- 1Department of Gastroenterology, Affiliated Hospital of Xuzhou Medical University, Xuzhou, China
- 2Institute of Digestive Diseases, Xuzhou Medical University, Xuzhou, China
- 3Zhejiang University Kunshan Biotechnology Laboratory, Zhejiang University Kunshan Innovation Institute, Kunshan, China
- 4State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, China
- 5Department of R&D, Suzhou VersaBio Technologies Co. Ltd., Kunshan, China
- 6Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China
Background: Aberrant DNA methylation has emerged as a class of promising biomarkers for early colorectal cancer (CRC) detection, but the performance of methylated C9orf50 and methylated KCNQ5 in stool DNA has never been evaluated.
Methods: Methylation specific quantitative PCR (qPCR) assays for methylated C9orf50 and methylated KCNQ5 were developed. The methylation levels of C9orf50 and KCNQ5 in 198 CRC patients, 20 advanced adenoma (AA) patients, 101 small polyp (SP) patients, and 141 no evidence of disease (NED) subjects were analyzed.
Results: The methylation levels of both KCNQ5 and C9orf50 genes were significantly higher in CRC and AA groups than those in SP and NED groups, but showed no significant difference among different stages of CRC. The sensitivities of methylated KCNQ5 and methylated C9orf50 for CRC detection were 77.3% (95% CI: 70.7–82.8%) and 85.9% (95% CI: 80.0–90.2%) with specificities of 91.5% (95% CI: 85.3–95.3%) and 95.0% (95% CI: 89.7–97.8%), respectively. When C9orf50 and methylated KCNQ5 were combined, the clinical performance for CRC detection was similar to that of methylated C9orf50 alone.
Conclusions: Stool DNA based methylated C9orf50 test has the potential to become an alternative approach for CRC screening and prevention.
Introduction
Colorectal cancer (CRC) is one of the most widespread and lethal malignancies globally. It ranked the third in incidence and the second in mortality worldwide in 2018, accounting for more than 1.8 million new cases and over 0.86 million deaths (1). According to the same study, its rankings for China were the second for incidence and the fifth for mortality for the same year, leading to nearly 517,000 new cases and over 245,000 deaths. Moreover, due to an aging population and lifestyle changes toward a more Western diet and activity pattern in recent years, CRC incidence rate has steadily increased and the age of first diagnosis has decreased significantly in China (2).
As it is generally believed that it takes 10 years or more for adenomas, the precancerous lesions, to develop into sporadic CRCs (3), there is an ample time window to identify and treat CRC at its early stages to reduce incidence and mortality rates. Indeed, results from randomized controlled trials and observational studies have provided compelling evidence that early screening could lead to significant reduction of CRC incidence and mortality (4–6), leading to a multitude of national and international guidelines for early CRC screening (7).
Multiple CRC screening approaches have been used in the clinics including fecal immunochemical test (FIT), guaiac-based fecal occult blood test (gFOBT), colonoscopy, flexible sigmoidoscopy, and stool DNA test, which have their distinct advantages and disadvantages. For example, the low sensitivity for detecting stage I CRCs and advanced adenomas (AA) of annual FIT or gFOBT test has limited their effectiveness as screening tools for early stage CRC detection (8, 9). On the other hand, colonoscopy, the gold standard of CRC screening, has demonstrated higher sensitivities for the detection of precancerous lesions and early stage CRC than FIT and gFOBT tests (10–12). However, its significantly higher cost, invasiveness, complicated bowel preparation, and potential complications (13–15) have prevented its wide acceptance by Chinese population (16). Besides, it is hardly a primary CRC screening method in developing countries with limited medical resources and personnel such as China. In contrast, recent application of blood- and stool-based molecular diagnostic assays for early CRC screening in the clinics has demonstrated the feasibility of these alternative approaches. Cologuard, the first stool-based CRC screening test approved by the US Food and Drug Administration (FDA) with relatively high sensitivity and specificity, includes hemoglobin, multiple genetic mutations, and BMP3 and NDRG4 methylation sites as biomarkers (8). Similar to colonoscopy, its high cost and complex procedure renders it unfriendly to low- and moderate-income countries like China. Plasma-based Epi proColon 2.0 test, another FDA-approved CRC screening test (17), utilizes a single SEPT9 methylation site as the biomarker. However, the sensitivities of SEPT9 methylation were much lower than those of colonoscopy and Cologuard test, especially for precancerous lesions and early stage CRCs (18–20).
Recently, aberrant DNA methylation has emerged as a class of promising biomarkers for cancer diagnosis (21–23), and their successful clinical application for CRC screening and prevention has been demonstrated by Cologuard and Epi proColon 2.0 tests. In addition to SEPT9, BMP3, and NDRG4 methylation biomarkers employed in the above two tests, aberrant methylation of other genes has been investigated as potential CRC biomarkers in the literature, including SDC2 (24, 25), SFRP1 (26–29), SFRP2 (29–31), GATA5 (32), ITGA4 (33–36), COL4A1 (33), COL4A2 (33), TLX2 (33), VIM (36–38), cg10673833 (39), GRIA4 (40), VIPR2 (41), EYA4 (42), MAP3K14-AS1 (42), MSC (42), CLIP4 (43), C9orf50 (43), and KCNQ5 (43). Some of these methylation biomarkers have been evaluated with both blood and stool samples for CRC detection but not methylated C9orf50 and methylated KCNQ5. Whereas both methylated C9orf50 and methylated KCNQ5 exhibited good performance for early CRC detection with blood samples with sensitivities of 50.0 and 87.5%, respectively, for stage I CRC (43), their performance with stool samples was lacking. The aim of this study was to address this unanswered question and thus to evaluate them as potential biomarkers for a low-cost, convenient, and more accurate screening method urgently needed to promote early CRC screening in China.
Materials and Methods
Sample Enrollment and Collection
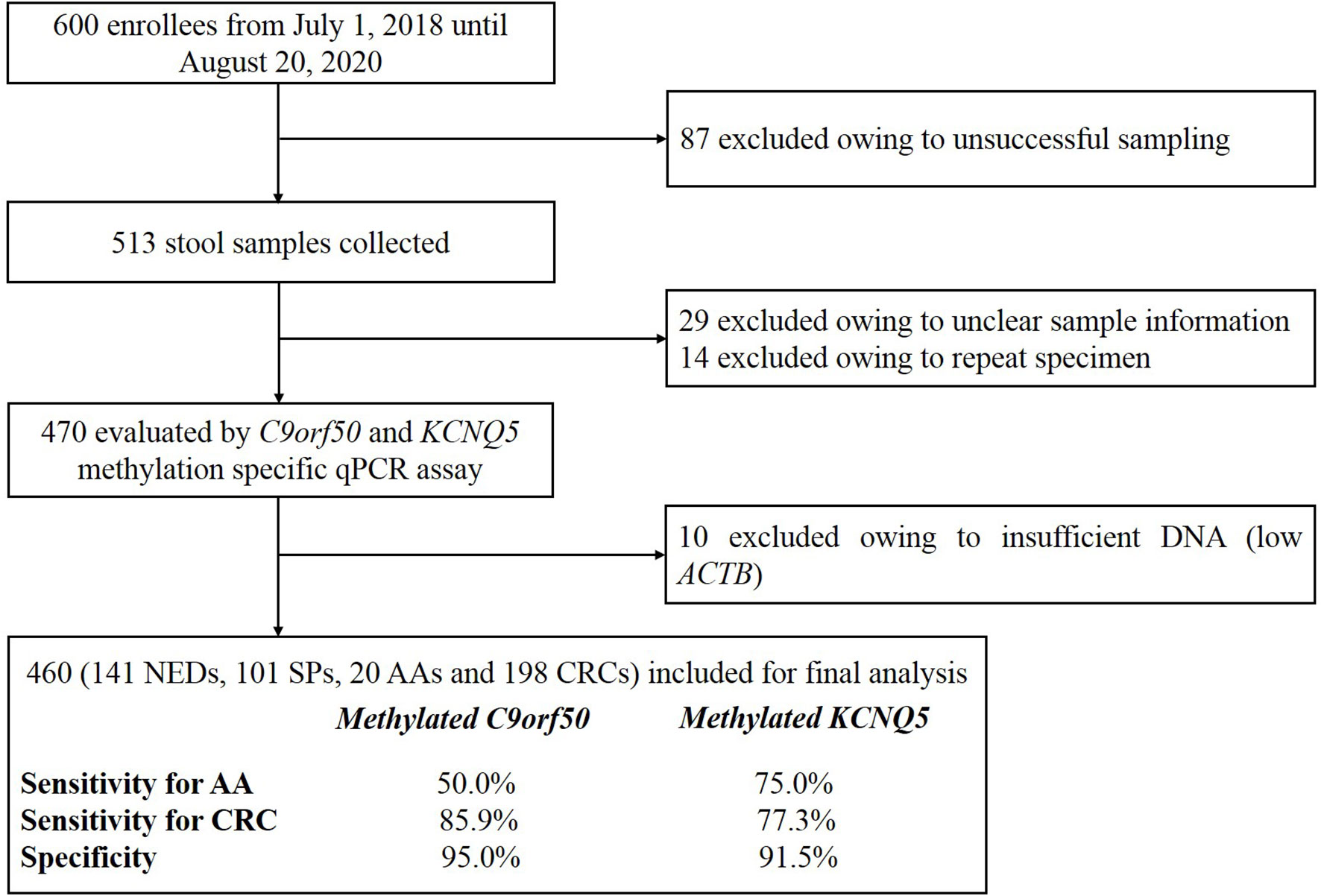
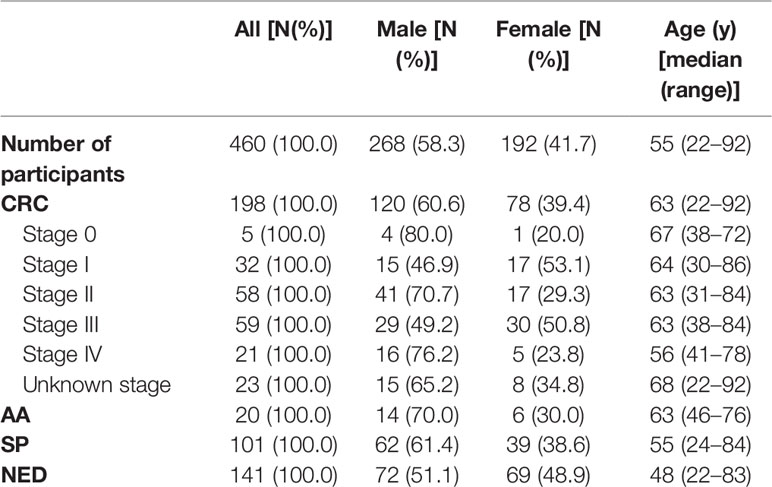
A total of 600 volunteers were recruited from July 1, 2018 until August 20, 2020 at the Affiliated Hospital of Xuzhou Medical University. All participants with colonoscopy results have acknowledged and signed the informed consent, and this study was performed according to the principles of the Helsinki Declaration and approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYFY2020-KL122). Eighty-seven participants were excluded owing to unsuccessful sampling, resulting in 513 stool samples collected (Figure 1). Forty-three samples were subsequently excluded before quantitative PCR (qPCR) test due to missing sample information or repeated sampling. Finally, after excluding 10 samples due to low human gDNA amounts reflected by ACTB Ct values, 460 samples with valid qPCR results for next step analysis included 198 from CRC patients, 20 from advanced adenoma (AA, an adenoma measuring ≥ 10 mm in size, with high-grade dysplasia, or with ≥ 25% villous features) (44, 45) patients, 101 from small polyp (SP, an adenoma < 10 mm in size without high-grade dysplasia and villous histologic features, or hyperplastic polyp < 10 mm in size) patients, and 141 from no evidence of disease (NED, control subjects with normal colonoscopy results) subjects. Stool samples were collected 1–5 day before routine bowel preparation for colonoscopy with single-use disposable sampling boxes (46), and approximately 5 g stool samples were transferred to 50ml collection tubes containing 25 ml of preservative buffer with sampling spoons. All stool samples were stored at room temperature no more than 7 days before being transferred to −80°C for long-term preservation and storage.
DNA Isolation and Methylation Testing
Stool human genomic DNA for each subject was isolated by VersaBio Human Genomic DNA FastPrep Kit (Suzhou VersaBio Technologies Co., Ltd., Kunshan, China) according to the previously published protocol (46). Briefly, the stool samples were homogenized at least for 1 min, and then centrifuged at 10,000 g for 20 min, and 150 μl supernatants were transferred into new 2 ml tubes for DNA extraction. Next, 500 μl preservative buffer was added to each supernatant and centrifuged at 20,000 g for 3 min. The resulting supernatant was then transferred to a new 2 ml tube, and 600 μl lysis buffer, and 20 μl proteinase K solution were added and subsequently incubated at 70°C for 10 min. Six hundred microliter ethanol was added into each sample and then loaded onto a spin column. After two washing steps, the stool human genomic DNA was eluted with 100 μl elution buffer. Bisulfite conversion of stool human genomic DNA and purification of the converted products followed the previously published procedure of a fast bisulfite conversion kit (Suzhou VersaBio Technologies Co., Ltd.) (46). One hundred and fifty microliter conversion buffer and 25 μl protection buffer were added to each purified DNA sample, and the resulting mixture was incubated at 80°C for 45 min. Next, 1 ml wash buffer A was added to each sample and loaded onto a spin column. After two washing steps, the converted DNA was eluted with 100μl elution buffer.
To analyze the methylation levels of KCNQ5 and C9orf50 genes in stool DNA, a triplex methylation specific qPCR (qMSP) assay based on Jensen S. et al. (43) was developed. All primers and probes for detecting methylated KCNQ5, methylated C9orf50, and internal control (ACTB) were synthesized by GENEWIZ BioTechnologies (Suzhou, China) and listed in Supplementary Table 1. VersaBio Multiplex Methylation Specific PCR Master Mix Kit (Suzhou VersaBio Technologies Co., Ltd.) was used. Thirty microliter reactions containing 15μl of bisulfite-converted stool DNA each were performed on ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA) under the following conditions: initial activation at 95°C for 20 min, followed by 50 cycles at 95°C for 10 s, 56°C for 30 s and 72°C for 10 s, and a final cooling to 40°C for 30 s. Three PCR replicates were performed simultaneously for each sample.
Data Analysis
ACTB Ct values were used to validate sample processing. The result for a stool sample was considered “invalid” if the ACTB mean Ct value of three qPCR reactions was greater than 41.0. The Ct values of methylated KCNQ5 and methylated C9orf50 were used to determine whether a stool sample was scored positive or negative for the corresponding methylation biomarker. The cutoff for methylated KCNQ5 was all three qPCR reactions producing amplification signals (3/3 algorithm) with a mean Ct of less than 35.0. The cutoff for methylated C9orf50 was all three qPCR reactions producing Ct values of less than 50.0 (3/3 algorithm). For two-biomarker combination of methylated KCNQ5 and methylated C9orf50, a stool sample would be scored positive when either methylated KCNQ5 or methylated C9orf50 was scored positive. Mean Ct value of each sample in CRC, AA, SP and no evidence of disease (NED) groups was used to represent its target methylation level. Those reactions without amplification signals were set a Ct value of 50 (the maximal number of PCR cycles) for the mean Ct analysis (47). The receiver operating characteristic (ROC) curves were plotted to calculate the area under curve (AUC) values. All statistical analyses were performed with IBM SPSS for Windows Version 22.0. Pearson chi-square test was used for sensitivity comparison among groups at a significance level of p < 0.05, the differences in methylation levels have been analyzed using the Mann–Whitney U test.
Results
A total of 513 stool samples were collected in this study, of which 43 were excluded due to insufficient sample information or repeated sampling. Four hundred and sixty samples returned valid data for methylated KCNQ5 and methylated C9orf50 qMSP assay. Among them, 198 were CRC patients, 20 were AA patients (Supplementary Table 2), 101 were patients with SPs, and 141 were NED subjects. For CRC group, 60.6% were male and the mean age was 61.7, including 5 stage 0, 32 stage I, 58 stage II, 59 stage III, 21 stage IV patients, and 23 patients of unknown stage (Table 1).
To determine whether the methylation levels of KCNQ5 and C9orf50 in stool DNA were capable of distinguishing CRC from other samples, the mean Ct values of each group was analyzed. As shown in Figures 2A, B, KCNQ5 and C9orf50 in CRC (p<0.0001), AA (p<0.0001) and SP (p<0.0001) groups all displayed significantly higher methylation levels than in NED group. The methylation levels of KCNQ5 and C9orf50 in CRC (p<0.0001) and AA (p<0.05 and p<0.01, respectively) groups were also significantly higher than those in SP group. C9orf50 showed significantly higher methylation levels in CRC (p<0.01) than in AA group, but KCNQ5 showed no significantly difference between CRC group and AA group (Figure 2A). In contrast, both KCNQ5 and C9orf50 showed no significant difference in the methylation levels among different stages of CRC (Figures 2C, D).

Figure 2 The methylation levels of KCNQ5 and C9orf50 in no evidence of disease (NED), small polyp (SP), advanced adenoma (AA), colorectal cancer (CRC) groups (A, B) and different stages of CRC (C, D). **p < 0.01; ***p < 0.0001, ns, no significant difference according to Student’s t-test. Red lines represent the median methylation levels of KCNQ5 or C9orf50. Mann–Whitney U test compared to methylation levels of KCNQ5 and C9orf50 in NED, SP, AA, CRC groups.
Methylated KCNQ5 was positive in 36.6% (37/101) of SP, 75.0% (15/20) of AA, 60.0% (3/5) of stage 0 CRC, 84.4% (27/32) of stage I CRC, 82.8% (48/58) of stage II CRC, 69.5% (41/59) of stage III CRC, 66.7% (14/21) of stage IV CRC, and 87.0% (20/23) of CRC of unknown stage. The sensitivity and the specificity of methylated KCNQ5 for detecting all stage CRC were 77.3% (95% CI: 70.7–82.8%) and 91.5% (95% CI: 85.3–95.3%). Methylated C9orf50 was positive in 25.7% (26/101) of SP, 50.0% (10/20) of AA, 60.0% (3/5) of stage 0 CRC, 90.6% (29/32) of stage I CRC, 87.9% (51/58) of stage II CRC, 84.7% (50/59) of stage III CRC, 85.7% (18/21) of stage IV CRC, and 82.6% (19/23) of CRC of unknown stage. The sensitivity and the specificity of methylated C9orf50 for detecting all stage CRC were 85.9% (95% CI: 80.0–90.2%) and 95.0% (95% CI: 89.7–97.8%). When methylated KCNQ5 and methylated C9orf50 were combined for CRC detection, the sensitivity and the specificity were 88.4% (95% CI: 82.9–92.3%) and 89.4% (95% CI: 82.8–93.7%), respectively (Figure 3A).

Figure 3 Sensitivities and specificities of methylated KCNQ5 and methylated C9orf50 in stool DNA for small polyp (SP), advanced adenoma (AA), and colorectal cancer (CRC) detection (A), and the receiver operating characteristic (ROC) curves of methylated KCNQ5 and methylated C9orf50 in stool DNA for CRC detection (B).
ROC analysis demonstrated the ability of methylated KCNQ5 alone, methylated C9orf50 alone, and the combination of methylated KCNQ5 and methylated C9orf50 to discriminate CRC from NEDs with AUC values of 0.846 (95% CI: 0.802–0.890), 0.904 (95% CI: 0.869–0.940), and 0.888 (95% CI: 0.849–0.928), respectively (Figure 3B). Sensitivities of methylated KCNQ5 and methylated C9orf50 in stool DNA for detecting CRC of different characteristics are summarized in Table 2. Methylated KCNQ5 showed no significant difference of sensitivities among different genders, age groups, stages, tumor locations, tumor sizes, and differentiation statuses, but sensitivities of methylated C9orf50 for detecting CRC of different locations showed statistically significant differences (p<0.05). Similar analysis was also performed on the 20 AA cases. As shown in Supplementary Table 3, both methylated KCNQ5 and methylated C9orf50 showed no significant sensitivity differences between different genders, polyp locations, and differentiation statuses. However, methylated C9orf50 but not methylated KCNQ5 exhibited significant sensitivity difference between different polyp sizes (p<0.05).
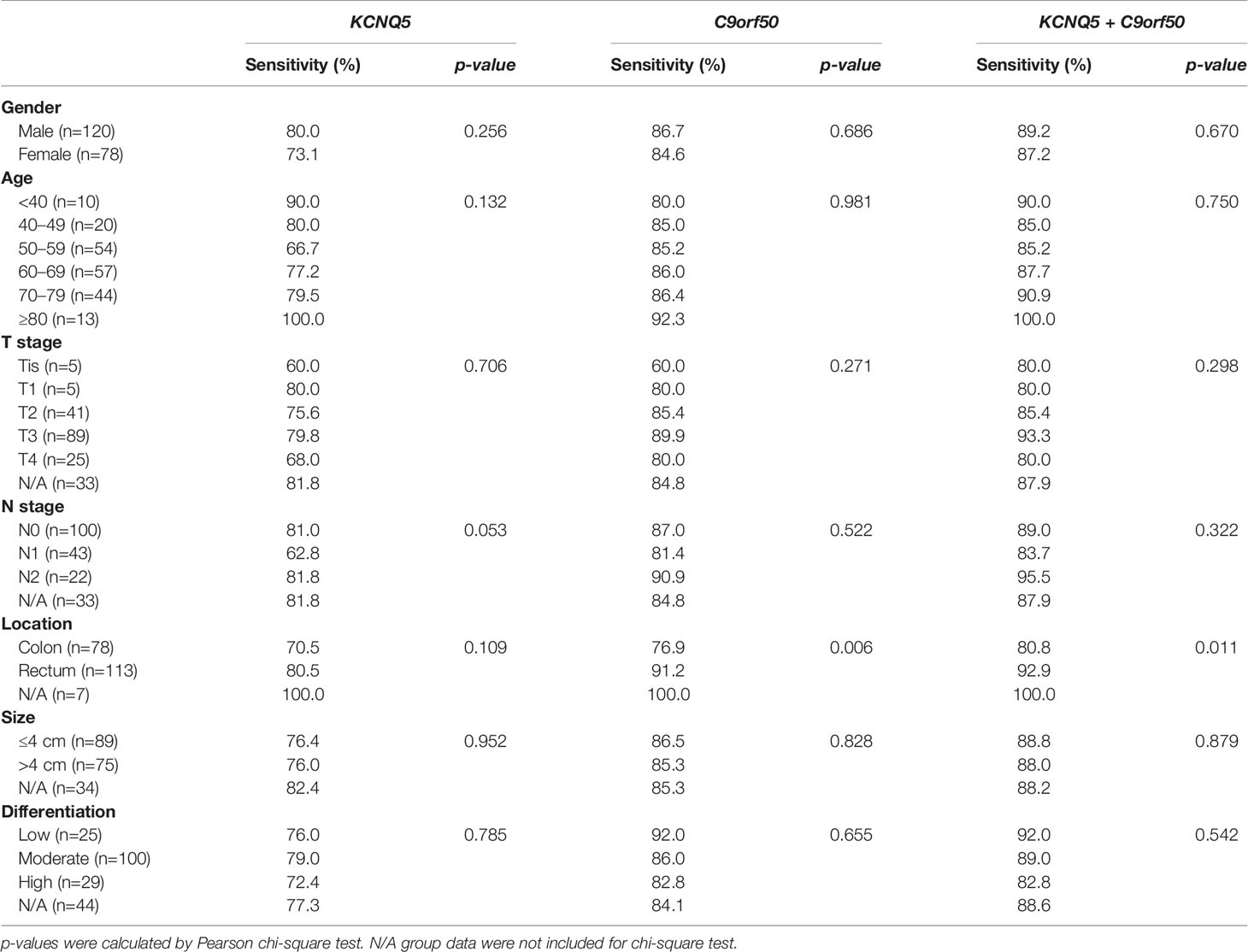
Table 2 Sensitivities of methylated KCNQ5 and methylated C9orf50 in stool DNA for detecting colorectal cancer (CRC) in different genders, age groups, stages, tumor locations, tumor sizes, and differentiation statuses.
Discussion
Colorectal cancer is one of the most prevalent and lethal malignancies globally and in China. The best approach to reduce its burden on individuals and societies is through prevention by early CRC screening, which has been shown to reduce CRC incidence and mortality rates (48). Current methods for early CRC screening all have disadvantages for developing countries including China, such as relatively poor diagnostic accuracy for low-cost FIT (8) and gFOBT tests (9) and high demand on limited medical resources and personnel for diagnostically more accurate colonoscopy, sigmoidoscopy, and stool-based Cologuard test (49). Therefore, development of low-cost, convenient, and more accurate early CRC screening methods will help promote widespread acceptance of early CRC screening by the population especially in developing countries.
In this study, we have examined the clinical performance of stool-based methylated C9orf50 and methylated KCNQ5 tests for CRC and precancerous lesion detection (Figure 3 and Table 3). Overall, the clinical performance of both methylation biomarkers for CRC detection in stool-based assays was similar to that in plasma-based assays (43) with two exceptions (Table 3). Stool methylated C9orf50 test showed a much higher sensitivity for stage I CRC than plasma-based test (90.6 vs. 50.0%). On the contrary, plasma methylated KCNQ5 test showed a much higher sensitivity for stage IV CRC than stool-based test (100.0 vs. 66.7%). One possible explanation could account for these differences between the results of stool test and plasma test (43). Stool DNA originated directly from cancer or polyp tissues in colon or rectum. However, after entering the blood stream, circulating tumor DNA were diluted by a person’s entire blood supply. Therefore, it is conceivable that tumor DNA was more concentrated in stool than in plasma, resulting in a higher sensitivity for CRC detection with stool samples. Such a phenomenon was also observed for methylated SEPT9 and methylated SDC2 in our previous studies (50, 56, 57).
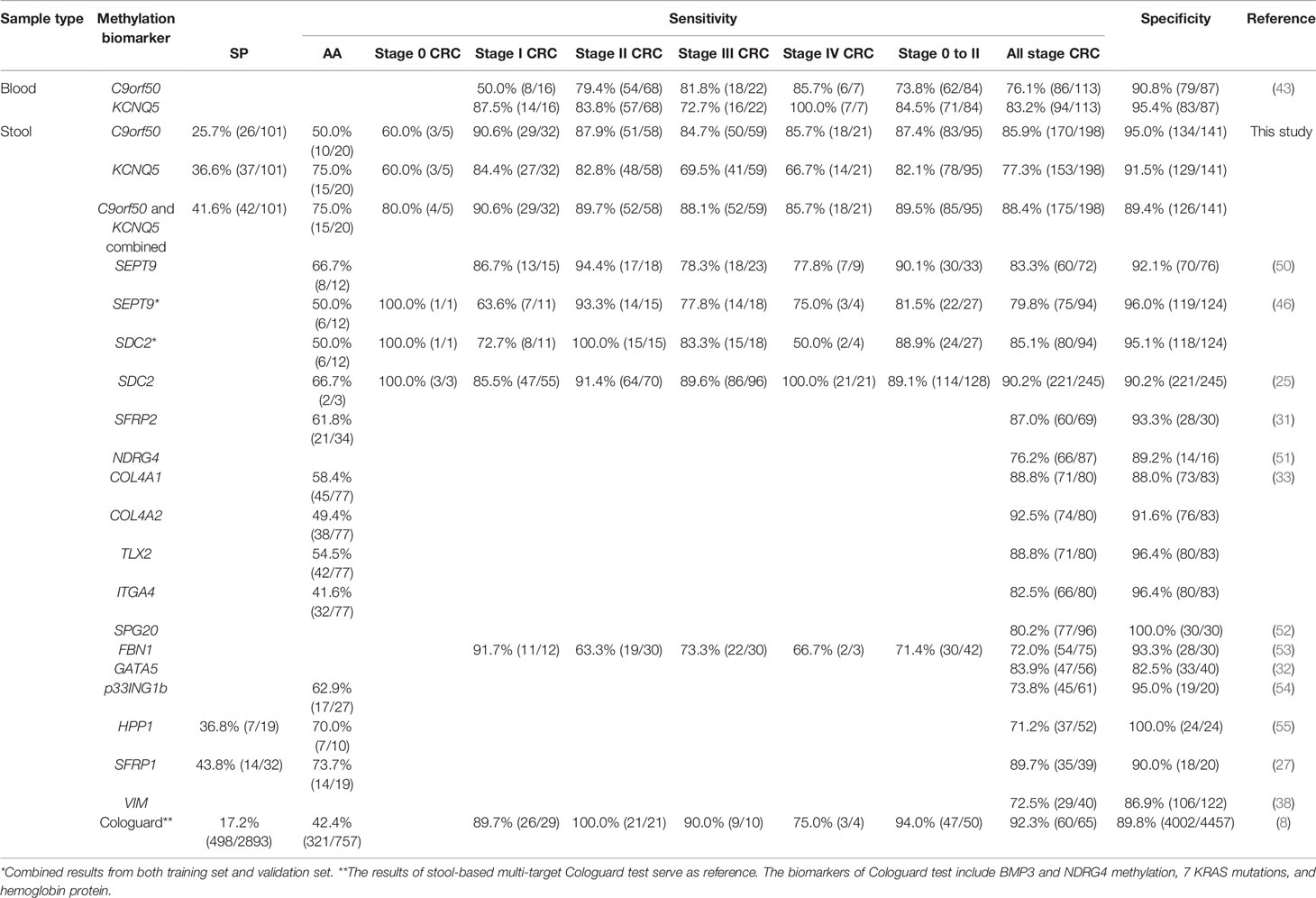
Table 3 Comparison of clinical performance of single methylation biomarkers for colorectal cancer (CRC) and precancerous lesion detection from representative studies.
When compared to other methylation biomarkers in stool-based tests, the sensitivities of methylated C9orf50 and methylated KCNQ5 for all stage CRC detection, 85.9 and 77.3% respectively, fell within the range of other markers from 71.2% for methylated HPP1 (55) to 92.5% for methylated COL4A2 (33). So did their specificities of 95.0% for methylated C9orf50 and 91.5% for methylated KCNQ5 in NED or healthy controls, when compared to the range observed for other markers from 82.5% for methylated GATA5 (32) to 100.0% for methylated SPG20 (52). As more detailed evaluations of the clinical performance for detecting different stages of CRC were not reported for most of the other markers, the sensitivities of methylated C9orf50 and methylated KCNQ5 for early stage CRC (stage 0 to II) could only be compared to those of methylated SEPT9 and methylated SDC2, which were very similar among all four methylation markers, 87.4% for methylated C9orf50, 82.1% for methylated KCNQ5, 90.1 and 81.5% for methylated SEPT9 (46, 50), and 88.9 and 89.1% for methylated SDC2 (25, 46). On the other hand, the sensitivities of most of the other methylation biomarkers for AA detection, from 41.6% for methylated ITGA4 (33) to 73.7% for methylated SFRP1 (27), were on par with those of methylated C9orf50 and methylated KCNQ5, 50.0 and 75.0% respectively.
When compared to the FDA-approved multi-target Cologuard test (8), the clinical performance of methylated C9orf50, 85.9% sensitivity for all stage CRC with 95.0% specificity, appeared to be similar to that of Cologuard, 92.3% sensitivity for all stage CRC with 89.8% specificity, whereas 77.3% sensitivity for all stage CRC with 91.5% specificity of methylated KCNQ5 seemed to be inferior to those of Cologuard. As far as precancerous lesions were concerned, the sensitivities of methylated C9orf50 for AA and SP, 50.0 and 25.7% respectively, were similar to those of Cologuard, 42.2 and 17.2%. In contrast, the sensitivities of methylated KCNQ5 for AA and SP, 75.0 and 36.6%, appeared significantly higher than those of Cologuard, suggesting a better performance in a screening setting where identification of precancerous lesions and early stage CRC was preferred.
Furthermore, when methylated C9orf50 and methylated KCNQ5 were combined, the clinical performance for CRC detection was similar to that of methylated C9orf50 alone, 88.4 vs. 85.9% for sensitivity and 89.4 vs. 95.0% for specificity. Thus, the performance improvement of two-marker combination appeared to be minimal, particularly for precancerous lesions and early stage CRC detection.
Whereas this study was the first to examine the clinical performance of stool-based methylated C9orf50 and methylated KCNQ5 tests for early CRC detection, it did have a few limitations. For example, the number of AA samples examined was relatively small, and the mean age of NED group was younger than CRC patients. Thus increasing the number of enrolled AA patients and comparable participant distribution in all groups should be considered in future studies. Meanwhile, stool methylated C9orf50 or methylated KCNQ5 test has not been directly compared with previous published methylation based CRC screening methods, such as plasma methylated SEPT9 test or Cologuard. Therefore, more validation studies in multiple clinical centers as well as a large prospective comparison study between different methods should be carried out in the future.
Conclusion
In the present study, we demonstrated that stool-based methylated C9orf50 test exhibited high sensitivities and specificity for the detection of precancerous lesions and all stage CRCs. Whereas stool-based methylated KCNQ5 test also demonstrated high sensitivities for the detection of precancerous lesions and early stage CRCs, its sensitivities for late stage CRCs were markedly lower than those of methylated C9orf50. These results suggested that stool-based methylated C9orf50 test has the potential to become an alternative approach for early diagnosis of CRC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYFY2020-KL122). All participants have acknowledged and signed the informed consent, and this study was performed according to the principles of the Helsinki Declaration and approved by the institutional review board of the Affiliated Hospital of Xuzhou Medical University (Ethics Committee reference number: XYFY2020-KL122). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YapC, GZ, MY, and YM performed the statistical analyses and drafted the manuscript. YanC, MY, XL, YanC, BM, SZ, and DL participated in sample collection and data analysis. GZ, SX, MZ, and SF conceived of the study and participated in the design and coordination of the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Suzhou Technology Entrepreneur Angel Project (grant CYTS2018051), Key Technologies R & D Program for Social Development of Jiangsu Province (grant BE2019688), Kunshan Leading Talent Project (grant 00311), Suzhou Innovation and Entrepreneurship Leading Talent Program (grant ZXL2020046), and Key Technologies R & D Program for Social Development of Xuzhou (grant KC17184).
Conflict of Interest
Authors GZ and SX were employed by Suzhou VersaBio Technologies Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.621295/full#supplementary-material
Supplementary Table 1 | Primers and probes used in this study.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin (2016) 66(2):115–32. doi: 10.3322/caac.21338
3. Engel C, Vasen HF, Seppälä T, Aretz S, Bigirwamungu-Bargeman M, de Boer SY, et al. No Difference in Colorectal Cancer Incidence or Stage at Detection by Colonoscopy Among 3 Countries With Different Lynch Syndrome Surveillance Policies. Gastroenterology (2018) 155(5):1400–9.e2. doi: 10.1053/j.gastro.2018.07.030
4. Brenner H, Stock C, Hoffmeister M. Effect of screening sigmoidoscopy and screening colonoscopy on colorectal cancer incidence and mortality: systematic review and meta-analysis of randomised controlled trials and observational studies. BMJ (2014) 348:g2467. doi: 10.1136/bmj.g2467
5. Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med (2013) 369(12):1106–14. doi: 10.1056/NEJMoa1300720
6. Pignone M, Rich M, Teutsch SM, Berg AO, Lohr KN. Screening for Colorectal Cancer in Adults at Average Risk: A Summary of the Evidence for the U.S. Prevent Serv Task Force Ann Internal Med (2002) 137(2):132–41. doi: 10.7326/0003-4819-137-2-200207160-00015
7. Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, et al. Colorectal cancer screening: a global overview of existing programmes. Gut (2015) 64(10):1637–49. doi: 10.1136/gutjnl-2014-309086
8. Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. New Engl J Med (2014) 370(14):1287–97. doi: 10.1056/NEJMoa1311194
9. Christian P. Colon cancer screening: which non-invasive filter tests? Digest Dis (2011) 29(s1):56–9. doi: 10.1159/000331127
10. Segnan N, Senore C, Andreoni B, Azzoni A, Bisanti L, Cardelli A, et al. Comparing attendance and detection rate of colonoscopy with sigmoidoscopy and FIT for colorectal cancer screening. Gastroenterology (2007) 132(7):2304–12. doi: 10.1053/j.gastro.2007.03.030
11. Quintero E, Castells A, Bujanda L, Cubiella J, Salas D, Lanas A, et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med (2012) 366(8):697–706. doi: 10.1056/NEJMoa1108895
12. Graser A, Stieber P, Nagel D, Schafer C, Horst D, Becker CR, et al. Comparison of CT colonography, colonoscopy, sigmoidoscopy and faecal occult blood tests for the detection of advanced adenoma in an average risk population. Gut (2009) 58(2):241–8. doi: 10.1136/gut.2008.156448
13. Swaroop VS, Larson MV. Colonoscopy as a Screening Test for Colorectal Cancer in Average-Risk Individuals. Mayo Clin Proc (2002) 77(9):951–6. doi: 10.4065/77.9.951
14. Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, et al. Colorectal Cancer Screening: Recommendations for Physicians and Patients From the U.S. Multi-Soc Task Force Colorectal Cancer Gastroenterol (2017) 153(1):307–23. doi: 10.1053/j.gastro.2017.05.013
15. Subramanian S, Tangka FKL, Hoover S, Royalty J, DeGroff A, Joseph D. Costs of colorectal cancer screening provision in CDC’s Colorectal Cancer Control Program: Comparisons of colonoscopy and FOBT/FIT based screening. Eval Program Plann (2017) 62:73–80. doi: 10.1016/j.evalprogplan.2017.02.007
16. Niu F, Wen J, Fu X, Li C, Zhao R, Wu S, et al. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol Biomarkers Prev (2017) 26(9):1411–9. doi: 10.1158/1055-9965.EPI-17-0153
17. Lamb YN, Dhillon S. Epi proColon((R)) 2.0 CE: A Blood-Based Screening Test for Colorectal Cancer. Mol Diagn Ther (2017) 21(2):225–32. doi: 10.1007/s40291-017-0259-y
18. Song L, Yu H, Jia J, Li Y. A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. Cancer Biomark (2017) 18(4):425–32. doi: 10.3233/CBM-160321
19. Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut (2014) 63(2):317–25. doi: 10.1136/gutjnl-2012-304149
20. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, Zou X, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health (2018) 6(5):e555–e67. doi: 10.1016/S2214-109X(18)30127-X
21. Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation.s. Expert Rev Mol Diagn (2014) 14(3):293–305. doi: 10.1586/14737159.2014.897610
22. Taryma-Lesniak O, Sokolowska KE, Wojdacz TK. Correction to: Current status of development of methylation biomarkers for in vitro diagnostic IVD applications. Clin Epigenet (2020) 12(1):107. doi: 10.1186/s13148-020-00902-9
23. Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci (2015) 2:13. doi: 10.3389/fmolb.2015.00013
24. Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, et al. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn (2013) 15(4):498–507. doi: 10.1016/j.jmoldx.2013.03.004
25. Han YD, Oh TJ, Chung TH, Jang HW, Kim YN, An S, et al. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenet (2019) 11(1):51. doi: 10.1186/s13148-019-0642-0
26. Pasha HF, Radwan MI, Yehia AM, Toam MM. Circulating methylated RUNX3 and SFRP1 genes as a noninvasive panel for early detection of colorectal cancer. Eur J Gastroenterol Hepatol (2019) 31(11):1342–9. doi: 10.1097/MEG.0000000000001532
27. Tang D, Wang D, Li H. Combination analysis of hypermethylated SFRP1 and SFRP2 gene in fecal as a novel epigenetic biomarker panel for colorectal cancer screening* *This work was supported by the grant from Programs of Science and Technology Commission Foundation of Jiangsu Province(NO. BS 2005 036). J Nanjing Med Univ (2008) 22(2):96–101. doi: 10.1016/S1007-4376(08)60020-9
28. Salehi R, Mohammadi M, Emami MH, Salehi AR. Methylation pattern of SFRP1 promoter in stool sample is a potential marker for early detection of colorectal cancer. Adv Biomed Res (2012) 1:87. doi: 10.4103/2277-9175.105169
29. Bartak BK, Kalmar A, Peterfia B, Patai AV, Galamb O, Valcz G, et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics (2017) 12(9):751–63. doi: 10.1080/15592294.2017.1356957
30. Yang Q, Huang T, Ye G, Wang B, Zhang X. Methylation of SFRP2 gene as a promising noninvasive biomarker using feces in colorectal cancer diagnosis: a systematic meta-analysis. Sci Rep (2016) 6:33339. doi: 10.1038/srep33339
31. Wang DR, Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol (2008) 14(4):524–31. doi: 10.3748/wjg.14.524
32. Lu H, Huang S, Zhang X, Wang D, Zhang X, Yuan X, et al. DNA methylation analysis of SFRP2, GATA4/5, NDRG4 and VIM for the detection of colorectal cancer in fecal DNA. Oncol Lett (2014) 8(4):1751–6. doi: 10.3892/ol.2014.2413
33. Liu X, Wen J, Li C, Wang H, Wang J, Zou H. High-Yield Methylation Markers for Stool-Based Detection of Colorectal Cancer. Dig Dis Sci (2020) 65(6):1710–9. doi: 10.1007/s10620-019-05908-9
34. Chang E, Park DI, Kim YJ, Kim BK, Park JH, Kim HJ, et al. Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepatogastroenterology (2010) 57(101):720–7.
35. Ausch C, Kim YH, Tsuchiya KD, Dzieciatkowski S, Washington MK, Paraskeva C, et al. Comparative analysis of PCR-based biomarker assay methods for colorectal polyp detection from fecal DNA. Clin Chem (2009) 55(8):1559–63. doi: 10.1373/clinchem.2008.122937
36. Ahmed D, Danielsen SA, Aagesen TH, Bretthauer M, Thiis-Evensen E, Hoff G, et al. A tissue-based comparative effectiveness analysis of biomarkers for early detection of colorectal tumors. Clin Transl Gastroenterol (2012) 3:e27. doi: 10.1038/ctg.2012.21
37. Song BP, Jain S, Lin SY, Chen Q, Block TM, Song W, et al. Detection of hypermethylated vimentin in urine of patients with colorectal cancer. J Mol Diagn (2012) 14(2):112–9. doi: 10.1016/j.jmoldx.2011.12.003
38. Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC,3, Sontag S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol (2007) 5(1):111–7. doi: 10.1016/j.cgh.2006.10.006
39. Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med (2020) 12(524):1–11. doi: 10.1126/scitranslmed.aax7533
40. Fadda A, Gentilini D, Moi L, Barault L, Leoni VP, Sulas P, et al. Colorectal cancer early methylation alterations affect the crosstalk between cell and surrounding environment, tracing a biomarker signature specific for this tumor. Int J Cancer (2018) 143(4):907–20. doi: 10.1002/ijc.31380
41. Vega-Benedetti AF, Loi E, Moi L, Orrù S, Zavattari P. Colorectal Cancer Early Detection in Stool Samples Tracing CpG Islands Methylation Alterations Affecting Gene Expression. Int J Mol Sci (2020) 21(12):4494. doi: 10.3390/ijms21124494
42. Barault L, Amatu A, Siravegna G, Ponzetti A, Nicolantonio FD. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut (2017) 67(11):1995–2005. doi: 10.1136/gutjnl-2016-313372
43. Jensen S, Øgaard N, Ørntoft MW, Rasmussen MH, Bramsen JB, Kristensen H, et al. Novel DNA methylation biomarkers show high sensitivity and specificity for blood-based detection of colorectal cancer-a clinical biomarker discovery and validation study. Clin Epigenet (2019) 11(1):158. doi: 10.1158/1538-7445.AM2018-5604
44. Morson BC. Evolution of cancer of the colon and rectum. Cancer (1974) 34(3):845–9. doi: 10.1002/1097-0142(197409)34:3+<845::AID-CNCR2820340710>3.0.CO;2-H
45. Winawer SJ, Zauber AG. The advanced adenoma as the primary target of screening. Gastrointest Endosc Clinics North America (2002) 12(1):1–9. doi: 10.1016/S1052-5157(03)00053-9
46. Zhao G, Liu X, Liu Y, Li H, Zheng M. Aberrant DNA Methylation of SEPT9 and SDC2 in Stool Specimens as an Integrated Biomarker for Colorectal Cancer Early Detection. Front Genet (2020) 11:643. doi: 10.3389/fgene.2020.00643
47. Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q, et al. Detection of Colorectal Cancer Using a Simplified SEPT9 Gene Methylation Assay Is a Reliable Method for Opportunistic Screening. J Mol Diagn JMD (2016) 18(4):535–45. doi: 10.1016/j.jmoldx.2016.02.005
48. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
49. Qian C-N. At-home cancer screening: a solution for China and other developing countries with a large population and limited number of healthcare practitioners. Chin J Cancer (2017) 36(1):68. doi: 10.1186/s40880-017-0235-2
50. Liu Y, Zhao G, Miao J, Li H, Ma Y, Liu X, et al. Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer. Front Genet (2020) 11:324. doi: 10.3389/fgene.2020.00324
51. Xiao W, Zhao H, Dong W, Li Q, Zhu J, Li G, et al. Quantitative detection of methylated NDRG4 gene as a candidate biomarker for diagnosis of colorectal cancer. Oncol Lett (2015) 9(3):1383–7. doi: 10.3892/ol.2014.2815
52. Zhang H, Song Y-C, Dang C-X. Detection of Hypermethylated Spastic Paraplegia-20 in Stool Samples of Patients with Colorectal Cancer. Int J Med Sci (2013) 10(3):230–4. doi: 10.7150/ijms.5278
53. Guo Q, Song Y, Zhang H, Wu X, Xia P, Dang C. Detection of hypermethylated fibrillin-1 in the stool samples of colorectal cancer patients. Med Oncol (2013) 30(4):695. doi: 10.1007/s12032-013-0695-4
54. He CG, Huang QY, Chen LS, Ling ZA, Wu HG, Deng HQ. p33(ING1b) methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. Oncol Lett (2014) 7(5):1639–44. doi: 10.3892/ol.2014.1923
55. Huang ZH, Li LH, Yang F, Wang JF. Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol (2007) 13(6):950–4. doi: 10.3748/wjg.v13.i6.950
56. Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S, et al. Aberrant DNA Methylation of SEPT9 and SDC2 in Stool Specimens as an Integrated Biomarker for Colorectal Cancer Early Detection. Front Genet (2020) 11:643. doi: 10.3389/fgene.2020.00643
Keywords: methylated C9orf50, methylated KCNQ5, stool DNA, colorectal cancer, early detection
Citation: Cao Y, Zhao G, Yuan M, Liu X, Ma Y, Cao Y, Miao B, Zhao S, Li D, Xiong S, Zheng M and Fei S (2021) KCNQ5 and C9orf50 Methylation in Stool DNA for Early Detection of Colorectal Cancer. Front. Oncol. 10:621295. doi: 10.3389/fonc.2020.621295
Received: 26 October 2020; Accepted: 14 December 2020;
Published: 29 January 2021.
Edited by:
Cornelis F. M. Sier, Leiden University, NetherlandsReviewed by:
Sarah Østrup Jensen, Aarhus University, DenmarkErin Leigh Symonds, Flinders Medical Centre, Australia
Patrizia Zavattari, University of Cagliari, Italy
Copyright © 2021 Cao, Zhao, Yuan, Liu, Ma, Cao, Miao, Zhao, Li, Xiong, Zheng and Fei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangmin Xiong, shangmin_xiong@hotmail.com; Minxue Zheng, minxue.zheng@sibet.ac.cn; Sujuan Fei, xyfyfeisj99@163.com
†These authors have contributed equally to this work
 Yaping Cao
Yaping Cao Guodong Zhao
Guodong Zhao Mufa Yuan
Mufa Yuan Xiaoyu Liu3
Xiaoyu Liu3 Shangmin Xiong
Shangmin Xiong