- 1Department of Biochemistry, Faculty of Chemistry, Philipps-University, Marburg, Germany
- 2Center for Synthetic Microbiology, Philipps-University, Marburg, Germany
Phytochrome activity is not only controlled by light but also by post-translational modifications, e. g. phosphorylation. One of the phosphatases responsible for plant phytochrome dephosphorylation and thereby increased activity is the phytochrome-associated protein phosphatase 5 (PAPP5). We show that PAPP5 recognizes phospho-site mimicking mutants of phytochrome B, when being activated by arachidonic acid (AA). Addition of AA to PAPP5 decreases the α-helical content as tracked by CD-spectroscopy. These changes correspond to conformational changes of the regulatory tetratricopeptide repeats (TPR) region as shown by mapping data from hydrogen deuterium exchange mass spectrometry onto a 3.0 Å crystal structure of PAPP5. Surprisingly, parts of the linker between the TPR and PP2A domains and of the so-called C-terminal inhibitory motif exhibit reduced deuterium uptake upon AA-binding. Molecular dynamics analyses of PAPP5 complexed to a phyB phosphopeptide show that this C-terminal motif remains associated with the TPR region in the substrate bound state, suggesting that this motif merely serves for restricting the orientations of the TPR region relative to the catalytic PP2A domain. Given the high similarity to mammalian PP5 these data from a plant ortholog show that the activation mode of these PPP-type protein phosphatases is highly conserved.
Introduction
During evolution plants evolved complex mechanisms to ensure their survival as sessile organisms. Given that sunlight is essential for survival and growth, plants employ a variety of photoreceptors and interconnected signaling cascades to optimize their photosynthetic potential and to synchronize their lifecycles with circadian and seasonal rhythms (Rockwell et al., 2006). As a classical plant photoreceptor phytochromes are photochromic toward red/far-red light and consist of a large (∼120 kDa) apoprotein with a covalently bound phytochromobilin (PΦB). This bilin chromophore enables plant phytochromes to absorb red/far-red light by cis-trans isomerization along the C15-C16 bond for triggering large structural changes within phytochrome dimers and distinguishing between the photon fluence rate, direction, duration and light quality. In addition, plant phytochromes like phytochrome B (phyB) act as temperature sensors (Jung et al., 2016; Legris et al., 2016), given their high activation energy for thermal dark reversion. Accordingly, these phytochromes are integrators of different environmental cues and trigger numerous developmental processes in planta such as seed germination, photomorphogenesis or floral induction (Rockwell et al., 2006; Franklin and Quail, 2010).
When plant phytochromes are photoconverted from their inactive, red-light sensitive Pr state to the active, far-red sensitive Pfr state, they are transported into the nucleus, where they can interact with various effectors such as phytochrome interaction factors (PIFs), a class of transcription factors, whose further modification and degradation is controlled by phytochromes (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Yamaguchi et al., 1999; Hisada et al., 2000; Park et al., 2004; Chen et al., 2005). However, PIFs are not the only interaction partners as phytochrome-dependent signal transduction pathways are complex and composed of an intricate network of numerous downstream-signaling components (Quail, 2002). Accordingly, phytochrome-dependent signaling is not only restricted to the cell nucleus, but also occurs in the cytoplasm. These cytosolic signaling routes presumably allow a faster mode of signal transduction, since relocalization of phytochromes into the nucleus requires up to 2 h (Parks and Spalding, 1999).
The N-terminal photosensory module (PSM) of plant phytochromes, which mediates light perception, resembles bacterial and fungal orthologs by consisting of a PAS (Per/Arndt/Sim), GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) and PHY (Phy-specific GAF) domain (Anders and Essen, 2015). Plant phytochromes have a unique composition for their C-terminal output module, comprising of two additional PAS domains and a histidine kinase-related domain. Furthermore, they possess an N-terminal extension (NTE) before the PSM, which is especially prominent in phyB and phyD with a length of ∼ 90 amino acids. This extension contains a serine- and glycine-rich region (Mathews, 2010) and is predicted to be largely unstructured. However, recent data indicate a light-dependent interaction with the PSM, as parts of the NTE exhibit light-switchable H/D exchange rates (Von Horsten et al., 2016). Interestingly, one of these parts (S84-K88) corresponds to a tandem of phosphorylation sites in phyB from Arabidopsis thaliana, S84 and S86 (Medzihradszky et al., 2013; Von Horsten et al., 2016). Phosphorylation of these sites shows an effect on the binding affinity of phytochromes toward some interaction partners and demonstrate the role of the NTE as a modulatory element for light-dependent phytochrome activity (Casal et al., 2002; Kim et al., 2004).
Whereas the kinase that phosphorylates plant phytochromes has not yet been discovered and an autophosphorylation activity appears to be present as well (Shin et al., 2016), four phosphatases have been identified to affect the phytochromes’ phosphorylation status (Bheri et al., 2020). The flower-specific phytochrome-associated protein phosphatase (FyPP) is named after its involvement in phytochrome-regulated flower formation and belongs to the family of type 6 protein phosphatases (Kim et al., 2002). The second, a type 7 phosphatase (PP7), controls phytochrome activity as well (Genoud et al., 2008). Two further phosphatases are the phytochrome-associated protein phosphatase 2c (PAPP2c), a phosphatase identified to interact with the PHY Domain of PhyA and to dephosphorylate the NTE of both AtPhyA and AtPhyB (Phee et al., 2008), and the type 5 serine/threonine protein phosphatase (PAPP5). The latter represents a unique type within the PPP family of serine/threonine phosphatases due to its N-terminal tetratricopeptide repeats (TPR) (Chen et al., 1994). The three repeats form a common α-helical bundle that is connected to the type 2A serine/threonine protein phosphatase (PP2A) domain via a long helical linker. A C-terminal inhibitory motif blocks together with the TPR region the active site of the PP2A domain, where two catalytic manganese ions are bound (Yang et al., 2005). PAPP5 is predominantly located in the nucleus but also present at lower levels in the cytoplasm, as the C-terminal motif of PAPP5 contains a putative nuclear localization signal (476–491) (Chen et al., 1994; Zeke et al., 2005). Furthermore, a second splice form of PAPP5 encoding mRNA yields a larger, membrane-localized isozyme of PAPP5 due to the presence of two additional transmembrane helices (De La Fuente Van Bentem et al., 2003). Studies by Ryu et al. have shown that the PP2A domain of PAPP5 dephosphorylates the NTE of AsPhyA and AtPhyB, whereas the TPR region binds to the C-terminal histidine kinase-related domain of AsPhyA (Ryu et al., 2005). The interaction between phytochromes and PAPP5 is thereby light-dependent and occurs preferentially, when the phytochrome adopts its activated Pfr state. Other functions of the soluble isoform of PAPP5 might be plantal signal transduction via plastid-derived tetrapyrroles (Barajas-Lopez Jde et al., 2013) or karrikins and the unknown endogenous KAI2 ligand (Struk et al., 2021).
Apparently, low in vitro activity is the reason for the late discovery of PAPP5 (Chen and Cohen, 1997). Variants of the human PP5 ortholog without the TPR region or the C-terminal inhibitory motif show an high increase in phosphatase activity. Mutagenesis studies have revealed that especially E76 (AtPAPP5: E61) of the TPR region (Kang et al., 2001) and the last 13 residues of the C-terminus are essential for blocking the active site (Chen and Cohen, 1997; Sinclair et al., 1999; Kang et al., 2001). Like mammalian PP5, PAPP5 is activated by the polyunsaturated fatty acid arachidonic acid (AA) (Ryu et al., 2005). As higher plants do not produce eicosapolyenoic acids, AA has been postulated to act in plants as an exogenous signaling molecule for stress and host defense (Savchenko et al., 2010).
Next to the absorption of light, phosphorylation/dephosphorylation presents a second pathway to fine-tune the activity of plant phytochromes. From the set of phosphatases responsible for dephosphorylating phytochromes, PAPP5 proves to be an interesting interaction partner since it interacts with both the N- and C-terminus of phytochrome A from Avena sativa and Arabidopsis thaliana (Ryu et al., 2005). In this report, we investigated AtPAPP5 and solved its crystal structure, which enabled a molecular understanding of the autoinhibition and the role of its C-terminal motif. Studies addressing the structural changes in AtPAPP5 upon AA-binding allowed us to create a general activation model for phosphatases of the PP5 subfamily.
Experimental Procedures
Cloning, Overexpression, and Purification of AtPAPP5, AtPhyB(1–651) and AtPhyB/S84D and AtPhyB/S86D Mutants
The DNA fragment encoding isoform 2 of papp5 from Arabidopsis thaliana (UniProtKB: Q84XU2-2; 1-484) was codon-optimized and flanked by NdeI and Sall restriction sites (Geneart, Invitrogen Life Technologies). Afterward, the fragment was subcloned into the pET-28a vector (Novagen) using the corresponding restriction sites.
The plasmid carrying the coding sequence for the photosensory module of Arabidopsis thaliana phytochrome B (UniProtKB: P14713-1; 1-651) was kindly provided by Andreas Zurbriggen (University Düsseldorf) and subcloned into the pCDF Duet-1 vector (Novagen) by polymerase chain reaction using primers, which introduced PciI and PstI restriction sites. S84D and S86D mutants were generated by QuickChange site-directed mutagenesis (Stratagene). All primers are listed in Supplementary Table 1. Sequences of the resulting plasmids were controlled by dideoxy sequencing (GATC).
Expression was performed in the E. coli BL21 Gold (DE3) strain (Novagen). Cultures for AtPAPP5 overproduction were grown in TB medium containing 35 mg/L kanamycin at 37°C to an OD595 of 0.6, the temperature was decreased to 18°C and expression was induced with 50 μM IPTG. After 22 h the cells were harvested by centrifugation (8,200 g, 15 min, 4°C), resuspended in Mn-buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 4 mM MnCl2, 0.1 mM β-mercaptoethanol), frozen in liquid nitrogen and stored at −80°C. Bacterial cells were lysed by a French Press cell (AMINCO) and the supernatant was separated by centrifugation (40,000 g, 30 min, 4°C), followed by Ni2+-affinity chromatography (HisTrapTM HP column, GE Healthcare) and eluted with TIS buffer (50 mM Tris, 250 mM imidazole pH 7.8, 100 mM NaCl, 4 mM MnCl2, 1 mM β-mercaptoethanol). A final purification step was done by size exclusion chromatography (Superdex 200 26/60, GE Healthcare) using 50 mM Tris pH 7.8, 1 mM EDTA, 100 mM NaCl, 4 mM MnCl2, 1 mM β-mercaptoethanol as buffer. Before usage of AtPAPP5 for assays the affinity tag was cleaved off by thrombin (overnight at 4°C) and removed by reversed Ni-NTA.
Overproduction and purification of wild-type AtPhyB(1-651) and the phosphomimic mutants harboring the PCB chromophore was performed as described before (Von Horsten et al., 2016). Prior to usage samples were centrifuged for 10 min at 4°C and 13,000 rpm.
Phosphatase Activity Assays
The assay was performed as described by Ryu et al. (2005). In 100 μL reaction mixture, 1 μg AtPAPP5 in kinase/phosphatase buffer (KP: 25 mM Tris-HCl pH 7.5, 5 mM MgCl2, 20 mM Mg(CH3COO)2, 0.2 mM EDTA, 0.2 EGTA) was used in the absence or presence of arachidonic acid/6% ethanol. The concentration of p-nitrophenyl phosphate (pNPP) was varied. After an incubation time of 15 min at 30°C the reaction was terminated by the addition of 900 μL 0.25 M NaOH and the absorbance at 410 nm was measured.
Microscale Thermophoresis Analysis of AtPAPP5-AtPhyB Interaction
Microscale thermophoresis was conducted to determine the AtPAPP5 dissociation constant toward the AtPhyB(1-651) mutants S84D and S86D by using a Monolith NT.115 instrument (NanoTemper). Freshly prepared AtPAPP5 (50 μM in labeling buffer) was labeled at 8°C overnight using the cysteine-reactive Monolith NT.115 Protein Labeling Kit GREEN MALEIMIDE (NanoTemper) to achieve a 1:1 molar ratio of labled protein to dye. A serial titration from 4 nM to 70 μM of S86D and 2 nM to 29 μM of S84D was prepared and labeled. AtPAPP5 was diluted to each sample until a final concentration of 50 nM. Afterward 100 μM AA was added and the mixture was incubated for 15 min before measurement. The assay was then transferred into premium coated capillaries (NanoTemper) and measured at 25°C. The LED power for each thermophoresis measurement was set to 80% and the laser power to 20%, the heating time was set to 30 s, followed by 5 s of cooling. Binding curves were obtained from the thermophoresis hot to cold ratio. Negative controls were performed without the addition of AA and with AtPhyB WT under the same conditions mentioned above. Each experiment was repeated in triplicate, analyzed with the NT Analysis software (NanoTemper) and the KD value was calculated with Origin 9.0 (OriginLab) using a 1:1 binding model.
Size Exclusion Chromatography
A 25 μM of AtPAPP5 in KP buffer (25 mM Tris-HCl pH 7.5, 5 mM MgCl2, 20 mM Mg(CH3COO)2, 0.2 mM EDTA, 0.2 EGTA) buffer was injected onto a Superdex 200 10/300 column using an Äkta purifier (GE Healthcare). The setup was repeated after adding 30 μM arachidonic acid. The runs were monitored at 280 nm. The apparent molecular masses of the eluted species were calculated by using a molecular weight standard curve based on a column calibration (LMW kit, GE Healthcare) performed previously.
Circular Dichroism Spectroscopy of AtPAPP5
A solution of 40 μM AtPAPP5 in HEPES buffer (10 mM HEPES pH 8.0, 100 mM NaCl, 4 mM MgCl2) with 0 μ, 25, 50, 75, 100, and 150 μM AA, using a 3.2 mM AA stock solution in EtOH were incubated at room temperature for 15 min and measured with a spectropolarimeter J-810 (Jasco) using a 1 mm cuvette. After a baseline correction with the corresponding buffer, which contained an equal concentration of ethanol, the data was averaged over three scans.
Crystallization and Structure Determination of AtPAPP5
AtPAPP5 was transferred into crystallization buffer (10 mM Tris pH 7.5, 100 mM NaCl, 5 mM MnCl2), concentrated to 15 mg/mL and crystallization attempts were pipetted with a Honeybee robot system (Zinsser Analytic) using commercially available screens (Qiagen) in a 96-well format. Crystals were grown at 18°C using the sitting-drop method and vapor-diffusion. After 2 weeks, small crystals were found growing in condition C10 from crystal screen Core I (Qiagen). To obtain bigger crystals optimizations were done using streak seeding with crushed crystals from the original hit. The crystallization drop for the optimizations consists of 1 μL protein solution and 1 μL of reservoir solution. For optimization, the hanging-drop vapor diffusion method was used. After 4 weeks, crystals appeared in a condition containing 0.1 M HEPES pH 7.0 and 50% (v/v) PEG4000. These were picked and incubated in cryoprotection buffer (reservoir buffer supplemented with 30% glycerol) prior to freezing in liquid nitrogen.
An AtPAPP5 dataset was collected at beamline 14.2 of BESSY-II (Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung, Berlin, Germany) and processed with the XDS (Kabsch, 2010) and CCP4 (Collaborative Computational Project, 1994) package. The structure for the dataset was solved by molecular replacement using PHENIX autosolve (Adams et al., 2010) and 1WAO as search model (Yang et al., 2005). Manual model building was conducted with COOT (Emsley and Cowtan, 2004) and refinement by PHENIX refine (Adams et al., 2010). Figures were created using PYMOL 1.6 (DeLano Scientific).
Hydrogen Deuterium Exchange-Mass Spectrometry of AtPAPP5
HDX-mass spectrometric analysis AtPAPP5 (60 μM) with and without arachidonic acid (75 μM) was carried out using a commercial HDX-automation setup (SYNAPT G2-Si, Waters) including a two-arm robotic autosampler (LEAP Technologies), an ACQUITY UPLC M-Class system (Waters) and HDX manager (Waters). AtPAPP5 samples were transferred by a PD-10 column into a low salt buffer (10 mM Tris pH 8.0, 100 mM NaCl, 4 mM MgCl2, 1 mM β-mercaptoethanol) and centrifuged for 10 min at 16,100 g and 4°C. Exchange reactions and analysis were performed as described before (Von Horsten et al., 2016).
Final assignment of deuterium incorporation was done with DynamX 3.0 (Waters). The minimum peak intensity was set to 103 counts and a peptide length between four and 15 was chosen. Moreover, tolerances of 0.5 min for the retention time and 25 ppm for m/z values were applied for the peptide assignment, generating an overall sequence coverage of 80.0%. 125 peptides were analyzed with an overall redundancy of 3.0 per amino acid. A standard deviation of 4 σ was used to quantify the amount of variation between the repetitions. A coverage map of all peptides is presented in Figure 1D. The results of the analysis were mapped onto the structure of AtPAPP5.
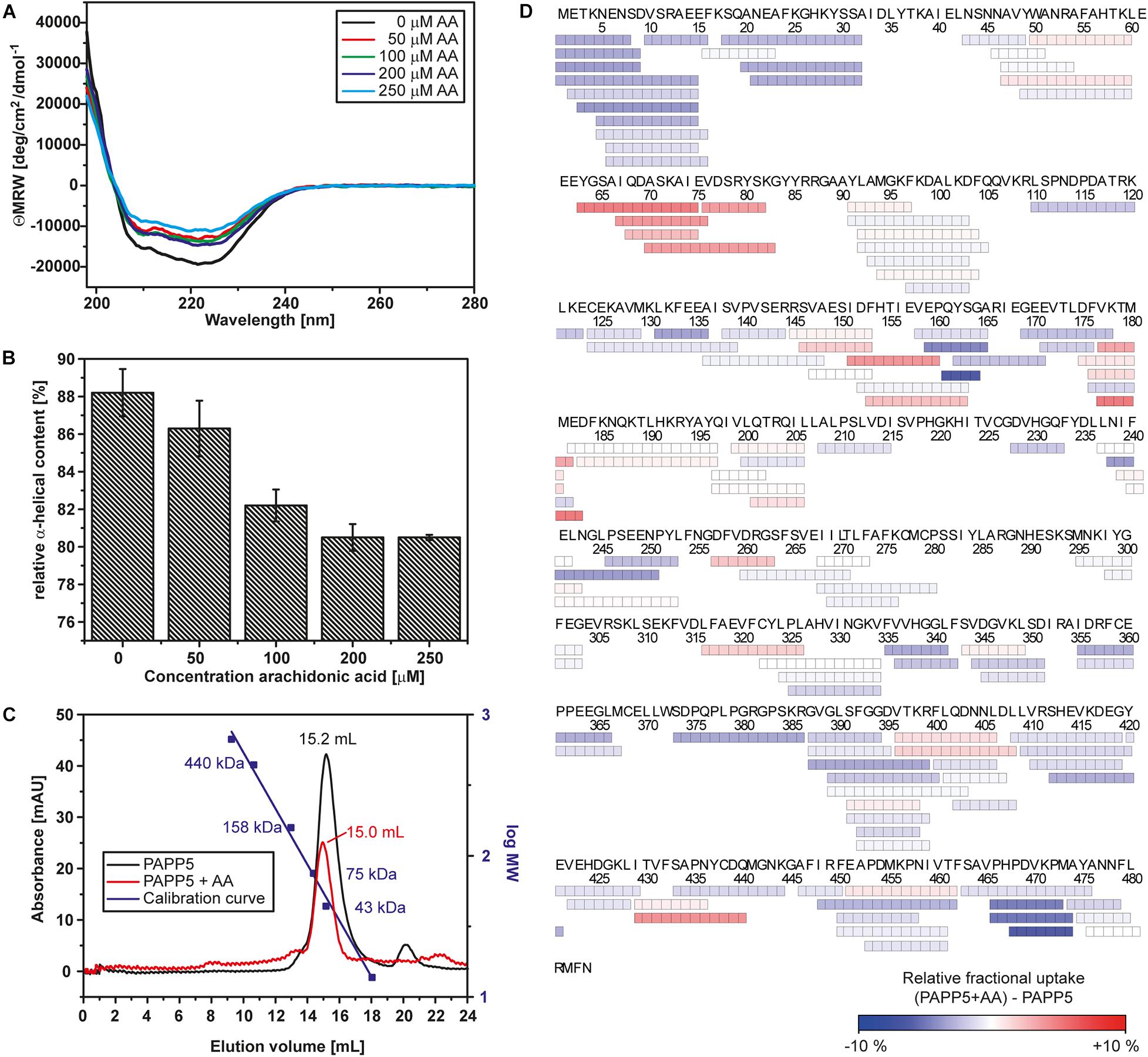
Figure 1. Arachidonic acid activates AtPAPP5 by altering its structure. (A) Far-UV CD-spectrum of AtPAPP5 in the absence or presence of various concentrations of arachidonic acid (AA). (B) Calculated relative alpha-helical contents from the CD measurements presented in a bar chart. (C) Size exclusion chromatogramm of AtPAPP5 in the presence or absence of AA. The addition of arachidonic acid alters the elution volume from 15.18 to 14.96 mL. The molecular masses of calibration standards are plotted using the second logarithmic ordinate. The absorbance was recorded at 280 nm, respectively. (D) Coverage map of the HDX-MS analysis for the difference between AtPAPP5 and AtPAPP5 with AA after 15 s deuterium incubation. Each box reflects one peptide and the coloring is according to the relative fractional deuterium uptake colored in blue-white-red presentation.
Molecular Dynamics Analysis of AtPAPP5•phyB Peptide Complexes
Molecular dynamics analyses of AtPAPP5 were done with the GPU-accelerated Amber18 suite (Lee et al., 2018) using the ff19sb force field for the protein (Tian et al., 2020) and the SPC/E water model. pKa values of ionizable residues of AtPAPP5 at pH 6.5 were estimated by H++. Using B3LYP/6-31G∗ model chemistry we calculated parameters for the μ-hydroxyl bridged di-Mn2+ center either in its free state or complexed to methyl-phosphate or phosphoserine-comprising phyB peptides by gaussian16 (Frisch et al., 2016) and MCPB.py (Li and Merz, 2016). As start geometry for these QM-calculations we used the di-Mn2+ center of human PP5 complexed to the Cdc37 substrate peptide (Oberoi et al., 2016). For MD simulations free AtPAPP5 was neutralized by 57 chloride and 58 sodium ions (∼0.15 M) and placed in a box with 31,718 water molecules, which equilibrated to box dimensions of 100.6 × 100.8 × 100.5 Å3 (Supplementary Table 2). AtPAPP5 ensembles were minimized, pre-equilibrated as NVT ensembles followed up for 5.5 ns as NpT ensembles at 300 K using 2 fs steps, 10 Å cutoff and a Monte-Carlo barostat. The systems were further equilibrated for 100 ns before generating projection trajectories with lengths of 180–1,000 ns. AtPAPP5 trajectories were processed and evaluated in jupyter notebooks using pytraj 2.0.5 (Roe and Cheatham, 2013),1 AmberTools20 and NGLview 2.7.7 (Nguyen et al., 2018). The total length of four independently generated production trajectories for AtPAPP5 in the unbound, closed and the phyB S80-T89 phosphorylated peptide states amount to 4 μs for each, for the complex between AtPAPP5 and the phyB D82-S86 peptide phosphorylated at S84 to 2.88 μs and the complex with phosphoserine to 3.50 μs (Supplementary Table 2). AtPAPP5-bound peptides were capped at their N- and C-termini by acetyl and methylamine groups, respectively.
Results
Activation of AtPAPP5 by Fatty Acids and Ethanol
For structural analyses, we used recombinant AtPAPP5 that was produced as histidine-tagged monomer by heterologous expression in E. coli. In contrast to previous reports (Ryu et al., 2005) we removed affinity tags for activity measurements.
Earlier reports showed that AA activates AtPAPP5 as well as the human PP5 ortholog (Chen and Cohen, 1997; Skinner et al., 1997; Ryu et al., 2005). To confirm the functionality of recombinant AtPAPP5, we measured the phosphatase activity by an assay based on para-nitrophenyl phosphate (pNPP) as chromogen. The amount of released phosphate (Pi) corresponds to the formation of para-nitrophenolate that is monitored at 405 nm. The assay was initially conducted for AtPAPP5 in the presence of 70 μM AA that was dissolved in ethanol. Control reactions were carried out with an equivalent volume of ethanol. Surprisingly, ethanol alone already activates AtPAPP5 by releasing the blocked active site (Figure 2A). In the presence of AA AtPAPP5 shows a Michaelis-Menten kinetics for varying pNPP concentrations with a KM-value of 92.2 mM and a Vmax value of 15 ± 1.1 μmol/mg/mL. These values resemble kinetic constants obtained by Ryu et al. (2005) for a GST-AtPAPP5 fusion with 100 μM AA (KM 160 mM; Vmax 22 μmol/mg/mL). Interestingly, addition of arachidonic acid caused only a doubled turnover rate relative to the ethanol-activated state (Vmax = 8.9 ± 1.9 μmol/min/mg) and a fourfold increase compared to wildtype without any AA and EtOH (Vmax = 3.6 ± 0.7 μmol/min/mg).
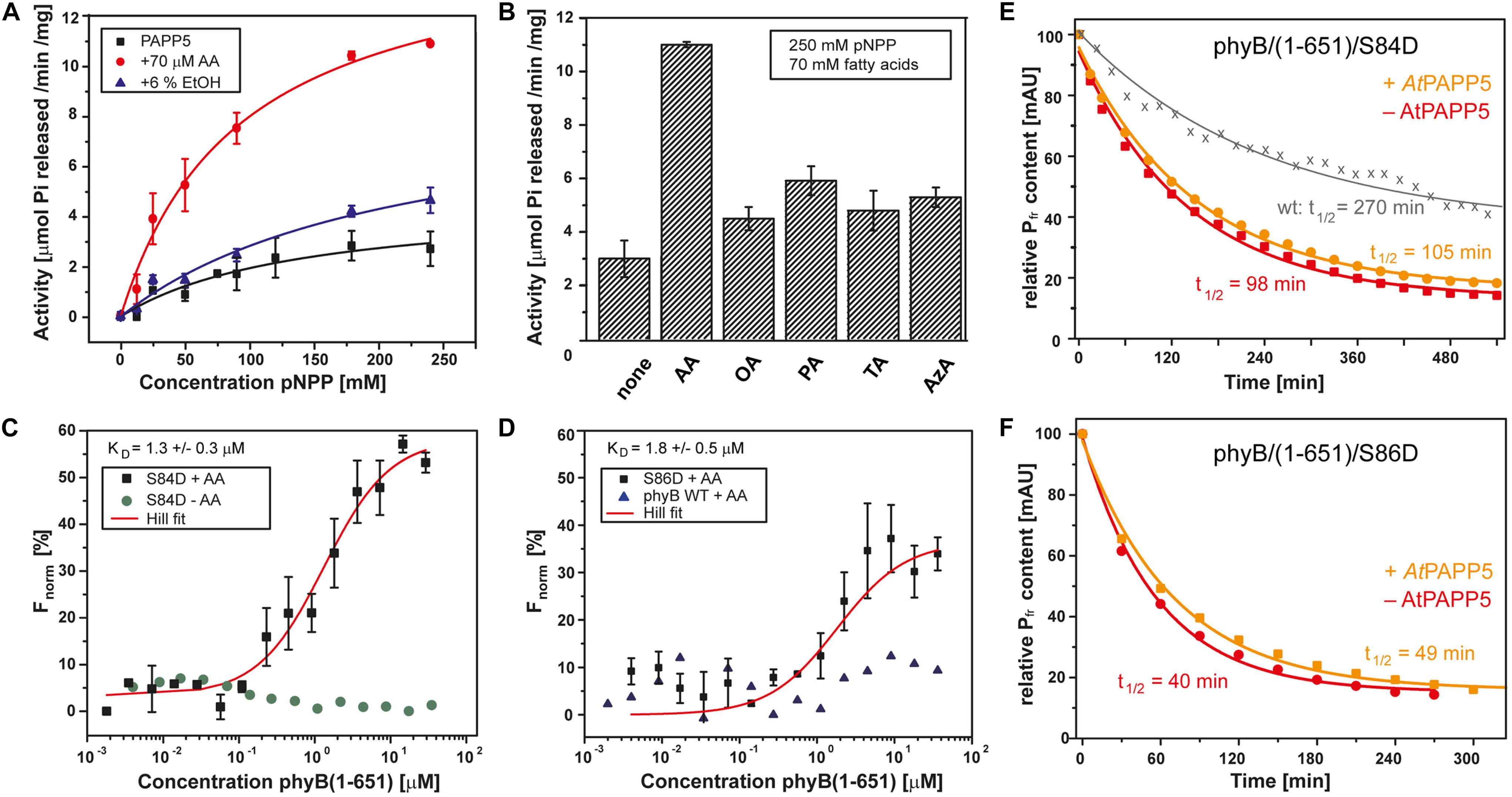
Figure 2. AtPAPP5 activity and interaction with the phosphomimic mutants S84D and S86D of phytochrome B. (A) Phosphatase activity (mean ± SD; n = 3) of AtPAPP5 in the presence or absence of 70 μM arachidonic acid (AA) or 6% ethanol (EtOH) and various concentrations of the artificial substrate, p-nitrophenyl phosphate (pNPP). (B) Influence of fatty acids (70 μM) on the phosphatase activity (mean ± SD; n = 3) of AtPAPP5 in the presence of 250 mM pNPP. The line marks the activity of ethanol alone. OA, oleic acid; PA, palmitic acid; TA, traumatic acid; AzA, azaleic acid. (C,D) Microscale thermophoresis experiments show binding (mean ± SD; n = 3) of AtPAPP5 toward AtPhyB/S86D and AtPhyB/S84D. Notably, the S84D achieved no saturation. Therefore, this value should only be regarded as approximation. As negative controls the measurement were done either without AA (C) or with wild-type AtPhyB(1-651) (D). The fitting of experimental data assumed a 1:1 binding model. (E,F) The dark conversion of the S84D and S86D variants (red), in the presence of AtPAPP5 and AA (orange) and the half-lives (t1/2) calculated therefrom.
We also checked PAPP5 activities in the presence of different fatty acids using saturating substrate concentrations, i.e., 250 mM pNPP (Figure 2B). None of the other fatty acids activated PAPP5 like AA. To further investigate the influence of ethanol on the phosphatase activity, ethanol concentrations were varied between 0 and 90% (v/v). Interestingly, these data show an up to 12-fold boost of PAPP5 activity with 75% ethanol (Supplementary Figure 1), which may reflect either partial unfolding of the TPR region or a disruption of TPR-PP2A domain interface.
AtPAPP5 Interacts With the N-terminal Extension of phyB
Structural studies on the catalytic domain of human PP5 complexed to a Cdc37-derived peptide showed that phosphomimetic S→D mutations serve as useful tools for analyzing phosphatase-phosphopeptide interactions (Oberoi et al., 2016). Accordingly, we proved direct interaction between AtPAPP5 and the NTE of AtPhyB by generating two phosphomimetic variants of AtPhyB(1-651), S84D, and S86D. Phosphorylation/dephosphorylation of S84 and S86 in the NTE phyB was shown to be strongly involved in the negative in vivo regulation of phyB (Medzihradszky et al., 2013; Nito et al., 2013). We performed sensitive interaction assays by microscale thermophoresis (MST) using AtPAPP5 that was labeled before with a green-fluorescent dye. After preincubation with AA for 15 min AtPAPP5 binding was measured against a concentration series with either AtPhyB/S84D or AtPhyB/S86D. Since our MST setup uses a green laser for fluorescence detection (λexc: 515–525 nm, λem: 650–685 nm), the phytochrome variants were kept in a photodynamical equilibrium with a prevalence of their activated Pfr state. AtPAPP5 showed a high affinity interaction to AtPhyB phosphomimic variants with an apparent KD of 1.3 ± 0.3 μM for S84D and 1.8 ± 0.5 μM for S86D, respectively (Figures 2C,D). To rule out non-specific binding, the measurements were repeated using recombinant wild-type AtPhyB(1–651) and resulted in signals not exceeding signal-to-noise ratios. Accordingly, non-specific binding to the photosensory module of AtPhyB in its non-phosphorylated Pfr state can be excluded. Furthermore, measurements in the absence of AA resulted in baselines showing the necessity of AtPAPP5 activation by AA (Figure 2C).
We analyzed the photochemical behavior of our phosphomimic mimics by UV/Vis spectroscopy. Compared to wild-type AtPhyB (Von Horsten et al., 2016) absorption maxima are bathochromically shifted, especially in the Pfr form with shifts of 8 nm for the S84D and 5 nm for the S86D variant (Supplementary Figure 2). Furthermore, both phytochrome B variants exhibit an accelerated dark conversion rate (t1/2 of S84D: 98 min; S86D: 40 min) compared to the photosensory module of wildtype phyB (t1/2: 270 min; Figures 2E,F; Von Horsten et al., 2016). Addition of AtPAPP5 and AA induced no further shift of the maxima according to UV/Vis spectra. However, there is a slightly reduced dark conversion rate with half-lives of 105 min for S84D and 49 min for S86D.
Closed State Structure of AtPAPP5
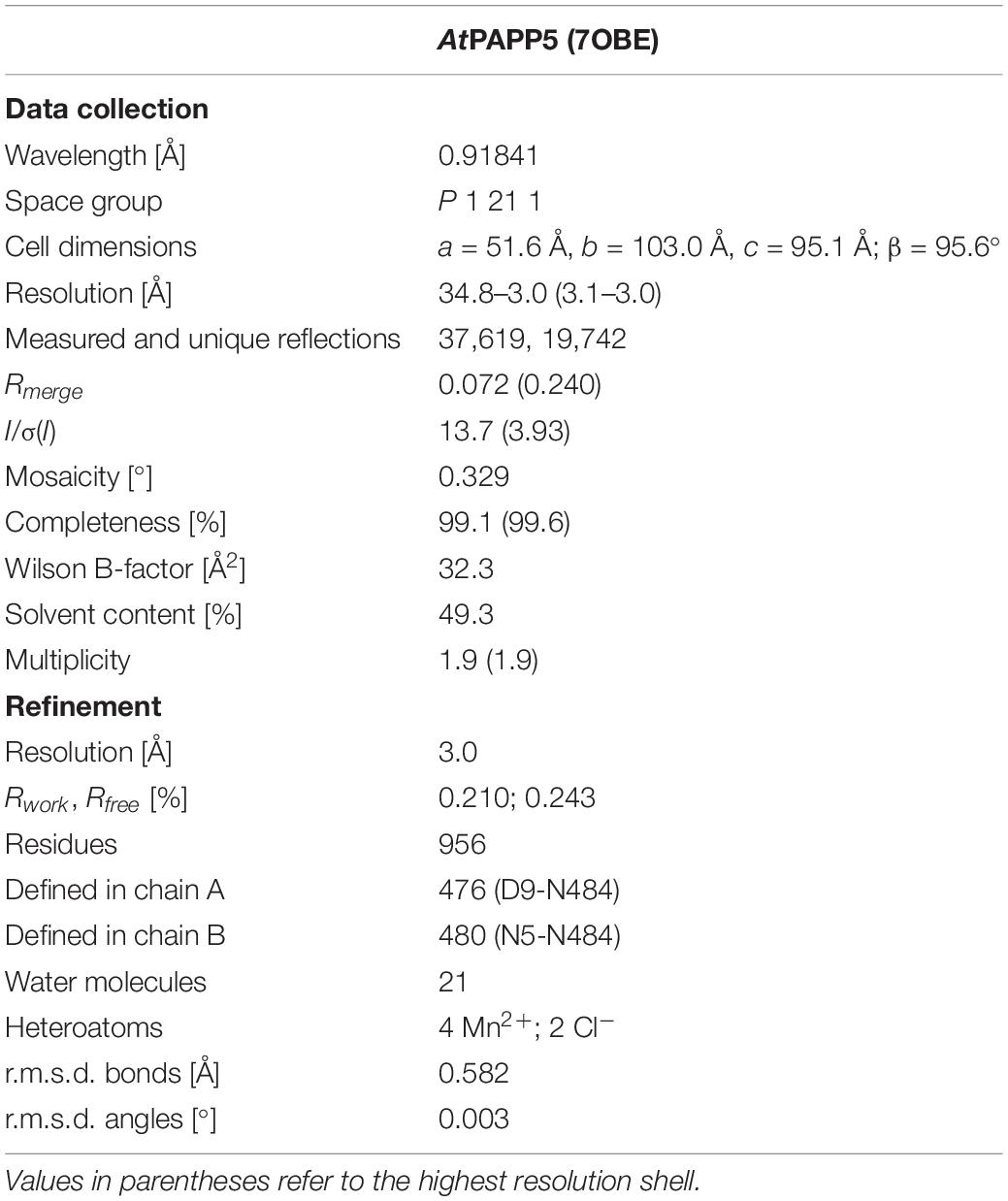
We crystallized AtPAPP5 in a monoclinic crystal form that resulted in a dataset at 3.0 Å resolution. Unfortunately, we could not treat these AtPAPP5 crystals with AA because they rapidly dissolved upon incubation. Molecular replacement using protein phosphatase 5 from Homo sapiens (HsPP5; PDB Code 1WAO) as search model (sequence identity: 56% for 484 amino acids) allowed structure determination with two molecules per asymmetric unit. Table 1 shows data collection and refinement statistics of our AtPAPP5 structure.
The AtPAPP5 structure shows the characteristic bilobal shape of a type 5 serine/threonine phosphatase with two manganese ions bound in the active site. Three TPR repeats (TPR1: V10-L42, TPR2: W50-V76 and TPR3: S81-L110) are linked via a long helical linker (P115-T173) to the PP2A domain (L174-N484). The TPR repeats of the N-terminal domain consist each of two α-helices as part of a helix-turn-helix motif; these motifs stack to each other causing formation of a superhelical tertiary structure. The last 14 amino acids of the PP2A domain form a separate motif, the C-terminal inhibitory motif that consists of a linker (K471-A476) and a short α-helical segment (αJ, N477-M482; Figure 3A). The strictly conserved PP2A domain adopts a compact α/β fold consisting of eleven α-helices and eleven β-strands, which form together the central β-sandwich (Figure 3E). This sandwich is flanked on one side by three short helices (α9-α11) and on the other side by six helices (α12-α17). The active site itself is embedded in the PP2A domain and covered by the TPR region and the C-terminal inhibitory motif. In its center two catalytic manganese ions are observed (Figures 3A,B).
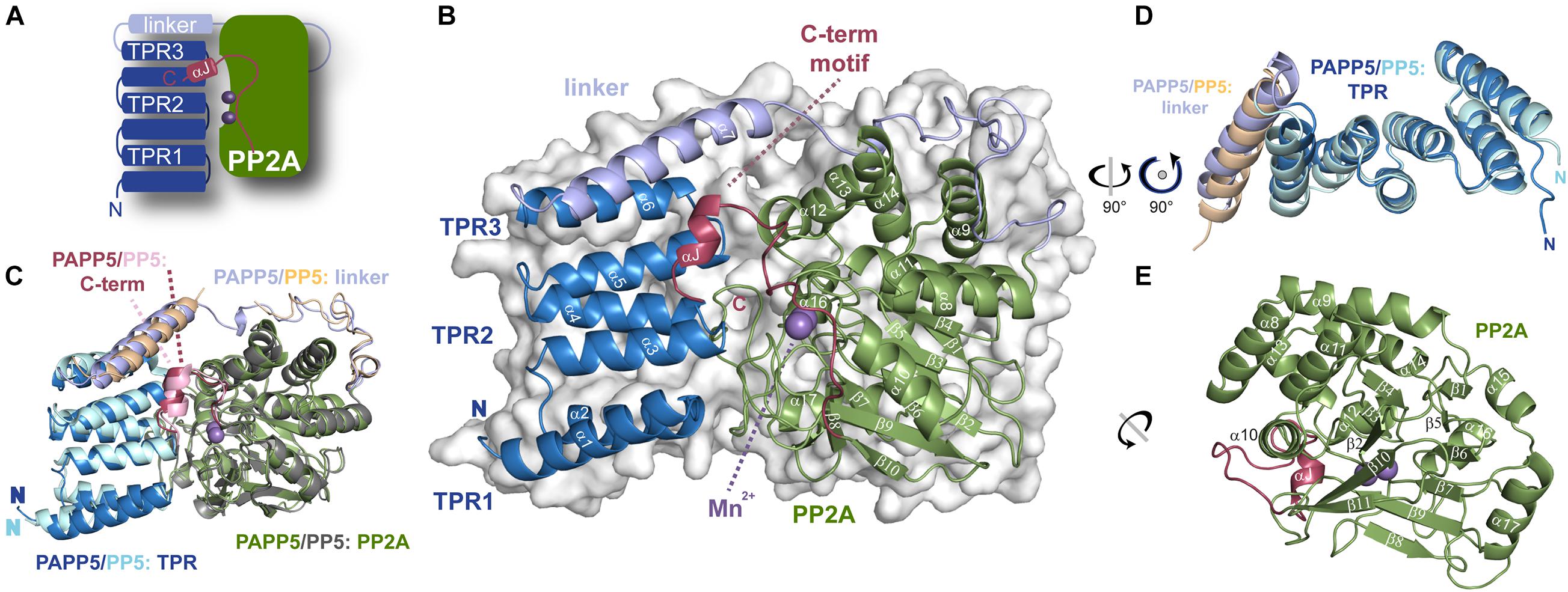
Figure 3. Crystal structure of AtPAPP5 phosphatase. (A) Schematic organization of AtPAPP5. (B) The TPR region (blue) is arranged in an α-helical bundle and connected via a long linker region (light blue) to the PP2A domain (green), in whose active site two Mn2+ ions (purple) are bound. The C-terminal inhibitory motif (raspberry) and the TPR region cover the active site. (C) Superposition of AtPAPP5 and HsPP5 structures (PDB code: 1WAO). (D) After superposition on the central TPR2 repeat, the superposed TPR regions show some deviations for the TPR3 repeat and the linker helix. (E) The catalytic PP2A domain consists of 11 β-strands arranged as a central β-sandwich that is surrounded by α-helices.
A superposition of AtPAPP5 with HsPP5 shows, that AtPAPP5 mimics the overall structure of the human phosphatase with an r.m.s.d. of 0.70 Å for 365 Cα-atoms (0.99 Å for 390 Cα -atoms). These structures represent the closed state of type 5 serine/threonine protein phosphatases, because the TPR region covers major parts of the active site region. Minor deviations are shown in the arrangement of the TPR region and the helix of the linker region (Figures 3C,D). In contrast to mammalian PP5 structures (Yang et al., 2005; Haslbeck et al., 2015) in our AtPAPP5 structure the linker region is completely defined by electron density.
The local environment of the active site is conserved between HsPP5 and AtPAPP5 as exemplified by residues D228, H230, D257, N289, H338 and H413 (numbering of amino acids corresponds to AtPAPP5), which are the coordination partners of the manganese ions. Here, Mn2+ ion 1 is tetra-valently coordinated by amino acids D257, N289, H338 and H413, whereas Mn2+ ion 2 interacts with amino acids D228, H230 and D257 (Figure 4A). For comparison, a crystal structure of the isolated human PP2A domain (PDB Code: 1S95) (Swingle et al., 2004) shows two additional water molecules. One of them acts like the oxygen atom of the opposing D257 as μ-bridge between the two Mn2+ ions thus forming slightly distorted octahedral coordination spheres around the catalytic Mn2+ ions (Figure 4B). These μ-bridging water ligands are missing in the AtPAPP5 structure due to low resolution. Instead, we observe a coordinated chloride ion in our AtPAPP5 structure (Figures 4A,C). This partly occupied chloride (molecule A: 59%, B: 65%) replaces a phosphate anion that was observed before in the active site of the isolated PP2A domain of HsPP5. Its five interacting ion partners are preserved in AtPAPP5 (R261, N289, H290, R386, and Y437) and require only some swiveling of the sidechains of R261 and R386 for phosphate binding.
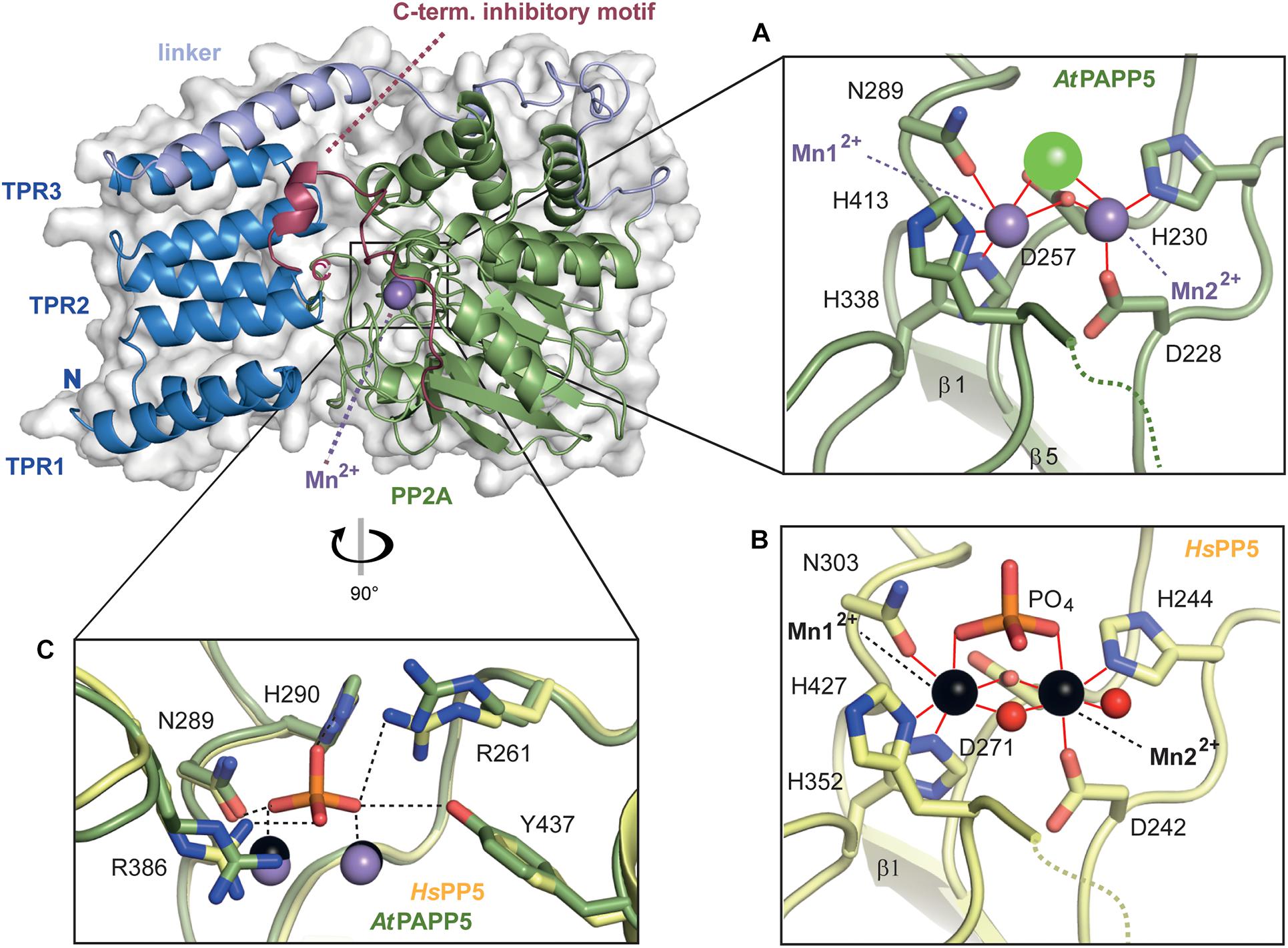
Figure 4. Close-up view of the active site of AtPAPP5. (A,B) Detailed presentation of the coordination spheres of the manganese ions in the phosphatase AtPAPP5 and HsPP5 (PDB Code: 1S95). Interactions of amino acids with the Mn2+ ions (AtPAPP5: purple, HsPP5: black) are shown in red. A green sphere represents a chloride ion and red spheres water molecules. (C) Superposition of the phosphate binding site of AtPAPP5 and HsPP5 (PDB code: 1S95). The interactions between the phosphate and the amino acids are shown as black dashed lines. The numbering of the amino acids refers to AtPAPP5.
The C-terminal Inhibitory Motif
The AtPAPP5 structure represents the autoinhibited state, in which both the TPR region as well as the C-terminal inhibitory motif (K471-N484) block the active site and deny access for putative substrate-like phosphopeptides (Figure 5). This mode of self-inhibition was already demonstrated by activity measurements of PP5-type deletion variants (Sinclair et al., 1999; Ryu et al., 2005).
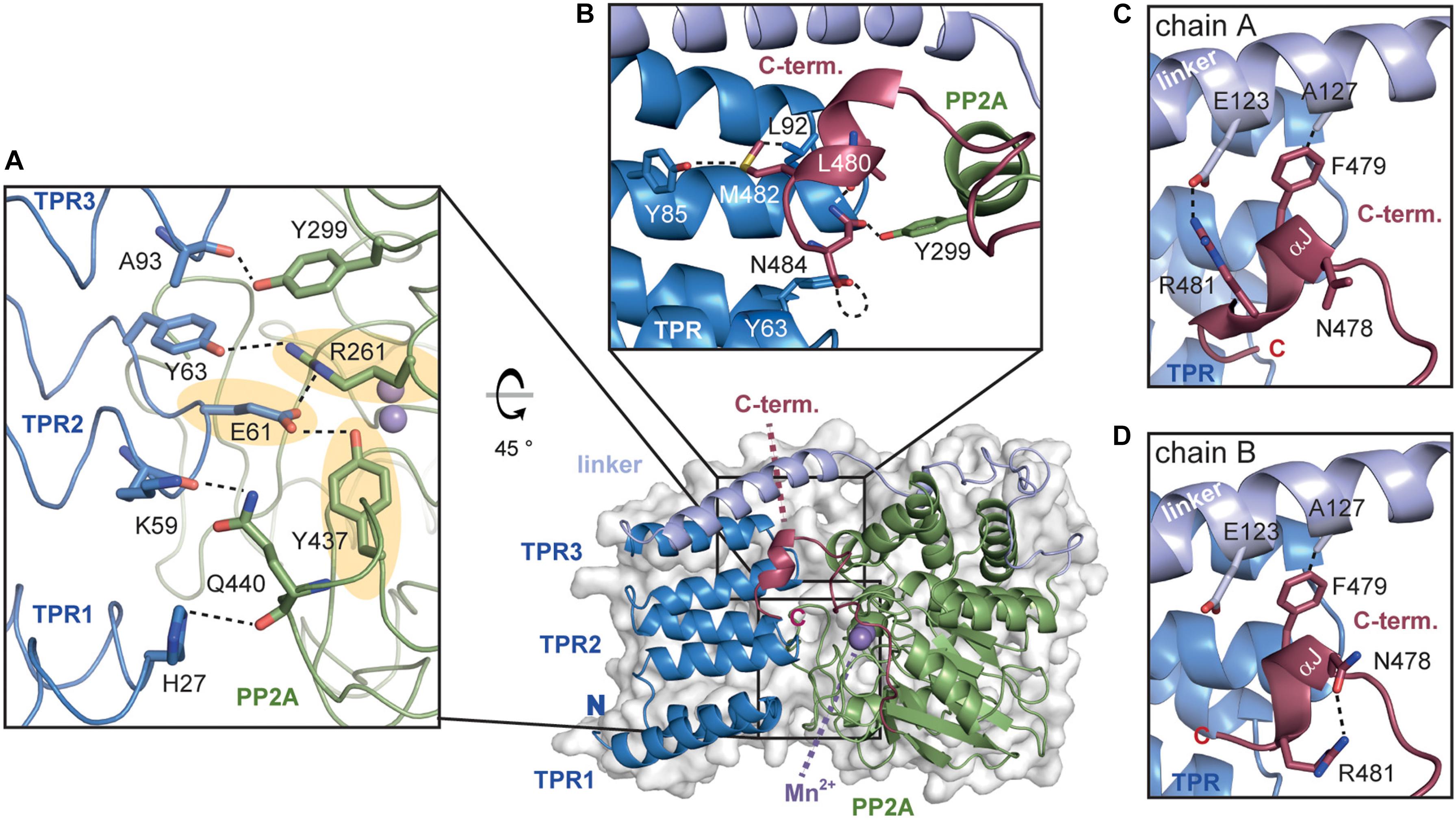
Figure 5. The TPR-PP2A interface of AtPAPP5. (A) The inhibition of the PP2A domain by the TPR region is based on a variety of direct interactions. The hydrogen bonds from E61 to R261 and Y437 (highlighted in yellow), which are also involved in phosphate binding, are depicted in dashed lines. (B) The C-terminal helical segment interacts with both the TPR and the PP2A domain. The hydrogen bonding network between C-terminal inhibitory motif and TPR2 as well as the PP2A domain is indicated with dashed lines. (C,D) The interactions of the C-terminal helical segment differ between chains A and B. The interactions of the amino acids are represented by dashed black lines.
An analysis of the interactions between the TPR and PP2A domain shows that all three TPR repeats form interactions with the PP2A domain (Figure 5A). Here, the prominent hydrogen bonds between the carboxylate group of E61 to Y437 and R261 are of particular interest, since both amino acids are involved in the coordination of the substrate’s phosphate group. Mutation of E61 in HsPP5 to alanine unblocked the active site and raised 10-fold the intrinsic phosphatase activity (Kang et al., 2001). Further stabilization of the inhibited state may be also provided by the interaction between the hydroxyl group of Y63 with R261. These interactions with the phosphate binding site as well as those between the amide oxygen of A93 and the hydroxyl group of Y299 are also conserved in the human and rat PP5 phosphatases. Finally, H27 from TPR1 and K59 from TPR2 form H-bonds with the main chain carbonyl group and the sidechain of Q440 in the PP2A domain, respectively.
The αJ-helix of the C-terminal inhibitory motif is indirectly involved in shielding the active site by stabilizing the closed-state interactions of the PP2A domain with the TPR region (Yang et al., 2005; Haslbeck et al., 2015). In AtPAPP5, the thioether group of M482 forms interactions with the carbonyl oxygen of Y85 as well as van-der-Waals interactions with L92 (Figure 5B). Unlike mammalian PP5, not only non-polar interactions are found in this region of AtPAPP5. One prominent interaction is formed by N484, which interacts via H-bonds with the carbonyl oxygen of Y63 on TPR1 and Y299 of the PP2A domain. The position of this amino acid is further stabilized by interactions with the amide group of L480. Overall, the polar amino acid N484 is found to bridge both the TPR and the PP2A domains.
The positioning of the αJ-helix is also supported by an interaction with the TPR-PP2A linker. Interestingly, in the two molecules of the asymmetric unit the αJ-helix interacts with different amino acids. Heterogeneity of this helix was also observed in the two PP5 structures from Rattus norvegicus (Haslbeck et al., 2015). While the hydrophobic interaction between A127 and the aromatic side chain of F479 is present in both molecules, the polar interactions occur only in molecule A (E123 and R481) and not in molecule B. In the latter, the side chains of R481 and N478 are in H-bonding distance (Figures 5C,D). This interaction pattern might be unique to plant phytochromes since mammalian PP5 lack the involved residues and show no comparable interactions for the C-terminal five amino acids in its structure.
Conformational Changes of the Tetratricopeptide Repeats Region Activate AtPAPP5
Heterogeneous interactions of the C-terminal inhibitory motif may indicate that the C-terminus only partly inhibits phosphatase activity and a dynamic equilibrium is formed between active and auto-inhibited AtPAPP5 forms. This might explain the small, but still measurable activity of the AtPAPP5 wildtype due to reversible breakage and reformation of interactions between the TPR and the PP2A domains. However, to achieve full phosphatase activity in vitro the addition of AA is needed. Size exclusion chromatography (SEC) shows an earlier elution of AtPAPP5 upon addition of AA, which indicates a larger hydrodynamic radius than the closed, non-activated state of AtPAPP5 (Figure 1C). To address this issue, we performed CD measurements in the presence of different AA concentrations (Figures 1A,B). These data reveal that the relative α-helical fraction decreases in the presence of AA thus implying partial unfolding of α-helical regions in AtPAPP5. In order to localize corresponding regions, we applied HDX-MS measurements to AtPAPP5 in the non-actived, closed state as well as in the AA-activated state. Generally, less folded regions are usually characterized by enhanced hydrogen deuterium exchange rates (Figure 1D).
A priori one may predict higher exchange rates for the TPR repeats in the presence of AA due to a disruption of the TPR-PP2A domain interface for opening up the active site of the PP2A domain. Interestingly, the HDX-MS data show only for TPR2 and the beginning of the TPR3 repeat increased exchange rates (Figure 6A). In the AA-activated state H27 of TPR1 experiences surprisingly a reduced H/D exchange rate, whereas its interaction partner within the closed state of AtPAPP5, Q440, shows increased H/D exchange. Y437 located on the same loop as Q440 shows an increased exchange rate as well. Furthermore, R261 experiences increased exchange so that it can be assumed that its interactions with E61 and Y63 are no longer formed in the AA-actived state. K385 shows a slightly reduced and K59 a slightly increased exchange. Interestingly, some of the interactions between the TPR3 repeat and the PP2A domain apparently remain intact, since Y299 and A93 show no significant changes. It is striking, that only D183 located on the α8-helix has an increased exchange rate. Accordingly, the interactions that D183 had formed to S163 of the linker and K188 of the PP2A domain in the closed state of AtPAPP5 seem to be broken by addition of AA (Figure 6A).
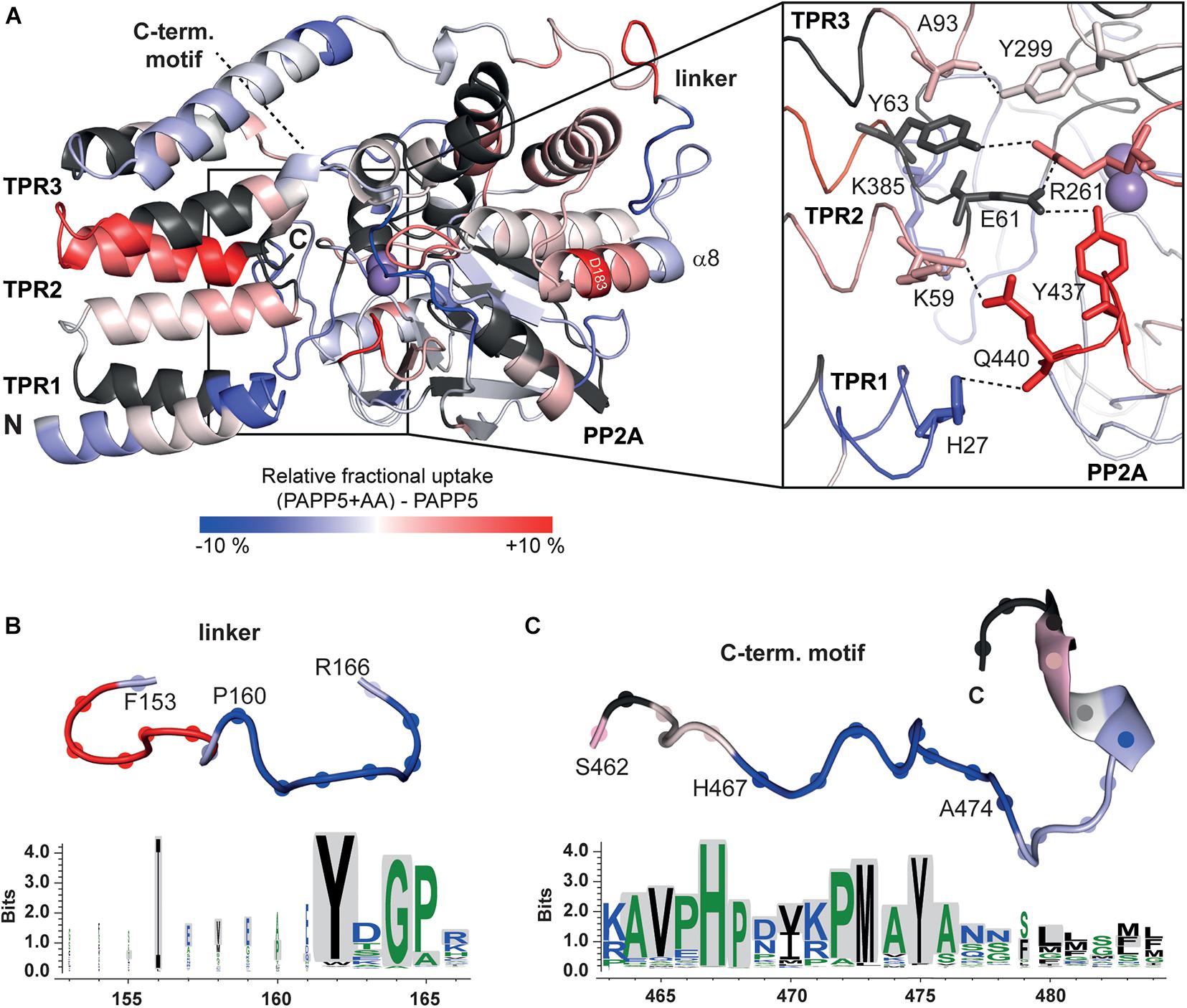
Figure 6. HDX-MS analysis of AtPAPP5 in blue-white-red representation. (A) The difference in deuterium uptake between AtPAPP5 alone and in the presence of arachidonic acid after 15 s. Regions in red show an increased uptake, while blue structural features represent a reduced exchange rate due to interaction with AA. (A) The magnification shows the residues involved in the interaction between TPR and PP2A domains colored according to their uptake. (B,C) The linker and the C-terminal motif show both a decreased deuterium uptake compared to the rest of the structure. The conservation of this regions within the PP5 family was generated using the server Weblogo 3.0 (Schneider and Stephens, 1990; Crooks et al., 2004) (using 60 ortholog sequences with a pairwise sequence identity of < 90%). The height of the letters represents the degree of conservation and gray shading marks the sequence of AtPAPP5.
Two further regions (P160-A165 and H467-A474) stand out due to their significantly reduced uptake rates in the AA-activated state (Figures 6B,C). The first one (P160-A165) is part of the linker between the TPR and PP2A domains. The second region with a reduced exchange rate is part of the C-terminal inhibitory motif (H467-A474). The αJ-helix of the C-terminal motif shows no significant changes in the presence of AA. The last three amino acids of the C-terminus that form both interactions with the TPR and the PP2A domain as well as the corresponding interaction partners are not defined by our HDX-MS analysis (Figure 6C). A comparison of the amino acid sequences of the PP5 phosphatases showed that the linker region contains only three conserved amino acids (Y162, G164, and P165). In contrast, the beginning of the C-terminal inhibitory motif is strictly conserved (Figure 6C).
In order to check for transferability of our AtPAPP5activation mechanism to other members of the PP5 family, sequence identities of the PP5 phosphatases from different organisms were aligned and analyzed for conservation by the Consurf server (Glaser et al., 2003; Landau et al., 2005). The active site and the interaction interface of the PP2A domain with the TPR region are strongly conserved, compared to the rest of the structure (Supplementary Figure 3). Interestingly, two of the three amino acids of the TPR region that interact with the C-terminal inhibitory motif (Y85 and L92) are conserved in all PP5 phosphatases, as well as E61, which binds in the active site. A complete conservation of the helices that unfold upon addition of AA might not be necessary, as only specific bonds are broken. Furthermore, both regions, which showed a decrease in HDX rate in the presence of AA, are conserved within the PP5 family indicating the general applicability of our activation mechanism. However, the nuclear localization signal is also localized in C-terminal inhibitory motif and could explain the high degree of conservation.
Although the HDX-MS analysis failed to identify unambiguously the AA binding of AtPAPP5, we can summarize (1) that AA binding induces a loss of structure only in the TPR2 repeat of the TPR region and (2) that the C-terminal inhibitory motif stays associated with the TPR region in the active state of AtPAPP5. One may tentatively suggest that H27 at the tip of TPR1 may represent an interaction partner of AA due to ionic interaction with the AA’s carboxylate group, as several hydrophobic, surface exposed side chains are located near H27 (F24, Y29, F55).
MD Analysis of a phyB-Phosphopeptide Bound Complex of AtPAPP5
As the rather local effects of AA-binding to PAPP5 are sufficient for activation of AtPAPP5, we wondered to what extent the TPR region has to unblock the active site for allowing substrate binding and catalysis. For that purpose, we performed a series of molecular-dynamics simulations, where we sequentially increased the size of the substrate (analog) bound to the di-manganese site in the active center. In the first step, we added a capped phosphoserine residue to the active site, equilibrated this system as NpT ensemble for 80 ns before extending the bound phosphoserine to the phyB D82-S86 peptide phosphorylated at S84. The orientation of the phyB was taken from the complex of human PP5 with a Cdc37 peptide (Oberoi et al., 2016). After further equilibration for 100 ns the AtPAPP5•peptide complex was elongated to the phyB S80-T89 peptide.
MD trajectories for the closed state of AtPAPP5 show only minor differences for the association of the TPR region and the PP2A domain when comparing to the crystal structure of AtPAPP5 and other PP5-type phosphatases (Figure 7). Only the interactions of Q440 with H27 and K59, which were observed in the crystal structure, are apparently weaker in the MD ensembles (Supplementary Table 4) due to intrusion of bridging water molecules. Docking of the short phosphoserine peptide N-acetyl-SEP-NMe into the active site of AtPAPP5 causes apparently no major structural adaptions of the closed state. The center-of-mass (COM) distances between the TPR region and the PP2A domain increases only slightly from ∼28 Å to 29–30 Å (Supplementary Figure 4). Nevertheless, the H-bonding interactions Y437/E61, Q440/K59 and Y299/Y63 of the substrate-free state are broken in the TPR-PP2A interface. Elongating the phosphopeptide to the S84-phosphorylated phyB peptide D82-S85 severely distorts the TPR-PP2A interface by triggering a rotation of the TPR-region by ∼40° relative to the PP2A domain and the linking helix (Figure 7) and an increase of the COM-COM distance of both domains to 32–34 Å. As a consequence, main chain interactions between G64-S65 from TPR2 and K385-R386, which are a feature of the closed states, are disrupted. Despite the large-scale conformational changes the C-terminal inhibitory motif remains associated during simulation time with the TPR region (Figure 8A). Given the mostly hydrophobic interactions of this motif with TPR3 residues, it is notable that its only salt bridge in the closed state between the C-terminal carboxyl group of N484 and K59, which is apparently only transiently formed during MD simulations of the closed state (Supplementary Table 4) and not defined at all for molecule B of the AtPAPP5 crystals, forms a new salt bridge to the side chain of K120 from the linker helix. Finally, in the complex with the capped phyB decapeptide S80-T89 there is a slight increase of the TPR-region rotation to ∼50° without a change of the COM-COM distance. The MD trajectories of this AtPAPP5 complex tend to form again more homogeneous ensembles in the now open state than the AtPAPP5•phyB/D82-S85 complexes, because the former exert a lesser degree of conformational variation for the TPR-PP2A domain arrangement (Figure 7 and Supplementary Figure 4).
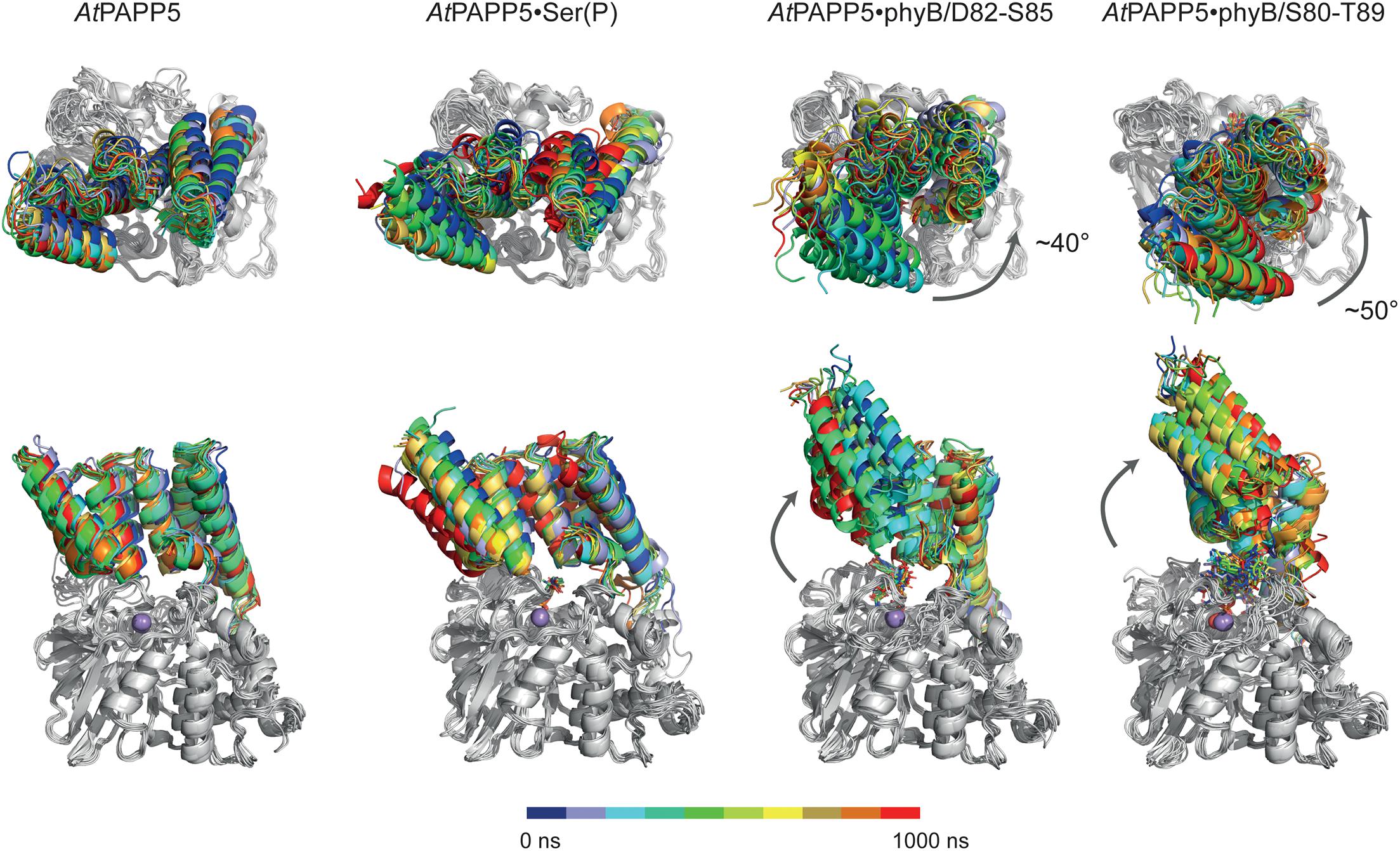
Figure 7. MD simulation of AtPAPP5 and its complexes to phosphorylated phyB peptides. Ten snapshots of a representative 1 μs trajectory are shown with the PP2A domains superposed on each other to highlight displacements of the TPR region. The upper row shows a view mostly along the linker helix between the TPR region and PP2A domain; here a counter-clockwise movement of the TPR region relative to the PP2A domain is apparent upon binding of phyB peptides. The lower row depicts a perpendicular view and shows partial off-swiveling of the TPR region to open up the peptide-binding channel of AtPAPP5.
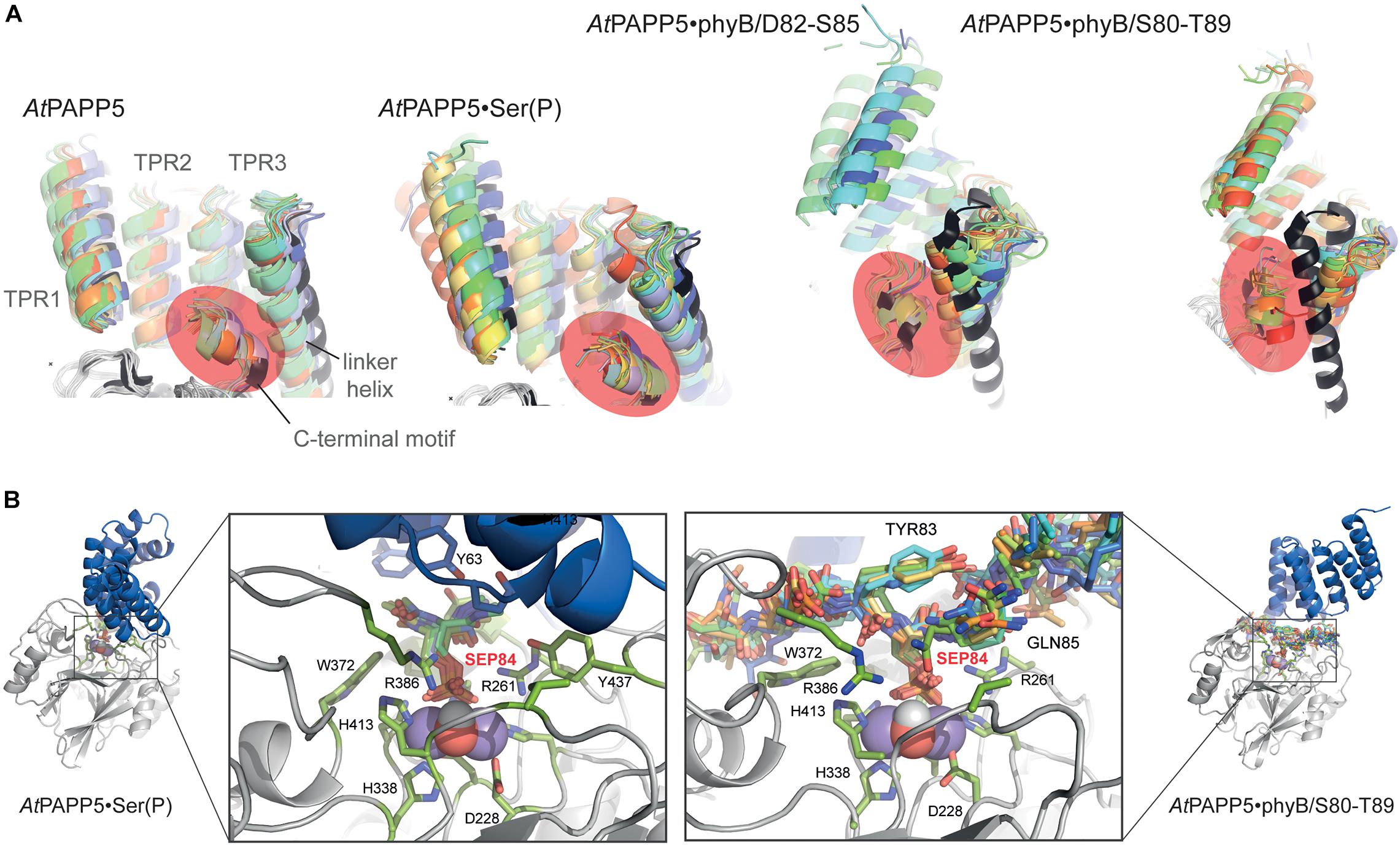
Figure 8. Effect of phosphopeptide binding to the TPR-PP2A domain arrangement in AtPAPP5. (A) The TPR regions of 10 MD snapshots (colored) are shown in comparison to the crystal structure of AtPAPP5 (black). The C-terminal inhibitory motif is highlighted in orange. In the phyB peptide complexes the motif moves closer to the N-terminal portion of the linker helix. (B) Active site shown for 10 MD snapshots of the AtPAPP5•N-acetyl-SEP-NMe (left) and the AtPAPP5•phyB/S80-T89 complexes.
The phosphoserine S84 of the phyB peptides is coordinated to the dimanganese center and forms ionic interactions with the sidechain of R261; the protonated H290 forms a hydrogen bond to the OG atom of the phosphoserine and apparently acts as general acid during catalysis (Figure 8B). Despite being sandwiched between the TPR and PP2A domains, the conformation of the bound phosphopeptides follows the in-out-in-out rule for peptide binding by PP5-type phosphatases as previously suggested (Oberoi et al., 2016), where the phosphoserine adopts the inward orientation toward the active site.
Discussion
Protein phosphorylation and dephosphorylation are commonly used as on-/off-switches in the regulation of cellular activities in almost all organisms. The transfer of a phosphate group to a protein often exerts profound effects on its structure and functional properties (Krebs, 1994). Accordingly, strict control of kinases and phosphatases often depends on regulatory domains or subunits, which undergo structural changes due to substrate binding (Uhrig et al., 2013). Studies with phosphatases of the PP5 family revealed that activation can be achieved by addition of AA or proteolytic digestion of their TPR region and 13 C-terminal amino acids (Kang et al., 2001). Our interaction studies with the AtPhyB variants mimicking the phosphorylated state of the NTE could show that the PSM alone is sufficient for the interaction and dephosphorylation by AtPAPP5 in the presence of AA. Additional interactions between AtPAPP5 and HKR of the C-terminal module may strengthen interactions between AtPhyB and AtPAPP5 but are not essential for the interaction with the phosphorylated state of AtPhyB.
Activity control by fatty acids is not restricted to phosphatases of the PP5 family. One example is the Rho kinase from Homo sapiens, which is blocked by a C-terminal autoinhibitory subunit, where addition of AA activates the kinase (Araki et al., 2001). Likewise, an inhibitory effect is known for the smooth muscle myosin light chain phosphatase (SM-PP1M). A dissociation of the catalytic subunit is triggered by AA, resulting in a reduced dephosphorylation efficiency (Gong et al., 1992; Somlyo and Somlyo, 1994). In plants, AA and other polyunsaturated fatty acids inhibit protein phosphatase MP2C (Baudouin et al., 1999), whereas in animals AA stimulate PP2Cα (Klumpp et al., 1998). Like for 2C-type phosphatases the binding mode and activation mechanism by AA are poorly understand for PP5 phosphatases. Previous data point to a disruption of the interaction between TPR and the active site of the PP2A domain (Skinner et al., 1997). Mutations of the amino acid E61 from TPR2, which interacts with the catalytic site and L480, which stabilizes the C-terminal helix, increase the phosphatase activity. Interestingly, the mutation of the amino acid N477 of the C-terminal inhibitory motif, which is not directly involved in interactions between the PP2A and TPR regions, shows a 50% decreased sensitivity to activation by AA (Kang et al., 2001). This contradicts the hypothesis that AA merely binds to the TPR region and directly effects a destabilization of the α-helical structure of the TPR region by interactions with its hydrophobic amino acids.
AA-driven structural transformations were initially indicated by soaking experiments of AtPAPP5 crystals, which caused rapid dissolution of the crystals. Our HDX-MS experiments show that after AA-addition accessibility changes occur indeed in the TPR region as well as in the linker and the C-terminal inhibitory motif. The end of the TPR2 repeat and the beginning of the TPR3 repeat possess increased exchange rates, indicating that interactions between the second and third repeat get broken and at least partial unfolding of this region takes place. Accordingly, the amino acids of the PP2A domain, which interact with the TPR1 and TPR2 repeats, show increased exchange rates, indicating a dissociation of these TPR-repeats from the active site surface of the PP2A domain as indicated by SEC due to the increased hydrodynamic radius of the AtPAPP5⋅AA complex. In contrast, the TPR3 repeat apparently still interacts with the PP2A domain as its H/D exchange rate is almost unaltered. Furthermore, the beginning of the C-terminal inhibitory motif and the end of the linker connecting the TPR and PP2A domains exert strongly reduced exchange rates upon addition of AA. Since these areas are the only ones in AtPAPP5 with decreased conformational dynamics in the AtPAPP5⋅AA complex, they may directly interact with each other. This notion is corroborated by our extensive MD simulations with docked phyB phosphopeptides. Here, we also observe dissocation of TPR1 from the PP2A domain and an increased exposure of TPR2 to solvent (Figure 7), whereas the C-terminal inhibitory motif still packs to TPR3 and the linker helix despite a swivel motion of 40–50° for the TPR region. Apparently, small-molecule activators of mammalian PP5 can act allosterically in an analogous manner as AA (Haslbeck et al., 2015). These synthetic compounds have been found to bind to the PP2A domain close to TPR1 and cause a slight swiveling motion of the TPR region. Although our HD/X MS-data could not unambiguously reveal the AA binding site of AtPAPP5, we found a significant decrease of accessibility for H27 of TPR1 upon AA addition. We suggest a scenario for PP5-type phosphatases, where allosteric activators like AA first weaken the TPR-PP2A interactions at the peripheral site of the closed state. The loss of TPR-PP2A interactions allows then a directed movement of the TPR domain relative to the PP2A domain. The resulting open state still provides parts of the TPR region, especially TPR3, for interaction with substrate peptides. In this scenario, the specificity for substrate phosphopeptides, such as the N-terminal, phosphorylated regions around S84 and S86 of phyB, would not only been conferred by the PP2A domain (Oberoi et al., 2016), but partly also by TPR3. Finally, during inactivation of the PP5-type phosphatase the C-terminal inhibitory motif may act then as a return spring for the back-positioning of the TPR region to the closed state conformation.
In order to prove the transferability of our activation mechanism for AtPAPP5 to other members of the PP5 family, sequence identities of the PP5 phosphatases from different organisms were aligned and analyzed for conservation by the Consurf server (Glaser et al., 2003; Landau et al., 2005). The active site and the interaction interface of the PP2A domain with the TPR region are strongly conserved, compared to the rest of the structure (Supplementary Figure 3). Interestingly, two of the three amino acids of the TPR region that interact with the C-terminal inhibitory motif (Y85 and L92) are conserved in all PP5 phosphatases, as well as E61, which binds in the active site. A complete conservation of the helices that unfold upon addition of AA might not be necessary, as only specific bonds are broken. Furthermore, both regions, which showed a decrease in HDX rate in the presence of AA, are conserved within the PP5 family indicating the general applicability of our activation mechanism. However, the nuclear localization signal is also localized in C-terminal inhibitory motif and could explain the high degree of conservation.
In plants the role of AA is still controversial. It has so far only been found in the plastids of algae and mosses, where it is produced by chain lengthening of the linoleic acid and subsequent desaturation. Interestingly, the exogenous treatment of plants with AA, a lipid that is usually derived from animals, triggers programmed cell death as well as plant defense mechanisms against insect pathologs (Bostock et al., 1981; Knight et al., 2001; García-Pineda et al., 2004).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.wwpdb.org/, 7OBE.
Author Contributions
SH performed the experiments. SH and L-OE analyzed the data, wrote the manuscript, and designed the research. Both authors reviewed the manuscript.
Funding
This work was supported by grants of the Deutsche Forschungsgemeinschaft (ES152/10, ES152/21) and the Center for Synthetic Microbiology.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Petra Gnau and Ralf Pöschke for help with molecular biology and crystallization, Uwe Linne and Andreas Mielcarek for HDX-MS measurements, the beamline staff of BESSY-II, Berlin, for support, Vitali Kalugin and Elisabeth Ignatz for data collection and Katrin Anders and Vitali Kalugin for support in structure refinement.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.733069/full#supplementary-material
Footnotes
References
Adams, P. D., Afonine, P. V., Bunkoczi, G., Chen, V. B., Davis, I. W., Echols, N., et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. doi: 10.1107/S0907444909052925
Anders, K., and Essen, L.-O. (2015). The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 35, 7–16. doi: 10.1016/j.sbi.2015.07.005
Araki, S., Ito, M., Kureishi, Y., Feng, J. H., Machida, H., Isaka, N., et al. (2001). Arachidonic acid-induced Ca2+ sensitization of smooth muscle contraction through activation of Rho-kinase. Pflugers Archiv Eur. J. Physiol. 441, 596–603. doi: 10.1007/s004240000462
Barajas-Lopez Jde, D., Kremnev, D., Shaikhali, J., Pinas-Fernandez, A., and Strand, A. (2013). PAPP5 is involved in the tetrapyrrole mediated plastid signalling during chloroplast development. PLoS One 8:e60305. doi: 10.1371/journal.pone.0060305
Baudouin, E., Meskiene, I., and Hirt, H. (1999). Short communication: unsaturated fatty acids inhibit MP2C, a protein phosphatase 2C involved in the wound-induced MAP kinase pathway regulation. Plant J. 20, 343–348. doi: 10.1046/j.1365-313X.1999.00608.x
Bheri, M., Mahiwal, S., Sanyal, S. K., and Pandey, G. K. (2020). Plant protein phosphatases: What do we know about their mechanism of action? FEBS J. 288, 756–785. doi: 10.1111/febs.15454
Bostock, R. M., Kuc, J. A., and Laine, R. A. (1981). Eicosapentaenoic and Arachidonic Acids from Phytophthora infestans Elicit Fungitoxic Sesquiterpenes in the Potato. Science 212, 67–69. doi: 10.1126/science.212.4490.67
Casal, J. J., Davis, S. J., Kirchenbauer, D., Viczian, A., Yanovsky, M. J., Clough, R. C., et al. (2002). The serine-rich N-terminal domain of oat phytochrome a helps regulate light responses and subnuclear localization of the photoreceptor. Plant Physiol. 129, 1127–1137. doi: 10.1104/pp.010977
Chen, M. X., and Cohen, P. T. (1997). Activation of protein phosphatase 5 by limited proteolysis or the binding of polyunsaturated fatty acids to the TPR domain. FEBS Lett. 400, 136–140. doi: 10.1016/S0014-5793(96)01427-5
Chen, M. X., Mcpartlin, A. E., Brown, L., Chen, Y. H., Barker, H. M., and Cohen, P. T. (1994). A novel human protein serine/threonine phosphatase, which possesses four tetratricopeptide repeat motifs and localizes to the nucleus. EMBO J. 13, 4278–4290. doi: 10.1002/j.1460-2075.1994.tb06748.x
Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15, 637–642. doi: 10.1016/j.cub.2005.02.028
Collaborative Computational Project (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763. doi: 10.1107/S0907444994003112
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
De La Fuente Van Bentem, S., Vossen, J. H., Vermeer, J. E. M., De Vroomen, M. J., Gadella, T. W. J., et al. (2003). The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol. 133, 702–712. doi: 10.1104/pp.103.026617
Emsley, P., and Cowtan, K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. doi: 10.1107/S0907444904019158
Franklin, K. A., and Quail, P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. doi: 10.1093/jxb/erp304
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., et al. (2016). ”Gaussian 16 Rev. C.01”. Wallingford, CT: Gaussian.
García-Pineda, E., Castro-Mercado, E., and Lozoya-Gloria, E. (2004). Gene expression and enzyme activity of pepper (Capsicum annuum L.) ascorbate oxidase during elicitor and wounding stress. Plant Sci. 166, 237–243. doi: 10.1016/j.plantsci.2003.09.013
Genoud, T., Santa Cruz, M. T., Kulisic, T., Sparla, F., Fankhauser, C., and Metraux, J. P. (2008). The protein phosphatase 7 regulates phytochrome signaling in Arabidopsis. PLoS One 3:e2699. doi: 10.1371/journal.pone.0002699
Glaser, F., Pupko, T., Paz, I., Bell, R. E., Bechor-Shental, D., Martz, E., et al. (2003). ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 19, 163–164. doi: 10.1093/bioinformatics/19.1.163
Gong, M. C., Fuglsang, A., Alessi, D., Kobayashi, S., Cohen, P., Somlyo, A. V., et al. (1992). Arachidonic acid inhibits myosin light chain phosphatase and sensitizes smooth muscle to calcium. J. Biol. Chem. 267, 21492–21498. doi: 10.1016/S0021-9258(19)36636-0
Haslbeck, V., Drazic, A., Eckl, J. M., Alte, F., Helmuth, M., Popowicz, G., et al. (2015). Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism. Biosci. Rep. 35, 1–11. doi: 10.1042/BSR20150042
Hisada, A., Hanzawa, H., Weller, J. L., Nagatani, A., Reid, J. B., and Furuya, M. (2000). Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12, 1063–1078. doi: 10.1105/tpc.12.7.1063
Jung, J. H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M. J., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889. doi: 10.1126/science.aaf6005
Kabsch, W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. D66, 125–132. doi: 10.1107/S0907444909047337
Kang, H., Sayner, S. L., Gross, K. L., Russell, L. C., and Chinkers, M. (2001). Identification of amino acids in the tetratricopeptide repeat and C-terminal domains of protein phosphatase 5 involved in autoinhibition and lipid activation. Biochemistry 40, 10485–10490. doi: 10.1021/bi010999i
Kim, D. H., Kang, J. G., Yang, S. S., Chung, K. S., Song, P. S., and Park, C. M. (2002). A phytochrome-associated protein phosphatase 2A modulates light signals in flowering time control in Arabidopsis. Plant Cell 14, 3043–3056. doi: 10.1105/tpc.005306
Kim, J. I., Shen, Y., Han, Y. J., Park, J. E., Kirchenbauer, D., Soh, M. S., et al. (2004). Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16, 2629–2640. doi: 10.1105/tpc.104.023879
Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., et al. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. doi: 10.1105/tpc.11.8.1445
Klumpp, S., Selke, D., and Hermesmeier, J. (1998). Protein phosphatase type 2C active at physiological Mg2+: stimulation by unsaturated fatty acids. FEBS Lett. 437, 229–232. doi: 10.1016/S0014-5793(98)01237-X
Knight, V. I., Wang, H., Lincoln, J. E., Lulai, E. C., Gilchrist, D. G., and Bostock, R. M. (2001). Hydroperoxides of fatty acids induce programmed cell death in tomato protoplasts. Physiol. Mol. Plant Pathol. 59, 277–286. doi: 10.1006/pmpp.2001.0366
Krebs, E. G. (1994). The growth of research on protein phosphorylation. Trends Biochem. Sci. 19:439. doi: 10.1016/0968-0004(94)90125-2
Landau, M., Mayrose, I., Rosenberg, Y., Glaser, F., Martz, E., Pupko, T., et al. (2005). ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302. doi: 10.1093/nar/gki370
Lee, T. S., Cerutti, D. S., Mermelstein, D., Lin, C., Legrand, S., Giese, T. J., et al. (2018). GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J. Chem. Inf. Model 58, 2043–2050. doi: 10.1021/acs.jcim.8b00462
Legris, M., Klose, C., Burgie, E. S., Rojas, C. C., Neme, M., Hiltbrunner, A., et al. (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900. doi: 10.1126/science.aaf5656
Li, P., and Merz, K. M. Jr. (2016). MCPB.py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model 56, 599–604. doi: 10.1021/acs.jcim.5b00674
Mathews, S. (2010). Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22, 4–16. doi: 10.1105/tpc.109.072280
Medzihradszky, M., Bindics, J., Adam, E., Viczian, A., Klement, E., Lorrain, S., et al. (2013). Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25, 535–544. doi: 10.1105/tpc.112.106898
Nguyen, H., Case, D. A., and Rose, A. S. (2018). NGLview-interactive molecular graphics for Jupyter notebooks. Bioinformatics 34, 1241–1242. doi: 10.1093/bioinformatics/btx789
Nito, K., Wong, C. C. L., Yates, J. R., and Chory, J. (2013). Tyrosine Phosphorylation Regulates the Activity of Phytochrome Photoreceptors. Cell Rep. 3, 1970–1979. doi: 10.1016/j.celrep.2013.05.006
Oberoi, J., Dunn, D. M., Woodford, M. R., Mariotti, L., Schulman, J., Bourboulia, D., et al. (2016). Structural and functional basis of protein phosphatase 5 substrate specificity. Proc. Natl. Acad. Sci. U S A. 113, 9009–9014. doi: 10.1073/pnas.1603059113
Park, E., Kim, J., Lee, Y., Shin, J., Oh, E., Chung, W. I., et al. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968–975. doi: 10.1093/pcp/pch125
Parks, B. M., and Spalding, E. P. (1999). Sequential and coordinated action of phytochromes A and B during Arabidopsis stem growth revealed by kinetic analysis. Proc. Natl. Acad. Sci. U S A. 96, 14142–14146. doi: 10.1073/pnas.96.24.14142
Phee, B. K., Kim, J. I., Shin, D. H., Yoo, J., Park, K. J., Han, Y. J., et al. (2008). A novel protein phosphatase indirectly regulates phytochrome-interacting factor 3 via phytochrome. Biochem. J. 415, 247–255. doi: 10.1042/BJ20071555
Quail, P. H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3, 85–93. doi: 10.1038/nrm728
Rockwell, N. C., Su, Y. S., and Lagarias, J. C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858. doi: 10.1146/annurev.arplant.56.032604.144208
Roe, D. R., and Cheatham, T. E. III (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 9, 3084–3095. doi: 10.1021/ct400341p
Ryu, J. S., Kim, J. I., Kunkel, T., Kim, B. C., Cho, D. S., Hong, S. H., et al. (2005). Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120, 395–406. doi: 10.1016/j.cell.2004.12.019
Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10, 859–868. doi: 10.1046/j.1365-313X.1996.10050859.x
Savchenko, T., Walley, J. W., Chehab, E. W., Xiao, Y., Kaspi, R., Pye, M. F., et al. (2010). Arachidonic acid: an evolutionarily conserved signaling molecule modulates plant stress signaling networks. Plant Cell 22, 3193–3205. doi: 10.1105/tpc.110.073858
Schneider, T. D., and Stephens, R. M. (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100. doi: 10.1093/nar/18.20.6097
Shin, A. Y., Han, Y. J., Baek, A., Ahn, T., Kim, S. Y., Nguyen, T. S., et al. (2016). Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat. Commun. 7:11545. doi: 10.1038/ncomms11545
Sinclair, C., Borchers, C., Parker, C., Tomer, K., Charbonneau, H., and Rossie, S. (1999). The tetratricopeptide repeat domain and a C-terminal region control the activity of Ser/Thr protein phosphatase 5. J. Biol. Chem. 274, 23666–23672. doi: 10.1074/jbc.274.33.23666
Skinner, J., Sinclair, C., Romeo, C., Armstrong, D., Charbonneau, H., and Rossie, S. (1997). Purification of a fatty acid-stimulated protein-serine/threonine phosphatase from bovine brain and its identification as a homolog of protein phosphatase 5. J. Biol. Chem. 272, 22464–22471. doi: 10.1074/jbc.272.36.22464
Somlyo, A. P., and Somlyo, A. V. (1994). Signal transduction and regulation in smooth muscle. Nature 372, 231–236. doi: 10.1038/372231a0
Struk, S., De Cuyper, C., Jacobs, A., Braem, L., Walton, A., De Keyser, A., et al. (2021). Unraveling the MAX2 Protein Network in Arabidopsis thaliana: Identification of the Protein Phosphatase PAPP5 as a Novel MAX2 Interactor. Mol. Cell Proteomics 20:100040. doi: 10.1074/mcp.RA119.001766
Swingle, M. R., Honkanen, R. E., and Ciszak, E. M. (2004). Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J. Biol. Chem. 279, 33992–33999. doi: 10.1074/jbc.M402855200
Tian, C., Kasavajhala, K., Belfon, K. A. A., Raguette, L., Huang, H., Migues, A. N., et al. (2020). ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 16, 528–552. doi: 10.1021/acs.jctc.9b00591
Uhrig, R. G., Labandera, A. M., and Moorhead, G. B. (2013). Arabidopsis PPP family of serine/threonine protein phosphatases: many targets but few engines. Trends Plant Sci. 18, 505–513. doi: 10.1016/j.tplants.2013.05.004
Von Horsten, S., Straß, S., Hellwig, N., Gruth, V., Klasen, R., Mielcarek, A., et al. (2016). Mapping light-driven conformational changes within the photosensory module of plant phytochrome B. Sci. Rep. 6:34366. doi: 10.1038/srep34366
Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S. A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. doi: 10.1038/srep34366
Yang, J., Roe, S. M., Cliff, M. J., Williams, M. A., Ladbury, J. E., Cohen, P. T., et al. (2005). Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 24, 1–10. doi: 10.1038/sj.emboj.7600496
Zeke, T., Morrice, N., Vazquez-Martin, C., and Cohen, P. T. W. (2005). Human protein phosphatase 5 dissociates from heat-shock proteins and is proteolytically activated in response to arachidonic acid and the microtubule-depolymerizing drug nocodazole. Biochem. J. 385, 45–56. doi: 10.1042/BJ20040690
Keywords: phytochrome, protein phosphatase, TPR domain, HDX-mass spectrometry, plantal photoreception
Citation: von Horsten S and Essen L-O (2021) Conformational Change of Tetratricopeptide Repeats Region Triggers Activation of Phytochrome-Associated Protein Phosphatase 5. Front. Plant Sci. 12:733069. doi: 10.3389/fpls.2021.733069
Received: 29 June 2021; Accepted: 21 September 2021;
Published: 14 October 2021.
Edited by:
András Viczián, Biological Research Centre, HungaryReviewed by:
Jeong-Il Kim, Chonnam National University, South KoreaWilliam Bryan Terzaghi, Wilkes University, United States
Copyright © 2021 von Horsten and Essen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lars-Oliver Essen, essen@chemie.uni-marburg.de
 Silke von Horsten1
Silke von Horsten1 Lars-Oliver Essen
Lars-Oliver Essen