Illumina-Based Analysis Yields New Insights Into the Fungal Contamination Associated With the Processed Products of Crataegi Fructus
- Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
Crataegi Fructus, a medicinal and edible herb in China, has been considered a popular dietary supplement globally. It is used for the treatment of dyspepsia and chronic heart failure according to the Chinese Pharmacopoeia (2020). However, fungal contamination in Crataegi Fructus affects its quality and safety, thus preventing its global promotion. In this study, we comprehensively studied the fungal community in processed products of Crataegi Fructus by high-throughput sequencing. A total of 21 Crataegi Fructus samples were collected from five provinces in China, and the samples were divided into five groups based on collection areas, as well as into three groups based on processing methods. We then targeted the internal transcribed spacer 2 sequence through the Illumina Miseq PE300 platform to investigate fungal composition and diversity. Results showed that all 21 samples were detected with fungal contamination, and Ascomycota was dominant at the phylum level. In the groups based on collection areas, Dothideomycetes, Pleosporaceae, and Alternaria were dominant at the class, family, and genus levels, respectively. In the groups based on processing methods, Dothideomycetes, Aspergillaceae, and Alternaria were the most abundant at the class, family, and genus levels, respectively. Differences in fungal communities between various groups were also observed. Furthermore, a total of 115 species were identified, among which seven were potential toxigenic, namely, Trichothecium roseum, Alternaria tenuissima, Aspergillus carbonarius, Penicillium brevicompactum, Aspergillus fumigatus, Rhizopus microspores, and Pichia fermentans. In conclusion, this study reveals great fungal richness and diversity of Crataegi Fructus, providing references for the prevention and control of fungal contamination of Crataegi Fructus in practical production.
Introduction
As a popular dietary supplement, hawthorn has been consumed worldwide. Hawthorn is a universal name, which represents all species in the Crataegus genus (1). This supplement is distributed widely in Asia, Europe, and North America. In Europe, it is made into canned fruits, jams, and jellies. Furthermore, based on the European Pharmacopeia (2017), hawthorn herbal materials are derived from Crataegus monogyna Jacq. (Lindm.) and Crataegus laevigata (Poir.) DC (syn. C. oxyacantha L.) (2). Hawthorn is also a medicinal and edible herb in China, and its products have been made into beverages and snacks. This herb was first recorded as Shan Zha (in Chinese) in Ben Cao Jing Ji Zhu in 536 AD (3). According to the recommendation of the Chinese Pharmacopoeia (2020), Crataegi Fructus (CF) can be mainly formulated using dried, roasted, and charred products, which are derived from Crataegus pinnatifida Bge. var. major N.E. and Crataegus pinnatifida Bge (4). Modern pharmacological studies have demonstrated that its extract is useful for the treatment of chronic heart failure (5). It also shows the treatment effect on high-calorie-diet-induced dyspepsia. Interestingly, compared with the effect of dried products, roasted and charred products have been demonstrated to possess a stronger curative effect (6). However, mycotoxin contamination in CF has been reported in recent years. Li et al. investigated the patulin contamination in hawthorn products, and results showed that six of 43 samples were detected to be positive, with contamination levels ranging from 19.8 to 206.88 microg/L. Meanwhile, the level of patulin in four samples exceeded the Chinese legal limit (50 microg/L) (7). Similarly, Zhou et al. also observed patulin contamination in one of 13 hawthorn samples by HPLC (8). Therefore, the quality and safety of CF have received extensive attention worldwide.
Fungal contamination in herbal materials is derived from the whole production chain, including cultivation, harvest, transport, processing, and storage. Among these procedures, processing is a crucial factor that affects the quality and safety of herbs. In order to enhance the curative effect or decrease toxicity, many herbs are processed before clinical application. Shen et al. compared the effect of stir-frying with sand and stir-frying on carbonized ginger. The result showed that the sand-fried ginger samples exhibited greater adsorption capacity than the stir-fried samples (9). Liu et al. indicated that processing methods, such as heating and water-washing, reduced the toxicity of Aconitum roots (10). In addition, processing procedures remarkably affect the fungal communities in herbs. He et al. studied the variation in fungal communities during the processing of Polygala tenuifolia roots. The result showed that the processing methods decreased the level of most fungal genera excluding Penicillium (11). Odongo et al. determined the influence of processing on the growth of fungi, indicating that cooking and fermentation methods inhibited fungal growth (12). A study performed by Guo et al. investigated the fungal community in raw and roasted Cassiae Semen samples. In comparison with raw samples, roasted samples had higher numbers of Penicillium and Periconia (13). Therefore, it is essential to assess the effect of processing methods on fungal contamination in herbs.
The development of high-throughput sequencing (HTS) provides new insights for revealing the role of microorganisms in human daily life. This method has been applied in different areas to analyze the diversity, composition, and function of microorganisms (14–16). The application of HTS for the investigation of fungal contamination in herbs exhibits some irreplaceable advantages. For example, overall fungal diversity and composition can be analyzed efficiently. In addition, some strains that cannot grow in a synthetic medium can be identified through HTS. It can also be applied to monitor the dynamic change of fungal communities in practical production (17–19). Thus, the analysis of fungal communities by HTS has become increasingly prevalent.
In this study, we firstly studied the fungal community in processed CF products using the HTS method and analyzed the differences in the fungal community between groups based on processing methods, as well as between groups based on collection areas. This study provides an efficient method for the analysis of fungal contamination in CF, thereby providing the scientific basis for its safe utilization.
Materials and Methods
Sampling and Treatments
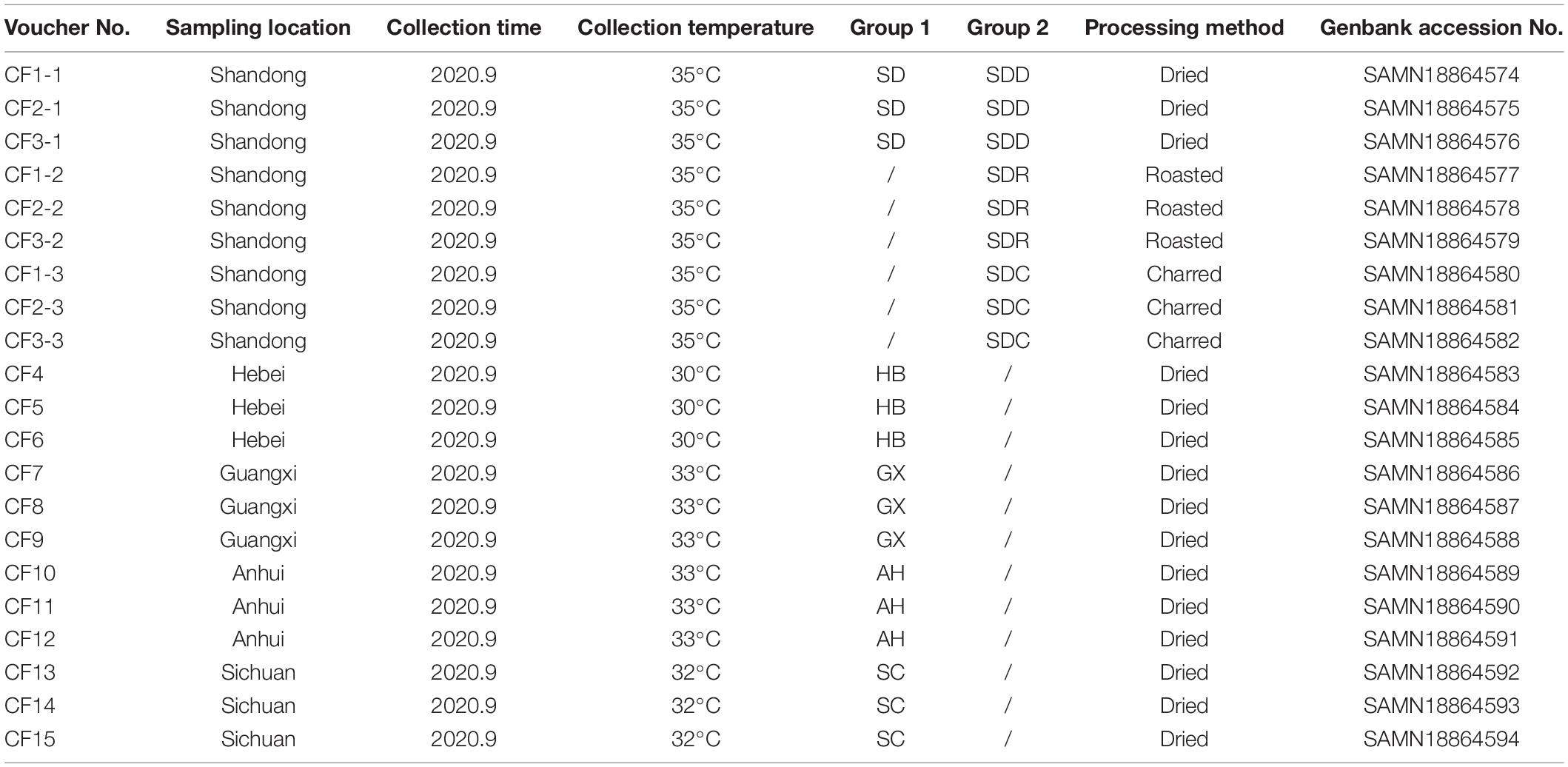
In this study, we collected 21 CF samples from herbal markets in Shandong, Hebei, Anhui, Guangxi, and Sichuan provinces in China (Figure 1). Among the 21 samples, 15 dried samples were divided into five groups, namely, the SD, HB, GX, AH, and SC groups based on collection areas. Meanwhile, nine samples from Shandong province were divided into three groups, namely, the SDD (dried), SDR (roasted), and SDC (charred) groups based on processing methods.
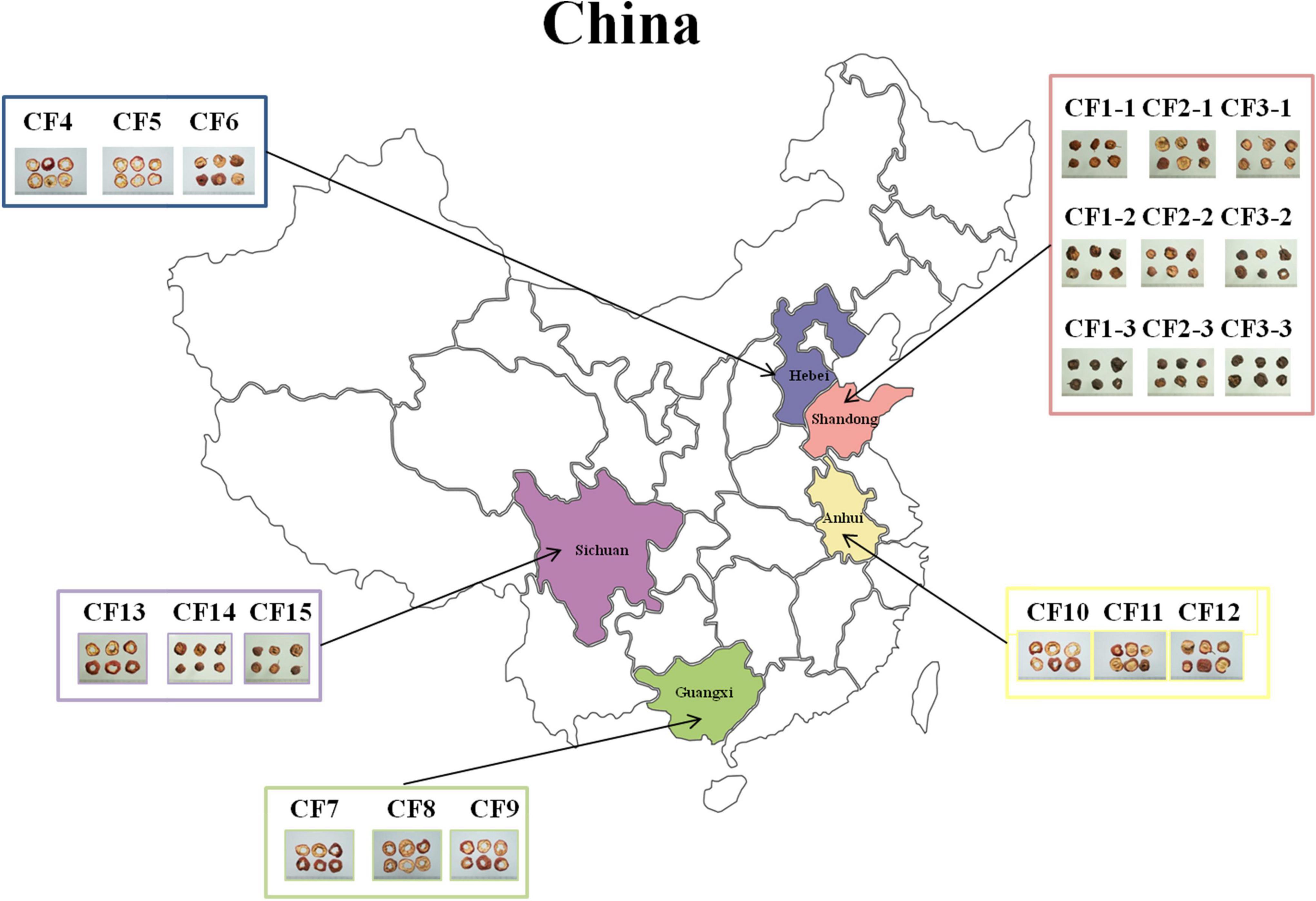
Figure 1. Overview of sampling in this study. A total of 21 Crataegi Fructus (CF) samples were collected from five provinces in China: Shandong (n = 9), Hebei (n = 3), Anhui (n = 3), Guangxi (n = 3), and Sichuan (n = 3) provinces.
The CF samples were processed as described by Chinese Pharmacopoeia (2020). Samples in the SDD group were dried with hot air or under the sun. The CF samples in the SDR group were processed by low-temperature heating (150°C) until they became brown. The CF samples in the SDC group were processed by medium-temperature heating (180°C) until they became brown and black. Each sample was collected with 500 g, and placed into 21 sterile paper bags, respectively. All the samples were assigned with voucher numbers and deposited in the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences (Table 1).
Total DNA Extraction and Polymerase Chain Reaction Amplification
Total community DNA was extracted from 2.7 g CF samples using an EZNA® soil DNA kit (Omega Bio-tek., Inc., Norcross, GA, United States) based on the manufacturer’s protocol. All the CF samples were transferred into a 50-ml sterilized centrifuge tube with 30 ml 1 × phosphate-buffered saline (PBS) (Beijing Solarbio Science and Technology Co., Ltd), and shaken with a cortex mixer for 5 min. In order to remove impurities, the mixture was filtered through double layers of sterile gauze. All filtrates were centrifugated at 12,000 rpm for 28 min to collect fungal strains for total DNA extraction. The DNA was stored at −20°C.
The ITS2 sequence was amplified using the universal primers of ITS3 (5′-GCATCGATGAAGAACGCAGC- 3′) and ITS4 (5′ -TCCTCCGCTTATTGATATGC-3′) (20). The Polymerase Chain Reaction (PCR) conditions were as follows: initial denaturation at 95°C for 3 min, 37 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 20 s, elongation at 72°C for 45 s, and final extension at 72°C for 10 min. The integrity and concentration of PCR products were verified by agarose (2%, W/V) gel electrophoresis.
High-Throughput Sequencing and Bioinformatics Analysis
Purified ITS2 amplicons were sequenced with an Illumina Miseq PE300 platform (Illumina, San Diego, CA, United States). Raw sequences were uploaded to the National Center for Biotechnology Information Sequence Read Archive database with accession numbers SAMN18864574- SAMN18864594.
The quality of demultiplexed reads was checked using Fastp software (v. 0.19.6).1 The reads were truncated with a minimum overlap of 10 bp and a PHRED score of at least 20 over a 50-bp sliding window. The reads were clustered into operational taxonomic units (OTUs, 97% similarity) using UPARSE (version 7.0.1090, http://www.drive5.com/uparse/) against the UNITE database (v. 8.0)2 in Quantitative Insights Into Microbial Ecology (QIIME, V. 1.9.1)3 (21, 22). Then, chimeric sequences were detected and removed by USEARCH (V. 7.0, 7.0).4 To ensure the accuracy of OTU annotation, we verified the taxonomical classification of all the OTUs by manual BLAST search in the International Nucleotide Sequence Database Collaboration. We constructed a rarefaction curve for normalization to even depths across samples in QIIME. All the OTUs were denominated at the kingdom, phylum, class, order, family, genus, and species levels. Alpha diversity indices involving Chao1, Good’s coverage, Simpson, ACE, and Shannon were measured through MOTHUR (v. 1.30.2).5 In order to estimate beta diversity, weighted UniFrac distance was visualized by principal coordinates analysis (PCoA) and non-metric multidimensional scaling (NMDS). Furthermore, the CF samples were hierarchically clustered by UPGMA. Partial least squares discriminant analysis (PLS-DA) was performed to study the differences between groups using the mixOmics package in the R software. We utilized the linear discriminant analysis effect size (LEFSe) algorithm to analyze the differences in fungal composition between various groups. A Circos graph was constructed using Circos software (23). Venn analysis, community barplot analysis, and heat mapping were conducted using the R software (24). Co-occurrence analysis was applied to reveal interactions among fungal communities at the genus level between different groups with the NetworkX package in Python (25).
Results
Diversity of Fungal Community in Crataegi Fructus Samples
A total of 1,525,316 ITS2 sequences were obtained from 21 CF samples after quality filtering. Rarefaction analysis revealed that the data to determine the depth for each sample was sufficient to estimate the fungal microbiome (Supplementary Figure 1). A total of 925 OTUs were obtained from the 21 CF samples. The distribution of OTUs in the 21 CF samples is listed in Supplementary Table 1. Based on Venn analysis (Figure 2), there were 421 shared OTUs between groups based on various collection areas. The numbers of OTUs in groups based on various collection areas are as follows: AH group 509 OTUs, SD group 378 OTUs, SC group 370 OTUs, HB group 255 OTUs, and GX group247 OTUs. Among the groups based on processing methods, 158 OTUs were shared between different products. The number of OTUs in the SDD group (378 OTUs) was the highest, followed by the SDR (222 OTUs) and SDC (95 OTUs) groups. Five alpha diversity indices, Chao1, Good’s coverage, Simpson, ACE, and Shannon, were measured to estimate the richness and diversity of the fungal community (Supplementary Table 2). High indices of Chao1 and ACE represent a large variation among species. Furthermore, the low Simpson and high Shannon indices demonstrate the high diversity of the fungal community in the samples. The result of Good’s coverage in all samples collected from various areas yielded an estimate of over 99.9%, indicating good overall sampling. Compared with those in the other samples, the Chao1 and ACE indices in the CF11 sample were highest, demonstrating the largest variation in it. In contrast, the highest index of Shannon was observed in the CF15 sample. Among the groups based on processing methods, the index of Good’s coverage in nine samples also yielded an estimate of over 99.9%. The Chao1, Shannon, and ACE indices in the CF3-1 sample were the highest, whereas the Chao1 and ACE indices in the CF1-3 sample were the lowest. Hence, the highest and lowest fungal diversity was observed in the CF3-1 and CF1-3 samples, respectively.
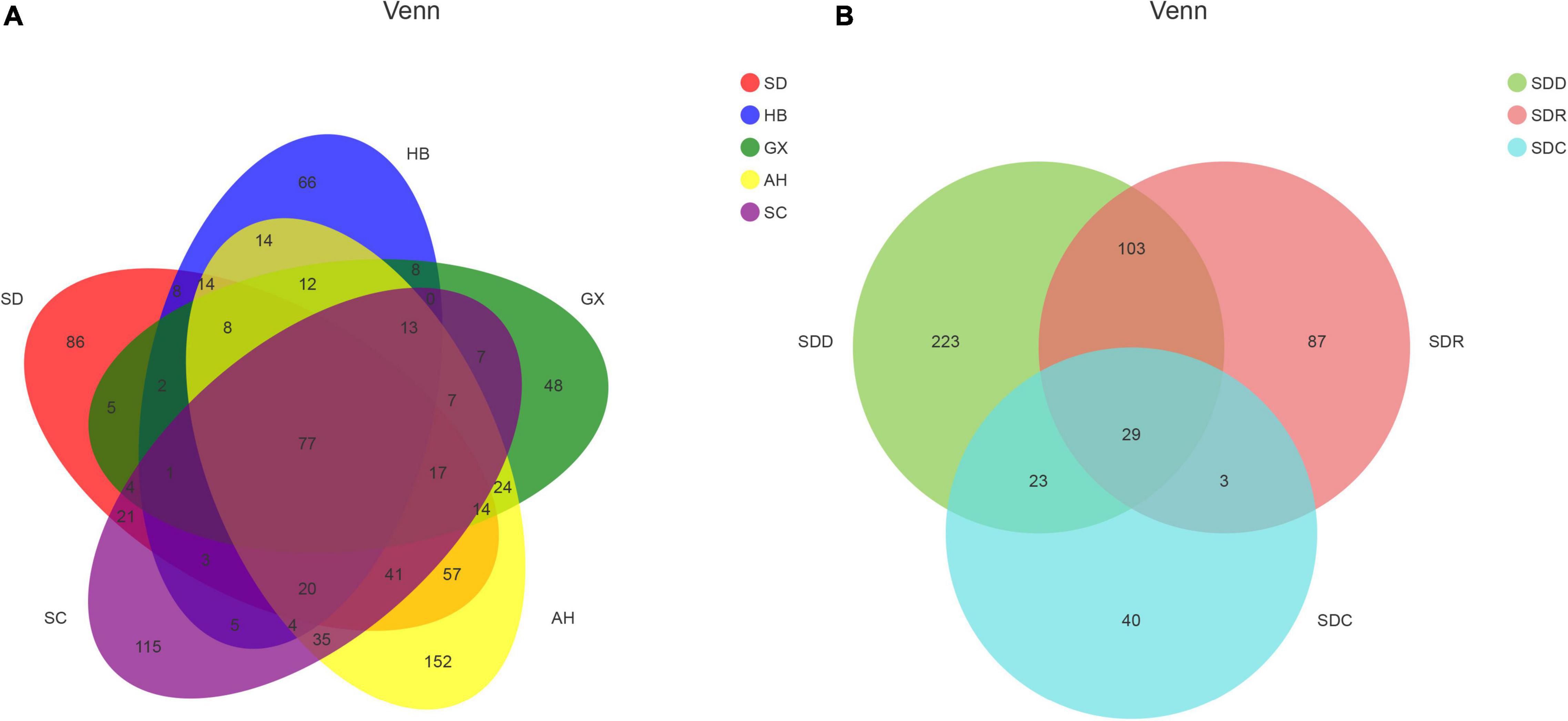
Figure 2. Venn diagram of operational taxonomic units (OTUs) in (A) samples from different collection areas and (B) samples based on processing methods.
Composition of Fungal Community in Crataegi Fructus Samples
All the 925 OTUs taxonomically spanned at least three phyla; some of the OTUs were classified as unclassified fungi or others. Ascomycota was dominant at the phylum level in all the samples that were collected from different areas, with a relative abundance of 64.25–98.09%. The relative abundance of Basidiomycota was 1.45–34.41% (Figure 3A). At the class level, Dothideomycetes (36.70–91.81%) was the most dominant among all the classes (Figure 3B). Furthermore, taxonomical classification at the family level showed that Pleosporaceae (9.35–67.17%) predominated among other families, followed by Didymellaceae (3.34–33.93%) and Cladosporiaceae (5.58–32.46%, Figure 3C). At the genus level, the three most abundant genera were Alternaria, Nothophoma, and Cladosporium, with relative abundances of 9.35–66.92%, 3.35–33.93%, and 5.57–32.46%, respectively (Figure 3D).
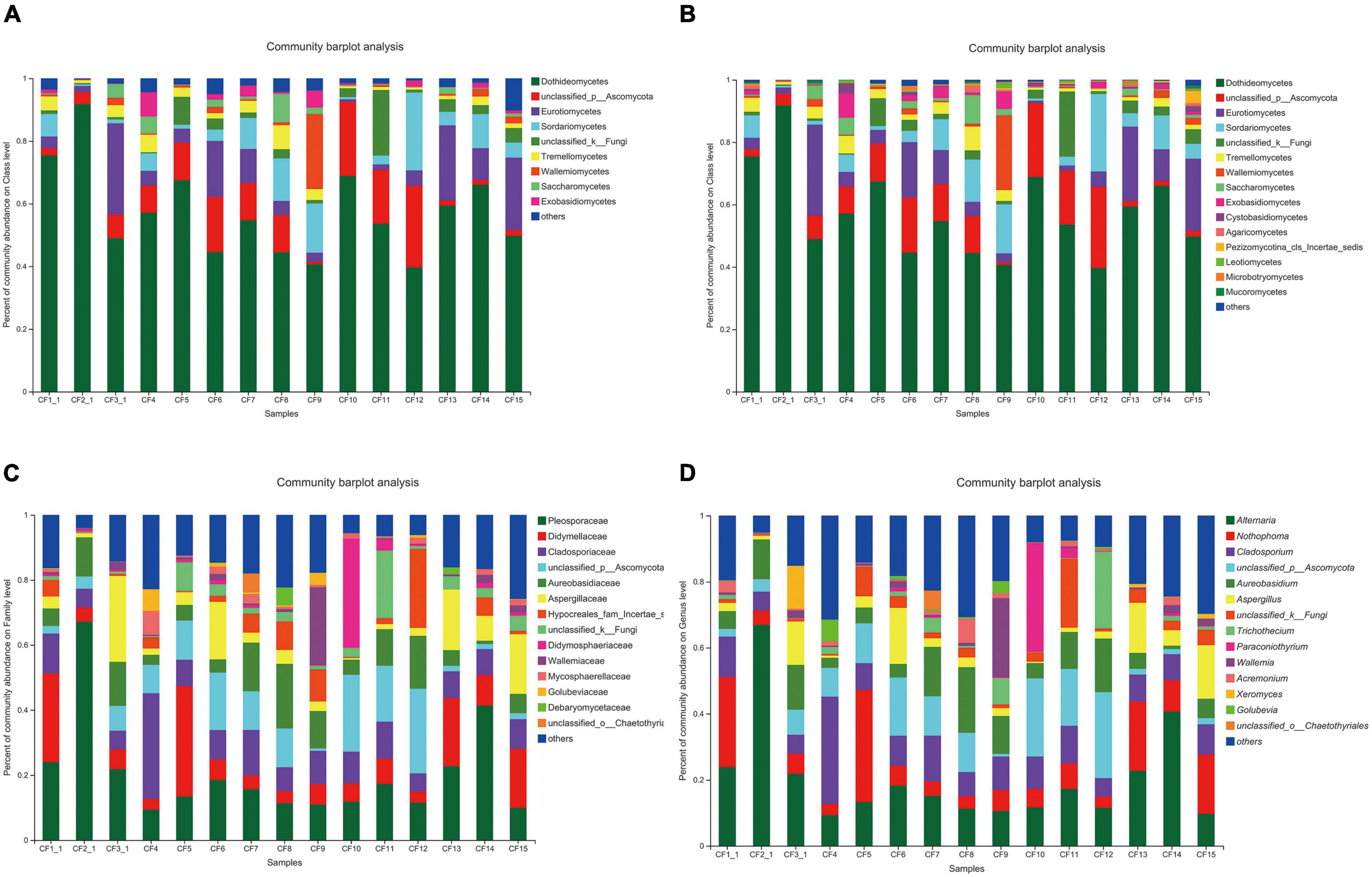
Figure 3. Fungal composition in CF samples from different collection areas at the (A) phylum level, (B) class level, (C) family level, and (D) genus level.
As for the groups based on processing methods, Ascomycota (69.44–98.09%) was also dominant at the phylum level in the SDD and SDR groups. However, the relative abundance of Basidiomycota (10.36–64.18%) was higher than that of Ascomycota (27.35–48.61%) in the SDC group (Figure 4A). Dothideomycetes (0–91.81%) was the most abundant at the class level (Figure 4B). Moreover, the abundances of Aspergillaceae, Pleosporaceae, and Microascaceae were highest at the family level, accounting for 1.29–29.74%, 0–67.17%, and 1.67–82.68% (Figure 4C). At the genus level, the relative abundance of Alternaria (0–66.92%) was highest among the 207 identifiable genera (Figure 4D).
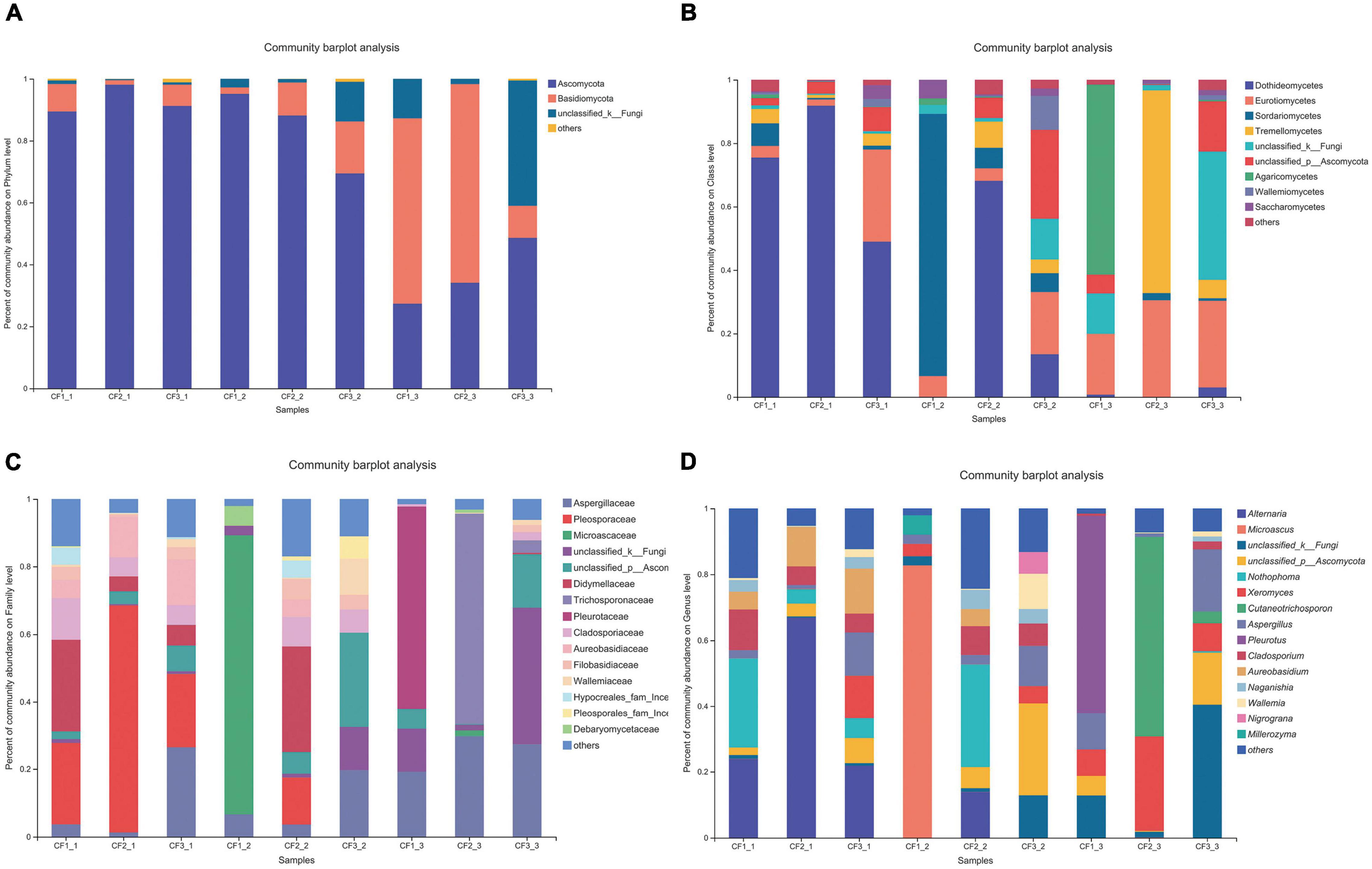
Figure 4. Fungal composition in various processed CF samples at the (A) phylum level, (B) class level, (C) family level, and (D) genus level.
In addition, among all the 925 OTUs, 135 could be identified at the species level by manual BLAST search. Seven potential mycotoxin-producing fungi were detected, T. roseum (detected in CF10, CF2-2, CF11, CF12, CF1-1, CF15, CF2-1, CF3-1, CF14, CF13, CF7, CF5, CF9, CF6, CF4, CF8, CF3-3, CF2-3, CF1-2, and CF1-3), A. tenuissima (detected in CF10, CF2-2, CF11, CF12, CF1-1, CF15, CF3-1, CF14, CF13, CF7, CF5, CF6, CF4, and CF8), A. carbonarius (detected in CF10, CF2-2, CF11, CF12, CF15, CF2-1, CF3-1, CF13, CF7, CF6, CF8, and CF2-3), P. brevicompactum (detected in CF2-2, CF12, CF15, and CF3-1), A. fumigatus (detected in CF11, CF15, and CF6), R. microspores (detected in CF2-2 and CF3-1), and P. fermentans (detected in CF3-1).
Comparison of Fungal Community in Crataegi Fructus From Different Collection Areas
We divided the 15 samples into five groups according to collection areas and compared the differences in fungal diversity and composition between different groups. For alpha diversity, the highest Shannon index was detected in the GX group, representing the greatest diversity of the fungal community in this group. Meanwhile, the highest indices of Chao 1 and Ace were observed in the AH group, indicating that this group had the greatest richnes. For beta diversity, we analyzed the diversity of fungal community by PCoA and NMDS analysis. The PCoA result showed that the SC, AH, and SD groups were significantly distinguishable with the exception of the GX and HB groups (ANOSIM, R = 0.5467, P = 0.001) (Figure 5A). However, differences between the AH and GX groups, and between the SD and HB groups were low based on the NMDS analysis (Figure 5B). Similarly, the PLS-DA result demonstrated that the difference in fungal composition between the SD and HB groups was low (Figure 5C). Furthermore, we compared the differences in fungal communities at various levels between groups (Figure 6A). At the phylum level, the relative abundance of Basidiomycota in the GX group was higher than that in the other groups. At the class level, some unclassified classes belonging to Ascomycota had the highest average percentage of community abundance in the AH group among the five groups. In comparison with the other groups, the average relative abundances of Teratosphaeriaceae, Coniothyriaceae, and some unclassified families belonging to Ascomycota in the AH group were higher. At the genus level, the relative abundance of Xeromyces in the SD group was higher than that in the other groups, while the relative abundance of Sphaerulina in the HB group was highest among all the groups. Meanwhile, the relative abundances of Coniothyrium, Fusicolla, and some unclassified genera belonging to Ascomycota were highest in the AH group. In addition, Figures 7A,B show the distribution and differences of the dominant genera with a relative abundance >5% in groups based on collection areas. The relative abundance of Alternaria, which was a dominant genus in CF, was higher in the SD and SC groups than in the other groups. We conducted a typing analysis at the genus level (Figure 5D), and the result showed that the relative abundance of Alternaria was the main factor that divided the 15 samples into two types, namely, type 1 (AH group, GX group, and CF4) and type 2 (SD group, SC group, CF5, and CF6).
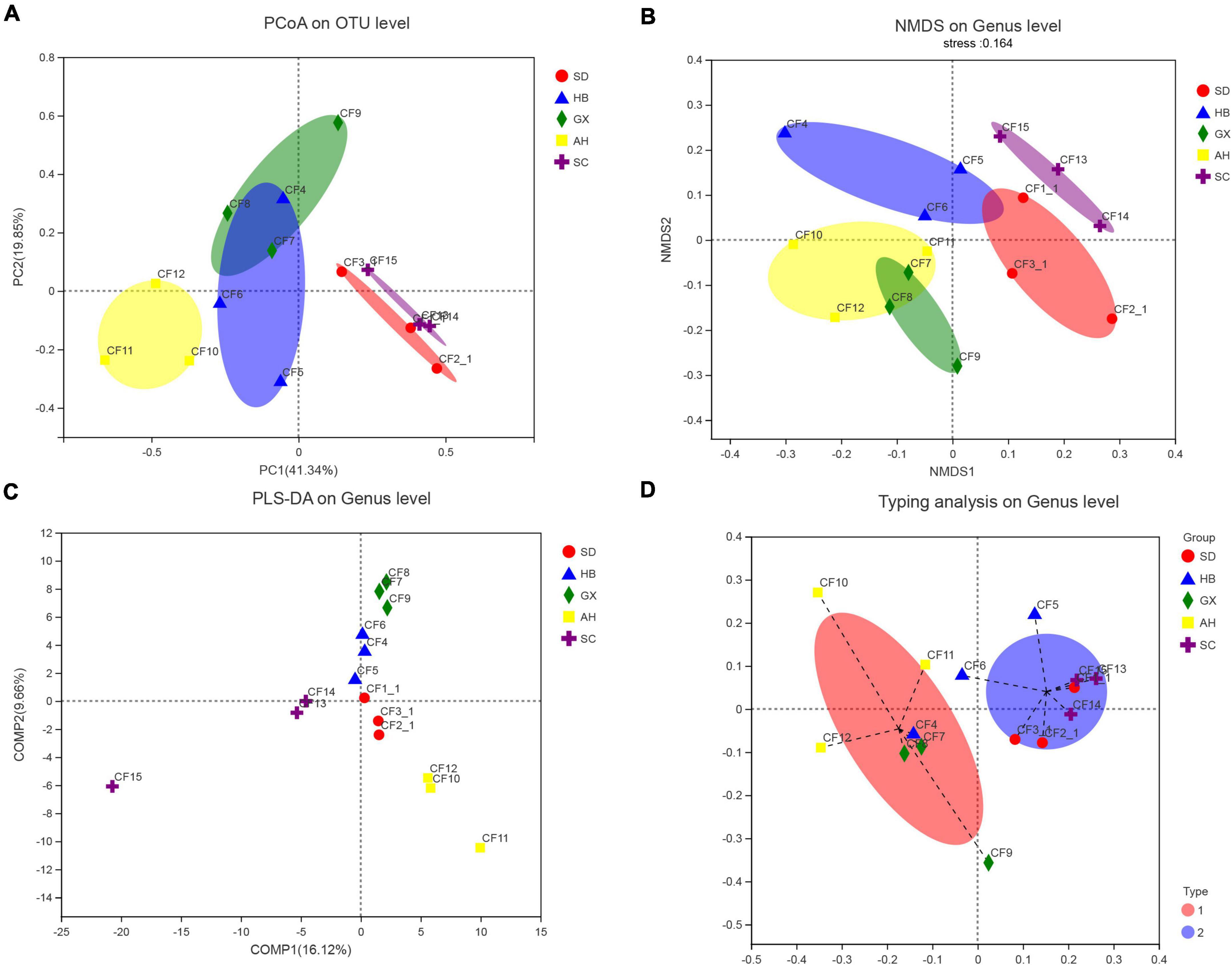
Figure 5. Analysis of the beta diversity of the fungal community in groups based on collection areas. (A) Principal coordinates analysis (PCoA) plot on OTU level, (B) non-metric multidimensional scaling (NMDS) plot on genus level, (C) Partial least squares discriminant analysis (PLS-DA) plot on genus level, and (D) typing analysis on genus level.
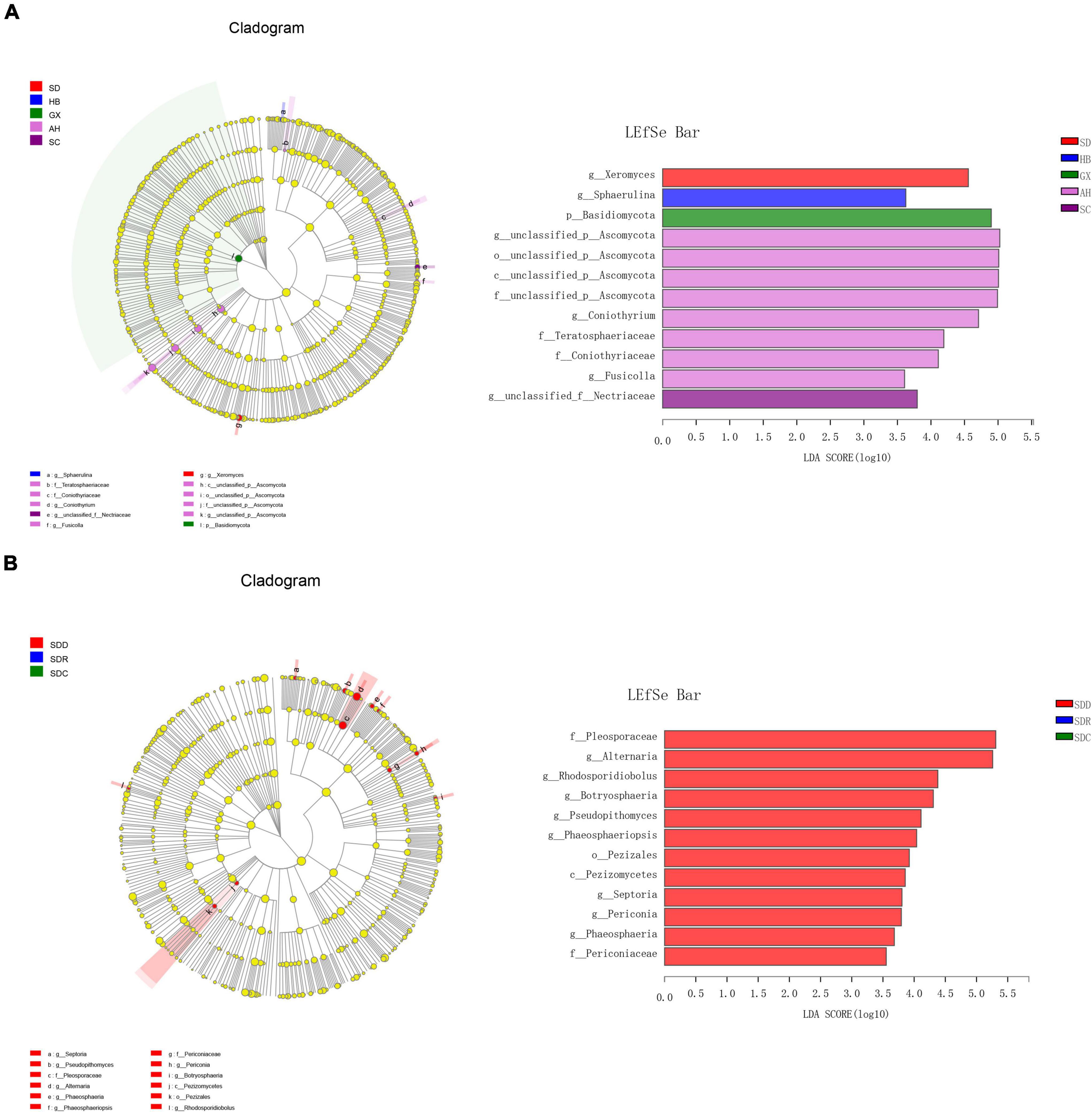
Figure 6. Fungal taxa with different abundances by linear discriminant analysis effect size (LEFSe) analysis in (A) groups based on collection areas and (B) groups based on processing methods.
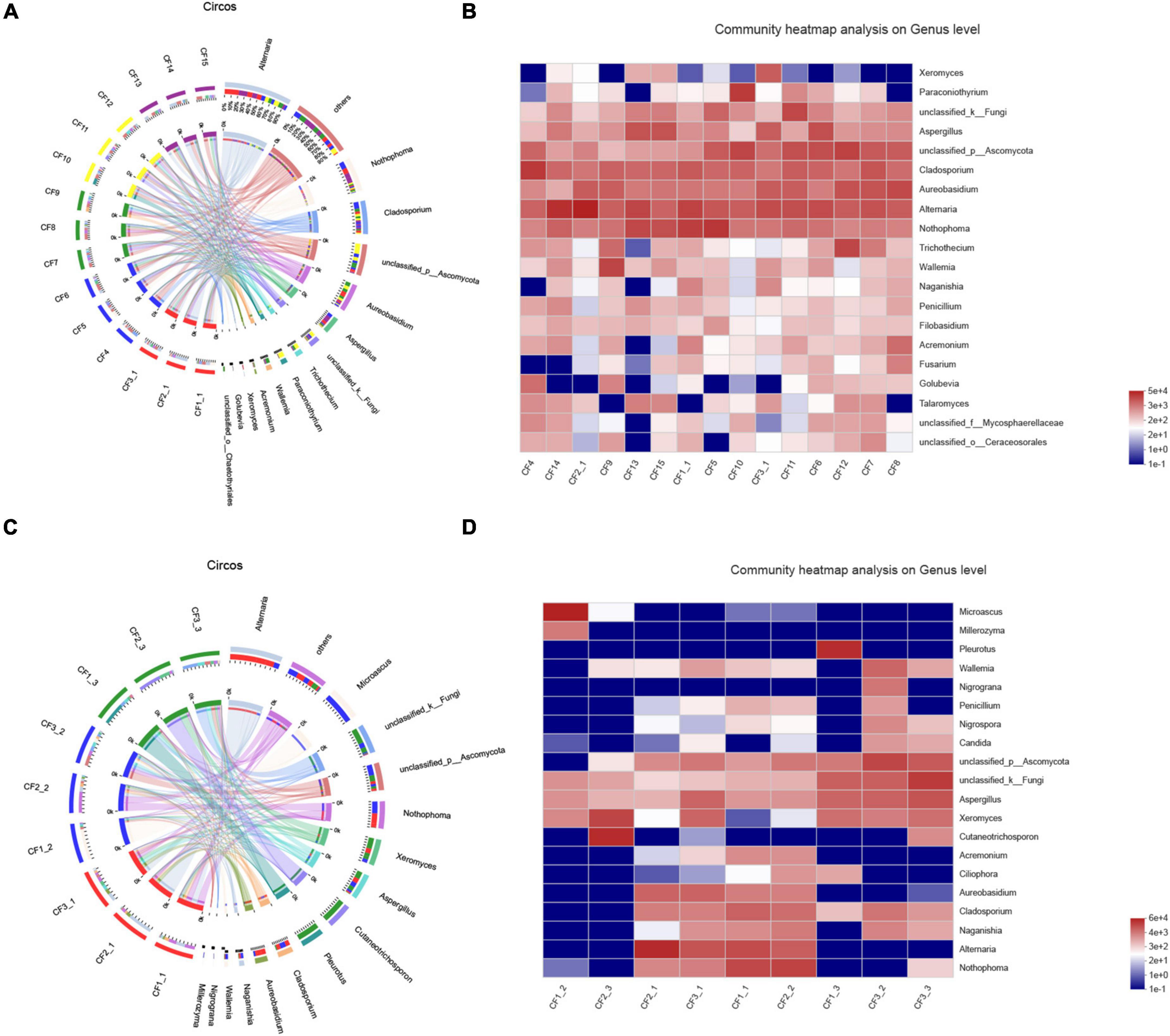
Figure 7. Distribution of fungal community at the genus level in (A) samples from different collection areas visualized by Circos, (B) a heatmap of the top 20 genera in samples from different collection areas, (C) in various processed CF samples, (D) a heatmap of the top 20 genera in samples based on processing methods.
Comparison of Fungal Community in Groups Based on Processing Methods
We collected nine samples and divided them into three groups (SDD, SDR, and SDC) based on processing methods. For alpha diversity, the average indices of Chao 1, Shannon, and ACE were observed to be highest in the SDD group, indicating its high richness and diversity. We also observed that the three groups showed various degrees of clustering based on the result of hierarchical clustering analysis (Figure 8A). Except for the SDR group, the two other groups were clustered into two branches. The results of NMDS analysis and PCoA were also consistent with hierarchical clustering analysis and showed that the SDR samples were assigned to other groups (Figures 8B,C). Based on the PLS-DA result, only the SDC group could be distinguished from the SDD and SDR groups (Figure 8D). With the exception of the SDC group, Ascomycota was the most abundant phylum in the SDD and SDR groups. However, the relative abundance of Basidiomycota was higher than that of Ascomycota in the SDC group. At the class level, the relative abundance of Dothideomycetes was highest in the SDD group, while Eurotiomycetes was the most abundant class in the SDC group. The relative abundance of Pezizales was higher in the SDD group than in the SDR and SDC groups. Based on LEfSe analysis (Figure 6B), the SDD group exhibited higher numbers of Pezizales at the order level and of Pleosporaceae and Periconiaceae at the family level than those in the SDR and SDC group. Meanwhile, Alternaria, Pseudopithomyces, Phaeosphaeriopsis, Septoria, Botryosphareia, and Phaeosphaeria were more common in the SDD group than in the SDR and SDC groups. The fungal composition in various groups showed differences at the genus level. Alternaria was dominant in the SDD group, while the relative abundance of Alternaria was low in the SDC group. On the contrary, the relative abundance of Xeromyces was higher in the SDC group than in the SDD and SDR groups (Figures 7C,D).
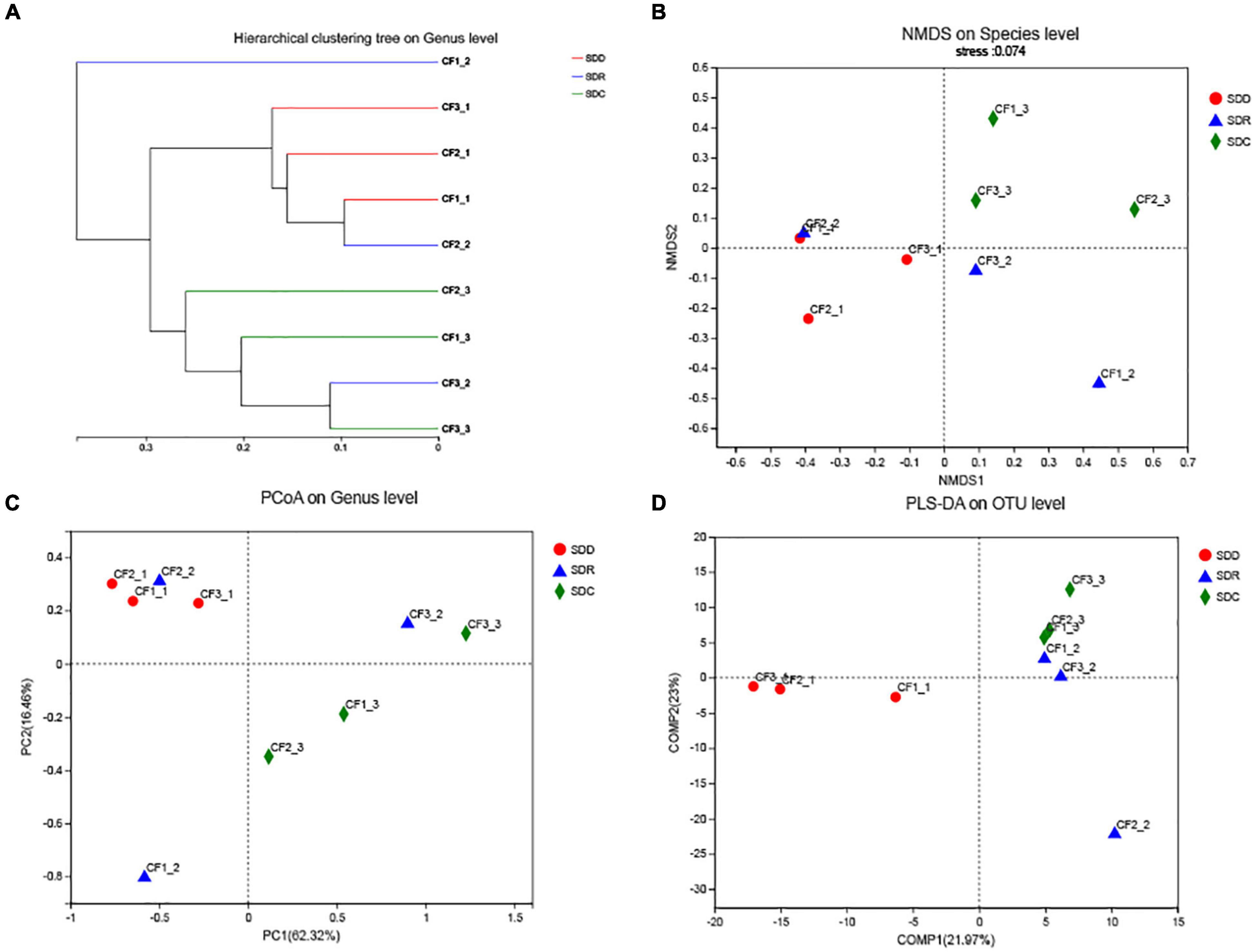
Figure 8. Analysis of the beta diversity of the fungal community in groups based on processing methods. (A) Hierarchical clustering tree on genus level, (B) NMDS plot on species level, (C) PCoA plot on genus level, and (D) PLS-DA plot on OTU level.
Co-occurrence Analysis
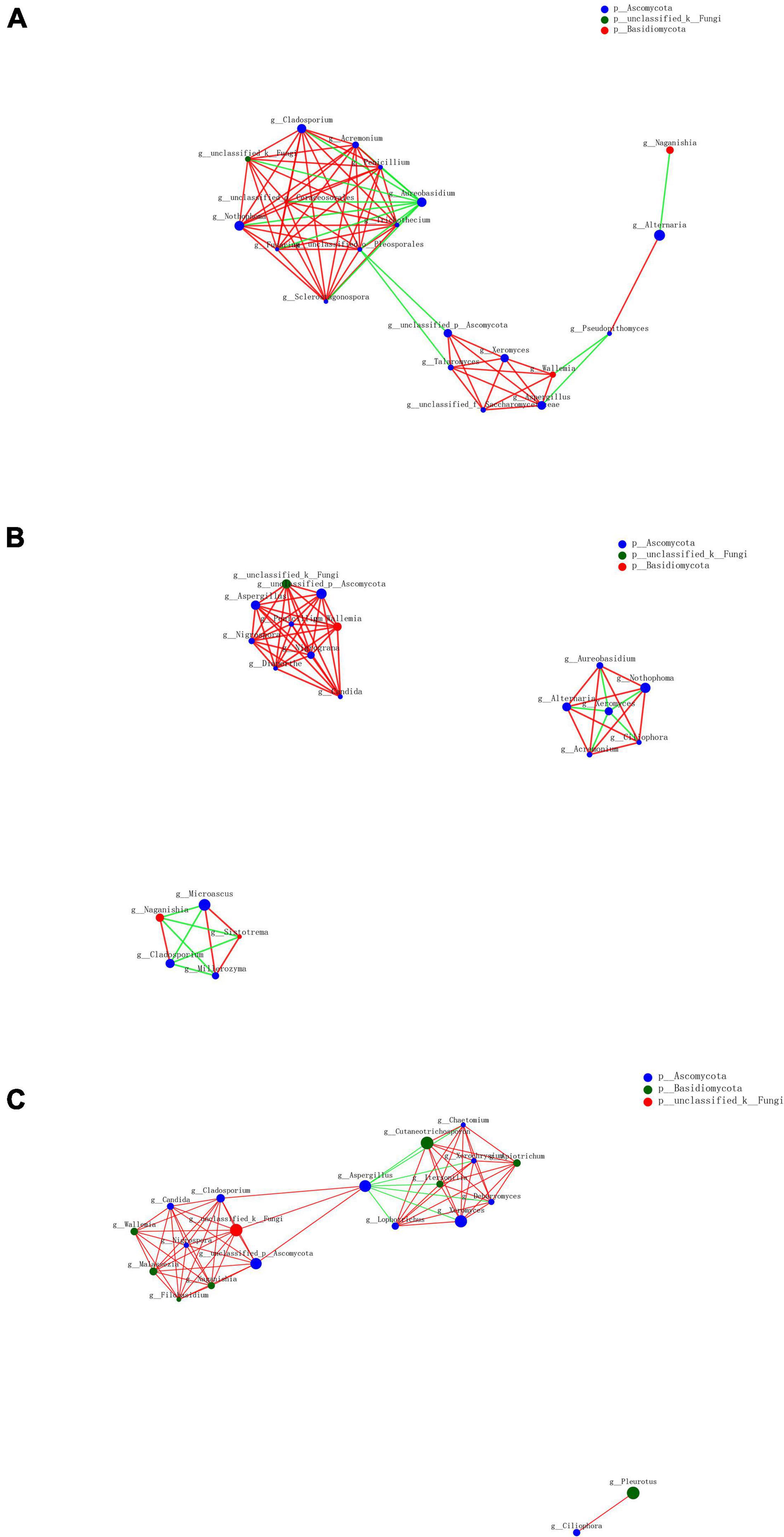
As a result of the diversity of fungal microbiome in different groups based on processing methods, we further analyzed the interactions of fungal genera among various groups. The top 20 genera were selected to reveal the microbiome relationship. The co-occurrence network analysis showed that the difference was distinguishable depending on different processing methods (Figure 9). A total of 55 positive and 14 negative correlations were recorded in the SDD group, while 51 positive and 11 negative correlations were recorded in the SDR group. Meanwhile, the SDC group had 63 positive and 7 negative correlations. The result showed that Alternaria, which was the dominant genus, was positively correlated with Pseudopithomyces but negatively correlated with Naganishia in the SDD group, while Alternaria was negatively correlated with Xeromyces and positively correlated with Aureobasidium, Nothophoma, Ciliophora, and Acremonium in the SDR group. Except for Alternaria, the correlations of Xeromyces differed among the three groups. In the SDD group, Xeromyces showed positive correlations with Wallemia, Aspergillus, Talaromyces, and some unclassified genera belonging to the Saccharomycetaceae family. However, Xeromyces exhibited negative correlations with Aureobasidium, Nothophoma, Ciliophora, Acremonium, and Alternaria. Furthermore, this genus was negatively correlated with Aspergillus and positively correlated with Lophotrichus, Itersonilia, Cutaneotrichosporon, Chaetomium, Xerochrysium, Apiotrichum, and Dearyomyces. In addition, Aspergillus exhibited the most correlations (10 correlations) in the SDC group, followed by the SDR (eight correlations) and SDD (six correlations) groups. For Wallemia, more correlations (eight correlations) were detected in the SDR group than in the SDC (seven correlations) and SDD (five correlations) groups. However, Cladosporium had a higher number of correlations in the SDD and SDC groups (nine correlations) than in the SDR group (four correlations).
Discussion
Fungal Contamination in Crataegi Fructus
Crataegi Fructus is considered a popular dietary supplement that has attracted global attention as a result of its edible and medicinal effects. However, exogenous contaminations, especially fungal contamination, in CF are inevitable. In this study, we detected fungal contamination in 21 CF samples that were collected in China. The result showed that all the samples were contaminated with fungi. At the phylum level, Ascomycota was dominant among most of the samples (except for CF1-3 and CF2-3). It is a common phylum that has high metabolite diversity (26). Meanwhile, this phylum also includes a wide range of plant and animal pathogens such as species from Alternaria, Aspergillus, Penicillium, and Fusarium. Contamination of foods and herbs by Ascomycota has been reported continuously. Zang et al. studied the dynamics of microbial community during fermentation of Suan yu (fermented fish) and found that Ascomycota was the predominant phylum in all stages of fermentation (27). In Saudi Arabia, Hashem and Alamri isolated 520 fungal strains from 15 spices, indicating that most of the species belonged to Ascomycota (28). In addition, it was observed that Alternaria contamination was detected in 16 out of 21 CF samples in this study, showing that Alternaria was the main fungal genus in the CF samples. According to a relevant study, Alternaria is a common and important fungal genus that is widely distributed worldwide. It is divided into 24 sections based on morphological and molecular identification (29). Many species in this genus have been considered as plant and post-harvest pathogens such as Alternaria bataticola, Alterneria porri, and Alterneria solani (30, 31). A previous study has reported that contamination by Alternaria species caused damage in Crataegus sp. tree leaves (32), while few studies focused on the contamination by Alternaria in CF fruits. Our result indicated that Alternaria was the most abundant genus among all the genera that contaminated CF. Similarly, Alternaria has also been reported as the main contaminated fungal genus in other herbs. The study performed by Pickova et al. showed that the quality of Milk Thistle, which is an herb used for treatment of liver disease, was affected by fungi in Alternaria and their mycotoxins (33). Zhao et al. also indicated that fungal strains in Alternaria severely threatened the industry of Dendrobium officinale (34). Furthermore, several Alternaria species were not only considered as spoilage agents for herbs but were also capable of synthesizing some mycotoxins such as alternariol, alternariol monomethyl ether, and altenuene, causing damage to human health (35). Thus, in order to identify the fungi in Alternaria efficiently and accurately, an identification method deserves to be developed. HTS provides an efficient method for the efficient identification of fungi that contaminate CF, guaranteeing safe utilization of CF.
Effect of Processing Method on the Fungal Community in Herb and Food
In order to improve efficacy and decrease toxicity, many herbs are processed before being distributed to the market. However, the processing methods also affect the fungal contamination in herbs. According to the Chinese Pharmacopoeia (2020), CF can be processed into dried, roasted, and charred products, and their efficacy shows degrees of differences in clinics. Moreover, few studies reported a comparison of the fungal microbiome in these products. In this study, we compared the difference in fungal composition and diversity between processed CF products. The result showed differences in the fungal community in the three groups. At the phylum level, Ascomycota was dominant in the SDD and SDR groups, while the relative abundance of Basidiomycota was highest in the SDC group. Furthermore, remarkable differences were observed at the genus level. According to the fungal composition analysis, the relative abundances of Aspergillus and Xeromyces were highest in the SDC group, followed by the SDR and SDD groups, and the relative abundances of Alternaria and Aureobasidium were highest in the SDD group, followed by the SDR and SDC groups. It can be concluded that the fungal community in the CF samples changes as the effect of processing temperature increases. Some fungal genera such as Aspergillus and Xeromyces become dominant from low abundance, while others such as Alternaria and Aureobasidium decrease significantly or are not even detected in the samples. During the CF processing procedure, temperature is an important factor that may affect the fungal community. Based on previous studies, temperature has a significant impact on fungal growth and mycotoxin production. Mellon et al. assessed the effect of temperature on the growth and aflatoxin production of A. flavus and A. parasiticus. The results showed that high temperature exhibited more inhibitory effect on the growth and aflatoxin production of these fungi than low temperature (36). Similarly, Cibelli et al. observed that the growth of A. alternata was remarkably inhibited under high-temperature conditions (37). Moreover, many studies have reported the impact of other processing methods on the fungal community. Cao et al. analyzed the variation of microbial community during the fermentation of Huafeng Dan Yaomu and found that the dominant fungal genera changed from Millerozyma and Saccharomycopsis to Pichia after 14 days of fermentation (38). In Nigeria, Omohimi et al. collected yam samples (including raw samples, chips, flakes, and flour) to compare the difference in fungal contamination. The result showed that the frequency of some strains in Aspergillus and Fusarium in processed samples was higher than that in dried samples (39). In addition to herbs, many foods will be processed to improve their taste or extend shelf life, such as hams. Mu et al. observed that the fungal community in Panxian Ham changed during the processing procedure (selection, salting, resting, and drying-ripening) (40), and this result was similar to the studies performed by Zhang et al. (41). In conclusion, the processing procedure influences the fungal community in herbs and foods significantly. Thus, it is important to study the relationship between processing methods and the fungal community, providing references for industrial processors to construct standard quality parameters.
Molecular Identification of Toxigenic and Non-toxigenic Strains
In this study, seven potential mycotoxin-producing fungi were detected in the CF samples, including T. roseum, A. tenuissima, A. carbonarius, P. brevicompactum, A. fumigatus, R. microspores, and P. fermentans. Previous studies reported that all these fungi were capable of producing mycotoxins, such as zearalenone, T-2 toxin, and deoxynivalenol (42–47). However, these fungi exhibit mycotoxin-producing ability inconsistently and are often non-toxigenic under some conditions. Both external and internal factors may influence the synthesis of mycotoxin (48–50). Therefore, the molecular identification of toxigenic and non-toxigenic fungi has attracted the attention of researchers. Kim et al. applied multiplex PCR to detect aflatoxigenic and non-aflatoxigenic fungi by designing two primer sets, primer set I (omtB/alfR/omtA) and primer set II (alfR/ver-1/omtA), indicating that only aflatoxigenic Aspergillus species could show three patterns in both primer sets (51). Similarly, Davari et al. characterized the aflatoxigenic A. flavus and Aspergillus parasiticus strains successfully using four sets of primers (specifically for nor1, ver1, omtA, and aflR) (52). Norlia et al. observed that all aflatoxin biosynthesis genes (aflR, aflP, aflD, aflM, and pksA) were amplified in aflatoxigenic A. flavus, and that at least one of these genes could not be amplified in non-aflatoxigenic A. flavus (53). Thus, a molecular method for efficient and accurate detection of toxigenic fungi should be developed to provide early warning and ensure the microbiological safety of herbs.
Conclusion
In this study, fungal contamination in CF was investigated using ITS2 amplicon sequencing. All the samples were detected to be positive for fungal contamination. A total of 115 fungal species were identified, among which seven were potential mycotoxin-producing fungi. Moreover, HTS has been demonstrated as a feasible method for efficient detection of fungal contamination in CF, serving as a consolidate reference for its safe application and quality improvement.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, SAMN18864574-SAMN18864594.
Author Contributions
XP: conceptualization, funding acquisition, resources, and validation. JY and MG: data curation. JY, MG, WJ, YD, and XP: formal analysis. JY and XP: methodology, writing (original draft), and writing (review and editing). All authors contributed to the article and approved the submitted version.
Funding
This study was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2021-I2M-1-071).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.883698/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves for operational taxonomic unit (OTU) in each Crataegi Fructus (CF) sample.
Supplementary Table 1 | Distribution of operational taxonomic units (OTUs) for 21 Crataegi Fructus (CF) samples.
Supplementary Table 2 | Alpha diversity indices in each CF sample.
Footnotes
- ^ https://github.com/OpenGene/fastp
- ^ https://unite.ut.ee/
- ^ http://qiime.org/install/index.html
- ^ http://www.drive5.com/usearch/
- ^ https://www.mothur.org/wiki/Download_mothur
References
2. European Directorate for the Quality of Medicines, Council of Europe. European Pharmacopaeia. 9th ed. Strasburg Cedex: EDQM (2017).
3. People’s Medical Publishing House.Zhong Yao Zhi. 2nd edn. (Vol. 3). Beijing: People’s Medical Publishing House (1984).
4. Press of Chemical Industry.Chinese Pharmacopeia Commission. Pharmacopoeia of People’s Republic of China (Part 1). Beijing: Press of Chemical Industry (2020). p. 33.
5. Pittler MH, Guo R, Ernst E. Hawthorn extract for treating chronic heart failure. Cochrane Db Syst Rev. (2008) 1:CD005312. doi: 10.1002/14651858.CD005312
6. Wang Y, Lv M, Wang T, Sun J, Wang Y, Xia M, et al. Research on mechanism of charred hawthorn on digestive through modulating “brain-gut” axis and gut flora. J Ethnopharmacol. (2019) 245:112166. doi: 10.1016/j.jep.2019.112166
7. Li F, Zhao S, Chin L, Li Y. Determination of patulin in apple and hawthorn beverages by solid-phase filtration column and liquid chromatography. J AOAC Int. (2007) 90:167–117. doi: 10.1093/jaoac/90.1.167
8. Zhou Y, Kong W, Li Y, Logrieco AF, Xu J, Yang M. A new solid-phase extraction and HPLC method for determination of patulin in apple products and hawthorn juice in China. J Sep Sci. (2012) 35:641–9. doi: 10.1002/jssc.201100919
9. Shen M, Jia-Li W, Hai-Pei S, Hui Y, Pei-Dong C, Wei-Feng Y, et al. Alternative processing technology for the preparation of carbonized Zingiberis Rhizoma by stir-frying with sand. Pharm Biol. (2020) 58:131–7. doi: 10.1080/13880209.2019.1711431
10. Liu S, Li F, Li Y, Li W, Xu J, Du H. A review of traditional and current methods used to potentially reduce toxicity of Aconitum roots in traditional Chinese medicine. J Ethnopharmacol. (2017) 207:237–50. doi: 10.1016/j.jep.2017.06.038
11. He L, Bai L, Shan L, Yu P, Zhang L, Dou X, et al. Variations in fungal microbiota and aflatoxin contamination during the processing of yuanzhi, a traditional chinese medicine. Ind Crops Prod. (2020) 152:112509. doi: 10.1016/j.indcrop.2020.112509
12. Odongo GA, Schlotz N, Baldermann S, Papadopoulou-Mourkidou E, Karaoglanidis GS. African Nightshade (Solanum scabrum Mill.): impact of cultivation and plant processing on its health promoting potential as determined in a human liver cell model. Nutrients. (2018) 10:E1532. doi: 10.3390/nu10101532
13. Guo M, Jiang W, Yang M, Dou X, Pang X. Characterizing fungal communities in medicinal and edible Cassiae Semen using high-throughput sequencing. Int J Food Microbiol. (2019) 16:108496. doi: 10.1016/j.ijfoodmicro.2019.108496
14. Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, García-Cobos S, et al. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol. (2017) 243:16–24. doi: 10.1016/j.jbiotec.2016.12.022
15. Lee S, An C, Xu S, Lee S, Yamamoto N. High-throughput sequencing reveals unprecedented diversities of Aspergillus species in outdoor air. Lett Appl Microbiol. (2016) 63:165–71. doi: 10.1111/lam.12608
16. Quijada NM, Hernández M, Rodríguez-Lázaro D. High-throughput sequencing and food microbiology. Adv Food Nutr Res. (2019) 91:275–300. doi: 10.1016/bs.afnr.2019.10.003
17. Yang F, Yang D, Liu S, Xu S, Wang F, Chen H, et al. Use of high-throughput sequencing to identify fungal communities on the surface of Citri Reticulatae Pericarpium during the 3-year aging process. Curr Microbiol. (2021) 78:3142–51. doi: 10.1007/s00284-021-02575-w
18. Hartmann P, Lang S, Zeng S, Duan Y, Zhang X, Wang Y, et al. Dynamic changes of the fungal microbiome in alcohol use disorder. Front Physiol. (2021) 12:699253. doi: 10.3389/fphys.2021.699253
19. Gschwend F, Hartmann M, Hug AS, Enkerli J, Gubler A, Frey B, et al. Long-term stability of soil bacterial and fungal community structures revealed in their abundant and rare fractions. Mol Ecol. (2021) 30:4305–20. doi: 10.1111/mec.16036
20. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: <snm>Innis</snm> <gnm>MA</gnm>, <snm>Gelfand</snm> <gnm>DH</gnm>, <snm>Sninsky</snm> <gnm>JJ</gnm>, <snm>White</snm> <gnm>TJ</gnm> editors. PCR Protocols a Guide to Methods and Applications. San Diego, CA: Academic Press (1990). p. 315–22. doi: 10.1016/b978-0-12-372180-8.50042-1
21. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. (2010) 26:2460–1. doi: 10.1093/bioinformatics/btq461
22. Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. (2013) 10:996–8. doi: 10.1038/nmeth.2604
23. Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. (2009) 19:1639–45. doi: 10.1101/gr.092759.109
24. Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. (2013) 7:1069–79. doi: 10.1038/ismej.2013.2
25. Hagberg AA, Schult DA, Swart PJ. Exploring network structure, dynamics, and function using NetworkX. In: <snm>Varoquaux</snm> <gnm>G</gnm>, <snm>Vaught</snm> <gnm>T</gnm>, <snm>Millman</snm> <gnm>J</gnm> editors. Proceedings of the 7th Python in Science Conference. Pasadena, CA: BibSonomy (2008). p. 11–5.
26. Hagee D, Abu Hardan A, Botero J, Arnone JT. Genomic clustering within functionally related gene families in Ascomycota fungi. Comput Struct Biotechnol J. (2020) 18:3267–77. doi: 10.1016/j.csbj.2020.10.020
27. Zang J, Xu Y, Xia W, Yu D, Gao P, Jiang Q, et al. Dynamics and diversity of microbial community succession during fermentation of Suan yu, a Chinese traditional fermented fish, determined by high throughput sequencing. Food Res Int. (2018) 111:565–73. doi: 10.1016/j.foodres.2018.05.076
28. Hashem M, Alamri S. Contamination of common spices in Saudi Arabia markets with potential mycotoxin-producing fungi. Saudi J Biol Sci. (2010) 17:167–75. doi: 10.1016/j.sjbs.2010.02.011
29. Woudenberg JH, Truter M, Groenewald JZ, Crous PW. Large-spored Alternaria pathogens in section Porri disentangled. Stud Mycol. (2014) 79:1–47. doi: 10.1016/j.simyco.2014.07.003
30. Ding S, Meinholz K, Gevens AJ. Spatiotemporal distribution of potato-associated alternaria species in wisconsin. Plant Dis. (2021) 105:149–55. doi: 10.1094/PDIS-11-19-2290-RE
31. Hay FS, Sharma S, Hoepting C, Strickland D, Luong K, Pethybridge SJ. Emergence of stemphylium leaf blight of onion in New York associated with fungicide tesistance. Plant Dis. (2019) 103:3083–92. doi: 10.1094/PDIS-03-19-0676-RE
32. Salazar-Cerezo S, Meneses-Sánchez MC, Martínez-Contreras RD, Martínez-Montiel N. Unraveling the fungal community associated with leaf spot on Crataegus sp. Microorganisms. (2020) 8:459. doi: 10.3390/microorganisms8030459
33. Pickova D, Ostry V, Toman J, Malir F. Presence of mycotoxins in milk thistle (Silybum marianum) food supplements: a review. Toxins. (2020) 12:782. doi: 10.3390/toxins12120782
34. Zhao W, Sun C, Wei L, Chen W, Wang B, Li F, et al. Detection and fitness of dicarboximide-resistant isolates of Alternaria alternata from Dendrobium officinale, a Chinese indigenous medicinal herb. Plant Dis. (2020) 105:2222–30. doi: 10.1094/PDIS-06-20-1246-RE
35. Pinto VE, Patriarca A. Alternaria species and their associated mycotoxins. Methods Mol Biol. (2017) 1542:13–32. doi: 10.1007/978-1-4939-6707-0_2
36. Mellon JE, Dowd MK, Beltz SB. Effects of temperature and medium composition on inhibitory activities of gossypol-related compounds against aflatoxigenic fungi. J Appl Microbiol. (2013) 115:179–86. doi: 10.1111/jam.12219
37. Cibelli F, Bevilacqua A, Raimondo ML, Campaniello D, Carlucci A, Ciccarone C, et al. Evaluation of fungal gowth on Olive-Mill wastewaters treated at high temperature and by high-pressure homogenization. Front Microbiol. (2017) 8:2515. doi: 10.3389/fmicb.2017.02515
38. Cao G, Ma F, Xu J, Zhang Y. Microbial community succession and toxic alkaloids change during fermentation of Huafeng Dan Yaomu. Lett Appl Microbiol. (2020) 70:318–25. doi: 10.1111/lam.13276
39. Omohimi C, Piccirillo C, Ferraro V, Roriz MC, Omemu MA, Santos SMD, et al. Safety of Yam-derived (Dioscorea rotundata) foodstuffs-chips, flakes and flour: effect of processing and post-processing conditions. Foods (2019) 8:12. doi: 10.3390/foods8010012
40. Mu Y, Su W, Mu Y, Jiang L. Combined application of high-throughput sequencing and metabolomics reveals metabolically active microorganisms during Panxian ham processing. Front Microbiol. (2020) 10:3012. doi: 10.3389/fmicb.2019.03012
41. Zhang XM, Dang XJ, Wang YB, Sun T, Wang Y, Yu H, et al. Diversity and composition of microbiota during fermentation of traditional Nuodeng ham. J Microbiol. (2021) 59:20–8. doi: 10.1007/s12275-021-0219-4
42. Böswald C, Engelhardt G, Vogel H, Wallnöfer PR. Metabolism of the Fusarium mycotoxins zearalenone and deoxynivalenol by yeast strains of technological relevance. Nat Toxins. (1995) 3:138–44. doi: 10.1002/nt.2620030304
43. Costa CLA, Cerqueira MBR, Garda-Buffon J. Kresoxim-methyl and famoxadone as activators of toxigenic potential of Aspergillus carbonarius. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. (2019) 36:1860–70. doi: 10.1080/19440049.2019.1670869
44. Guruceaga X, Perez-Cuesta U, Abad-Diaz de Cerio A. Fumagillin, a mycotoxin of Aspergillus fumigatus: biosynthesis, biological activities, detection, and applications. Toxins. (2019) 12:7. doi: 10.3390/toxins12010007
45. Jennessen J, Nielsen KF, Houbraken J. Secondary metabolite and mycotoxin production by the Rhizopus microsporus group. J Agric Food Chem. (2005) 53:1833–40. doi: 10.1021/jf048147n
46. Ntasiou P, Myresiotis C, Konstantinou S, Papadopoulou-Mourkidou E, Karaoglanidis GS. Identification, characterization and mycotoxigenic ability of Alternaria spp. causing core rot of apple fruit in Greece. Int J Food Microbiol. (2015) 197:22–9. doi: 10.1016/j.ijfoodmicro.2014.12.008
47. Tang Y, Xue H, Bi Y, Li Y, Wang Y, Zhao Y, et al. A method of analysis for T-2 toxin and neosolaniol by UPLC-MS/MS in apple fruit inoculated with Trichothecium roseum. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. (2015) 32:480–7. doi: 10.1080/19440049.2014.968884
48. Amaike S, Keller NP. Aspergillus flavus. Annu Rev Phytopathol. (2011) 49:107–33. doi: 10.1146/annurev-phyto-072910-095221
49. Camardo Leggieri M, Decontardi S, Bertuzzi T, Pietri A, Battilani P. Modeling growth and toxin production of toxigenic fungi signaled in cheese under different temperature and water activity regimes. Toxins. (2016) 9:9010004. doi: 10.3390/toxins9010004
50. Casquete R, Benito MJ, Córdoba MG. The growth and aflatoxin production of Aspergillus flavus strains on a cheese model system are influenced by physicochemical factors. J Dairy Sci. (2017) 100:6987–96. doi: 10.3168/jds.2017-12865
51. Kim DM, Chung SH, Chun HS. Multiplex PCR assay for the detection of aflatoxigenic and non-aflatoxigenic fungi in meju, a Korean fermented soybean food starter. Food Microbiol. (2011) 28:1402–8. doi: 10.1016/j.fm.2011.06.017
52. Davari E, Mohsenzadeh M, Mohammadi G, Rezaeian-Doloei R. Characterization of aflatoxigenic Aspergillus flavus and A. parasiticus strain isolates from animal feedstuffs in northeastern Iran. Iran J Vet Res. (2015) 16:150–5.
Keywords: Crataegi Fructus, high-throughput sequencing, ITS2, fungal contamination, collection areas, processing methods
Citation: Yu J, Guo M, Jiang W, Dao Y and Pang X (2022) Illumina-Based Analysis Yields New Insights Into the Fungal Contamination Associated With the Processed Products of Crataegi Fructus. Front. Nutr. 9:883698. doi: 10.3389/fnut.2022.883698
Received: 25 February 2022; Accepted: 13 April 2022;
Published: 12 May 2022.
Edited by:
Sara Panseri, University of Milan, ItalyReviewed by:
Pradeep Kumar, North Eastern Regional Institute of Science and Technology, IndiaFuguo Xing, Institute of Food Science and Technology (CAAS), China
Copyright © 2022 Yu, Guo, Jiang, Dao and Pang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohui Pang, xhpang@implad.ac.cn
 Jingsheng Yu
Jingsheng Yu Mengyue Guo
Mengyue Guo Wenjun Jiang
Wenjun Jiang  Xiaohui Pang
Xiaohui Pang