- 1Ecological Science Group, The James Hutton Institute, Dundee, United Kingdom
- 2Low and Medium Energy Physics, Jožef Stefan Institute, Ljubljana, Slovenia
- 3Future Food Beacon of Excellence and School of Biosciences, University of Nottingham, Loughborough, United Kingdom
- 4Distinguished Scientist Fellowship Program, King Saud University, Riyadh, Saudi Arabia
Shoot zinc (Zn) concentration in Brassica oleracea is affected by soil Zn and phosphorus (P) supply. Most problematic is the negative impact of P fertilizers on Zn concentrations in crops, which makes balancing yield and mineral quality challenging. To evaluate early molecular mechanisms involved in the accumulation of large shoot Zn concentrations regardless of the P supply, two B. oleracea accessions differing in root architecture and root exudates were grown hydroponically for two weeks with different combinations of P and Zn supply. Ionome profiling and deep RNA sequencing of roots revealed interactions of P and Zn in planta, without apparent phenotypic effects. In addition, increasing P supply did not reduce tissue Zn concentration. Substantial changes in gene expression in response to different P and/or Zn supplies in roots of both accessions ensured nutritionally sufficient P and Zn uptake. Numerous genes were differentially expressed after changing Zn or P supply and most of them were unique to only one accession, highlighting their different strategies in achieving nutrient sufficiency. Thus, different gene networks responded to the changing P and Zn supply in the two accessions. Additionally, enrichment analysis of gene ontology classes revealed that genes involved in lipid metabolism, response to starvation, and anion transport mechanisms were most responsive to differences in P and Zn supply in both accessions. The results agreed with previously studies demonstrating alterations in P and Zn transport and phospholipid metabolism in response to reduced P and Zn supply. It is anticipated that improved knowledge of genes responsive to P or Zn supply will help illuminate the roles in uptake and accumulation of P and Zn and might identify candidate genes for breeding high-yield-high-Zn brassicas.
Introduction
Plants have an extraordinary ability to survive in a fluctuating environment. When facing scarcity in essential mineral elements (nutrients) plants employ a plethora of morphological, physiological and metabolic acclimatory mechanisms to enhance nutrient acquisition and/or their within-plant utilisation (White and Brown, 2010; White et al., 2013b). In the case of phosphorus (P) deficiency, enhanced phosphate (Pi) acquisition is delivered by enhancing the mobilisation of Pi from soils, increasing the volume of soil explored by the root system to capture solubilized Pi and increasing the rate of Pi uptake across the root rhizodermis (Rose et al., 2013; Hunter et al., 2014; Wissuwa et al., 2016). Within-plant mechanisms reducing plant P-requirements include alterations in P translocation from younger to older tissues, utilisation of P-sparing metabolic pathways, reducing the abundance of nucleic acids, and recycling P through lipid remodelling (Veneklaas et al., 2012; Nakamura, 2013; Pant et al., 2015; Siebers et al., 2015; Tawaraya et al., 2018). In the case of zinc (Zn) deficiency, increased Zn uptake is achieved mainly by increasing Zn uptake capacity through a tightly regulated network of molecular interactions in which the coordinated expression of Zn transporters plays a major role in Zn acquisition from soil, in movement between organs and tissues and in intracellular sequestration of Zn (Assunção et al., 2010), although chemical modification of the rhizosphere to increase Zn phytoavailability and greater exploration of the root volume are also beneficial (White et al., 2013a). Adaptive traits involved in plant responses to Pi deficiency will also improve Zn acquisition (Rose et al., 2013). Thus, although the requirements for P and Zn differ by orders of magnitude, i.e. 2,000–5,000 µg P g-1 dry weight (DW) and 15–30 µg Zn g-1 DW in most crops (White and Brown, 2010) and their roles in the plants are contrasting, they are both poorly mobile in soil and as such, traits involved in the increased acquisition of one can improve the acquisition of the other.
Phosphorus is one of the key yield-limiting nutrients for crop production and responses to Pi deficiency and the optimisation of P-use-efficiency (PUE) have achieved significantly more attention than has improving Zn-use efficiency (ZnUE). This is unfortunate, since Zn deficiency in soils is a global problem and improving Zn nutrition of crops and increasing Zn concentration in seeds can have several beneficial effects including higher yields, better seed viability and seedling vigour, smaller seeding rate and improved tolerance to abiotic and biotic stress (Broadley et al., 2007; Cakmak, 2008; White and Veneklaas, 2012). In addition, Zn malnutrition (i.e. insufficient intake of Zn in animals and humans) remains a significant health problem, particularly in children (White and Broadley, 2009; Bouis and Saltzman, 2017; Cakmak and Kutman, 2018).
Recent findings suggest that there is a molecular coordination between P and Zn homeostasis (Huang et al., 2000; Bouain et al., 2014; Khan et al., 2014; Briat et al., 2015; Kisko et al., 2018) which may help interpret observations of P-Zn interactions in different plants. Three different types of P-Zn interactions have been reported: (1) P-induced Zn deficiency (Stukenholtz et al., 1966; Cakmak and Marschner, 1986; Li et al., 2003; Zhang et al., 2012), (2) Zn-deficiency-induced P overaccumulation (Cakmak and Marschner, 1986; Marschner and Cakmak, 1986; Huang et al., 2000; Bouain et al., 2014; Santos et al., 2018) and (3) P-deficiency-induced Zn overaccumulation (Haldar and Mandal, 1981; Misson et al., 2005; Barben et al., 2010; Pedas et al., 2011). The consequences of these P-Zn interactions are negative effects on the yield, quality and longevity of crops and their products with potential negative effects on mineral element composition of produce. This indicates that agricultural management strategies should seek for balanced mineral nutrition to optimize yields and resource use efficiencies.
A prerequisite for the development of crops with greater PUE without reduced Zn concentrations in edible parts is an understanding of the regulatory mechanisms underlying P and Zn deficiency responses and their within-plant interactions. Since P deficiency is perceived by the root tip (Ham et al., 2018) and the subsequent biochemical and morphological adjustments made by plants to increase both Pi and Zn acquisition take place in roots, roots have been the focus of research on the molecular responses to P and Zn deficiency. Since acclimation to low Pi or Zn supply relies fundamentally on changes in gene expression, recent technical advances in “bottom-up” omics technologies have allowed novel and integrative insights into these processes (Lan et al., 2015). However, many inconsistent observations still need to be resolved before meaningful conclusions can be drawn. For example, although transcriptomic studies have uncovered a surprisingly large number of Pi starvation inducible (PSI) genes, these do not respond similarly in all studies, which might be attributed to stochastic fluctuations in gene expression, differences in growth conditions, plant growth stage or tissue sampled, plant nutritional status, differences in sample handling or experimental design, plant species studied, or combination of these factors (Lan et al., 2015). Nonetheless, by combining four studies (Misson et al., 2005; Morcuende et al., 2007; Lan et al., 2012; Woo et al., 2012) of a genetic model plant Arabidopsis thaliana (L.) Heynh., 95 core PSI genes were identified, with genes related to lipid metabolism and galactolipid biosynthesis representing the largest group, emphasizing the importance of these processes for P recycling (Lan et al., 2015). Indeed, use of the promoter for SQD1, a gene involved in sulfolipid synthesis, was suggested for the development of “smart plants” to monitor plant P status (Hammond et al., 2003). Further important PSI genes were SPX domain-containing transcription factors, Pi transporters of the PHT family and the PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR (Lan et al., 2015). Similarly, these groups of genes have been observed in responses of other plants to Pi deficiency for example in Brassica rapa L. (Hammond et al., 2011), rice (Orzya sativa L.; Secco et al., 2013), maize (Zea mays L.; Sun et al., 2016) and poplar (Populus x canescens (Aiton) Sm.; Kavka and Polle, 2017) indicating some consensus can be achieved.
In a species-wide study of Brassica oleracea L., which has been bred into a wide range of crops such as broccoli, cabbage, kale and cauliflower that are among the most commonly consumed and economically important vegetables in the world (Thomson et al., 2007), large variability in PUE was found that correlated with root development and architecture (Hammond et al., 2009). Concurrently, large genetic variation in shoot Zn concentrations was observed and shoot Zn concentration was found to be increased by increasing soil Zn concentration and reduced by increasing soil P concentrations (Broadley et al., 2010). In contrast, increasing soil P concentration increased shoot calcium (Ca) and magnesium (Mg) concentrations (Broadley et al., 2008). There was no correlation between shoot Zn concentration and shoot DW (i.e. no dilution effect), which indicated that shoot Zn concentration in leafy Brassica crops could be improved through cultivar selection. This was recently confirmed for cabbage and broccoli (White et al., 2018). However, a negative correlation between shoot P concentration and shoot DW was observed (Broadley et al., 2010), which indicated that optimal yield required fine tuning P-fertilizer application. Since Brassica vegetables are widely consumed and have relatively large Zn concentrations but small concentrations of phytate, which inhibits Zn absorption in the human intestine, they are promising candidates for delivering more bioavailable Zn to human and animal diets (White and Broadley, 2011; White et al., 2018).
In order to understand the early responses of plants to low P and/or Zn supply, differences in gene expression were studied in young roots of two B. oleracea accessions grown in a hydroponic system. Plants were analyzed at a time point at which neither ionomic changes nor growth parameters were greatly affected by the treatments. The aim of the study was to identify root-specific molecular networks and genes involved in the responses of the two B. oleracea accessions to contrasting P and Zn supply.
Material and Methods
Plant Material, Experimental Setup and Ionome Measurements
Seeds of eight Brassica oleracea accessions (Table S1) with contrasting PUE and shoot Zn concentration (Hammond et al., 2009; Broadley et al., 2010) were obtained from the Genetic Resource Unit, Warwick Crop Centre of the University of Warwick. The accessions showed 3.9-fold variability in PUE (inferred as physiological P efficiency ratio (PPUE) calculated by dividing shoot yield by the shoot P concentration) and 8-fold variability in shoot Zn concentration when plants were grown on a peat-based compost with low P (5.25 mg L-1) amendments (Hammond et al., 2009; Broadley et al., 2010). When the same accessions were grown on a peat-based compost with high P (15.75 mg L-1) amendments the PPUE varied 14-fold and shoot Zn varied 2.9-fold (Hammond et al., 2009; Broadley et al., 2010). Accessions were grown to maturity in a glasshouse to multiply seeds for subsequent experiments to ensure comparable seed characteristics.
Two independent hydroponic experiments were performed. The phytoavailability of Zn and P in a hydroponic system is generally controlled by fewer factors than in solid substrates, making it the method of choice to study Zn-P interactions (Santos et al., 2018), especially when roots must be harvested (Nguyen et al., 2016). For both experiments, seeds were germinated on the surface of filter paper moistened with deionized water in Petri dishes placed at 16°C in the dark for five days. Six 5-day-old seedlings per accession were transferred to a 5-L bucket holding a solution containing 2 mM Ca(NO3)2, 2 mM NH4NO3, 0.75 mM MgSO4, 0.5 mM KOH, 0.1 mM FeNaEDTA, 30 µM H3BO3, 25 µM CaCl2, 10 µM MnSO4, 3µM CuSO4, and 0.5 µM Na2MoO4, and either 0.25 mM KH2PO4 (High P treatment) or 0.025 mM KH2PO4 plus 0.225 mM KCl (Low P treatment). Either 1 µM ZnSO4 (High Zn) or no ZnSO4 (Low Zn) was added to each P treatment. Plants were, therefore, subjected to four treatments (Low P & Low Zn, Low P & High Zn, High P & Low Zn, and High P & High Zn). The pH was adjusted to 6 in all treatments by addition of H2SO4. Experiments were performed in a glasshouse at the James Hutton Institute (Invergowrie, Scotland, UK, latitude 56°27’22.7” N, longitude 03°04’09.9” W) and lasted two weeks, with one change of the nutrient solution, which was continuously aerated. Buckets were set in a complete randomized design on a single bench. Daytime and night time temperatures were maintained at 24°C and 18°C, respectively. Daily irradiance (>200 W m-2 for 16 h) was obtained by artificial illumination (MASTER SON-T PIA Green Power lamps, Philips, Guildford, UK).
When the first experiment, which comprised eight B. oleracea accessions grown under the said four treatments, was harvested, roots and shoots were separated and dried in an oven at 70°C for three days, before their dry weight (DW) was determined. Six individuals of each accession were subjected to each treatment. Because the root DW was small, roots of 2-3 individuals had to be pooled to obtain one sample for ionome assessment.
For the second experiment, two accessions (C6 and F103) were selected because they have different root architecture and they differ in the amount and the number of root exudates (Pongrac et al. unpublished results). Twenty-four individuals per accession were grown with each of the aforementioned treatments. At harvest, roots of three individuals per accession and per treatment were frozen in liquid nitrogen for RNA extraction yielding three replicates for gene expression analysis. Shoots of these three plants were dried in an oven at 70°C for three days to determine shoot DW. The remaining individuals were photographed (Figure S1), root length was determined using a ruler, roots were separated from shoots, dried in an oven at 70°C for three days and weighed. Because the root DW was small, roots of 2-3 individuals were pooled for ionome assessment, yielding 5 to 9 replicates for ionome analysis.
Concentrations of sodium (Na), Mg, P, sulphur (S), chlorine (Cl), potassium (K), Ca, manganese (Mn), nickel (Ni), copper (Cu), iron (Fe), and Zn were determined in plant material using inductively-coupled-plasma-mass spectrometry following microwave-assisted acid digestion of dried roots and shoots as described by White et al. (2012).
RNA Extraction and RNAseq
Frozen root samples were ground to powder using a pre-cooled pestle and mortar, after which the powdered material was transferred to a 2 ml Eppendorf tube. One mL of Trizol® was added to each sample, shaken thoroughly and incubated at room temperature for 5 min. Subsequently 200µl chloroform was added to each sample after which they were again shaken and incubated for 5 min at room temperature. The samples were centrifuged at 12000×g for 15 min at 4°C and 600µl supernatant was transferred to a new 2 ml Eppendorf tube to which 600µl of isopropanol was added. The Eppendorf tubes were inverted for mixing after which samples were centrifuged at 12000×g for 15 min at 4°C. The supernatant was removed and the pellet was rinsed in 700µl 70% ethanol and dried at room temperature for 10 min before being resuspended in 100 µl of MiliQ water. The RNA was further purified using the ENZO kit (Zymo Research, Irvine, California, USA) following the manufacturer’s instructions. An “On column DNAse treatment” was included and DNA was eluted twice with 100µl of pre-warmed MilliQ water. Library preparation, single-end reads of 50 bases (SE50) sequencing, data filtering and cleaning, alignments with the reference database (ftp://ftp.ensemblgenomes.org/pub/plants/release-35/fasta/brassica_oleracea/dna/) and gene expression quantification as well as detection of differential gene expression was performed by BGI (https://www.bgi.com/global/) according to their standard analysis pipeline (Figure S2). In brief, mRNA, selected via polyA tails, was used for cDNA library construction on which SE50 sequencing was performed. Raw reads were filtered to remove adaptor sequences, low quality reads and reads with high content of unknown bases. Sequences were then aligned using Bowtie2 and HISAT tools. Gene expression was quantified as FPKM, i.e. values which consider the gene length and total number of mapped reads and therefore eliminate the influence of gene size differences and sequencing discrepancy on gene expression, using RSEM package. Differentially expressed genes (DEGs) were defined using the NOISeq method (Tarazona et al., 2011) and were identified by comparing the gene expression in the each of the three treatments with the gene expression in the Low P & Low Zn treatment. NIOSeq uses gene expression values (FPKM) to calculate log2 (fold change) and absolute differences between all comparisons to build a noise distribution model. The average expression in all three replicates was then used to assess if the average fold change and average absolute difference value are above the noise and, if so, the gene was defined as a DEG. A filter of fold change >2 and a diverge probability greater than or equal to 80% were applied. Differentially expressed genes were subsequently mapped to gene ontology (GO) terms (http://www.geneontology.org). In the standard analysis pipeline of BGI a hypergeometric test was used to find significantly enriched GO terms based on “GO Term Finder” (http://www.yeastgenome.org/help/analyze/go-term-finder). The calculated p-values took into account the number of genes belonging to a GO term, the number of DEGs in total, the number of genes for the GO term in the Brassica genome and the overall number of GO annotatable genes. The resulting value was thus scaled for the GO-group size. Additionally, the p-values were Bonferroni corrected for multiple comparisons. Gene ontology terms with p ≤ 0.05 were selected for interpretation.
Statistical Analysis
The experiments followed a completely randomized design with four treatments and eight accessions in the first experiment and four treatments and two accessions in the second experiment. Figures were created and statistical analyses were performed in RStudio Version 1.0.143 (www.rstudio.com) in combination with R version 3.4.0 (2017-04-21); for graphics the ggplot2 package was used. Using the lsmeans package a two-way analysis of variance (ANOVA) was performed with accession and treatment as independent variables followed by a Tukey post-hoc test at p < 0.05 to assess the effect of accessions and P and Zn supply on biomass and ionome of roots and shoot. For the data in the first experiment, one-way ANOVA followed by a Tukey post-hoc test at p < 0.05 was used to detect significant treatment effects for each of the eight accessions. For two-group comparisons, Student t-test at p < 0.05 was performed. Correlations between shoot dry weight, P and Zn concentration were evaluated using the geom_smooth function of the ggplot2 package method and applying the lm (linear mixed model) method. The ggpmisc package was used to calculate and add p-values and R2 values to the graph using the functions stat_poly_eq and stat_fit_glance. Principal component analysis (PCA) was performed using the R packages FactoMineR and factoextra. Data were centred and scaled, and normal distribution confirmed, before principal components (PCs) were computed. Contribution of variables to the computed PCs was depicted graphically using the fviz_contrib function. The individual data points were plotted using the fviz_pca_ind function. Hierarchical clustering was performed using the hcut function of the factoextra package.
Results
Growth and Ionome of Brassica oleracea Accessions Receiving Contrasting Combinations of P and Zn Supply in Hydroponics
Biomass production and element concentrations of roots and shoots were evaluated in eight B. oleracea accessions grown hydroponically in solutions with contrasting combinations of P and Zn supply (Figure 1). Considering genotype and treatment as independent variables, two-way ANOVA (Table 1) revealed that, for the eight accessions studied: (1) genotype affected root length and shoot P concentration, but treatment and genotype × treatment interactions did not, (2) both genotype and treatment affected root and shoot DW, but their interaction did not, (3) genotype, treatment and their interaction affected root P concentration and shoot Zn concentration, and (4) treatment affected root Zn concentration, but genotype and genotype × treatment interaction did not. Since genotype × treatment interactions were significant only for some responses, one-way ANOVA was conducted to depict treatment effects for each accession separately. No treatment effects were observed on root or shoot biomass (Figures 1A, B). No apparent effects of P supply on root and shoot Zn concentrations were found in the B. oleracea accessions studied (Figures 1E, F). Zinc concentration increased with increasing Zn supply in both roots and shoots of all accessions grown hydroponically in the experiments reported here (Figures 1E, F).
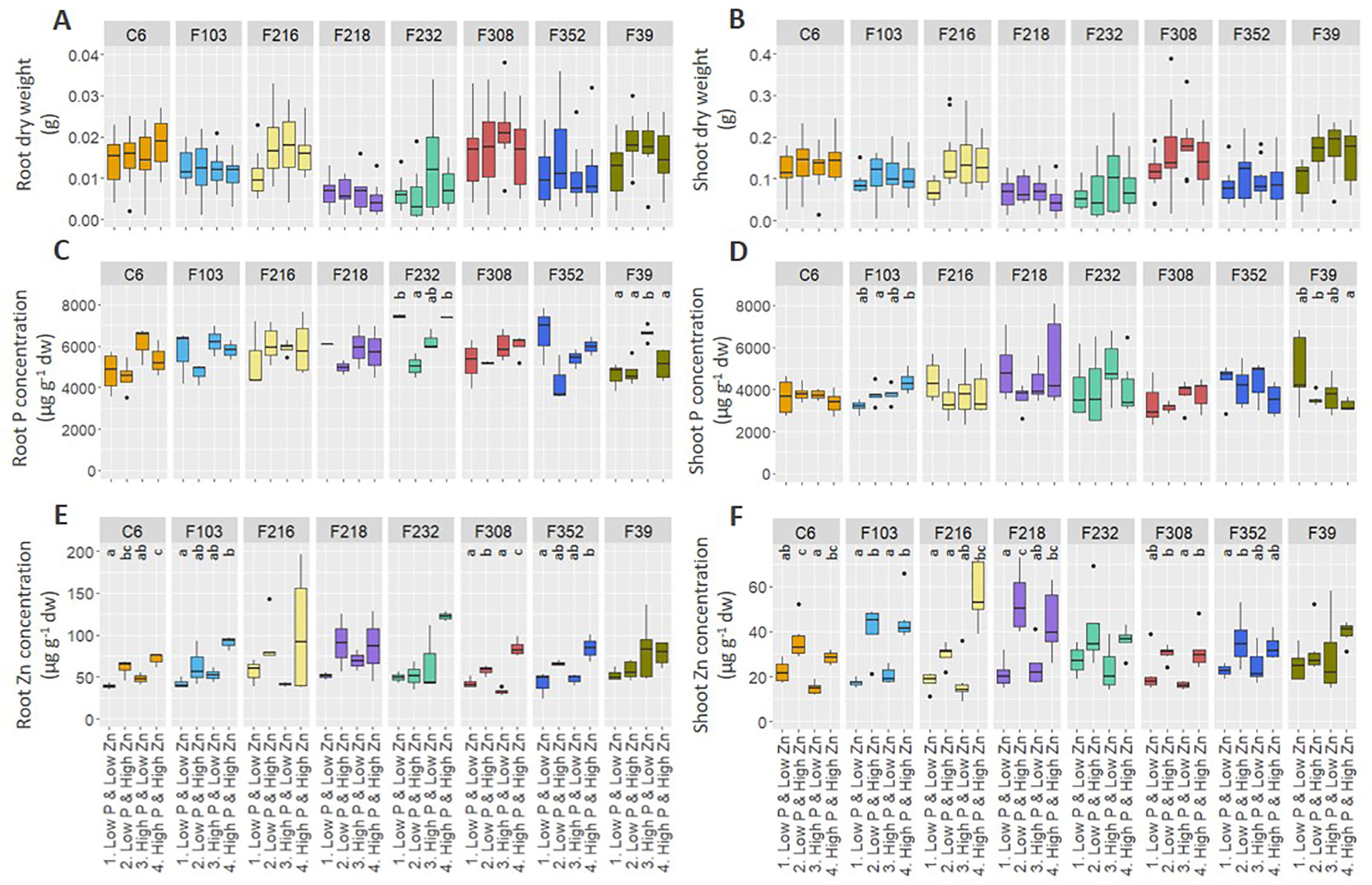
Figure 1 Effect of contrasting combinations of P and Zn supply on eight B. oleracea accessions. Plants were grown hydroponically for two weeks after which biomass (A, B) and P (C, D) and Zn (E, F) concentration were determined in roots (A, C, E) and shoots (B, D, F). Shown are Tukey boxplots representing 25th and 75th percentile of the data with the centerline representing the median, whiskers representing the 5th and 95th percentile and the black dots representing outliers (n = 4-6 for each accession and each treatment). Significant differences between treatments for particular accession are indicated by different letters above the box-plots (one-way analysis of variance followed by Tukey post-hoc test at p < 0.05). dw, dry weight.
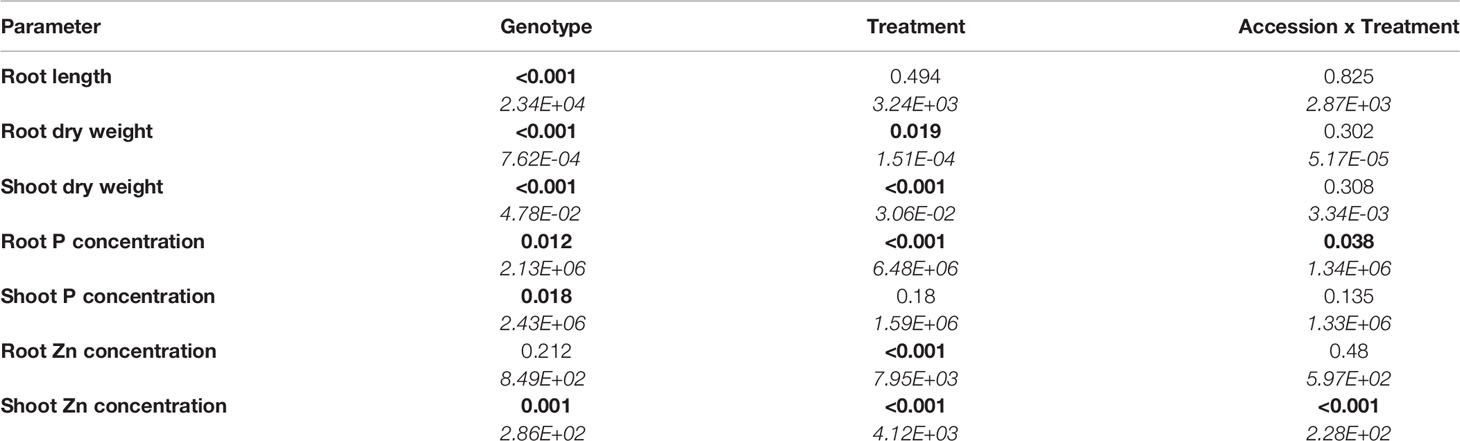
Table 1 Two-way analysis of variance table with p-values (those less than 5% are highlighted in bold) and means squares (in italics) for response variables in eight B. oleracea accessions grown in hydroponics at Low P and High P each with High Zn or Low Zn treatments for two weeks (n = 4-6 for each accession and each treatment).
Both shoot P concentration and shoot Zn concentration were negatively correlated with shoot DW, while there was a positive correlation between shoot P concentration and shoot Zn concentration (Figure 2).
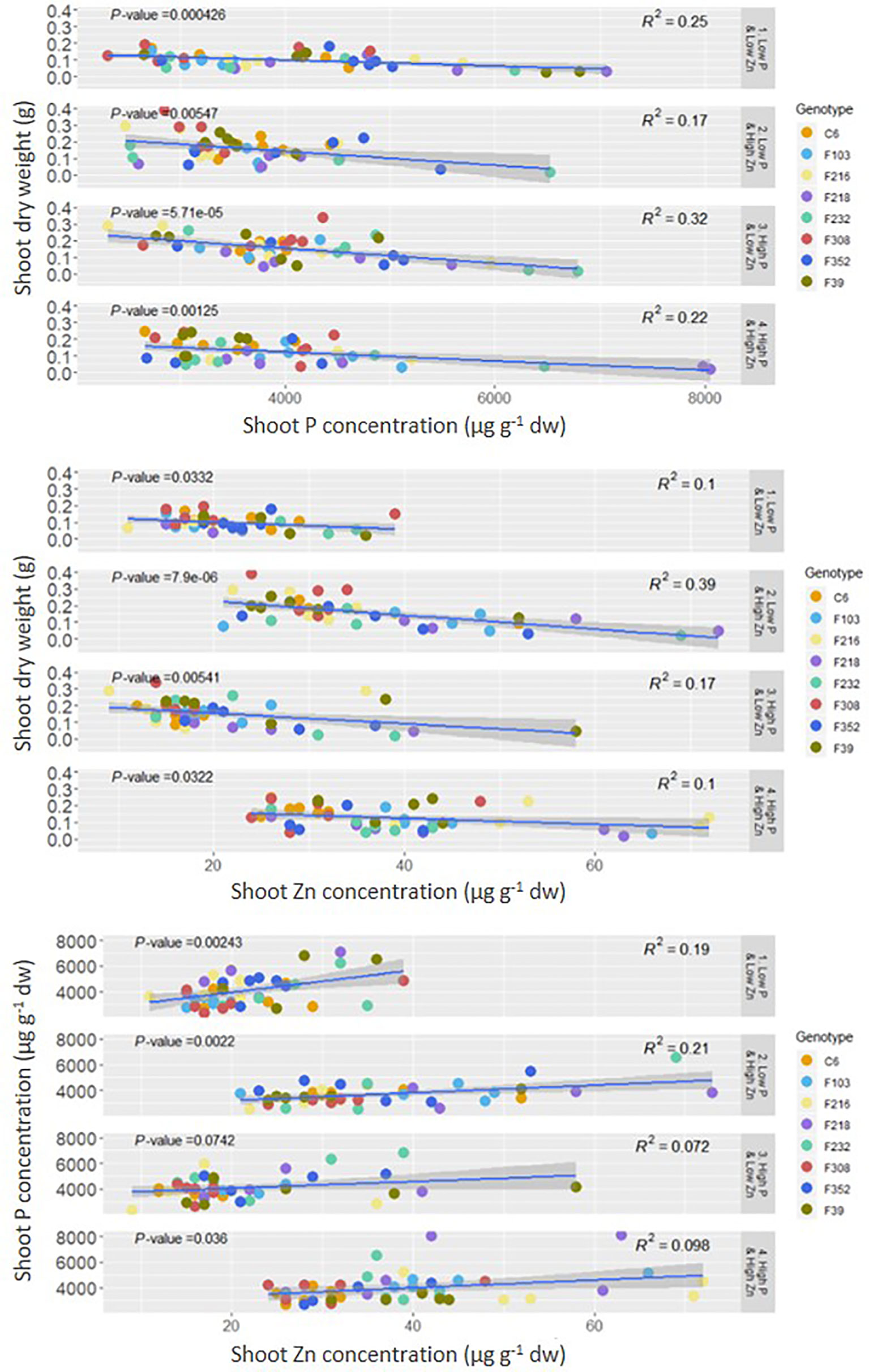
Figure 2 Correlation between shoot P or Zn concentration and shoot dry weight and between shoot P and Zn concentration in eight B. oleracea accessions grown hydroponically for two weeks with contrasting combinations of P and Zn supply. Significant correlations are indicated by p-value < 0.05 dw, dry weight.
Responses of the C6 and the F103 Accessions Receiving Combinations of P and Zn Supply in Hydroponics
The F103 accession was selected for further detailed analysis because of its marked response to the nutritional treatments in the first experiment (i.e. treatment effects were observed for shoot P and Zn concentrations and root Zn concentrations, Figures 1D–F). The C6 accession was selected because its shoot Zn concentration, but not its shoot P concentration, was affected by the nutritional treatments in the first experiment (Figures 1C–F). In addition, these two accessions differed in their root architecture with the F103 accession having a larger number of lateral roots, which were also longer than in the C6 accession, and in their root exudation with the C6 accession releasing a larger quantity and number of polar compounds than the F103 accession (Pongrac et al., unpublished results).
Root length, DW and ionome (concentrations of P, Zn, Mg, S, Cl, K, Ca, Mn, Fe, Ni, Cu, and Na) of roots and shoots, and root gene expression were evaluated in the two selected accessions. Across all treatments, roots of B. oleracea accumulated less biomass (Figure 3) and had smaller concentrations of Mg, S, Cl, K, Ca, Mn, and Ni than shoots (Figures S3 and S4). The two-way ANOVA, considering the genotype and treatment as independent variables (Table 2), revealed that the genotype affected root and shoot DW, while the treatment and genotype × treatment interactions did not. In agreement with the first experiment, the C6 accession had, on average across all treatments, significantly larger roots and shoots than the F103 accession, but no treatment-specific differences were observed for root and shoot DW of the two accessions (Figures 3A, B). Only the treatment, but not the genotype or genotype × treatment interactions, affected root and shoot P concentrations and shoot Zn concentration (Table 2). An increase in P supply increased root P concentration in both accessions, while in shoots no significant effects of P and Zn supply were seen (Figures 3C, D). An increase in Zn supply increased root Zn concentrations, except in the C6 accession at Low P supply (Figure 3E). The largest root Zn concentration was found in the High P & High Zn treatment (Figure 3E) in both accessions. In shoots, Zn concentrations increased significantly with increasing Zn supply in both accessions at High P, but not at Low P (Figure 3F).
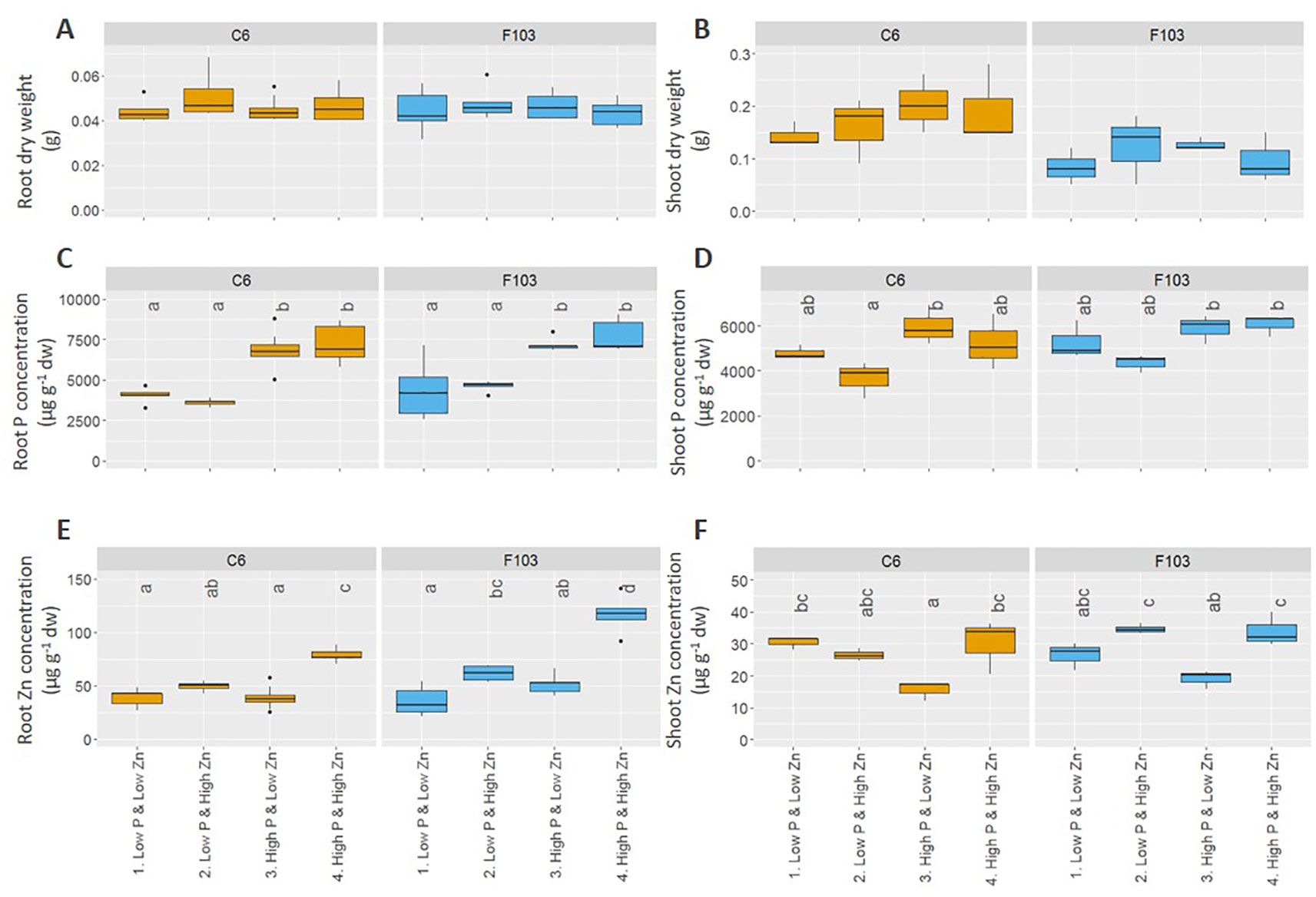
Figure 3 Effect of contrasting combinations of P and Zn supply on two B. oleracea accessions (C6 and F103). Plants were grown hydroponically for two weeks after which biomass (A, B) and P (C, D) and Zn (E, F) concentration were determined in roots (A, C, E) and shoots (B, D, F). Shown are Tukey boxplots representing 25th and 75th percentile of the data with the centerline representing the median and whiskers representing the 5th and 95th percentile (n = 5-9 for roots and n = 3 for shoots for each accession and each treatment). Significant differences between accessions and treatments are indicated by different letters above the box-plots (Two-way analysis of variance followed by Tukey post-hoc test at p < 0.05). dw, dry weight.
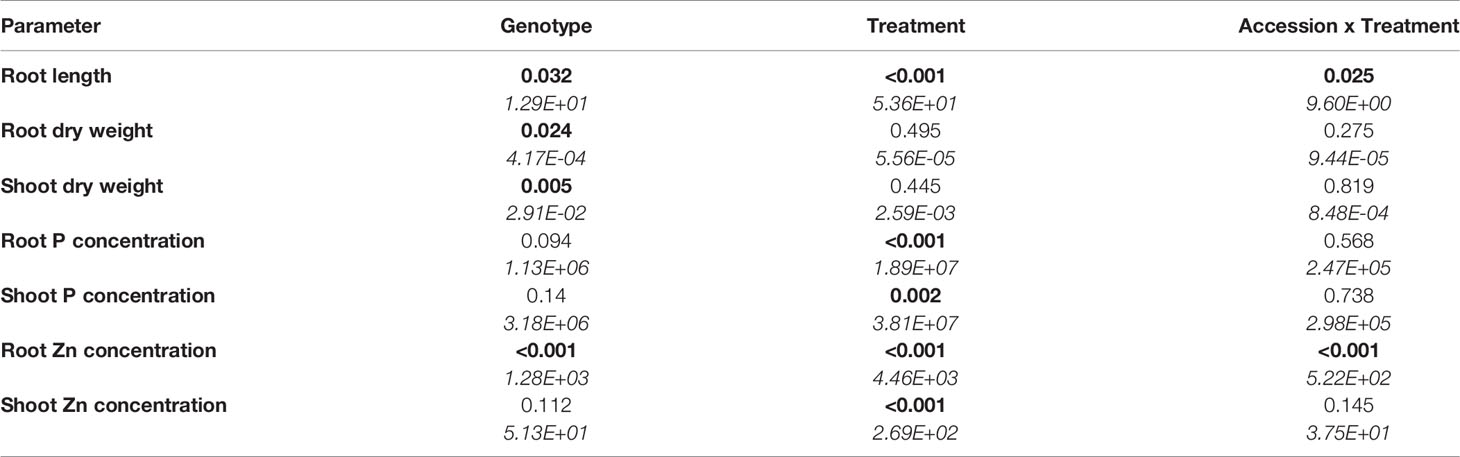
Table 2 Two-way analysis of variance table with p-values (those less than 5% are highlighted in bold) and means squares (in italics) for response variables in two B oleracea accessions (the C6 and the F103) grown in hydroponics at Low P and High P each with High Zn or Low Zn treatments for two weeks (n = 3 for each accession and each treatment).
A PCA was used to visualize the major sources of variation in root and shoot ionomes of the two accessions. Root and shoot samples separated clearly and a greater spread of data were observed in the root ionome than in the shoot ionome (Figure S5). A detailed PCA of the root ionome based on DW, P and Zn concentrations (Figures 3C–F), Mg, S, Cl, K and Ca concentrations (Figure S3) and Mn, Fe, Ni, Cu and Na concentrations (Figure S4) revealed that the first two PCs accounted for 61.4% of the variance (Figure 4A). In the PCA plot of the roots of individual plants a clear separation of the ionome was seen between Low P and High P treatments in the treatments with High Zn. By contrast, in the treatments with Low Zn the P supply did not lead to a clear effect on the root ionome. Variables with significant contributions to the first PC were root Fe, P, Ni, Mg, Cu and Ca concentrations (Figure 4B), while variables with significant contribution to the second PC were root K, S, Zn and Cu concentrations (Figure 4C). Root P and Fe concentrations were significantly smaller in plants receiving a Low P supply than those receiving a High P supply (Figures 3C and S4C).
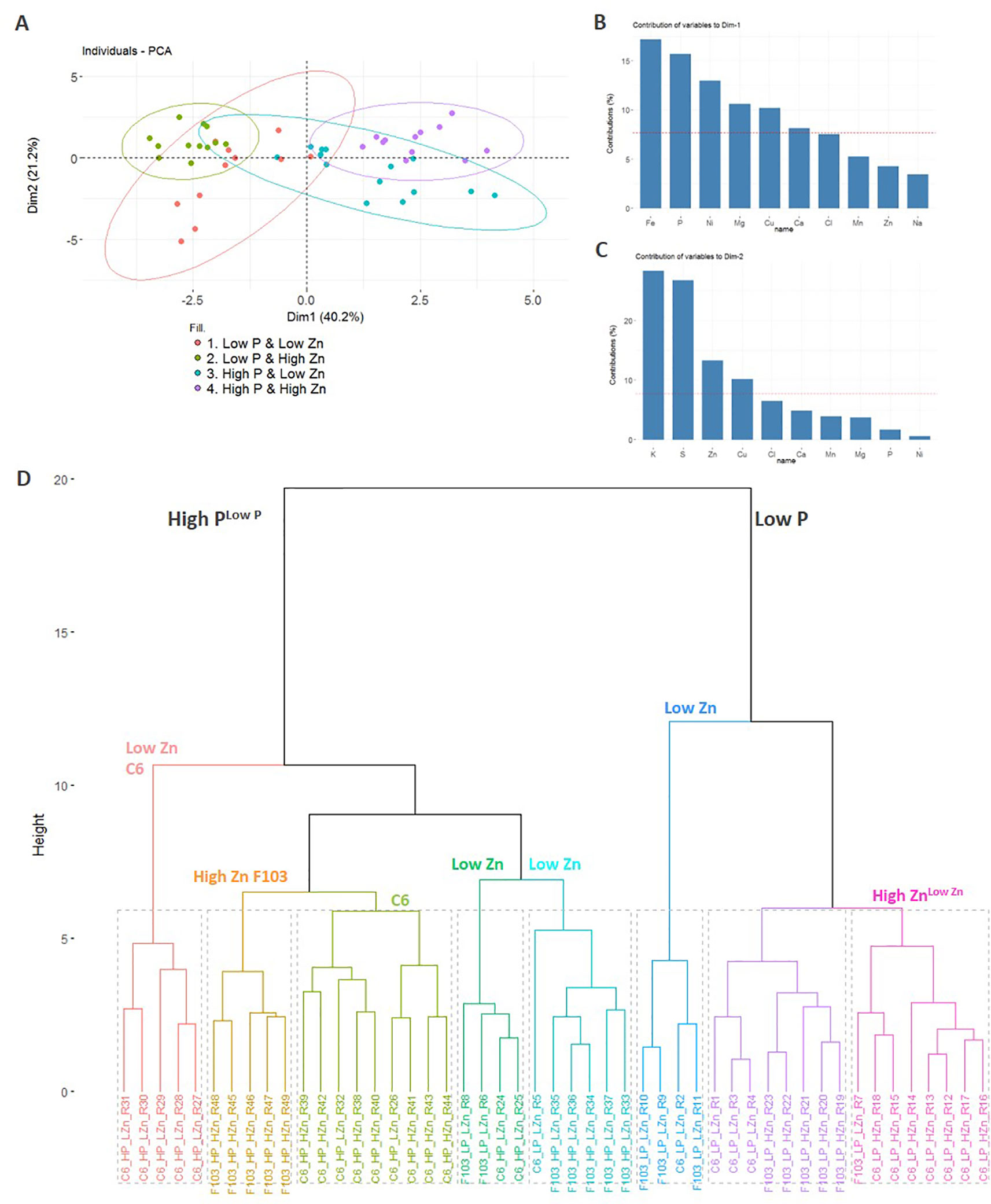
Figure 4 Principal component analysis (PCA) of the root ionome of two B. oleracea accessions (C6 and F103). The root dry weight and concentrations of Na, Mg, P, S, Cl, K, Ca, Mn, Fe, Cu, Zn and Ni in roots of the C6 and F103 accessions grown hydroponically for two weeks at different P and Zn supply were included in the PCA to detect major effects on variations observed. The PCA plot depicts the first two PCs (Dim1 and Dim2) for response variables grouped by treatments (A). The bar plots show the contribution of ten variables to PC1 (B) and PC2 (C). Cluster dendrogram of all replicates (n = 4-9 for each accession and each treatment; (D). In (B and C), the red dashed line at 7.7% indicates the value at which uniform contribution of all variables tested occurs.
Independently, in a hierarchical clustering analysis of the root ionome, samples separated clearly depending on the P supply (Figure 4D). Within the High P cluster, the majority of samples were grouped based on the Zn supply. Within the High P cluster, samples of the F103 accession from the High Zn treatment formed a group, as did those from the Low Zn treatment, indicating a genotypic effect. Notably, the P supply had a stronger influence on the clustering of samples than the Zn supply, which in turn had a greater effect on clustering than did the genotype, whose effects were not consistent. In order to link these observations and expand our understanding of P-Zn interactions in B. oleracea gene expression and regulatory networks in roots were studied in greater detail.
Gene Expression of the Brassica oleracea C6 and F103 Accessions Receiving Combinations of P and Zn Supply in Hydroponics
Roots of the C6 and the F103 accessions receiving four combinations of P and Zn supply for two weeks in hydroponics were analyzed via RNAseq in triplicates (all genes can be found in Table S2). Raw data of the RNAseq experiment can be downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) database, accession number GSE127467. Sequencing of 24 samples returned on average 23,569,302 clean reads per sample (72.2%) which were then mapped to the reference genome (ftp://ftp.ensemblgenomes.org/pub/plants/release-35/fasta/brassica_oleracea/dna/), while the average genome mapping ratio was 91.78%, showing that on average 91% of all reads could be mapped to the reference genome. On average the expression of 38,103 genes was detected.
Treatment Effects on the Patterns in Gene Expression in the Two B. oleracea Accessions
To understand the way by which contrasting combinations of P and Zn supply affect gene expression in B. oleracea, differentially expressed genes (DEGs) were identified. The expression of genes in roots of plants receiving Low P & High Zn, High P & Low Zn or High P & High Zn supply was compared to that in roots of plants receiving Low P & Low Zn (Figure 5A, comparison 1 through 3). All details regarding the comparisons are provided in Table S3. In general, a larger number of DEGs were found in the C6 accession than in the F103 accession (Figure 5B).
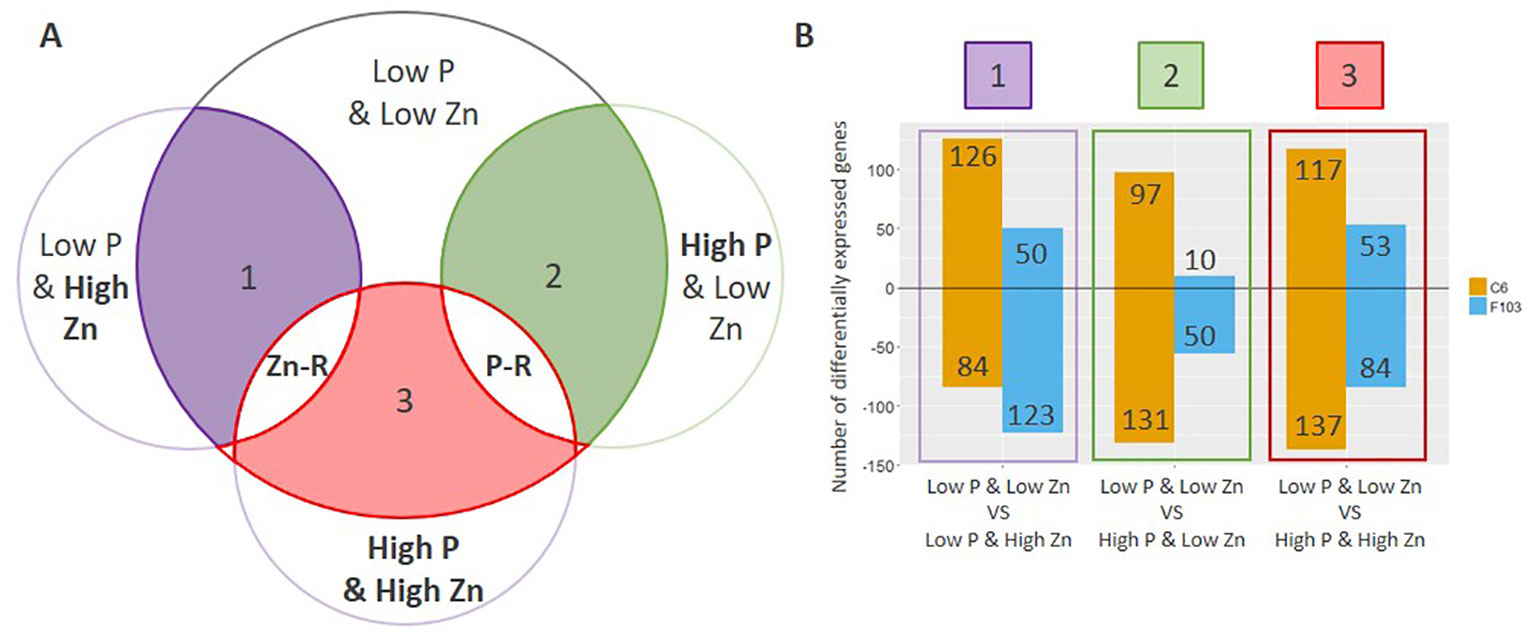
Figure 5 Comparative analysis of differentially expressed genes (DEGs) in roots of B. oleracea accessions C6 and F103 grown hydroponically for two weeks with contrasting combinations of P and Zn supply. Differentially expressed genes were defined by comparing the Low P & Low Zn treatment with the three other treatments as schematically represented in (A), where the contrasting supply (Low vs. High) is highlighted in bold. The number of DEGs (up-regulated are positive values and down-regulated are negative values) in each of these three comparisons for each accession is displayed in (B). Genes that were either consistently up-regulated or down-regulated in response to either Zn or P supply in both or only in one of the accessions were defined as Zn-responsive (Zn-R) and P-responsive (P-R).
Following the selection of DEGs in different treatments, P-responsive and Zn-responsive (P-R and Zn-R, respectively) genes were defined (Figure 5A). A gene was considered to be P-responsive if it responded to High P treatment (expression in the Low P & Low Zn treatment compared to expression in the High P & Low Zn treatment and compared to expression in the High P & High Zn treatment) with either an increase (up-regulated) or decrease (down-regulated) in expression. Likewise, Zn-responsive genes were defined as those responding to increased Zn supply (expression in the Low P & Low Zn treatment compared to expression in the Low P & High Zn treatment and compared to expression in High P & High Zn). Twenty-seven P-responsive genes were detected in the two accessions. Of these 26 were down-regulated and one was up-regulated. Among the down-regulated P-responsive genes was a putative Pi-transporter (Bo6g120500, orthologue of AT1G20860: Probable inorganic phosphate transporter 1-8), whose expression was 6 to 16-fold smaller in the High P treatments (Figure 6A). Another gene down regulated in both treatments with High P was a predicted AtHAK5 homologue (Figure 6B), a high-affinity K-transporter (White, 2013), whose expression changes were not reflected in changes in root or shoot K concentrations (Figures S3G, H). Also common to both accessions responding to increased Zn supply was the down-regulation of four Zn-responsive genes, one of which was a predicted Zn transporter from B. oleracea var. oleracea (Parkin et al., 2014), whose expression was repressed under High Zn (Figure 6C).
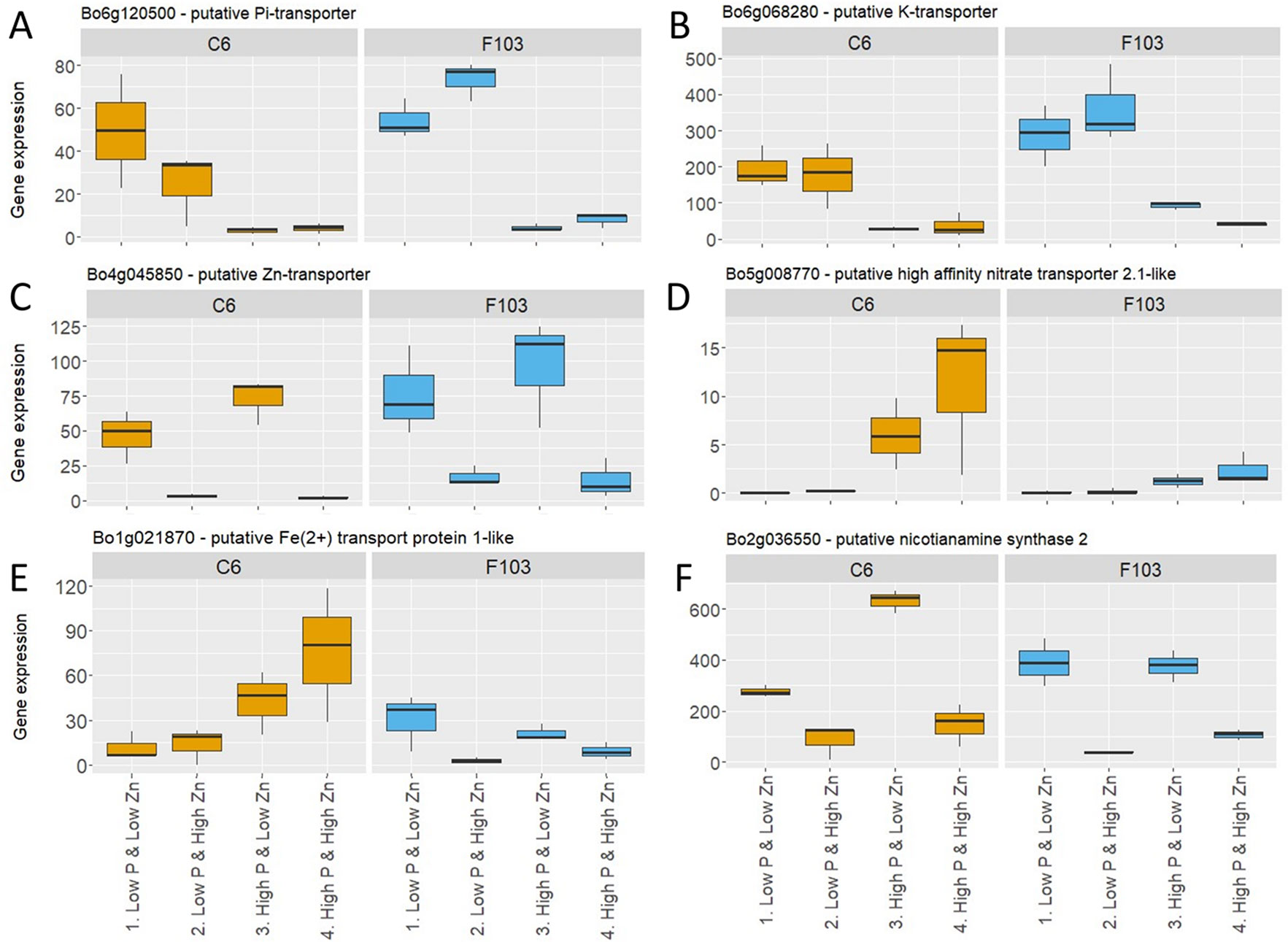
Figure 6 Expression of selected P-responsive and Zn-responsive genes in roots of B. oleracea accessions C6 and F103 grown hydroponically for two weeks with contrasting combinations of P and Zn supply. These genes were either P-responsive in both accessions: (A) Pi-transporter (Bo6g120500) and (B) K-transporter (Bo6g068280), Zn-responsive in both accessions: (C) putative Zn-transporter (Bo4g045850), P-responsive in the C6 accession: (D) predicted high-affinity nitrate transporter 2.1-like (Bo5g008770) and (E) putative iron2+ transport protein1-like (Bo5g008770), or Zn-responsive in the F103 accession: (F) predicted nicotianamine synthase 2 (Bo2g036550).
A larger number of P- and Zn-responsive genes were unique to only one accession. Ninety-two P-responsive genes, 55 of which were down-regulated and 37 up-regulated by increasing P supply, were found in the C6 accession, while in the F103 accession only six P-responsive genes were found, of which two were down-regulated and four were up-regulated by increasing P supply. An example of a P-responsive gene in the C6 accession encoded a predicted high-affinity nitrate transporter 2.1-like protein. This gene was induced at High P & Low Zn and at High P & High Zn when compared to Low P & Low Zn (Figure 6D). A second example of a P-responsive gene in the C6 accession only was a putative iron2+ transport protein1-like, whose expression was induced in treatments with High P supply compared to the treatments with Low P supply (Figure 6E). Thirty-four Zn-responsive genes were found in the C6 accession, of which seven were down-regulated and 27 were up-regulated by increased Zn supply, while in the F103 accession 28 Zn-responsive genes were found of which 21 were down regulated and seven were up-regulated by increased Zn supply. An example of a Zn-responsive gene in the F103 accession encoded a predicted nicotianamine synthase (NAS) 2, whose expression was reduced by increased Zn supply (Figure 6F).
Gene Ontology Analysis
To capture the physiological processes involved in responses to contrasting combinations of Zn and P supply, a GO term enrichment analysis was performed. For ease of discussion, GO term enrichments among DEGs were labeled as follows: responsive to Zn supply (comparisons 1 and 3) and responsive to P supply (comparisons 2 and 3) in either the C6 or the F103 accession. All GO terms with significant p-values are shown in Table S4. Thirty-nine significantly enriched GO term categories were found, which belonged to groups annotated to biological processes, molecular function or cellular compartment (Figure 7). In response to Zn supply there was a GO-term enrichment in “response to oxygen containing compound” in the C6 accession, while there was a GO-term enrichment in “cellular response to starvation,” “glycolipid biosynthetic process,” “cellular trivalent inorganic anion homeostasis,” “cation homeostasis” and “response to reactive oxygen species (ROS)” in the F103 accession. None of the enrichments were common to both accessions in response to Zn supply, while five processes (“cellular response to starvation,” “glycolipid biosynthetic process,” “cellular trivalent inorganic anion homeostasis,” “potassium symporter activity” and “phosphoric ester hydrolase activity”) were enriched in both accessions as a response to changing P supply (Figure 7), showing a conserved transcriptional response to vagaries in P supply. In addition, accession-specific GO enrichments were found. Only one process (“response to oxygen-containing compound”) was common to both Zn and P responses (significant enrichment in all three comparisons) in the C6 accession while four such processes (“cellular response to starvation,” “glycolipid biosynthetic process,” “cellular trivalent inorganic anion homeostasis” and “cation homeostasis”) were common to both Zn and P responses in the F103 accession.
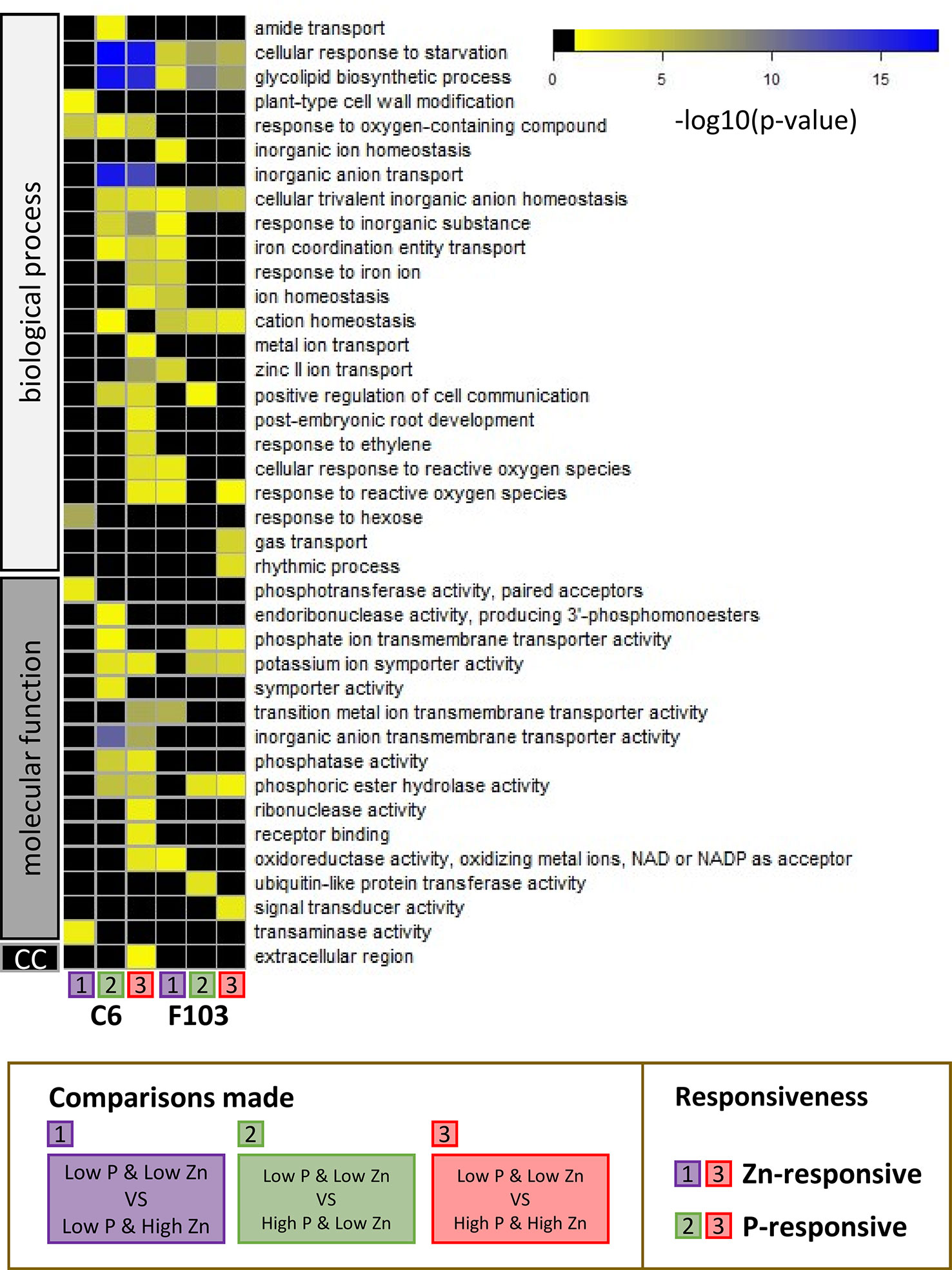
Figure 7 Gene ontology (GO) analysis of enriched functional categories in roots of B. oleracea accessions C6 and F103 grown hydroponically for two weeks with contrasting combinations of P and Zn supply. Differentially expressed genes in the C6 and the F103 accessions were defined between the treatments Low P & Low Zn with Low P & High Zn [1], with High P & Low Zn [2] and with High P & High Zn [3] for each accession. See Figure 5A for schematic representation of comparisons. Among these DEGs the GO-term enrichments were calculated and scored by Bonferroni corrected p-values as indicated by the color legend (in black: GO-term with p-values above 0.05, yellow to blue: increasing significance for GO-enrichment). Gene ontology term enrichments in the GO-groups: biological process, molecular function and cellular compartment (cc) were detected. Where child and ancestral GO-categories where enriched, only the most defined child categories were selected.
Discussion
Growth and Ionome Responses of B. oleracea Accession Receiving Combinations of P and Zn Supply in Hydroponics
To understand the molecular networks and genes involved in the early responses of B. oleracea to contrasting P and Zn supply, gene expression in roots of young plants was profiled in a hydroponic system. Two B. oleracea accessions, C6 and F103, which responded significantly to the contrasting P and Zn supply (Figure 1) and differ in their root architecture and profiles of root exudates (Pongrac et al., unpublished results) were selected from a set of eight accessions (Table S1) for further analysis. The hydroponic experiments were designed to capture early root responses at a molecular level, with minimal effects on plant growth and tissue ionomes. There were no interactions of treatments and genotypes for shoot biomass or shoot P concentration (Tables 1 and 2) and the shoot P concentration (Figures 1D and 3D) was above the shoot P concentration reported to be sufficient (2,000 mg P kg-1 DW) in B. oleracea (Campbell, 2000). Purple anthocyanin pigmentation, which is indicative of systemic P deficiency, was also absent (Figure S1; Jiang et al., 2007). Additionally, in plants grown in treatments with Low Zn supply, shoot Zn concentrations were at the lower limit of Zn sufficiency, which is reported to be 20 mg Zn kg-1 DW for B. oleracea (Campbell, 2000), but were never significantly below this value (Figures 1F and 3F). Two explanations for the lack of deficiency symptoms observed in these young Brassica plants can be offered. The first is that seed reserves, which may provide sufficient P supply from one to four weeks (White and Veneklaas, 2012), could have provided adequate nutrition of plants. A delay in reducing growth in young plants challenged with P and Zn deficiencies has been reported previously for barley (Huang et al., 2000). The second explanation is that differences at the molecular level in roots of plants grown in different treatments, enable the activation of regulatory networks compensating for insufficient P or Zn supply. Large differences in gene expression were observed in roots of both B. oleracea accessions in response to contrasting P and Zn supply in the hydroponic system (Figures 5–7). Thus, studying young plants receiving sub-optimal P and Zn supply that do not exhibit visual deficiency symptoms allowed us to capture key compensatory mechanisms enabling optimal growth of these plants at the molecular level.
Plants in the hydroponic system grew well (Figure S1) and had similar shoot P concentrations (Figure 1D) as B. oleracea plants grown on compost (Hammond et al., 2009; Broadley et al., 2010; Table S1). By contrast, shoot Zn concentrations (Figure 1F) were less when grown in the hydroponic system, even with an adequate Zn supply, than shoot Zn concentrations observed when these accessions grown in compost (Hammond et al., 2009; Broadley et al., 2010; Table S1). These results suggest that shoot P concentration is less variable than shoot Zn concentration across growth environments. Similarly, shoot P concentration in cabbage (B. oleracea var. capitata) showed 3-fold variation, while shoot Zn concentration varied 15-fold, in plants grown on three unamended soil types (Pongrac et al., 2019). Such differences in variation between concentrations of macro- and micronutrients were also reported for A. thaliana (Berezin et al., 2012). Noteworthy was a negative correlation between shoot P and Zn concentrations with shoot dry weight, but a positive correlation between shoot P concentration and shoot Zn concentration, among the B. oleracea accessions grown in the hydroponic system (Figure 2). In line with similar observations in B. oleracea grown in compost (Broadley et al., 2010) and in cereals (Murphy et al., 2008; Zhao et al., 2009), these observations reflect the delicate balance between yield and mineral element composition that must be considered carefully in breeding and biofortification programs. Overall, these results indicate the suitability of the hydroponic system for experiments to study gene expression in roots and identify early molecular changes in response to contrasting nutrient supply.
The root system is the plant organ that first responds to changes in the rhizosphere. Therefore, it was not surprising to observe larger variation in the root than in the shoot ionome (Figure S5) and a larger number of significant differences between treatments in the concentrations of elements in roots than in shoots (Figures 3, S3, and S4). Most prominent was the greater root Fe concentrations in treatments receiving High P supply than those receiving Low P supply (Figure S4C), while shoot Fe concentrations were hardly affected (Figure S4D). This highlights an interesting connection between P and Fe homeostasis that has also been observed in other plant species. In roots of strawberry (Fragaria × ananassa (Duchesne ex Weston) Duchesne ex Rozier), for example, Fe concentration was strongly reduced upon P starvation (Valentinuzzi et al., 2015) as was the Fe concentration in rice (Oryza sativa) roots (Saenchai et al., 2016) and maize (Zea mais) roots (Nichols et al., 2012). By contrast, roots of P-starved A. thaliana plants had significantly larger Fe concentrations than did roots of P-replete plants in two different studies (Misson et al., 2005; Baxter et al., 2008; Raghothama et al., 2008), as did roots of bush bean (Phaseolus vulgaris L.; Wallace et al., 1974) and rice seedlings (Zheng et al., 2009). No effect of P supply was observed on Fe concentrations in roots of maize (Prabhakaran Nair and Rabu, 1975) or poplar (Kavka and Polle, 2017). These contradicting observations preclude consensus on the interactions between P deficiency and Fe nutrition, but they do indicate crosstalk between Fe and P nutrition which merits further investigation (Briat et al., 2015; Bouain et al., 2019), especially since Fe deficiency in humans is of great concern and Fe fortified food would benefit many of the world’s population (White and Broadley, 2009).
In order to evaluate the source of the variation in the root ionome two independent techniques were used: PCA and cluster analysis. They both showed (Figure 4) that P supply had a larger effect on the root ionome than did Zn supply, possibly reflecting a greater importance of P for metabolic processes and plant growth than Zn. Even though the B. oleracea accessions studied in the hydroponic system did not show different growth rates in response to contrasting P supply, these would be expected subsequently (Dechassa et al., 2003). Additionally, the PCA analysis revealed root Fe concentration as the strongest contributor (explaining 40.2% of the variation in the root ionome) to PC1, which further stresses the connection between Fe and P homeostasis in plants.
Gene Expression in Roots of B. oleracea Accessions Receiving Different P and Zn Supplies
Significant differences in gene expression in roots of the two B. oleracea accessions were observed depending upon P and Zn supply, as reflected by the large number of DEGs (Figure 5B). A greater number of DEGs responding to P and Zn supply were observed in the C6 accession than in the F103 accession, which might be linked with the observation that C6 releases a larger number and greater amounts of root exudates than the F103 accession (Pongrac et al., unpublished results). To uncover DEGs responding most significantly P and Zn supply, P-responsive and Zn-responsive genes were defined (Figure 5A). Of the 27 P-responsive genes detected in the two accessions, 26 were down-regulated and one was up-regulated. These genes represent the constitutive response of the two B. oleracea accessions to P supply. Orthologues of fourteen B. oleracea P-responsive genes found in our study could be found within a set of 95 core PSI genes in A. thaliana (Lan et al., 2015), illustrating a common genetic response to Low P across different species. Among these fourteen genes were two genes encoding Pi transporters Bo6g120500 and Bo6g120510, orthologues of At1g76430 or PHT1;9 (Pi transporter 1;9), and Bo3g134470, an orthologue of At3g47420 or G3Pp1 (Pi starvation-induced gene 3). PHT1;9, together with PHT1;8, plays an important role in Pi acquisition during P-starvation (Remy et al., 2012) and G3Pp1 is a member of a gene family of Glycerol-3-phosphate permeases, likely to be involved in the mobilization of G3P (Glycerol -3- phosphate) during P-starvation (Misson et al., 2005). Members of the latter gene family are induced greatly upon P starvation and A. thaliana mutants in G3Pp1 show longer lateral roots than wild type indicative of a P-starvation phenotype even under sufficient P supply (Ramaiah et al., 2011).
Among the down-regulated P-responsive genes was a putative Pi-transporter (Bo6g120500, orthologue of AT1G20860: Probable inorganic phosphate transporter 1-8), whose repressed expression in the High P treatments (Figure 6A) suggested that this was a high affinity P transporter required for Pi uptake at low external Pi concentrations. These have been shown to be repressed by an increase in P supply in A. thaliana (Mudge et al., 2002; Bayle et al., 2011; Remy et al., 2012) and also to be induced by Zn deficiency in barley (Hordeum vulgare L.; Huang et al., 2000).
Both accessions responded to increased Zn supply by down-regulation of four Zn-responsive genes, one of which (Bo4g045850) was a predicted Zn transporter from B. oleracea var. oleracea, whose expression was repressed under High Zn (Figure 6C). Bo4g045850 is an orthologue AtZIP3 (Zn transporter precursor), a Zn transporter which is induced by Zn deficiency in A. thaliana (Inaba et al., 2015) but requires further investigation to uncover its mode of action. Other members of the ZIP family have been studied in A. thaliana and their regulation under Zn deficiency has been demonstrated. ZIP9 and ZIP12 for example play a role in Zn uptake with mutants lacking these transporters showing reduced Zn concentrations (Inaba et al., 2015). This Zn transporter (Bo4g045850) might provide a molecular target for breeding cultivars with larger tissue Zn concentrations, however detailed understanding of its regulation will be needed to ensure its optimal expression for significant increase in Zn uptake and larger shoot Zn concentrations.
Another P-responsive gene in roots of the C6 accession was Bo5g008770, a putative high affinity nitrate transporter and orthologue to AtNRT2.1 (nitrate transporter 2.1). In A. thaliana, AtNRT2.1 is induced by N starvation and repressed by treatment with ethylene (Zheng et al., 2013). It has also been implicated in lateral root initiation, which results in more root branching when N supply limits plant growth (Zheng et al., 2013). It was reported that P deficiency can induce ethylene production (Borch et al., 1999), which may explain the observed induction of Bo5g008770 by increased P supply in the C6 accession (Figure 6D). It is likely that increasing P supply reduces ethylene production which then allows the induction of Bo5g008770 expression. Thus, the uptake of P and N might be balanced to provide an optimal N:P ratio in the plant (Raven, 2015). Another example of a P-responsive gene in roots of the C6 accession is Bo1g021870, which encodes a solute carrier family 39 (zinc transporter) protein (Figure 6E). The A. thaliana orthologue is AtIRT1 (Iron-Regulated Transporter 1) which is induced by Fe deficiency and in the presence of Zn, is able to affect the accumulation of different divalent cations such as Fe2+, Zn2+, Mn2+, Cd2+ and Cu2+ and acts together with IRT2 to maintain Fe homeostasis (Vert et al., 2009; Shanmugam et al, 2011). Since Bo1g021870 was up regulated in roots of accession C6, but not in roots of accession F103, in the High P treatment, this might explain in part the large root Fe concentration of the C6 accession under these conditions (Figure S4C).
A Zn-responsive gene unique to the F103 accession was Bo2g036550: a predicted nicotianamine synthase and orthologue to AtNAS2, whose expression was reduced by increased Zn supply (Figure 6F). It could be linked to the role of nicotianamine in transporting Zn within the plant (White and Broadley, 2011; Deinlein et al., 2012) or in solubilizing Zn in the soil through the synthesis and release of phytosiderophores (Clemens et al., 2013). Just like its A. thaliana counterpart, BoNAS2 could have played a role in increasing shoot Zn concentration in plants from the Low Zn treatments when its expression was high. An interaction between plant P, Zn and Fe nutrition has been proposed to act via PHR1 (Phosphate Starvation Response 1) and PHL1 (PHR LIKE 1) associations with the promoter of Fer1 (Ferritin 1), which allow the expression of Fer1 to be regulated in an Fe-independent manner (Briat et al., 2015). The smaller root Fe concentration in plants from the Low P treatment compared to those from the High P treatment (Figure S4C) might, therefore, be linked to changes in the expression of genes connected with Fe homeostasis, such as BoNAS2 (Bo2g036550). Overexpression of NAS genes has been associated with Fe-deficiency tolerance in several plant species (Nozoye, 2018). NAS1, was one of only two genes consistently downregulated in response to Pi deficiency in A. thaliana according to four studies analyzing expression in roots or whole seedlings (Lan et al., 2015).
Gene Ontology Analysis
The greater number of the P-responsive genes in roots of the C6 accession than in roots of the F103 accession suggest that these two accessions might employ different genetic strategies when challenged by an insufficient P supply. The GO enrichment analysis highlighted these differences (Figure 7). For example, it revealed an enrichment in “inorganic anion transmembrane transporter activity” in the C6 accession but not in the F103 accession. This could indicate a greater flexibility in ion transport capabilities of the C6 accession. Interestingly, it was observed that GO-terms are less conserved among Zn-responsive genes and depend much more on genotype and P supply (Figure 7). This might suggest that the response to Zn supply is less conserved and more varied than the response to P supply. Notably, “Zinc II ion transport” processes were enriched among genes whose expression was changed by Zn supply in the C6 accession only at High P, while in the F103 accession this occurred at Low P supply.
Other important pathways were the “cellular response to starvation” and “glycolipid biosynthetic process” which were enriched in both accessions under almost all conditions assayed, highlighting the robust changes in response to contrasting combinations of P or Zn supply (Figure 7). As has been reported previously, a lack of P leads to remodelling of lipids, which is enabled by changes in gene expression (Nakamura, 2013; Tawaraya et al., 2014; Siebers et al., 2015), particularly through PHR1 in A. thaliana (Pant et al., 2015). Phosphate deficiency results in alterations in the membrane lipid composition of plants, because phospholipids are replaced with non-phosphorus glycolipids and sulfolipids (Li et al., 2006). Lipids play crucial roles in the ability of plants to survive and acclimatize to environmental changes. The lipid response during Pi deprivation differs between leaves and roots, as leaves contain large amounts of galactolipids (Siebers et al., 2015). Membrane phospholipids constitute about 20% of the total amount of P in the leaves of Pi-replete plants, and thus represent a large pool of P that can be mobilized when internal Pi levels are insufficient to match the demand of the plant (Veneklaas et al., 2012). In many plants, as exemplified by poplar, changes in P availability affect the expression of genes belonging to the functional category “galactolipid synthesis” (Kavka and Polle, 2017).
Taking a closer look at GO category “cellular response to reactive oxygen species,” which was enriched in the P-responsive genes of the F103 accession and also among genes responsive to a combination of P and Zn supply in the C6 accession, suggests that the sensing of P-withdrawal might occur through the formation of ROS, in agreement with previous reports (Ham et al., 2018). Interplay between signaling networks for P deficiency and Zn deficiency through ROS might enable the C6 accession to integrate both signals and ensure an appropriate response to precise environmental conditions.
Taking into consideration that all analysis performed was limited by the quality of the gene annotation of the B. oleracea genome, the value of the dataset is likely to increase in the future when more information become available.
Summary
Early responses to insufficient P and Zn supply were assessed by analyzing gene expression in roots of young B. oleracea plants, not yet exhibiting visible deficiency symptoms. A number of Zn and P-responsive genes were detected. Their expression changes were related to acclimation to low-nutrient environments and contributed to maintaining balanced nutrition. The gene expression changes agreed with previous studies demonstrating alterations in Pi transport and phospholipid metabolism in response to reduced Pi supply. It is anticipated that improved knowledge of genes responsive to P or Zn supply will help illuminate their roles in uptake and accumulation of P and Zn and might identify candidate genes for breeding high-yield-high-Zn brassicas.
Data Availability Statement
The datasets generated for this study can be found in Gene Expression Omnibus, GSE127467.
Author Contributions
PP and PW conceived and designed the research. PP, JT, GW, and SF conducted experiments. PP and SF analyzed data. PP, SF, and PW wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Rural and Environment Science and Analytical Services Division of the Scottish Government, an EU Marie Curie Intra-European Fellowship (REA grant agreement n°623305) to PP who also acknowledges financial support from the Slovenian Research Agency (P1-0212 and P1-0112 programs and N7-077 project), and a DFG Postdoctoral Fellowship to SF (FI 2152/1-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank David E. Salt at the University of Nottingham for his mentorship of SF.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01645/full#supplementary-material
References
Assunção, A. G. L., Schat, H., Aarts, M. G. M. (2010). Regulation of the adaptation to zinc deficiency in plants. Plant Signal. Behav. 5, 1553–1555. doi: 10.4161/psb.5.12.13469
Barben, S. A., Hopkins, B. G., Jolley, V. D., Webb, B. L., Nichols, B. A. (2010). Phosphorus and zinc interactions in chelator-buffered solution grown russet burbank potato. J. Plant Nutr. 33, 587–601. doi: 10.1080/01904160903506308
Baxter, I. R., Vitek, O., Lahner, B., Muthukumar, B., Borghi, M., Morrissey, J., et al. (2008). The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc. Natl. Acad. Sci. 105, 12081–12086. doi: 10.1073/pnas.0804175105
Bayle, V., Arrighi, J.-F., Creff, A., Nespoulous, C., Vialaret, J., Rossignol, M., et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23, 1523–1535. doi: 10.1105/tpc.110.081067
Berezin, I., Elazar, M., Gaash, R., Avramov-Mor, M., Shaul, O. (2012). “The use of hydroponic growth systems to study the root and shoot ionome of Arabidopsis thaliana,” in Hydroponics - A Standard Methodology for Plant Biological Researches. Ed. Asao, T. (Rijeka: InTech), 135–152. doi: 10.5772/36558
Borch, K., Bouma, T. J., Lynch, J. P., Brown, K. M. (1999). Ethylene: a regulator of root architectural responses to soil phosphorus availability. Plant Cell Environ. 22, 425–431. doi: 10.1046/j.1365-3040.1999.00405.x
Bouain, N., Kisko, M., Rouached, A., Dauzat, M., Lacombe, B., Belgaroui, N., et al. (2014). Phosphate/zinc interaction analysis in two lettuce varieties reveals contrasting effects on biomass, photosynthesis, and dynamics of Pi transport. BioMed. Res. Int. 2014, 1–9. doi: 10.1155/2014/548254
Bouain, N., Krouk, G., Lacombe, B., Rouached, H. (2019). Getting to the root of plant mineral nutrition: combinatorial nutrient stresses reveal emergent properties. Trends Plant Sci. 24, 542–552. doi: 10.1016/j.tplants.2019.03.008
Bouis, H. E., Saltzman, A. (2017). Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Sec. 12, 49–58. doi: 10.1016/j.gfs.2017.01.009
Briat, J.-F., Rouached, H., Tissot, N., Gaymard, F., Dubos, C. (2015). Integration of P, S, Fe, and Zn nutrition signals in Arabidopsis thaliana: potential involvement of phosphate starvation response 1 (PHR1). Front. Plant Sci. 6, 290. doi: 10.3389/fpls.2015.00290
Broadley, M. R., White, P. J., Hammond, J. P., Zelko, I., Lux, A. (2007). Zinc in plants. New Phytol. 173, 677–702. doi: 10.1111/j.1469-8137.2007.01996.x
Broadley, M. R., Hammond, J. P., King, G. J., Astley, D., Bowen, H. C., Meacham, M. C., et al. (2008). Shoot calcium and magnesium concentrations differ between subtaxa, are highly heritable, and associate with potentially pleiotropic loci in Brassica oleracea. Plant Physiol. 146, 1707–1720. doi: 10.1104/pp.107.114645
Broadley, M., Lochlainn, S., Hammond, J., Bowen, H., Cakmak, I., Eker, S., et al. (2010). Shoot zinc (Zn) concentration varies widely within Brassica oleracea L. and is affected by soil Zn and phosphorus (P) levels. J. Hortic. Sci. Biotechnol. 85, 375–380. doi: 10.1080/14620316.2010.11512683
Cakmak, I., Kutman, U. B. (2018). Agronomic biofortification of cereals with zinc: a review. Eur. J. Soil Sci. 69, 172–180. doi: 10.1111/ejss.12437
Cakmak, I., Marschner, H. (1986). Mechanism of phosphorus-induced zinc deficiency in cotton. I. zinc deficiency-enhanced uptake rate of phosphorus. Physiol. Plant 68, 483–490. doi: 10.1111/j.1399-3054.1986.tb03386.x
Cakmak, I. (2008). Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 302, 1–17. doi: 10.1007/s11104-007-9466-3
Campbell, C. R. (2000). “Reference sufficiency ranges for plant analysis in the southern region of the United States,” in South. Coop. Ser. Bull. North Carolina Digital Collections, State Library of North Carolina. 394, 122.
Clemens, S., Deinlein, U., Ahmadi, H., Höreth, S., Uraguchi, S. (2013). Nicotianamine is a major player in plant Zn homeostasis. BioMetals 26, 623–632. doi: 10.1007/s10534-013-9643-1
Dechassa, N., Schenk, M. K., Claassen, N., Steingrobe, B. (2003). Phosphorous efficiency of cabbage, carrot and potato. Plant Soil 250, 215–224. doi: 10.1023/A:1022804112388
Deinlein, U., Weber, M., Schmidt, H., Rensch, S., Trampczynska, A., Hansen, T. H., et al. (2012). Elevated nicotianamine levels in Arabidopsis halleri roots play a key role in zinc hyperaccumulation. Plant Cell 24, 708–723. doi: 10.1105/tpc.111.095000
Haldar, M., Mandal, L. N. (1981). Effect of phosphorus and zinc on the growth and phosphorus, zinc, copper, iron and manganese nutrition of rice. Plant Soil 59, 415–425. doi: 10.1007/BF02184546
Ham, B.-K., Chen, J., Yan, Y., Lucas, W. J. (2018). Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49, 1–9. doi: 10.1016/j.copbio.2017.07.005
Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C., May, S. T., et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132, 578–596. doi: 10.1104/pp.103.020941
Hammond, J. P., Broadley, M. R., White, P. J., King, G. J., Bowen, H. C., Hayden, R., et al. (2009). Shoot yield drives phosphorus use efficiency in Brassica oleracea and correlates with root architecture traits. J. Exp. Bot. 60, 1953–1968. doi: 10.1093/jxb/erp083
Hammond, J. P., Mayes, S., Bowen, H. C., Graham, N. S., Hayden, R. M., Love, C. G., et al. (2011). Regulatory hotspots are associated with plant gene expression under varying soil phosphorus supply in Brassica rapa. Plant Physiol. 156, 1230–1241. doi: 10.1104/pp.111.175612
Huang, C., Barker, S. J., Langridge, P., Smith, F. W., Graham, R. D. (2000). Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiol. 124, 415–422. doi: 10.1104/pp.124.1.415
Hunter, P., Teakle, G., Bending, G. (2014). Root traits and microbial community interactions in relation to phosphorus availability and acquisition, with particular reference to Brassica. Front. Plant Sci. 5, 27. doi: 10.3389/fpls.2014.00027
Inaba, S., Kurata, R., Kobayashi, M., Yamagishi, Y., Mori, I., Ogata, Y., et al. (2015). Identification of putative target genes of bZIP19, a transcription factor essential for Arabidopsis adaptation to Zn deficiency in roots. Plant J. 84, 323–334. doi: 10.1111/tpj.12996
Jiang, C., Gao, X., Liao, L., Harberd, N. P., Fu, X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145, 1460–1470. doi: 10.1104/pp.107.103788
Kavka, M., Polle, A. (2017). Dissecting nutrient-related co-expression networks in phosphate starved poplars. PloS One 12, e0171958–e0171958. doi: 10.1371/journal.pone.0171958
Khan, G. A., Bouraine, S., Wege, S., Li, Y., de Carbonnel, M., Berthomieu, P., et al. (2014). Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 65, 871–884. doi: 10.1093/jxb/ert444
Kisko, M., Bouain, N., Safi, A., Medici, A., Akkers, R. C., Secco, D., et al. (2018). LPCAT1 controls phosphate homeostasis in a zinc-dependent manner. eLife 7, e32077. doi: 10.7554/eLife.32077
Lan, P., Li, W., Schmidt, W. (2012). Complementary proteome and transcriptome profiling in phosphate-deficient Arabidopsis roots reveals multiple levels of gene regulation. Mol. Cell. Proteomics 11, 1156–1166. doi: 10.1074/mcp.M112.020461
Lan, P., Li, W., Schmidt, W. (2015). “‘Omics’ approaches towards understanding plant phosphorus acquisition and use,” in Annual Plant Reviews Volume 48 (Hoboken, NJ, USA: John Wiley & Sons, Inc.), 65–97. doi: 10.1002/9781118958841.ch3
Li, H.-Y., Zhu, Y.-G., Smith, S. E., Smith, F. A. (2003). Phosphorus–zinc interactions in two barley cultivars differing in phosphorus and zinc efficiencies. J. Plant Nutr. 26, 1085–1099. doi: 10.1081/PLN-120020077
Li, M., Welti, R., Wang, X. (2006). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. roles of phospholipases d 1 and d 2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761. doi: 10.1104/pp.106.085647
Marschner, H., Cakmak, I. (1986). Mechanism of phosphorus-induced zinc deficiency in cotton. II. Evidence for impaired shoot control of phosphorus uptake and translocation under zinc deficiency. Physiol. Plant 68, 491–496. doi: 10.1111/j.1399-3054.1986.tb03387.x
Misson, J., Raghothama, K. G., Jain, A., Jouhet, J., Block, M. A., Bligny, R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. 102, 11934–11939. doi: 10.1073/pnas.0505266102
Morcuende, R., Bari, R., Gibon, Y., Zheng, W., Pant, B. D., Blasing, O., et al. (2007). Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 30, 85–112. doi: 10.1111/j.1365-3040.2006.01608.x
Mudge, S. R., Rae, A. L., Diatloff, E., Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Murphy, K. M., Reeves, P. G., Jones, S. S. (2008). Relationship between yield and mineral nutrient concentrations in historical and modern spring wheat cultivars. Euphytica 163, 381–390. doi: 10.1007/s10681-008-9681-x
Nakamura, Y. (2013). Phosphate starvation and membrane lipid remodeling in seed plants. Prog. Lipid Res. 52, 43–50. doi: 10.1016/j.plipres.2012.07.002
Nguyen, N. T., McInturf, S. A., Mendoza-Cózatl, D. G. (2016). Hydroponics: a versatile system to study nutrient allocation and plant responses to nutrient availability and exposure to toxic elements. J. Vis. Exp. 113, 54317. doi: 10.3791/54317
Nichols, B. A., Hopkins, B. G., Jolley, V. D., Webb, B. L., Greenwood, B. G., Buck, J. R. (2012). Phosphorus and zinc interactions and their relationships with other nutrients in maize grown in chelator-buffered nutrient solution. J. Plant Nutr. 35, 123–141. doi: 10.1080/01904167.2012.631672
Nozoye, T. (2018). The nicotianamine synthase gene is a useful candidate for improving the nutritional qualities and Fe-deficiency tolerance of various crops. Front. Plant Sci. 9, 340. doi: 10.3389/fpls.2018.00340
Pant, B. D., Burgos, A., Pant, P., Cuadros-Inostroza, A., Willmitzer, L., Scheible, W.-R. (2015). The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 66, 1907–1918. doi: 10.1093/jxb/eru535
Parkin, I. A. P., Koh, C., Tang, H., Robinson, S. J., Kagale, S., Clarke, W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 15, R77. doi: 10.1186/gb-2014-15-6-r77
Pedas, P., Husted, S., Skytte, K., Schjoerring, J. K. (2011). Elevated phosphorus impedes manganese acquisition by barley plants. Front. Plant Sci. 2, 37. doi: 10.3389/fpls.2011.00037
Pongrac, P., McNicol, J. W., Lilly, A., Thompson, J. A., Wright, G., Hillier, S., et al. (2019). Mineral element composition of cabbage as affected by soil type and phosphorus and zinc fertilisation. Plant Soil 434, 151–165. doi: 10.1007/s11104-018-3628-3
Prabhakaran Nair, K. P., Rabu, G. R. (1975). Zinc–phosphorus–iron interaction studies in maize. Plant Soil 42, 517–536. doi: 10.1007/BF00009940
Raghothama, K. G., Salt, D. E., Ward, J. T., Lahner, B., Yakubova, E. (2008). The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147, 1181–1191. doi: 10.1104/pp.108.118562
Ramaiah, M., Jain, A., Baldwin, J. C., Karthikeyan, A. S., Raghothama, K. G. (2011). Characterization of the phosphate starvation-induced glycerol-3-phosphate permease gene family in Arabidopsis. Plant Physiol. 157, 279–291. doi: 10.1104/pp.111.178541
Raven, J. A. (2015). “Interactions between Nitrogen and Phosphorus metabolism,” in Annual Plant Reviews, (Hoboken, NJ, USA: John Wiley & Sons, Inc.) 48, 187–214. doi: 10.1002/9781118958841.ch7
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sá-Correia, I., Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Rose, T. J., Impa, S. M., Rose, M. T., Pariasca-Tanaka, J., Mori, A., Heuer, S., et al. (2013). Enhancing phosphorus and zinc acquisition efficiency in rice: a critical review of root traits and their potential utility in rice breeding. Ann. Bot. 112, 331–345. doi: 10.1093/aob/mcs217
Saenchai, C., Bouain, N., Kisko, M., Prom-u-thai, C., Doumas, P., Rouached, H. (2016). The involvement of OsPHO1;1 in the regulation of iron transport through integration of phosphate and zinc deficiency signaling. Front. Plant Sci. 7, 1–9. doi: 10.3389/fpls.2016.00396
Santos, E. F., Pongrac, P., Reis, A. R., White, P. J., Lavres, J. (2018). Phosphorus-zinc interactions in cotton: consequences for biomass production and nutrient-use efficiency in photosynthesis. Physiol. Plant 166, 996–1007. doi: 10.1111/ppl.12867
Secco, D., Jabnoune, M., Walker, H., Shou, H., Wu, P., Poirier, Y., et al. (2013). Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25, 4285–4304. doi: 10.1105/tpc.113.117325
Shanmugam, V., Lo, J. C., Wu, C. L., Wang, S. L., Lai, C. C., Connolly, E. L., et al. (2011). Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana - the role in zinc tolerance. New Phytol. 190, 125–137. doi: 10.1111/j.1469-8137.2010.03606.x
Siebers, M., Dörmann, P., Hölzl, G. (2015). “Membrane remodelling in phosphorus-deficient plants,” in Annual Plant Reviews, vol. 48. (Hoboken, NJ, USA: John Wiley & Sons, Inc.), 237–263. doi: 10.1002/9781118958841.ch9
Stukenholtz, D. D., Olsen, R. J., Gogan, G., Olson, R. A. (1966). On the mechanism of phosphorus-zinc interaction in corn nutrition. Soil Sci. Soc. Am. J. 30, 759–763. doi: 10.2136/sssaj1966.03615995003000060029x
Sun, Y., Mu, C., Chen, Y., Kong, X., Xu, Y., Zheng, H., et al. (2016). Comparative transcript profiling of maize inbreds in response to long-term phosphorus deficiency stress. Plant Physiol. Biochem. 109, 467–481. doi: 10.1016/j.plaphy.2016.10.017
Tarazona, S., García-Alcalde, F., Dopazo, J., Ferrer, A., Conesa, A. (2011). Differential expression in RNA-seq: a matter of depth. Genome Res. 21, 2213–2223. doi: 10.1101/gr.124321.111
Tawaraya, K., Horie, R., Saito, S., Wagatsuma, T., Saito, K., Oikawa, A. (2014). Metabolite profiling of root exudates of common bean under phosphorus deficiency. Metabolites 4, 599–611. doi: 10.3390/metabo4030599
Tawaraya, K., Honda, S., Cheng, W., Chuba, M., Okazaki, Y., Saito, K., et al. (2018). Ancient rice cultivar extensively replaces phospholipids with non-phosphorus glycolipid under phosphorus deficiency. Physiol. Plant 163, 297–305. doi: 10.1111/ppl.12699
Thomson, C. A., Newton, T. R., Graver, E. J., Jackson, K. A., Reid, P. M., Hartz, V. L., et al. (2007). Cruciferous vegetable intake questionnaire improves cruciferous vegetable intake estimates. J. Am. Diet. Assoc. 107, 631–643. doi: 10.1016/j.jada.2007.01.016
Valentinuzzi, F., Pii, Y., Vigani, G., Lehmann, M., Cesco, S., Mimmo, T. (2015). Phosphorus and iron deficiencies induce a metabolic reprogramming and affect the exudation traits of the woody plant Fragaria×ananassa. J. Exp. Bot. 66, 6483–6495. doi: 10.1093/jxb/erv364
Veneklaas, E. J., Lambers, H., Bragg, J., Finnegan, P. M., Lovelock, C. E., Plaxton, W. C., et al. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195, 306–320. doi: 10.1111/j.1469-8137.2012.04190.x
Vert, G., Barberon, M., Zelazny, E., Séguéla, M., Briat, J. F., Curie, C. (2009). Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta 229, 1171–1179. doi: 10.1007/s00425-009-0904-8
Wallace, A., Elgazzar, A. A., Cha, J. W., Alexander, G. V. (1974). Phosphorus levels versus concentrations of zinc and other elements in bush bean plants. Soil Sci. 117, 347–351. doi: 10.1097/00010694-197406000-00008
White, P. J., Broadley, M. R. (2009). Biofortification of crops with seven mineral elements often lacking in human diets - iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 182, 49–84. doi: 10.1111/j.1469-8137.2008.02738.x
White, P. J., Broadley, M. R. (2011). Physiological limits to zinc biofortification of edible crops. Front. Plant Sci. 2, 80. doi: 10.3389/fpls.2011.00080
White, P. J., Brown, P. H. (2010). Plant nutrition for sustainable development and global health. Ann. Bot. 105, 1073–1080. doi: 10.1093/aob/mcq085
White, P. J., Veneklaas, E. J. (2012). Nature and nurture: the importance of seed phosphorus content. Plant Soil 357, 1–8. doi: 10.1007/s11104-012-1128-4
White, P. J., Broadley, M. R., Thompson, J. A., McNicol, J. W., Crawley, M. J., Poulton, P. R., et al. (2012). Testing the distinctness of shoot ionomes of angiosperm families using the Rothamsted Park Grass Continuous Hay Experiment. New Phytol. 196, 101–109. doi: 10.1111/j.1469-8137.2012.04228.x
White, P. J., George, T. S., Dupuy, L. X., Karley, A. J., Valentine, T. A., Wiesel, L., et al. (2013a). Root traits for infertile soils. Front. Plant Sci. 4, 1–7. doi: 10.3389/fpls.2013.00193
White, P. J., George, T. S., Gregory, P. J., Bengough, A. G., Hallett, P. D., McKenzie, B. M. (2013b). Matching roots to their environment. Ann. Bot. 112, 207–222. doi: 10.1093/aob/mct123
White, P. J., Pongrac, P., Sneddon, C. C., Thompson, J. A., Wright, G. (2018). Limits to the biofortification of leafy brassicas with zinc. Agriculture 8, 1–14. doi: 10.3390/agriculture8030032
White, P. J. (2013). Improving potassium acquisition and utilisation by crop plants. J. Plant Nutr. Soil Sci. 176, 305–316. doi: 10.1002/jpln.201200121
Wissuwa, M., Kretzschmar, T., Rose, T. J. (2016). From promise to application: root traits for enhanced nutrient capture in rice breeding. J. Exp. Bot. 67, 3605–3615. doi: 10.1093/jxb/erw061
Woo, J., MacPherson, C. R., Liu, J., Wang, H., Kiba, T., Hannah, M. A., et al. (2012). The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 12, 62. doi: 10.1186/1471-2229-12-62
Zhao, F. J., Su, Y. H., Dunham, S. J., Rakszegi, M., Bedo, Z., McGrath, S. P., et al. (2009). Variation in mineral micronutrient concentrations in grain of wheat lines of diverse origin. J. Cereal Sci. 49, 290–295. doi: 10.1016/j.jcs.2008.11.007
Zheng, D., Han, X., An, Y., Guo, H., Xia, X., Yin, W. (2013). The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ. 36, 1328–1337. doi: 10.1111/pce.12062
Keywords: mineral nutrition, kale, broccoli, element interaction, RNAseq, gene ontology
Citation: Pongrac P, Fischer S, Thompson JA, Wright G and White PJ (2020) Early Responses of Brassica oleracea Roots to Zinc Supply Under Sufficient and Sub-Optimal Phosphorus Supply. Front. Plant Sci. 10:1645. doi: 10.3389/fpls.2019.01645
Received: 28 February 2019; Accepted: 21 November 2019;
Published: 09 January 2020.
Edited by:
Karl H. Mühling, University of Kiel, GermanyReviewed by:
Marta R. M. Lima, University of New Hampshire, United StatesAsif Naeem, Nuclear Institute for Agriculture and Biology, Pakistan
Copyright © 2020 Pongrac, Fischer, Thompson, Wright and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paula Pongrac, paula.pongrac@ijs.si
†These authors have contributed equally to this work
 Paula Pongrac
Paula Pongrac Sina Fischer
Sina Fischer Jacqueline A. Thompson1
Jacqueline A. Thompson1 Philip J. White
Philip J. White