Metagenomic Analysis Identified Stenotrophomonas maltophilia Pneumonia in an Infant Suffering From Unexplained Very Severe Pneumonia
- Department of Pediatrics, Tangdu Hospital, Air Force Medical University, Xi'an, China
Pneumonia poses a significant global morbidity and mortality burden on children. Etiological diagnosis and matched anti-microbial therapy are particularly important for very severe pneumonia. Although great advances have been achieved in diagnostic approaches, it remains challenging to identify pathogens in unexplained pneumonia (UP) cases. In this study, we applied next-generation sequencing (NGS) technology and a metagenomic approach to detect and characterize respiratory bactiera in an UP case in infant. Stenotrophomonas maltophilia was the only bacterial pathogen detected in blood. Metagenomic sequencing also provided bacteria genomic sequences, which could be used to evaluate the role of this pathogen in the disease. This NGS method has the potential to improve the identification of causative organisms in patients with pneumonia and the delivery of appropriate, pathogen-directed antibiotic therapy.
Introduction
Globally, pneumonia is the top cause of death in children with age standardized mortality rates of 38.9/100,000 children. Furthermore, pneumonia remained one of the leading causes of disability-adjusted life years in children (1, 2). The World Health Organization (WHO) guideline defines “very severe pneumonia” as children with tachypnea and general danger signs (e.g., severe chest in-drawing, cyanosis, difficulty breastfeeding, unconsciousness) (3). With the development of detecting techniques, we have more detection methods to identify pathogens (e.g., immunofluorescence technique or polymerase chain reaction of respiratory secretion), comparing with traditional respiratory secretion culture and blood culture. But it remains challenging to identify pathogens in unexplained pneumonia (UP) cases. We present here a case of Stenotrophomonas maltophilia severe pneumonia in an infant diagnosed by Next-generation sequencing method. Furthermore, metagenomic sequencing afforded information regarding the quantity of each pathogens in the sample, which could be used to evaluate the role of each pathogen in the disease. Our study highlights the potential of metagenomic sequencing for pathogen identification in UP cases.
Case Report
A 10-week-old extremely preterm girl was admitted to our hospital for severe paroxysmal cyanosis and tachypnea. The infant was the product of a 25-week-and-4-day gestation and was a test-tube baby, from a gestational diabetes mother. The pregnancy was otherwise uneventful. A vaginal delivery was performed. The infant was 890 g with Apgar scores of 5, 9, and 9 at 1, 5, and 10 min, respectively. The infant was independent on the oxygen when she was discharged after 58 days of hospitalization with 2,020 g.
At day of life 70, the infant presented with symptoms of poor response and appetite, but her parents did not notice these symptoms. At day of life 72, the infant has symptoms of paroxysmal cyanosis and tachypnea, then was subsequently admitted to our hospital. Upon arrival to our hospital, vital signs were notable for respiratory rate of 70 per minute and heart rate of 170 beats per minute. Physical exam revealed coarse breath sounds and pulmonary moist rales. The infant was diagnosed with severe pneumonia and we immediately gave CPAP assisted ventilation (FiO2 50% PEEP 5 cmH2O). Meanwhile, we actively carried out blood culture, sputum culture, throat swab, respiratory virus (respiratory syncytial virus, influenza viruses, parainfluenza viruses, adenovirus, rhinovirus) and mycoplasma detection from blood and galactomannan detection. Then intravenous fluids and meropenem were administered (Figure 1). Chest X-ray showed high-density shadow in the left middle lobe lung on Day 1. But her general condition progressively worsened after 3 days. Her white blood cell count increased from 5.71 × 109/L (with 52% neutrophils and 27.5% lymphocytes) to 16.37 × 109/L (with 74.9% neutrophils and 16.7% lymphocytes), whereas her C-reactive protein and procalcitonin levels were normal. Pulmonary atelectasis in the right upper lobe and bilateral alveolar consolidation by chest computed tomography on Day 6 after the onset of symptoms (Figure 2A). Meanwhile repeated blood and sputum secretions were negative for bacteria, respiratory virus, tuberculosis, and fungi in cultures, and G-test (1,3 -beta-D-glucan test) and GM-test (galactomannan test), respectively. Her oxygenation index was 125 at admission, indicating a bad oxygenation status. And her echocardiography and blood pro-brain natriuretic peptide readings were normal. During this period, the infant showed polypnea, paroxysmal dysphoria, cough, and abundant yellow sticky respiratory secretions. Given the concern for a severe infection, empiric broad-spectrum antibiotics consisting of meropenem, vancomycin, and voriconazole to cover typical bacterial pathogens and fungal infection after a second infectious workup was performed including repeated blood culture, sputum culture (Figure 1).
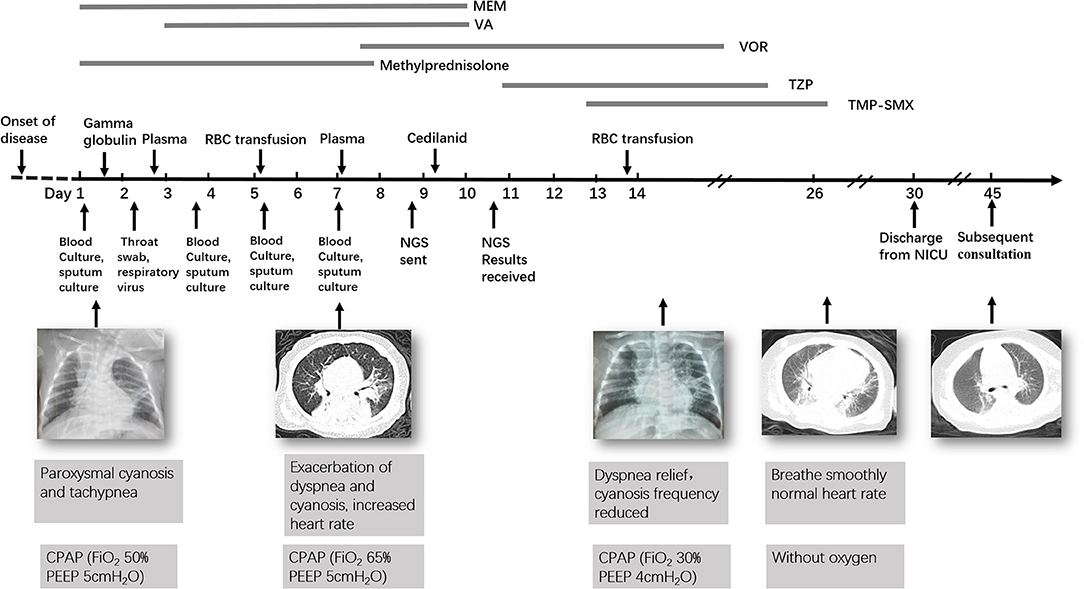
Figure 1. Timeline of the patient's clinical course and outcome. MEM, Meropenem; VA, Vancomycin; VOR, Voriconazole; TZP, Tazobactam and Piperacillin; TMP-SMX, Trimethoprim/Sulfamethoxazole.
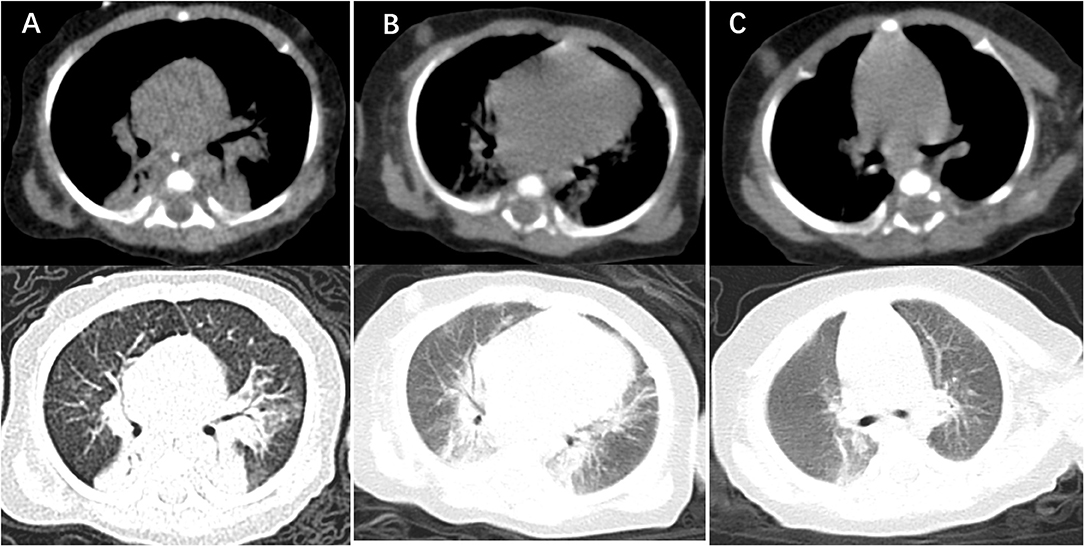
Figure 2. Computer tomography imaging showing lung lesion. (A) CT of the chest obtained around the time of clinical presentation most severely, showing bilateral alveolar multiple patchy shadows and local consolidation, more prominent on the left lung than right. (B) CT of the chest obtained after treatment 14 days with TMP-SMX, showing absorption multiple patchy and local consolidation. (C) CT of the chest obtained around the time of 2 weeks after discharge from NICU, showing that dual markings slightly increased, the lung parenchyma was no significant consolidation.
At day 10, the microbial species were then assessed by metagenomic analysis. Deoxyribonucleic acid libraries for Next-generation sequencing (NGS) were prepared as previously described (4, 5). Sequencing was performed on an Illumina MiSeq instrument. Total numbers of raw reads were 13849671, and non-human reads accounted for 5.92%. Stenotrophomonas maltophilia reads (428) were the only bacterial pathogen in blood (Figure 3A). In addition, we also detected several fungi pathogens in blood, including Penicillium chrysogenum reads (404), Pichia anomala reads (134), and Candida parapsilosis reads (6) (Figures 3B–D). The infant was then treated with Tazobactam and Piperacillin (TZP) and trimethoprim/sulfamethoxazole (TMP-SMX) successively, subjected to non-invasive positive pressure ventilation. Her oxygenation status and chest computed tomography showed significant improvement after treatments (Figure 2B), and the infant was discharged on Day 30 after admission. Two weeks after discharge, the infant appeared good responses and appetite, and her chest computed tomography showed distribution of the two slightly thickened lung markings, clear lung fields and no substantive changes (Figure 2C).
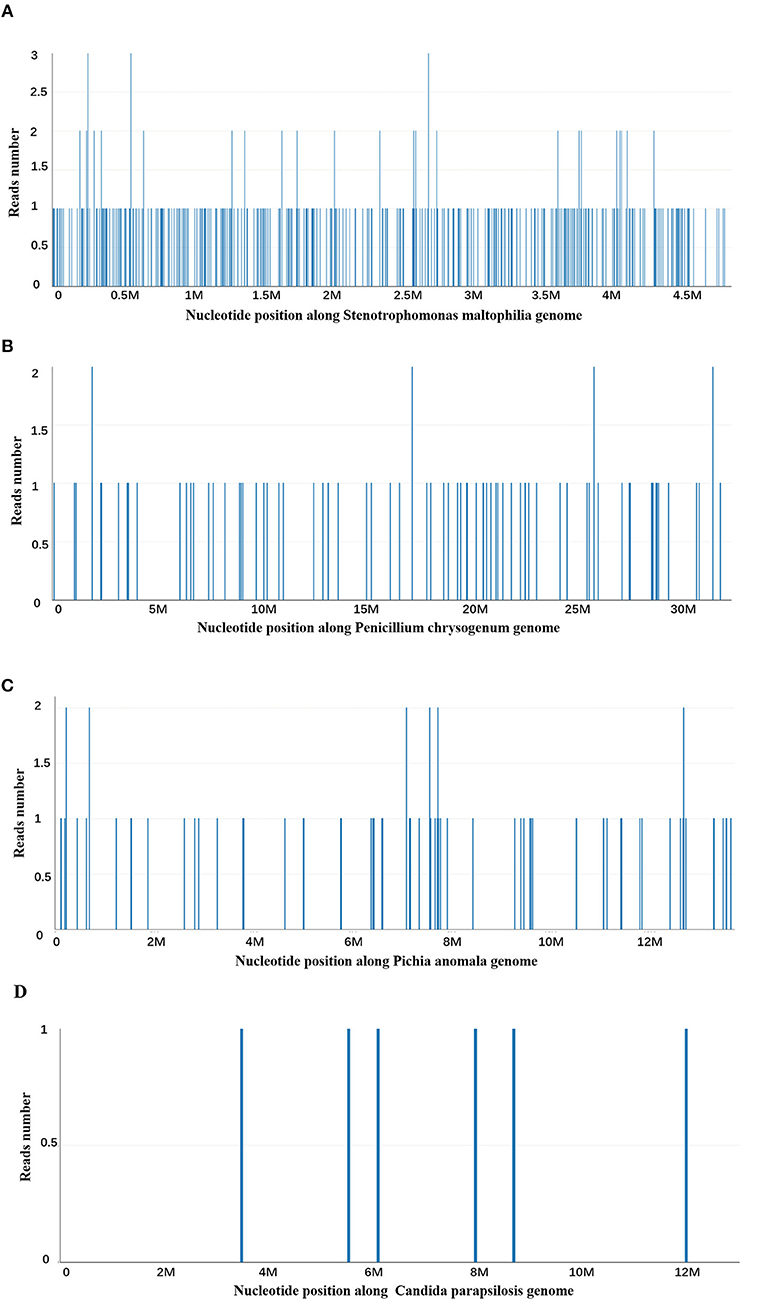
Figure 3. Next-generation sequencing and a metagenomic analysis. (A) Stenotrophomonas maltophilia specific reads and its nucleotide position along Stenotrophomonas maltophilia genome. (B) Penicillium chrysogenum specific reads and its nucleotide position along Penicillium chrysogenum genome. (C) Pichia anomala specific reads and its nucleotide position along Pichia anomala genome. (D) Candida parapsilosis specific reads and its nucleotide position along Candida parapsilosis genome.
Discussion
Bacteria, virus, and fungi are the major pathogens of human respiratory infections. However, pathogens remain undetected by routine clinical testing in ~30% of respiratory cases (6). Therefore, for these patients, we need to choose more sensitive and effective ways to detect pathogens for etiological diagnosis and reasonable treatment. In this study, we report a case of unexplained very severe pneumonia due to S. maltophilia that was diagnosed by plasma whole-genome NGS method.
Stenotrophomonas maltophilia is a commensal and an emerging pathogen earlier noted in broad-spectrum life-threatening infections among the vulnerable, but more recently as a pathogen in immunocompetent individuals (7). In addition, S. maltophilia has emerged as an important pathogen that induces nosocomial infections (8). S. maltophilia is a non-fermentative, gram-negative bacillus and causes severe infectious diseases, such as pneumonia, bacteremia, skin and soft-tissue infection, urinary tract infection, and meningitis (9–12). In recent times, the consistency of infection by the organism as reported worldwide is quite alarming. S. maltophilia accounts for about 3.7% (n = 10,000) in hospital discharges (13–15). A recovery rate of 3.3% in S. maltophilia infections was reported in the United States (16). And the mortality rate due to S. maltophilia infection was 36.6% rate found in a large scale study (17) and 25–51% in a multi-center study (18). Pathogenesis is by colonization, rather than infection, which is often accompanied by tissue invasion (19). The most important risk factors for S. maltophilia infection in neonates and infants are invasive procedures; previous exposure to antibiotics, such as carbapenem and aminoglycoside; and prolonged hospitalization (20). The duration of hospitalization of some patients before the onset of the Stenotrophomonas related clinical features and/or diagnosis is an important factor in nosocomial infection. Exemplifying case studies considered the duration of hospitalization before the onset of S. maltophilia bacteremia, which ranged from 11.5 to 24 days (21, 22). Neonate and infants have low immunity, and often have severe and uncontrollable symptoms after infection. S. maltophilia is intrinsically resistant to lots of antibiotics, including carbapenem and aminoglycosides which are used empirically for nosocomial infection (8). Therefore, early identification and appropriate treatment are important.
In principle, conventional cultural methods on nutrient agar support the growth of S. maltophilia, although certain strains require methionine (23, 24). However, we did not find any other pathogenic bacteria in conventional blood culture and sputum culture, which brought great difficulties to diagnosis and treatment. The reason may be inappropriate culture media or low bacterial counts but high virulent. We treated the children with empirical broad-spectrum antibiotics but her symptoms gradually worsened. Therefore, we choose microbial sequencing research to further explore pathogens. Common methods include 16S rDNA sequencing and metagenomic sequencing. Metagenomic sequencing has great potential utility in the diagnosis of infectious disease since it can obtain all kinds of pathogen information, including bacteria, viruses, fungi, and parasites, etc., and simultaneously interrogate host responses. While 16S rDNA refers to the identification of bacteria species by using the method of 16S rDNA sequence, which can only detect bacteria-like pathogens and that cannot fully cover the source of infection in clinical samples. In addition, metagenomic sequencing has multiple advantages compared with the 16S rDNA sequencing method including increased detection of diversity, increased prediction of genes and improved the accuracy of species detection (25). In this case, we did not find any other pathogens in repeated blood culture, sputum culture and other etiological examination, and her C-reactive protein and procalcitonin levels were normal during whole clinical course of the pneumonia. Therefore, we need total metagenomics to cover a broader range of pathogens.
In this study, we applied next-generation sequencing (NGS) technology and a metagenomic approach to detect S. maltophilia infection in infant. Metagenomics provides a means of assessing the total genetic pool of all the microbes in a sample, in a culture-independent manner (26). Firstly, it only takes 48 h to have the identification, which is faster than the traditional bacterial culture. Secondly, these approaches have the benefit of increasing the number and proportion of pathogen reads in the sequence data, which can increase the detection sensitivity for microorganisms (26–28). In addition, key advantage of NGS approaches is that the sequencing data can potentially be leveraged or additional analyzed beyond the mere identification of a causative pathogen, such as microbiome characterization and parallel analyses of human host responses through transcriptome profiling by RNA sequencing (RNA-seq). Thus, the clinical utility of NGS in diagnosis may be in the most difficult-to-diagnose cases or for immunocompromised patients, in whom the spectrum of potential pathogens is greater (29). In principle, the sensitivity, specificity and credibility of the body fluids or tissue samples from the infected site are higher, such as bronchoalveolar lavage fluid (BALF) for pneumonia. However, due to the young age and serious condition of the infant, BALF was not suitable, we selected blood samples for testing. Due to the abundant blood vessels in the lungs of small infants, pathogens or toxins produced by them can enter the blood, which makes the detection rate of pathogens in the blood significantly increased. NGS combines molecular biology techniques and informatics to filter human sequences and identify any microorganism sequences directly from patient plasma (30). Moreover, metagenomic sequencing could provide detailed genomic information of the pathogens presented in samples, which could be used for genotype characterization and phylogenetic analysis (31).
However, a positive pathogen test is not always meaning an infectious pathogen. To determine whether it is contaminated bacteria, colonizing bacteria or pathogenic bacteria, it requires a comprehensive analysis according to the infection site, clinical manifestations of patients, characteristics of medical history, environmental conditions, immune status, imaging, hematological examination, and response to empirical treatment. In this case, the peripherally inserted central venous catheters (PICC) was established 1 week after birth, and the PICC was removed on the day before her first discharge, which was used for 50 days. After the first admission, the infant was treated with meropenem, vancomycin, and cefoperazone sodium sulbactam successively, and the final exposure to broad-spectrum antibiotics was at 18 days before first discharge with cefoperazone sodium and sulbactam sodium. Infants, particularly those born prematurely, receive extensive antibiotic therapy during the first weeks of life may have S. maltophilia colonization in the respiratory tract (20). The infant was the extremely preterm of a 25-week-and-4-day gestation and had the most important risk factors for S. maltophilia infection, such as PICC, previous exposure to antibiotics in her early life and prolonged hospitalization. Futhermore, clinical manifestations of this infant and responses to empirical treatment were consistent with the characteristics of S. maltophilia infection. Therefore, we selected the antibiotic, TZP and TMP/SMX, which are sensitive to S. maltophilia. In addition, we also detected several fungal pathogens in blood, including Penicillium chrysogenum reads (404), Pichia anomala reads (134), and Candida parapsilosis reads (6). Due to the negative fungal tests (G-test, GM-test, and Fungal culture) and her clinical manifestations, we did not consider that this infant suffered severe fungal infection. Furthermore, the infant was prophylactic treated with VOR that can cover these fungal pathogens before NGS results received.
After receiving the NGS results, we firstly considered stopping meropenem and vancomycin because of their natural resistance to S. maltophilia. In addition, we cannot confirm the infection of S. maltophilia or the possibility of contamination, so we chose broad-spectrum antibiotics, TZP, that S. maltophilia were sensitive to at the same time. But there was no significant improvement in symptoms after 2 days' treatment in this case. Previous studies have shown that TZP can be a candidate for combination therapy in treating S. maltophilia because isolates showed a high susceptibility rate to this combination. However, TZP are not suitable for monotherapeutic strategy for treating S. maltophilia because this microorganism has a high intrinsic resistance to most penicillins and cephalosporins, as well as to all carbapenems (32). Thus, we added TMP-SMX combined treatment considering the severity of the disease and the possible insensitivity to TZP. TMP/SMX, ticarcillin-clavulanate, fluoroquinolone, colistin, and tigecycline are candidates that show consistent therapeutic activity against S. maltophilia. Although increasing resistance of S. maltophilia to TMP-SMX has been reported (8), the present study suggests that TMP-SMX may still be suitable as first-line treatment (7), and be recommended for being most potent antibiotics against S. maltophilia (22). In addition, TMP-SMX has been recommended by a number of researchers as initial therapeutic option for serious S. maltophilia infection (33). Wang et al. showed that clinical success rates of monotherapy with TMP-SMX was 61% (34). In this case, we have treated infant with TMP/SMX showing benefit outcomes. Studies showed that treatment of S. maltophilia infection with a combination of two or three antimicrobials can be considered in the current practice because S. maltophilia has a high resistance rate (34, 35). And in this case, it may be the synergistic effects of the two antibiotics.
In conclusion, S. maltophilia should be considered as a possible infectious agent in pediatric patients with high Stenotrophomonas infection risk factors. For unexplained very severe pneumonia, NGS will enhance the diagnostic rate for “unknown” pathogens in pediatrics and facilitate the identification of new pathogens. Ongoing technological and bioinformatic innovations, such as the NGS diagnostic tools, will hopefully apply to the clinic for timely, targeted, and precise antibiotic use in the ICU.
Data Availability Statement
The raw data can be accessed from EBI metagenomics (Accession: PRJEB32856).
Ethics Statement
Written informed consent was obtained from the individual(s) and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YL, B-XW, and XJ conceptualized and designed the study, drafted the initial manuscript, and reviewed. YL and N-NZ reviewed and revised the manuscript. YL, LZ, Z-BG, and JT designed the data collection instruments, collected data, carried out the initial analyses. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from the Innovation Foundation of Tangdu Hospital of Fourth Military Medical University (2016JCYJ010) and Shannxi Provincial Science Foundation of China (2018JM7052).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Yun Yan for reviewing our manuscript.
References
1. Rudan I, Boschi-Pinto C, Biloglav Z, Mulholland K, Campbell H. Epidemiology and etiology of childhood pneumonia. Bull World Health Organ. (2008) 86:408–16. doi: 10.2471/blt.07.048769
2. Kyu HH, Pinho C, Wagner JA, Brown JC, Bertozzi-Villa A, Charlson FJ, et al. Global and national burden of diseases and injuries among children and adolescents between 1990 and 2013: findings from the global burden of disease 2013 study. JAMA Pediatr. (2016) 170:267–87. doi: 10.1001/jamapediatrics.2015.4276
3. Revised WHO Classification and Treatment of Pneumonia in Children at Health Facilities: Evidence Summaries. Geneva: World Health Organization (2014).
4. De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. (2013) 155:1178–87. doi: 10.1016/j.cell.2013.10.034
5. De Vlaminck I, Martin L, Kertesz M, Patel K, Kowarsky M, Strehl C, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci USA. (2015) 112:13336–41. doi: 10.1073/pnas.1517494112
6. Heikkinen T, Jarvinen A. The common cold. Lancet. (2003) 361:51–9. doi: 10.1016/s0140-6736(03)12162-9
7. Adegoke AA, Stenstrom TA, Okoh AI. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: looking beyond contemporary antibiotic therapy. Front Microbiol. (2017) 8:2276. doi: 10.3389/fmicb.2017.02276
8. Looney WJ, Narita M, Muhlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis. (2009) 9:312–23. doi: 10.1016/s1473-3099(09)70083-0
9. Furuichi M, Ito K, Miyairi I. Characteristics of Stenotrophomonas maltophilia bacteremia in children. Pediatr Int. (2016) 58:113–8. doi: 10.1111/ped.12745
10. Waters V, Atenafu EG, Salazar JG, Lu A, Yau Y, Matukas L, et al. Chronic Stenotrophomonas maltophilia infection and exacerbation outcomes in cystic fibrosis. J Cyst Fibros. (2012) 11:8–13. doi: 10.1016/j.jcf.2011.07.008
11. Flores-Trevino S, Gutierrez-Ferman JL, Morfin-Otero R, Rodriguez-Noriega E, Estrada-Rivadeneyra D, Rivas-Morales C, et al. Stenotrophomonas maltophilia in Mexico: antimicrobial resistance, biofilm formation and clonal diversity. J Med Microbiol. (2014) 63:1524–30. doi: 10.1099/jmm.0.074385-0
12. Pompilio A, Crocetta V, Ghosh D, Chakrabarti M, Gherardi G, Vitali LA, et al. Stenotrophomonas maltophilia phenotypic and genotypic diversity during a 10-year colonization in the lungs of a cystic fibrosis patient. Front Microbiol. (2016) 7:1551. doi: 10.3389/fmicb.2016.01551
13. Huang YW, Hu RM, Chu FY, Lin HR, Yang TC. Characterization of a major facilitator superfamily. (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J Antimicrob Chemother. (2013) 68:2498–505. doi: 10.1093/jac/dkt250
14. Garcia-Leon G, Ruiz de Alegria Puig C, Garcia de la Fuente C, Martinez-Martinez L, Martinez JL, Sanchez MB. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin Microbiol Infect. (2015) 21:464–7.doi: 10.1016/j.cmi.2015.01.007
15. Reynaud Q, Weber E, Gagneux-Brunon A, Suy F, Romeyer-Bouchard C, Lucht F, et al. Late Stenotrophomonas maltophilia pacemaker infective endocarditis. Med Mal Infect. (2015) 45:95–7. doi: 10.1016/j.medmal.2015.01.016
16. Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. (2010) 51(Suppl. 1):S81–7. doi: 10.1086/653053
17. Jeon YD, Jeong WY, Kim MH, Jung IY, Ahn MY, Ann HW, et al. Risk factors for mortality in patients with Stenotrophomonas maltophilia bacteremia. Medicine. (2016) 95:e4375. doi: 10.1097/md.0000000000004375
18. Ebara H, Hagiya H, Haruki Y, Kondo E, Otsuka F. Clinical characteristics of Stenotrophomonas maltophilia bacteremia: a regional report and a review of a Japanese case series. Intern Med. (2017) 56:137–42.doi: 10.2169/internalmedicine.56.6141
19. Juhasz E, Krizsan G, Lengyel G, Grosz G, Pongracz J, Kristof K. Infection and colonization by Stenotrophomonas maltophilia: antimicrobial susceptibility and clinical background of strains isolated at a tertiary care centre in Hungary. Ann Clin Microbiol Antimicrob. (2014) 13:333. doi: 10.1186/s12941-014-0058-9
20. Mutlu M, Yilmaz G, Aslan Y, Bayramoglu G. Risk factors and clinical characteristics of Stenotrophomonas maltophilia infections in neonates. J Microbiol Immunol Infect. (2011) 44:467–72. doi: 10.1016/j.jmii.2011.04.014
21. Lai CH, Chi CY, Chen HP, Chen TL, Lai CJ, Fung CP, et al. Clinical characteristics and prognostic factors of patients with Stenotrophomonas maltophilia bacteremia. J Microbiol Immunol Infect. (2004) 37:350–8.
22. Friedman ND, Korman TM, Fairley CK, Franklin JC, Spelman DW. Bacteraemia due to Stenotrophomonas maltophilia: an analysis of 45 episodes. J Infect. (2002) 45:47–53. doi: 10.1053/jinf.2002.0978
23. O'Malley CA. Infection control in cystic fibrosis: cohorting, cross-contamination, and the respiratory therapist. Respir Care. (2009) 54:641–57.
24. Pinot C, Deredjian A, Nazaret S, Brothier E, Cournoyer B, Segonds C, et al. Identification of Stenotrophomonas maltophilia strains isolated from environmental and clinical samples: a rapid and efficient procedure. J Appl Microbiol. (2011) 111:1185–93. doi: 10.1111/j.1365-2672.2011.05120.x
25. Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. (2016) 469:967–77. doi: 10.1016/j.bbrc.2015.12.083
26. Lefterova MI, Suarez CJ, Banaei N, Pinsky B. A. Next-generation sequencing for infectious disease diagnosis and management: a report of the association for molecular pathology. J Mol Diagn. (2015) 17:623–34. doi: 10.1016/j.jmoldx.2015.07.004
27. Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, et al. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. (2014) 370:2408–17. doi: 10.1056/NEJMoa1401268
28. Dunne WM Jr. Westblade LF, Ford B. Next-generation and whole-genome sequencing in the diagnostic clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis. (2012) 31:1719–26. doi: 10.1007/s10096-012-1641-7
29. Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. (2019) 20:341–55. doi: 10.1038/s41576-019-0113-7
30. Abril MK, Barnett AS, Wegermann K, Fountain E, Strand A, Heyman BM, et al. Diagnosis of Capnocytophaga canimorsus sepsis by whole-genome next-generation sequencing. Open Forum Infect Dis. (2016) 3:ofw144. doi: 10.1093/ofid/ofw144
31. Zou X, Tang G, Zhao X, Huang Y, Chen T, Lei M, et al. Simultaneous virus identification and characterization of severe unexplained pneumonia cases using a metagenomics sequencing technique. Sci China Life Sci. (2017) 60:279–86. doi: 10.1007/s11427-016-0244-8
32. Rhee JY, Song JH, Ko KS. Current situation of antimicrobial resistance and genetic differences in Stenotrophomonas maltophilia complex isolates by multilocus variable number of tandem repeat analysis. Infect Chemother. (2016) 48:285–93. doi: 10.3947/ic.2016.48.4.285
33. Lo WT, Wang CC, Lee CM, Chu ML. Successful treatment of multi-resistant Stenotrophomonas maltophilia meningitis with ciprofloxacin in a pre-term infant. Eur J Pediatr. (2002) 161:680–2. doi: 10.1007/s00431-002-1095-5
34. Wang YL, Scipione MR, Dubrovskaya Y, Papadopoulos J. Monotherapy with fluoroquinolone or trimethoprim-sulfamethoxazole for treatment of Stenotrophomonas maltophilia infections. Antimicrob Agents Chemother. (2014) 58:176–82. doi: 10.1128/aac.01324-13
Keywords: severe pneumonia, Stenotrophomonas maltophilia, infant, next-generation sequencing, metagenomics
Citation: Lin Y, Wang B-X, Zhang N-N, Zhang L, Gao Z-B, Tian J and Jiang X (2019) Metagenomic Analysis Identified Stenotrophomonas maltophilia Pneumonia in an Infant Suffering From Unexplained Very Severe Pneumonia. Front. Pediatr. 7:380. doi: 10.3389/fped.2019.00380
Received: 30 May 2019; Accepted: 03 September 2019;
Published: 19 September 2019.
Edited by:
Anne B. Chang, Charles Darwin University, AustraliaReviewed by:
Jose L. Martinez, Spanish National Research Council (CSIC), SpainBülent Taner Karadag, Marmara University, Turkey
Copyright © 2019 Lin, Wang, Zhang, Zhang, Gao, Tian and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Lin, yiyilin19800823@126.com; Xun Jiang, jiangx@fmmu.edu.cn
†These authors have contributed equally to this work
 Yan Lin
Yan Lin Bao-Xi Wang
Bao-Xi Wang Ni-Ni Zhang
Ni-Ni Zhang Lei Zhang
Lei Zhang Zhi-Bo Gao
Zhi-Bo Gao Jiao Tian
Jiao Tian  Xun Jiang
Xun Jiang