- 1Central Laboratory, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, China
- 2Department of Oral Implantology, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, China
Preclinical animal models of oral squamous cell carcinoma (OSCC) have been extensively studied in recent years. Investigating the pathogenesis and potential therapeutic strategies of OSCC is required to further progress in this field, and a suitable research animal model that reflects the intricacies of cancer biology is crucial. Of the animal models established for the study of cancers, mouse tumor-bearing models are among the most popular and widely deployed for their high fertility, low cost, and molecular and physiological similarity to humans, as well as the ease of rearing experimental mice. Currently, the different methods of establishing OSCC mouse models can be divided into three categories: chemical carcinogen-induced, transplanted and genetically engineered mouse models. Each of these methods has unique advantages and limitations, and the appropriate application of these techniques in OSCC research deserves our attention. Therefore, this review comprehensively investigates and summarizes the tumorigenesis mechanisms, characteristics, establishment methods, and current applications of OSCC mouse models in published papers. The objective of this review is to provide foundations and considerations for choosing suitable model establishment methods to study the relevant pathogenesis, early diagnosis, and clinical treatment of OSCC.
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common human malignancies and endangers human health, and it accounts for 40% of all head and neck squamous cell carcinoma (HNSCC) cases (1, 2). Although cancer treatments have developed rapidly, the 5-year survival rate for OSCC is still only remained at 50% over the last few decades (3–5). Given the high incidence and poor prognosis of OSCC, an increasing number of researchers are conducting in-depth investigations into the pathogenesis and potential therapeutic targets of OSCC (6–10).
Researches on OSCC can be performed both in vitro and in vivo. Although experiments in vitro have the advantages of relative simplicity, species specificity, convenience, and automation, the extrapolation of in vitro results to predict the behavior of tumors in intact organisms is challenging (11). By contrast, experiments in vivo using animal models are representative of whole organisms and the use of animal models can avoid issues related to safety, ethics, and extended research cycles that arise in human experiments. Moreover, animal models can accurately reflect the tumor microenvironment—which includes a variety of cytokines, infiltrating immune cells, tumor stroma, and blood vessels—and significant tumor biological behaviors, such as invasion and metastasis. Therefore, a suitable animal model is a prerequisite for clarifying the initiation and progression of OSCC.
Previous researches have revealed that several kinds of animals can be used to establish OSCC models, including hamsters, rats, mice, dogs, and cats (12–17). As early as 1954, squamous cell carcinoma (SCC) of the cheek was induced in hamsters by 9,10-dimethyl-1,2-benzanthracene (DMBA), and hamster SCC exhibits many similarities to human OSCC, including the morphology, histology, infiltration and metastasis, expression of biomarkers, and genetic and epigenetic alterations (18–24). Nevertheless, this model also has its shortcomings, for example, the humans lack cheek pouches, and hamster cheek pouches have inadequate lymphatic drainage (25). Moreover, non-murine models have also been used for the research of OSCC etiology, treatment, and tumor–immune system interactions (17). For example, the metastasis and bone invasion of OSCC has been studied in cats, which can be used to mimic highly malignant OSCC (16, 26, 27), and OSCC has also been researched using dogs for identifying risk factors associated with survival in dogs with non-tonsillar OSCC (15). However, there are fewer reagents available to study dogs and cats, and species-specific drug metabolism and solubility issues are challenging to solve with these models (28).
Rodents other than hamsters, especially rats and mice, are the most commonly used animals in OSCC modeling. The rats are larger but correspondingly more expensive, and there are few immunodeficient or genetically engineered rats. The mouse is characterized by small size, a propensity to breed in captivity, a lifespan of 3 years, extensive physiological and molecular similarities to humans, and an entirely sequenced genome (29). The abundance of particular types of mice, such as immunodeficient mice, genetically engineered mice, and humanized mice, provides a variety of new platforms for the establishment of OSCC models. Therefore, OSCC mouse models have attracted increasing attention from numerous scholars in the OSCC research field.
This review focuses on several aspects of OSCC mouse models and is organized by the three main methods by which these models are established: chemical carcinogen-induced, transplanted, and genetically modified OSCC mouse models (Figure 1). Details are provided on the selection and establishment of OSCC mouse models and differences in their mechanisms, characteristics, establishment methods, and applications. This review aims to provide a reference and direction for researchers who are working toward conquering OSCC.
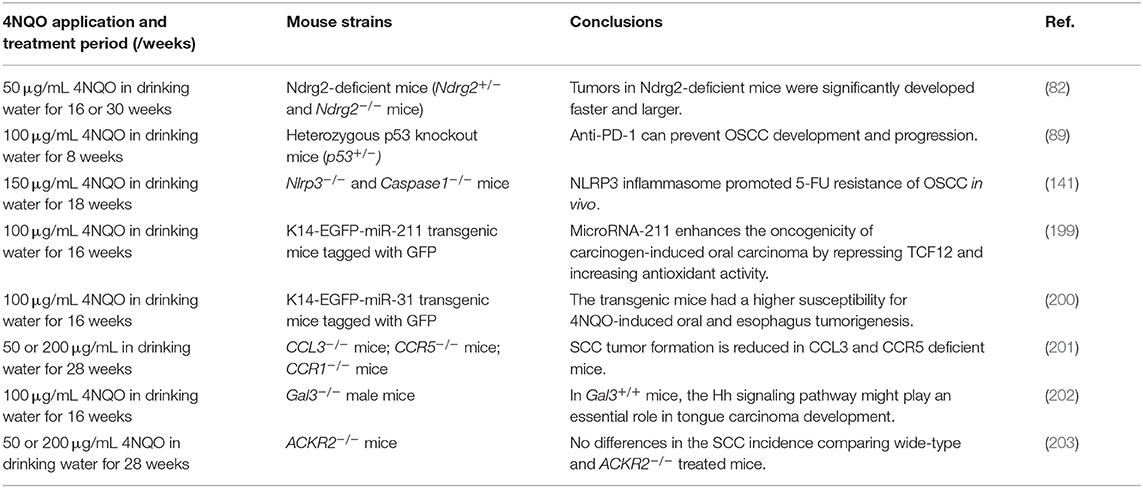
Figure 1. Current mouse models of OSCC. Various methods of establishing OSCC mouse models can be divided into three categories: chemical carcinogen-induced, transplanted, and genetically engineered mouse models. Adhesions: OSCC, oral squamous cell carcinoma; 4NQO, 4-nitroquinoline-1-oxide; B[a]P, Benzo[a]pyrene; DB[a,l]P, Dibenzo[a,l]pyrene; NNN, N'-nitrosonornicotine; CDX, cell-derived xenograft; PDX, patient-derived xenograft; GEMMs, genetically engineered models; SCC, squamous cell carcinoma.
Chemical Carcinogen-Induced Mouse Models
The main risk factors for OSCC include tobacco, alcohol, long-term chewing of betel quid, human papillomavirus (HPV), and oral lichen planus (especially the erosive form) (30, 31). There are more than 60 carcinogens in cigarette smoke and 16 in unburned tobacco, including tobacco-specific nitrosamines [such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and N'-nitrosonornicotine (NNN)], polycyclic aromatic hydrocarbons (PAHs, such as benzo[a]pyrene), and aromatic amines (such as 4-aminobiphenyl) (32). These chemicals have been reported to induce DNA adducts and correlate with a predisposition to cancer. DNA adducts—the result of covalent binding between DNA and carcinogens, related substances, or their metabolites—are central in the carcinogenesis caused by these agents (32, 33).
Knowing the above-mentioned risk factors for OSCC, researchers have induced murine oral carcinogenesis using a variety of chemical carcinogens, such as 4-nitroquinoline-1-oxide (4NQO) (34), Benzo[a]pyrene (B[a]P) (35), Dibenzo[a,l]pyrene (DB[a,l]P) (36, 37), NNN (37), and combination of 4NQO and arecoline or ethanol (38, 39). The exposure of these chemical carcinogens to the oral cavity of mice can naturally produce a primary OSCC mouse model.
4NQO-Induced OSCC Mouse Model
Mechanism of 4NQO-Induced OSCC Mouse Model
The compound 4NQO is an aromatic amine heterocyclic compound and a precursor carcinogen that is typically manufactured for research purposes. Currently, it is the most recognized and frequently used chemical carcinogen for establishing chemical carcinogen-induced OSCC mouse models (40). Researches have proved that 4NQO mimics tobacco and plays a carcinogenic role by causing intracellular oxidative stress, DNA adduction, mutagenesis, and tumor induction (Figure 2) (41–43). It has been revealed as a potent inducer of intracellular oxidative stress by generating reactive oxygen species (ROS) (44, 45), which is related to carcinogenesis that is caused by DNA damage (46–49). Although 4NQO is not a direct carcinogen, it is an electrophilic species produced by metabolism in the body and undergoes an irreversible reaction with the nucleophilic part of DNA, which ultimately introduce mutations. The carcinogenic action of 4NQO is initiated by the enzymatic reduction of its nitro group: first, 4NQO is reduced to 4-hydroxy amino quinoline-1-oxide (4HAQO) by NADH and NAD(P)H, which thus act as 4NQO nitroreductase and quinone reductase, respectively (50). Then, 4HAQO can be acetylated by seryl-tRNA synthetase to form a seryl-AMP enzyme complex (51). The resultant 4HAQO and seryl-AMP enzyme complex are carcinogenic metabolites that induce the formation of DNA adducts. The cellular detoxification of 4NQO is carried out by multidrug resistance protein (MRP) and glutathione S-transferase P (GSTP1-1), which may play essential roles in preventing the initiation and progression events of carcinogenesis (52). The imbalance between the two pathways is the leading cause of tumor induction by 4NQO. Nuclear magnetic resonance (NMR) studies and in vivo experiments have revealed that 4HAQO binds preferentially to G residues of DNA (53, 54). Moreover, the third and fourth position of the acetylated metabolite of 4NQO can react with the N2 and C8 positions of guanine (55). Mutations caused by these DNA adducts result in guanine to pyrimidine substitution (56, 57).
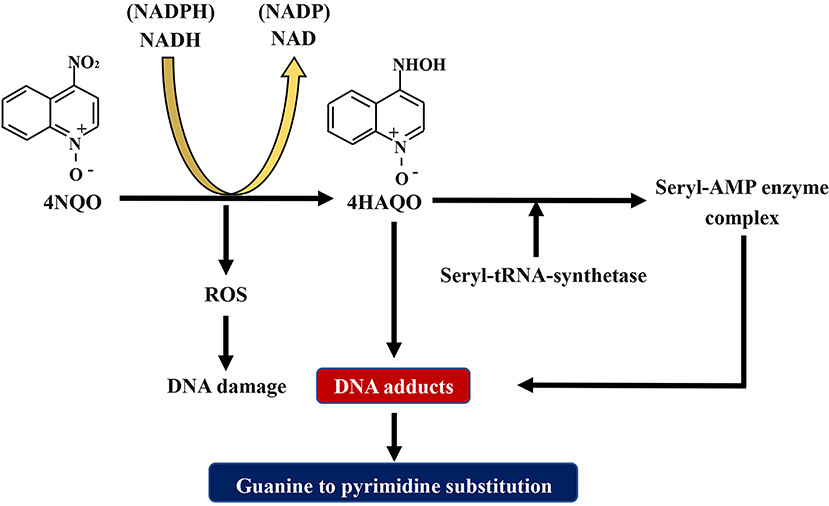
Figure 2. Schematic diagram about the mechanism of 4NQO carcinogenesis. 4NQO is firstly reduced to 4HAQO by NADH and NAD(P)H, while 4HAQO can be acetylated by seryl-tRNA synthetase to form a seryl-AMP enzyme complex. The resultant 4HAQO and seryl-AMP enzyme complex are carcinogenic metabolites that induce the formation of DNA adducts. Mutations caused by these DNA adducts lead to guanine-to-pyrimidine substitution. Adhesions: 4NQO, 4-nitroquinoline-1-oxide; 4HAQO, 4-hydroxy amino quinoline-1-oxide.
Characteristics of the 4NQO-Induced OSCC Mouse Model
The 4NQO-induced mouse model reflects the multistage dynamic carcinogenicity of human OSCC, from dysplasia to invasion. Researchers using this model reported that the gross morphology of lesions changed from mild to severe dysplasia (Figure 3A), and the pathological stages of tongue lesions underwent hyperplasia, dysplasia (mild, moderate, and severe), in situ carcinoma, and invasive squamous cell carcinoma (SCC) (Figure 3B) (58). One of the characteristics of this model is that not all mice develop the same lesions at the same time, and a variety of lesions can be seen on the tongue of a single mouse (Figure 3C). In a study using the 4NQO-induced oral cancer pain model, adult female C57BL/6 mice were given 4NQO (100 μg/mL) dissolved in propylene glycol for 16 weeks. The treatment was then switched to regular drinking water for 16–28 weeks. By 28 weeks, pathological changes in the tongue were observed in all mice, of which 20.8% had moderate atypical hyperplasia, 45.9% developed severe atypical hyperplasia and carcinoma in situ, and 33.3% progressed to squamous cell carcinoma (61). But the OSCC mouse model established by this method had a low degree of malignancy and rarely underwent metastasis and bone invasion (40).
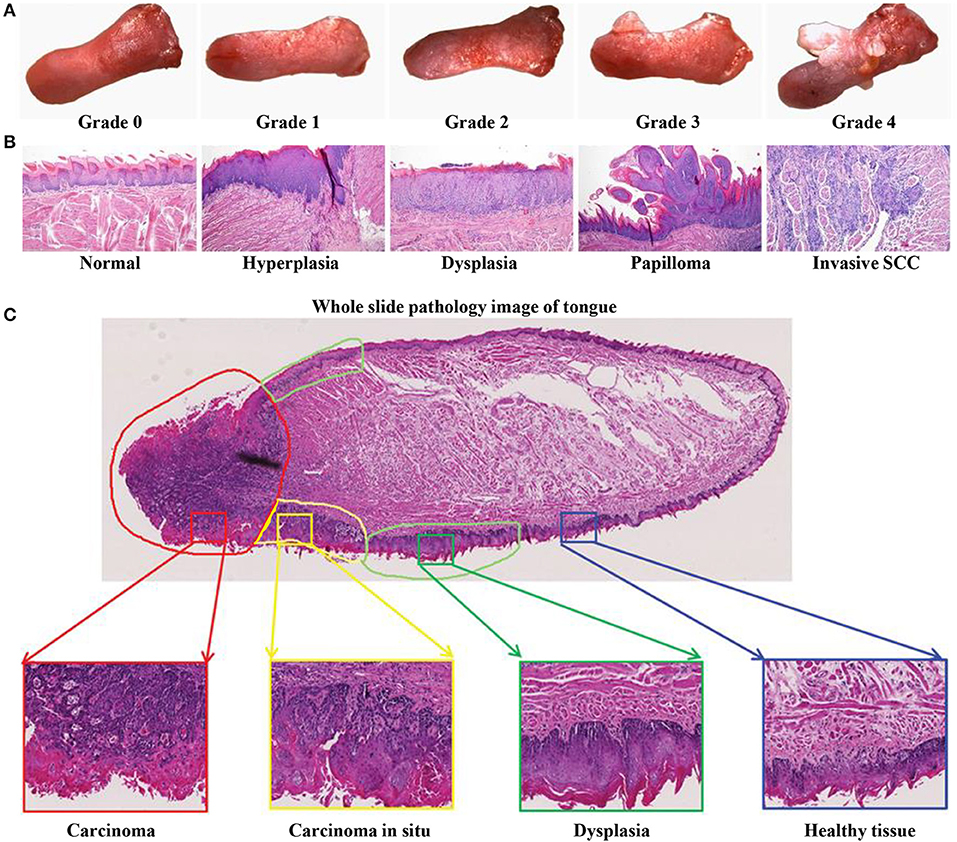
Figure 3. 4NQO-induced OSCC mouse model. (A–C) Representative gross morphology and pathological stages of the tongue from 4NQO-induced mice. (A) The gross tongue lesion grading system (8×), and the severity of the lesions gradually increased from 0 to 4. (B) Representative pathological stages of tongue lesions. (A,B) Reproduced and modified with permission (59). Copyright © 2014, National Academy of Sciences. (C) A whole slice image of tongue specimen contains tongue lesions of dysplasia, carcinoma in situ, and carcinoma. Reproduced and modified with permission (60). Copyright © 2017, Wiley Online Library.
The molecular events showing in the 4NQO-induced mouse model closely resemble those observed in human HNSCC patients (62, 63). Moreover, the 4NQO-induced OSCC mouse model has been proved effective because it is very similar to the occurrence of human OSCC at the genetic and molecular level. A review of molecular alterations at various stages of 4NQO-induced oral carcinogenesis had been accomplished by Kanojia and Vaidya, which covered apoptosis-related proteins, cell cycle-related proteins, proteins of cell-cell interactions, and cytoskeletal proteins (40). To identify the genetic alterations of 4NQO-induced C57BL/6J mice during the development of lingual SCC, Liu et al. (64) induced C57BL/6J mice with 4NQO (50 mg/L in drinking water) and then harvested lingual mucosa samples from different stages, namely normal tissue (0 week) and early-stage (12 week) and advanced-stage (28 week), respectively. Through microarray and methylated DNA immunoprecipitation sequencing, the result of bioinformatics analysis revealed significant alterations in 63 hub genes and promoter methylation, including Tbp, Smad1, Smad4, Pdpk1, Camk2, Atxn3, and Cdh2. Of the 63 human orthologous genes, 100% were reported to be associated with human cancer, and up to 55.5% were relevant to human oral cancer. Schoop et al. (65) revealed that the immunopathology of the murine oral tumor was similar to that of the described human oral tumor through immunohistochemistry labeled with cyclin D1 and E-cadherin.
The carcinogenic effect of 4NQO starts with its reduction by 4NQO reductase, so the distribution and quantity of 4NQO reductase in different tissues affect the tissue specificity of 4NQO carcinogenesis (66). 4NQO can induce carcinogenesis in many parts of the oral cavity, such as the dorsal tongue, ventral tongue, and palate (67, 68). Additionally, 4NQO passes from the oral cavity through the entire digestive tract, and the esophagus also contains a large amount of 4NQO reductase; therefore, this model can also form esophageal cancer (67, 69).
Methods for 4NQO-Induced OSCC Mouse Model
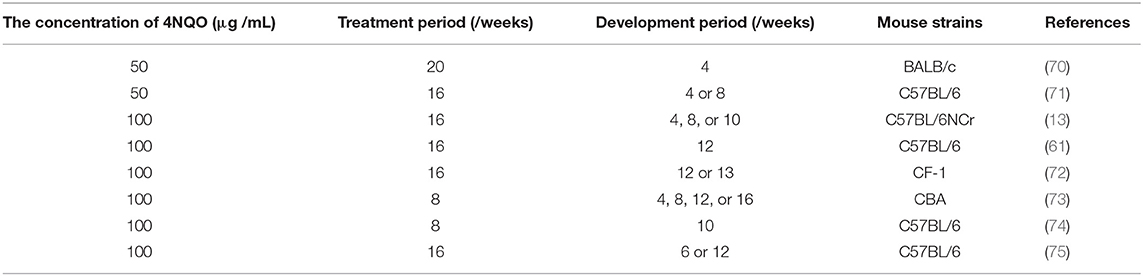
The strains of mice used for the 4NQO-induced OSCC mouse model include immunocompetent C57BL/6, BALB/c, CF-1, and CBA mice (Table 1). Most studies that have applied this method used adult mice aged 6–8 weeks, but Vincent-Chong et al. (76) observed that 92% of old mice (aged 65–70 weeks) developed severe dysplasia/invasive squamous cell carcinoma, while the incidence in young mice (aged 7–12 weeks) was 69%. CBA mice have been reported to be more susceptible to 4NQO induction than C57BL/6 mice (67, 77). Tang et al. (67) used 4NQO in drinking water to induce C57BL/6 and CBA mice and found that both strains developed a variety of precancerous lesions and carcinogenesis in the tongue and esophagus. CBA mice were treated with 50 and 100 μg/mL 4NQO for 8 weeks, and 100% of them developed tongue lesions, and 100 μg/mL 4NQO treatment for 8 or 16 weeks resulted in carcinogenesis of the tongue and esophagus. After 16 weeks of treatment with 100 μg/mL 4NQO, tongue lesions were observed in 100% of C57BL/6 mice, but visible gross lesions were not seen until the following 4–12 weeks.
Administration of 4NQO can be carried out by topical application or the addition to drinking water, and both methods have successfully established the OSCC mouse model (65, 76, 78–81). Initially, researchers used localized smearing of 4NQO to induce OSCC, which was the same approach used to induce SCC in hamster pouches by DMBA (65, 80). Schoop et al. (65) smeared the tongues of male CBA mice with 4NQO (5 mg/mL) dissolved in propylene glycol for 16 weeks, with three treatments every week. From 24 to 40 weeks, the researchers detected the continuous development of hyperplasia; mild, moderate, and severe dysplasia; and squamous cell carcinoma. The smear method ensures that 4NQO acts on the oral cavity to a great extent and reduces the 4NQO burden in the digestive tract as much as possible. However, some researchers have recently used drinking water containing 4NQO to establish the OSCC model since it is a more natural means of administration, faster to model, and less painful to the mice than the topical application method (76, 78, 79, 81).
As shown in Table 1, the dosage and administration time of 4NQO used by the drinking water method in different studies are diverse. The most commonly used 4NQO concentrations in drinking water to induce OSCC are 50 and 100 μg/mL (61, 70). In general, a higher concentration means the establishment of OSCC is faster. Typically, the mice are exposed to 4NQO in the drinking water for about 16 weeks (treatment period) and then provided with normal drinking water for an additional 6–16 weeks (development period) (76, 78, 79, 81). There is no unified standard for the establishment of 4NQO-induced OSCC mouse models at present. Researchers can adjust these factors according to the purpose of the study. Tang et al. (67) administered different concentrations of 4NQO in drinking water to CBA mice: 100 μg/mL 4NQO for 16 weeks, 100 μg/mL for 8 weeks, 50 μg/mL for 8 weeks, and 20 μg/mL for 8 weeks. Of the four groups, 100% of the first three groups developed oral cavity lesions; the values of the bromodeoxyuridine (BrdUrd) labeling index, which can be used to reflect the degree of malignancy of oral cavity lesions, were 21.0 ± 0.9%, 15.1 ± 0.8%, 10.9 ± 0.8%, and 11.1 ± 0.5%, respectively, for the four groups.
Usage of 4NQO-Induced OSCC Mouse Model
Since the similarities of the 4NQO-induced OSCC mouse model in pathogenesis, pathological changes, and host immune activity, and molecular level to those of humans, this model is widely used to study OSCC, especially in developing the biomarkers for early diagnosis and the transformation of the epithelium. Because 4NQO simulates tobacco-related gene mutation and can induce primary OSCC, this model has been used to explore the pathogenesis of OSCC (82). A spontaneous tumor has a more natural tumor microenvironment (76, 83) and can also be used in the study of cytokines (61), mesenchymal stem cells (84), natural killer (NK) cells (85, 86), microbiomes (7), and angiogenesis (87). The murine lesions induced by 4NQO constitute a dynamic and continuous process and can be used to study the early detection and prevention of precancerous OSCC lesions (88–90). The immune activity of this model renders it suitable for researching the changes in tumor immunology in the process and development of tumorigeneses, as well as related subjects such as immunosuppression (91–94) and immunotherapy (34, 95, 96). In conclusion, the 4NQO-induced OSCC mouse model appears to be the best available model for research the diagnostic and prognostic markers for OSCC. However, this model has limited application for studies on the invasion and metastasis of malignant tumors because of its low probability of metastasis and bone invasion.
Other Chemical Carcinogen-Induced OSCC Mouse Models
Tobacco-Related Chemical Carcinogens-Induced OSCC
Benzo[a]pyrene (B[a]P)
B[a]P, a member of the PAHs family and a procarcinogen, is widely distributed in tobacco smoke, charcoal-grilled foods, contaminated water, engine exhaust, and soil (97). In a 2-year bioassay of female B6C3F1 mice, Culp et al. (35) compared the effect of coal tar and B[a]P on tumor induction and discovered that the mice fed with 100 ppm B[a]P developed tongue lesions (papillomas and carcinomas) with an incidence of 23/48. However, the main target of B[a]P is the forestomach. Although high doses of B[a]P can induce tongue cancer, the toxicity of B[a]P is high, and the low carcinogenic specificity of B[a]P will produce lesions in other parts of the body, including the liver, lung, forestomach, esophagus, and larynx, which limit its application.
Dibenzo[a,l]pyrene (DB[a,l]P)
DB[a,l]P is a potent carcinogen produced in cigarette smoke and has been used to induce carcinogenesis of the lung, skin, mammary gland, and oral cavity in mice and rats (36, 98–100). Guttenplan et al. (36) revealed that a 24 nmol dose of DB[a,l]P by topical application to the oral cavity resulted in neoplasia in 31% of B6C3F1 mice, accompanied by the elevated expression of p53 and COX-2 protein. Furthermore, DB[a,l]P induced ovarian tumors.
DB[a,l]P and N'-nitrosonornicotine (NNN)
Guttenplan et al. (37) compared the carcinogenesis and mutations resulting from the administration of DB[a,l]P, NNN, and both NNN and DB[a,l]P. DB[a,l]P (0.16 μmol) was topically applied on the tongue of lacI mice three times per week, while NNN (8.46 μmol) was applied two times per week. After induction for 5 weeks and development for 4 weeks, the mice were euthanized. This study revealed that DB[a,l]P + NNN caused the highest percentage of genetic mutations.
4NQO Combined With Other Chemical Carcinogens
4NQO and arecoline
Arecoline is an alkaloid extracted from betel nut and plays an essential role in the progression of oral cancer (101, 102). Chang et al. (38) treated C57BL/6JNarl mice with arecoline, 4NQO, or both arecoline and 4NQO for 8 weeks to induce oral carcinogenesis. They found that 100% of the mice induced by both 4NQO (200 μg/mL) and arecoline (500 μg/mL) developed tongue tumors, while 57 and 0% of the mice exposed to only 4NQO or arecoline developed tongue tumors, respectively. Similar to the results in human studies (103), immunohistochemical analysis revealed the expression of αB-crystallin and Hsp27 in this mouse tumor model. In this model, the mouse oral cavity was exposed to 4NQO and arecoline at the same time, simulating the etiology for oral cancer associated with betel nut.
4NQO and ethanol
Alcohol is another risk factor for oral cancer. Guo et al. (39) treated male wild-type C57BL/6J mice and 5-Lox knockout mice (Alox5tm1Fun/J) with 100 μg/mL 4NQO in drinking water for 8 weeks, and then in the following 16 weeks, 8% ethanol was provided ad libitum as the sole drink water. The incidence of OSCC increased from 20 (no ethanol) to 43% (8% ethanol) in wild-type mice, while fewer cancers were induced in the 5-Lox knockout mice. This study revealed that ethanol could promote 4NQO-induced oral carcinogenesis in mice by activating the 5-LOX pathway of arachidonic acid metabolism.
Transplanted OSCC Mouse Models
Transplanted mouse models can be more accurately named “animal cultures” because tumor cells or tumor tissues are transferred to mice for culture. As early as 1969, Rygaard (104) successfully transplanted human malignant tumors into nude mice for the first time, which offered a new prospect in the research and application of transplanted mouse models.
Xenograft OSCC Mouse Models
Classifications of Xenograft OSCC Mouse Models
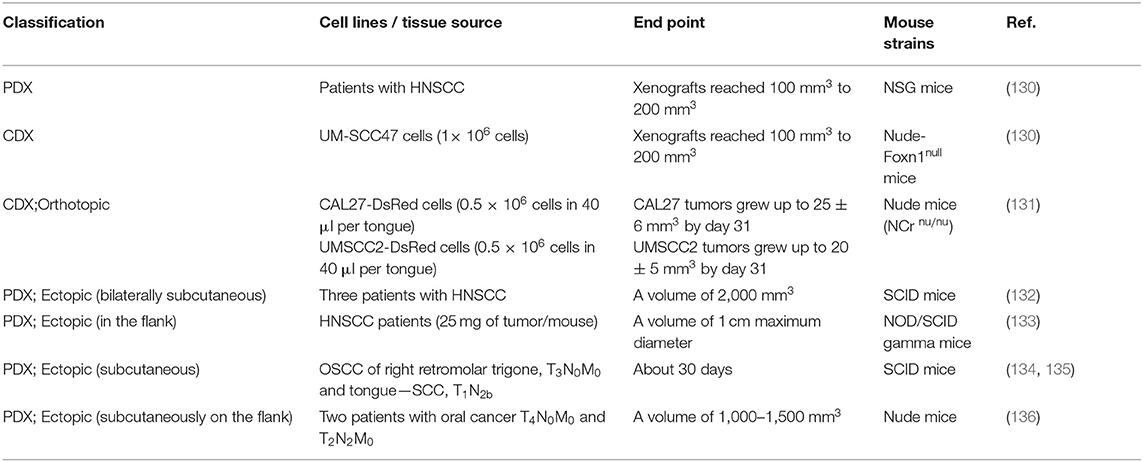
In tumor xenograft mice, tumor tissue or cell lines from one species are propagated in ectopic or orthotopic sites in immunodeficient mice (29). As shown in Table 2, depending on the source of the graft, xenograft mouse models can be divided into cell-derived xenografts (CDX) and patient-derived xenografts (PDX), which are derived from tumor cells cultured in vitro and fresh tumor tissues from patients, respectively. Xenograft mouse models can also be divided into ectopic or orthotopic mouse models depending on the tumor location. In ectopic mouse models, tumor cells are subcutaneously injected into the flank or back, while in orthotopic mice, tumor cells are typically transplanted to the tongue of mice.
Characteristics of Xenograft OSCC Mouse Models
A xenograft can be continuously transplanted in the same species or the same strain of animal, and the characteristics of easy replication, a high modeling success rate, and high stability facilitates the generation of abundant experimental mice.
The prominent characteristic of CDX models is high tumor consistency: after inoculating a certain number of tumor cells into mice, the volume and growth rate of the tumor are consistent, the inter-individual difference is low, and the effects on the hosts are identical (Figures 4A–D). But the transplanted tumor microenvironment lacks surrounding tissues, especially the stromal cells, vascular and lymphatic circulation, and immune cells of human origin (Figure 4D) (105, 106). Because different immunodeficient mice are used, the xenograft tissue lacks the structure of a typical immune system, although the immune cells are not absent. HE staining of HSC2-bearing tongues, dissected from athymic nude mice injected with HSC2 cells, revealed an immune reaction that included neutrophils, lymphocytes, and macrophages (107). CDX models can be considered PDX models with too many generations to be traceable. After multiple generations, the heterogeneity in the genetic, histological, and phenotypic characteristics of the tumors causes gradual differences from the original tumor tissue (108). For this reason, the US National Cancer Institute (NCI) retired the NCI-60 panel−60 human tumor cell lines established by NCI—from the drug-screening program (109). PDX models can more accurately reflect the mechanism of tumor occurrence and development in patients. In contrast to the traditional CDX models, PDX models preserve the genotypic and phenotypic diversity of tumor tissue to reflect the characteristics of the original tumor genuinely. PDX models also maintain tumor stromal cells and tumor microenvironment (Figures 4F–H). In a previous study, HE and immunohistochemistry (IHC) staining revealed that the histopathology and IHC were highly consistent between the PDX models and the corresponding patients (112). However, after the secondary transfer of the PDX tumor, the tumor stroma disappeared rapidly and was replaced by mouse interstitial cells (113).
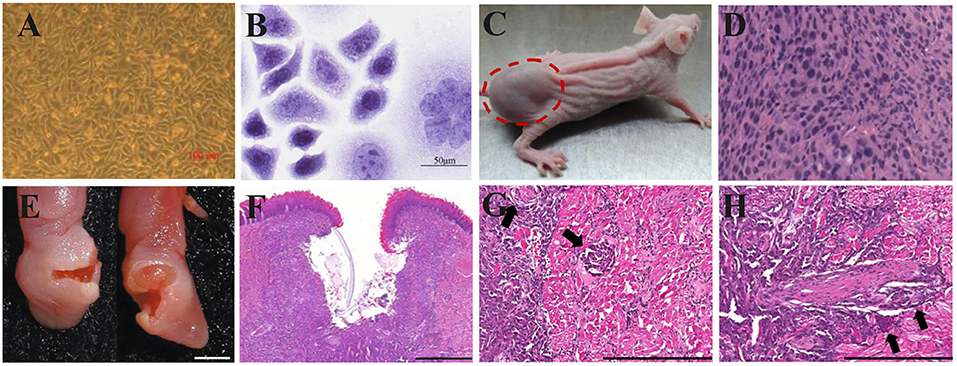
Figure 4. Xenograft OSCC mouse models. (A–D) An ectopic mouse model of OSCC. (A) OSCC-BD cells overlapped and lost contact inhibition. (B) HE staining showed that OSCC-BD cells had malignant characters. (C) The nude mice were subcutaneously injected with OSCC-BD cells. Tumor formation was observed, and neoplasms were about 1.0± 0.2 cm in size. (D) HE staining showed the neoplasm was typical squamous cell carcinoma. Scale bars, 100 μm (A) and 50 μm (B). Reproduced and modified with permission (110). Copyright © 2015, Springer Nature. (E–H) An orthotopic mouse model injected into the lateral border of the tongue of non-obese diabetes–severe combined immune deficiency (NOD-SCID) mice via HSC-3 cells. The HSC-3 bearing tumors have (E,F) ulcers, (G) neural, and (H) vascular invasion (arrows). Scale bars, 0.2 cm (E), 400 μm (F), and 200 μm (G,H). Reproduced and modified with permission (111). Copyright © 2018, Springer Nature.
The ectopic OSCC model can be established by subcutaneous injection with little effort; the operation is simple, and the observation of ectopic tumors and the measurement of tumor volume are also intuitive. Furthermore, the growth of subcutaneous tumors is not limited by the oral cavity size of mice (Figure 4C). However, the anatomical structures of the oral cavity and the subcutaneous tissue have some differences, such as the vascular distribution, lymphatic reflux, and bone adjacency. The orthotopic model can provide an experimental environment more similar to the original tumor environment than the ectopic model (28). Besides, more aggressive behaviors of regional and distant metastasis were discovered in the orthotopic models (114, 115). However, the growth of intra-oral tumors in the orthotopic model will not only cause pain but also hinder the diet of mice, which is against humanitarianism. Fitch et al. (116) firstly established an orthotopic model of human OSCC cell lines. Tumor cells were extracted from tumors growing subcutaneously and then injected into the tongue of nude mice. Equal tumorigenicity of tumor cells was finally detected in oral and subcutaneous tissues. In contrast, Myers et al. (117) compared the tumorigenicity of three human OSCC cell lines, Tu159, Tu167, and MDA1986, in the orthotopic tongue and ectopic subcutaneous tissues. It was found that all three types of SCC were more likely to develop in orthotopic than ectopic. The local tumor growth, regional lymph node metastases, and distant visceral metastases of the established orthotopic nude mouse model showed similar histopathology and biological characteristics to the patient's primary tumor.
Methods of Xenograft OSCC Mouse Models
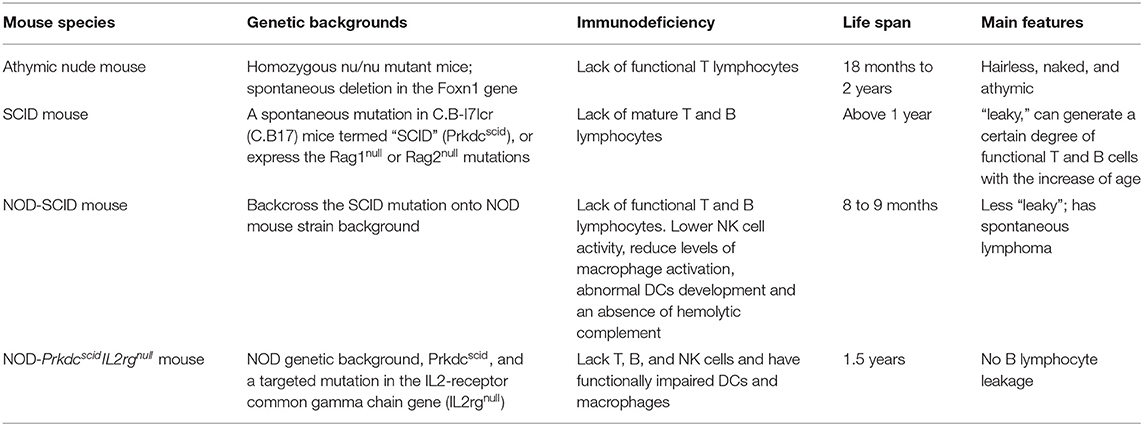
The mice used in this model need to be immunodeficient because xenografts can otherwise trigger an immune response and cause a rejection reaction and graft-vs.-host disease (GvHD). The commonly used immunodeficient mice are athymic nude mice, severe combined immune deficiency (SCID) mice, non-obese diabetes–severe combined immune deficiency (NOD-SCID) mice, and NOD-PrkdcscidIL2rgnull mice, such as NOD.Cg-PrkdcscidIl2rgtm1Sug/ShiJic mouse (NOG) and NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mouse (NSG). Different gene mutations cause varying degrees of immunodeficiency in the four kinds of mice. Comparisons by various researchers have revealed that the mice with the highest potential for successfully engrafting and investigating human cancers are the immunodeficient IL2rgnull mice, followed by NOD-SCID mice, SCID mice, and athymic nude mice, in descending order of efficacy (Table 3) (118–120).
In ectopic mouse models, tumor cells are subcutaneously injected into the flank or back, while in orthotopic mice, tumor cells are mainly transplanted into the tongue of mice. Moreover, the orthotopic OSCC mouse model can also be established at the floor-of-mouth (FOM) region (126–128). Transplanting tumor cells into the FOM region allowed the tumor to occur in the submental region of the jaw rather than in the mouth, in order to create a tumor that was easier to surgically remove and avoided inevitable complications if a trans-oral resection was attempted (127).
CDX models are developed using established human-derived cell lines, such as the human oral carcinoma cell lines SCC9 (1 × 107 cells) (129), UM-SCC47 (1 × 106 cells) (130), CAL27-DsRed (0.5 × 106 cells in 40 μL per tongue) (131), and UMSCC2-DsRed (0.5 × 106 cells in 40 μL per tongue) (131) (Table 4). The construction of PDX models has four phases (P): surgical removal of the tumor (P0), engraftment (P1), expansion (P2), and treatment (P3…Pn). The tumor tissue obtained from surgery is cut into fragments (diameter of about 1 mm), mixed with Matrigel, and then transplanted into immunodeficient mice. When the tumor grows to a specific size, the tumor is removed and then re-transplanted for tissue expansion. The growth rate of each cell line or tumor tissue derived from different patients varies, and the time required for the successful construction of the transplanted tumor has differed among studies. Therefore, the endpoint of tumor growth is evaluated by the volume of the tumor (Table 4).
Usage of Xenograft OSCC Mouse Models
The development of xenograft mouse models has enabled the study of human tumor tissues and cell lines in vivo in immunodeficient mice. In light of the advantages as mentioned above of “animal cultures” of tumor cells or tissues in vivo, most antineoplastic drugs have been tested using xenograft mouse models in the preclinical stage. Compared with CDX models, PDX models are more advanced preclinical oncology models for new drug development and can achieve better preclinical drug efficacy testing and analysis. As the tumor model closest to the actual conditions of the human body, the PDX model can provide personalized drug guidance for a single patient and realize precision therapy for a tumor. In the field of OSCC research, xenograft OSCC mouse models are used for screening drugs (137–139) and studying drug resistance, such as cisplatin (9, 140), 5-fluorouracil (141), and cetuximab (142). Potential therapeutic targets of OSCC can also be verified in xenograft mouse models (143, 144). Researchers have also focused on invasion and metastasis (145–149), new techniques for detecting tumors (138), postoperative recurrence (127, 128), and comparisons with other models or primary tumors (132, 150). Nevertheless, xenograft mouse models are incapable of simulating the tumor microenvironment and cannot be used to study the interaction between tumors and host immunity because of the use of immunodeficient mice.
Syngeneic OSCC Mouse Models
Syngeneic OSCC mouse models are derived from the allografts of immortalized mouse tumor cell lines, chemical carcinogen-induced spontaneous mouse tumors, or genetically engineered mouse models (GEMMs). Syngeneic mice can effectively avoid tissue rejection and GVHD caused by the transplantation of tumor cells into allogeneic mice. The models also have the inherent advantages of transplanted tumor models, such as the high speed of establishment, high consistency, and high stability. These models can better reflect the tumor microenvironment and more comprehensively simulate the complexity of the tumor. Therefore, they are widely used in the study of oncoimmunology and show great potential in the development of novel therapy for OSCC, especially immunotherapy.
Syngeneic Models Derived From Murine OSCC Cell Lines
The principle of syngeneic models derived from murine cell lines is similar to that of the CDX model. Various murine cell lines have been used to establish syngeneic OSCC mouse models, including murine squamous cell carcinoma SCC7 (151, 152), murine oral cancer (MOC) cell lines (153), and MOC1 (154), MOC2 (155), and murine OSCC Sq-1979 cells (156). Moroishi et al. (157) subcutaneously transplanted 1 × 105 SCC7 cells into both back flanks of C3H/HeOu mice and discovered that the tumor growth was aggressive, while all of the mice transplanted with LATS1/2 dKO SCC7 cells were tumor-free. Similarly, Dong et al. (152) subcutaneously injected 1 × 106 SCC7 cells into the right abdomen of C3H/HeJ mice to assess the therapeutic effect of the tumor-derived autophagosome vaccine (DRibble). To evaluate the antineoplastic effect of near-infrared photoimmunotherapy (NIR-PIT) produced by conjugating IR700DX (a photo-absorber) with anti-CD44 monoclonal antibodies, Nagaya et al. (153) built syngeneic models by subcutaneously injecting immunogenic MOC1 cells (2.0 × 106), moderately immunogenic MOC2-luc cells (1.5 × 105), and poorly immunogenic MOC2 mKate2 cells (1.5 × 105) into C57BL/6 mice. The authors ultimately demonstrated that NIR-PIT could significantly inhibit tumor growth in the three models. Similarly, Adachi et al. (156) subcutaneously injected 1 × 107 Sq-1979 cells into the posterior neck area of C3H/HeN mice to examine the genetic changes during the development of OSCC. In summary, these different murine OSCC cell lines were used to generate stable and straightforward syngeneic OSCC models successfully.
Syngeneic Models Derived From 4NQO-Induced Murine OSCC
Similar to the construction of the PDX model, Chen et al. (158) induced C57BL/6 mice with 4NQO (100 μg/mL in drinking water) for 16 weeks, and the mice were sacrificed at the 28th week to establish the mouse tongue squamous cell carcinoma cell lines MTCQ1 and MTCQ2. After that, the cells were injected into the flanks and the tongues of C57BL/6 mice to establish ectopic and orthotopic mouse models, respectively. Compared with the human SAS tongue SCC cell line, the proliferation of MTCQ cells was lower, but the migration and invasion abilities were much higher than those of SAS cells. A subclone of GFP-labeled MTCQ1 cells was identified by immunostaining and fluorescence imaging, and extensive cervical lymph node metastasis and lung metastasis were discovered. Some treatment methods, such as miRNAs (particularly miR-134), cisplatin treatment, and anti-PD-L1 immunotherapy, also had therapeutic effects on this model. Besides, Chen et al. (159) used 4NQO-induced OSCC transgenic mice to establish a syngeneic model. K14-EGFP-miR-211 transgenic mice were induced by 4NQO (100 μg/mL in drinking water) for 16 weeks and then sacrificed at set times. After that, the cell lines, designated MOC-L1 to MOC-L4, were established from the dissections of OSCC lesions on the dorsal tongue surface. The cell lines were used to obtain orthotopic xenografts and perform real-time in vivo tumor imaging by injecting 5 × 106 cells into the central tongue portion of C57BL/6 mice. Additionally, the cells were used to analyze distant metastasis and assess the therapeutic efficacy of cisplatin.
The immune system is a powerful weapon against a tumor, and the emergence and development of oncoimmunology have made conquering cancer an achievable goal. Syngeneic OSCC mouse models are feasible tools for oncoimmunology, but the major issue is that the models represent oral cancer in mice and form murine tumors with murine targets. There are significant differences in the composition and reaction mechanisms between mice and humans, and some targets in humans do not exist or respond in mice.
Humanized Mouse Models
The humanized mouse, which is developed by transplanting functional human cells or tissues into immunodeficient Il2rgnull mice, is an emerging preclinical model for studying human disease (160). For the study of tumors, current research focuses on [1] the establishment of novel tumor models (such as humanized CDX tumor models and humanized PDX tumor models) that are associated with human tumor-immune system interactions and [2] the exploration of therapy (including NK cell therapy, T cell editing, cytokine therapy, co-stimulatory enhancement, and checkpoint inhibitors) (161).
Morton et al. (162) reconstructed the tumor microenvironment in a humanized PDX mouse model of HNSCC (Figure 5). Tumors of a xenochimeric mouse (XactMice) presented human cells derived from humanized bone marrow, infiltration of human T and B cell populations, lymphangiogenesis, cytokine expression, and a dynamic microenvironment. Therefore, the XactMice system accurately recapitulates the growth of the original tumor in vivo. The humanized mouse model of HNSCC was optimized in another study by Morton et al. (163). After the dual infusion of human hematopoietic stem and progenitor cells (HSPCs) and mesenchymal stem cells (MSCs), the percentage of human immune cells in the bone marrow of established mice was almost twice that in mice implanted with HSPCs alone, and mature peripheral human immune cells were 9–38-fold more abundant. The dually engrafted mice also had more regulatory T cells, cytotoxic T cells, and MSCs. Thus, the dual infusion of HSPCs and MSCs resulted in a higher degree of humanization, which further increased the accuracy of the model.
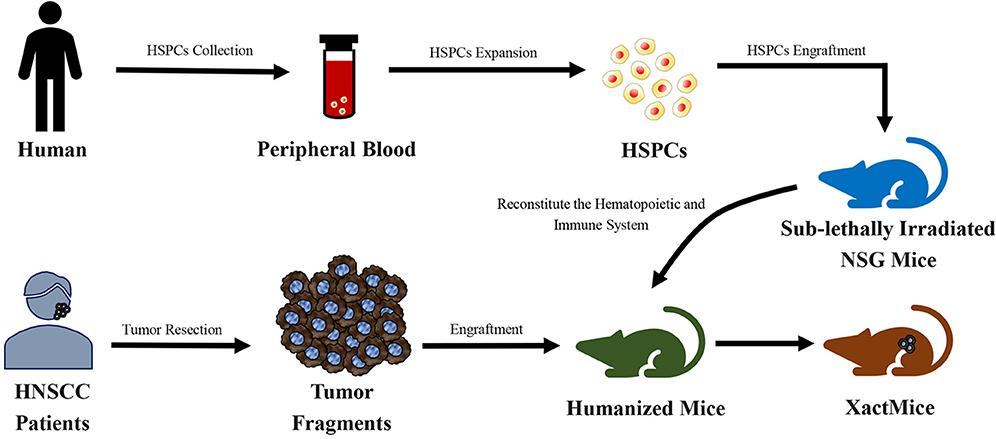
Figure 5. Schematic diagram of the generation of xenochimeric mice (XactMice) (162). After the cells are harvested from either cord blood or Granulocyte Colony-stimulating Factor (G-CSF) mobilized adult peripheral blood, the human hematopoietic stem and progenitor cells (HSPCs), which contain hematopoietic stem cells (HSCs), were expanded by an ex vivo technique and injected into sub-lethally irradiated NOD/SCID/IL2rg−/− (NSG) mice to reconstitute the hematopoietic and immune system. Subsequently, tumor tissue from head and neck squamous cell carcinoma (HNSCC) patients were engrafted into NSG mice to generate the XactMice.
As a new model, the technology of humanized mice is relatively immature, and the existing humanized immune mouse model system undeniably has many shortcomings. For instance, the immune response in the humanized mouse may be the result of tissue incompatibility (164). The transplanted human immune cells, mainly T cells, will produce GvHD and an immune attack on recipient mice and subsequently cause their death, so the operating window for conducting experiments is short (165). Nevertheless, the humanized mouse is entirely in line with the need to build a copy of the disease, so it remains a positive direction for the development of mouse models in the future.
Transplanted Mouse Models of HPV-Related Oral Cancer
The HPV infection is one of the risk factors for HNSCC, and the incidence of HPV-related oropharyngeal squamous cell carcinoma (OPSCC) increases sharply in men under the age of 50 (166). Within 20 years, the percentage of HPV-positive OPSCC in the United States and some European countries has risen from <20% to more than 70% (166). Given the transfer of the incidence of HNSCC to the HPV-positive population, HPV-positive oral cancer has aroused the interest of researchers, and the corresponding mouse models have been gradually developed. Currently, mouse models of HPV-positive oral cancer can be classified as transplanted models and genetically engineered mouse models (GEMMs) (Figure 6), the latter of which will be introduced in section GEMMs of HPV-related oral cancer.
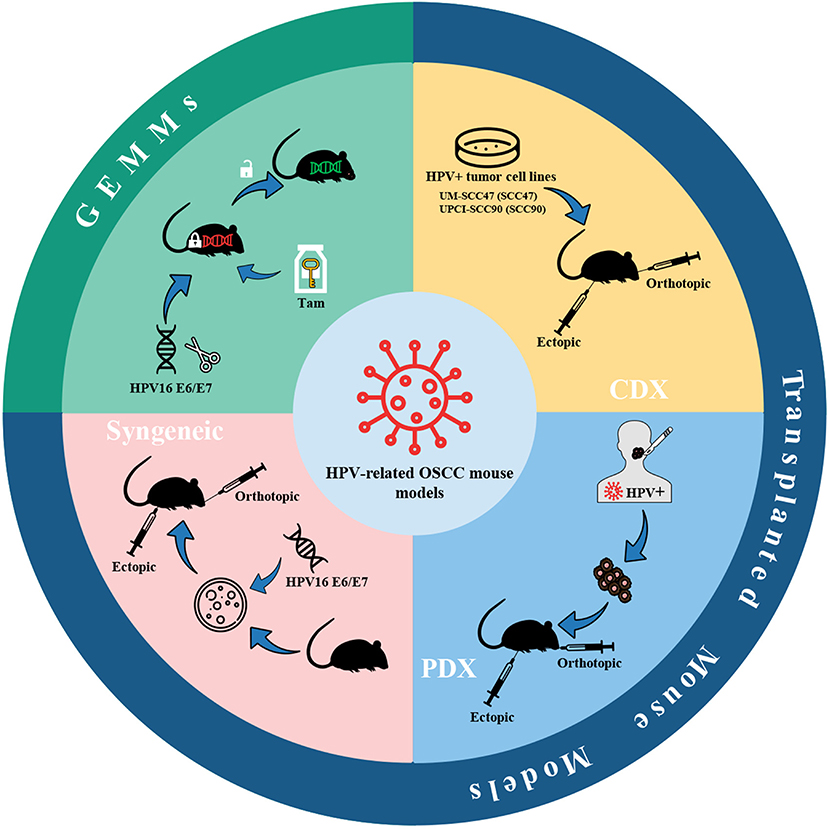
Figure 6. HPV-related OSCC mouse models. Mouse models of HPV-positive oral cancer can be generated by injecting HPV-positive HNSCC cell lines (CDX) or transplanting tumor fragments from HPV-positive HNSCC patients (PDX) or engrafting syngeneic HPV-positive cell lines, e.g., HPV16 E6/E7-expressing MTECs. Also, conditional GEMMs can be utilized to establish HPV-related OSCC mouse models. Adhesions: HPV, human papillomavirus; CDX, cell-derived xenograft; PDX, patient-derived xenograft; MTECs, mouse tonsil epithelial cells; GEMMs, genetically engineered models; Tam, tamoxifen.
The high-risk subtypes of HPV are mainly HPV16 and HPV18, while the former is considered to be associated with 90% of HPV-related cancers (167). The two proteins E6 and E7 in the early coding region of the HPV genome are highly conserved in high-risk HPV subtypes and are the most critical viral-encoded proteins involved in cancer (168). Thus, the current mouse models of HPV-related oral cancer mainly focus on the construction of cell lines or mouse strains expressing HPV16 E6/E7.
Brand et al. (169) established an HPV-related CDX model by bilaterally injecting HPV-positive HNSCC cell lines UM-SCC47 (SCC47) and UPCI-SCC90 (SCC90) into the athymic nude mice. The tumor fragments of HNSCC patients were also utilized to implant into NSG mice bilaterally to establish the PDX model. Keysar et al. (170) developed a model covering a total of 25 strains of HNSCC clinical spectrum, in which primary and recurrent tumors (including HPV-positive and HPV-negative) from HNSCC patients were implanted on mice using a modified floor-of-the-mouth (FOM) or base-of-tongue (BOT) implantation protocol to produce orthotopic tumors. Through the continuous passage of these models, the tumors maintained their original morphology, genetic characteristics, and drug susceptibilities. The gene characteristics of these tumors were following the known mutation frequencies of TP53, PI3KCA, NOTCH1, and NOTCH2. In addition, Facompre et al. (171) have also verified an elevated level of expression of p16INK4A and E6/E7 virus oncogene transcripts. Meanwhile, the barriers of HPV-related HNSCC PDX models were also summarized in the research of Facompre et al. including a low engrafted rate and the formation of Epstein-Barr virus-positive (EBV+) human large B-cell lymphomas. The biological characteristics of HPV-positive HNSCC are low degree of malignancy, low invasiveness, and weak growth potential of tumor cells in vitro, which lead to the paucity of available HPV-positive HNSCC cell lines (172). Accordingly, selection biases against the more typical molecular characteristics of HPV-positive tumors may occur during the experiments. Likewise, using less invasive tumors as xenografts may have adverse growth characteristics and further prevent PDX from remaining stable during continuous passage in vivo (171).
Another HPV-related mouse oral cancer model is generated by syngeneic HPV-positive cell lines. John Lee et al. (173) transferred retroviruses expressing HPV16 E6, E7, H-Ras, and an empty control vector into mouse tonsil epithelial cells (MTECs) to construct stable murine HPV-related cell lines in vitro. After that, 5 × 105 cells were injected into the tongue of C57BL/6 mice, or 1 × 106 cells were injected into the subcutaneous tissue of the upper back near the spine. It turned out that only HPV16 E6/H-Ras MTECs and HPV16 E6/E7/H-Ras MTECs formed invasive tumors in syngeneic C57BL/6 mice at orthotopic and ectopic sites. On this basis, in order to investigate whether HPV-specific immune mechanisms can result in tumor clearance, Williams et al. (174) developed preclinical models by injecting 1 × 106 HPV16 E6/E7/H-ras MTECs or HPV-negative cells subcutaneously on the right flank of C57BL/6 or SCID mice. Moreover, Mermod et al. (175) developed a novel HPV-positive HNSCC mouse model in which 1 × 105 HPV16 E6/E7-expressing MTECs were transplanted into the submental region of the FOM. After that, the tumor was surgically removed to investigate the progress of postoperative primary tumor recurrence and regional lymph node metastasis. Similarly, Paolini et al. (176) established HPV16 E7 expressing mouse OSCC AT-84 cells (AT-84 E7 cells) and AT-84-E7 luminescent cells. Then 6 × 105 AT-84 cells were injected into the FOM to obtain orthotopic tumors. Despite widely used, a major concern in syngeneic mouse models is the representativeness of mouse-derived tumor cells to human-derived tumor cells.
Although transplanted mouse models are currently the most commonly used animal models of HPV-related oral cancer, and their successful establishment has been documented in several publications, the lack of an appropriate HPV-positive mouse model—remaining stable during continuous passage in vivo and possessing more molecular characteristics with HPV-positive tumor cells of human origin—is still an obstacle to preclinical evaluation and treatment.
Genetically Modified Mouse Models
Genetically modified mouse models, also known as genetically engineered mouse models (GEMMs), are intricate and novel animal models that have been a beneficial outcome of the development of genetic engineering technology. A review of GEMMs of OSCC was published by Ishida et al. (177), so we mainly provide an overview of GEMMs.
An Overview of GEMMs
Classifications of GEMMs
GEMMs can be classified as either loss of function or gain of function. Loss of function entails gene knockout or knockdown, in which the expression of target genes is depleted or silenced. In the field of oncology, the blocked genes are typically oncogenes, tumor-suppressor genes, and metabolic genes (178). Gain-of-function studies use knock-in models of oncogene overexpression to study the function of an oncogene in vivo. According to the specificity, GEMMs can be classified as either conventional GEMMs or conditional GEMMs. Conventional GEMMs alter the gene of interest in every cell in the body, which is inconsistent with the reality where multiple mutational gene sites in a single cell suppress cell apoptosis and promote proliferation, resulting in tumorigenesis (179). Because of the spatiotemporal specificity of conditional GEMMs, their genes can be altered in different tissues or periods using conditional genetically modified techniques.
Characteristics of GEMMs
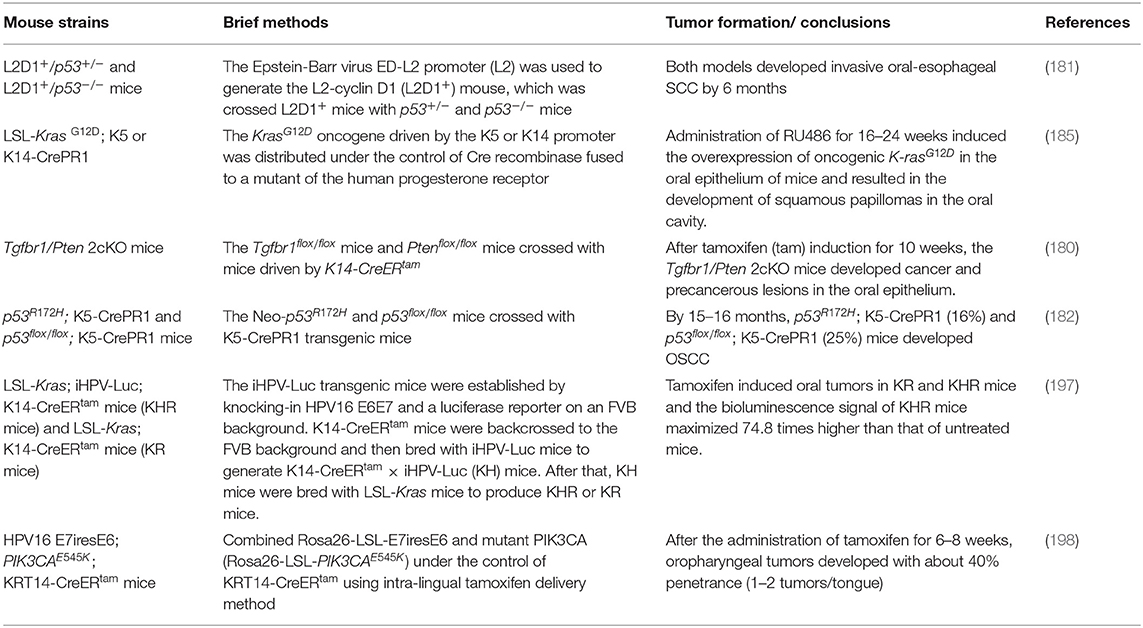
GEMMs enable the editing of specific genes through the activation or overexpression of oncogenes and the inactivation or silencing of tumor-suppressor genes. The pathological changes in these GEMMs, including hyperkeratosis, abnormal hyperplasia, carcinoma in situ, and invasive carcinoma (180–184), are similar to those of primary OSCC in humans and 4NQO-induced OSCC in mice. By the age of 5–6 months, histologic examination of L2D1+/p53+/− and L2D1+/p53−/− mice revealed hyperkeratosis, hyperplasia, severe epithelial dysplasia, and even cancer (181). Importantly, tumors of GEMMs grow in an environment with full immunity, so GEMMs are especially suitable for oncoimmunology study.
However, as a novel and promising model, there are still several barriers to the full application of GEMMs. Firstly, although the introduction of mice expressing Cre recombinase is driven by oral mucosa-specific promoters such as K5 or K14, the tumors generated by GEMMs have low specificity and appear in sites other than the oral cavity, such as the skin, tongue, esophagus, and forestomach (180, 181, 185, 186). Secondly, because of the full range of genetic changes caused by gene editing, GEMMs have a short lifespan and a high mortality rate, and highly malignant OSCC may not develop. For example, at 5 months, p53−/− and L2D1+/p53−/− mice had to be euthanized as a result of the morbidity caused by systemic lymphomas or sarcomas (181). Thirdly, the development of the human tumor is caused by mutations in a small number of cells surrounded by normal cells, while the introduction of exogenous genes or the knockout of endogenous genes in GEMMs will occur in all (conventional GEMMs) or most (conditional GEMMs) of the cells. Human tumors are usually accompanied by multiple mutations and metastases, which is not the case with GEMMs (187). Finally, the role of altered genes is questionable in the occurrence and development of OSCC. Kras, one of the most frequently used mutant genes in GEMMs of OSCC, has a low mutation frequency in human HNSCC, while the ras family is accounting for only 5% (188, 189). Almost all HPV-positive oral cancer patients are p53 wild type (190), which indicates that GEMMs of OSCC generated by p53 gene mutation are not representative of HPV-positive oral cancer.
Methods of GEMMs
C57BL/6 is the first mouse strain to have its complete genome sequenced and is regarded as a “standard” inbred line, which can provide a genetic background for many mutant genes. Therefore, C57BL/6 is widely used as a transgenic mouse model to mimic human genetic defects in genetic experiments. Additionally, Balb/c mice can also be used as the genetic background of GEMMs (154).
The specificity of conditional GEMMs is controlled by the appearance of a recombinase, for example, the Cre-loxP recombinase system (Figure 7) (191, 192). The temporal specificity of conditional GEMMs is achieved through inducible promoters, which are regulated by exogenous chemicals, such as tamoxifen (180) and RU486 (185, 193). When the chemicals are present or removed, the promoters are activated, and then the expression of downstream target genes is changed (194, 195). The spatial specificity or tissue specificity in conditional GEMMs is determined by Cre transgenes that are expressed from tissue-specific promoters (29). For the oral cavity, the known optimal promoters are the keratin 5 (K5) and keratin 14 (K14) promoters (177). K5 is expressed within the basal layer of the tongue and forestomach stratified squamous epithelia, while K14 has been found to be expressed in the basal layer of the oral mucosa and tongue (185). Furthermore, a promoter of the Epstein–Barr virus, ED-L2, has been shown to target genes in oral and esophageal squamous epithelial cells (181, 196).
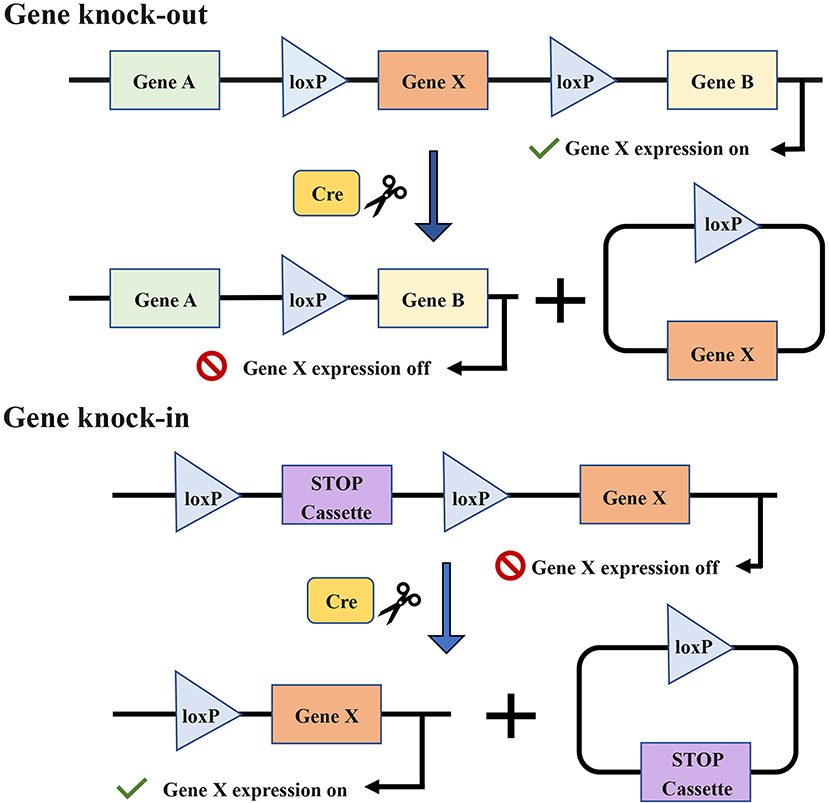
Figure 7. The target gene X is knocked out or knocked in via the Cre-loxP recombinase system. Cre enzymes can respectively catalyze recombination between loxP sites oriented in the same direction and flank the target gene. The introduction of Cre recombinase can cause the knock-out of the gene X. The conditional knock-in of the gene X requires another sequence named strong translational and transcriptional termination (STOP), which can terminate the expression of gene X. When Cre recombinase is present, the STOP cassette can be removed and then gene X is expressed.
As shown in Table 5, the current GEMMs of OSCC are mainly conditional, the implementation of which mostly depends on the hybridization of the Cre mouse and gene-floxed mouse. Different genetic changes cause different cancer rates, and the carcinogenic time ranges from 10 weeks to 16 months. Generally, the activation of oncogenes or inhibition of tumor-suppressor genes accelerates the progression of carcinogenesis, while the activation of tumor-suppressor genes or the knockout of oncogenes will slow down or even halt carcinogenesis.
Usage of GEMMs
At present, several studies have induced oral carcinogenesis by changing the expression of oncogenes or tumor-suppressor genes and identified the links between genes and cancer. The simple GEMMs can be used to reveal novel mechanisms of carcinogenesis and will be an essential tool in the study of oral cancer-related gene mutations or proteins in the future. However, the high mortality caused by unexpected primary tumors limits its application in highly malignant OSCC.
4NQO-Induced Combined With GEMMs
Given that mice induced by 4NQO can develop spontaneous OSCC and precancerous lesions, some studies have combined 4NQO induction with GEMMs. This combined model integrates the advantages of both methods. Wild-type mice were used as a control in the study of this combined model. The wild-type and genetically modified mice were induced by 4NQO (50, 100, or 150 μg/mL in drinking water) for 8–30 weeks (Table 6). The effect of 4NQO on both mice reflected the effect of gene changes on OSCC, such as susceptibility and resistance.
In the combined model, in contrast to simple 4NQO induction, specific gene deletion or overexpression can be studied; compared with simple GEMMs, more specific OSCC can be induced, and genetic changes that are not directly carcinogenic can be investigated. This model can be used to study the role of miRNA (199, 200), proteins (82, 184, 203), and pathways (201, 202) involved in the carcinogenesis of OSCC. Furthermore, the knock-in of green fluorescent protein can be used to label cells with specific gene changes (158, 199).
GEMMs of HPV-Related Oral Cancer
By utilizing the tamoxifen-regulated Cre recombinase system, Zhong et al. (197) targeted HPV-16 oncogenes E6 and E7 and a luciferase reporter gene (iHPV-Luc) in epithelial cells of transgenic mice and established a preclinical model of autologous HPV-positive oral tumors (Table 5). Tamoxifen treatment in this model generated the development of oral tumors that detected the expression of HPV biomarkers p16 and MCM7, PIK3CA, and PTEN mutations following HNSCC sequencing data and an active mTOR-PI3K pathway. However, this GEMM still has some limitations in accurately reflecting the clinical conditions of HNSCC patients (197). First, Mutant Kras was required for the development of HPV-positive tumors, while the ras family mutations account for only about 5% of HNSCC cases. Second, the expression of HPV oncogene was artificially driven by a cytomegalovirus promoter and did not reflect human HPV positive tumors. Finally, HPV-positive mice developed oral tumors rather than oropharyngeal cancers commonly seen in HNSCC patients. Caper et al. (198) established a novel inducible GEMM of HPV-related OPSCC, in which the HPV16 oncogenes were activated in a tissue-specific and temporal manner in vivo (Table 5). The expression of HPV16 E6/E7 and the tissue-specific expression of mutant PIK3CAE545K were induced by intra-lingual tamoxifen delivery, which replicated the histological and molecular characteristics of human HPV-positive OPSCC, including robust immune cell infiltration. Meanwhile, oropharyngeal lesions were accompanied by robust S6 phosphorylation at Ser235/236 in oropharyngeal tumors and low-level ERK1/2 activation (Thr202/Tyr204 phosphorylation).
Conclusions
In conclusion, mouse tumor-bearing models provide a preclinical study platform and make a significant contribution to the development of a cure for OSCC. A comprehensive understanding of model establishment is crucial for thoroughly studying OSCC. While it is impossible to replicate the actual situation of OSCC absolutely, mouse models can come very close. Thus, each model should be considered on the premise of its shortcomings, and the appropriate model should be chosen according to their overall advantages. Furthermore, choosing the appropriate model based on the specific research purpose can improve research efficiency. In a single research study, several types of models can be mixed according to the research purpose to ensure the scientific robustness and correctness of the results.
Author Contributions
QL, HD, YM, and YN: conceptualization. QL, HD, and GY: literature search. QL and HD: writing. HD: figure rendering. QL: table editing. QL, HD, GY, YS, YM, and YN: manuscript correction. All authors read and approved the final manuscript.
Funding
This review was funded by the National Natural Sciences Foundation of China (Nos. 81772880 to YN, 81371680 and 81571800 to YM), Development of Science and Technology of Nanjing (No. 201803036 to YM), Jiangsu Provincial Medical Talent (No. ZDRCC2016016 to YM), Nanjing Medical Science and Technique Development Foundation (QRX17083, ZKX18035), and Nanjing Municipal Key Medical Laboratory Constructional Project Funding.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the original authors of Figures 3, 4 and all references for their significant researches contributing to this manuscript. Besides, the authors wish to thank MDPI English editing for their editing service.
References
1. Park J, Zhang X, Lee SK, Song NY, Son SH, Kim KR, et al. CCL28-induced RARβ expression inhibits oral squamous cell carcinoma bone invasion. J Clin Invest. (2019) 129:5381–99. doi: 10.1172/JCI125336
2. Fan H, Tian H, Cheng X, Chen Y, Liang S, Zhang Z, et al. Aberrant Kank1 expression regulates YAP to promote apoptosis and inhibit proliferation in OSCC. J Cell Physiol. (2020) 235:1850–65. doi: 10.1002/jcp.29102
3. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. (2011) 61:69–90. doi: 10.3322/caac.20107
4. Sahu N, Grandis JR. New advances in molecular approaches to head and neck squamous cell carcinoma. Anticancer Drugs. (2011) 22:656–64. doi: 10.1097/CAD.0b013e32834249ba
5. Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. (2007) 26:645–62. doi: 10.1007/s10555-007-9082-y
6. Gao P, Liu S, Yoshida R, Shi CY, Yoshimachi S, Sakata N, et al. Ral GTPase activation by downregulation of RalGAP enhances oral squamous cell carcinoma progression. J Dent Res. (2019) 98:1011–9. doi: 10.1177/0022034519860828
7. Stashenko P, Yost S, Choi Y, Danciu T, Chen T, Yoganathan S, et al. The oral mouse microbiome promotes tumorigenesis in oral squamous cell carcinoma. mSystems. (2019) 4:e00323–19. doi: 10.1128/mSystems.00323-19
8. Yang Z, Liang X, Fu Y, Liu Y, Zheng L, Liu F, et al. Identification of AUNIP as a candidate diagnostic and prognostic biomarker for oral squamous cell carcinoma. EBioMed. (2019) 47:44–57. doi: 10.1016/j.ebiom.2019.08.013
9. Khoo XH, Paterson IC, Goh BH, Lee WL. Cisplatin-resistance in oral squamous cell carcinoma: regulation by tumor cell-derived extracellular vesicles. Cancers. (2019) 11:1166. doi: 10.3390/cancers11081166
10. Kim SY, Han YK, Song JM, Lee CH, Kang K, Yi JM, et al. Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma. (OSCC). Clin Epigenet. (2019) 11:116. doi: 10.1186/s13148-019-0715-0
11. Yoon M, Campbell JL, Andersen ME, Clewell HJ. Quantitative in vitro to in vivo extrapolation of cell-based toxicity assay results. Critical Rev Toxicol. (2012) 42:633–52. doi: 10.3109/10408444.2012.692115
12. Warner BM, Casto BC, Knobloch TJ, Accurso BT, Weghorst CM. Chemoprevention of oral cancer by topical application of black raspberries on high at-risk mucosa. Oral Surg Oral Med Oral Pathol Oral Radiol. (2014) 118:674–83. doi: 10.1016/j.oooo.2014.09.005
13. Vincent-Chong VK, DeJong H, Attwood K, Hershberger PA, Seshadri M. Preclinical prevention trial of calcitriol: impact of stage of intervention and duration of treatment on oral carcinogenesis. Neoplasia. (2019) 21:376–88. doi: 10.1016/j.neo.2019.02.002
14. Khiavi MM, Abdal K, Abbasi MM, Hamishehkar H, Aghbali AA, Salehi R, et al. Comparison of injectable doxorubicin & its nanodrug complex chemotherapy for the treatment of 4-nitroquinoline-1-oxide induced oral squamous cell carcinoma in rats. Indian J Med Res. (2017) 145:112–7. doi: 10.4103/ijmr.IJMR_542_14
15. Fulton AJ, Nemec A, Murphy BG, Kass PH, Verstraete FJ. Risk factors associated with survival in dogs with nontonsillar oral squamous cell carcinoma 31 cases. (1990–2010). J Am Vet Med Assoc. (2013) 243:696–702. doi: 10.2460/javma.243.5.696
16. Soltero-Rivera MM, Krick EL, Reiter AM, Brown DC, Lewis JR. Prevalence of regional and distant metastasis in cats with advanced oral squamous cell carcinoma: 49 cases. (2005–2011). J Feline Med Surg. (2014) 16:164–9. doi: 10.1177/1098612X13502975
17. Rossa C, D'Silva NJ. Non-murine models to investigate tumor-immune interactions in head and neck cancer. Oncogene. (2019) 38:4902–14. doi: 10.1038/s41388-019-0776-8
18. Manimaran A, Manoharan S. Tumor preventive efficacy of emodin in 7,12-dimethylbenz[a]Anthracene-induced oral carcinogenesis: a histopathological and biochemical approach. Pathol Oncol Res. (2018) 24:19–29. doi: 10.1007/s12253-017-0205-7
19. Chen YK, Hsue SS, Lin LM. Correlation between inducible nitric oxide synthase and p53 expression for DMBA-induced hamster buccal-pouch carcinomas. Oral Dis. (2003) 9:227–34. doi: 10.1034/j.1601-0825.2003.02878.x
20. Li N, Sood S, Wang S, Fang M, Wang P, Sun Z, et al. Overexpression of 5-lipoxygenase and cyclooxygenase 2 in hamster and human oral cancer and chemopreventive effects of zileuton and celecoxib. Clin Cancer Res. (2005) 11:2089–96. doi: 10.1158/1078-0432.CCR-04-1684
21. Vidya Priyadarsini R, Kumar N, Khan I, Thiyagarajan P, Kondaiah P, Nagini S. Gene expression signature of DMBA-induced hamster buccal pouch carcinomas: modulation by chlorophyllin and ellagic acid. PLoS ONE. (2012) 7:e34628. doi: 10.1371/journal.pone.0034628
22. Shklar G. Development of experimental oral carcinogenesis and its impact on current oral cancer research. J Dent Res. (1999) 78:1768–72. doi: 10.1177/00220345990780120101
23. Mognetti B, Di Carlo F, Berta GN. Animal models in oral cancer research. Oral Oncol. (2006) 42:448–60. doi: 10.1016/j.oraloncology.2005.07.014
24. Salley JJ. Experimental carcinogenesis in the cheek pouch of the Syrian hamster. J Dent Res. (1954) 33:253–62. doi: 10.1177/00220345540330021201
25. Smith LP, Thomas GR. Animal models for the study of squamous cell carcinoma of the upper aerodigestive tract: a historical perspective with review of their utility and limitations. Part A. Chemically-induced de novo cancer, syngeneic animal models of HNSCC, animal models of transplanted xenogeneic human tumors. Int J Cancer. (2006) 118:2111–22. doi: 10.1002/ijc.21694
26. Ballegeer EA, Madrill NJ, Berger KL, Agnew DW, McNiel EA. Evaluation of hypoxia in a feline model of head and neck cancer using. (6)(4)Cu-ATSM positron emission tomography/computed tomography. BMC Cancer. (2013) 13:218. doi: 10.1186/1471-2407-13-218
27. Rathore K, Alexander M, Cekanova M. Piroxicam inhibits Masitinib-induced cyclooxygenase 2 expression in oral squamous cell carcinoma cells in vitro. Transl Res. (2014) 164:158–68. doi: 10.1016/j.trsl.2014.02.002
28. Supsavhad W, Dirksen WP, Martin CK, Rosol TJ. Animal models of head and neck squamous cell carcinoma. Vet J. (2016) 210:7–16. doi: 10.1016/j.tvjl.2015.11.006
29. Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. (2007) 7:645–58. doi: 10.1038/nrc2192
30. Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. (2002) 52:195–215. doi: 10.3322/canjclin.52.4.195
31. Johnson N. Tobacco use and oral cancer: a global perspective. J Dental Educat. (2001) 65:328–39. Available online at: http://www.jdentaled.org/content/65/4/328.long
32. Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. (2003) 3:733–44. doi: 10.1038/nrc1190
33. Jin F, Thaiparambil J, Donepudi SR, Vantaku V, Piyarathna DWB, Maity S, et al. Tobacco-specific carcinogens induce hypermethylation, DNA adducts, and DNA damage in bladder cancer. Cancer Prev Res. (2017) 10:588–97. doi: 10.1158/1940-6207.CAPR-17-0198
34. Ludwig S, Hong CS, Razzo BM, Fabian KPL, Chelvanambi M, Lang S, et al. Impact of combination immunochemotherapies on progression of 4NQO-induced murine oral squamous cell carcinoma. Cancer Immunol Immunother. (2019) 68:1133–41. doi: 10.1007/s00262-019-02348-2
35. Culp SJ, Gaylor DW, Sheldon WG, Goldstein LS, Beland FA. A comparison of the tumors induced by coal tar and benzo[a]pyrene in a 2-year bioassay. Carcinogenesis. (1998) 19:117–24. doi: 10.1093/carcin/19.1.117
36. Guttenplan JB, Kosinska W, Zhao ZL, Chen KM, Aliaga C, DelTondo J, et al. Mutagenesis and carcinogenesis induced by dibenzo[a,l]pyrene in the mouse oral cavity: a potential new model for oral cancer. Int J Cancer. (2012) 130:2783–90. doi: 10.1002/ijc.26344
37. Guttenplan JB, Chen KM, Sun YW, Shalaby NAE, Kosinska W, Desai D, et al. The effects of the tobacco carcinogens N'-nitrosonornicotine and dibenzo[a,l]pyrene individually and in combination on DNA damage in human oral leukoplakia and on mutagenicity, and mutation profiles in lacI mouse tongue. Chem Res Toxicol. (2019) 32:1893–9. doi: 10.1021/acs.chemrestox.9b00257
38. Chang NW, Pei RJ, Tseng HC, Yeh KT, Chan HC, Lee MR, et al. Co-treating with arecoline and 4-nitroquinoline 1-oxide to establish a mouse model mimicking oral tumorigenesis. Chem Biol Interact. (2010) 183:231–7. doi: 10.1016/j.cbi.2009.10.005
39. Guo Y, Wang X, Zhang X, Sun Z, Chen X. Ethanol promotes chemically induced oral cancer in mice through activation of the 5-lipoxygenase pathway of arachidonic acid metabolism. Cancer Prev Res. (2011) 4:1863–72. doi: 10.1158/1940-6207.CAPR-11-0206
40. Kanojia D, Vaidya MM. 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral Oncol. (2006) 42:655–67. doi: 10.1016/j.oraloncology.2005.10.013
41. Kartasova T, Cornelissen BJ, Belt P, van de Putte P. Effects of UV, 4-NQO and TPA on gene expression in cultured human epidermal keratinocytes. Nucleic Acids Res. (1987) 15:5945–62. doi: 10.1093/nar/15.15.5945
42. Kondo S. A test for mutation theory of cancer: carcinogenesis by misrepair of DNA damaged by 4-nitroquinoline 1-oxide. Br J Cancer. (1977) 35:595–601. doi: 10.1038/bjc.1977.93
43. Kao YY, Tu HF, Kao SY, Chang KW, Lin SC. The increase of oncogenic miRNA expression in tongue carcinogenesis of a mouse model. Oral Oncol. (2015) 51:1103–12. doi: 10.1016/j.oraloncology.2015.10.007
44. Koike R, Uchiyama T, Arimoto-Kobayashi S, Okamoto K, Negishi T. Increase of somatic cell mutations in oxidative damage-sensitive drosophila. Genes Environ. (2018) 40:3. doi: 10.1186/s41021-017-0090-z
45. Arima Y, Nishigori C, Takeuchi T, Oka S, Morimoto K, Utani A, et al. 4-Nitroquinoline 1-oxide forms 8-hydroxydeoxyguanosine in human fibroblasts through reactive oxygen species. Toxicol Sci. (2006) 91:382–92. doi: 10.1093/toxsci/kfj161
46. Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. (2018) 80:50–64. doi: 10.1016/j.semcdb.2017.05.023
47. Yang J, Zhao X, Tang M, Li L, Lei Y, Cheng P, et al. The role of ROS and subsequent DNA-damage response in PUMA-induced apoptosis of ovarian cancer cells. Oncotarget. (2017) 8:23492–506. doi: 10.18632/oncotarget.15626
48. Tsai CH, Hung AC, Chen YY, Chiu YW, Hsieh PW, Lee YC, et al. 3'-hydroxy-4'-methoxy-β-methyl-β-nitrostyrene inhibits tumorigenesis in colorectal cancer cells through ROS-mediated DNA damage and mitochondrial dysfunction. Oncotarget. (2017) 8:18106–17. doi: 10.18632/oncotarget.14996
49. El-Hussein A, Hamblin MR. ROS generation and DNA damage with photo-inactivation mediated by silver nanoparticles in lung cancer cell line. IET Nanobiotechnol. (2017) 11:173–8. doi: 10.1049/iet-nbt.2015.0083
50. Benson AM. Conversion of 4-nitroquinoline 1-oxide. (4NQO) to 4-hydroxyaminoquinoline 1-oxide by a dicumarol-resistant hepatic 4NQO nitroreductase in rats and mice. Biochem Pharmacol. (1993) 46:1217–21. doi: 10.1016/0006-2952(93)90470-H
51. Friedberg EC. A history of the DNA repair and mutagenesis field: I. The discovery of enzymatic photoreactivation. DNA Repair (Amst). (2015) 33:35–42. doi: 10.1016/j.dnarep.2015.06.007
52. Morrow CS, Diah S, Smitherman PK, Schneider E, Townsend AJ. Multidrug resistance protein and glutathione S-transferase P1–1 act in synergy to confer protection from 4-nitroquinoline 1-oxide toxicity. Carcinogenesis. (1998) 19:109–15. doi: 10.1093/carcin/19.1.109
53. Tada M, Tada M. Main binding sites of the carcinogen, 4-nitroquinoline 1-oxide in nucleic acids. Biochim Biophys Acta. (1976) 454:558–66. doi: 10.1016/0005-2787(76)90281-1
54. Winkle SA, Tinoco I Jr. Interactions of 4-nitroquinoline 1-oxide with four deoxyribonucleotides. Biochemistry. (1978) 17:1352–6. doi: 10.1021/bi00600a033
55. Galiegue-Zouitina S, Bailleul B, Loucheux-Lefebvre MH. Adducts from in vivo action of the carcinogen 4-hydroxyaminoquinoline 1-oxide in rats and from in vitro reaction of 4-acetoxyaminoquinoline 1-oxide with DNA and polynucleotides. Cancer Res. (1985) 45:520–5.
56. Fronza G, Campomenosi P, Iannone R, Abbondandolo A. The 4-nitroquinoline 1-oxide mutational spectrum in single stranded DNA is characterized by guanine to pyrimidine transversions. Nucleic Acids Res. (1992) 20:1283–7. doi: 10.1093/nar/20.6.1283
57. Galiegue-Zouitina S, Daubersies P, Loucheux-Lefebvre MH, Bailleul B. Mutagenicity of N2 guanylarylation is SOS functions dependent and reminiscent of the high mutagenic property of 4NQO. Carcinogenesis. (1989) 10:1961–6. doi: 10.1093/carcin/10.10.1961
58. Li J, Liang F, Yu D, Qing H, Yang Y. Development of a 4-nitroquinoline-1-oxide model of lymph node metastasis in oral squamous cell carcinoma. Oral Oncol. (2013) 49:299–305. doi: 10.1016/j.oraloncology.2012.10.013
59. Tang XH, Osei-Sarfo K, Urvalek AM, Zhang T, Scognamiglio T, Gudas LJ. Combination of bexarotene and the retinoid CD1530 reduces murine oral-cavity carcinogenesis induced by the carcinogen 4-nitroquinoline 1-oxide. Proc Natl Acad Sci USA. (2014) 111:8907–12. doi: 10.1073/pnas.1404828111
60. Lu G, Wang D, Qin X, Muller S, Wang X, Chen AY, et al. Detection and delineation of squamous neoplasia with hyperspectral imaging in a mouse model of tongue carcinogenesis. J Biophoton. (2018) 11:e201700078. doi: 10.1002/jbio.201700078
61. Scheff NN, Ye Y, Bhattacharya A, MacRae J, Hickman DN, Sharma AK, et al. Tumor necrosis factor alpha secreted from oral squamous cell carcinoma contributes to cancer pain and associated inflammation. Pain. (2017) 158:2396–409. doi: 10.1097/j.pain.0000000000001044
62. Ye Y, Ono K, Bernabe DG, Viet CT, Pickering V, Dolan JC, et al. Adenosine triphosphate drives head and neck cancer pain through P2X2/3 heterotrimers. Acta Neuropathol Commun. (2014) 2:62. doi: 10.1186/2051-5960-2-62
63. Yuan B, Oechsli MN, Hendler FJ. A region within murine chromosome 7F4, syntenic to the human 11q13 amplicon, is frequently amplified in 4NQO-induced oral cavity tumors. Oncogene. (1997) 15:1161–70. doi: 10.1038/sj.onc.1201269
64. Liu H, Li J, Yang Y, Liu L, Yu L, Tu M, et al. Alterations of 63 hub genes during lingual carcinogenesis in C57BL/6J mice. Sci Rep. (2018) 8:12626. doi: 10.1038/s41598-018-31103-3
65. Schoop RA, Noteborn MH, Baatenburg de Jong RJ. A mouse model for oral squamous cell carcinoma. J Mol Histol. (2009) 40:177–81. doi: 10.1007/s10735-009-9228-z
66. Booth DR. A relationship found between intra-oral sites of 4NQO reductase activity and chemical carcinogenesis. Cell Tissue Kinet. (1990) 23:331–40. doi: 10.1111/j.1365-2184.1990.tb01129.x
67. Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. (2004) 10:301–13. doi: 10.1158/1078-0432.CCR-0999-3
68. Vered M, Yarom N, Dayan D. 4NQO oral carcinogenesis: animal models, molecular markers and future expectations. Oral Oncol. (2005) 41:337–9. doi: 10.1016/j.oraloncology.2004.07.005
69. Hasina R, Mollberg N, Kawada I, Mutreja K, Kanade G, Yala S, et al. Critical role for the receptor tyrosine kinase EPHB4 in esophageal cancers. Cancer Res. (2013) 73:184–94. doi: 10.1158/0008-5472.CAN-12-0915
70. Maji S, Samal SK, Pattanaik L, Panda S, Quinn BA, Das SK, et al. Mcl-1 is an important therapeutic target for oral squamous cell carcinomas. Oncotarget. (2015) 6:16623–37. doi: 10.18632/oncotarget.3932
71. Jiang L, Huang R, Wu Y, Diao P, Zhang W, Li J, et al. Overexpression of CDK7 is associated with unfavourable prognosis in oral squamous cell carcinoma. Pathology. (2019) 51:74–80. doi: 10.1016/j.pathol.2018.10.004
72. Droguett D, Castillo C, Leiva E, Theoduloz C, Schmeda-Hirschmann G, Kemmerling U. Efficacy of quercetin against chemically induced murine oral squamous cell carcinoma. Oncol Lett. (2015) 10:2432–8. doi: 10.3892/ol.2015.3598
73. Foy JP, Tortereau A, Caulin C, Le Texier V, Lavergne E, Thomas E, et al. The dynamics of gene expression changes in a mouse model of oral tumorigenesis may help refine prevention and treatment strategies in patients with oral cancer. Oncotarget. (2016) 7:35932–45. doi: 10.18632/oncotarget.8321
74. Wu JS, Zheng M, Zhang M, Pang X, Li L, Wang SS, et al. Porphyromonas gingivalis promotes 4-nitroquinoline-1-oxide-induced oral carcinogenesis with an alteration of fatty acid metabolism. Front Microbiol. (2018) 9:2081. doi: 10.3389/fmicb.2018.02081
75. Liu YC, Ho HC, Lee MR, Yeh CM, Tseng HC, Lin YC, et al. Cortactin is a prognostic marker for oral squamous cell carcinoma and its overexpression is involved in oral carcinogenesis. Environ Toxicol. (2017) 32:799–812. doi: 10.1002/tox.22280
76. Vincent-Chong VK, DeJong H, Rich LJ, Patti A, Merzianu M, Hershberger PA, et al. Impact of age on disease progression and microenvironment in oral cancer. J Dent Res. (2018) 97:1268–76. doi: 10.1177/0022034518775736
77. Vitale-Cross L, Czerninski R, Amornphimoltham P, Patel V, Molinolo AA, Gutkind JS. Chemical carcinogenesis models for evaluating molecular-targeted prevention and treatment of oral cancer. Cancer Prev Res. (2009) 2:419–22. doi: 10.1158/1940-6207.CAPR-09-0058
78. Bisetto S, Whitaker-Menezes D, Wilski NA, Tuluc M, Curry J, Zhan T, et al. Monocarboxylate transporter 4. (MCT4) knockout mice have attenuated 4NQO induced carcinogenesis; a role for MCT4 in driving oral squamous cell cancer. Front Oncol. (2018) 8:324. doi: 10.3389/fonc.2018.00324
79. Chen Y, Jiang Y, Liao L, Zhu X, Tang S, Yang Q, et al. Inhibition of 4NQO-induced oral carcinogenesis by dietary oyster shell calcium. Integr Cancer Ther. (2016) 15:96–101. doi: 10.1177/1534735415596572
80. Fujino H, Chino T, Imai T. Experimental production of labial and lingual carcinoma by local application of 4-nitroquinoline N-oxide. J Natl Cancer Inst. (1965) 35:907–18.
81. Khandelwal AR, Moore-Medlin T, Ekshyyan O, Gu X, Abreo F, Nathan CO. Local and systemic curcumin C3 complex inhibits 4NQO-induced oral tumorigenesis via modulating FGF-2/FGFR-2 activation. Am J Cancer Res. (2018) 8:2538–47.
82. Tamura T, Ichikawa T, Nakahata S, Kondo Y, Tagawa Y, Yamamoto K, et al. Loss of NDRG2 expression confers oral squamous cell carcinoma with enhanced metastatic potential. Cancer Res. (2017) 77:2363–74. doi: 10.1158/0008-5472.CAN-16-2114
83. Wu T, Hong Y, Jia L, Wu J, Xia J, Wang J, et al. Modulation of IL-1β reprogrammes the tumor microenvironment to interrupt oral carcinogenesis. Sci Rep. (2016) 6:20208. doi: 10.1038/srep20208
84. Chen Y, Wang X, Fang J, Song J, Ma D, Luo L, et al. Mesenchymal stem cells participate in oral mucosa carcinogenesis by regulating T cell proliferation. Clin Immunol. (2019) 198:46–53. doi: 10.1016/j.clim.2018.12.001
85. Young MR. Use of carcinogen-induced premalignant oral lesions in a dendritic cell-based vaccine to stimulate immune reactivity against both premalignant oral lesions and oral cancer. J Immunother. (2008) 31:148–56. doi: 10.1097/CJI.0b013e31815bdbf5
86. Li J, Qiu G, Fang B, Dai X, Cai J. Deficiency of IL-18 aggravates esophageal carcinoma through inhibiting IFN-γ production by CD8(+)T cells and NK cells. Inflammation. (2018) 41:667–76. doi: 10.1007/s10753-017-0721-3
87. Ludwig N, Yerneni SS, Razzo BM, Whiteside TL. Exosomes from HNSCC promote angiogenesis through reprogramming of endothelial cells. Mol Cancer Res. (2018) 16:1798–808. doi: 10.1158/1541-7786.MCR-18-0358
88. Ohnishi Y, Fujii T, Ugaki Y, Yasui H, Watanabe M, Dateoka S, et al. Usefulness of a fluorescence visualization system for the detection of oral precancerous and early cancerous lesions. Oncol Rep. (2016) 36:514–20. doi: 10.3892/or.2016.4776
89. Wang J, Xie T, Wang B, William WN Jr, Heymach JV, El-Naggar AK, et al. PD-1 Blockade prevents the development and progression of carcinogen-induced oral premalignant lesions. Cancer Prev Res. (2017) 10:684–93. doi: 10.1158/1940-6207.CAPR-17-0108
90. Chen Y, Li Q, Li X, Ma D, Fang J, Luo L, et al. Blockade of PD-1 effectively inhibits in vivo malignant transformation of oral mucosa. Oncoimmunology. (2018) 7:e1388484. doi: 10.1080/2162402X.2017.1388484
91. Chu M, Su YX, Wang L, Zhang TH, Liang YJ, Liang LZ, et al. Myeloid-derived suppressor cells contribute to oral cancer progression in 4NQO-treated mice. Oral Dis. (2012) 18:67–73. doi: 10.1111/j.1601-0825.2011.01846.x
92. Chen WC, Lai CH, Chuang HC, Lin PY, Chen MF. Inflammation-induced myeloid-derived suppressor cells associated with squamous cell carcinoma of the head and neck. Head Neck. (2017) 39:347–55. doi: 10.1002/hed.24595
93. Oghumu S, Knobloch TJ, Terrazas C, Varikuti S, Ahn-Jarvis J, Bollinger CE, et al. Deletion of macrophage migration inhibitory factor inhibits murine oral carcinogenesis: Potential role for chronic pro-inflammatory immune mediators. Int J Cancer. (2016) 139:1379–90. doi: 10.1002/ijc.30177
94. Wu JS, Li L, Wang SS, Pang X, Wu JB, Sheng SR, et al. Autophagy is positively associated with the accumulation of myeloidderived suppressor cells in 4nitroquinoline1oxideinduced oral cancer. Oncol Rep. (2018) 40:3381–91. doi: 10.3892/or.2018.6747
95. Wen L, Lu H, Li Q, Li Q, Wen S, Wang D, et al. Contributions of T cell dysfunction to the resistance against anti-PD-1 therapy in oral carcinogenesis. J Exp Clin Cancer Res. (2019) 38:299. doi: 10.1186/s13046-019-1185-0
96. De Costa AM, Justis DN, Schuyler CA, Young MR. Administration of a vaccine composed of dendritic cells pulsed with premalignant oral lesion lysate to mice bearing carcinogen-induced premalignant oral lesions stimulates a protective immune response. Int Immunopharmacol. (2012) 13:322–30. doi: 10.1016/j.intimp.2012.05.004
97. Phillips DH. Polycyclic aromatic hydrocarbons in the diet. Mutat Res. (1999) 443:139–47. doi: 10.1016/S1383-5742(99)00016-2
98. Prahalad AK, Ross JA, Nelson GB, Roop BC, King LC, Nesnow S, et al. Dibenzo[a,l]pyrene-induced DNA adduction, tumorigenicity, and Ki-ras oncogene mutations in strain A/J mouse lung. Carcinogenesis. (1997) 18:1955–63. doi: 10.1093/carcin/18.10.1955
99. Cavalieri EL, Rogan EG, Higginbotham S, Cremonesi P, Salmasi S. Tumor-initiating activity in mouse skin and carcinogenicity in rat mammary gland of dibenzo[a]pyrenes: the very potent environmental carcinogen dibenzo[a, l]pyrene. J Cancer Res Clin Oncol. (1989) 115:67–72. doi: 10.1007/BF00391602
100. Higginbotham S, RamaKrishna NV, Johansson SL, Rogan EG, Cavalieri EL. Tumor-initiating activity and carcinogenicity of dibenzo[a,l]pyrene versus 7,12-dimethylbenz[a]anthracene and benzo[a]pyrene at low doses in mouse skin. Carcinogenesis. (1993) 14:875–8. doi: 10.1093/carcin/14.5.875
101. Tu HF, Chen MY, Lai JC, Chen YL, Wong YW, Yang CC, et al. Arecoline-regulated ataxia telangiectasia mutated expression level in oral cancer progression. Head Neck. (2019) 41:2525–37. doi: 10.1002/hed.25718
102. Kuo TM, Nithiyanantham S, Lee CP, Hsu HT, Luo SY, Lin YZ, et al. Arecoline N-oxide regulates oral squamous cell carcinoma development through NOTCH1 and FAT1 expressions. J Cell Physiol. (2019) 234:13984–93. doi: 10.1002/jcp.28084
103. Lo WY, Tsai MH, Tsai Y, Hua CH, Tsai FJ, Huang SY, et al. Identification of over-expressed proteins in oral squamous cell carcinoma. (OSCC) patients by clinical proteomic analysis. Clin Chim Acta. (2007) 376:101–7. doi: 10.1016/j.cca.2006.06.030
104. Rygaard J, Povlsen CO. Heterotransplantation of a human malignant tumour to “Nude” mice. Acta Pathol Microbiol Scand. (1969) 77:758–60. doi: 10.1111/j.1699-0463.1969.tb04520.x
105. Sikder H, Huso DL, Zhang H, Wang B, Ryu B, Hwang ST, et al. Disruption of Id1 reveals major differences in angiogenesis between transplanted and autochthonous tumors. Cancer Cell. (2003) 4:291–9. doi: 10.1016/S1535-6108(03)00245-9
106. Lwin TM, Hoffman RM, Bouvet M. Advantages of patient-derived orthotopic mouse models and genetic reporters for developing fluorescence-guided surgery. J Surg Oncol. (2018) 118:253–64. doi: 10.1002/jso.25150
107. Chodroff L, Bendele M, Valenzuela V, Henry M, Ruparel S. EXPRESS: BDNF signaling contributes to oral cancer pain in a preclinical orthotopic rodent model. Mol Pain. (2016) 12:1–17. doi: 10.1177/1744806916666841
108. Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. (2001) 84:1424–31. doi: 10.1054/bjoc.2001.1796
109. Ledford H. US cancer institute to overhaul tumour cell lines. Nature. (2016) 530:391. doi: 10.1038/nature.2016.19364
110. Dong Y, Zhao Q, Ma X, Ma G, Liu C, Chen Z, et al. Establishment of a new OSCC cell line derived from OLK and identification of malignant transformation-related proteins by differential proteomics approach. Sci Rep. (2015) 5:12668. doi: 10.1038/srep12668
111. Rivera C, Zandonadi FS, Sanchez-Romero C, Soares CD, Granato DC, Gonzalez-Arriagada WA, et al. Agrin has a pathological role in the progression of oral cancer. Br J Cancer. (2018) 118:1628–38. doi: 10.1038/s41416-018-0135-5
112. Sun S, Zhang Z. Patient-derived xenograft platform of OSCC: a renewable human bio-bank for preclinical cancer research and a new co-clinical model for treatment optimization. Front Med. (2016) 10:104–10. doi: 10.1007/s11684-016-0432-4
113. Maykel J, Liu JH, Li H, Shultz LD, Greiner DL, Houghton J. NOD-scidIl2rg. (tm1Wjl) and NOD-Rag1. (null) Il2rg. (tm1Wjl): a model for stromal cell-tumor cell interaction for human colon cancer. Digest Dis Sci. (2014) 59:1169–79. doi: 10.1007/s10620-014-3168-5
114. Sumareva R, Ukrainsky G, Kiremidjian-Schumacher L, Roy M, Wishe HI, Steinfeld AD, et al. Effect of combined adoptive immunotherapy and radiotherapy on tumor growth. Radiation Oncol Invest. (1999) 7:22–9. doi: 10.1002/(SICI)1520-6823(1999)7:1<22::AID-ROI3>3.0.CO;2-6
115. Mery B, Rancoule C, Guy JB, Espenel S, Wozny AS, Battiston-Montagne P, et al. Preclinical models in HNSCC: a comprehensive review. Oral Oncol. (2017) 65:51–6. doi: 10.1016/j.oraloncology.2016.12.010
116. Fitch KA. The development of a head and neck tumor model in the nude mouse. Head Neck Oncol Res. (1988) 187–90.
117. Myers JN, Holsinger FC, Jasser SA, Bekele BN, Fidler IJ. An orthotopic nude mouse model of oral tongue squamous cell carcinoma. Clin Cancer Res. (2002) 8:293–8. Available online at: https://clincancerres.aacrjournals.org/content/8/1/293.long
118. Agliano A, Martin-Padura I, Mancuso P, Marighetti P, Rabascio C, Pruneri G, et al. Human acute leukemia cells injected in NOD/LtSz-scid/IL-2Rgamma null mice generate a faster and more efficient disease compared to other NOD/scid-related strains. Int J Cancer. (2008) 123:2222–7. doi: 10.1002/ijc.23772
119. Takenaka K, Prasolava TK, Wang JC, Mortin-Toth SM, Khalouei S, Gan OI, et al. Polymorphism in Sirpa modulates engraftment of human hematopoietic stem cells. Nat Immunol. (2007) 8:1313–23. doi: 10.1038/ni1527
120. Phillips RA, Jewett MA, Gallie BL. Growth of human tumors in immune-deficient scid mice and nude mice. Curr Top Microbiol Immunol. (1989) 152:259–63. doi: 10.1007/978-3-642-74974-2_31
121. Szadvari I, Krizanova O, Babula P. Athymic nude mice as an experimental model for cancer treatment. Physiol Res. (2016) 65(Suppl 4):S441–S53. Available online at: http://www.biomed.cas.cz/physiolres/pdf/65/65_S441.pdf
122. Bosma MJ, Carroll AM. The SCID mouse mutant: definition, characterization, and potential uses. Annual Rev Immunol. (1991) 9:323–50. doi: 10.1146/annurev.iy.09.040191.001543
123. Prochazka M, Gaskins HR, Shultz LD, Leiter EH. The nonobese diabetic scid mouse: model for spontaneous thymomagenesis associated with immunodeficiency. Proc Natl Acad Sci USA. (1992) 89:3290–4. doi: 10.1073/pnas.89.8.3290
124. Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. (2002) 100:3175–82. doi: 10.1182/blood-2001-12-0207
125. Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. (2005) 174:6477–89. doi: 10.4049/jimmunol.174.10.6477
126. Dinesman A, Haughey B, Gates GA, Aufdemorte T, Von Hoff DD. Development of a new in vivo model for head and neck cancer. Otolaryngol Head Neck Surg. (1990) 103:766–74. doi: 10.1177/019459989010300517
127. Behren A, Kamenisch Y, Muehlen S, Flechtenmacher C, Haberkorn U, Hilber H, et al. Development of an oral cancer recurrence mouse model after surgical resection. Int J Oncol. (2010) 36:849–55. doi: 10.3892/ijo_00000562
128. Roh V, Abramowski P, Hiou-Feige A, Cornils K, Rivals JP, Zougman A, et al. Cellular barcoding identifies clonal substitution as a hallmark of local recurrence in a surgical model of head and neck squamous cell carcinoma. Cell Rep. (2018) 25:2208–22.e7. doi: 10.1016/j.celrep.2018.10.090
129. Li J, Huang J, Jeong JH, Park SJ, Wei R, Peng J, et al. Selective TBK1/IKKi dual inhibitors with anticancer potency. Int J Cancer. (2014) 134:1972–80. doi: 10.1002/ijc.28507
130. Brand TM, Hartmann S, Bhola NE, Li H, Zeng Y, O'Keefe RA, et al. Cross-talk Signaling between HER3 and HPV16 E6 and E7 mediates resistance to PI3K inhibitors in head and neck cancer. Cancer Res. (2018) 78:2383–95. doi: 10.1158/0008-5472.CAN-17-1672
131. Bais MV, Kukuruzinska M, Trackman PC. Orthotopic non-metastatic and metastatic oral cancer mouse models. Oral Oncol. (2015) 51:476–82. doi: 10.1016/j.oraloncology.2015.01.012
132. Pearson AT, Finkel KA, Warner KA, Nor F, Tice D, Martins MD, et al. Patient-derived xenograft. (PDX) tumors increase growth rate with time. Oncotarget. (2016) 7:7993–8005. doi: 10.18632/oncotarget.6919
133. Li H, Wheeler S, Park Y, Ju Z, Thomas SM, Fichera M, et al. Proteomic characterization of head and neck cancer patient-derived xenografts. Mol Cancer Res. (2016) 14:278–86. doi: 10.1158/1541-7786.MCR-15-0354
134. Bothwell KD, Shaurova T, Merzianu M, Suresh A, Kuriakose MA, Johnson CS, et al. Impact of short-term 1,25-dihydroxyvitamin D3 on the chemopreventive efficacy of erlotinib against oral cancer. Cancer Prev Res. (2015) 8:765–76. doi: 10.1158/1940-6207.CAPR-14-0454
135. Seshadri M, Merzianu M, Tang H, Rigual NR, Sullivan M, Loree TR, et al. Establishment and characterization of patient tumor-derived head and neck squamous cell carcinoma xenografts. Cancer Biol Ther. (2009) 8:2275–83. doi: 10.4161/cbt.8.23.10137
136. Lu Z, He Q, Liang J, Li W, Su Q, Chen Z, et al. miR-31–5p Is a potential circulating biomarker and therapeutic target for oral cancer. Mol Ther Nucleic Acids. (2019) 16:471–80. doi: 10.1016/j.omtn.2019.03.012
137. Kerk SA, Finkel KA, Pearson AT, Warner KA, Zhang Z, Nor F, et al. 5T4-targeted therapy ablates cancer stem cells and prevents recurrence of head and neck squamous cell carcinoma. Clin Cancer Res. (2017) 23:2516–27. doi: 10.1158/1078-0432.CCR-16-1834
138. Rich LJ, Seshadri M. Photoacoustic monitoring of tumor and normal tissue response to radiation. Sci Rep. (2016) 6:21237. doi: 10.1038/srep21237
139. Zhang L, Nomie K, Zhang H, Bell T, Pham L, Kadri S, et al. B-cell lymphoma patient-derived xenograft models enable drug discovery and are a platform for personalized therapy. Clin Cancer Res. (2017) 23:4212–23. doi: 10.1158/1078-0432.CCR-16-2703
140. Fang Z, Zhao J, Xie W, Sun Q, Wang H, Qiao B. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. (2017) 6:2897–908. doi: 10.1002/cam4.1253
141. Feng X, Luo Q, Zhang H, Wang H, Chen W, Meng G, et al. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res. (2017) 36:81. doi: 10.1186/s13046-017-0553-x
142. Ozawa H, Ranaweera RS, Izumchenko E, Makarev E, Zhavoronkov A, Fertig EJ, et al. SMAD4 loss is associated with cetuximab resistance and induction of MAPK/JNK activation in head and neck cancer cells. Clin Cancer Res. (2017) 23:5162–75. doi: 10.1158/1078-0432.CCR-16-1686
143. Ma J, Ren Y, Zhang L, Kong X, Wang T, Shi Y, et al. Knocking-down of CREPT prohibits the progression of oral squamous cell carcinoma and suppresses cyclin D1 and c-Myc expression. PLoS ONE. (2017) 12:e0174309. doi: 10.1371/journal.pone.0174309
144. Wang Y, Zhu Y, Wang Q, Hu H, Li Z, Wang D, et al. The histone demethylase LSD1 is a novel oncogene and therapeutic target in oral cancer. Cancer Lett. (2016) 374:12–21. doi: 10.1016/j.canlet.2016.02.004
145. Tohyama R, Kayamori K, Sato K, Hamagaki M, Sakamoto K, Yasuda H, et al. Establishment of a xenograft model to explore the mechanism of bone destruction by human oral cancers and its application to analysis of role of RANKL. J Oral Pathol Med. (2016) 45:356–64. doi: 10.1111/jop.12376
146. Wang Y, Guo W, Li Z, Wu Y, Jing C, Ren Y, et al. Role of the EZH2/miR-200 axis in STAT3-mediated OSCC invasion. Int J Oncol. (2018) 52:1149–64. doi: 10.3892/ijo.2018.4293
147. Wang H, Luo Q, Feng X, Zhang R, Li J, Chen F. NLRP3 promotes tumor growth and metastasis in human oral squamous cell carcinoma. BMC Cancer. (2018) 18:500. doi: 10.1186/s12885-018-4403-9
148. Jia L, Wang J, Wu T, Wu J, Ling J, Cheng B. In vitro and in vivo antitumor effects of chloroquine on oral squamous cell carcinoma. Mol Med Rep. (2017) 16:5779–86. doi: 10.3892/mmr.2017.7342
149. Sasabe E, Tomomura A, Tomita R, Sento S, Kitamura N, Yamamoto T. Ephrin-B2 reverse signaling regulates progression and lymph node metastasis of oral squamous cell carcinoma. PLoS ONE. (2017) 12:e0188965. doi: 10.1371/journal.pone.0188965
150. Campbell KM, Lin T, Zolkind P, Barnell EK, Skidmore ZL, Winkler AE, et al. Oral cavity squamous cell carcinoma xenografts retain complex genotypes and intertumor molecular heterogeneity. Cell Rep. (2018) 24:2167–78. doi: 10.1016/j.celrep.2018.07.058
151. Su H, Luo Q, Xie H, Huang X, Ni Y, Mou Y, et al. Therapeutic antitumor efficacy of tumor-derived autophagosome. (DRibble) vaccine on head and neck cancer. Int J Nanomed. (2015) 10:1921–30. doi: 10.2147/IJN.S74204
152. Dong H, Su H, Chen L, Liu K, Hu HM, Yang W, et al. Immunocompetence and mechanism of the DRibble-DCs vaccine for oral squamous cell carcinoma. Cancer Manage Res. (2018) 10:493–501. doi: 10.2147/CMAR.S155914
153. Nagaya T, Nakamura Y, Okuyama S, Ogata F, Maruoka Y, Choyke PL, et al. Syngeneic mouse models of oral cancer are effectively targeted by anti-CD44-based NIR-PIT. Mol Cancer Res. (2017) 15:1667–77. doi: 10.1158/1541-7786.MCR-17-0333
154. Chung MK, Jung YH, Lee JK, Cho SY, Murillo-Sauca O, Uppaluri R, et al. CD271 confers an invasive and metastatic phenotype of head and neck squamous cell carcinoma through the upregulation of slug. Clin Cancer Res. (2018) 24:674–83. doi: 10.1158/1078-0432.CCR-17-0866
155. Judd NP, Allen CT, Winkler AE, Uppaluri R. Comparative analysis of tumor-infiltrating lymphocytes in a syngeneic mouse model of oral cancer. Otolaryngol Head Neck Surg. (2012) 147:493–500. doi: 10.1177/0194599812442037
156. Adachi M, Mizuno-Kamiya M, Takayama E, Kawaki H, Inagaki T, Sumi S, et al. Gene expression analyses associated with malignant phenotypes of metastatic sub-clones derived from a mouse oral squamous cell carcinoma Sq-1979 cell line. Oncol Lett. (2018) 15:3350–6. doi: 10.3892/ol.2017.7648
157. Moroishi T, Hayashi T, Pan WW, Fujita Y, Holt MV, Qin J, et al. The hippo pathway kinases LATS1/2 suppress cancer immunity. Cell. (2016) 167:1525–39e17. doi: 10.1016/j.cell.2016.11.005
158. Chen YF, Chang KW, Yang IT, Tu HF, Lin SC. Establishment of syngeneic murine model for oral cancer therapy. Oral Oncol. (2019) 95:194–201. doi: 10.1016/j.oraloncology.2019.06.026
159. Chen YF, Liu CJ, Lin LH, Chou CH, Yeh LY, Lin SC, et al. Establishing of mouse oral carcinoma cell lines derived from transgenic mice and their use as syngeneic tumorigenesis models. BMC Cancer. (2019) 19:281. doi: 10.1186/s12885-019-5486-7
160. De La Rochere P, Guil-Luna S, Decaudin D, Azar G, Sidhu SS, Piaggio E. Humanized mice for the study of immuno-oncology. Trends Immunol. (2018) 39:748–63. doi: 10.1016/j.it.2018.07.001
161. Walsh NC, Kenney LL, Jangalwe S, Aryee KE, Greiner DL, Brehm MA, et al. Humanized mouse models of clinical disease. Annu Rev Pathol. (2017) 12:187–215. doi: 10.1146/annurev-pathol-052016-100332
162. Morton JJ, Bird G, Keysar SB, Astling DP, Lyons TR, Anderson RT, et al. XactMice: humanizing mouse bone marrow enables microenvironment reconstitution in a patient-derived xenograft model of head and neck cancer. Oncogene. (2016) 35:290–300. doi: 10.1038/onc.2015.94
163. Morton JJ, Keysar SB, Perrenoud L, Chimed TS, Reisinger J, Jackson B, et al. Dual use of hematopoietic and mesenchymal stem cells enhances engraftment and immune cell trafficking in an allogeneic humanized mouse model of head and neck cancer. Mol Carcinog. (2018) 57:1651–63. doi: 10.1002/mc.22887
164. Morton JJ, Bird G, Refaeli Y, Jimeno A. Humanized mouse xenograft models: narrowing the tumor-microenvironment gap. Cancer Res. (2016) 76:6153–8. doi: 10.1158/0008-5472.CAN-16-1260
165. Greenblatt MB, Vrbanac V, Tivey T, Tsang K, Tager AM, Aliprantis AO. Graft versus host disease in the bone marrow, liver and thymus humanized mouse model. PLoS ONE. (2012) 7:e44664. doi: 10.1371/journal.pone.0044664
166. Sathish N, Wang X, Yuan Y. Human papillomavirus. (HPV)-associated oral cancers and treatment strategies. J Dental Res. (2014) 93(7 Suppl):29–36s. doi: 10.1177/0022034514527969
167. zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. (2002) 2:342–50. doi: 10.1038/nrc798
168. Ghittoni R, Accardi R, Hasan U, Gheit T, Sylla B, Tommasino M. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes. (2010) 40:1–13. doi: 10.1007/s11262-009-0412-8
169. Brand TM, Hartmann S, Bhola NE, Peyser ND, Li H, Zeng Y, et al. Human papillomavirus regulates HER3 expression in head and neck cancer: implications for targeted HER3 therapy in HPV(+) patients. Clin Cancer Res. (2017) 23:3072–83. doi: 10.1158/1078-0432.CCR-16-2203
170. Keysar SB, Astling DP, Anderson RT, Vogler BW, Bowles DW, Morton JJ, et al. A patient tumor transplant model of squamous cell cancer identifies PI3K inhibitors as candidate therapeutics in defined molecular bins. Mol Oncol. (2013) 7:776–90. doi: 10.1016/j.molonc.2013.03.004
171. Facompre ND, Sahu V, Montone KT, Harmeyer KM, Nakagawa H, Rustgi AK, et al. Barriers to generating PDX models of HPV-related head and neck cancer. Laryngoscope. (2017) 127:2777–83. doi: 10.1002/lary.26679
172. Taberna M, Mena M, Pavon MA, Alemany L, Gillison ML, Mesia R. Human papillomavirus-related oropharyngeal cancer. Annal Oncol. (2017) 28:2386–98. doi: 10.1093/annonc/mdx304
173. Hoover AC, Spanos WC, Harris GF, Anderson ME, Klingelhutz AJ, Lee JH. The role of human papillomavirus 16 E6 in anchorage-independent and invasive growth of mouse tonsil epithelium. Arch Otolaryngol. (2007) 133:495–502. doi: 10.1001/archotol.133.5.495
174. Williams R, Lee DW, Elzey BD, Anderson ME, Hostager BS, Lee JH. Preclinical models of HPV+ and HPV- HNSCC in mice: an immune clearance of HPV+ HNSCC. Head Neck. (2009) 31:911–8. doi: 10.1002/hed.21040
175. Mermod M, Hiou-Feige A, Bovay E, Roh V, Sponarova J, Bongiovanni M, et al. Mouse model of postsurgical primary tumor recurrence and regional lymph node metastasis progression in HPV-related head and neck cancer. Int J Cancer. (2018) 142:2518–28. doi: 10.1002/ijc.31240
176. Paolini F, Massa S, Manni I, Franconi R, Venuti A. Immunotherapy in new pre-clinical models of HPV-associated oral cancers. Hum Vaccines Immunother. (2013) 9:534–43. doi: 10.4161/hv.23232
177. Ishida K, Tomita H, Nakashima T, Hirata A, Tanaka T, Shibata T, et al. Current mouse models of oral squamous cell carcinoma: genetic and chemically induced models. Oral Oncol. (2017) 73:16–20. doi: 10.1016/j.oraloncology.2017.07.028
178. Walrath JC, Hawes JJ, Van Dyke T, Reilly KM. Genetically engineered mouse models in cancer research. Adv Cancer Res. (2010) 106:113–64. doi: 10.1016/S0065-230X(10)06004-5
179. Lampreht Tratar U, Horvat S, Cemazar M. Transgenic mouse models in cancer research. Front Oncol. (2018) 8:268. doi: 10.3389/fonc.2018.00268
180. Bian Y, Hall B, Sun ZJ, Molinolo A, Chen W, Gutkind JS, et al. Loss of TGF-β signaling and PTEN promotes head and neck squamous cell carcinoma through cellular senescence evasion and cancer-related inflammation. Oncogene. (2012) 31:3322–32. doi: 10.1038/onc.2011.494
181. Opitz OG, Harada H, Suliman Y, Rhoades B, Sharpless NE, Kent R, et al. A mouse model of human oral-esophageal cancer. J Clin Invest. (2002) 110:761–9. doi: 10.1172/JCI0215324
182. Li Z, Gonzalez CL, Wang B, Zhang Y, Mejia O, Katsonis P, et al. Cdkn2a suppresses metastasis in squamous cell carcinomas induced by the gain-of-function mutant p53(R172H). J Pathol. (2016) 240:224–34. doi: 10.1002/path.4770
183. Ambatipudi S, Bhosale PG, Heath E, Pandey M, Kumar G, Kane S, et al. Downregulation of keratin 76 expression during oral carcinogenesis of human, hamster and mouse. PLoS ONE. (2013) 8:e70688. doi: 10.1371/journal.pone.0070688
184. Chen W, Kang KL, Alshaikh A, Varma S, Lin YL, Shin KH, et al. Grainyhead-like 2. (GRHL2) knockout abolishes oral cancer development through reciprocal regulation of the MAP kinase and TGF-β signaling pathways. Oncogenesis. (2018) 7:38. doi: 10.1038/s41389-018-0047-5
185. Caulin C, Nguyen T, Longley MA, Zhou Z, Wang XJ, Roop DR. Inducible activation of oncogenic K-ras results in tumor formation in the oral cavity. Cancer Res. (2004) 64:5054–8. doi: 10.1158/0008-5472.CAN-04-1488
186. El-Bayoumy K, Chen KM, Zhang SM, Sun YW, Amin S, Stoner G, et al. Carcinogenesis of the oral cavity: environmental causes and potential prevention by black raspberry. Chem Res Toxicol. (2017) 30:126–44. doi: 10.1021/acs.chemrestox.6b00306
187. Frijhoff AF, Conti CJ, Senderowicz AM. Advances in molecular carcinogenesis: current and future use of mouse models to screen and validate molecularly targeted anticancer drugs. Mol Carcinogenesis. (2004) 39:183–94. doi: 10.1002/mc.20013
188. Munirajan AK, Mohanprasad BK, Shanmugam G, Tsuchida N. Detection of a rare point mutation at codon 59 and relatively high incidence of H-ras mutation in Indian oral cancer. Int J Oncol. (1998) 13:971–4. doi: 10.3892/ijo.13.5.971
189. Saranath D, Chang SE, Bhoite LT, Panchal RG, Kerr IB, Mehta AR, et al. High frequency mutation in codons 12 and 61 of H-ras oncogene in chewing tobacco-related human oral carcinoma in India. Br J Cancer. (1991) 63:573–8. doi: 10.1038/bjc.1991.133
190. Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. (2018) 18:269–82. doi: 10.1038/nrc.2018.11
191. Ma Y, Zhang L, Huang X. Building Cre knockin rat lines using CRISPR/Cas9. Methods Mol Biol. (2017) 1642:37–52. doi: 10.1007/978-1-4939-7169-5_3
192. Hubbard EJ. FLP/FRT and Cre/lox recombination technology in C. elegans. Methods. (2014) 68:417–24. doi: 10.1016/j.ymeth.2014.05.007
193. Kellendonk C, Tronche F, Monaghan AP, Angrand PO, Stewart F, Schutz G. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. (1996) 24:1404–11. doi: 10.1093/nar/24.8.1404
194. Chanphai P, Thomas TJ, Tajmir-Riahi HA. Design of functionalized folic acid-chitosan nanoparticles for delivery of tetracycline, doxorubicin, and tamoxifen. J Biomol Struct Dyn. (2019) 37:1000–6. doi: 10.1080/07391102.2018.1445559
195. Dominguez-Monedero A, Davies JA. Tamoxifen- and mifepristone-inducible versions of CRISPR effectors, Cas9 and Cpf1. ACS Synthet Biol. (2018) 7:2160–9. doi: 10.1021/acssynbio.8b00145
196. Nakagawa H, Inomoto T, Rustgi AK. A CACCC box-like cis-regulatory element of the Epstein-Barr virus ED-L2 promoter interacts with a novel transcriptional factor in tissue-specific squamous epithelia. J Biol Chem. (1997) 272:16688–99. doi: 10.1074/jbc.272.26.16688
197. Zhong R, Pytynia M, Pelizzari C, Spiotto M. Bioluminescent imaging of HPV-positive oral tumor growth and its response to image-guided radiotherapy. Cancer Res. (2014) 74:2073–81. doi: 10.1158/0008-5472.CAN-13-2993
198. Carper MB, Troutman S, Wagner BL, Byrd KM, Selitsky SR, Parag-Sharma K, et al. An Immunocompetent mouse model of HPV16(+) head and neck squamous cell carcinoma. Cell Rep. (2019) 29:1660–74.e7. doi: 10.1016/j.celrep.2019.10.005
199. Chen YF, Yang CC, Kao SY, Liu CJ, Lin SC, Chang KW. MicroRNA-211 enhances the oncogenicity of carcinogen-induced oral carcinoma by repressing TCF12 and increasing antioxidant activity. Cancer Res. (2016) 76:4872–86. doi: 10.1158/0008-5472.CAN-15-1664
200. Tseng SH, Yang CC, Yu EH, Chang C, Lee YS, Liu CJ, et al. K14-EGFP-miR-31 transgenic mice have high susceptibility to chemical-induced squamous cell tumorigenesis that is associating with Ku80 repression. Int J Cancer. (2015) 136:1263–75. doi: 10.1002/ijc.29106
201. da Silva JM, Moreira Dos Santos TP, Sobral LM, Queiroz-Junior CM, Rachid MA, Proudfoot AEI, et al. Relevance of CCL3/CCR5 axis in oral carcinogenesis. Oncotarget. (2017) 8:51024–36. doi: 10.18632/oncotarget.16882
202. de Oliveira Santos D, Loyola AM, Cardoso SV, Chammas R, Liu FT, de Faria PR. Hedgehog signaling pathway mediates tongue tumorigenesis in wild-type mice but not in Gal3-deficient mice. Exp Mol Pathol. (2014) 97:332–7. doi: 10.1016/j.yexmp.2014.09.018
Keywords: mouse models, OSCC, chemical carcinogen-induced, transplanted, xenograft, syngeneic, HPV, genetically engineered models
Citation: Li Q, Dong H, Yang G, Song Y, Mou Y and Ni Y (2020) Mouse Tumor-Bearing Models as Preclinical Study Platforms for Oral Squamous Cell Carcinoma. Front. Oncol. 10:212. doi: 10.3389/fonc.2020.00212
Received: 23 September 2019; Accepted: 06 February 2020;
Published: 25 February 2020.
Edited by:
Christian Simon, Lausanne University Hospital (CHUV), SwitzerlandReviewed by:
Ruud Brakenhoff, VU University Medical Center, NetherlandsVincent Roh, Lausanne University Hospital (CHUV), Switzerland
Copyright © 2020 Li, Dong, Yang, Song, Mou and Ni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongbin Mou, yongbinmou@163.com; Yanhong Ni, niyanhong12@163.com
†These authors have contributed equally to this work
 Qiang Li
Qiang Li Heng Dong
Heng Dong Guangwen Yang
Guangwen Yang Yuxian Song
Yuxian Song Yongbin Mou
Yongbin Mou Yanhong Ni
Yanhong Ni