- 1Gujarat Biotechnology Research Centre, Department of Science and Technology, Government of Gujarat, Gandhinagar, India
- 2Gujarat Cancer Research Institute, Civil Hospital, Ahmedabad, India
- 3Clinical Genetics, ONE-Centre for Rheumatology and Genetics, Vadodara, India
Background: Breast and ovarian cancers are the most prevalent cancers and one of the leading causes of death in Indian women. The healthcare burden of breast and ovarian cancers and the rise in mortality rate are worrying and stress the need for early detection and treatment.
Methods: We performed amplicon sequencing of 144 cases who had breast/ovarian cancer disease (total 137 cases are patients and seven are tested for BRCA1/2 carrier) Using our custom designed gene panel consisting of 14 genes, that are associated with high to moderate risk of breast and ovarian cancers. Variants were called using Torrent Variant Caller and were annotated using ThermoFisher’s Ion Reporter software. Classification of variants and their clinical significance were identified by searching the variants against ClinVar database.
Results: From a total of 144 cases, we were able to detect 42 pathogenic mutations in [40/144] cases. Majority of pathogenic mutations (30/41) were detected in BRCA1 gene, while (7/41) pathogenic mutations were detected in BRCA2 gene, whereas, (2/41) pathogenic mutations were detected in TP53 gene and BRIP1, PALB2, and ATM genes respectively. So, BRCA genes contributed 88.09% of pathogenic mutations, whereas non-BRCA genes contributed 11.91% of pathogenic mutations. We were also able to detect 25 VUS which were predicted to be damaging by in silico prediction tools.
Conclusion: Early detection of cancers in the Indian population can be done by genetic screening using customized multi-gene panels. Indications of our findings show that in the Indian population, apart from the common BRCA genes, there are other genes that are also responsible for the disease. High frequency mutations detected in the study and variants of uncertain significance predicted to be damaging by in silico pathogenicity prediction tools can be potential biomarkers of hereditary breast and ovarian cancer in Indian HBOC patients.
Introduction
Breast and ovarian cancer are the most prevalent cancer and leading cause of death in Indian women. The incidence of breast cancer cases in India in 2018 was estimated to be 162,468, which is 27.6% of all cancer cases in females. Whereas, for ovarian cancers, it was estimated to be 36,170, which is 6.2% of all cancer cases in females (1). Gujarat ranks first in deaths due to breast cancer and seventh in deaths due to ovarian cancer among other states of India (2). Moreover, the ratio of number of deaths to new cases in case of breast cancer is increased to 56.30% (87090/162648) in India which is much higher as compared to F.6% (131,347/458,718) in Europe and 19% (48,850/256,222) in the US (1, 3, 4). In case of ovarian cancer burden in India, the ratio of number of deaths to new cases is 66.39% (24015/36170) which is also higher in comparison to Europe 65.77% (44576/67771) and US 57.24% (14008/24469) (1, 3, 4).
Hereditary breast and ovarian cancer (HBOC) is an inherited disorder in which the risk of breast and ovarian cancers is higher than normal BOC. About 5–10% of breast cancers and 10–15% of ovarian cancers can be attributed to HBOC (5). HBOC is characterized by bilateral cancer with a family history of breast or ovarian cancer in relatives. BRCA1 and BRCA2 (BRCA1/2) genes are maximally associated with predisposition to HBOC; however, in addition to the BRCA1/2 genes, the National Comprehensive Cancer Network (NCCN) guidelines have been expanded to incorporate non-BRCA genes into gene panels for increased medical management (6). According to the 2014 NCCN guidelines for multi-gene testing, women with mutations in the non-BRCA genes, such as TP53, PTEN, CDH1, ATM, CHEK2, or PALB2 tumor suppressor genes and others also have an increased risk of breast and ovarian cancer (2, 6). In one of the studies, in individuals who underwent multigene testing, the researchers found that 3.8% of BRCA1/2 mutation-negative individuals harbored deleterious mutations in other hereditary cancer predisposition genes. 40–50% more individuals with mutations in these cancer susceptibility genes could be identified by multigene panel testing as compared to testing for BRCA1 and BRCA2 genes only (7). A high rate of mortality in India due to cancers is mainly because of late diagnosis and delayed treatment (8). Thus, it is suggested that non-BRCA genes must also be tested so that deleterious non-BRCA mutations can also be covered and presentation of HBOC related mutations in Indian population can be achieved. This would enable us to gather clinically useful data for HBOC risk assessment and diagnosis in Indian population. For this, gene panels that cover both BRCA1/2 and non-BRCA genes should be incorporated so that a detailed picture of mutations contributed by both BRCA and non-BRCA genes can be obtained (2, 9–12). In our study we used 14 gene panels, in accordance with 2014 NCCN guidelines and other literature, for determining the contribution of BRCA and non-BRCA genes in HBOC in the Indian population (6, 13). We also sought to find out mutations specific/prevalent to the Indian cohort, so that they can be used as hotspots for readily detecting HBOC in India. For this, we used next generation sequencing (amplicon sequencing) based approach to screen 14 high to moderate risk genes associated with breast and ovarian cancers in a cohort of 144 Indians, related and unrelated individuals, with at least a 1st degree family history of breast and/or ovarian cancer (6).
The percentage of detection of pathogenic mutations in non-BRCA genes in patients with breast and ovarian cancers is 17.64%, which indicates that, testing of non-BRCA genes in combination of BRCA1/2 genes is equally necessary for a complete risk assessment and for developing a more accurate method for early detection, diagnosis and treatment (14, 15).
Materials and Methods
Patient Selection
The study was conducted and approved in accordance with the Gujarat Cancer Research Institute’s Institutional Review Board Committee at GCRI, Civil Hospital, Ahmedabad, Gujarat, India. Patients undergoing treatment or treated earlier and having a family history of breast or ovary cancer in the first or second degree relative were selected for the study. Inclusion of patients was made according to the ICMR guidelines (16). Written informed consent was also obtained from each participant of the study. Clinical and pathologic details were retrieved from the medical records. Genetic counseling of breast and ovary cancer patients was performed prior to patient’s selection, through cancer camps held at Gujarat Cancer and Research Institute. Out of 144 cases, 108 patients were having breast cancer, and 23 patients were having ovarian cancer, three patients were having breast and cervical cancer whereas three patients had breast and ovarian cancer. Seven carrier cases were enrolled as relatives of two different patients Figure 1. Majority of the patients selected (64/144) were below 45 years with a mean age of 45 years at the time of diagnosis Figure 2. Out of 144 cases, 18 cases were from six different families; hence they were interrelated, and 126 cases were unrelated individuals from singular families.
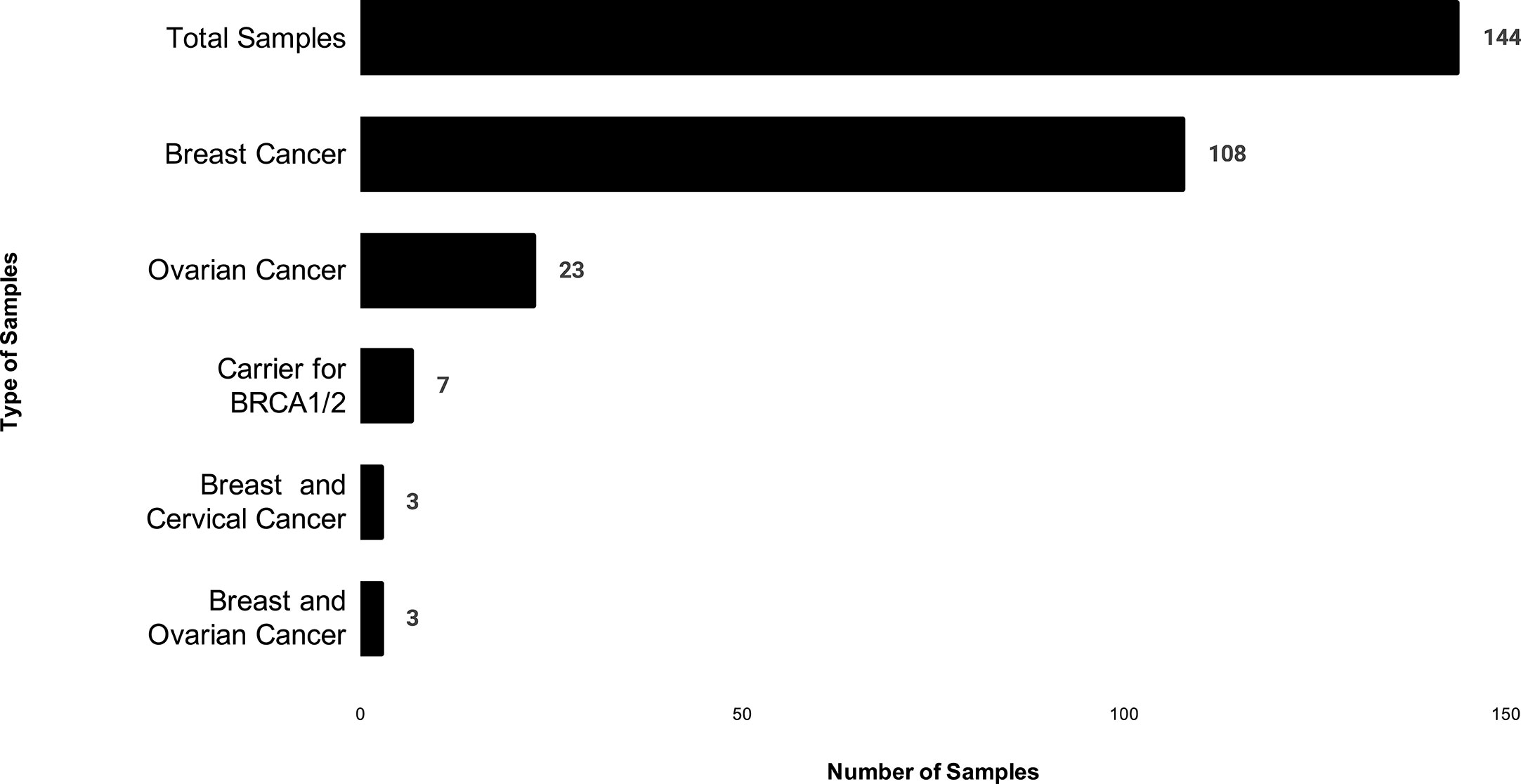
Figure 1 The distribution of patients based on cancer type in which X-axis shows number of patients, and Y- axis shows type of cancer in our study.
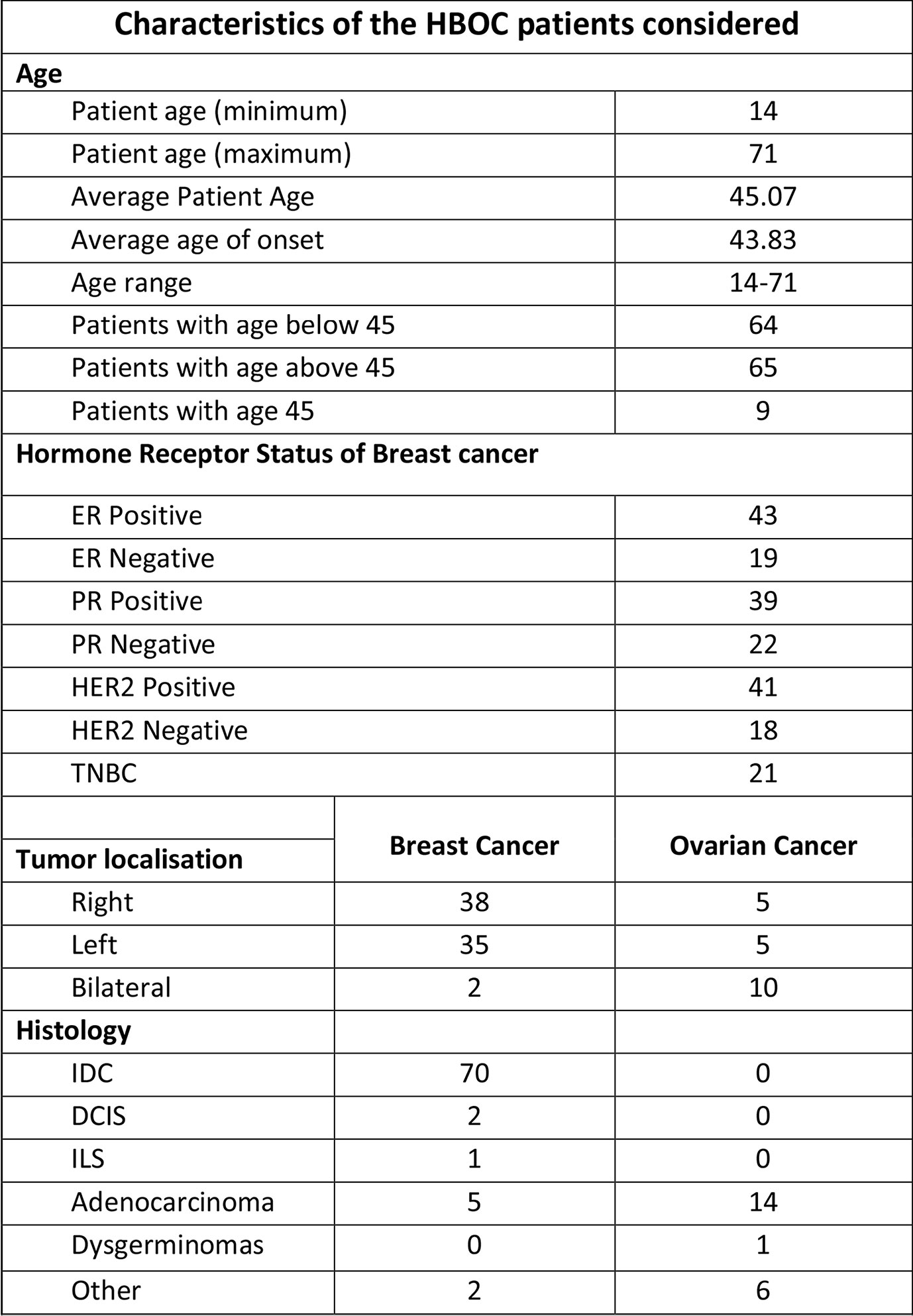
Figure 2 Characteristics of HBOC patients considered for the study including patients’ age, tumor localization and histology.
Amplicon Sequencing
Customized Multi-Gene Panel
In this study, a custom Ion AmpliSeq™ Panel of 14 HBOC associated high-penetrance and moderate risk genes as mentioned in the NCCN guidelines and other literature was considered for mutational profiling of Indian HBOC patients (6, 13). The 14 HBOC genes, BRCA1 (NM_007300.3), BRCA2 (NM_000059.3), TP53 (NM_000546.4), PTEN (NM_000314.4), CDH1 (NM_004360.3), STK11 (NM_000455.4), ATM (NM_000051.3), BARD1 (NM_000465.3), BRIP1 (NM_032043.2), CHEK2 (NM_007194.3), ERBB2 (NM_004448.3), NBN (NM_002485.4), PALB2 (NM_024675.3), RAD51C (NM_058216.1), were sequenced using 965 amplicons in two pools.
Sample Preparation
Genomic DNA was extracted from blood samples using QIAamp DNA Blood Mini kit (QIAGEN, Germany). The concentration of DNA was determined using Qubit Fluorometer 2.0 and 4.0®(ThermoFisher), and purity of DNA was determined using QIAxpert (QIAGEN).
Sequencing
Multiplex PCR was performed using 50–100 ng genomic DNA with a premixed primer pool and Ion AmpliSeq™ HiFi master mix (Ion AmpliSeq™ Library Kit 2.0) for 2 min at 99°C, followed by 16 cycles of 99°C for 15 s and 60°C for 4 min, ending with a holding period at 10°C. The PCR amplicons were treated with 2 µl FuPa reagent to partially digest primer sequences and phosphorylate the amplicons at 50°C for 10 min, followed by 55°C for 10 min, then 60°C for 20 min. The amplicons were ligated to adapters with the diluted barcodes of the Ion Xpress™ Barcode Adapters kit (ThermoFisher) for 30 min at 22°C then 68°C for 5 min followed by 72°C for 5 min. Adaptor ligated amplicon libraries were purified using Agencourt® AMPure® XP reagents (Beckman Coulter, Tokyo, Japan). The library concentration was determined using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), then each library was diluted to 100 pmol, and the same amount of libraries was pooled for one sequencing reaction. Next, emulsion PCR was carried out using the Ion OneTouch™ 2 System (ThermoFisher) according to the manufacturer’s instructions. Template-positive Ion Sphere™ Particles were then enriched with Dynabeads® MyOne™ Streptavidin C1 Beads (ThermoFisher) using an Ion OneTouch™ ES system (ThermoFisher). Massively parallel sequencing was carried out on Ion Proton and Ion S5™ using the Ion Proton Sequencing 200 Kit and Ion S5 540 and Ion S5 520 and 530 OT2 and sequencing kit according to the manufacturer’s instructions.
Data Processing
The raw sequence data was processed using standard Ion Torrent Suite™ Software running on the Torrent Server. Raw signal data were analyzed using Torrent Suite™. The pipeline includes signal processing, base calling, quality score assignment, adapter trimming, PCR duplicate removal, read alignment to human genome 19 reference (hg19), quality control of mapping quality, coverage analysis Figure 2. Identification of sequence variants was carried out via Torrent Variant Caller Plugin software (ThermoFisher), and coverage of each amplicon was obtained by the Coverage Analysis Plugin software (ThermoFisher). The variant call parameter setting was germline high stringency. Mutations with low quality, occurring intronic regions were discarded from the datasets. Following this, annotation of the remaining high quality single-nucleotide variants, insertions, deletions, and splice site alterations was performed using Ion Reporter™ Server System (ThermoFisher). Mutations with known clinical significance were identified by searching variants against ClinVar database. Functional consequences of variants which are novel or which were under the category of variants of uncertain significance or those which could not be classified as pathogenic or likely pathogenic by prior reports were assessed using 17 in silico pathogenicity prediction tools which were incorporated in the search engine named Varsome: The Human Genomics Community (17).
Variant Classification
Classification of variants was done according to the American College of Medical Genetics and Genomics (ACMG) recommendations for standard interpretation and reporting of sequence variations (18). The variants were classified into five categories, such as pathogenic, likely pathogenic, variant of uncertain significance (VUS), likely benign, and benign.
Mutational Pattern of Indian Cohort Assessment
In order to check the differences in mutational pattern of Indian HBOC patients with that of other populations, we compared the minor allele frequencies of clinically significant mutations from the Indian HBOC patients with that of South Asian, European (Finnish and non-Finnish), Ashkenazi, Latino, East Asian, and African populations using the GnomAD browser (19).
Results
Amplicon sequencing targeting 14 genes in 144 samples generated 42.76 GB of sequences with 428 million high quality sequencing reads. Mean sequencing depth of high quality reads was 3,602×, and >95% reads were mapped on to the target regions. Alignment of sequencing reads was done on to the targeted regions of reference genome hg19 which produced a total of 7,626 SNVs and INDELs in 144 samples, of which 3,488 variants were observed to be occurring in exonic region, 3,936 in intronic region, 11 in splice sites, and 191 in untranslated regions. Introns, synonymous variants, and variants below 100× coverage were removed to finally obtain 1,770 variants.
Distribution of Variants in Patients Across 14 Genes
To start with, we constructed a heat map to obtain the distribution of Variants among all 144 cases across all 14 genes included in the multi-gene panel. Mutations were detected in all 14 genes tested: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EBB2, NBN, PALB2, PTEN, RAD51C, STK11, TP53. Distribution of variants revealed that the majority 30.50% was in the BRCA1/2 genes as compared to the 12 non-BRCA genes altogether 69.5%. Of the BRCA mutations, the majority 14.96% were detected in BRCA1 and 15.53% of mutations were detected in BRCA2. In the non-BRCA genes, the mutation detection rate was as follows: NBN [12.20%], ATM (11.36%), BARD1 (10.11%), ERBB2 (8.14%), BRIP1 (6.81%), TP53 (6.74%), STK11 (4.31%), CDH1 (4.06%), CHEK2 (1.84%), PALB2 (1.23%), PTEN (1.04%) and RAD51C [1.57%] Figure 3.
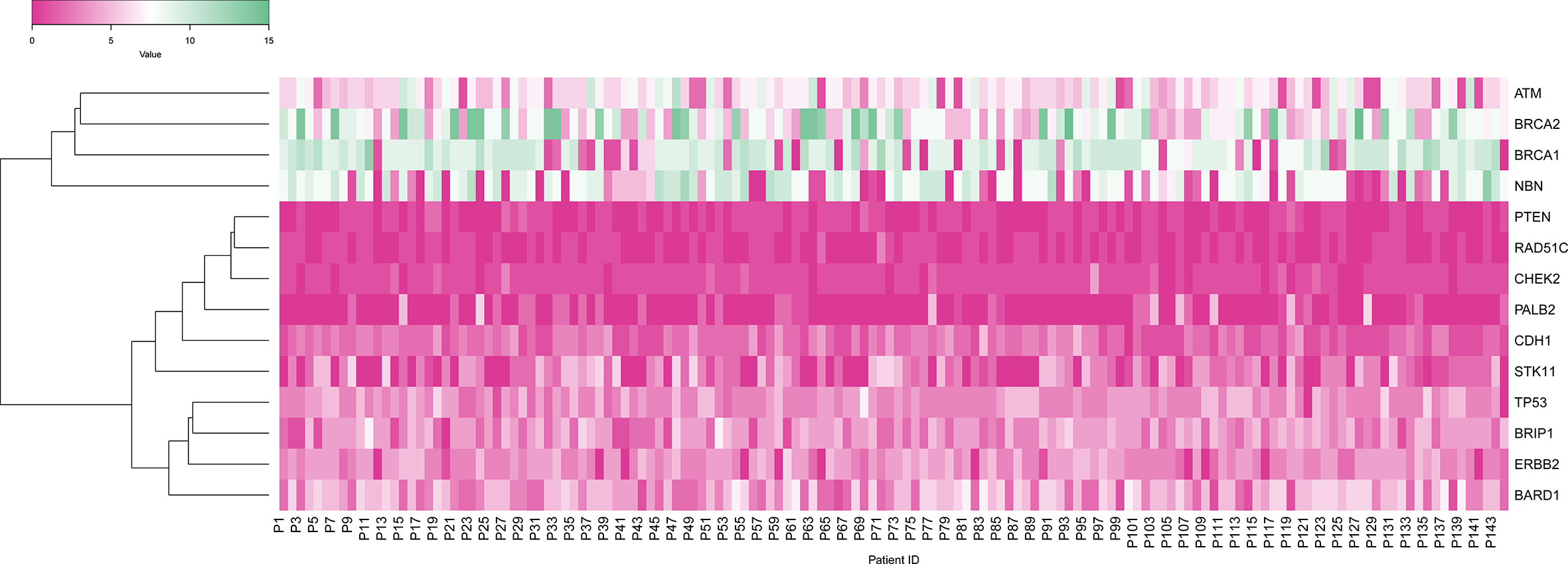
Figure 3 Heat map showing gene-wise distribution of variants in 144 patients in which X-axis shows patients, and Y-axis shows the gene variants.
Categorization of All Mutations Into ClinVar Categories
After filtration, 1,770 variants obtained were classified based on searching each variant against the ClinVar database, and according to their clinical significance, the variants were categorized into different categories, such as, Pathogenic variants, Likely Pathogenic variants, Variants of Uncertain Significance and Benign/Likely benign variants. 2.37% variants were placed into the category of Pathogenic variants, 1.41% variants were placed into the category of variants of uncertain significance (VUS), 96.29% variants were placed into the category of Benign/Likely benign variants.
Pathogenic Mutations
From total 42 pathogenic mutations, 30 (71.42%) mutations were detected in BRCA1 gene, seven (16.66%) mutations were detected in BRCA2 gene, two (4.76%) mutations were detected in TP53 gene and one (2.38%) mutation each was detected in PALB2, BRIP1, and ATM genes respectively Figures 4A, B. The details of the mutations with clinical information of patients have been summarized in the Table 1. One pathogenic mutation c.5137+1G>A in BRCA1 gene was found to be present in 7/144 cases. Other two pathogenic mutations, which are c.5098delC and c.68_69delAG in BRCA1 gene, were found to be present in 4/144 and 3/144 cases respectively. Moreover, five pathogenic mutations, which are, c.1016delA in BRCA1 gene, c.1907C>G in BRCA2 gene, c.3331C>T in BRCA1 gene, c.5566C>T in BRCA1 gene and c.682-2A>G in BRCA2 gene were found to be present in 2/144 cases. Whereas, other pathogenic mutations were found to be present in 1/144 cases, Table 1. While in case of 18 cases from total six different families, pathogenic mutation was detected in five families. Three relatives of a cancer patient (P24) were found to have the same pathogenic mutation in in BRCA1 (c.5098delC) gene as the patient, and thus they might show symptoms of HBOC in future. None of the four relatives of other cancer patients (P8) were found to have any HBOC related mutations suggesting that they did not inherit the mutations. While from two sisters, P4 and P5, pathogenic mutation was only detected in BRCA1 (c.1016delA) in patient P5. Moreover, in the screening of the other two sisters P41 and P42, the same pathogenic mutation in BRCA2 (c.1907C>G) was identified. Further in two relatives of the patient P100, pathogenic mutation was detected in P136 relative in BRCA1 (c.5137+1G>A) gene, the same as patient P100.
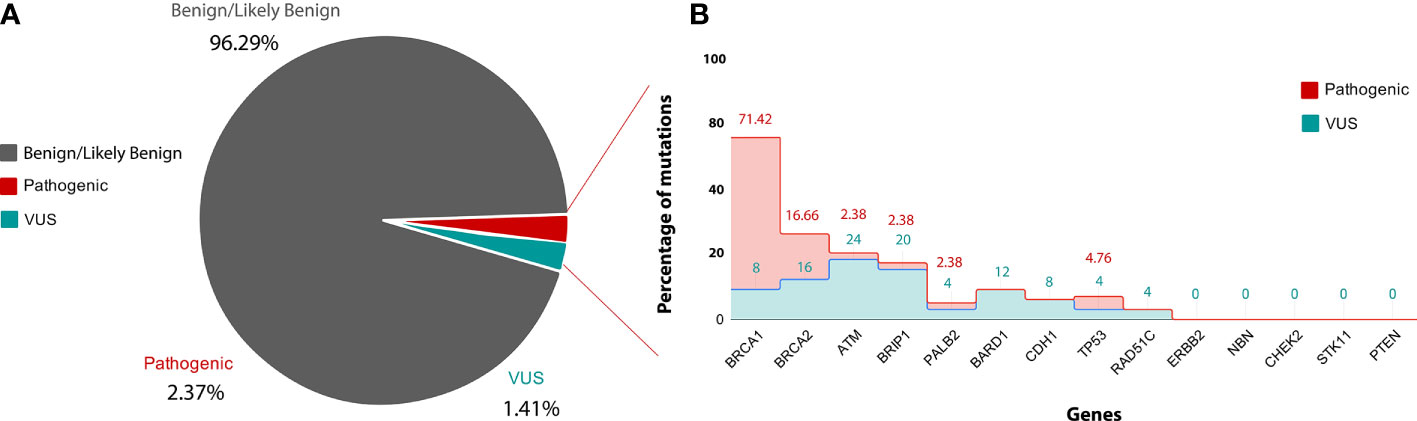
Figure 4 (A) is showing classification of variants into clinvar categories: Benign/likely Benign (Grey), VUS (blue) and pathogenic (red) according to ACMG guideline. (B) is showing gene-wise percentage of pathogenic and VUS mutation. X-axis showing name of genes used in the study and Y-axis showing percentage of pathogenic (red) and VUS (blue) mutation.
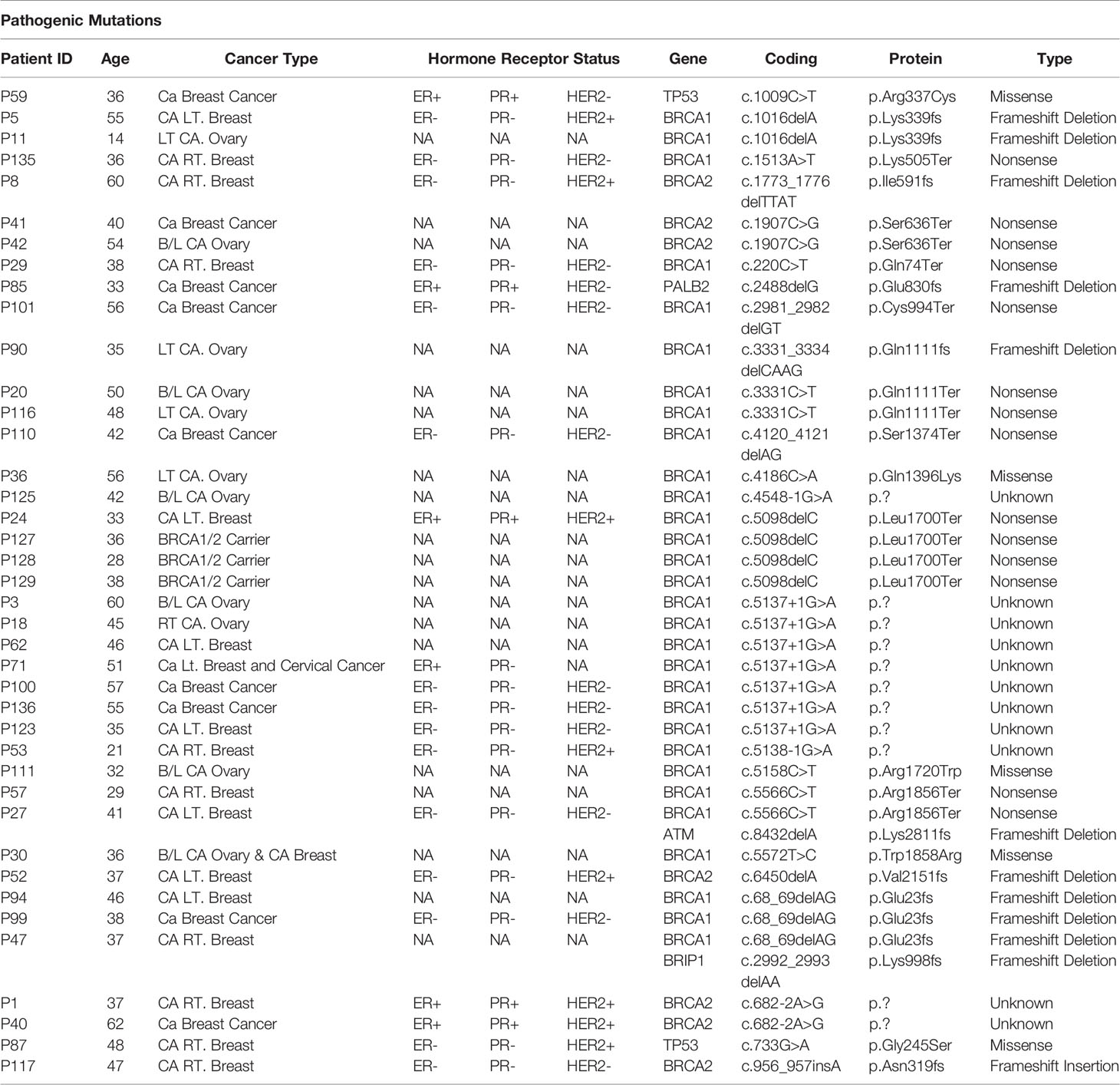
Table 1 Showing the details of ClinVar pathogenic mutations and clinical details of patients in the study.
VUS
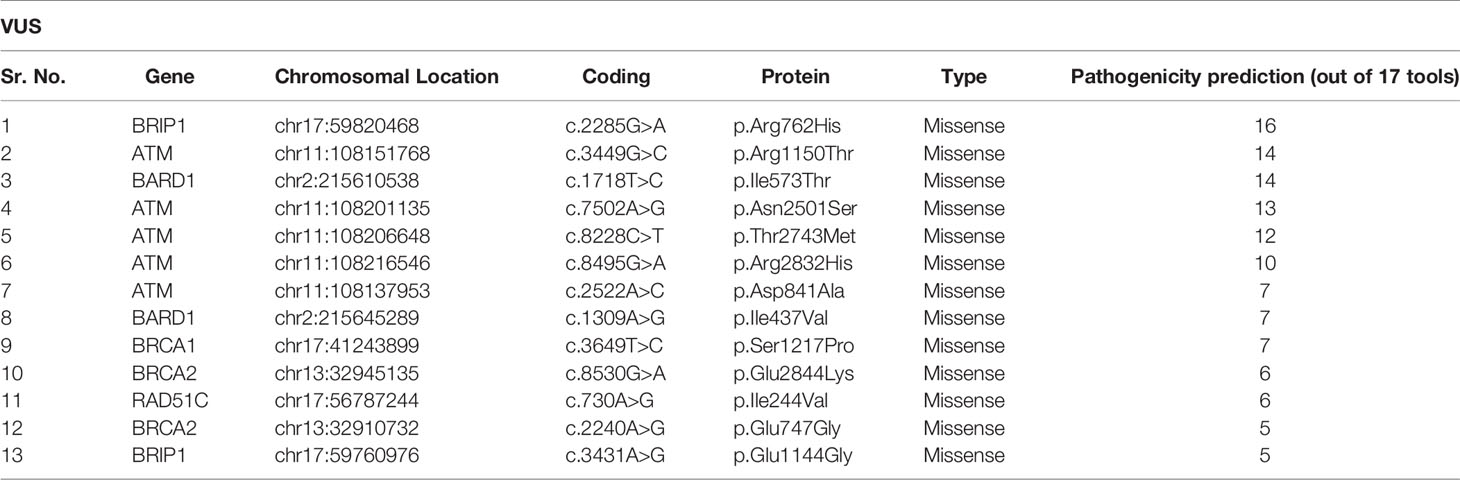
In this study, we also detected 25 missense variants Table 2 that are clinically placed in the category of “variants of uncertain significance-VUS” by ClinVar (20). Clinical significance of these VUS was evaluated by Varsome which subjects each mutation to 17 in silico pathogenicity prediction tools including Mutation Taster®, Mutation Assessor, FATHMM, FATHMM-MKL, FATHMM-XF, LRT, Eigen, Eigen PC, SIFT, SIFT 4G, PROVEAN, MVP, REVEL, PrimateAI, MetaSVM, MetalR, and DANN. Varsome is the human genome variant search engine for prediction of pathogenicity by multiple tools (17). Out of these 25 VUS, c.2285G>A in BRIP1 was predicted to be pathogenic by 16 of the 17 pathogenicity prediction tools, c.3449G>C in ATM and c.1718T>C in BARD1 were predicted to be pathogenic by 14 of the 17 pathogenicity prediction tools, c.7502A>G in ATM was predicted to be pathogenic by 13 of the 17 tools, while c.8228C>T in ATM was predicted to be pathogenic by 12 out of the 17 tools, c.8495G>A and c.2522A>C in ATM was predicted to be pathogenic by 10 and 7 tools respectively Figure 5.
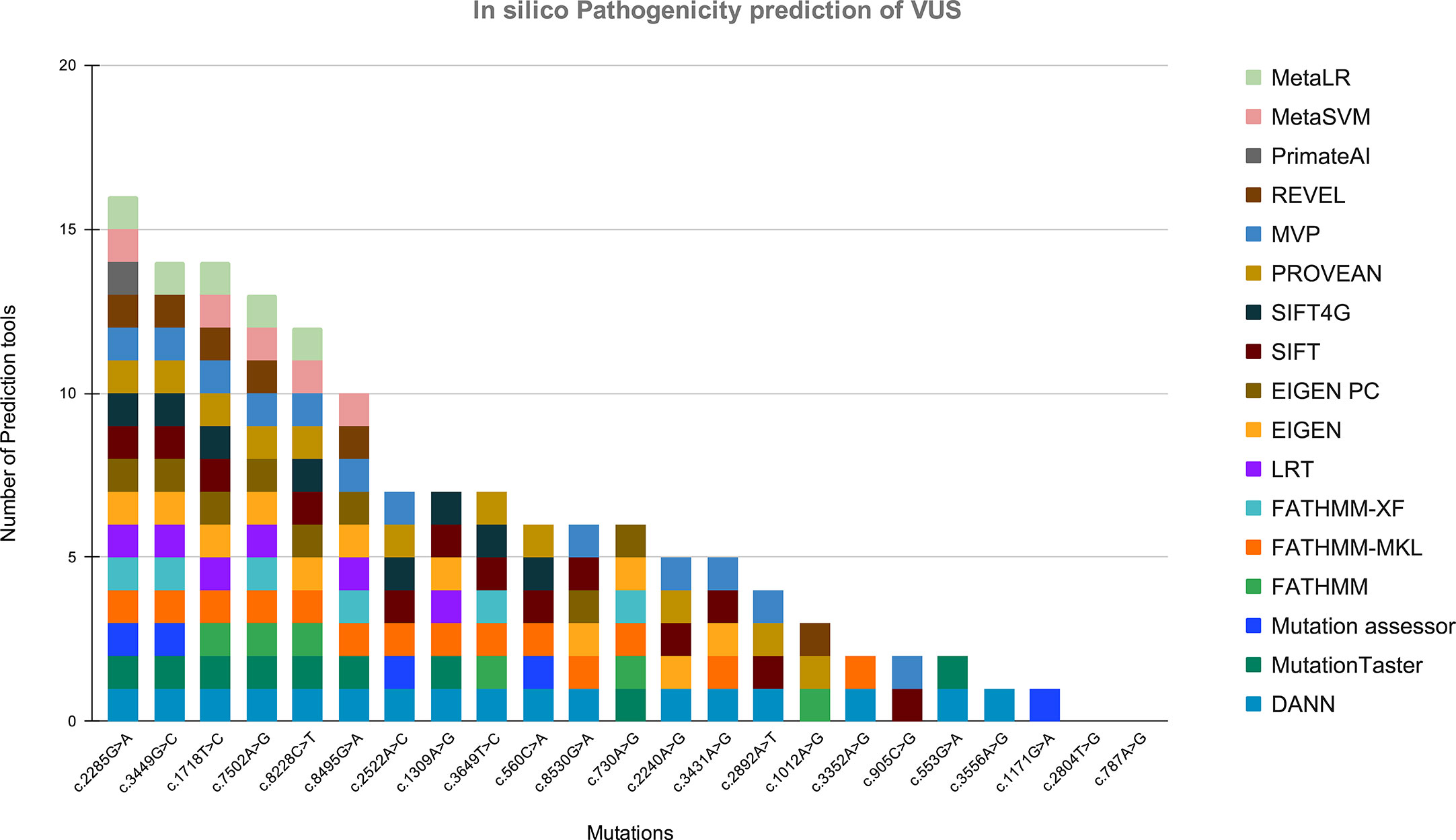
Figure 5 Showing the in silico pathogenicity prediction of VUS. X-axis shows the mutation found in our study, and Y-axis shows the number of tools predicting the variant as a pathogenic mutation.
Comparison of Mutational Profile of Indian HBOC Patients With Other Populations
When the pathogenic mutations and VUS found in our data were compared with the GnomeAD database, we found that mutational frequency distribution differs in Indian patients from other populations. On comparing the minor allele frequencies of clinically significant mutations, including pathogenic and VUS from Indian HBOC patients with that of other populations such as South Asian, East Asian, Latino, Ashkenazi, European-Finnish, European-non-Finnish and African populations, a separation of mutational frequencies of Indian cohort with other global population was observed, suggesting a different mutational frequency profile of Indian HBOC patients. We first compared the minor allele frequencies of pathogenic mutations detected in our study with that of other populations mentioned in the additional file (see Supplementary Table 1). Secondly, we compared the minor allele frequencies of variants of uncertain significance (VUS) detected in our study with that of other populations mentioned in the additional file (see Supplementary Table 2). It was found that 11 out of 25 VUS detected in our study namely, c.3431A>G in BRIP1, c.553G>A in BRIP1, c.3556A>G in BRIP1, c.787A>G in TP53, c.8530G>A in BRCA2, c.2240A>G in BRCA2, c.2892A>T in BRCA2, c.905C>G in BRCA2, c.3449G>C in ATM, c.2522A>C in ATM, and c.3352A>G in ATM were only detected in South Asian population out of all the populations considered in the comparison study.
Discussion
Population Specific Scenario in Genetic Predisposition
In the present, study Amplicon sequencing of 14 potential high risk genes was performed to uncover mutational profiles of 144 Indian patients having a family history of breast and or ovarian cancer.
Distribution of Mutations in Non-BRCA Genes
The study indicates that in the Indian population, mutations in the BRCA1/2 genes are the major contributors 37/144 (25.69%) for hereditary breast and/or ovarian cancers. Whereas the non-BRCA genes contribute 5/144 (3.47%) for hereditary breast and/or ovarian cancers; two mutations were detected in TP53 gene, and one mutation each was detected in PALB2, BRIP1, and ATM genes respectively. The contribution of pathogenic mutations by non-BRCA genes is 3.47% which is lower as compared to the other Indian studies involving multi-gene panel which might be due to lower number of cases in our study (12). Multigene panels ranging from 13 genes to 29 genes have been used in different studies for hereditary breast and ovarian cancer in different population and pathogenic variants in other non-BRCA genes have been consistently reported (21, 22)
Clinically Significant Mutations in Indian Patients
In our study, two recurrent BRCA1 pathogenic mutations were observed in more than three cases, and these mutations are c.5137+1G>As and c.5098delC (12, 23–25), out of which the most frequent pathogenic mutation c.5137+1G>A has also been reported in another Indian study (12). While, c.5098delC, has not been reported in any other Indian study as per our best knowledge. It was also found that a pathogenic mutation, c.5137+1G>A in BRCA1, is found nowhere in the other populations considered for comparison except in South Asian population (MAF: 3.27E-05), suggesting it to be a possible biomarker for HBOC in Indian population. Other pathogenic mutations which were detected in 2/144 cases each (MAF:1.39E-02) in our study are c.3331C>T and c.5566C>T in BRCA1, out of which, c.3331C>T is found to be detected only in East Asian population (MAF: 5.44E-05), while c.5566C>T is found to be detected only in Latino population (MAF: 2.89E-05) and South Asian population (MAF: 6.53E05). Another pathogenic mutation, c.4548-1G>A in BRCA1 (MAF: 6.94E-03) is found to be detected in only the South Asian population (MAF: 9.80E-05). The findings obtained suggest that the recurrent pathogenic mutations that are detected in our study and are not found to be detected in any other population except the South Asian population can be possible biomarkers for HBOC in the Indian population. Other recurrent mutations detected in our study such as c.5098delC in BRCA1, c.682-2A>G in BRCA2, c.1907C>G in BRCA2 and c.1016delA in BRCA1 were not available in the GnomAD database for comparison of their frequencies in different populations. Another pathogenic mutation, c.68_69delAG in BRCA1 (MAF: 2.08E-02) is also found to be highly frequent in Ashkenazi population (MAF: 4.05E-03) and is also reported as a founder mutation in the Ashkenazi Jewish population suggesting that the Ashkenazi founder mutation is not only present in the Ashkenazi descent but also present in non-Ashkenazi descent in Israel, Spain, Poland, India, and other countries in Central and Eastern Europe (26–29). The frequency range of this mutation in the Indian population is reported to be 0.5–4.1% (27). The frequency of this mutation in the present study cohort was 2.08% (3/144), which falls under the reported range of 0.5–4.1%. Also, 11 VUS coincide only with the South Asian population and are also predicted to be disease-causing by prediction tools; it suggests that these VUS could be possible biomarkers for HBOC in the Indian population. Observation of differences in mutational spectrum of Indian cohort and other global populations supports the need of generating population specific mutational profiles, which will help in designing more accurate, effective strategies in HBOC risk assessment/screening, diagnosis and therapy.
Important Genes for Indian HBOC Patients
Clinically important genes have been found to have mutations in many genes from the 14 genes included in this study. We found pathogenic mutations in six genes including BRCA1, BRCA2, BRIP1, TP53, ATM, and PALB2. We also found VUS predicted to be pathogenic in six genes including BRCA1, BRCA2, BRIP1, ATM, RAD51C, and BARD1. Hence, it is suggested that apart from the BRCA genes, other non-BRCA genes such as ATM, PALB2, and TP53 also contribute to HBOC in the Indian population. The findings of the current study strongly emphasize incorporation of multi-gene panels, including high-penetrance genes such as BRCA1/2 and moderate-risk or non-BRCA genes in genetic testing of Indian HBOC patients. These findings will help in better understanding of the contribution of high-risks and moderate-risks genes in onset, prevention, and management of hereditary breast and ovarian cancer in Indian patients. The above findings can be further validated on much wider populations for developing future risk assessment and screening programs of HBOC in India.
Conclusions
Mutational frequency patterns of Indian hereditary breast and ovarian cancer patients are different from the other global populations. High frequency clinically significant pathogenic mutations, recurrent novel mutations, and variants of uncertain significance, which were predicted to be disease-causing by majority of in silico tools, are suggested to be biomarkers for screening and risk assessment strategies of Indian HBOC patients. Five variants of uncertain significance that were predicted to be pathogenic or disease-causing by more than 10 of 17 pathogenicity prediction tools can be further studied and validated on more numbers of samples to understand their role in HBOC. Further validation of these mutations on more number of Indian HBOC patients shall strengthen their role and associations with disease development and progression, which in turn will also help in designing strategies in the prevention, diagnosis, and better disease management.
Data Availability Statement
We have deposited our sequence data in NCBI and our submission Bioproject Accession number is PRJNA675417.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board, Gujarat Cancer Research Institute Registration no.: ECR/41/Inst/GJ/2013/RR-16, date of registration: 16th April 2016. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CJ, MJ, PB, and IS were responsible for the conception and design. FS, NB, BT, IS, MK, and KP were responsible for the acquisition of samples and filling out of the consent forms. MK, KP, and MS were involved in interpretation of the data. MK and KP contributed to the manuscript writing. KS was responsible for the genetic counseling of patients. All authors contributed to the article and approved the submitted version.
Funding
Gujarat Biotechnology Research Centre (GBRC) and Gujarat Cancer Research Institute (GCRI) were funded by Gujarat State Biotechnology Mission (GSBTM), Department of Science and Technology, Government of Gujarat (21WU41).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to the patients and their families for registering themselves and providing consent for participation in our study. We also thank Gujarat State Biotechnology Mission (GSBTM) for funding this study. We would also like to thank our lab mate Ms. Mital Patel for helping us in successful conduction of camps. We express our deep gratitude to all the mentors, especially Mr. Apurvasinh Puvar who helped us get over any obstacles we faced during different phases of the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.568786/full#supplementary-material
Abbreviations
ATM, ataxia-telangiectasia mutated; BARD1, BRCA1 Associated RING Domain 1; BLAST, Basic Local Alignment Search Tool; BOC, Breast and Ovarian Cancer; BRCA1, Breast cancer type 1 susceptibility protein; BRCA2, Breast cancer type 2 susceptibility protein; BRIP1, BRCA1 interacting protein 1 gene; CHEK2, checkpoint kinase 2 gene; ERBB2, Erb-B2 Receptor Tyrosine Kinase 2; HBOC, Hereditary Breast and Ovarian Cancer; MAF, Minor Allele Frequency; MVP, Missense Variant Pathogenicity; NBN, Nibrin; NCCN, National Comprehensive Cancer Network; NGS, Next Generation Sequencing; PALB2, Partner And Localizer Of BRCA2; PTEN, Phosphatase And Tensin Homolog; RAD51C, RAD51 Paralog C; STK11 Serine/Threonine Kinase 11; TP53, Tumor Protein P53; VUS, Variants of Uncertain Significance.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68:394–424. doi: 10.3322/caac.21492
2. Hereditary Breast and Ovarian Cancer(2012). Available at: https://www.cancer.net/cancer-types/hereditary-breast-and-ovarian-cancer (Accessed April 9, 2020).
3. Globocan 2018. India factsheet - India Against Cancer(2018). Available at: http://cancerindia.org.in/globocan-2018-india-factsheet/ (Accessed April 9, 2020).
4. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer J Clin (2015) 65:87–108. doi: 10.3322/caac.21262
5. Available at: https://www.jax.org/education-and-learning/clinical-and-continuing-education/cancerresources/hereditary-breast-and-ovarian-cancer-syndrome-factsheet (Accessed April 9, 2020).
6. National Comprehensive Cancer Network (NCCN). NCCN Clinical Practice Guidelines in Oncology. Genetic/Familial High-Risk Assessment: Breast and Ovarian. (2016). https://www.nccn.org/professionals/physician_gls/PDF/genetics_screening.pdf.
7. Desmond A, Kurian AW, Gabree M, Mills MA, Anderson MJ, Kobayashi Y, et al. Clinical Actionability of Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Risk Assessment. JAMA Oncol (2015) 1:943–51. doi: 10.1001/jamaoncol.2015.2690
8. Banavali SD. Delivery of cancer care in rural India: Experiences of establishing a rural comprehensive cancer care facility. Indian J Med Paediatr Oncol (2015) 36:128–31. doi: 10.4103/0971-5851.158848
9. Antoniou A, Pharoah PDP, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average Risks of Breast and Ovarian Cancer Associated with BRCA1 or BRCA2 Mutations Detected in Case Series Unselected for Family History: A Combined Analysis of 22 Studies. Am J Hum Genet (2003) 72:1117–30. doi: 10.1086/375033
10. Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol (2007) 25:1329–33. doi: 10.1200/JCO.2006.09.1066
11. Lincoln SE, Kobayashi Y, Anderson MJ, Yang S, Desmond AJ, Mills MA, et al. A Systematic Comparison of Traditional and Multigene Panel Testing for Hereditary Breast and Ovarian Cancer Genes in More Than 1000 Patients. J Mol Diagn (2015) 17:533–44. doi: 10.1016/j.jmoldx.2015.04.009
12. Singh J, Thota N, Singh S, Padhi S, Mohan P, Deshwal S, et al. Screening of over 1000 Indian patients with breast and/or ovarian cancer with a multi-gene panel: prevalence of BRCA1/2 and non-BRCA mutations. Breast Cancer Res Treat (2018) 170:189–96. doi: 10.1007/s10549-018-4726-x
13. Available at: https://www.pharmamedtechbi.com/~/media/Supporting%20Documents/The%20Gray%20Sheet/39/37/mm_0001_coveragepositioncriteria_genetic_testing_for_breast_and_ovarian_cancer.pdf (Accessed April 9, 2020).
14. Eliade M, Skrzypski J, Baurand A, Jacquot C, Bertolone G, Loustalot C, et al. The transfer of multigene panel testing for hereditary breast and ovarian cancer to healthcare: What are the implications for the management of patients and families? Oncotarget (2017) 8:1957–71. doi: 10.18632/oncotarget.12699
15. Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, et al. Frequency of mutations in individuals with breast cancer referred forBRCA1andBRCA2testing using next-generation sequencing with a 25-gene panel. Cancer (2015) 121:25–33. doi: 10.1002/cncr.29010
16. Available at: https://www.icmr.nic.in/sites/default/files/guidelines/ICMR_Ethical_Guidelines_2017 (Accessed April 9, 2020).
17. Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Aguilera MA, Meyer R, et al. VarSome: the human genomic variant search engine. Bioinformatics (2019) 35:1978–80. doi: 10.1093/bioinformatics/bty897
18. Richards S, on behalf of the ACMG Laboratory Quality Assurance Committee, Aziz N, Bale S, Bick D, Das S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17:405–23. doi: 10.1038/gim.2015.30
19. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature (2020) 581(7809):434–43. doi: 10.1101/531210
20. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res (2014) 42:D980–5. doi: 10.1093/nar/gkt1113
21. Wong MW, Nordfors C, Mossman D, Pecenpetelovska G, Avery-Kiejda KA, Talseth-Palmer B, et al. BRIP1, PALB2, and RAD51C mutation analysis reveals their relative importance as genetic susceptibility factors for breast cancer. Breast Cancer Res Treat (2011) 127:853–9. doi: 10.1007/s10549-011-1443-
22. Li J, Meeks H, Feng B-J, Healey S, Thorne H, Makunin I, et al. Targeted massively parallel sequencing of a panel of putative breast cancer susceptibility genes in a large cohort of multiple-case breast and ovarian cancer families. J Med Genet (2016) 53:34–42. doi: 10.1136/jmedgenet-2015-103452
23. Ashton-Prolla P, Vargas FR. Prevalence and impact of founder mutations in hereditary breast cancer in Latin America. Genet Mol Biol (2014) 37:234–40. doi: 10.1590/s1415-47572014000200009
24. Janavičius R. Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J (2010) 1:397–412. doi: 10.1007/s13167-010-0037-y
25. Founder mutation - SNPedia. Available at: https://www.snpedia.com/index.php/Founder_mutation (Accessed April 9, 2020).
26. Levy-Lahad E, Catane R, Eisenberg S, Kaufman B, Hornreich G, Lishinsky E, et al. Founder BRCA1 and BRCA2 mutations in Ashkenazi Jews in Israel: frequency and differential penetrance in ovarian cancer and in breast-ovarian cancer families. Am J Hum Genet (1997) 60:1059–67.
27. Mannan AU, Singh J, Lakshmikeshava R, Thota N, Singh S, Sowmya TS, et al. Detection of high frequency of mutations in a breast and/or ovarian cancer cohort: implications of embracing a multi-gene panel in molecular diagnosis in India. J Hum Genet (2016) 61:515–22. doi: 10.1038/jhg.2016.4
28. Pathak A, Rathore A, Ranjan S, Vishwanath S, Dubey AP, Nikhil P. BRCA 1 and 2 mutations in carcinoma breast: An Indian study. Clin Cancer Invest J (2018) 7:97. doi: 10.4103/ccij.ccij_9_18
Keywords: genetic testing, next generation sequencing, amplicon sequencing, non-BRCA genes, customized multi-gene panel, hereditary breast and ovarian cancer, BRCA1 and BRCA2
Citation: Kadri MSN, Patel KM, Bhargava PA, Shah FD, Badgujar NV, Tarapara BV, Patel PS, Shaikh MI, Shah K, Patel A, Pandya S, Vora H, Joshi CG and Joshi MN (2021) Mutational Landscape for Indian Hereditary Breast and Ovarian Cancer Cohort Suggests Need for Identifying Population Specific Genes and Biomarkers for Screening. Front. Oncol. 10:568786. doi: 10.3389/fonc.2020.568786
Received: 24 June 2020; Accepted: 02 December 2020;
Published: 21 January 2021.
Edited by:
Edenir Inez Palmero, Barretos Cancer Hospital, BrazilReviewed by:
Ian Campbell, Peter MacCallum Cancer Centre, AustraliaJiheng Xu, New York University, United States
Copyright © 2021 Kadri, Patel, Bhargava, Shah, Badgujar, Tarapara, Patel, Shaikh, Shah, Patel, Pandya, Vora, Joshi and Joshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Madhvi N. Joshi, madhvimicrobio@gmail.com
†These authors share first authorship
 Mohammed Shaad N. Kadri
Mohammed Shaad N. Kadri Komal M. Patel
Komal M. Patel Poonam A. Bhargava
Poonam A. Bhargava Franky D. Shah
Franky D. Shah Nutan V. Badgujar
Nutan V. Badgujar Bhoomi V. Tarapara
Bhoomi V. Tarapara Prabhudas S. Patel
Prabhudas S. Patel Mohammed Inayatullah Shaikh
Mohammed Inayatullah Shaikh Krati Shah
Krati Shah Apurva Patel
Apurva Patel Shashank Pandya
Shashank Pandya Hemangini Vora
Hemangini Vora Chaitanya G. Joshi
Chaitanya G. Joshi Madhvi N. Joshi
Madhvi N. Joshi