- 1Qilu Medical University, Zibo, Shandong, China
- 2Shandong Benon Biological Technology Co., Ltd., Jinan, Shandong, China
- 3Shandong Engineering Research Center of Precision Nutrition and Healthy Aging, Qilu Medical University, Zibo, Shandong, China
- 4Research Institute of Natural Products and Health Industry Innovation, Qilu Medical University, Zibo, Shandong, China
Introduction: This study employs an integrated approach combining multi-omics analysis, network pharmacology, machine learning, and experimental validation to elucidate the molecular mechanisms of Fructus Xanthii (Chinese name: Cang-Er-Zi) in treating asthma.
Methods: Asthma-related differentially expressed genes (DEGs) were identified from GEO datasets (GSE63142 and GSE14787). Weighted gene co-expression network analysis (WGCNA) was performed on GSE14787. Active ingredient targets of Fructus Xanthii were predicted using TCMSP and SwissTargetPrediction. Integration included machine learning (RF, SVM, XGB), PPI network analysis, GO/KEGG enrichment, immune infiltration (CIBERSORT), molecular docking, molecular dynamics simulations, and in vivo validation in an ovalbumin-induced murine asthma model.
Results: Yielded 3,755 DEGs and the MEblack module correlated with asthma. Identified 1,317 potential targets, with 100 intersecting DEGs. Hub targets included HSP90AB1, CCNB1, CASP9, CDK6, NR3C1, ERBB2, and CCK. Strong binding affinities (e.g., carboxyatractyloside with HSP90AB1 at −10.09 kcal/mol) and stable complexes were confirmed. Immune profiling showed altered cell populations. In vivo, Fructus Xanthii extract reduced inflammation, cytokines (TNF-α, IL-6, IL-1β, IL-5), and hub gene expression.
Discussion: Fructus Xanthii exerts anti-asthmatic effects by modulating HSP90AB1/IL6/TNF and PI3K-AKT pathways, regulating inflammation, cell cycle, apoptosis, and immune homeostasis, providing empirical support for multi-target traditional Chinese medicine strategies in asthma management.
1 Introduction
Asthma, a chronic respiratory condition characterized by airway hyperresponsiveness, inflammation, excessive mucus production, and structural alterations, commonly results in recurrent episodes and compromised pulmonary function (Porsbjerg et al., 2023). While conventional therapies such as inhaled corticosteroids and beta2-agonists have significantly improved disease control, their limitations, including side effects and incomplete symptom resolution, have spurred interest in alternative therapeutic strategies (Busse, 2019). Traditional Chinese Medicine (TCM) has emerged as a promising complementary approach due to its holistic perspective on health and its emphasis on restoring systemic balance through multi-component herbal formulations (Chan and Ng, 2020). In TCM philosophy, asthma is conceptualized as a manifestation of vital energy (Qi) imbalance, particularly involving deficiencies within the lung, spleen, and kidney systems, compounded by interference from pathogenic factors such as wind or heat (Zhou et al., 2019). Historically, TCM therapeutic interventions have been developed according to the principles of “emperor, minister, assistant, and messenger” herbs, which function synergistically to reestablish homeostatic balance and facilitate effective Qi circulation (Liu et al., 2021).
Fructus Xanthii, a herb traditionally utilized in Chinese medicine, has long been applied in treatments targeting diverse inflammatory and allergic conditions. In traditional Chinese medicine (TCM) practice, this botanical agent is highly regarded for its “wind-dispelling” properties and efficacy in treating exterior patterns, demonstrating therapeutic benefits for rhinosinusitis-related symptoms, cephalgia, and select musculoskeletal discomfort (Luo et al., 2020; Luo et al., 2021; Wang et al., 2023). While systematic clinical trials specifically addressing asthma remain sparse, its integration within multi-herb respiratory formulas implies a synergistic mechanism in mitigating airway inflammatory responses and alleviating associated symptoms through combined pharmacological actions (Barnes et al., 2019; Finotto, 2019; Zinellu et al., 2019).
Fructus Xanthii exhibits a complex array of phytochemicals, with sesquiterpene lactones serving as its principal active components. Studies on related Xanthium species have identified key bioactive molecules, including xanthinosin, xanthatin, and structurally diverse mogolides (including Mogolide A, D, E), which are isolated through advanced chromatographic techniques. These compounds are implicated in mediating the anti-inflammatory mechanisms observed in preclinical evaluations. In addition to sesquiterpene lactones, Fructus Xanthii exhibits a complex phytochemical profile comprising phenylpropanoids, lignans, coumarin derivatives, and multifarious flavonoids, collectively contributing to its multifaceted therapeutic properties (Han et al., 2023). Mechanistically, the pharmacological effects of Fructus Xanthii derive predominantly from its ability to modulate key inflammatory signaling pathways. Notably, the sesquiterpene lactones have been associated with inhibition of nuclear factor-kappa B (NF-κB) nuclear translocation and modulation of mitogen-activated protein kinase (MAPK) signaling cascades. This molecular intervention effectively suppresses downstream transcriptional activation, thereby curtailing synthesis and secretion of pro-inflammatory mediators, including cytokines and chemokines. Consequently, the therapeutic agents mitigate pathological recruitment of immune cells to inflamed tissue microenvironments, establishing a critical mechanism for their anti-inflammatory effects (Han et al., 2023). The ability of Fructus Xanthii to modulate inflammatory mediators positions it as a promising therapeutic candidate for chronic inflammatory disorders typified by asthma, particularly given its impact on hallmark pathological features such as airway hyperresponsiveness and hypersecretion of mucins (Athari, 2019; Peters et al., 2019). Notably, its proposed immunomodulatory activity involves recalibrating Th1/Th2 immune responses (Han et al., 2022). Despite these insights, comprehensive elucidation of the molecular pathways linking Fructus Xanthii’s bioactive constituents to asthma pathophysiology remains an unmet research objective.
In the present study, we employed an integrated multi-omics approach, encompassing differential gene expression analysis from GEO datasets (GSE63142 and GSE14787) and weighted gene co-expression network analysis (WGCNA), to identify asthma-related differentially expressed genes (DEGs) and core modules. This was complemented by a network pharmacology framework to predict active ingredients and targets of Fructus Xanthii, constructing a “disease-drug-ingredient-target” network and performing protein-protein interaction (PPI) analysis. Pivotal therapeutic targets were systematically mapped through the integration of machine learning algorithms (random forest, support vector machine, and extreme gradient boosting) with PPI network topology. Computational validation was achieved via Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, immune cell infiltration profiling using CIBERSORT and ESTIMATE algorithms, molecular docking, and molecular dynamics simulations, confirming stable binding affinities and complex dynamics for key compounds such as carboxyatractyloside with hub targets (e.g., HSP90AB1, CCNB1, and CASP9). These in silico findings were substantiated by in vivo experimental validation in an ovalbumin-induced murine asthma model, demonstrating dose-dependent anti-inflammatory effects of Fructus Xanthii aqueous extract, including reduced cytokine levels (TNF-α, IL-6, IL-1β, IL-5), ameliorated lung histopathology, and downregulated expression of hub genes (HSP90AB1, CCNB1, CASP9, PI3K, AKT1). Collectively, our results elucidate the multi-target anti-asthmatic mechanisms of Fructus Xanthii, involving modulation of inflammation, cell cycle progression, apoptosis, and immune balance through pathways such as HSP90AB1/IL6/TNF and PI3K-AKT, thereby providing robust scientific evidence for the application of traditional Chinese medicine in asthma management.
2 Materials and methods
2.1 Prediction of active ingredients and targets of Fructus Xanthii
This study employed a multi-platform integrated analysis strategy to dissect the pharmacological mechanisms of Fructus Xanthii. Initially, chemical constituents of the herb were extracted through the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://www.tcmsp-e.com/). We then identified qualified active ingredients based on pharmacokinetic (ADME) parameters, retaining molecules with oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18. Subsequently, SMILES structural formulas of the target components were acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and imported into the SwissTarget Prediction platform (https://www.swisstargetprediction.ch/) for virtual screening of potential therapeutic targets. The target gene names were then standardized through the UniProt database (https://www.uniprot.org/) (Xing et al., 2018).
2.2 Collection of asthma dataset
We retrieved asthma-related gene expression datasets from the Gene Expression Omnibus (GEO) database using the keywords “asthma” and “Homo sapiens”. For differential gene analysis, we selected GSE63142 (27 healthy controls vs. 56 severe asthma samples) and GSE14787 (13 healthy controls vs. 60 severe asthma samples). After filtering, 27 normal and 56 severe asthma samples from GSE63142 were used for DEG identification. The GSE14787 dataset was integrated to construct immune infiltration and weighted gene co-expression networks (WGCNA) (Zhou et al., 2025).
2.3 Building a weighted Co-expression network
We further processed the GSE14787 dataset through weighted gene co-expression network analysis (WGCNA) using R version 4.1.2 to identify co-expressed gene modules. Initially, we performed hierarchical clustering on the research samples to detect and eliminate outlier samples. Subsequently, we constructed a scale-free network and utilized the pick Soft Threshold function to select an appropriate soft threshold. Following this, we established an adjacency matrix and transformed it into a topological overlap matrix (TOM), employing various degrees to generate a gene tree diagram with module colors. Finally, we calculated the correlation between each module and the differential samples (Liu et al., 2025).
2.4 Identification of differentially expressed genes in asthma
Subsequently, we standardized the GSE63142 dataset using the “Sva” package in R 4.1.2. Differentially expressed genes (DEGs) were identified using the “limma” package with selection criteria of absolute log2 fold change >1 and adjusted P-value <0.05. Heatmaps and volcano plots visualizing the DEGs were generated using the “pheatmap” and “ggplot2” packages in R 4.1.2 (Feng et al., 2025).
2.5 Intersection target acquisition of Fructus Xanthii for asthma treatment
We utilized the SRplot (https://www.bioinformatics.com.cn/) to generate Venn diagrams. The identified asthma-related genes, target genes, and active ingredients of Fructus Xanthii were imported into Cytoscape 3.8.1 for visual data analysis to construct a “disease-drug-active ingredient-target” network. The overlapping target genes identified through this analysis were predicted as potential therapeutic targets of Fructus Xanthii in asthma treatment (Guo et al., 2025).
2.6 Construction of protein interaction network
The intersected target dataset was uploaded to the STRING database (https://string-db.org/) with Homo sapiens as the organism and an interaction score threshold ≥0.4 to construct a refined protein-protein interaction (PPI) network. Network visualization and functional analysis were performed using Cytoscape v3.10.2. Key analyses included.
1. Topological profiling via CytoNCA plugin to evaluate node centrality;
2. Hub identification using Cytohuba with MCC, DMNC, MNC, and EPC algorithms;
3. Functional clustering via MCODE to detect modules. This workflow systematically characterized network architecture and the biological significance of key components.
2.7 GO and KEGG analysis
To systematically characterize the functional annotations of key target genes, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses employing a comprehensive bioinformatics workflow. This analysis utilized multiple R packages (version 4.1.2), including org. Hs. eg.db, colorspace, stringi, DOSE, clusterProfiler, pathview, ggplot2, and limma, with a stringent significance threshold of P < 0.05. The GO enrichment analysis was conducted across three ontological domains: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). For result visualization and interpretation, we leveraged the Metware Cloud platform (https://cloud.metware.cn/) to generate publication-ready graphics. Notably, the top 10 most significantly enriched terms (P < 0.05) within each GO category and KEGG pathway were prioritized for detailed representation, facilitating biological insight extraction.
2.8 Molecular docking
Active ingredients from Fructus Xanthii were selected and subjected to molecular docking against the predicted core targets. The three-dimensional (3D) structures of these bioactive compounds were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov), while the structural data for target proteins were obtained from the Protein Data Bank (PDB, https://www.rcsb.org). Molecular docking simulations and visual analysis were carried out using AutoDock Tools 1.5.7 and PyMOL molecular visualization software (Huang et al., 2024b; Huang et al., 2024b). To assess the stability of protein-ligand interactions, molecular dynamics (MD) simulations were applied to the screened potential targets in complex with carboxyatractyloside. Molecular dynamics were performed in GROMACS 2023.2 with CHARMM36/TIP3P; triplicate 100-ns runs per complex. The resulting MD trajectory data were visualized through Xmgrace software version 5.1.25.
2.9 Machine learning screening hub genes
We further developed a machine learning model based on the intersection gene expression matrix in the GSE14787 dataset using “caret” (R 4.1.2), encompassing Random Forest (RF), Support Vector Machine (SVM), Generalized Linear Model (GLM), and Extreme Gradient Boosting (XGB). Subsequent screening of intersecting target genes was performed to identify potential hub genes for the treatment of asthma with Fructus Xanthii. Models were trained with 10-fold cross-validation; feature importance was summarized by mean decrease accuracy (RF) and SHAP values (XGBoost). GLM residuals were higher, consistent with linear model limitations in small-sample, nonlinear settings. The validity of the models was assessed through residual analysis and ROC evaluation, resulting in the identification of the top 10 variables as hub genes.
2.10 Immune infiltration analysis
To investigate potential links between key hub genes and immune microenvironment dynamics in asthma, we utilized the CIBERSORT tool (https://cibersortx.stanford.edu/) for immunophenotyping of 22 cellular subsets within GSE14787 asthma samples. Subsequent application of the ESTIMATE algorithm enabled quantification of tissue immune infiltration characteristics, while Spearman rank correlation analysis systematically assessed gene-immune cell association patterns.
2.11 Animal experiment
This study utilized 4- to 6-week-old specific pathogen-free (SPF) female BALB/c mice (weight range: 16–18 g), sourced from Beijing Vital River Laboratory Animal Technology Co., Ltd., with eight mice per experimental group. The asthma model was induced using ovalbumen (OVA, Cat: A5503, Sigma-Aldrich) as the allergen. Following a 3-day acclimatization period, mice were randomly assigned to the following groups: Control group, Model group, Dexamethasone treatment group (1 mg/kg), and Fructus Xanthii aqueous extract treatment groups at low (10 mg/kg), medium (20 mg/kg), and high (30 mg/kg) doses. Sensitization was performed on days 0 and 7 by intraperitoneal (i.p.) injection of 20 μg OVA emulsified in 100 μL of 4% aluminum hydroxide adjuvant (Cat: BS010, ZHHC, China) diluted in 100 μL of sterile physiological saline. From days 15–19, mice received daily oral administration of Fructus Xanthii aqueous extract or dexamethasone, administered 2 h prior to allergen challenge. Subsequently, for five consecutive days (days 15–19), mice were subjected to 30-min nebulization with 5% OVA to provoke airway inflammation, as outlined in Figure 8A. After the model was established, the mice were anesthetized by intraperitoneal injection of tribromoethanol (0.2 mL/10 g) to facilitate tissue collection. Blood samples were collected via cardiac puncture, and serum was isolated by centrifugation at 850 × g for 10 min at 4 °C, while lung tissues were snap-frozen and stored at −80 °C for subsequent analyses. Bronchoalveolar lavage fluid (BALF) was harvested by exposing the trachea of each mouse, inserting a venous catheter into the cervical trachea, and securing it with sutures. The lungs were lavaged three times with 0.4 mL, 0.5 mL, and 0.6 mL of phosphate-buffered saline (PBS), respectively, and the instilled fluid was gently aspirated using a syringe for downstream immunological and biochemical assessments. Followed by cervical dislocation to confirm death. This method ensures rapid and humane euthanasia, minimizing animal distress. All procedures adhered to ethical guidelines for animal experimentation, ensuring minimal distress and compliance with institutional protocols. All experimental procedures were strictly carried out by the guiding principles of the Animal Care and Use Committee of Qilu Medical University, and the experimental plans were approved by the Laboratory Animal Ethics Committee of Qilu Medical University (YXLL2025R032).
2.12 Preparation of Fructus Xanthii
Commercial Fructus Xanthii was purchased from Omniherb (Daegu, Korea). The material (voucher specimen No. 21032) was authenticated according to the Korean Ministry of Food and Drug Safety (KFDA) guidelines. Fructus Xanthii was extracted using hot water to prepare the aqueous extract. The aqueous extract was prepared using a standardized hot water extraction method (1:10 w/v, 100 °C, 2 h × 2), where the yield was approximately 18.5%. The HPLC fingerprint is consistent with published profiles of Fructus Xanthii aqueous extracts, showing major peaks corresponding to xanthatin and phenolic acids (Luo et al., 2021; Han et al., 2022).
2.13 qPCR
Primers for mouse Hsp90ab1, CCNB1, CASP9, PI3K, AKT1, and the reference gene GAPDH were designed using Primer-BLAST (NCBI, https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer sequences were: Hsp90ab1-F: 5′- GCTGAAGATGACCGACTATGAC-3′, Hsp90ab1-R: 5′- AGTCCAGCTCCATGATGTTGAC-3′; AKT1-F: 5′-ATGGACTACCTGCACTCGGA-3′, AKT1-R: 5′-GATGACCGTGTAGCCATTGT-3′; GAPDH-F: 5′-AGGTCGGTGTGAACGGATTTG-3′, GAPDH-R: 5′-TGTAGACCATGTAGTTGAGGTCA-3′; CCNB1-F: 5′- AGAGGTGGAACTTGCTGAGCCT-3′, CCNB1-R: 5′- GCACATCCAGATGTTTCCATCGG-3′; CASP9-F: 5′- GCTGTGTCAAGTTTGCCTACCC-3′, CASP9-R: 5′- CCAGAATGCCATCCAAGGTCTC-3′; PI3k-F: 5′- CTCTCCTGTGCTGGCTACTGT-3′, PI3K-R: 5′- GCTCTCGGTTGATTCCAAACT-3′. Primer specificity was validated by BLAST analysis. Each 20 µL reaction contained 10 µL SYBR Green Master Mix, 0.4 µL forward primer (10 µM), 0.4 µL reverse primer (10 µM), 2 µL cDNA, and 7.2 µL nuclease-free water. Cycling conditions were: initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min, with a melt curve stage (95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s) to verify amplicon specificity. Samples were run in triplicate, with no-template controls (NTCs) to detect contamination. GAPDH was used for normalization, and relative gene expression was calculated using the 2−ΔΔCt method (ΔCt = Ct (target gene) - Ct (GAPDH), ΔΔCt = ΔCt (sample) - ΔCt (control)). Amplification efficiency was validated by standard curve analysis (95%–105%, R2 > 0.99). Data were analyzed using GraphPad Prism 9.0, expressed as mean ± standard deviation (SD).
3 Results
3.1 Acquisition of the disease targets
We downloaded the GSE63142 dataset from the GEO database and conducted data organization and analysis using the R language. A total of 3,755 differentially expressed genes were identified under the criteria of log2|FC| ≥ 1 and P < 0.05. Based on these screening results, we generated a volcano plot (Figure 1A) illustrating the differentially expressed genes in asthma. Additionally, we selected upregulated and downregulated genes to create a heatmap (Figure 1B). Among the differentially expressed genes, 1946 were upregulated, including TFF1, FKBP5, AKR1B10, CXCL14, CST1, TPRXL, NOS2, CPA3, KRT6A, PHACTR3, TCN1, CLCA1, and CEACAM5, among others. Conversely, 1809 genes were downregulated, including LOC100132287, C7orf26, LTF, GPR156, SCGB3A1, SCGB1A1, CSH1, TMEM45A, C20orf46, FHOD3, C3, LYPD2, among others. These differentially expressed genes are involved in various signaling pathways (Figure 1C).
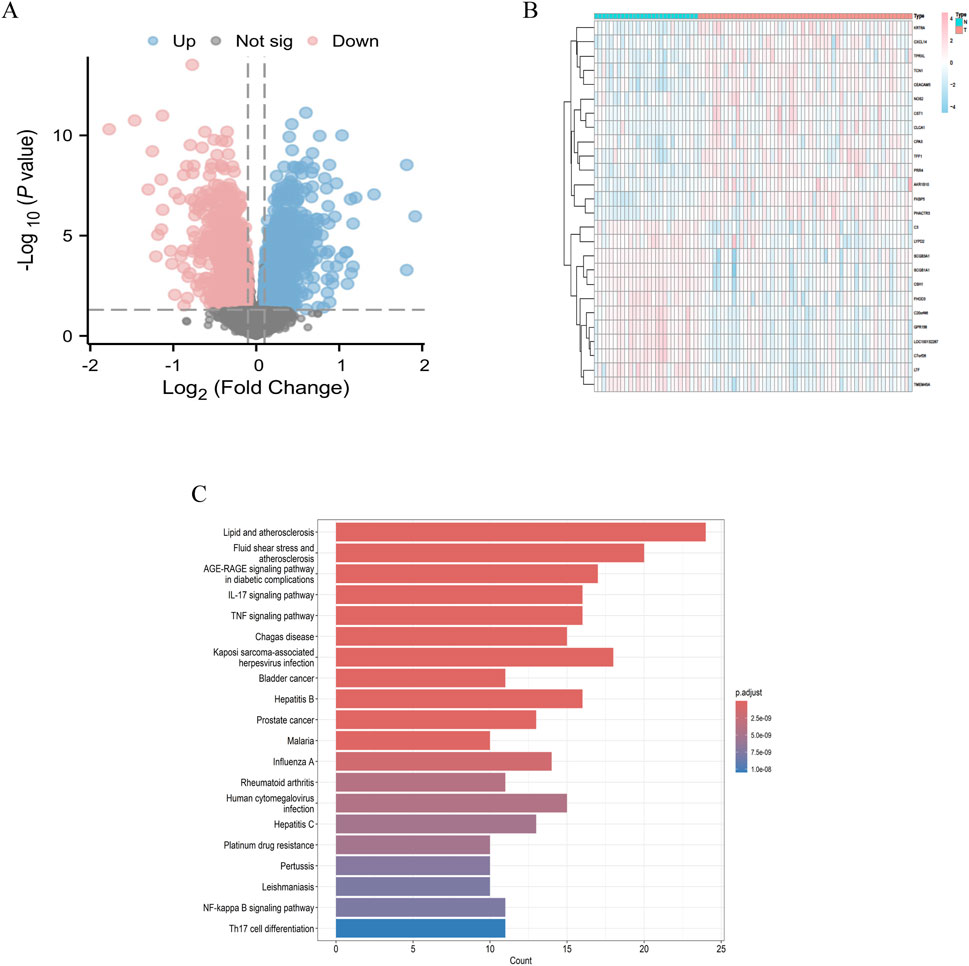
Figure 1. Differential genes in asthma (A) Volcano plot depicting differentially expressed genes in asthma; (B) Heatmap visualization of differentially expressed genes in asthma; (C) Enrichment analysis of signaling pathways for differences.
3.2 Asthma-related core gene clusters
We employed the Weighted Gene Co-expression Network Analysis (WGCNA) methodology to analyze the GSE14787 dataset and construct a gene co-expression network for identifying key gene modules associated with asthma. Initially, we detected and eliminated outliers from abnormal samples. Subsequently, we constructed a scale-free network and utilized the pickSoftThreshold function to select the optimal soft power β = 18 (R2 = 0.962) (Figure 2A). We then established an adjacency matrix, converted it into a topological overlap matrix (TOM), and applied varying degrees to generate a gene tree diagram with corresponding module colors (Figures 2B–D). Our analysis revealed that MEblack and MEblue modules exhibited significant positive correlations with asthma samples, with correlation coefficients of 0.42 and 0.23, respectively. Consequently, the MEblack gene cluster was selected for further investigation (Figure 2E). Genes within each module were filtered based on their significance and module correlation, with thresholds set at geneSigFilter = 0.5 and moduleSigFilter = 0.5. The results identified MEblack as a cluster containing 741 asthma-related core genes, including POMP, GUSBL2, RBM39, PSMA6, RFX1, TCEB1, PSMD6, and others.
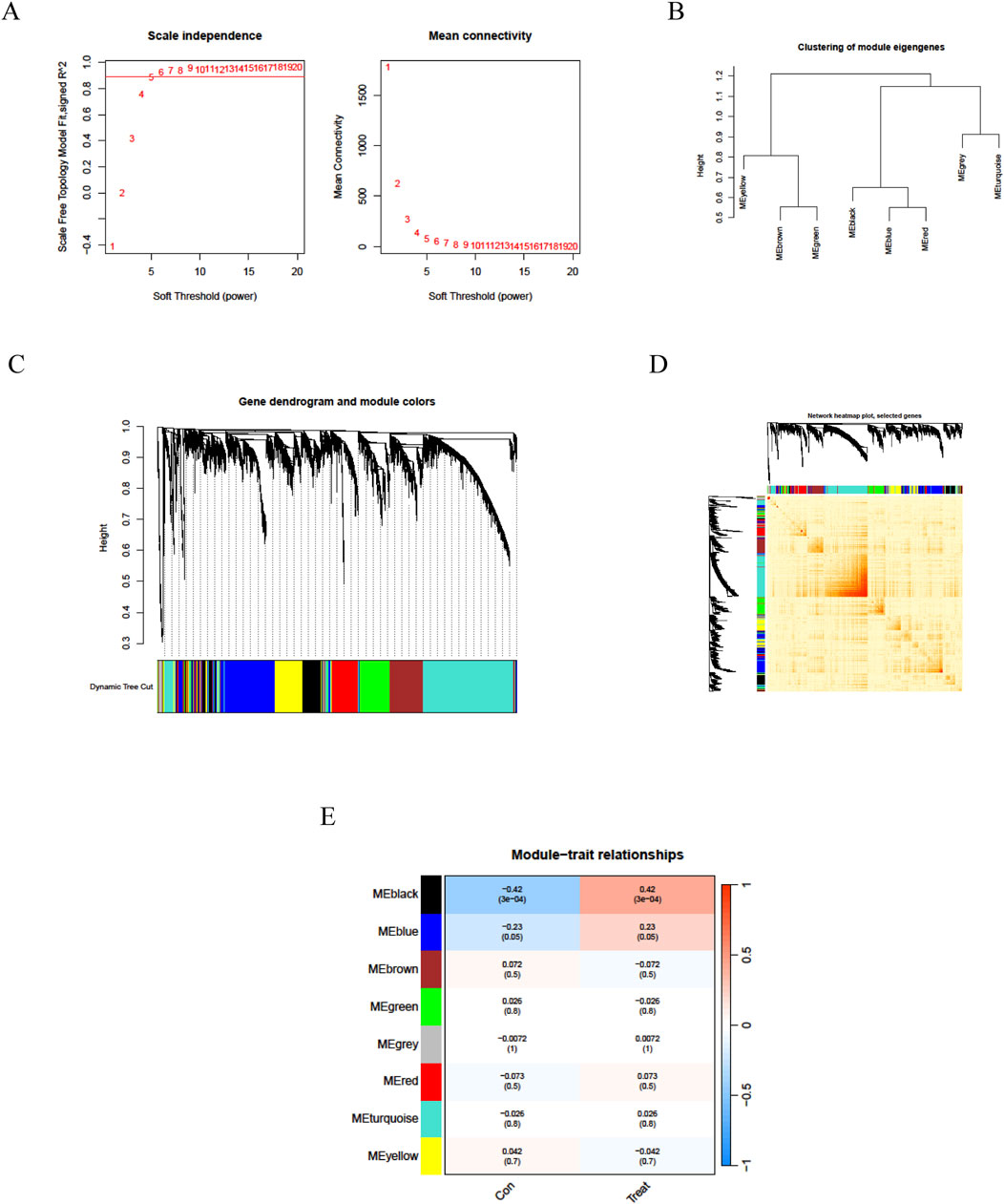
Figure 2. Weighted gene co-expression network analysis of asthma. (A) WGCNA soft threshold power selection plot; (B) Gene dendrogram based on WGCNA clustering; (C) Topological Overlap Matrix (TOM) visualization; (D) WGCNA Gene heatmap. (E) Heatmap depicting module-trait relationships with associated correlation values and significance levels (red indicates positive correlation, blue indicates negative correlation; values in cells are correlation coefficients with p-values in parentheses).
3.3 Target prediction of Fructus Xanthii in the treatment of asthma
Through the application of TCMSP and subsequent searches in the SwissTarget Prediction database, we identified the chemical composition and potential targets of Fructus Xanthii, yielding a total of 1,317 potential targets. By intersecting these targets with 741 asthma gene clusters and 3,755 asthma DEGs, we identified 100 specific targets through which Fructus Xanthii may exert therapeutic effects on asthma (Figure 3A).
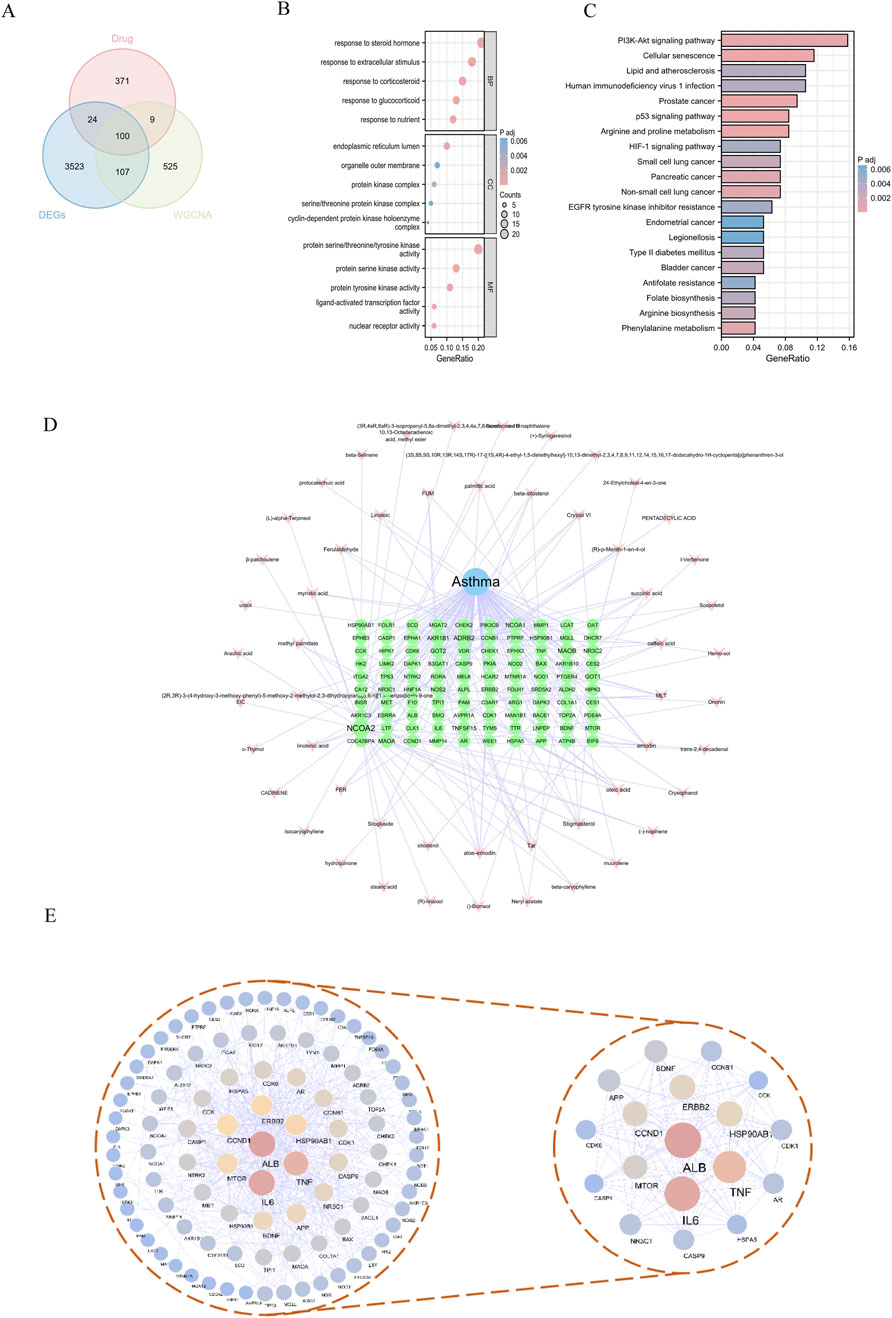
Figure 3. Target Prediction of Fructus Xanthii in the treatment of asthma (A) Venn diagram illustrating the intersection targets of Fructus Xanthii in asthma treatment; (B) GO enrichment analysis therapeutic effects of Fructus Xanthii on asthma; (C) KEGG pathway enrichment analysis of Fructus Xanthii in asthma treatment; (D) Network visualization of disease-drug-active ingredient-target interactions; (E) Protein-protein interaction network revealing therapeutic mechanisms of Fructus Xanthii in asthma.
To elucidate the functional roles of the 100 overlapping genes, a comprehensive functional enrichment analysis was performed. Gene Ontology (GO) annotation revealed 1,275 significantly enriched terms across three ontological categories—biological processes, cellular components, and molecular functions—in the context of Fructus Xanthii’s therapeutic intervention in asthma. Biological process terms dominated the enrichment landscape (1,153 entries), with prominent clusters including steroid hormone response, glucocorticoid/corticosteroid responsiveness, nutrient/extracellular stimulus adaptation, and related signaling cascades. Cellular component annotations (31 entries) highlighted endoplasmic reticulum subcompartments, protein kinase complexes (e.g., cyclin-dependent holoenzymes, serine/threonine kinase complexes), and organelle membrane structures. Molecular function enrichment (91 entries) prioritized kinase activities (serine/threonine/tyrosine-specific), nuclear receptor signaling, and ligand-dependent transcription factor regulation (Figure 3B).
KEGG functional enrichment analysis identified 102 related pathways involved in the intervention of Fructus Xanthii in asthma, mainly including Arginine and proline metabolism, Prostate cancer, Cellular senescence, p53 signaling pathway, and PI3K-Akt signaling pathway (Figure 3C). The intersecting targets were imported into Cytoscape 3.8.1 software to construct a “disease-drug-active ingredient-target” network (Figure 3D). Additionally, a protein interaction network was constructed using the intersecting genes, with nodes assigned blue-yellow-wine red gradients based on degree values. The network analysis revealed 96 nodes with 503 interrelationships. The highest-ranked targets in terms of degree were ALB, IL6, TNF, HSP90AB1, ERBB2, and CCND1, among others (Table 1). By screening for core targets with degree values greater than twice the median, a network comprising 18 nodes and 120 interrelationships was identified (Figure 3E).
3.4 Identification of target hub genes with machine learning
To further investigate the core genes within the network, we identified a network comprising 16 targets as the core by applying a threshold of degree values exceeding twice the median. We developed a machine learning model using the “caret” package in R and assessed its validity through residual analysis and ROC evaluation. The results demonstrated that the Random Forest (RF), Support Vector Machine (SVM), and XGBoost (XGB) machine learning methods yielded small residuals, whereas the Generalized Linear Model (GLM) produced larger residuals (Figures 4A,B). Concurrently, ROC evaluation revealed that RF, SVM, and XGB methods all achieved values greater than 0.7, with XGBoost exhibiting the highest ROC value of 0.883 (Figure 4C). Based on a comprehensive analysis of both residuals and ROC performance, we selected the RF, SVM, and XGB models for subsequent analysis and identified the top 10 variables from each model as central genes (Figure 4D). The intersection of these three machine learning approaches revealed 7 core targets (Figure 4E): HSP90AB1, CCNB1, CCK, CDK6, CASP9, NR3C1, and ERBB2.
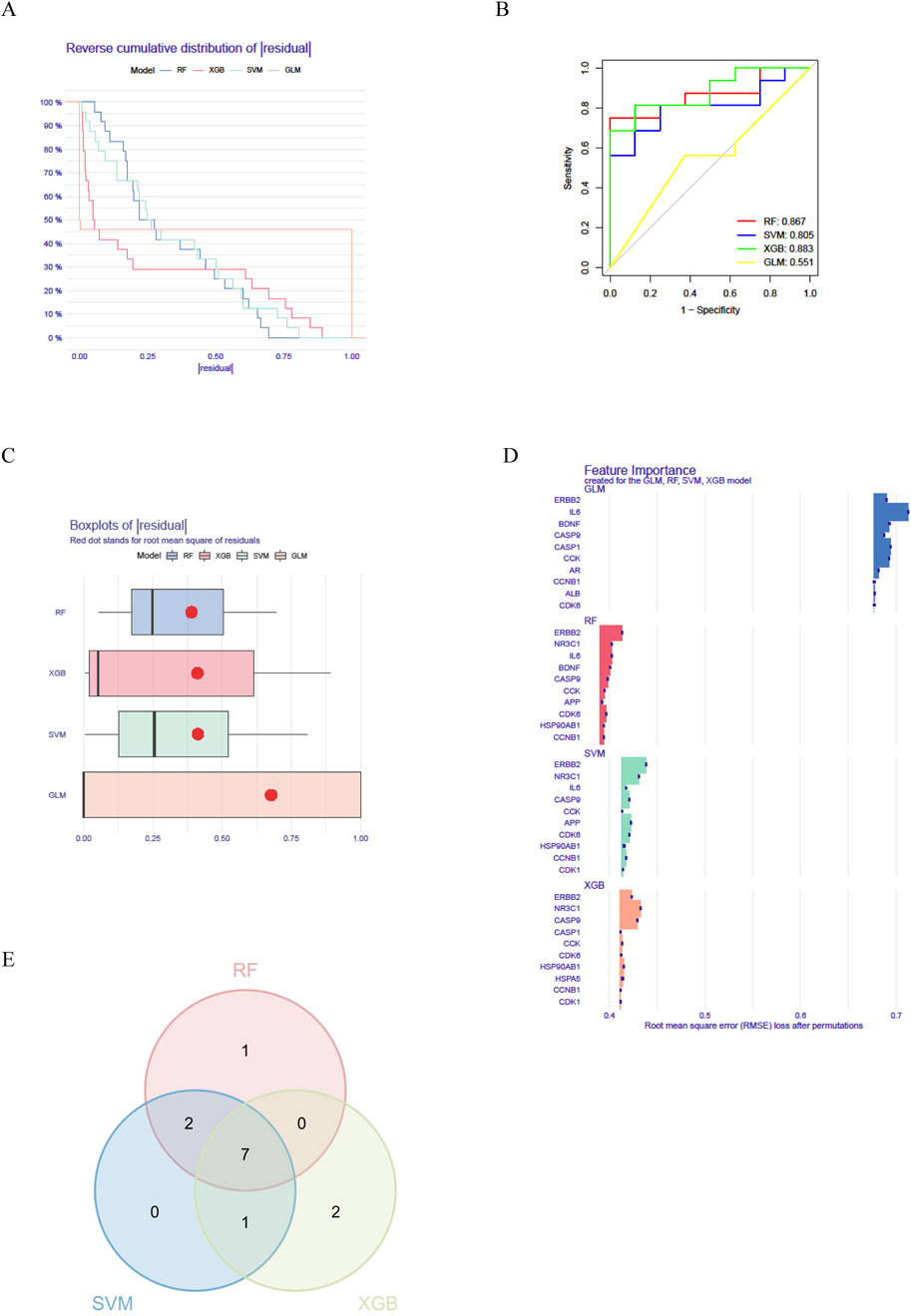
Figure 4. Identification of key genes involved in the treatment of asthma with Fructus Xanthii (A) Machine learning residual evaluation; (B) Machine learning residual evaluation; (C) Machine learning ROC evaluation; (D) Identification of key genes; (E) Three intersections of machine learning.
3.5 Molecular docking virtual verification
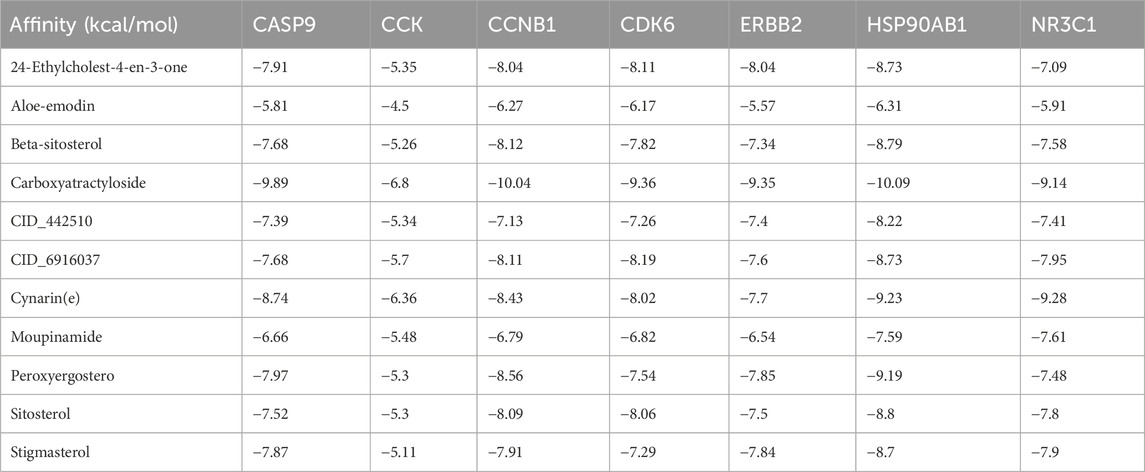
Molecular docking analysis was conducted to evaluate the binding affinities of eleven compounds against seven key target genes (HSP90AB1, CCNB1, CCK, CDK6, CASP9, NR3C1, ERBB2), with interaction scores ranging from −5.3 kcal/mol to −10.09 kcal/mol (see Table 1). A lower docking score correlates with a stronger binding affinity, with scores below −5.0 kcal/mol indicating potential interactions and those below −7.0 kcal/mol suggesting robust binding affinities. Among the targets, Carboxyatractyloside demonstrated the highest binding affinity for HSP90AB1. The binding interactions of the five key targets with their respective ligands are shown in Figure 5. The binding pockets of HSP90AB1, CCNB1, CDK6, CASP9, and ERBB2 were effectively occupied by the selected ligands, with stability provided through hydrogen bonds and hydrophobic interactions. In particular, Carboxyatractyloside formed hydrogen bonds with LYS53 of HSP90AB1 at a distance of 3.00 Å (see Figure 5A). The binding sites and affinities for the other proteins and ligands are presented in Figures 5B–E. These results highlight the effective binding of the five compounds to the core targets through hydrogen bonds and hydrophobic interactions.
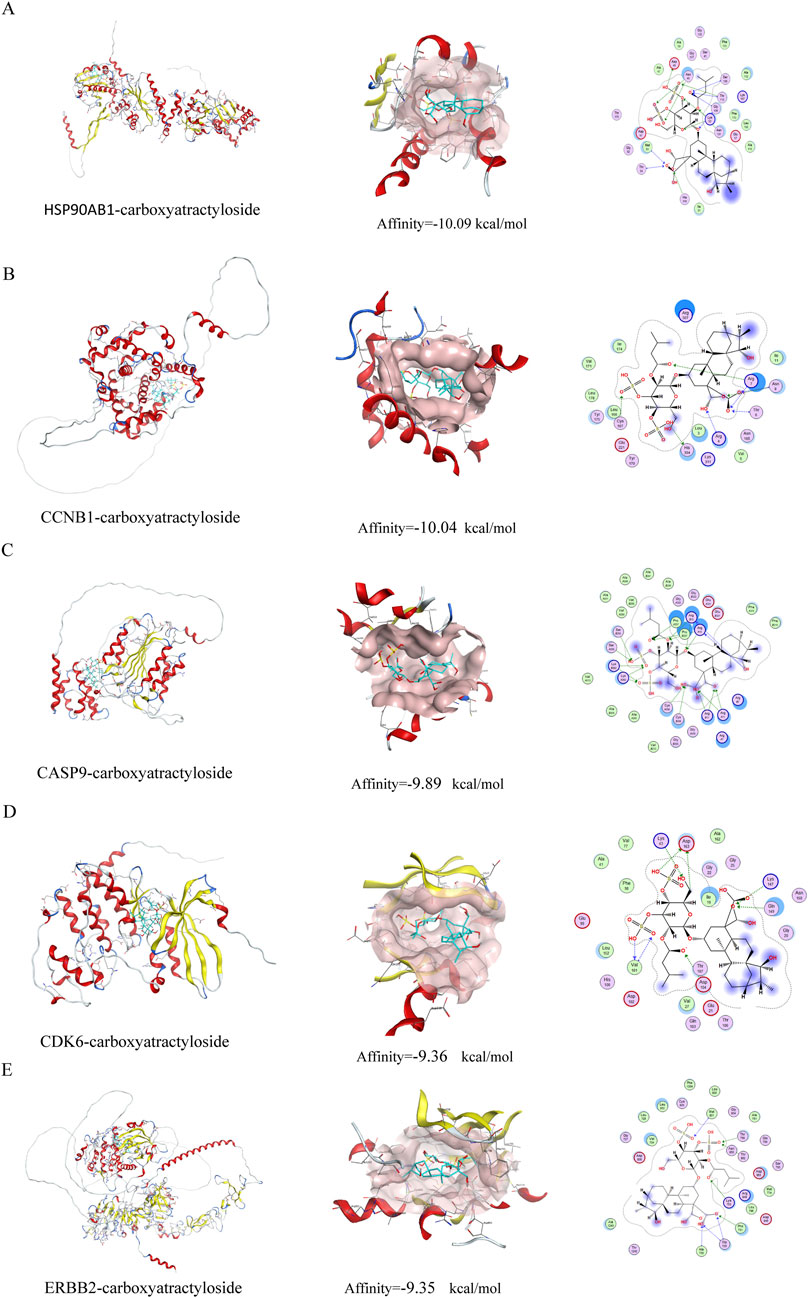
Figure 5. Schematic diagram of molecular docking. (A) HSP90AB1-carboxyatractyloside; (B) CCNB1-carboxyatractyloside; (C) CASP9-carboxyatractyloside; (D) CDK6-carboxyatractyloside; (E) ERBB2-carboxyatractyloside.
3.6 Molecular dynamics simulations
To characterize the dynamic stability of the top 3 protein-ligand complexes (HSP90AB1/CCNB1/CASP9-Carboxyatractyloside), we performed 100-ns MD simulations with triplicate sampling. Structural equilibration was verified through root mean square deviation (RMSD) trajectories, where values below 3 nm (Figure 6A) confirmed physiological stability across all systems. Post-equilibration RMSD plateaued at 1.16 ± 0.13 Å (HSP90AB1) to 2.85 ± 0.18 Å (CCNB1), indicating ligand-specific stabilization patterns. Atomic positional fluctuations were quantified via root mean square fluctuation (RMSF) analysis (Figure 6B), revealing constrained molecular motions with values ranging from 0.19 ± 0.06 Å (CCNB1) to 0.47 ± 0.23 Å (HSP90AB1). Compactness metrics derived from radius of gyration (Rg) calculations (Figure 6C) showed sustained structural integrity throughout simulations (2.18 ± 0.11 nm [CCNB1] - 3.26 ± 0.15 nm [HSP90AB1]). The SASA values of the three complexes also demonstrated stability, reaching average values of 420.87 ± 3.79 nm2, 241.87 ± 8.62 nm2, and 215.76 ± 3.49 nm2, respectively.
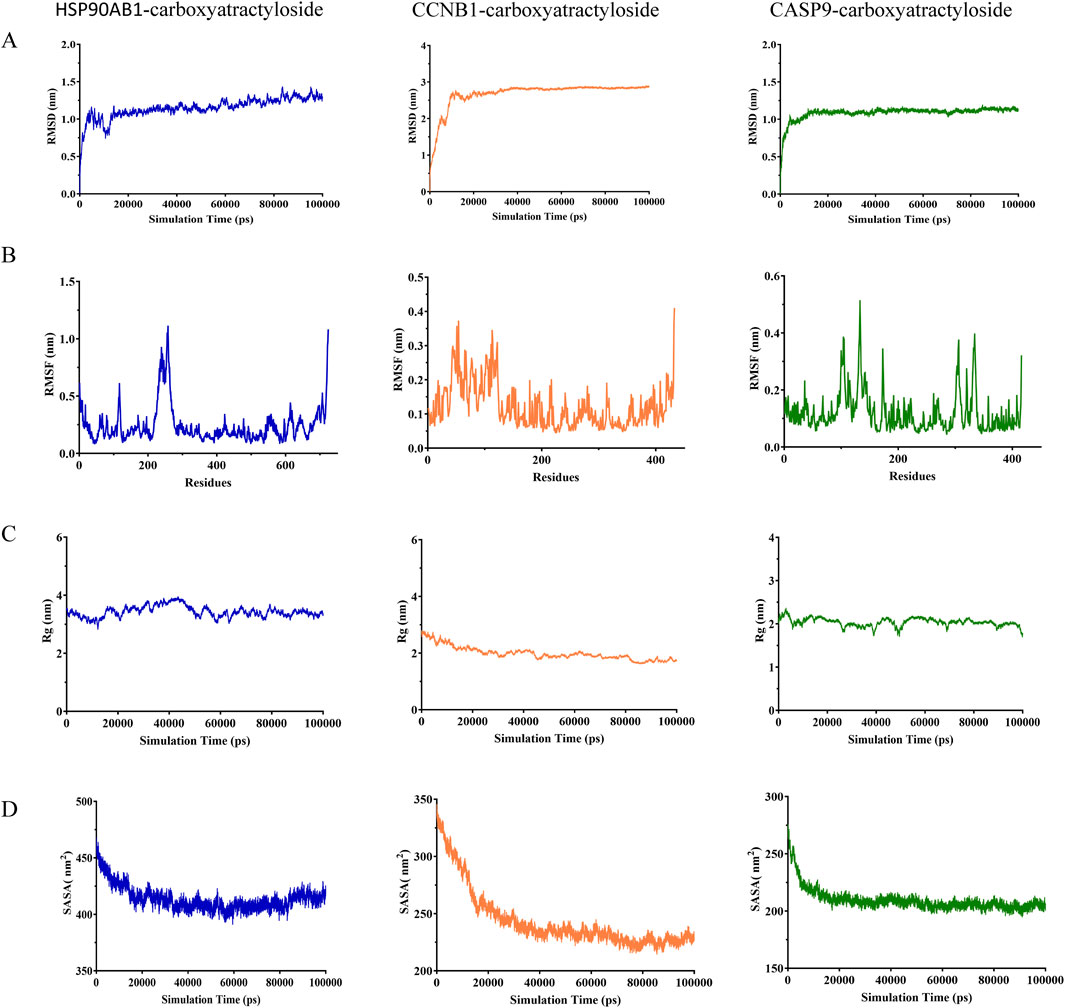
Figure 6. Molecular dynamics simulation and binding free energy calculation (A) RMSD values of the three complexes. (B) RMSF values of the three complexes. (C) Radius of gyration (Rg) values of the three complexes. (D) Solvent-accessible surface area (SASA) values of the three complexes.
3.7 Immune infiltration analysis
We initially employed the CIBERSORT algorithm to characterize immune cell composition across 22 distinct populations in GSE14787 samples (Figures 5A,B). Comparative analysis between asthmatic and non-asthmatic tissues revealed significant alterations in immune cell profiles (Figure 5C). The findings revealed (Figures 7C,D) that M2 macrophages and memory B cells were significantly decreased in asthma samples, whereas plasma cells were significantly elevated, suggesting the importance of focusing on macrophages, plasma cells, and memory B cells in future investigations. To explore functional relationships, we subsequently investigated correlations between the expression of key hub genes and immune cell infiltration profiles (Figure 5D), providing insights into potential regulatory networks linking gene expression to immune microenvironment composition. The heatmap reveals distinct immunomodulatory roles for core asthma targets: NR3C1 exhibits strong positive correlations with regulatory immune cells—notably memory B cells (r ≈ 0.64) and T follicular helper cells (r ≈ 0.48)—suggesting its role in enhancing immunosuppressive responses, consistent with its function as the glucocorticoid receptor. Conversely, HSP90AB1 shows divergent associations: positive with pro-inflammatory M1 macrophages (r ≈ 0.38) but negative with anti-inflammatory M2 macrophages (r ˜ −0.41), indicating its involvement in macrophage polarization imbalance. ERBB2 displays broad negative correlations with lymphocyte populations, including memory B cells (r ˜ −0.71) and plasma cells (r ˜ −0.43), aligning with its known role in mucus hypersecretion and adaptive immune suppression. Critically, NR3C1, HSP90AB1, and ERBB2 collectively suppress plasma cells (unified negative correlations), implicating synergistic control of humoral immunity dysregulation in asthma. CDK6 and CCNB1 show moderate negative associations with neutrophils and mast cells, while CCK exhibits no significant immune correlations (p > 0.05), implying non-immune mechanistic pathways.

Figure 7. Immune infiltration analysis of target hub genes (A) Proportional histograms depicting the distribution of 22 immune cell types; (B) Heatmap illustrating the expression abundance of 22 immune cell populations; (C) Box plots showing differential expression of 22 immune cell types between normal and DN groups; (D) Correlation heatmap of immune cell populations. (E) Heat map of the correlation between immune infiltration and the hub genes’ expression.
3.8 Fructus Xanthii aqueous extract enhanced pulmonary function and mitigated inflammatory infiltration of type II immune response in an OVA-Induced murine asthma model
Figure 8 provides a comprehensive evaluation of the therapeutic efficacy of Fructus Xanthii aqueous extract in mitigating ovalbumen (OVA)-induced allergic asthma in a murine model, integrating the experimental design, physiological responses, histopathological observations, inflammatory metrics, cytokine profiles, and molecular gene expression analyses in BALB/c mice subjected to OVA sensitization and challenge. The study protocol (A) outlines a 1-week adaptation period, followed by a 2-week OVA sensitization phase via intraperitoneal (i.p.) injection to establish the asthma model, and a subsequent 1-week treatment phase where groups received either vehicle (for control and model groups), low (L), medium (M), or high (H) concentrations of Fructus Xanthii aqueous extract, or dexamethasone as a positive control drug. Final assessments included body weight monitoring, bronchoalveolar lavage fluid (BALF) collection, histopathological examination using hematoxylin and eosin (H&E) staining, inflammation scoring, enzyme-linked immunosorbent assay (ELISA) for cytokine quantification, and real-time quantitative polymerase chain reaction (qPCR) for gene expression profiling. Body weight dynamics (B) reveal a significant time-dependent decline in the Model group, reflecting systemic inflammation or anaphylactic stress induced by OVA, which was progressively alleviated by Fructus Xanthii extract in a dose-dependent manner, with high concentrations nearing the protective effect of dexamethasone, suggesting potential anti-cachectic or immunomodulatory benefits. Histopathological analysis (C) showcases pristine alveolar structures in the Control group, contrasted by severe asthmatic features in the Model group—such as thickened alveolar septa, dense peribronchial and perivascular leukocyte infiltration (predominantly eosinophils, lymphocytes, and macrophages), bronchial epithelial hypertrophy, goblet cell metaplasia with excessive mucus, and luminal narrowing—while dexamethasone and Fructus Xanthii extract treatments, particularly at medium and high doses, restored alveolar integrity and reduced inflammatory cell aggregation in a concentration-dependent manner, indicating glucocorticoid-like anti-inflammatory activity (scale bars: 100 μm upper panels, 50 μm lower panels). Inflammation scores (D) derived from H&E histopathology highlight a marked elevation in the Model group (***p < 0.001 versus Control), indicative of robust Th2-driven airway inflammation, which was significantly attenuated by dexamethasone and higher doses of Fructus Xanthii extract (**p < 0.01 or *p < 0.05 versus Model), underscoring effective suppression of inflammatory cascades (mean ± SEM, n = 8). Cytokine quantification in BALF (E) demonstrates substantial increases in tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), interleukin-1 beta (IL-1β), and interleukin-5 (IL-5) in the Model group (***p < 0.001 versus Control), reflecting enhanced eosinophilic recruitment, innate immune activation, and pleiotropic inflammation, which were dose-dependently reduced by Fructus Xanthii extract, with high-dose effects matching dexamethasone’s potent inhibition (**p < 0.01 or *p < 0.05 versus Model), suggesting a therapeutic modulation of cytokine networks. Molecular analysis via qPCR (F) reveals significant upregulation of heat shock protein 90ab1 (Hsp90ab1), cyclin B1 (CCNB1), caspase-9 (CASP9), phosphoinositide 3-kinase (PI3K), and protein kinase B isoform 1 (AKT1) in the Model group (***p < 0.001 versus Control), implicating heightened chaperone activity, cell cycle progression, apoptosis, and PI3K/AKT-mediated survival signaling in asthma-aggravated lung injury; these were dose-dependently downregulated by Fructus Xanthii extract, with high-dose effects paralleling dexamethasone (**p < 0.01 or *p < 0.05 versus Model), potentially through interference with stress responses, cell cycle arrest, apoptosis induction, or PI3K/AKT inhibition to limit inflammatory hyperplasia. All data are presented as mean ± SEM (n = 8), with statistical significance assessed by one-way ANOVA followed by Tukey’s multiple comparisons test, collectively affirming the concentration-dependent anti-inflammatory and anti-asthmatic properties of Fructus Xanthii aqueous extract, comparable to dexamethasone. Western blot analysis further validated these findings at the protein level, showing increased expression of HSP90AB1 and AKT1 in the Model group, with normalization in treated groups, using GAPDH as the loading control (Figure 8G). All data are presented as mean ± SEM, with statistical significance determined by one-way ANOVA followed by Tukey’s multiple comparisons test. These results collectively demonstrate the concentration-dependent anti-inflammatory and anti-asthmatic effects of Fructus Xanthii aqueous extract, comparable to dexamethasone, through modulation of inflammatory cytokines and key signaling pathways.
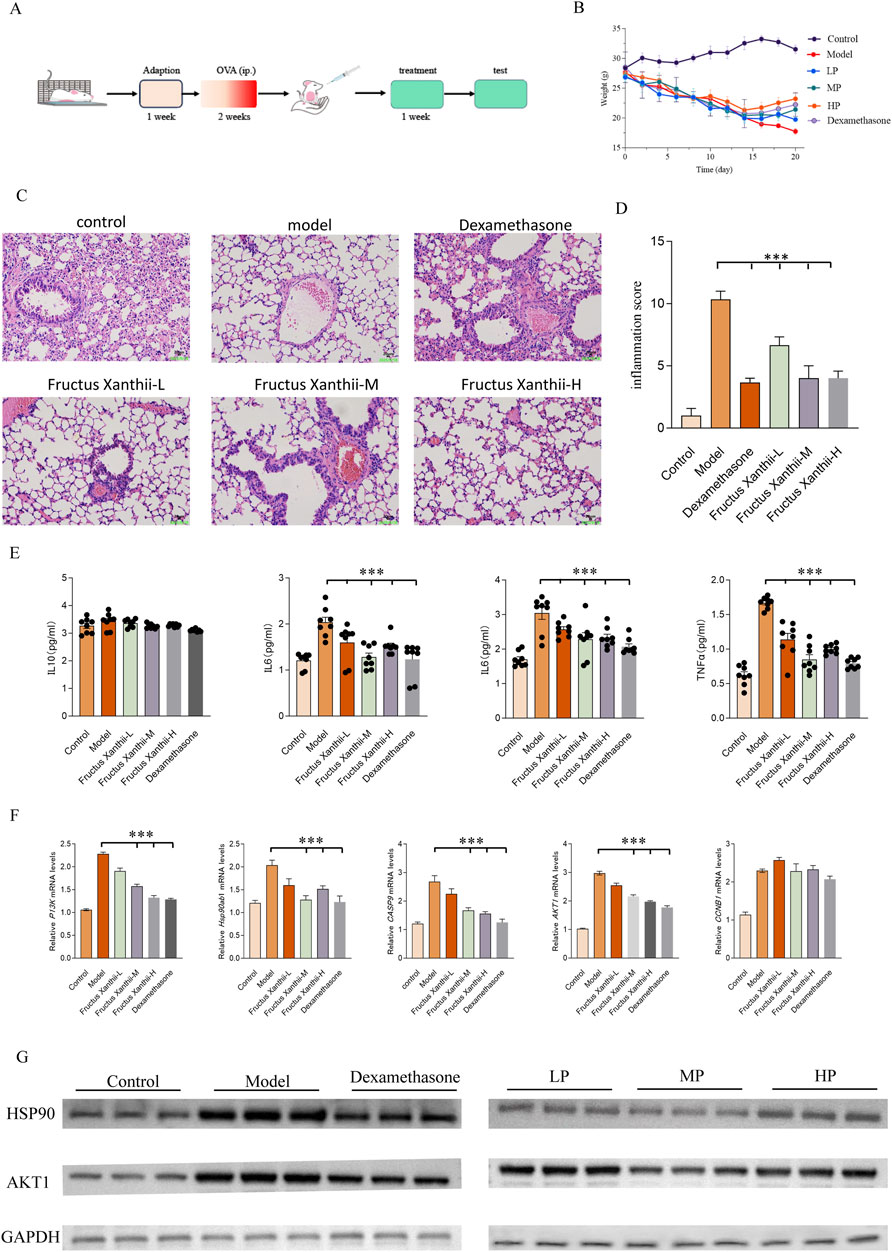
Figure 8. Comprehensively evaluates the therapeutic potential of Fructus Xanthii aqueous extract in an ovalbumin (OVA)-induced allergic asthma murine model, detailing the experimental timeline (A), body weight dynamics (B), histopathological lung alterations via H&E staining (C), inflammation scores (D), cytokine profiles in bronchoalveolar lavage fluid (BALF) via ELISA (E), and mRNA expression of key genes (Hsp90ab1, CCNB1, CASP9, PI3K, AKT1) via qPCR (F) in BALB/c mice. The protocol involved a 1-week adaptation, 2-week OVA sensitization, and 1-week treatment with low (L), medium (M), or high (H) doses of Fructus Xanthii extract or dexamethasone, followed by multifaceted assessments. Body weight declined in the Model group but was mitigated dose-dependently by Fructus Xanthii and dexamethasone (B). Histopathology revealed severe inflammation in the Model group, with progressive amelioration in treated groups, particularly at higher Fructus Xanthii doses and dexamethasone (C), corroborated by reduced inflammation scores (D). Cytokine levels (TNF-α, IL-6, IL-1β, IL-5) were significantly elevated in the Model group, with notable suppression by Fructus Xanthii and dexamethasone (E). Gene expression analysis indicated upregulated Hsp90ab1, CCNB1, CASP9, PI3K, and AKT1 in the Model group, with dose-dependent downregulation by treatments, suggesting modulation of stress response, cell cycle, apoptosis, and survival pathways (F). Data, presented as mean ± SEM (n = 8), highlight Fructus Xanthii’s concentration-dependent anti-inflammatory and anti-asthmatic efficacy, comparable to dexamethasone, with statistical significance (***p < 0.001) established via ANOVA with Tukey’s test. (G) Western blot analysis of HSP90AB1 and AKT1 protein levels in lung tissues, with GAPDH as the loading control, showing increased expression in the Model group and normalization in treated groups. All data are expressed as mean ± SEM (n = 8 per group); statistical significance was assessed by one-way ANOVA followed by Tukey’s multiple comparisons test.
4 Discussion
Our integrated analysis demonstrates that asthma involves complex transcriptional dysregulation, as evidenced by the identification of 3,755 differentially expressed genes (DEGs). Specifically, WGCNA highlighted the MEblack module (741 genes), enriched in proteostasis regulators (e.g., POMP, PSMA6) and immune signaling components, as a key driver of asthma pathogenesis (Dong et al., 2017). This suggests a dual-pronged disease mechanism involving both immune activation and impaired epithelial stress responses, a paradigm that extends beyond traditional inflammatory models. By integrating Fructus Xanthii’s 1,317 predicted targets with the asthma-related genes, we identified 100 overlapping targets, which were significantly enriched in pathways related to steroid hormone response (e.g., glucocorticoid receptor NR3C1), kinase activity (e.g., HSP90AB1), and immune modulation. Crucially, KEGG analysis revealed the PI3K-AKT and p53 signaling pathways, central regulators of airway remodeling and cellular senescence, respectively, as key targets of Fructus Xanthii. Further refinement using machine learning (RF/SVM/XGBoost) pinpointed seven hub genes: HSP90AB1, CCNB1, CASP9, CDK6, NR3C1, ERBB2, and CCK. Of particular interest is the HSP90AB1-NR3C1 axis. Molecular docking revealed that Carboxyatractyloside, a key compound in Fructus Xanthii, binds with high affinity (−10.09 kcal/mol) to LYS53 of HSP90AB1, potentially stabilizing its ATPase domain and preventing NR3C1 degradation. This mechanism could directly address glucocorticoid resistance, a hallmark of severe asthma. Molecular dynamics simulations confirmed the stable binding (RMSD <3 Å) of Carboxyatractyloside to HSP90AB1, CCNB1, and CASP9, demonstrating compact and stable complex dynamics (Rg: 2.18–3.26 nm). Immune infiltration analysis further supported the relevance of these targets, linking HSP90AB1 and ERBB2 expression to M2 macrophage polarization and highlighting asthma-specific immune dysregulation. The GEO datasets (GSE63142 and GSE14787) predominantly comprised samples from patients with severe asthma (Porsbjerg et al., 2023). While this focused enabled robust identification of disease-associated signals, generalizability to mild or intermittent asthma phenotypes remained uncertain, as these subgroups might exhibit distinct molecular signatures and inflammatory profiles (Kuo CS, et al. J Allergy Clin Immunol 2017; 139:1797–1807). Second, carboxyatractyloside, despite demonstrating the strongest binding affinity to HSP90AB1 (−10.09 kcal/mol), is a known inhibitor of the mitochondrial ADP/ATP translocase and has been associated with hepatotoxicity and nephrotoxicity in preclinical models (Obatomi and Bach, 1998). Its clinical translation would require comprehensive tiered toxicological profiling (acute, sub-chronic, and genotoxicity studies) prior to any therapeutic consideration. Third, the current study relies entirely on computational predictions and in vivo observations without in vitro mechanistic validation. Functional consequences of modulating hub genes via siRNA-mediated knockdown or lentiviral overexpression in human bronchial epithelial cells or macrophage models have not been evaluated. Such experiments are essential to confirm target engagement, downstream pathway modulation, and phenotypic outcomes. Future studies should include cell-based knockdown/overexpression and systematic toxicology to substantiate translational relevance. In vivo experiments in an OVA-sensitized murine model corroborated these findings, demonstrating Fructus Xanthii aqueous extract’s dose-dependent efficacy: attenuated body weight loss, restored lung histopathology (reduced peribronchial infiltration, epithelial hypertrophy, and mucus hypersecretion via H&E), lowered inflammation scores, suppressed BALF cytokines (TNF-α, IL-6, IL-1β, IL-5), and downregulated hub gene expression (HSP90AB1, CCNB1, CASP9, PI3K, AKT1), rivaling dexamethasone. This validates Fructus Xanthii’s role in blocking NF-κB/MAPK cascades, inhibiting extracellular HSP90α-mediated RhoA/MLC signaling, and preserving epithelial barrier integrity, as extracellular HSP90α exacerbates bronchial dysfunction in asthma.
Acute asthma exacerbations are strongly correlated with inflammatory mediators released by infiltrating immune cells in the airways. These mediators can induce epithelial cell damage and programmed cell death, disrupting airway mucosal integrity and function. Notably, damaged epithelial tissue may further amplify the innate immune response by persistently releasing endogenous danger signals, including damage-associated molecular patterns (DAMPs), which act as triggers for immune activation (Méndez-Enríquez and Hallgren, 2019; Miller et al., 2021). This persistent immune activation triggered by cell damage eventually leads to abnormal airway structure remodeling and promotes the chronic process of the disease. Based on this pathological mechanism, modern asthma treatment strategies should focus on targeting and regulating the key nodes in the inflammatory cascade reaction, while exploring new therapeutic approaches to block the abnormal death pathways of epithelial cells. Hsp90 is compositively expressed in eukaryotes and upregulated under stress. Its subtype Hsp90α (HSP90AB1) is distributed in the cytoplasm and the surface of some cells, and can be actively secreted into the intercellular space, participating in wound healing, inflammation regulation, and cell migration. Airway epithelial barrier injury is associated with asthma. Secretory Hsp90α intensifies the pathological process through pro-inflammatory effects, while Hsp90 inhibitors can effectively alleviate asthma-induced epithelial barrier dysfunction (Ye et al., 2019; Huang et al., 2024a). Extracellular HSP90α (eHSP90α) can activate the RhoA/myosin light chain (MLC) signal transduction pathway, leading to continuous phosphorylation of MLC and cytoskeletal remodeling, disrupting the bronchial epithelial barrier and resulting in the deficiency of two essential adhesion connexin proteins, E-cadherin and β-catenin. The disruption of the integrity of this epithelial barrier can lead to increased permeability of the bronchial epithelium, intensifying the inflammatory response and the development of asthma (Dong et al., 2017). In addition, studies have found that the glucocorticoid receptor (GR) encoded by the NR3C1 gene, as a core regulator of immune and inflammatory response, is a therapeutic target in asthma and other chronic inflammatory diseases. Corticosteroid drugs (such as hydrocortisone, prednisone, etc.) exert anti-inflammatory effects precisely by activating GR (Panek et al., 2015; Pan et al., 2022). The enrichment of NR3C1 in “response to corticosteroid” pathways, downstream of HSP90AB1, suggests that Fructus Xanthii may restore glucocorticoid sensitivity by stabilizing GR through HSP90AB1 chaperone function. ERBB2, often overexpressed in asthmatic epithelium, drives mucus hypersecretion. Its inhibition by Fructus Xanthii components could further mitigate airway remodeling (Hough et al., 2020).
While our bioinformatics analysis of public datasets provides valuable insights into the potential therapeutic mechanisms of Fructus Xanthii in asthma, the computational predictions, including molecular docking and machine learning results, require experimental validation. Future in vitro and in vivo studies are needed to confirm predicted target interactions (e.g., Carboxyatractyloside binding) and assess pharmacological efficacy in diverse patient populations. Furthermore, rigorous evaluation of Carboxyatractyloside’s bioavailability, potential toxicity (especially hepatotoxicity), and therapeutic efficacy is crucial before clinical translation. Given the complexity of Fructus Xanthii’s chemical composition, further studies should focus on isolating and characterizing the specific active constituents. Finally, while our immune infiltration analysis provides valuable insights, it relies on computational deconvolution of bulk RNA-seq data. Single-cell RNA sequencing would provide a more granular understanding of the immune landscape in asthma and the cell-type-specific effects of Fructus Xanthii.
Despite these limitations, our study identifies seven core targets (HSP90AB1, CCNB1, CCK, CDK6, CASP9, NR3C1, and ERBB2) that offer promising avenues for developing novel asthma therapies. Through the integration of network pharmacological analysis, machine learning, and molecular dynamics simulation technology, we screened the potential targets of Fructus Xanthii for the treatment of asthma. These key molecular targets identified by these screens are potential intervention nodes for core biological processes such as mediating inflammatory cascades, cell cycle regulation, and immune homeostasis maintenance. Further validation experiments targeting these potential targets will be carried out in subsequent studies.
In conclusion, several potential core targets of Fructus Xanthii for the treatment of asthma were screened out in this study. They may serve as potential biomarkers for the diagnosis of asthma and the monitoring of disease severity. In addition, targeted therapy against these molecules may also bring more effective treatment strategies to asthma patients. Finally, the differences in different immune cells found in this study between asthma patients and non-asthma patients need further research. By exploring the differential expression of the potential targets of Fructus Xanthii in different immune cells, it is helpful to understand the therapeutic potential of Fructus Xanthii and provide new ideas for the development of innovative asthma treatment methods.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal study was approved by all procedures adhered to ethical guidelines for animal experimentation, ensuring minimal distress and compliance with institutional protocols. All experimental procedures were strictly carried out by the guiding principles of the Animal Care and Use Committee of Qilu Medical University, and the experimental plans were approved by the Laboratory Animal Ethics Committee of Qilu Medical University (YXLL2025R032). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YD: Data curation, Conceptualization, Methodology, Writing – original draft, Formal Analysis. ZF: Writing – original draft, Methodology, Data curation, Conceptualization, Formal Analysis. XL: Methodology, Data curation, Writing – original draft, Supervision. YM: Data curation, Writing – original draft, Formal Analysis, Methodology. DW: Methodology, Data curation, Formal Analysis, Conceptualization, Writing – original draft. YS: Supervision, Writing – original draft, Formal Analysis, Resources, Funding acquisition, Writing – review and editing. YF: Supervision, Formal Analysis, Methodology, Writing – original draft, Resources, Funding acquisition, Writing – review and editing.
Funding
The authors declare that financial support was received for the research and/or publication of this article. Special Research Start-up Fund for the Introduction of High-level Talents of Qilu Medical University (004080341); Open Project of Basic Medicine of Qilu Medical University (JCYXKFKC202407).
Conflict of interest
Author ZF was employed by Shandong Benon Biological Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Athari, S. S. (2019). Targeting cell signaling in allergic asthma. Sig Transduct. Target Ther. 4, 45. doi:10.1038/s41392-019-0079-0
Barnes, P. J., Szefler, S. J., Reddel, H. K., and Chipps, B. E. (2019). Symptoms and perception of airway obstruction in asthmatic patients: clinical implications for use of reliever medications. J. Allergy Clin. Immunol. 144, 1180–1186. doi:10.1016/j.jaci.2019.06.040
Busse, W. W. (2019). Biological treatments for severe asthma: a major advance in asthma care. Allergol. Int. 68, 158–166. doi:10.1016/j.alit.2019.01.004
Chan, H. H. L., and Ng, T. (2020). Traditional Chinese Medicine (TCM) and allergic diseases. Curr. Allergy Asthma Rep. 20, 67. doi:10.1007/s11882-020-00959-9
Dong, H., Le, Y., Wang, Y., Zhao, H., Huang, C., Hu, Y., et al. (2017). Extracellular heat shock protein 90α mediates HDM-induced bronchial epithelial barrier dysfunction by activating RhoA/MLC signaling. Respir. Res. 18, 111. doi:10.1186/s12931-017-0593-y
Feng, X., Peng, D., Qiu, Y., Guo, Q., Zhang, X., Li, Z., et al. (2025). Identification and validation of Aging- and endoplasmic reticulum stress-related genes in periodontitis using a competing endogenous RNA network. Inflammation 48, 1326–1349. doi:10.1007/s10753-024-02124-0
Finotto, S. (2019). Resolution of allergic asthma. Semin. Immunopathol. 41, 665–674. doi:10.1007/s00281-019-00770-3
Guo, Q., Zhong, X., Dang, Z., Zhang, B., and Yang, Z. (2025). Identification of GBN5 as a molecular biomarker of pan-cancer species by integrated multi-omics analysis. Discov. Oncol. 16, 85. doi:10.1007/s12672-025-01840-9
Han, J., Zhang, S., Jiang, B., Wang, J., Ge, X., Wu, B., et al. (2022). Sesquiterpene lactones from Xanthium sibiricum Patrin alleviate asthma by modulating the Th1/Th2 balance in a murine model. Phytomedicine 99, 154032. doi:10.1016/j.phymed.2022.154032
Han, J., Wang, J., Wang, Y., Zhu, Z., Zhang, S., Wu, B., et al. (2023). Sesquiterpene lactones-enriched fractions from Xanthium mongolicum Kitag alleviate RA by regulating M1 macrophage polarization via NF-κB and MAPK signaling pathway. Front. Pharmacol. 14, 1104153. doi:10.3389/fphar.2023.1104153
Hough, K. P., Curtiss, M. L., Blain, T. J., Liu, R.-M., Trevor, J., Deshane, J. S., et al. (2020). Airway remodeling in asthma. Front. Med. 7, 191. doi:10.3389/fmed.2020.00191
Huang, H., Qiao, Y., Chu, L., Ye, C., Lin, L., Liao, H., et al. (2024a). Up-regulation of HSP90α in HDM-induced asthma causes pyroptosis of airway epithelial cells by activating the cGAS-STING-ER stress pathway. Int. Immunopharmacol. 131, 111917. doi:10.1016/j.intimp.2024.111917
Huang, W., Liu, X., Li, X., Zhang, R., Chen, G., Mao, X., et al. (2024b). Integrating network pharmacology, molecular docking and non-targeted serum metabolomics to illustrate pharmacodynamic ingredients and pharmacologic mechanism of Haizao Yuhu Decoction in treating hyperthyroidism. Front. Endocrinol. 15, 1438821. doi:10.3389/fendo.2024.1438821
Liu, J.-X., Zhang, Y., Yuan, H.-Y., and Liang, J. (2021). The treatment of asthma using the Chinese materia medica. J. Ethnopharmacol. 269, 113558. doi:10.1016/j.jep.2020.113558
Liu, X., Li, X., Huang, W., Cui, Y., Cheng, F., Chen, G., et al. (2025). Elucidating the pharmacological foundations and mechanisms of the sihai Shuyu formula in treating Graves’ disease through integrated serum metabolomics and network pharmacology with molecular docking techniques. Front. Endocrinol. 16, 1511808. doi:10.3389/fendo.2025.1511808
Luo, Y., Wang, C.-Z., Sawadogo, R., Tan, T., and Yuan, C.-S. (2020). Effects of herbal medicines on pain management. Am. J. Chin. Med. 48, 1–16. doi:10.1142/S0192415X20500019
Luo, W., Wu, B., Tang, L., Li, G., Chen, H., and Yin, X. (2021). Recent research progress of cirsium medicinal plants in China. J. Ethnopharmacol. 280, 114475. doi:10.1016/j.jep.2021.114475
Méndez-Enríquez, E., and Hallgren, J. (2019). Mast cells and their progenitors in allergic asthma. Front. Immunol. 10, 821. doi:10.3389/fimmu.2019.00821
Miller, R. L., Grayson, M. H., and Strothman, K. (2021). Advances in asthma: new understandings of asthma’s natural history, risk factors, underlying mechanisms, and clinical management. J. Allergy Clin. Immunol. 148, 1430–1441. doi:10.1016/j.jaci.2021.10.001
Obatomi, D. K., and Bach, P. H. (1998). Biochemistry and toxicology of the diterpenoid glycoside atractyloside. Food Chem. Toxicol. 36, 335–346. doi:10.1016/s0278-6915(98)00002-7
Pan, C., Chen, Y., Dong, Z., and Liu, H. (2022). Association of the glucocorticoid receptor gene polymorphisms with TGF-β1 mRNA expression in childhood asthma. Iran. J. Pediatr. 32. doi:10.5812/ijp-114153
Panek, M., Pietras, T., Fabijan, A., Zioło, J., Wieteska, Ł., Małachowska, B., et al. (2015). The NR3C1 glucocorticoid receptor gene polymorphisms may modulate the TGF-beta mRNA expression in asthma patients. Inflammation 38, 1479–1492. doi:10.1007/s10753-015-0123-3
Peters, M. C., Kerr, S., Dunican, E. M., Woodruff, P. G., Fajt, M. L., Levy, B. D., et al. (2019). Refractory airway type 2 inflammation in a large subgroup of asthmatic patients treated with inhaled corticosteroids. J. Allergy Clin. Immunol. 143, 104–113.e14. doi:10.1016/j.jaci.2017.12.1009
Porsbjerg, C., Melén, E., Lehtimäki, L., and Shaw, D. (2023). Asthma. Lancet 401, 858–873. doi:10.1016/S0140-6736(22)02125-0
Wang, J., Wang, D., Feng, L., Li, X., Gong, Y., Wang, Z., et al. (2023). Eremophilane-type and xanthanolide-type sesquiterpenes from the aerial parts of Xanthium sibiricum and their anti-inflammatory activities. Phytochemistry 208, 113603. doi:10.1016/j.phytochem.2023.113603
Xing, X., Chen, S., Li, L., Cao, Y., Chen, L., Wang, X., et al. (2018). The active components of Fuzheng Huayu formula and their potential mechanism of action in inhibiting the hepatic stellate cells viability – a network pharmacology and transcriptomics approach. Front. Pharmacol. 9, 525. doi:10.3389/fphar.2018.00525
Ye, C., Huang, C., Zou, M., Hu, Y., Luo, L., Wei, Y., et al. (2019). The role of secreted Hsp90α in HDM-induced asthmatic airway epithelial barrier dysfunction. BMC Pulm. Med. 19, 218. doi:10.1186/s12890-019-0938-z
Zhou, W., Chen, Z., Li, W., Wang, Y., Li, X., Yu, H., et al. (2019). Systems pharmacology uncovers the mechanisms of anti-asthma herbal medicine intervention (ASHMI) for the prevention of asthma. J. Funct. Foods 52, 611–619. doi:10.1016/j.jff.2018.11.048
Zhou, L., Zhou, Q., Guo, Q., Lai, P., Rui, C., Li, W., et al. (2025). Dual role of cathepsin S in cutaneous melanoma: insights from mendelian randomization and bioinformatics analysis. BMC Cancer 25, 104. doi:10.1186/s12885-025-13481-w
Keywords: Fructus Xanthii, asthma, network pharmacology, machine learning, molecular docking, immune infiltration
Citation: Dong Y, Fan Z, Li X, Ming Y, Wang D, Song Y and Fan Y (2025) Integrating multi-omics, network pharmacology, and experimental validation to unveil the molecular mechanisms of Fructus Xanthii in treating asthma. Front. Pharmacol. 16:1722150. doi: 10.3389/fphar.2025.1722150
Received: 10 October 2025; Accepted: 18 November 2025;
Published: 01 December 2025.
Edited by:
Shaoqiu Chen, University of Hawaii at Mānoa, United StatesReviewed by:
Bao Wang, Jilin Cancer Hospital, ChinaMengfei Long, Southwest University, China
Mingyang Jiang, National University of Singapore, Singapore
Copyright © 2025 Dong, Fan, Li, Ming, Wang, Song and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanda Song, eXNvbmdAc2R1dC5lZHUuY24=; Youpeng Fan, ZmFueW91cGVuZzE5OTRAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yiqun Dong
Yiqun Dong Zhaosen Fan2†
Zhaosen Fan2† Yuanda Song
Yuanda Song Youpeng Fan
Youpeng Fan