- 1Key Laboratory of Beijing for Identification and Safety Evaluation of Chinese Medicine, Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 2College of Pharmacy, Hubei University of Chinese Medicine, Wuhan, China
- 3Institute of Disease Control and Prevention, Academy of Military Medical Sciences, Beijing, China
Mu-tong (Akebiae Caulis) is a traditional Chinese medicine commonly used as a diuretic and antiphlogistic. A common adulterant of Mu-tong is Guan-mu-tong (Aristolochiae Manshuriensis Caulis), which is derived from the stem of Aristolochia manshuriensis Komarov, and contains carcinogenic aristolochic acids. We used an alternative technique, loop-mediated isothermal amplification (LAMP), to differentiate Mu-tong from Guan-mu-tong because LAMP is quick, highly sensitive, and specific. We designed a set of four common primers (G-F3, G-B3, G-FIP, and G-BIP) and a loop primer (G-LB) for LAMP based on the internal transcribed spacer 2 sequence of Ar. manshuriensis. We successfully amplified the LAMP assays and visual detection occurred within 60 min at isothermal conditions of 65°C. The LAMP reaction exhibited a tenfold increase in detection (4.22 pg/μl DNA) over conventional polymerase chain reaction demonstrating that LAMP is a useful technique to detect Guan-mu-tong. We conclude that the LAMP technique is a potentially valuable safety control method for simple and efficient discrimination of Mu-tong from its adulterant Guan-mu-tong.
Introduction
The use of traditional herbal medicine as dietary supplements has become increasingly popular in many countries due to the consumer’s belief that herbal medicine is both natural and harmless. In order to establish proper regulatory oversight for consumer safety, a procedure is needed that not only identifies the herbal plant in question, but also detects possible adulterants. One such case is that of Mu-tong (Akebiae Caulis), a common traditional Chinese medicine used as a diuretic and antiphlogistic (Kawata et al., 2007) which may be adulterated by an aristolochic acid containing herb, Guan-mu-tong (Aristolochiae Manshuriensis Caulis) derived from the stem of Aristolochia manshuriensis Komarov (Aristolochiaceae) (But and Ma, 1999; Lord et al., 1999; Ma and Zhang, 2002). Guan-mu-tong contains nephrotoxic and carcinogenic aristolochic acids (AAs), which cause aristolochic acid nephropathy (AAN) and upper tract urothelial carcinomas (UTUC) (Grollman et al., 2007; Hoang et al., 2013; Poon et al., 2013), with the inadvertent use of adulterated Guan-mu-tong by consumers leading to possible renal failure (But and Ma, 1999; Lord et al., 1999).
According to the Pharmacopoeia of the People’s Republic of China 2015 (State Pharmacopoeia Committee, 2015), Mu-tong comes from the stems of Akebia quinata (Houtt.) Decne., Ak. trifoliata (Thunb.) Koidz. and Ak. trifoliata (Thunb.) Koidz. var. australis (Diels) T. Shimizu. Not only is Mu-tong believed to have diuretic and anti-inflammatory properties, but it is also commonly used for weight loss and one species of Mu-tong (Ak. quinata) is an important ingredient used in a traditional Korean medicine. With seemingly similar common names, Mu-tong and Guan-mu-tong are commonly confused, and adding to the confusion is the post-harvest process, which makes proper identification of the dried stems difficult despite differing morphological characteristics of the plants (Figure 1). Currently, high performance liquid chromatography (HPLC) and HPLC–MS are able to differentiate between Aristolochiaceae plant material and non-Aristolochiaceae material through the analysis of aristolochic acid I, II, and other analogs (Hashimoto et al., 1999; Wei et al., 2005). However, both the HPLC and HPLC–MS methods may be hindered by climatic and altitudinal effects, which can cause chemical constituent changes. In another approach, DNA sequencing and DNA barcoding approaches are used for authentication of Aristolochiaceae and non-Aristolochiaceae plant material, but these procedures are time consuming (Han et al., 2005; Li et al., 2014; Wu et al., 2015). Due to these limitations, a rapid and effective identification method is needed for detecting Guan-mu-tong, in adulterated samples of Mu-tong.
Loop-mediated isothermal amplification (LAMP) is a quick, yet effective technique for amplifying DNA under isothermal conditions, which shows both high sensitivity and specificity. Due to LAMP’s simplicity, ease of use, prompt results, and straight-forward visual interpretation (Notomi et al., 2000; Mori et al., 2001; Nagamine et al., 2001; Mori and Notomi, 2009), this technique has been used for diagnosis of viral diseases (Mansour et al., 2015; Mongan et al., 2015; Nyan and Swinson, 2016; Venkatesan et al., 2016). In addition, LAMP has been used for the detection of: fungi (Niessen, 2015), milk pathogens (Cornelissen et al., 2016), tumors (Horibe et al., 2007), and recently herbal medicine adulterants. For example, the RT-LAMP assay was able to detect eastern or western strains of bluetongue virus in India in 60–90 min with the sensitivity equal to real-time RT-PCR but greater than conventional RT-PCR (Maan et al., 2016). In another example, the LAMP assay was used to rapidly and precisely discriminate the feces of sika deer (Cervus nippon) from Japanese serow (Capricornis crispus) within 60 min at 63°C (Aikawa et al., 2015). Furthermore, Bosward et al. (2016) reported a sensitivity of 20 pg of extracted DNA/reaction for a LAMP assay designed to detect Streptococcus agalactiae in bovine milk. Although previous studies have demonstrated the successful applicability of the LAMP technique in authenticating traditional Chinese medicinal materials such as: Taraxacum formosanum (Lai et al., 2015), Catharanthus roseus (Chaudhary et al., 2012), Panax ginseng (Sasaki et al., 2008), and Hedyotis diffusa (Li et al., 2013); to our knowledge, LAMP has not been applied for discriminating between Mu-tong and its toxic adulterant Guan-mu-tong. DNA barcoding is becoming a universal molecular technique used for effective species identification. Currently, due to high species resolution and moderate sequence length, the internal transcribed spacer 2 (ITS2) region has been widely used to authenticate plant species, especially medicinal plants (Lv et al., 2015; Xin et al., 2015; Zhao et al., 2015; Michel et al., 2016). Therefore, the ITS2 sequence is a suitable target-gene choice for designing primers for the LAMP assay. The aim of this study is to develop a sensitive and specific LAMP method based on the ITS2 sequence to rapidly and accurately identify Mu-tong and its toxic adulterant Guan-mu-tong.
Materials and Methods
Plant Materials
We collected three samples of each Mu-tong species (Ak. quinata, Ak. trifoliata and Ak. trifoliata var. australis) as well as three samples of the adulterant Guan-mu-tong (Ar. manshuriensis) for testing. The samples were collected from Jilin, Shanxi and Beijing (Table 1) and were identified by Junlin Yu at Tonghua Normal University, College of Pharmaceutical and Food Science and also Wei Sun at the Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences. All materials were stored in the Herbarium of the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Beijing, China.
DNA Isolation and Quantification
We used approximately 30 mg of fresh leaves or stems for DNA isolation utilizing the Plant Genomic DNA Kit (Tiangen Biotech Co., China) and we determined total DNA concentrations using Qubit® 2.0 (Life Tech, Invitrogen, USA). We validated the identification of all plant material specimens using PCR-based sequences.
Primer Design
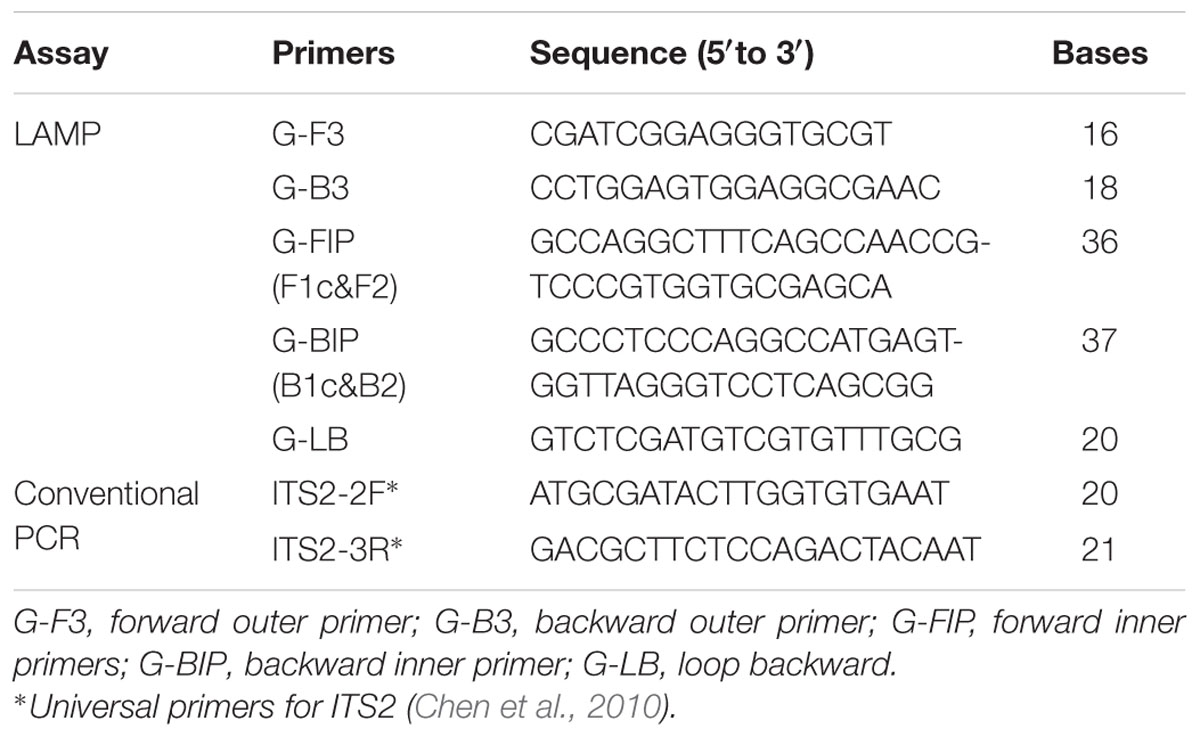
DNA barcoding is a useful and effective tool, which enables the authentication of plant species accurately by using the ITS2 sequence, which is considered suitable for the identification of medicinal plants. We obtained ITS2 sequences of Ar. manshuriensis, Ak. quinata, Ak. trifoliata and Ak. trifoliata var. australis using DNA barcoding methods and based on these sequences, we designed LAMP primers for Ar. manshuriensis. The sequence alignment was carried out by MUSCLE in software MEGA 5.0. The variation of the ITS2 sequence within Ar. manshuriensis, Ak. quinata, Ak. trifoliata and Ak. trifoliata var. australis was aligned as illustrated in Figure 2. By utilizing PrimerExplorerV41, we designed four common primers (G-F3, G-B3, G-FIP, and G-BIP) and a loop primer (G-LB) from ITS2 sequence of Ar. manshuriensis and are as follows: the forward outer primer G-F3 (5′-CGATCGGAGGGTGCGT-3′), backward outer primer G-B3 (5′-CCTGGAGTGGAGGCGAAC-3′), forward inner primers G-FIP (5′-GCCAGGCTTTCAGCCAACCGTCCCGTGGTGCGAGCA-3′), backward inner primer G-BIP (5′-GCCCTCCCAGGCCATGAGTGGTTAGGGTCCTCAGCGG-3′) and loop backward G-LB (5′-GTCTCGATGTCGTGTTTGCG-3′) (Table 2).
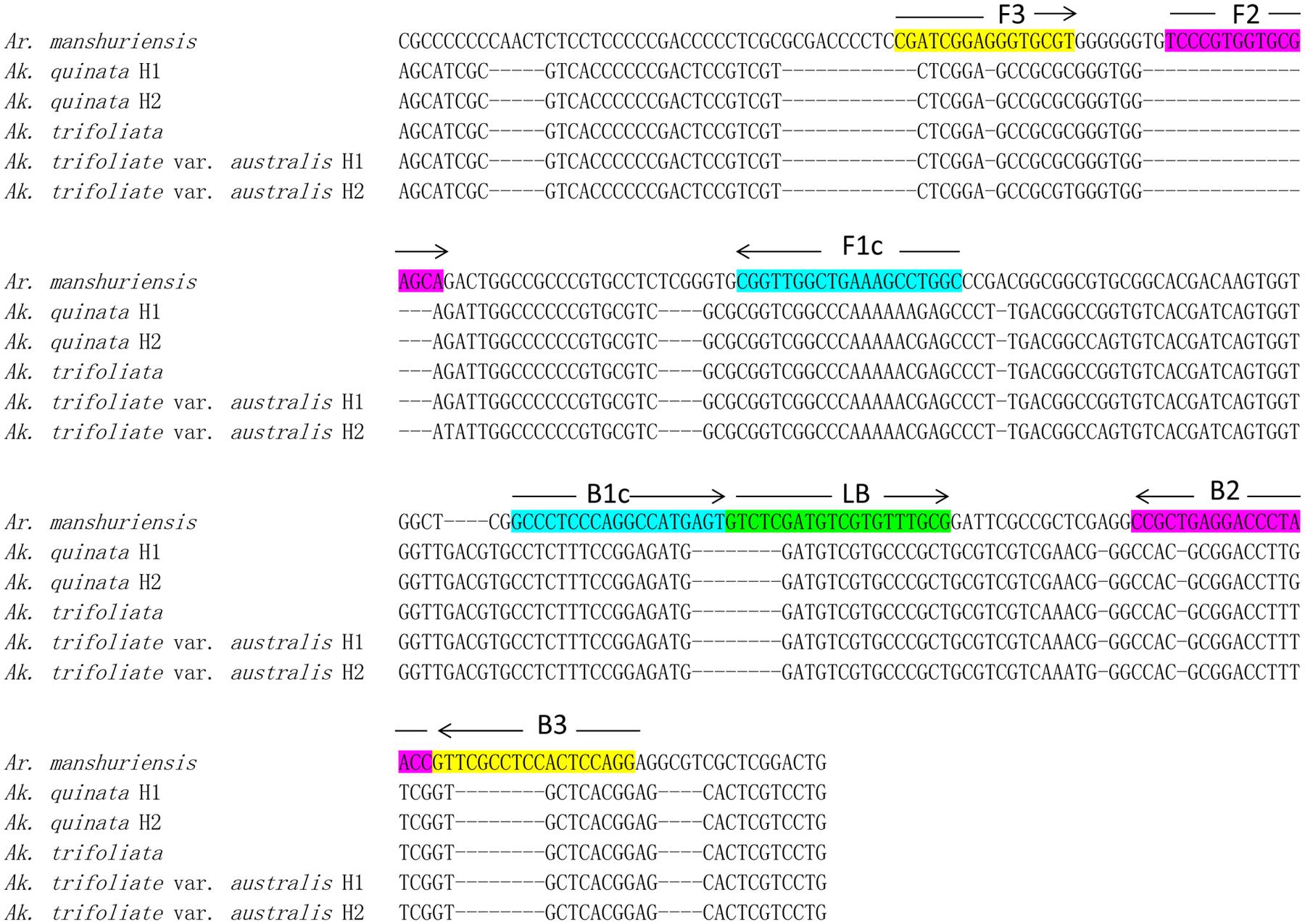
FIGURE 2. Sequence alignment of the internal transcribed spacer 2 (ITS2) of Aristolochiae Manshuriensis Caulis and Akebiae Caulis. Hyphens (-) denote alignment gaps. The specific primer and loop primer were designed from seven loci (F3, F2, FIc, B1c, B2, B3, and LB) as indicated in color boxes. Arrows represent the direction of 5′-3′ amplification in loop-mediated isothermal amplification (LAMP). H1 and H2 indicate different haplotypes.
LAMP Reaction
All LAMP reactions occurred in a 25 μl mixture (DNA amplification kit; Loopamp, Eiken China Co., Ltd., Shanghai, China), which contained 2 × Reaction Mix, 1 μl Bst DNA polymerase, 0.2 μM of each outer primer (F3 and B3), 1.6 μM of each inner primer (FIP and BIP), 0.8 μM of loop primer (LB), 1 μl DNA template. We performed the LAMP reaction in an isothermal amplification system at 65°C for 60 min followed by inactivation at 80°C for 5 min. Additionally, EvaGreen fluorescent dye (Biotium, Hayward, CA) (20 × in water) was added to give a final concentration of 0.5×, when the LAMP reaction was carried out in a real-time PCR machine (Rotor-Gene Q, QIAGEN, Germany).
Detection of LAMP Products
We utilized three different methods to detect LAMP products. First, we monitored real-time amplification of LAMP assays using a Loopamp real-time turbidimeter (LA-230; Eiken Chemical Co., Ltd., Tochigi, Japan) to monitor turbidity by recording the optical density at 400 nm every 6 s. Second, we employed visual inspection where we observed color changes in the LAMP reaction mixtures, caused by the addition of 1 μl of calcein (Fluorescence Detection Reagent; Loopamp, Eiken China Co., Ltd., Shanghai, China) before the LAMP reaction. Third, we monitored fluorescence of the EvaGreen dye in a Rotor-Gene Q. The reaction mixtures were conducted at 65°C for 60 min. Real-time fluorescence data was acquired every 60 s (a total of 60 cycles) on the Green channel (excitation at 470 nm and detection at 510 nm), followed by melting curve analysis from 65 to 99°C using 1°C steps. Real-time LAMP results were analyzed by Tt (time threshold) values.
Specificity and Sensitivity of LAMP
The samples of Ar. manshuriensis (Guan-mu-tong) were used to assess the specificity of the LAMP test, which we conducted by preparing genomic DNA from Ar. manshuriensis in serial 10-fold dilutions to produce concentrations ranging from 42.2 ng/μl to 42.2 fg/μl. We amplified both LAMP and conventional PCR for sensitivity testing.
Conventional PCR and Real Time PCR
We performed conventional PCR methodology by using a 25 μl mixture containing 12.5 μl Taq PCR Mix (Beijing Aidlab Biotech Co., China), 1 μl of each 2F and 3R primers (2.5μM) (Table 2), and 1 μl DNA template. We implemented the PCR reaction at 94°C for 5 min; followed by 40 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s; and a final extension step at 72°C for 10 min. We inspected the amplification products by 1% agarose gel electrophoresis. Furthermore, real-time PCR was carried out in Rotor-Gene Q (QIAGEN, Germany) by using 12.5 μL 2 × HRM PCR master mix (Type-it® HRMTM PCR Kit, QIAGEN, Germany). The reaction was performed as described above and followed by a melting analysis from 65 to 99°C with increment 1°C/s. We analyzed the real-time PCR results in terms of Tt (time threshold) values.
Detection of Mixed Sample
To detect Ar. manshuriensis within a mixed sample, we combined Ar. manshuriensis and Ak. quinata plants into mixed proportioned samples which were then used for the contamination test. The proportioned samples of Ar. manshuriensis and Ak. quinata were mixed in the ratios of 1:99, 3:97, 5:95, 7:93, 10:90, 20:80, 30:70, 40:60, 50:50, 60:40, 70:30, 80:20, 90:10, and 99:1, respectively. We used DNA extraction to obtain the genomic DNA. After LAMP amplification, we observed results using visual inspection and real-time fluorescence curve.
Results
Specificity of LAMP
To test the specificity of LAMP to detect Guan-mu-tong in adulterated samples of Mu-tong, we used the samples of Ar. manshuriensis as the positive samples, the three Akebia species as the negative samples and double-distilled water as the negative control. As depicted in Figure 3A, Guan-mu-tong (Gmt-1–Gmt-3) was detected by LAMP within 25–30 min and reached saturation at 45–60 min. In contrast, LAMP of the three Akebia species (Mt-1–Mt-9) and double-distilled water showed negative throughout the 60 min (Figure 3A) reaction, suggesting that the primer group could be used to detect Guan-mu-tong. We obtained similar results by directly observing the color change caused by the addition of calcein to the LAMP samples. Upon completion of the LAMP assay, positive reactions (Gmt-1–Gmt-3) changed from orange to green, whereas the negative reactions (Mt-1–Mt-9) and negative control (double-distilled water) remained orange (Figure 3B). Thus, these two detection methods showed the same sensitivity, indicating that the LAMP method developed in this study is able to promptly discriminate Guan-mu-tong from Mu-tong within 60 min under isothermal conditions.
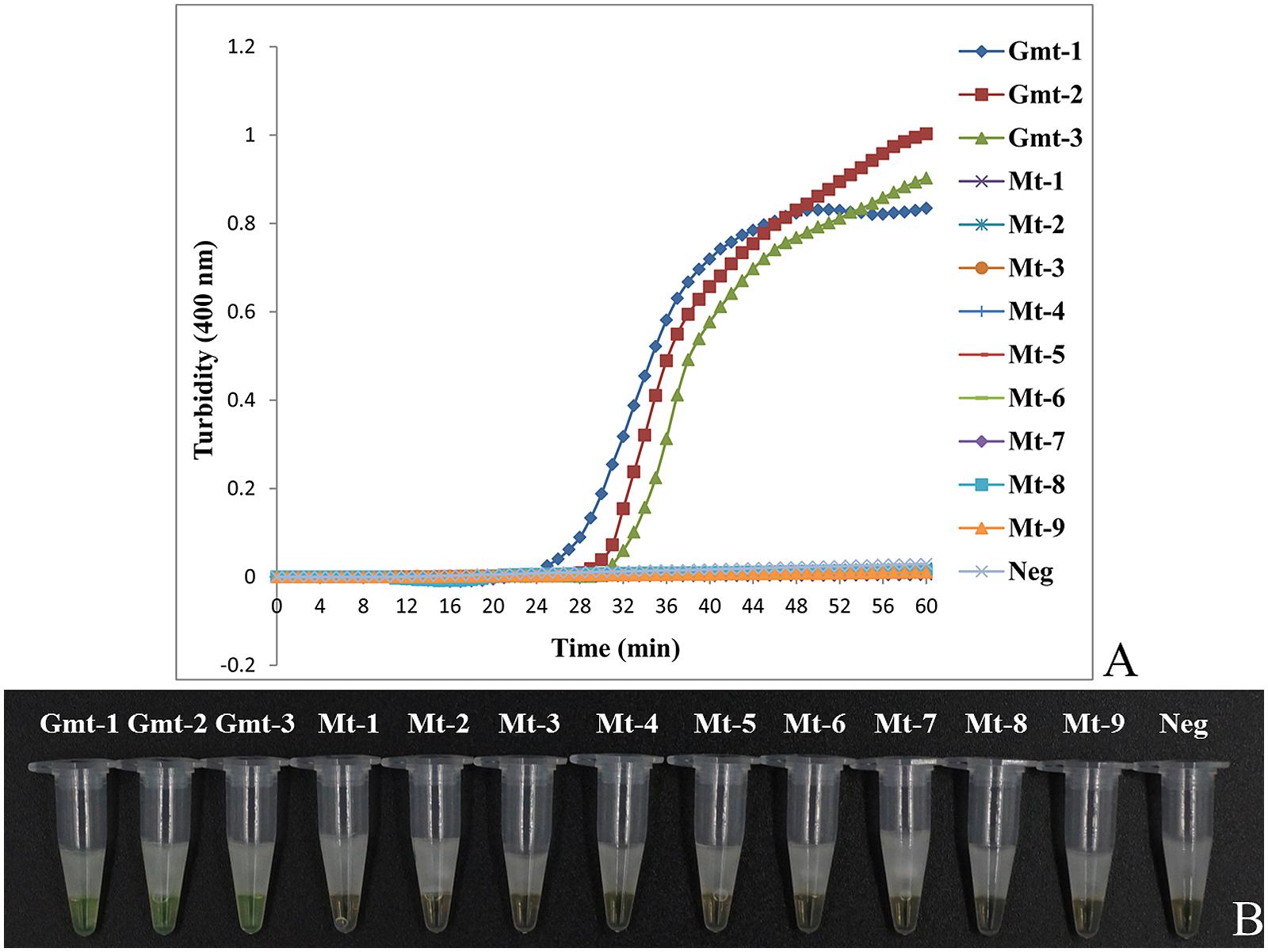
FIGURE 3. Differentiation of Akebiae Caulis from Aristolochiae Manshuriensis Caulis using LAMP. Gmt-1 to Gmt-3, Aristolochia manshuriensis; Mt-1 to Mt-3, Akebia trifoliata; Mt-4 to Mt-6, Ak. trifoliata var. australis; Mt-7 to Mt-9, Ak. quinata; Neg, negative control (double-distilled water). (A) Turbidity was monitored by a Loopamp real-time turbidimeter at 400 nm every 6 s, amplification was performed at 65°C for 60 min; (B) A visual color change detection method was compared. 1 μl of calcein (fluorescent detection reagent) was added to 25 μl of LAMP reaction mixture before the LAMP reaction.
Sensitivity of LAMP
Our results show that the turbidity-based real-time LAMP assay of Ar. manshuriensis was detectable with 42.2 ng/μl to 4.22 pg/μl of DNA template (Figure 4A). Similarly, results from the direct visual method showed that the positive reactions (1–5) turned green, while the negative samples and negative control (6–8) remained orange (Figure 4B). We were able to confirm the sensitivity of the reactions by using the fluorescence-based real-time LAMP platform, with DNA templates ranging in concentration from 42.2 ng/μl to 4.22 pg/μl per reaction tube, and Tt values ranging from 17.28 to 43.51 min (Additional File: Supplementary Figure S1A). No amplification was acquired for the 422 and 42.2 fg/μl templates. Thus, the detection limit of the LAMP assay for authentication Ar. manshuriensis developed in this study was 4.22 pg/μl (Figures 4A,B; Additional File: Supplementary Figure S1A). For comparison purposes, we also performed conventional PCR using the universal primers for ITS2 and found that the detection limit for conventional PCR was 42.2 pg/μl (Figure 4C). We also found that in the real-time PCR platform, the Tt values increased from 33.16 to 45.99 min for templates ranging from 42.2 ng/μl to 42.2 pg/μl per reaction tube (Additional File: Supplementary Figure S1B). As to its sensitivity, however, no amplification was observed when the concentration of template ranged from 4.22 pg/μl to 42.2 fg/μl, indicating that the sensitivity is lower than that of LAMP. These results suggest that LAMP is a highly sensitive method for detection of Ar. manshuriensis genomic DNA.
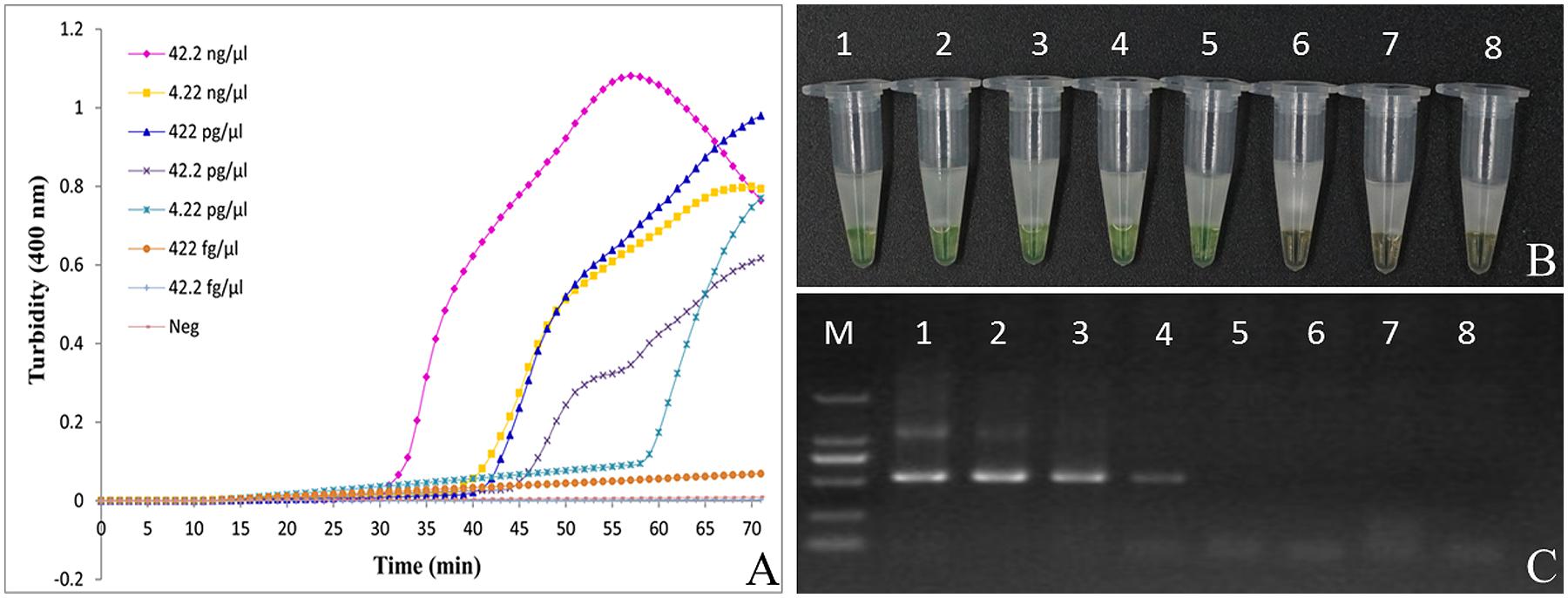
FIGURE 4. Comparison of sensitivity between the LAMP reaction and conventional PCR for detection of Aristolochiae Manshuriensis Caulis. The pure genomic DNA extracted from Aristolochia manshuriensis was diluted in a serial 10-fold dilution. Both LAMP reaction (A) and (B) and conventional PCR (C) were carried out in duplicate for each dilution point. Tubes and lanes: 1, 42.2 ng/μl; 2, 4.22 ng/μl; 3, 422 pg/μl; 4, 42.2 pg/μl; 5, 4.22 pg/μl; 6, 422 fg/μl; 7, 42.2 fg/μl; 8, Neg, negative control (double-distilled water). (A) Turbidity was monitored by a Loopamp real-time turbidimeter at 400 nm every 6 s; (B) A visual color change detection method was compared. 1μl of calcein (fluorescent detection reagent) was added to 25 μl of LAMP reaction mixture before the LAMP reaction; (C) The PCR products were detected by 1% agarose gel electrophoresis.
Detection of Ar. manshuriensis within a Mixed Sample
After LAMP amplification, results from the direct visual method showed that all mixed samples in this study changed color from orange to green, indicating positive results in all mixed samples of different ratios (Figure 5A), with the exception of samples 1 and 2 (negative sample and double-distilled water, respectively), which remained orange. Additionally, we observed from the fluorescence-based real-time LAMP assay that the amplification curve appeared for all the mixed proportioned samples (Figure 5B); the Tt values increased from 17.72 to 24.30 min for all mixed ratios (1:99 to 99:1) of Ar. manshuriensis (Figure 5B). No amplification curve was obtained from either the negative sample or the double-distilled water (samples 1 and 2). These results demonstrate that Ar. manshuriensis within a mixed sample can be detected using the LAMP method when its ratio greater than or equal to 1% in the mixed sample. As a result, the developed LAMP method for Ar. manshuriensis detection is prompt and reliable for situations where Mu-tong may be contaminated by Guan-mu-tong.
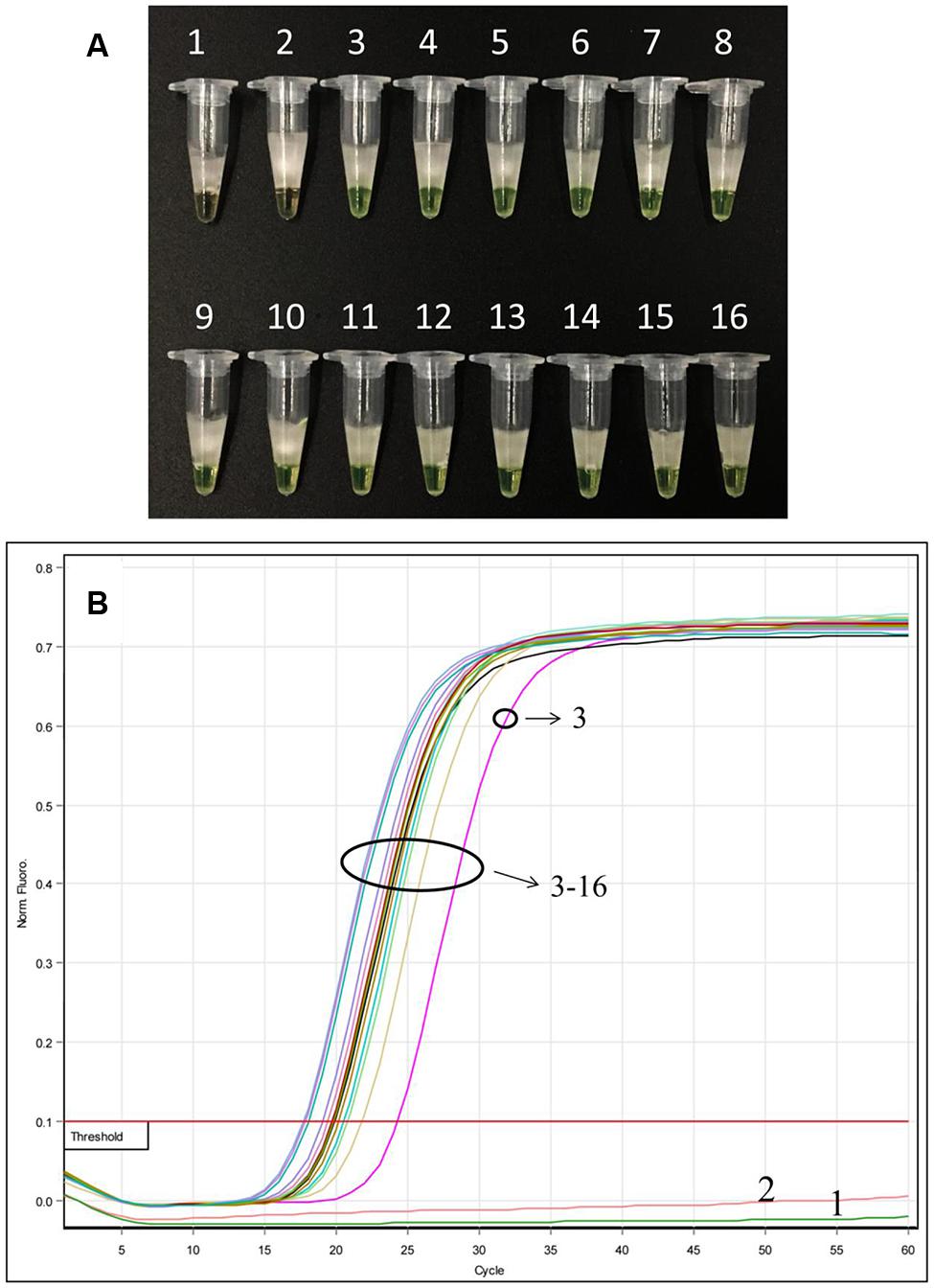
FIGURE 5. Detection of Ar. manshuriensis within a mixed sample in various ratios using the LAMP method. The sample powder of Ar. manshuriensis and Ak. quinata were mixed in the ratio 1:99, 3:97, 5:95, 7:93, 10:90, 20:80, 30:70, 40:60, 50:50, 60:40, 70:30, 80:20, 90:10, and 99:1, respectively. 1: Ak. quinata (negative sample); 2: double-distilled water (negative control); 3: 1:99; 4: 3:97; 5: 5:95; 6: 7:93; 7: 10:90; 8: 20:80; 9: 30:70; 10: 40:60; 11: 50:50; 12: 60:40; 13: 70:30; 14: 80:20; 15: 90:10; and 16: 99:1. (A) A visual color change detection method was compared. 1 μl of calcein (fluorescent detection reagent) was added to 25 μl of LAMP reaction mixture before the LAMP reaction; (B) In real time LAMP reaction, we monitored fluorescence in a Rotor-Gene Q.
Discussion
In this study, we investigated the usefulness of the LAMP method as a prompt and effective way of detecting Guan-mu-tong, as a common adulterant of Mu-tong. Currently, few methods are available for the identification Akebiae Caulis and its toxic adulterant Aristolochiae Manshuriensis Caulis (Ar. manshuriensis). Some previous studies have shown that both the PCR-based assay (Han et al., 2005; Li et al., 2014; Wu et al., 2015) and chemical method (Hashimoto et al., 1999; Wei et al., 2005) can also be used to authenticate Akebiae Caulis and Aristolochiae Manshuriensis Caulis. While these methods are able to accurately discriminate between species, they require complicated and time-consuming procedures, delicate instruments, and professional statistical methodologies.
The basic principles of PCR-based assay are: DNA fragment amplification and fragment detection via agarose gel electrophoresis. During conventional PCR procedures, at least three steps have high probabilities of introducing undesirable effects such as denaturation, annealing, and extension. In general, to conduct a PCR assay it takes approximately 2–3 h, depending on parameters, but this timeframe may be considered too time consuming for some situations. In contrast, the LAMP assay is performed in isothermal conditions (60–65°C) because LAMP requires Bst DNA polymerase, which has unique strand displacement activity. This characteristic allows not only for the LAMP reactions to occur faster, but also reduces the possibility of denaturation during the procedure. For example, in this study, we show that the developed LAMP method can detect Ar. manshuriensis from Akebiae Caulis at 65°C within 60 min. In evaluating the detection of Guan-mu-tong from mixed samples, we show that contamination can be detected at 65°C within 60 min given that the ratio of the contaminant, Ar. manshuriensis is at least 1%. The simplicity of the procedure suggests that it will be a significant, useful, and convenient method for identification of toxic herbal contaminants from tainted nontoxic herbal plant samples.
Moreover, PCR requires high-precision instrumentation due to the temperature cycling environment needed, whereas LAMP can be performed in constant temperature conditions decreasing the need for expensive equipment. Other detection procedures such as chemical detection (i.e., U/HPLC and U/HPLC–MS) also require highly specialized technology such as chromatographic instruments. In contrast, the LAMP method needs no such expensive instrumentation; only a water bath or heating block is enough. Additionally, agarose gel electrophoresis is required after amplification in conventional PCR to determine results, but the LAMP results can be detected by several methods. These detection methods include a simplified methodology that allows an individual to obtain LAMP results by use of the naked eye when an additional metal indicator such as calcein (Parida et al., 2008; Tomita et al., 2008), hydroxy naphthol blue (HNB) (Goto et al., 2009), or dye SYBR green (Parida et al., 2005) is added to the sample. Where SYBR green is added after LAMP amplification; calcein or HNB can be added at the beginning of the reaction to reduce the chance of contamination. A second method of detecting LAMP amplification products can be accomplished by monitoring with a real-time turbidimeter. Lastly, LAMP results are also detectable through agarose gel electrophoresis, similar to conventional PCR procedures, but we suggest avoiding this method because open-tube operation increases the risk of contamination.
Despite the many advantages of the LAMP reaction, several drawbacks to this procedure do exist. First, we acknowledge the occurrence of false positives due to high amplification efficiency as well as contamination issues. To address contamination during the LAMP procedure, we added low-melting-point paraffin wax to the reaction tubes to prevent contaminants, which previous studies show to be effective for the LAMP technique (Li et al., 2012; Liu et al., 2012). In addition, spatial separation of the reagent and test samples is needed in order to reduce the risk of contamination. Second, frequently used barcodes are not always suitable for designing LAMP primers. The primer design is an important step in the LAMP procedure and selecting a proper barcode marker is the basis of the primer design. The barcode sequence must be able to distinguish species effectively and include all regions adapted for the design of LAMP primers. Thus, we suggest try to design some groups of LAMP primers as candidates based on common barcode markers like ITS2, matK, or psbA-trnH etc. The best group of primers can be obtained by screening candidate primers in amplification efficiency.
The application of DNA barcoding in the identification of herbal materials is widely used, with ITS2 proposed as a core marker and psbA-trnH as a supplementary marker for differentiation of medicinal plants (Chen et al., 2010, 2014; Li et al., 2015). The wide use of the ITS2 sequence is due in great part to the successful identification of medicinal plant material by previous studies (Gu et al., 2013; Xin et al., 2013, 2015; Zhao et al., 2015) as well as its easy amplification. The use of the psbA-trnH sequence is less effective than ITS2 as shown both by our previous work with Aristolochiaceous plant materials (Wu et al., 2015), as well as this current study, which found that the psbA-trnH sequence is not suitable for obtaining LAMP primers. No primers were obtained when we uploaded the psbA-trnH sequence to PrimerExplorerV4 to design a LAMP primer. However, two sets of LAMP primers (each containing four primers: FIP, BIP, F3, and B3) were obtained from PrimerExplorerV42 based on the ITS2 sequence of Ar. manshuriensis. After we acquired the two LAMP primer sets, we tested both for usability and determined that only one set was useable (data not shown). In this study, the design of LAMP primers was based on the ITS2 sequence, which is a useful DNA barcode in Aristolochia species as well as its adulterant (Wu et al., 2015). Four primers (G-F3, G-FIP, G-BIP, and G-B3) and one loop primer (G-LB) were designed from seven loci in the ITS2 sequence of Ar. manshuriensis (Figure 2) which were specific to Guan-mu-tong (Ar. manshuriensis), but not the three Akebia (Mu-tong) species (Figure 2). The LAMP technique is a highly sensitive method, with DNA template amplification at low concentrations (six copies) (Notomi et al., 2000). We found that the LAMP assay for Ar. manshuriensis detection was tenfold more sensitive than conventional PCR assay sensitivity.
Although several methods were introduced to identify Akebiae Caulis and Aristolochiae Manshuriensis in previous studies, none of them could: (i) provide an on-site and rapid method for exact and effective authentication, nor (ii) provide results using simple, readily available heating devices. Compared to the several methods used to authenticate Akebiae Caulis from Aristolochiae Manshuriensis, results indicate that the LAMP assay is a superior process. Our study introduces a novel method for the detection of toxic Guan-mu-tong from widely used Mu-tong. With the increase of widespread attention of governmental agencies on the safety of traditional medicines, our results indicate that the LAMP method is an effective technique to test for the safety and quality of medicinal plant specimens due to its simplicity, rapidness, cost-effectiveness, and high sensitivity.
Author Contributions
LW and BW performed the experiments. MZ collected and analyzed data. LW wrote the manuscript. WS, PW, WL, PZ, YS, CX and SC conceived the study design and read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Major Scientific and Technological Special Project for “Significant New Drugs Creation” (No. 2014ZX09304307) and “National High-tech R&D Program of China (863 Program)” (No. 2012AA021602). Thanks Tianyuan Zhang for taking photos in this study.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00887
FIGURE S1 | Comparison of sensitivity between fluorescence-based real-time LAMP reaction and real-time PCR for detection of Aristolochiae Manshuriensis Caulis. The pure genomic DNA extracted from Aristolochia manshuriensis was diluted in a serial 10-fold dilution. Both real time LAMP (A) and real time PCR (B) were carried out in duplicate for each dilution point. Tubes and lanes: 1, 42.2 ng/μl; 2, 4.22 ng/μl; 3, 422 pg/μl; 4, 42.2 pg/μl; 5, 4.22 pg/μl. (A) In real time LAMP reaction, we monitored fluorescence in a Rotor-Gene Q; (B) In real time PCR reaction, we monitored fluorescence in a Rotor-Gene Q.
Footnotes
References
Aikawa, T., Horino, S., and Ichihara, Y. (2015). A novel and rapid diagnostic method for discriminating between feces of sika deer and Japanese serow by loop-mediated isothermal amplification. Mamm. Genome 26, 355–363. doi: 10.1007/s00335-015-9572-0
Bosward, K. L., House, J. K., Deveridge, A., Mathews, K., and Sheehy, P. A. (2016). Development of a loop-mediated isothermal amplification assay for the detection of Streptococcus agalactiae in bovine milk. J. Dairy Sci. 99, 2142–2150. doi: 10.3168/jds.2015-10073
But, P. P., and Ma, S. C. (1999). Chinese-herb nephropathy. Lancet 354, 1731–1732. doi: 10.1016/S0140-6736(05)76720-9
Chaudhary, A. A., Mohsin, M., and Ahmad, A. (2012). Application of loop-mediated isothermal amplification (LAMP)-based technology for authentication of Catharanthus roseus (L.) G. Don. Protoplasma 249, 417–422. doi: 10.1007/s00709-011-0293-2
Chen, S. L., Pang, X. H., Song, J. Y., Shi, L. C., Yao, H., Han, J. P., et al. (2014). A renaissance in herbal medicine identification: from morphology to DNA. Biotechnol. Adv. 32, 1237–1244. doi: 10.1016/j.biotechadv.2014.07.004
Chen, S. L., Yao, H., Han, J. P., Liu, C., Song, J. Y., Shi, L. C., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5:e8613. doi: 10.1371/journal.pone.0008613
Cornelissen, J. B., De Greeff, A., Heuvelink, A. E., Swarts, M., Smith, H. E., Van der Wal, F. J., et al. (2016). Rapid detection of Streptococcus uberis in raw milk by loop-mediated isothermal amplification. J. Dairy Sci. 99, 4270–4281. doi: 10.3168/jds.2015-10683
Goto, M., Honda, E., Ogura, A., Nomoto, A., and Hanaki, K.-I. (2009). Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxyl naphthol blue. Biotechniques 46, 167–172. doi: 10.2144/000113072
Grollman, A. P., Shibutani, S., Moriya, M., Miller, F., Wu, L., Moll, U., et al. (2007). Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. U.S.A. 104, 12129–12134. doi: 10.1073/pnas.0701248104
Gu, W., Song, J. Y., Cao, Y., Sun, Q. W., Yao, H., Wu, Q. A., et al. (2013). Application of the ITS2 region for barcoding medicinal plants of Selaginellaceae in Pteridophyta. PLoS ONE 8:e67818. doi: 10.1371/journal.pone.0067818
Han, S., Kim, S., Yang, C. H., and Seo, J. (2005). Genetical identification of Akebia and Aristolochia species by using Pyrosequencing. J. Biotechnol. 120, 360–363. doi: 10.1016/j.jbiotec.2005.06.030
Hashimoto, K., Higuchi, M., Makino, B., Sakakibara, I., Kubo, M., Komatsu, Y., et al. (1999). Quantitative analysis of aristolochic acids, toxic compounds, contained in some medicinal plants. J. Ethnopharmacol. 64, 185–189. doi: 10.1016/S0378-8741(98)00123-8
Hoang, M. L., Chen, C.-H., Sidorenko, V. S., He, J., Dickman, K. G., Yun, B. H., et al. (2013). Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med. 5, ra102–ra197. doi: 10.1126/scitranslmed.3006200
Horibe, D., Ochiai, T., Shimada, H., Tomonaga, T., Nomura, F., Gun, M., et al. (2007). Rapid detection of metastasis of gastric cancer using reverse transcription loop-mediated isothermal amplification. Int. J. Cancer 120, 1063–1069. doi: 10.1002/ijc.22397
Kawata, J., Kameda, M., and Miyazawa, M. (2007). Constituents of essential oil from the dried fruits and stems of Akebia quinata (THUNB.) DECNE. J. Oleo Sci. 56, 59–63. doi: 10.5650/jos.56.59
Lai, G.-H., Chao, J., Lin, M.-K., Chang, W.-T., Peng, W.-H., Sun, F.-C., et al. (2015). Rapid and sensitive identification of the herbal tea ingredient Taraxacum formosanum using loop-mediated isothermal amplification. Int. J. Mol. Sci. 16, 1562–1575. doi: 10.3390/ijms16011562
Li, M., Au, K.-Y., Lam, H., Cheng, L., But, P. P.-H., and Shaw, P.-C. (2014). Molecular identification and cytotoxicity study of herbal medicinal materials that are confused by Aristolochia herbs. Food Chem. 147, 332–339. doi: 10.1016/j.foodchem.2013.09.146
Li, M., Wong, Y.-L., Jiang, L.-L., Wong, K.-L., Wong, Y.-T., Bik-San Lau, C., et al. (2013). Application of novel loop-mediated isothermal amplification (LAMP) for rapid authentication of the herbal tea ingredient Hedyotis diffusa Willd. Food Chem. 141, 2522–2525. doi: 10.1016/j.foodchem.2013.05.085
Li, X. L., Liu, W., Wang, J., Zou, D. Y., Wang, X. S., Yang, Z., et al. (2012). Rapid detection of Trichinella spiralis larvae in muscles by loop-mediated isothermal amplification. Int. J. Parasitol. 42, 1119–1126. doi: 10.1016/j.ijpara.2012.09.011
Li, X. W., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y. T., and Chen, S. L. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. doi: 10.1111/brv.12104
Liu, W., Zou, D. Y., Li, Y., Wang, X. S., He, X., Wei, X., et al. (2012). Sensitive and rapid detection of the new Delhi metallo-beta-lactamase gene by loop-mediated isothermal amplification. J. Clin. Microbiol. 50, 1580–1585. doi: 10.1128/JCM.06647-11
Lord, G. M., Tagore, R., Cook, T., Gower, P., and Pusey, C. D. (1999). Nephropathy caused by Chinese herbs in the UK. Lancet 354, 481–482. doi: 10.1016/S0140-6736(99)03380-2
Lv, T., Teng, R., Shao, Q., Wang, H., Zhang, W., Li, M., et al. (2015). DNA barcodes for the identification of Anoectochilus roxburghii and its adulterants. Planta 242, 1167–1174. doi: 10.1007/s00425-015-2353-x
Ma, H. M., and Zhang, B. L. (2002). Comparison among families of Mutong. China J. Chin. Mater. Med. 27, 412–418. doi: 10.3321/j.issn:1001-5302.2002.06.004
Maan, S., Maan, N. S., Batra, K., Kumar, A., Gupta, A., Rao, P. P., et al. (2016). Reverse transcription loop-mediated isothermal amplification assays for rapid identification of eastern and western strains of bluetongue virus in India. J. Virol. Methods 234, 65–74. doi: 10.1016/j.jviromet.2016.04.002
Mansour, S. M., Ali, H., Chase, C. C., and Cepica, A. (2015). Loop-mediated isothermal amplification for diagnosis of 18 World Organization for Animal Health (OIE) notifiable viral diseases of ruminants, swine and poultry. Anim. Health Res. Rev. 16, 89–106. doi: 10.1017/S1466252315000018
Michel, C.-I., Meyer, R. S., Taveras, Y., and Molina, J. (2016). The nuclear internal transcribed spacer (ITS2) as a practical plant DNA barcode for herbal medicines. J. Appl. Res. Med. Arom. Plants (in press). doi: 10.1016/j.jarmap.2016.02.002
Mongan, A. E., Yusuf, I., Wahid, I., and Hatta, M. (2015). The evaluation on molecular techniques of reverse transcription loop-mediated isothermal amplification (RT-LAMP), reverse transcription polymerase chain reaction (RT-PCR), and their diagnostic results on MinION TM nanopore sequencer for the detection of dengue virus serotypes. Amer. J. Microbiol. Res. 3, 118–124. doi: 10.12691/ajmr-3-3-4
Mori, Y., Nagamine, K., Tomita, N., and Notomi, T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289, 150–154. doi: 10.1006/bbrc.2001.5921
Mori, Y., and Notomi, T. (2009). Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 15, 62–69. doi: 10.1007/s10156-009-0669-9
Nagamine, K., Watanabe, K., Ohtsuka, K., Hase, T., and Notomi, T. (2001). Loop-mediated isothermal amplification reaction using a nondenatured template. Clin. Chem. 47, 1742–1743.
Niessen, L. (2015). Current state and future perspectives of loop-mediated isothermal amplification (LAMP)-based diagnosis of filamentous fungi and yeasts. Appl. Microbiol. Biotechnol. 99, 553–574. doi: 10.1007/s00253-014-6196-3
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucl. Acids Res. 28:e63. doi: 10.1093/nar/28.12.e63
Nyan, D.-C., and Swinson, K. L. (2016). A method for rapid detection and genotype identification of hepatitis C virus 1-6 by one-step reverse transcription loop-mediated isothermal amplification. Int. J. Infect. Dis. 43, 30–36. doi: 10.1016/j.ijid.2015.12.002
Parida, M., Horioke, K., Ishida, H., Dash, P. K., Saxena, P., Jana, A. M., et al. (2005). Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43, 2895–2903. doi: 10.1128/JCM.43.6.2895-2903.2005
Parida, M., Sannarangaiah, S., Dash, P. K., Rao, P., and Morita, K. (2008). Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 18, 407–421. doi: 10.1002/rmv.593
Poon, S. L., Pang, S.-T., McPherson, J. R., Yu, W., Huang, K. K., Guan, P., et al. (2013). Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med. 5, ra101–ra197. doi: 10.1126/scitranslmed.3006086
Sasaki, Y., Komatsu, K., and Nagumo, S. (2008). Rapid detection of Panax ginseng by loop-mediated isothermal amplification and its application to authentication of ginseng. Biol. Pharm. Bull. 31, 1806–1808. doi: 10.1248/bpb.31.1806
State Pharmacopoeia Committee (2015). Chinese Pharmacopoeia. Beijing: Medical Science and Technology Press.
Tomita, N., Mori, Y., Kanda, H., and Notomi, T. (2008). Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 3, 877–882. doi: 10.1038/nprot.2008.57
Venkatesan, G., Balamurugan, V., Bhanuprakash, V., Singh, R. K., and Pandey, A. B. (2016). Loop-mediated isothermal amplification assay for rapid and sensitive detection of sheep pox and goat pox viruses in clinical samples. Mol. Cell. Probes 30, 174–177. doi: 10.1016/j.mcp.2016.02.004
Wei, F., Cheng, X. L., Ma, L. Y., Jin, W. T., Schaneberg, B. T., Khan, I. A., et al. (2005). Analysis of aristolochic acids and analogues in medicinal plants and their commercial products by HPLC-PAD-ESI/MS. Phytochem. Anal. 16, 222–230. doi: 10.1002/pca.849
Wu, L., Sun, W., Wang, B., Zhao, H. Y., Li, Y. L., Cai, S. Q., et al. (2015). An integrated system for identifying the hidden assassins in traditional medicines containing aristolochic acids. Sci. Rep. 5:11318. doi: 10.1038/srep11318
Xin, T. Y., Li, X. J., Yao, H., Lin, Y. L., Ma, X. C., Cheng, R. Y., et al. (2015). Survey of commercial Rhodiola products revealed species diversity and potential safety issues. Sci. Rep. 5:8337. doi: 10.1038/srep08337
Xin, T. Y., Yao, H., Gao, H. H., Zhou, X. Z., Ma, X. C., Xu, C. Q., et al. (2013). Super food Lycium barbarum (Solanaceae) traceability via an internal transcribed spacer 2 barcode. Food Res. Int. 54, 1699–1704. doi: 10.1016/j.foodres.2013.10.007
Keywords: loop-mediated isothermal amplification (LAMP), Akebia, Aristolochia manshuriensis, internal transcribed spacers 2 (ITS2), rapid authentication, visual detection
Citation: Wu L, Wang B, Zhao M, Liu W, Zhang P, Shi Y, Xiong C, Wang P, Sun W and Chen S (2016) Rapid Identification of Officinal Akebiae Caulis and Its Toxic Adulterant Aristolochiae Manshuriensis Caulis (Aristolochia manshuriensis) by Loop-Mediated Isothermal Amplification. Front. Plant Sci. 7:887. doi: 10.3389/fpls.2016.00887
Received: 17 February 2016; Accepted: 06 June 2016;
Published: 20 June 2016.
Edited by:
Basil J. Nikolau, Iowa State University, USAReviewed by:
Ankush Prasad, Tohoku Institute of Technology, JapanYuhui Chen, The Samuel Roberts Noble Foundation, USA
Copyright © 2016 Wu, Wang, Zhao, Liu, Zhang, Shi, Xiong, Wang, Sun and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Sun, wsun@icmm.ac.cn; Shilin Chen, slchen@icmm.ac.cn
 Lan Wu
Lan Wu Bo Wang1
Bo Wang1 Wei Liu
Wei Liu Wei Sun
Wei Sun Shilin Chen
Shilin Chen