- 1Department of Soil, Plant and Food Science, University of Bari “Aldo Moro”, Bari, Italy
- 2Department of Biological and Environmental Sciences and Technologies, University of Salento, Lecce, Italy
- 3Department of Agriculture, Food, Natural Resources and Engineering, University of Foggia, Foggia, Italy
The recent outbreak of the Olive Quick Decline Syndrome (OQDS), caused by Xylella fastidiosa subsp. pauca (Xf), is dramatically altering ecosystem services in the peninsula of Salento (Apulia Region, southeastern Italy). Here we report the accomplishment of several exploratory missions in the Salento area, resulting in the identification of thirty paucisymptomatic or asymptomatic plants in olive orchards severely affected by the OQDS. The genetic profiles of such putatively resistant plants (PRPs), assessed by a selection of ten simple sequence repeat (SSR) markers, were compared with those of 141 Mediterranean cultivars. Most (23) PRPs formed a genetic cluster (K1) with 22 Italian cultivars, including ‘Leccino’ and ‘FS17’, previously reported as resistant to Xf. The remaining PRPs displayed relatedness with genetically differentiated germplasm, including a cluster of Tunisian cultivars. Markedly lower colonization levels were observed in PRPs of the cluster K1 with respect to control plants. Field evaluation of four cultivars related to PRPs allowed the definition of partial resistance in the genotypes ‘Frantoio’ and ‘Nocellara Messinese’. Some of the PRPs identified in this study might be exploited in cultivation, or as parental clones of breeding programs. In addition, our results indicate the possibility to characterize resistance to Xf in cultivars genetically related to PRPs.
Introduction
Xylella fastidiosa (Xf) is a Gram-negative bacterium affecting more than 350 plant species, including tree crops of major economic importance (Zarco-Tejada et al., 2018). Disease symptoms include leaf scorching, leaf and twig wilting and, in a second phase, tree die-back, due to the formation of bacterial biofilms in xylematic vessels (Purcell and Hopkins, 1996). In the peninsula of Salento (Apulia Region, southeastern Italy), Xf subsp. pauca is responsible of the outbreak of the Olive Quick Decline Syndrome (OQDS), resulting in a massive socio-economic impact in the last 10 years (Loconsole et al., 2014; Saponari et al., 2019). Recent data indicate the occurrence of about 5 million unproductive, heavily infected or dead, olive trees in Salento, leading to a loss of about 10% of Italian olive oil (White et al., 2020). According to spatially explicit economic models, Xf may impact Italian olive growers from 1.5 to 5.9 billion euros in the next 50 years and cause further economic losses by affecting cultural heritage and landscape values (Schneider et al., 2020).
Xf can be limited by agronomic practices affecting the vector Philaenus spumarius (Liccardo et al., 2020; Schneider et al., 2020) and, possibly, by acting on the soil microbiome (Fernández-González et al., 2019). However, replanting with resistant genotypes appears as the most feasible and promising strategy to control the bacterium. Here, we use the term ‘resistance’ in a broad sense, including both resistance sensu stricto, i.e., the ability to limit pathogen proliferation, and tolerance, i.e., the capacity to limit the development of symptoms regardless of the level of pathogen colonization (Pagán and García-Arenal, 2018). So far, the cultivars ‘Leccino’ and ‘FS17’ (also referred to as ‘Favolosa’) were shown to display resistance to Xf (Baù et al., 2017; Boscia et al., 2017). Therefore, the current regulatory measures only allow ‘Leccino’ and ‘FS17’ for olive reconversion in infected areas. It was shown that resistance in ‘Leccino’ and ‘FS17’ is associated with reduced levels of bacterial growth in xylematic vessels (Saponari et al., 2016; Giampetruzzi et al., 2020).
Screening olive biodiversity for response to Xf may lead to the identification of resistant genotypes in addition to ‘Leccino’ and ‘FS17’. In turn, this would represent the basis for the reconversion of infected areas into more complex, resilient, agro-ecosystems, and the selection of cultivars merging resistance with other economically important traits, such as agronomic performance and oil quality. So far, screenings were limited to a few Italian cultivars (Baù et al., 2017; Boscia et al., 2017). None of them displayed resistance levels comparable to those of ‘Leccino’ and ‘FS17’; however, a wide range of variation was detected. Intermediate resistance was reported for ‘Frantoio’, ‘Toscanina’, ‘Termite di Bitetto’, ‘Maiatica’, ‘Dolce di Cassano’, ‘Oliastro’, ‘Nociara’, and ‘Nocellara Etnea’ (Baù et al., 2017; Boscia et al., 2017).
In Salento, about 85% of the olive trees refer to two cultivars, ‘Cellina di Nardò’ and ‘Ogliarola Salentina’, both extremely susceptible to Xf (Luvisi et al., 2017). In heavily infected orchards, this creates a spooky scenario of desiccated trees, in which putatively resistant genotypes, asymptomatic or paucisymptomatic, stand out. Here, we present the results of a study mainly aimed to: (i) identify genotypes putatively resistant to Xf, through exploratory missions conducted in Salento; (ii) establish genetic relationships between putatively resistant genotypes and a large panel of cultivars occurring in the Mediterranean region.
Materials and Methods
Exploratory Missions and Characterization of Putatively Resistant Plants
Exploratory missions were carried out in the Autumns of 2019 and 2020 upon report, by local growers, of asymptomatic or paucisymptomatic plants (reported plants or RPs) in olive orchards heavily affected by Xf. RPs were evaluated for disease severity (S) using the pathometric scale previously described by Luvisi et al. (2017), ranging from 0 (no symptoms) to 3 (canopy with uniformly distributed desiccated branches). Three control plants (CPs) randomly chosen in the proximity of each RP were also evaluated for disease severity. A RP was deemed as a putatively resistant plant (PRP) if SCP − SRP (ΔS) ≥ 1.5, with SCP being the average value of S calculated from its three CPs.
For bacterial quantification, 1-year-old branches (8–10), with attached leaves, were collected from the apical, median, and basal portions of the canopy of PRPs and CPs. Branches from each triad of CPs were pooled. Approximately one gram of leaf petioles was transferred into an extraction bag (BIOREBA, Switzerland) and processed as described by Sabella et al. (2018). DNA was extracted according to Edwards et al. (1991) and used as template for Xf detection by the TaqMan real-time PCR protocol with the XF-F/R primers and the XF-P probe (Harper et al., 2010). Each reaction consisted of 5 μL from a 20 ng μL−1 dilution of DNA, 12.5 μL of TaqMan Environmental Master Mix 2.0 (Applied Biosystems, CA, United States), 0.4 μM of each primer, 0.2 μM of TaqMan probe, and ultrapure DNase/RNase-free water (Carlo Erba Reagents, Italy) in a total volume of 25 μL. The cycling conditions were those described by Harper et al. (2010). Xf concentration (C), expressed as bacterial cfu ml–1, was inferred from Cq values using a standard curve with dilutions ranging from 102 to 107 cfu ml–1, as described by D’Attoma et al. (2019). The minimum value of 102 cfu ml–1 was assigned to negative samples as detection limit, whereas the maximum value of 107 cfu ml–1 was assigned to samples in which the bacterium could not be detected due to extremely advanced plant desiccation. The parameter ΔC, given by CCP – CPRP, was finally calculated for each PRP.
Disease symptoms and bacterial concentrations recorded for PRPs falling in the genetic cluster K1, which was identified by hierarchical clustering, were compared with those of respective controls, using the Wilcoxon signed-rank test.
SSR Marker Analysis and Assessment of Genetic Relationships With Known Cultivars
DNA of PRPs was used as template to perform Simple Sequence Repeat (SSR) marker analysis, using a set of 10 primer combinations previously reported to be effective in differentiating olive genetic resources (Supplementary Table 1; Sefc et al., 2000; Carriero et al., 2002; De La Rosa et al., 2002; Pasqualone et al., 2016). PCR reactions were performed following the protocol described by Spadoni et al. (2019). Amplicons were resolved with an internal size standard (GeneScan 500 LIZ; Applied Biosystems) by capillary electrophoresis on an ABI 3130 Genetic Analyzer (Applied Biosystems, CA, United States). SSR allele sizes were estimated using the software GeneMapper v.3.7. The informativeness of each SSR marker was estimated through the polymorphic information content (PIC) index, which was calculated using the software Cervus v.3.0.7 (Marshall et al., 1998).
Marker profiles were merged with those of Mediterranean cultivars, available from previous studies (Alba et al., 2009; Boucheffa et al., 2017; di Rienzo et al., 2018; Miazzi et al., 2020; Debbabi et al., 2021). UPGMA hierarchical clustering was performed using the Nei’s distance and the ape R package (Paradis et al., 2004). Parametric clustering was performed using the software STRUCTURE v.2.3.4. (Pritchard et al., 2000) for a number of sub-populations (K) ranging from 1 to 10, using 10 runs for each K, a burn-in period of 50,000, and 100,000 Markov chain Monte Carlo iterations. The software Structure Harvester (Earl and vonHoldt, 2012) was used to infer the value of K best describing genetic structure, based on the ΔK statistics (Evanno et al., 2005).
Characterization of Cultivars for Response to Xf
Four cultivars displaying genetic similarity with PRPs (‘Nocellara Messinese’, ‘Frantoio’, ‘Bella di Spagna’, ‘Pendolino’) were represented at the experimental orchard ‘Murrone’, located in Caprarica di Lecce (Province of Lecce, Italy, 40°15′59″N 18°14′52″E), together with the susceptible controls ‘Cellina di Nardò’ and ‘Ogliarola Salentina’, and the resistant control ‘Leccino’. The orchard, planted in 2015, is heavily affected by Xf. Disease severity and bacterial quantification were assessed in Autumn 2020 on eight biological replicates for each cultivar, following the methodologies described above. Significant differences among cultivars were assessed using the Wilcoxon rank-sum test and the false discovery rate (FDR) correction to control for Type I error.
Results
Exploratory Missions and Characterization of Putatively Resistant Plants
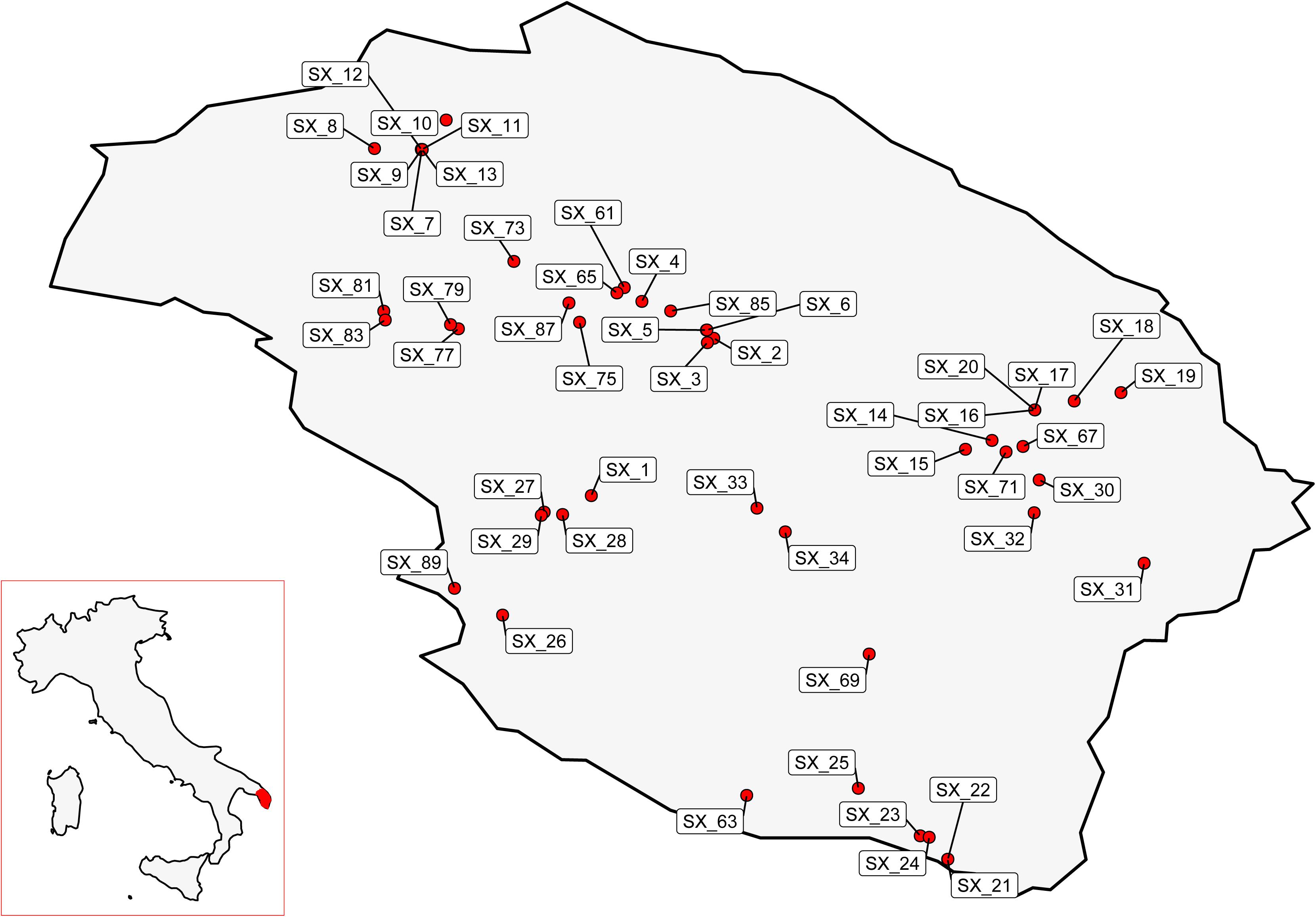
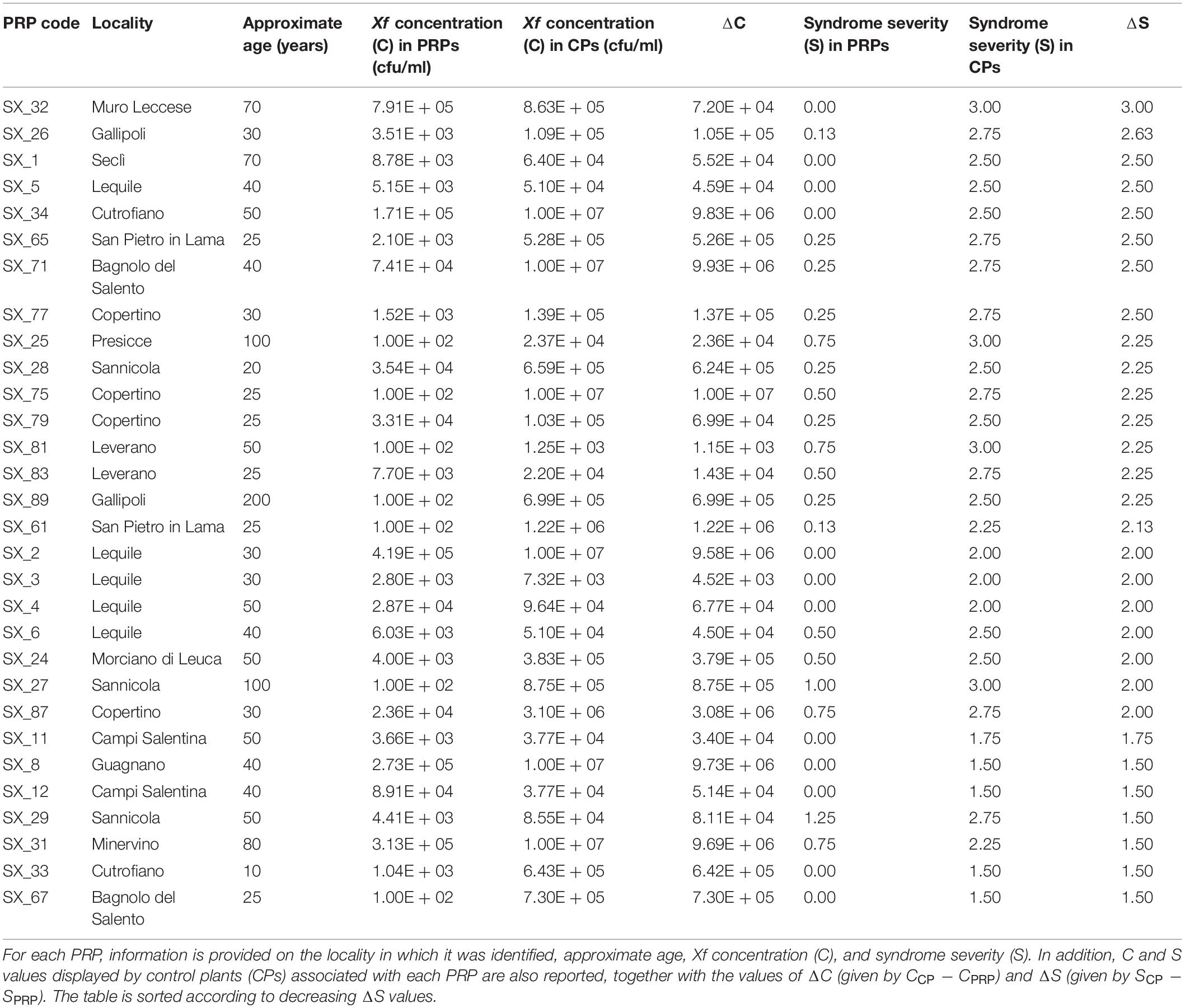
Exploratory missions were carried out in the Province of Lecce (Salento) for a periodical screening of 49 plants reported by local farmers to stand out in orchards heavily infected by Xf (Figure 1). For each plant, difference in disease severity with neighboring control plants (ΔS) was assessed (Figure 2). Thirty putative resistant plants (PRPs) were selected, which were associated, compared to control plants (CPs), with a difference in disease severity (ΔS) ≥ 1.5. Twelve of them also appeared asymptomatic (S = 0) (Table 1). The distribution of S and C values associated with PRPs and CPs are reported in Supplementary Figure 1.
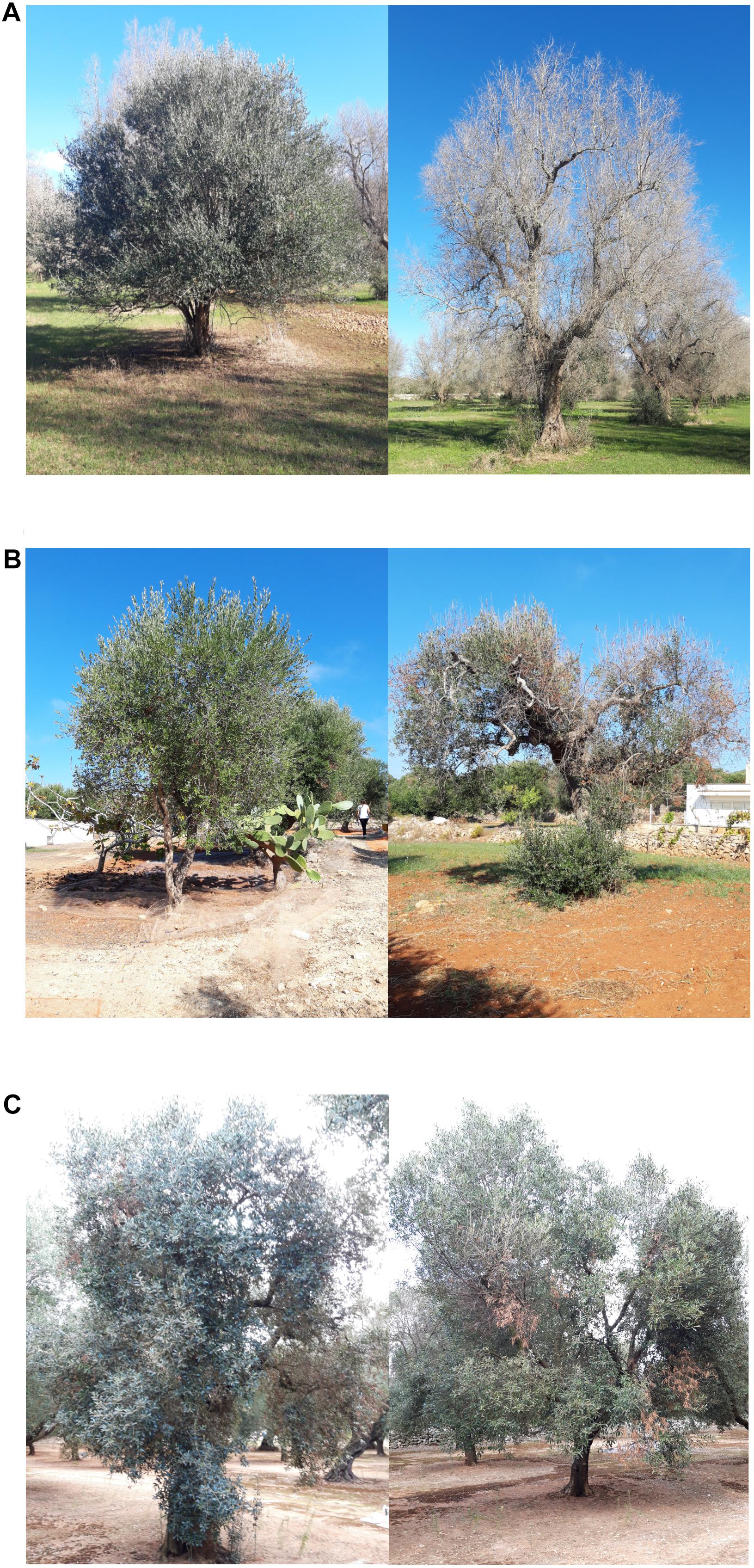
Figure 2. Phenotypes of plants monitored in this study (left) and respective controls (right) for (A) ΔS = 3, (B) ΔS = 2, and (C) ΔS = 1.
PRP Fingerprinting and Genetic Relationships With Known Cultivars
The thirty identified PRPs were genotyped with ten short sequence repeat (SSR) markers previously reported as highly polymorphic in olive germplasm (Sefc et al., 2000; Carriero et al., 2002; De La Rosa et al., 2002; Pasqualone et al., 2016). Identical DNA fingerprints were found for the two PRPs SX_77 and SX_79, detected in the same locality (Copertino), and for the three PRPs SX_65, SX_81, and SX_89), detected in different localities (San Pietro in Lama, Leverano, and Gallipoli) (Table 1).
SSR fingerprints obtained for the PRPs were compared with those of 141 cultivars from several Mediterranean Countries, taking profit of SSR profiles available from previous studies (Alba et al., 2009; Boucheffa et al., 2017; di Rienzo et al., 2018; Miazzi et al., 2020; Debbabi et al., 2021). A total of 124 alleles was detected (Supplementary Table 2) with an average of 12.4 alleles per locus. PIC index values ranged from 0.49 to 0.90 with an average of 0.73, indicating high informativeness on DNA polymorphism of the selected SSR marker loci (Supplementary Table 2). None of the PRPs displayed a marker profile identical to the resistant cultivars ‘Leccino’ and ‘FS17’, or to the susceptible cultivars ‘Cellina di Nardò’ and ‘Ogliarola Salentina’.
Hierarchical clustering (Figure 3) revealed the occurrence of a genetic cluster (K1) grouping most (23) PRPs with 22 Italian cultivars, including the resistant cultivars ‘Leccino’ and ‘FS17’. The PRPs SX_2, SX_8, SX_31, SX_61, SX_67, and SX_83, together with the two groups of putatively identical PRPs SX_77_79 and SX_65_81_89, formed a sub-cluster (K1/L) of genotypes closely related with ‘Leccino’, thus suggesting the occurrence of clonal variation around this cultivar. Five PRPs (SX_25, SX_26, SX_27, SX_29, and SX_34) formed another sub-cluster (K1/C) of genotypes closely related to the cultivar ‘Ciciulara’. PRPs not included in K1 were scattered throughout the dendrogram. Notably, the PRP SX_32, associated with the highest ΔS (3), formed a cluster (K2) with several Tunisian cultivars (‘Regueb’, ‘Chemlali Sfax’, ‘Sayali’, and ‘Tamri Douiret’), thus suggesting that this plant might have an allochtonous origin.
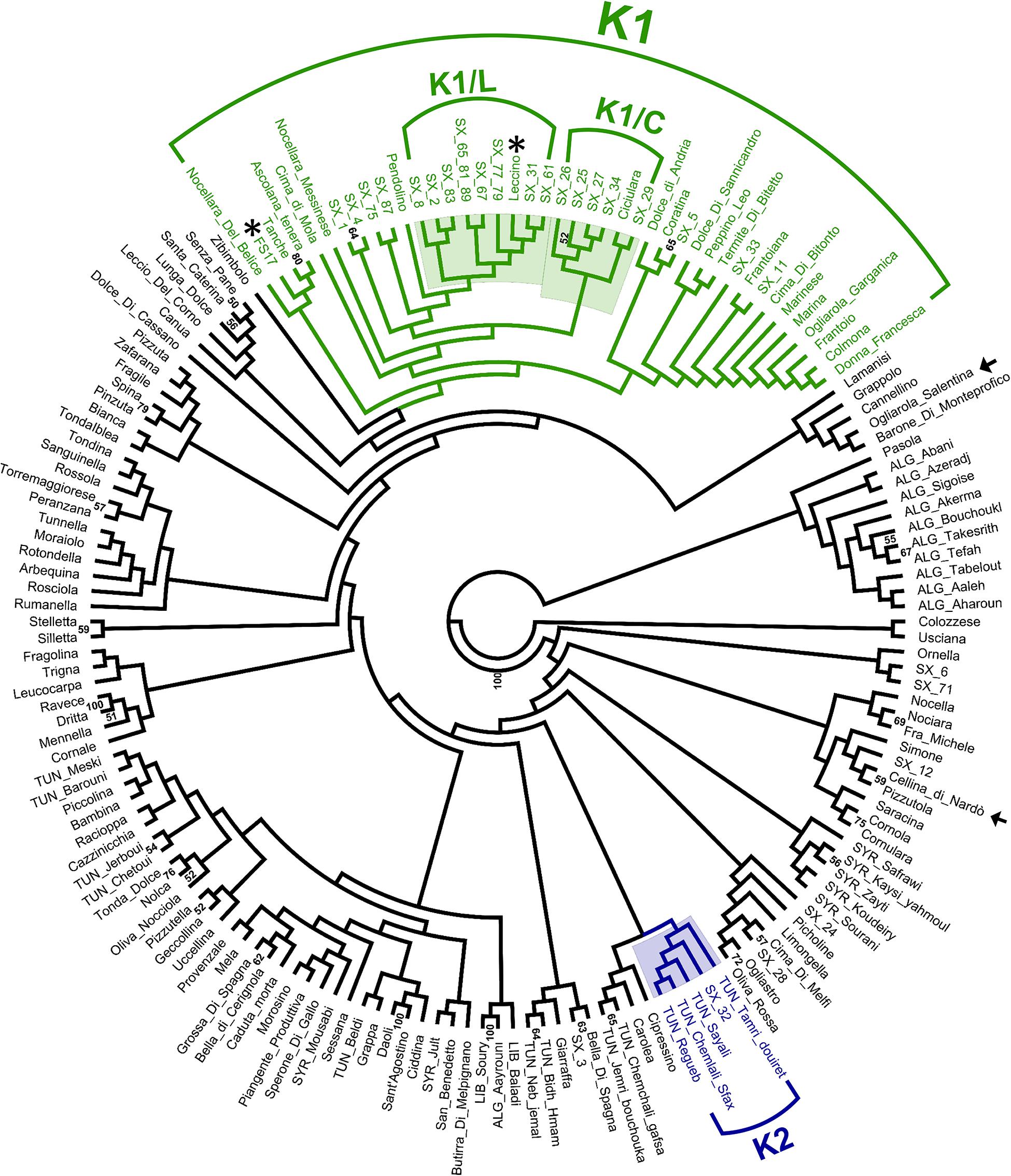
Figure 3. UPGMA clustering based on short sequence repeat (SSR) DNA fingerprints of 30 putatively resistant plants and 141 olive cultivars. The cluster K1 (with the sub-clusters K1/L and K1/C) and the cluster K2 are highlighted. Bootstrap values > 50 are shown. The resistant and susceptible control cultivars are indicated with asterisks and arrows, respectively.
Parametric analysis indicated a model with three sub-populations (Subpop_1–3) as the most appropriate to describe genetic structure (Supplementary Figure 2). In accordance with the results of hierarchical clustering, most individuals grouped in K1 and K2 were mainly referable to Subpop_1 and Subpop_2, respectively (Supplementary Table 3).
PRPs included in K1 displayed significantly lower bacterial colonization than respective controls (p = 2.38 ∗ 10–7) (Supplementary Figure 3). As for the remaining PRPs, they were characterized by highly variable ΔC values, ranging from –5.14∗104 (SX_12) to 9.93∗106 (SX_71) (Table 1).
Response of Four Cultivars Displaying Genetic Similarity With PRPs
Among the cultivars showing genetic similarity with PRPs, four were replicated in a 5-year-old experimental orchard heavily infested by Xf, and therefore could be evaluated for response to the bacterium: ‘Pendolino’ and ‘Frantoio’, and ‘Nocellara Messinese’, included in the cluster K1; ‘Bella di Spagna’, related to the plant SX_3. Disease severity was significantly lower in ‘Nocellara Messinese’ and ‘Frantoio’ than in the susceptible control cultivars ‘Ogliarola Salentina’ and ‘Cellina di Nardò’ (Figure 4A). Significant differences were also found between the resistant control ‘Leccino’, displaying the lowest level of symptoms, and all the other cultivars (Figure 4A). ‘Leccino’ also exhibited the lowest level of bacterial colonization (Figure 4B).
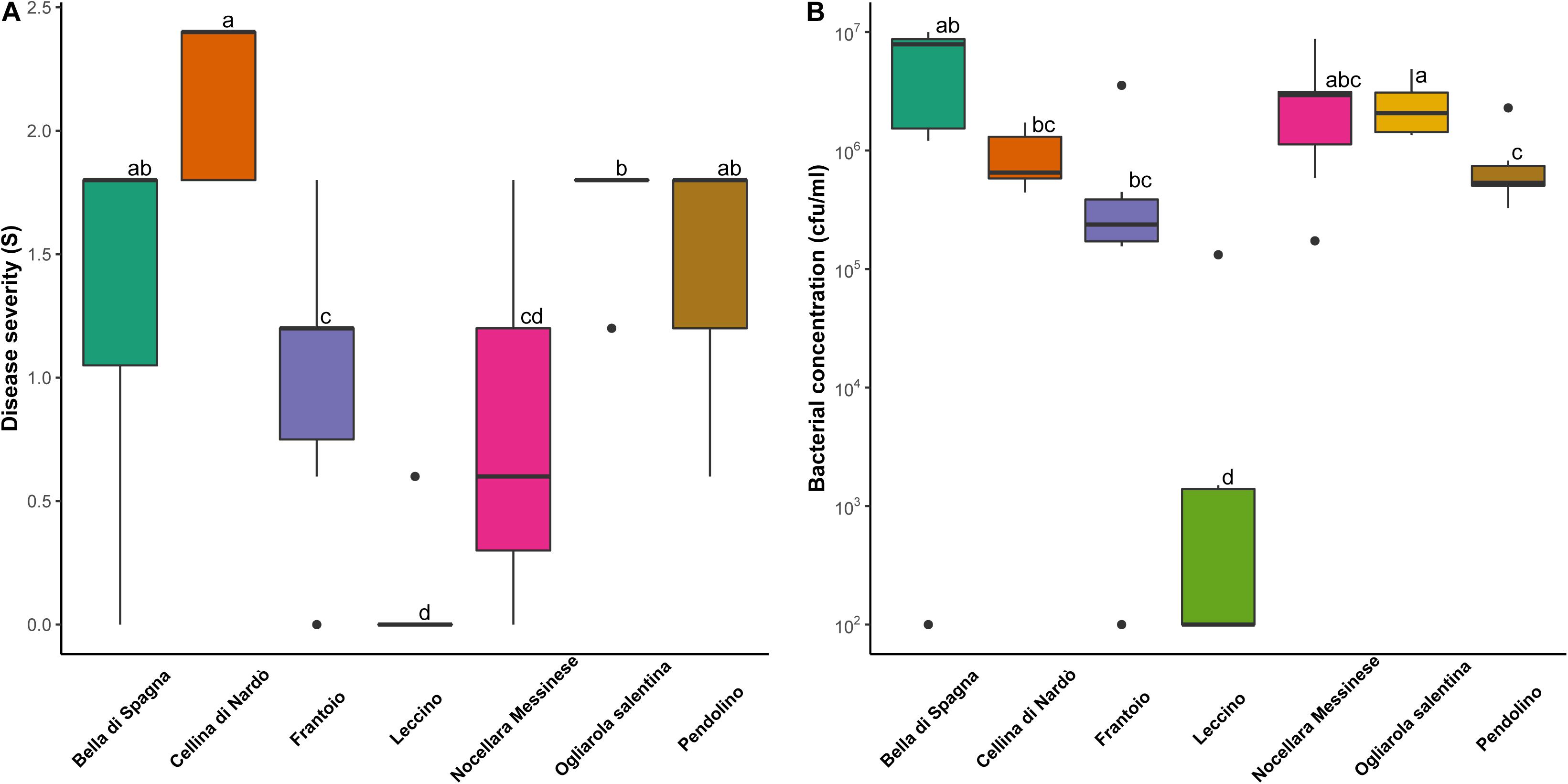
Figure 4. Distribution of (A) disease severity and (B) bacterial concentration observed for eight biological replicates of the following olive cultivars: ‘Bella di Spagna’, ‘Cellina di Nardo’, ‘Frantoio’, ‘Leccino’, ‘Nocellara Messinese’, ‘Ogliarola Salentina’, and ‘Pendolino’. Different letters indicate significant differences following the application of the Wilcoxon rank-sum test (p < 0.05).
Correlation was found between the level of symptoms and bacterial concentration (p < 0.05) in individual trees. Consistently, the two outlier individuals within the cultivars ‘Bella di Spagna’ and ‘Frantoio’, displaying no visible symptoms, were also characterized by undetectable Xf colonization.
Discussion
There is an urgent need to reconvert olive orchards exposed to Xf with resistant germplasm. So far, only two cultivars, ‘Leccino’ and ‘FS17’, were reported as resistant to Xf (Boscia et al., 2017). Although to a low extent, both of them support bacterial colonization and may display disease symptoms (Boscia et al., 2017; Giampetruzzi et al., 2020); in addition, their performance under pathogen pressure, with respect to agronomic, qualitative and technological traits, should be further investigated. In this context, the identification of further sources of resistance to Xf may allow the selection of cultivars increasing the complexity, and thus the resilience, of olive agro-ecosystems, and displaying superior features.
The screening of olive germplasm for response to Xf may be accomplished through the set-up of experimental orchards in areas heavily infected by the bacterium. This approach, besides being relatively costly, requires several months or years for the emergence of symptoms after exposure to the bacterium. In this study, we followed an alternative and rapid approach for the identification of germplasm resistant to Xf, based on the detection, in existing orchards, of individuals standing out among heavily symptomatic or dead trees. Some of the thirty PRPs identified in this study might deserve direct exploitation in cultivation. Therefore, we are currently proceeding with their clonal propagation and thoroughly characterization in replicated trials with respect to response to Xf and other main economic traits.
SSR profiles obtained for PRPs were compared with those available from previous studies, referring to a large panel of cultivars occurring in Italy and the Mediterranean area. Most (23) of the PRPs were grouped in the cluster K1, including the resistant cultivars ‘Leccino’ and ‘FS17’ (Figure 3). This indicates that the screening of cultivars in K1 might represent a valuable strategy for the identification of germplasm resistant to Xf.
Eight different genotypes, associated with 11 PRPs, formed the sub-cluster K1/L together with the cultivar ‘Leccino’. Noteworthy, none of these genotypes fully matched the one of ‘Leccino’, suggesting that wide clonal variation exists within germplasm identified as ‘Leccino’. Screening of such variation might lead to the selection of clones displaying higher levels of resistance and/or superior attributes with respect to other economic traits. The cultivar ‘Ciciulara’, forming another sub-cluster (K1/C) with five closely related PRPs, was previously characterized in Salento in the framework of a Regional Project aimed at the recovery of minor olive germplasm (Miazzi et al., 2020). Interestingly, ‘Ciciulara’ is clearly distinct at the morphological level from ‘Leccino’, as it displays very large drupes that do not turn completely black at maturity.
All the cultivars displaying similarity with the PRPs identified in this study represent obvious targets for future investigations aiming to the characterization of new sources of resistance to Xf. Here, we assessed the response of three other cultivars of this kind, namely ‘Frantoio’, ‘Nocellara Messinese’, and ‘Pendolino’, included in K1, and ‘Bella di Spagna’, related to SX_3. Both ‘Nocellara Messinese’ and ‘Frantoio’ displayed significantly lower symptoms than the susceptible controls ‘Ogliarola Salentina’ and ‘Cellina di Nardò’, although none of them displayed the same level of resistance of ‘Leccino’. This is consistent with preliminary studies indicating partial resistance in ‘Frantoio’ (Boscia et al., 2017; Luvisi et al., 2017).
Of main interest it would be the characterization of the response of Tunisian cultivars closely related to the plant SX_32, which displayed no symptoms and the highest possible ΔS value (3). In general, the exploration of olive gene pools occurring outside Italy might lead to the identification of new sources of resistance to Xf. In this respect, we highlight previous studies reporting regional stratification of olive biodiversity in the Mediterranean region (Belaj et al., 2012; El Bakkali et al., 2013) and the occurrence of hotspots of genetic diversity in specific areas (Hosseini-Mazinani et al., 2014; Deddabi et al., 2020).
Based on Xf quantification in xylematic vessels, we showed that reduced symptoms in ‘Leccino’ and PRPs included in the cluster K1 might be due to (active or passive) defense mechanisms limiting bacterial proliferation. This is consistent with preliminary studies indicating partial resistance in ‘Frantoio’ (Boscia et al., 2017). In contrast, tolerance to the bacterium might be at the basis of the response of ‘Nocellara Messinese’, which exhibits markedly lower symptoms than ‘Ogliarola Salentina’ and ‘Cellina di Nardò’, but similar levels of Xf colonization. Similarly, tolerance might characterize SX_32, displaying a similar level of bacterial colonization with respect to the control (Table 1).
Overall, the results of this study may guide future efforts aimed to the selection of cultivars displaying resistance to Xf. High-throughput genotyping by whole or partial genome sequencing (Taranto et al., 2018; Pavan et al., 2020) might allow the collection of more comprehensive fingerprint data on the clones considered in this work, and the characterization of loci associated with resistance to Xf by means of conventional or extreme phenotype genome-wide association studies (Yang et al., 2015).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, upon reasonable request to the corresponding author.
Author Contributions
SP and AL conceived the research. MV, FNic, ES, AA, and CN performed exploratory missions and identified putatively resistant plants. VM, LS and FNig contributed to the evaluation of cultivars for response to Xylella fastidiosa. VF, MS, and CM contributed to SSR analysis. SP and CD performed data analysis. SP wrote the first draft of the manuscript. AL, LD, CL, CM, LR, and FNig critically revised the manuscript. All authors have read and approved the final manuscript.
Funding
This research was carried out within the framework of the following projects: Screen-X (Screening del germoplasma olivicolo salentino per l’individuazione di accessioni resistenti a Xylella fastidiosa), funded by the Apulian Regional Government; RE.D.O.XY (Valutazione del germoplasma olivicolo pugliese e miglioramento genetico per la resistenza a Xylella fastidiosa) funded by the Apulian Regional Government; APPROCCI (Approcci di Next generation sequencing per l’analisi di variabilità e di espressione genica in genotipi di olivo autoctoni pugliesi) funded by the Apulian Regional Government; Rigenerazione sostenibile dell’agricoltura nei territori colpiti da Xylella fastidiosa, funded by the Italian Ministry of Agricultural, Food and Forestry Policies.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge Mr. Antonio Ceglie and Dr. Nicola Murrone for the management of experimental orchard ‘Murrone’.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.723879/full#supplementary-material
Supplementary Figure 1 | Distribution of (A) disease severity scores (S) and (B) bacterial concentration (C) in thirty putatively resistant plants (PRPs) and respective control plants (CPs).
Supplementary Figure 2 | Parametric study of genetic structure. (A) Distribution of the ΔK parameter. (B) Genetic structure for three subpopulations (K = 3). Each individual is represented by a vertical line, which is partitioned into colored segments whose length is proportional to the estimated membership fraction (qi) in each subpopulation.
Supplementary Figure 3 | Bacterial colonization levels in PRPs included in K1 with respect to control plants.
Supplementary Table 1 | Features of the SSR loci used in this study, including the repeat motif, forward and reverse sequences, and reference in scientific literature.
Supplementary Table 2 | Summary statistics for the SSR analysis. For each SSR locus, the number of alleles, allele sizes, and polymorphic information content (PIC), are indicated.
Supplementary Table 3 | Membership coefficients (qi) of the individuals genotyped in this study for the three subpopulations identified by STRUCTURE analysis. Individuals grouped in the clusters K1 and K2 identified by hierarchical clustering are indicated.
References
Alba, V., Sabetta, W., Blanco, A., Pasqualone, A., and Montemurro, C. (2009). Microsatellite markers to identify specific alleles in DNA extracted from monovarietal virgin olive oils. Eur. Food Res. Technol. 229, 375–382.
Baù, A., Delbianco, A., Stancanelli, G., and Tramontini, S. (2017). Susceptibility of Olea europaea L. varieties to Xylella fastidiosa subsp. pauca ST53: systematic literature search up to 24 March 2017. EFSA J. 15:e04772. doi: 10.2903/j.efsa.2017.4772
Belaj, A., Dominguez-García, M. D. C., Atienza, S. G., Martín Urdíroz, N., de la Rosa, R., Satovic, Z., et al. (2012). Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 8, 365–378.
Boscia, D., Altamura, G., Ciniero, A., Di Carolo, M., Dongiovanni, C., Fumarola, G., et al. (2017). Resistenza a Xylella fastidiosa in diverse cultivar di olivo. Inf. Agrar. 11, 59–63.
Boucheffa, S., Miazzi, M. M., di Rienzo, V., Mangini, G., Fanelli, V., Tamendjari, A., et al. (2017). The coexistence of oleaster and traditional varieties affects genetic diversity and population structure in Algerian olive (Olea europaea) germplasm. Genet. Resour. Crop Evol. 64, 379–390.
Carriero, F., Fontanazza, G., Cellini, F., and Giorio, G. (2002). Identification of simple sequence repeats (SSRs) in olive (Olea europaea L.). Theor. Appl. Genet. 104, 301–307.
D’Attoma, G., Morelli, M., Saldarelli, P., Saponari, M., Giampetruzzi, A., Boscia, D., et al. (2019). Ionomic differences between susceptible and resistant olive cultivars infected by Xylella fastidiosa in the outbreak area of salento, Italy. Pathogens 8:272. doi: 10.3390/pathogens8040272
De La Rosa, R., James, C. M., and Tobutt, K. R. (2002). Isolation and characterization of polymorphic microsatellites in olive (Olea europaea L.) and their transferability to other genera in the Oleaceae. Mol. Ecol. Notes 2, 265–267.
Debbabi, O. S., Mnasri, S. R., Amar, F. B., Naceur, M. B., Montemurro, C., and Miazzi, M. M. (2021). Applications of microsatellite markers for the characterization of olive genetic resources of Tunisia. Genes 12:286. doi: 10.3390/genes12020286
Deddabi, O. S., Montemurro, C., Maachia, S. B., Amar, F. B., Fanelli, V., Gadaleta, S., et al. (2020). A hot spot of olive biodiversity in the Tunisian oasis of degache. Diversity 12:358. doi: 10.3390/genes12020286
di Rienzo, V., Sion, S., Taranto, F., D’Agostino, N., Montemurro, C., Fanelli, V., et al. (2018). Genetic flow among olive populations within the Mediterranean basin. PeerJ. 7:e5260.
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361.
Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19:1349.
El Bakkali, A., Haouane, H., Moukhli, A., Costes, E., Van Damme, P., and Khadari, B. (2013). Construction of core collections suitable for association mapping to optimize use of Mediterranean olive (Olea europaea L.) genetic resources. PLoS One 8:e61265. doi: 10.1371/journal.pone.0061265
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620.
Fernández-González, A. J., Villadas, P. J., Gómez-Lama Cabanás, C., Valverde-Corredor, A., Belaj, A., Mercado-Blanco, J., et al. (2019). Defining the root endosphere and rhizosphere microbiomes from the World olive germplasm collection. Sci. Rep. 9:20423. doi: 10.1038/s41598-019-56977-9
Giampetruzzi, A., Baptista, P., Morelli, M., Cameirão, C., Lino Neto, T., Costa, D., et al. (2020). Differences in the endophytic microbiome of olive cultivars infected by Xylella fastidiosa across seasons. Pathogens 9:723. doi: 10.3390/pathogens9090723
Harper, S. J., Ward, L. I., and Clover, G. R. G. (2010). Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology 100, 1282–1288. doi: 10.1094/PHYTO-06-10-0168
Hosseini-Mazinani, M., Mariotti, R., Torkzaban, B., Sheikh-Hassani, M., Ataei, S., Cultrera, N. G. M., et al. (2014). High Genetic Diversity Detected in Olives beyond the Boundaries of the Mediterranean Sea. PLoS One 9:e93146. doi: 10.1371/journal.pone.0093146
Liccardo, A., Fierro, A., Garganese, F., Picciotti, U., and Porcelli, F. (2020). A biological control model to manage the vector and the infection of Xylella fastidiosa on olive trees. PLoS One 15:e0232363. doi: 10.1371/journal.pone.0232363
Loconsole, G., Potere, O., Boscia, D., Altamura, G., Djelouah, K., Elbeaino, T., et al. (2014). Detection of Xylella fastidiosa in olive trees by molecular and serological methods. Eur. J. Plant Pathol. 96, 7–14.
Luvisi, A., Aprile, A., Sabella, E., Vergine, M., Nicolì, F., Nutricati, E., et al. (2017). Xylella fastidiosa subsp. pauca (CoDiRO strain) infection in four olive (Olea europaea L.) cultivars: profile of phenolic compounds in leaves and progression of leaf scorch symptoms. Phytopathol. Mediterr. 56, 259–273.
Marshall, T. C., Slate, J., Kruuk, L. E. B., and Pemberton, J. M. (1998). Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 7, 639–655. doi: 10.1046/j.1365-294x.1998.00374.x
Miazzi, M. M., di Rienzo, V., Mascio, I., Montemurro, C., Sion, S., Sabetta, W., et al. (2020). Re.Ger.O.P.: an Integrated Project for the Recovery of Ancient and Rare Olive Germplasm. Front. Plant Sci. 11:73. doi: 10.3389/fpls.2020.00073
Pagán, I., and García-Arenal, F. (2018). Tolerance to plant pathogens: theory and experimental evidence. Int. J. Mol. Sci. 19:810.
Paradis, E., Claude, J., and Strimmer, K. (2004). APE: an R package for analyses of phylogenetics and evolution. Bioinformatics 20, 289–290. doi: 10.1093/bioinformatics/btg412
Pasqualone, A., Montemurro, C., Di Rienzo, V., Summo, C., Paradiso, V. M., and Caponio, F. (2016). Evolution and perspectives of cultivar identification and traceability from tree to oil and table olives by means of DNA markers. J. Sci. Food Agric. 96, 3642–3657. doi: 10.1002/jsfa.7711
Pavan, S., Delvento, C., Ricciardi, L., Lotti, C., Ciani, E., and D’Agostino, N. (2020). Recommendations for choosing the genotyping method and best practises for quality control in crop genome-wide association studies. Front. Genet. 11:447. doi: 10.3389/fgene.2020.00447
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Purcell, A. H., and Hopkins, D. L. (1996). Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34, 131–151.
Sabella, E., Luvisi, A., Aprile, A., Negro, C., Vergine, M., Nicolì, F., et al. (2018). Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 220, 60–68. doi: 10.1016/j.jplph.2017.10.007
Saponari, M., Boscia, D., Altamura, G., D’Attoma, G., Cavalieri, V., Loconsole, G., et al. (2016). Pilot project on Xylella fastidiosa to reduce risk assessment uncertainties. EFSA Support. Publ. 1013:60. doi: 10.1016/j.jplph.2017.10.007
Saponari, M., Giampetruzzi, A., Loconsole, G., Boscia, D., and Saldarelli, P. (2019). Xylella fastidiosa in olive in apulia: where we stand. Phytopathol 109, 175–186. doi: 10.1094/PHYTO-08-18-0319-FI
Schneider, K., van der Werf, M., Cendoya, M., Mourits, M., Navas-Cortés, J. A., Vicent, A., et al. (2020). Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. U. S. A. 117, 9250–9259.
Sefc, K. M., Lopes, M. S., Mendonca, D., Santos, M. R. D., Da Camara Machado, M., and Da Camara Machado, A. (2000). Identification of microsatellites loci in olive (Olea europaea) and their characterization in Italian and Iberian olive trees. Mol. Ecol. 9, 1171–1173. doi: 10.1046/j.1365-294x.2000.00954.x
Spadoni, A., Sion, S., Gadaleta, S., Savoia, M., Piarulli, L., Fanelli, V., et al. (2019). A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agric. Sci. Technol. 21, 1215–1226.
Taranto, F., Nicolia, A., Pavan, S., De Vita, P., and D’Agostino, N. (2018). Biotechnological and digital revolution for climate-smart plant breeding. Agronomy 8:277.
White, S. M., Navas-Cortés, J. A., Bullock, J. M., Boscia, D., and Chapman, D. S. (2020). Estimating the epidemiology of emerging Xylella fastidiosa outbreaks in olives. Plant Pathol. 69, 1403–1413.
Yang, J., Jiang, H., Yeh, C. T., Yu, J., Jeddeloh, J. A., Nettleton, D., et al. (2015). Extreme-phenotype genome-wide association study (XP-GWAS): a method for identifying trait-associated variants by sequencing pools of individuals selected from a diversity panel. Plant J. 84, 587–596. doi: 10.1111/tpj.13029
Keywords: olive, Xylella fastidiosa, genetic diversity, DNA markers, resistance
Citation: Pavan S, Vergine M, Nicolì F, Sabella E, Aprile A, Negro C, Fanelli V, Savoia MA, Montilon V, Susca L, Delvento C, Lotti C, Nigro F, Montemurro C, Ricciardi L, De Bellis L and Luvisi A (2021) Screening of Olive Biodiversity Defines Genotypes Potentially Resistant to Xylella fastidiosa. Front. Plant Sci. 12:723879. doi: 10.3389/fpls.2021.723879
Received: 11 June 2021; Accepted: 23 July 2021;
Published: 16 August 2021.
Edited by:
Marcello Mastrorilli, Council for Agricultural and Economics Research (CREA), ItalyReviewed by:
Riccardo Aversano, University of Naples Federico II, ItalyPanagiotis Kalaitzis, Mediterranean Agronomic Institute of Chania, Greece
Copyright © 2021 Pavan, Vergine, Nicolì, Sabella, Aprile, Negro, Fanelli, Savoia, Montilon, Susca, Delvento, Lotti, Nigro, Montemurro, Ricciardi, De Bellis and Luvisi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Pavan, stefano.pavan@uniba.it
 Stefano Pavan
Stefano Pavan Marzia Vergine
Marzia Vergine Francesca Nicolì
Francesca Nicolì Erika Sabella
Erika Sabella Alessio Aprile
Alessio Aprile Carmine Negro
Carmine Negro Valentina Fanelli
Valentina Fanelli Michele Antonio Savoia
Michele Antonio Savoia Vito Montilon
Vito Montilon Leonardo Susca
Leonardo Susca Chiara Delvento
Chiara Delvento Concetta Lotti
Concetta Lotti Franco Nigro
Franco Nigro Cinzia Montemurro
Cinzia Montemurro Luigi Ricciardi1
Luigi Ricciardi1 Luigi De Bellis
Luigi De Bellis Andrea Luvisi
Andrea Luvisi