- 1College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
- 2Zhejiang Provincial Key Laboratory of Horticultural Plant Integrative Biology, Zhejiang University, Hangzhou, China
- 3The State Agriculture Ministry Laboratory of Horticultural Plant Growth, Development and Quality Improvement, Zhejiang University, Hangzhou, China
- 4Plant and Crop Sciences Division, School of Biosciences, University of Nottingham, Loughborough, UK
Lignin is important for plant secondary cell wall formation and participates in resistance to various biotic and abiotic stresses. Loquat undergoes lignification not only in vegetative tissues but also in flesh of postharvest fruit, which adversely affects consumer acceptance. Thus, researches on lignin biosynthesis and regulation are important to understand loquat fruit lignification. In loquat, a gene encoding an enzyme in the lignin biosynthesis pathway, Ej4CL1, was reported to be regulated by transcription factors, including EjMYB1, EjMYB2, EjMYB8, and EjAP2-1, knowledge of this process is still limited. With the aim of identifying novel transcriptional factors controlling lignin biosynthesis in loquat, the promoter of Ej4CL1 was utilized to screen a cDNA library by yeast one hybrid assay. A novel R2R3 MYB, named EjODO1, was identified. Real-time PCR analyses indicated that EjODO1 is highly expressed in lignified stems and roots. During fruit development, expression of EjODO1 decreased along with the reduction of lignin content and became undetectable in mature ripe fruit. Thus, EjODO1 is likely to be involved in lignification of vegetative organs and early fruit development but not in mature fruit or postharvest lignification. Dual-luciferase assay indicated that EjODO1 could trans-activate promoters of lignin biosynthesis genes, such as EjPAL1, Ej4CL1, and Ej4CL5 and transient overexpression of EjODO1 triggered lignin biosynthesis. These results indicate a role for EjODO1 in regulating lignin biosynthesis in loquat which is different from the previously characterized transcription factors.
Introduction
Lignin is a complex phenylpropanoid polymer that constitutes a vital component of plant secondary cell walls, and imparts ‘waterproofing’ capacity as well as mechanical strength, rigidity, and environmental protection. The classical building blocks of lignin polymer comprise p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) (Boerjan et al., 2003; Rogers and Campbell, 2004; Vanholme et al., 2010). Various enzymes are involved in the lignin biosynthesis pathway, and most of them have been targeted for down-regulation, generating plants with reduced lignin levels or altered ratio of lignin monomers (Nicholas and Clint, 2010; Vanholme et al., 2013).
In addition to the biosynthetic pathway, a complex network of transcription factors has been reported regulating lignin biosynthesis. MYB transcription factors have been widely investigated and AtMYB58, AtMYB63, and AtMYB85 specifically and directly activated the genes encoding the monolignol biosynthetic enzymes by binding to a conserved AC element in their promoters, but not genes for the cellulose and hemicellulose biosynthetic pathways (Zhong et al., 2008; Zhou et al., 2009). Similar MYB transcription factors have also been reported in other plants, such as PtMYB1 and PtMYB4 in Pinus taeda (Patzlaff et al., 2003a,b), PtoMYB216 in Populus spp. (Tian et al., 2013), and ZmMYB31in Zea mays (Fornalé et al., 2010). On contrast, AtMYB4, an active repressor, negatively regulated the expression AtC4H (Jin et al., 2000). AtMYB7 and AtMYB32, homologs of AtMYB4, also repressed lignin biosynthesis genes, such as AtPAL, AtC4H, At4CL1, and AtCOMT (Jin et al., 2000; Preston et al., 2004; Fornalé et al., 2014).
Lignin accumulation in perennial woody trees but rarely occurs in flesh fruit and has mainly been reported in pear (stone cells, Lu et al., 2015; Yang et al., 2015), mangosteen (pericarp, Ketsa and Atantee, 1998; Kamdee et al., 2014), and loquat (Cai et al., 2006c). In loquat fruit, lignin content increasing during postharvest storage in flesh (edible layer), which was associated with an increase in firmness and reduction of juice yield (Cai et al., 2006b), subsequently adversely affecting fruit edible quality. More worse, loquat is chilling sensitive fruit, as flesh lignification of loquat fruit can also be accelerated by low temperature (0°C, Cai et al., 2006d). Chill injury induced lignification can be alleviated by low temperature conditioning (LTC), acetylsalicylic acid (ASA), 1-methylcyclopropene (1-MCP), methyl jasmonate (MeJA), heat treatment (HT), etc (Cai et al., 2006a,b,d; Cao et al., 2008; Xu et al., 2014). Due to the significance of lignification and the existence of numerous effective treatments, loquat fruit was widely used as a model fruit to understand flesh lignification.
Investigations of the mechanisms underlying loquat lignification have been at both the biochemical- and molecular- level. Analysis of most of the biosynthetic enzymes, including phenylalanine ammonia lyase (PAL), 4-coumarate: coenzyme A ligase (4CL) and cinnamyl alcohol dehydrogenase (CAD), showed a correlation with fruit lignin content (Cai et al., 2006c; Shan et al., 2008) and their coding genes have also been analyzed. EjCAD1 was first identified as a key candidate in regulated chilling injury-related lignification (Shan et al., 2008); the expression of other genes, such as EjCcoAOMT, also showed some positive correlation with lignin content in loquat fruit (Liu et al., 2015) and Ej4CL1 was significantly inhibited by LTC and HT, which indicated Ej4CL1 also involved in loquat fruit chilling injury induced lignification (Unpublished data). Recently two EjMYB genes, i.e., EjMYB1 and EjMYB2, were characterized (Xu et al., 2014) and shown to be capable of binding to the promoters of lignin biosynthetic genes, but they manifested opposite roles in regulating lignification, with EjMYB1 acting as an activator and EjMYB2 as a repressor (Xu et al., 2014). Subsequently, an AP2/ERF gene, EjAP2-1, was discovered to be able to interact with EjMYB at the protein level (Zeng et al., 2015) and EjAP2-1 transcriptionally suppressed the Ej4CL1 promoter, involving the EAR motif in EjAP2-1 acting via EjMYB (Zeng et al., 2015). Meanwhile, EjNAC1, homolog of AtVND6 and AtVND7, was also shown to act as a positive upstream regulator of loquat fruit lignification and could trigger lignin accumulation in tobacco leaves (Xu et al., 2014). All these results indicated that fruit lignification can be regulated by a range of transcription factors (Xie et al., 2016).
Interestingly, among the promoters of structural genes in the lignin pathway that were tested, changes in Ej4CL1 promoter activity showed the most significant responses to these transcription factors when tested, including three EjMYB (Xu et al., 2014; Wang et al., 2016), EjAP2-1 (Zeng et al., 2015), EjHSF3 (Zeng et al., 2016), and EjNAC1 (Xu et al., 2015). These findings led us to conduct the present research leading to the identification of, a R2R3 type MYB transcription factor, EjODORANT1 (EjODO1), which is a homolog of petunia volatile related PhODO1 (Spitzer-Rimon et al., 2012) and was obtained as the result of yeast one hybrid screening by using the Ej4CL1 promoter as bait. The regulatory mechanisms of EjODO1 was analyzed using real-time PCR, dual-luciferase, and transient over-expression systems.
Materials and Methods
Plant Materials
Red-flesh loquat ‘Luoyangqing’ (LYQ) fruits were collected from an orchard in Luqiao (Zhejiang province, China) at six different stages during fruit development: S1, fruitlet, 60 days after full bloom (DAFB); S2, immature green, 75 DAFB; S3, mature green 90 DAFB; S4, breaker, 100 DAFB; S5, half ripe 108 DAFB; S6, fully ripe 115 DAFB. Fruit harvested at 115 DAFB reached commercial maturity. Fruit without visible disease and mechanical wounding were selected for the study and three replicates were set for all sampling points. To test tissue specificity of the expression of EjODO1, vegetative tissues, including roots, stems and leaves, were harvested from germinated seedlings from seeds of loquat fruit, flowers were harvested at full bloom from orchard trees. Samples for all materials were frozen in liquid nitrogen and stored at -80°C.
RNA Extraction and Real-Time Quantitative PCR
Total RNA and cDNA for different tissues and developmental stages of loquat were prepared according to the protocol described by Zeng et al. (2015). For Real-time PCR, gene specific primers were designed using Primer3 (vision 0.4.0)1. The specificity of these primers was determined by examining the melting curve and product resequencing. All reactions were normalized using the Ct value corresponding to the loquat actin gene EjACT (Fu et al., 2012). Primers for EjODO1 were as follow: forward 5′- ATTCCCCAAGCAATGAGTCTCAG-3′; reverse 5′- TGCTAAGCTATTCTCCTCCGTTGG -3′. Real-time PCR analysis was performed with a LightCycler 1.5 instrument (Roche) using a mixture (10 μl total volume) comprising 2 μl of 5 × LightCycler FastStart DNA MasterPLUS SYBR Green I Master Mix (Roche), 0.5 μl of each primer (10 μM), 1 μl of diluted cDNA and 6 μl PCR-grade H2O. The PCR conditions included an initial denaturation for 5 min at 95°C, followed by 45 cycles of 95°C for 5 s, 60°C for 5 s, and 72°C for 10 s, and completed with a melting- curve analysis program. Three biological replicates were included for each sampling point or tissue.
Yeast One Hybrid Library Screening and Confirmation
Yeast one hybrid library screening was conducted with the MatchmakerTM Gold Yeast One-Hybrid Library Screening System (Clontech, Mountain View, CA, USA). A cDNA library was constructed with different loquat tissues (including root, stem, leaf, flower stalk, pericarp, and pulp) and used as prey and the promoter of Ej4CL1 was used as bait for library screening. The construct of the Ej4CL1 promoter was described in our previous report (Xu et al., 2014). All colonies were sequenced and blasted to GenBank for annotation. In order to verify the interaction between transcription factors and Ej4CL1 promoter, 11 full-length specific transcription factors (excluding previously characterized EjMYB1, EjMYB2, Xu et al., 2014; EjMYB8, Wang et al., 2016) were sub-cloned into pGADT7 vector, the corresponding pGADT7 clone and pGADT7 empty vector was, respectively, transformed into Y1HGold[pEj4CL1/AbAi]. AD-p53 was transformed into Y1HGold[p53/AbAi]. Yeast one-hybrid assays were conducted on SD medium with aureobasidin A and without leucine (SD/Leu + AbA) at 30°C for 3–5 days to test the interaction.
Alignment of EjODO1 with R2R3 EjMYB in both loquat and Arabidopsis was performed using the neighbour-joining (NJ) method in ClustalX (v.1.81), and a phylogenetic tree was constructed with FigTree (v.1.3.1).
Dual-Luciferase Assay
Dual-luciferase assay was carried out to investigate the transactivation activity of transcription factors to target promoters. Full-length EjODO1 was amplified and inserted into NotI and SpeI sites of pGreen II 0029 62-SK vector, while pGreen II 0800-LUC carrying promoters of lignin biosynthesis were constructed by Xu et al. (2014). All constructs were electroporated into Agrobacterium tumefaciens GV3101, using Gene PulserXcellTM Electroporation Systems (Bio-Rad). Dual luciferase assays were performed according to our previous reports (Zeng et al., 2015). The empty vector mixed with the promoters was set as 1 and the analysis was carried out with at least three replicates.
Transient Over-Expression Analysis of EjODO1 in Nicotiana tabacum leaves
In order to verify the role of EjODO1 in the regulation of lignin biosynthesis (based on present results) and fragrance biosynthesis (ODO genes were previously linked to volatiles, Verdonk et al., 2005), a transient expression transformation system was adopted in N. tabacum leaves.
Agrobacterium cultures with EjODO1 or empty vector pGreen II 0029 62-SK (SK) were suspended in the same infiltration buffer used for dual luciferase assay and OD600 were adjusted to 0.75. Target gene (EjODO1) and negative control (SK) were infiltrated on two separate sides of the fifth true leave of N. tabacum. Five days after infiltration, tissue from each of the infiltrated leaves was taken for lignin and volatile compounds analysis.
Lignin content of tobacco leaves was measured according to the methods described by Xu et al. (2014). Data were expressed on a fresh weight basis, and all measurements were done in triplicate.
Volatiles were collected by placing 0.2 g frozen tissues powder into a 4 ml headspace vial containing 1 ml saturated sodium chloride solution and 50 μl 1-hexanol (0.1%, v/v) was added as an internal standard. The samples were incubated at 40°C for 30 min with continuous agitation (600 rpm) after fully vortexing for a few seconds. A SPME fiber coated with 50/30 μm Divinylbenzene/Carboxen/Polydimethylsiloxane (DVB/CAR/PDMS; Supelco Co., Bellefonte, PA, USA) was used for the extract of volatiles under the same conditions (40°C, 600 rpm). Volatile compounds were determined as described by Shen et al. (2016). The internal standards were used for compensating differences between samples, and the abundance of each volatile was calculated based on its peak area.
Results
Yeast One Hybrid Screening for Proteins Interacting with the Promoter of Ej4Cl1
In our previous studies, Ej4CL1 was shown to be regulated by EjMYB1, EjMYB2, EjMYB8, EjAP2-1, and EjHSF3 (Xu et al., 2014; Zeng et al., 2015, 2016). In order to obtain further information about the transcriptional regulatory mechanism controlling lignin biosynthesis in loquat, yeast one-hybrid screening was employed to look for novel transcription factors, using the Ej4CL1 promoter as a bait. A total of 173 PCR products were obtained (data not shown), among which were 11 putative transcription factors including three MYBs (the previously characterized EjMYB genes were disregarded), three Zinc finger proteins, three AP2/ERFs, one bHLH, and one NAC (Supplementary Figure S1). However, only EjODO1 (Genbank no. KX347550) showed high binding affinity for Ej4CL1 promoter, unlike the other 10 putative transcription factors (Figure 1; Supplementary Figure S1).
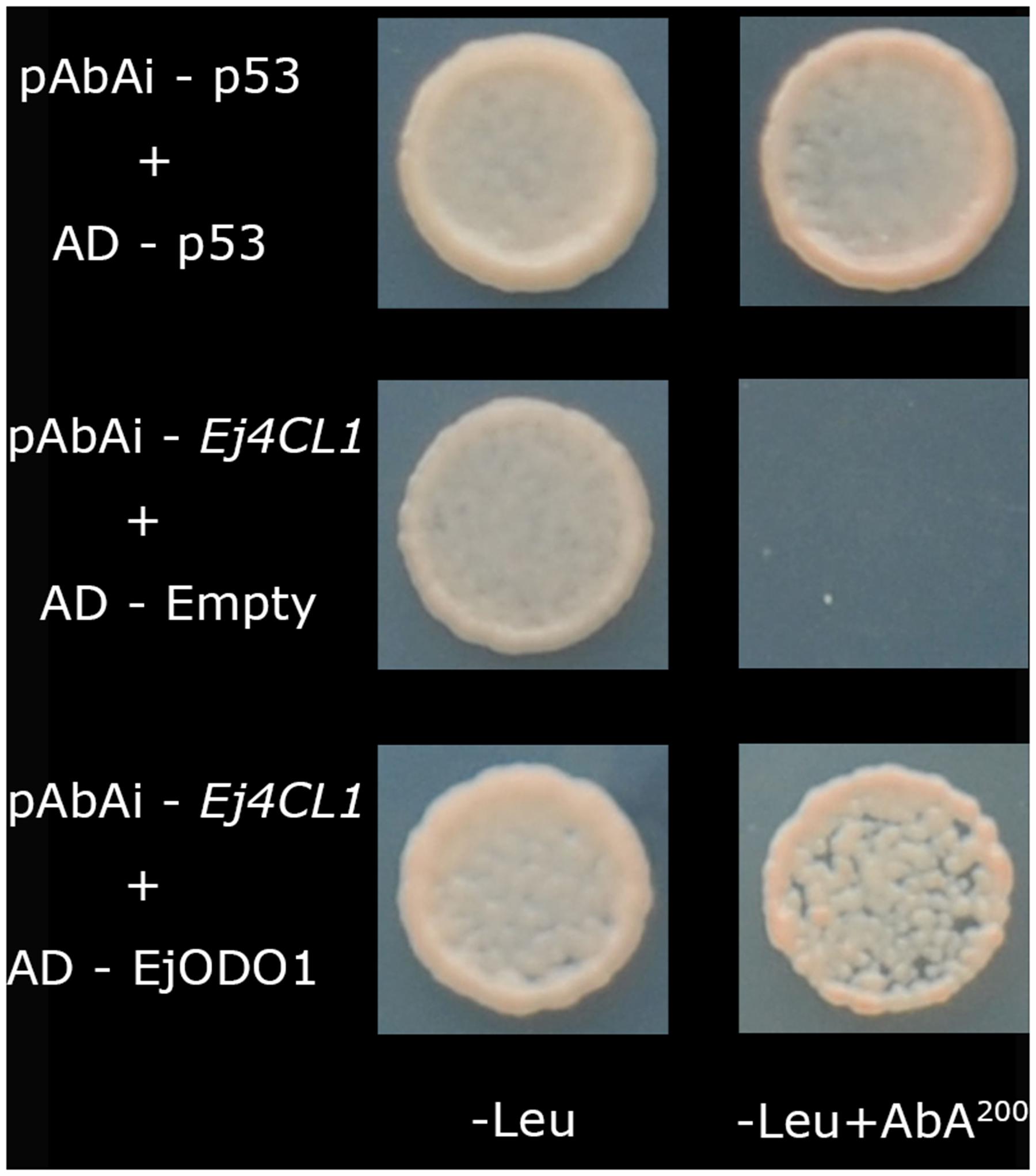
FIGURE 1. Protein–DNA interaction between EjODO1 and the promoter of Ej4CL1 using yeast one hybrid analysis. Interaction was determined on SD medium lacking Leu in the presence of aureobasidin A (-Leu + AbA200). AD-p53 and pAbAi-p53 were used as a positive control; AD-empty and pAbAi-pEj4CL1 were used as a negative control.
Phylogenetic Analysis of EjODO1
EjODO1 is an R2R3 type MYB transcription factor with conserved R2R3 domain (Figure 2). Further phylogenetic analysis indicated that EjODO1 is different from previously characterized EjMYB genes (Figure 3), and it was closest to PhODO1, a transcriptional regulator of volatile benzenoids in petunia flowers (Verdonk et al., 2005) and two lignin biosynthesis and secondary cell-wall related transcription factors (AtMYB85 and AtMYB42; Zhong et al., 2008).
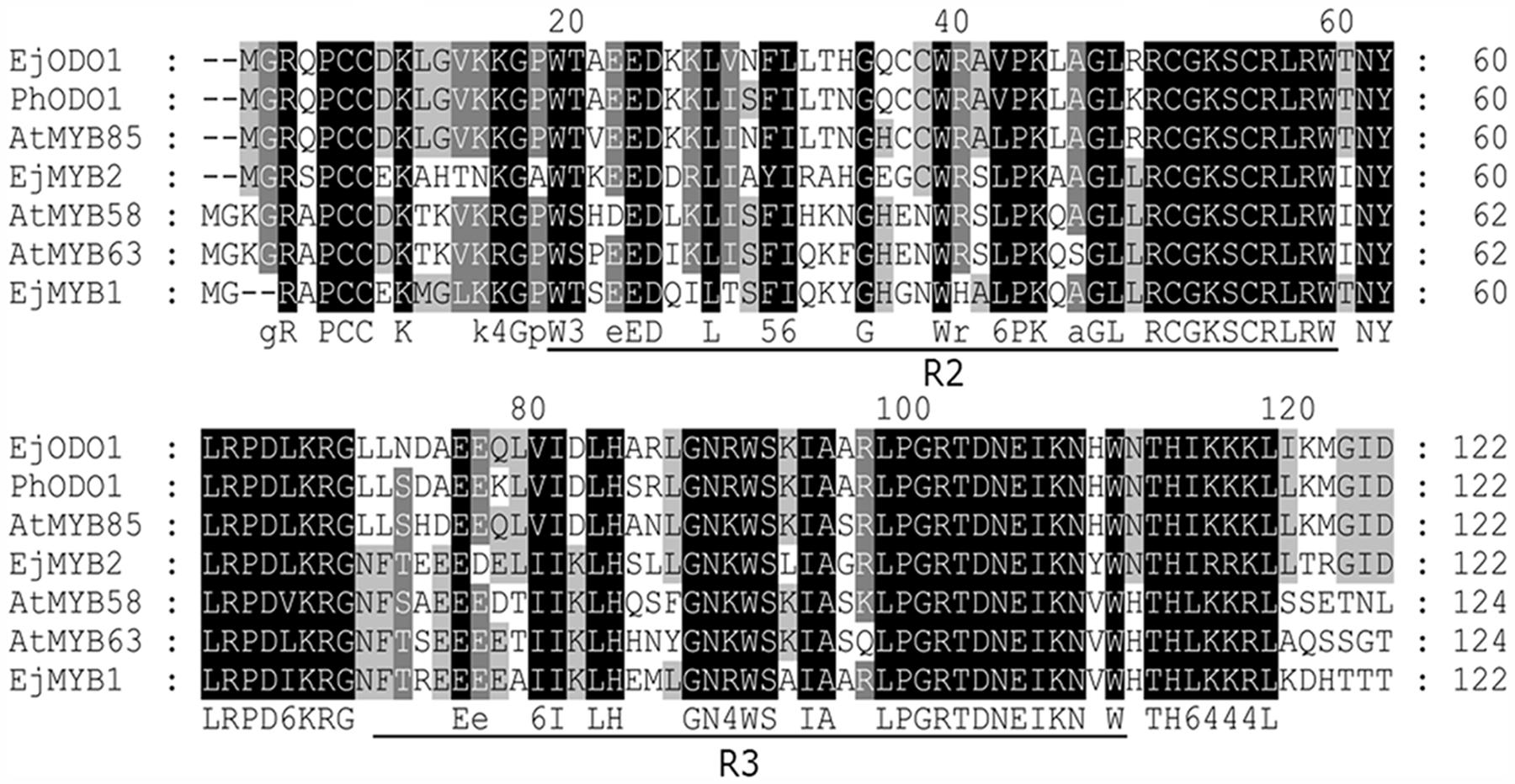
FIGURE 2. Alignment of amino acid sequences of the R2R3 domains of EjODO1, PhODO1, and other MYBs. Identical amino acid residues were shaded in black, and similar in gray. The R2 and R3 MYB conserved amino acid sequence among the different genes were underlined and refer to two repeats of the MYB DNA-binding domain.
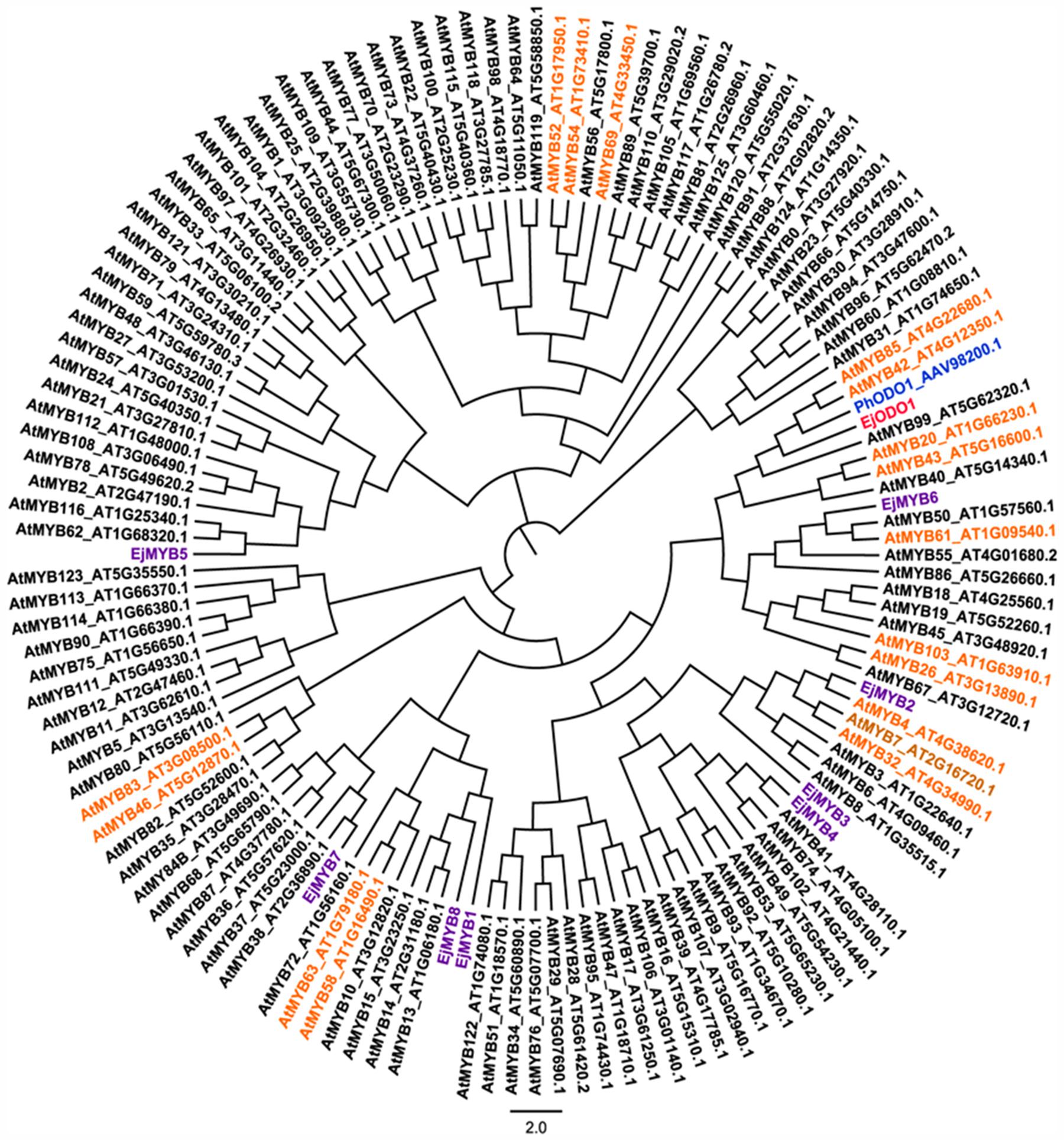
FIGURE 3. Phylogenetic analysis of EjODO1, PhODO1, and R2R3 MYB members from loquat and Arabidopsis. Fragrance biosynthesis related PhODO1 was highlighted in blue, the MYBs that involved in the regulation of lignin biosynthesis or secondary wall metabolism in Arabidopsis were indicated in orange and previously reported loquat EjMYB were highlighted in purple. Full length MYB protein sequences from Arabidopsis were downloaded from The Arabidopsis Information Resource (TAIR), and their accession number were embedded in the diagram.
Association between EjODO1 and Lignin Content in Different Loquat Tissues and Fruit Developmental Stages
Tissue specificity analysis indicated that EjODO1 was most highly expressed in stems and at a much lower level in roots, leaves and flowers, and was undetectable in the flesh of commercial mature loquat fruit (Figure 4). In order to investigate whether EjODO1 is either not expressed at all, or only at a specific stage in fruit development, levels were examined in a loquat fruit developmental series. As shown in Figure 5, lignin content in loquat flesh gradually decreased during fruit development, from 37.57 × 103A280kg-1FW-1 at S1 stage to 1.06 × 103 A280kg-1FW-1at S6 stage (commercial maturity). Meanwhile, the expression level of EjODO1 gradually decreased during fruit developmental, reaching an extremely low level at S5 stage (with approximate 0.0065% of S1 stage fruit) and was non-detectable at the S6 (ripe) stage (Figure 5). EjODO1 also could not be detected in low temperature-induced lignified loquat fruit (data not shown).
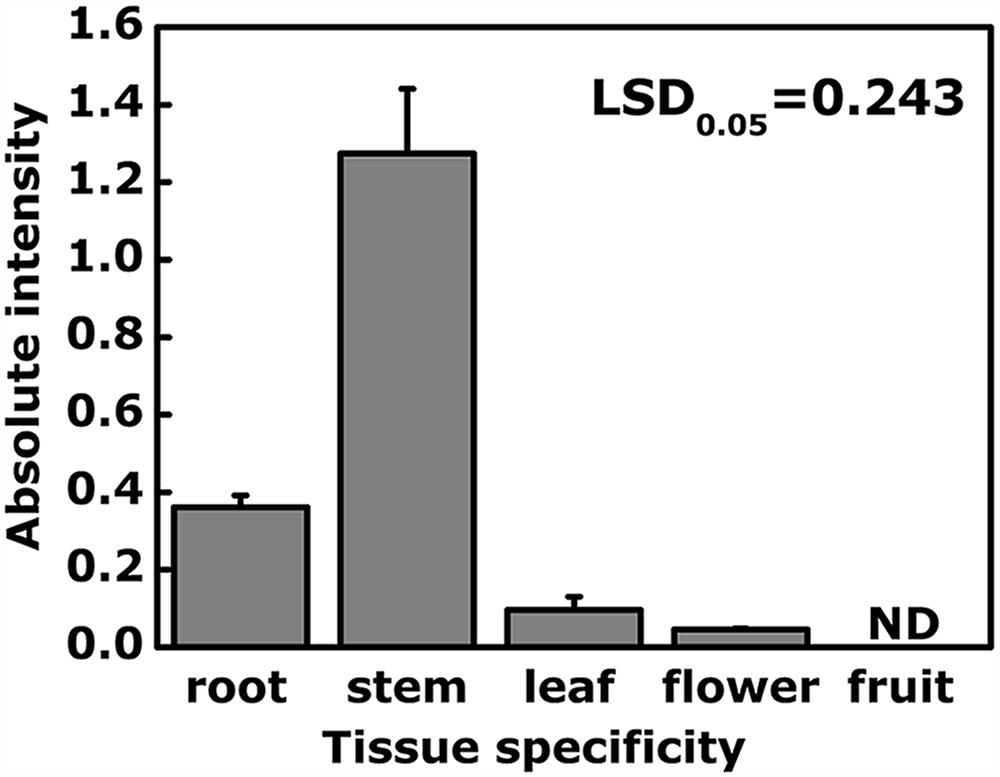
FIGURE 4. Analysis of expression of EjODO1 in different tissues of loquat. Five tissues were used for gene expression of EjODO1, inculding vegetative tissues (roots, stems, and leaves of seedlings) and reproductive tissues (flowers, harvested at full bloom; fruit, mature fruit at harvest). Gene expression was expressed as absolute intensity compared to EjACT. Data are reported as means + SE. LSDs represent least significantdifference at 0.05. ND, not detectable in the pulp.
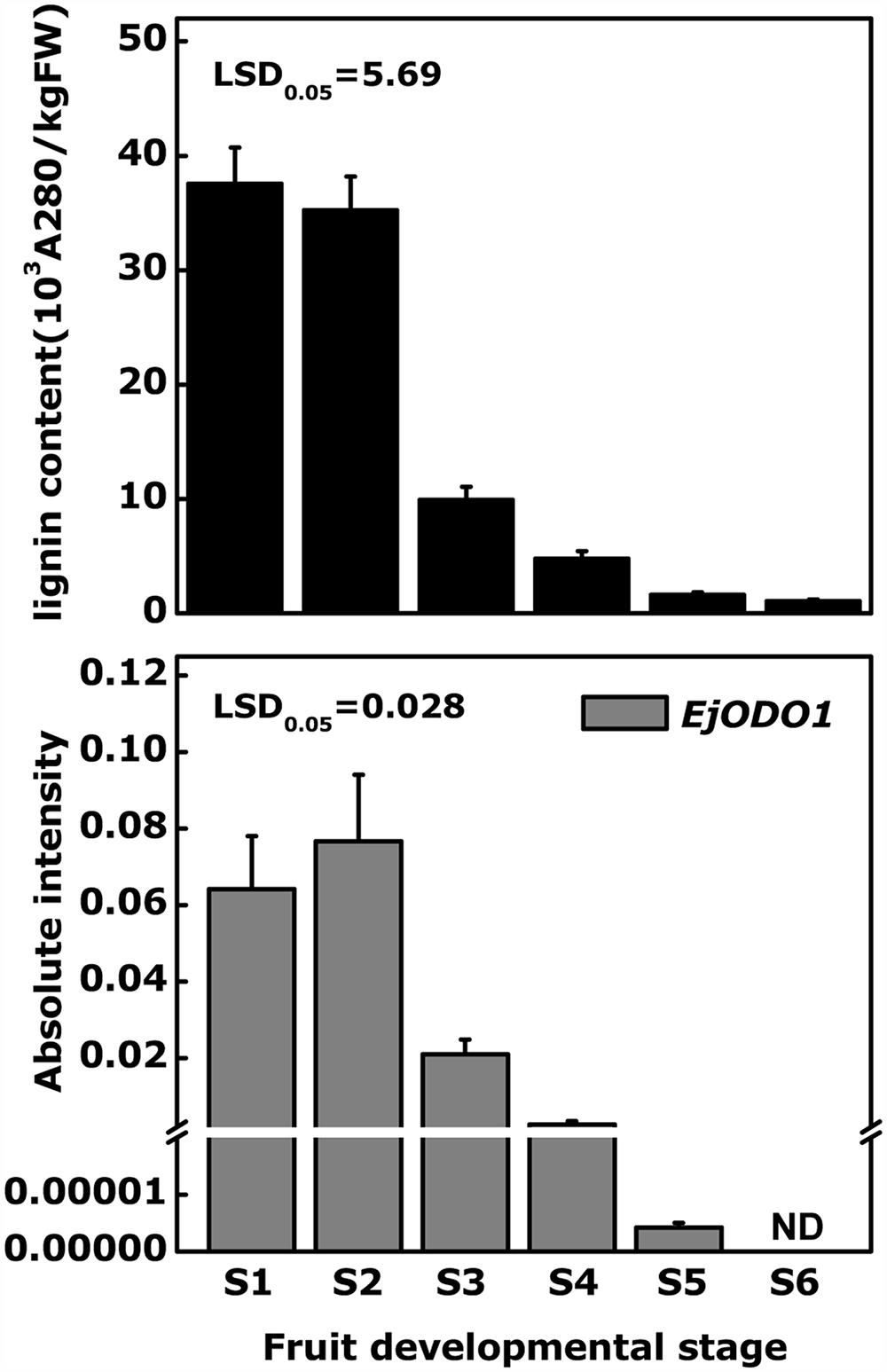
FIGURE 5. Comparison of changes in lignin content and expression of EjODO1 at different developmental stages of loquat fruit. Red-flesh loquat ‘LYQ’ fruits were harvested at six different stages during fruit development:S1, 60 days afterfull bloom (DAFB); S2, 75 DAFB; S3,90 DAFB; S4, 100 DAFB; S5, 108 DAFB; S6, 115 DAFB, respectively. Fruit reach commercial mature at stage S6. Gene expression was expressed as absolute intensity compared to EjACT. Lignin content was expressed as A280 absorbance per kilogram of fresh weight (103 A280 kg-1 of fresh weight). ND, not detected. Error bars indicate SEs from three biological replicates.
In vivo Interaction of EjODO1 and Promoters of Loquat Lignin Biosynthesis Genes
Using the dual-luciferase assay, the trans-activation activities of EjODO1 on the promoters of the EjPAL, Ej4CL, and EjCAD genes from loquat were investigated. The results indicated that EjODO1 trans-activated the promoters of EjPAL1, Ej4CL1, and Ej4CL5, reached approximately 5.5, 11.5, and 5.7 fold, respectively (Figure 6). In order to further test the relationship between EjODO1 and lignin biosynthesis, 13 genes within the phenylpropanoid pathway of Arabidopsis were selected for further analysis. The results showed that EjODO1 could interact with many of the promoters of these lignin-related genes, including AtPAL1, AtPAL2, AtC4H, At4CL1, At4CL2, AtHCT, AtC3H1, AtCCoAOMT1, AtCCR1, and AtCAD5 (Figure 6).
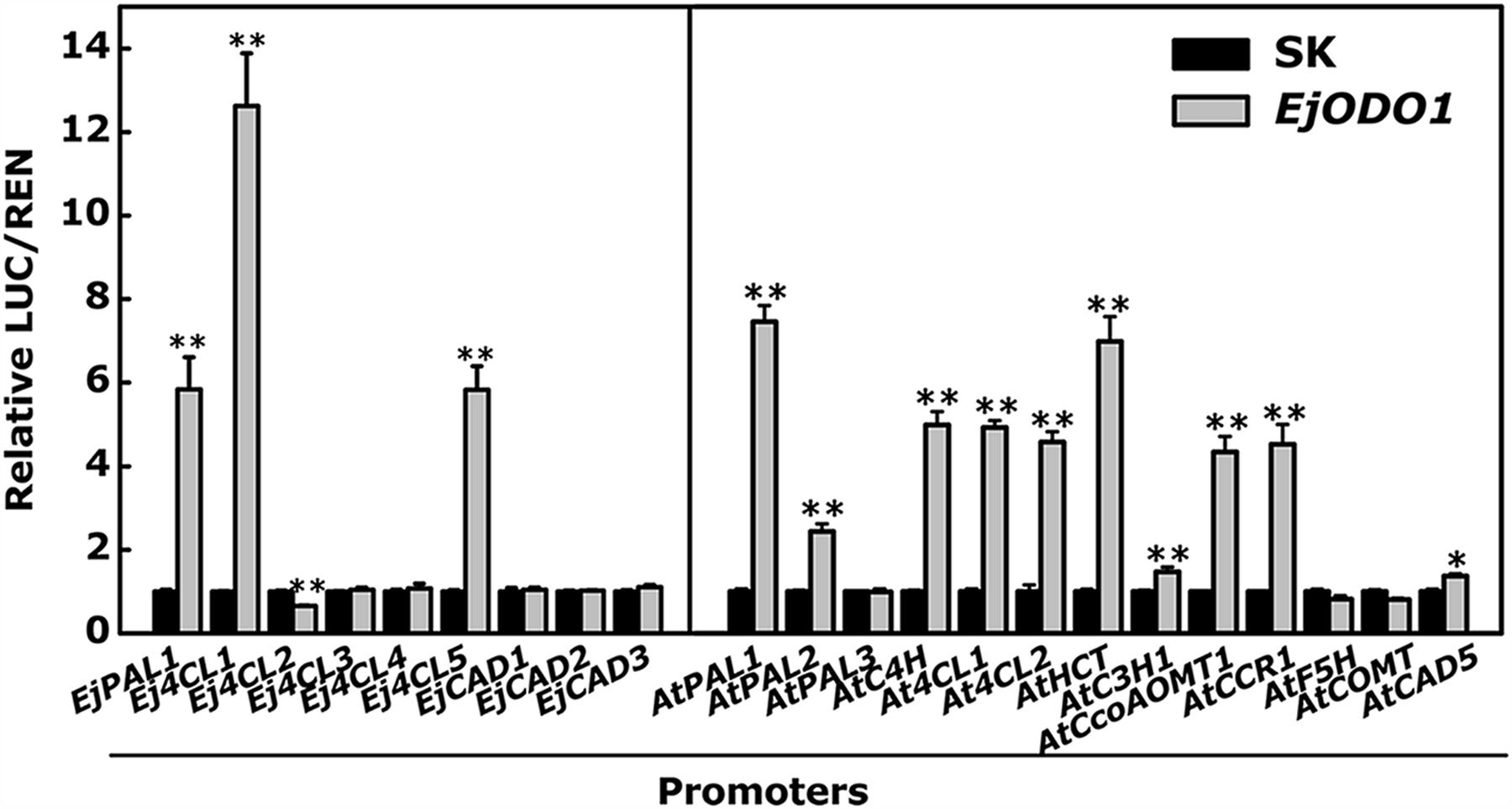
FIGURE 6. In vivo interaction of EjODO1 with promoters of lignin biosynthesis genes from loquat and Arabidopsis. 9 promoters of lignin biosynthesis from loquat and 13 promoters within the phenylpropanoid pathway from Arabidopsis were used for dual luciferase assey. The ratio of LUC/REN of the empty vector (SK) plus promoter was used as calibrator (set as 1). Error bars indicate SE from at least three replicates. Statistical analyses were performed using Student’s t-test: ∗P < 0.05, ∗∗P < 0.01.
Transient Expression of EjODO1 and its Role in Lignin Biosynthesis
In order to verify the significance of the results obtained from yeast one hybrid and dual-luciferase assay, transient over-expression was performed for functional analysis of EjODO1 in tobacco leaves. EjODO1, driven by the CaMV 35S promoter in the pGreen II 0029 62-SK vector, was introduced into N. tabacum leaves using Agrobacterium. The results indicated that EjODO1 significantly increased lignin content to 1.97 × 103A280kg-1FW-1(P < 0.01), compared with 1.33 × 103A280kg-1FW-1 for tobacco leaves expressing the empty vector (Figure 7A). With regard to the volatile analysis, two volatile compounds benzaldehyde and benzyl alcohol, products of the phenyl propanoid pathway were detected, but there were no differences in volatiles between the control (SK) and EjODO1 treatments (Figure 7B).
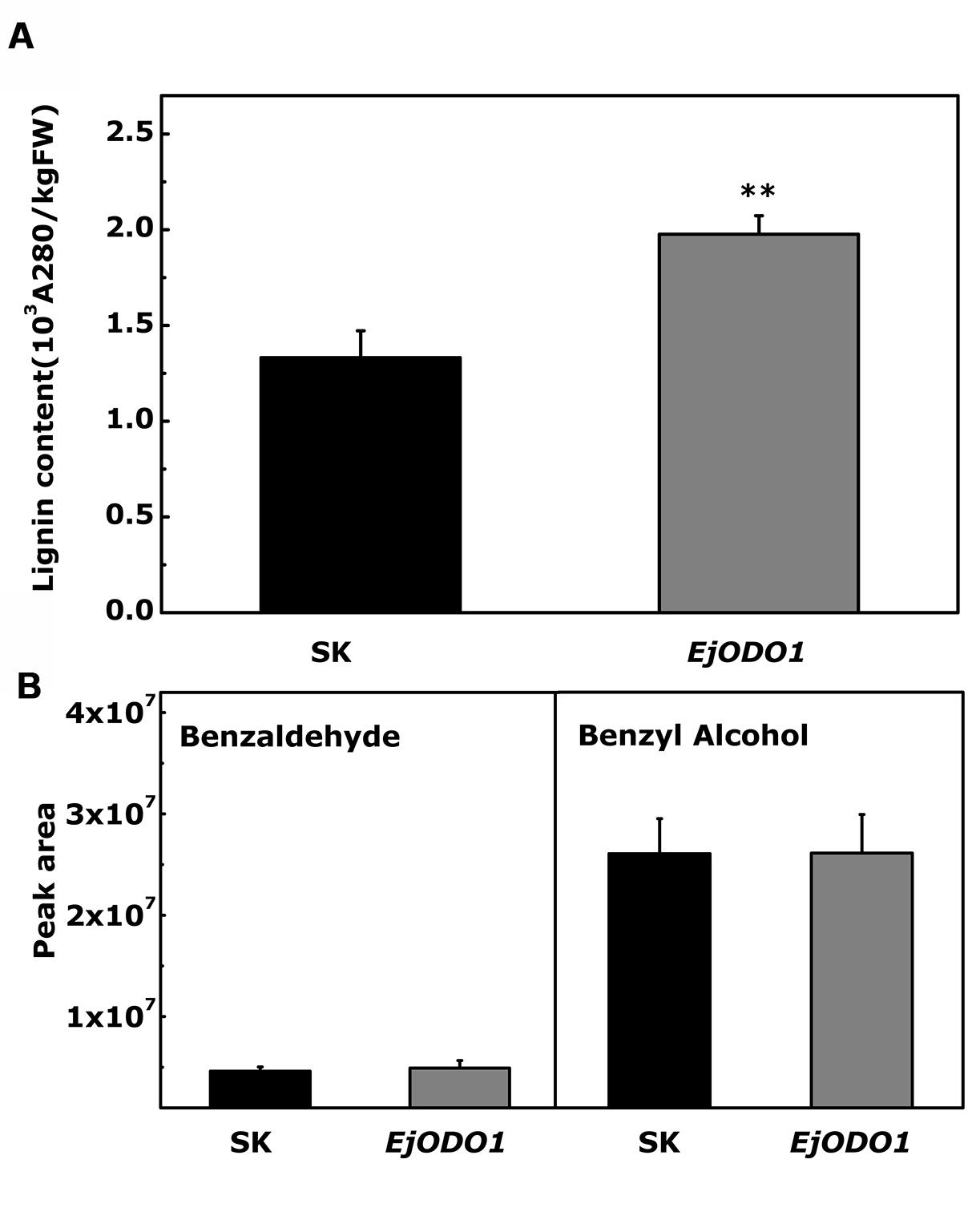
FIGURE 7. Transient over-expression of EjODO1 in Nicotiana tabacum leaves. The transient over-expression experiments were conducted with empty vector pGreen II 0029 62-SK (SK) and EjODO1 on two separate sides of the fifth leaf. Tissues from each of the infiltrated leaves were taken to measure both lignin content (A) and volatile compotents (B) after 5 days of infiltration. Lignin content was measured based on precipitation of lignin thioglycolic acid. Volatile compounds were determined by HS SPME GC-MS. Error bars indicate SEs from three biological replicates. Statistical analyses were performed using Student’s t-test: ∗∗P < 0.01.
Discussion
Lignin biosynthesis and the process of lignification is regulated by many transcription factors, such as EjMYB1, EjMYB2, EjMYB8, EjNAC1, and EjAP2-1, which have been investigated in relation to the regulation of fruit lignification. Among these transcription factors, EjMYB1, EjMYB2, and EjMYB8 could directly bind to and transcriptionally regulate promoters of lignin biosynthetic genes (Xu et al., 2014; Wang et al., 2016), while EjAP2-1 forms a protein-protein complex with EjMYB (Zeng et al., 2015), and the regulatory mechanism of EjNAC1 remains unclear (Xu et al., 2015).
Here, a previously characterized fruit lignification-related gene, Ej4CL1, was selected for cDNA library screening, using yeast one hybrid assay, and EjODO1 was identified. EjODO1 has a conserved R2R3 domain and belongs to a member of the MYB family, which is similar to EjMYB1, EjMYB2, and EjMYB8 (Wang et al., 2016; Xu et al., 2014). MYB proteins function as key regulators of the synthesis of phenylpropanoid-derived compounds, and some R2R3-MYB transcription factors regulate the synthesis of more than one class of phenylpropanoid-derived metabolites. Based on phylogenetic analysis, EjODO1 clustered with PhODO1 in petunia flowers, which is related to volatile benzenoids (Verdonk et al., 2005; Spitzer-Rimon et al., 2012) and AtMYB85, which was considered as lignin specific activator (Zhong et al., 2008). Overexpression of VvMYB5a in tobacco affected the metabolism of anthocyanins, flavonols, tannins, and lignins (Deluc et al., 2006). Moreover, AtMYB75/PAP1 regulated a series of phenylpropanoid-derived compounds, including monolignol, anthocyanin, proanthocyanidin, flavonols, and phenolic acid (Deluc et al., 2006; Zuluaga et al., 2008). All these findings indicated the dual potential for EjODO1 in lignin regulation and fragrance biosynthesis.
The expression pattern showed that EjODO1 was highly and preferentially expressed in stems and roots of loquat, indicating that it may play a significant role in processes involved in formation of these actively lignifying tissues. These results were similar to some other previous observations with known MYB transcription factors, such as AtMYB103, which is only expressed in stems and influences the biosynthesis of S lignin in Arabidopsis stems (Zhong et al., 2008; Ohman et al., 2013); AtMYB75 showed the highest expression in the lower part of the inflorescence stem, and lower levels of AtMYB75 expression could be detected in flowers, leaves, and siliques, but none could be detected in roots (Bhargava et al., 2010). Moreover, the expression level of EjODO1 diminished as fruit ripening proceeded, speculated to be involved in lignin biosynthesis during fruit development, especially for young fruit. EjODO1 is not detectable in mature fruit and its subsequent storage period, which is different from previously characterized transcription factors, such as EjMYB1, EjMYB2, and EjMYB8, as these transcription factors were generally characterized as related to postharvest lignification in loquat fruit (Xu et al., 2014; Wang et al., 2016).
Further analysis indicated EjODO1 is a transcriptional activator of lignin biosynthesis. Dual-luciferase assay revealed that EjODO1 induced promoter activities of lignin biosynthesis genes in both loquat and Arabidopsis, which was similar to the transcriptional activator EjMYB1 and functionally opposite to the transcriptional repressor EjMYB2 (Xu et al., 2014). Other lignin-specific transcription factors, such as AtMYB58, AtMYB63, and AtMYB85 are able to specifically induce expression driven by promoters of lignin biosynthesis genes, including promoters of PAL and 4CL (Zhong et al., 2008; Zhou et al., 2009). Yeast one hybrid assay indicated that EjODO1 could directly bind to the promoter of Ej4CL1 and then promote trans-activation of the target genes. Transient overexpression of EjODO1 was conducted to determine the lignin content and volatile substances, and the results indicated that EjODO1 could promote the accumulation of lignin without any change in volatiles between the control (SK) and EjODO1 expressers. Thus, it appears that in loquat, ODORANT1 activity leads to transcriptional regulation of lignin production but not volatile benzenoid biosynthesis, which differ to its homolog in petunia PhODO1 (Verdonk et al., 2005; Spitzer-Rimon et al., 2012).
Conclusion
The present study has identified a novel activator of loquat lignin biosynthesis, EjODO1, by yeast one-hybrid screening, which, unlike the previously characterized transcription factors, appears to be a regulator of lignin biosynthesis in vegetative organs and during early fruit development.
Author Contributions
JZ, X-rY, and K-sC designed the experiment. JZ, HG, and CZ performed the experiment. JZ and X-rY wrote the first draft of the manuscript. X-rY, XL, DG, and K-sC conducted analysis of data. X-rY, XL, DG, and K-sC contributed substantially to the revisions.
Funding
This research was supported by the National Key Research and Development Program (SQ2016YFNC010081); National Natural Science Foundation of China (no. 31630067); Program of International Science and Technology Cooperation (2011DFB31580); the Natural Science Foundation of Zhejiang Province, China (LR16C150001).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01360
FIGURE S1 | Protein-DNA interaction between the screened proteins and the promoter of Ej4CL1 using yeast one hybrid analysis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Bhargava, A., Mansfield, S. D., Hall, H. C., Douglas, C. J., and Ellis, B. E. (2010). MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 154, 1428–1438. doi: 10.1104/pp.110.162735
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Cai, C., Chen, K. S., Xu, W. P., Zhang, W. S., Li, X., and Ferguson, I. (2006a). Effect of 1-MCP on postharvest quality of loquat fruit. Postharvest Biol. Technol. 40, 155–162. doi: 10.1016/j.postharvbio.2005.12.014
Cai, C., Li, X., and Chen, K. S. (2006b). Acetylsalicylic acid alleviates chilling injury of postharvest loquat (Eriobotrya japonica Lindl.) fruit. Eur. Food Res. Technol. 223, 533–539. doi: 10.1007/s00217-005-0233-5
Cai, C., Xu, C. J., Li, X., Ferguson, I. B., and Chen, K. S. (2006c). Accumulation of lignin in relation to change in activities of lignification enzymes in loquat fruit flesh after harvest. Postharvest Biol. Technol. 40, 163–169. doi: 10.1016/j.postharvbio.2005.12.009
Cai, C., Xu, C. J., Shan, L. L., Li, X., Zhou, C. H., Zhang, W. S., et al. (2006d). Low temperature conditioning reduces postharvest chilling injury in loquat fruit. Postharvest Biol. Technol. 41, 252–259. doi: 10.1016/j.postharvbio.2006.04.015
Cao, S. F., Zheng, Y. H., Yang, Z. F., Tang, S. S., Jin, P., Wang, K. T., et al. (2008). Effect of methyl jasmonate on the inhibition of Colletotrichum acutatum infection in loquat fruit and the possible mechanisms. Postharvest Biol. Technol. 49, 301–307. doi: 10.1016/j.postharvbio.2007.12.007
Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140, 499–511. doi: 10.1104/pp.105.067231
Fornalé, S., Lopez, E., Salazar-Henao, J. E., Fernández-Nohales, P., Rigau, J., and Caparros-Ruiz, D. (2014). AtMYB7, a new player in the regulation of UV- sunscreens in Arabidopsis thaliana. Plant Cell Physiol. 55, 507–516. doi: 10.1093/pcp/pct187
Fornalé, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64, 633–644. doi: 10.1111/j.1365-313X.2010.04363.x
Fu, X. M., Kong, W. B., Peng, G., Zhou, J. Y., Azam, M., Xu, C. J., et al. (2012). Plastid structure and carotenogenic gene expression in red- and white-fleshed loquat (Eriobotrya japonica) fruits. J. Exp. Bot. 63, 341–354. doi: 10.1093/jxb/err284
Jin, H. L., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Kamdee, C., Imsabai, W., Kirk, R., Allan, A. C., Ferguson, I. B., and Ketsa, S. (2014). Regulation of lignin biosynthesis in fruit pericarp hardening of mangosteen (Garcinia mangostana L.) after impact. Postharvest Biol. Technol. 97, 68–76. doi: 10.1016/j.postharvbio.2014.06.004
Ketsa, S., and Atantee, S. (1998). Phenolics, lignin, peroxidase activity and increased firmness of damaged pericarp of mangosteen fruit after impact. Postharvest Biol. Technol. 14, 117–124. doi: 10.1016/S0925-5214(98)00026-X
Liu, Y. X., Zou, D. M., Wu, B. S., Lin, D. H., Zhang, Z. H., and Wu, J. C. (2015). Cloning and expression analysis of a CCoAOMT homolog in loquat fruit in response to low-temperature storage. Postharvest Biol. Technol. 105, 45–50. doi: 10.1016/j.postharvbio.2015.03.008
Lu, G. L., Li, Z. J., Zhang, X. F., Wang, R., and Yang, S. L. (2015). Expression analysis of lignin-associated genes in hard end pear(Pyrus pyrifolia Whangkeumbae) and its response to calcium chloride treatment conditions. J. Plant Growth Regul. 34, 251–262. doi: 10.1007/s00344-014-9461-x
Nicholas, D. B., and Clint, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/annurev-genet-102209-163508
Ohman, D., Demedts, B., Kumar, M., Gerber, L., Gorzsás, A., Goeminne, G., et al. (2013). MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 73, 63–76. doi: 10.1111/tpj.12018
Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L. J., Smith, C., et al. (2003a). Characterisation of a pine MYB that regulates lignification. Plant J. 36, 743–754. doi: 10.1046/j.1365-313X.2003.01916.x
Patzlaff, A., Newman, L. J., Dubos, C., Whetten, R., Smith, C., McInnis, S., et al. (2003b). Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 53, 597–608. doi: 10.1023/B:PLAN.0000019066.07933.d6
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
Rogers, L. A., and Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Shan, L. L., Li, X., Wang, P., Cai, C., Zhang, B., Sun, C. D., et al. (2008). Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. doi: 10.1007/s00425-008-0696-2
Shen, S. L., Yin, X. R., Zhang, B., Xie, X. L., Jiang, Q., Grierson, D., et al. (2016). CitAP2.10 activation of CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 67, 4105–4115. doi: 10.1093/jxb/erw189
Spitzer-Rimon, B., Farhi, M., Albo, B., Can’ani, A., Ben Zvi, M. M., Masci, T., et al. (2012). The R2R3-MYB-Like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24, 5089–5105. doi: 10.1105/tpc.112.105247
Tian, Q. Y., Wang, X. Q., Li, C. F., Lu, W. X., Yang, L., Jiang, Y. Z., et al. (2013). Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS ONE 8:e76369. doi: 10.1371/journal.pone.0076369
Vanholme, R., Cesarino, I., Rataj, K., Xiao, Y., Sundin, L., Goeminne, G., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341, 1103–1106. doi: 10.1126/science.1241602
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Verdonk, J. C., Haring, M. A., van Tunen, A. J., and Schuurink, R. C. (2005). ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell 17, 1612–1624. doi: 10.1105/tpc.104.028837
Wang, W. Q., Zhang, J., Ge, H., Li, S. J., Li, X., Yin, X. R., et al. (2016). EjMYB8 transcriptionally regulates flesh lignification in loquat fruit. PLoS ONE 11:e0154399. doi: 10.1371/journal.pone.0154399
Xie, X. L., Yin, X. R., and Chen, K. S. (2016). Roles of APETALA2/Ethylene responsive factors in regulation of fruit quality. Crit. Rev. Plant Sci. 35, 120–130. doi: 10.1080/07352689.2016.1213119
Xu, Q., Wang, W. W., Zeng, J. K., Zhang, J., Grierson, D., Li, X., et al. (2015). A NAC transcription factor, EjNAC1, affects lignification of loquat fruit by regulating lignin. Postharvest Biol. Technol. 102, 25–31.
Xu, Q., Yin, X. R., Zeng, J. K., Ge, H., Song, M., Xu, C. J., et al. (2014). Activator- and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J. Exp. Bot. 65, 4349–4359. doi: 10.1093/jxb/eru208
Yang, S. L., Zhang, X. N., Lu, G. L., Wang, C. R., and Wang, R. (2015). Regulation of gibberellin on gene expressions related with the lignin biosynthesis in ‘Wangkumbae’ pear(Pyrus pyrifolia Nakai) fruit. Plant Growth Regul. 76, 127–134. doi: 10.1007/s10725-014-9982-0
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi: 10.1111/pbi.12351
Zeng, J. K., Li, X., Zhang, J., Ge, H., Yin, X. R., and Chen, K. S. (2016). Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell Environ. 39, 1780–1789. doi: 10.1111/pce.12741
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., and Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. doi: 10.1105/tpc.108.061325
Zhou, J., Lee, C., Zhong, R., and Ye, Z.-H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. doi: 10.1105/tpc.108.063321
Keywords: EjODO1, lignification, loquat, MYB, transcriptional regulation
Citation: Zhang J, Ge H, Zang C, Li X, Grierson D, Chen K-s and Yin X - r (2016) EjODO1, a MYB Transcription Factor, Regulating Lignin Biosynthesis in Developing Loquat (Eriobotrya japonica) Fruit. Front. Plant Sci. 7:1360. doi: 10.3389/fpls.2016.01360
Received: 06 June 2016; Accepted: 26 August 2016;
Published: 16 September 2016.
Edited by:
Maoteng Li, Huazhong University of Science and Technology, ChinaReviewed by:
Yan Long, Chinese Academy of Agricultural Sciences, ChinaLiezhao Liu, Southwest University, China
Dengfeng Hong, Huazhong Agricultural University, China
Copyright © 2016 Zhang, Ge, Zang, Li, Grierson, Chen and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun-song Chen, akun@zju.edu.cn Xue-ren Yin, xuerenyin@zju.edu.cn
 Jing Zhang1,2,3
Jing Zhang1,2,3 Donald Grierson
Donald Grierson Kun-song Chen
Kun-song Chen Xue-ren Yin
Xue-ren Yin