- 1Department of Frontier Life Sciences, Faculty of Life Sciences, Kyoto Sangyo University, Kyoto, Japan
- 2Center for Plant Sciences, Kyoto Sangyo University, Kyoto, Japan
The chloroplast-localized cystathionine β-synthase X (CBSX) proteins CBSX1 and CBSX2 have been proposed as modulators of thioredoxins (Trxs). In this study, the contribution of CBSX proteins to the redox regulation of thiol enzymes in the chloroplast Trx system was evaluated both in vitro and in vivo. The in vitro biochemical studies evaluated whether CBSX proteins alter the specificities of classical chloroplastic Trx f and Trx m for their target proteins. However, addition of CBSX proteins did not alter the specificities of Trx f and Trx m for disulfide bond reduction of the photosynthesis-related major thiol enzymes, FBPase, SBPase, and NADP-MDH. In vivo analysis showed that CBSX-deficient mutants grew similarly to wild type plants under continuous normal light conditions and that CBSX deficiency did not affect photo-reduction of photosynthesis-related thiol enzymes by Trx system at several light intensities. Although CBSX proteins have been suggested as modulators in the chloroplast Trx system, our results did not support this model, at least in the cases of FBPase, SBPase, and NADP-MDH in leaves. However, fresh weights of the cbsx2 mutants were decreased under short day. Since Trxs regulate many proteins participating in various metabolic reactions in the chloroplast, CBSX proteins may function to regulate other chloroplast Trx target proteins, or serve as modulators in non-photosynthetic plastids of flowers. As a next stage, further investigations are required to understand the modulation of Trx-dependent redox regulation by plastidal CBSX proteins.
Introduction
Proteins belonging to the chloroplastic thioredoxin (Trx) family are reduced via ferredoxin and ferredoxin-thioredoxin reductase, using electrons from the photosynthetic electron transport pathway in light conditions (Schurmann and Buchanan, 2008; Geigenberger and Fernie, 2014; Buchanan, 2016). The reduced forms of Trx proteins regulate the activity of thiol enzymes, such as those in the Calvin-Benson cycle, by reduction of their disulfide bonds (Michelet et al., 2013). This Trx-dependent regulation of thiol enzymes is known as chloroplast redox regulation; the NADPH-dependent Trx system in the chloroplast is also known as the NTRC system. NTRC proteins uniquely contain both thioredoxin reductase and thioredoxin domains (Serrato et al., 2004; Perez-Ruiz et al., 2006). NTRC system involvement in plastidial redox-regulation processes has also been suggested (Nikkanen and Rintamaki, 2014; Buchanan, 2016; Cejudo et al., 2019).
Typical Trx family proteins in the chloroplast stroma of Arabidopsis thaliana are categorized into five types: two f-type isoforms (Trx f1 and f2); four m-type isoforms (Trx m1, m2, m3, and m4); one x-type isoform (Trx x); two y-type isoforms (Trx y1 and y2); and one z-type isoform (Trx z; Michelet et al., 2013; Serrato et al., 2013; Balsera et al., 2014). Trx f and Trx m were originally identified as regulators of thiol enzymes involved in photosynthetic carbon fixation and related processes in the chloroplast (Jacquot et al., 1978; Wolosiuk et al., 1979). Furthermore, using in vitro biochemical approaches, Trx specificities for their target thiol enzymes were well characterized (Schurmann and Buchanan, 2008; Geigenberger and Fernie, 2014). Calvin-Benson cycle enzymes, including fructose-1,6-bisphosphatase (FBPase; Wolosiuk and Buchanan, 1977; Collin et al., 2003) and sedoheptulose-1,7-bisphosphatase (SBPase; Breazeale et al., 1978; Nishizawa and Buchanan, 1981), and the γ subunit of ATP synthase (Schwarz et al., 1997) were mainly regulated by Trx f. NADP malate dehydrogenase (MDH) was characterized as an enzyme regulated by Trx m (Issakidis et al., 1992), although this enzyme was also later shown to be efficiently activated by Trx f (Collin et al., 2003; Yoshida et al., 2015). However, little analysis of the physiological role of the Trx f and Trx m proteins was available for a long time. Recently, several groups characterized Trx f- and Trx m-deficient mutants using Arabidopsis T-DNA insertion lines and RNA interference. Trx m-deficient mutants were impaired in photoreduction of photosynthesis-related enzymes including FBPase, SBPase, and MDH, and showed severe growth defects as a result (Wang et al., 2013; Okegawa and Motohashi, 2015a; Da et al., 2018). In contrast, Trx f-deficient mutants did not exhibit substantial defects (Yoshida et al., 2015; Naranjo et al., 2016; Thormahlen et al., 2017), although the in vitro biochemical analysis clearly showed that the FBPase (Wolosiuk and Buchanan, 1977) and SBPase (Breazeale et al., 1978) activities were regulated by Trx f. These results suggest that in vivo, Trx m is a more significant redox regulator than Trx f in the activation of FBPase, SBPase, and NADP-MDH. Further research is needed to explain the discrepancy between the in vitro and in vivo studies.
As a regulator of Trx proteins, thioredoxin-interacting protein (TXNIP) was first identified in mammals (Nishiyama et al., 1999). TXNIP interacts with the active site of Trx and inhibits its disulfide reduction activity (Spindel et al., 2012). In plants, CBSX proteins were identified as candidate Trx regulators (Yoo et al., 2011; Jung et al., 2013). CBSX proteins can enhance Trx disulfide reduction activity, in contrast to TXNIP (Yoo et al., 2011). In cyanobacteria, fusion proteins containing a CBS domain and CP12 component were found (Stanley et al., 2013). Plant CP12 protein forms complexes with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and phosphoribulokinase (PRK), which are also well known redox-regulated Calvin-Benson cycle enzymes. It thus functions as a regulator in the Calvin-Benson cycle (Michelet et al., 2013). The discovery of CBS-CP12 fusion proteins in cyanobacteria implies that the CBS-domain may act as a regulator in the redox regulation system (Lopez-Calcagno et al., 2014). Arabidopsis CBSX1 and CBSX2 were indeed proposed as Trx regulators in chloroplasts (Yoo et al., 2011; Jung et al., 2013).
In this study, we evaluated the possibility that CBSX1 and CBSX2 regulate Trx f and Trx m in the chloroplast redox regulation system using in vitro and in vivo experiments. Specifically, we investigated whether CBSX1 and CBSX2 modulates the target specificity of Trx f and Trx m in redox regulation of the photosynthesis-related thiol enzymes FBPase, SBPase, and NADP-MDH. However, we could not find any evidence that support this possibility. CBSX1 and CBSX2 may function not as a Trx regulator for Calvin-Benson cycle enzyme activation, but rather as a regulator for other thiol enzymes in flowers and leaves.
Materials and Methods
Construction of Expression System for Arabidopsis CBSX1, CBSX2, FBPase, SBPase, and NADP-MDH
The mature forms of Arabidopsis chloroplast proteins were predicted using TargetP 1.1 (Emanuelsson et al., 2007). The mature forms of the CBSX1 (AT4G36910), CBSX2 (AT4G34120), FBPase (AT3G54050), SBPase (AT3G55800), and NADP-MDH (AT5G58330) genes were obtained by PCR amplification from an Arabidopsis cDNA library (Yamazaki et al., 2004). Primers for cloning and vector constructs are described in Supplementary Table S1. The amplified CBSX1 and CBSX2 genes were directly inserted by seamless cloning into the NdeI and BamHI sites of pET23a and the NcoI and EcoRI sites of pET23d, respectively (vectors from Merck Millipore, Darmstadt, Germany), using seamless ligation cloning extract (SLiCE) from a laboratory Escherichia coli JM109 strain (Okegawa and Motohashi, 2015b; Motohashi, 2017b). The amplified FBPase and SBPase genes were cloned into the NdeI and EcoRI sites of pET23a, and the amplified NADP-MDH gene into the NcoI and BamHI sites of pET23d, by the conventional restriction enzyme method. The length of cloned DNA inserts was confirmed by colony PCR (Okegawa et al., 2016; Motohashi, 2017a), using T7 promoter and T7 terminator primers (Okegawa and Motohashi, 2015c). Plasmids were prepared from colony PCR-positive clones and the inserted DNA sequences of plasmids were confirmed by DNA sequencing (Sanger et al., 1977).
Preparation of Arabidopsis CBSX1, CBSX2, FBPase, SBPase, NADP-MDH, Trx f1, Trx m1, and Trx m4 Proteins
Arabidopsis proteins were overexpressed in E. coli BL21 (DE3; Motohashi et al., 2009).
Arabidopsis CBSX1, CBSX2, FBPase, and SBPase proteins were purified as follows. Escherichia coli cells overexpressing Arabidopsis proteins were suspended in 25 mM Tris-HCl (pH 7.5) and disrupted by sonication (Sonifier® 250, Branson) at 4°C. The disrupted cells were centrifuged at 100,000 g for 40 min. The supernatant (crude extract) was applied to a Toyopearl® DEAE-650 M column (Tosoh) and eluted using a linear gradient of NaCl (0–300 mM) in 25 mM Tris-HCl (pH 7.5). Peak fractions containing each Arabidopsis protein target were collected, and solid ammonium sulfate was added to obtain a final concentration of 1.4 M. The solution was then loaded into a Toyopearl® Butyl-650 M column (Tosoh) and eluted using an inverse gradient of ammonium sulfate (1.4–0 M) in 25 mM Tris-HCl (pH 7.5). Peak fractions containing each Arabidopsis protein were collected. Purified CBSX1 and SBPase were concentrated using Amicon® Ultra-15 (10,000 NMWL membrane, Merck Millipore); the buffer in the protein solution was exchanged with 25 mM Tris-HCl (pH 7.5). Purified CBSX1 and SBPase were stored at −80°C in 16% (v/v) glycerol. The protein fractions of CBSX2 and FBPase underwent further column chromatography for additional purification. CBSX2 collected fractions were dialyzed into 10 mM sodium phosphate buffer (pH 6.8). The solution was then applied to an HA-Ultrogel (Pall) and eluted using a sodium phosphate buffer gradient (pH 6.8, 10–200 mM). His-tagged FBPase collected fractions were dialyzed into 25 mM Tris-HCl (pH 7.5). The solution was then applied to the TALON® metal affinity resin (Takara Bio USA, Inc., Mountain View, CA, United States) equilibrated with 300 mM NaCl and 25 mM Tris-HCl (pH 7.5) and eluted with the buffer, which contained 150 mM imidazole. Purified CBSX2 and FBPase were concentrated and stored at −80°C in 16% (v/v) glycerol.
Arabidopsis NADP-MDH was purified without an affinity tag in the absence of dithiothreitol (DTT) as described previously (Okegawa and Motohashi, 2016).
Arabidopsis Trx f1 (AT3G02730) was purified without an affinity tag as follows (Motohashi and Hisabori, 2006). Escherichia coli cells overexpressing Arabidopsis Trx f1were suspended in 25 mM Tris-HCl (pH 8.1) and disrupted by sonication (Sonifier® 250, Branson) at 4°C. The disrupted cells were centrifuged at 100,000 g for 40 min. The supernatant (crude extract) was applied to a Toyopearl® QAE-550C column (Tosoh) and eluted using a linear gradient of NaCl (0–300 mM) in 25 mM Tris-HCl (pH 8.1). Peak fractions containing Trx f1 were collected, and solid ammonium sulfate was added to obtain a final concentration of 1.4 M. The solution was then loaded into a Toyopearl® Butyl-650 M column (Tosoh) and eluted using an inverse gradient of ammonium sulfate (1.4–0 M) in 25 mM Tris-HCl (pH 8.1). Peak fractions containing Trx f1 were collected and dialyzed into 25 mM MES-NaOH (pH 6.1). The solution was loaded into a Toyopearl® SP-650M column (Tosoh) and eluted using a linear gradient of NaCl (0-300 mM) in 25 mM MES-NaOH (pH6.1). Peak fractions containing Trxf1 were collected and then concentrated using Amicon® Ultra-15 (10,000 NMWL membrane); the buffer in the protein solution was exchanged with 25 mM Tris-HCl (pH 7.5). Purified Trx f1 was stored at −80°C in 16% (v/v) glycerol.
Trx m1 (AT1G03680; Motohashi et al., 2003, 2009) and Trx m4 (AT3G15360; Motohashi et al., 2009; Motohashi and Okegawa, 2014) were purified by conventional column chromatography, as described previously, without DTT. Deduced amino acid sequences of Trx m1, m2, and m4 proteins were aligned by ClustalW (Thompson et al., 1994) and formatted by ESPript 3.0 (Robert and Gouet, 2014).
Preparation of Antibodies
Polyclonal antibodies were prepared from rabbits immunized with CBSX1, CBSX2, FBPase, and SBPase (Okegawa and Motohashi, 2015a), and NADP-MDH (Okegawa and Motohashi, 2016) recombinant proteins. Purified Trx f1 and Trx m1 were also used as antigens for polyclonal antibodies production. Polyclonal antibodies for PrxA (AT3G11630) and PrxB (AT5G06290) were prepared as peptide antibodies against C-terminal consensus amino acid sequence between PrxA and PrxB (Cys-SMKPDPKLSKEYFSAI). Polyclonal antibodies for PetA (ATCG00540) and PetC (AT4G03280) were prepared from rabbits immunized with partially purified inclusion bodies from recombinant expression proteins (Supplementary Table S1).
Reduction Assay of Trx-Target Proteins in vitro
Thioredoxins-dependent redox-regulated proteins (FBPase, SBPase, and NADP-MDH; 3 μM) were incubated in the presence of DTT (0–1 mM) in 3 mM MgCl2 and 25 mM Tris-HCl (pH 7.5) for 30 min at 25°C. Each enzyme was incubated with either Trx-f or Trx-m alone (total 5 μM), and with one of these plus 3 mM adenosine 5'-monophosphate (AMP) and each of CBSX1 or CBSX2 (total 5 μM). Reactions were terminated by precipitation with trichloroacetic acid (final 10%). The precipitants were washed with ice-cold acetone and dissolved in buffer containing 125 mM Tris-HCl (pH 6.8), 4% sodium dodecyl sulfate (SDS), 20% glycerol, and 2 mM 4-acetamido-4'-maleimidylstilbene-2,2'-disulfonic acid (AMS). The samples were incubated for 30 min at 25°C with vigorous mixing, and then denatured for 2 min at 95°C. Reduced and oxidized proteins were separated by non-reducing SDS-PAGE and stained by Coomassie Brilliant Blue (CBB) R-250.
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia-0 was used as the wild type. The T-DNA insertion line GABI_050D12 (cbsx1) was obtained from the Nottingham Arabidopsis stock center (NASC, United Kingdom). The T-DNA insertion line SALK_136934C (cbsx2; Jung et al., 2013) was obtained from the Salk Institute Genomic Analysis Laboratory (SIGnAL, CA, United States). Plants were grown in soil, or in petri dishes containing Murashige Skoog (MS) medium (Murashige and Skoog, 1962) with 1.2% (w/v) agar and 1% (w/v) sucrose in the growth-chambers (50 μmol photons m−2 s−1, continuous light, 23°C). Agar-sown seeds were first surface sterilized by soaking in 5% (v/v) sodium hypochlorite for 5 min. To generate double mutants, cbsx1 and cbsx2 T-DNA single mutants were crossed. The cbsx1 and cbsx2 double mutant was selected from F2 plants. The presence of T-DNA insertions was confirmed by PCR (for primer, see Supplementary Table S2) and DNA agarose gel electrophoresis (Motohashi, 2019).
Analysis of mRNA Expression in cbsx Mutants by RT-PCR
The mRNA from wild type and cbsx mutants was prepared using Sepasol®-RNA I Super G (Nakalai tesque, Kyoto, Japan), and cDNA samples were synthesized for 20 min at 42°C by ReverTra Ace® (Toyobo, Osaka, Japan), using oligo(dT)20. The cDNAs from wild type and cbsx mutants were amplified by KAPATaq EXtra (KAPA Biosystems, Wilmington, MA, United States), with the following cycle conditions: 94°C for 2 min, (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min) × 25 cycles. The primers used are listed in Supplementary Table S2. Amplified samples were electrophoresed and visualized by a combination of a black light and a longpass emission-filter (SC-46, Fujifilm, Tokyo, Japan; Motohashi, 2019).
Expression Analysis of CBSX1, CBSX2, and Redox Regulation-Related Proteins by Western Blotting
To analyze protein expression in Arabidopsis, leaves and flowers of wild type and cbsx mutants were frozen and ground in liquid nitrogen; total proteins were extracted in 1 × SDS sample buffer [2% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 10% (v/v) glycerol, and 62.5 mM Tris-HCl (pH 6.8)]. Extracted proteins (30 or 90 μg) were loaded on SDS-PAGE [15% (w/v) acrylamide] and detected by specific antibodies. Immunoblot signal was detected with a LAS-3000UVmini lumino-image analyzer (Fujifilm, Tokyo, Japan). Uncropped original images were indicated in Supplementary Figure S1.
Chlorophyll Fluorescence Analysis
Chlorophyll fluorescence was measured with a Mini-pulse-amplitude modulation (PAM) portable chlorophyll fluorometer (Walz, Germany). Minimum fluorescence (Fo) was obtained with open PSII centers in the dark-adapted state by a low-intensity measuring light (wavelength 650 nm, 0.05–0.1 μmol photons m−2 s−1). A saturating pulse of white light (800 ms, 3,000 μmol photons m−2 s−1) was applied to determine the maximum fluorescence with closed PSII centers in the dark-adapted state (Fm) and during actinic light (AL) illumination (Fm'). The steady-state fluorescence level (Fs) was recorded during AL illumination (25-828 μmol photons m−2 s−1). The quantum yield of PSII (ΦPSII) was calculated as (Fm'−Fs)/Fm' (Genty et al., 1989). The relative rate of electron transport through PSII (ETR) was calculated as ΦPSII × light intensity (μmol photons m−2 s−1). Non-photochemical quenching (NPQ) was calculated as (Fm−Fm')/Fm'. For the analysis of light-intensity dependence of fluorescence parameters, AL intensity was increased in a stepwise manner every 2 min after the application of a saturating pulse.
Photo-Reduction Assay of Trx-Target Proteins in vivo
Photo-reduction of Trx target enzymes in seedlings was determined using the free thiol-specific modifying reagent, 4-acetamido-4'-maleimidylstilbene-2,2'-disulfonic acid (AMS; Thermo Fisher Scientific, Carlsbad, CA, United States), as described by (Okegawa and Motohashi, 2015a), with minor modifications. Seedlings were incubated in the dark for 3 h and then illuminated at three different light intensities for 1 h (50, 200, or 800 μmol photons m−2 s−1). Seedlings were collected thereafter and frozen in liquid nitrogen. Frozen samples were ground using 5 mm tungsten beads and incubated in 10 mM AMS, 10 mM EDTA, 4% (w/v) SDS, 8 M urea, and 125 mM Tris-HCl (pH 6.8) for 2 h at 25°C to complete the labeling of thiol groups with AMS. Hydrophilic proteins were separated from the lipid fraction by the addition of an equal volume of chloroform to the sample. The aqueous portion was collected by centrifugation at 15,000 g for 10 min). Reduced and oxidized proteins were separated by non-reducing SDS-PAGE [12.5% (w/v) acrylamide] and detected by western blotting. Immunoblot signal was detected with a LAS-3000UVmini lumino-image analyzer (Fujifilm, Tokyo, Japan). The reduction level was quantified using Multi Gauge 3.1 software (Fujifilm, Tokyo, Japan) and the ratio of the reduced form to total protein was determined. Uncropped original images were indicated in Supplementary Figure S1.
Results
Contribution of CBSX Proteins to Reduction of Photosynthesis-Related Thiol Enzymes, in vitro
Previous work showed that CBSX1 and CBSX2 proteins interact with Trxs (Yoo et al., 2011; Jung et al., 2013), and that CBSX1 protein directly binds with both Trx f and Trx m proteins in vitro (Yoo et al., 2011). Photosynthesis-related thiol enzymes, including several Calvin-Benson cycle enzymes, are regulated by Trx proteins in the light. Published in vitro studies indicate preferential reduction by Trx f of FBPase (Wolosiuk and Buchanan, 1977; Collin et al., 2003) and SBPase (Breazeale et al., 1978; Nishizawa and Buchanan, 1981), whereas NADP-MDH is reduced by both Trx f and Trx m proteins (Issakidis et al., 1992; Collin et al., 2003). In contrast, in vivo, levels of photo-reduction in thiol enzymes such as FBPase, SBPase, and NADP-MDH were significantly decreased in the Trx m-deficient mutants (Okegawa and Motohashi, 2015a). If chloroplast CBSX1 and CBSX2 proteins function as modulators of the chloroplast Trx system, the discrepancy between in vitro and in vivo results may be explained. To evaluate this possibility, we firstly evaluated the contribution of CBSX proteins to the reduction of thiol enzymes by Trx f and Trx m proteins, using purified recombinant proteins in vitro. Our results were consistent with previous reports described above. FBPase was efficiently reduced in vitro by Trx f1, not Trx m1 (Figure 1A, Trx-only elements), and SBPase was reduced by both Trx f1 and Trx m1, but Trx f1 could reduce more efficiently than Trx m1 (Figure 1B, Trx only elements). On the other hand, NADP-MDH was reduced to the same extent by both Trx f1 and Trx m1 (Figure 1C, Trx only elements). Given this, the effects of addition of CBSX1 or CBSX2 on Trx-dependent reduction of the thiol enzymes by Trx f1 and m1 were evaluated, in vitro. Since CBSX proteins are known to function with AMP as a ligand (Ereno-Orbea et al., 2013), 3 mM of AMP was added to each reaction medium. Under these reaction conditions, both Trx f1 and Trx m1 were reduced in a DTT concentration-dependent manner (Supplementary Figure S2). Reduction of FBPase and SBPase by Trx f1 and Trx m1 was not affected by the addition of CBSX1 (Figure 1, CBSX1 elements) or CBSX2 (Figure 1, CBSX2 elements) with AMP. Trx f1 reduced FBPase and SBPase disulfide bonds as efficiently in the presence of CBSXs and AMP as with Trx f1 alone (Figures 1A,B). The addition of CBSX1 or CBSX2 did not also affect reduction of NADP-MDH by Trx f1 and Trx m1 with AMP; NADP-MDH was equally reduced by Trx f1 and Trx m1 in the presence of CBSXs and AMP (Figure 1C). In addition to Trx m1, Arabidopsis has abundant Trx m4 protein in its chloroplasts (Okegawa and Motohashi, 2015a). As with Trx m1, reduction patterns of the three thiol enzymes by Trx m4 in the presence of CBSX1 or CBSX2 were not affected [Figure 1 (Trx m1) and Supplementary Figure S3]. These in vitro experiments did not indicate any contribution of CBSX proteins to Trx-dependent reduction of three photosynthesis-related thiol enzymes under studied conditions.
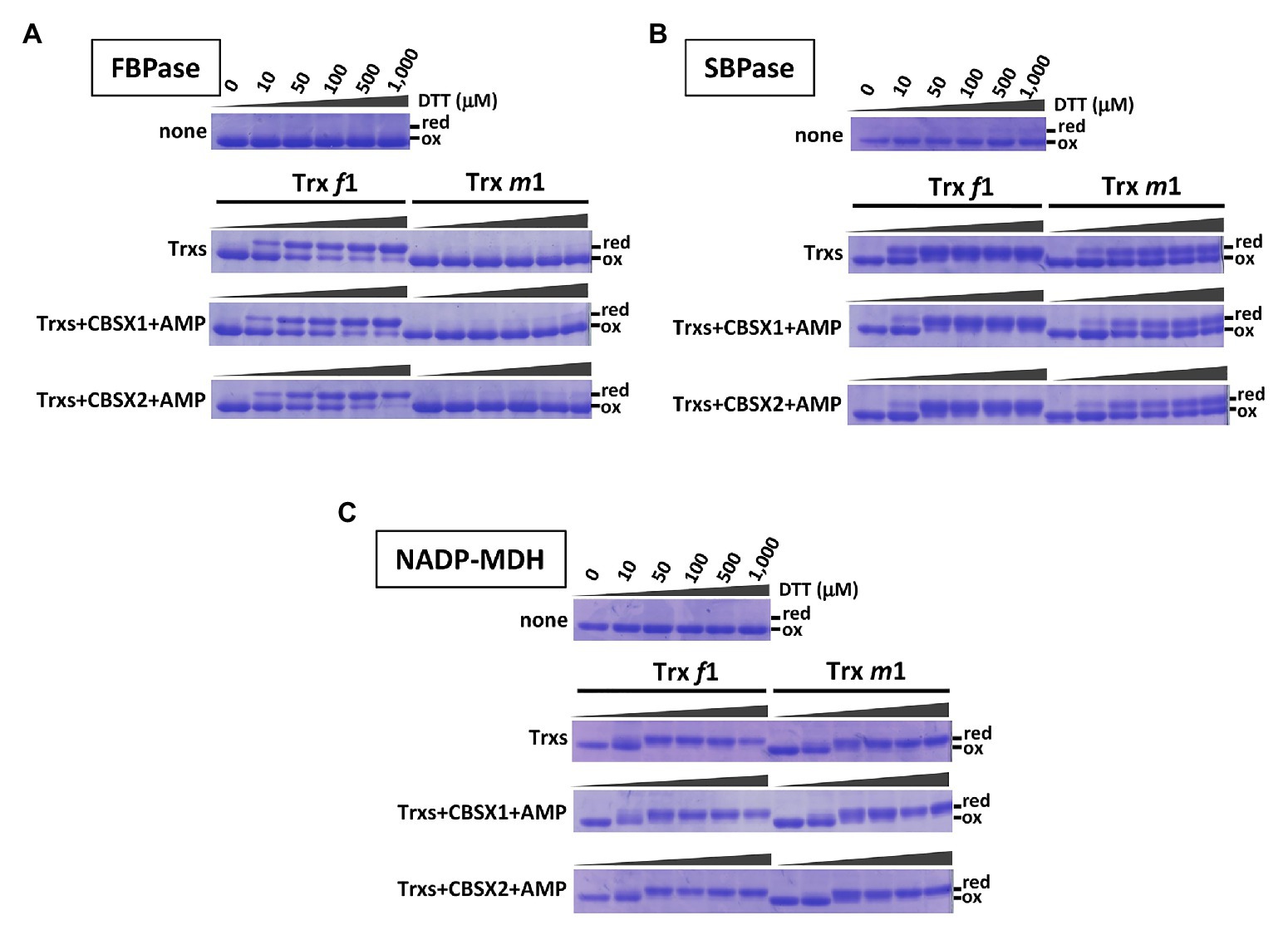
Figure 1. Contribution of cystathionine β-synthase X (CBSX) proteins to thioredoxins (Trx)-dependent reduction of thiol-enzymes, in vitro. Thiol-enzyme proteins (3 μM) were reduced in each dithiothreitol (DTT) concentration for 30 min at 25°C. DTT concentrations in each result shown are the same as for the first element. None: contained thiol-enzyme only; Trxs: thiol-enzyme with 5 μM of the indicated Trx; Trxs+CBSXs+AMP: thiol-enzyme with 5 μM indicated Trx, 5 μM CBSX1 or CBSX2 as indicated, and 3 mM adenosine 5'-monophosphate (AMP). Reduced (red) and oxidized (ox) forms of thiol-enzyme were detected by CBB staining. (A): FBPase; (B): SBPase; and (C): NADP-MDH.
Arabidopsis cbsx1 and cbsx2 Mutants Did Not Show Obvious Visible Defects
Arabidopsis has two chloroplast-localized CBSX proteins, namely CBSX1 and CBSX2. The deduced amino acid sequences of these proteins were highly conserved, showing 91% identity. As T-DNA insertion mutants of CBSXs, the cbsx1 (GABI_050D12), and cbsx2 (SALK_136934C) mutants were used to investigate the proteins’ physiological significance (Figure 2A). The cbsx1 and cbsx2 mutants did not express the mRNAs of CBSX1 and CBSX2, respectively (Figure 2B). To obtain the double mutant cbsx1 cbsx2, the cbsx1 and cbsx2 single mutants were crossed. The cbsx mutants did not show obvious phenotypic defects compared with the wild type under normal growth conditions in soil for 5 weeks (Figure 2C). We could not observe morphological abnormalities (Figure 2C) and differences of chlorophyll content of seedlings (Figures 2D,E) between wild type and the cbsx mutants under two different growth conditions. However, the fresh weight of the cbsx mutants was gradually decreased in the order of wild type, single mutants, and double mutant in soil under short day condition (Figure 2F). The fresh weight of the cbsx2 and cbsx double mutant grown under short day condition were decreased to ~74% and ~65% of that of wild type (Figure 2F), although there were no obvious differences in the fresh weights between mutants and wild type shoots when these were grown on MS medium agar plates for 3-week-old (Figure 2G).
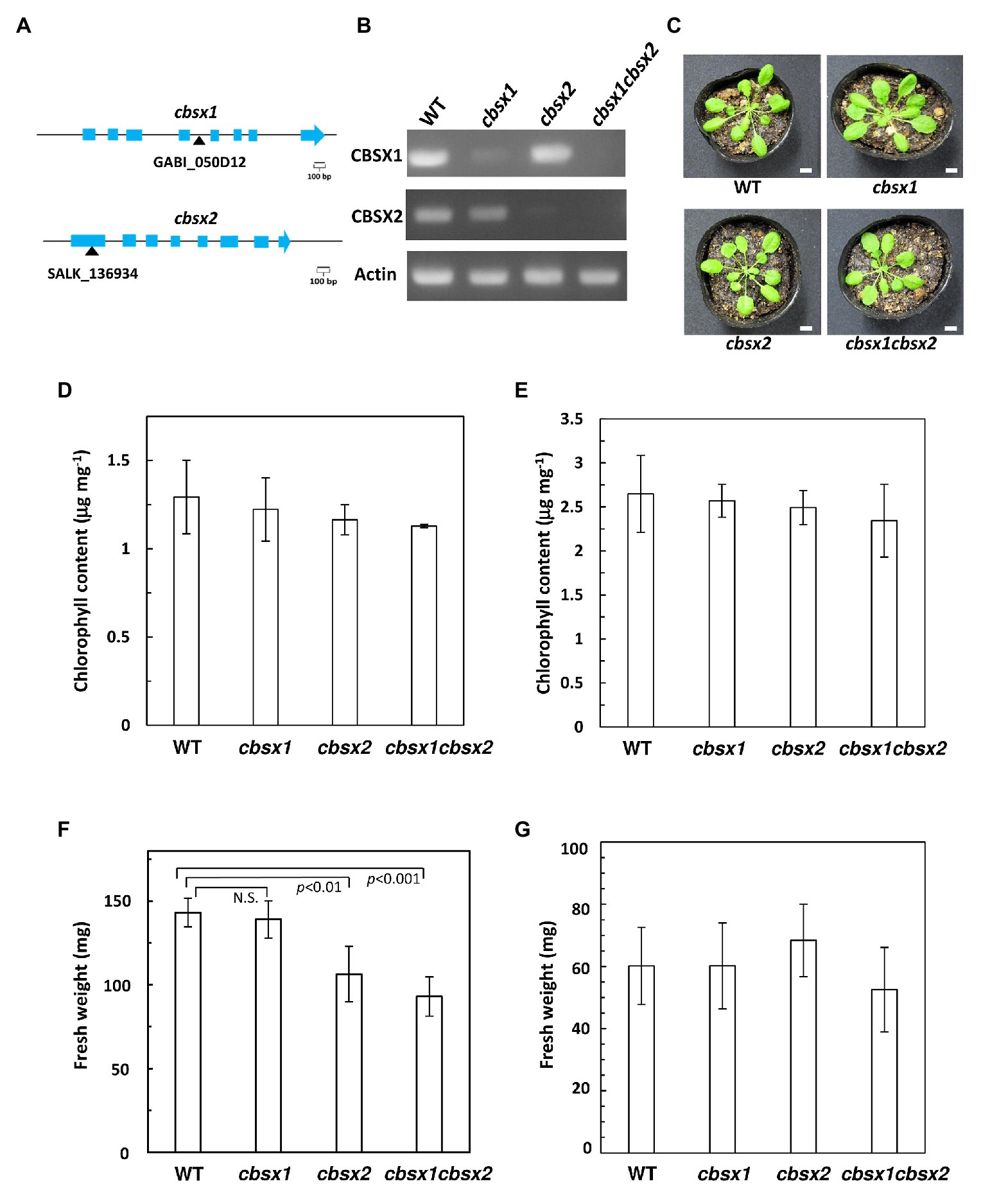
Figure 2. Growth phenotype in Arabidopsis cbsx mutants. (A) Schematic diagram of CBSX1 and CBSX2 gene structures. The T-DNA insertion sites are indicated with black triangles; light blue boxes indicate coding regions. (B) Analysis of mRNA expression in Arabidopsis cbsx mutants by RT-PCR. (C) Phenotype of cbsx mutants grown in soil for 35 days in growth chambers [50 μmol photons m−2 s−1, short day (8 h light/16 h dark), 23°C]. White bars indicate 10 mm. (D) Chlorophyll content of seedlings [50 μmol photons m−2 s−1, short day (8 h light/16 h dark), 23°C], per unit fresh weight (n = 3). (E) Chlorophyll content of seedlings (50 μmol photons m−2 s−1, continuous light, 23°C), per unit fresh weight (n = 3). (F) Fresh weight of seedlings (n = 10) grown on soil for 35 days in growth-chambers [50 μmol photons m−2 s−1, short day (8 h light/16 h dark), 23°C]. Statistical analyses were performed using the Student’s t-test. N.S., not significant. (G) Fresh weight of seedlings (n = 10) grown on Murashige Skoog (MS) medium for 22 days in growth-chambers (50 μmol photons m−2 s−1, continuous light, 23°C). Each value is shown as the mean ± SD of independent plants.
Arabidopsis CBSX1 and CBSX2 Proteins Are Present in Leaves and Flowers
According to the Arabidopsis eFP browser 2.0 database for gene expression in Arabidopsis (Winter et al., 2007), the mRNA of CBSX genes is expressed in leaves, flowers, and seeds. To detect proteins in Arabidopsis tissues, we generated specific antibodies for CBSX1 and CBSX2 proteins. The mature forms of the CBSX1 and CBSX2 proteins were predicted to be ~18 kDa proteins, using ChloroP 1.1 (Emanuelsson et al., 1999), and were detected as ~20 kDa proteins in wild type Arabidopsis, using antibodies obtained as described above. The specificities of the antibodies for CBSX1 and CBSX2 were confirmed by loss of signal in each deficient mutant (Figure 3). The CBSX1 and CBSX2 proteins were present in leaves (Figure 3A, WT) and flowers (Figure 3B, WT), as suggested by the Arabidopsis eFP browser. Western blots detected two bands for CBSX2 protein. We evaluated the two bands as resulting from distinct processing of chloroplast transit sequence elements, not as distinct redox forms, because the CBSX1 and CBSX2 proteins had no cysteine residue in the mature forms. In the cbsx deficient mutants, chloroplast Trxs (Trx f1 and m1), Trx-regulated thiol enzymes (FBPase, SBPase, and NADP-MDH), and PrxA and PrxB that were supplied reducing equivalents by Trxs, were normally detected in leaves (Figure 3A; Supplementary Figure S4). These results indicated that the disappearance of CBSX proteins did not affect the abundance of Trx-related proteins in Arabidopsis photosynthetic leaves.
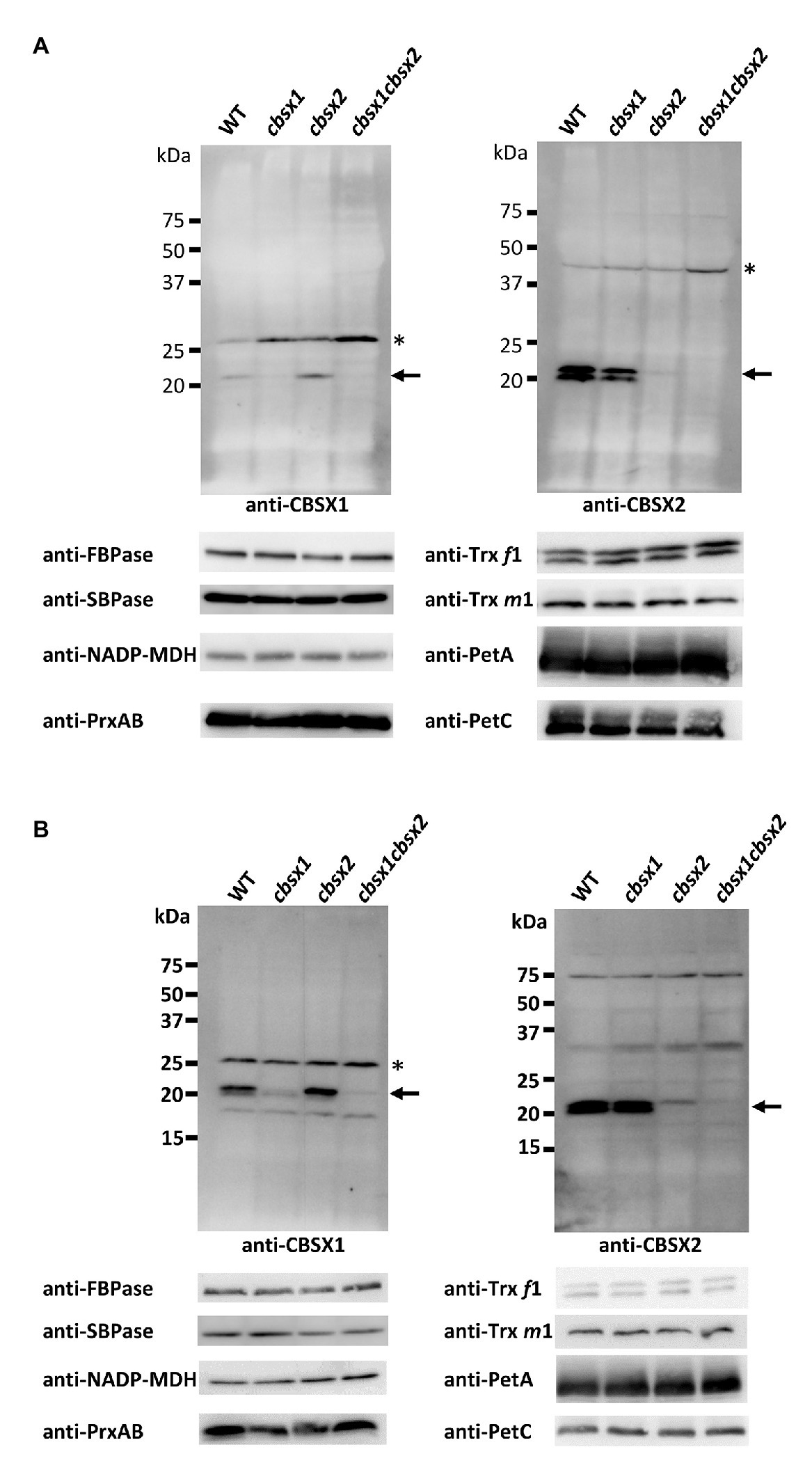
Figure 3. Protein accumulation of CBSX proteins and redox regulation-related proteins in the leaves (A) and flowers (B) of cbsx mutants. Thirty or ninety micrograms of protein were loaded in each lane. Arrows indicate position of CBSX1 or CBSX2; asterisks indicate positions of non-specific bands.
The cbsx Single and Double Mutants Showed Normal Electron Transport Activities in Leaves
CBSX1 and CBSX2 proteins are suggested to function as modulators of Trxs in leaves, which regulate their target enzyme activity (Yoo et al., 2011; Lopez-Calcagno et al., 2014). Several Calvin cycle enzymes are Trx-regulated proteins. If CBSX proteins are functional regulators for Trxs, photosynthetic activity in leaves may be affected in cbsx mutants. To characterize photosynthetic electron transport activity in the mutants, their chlorophyll fluorescence parameters were analyzed (Figure 4). The maximum quantum yield of PSII (Fv/Fm) reflects the intactness of PSII. The three cbsx mutants exhibited the same level of Fv/Fm as the wild type (Figure 4A). The light-intensity dependence of the electron transport rate (ETR) is an indicator of the relative flow rate of electrons through PSII during steady-state photosynthesis, and the NPQ of chlorophyll fluorescence mainly reflects the level of thermal dissipation of excess light energy. No differences in ETR and NPQ were observed between the wild type and the cbsx mutants (Figures 4B,C). These results showed that a deficiency of CBSX did not affect these photosynthetic pathways.
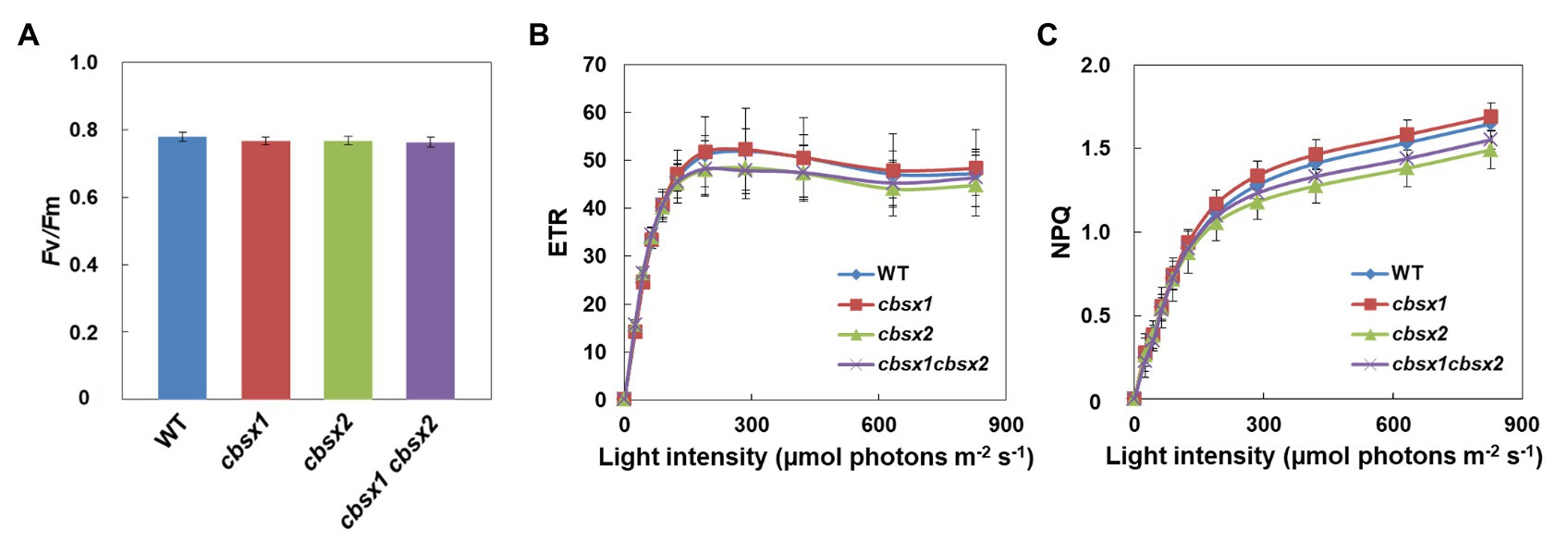
Figure 4. Electron transport activity in the wild type (WT), cbsx1, cbsx2, and cbsx1 cbsx2 mutants. (A) The maximum quantum yield of PSII (Fv/Fm). Each value is the mean ± SD of 10 independent replicates. (B) Light-intensity dependence of the electron transport rate (ETR). The ETR was calculated as ΦPSII × light intensity (μmol photons m−2 s−1). Each value is the mean ± SD of five independent replicates. (C) Light-intensity dependence of the non-photochemical quenching (NPQ) of chlorophyll fluorescence. Each value is the mean ± SD of five independent replicates. Seedlings were grown in soil for 34 days in growth-chambers (50 μmol photons m−2 s−1, continuous light, 23°C).
Normal Photo-Reduction of Photosynthesis-Related Thiol Enzymes in cbsx Mutants
We next investigated the contributions of CBSX1 and CBSX2 to Trx-dependent redox regulation in chloroplasts, using the cbsx1, cbsx2, and cbsx1 cbsx2 mutants. Photosynthesis-related thiol enzymes, including several Calvin-Benson cycle enzymes, are reduced by Trx, using electrons from photosynthetic electron transport, and the enzymes are thus activated in light conditions (Hisabori et al., 2007; Michelet et al., 2013; Geigenberger and Fernie, 2014; Buchanan, 2016). If CBSX proteins modulate the activity of photosynthesis-related thiol enzymes via a Trx-dependent system, the photo-reduction levels of thiol enzymes should be altered in the cbsx mutants. FBPase, SBPase, and NADP-MDH disulfide bonds were reduced by Trx in a light intensity-dependent manner in the wild type (Figure 5, WT). The photo-reduction of FBPase, SBPase, and NADP-MDH in the cbsx mutants was compared to that in the wild type (Figure 5, upper elements). At all light intensities studied, the photo-reduction levels of FBPase, SBPase, and NADP-MDH did not differ between the wild type and cbsx mutants (Figure 5, lower elements). Additionally, Trxs (Trx f and Trx m) and Trx-related proteins [FBPase, SBPase, NADP-MDH, 2-Cys peroxiredoxin A (PrxA), and PrxB] were normally present in leaves of the three cbsx mutants (Figure 3A). This in vivo analysis thus showed no observable contribution of CBSXs to redox regulation of FBPase, SBPase, and NADP-MDH under the studied conditions.
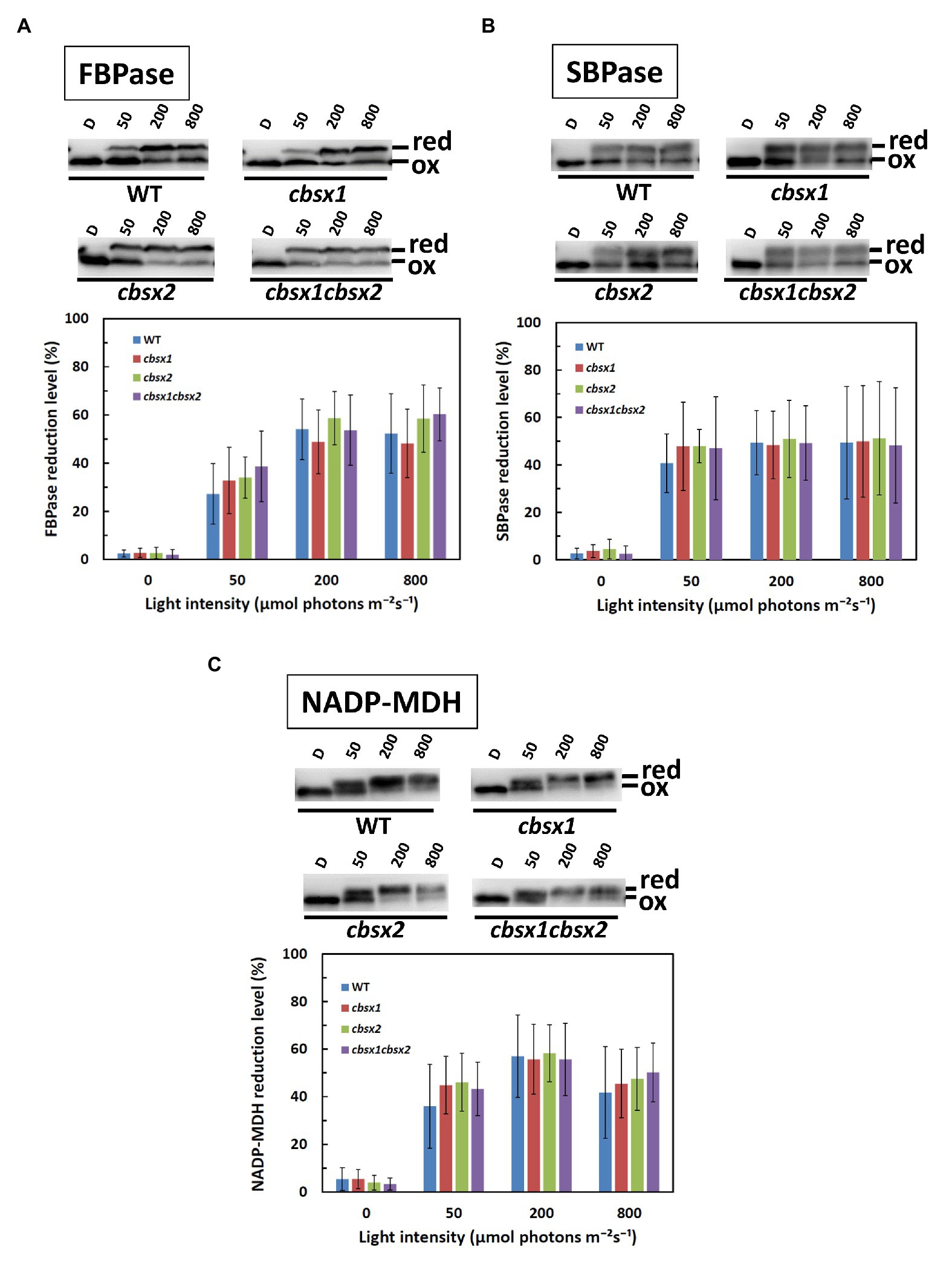
Figure 5. Photo-reduction of photosynthesis-related thiol enzymes in the wild type (WT) and the cbsx mutants. (A): FBPase; (B): SBPase; and (C): NADP-MDH. Seedlings were left in darkness for 3 h and illuminated at 50, 200, and 800 μmol photons m−2 s−1 steps for 1 h each. Samples were collected at each of these light intensities and modified with 4-acetamido-4'-maleimidylstilbene-2,2'-disulfonic acid (AMS). Ninety micrograms of protein samples were loaded on non-reducing SDS-PAGE. Reduced (red) and oxidized (ox) forms were detected by western blotting (upper element of each panel), and reduction levels of thiol enzymes were determined as described in Materials and Methods (lower element of each panel). Each value is the mean ± SD (independent-biological replicates, FBPase: n = 9; SBPase: n = 8; and NADP-MDH: n = 7).
Discussion
Arabidopsis CBSX proteins are proposed as modulators of Trx activity in previous works (Yoo et al., 2011; Jung et al., 2013). Although direct interactions between Trxs and CBSXs have been reported (Yoo et al., 2011), their regulatory mechanism is still unknown. CP12 protein forms the PRK/GAPDH/CP12 regulatory complex in Trx-dependent redox regulation. In cyanobacteria, a fused CP12-CBSX gene was also found (Stanley et al., 2013). These fusion genes imply that CBSX proteins may function as a modulator that cooperates with CP12 in Trx-dependent redox regulation. Lopez-Calcagno et al. (2014) suggested that CBSX proteins are modulators in the redox regulation of Trx-dependent thiol-enzymes in Arabidopsis and cyanobacteria (Lopez-Calcagno et al., 2014). However, few biochemical and physiological analyses of CBSX proteins in Trx-dependent redox regulation have been performed. In particular, their contribution to photosynthesis-related Calvin cycle thiol enzyme reduction in leaves has not been tested. In this study, we evaluated the contributions of CBSX1 and CBSX2 proteins to the redox regulation of photosynthesis-related thiol enzymes by chloroplast Trx f and Trx m, in vitro and in vivo.
In vitro, we evaluated whether CBSX proteins can alter the target specificity of Trx f and Trx m, using photosynthesis-related thiol enzymes. We could not demonstrate that CBSX1 and CBSX2 proteins modulated the Trx f- or Trx m-dependent reduction of thiol enzymes such as FBPase, SBPase, and NADP-MDH. CBSX proteins may contribute to the activation of thiol enzymes other than these, because the chloroplast Trx system does function in various redox regulation pathways (Motohashi et al., 2001; Buchanan and Balmer, 2005). Alternatively, other ligands for CBS-domain proteins may be required. CBS-domain proteins are known in wide variety of organisms, and require AMP as a ligand for activation (Baykov et al., 2011). However, several other ligands, including ADP, ATP, and NADH, have been also suggested for CBSXs (Ereno-Orbea et al., 2013). In this study, we used AMP, as the best-characterized ligand. Under these conditions, CBSX proteins had no observable effect on Trx f or Trx m-dependent activation of photosynthesis-related proteins, but other compounds may be bound as ligands to Arabidopsis CBSX1 and CBSX2 proteins in chloroplast redox regulation. Arabidopsis has also Trx m2 protein, in addition to Trx m1 and m4, as a major m-type Trx. Trx m2 is closely related to Trx m1 (81% identity in their deduced amino acid sequences, Supplementary Figure S5) and has the same redox potential value (−335 mV) and similar enzyme kinetic parameters (Yoshida and Hisabori, 2017), although Trx m4 has lower similarity to Trx m1 (60% identity of their deduced amino acid sequences, Supplementary Figure S5) and a different redox potential value (−312 mV). From these facts, we concluded that Trx m2 has similar properties to Trx m1 proteins and is unlikely to be changed due to specificity by CBSX proteins.
In vivo, the cbsx1, cbsx2, and cbsx1 cbsx2 mutants did not exhibit obvious morphological defects compared to the wild type (Figure 2C). Consistent with their phenotype, the photosynthetic parameters of the three cbsx mutants did not differ from those of the wild type (Figure 4). Deficiencies of the CBSX proteins did not affect photosynthetic activity. Moreover, we analyzed photoreduction of three thiol enzymes, FBPase, SBPase, and NADP-MDH, in three cbsx mutants. The photo-reduction rates of these enzymes in the cbsx mutants were similar to those of the wild type (Figure 5). For these photosynthesis-related enzymes, the CBSX proteins may not be required for regulation of their activities by Trx f or Trx m under the light conditions used in this study. However, fresh weight of the cbsx single and double mutants was gradually decreased in soil under short day condition (Figure 2F). Particularly, fresh weight of the cbsx double mutant was decreased to ~65% of that of WT during the condition (Figure 2F), although obvious difference in growth of the cbsx mutants could not be observed on MS plates under continuous light (Figures 2E,G). Interestingly, regarding other redox-related gene deficient mutants, Lepistö et al. reported that the ntrc mutants displayed no phenotype when grown under continuous light, but exhibited a strong growth suppression under short day condition (Lepisto et al., 2009). The ntrc mutant grown under short day condition had smaller mesophyll cells and contained fewer chloroplasts than wild type leaves. Similar mechanism may also function in the cbsx mutants.
From the in vitro and in vivo analyses, we concluded that CBSX1 and CBSX2 proteins did not function as modulators of Trx f and Trx m in redox regulation of the three major thiol enzymes FBPase, SBPase, and NADP-MDH in leaves, at least under the studied conditions. In the last 20 years, a large number of Trx target proteins have been newly identified (Geigenberger et al., 2017). More recently, new target proteins have also been identified (Okegawa and Motohashi, 2020; Yoshida et al., 2020). Although Arabidopsis CBSX proteins are suggested to modulate Trx-activity (Yoo et al., 2011; Jung et al., 2013), it is unclear which Trx-dependent pathways do CBSX proteins regulate. There are several possibilities other than the possibility of Trx-related pathways, including Calvin cycle enzymes in the leaves. CBSX proteins may modulate other unknown redox-related pathways, particularly concern to the growth delay of the cbsx mutants grown in soil under short day condition. Expression of CBSX mRNA was seen in leaves, flowers, and seeds in the Arabidopsis eFP Browser 2.0 (Winter et al., 2007) and we detected accumulation of CBSX proteins in leaves and flowers by western blotting, using anti-CBSX1 and anti-CBSX2. Given this, CBSX proteins may function as modulators for thiol enzymes other than those involved in photosynthesis in leaves and flowers. In particular, redox regulation by Trx proteins in flowers remains largely uninvestigated. To elucidate the functions of CBSX proteins, the effects of Trx proteins including Trx f, Trx m, and other chloroplast Trxs on thiol enzymes in leaves and flowers should form the next stage of investigation.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
YO and KM contributed conception and design of the study. RM, YO, NS, and KM carried out experiments. RM, YO, NS, and KM performed the statistical analysis. RM and KM wrote the first draft of the manuscript. YO and KM wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the MEXT-Supported Program for the Strategic Research Foundation at Private Universities grant number S1511023 (to KM) and the Institute for Fermentation (Osaka) grant number G-2019-2-067 (to KM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Akira Kawabe (Kyoto Sangyo University) for the discussion on statistical analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.530376/full#supplementary-material
References
Balsera, M., Uberegui, E., Schurmann, P., and Buchanan, B. B. (2014). Evolutionary development of redox regulation in chloroplasts. Antioxid. Redox Signal. 21, 1327–1355. doi: 10.1089/ars.2013.5817
Baykov, A. A., Tuominen, H. K., and Lahti, R. (2011). The CBS domain: a protein module with an emerging prominent role in regulation. ACS Chem. Biol. 6, 1156–1163. doi: 10.1021/cb200231c
Breazeale, V. D., Buchanan, B. B., and Wolosiuk, R. A. (1978). Chloroplast sedoheptulose 1,7-bisphosphatase: evidence for regulation by the ferredoxin/thioredoxin system. Z. Naturforsch. C 33, 521–528. doi: 10.1515/znc-1978-7-812
Buchanan, B. B. (2016). The path to thioredoxin and redox regulation in chloroplasts. Annu. Rev. Plant Biol. 67, 1–24. doi: 10.1146/annurev-arplant-043015-111949
Buchanan, B. B., and Balmer, Y. (2005). Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56, 187–220. doi: 10.1146/annurev.arplant.56.032604.144246
Cejudo, F. J., Ojeda, V., Delgado-Requerey, V., Gonzalez, M., and Perez-Ruiz, J. M. (2019). Chloroplast redox regulatory mechanisms in plant adaptation to light and darkness. Front. Plant Sci. 10:380. doi: 10.3389/fpls.2019.00380
Collin, V., Issakidis-Bourguet, E., Marchand, C., Hirasawa, M., Lancelin, J. M., Knaff, D. B., et al. (2003). The Arabidopsis plastidial thioredoxins: new functions and new insights into specificity. J. Biol. Chem. 278, 23747–23752. doi: 10.1074/jbc.M302077200
Da, Q., Sun, T., Wang, M., Jin, H., Li, M., Feng, D., et al. (2018). M-type thioredoxins are involved in the xanthophyll cycle and proton motive force to alter NPQ under low-light conditions in Arabidopsis. Plant Cell Rep. 37, 279–291. doi: 10.1007/s00299-017-2229-6
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Emanuelsson, O., and Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Ereno-Orbea, J., Oyenarte, I., and Martinez-Cruz, L. A. (2013). CBS domains: ligand binding sites and conformational variability. Arch. Biochem. Biophys. 540, 70–81. doi: 10.1016/j.abb.2013.10.008
Geigenberger, P., and Fernie, A. R. (2014). Metabolic control of redox and redox control of metabolism in plants. Antioxid. Redox Signal. 21, 1389–1421. doi: 10.1089/ars.2014.6018
Geigenberger, P., Thormahlen, I., Daloso, D. M., and Fernie, A. R. (2017). The unprecedented versatility of the plant thioredoxin system. Trends Plant Sci. 22, 249–262. doi: 10.1016/j.tplants.2016.12.008
Genty, B., Briantais, J. M., and Baker, N. R. (1989). The relationship between the quantum yield of photosynthetic electron-transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990, 87–92. doi: 10.1016/S0304-4165(89)80016-9
Hisabori, T., Motohashi, K., Hosoya-Matsuda, N., Ueoka-Nakanishi, H., and Romano, P. G. (2007). Towards a functional dissection of thioredoxin networks in plant cells. Photochem. Photobiol. 83, 145–151. doi: 10.1562/2006-02-27-IR-816
Issakidis, E., Miginiac-Maslow, M., Decottignies, P., Jacquot, J. P., Cretin, C., and Gadal, P. (1992). Site-directed mutagenesis reveals the involvement of an additional thioredoxin-dependent regulatory site in the activation of recombinant sorghum leaf NADP-malate dehydrogenase. J. Biol. Chem. 267, 21577–21583. doi: 10.1016/S0021-9258(19)36649-9
Jacquot, J. -P., Vidal, J., Gadal, P., and Schürmann, P. (1978). Evidence for the existence of several enzyme-specific thioredoxins in plants. FEBS Lett. 96, 243–246. doi: 10.1016/0014-5793(78)80410-4
Jung, K. W., Kim, Y. Y., Yoo, K. S., Ok, S. H., Cui, M. H., Jeong, B. C., et al. (2013). A cystathionine-beta-synthase domain-containing protein, CBSX2, regulates endothecial secondary cell wall thickening in another development. Plant Cell Physiol. 54, 195–208. doi: 10.1093/pcp/pcs166
Lepisto, A., Kangasjarvi, S., Luomala, E. M., Brader, G., Sipari, N., Keranen, M., et al. (2009). Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiol. 149, 1261–1276. doi: 10.1104/pp.108.133777
Lopez-Calcagno, P. E., Howard, T. P., and Raines, C. A. (2014). The CP12 protein family: a thioredoxin-mediated metabolic switch? Front. Plant Sci. 5:9. doi: 10.3389/fpls.2014.00009
Michelet, L., Zaffagnini, M., Morisse, S., Sparla, F., Perez-Perez, M. E., Francia, F., et al. (2013). Redox regulation of the Calvin-Benson cycle: something old, something new. Front. Plant Sci. 4:470. doi: 10.3389/fpls.2013.00470
Motohashi, K. (2017a). Evaluation of the efficiency and utility of recombinant enzyme-free seamless DNA cloning methods. Biochem. Biophys. Rep. 9, 310–315. doi: 10.1016/j.bbrep.2017.01.010
Motohashi, K. (2017b). Seamless ligation cloning extract (SLiCE) method using cell lysates from laboratory Escherichia coli strains and its application to SLiP site-directed mutagenesis. Methods Mol. Biol. 1498, 349–357. doi: 10.1007/978-1-4939-6472-7_23
Motohashi, K. (2019). Development of highly sensitive and low-cost DNA agarose gel electrophoresis detection systems, and evaluation of non-mutagenic and loading dye-type DNA-staining reagents. PLoS One 14:e0222209. doi: 10.1371/journal.pone.0222209
Motohashi, K., and Hisabori, T. (2006). HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 281, 35039–35047. doi: 10.1074/jbc.M605938200
Motohashi, K., Kondoh, A., Stumpp, M. T., and Hisabori, T. (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. U. S. A. 98, 11224–11229. doi: 10.1073/pnas.191282098
Motohashi, K., Koyama, F., Nakanishi, Y., Ueoka-Nakanishi, H., and Hisabori, T. (2003). Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. J. Biol. Chem. 278, 31848–31852. doi: 10.1074/jbc.M304258200
Motohashi, K., and Okegawa, Y. (2014). Method for enhancement of plant redox-related protein expression and its application for in vitro reduction of chloroplastic thioredoxins. Protein Expr. Purif. 101, 152–156. doi: 10.1016/j.pep.2014.07.001
Motohashi, K., Romano, P. G., and Hisabori, T. (2009). Identification of thioredoxin targeted proteins using thioredoxin single cysteine mutant-immobilized resin. Methods Mol. Biol. 479, 117–131. doi: 10.1007/978-1-59745-289-2_8
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Naranjo, B., Diaz-Espejo, A., Lindahl, M., and Cejudo, F. J. (2016). Type-f thioredoxins have a role in the short-term activation of carbon metabolism and their loss affects growth under short-day conditions in Arabidopsis thaliana. J. Exp. Bot. 67, 1951–1964. doi: 10.1093/jxb/erw017
Nikkanen, L., and Rintamaki, E. (2014). Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 369:20130224. doi: 10.1098/rstb.2013.0224
Nishiyama, A., Matsui, M., Iwata, S., Hirota, K., Masutani, H., Nakamura, H., et al. (1999). Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J. Biol. Chem. 274, 21645–21650. doi: 10.1074/jbc.274.31.21645
Nishizawa, A. N., and Buchanan, B. B. (1981). Enzyme regulation in C4 photosynthesis. Purification and properties of thioredoxin-linked fructose bisphosphatase and sedoheptulose bisphosphatase from corn leaves. J. Biol. Chem. 256, 6119–6126. doi: 10.1016/S0021-9258(19)69136-2
Okegawa, Y., Koshino, M., Okushima, T., and Motohashi, K. (2016). Application of preparative disk gel electrophoresis for antigen purification from inclusion bodies. Protein Expr. Purif. 118, 77–82. doi: 10.1016/j.pep.2015.10.008
Okegawa, Y., and Motohashi, K. (2015a). Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. Plant J. 84, 900–913. doi: 10.1111/tpj.13049
Okegawa, Y., and Motohashi, K. (2015b). Evaluation of seamless ligation cloning extract preparation methods from an Escherichia coli laboratory strain. Anal. Biochem. 486, 51–53. doi: 10.1016/j.ab.2015.06.031
Okegawa, Y., and Motohashi, K. (2015c). A simple and ultra-low cost homemade seamless ligation cloning extract (SLiCE) as an alternative to a commercially available seamless DNA cloning kit. Biochem. Biophys. Rep. 4, 148–151. doi: 10.1016/j.bbrep.2015.09.005
Okegawa, Y., and Motohashi, K. (2016). Expression of spinach ferredoxin-thioredoxin reductase using tandem T7 promoters and application of the purified protein for in vitro light-dependent thioredoxin-reduction system. Protein Expr. Purif. 121, 46–51. doi: 10.1016/j.pep.2016.01.005
Okegawa, Y., and Motohashi, K. (2020). M-type thioredoxins regulate the PGR5/PGRL1-dependent pathway by forming a disulfide-linked complex with PGRL1. Plant Cell 32, 3866–3883. doi: 10.1105/tpc.20.00304
Perez-Ruiz, J. M., Spinola, M. C., Kirchsteiger, K., Moreno, J., Sahrawy, M., and Cejudo, F. J. (2006). Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell 18, 2356–2368. doi: 10.1105/tpc.106.041541
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. doi: 10.1093/nar/gku316
Sanger, F., Nicklen, S., and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74, 5463–5467.
Schurmann, P., and Buchanan, B. B. (2008). The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid. Redox Signal. 10, 1235–1274. doi: 10.1089/ars.2007.1931
Schwarz, O., Schurmann, P., and Strotmann, H. (1997). Kinetics and thioredoxin specificity of thiol modulation of the chloroplast H+-ATPase. J. Biol. Chem. 272, 16924–16927. doi: 10.1074/jbc.272.27.16924
Serrato, A. J., Fernandez-Trijueque, J., Barajas-Lopez, J. D., Chueca, A., and Sahrawy, M. (2013). Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front. Plant Sci. 4:463. doi: 10.3389/fpls.2013.00463
Serrato, A. J., Perez-Ruiz, J. M., Spinola, M. C., and Cejudo, F. J. (2004). A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J. Biol. Chem. 279, 43821–43827. doi: 10.1074/jbc.M404696200
Spindel, O. N., World, C., and Berk, B. C. (2012). Thioredoxin interacting protein: redox dependent and independent regulatory mechanisms. Antioxid. Redox Signal. 16, 587–596. doi: 10.1089/ars.2011.4137
Stanley, D. N., Raines, C. A., and Kerfeld, C. A. (2013). Comparative analysis of 126 cyanobacterial genomes reveals evidence of functional diversity among homologs of the redox-regulated CP12 protein. Plant Physiol. 161, 824–835. doi: 10.1104/pp.112.210542
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal-W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Thormahlen, I., Zupok, A., Rescher, J., Leger, J., Weissenberger, S., Groysman, J., et al. (2017). Thioredoxins play a crucial role in dynamic acclimation of photosynthesis in fluctuating light. Mol. Plant 10, 168–182. doi: 10.1016/j.molp.2016.11.012
Wang, P., Liu, J., Liu, B., Feng, D., Da, Q., Shu, S., et al. (2013). Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol. 163, 1710–1728. doi: 10.1104/pp.113.228353
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Wolosiuk, R. A., and Buchanan, B. B. (1977). Thioredoxin and glutathione regulate photosynthesis in chloroplasts. Nature 266, 565–567. doi: 10.1038/266565a0
Wolosiuk, R. A., Crawford, N. A., Yee, B. C., and Buchanan, B. B. (1979). Isolation of three thioredoxins from spinach leaves. J. Biol. Chem. 254, 1627–1632. doi: 10.1016/S0021-9258(17)37818-3
Yamazaki, D., Motohashi, K., Kasama, T., Hara, Y., and Hisabori, T. (2004). Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 45, 18–27. doi: 10.1093/pcp/pch019
Yoo, K. S., Ok, S. H., Jeong, B. C., Jung, K. W., Cui, M. H., Hyoung, S., et al. (2011). Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 23, 3577–3594. doi: 10.1105/tpc.111.089847
Yoshida, K., Hara, S., and Hisabori, T. (2015). Thioredoxin selectivity for thiol-based redox regulation of target proteins in chloroplasts. J. Biol. Chem. 290, 14278–14288. doi: 10.1074/jbc.M115.647545
Yoshida, K., and Hisabori, T. (2017). Distinct electron transfer from ferredoxin-thioredoxin reductase to multiple thioredoxin isoforms in chloroplasts. Biochem. J. 474, 1347–1360. doi: 10.1042/BCJ20161089
Keywords: chloroplast, cystathionine β-synthase X, photo-reduction, photosynthesis, redox regulation, thiol-enzyme, thioredoxin
Citation: Murai R, Okegawa Y, Sato N and Motohashi K (2021) Evaluation of CBSX Proteins as Regulators of the Chloroplast Thioredoxin System. Front. Plant Sci. 12:530376. doi: 10.3389/fpls.2021.530376
Edited by:
Nicolas Rouhier, Université de Lorraine, FranceReviewed by:
Pascal Rey, Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), FranceFrancisco Javier Cejudo, Sevilla University, Spain
Copyright © 2021 Murai, Okegawa, Sato and Motohashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ken Motohashi, motohas@cc.kyoto-su.ac.jp
†These authors have contributed equally to this work
 Ryota Murai1
†
Ryota Murai1
† Yuki Okegawa
Yuki Okegawa Ken Motohashi
Ken Motohashi