- 1Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, China
- 2Yunnan Provincial Key Laboratory of Panax notoginseng, Kunming, China
Pathogenesis-related proteins (PRs) are a class of proteins that accumulate in response to biotic and abiotic stresses to protect plants from damage. In this study, a gene encoding a PR-like protein (PnPR-like) was isolated from Panax notoginseng, which is used in traditional Chinese herbal medicines. An analysis of gene expression in P. notoginseng indicated that PnPR-like was responsive to an infection by the root rot pathogen Fusarium solani. The expression of this gene was induced by several signaling molecules, including methyl jasmonate, ethephon, hydrogen peroxide, and salicylic acid. The PnPR-like-GFP fusion gene was transiently expressed in onion (Allium cepa) epidermal cells, which revealed that PnPR-like is a cytoplasmic protein. The purified recombinant PnPR-like protein expressed in Escherichia coli had antifungal effects on F. solani and Colletotrichum gloeosporioides as well as inhibited the spore germination of F. solani. Additionally, the in vitro ribonuclease (RNase) activity of the recombinant PnPR-like protein was revealed. The PnPR-like gene was inserted into tobacco (Nicotiana tabacum) to verify its function. The gene was stably expressed in T2 transgenic tobacco plants, which exhibited more RNase activity and greater disease resistance than the wild-type tobacco. Moreover, the transient expression of hairpin RNA targeting PnPR-like in P. notoginseng leaves increased the susceptibility to F. solani and decreased the PnPR-like expression level. In conclusion, the cytoplasmic protein PnPR-like, which has RNase activity, is involved in the P. notoginseng defense response to F. solani.
Introduction
Plants are sessile organisms that are usually subjected to a variety of biotic and abiotic stresses during growth. Specifically, fungal diseases account for 70–80% of all plant diseases. Effector-triggered immunity (ETI) is an important part of plant innate immunity that protects against pathogen infections (Zhang J. et al., 2018). In plants, ETI is established via the recognition of virulence effectors by the corresponding receptor resistance (R) proteins in a specific gene-to-gene manner (Zhang W. et al., 2018). During a pathogen attack, the ETI induction mechanism in plants is activated, leading to the accumulation of pathogenesis-related proteins (PRs) (Mazumder et al., 2013). The accumulated PRs help plants prevent reinfections, resulting in the development of systemic acquired resistance (Zhang et al., 2013).
On the basis of the similarity of their amino acid sequences, structures, serological relationships, and biological activities, the PRs have been grouped into 17 families (Kaur et al., 2017). These PRs have diverse functions, contributing to cell wall rigidification, signal transduction, and antimicrobial activities, and they are mainly expressed in plants as chitinases, glucanases, and thaumatin-like proteins (Farrakh et al., 2018). Many PRs are distributed in plant cell gaps and vacuoles, and their distribution is related to their isoelectric point and the exposure to stress (Jo et al., 2020). The PRs are usually small (10–40 kDa) and mostly acidic. Therefore, they are able to accumulate in intracellular and intercellular spaces.
In plant cells, specific signaling pathways stimulate the defense system, making plant signaling molecules, including some hormones, crucial for the regulation of defense gene expression. Specially, some PR genes are considered as the signature genes of salicylic acid (SA) and jasmonic acid (JA) pathways in model and many crop plants, in which the biotrophic pathogen activates the SA pathway, whereas the necrotrophic pathogen stimulates the JA pathway (Ali et al., 2017). Both abscisic acid and JA up-regulate the expression of ginseng (Panax ginseng C.A. Mey) PR10-1 and PR10-2 genes to varying degrees (Lee et al., 2012). The moss (Physcomitrella patens) PpPR-10 genes are highly expressed in response to SA (Castro et al., 2016). The application of exogenous methyl jasmonate (MeJA), SA, and ethylene can increase the NtPR1a expression level in tobacco (Nicotiana tabacum) (Liu Y. et al., 2019). The ScPR10 transcription level in sugarcane (Saccharum spp. hybrid) is reportedly continuously up-regulated in the first 12 h after a MeJA treatment, and ScPR10 expression is highly induced by SA and MeJA (Peng et al., 2017). In Chilean strawberry (Fragaria chiloensis), the relative expression levels of β-1,3-glucanase genes (FcBG2-1, FcBG2-2, and FcBG2-3) and chitinase genes (FcCHI2-2 and FcCHI3-1) are up-regulated by MeJA (Macedo et al., 2017).
Many PRs have antifungal activities. The broad-spectrum antifungal activities of purified maize (Zea mays) PR10 can disrupt the conidial germination and hyphal growth of various species, including Botrytis cinerea, Sclerotinia sclerotiorum, Fusarium oxysporum, Verticillium dahliae, and Alternaria solani (Zandvakili et al., 2017). The in vitro antimicrobial activities of most PRs are due to their hydrolytic effects on cell walls, contact toxicity and perhaps their involvement in defense signaling. Moreover, some PRs may function as RNase, suggesting they may degrade fungal RNA during infections (Chen et al., 2013). For example, the recombinant LrPR4 protein expressed in Escherichia coli shows RNase activity toward hydrolyzing RNA from Lycoris radiata bulbs and has antifungal activity against rice (Oryza sativa) blast Magnaporthe grisea (Li et al., 2009). The in vitro antifungal activity of wheatwin1, a wheat (Triticum aestivum) PR4, is due to its RNase ability (Caporale et al., 2004; Bertini et al., 2009). The RNase activity of jelly fig (Ficus awkeotsang Makino) PR-4s is correlated to its inhibitory effect on fungal growth (Lu et al., 2012). An earlier investigation confirmed the in vitro RNase activity of soybean (Glycine max) GmPRP, which is a PR that can inhibit Phytophthora sojae mycelial growth (Xu et al., 2014). Another study proved that GbPR10-1 from sea island cotton (Gossypium barbadense) adversely affects V. dahliae mycelial growth and shows in vitro RNase activity (Chen et al., 2013).
Panax notoginseng (Burk) F.H. Chen is a perennial herb in the family Araliaceae. Dried P. notoginseng roots are important medicinal materials for producing drugs useful for treating cardiovascular diseases. As a component of traditional Chinese herbal medicines, P. notoginseng is widely cultivated in Yunnan Province in China. However, the humid environments in which it is grown are suitable for pests and diseases, especially root rot, which is problematic for the sustainable commercial production of P. notoginseng. Root rot, which substantially affects the quality and yield of P. notoginseng medicinal materials, is mainly caused by the pathogenic fungus F. solani (Chen et al., 2015). In an earlier study, we proved that treating P. notoginseng roots with exogenous MeJA can significantly improve the resistance of plants to F. solani, and the expression levels of many P. notoginseng PR genes changed in response to an exogenous MeJA treatment and F. solani infection through transcriptome sequencing (Liu D. et al., 2019). To clarify the PR functions that protect plants from F. solani, a PR-like gene was isolated from P. notoginseng. The full-length cDNA sequence of this gene was obtained and named PnPR-like (GenBank accession number MT515437). Additionally, the PnPR-like expression profile as well as the expression of PnPR-like in a prokaryotic system and the subcellular localization of the encoded protein were analyzed. To functionally validate PnPR-like, it was overexpressed in tobacco and the hairpin RNA targeting PnPR-like was transiently expressed in P. notoginseng leaves through the RNAi-mediated gene silencing.
Materials and Methods
Plant and Fungal Materials
Sterile tobacco seedlings were cultured in a climate-controlled cabinet and used for genetic transformations. Additionally, 1-year-old P. notoginseng plants grown in a greenhouse under double-layered shade nets were used for gene cloning and expression pattern analyses. Prior to their use in experiments, F. solani, and Colletotrichum gloeosporioides mycelia were maintained on potato dextrose agar (PDA) medium at 4°C.
Chemical Treatments and Fungal Inoculation
The roots of healthy 1-year-old P. notoginseng plants were wounded with scissors, after which the root-dipping method was used to treat plants with SA (200 μM), MeJA (100 μM), ethephon (ETH, 1 mM), or H2O2 (1 mM) for 30 min. The P. notoginseng roots treated with sterile water were used as control samples. The roots were collected at 0, 4, 12, 24, 48, and 72 h after the treatments. Regarding the fungal inoculations, the wounded roots of healthy 1-year-old P. notoginseng seedlings were treated with a MeJA solution (100 μM; treatment group) or sterile water (control group) for 30 min. After 24 h, the roots were inoculated with a fresh F. solani spore suspension (106 spores mL–1) for 30 min. For the two P. notoginseng groups, the roots were collected at 0, 4, 12, 24, 48, and 72 h post-inoculation. All samples were immediately frozen in liquid nitrogen and stored at −80°C before the subsequent nucleic acid extraction.
Rapid Amplification of cDNA Ends (RACE) and Bioinformatics Analyses
The PnPR-like unigene sequence determined from the P. notoginseng transcriptome sequencing data included the 5′ untranslated region (UTR) and the start codon. Therefore, a 3′ RACE was performed to obtain the full-length cDNA sequence. The gene-specific primers used for the 3′ RACE were designed based on the unigene sequence (Supplementary Table 1). The mRNA was isolated from 100 μg P. notoginseng total RNA using the NucleoTrap®, mRNA Midi Kit (Macherey-Nagel, Germany). The SMART RACE cDNA Amplification Kit (Clontech, United States) was used to synthesize cDNA and complete the RACE-PCR. The PCR product was cloned into the pMD-18T vector (TaKaRa, Japan) and the resulting recombinant plasmid was inserted into competent E. coli DH5α cells. The recombinant clones were analyzed by sequencing. The GenBank blastn tools1 were used to assemble the unigene and RACE products. The ORFfinder online tool2 was used to search for the open reading frame (ORF) in the assembled sequence. Bioinformatics analyses, including the sequence analysis, multiple sequence alignment, and cluster analysis, were completed as described by Taif et al. (2020).
Quantitative Real-Time PCR
The PnPR-like gene expression patterns in P. notoginseng roots during an infection by F. solani as well as after a treatment with SA, ETH, MeJA, or H2O2 were analyzed in a quantitative real-time PCR (qRT-PCR) assay. A gene-specific primer pair was designed based on the PnPR-like cDNA sequence (Supplementary Table 1). The P. notoginseng actin 2 gene (PnACT2; GenBank accession number KF815706.2) was used as an internal reference gene. The qRT-PCR was performed using the ABI Prism 7,500 Sequence Detection System (Applied Biosystems, United States). The qRT-PCR conditions in this study were the same as those described by Liu et al. (2012). The PnPR-like expression level in each sample was calculated using the 2–Δ Δ Ct method. The qRT-PCR analysis was completed with three replications.
Subcellular Localization
The PnPR-like ORF was cloned into the pGEM-T Easy vector (Promega, United States) to produce the pGEM-T Easy-PnPR-like plasmid, which was digested with NdeI and BamHI. The same restriction enzymes were used to digest the binary expression vector pBIN m-gfp5-ER, which includes the GFP (green fluorescent protein) gene. The PnPR-like ORF was incorporated into pBIN m-gfp5-ER using T4 DNA ligase (Promega, United States) to generate the pBIN m-gfp5-ER-PnPR-like recombinant plasmid, which included the PnPR-like-GFP fusion gene. This plasmid was inserted into Agrobacterium tumefaciens EHA105 cells using the CaCl2 freeze–thaw method. The transient expression of the PnPR-like-GFP fusion gene in onion epidermal cells was analyzed as described by Liu et al. (2018).
Expression and Purification of the Recombinant PnPR-Like Protein in E. coli
The PnPR-like ORF was removed from the pGEM-T Easy-PnPR-like plasmid by restriction enzyme digestion and then subcloned into the pET-32a vector. The resulting pET-32a-PnPR-like plasmid was inserted into competent E. coli BL21 cells. The transformed E. coli cells were cultured in liquid Luria-Bertani (LB) medium supplemented with ampicillin (50 mg L–1) for 12 h at 37°C. When the optical density (at 600 nm) of the bacterial culture reached 0.6, isopropyl β-D-1-thiogalactopyranoside (IPTG, 1 mM final concentration) was added, after which the bacterial culture was incubated at 25°C in a shaker to induce PnPR-like expression. Aliquots of the bacterial culture were collected at 2, 4, 6, and 8 h after the E. coli cells were induced. The bacterial cells were lysed with lysozyme and then centrifuged. The supernatant and the precipitate were collected and analyzed by SDS-PAGE. The polyhistidine-tagged recombinant PnPR-like protein was purified using a Ni-NTA column (Sangon Biotech, China) as described by Wang et al. (2019).
Analyses of the RNase and Antifungal Activities of the Recombinant PnPR-Like Protein
The recombinant PnPR-like protein concentration was determined with the Bradford Protein Assay Kit (Sangon Biotech). To further evaluate the PnPR-like activity, fresh F. solani and C. gloeosporioides mycelia were activated on agar-solidified PDA medium. When the diameter of the fungal colonies reached 2 cm, sterile filter paper disks (0.6 mm in diameter) containing 5, 10, and 20 μg recombinant PnPR-like protein were added around the fungal colonies. As controls, sterile filter paper disks moistened with ddH2O and buffer were also placed around the fungal colonies. After a 3-day incubation at 28°C, the in vitro antifungal effects of the recombinant PnPR-like protein on F. solani and C. gloeosporioides were observed. Specifically, three replications of the antifungal assay were analyzed with Photoshop 7.0 to calculate the average mycelial growth inhibition areas (mm2) for the two pathogenic fungi. To further observe the inhibitory effect of PnPR-like on the growth of F. solani, the spore growth inhibition was also tested. The F.solani spores were grown in the presence of recombinant PnPR-like proteins (40 μg/ml) using glucose solution as control. Inhibition of spore germination was evaluated after 8 h at 21°C and was examined under an optical microscopy (Nikon, JPN). Images acquired through the digital photo camera were sized and optimized for contrast and brightness using Photoshop 7.0. The experiment was carried out at least three times independently using the same conditions.
RNase activity of the recombinant PnPR-like was performed using 20 μg protein incubated with 5 μg of extracted RNA from F. solani. The reaction was incubated at 37°C for 30 min, the proteins were removed by phenol–chloroform (1:1) extraction and the results were observed on 1.4% agarose gel. RNase Inhibitor (40 U; promega, United States) was used as a control. The value of RNase activity of the recombinant PnPR-like protein was measured by spectrophotometric assay. The reaction mixture contained 20 μg of F.solani RNA and 1 μg of recombinant protein. After 30 min incubation at 37°C, the residual not degraded RNA was recovered by an equal volume of 4 M LiCl at 4°C for 3 h, whereas the free oligonucleotides remained in the supernatant. The reaction mixture without incubation contained the same content of F.solani RNA, recombinant protein, and 4M LiCl was as a control. The absorbance of the supernatant at 260 nm was read in comparison with the absorbance of control RNA treated as above. One unit of RNase activity (U/mg) was defined as one OD260 value adding from the digested RNA by 1 mg protein.
Generation and Screening of Transgenic Tobacco Plants
The PnPR-like ORF fragment was obtained by digesting the pGEM-T-PnPR-like plasmid with BamHI and EcoR I. The PnPR-like fragment was then inserted into the pCAMBIA2300s plant expression vector digested with the same two restriction enzymes. Competent E. coli DH5α cells were transformed with pCAMBIA2300s-PnPR-like. The transformation was confirmed by PCR. The recombinant plasmid was then inserted into A. tumefaciens LBA4404 cells using a CaCl2 freeze–thaw method, with transformants detected by PCR. The A. tumefaciens cells carrying pCAMBIA2300s-PnPR-like were used to transform N. tabacum L. cv “Xanthi” leaf disks (Horsch et al., 1985). The T0 generation transgenic tobacco plants verified by PCR were grown in a greenhouse to produce the T1 and T2 generations.
Analysis of Transgene Expression and Evaluation of the Disease Resistance of T2 Transgenic Tobacco
The PnPR-like transcription level in T2 tobacco lines was analyzed by qRT-PCR. The PnACT2 gene was used as an internal reference control. Additionally, wild-type (WT) tobacco served as a negative control, whereas the transgenic tobacco line with the highest Ct value was used as a positive control (The expression level in this line was set as 1). In addition, the RNase activity of total protein of PnPR-like transgenic tobacco lines was tested. The 0.5 g tobacco leaves of transgenic tobacco and WT were quickly ground into powder with liquid nitrogen, and then 500 μL of protein extract was added with sufficient grinding, which was following the incubation at 4°C for overnight. After centrifuging at 4°C and 12,000 g for 30 min, the supernatant was obtained to test the RNase activity by spectrophotometric assay as previously described.
To further functionally characterize PnPR-like, four transgenic tobacco lines in which PnPR-like was expressed at relatively high levels were examined regarding their resistance to fungal infections. The WT plants were used as controls. Fully expanded tobacco leaves of a uniform size were wounded and then inoculated with 100 μL F. solani spore suspension (106 spores mL–1). The inoculated leaves were placed on sterile water-soaked filter paper in a foam box, which was then sealed with plastic wrap and maintained under light in an incubator set at 25°C for 1 week. The leaves were collected and the lesions due to the F. solani infection were examined. Additionally, the roots of sterile WT and transgenic tobacco plants were wounded and then immersed in the F. solani spore suspension (106 spores mL–1) for 30 min. The inoculated tobacco plants were grown in Hoagland nutrient solution under light in an incubator set at 25°C for 1 week. The disease symptoms induced by the F. solani infection were analyzed.
Transient Expression of Hairpin RNA Targeting PnPR-Like in P. notoginseng
Primers specific for PnPR-like were designed and then modified at the 5′ end by the addition of the attB adapter sequence (Supplementary Table 1). The PCR product with the attB adapter sequence as well as the RNAi vector pHellsgate 2 were subjected to a BP recombination reaction using the BP ClonaseTM II enzyme mix (Invitrogen, United States). The reaction product was inserted into competent E. coli DH10B cells, with the transformants selected on agar-solidified LB medium containing spectinomycin (90 mg L–1). The recombinant plasmid was subsequently digested with XbaI and XhoI to confirm the exogenous gene fragments were correctly recombined.
The pHellsgate 2-PnPR-like recombinant plasmid and an empty pHellsgate two vector were inserted into separate A. tumefaciens EHA105 cells. Positive clones were selected on agar-solidified LB medium supplemented with kanamycin (90 mg L–1) and rifampicin (20 mg L–1). The cells carrying the empty pHellsgate two vector were used as controls. The A. tumefaciens EHA105 clones were added to MGL medium and incubated for 5 h at 28°C in a constant temperature shaker (150 rpm). Young P. notoginseng leaves were wounded and placed on filter paper moistened with water. The A. tumefaciens cells containing the RNAi vector were added to the P. notoginseng leaves at the wound site. The hairpin RNA from pHellsgate 2-PnPR-like was transiently expressed in P. notoginseng leaves for approximately 24 h in a climate-controlled cabinet set at 25°C, after which a fresh F. solani spore suspension (106 spores mL–1) was used to inoculate the leaves at the wound site. The infected leaves were collected at 72 h after the inoculation for an examination of the lesions caused by F. solani. The samples were frozen in liquid nitrogen and stored at −80°C. Finally, the PnPR-like expression level after the transient expression of the hairpin RNA used for RNAi in P. notoginseng leaves was analyzed by qRT-PCR.
Statistical Analyses
Data regarding the relative PnPR-like expression levels, the inhibition of fungal growth by the recombinant PnPR-like protein, RNase activity, and the lesion areas are presented herein as the mean ± standard deviation. The data were analyzed with the SPSS software (version 17.0), using Student’s t-test to determine the significance of the differences between the treated or inoculated and control samples as well as between the WT and T2 transgenic lines.
Results
Isolation and Analysis of a PnPR-Like Gene From P. notoginseng
In this study, a gene encoding a PR-like protein was isolated from P. notoginseng. The full-length PnPR-like cDNA is 976 bp in length, with a 717-bp ORF, a 96-bp 5′ UTR, and a 163-bp 3′ UTR. The PnPR-like ORF encodes a protein with 238 amino acid residues, with a molecular mass of approximately 26.64 kDa and an isoelectric point of about 5.69. A BLASTp analysis indicated that the protein sequence encoded by PnPR-like is very similar to the sequences of many PR-like proteins and PRs, including DcPR-like (XP_017223871.1) in carrot (Daucus carota), CsPR-like (XP_028067451.1) in Camellia sinensis, InPR-like (XP_019190454.1) in Ipomoea nil, PmPR-like (XP_008237645.1) in Prunus mume, JrPR-like (XP_018849219.1) in Juglans regia, MnPR (XP_010110490.1) in Morus notabilis, and PtPR (XP_002306682.2) in Populus trichocarpa. A multiple sequence alignment of PnPR-like and the homologous sequences confirmed the high homology among these protein sequences (Figure 1A).
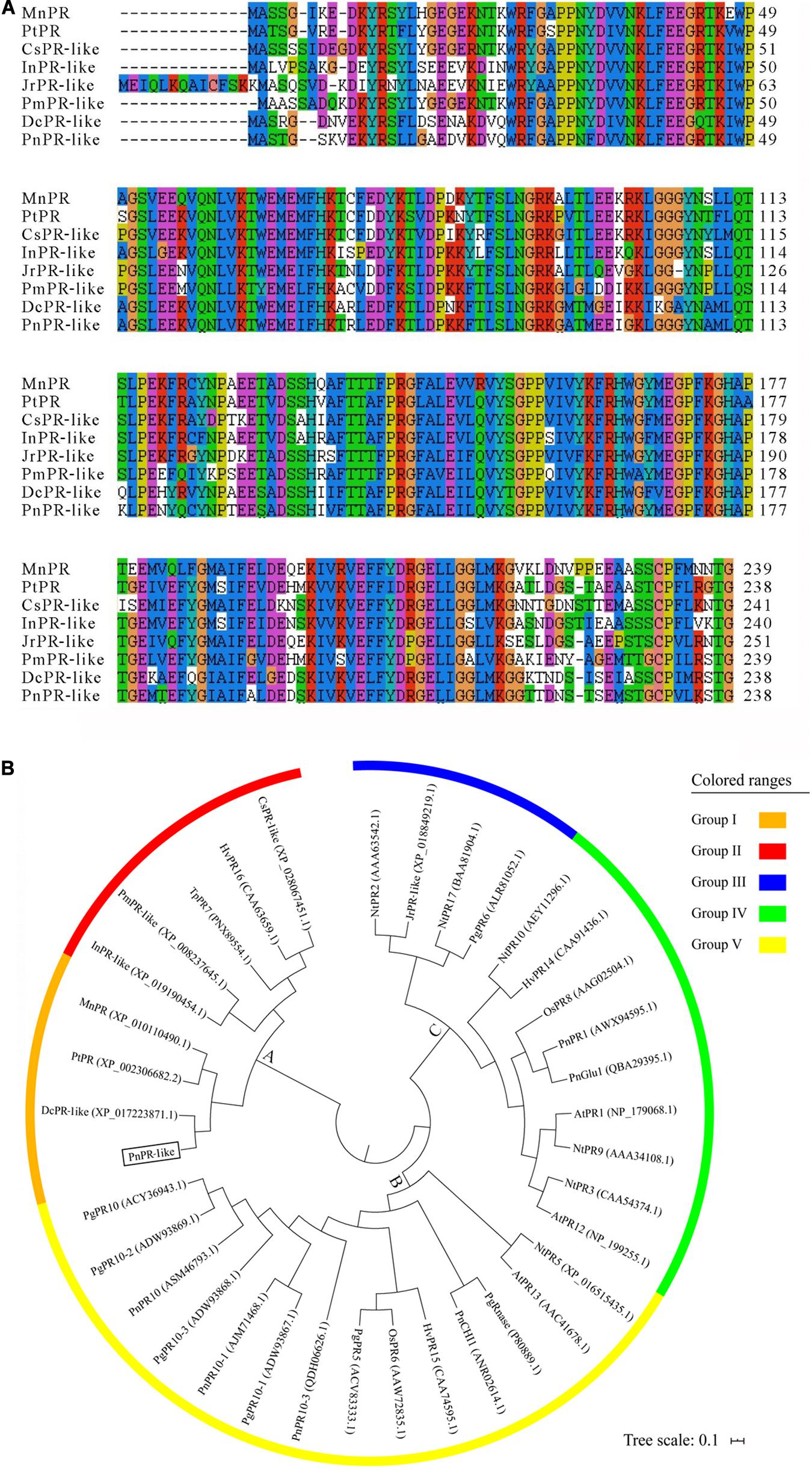
Figure 1. Multiple sequence alignment and phylogenetic tree of PnPR-like and some known PRs (like). (A) The multiple alignment of the deduced amino acid sequences of PnPR-like and seven homologous sequences were performed with the ClustalW. (B) The phylogenetic tree of PnPR-like and 35 known PRs was constructed with Mega seven. The scale bar equals a distance of 10 changes per 100 amino acid positions. The protein sequences are from M. notabilis (MnPR, XP_010110490.1), P. trichocarpa (PtPR, XP_002306682.2), C. sinensis (CsPR-like, XP_028067451.1), I. nil (InPR-like, XP_019190454.1), J. regia (JrPR-like, XP_018849219.1), P. mume (PmPR-like, XP_008237645.1), carrot (DcPR-like, XP_017223871.1), N. tabacum (AtPR1, NP_179068.1), N. tabacum (NtPR2, AAA63542.1), N. tabacum (NtPR3, CAA54374.1), N. tabacum (NtPR5, XP_016515435.1), rice (OsPR6, AAW72835.1), T. pretense (TpPR7, PNX89554.1), rice (OsPR8, AAG02504.1), N. tabacum (NtPR9, AAA34108.1), N. tabacum (NtPR10, AEY11296.1), A. thaliana (AtPR12, NP_199255.1), A. thaliana (AtPR13, AAC41678.1), wheat (HvPR14, CAA91436.1), wheat (HvPR15, CAA74595.1), wheat (HvPR16, CAA63659.1), N. tabacum (NtPR17, BAA81904.1), P. ginseng (PgPR5, ACV83333.1), P. ginseng (PgPR6, ALR81052.1), P. ginseng (PgPR10-1, ADW93867.1), P. ginseng (PgPR10-2, ADW93869.1), P. ginseng (PgPR10-3, ADW93868.1), P. ginseng (PgRnase, P80889.1), P. notoginseng (PnPR1, AWX94595.1), P. notoginseng (PnPR10, ASM46793.1), P. notoginseng (PnPR10-1, AJM71468.1), P. notoginseng (PnCHI1, ANR02614.1), P. notoginseng (PnGlu1, QBA29395.1), P. notoginseng (PnPR10, ASM46793.1), and P. notoginseng (PnPR10-3, MK238491), respectively.
In addition to PnPR-like and the above-mentioned 7 homologous proteins, 13 PRs from Panax species and 15 other PR family proteins were used to construct a phylogenetic tree. The PRs and PR-like proteins were divided into three clusters (A–C) and further subdivided into five groups. Cluster A included PnPR-like, the two homologous PRs and four PR-like proteins, TpPR7, and HvPR16 (Figure 1B), implying that PnPR-like is highly homologous to PRs. Additionally, PnPR-like, MnPR, PtPR, and DcPR-like formed Group I. The 13 PRs from Panax species were unequally distributed among the three clusters. Specifically, the PR10s, including PnPR10, PgPR10, PnPR10-1, PnPR10-3, PgPR10-1, PgPR10-2, and PgPR10-3, as well as PgPR5, PgRnase, and PnCHI1 belonged to Cluster B, whereas PgPR6, PnPR1, and PnGlu1 were included in Cluster C. These phylogenetic relationships suggest that PnPR-like encodes a P. notoginseng PR.
PnPR-Like Expression Is Responsive to Several Signaling Molecules and Is Up-Regulated During the F. solani Infection
The qRT-PCR data revealed the up-regulated expression of PnPR-like following treatments with four signaling molecules (Figures 2A–D). Relative to the control level, the PnPR-like expression level was highest in the roots after the MeJA treatment, followed by the SA, H2O2, and ETH treatments. The PnPR-like gene was most highly expressed (i.e., 5.2-times higher than the control level) at 24 h after the treatments with MeJA. Regarding the SA treatment, the PnPR-like expression level was highest at 24 h (i.e., 5-times higher than the control level). The highest PnPR-like expression levels in response to the H2O2 and ETH treatments (i.e., 3.2- and 2.3-times higher than the control level, respectively) were detected at the 12-h time-point.
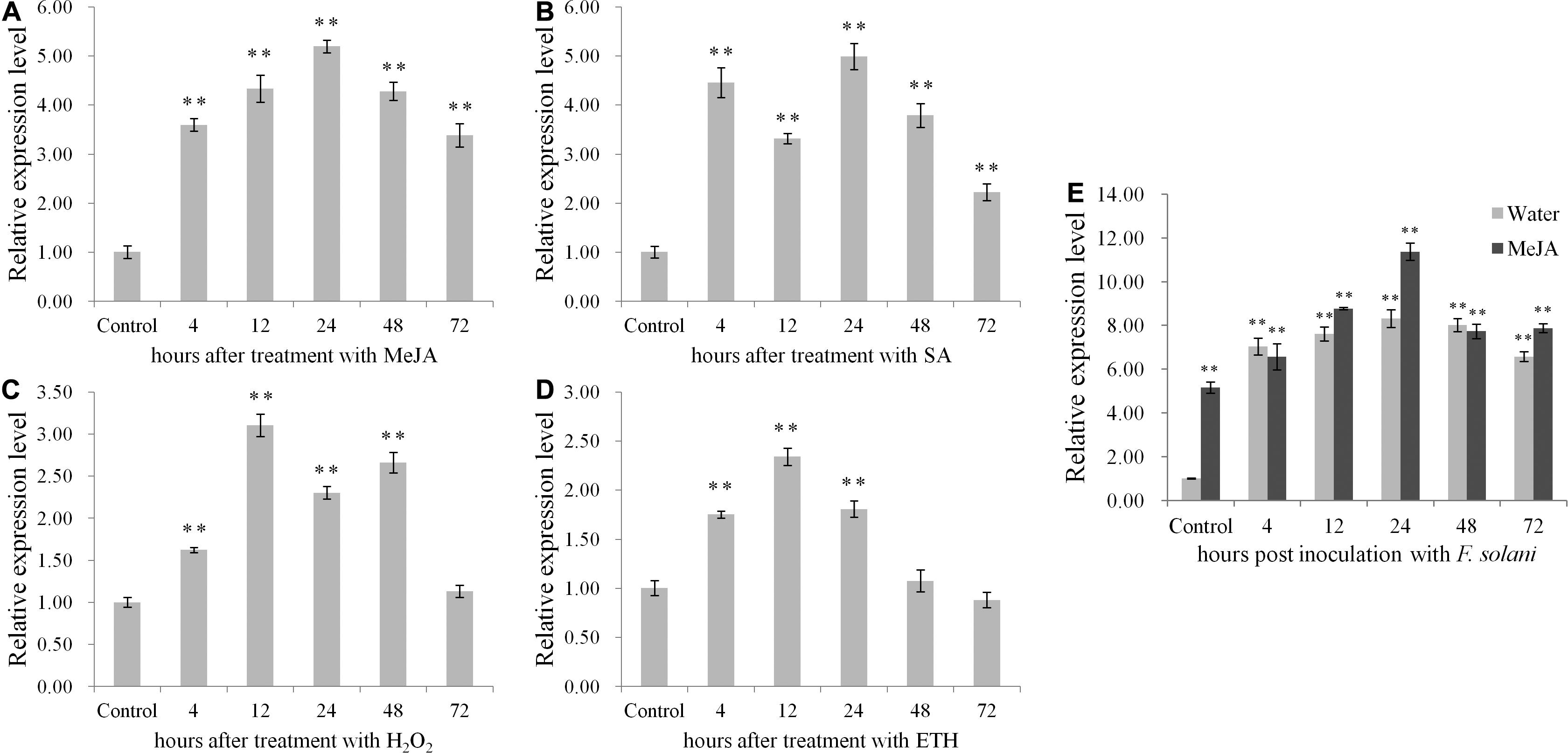
Figure 2. The expression profiles of PnPR-like in P. notoginseng roots. (A–E) The relative expression levels of PnPR-like after treatment with four signal molecules including MeJA (A), SA (B), H2O2 (C), and ETH (D) as well as after inoculation with F. solani (E) were analyzed by qRT-PCR. The roots were collected at 4-, 12-, 24-, 48-, and 72-h post-inoculation, respectively. The P. notoginseng roots respectively pretreated with MeJA and sterile water were inoculated with F. solani spore’s suspension. Results are showed with the average values calculated from three replicates, and the statistical differences between the treatments or inoculation and the control were analyzed by Student’s t-test (**p < 0.01).
After the inoculation with F. solani, PnPR-like expression increased immediately in the P. notoginseng roots pre-treated with MeJA, with an expression level about 5.1-times higher than that in the plants pre-treated with water (Figure 2E). Moreover, the peak PnPR-like expression levels in the two groups of P. notoginseng roots were detected at 24 h; the expression level in the roots pre-treated with MeJA was about 1.4-times higher than that in the roots pre-treated with water and 11.2-times higher than that in the control roots (uninoculated roots pre-treated with sterile water).
The PnPR-Like Protein Is Localized in the Cytoplasm
The deduced PnPR-like protein lacks a predicted signal peptide and may be an endocrine protein. To investigate the subcellular localization of PnPR-like, a PnPR-like-GFP expression cassette was constructed and transiently expressed in onion epidermal cells following A. tumefaciens-mediated transformation. An examination with a confocal laser scanning microscope indicated that the GFP signal was specifically distributed in the cytoplasm of onion epidermal cells in which the PnPR-like-GFP fusion gene was expressed, whereas it was detected throughout the cells transformed with the empty GFP vector (Figure 3). Thus, PnPR-like appears to be a cytoplasmic protein.
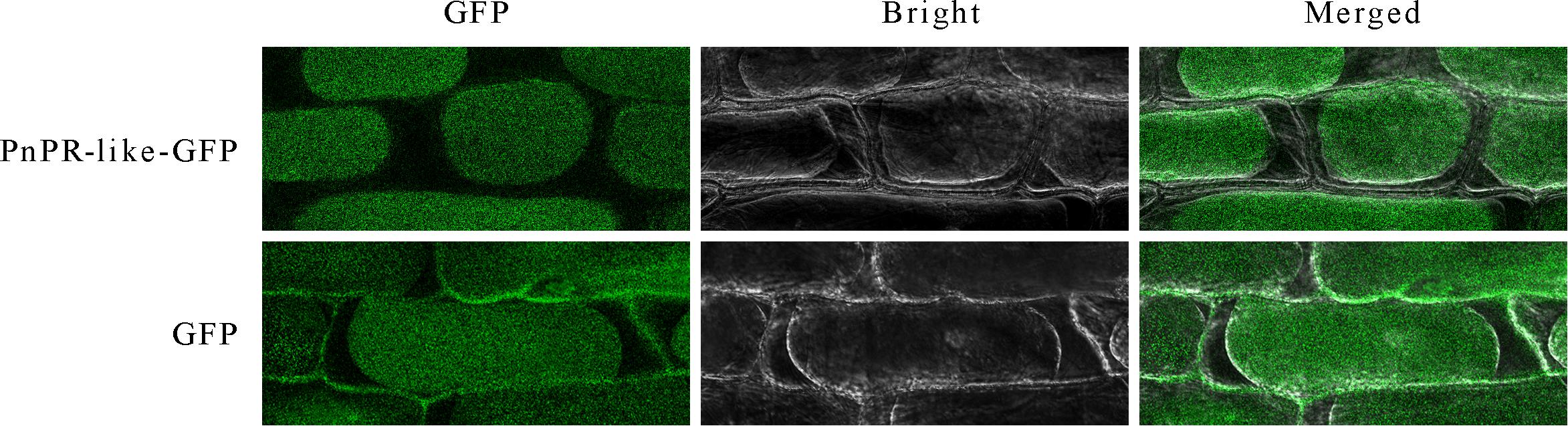
Figure 3. Subcellular analysis revealed the location of PnPR-like in the cytoplasm through the transient expression of the PnPR-like-GFP fusion gene in the onion epidermal cells. The PnPR-like-GFP fusion gene and GFP (control) were transiently expressed in onion epidermal cells mediated by A. tumefaciens. The onion epidermal tissues were treated with 20% sucrose solution to induce plasmolysis, and then a confocal laser microscopy was used to display GFP (green fluorescence), bright field (bright), and combination (merged) views, respectively.
The PnPR-Like Recombinant Protein Expressed in E. coli Has RNase and Antifungal Activities
The SDS-PAGE analysis revealed that the recombinant PnPR-like protein expressed in E. coli is about 43 kDa, which is consistent with the expected size. The recombinant protein was purified with a Ni-NTA column and then concentrated with a Millipore ultrafiltration tube. In the subsequent antifungal assay, the recombinant PnPR-like protein significantly inhibited C. gloeosporioides and F. solani mycelial growth (Figures 4A,B). For both fungi, the mycelial growth inhibition increased as the amount of recombinant PnPR-like protein increased. Moreover, the inhibitory effect of the recombinant protein was greater for F. solani than for C. gloeosporioides (Figure 4C). To further observe the inhibitory effect of PnPR-like on the growth of F. solani, spore growth inhibition was also tested. The Figure 4D showed the growth of F. solani in the absence of PnPR-like protein, whereas the effect of PnPR-like is reported in Figure 4E. It is clear that the PnPR-like protein obviously inhibited the germination of F. solani spores and the mycelium growth. Moreover, the recombinant PnPR-like protein exhibited in vitro RNase activity (Figure 4F), and the tested enzymatic activity was 325 U mg–1. RNA degradation caused by 20 μg of PnPR-like protein was clearly visible (lane two, Figure 4F). Meanwhile, its RNase activity was proved to be sensitive to the RNase inhibitor treatment, and no degradation was observed in presence of 20 μg of PnPR-like plus RNase inhibitor (lane three, Figure 4F).
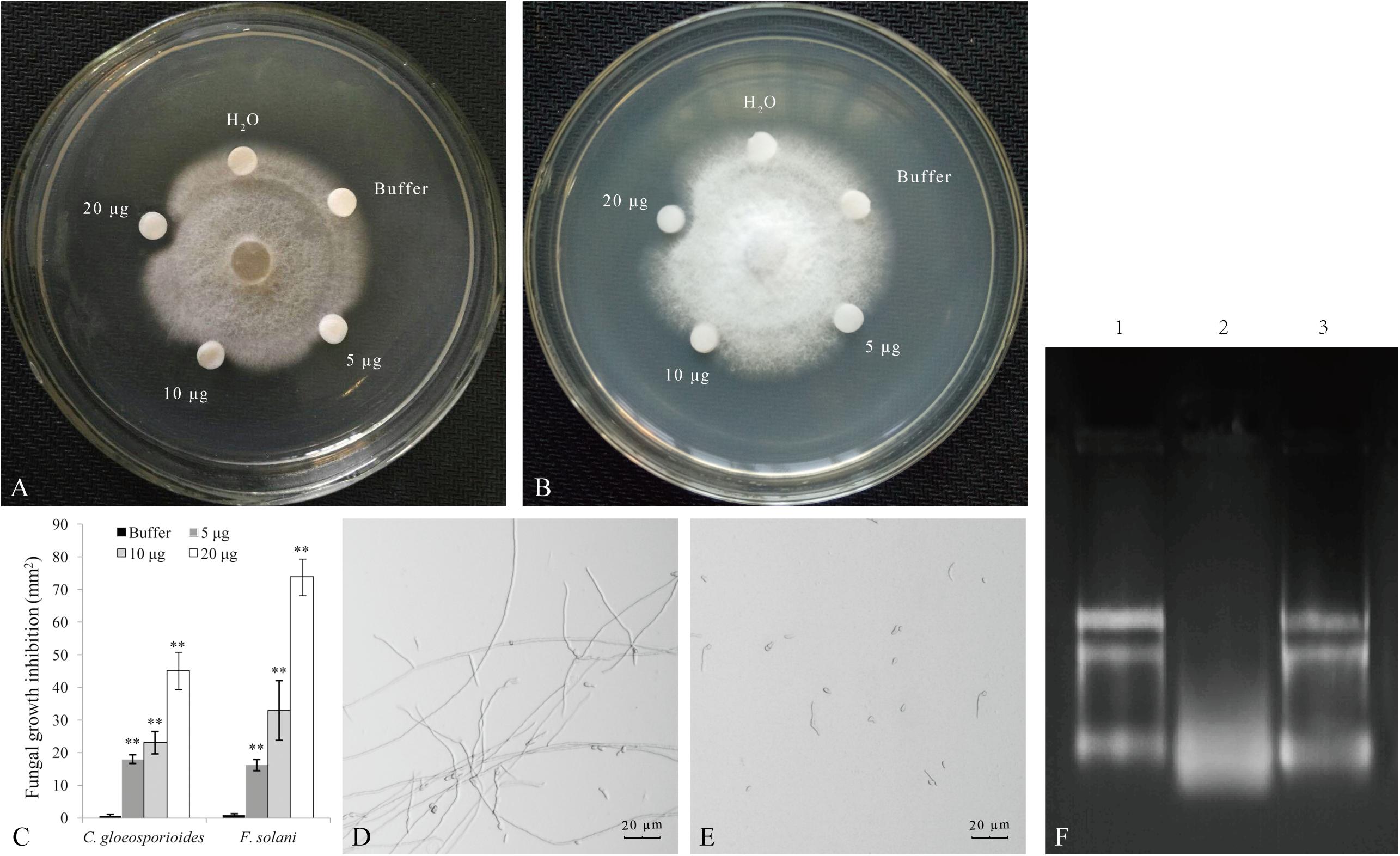
Figure 4. The antifungal assay of the recombinant PnPR-like protein expressed in E. coli. (A,B) The recombinant PnPR-like protein has evident antifungal activity to C. gloeosporioides (A) and F. solani (B). (C) The fungal growth inhibition analysis shows the antifungal activities are increased with the increase of recombinant PnPR-like protein. (D,E) Antifungal effect of PnPR-like on F. solani spore germination revealed by optical microscopy. F. solani spores germinated in the presence of glucose solution (D) and PnPR-like (E), respectively. (F) RNase activity of the recombinant PnPR-like protein tested against F. solani total RNA. The F. solani total RNA (lane 1) was incubated with PnPR-like for 30 min (lane 2); The F. solani total RNA was incubated with PnPR-like and RNase inhibitor for 30 min (lane 3). Results are showed with the average values calculated from three replicates, and the statistical differences are analyzed by Student’s t-test (**p < 0.01).
The Overexpression of PnPR-Like in Tobacco Confers a High Level of Resistance to F. solani
The PnPR-like gene was inserted into tobacco plants to investigate its biological function related to defense responses. The T0 generation transgenic tobacco plants were obtained via a leaf disk transformation method, with 32 positive PnPR-like transgenic tobacco plants identified by a PCR analysis. Twelve transgenic tobacco plants were randomly selected to develop T2 lines by self-crossing for two generations. There were no visible phenotypic differences between the T2 tobacco lines and the WT control. Additionally, qRT-PCR data indicated that PnPR-like was stably expressed in 12 analyzed T2 transgenic tobacco lines (Figure 5A). More specifically, the PnPR-like expression levels were highest in transgenic lines Pl-10, Pl-11, Pl-12, and Pl-16 (Figure 5A). Accordingly, these four lines were used for testing the RNase activity and evaluating the resistance to fungal infections. The total protein of WT and PnPR-like transgenic tobacco showed the RNase activity, but the RNase activity of the PnPR-like transgenic lines was significantly higher than the WT (Figure 5B).

Figure 5. The expression levels, RNase activities, and resistance analyses of T2 generation PnPR-like transgenic tobacco. (A) The expression levels of PnPR-like in the T2 generation of transgenic tobacco were by detected through qRT-PCR, and PnPR-like was expressed in all transgenic tobacco plants. The Pl-1, Pl-3, Pl-4, Pl-5, Pl-6, Pl-9, Pl-10, Pl-11, Pl-12, Pl-16, Pl-17, and Pl-19 are the T2 generation transgenic tobacco lines, and WT is the wild-type tobacco. (B) The RNase activity of total protein of PnPR-like transgenic tobacco and WT tested against F. solani total RNA. (C) The roots inoculation assay reveals the enhanced resistance of PnPR-like transgenic tobacco lines against F. solani. (D) The leave inoculation assay shows the enhanced resistance of PnPR-like transgenic tobacco lines against F. solani. (E) The lesion sizes in leaves of PnPR-like transgenic tobacco lines and WT during F. solani infection. The results show the mean of three replicate calculations and statistical differences were analyzed by Student’s t-test (**p < 0.01).
At 7 days after the inoculation with F. solani, the roots of WT tobacco plants were blackened and the leaf chlorosis was severe. In contrast, the roots of the four PnPR-like transgenic lines were growing normally, with no blackening or rotting (Figure 5C). Furthermore, the F. solani infection resulted in mild chlorosis around the inoculation site of the transgenic tobacco leaves, whereas the WT tobacco leaves had severe chlorotic lesions and were rotting (Figure 5D). The lesions were almost 12-times larger on the WT leaves than on the transgenic tobacco leaves (Figure 5E) at 7 days after the inoculation with F. solani. The inoculation assay results suggest that the transgenic tobacco plants were more resistant to F. solani than the WT plants. Thus, the overexpression of PnPR-like in tobacco confers strong resistance to F. solani.
The Transient Expression of the Hairpin RNA Targeting PnPR-Like in P. notoginseng Leaves Increases the Susceptibility to F. solani
The pHellsgate 2-PnPR-like and the pHellsgate two empty vector were inserted into separate P. notoginseng leaves, which were then inoculated with F. solani. At 3 days post-inoculation, the infected leaves were withered, yellow, and decayed around the inoculation site (Figure 6A). However, the lesions were significantly larger on the leaves transformed with pHellsgate 2-PnPR-like than on the leaves transformed with the empty vector. The average leaf lesion area was 2.5 cm2 for the PnPR-like RNAi samples, whereas it was less than 1 cm2 for the empty vector control samples (Figure 6B). Additionally, the PnPR-like expression level in the control leaves was about five-times higher than that in the leaves transformed with pHellsgate 2-PnPR-like (Figure 6C). Thus, the transient expression of the hairpin RNA targeting PnPR-like increased the susceptibility of P. notoginseng leaves to F. solani.
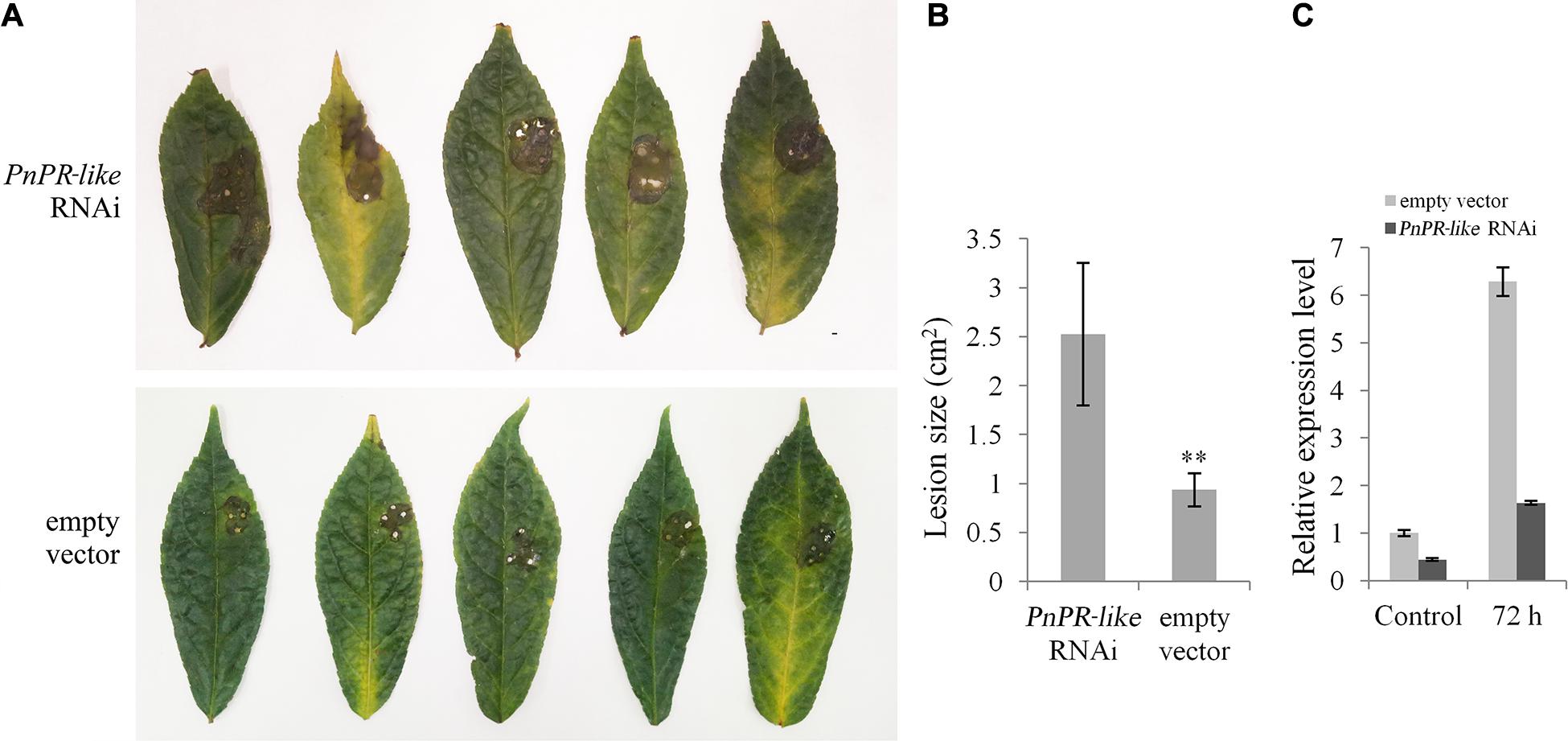
Figure 6. Analysis of the hairpin RNA targeting PnPR-like transiently expressed in the P. notoginseng leaves. (A) The symptoms of P. notoginseng leaves after inoculation with F. solani, in which the PnPR-like RNAi vector and the empty vector was expressed, respectively. (B) The lesion size in P. notoginseng leaves caused by F. solani infection using Student’s t-test (**p < 0.01). The transient expression of the hairpin RNA targeting PnPR-like increased the susceptibility of P. notoginseng leaves to F. solani. (C) The relative expression levels of PnPR-like in the two group of P. notoginseng leaves evaluated by qRT-PCR. After inoculation with F. solani for 72 h, the PnPR-like expression level in the control leaves was about 5-times higher than that in the leaves transformed with pHellsgate 2-PnPR-like, which indicates that the transient expression of RNAi construct decreased the expression level of PnPR-like in P. notoginseng leaves. Results are showed with the average values calculated from three replicates, and the Student’s t-test was used to analyze the statistical differences (**p < 0.01).
Discussion
Pathogenesis-related proteins are widely distributed in higher plants, wherein they are important for defense responses that protect plant cells from diverse stresses. In this study, a new PR gene, PnPR-like, was isolated from P. notoginseng. The deduced PnPR-like amino acid sequence is highly similar to the sequences of some known plant PR-like proteins. The PRs are mainly distributed in the intercellular space. However, the deduced PnPR-like protein appears to lack a signal peptide, and its subcellular localization suggests PnPR-like may be an intracellular protein. In the current study, the PnPR-like-GFP fusion protein was detected exclusively in the cytoplasm of onion epidermal cells, implying PnPR-like encodes a cytoplasmic protein. In the PR family, PR10 is a class of typical cytoplasmic proteins with no signal peptide (Han et al., 2017). Previous studies proved that GmPR10 (Xu et al., 2014), ZmPR10.1 (Xie et al., 2010), JcPR10a (Agarwal et al., 2012), and P. notoginseng PR10-3 (Tang et al., 2019) are localized in the cytoplasm. These intracellular proteins are synthesized by free ribosomes and help mediate cytosolic reactions associated with mitochondria, nuclei, and peroxidases. Some intracellular PRs can bind to ligands, including hormones, biological macromolecules, and active substances with important biological functions, to regulate growth and development as well as defense responses (Mogensen et al., 2002).
The expression of many PR genes is regulated by specific hormone signaling pathways. In Arabidopsis thaliana, SA treatments significantly up-regulate PR1, PR2, and PR5 expression, but have the opposite effect on the expression of PR3, PR12, and PR13 (Seo et al., 2008). The expression of Brassica juncea PR genes is responsive to JA. The BjPR3, BjPR4, and BjPR12 expression levels reportedly increase following a JA treatment, whereas an abscisic acid treatment down-regulates the transcription of BjPR1, BjPR2, and BjPR5 (Ali et al., 2017). An earlier investigation of Lilium regale indicated that the stress-related signaling molecules JA, SA, ETH, and H2O2 up-regulate PR10-1/-2/-5/-6/-7 expression, but down-regulate the expression of PR10-3/-4/-8/-9 (He et al., 2014). In the current study, PnPR-like expression was up-regulated by MeJA, ETH, H2O2, and SA to varying degrees. Moreover, PnPR-like expression was also induced by an infection of F. solani. To effectively defend against pathogens, plants have evolved sophisticated defense mechanisms that “sense” pathogen attacks and activate appropriate defense responses (Ji et al., 2014; Gupta et al., 2015). Additionally, cross-communicating signal transduction networks are formed in plants to regulate the induced defense responses. Specifically, necrotrophic pathogen F. solani mainly stimulates JA pathway, whereas the cross-talk among SA, JA, and ETH signaling pathways helps control the defense response of P. notoginseng to F. solani (Ali et al., 2017; Liu Y. et al., 2019). Moreover, the MeJA regulates the expression of genes encoding PnPR10-3, osmotin-like protein, and β-1,3-glucanase following an F. solani infection (Tang et al., 2017; Taif et al., 2020). The activation of P. notoginseng PRs, including PnPR-like, is part of a regulatory network mediated by the cross-talk among hormone signaling pathways during an F. solani infection; however, the precise regulatory mechanism will need to be more thoroughly characterized.
The PRs are an important part of plant defense systems. Some PRs are hydrolytic enzymes, such as chitinases (NtPR-Q, VpPR4-1, and EuCHIT2) and β-1,3-glucanases (PnGlu1 and HbPR2), that hydrolyze β-1,3-glucans and chitin, which are cell wall components in most higher fungi (Dai et al., 2016; Sunpapao and Pornsuriya, 2016; Dong et al., 2017; Tang et al., 2017). The antifungal activities of PR5 proteins, including thaumatin-like and osmotin-like proteins, result in permeabilized plasma membranes, weakened cell walls, and plasmolysis (Rather et al., 2015). The Ypr6 (proteinase inhibitor), Ypr12 (plant defensin), Ypr13 (plant thionin), and Ypr14 (lipid transfer protein) antimicrobial peptides are usually cysteine-rich peptides with broad-spectrum antifungal activities (Loon et al., 2006; Nawrot et al., 2013). Many PR10 proteins and PR4 protrins have been shown to have RNase activity (Xu et al., 2010; Wang et al., 2011; Yang et al., 2018). The recombinant Theobroma cacao PR-10 protein has antifungal activities against the basidiomycetous pathogen Moniliophthora perniciosa and the yeast Saccharomyces cerevisiae, while also exhibiting in vitro and in vivo RNase activities (Pungartnik et al., 2009). Two PR10 proteins from ginseng (PgPR10s) have been characterized as RNases (Lee et al., 2012). A recent study proved that recombinant PnPR10-3 functions as an RNase in vitro has clear antifungal effects on Fusarium species (F. oxysporum, F. solani, and F. verticillioides) (Tang et al., 2019). The RNase activity in Capsicum chinense PR-4 protein plays a protective role by degrading RNA of invasive pathogens (Guevara-Morato et al., 2010). The antifungal activity of T. cacao PR-4b is directly dependent of its RNase activity (Menezes et al., 2014). In the current study, the recombinant PnPR-like protein had RNase activity in vitro and was able to inhibit C. gloeosporioides and F. solani mycelial growth, meanwhile, the spore germination of F. solani was significantly inhibited by the recombinant PnPR-like protein. Thus, in addition to the PR10 and PR4 proteins, some other PRs have RNase activities related to plant defense responses during pathogen infections.
The application of reverse genetics technology involving gene overexpression and gene silencing (e.g., RNAi) has enabled the rapid functional characterization of PR genes. For example, overexpressing the sugarcane PR10 gene in tobacco enhances the resistance of transgenic plants to Pseudomonas solanacearum and F. solani (Peng et al., 2017). Transgenic grape (Vitis vinifera) lines overexpressing VpPR10.1 are more resistant to downy mildew caused by Plasmopara viticola than WT plants (Ma et al., 2018). Constitutive overexpression of wheat PR4a and PR4b in transgenic tobacco plants decreased Phytophthora nicotianae susceptibility, and the T4 generation plants were found to be significantly more resistant against P. nicotianae (Fiocchetti et al., 2007). Overexpression of VpPR4-1 increased powdery mildew resistance of grape (Dai et al., 2016). Additionally, transgenic tobacco plants overexpressing the L. regale PR10-5 gene exhibit increased resistance to F. oxysporum (Chen et al., 2017). The RNAi-mediated silencing of PR5 expression was observed to decrease the resistance of cherry tomato (Lycopersicum esculentum) to Alternaria alternata (Zhai et al., 2018). In another study, fungal colonization significantly increased in mature PR10-RNAi transgenic maize kernels inoculated with Aspergillus flavus (Chen et al., 2010). In the present study, the F. solani infection rate was significantly lower for PnPR-like transgenic tobacco than for the WT plants, reflecting the increase in disease resistance induced by the overexpression of PnPR-like. To further elucidate the PnPR-like function, PnPR-like expression was suppressed via RNAi, which resulted in P. notoginseng leaves that were more susceptible to F. solani than the WT leaves in vitro. These findings clearly indicate that PnPR-like is a defense response gene in P. notoginseng that protects plants against F. solani infections. Fusarium species, including F. oxysporum and F. solani, cause serious diseases in many crops, ornamental plants, and medicinal plants. Therefore, they are very important agricultural pathogens. The best way to sustainably and effectively control plant diseases involves developing disease-resistant cultivars (Moscou and van Esse, 2017). A deeper understanding of the molecular basis of plant disease resistance may enhance the genetic engineering of crop species to overcome the limitations of traditional breeding strategies. The P. notoginseng genes encoding PRs, including PnPR-like, are excellent candidate genes for the genetic engineering of plants with increased disease resistance.
Conclusion
The PnPR-like gene isolated from P. notoginseng in this study encodes a cytoplasmic protein. Additionally, PnPR-like expression is responsive to F. solani infections and is induced by four signaling molecules, including MeJA. The recombinant PnPR-like protein has significant in vitro antifungal and RNase activities. Moreover, the overexpression of PnPR-like results in transgenic tobacco plants that is highly resistant to F. solani. In contrast, the RNAi-mediated down-regulation of PnPR-like expression increases the susceptibility of P. notoginseng plants to F. solani. Therefore, PnPR-like plays an important role in the P. notoginseng defense response to the root rot pathogen. The regulatory effects of hormone signaling pathways on PnPR-like expression and the mechanism underlying the PnPR-like RNase activity in defense responses to root rot will be explored more precisely in future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI: accession number MT515437.
Author Contributions
SL: methodology, data curation, experiment execution, and writing-original draft preparation. ZW: methodology and material collection. BT: investigation and software. LZ: validation. HC: conceptualization. XC: supervision. FG: methodology. DL: methodology, supervision, writing-reviewing, and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by three grants received from National Natural Sciences Foundation of China (81560610 and 82060693) and Young and Elite Talents Project of Yunnan Ten Thousand Talents Plan, respectively.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.610176/full#supplementary-material
Footnotes
References
Agarwal, P., Bhatt, V., Singh, R., Das, M., Sopory, S. K., and Chikara, J. (2012). Pathogenesis-related gene, JcPR-10a from Jatropha curcas exhibit RNase and antifungal activity. Mol. Biotechnol. 54, 412–425. doi: 10.1007/s12033-012-9579-7
Ali, S., Mir, Z. A., Tyagi, A., Bhat, J. A., Chandrashekar, N., Papolu, P. K., et al. (2017). Identification and comparative analysis of Brassica juncea pathogenesis-related genes in response to hormonal, biotic and abiotic stresses. Acta Physiol. Plant 39:268. doi: 10.1007/s11738-017-2565-8
Bertini, L., Caporale, C., Testa, M., Proietti, S., and Caruso, C. (2009). Structural basis of the antifungal activity of wheat PR4 proteins. FEBS Lett. 583, 2865–2871. doi: 10.1016/j.febslet.2009.07.045
Caporale, C., Berardino, I., Leonardi, L., Bertini, L., Cascone, A., Buonocore, V., et al. (2004). Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Lett. 575, 71–76. doi: 10.1016/j.febslet.2004.07.091
Castro, A., Vidal, S., and Ponce de León, I. (2016). Moss pathogenesis-related-10 protein enhances resistance to Pythium irregulare in Physcomitrella patens and Arabidopsis thaliana. Front. Plant Sci. 7:580. doi: 10.3389/fpls.2016.00580
Chen, J. L., Liu, K., Miao, C. P., Sun, S., Chen, Y. W., Xu, L. H., et al. (2015). Salt tolerance of endophytic Trichoderma koningiopsis YIM PH30002 and its volatile organic compounds (VOCs) allelopathic activity against phytopathogens associated with Panax notoginseng. Ann. Microbiol. 66, 981–990. doi: 10.1007/s13213-015-1171-5
Chen, L., Sun, N., Wang, J., Ling, H., Zhang, L., and Zuo, K. (2013). Functional analysis of a wilt fungus inducible PR10-1 gene from cotton. Am. J. Sci. 04, 417–426. doi: 10.4236/ajps.2013.42A054
Chen, R., He, H., Yang, Y., Qu, Y., Ge, F., and Liu, D. (2017). Functional characterization of a pathogenesis-related protein family 10 gene, LrPR10-5, from Lilium regale Wilson. Australas Plant Path. 46, 251–259. doi: 10.1007/s13313-017-0485-0
Chen, Z. Y., Brown, R. L., Damann, K. E., and Cleveland, T. E. (2010). PR10 expression in maize and its effect on host resistance against Aspergillus flavus infection and aflatoxin production. Mol. Plant Pathol. 11, 69–81. doi: 10.1111/j.1364-3703.2009.00574.x
Dai, L., Wang, D., Xie, X., Zhang, C., Wang, X., Xu, Y., et al. (2016). The novel gene VpPR4-1 from Vitis pseudoreticulata increases powdery mildew resistance in transgenic Vitis vinifera L. Front. Plant Sci. 7:695. doi: 10.3389/fpls.2016.00695
Dong, X., Zhao, Y., Ran, X., Guo, L., and Zhao, D. G. (2017). Overexpression of a new chitinase gene EuCHIT2 enhances resistance to Erysiphe cichoracearum DC in tobacco plants. Int. J. Mol. Sci. 18:2361. doi: 10.3390/ijms18112361
Farrakh, S., Wang, M., and Chen, X. (2018). Pathogenesis-related protein genes involved in race-specific all-stage resistance and non-race specific high-temperature adult-plant resistance to Puccinia striiformis f. sp. tritici in wheat. J. Integr. Agr. 17, 2478–2491. doi: 10.1016/s2095-3119(17)61853-7
Fiocchetti, F., D’Amore, R., Palma, M., Bertini, L., Caruso, C., Caporale, C., et al. (2007). Constitutive over-expression of two wheat pathogenesis related genes enhances resistance of tobacco to Phytophthora nicotianae. Plant Cell Tiss. Org. 92, 73–84. doi: 10.1007/s11240-007-9306-4
Guevara-Morato, M. Á, García de Lacoba, M., García-Luque, I., and Serra, M. T. (2010). Characterization of a pathogenesis-related protein 4 (PR-4) induced in Capsicum chinense L3 plants with dual RNase and DNase activities. J. Exp. Bot. 61, 3259–3271. doi: 10.1093/jxb/erq148
Gupta, R., Lee, S. E., Agrawal, G. K., Rakwal, R., Park, S., Wang, Y., et al. (2015). Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front. Plant Sci. 6:352. doi: 10.3389/fpls.2015.00352
Han, X., He, X., Qiu, W., Lu, Z., Zhang, Y., Chen, S., et al. (2017). Pathogenesis-related protein PR10 from Salix matsudana Koidz exhibits resistance to salt stress in transgenic Arabidopsis thaliana. Environ. Exp. Bot. 141, 74–82. doi: 10.1016/j.envexpbot.2017.07.008
He, H., Liu, D., Zhang, N., Zheng, W., Han, Q., Ji, B., et al. (2014). The PR10 gene family is highly expressed in Lilium regale Wilson during Fusarium oxysporum f. sp. lilii infection. Genes Genom. 36, 497–507. doi: 10.1007/s13258-014-0185-x
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Eichholtz, D., Rogers, S. G., and Fraley, R. T. (1985). A simple and general method for transferring genes into plants. Science 227, 1229–1231. doi: 10.1126/science.227.4691.1229
Ji, S. H., Gururani, M. A., and Chun, S. C. (2014). Expression analysis of rice pathogenesis-related proteins involved in stress response and endophytic colonization properties of gfp-tagged Bacillus subtilis CB-R05. Appl. Biochem. Biotech. 174, 231–241. doi: 10.1007/s12010-014-1047-3
Jo, B. R., Yu, J. M., Jang, S., Ahn, J. W., Kim, H. S., Seoung, E. A., et al. (2020). Cloning, expression, and purification of a pathogenesis-related protein from Oenanthe javanica and its biological properties. Biol. Pharm. Bull. 43, 158–168. doi: 10.1248/bpb.b19-00801
Kaur, A., Pati, P. K., Pati, A. M., and Nagpal, A. K. (2017). In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PLoS One 12:9. doi: 10.1371/journal.pone.0184523
Lee, O. R., Kim, Y. J., Balusamy, S. R. D., Khorolragchaa, A., Sathiyaraj, G., Kim, M. K., et al. (2012). Expression of the ginseng PgPR10-1 in Arabidopsis confers resistance against fungal and bacterial infection. Gene 506, 85–92. doi: 10.1016/j.gene.2012.06.039
Li, X., Xia, B., Jiang, Y., Wu, Q., Wang, C., He, L., et al. (2009). A new pathogenesis-related protein, LrPR4, from Lycoris radiata, and its antifungal activity against Magnaporthe grisea. Mol. Biol. Rep. 37, 995–1001. doi: 10.1007/s11033-009-9783-0
Liu, D., He, X., Li, W., Chen, C., and Ge, F. (2012). A β-1,3-glucanase gene expressed in fruit of Pyrus pyrifolia enhances resistance to several pathogenic fungi in transgenic tobacco. Eur. J. Plant Pathol. 135, 265–277. doi: 10.1007/s10658-012-0083-5
Liu, D., Zhao, Q., Cui, X., Chen, R., Li, X., Qiu, B., et al. (2019). A transcriptome analysis uncovers Panax notoginseng resistance to Fusarium solani induced by methyl jasmonate. Genes Genom. 41, 1383–1396. doi: 10.1007/s13258-019-00865-z
Liu, D. Q., Han, Q., Shah, T., Chen, C. Y., Wang, Q., Tang, B. F., et al. (2018). A hybrid proline-rich cell-wall protein gene JsPRP1 from Juglans sigillata Dode confers both biotic and abiotic stresses in transgenic tobacco plants. Trees 32, 1199–1209. doi: 10.1007/s00468-018-1702-3
Liu, Y., Liu, Q., Tang, Y., and Ding, W. (2019). NtPR1a regulates resistance to Ralstonia solanacearum in Nicotiana tabacum via activating the defense-related genes. Biochem. Bioph. Res. Co. 508, 940–945. doi: 10.1016/j.bbrc.2018.12.017
Loon, L. C., Rep, M., and Pieterse, C. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Lu, H. C., Lin, J. H., Chua, A. C. N., Chung, T. Y., Tsai, I. C., Tzen, J. T. C., et al. (2012). Cloning and expression of pathogenesis-related protein 4 from jelly fig (Ficus awkeotsang Makino) achenes associated with ribonuclease, chitinase and anti-fungal activities. Plant Physiol. Bioch. 56, 1–13. doi: 10.1016/j.plaphy.2012.04.004
Ma, H., Xiang, G., Li, Z., Wang, Y., Dou, M., Su, L., et al. (2018). Grapevine VpPR10.1 functions in resistance to Plasmopara viticola through triggering a cell death-like defence response by interacting with VpVDAC3. Plant Biotechnol. J. 16, 1488–1501. doi: 10.1111/pbi.12891
Macedo, R., Sales, L. P., Yoshida, F., Silva-Abud, L. L., and Lobo, M. (2017). Potential worldwide distribution of Fusarium dry root rot in common beans based on the optimal environment for disease occurrence. PLoS One 12:11. doi: 10.1371/journal.pone.0187770
Mazumder, M., Das, S., Saha, U., Chatterjee, M., Bannerjee, K., and Basu, D. (2013). Salicylic acid-mediated establishment of the compatibility between Alternaria brassicicola and Brassica juncea is mitigated by abscisic acid in Sinapis alba. Plant Physiol. Bioch. 70, 43–51. doi: 10.1016/j.plaphy.2013.04.025
Menezes, S., Silva, E., Lima, E., Sousa, A., Andrade, B., Lemos, L., et al. (2014). The pathogenesis-related protein PR-4b from Theobroma cacao presents RNase activity, Ca2+ and Mg2+ dependent-DNase activity and antifungal action on Moniliophthora perniciosa. BMC Plant Biol. 14:161. doi: 10.1186/1471-2229-14-161
Mogensen, J. E., Wimmer, R., Larsen, J. N., Spangfort, M. D., and Otzen, D. E. (2002). The major birch allergen, Bet v 1, shows affinity for a broad spectrum of physiological ligands. J. Biol. Chem. 277, 23684–23692. doi: 10.1074/jbc.M202065200
Moscou, M., and van Esse, P. (2017). The quest for durable resistance. Science 358, 1541–1542. doi: 10.1126/science.aar4797
Nawrot, R., Barylski, J., Nowicki, G., Broniarczyk, J., Buchwald, W., and Goździcka-Józefiak, A. (2013). Plant antimicrobial peptides. Folia Microbiol. 59, 181–196. doi: 10.1007/s12223-013-0280-4
Peng, Q., Su, Y., Ling, H., Ahmad, W., Gao, S., Guo, J., et al. (2017). A sugarcane pathogenesis-related protein, ScPR10, plays a positive role in defense responses under Sporisorium scitamineum, SrMV, SA, and MeJA stresses. Plant Cell Rep. 36, 1427–1440. doi: 10.1007/s00299-017-2166-4
Pungartnik, C., Silva, A., Melo, S., Gramacho, K., Cascardo, J., Brendel, M., et al. (2009). High-affinity copper transport and Snq2 export permease of Saccharomyces cerevisiae modulate cytotoxicity of PR-10 from Theobroma cacao. Mol. Plant Microbe 22, 39–51. doi: 10.1094/MPMI-22-1-0039
Rather, I. A., Awasthi, P., Mahajan, V., Bedi, Y. S., Vishwakarma, R. A., and Gandhi, S. G. (2015). Molecular cloning and functional characterization of an antifungal PR-5 protein from Ocimum basilicum. Gene 558, 143–151. doi: 10.1016/j.gene.2014.12.055
Seo, P. J., Lee, A. K., Xiang, F., and Park, C. M. (2008). Molecular and functional profiling of Arabidopsis pathogenesis-related genes: insights into their roles in salt response of seed germination. Plant Cell Physiol. 49, 334–344. doi: 10.1093/pcp/pcn011
Sunpapao, A., and Pornsuriya, C. (2016). Overexpression of β-1,3-glucanase gene in response to Phytophthora palmivora infection in leaves of Hevea brasiliensis clones. Walailak J. Sci. Technol. 13, 35–43. doi: 10.14456/wjst.2016.4
Taif, S., Zhao, Q., Pu, L., Li, X., Liu, D., and Cui, X. (2020). A ß-1,3-glucanase gene from Panax notoginseng confers resistance in tobacco to Fusarium solani. Ind. Crop Prod. 143:111947. doi: 10.1016/j.indcrop.2019.111947
Tang, B., Li, X., Pu, L., Zhao, Q., Cui, X., Ge, F., et al. (2019). A pathogenesis-related protein 10 gene PnPR10–3 was involved in molecular interaction between Panax notoginseng and Fusarium solani. Australas Plant Path. 48, 447–456. doi: 10.1007/s13313-019-00644-0
Tang, Y., Liu, Q., Liu, Y., Zhang, L., and Ding, W. (2017). Overexpression of NtPR-Q up-regulates multiple defense-related genes in Nicotiana tabacum and enhances plant resistance to Ralstonia solanacearum. Front. Plant Sci. 8:1963. doi: 10.3389/fpls.2017.01963
Wang, N., Xiao, B., and Xiong, L. (2011). Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 168, 2212–2224. doi: 10.1016/j.jplph.2011.07.013
Wang, Q., Qiu, B. L., Li, S., Zhang, Y. P., Cui, X. M., Ge, F., et al. (2019). A methyl jasmonate induced defensin like protein from Panax notoginseng confers resistance against Fusarium solani in transgenic tobacco. Biol. Plant. 63, 797–807. doi: 10.32615/bp.2019.123
Xie, Y. R., Chen, Z. Y., Brown, R. L., and Bhatnagar, D. (2010). Expression and functional characterization of two pathogenesis-related protein 10 genes from Zea mays. J. Plant Physiol. 167, 121–130. doi: 10.1016/j.jplph.2009.07.004
Xu, P., Jiang, L., Wu, J., Li, W., Fan, S., and Zhang, S. (2014). Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 41, 4899–4909. doi: 10.1007/s11033-014-3356-6
Xu, Y., Yu, H., He, M., Yang, Y., and Wang, Y. (2010). Isolation and expression analysis of a novel pathogenesis-related protein 10 gene from Chinese wild Vitis pseudoreticulata induced by Uncinula necator. Biologia 65, 653–659. doi: 10.2478/s11756-010-0056-0
Yang, D., Jiang, L., Bao, Y., Liu, W., Zhang, H., Chen, L., et al. (2018). Molecular cloning, bacterial expression and promoter analysis of pathogenesis-related protein 10 gene from Panax notoginseng. Physiol. Mol. Plant P. 104, 135–140. doi: 10.1016/j.pmpp.2018.10.003
Zandvakili, N., Zamani, M., Motallebi, M., and Moghaddassi Jahromi, Z. (2017). Cloning, overexpression and in vitro antifungal activity of Zea Mays PR10 Protein. Iran J. Biotechnol. 15, 42–49. doi: 10.15171/ijb.1357
Zhai, X., Kong, Q., An, P., and Ren, X. (2018). The function and mechanism of pathogenesis-related 5 protein resistance in cherry tomato in response to Alternaria alternata. Food Biotechnol. 32, 178–190. doi: 10.1080/08905436.2018.1481088
Zhang, J., Wang, F., Liang, F., Zhang, Y., Ma, L., Wang, H., et al. (2018). Functional analysis of a pathogenesis-related thaumatin-like protein gene TaLr35PR5 from wheat induced by leaf rust fungus. BMC Plant Biol. 18:76. doi: 10.1186/s12870-018-1297-2
Zhang, W., Zhao, F., Jiang, L., Chen, C., Wu, L., and Liu, Z. (2018). Different pathogen defense strategies in Arabidopsis: more than pathogen recognition. Cells 7:12. doi: 10.3390/cells7120252
Keywords: Fusarium solani, Panax notoginseng, RNase activity, overexpression, RNAi, defense response
Citation: Li S, Wang Z, Tang B, Zheng L, Chen H, Cui X, Ge F and Liu D (2021) A Pathogenesis-Related Protein-Like Gene Is Involved in the Panax notoginseng Defense Response to the Root Rot Pathogen. Front. Plant Sci. 11:610176. doi: 10.3389/fpls.2020.610176
Received: 25 September 2020; Accepted: 17 December 2020;
Published: 13 January 2021.
Edited by:
Maria Del Mar Jimenez-Gasco, Pennsylvania State University (PSU), United StatesReviewed by:
Jia-He Wu, Chinese Academy of Sciences, ChinaCarla Caruso, University of Tuscia, Italy
Copyright © 2021 Li, Wang, Tang, Zheng, Chen, Cui, Ge and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Diqiu Liu, Diqiuliu@126.com
 Shan Li
Shan Li Zie Wang1,2
Zie Wang1,2 Feng Ge
Feng Ge