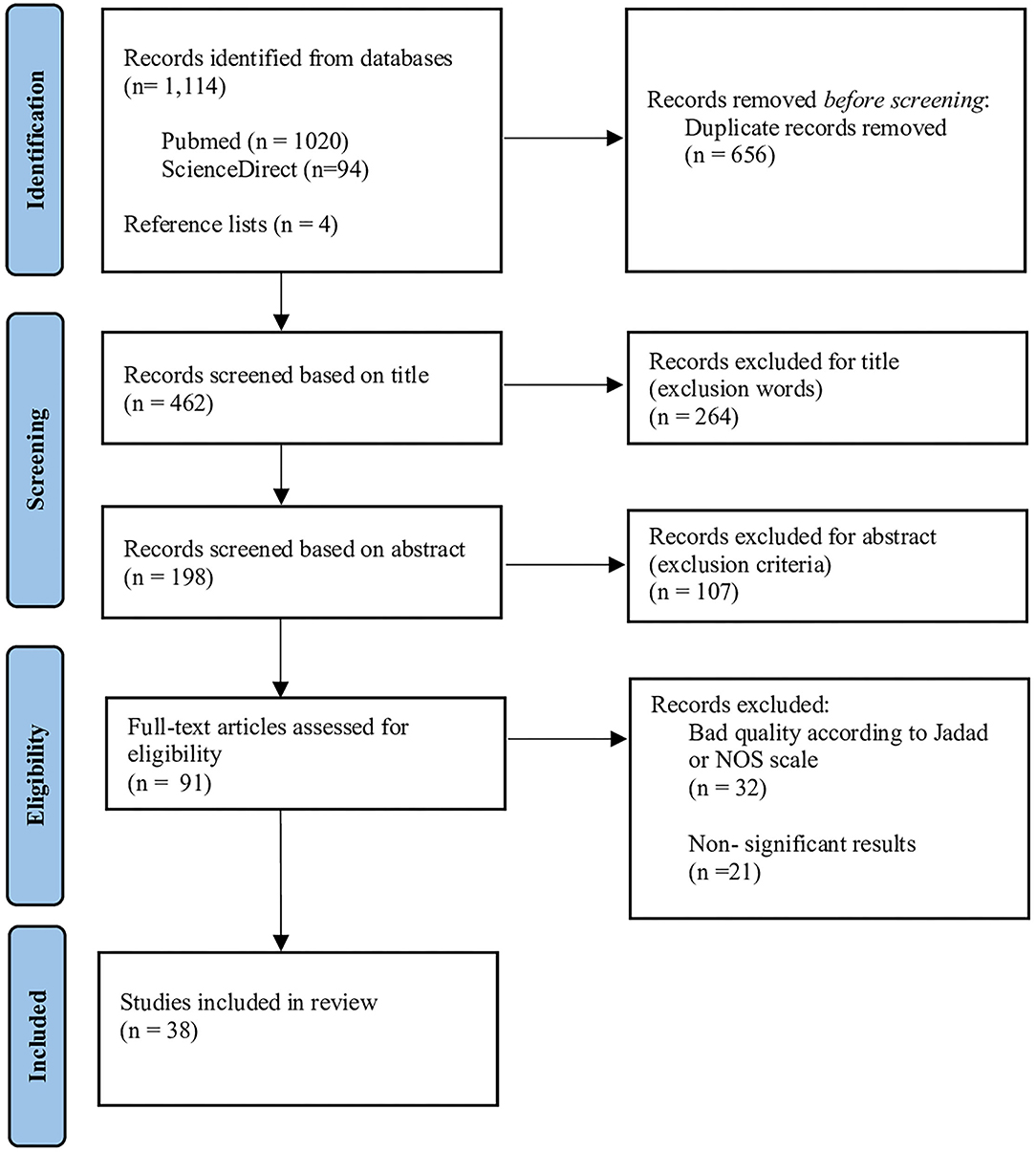
Figure 1 summarizes the search and study selection process using the PRISMA flow diagram. An initial sample of 1,118 articles was identified, 1,114 from databases, and 4 articles by reference lists. 656 duplicates were subsequently excluded, the remaining 462 articles were screened for title and abstract, based on exclusion word and criteria, previously mentioned, resulting in the removal of 371 articles. Then, 91 articles were full-text analyzed. A total of 32 articles were excluded, i.e., 21 articles did not present significant results in terms of a decrease in lipid parameters after treatment or no gene-diet interaction identified, and 32 articles were excluded because the methodologies were recognized as weak, according to quality assessment scales (NOS and Jadad). Finally, 38 articles were selected for the systematic review, of which 29 were randomized controlled trials (RCT), 2 cohorts and 7 cross-sectional studies. The quality and the level of evidence of these included articles are provided in detail in Supplementary Table S3. Results showed that 29 RCT evaluated with the Jadad scale had adequate quality (score ≥ 8 points) and level I of evidence (AHRQ scores). Regarding the items evaluated by the scale, only 5 double-blinded studies were identified; the rest were single-blinded and unblinded. All articles clearly defined the objectives of the study, the inclusion and exclusion criteria, and the statistical methods used. All studies had at least one control group or comparison group, and it is highlighted that 24 articles present in their methodology the justification of the sample size used (statistical power calculation).
Cohort and cross-sectional studies evaluated with the Newcastle-Ottawa Scale (NOS) had scores ranging from 7 to 9 (good and excellent quality). It should be noted that all the articles fulfilled more than 98% of all items evaluated by the NOS scale, such as satisfactory representativeness of the sample, justification or the sample size, exposure verification, comparability between subjects from different groups, evaluation of the results and precise description of the statistical tests used. See Supplementary Tables S4, S5 for detailed information.
The included studies showed significant heterogeneity regarding sample size, population characteristics, type of dietary intervention, genes and polymorphisms evaluated, allele frequency, and experimental analyses. Most of the studies included derived from major clinical trials like PREDIMED (Prevención con Dieta Mediterránea), CORDIOPREV (CORonary Diet Intervention with Olive oil and cardiovascular PREVention study), RISCK, POUNDS lost, and Food4Me. All of the studies were performed in adults (19–85 y/o) males or females who met all inclusion criteria, with a total sample size of n = 47,956 and an average sample size of n = 1,262 for all 38 studies included.
A large part of the population evaluated in the included studies were from Spain (n = 11) and China (n = 6), and a majority of the studies included Caucasians since they were carried out in countries such as the United Kingdom (n = 3), the United States (n = 3), Canada (n = 2), and other countries such as The Netherlands and New Zealand (n = 5). The rest of the studies included populations from Mexico, Brazil, Korea, and the Inuit population. The list of genes that predominated in this review included: CETP, AGT, CYP7A1, PPAR, APOB, APOE, APOA, APOC, LPL, ABCA1, LIPC, FADS1, FASN, FAAH, and SREBP-1C. There are several genes whose participation indirectly alters lipid metabolism (FTO, GHRELIN, LEP, POMC, among others); their influence is significant and remarkable, but this review considered genes directly involved in lipid metabolism.
Gene-diet interaction with nutrients or diet components identified in included articles were varied but were clearly categorized into four major groups, developed as main points throughout this review: studies that approached dietary fat intake recommendations and gene variants (n = 26), studies that addressed dietary recommendations related to gene variants and carbohydrate or dietary fiber intake (n = 6), dietary recommendations related to gene variants and nutraceuticals and dietary supplements intake (n = 7) and studies about dietary recommendations related to gene variants and protein intake (n = 2).
Dietary Fat Intake Recommendations and Gene Variants Related to Fat
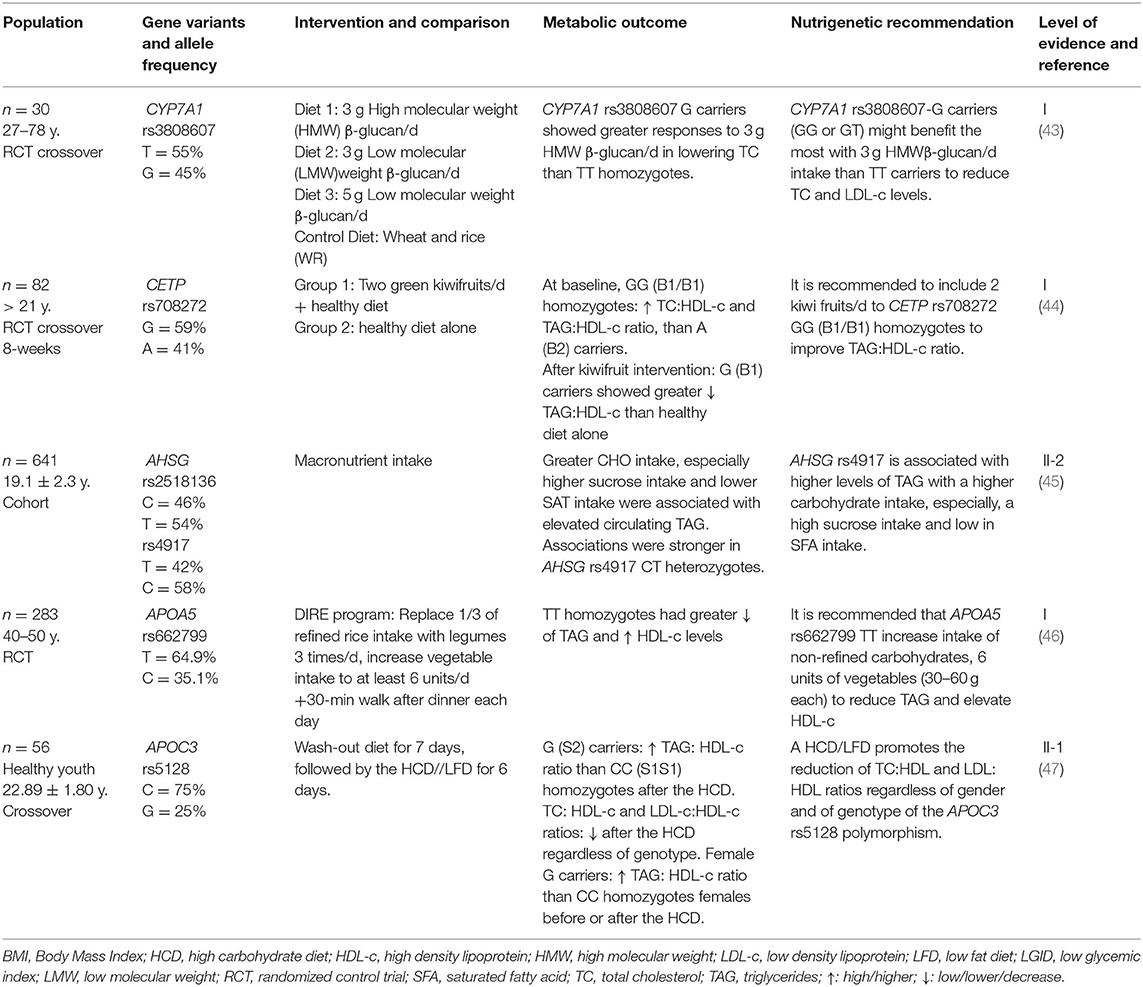
The official recommendation for achieving dietary fat for the healthy adult population is 20–35% of total calorie intake. However, it is equally important to emphasize the quality of fats in this percentage. A low-fat diet (LFD) could be considered when fat provides <20% total calorie intake, a moderate-fat diet 20–35%, and a high-fat diet (HFD) > 35% (57). Convincing evidence has shown that maintaining a healthy unsaturated/saturated fat balance might be more beneficial for reducing the risk of chronic diseases (58). Nevertheless, these recommendations do not consider the genetic background of different individuals. The findings regarding the interaction between gene variants and dietary fat intake are broken down below (Table 1).
Cholesteryl Ester Transfer Protein
CETP is a protein synthesized in the liver, and its function is to mediate the transfer of cholesterol esters from HDL (High-density lipoprotein) to VLDL (Very low-density lipoprotein), chylomicrons, LDL (Low-density lipoprotein), and the transfer of triacylglycerols (TAG) from VLDL and chylomicrons to HDL. When this protein is inhibited, increased HDL-c and decreased LDL-c occur (59).
With attention to the CETP gene, Garcia-Rios et al. (18) evaluated 424 adults with metabolic syndrome in an RCT that compared the Mediterranean diet (MedDiet) with a low-fat diet (LFD). CETP rs3764261 T-carriers (TT and TG) displayed higher HDL-c plasma concentrations (p = 0.021). TT and TG had lower TAG concentrations following the same diet than GG (p = 0.020). In this manner, the final recommendation for CETP rs376461 TT and TG is to increase the intake of monounsaturated (MUFAs) (22%) and polyunsaturated fats (PUFAs) (6%) and reduce intake of saturated fats (SFA) (>7%).
On the other hand, Rudkowska et al. (19) also evaluated CETP polymorphism in a cohort study in an Inuit population where CETP Ile405Val rs5882 TT was associated with elevated TC in subjects with a high fat intake (p = 0.0460). Therefore, this genotype might benefit from an LFD (19). It is essential to state that this recommendation might be specific only for the Inuit population.
Apolipoprotein E
APOE is perhaps one of the most studied genes for gene-diet interactions due to its importance as an essential regulatory protein in cholesterol homeostasis. It is a transporter for TAG-rich lipoproteins and a major ligand for transport to the liver by receptor-mediated endocytosis (21). APOE is a crucial component of plasma lipoproteins because it is involved in their production, conversion, and clearance. Specifically, APOE associates with chylomicrons and its remnants, VLDL, IDL, and shows preferential binding to HDL-c. It also binds with other receptors like the LDL and VLDL receptors. It is estimated that APOE E3/E4 and E4/E4 variants are associated with higher risks (22 and 45%, respectively for developing cardiovascular disease than E3/E3) (60).
Shatwan et al. (20) studied various APOE polymorphisms. They found a specific gene-diet interaction with APOE rs1064725 TT homozygotes, as they had significantly lower total cholesterol (TC) concentrations after a high MUFA (monounsaturated fatty acids) diet compared with a high SFA (saturated fatty acids) diet and ω-6 PUFA (polyunsaturated fatty acids) diet (p = 0.003 and p = 0.004). According to the authors, a possible explanation for this gene-diet interaction is that a high MUFA diet could shorten the residence time of very-low-density lipoprotein (VLDL) particles in the circulation, therefore, regulating and increasing TAG-rich lipoproteins clearance rate among TT homozygotes of APOE rs1064725.
21 investigated the effect of replacing saturated fatty acids (SFA) with either MUFAs or carbohydrates of high or low glycemic index (GI) in five different possible pattern combinations of APOE rs429358 and rs7412. After 24 weeks of dietary intervention, the five patterns resulted in different significant interactions between E2, E3/E3, and E4 carriers; APOE E3/E3 carriers benefit from a high MUFA/high GI diet in order to reduce TC (p = 0.03); E4 carriers benefit the most from a low fat/low GI diet (p = 0.03), and E2 carriers benefit from an LFD to maintain healthy circulating levels of TAG (p = 0.02). The article mentions that the isoforms of APOE alter the physical structure of the lipoprotein itself. E2 isoform, for example, reduces the affinity of the particle to bind to LDL receptors. This difference explains why E2 transport is associated with low levels of LDL-c.
On the other hand, there may be less competition in uptake for the E2 isoform and its content. The association of E4 carriers with dyslipidemia is not so clear; this isoform is preferentially incorporated into VLDL, and this could compete with the uptake of LDL by the LDL receptor, thus decreasing the hepatic LDL uptake and prolonging its presence in circulation. This mechanism is supported by other different in vitro studies and may explain why E4 carriers express a greater sensitivity to dietary fat reduction, specifically SFAs (20, 61).
Fallaize et al. (22) studied APOE rs429358 and rs7412 in an RCT and found that through different levels of gene-personal nutrition. Gene-based personalized nutrition was associated with a significant reduction in total fat and SFA intake compared to a not gene-based personalized nutrition approach in E4 carriers. There was also a significant impact of genotype on the change in TC concentrations after dietary advice intervention (total fat, p = 0.016; SFAs, p = 0.025; MUFAs, p = 0.019; PUFAs, p = 0.024). The study advocates for dietary information based on personal genetics as much more understandable and valuable than general dietary guidelines and suggests that it could help to increase the motivation for effective dietary changes. Nevertheless, this finding could be exclusive to this study and does not recount to personalized recommendations for individual health benefits.
In this context, Gomez-Delgado et al. (61) evaluated patients with coronary heart disease from the CORDIOPREV RCT study, which compared the MedDiet (high in MUFAs) and an LFD (<30%). After 3 years, T carriers in the MedDiet group had a significantly larger reduction in postprandial TAG levels than CC (homozygous wild type) (p = 0.03). So, it is recommended that APOE rs439401 T carriers follow a MedDiet pattern to decrease TAG levels. These results add up to a final recommendation for APOE rs439401 and rs7412 benefit from the MedDiet pattern with a moderate fat intake high in MUFAs and low in SFA. This article provides an explanation for these findings based on the fact that a diet rich in MUFAs increases the secretion of VLDL-c (that contains APOE) and IDL-c, leading to faster elimination of TAG from the bloodstream, thus preventing its conversion to LDL-c and, possibly, reducing its atherogenic effect.
Carvalho-Wells et al. (24) found that APOE rs7412 and rs429358 E4/E4 and E3/E4 experienced more significant reductions in TAG with a high SFA diet and 3.45 g ω-3 supplementation, than with the LFD (p = 0.033). These findings can be interpreted as an additional therapeutic effect of ω-3 for these polymorphisms.
Apolipoprotein A1
APOA1 gene encodes a protein that plays a fundamental role as a component of HDL. It plays a pivotal role in reverse cholesterol transport as it promotes cholesterol transportation from tissues to the liver for excretion. It is also a cofactor for cholesterol acyltransferase (LCAT), the enzyme responsible for most plasma cholesteryl esters' formation (62).
De Luis et al. (28) evaluated 282 adults with obesity in an RCT for 12 weeks and analyzed APOA1 rs670. All participants were randomized into an LFD (27% fats) or an HFD (38%), both diets with a caloric restriction of 500 kcal based on individual estimates. A carriers with LFD showed increased levels of HDL-c after the LFD (p = 0.03). It is essential to mention that A carriers already had elevated HDL-c before the dietary intervention; previous studies had made similar observations. On the other hand, Rudkowska et al. (19) reported that APOA1 rs670 G in an Inuit population cohort study is associated with an increase in LDL-c with an HFD, yet decreased levels of HDL-c were noticed with a diet low in saturated fat. These two studies support the recommendation for APOA1 rs670 to follow an LFD. However, both studies share the limitation of not specifying the type of unsaturated fats used in the intervention, such as MUFAs and PUFAs. They only refer to the total percentage of fat, so the potential interactions between these types of fat may have been overlooked.
Apolipoprotein A5
Apolipoprotein A5 (APOA5) is produced in the liver, and its primary lipoprotein associations are VLDL-c, chylomicrons, and HDL-c. Its primary function is promoting lipoprotein lipase (LPL) activity on TAG lipolysis (59). For this reason, APOA5 plays a pivotal role in regulating plasma TAG, a risk factor for cardiovascular disease (63). Genetic studies have reported that polymorphisms in APOA5 are among the most determinant factors for developing elevated TAG levels. APOA5 CC homozygotes and TC heterozygotes have been associated with increased cardiovascular disease in various populations (64).
Zhang et al. (26) studied 734 overweight or obese adults who were randomly assigned to four different diets with different percentages of energy derived from fat, protein, and carbohydrate, and found that APOA5 rs964184 CG and GG benefit from LFD (<20% energy from fat) to reduce TC and LDL-c. This is concurrent with Sánchez-Moreno et al. (27) found with APOA5 rs662799 in a cross-sectional study in adults with a BMI > 26 kg/m2, where TC heterozygotes seemed to benefit from an LFD for controlling VLDL and TAG levels. These coincident results and recommendations suggest that this gene-diet interaction could be a trend in obese individuals; however, it is also possible that an impaired efficiency of ribosomal translation in the −1131C allele may lead to a reduced LPL-mediated TAG uptake into adipocytes. Another possible mechanism is linked to the regulation of APOA5 by thyroid hormones, or peroxisome proliferator-activated receptors (PPARs) (20, 65).
Peroxisome Proliferator-Activated Receptors
The PPAR family encompasses critical transcriptional regulators of lipid and carbohydrate metabolism. The isotypes of PPAR include PPARα, PPARδ, and PPARγ, and these codify a great variety of genes responsible for glucose and carbohydrate homeostasis as well as cholesterol metabolism. Any alteration in these genes can lead to obesity and cardiovascular disease. For example, the C allele of PPARα is a crucial regulator of lipid metabolism and has been known to have cardioprotective effects because it associates with higher levels of HDL-c and reduced levels of TAG and VLDL-c (66).
PPARs are known as major transcriptional regulators of lipid and carbohydrate metabolism. Two studies based on the same RCT “RISCK Study” (29, 30) evaluated PPARγ rs1801282 (Pro12Ala) and PPARα rs1800206 (Leu162Val) in the same intervention, where the participants were randomly assigned to one of 3 diets to be consumed for 24 weeks: reference diet (high SFA), high MUFA diet and low-fat diet, all with equivalent PUFA intake. In the first study, 415 subjects were included, and only significant interaction between dietary PUFA:SFA ratio and genotype as a determinant of plasma concentrations of TC (p = 0.02), LDL-c (p = 0.002), and TAG (p = 0.02) was found; at a low PUFA:SFA ratio (0.33) mean TC and LDL-c concentrations in G (Ala12) carriers was significantly higher relative to non-carriers. There was also a significant trend in reducing plasma TAG (-50%) in G (Ala12) carriers as the PUFA:SFA ratio increased from 0.34 to 0.65 (p = 0.002). In the second study, also by AlSaleh et al. (29), both Pro12Ala PPARγ and Leu162Val PPARα had 45.7% lower levels of LDL-c after a high MUFA diet, compared to those with a low-fat diet (p = 0.005). Furthermore, PPARγ G carriers in the reference diet group (high in SFA) had significantly higher TC and LDL-c than C (Pro12) carriers.
Even though this gene-diet interaction has not been described before, the study also concluded the results were related to an increased PUFA intake rather than the SFA intake. In the second study, several SNPs were genotyped in 466 subjects, and it was found that co-carriers of the PPARγ rs1801282 G (Ala12) and PPARα rs1800206 V (Val162) alleles had 45.7% lower LDL-c and 53.5% lower sdLDL (small dense LDL-c) as a proportion of LDL, after the high MUFA diet, compared to LFD.
The results discussed above complement what Rudkowska et al. (19) found in the Inuit cohort. Participants with C allele (wild type) of PPARγ2 rs10856710 had higher TC and LDL-c levels (p = 0.0175) when consuming a higher-TotFat and SatFat diet. These findings indicate that PPARγ2 rs10856710 C carriers benefit from a low SFA intake to lower TC and LDL-c levels.
Leptin Receptor
Leptin Receptor (LEPR) is responsible for coding the receptor for leptin, a hormone that regulates appetite and body weight. LEPR is also involved in other mechanisms like the regulation of fat metabolism. Mutations in this gene are associated with obesity (67).
Domínguez-Reyes et al. (25) analyzed the dietary fat intake of 100 subjects with normal weight and obesity using a frequency food consumption questionnaire. This study found that LEPR rs1137101 G carriers had two times higher risk of total cholesterol levels ≥200 mg/dl (OR = 2.1, 95 % CI 1.15–3.8; p = 0.008), AG/GG had a high risk of hypercholesterolemia (OR = 9.4, 95 % CI 2.1–41.5; p = 0.003). Furthermore, AG/GG of the rs1137101 polymorphism in the LEPR gene with an intake ≥12 g/day of saturated fatty acids have 3.8 times higher risk of cholesterol levels ≥200 mg/dl (p = 0.002) and 2.4 times higher risk of triglyceride levels ≥150 mg/dl (p = 0.02) than those with an intake <12 g/day SFA intake. Therefore LEPR rs1137101 AG/GG might benefit from a low SFA intake (<12 g/day SFA).
A possible explanation for this interaction is that when leptin signaling is defective in the liver, this may induce suboptimal protein expression levels of the lipolysis stimulator receptor, which may contribute to elevated postprandial lipemia (25, 68).
Hepatic Lipase (LIPC)
LIPC is a lipolytic enzyme that hydrolyzes TAG and phospholipids present in plasmatic lipoproteins like chylomicrons, LDL, and HDL lipoproteins, releasing free fatty acids and smaller lipoprotein particles (69).
Xu et al. (31) tested the effect of rs2070895 variant in LIPC on changes in blood lipids in response to various weight-loss diets: 2 diets were low fat (20%), and 2 diets were high fat (40%), or 2 diets were average protein (15%), and 2 diets were high protein (25%). After 2 years of intervention, they observed a long-term, cumulative pattern. Dietary fat significantly modified the genetic effects of rs2070895 on changes in serum TC, LDL cholesterol, and HD-c (p < 0.05). Data suggest that A carriers might benefit more in terms of lipid profile by eating a low-fat (20%), high-carbohydrate (55–65%) diet. A possible explanation for these findings comes from in animal studies showing that LIPC activity can be suppressed by a high-fat diet (70).
A similar interaction was found in the cohort study by Rudkowska et al. (19), where TT homozygotes of LIPC C-514T rs1800588 had higher TAG levels with a higher fat intake (p = 0.007).
Finally, Smith et al. (32) investigated dietary modulation of LIPC rs1800588 (-514 C/T) for lipids, comparing a high-fat Western diet and a low-fat traditional Hispanic diet consumption. The authors claim that the dietary patterns differed in total fat and fat composition (e.g., greater MUFAs contents in the Hispanic diet). In major allele carriers (CC/CT), HDL-c levels were higher following the Western diet than the Hispanic diet. Only major allele carriers benefited from the higher fat diet for HDL-c. However, in C carriers but not in T carriers, the Western diet was associated with higher LDL-c and higher TC in both phases of experimentation (high-fat Western diet and low-fat traditional Hispanic diet). LIPC rs1800588 CC/CT might benefit from an HFD (39% total fat) to raise HDL-c levels but may also raise LDL-c and TC levels. This result is somewhat controversial compared to Rudkowska et al. (19) and Xu et al. (31). It would be necessary to differentiate and prioritize between the benefits of an elevated HDL-c and an elevated LDL-c and their proportional contribution to total cholesterol.
Lipoprotein Lipase
Lipoprotein Lipase (LPL) is a crucial enzyme for lipid metabolism. It catalyzes the hydrolysis of triacylglycerols of chylomicrons and VLDL-c to provide free fatty acids for oxidation and, later, their use in the heart and other tissues and their storage in adipose tissue (71). In order to understand the relationship between rs13702T > C polymorphism in the 3'untranslated region of LPL and the fat intake on TAG, Corella et al. (33) studied 7,187 participants in the PREDIMED randomized trial that tested a MedDiet intervention (2 groups: one supplemented with extra-virgin olive oil and the other supplemented with nuts) compared with a low-fat diet control group. The rs13702T > C polymorphism was strongly associated with lower TAG in C carriers and interacted synergistically with dietary monounsaturated (p = 0.038) and unsaturated fat intake (p = 0.037), decreasing TAG at baseline. By 3 years, a gene-diet interaction was found in which the C allele was associated with a greater reduction in TAG after intervention with MedDiet, high in unsaturated fats (p = 0.025). The rs13702 C allele, which disrupts a microRNA-410 recognition element seed site resulting in a gain of function, was strongly associated with lower TAG levels, meaning that LPL rs13702 C carriers benefit from a high-unsaturated fat (30 g/day) MedDiet intervention in order to reduce TAG. According to Corella et al. (33), a possible explanation for this observation is that this interaction could result from the decrease in cellular oxidative stress associated with the consumption and content of unsaturated fats.
Fatty Acid-Binding Protein 2
The Fatty Acid-Binding Protein 2 (FABP2) gene encodes the intestinal fatty acid-binding protein (FABP), which is found in enterocytes and is involved in the intracellular transport of long-chain fatty acids. The Thr54 isoform (T allele) has been shown to have a two-fold greater binding affinity for the long-chain fatty acids. The Thr54 isoform has been associated with hypertriglyceridemia, increased body mass index (BMI), hyperinsulinemia, and insulin resistance.
De Luis et al. (35) showed that FABP2 rs1799883 T carriers had a better metabolic response than obese patients with GG (Ala54Ala), with a decrease of total cholesterol, LDL-c, insulin levels, leptin levels, and HOMA-IR after weight loss with a high PUFA-hypocaloric diet (SAT = 21.8%, MUFAs: 55.5%, PUFAs = 22.7%, 7-g/day ω-6 fatty acids, 2-g/day ω-3fatty acids, and a ratio ω-3/ω-6 of 3.5). On the other hand, Martinez-Lopez et al. (34) showed that a moderate-fat diet (Fat:30%, SAT < 7%, MUFAs: 10–15%, and PUFAs: 10%, protein: 15%, CHO: 55%) had a positive effect on anthropometric and biochemical variables in overweight or obese subjects. This study showed that the T carriers responded better to a moderate-fat diet. In this way, Martinez-Lopez et al. (34) and De Luis et al. (35) suggested that high ingestion of PUFAs and MUFAs and a limiting dietary intake of SFA was beneficial for T carriers since the effects of the T allele and the diet contributed to the decrease of metabolic parameters as a result of the changes in the amount and type of fat intake. Under this condition, the isoform Thr54-I-FABP captures more “good” substrates (MUFAs and PUFAs) instead of saturated fats. Consequently, the Thr54 isoform (T allele) could be contributing indirectly to the expression of genes influenced by the peroxisome proliferator-activated receptor (72).
Fatty Acid Desaturase
Genes in the fatty acid desaturase (FADS) family of genes control the fatty acid metabolism pathway in the human body. This gene encodes a protein (desaturase enzymes) that controls and regulates the unsaturation of fatty acids by introducing double bonds between the defined carbons of the chain of the respective fatty acid. The protein encoded by the FADS1 gene, for example, desaturates the PUFAs ω-3 and ω-6, acting as d-5 desaturase, so that EPA and arachidonic acid are formed (73).
Ching et al. (36) aimed to determine the interaction of ω-3 and ω-6 PUFAs with rs174547 in the FADS1 gene in metabolic syndrome (MS). Chinese and Indian vegetarians were classified according to ω-3 and ω-6 intake. Results showed that middle LA (linoleic acid) and ALA (alfa-linoleic acid) groups with a middle intake of LA (5.87–8.19 g/day) had lower HDL-c levels (p < 0.05).
A possible explanation for this is that over-consumption of ω-6 may inhibit the production of ω-3 PUFAs, which reduces HDL-c level among those with TT homozygotes of rs174547 in the FADS1 gene. Dietary intake of LA may interact with rs174547 in the FADS1 gene to affect HDL-c level, which warrants the need to monitor the amount of LA and ALA in the regular diet of vegetarians. The health benefit of specific PUFAs may depend on individual FADS1 genotypes or δ-5 desaturase activity, which is a rate-limiting enzyme in PUFAs biosynthesis encoded by the FADS1 and FADS2 genes (36, 74).
Lu et al. (37) examined whether genetic variations in the FADS gene cluster region interact with dietary intakes of ω-3 and ω-6 PUFAs to affect plasma total, HDL and non-HDL-cholesterol concentrations, and their relation with rs174546, rs482548, and rs174570 in the FADS gene cluster region, in 3575 participants in the second survey of the Doetinchem Cohort Study. Results showed significant associations between rs174546 genotypes and total and non-HDL-c concentrations in the group with a high intake of ω-3 but not in the low-intake group (p = 0.02). A significant gene-diet interaction was found as TC differed significantly according to rs175456 genotype (p = 0.02). The C carriers had higher TC concentrations, especially and only statistically significant in subjects with a high ω-3 PUFA intake (p = 0.006). Moreover, ω-6 PUFA intake also modified rs174546 genotype as the C allele was associated with higher HDL-c concentrations in subjects with a high intake (p = 0.004). No difference was seen for low ω-6 intake.
Although these studies do not provide the highest level of evidence due to their design, both results are congruent and show that polymorphisms in the FADS1 and FADS2 genes can affect non-HDL concentrations, leading to the conclusion that these polymorphisms benefit from a diet high in PUFAs, specifically, high in ω-6. The potential mechanism underlying this interaction between elevated desaturase activity and elevated CT or LDL-c levels is not yet entirely clear.
Fatty Acid Synthase
FASN codifies for a multifunctional enzyme that catalyzes palmitate synthesis from acetyl-CoA and malonyl-CoA into long-chain SFA (75).
Dolley et al. (38) hypothesized that positional candidate FASN polymorphisms and dietary fat might modulate LDL particle size. A Quebec Family Study (QFS) sample was studied, and five single nucleotide FASN polymorphisms were genotyped. Results showed that FASN rs4246444 was associated with LDL peak particle diameter (LDL-PPD) when fat intake was taken into account (p = 0.001). A carriers showed smaller LDL-PPD only when consuming a high amount of fat >35% total energy compared to the C carriers. The outcomes suggest that dietary fat intake may modify the effect of the FASN rs4246444 on LDL-PPD since FASN rs4246444-A carriers benefited from an LFD (<35% TE) in order to avoid LDL-PPD decrease and increase the number of LDL particles.
The diameter of small LDL-PPD is relevant because small LDL particles are considered more atherogenic as they can filter into vascular tissue and deposit in the subendothelial layer, contributing to the formation of atherogenic plaque and its consequences (76).
Tumor Necrosis Factor (TNF-α)
The superfamily of Tumor Necrosis Factor (TNF) genes codify multifunctional pro-inflammatory cytokines. One of its most studied variants, TNF-α, is a cytokine secreted by macrophages. This cytokine is implicated in multiple diseases, including those related to lipid metabolism, insulin resistance, arthritis, cancer, and others (77). Its increased expression in the adipose tissue of humans has been correlated with the degree of adiposity and associated with insulin resistance (39).
de Luis et al. (39) investigate the influence of G-308, rs1800629 of TNF-α on metabolic changes and weight loss secondary to high MUFA (Diet M) vs. high PUFA (Diet P) hypocaloric diets. In obese subjects, randomly allocated during 3 months to one of these diets for 3 months. Diet P resulted in reduced total cholesterol levels (p = 0.01), LDL cholesterol levels (p = 0.008) and TAG (p = 0.02). Therefore, G carriers (promoter variant of TNF-α gene) have a better metabolic response than obese carriers of A-allele consuming a high PUFA hypocaloric diet.
Gomez-Delgado et al. (23) obtained different results when examining whether the long-term consumption of a MedDiet, enriched in olive oil, compared with a low-fat diet, interacts with rs1800629 and rs1799964 SNPs at the TNF-α gene. In order to improve TAG, glycemic control, and inflammation markers. After 12 months of MedDiet, the decrease in TAG was statistically significant in GG subjects but not in A carriers (p = 0.005). Thus, the authors demonstrated that postprandial abnormalities associated with MS could be attenuated with high MUFA diets. One hypothesis to explain these findings could be related to the phenotypic flexibility associated with GG homozygotes MUFAs play a crucial role in maintaining phenotypic flexibility through TAG and inflammation homeostasis.
Max-Like Protein X Interacting Protein-Like
The MLXIPL gene encodes a transcription factor that, with the help of glucose, binds and activates elements for the response to carbohydrates in the promoters of TAG synthesis genes (78).
The effect of the MedDiet was analyzed by Ortega-Azorín et al. (40) in rs3812316 of the MLXIPL (Max-like protein X interacting protein-like) gene. A MedDiet intervention vs. a control diet for cardiovascular prevention was tested. The MLXIPL rs3812316 was associated with lower baseline TAG (p < 0.0001) and lower hypertriglyceridemia (odds ratio, 0.73 in G-carriers vs. CC homozygotes). This association was modulated by baseline adherence to MedDiet. When adherence to MedDiet was high, the protection was stronger and vice versa. Both the MLXIPL rs3812316 (p = 0.0001) and the MedDiet intervention (p = 0.030) were significantly associated with decreased TAG throughout the follow-up. Results suggest that MedDiet enhances the TAG-lowering effect of the MLXIPL rs381231.
The study was one of the first to report this gene-diet interaction. However, although the regulation of the MLXIPL remains unknown, several animal models have reported that the diet modulates the expression of this gene, thus affecting the metabolism of TAGs, which matches the outcome of the above-described study.
Transcription Factor 7 Like 2
The TCF7L2 gene or transcription factor 7-like-2 encodes a protein that acts as a transcription factor with multiple functions and multiple polymorphisms. This gene is so broad that it is known as a pleiotropic gene (79).
Corella et al. (80) investigated the Transcription factor 7-like 2 (TCF7L2) rs7903146 polymorphism associations with type 2 diabetes, glycemia, blood lipids, and cardiovascular disease incidence. A randomized trial (two MedDiet interventions: group MedDiet + extra virgin olive oil (50 ml/d), group MedDiet + nuts (30 g/day), and a control group) with PREDIMED study sample was undertaken. Data were analyzed at baseline and after a median follow-up of 4.8 years. When adherence to the MedDiet was low, TT homozygotes had higher TC, LDL-c, and TAG (p = 0.005, p = 0.003, p = 0.046, respectively). Nevertheless, this increase was not observed (p = 0.605). Authors suggest that the overall MedDiet pattern rather than specific foods contribute to maintaining adequate TC, LDL-c, and TAG; therefore, TCF7L2 rs7903146 TT homozygotes recommendation is to follow a MedDiet pattern to maintain adequate levels of TC, LDL-c and TAG.
Fatty Acid Amide Hydrolase
Fatty Acid Amide Hydrolase (FAAH) gene encodes an integral membrane protein whose function is to hydrolyze bioactive amides to free fatty acids and ethanolamine (81).
Deluis et al. (41) investigated the role of FAAH rs324420 polymorphism on metabolic changes secondary to the administration of two kinds of diets. Basal and final measurements of 248 patients with obesity were compared after a 3-month period in which they received either an LFD or an LCD (low carbohydrate diet). With the LFD, TC and TAG decreased in the CC (wild-type homozygotes) and only decreased TC in AA homozygotes (p < 0.05). FAAH rs324420 CC benefits from consuming either an LFD to reduce TC and TAG or an LCD to decrease TC and LDL-c levels. The authors argue that a possible mechanism for these effects is that dysregulation of FAAH can lead to a higher hepatic and central endocannabinoids concentration, consequently leading to an increase in energy storage. With a hypocaloric diet, this regulation decreases and results in the observed improvement of the lipid profile.
Sterol Regulatory Element-Binding Transcription Factor 1
The SREBP-1c gene, as its name implies, encodes a transcription factor that binds to the sterol regulatory element (SRE1), which is one reason why it is found in the promoter of the LDL-c receptor gene and other genes involved in sterol biosynthesis. These genes have different transcription variants, including SREBP-1a and SREBP-1c (Genecards Org, 2021).
Zhang et al. (42) investigated the possible association between SREBP-1c rs2297508 polymorphism and lipid traits changes following a high-CHO/low-fat diet (15% fat and 70% CHO) for 6 days. Gender-specific changes in lipid profile emerged after treatment: levels of TC in males with GG homozygotes and C carriers decreased after HC/LF diet while female GG homozygotes also decreased levels of TC but increased TAG levels (p < 0.05). An HC/LFD might benefit SREBP-1c rs2297508 male and female GG homozygotes regarding the reduction of TC levels but may increase triglyceridemia in females.
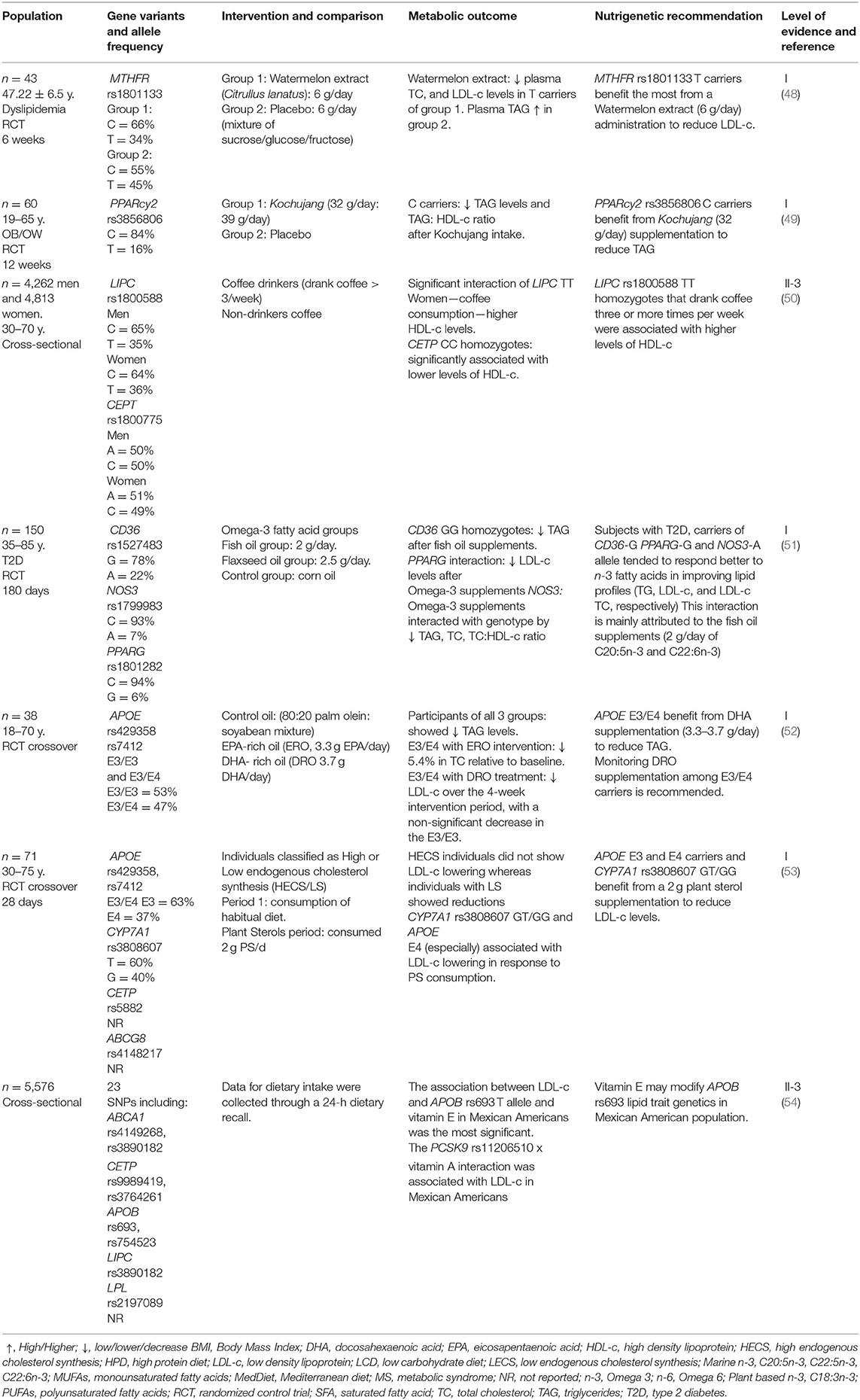
Dietary Recommendations Related to Gene Variants and Carbohydrate or Dietary Fiber Intake
Carbohydrates should provide 40–60% of the total calorie intake in a balanced diet. Dietary fiber and sugars form carbohydrates, but their recommended intakes are specific and separate from carbohydrates. Dietary fiber consists mainly of structural polysaccharides that cannot be digested in the gastrointestinal tract of humans. There is ample evidence supporting that an adequate intake of fiber (20–35 g/day) helps to reduce blood cholesterol, prevent cardiovascular disease, and prevent constipation; there is also evidence about limiting the intake of free sugars to less than 10% of total energy intake, and even further reduction to 5%, as aiding measures for additional health benefits (58). Six studies were found in this systematic review that demonstrated a gene-diet interaction with carbohydrates or fiber (Table 2).
Cytochrome P450 Family 7 Subfamily A Member 1
The cytochrome P450 proteins are monooxygenases that catalyze many reactions in drug metabolism and the synthesis of cholesterol, steroids, and other lipids. This endoplasmic reticulum membrane protein catalyzes the first reaction in the cholesterol catabolic pathway of extrahepatic tissues, converting cholesterol to bile acids (82). Wang et al. (43) studied the effect of two different molecular weight β-glucan preparations on LDL-c and TC, in three groups of volunteers: 3 g/day high molecular weight (HMW), 3 g/day low molecular weight (LMW), and 5 g/day LMW, in mildly hypercholesterolemic adults, typified for CYP7A1 rs3808607 G carriers. After a 5-week intervention, GG homozygotes seemed to benefit the most with 3 g HMW β-glucan/d administration and, more importantly, than TT homozygotes concerning TC and LDL-c levels (p < 0.05). This study provided knowledge regarding the influence of the molecular weight of β-glucans on their physiological effects, highlighting the importance of supplementing the diet with the polysaccharide and controlling its quality and molecular weight.
Apolipoprotein A5
Jang et al. (46) studied a dietary intervention program in APOA5 rs662799 adult carriers with hypertriglyceridemia that consisted in replacing 1/3 of refined rice intake with legumes three times per day and an increased vegetable intake (6 units, 30–70 g/unit). A 12-week intervention significantly decreased TAG and increased HDL-c regardless of genotype (p = 0.002), although significantly lower TAG and higher HDL-c levels were recorded in APOA5−1121TT homozygotes compared with C carriers (p = 0.044). It is recommended that APOA5 rs662799 TT homozygotes increase the intake of non-refined carbohydrates, 6 units of vegetables (30–60 g each) to reduce TAG and elevate HDL-c. APOA5 was found to activate LPL in vitro and to accelerate TAG catabolism in animal studies. Therefore, reduced TAG levels in C carriers may be explained based on their reportedly reduced APOA5 activity, which may result in decreased lipoprotein lipase (LPL) activity (46).
Cholesteryl Ester Transfer Protein
Gammon et al. (44) compared 82 hypercholesterolemic adults who were Cholesteryl ester transfer protein (CETP) Taq1B rs708272 carriers with two groups: a healthy diet and healthy diet plus two green kiwifruits per day. CETP Taq1B CC homozygotes (B1/B1) decreased TAG/HDL-c ratio after the kiwifruit intervention compared to the control group (p = 0.001). So, the final recommendation was to include two kiwi fruits per day to CETP rs708272 C carriers to improve TAG/HDL-c ratio. Elevated CETP activity results in enhanced TAG enrichment of HDL-c and LDL-c. CETP mass or activity was not measured in this intervention, but the difference in C carriers and T carriers (B2) could have been mediated by kiwifruit polyphenols decreasing CETP activity (83).
Alpha 2 Heremans-Schmid Glycoprotein
Similarly, Robinson et al. (45) performed another study involving 641 young Mexican healthy college students, where Alpha 2 Heremans-Schmid glycoprotein (AHSG) rs4917 carriers had higher levels of TAG with a higher carbohydrate intake (p = 0.004), specifically high in sugars (p = 0.05) and low in saturated fats (p = 0.01). AHSG is a gene that codes for Fetuin-A (FetA), a hepatokine and an adipokine that inhibits insulin receptor tyrosine kinase and thus reduces insulin sensitivity via decreased translocation of glucose transporter type 4; FetA has also been shown to bind saturated fatty acids and stimulate inflammatory cytokine release via toll-like receptor 4 (84). In this sense, Robinson et al. (45) detected that T carriers were associated with higher TAG levels in this cohort. However, the association was also strongly correlated with BMI. On the contrary, Song et al. (47) found in a group of Chinese healthy young college students that an intervention with a high carbohydrate (70% total calorie intake) LFD (15% total calorie intake) had favorable effects on TC, LDL-c, and HDL-c ratios, regardless of gender or Apolipoprotein C3 (APOC3) SstI polymorphism (p = 0.001).
Dietary Recommendations Related to Gene Variants and Nutraceuticals and Dietary Supplements Intake
Nutraceuticals are defined as food or part of a food that provides health benefits, including preventing and treating disease beyond essential nutritional functions. Most nutraceuticals can be found in foods of plant origin; nevertheless, some of them can be found in animal foods (e.g., ω-3 fatty acids in fish). Besides, nutraceuticals come in different forms (whole foods, dietary supplements, or isolated compounds. In this way, dietary supplements are defined as “concentrated sources of nutrients (macronutrients, vitamins, minerals, enzymes, amino acids) or other substances with a physiological or nutritional effect to complement the diet” (85).
Both nutraceuticals and dietary supplements might positively impact consumer lipid traits when considering their genetic background.
Methylenetetrahydrofolate Reductase
The methylenetetrahydrofolate reductase (MTHFR) is an essential enzyme for folate metabolism, a necessary process for cell metabolism involving DNA, RNA, and protein methylation. It has also been shown that MTHFR C677T polymorphism is associated with hyperhomocysteinemia, hypertriglyceridemia, and elevated TC and LDL-c. In this context, Massa et al. (48) investigated the effect of a watermelon extract (Citrullus lanatus) supplementation (6 g/day) in adults carriers of MTHFR rsC677T or rs1801133 with diagnosed with dyslipidemia and found that total cholesterol and LDL-c levels were reduced after 6 weeks (p <0.01). The most significant reduction was observed in CT and TT hetero and homozygotes. The effect may be related to the action of antioxidant compounds present in this fruit, such as lycopene, citrulline, and arginine (48).
Peroxisome Proliferator-Activated Receptor Gamma
Peroxisome proliferator-activated receptor gamma (PPARγ) agonists improve adipogenesis and insulin resistance, as well as dyslipidemia, without reducing food intake, and mutations in the PPARγ are associated with obesity-related phenotypes. Lee et al. (49) analyzed the effects of Kochujang (Korean fermented soybean-based red pepper paste) consumed for 12 weeks (32 g/day: 39 g/day wet weight) by Korean subjects carriers of PPARγ2 C1431T rs3856806, with overweight or obesity. Significant plasma TAG reduction with Kochujang supplementation was not shown in the T allele rather than in the C carriers. Otherwise, the TAG/HDL ratio was lower with Kochujang supplementation in T carriers compared with the C carriers, since the plasma HDL-c and apoA-1 were also reduced in T carriers. These observations suggest a beneficial effect of Kochujang supplementation on the TAG and TAG/HDL weakened by the T mutant allele of the PPARγ C1431T. Despite the concern of the high sodium content of Kochujang, it may have the potential to be developed as a functional supplement for the obese population (49).
Hepatic Lipase and Cholesteryl Ester Transfer Protein
Certain foods and beverages, like coffee, have been associated with beneficial health effects due to their phytochemical compounds, such as caffeine, chlorogenic acids, and trigonelline (86). Hsu et al. (50) analyzed 4,262 men and 4,813 women from the Taiwan Biobank, with polymorphisms in the genes of Hepatic lipase (LIPC) rs1800588 and Cholesteryl ester transfer protein (CETP) rs18007, both of which participate in lipoprotein metabolism. The cohort was divided into coffee drinkers (drank coffee > 3 times/week) vs. non-coffee drinkers. Only female coffee drinkers were significantly associated with higher HDL-c levels after adjusting for confounders, including rs1800588 (LIPC) and rs1800775 (CETP) variants. In this way, consumption of coffee and other caffeinated beverages may be beneficial for plasma levels of estrogen, sex hormone-binding globulin, and estradiol in women. However, information on the type of coffee consumed or drinking style was unavailable in the Taiwan Biobank database.
Cluster of Differentiation 36, Nitric Oxide Synthase 3, and Peroxisome Proliferator-Activated Receptor Gamma
Corella and Ordovás (87) suggested that CD36, NOS3, and PPARγ genes have interactions with omega-3 (ω-3) fatty acids affecting the levels of blood lipids in intervention studies. Therefore, Zheng et al. (51) performed an RCT to study the gene-diet interaction of CD36 (rs1527483), NOS3 (rs1799983), and PPARγ (rs1801282) in response to supplements of omega-3 fatty acids (from fish oil: 2 g/day of C20:5ω-3 and C22:6ω-3, or flaxseed oil: 2.5 g/day of C18:3ω-3) in blood lipids of adults with T2D. After 180 days of treatment, omega-3 supplements from both sources interacted with CD36 on TAG (p = 0.042) and with PPARγ rs1801282 to reduce LDL-c levels (p = 0.02). These interactions were more pronounced with fish oil supplements, so the administration of 2 g/day of omega-3 (C20:5ω-3 and C22:6ω-3) is beneficial to reduce TAG or LDL-c in CD36 (rs1527483) and PPARγ rs1801282) carriers.
Polymorphism and Oil Supplements
Olano-Martin et al. (52) investigated the response of the plasma lipoprotein profile to eicosapentaenoic acid (EPA) vs. docosahexaenoic acid (DHA) in healthy adults with APOE rs429358, rs7412, E3/E3, and E3/E4. Four weeks of EPA-rich oil (ERO: 3.3 g EPA/day) and DHA-rich oil (DRA: 3.7 g DHA/day) supplementation resulted in significant TAG reductions (28 and 19%, respectively). The authors attribute this effect to an EPA/DHA mediated suppression of genes involved in the fatty acid synthesis and upregulation of those controlling beta-oxidation, resulting in an overall reduction in fatty acids available for TAG synthesis. In E4 carriers, DRO treatment resulted in a 10% increase in LDL-c (p = 0.029). For this reason, it is essential to monitor DRO supplementation in APOE rs429358 E3/E4. The findings generally agree with the greater VLDL to LDL conversion described in biokinetic studies in E4 carriers (70–80% vs. 50–60% in wild type E3/E3) (88). In this context, Carvalho-Wells et al. (24) determined the effect of dietary fat quantity and composition on both lipid and non-lipid cardiovascular disease biomarkers according to APOE genotype. The study involved 88 adults with an average BMI of 26 that were randomized into three isoenergetic diets: a low-fat diet, a high-fat/high SFA diet, and a high SFA supplemented with 3.45 g of DHA (HSF-DHA) per day. An overall diet effect was evident for all cholesterol fractions (p < 0.01), with no significant genotype x diet interaction. A genotype x diet interaction (p = 0.033) was evident for plasma TAG, with 17% and 30% decreases in APOE E3/E3 and APOE E3/E4 individuals, respectively, after the HSF-DHA diet compared with the low-fat diet. These observations indicate that E3/E3 and E3/E4 subjects benefit from 3.45 g DHA supplementation, even with a high SFA diet, in order to reduce TAG levels. APOE acts as a high-affinity ligand for the removal of VLDL remnants by the liver. The selective affinity of the apoE4 protein isoform for VLDL in contrast with the E2 and E3 isoforms, which have a preference for more lipid-poor large HDLs, may help explain the greater TAG lowering in APOE E4 carriers (89).
The efficacy of plant sterols (PS) as cholesterol-lowering agents is limited by a large heterogeneity across individuals, potentially because of genetic polymorphisms. MacKay et al. (53) evaluated the effect of ingesting 2 g of plant sterols/d or a placebo in mildly hypercholesterolemic adults preselected as exhibiting either high endogenous cholesterol synthesis or low endogenous cholesterol synthesis, as well as the presence of some polymorphisms: (Apolipoprotein E) APOE E3/E4, Cytochrome P450 Family 7 Subfamily A Member 1 (CYP7A1) rs3808607, CETP rs5882 and ATP-binding cassette sub-family G member 8 (ABCG8) rs4148217. Only CYP7A1-rs3808607 GT (p = 0.0006), GG (p = 0.0009), APOE E3 (p = 0.037) and especially E4 (p < 0.0001) were associated with LDL-c lowering in response to PS consumption, this specific finding could serve as potential predictive genetic markers to identify individuals who would derive maximum LDL-c lowering with PS consumption.
As it has been described throughout this review, both environmental and genetic factors impact lipid traits. Dumitrescu et al. (54) genotyped 23 lipid-associated variants in non-Hispanic white, non-Hispanic black, and Mexican-American subjects collected from the National Health and Nutrition Examination Surveys (NHANES). Gene-environment interactions between vitamin A or E and variants previously associated with HDL-c, LDL-c, and TAG levels were modeled. The most significant interaction was Apolipoprotein B (APOB) rs693 x vitamin E (p < 0.05) for LDL-c levels and proprotein convertase subtilisin/kexin type 9 (PCSK9) rs11206510 x vitamin A associated with LDL-c, both among Mexican-Americans. PCSK9 is an enzyme that binds to low-density lipoprotein receptors (LDL receptors), which stop LDL-c from being removed from the blood, leading to an increase in blood levels of LDL-c. Thus, understanding the mechanism of the interaction between these lipid-associated variants and nutritional factors, such as serum vitamin E and A levels, is imperative to determining the etiology of a poor lipid profile and could, consequently, have implications in clinical care.
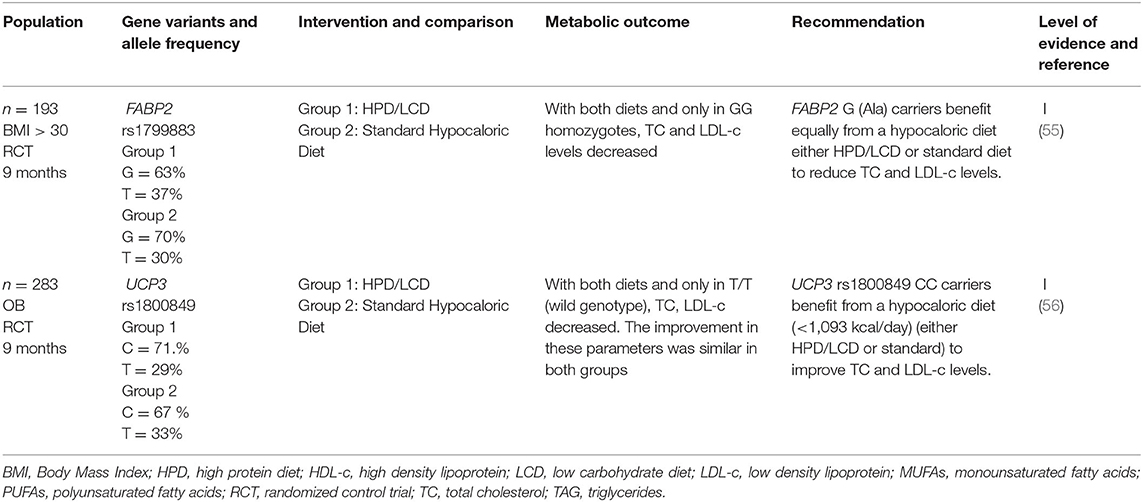
Dietary Recommendations Related to Gene Variants and Protein Intake
Fatty Acid-Binding Protein 2
Recent studies have suggested metabolic responses differences between the effects of various dietary approaches, including high protein diets.
As mentioned before, the Thr54 isoform (T allele) of FABP2 rs1799883 has been associated with hypertriglyceridemia, and increased body mass index (BMI), hyperinsulinemia, and insulin resistance (90, 91).
de Luis et al. (55) investigated the influence of the variation Ala54Thr in the intestinal fatty acid-binding protein 2 gene (FABP2) on metabolic response secondary to the intake of a high-protein/low-carbohydrate (HP) vs. standard hypocaloric diet in subjects with obesity during 9 months. The two diets resulted in decreased anthropometric parameters in both genotype groups, but the improvement in most parameters was greater with HP than with the standard diet. With both diets and only in the wild genotype (GG homozygotes), TC and LDL-c levels decreased. Therefore, T carriers (Thr54) have a different metabolic response after weight loss than obese G carriers, with no effect on LDL-c or other metabolic parameters.
Uncoupling Protein 3
Uncoupling protein 3 (UCP3) belongs to a family of mitochondrial transporters that could uncouple oxidative phosphorylation by increasing the proton leak of the inner mitochondrial membrane. Decreased expression or function of UCP3 could reduce energy expenditure and increase energy storage as fat (92).
The effect of rs1800849 in the UCP3 promotor (−55C– > T) on cardiovascular risk factors and weight loss after a high protein/low carbohydrate vs. standard hypocaloric diet was studied as well by de Luis et al. (56). Results showed that with both diets and only in C carriers, total and LDL- c levels decreased. T carriers (mutant type) have a different response secondary to a standard hypocaloric diet than wild-type obese patients, and the authors hypothesized that the distribution of macronutrient and type of dietary fat could be involved in this different metabolic response. From this standpoint, they concluded that UCP3 rs1800849 CC homozygotes benefit from a hypocaloric diet (either high protein/low carbohydrate or standard diet) to improve TC and LDL-c.
Discussion
To the best of our knowledge, this is the first systematic review that collects information about the interaction of genetic variants with diet, seeking to identify nutrigenetic recommendations as potential tools to improve health through precision nutrition.
The objective of identifying and understanding gene-diet interactions is to enable guiding to feeding patterns based on individual and population genetic susceptibilities in contrast to the traditional “one size fits all” dietary recommendations. Although the concept of gene-diet interaction is attractive and its progress in recent years is encouraging, the lack of enough clinical interventions and replication is still a significant barrier limiting the acceleration of the field growth and its translation into practice.
The primary outcome of this review was the identification of potential nutrigenetic recommendations that demonstrate a strong interaction between gene-diet and circulating lipid variations. The difference between a recommendation and a requirement lies in the principle that a recommendation is population-based, and a requirement addresses the particularities of an individual, hence, making a dietetic treatment more appropriate and effective. The information identified in this review differentiates from generic or conventional recommendations for the dietary approach to dyslipidemias because the nutrigenetic recommendations are based on significant gene-diet interactions.
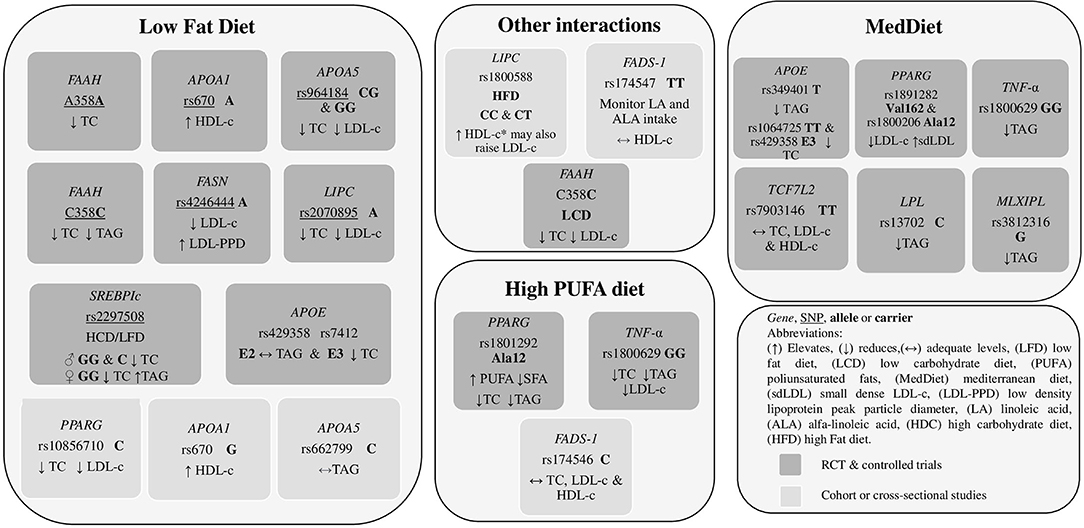
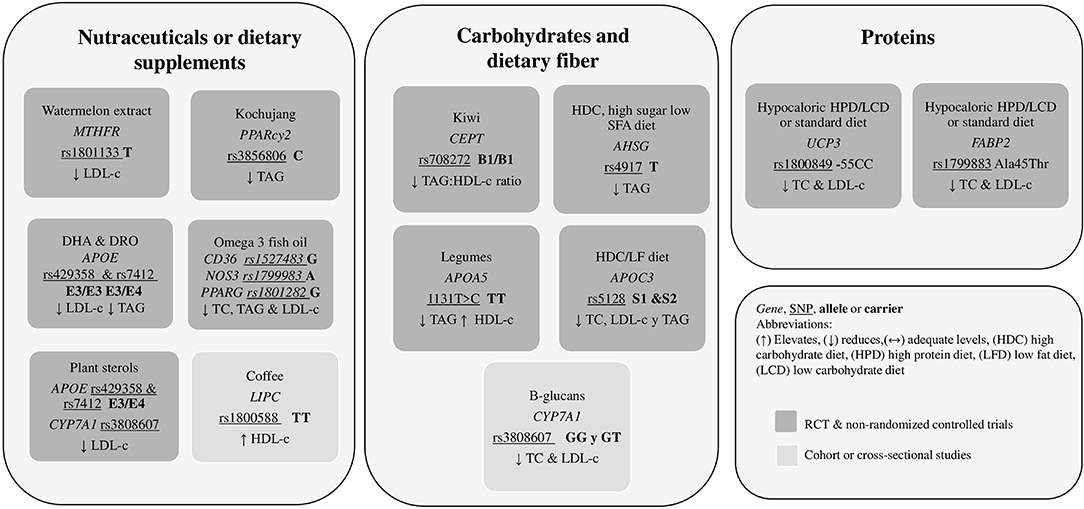
It was observed that the distribution of macronutrients intake is a crucial point in the interaction between body weight and metabolic response, considering the lipid profile or other biochemical markers. Dietary fat significantly modified the genetic effects of polymorphisms on changes in serum TC, LDL-c, and HDL-c. It was identified that the types and dosage of fatty acids included in the diet, whether they are PUFAS, MUFAS, or SFA, play a fundamental role in lipid metabolism. It should be noted that the dietary recommendations that benefit individuals with a particular genotype do not work in the same way for individuals with a different genotype. That is why genotyping individuals and populations is very relevant. Results of studies allowed us to observe how, through the intake of specific amounts and kinds of fats, carbohydrates/fiber, proteins, nutraceuticals or through the administration of precise dietary patterns made according to the anthropometric, physiological, and genetic needs of people, the decrease in biochemical parameters, mainly plasma lipids, was significant. Figure 2 summarizes the dietary recommendations related to gene variants and dietary fat intake, and Figure 3 summarizes the dietary recommendations related to gene variants and diet components identified throughout this review. The information collected in this review constitutes a finding that encourages and leads to creating nutritional strategies for preventing and controlling cardiometabolic diseases, a latent public health problem worldwide.
Concerning the consumption of carbohydrates, it is evident that the nutritional quality of these complex carbohydrate-rich and low glycemic index diets has a significant influence. The already known lipid-lowering effect of dietary fiber is reaffirmed. However, more research is needed to establish and understand the interaction of lipid genes with carbohydrates/fiber intake. It is also necessary to bear in mind that the general eating pattern helps maintain adequate levels of lipid traits and reduce cardiovascular risk.
Nevertheless, all recommendations can stand out from this review and highlight the importance of personalized nutrition. For example, gene-diet interactions with FADS genes were particularly appealing because there is a rise in current ω-3 y ω-6 vegetable sources like chia and flaxseed. However, gene-diet interactions found in Ching et al. (36) and Lu et al. (37) prove that these recommendations may not be adequate for people with polymorphisms in the FADS gene cluster family. Olano-Martin et al. (52) findings also highlight relevant gene-diet interactions with another famous and generic recommendation, fish oil supplementation with APOE and EPA-DHA administration, in which APOE rs429358 E3/E4 had an increase in LDL-c levels with DRO treatment which proves that this recommendation may not be adequate for everyone.
One of the strengths of this systematic review is that it addresses the evidence from studies that investigate the combined effect of genotype plus diet on health outcomes, which is essential for establishing evidence-based nutrigenetic advice. A limitation of this review was that due to the heterogeneity of the information obtained from the included articles, it was difficult to compare studies in terms of specific results and dietary recommendations suggested by the different authors. Another limitation was the non-inclusion of information since it came from articles that were evaluated with low methodological quality. However, the information transmitted can be considered enriching, recognizing that it is a pioneering review. Although the nutrigenetic area is still in its infancy, the available information is still considered scarce, heterogeneous and some findings are based on semi-quantitative data. It is crucial to stimulate debate on their usefulness and continue generating research as the knowledge on the subject increases.
The scientific community and community at large are facing the fact that nutrigenetics is unregulated, with few defined standards, there is a lack of educational resources or guidance for implementing the outcomes of nutrigenetic research. In this sense, it is considered that the proposed guidelines to evaluate scientific validity and evidence for genotype-based dietary advice by Grimaldi et al. (93) could be an effective tool to subsequently and individually evaluate each of the possible nutrigenetic recommendations included in this study as it is essential to consider aspects such as the study quality rating, type of gene x diet interaction (direct phenotype, intermediate phenotype, indirect phenotype), biological plausibility among others, to avoid misuse and to protect the public, personalized nutrigenetic advice and information should be based on clear evidence.
A proposal for further research is that the evidence obtained in this review could be a starting point for the future elaboration of “nutrition gene cards” that assess the evidence supporting specific gene × diet interactions and their relationship with a specific health outcome.
Conclusion
The nutrigenetic recommendations identified may serve as dietary tools for the preventive treatment and control alterations in lipid metabolism. They are also potential tools for future research and a basis for creating nutrigenetic patterns and nutrigenetic portfolios containing specific dietary material for populations with different genetic and physiological characteristics. They can also be considered in potential strategies to treat public health problems related to dyslipidemias and several chronic degenerative diseases.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author Contributions
YP-B: conceptualization, data curation, and writing—original draft preparation. IR-I and KG-B: conceptualization, visualization, methodology, data curation, and writing—review and editing. NP-N: writing—original draft. JT: writing—review and editing. SS-A: writing—review and editing and supervision. EM: conceptualization, visualization, writing—review and editing, supervision, funding acquisition, and project administration. All authors contributed to the article and approved the submitted version.
Funding
The State Council of Science and Technology (COECYTJAL) funded this project with the Grant Number FODECIJAL-2019-8224.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge the contribution of students from the ITESO Programa de Aplicación Profesional Innovación para la salud, and UDG CUCS Prácticas Profesionales en Nutrición. As well as COECYTJAL for funding this research (FODECIJAL-2019-8224).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.830283/full#supplementary-material
Abbreviations
ABCG8, ATP-binding cassette sub-family G member 8; AHSG, alpha2 Heremans-Schmid glycoprotein; APOA5, apolipoprotein A5; APOB, apolipoprotein B; APOC3, apolipoprotein C3; APOE, apolipoprotein E; CD36, cluster of differentiation 36,; CETP, cholesteryl ester transfer protein; CVD, cardiovascular diseases; CYP7A1, cytochrome P450 Family 7 Subfamily A Member 1; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; FAAH, fatty acid amide hydrolase; FABP2, fatty acid binding protein 2; FADS, fatty acid desaturase; FetA, Fetuin-A; GI, glycemic index; GWAS, genome-wide association studies; HDL, high-density lipoprotein; HFD, high fat diet; IDL, intermediate density lipoprotein; LA, linoleic acid; LDF, low-fat diet; LDL, low density lipoprotein; LDL-PPD, LDL peak particle diameter; LIPC, hepatic lipase; LPL, lipoprotein lipase; MeSH, medical subject headings; MLXIPL, Max-like protein X interacting protein-like; MS, metabolic syndrome; MTHFR, methylenetetrahydrofolate reductase; MUFAs, monounsaturated fatty acids; NCBI, National Center for Biotechnology Information; NOS, Newcastle-Ottawa Scale; NOS3, nitric oxide synthase 3; PCSK9, proprotein convertase subtilisin/kexin type 9; PPARs, peroxisome proliferator activated receptors; PPARɤ2, peroxisome proliferator activated receptor gamma C1431T polymorphism; PRISMA-P, preferred reporting items for systematic reviews and meta-analyses protocol; PS, plant sterols; PUFAs, polyunsaturated fatty acids; RCT, randomized control trial; SFA, saturated fatty acids; SNP, single nucleotide polymorphism; SREBP-1c, sterol regulatory element binding transcription factor 1; TAG, triacylglycerols; T2D, type 2 diabetes; TC, total cholesterol; TCF7L2, transcription factor 7 like 2; TNF- a, tumor necrosis factor; UCP3, uncoupling protein 3; US NLM, United States National Library of Medicine.
References
1. Vos T, Allen C, Arora M, Barber RM, Bhutta ZA, Brown A, et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. (2016) 388:1545–602. doi: 10.1016/S0140-6736(16)31678-6
2. Gadde KM, Martin CK, Berthoud HR, Heymsfield SB. Obesity: pathophysiology and management. J Am Coll Cardiol. (2018) 71:69–84. doi: 10.1016/j.jacc.2017.11.011
3. Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ. ACC/AHA guideline on the primary prevention of cardiovascular disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. (2019) 140:596–646. doi: 10.1161/CIR.0000000000000725
4. Hannon BA, Khan NA, Teran-Garcia M. Nutrigenetic contributions to dyslipidemia: A focus on physiologically relevant pathways of lipid and lipoprotein metabolism. Nutrients. (2018) 10:1404. doi: 10.3390/nu10101404
5. de Luis DA, Romero E, Izaola O, Primo D, Aller R. Cardiovascular risk factors and insulin resistance after two hypocaloric diets with different fat distribution in obese subjects: Effect of the rs10767664 gene variant in brain-derived neurotrophic factor. Lifestyle Genomics. (2017) 10:163–71. doi: 10.1159/000485248
6. Barrea L, Annunziata G, Bordoni L, Muscogiuri G, Colao A, Savastano S. Nutrigenetics—personalized nutrition in obesity and cardiovascular diseases. Int J Obes Suppl. (2020) 10:1–13. doi: 10.1038/s41367-020-0014-4
7. Vazquez-Vidal I, Desmarchelier C, Jones PJH. Nutrigenetics of blood cholesterol concentrations: towards personalized nutrition. Curr Cardiol Rep. (2019) 21:3–7. doi: 10.1007/s11886-019-1124-x
8. Fall T, Mendelson M, Speliotes EK. Recent advances in human genetics and epigenetics of adiposity: pathway to precision medicine? Gastroenterology. (2017) 152:1695–706. doi: 10.1053/j.gastro.2017.01.054
9. Romero-Cortes T, Lopez-Perez PA, Toledo AKM, Perez-Espana VH, Aparicio-Burgos JE, Cuervo-Parra JA. Nutrigenomics and nutrigenetics in functional foods. Int J Bio-resource Stress Manag. (2018) 9:661–72. doi: 10.23910/IJBSM/2018.9.6.1922
10. Beckett EL, Jones PR, Veysey M, Lucock M. Nutrigenetics—personalized nutrition in the genetic age. Explor Res Hypothesis Med. (2017) 2:1–8. doi: 10.14218/ERHM.2017.00027
11. Toro-Martín D, Arsenault BJ, Després JP, Vohl MC. Precision nutrition: a review of personalized nutritional approaches for the prevention and management of metabolic syndrome. Nutrients. (2017) 9:1–28. doi: 10.3390/nu9080913
12. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int J Surg. (2021) 88:1–11. doi: 10.1016/j.ijsu.2021.105906
13. Richardson WS, Wilson MC, Nishikawa J, Hayward RSA. The well-built clinical question: a key to evidence-based decisions. ACP J Club. (1995) 123:A12. doi: 10.7326/ACPJC-1995-123-3-A12
14. Jadad AR, Moore RA, Carroll D, Jenkinson C, Reynolds DJM, Gavaghan DJ. Assessing the quality of reports of randomized clinical trials: Is blinding necessary? Control Clin Trials. (1996) 17:1–12. doi: 10.1016/0197-2456(95)00134-4
15. Wells G, Shea B, Higgins J, Sterne J, Tugwell P, Reeves B. Checklists of methodological issues for review authors to consider when including non-randomized studies in systematic reviews. Res Synth Methods. (2013) 4:63–77. doi: 10.1002/jrsm.1077
16. Stang A Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. (2010) 25:603–5. doi: 10.1007/s10654-010-9491-z
17. AHRQ. US Preventive Services Force Guides to Clinical Preventive Services. Rock- ville: Agency for Healthcare Research and Quality (1996).
18. Garcia-Rios A, Alcala-Diaz JF, Gomez-Delgado F, Delgado-Lista J, Marin C, Leon-Acuña A. Beneficial effect of CETP gene polymorphism in combination with a Mediterranean diet influencing lipid metabolism in metabolic syndrome patients: CORDIOPREV study. Clin Nutr. (2018) 37:229–34. doi: 10.1016/j.clnu.2016.12.011
19. Rudkowska I, Dewailly E, Hegele RA, Boiteau V, Dubé-Linteau A, Abdous B. Gene-diet interactions on plasma lipid levels in the Inuit population. Br J Nutr. (2013) 109:953–61. doi: 10.1017/S0007114512002231
20. Shatwan IM, Weech M, Jackson KG, Lovegrove JA, Vimaleswaran KS. Apolipoprotein e gene polymorphism modifies fasting total cholesterol concentrations in response to replacement of dietary saturated with monounsaturated fatty acids in adults at moderate cardiovascular disease risk. Lipids Health Dis. (2017) 16:1–9. doi: 10.1186/s12944-017-0606-3
21. Griffin BA, Walker CG, Jebb SA, Moore C, Frost GS, Goff L. APOE4 genotype exerts greater benefit in lowering plasma cholesterol and apolipoprotein B than wild type (E3/E3), after replacement of dietary saturated fats with low glycaemic index carbohydrates. Nutrients. (2018) 10:1–13. doi: 10.3390/nu10101524
22. Fallaize R, Celis-Morales C. MacReady AL, Marsaux CFM, Forster H, O'Donovan C, et al. The effect of the apolipoprotein E genotype on response to personalized dietary advice intervention: Findings from the Food4Me randomized controlled trial. Am J Clin Nutr. (2016) 104:827–36. doi: 10.3945/ajcn.116.135012
23. Gomez-Delgado F, Alcala-Diaz JF, Garcia-Rios A, Delgado-Lista J, Ortiz-Morales A, Rangel-Zuñiga O. Polymorphism at the TNF-alpha gene interacts with Mediterranean diet to influence triglyceride metabolism and inflammation status in metabolic syndrome patients: From the CORDIOPREV clinical trial. Mol Nutr Food Res. (2014) 58:1519–27. doi: 10.1002/mnfr.201300723
24. Carvalho-Wells AL, Jackson KG, Lockyer S, Lovegrove JA, Minihane AMAPOE. genotype influences triglyceride and C-reactive protein responses to altered dietary fat intake in UK adults. Am J Clin Nutr. (2012) 96:1447–53. doi: 10.3945/ajcn.112.043240
25. Domínguez-Reyes T, Astudillo-López CC, Salgado-Goytia L, Muñoz-Valle JF, Salgado-Bernabé AB, Guzmán-Guzmán IP. Interaction of dietary fat intake with APOA2, APOA5 and LEPR polymorphisms and its relationship with obesity and dyslipidemia in young subjects. Lipids Health Dis. (2015) 14:1–10. doi: 10.1186/s12944-015-0112-4
26. Zhang X, Bray QQ, Hu GA, Sacks FB, Qi FML. APOA5 genotype modulates 2-y changes in lipid profile in response to weight-loss diet intervention: The Pounds Lost Trial. Am J Clin Nutr. (2012) 96:917–22. doi: 10.3945/ajcn.112.040907
27. Sánchez-Moreno C, Ordovas J, Smith C, Baraza J, Lee Y-C, Garaulet M. APOA5 gene variation interacts with dietary fat intake to modulate obesity and circulating triglycerides in a mediterranean population. J Nutr. (2011) 141:380–5. doi: 10.3945/jn.110.130344
28. de Luis D, Izaola O, Primo D, Aller R. Role of rs670 variant of APOA1 gene on metabolic response after a high fat vs. a low fat hypocaloric diets in obese human subjects. J Diabetes Complications. (2019) 33:249–54. doi: 10.1016/j.jdiacomp.2018.10.015
29. AlSaleh A, O'Dell SD, Frost GS, Griffin BA, Lovegrove JA, Jebb SA, et al. Interaction of PPARG Pro12Ala with dietary fat influences plasma lipids in subjects at cardiometabolic risk. J Lipid Res. (2011) 52:2298–303. doi: 10.1194/jlr.P019281
30. Alsaleh A, Frost GS, Griffin BA, Lovegrove JA, Jebb SA, Sanders TAB. PPARγ2 gene Pro12Ala and PPARα gene Leu162Val single nucleotide polymorphisms interact with dietary intake of fat in determination of plasma lipid concentrations. J Nutrigenet Nutrigenomics. (2012) 4:354–66. doi: 10.1159/000336362
31. Xu M, Ng SS, Bray GA, Ryan DH, Sacks FM, Ning G. Dietary fat intake modifies the effect of a common variant in the LIPC gene on changes in serum lipid concentrations during a long-term weight-loss intervention trial. J Nutr. (2015) 145:1289–94. doi: 10.3945/jn.115.212514
32. Smith C, Van Rompay M, Mattei J, Garcia J, Garcia-Bailo B, Lichtenstein A. Dietary fat modulation of hepatic lipase variant−514 C/T for lipids: a crossover randomized dietary intervention trial in Caribbean Hispanics. Physiol Genomics. (2017) 49:592–600. doi: 10.1152/physiolgenomics.00036.2017
33. Corella D, Sorlí JV, Estruch R, Coltell O, Ortega-Azorín C, Portolés O. MicroRNA-410 regulated lipoprotein lipase variant rs13702 is associated with stroke incidence and modulated by diet in the randomized controlled PREDIMED trial. Am J Clin Nutr. (2014) 100:719–31. doi: 10.3945/ajcn.113.076992
34. Martinez-Lopez E, Garcia-Garcia MR, Gonzalez-Avalos JM, Maldonado-Gonzalez M, Ruiz-Madrigal B, Vizmanos B. Effect of Ala54Thr polymorphism of FABP2 on anthropometric and biochemical variables in response to a moderate-fat diet. Nutrition. (2013) 29:46–51. doi: 10.1016/j.nut.2012.03.002
35. De Luis D, Aller R, Izaola O, Gonzalez Sagrado M, de La Fuente B, Conde R, et al. Effect of fatty acid-binding protein 2 Ala54Thr genotype on weight loss and cardiovascular risk factors after a high-polyunsaturated fat diet in obese patients. J Investig Med. (2012) 60:1194–8. doi: 10.2310/JIM.0b013e318271fb25
36. Ching YK, Chin YS, Appukutty M, Ramanchadran V, Ang Y. Interaction of dietary linoleic acid and α-linolenic acids with rs174547 in FADS1 gene on metabolic syndrome components among vegetarians. Nutrients. (2019) 11:1–16. doi: 10.3390/nu11071686
37. Lu Y, Feskens EJM, Dollé MET, Imholz S, Verschuren WMM, Müller M. Dietary n-3 and n-6 polyunsaturated fatty acid intake interacts with FADS1 genetic variation to affect total and HDL-cholesterol concentrations in the Doetinchem Cohort Study. Am J Clin Nutr. (2010) 92:258–65. doi: 10.3945/ajcn.2009.29130
38. Dolley G, Boisclair ME, Lamarche B, Després JP, Bouchard C, Pérusse L. Interactions between dietary fat intake and FASN genetic variation influence LDL peak particle diameter. J Nutrigenet Nutrigenomics. (2011) 4:137–45. doi: 10.1159/000327778
39. de Luis DA, Aller R, Izaola O, Sagrado MG, Conde R. Role of G308 promoter variant of tumor necrosis factor alpha gene on weight loss and metabolic parameters after a high monounsaturated versus a high polyunsaturated fat hypocaloric diets. Med Clin. (2013) 141:189–93. doi: 10.1016/j.medcli.2012.12.021
40. Ortega-Azorín C, Sorlí JV, Estruch R, Asensio EM, Coltell O, González JI. Amino acid change in the carbohydrate response element binding protein is associated with lower triglycerides and myocardial infarction incidence depending on level of adherence to the mediterranean diet in the PREDIMED trial. Circ Cardiovasc Genet. (2014) 7:49–58. doi: 10.1161/CIRCGENETICS.113.000301
41. Deluis DA, Sagrado MG, Aller R, Izaola O, Conde R. Effects of C358A missense polymorphism of the degrading enzyme fatty acid amide hydrolase on weight loss, adipocytokines, and insulin resistance after 2 hypocaloric diets. Metabolism. (2010) 59:1387–92. doi: 10.1016/j.metabol.2009.12.029
42. Zhang Z, Gong RR, Du J, Xiao LY, Duan W, Zhou XD. Associations of the SREBP-1c gene polymorphism with gender-specific changes in serum lipids induced by a high-carbohydrate diet in healthy chinese youth. Appl Physiol Nutr Metab. (2011) 36:226–32. doi: 10.1139/h11-005
43. Wang Y, Harding SV, Eck P, Thandapilly SJ, Gamel TH, Abdel-Aal ESM. High-molecular-weight β-glucan decreases serum cholesterol differentially based on the CYP7A1 rs3808607 polymorphism in mildly hypercholesterolemic adults. J Nutr. (2016) 146:720–7. doi: 10.3945/jn.115.223206
44. Gammon CS, Minihane AM, Kruger R, Conlon CA, Von Hurst PR, Jones B. TaqIB polymorphism in the cholesteryl ester transfer protein (CETP) gene influences lipid responses to the consumption of kiwifruit in hypercholesterolaemic men. Br J Nutr. (2014) 111:1077–84. doi: 10.1017/S0007114513003437
45. Robinson KN, Vazquez-Vidal I, Marques C, Andrade FCD, Aradillas-Garcia C, Teran-Garcia M. Circulating triglycerides and the association of triglycerides with dietary intake are altered by alpha-2-heremans-schmid glycoprotein polymorphisms. J Nutrigenet Nutrigenomics. (2017) 10:75–83. doi: 10.1159/000478657
46. Jang Y, Chae JS, Kim OY, Park HJ, Kim JY, Paik JK. APOA5-1131T > C genotype effects on apolipoprotein A5 and triglyceride levels in response to dietary intervention and regular exercise (DIRE) in hypertriglyceridemic subjects. Atherosclerosis. (2010) 211:512–9. doi: 10.1016/j.atherosclerosis.2010.03.014
47. Song YY, Gong RR, Zhang Z, Li YH, Xiao LY, Zhou XD, Fang DZ. A high-carbohydrate diet enhances the adverse effect of the S2 allele of APOC3 SstI polymorphism on the TG/HDL-C ratio only in young Chinese females. Brazilian J Med Biol Res. (2011) 44:524–30. doi: 10.1590/S0100-879X2011007500065
48. Massa NM, Silva AS, de Oliveira CV, Costa MJ, Persuhn DC, Barbosa CV, et al. Supplementation with watermelon extract reduces total cholesterol and ldl cholesterol in adults with dyslipidemia under the influence of the MTHFR C677T Polymorphism. J Am Coll Nutr. (2016) 35:514–20. doi: 10.1080/07315724.2015.1065522
49. Lee Y, Cha YS, Park Y, Lee M. PPARγ2 C1431T polymorphism interacts with the antiobesogenic effects of Kochujang, a Korean fermented, soybean-based red pepper paste, in overweight/obese subjects: a 12-week, double-blind randomized clinical trial. J Med Food. (2017) 20:610–7. doi: 10.1089/jmf.2016.3911
50. Hsu T, Tantoh DM, Lee K, Ndi ON, Lin L, Chou M. Genetic and non-genetic factor-adjusted association between coffee drinking and high-density lipoprotein cholesterol in Taiwanese Adults: stratification by sex. Nutrients. (2019) 11:1–11. doi: 10.3390/nu11051102
51. Zheng JS, Chen J, Wang L, Yang H, Fang L, Yu Y. Replication of a gene-diet interaction at cd36, nos3 and pparg in response to omega-3 fatty acid supplements on blood lipids: a double-blind randomized controlled trial. EBioMed. (2018) 31:150–6. doi: 10.1016/j.ebiom.2018.04.012
52. Olano-Martin E, Anil E, Caslake MJ, Packard CJ, Bedford D, Stewart G. Contribution of apolipoprotein E genotype and docosahexaenoic acid to the LDL-cholesterol response to fish oil. Atherosclerosis. (2010) 209:104–10. doi: 10.1016/j.atherosclerosis.2009.08.024
53. MacKay DS, Eck PK, Gebauer SK, Baer DJ, Jones PJH. CYP7A1-rs3808607 and APOE isoform associate with LDL cholesterol lowering after plant sterol consumption in a randomized clinical trial. Am J Clin Nutr. (2015) 102:951–7. doi: 10.3945/ajcn.115.109231
54. Dumitrescu L, Goodloe R, Brown-Gentry K, Mayo P, Allen M, Jin H. Serum vitamins A and e as modifiers of lipid trait genetics in the National Health and Nutrition Examination Surveys as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. Hum Genet. (2012) 131:1699–708. doi: 10.1007/s00439-012-1186-y
55. de Luis DA, Izaola O, de la Fuente B, Primo D, Romero E. Role of fatty acid-binding protein 2 Ala54Thr genotype on weight loss and cardiovascular risk factors after a high-protein/low-carbohydrate versus a standard hypocaloric diet during 9 months. Ann Nutr Metab. (2015) 67:81–6. doi: 10.1159/000438947
56. de Luis DA, Aller R, Izaola O, Romero E. Effect of-55CT polymorphism of UCP3 on insulin resistance and cardiovascular risk factors after a high protein/low carbohydrate versus a standard hypocaloric diet. Ann Nutr Metab. (2016) 68:157–63. doi: 10.1159/000444150
57. Vannice G, Rasmussen H. Position of the academy of nutrition and dietetics: dietary fatty acids for healthy adults. J Acad Nutr Diet. (2014) 114:136–53. doi: 10.1016/j.jand.2013.11.001
58. Academy of Nutrition and Dietetics. Position of the Academy of Nutrition and Dietetics: total diet approach to healthy eating. J Acad Nutr Dietet. (2013) 113:307–17. doi: 10.1016/j.jand.2012.12.013
59. Feingold K, Anawalt B, Boyce AE. Introduction to Lipids and Lipoproteins. South Dartmouth (MA): Endotext, MDText.com, Inc. (2000).
60. Xu M, Zhao J, Zhang Y, Ma X, Zhi DQ. Apolipoprotein e Gene variants and risk of coronary heart disease: a meta-analysis. Biomed Res Int. (2016) 2016:3912175. doi: 10.1155/2016/3912175
61. Gomez-Delgado F, Alcala-Diaz JF, Leon-Acuña A, Lopez-Moreno J, Delgado-Lista J, Gomez-Marin B. Apolipoprotein E genetic variants interact with Mediterranean diet to modulate postprandial hypertriglyceridemia in coronary heart disease patients: CORDIOPREV study. Eur J Clin Invest. (2019) 49:1–9. doi: 10.1111/eci.13146
62. GeneCards Org,. Apolipoprotein A1 [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=APOA1&keywords=APOA1 (accessed July 28, 2021).
63. Paththinige CS, Sirisena ND, Dissanayake VHW. Genetic determinants of inherited susceptibility to hypercholesterolemia—a comprehensive literature review. Lipids Health Dis. (2017) 16:1–22. doi: 10.1186/s12944-017-0488-4
64. Lin YC, Nunez V, Johns R, Shiao SPK. APOA5 gene polymorphisms and cardiovascular diseases: metaprediction in global populations. Nurs Res. (2017) 66:164–74. doi: 10.1097/NNR.0000000000000207
65. Prieur X, Huby T, Coste H, Schaap FG, Chapman MJ, Rodríguez JC. Thyroid hormone regulates the hypotriglyceridemic gene APOA5. J Biol Chem. (2005) 280:27533–43. doi: 10.1074/jbc.M503139200
66. Rashid A, Jafar S, Yaqub RK. Role of peroxisome proliferator-activated receptor (PPAR)-α gene in dyslipidemia. Rawal Med J. (2020) 45:54–7.
67. GeneCards Org. LEPR Gene [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=LEPR (accessed July 28, 2021).
68. Stenger C, Hanse M, Pratte D, Mbala M, Akbar S, Koziel V. Up-regulation of hepatic lipolysis stimulated lipoprotein receptor by leptin: a potential lever for controlling lipid clearance during the postprandial phase. FASEB J. (2010) 24:4218–28. doi: 10.1096/fj.10-160440
69. GeneCards Org,. LIPC Gene [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=LIPC&keywords=lipc (accessed July 28, 2021).
70. Cahova M, Dankova H, Palenickova E, Papackova Z, Kazdova L. The opposite effects of high-sucrose and high-fat diet on fatty acid oxidation and very low density lipoprotein secretion in rat model of metabolic syndrome. J Nutr Metab. (2012). doi: 10.1155/2012/757205
71. Xie L, Li YM. Lipoprotein lipase (LPL) polymorphism and the risk of coronary artery disease: a meta-analysis. Int J Environ Res Public Health. (2017) 14:84. doi: 10.3390/ijerph14010084
72. Liao F-H, Liou T-H, Chiu W-C, Shieh M-J, Chien Y-W. Differential effects of high MUFA with high or low P/S ratio (polyunsaturated to saturated fatty acids) on improving hepatic lipolytic enzymes and mediating PPARγ related with lipoprotein lipase and hormone-sensitive lipase of white adipose tissue in diet. Int J Obes. (2010) 34:1608–17. doi: 10.1038/ijo.2010.88
73. GeneCards Org. FADS1 Gene [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FADS1&keywords=fads (accessed July 7, 2021).
74. Dumont J, Goumidi L, Grenier-Boley B, Cottel D, Marécaux N, Montaye M. Dietary linoleic acid interacts with FADS1 genetic variability to modulate HDL-cholesterol and obesity-related traits. Clin Nutr. (2018) 37:1683–9. doi: 10.1016/j.clnu.2017.07.012
75. GeneCards Org. FASN Gene [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FASN&keywords=fasn (accessed July 7, 2021).
76. Kaitosaari T, Rönnemaa T, Viikari J, Leino A, Jokinen E, Simell O. Low-density lipoprotein (LDL) particle size in healthy prepubertal children: The STRIP study. Acta Paediatr Int J Paediatr. (2006) 95:1668–73. doi: 10.1080/08035250600746294
77. NCBI. TNF Tumor Necrosis Factor [Homo sapiens (Human)] [Internet]. NCBI Reference Sequences (RefSeq) (2021). Available online at: https://www.ncbi.nlm.nih.gov/gene/7124 (accessed July 28, 2021).
78. NCBI. No MLXIPL MLX Interacting Protein Like [Homo sapiens (Human)] [Internet]. NCBI Reference Sequences (RefSeq). (2021). Available online at: https://www.ncbi.nlm.nih.gov/gene/51085 (accessed July 28, 2021).
79. GeneCards Org. No TCF7L2 Gene (Protein Coding) [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TCF7L2&keywords=TCF7L2 (accessed July 28, 2021).
80. Corella D, Carrasco P, Sorlí JV, Estruch R, Rico-Sanz J, Martínez-González MÁ. Mediterranean diet reduces the adverse effect of the TCF7L2-rs7903146 polymorphism on cardiovascular risk factors and stroke incidence: a randomized controlled trial in a high-cardiovascular-risk population. Diabetes Care. (2013) 36:3803–11. doi: 10.2337/dc13-0955
81. GeneCards Org. FAAH Gene Fatty Acid Amide Hydrolase [Internet]. (2021). Available online at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAAH&keywords=faah (accessed July 30, 2021).
82. Inamine T, Higa S, Noguchi F, Kondo S, Omagari K, Yatsuhashi H. Association of genes involved in bile acid synthesis with the progression of primary biliary cirrhosis in Japanese patients. J Gastroenterol. (2013) 48:1160–70. doi: 10.1007/s00535-012-0730-9
83. Lam CK, Zhang Z, Yu H, Tsang SY, Huang Y, Chen ZY. Apple polyphenols inhibit plasma CETP activity and reduce the ratio of non-HDL to HDL cholesterol. Mol Nutr Food Res. (2008) 52, 950–8. doi: 10.1002/mnfr.200700319
84. Pal D, Dasgupta S, Kundu R, Maitra S, Das G, Mukhopadhyay S, et al. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat Med [Internet]. (2012) 18:1279–85. doi: 10.1038/nm.2851
85. Hardy G. Nutraceuticals and functional foods: introduction and meaning. Nutrition. (2000) 16:688–9. doi: 10.1016/S0899-9007(00)00332-4
86. Montenegro J, Silva dos Santos L, Gonçalves Gusmão de Souza R, Barbosa Lima LG, Santos Mattos D, Prunes B, et al. Bioactive compounds, antioxidant activity and antiproliferative effects in prostate cancer cells of green and roasted coffee extracts obtained by microwave-assisted extraction (MAE). Food Res Int. (2021) 140:110014. doi: 10.1016/j.foodres.2020.110014
87. Corella D, Ordovás JM. Interactions between dietary n-3 fatty acids and genetic variants and risk of disease. Br J Nutr. (2012) 107(SUPPL. 2):S271–S283. doi: 10.1017/S0007114512001651
88. Welty F, Lichtenstein A, Barrett P, Jenner J, Dolnikowski G, Schaefer E. Effects of ApoE genotype on ApoB-48 and ApoB-100 kinetics with stable isotopes in humans. Arterioscler Thromb Vasc Biol. (2000) 20:1807–10. doi: 10.1161/01.ATV.20.7.1807
89. Dong L, Weisgraber K. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. (1996) 271:19053–7. doi: 10.1074/jbc.271.32.19053
90. Baier LJ, Sacchettini JC, Knowler WC, Eads J, Paolisso G, Tataranni PA. An amino acid substitution in the human intestinal fatty acid binding protein is associated with increased fatty acid binding, increased fat oxidation, and insulin resistance. J Clin Invest. (1995) 95:1281–7. doi: 10.1172/JCI117778
91. Dworatzek PDN, Hegele RA, Wolever TMS. Postprandial lipemia in subjects with the threonine 54 variant of the fatty acid-binding protein 2 gene is dependent on the type of fat ingested. Am J Clin Nutr. (2004) 79:1110–7. doi: 10.1093/ajcn/79.6.1110
92. Kopecký J, Rossmeisl M, Flachs P, Bardová K, Brauner P. Mitochondrial uncoupling and lipid metabolism in adipocytes. Biochem Soc Trans. (2001) 29(Pt 6):791-−7. doi: 10.1042/bst0290791
Keywords: nutritional genetics, nutrigenomics, dyslipidemia, obesity, polymorphism, gene-diet interaction
Citation: Pérez-Beltrán YE, Rivera-Iñiguez I, Gonzalez-Becerra K, Pérez-Naitoh N, Tovar J, Sáyago-Ayerdi SG and Mendivil EJ (2022) Personalized Dietary Recommendations Based on Lipid-Related Genetic Variants: A Systematic Review. Front. Nutr. 9:830283. doi: 10.3389/fnut.2022.830283
Received: 06 December 2021; Accepted: 16 February 2022;
Published: 21 March 2022.
Edited by:
Annalisa Terranegra, Sidra Medicine, QatarReviewed by:
Margarita Teran-Garcia, University of Illinois at Urbana-Champaign, United StatesRosaura Leis, University of Santiago de Compostela, Spain
Mahmut Cerkez Ergoren, Near East University, Cyprus
Copyright © 2022 Pérez-Beltrán, Rivera-Iñiguez, Gonzalez-Becerra, Pérez-Naitoh, Tovar, Sáyago-Ayerdi and Mendivil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Edgar J. Mendivil, edgarjmendivil@gmail.com
 Yolanda E. Pérez-Beltrán
Yolanda E. Pérez-Beltrán Ingrid Rivera-Iñiguez
Ingrid Rivera-Iñiguez Karina Gonzalez-Becerra
Karina Gonzalez-Becerra Naomi Pérez-Naitoh
Naomi Pérez-Naitoh Juscelino Tovar
Juscelino Tovar Sonia G. Sáyago-Ayerdi
Sonia G. Sáyago-Ayerdi Edgar J. Mendivil
Edgar J. Mendivil