- 1Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, China
- 2College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
Gibberellins (GAs) are plant hormones that play fundamental roles in plant growth and development. Gibberellin 2-oxidase (GA2ox) plays a direct role in determining the levels of bioactive GAs by catalyzing bioactive GAs or their immediate precursors to inactive forms. In this study, a GA2ox gene, designated JcGA2ox6, was isolated from Jatropha curcas. JcGA2ox6 is expressed in all tissues of adult Jatropha, with the highest expression level in male flowers and the lowest expression level in young leaves. Overexpression of JcGA2ox6 in Arabidopsis resulted in a typical dwarf phenotype, along with late flowering, smaller leaves and flowers, shorter siliques and smaller seeds. Similarly, when JcGA2ox6 was overexpressed in Jatropha, the transgenic plants exhibited a dwarf phenotype with dark-green leaves and smaller inflorescences, flowers, fruits and seeds. However, the flowering time of Jatropha was not affected by overexpression of JcGA2ox6, unlike that in the transgenic Arabidopsis. Moreover, the number of flowers per inflorescence, the weight of 10 seeds and the seed oil content were significantly decreased in transgenic Jatropha. The results indicated that overexpression of JcGA2ox6 had a great impact on the vegetative and reproductive growth of transgenic Jatropha. Furthermore, we found that the dwarf phenotype of transgenic Jatropha was caused by a decrease in endogenous bioactive GA4, which was correlated with the degree of dwarfism.
Introduction
Physic nut (Jatropha curcas L.) is a perennial woody plant that belongs to the Euphorbiaceae family and is monoecious, with male and female flowers borne on the same inflorescence (Divakara et al., 2010). Jatropha seeds contain a high amount of oil and therefore represent a promising feedstock for renewable biodiesel production (King et al., 2009; Abdulla et al., 2011; Sato et al., 2011; Chakrabarti and Prasad, 2012) and bio-jet fuel (Li et al., 2010). However, the potential of Jatropha as a biofuel plant is limited by its low seed production, which results from excessive vegetative growth and erratic flowering and fruiting (Ghosh et al., 2010; Ye et al., 2014). Previous studies showed that soil application of the gibberellin (GA) biosynthesis inhibitor paclobutrazol (PAC) in Jatropha promoted the transition of shoot growth from the vegetative to reproductive phase, which resulted in significant increases in the numbers of inflorescence and infructescence per plant and, therefore, a higher seed yield (Ghosh et al., 2010; Song et al., 2013). Similar to Jatropha, PAC treatment reduced vegetative vigor and improved flowering in mangoes (Winston, 1992). In young macadamia trees, uniconazole, another type of GA biosynthesis inhibitor, was applied as a soil drench, resulting in flower initiation (Nagao et al., 1999). Reduced levels of endogenous GA have been correlated with flowering in citrus (Koshita et al., 1999) and lychee plants (Chen, 1990). These studies suggest that GA suppresses floral initiation in perennial woody plants, which is contrary to the role of GA in promoting flowering in most herbaceous plants such as Arabidopsis (Blazquez et al., 1998), maize (Evans and Poethig, 1995), and chrysanthemums (Dong et al., 2017). Therefore, it is valuable to identify the functional genes responsible for reducing the content of endogenous GA to promote the floral initiation of Jatropha.
Gibberellins are plant hormones that control diverse aspects of plant growth and development, such as seed germination, shoot elongation, leaf expansion, flower initiation and fruit development (Harberd et al., 1998; Fleet and Sun, 2005; Yamaguchi, 2008). Three major oxidase gene families of GA 20-oxidase (GA20ox), GA 3-oxidase (GA3ox) and GA 2-oxidase (GA2ox) participate in GA synthesis and degradation by a series of conversions from geranylgeranyl diphosphate (Hedden and Phillips, 2000). The GA2oxs are pivotal genes that reduce the endogenous bioactive GA content of plants with dwarfism (Lee and Zeevaart, 2005; Schwechheimer, 2008; Yamaguchi, 2008; Zhou et al., 2012). The functions of GA2ox genes have been characterized in various plant species. Overexpression of AtGA2ox7 or AtGA2ox8 in Arabidopsis decreased GA levels and flowering time was delayed in transgenic plants (Schomburg et al., 2003), and GA4 had been found to promote the transition from the vegetative to the reproductive phase (Eriksson et al., 2006; Yamaguchi et al., 2014). In rice, the transgenic plants carrying Actin:OsGA2ox1 showed late flowering with low endogenous levels of GA1 (Sakamoto et al., 2003), and ectopic expression of GA2ox6 can increase grain yield by 10–30% (Lo et al., 2017). However, in woody plant poplars, overexpression of PtGA2ox causes the dwarf trait and early flowering (Zawaski et al., 2011). These results led us to genetically inhibit vegetative growth and promote reproductive growth in Jatropha by overexpressing Jatropha GA2ox genes.
Based on a previous study, there are five GA2-oxidase homologue genes in Jatropha (Gao et al., 2015), designated as JcGA2ox2 (GenBank accession No. KDP37976), JcGA2ox4 (GenBank accession No. KDP27967), JcGA2ox6 (GenBank accession No. KDP28294), JcGA2ox7 (GenBank accession No. KDP39055), and JcGA2ox8 (GenBank accession No. KDP30016). In this study, we isolated JcGA2ox6 and analyzed its function in transgenic Arabidopsis and Jatropha, and we found that overexpression of JcGA2ox6 had a significant impact on plant growth and development. The transgenic plants exhibited a typical dwarf phenotype with darker green leaves and smaller reproductive organs.
Materials and Methods
Plant Materials and Growth Conditions
Jatropha curcas plants cultivated in Xishuangbanna, Yunnan Province, China, were used in this study as described previously (Pan and Xu, 2011). The Arabidopsis thaliana ecotype Columbia (Col-0) and the transgenic lines were grown in plant growth chambers at 22 ± 2°C under long-day (LD, 16 h light/8 h dark) or short-day (SD, 8 h light/16 h dark) conditions. Phenotype analysis was performed on homozygous (T3) Arabidopsis plants and heterozygous (T0) Jatropha plants.
Cloning of JcGA2ox6 cDNA
Total RNA was extracted from the leaves of flowering Jatropha plants using the protocol described by Ding et al. (2008). First-strand cDNA was synthesized using M-MLV reverse transcriptase from TAKARA (Dalian, China) according to the manufacturer’s instructions. A full-length JcGA2ox6 CDS was amplified by PCR using the primers XA579 and XA580 (Supplementary Table 1), which introduced BamHI and SalI restriction sites at the ends of the JcGA2ox6 CDS fragment, respectively. The PCR products were subsequently cloned into the pGEM-T vector (Promega Corporation, Madison, WI, United States) and sequenced. All primers used in this research are listed in Supplementary Table 1.
Sequences and Phylogenetic Analyses of JcGA2ox6
The JcGA2ox6 amino acid sequence was deduced according to the coding sequence (GenBank accession No. KDP28294). Related sequences were identified through a BLAST search1. To determine the amino acid identities, the alignment results were subjected to pairwise comparisons using DNAMAN 6.0. A phylogenetic tree was constructed based on the protein sequences with MEGA 5.0.2 A neighbor-joining phylogenetic tree was generated with MEGA 5.0 using the Poisson model, with gamma-distributed rates and 1,000 bootstrap replicates.
Construction of Plant Expression Vectors and Transformation of Arabidopsis and Jatropha
To construct the plant overexpression vector 35S:JcGA2ox6, the JcGA2ox6 sequence was excised from the pGEM-T vector (Promega, Madison, WI, United States) using the restriction enzymes BamHI and SalI. Then, JcGA2ox6 was cloned into the pOCA30 vector containing the CaMV35S promoter and the 35S enhancer (Chen and Chen, 2002). The JcUEP promoter (Tao et al., 2015) was obtained by PCR from the Jatropha genomic DNA using the primers XB348 and XB349 (Supplementary Table 1), which introduced HindIII and SacI restriction sites, respectively. The PCR products were cloned into pGEM-T and sequenced. To construct the JcUEP:JcGA2ox6 plasmid, the CaMV35S promoter of the 35S:JcGA2ox6 vector was replaced with the JcUEP promoter using the restriction enzymes HindIII and SacI.
Transformation of Arabidopsis with the Agrobacterium strain EHA105 carrying the 35S:JcGA2ox6 construct was performed using the floral dip method (Clough and Bent, 1998). Transformation of Jatropha with the Agrobacterium strain EHA105 carrying the 35S:JcGA2ox6 and JcUEP:JcGA2ox6 constructs was performed according to the protocol described by Pan et al. (2010) and Fu et al. (2015).
Expression Analysis by Quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from frozen Jatropha tissues as described by Ding et al. (2008). Arabidopsis total RNA was extracted from frozen tissues using TRIzol reagent (Transgene, China). First-strand cDNA was synthesized using the PrimeScript® RT Reagent Kit with gDNA Eraser (TAKARA, Dalian, China). qRT-PCR was performed using SYBR® Premix Ex TaqTM II (TAKARA) on a Roche 480 Real-Time PCR Detection System (Roche Diagnostics). qRT-PCR was performed using three independent biological replicates and three technical replicates for each sample. Data were analyzed using the 2-ΔΔCT method as described by Livak and Schmittgen (2001). The transcript levels of specific genes were normalized using Jatropha JcActin1 or Arabidopsis AtActin2. The primers used for qRT-PCR are listed in Supplementary Table 1.
Chlorophyll Content Measurement
Three 1-cm2 leaf segments were removed from mature leaves of wild-type (WT) and transgenic Jatropha using a hole punch. The total chlorophyll, chlorophyll a and chlorophyll b contents were measured following the protocol described by Arnon (1949). Each measurement was repeated three times.
Quantification of Endogenous GAs
The WT and T1 transgenic Jatropha (lines L43 and L27) were grown in soil for 6 weeks in a growth chamber at 28°C under a 16 h light/8 h dark photoperiod (Figure 8A). Their young stems (from shoot tip to node 2) (Figure 8B) were collected for GA quantification. The GA contents were determined by the Wuhan Greensword Creation Technology Company, and the analysis was performed as described previously (Chen et al., 2012). Three independent biological replicates and three technical replicates were measured for each sample.
Results
Cloning and Sequence Analysis of JcGA2ox6
A combined reverse transcriptase-polymerase chain reaction (RT-PCR) strategy was used to isolate JcGA2ox6 cDNA from Jatropha. The JcGA2ox6 coding sequence (CDS) (GenBank accession no. KDP28294) comprises 1002 bp and encodes a 333-amino-acid protein. A multiple alignment was performed using the JcGA2ox6 sequence and the sequences of GA2ox homologs from other species (Supplementary Figure 1A). JcGA2ox6 showed 84, 74, 71, and 57% sequence identity with Ricinus communis GA2ox5 (RcGA2ox5), Populus trichocarpa GA2ox2 (PtGA2ox2), Vitis vinifera GA2ox5 (VvGA2ox5) and Arabidopsis thaliana GA2ox6 (AtGA2ox6), respectively.
To better characterize the JcGA2ox genes within the large GA2ox family, a phylogenetic analysis was performed with amino acid sequences of the GA2oxs in some plant species (Supplementary Figure 1B). The analysis showed that JcGA2ox2, JcGA2ox4, and JcGA2ox6 have the highest identity with RcGA2ox2, RcGA2ox4, and RcGA2ox5 protein from Ricinus communis, respectively, and that they clustered with the C19-GA2ox group of Arabidopsis. This result suggests that all of them encode this type of enzyme. In contrast, JcGA2ox7 and JcGA2ox8 clustered with the C20-GA2oxs group of Arabidopsis.
Expression Pattern of JcGA2ox6 in Jatropha
To investigate the expression pattern of JcGA2ox6 in Jatropha, we performed a qRT-PCR analysis with the total RNAs extracted from various tissues of adult plants, including the roots, stems, young and mature leaves, shoot tips, inflorescences, male and female flowers, fruits at 10 days after pollination (DAP), pericarps at 30 DAP and seeds at 30 DAP. The expression profile showed that JcGA2ox6 was almost constitutively expressed in adult Jatropha (Figure 1). The primary expression levels were detected in the roots, stems, flowers and pericarps at 30 DAP, with the highest level found in male flowers. Very low expression levels were detected in the mature leaves, shoot tips, inflorescences, fruits and seeds at 30 DAP, especially the young leaves.
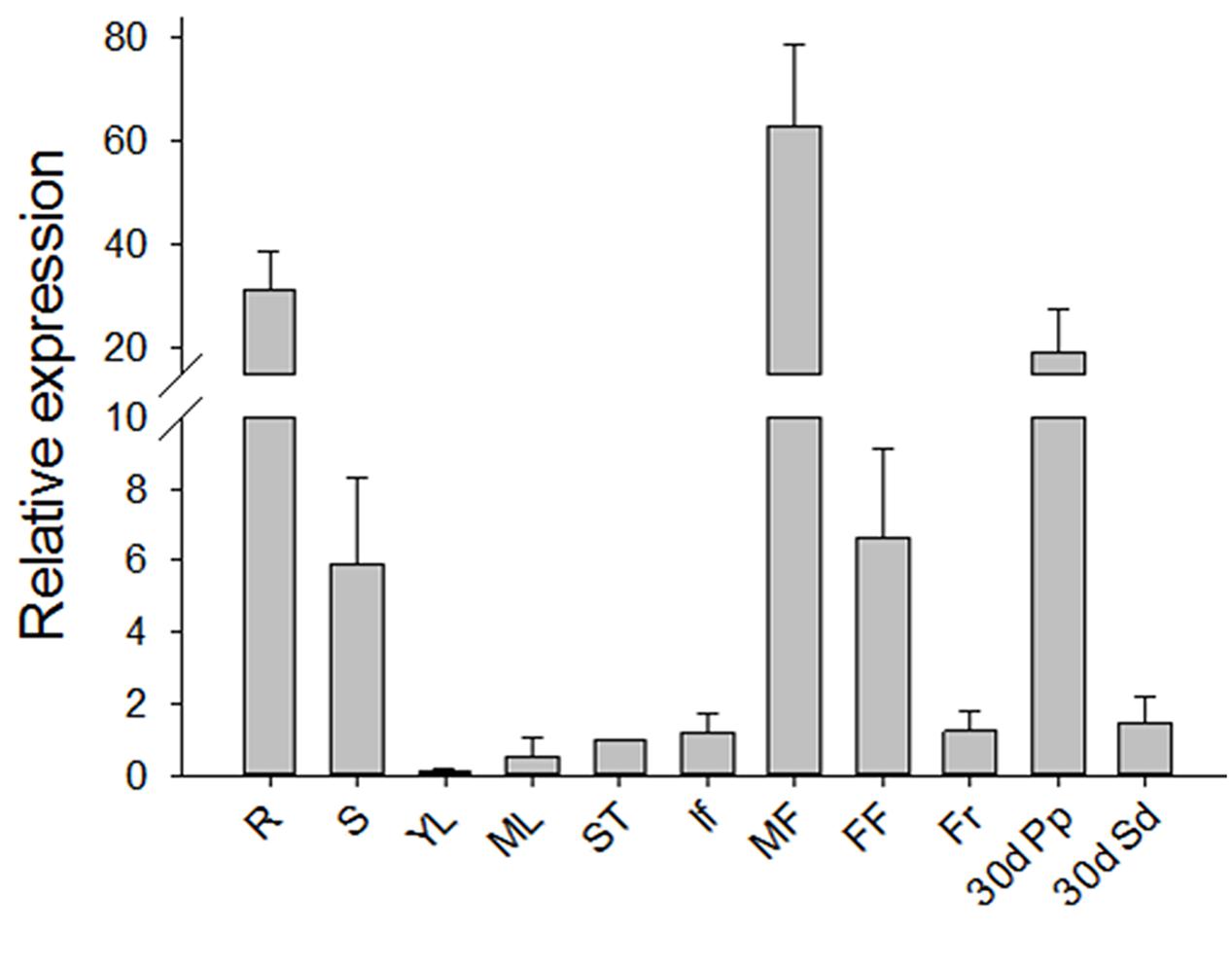
FIGURE 1. Expression profile of JcGA2ox6 in three-year-old adult Jatropha. The qRT-PCR results were obtained from three independent biological replicates and three technical replicates for each sample. R, roots; S, stems; YL, young leaves; ML, mature leaves; ST, shoot tips; If, inflorescences; MF, male flowers; FF, female flowers; Fr, fruits at 10 days after pollination (DAP); 30d Pp, pericarps at 30 DAP; 30d Sd, seeds at 30 DAP. The levels of the detected amplicons were normalized using the amplified products of the JcActin1. The mRNA level in the shoot tips was set as the standard, with a value of 1. The values are presented as the means ± standard deviation.
Overexpression of JcGA2ox6 in Arabidopsis Caused the Dwarf Phenotype with Late Flowering, Smaller Flowers, Shorter Siliques and Smaller Seeds
To determine the roles of JcGA2ox6 in plant growth and development, 35S:JcGA2ox6 was transformed into Arabidopsis for preliminary analysis. WT Arabidopsis under the same growth conditions was used as a control. Transgenic plants were confirmed by qRT-PCR analysis of JcGA2ox6 expression using rosette leaves. More than twenty independent T1 transgenic lines were generated with the 35S:JcGA2ox6 construct. Transgenic plants showed high JcGA2ox6 expression levels (Figure 2E). Under LD conditions, most transgenic lines showed a dwarf phenotype with late flowering.
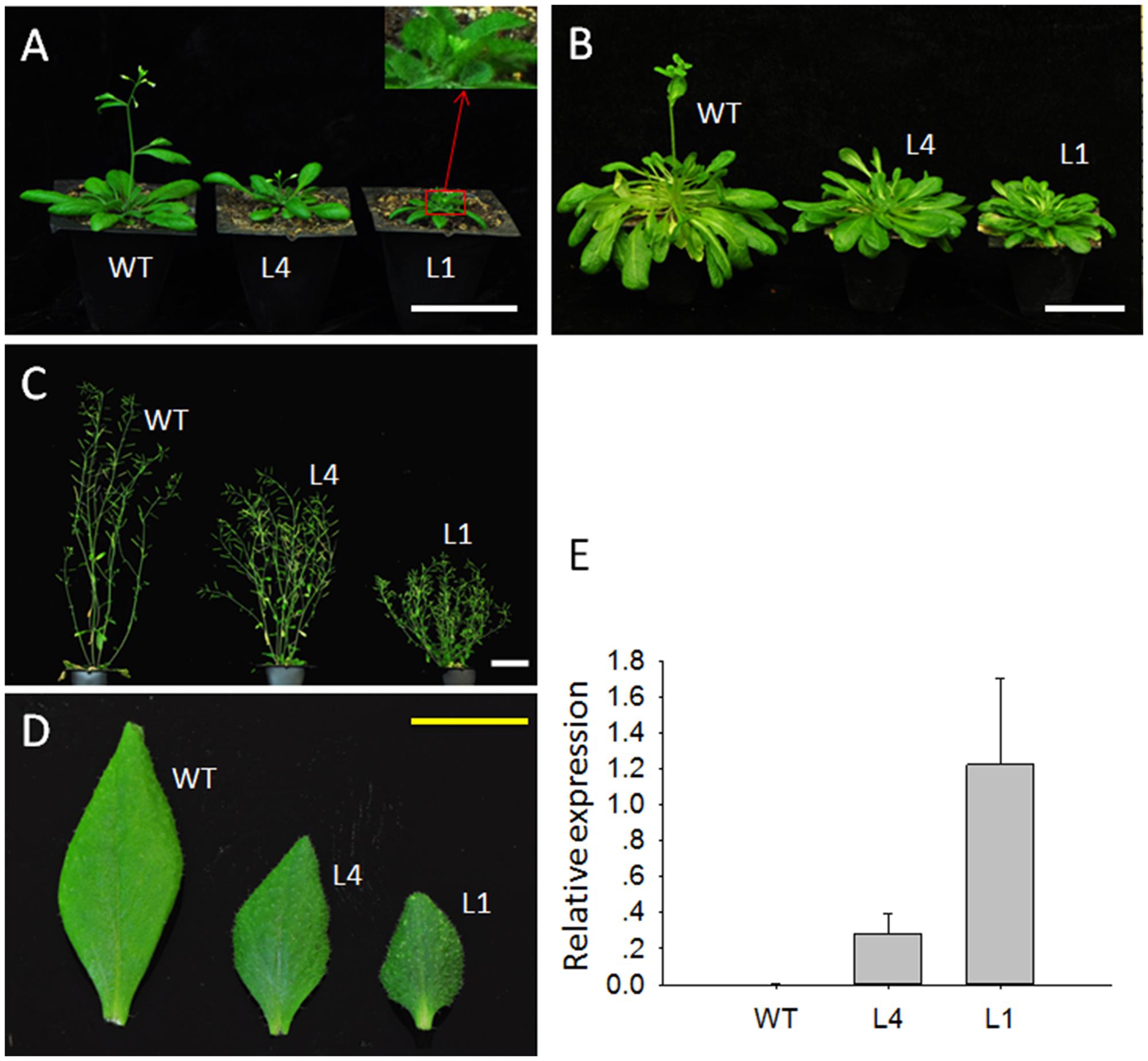
FIGURE 2. Ectopic expression of JcGA2ox6 causes the dwarf phenotype with small leaves and late flowering in transgenic Arabidopsis. (A) Thirty-five-day-old plants grown under LD conditions. (B) One-hundred-twenty-five-day-old plants grown under SD conditions. (C) Seventy-five-day-old plants grown under LD conditions. (D) The first cauline leaves of WT and transgenic Arabidopsis plants under LD conditions. (E) qRT-PCR analysis of JcGA2ox6 in WT and transgenic Arabidopsis plants. The levels of the detected amplicons were normalized using the amplified products of AtActin2. The values are presented as the means ± standard deviation. WT, wild type; L4 and L1 represent independent 35S:JcGA2ox6 transgenic Arabidopsis lines. White bars = 5 cm, yellow bar = 1 cm.
We selected two independent homozygous lines (L4 and L1) in the T3 generation to examine the phenotypes. Compared with the WT plants, Arabidopsis overexpressing JcGA2ox6 were 12.1–23 cm shorter (Figure 2C and Table 1) and produced smaller rosette and cauline leaves (Figures 2A,D) under LD conditions. Furthermore, the transgenic lines bolted later under both LD and SD conditions, but there was no significant difference in rosette leaf number (Figures 2A,B and Tables 1, 2). The phenotypes of the transgenic Arabidopsis were similar to that of the GA-deficient mutant ga1–3 (Koornneef and Vanderveen, 1980), which suggested that overexpression of JcGA2ox6 reduced the endogenous GA levels. Furthermore, flower development was also affected, as smaller sepals, petals, stamens and pistils were noted (Figures 3A,B). However, the transgenic plants were fertile, producing shorter siliques and smaller seeds (Figures 3C–E).
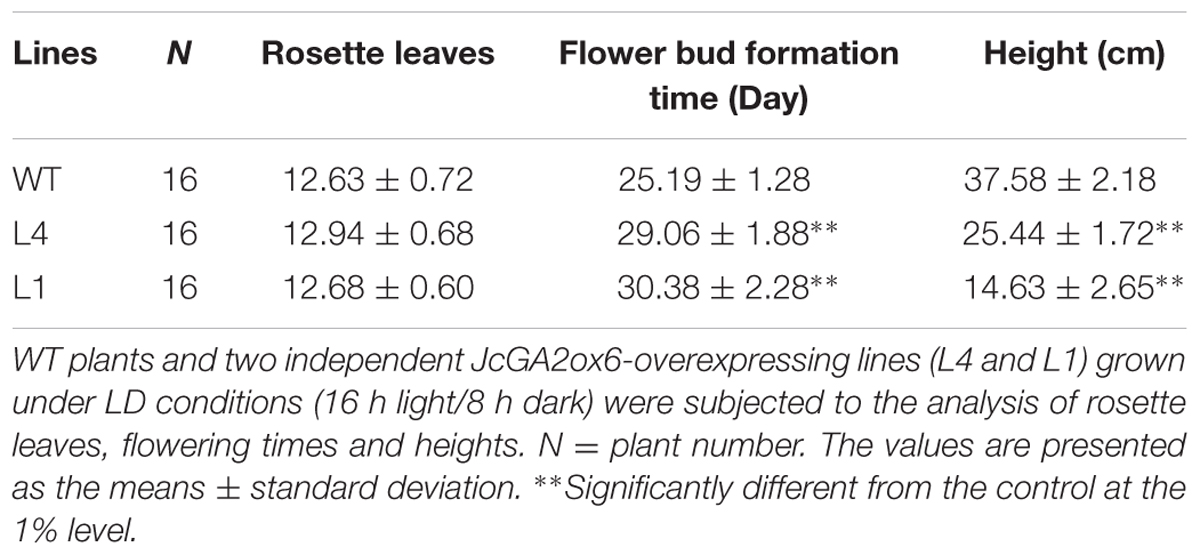
TABLE 1. Overexpression of JcGA2ox6 caused dwarf phenotype with late flowering in transgenic Arabidopsis under LD conditions.
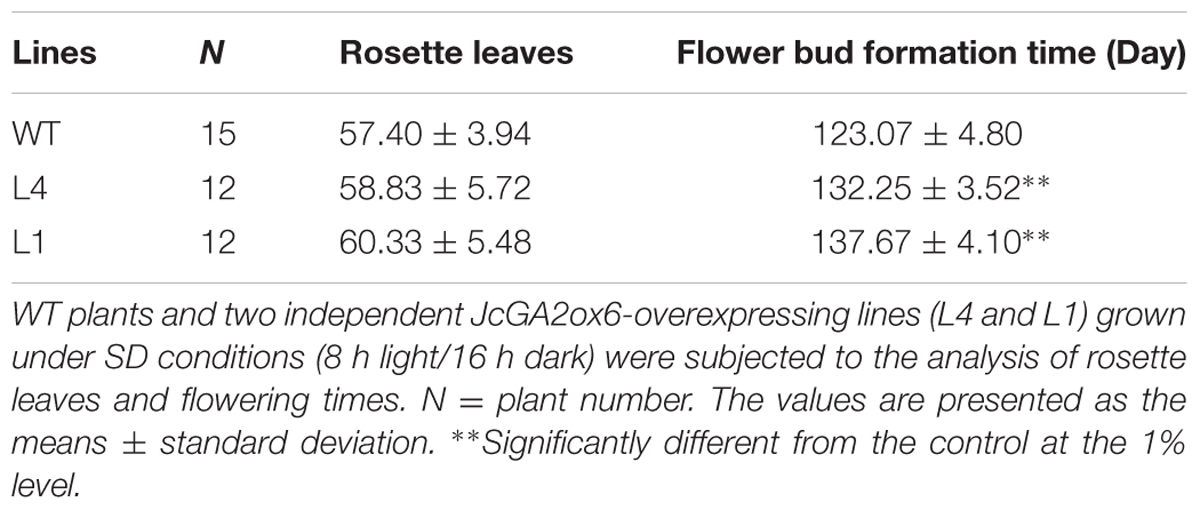
TABLE 2. Overexpression of JcGA2ox6 causes late flowering in transgenic Arabidopsis under SD conditions.
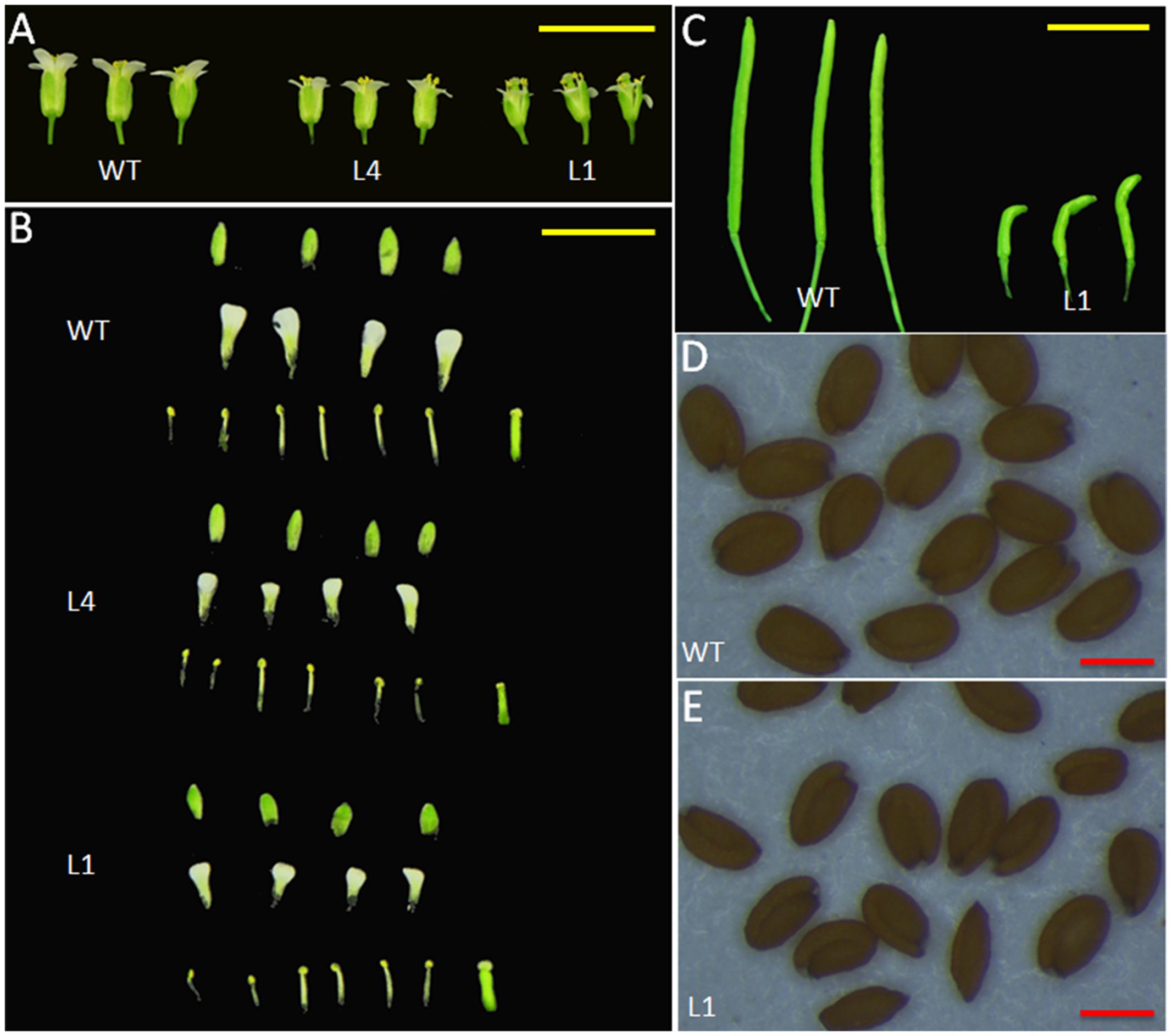
FIGURE 3. Morphological changes in the reproductive organs of 35S:JcGA2ox6 transgenic Arabidopsis. (A) Flowers from WT and transgenic L4 and L1 lines. (B) Anatomy of the flowers. (C) Siliques from the WT and transgenic line L1. (D,E) Seeds from the WT and transgenic line L1. Yellow bars = 5 mm, red bars = 0.3 mm.
Overexpression of JcGA2ox6 in Jatropha Caused a Dwarf Phenotype with Small Dark-Green Leaves
To further test whether JcGA2ox6 behaves accordingly in Jatropha, the transgenic Jatropha overexpressing JcGA2ox6 was generated. Transgenic shoots overexpressing JcGA2ox6 under the control of the CaMV35S promoter showed abnormal phenotypes of severely crimpled leaves and no obvious stems (Supplementary Figure 2). Moreover, these shoots were unable to generate roots. Therefore, we grafted these shoots onto WT rootstocks, but they hardly grew. These observations prompted us to replace the CaMV35S promoter with a weaker, JcUEP promoter, which is an alternative to the CaMV35S promoter for driving constitutive overexpression of transgenes in Jatropha (Tao et al., 2015). We successfully generated more than 30 transgenic Jatropha lines carrying the JcUEP:JcGA2ox6 transgene.
When all plants grew in pots for 2 months, the JcUEP:JcGA2ox6 transgenic plants exhibited a dwarf phenotype with small dark-green leaves (Figure 4A). The average height of the transgenic Jatropha plants was decreased by 74% compared with that of the WT plants (Figure 4B). Since the leaves of transgenic plants turned dark green, the mature leaves (Figure 4C) were collected to measure the chlorophyll content. The average chlorophyll a, chlorophyll b and total chlorophyll contents in the transgenic plants were 1.9-, 1.3-, and 1.6-fold higher than those of the WT plants, respectively (Figure 4D). This result is consistent with studies in for example transgenic potato (Jackson and Prat, 1996; Martínez-García et al., 2001) and soybean (Suo et al., 2012), which showed that the chlorophyll concentration was inversely correlated with GA1 or GA1 and GA4 contents. In Solanum species, overexpression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. produced darker green leaves with higher concentrations of chlorophylls (Dijkstra et al., 2008). After 5 months of plant growth in the field, the transgenic plants were still dwarfed (Figures 5A–C). In addition, shoot branching was not affected by this transgene. The numbers of branches in WT and transgenic Jatropha were almost the same. We examined the phenotypes of two independent transgenic Jatropha lines L43 and L27, which exhibited intermediate and high expression levels of JcGA2ox6, respectively (Figure 5D). Accordingly, the heights (from the soil to the highest part of the main stem) of L43 and L27 lines were decreased by 37 and 60%, respectively, compared with that of the WT plant (Figure 5E). The retarded vegetative growth was similar to that of transgenic Arabidopsis (Figure 2C and Table 1).
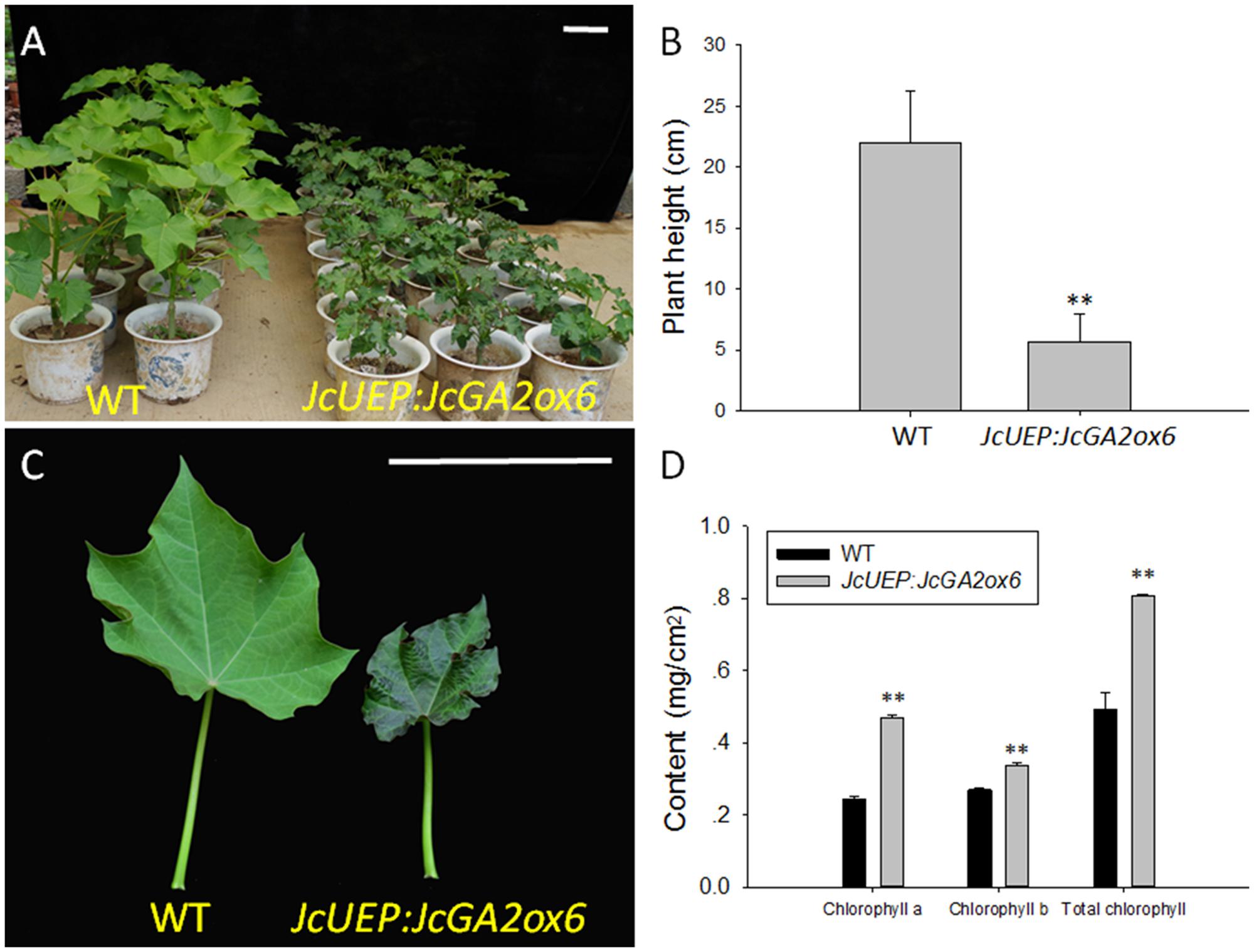
FIGURE 4. Phenotypic analysis of JcUEP:JcGA2ox6 transgenic Jatropha plants grown in pots for 2 months. (A,B) A height comparison between WT and transgenic Jatropha. (C) The mature leaves from the WT and transgenic Jatropha plants. (D) Chlorophyll a, chlorophyll b and total chlorophyll contents of the mature leaves. The values are presented as the means ± standard deviation. Bars = 10 cm. ∗∗Significantly different from the control at the 1% level.
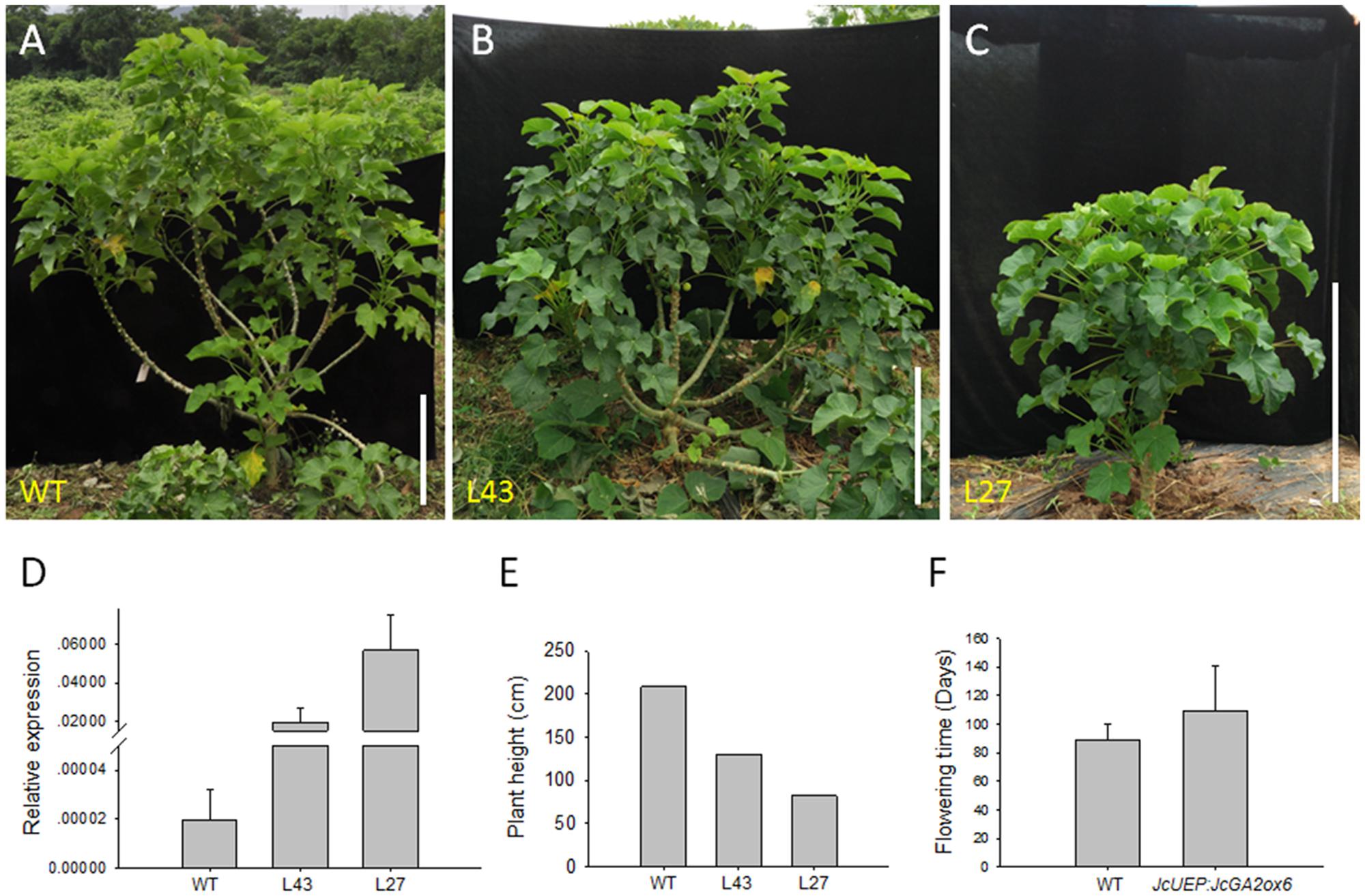
FIGURE 5. Dwarf phenotype of JcUEP:JcGA2ox6 transgenic Jatropha grown in the field for 5 months. (A) WT. (B,C) Transgenic Jatropha (L43 and L27). (D) qRT-PCR analysis of JcGA2ox6 in the WT and transgenic Jatropha plants. The levels of the detected amplicons were normalized using the amplified products of JcActin1. The values are presented as the means ± standard deviation. (E) Comparison of the heights of the WT and transgenic Jatropha plants. (F) Comparison of the flowering time of the WT and transgenic Jatropha plants. The values are presented as the means ± standard deviation. Bars = 50 cm.
Overexpression of JcGA2ox6 in Jatropha Affected Flower, Fruit and Seed Development
When WT and transgenic plants were grown in the field for approximately 3 months, inflorescence buds emerged in both plants (Figure 5F), suggesting that the flowering time of Jatropha was not affected by overexpression of JcGA2ox6, unlike the late-flowering noted in transgenic Arabidopsis. The inflorescence numbers in the transgenic lines, contrary to our expectation, were lower than those in the WT plants (Figure 6D). Additionally, each inflorescence produced fewer male and female flowers (Figure 6E). Moreover, the inflorescences size was reduced in the transgenic Jatropha lines (Figure 6A) because of the shorter inflorescence stalks (Supplementary Figure 3A) and smaller male and female flowers (Figures 6B,C and Supplementary Figures 3B–E). In addition, similar to transgenic Arabidopsis (Figure 3B), the floral organs remained intact in transgenic Jatropha (Figures 6F,G) and plants were fertile.
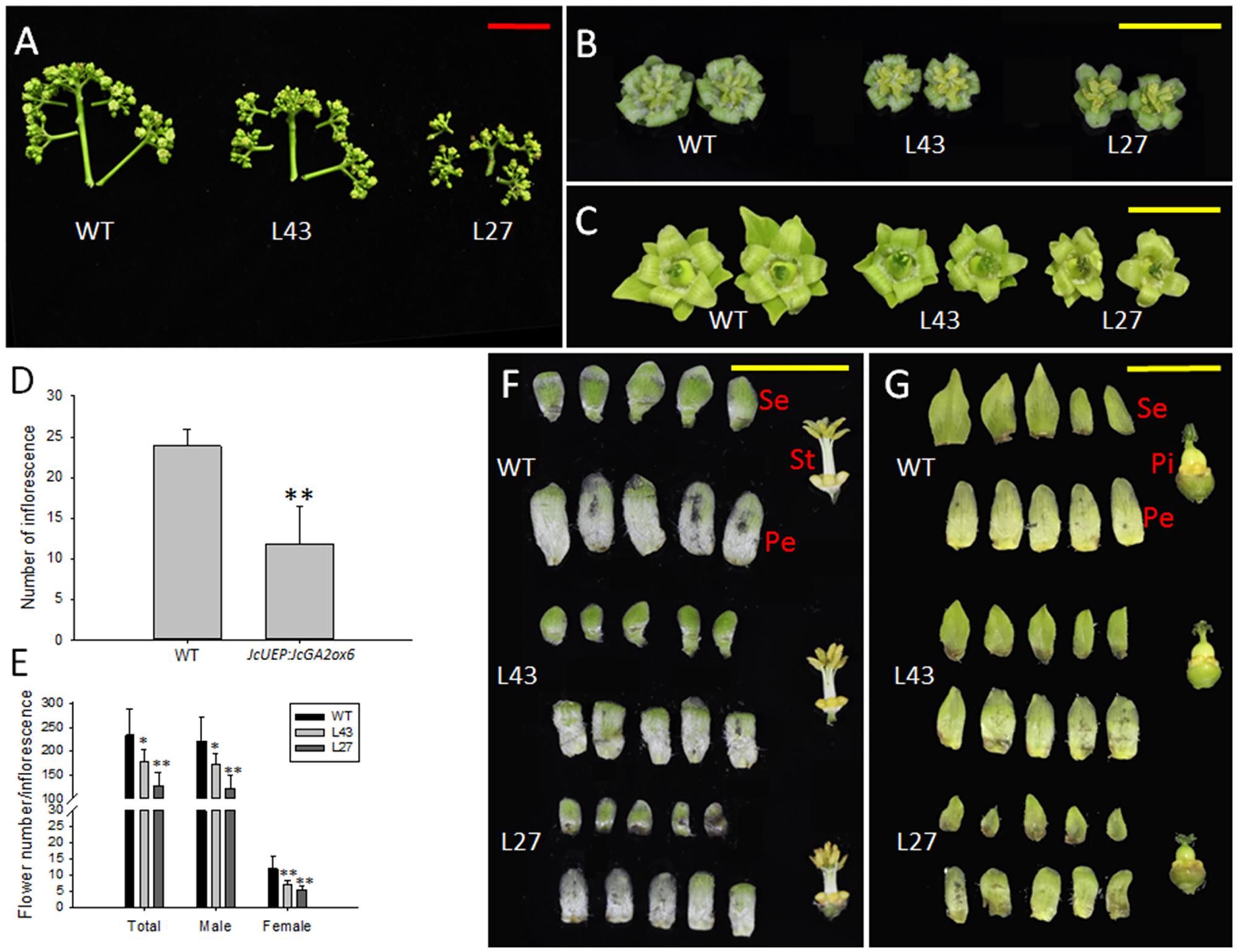
FIGURE 6. Inflorescence and flower development in JcUEP:JcGA2ox6 transgenic Jatropha. (A) Inflorescences of WT and transgenic Jatropha (L43 and 27). (B) Male flowers of WT and transgenic Jatropha (L43 and 27). (C) Female flowers of WT and transgenic Jatropha (L43 and 27). (D) The inflorescence numbers in WT and transgenic Jatropha. Six WT plants and eight transgenic plants were analyzed. (E) The flower numbers per inflorescence in WT and transgenic Jatropha plants. The values are presented as the means ± standard deviation; ∗significantly different from the control at the 5% level, ∗∗significantly different from the control at the 1% level. (F) Anatomy of male flowers from WT and transgenic Jatropha (L43 and 27). (G) Anatomy of female flowers from WT and transgenic Jatropha (L43 and 27). Se, sepals; Pe, petals; St, stamens; Pi, pistils. Red bar = 5 cm, yellow bars = 1 cm.
Subsequently, fruit and seed development was affected as well by this transgene. Compared with WT plants, transgenic Jatropha lines L43 and L27 exhibited smaller fruits (Figure 7A), as the fruit lengths were reduced by 13.04 and 31.59%, respectively (Figure 7B), while the widths were almost the same (Figure 7C). Consequently, the seeds from the L43 and L27 lines were also small, with 12.90 and 22.04% shorter lengths, respectively, compared with the seed length in WT plants (Figures 7D–G). The results indicated that overexpressing JcGA2ox6 in transgenic Jatropha inhibited the elongation of fruits and seeds other than the widths. It is known that GA induces cell elongation (Kende and Zeevaart, 1997). We supposed a reduced GA content caused by overexpression of JcGA2ox6 would repress cell elongation in fruits and seeds. Consistent with this, in liliaceous Tricyrtis sp., most transgenic plants overexpressing TfGA2ox2 showed no significant differences in cell widths, but the lengths significantly decreased compared with those of control plants (Otani et al., 2013). Furthermore, we analyzed the seed weights and seed oil contents in the L43 and L27 lines. The oil contents were measured using a mini-spec mq-one Seed Analyzer (Bruker Optik, Ettlingen, Germany) (Pan and Xu, 2011). The results showed that the average weights of 10 seeds and the seed oil contents in transgenic lines were significantly decreased (Figures 7H,I).
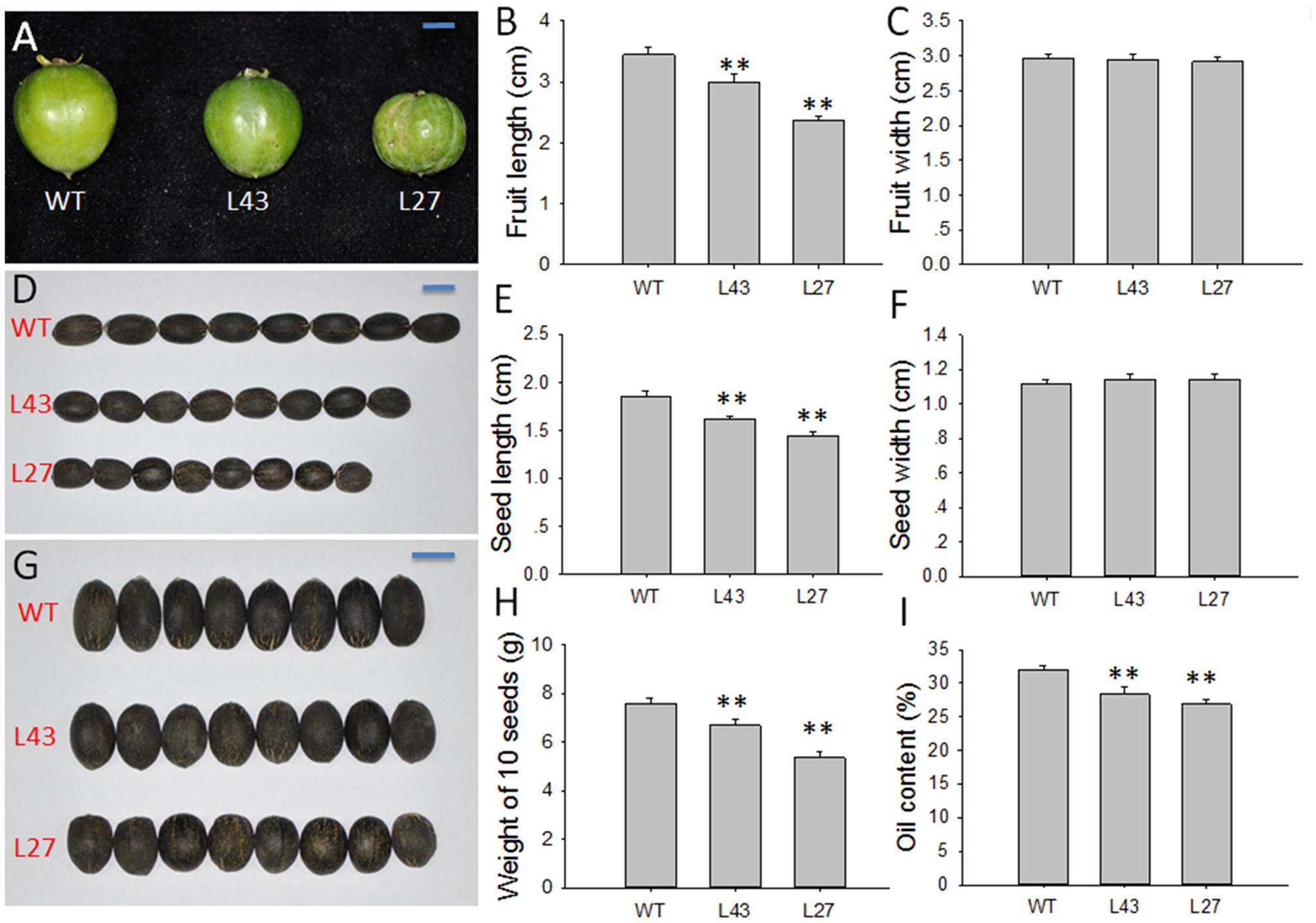
FIGURE 7. Agronomic traits of the fruits and seeds in WT and JcUEP:JcGA2ox6 transgenic Jatropha. (A) Fruits of WT and transgenic Jatropha (L43 and 27). (B,C) Fruit length and width of WT and transgenic Jatropha (L43 and 27). (D,G) Seeds of WT and transgenic Jatropha (L43 and 27). (E,F) Seed length and width of WT and transgenic Jatropha (L43 and 27). (H) Weight of 10 seeds in WT and transgenic Jatropha (L43 and 27). (I) Oil content of WT and transgenic Jatropha (L43 and 27). The values are presented as the means ± standard deviation. Bars = 1 cm. ∗∗Significantly different from the control at the 1% level.
Effect of Overexpression of JcGA2ox6 on the Endogenous GA Contents
To determine whether the endogenous GA contents in transgenic Jatropha (L43 and L27) were affected by JcGA2ox6, the non-13-hydroxylated GAs (GA12, GA9, GA7, GA4 and GA34) and the 13-hydroxylated GAs (GA53, GA20, GA3, GA1 and GA8) (Figure 8D) were determined in 6-week-old T1 and WT seedlings (Figure 8A). The results (Figure 8C) showed that the levels of bioactive GA4 were significantly decreased in transgenic lines and were correlated with the degree of dwarfism (Figure 8A). However, the levels of GA34, the deactivated product of GA4, did not differ significantly between the WT and transgenic lines. Contrary to GA4, the bioactive GA3 levels were increased in transgenic lines. Therefore, we supposed that the reduced GA4 levels caused negative feedback control of GA biosynthetic gene expression, resulting in an increase in GA3 levels. Consequently, elevated expression levels of JcGA20ox1, JcGA20ox3, JcGA3ox1, JcGA3ox2 and JcGA3ox3 were detected in transgenic lines (Supplementary Figure 4). This effect of negative feedback regulation was previously confirmed in other plant species (Cowling et al., 1998; Suo et al., 2012). Another two bioactive GAs, GA1 and GA7, were not detected in either the WT or transgenic lines. However, GA8, the deactivated product of GA1 (Yamaguchi, 2008), accumulated in both lines, and there was a significant increase in its levels in the transgenic lines, implying that elevated expression levels of GA biosynthetic genes caused higher GA1 levels than WT plants. This result indicated the GA1 in Jatropha seedlings was completely deactivated by JcGA2ox. Taken together, it suggests that the dwarf phenotype of transgenic Jatropha resulted from a decrease in the endogenous bioactive GA4. In addition, the transgenic lines displayed decreased levels of GA20, GA12 and GA9, the immediate precursors of GA1 and GA4 (Figure 8D), while GA53 did not change significantly.
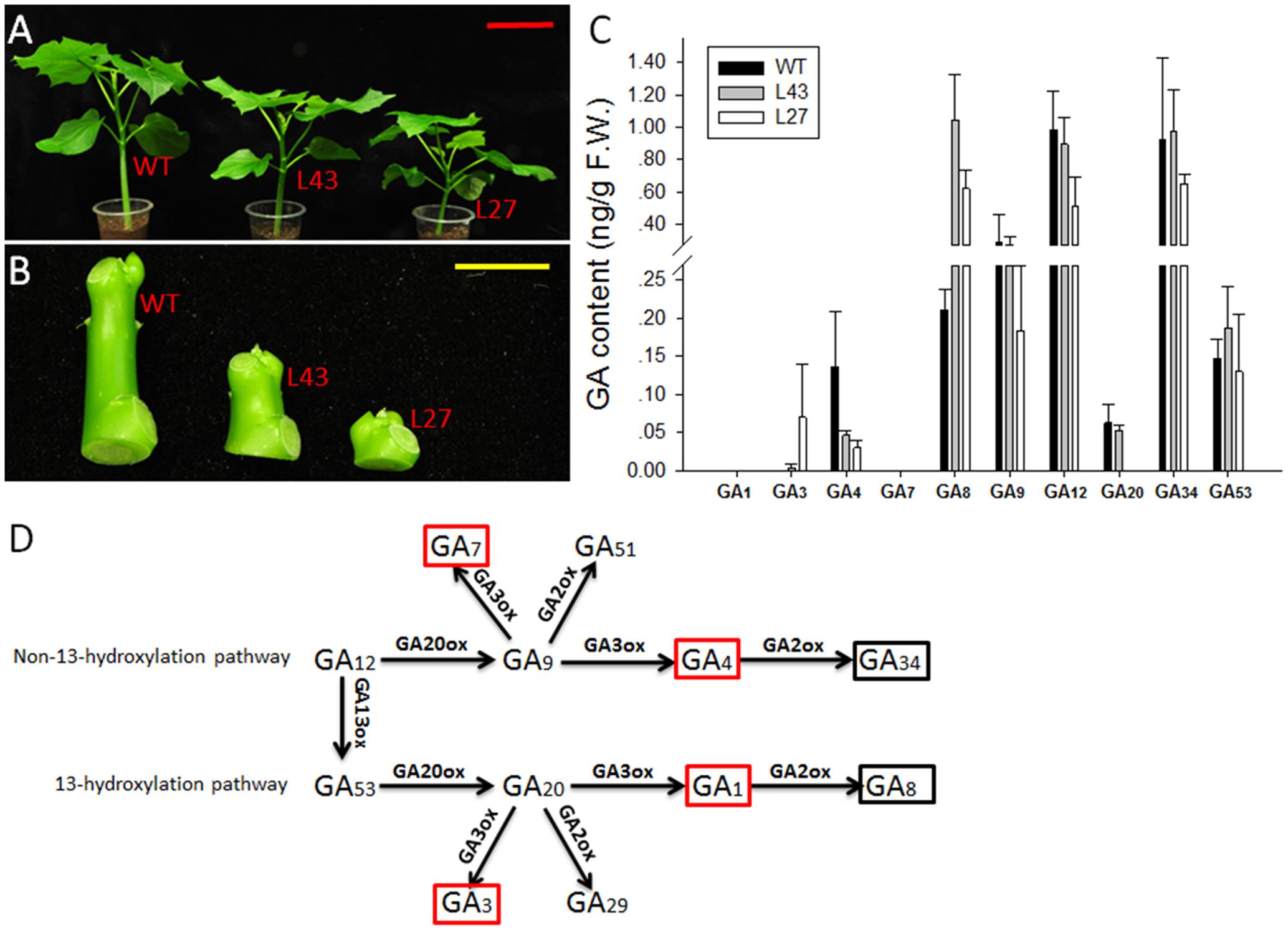
FIGURE 8. Analysis of GA contents in 6-week-old seedlings from WT and JcUEP:JcGA2ox6 transgenic Jatropha (T1). (A) Six-week-old seedlings from WT and transgenic Jatropha (L43 and 27). (B) Samples of young stems (from shoot tip to node 2) were collected for GA quantification. (C) GA contents of WT and transgenic Jatropha (L43 and 27). The values are presented as the means ± standard deviation. (D) The schematic representation of the principal pathways of GA metabolism in higher plants. Red box, bioactive GA; Black box, inactive GA. Red bar = 10 cm, yellow bar = 1 cm.
Discussion
In the present study, we identified a GA2-oxidase homologue gene in Jatropha, JcGA2ox6. The transgenic Arabidopsis and Jatropha overexpressing JcGA2ox6 demonstrated the GA-deficient phenotype of dwarfism. By analyzing the contents of the endogenous GAs in transgenic Jatropha seedlings, the levels of bioactive GA4 significantly decreased in transgenic lines (Figure 8C) and were correlated with the degree of dwarfism (Figure 8A). However, there was no equivalent increase in the amount of GA34, the deactivated product of GA4. This result is probably due to a rapid turnover of GA34 to its catabolite (Coles et al., 1999; Thomas et al., 1999). Though GA4 was downregulated in transgenic Jatropha, GA3 was upregulated. This may suggest that the non-13-hdroxylation pathway was redirecting synthesis toward the 13-hydroxylation pathway. Thus, instead of conversion of GA12 to GA9, GA4, flow was toward GA53, GA20, and then GA3. In addition, the bioactive GA1 and GA7 were not detected in either the WT or transgenic lines, suggesting that they did not function as a promoter in the stem elongation of Jatropha seedlings, not to mention GA3 which was elevated in the transgenic lines. Obviously, GA4 is the major bioactive GA that regulates Jatropha stem elongation. Similarly, in Arabidopsis and hybrid aspen, the GA4 plays a pivotal role in controlling shoot elongation (Xu et al., 1997; Cowling et al., 1998; Israelsson et al., 2004). Perhaps GA1, GA3 and GA7 are required in other aspects of Jatropha growth and development, as the major bioactive GA is different in the diverse tissue development stages of some plant species. In rice, the GA4 levels were undetectable in the uppermost internodes and GA1 was the major bioactive GA in vegetative tissues of rice (Zhu et al., 2006). In contrast, an extremely high amount of GA4 accumulated in pollen (Kobayashi et al., 1988). Because drastic GA-dependent reactions occur during the reproductive stage, such as rapid elongation of the heading stem and pollen tube, rice may use the most effective GA at this stage and use GA1 for ordinary GA-dependent reactions at the vegetative stage (Ueguchi-Tanaka et al., 2007). In Populus, the GA3 and GA4 regulated different glucan hydrolase family 17 genes (GH17s) to govern dormancy cycling at the shoot apex (Rinne et al., 2011).
In addition to the dwarf phenotype, transgenic Arabidopsis and Jatropha overexpressing JcGA2ox6 also displayed darker green leaves (Figures 2D, 4C) and smaller reproductive organs (Figures 3, 6A–C,F,G, 7). It indicated the transgene had similar effects on some aspects of Arabidopsis and Jatropha growth and development, including stem elongation, leaf expansion, and flower and fruit development. However, the effects of the transgene on the flowering time differed between the transgenic Arabidopsis and Jatropha. Overexpression of JcGA2ox6 delayed the flowering time in transgenic Arabidopsis (Figures 2A,B) with no significant difference in the number of rosette leaves. It suggested that the reduced growth rate caused late flowering. In transgenic Jatropha, however, the flowering time was not significantly different from that of the WT plant (Figure 5F). Similarly, transgenic plants of liliaceous Tricyrtis sp. overexpressing the TfGA2ox gene from T. fournieri exhibited the dwarf phenotype, but no apparent alteration in the flowering time was observed (Otani et al., 2013). However, overexpression of PtGA2ox caused early-flowering in the poplar (Zawaski et al., 2011). These findings suggest that the GA regulation of flowering differs between plant species.
In this study, we expected to promote reproductive growth by reducing the endogenous GA content in Jatropha. However, transgenic Jatropha showed an unexpected phenotype of fewer inflorescences and flowers. Even the seed weights and oil contents were decreased. The possible reason for this result is the usage of a constitutive JcUEP promoter, which is active throughout the entire growth phase of the plant. Because the vegetative growth of transgenic Jatropha is partially suppressed (Figure 4), there is not enough nutrition provided for blossoming and fructification. In rice, when OsGA2ox1 was constitutively expressed under the direction of the rice actin promoter, transgenic rice showed severe dwarfism but failed to set grain (Sakamoto et al., 2003). In contrast, the expression of OsGA2ox1 under the control of the promoter of OsGA3ox2 resulted in a semi-dwarf phenotype that is normal in flowering and grain development (Sakamoto et al., 2003). To guarantee normal vegetative growth, tissue-specific or inducible promoters that are active in the transition from vegetative to reproductive growth in Jatropha should be used to drive JcGA2ox6. Although the expected phenotype of vigorous reproductive growth was not exhibited in transgenic Jatropha, the dwarf trait may allow for dense field plantation and increase the efficiency of fruit collection in Jatropha plantations. For example, during close planting in narrow rows, the suitable density for the thick planting of a new variety of semi-dwarf soybean ranged from 500 thousand plants to 600 thousand plants, producing a high yield of 5467.95 kg/hm2 (Zheng et al., 2013). In relay intercropped cotton, the light use efficiency and yield were both increased linearly with plant density (Mao et al., 2014). Propagation of transgenic Jatropha overexpressing JcGA2ox6 for dense plantation is in progress.
Author Contributions
Y-XH, Y-BT, and Z-FX designed the experiments. Y-XH performed the experiments and analyzed the data. Y-XH, Y-BT, and Z-FX wrote the paper.
Funding
This work was supported by funding from the Natural Science Foundation of Yunnan Province (2016FB048), the National Natural Science Foundation of China (31370595), the Program of Chinese Academy of Sciences (ZSZC-014, 2017XTBG-T02), and the West Light Foundation of Chinese Academy of Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Ms. Zhifang Zhou, Ms. Meili Zhao, Mr. Xin Ming, Mr. Li Cai, and Mr. Zhiyu Pu for transplanting and maintaining transgenic Jatropha plants. The authors gratefully acknowledge the Central Laboratory of the Xishuangbanna Tropical Botanical Garden for providing the research facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02103/full#supplementary-material
Footnotes
References
Abdulla, R., Chan, E. S., and Ravindra, P. (2011). Biodiesel production from Jatropha curcas: a critical review. Crit. Rev. Biotechnol. 31, 53–64. doi: 10.3109/07388551.2010.487185
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts - polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/Pp.24.1.1
Blazquez, M. A., Green, R., Nilsson, O., Sussman, M. R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10, 791–800. doi: 10.1105/tpc.10.5.791
Chakrabarti, P. P., and Prasad, R. B. N. (2012). “Biodiesel production from Jatropha curcas oil,” in Jatropha, Challenges for a New Energy Crop, eds B. Bahadur, M. Sujatha, and N. Carels (New York, NY: Springer), 463–490. doi: 10.1007/978-1-4614-4806-8_25
Chen, C. H., and Chen, Z. X. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. doi: 10.1104/pp001057
Chen, M. L., Fu, X. M., Liu, J. Q., Ye, T. T., Hou, S. Y., Huang, Y. Q., et al. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905, 67–74. doi: 10.1016/j.jchromb.2012.08.005
Chen, W. S. (1990). Endogenous growth-substances in xylem and shoot tip diffusate of lychee in relation to flowering. HortScience 25, 314–315.
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coles, J. P., Phillips, A. L., Croker, S. J., Garcia-Lepe, R., Lewis, M. J., and Hedden, P. (1999). Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 17, 547–556.
Cowling, R. J., Kamiya, Y., Seto, H., and Harberd, N. P. (1998). Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis. Plant Physiol. 117, 1195–1203. doi: 10.1104/pp.117.4.1195
Dijkstra, C., Adams, E., Bhattacharya, A., Page, A. F., Anthony, P., Kourmpetli, S., et al. (2008). Over-expression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Rep. 27, 463–470. doi: 10.1007/s00299-007-0471-z
Ding, L. W., Sun, Q. Y., Wang, Z. Y., Sun, Y. B., and Xu, Z. F. (2008). Using silica particles to isolate total RNA from plant tissues recalcitrant to extraction in guanidine thiocyanate. Anal. Biochem. 374, 426–428. doi: 10.1016/j.ab.2007.11.030
Divakara, B. N., Upadhyaya, H. D., Wani, S. P., and Gowda, C. L. L. (2010). Biology and genetic improvement of Jatropha curcas L.: a review. Appl. Energy 87, 732–742. doi: 10.1016/j.apenergy.2009.07.013
Dong, B., Deng, Y., Wang, H. B., Gao, R., Stephen, G. K., Chen, S. M., et al. (2017). Gibberellic acid signaling is required to induce flowering of chrysanthemums grown under both short and long days. Int. J. Mol. Sci. 18:E1259. doi: 10.3390/ijms18061259
Eriksson, S., Bohlenius, H., Moritz, T., and Nilsson, O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18, 2172–2181. doi: 10.1105/tpc.106.042317
Evans, M. M. S., and Poethig, R. S. (1995). Gibberellins promote vegetative phase-change and reproductive maturity in maize. Plant Physiol. 108, 475–487. doi: 10.1104/Pp.108.2.475
Fleet, C. M., and Sun, T. P. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85. doi: 10.1016/j.pbi.2004.11.015
Fu, Q. T., Li, C. Q., Tang, M. Y., Tao, Y. B., Pan, B. Z., Zhang, L., et al. (2015). An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnol. Rep. 9, 405–416. doi: 10.1007/s11816-015-0377-0
Gao, C. C., Ni, J., Chen, M. S., and Xu, Z. F. (2015). Characterization of genes involved in gibberellin metabolism and signaling pathway in the biofuel plant Jatropha curcas. Plant Divers. Resour. 37, 157–167.
Ghosh, A., Chikara, J., Chaudhary, D. R., Prakash, A. R., Boricha, G., and Zala, A. (2010). Paclobutrazol arrests vegetative growth and unveils unexpressed yield potential of Jatropha curcas. J. Plant Growth Regul. 29, 307–315.
Harberd, N. P., King, K. E., Carol, P., Cowling, R. J., Peng, J. R., and Richards, D. E. (1998). Gibberellin: inhibitor of an inhibitor of...? Bioessays 20, 1001–1008.
Hedden, P., and Phillips, A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530. doi: 10.1016/S1360-1385(00)01790-8
Israelsson, M., Mellerowicz, E., Chono, M., Gullberg, J., and Moritz, T. (2004). Cloning and overproduction of gibberellin 3-oxidase in hybrid aspen trees. Effects of gibberellin homeostasis and development. Plant Physiol. 135, 221–230. doi: 10.1104/pp.104.038935
Jackson, S. D., and Prat, S. (1996). Control of tuberisation in potato by gibberellins and phytochrome B. Physiol. Plant. 98, 407–412. doi: 10.1034/j.1399-3054.1996.980224.x
Kende, H., and Zeevaart, J. A. D. (1997). The five ”classical” plant hormones. Plant Cell 9, 1197–1210. doi: 10.1105/Tpc.9.7.1197
King, A. J., He, W., Cuevas, J. A., Freudenberger, M., Ramiaramanana, D., and Graham, I. A. (2009). Potential of Jatropha curcas as a source of renewable oil and animal feed. J. Exp. Bot. 60, 2897–2905. doi: 10.1093/jxb/erp025
Kobayashi, M., Yamaguchi, I., Murofushi, N., Ota, Y., and Takahashi, N. (1988). Fluctuation and localization of endogenous gibberellins in rice. Agric. Biol. Chem. 52, 1189–1194. doi: 10.1271/bbb1961.52.1189
Koornneef, M., and Vanderveen, J. H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58, 257–263. doi: 10.1007/Bf00265176
Koshita, Y., Takahara, T., Ogata, T., and Goto, A. (1999). Involvement of endogenous plant hormones (IAA, ABA, GAs) in leaves and flower bud formation of satsuma mandarin (Citrus unshiu Marc.). Sci. Hortic. 79, 185–194. doi: 10.1016/S0304-4238(98)00209-X
Lee, D. J., and Zeevaart, J. A. D. (2005). Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 138, 243–254. doi: 10.1104/pp.104.056499
Li, L. X., Coppola, E., Rine, J., Miller, J. L., and Walker, D. (2010). Catalytic hydrothermal conversion of triglycerides to non-ester biofuels. Energy Fuels 24, 1305–1315. doi: 10.1021/ef901163a
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, S. F., Ho, T. H. D., Liu, Y. L., Jiang, M. J., Hsieh, K. T., Chen, K. T., et al. (2017). Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice. Plant Biotechnol. J. 15, 850–864. doi: 10.1111/pbi.12681
Mao, L. L., Zhang, L. Z., Zhao, X. H., Liu, S. D., van der Werf, W., Zhang, S. P., et al. (2014). Crop growth, light utilization and yield of relay intercropped cotton as affected by plant density and a plant growth regulator. Field Crops Res. 155, 67–76. doi: 10.1016/j.fcr.2013.09.021
Martínez-García, J. F., García-Martínez, J. L., Bou, J., and Prat, S. (2001). The interaction of gibberellins and photoperiod in the control of potato tuberization. J. Plant Growth Regul. 20, 377–386. doi: 10.1007/s003440010036
Nagao, M. A., Ho-a, E. B., and Yoshimoto, J. R. (1999). Uniconazole retards growth and increases flowering of young macadamia trees. HortScience 34, 104–105.
Otani, M., Meguro, S., Gondaira, H., Hayashi, M., Saito, M., Han, D. S., et al. (2013). Overexpression of the gibberellin 2-oxidase gene from Torenia fournieri induces dwarf phenotypes in the liliaceous monocotyledon Tricyrtis sp. J. Plant Physiol. 170, 1416–1423. doi: 10.1016/j.jplph.2013.05.002
Pan, B. Z., and Xu, Z. F. (2011). Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 30, 166–174. doi: 10.1007/s00344-010-9179-3
Pan, J. L., Fu, Q. T., and Xu, Z. F. (2010). Agrobacterium tumefaciens-mediated transformation of biofuel plant Jatropha curcas using kanamycin selection. Afr. J. Biotechnol. 9, 6477–6481.
Rinne, P. L. H., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Sakamoto, T., Morinaka, Y., Ishiyama, K., Kobayashi, M., Itoh, H., Kayano, T., et al. (2003). Genetic manipulation of gibberellin metabolism in transgenic rice. Nat. Biotechnol. 21, 909–913. doi: 10.1038/nbt847
Sato, S., Hirakawa, H., Isobe, S., Fukai, E., Watanabe, A., Kato, M., et al. (2011). Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 18, 65–76. doi: 10.1093/dnares/dsq030
Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A. D., and Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell 15, 151–163. doi: 10.1105/tpc.005975
Schwechheimer, C. (2008). Understanding gibberellic acid signaling - are we there yet? Curr. Opin. Plant Biol. 11, 9–15. doi: 10.1016/j.pbi.2007.10.011
Song, J., Chen, M. S., Li, J. L., Niu, L. J., and Xu, Z. F. (2013). Effects of soil-applied paclobutrazol on the vegetative and reproductive growth of biofuel plant Jatropha curcas. Plant Divers. Resour. 35, 173–179.
Suo, H., Ma, Q., Ye, K., Yang, C., Tang, Y., Hao, J., et al. (2012). Overexpression of AtDREB1A causes a severe dwarf phenotype by decreasing endogenous gibberellin levels in soybean [Glycine max (L.) Merr.]. PLOS ONE 7:e45568. doi: 10.1371/journal.pone.0045568
Tao, Y. B., He, L. L., Niu, L. J., and Xu, Z. F. (2015). Isolation and characterization of an ubiquitin extension protein gene (JcUEP) promoter from Jatropha curcas. Planta 241, 823–836. doi: 10.1007/s00425-014-2222-z
Thomas, S. G., Phillips, A. L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. U.S.A. 96, 4698–4703. doi: 10.1073/pnas.96.8.4698
Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., et al. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19, 2140–2155. doi: 10.1105/tpc.106.043729
Winston, E. C. (1992). Evaluation of paclobutrazol on growth, flowering and yield of mango cv Kensington pride. Aust. J. Exp. Agric. 32, 97–104. doi: 10.1071/Ea9920097
Xu, Y. L., Gage, D. A., and Zeevaart, J. A. D. (1997). Gibberellins and stem growth in Arabidopsis thaliana - effects of photoperiod on expression of the GA4 and GA5 loci. Plant Physiol. 114, 1471–1476. doi: 10.1104/pp.114.4.1471
Yamaguchi, N., Winter, C. M., Wu, M. F., Kanno, Y., Yamaguchi, A., Seo, M., et al. (2014). Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344, 638–641. doi: 10.1126/science.1250498
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Ye, J., Geng, Y., Zhang, B., Mao, H., Qu, J., and Chua, N.-H. (2014). The Jatropha FT ortholog is a systemic signal regulating growth and flowering time. Biotechnol. Biofuels 7:91.
Zawaski, C., Kadmiel, M., Pickens, J., Ma, C., Strauss, S., and Busov, V. (2011). Repression of gibberellin biosynthesis or signaling produces striking alterations in poplar growth, morphology, and flowering. Planta 234, 1285–1298. doi: 10.1007/s00425-011-1485-x
Zheng, W., Du, Z. M., Guo, T., Liu, Z. T., Wang, Z. X., Li, C. D., et al. (2013). Breed semi-dwarf, resistant high density and super high-yielding soybean variety ‘henong 60’ using American dwarf source as hybridization paternal. J. Agric. 3, 27–30.
Zhou, B., Lin, J. Z., Peng, W. S., Peng, D., Zhuo, Y. H., Zhu, D. F., et al. (2012). Dwarfism in Brassica napus L. induced by the over-expression of a gibberellin 2-oxidase gene from Arabidopsis thaliana. Mol. Breed. 29, 115–127. doi: 10.1007/s11032-010-9530-1
Keywords: Jatropha, Arabidopsis, gibberellin, GA2-oxidase, dwarf
Citation: Hu Y-X, Tao Y-B and Xu Z-F (2017) Overexpression of Jatropha Gibberellin 2-oxidase 6 (JcGA2ox6) Induces Dwarfism and Smaller Leaves, Flowers and Fruits in Arabidopsis and Jatropha. Front. Plant Sci. 8:2103. doi: 10.3389/fpls.2017.02103
Received: 17 September 2017; Accepted: 27 November 2017;
Published: 12 December 2017.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Christiaan Van Der Schoot, Norwegian University of Life Sciences, NorwayHak Soo Seo, Seoul National University, South Korea
Copyright © 2017 Hu, Tao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Bin Tao, taoyanbin@xtbg.ac.cn Zeng-Fu Xu, zfxu@xtbg.ac.cn
 Ying-Xiong Hu1,2
Ying-Xiong Hu1,2 Zeng-Fu Xu
Zeng-Fu Xu