Metataxonomic Analysis of Milk Samples From SARS-CoV-2-Positive and SARS-CoV-2-Negative Women
- 1Department of Nutrition and Food Science, Complutense University of Madrid, Madrid, Spain
- 2Department of Neonatology, Biomedical Research Foundation-IDIPAZ, La Paz University Hospital, Madrid, Spain
- 3Department of Neonatology, Quirónsalud Madrid University Hospital and Quirónsalud San José Hospital, Biomedical Research Foundation-IDIPAZ, La Paz University Hospital, Madrid, Spain
- 4Department of Neonatology, Quirónsalud Madrid University Hospital and Quiroónsalud San José Hospital, Madrid, Spain
- 5Department of Neonatology, 12 de Octubre University Hospital, Madrid, Spain
- 6Department of Neonatology, Regional Human Milk Bank, 12 de Octubre University Hospital, Madrid, Spain
Objective: To assess the impact of SARS-CoV-2 viral infection on the metataxonomic profile and its evolution during the first month of lactation.
Methods: Milk samples from 37 women with full-term pregnancies and mild SARS-CoV-2 infection and from 63 controls, collected in the first and fifth postpartum weeks, have been analyzed. SARS-CoV-2 RNA was assessed by reverse transcription polymerase chain reaction (RT-PCR) both in cases and controls. After DNA extraction, the V3-V4 hypervariable region of the gene 16S rRNA was amplified and sequenced using the MiSeq system of Illumina. Data were submitted for statistical and bioinformatics analyses after quality control.
Results: All the 1st week and 5th week postpartum milk samples were negative for SARS-CoV-2 RNA. Alpha diversity showed no differences between milk samples from the study and control group, and this condition was maintained along the observation time. Analysis of the beta-diversity also indicated that the study and control groups did not show distinct bacterial profiles. Staphyloccus and Streptococcus were the most abundant genera and the only ones that were detected in all the milk samples provided. Disease state (symptomatic or asymptomatic infection) did not affect the metataxonomic profile in breast milk.
Conclusion: These results support that in the non-severe SARS-CoV-2 pregnant woman infection the structure of the bacterial population is preserved and does not negatively impact on the human milk microbiota.
Introduction
The possibility of mother-to-infant transmission of SARS-CoV-2 during pregnancy and breastfeeding was a matter of concern at the start of the COVID-19 pandemic. The first question to address about the impact of SARS-CoV-2 infection on breastfeeding was if the virus was present or not in human colostrum or mature milk samples, with a few studies finding positive samples (1) but most of them showing negative results (2–8). Second, the research focused on testing whether these biological fluids carry anti-SARS-CoV-2 antibodies with the property of protecting nursing babies from infection (4, 9, 10).
SARS-CoV-2 initially interacts with the mucosal surface of the upper respiratory tract (11, 12) and with that of the gastrointestinal tract (13), which are associated with a highly complex microbiota. SARS-CoV-2 infection pathophysiology is related to the angiotensin-converting enzyme receptor 2 (ACE2), whose activity is influenced and, in turn, influences the microbiota (14). These facts suggest an implication of the host microbiota in the modulation of SARS-CoV-2 infection (15). Some studies have already found that the presence of key immunomodulatory species in the gut microbiota, such as bifidobacteria, Faecalibacterium prausnitzii, or Eubacterium rectale, is reduced in COVID-19 patients (16, 17), and the magnitude of this change is closely associated to disease severity (18, 19). It has been reported that the gut dysbiosis usually persists beyond disease state resolution and may contribute to the long-lasting COVID-19 sequelae (18, 19). The modulatory role of the microbiota has already been observed with other respiratory viruses, including other coronaviruses and the respiratory syncytial virus (20–22).
Breast milk contains a site-specific microbiota (23), which is dominated by species of the genera Staphylococcus, Streptococcus, Corynebacterium, and Cutibacterium, although lactic acid bacteria and bifidobacteria can also be isolated. The microbiome of human milk seems to be a dynamic and complex ecosystem which is not randomly assembled but forms well-organized bacterial consortia and networks (24). This microbiome and its evolution along the lactation period are relevant from both the scientific and biomedical points of view since human milk-associated microbes may determine, at least partly, the pattern of gastrointestinal colonization during early life (25, 26). Breast milk bacteria not only drive infant gut colonization but, also, they may play other important biological functions, such as protecting against pathogens, influencing the development of the mucosal immune system, and helping digestion and nutrients’ absorption (27, 28). However, there are many maternal, neonatal and environmental factors that can affect the composition of the milk microbiome over time (29–31). Since colostrum and mature milk microbes are among the first colonizers of the human gut, any factor disturbing the microbiome profile of these biological fluids may potentially impact infant gut colonization and future health. Interestingly, DNA from some gut-associated strict anaerobes which presence is reduced in COVID-19 patients (Faecalibacterium or Eubacterium, among others) has also been detected in human milk using culture-independent techniques (32–36).
On April 2020, a multidisciplinary group designed a research plan to foster the knowledge on breastfeeding and SARS-CoV-2 infection, with the aims of providing data on its safety, i.e., exclusion of human milk as a source of virus spread, and its efficacy in the transfer of protection factors against SARS-CoV-2 infection. The purpose of this work is to report on the impact of SARS-CoV-2 viral infection on the breast milk metataxonomic profile and its evolution during the first month of lactation.
Materials and Methods
A multicenter, prospective case and control study was conducted in four maternity hospitals of Madrid, which gather a total number of deliveries per year above 13.000. Since March 2020, all pregnant women underwent routine nasopharyngeal SARS-CoV-2 Reverse Transcription Polymerase Chain Reaction (RT-PCR) test as screening upon admission prior delivery. Recruitment was completed between April and July 2020. The study was approved by the referral Clinical Research Ethics Committee (La Paz University Hospital). Informed consent was obtained from mothers before enrollment. Every mother-infant’ information was treated anonymously.
Study Procedures
Term pregnant women with confirmed SARS-CoV-2 infection at the time of delivery were approached, providing they were in good clinical condition and had a decision to breastfeed their healthy babies (study group; n = 45). Consecutive sample (1case:2controls) of SARS-CoV-2 negative pregnant women, who were in identical conditions, was obtained (control group; n = 96). Prospective clinical data of participant mothers and their infants were recorded.
Women were instructed on how to extract and store milk to avoid contaminations. Milk samples were immediately frozen at −20°C and shipped on dry ice (−78.5°C) to the Department of Nutrition and Food Science, Complutense University of Madrid, for further processing. Breast milk samples collected during the first- and fifth-week postpartum were used for analyses.
Reverse Transcription Polymerase Chain Reaction Assays
RNA extraction from the milk samples (200 μL) was carried out using the KINGFISHER FLEX 96 extraction robot (Thermo Fisher Scientific), the MagMax_Core_Flex extraction program and the MagMAX Viral/Pathogen II Nucleic Acid Isolation kit (Applied Biosystems, Thermo Fisher Scientific). For the detection of SARS-CoV-2, the TaqPath COVID-19 CE-IVD RT-PCR kit (Applied Biosystems, Thermo Fisher Scientific) was used in a 384-well format with the QuantStudio 7 Flex System equipment (Applied Biosystems). In this kit, probes anneal to three target sequences (ORF1ab, N Protein and S Protein genes) that are specific to SARS-CoV-2. All lab kits were used following the manufacturer’s instructions.
DNA Extraction, Metataxonomic and Bioinformatic Analysis
DNA extraction from the milk samples (1 mL) was performed as described by Lackey et al. (37). The V3-V4 hypervariable region of the gene 16S rRNA was amplified and sequenced using the MiSeq system of Illumina at the facilities of Parque Científico de Madrid (Tres Cantos, Spain) with the Illumina MiSeq 2 × 300 bp pair-end protocol (Illumina Inc., San Diego, CA, United States) (38).
Raw sequences data were demultiplexed and quality filtered with the Illumina MiSeq Reporter analysis software. Bioinformatic analysis was performed with QIIME 2 2019.1 (39) pipelines. The denoising step was performed with DADA2 (40). The forward reads were truncated at position 295 and trimming the first 12 nucleotides, while the reverse sequences were truncated at the 275 nucleotides and trimming the first 9 nucleotides, in order to discard nucleotides which median quality were Q20 or below. Also, all amplicon sequence variants (ASVs) were classified in a feature count table. Taxonomy was assigned to each ASVs with the q2-feature-classifier (41) classify-sklearn, naïve Bayes taxonomy classifier, using the SILVA 138 reference database (42). Subsequent bioinformatic analysis was conducted using R version 3.5.1 (R Core Team, 2013).1 The decontam package (43) was used in order to identify, visualize and remove contaminating DNA.
ASVs, genera and phyla sequence count tables per sample were generated, and bacterial taxa abundances were normalized to the total number of sequences in each sample (relative abundance). Alpha diversity was assessed using the Shannon and Simpson diversity indexes. For the beta diversity studies, a quantitative (relative abundance) and a qualitative (presence/absence) analysis, for the bacterial profiles, were performed with the Bray-Curtis dissimilarity and binary Jaccard indices, respectively. Principal Coordinates Analysis (PCoA) was performed in order to plot patterns of bacterial profiles through the Bray-Curtis and binary Jaccard distance matrices.
A set of “core” genera were characterized for each group. To be included in the core taxa, a genus must have been present in ≥ 90% of the samples with a representability ≥ 0.1% of the relative abundance of each sample, for one or two groups. The 5 most-abundant phyla from all the milk samples were selected as most abundance phyla and the rest were included in the “minor_phyla” group. The 19 most-abundant genera from all the milk samples were selected as most abundance genera and the rest were included in the “minor_genera” group.
Statistical Analysis
Demographic data were expressed as mean and standard deviation (SD) and differences between study and control groups were assessed using the Fisher exact (or Chi-squared) and the t-test for categorical and quantitative variables, respectively.
Quantitative data on microbiota were expressed as median and interquartile range (IQR). Differences between study and control groups were assessed using the Wilconxon rank test. PCoA and the PERMANOVA analysis with 999 permutations were performed to examine for similarities in the bacterial profile between groups. Statistical analysis and plotting were performed in the R environment using R version 3.5.1 (R Core Team, 2013) (see text footnote 1) with library ggplot2 (44). Differences were considered statistically significant at p < 0.05.
Results
Forty-five term pregnant women with confirmed non-severe SARS-CoV-2 infection and uncomplicated delivery (study group), and 96 SARS-CoV-2 negative women in identical conditions (control group) were approached. Of them, 37 study group and 63 control group women were included in the final analyses. All women had normal weight (body mass index: 19–24) with only one exception (one woman of the study group was overweighed). Details on participants’ chart flow and reasons for exclusion are described on Figure 1.
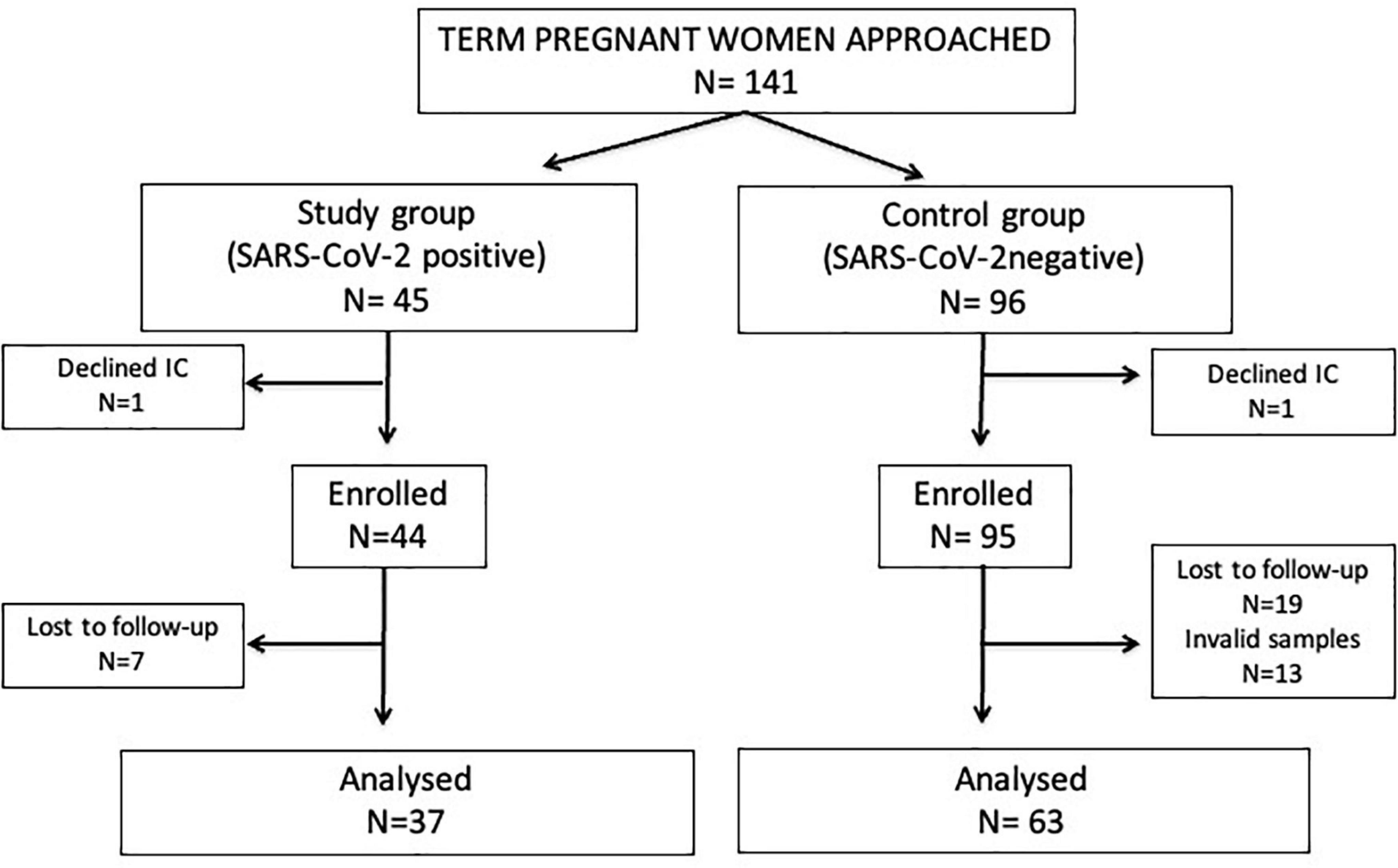
Figure 1. Flow diagram of the study participants. IC, informed consent. Invalid samples: non-availability, inadequate patient identification or inadequate handling or storage.
No differences between the study and control group were found regarding maternal age (33.9 ± 5.4 years vs. 34.5 ± 4.4 years, p = 0.56), prevalence of previous maternal health problems [6 (16%) vs. 10 (16%), p = 0.99; including the case of overweight in the study group], rates of vaginal delivery [26 (70%) vs. 48 (76%), p = 0.85], gestational age (39.1 ± 1.7 weeks vs. 38.9 ± 1.9 weeks, p = 0.64) and birth weight (3,187 ± 543 g vs. 3,179 ± 536 g, p = 0.94) of infants.
Mild SARS-CoV-2 infection related symptoms were present in 21 (56.8%) of study group women, including fever (48%), anosmia (48%), cough (43%), ageusia (14%), odynophagia (10%), myalgia (10%), diarrhea (10%), or headache (5%); and 19 out of the 21 symptomatic women received medication (anticoagulation, antibiotics, hydroxychloroquine, oxygen therapy) around labor. None of the infants of mothers in the study or the control group presented clinical signs associated to SARS-COV-2 infection within the first month of life. By hospital protocol, nasopharyngeal RT-PCR was performed in neonates of positive SARS-COV-2 mothers, resulting negative in all cases.
All the 1st week and 5th week postpartum samples collected and processed were negative for SARS-CoV-2 RNA (study group: 36 samples at 1st week and 37 samples at 5th week postpartum; control group: 63 samples at 1st week and 63 samples at 5th week postpartum), that were submitted to metataxonomic analysis. Among them, 197 milk samples (study group: 36 at 1st week, 37 at 5th week; control group: 62 at 1st and 5th week) showed a distinct single electrophoretic band after the first PCR round; the size of these amplicons matched the exact size of the V3-V4 hypervariable region and, therefore, they were selected for the second PCR round and sequencing. The 16S rRNA gene sequencing yielded 12,471,894 high quality filtered sequences, ranging from 21,862 to 183,796 reads per sample [median sequence per sample 60,067 (51,499–70,938)] with a total of 5,003 ASVs.
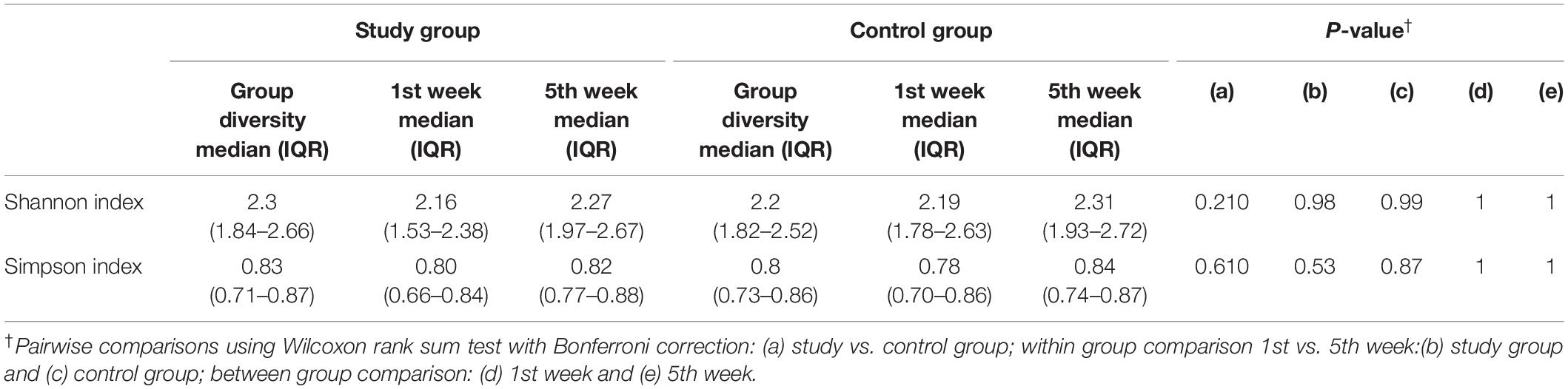
Alpha diversity showed no differences between milk samples from the study and control group, and this condition was maintained throughout the observation period (Table 1).
The overall analysis of the beta diversity indicated that the profiles of bacterial genera of the study and control groups did not show distinct bacterial profiles. The PCoA plots of the Bray-Curtis distance matrix (relative abundance) yielded no differences between groups (p = 0.163), while the binary Jaccard distance matrix revealed small but statistically significant differences (p = 0.016) in the presence/absence of bacterial taxa between the study and the control groups (Figure 2).
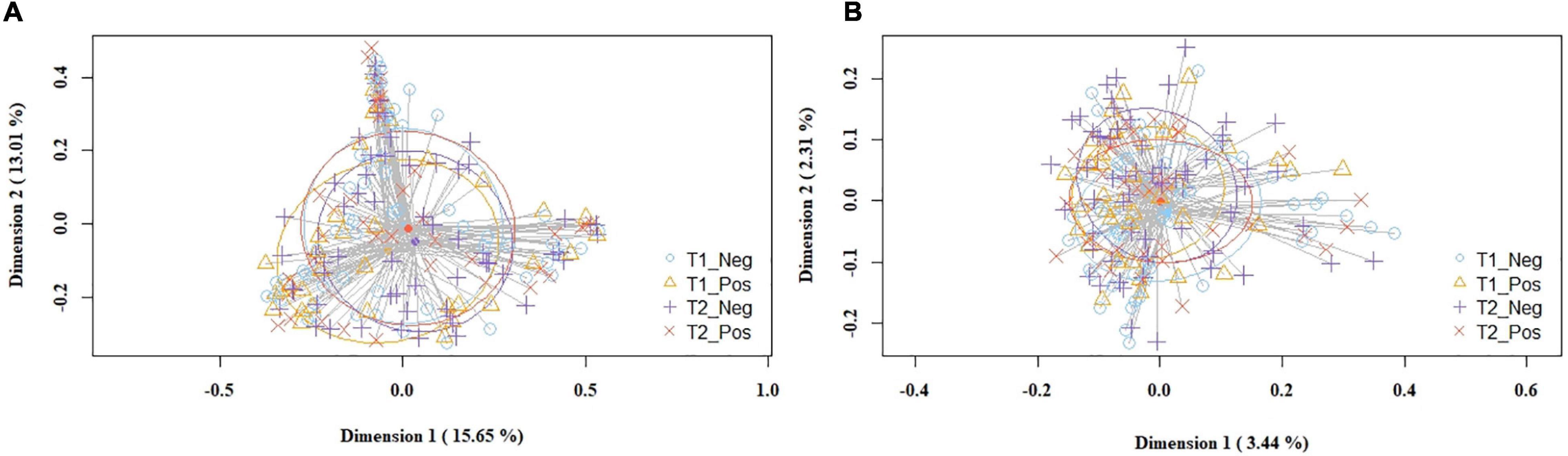
Figure 2. PCoA plots of bacterial profiles (at the ASVs level) of milk samples according to SARS-CoV-2 status (study and control group) and study time point, based on the Bray-Curtis similarity analysis (relative abundance) (A), and Jaccard’s coefficient for binary data (presence/absence) (B). Where blue circles (T1_Neg) refer to 1st week samples from the control group, yellow triangles (T1_Pos) to 1st week samples from the study group, purple crosses (T2_Neg) to 5th week samples from the control group, and orange × s (T2_Pos) to 5th week samples from the study group. The value given on each axis label represents the percentage of the total variance explained by that axis.
The frequency of the most abundant bacterial phyla and genera detected in milk samples according to group and observation time is displayed on Table 2, indicating a high similarity in the relative abundance between groups, with a few exceptions; among them, the relative abundance of the genus Gemella was higher among the samples of the control group. Staphylococcus and Streptococcus were the most abundant genera and the only ones that were detected in all the milk samples processed. No differences regarding diversity or any taxonomic level were found in relation to disease expression in SARS-CoV-2-positive women (symptomatic vs. asymptomatic infected women).
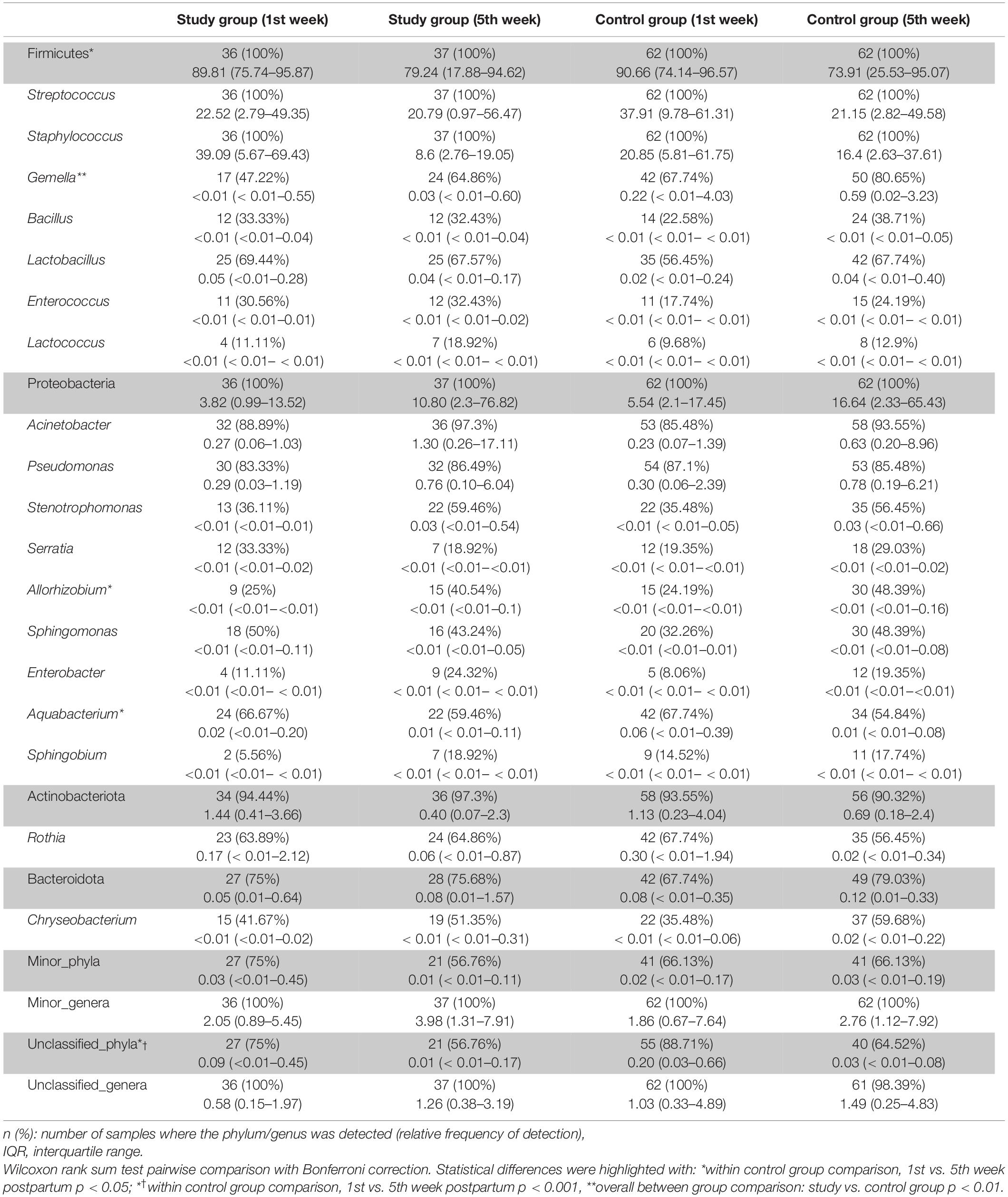
Table 2. Frequency of the most abundant bacterial phyla and genera detected in milk samples according to group and study time point.
Subsequently, the potential variation of the milk bacteriome composition over time was assessed. For this purpose, all the samples obtained at 1st week were compared with those collected at 5th week postpartum. This approach revealed that alpha diversity assessed by Simpson index increased [1st week 0.79 (0.67–0.85); 5th week 0.83 (0.74–0.87); p = 0.021]. Non-significant trends were observed in alpha diversity assessed by the Shannon index [1st week 2.19 (1.76–2.55); 5th week 2.3 (1.94–2.7); p = 0.210]. Beta diversity did not vary over time, neither using the Bray-Curtis distance matrix (p = 0.789) nor the binary Jaccard distance matrix (p = 0.934) (Figure 3). However, the analysis of specific bacterial taxonomic groups revealed a decrease in abundance of the phylum Firmicutes and the genus Staphylococcus over time and the opposite trends for several genera in the phylum Proteobacteria and other unclassified phyla (Table 3).
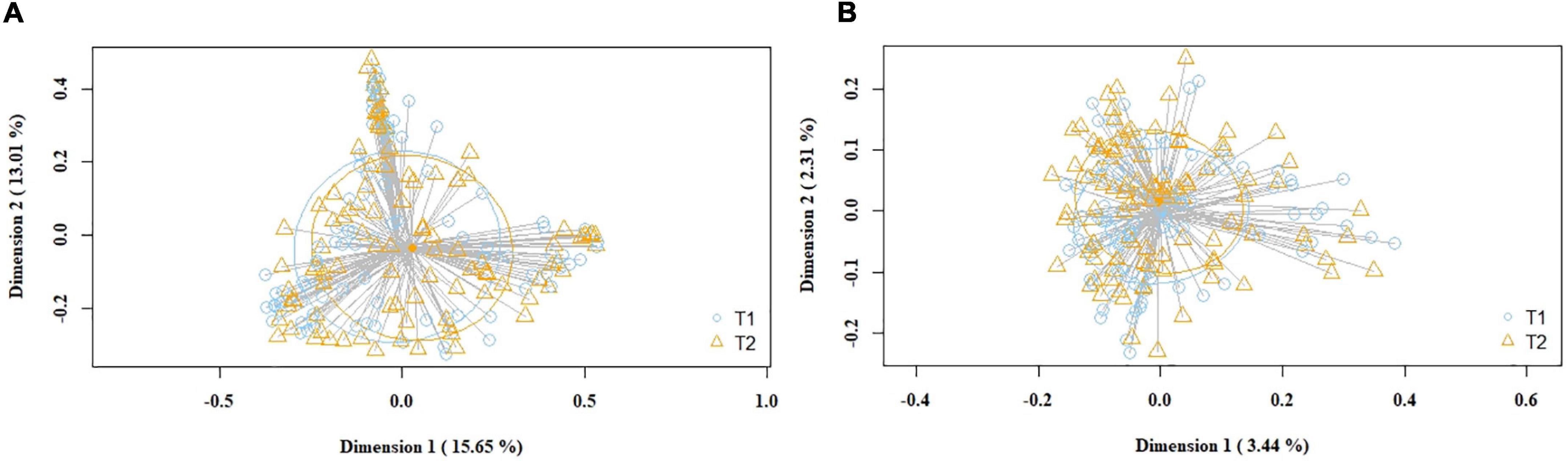
Figure 3. PCoA plots of bacterial profiles (at the ASVs level) of pooled (study and control group) milk samples according to time, based on the Bray-Curtis similarity (relative abundance) (A) and Jaccard’s coefficient for binary data (presence/absence) (B) analyses. Where blue circles (T1) refer to 1st week samples, and yellow triangles (T2), to 5th week samples. The value given on each axis label represents the percentage of the total variance explained by that axis.
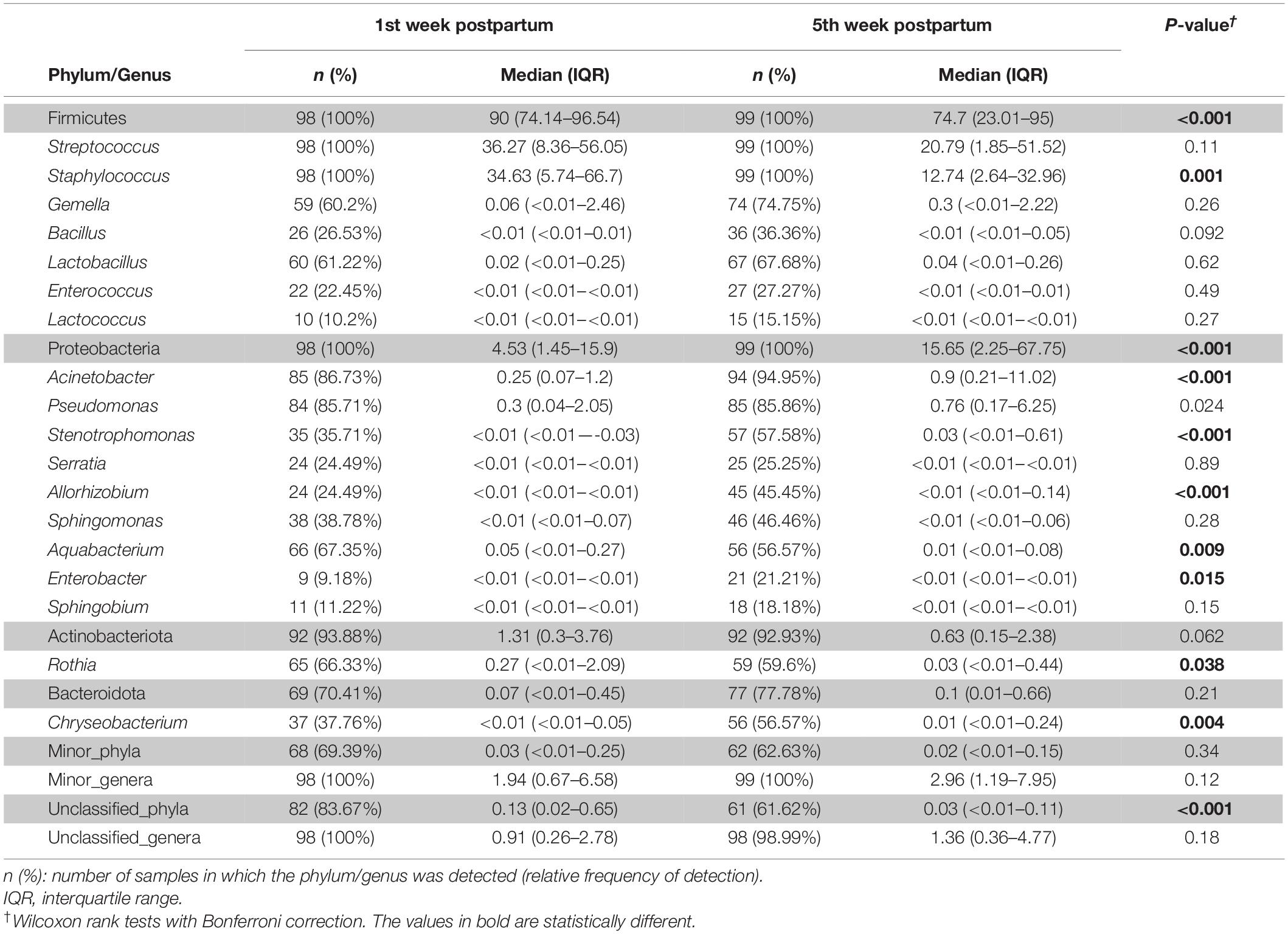
Table 3. Frequency of the most abundant bacterial phyla (shadowed boxes) and genera detected in the milk samples according to time.
When the samples were compared according to mode of labor completion (vaginal delivery vs. cesarean section), no statistical differences were found in relation to the biodiversity indices [Shannon and Simpson indices of 2.22 (1.78–2.53) and 0.78 (0.69–0.86), respectively, for the vaginal delivery group; Shannon and Simpson indices of 2.12 (1.6–2.56) and 0.80 (0.67–0.84), for the cesarean group]. Similarly, no statistical differences were observed concerning beta diversity neither using the Bray-Curtis distance matrix (p = 0.7413) nor the binary Jaccard distance (p = 0.526) (Figure 4). Analysis of the relative abundance of specific bacterial taxonomic groups showed a higher relative abundance of phylum Proteobacteria (p = 0.036) and genus Pseudomonas, Stenotrophomonas (p < 0.001) and Serratia (p = 0.002) among women who had cesarean section, and a higher abundance of the genus Lactobacillus among women with vaginal delivery (Table 4).
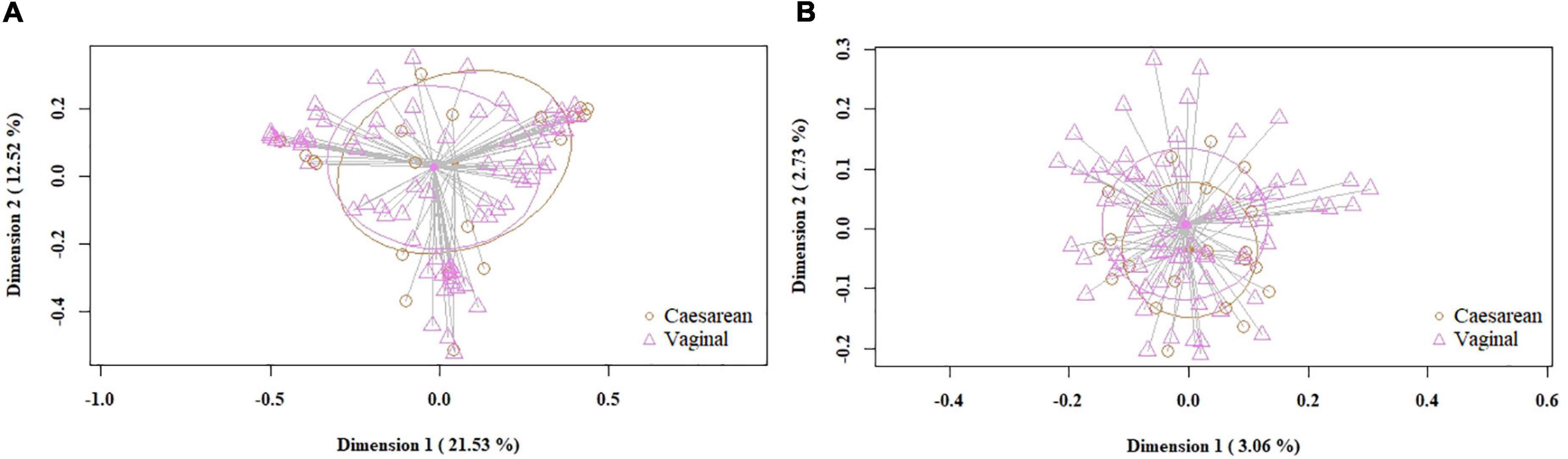
Figure 4. PCoA plots of bacterial profiles (at the ASVs level) of milk samples according to mode of labor completion, based on the Bray-Curtis similarity analysis (relative abundance) (A) and Jaccard’s coefficient for binary data (presence/absence) (B). Where orange circles refer to samples from women who had cesarean section, and purple triangles, to those who had vaginal delivery. The value given on each axis label represents the percentage of the total variance explained by that axis.
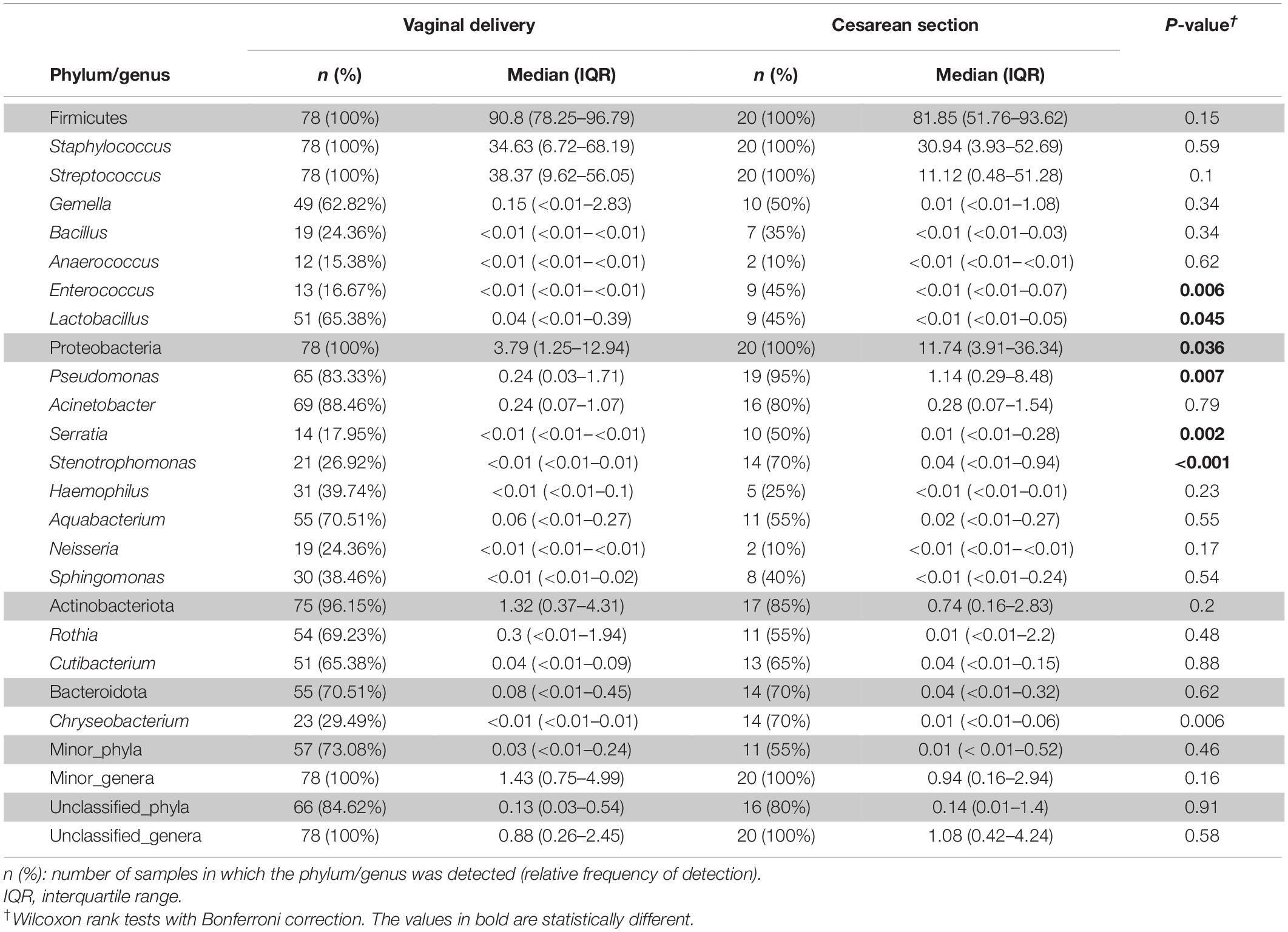
Table 4. Frequency of the most abundant bacterial phyla (shadowed boxes) and genera detected in the milk samples according to mode of delivery.
Discussion
This study aimed to address potential differences on breast milk microbiota according to mother’s SARS-CoV-2 status, by means of the 16S RNA gene-based metataxonomic analysis. Overall, the results have shown great similarity in the structure of the bacterial populations in milk samples collected at first week and at fifth week of lactation, between the infected and the non-infected women. This suggests that peri-partum maternal SARS-CoV-2 infection does not have an impact on the human milk microbiota. On a recent report conducted on the same cohort of pregnant women (45), we have documented a distinct immunological profile in the human milk of SARS-CoV-2 infected mothers, consisting of higher concentrations of cytokines, chemokines and growth factors compared to that of negative women. Concentrations of most of the immune factors analyzed remained stable over time in SARS-CoV-2-positive women milk samples, but most of these compounds significantly decreased from the first to the fifth week postpartum in negative women. The results of both studies, together with the evidence of passive transfer of SARS-CoV-2-specific antibodies (4, 9, 10) and the lack of detection of this virus in most studies targeting human milk (2–4, 6, 45–48), reinforces the notion of safety and strengthens the current recommendation of breastfeeding in SARS-CoV-2-positive mothers.
Staphylococcus and Streptococcus were the most abundant bacterial genera and the only ones detected in all the milk samples. Such abundance of these two genera is in accord with previous studies (32, 35, 37, 49, 50), therefore supporting these two species as the hard core of the human milk bacteriome. Several studies have shown that coagulase-negative staphylococci are very abundant in colostrum or feces of breast-fed infants during the first days after birth (51), and that their levels tend to decrease in the following weeks (49, 52–54), usually coinciding with a rise in enterobacteria detection (54). Although few significant differences were also found at the phylum or genus level, this can be due to the relatively high degree of interindividual variability that characterizes the human milk bacteriome (50, 55–59).
Human milk is a relevant source of bacteria to the infant gut (60–62), that have a strong and perdurable impact on infant gut colonization (26). Therefore, the finding that the bacteriome structure evolution in the samples obtained during the first month postpartum in the SARS-CoV-2-positive women mirrors that of the negative ones is of utmost importance. So far, studies addressing the impact of maternal infections on the microbiological composition of milk are very scarce. It has been described that human papilloma virus (HPV) infection does not modify the bacterial composition of milk (63), although the authors declared that their results were not conclusive because of the low sample size (only 3 out of 35 samples analyzed were from HPV positive mothers). Breastfeeding has been repeatedly acknowledged as providing a certain degree of protection against mother-to-infant transmission of some viruses, including HIV (64). Genome analysis of a Ligilactobacillus salivarius strain with a high ability to inhibit in vitro HIV infectivity (65) revealed the existence of a mechanism by which such strain could interfere with gp120 attachment to immune cells (66). Interestingly, such strain had been isolated from human milk of a HIV-positive woman (with a high HIV titer in milk) whose breastfed infant remained uninfected during exclusive breastfeeding period.
The potential roles of the human milk microbiota in infants’ protection against life-viral infections (HIV, Zika, Ebola, cytomegalovirus…) or in minimizing their impact on health remains yet unexplored. It has been suggested, however, that the outcomes after suffering neonatal viral infections may be influenced by the interactions established between human milk oligosaccharides and the human milk and infant gut microbiomes (67).
Regardless of maternal SARS-CoV-2 status, our study has shown that the bacteriome composition changed from the 1st week to the 5th week postpartum milk, which confirms previous observations (68, 69). Different factors have been suggested as drivers of these differences, including an increased permeability of the mammary tight junctions during the first days after birth, or the impact of accumulating exposures to the infant microbiota over time. The decrease in the relative abundance of Staphylococcus from colostrum to mature milk samples has also been described before (70).
In contrast, only small differences in the bacteriome composition and structure were found when women were compared depending on the mode of labor completion (vaginal delivery vs. cesarean section). Previous studies have reported contradictory results when addressing the influence of mode of delivery on the composition of the human milk bacteriome, ranging from non-significant differences in the bacteriome profile (71) to significant differences at a variety of taxon levels (57, 72–75). In our study, cesarean section was associated to an increased presence of Pseudomonas, which is in accord with previous reports (76), but also to Serratia and Stenotrophomonas. All these bacterial genera are very relevant in the Neonatal Intensive Care Unit setting; therefore, this additional finding of our study also arouses interest from the perspective of increasing knowledge of the risk factors for perinatal infection.
This study faces some limitations. First, although the sample size was enough to reach statistical significance, it was relatively small, making difficult to extrapolate the results. Second, mothers with severe SARS-CoV-2 infection were not included. In addition, some factors known to modify the breast milk microbiota, such as overweight and obesity, could not be assessed since only one overweighed woman (study group) was included in this study. On the other hand, there was no comparison between the microbiome of the breast milk samples and the fecal microbiome of the respective infants (since the later samples were not available). In addition, the analysis of the milk microbiome was limited to two samples from each woman, with an interval of approximately 1 month. This decision was taken after confirmation of the lack of viral RNA in any of the milk samples collected over time (45). We considered that this observation period was adequate to elucidate an eventual distinct evolution of the bacterial structure related to mother’s infection status and/or to the time passed after birth. On the other hand, a strength of this work is the systematic approach to both SARS-CoV-2 documented infection and control women.
In summary, the results of this study indicate that the bacterial structure and composition of the human milk is, overall, well preserved among the SARS-CoV-2-positive women, providing additional support to foster breastfeeding in this population. The finding is relevant in terms of safety and efficacy of breastfeeding in this context, given the important role of colostrum and mature milk microbes as one of the first colonizers of the human gut, with potential impact on long-term health outcomes of infants.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, BioProject, PRJNA795855.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethical Committee of Clinical Research of La Paz University Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
NG-T, IC, RA, and CA participated in sampling management and analysis, drafted the initial manuscript, reviewed, and approved the final version. LS-G participated in designing the study, patient’s enrollment, data gathering and analysis, drafted the initial manuscript, reviewed, and approved the final version. AP and JR conceptualized, designed the study, participated in data analyses, drafted the initial manuscript, reviewed, and approved the final version. FC conceptualized, designed the study, reviewed, and approved the final version of the manuscript. RG-S, ML-A, MM-P, DE-V, and EC-A participated in patient’s enrollment, data gathering, reviewed, and approved the final version of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Instituto de Salud San Carlos III (COV20/01046) and RETICS “Maternal and Child Health and Development Network” (SAMID Network), funded by the PN I+D+I 2013–2016 Spain, ISCIII-Sub-Directorate General for Research Assessment and Promotion and the European Regional Development Fund (ERDF) (RD16/0022). IC was the recipient of a postdoctoral grant (BES-2017-080713) from the Spanish Ministry of Science, Innovation and Universities.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank all the women that participated in the study.
Footnotes
References
1. Groß R, Conzelmann C, Müller JA, Stenger S, Steinhart K, Kirchhoff F, et al. Detection of SARS-CoV-2 in human breastmilk. Lancet. (2020) 395:1757–8. doi: 10.1016/S0140-6736(20)31181-8
2. Marín Gabriel MA, Cuadrado I, Álvarez Fernández B, González Carrasco E, Alonso Díaz C, Llana Martín I, et al. Multicentre Spanish study found no incidences of viral transmission in infants born to mothers with COVID-19. Acta Paediatr. (2020) 109:2302–8. doi: 10.1111/apa.15474
3. Chambers CD, Krogstad P, Bertrand K, Contreras D, Tobin NH, Bode L, et al. Evaluation of SARS-CoV-2 in breastmilk from 18 infected women. JAMA. (2020) 324:1347–8. doi: 10.1001/jama.2020.15580
4. Pace RM, Williams JE, Järvinen KM, Belfort MB, Pace CD, Lackey KA, et al. Characterization of SARS-CoV-2 RNA, antibodies and neutralizing capacity in milk produced by women with COVID-19. mBio. (2021) 12:e3192–3120. doi: 10.1128/mBio.03192-20
5. Bertino E, Moro GE, De Renzi G, Viberti G, Cavallo R, Coscia A, et al. Detection of SARS-CoV-2 in milk from COVID-19 positive mothers and follow-up of their infants. Front Pediatr. (2020) 8:597699. doi: 10.3389/fped.2020.597699
6. Martins-Filho PR, Santos VS, Santos HP. Jr To breastfeed or not to breastfeed? Lack of evidence on the presence of SARS-CoV-2 in breastmilk of pregnant women with COVID-19. Rev Panam Salud Publica. (2020) 44:e59. doi: 10.26633/RPSP.2020.59
7. Cheema R, Partridge E, Kair LR, Kuhn-Riordon KM, Silva AI, Bettinelli ME. Protecting breastfeeding during the COVID-19 pandemic. Am J Perinatol. (2020). doi: 10.1055/s-0040-1714277
8. Bäuerl C, Randazzo W, Sánchez G, Selma-Royo M, García Verdevio E, Martínez L, et al. SARS-CoV-2 RNA and antibody detection in breast milk from a prospective multicentre study in Spain. Arch Dis Child Fetal Neonatal Ed. (2021) 107:216–21. doi: 10.1136/archdischild-2021-322463
9. Demers-Mathieu V, Do DM, Mathijssen GB, Sela DA, Seppo A, Järvinen KM, et al. Difference in levels of SARS-CoV-2 S1 and S2 subunits – and nucleocapsid protein-reactive SIgM/IgM. IgG and SIgA/IgA antibodies in human milk. J Perinatol. (2020) 1:1–10. doi: 10.1038/s41372-020-00805-w
10. Fox A, Marino J, Amanat F, Krammer F, Hahn-Holbrook J, Zolla-Pazner S, et al. Robust and specific secretory IgA against SARS-CoV-2 detected in human milk. iScience. (2020) 23:101735. doi: 10.1016/j.isci.2020.101735
11. Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang L, et al. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. (2020) 71:713–20. doi: 10.1093/cid/ciaa203
12. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. (2020) 81:465–9. doi: 10.1038/s41586-020-2196-x
13. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. (2020) 158:1831−1833.e3. doi: 10.1053/j.gastro.2020.02.055
14. Yang T, Chakraborty S, Saha P, Mell B, Cheng X, Yeo JY, et al. Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS-CoV-2 infectivity. Hypertension. (2020) 76:e1–3. doi: 10.1161/hypertensionaha.120.15360
15. He Y, Wang J, Li F, Shi Y. Main clinical features of COVID-19 and potential prognostic and therapeutic value of the microbiota in SARS-CoV-2 infections. Front Microbiol. (2020) 11:1302. doi: 10.3389/fmicb.2020.01302
16. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with COVID-19 or H1N1 influenza. Clin Infect Dis. (2020) 71:2669–78. doi: 10.1093/cid/ciaa709
17. Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in gut microbiota of patients With COVID-19 during time of hospitalization. Gastroenterology. (2020) 159:944-955.e8. doi: 10.1053/j.gastro.2020.05.048
18. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. (2021) 70:698–706. doi: 10.1136/gutjnl-2020-323020
19. Zuo T, Liu Q, Zhang F, Lui GC, Tso EY, Yeoh YK, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. (2021) 70:276–84. doi: 10.1136/gutjnl-2020-322294
20. McGillivary G, Mason KM, Jurcisek JA, Peeples ME, Bakaletz LO. Respiratory syncytial virus-induced dysregulation of expression of a mucosal beta-defensin augments colonization of the upper airway by non-typeable Haemophilus influenzae. Cell Microbiol. (2009) 11:1399–408. doi: 10.1111/j.1462-5822.2009.01339.x
21. Hasegawa K, Mansbach JM, Ajami NJ, Espinola JA, Henke DM, Petrosino JF, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. Eur Respir J. (2016) 48:1329–39. doi: 10.1183/13993003.00152-2016
22. de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. (2016) 194:1104–15. doi: 10.1164/rccm.201602-0220OC
23. Fernández L, Pannaraj PS, Rautava S, Rodríguez JM. The microbiota of the human mammary ecosystem. Front Cell Infect Microbiol. (2020) 10:586667. doi: 10.3389/fcimb.2020.586667
24. Ma Z, Guan Q, Ye C, Zhang C, Foster JA, Forney LJ. Network analysis suggests a potentially “evil” alliance of opportunistic pathogens inhibited by a cooperative network in human milk bacterial communities. Sci Rep. (2015) 5:8275. doi: 10.1038/srep08275
25. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. (2017) 81:e36–17. doi: 10.1128/MMBR.00036-17
26. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690-703. doi: 10.1016/j.chom.2015.04.004
27. Jeurink PV, van Bergenhenegouwen J, Jiménez E, Knippels LM, Fernández L, Garssen J, et al. Human milk: a source of more life than we imagine. Benef Microbes. (2013) 4:17–30. doi: 10.3920/BM2012.0040
28. Rodríguez JM, Fernández L, Verhasselt V. The gut?breast axis: programming health for life. Nutrients. (2021) 13:606. doi: 10.3390/nu13020606
29. Pace RM, Williams JE, Robertson B, Lackey KA, Meehan CL, Price WJ, et al. Variation in human milk composition is related to differences in milk and infant fecal microbial communities. Microorganisms. (2021) 9:1153. doi: 10.3390/microorganisms9061153
30. Mantziari A, Rautava S. Factors in incluencing the microbial composition of human milk. Semin Perinatol. (2021) 45:151507. doi: 10.1016/j.semperi.2021.151507
31. Consales A, Cerasani J, Sorrentino G, Morniroli D, Colombo L, Mosca F, et al. The hidden universe of human milk microbiome: origin, composition, determinants, role, and future persperctives. Eur J Pediatri. (2022). doi: 10.1007/s00431-022-04383-1
32. Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. (2012) 96:544–51. doi: 10.3945/ajcn.112.037382
33. Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. (2013) 110:1253–62. doi: 10.1017/S0007114513000597
34. Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. (2014) 16:2891–904. doi: 10.1111/1462-2920.12238
35. Jiménez E, de Andrés J, Manrique M, Pareja-Tobes P, Tobes R, Martínez-Blanch JF, et al. Metagenomic analysis of milk of healthy and mastitis-suffering women. J Hum Lact. (2015) 31:406–15. doi: 10.1177/0890334415585078
36. Patel SH, Vaidya YH, Patel RJ, Pandit RJ, Joshi CG, Kunjadiya AP. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep. (2017) 7:7804. doi: 10.1038/s41598-017-08451-7
37. Lackey KA, Williams JE, Meehan CL, Zachek JA, Benda ED, Price WJ, et al. What’s normal? Microbiomes in human milk and infant feces are related to each other but vary geographically: the INSPIRE study. Front Nutr. (2019) 6:45. doi: 10.3389/fnut.2019.00045
38. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. (2013) 41:e1. doi: 10.1093/nar/gks808
39. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
40. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
41. Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. (2018) 6:90. doi: 10.1186/s40168-018-0470-z
42. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
43. Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. (2018) 6:226. doi: 10.1186/s40168-018-0605-2
44. Wickham H. Ggplot2 Elegant Graphics for Data Analysis. Cham: Springer International Publishing (2016). doi: 10.1007/978-3-319-24277-4
45. Sánchez García L, Gómez-Torres N, Cabañas F, González-Sánchez R, López-Azorín M, Moral-Pumarega MT, et al. Immediate pre-partum SARS-CoV-2 status and immune profiling of breastmilk: a case-control study. Front Immunol. (2021) 12:720716. doi: 10.3389/fimmu.2021.720716
46. Lackey KA, Pace RM, Williams JE, Bode L, Donovan SM, Järvinen KM, et al. SARS-CoV-2 and human milk: what is the evidence? Matern Child Nutr. (2020) 16:e13032. doi: 10.1111/mcn.13032
47. Yang N, Che S, Zhang J, Wang X, Tang Y, Wang J, et al. COVID-19 evidence and recommendations working group. breastfeeding of infants born to mothers with COVID-19: a rapid review. Ann Transl Med. (2020) 8:618. doi: 10.21037/atm-20-3299
48. Chen L, Li Q, Zheng D, Jiang H, Wei Y, Zou L, et al. Clinical characteristics of pregnant women with Covid-19 in Wuhan. China N Engl J Med. (2020) 382:e100. doi: 10.1056/NEJMc2009226
49. Jiménez E, Delgado S, Maldonado A, Arroyo R, Albujar M, García N, et al. Staphylococcus epidermidis: a differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. (2008) 8:143. doi: 10.1186/1471-2180-8-143
50. Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. (2011) 6:e21313. doi: 10.1371/journal.pone.0021313
51. Jiménez E, Delgado S, Fernández L, García N, Albújar M, Gómez A, et al. Assessment of the bacterial diversity of human colostrum and screening of Staphylococcal and Enterococcal populations for potential virulence factors. Res Microbiol. (2008) 159:595–601. doi: 10.1016/j.resmic.2008.09.001
52. Lundequist B, Nord CE, Winberg J. The composition of the faecal microflora in breastfed and bottle-fed infants from birth to eight weeks. Acta Pediatr Scan. (1985) 74:45–51. doi: 10.1111/j.1651-2227.1985.tb10919.x
53. Borderon JC, Lionnet C, Rondeau C, Suc AI, Laugier J, Gold F. Current aspects of fecal flora of the newborn without antibiotherapy during the first 7 days of life: Enterobacteriaceae, enterococci, staphylococci. Pathol Biol. (1996) 44:416–22.
54. Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegård IL, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. (2006) 59:96–101. doi: 10.1203/01.pdr.0000191137.12774.b2
55. Martín R, Heilig GHJ, Zoetendal EG, Jiménez E, Fernández L, Smidt H, et al. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res Microbiol. (2007) 158:31–7. doi: 10.1016/j.resmic.2006.11.004
56. Boix-Amorós A, Collado MC, Mira A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front Microbiol. (2016) 7:492. doi: 10.3389/fmicb.2016.00492
57. Cabrera-Rubio R, Mira-Pascual L, Mira A, Collado MC. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. (2016) 7:54–60. doi: 10.1017/S2040174415001397
58. Chen PW, Lin YL, Huang MS. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J Food Drug Anal. (2018) 26:1235–44. doi: 10.1016/j.jfda.2018.03.004
59. Avershina E, Angell IL, Simpson M, Storrø O, Øien T, Johnsen R, et al. Low maternal microbiota sharing across gut, breast milk and vagina, as revealed by 16S rRNA gene and reduced metagenomic sequencing. Genes (Basel). (2018) 9:E231. doi: 10.3390/genes9050231
60. Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. (2017) 171:647–54. doi: 10.1001/jamapediatrics.2017.0378
61. Murphy K, Curley D, O’Callaghan TF, O’Shea CA, Dempsey EM, O’Toole PW, et al. The composition of human milk and infant fecal microbiota over the first three months of life: a pilot study. Sci Rep. (2017) 7:40597. doi: 10.1038/srep40597
62. Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armaninni F, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomics profiling. mSystems. (2017) 2:e00164. doi: 10.1128/mSystems.00164-16
63. Tuominen H, Rautava S, Collado MC, Syrjänen S, Rautava J. HPV infection and bacterial microbiota in breast milk and infant oral mucosa. PLoS One. (2018) 13:e0207016. doi: 10.1371/journal.pone.0207016
64. World Health Organization.Guideline: Updates on HIV and Infant Feeding: the Duration of Breastfeeding, and Support from Health Services to Improve Feeding Practices Among Mothers Living with HIV. (2016). Available online at: https://www.who.int/publications/i/item/9789241549707 (accessed January 1, 2016).
65. Martín V, Maldonado A, Fernández L, Rodríguez JM, Connor RI. Inhibition of human immunodeficiency virus type 1 by lactic acid bacteria from human breast milk. Breastfeed Med. (2010) 5:153–8. doi: 10.1089/bfm.2010.0001
66. Langa S, Maldonado-Barragán A, Delgado S, Martín R, Martín V, Jiménez E, et al. Characterization of Lactobacillus salivarius CECT 5713, a strain isolated from human milk: from genotype to phenotype. Appl Microbiol Biotechnol. (2012) 94:1279–87. doi: 10.1007/s00253-012-4032-1
67. Ramani S, Stewart CJ, Laucirica DR, Ajami NJ, Robertson B, Autran CA, et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat Commun. (2018) 9:5010. doi: 10.1038/s41467-018-07476-4
68. Hoashi M, Meche L, Mahal LK, Bakacs E, Nardella D, Naftolin F, et al. Human milk bacterial and glycosylation patterns differ by delivery mode. Reprod Sci. (2016) 23:902–7. doi: 10.1177/1933719115623645
69. Drago L, Toscano M, De Grandi R, Grossi E, Padovani EM, Peroni DG. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. (2017) 11:875–84. doi: 10.1038/ismej.2016.183
70. Simpson MR, Avershina E, Storrø O, Johnsen R, Rudi K, ?jen T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J Dairy Sci. (2018) 101:889–99. doi: 10.3168/jds.2017-13411
71. Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. (2016) 4:1. doi: 10.1186/s40168-015-0145-y
72. Khodayar-Pardo P, Mira-Pascual L, Collado MC, Martínez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. (2014) 34:599–605. doi: 10.1038/jp.2014.47
73. Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, et al. Distinct patterns in human milk microbiota and fatty acid profiles across specific geographic locations. Front Microbiol. (2016) 7:1619. doi: 10.3389/fmicb.2016.01619
74. Ding M, Qi C, Yang Z, Jiang S, Bi Y, Lai J, et al. Geographical location specific composition of cultured microbiota and Lactobacillus occurrence in human breast milk in China. Food Funct. (2019) 10:554–64. doi: 10.1039/c8fo02182a
75. Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr. (2019) 6:4. doi: 10.3389/fnut.2019.00004
Keywords: SARS-CoV-2, human milk, microbiota, metataxonomic analysis, infant gut colonization
Citation: Gómez-Torres N, Sánchez-García L, Castro I, Arroyo R, Cabañas F, González-Sánchez R, López-Azorín M, Moral-Pumarega MT, Escuder-Vieco D, Cabañes-Alonso E, Rodríguez JM, Alba C and Pellicer A (2022) Metataxonomic Analysis of Milk Samples From SARS-CoV-2-Positive and SARS-CoV-2-Negative Women. Front. Nutr. 9:853576. doi: 10.3389/fnut.2022.853576
Received: 12 January 2022; Accepted: 24 February 2022;
Published: 18 March 2022.
Edited by:
Yao-Tsung Yeh, Fooyin University, TaiwanReviewed by:
Alba Pérez-Cataluña, Institute of Agrochemistry and Food Technology (CSIC), SpainSercan Karav, Çanakkale Onsekiz Mart University, Turkey
Copyright © 2022 Gómez-Torres, Sánchez-García, Castro, Arroyo, Cabañas, González-Sánchez, López-Azorín, Moral-Pumarega, Escuder-Vieco, Cabañes-Alonso, Rodríguez, Alba and Pellicer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Sánchez-García, laurasg.hulp@gmail.com; Claudio Alba, c.alba@ucm.es
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
 Natalia Gómez-Torres
Natalia Gómez-Torres Laura Sánchez-García
Laura Sánchez-García Irma Castro1
Irma Castro1  Fernando Cabañas
Fernando Cabañas Raquel González-Sánchez
Raquel González-Sánchez Manuela López-Azorín
Manuela López-Azorín M. Teresa Moral-Pumarega
M. Teresa Moral-Pumarega Diana Escuder-Vieco
Diana Escuder-Vieco Juan Miguel Rodríguez
Juan Miguel Rodríguez Claudio Alba
Claudio Alba Adelina Pellicer
Adelina Pellicer