- 1Microbial Genetics, Institute of Applied Microbiology, RWTH Aachen University, Aachen, Germany
- 2Genetics, Matthias-Schleiden-Institute, Friedrich-Schiller-University Jena, Jena, Germany
- 3Plant Biotechnology, National Research Centre, Cairo, Egypt
- 4Molecular Biology of Plant-Microbe Interactions, Albrecht-von-Haller Institute of Plant Sciences, Schwann-Schleiden Research Center, Georg-August-University Göttingen, Göttingen, Germany
Sporisorium reilianum f. sp. zeae (SRZ) is a biotrophic fungus causing head smut in maize. Maize infection with SRZ leads to very little cell death suggesting the presence of cell-death suppressinpg effectors. Several hundred effector proteins have been predicted based on genome annotation, genome comparison, and bioinformatic analysis. For only very few of these effectors, an involvement in virulence has been shown. In this work, we started to test a considerable subset of these predicted effector proteins for a possible function in suppressing cell death. We generated an expression library of 62 proteins of SRZ under the control of a strong constitutive plant promoter for delivery into plant cells via Agrobacterium tumefaciens-mediated transient transformation. Potential apoplastic effectors with high cysteine content were cloned with signal peptide while potential intracellular effectors were also cloned without signal peptide to ensure proper localization after expression in plant cells. After infiltration of Nicotiana benthamiana leaves, infiltration sites were evaluated for apparent signs of hypersensitive cell death in absence or presence of the elicitin INF1 of Phytophthora infestans. None of the tested candidates was able to induce cell death, and most were unable to suppress INF1-induced cell death. However, the screen revealed one predicted cytoplasmic effector (sr16441) of SRZ that was able to reliably suppress INF1-induced cell death when transiently expressed in N. benthamiana lacking its predicted secretion signal peptide. This way, we discovered a putative function for one new effector of SRZ.
Introduction
The phytopathogenic biotrophic basidiomycete Sporisorium reilianum f. sp. zeae (SRZ) is the causative agent of head smut disease of maize. The disease can cause great damage and leads to complete harvest loss of affected individual plants. In nature, the disease is transmitted by soil-borne diploid teliospores. Under favorable environmental conditions, teliospores germinate, undergo meiosis, and generate haploid sporidia of different mating types (Hanna, 1929; Halisky, 1963; Martinez et al., 2002). Prior to plant infection, compatible haploid sporidia form conjugation hyphae that grow toward each other and fuse at their tips (Schirawski et al., 2005). After mating, the fungus then grows as dikaryotic hyphae that penetrate and colonize the plant initially without causing severe symptoms (Martinez et al., 1999; Prom et al., 2011; Poloni and Schirawski, 2016). Symptoms become evident only at the flowering time when spore formation and phyllody occur in the inflorescences (Wilson and Frederiksen, 1970; Martinez et al., 1999; Ghareeb et al., 2011; Poloni and Schirawski, 2016).
For host plant colonization, pathogens have to overcome several lines of plant defense. Plant defense mechanisms allow perception of pathogen attack and activation of pre- and postinvasion defense responses to minimize damages imposed by destructive invaders (Dangle and Jones, 2001). Plant pathogens acquired the ability to defeat plant immunity responses, resulting in a co-evolutionary arms race for resistance or susceptibility. The first line of defense of the plant's innate immune system is provided by pattern recognition receptors (PRRs) that recognize conserved microbial- or pathogen-associated molecular patterns (MAMPs or PAMPs) and trigger the so-called PAMP-triggered immunity (PTI) response (Jones and Dangl, 2006; Coll et al., 2011). Pathogens can overcome PTI-based defenses by expressing specific effectors that suppress PTI and lead to effector-triggered-susceptibility (ETS) (Coll et al., 2011; Asai and Shirasu, 2015).
Effector proteins secreted by successful plant pathogens modulate and reprogram the defense systems of the host. Two types of secreted effector proteins are known: Apoplastic effectors that are targeted to the plant extracellular space, and cytoplasmic effectors that are delivered inside the plant cell and target different subcellular compartments (Bos et al., 2006; Kamoun, 2006; Asai and Shirasu, 2015). Effectors may be recognized by plant disease resistance (R) proteins, which may result in hypersensitive response (HR), a form of programmed cell death (PCD). Host cell death does not always have a negative impact on the plant. Targeted destruction of specific plant cells can be a powerful mechanism of defense against biotrophic plant pathogens that rely on living host cells to colonize and complete their infection cycles (Lam, 2004; Reape et al., 2007). The infection success of biotrophic pathogens is therefore determined by the ability of the pathogen to suppress the induction of plant defense responses leading to programmed cell death. Suppression of PCD through secretion of specific effectors delivered into host cells has been shown for many different systems. The effector protein AvrPiz-t secreted by Magnaporthe oryzae suppresses the mouse BAX-induced programmed cell death in Nicotiana benthamiana leaves (Li et al., 2009), while MoHEG13 antagonizes cell death induced by M. oryzae Necrosis-and ethylene-inducing-protein-1 (Nep1)-like proteins in N. benthamiana (Mogga et al., 2016). The Avr3a effector of Phytophthora infestans interacts with the potato U-box E3 ubiquitin ligase CMPG1 and stabilizes it to suppress Infestin1 (INF1)-mediated cell death (Bos et al., 2010; Derevnina et al., 2016). INF1 is a P. infestans elicitin inducing HR cell death. Elicitins are highly conserved extracellular proteins secreted by phytopathogenic microorganisms that have features of pathogen-associated molecular patterns (PAMPs) and trigger defenses in a variety of plant species (Du et al., 2015; Derevnina et al., 2016). Expression of P. infestans INF1 is largely used in N. benthamiana to screen for effectors that function as immunosuppressants. The effector AVR3a-KI, a P. infestans host-translocated (cytoplasmic) effector, suppresses the HR cell death triggered by INF1 (Bos et al., 2006).
To successfully infect maize, S. reilianum f. sp. zeae is predicted to secrete hundreds of effector proteins (Schirawski et al., 2010; Schuster et al., 2018; Schweizer et al., 2018; Ghareeb et al., 2019) that facilitate modulation of plant innate immunity and colonization of the host tissue (Ghareeb et al., 2019), supposedly by suppressing plant innate immune responses. For only very few effectors, an involvement in virulence has been shown (Ghareeb et al., 2015; Schweizer et al., 2018; Ghareeb et al., 2019). Since plant penetration is followed by a long phase of fungal proliferation within the plant tissue but without prominent disease symptoms, some effectors likely function in suppressing cell death.
In the present study, we aimed to assign a function to more effector proteins by identifying effectors that could suppress cell death. We selected a set of bioinformatically predicted small secreted proteins from SRZ, created an expression library of 62 constructs and expressed them in Nicotiana benthamiana via Agrobacterium-mediated transient gene expression, under the control of a strong constitutive plant promoter. We evaluated their ability to induce hypersensitive cell death (PCD) or suppress PCD triggered by the elicitin INF1. Potential apoplastic effectors with high cysteine content were cloned with signal peptide while potential intracellular effectors were also cloned without signal peptide to ensure proper localization after expression in plant cells. This way, one candidate effector (sr16441) was identified that is able to suppress cell death induced by the elicitin INF1.
Materials and Methods
Cloning of Candidate Effector Genes
Effector candidates were mined from a large collection of SRZ proteins predicted to be secreted and lacking functional annotation (Schirawski et al., 2010; Schweizer et al., 2018). SignalP 5.0 (Armenteros et al., 2019) was used to predict the location of putative secretion signal peptides. Amino acid sequences of mature (i.e. lacking their signal peptide) putative effector proteins were analyzed for their cysteine content using the webserver DIANNA (Ferrè and Clote, 2005). We selected 56 effector candidates for cloning, with, without, or both with and without predicted signal peptides, totaling 62 constructs (see Table 1). Gene-specific primer pairs (Table S1) were used in PCR amplification reactions with genomic DNA from SRZ isolates as template. Amplified genes were cloned into the binary plasmid pHG44-GWY. pHG44-GWY is a derivative of pHG44 that was modified by Gibson assembly (Gibson et al., 2009). The plasmid pHG44 in turn was generated from the plasmid pP35S:GFP-SAD1ΔSP-T35 (Ghareeb et al., 2015) by digesting it with AscI to produce a 6.5 kb fragment, which then was dephosphorylated using calf intestinal alkaline phosphatase (NEB). pP35S:mCherry-T35 plasmid was digested using BssHII and MluI to generate a 1.6 kb fragment containing the P35S:mCherry-T35 construct. Ligation of the two aforementioned fragments resulted in the pP35S:GFP-SAD1ΔSP-T35-P35S:mCherry-T35 expression vector (pHG44). A pair of primers containing 20 bp overhangs (Table S1) was used to amplify a segment of about 3 kb from pHG44 consisting of left and right border repeats from nopaline C58 T-DNA, the resistance cassette (AmpR) and origin of replication (ori and oriV). The amplified segment was then ligated with a 3 kb BsrBI fragment of the plasmid HBT-sGFP(S65T)-NOS supplied by Jen Sheen, Boston, USA (Cheng et al., 2001) containing the recombination sites attR1 and attR2 suited for Gateway cloning along with the 35S-PPDK hybrid promoter and nos terminator (Figure 1).
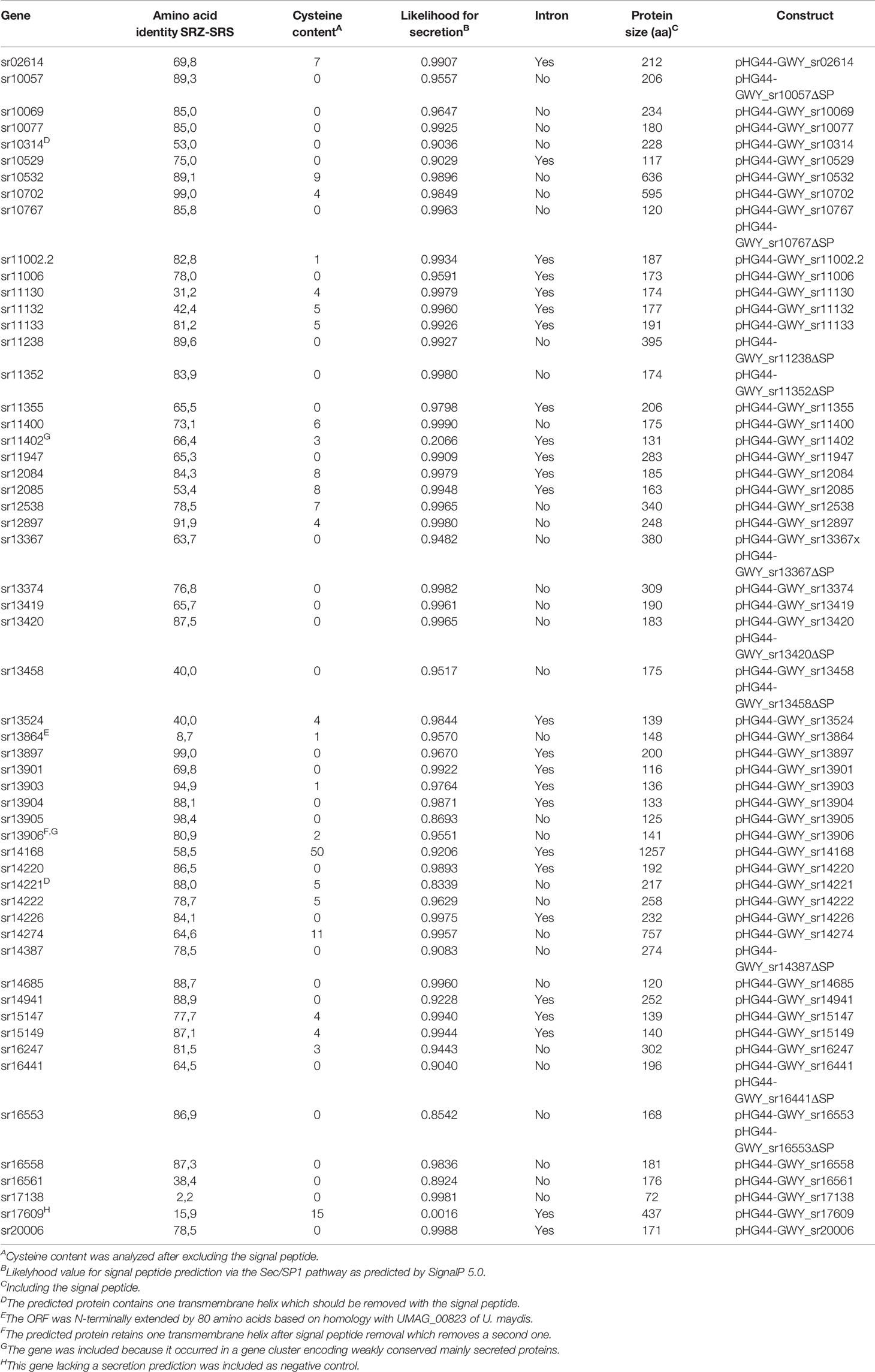
Table 1 SRZ genes selected to be tested for programmed cell death (PCD) suppression in N. benthamiana in this study. Fifty-six effector candidates were cloned from genomic DNA, some of them carry introns.

Figure 1 Principal elements of the Gateway-modified gene expression vector pHG44-GWY. The binary vector pHG44 was modified to carry a Gateway cloning cassette with the ccdB suicide gene and attR recombination sites along with the promoter 35SPPDK and nos terminator between its left and right border repeats from nopaline C58 T-DNA.
All constructs were verified first by restriction digest using several enzymes, then by sequencing. A total of 25 out of 46 constructs cloned with signal peptide contain introns (Table 1). A total of 49 constructs (see Figure S2) were tested in TSL, Norwich, UK, for their capacity to induce or suppress cell death induced by INF1, under the supervision of Sophien Kamoun, using p35S_ ΔGFP (Chaparro-Garcia et al., 2015) as a negative control and pBIN-plus-Avr3a-KI (Chaparro-Garcia et al., 2015) as a positive control. The remaining constructs were evaluated at the RWTH Aachen University, where pHG44-GWY_GFP was used as negative control, and pBINplus-Avr3a-KI was used as a positive control. A. tumefaciens strains GV3101 containing the constructs pGR106-INF1 (Huitema et al., 2005) and pBINplus-Avr3a-KI (Chaparro-Garcia et al., 2015) were kindly provided by Sophien Kamoun. INF1 was amplified using the primer pairs indicated in Table S1 and cloned in pHG44-GWY to test the efficiency of this plasmid.
Growth of Microbial Strains and Cultivation of Plants
Recombinant binary plasmids were maintained and propagated in Escherichia coli, strain Top10, grown in lysogeny broth (LB) media supplemented with 50 µg/ml carbenicillin. Basic molecular cloning techniques were used (Maniatis et al., 1988).
Recombinant A. tumefaciens genotype GV3101pMP90RK carrying constructs in the pHG44-GWY vector were routinely grown in LB media with appropriate antibiotics (rifampicin 100 µg/ml, gentamycin 50 µg/ml, kanamycin 50 µg/ml, carbenicillin 50 µg/ml) at 28°C with shaking at 180 rpm overnight. Agrobacterium strains were transformed with plasmid vectors by heat shock using a protocol provided by DNA Cloning Service e. K. available online (http://www.dna-cloning.com/agrobacterium). Transformed cells were grown, collected by centrifugation (5000 rpm, 5 minutes, at room temperature), re-suspended in infiltration buffer (10 mM MgCl2, 10 mM MES, pH 5.6, and 200 mM acetosyringone), and incubated at room temperature for 1–2 hours before infiltration.
N. benthamiana plants were cultivated and maintained throughout the experiments in a plant growth chamber or greenhouse under 16/8 hour light/dark photoperiod at 22°C and high light intensity. The experiments were performed using leaves of 4 to 6 week-old plants.
Cell Death and Cell Death Suppression Assays
All constructs were first tested whether they induce cell death. The known PCD elicitor psojNIP (Qutob et al., 2002) or pGR106-INF1 (Huitema et al., 2005) was used as a control. A. tumefaciens strains expressing the SRZ effectors or controls were grown to a final OD600 of 0.5 and used to infiltrate the abaxial leaf side of 4 to 6 week-old N. benthamiana plants using a 1-ml syringe. Induction of PCD was visually assessed at 3, 4, and 5 days after infiltration. For cell death suppression assays, the infiltration sites were challenged again after 24 hours with recombinant A. tumefaciens carrying pGR106-INF1 at a final OD600 of 0.2 as previously described (Huitema et al., 2004; Bos et al., 2006); GFP and Avr3a-KI served as negative and positive control, respectively. A suppressor of posttranscriptional gene silencing from Tomato bushy stunt virus (P19) known to increase gene expression in the agroinfiltration assay (Oh et al., 2009) was used to improve the expression of 13 constructs (see Figures 3C, E). Strains carrying the plasmid pBIN61-P19 (Voinnet et al., 1999) were mixed in induction buffer with strains carrying the candidate effectors in a ratio of 1:1 (final OD600 of 1) and coinfiltrated. Symptom development and possible suppression of PCD was monitored at 3, 4, and 5 days after the second infiltration (Bos et al., 2006; Oh et al., 2009). The degree of PCD of leaves (HR index) was scored on a previously described seven-point scale according to the size of the necrotic area (grade 0 when no necrosis is observed, grade 7 when necrosis is confluent) (Wu et al., 2017). Each treatment was assayed on two plants with three leaves for each plant. Therefore, at least five infiltration sites were evaluated for each treatment. The experiment done in Aachen was conducted at least three times, two times without pBIN61-P19. The number of infiltration sites showing PCD was counted for each construct. One-way ANOVA with post-hoc Tukey test or independent-samples t-tests was used for statistical analysis conducted in SPSS (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.). The graphs (except those in the Supplemental Material available online) display results of the experiments when pBIN61-P19 was used.
Results
We randomly selected a total of 82 putative effector proteins for PCR amplification of the respective open reading frames. After excluding genes with weak or unsuccessful PCR-amplification, we finally generated a library of 62 constructs using primers corresponding to 56 ORFs that were either cloned with (46 genes), without (four genes), or with and without (six genes) their predicted signal peptides (Table 1). Since apoplastic effectors often contain multiple cysteine residues (Stergiopoulos and de Wit, 2009), we made sure that the ten ORFs selected for cloning without signal peptide (and thus ending up within the plant cell after heterologous expression) encoded proteins with no or only one cysteine residue (Table 1). The amplicons were cloned under the control of the constitutive hybrid 35S-PPDK promoter suited for gene expression in N. benthamiana (Figure 1) and were sequenced prior to use. Of the 56 ORFs that were cloned, 53 had less than 95% amino acid conservation to the respective orthologs of S. reilianum f. sp. reilianum, 46 had less than 300 amino acids, and one did not have a secretion prediction and served as negative control (Table 1).
To determine whether diverse S. reilianum f. sp. zeae effectors perturb host cellular processes, the 62 constructs were expressed in N. benthamiana using Agrobacterium-mediated transient transformation. This method has been shown to be a valuable initial screening tool to determine whether particular genes induce or suppress defense-associated PCD (Huitema et al., 2004; Oh et al., 2009). To verify that none of the candidate effectors induces cell death, N. benthamiana leaves were first infiltrated with A. tumefaciens strains carrying each of the 62 constructs or the controls psojNIP (Qutob et al., 2002) or pGR106-INF1 (Huitema et al., 2005) that encode known cell death-inducing elicitins. The infiltrated sites were visually evaluated at 3, 4, and 5 days after infiltration for signs of cell death. Phenotypic evaluation of the infiltrated sites revealed that only the known elicitins NIP of P. sojae and INF1 of P. infestans but none of the 62 constructs led to cell death induction (Figure 2; Figure S1, see Supplemental Material available online).

Figure 2 PCD induction assay. Agrobacterium strains carrying constructs with Sporisorium reilianum f. sp. zeae (SRZ) effectors were infiltrated in N. benthamiana. The results were evaluated at 3, 4, and 5 days after infiltration and compared to that of Infestin1 (INF1). None of the constructs could induce cell death under our experimental conditions. Pictures were taken at 4 days after infiltration.
We wondered whether a lack of cell-death induction activity of the tested constructs was a result of non-sufficient mRNA generation or wrong splicing of constructs that were cloned with introns. Therefore, we isolated total RNA of N. benthamiana leaves 4 days after infiltration with A. tumefaciens strains carrying one of seven constructs for expression of selected effectors, either with or without intron and either with or without signal peptide. Using RT-PCR, we could show that all seven constructs were expressed and that the three tested constructs with intron were correctly spliced (Figure S2, see Supplemental Material available online). Since all effector constructs were cloned in their native form without any tag that could be used in Western blot experiments, we decided to check protein expression by cloning the INF1 gene in the effector delivery vector pHG44-GWY. We compared the cell-death inducing activity of INF1 when expressed from pGR106-INF1 or from pHG44-GWY_INF1. Both constructs clearly induced necrosis of the infiltrated area (Figure 3A). Although necrosis induction by pHG44-GWY_INF1 was slightly weaker, this experiment showed that expression of pHG44-GWY led to protein expression strong enough to induce a cell-death response. Hence the pHG44-GWY vector was found to be suitable for the conducted experiment, suggesting that none of the tested constructs was able to induce cell death in N. bentamiana.
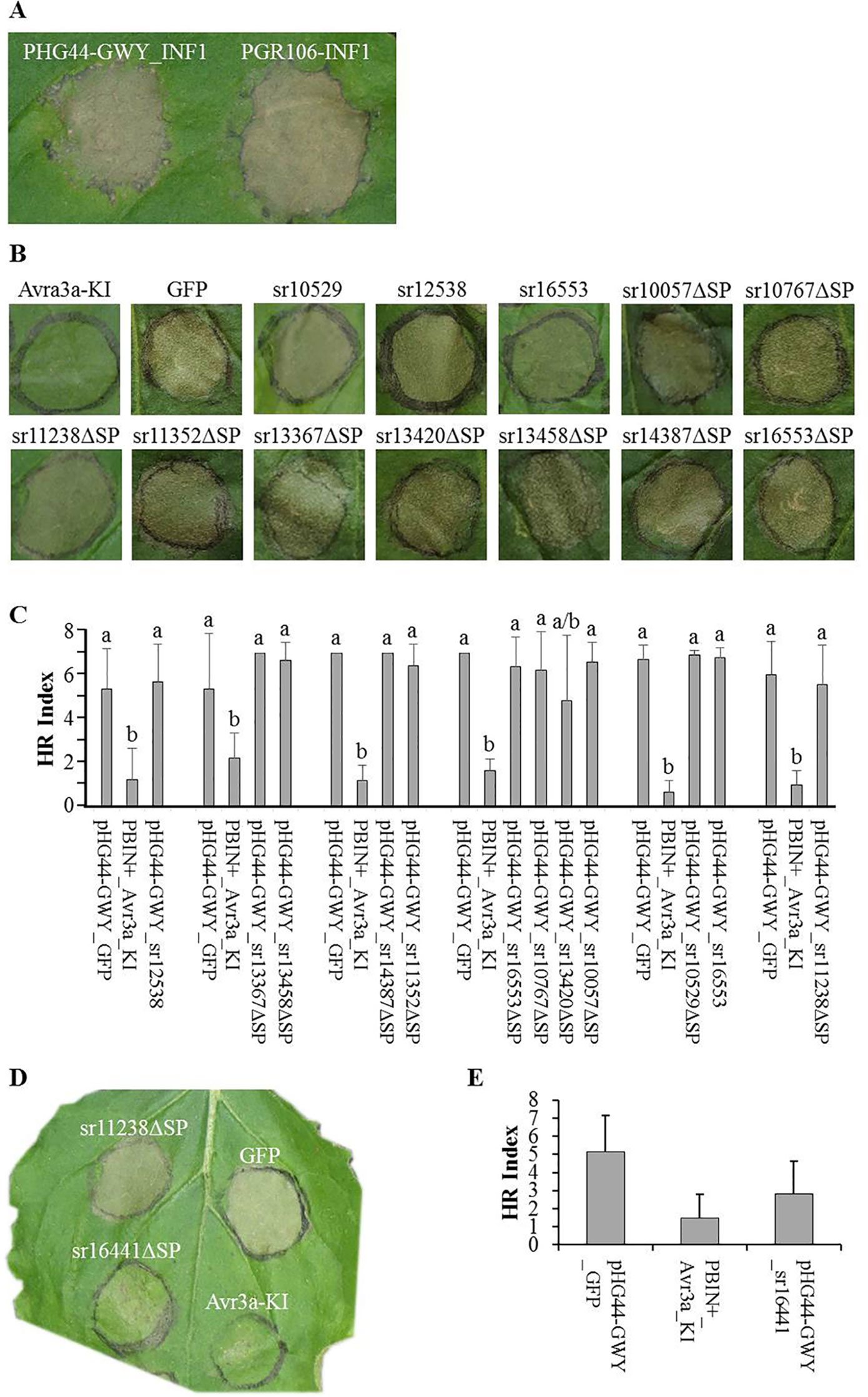
Figure 3 Cell death suppression assays. Agrobacterium strains carrying effectors were infiltrated in N. benthamiana. Candidate effectors from Sporisorium reilianum f. sp. zeae (SRZ) were infiltrated along with GFP (negative control) and Avr3a-KI (positive control), one day later the infiltration sites were challenged with the elicitin Infestin1 (INF1). The infiltration sites were evaluated after 3, 4, and 5 days. Pictures were taken at day 4. (A) Transient expression of both, pHG44-GWY-INF1 and pGR106-INF1 induced necrosis of the infiltrated area. Although necrosis induction by pHG44-GWY_INF1 was slightly weaker, we could show that expression of pHG44-GWY led to an expression strong enough to induce a cell-death response. (B) Leaves infiltrated with GFP and challenged with INF1 showed cell death, while leaves infiltrated with Avr3a-KI and challenged with INF1 did not show cell death. Furthermore, most of the constructs carrying Sporisorium reilianum f. sp. zeae (SRZ) effectors could not suppress INF1-induced cell death. (C) Quantitative comparison of the means of HR indexes of the infiltration sites of SRZ candidate effectors (co-infiltrated with pBIN61-P19) shows that they were statistically significantly higher then that from pBINplus-AVR3a-KI and had no significant difference to the means of pHG44-GWY-GFP, indicating that those candidate effectors are not able to suppress INF1-induced cell death. The experiment was conducted three times, two times without pBIN61-P19. Each column shows the mean and standard deviation. The letters above each column indicate statistically significant differences of the HR index (P < 0.01). (D) The strain carrying the predicted cytoplasmic effector sr16441ΔSP could suppress INF1-induced cell death. (E) Quantitative comparison shows that the mean of HR index of pHG44-GWY-sr16441ΔSP was statistically significantly lower than that of the pHG44-GWY-GFP control and was as low as that of pBINPLUS-AVR3a-KI (P < 0.01), indicating that sr16441ΔSP was able to suppress INF1-induced cell death. The experiments were repeated at least three times. The percentage was calculated from 30 infiltration sites. Columns show the mean and standard deviation. Different letters above each column indicate statistically significant differences of the HR index (P < 0.01).
To test whether any of the cloned SRZ effectors had a function in suppressing INF1-induced cell death, we first infiltrated leaves of N. benthamiana with Agrobacteria delivering the effector constructs, and challenged the same area at 24 hours with Agrobacteria delivering the construct for expression of INF1. When first infiltrating the leaves with Agrobacteria delivering a construct for expression of AVR3a-KI, an effector of P. infestans that was shown to suppress INF1-induced cell death (Bos et al., 2006), a challenge with INF1-expressing Agrobacteria did not lead to cell death. In contrast, when first infiltrating the leaves with Agrobacteria delivering a construct for expression of GFP, challenging with INF1-expressing Agrobacteria led to clearly visible cell death (Figure 3B). Of the 62 tested constructs, 61 were not able to suppress INF1-induced cell death (Figures 3B, C; Figure S3, see Supplemental Material available online).
In contrast, delivery of the sr16441ΔSP expression construct efficiently prevented INF1-induced cell death (Figures 3D, E). We quantified the amount of induced necrosis within the infiltrated leaf area (Wu et al., 2017). The necrosis ratio of pHG44-GWY-sr16441ΔSP was significantly lower than that of the pHG44-GWY-GFP control and was as low as that of pBINplus-AVR3a-KI (P < 0.01) (Figure 3E). We verified the cell-death suppression activity of sr16441ΔSP by infiltration of pHG44-GWY-sr16441ΔSP together with p19 (Oh et al., 2009), which led to even clearer cell death suppression response (not shown). Interestingly, using pHG44-GWY-sr16441, which leads to expression of sr16441 including its putative signal peptide, did not lead to suppression of INF1-induced cell death (Figure S3, see Supplemental Material available online). This result suggests that cytoplasmically expressed sr16441ΔSP can consistently suppress PCD induced by INF1 in N. benthamiana.
Discussion
In this study, we evaluated the ability of 62 expression constructs for putative effectors of SRZ to induce cell death or suppress INF1-induced cell death in N. benthamiana. None of the tested constructs induced cell death, but sr16441 suppressed the cell death induced by INF1 when expressed without signal peptide.
Suppression of plant innate immunity is an important function of plant pathogens during plant cell invasion (Hogenhout et al., 2009). AVR3a-KI, a P. infestans host-translocated (cytoplasmic) effector, deregulates plant immune signaling leading to suppression of the cell death triggered by several pathogen molecules, among them the PAMP-like elicitin INF1 of P. infestans (Bos et al., 2006; Bos et al., 2010; Gilroy et al., 2011). To suppress PCD triggered by INF1, Avr3a-KI interacts with and stabilizes the host ubiquitin E3 ligase CMPG1, which is required for INF1-dependent cell death. Stabilization of CMPG1 by AVR3a consists of modifying CMPG1 activity, preventing the normal 26S proteasome-dependent degradation of itself and potentially of its protein substrates in the host cell. Thus, AVR3a blocks signal transduction cascades initiated at the plasma membrane after pathogen perception (Bos et al., 2010). Currently, over 30 effectors from four different oomycete species are known to suppress INF1-triggered responses, however, knowledge on how elicitin-triggered responses are suppressed is currently limited to AVR3a-KI (Derevnina et al., 2016).
Cell death-inducing effectors have been identified in the Ustilago hordei–barley pathosystem that is genetically ruled by a “gene-for-gene” interaction. One of the avirulence proteins, UhAvr1, induces local cell death during an incompatible interaction with barley, in the presence of Ruh1 (Ali et al., 2014). Incompatible interaction between SRZ and sorghum leads to induction of phytoalexins, which culminates in cell death at the site of infection (Zuther et al., 2012). Hence, the existence of avirulence effectors in SRZ that would induce cell death in N. benthamiana would be plausible. It is possible that cell death-inducing effectors are among the majority of effectors that were not tested, or that the used constructs did not lead to protein expression. Also, the used method would only identify cell death effectors targeting conserved host proteins, while most effectors may induce cell death only in one particular plant species. Therefore, testing the selected effectors for cell death induction in maize or sorghum might prove rewarding.
The putative effector sr16441 that we identified as suppressing INF1-induced cell death in N. benthamiana, is a predicted small protein of 196 amino acids excluding its signal peptide. The protein shows only 65% identity to its closest homolog SRS1_16441 of Sporisorium reilianum f. sp. reilianum. The protein has weakly conserved homologs also in Sporisorium scitamineum (SPSC_01549 and SPSC_1550) and Sporisorium graminicola (EX895_003212). In how far the proteins fulfill the same or a similar function in their respective host plants needs to be elucidated. The candidate effector sr16441 does not have any recognizable domains but has a clear prediction for secretion. In N. benthamiana, it was able to suppress INF1-induced cell death only when expressed inside the plant cell, i.e. without its signal peptide. This indicates that its signal peptide is functional and that for INF1-induced cell death suppression, an intracellular localization is necessary. Supposing that sr16441 also functions in cell death suppression in maize when secreted by S. reilianum, the protein would need to be taken up by the plant cell and target a factor that is conserved between N. benthamiana and Maize.
Further investigations are needed to verify whether sr16441 contributes to virulence, host selection, or symptom formation of S. reilianum on maize and sorghum. Gene expression analysis, localization of the protein after secretion by the fungus, and a possible function in suppressing cell death in sorghum or maize should be tested. However, a possible screening system would depend on the identification of a factor that reliably induces cell death in sorghum or maize. Although there is still a lot to do to elucidate the function of sr16441 in its natural system, the conducted assay revealed a possible function for this candidate effector that can now be elucidated in detail.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
JS and DD conceived the study. DD did the cloning, cell-death and cell death-suppression experiments. NA did the effector expression control. DD and JS wrote the manuscript and prepared the figures. HG generated pHG44. All authors read and approved the submitted manuscript version.
Funding
DD was funded by the mobility program for Brazilian students “Science without Borders” (Ciência sem Fronteiras). NA was supported by the German Academic Exchange Service (DAAD). JS received funding from the German Research Foundation (DFG), as well as institutional support by the RWTH Aachen University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Sophien Kamoun, Lida Derevnina and Joe Win (The Sainsbury Laboratory, UK) for the guidance and for sharing material, and to Lars Blank (RWTH Aachen University) for the support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00095/full#supplementary-material
References
Ali, S., Laurie, J. D., Linning, R., Cervantes-Chávez, J. A., Gaudet, D., Bakkeren, G. (2014). An immunity-triggering effector from the barley smut fungus Ustilago hordei resides in an Ustilaginaceae-specific cluster bearing signs of transposable element-assisted evolution. PLoS Pathog. 10 (7), e1004223. doi: 10.1371/journal.ppat.1004223
Armenteros, J. J. A., Tsirigos, K. D., Sønderby, C. K., Petersen, T. N., Winther, O., Brunak, S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37 (4), 420–423. doi: 10.1038/s41587-019-0036-z
Asai, S., Shirasu, K. (2015). Plant cells under siege: Plant immune system versus pathogen effectors. Curr. Opin. Plant Biol. 28, 1–8. doi: 10.1016/j.pbi.2015.08.008
Bos, J. I. B., Kanneganti, T. D., Young, C., Cakir, C., Huitema, E., Win, J., et al. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48 (2), 165–176. doi: 10.1111/j.1365-313X.2006.02866.x
Bos, J. I. B., Armstrong, M. R., Gilroy, E. M., Boevink, P. C., Hein, I., Taylor, R. M., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. 107 (21), 9909–9914. doi: 10.1073/pnas.0914408107
Chaparro-Garcia, A., Schwizer, S., Sklenar, J., Yoshida, K., Petre, B., Bos, J. I. B., et al. (2015). Phytophthora infestans RXLR-WY effector AVR3a associates with dynamin-related protein 2 required for endocytosis of the plant pattern recognition receptor FLS2. PLoS One 10 (9), e0137071. doi: 10.1371/journal.pone.0137071
Cheng, S.-H., Sheen, J., Gerrish, C., Bolwell, G. P. (2001). Molecular identification of phenylalanine ammonia-lyase as a substrate of a specific constitutively active Arabidopsis CDPK expressed in maize protoplasts. FEBS Lett. 503 (2-3), 185–188. doi: 10.1016/s0014-5793(01)02732-6
Coll, N. S., Epple, P., Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18 (8), 1247–1256. doi: 10.1038/cdd.2011.37
Dangle, J. L., Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 (6839), 826–833. doi: 10.1038/35081161
Derevnina, L., Dagdas, Y. F., De la Concepcion, J. C., Bialas, A., Kellner, R., Petre, B., et al. (2016). Nine things to know about elicitins. New Phytol. 212 (12), 888–895. doi: 10.1111/nph.14137
Du, J., Verzaux, E., Chaparro-Garcia, A., Bijsterbosch, G., Keizer, L. C. P., Zhou, J., et al. (2015). Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 1 (3), 1–5. doi: 10.1038/nplants.2015.34
Ferrè, F., Clote, P. (2005). DiANNA: a web server for disulfide connectivity prediction. Nucleic Acids Res. 33 (Web Server Issue), W230–W232. doi: 10.1093/nar/gki412
Ghareeb, H., Becker, A., Iven, T., Feussner, I., Schirawski, J. (2011). Sporisorium reilianum infection changes inflorescence and branching architectures of maize. Plant Physiol. 156 (4), 2037–2052. doi: 10.1104/pp.111.179499
Ghareeb, H., Drechsler, F., Löfke, C., Teichmann, T., Schirawski, J. (2015). Suppressor of apical dominance 1 of Sporisorium reilianum modulates inflorescence branching architecture in maize and Arabidopsis. Plant Physiol. 169 (4), 2789–2804. doi: 10.1104/pp.15.01347
Ghareeb, H., Zhao, Y., Schirawski, J. (2019). Sporisorium reilianum possesses a pool of effector proteins that modulate virulence on maize. Mol. Plant Pathol. 20 (1), 124–136. doi: 10.1111/mpp.12744
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6 (5), 343–345. doi: 10.1038/nmeth.1318
Gilroy, E. M., Taylor, R. M., Hein, I., Boevink, P., Sadanandom, A., Birch, P. R. J. (2011). CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190 (3), 653–666. doi: 10.1111/j.1469-8137.2011.03643.x
Halisky, P. M. (1963). Head smut of sorghum, sudangrass, and corn, caused by Sphacelotheca reiliana (Kühn) Clint. Hilgardia 34 (8), 287–304. doi: 10.3733/hilg.v34n08p287
Hanna, W. (1929). Studies in the physiology and cytololgy of Ustilago zeae and Sorosporium reilianum. Phytopathology 19 (5), 415–442.
Hogenhout, S. A., Van der Hoorn, R. A. L., Terauchi, R., Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant-Microbe Interact. 22 (2), 115–122. doi: 10.1094/mpmi-22-2-0115
Huitema, E., Bos, J. I. B., Tian, M., Win, J., Waugh, M. E., Kamoun, S. (2004). Linking sequence to phenotype in Phytophthora-plant interactions. Trends Microbiol. 12 (4), 193–200. doi: 10.1016/j.tim.2004.02.008
Huitema, E., Vleeshouwers, V. G. A. A., Cakir, C., Kamoun, S., Govers, F. (2005). Differences in intensity and specificity of hypersensitive response induction in Nicotiana spp. by INF1, INF2A, and INF2B of Phytophthora infestans. Mol. Plant-Microbe Interact. 18 (3), 183–193. doi: 10.1094/MPMI-18-0183
Jones, J. D. G., Dangl, ,. J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44 (1), 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Lam, E. (2004). Controlled cell death, plant survival and development. Nat. Rev. Mol. Cell Biol. 5 (4), 305–315. doi: 10.1038/nrm1358
Li, W., Wang, B., Wu, J., Lu, G., Hu, Y., Zhang, X., et al. (2009). The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 22 (4), 411–420. doi: 10.1094/mpmi-22-4-0411
Maniatis, T., Fritsch, E. F., Sambrook, J. (1988). Molecular cloning: a laboratory manual. 2nd edition. Cold Spring Harbor New York.
Martinez, C., Roux, C., Dargent, R. (1999). Biotrophic development of Sporisorium reilianum f. sp. zeae in vegetative shoot apex of maize. Phytopathology 89 (3), 247–253. doi: 10.1094/PHYTO.1999.89.3.247
Martinez, C., Roux, C., Jauneau, A., Dargent, R. (2002). The biological cycle of Sporisorium reilianum f.sp. zeae: an overview using microscopy. Mycologia 94 (3), 505–514. doi: 10.1080/15572536.2003.11833215
Mogga, V., Delventhal, R., Weidenbach, D., Langer, S., Bertram, P. M., Andresen, K., et al. (2016). Magnaporthe oryzae effectors MoHEG13 and MoHEG16 interfere with host infection and MoHEG13 counteracts cell death caused by Magnaporthe-NLPs in tobacco. Plant Cell Rep. 35 (5), 1169–1185. doi: 10.1007/s00299-016-1943-9
Oh, S.-K., Young, C., Lee, M., Oliva, R., Bozkurt, T. O., Cano, L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21 (9), 2928–2947. doi: 10.1105/tpc.109.068247
Poloni, A., Schirawski, J. (2016). Host specificity in Sporisorium reilianum is determined by distinct mechanisms in maize and sorghum. Mol. Plant Pathol. 17 (5), 741–754. doi: 10.1111/mpp.12326
Prom, L. K., Perumal, R., Erattaimuthu, S. R., Erpelding, J. E., Montes, N., Odvody, G. N., et al. (2011). Virulence and molecular genotyping studies of Sporisorium reilianum isolates in sorghum. Plant Dis. 95 (5), 523–529. doi: 10.1094/pdis-10-10-0720
Qutob, D., Kamoun, S., Gijzen, M. (2002). Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 32 (3), 361–373. doi: 10.1046/j.1365-313X.2002.01439.x
Reape, T. J., Molony, E. M., McCabe, P. F. (2007). Programmed cell death in plants: Distinguishing between different modes. J. Exp. Bot. 59 (3), 435–444. doi: 10.1093/jxb/erm258
Schirawski, J., Heinze, B., Wagenknecht, M., Kahmann, R. (2005). Mating type loci of Sporisorium reilianum: novel pattern with three a and multiple b specificities. Eukaryotic Cell. 4 (8), 1317–1327. doi: 10.1128/EC.4.8.1317-1327.2005
Schirawski, J., Mannhaupt, G., Münch, K., Brefort, T., Schipper, K., Doehlemann, G., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330 (6010), 1546–1548. doi: 10.1126/science.1195330
Schuster, M., Schweizer, G., Kahmann, R. (2018). Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes. Fungal Gen. Biol. 112, 21–30. doi: 10.1016/j.fgb.2016.12.003
Schweizer, G., Münch, K., Mannhaupt, G., Schirawski, J., Kahmann, R., Dutheil, J. Y. (2018). Positively selected effector genes and their contribution to virulence in the smut fungus Sporisorium reilianum. Genome Biol. Evol. 10 (2), 629–645. doi: 10.1093/gbe/evy023
Stergiopoulos, I., de Wit, P. J. G. M. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Voinnet, O., Pinto, Y. M., Baulcombe, D. C. (1999). Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. 96 (24), 14147–14152. doi: 10.1073/pnas.96.24.14147
Wilson, J. M., Frederiksen, R. A. (1970). Histopathology of the interaction of Sorhum bicolor and Sphacelotheca reiliana. Phytopathology 60, 828–832. doi: 10.1094/Phyto-60-828
Wu, C. H., Abd-El-Haliem, A., Bozkurt, T. O., Belhaj, K., Terauchi, R., Vossen, J. H., et al. (2017). NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U. S. A. 114 (30), 8113–8118. doi: 10.1073/pnas.1702041114
Keywords: Sporisorium reilianum, plant pathogen, hypersensitive response, effector proteins, Nicotiana benthamiana, agroinfiltration, INF1 elicitin, cell death suppression
Citation: Dutra D, Agrawal N, Ghareeb H and Schirawski J (2020) Screening of Secreted Proteins of Sporisorium reilianum f. sp. zeae for Cell Death Suppression in Nicotiana benthamiana. Front. Plant Sci. 11:95. doi: 10.3389/fpls.2020.00095
Received: 27 September 2019; Accepted: 22 January 2020;
Published: 19 February 2020.
Edited by:
Sébastien Duplessis, INRA Centre Nancy-Lorraine, FranceReviewed by:
Steven Whitham, Iowa State University, United StatesTobias Link, University of Hohenheim, Germany
Copyright © 2020 Dutra, Agrawal, Ghareeb and Schirawski. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jan Schirawski, jan.schirawski@uni-jena.de
†Present Address: Hassan Ghareeb, Plant Cell Biology, Albrecht-von-Haller Institute of Plant Sciences, Georg-August-University Göttingen, Göttingen, Germany
 Deiziane Dutra1
Deiziane Dutra1 Nisha Agrawal
Nisha Agrawal Hassan Ghareeb
Hassan Ghareeb Jan Schirawski
Jan Schirawski