- 1Department of Orthodontics, The Affiliated Stomatology Hospital of Southwest Medical University, Luzhou, China
- 2Department of Oral Maxillofacial Surgery, Shenzhen Hospital, Southern Medical University, Shenzhen, China
- 3Chongqing Key Laboratory for Oral Diseases and Biomedical Sciences, Chongqing Municipal Key Laboratory of Oral Biomedical Engineering of Higher Education, College of Stomatology, Chongqing Medical University, Chongqing, China
Objective: Tooth morphogenesis and the formation of hard tissues have been reported to be closely related to circadian rhythms. This study investigates the spatiotemporal expression and relationship of p75NTR with core clock genes, mineralization-related or odontogenesis-related genes, and aims to derive the potential role of p75NTR in regulating circadian rhythm and incrementality growth line formation during tooth development.
Materials and methods: The dynamic morphology of the rat dental germ was observed at seven stages (E14.5 d, E16.5 d, E18.5 d, P.N. 4 d, P.N. 7 d, P.N. 10 d, and P.N. 15 d). Next, the expressions of p75NTR and other target factors were traced. The ectomesenchymal stem cells (EMSCs) were isolated from the E18.5d rat dental germs and synchronized using 50% of fetal bovine serum. Then, they were cultured in light/light (L.L.), dark/dark (D.D.), and light/dark (L.D.) conditions for 48 h. The total RNA was collected every 4 h, and the circadian rhythm dynamics of target factors were observed. To reveal the mechanism further, p75NTR was down-regulated in p75NTRExIII−/− mice and up-regulated in immortalized mouse dental apical papilla progenitor cells. The change tendencies of other target factors were also detected.
Results: The clock genes Bmal1, Clock, Per1, and Per2 were all expressed in tooth germs before the formation of dental hard tissues and demonstrated a regular oscillating expression pattern in EMSCs from dental germs. Their expression was affected by the L.D. stimulus, and most of them were promoted by D.D. conditions. p75NTR presented a similar expression pattern and a positive or negative relationship with most clock genes, mineralization-related and odontogenesis-related factors, such as brain and muscle ARNT-like protein-1 (Bmal1), runt-related transcription factor 2 (Runx2), alkaline phosphatase (ALP), MSH-like 1 (MSX1), dentin matrix acidic phosphoprotein 1 (Dmp1), and dentin sialophosphoprotein (Dspp). Moreover, the arrangement, morphology, and even boundary in pre-odontoblast/pre-ameloblast layers were disordered in the p75NTRExIII−/− mice.
Conclusion: Circadian rhythm was found to affect tooth development. p75NTR might play a crucial role in regulating clock genes in the mineralization and formation of the dental hard tissues. p75NTR is actively involved in the odontoblast-ameloblast junction and cell polarity establishment during tooth morphogenesis.
Introduction
Most physiological and behavioral processes, such as hormone secretion, metabolism, growth, sleep, among others, are governed by circadian rhythms, which are managed by internal biological clocks (Richards and Gumz, 2013; Neumann et al., 2019). The suprachiasmatic nucleus (SCN), located in the brain’s anterior hypothalamus, is generally considered the “master clock” controlling the circadian rhythms (Weaver, 1998). Moreover, peripheral clocks have been discovered in several body tissues and found to be regulated by the SCN via a transcriptional-translational feedback loop (TTFL) (Okawa et al., 2020; Allada and Bass, 2021). Importantly, circadian locomotor output cycles kaput (Clock) and brain and muscle ARNT-like protein-1 (Bmal1) serve as positive feedback signals, form heterodimers, and activate the transcription of period (Per) and cryptochrome (Cry) genes by binding to a cis-regulatory enhancer sequence known as the E-box element on the target gene promoter (Gekakis et al., 1998; Shearman et al., 2000). After reaching a certain concentration, Per and Cry proteins are phosphorylated and translocated into the nucleus to inhibit the transcriptional activation of Clock/Bmal1 by competitively binding to the E-box element (Brown et al., 2012). This negative TTFL acts as a core circadian regulator to maintain the 24-h rhythm.
Clock genes play an essential role in tooth development (Nirvani et al., 2017; Papakyrikos et al., 2020). Clock, Bmal1, Per, and Cry are expressed in dental tissues, especially in dental hard tissues during tooth development (Zheng et al., 2011; Lacruz et al., 2012). The phenomenon of regular incremental growth lines (e.g., daily Retzius’s lines in enamel, von Ebner’s lines in dentine) implies that the formation of dental hard tissues characterizes the circadian rhythm and is tightly controlled by the time (Antoine et al., 2009; Lacruz et al., 2012). The collagen production in dentin follows a 12 hour-pattern, with twice as much collagen secreted during the 12 h of daylight than the 12 h of nighttime (Lopez Franco et al., 2006). Previous studies have demonstrated that tooth morphogenesis and hard tissue formation have been closely related to circadian rhythms (Lacruz et al., 2012). However, how the core clock genes affect tooth development is still unclear.
p75 neurotrophin receptor (p75NTR) demonstrated a strong expression in epithelial-mesenchymal interaction, dental papilla, and dental follicle during tooth development, and positively regulated the mineralization in ectomesenchymal stem cells (EMSCs) (Wen et al., 2012; Li et al., 2017). Further research revealed that the incisors’ daily mineralization speed and the incremental growth line width were significantly lower in p75NTR knockout mice than in wild-type mice (Wang et al., 2020; Zhao et al., 2020). There is also evidence that p75NTR regulates tooth morphogenesis and mineralization along with the circadian rhythm and incremental growth line formation during tooth development. p75NTR, a member of the tumor necrosis factor receptor superfamily, has been reported to manage a wide range of biological functions via multiple intracellular signaling pathways. Recently, p75NTR was reported to be controlled by Clock/Bmal1 by bonding to the E-box element and was considered a clock gene–regulating oscillatory component of circadian rhythms (Baeza-Raja et al., 2013). Therefore, p75NTR might act as a clock-controlled gene in tooth development and regulate periodic mineralization during incremental growth line formation.
The aims of this study are to investigate the spatiotemporal expression and relationship of p75NTR with core clock genes, mineralization-related or odontogenesis-related genes, and further to reveal the potential role of p75NTR in regulating the circadian rhythms and incremental growth line formation during tooth development via the in vivo experiment of p75NTRExIII knockout mice and in vitro experiment of EMSCs cultured under light/light (L.L.), dark/dark (D.D.), and light/dark (L.D.) conditions, which would contribute to illustrate the circadian rhythm and biomineralization in tooth development.
Materials and methods
Experimental animals
Sprague–Dawley (S.D.) rats were provided by the Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences, Chongqing Medical University. p75NTR knockout mice used in this study were gifted by the Jackson Laboratory (#:031,162). In 1992, it was reported that these mutant mice have a targeted deletion of exon III of the p75NTR (p75NTRExIII−/−) and could not express functional full-length p75NTR (Lee et al., 1992). The presence of a vaginal plug is considered embryonic day 0.5 (E 0.5 d), and the day of littermate birth is regarded as post-natal day 1(PN1 d). All procedures were approved by the Medical Ethics Committee of the Chongqing Medical University.
H.E. and immunohistochemistry staining
The first molars were dissected from the E14.5 d, E16.5 d, E18.5 d, P.N.4 d, P.N.7 d, P.N.10 d, and P.N.15 d rats (Figures 1, 2) and the E16.5d p75NTRExIII−/− and p75NTRExIII+/+ mice (Figure 4), fixed in 4% paraformaldehyde, decalcified with 10% EDTA, and embedded in paraffin. The 6-μm sections of tissue specimens were obtained for H.E. and immunohistochemistry staining. The primary antibodies were used in this study are as follows: rabbit anti-rat p75NTR (1:1,500; Abcam, Cambridge, MA, United States, ab245134, monoclonal), rabbit anti-rat BMAL1 (1:1,000; Abcam, Cambridge, MA, United States, ab230822, monoclonal), rabbit anti-rat CLOCK (1:1,000; Abcam, Cambridge, MA, United States, ab3517, polyclonal), rabbit anti-rat PER1 (1:500; Bioss, Beijing, China,bs-2350R, polyclonal), rabbit anti-rat CRY1 (1:500; Bioss, Beijing, China bs-11441R, polyclonal), rabbit anti-rat ALP (1:500; Bioss, Beijing, China, bs-2928R, polyclonal), rabbit anti-rat COL1 (1:500; Bioss, Beijing, China, bs-0578R, polyclonal). These specimens were treated with the DAB Detection Kit Streptavidin-Biotin (ZSGB-BIO, Beijing, China) and Hematoxylin and Eosin Staining Kit (Beyotime, Shanghai, China) according to the manufacturer’s protocols, followed by visualization under phase-contrast microscopy.
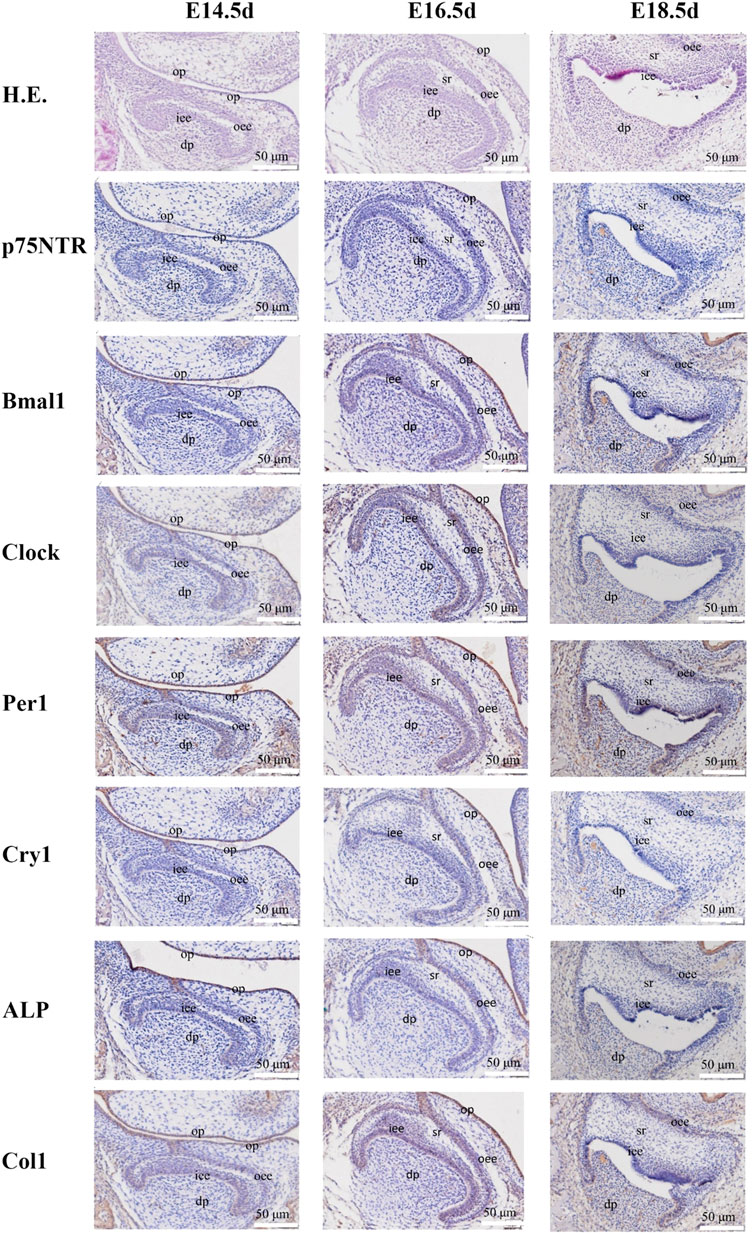
FIGURE 1. The images of H.E. staining and immunohistochemistry staining for prenatal rat dental germs. H.E. staining demonstrated that the rat dental germs entered the early cap stage at E14.5 d, the cap stage and early bell stage at E16.5 d, and the bell stage at E18.5 d. The separation between the pre-odontoblast and pre-ameloblast layers occurred in all E18.5 d species. Immunohistochemistry staining revealed that Clock, Per1, and Col1 were detected in the epithelial-mesenchymal interaction area, dental follicle, and dental papilla at E14.5 d, and became stronger at E16.5 d when p75NTR, Bmal1, and ALP were detected. All the factors were expressed at E18.5 d, but Cry1 showed the weakest expression. All experiments were repeated three times independently. op: oral epithelium; dp: dental papilla; iee: inner enamel epithelium; oee: outer enamel epithelium; sr: stellate reticulum. The scale bar represents 50 μm, respectively.
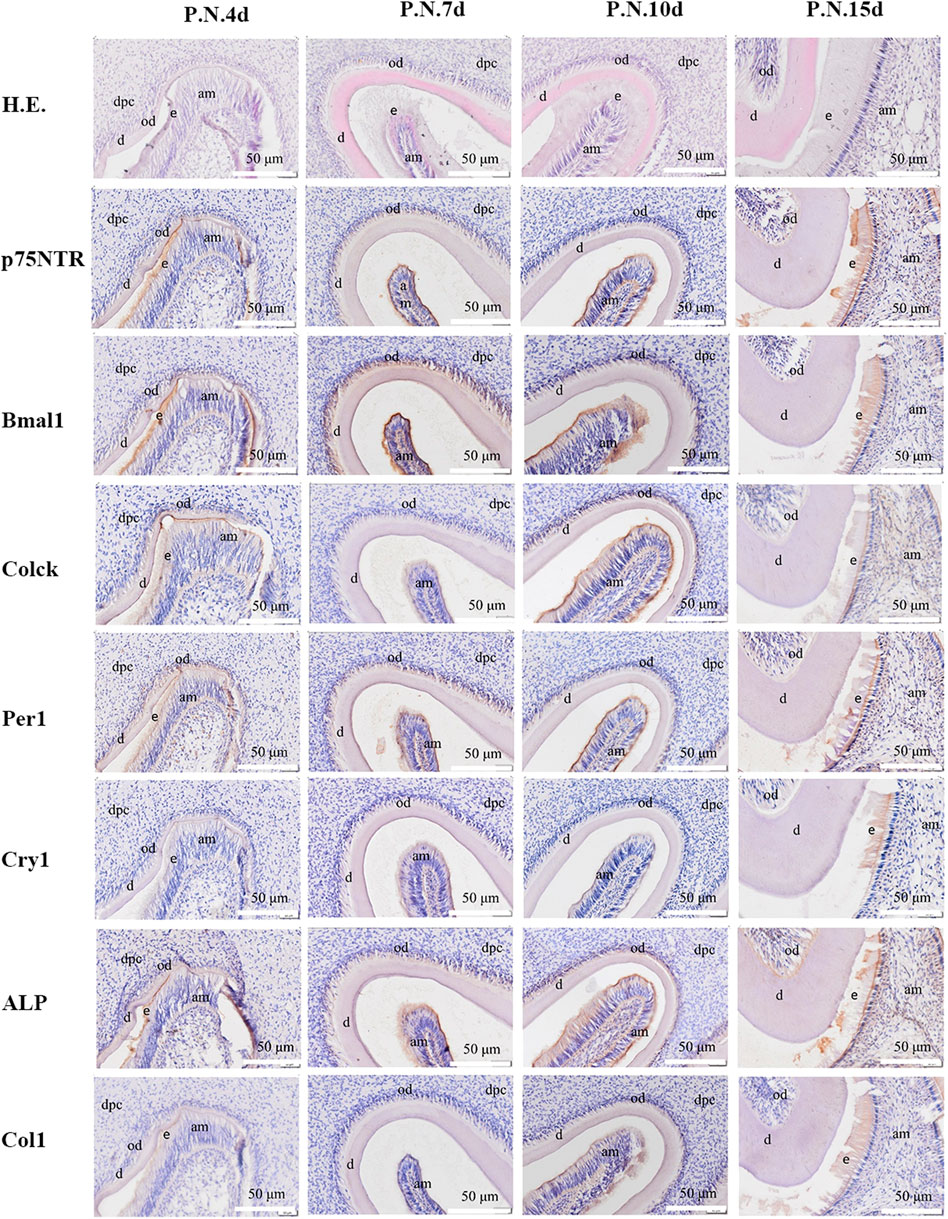
FIGURE 2. The images of H.E. staining and immunohistochemistry staining for post-natal rat dental germs. H.E. staining revealed that the morphogenesis of molar cusps was completed, and hard tissues (enamel and dentine) began to form at PN4 d. The hard tissues gradually thickened at PN7 d. The tooth roots began to shape at PN10 d and PN15 d. The separation continued between the inner enamel epithelium and enamel in all the post-natal species. Immunohistochemistry staining showed that all the factors were detected and distributed in odontoblast and ameloblast layers, and then in dental papilla in the post-natal species. All experiments were repeated three times independently. dpc: dental papilla cells; od: odontoblast; am: ameloblast; e: enamel; d: dentin. The scale bar represents 500 and 50 μm, respectively.
Isolation and culture of EMSCs from dental germs
The first molars of the upper and lower jaws were dissected from seven to ten embryos of E18.5d rats. The minced tissue was mixed with 1% trypsin/1 mM of the EDTA solution (Sigma, St. Louis, MO, United States) at 37°C for 10 min and neutralized with Dulbecco’s modified Eagle’s medium/Ham’s F12 (DMEM/F12) (Sigma, St. Louis, MO, United States) containing 10% of FBS (Gibco Waltham, MA United States). Next, the suspension was centrifuged at 800 rpm for 5 min. The cell pellet was resuspended in DMEM/F12 supplemented with 10% FBS and antibiotics (100 μg/ml of penicillin and 100 μg/ml of streptomycin) at a density of 2 * 105/ml and then cultured at 37°C in a 5% CO2 humidified incubator. The culture medium was changed every 2 days and cells were passaged when the cell density was fused to 70 %–80%. E18.5 d rat EMSCs at passage three were used in the following experiment.
Clock synchronization experiment of EMSCs under three conditions
E18.5 d rat EMSCs were seeded into 6-well plates at a density of 1 × 105 cells per well and incubated in the routine medium as mentioned above until confluency. They were synchronized using 50% of the horse serum (Gibco Waltham, MA,United States), and total RNA was collected every 4 hours over a circadian cycle at the following zeitgeber time (Z.T.) after synchronization: ZT0, 4, 8, 12, 16, 20, 24, 28, 32, 36, 40, 44, and 48. The cells were exposed to the serum for 2 h and then incubated under three conditions, namely L.L.-cycle, D.D.-cycle, and 12-h L.D.-cycle (12 h dark and 12 h light), until sample collection.
Genotype identification of p75NTR-knockout mice
All mice were housed under specific pathogen-free conditions (22°C, 12/12-h L.D., 50%–55% humidity) in the Chongqing Key Laboratory for Oral Diseases and Biomedical Sciences. The p75NTR knockout (p75NTRExIII−/−) and wild-type (p75NTRExIII+/+) littermates used in this research were obtained from mating between heterozygous (p75NTRExIII+/−) females and males. The tail DNA was extracted and used to determine their genotypes by PCR analysis (Zhang et al., 2015).
Wild-type mice feeding under D.D. And 12-h D.L. conditions
p75NTRExIII+/+ mice were fed and mated under D.D.-cycle and D.L.-cycle (natural day and night), respectively, at PN7 d, four wild-type mice were killed at 7:30 a.m. and 7:30 p.m. The maxilla and mandible of five mice were separated under sterile conditions, and the first molars of mice were removed under the anatomical microscope and then divided into enamel organs and dental papilla. All experiments were repeated at least three times independently. Total RNA was extracted from the isolated dental papilla with the steady pure universal RNA Extraction Kit II and subsequently analyzed quantitatively by RT qPCR.
p75NTR over-expression plasmid constructs and transfection
The coding region of p75NTR was amplified from the cDNAs of mice and then cloned into vector GV492. The plasmid GV492-p75NTR was co-transfected with lentivirus helper plasmids (Helper 1.0 and Helper 2.0) into HEK-293T cells using Lipofectamine 2000 (Invitrogen, United States) according to the manufacturer’s protocol, the negative control carried corresponding fluorescent markers and contain resistant genes, but do not express other target genes. Supernatants containing the virus were collected 72 h following transfection and then infected the immortalized mouse dental apical papilla progenitor cells. iSCAP (Using the previously characterized reversible immortalization system, which expresses SV40 T antigenflanked with Cre/loxP sites, Wang et al. (2014) demonstrated that the mouse SCAPs can be effectively immortalized with an enhanced proliferative activity) was gifted by Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences. The cells were selected with 2 μg/ml of puromycin 48 h later. p75NTR over-expression iSCAP and negative control iSCAP were seeded into 6-well plates at a density of 1 × 105 cells per well and incubated in the routine medium until confluency. All experiments were repeated at least three times independently.
Quantitative RT qPCR
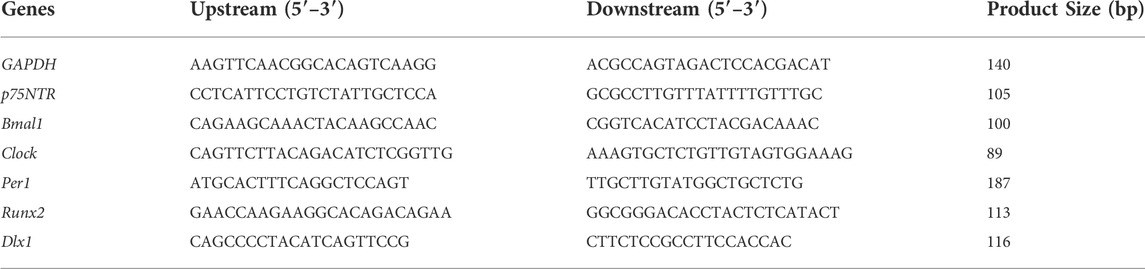
According to the manufacturer’s specifications, the total RNA was extracted using Steady Pure Universal RNA Extraction Kit II. The total RNA was determined using nanodrop spectroscopy before cDNA synthesis using Evo M-MLV Mix Kit with gDNA Clean for qPCR, with Oligo dT at 37°C for 15 min in a 20-μL reaction. Real-time RT-PCR was conducted on 0.01ug per well cDNA samples with SYBR Green PCR SuperMix (Biorad) using the CFX Connect™ Real-Time PCR Detection System (BioRad) under the following cycling conditions: 95°C for 3 min; 40 cycles of 95°C for 5 s; and 60°C for 30 s. Results were normalized relative to a housekeeping gene’s GAPDH expression. Primers were designed against the following genes: glyceraldehyde 3-phosphate dehydrogenase (GAPDH), low-affinity neurotrophin receptor p75NTR (p75NTR), MAGE family member D1 (Mage-D1), aryl hydrocarbon receptor nuclear translocator-like (ARNTL or Bmal1), clock circadian regulator (Clock), period circadian clock 1 (Per1), period circadian clock 2 (Per2), runt-related transcription factor 2 (Runx2), alkaline phosphatase (ALP), collagen type I (Col1), Msh Homeobox 1 (Msx1), distal-less homeobox 1 (Dlx1), dentin matrix acidic phosphoprotein 1 (DMP1) and dentin sialophosphoprotein (Dspp) (Tables 1, 2 enlist the primer sequences used in this study). The specificity of all primers was tested by BLAST, and the melting curve of the RT-PCR result again proved its specificity.
Statistical analyses
Following the determination of normal distribution by F-test, normally distributed data were analyzed by t test and non-normally distributed data were analyzed by Mann-Whitney test. All the data are expressed as the means ± standard deviations (S.D.). Statistical significance was assessed using the Prism 8.0 software (GraphPad Software, San Diego, CA, United States). p-values < 0.05 were considered statistically significant. All experiments were repeated thrice.
Results
Dynamic histological observation of rat tooth development
H.E. staining demonstrated that the rat dental germs entered the early cap stage at E14.5 d, the cap stage and early bell stage at E16.5 d, and the bell stage at E18.5 d (Figure 1 and Supplementary Figure S1). In the post-natal species, the morphogenesis of molar cusps was completed, and hard tissues (enamel and dentine) began to be detected at PN4 d (Figure 2 and Supplementary Figure S2). Next, the hard tissues gradually thickened at PN7 d. The tooth roots began to shape at PN10 d and PN15 d. Interestingly, the inner enamel epithelium was easy to separate from odontoblast layers in the E18.5 d species during tissue sectioning, indicating that the adhesion between odontoblasts and ameloblasts would weaken at E18.5 d. This separation between the inner enamel epithelium and enamel continued to occur in all the species in the following four post-natal stages.
Immunohistochemistry staining illustrated that Clock, Per1, and Col1 were expressed in the epithelial-mesenchymal interaction area, dental follicle, and dental papilla at E14.5 d (Figure 1 and Supplementary Figure S1). The other p75NTR, Bmal1, Cry1, and ALP factors were either little or no expressed. At E16.5 d, the expressions of Clock, Per1, and Col1 became stronger, and the expressions of p75NTR, Bmal1, and ALP were detected. All the factors were expressed at E18.5 d, but Cry1 showed the weakest expression. In the post-natal species (Figure 2 and Supplementary Figure S2), all the factors were detected and distributed in the odontoblast and ameloblast layers, and further in the dental papilla. Of the eight factors detected in this assay, Bmal1 showed the strongest expression, Per1, Clock, and p75NTR demonstrated a moderate expression, ALP and Col1 had an increased expression, and Cry1 had the weakest expression throughout.
In vitro observation of the circadian rhythm dynamics in rat EMSCs
Quantitative RT-PCR analyses demonstrated the temporal expression profiles of p75NTR, clock, mineralization-related and odontogenesis-related genes in E18.5 d EMSCs collected every 4 h during the ZT0 h–ZT48 h following serum synchronization (Figure 3). The relative expression levels of p75NTR mRNA were significantly higher at ZT4, ZT24, and ZT36-44 and were lower at ZT16, ZT32, and ZT48 in the L.L. condition. The peak times of p75NTR mRNA expression in the D.D. and L.D. conditions were similar to that in the L.L. condition. Still, the amplitudes in the D.D. condition were the biggest (the Y-axis unit in D.D. is more than twice or three times that in the L.L. and L.D. conditions). A supramaximal peak at ZT4 was also detected. The results revealed that the mRNA expression of p75NTR characterized the prominent oscillation in a day and was affected by light stimulus.
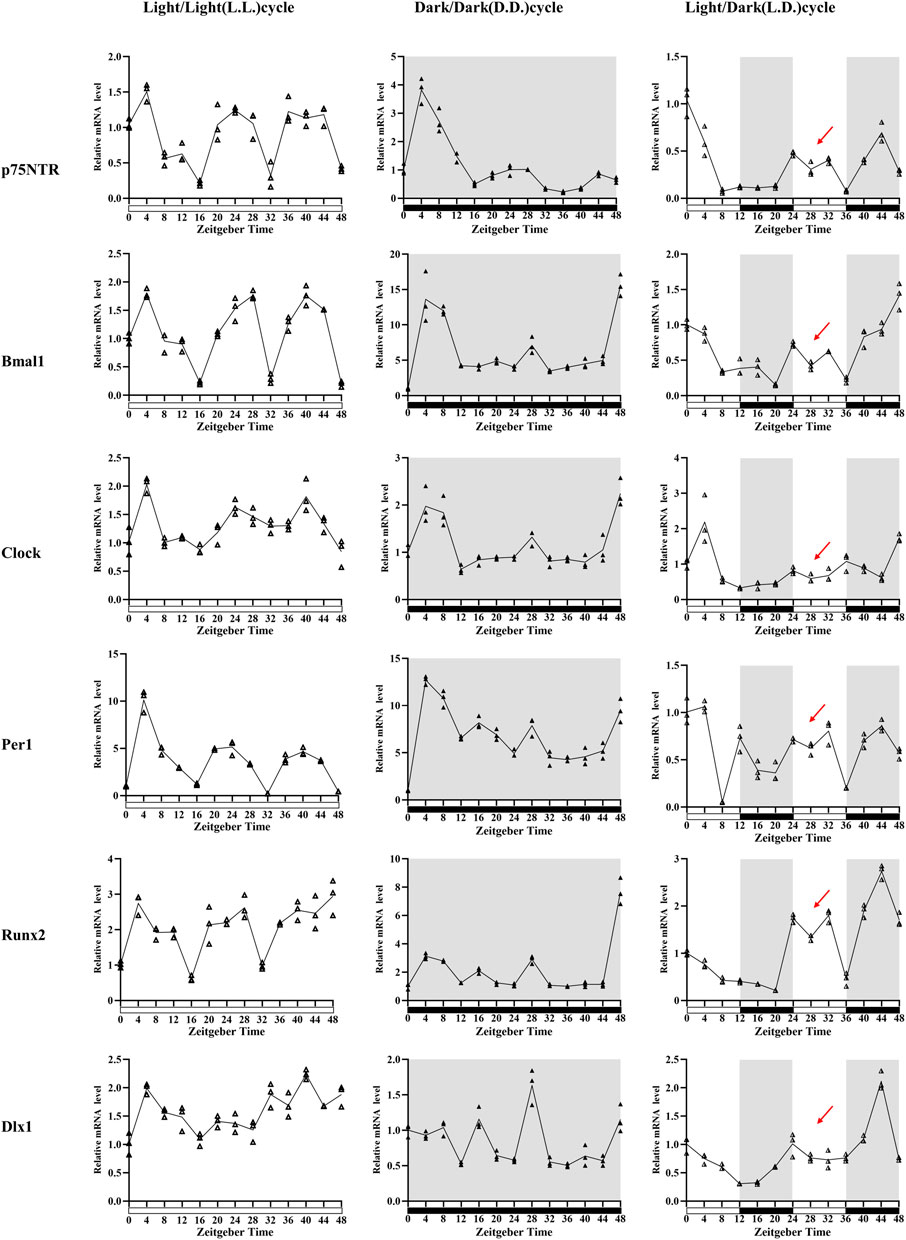
FIGURE 3. The in vitro observation of circadian rhythm dynamics in rat E18.5 d EMSCs. Quantitative RT-PCR analyses depicted that p75NTR mRNA expression patterns were similar in three conditions: higher at about ZT4, ZT24, and ZT36-44 and lower at about ZT16, ZT32, and ZT48. However, the patterns had highest amplitudes, and a supramaximal peak was detected at ZT4 in the D.D. condition. Of the three clock genes, Bmal1 and Clock showed a similar mRNA expression pattern to p75NTR in three conditions. Per1 was similar in the L.L. condition, but four, not three, peak times presented in the L.D. and D.D. conditions. The mineralization-related factors Runx2 also showed a similar mRNA expression pattern to p75NTR in L.L. and D.D. conditions, but four peak times were observed in the L.D. condition: ZT0, ZT24, ZT32, and ZT44. The odontogenesis-related factor Dlx1 showed irregular oscillating expressions: ZT4, ZT20, ZT32, and ZT40 in the L.L. condition; ZT8, ZT16, ZT28, ZT40, and ZT40 in the D.D. condition; and ZT0, ZT24, and ZT44 in the L.D. condition. Besides, the peak at ZT28 in all the six detected factors in D.D. condition, but it was replaced by a V-shape (as indicated by the red arrows) in the L.D. condition when the cells were exposed to the second 12-h light stimulus. All experiments were repeated three times independently. Data are expressed as mean ± S.D. n = 13 per group.
Of the three clock genes detected in this study, Bmal1 and Clock depicted a similar mRNA expression pattern and tendency to p75NTR in the three conditions, indicating a possible relation of p75NTR with Bmal1 and Clock in tooth development. The Per1 mRNA expression pattern in the L.L. condition was similar to that of p75NTR and showed the three peak times. However, four peak times were observed in the other two conditions. Besides, Bmal1 amplitudes were considerably higher in the D.D. condition (the Y-axis maximum unit is 20), whereas Per1 amplitudes were considerably higher in L.L. and D.D. conditions (the Y-axis maximum unit is 15). These results indicated that light stimulus would play a crucial role in the temporal expression and oscillation of clock genes in rat EMSCs.
The mineralization-related factor Runx2 revealed the three mRNA expression peaks at ZT4, ZT28, and ZT48 in both L.L. and D.D. conditions, which were similar to that of p75NTR, Bmal1, and Clock, indicating a circadian rhythm oscillation in rat EMSCs. However, four peak times were observed in the L.D. condition: ZT0, ZT24, ZT32, and ZT44. The peak times of odontogenesis-related factor Dlx1 showed irregular oscillating expressions, namely ZT4, ZT20, ZT32, and ZT40 in the L.L. condition; ZT8, ZT16, ZT28, ZT40, and ZT40 in the D.D. condition; and ZT0, ZT24, and ZT44 in the L.D. condition.
There are two variables in this experiment: light stimulus and time. The D.D. condition considerably promoted the mRNA expression of Bmal1 and Runx2, then p75NTR. In contrast, the L.L. condition significantly promoted the mRNA expression of Per1. Interestingly, a peak was found at ZT28 in all the six detected factors under the D.D. condition, but this peak became a V-shape (as indicated by the red arrows in Figure 3) under the L.D. condition when the cells were exposed to the second 12-h light stimulus. The results suggested that light stimulus affected the mRNA expression oscillation of rat EMSCs. The peak-to-peak periods of p75NTR, Bmal1, Clock, and Runx2 were maintained for about 24 h, and the peak times were nearly consistent between L.L. and D.D. conditions.
Identification of p75NTR knockout mice and the histological observation of dental germs
The genotypes of mice were determined by RT-PCR (Figure 4A). Of the six detected littermates, three with two bands of 280 bp and 345 bp were identified as heterozygous mice, two with one band of 345 bp were identified as wild-type mice, and one with one band of 280 bp was identified as the knockout mice. Immunohistochemical staining was performed on the wild-type mice and p75NTR knockout mice to further confirm the knockout of p75NTR. The results showed that p75NTR was almost not expressed in the tooth germs of the knockout mice (Figure 4C).
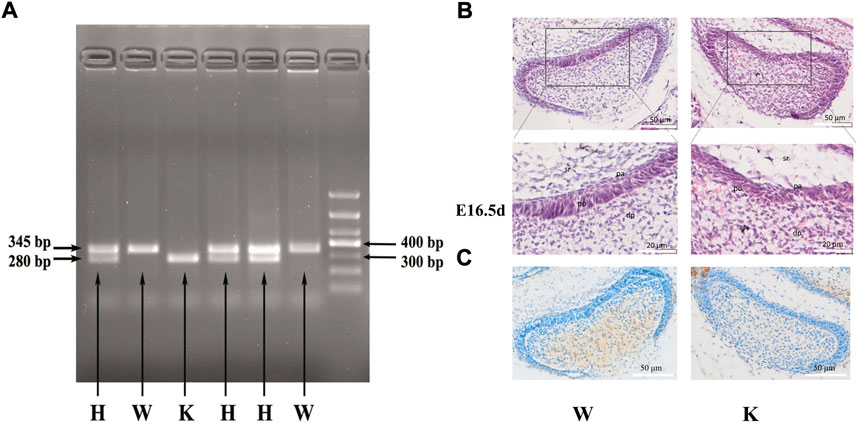
FIGURE 4. The genotype identification and dental germ morphology of p75NTR knockout mice. Of the six littermates detected, three with two bands of 280 bp and 345 bp were identified as heterozygous mice, two with one band of 345 bp were identified as wild-type mice, and one with one band of 280 bp was identified as knockout mice (A). H.E. staining revealed that the pre-odontoblast and pre-ameloblast layers tightly adhered to each other in the E16.5 d mouse dental germ (B). The long columnar pre-odontoblasts and short columnar pre-ameloblasts were regularly arranged with apparent polarity and boundary in the wild-type mice. In contrast, the regular shape, polarity, and boundary disappeared in the knockout mice. (C) Immunohistochemical staining was performed on the wild-type mice and p75NTR knockout mice. The results showed that p75NTR was almost not expressed in the tooth germs of the knockout mice. dp: dental papilla; po: pre-odontoblast; pa: pre-ameloblast; sr: stellate reticulum. The scale bar represents 50 and 20 μm, respectively. Wild-type mice, n = 3; p75NTR knockout mice, n = 2.
H.E. staining revealed that the dental germs of E16.5 d mouse were present at the early bell stage (Figure 4B). The pre-odontoblast and pre-ameloblast layers tightly adhered to each other in wild-type and knockout mice. Interestingly, long columnar pre-odontoblasts and short columnar pre-ameloblasts in wild-type mice were regularly arranged with apparent polarity and boundary. In contrast, the pre-odontoblast and pre-ameloblast layers in the knockout mice were not observed their regular shape and polarity. The boundary between pre-odontoblast and pre-ameloblast layers also disappeared. The results indicated that p75NTR might play a critical role in the shape and the polarity of odontoblasts and ameloblasts during tooth development.
In vivo observation of circadian rhythm dynamics in the dental papilla of the model mice
The results of PN7 d wild-type mice indicated that the gene expression of dental papilla was not only related to D.D. and L.D. conditions, but also related to sampling time (Figure 5). p75NTR mRNA presented a significant difference between D.D. and L.D. conditions (p < 0.05). However, the change tendencies at two sampling times were reversed. p75NTR mRNA expression in the L.D. condition was significantly higher at 7:30 a.m. and significantly lower at 7:30 p.m. than in the D.D. condition (p < 0.05). The results demonstrated that p75NTR mRNA expression fluctuated in a day and was significantly affected by light stimulus, further confirming that p75NTR might be involved in regulating the circadian rhythms of dental papilla. The expression pattern of Mage-D1 was similar to that of p75NTR. Unlike p75NTR and Mage-D1, all the four clock genes were significantly more expressed in the D.D. condition than in the L.D. condition (p < 0.05). Except for Bmal1, which showed the same big change at both two sampling times of 7:30 a.m. and 7:30 p.m., the changes of Clock, Per1, and Per2 were small at 7:30 a.m. and much larger at 7:30 p.m. The results demonstrated that the clock gene expression in dental papilla fluctuated in a day and greatly increased under the D.D. condition.
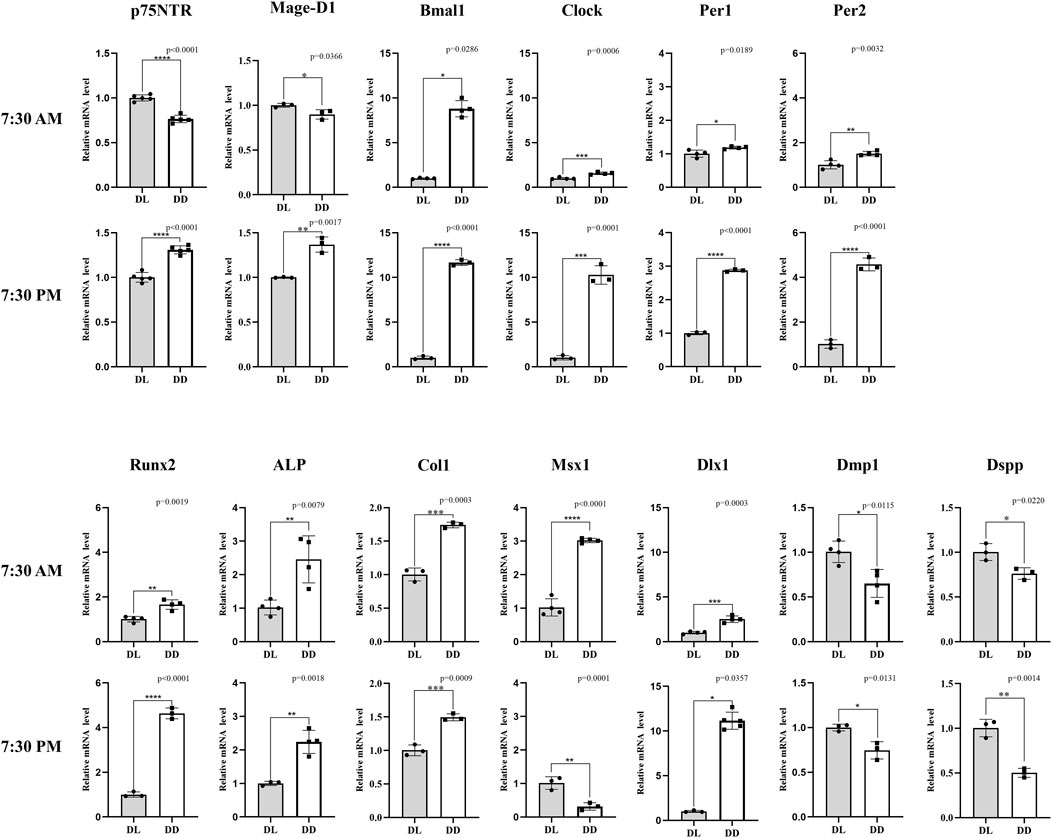
FIGURE 5. The in vivo observation of circadian rhythm dynamics in PN7 d dental germs of wild-type mice. p75NTR mRNA showed a significant difference between D.D. and L.D. conditions (p < 0.05), but the change tendencies at two sampling times were opposite: p75NTR mRNA expression in the L.D. condition was significantly higher at 7:30 a.m. and significantly lower at 7:30 p.m. in the D.D. condition (p < 0.05). The expression pattern of Mage-D1 was similar to that of p75NTR. Most factors (Bmal1, Clock, Per1, Per2, Runx2, ALP, Col1, and Dlx1) showed the same change tendency at the two sampling times: mRNA expression was significantly higher in the D.D. condition than that in the L.D. condition (p < 0.05). The change amplitudes of Clock, Per1, Per2, Runx2, and Dlx1 were significantly larger at the sampling time of 7:30 PM. Contrary to p75NTR, Msx1 mRNA expression in the L.D. condition was significantly lower at 7:30 a.m. and significantly higher at 7:30 p.m. compared with the D.D. condition (p < 0.05). Dmp1and Dspp mRNA expression in the D.D. condition was significantly lower than that in L.D. condition at both sampling times (p < 0.05). All experiments were repeated at least three times independently. Data are expressed as mean ± S.D. L. D. Condition group, n = 11; D. D. Condition group, n = 5.
The mineralization-related factors of Runx2 ,ALP, and Col1 revealed the same tendency that their mRNA expressions under the D.D. condition were significantly higher than that under the L.D. condition (p < 0.05), but the change of Runx2 between 7:30 a.m. and 7:30 p.m. was obviously bigger. All the factors of Msx1, Dlx1, Dmp1 and Dspp, which have been recognized as the key factors related to tooth development, revealed a significant mRNA expression between D.D. and L.D. conditions (p < 0.05), but the change tendencies were inconsistent. The Dlx1 mRNA expression pattern was similar to that of Clock, Per1, Per2, and Runx2. Notably, the same tendency that the Dlx1 mRNA expression under the D.D. condition was higher than that under the L.D. condition was shown at both sampling times, but the change was much more prominent at 7:30 p.m.. In contrast to Dlx1, Dmp1, and Dspp showed a reverse tendency that its mRNA expression under the D.D. condition was lower than that under the L.D. condition at both sampling times (p < 0.05). Unlike all the factors detected in this assay, the Msx1 mRNA expression in the L.D. condition was significantly lower at 7:30 a.m. but significantly higher at 7:30 p.m. than under the D.D. condition. This expression pattern was quite the opposite of p75NTR, indicating that p75NTR might be negatively related to Msx1 in the circadian rhythm of tooth development. Most of the mineralization-related and odontogenesis-related factors detected in this study revealed similar expression patterns to core clock genes, implying that the circadian rhythm would greatly affect the formation of dental hard tissues. These might be the possible reason for the formation of incremental growth lines, such as daily Retzius’s lines in enamel and von Ebner’s lines in dentine.
Effect of p75NTR knockout or over-expression on tooth circadian rhythm and mineralization
The effect of p75NTR knockout was determined by the in vivo experiment of the model mice. Figure 6 shows the mRNA expression difference between knockout and wild-type mice. As expected, p75NTR mRNA in the knockout mice was significantly lower than in wild-type mice (p < 0.05). However, the mRNA expression of Mage-D1 was not significantly reduced in the knockout mice (p > 0.05). Interestingly, of the four core clock genes detected, Bmal1 demonstrated the exact noticeable change and tendency as p75NTR, which indicated that Bmal1 mRNA expression might be closely related to p75NTR. The other three clock genes depicted the reverse tendency. Per1 mRNA was statistically higher in the knockout mice (p > 0.05). Clock and Per2 mRNA were slightly higher in the knockout mice but showed no significant difference (p > 0.05). Of the three mineralization-related factors, Runx2 mRNA expression in the knockout mice was significantly higher than in the wild-type mice (p < 0.05). In comparison, ALP and Col1 mRNA expression in the knockout mice was significantly lower than in the wild-type mice. Dlx1 showed no significant difference among the four odontogenesis-related factors (p > 0.05). Msx1, Dmp1, and Dspp were significantly lower in the knockout mice than in the wild-type mice (p < 0.05).
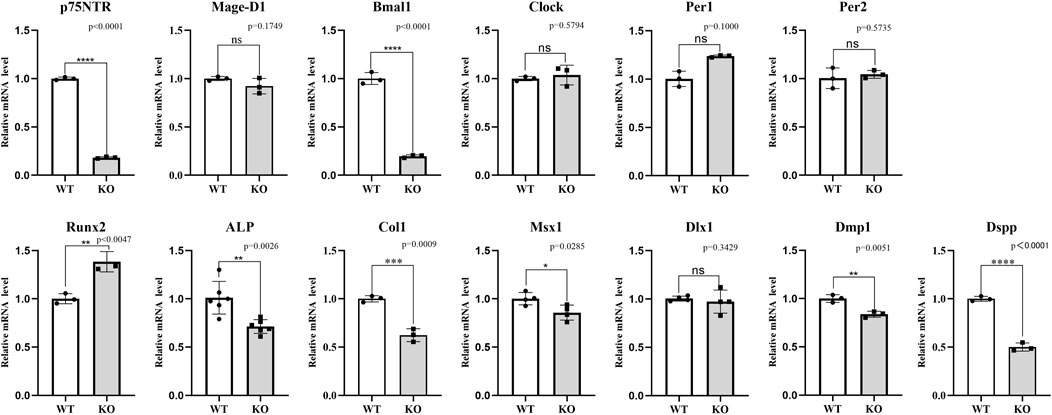
FIGURE 6. The in vivo assays for the effect of p75NTR knockout on the mRNA expression in dental germs at PN7d. As expected, p75NTR mRNA in the knockout mice was significantly lower than in THE wild-type mice (p < 0.05). Mage-D1, Bmal1, ALP, Col1, Msx1, Dmp1, and Dspp showed the same change in the p75NTR knockout mice, indicating a positive relationship with p75NTR. In contrast, Runx2 showed the reverse change, presenting a negative relationship with p75NTR. Clock, Per1, Per2, and Dlx1 showed no significant difference between p75NTR knockout and wild-type mice (p > 0.05). All experiments were repeated at least three times independently. Data are expressed as mean ± S.D. L. D. Condition group, n = 6; D. D. Condition group, n = 6.
The effect of p75NTR over-expression was determined by the in vitro experiment of iSCAP. Figure 7 shows that p75NTR mRNA expression was considerably higher in the over-expression group than in the control group (p < 0.05). Seven factors presented a significant change (p < 0.05): six positive relationships (Mage-D1, Bmal1, Clock, Runx2, Col1, and Msx1) and one negative relationships (Dspp) with p75NTR. The other five factors (Per1, Per2, ALP, Dlx1, and Dmp1) showed no significant change (p > 0.05). Compared with the results presented above, the change tendency of Bmal1, Per1, Per2, Col1, Msx1, and Dlx1 with p75NTR knockout and over-expression were consistent in the in vivo model mice and in vitro cell experiment. However, the change tendency of Runx2 and Dspp was reversed. The Mage-D1 and Clock showed no significant difference in the model mice experiment but exhibited a positive relationship with p75NTR over-expression in the cells experiment.
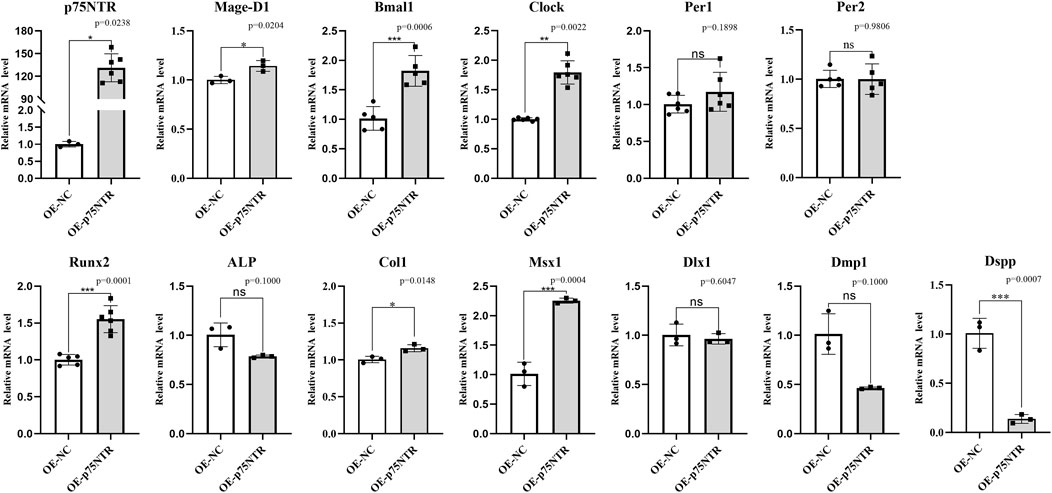
FIGURE 7. The in vitro assays for the effect of p75NTR over-expression on the mRNA expression in immortalizing stem cells from dental apical papilla (iSCAP). p75NTR was expected to be significantly higher expressed in the over-expression group than that in the control group (p < 0.05). Mage-D1, Bmal1, Clock, Runx2, Col1, and Msx1 showed an apparent positive relationship when p75NTR was significantly up-regulated. In contrast, ALP, Dmp1, and Dspp showed a negative relationship with p75NTR. Per1, Per2, ALP, Dlx1, and Dmp1 showed no significant difference between the over-expression group and control group (p > 0.05) with lower than that in wild-type mice (p < 0.05). All experiments were repeated at least three times independently. Data are expressed as mean ± S.D. Overexpression negative control group, n = 3; p75NTR over-expression group, n = 3.
Discussion
Teeth are an essential model for gaining insights into the general processes of biomineralization (Jussila and Thesleff, 2012; Bakhit et al., 2018; Pagella et al., 2020). It comprises three mineralized tissues, namely dentine, cementum, and enamel, of which the biomineralization is specific for each dental hard tissue. Moreover, they are unique in the body, involving specific proteins, such as collagen and non-collagenic matrix proteins, not found elsewhere. Therefore, the mechanism of tooth biomineralization is still unclear. The evidence exhibited regular incremental growth lines in all three dental hard tissues mentioned above, indicating that tooth biomineralization was controlled by time and is a characteristic of circadian rhythm (Zheng et al., 2011; He et al., 2019; Papakyrikos et al., 2020). In this study, the clock genes were detected to be expressed in dental germs during tooth development, and p75NTR was found to play an essential role in regulating the circadian rhythm during the formation of dental hard tissues. In order to reveal the mechanism of p75NTR in regulating clock genes in the mineralization and formation of the dental hard tissues, we used four models in this study. The model of rats was used to observe the dynamic morphogenesis of the first molar and the expression of p75NTR and clock factor protein. The model of EMSCs were used to explore the possible oscillation relationship between p75NTR and core clock factors. The p75NTR knockout mouse is a model for downregulation of p75NTR expression, and overexpression of p75NTR in iSCAP cells is a model for upregulation of p75NTR expression.
The tooth morphogenesis is triggered by the sequential and reciprocal interactions between ectomesenchyme generated from the cranial neural crest and dental epithelium (Jussila and Thesleff, 2012; Bakhit et al., 2018). The dynamic histological observation of this study revealed that the rat molar germs entered the early cap stage at E14.5 d, the cap stage and early bell stage at E16.5 d, and the bell stage at E18.5 d. The morphogenesis of molar cusps was completed at PN4 d. These results were consistent with that of previous studies (Taniguchi et al., 1999; Yang et al., 2020). Interestingly, the separations between pre-odontoblast and pre-ameloblast layers were found to occur in all the E18.5 d rat first molar, implying a significant decrease in the cell adhesion between odontoblasts and ameloblasts at this stage. This finding indicated that the cell crosstalk of epithelial-mesenchymal interactions might primarily occur during the early tooth morphogenesis. In the post-natal rat first molar, the dental hard tissues began to be detected at PN4 d, and the inner enamel epithelium was also found easy to separate from enamel. It might be the structure of dentine tubules that enhanced the adhesion of odontoblasts to dentine.
The clock genes of Bmal1, Clock, Per1, and Cry1 were detected in this study. Dynamic histological observation demonstrated that Clock and Per1 were apparently expressed in the rat dental germs at E14.5 d, Bmal1, and Cry1 began to be represented at E16.5 d. Although Cry1 was weakly expressed throughout, the other three displayed an increased expression in the following rat first molar germs. The expression’s initial times were largely consistent with the report by Zheng et al. [12] that Bmal1, Clock, Per1, and Per2 were not detected at E14 d or 15 d and began to be expressed in the mouse dental germs at E17 d. The expression distributions were completely consistent with the previous studies (Polly, 2015; Allada and Bass, 2021). The expressions of Bmal1, Clock, Per1, and Cry1 were initially detected in the pre-odontoblast and pre-ameloblast layers and then detected in the dental papilla cells, stratum intermedium, and stellate reticulum. These results indicated that the clock genes might participate in the complex epithelial-mesenchymal crosstalk networks and mainly regulate dental hard tissue formation during tooth development (Tao et al., 2016; Pincha et al., 2022).
p75NTR, a well-conserved transmembrane neurotrophin receptor, was shown to play a critical role in tooth morphogenesis and mineralization in the previous studies by this study’s researchers (Wen et al., 2012; Li et al., 2017; Wang et al., 2020; Zhao et al., 2020). Baeza-Raja et al. (2013) reported that p75NTR expression oscillated via the direct binding of Clock/Bmal1 to noncanonical E-box elements present in the p75NTR promoter. p75NTR might be a novel clock gene–regulating oscillatory components of circadian rhythms. Therefore, p75NTR was selected and investigated in this study to determine its potential role in the circadian rhythm and incremental growth line formation during tooth development. The dynamic expression of p75NTR was similar to that of clock genes during the development of rat dental germs, indicating its relationship with the circadian rhythm. Moreover, p75NTR knockout mice exhibited the disorder of pre-odontoblast/pre-ameloblast arrangement and morphology shape and even the disappearance of boundaries between pre-odontoblast and pre-ameloblast layers. There is another possibility that these phenotypes are delayed because p75NTR is knocked out. In general, this finding indicated that p75NTR might regulate cell polarity during tooth development. Cell polarity is critical in cellular processes ranging from cell migration to asymmetric cell division and axon and dendrite specification (Piroli et al., 2019). p75NTR was reported to directly interact with the polarity protein Par-3 and recruited to regulate the axon-glial junction, forming a complex that points to a critical role in establishing the cell polarity for myelination (Chan et al., 2006). As a transmembrane receptor, p75NTR might participate in the odontoblast-ameloblast junction and cell polarity establishment during tooth morphogenesis.
The ablation of SCN was reported to result in a disrupted patterning of rat’s incremental lines in dentine and supported the involvement of the circadian clock in tooth development (Ohtsuka-Isoya et al., 2001). The opinion was confirmed by the report that Bmal1−/− mice showed the fainter daily lines in dentine than Bmal1+/+ and Bmal1+/− mice (He et al., 2019). The phenomenon of incremental lines might be related to the disorder of pre-odontoblast/pre-ameloblast polarity and the arrangement found in p75NTRExIII−/− mice of this study. p75NTR was speculated to play a crucial role in regulating clock genes during the formation of dental hard tissues. The recent research certainly supported this speculation that the incisors’ daily mineralization speed and incremental growth line width were significantly lower in p75NTRExIII−/− mice than in p75NTRExIII+/+ mice (Wang et al., 2020; Zhao et al., 2020). Importantly, further studies are needed to reveal the signaling networks under this process.
Previous studies reported the detection of clock genes in dental germs and depicted a regular oscillation expression pattern, indicating that biological clocks affect tooth development (Nirvani et al., 2017; Papakyrikos et al., 2020; Huang et al., 2021). The 48-h circadian rhythm dynamics in rat EMSCs showed that the clock genes Bmal1, Clock, and Per1 presented a regular oscillation in mRNA expression. A similar oscillation was observed in p75NTR and Runx2 mRNA expression but not in Dlx1 mRNA expression, indicating that p75NTR and mineralization-related factor Runx2 might be involved in the circadian rhythm of tooth development. Previous studies support this finding that clock genes (Bmal1, Clock, Per1, and Per2) and two markers of ameloblast differentiation (amelogenin and kallikrein-related peptidase 4) have regular oscillations in ameloblasts (Zheng et al., 2013; Huang et al., 2021). Moreover, this oscillation was affected by L.D. stimulus. The D.D. condition significantly increased the mRNA expression of Bmal1, p75NTR, and Runx2 in this study. The light stimulus disturbed the oscillation peaks of all the detected factors. These results were confirmed by the in vivo experiment in this study. The clock genes (Bmal1, Clock, Per1, and Per2), mineralization-related factors (Runx2, ALP, and Col1), and odontogenesis-related (Dlx1) in dental germs presented a similar tendency that the mRNA expression was considerably higher in the D.D. condition than in L.D. Moreover, this change was much more prominent in most of them at sampling times of 7:30 p.m. These results indicated that the D.D. condition promoted the dental mineralization, which was consistent with the previous reports that the mineralization of dental hard tissues increased in the night during the day (Lacruz et al., 2012; Satou et al., 2019).
The data in this study further confirm that circadian rhythms are involved in tooth development. However, the molecular mechanisms of the effects of clock genes in tooth development and incremental growth lines are unclear. p75NTR was reported to directly bond to the Clock/Bmal1 heterodimer via the E-box element and participated in regulating circadian rhythms (Baeza-Raja et al., 2013). In a recent study by Zhao et al. (2020) the calcein fluorescence assay showed that the distance between the calcein fluorescence bands was significantly lower than that in wild-type and heterozygous mice, indicating that p75NTR would regulate the daily mineralization speed and incremental growth line width during tooth development. It was also proposed that p75NTR might participate in regulating circadian rhythm during dental incremental line formation and Mage-D1 might play an underlying role in this process. In this study, Mage-D1 expression was positively correlated with p75NTR except that Mage-D1 was not significantly reduced with p75NTR knockout, implying that Mage-D1 expression is not completely regulated by p75NTR. Wang et al. (2020) believed that p75NTR can also regulate tooth mineralization via enhancing the PI3K/Akt/β-catenin pathway, further illustrating that Mage-D1 is not the only bridge factor for p75NTR to regulate tooth periodic development and rhythmic mineralization. To further reveal its effects and mechanisms in circadian rhythms during tooth development, p75NTR knockout or over-expression was performed in this study. Bmal1 and Msx1 showed a positive relationship with p75NTR in both in vivo model mice and in vitro cell experiments. Per1, Per2, and Dlx1 showed no significant change when p75NTR was up- and down-expressed. In the cell experiment, the Mage-D1 and Clock was positively related with p75NTR but uncorrelated with p75NTR in the model mice. Interestingly, Runx2 was positively related to p75NTR in the cell experiment and negatively related in the model mice. This contradictory conclusion is attributed to the diverse biological functions of p75NTR and complex signal regulation mechanism. Because of the formation of bone tissue, another process of mineral deposition in vivo, our previous research showed that the mineralized development of mouse femur was inhibited after p75NTR knock (Zhao et al., 2020). However, Wang et al. (2020) found that loss of P75NTR upregulated Runx2 expression, thereby promoting BMSCs mineralization in vitro. Thus, the signaling network related to the regulation of biological mineralization by p75NTR is more complex than initially understood, and further research is still needed in the future. In contrast, ALP, Dmp1, and Dspp were negatively related to p75NTR in the cell experiment and in model mice. These reverse tendencies might be the different cell niches between in vitro cell experiments and in vivo model mice. The data in this study indicated that p75NTR might participate in regulating circadian rhythms during tooth development via clock genes Bmal1 and Clock, especially in mineralization and dental hard tissue formation. This is consistent with the previous studies that the clock gene Bmal1 was involved in the up-regulation of mineralization in mouse bone marrow stromal cells (Chen et al., 2012; Samsa et al., 2016). Fu et al. (2005) and Min et al. (2016) also reported that Bmal1 promoted the expression of mineralization-related factors Runx2 and OCN, which was inhibited by Per1/Per2 and Cry1/Cry2. Therefore, it was speculated that the core circadian regulator of negative TTFL was involved in cell mineralization, and p75NTR might be crucial in this process.
Conclusion
The clock genes Bmal1, Clock, Per1, and Per2 were all detected in tooth germs before the formation of dental hard tissues and showed a regular oscillating expression pattern in EMSCs from dental germs. Their expression was affected by L.D. stimulus, and most of them were promoted by D.D. conditions. p75NTR showed a close relationship with majority of the clock genes, mineralization-related and odontogenesis-related factors detected in this study, such as Bmal1, Runx2, ALP, Msx1, and Dmp1. It was speculated that p75NTR might participate in cell mineralization and dental hard tissue formation via the clock genes Bmal1 and Clock. Moreover, p75NTR plays a potential role in the odontoblast-ameloblast junction and cell polarity establishment during tooth morphogenesis. Of course, the limitations are also present in this study. Some results in the in vitro experiment did not match and were even contradictory to those in the in vivo model mice. For example, Runx2 was positively related to p75NTR in the in vivo experiment and negatively related to p75NTR in the in vivo model mice. The in vivo cell niche and the signaling networks regulating tooth development are more complex than initially understood. Further studies are still needed in the future.
Data availability statement
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by The Medical Ethics Committee of the Chongqing Medical University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
All authors contributed to the study concepts and design. HY, BX, XY, and CL carried out in vivo experiments on p75NTR knockout mice. HY, BX, ML, and YZ carried out in vitro experiments on ectomesenchymal stem cells. HY, CL, and XZ carried out H.E. and immunohistochemistry staining. HY, BX collected the data. HY, BX, MZ, and XW performed the analysis. HY, BX, MZ, and XW drafted the manuscript. All authors reviewed and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81970906; 82000997) and Shenzhen Science and Technology Program (JCYJ20180306174437246).
Acknowledgments
Thanks to the platform and help provided by Chongqing Key Laboratory of oral diseases and biomedicine.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.981311/full#supplementary-material
References
Allada R., Bass J. (2021). Circadian mechanisms in medicine. N. Engl. J. Med. 384, 550–561. doi:10.1056/NEJMra1802337
Antoine D., Hillson S., Dean M. C. (2009). The developmental clock of dental enamel: A test for the periodicity of prism cross-striations in modern humans and an evaluation of the most likely sources of error in histological studies of this kind. J. Anat. 214, 45–55. doi:10.1111/j.1469-7580.2008.01010.x
Baeza-Raja B., Eckel-Mahan K., Zhang L., Vagena E., Tsigelny I. F., Sassone-Corsi P., et al. (2013). p75 neurotrophin receptor is a clock gene that regulates oscillatory components of circadian and metabolic networks. J. Neurosci. 33, 10221–10234. doi:10.1523/JNEUROSCI.2757-12.2013
Bakhit A., Kawashima N., Hashimoto K., Noda S., Nara K., Kuramoto M., et al. (2018). Strontium ranelate promotes odonto-/osteogenic differentiation/mineralization of dental papillae cells in vitro and mineralized tissue formation of the dental pulp in vivo. Sci. Rep. 8, 9224. doi:10.1038/s41598-018-27461-7
Brown S. A., Kowalska E., Dallmann R. (2012). (Re)inventing the circadian feedback loop. Dev. Cell 22, 477–487. doi:10.1016/j.devcel.2012.02.007
Chan J. R., Jolicoeur C., Yamauchi J., Elliott J., Fawcett J. P., Ng B. K., et al. (2006). The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science 314, 832–836. doi:10.1126/science.1134069
Chen Y., Xu X., Tan Z., Ye C., Zhao Q. (2012). Age-related BMAL1 change affects mouse bone marrow stromal cell proliferation and osteo-differentiation potential. Arch. Med. Sci. 8, 30–38. doi:10.5114/aoms.2012.27277
Fu L., Patel M. S., Bradley A., Wagner E. F., Karsenty G. (2005). The molecular clock mediates leptin-regulated bone formation. Cell 122, 803–815. doi:10.1016/j.cell.2005.06.028
Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., et al. (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569. doi:10.1126/science.280.5369.1564
He L., Hao Y., Zhen L., Liu H., Shao M., Xu X., et al. (2019). Biomineralization of dentin. J. Struct. Biol. 207, 115–122. doi:10.1016/j.jsb.2019.05.010
Huang W., Zheng X., Yang M., Li R., Song Y. (2021). PER2-mediated ameloblast differentiation via PPARγ/AKT1/β-catenin axis. Int. J. Oral Sci. 13, 16. doi:10.1038/s41368-021-00123-7
Jussila M., Thesleff I. (2012). Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 4, a008425. doi:10.1101/cshperspect.a008425
Lacruz R. S., Hacia J. G., Bromage T. G., Boyde A., Lei Y., Xu Y., et al. (2012). The circadian clock modulates enamel development. J. Biol. Rhythms 27, 237–245. doi:10.1177/0748730412442830
Lee K. F., Li E., Huber L. J., Landis S. C., Sharpe A. H., Chao M. V., et al. (1992). Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 69, 737–749. doi:10.1016/0092-8674(92)90286-l
Li G., Liu J., Wang Y., Yang K., Zhao M., Xiao Y., et al. (2017). LNGFR targets the Wnt/β-catenin pathway and promotes the osteogenic differentiation in rat ectomesenchymal stem cells. Sci. Rep. 7, 11021. doi:10.1038/s41598-017-11555-9
Lopez Franco G. E., Huang A., Stone D. S., Blank R. D. (2006). Increased Young's modulus and hardness of Col1a2oim dentin. J. Dent. Res. 85, 1032–1036. doi:10.1177/154405910608501111
Min H. Y., Kim K. M., Wee G., Kim E. J., Jang W. G. (2016). Bmal1 induces osteoblast differentiation via regulation of BMP2 expression in MC3T3-E1 cells. Life Sci. 162, 41–46. doi:10.1016/j.lfs.2016.08.002
Neumann A. M., Schmidt C. X., Brockmann R. M., Oster H. (2019). Circadian regulation of endocrine systems. Auton. Neurosci. 216, 1–8. doi:10.1016/j.autneu.2018.10.001
Nirvani M., Khuu C., Utheim T. P., Hollingen H. S., Amundsen S. F., Sand L. P., et al. (2017). Circadian rhythms and gene expression during mouse molar tooth development. Acta Odontol. Scand. 75, 144–153. doi:10.1080/00016357.2016.1271999
Ohtsuka-Isoya M., Hayashi H., Shinoda H. (2001). Effect of suprachiasmatic nucleus lesion on circadian dentin increment in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280, R1364–R1370. doi:10.1152/ajpregu.2001.280.5.R1364
Okawa H., Egusa H., Nishimura I. (2020). Implications of the circadian clock in implant dentistry. Dent. Mat. J. 39, 173–180. doi:10.4012/dmj.2019-291
Pagella P., Porcheri C., Mitsiadis T. A. (2020). Exploiting teeth as a model to study basic features of signaling pathways. Biochem. Soc. Trans. 48, 2729–2742. doi:10.1042/BST20200514
Papakyrikos A. M., Arora M., Austin C., Boughner J. C., Capellini T. D., Dingwall H. L., et al. (2020). Biological clocks and incremental growth line formation in dentine. J. Anat. 237, 367–378. doi:10.1111/joa.13198
Pincha N., Marangoni P., Haque A., Klein O. D. (2022). Parallels in signaling between development and regeneration in ectodermal organs. Curr. Top. Dev. Biol. 149, 373–419. doi:10.1016/bs.ctdb.2022.02.006
Piroli M. E., Blanchette J. O., Jabbarzadeh E. (2019). Polarity as a physiological modulator of cell function. Front. Biosci. 24, 451–462. doi:10.2741/4728
Polly P. D. (2015). Gene networks, occlusal clocks, and functional patches: New understanding of pattern and process in the evolution of the dentition. Odontology 103, 117–125. doi:10.1007/s10266-015-0208-3
Richards J., Gumz M. L. (2013). Mechanism of the circadian clock in physiology. Am. J. Physiol. Regul. Integr. Comp. Physiol. 304, R1053–R1064. doi:10.1152/ajpregu.00066.2013
Samsa W. E., Vasanji A., Midura R. J., Kondratov R. V. (2016). Deficiency of circadian clock protein BMAL1 in mice results in a low bone mass phenotype. Bone 84, 194–203. doi:10.1016/j.bone.2016.01.006
Satou R., Shibukawa Y., Kimura M., Sugihara N. (2019). Light conditions affect rhythmic expression of aquaporin 5 and anoctamin 1 in rat submandibular glands. Heliyon 5, e02792. doi:10.1016/j.heliyon.2019.e02792
Shearman L. P., Sriram S., Weaver D. R., Maywood E. S., Chaves I., Zheng B., et al. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288, 1013–1019. doi:10.1126/science.288.5468.1013
Taniguchi K., Okamura K., HayashiM. , Funakoshi T., MotokaWa W. (1999). The effect of mechanical trauma on the tooth germ of rat molars at various developmental stages: A histopathological study. Endod. Dent. Traumatol. 15, 17–25. doi:10.1111/j.1600-9657.1999.tb00743.x
Tao J., Zhai Y., Park H., Han J., Dong J., Xie M., et al. (2016). Circadian rhythm regulates development of enamel in mouse mandibular first molar. PLoS One 11, e0159946. doi:10.1371/journal.pone.0159946
Wang J., Zhang H., Zhang W., Huang E., Wang N., Wu N., et al. (2014). Bone morphogenetic protein-9 effectively induces osteo/odontoblastic differentiation of the reversibly immortalized stem cells of dental apical papilla. Stem Cells Dev. 23, 1405–1416. doi:10.1089/scd.2013.0580
Wang Y., Yang K., Li G., Liu R., Liu J., Li J., et al. (2020). p75NTR(-/-) mice exhibit an alveolar bone loss phenotype and inhibited PI3K/Akt/β-catenin pathway. Cell Prolif. 53, e12800. doi:10.1111/cpr.12800
Weaver D. R. (1998). The suprachiasmatic nucleus: A 25-year retrospective. J. Biol. Rhythms 13, 100–112. doi:10.1177/074873098128999952
Wen X., Liu L., Deng M., Zhang L., Liu R., Xing Y., et al. (2012). Characterization of p75(+) ectomesenchymal stem cells from rat embryonic facial process tissue. Biochem. Biophys. Res. Commun. 427, 5–10. doi:10.1016/j.bbrc.2012.08.109
Yang J., Lu X., Liu S., Zhao S. (2020). The involvement of genes related to bile secretion pathway in rat tooth germ development. J. Mol. Histol. 51, 99–107. doi:10.1007/s10735-020-09861-0
Zhang L., Chen P., Chen L., Weng T., Zhang S., Zhou X., et al. (2015). Inhibited Wnt signaling causes age-dependent abnormalities in the bone matrix mineralization in the Apert syndrome FGFR2(S252W/+) mice. PLoS One 10, e112716. doi:10.1371/journal.pone.0112716
Zhao M., Wang Y., Li G., Li J., Yang K., Liu C., et al. (2020). The role and potential mechanism of p75NTR in mineralization via in vivo p75NTR knockout mice and in vitro ectomesenchymal stem cells. Cell Prolif. 53, e12758. doi:10.1111/cpr.12758
Zheng L., Papagerakis S., Schnell S. D., Hoogerwerf W. A., Papagerakis P. (2011). Expression of clock proteins in developing tooth. Gene Expr. Patterns 11, 202–206. doi:10.1016/j.gep.2010.12.002
Keywords: p75 neurotrophin receptor, circadian rhythm, mineralization, cell polarity, tooth development
Citation: Yuan H, Xie B, Yu X, Lin C, Li M, Zhang Y, Zou X, Lu M, Zhao M and Wen X (2022) A potential role of p75NTR in the regulation of circadian rhythm and incremental growth lines during tooth development. Front. Physiol. 13:981311. doi: 10.3389/fphys.2022.981311
Received: 29 June 2022; Accepted: 09 September 2022;
Published: 23 September 2022.
Edited by:
Ruifeng Ray Cao, University of Minnesota Twin Cities, United StatesReviewed by:
Michel Goldberg, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceJuliane Isaac, Université Paris Cité, France
Copyright © 2022 Yuan, Xie, Yu, Lin, Li, Zhang, Zou, Lu, Zhao and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiujie Wen, wenxiujie@tom.com; Manzhu Zhao, 501252@hospital.cqmu.edu.cn
†These authors have contributed equally to this work
 Hongyan Yuan
Hongyan Yuan Bo Xie
Bo Xie Xia Yu1
Xia Yu1