- 1Government Department of School Education, Jammu, India
- 2Vatsalya Clinic, New Delhi, India
The influence of nutrition and environment on human health has been known for ages. Phytonutrients (7,000 flavonoids and phenolic compounds; 600 carotenoids) and pro-health nutrients—nutraceuticals positively add to human health and may prevent disorders such as cancer, diabetes, obesity, cardiovascular diseases, and dementia. Plant-derived bioactive metabolites have acquired an imperative function in human diet and nutrition. Natural phytochemicals affect genome expression (nutrigenomics and transcriptomics) and signaling pathways and act as epigenetic modulators of the epigenome (nutri epigenomics). Transcriptomics, proteomics, epigenomics, miRNomics, and metabolomics are some of the main platforms of complete omics analyses, finding use in functional food and nutraceuticals. Now the recent advancement in the integrated omics approach, which is an amalgamation of multiple omics platforms, is practiced comprehensively to comprehend food functionality in food science.
Introduction
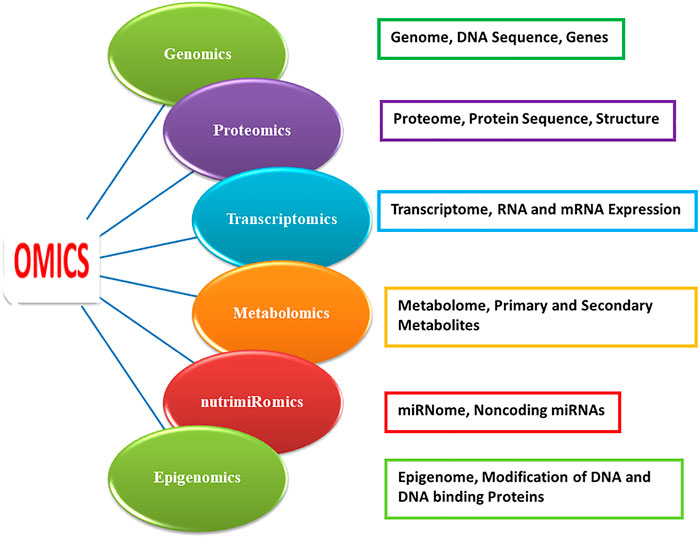
The term “nutraceuticals” was coined by Dr. Stephen (1989). Nutraceuticals comprise “any nontoxic food extract supplement that has scientifically proven health benefits for both disease treatment and prevention” (DeFelice, 1995). Nutraceuticals combines the two disciplines of nutrition and pharmaceuticals (Figure 1). But, after the act of the dietary supplement health and education (1994) came into force, the designation of nutraceuticals stretched to include minerals, vitamins, herbs, extra-botanicals, amino acids, and dietary substances for usage as a diet supplement by humans (Stauffer, 1999). Instead of the nutraceutical term, the term dietary supplement is well-accepted globally and in the regulatory systems. About 470 nutraceutical and functional food products were accessible in 1999 with recognized fitness profits (Brower, 1999). Functional food or medicinal foods is a name entitled for edible foodstuffs with improved content of bioactive compounds by breeding, ecological influences, or genomic engineering and fortified foods that deliver health benefits, besides elementary nutrition. The term “functional food” was coined in Japan (1980) (Arihara 2014). Roberfroid (1999) defined functional foods as “products that have a relevant effect on well-being and health or result in reducing the risk of diseases.” However, an internationally accepted definition of functional food and nutraceuticals is not available (Arihara 2014; Télessy et al., 2019). Functional food is mainly referred to as “processed foods having disease-preventing and/or health-promoting benefits in addition to their nutritive value” (Arihara 2014). Functional foods cover nutraceuticals, pharma foods, probiotics, designer foods, medical foods, and vita foods (Arihara 2014). “Nutraceutical” is a food or food part that provides health benefits and encompasses disease prevention and treatment. Products as diverse as isolated nutrients, dietary supplements, and diets to “designer” foods, synthetic products, herbal products, and processed foods fall under the umbrella of nutraceuticals. Nutraceuticals are beyond food but subordinate to pharmaceuticals (Dudeja and Gupta 2017; Mohanty and Singhal, 2018; Télessy et al., 2019). Functional foods and nutraceuticals (FFNs) possess numerous health benefits. Besides essential nutrients, for example, fatty acids, carbohydrates, proteins, vitamins, and minerals, non-essential nutritional and bioactive food components such as folates, phenolics, polyamines, flavonoids, anthocyanin and nonflavonoid condensed tannins (ellagitannins), and carotenoids modulate various cellular processes (Abbasi et al., 2015; Kumar et al., 2018; Islam et al., 2021; Nayak et al., 2021). FFNs provide protection against disorders, for instance, cancer, obesity, high blood pressure, cardiovascular diseases (CVDs), gastrointestinal tract disorder, type II diabetes, inflammation, microbial, viral and parasitic infections, psychotic diseases, spasmodic disorders, ulcers, etc. (Abbasi et al., 2015; Kumar et al., 2018; Hamid and Hamid, 2019; Islam et al., 2021; Nayak et al., 2021; Salunkhe et al., 1983). Different omics platforms (Figure 1) will help in deepening our knowledge of food–body interactions, the influence of functional foods on consumption and the mechanism of their actions, safety issues, improvement of the nutritional value of staple foods, efficient use of nutrients, and characterization and development of new functional foods from traditional medicines and nutritional security (Kato et al., 2011; Abbott, 2014; Tian et al., 2016; Pazhamala et al., 2021). This review is an elaborate and informative type of review that describes the current status and efficient progress of various omics technologies in the arena of nutraceutical and functional foods. This review which discusses compiled data of multi-omics studies in nutraceutical and functional foods will enlighten our understanding of the potential of efficient foods and nutraceuticals for the betterment of human health.
Genomics
Earlier, nutrition or FFN investigations were piloted principally for reviewing the significance of functional food or nutrient by its deficiency expressed as health-linked complications. Intermingled concepts of diet and health bonds were strongly recognized, though, only after the wealth of genomic information in form of three billion bases in every human cell was produced by Human Genome Project (HGP) in the year 2003, the nutritional research area developed to emphasize on the direct communication between nutrient and human genome (Venter et al., 2001; Austin, 2004).Single-nucleotide polymorphisms (SNPs) within human populations revealed variance in rejoinder to dietary nutrients (Subbiah, 2007). Therefore, nutritional genomics has developed due to the revolution of human genomics. Nutritional genomics consists of nutrigenetics and nutrigenomics:
Nutrigenetics
Nutrigenetics estimates regulation of gene nutrition by presenting exactly how SNPs within persons influence their responses to nutritional constituents.
Nutrigenomics
Alternatively, nutrigenomics scrutinizes nutrition and regulation of gene expression by indicating how dietary nutrients interconnect with human genomic sequences and change their gene expression and gene transcripts. It provides authorization for nutrition–gene regulation owing to advanced omics approaches (Kato, 2008). Nutrigenomics also denoted as Nutri-Omics or nutritional genomics is a sub-discipline of food and nutrition research via the application of various highly effective high-throughput genomics (McGuire et al., 2020; Müller and Kersten, 2003) and omics-based approaches including transcriptomics, proteomics, epigenomics, nutrimiRomics, metabolomics, and bioinformatics (Pazhamala et al., 2021) (Figure 1). Cominetti et al. (2017) referred to nutrigenomics as the investigation of communications between food nutrient ingredients and human genome and evaluation of genomic expression and metabolic functions after nourishing. The prompt progression of technologies, for example, high-throughput sequencing of the genome (DNA) and transcriptome (RNA), proteome sequencing by mass spectrometry (MS), and upgraded outcome of DNA microarrays has contributed greatly to expanded applicability of omics platforms in the area of food science and directed field of nutri-omics to the right direction (Fu et al., 2010; Gehlenborg et al., 2010). Today different nutri-omics platforms are prerequisites to gain deeper insights into and knowledge of the influence of dietary food components on humans along with the mechanism of their actions. The overall goal of nutrigenomics appreciates in what manner nutrition impacts cell metabolic pathways and homeostatic regulation. Furthermore, how this regulation interrupts the initial stage of a lifestyle diet–linked ailment depends on the genotypes of the individual (Müller and Kersten, 2003). The bond between nutrition and human health is essentially affected by the interaction between the nutrient and genes. The functional sensitivity of genes to nutrients delivers the central foundation of nutrigenomics. Both while in the uterus of the pregnant mother and through the primary years of life, under and over-nourished mother–child components imprint gene modifications causing chronic metabolic complications in the future life of humans (Chávez and Muñoz de Chávez, 2003).
Transcriptomics
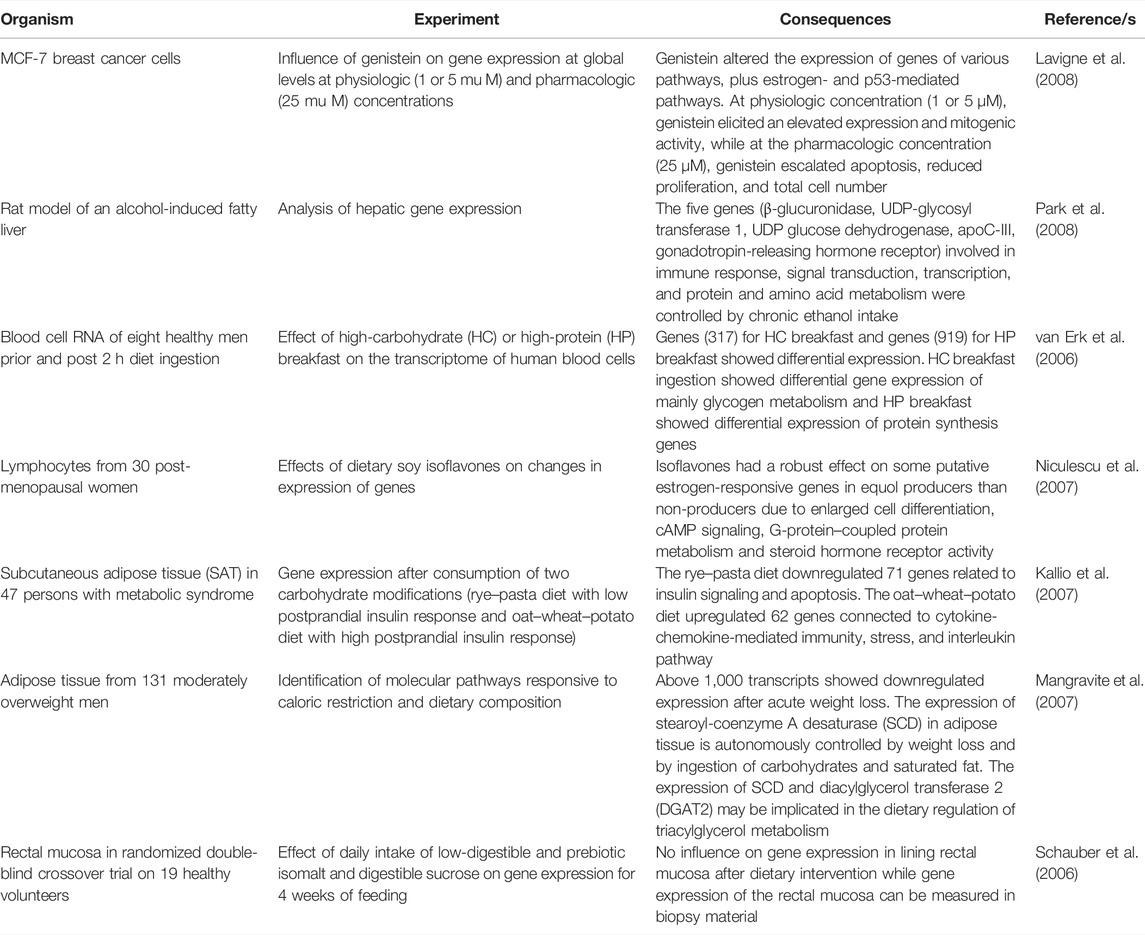
Among all omics platforms, transcriptomics appears to be the utmost efficacious and extensively used technology in the field of nutrigenomic studies due to its efficiency and high data representation (Kato, 2008). Transcriptome includes the whole mRNA or transcript complements transcribed or expressed from genes. So, transcriptomics is a potent platform for expression profiling of various genes in a genome (Lowe et al., 2017). Diverse bioactive constituents of food can affect the gene expression in primary to changed cellular biological processes along with cell metabolism, cell proliferation, and tissue differentiation, and the death of the cell and their imbalance may cause syndromes such as diabetes or cancer. Owing to that, gene expression interrogation at the genome level influenced by dietary functional nutrients is predominantly significant in nutrigenomics studies. Transcriptomics for investigating the influence of food components on the expression of genes has been adopted in diverse investigational paradigms together with animal cell cultures, animal models, and human beings. With transcriptomics technology, clinical trials on obese humans with dietary intervention have been carried out to check the influence of energy-restricted diets on the expression of genes present in adipose tissue (ClÉment et al., 2004; Dahlman et al., 2005). The human transcriptomic study assessed duodenal mucosa for the gene expression profiles of Lactobacillus G Gon (DiCaro et al., 2005). The transcriptomic investigations for understanding the variations in global gene expression due to various dietary interventions, for example, deficiency of nutrients, fasting, ingestion of disproportionate nutrients, and specific food factors have been performed (Endo et al., 2002; Matsuzaki et al., 2005; Kamei et al., 2010; Nakai et al., 2008; Saito et al., 2010; Ohta et al., 2006; Tachibana et al., 2005). The transcriptomic investigation of rat liver that was subjected to mild caloric restriction (5–30 percent lesser amount of food for 1 week or 1 month) (Saito et al., 2010) was carried out to examine food functionality by discriminating direct consequences of food nutrients and secondary effects produced by alterations in food ingestion behavior. The cyp4a14 gene expression revealed restriction level–dependent changes, so it can be used as a biomarker for the beneficial effects of functional food nutrients on energy metabolism. Microarray technology allows simultaneous quantification of thousands of mRNA before and after exposure to bioactive dietary components (Kato, 2008; Garosi et al., 2005). The DNA microarray tool has been used in in vivo conditions for documentation of cellular responses to food components and their cellular targets at molecular levels in various investigations, for instance, green tea catechins (McLoughlin et al., 2004; Vittal et al., 2004), vitamin D and vitamin E (Johnson and Manor, 2004; Lin et al., 2002), polyunsaturated fatty acids (Lapillonne et al., 2004; Kitajka et al., 2004; Narayanan et al., 2003), soy isoflavones (Herzog et al., 2004), quercetin (Murtaza et al., 2006), anthocyanins (Tsuda et al., 2006), arginine (Leong et al., 2006) and hypoallergenic wheat flour (Narasaka et al., 2006). Some other transcriptome expression studies involving the use of microarray are listed in Table 1. Apart from expression studies, transcriptomics has also been practiced to evaluate the safety of food (Kato, 2008). The swift accretion of nutri-transcriptomic microarray data stimulated the formation of an integrated open-source web-based database which guarantees efficient organization, storage, and investigation of the huge volume of microarray data produced from all nutri-transcriptomic research studies (Saito et al., 2005).
Proteomics
Proteomics involves high-throughput analysis of proteomes in cells, tissues, or biological fluids which are being expressed by the genome (Thongboonkerd, 2007; Husi and Albalat, 2014; Hixson et al., 2017) and also facilitates novel protein discovery. The proteome is the complete complement of proteins expressed from a set of specific genes in any biological organism at a given point of time and specific environment (Trayhurn, 2000; Hixson et al., 2017). The proteome is dynamic, continuously changes in line with cell type and cell functional state, and extra complex than the genome (Thongboonkerd, 2007; Hixson et al., 2017). From the genome of humans, nearly 25,000 functional genes are encoded, whereas the proteome includes about 250,000 proteins because of alternative splicing and posttranslational modifications (Kussmann and Affolter, 2006). The multifaceted proteome can be evaluated by proteomics tools. Proteomics covers protein investigation by protein separation, protein quantification, and protein identification (Kussmann et al., 2008) and has promptly proceeded from gel-based techniques such as 2-DE (two-dimensional) gel electrophoresis to technologies such as mass spectrometry (MS) by evaporation of peptides and proteins by MALDI (matrix-assisted laser desorption/ionization) and ESI (electrospray ionization), multiple reaction monitoring, and multiplexed immunoassays (Swatton et al., 2004; de Roos and McArdle, 2008; Zhang et al., 2014). Nutritional proteomics provides knowledge about the complex interaction of nutrition-protein regulation, identification of new biomarkers for dietary status, and develops novel stratagems for diet-related avoidance and interference of ailments (Ovesná et al., 2008). Nutritional proteomics, which is an essential fragment of nutrigenomics, studies the influences of functional food nutrients on the expression of proteins and delivers prospectives for differentiating biomarkers that show sensitivity to dietary interventions (Fuchs et al., 2005a). Limited nutritional proteomics analysis in humans involved cell culture (colon cancer cell and endothelial cell) studies which confirmed the effects of food components (butyrate, flavonoid, and genistein) on protein profiles and identified the cellular molecular target proteins of components of the diet (Tan et al., 2002; Herzog et al., 2004; Wenzel et al., 2004; Fuchs et al., 2005b; Fuchs et al. 2005c; Fuchs et al. 2005d). Exponentially cumulative publications on proteomics in nutritional research showed its prospective (de Roos et al., 2005; Breikers et al., 2006; Griffiths and Grant, 2006; Kim et al., 2006). Proteomics of liver tissues of two mice strains that were susceptible and resistant to atherosclerosis led to the identification of 30 differentially expressed proteins of oxidative stress and lipid metabolism which were significantly altered in response to an atherogenic diet proposing that the identified proteins add to variances in atherogenesis susceptibility (Park et al., 2004). With the protein microarray, the cholesterol diet–induced expression of proteins revealed an altered pattern of phosphorylation (Puskas et al., 2006). The 2D gel and MALDI-TOF-MS proteomic analysis techniques identified and characterized novel prospective allergens from transgenic soybean and nontransgenic soybean (Batista et al., 2007), allergens from wheat flour, and it was discovered that the nine glutenin subunits are utmost principal IgE-binding antigens (Akagawa et al., 2007) and 15 host defense proteins were identified (Smolenski et al., 2007). After intervention with flaxseed diet in seven humans, the PBMC—peripheral blood mononuclear cell proteome showed significant influence on steady-state levels of sixteen proteins and improved peroxiredoxin and decreased long-chain fatty acid beta-oxidation multienzyme complex and glycoprotein IIIa/II levels (Fuchs et al., 2007). There is a public 2-DE database for proteome of human PBMCs (peripheral blood mononuclear cells), with the potential to examine the proteomics alterations concomitant to interventions of diet or drugs (Vergara et al., 2008). Comparative proteomics between rat livers subjected to 30% food restriction and control rats led to identification of nine proteins showing upregulation and nine proteins with downregulation. The 10% food restriction caused nine upregulated and two downregulated proteins with the 2-DE technique. The prohibitin which regulates longevity was also upregulated (Artal-Sanz and Tavernarakis, 2009) and can prove to be a highly promising and efficient proteomic biomarker for positive results of food components (Kussmann et al., 2010a). In the food and nutrition proteomics research approach, the influence of trivial caloric restriction was also investigated (Takahashi et al., 2011). The rats were fed on a GSE—grape seed extract–supplemented diet. Their homogenates of brains when subjected to proteomics investigation lead to the identification of thirteen candidate proteins (Deshane et al., 2004). Some of these proteins were under regulation by supplementation of grape seed extract in opposed order from earlier studies for the identified proteins in mouse models of Alzheimer’s neurodegeneration syndrome signifying their modulation by GSE as a neuroprotective agent (Deshane et al., 2004). With mass spectrometry (MALDI-TOF MS) serum protein B-chain of α2-HS glycoprotein diet–related biomarkers with a role in insulin resistance and immune function were isolated from human subjects fed on a cruciferous-supplemented diet in contrast to control human subjects (Mitchell et al., 2005). Mingling of transcriptomics with proteomics leads to the identification of enzymes and transporters involved in fatty acid metabolism, sequestration, and transcriptional regulation in zinc-deficient models. The hepatic lipid accretion pathways were designed (tom Dieck et al., 2005).
Metabolomics
Nutritional metabolomics is a unique modern omics know-how in research (Rochfort, 2005) involving the investigation of global metabolite profiles in any organism in precise ecological scenarios. Metabolomics provides the perception of biochemical deviations after the intervention of diet and impacts safety issues of GMO—genetically modified food (Dixon et al., 2006). The metabolome comprises a complete set of metabolites biosynthesized in any biological organism, whereas the metabolites are final products of biological reactions of metabolism, signifying the genome–environ interaction (Rochfort, 2005). Beyond 10,000 categories of key metabolites being existent in animals, the figure for proteins is assumed to drive beyond 100,000. This metabolite feature probably results in extra-broad features of metabolomics compared to proteomics. Metabolite analysis is challenging and frequently needs the usage of high-level techniques owing to much greater diversity and abundance of metabolites than mRNA transcripts and proteins. In spite of these technical hitches, metabolomics is a potent technique in neutraceuticals and food nutrition (Oresic, 2009; Zivkovic and German, 2009) and uses NMR—nuclear magnetic resonance, MS—mass spectrometry (gas chromatography (GC)-MS and liquid chromatography (LC)-MS and capillary electrophoresis (CE)-MS (Prakash et al., 2018; Kumar et al., 2019). NMR and MS in conjunction with potent bioinformatics podiums significantly boost metabolomics in the investigation of nutrition (German et al., 2003; Trujillo et al., 2006; Hall et al., 2008). The nutrition metabolomics study performed in five fit premenopausal women under controlled state of affairs determined the biochemical changes subsequent to clear intervention of diet with soy isoflavones and showed that soy-induced isoflavones brought modifications in plasma components such as plasma lipoprotein, amino acid, and carbohydrates with a role in energy metabolism (Solanky et al., 2003). Other examples include deducing the metabolic consequences of vitamin E supplements in in vitro conditions in a mouse model of motor neuro-degeneration, evaluation of human biological reaction to various diets, for example., chamomile tea or vegan, low- and high-meat diets, the characterization of the variations in metabolic profile attributable to dissimilar populations such as Chinese, American, and Japanese or Swedish and British inhabitants (Dumas et al., 2006; Griffin et al., 2002; Lenz et al., 2004; Wang et al., 2005; Stella et al., 2006). Metabolite profiles of chamomile tea consumption in humans estimated with chemometric methods and 1HNMR spectroscopy exhibited a high degree of variation with clear discrepancy in excretion of urine, signifying augmented hippurate and glycine and reduced creatinine levels (Wang et al., 2005). The influence of green tea ingestion in humans showed a robust upsurge in intermediates of citric acid cycle urinary metabolites suggestive of the influence of flavanols present in green tea on oxidative energy metabolism (van Dorsten et al., 2006). The metabolic response of rats fed with whole-grain flour and refined wheat flour showed that some intermediates of the TCA(tri carboxylic acid) cycle, aromatic amino acids, and hippurate significantly escalated in the urine of rats nourished on whole-grain flour (Fardet et al., 2007). The proton NMR-based metabolomics approach investigated and identified the overall biochemical consequences of short-term high consumption of animal milk protein or meat protein on 8-year-old boys and found that milk diet augmented excretion of hippurate and marginally altered serum lipid metabolic profile, while the meat diet amplified excretion of histidine, creatine, and urea in urine with no influence on the serum profile (Bertram et al., 2007). Nutimetabolomics contributed to the revolution of omics. Human food constituent–induced changes in nutritional metabolic profiles are not well-understood, so the global picture of human metabolism is not imaginable. Technological advancements can overcome future challenges of using metabolomics in nutrition research (Ovesná et al., 2008).
Epigenomics
Epigenomics is the investigation of the epigenome–plastic variations at the level of epigenetics, for example, cytosine DNA methylation and histone modifications devoid of variations in DNA nucleotide sequence and small RNA–mediated methylation (Kussmann et al., 2010b; Yadav et al., 2018; Lloyd and Lister, 2022). Epigenetics modifications modulate gene expression at respective locations (Yadav et al., 2018). The pregnant mother’s nutrition during the development of the fetus influences the acquired predisposition to lifestyle-linked disorders such as hypertension, obesity, diabetes, cardiovascular diseases, etc in future (Lusis et al., 2008) on the basis of epigenetic alterations. NGS—next-generation sequencing and genome-wide analysis technologies may throw light on the epigenomic modification in nutritional omics (Hawkins et al., 2010). Effect of nutrition on initial life stages, for example, fetal, suckling, and growing age on health in advanced stages of life may be allocated by chromatin modifications in future.
Nutri miRomics
A new emergent area of omics platforms in functional foods is associated with RNA transcripts not translating into proteins. MicroRNAs (miRNAs) are small, single-stranded, endogenously encoded, 18–25 nucleotide conserved, non-coding RNAs that regulate expression of the protein-coding genes by PTGS—posttranscriptional gene silencing (Pandita, 2018a) and possess roles in nutrient homeostasis, hormone homeostasis, signaling pathways, immune response, human disorders, etc. (Pandita, 2018b; Pandita and Wani, 2019; Pandita 2021). A non-coding gene previously referred to as “JUNK” under the spotlight as “just unrevealed new-fangled know-how” of future treasure veiled in genomes that produce functional RNA molecules for internal signals and hold potential in medicine at personalized levels (Pandita and Pandita, 2016). Since the accruing confirmations support the significance of miRNA in disease development (Pandita, 2018a) and health maintenance, the know-how on miRNA eminence is, indeed, indispensable for understanding the interaction between functional food constituents and the human body. Nutrient such as phosphate helps in the processes of replication of DNA, development of phospholipid bilayer, and biosynthesis of ATP molecules, whereas the deficiency of P causes upregulation and downregulation of miRNAs. Global microRNA investigation can be performed by high-throughput tools of microRNA sequencing and microRNA microarray (Pandita, 2019). RNAi triggered by short double-stranded small interfering RNA (siRNA) has been used in commercial crops such as cassava plants deprived of cyanogenic linamarin, tomato plants with few allergens, dietary antioxidant–rich fortified tomatoes, Flavr Savr tomato, and ring spot–resistant cultivars of papaya plant (Pandita, 2018b). Nutrimiromics investigates the influence of food on gene expression modifications because of microRNA epigenetics affecting the jeopardy of chronic disease development. Nutrimiromics comprehends the association between food components and the reaction of microRNAs in particular body parts. Nutritional supplements are prospectively prized army to fight against obesity—the imbalance between food consumption and energy outflow (Downs et al., 2005; Lau et al., 2008; Li et al., 2008). Whole-genome screening approaches enabled the identification of coding genes sensitive to nutritional supplements. The hypotheses can be formulated by explaining the mode of action of the supplement under interrogation (Roy et al., 2004; Roy et al., 2007). Expression profiling has provided innumerable new adipose tissue genes for the regulation of obesity. Assimilating expression patterns with genome-wide linkage and association maps can recognize novel obesity susceptible genes (Dahlman and Arner, 2007). MicroRNAs induced during adipogenesis fast-track development of fat cells and show downregulation in obesity. Ectopic expressions of microRNA-103 or microRNA-143 present in preadipocytes speed up adipogenesis, proving that miRNA plays a crucial function in obesity (Xie et al., 2009). Angiogenesis helps in adipose tissue growth and is controlled by miRNAs (Sen et al., 2009). Nutritional supplements against obesity need to check for adipose tissue miRNA regulation for speeding up the catabolism of fat at cellular and tissue levels. Nutri miRomics represents a potent tool and will probably emerge as a key pilot of forthcoming nutritional supplement trade. Resolvins—DHA and EPA metabolites develop due to injury of tissues that back inflammation homeostasis by NF-kB downregulation (Zárate et al., 2017). The microRNA-21, microRNA-146b, microRNA-142 family, microRNA-203, microRNA-208a, microRNA-219, and microRNA-302d show temporal and differential expression in tissue exudates and resolvin D1 biosynthesized in resolution controls microRNA-21, microRNA-146b, microRNA-208a, and microRNA-219 (Recchiuti, et al., 2011). In HepG2 cells, EGCG downregulates microRNA-30b, microRNA -453, microRNA-520-e, microRNA-629, and microRNA-608 which are involved in glycolysis and gluconeogenesis, inflammatory pathways, insulin signaling pathway, oxidative phosphorylation, peroxisome proliferator–activated receptors (PPARs), signaling pathway, and glutathione metabolism (Arola-Arnal and Bladé, 2011). EGCG upregulated the miR-let-7b in melanoma cell lines and metastatic melanoma tumors and suppresses melanoma tumor growth by activation of the intercellular signaling pathway, cAMP/protein kinase A (PKA)/protein phosphatase 2A (PP2A) (Yamada et al., 2016). From the blood of mice nourished on 10 g of rapeseed bee pollen/kg of body weight, 132 plant-derived microRNAs (miR-166a and miR-159a were profuse) were identified (Chen et al., 2016). In the plasma samples of four healthy adult humans who ingested 102 g of broccoli, no variation was detected in broccoli-derived microRNA-824 and microRNA-167a after 4 h of broccoli ingestion (Baier et al., 2014). Table 2 lists some other studies where the principal miRNAs are modulated by the respective nutrient and bioactive compounds.
Concluding Remarks
Plants are the picture-perfect cradle of FFNs owing to the presence of thousands of naturally valuable bioactive metabolites and phytochemical food compounds which can be augmented by genetic improvement. The biotechnological omics tool goals in nutrigenomics research range widely from identification, isolation, and innovation of new functionality, manipulations, revelation of the mode of action and security concerns of specific health-stimulating metabolites and phyto-compounds or their combination for designing and developing FFNs which can prove as a key stratagem in complementary medicine. The ultimatum of omics-based approaches will endure for growth and development in food and nutrition sciences. The extensive investigation of one specific omics approach will expand the understanding of the communication between essential food constituents and the human body. The permutation and unification of various omics data will arrange for supplementary concrete statistics on what takes place within the body in reaction to food consumption. Furthermore, the integration of various omics approaches or the integrated omics may intensify the significance of omics research.
Author Contributions
DP conceptualized the review. DP and AP wrote the review article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbasi A. M., Shah M. H., Khan M. A. (2015). “Phytochemicals and Nutraceuticals,” in Wild Edible Veetables of Lesser Himalayas (Cham: Springer), 31–65. doi:10.1007/978-3-319-09543-1_3
Abbott R. (2014). “Documenting Traditional Medical Knowledge,” in Ryan Abbott, Documenting Traditional Medical Knowledge (Geneva, Switzerland: World Intellectual Property Organization), 1–48.
Akagawa M., Handoyo T., Ishii T., Kumazawa S., Morita N., Suyama K. (2007). Proteomic Analysis of Wheat Flour Allergens. J. Agric. Food Chem. 55 (17), 6863–6870. doi:10.1021/jf070843a
Al-Ansari M. M., Aboussekhra A. (2015). miR-146b-5p Mediates P16-dependent Repression of IL-6 and Suppresses Paracrine Procarcinogenic Effects of Breast Stromal Fibroblasts. Oncotarget 6 (30), 30006–30016. doi:10.18632/oncotarget.4933
Arihara K. (2014). “FUNCTIONAL FOODS,” in Michael Dikeman, Carrick Devine, Encyclopedia of Meat Sciences. 2nd Edn (Cambridge, United States: Academic Press), 32–36. doi:10.1016/B978-0-12-384731-7.00172-0
Arola-Arnal A., Bladé C. (2011). Proanthocyanidins Modulate microRNA Expression in Human HepG2 Cells. PLoS ONE 6 (10), e25982. doi:10.1371/journal.pone.0025982
Artal-Sanz M., Tavernarakis N. (2009). Prohibitin Couples Diapause Signalling to Mitochondrial Metabolism during Ageing in C. elegans. Nature 461 (7265), 793–797. doi:10.1038/nature08466
Austin C. P. (2004). The Impact of the Completed Human Genome Sequence on the Development of Novel Therapeutics for Human Disease. Annu. Rev. Med. 55 (1), 1–13. doi:10.1146/annurev.med.55.091902.104426
Baier S. R., Nguyen C., Xie F., Wood J. R., Zempleni J. (2014). MicroRNAs Are Absorbed in Biologically Meaningful Amounts from Nutritionally Relevant Doses of Cow Milk and Affect Gene Expression in Peripheral Blood Mononuclear Cells, HEK-293 Kidney Cell Cultures, and Mouse Livers. J. Nutr. 144 (10), 1495–1500. doi:10.3945/jn.114.196436
Batista R., Martins I., Jenö P., Ricardo C. P., Oliveira M. M. (2007). A Proteomic Study to Identify Soya Allergens - the Human Response to Transgenic versus Non-transgenic Soya Samples. Int. Arch. Allergy Immunol. 144 (1), 29–38. doi:10.1159/000102611
Bertram H. C., Hoppe C., Petersen B. O., Duus J. Ø., Mølgaard C., Michaelsen K. F. (2007). An NMR-Based Metabonomic Investigation on Effects of Milk and Meat Protein Diets Given to 8-Year-Old Boys. Br. J. Nutr. 97 (4), 758–763. doi:10.1017/s0007114507450322
Bigagli E., Cinci L., Paccosi S., Parenti A., D'Ambrosio M., Luceri C. (2017). Nutritionally Relevant Concentrations of Resveratrol and Hydroxytyrosol Mitigate Oxidative Burst of Human Granulocytes and Monocytes and the Production of Pro-inflammatory Mediators in LPS-Stimulated RAW 264.7 Macrophages. Int. Immunopharmacology 43, 147–155. doi:10.1016/j.intimp.2016.12.012
Boesch-Saadatmandi C., Wagner A. E., Wolffram S., Rimbach G. (2012). Effect of Quercetin on Inflammatory Gene Expression in Mice Liver In Vivo - Role of Redox Factor 1, miRNA-122 and miRNA-125b. Pharmacol. Res. 65 (5), 523–530. doi:10.1016/j.phrs.2012.02.007
Breikers G., van Breda S. G. J., Bouwman F. G., van Herwijnen M. H. M., Renes J., Mariman E. C. M., et al. (2006). Potential Protein Markers for Nutritional Health Effects on Colorectal Cancer in the Mouse as Revealed by Proteomics Analysis. Proteomics 6 (9), 2844–2852. doi:10.1002/pmic.200500067
Brower V. (1999). Nutraceuticals: Poised for a Healthy Slice of the Healthcare Market? Nat. Biotechnol. 16, 728–731. doi:10.1038/nbt0898-728
Chávez A., Muñoz de Chávez M. (2003). Nutrigenomics in Public Health Nutrition: Short-Term Perspectives. Eur. J. Clin. Nutr. 57 (Suppl. 1), S97–S100. doi:10.1038/sj.ejcn.1601809
Chen X., Dai G. H., Ren Z. M., Tong Y. L., Yang F., Zhu Y. Q. (2016). Identification of Dietetically Absorbed Rapeseed (Brassica Campestris L.) Bee Pollen microRNAs in Serum of Mice. Biomed. Res. Int. 2016 1–5. doi:10.1155/2016/5413849
ClÉment K., Viguerie N., Poitou C., Carette C., Pelloux V., Curat C. A., et al. (2004). Weight Loss Regulates Inflammation‐related Genes in white Adipose Tissue of Obese Subjects. FASEB j. 18, 1657–1669. doi:10.1096/fj.04-2204com
Cominetti C., Horst M. A., Rogero M. M. (2017). Brazilian Society for Food and Nutrition Position Statement: Nutrigenetic Tests. Nutrire 42, 10. doi:10.1186/s41110-017-0033-2
Dahlman I., Arner P. (2007). Obesity and Polymorphisms in Genes Regulating Human Adipose Tissue. Int. J. Obes. 31, 1629–1641. doi:10.1038/sj.ijo.0803657
Dahlman I., Linder K., Arvidsson Nordström E., Andersson I., Lidén J., Verdich C., et al. (2005). Changes in Adipose Tissue Gene Expression with Energy-Restricted Diets in Obese Women. Am. J. Clin. Nutr. 81, 1275–1285. doi:10.1093/ajcn/81.6.1275
de Roos B., Duivenvoorden I., Rucklidge G., Reid M., Ross K., Lamers R. J., et al. (2005). Response of Apolipoprotein E*3-Leiden Transgenic Mice to Dietary Fatty Acids: Combining Liver Proteomics with Physiological Data. FASEB J. 19 (3), 813–815. doi:10.1096/fj.04-2974fje
de Roos B., McArdle H. J. (2008). Proteomics as a Tool for the Modelling of Biological Processes and Biomarker Development in Nutrition Research. Br. J. Nutr. 99 (Suppl. 3), S66–S71. doi:10.1017/S0007114508006909
DeFelice S. L. (1995). The Nutraceutical Revolution: its Impact on Food Industry R&D. Trends Food Sci. Technology 6 (2), 59–61. doi:10.1016/s0924-2244(00)88944-x
Deshane J., Chaves L., Sarikonda K. V., Isbell S., Wilson L., Kirk M., et al. (2004). Proteomics Analysis of Rat Brain Protein Modulations by Grape Seed Extract. J. Agric. Food Chem. 52, 7872–7883. doi:10.1021/jf040407d
Dicaro S., Tao H., Grillo A., Elia C., Gasbarrini G., Sepulveda A., et al. (2005). Effects of on Genes Expression Pattern in Small Bowel Mucosa. Dig. Liver Dis. 37, 320–329. doi:10.1016/j.dld.2004.12.008
Dieck H. t., Döring F., Fuchs D., Roth H.-P., Daniel H. (2005). Transcriptome and Proteome Analysis Identifies the Pathways that Increase Hepatic Lipid Accumulation in Zinc-Deficient Rats. J. Nutr. 135, 199–205. doi:10.1093/jn/135.2.199
Dixon R. A., Gang D. R., Charlton A. J., Fiehn O., Kuiper H. A., Reynolds T. L., et al. (2006). Applications of Metabolomics in Agriculture. J. Agric. Food Chem. 54, 8984–8994. doi:10.1021/jf061218t
Downs B. W., Bagchi M., Subbaraju G. V., Shara M. A., Preuss H. G., Bagchi D. (2005). Bioefficacy of a Novel Calcium-Potassium Salt of (−)-hydroxycitric Acid. Mutat. Research/Fundamental Mol. Mech. Mutagenesis 579, 149–162. doi:10.1016/j.mrfmmm.2005.02.021
Dudeja P., Gupta R. K. (2017). “Nutraceuticals,” in Rajul Kumar Gupta, Dudeja, Singh Minhas, Food Safety in the 21st Century (Cambridge, United States: Academic Press), 491–496. doi:10.1016/B978-0-12-801773-9.00040-6
Dumas M.-E., Maibaum E. C., Teague C., Ueshima H., Zhou B., Lindon J. C., et al. (2006). Assessment of Analytical Reproducibility of 1H NMR Spectroscopy Based Metabonomics for Large-Scale Epidemiological Research: the INTERMAP Study. Anal. Chem. 78 (7), 2199–2208. doi:10.1021/ac0517085
Endo Y., Fu Z., Abe K., Arai S., Kato H., Kato H. (2002). Dietary Protein Quantity and Quality Affect Rat Hepatic Gene Expression. J. Nutr. 132, 3632–3637. doi:10.1093/jn/132.12.3632
Fardet A., Canlet C., Gottardi G., Lyan B., Llorach R., Rémésy C., et al. (2007). Whole-Grain and Refined Wheat Flours Show Distinct Metabolic Profiles in Rats as Assessed by a 1H NMR-Based Metabonomic Approach. J. Nutr. 137 (4), 923–929. doi:10.1093/jn/137.4.923
Fu W. J., Stromberg A. J., Viele K., Carroll R. J., Wu G. (2010). Statistics and Bioinformatics in Nutritional Sciences: Analysis of Complex Data in the Era of Systems Biology☆. J. Nutr. Biochem. 21, 561–572. doi:10.1016/j.jnutbio.2009.11.007
Fuchs D., de Pascual-Teresa S., Rimbach G., Virgili F., Ambra R., Turner R., et al. (2005a). Proteome Analysis for Identification of Target Proteins of Genistein in Primary Human Endothelial Cells Stressed with Oxidized LDL or Homocysteine. Eur. J. Nutr. 44, 95–104. doi:10.1007/s00394-004-0499-6
Fuchs D., Erhard P., Rimbach G., Daniel H., Wenzel U. (2005b). Genistein Blocks Homocysteine-Induced Alterations in the Proteome of Human Endothelial Cells. Proteomics 5, 2808–2818. doi:10.1002/pmic.200401174
Fuchs D., Erhard P., Turner R., Rimbach G., Daniel H., Wenzel U. (2005c). Genistein Reverses Changes of the Proteome Induced by Oxidized-LDL in EA·hy 926 Human Endothelial Cells. J. Proteome Res. 4, 369–376. doi:10.1021/pr049820r
Fuchs D., Piller R., Linseisen J., Daniel H., Wenzel U. (2007). The Human Peripheral Blood Mononuclear Cell Proteome Responds to a Dietary Flaxseed-Intervention and Proteins Identified Suggest a Protective Effect in Atherosclerosis. Proteomics 7, 3278–3288. doi:10.1002/pmic.200700096
Fuchs D., Winkelmann I., Johnson I. T., Mariman E., Wenzel U., Daniel H. (2005d). Proteomics in Nutrition Research: Principles, Technologies and Applications. Br. J. Nutr. 94, 302–314. doi:10.1079/bjn20051458
Garosi P., De Filippo C., van Erk M., Rocca-Serra P., Sansone S.-A., Elliott R. (2005). Defining Best Practice for Microarray Analyses in Nutrigenomic Studies. Br. J. Nutr. 93, 425–432. doi:10.1079/bjn20041385
Gehlenborg N., O'Donoghue S. I., Baliga N. S., Goesmann A., Hibbs M. A., Kitano H., et al. (2010). Visualization of Omics Data for Systems Biology. Nat. Methods 7, S56–S68. doi:10.1038/nmeth.1436
German J. B., Roberts M.-A., Watkins S. M. (2003). Genomics and Metabolomics as Markers for the Interaction of Diet and Health: Lessons from Lipids. J. Nutr. 133, 2078S–2083S. doi:10.1093/jn/133.6.2078s
Griffin J. L., Muller D., Woograsingh R., Jowatt V., Hindmarsh A., Nicholson J. K., et al. (2002). Vitamin E Deficiency and Metabolic Deficits in Neuronal Ceroid Lipofuscinosis Described by Bioinformatics. Physiol. Genomics 11 (3), 195–203. doi:10.1152/physiolgenomics.00100.2002
Griffiths H. R., Grant M. M. (2006). The Use of Proteomic Techniques to Explore the Holistic Effects of Nutrients In Vivo. Nutr. Res. Rev. 19 (2), 284–293. doi:10.1017/s0954422407249698
Hall R. D., Brouwer I. D., Fitzgerald M. A. (2008). Plant Metabolomics and its Potential Application for Human Nutrition. Physiol. Plant 132, 162–175. doi:10.1111/j.1399-3054.2007.00989.x
Hamid S. B., Hamid A. F. A. (2019). “Roles of Nutraceuticals and Functional Food in Prevention of Cardiovascular Disease,” in Nutraceutical and Functional Foods in Disease Prevention (Hershey, PA: IGI Global), 126–165. doi:10.4018/978-1-5225-3267-5.ch005
Hawkins R. D., Hon G. C., Ren B. (2010). Next-generation Genomics: an Integrative Approach. Nat. Rev. Genet. 11, 476–486. doi:10.1038/nrg2795
Herzog A., Kindermann B., Döring F., Daniel H., Wenzel U. (2004). Pleiotropic Molecular Effects of the Pro-apoptotic Dietary Constituent Flavone in Human colon Cancer Cells Identified by Protein and mRNA Expression Profiling. Proteomics 4, 2455–2464. doi:10.1002/pmic.200300754
Hixson K. K., Lopez-Ferrer D., Robinson E. W., Paša-Tolić L., Lindon John. C., Tranter George. E., et al. (2017). “Proteomics,” in Encyclopedia of Spectroscopy and Spectrometry. 3rd Edn (Cambridge, United States: Academic Press), 766–773. doi:10.1016/b978-0-12-803224-4.00061-3
Husi H., Albalat A. (2014). “Proteomics,” in Sandosh Padmanabhan, Handbook of Pharmacogenomics and Stratified Medicine (Cambridge, United States: Academic Press), 147–179. doi:10.1016/b978-0-12-386882-4.00009-8
Islam S. U., Ahmed M. B., Ahsan H., Lee Y.-S. (2021). Recent Molecular Mechanisms and Beneficial Effects of Phytochemicals and Plant-Based Whole Foods in Reducing LDL-C and Preventing Cardiovascular Disease. Antioxidants 10 (5), 784. doi:10.3390/antiox10050784
Johnson A., Manor D. (2004). The Transcriptional Signature of Vitamin E. Ann. NY Acad. Sci. 1031, 337–338. doi:10.1196/annals.1331.037
Kallio P., Kolehmainen M., Laaksonen D. E., Kekäläinen J., Salopuro T., Sivenius K., et al. (2007). Dietary Carbohydrate Modification Induces Alterations in Gene Expression in Abdominal Subcutaneous Adipose Tissue in Persons with the Metabolic Syndrome: the FUNGENUT Study. Am. J. Clin. Nutr. 85, 1417–1427. doi:10.1093/ajcn/85.5.1417
Kamei A., Watanabe Y., Ishijima T., Uehara M., Arai S., Kato H., et al. (2010). Dietary Iron-Deficient Anemia Induces a Variety of Metabolic Changes and Even Apoptosis in Rat Liver: a DNA Microarray Study. Physiol. Genomics 42, 149–156. doi:10.1152/physiolgenomics.00150.2009
Kato H. (2008). Nutrigenomics: the Cutting Edge and Asian Perspectives. Asia Pac. J. Clin. Nutr. 17 (Suppl. 1), 12–15.
Kato H., Takahashi S., Saito K. (2011). Omics and Integrated Omics for the Promotion of Food and Nutrition Science. J. Traditional Complement. Med. 1 (1), 25–30. doi:10.1016/S2225-4110(16)30053-0
Kim H., Deshane J., Barnes S., Meleth S. (2006). Proteomics Analysis of the Actions of Grape Seed Extract in Rat Brain: Technological and Biological Implications for the Study of the Actions of Psychoactive Compounds. Life Sci. 78 (18), 2060–2065. doi:10.1016/j.lfs.2005.12.008
Kitajka K., Sinclair A. J., Weisinger R. S., Weisinger H. S., Mathai M., Jayasooriya A. P., et al. (2004). Effects of Dietary omega-3 Polyunsaturated Fatty Acids on Brain Gene Expression. Proc. Natl. Acad. Sci. U.S.A. 101 (30), 10931–10936. doi:10.1073/pnas.0402342101
Kronski E., Fiori M. E., Barbieri O., Astigiano S., Mirisola V., Killian P. H., et al. (2014). miR181b Is Induced by the Chemopreventive Polyphenol Curcumin and Inhibits Breast Cancer Metastasis via Down-Regulation of the Inflammatory Cytokines CXCL1 and -2. Mol. Oncol. 8, 581–595. doi:10.1016/j.molonc.2014.01.005
Kumar A., Kujur A., Singh P. P., Prakash B. (2019). Nanoencapsulated Plant-Based Bioactive Formulation against Food-Borne Molds and Aflatoxin B1 Contamination: Preparation, Characterization and Stability Evaluation in the Food System. Food Chem. 287, 139–150. doi:10.1016/j.foodchem.2019.02.045
Kumar A., Mosa K. A., Ji L., Kage U., Dhokane D., Karre S., et al. (2018). Metabolomics-assisted Biotechnological Interventions for Developing Plant-Based Functional Foods and Nutraceuticals. Crit. Rev. Food Sci. Nutr. 58 (11), 1791–1807. doi:10.1080/10408398.2017.1285752
Kussmann M., Affolter M. (2006). Proteomic Methods in Nutrition. Curr. Opin. Clin. Nutr. Metab. Care 9, 575–583. doi:10.1097/01.mco.0000241667.23137.a2
Kussmann M., Krause L., Siffert W. (2010a). Nutrigenomics: where Are We with Genetic and Epigenetic Markers for Disposition and Susceptibility? Nutr. Rev. 68, S38–S47. doi:10.1111/j.1753-4887.2010.00326.x
Kussmann M., Panchaud A., Affolter M. (2010b). Proteomics in Nutrition: Status Quo and Outlook for Biomarkers and Bioactives. J. Proteome Res. 9, 4876–4887. doi:10.1021/pr1004339
Kussmann M., Rezzi S., Daniel H. (2008). Profiling Techniques in Nutrition and Health Research. Curr. Opin. Biotechnol. 19, 83–99. doi:10.1016/j.copbio.2008.02.003
Lapillonne A., Clarke S. D., Heird W. C. (2004). Polyunsaturated Fatty Acids and Gene Expression. Curr. Opin. Clin. Nutr. Metab. Care 7 (2), 151–156. doi:10.1097/00075197-200403000-00008
Lau F. C., Bagchi M., Sen C. K., Bagchi D. (2008). Nutrigenomic Basis of Beneficial Effects of Chromium(III) on Obesity and Diabetes. Mol. Cel Biochem 317, 1–10. doi:10.1007/s11010-008-9744-2
Lavigne J. A., Takahashi Y., Chandramouli G. V. R., Liu H., Perkins S. N., Hursting S. D., et al. (2008). Concentration-dependent Effects of Genistein on Global Gene Expression in MCF-7 Breast Cancer Cells: an Oligo Microarray Study. Breast Cancer Res. Treat. 110 (1), 85–98. doi:10.1007/s10549-007-9705-6
Lenz E. M., Bright J., Wilson I. D., Hughes A., Morrisson J., Lindberg H., et al. (2004). Metabonomics, Dietary Influences and Cultural Differences: a 1H NMR-Based Study of Urine Samples Obtained from Healthy British and Swedish Subjects. J. Pharm. Biomed. Anal. 36 (4), 841–849. doi:10.1016/j.jpba.2004.08.002
Leong H. X., Simkevich C., Lesieur-Brooks A., Lau B. W., Fugere C., Sabo E., et al. (2006). Short-term Arginine Deprivation Results in Large-Scale Modulation of Hepatic Gene Expression in Both normal and Tumor Cells: Microarray Bioinformatic Analysis. Nutr. Metab. (Lond) 3, 37. doi:10.1186/1743-7075-3-37
Li H., Jia Z., Li A., Jenkins G., Yang X., Hu J., et al. (2013). Resveratrol Repressed Viability of U251 Cells by miR-21 Inhibiting of NF-Κb Pathway. Mol. Cel Biochem 382, 137–143. doi:10.1007/s11010-013-1728-1
Li J.-J., Huang C. J., Xie D. (2008). Anti-obesity Effects of Conjugated Linoleic Acid, Docosahexaenoic Acid, and Eicosapentaenoic Acid. Mol. Nutr. Food Res. 52, 631–645. doi:10.1002/mnfr.200700399
Lin R., Nagai Y., Sladek R., Bastien Y., Ho J., Petrecca K., et al. (2002). Expression Profiling in Squamous Carcinoma Cells Reveals Pleiotropic Effects of Vitamin D3 Analog EB1089 Signaling on Cell Proliferation, Differentiation, and Immune System Regulation. Mol. Endocrinol. 16, 1243–1256. doi:10.1210/mend.16.6.0874
Lloyd J. P. B., Lister R. (2022). Epigenome Plasticity in Plants. Nat. Rev. Genet. 23, 55–68. doi:10.1038/s41576-021-00407-y
Lowe R., Shirley N., Bleackley M., Dolan S., Shafee T. (2017). Transcriptomics Technologies. Plos Comput. Biol. 13 (5), e1005457. doi:10.1371/journal.pcbi.1005457
Lusis A. J., Attie A. D., Reue K. (2008). Metabolic Syndrome: from Epidemiology to Systems Biology. Nat. Rev. Genet. 9, 819–830. doi:10.1038/nrg2468
Mangravite L. M., Dawson K., Davis R. R., Gregg J. P., Krauss R. M. (2007). Fatty Acid Desaturase Regulation in Adipose Tissue by Dietary Composition Is Independent of Weight Loss and Is Correlated with the Plasma Triacylglycerol Response. Am. J. Clin. Nutr. 86 (3), 759–767. doi:10.1093/ajcn/86.3.759
Matsuzaki K., Kato H., Sakai R., Toue S., Amao M., Kimura T. (2005). Transcriptomics and Metabolomics of Dietary Leucine Excess. J. Nutr. 135, 1571S–1575S. doi:10.1093/jn/135.6.1571s
McGuire A. L., Gabriel S., Tishkoff S. A., Wonkam A., Chakravarti A., Furlong E. E. M., et al. (2020). The Road Ahead in Genetics and Genomics. Nat. Rev. Genet. 21 (10), 581–596. doi:10.1038/s41576-020-0272-6
McLoughlin P., Roengvoraphoj M., Gissel C., Hescheler J., Certa U., Sachinidis A. (2004). Transcriptional Responses to Epigallocatechin-3 Gallate in HT 29 colon Carcinoma Spheroids. Genes to Cells 9 (7), 661–669. doi:10.1111/j.1356-9597.2004.00754.x
Mitchell B. L., Yasui Y., Lampe J. W., Gafken P. R., Lampe P. D. (2005). Evaluation of Matrix-Assisted Laser Desorption/ionization-Time of Flight Mass Spectrometry Proteomic Profiling: Identification of Alpha 2-HS Glycoprotein B-Chain as a Biomarker of Diet. Proteomics 5, 2238–2246. doi:10.1002/pmic.200401099
Mohanty S., Singhal K. (2018). “Functional Foods as Personalised Nutrition: Definitions and Genomic Insights,” in Functional Food and Human Health (Berlin, Germany: Springer), 513–535. doi:10.1007/978-981-13-1123-9_22
Müller M., Kersten S. (2003). Nutrigenomics: Goals and Strategies. Nat. Rev. Genet. 4, 315–322. doi:10.1038/nrg1047
Murtaza I., Marra G., Schlapbach R., Patrignani A., Künzli M., Wagner U., et al. (2006). A Preliminary Investigation Demonstrating the Effect of Quercetin on the Expression of Genes Related to Cell-Cycle Arrest, Apoptosis and Xenobiotic Metabolism in Human CO115 colon-adenocarcinoma Cells Using DNA Microarray. Biotechnol. Appl. Biochem. 45, 29–36. doi:10.1042/BA20060044
Nakai Y., Hashida H., Kadota K., Minami M., Shimizu K., Matsumoto I., et al. (2008). Up-Regulation of Genes Related to the Ubiquitin-Proteasome System in the Brown Adipose Tissue of 24-H-Fasted Rats. Biosci. Biotechnol. Biochem. 72, 139–148. doi:10.1271/bbb.70508
Narasaka S., Endo Y., Fu Z., Moriyama M., Arai S., Abe K., et al. (2006). Safety Evaluation of Hypoallergenic Wheat Flour by Using a DNA Microarray. Biosci. Biotechnol. Biochem. 70 (6), 1464–1470. doi:10.1271/bbb.60002
Narayanan B. A., Narayanan N. K., Simi B., Reddy B. S. (2003). Modulation of Inducible Nitric Oxide Synthase and Related Proinflammatory Genes by the omega-3 Fatty Acid Docosahexaenoic Acid in Human colon Cancer Cells. Cancer Res. 63 (5), 972–979.
Nayak S. N., Aravind B., Malavalli S. S., Sukanth B. S., Poornima R., Bharati P., et al. (2021). Omics Technologies to Enhance Plant Based Functional Foods: An Overview. Front. Genet. 12. doi:10.3389/fgene.2021.742095
Niculescu M. D., Pop E. A., Fischer L. M., Zeisel S. H. (2007). Dietary Isoflavones Differentially Induce Gene Expression Changes in Lymphocytes from Postmenopausal Women Who Form Equol as Compared with Those Who Do Not. J. Nutr. Biochem. 18, 380–390. doi:10.1016/j.jnutbio.2006.06.002
Ohta Y., Sami M., Kanda T., Saito K., Osada K., Kato H. (2006). Gene Expression Analysis of the Anti-obesity Effect by Apple Polyphenols in Rats Fed a High Fat Diet or a normal Diet. J. Oleo Sci. 55, 305–314. doi:10.5650/jos.55.305
Oresic M. (2009). Metabolomics, a Novel Tool for Studies of Nutrition, Metabolism and Lipid Dysfunction. Nutr. Metab. Cardiovasc. Dis. 19, 816–824. doi:10.1016/j.numecd.2009.04.018
Ortega F. J., Cardona-Alvarado M. I., Mercader J. M., Moreno-Navarrete J. M., Moreno M., Sabater M., et al. (2015). Circulating Profiling Reveals the Effect of a Polyunsaturated Fatty Acid-Enriched Diet on Common microRNAs. J. Nutr. Biochem. 26, 1095–1101. doi:10.1016/j.jnutbio.2015.05.001
Ovesná J., Slabý O., Toussaint O., Kodícek M., Marsík P., Pouchová V., et al. (2008). High Throughput 'omics' Approaches to Assess the Effects of Phytochemicals in Human Health Studies. Br. J. Nutr. 99 (Suppl. 1), ES127–34. doi:10.1017/S0007114508965818
Pandita D. (2018a). Plant miRNAs: Micro Structure and Macro Character. Res. Rev. J. Agric. Allied Sci. 7, 83–84.
Pandita D. (2021). “Role of miRNAi Technology and miRNAs in Abiotic and Biotic Stress Resilience,” in Plant Perspectives to Global Climate Changes. Editors T. Aftab, and A. Roychoudhury, 303–330. doi:10.1016/B978-0-323-85665-2.00015-7
Pandita D., Pandita A. (2016). Jumping Genes-"The Other Half of the Human Genome" and the Missing Heritability Conundrum of Human Genetic Disorders. Bbj 11 (3), 1–18. doi:10.9734/bbj/2016/13904
Pandita D. (2019) Plant MIRnome: miRNA Biogenesis and Abiotic Stress Response, 449, 474. In: M. Hasanuzzaman, K. Hakeem, K. Nahar, and H. Alharby editions Plant Abiotic Stress Tolerance. (Cham: Springer). doi:10.1007/978-3-030-06118-0_18
Pandita D., Wani S. H. (2019). “MicroRNA as a Tool for Mitigating Abiotic Stress in Rice (Oryza Sativa L.),” in Recent Approaches in Omics for Plant Resilience to Climate Change. Editor S. Wani (Cham: Springer), 109–133. doi:10.1007/978-3-030-21687-0_6
Park J. Y., Seong J. K., Paik Y.-K. (2004). Proteomic Analysis of Diet-Induced Hypercholesterolemic Mice. Proteomics 4, 514–523. doi:10.1002/pmic.200300623
Park S.-H., Choi M.-S., Park T. (2008). Changes in the Hepatic Gene Expression Profile in a Rat Model of Chronic Ethanol Treatment. Food Chem. Toxicol. 46 (4), 1378–1388. doi:10.1016/j.fct.2007.08.029
Pazhamala L. T., Kudapa H., Weckwerth W., Millar A. H., Varshney R. K. (2021). Systems Biology for Crop Improvement. Plant Genome 14, 1–23. doi:10.1002/tpg2.20098
Prakash B., Kujur A., Yadav A., Kumar A., Singh P. P., Dubey N. K. (2018). Nanoencapsulation: An Efficient Technology to Boost the Antimicrobial Potential of Plant Essential Oils in Food System. Food Control 89, 1–11. doi:10.1016/j.foodcont.2018.01.018
Puskas L., Menesi D., Feher L., Kitajka K. (2006). High-throughput Functional Genomic Methods to Analyze the Effects of Dietary Lipids. Cpb 7 (6), 525–529. doi:10.2174/138920106779116801
Recchiuti A., Krishnamoorthy S., Fredman G., Chiang N., Serhan C. N. (2011). MicroRNAs in Resolution of Acute Inflammation: Identification of Novel Resolvin Dl‐miRNA Circuits. FASEB j. 25, 544–560. doi:10.1096/fj.10-169599
Roberfroid M. B. (1999). Concepts in Functional Foods: the Case of Inulin and Oligofructose. J. Nutr. 129 (7 Suppl. l), 1398S–401S. doi:10.1093/jn/129.7.1398S
Rochfort S. (2005). Metabolomics Reviewed: a New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 68, 1813–1820. doi:10.1021/np050255w
Roessler C., Kuhlmann K., Hellwing C., Leimert A., Schumann J. (2017). Impact of Polyunsaturated Fatty Acids on miRNA Profiles of Monocytes/macrophages and Endothelial Cells-A Pilot Study. Ijms 18, 284. doi:10.3390/ijms18020284
Roy S., Rink C., Khanna S., Phillips C., Bagchi D., Bagchi M., et al. (2004). Body Weight and Abdominal Fat Gene Expression Profile in Response to a Novel Hydroxycitric Acid-Based Dietary Supplement. Gene Expr. 11, 251–262. doi:10.3727/000000003783992289
Roy S., Shah H., Rink C., Khanna S., Bagchi D., Bagchi M., et al. (2007). Transcriptome of Primary Adipocytes from Obese Women in Response to a Novel Hydroxycitric Acid-Based Dietary Supplement. DNA Cel Biol. 26, 627–639. doi:10.1089/dna.2007.0617
Ryu M.-S., Langkamp-Henken B., Chang S.-M., Shankar M. N., Cousins R. J. (2011). Genomic Analysis, Cytokine Expression, and microRNA Profiling Reveal Biomarkers of Human Dietary Zinc Depletion and Homeostasis. Proc. Natl. Acad. Sci. U.S.A. 108, 20970–20975. doi:10.1073/pnas.1117207108
Saito K., Arai S., Kato H. (2005). A Nutrigenomics Database - Integrated Repository for Publications and Associated Microarray Data in Nutrigenomics Research. Br. J. Nutr. 94, 493–495. doi:10.1079/bjn20051536
Saito K., Ohta Y., Sami M., Kanda T., Kato H. (2010). Effect of Mild Restriction of Food Intake on Gene Expression Profile in the Liver of Young Rats: Reference Data for In Vivo Nutrigenomics Study. Br. J. Nutr. 104, 941–950. doi:10.1017/s0007114510001625
Salunkhe D. K., Jadhav S. J., Kadam S. S., Chavan J. K., Luh B. S. (1983). Chemical, Biochemical, and Biological Significance of Polyphenols in Cereals and Legumes. C R. C Crit. Rev. Food Sci. Nutr. 17 (3), 277–305. doi:10.1080/10408398209527350
Schauber J., Weiler F., Gostner A., Melcher R., Kudlich T., Lührs H., et al. (2006). Human Rectal Mucosal Gene Expression after Consumption of Digestible and Non-digestible Carbohydrates. Mol. Nutr. Food Res. 50, 1006–1012. doi:10.1002/mnfr.200600084
Sen C. K., Gordillo G. M., Khanna S., Roy S. (2009). Micromanaging Vascular Biology: Tiny micrRNAs Play Big Band. J. Vasc. Res. 46 (6), 527–540. doi:10.1159/000226221
Smolenski G., Haines S., Kwan F. Y.-S., Bond J., Farr V., Davis S. R., et al. (2007). Characterisation of Host Defence Proteins in Milk Using a Proteomic Approach. J. Proteome Res. 6 (1), 207–215. doi:10.1021/pr0603405
Solanky K. S., Bailey N. J. C., Beckwith-Hall B. M., Davis A., Bingham S., Holmes E., et al. (2003). Application of Biofluid 1H Nuclear Magnetic Resonance-Based Metabonomic Techniques for the Analysis of the Biochemical Effects of Dietary Isoflavones on Human Plasma Profile. Anal. Biochem. 323 (2), 197–204. doi:10.1016/j.ab.2003.08.028
Song J., Jun M., Ahn M.-R., Kim O. Y. (2016). Involvement of miR-Let7A in Inflammatory Response and Cell Survival/apoptosis Regulated by Resveratrol in THP-1 Macrophage. Nutr. Res. Pract. 10, 377–384. doi:10.4162/nrp.2016.10.4.377
Subbiah M. T. R. (2007). Nutrigenetics and Nutraceuticals: the Next Wave Riding on Personalized Medicine. Translational Res. 149, 55–61. doi:10.1016/j.trsl.2006.09.003
Swatton J. E., Prabakaran S., Karp N. A., Lilley K. S., Bahn S. (2004). Protein Profiling of Human Postmortem Brain Using 2-dimensional Fluorescence Difference Gel Electrophoresis (2-D DIGE). Mol. Psychiatry 9, 128–143. doi:10.1038/sj.mp.4001475
Tachibana N., Matsumoto I., Fukui K., Arai S., Kato H., Abe K., et al. (2005). Intake of Soy Protein Isolate Alters Hepatic Gene Expression in Rats. J. Agric. Food Chem. 53, 4253–4257. doi:10.1021/jf0481103
Takahashi K., Sasano T., Sugiyama K., Kurokawa J., Tamura N., Soejima Y., et al. (2016). High-fat Diet Increases Vulnerability to Atrial Arrhythmia by Conduction Disturbance via miR-27b. J. Mol. Cell Cardiol. 90, 38–46. doi:10.1016/j.yjmcc.2015.11.034
Takahashi S., Masuda J., Shimagami H., Ohta Y., Kanda T., Saito K., et al. (2011). Mild Caloric Restriction Up-Regulates the Expression of Prohibitin: A Proteome Study. Biochem. Biophysical Res. Commun. 405, 462–467. doi:10.1016/j.bbrc.2011.01.054
Tan S., Seow T. K., Liang R. C. M. Y., Koh S., Lee C. P. C., Chung M. C. M., et al. (2002). Proteome Analysis of Butyrate-Treated Human colon Cancer Cells (HT-29). Int. J. Cancer 98, 523–531. doi:10.1002/ijc.10236
Télessy I. G., Singh Ram. B., Ross Watson Ronald., Takahashi Toru. (2019). “Nutraceuticals,” in The Role of Functional Food Security in Global Health (Cambridge, United States: Academic Press), 409–421. doi:10.1016/B978-0-12-813148-0.00024-4
Tian J., Bryksa B. C., Yada R. Y. (2016). Feeding the World into the Future - Food and Nutrition Security: the Role of Food Science and Technology. Front. Life Sci. 9 (3), 155–166. doi:10.1080/21553769.2016.1174958
Tili E., Michaille J.-J., Adair B., Alder H., Limagne E., Taccioli C., et al. (2010). Resveratrol Decreases the Levels of miR-155 by Upregulating miR-663, a microRNA Targeting JunB and JunD. Carcinogenesis 31, 1561–1566. doi:10.1093/carcin/bgq143
Tomé-Carneiro J., Larrosa M., Yánez-Gascón M. J., Dávalos A., Gil-Zamorano J., Gonzálvez M., et al. (2013). One-year Supplementation with a Grape Extract Containing Resveratrol Modulates Inflammatory-Related microRNAs and Cytokines Expression in Peripheral Blood Mononuclear Cells of Type 2 Diabetes and Hypertensive Patients with Coronary Artery Disease. Pharmacol. Res. 72, 69–82. doi:10.1016/j.phrs.2013.03.011
Trayhurn P. (2000). Proteomics and Nutrition - A Science for the First Decade of the New Millennium. Br. J. Nutr. 83, 1–2. doi:10.1017/s0007114500000015
Trujillo E., Davis C., Milner J. (2006). Nutrigenomics, Proteomics, Metabolomics, and the Practice of Dietetics. J. Am. Diet. Assoc. 106, 403–413. doi:10.1016/j.jada.2005.12.002
Tsuda T., Ueno Y., Yoshikawa T., Kojo H., Osawa T. (2006). Microarray Profiling of Gene Expression in Human Adipocytes in Response to Anthocyanins. Biochem. Pharmacol. 71 (8), 1184–1197. doi:10.1016/j.bcp.2005.12.042
van Dorsten F. A., Daykin C. A., Mulder T. P. J., van Duynhoven J. P. M. (2006). Metabonomics Approach to Determine Metabolic Differences between green tea and Black tea Consumption. J. Agric. Food Chem. 54, 6929–6938. doi:10.1021/jf061016x
van Erk M. J., Blom W. A., van Ommen B., Hendriks H. F. (2006). High-protein and High-Carbohydrate Breakfasts Differentially Change the Transcriptome of Human Blood Cells. Am. J. Clin. Nutr. 84, 1233–1241. doi:10.1093/ajcn/84.5.1233
Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., et al. (2001). The Sequence of the Human Genome. Science 291, 1304–1351. doi:10.1126/science.1058040
Vergara D., Chiriacò F., Acierno R., Maffia M. (2008). Proteomic Map of Peripheral Blood Mononuclear Cells. Proteomics 8 (10), 2045–2051. doi:10.1002/pmic.200700726
Vinciguerra M., Sgroi A., Veyrat-Durebex C., Rubbia-Brandt L., Buhler L. H., Foti M. (2009). Unsaturated Fatty Acids Inhibit the Expression of Tumor Suppressor Phosphatase and Tensin Homolog (PTEN) via microRNA-21 Up-Regulation in Hepatocytes. Hepatology 49, 1176–1184. doi:10.1002/hep.22737
Vittal R., Selvanayagam Z. E., Sun Y., Hong J., Liu F., Chin K. V., et al. (2004). Gene Expression Changes Induced by green tea Polyphenol (-)-Epigallocatechin-3-Gallate in Human Bronchial Epithelial 21BES Cells Analyzed by DNA Microarray. Mol. Cancer Ther. 3 (9), 1091–1099.
Wang Y., Tang H., Nicholson J. K., Hylands P. J., Sampson J., Holmes E. (2005). A Metabonomic Strategy for the Detection of the Metabolic Effects of Chamomile (Matricaria Recutita L.) Ingestion. J. Agric. Food Chem. 53 (2), 191–196. doi:10.1021/jf0403282
Wang Z., Filgueiras L. R., Wang S., Serezani A. P. M., Peters-Golden M., Jancar S., et al. (2014). Leukotriene B4 Enhances the Generation of Proinflammatory microRNAs to Promote MyD88-dependent Macrophage Activation. J.I. 192, 2349–2356. doi:10.4049/jimmunol.1302982
Wenzel U., Herzog A., Kuntz S., Daniel H. (2004). Protein Expression Profiling Identifies Molecular Targets of Quercetin as a Major Dietary Flavonoid in Human colon Cancer Cells. Proteomics 4, 2160–2174. doi:10.1002/pmic.200300726
Xie H., Lim B., Lodish H. F. (2009). MicroRNAs Induced during Adipogenesis that Accelerate Fat Cell Development Are Downregulated in Obesity. Diabetes 58, 1050–1057. doi:10.2337/db08-1299
Xing Y., Liu Z., Yang G., Gao D., Niu X. (2015). MicroRNA Expression Profiles in Rats with Selenium Deficiency and the Possible Role of the Wnt/β-Catenin Signaling Pathway in Cardiac Dysfunction. Int. J. Mol. Med. 35, 143–152. doi:10.3892/ijmm.2014.1976
Yadav C. B., Pandey G., Muthamilarasan M., Prasad M. (2018). Epigenetics and Epigenomics of Plants. Adv. Biochem. Eng. Biotechnol. 164, 237–261. doi:10.1007/10_2017_51
Yamada S., Tsukamoto S., Huang Y., Makio A., Kumazoe M., Yamashita S., et al. (2016). Epigallocatechin-3-O-gallate Up-Regulates microRNA-Let-7b Expression by Activating 67-kDa Laminin Receptor Signaling in Melanoma Cells. Sci. Rep. 6, 19225. doi:10.1038/srep19225
Yang W.-M., Jeong H.-J., Park S.-Y., Lee W. (2014). Induction of miR-29a by Saturated Fatty Acids Impairs Insulin Signaling and Glucose Uptake through Translational Repression of IRS-1 in Myocytes. FEBS Lett. 588, 2170–2176. doi:10.1016/j.febslet.2014.05.011
Zárate R., El Jaber-Vazdekis N., Tejera N., Pérez J. A., Rodríguez C. (2017). Significance of Long Chain Polyunsaturated Fatty Acids in Human Health. Clin. Transl Med. 6 (1), 25.
Zhang Z., Wu S., Stenoien D. L., Paša-Tolić L. (2014). High-throughput Proteomics. Annu. Rev. Anal. Chem. 7, 427–454. doi:10.1146/annurev-anchem-071213-020216
Keywords: functional food, nutraceuticals, nutrigenomics, transcriptomics, proteomics, metabolomics, epigenomics, miRNomics
Citation: Pandita D and Pandita A (2022) Omics Technology for the Promotion of Nutraceuticals and Functional Foods. Front. Physiol. 13:817247. doi: 10.3389/fphys.2022.817247
Received: 17 November 2021; Accepted: 23 March 2022;
Published: 13 May 2022.
Edited by:
Monica Colitti, University of Udine, ItalyReviewed by:
Dipayan Sarkar, North Dakota State University, United StatesEswar Shankar, The Ohio State University, United States
Copyright © 2022 Pandita and Pandita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deepu Pandita, deepupandita@gmail.com
 Deepu Pandita
Deepu Pandita Anu Pandita2
Anu Pandita2