Pediatric obstructive sleep apnea diagnosis: leveraging machine learning with linear discriminant analysis
- 1Department of Child Health Care, Children’s Hospital Capital Institute of Pediatrics, Chinese Academy of Medical Sciences & Peking Union Medical College, Capital Institute of Pediatrics, Beijing, China
- 2Pharmacovigilance Research Center for Information Technology and Data Science, Cross-strait Tsinghua Research Institute, Xiamen, China
- 3Department of Otolaryngology, Head and Neck Surgery, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing, China
- 4Respiratory Department, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing, China
- 5Faculty of Information Technology, Beijing University of Technology, Beijing, China
- 6Department of Otolaryngology, Head and Neck Surgery, Children’s Hospital Capital Institute of Pediatrics, Beijing, China
- 7Department of Automation, Tsinghua University, Beijing, China
Objective: The objective of this study was to investigate the effectiveness of a machine learning algorithm in diagnosing OSA in children based on clinical features that can be obtained in nonnocturnal and nonmedical environments.
Patients and methods: This study was conducted at Beijing Children's Hospital from April 2018 to October 2019. The participants in this study were 2464 children aged 3–18 suspected of having OSA who underwent clinical data collection and polysomnography(PSG). Participants’ data were randomly divided into a training set and a testing set at a ratio of 8:2. The elastic net algorithm was used for feature selection to simplify the model. Stratified 10-fold cross-validation was repeated five times to ensure the robustness of the results.
Results: Feature selection using Elastic Net resulted in 47 features for AHI ≥5 and 31 features for AHI ≥10 being retained. The machine learning model using these selected features achieved an average AUC of 0.73 for AHI ≥5 and 0.78 for AHI ≥10 when tested externally, outperforming models based on PSG questionnaire features. Linear Discriminant Analysis using the selected features identified OSA with a sensitivity of 44% and specificity of 90%, providing a feasible clinical alternative to PSG for stratifying OSA severity.
Conclusions: This study shows that a machine learning model based on children's clinical features effectively identifies OSA in children. Establishing a machine learning screening model based on the clinical features of the target population may be a feasible clinical alternative to nocturnal OSA sleep diagnosis.
1 Introduction
Obstructive sleep apnea (OSA) is a severe type of sleep-disordered breathing (SDB) characterized by recurrent events, including instances of partial or complete obstruction of the upper airway and disruptions in normal oxygenation, ventilation, and sleep patterns (1). In children, the estimated prevalence of OSA is 1%–5% (2). However, it is notably higher, ranging from 33% to 61%, among youths who are diagnosed with obesity (3). Untreated OSA has been linked to disturbances in both the cardiovascular and metabolic systems, as well as neurocognitive and behavioral dysfunction (4). Additionally, it is associated with hypertension (5), diminished school performance (6), growth failure (7), and a decline in overall quality of life (8).
Polysomnography (PSG) is the gold standard for diagnosing and characterizing OSA (2). However, it has drawbacks, such as being costly, time-consuming, and requiring specialized facilities and staff. Data collection of PSG requires an overnight hospital stay in a specially equipped sleeping suite involving more than 15 measurement channels (9). Based on these limitations, previous research indicates that over 80% of individuals with OSA are estimated to remain undiagnosed (10), and only 5%–10% of children receive PSG before adenotonsillectomy, leading to potential overuse of surgeries and postoperative risks (11). There is a view that developing a practical and affordable OSA diagnostic model would greatly benefit children in areas with limited sleep laboratory facilities (12).
Machine Learning (ML) methods represent an evolving approach capable of simultaneously and autonomously processing substantial volumes of data, continually refining their classification performance through previous experiences (13). Particularly adept at discerning patterns within data featuring numerous variables (14), ML methods harness extensive clinical datasets to craft practical diagnostic tools. In recent years, promising results have been reported in studies involving ML methods that facilitate OSA diagnosis using children's clinical features to develop diagnostic tools (15, 16), even to classify sleep stages in children with OSA (17). Most OSA diagnosis studies using ML methods rely on nocturnal biological signals such as Electrocardiogram (ECG), Electroencephalogram (EEG), Oxygen saturation (SpO2), and airflow signals to build diagnostic models (18). Compared with PSG, this approach dramatically reduces the number of biomarkers collected but still requires specialized electronic equipment for data collection.
Using clinical features and questionnaires is a cost-effective approach for identifying children with OSA, circumventing the need for specialized laboratory and sleep monitoring equipment (19–23). Notably, a study has enhanced the efficacy of OSA screening by using Selected Features and optimizing existing ones, demonstrating improved performance after eliminating redundancy features (24). However, these studies do not use cross-validation or test sets to verify classification performance. Given these considerations, our hypothesis posits that a machine learning approach, integrating multiple clinical variables, can more effectively identify pediatric OSA than standalone questionnaires. Consequently, this study pursues dual objectives: (1) Construction of a machine learning model solely relying on non-invasive clinical features for pediatric OSA diagnosis, and (2) Assessment and comparison of its performance against models derived from established sleep questionnaires.
2 Materials and methods
2.1 Dataset and inclusion and exclusion criteria
The study received approval from the Ethics Committee of Beijing Children's Hospital, Capital Medical University [(2022)-E-111-R]. Data collection occurred between April 2018 and October 2019 and involved children aged 3–18 years suspected of having OSA. Written informed consent was obtained from the parent or guardian of each participating child. A specialized physician measured the children's height, weight, neck, waist, and hip circumference before PSG. Additionally, the parents or guardians filled out an information collection form to provide the necessary features for the study.
Exclusion criteria included missing demographic information, presence of other disease-causing disordered breathing (e.g., neuromuscular diseases, craniofacial dysplasia, Down syndrome), chronic lung diseases, previous adenoidectomy and/or tonsillectomy, respiratory infection within the last three weeks, total sleep time less than 5 h, and non-completion of the questionnaire. The study adhered to the reporting guidelines of the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) statement (25).
2.2 Clinical feature data collection
The features used in this study were collected from no wearable devices or other overnight biomarker recordings. (1) Demographic and anthropometric data, including age, sex, neck circumference, abdominal circumference, hip circumference, and Body mass index (BMI), were collected. Overweight status was assessed according to the BMI standard for children aged 2–18 in China (26). The ratios of neck circumference to height, waist circumference to height, hip circumference to height, and waist to hip ratio were also calculated. (2) Children's symptoms and living habits were determined based on existing questionnaires such as the Pediatric Sleep Questionnaire (PSQ) (27), the Obstructive Sleep Apnea-18 (OSA-18) (28), and the Hong Kong Children's Sleep Questionnaire (HK-CSQ) (29). Some questions were refined to provide more information. In addition, we also collected information about family members, including the prevalence of snoring, OSA, and smoking, as well as the educational level of the parents. A total of 102 features have been collected, and a list of these features is shown in Supplementary Material 1.
Specialized doctors collected the above data using a questionnaire. The questionnaire was designed to gather clinical features, not as a screening test, and was formulated in simple language consistent with the local culture. Any confusing questions were clarified by a doctor.
2.3 Polysomnography
All patients underwent standard nocturnal PSG at Beijing Children's Hospital of Capital Medical University. The Alice 5 PSG device (Philips, Amsterdam, The Netherlands) was used for data collection. The data were manually scored according to the American Academy of Sleep Medicine (AASM) 2012 scoring criteria (30). Obstructive apnea was defined as a greater than 90% reduction in oronasal flow for at least two respiratory cycles, accompanied by respiratory efforts throughout the event. Hypopnea is defined as a reduction in airflow of at least ≥30%, accompanied by event-related arousal or oxygen desaturations of >3%, which persists for a minimum of two respiratory cycles. The apnea hypopnea index (AHI) was defined as the mean number of obstructive apnea and hypopnea events per hour during sleep. In this study, AHI ≥5 and AHI ≥10 were used as grouping criteria. The statistical differences between these groups are shown in Supplementary Material 1.
2.4 Data preprocessing
2.4.1 Raw datasets
The study included 2,464 patients (1,665 boys, 799 girls). Each participant in the raw dataset had 102 features and one label column. The label values were encoded as 1 for the positive class and 0 for the negative class. Python 3.6 and the open-source Python automated machine learning library, PyCaret 2.3.10, were used for data preprocessing, modeling, evaluation, statistical analysis, and feature analysis33 (31).
2.4.2 Raw data preprocessing
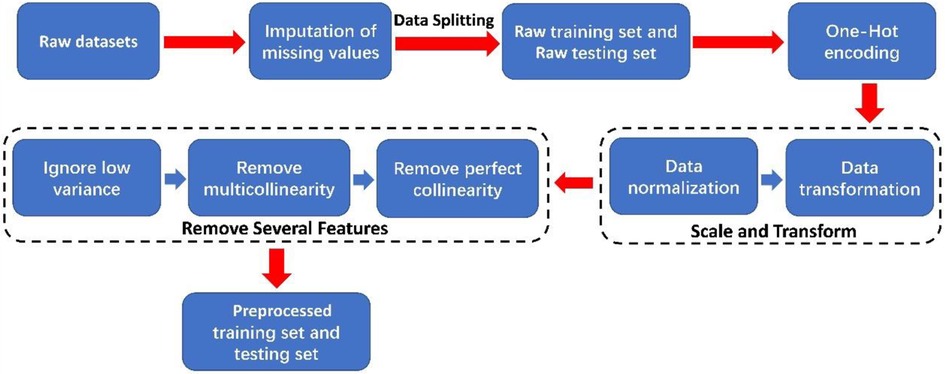
In this paper, the process of preprocessing the raw dataset is shown in Figure 1. Initially, missing values in categorical features were imputed with “NaN”. The dataset was then randomly split into a training set (1,971 samples) and a testing set (493 samples) with an 8:2 ratio. Afterward, one-hot encoding was applied to categorical features. Subsequently, we used the “z-score” normalization methods to perform data normalization and employed the “Yeo-Johnson” transformation methods to perform data transformation on continuous features (32). The “z-score” normalization method is used to find out the mean and standard deviation of the sample feature “x” and then use “(x-mean)/std” to replace the original feature value. The mean of each feature after “z-score” standardization is 0, and the standard deviation is 1. The “Yeo-Johnson” transformation methods bring the shape of the probability density function of the features closer to a normal distribution.
2.5 Machine learning methods
Instead of complex models with limited interpretability, six common ML methods were used for data modeling and evaluated for pediatric OSA binary classification performance using actual clinical data during cross-validation (33) as reported in previous studies (34–36). ML methods included logistic regression (LR) (37), linear discriminant analysis (LDA) (38), radial basis function kernel support vector machine (RBF-SVM) (39–41), CatBoost (42), AdaBoost (43), and random forest classifiers (RF) (44).
Several common metrics were implemented for the performance evaluation of each ML algorithm on the training and testing sets, including accuracy, balanced accuracy (BA), the area under the receiver operating characteristic curve (AUC) (45), positive predictive value (PPV), negative predictive value (NPV), sensitivity and specificity. Accuracy refers to the proportion of correctly identified samples in all samples. Balanced accuracy is a corrected measure of accuracy used for comparing datasets with imbalances in sample size. Higher AUC values indicate better classification performance of the model. The probability threshold was set to 0.5.
2.6 The procedure of data modeling
The data modeling procedure, as shown in Figure 2, was divided into three stages: model training, hyperparameter tuning of the selected model, and model evaluation. Hyperparameter tuning was performed using a random grid search, and the highest AUC value was used as the target. Five random seed values were utilized for data splitting during preprocessing to reduce bias (46). Six metrics were obtained from each hyperparameter-tuned ML model on the testing datasets. The average AUC value was calculated from five repetitions of the data modeling process, and other evaluation metrics were obtained similarly.
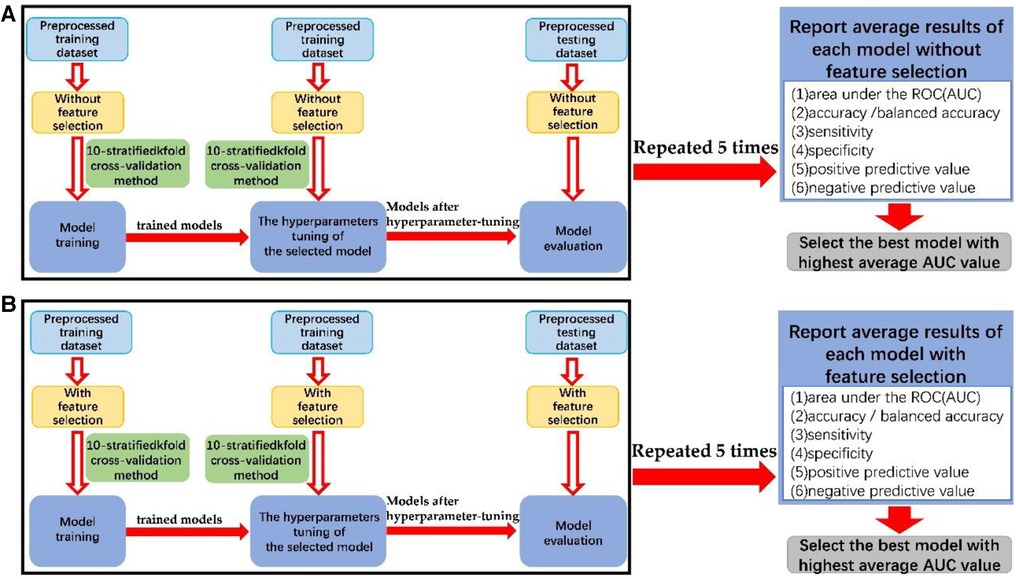
Figure 2. The block diagram of the process of data modeling (A) without feature selection and (B) with feature selection.
The preprocessed training datasets underwent model training and optimization using 10-stratified k-fold cross-validation. Overfitting was assessed by comparing results between training and testing datasets, and the selected hyperparameters were evaluated by comparing the trained model with the hyperparameter-tuned model. More reliable estimates of model performance were obtained through 10-stratified k-fold cross-validation in each repetition, ensuring consistent class distribution in each fold. The hyperparameters tuned through cross-validation of each machine learning algorithm is shown in Supplementary Material 5.
In addition, the best model was then trained and tested on preprocessed datasets that included 22 features of the PSQ questionnaire to predict the severity of OSA.
2.7 The procedure of feature selection
To select predictive features for OSA samples and create a concise model, we employed the Elastic net method as the feature selection algorithm (47). This method applies L1 and L2 penalties during training to shrink coefficients of unimportant features to zero. The Elastic net was implemented on all preprocessed datasets with two optimal parameters (alpha and L1_ratio) using the ElasticNet and ElasticNetCV functions (48). Features with coefficients having an absolute value greater than zero were retained, while those with a coefficient of zero were eliminated.
The feature selection procedure and optimal alpha and L1_ratio values are demonstrated in Supplementary Material 2. Finally, we performed a paired t test to compare the predictive performance of the best model based on PSQ and our selected feature set when an AHI greater than 5 and 10 events/h were the classification criteria. P < 0.05 was considered significant.
3 Results
After feature selection, 31 features were retained for a binary classification threshold of AHI ≥10 events/h and 47 features for AHI ≥5 events/h. The selected features are listed in Supplementary Material 3, and the details of the feature coefficients’ absolute values are reported in Supplementary Material 4. Twenty-seven features are simultaneously selected by machine learning algorithms for diagnostic tasks with AHI thresholds of 5 and 10. Among them, Sex has been proven to be a risk factor for OSA in children (49). Five features associated with Children's performance during bedtime had the highest feature importance values, indicating that these five features best-discriminated patients with vs. without moderate to severe pediatric OSA. The five features are A2. Snore more than half the time; A3. Always snore; A4. Snore loudly; A6. Have trouble breathing or struggle to breathe; A7. Stop breathing during the night; Q4. Mouth breathing during sleep. Based on the result of the feature coefficients’ absolute values, four features associated with Children's body measurement data and one feature associated with characteristics of family members were also used in the final model, e.g., Neck circumference, Hip circumference, Neck/height_ratio, Waist/hip_ratio, the educational level of the mother.
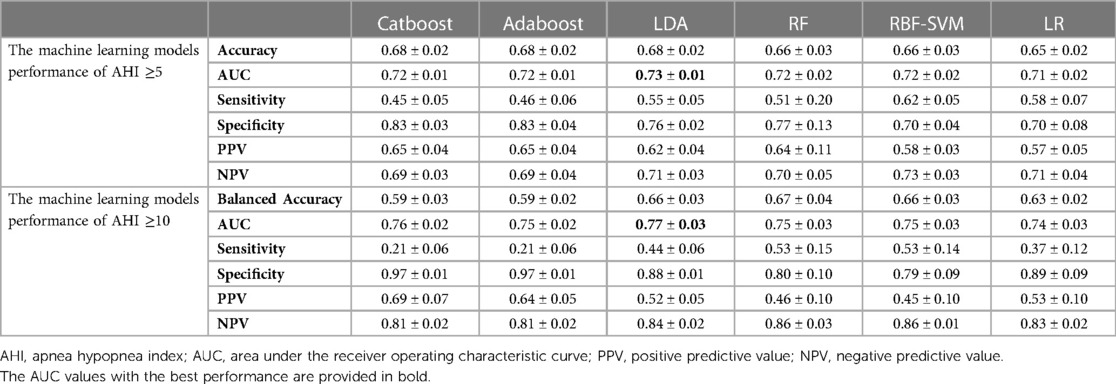
We compared the performance of six machine learning models for predicting OSA. Table 1 summarizes their average performance, obtained by repeating the data modeling process five times on the test datasets with all features. The performance was evaluated using six metrics, expressed as mean ± standard deviation. The results showed that the LDA classifier outperformed other models with the highest average AUC value of 0.73 and a mean accuracy rate of 68% when using an AHI of 5 events/h as the binary classification threshold. For the AHI of 10 events/h threshold, the LDA classifier remained the best model with the highest average AUC value of 0.77 and a mean balanced accuracy rate of 66%.
The performance of the selected features obtained by repeating the data modeling process five times on the test datasets is shown in Table 2. The LDA classifier achieved the highest average AUC value of 0.73 and a mean accuracy rate of 68% for the AHI ≥5 group. Additionally, it had the highest average AUC value of 0.78 and a mean balanced accuracy rate of 67% for the AHI ≥10 group.
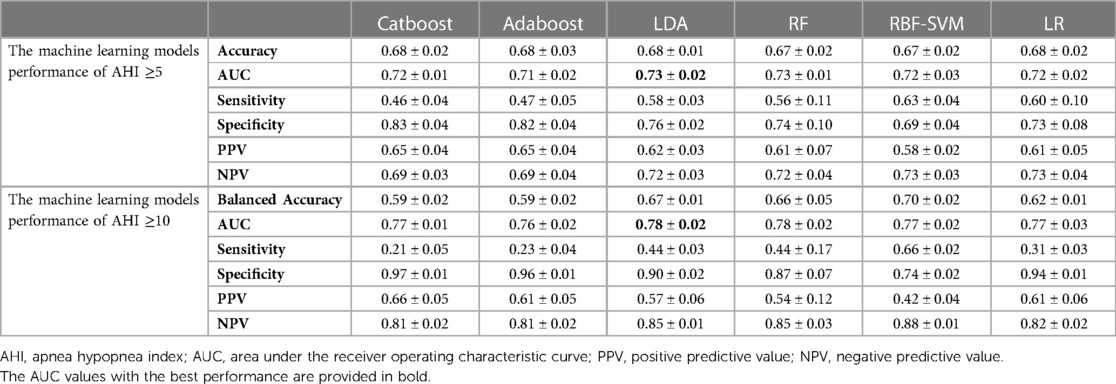
Table 2. Performance of each machine learning model using selected features on test dataset (mean ± SD).
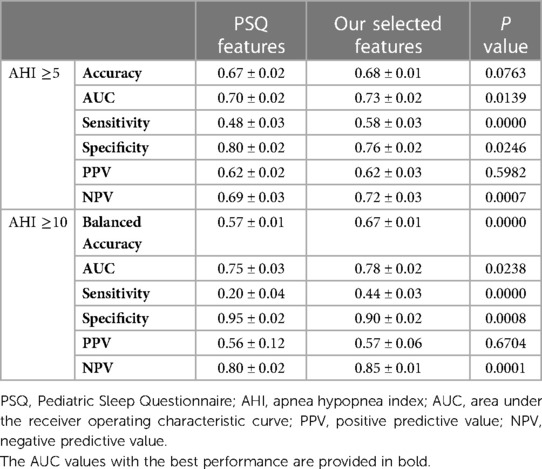
Furthermore, we compared the diagnostic ability of feature selection with the PSQ questionnaire using the LDA algorithm. The results, presented in Table 3, demonstrate that the LDA model of feature selection had a higher AUC value, sensitivity, and NPV than PSQ for classifying both the AHI ≥5 and AHI ≥10 groups (P < 0.05). These findings demonstrate that our selected features, when analyzed using the LDA algorithm, offer superior diagnostic accuracy and classification ability compared to the traditional PSQ features, particularly in distinguishing between different severity levels of pediatric OSA. The improvements in AUC, sensitivity, and specificity underscore the potential clinical utility of the feature selection approach in enhancing the accuracy of OSA diagnosis.
4 Discussion
The diagnostic ability of six ML methods based on clinical features in children with OSA was assessed in our study. The feature data used in this study can be collected during the day to avoid the burden of physiological signal collection throughout the night on children. The performance of each model was evaluated by the average accuracy rate and AUC value, which were obtained after five repetitions of the data modeling process. The ML models using the dataset with selected features showed slightly better performance compared to those using the dataset with all features. Among the ML models using the selected feature dataset, LDA obtained the best performance with an AUC value of 0.73 and an accuracy rate of 68% for the AHI ≥5 group and an AUC value of 0.78 and an accuracy rate of 67% for the AHI ≥10 group. Classical LDA projects the data onto a lower-dimensional vector space by the projection hyperplane that minimizes the interclass variance and maximizes the distance between the projected means of the classes (38). One possible explanation is the low feature dimension in our study, which gives the LDA model advantages in classification tasks. After feature selection further reduces the number of features, the performance of LDA is further improved.
Sleep questionnaires are the primary method for the daytime diagnosis of OSA in children. Improving its diagnostic performance is a concern of researchers. A recent meta-analysis reported the performance of the PSQ in predicting OSA in children and showed that the accuracy of predicting children with moderate OSA was 62.45%, the sensitivity was 0.79 [95% CI 0.69, 0.86], and the specificity was 0.47 [95% CI 0.28, 0.67]. Therefore, the reliability of the PSQ in real-world clinical populations is still uncertain (50). A recent study used clinical features to identify adult OSA and obtained an AUC value of 0.92, which has an advantage over our predictive model. One possible reason is that part of the predictive characteristics of children come from parents’ description of their children's sleep, which may reduce the reliability of the features (51).
In this study, we observed that specific questions in the original questionnaire were prone to ambiguity, potentially leading to less specific responses from parents. For example, question A3 from the Pediatric Sleep Questionnaire (PSQ), which asks about the frequency of snoring with the descriptor “Always,” might be challenging for parents to interpret accurately. The term “Always” is inherently vague, and parental subjective interpretations may vary, introducing potential inaccuracies in the data. We refined the answers to such questions to address this issue and enhance the precision of responses. Specifically, we focused on the A3 question regarding “Always snore” and introduced a new labeling scheme: Label “0” signifies a negative response, Label “1” corresponds to less than one time per month, Label “2” represents 1–2 times per month, Label “3” indicates 1 to 3 times per week, Label “4” denotes more than three times per week, and Label “5” designates an unclear response. Our research team conducted This refinement process internally, and no direct communication with the questionnaire developers was pursued. While we acknowledge that the original questionnaire designers may have had specific considerations, these adjustments were made within our research team to align with the goals of our study. Through these refinements, we aim to improve the quality and accuracy of the data, ensuring that machine learning algorithms can better comprehend and leverage this information for a more effective classification of pediatric OSA.
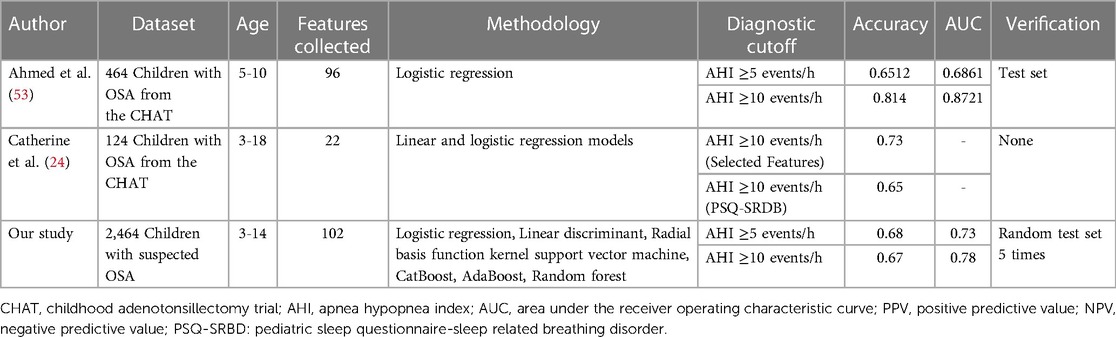
Table 4 compares previous studies using machine learning methods to predict OSA in children based on clinical features. Some of these studies have demonstrated that diagnostic models based on machine learning methods and self-built features can offer superior diagnostic accuracy compared to OSA screening questionnaires. For example, Catherine et al. compared the use of self-built features with the PSQ questionnaire in their study and reported the performance of both methods in children with severe OSA. The prediction accuracy of the self-built features was 0.73 (0.64–0.80) compared to that PSQ questionnaire, 0.63 (0.54–0.72) (24). Similarly, Ahmed et al. also used and compared self-built features with five different screening questionnaires, with the self-built model achieving better performance in children with severe OSA (23). The above studies all used data from the Childhood Adenotonsillectomy Trial (CHAT), a randomized controlled trial that compared early outcomes of T&A (52). Our study found similar findings in children with a suspected diagnosis of OSA who went to the outpatient clinic, using an ML algorithm based on 22 features of the PSQ to predict children's level of OSA, and its diagnostic performance was weaker than that of the diagnostic model after feature selection. One possible reason is that the features in the OSA screening questionnaire do not provide enough valid diagnostic information.
In our study, as shown in Supplementary Material 1, when an AHI of 5 events/h was used as the cutoff value, only 11 of the 22 features of the PSQ questionnaire showed significant differences between individuals with AHI greater than or less than 5 events/h (P < 0.05), which meant that half of the features could not benefit diagnosis. In addition, 29 and 34 clinical features from our questionnaire showed significant differences between groups when the AHI cutoff values were 5 and 10 events/h, respectively. Our feature list greatly increased the number of potentially effective features compared with the PSQ questionnaire. We believe it is important to consider the features’ effectiveness when establishing a simple diagnostic tool for OSA based on artificial intelligence. Additionally, due to differences in language and culture, finding effective feature combinations specific to the target population and establishing a diagnostic model may be more suitable than directly using the existing OSA diagnostic questionnaire. Our study is based on non-invasive clinical features. It has a significant cost advantage over traditional sleep monitoring (PSG), which creates the possibility of providing efficient OSA screening tools for children in a resource-limited medical environment. In this context, our model may provide a rapid and economical means for medical teams to identify children with OSA, leading to early intervention and treatment. However, we emphasize that more in-depth research and verification are needed before this method is put into clinical application, and its generalization in different populations, cultures, and medical practices still needs further investigation. We encourage future research teams to conduct more extensive, multicenter research to assess this method's applicability and robustness more comprehensively.
There are some limitations in this study. The AHI cut-off in this study does not include 1 event per hour, which is usually referred to as mild OSA. It should be noted that mild OSA children with persistent symptoms should be recommended for treatment. Although we collected 102 features, we believe not all OSA-related features were collected, for example, other OSA-related features like genetics and environment. Considering other factors affecting OSA, such as genetics and environment, more data types should be added in the future to increase clinical features. In addition, single-center studies, data that may be missing, retrospective design, the age of the participants and the lack of universality of the country may all contribute to data source bias. Although the sample size was larger than that of previous studies, there is still a risk of over-fitting using the same population data for result verification, and future external verification queues are needed to solve this problem. In the machine learning algorithms used in this study, LDA exhibited better performance. One possible reason is that LDA is simple and computationally efficient, which may make it more robust and less prone to overfitting. Nevertheless, we recognize the need for further research to fully elucidate the underlying reasons for the observed performance differences among these classifiers. In addition, more excellent machine learning models such as Bayesian networks should be considered as one of the classification models. In this study, we only focused on traditional feature-based methods for disease classification without exploring the potential of deep learning methods for processing high-dimensional features like images and sounds. With the advantages of deep learning in sleep research (54), introducing deep learning methods into studying OSA disease classification may have potential benefits. Future work can explore deep learning models such as convolution neural networks (CNN) to improve the modeling ability of complex feature relationships.
5 Conclusions
In this study, we used ML algorithms to analyze the clinical features of children with a suspected diagnosis of OSA. We found that using ML to investigate clinical features is an effective method to identify OSA in children, and ML models based on clinical features had better predictive ability than ML models based on the PSQ questionnaire. Using the features of nonnocturnal biological signals to stratify the severity of children's OSA is an essential diagnostic supplement to PSG. It provides references for children in areas where PSG is unavailable and cannot be used to determine the severity of OSA. Future research should search for additional practical features that can improve the prediction performance of ML algorithms.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
HQ: Writing – original draft. LZ: Software, Writing – original draft. XL: Data curation, Writing – original draft. YQ: Resources, Writing – review & editing. ZX: Project administration, Writing – review & editing. JZ: Investigation, Writing – review & editing. SW: Methodology, Writing – review & editing. LZ: Data curation, Writing – review & editing. TJ: Supervision, Writing – review & editing. LM: Validation, Writing – review & editing. YK: Visualization, Writing – review & editing. XJ: Resources, Writing – review & editing. YL: Software, Writing – review & editing. YQ: Writing – review & editing. JJ: Project administration, Writing – review & editing. XN: Supervision, Writing – review & editing. QW: Project administration, Writing – review & editing. JT: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This work was supported by the Capital’s Funds for Health Improvement and Research (2022-2-1132); Beijing Hospitals Authority’s Ascent Plan (DFL20221102); National natural science foundation of China (81970900); Public service development and reform pilot project of Beijing Medical Research Institute (BMR2021-3); Beijing Hospitals Authority Youth Programme (QML20201206); National Natural Science Foundation of China (82000991).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2024.1328209/full#supplementary-material
References
1. Kaditis AG, Alonso Alvarez ML, Boudewyns A, Alexopoulos EI, Ersu R, Joosten K, et al. Obstructive sleep disordered breathing in 2- to 18-year-old children: diagnosis and management. Eur Respir J. (2016) 47:69–94. doi: 10.1183/13993003.00385-2015
2. Marcus CL, Brooks LJ, Draper KA, Gozal D, Halbower AC, Jones J, et al. Diagnosis and management of childhood obstructive sleep apnea syndrome. Pediatrics. (2012) 130:e714–755. doi: 10.1542/peds.2012-1672
3. Roche J, Isacco L, Masurier J, Pereira B, Mougin F, Chaput JP, et al. Are obstructive sleep apnea and sleep improved in response to multidisciplinary weight loss interventions in youth with obesity? A systematic review and meta-analysis. Int J Obes (Lond). (2020) 44:753–70. doi: 10.1038/s41366-019-0497-7
4. Tan HL, Gozal D, Kheirandish-Gozal L. Obstructive sleep apnea in children: a critical update. Nat Sci Sleep. (2013) 5:109–23. doi: 10.2147/NSS.S51907
5. Kang KT, Chiu SN, Weng WC, Lee PL, Hsu WC. Comparisons of office and 24-hour ambulatory blood pressure monitoring in children with obstructive sleep apnea. J Pediatr. (2017) 182:177–183.e2. doi: 10.1016/j.jpeds.2016.11.032
6. Galland B, Spruyt K, Dawes P, McDowall PS, Elder D, Schaughency E. Sleep disordered breathing and academic performance: a meta-analysis. Pediatrics. (2015) 136:e934–946. doi: 10.1542/peds.2015-1677
7. Bonuck KA, Freeman K, Henderson J. Growth and growth biomarker changes after adenotonsillectomy: systematic review and meta-analysis. Arch Dis Child. (2009) 94:83–91. doi: 10.1136/adc.2008.141192
8. Rosen CL, Palermo TM, Larkin EK, Redline S. Health-related quality of life and sleep-disordered breathing in children. Sleep. (2002) 25:657–66. doi: 10.1093/sleep/25.6.648
9. Ghaemmaghami H, Abeyratne UR, Hukins C. Normal probability testing of snore signals for diagnosis of obstructive sleep apnea. Annu Int Conf IEEE Eng Med Biol Soc. (2009) 2009:5551–4. doi: 10.1109/IEMBS.2009.5333733
10. Finkel KJ, Searleman AC, Tymkew H, Tanaka CY, Saager L, Safer-Zadeh E, et al. Prevalence of undiagnosed obstructive sleep apnea among adult surgical patients in an academic medical center. Sleep Med. (2009) 10:753–8. doi: 10.1016/j.sleep.2008.08.007
11. Garde A, Hoppenbrouwer X, Dehkordi P, Zhou G, Rollinson AU, Wensley D, et al. Pediatric pulse oximetry-based OSA screening at different thresholds of the apnea-hypopnea index with an expression of uncertainty for inconclusive classifications. Sleep Med. (2019) 60:45–52. doi: 10.1016/j.sleep.2018.08.027
12. Roebuck A, Clifford GD. Comparison of standard and novel signal analysis approaches to obstructive sleep apnea classification. Front Bioeng Biotechnol. (2015) 3:114. doi: 10.3389/fbioe.2015.00114
13. Akkoyunlu B, Soylu MY. A study of student's perceptions in a blended learning environment based on different learning styles[J]. J Educ Techno Soc. (2008) 11(1):183–93.
14. Passos IC, Mwangi B, Kapczinski F. Big data analytics and machine learning: 2015 and beyond. Lancet Psychiatry. (2016) 3:13–5. doi: 10.1016/S2215-0366(15)00549-0
15. Ramachandran A, Karuppiah A. A survey on recent advances in machine learning based sleep apnea detection systems. Healthcare (Basel). (2021) 9(7):914. doi: 10.3390/healthcare9070914
16. Mostafa SS, Mendonça F, Ravelo-García AG, Morgado-Dias F. A systematic review of detecting sleep apnea using deep learning. Sensors (Basel). (2019) 19(22):4934. doi: 10.3390/s19224934
17. Vaquerizo-Villar F, Alvarez D, Gutierrez-Tobal GC, Del Campo F, Gozal D, Kheirandish-Gozal L, et al. A deep learning model based on the combination of convolutional and recurrent neural networks to enhance pulse oximetry ability to classify sleep stages in children with sleep apnea. Annu Int Conf IEEE Eng Med Biol Soc. (2023) 2023:1–4. doi: 10.1109/EMBC40787.2023.10341100
18. Gutiérrez-Tobal GC, Álvarez D, Kheirandish-Gozal L, Del Campo F, Gozal D, Hornero R. Reliability of machine learning to diagnose pediatric obstructive sleep apnea: systematic review and meta-analysis. Pediatr Pulmonol. (2022) 57:1931–43. doi: 10.1002/ppul.25423
19. Tanphaichitr A, Chuenchod P, Ungkanont K, Banhiran W, Vathanophas V, Gozal D. Validity and reliability of the Thai version of the pediatric obstructive sleep apnea screening tool. Pediatr Pulmonol. (2021) 56:2979–86. doi: 10.1002/ppul.25534
20. Maggio ABR, Beghetti M, Cao Van H, Grasset Salomon C, Barazzone-Argiroffo C, Corbelli R. Home respiratory polygraphy in obstructive sleep apnea syndrome in children: comparison with a screening questionnaire. Int J Pediatr Otorhinolaryngol. (2021) 143:110635. doi: 10.1016/j.ijporl.2021.110635
21. Pires PJS, Mattiello R, Lumertz MS, Morsch TP, Fagondes SC, Nunes ML, et al. Validation of the Brazilian version of the pediatric obstructive sleep apnea screening tool questionnaire. J Pediatr (Rio J). (2019) 95:231–7. doi: 10.1016/j.jped.2017.12.014
22. Jordan L, Beydon N, Razanamihaja N, Garrec P, Carra MC, Fournier BP, et al. Translation and cross-cultural validation of the French version of the sleep-related breathing disorder scale of the pediatric sleep questionnaire. Sleep Med. (2019) 58:123–9. doi: 10.1016/j.sleep.2019.02.021
23. Abumuamar AM, Chung SA, Kadmon G, Shapiro CM. A comparison of two screening tools for paediatric obstructive sleep apnea. J Sleep Res. (2018) 27:e12610. doi: 10.1111/jsr.12610
24. Kennedy CL, Onwumbiko BE, Blake J, Pereira KD, Isaiah A. Prospective validation of a brief questionnaire for predicting the severity of pediatric obstructive sleep apnea. Int J Pediatr Otorhinolaryngol. (2022) 153:111018. doi: 10.1016/j.ijporl.2021.111018
25. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Br Med J. (2015) 350:g7594. doi: 10.1136/bmj.g7594
26. Li H, Zong XN, Ji CY, Mi J. Body mass index cut-offs for overweight and obesity in Chinese children and adolescents aged 2-18 years. Zhonghua liu Xing Bing xue za zhi= Zhonghua Liuxingbingxue Zazhi. (2010) 31:616–20. doi: 10.3760/cma.j.issn.0253-9624.2018.11.010
27. Chervin RD, Hedger K, Dillon JE, Pituch KJ. Pediatric sleep questionnaire (PSQ): validity and reliability of scales for sleep-disordered breathing, snoring, sleepiness, and behavioral problems. Sleep Med. (2000) 1:21–32. doi: 10.1016/S1389-9457(99)00009-X
28. Franco RA Jr., Rosenfeld RM, Rao M. First place–resident clinical science award 1999. Quality of life for children with obstructive sleep apnea. Otolaryngol Head Neck Surg. (2000) 123:9–16. doi: 10.1067/mhn.2000.105254
29. Li AM, Cheung A, Chan D, Wong E, Ho C, Lau J, et al. Validation of a questionnaire instrument for prediction of obstructive sleep apnea in Hong Kong Chinese children. Pediatr Pulmonol. (2006) 41:1153–60. doi: 10.1002/ppul.20505
30. Berry RB, Budhiraja R, Gottlieb DJ, Gozal D, Iber C, Kapur VK, et al. Rules for scoring respiratory events in sleep: update of the 2007 AASM manual for the scoring of sleep and associated events. Deliberations of the sleep apnea definitions task force of the American academy of sleep medicine. J Clin Sleep Med. (2012) 8:597–619. doi: 10.5664/jcsm.2172
31. Ali M. PyCaret: An Open Source, Low-Code Machine Learning Library in Python, pyCaret Version 1.0. 0 (2020).
32. Yeo I-K, Johnson RA. A new family of power transformations to improve normality or symmetry. Biometrika. (2000) 87:954–9. doi: 10.1093/biomet/87.4.954
33. Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI (U S). (1995) 14:1137–45.
34. Hajipour F, Jozani MJ, Moussavi ZJM, Engineering B. Computing. A comparison of regularized logistic regression and random forest machine learning models for daytime diagnosis of obstructive sleep apnea. Med Biol Eng Comput. (2020) 58:2517–29. doi: 10.1007/s11517-020-02206-9
35. Bozkurt S, Bostanci A, Turhan M. Can statistical machine learning algorithms help for classification of obstructive sleep apnea severity to optimal utilization of polysomno graphy resources? Methods Inf Med. (2017) 56:308–18. doi: 10.3414/ME16-01-0084
36. Mencar C, Gallo C, Mantero M, Tarsia P, Carpagnano GE, Foschino Barbaro MP, et al. Application of machine learning to predict obstructive sleep apnea syndrome severity. Health Informatics J. (2020) 26:298–317. doi: 10.1177/1460458218824725
37. Menard S. Six approaches to calculating standardized logistic regression coefficients. Am Stat. (2004) 58:218–23. doi: 10.1198/000313004X946
39. Boser BE, Guyon IM, Vapnik VN. A training algorithm for optimal margin classifiers. Proceedings of the Fifth Annual Workshop on Computational Learning Theory (1992). p. 144–52
40. Vapnik V. The Nature of Statistical Learning Theory[M]. Springer Science & Business Media (2013).
41. Govindarajan M. A hybrid RBF-SVM ensemble approach for data mining applications. Int J Intell Syst. (2014) 6:84–95. doi: 10.5815/ijisa.2014.03.09
42. Freund Y, Schapire R, Abe N. A short introduction to boosting. J Japanese Soc Artif Intel. (1999) 14:1612.
43. Odegua R. An empirical study of ensemble techniques (bagging boosting and stacking). Proceedings Conf.: Deep Learn. IndabaXAt (2019).
45. Fawcett T. An introduction to ROC analysis. Pattern Recog Lett. (2006) 27:861–74. doi: 10.1016/j.patrec.2005.10.010
47. Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Series B Stat Methodol. (2005) 67:301–20. doi: 10.1111/j.1467-9868.2005.00503.x
48. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. (2010) 33:1. doi: 10.18637/jss.v033.i01
49. Xu Z, Wu Y, Tai J, Feng G, Ge W, Zheng L, et al. Risk factors of obstructive sleep apnea syndrome in children. J Otolaryngol Head Neck Surg. (2020) 49:11. doi: 10.1186/s40463-020-0404-1
50. Parenti SI, Fiordelli A, Bartolucci ML, Martina S, D'Antò V, Alessandri-Bonetti G. Diagnostic accuracy of screening questionnaires for obstructive sleep apnea in children: a systematic review and meta-analysis. Sleep Med Rev. (2021) 57:101464. doi: 10.1016/j.smrv.2021.101464
51. Maniaci A, Riela PM, Iannella G, Lechien JR, La Mantia I, De Vincentiis M, et al. Machine learning identification of obstructive sleep apnea severity through the patient clinical features: a retrospective study. Life (Basel). (2023) 13(3):702. doi: 10.3390/life13030702
52. Marcus CL, Moore RH, Rosen CL, Giordani B, Garetz SL, Taylor HG, et al. A randomized trial of adenotonsillectomy for childhood sleep apnea. New Engl J Med. (2013) 368:2366–76. doi: 10.1056/NEJMoa1215881
53. Ahmed S, Hasani S, Koone M, Thirumuruganathan S, Diaz-Abad M, Mitchell R, et al. An empirical study of questionnaires for the diagnosis of pediatric obstructive sleep apnea. Annu Int Conf IEEE Eng Med Biol Soc. (2018) 2018:4097–100. doi: 10.1109/EMBC.2018.8513389
54. Vaquerizo-Villar F, Gutiérrez-Tobal GC, Calvo E, Álvarez D, Kheirandish-Gozal L, Del Campo F, et al. An explainable deep-learning model to stage sleep states in children and propose novel EEG-related patterns in sleep apnea. Comput Biol Med. (2023) 165:107419. doi: 10.1016/j.compbiomed.2023.107419
Keywords: obstructive sleep apnea, machine learning, artificial intelligence, computer-aided diagnosis, children
Citation: Qin H, Zhang L, Li X, Xu Z, Zhang J, Wang S, Zheng L, Ji T, Mei L, Kong Y, Jia X, Lei Y, Qi Y, Ji J, Ni X, Wang Q and Tai J (2024) Pediatric obstructive sleep apnea diagnosis: leveraging machine learning with linear discriminant analysis. Front. Pediatr. 12:1328209. doi: 10.3389/fped.2024.1328209
Received: 26 October 2023; Accepted: 30 January 2024;
Published: 14 February 2024.
Edited by:
Gonzalo C. Gutiérrez-Tobal, University of Valladolid, SpainReviewed by:
Daniela Ferreira-Santos, University of Porto, PortugalAntonino Maniaci, Kore University of Enna, Italy
© 2024 Qin, Zhang, Li, Xu, Zhang, Wang, Zheng, Ji, Mei, Kong, Jia, Lei, Qi, Ji, Ni, Wang and Tai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Ni nixin@bch.com.cn Qing Wang 8010020958@189.cn Jun Tai trenttj@163.com
†These authors have contributed equally to this work
 Han Qin1,†
Han Qin1,†  Liping Zhang
Liping Zhang Xiaodan Li
Xiaodan Li Jie Zhang
Jie Zhang Tingting Ji
Tingting Ji Yaru Kong
Yaru Kong Xinbei Jia
Xinbei Jia Yi Lei
Yi Lei Yuwei Qi
Yuwei Qi Jie Ji
Jie Ji Xin Ni
Xin Ni Jun Tai
Jun Tai