- 1School of Biology and Basic Medical Sciences, Medical College, Soochow University, Suzhou, China
- 2Institute of Agricultural Biotechnology and Ecology (IABE), Soochow University, Suzhou, China
Circadian clock system disorders can lead to uncontrolled cell proliferation, but the molecular mechanism remains unknown. We used a Bombyx mori animal model of single Period gene (BmPer) expression to investigate this mechanism. A slow growing developmental cell model (Per-KD) was isolated from a B. mori ovarian cell line (BmN) by continuous knock down of BmPer expression. The effects of BmPer expression knockdown (Per-KD) on cell proliferation and apoptosis were opposite to those of m/hPer1 and m/hPer2 in mammals. The knockdown of BmPer expression led to cell cycle deceleration with shrinking of the BmN cell nucleus, and significant inhibition of nuclear DNA synthesis and cell proliferation. It also promoted autophagy via the lysosomal pathway, and accelerated apoptosis via the caspase pathway.
Introduction
The circadian clock system and cell cycle are closely related (Shostak et al., 2016), and cell cycle progression occurs at a specific time in the circadian rhythm (Scheving et al., 1983; Bjarnason and Jordan, 2000; Garcia et al., 2001; Smaaland et al., 2002). Some key proteins in cell cycle, such as Cyclin D1, Cyclin B1, Cyclin E, Cyclin A, P53, WEE1, C-MYC, P21, Mdm2, ATM, ATR and Gadd45, exhibit circadian rhythm-dependent expression (Pines, 1999; Delaunay and Laudet, 2002; Duffield, 2003; Sato et al., 2003). Regulation of the cell cycle by the circadian clock system has been observed in mouse hepatic cells (Matsuo et al., 2003) and mouse embryonic fibroblasts (MEFs) NIH3T3 (Bieler et al., 2014; Feillet et al., 2014), respectively. Altered expression of clock genes has been seen in cells with abnormal cell cycle kinetics or in rapidly dividing cancer cells (Fu and Kettner, 2013; Cadenas et al., 2014).
Circadian clock disorders or impairment have been reported to lead to abnormal cell proliferation and promotion of tumor growth in mouse models (Filipski et al., 2002, 2004; EI Cheikh et al., 2014). Knockout of the circadian clock genes mCry1 and mCry2 has been shown to accelerate the cell cycle in MEFs (Destici et al., 2011). Circadian clock disorders can directly inhibit the cell cycle, and the apoptosis of breast cancer cells (Blakeman et al., 2016). The results of some studies are inconsistent or contradictory. For example, knockout of the circadian clock gene mClk was reported to arrest the cell cycle and promote apoptosis in embryonic stem cells (Lu et al., 2016). We previously reported that knocking down the expression of the silkworm circadian clock Period gene (BmPer) inhibited the cell proliferation by repressing glycometabolism in Bombyx mori ovarian (BmN) cells (Tao et al., 2017). The mutual regulation of the circadian clock and cell cycle generates conflicting cellular signals and indicate that further analysis of the mechanism of circadian clock regulation of cell proliferation is necessary.
Period (Per) is a core component of the circadian clock transcriptional regulatory network (Dunlap, 1999; Takahashi et al., 2008). In mammals, mPER1 regulates multiple cell cycle proteins (Gery et al., 2006; Yang et al., 2009). mPER1 and mPER2 proteins have been shown to inhibit breast cancer in vivo by inducing cancer cell apoptosis (Fu et al., 2002; Gery et al., 2006; Blakeman et al., 2016). However, mammalian mPer has multiple subtypes with distinct temporal and spatial expression of functional protein products (Shearman et al., 2000; Bae et al., 2001; Cermakian et al., 2001; Zheng et al., 2001).
In this study, an animal model with a single BmPer gene product was selected to investigate the effect of Per-KD on the cell cycle and avoid the interaction of multiple Per expression products. There have been no previous reports of cell cycle changes after simultaneous knockdown or knockout of all mPer genes. A slow growing developmental model expressing a single BmPer gene that was continually knocked down in BmN cells (Per-KD) was used in this study. The BmN cells were free of endocrine influences. We compared cell proliferation and programmed cell death (PCD) and investigated the regulatory mechanisms in mutant and wild-type BmN cells.
Materials and Methods
Cell Preparation
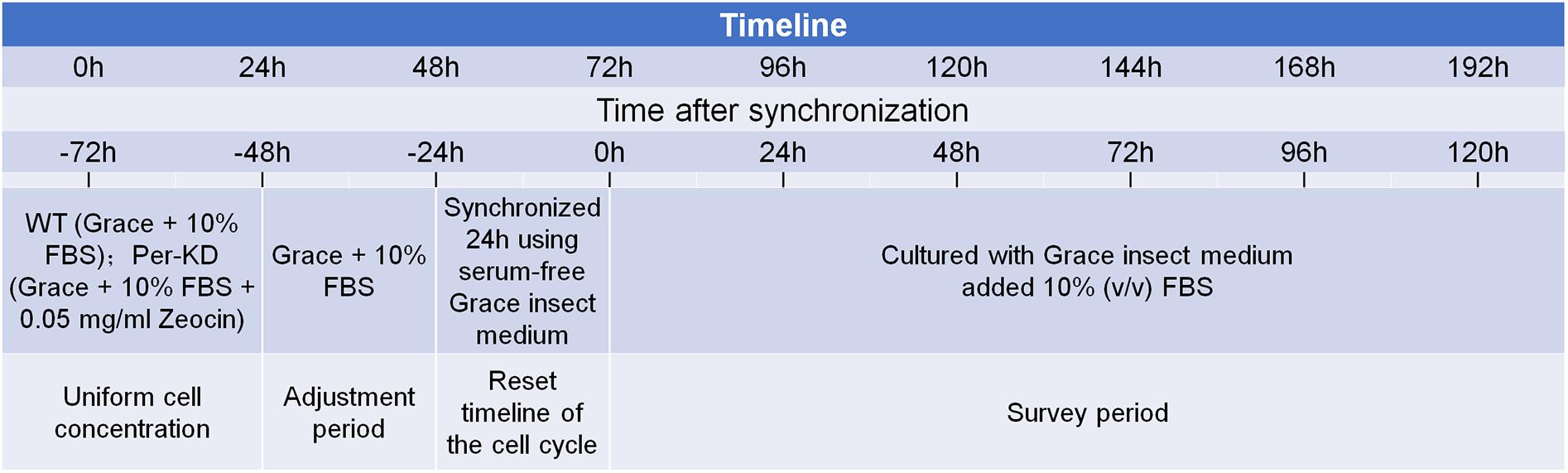
A wild-type (WT) B. mori ovary cell line (BmN) and a mutant line with stable interference of the BmPer gene (Per-KD) (Tao et al., 2017), were maintained in our laboratory and cultured in Grace insect medium (11605094, GIBCO, United States) with 10% (v/v) fetal bovine serum (FBS) (04-121-1A; Biological Industries, United States) at 26°C in the dark. The medium for culture of Per-KD cells included 0.05 mg/mL Zeocin (R25001, Invitrogen, United States). As shown in Figure 1, cell lines were synchronized by 24 h culture in serum-free Grace insect medium. The medium was then replaced with Grace insect medium with 10% FBS (v/v). The cells were counted and adjusted to the desired concentration. The time at which the synchronization process ended was recorded as time 0 h after synchronization.
Cell Proliferation Assay
After synchronization, the rate of cell division was determined at 0, 24, 48, 72, 96, and 120 h of growth in Grace insect medium with 10% (v/v) FBS with a methyl thiazolyl tetrazolium (MTT) assay (C0009, Beyotime, China). The cells (100 μL, 1 × 105 cells/mL) were incubated for 4 h in 96-well plates at 26°C in the dark and additional 4 h at 37°C in the dark after adding 100 μL formazan. The absorbance at 570 nm was measured with an Eon microplate reader (BioTek, VT, United States). The measurement was repeated in five culture wells.
Staining Methods
Synchronized BmN cells (1000 μL, 1.5 × 105 cells/mL) were cultured in Grace insect medium with 10% (v/v) FBS. The cells were stained with using Click-iTTM EdU Alexa FluorTM 488 Imaging Kits (C10337, Invitrogen, United States) following the manufacturer’s instructions (Salic and Mitchison, 2008; Ning et al., 2013), diamidino-phenyl-indole (DAPI; C1006, Beyotime, China) and TdT-mediated dUTP nick end labeling (TUNEL; 11684795910, Roche, Switzerland) as previously described (Liu et al., 2014; Li et al., 2017), monodansylcadaverine (MDC; G0170, Solarbio, China) as described by Biederbick et al. (1995), and Lyso-Tracker Red (C1046, Beyotime, China) as described by Yan et al. (2016). Immunohistochemical staining was performed using an anti-human cleaved-caspase-3 primary antibody (1:200, 9661s, CST, United States) and an Alexa Fluor 594-conjugated goat anti-mouse IgG (H+L) secondary antibody (1:300, AS054, ABclonal, China) as described by Ji et al. (2013).
Flow Cytometry
Synchronized BmN cells (1000 μL,1 × 106 cells/mL) were transferred to Eppendorf tubes containing Grace insect medium with 10% (v/v) FBS. Cell cycle and apoptosis assays were conducted simultaneously at 0, 24, 48, 72, 96, and 120 h. Cells were harvested by low speed centrifugation (4°C, 1000 rpm for 10 min), washed twice in precooled phosphate buffered saline (PBS; SH30256.01, HyClone, United States), resuspended in 1 mL PBS, and then fixed overnight at 4°C after adding 0.5 mL precooled 70% ethanol. Centrifugation and washing were repeated, and the cells were resuspended in 500 μL PBS. For flow cytometry, 5 μL RNase A and 5 μL 1 mg/mL propidium iodide (PI) were added to 100 μL of cell suspension and placed in direct sunlight for 30 min at 4°C before adding 400 μL PBS. The cell cycle assay was performed with an FC500 series flow cytometer (Beckman Coulter, Atlanta, GA, United States). Apoptosis was assayed with an annexin V-FITC/PI kit (AD10, Dojindo, Japan) following the manufacturer’s instructions.
Real-Time Polymerase Chain Reaction (qRT-PCR)
The mRNA expression of marker genes was assayed by qRT-PCR using the primers shown in Supplementary Table S1 as described by Tao et al. (2017). Total RNA was extracted from WT and Per-KD BmN cells using RNAiso Plus kits (9109, TaKaRa, China), and DNA in the total RNA samples was digested using RNase-free DNase I (2270A, TaKaRa, China). cDNA was synthesized using PrimeScript RT reagent kits (RR037A, TaKaRa, China) following the manufacturer’s instructions, RT-qPCR was performed with Premix Ex Taq (RR420A, TaKaRa, China) using an ABI StepOne Plus (Life technologies, Marsiling Industrial Estate, Singapore) and the fluorescent dye SYBR in a total reaction volume of 20 μL. The reaction conditions were 95°C preinitiation heating for 30 s followed by 40 cycles at 95°C for 5 s and 60°C for 30 s. Melting curve analysis was used to confirm the amplification of specific products. The BmRp49 gene was used to normalize the qRT-PCR results and a standard curve was used to determine the expression levels of the samples. All assays were performed in triplicate.
Caspase-3 Activity Assay
WT and Per-KD cells (1.96 × 106 cells) were cultured several days in Grace insect medium with 10% (v/v) FBS. Cells were collected by centrifugation (4°C, 1000 g, 10 min) at 24, 72, and 120 h after synchronization and then resuspended in 150 μL RIPA lysis buffer (P0013B, Beyotime, China) with 1% phenylmethanesulfonyl fluoride (PMSF; ST506, Beyotime, China). The suspended cells were lysed with an ultrasonic disruptor (Q700; Qsonica, Newtown, CT, United States) at 300 W, pulse on 3 s, pulse off 10 s, for 40 cycles. The lysate was centrifuged at 4°C at 12 000 rpm for 10 min. Caspase-3 activity in the supernatant was assayed with a colorimetric assay (BC3830; Solarbio, China) following the kit manufacturer’s instructions. The reaction system included 70 μL buffer solution, 25 μL sample and 5 μL acetyl-Asp-Glu-Val-Asp p-nitroanilide (Ac-DEVD-pNA). Buffer solution was used as the negative control. The assay was conducted for 2.5 h at 37°C in the dark. The absorbance at 405 nm was read with an Eon microplate reader (BioTek, VT, United States). Caspase-3 activity was estimated by comparison to a standard curve.
Results
Inhibition of BmPer Expression Causes Inhibition of the Cell Cycle
We previously achieved continuous knockdown of BmPer gene expression in BmN cells and generated a Per-KD B. mori model with developmental disorders. The qPCR and western blotting results showed that BmPer gene transcription in Per-KD cells was downregulated by about 40% compared with WT cells and BmPER protein expression was significantly reduced by about 80%. BmPer gene knockdown thus significantly reduced both gene and protein expression of Per (Tao et al., 2017). In this study, DAPI staining of synchronized cell lines after culture for 24–120 h (Figure 2A) revealed that Per-KD nuclei were significantly smaller than WT nuclei (p < 0.001, Figure 2B). The difference in the number of DAPI-stained nuclei revealed that the proliferation rate of Per-KD cells was significantly lower than that of WT cells and that the difference increased with time (p < 0.001, Supplementary Figure S1).
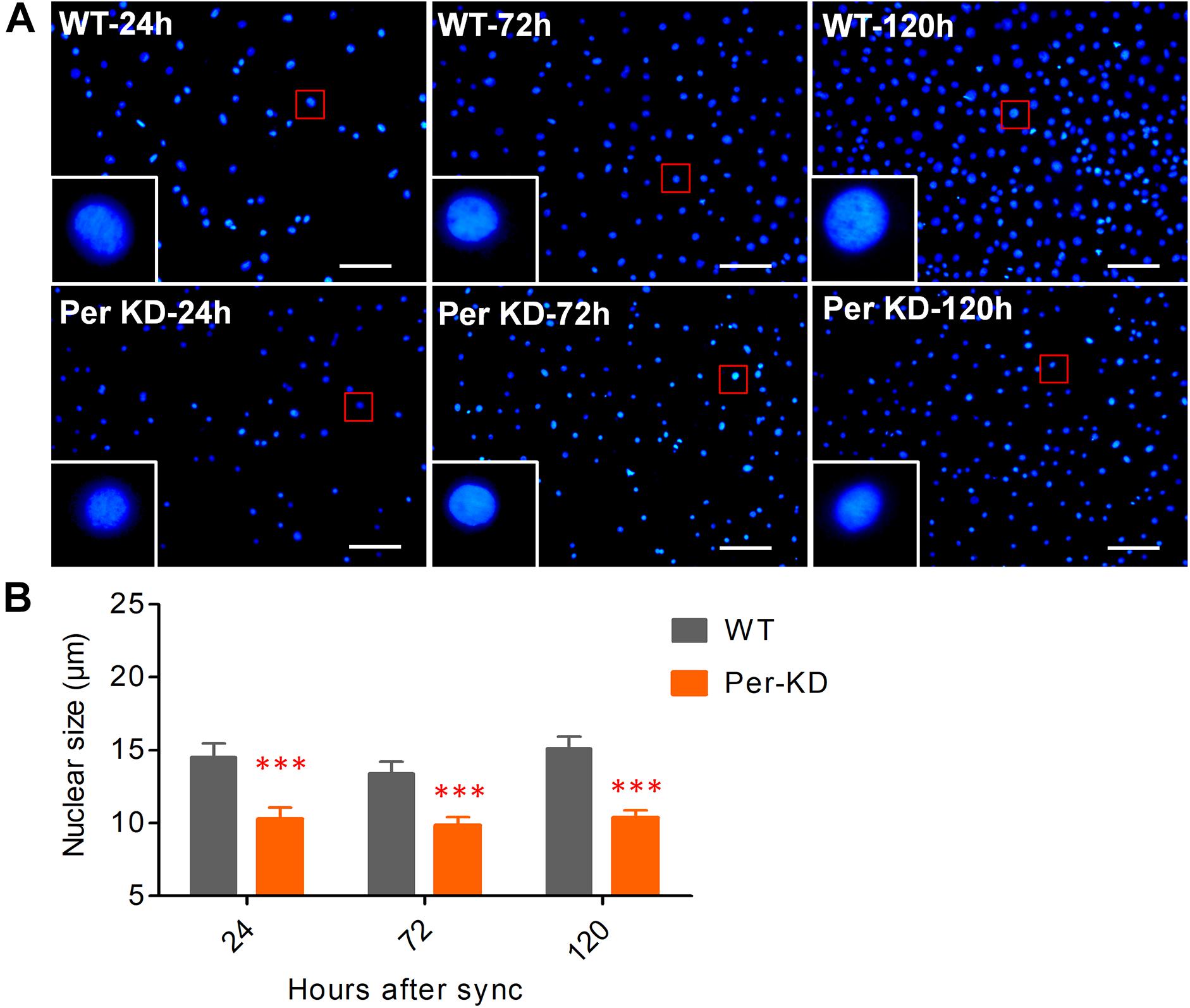
Figure 2. BmPer gene knockdown changes nuclear size. Cell lines were synchronized for 24 h in serum-free Grace insect medium. The nuclei were stained after 24, 72, and 120 h of culture in medium with 10% (v/v) FBS. (A) Cell nuclei were stained with 4’-6-diamidino-2-phenylindole (DAPI). Bar = 100 μm. (B) Nuclear size. ∗∗∗p < 0.001 (n = 3 plates with size measurement of 120 nuclei). WT, wild-type BmN cells; Per-KD, BmPer knockdown BmN cells.
BmPer knockdown affects the proliferation of BmN cells by inhibiting nuclear DNA synthesis. The effect of knockdown of BmPer expression on BmN cell proliferation was assayed by the amount of nuclear EdU incorporation over 8h. The difference in the percentage of EdU-positive cells indicated that the proliferation rate was significantly lower in Per-KD than in WT cells (p < 0.001, Figures 3A,B). Cell proliferation at 0–120 h after synchronization was also assayed by MTT. Compared with WT cells (Figure 3C), Per-KD cell proliferation began to slow at 72 h, reaching and became significantly different 120 h (p < 0.01).
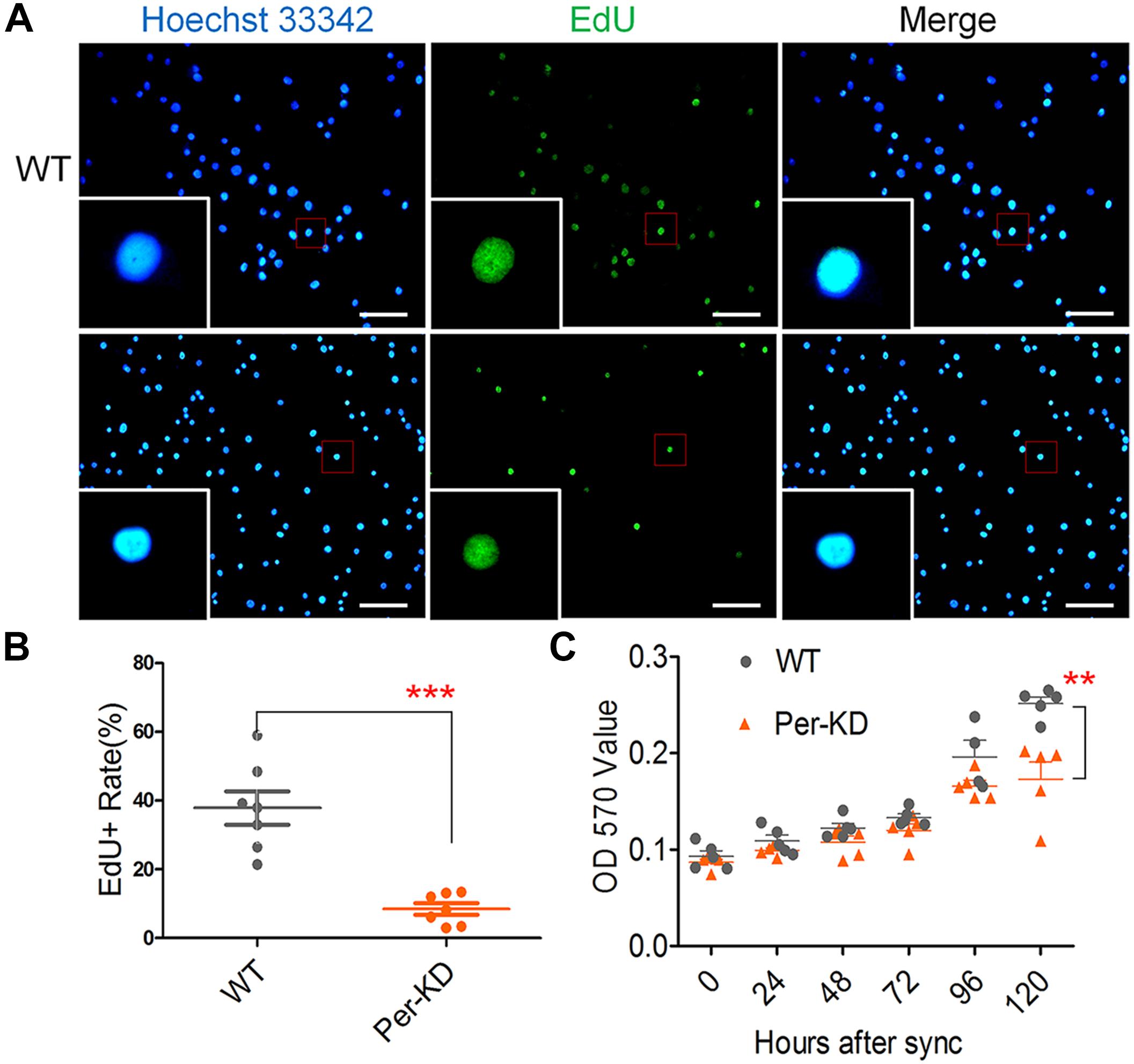
Figure 3. Changes in the proliferation of Per-KD cells following inhibition of nuclear DNA synthesis. Cells were synchronized for 24 h in serum-free Grace insect medium. The medium was replaced by Grace insect medium with 10% (v/v) FBS. After 8 h incubation with EdU (A) nuclei with green fluorescence indicate DNA synthesis. All nuclei show Blue Hoechst 33342 staining. Bar = 100 μm. (B) EdU-positive staining percentage (n = 7). (C) After synchronization and culture in Grace insect medium with 10% (v/v) FBS, the rate of cell division was determined with an MTT assay (n = 5). ∗∗p < 0.01, ∗∗∗p < 0.001.
Cell Cycle Slowing in Per-KD Cells
The knockdown of BmPer expression resulted in deceleration of BmN cells in the G0/G1 phase. Flow cytometry analysis of the cell cycle distribution in PI-stained Per-KD and WT cells found that the Per-KD cell cycle decelerated and the percentage of cells in the G0/G1 phase increased with the time in culture (Figure 4A). The percentage of cells in the G0/G1 phase was higher in the Per-KD group than in the WT group at 24, 48, 72, and 120 h after synchronization (Figure 4B). The percentage of cells in S phase was lower in Per-KD than in WT cells at each assay time (p < 0.001, Figure 4C). The percentage in G2/M was higher in Per-KD than in WT cells at 24 and 48 h after synchronization (p < 0.001); but then decreased, and at 120 h the percentage in G2/M was lower in Per-KD than in WT cells (p < 0.01, Figure 4D). The cell cycle distribution data showed that WT cells cultured for 24–120 h after cell cycle synchronization were mainly distributed in the S phase, and cells in G0/G1 phase gradually decreased in number. The Per-KD cells in S phase increase as those in G2/M decrease. (Figure 4E).
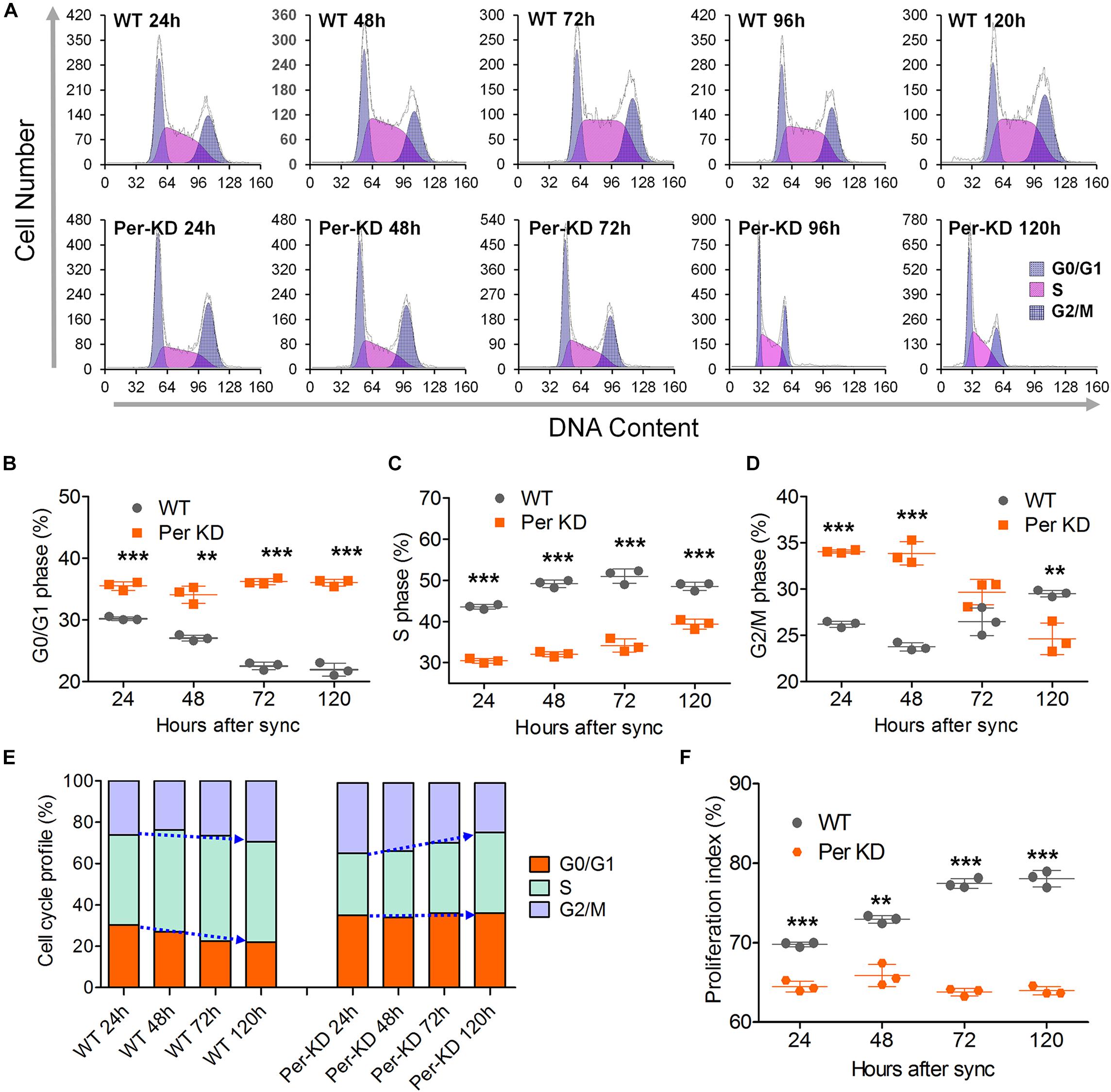
Figure 4. Cell cycle changes induced by BmPer gene knockdown. WT and BmPer knockdown BmN cells (Per-KD) were assayed 24, 48, 72, and 120 h after synchronization. (A) PI-FCM (Propidium Iodide-flow cytometry) results. (B–D) Percentages of cells in G0/G1 phase, S phase and G2/M phase. (E) Cell cycle profile. (F) Proliferation index = (S+G2/M)/(G0/G1+S+G2/M) × 100%. ∗∗p < 0.01, ∗∗∗p < 0.001 (n = 3).
Flow cytometry of PI-labeled cells showed that the proliferation index of Per-KD cells was lower than that of WT cells at 24 (p < 0.001) and 120 h (p < 0.01) after cell synchronization. The proliferation index remained low in Per-KD cells and did not increase with prolonged culture time as it did in WT cells (Figure 4F). RT-qPCR assays of the transcription of five cell cycle related genes shown in Supplementary Figure S2 are consistent with the flow cytometry results indicative of a blocked cell cycle shown in Figure 4E. Knockdown of BmPer gene expression was shown to result in slowed BmN cell proliferation, which is consistent with the observations shown in Figures 2, 3.
The Process of PCD Is Accelerated in Per-KD Cells
These results suggest that knockdown of BmPer expression slowed cell proliferation by slowing the cell cycle, but that accelerated cell death led to a decrease in cell number. Therefore, we investigated changes in the process of PCD in WT and Per-KD cells. Autophagy assay by MDC and Lyso-Tracker Red staining showed that more Per-KD than WT cells included green MDC or red Lyso-Tracker autophagosomes 120 h after developmental synchronization (Figures 5A,C), indicating a significant increase in the autophagy of Per-KD compared with WT cells (p < 0.001, Figures 5B,D). Assays of the transcription of autophagy associated genes 6 and 8 (BmAtg6 and BmAtg8) revealed that both were upregulated in Per-KD compared with WT cells at 24, 72, and 96 h (Figures 5E,F).
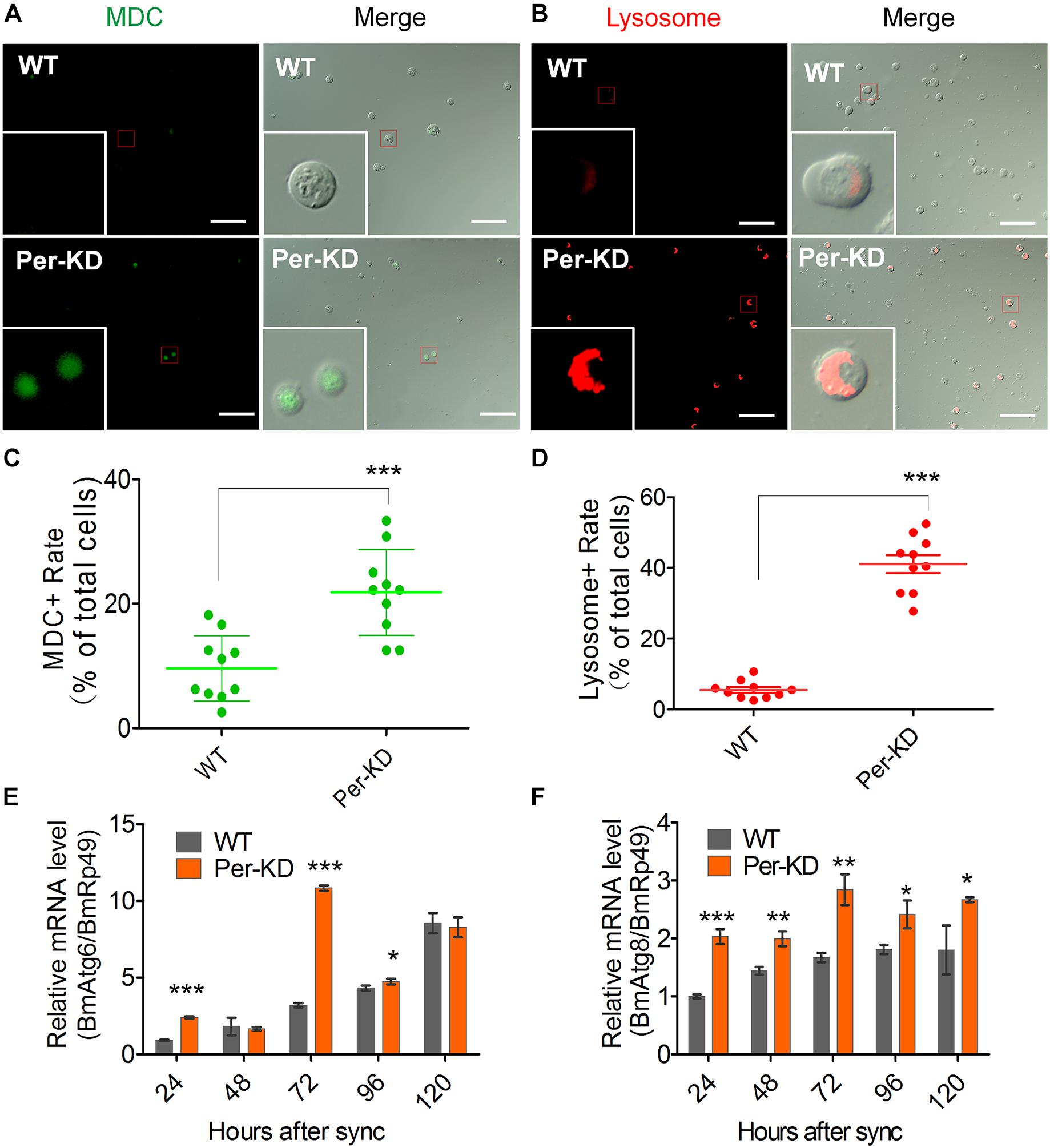
Figure 5. Autophagy was induced by BmPer gene knockdown. Autophagy was assayed in WT and Per-KD cells at 24, 48, 72, 96, and 120 h after synchronization. Cell dyeing were performed at 120 h after the synchronization process. (A) Monodansylcadaverine (MDC) staining is visible as green fluorescence in autophagosomes and (B) Lyso-Tracker Red fluorescence staining shows lysosomes in autophagic cells at 120 h. (C) Percentage of positive MDC staining. (D) Percentage of positive lysosome staining. (E,F) qRT-PCR assay of relative BmAtg6 (E) and BmAtg8 gene transcription (F) after synchronization. Bar = 100 μm. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, (n = 3).
Apoptosis and necrosis were assayed by annexin V-FITC and PI flow cytometry (Figure 6A). The results of the annexin V-FITC single-positive cell assay showed increased early apoptosis of Per-KD compared with WT cells at 48 (p < 0.001), 72 (p < 0.001), and 120 h (p < 0.01). The results at 24 and 96 h were not significantly different (Figure 6B). The PI and annexin V-FITC double-positive assay showed that apoptosis was increased in Per-KD cells at 24, 48 (p < 0.05), 72 (p < 0.001), and 120 h (p < 0.01), but the opposite result (p < 0.05) was observed at 96 h (p < 0.05, Figure 6C). The results of the PI single-marker assay of cell found that necrosis was lower in Per-KD than in WT cells at 72 and 120 h (both p < 0.001). No significant difference was observed at other times (Figure 6D).
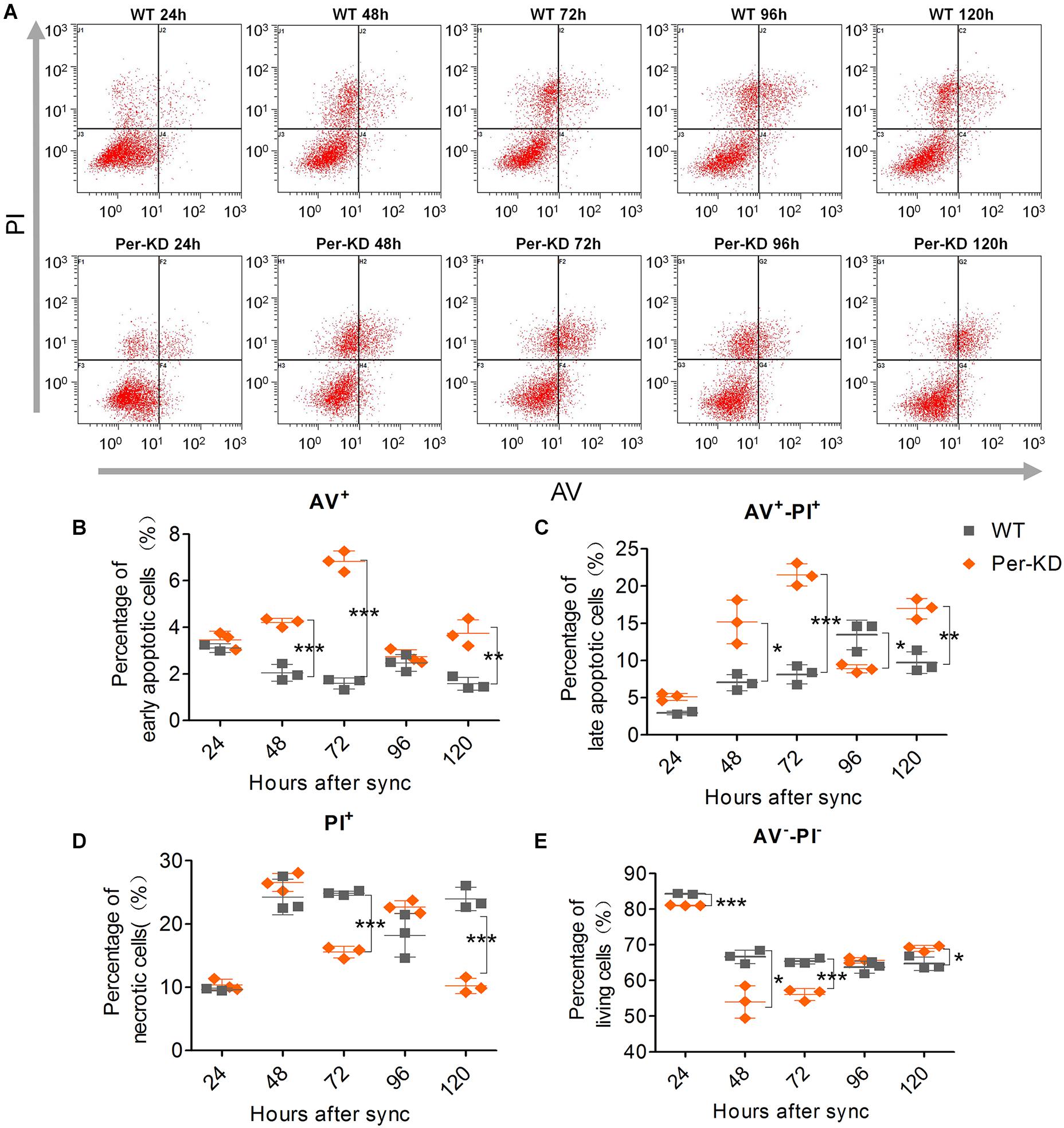
Figure 6. Cell apoptosis and cytoclasis assayed by Annexin V-FITC/PI-FCM. Apoptosis and necrosis were assayed in WT and Per-KD cells every 24 h after synchronization. (A) Annexin V-FITC/PI flow cytometry assay of apoptotic cells. (B) Annexin V-FITC single-positive apoptotic cells. (C) Double-positive Annexin V-FITC/PI positive nonviable apoptotic cells. (D) PI single-positive necrotic cells. (E) Double-negative Annexin V-FITC living cells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, (n = 3).
Flow cytometry found that the percentage of viable Per-KD cells was lower than that of WT cells at 24 and 72 h (both p < 0.001), but higher after 120 h (p < 0.05, Figure 6E). The effects of knockdown of BmPer expression on apoptosis and necrosis were greater soon after synchronization (24–72 h) than at later stages (120 h). This is inconsistent with the observations shown in Figure 5 of the effects of slowing the cell cycle on proliferation. Therefore, the decrease in the proportion of viable cells with Per gene expression knockdown that occurred shortly after synchronization, resulted from apoptosis rather than necrosis.
The promotion of apoptosis of BmN cells following knockdown of BmPer expression was also assayed by TUNEL staining. Positive TUNEL staining was increased in Per-KD cells compared with WT cells 120 h after synchronization (p < 0.01, Figure 7A and Supplementary Figure S3A). qRT-PCR showed that the expression of BmDronc mRNA, an apoptosis marker, was higher in Per-KD than in WT cells in the 24–120 h after synchronization (Figure 7B). An immunofluorescence assay found no significant differences in BmCaspase-3 protein expression in Per-KD and WT cells at 24 and 72 h after cell synchronization, but found significantly increased expression in Per-KD cells at 120 h (p < 0.01, Figure 7C and Supplementary Figure S3B). BmCaspase-3 activity was significantly higher in Per-KD cells than in WT cells (p < 0.01) in the 24–120 h after cell synchronization (Figure 7D). The results indicate that the knockdown of BmPer expression induced apoptosis by the caspase pathway.
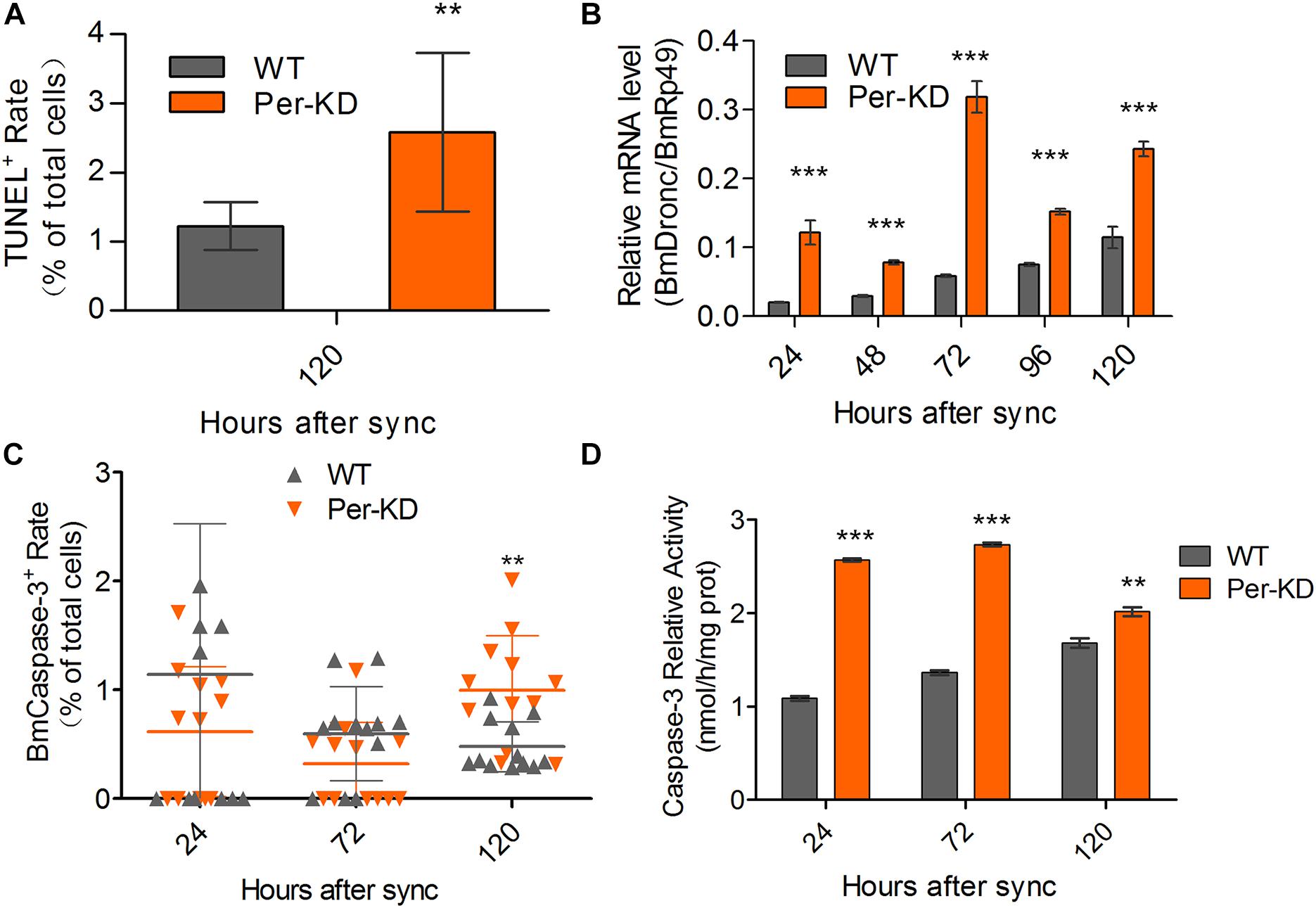
Figure 7. Cell apoptosis induced by BmPer gene knockdown. Apoptosis was assayed in WT and Per-KD cells every 24 h after synchronization. (A) Percentage of single-positive stained TUNEL cells (n = 12). (B) BmDronc mRNA expression assayed by qRT-PCR. The reference gene was BmRp49 (n = 3). (C) BmCaspase-3 immunofluorescence staining (n = 12). (D) Caspase-3 activity (n = 3). Nuclei were visualized by blue DAPI fluorescence. ∗∗p < 0.01 and ∗∗∗p < 0.001.
Discussion
A Possible Mechanism of Inhibition of Cell Growth and Proliferation by Knockdown of BmPer Gene Expression
The core components of circadian clock signal input negative feedback loop can promote or inhibit cell growth and proliferation (Filipski et al., 2002, 2004; EI Cheikh et al., 2014; Blakeman et al., 2016; Lu et al., 2016; Tao et al., 2017). The mechanisms have not been described. In mammals with multiple Per subtypes, the temporal and spatial expression of homologous genes, and the activities of their protein products, are very different (Shearman et al., 2000; Bae et al., 2001; Cermakian et al., 2001; Zheng et al., 2001). Regulation of cell proliferation by the circadian clock system is not conducted via a single pathway.
This study evaluated the effects of Per knocking-down on cell proliferation using a cell model of a Period gene disorder, Per-KD, which was constructed previously with a silkworm ovary cell line (Tao et al., 2017). Comparison of the regulation of cell proliferation and PCD between mutant Per-KD and WT BmN cells revealed that BmPer knockdown led to shrinkage of BmN cell nuclei, decelerated cell cycling, increased autophagy and apoptosis, and decreased cell proliferation. The proposed mechanism is shown in Figure 8.
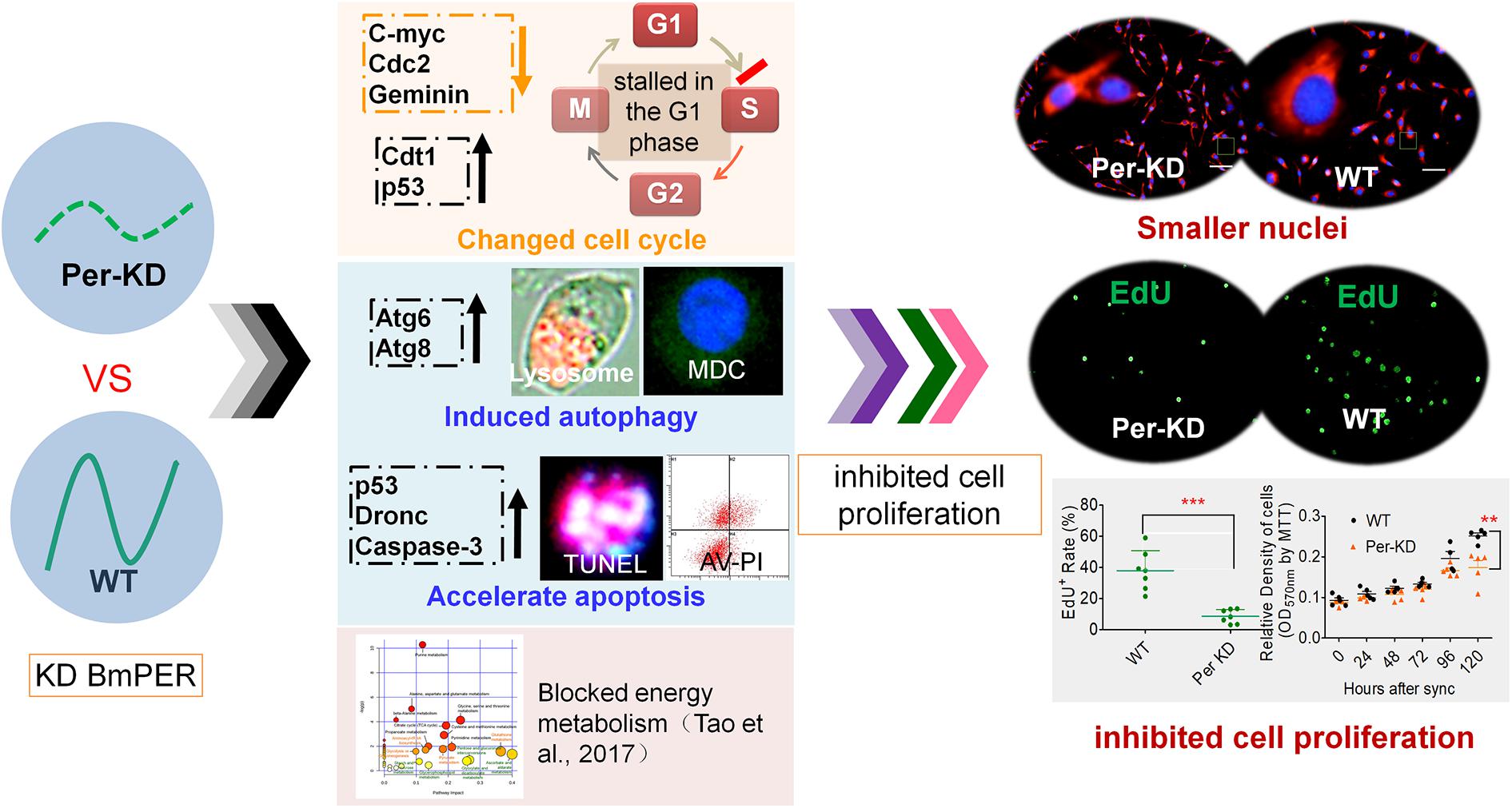
Figure 8. A proposed mechanism of inhibition of the cell cycle associated with knockdown of BmPer expression. Knockdown of the BmPer gene causes cell cycle deceleration, increased autophagy and apoptosis, or other previously reported changes in cellular metabolism (Tao et al., 2017), ultimately lead to cell shrinkage and inhibition of cell proliferation.
The size of the daughter cells produced by cytokinesis may influence cell cycle progression, with large daughter cells accelerating G1 and G2 and promoting cell cycle progression; small daughter cells have the opposite effects (Barnum and O’Connell, 2014). In this study, the Per-KD cells had smaller nuclei, decreased nuclear DNA synthesis in dividing cells, and a delayed cell cycle with cell development blocked in G0/G1. The observed effects of BmPer gene knockdown increased with prolonged culture after synchronization.
Previous studies have shown that a defective circadian clock system can disrupt the cell cycle and affect apoptosis in addition to influencing cell metabolism (Blakeman et al., 2016). BmPer knockdown in the silkworm has been reported to suppress cellular glucose metabolism and was correlated with changes in amino acid metabolism and inhibition of cell growth (Tao et al., 2017). Inhibition of cell proliferation has been associated with accelerated apoptosis with impairment of the circadian clock system by knockdown of mClk expression in mESCs (Lu et al., 2016). In this study, the observed effects on the regulation of PCD in Per-KD BmN cells included induction of autophagy by the lysosomal pathway and apoptosis by the caspase pathway.
Effects of the Period Gene on the Proliferation Differ in Mammalian and Silkworm Cells
The effects of changes of Per gene expression on cell proliferation have not been reported in animal models or normal cells, but a positive correlation of m/hPer1 and m/hPer2 gene expression with promotion of cell proliferation and reduction of cell apoptosis has been shown in mammalian tumor models. Mice lacking mPer2 expression develop lymphoma (Fu et al., 2002), and hPer2 expression is downregulated in human lymphoma and myeloid malignancy cells (Gery et al., 2005). Downregulation of hPer1 expression in mouse mammary carcinoma cells or human oral squamous cell carcinoma can alter cell cycle distribution, promote cell cycling, and reduce apoptosis (Borgs et al., 2009; Yang et al., 2009; Soták et al., 2014; Zhao et al., 2016). Downregulation of Per2 expression in mammals promotes expression of the cell proliferation-associated proteins Cyclin A, Cyclin D1, Mdm2, C-MYC, β-Catenin and Cyclin E, thereby promoting cell proliferation (Fu et al., 2002; Rosbash and Takahashi, 2002; Lee, 2006; Yang and Stockwell, 2008).
In addition, hPER1 is known to be an anti-apoptosis factor in human pancreatic, liver, and gingival cancer (Sato et al., 2009; Sato et al., 2011). Overexpression of hPer1 inhibits cell proliferation (Gery et al., 2006, 2007) and reduces the expression of Cyclin B1 and WEE1 proteins, promotes apoptosis, and inhibits cell proliferation in HCT116 human colonic cancer cells and in human prostate cancer cells (Gery et al., 2006; Cao et al., 2009). Overexpression of human hPer2 has been shown to inhibit the growth of the K562 and U937 myeloid leukemia cell, leading to K562 cell proliferation arrest, loss of clonality, and apoptosis (Gery et al., 2005). Overexpression of the mouse mPer2 gene has been shown to inhibit both tumor formation, and cell proliferation by decreasing Cyclin B1 and Cyclin A2 expression, while promoting p53 transcription to accelerate apoptosis (Hua et al., 2006; Lee, 2006; Hua et al., 2007; Wood et al., 2008; Yang and Stockwell, 2008). In this study, knockdown of BmPer gene expression in BmN cells induced autophagy and apoptosis, and inhibited cell proliferation. This is opposite to the effects of inhibition of m/hPer1 and m/hPer2 expression in mammalian cells.
It has been reported that the circadian clock and cell cycle are coordinated via MYC, WEE1, P16, P53 and other coupling factors (Shostak, 2017), the loss of mPer2 function results in the increased expression of c-Myc and partially impairs p53-mediated apoptosis (Fu et al., 2002). The expression products of the cellular-oncogene c-Myc and cell division cycle 2 gene (Cdc2) directly regulated the proliferation of cultured cells, and that C-MYC protein accelerated the progression of cells through G1 and S (Herrera-Merchan et al., 2010). Cdc2 was shown to regulate cell differentiation from G2 to M (Morgan, 1995). The transcription level of the Geminin and Cdc10-dependent transcript 1 gene (Cdt1) oscillates throughout the cell cycle and influences nuclear replication and cell division. Geminin is not transcribed during the G1 phase and is significantly upregulated during the S, G2, and M phases (Suchyta et al., 2015). Cdt1 transcription is low in the S phase and high in G1 (Nishitani et al., 2001). The tumor suppressor gene p53 is a cell cycle suppressor and an important regulator of G1/S checkpoints (Fu et al., 2016). Activation of p53 gene expression induces cell cycle arrest, promotes apoptosis, and transcription of senescence genes in response to stress, such as DNA damage (Herrera-Merchan et al., 2010). It can thus be seen that the mPer gene regulates the cell cycle by its effects on C-MYC, CDC2, P53, and other coupling factors in mammalian cells.
We investigated the transcription of these five genes in a silkworm ovary cell line with BmPer expression knockdown (Supplementary Figure S2). Downregulation of Bmc-Myc, BmCdc2, and BmGeminin at 96 and 120 h after synchronization of cell proliferation, and upregulation of BmCdt1 and Bmp53 expression, with decreased cell proliferation and increased apoptosis (Figure 8). It has been reported that knockdown of the mPer2 gene in mice results in upregulation of c-Myc and downregulation of p53 gene expression as well as promotion of cell proliferation and reduction of cell apoptosis (Fu et al., 2002). These effects differ in mammalian and silkworm cells and can be explained as follows.
(1) Effects of the BmPer and mPer genes on cell proliferation may depend on species differences in PER protein function in the circadian clock transcription–translation feedback loop (TTFL). However, we believe that BmPer or mPer knockdown affects cell proliferation and apoptosis of both mice and BmN cells via mediating C-MYC, P53 and other factors. In mammals, the most widely accepted explanation of the TTFL is that mPER independently or together with mCRYs (mCRYs/mPERs) binds CLOCK/BMAL1, with feedback inhibition of mCry and mPer genes transcription (Reppert and Weaver, 2001; Ko and Takahashi, 2006). On the other hand, some reports indicate that mPER2 can independently promote gene transcription at some time in the TTFL (Akashi et al., 2014; Ye et al., 2014). The available evidence indicates that mPER may have dual functions in different situations. The Danaus plexippus and Bombyx mori are both members of order Lepidoptera, in which DpCRY2 shuts down CLOCK-CYCLE-mediated transcription. DpPER likely helps to transport DpCRY2 from the cytoplasm to nucleus, but has no inhibitory feedback effect on DpPER (Zhu et al., 2008).
Bombyx mori has only a single Period gene and protein, but there are multiple mPer gene subtypes in mice. It has been reported that mPER1 and mPER2 proteins have distinct and complementary roles in the mouse clock mechanism (Bae et al., 2001; Zheng et al., 2001). The roles of the mPer2 and mPer1 genes on cell proliferation and apoptosis have not been investigated after knockout of their complementary genes (mPer1 or mPer2). Consequently, the effects of mPer1 or mPer2 on cell proliferation and apoptosis may not reflect the influence of mPer genes.
(2) Metabolism affects cell proliferation in complex ways. Previous studies have shown that fast-dividing pluripotent stem cells are usually highly glycolytic, with the energy provided by glycolysis as key driver of cell proliferation (Folmes et al., 2012; Zhang et al., 2012). Mice deficient in mPer2 have significantly increased circulating insulin levels compared with WT mice, and that can lead to upregulation of glucose metabolism (Zhao et al., 2012). In mouse models, the anterior tibialis muscle in mPer2 knockout mice has increased levels of glycolytic enzymes such as triose phosphate isomerase and enolase (Bae et al., 2006). However, double knockout of mPer1 and mPer2 has the opposite effect on blood insulin levels (Lamia et al., 2008). We previously assayed cell metabolomics by gas chromatography/liquid chromatography-mass spectrometry (GC/LC-MS) and found that knockdown of BmPer gene expression resulted in significant inhibition of glycometabolism, the amino acids that used glucose metabolites as a source were also downregulated. The activities of key glycolytic enzymes, such as hexokinase (BmHK), phosphofructokinase (BmPFK), and citrate synthase (BmCS), were significantly decreased and transcription of their encoding genes, as well as that of pyruvate kinase, were significantly downregulated (Tao et al., 2017). The results clearly showed that inhibition of the circadian clock gene BmPer repressed glycometabolism. The results differ from the increase in glycometabolism caused by knockout of the mPer2 gene (Bae et al., 2006; Zhao et al., 2012), but are consistent with changes in the glycometabolism in mPer1/mPer2 double knockout mice (Lamia et al., 2008). Species differences in the activities of gene products may thus help explain the opposite effects of BmPer knockdown and mPer1 or mPer2 knockdown or deletion on cell proliferation.
Conclusion
Knockdown of BmPer expression in Bombyx mori blocked the BmN cell cycle, decreased nuclear size, accelerated apoptosis and autophagy, and inhibited cell proliferation.
Author Contributions
S-QX and J-FQ conceived this project. XL, J-FQ, W-ZC, X-FL, HT, and Y-HS performed the research. XL, J-FQ, and S-QX analyzed the data and wrote the manuscript. All authors participated in the revision of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31672492), the Provincial Key R&D Program of Jiangsu (Grant No. BE2015317), the China Agriculture Research System (CARS) (Grant No. CARS-22), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), Postgraduate Research and Practice Innovation Program of Jiangsu Province (Grant No. KYCX18_2521). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The manuscript was edited by a native English speaker of International Science Editing Scientific Services.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00537/full#supplementary-material
References
Akashi, M., Okamoto, A., Tsuchiya, Y., Todo, T., Nishida, E., and Node, K. (2014). A positive role for PERIOD in mammalian circadian gene expression. Cell Rep. 7, 1056–1064. doi: 10.1016/j.celrep.2014.03.072
Bae, K., Jin, X., Maywood, E. S., Hastings, M. H., Reppert, S. M., and Weaver, D. R. (2001). Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30, 525–536. doi: 10.1016/s0896-6273(01)00302-6
Bae, K., Lee, K., Seo, Y., Lee, H., Kim, D., and Choi, I. (2006). Differential effects of two period genes on the physiology and proteomic profiles of mouse anterior tibialis muscles. Mol. Cells 22, 275–284.
Barnum, K. J., and O’Connell, M. J. (2014). Cell cycle regulation by checkpoints. Methods Mol. Biol. 1170, 29–40. doi: 10.1007/978-1-4939-0888-2_2
Biederbick, A., Kern, H. F., and Elsässer, H. P. (1995). Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur. J. Cell Biol. 66, 3–14.
Bieler, J., Cannavo, R., Gustafson, K., Gobet, C., Gatfield, D., and Naef, F. (2014). Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 10:739. doi: 10.15252/msb.20145218
Bjarnason, G. A., and Jordan, R. (2000). Circadian variation of cell cycle and cell cycle protein expression in man: clinical implications. Prog. Cell Cycle Res. 4, 193–206. doi: 10.1007/978-1-4615-4253-7_17
Blakeman, V., Williams, J. L., Meng, Q. J., and Streuli, C. H. (2016). Circadian clocks and breast cancer. Breast Cancer Res. 18:89. doi: 10.1186/s13058-016-0743-z
Borgs, L., Beukelaers, P., Vandenbosch, R., Belachew, S., Nguyen, L., and Malgrange, B. (2009). Cell circadian cycle: new role for mammalian core clock genes. Cell Cycle 8, 832–837. doi: 10.4161/cc.8.6.7869
Cadenas, C., van de Sandt, L., Edlund, K., Lohr, M., Hellwig, B., Marchan, R., et al. (2014). Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle 13, 3282–3291. doi: 10.4161/15384101.2014.954454
Cao, Q., Gery, S., Dashti, A., Yin, D., Zhou, Y., Gu, J., et al. (2009). A role for the clock gene per1 in prostate cancer. Cancer Res. 69, 7619–7625. doi: 10.1158/0008-5472.CAN-08-4199
Cermakian, N., Monaco, L., Pando, M. P., Dierich, A., and Sassone-Corsi, P. (2001). Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the period1 gene. EMBO J. 20, 3967–3974. doi: 10.1093/emboj/20.15.3967
Delaunay, F., and Laudet, V. (2002). Circadian clock and microarrays: mammalian genome gets rhythm. Trends Genet. 18, 595–597. doi: 10.1016/s0168-9525(02)02794-4
Destici, E., Oklejewicz, M., Saito, S., and van der Horst, G. T. (2011). Mammalian cryptochromes impinge on cell cycle progression in a circadian clock-independent manner. Cell Cycle 10, 3788–3797. doi: 10.4161/cc.10.21.17974
Duffield, G. E. (2003). DNA microarray analyses of circadian timing: the genomic basis of biological time. J. Neuroendocrinol. 15, 991–1002. doi: 10.1046/j.1365-2826.2003.01082.x
Dunlap, J. C. (1999). Molecular bases for circadian clocks. Cell 96, 271–290. doi: 10.1016/s0092-8674(00)80566-8
EI Cheikh, R., Bernard, S., and Ei Khatib, N. (2014). Modeling circadian clock-cell cycle interaction effects on cell population growth rates. J. Theor. Biol. 363, 318–331. doi: 10.1016/j.jtbi.2014.08.008
Feillet, C., Krusche, P., Tamanini, F., Janssens, R. C., Downey, M. J., Martin, P., et al. (2014). Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc. Natl. Acad. Sci. U.S.A. 111, 9828–9833. doi: 10.1073/pnas.1320474111
Filipski, E., Delaunay, F., King, V. M., Wu, M. W., Claustrat, B., Gréchez-Cassiau, A., et al. (2004). Effects of chronic jet lag on tumor progression in mice. Cancer Res. 64, 7879–7885. doi: 10.1158/0008-5472.can-04-0674
Filipski, E., King, V. M., Li, X., Granda, T. G., Mormont, M. C., Liu, X., et al. (2002). Host circadian clock as a control point in tumor progression. J. Natl. Cancer Inst. 94, 690–697. doi: 10.1093/jnci/94.9.690
Folmes, C. D., Dzeja, P. P., Nelson, T. J., and Terzic, A. (2012). Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11, 596–606. doi: 10.1016/j.stem.2012.10.002
Fu, L., and Kettner, N. M. (2013). The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 119, 221–282. doi: 10.1016/B978-0-12-396971-2.00009-9
Fu, L., Pelicano, H., Liu, J., Huang, P., and Lee, C. (2002). The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111, 41–50. doi: 10.1016/s0092-8674(02)00961-3
Fu, X.-J., Li, H.-X., Yang, K., Chen, D., and Tang, H. (2016). The important tumor suppressor role of PER1 in regulating the cyclin-CDK-CKI network in SCC15 human oral squamous cell carcinoma cells. Onco Targets Ther. 9, 2237–2245. doi: 10.2147/OTT.S100952
Garcia, M. N., Barbeito, C. G., Andrini, L. A., and Badrán, A. F. (2001). Circadian rhythm of DNA synthesis and mitotic activity in tongue keratinocytes. Cell Biol. Int. 25, 179–183. doi: 10.1006/cbir.2000.0585
Gery, S., Gombart, A. F., Yi, W. S., Koeffler, C., Hofmann, W. K., and Koeffler, H. P. (2005). Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood 106, 2827–2836. doi: 10.1182/blood-2005-01-0358
Gery, S., Komatsu, N., Baldjyan, L., Yu, A., Koo, D., and Koeffler, H. P. (2006). The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 22, 375–382. doi: 10.1016/j.molcel.2006.03.038
Gery, S., Komatsu, N., Kawamata, N., Miller, C. W., Desmond, J., Virk, R. K., et al. (2007). Epigenetic silencing of the candidate tumor suppressor gene Per1 in non-small cell lung cancer. Clin. Cancer Res. 13, 1399–1404. doi: 10.1158/1078-0432.ccr-06-1730
Herrera-Merchan, A., Cerrato, C., Luengo, G., Dominguez, O., Piris, M. A., Serrano, M., et al. (2010). miR-33-mediated downregulation of p53 controls hematopoietic stem cell self-renewal. Cell Cycle 9, 3277–3285. doi: 10.4161/cc.9.16.12598
Hua, H., Wang, Y., Wan, C., Liu, Y., Zhu, B., Wang, X., et al. (2007). Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 14, 815–818. doi: 10.1038/sj.cgt.7701061
Hua, H., Wang, Y., Wan, C., Liu, Y., Zhu, B., Yang, C., et al. (2006). Circadian gene mPer2, overexpression induces cancer cell apoptosis. Cancer Sci. 97, 589–596. doi: 10.1111/j.1349-7006.2006.00225.x
Ji, M. M., Liu, A. Q., Gan, L. P., Xing, R., Wang, H., Sima, Y. H., et al. (2013). Functional analysis of 30K proteins during silk gland degeneration by a caspase-dependent pathway in Bombyx. Insect Mol. Biol. 22, 273–283. doi: 10.1111/imb.12019
Ko, C. H., and Takahashi, J. S. (2006). Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15, R271–R277.
Lamia, K. A., Storch, K. F., and Weitz, C. J. (2008). Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177. doi: 10.1073/pnas.0806717105
Lee, C. C. (2006). Tumor suppression by the mammalian period genes. Cancer Causes Control. 17, 525–530. doi: 10.1007/s10552-005-9003-8
Li, K. L., Zhang, Y. H., Xing, R., Zhou, Y. F., Chen, X. D., Wang, H., et al. (2017). Different toxicity of cadmium telluride, silicon, and carbon nanomaterials against hemocytes in silkworm, Bombyx mori. RSC Adv. 7, 50317–50327. doi: 10.1039/c7ra09622d
Liu, T., Xing, R., Zhou, Y. F., Zhang, J., Su, Y. Y., Zhang, K. Q., et al. (2014). Hematopoiesis toxicity induced by CdTe quantum dots determined in an invertebrate model organism. Biomaterials 35, 2942–2951. doi: 10.1016/j.biomaterials.2013.12.007
Lu, C., Yang, Y., Zhao, R., Hua, B., Xu, C., Yan, Z., et al. (2016). Role of circadian gene clock during differentiation of mouse pluripotent stem cells. Protein Cell. 7, 820–832. doi: 10.1007/s13238-016-0319-9
Matsuo, T., Yamaguchi, S., Mitsui, S., Emi, A., Shimoda, F., and Okamura, H. (2003). Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259. doi: 10.1126/science.1086271
Ning, H., Albersen, M., Lin, G., Lue, T. F., and Lin, C. S. (2013). Effects of EdU labeling on mesenchymal stem cells. Cytotherapy 15, 57–63. doi: 10.1016/j.jcyt.2012.10.010
Nishitani, H., Taraviras, S., Lygerou, Z., and Nishimoto, T. (2001). The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 276, 44905–44911. doi: 10.1074/jbc.M105406200
Reppert, S. M., and Weaver, D. R. (2001). Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol. 63, 647–676.
Rosbash, M., and Takahashi, J. S. (2002). Circadian rhythms: the cancer connection. Nature 420, 373–374.
Salic, A., and Mitchison, T. J. (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 2415–2420. doi: 10.1073/pnas.0712168105
Sato, F., Nagata, C., Liu, Y., Suzuki, T., Kondo, J., Morohashi, S., et al. (2009). PERIOD1 is an anti-apoptotic factor in human pancreatic and hepatic cancer cells. J. Biochem. 146, 833–838. doi: 10.1093/jb/mvp126
Sato, F., Wu, Y., Bhawal, U. K., Liu, Y., Imaizumi, T., Morohashi, S., et al. (2011). PERIOD1 (PER1) has anti-apoptotic effects, and PER3 has pro-apoptotic effects during cisplatin (CDDP) treatment in human gingival cancer CA9-22 cells. Eur. J. Cancer 47, 1747–1758. doi: 10.1016/j.ejca.2011.02.025
Sato, T. K., Panda, S., Kay, S. A., and Hogenesch, J. B. (2003). DNA arrays: applications and implications for circadian biology. J. Biol. Rhythms 18, 96–105. doi: 10.1177/0748730403252245
Scheving, L. E., Tsai, T. H., and Scheving, L. A. (1983). Chronobiology of the intestinal tract of the mouse. Am. J. Anat. 168, 433–465. doi: 10.1002/aja.1001680405
Shearman, L. P., Jin, X., Lee, C., Reppert, S. M., and Weaver, D. R. (2000). Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol. Cell. Biol. 20, 6269–6275. doi: 10.1128/mcb.20.17.6269-6275.2000
Shostak, A. (2017). Circadian clock, cell division, and cancer: from molecules to organism. Int. J. Mol. Sci. 18, E873. doi: 10.3390/ijms18040873
Shostak, A., Ruppert, B., Ha, N., Bruns, P., Toprak, U. H., Seq Project, M. M. M. L. -, et al. (2016). MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat. Commun. 7:11807. doi: 10.1038/ncomms11807
Smaaland, R., Sothern, R. B., Laerum, O. D., and Abrahamsen, J. F. (2002). Rhythms in human bone marrow and blood cells. Chronobiol. Int. 19, 101–127. doi: 10.1081/cbi-120002594
Soták, M., Sumová, A., and Pácha, J. (2014). Cross-talk between the circadian clock and the cell cycle in cancer. Ann. Med. 46, 221–232. doi: 10.3109/07853890.2014.892296
Suchyta, M., Miotto, B., and McGarry, T. J. (2015). An inactive geminin mutant that binds cdt1. Genes 6, 252–266. doi: 10.3390/genes6020252
Takahashi, J. S., Hong, H. K., Ko, C. H., and McDearmon, E. L. (2008). The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9, 764–775. doi: 10.1038/nrg2430
Tao, H., Li, X., Qiu, J. F., Cui, W. Z., Sima, Y. H., and Xu, S. Q. (2017). Inhibition of expression of the circadian clock gene Period causes metabolic abnormalities including repression of glycometabolism in Bombyx mori cells. Sci. Rep. 7:46258. doi: 10.1038/srep46258
Wood, P. A., Yang, X., Taber, A., Oh, E. Y., Ansell, C., Ayers, S. E., et al. (2008). Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol. Cancer Res. 6, 1786–1793. doi: 10.1158/1541-7786.MCR-08-0196
Yan, S. Q., Xing, R., Zhou, Y. F., Li, K. L., Su, Y. Y., Qiu, J. F., et al. (2016). Reproductive toxicity and gender differences induced by cadmium telluride quantum dots in an invertebrate model organism. Sci. Rep. 6:34182. doi: 10.1038/srep34182
Yang, W. S., and Stockwell, B. R. (2008). Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 9:R92. doi: 10.1186/gb-2008-9-6-r92
Yang, X., Wood, P. A., Ansell, C. M., Quiton, D. F., Oh, E. Y., Du-Quiton, J., et al. (2009). The circadian clock gene Per1 suppresses cancer cell cycle and tumor growth at specific times of day. Chronobiol. Int. 26, 1323–1339. doi: 10.3109/07420520903431301
Ye, R., Selby, C. P., Chiou, Y. Y., Ozkan-Dagliyan, I., Gaddameedhi, S., and Sancar, A. (2014). Dual modes of CLOCK:BMAL1 inhibition mediated by Cryptochrome and period proteins in the mammalian circadian clock. Genes Dev. 28, 1989–1998. doi: 10.1101/gad.249417.114
Zhang, J., Nuebel, E., Daley, G. Q., Koehler, C. M., and Teitell, M. A. (2012). Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 11, 589–595. doi: 10.1016/j.stem.2012.10.005
Zhao, Q., Zheng, G., Yang, K., Ao, Y. R., Su, X. L., Li, Y., et al. (2016). The clock gene PER1 plays an important role in regulating the clock gene network in human oral squamous cell carcinoma cells. Oncotarget 7, 70290–70302. doi: 10.18632/oncotarget.11844
Zhao, Y., Zhang, Y., Zhou, M., Wang, S., Hua, Z., and Zhang, J. (2012). Loss of mPer2 increases plasma insulin levels by enhanced glucose-stimulated insulin secretion and impaired insulin clearance in mice. FEBS Lett. 586, 1306–1311. doi: 10.1016/j.febslet.2012.03.034
Zheng, B., Albrecht, U., Kaasik, K., Sage, M., Lu, W., Vaishnav, S., et al. (2001). Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694. doi: 10.1016/s0092-8674(01)00380-4
Keywords: Period gene, cell cycle, apoptosis, autophagy, ovarian cell line, Bombyx mori
Citation: Qiu J-F, Li X, Cui W-Z, Liu X-F, Tao H, Yang K, Dai T-M, Sima Y-H and Xu S-Q (2019) Inhibition of Period Gene Expression Causes Repression of Cell Cycle Progression and Cell Growth in the Bombyx mori Cells. Front. Physiol. 10:537. doi: 10.3389/fphys.2019.00537
Received: 26 January 2019; Accepted: 15 April 2019;
Published: 03 May 2019.
Edited by:
Elzbieta M. Pyza, Jagiellonian University, PolandReviewed by:
Kenji Tomioka, Okayama University, JapanGiorgio F. Gilestro, Imperial College London, United Kingdom
Copyright © 2019 Qiu, Li, Cui, Liu, Tao, Yang, Dai, Sima and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shi-Qing Xu, szsqxu@suda.edu.cn
†These authors have contributed equally to this work
 Jian-Feng Qiu
Jian-Feng Qiu Xue Li
Xue Li Wen-Zhao Cui
Wen-Zhao Cui Xiao-Fei Liu
Xiao-Fei Liu Hui Tao
Hui Tao Kun Yang
Kun Yang Tai-Ming Dai
Tai-Ming Dai Yang-Hu Sima
Yang-Hu Sima Shi-Qing Xu
Shi-Qing Xu