- 1Agricultural Biotechnology Division, National Institute for Biotechnology and Genetic Engineering, Constituent College of Pakistan Institute of Engineering and Applied Sciences, Faisalabad, Pakistan
- 2Department of Biotechnology, Balochistan University of Information Technology, Engineering and Management Sciences, Quetta, Pakistan
- 3Soil and Environmental Biotechnology Division, National Institute for Biotechnology and Genetic Engineering, Constituent College of Pakistan Institute of Engineering and Applied Sciences, Faisalabad, Pakistan
Basmati rice is famous around the world for its flavor, aroma, and long grain. Its demand is increasing worldwide, especially in Asia. However, its production is threatened by various problems faced in the fields, resulting in major crop losses. One of the major problems is bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo). Xoo hijacks the host machinery by activating the susceptibility genes (OsSWEET family genes), using its endogenous transcription activator like effectors (TALEs). TALEs have effector binding elements (EBEs) in the promoter region of the OsSWEET genes. Out of six well-known TALEs found to have EBEs in Clade III SWEET genes, four are present in OsSWEET14 gene’s promoter region. Thus, targeting the promoter of OsSWEET14 is very important for creating broad-spectrum resistance. To engineer resistance against bacterial blight, we established CRISPR-Cas9 mediated genome editing in Super Basmati rice by targeting 4 EBEs present in the promoter of OsSWEET14. We were able to obtain four different Super Basmati lines (SB-E1, SB-E2, SB-E3, and SB-E4) having edited EBEs of three TALEs (AvrXa7, PthXo3, and TalF). The edited lines were then evaluated in triplicate for resistance against bacterial blight by choosing one of the locally isolated virulent Xoo strains with AvrXa7 and infecting Super Basmati. The lines with deletions in EBE of AvrXa7 showed resistance against the Xoo strain. Thus, it was confirmed that edited EBEs provide resistance against their respective TALEs present in Xoo strains. In this study up to 9% editing efficiency was obtained. Our findings showed that CRISPR-Cas9 can be harnessed to generate resistance against bacterial blight in indigenous varieties, against locally prevalent Xoo strains.
Introduction
In plants, different genome editing strategies have been exploited including zinc finger nucleases, transcription activator-like effector nucleases (TALEs), and clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated (Cas) nucleases. Among these tools, the RNA guided CRISPR-Cas9 system has become the method of choice for genome editing because of its simplicity, ease of performing, and versatility. This system exploits the complementary base pairing mechanism of DNA to guide site-specific Cas9 endonuclease to the target site. The guide RNA (gRNA) screens the template, recognizes the specific complementary target sequence, and signals to Cas9 to introduce a double stranded break (DSB) at the target site. A triplet of nucleotides (NGG) at the 3′ end of the target site, also known as protospacer-associated motif (PAM), is essential for Cas9 to introduce a double stranded break (DSB) 3 bp upstream of the PAM sequence. These DSBs are repaired either via imprecise non-homologous end joining (NHEJ) or template directed precise homology directed repair (HDR) (Barrangou et al., 2007; Cong et al., 2013; Mali et al., 2013; Hsu et al., 2014; Wright et al., 2016). Until today, this technology has been successfully used to engineer resistance against various pathogens and for agronomic trait enhancement.
Rice is a staple food crop which belongs to the family Poaceae, genus Oryza. It has been cultivated for more than 10,000 years and is the second most commonly cultivated cereal in the world (Sasaki, 2001; Pazuki and Sohani, 2013). For Asian countries, it contributes 50–80% of daily calories and serves more than 90% of the population (Khush, 2005; Zeigler and Barclay, 2008). Among different varieties of rice, Basmati rice is famous around the globe for its flavor, aroma, and long grain. Due to food insecurity, rice demand is increasing every day (Shobarani et al., 2010). However, Basmati rice production is threatened by various problems in the field resulting in major crop losses. One of the grave problems is bacterial blight caused by Xanthomonas oryzae pv. Oryzae (Xoo). It is the most destructive and deadly bacterial disease and can cause up to 75% crop loss. Xoo hijacks the host machinery by activating the susceptibility genes (SWEET family genes), using its endogenous transcription activator like effectors (TALE). TALEs have their effector binding elements (EBEs) in the promoter region of OsSWEET genes. These effectors divert sugars from the plant cell to fulfill the pathogen’s nutritional needs (Chen et al., 2012). Most of the geographically distinct Xoo strains target OsSWEET14, which encodes the sucrose-efflux transporter family (Chen et al., 2010). There are six known TALEs which target promoter regions of OsSWEET genes, and the EBEs for four different TALEs, PthXo3, AvrXa7, TalC, and TalF (previously known as Tal5) are present in OsSWEET14 (Antony et al., 2010; Yu et al., 2011; Streubel et al., 2013; Oliva et al., 2019) (Figure 1A).
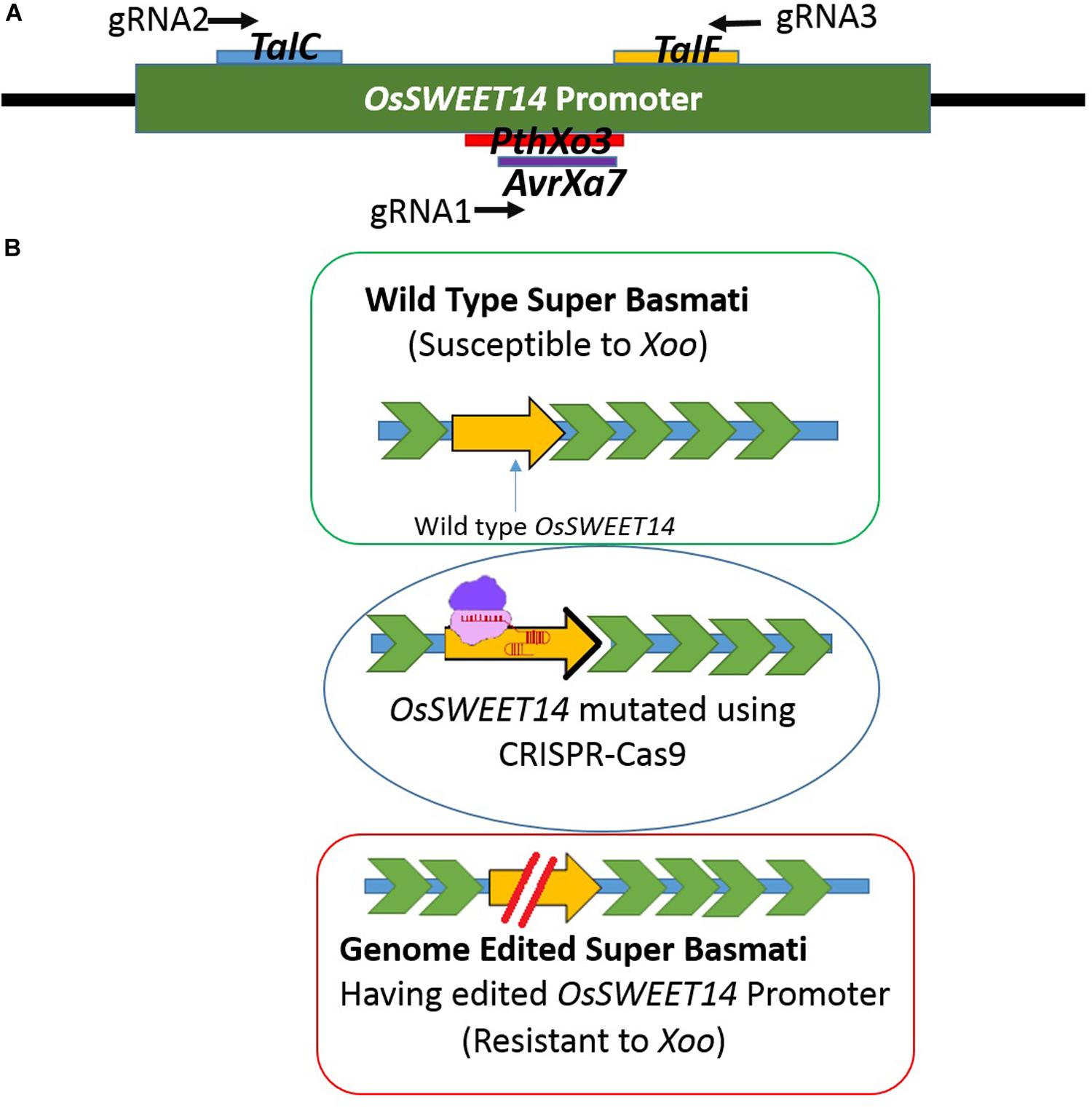
Figure 1. (A) Schematic figure of the promoter showing EBEs location on the promoter against which three gRNAs were designed. gRNA1 was designed to target PthXo3 and AvrXa7, gRNA2 for TalC and gRNA3 targeted TalF. (B) Schematic figure of developing resistance by editing the OsSWEET14 gene. The intact promoter was susceptible to Xoo whereas the EBE edited promoter can be resistant to Xoo.
Rice has been used extensively as a model for performing genome editing studies via CRISPR-Cas9 due to its diploid nature (Jiang et al., 2013; Hu et al., 2016; Abe et al., 2018). There are some prior studies where genome editing has been done to develop resistance against bacterial blight using different genome editing platforms including CRISPR-Cas9 (Blanvillain-Baufumé et al., 2017; Oliva et al., 2019; Xu et al., 2019). Customization of the CRISPR-Cas9 tool offers an avenue to target and mutate TALE binding elements in the promoter region of OsSWEET14 to resist the spread of bacterial blight (Jiang et al., 2013). Different types of natural mutations have been reported in OsSWEET14 genes in different rice cultivars which provide immunity against Xoo strains (Hutin et al., 2015b). However, no natural mutations have been reported in Super Basmati rice (Zaka et al., 2018). Here, we have employed CRISPR-Cas9 technology to engineer resistance against bacterial blight in Super Basmati rice for the first time by targeting the promoter region of the OsSWEET14 gene (Figure 1B).
Materials and Methods
gRNAs Design and Construct Development
The genome sequence of Super Basmati rice was not available in nucleotide databases. To know the exact sequence of the OsSWEET14 gene promoter and to design gRNAs, different primer sets (OsP-F: 5′ ATTGGCACTTTCTGTCATGCATG 3′ and OsP-R: 5′ GCAAGATCTTGATTAACTAGCTAGC 3′) were first designed on the OsSWEET14 gene promoter based on the sequences of other rice varieties available in the database. The genomic DNA of Super Basmati rice was extracted using the cetyl trimethylammonium bromide (CTAB) method (Stewart, 1993) followed by polymerase chain reaction (PCR). An amplicon of 457 bp in length was cloned into a pTZ57 R/T (Thermo Fisher Scientific, United States) vector and sent for sequencing. All gRNAs were designed manually to target their respective EBEs (Table 1). The overhangs of GGCA and AAAC were added a 5′ end of forward and reverse gRNA respectively for cloning at the BsaI site (Table 2). The gRNAs were screened for potential off-targets using the online tool Cas-OFFinder1. Analysis revealed that there were no off-targets. The gRNAs were synthesized, annealed, and cloned at the BsaI site in pRGEB32 (Addgene plasmid # 63142) under the rice U3 promoter (Xie et al., 2015). The pRGEB32 already has rice codon optimized Cas9 expressed under the Ubi promoter. The constructs with the Cas9 gene and gRNA cassette (shown in Figure 2) were confirmed by PCR and restriction followed by Sanger sequencing. The Cas9 gene was confirmed by PCR using specific primers (Cas9-F 5′ AGCATCGGCCTGGACATCGGC 3′ and Cas9 R- 5′ CCGGAACTTGATCATGTGGG 3′). The full length Cas9 was also confirmed in constructs using primer set 3-full Ca9 (Supplementary Table S1).

Figure 2. Schematic diagram of the construct used for genome editing. All three gRNAs were cloned in the same way. The construct was expressing gRNA under the OsU3 promoter. The rice codon-optimized Cas9 was expressing under the Ubi promoter.
Plant Material
Super Basmati (Pakistan’s indigenous rice variety) was used to establish genome editing against bacterial blight. The rice variety IR24 was used as a susceptible control. Rice seeds were obtained from DNA Markers and the Applied Genomics Lab of National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan. The seeds were manually de-husked, and surface sterilized with 70% ethanol followed by dipping in 50% (v/v) commercial bleach (having 5.25% sodium hypochlorite). After washing with distilled water, seeds were placed on MS media (Murashige and Skoog, 1962) supplemented with vitamins and 2,4-Dichlorophenoxyacetic acid (2,4-D) for callus formation.
Rice Transformation and Growth Conditions
The 28 days old embryogenic calli were selected and placed on an osmotic medium with 0.25 M mannitol. The constructs with gRNA and Cas9 were confirmed again and coated on 1 μm gold particles (Bio-Rad, United States) for biolistic transformation. The rice calli were transformed through bombardment by a gene gun (PDS-1000/Bio-Rad, United States) following optimized protocol for biolistic transformation of Super Basmati rice (Mukhtar and Hasnain, 2018). For each gRNA approximately 4,000 calli were transformed. The transformed calli were placed at 28–30°C for 24 h in a dark room under sterile conditions. After 24 h, the transformed calli were shifted to the selection medium with 50 mg/L hygromycin. After 15 days they were again shifted to fresh selection medium. After completing 30 days on the selection medium, the surviving calli were shifted to pre-regeneration and then to regeneration media. The regenerated plantlets were shifted to rooting media. After the development of roots, they were transferred to soil and kept in the greenhouse under controlled conditions. After 15 days the plants were shifted to large pots and allowed to grow to maturity and the seeds were then harvested.
Confirmation of Transgenic Rice
To confirm the presence of Cas9 and gRNA cassette, DNA was extracted from all plants that were developed through tissue culture, using the CTAB method (Stewart, 1993). The construct was confirmed by PCR using forward primer at vector backbone (OsPRGEB32 F 5′ GGTGCTACCAGCAAATGC TGGAAGCCG3′) and reverse primer designed on gRNA (OsPgRNA1-R: 5′-AAACGGTTGGAGGGGGTTTATATC-3′, OsPgRNA2-R: 5′-AAAC GCTGCTGACATGCATGCCC-3′ and OsPgRNA3-R: 5′-AAAC TTCAAGCAAAGCAAACTCAC-3′ for gRNA 1, 2, and 3 constructs, respectively). The 273 bp amplicon confirmed the presence of construct (Supplementary Figure S2). Gene specific primers were used to confirm the presence of Cas9 (Table 1- primer set 2 (partial Cas9) and 3 (full Cas9). Promoter fragments of the OsSWEET14 gene were amplified using primers (Supplementary Table S1- primer set 1) from all the plants and PCR products were sent for Sanger sequencing.
T7 Endonuclease Assay
For the T7 endonuclease assay, the genomic DNA was extracted from T0 plants and the flanking target region was amplified using primer set 1 (Supplementary Table S1- OsP-F and OsP-R) to amplify the promoter (Supplementary Table S1). The purified PCR amplicons were denatured and renatured then subjected to T7 endonuclease I (NEB, M0302) following the manufacturer’s instructions. The products were resolved on 2% agarose gel and then stained with ethidium bromide.
Screening Against Bacterial Blight Resistance
The screening of edited lines against bacterial blight was performed under containment glasshouse conditions. The 1st round of screening was performed on T0 plants. Two plants from each line were selected for inoculation. T1 seeds were then collected from primary transformants. Seeds from each line were sown in separate pots containing soil (Excluding SB-E1 because seed filling was disturbed for this line). When the plants reached the three leaf stage, they were arranged in different batches for screening against bacterial blight. The experiment was conducted in three batches. There were six pots in each batch. Each pot had three plants of wild type Super Basmati, SB-E2, SB-E3, SB-E4, IR-4 (susceptible check), and a negative control. So, the edited plants were arranged along with their negative, susceptible, and wild type controls. There were three pots for each line, so a total of nine plants were inoculated for each line. The wild type Super Basmati has no editing, and the IR-24 line is susceptible to almost all kinds of Xoo strains, and was therefore used as a susceptible control. The negative control is the plant for which scissors were dipped in distilled water, without any Xoo strain, to check the lesion length introduced due to scissor injury. All of these lines were screened for bacterial blight resistance using a local virulent Xoo strain. This strain was selected on the basis that one of the four EBEs of OsSWEET14 is AvrXa7 which was present in the strain. The inoculum was prepared as described by Tu et al. (2000). All the lines were maintained in a uniform environment in the glasshouse at 30°C and 85% relative humidity. For leaf clip inoculation, 45 days old plants were used. For each plant three to four leaves were uniformly inoculated with scissors dipped in bacterial suspension (Kauffman, 1973). At 14 days post-inoculation (dpi) the plant’s responses were observed and data was recorded by measuring the full length of the leaf and lesion length in centimeters (cm). The percentage disease leaf area (%DLA) was then calculated using the below formula.
RNA Extraction, cDNA Synthesis and RT-qPCR
The 30 days old rice seedlings were inoculated with scissors dipped in Xoo suspension. The total RNA was extracted after 24 h post-inoculation (hpi) from rice leaves using TRIzol reagent following the manufacturer’s instructions (Invitrogen, United States). The isolated RNA was treated with DNaseI as per the manufacturer’s instructions (Thermo Fisher Scientific, United States). The complementary DNA (cDNA) was synthesized from RNA using a RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific, United States) following instructions given by the manufacturer. The induction of OsSWEET14 by Xoo was determined in wild type as well as in edited plants by RT-qPCR. The OsSWEET14 transcripts were amplified using OsSWEET14-specific primers OsSWEET14-RT-F/OsSWEET14-RT-R (Supplementary Table S1). A volume of 25 μL reaction mixture was made using 12.5 μL SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific, United States), 0.1 pmole of forward and reverse primers, 2.5 μL cDNA (∼25 ng), and 9.5 μL water. The conditions were optimized and finally the reaction was performed using a Bio-Rad iQ5 thermal cycler (Bio-Rad, United States). The expression of sucrose phosphate synthase (SPS) was used as an internal control and was amplified using primers SPS-F/SPS-R (Supplementary Table S1). The quantification results were analyzed using the 2–ΔΔΔCt method (Livak and Schmittgen, 2001). Each reaction was performed in triplicate.
Phenotyping
The plants were observed visually during growth. After harvesting the seeds from edited plants, the germination of the seeds was checked. The seeds were dehusked, sterilized and placed in six-well plates in water and the rate of germination was recorded. This was done for all the lines except SB-E1 because we were unable to obtain seeds from this line. The root and shoot lengths were measured by growing three to four seeds from each line vertically on square plates with ½ MS media (no sucrose was added). After 1 week, the root and shoot lengths were again measured and compared with the wild type.
Results
Development of Constructs
The sequence of the OsSWEET14 gene promoter was amplified using the primer set OsP-F/OsP-R (Supplementary Table S1). This primer set was designed on the sequences of other rice varieties available in the database. Upon amplification a PCR, a product of 457 bp, was amplified which was cloned and sequenced. The sequenced region of the OsSWEET14 promoter from Super Basmati rice was submitted to Genbank and is available online under Accession No. MK791135.1. Three different gRNAs (Table 2) were designed to target EBEs (Table 1) of AvrXa7, PthXo3, TalF, and TalC in the promoter region. Due to overlapping EBEs for AvrXa7 and PthXo3, the first gRNA (gRNA1) was designed to target both EBEs simultaneously. Similarly, gRNA2 and gRNA3 were designed to target TalC and TalF, EBEs, respectively.
The constructs containing the Cas9 gene and gRNA cassette were initially confirmed by PCR. Gene specific primers were used to confirm the Cas9 gene with an amplicon length of 490 bp (Supplementary Figure S3A). In the final constructs, full-length Cas9 (4100 bp) was confirmed (Supplementary Figure S3B). The gRNA cassette was confirmed using the forward primer (OsPRGEB32 F GGTGCTACCAGCAAATGCTGGAAGCCG) and reverse primer of the respective gRNA (Table 2). The 273 bp product confirmed the presence of gRNA along with its scaffold in the pRGEB32 vector (Supplementary Figure S1A). The presence of gRNAs in constructs was also confirmed by restriction analysis. gRNA insertion in pRGEB32 results in disruption of the BsaI site. Due to this disruption, restriction with BsaI and HindIII enzymes did not release 400 bp fragments in positive clones. Thus, the absence of 400 bp confirmed that all the gRNAs were successfully cloned (Supplementary Figures S1B,C). Finally, all constructs were confirmed by Sanger sequencing (Supplementary Figure S1D).
Transgenic Plants Development
The process of transgenic plant development and screening against bacterial blight is shown in Supplementary Figures S4B,C. Cas9 expressing transgenic rice lines were developed under the Ubi promoter. All the three gRNAs (designated as gRNA1, gRNA2, and gRNA3) were expressed under rice U3 promoter in a pRGEB32 vector with rice codon-optimized Cas9. A total of 48 transgenic lines were obtained for gRNA1, 35 lines for gRNA2, and 51 lines for gRNA3. Both edited and wild type lines were maintained at 30°C in a containment glasshouse.
Mutation Detection
To determine whether CRISPR-Cas9 was able to cleave and generate DSBs at the target site and repaired it via NHEJ, total genomic DNA was isolated using the CTAB method. Target regions were amplified using PCR and subjected to T7 Endonuclease I (T7EI) mutation detection analysis which degrades single-stranded regions of non-complementarity resulting from indels; these regions are then detected by sequencing. The 457 bp target region from T0 was digested with T7E1 and the expected bands (200 and 250 bp approximately) were observed in all five edited lines (Figure 3C). The wild type plants did not show these bands and thus confirmed editing in five lines. Moreover, to further confirm the editing, a 457 bp promoter fragment of the OsSWEET14 gene from all transgenic plants was sent for Sanger sequencing. We were successful in obtaining two transgenic lines with a mutated promoter fragment for gRNA1 (targeting AvrXa7 and PthXo3). Out of these two lines, one line had 24 bp deletion while the second line showed 4 bp deletion at the target site (Figure 3A). We were unable to obtain transgenic lines for gRNA2. All those lines that were edited with gRNA2 had intact EBE of TalC. Out of 51 transgenic lines for gRNA3, we were able to obtain three lines with 4, 5, and 18 bp deletions at the target site. Thus, in two of the three lines (having 4 and 18 bp deletions), we successfully disrupted the EBE of TalF. In the third line, 5 bp deletion was not able to disrupt the EBE of TalF (Figure 3B). These results showed that CRISPR-Cas9 was able to do editing at the target site while no such activity was observed in the control plants. The nature of mutations was also observed in edited lines. We obtained only one biallelic mutation in SB-E1 while the rest of the mutations were mono-allelic (Supplementary Table S2). However, the edits were transmitted successfully to their progeny.
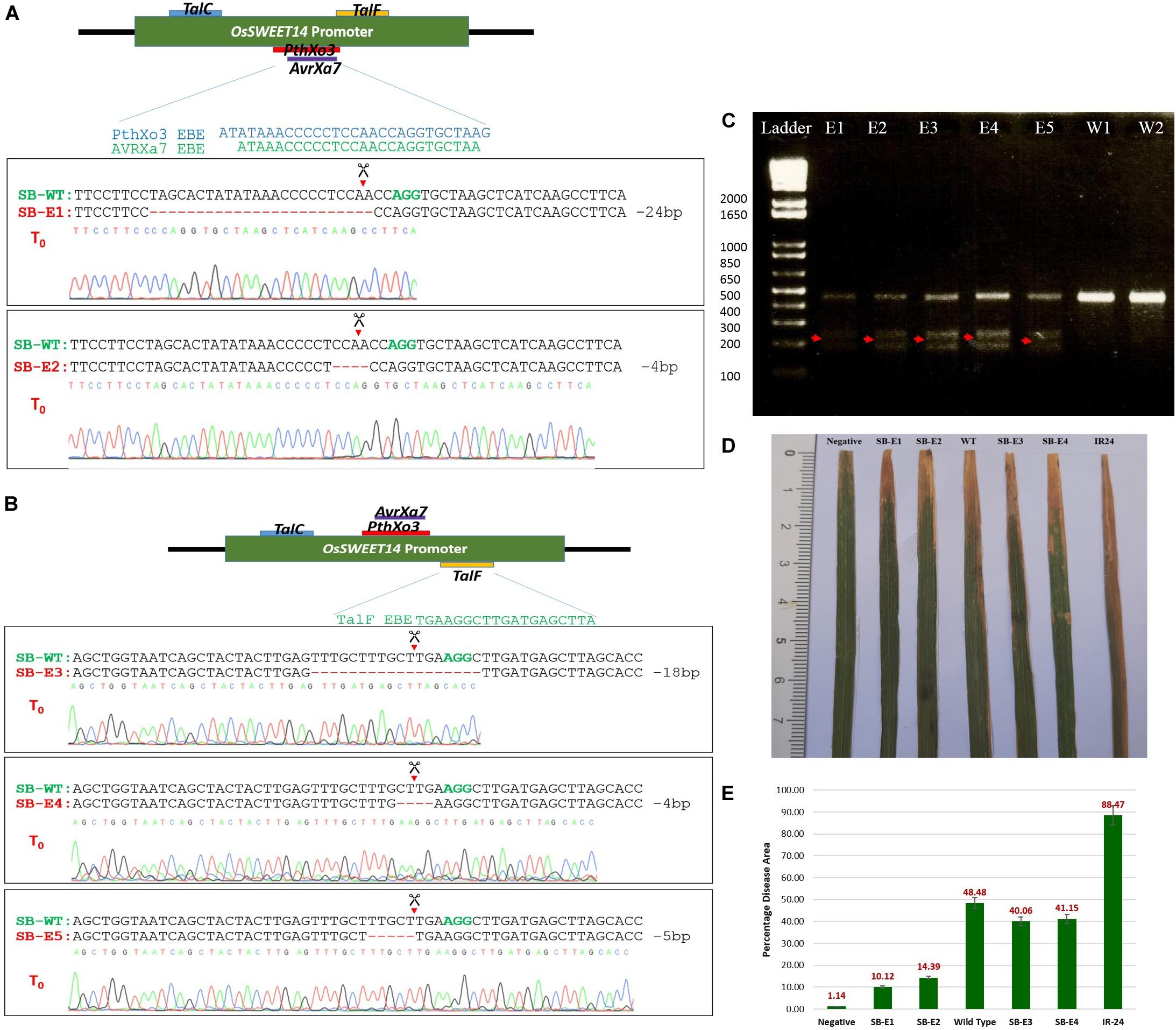
Figure 3. (A) Sanger sequencing of AvrXa7 and PthXo3 EBE edited plants. The plants were named as SB-WT (Super Basmati wild type), SB-E1 (Super Basmati edited 1) and SB-E2 (Super Basmati edited 2). Green colored bases AGG represents the PAM sequence and red dashes symbolize the deletion at the target site. SB-E1 and SB-E2 have 24 and 4 bp deletion at the target site, respectively. (B) Sanger sequencing of TalF EBE edited plants. The plants were named SB-WT (Super Basmati wild type), SB-E3 (Super Basmati edited 3), SB-E4 (Super Basmati edited 4), and SB-E5 (Super Basmati edited 5). SB-E3 and SB-E4 have 18 and 4 bp deletion at the target site, respectively, disrupting TalF EBE, whereas SB-E5 had 5 bp deletion at the target site but TalF EBE remained intact. (C) T7 Endonuclease assay of T0 edited and wild type plants. Red arrows showed the bands from edited plants that are absent in wild type plants (E1 = SB-E1, E2 = SB-E2, E3 = SB-E3, E4 = SB-E4, E5 = SB-E5, W1 and W2 = Wild Type Super Basmati). (D) Lesions induced by Xoo after 14 days post-inoculation on edited and control plants. SB-E1 and SB-E2 has reduced lesion length as compared to control plants. Negative = Plant was inoculated with scissors dipped in water. All the other lines were inoculated with scissors dipped in Xoo suspension. (E) Average of Percentage disease leaf area (% DLA) of the edited and control plants. The lowest DLA 10.12% was observed in the case of SB-E1 plants where 24 bp deletion was detected. The highest DLA 88.47% was present in IR-24 which was used as a susceptible check.
Bacterial Blight Resistance Assays
The edited lines were challenged with a prevalent Xoo strain via the leaf clip inoculation method. The strain has AvrXa7 TALE. The presence of AvrXa7 was not only confirmed by sequencing but the same strain was screened against a rice cultivar (IRBB7) which showed resistance against this strain. IRBB7 has Xa7 gene, which provides resistance against Xoo isolates with TALE AvrXa7 (Yang et al., 2000). This strain was tested to induce the OsSWEET14 gene. It was able to induce OsSWEET14. Therefore, this strain was selected for inoculating edited lines. The wild type Super Basmati rice was used as the control and IR24 (IRRI Line) as a susceptible check. All the inoculated lines were kept at 30°C in a greenhouse with 85% humidity. Two plants from the control and T0 edited lines, whereas nine plants from control and T1 edited lines were inoculated. The percentage disease leaf area (%DLA) for three inoculated leaves from each plant was calculated (Figure 3E and Supplementary Tables S3, S4). After 14 days post-inoculation, plants were observed for bacterial blight infection. There were significant differences in the rate of infection among edited and control lines. The disease area among edited and control lines were compared in terms of %DLA covered by infection. In AvrXa7/PthXo3 edited lines, the incidence of infection was very low as compared to the wild type Super Basmati and IR24 lines (Figure 3D). The lowest DLA (10.12%) covered by infection was seen in the case of Super Basmati edited line 1 (SB-E1) in which 24 bp deletion in overlapping EBEs of AvrXa7 and PthXo3 occurred. The other variant, SB-E2, showed 14.4% DLA, which was also lower than the control plants. SB-E3 and SB-E4, with disrupted TalF EBE and intact AvrXa7/PthXo3 EBE, showed 40 and 41% DLA, respectively, when inoculated with the same strain (possessing only AvrXa7). 48 and 88% disease incidence were observed for wild type Super Basmati and IR24, respectively. This confirmed our hypothesis that editing EBEs in the promoter of the OsSWEET genes provides resistance against corresponding TALEs.
Relative OsSWEET14 Induction by Xoo Strain
Xoo strains encode different TALEs to activate endogenous SWEET genes for a successful infection. The OsSWEET14 activation by the Xoo strain in wild type Super Basmati Rice and edited lines were determined using real-time quantitative PCR. Expression of OsSWEET14 was induced by inoculating both wild type and edited lines by the Xoo strain. Five lines [(1) Non-inoculated wild type Super Basmati, (2) Inoculated wild type Super Basmati, (3) Inoculated edited line SB-E2, (4) Inoculated SB-E3, and (5) Inoculated SB-E4] were selected to compare OsSWEET14 activation by Xoo. Upon infiltration with the Xoo strain, the expression of OsSWEET14 was considerably high in WT-SB as compared to non-inoculated WT-SB (Figure 4). The edited line (SB-E2) carrying a mutation for AvrXa7, showed very low expression of OsSWEET14. Whereas in the SB-E3 and SB-E4 lines (carrying edited TalF but intact AvrXa7), the strain was still able to induce OsSWEET14. This clearly indicates that the Xoo strain was able to induce OsSWEET14 expression to establish successful infection in Super Basmati rice. In addition, the induction of OsSWEET14 expression in SB-E3 and SB-E4 edited lines clearly showed that the Xoo strain contained AvrXa7 TALE to infect Super Basmati rice. As SB-E2 edited line has mutated EBE for AvrXa7 and the strain was unable to induce OsSWEET14 expression. As a result, relative expression of OsSWEET14 was significantly lower in the SB-E2 edited line as compared to other inoculated lines.
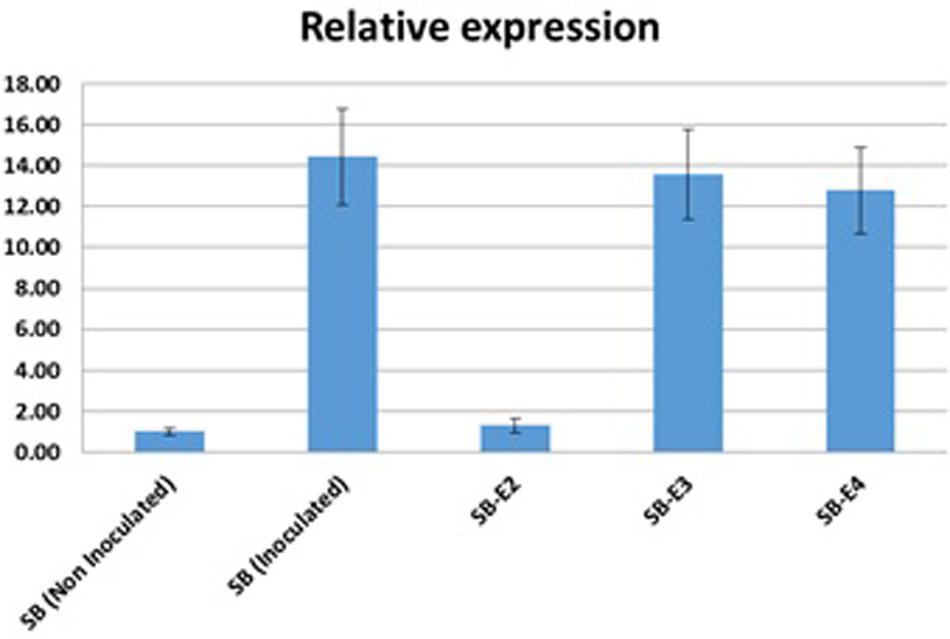
Figure 4. OsSWEET14 Induction by Xoo strain: Relative mRNA levels (2–ΔΔCt) of OsSWEET14 in leaves of wild type and edited rice lines were compared. qRT-PCR was conducted in wild type Super Basmati and edited lines (SB-E2, SB-E3, SB-E4). Samples were harvested after 24 h of Inoculation with a locally virulent Xoo strain (mean ± s.e.m., n = 3 leaf samples from biological replicates) with expression normalized to rice SPS levels; repeated independently three times with comparable results.
Phenotyping
The plants were carefully observed for any phenotypic changes. All the edited plants showed a normal phenotype and were fertile just like the wild type plants. The harvested seeds also showed a normal phenotype. The germination was checked for seeds from all the edited lines (Except SB-E1) and seeds showed normal germination (Supplementary Figure S5). The shoot and root lengths were also normal for the edited lines (Figure 5).
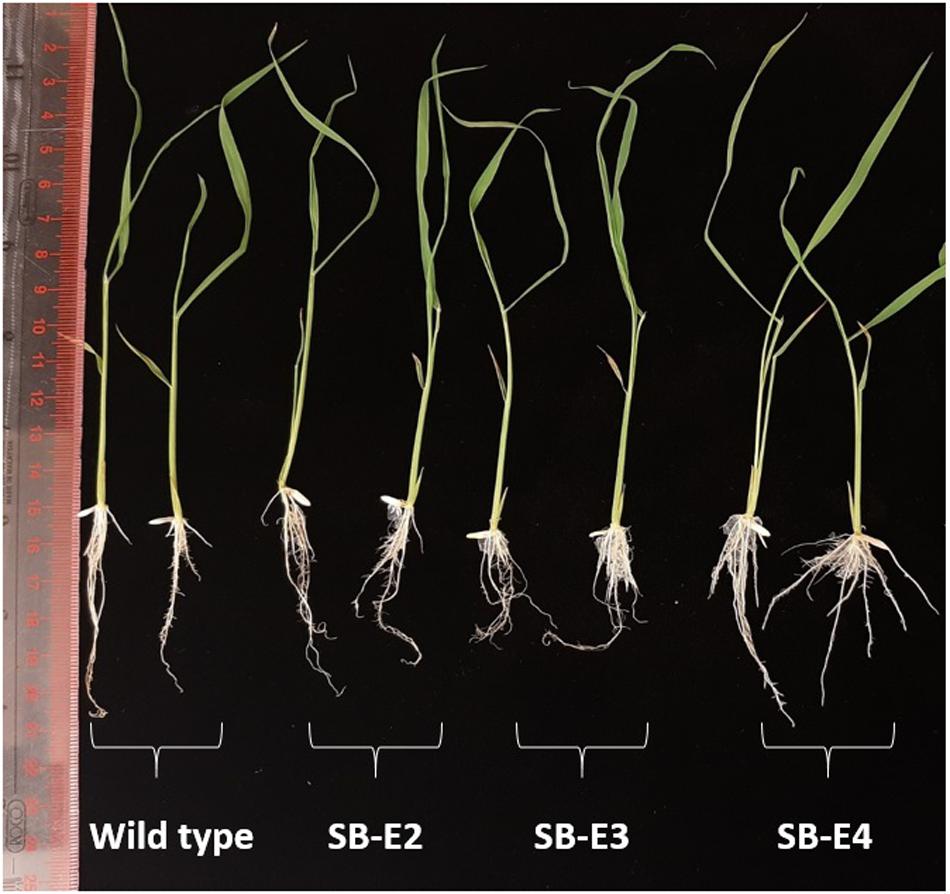
Figure 5. Root length and shoot length of wild type and edited plants. Edited plants show normal root and shoot length just like wild type plants (two representative plants are shown in figure out of three).
Discussion
In a global scenario, bacterial blight is a destructive disease of rice, and in Asia specifically, it severely damages crop yield. Traditionally, various approaches have been used to develop resistance in rice cultivars which include the introgression of resistance genes (R genes), host-derived resistance, and loss of susceptibility through mutation in recessive R genes (Dou and Zhou, 2012; Kim et al., 2015; Busungu et al., 2016; Li et al., 2019). Despite the effectiveness of R genes, the constant introduction of R-genes in rice breeding programs may result in the emergence of new Xoo strains, which can overcome R gene-mediated resistance (Ji et al., 2016). With the advent of new technologies like genome editing, various strategies have been employed, e.g., TALENs and CRISPR-Cas9 to develop disease resistance (Khan et al., 2014). Here in this study we used CRISPR-Cas9 technology to develop disease resistance in Basmati rice.
One approach which can be used to remove disease predisposition is to mutate the susceptible portion of the genome (Iyer-Pascuzzi and McCouch, 2007; Hutin et al., 2015a; Kourelis and van der Hoorn, 2018; Zaidi et al., 2018). The idea to mutate the bacterial blight susceptible region of the rice genome sparked from the presence of three naturally occurring recessive R genes, i.e., xa13, xa25, and xa41 (t). These are mutated alleles of OsSWEET11, OsSWEET13, and OsSWEET14, respectively, which offers resistance against their respective TALEs present in the Xoo strain (Chu et al., 2006; Yang et al., 2006; Yuan et al., 2009; Hutin et al., 2015b). It has been reported that Xa41(t) (having 18 bp deletion) offers broad-spectrum resistance against a large collection of Xoo strains (Hutin et al., 2015b). Recently it was reported that some Xoo strains still cause infection in rice despite R genes (Carpenter et al., 2018; Doucouré et al., 2018). These failures have prompted plant genome engineers to develop resistance against this rapidly evolving pathogen by mutating susceptibility genes. TALENs have been employed in the past, but extensive protein engineering makes developing broad-spectrum immunity against bacterial blight difficult (Li et al., 2012). The prokaryotic immune system (CRISPR-Cas9) was customized and used for editing susceptibility genes in rice (Jiang et al., 2013). In the current study this customized tool was used for editing the OsSWEET14 gene’s promoter of Basmati, to obtain resistance against bacterial blight.
Basmati rice is famous for its long grain and aroma and none of the natural mutations are reported in OsSWEET14 for Super Basmati rice. For the OsSWEET13 gene, one deletion and substitutions are naturally present in the promoter which disrupts all the EBE variants of PthXo2 TALE, i.e., PthXo2A, PthXo2B, and PthXo2C (Zaka et al., 2018; Oliva et al., 2019). It was reported that in the majority of cases, Xoo strains induce the expression of OsSWETT14 for the onset of infection because it has the EBEs of four different TALEs. OsSWEET14 was targeted previously as a single target as well as in combination with other OsSWEET genes to create broad-spectrum resistance in different rice cultivars (Blanvillain-Baufumé et al., 2017; Oliva et al., 2019; Xu et al., 2019). They have developed different lines with edited EBEs of the OsSWEET14 gene’s promoter. Each EBE edited line provided resistance against specific TALE. In the present study, the OsSWEET14 gene’s promoter was selected to modify the respective EBEs of four TALEs via CRISPR-Cas9. We have successfully mutated EBEs of three TALEs (AvrXa7, PthXo3, and TalF). The edited rice lines (SB-E1, SB-E2, SB-E3, and SB-E4) have modified alleles of OsSWEET14. Among these four edited lines, SB-E1 and SB-E2 did not respond to AvrXa7/PthXo3 to establish a successful infection. The new germplasm created in this way showed resistance against a locally virulent Xoo strain. These results indicated that CRISPR-Cas9 technology can also be employed on Basmati rice to create resistance against bacterial blight.
Blanvillain-Baufumé et al. (2017) created the allele library of OsSWEET14 for developing resistance against bacterial blight. The OsSWEET14 gene’s promoter was targeted using TALENS, and they observed 51% editing efficiency. In the current study the EBEs of the OsSWEET14 gene’s promoter were targeted using CRISPR-Cas9 and we were able to obtain only 9% editing efficiency. All the editing events were in close proximity to the predicted cleavage site by Cas9 which is three base pairs upstream of the PAM sequence. This indicates the specificity of this editing system. However, we observed low editing efficiency as compared to the previous report of editing the OsSWEET14 gene’s promoter. The reason for low editing efficiency could be the difference in the editing tool (TALENs) and rice variety as they have used a non-Basmati background. The editing efficiency can be improved by using Cas9 fusion with chromatin-modulating peptides (CMPs), derived from high mobility group proteins. This fusion exhibited many folds improved activity (Ding et al., 2019). Thus, such type of improved systems can be tested in elite varieties, like Super Basmati, to check improvement in editing efficiency. Blanvillain-Baufumé et al. (2017) obtained mixed events (insertion/deletion) at the target sites including the insertion of 22 bp and deletions of up-to 51 bp, whereas another study by Jiang et al. (2013) reported only deletions at the target site. In our case, we were only able to obtain deletions at the target sites, because the repairing of DSB (introduced by CRISPR-Cas9) was done through a random NHEJ process. Therefore, obtaining deletions, insertions, or point mutations at the target site is solely dependent on the host repair machinery.
The diversity of TALEs present in different Xoo strains was studied previously by analyzing Xoo genome sequence data. Most of the Asian strains had AvrXa7 (OsSWEET14) and PthXo2 (SWEET13) (Oliva et al., 2019). Sequencing data of the most prevalent Xoo strain infecting Basmati showed that this strain had AvrXa7 for infecting Basmati. The induction of OsSWEET14 was also checked by the tested Xoo strain and RT-qPCR results confirmed OsSWEET14 induction by the tested Xoo strain. So, targeting the promoter of OsSWEET14 in Super Basmati is very effective in generating broad-spectrum resistance. However, this strategy can be modified depending on the TALEs present in Xoo strains and their respective EBEs in rice cultivars. More work is still required, however, in order to deal with bacterial blight by characterizing maximum Xoo isolates. Some Xoo TALEs also target OsSWEET13 and OsSWEET11 for the onset of infection. But the isolates which we have characterized were mostly targeting OsSWEET14, which agrees with studies reported by Blanvillain-Baufumé et al. (2017).
In a previous report it was shown that Asian Xoo strains did not have TalF (Oliva et al., 2019). Our study also supports their finding because our locally isolated strain did also not possess TalF. The selected Xoo strain was inoculated on all the edited lines and disease development was recorded. The symptoms were reduced on AvrXa7 edited lines. When TalF-edited lines were challenged with the same Xoo strain, the symptoms were not reduced. This was further confirmed by RT-qPCR of the control and edited lines. The relative expression of OsSWEET14 was similar in wild type SB and TalF edited lines (SB-E3, SB-E4), whereas the strain was unable to induce OsSWEET14 in the AvrXa7 edited line (SB-E2). This showed that the strain has AvrXa7 to infect Super Basmati. These findings confirmed our hypothesis that targeting EBEs in the promoter region only provides specific resistance to their corresponding TALE present in the Xoo strain.
Mutations in susceptibility genes can also have side effects on normal plant physiology (van Schie and Takken, 2014). In a previous study, rice plants with a OsSWEET14 TDNA insertion mutant had smaller seeds as compared to wild type plants, although it showed resistance against Xoo strains (Antony et al., 2010). In contrast, OsSWEET14 EBE edited rice plants were no longer susceptible to Xoo and showed normal growth (Li et al., 2012). In the current study the edited lines were visually observed during the growth period and after harvest. All the edited lines showed no detectable growth defects in greenhouse conditions except for SB-E1. In SB-E1 seed filling was disturbed. The reason could be attributed to a large deletion in the promoter region. This also indicates the role of OsSWEET genes in seed filling (Sosso et al., 2015; Yang et al., 2018; Hu et al., 2019). However, further investigation is required in this regard. Thus, in comparison to former studies, we came across both types of results, and out of four EBE mutated lines we observed abnormality in only one edited line.
There are some previous reports of CRISPR-Cas9 mediated genome editing of rice to develop resistance against bacterial blight. Editing in the promoter fragment of OsSWEET genes was reported using CRISPR-Cas9 to develop resistance against bacterial blight (Oliva et al., 2019; Xu et al., 2019). But the majority of previous genome editing was performed in rice cultivar kitake (japonica) and cannot be used for breeding programs of Basmati rice. Recently, genome editing via CRISPR-Cas9 was performed to mutate EBEs of OsSWEET11, OsSWEET13, and OsSWEET14 to create resistance in Indica cultivar (IR-64 and Ciherang-Sub1). These cultivars can be used by breeders of Asia and Africa in breeding programs but there was no report on creating resistance in the Basmati cultivars. Therefore, we performed genome editing in the elite Super Basmati rice cultivar to be incorporated into breeding programs. The present study was designed to establish CRISPR-Cas9 mediated genome editing in Basmati rice. To the best of our knowledge, this is the first report of genome editing in Basmati rice using the CRISPR-Cas9 toolbox.
This study shows the potential of CRISPR-Cas9 based genome editing in elite Super Basmati rice. However, there is still more work needed to deal with bacterial blight, by characterizing the maximum of local Xoo isolates, because some Xoo TALEs also target OsSWEET13 and OsSWEET11. So, to create broad-spectrum resistance against all the native Xoo strains, multiplexing can also be done in Basmati. Further, a study can be planned to establish multiplex genome editing against bacterial blight by simultaneously targeting multiple EBEs in the promoter regions of OsSWEET genes, which will ultimately result in reducing bacterial blight. This approach of dealing with bacterial blight by targeting the promoter of OsSWEET genes will not prevent the adaptation of pathogens. The strength of this method also depends on the ability of local Xoo strains to acclimate to recessive resistance alleles. To develop durable resistance against pathogens, it is better to create major changes in EBEs of the OsSWEET gene’s promoter. Combining these edited alleles with locally effective resistance genes can be a more effective way to reduce disease pressure. In conclusion, by understanding TALE interaction with EBEs of OsSWEET14 genes, we were able to create resistance against a corresponding Xoo strain.
Genome editing can have off-target effects. The gRNAs used in this study were analyzed for off-targeting against the rice genome available in the database. All the gRNAs did not have any off-targets. Furthermore, if any off-target mutations are still present as a result of genome editing or tissue culturing, they will be eliminated during crossing. Full genome sequencing will be required to confirm any off-targeting. Finally, our initial data shows the potential of CRISPR-Cas9 based genome editing in elite Basmati cultivar. However, the establishment of a transgene-free genome editing protocol for targeting multiple EBEs simultaneously is still needed in Super Basmati rice to create broad-spectrum resistance against a large collection of Xoo strains. Such transgene-free genome-edited elite lines created in this way can be added in breeding programs.
Conclusion
Our results show that targeting EBEs of respective TALEs, employing the CRISPR-Cas9 approach, can provide highly selective and promising immunity against bacterial blight.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
SM planned the work. KZ, MK, and IA planned the experiments. KZ performed all the experiments and wrote the manuscript. MK and IA edited and finalized the manuscript. ZM helped establishing tissue culture of Super Basmati. SY and KE performed the resistance assays against Xoo. MA provided the seeds for this study and give critical suggestions.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00575/full#supplementary-material
Footnotes
References
Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. (2018). Production of high oleic/low linoleic rice by genome editing. Plant Physiol. Biochem. 131, 58–62. doi: 10.1016/j.plaphy.2018.04.033
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864–3876. doi: 10.1105/tpc.110.078964
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Blanvillain-Baufumé, S., Reschke, M., Solé, M., Auguy, F., Doucoure, H., Szurek, B., et al. (2017). Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET 14−inducing TAL effectors. Plant Biotechnol. J. 15, 306–317.
Busungu, C., Taura, S., Sakagami, J.-I., and Ichitani, K. (2016). Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 66, 636–645. doi: 10.1270/jsbbs.16062
Carpenter, S. C., Mishra, P., Ghoshal, C., Dash, P. K., Wang, L., Midha, S., et al. (2018). A strain of an emerging Indian pv. oryzae pathotype defeats the rice bacterial blight resistance gene without inducing a clade III gene and is nearly identical to a recent Thai isolate. Front. Microbiol. 9:2703. doi: 10.3389/fmicb.2018.02703
Chen, L.-Q., Hou, B.-H., Lalonde, S., Takanaga, H., Hartung, M. L., Qu, X.-Q., et al. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532. doi: 10.1038/nature09606
Chen, L.-Q., Qu, X.-Q., Hou, B.-H., Sosso, D., Osorio, S., Fernie, A. R., et al. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211. doi: 10.1126/science.1213351
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Ding, X., Seebeck, T., Feng, Y., Jiang, Y., Davis, G. D., and Chen, F. (2019). Improving CRISPR-Cas9 genome editing efficiency by fusion with chromatin-modulating peptides. CRISPR J. 2, 51–63. doi: 10.1089/crispr.2018.0036
Dou, D., and Zhou, J.-M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Doucouré, H., Pérez-Quintero, A. L., Reshetnyak, G., Tekete, C., Auguy, F., Thomas, E., et al. (2018). Functional and genome sequence-driven characterization of TAL effector gene repertoires reveals novel variants with altered specificities in closely related Malian Xanthomonas oryzae pv. oryzae strains. Front. Microbiol. 9:1657. doi: 10.3389/fmicb.2018.01657
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, B., Wu, H., Huang, W., Song, J., Zhou, Y., and Lin, Y. (2019). SWEET gene family in Medicago truncatula: genome-wide identification, expression and substrate specificity analysis. Plants 8:338. doi: 10.3390/plants8090338
Hu, X., Wang, C., Fu, Y., Liu, Q., Jiao, X., and Wang, K. (2016). Expanding the range of CRISPR/Cas9 genome editing in rice. Mol. Plant 9, 943–945. doi: 10.1016/j.molp.2016.03.003
Hutin, M., Pérez-Quintero, A. L., Lopez, C., and Szurek, B. (2015a). MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front. Plant Sci. 6:535. doi: 10.3389/fpls.2015.00535
Hutin, M., Sabot, F., Ghesquière, A., Koebnik, R., and Szurek, B. (2015b). A knowledge−based molecular screen uncovers a broad−spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042
Iyer-Pascuzzi, A. S., and McCouch, S. R. (2007). Recessive resistance genes and the Oryza sativa-Xanthomonas oryzae pv. oryzae pathosystem. Mol. Plant Microbe Interact. 20, 731–739.
Ji, Z., Ji, C., Liu, B., Zou, L., Chen, G., and Yang, B. (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 7:13435. doi: 10.1038/ncomms13435
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41:e188. doi: 10.1093/nar/gkt780
Kauffman, H. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Khan, M. A., Naeem, M., and Iqbal, M. (2014). Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol. 139, 27–37. doi: 10.1007/s10658-014-0377-x
Khush, G. S. (2005). What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 59, 1–6. doi: 10.1007/s11103-005-2159-5
Kim, S.-M., Suh, J.-P., Qin, Y., Noh, T.-H., Reinke, R. F., and Jena, K. K. (2015). Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theor. Appl. Genet. 128, 1933–1943. doi: 10.1007/s00122-015-2557-2
Kourelis, J., and van der Hoorn, R. A. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Li, C., Li, W., Zhou, Z., Chen, H., Xie, C., and Lin, Y. (2019). A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene−free bacterial blight−resistant rice. Plant Biotechnol. J. 18, 313–315. doi: 10.1111/pbi.13217
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., Dicarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Mukhtar, Z., and Hasnain, S. (2018). Optimization of particle bombardment conditions for rice (Oryza sativa L.) transformation. Pak. J. Agric. Sci. 55, 271–278. doi: 10.21162/pakjas/18.3678
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
Pazuki, A., and Sohani, M. M. (2013). Phenotypic evaluation of scutellum-derived calluses in ‘Indica’rice cultivars. Acta Agric. Slov. 101, 239–247.
Sasaki, T. (2001). Rice genome analysis to understand the rice plant as an assembly of genetic codes. Photosynth. Res. 70, 119–127.
Shobarani, N., Prasad, G., Prasad, A., Sailaja, B., Muthuraman, P., Numeera, S., et al. (2010). Rice Almanac–India. DRR Tech. Bull. 5, 6–7.
Sosso, D., Luo, D., Li, Q.-B., Sasse, J., Yang, J., Gendrot, G., et al. (2015). Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 47, 1489–1493. doi: 10.1038/ng.3422
Stewart, C. N. Jr. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14, 748–750.
Streubel, J., Pesce, C., Hutin, M., Koebnik, R., Boch, J., and Szurek, B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector−mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200, 808–819. doi: 10.1111/nph.12411
Tu, J., Datta, K., Khush, G., Zhang, Q., and Datta, S. (2000). Field performance of Xa21 transgenic indica rice (Oryza sativa L.), IR72. Theor. Appl. Genet. 101, 15–20. doi: 10.1007/s001220051443
van Schie, C. C., and Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Wright, A. V., Nuñez, J. K., and Doudna, J. A. (2016). Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell 164, 29–44. doi: 10.1016/j.cell.2015.12.035
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12, 1434–1446. doi: 10.1016/j.molp.2019.08.006
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yang, B., Zhu, W., Johnson, L. B., and White, F. F. (2000). The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 97, 9807–9812. doi: 10.1073/pnas.170286897
Yang, J., Luo, D., Yang, B., Frommer, W. B., and Eom, J. S. (2018). SWEET11 and 15 as key players in seed filling in rice. New Phytol. 218, 604–615. doi: 10.1111/nph.15004
Yu, Y., Streubel, J., Balzergue, S., Champion, A., Boch, J., Koebnik, R., et al. (2011). Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant Microbe Interact. 24, 1102–1113. doi: 10.1094/MPMI-11-10-0254
Yuan, M., Chu, Z., Li, X., Xu, C., and Wang, S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 50, 947–955. doi: 10.1093/pcp/pcp046
Zaidi, S. S.-E.-A., Mukhtar, M. S., and Mansoor, S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906. doi: 10.1016/j.tibtech.2018.04.005
Zaka, A., Grande, G., Coronejo, T., Quibod, I. L., Chen, C.-W., Chang, S.-J., et al. (2018). Natural variations in the promoter of OsSWEET13 and OsSWEET14 expand the range of resistance against Xanthomonas oryzae pv. oryzae. PLoS One 13:e0203711. doi: 10.1371/journal.pone.0203711
Keywords: genome editing, CRISPR-Cas9, rice improvement, bacterial blight, Xanthomonas oryzae
Citation: Zafar K, Khan MZ, Amin I, Mukhtar Z, Yasmin S, Arif M, Ejaz K and Mansoor S (2020) Precise CRISPR-Cas9 Mediated Genome Editing in Super Basmati Rice for Resistance Against Bacterial Blight by Targeting the Major Susceptibility Gene. Front. Plant Sci. 11:575. doi: 10.3389/fpls.2020.00575
Received: 03 February 2020; Accepted: 17 April 2020;
Published: 12 June 2020.
Edited by:
Vladimir Nekrasov, Rothamsted Research, United KingdomReviewed by:
Bing Yang, University of Missouri, United StatesJana Streubel, Leibniz University Hannover, Germany
Copyright © 2020 Zafar, Khan, Amin, Mukhtar, Yasmin, Arif, Ejaz and Mansoor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shahid Mansoor, shahidmansoor7@gmail.com
 Kashaf Zafar
Kashaf Zafar Muhammad Zuhaib Khan
Muhammad Zuhaib Khan Imran Amin
Imran Amin Zahid Mukhtar
Zahid Mukhtar Sumera Yasmin
Sumera Yasmin Muhammad Arif
Muhammad Arif Khansa Ejaz
Khansa Ejaz Shahid Mansoor
Shahid Mansoor
