- 1The National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Plant Pathology, China Agricultural University, Beijing, China
- 3Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 4College of Chemistry and Environment Engineering, Pingdingshan University, Pingdingshan, China
Powdery mildew resistance gene Pm4b, originating from Triticum persicum, is effective against the prevalent Blumeria graminis f. sp. tritici (Bgt) isolates from certain regions of wheat production in China. The lack of tightly linked molecular markers with the target gene prevents the precise identification of Pm4b during the application of molecular marker-assisted selection (MAS). The strategy that combines the RNA-Seq technique and the bulked segregant analysis (BSR-Seq) was applied in an F2:3 mapping population (237 families) derived from a pair of isogenic lines VPM1/7∗Bainong 3217 F4 (carrying Pm4b) and Bainong 3217 to develop more closely linked molecular markers. RNA-Seq analysis of the two phenotypically contrasting RNA bulks prepared from the representative F2:3 families generated 20,745,939 and 25,867,480 high-quality read pairs, and 82.8 and 80.2% of them were uniquely mapped to the wheat whole genome draft assembly for the resistant and susceptible RNA bulks, respectively. Variant calling identified 283,866 raw single nucleotide polymorphisms (SNPs) and InDels between the two bulks. The SNPs that were closely associated with the powdery mildew resistance were concentrated on chromosome 2AL. Among the 84 variants that were potentially associated with the disease resistance trait, 46 variants were enriched in an about 25 Mb region at the distal end of chromosome arm 2AL. Four Pm4b-linked SNP markers were developed from these variants. Based on the sequences of Chinese Spring where these polymorphic SNPs were located, 98 SSR primer pairs were designed to develop distal markers flanking the Pm4b gene. Three SSR markers, Xics13, Xics43, and Xics76, were incorporated in the new genetic linkage map, which located Pm4b in a 3.0 cM genetic interval spanning a 6.7 Mb physical genomic region. This region had a collinear relationship with Brachypodium distachyon chromosome 5, rice chromosome 4, and sorghum chromosome 6. Seven genes associated with disease resistance were predicted in this collinear genomic region, which included C2 domain protein, peroxidase activity protein, protein kinases of PKc_like super family, Mlo family protein, and catalytic domain of the serine/threonine kinases (STKc_IRAK like super family). The markers developed in the present study facilitate identification of Pm4b during its MAS practice.
Introduction
In wheat (Triticum aestivum L.), powdery mildew is caused by the biotrophic fungus Blumeria graminis f. sp. tritici (Bgt) (Green et al., 2014). The epidemics of powdery mildew often occur in the wheat producing regions with cool and humid climates (Cowger et al., 2012). In China, this foliar disease is endangering most regions of winter wheat and spring wheat productions. In the last decade, the proper management measures can retrieve about 1.4984 million metric tons of yield losses, and the actual annual grain loss caused by powdery mildew was limited to 0.3045 million metric tons (Liu et al., 2016).
The use of host resistance is a commonly recognized means to reduce the economic losses and to control the epidemics of diseases (Hulbert et al., 2001). The development of powdery mildew-resistant wheat cultivars requires the availability of resistance genes. At present, 78 permanently designated and many other temporally designated powdery mildew resistance genes or alleles have been documented1. Some of them have single alleles, while others have multiple alleles (e.g., Pm1, Pm2, Pm3, Pm4, Pm5, and Pm24 loci) (Hsam et al., 1998; Singrün et al., 2003; Hao et al., 2008, 2015; Bhullar et al., 2009, 2010; Xie et al., 2012; Zhang et al., 2016). The French cultivar VPM1 resistant to powdery mildew was developed from a complicated interspecific cross involving Aegilops ventricosa (Zhuk.) Chennav, and T. persicum Vav. (syn. T. turgidum var. carthlicum Nevski.) (Doussinault et al., 1983). A powdery mildew resistance gene in VPM1 was localized on a T. persicum chromosomal segment that was translocated onto the long arm of wheat chromosome 2A, and proved to be an allele in locus Pm4, designated Pm4b (Bariana and McIntosh, 1994). Even though it was identified over 30 years ago, Pm4b is still effective in certain regions of China and the United States (Wang et al., 2005; Parks et al., 2008; Zeng et al., 2014). It was also used to enhance powdery mildew resistance in triticale (× Triticosecale Wittmack) (Kowalczyk et al., 2011).
Great efforts have been taken to tag Pm4b and other Pm4 alleles. Due to the alien origin nature and low abundance of the markers used, e.g., restriction fragment length polymorphism (RFLP) and its conversion of sequence-tagged site (STS), random amplified polymorphic DNA (RAPD), simple sequence repeats (SSRs), and sequence-related amplified polymorphism (SRAP), previous works on the molecular mapping of Pm4b did not generate closely linked molecular markers. Based on the RFLP marker BCD1231 linked to Pm4a, Chen et al. (2002) designed an STS marker STS470 and mapped it 3.0 cM away from Pm4b. Another STS marker STS-241 developed from a cloned RAPD fragment was 4.9 cM from Pm4b (Yi et al., 2008). In that work, the SRAP marker Me8/Em7-220 was mapped 7.1 cM away from Pm4b. In an attempt to map Pm4c, Hao et al. (2008) also tested the Pm4-linked markers using a mapping population derived from the cross between VPM1 and the susceptible wheat Chancellor. An SSR marker Xbarc122 (2.0 cM from the target gene) was closer to Pm4b than other existing molecular markers. Using the near-isogenic line (NIL) CI 14124, Ma et al. (2004) mapped Pm4a by screening 46 pairs of microsatellite primers, and determined that the genetic distance between SSR marker Xgwm356 and Pm4a was 4.8 cM. An allele of Pm4, designated Pm4d, was derived from einkorn wheat (T. monococcum L.) and flanked by SSR markers Xgwm526 and Xbarc122 at genetic distances of 3.4 and 1.0 cM, respectively (Schmolke et al., 2012). Because of their less close association with the target gene, these markers may not effectively detect Pm4b. Recently, Li et al. (2017) reported a new allele in the Pm4 locus in the common wheat line D29, designated Pm4e, which was flanked by SSR markers Xgdm93 and Xhbg327 and co-segregated with STS markers Xsts_bcd1231 and TaAetPR5. However, the relationship between these markers and Pm4b was not clear.
Common wheat is a hexaploid species (2n = 6x = 42; AABBDD genomes) with a large sized genome (∼17 Gbp) and ∼90% repetitive sequences (Gupta et al., 2008; Shewry, 2009), so traditional classes of molecular markers, such as RFLP, RAPD (Fabritius et al., 1997), SSR (Duan et al., 2003; Cheema et al., 2008; Tsilo et al., 2009), amplified fragment length polymorphism (AFLP) (Cai et al., 2003; Asnaghi et al., 2004), as well as cleaved amplified polymorphic sequence (CAPS) for detecting single nucleotide polymorphism (SNP) (Lambreghts et al., 2009), cannot meet the demand for identifying closely linked markers due to inadequate density and high levels of duplication. Bulked segregant analysis-RNA-Seq (BSR-Seq) is a new genetic mapping strategy that combines the power of bulked segretant analysis (BSA) (Michelmore et al., 1991) and the ease of RNA-Seq technique. Using this strategy, Liu et al. (2012) mapped the genes in the population for which even no polymorphic markers were previously identified, resulting in cloning of glossy3 (gl3) gene from maize (Zea mays L.). Trick et al. (2012) sequenced mRNA from the NILs spanning a ∼30 cM interval including the GPC-B1 locus and two bulked samples that consisted of homozygous recombinant lines contrasting for their grain protein content (GPC) phenotypes. After discriminating for SNPs from the RNA-Seq data between the two NILs, they identified 39 new SNP markers, corresponding to 67% of the validated SNPs, mapped across a 12.2 cM interval including GPC-B1, and defined this gene to an interval containing 13–18 genes in the syntenic cereal genomes within a 0.4 cM interval of wheat. Ramirez-Gonzalez et al. (2015) combined BSA and the next generation sequencing technique to construct a high density genetic map of wheat and localized the stripe rust (caused by Puccinia striiformis West.) resistance gene Yr15 to a 0.77 cM interval. In a recent study, YrMM58 and YrHY1 for resistance to stripe rust were mapped in the distal ∼16 Mb region on chromosome 2AS (Wang et al., 2017). These studies have demonstrated that BSR-Seq is effective to identify SNP markers for fine-mapping and even cloning target genes, especially in the genome regions with low polymorphism.
The objectives of this study were to characterize the resistance of Pm4b to different Bgt isolates that were collected from China and to develop closely linked markers that can be used to detect Pm4b using BSR-Seq technique and comparative genomics approach. Results of this study will be useful for marker-assisted breeding and pyramiding Pm4b with other resistance genes for the improvement of wheat against powdery mildew.
Materials and Methods
Plant Materials
In 2005, Pm4b was transferred from VPM1 [pedigree: Aegilops ventricosa/T. turgidum L. var. carthlicum (T. persicum)//3∗ T. aestivum cv. Marne] to the Chinese winter wheat cultivar Bainong 3217 resulting in the production of the Pm4b NIL VPM1/7∗Bainong 3217 F4 (Zhou et al., 2005). The susceptible recurrent parent Bainong 3217, with the pedigree of [(Funo × Neixiang 5) F1 × Xiannong 39] F2 × (Xinong 64(4)43 line 2 × Yanda 24) F1, was widely grown in the Huang and Huai Rivers Valley Winter Wheat Zone (Huang et al., 1982). In 2013, line VPM1/7∗Bainong 3217 F4 was crossed with Bainong 3217 to produce F1, F2, and F2:3 populations to be used in the genetic analysis and molecular mapping of Pm4b. A set of 23 differential wheat cultivars or lines that carry known powdery mildew resistance genes was used to differentiate the Bgt isolates. Twenty-seven wheat entries carrying known powdery mildew resistance genes and 46 wheat cultivars were used to validate the Pm4b-lined markers developed in the present study. Winter wheat cultivars Zhongzuo 9504 was used as the susceptible control in the assessments of powdery mildew resistance.
Evaluation of Resistance to Powdery Mildew at the Seedling Stage
Genetic analysis and molecular mapping of the target resistance gene were conducted using the F1, F2, and F2:3 populations derived from the cross VPM1/7∗Bainong 3217 F4 × Bainong 3217. The parents and the mapping populations were grown in plastic trays with 5 × 10 wells (5 cm × 5 cm × 5 cm in dimension). At least 15 plants were tested for each F2:3 family. The conidiospores of Bgt isolate E20 freshly increased on the susceptible cultivar Zhongzuo 9504 were dusted on the tested seedlings at one-leaf stage. The inoculated plants were grown in a greenhouse at 20°C/14°C (day/night) with a photoperiod of 16 h light/8 h dark. Fifteen days after inoculation when the susceptible control Zhongzuo 9504 plants were heavily diseased, infection type (IT) of each plant was visually rated on a 0–4 scale as described by Liu et al. (1999). Plants were classified into the resistant group when the ITs were 0–2 or the susceptible group when the ITs were 3–4. Forty-six Bgt isolates were used to determine the effectiveness of Pm4b against powdery mildew using the same method as described above. These isolates were purified at least three times by the single colony method after they were collected from different wheat fields located in the provinces of Hebei, Shandong, Henan, Shanxi, Beijing, Jiangsu, Yunnan, and Guizhou in China.
Genotyping of F2:3 Lines Using BSR-Seq Analysis
The BSR-Seq approach was performed on selected F2:3 families from the mapping population of VPM1/7∗Bainong 3217 F4 × Bainong 3217 cross. The representative plants from each F2:3 family with known Bgt-resistant/susceptible phenotypes were grown in a Bgt-free growth chamber. The phenotypically contrasting bulks of leaf samples were created by pooling equal size of the primary leaf from each representative plant two-leaf-old of 50 homozygous resistant and 50 homozygous susceptible F2:3 families. Total RNA of the two bulks of leaf samples was separately extracted using the Illumina TruSeq RNA Sample Prep Kit (Illumina, Inc., San Diego, CA, United States) to be used in RNA-Seq analysis using the platform of Illumina HiSeq 4000 (Beijing Southern Genome Research Technology Co., Ltd., Beijing, China). The raw sequencing reads generated were quality controlled using software Trimmomatic v0.36 (Bolger et al., 2014) with the default parameters. Using software STAR v2.5.1b (Dobin et al., 2013), the clean reads were aligned to the Chinese Spring whole genome assembly sequences (IWGSC WGS v1, NRGene DeNovoMAGIC, Seq Repository of Wheat Portal on URGI, INRA, France2 with the mismatch rate of less than 5%. The uniquely mapped read pairs were used in further analysis. The read alignments were masked for PCR duplications and split for reads spanning introns before they were used to call SNPs and InDels using module “HaplotypeCaller” software GATK v3.6 (McKenna et al., 2010). The resulting SNPs and InDels with sequencing depth less than 6 were discarded, and the remaining ones were applied to BSA. Only variants with allele frequency difference (AFD) > 0.6 and P-value of Fisher’s exact test on read count data < 1e-8 were classified as resistance-associated variants and used as templates for marker development.
Development of SNP and SSR Markers
The SNPs associated with the powdery mildew resistance identified by BSR-Seq analysis were selected for marker development. The flanking sequences approximately 3 kb of the candidate SNPs were used as templates for designing PCR primers using the web-based program available at GSP website3. The closest SNP markers were used as queries for BLAST against the Chinese Spring whole genome assembly sequences (IWGSC WGS v1, NRGene DeNovoMAGIC, Seq Repository of Wheat Portal on URGI, INRA, France). The genomic sequences located downstream of the Pm4b-linked SNP marker (2AL71) developed in this study were used as templates to design SSR primers using batchprimer34. Polymorphic SSR markers between the parents and the contrasting DNA bulks were used to construct the genetic linkage map of Pm4b.
DNA Amplification and Electrophoresis
Genomic DNA was extracted from the fresh leaf tissues from each family of the F2:3 mapping population, following the cetyltrimethylammonium ammonium bromide (CTAB) method (Saghai-Maroof et al., 1984). The resistant and susceptible DNA pools were created by separately bulking equal amount of DNA from 16 resistant and 16 susceptible F2:3 families for detecting the polymorphism of SNPs and SSR markers. PCR was performed in a Biometra T3000 Thermocycler (ABI, New York, NY, United States). A reaction mixture (10 μL) consisted of 50–100 ng of template DNA, 0.4 μM each of the forward and reverse primers, 1 U of Taq polymerase, 0.4 mM dNTPs, and 2 μL 10× buffer with 20 mM Mg2+. Amplification of DNA was programmed at 94°C for 4 min; 35 cycles of 94°C for 45 s, 52–60°C for 45 s, and 72°C for 1 min. The reaction was terminated after an extension at 72°C for 10 min. The resulting PCR products were mixed with 2 μL loading buffer (98% formamide, 10 mM EDTA, 0.25% bromophenol blue, and 0.25% xylene cyanol) prior to separation on 1–2% agarose gel or 8% non-denaturing polyacrylamide gel (Acr:Bis = 19:1 or 39:1).
Physical Mapping and Comparative Genomics Analysis
The sequences of SSR markers Xics13 and Xics43 that flanked Pm4b were used to search against the genomic regions of the Chinese Spring whole genome assembly sequences (IWGSC WGS v1, NRGene DeNovoMAGIC, Seq Repository of Wheat Portal on URGI, INRA, France) and the Chinese Spring cDNA sequence information was used to obtain the genes that were included in the interval of the two Pm4b-flanking markers. Then, these genes were annotated by the online programs EnsemblPlants5 and NCBI6. These online databases provide the annotation information for the genes of T. aestivum and the homologous genes of Brachypodium distachyon (L.), rice (Oryza sativa L.), and sorghum (Sorghum bicolor L.).
Statistical Analysis and Linkage Map Construction
The Chi-squared test (χ2) for the goodness of fit was performed to determine the deviations of observed data from the expected segregation ratios using SAS 8.0 statistical analysis package (SAS Institute, Cary, NC, United States). Linkage between markers and the target resistance gene was established with the software Mapmaker/Exp Version 3.0b (Lincoln et al., 1993). Genetic distances were determined using the Kosambi function. The logarithm of the odds ratio (LOD) threshold score was set at 3.0 and the maximum distance allowed between markers was set at 50.0 cM.
Results
Characterization of Pm4b Resistance to Different Bgt Isolates
Forty-six Bgt isolates were used to examine the virulence spectrum against Pm4b in line VPM1/7∗Bainong 3217 F4. These isolates produced different ITs on the differential wheat cultivars or lines with known powdery mildew resistance genes (Supplementary Table S1). Line VPM1/7∗Bainong 3217 F4 was resistant to 52.2% of the isolates tested, while Khapli/8∗Cc carrying Pm4a was resistant to 39.1% of them. Line VPM1/7∗Bainong 3217 F4 was resistant to 72.7% of isolates that were collected from Hebei province, and it was effective against half of the isolates from Henan, and Shandong provinces. Bainong 3217 was as susceptible as the control Zhongzuo 9504 to all the Bgt isolates tested.
Inheritance of Resistance to Powdery Mildew in Line VPM1/7∗Bainong 3217 F4
The pair of NILs VPM1/7∗Bainong 3217 F4 and Bainong 3217 differed in their reactions to Bgt isolate E20 (Table 1). Line VPM1/7∗Bainong 3217 F4 was highly resistant with an IT of 0, while the recurrent parent Bainong 3217 was highly susceptible with an IT of 4. The F1 plants derived from the cross between VPM1/7∗Bainong 3217 F4 and Bainong 3217 produced ITs that were similar to the resistant line VPM1/7∗Bainong 3217 F4. The F2 plants exhibited a 3:1 segregation ratio for the resistant and susceptible plants and the F2:3 population showed a 1:2:1 segregating ratio for the homozygously resistant, heterozygous, and homozygously susceptible families (Table 1). All these results demonstrate that the resistance to Bgt isolate E20 in line VPM1/7∗Bainong 3217 F4 inherits in a mode of single dominant gene.
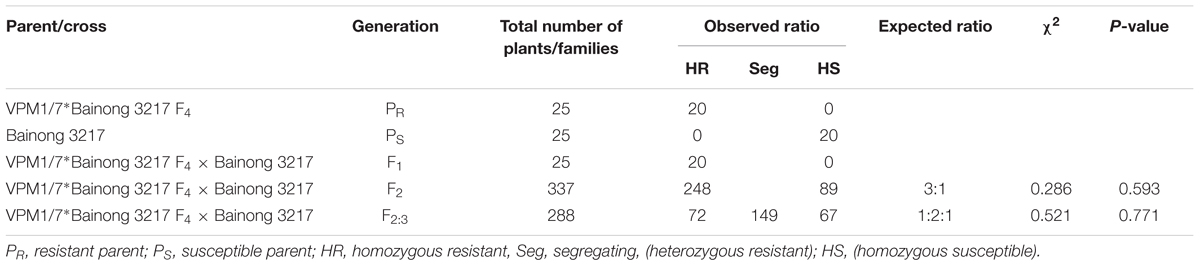
TABLE 1. Classification of responses to the Blumeria graminis f. sp. tritici isolate E20 for the F1, F2, and F2:3 progenies derived from VPM1/7∗Bainong 3217 F4 × Bainong 3217 cross.
RNA-Seq Analysis of the Bulked RNA Pools with Distinct Reactions to Powdery Mildew
The RNA-Seq analysis generated 22,700,130 and 25,870,804 raw read pairs for the powdery mildew resistant and susceptible bulks, respectively. After quality control process, the resistant bulk remained 20,745,939 high-quality read pairs and 18,217,476 (82.8%) of them were uniquely mapped to the Chinese Spring wheat whole genome draft assembly. There were 25,867,480 high-quality and 20,741,967 (80.2%) uniquely mapped reads for the susceptible bulk. Variant calling identified 283,866 raw SNPs and InDels between the two bulks, and 101,835 of them had a depth > 6. The SNPs with high association level focused on chromosome 2AL (Figures 1A,B). Results of BSA revealed that 84 variants were potentially associated with the target powdery mildew resistance gene. These SNPs mainly distributed on chromosome 2AL, which indicates that the resistance gene is located on this chromosome arm. Forty-six SNPs were enriched in an about 25 Mb region in the distal part of chromosome 2AL (Supplementary Table S2).
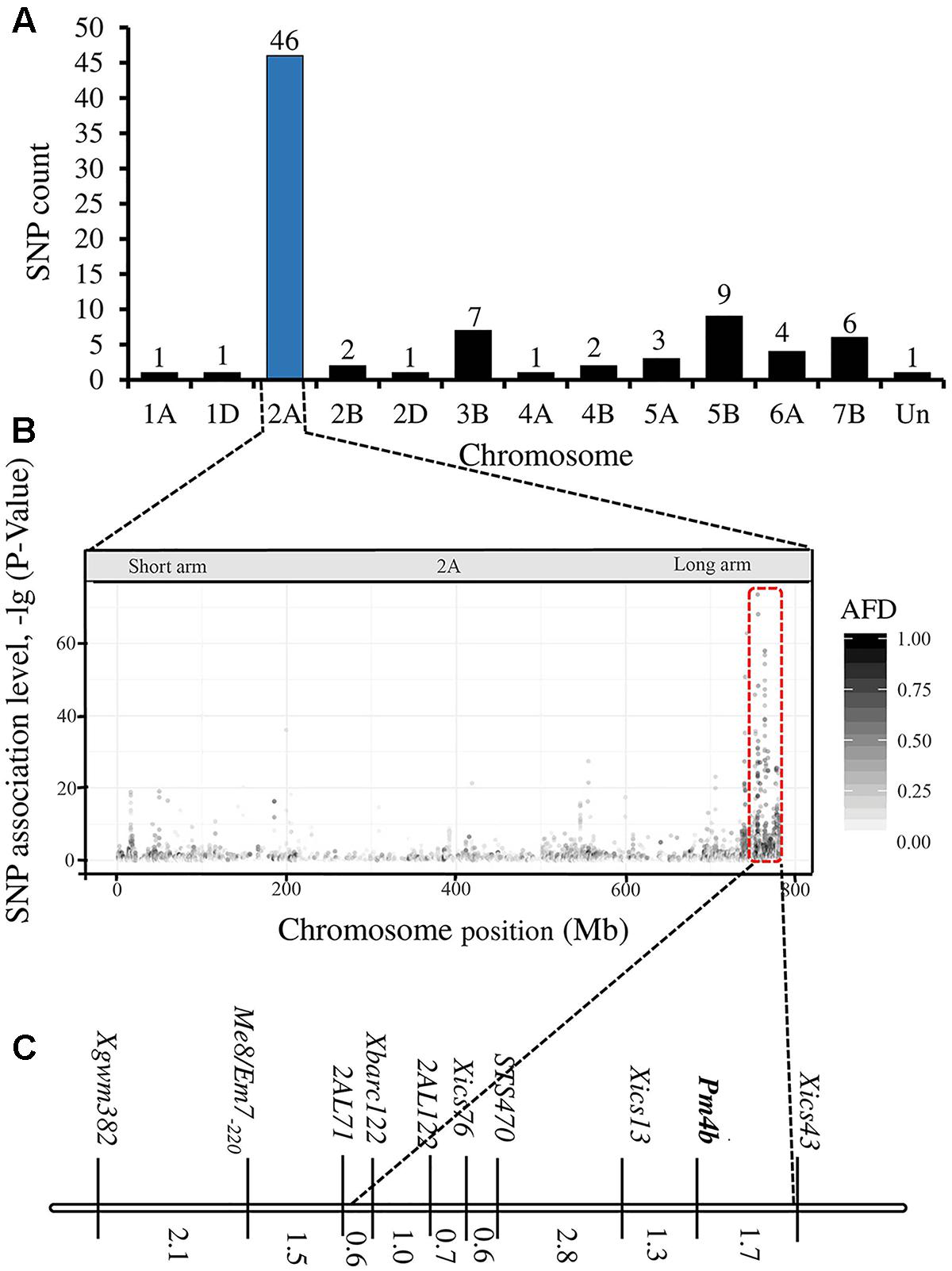
FIGURE 1. Overview of analyses. Number of polymorphic SNPs distributed on the different chromosomes (A) (Un means the unmapped SNPs), distribution of single nucleotide variation on wheat chromosomes (B), and linkage map of Pm4b from this study (C).
Polymorphic Analysis of Specific Primers Designed Based on the SNP Calling
Using the 46 SNP-containing sequences Blast analysis resulted in 14 homologous scaffolds from the Chinese Spring genomic sequence. These scaffolds were used as templates for designing 53 pairs of SNP primers on the GSP website, and 12 of them produced specific primers (Supplementary Table S3). These primer pairs amplified 700–900 bp nucleic acid sequences containing the SNP variants in line VPM1/7∗Bainong 3217 F4, Bainong 3217 and the two contrasting bulked F2:3 families. Sequence analysis of the resulting amplicons confirmed the consistency of polymorphisms for the SNP variants between the parents and the two contrasting DNA bulks. The SNP markers 2AL43, 2AL83, 2AL71, and 2AL122 were polymorphic between the parents and two contrasting bulked F2:3 families (Figure 2), indicating that they were possibly linked to Pm4b.
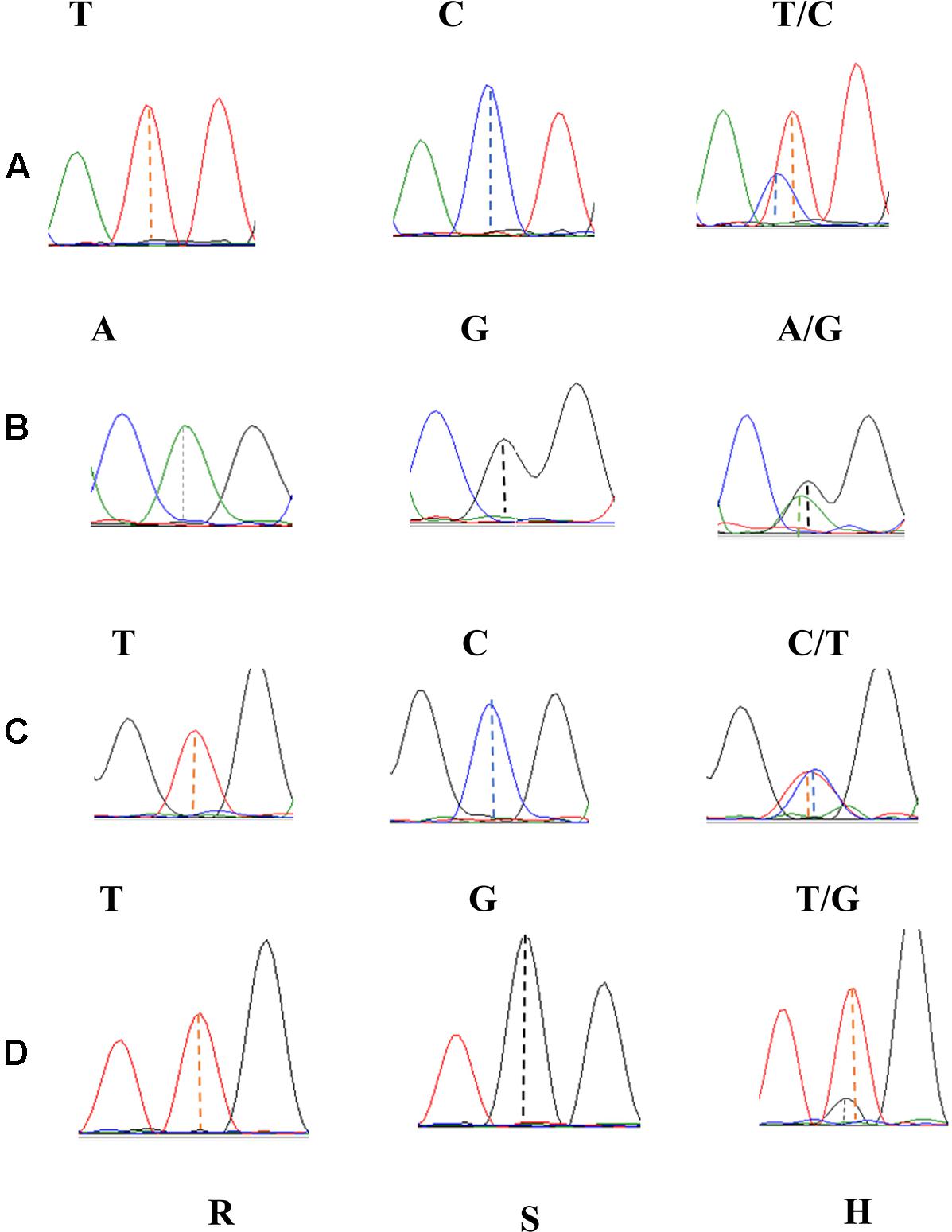
FIGURE 2. Sanger sequencing profiles of SNP markers 2AL43 (A), 2AL71 (B), 2AL83 (C), and 2AL122 (D) in homozygous resistant (R), homozygous susceptible (S), and heterozygous F2:3 families (H).
Development of SSR Markers and Construction of Genetic Linkage Map
Based on the linkage analysis, the SNP markers 2AL43, 2AL83, 2AL71, and 2AL122 were located on the same side of Pm4b (Table 2), and the corresponding physical locations in the Chinese Spring reference sequences were 755,793,708, 770,411,718, 772,336,969, and 774,673,558 on the distal end of chromosome arm 2AL, respectively. Based on the information on the sites of these SNPs, Pm4b was located at a position between marker 2AL122 and the end of chromosome 2AL. Then, the genome sequence from the corresponding location of 2AL71 to the end of chromosome arm 2AL was used as template to design SSR primers (Supplementary Table S4). Among the 98 pairs of SSR primers designed, twelve were polymorphic between the two parents. Markers Xics13, Xics43, and Xics76 were polymorphic between the contrasting DNA bulks, indicating their possible linkage to Pm4b. Based on their amplification patterns, Xics13 and Xics43 were co-dominant. Xics13 amplified 264 bp and 252 bp bands, and Xics43 produced 201 bp and 217 bp bands in the resistant and susceptible individuals of the mapping population, respectively (Figure 3). The dominant marker Xics76 produced a 167 bp fragment from the resistant individuals and null from the susceptible individuals. In addition, the polymorphism of the published Pm4-linked markers was examined using the F2:3 mapping population. Four markers STS470, Xbarc122, Me8/Em7-220, and Xgwm382 were polymorphic between the parents and the bulks (Table 3). Xgwm356 specific for Pm4a was not incorporated into the new genetic linkage map of Pm4b because it was not polymorphic between the bulks. The other molecular markers that were previously linked to Pm4b, Pm4c, Pm4d, and Pm4e were not polymorphic between neither the parents nor the bulks of the current mapping population (Table 3). Therefore, the newly developed polymorphic SNP markers 2AL71 and 2AL122 and SSR markers Xics13, Xics43, and Xics76, together with the four polymorphic Pm4-linked markers, were used to construct the genetic linkage map after genotying the F2:3 mapping population (Figure 1C). In this linkage map, the SNP markers 2AL122 and 2AL71 were closer to Pm4b than the previously identified markers, except for STS470. The newly designed SSR markers Xics76, Xics13, and Xics43 were located on the distal side to the SNP markers and closer to the target gene. Pm4b was flanked by markers Xics13 and Xics43 with genetic distances of 1.3 and 1.7 cM, respectively.
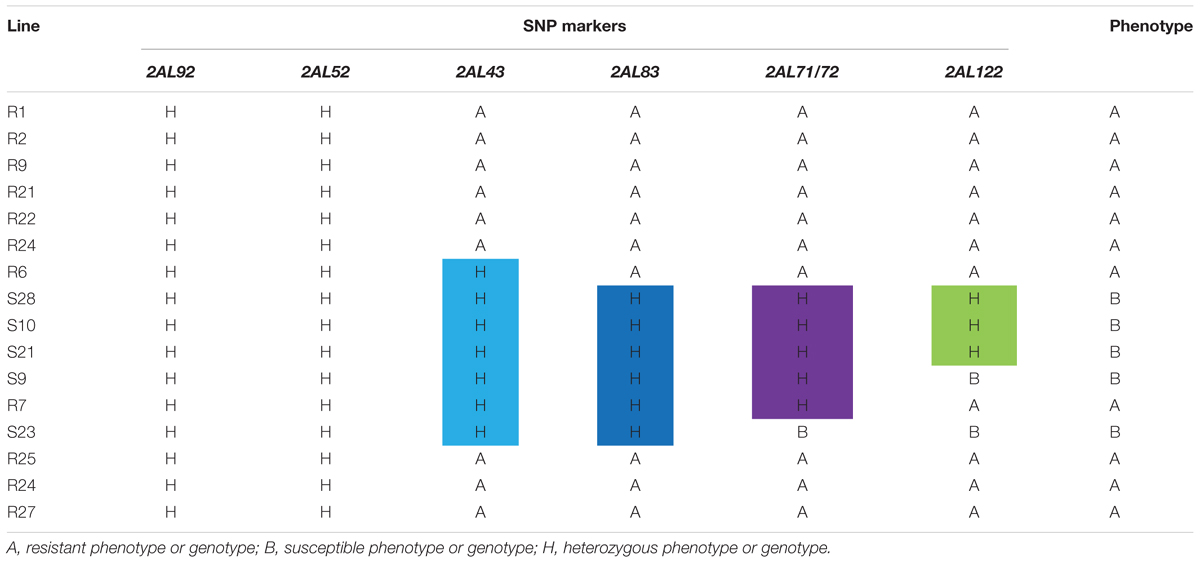
TABLE 2. Graphical genotypes of the SNP markers for the 16 F2:3 individuals used to develop the markers closely linked to Pm4b.
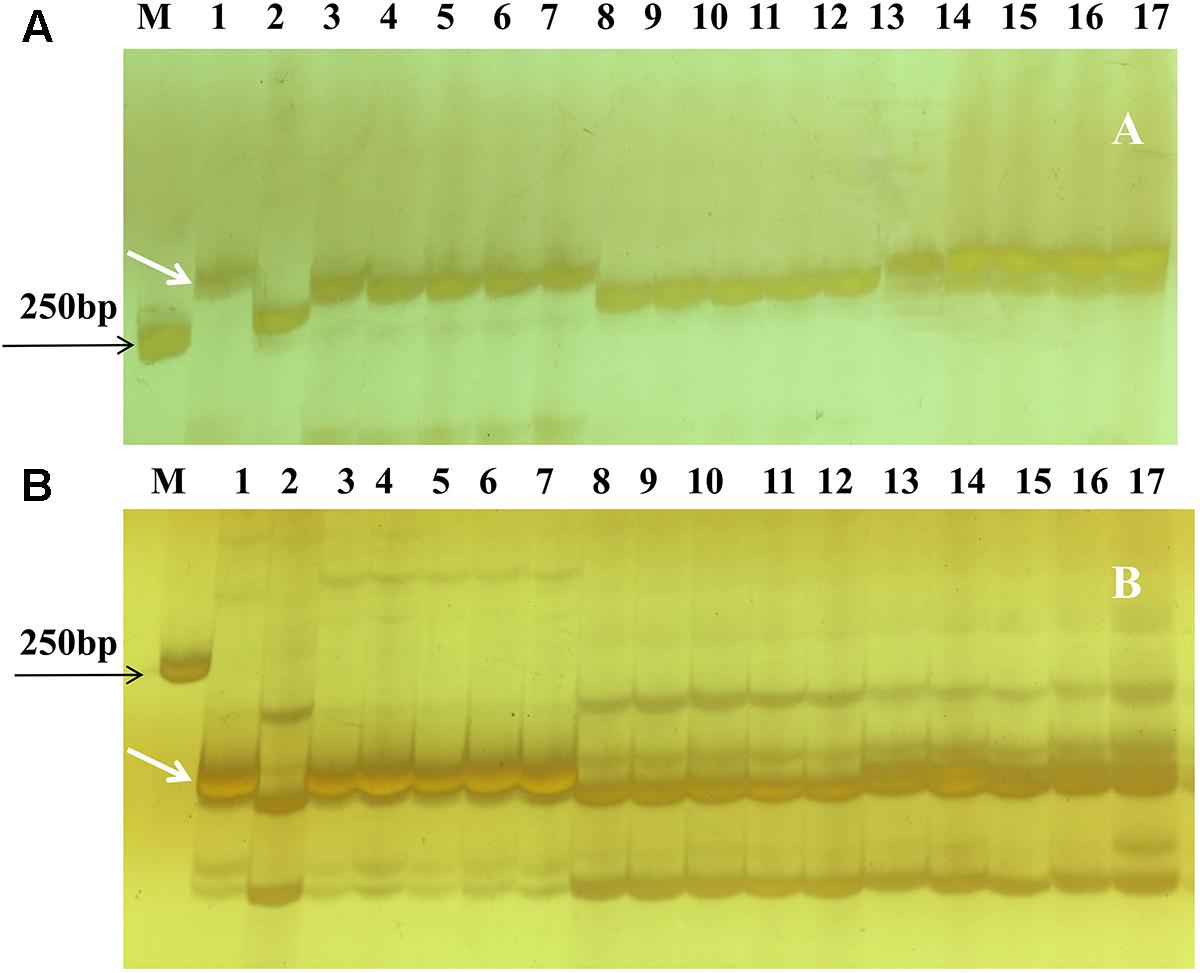
FIGURE 3. Amplification patterns of Pm4b-linked polymorphic SSR markers Xics13 (A) and Xics43 (B) in the parents and selected F2:3 Families of VPM1/7∗Bainong 3217 F4 × Bainong 3217 in 8% silver-stained non-denaturing polyacrylamide gels. Lane M, DL2000; lane 1: VPM1/7∗Bainong 3217 F4; lane 2: Bainong 3217; lanes 3–7: homozygous resistant F2:3 families; lanes 8–12, homozygous susceptible F2:3 families; and lanes 13–17: heterozygous F2:3 families. White arrows indicate the polymorphic bands specific for Pm4b.
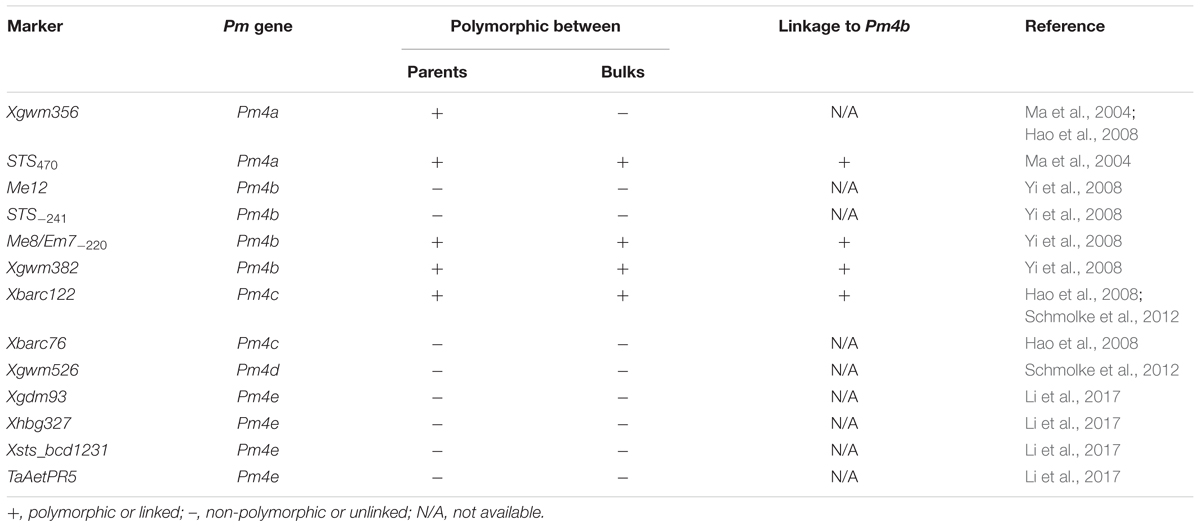
TABLE 3. Analysis of polymorphism in the mapping population of VPM1/7∗Bainong 3217 F4 × Bainong 3217 with the markers located on Pm4 locus on chromosome 2AL.
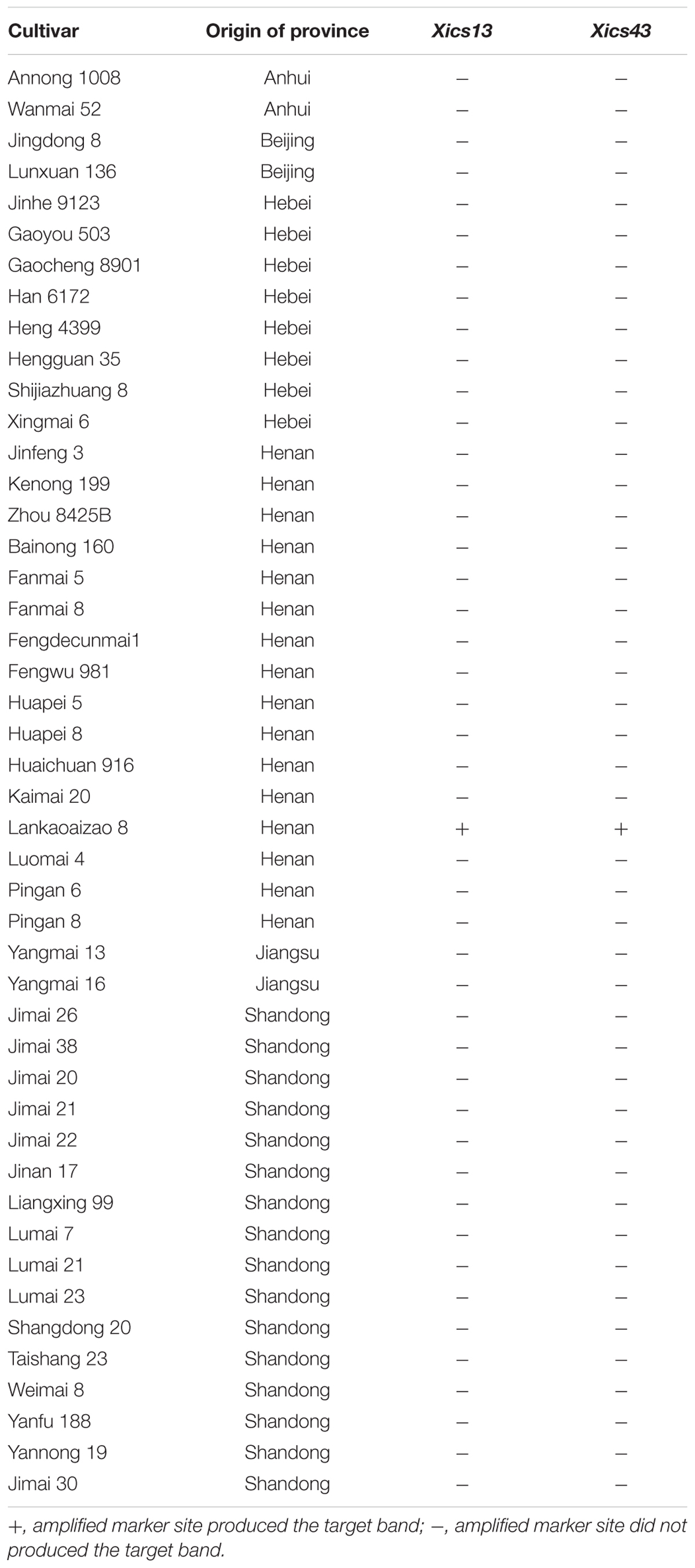
Validation of Markers Xics13 and Xics43
To validate the Pm4b-flanking markers, Xics13 and Xics43 were used to amplify 27 wheat differential cultivars or lines that carry known Pm genes. The target bands were amplified only from line VPM1/7∗Bainong 3217 F4, but not from the other 26 wheat accessions (Table 4). Meanwhile, markers Xics13 and Xics43 were used to genotype Pm4b in a panel of 46 wheat cultivars. Lankaoaizao 8 showed the same banding patterns as line VPM1/7∗Bainong 3217 F4, indicating that it may carry Pm4b (Table 5). However, the other 45 cultivars showed the same banding patterns as Bainong 3217, indicating the absence of Pm4b. This indicates that Xics13 and Xics43 are diagnostic molecular markers linked to Pm4b, which can be used in the marker-assisted selection (MAS) program for detecting and pyramiding Pm4b with other genes in the breeding program.
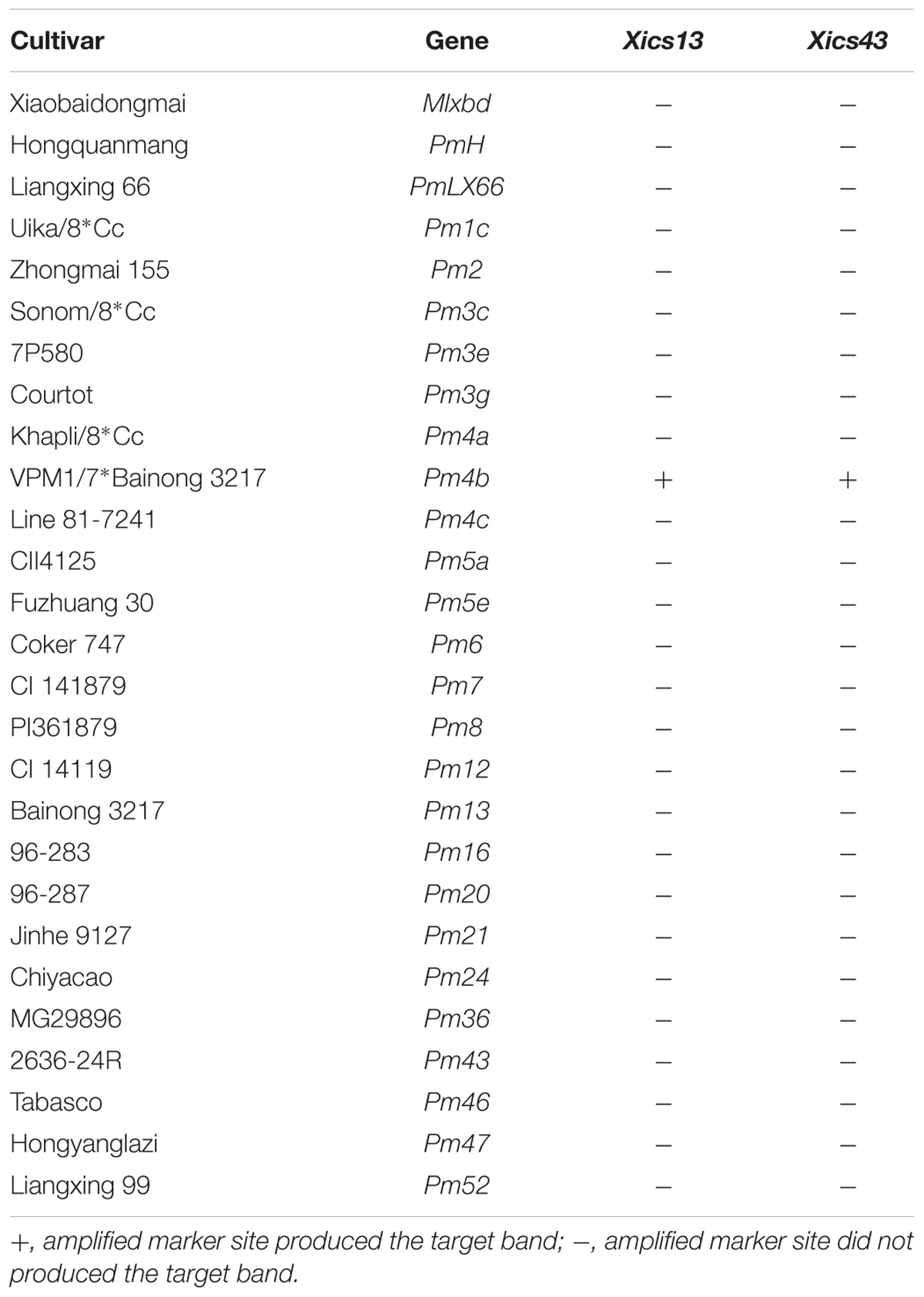
TABLE 4. Validation of Pm4b-linked SSR marker Xics13 and Xics43 on 27 Chinese wheat cultivars or lines with known powdery mildew resistance genes.
Physical Mapping and Comparative Genomics Analysis
The DNA sequences associated with the Pm4b-flanking markers Xics13 and Xics43 were positioned on the Chinese Spring reference genome sequence. The existing cDNA sequence information of the Chinese Spring database was used to enucleate the genes in the genomic regions between the two Pm4b-flanking markers. This region spanned a physical interval of about 6.7 Mb (773,680,429–780,760,825) on the Chinese Spring chromosome 2AL and contained 101 predicted genes. This region displayed a collinear relationship with B. distachyon in a 0.99 Mb (26,657,763–27,645,510) genomic region containing 49 predicted genes on chromosome 5, a 1.20 Mb (33,028,549–34,234,325) genomic region containing 47 predicted genes on rice chromosome 4, and a 0.61 Mb (60,898,697–61,519,884) genomic region containing 36 predicted genes on sorghum chromosome 6 (Figure 4). Seven transcripts, which encode for disease resistance-associated proteins, such as C2 domain, peroxidase activity protein, protein kinases of PKc_like super family, Mlo family protein, and catalytic domain of the serine/threonine kinases (STKc_IRAK like super family), were identified in this collinear genomic regions (Table 6).
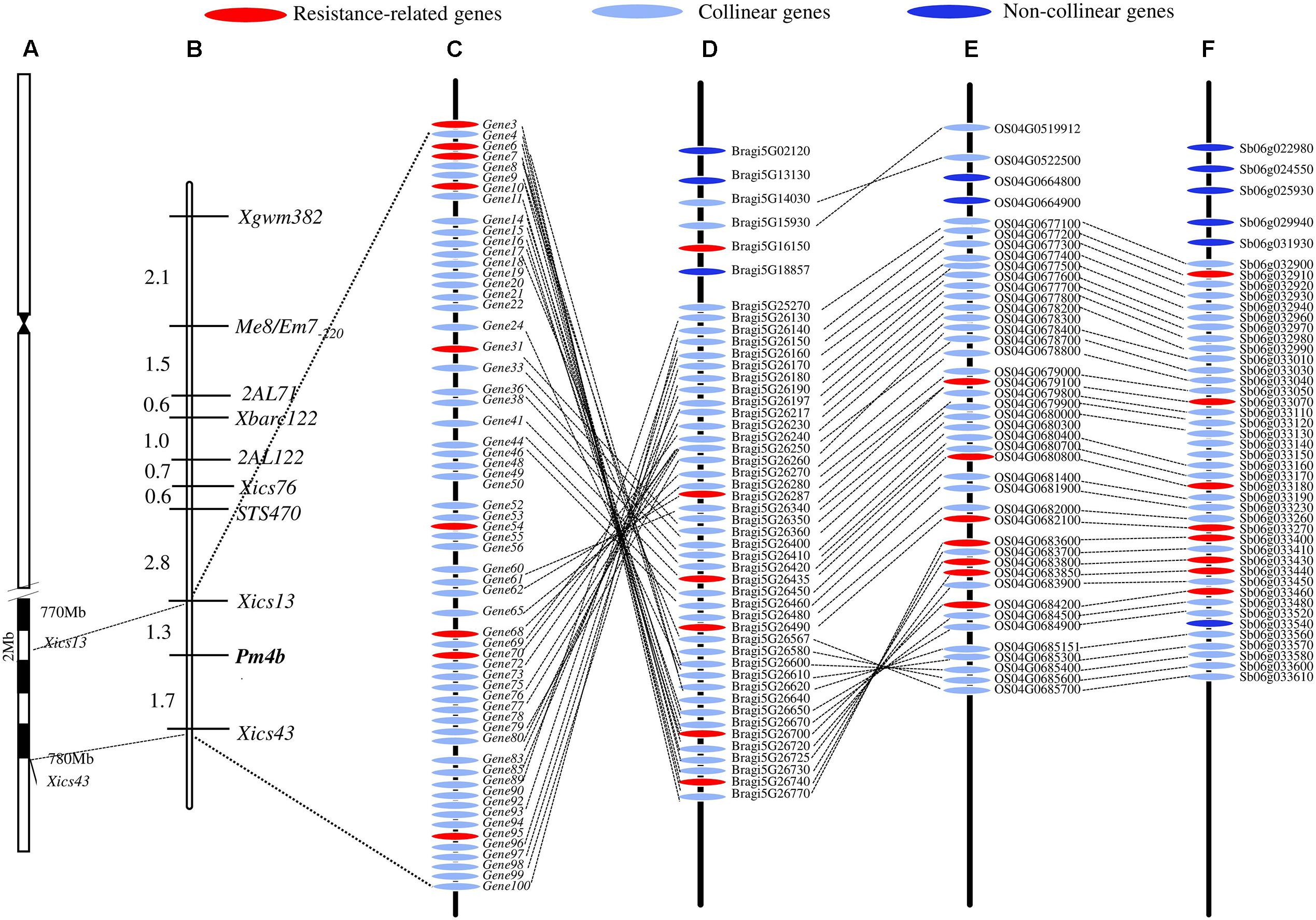
FIGURE 4. Schematic diagram of Pm4b physical interval on the chromosome position (A), genetic linkage map (B), and synteny genes among T. aestivum cv. Chinese Spring (C) and Brachypodium distachyon (D), rice (E), and sorghum (F) in the Pm4b corresponding genomic interval. Pm4b is located in an about 6.7 Mb physical interval on the end of chromosome 2AL, and the collinear genes and resistance-related genes among T. aestivum cv. Chinese Spring, B. distachyon, rice and sorghum in the Pm4b corresponding genomic interval are shown.
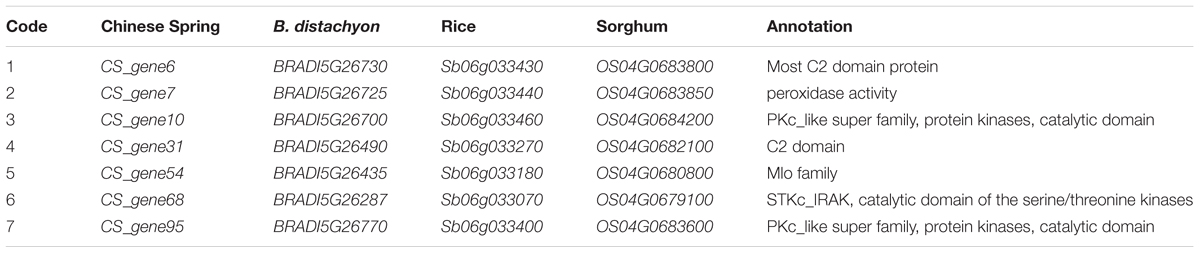
TABLE 6. Disease resistance genes identified in the Pm4b collinear genomic regions in Triticum aestivum cv. Chinese Spring, Brachypodium distachyon, rice, and sorghum.
Discussion
Pm4b was effective in certain areas of wheat producing fields in China. Results of BSR-Seq analysis demonstrated that Pm4b was located on the distal end of chromosome arm 2AL, which is consistent with its localization in previous study (Bariana and McIntosh, 1993). Three newly developed SSR markers, Xics13, Xics43, and Xics76, were incorporated into the genetic linkage map of Pm4b. These markers were able to produce the diagnostic banding patterns for clearly distinguishing Pm4b from other known Pm genes. The identification of these markers will facilitate molecular detection of Pm4b in MAS programs or pyramiding it with other effective genes for providing a broader spectrum of resistance against powdery mildew.
Pm4b was derived from a T. persicum chromosomal segment that was introgressed onto wheat chromosome 2AL (Bariana and McIntosh, 1993). The lack of homeologous chromosome pairing may prevent the recombination between this T. persicum chromosomal segment and the corresponding wheat chromosome. Moreover, the poor abundance of markers applied in previous works impeded the identification of molecular markers closely linked to Pm4b (Hao et al., 2008; Yi et al., 2008). Based on the linkage analysis in the current study, all the previously identified gene-linked markers were located on the proximal side of Pm4b using the mapping population in this study (Figure 1C).
To develop more gene-linked markers, the BSR-Seq technique was applied with the mapping population derived from the Pm4b NILs. This technique resulted in the identification of four polymorphic SNP markers, which were anchored in the distal region of chromosome 2AL of the Chinese Spring wheat genome sequence. An interval (∼20 Mb) of the genomic sequence that flanked these SNP markers was used to develop SSR markers associated with the target gene. Among the 98 pairs of SSR primers designed from the sequence of this interval, three polymorphic markers were detected in the F2:3 mapping population, which were incorporated in the genetic linkage map of Pm4b. The transferability of SSRs between common wheat and the relative species varied (Kuleung et al., 2004). The low efficiency of polymorphic markers designed based on the Chinese Spring genome sequence may attribute to the origin of Pm4b from T. persicum. The emergence of new strategies provides a great potential to identify candidate genes of wheat. In the study of cloning Pm21 gene that was transferred from chromosome 6VS of Haynaldia villosa L. to wheat chromosome 6AL, Xing et al. (unpublished) combined the strategies of development of cytogenetic stocks, mutagenesis, RenSeq and PacBio and identified an NBS-LRR type gene NLR1-V from the Pm21 locus. He et al. (2017) identified Pm21 as a typical coiled-coil-nucleotide-binding site, leucine-rich repeat (CC-NBS-LRR) gene by an integrated strategy of resistance gene analog (RGA)-based cloning via comparative genomics, physical and genetic mapping. Based on the flow sorting and sequencing of mutant chromosomes technique, Sánchez-Martín et al. (2016) developed the MutChromSeq technique to recognize induced mutations and isolated Pm2 gene that was derived from Ae. tauschii Coss.
RNA-Seq is a way to look for the alien pieces with de novo assembly of the transcriptome data (Liu et al., 2012). For the species without the reference genome sequence, however, it may increase the difficulty to obtain effective Single Nucleotide Variants (SNVs), which prevents the identification of enough variants associated with the traits of interest. In such cases, the genome sequences of the homoeologous species are often used as the reference genomes. The challenges in de novo assembly of pooled RNA-Seq data in a huge genome of a hexaploid species such as wheat using only the mapped reads into high-quality and full-length transcripts may hinder the finding of introduced genes from other related species. Also, it may miss the expressed sequences in the gaps of the Chinese Spring reference genome sequences, the highly homologous and homoeologous sequences, the sequences dislike the Chinese Spring reference genome sequences, and the unique sequences from the related species. The technique of BSR-Seq often cannot obtain the induced expression information of disease-resistance genes taking into consideration that the RNA samples are extracted from leaves uninoculated with any Bgt isolates. Moreover, the effective use of this methodology is largely associated with the sequencing depth. The limited sequencing depth may be inadequate for calling reliable variants from low expressed genes for the purpose of association analysis. Sequencing in higher depth and longer length, improving the BSR-Seq in de novo assembly of low expressed genes, and optimizing algorithms of variant calling and allele frequency estimations in pooled RNA-Seq samples would be helpful to solve this problem.
Physical mapping of the Xics13 and Xics43 markers that were linked to Pm4b to the Chinese Spring reference genome sequence enabled the identification of candidate genes for disease resistance, for example, C2 domain, peroxidase activity protein, protein kinases of PKc_like super family, Mlo family protein, and catalytic domain of the serine/threonine kinases (STKc_IRAK like super family) (Table 5). Up to now, four wheat powdery mildew resistance genes, Pm3b, Pm21, Pm2, and Pm60, have been cloned in wheat. Pm3b is a member of the CC-NBS-LRR type of disease resistance genes (Yahiaoui et al., 2004). Stpk-V, a serine/threonine protein kinase gene, was shown to be a member of Pm21 (Cao et al., 2011). In the most recent studies, Pm21 proved to be the CC-NBS-LRR gene NLR1-V (He et al., 2017; Xing et al., unpublished). Pm2 was also identified as a CC-NB-ARC-LRR resistance gene (Sánchez-Martín et al., 2016). Pm60, originating from T. urartu Thumanjan ex Gandilyan, is also a NB-LRR gene (Zou et al., 2017). The wild-type Mlo gene is a negative regulator of resistance to powdery mildew in barley (Hordeum vulgare L.) (Büschges et al., 1997). Currently, over 100 R genes have been cloned from various species, and the NBS-LRR proteins are the most abundant class of disease resistance genes in plants (Yang et al., 2013). Based on the features of their N-terminal structures, this protein family includes two major subfamilies: the Toll-interleukin (TIR-NBS-LRR) subfamily and the coiled-coil (CC-NBS-LRR) subfamily (Krattinger and Keller, 2016). In the present study, no NBS-LRR type of resistance gene was predicted in the target genomic region. There is a need to fine map Pm4b to narrow the genomic region that ensures the precisely identification of the candidate gene of Pm4b.
Zeng et al. (2014) reported that the mean frequency of virulent isolates on Pm4b was 42.5% out of 1082 Bgt isolates from the major wheat-growing regions of China, with the lowest virulence frequency of 16.7% for the isolates from the mid-Valley of the Yangtze River. Results of the present study also demonstrated the effectiveness of Pm4b in some provinces in northern part of China. Also, pyramiding multiple resistance genes is another effective means to improve disease resistance. Mwale et al. (2017) detected 24 cultivars that carried Pm4b gene among 60 wheat cultivars from China using the gene-linked molecular markers. Based on that study, the combination of genes Pm2+Pm4b+Pm8 was possibly present in cultivars Xinxuan 2039, Lankao 008 and Zhengmai 366, and Yumai 368 may possess Pm2+Pm4b+Pm6. Using the gene-linked markers, Zhang et al. (2002) identified 11 wheat lines that pyramided Pm4b, Pm13, and Pm21 genes. The lines with multiple genes provided better resistance to powdery mildew than the single gene. Line VPM1/7∗Bainong 3217 F4, which was developed using VPM1 as the donor parent and Bainong 3217 as the recurrent parent, has promising agronomic traits in addition to the resistance to powdery mildew (Zhou et al., 2005). This ensures its direct application in the breeding programs in China. The development of the breeder-friendly PCR markers can facilitate the effective identification of Pm4b in the breeding populations.
In summary, four SNP and three SSR markers were developed by means of the BSR-Seq technique, which mapped Pm4b gene in a 3.0 cM genetic interval corresponding to a 6.7 Mb genomic region. The putative genes in this interval were annotated by the web-based programs EnsemblPlants and NCBI. This interval had a good collinearity with certain genomic regions of B. distachyon (chromosome 5), rice (chromosome 4) and sorghum (chromosome 6). The collinear genomic region contained seven disease resistance genes. Xics13 and Xics43 can be used as the diagnostic molecular markers for identifying Pm4b during its marker-assisted selection.
Author Contributions
HjL and PW conceived and designed the study. PW, JH, and DQ conducted the experiments. PW, JX, JL, ML, and ZL analyzed the data. HZ, LY, and HwL performed the phenotypic tests and other works involved in this study. PW and HjL wrote the manuscript with the contributions of YZ, ZL, and ZZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The financial supports provided by the National Key Research and Development Program of China (2016YFD0101004, 2017YFD0101000, and 2017YFD0101600), the National Natural Science Foundation of China (31471491), the CAAS Innovation Team and the National Engineering Laboratory of Crop Molecular Breeding are gratefully appreciated.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00095/full#supplementary-material
Footnotes
- ^https://shigen.nig.ac.jp/wheat/komugi/genes/download.jsp
- ^https://wheat-urgi.versailles.inra.fr/Seq-Repository/News
- ^https://probes.pw.usda.gov/GSP/
- ^https://wheat.pw.usda.gov/demos/BatchPrimer3/
- ^http://plants.ensembl.org/index.html
- ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
References
Asnaghi, C., Roques, D., Ruffel, S., Kaye, C., Hoarau, J. Y., Télismart, H., et al. (2004). Targeted mapping of a sugarcane rust resistance gene (Bru1) using bulked segretant analysis and AFLP markers. Theor. Appl. Genet. 108, 759–764. doi: 10.1007/s00122-003-1487-6
Bariana, H. S., and McIntosh, R. A. (1993). Cytogenetic studies in wheat. XV. Location of rust resistance genes in VPMl and their genetic linkage with other disease resistance genes in chromosome 2. Genome 36, 476–482. doi: 10.1139/g93-065
Bariana, H. S., and McIntosh, R. A. (1994). Characterisation and origin of rust and powdery mildew resistance genes in VPM1 wheat. Euphytica 76, 53–61. doi: 10.1007/BF00024020
Bhullar, N. K., Street, K., Mackay, M., Yahiaoui, N., and Keller, B. (2009). Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. U.S.A. 106, 9519–9524. doi: 10.1073/pnas.0904152106
Bhullar, N. K., Zhang, Z. Q., Wicker, T., and Keller, B. (2010). Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project. BMC Plant Biol. 10:88. doi: 10.1186/1471-2229-10-88
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Büschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., et al. (1997). The barely Mlo gene: a novel control element of plant pathogen resistance. Cell 88, 695–705. doi: 10.1016/S0092-8674(00)81912-1
Cai, H. W., Gao, Z. S., Yuyama, N., and Ogawa, N. (2003). Identification of AFLP markers closely linked to the rhm gene for resistance to Southern Corn Leaf Blight in maize by using bulked segretant analysis. Mol. Genet. Genomics 269, 299–303. doi: 10.1007/s00438-003-0837-z
Cao, A., Xing, L., Wang, X., Yang, X., Wang, W., Sun, Y., et al. (2011). Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. U.S.A. 108, 7727–7732. doi: 10.1073/pnas.1016981108
Cheema, K. K., Grewal, N. K., Vikal, Y., Sharma, R., Lore, J. S., Das, A., et al. (2008). A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4L and transferred to Oryza sativa L. Genet. Res. 90, 397–407. doi: 10.1017/S0016672308009786
Chen, S. B., Cai, Y. L., Zhou, R. H., and Jia, J. Z. (2002). An STS marker for the wheat powdery mildew resistance gene Pm4 (In Chinese). J. Southwest Agric. Univ. 24, 231–234.
Cowger, C., Miranda, L., Griffey, C., Hall, M., Murphy, J. P., and Maxwell, J. (2012). “Wheat powdery mildew,” in Disease Resistance in Wheat, ed. I. Sharma (Oxfordshire: CABI), 84–119. doi: 10.1079/9781845938185.0084
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Doussinault, G., Delibes, A., Sanchez-Monge, R., and Garcia-Olmedo, F. (1983). Transfer of a dominant gene for resistance to eyespot disease from a wild grass to hexaploid wheat. Nature 303, 698–700. doi: 10.1038/303698a0
Duan, Y., Li, W., Wu, W., Pan, R., Zhou, Y., Qi, J., et al. (2003). Genetic analysis and mapping of gene fzp(t) controlling spikelet differentiation in rice. Sci. China C Life Sci. 46, 328–334. doi: 10.1360/03yc9035
Fabritius, A. L., Shattock, R. C., and Judelson, H. S. (1997). Genetic analysis of metalaxyl insensitivity loci in Phytophthora infestans using linked DNA markers. Phytopathology 87, 1034–1040. doi: 10.1094/PHYTO.1997.87.10.1034
Green, A. J., Berger, G., Griffey, C. A., Pitman, R., Thomason, W., and Balota, M. (2014). Genetic resistance to and effect of leaf rust and powdery mildew on yield and its components in 50 soft red winter wheat cultivars. Crop Prot. 64, 177–186. doi: 10.1016/j.cropro.2014.06.023
Gupta, P. K., Mir, R. R., Mohan, A., and Kumar, J. (2008). Wheat genomics: present status and future prospects. Int. J. Plant Genomics 2008:896451. doi: 10.1155/2008/896451
Hao, Y. F., Liu, A. F., Wang, Y. H., Feng, D. S., Gao, J. R., Li, X. F., et al. (2008). Pm23: a new allele of Pm4 located on chromosome 2AL in wheat. Theor. Appl. Genet. 117, 1205–1212. doi: 10.1007/s00122-008-0827-y
Hao, Y. F., Parks, R., Cowger, C., Chen, Z. B., Wang, Y. Y., Bland, D., et al. (2015). Molecular characterization of a new powdery mildew resistance gene Pm54 in soft red winter wheat. Theor. Appl. Genet. 128, 465–476. doi: 10.1007/s00122-014-2445-1
He, H., Zhu, S., Ji, Y., Jiang, Z., Zhao, R., and Bie, T. (2017). Map-based cloning of the gene Pm21 that confers broad spectrum resistance to wheat powdery mildew. arXiv:1708.05475.
Hsam, S. L. K., Huang, X. Q., Ernst, F., Hartl, L., and Zeller, F. J. (1998). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). Alleles at the Pm1 locus. Theor. Appl. Genet. 96, 1129–1134. doi: 10.1007/BF00225724
Huang, G. Z., Zhu, M. Z., Wang, S. J., and Wu, Z. H. (1982). Development of the wheat cultivar Bainong 3217 and analysis of its high and stable yield (In Chinese). J. Baiquan Agric. Coll. 1, 1–10.
Hulbert, S. H., Webb, C. A., Smith, S. M., and Sun, Q. (2001). Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. doi: 10.1146/annurev.phyto.39.1.285
Kowalczyk, K., Gruszecka, D., Nowak, M., and Lesniowska-Nowak, J. (2011). Resistance of triticale hybrids with Pm4b and Pm6 genes to powdery mildew. Acta Biol. Crac. Ser. Bot. 53, 57–62. doi: 10.2478/v10182-011-0008-1
Krattinger, S. G., and Keller, B. (2016). Molecular genetics and evolution of disease resistance in cereals. New Phytol. 212, 320–332. doi: 10.1111/nph.14097
Kuleung, C., Baenziger, P. S., and Dweikat, I. (2004). Transferability of SSR markers among wheat, rye, and triticale. Theor. Appl. Genet. 108, 1147–1150. doi: 10.1007/s00122-003-1532-5
Lambreghts, R., Shi, M., Belden, W. J., Decaprio, D., Park, D., Henn, M. R., et al. (2009). A high-density single nucleotide polymorphism map for Neurospora crassa. Genetics 181, 767–781. doi: 10.1534/genetics.108.089292
Li, N., Jia, H. Y., Kong, Z. X., Tang, W. B., Ding, Y. X., Liang, J. C., et al. (2017). Identification and marker-assisted transfer of a new powdery mildew resistance gene at the Pm4 locus in common wheat. Mol. Breed. 37, 79. doi: 10.1007/s11032-017-0670-4
Lincoln, S. E., Daly, M. J., and Lander, E. S. (1993). Constructing Linkage Maps with MAPMAKER/Exp Version 3.0: A Tutorial Reference Manual, 3rd Edn. Cambridge, MA: Whitehead Institute for Medical Research.
Liu, S. Z., Yeh, C. T., Tang, H. M., Nettleton, D., and Schnable, P. S. (2012). Gene mapping via bulked segretant RNA-Seq (BSR-Seq). PLOS ONE 7:e36406. doi: 10.1371/journal.pone.0036406
Liu, W. C., Liu, Z. D., Huang, C., Lu, M. H., Liu, J., and Yang, Q. P. (2016). Statistics and analysis of crop yield losses caused by main disease and insect pests in recent 10 years (In Chinese). Plant Prot. 42, 1–9.
Liu, Z. Y., Sun, Q. X., Ni, Z. F., and Yang, T. (1999). Development of SCAR markers linked to the Pm21 gene conferring resistance to powdery mildew in common wheat. Plant Breed. 118, 215–219. doi: 10.1046/j.1439-0523.1999.118003215.x
Ma, Z. Q., Wei, J. B., and Cheng, S. H. (2004). PCR-based markers for the powdery mildew resistance gene Pm4a in wheat. Theor. Appl. Genet. 109, 140–145. doi: 10.1007/s00122-004-1605-0
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segretant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. U.S.A. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Mwale, M. M., Tang, X. L., and Chilembwe, E. (2017). Molecular detection of disease resistance genes to powdery mildew (Blumeria graminis f. sp. tritici) in wheat (Triticum aestivum) cultivars. Afr. J. Biotechnol. 16, 22–31. doi: 10.5897/AJB2016.15720
Parks, R., Carbone, I., Murphy, J. P., Marshall, D., and Cowger, C. (2008). Virulence structure of the eastern US wheat powdery mildew population. Plant Dis. 92, 1074–1082. doi: 10.1094/PHYTO-99-7-0840
Ramirez-Gonzalez, R. H., Segovia, V., Bird, N., Fenwick, P., Holdgate, S., Berry, S., et al. (2015). RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol. J. 13, 613–624. doi: 10.1111/pbi.12281
Saghai-Maroof, M. A., Soliman, K. M., Jorgensen, R. A., and Allard, R. W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. U.S.A. 81, 8014–8018. doi: 10.1073/pnas.81.24.8014
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17:221. doi: 10.1186/s13059-016-1082-1
Schmolke, M., Mohler, V., Hartl, L., Zeller, F. J., and Hsam, S. L. K. (2012). A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol. Breed. 29, 449–456. doi: 10.1007/s11032-011-9561-2
Singrün, C. H., Hsam, S. L. K., Hartl, L., Zeller, F. J., and Mohler, V. (2003). Powdery mildew resistance gene Pm22 in cultivar Virest is a member of the complex Pm1 locus in common wheat (Triticum aestivum L. em Thell.). Theor. Appl. Genet. 106, 1420–1424. doi: 10.1007/s00122-002-1187-7
Trick, M., Adamski, N. M., Mugford, S. G., Jiang, C. C., Febrer, M., and Uauy, C. (2012). Combining SNP discovery from next-generation sequencing data with bulked segretant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol. 12:14. doi: 10.1186/1471-2229-12-14
Tsilo, T. J., Chao, S., Jin, Y., and Anderson, J. A. (2009). Identification and validation of SSR markers linked to the stem rust resistance gene Sr6 on the short arm of chromosome 2D in wheat. Theor. Appl. Genet. 118, 515–524. doi: 10.1007/s00122-008-0917-x
Wang, Y., Zhang, H., Xie, J., Guo, B. M., Chen, Y. X., Zhang, H. Y., et al. (2017). Mapping stripe rust resistance genes by BSR-Seq: YrMM58 and YrHY1 on chromosome 2AS in Chinese wheat lines Mengmai 58 and Huaiyang 1 are Yr17. Crop J. (in press). doi: 10.1016/j.cj.2017.03.002
Wang, Z. L., Li, L. H., He, Z. H., Duan, X. Y., Zhou, Y. L., Chen, X. M., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/PD-89-0457
Xie, W., Ben-David, R., Zeng, B., Dinoor, A., Xie, C., Sun, Q., et al. (2012). Suppressed recombination rate in 6VS/6AL translocation region carrying the Pm21 locus introgressed from Haynaldia villosa into hexaploid wheat. Mol. Breed. 29, 399–412. doi: 10.1007/s11032-011-9557-y
Yahiaoui, N., Srichumpa, P., Dudler, R., and Keller, B. (2004). Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 37, 528–538. doi: 10.1046/j.1365-313X.2003.01977.x
Yang, S. H., Li, J., Zhang, X. H., Zhang, Q. J., Huang, J., Chen, J. Q., et al. (2013). Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc. Natl. Acad. Sci. U.S.A. 110, 18572–18577. doi: 10.1073/pnas.1318211110
Yi, Y. J., Liu, H. Y., Huang, X. Q., An, L. Z., Wang, F., and Wang, X. L. (2008). Development of molecular markers linked to the wheat powdery mildew resistance gene Pm4b and marker validation for molecular breeding. Plant Breed. 127, 116–120. doi: 10.1111/j.1439-0523.2007.01443.x
Zeng, F. S., Yang, L. J., Cong, S. J., Shi, W. Q., Zhang, X. J., Wang, H., et al. (2014). Virulence and diversity of Blumeria graminis f. sp. tritici population in China. J. Integr. Agric. 13, 2424–2437. doi: 10.1007/s00122-011-1651-3
Zhang, R., Sun, B., Chen, J., Cao, A. Z., Xing, L. P., Feng, Y. G., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129, 1975–1984. doi: 10.1007/s00122-016-2753-8
Zhang, Z. Y., Chen, X., Zhang, C., Xin, Z. Y., and Chen, X. M. (2002). Selecting the pyramids of powdery mildew resistance genes Pm4b, Pm13 and Pm21 in wheat assisted by molecular marker (In Chinese). Sci. Agric. Sin. 35, 789–793.
Zhou, R. H., Zhu, Z. D., Kong, X. Y., Huo, N. X., Tian, Q. Z., Li, P., et al. (2005). Development of wheat near-isogenic lines for powdery mildew resistance. Theor. Appl. Genet. 110, 640–648. doi: 10.1007/s00122-004-1889-0
Keywords: powdery mildew, Pm4b, BSR-Seq, RNA-Seq, SNP, SSR marker
Citation: Wu P, Xie J, Hu J, Qiu D, Liu Z, Li J, Li M, Zhang H, Yang L, Liu H, Zhou Y, Zhang Z and Li H (2018) Development of Molecular Markers Linked to Powdery Mildew Resistance Gene Pm4b by Combining SNP Discovery from Transcriptome Sequencing Data with Bulked Segregant Analysis (BSR-Seq) in Wheat. Front. Plant Sci. 9:95. doi: 10.3389/fpls.2018.00095
Received: 06 November 2017; Accepted: 17 January 2018;
Published: 14 February 2018.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Morten Lillemo, Norwegian University of Life Sciences, NorwayPer Langkjaer Gregersen, Aarhus University, Denmark
Copyright © 2018 Wu, Xie, Hu, Qiu, Liu, Li, Li, Zhang, Yang, Liu, Zhou, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongjie Li, lihongjie@caas.cn Zhongjun Zhang, zhangzj@cau.edu.cn
 Peipei Wu1,2
Peipei Wu1,2 Jingzhong Xie
Jingzhong Xie Jinghuang Hu
Jinghuang Hu Zhiyong Liu
Zhiyong Liu Jingting Li
Jingting Li Zhongjun Zhang
Zhongjun Zhang Hongjie Li
Hongjie Li