Corrigendum: A Tree Peony Trihelix Transcription Factor PrASIL1 Represses Seed Oil Accumulation
- 1College of Landscape Architecture and Arts, Northwest A&F University, Yangling, China
- 2Oil Peony Engineering Technology Research Center of National Forestry Administration, Yangling, China
- 3Department of Biological Sciences, East Tennessee State University, Johnson City, TN, United States
- 4Academy of Medical Sciences, Zhengzhou University, Zhengzhou, China
In many higher plants, seed oil accumulation is governed by complex multilevel regulatory networks including transcriptional regulation, which primarily affects fatty acid biosynthesis. Tree peony (Paeonia rockii), a perennial deciduous shrub endemic to China is notable for its seed oil that is abundant in unsaturated fatty acids. We discovered that a tree peony trihelix transcription factor, PrASIL1, localized in the nucleus, is expressed predominantly in developing seeds during maturation. Ectopic overexpression of PrASIL1 in Nicotiana benthamiana leaf tissue and Arabidopsis thaliana seeds significantly reduced total fatty acids and altered the fatty acid composition. These changes were in turn associated with the decreased expression of multitudinous genes involved in plastidial fatty acid synthesis and oil accumulation. Thus, we inferred that PrASIL1 is a critical transcription factor that represses oil accumulation by down-regulating numerous key genes during seed oil biosynthesis. In contrary, up-regulation of oil biosynthesis genes and a significant increase in total lipids and several major fatty acids were observed in PrASIL1-silenced tree peony leaves. Together, these results provide insights into the role of trihelix transcription factor PrASIL1 in controlling seed oil accumulation. PrASIL1 can be targeted potentially for oil enhancement in tree peony and other crops through gene manipulation.
Introduction
In many higher plants, the seed storage reserves include oils as triacylglycerols (TAGs), carbohydrates, and storage proteins. Seed oils not only provide an essential energy source to support post-germinative growth and subsequent seedling development but also serve as the main source of nourishment for humans and farm animal (Li et al., 2006; Graham, 2008). Currently, oils are utilized by a broad variety of industries as well, such as raw materials for the manufacture of pharmaceuticals and biofuels (Durrett et al., 2008; Egert et al., 2014). As such, knowing the regulatory function of critical genes in seed oil accumulation is of significant basic and strategic benefits.
The accumulation of TAG is a complex biochemical process involving two subcellular organelles (Bates et al., 2013). The synthesis of fatty acids (FA) is initiated in the plastid in a multistep process with the involvement of many key enzymes. Plastidial pyruvate dehydrogenase complex (PDHC) first catalyzes pyruvic acid transformation to acetyl-CoA, and then 3-ketoacyl-ACP synthases (KAS catalyzes the condensation reactions (Johnston et al., 1997; Pidkowich et al., 2007). Subsequently, FAs are transferred to the endoplasmic reticulum as acyl-coenzyme A (FA-CoA), and some undergo desaturation catalyzed by oleate desaturase (FAD2) and linoleate desaturase (FAD3) (Browse et al., 1993; Okuley et al., 1994). Finally, FA-CoAs are esterified with glycerol-3-phosphate (G3P) to complete the assembly of TAG. This process is accomplished by successive catalyzation by glycerol 3-phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAAT), phosphatidic acid phosphatase (PAP), and diacylglycerol acyltransferase (DGAT). Additionally, phospholipid: diacylglycerol acyltransferase (PDAT) can also catalyze an acyl group from the sn-2 site of phosphatidylcholine (PC) to DAG to synthesize TAG (Baud and Lepiniec, 2010).
Transcriptional regulation is an important means to control gene expression for seed oil accumulation. Some of the transcription factors (TFs), referred to as master regulators, besides regulating the expression profile of biosynthetic genes, also the action of other TFs (Li et al., 2017). The master regulators such as WRINKLED1 (WRI1), FUSCA3 (FUS3), LEAFY COTYLEDON1 (LEC1), LEC2, and ABSCISIC ACID INSENSITIVE3 (ABI3) govern the seed maturity as well as oil synthesis process (Weselake et al., 2009; Baud and Lepiniec, 2010; Troncoso Ponce et al., 2011). Primarily, WRI1 alters the oil content by upregulating glycolytic and plastidial FA biosynthesis genes (Cernac and Benning, 2004; Baud et al., 2009; Kong and Ma, 2018; Kong et al., 2020b). Deletion of WRI1 led to an 80% decrease in Arabidopsis seed lipid level (Focks and Benning, 1998), and conversely, overexpression of WRI1 significantly increased the oil level (Maeo et al., 2009; Sanjaya et al., 2011; Adhikari et al., 2016). ABI3, FUS3, and LEC2 (AFL) as members of the B3 domain family, which have a B3 DNA-binding domain (Luerssen et al., 1998; Stone et al., 2001; Santos-Mendoza et al., 2008). These master regulators bind to the RY elements in the promoter region of the target genes involved in FA biosynthesis, and TAG storage protein synthesis to promote lipid yield (Reinders et al., 2002; Baud et al., 2016; Zhang et al., 2016). Activation of LEC1 also upregulates a suite of genes contributed to glycolysis, and FA biosynthesis, elongation and desaturation (Mu et al., 2008). Both LEC1 and LEC2 are positive modulators of WRI1, FUS3, and ABI3 upstream (Kagaya et al., 2005; Weselake et al., 2009). Furthermore, there is a positive feedback loop among ABI3 and FUS3 that ensures continuous expression of themselves and each other (To et al., 2006). Moreover, some positive regulators of FA synthesis and oil accumulation, including MYB92, MYB96, and tandem CCCH zinc finger protein, GmZF351 and GmZF392 were uncovered (Lee et al., 2018; To et al., 2020; Lu et al., 2021). In addition to the positive regulators, there are also several TFs implicated in the inhibition of seed lipid synthesis. A group of B3 domain family TFs including VIVIPAROUS1/ABI3-LIKE1 (VAL1), VAL2, and VAL3 repress oil accumulation by downregulating the AFL genes and their target genes of the network (Tsukagoshi et al., 2007). The CHD3 chromatin remodeling factor PICKLE (PKL) also can downregulate the members of the AFL network, thereby contributing to the shutdown of the seed maturation program (Ogas et al., 1999; Zhang and Ogas, 2009). The Arabidopsis 6b interacting protein 1-like 1 (ASIL1) inhibits the transcription of embryonic maturation genes in Arabidopsis seedlings and acts downstream of the miRNA to restrict seed maturation (Gao et al., 2009; Willmann et al., 2011). MYB89 and MYB76 were also investigated as two inhibitors of seed oil accumulation. Recently, a novel FA synthesis repressor, TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4, was investigated to inhibit the expression of fatty acid biosynthetic genes by interacting with WRI1. These finding is of great importance for advancing the study of transcriptional regulatory networks of seed oil accumulation (Duan et al., 2017; Li et al., 2017; Kong et al., 2020a).
Tree peony (Paeonia section Moutan DC.), a perennial deciduous shrub endemic to China, has been cultivated for ornamental and medicinal usage for over 2000 years (Zhou et al., 2014). Recently, tree peony seed oil gained prominence as a rich resource for unsaturated fatty acids (UFAs > 90%) such as linoleic acid (LA, ∼25%) and α-linolenic acid (ALA, ∼45%), which are essential to the human body (Li et al., 2015; Zhang et al., 2018). Thus, tree peony became an emerging woody oil crop in China and its seed oil has been authorized for human consumption since 2011 (Zhao et al., 2020). Consequently, tree peony cultivation has been promoted in China and attracted extensive attention as a novel oil crop.
Recent studies unveiled a host of genes engaged in seed oil biosynthesis and accumulation in tree peony (Yin et al., 2018; Zhang et al., 2019; Zhao et al., 2020). However, the role of TFs regulating oil accumulation in tree peony seeds is poorly understood. Previous transcriptome data revealed that a trihelix TF, PrASIL1 is highly expressed in seeds during early seed development and maturation in tree peony. Although implicated in the temporal regulation of seed filling and maturation in Arabidopsis, its role in seed oil biosynthesis is not well-studied. In this study, we show that PrASIL1 acts as a negative regulator and its reduced expression is associated with upregulation of oil biosynthesis genes and oil accumulation in seeds until maturation in tree peony.
Materials and Methonds
Plant Materials and Growth Conditions
Paeonia rockii was grown at the wild tree peony germplasm repository of Northwest Agronomy and Forestry University, Shaanxi Province, China (34°16′ N, 108°4′ E). The seeds at 20, 40, 60, 80, and 100 days after pollination (DAP) and other organs, such as roots, stems, leaves, calyxes, petals, stamens, and pistils of P. rockii were collected into liquid nitrogen and then stored at −80°C. Additionally, 2-year-old seedlings at 4 weeks post-germination were selected for virus-induced gene silencing (VIGS). Both plants, Nicotiana benthamiana for transient expression studies and Arabidopsis thaliana (Columbia-0; wild type) for ectopic overexpression experiment were grown in the same climate chamber with 16h light/8h dark and 65% relative humidity at 22°C.
Gene Cloning and Plasmid Construction
RNA was extracted from seeds or other organs using the RNA Prep Pure Plant kit (TIANGEN) and reverse transcribed using PrimeScript™ RT reagent Kit (TaKaRa). Specific forward and reverse primers (PrASIL1-F/PrASIL1-R) were used to amplify coding region of PrASIL1. The PCR products were purified by DNA Gel Extraction Kit (Sangon) and cloned into pMD19-T vector. The construct 35S:PrASIL1-GFP for subcellular localization assay was generated by inserting the CDS of PrASIL1 without the stop codon into pCAMBIA2300-GFP vector. The plasmids 2 × 35S:PrASIL1 used for transient expression assay in N. benthamiana and 35S:PrASIL1 used for stable overexpression experiment in Arabidopsis thaliana were constructed by inserting the CDS of PrASIL1 into pB110 and pCAMBIA2300, respectively. To obtain the TRV2-PrASIL1 construct for VIGS assay, the silencing fragment was amplified and connected to TRV2. The primers used for gene cloning and plasmid construction in this study which were designed by Oligo 6.0 software are listed in Supplementary Table 1.
Phylogenetic Analysis and Conserved Motif Analysis
Previously, thirty trihelix genes of A. thaliana were reviewed in detail (Kaplan-Levy et al., 2012). Of these, protein sequences of nineteen representative trihelix genes of A. thaliana and PrASIL1 were selected for multiple sequence alignment using Clustalx2.11.1 The sequences of Arabidopsis trihelix proteins were obtained from the TAIR database.2 A phylogenetic tree was generated in MEGA7.0 using the neighbor-joining (NJ) method with Poisson correction and 1000 replicates for bootstrap analysis. Furthermore, the MEME tool3 (Bailey et al., 2015) was used to identify and compare conserved motifs among PrASIL1 P. rockii and trihelix proteins of A. thaliana. The analysis parameters were set to the maximum number of motifs as 10 and the motif width of 6–50 aa.
Subcellular Localization
35S:PrASIL1-GFP, as described previously was used for localization studies. 35S:NbWRKY8-mCherry was used as a positive nucleus marker (Ishihama et al., 2011). They were transformed into onion epidermal cells by particle bombardment, in which gold powder embedding was performed according to the BIO-RAD Biolistic PDS-1000/He system (BIO-RADCA, United States) (Huang and Liu, 2006). Onion epidermis (approximately 1.5 cm × 1.5 cm) was placed on MS solid medium and incubated at 22°C for 24 hours. Subsequently, onion epidermal cells were bombarded using a particle bombardment with a rupture disk pressure of 1.1 kPa and distanced at 5cm. After incubation for at least 15 h at room temperature in the dark, the onion epidermal cells were then observed under a confocal laser scanning microscope (UltraVIEW VoX, PerkinElmerafter). GFP was excited by a 488 nm laser, the emission was obtained in 560–620 nm. The mCherry was excited by a 552 nm laser and acquires emission at 600–650 nm. The dimensions of the visual field were 237.42 × 237.42 μm2.
Transient Overexpression in Nicotiana benthamiana Leaves
The Agrobacterium tumefaciens strain LBA4404 was transformed with pB110 binary vector carrying 35S:PrASIL1 using the freeze-thaw method (Behera et al., 2021), and positive colonies were selected on solidYEP medium with kanamycin (50 μg/mL) and rifampicin (50 μg/mL). The positive colonies were then subcultured in YEP liquid medium with the same antibiotics and culture d at 28°C with constant shaking overnight. The bacterial cells were then harvested by centrifugation at 400xg for 10 min and suspended in the infiltration buffer (300 μM acetosyringone, 10 mM MES, 10 mM MgCl2, pH 5.7) to a final OD600 of 0.6. The suspension was maintained in the dark for 3 h at 28°C before infiltration. To enhance the PrASIL1 expression in the leaf tissue of N. benthamiana, suspension of Agrobacterium with P19 silencing inhibitor and suspension of Agrobacterium containing 35S:PrASIL1 were mixed in a ratio of 1:1 and then infiltrated into 4-week-old tobacco leaves using a needleless syringe. Untreated leaves and leaves injected with Agrobacterium containing P19 served as the control group.
Visualization of Lipid Droplets
To visualize lipid droplets (LDs) in N. benthamiana leaf tissues, LDs were stained with 2 μg/mL Nile Red (YUANYE) in 0.01 mM phosphate buffer saline (pH 7.2). Confocal images were obtained utilizing a laser scanning confocal microscope (SP8; Leica). Nile Red was excited by a 488 nm laser, the emission was obtained in 560–620 nm. The dimensions of each visual field were 232.50 × 232.50 μm2. Number of LDs counted per image area using ImageJ software. Three biological replicates were performed to visualize LDs in N. benthamiana leaf tissues.
Stable Transformation of Arabidopsis thaliana
The P. rockii 35S:PrASIL1 overexpression plasmid was constructed by pCAMBIA2300 and transformed into A. tumefaciens strain GV3101. Subsequently, GV3101 expressing 35S:PrASIL1 was transformed into wild-type Arabidopsis plants using the floral dip method (Clough and Bent, 1998). The T0 seeds were germinated on 1/2 MS media with kanamycin (500 μg/mL) to select transformed lines. The positive lines were confirmed by amplifying the full length of PrASIL1 using reverse transcription-PCR (RT-PCR) and their zygosity was determined if the independent lines of T1 generation showed 3:1 segregation on selection medium. These transgenic lines were grown up to T3 generation to obtain homozygous lines.
Mature Arabidopsis seeds were collected from the main inflorescence, especially from siliques that grow at the basal position. The seeds from several lines were randomly chosen and photographed utilizing a Nikon Eclipse 50i upright microscope.
Virus-Induced Gene Silencing of PrASIL1
The TRV vectors TRV1 and TRV2 have been described previously (Liu et al., 2002). The TRV2:GFP and TRV2:PrASIL1 (containing a 290 bp fragment of PrASIL1) constructs were transformed into GV3101 for VIGS assay. The transformed GV3101 were grown in YEP medium with appropriate antibiotics at 28°C with constant shaking overnight. The bacterial cells were harvested by centrifugation at 4,000 × g and resuspended in infiltration buffer as described previously (Xie et al., 2019), and kept in darkness for 4–6 h at 28°C. The A. tumefaciens infiltration buffer containing TRV1 and TRV2 vectors were mixed in a ratio of 1:1 to a final OD600 of 0.8 before infiltration into leaves of P. rockii seedlings. The leaves of WT and infiltrated with TRV2:GFP were used as controls. Six days post infiltration (DAI), the leaves were used for GFP expression analysis. At 14 DAI, the leaf samples from ten individual plants were collected for the determination of oil content.
Fatty Acids Quantification
To quantify the FA content, 8 mg of Arabidopsis seeds or 50 mg of dried leaves of N. benthamiana and tree peony were used for each biological replicate. The extraction and analysis of FAs in leaf tissues (Li et al., 2012; Ji et al., 2018), and seeds were carried out as previously described (Chen et al., 2012). Briefly, total FAs were converted to FA methyl esters in a methanol solution containing 1 M HCl for 2 h at 80°C. FAs from leaves or seeds were subsequently measured utilizing a gas chromatograph (8890; Agilent). A capillary column (HP-INNOWax; 60 m × 0.25 mm I.D., 0.25 μm) with nitrogen as carrier gas (flow rate of 1.0 mL min–1) was used. The initiating temperature was 170°C and kept for 5 min, then raised to 210°C at a rate of 2°C min–1. The peaks of each FA species were identified corresponding to the FA methyl ester analytical standard (catalog number: CDAA-252795; ANPEL). Concentrations of FA species were normalized against the internal control heptadecanoic acid (ANPEL). Experiments were performed with three biological replicates.
Expression Analysis
The expression of the transformed cDNA in transgenic plants was confirmed at the transcription level using reverse transcription-PCR (RT-PCR). Quantitative RT-PCR (qRT-PCR) was carried out for three biological replicates using SYBR® Premix Ex Taq™ kit (TaKaRa). Three independent biological replicas are used for expression analysis. The 18S-26S internal transcribed spacer (18S-26S ITS) gene of P. rockii, NbL23 of N. benthamiana, and Atactin7 of Arabidopsis were used as an internal control to normalize the gene expression. The 2–△△CT values were adopted to represent the relative expression levels (Livak and Schmittgen, 2001). The primers used for RT-PCR and qRT-PCR are listed in Supplementary Table 2.
Results
PrASIL1 Expression in Tree Peony Seeds Is Reciprocal to Oil Accumulation
Previously, we performed RNA-seq analysis of tree peony seeds at three developmental stages and found that the transcript levels of genes related to plastid fatty acid synthesis, desaturation, and triacylglycerol assembly were high (Zhang et al., 2019). Subsequently, we also identified the function of PrFAD2 and PrFAD3 genes in ALA synthesis (Zhang et al., 2019).
Transcriptomics also revealed that a trihelix TF PrASIL1 of tree peony was highly expressed in the early and late stages of seed development suggesting its association with seed-related traits (Figure 1A). Transcripts for additional paralogs of PrASIL1, named PrASIL1-2, PrASIL1-3, and PrASIL1-4 with roughly similar expression profiles as PrASIL1 were also identified (Supplementary Figure 1). Although, the expression levels for all PrASIL1 paralogs were relatively high at 20 DAP, they decreased progressively with seed development (60 DAP). Among the paralogs, more than 50% of the transcripts expressed were contributed by PrASIL1 (Figure 1B), suggesting a limited role for the remaining in seed development and oil accumulation. Therefore, we primarily selected PrASIL1 as the target to further investigate its function in tree peony seed oil accumulation.
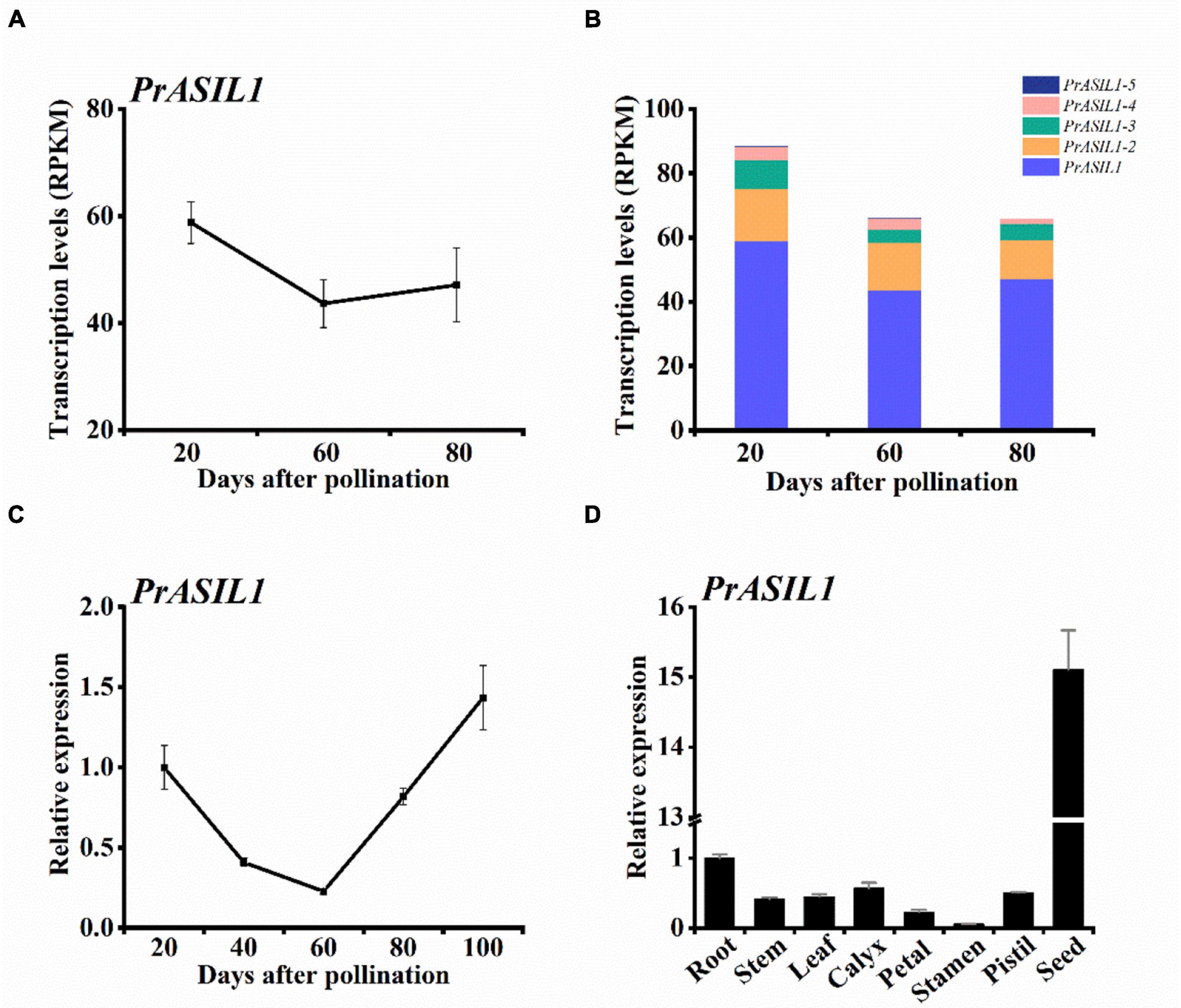
Figure 1. PrASIL1 is highly expressed in developing seeds of P. rockii. (A) PrASIL1 transcript levels in developing seeds. (B) Comparison of transcript levels of all PrASIL1 paralogs in developing seeds. (C) Expression pattern of PrASIL1 in developing seeds of P. rockii and (D) PrASIL1 expression in various tissues of P. rockii. Values are mean ± SD (n = 3).
We further examined the expression pattern of PrASIL1 in various developmental stages of tree peony seeds by utilizing qRT-PCR. The expression of PrASIL1 at the 20 DAP and 100 DAP was higher that levels at 60 DAP (Figure 1C). This dynamic expression pattern of PrASIL1 is reciprocally associated with the rate of fatty acid synthesis, which is at its highest during early to mid-developmental phase of the seed and plateaus by 100 DAP (Zhang et al., 2018). The transition of early to mid-maturation phase is marked by an increase in the storage lipid synthesis and thus a low expression of PrASIL1 is desired. In contrast, the transition from mid to late seed maturation phase is accompanied with a metabolic switch in the seed tissue to increase the energy molecules such as sucrose biosynthesis to prepare the seed for germination. Thus, it is likely that PrASIL1 plays a part in the early and mature stages of seed development that is likely associated with initiation and termination of seed oil synthesis (Willmann et al., 2011).
We also examined the expression of PrASIL1 in different organs including seeds at 100 DAP. Interestingly, a low-level expression of PrASIL1 was noted in all the organs examined and relatively the expression was most abundant in matured seeds (Figure 1D). We predict that since the organs other than seed do not accumulate oil, PrASIL is not expected to play any regulatory role and thus its expression is likely maintained at a low level. It is also possible that other trans-regulatory factors might be suppressing the expression of ASIL1, including the positive regulators of lipid biosynthesis, because of the low oil content in these organs. In contrast, PrASIL1 expression is dynamic in seeds as well as varying with development and oil biosynthesis.
Molecular Characterization of PrASIL1
The open reading frame of PrASIL1 is 858bp and encodes for 285aa protein with one trihelix conserved domain and belongs to the trihelix family (Supplementary Figure 2 and Figure 2A). The trihelix family is divided into five sub-families based on the differences in their conserved domains (Kaplan-Levy et al., 2012). Phylogenetic analysis with select Arabidopsis trihelix TFs showed that PrASIL1 clustered into SIP1 subfamily. Comparison of the PrASIL1 with SIP1 TFs indicated four conserved motifs between them. Among them, only one conserved motif is located toward the C-terminal region, but the other three are located toward their N-terminal region. Notably, the conserved motifs of the trihelix TFs are mostly located at the N-terminal, suggesting it as the most conserved region (Figure 2B and Supplementary Figure 3).
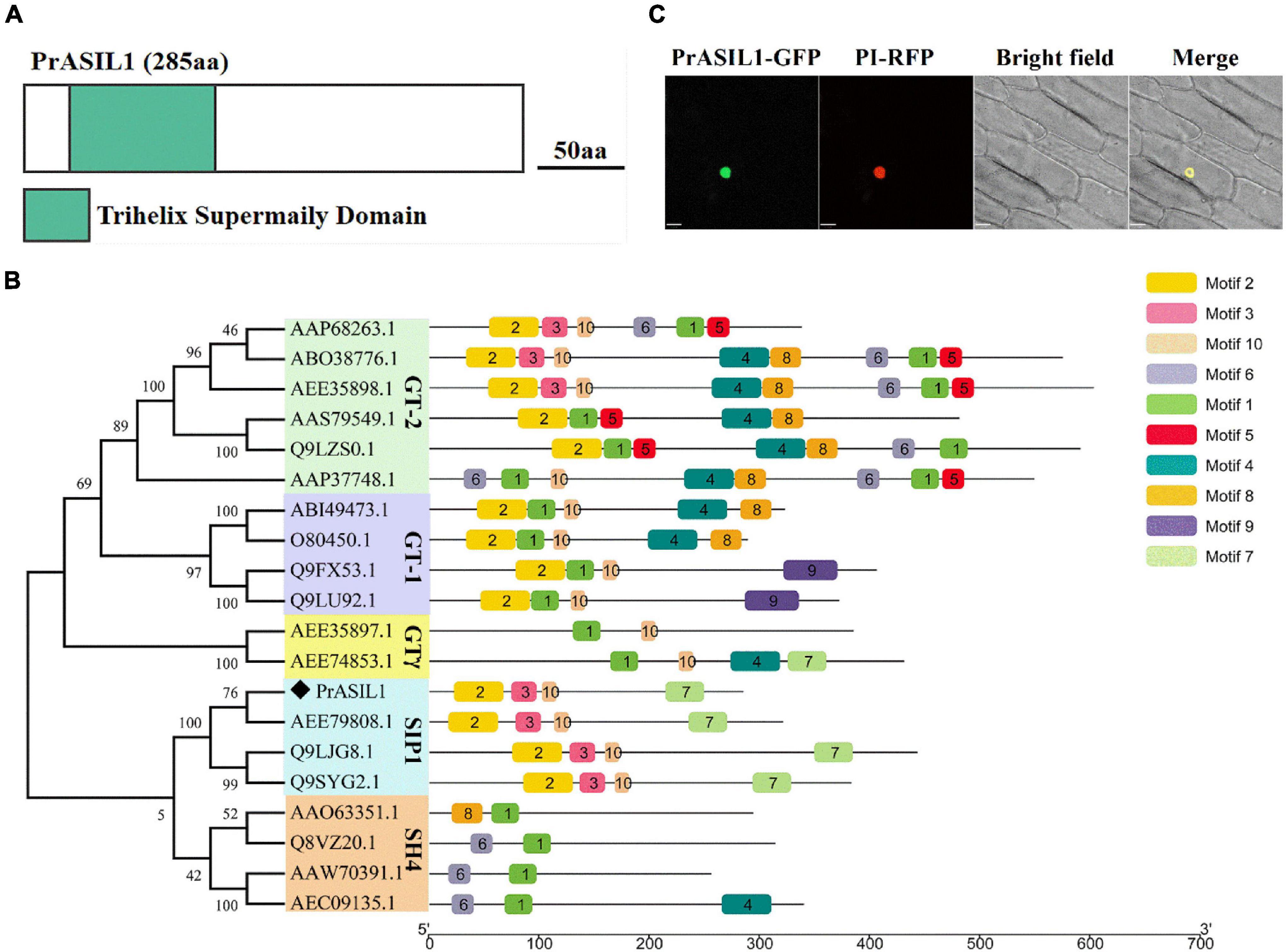
Figure 2. Molecular characterization of PrASIL1. (A) Schematic representation of PrASIL1 structure. aa, Amino acids. (B) Phylogenetic tree and conserved motifs of the PrASIL1. (C) Subcellular localization of the PrASIL1 protein fused with GFP (35S:PrASIL1-GFP) in onion epidermal cells. Bars = 33 μm.
To determine the subcellular localization of PrASIL1, a GFP fusion vector 35S:PrASIL1-GFP was constructed and transformed into onion epidermal cells. The results illustrate that PrASIL1-GFP co-localized with 35S:NbWRKY8-mCherry, a nuclear transcription factor, indicating that PrASIL1 is nuclear localized (Figure 2C) and likely to function as a TF.
Transient Overexpression of PrASIL1 Reduces Oil Content and Changes Fatty Acids Profile in Nicotiana benthamiana Leaf Tissue
The Agrobacterium-mediated transient expression is an advantageous system to study gene expression due to time-efficiency compared to stable transformation systems (Wroblewski et al., 2005). Additionally, several studies have successfully demonstrated that N. benthamiana leaf tissue can be used for transient expression of genes involved in FA synthesis and TAG accumulation to affect their metabolism (Grimberg et al., 2015; Ma et al., 2015, 2016; An et al., 2017; Snell et al., 2019; Behera et al., 2021). Here, we transiently expressed PrASIL1 under the control of dual CaMV 35S promoters in N. benthamiana leaves. The coding sequence for viral protein P19, an inhibitor of ectopic gene silencing, was also co-transformed. The expression of the PrASIL1 was confirmed by RT-PCR, 6 days after leaf infiltration (Figure 3A). The overexpression of PrASIL1 significantly decreased the number of lipid droplets (LDs), relative to the mock- and P19-transformed control leaves (Figures 3B,C). More specifically, overexpression of PrASIL1 in leaves caused a considerable decrease in the numbers of small-, medium-, and large-sized LDs, compared to P19 control, by 58, 75, and 100%, respectively (Figure 3D).
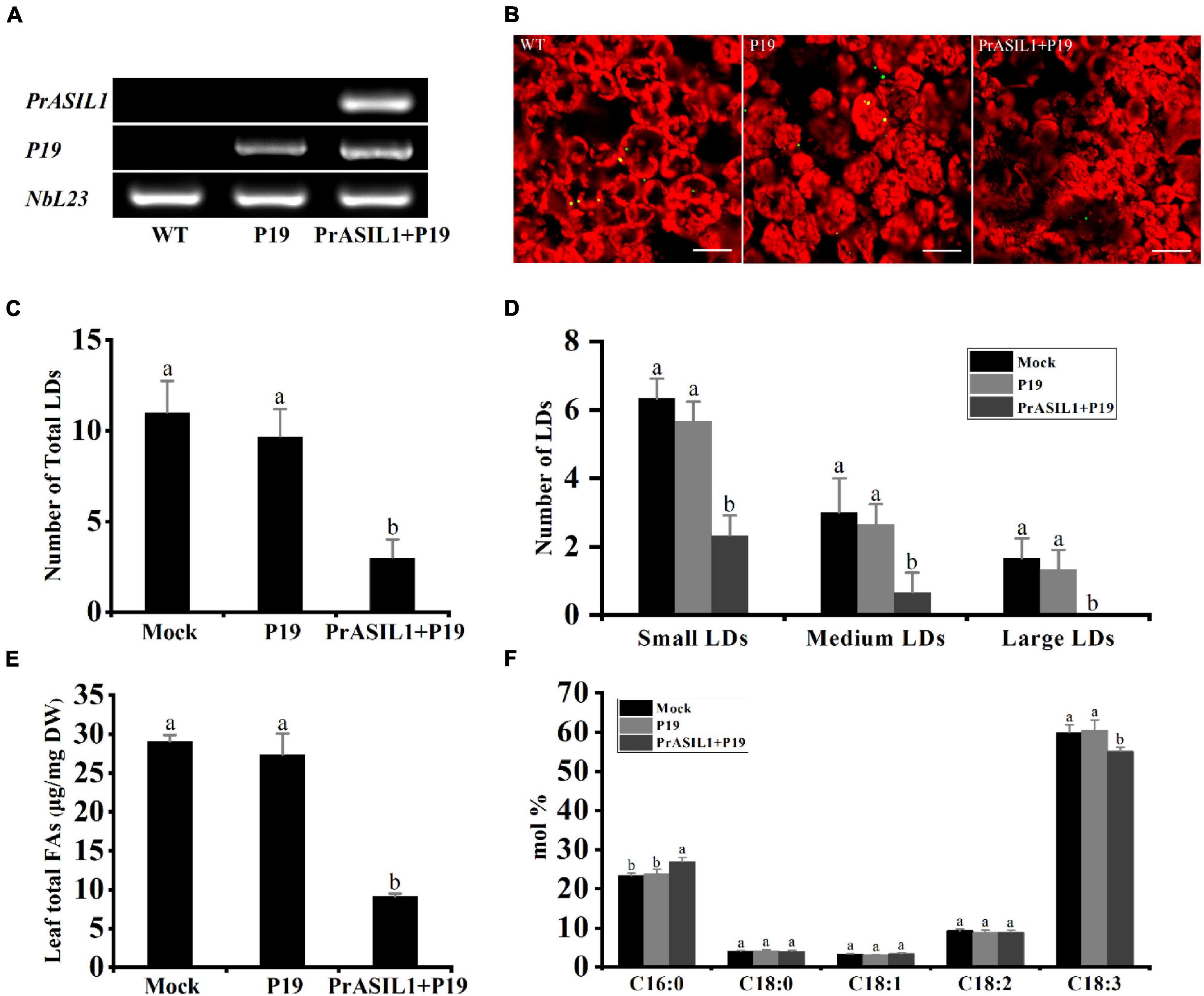
Figure 3. PrASIL1 reduces oil content and changes FA profiles in transformed N. benthamiana leaf tissue. (A) RT-PCR analysis of PrASIL1 expressed in tobacco leaf tissue. (B) Representative confocal images of LDs in N. benthamiana leaf tissue. Green color shows LDs and red color shows chloroplast. Images are shown a 232.50 × 232.50 μm2 field of the leaf tissue. Bars = 40 μm. (C) Number of total LDs per image area in PrASIL1-transformed, mock, and P19-transformed N. benthamiana leaf tissue. (D) Number of LDs in different size categories per image area. Small LDs: Nile Red-stained lipid area < 3 μm2, Medium LDs: 3–6 μm2, Large LDs: 6–10 μm2. (E) Comparison of total FA content (μg/mg) among PrASIL1-transformed, mock, and P19-transformed N. benthamiana leaf tissue. (F) Fatty acid composition of PrASIL1-transformed, mock, and P19-transformed N. benthamiana leaf tissue. DW, Dry weight. Values are mean ± SD (n = 3). Different letters indicate significant difference at P < 0.05, as confirmed by one-way ANOVA with Tukey’s post-test.
Consistent with the number of LDs, the total FA content was also remarkably lower in the leaves transiently overexpressing PrASIL1 than in the mock- and P19-transformed controls, suggesting a negative correlation between PrASIL1 expression and oil biosynthesis (Figure 3E). Previous studies have shown that the change of oil content in tobacco leaf tissues was accompanied by an alteration of the FA composition (Vanhercke et al., 2014; Cai et al., 2015). Here, the expression of PrASIL1 resulted in the change of FA composition (Figure 3F). Specifically, the FA composition of C18:3 in PrASIL1-overexpressing lines was decreased to ∼89%, while the C16:0 was significantly enhanced, relative to the P19-transformed controls (Figure 3F). This reduction might be due to the inhibition of genes involved in FA biosynthesis and TAG deposition by overexpression of PrASIL1.
Transient Overexpression of PrASIL1 in Leaf Tissue of Nicotiana benthamiana Downregulates the Genes Involved in Fatty Acids and Triacylglycerol Biosynthesis
To investigate how PrASIL1 resulted in reduced oil accumulation, qRT-PCR was performed to quantitatively analyze the expression levels of several genes related to FA and TAG biosynthesis in the N. benthamiana leaves. Previous studies showed that the expression of AtLEC2 in N. benthamiana leaves increased the transcription level of NbWRI1, a positive master regulator of FA biosynthesis (Nookaraju et al., 2014). Here, overexpression of PrASIL1 decreased the expression level of NbWRI1 (Figure 4). We further analyzed the expression levels of genes regulated by WRI1, including SUCROSE SYNTHASE (SUS), PHOSPHOENOLPYRUVATE ENOLASE 1 (ENO1), PYRUVATE KINASE ALPHA SUBUNIT (PKpα), PLASTIDIAL PYRUVATE KINASE BETA SUBUNIT 1 (PKpβ1), PDH E1 COMPONENT ALPHA SUBUNIT (PDH-E1α), BIOTIN CARBOXYL CARRIER PROTEIN 2 (BCCP2), KAS1 and ACYL CARRIER PROTEIN 5 (ACP5). Except for PKpα, the expression levels of the other genes in PrASIL1-transformed leaves were significantly lower than those in P19-transformed leaves; the expression level of NbBCCP2 was reduced the most by 70% (Figure 4). The expression levels of several other FA and TAG biosynthesis genes were also down-regulated in PrASIL1-transformed leaves compared with the control leaves; these include KETOACYL-ACP REDUCTASE (KAR), LIPOAMIDE DEHYDROGENASE1 (LPD1), ENOYL-ACYL CARRIER PROTEIN REDUCTASE (MOD1), NAD-DEPENDENT G3P DEHYDROGENASE (GPDH), FAD2, FAD3, GPAT9, and DGAT1 but not PDAT2 (Figure 4). These results confirm that the expression of PrASIL1 induced down-regulation of several genes involved in FA and TAG biosynthesis.
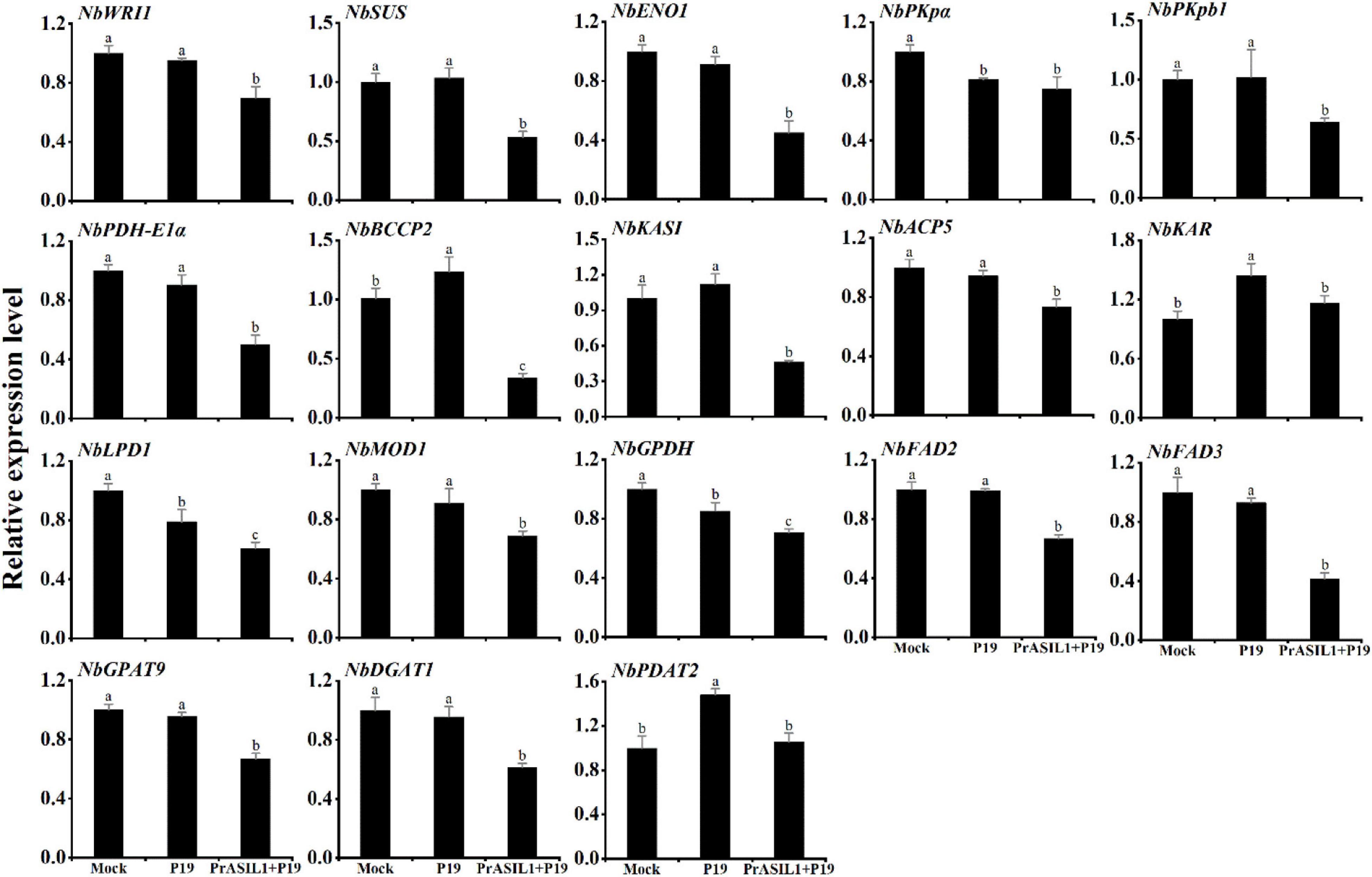
Figure 4. Comparison of the expression levels of genes involved in FA biosynthesis to TAG accumulation among PrASIL1-transformed, mock, and P19-transformed N. benthamiana leaf tissue. Results were normalized to the expression of NbL23 and calibrated to the levels of mock tobacco leaves. Values are means ± SD (n = 3). Different letters indicate significant difference at P < 0.05, as confirmed by one-way ANOVA with Tukey’s post-test.
PrASIL1 Represses Seed Oil Accumulation and Alters Fatty Acids Composition in Transgenic Arabidopsis
Since PrASIL1 is mostly expressed in the seeds, we investigated the role of PrASIL1 in seed oil accumulation by stable expression of PrASIL1 in Arabidopsis. We transformed the PrASIL1-overexpression construct (35S:PrASIL1) into wild-type Arabidopsis (Columbia-0). The independent T3 homozygous transgenic lines were identified by RT-PCR, and three lines with relatively high expression of PrASIL1 (OE-4, 5, and 7) were selected for further analysis (Figure 5A).

Figure 5. Overexpression of PrASIL1 reduces seed oil content and alters FA composition in transgenic Arabidopsis. (A) RT-PCR analysis of PrASIL1 expressed in transgenic Arabidopsis seed. (B) Microscopic observation of mature seeds randomly collected from wild-type (Col-0) and PrASIL1 transgenic plants. Bars = 500μm. (C) Quantitative comparison of seed size (length and width) between the wild type (Col-0) and PrASIL1 transgenic plants. (D) Quantitative comparison of the dry weight of seeds between the wild type (Col-0) and PrASIL1 transgenic plants. (E) Comparison of total FA content (μg/mg) between the wild type (Col-0) and PrASIL1 transgenic plants. (F) Comparison of total FA content (μg/dry seed) between the wild type (Col-0) and PrASIL1 transgenic plants. (G) Fatty acid composition of wild type (Col-0) and PrASIL1 transgenic plants. DW, Dry weight. Values are means ± SD (n = 3). Asterisks indicate significant differences in the seed size (C), dry weight of seed (D), seed total FA content (E,F) compared to that in the wild type (two-tailed paired Student’s t-test, P < 0.05). Different letters indicate significant difference in the contents of major seed FA compositions (G) compared to that in the wild type (one-way ANOVA with Tukey’s post-test, P < 0.05).
The overexpression of PrASIL1 did not show any phenotypic variation during its development in any of the tissues even though it was expressed under a strong constitutive promoter CAMV35S, except in seeds. In the three OE lines, the size and weight of mature seed was reduced, compared to the wild-type Arabidopsis seeds (Figures 5B–D). The total and per seed FA content was also significantly less in OE lines than in wild-type (Figures 5E,F). Furthermore, the increase in total FA content was accompanied by an alteration in FA composition. Compared to the wild type, the composition of C18:1 and C18:3 was significantly lower in OE seeds, while the proportion of C16:0 was significantly elevated (Figure 5G). Interestingly, lack of any significant phenotypic differences in the OE seedlings or their development compared to the wild-type suggests a less effective involvement of ASIL1 in other physiological processes. Overall, these results suggest that the overexpression of PrASIL1 led to reduced seed oil accumulation and altered FA composition in mature seeds.
PrASIL1 Represses the Expression of Fatty Acids and Oil Synthetic Genes Including Upstream Positive Regulators During Seed Maturation in Transgenic Arabidopsis
To identify the genes regulated by PrASIL1 in Arabidopsis, we examined the expression of various FA and TAG biosynthesis genes in seeds by qRT-PCR. First, we analyzed the expression levels of WRI1, LEC1, LEC2, and FUS3, the most important positive regulators involved in seed oil accumulation. The transcript levels of all these genes in transgenic seeds (12 DAP) were remarkably lower compared with wild-type, with subtle differences among the three transgenic lines (Figure 6). Second, the expression levels of 15 key FA biosynthetic genes were quantified. The transcript levels of SUS2, PKp-β1, PDH-E1β, ACETYL CO-ENZYME A CARBOXYLASE BIOTIN CARBOXYLASE SUBUNIT 2 (CAC2), BCCP2, MALONYL COA-ACP MALONYLTRANSFERASE (MCAMT), KAR, KASI, MOD, KASII, ACYL CARRIER PROTEIN DESATURASE 5 (AAD5), FATTY ACYL-ACP THIOESTERASES A (FATA), ACP5, FAD2, and FAD3 in transgenic seeds were significantly lower than those of wild type, except that PDH-E1β, HAD, FATA, and ACP5 in OE-5 or OE-7 were not significantly lower than those of wild type (Figure 6). Third, six key TAG synthetic genes, including GPDH, GPAT9, LPAAT1, DGAT1, PDAT1, and OLEOSIN (OLEO3) were selected to analyze their transcript levels. Compared to wild-type seeds, their transcript levels in transgenic seeds were significantly lower, except the GPDH in OE-7 (Figure 6). Taken together, these findings also suggest that PrASIL1 inhibits the oil accumulation by repressing the expression of several FA and TAG synthetic genes including their upstream regulators, during seed maturation.
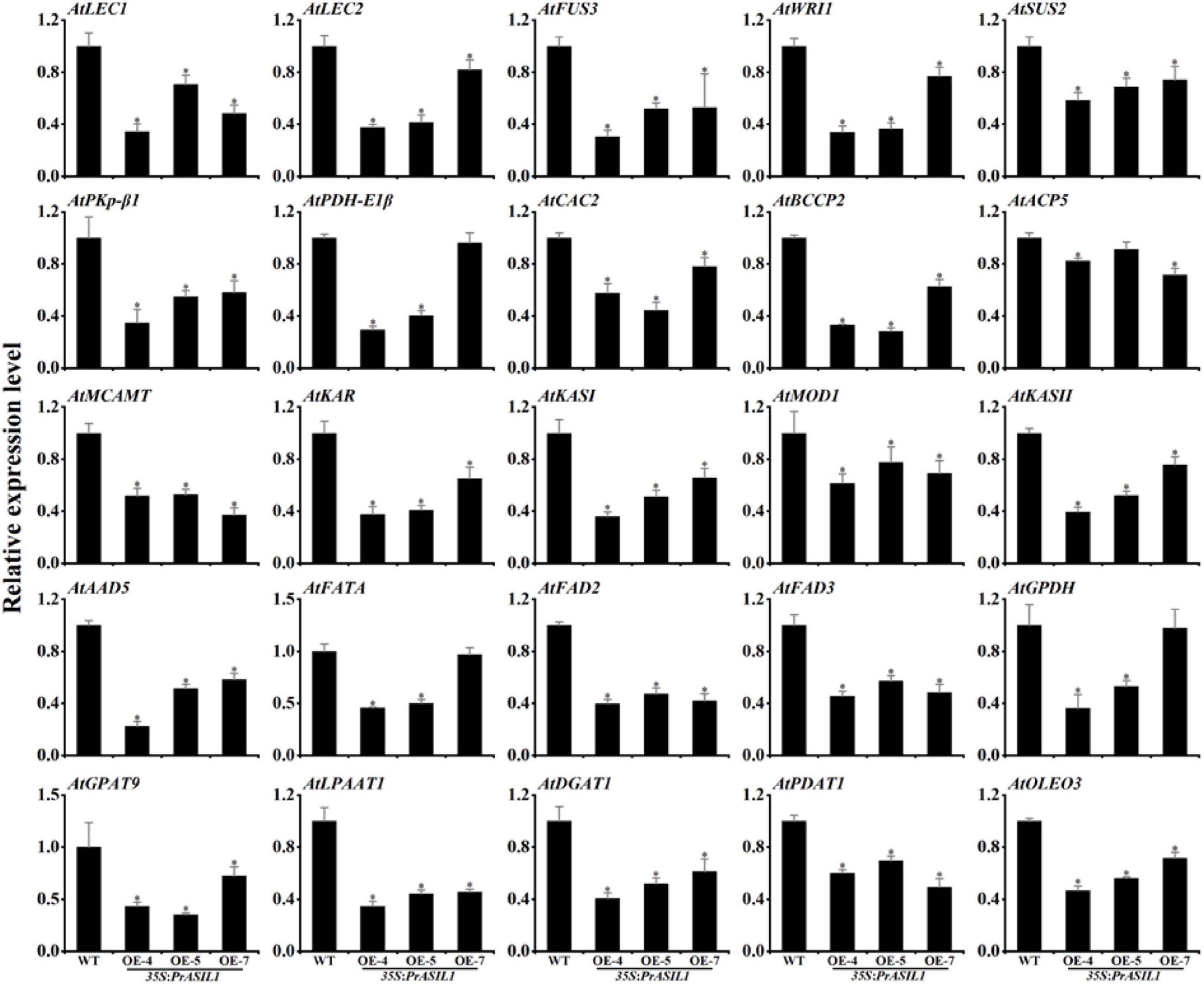
Figure 6. Comparison of the expression levels of genes involved in FA biosynthesis, modification, and TAG accumulation between the seed of wild type (Col-0) and PrASIL1 transgenic plants. Results were normalized to the expression of AtACTIN7 and calibrated to the levels of wild type. Values are mean ± SD (n = 3). Asterisks indicate significant differences in the expression levels compared to that in the wild type (two-tailed paired Student’s t-test, P < 0.05).
Reduced Expression of PrASIL1 in Paeonia rockii Enhances the Oil Content and Affects Fatty Acids Composition
To further validate the role of PrASIL1 in the regulation of oil accumulation in tree peony, we suppressed its endogenous expression in the leaves utilizing virus-induced gene silencing (VIGS) construct TRV2. GFP was used as a reporter and many green fluorescent spots were observed in the leaves inoculated with TRV2:GFP and TRV2:PrASIL1 at 6 dpi, but not in the WT, under the excitation by blue light (Figure 7A) or using confocal microscopy (Figure 7B). The expression of TRV1 and TRV2 RNA was confirmed in the leaves of the inoculated line through RT-PCR (Figure 7C). Using qRT-PCR, we showed that the transcript level of endogenous PrASIL1 in leaves inoculated with TRV2:PrASIL1 was significantly lower than that of the WT and TRV2:GFP lines, and there was no significant difference between the latter two lines (Figure 7D).
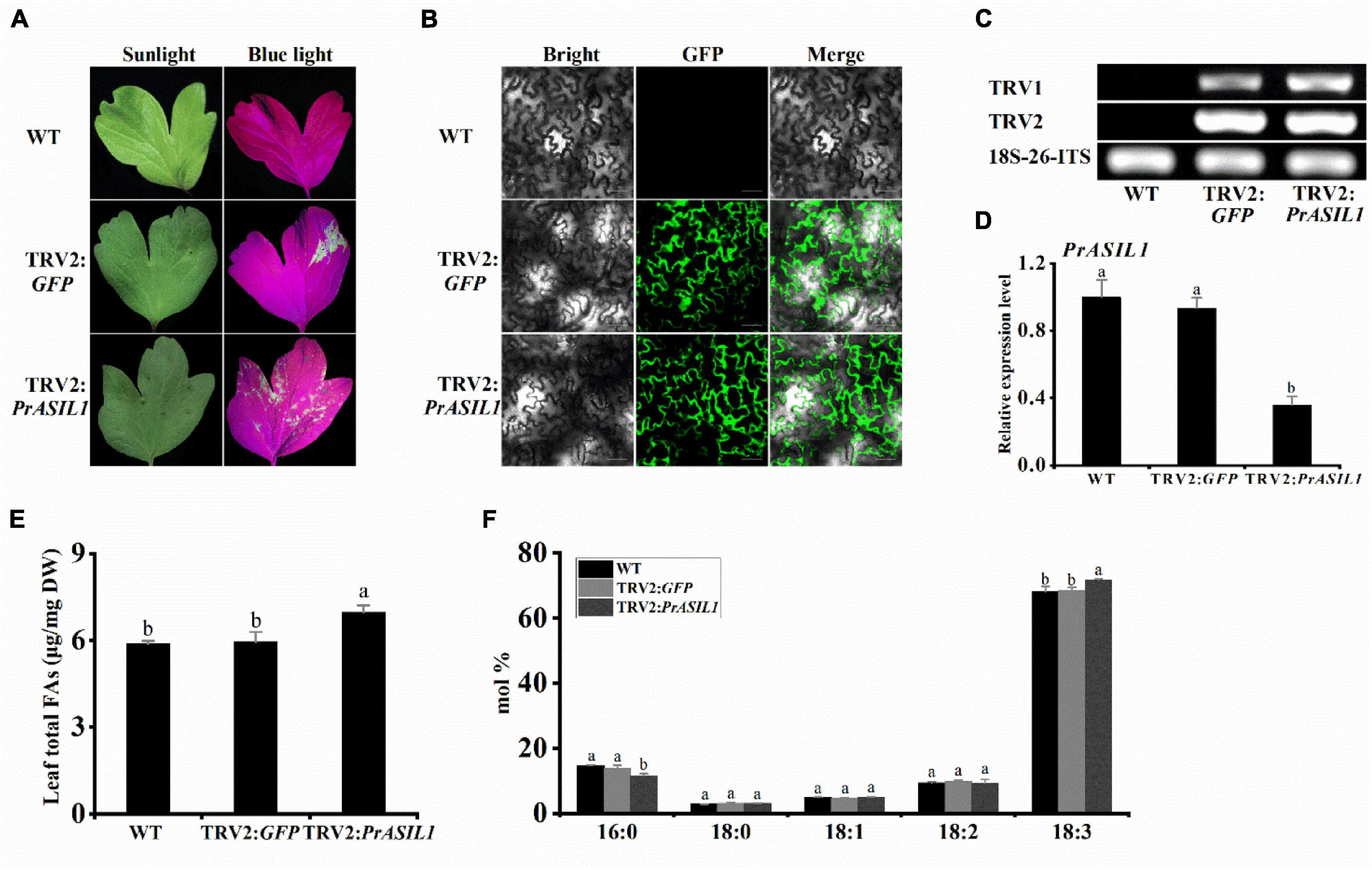
Figure 7. Reduced expression of PrASIL1 in Paeonia rockii enhances the oil content and affects FA composition. (A) Image of P. rockii leaves infected with TRV2-GFP or TRV2-PrASIL1 at 6 days post-infiltration under blue light. (B) Confocal microscopy image of P. rockii leaf tissue infected with TRV2-GFP or TRV2-PrASIL1 at 6 days post-infiltration. Bars = 30μm. (C) RT-PCR analysis of TRV1 and TRV2 accumulation levels in P. rockii leaf tissue at 2 weeks post-infiltration. (D) Silencing efficiency of PrASIL1 determined by qRT-PCR analysis. Relative expression of PrASIL1 was normalized to 18S-26S ITS and calibrated to the levels of WT leaves. (E) Comparisons of total FA content (μg/mg) among the WT, TRV2:GFP control, and PrASIL1-silenced plants. (F) Fatty acid composition of WT, TRV2:GFP control, and PrASIL1-silenced plants. DW, Dry weight. Values are mean ± SD (n = 3). Different letters indicate significant difference at P < 0.05, as confirmed by one-way ANOVA with Tukey’s post-test.
Further, it is worth noting that the total FA content in PrASIL1-silenced leaves was much higher as the endogenous PrASIL1 transcript levels decreased, relative to theTRV2:GFP control and the WT leaves (Figure 7E). Biochemical analyses revealed that the C16:0 composition was considerably lower, while the proportion of C18:3 was significantly increased, in the leaves of PrASIL1-silenced plants than those of the other two controls (Figure 7F). These results further confirmed that the expression level of PrASIL1 can negatively affect the oil content and FA composition.
PrASIL1 Silencing in Tree Peony Increases the Transcript Abundance of Fatty Acids and Triacylglycerol Biosynthesis Genes
From the above studies, it is clear that the overexpression of PrASIL1 in N. benthamiana leaves and Arabidopsis seeds decreased the expression of FA and oil synthesis-related genes. Conversely, we studied the effects of silencing PrASIL1 on genes contributing to FA and oil synthesis, which was opposite to its overexpression. The transcripts of FA biosynthesis genes, including PDH-E1α, BCCP2, MOD1, KASII, SAD, FATA, FATB, FAD2, and FAD3, were higher in the PrASIL1-silenced plants, relative to TRV2:GFP control and the WT lines (Figure 8). Similarly, genes involved in TAG assembly and accumulation, LPAAT4, DGAT1, PDAT2, and OLEO were also higher in PrASIL1-silenced plants compared to the controls; specifically, the expression level of LPAAT4 was increased by three times and FAD3 expression was higher than FAD2 (Figure 8). All together, these results corroborate that the expression of PrASIL1 is reciprocally associated with in FA and oil synthesis by affecting their gene expression.
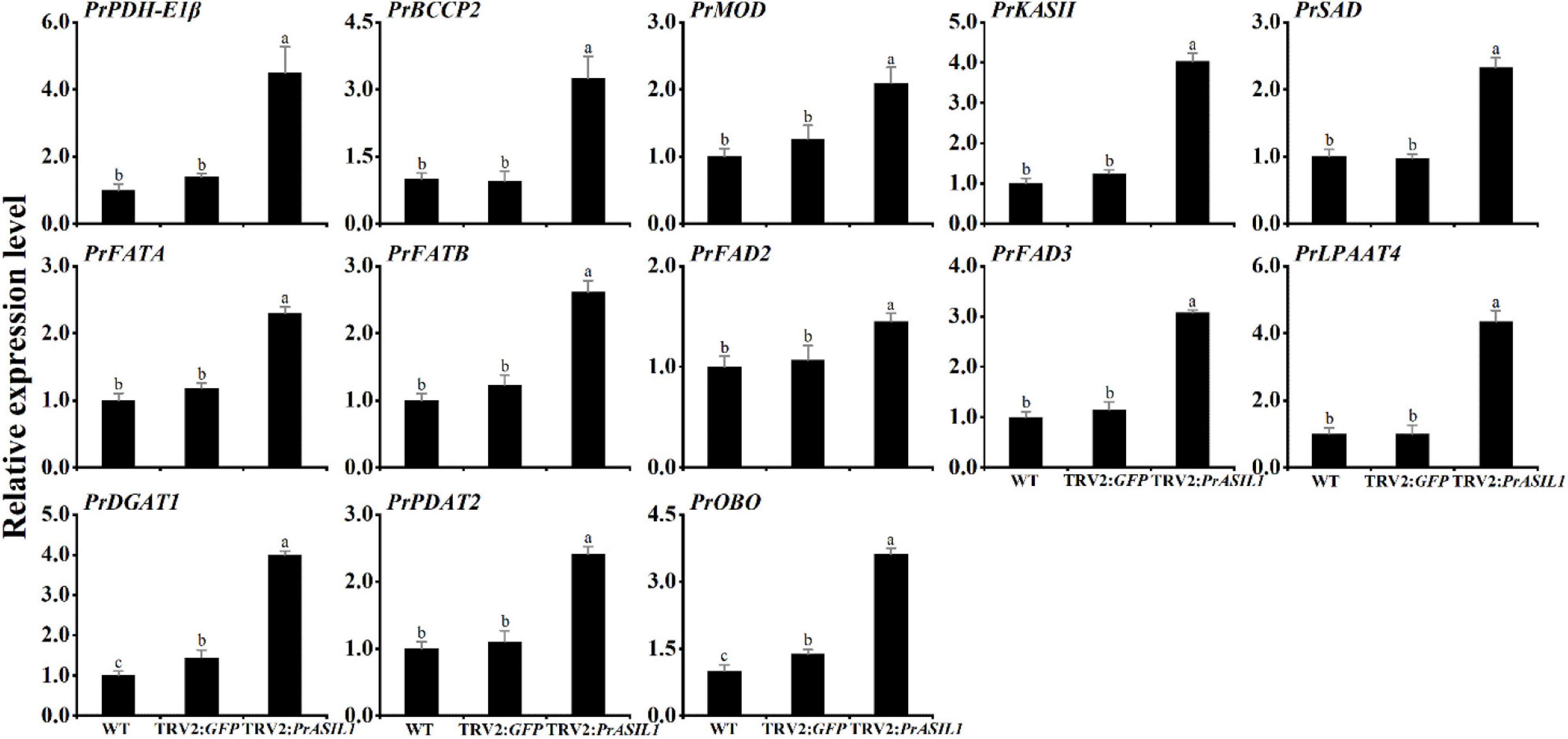
Figure 8. Comparison of the expression levels of FA and TAG biosynthetic genes among the WT, TRV2:GFP control, and PrASIL1-silenced plants. Relative expression of PrASIL1 was normalized to 18S-26S ITS and calibrated to the levels of WT leaves. Values are mean ± SD (n = 3). Different letters indicate significant difference at P < 0.05, as confirmed by one-way ANOVA with Tukey’s post-test.
Discussion
The transcriptional regulation of the biosynthetic genes is a major factor affecting the supply of FA in TAG biosynthesis. Over the past decade, the transcriptional regulation of oil accumulation in many oil crops has been revealed. Overexpression of the endogenous ZmLEC1 and ZmWRI1 in maize markedly increased seed oil content (Shen et al., 2010). LEC2 from both castor and cocoa exhibits a function that enhance oil level (Kim et al., 2014; Zhang et al., 2014). Both BnLEC1 and BnWRI1 overexpression in Brassica napus significantly increased seed oil levels, While deletion of the BnFUS3 resulted in reduced seed oil accumulation and affected the expression of BnLEC1, BnLEC2, BnABI3, and BnWRI1 (Elahi et al., 2015, 2016; Li et al., 2015). Recently, a transcriptional regulatory network of soybean oil accumulation consisting of GmLEC1, GmLEC2, GmNFYA, GmZF392, GmZF351, GmLEC1, and GmWRI1 was identified, which involves multiple reactions of glycolysis, de novo FA synthesis, TAG assembly, and oil deposition to ultimately promotes oil accumulation (Lu et al., 2021). A regulatory network was also shown in oil palm, in which EgWRI1-1 is activated by three ABA-responsive transcription factors, EgNF-YA3, EgNF-YC2, and EgABI5, thereby promoting FA synthesis (Yeap et al., 2017).
In this present study, We found that a trihelix TF, PrASIL1, is highly expressed during early and late stage of seed maturation in tree peony (Figure 1C). This temporal expression coincides with the low rate of FA biosynthesis in the early and late stages of seed maturation (Zhang et al., 2018). Interestingly, the low PrASIL1 expression in all the tissues examined, including those that do not accumulate much oil suggest that perhaps PrASIL1 is expressed constitutively and associated primary FA metabolism and/or other functions. Previous studies have shown that trihelix TFs have multiple functions, such as in response to light and biotic and abiotic stresses (Gilmartin et al., 1992; Perisic and Lam, 1992; Osorio et al., 2012), plant morphogenesis, growth, and development (Tzafrir et al., 2004; Sun et al., 2015; Shibata et al., 2018). In Arabidopsis, AtASIL1 regulates a number of embryonic maturation genes and acts downstream of miRNAs to inhibit seed maturation (Gao et al., 2009; Willmann et al., 2011). It was inferred that ASIL1 might indirectly inhibit positive regulators of FA biosynthesis, such as LEC1, LEC2, FUS3, and ABI3 (Weselake et al., 2009). Phylogenetic analysis with Arabidopsis trihelix TFs showed that PrASIL1 is closest to AtASIL1, and there are four conserved motifs shared between them (Figure 2B), and are likely functionally conserved. But noteworthy, previous reports have shown that ASIL1 may repress the expression of LEC1, LEC2, FUS2, and ABI3 in Arabidopsis. In our study, overexpression of PrASIL1 was shown to reduce the expression of genes related to oil metabolism globally, including those derived from sucrose catabolism, de novo FA biosynthesis and modification, TAG assembly, and storage into the oil body as well as several master positive regulators.
Both transient and stable overexpression of PrASIL1 induced a remarkable decrease of the total FAs and altered their compositions (Figures 3E,F, 5). Furthermore, stable expression of PrASIL1 in Arabidopsis also resulted in a significant reduction in mature seed size, seed weight (Figure 5). Additionally, reverse genetics approach revealed that the silencing of PrASIL1 leads to an increase in the levels of the total FAs and affected the composition of several major FAs (Figures 7E,F). In all the cases, the level of PUFA, especially C18:3, and FAD3 expression over FAD2 are highly affected. Thus, it is clear that a global shift in the expression of oil synthesis-related genes was induced by PrASIL1, which can impact both FA content and composition, specifically associated with PUFAs.
The coordinated expression of genes engaged in the oil biosynthesis is crucial for seed oil accumulation. The overexpression or silencing of PrASIL1 affected the expression of several critical genes involved in multiple biological processes, such as glycolysis, FA synthesis and modification, and TAG assembly and accumulation, and thereby the oil accumulation. For example, SUS2 is a key enzyme for carbon metabolism in plant tissues, and directly participates in the sucrose catalytic reaction (Baroja-Fernandez et al., 2012). Similarly, ENO1 and PKp-β1 are functionally important in converting sucrose to acetyl-CoA, a precursor of the FA synthesis. Thus, the decreased expression of SUS2, ENO1, and PKp-β1 in PrASIL1-overexpressed lines is likely to attenuate the production and distribution of photosynthetic products, thereby reducing the carbon source for oil accumulation.
Pyruvate produced by glycolytic metabolism in most oil-synthesizing tissues is converted into acetyl-CoA under the catalysis of PDHC, which is encoded by PDH-E1α, PDH-E1β, and LPD1 (McGlew et al., 2015). Both PDH-E1α and LPD1 were upregulated in AtLEC1- and AtWRI1-overexpressing plants, respectively, and were accompanied by an increase in lipid content (Mu et al., 2008; To et al., 2020). Similarly, the overexpression of PrASIL1 decreased the expression of NbPDH-E1α and NbLPD1 in tobacco and AtPDH-E1β in Arabidopsis, while the PrPDH-E1β was upregulated in PrASIL1-silenced tree peony. Next, the acetyl-CoA is catalyzed by ACCase to produce malonyl-CoA, a key step that determines the flow of FA biosynthesis in plastids (Mu et al., 2008). The transcripts for ACCase subunits, CAC2 and BCCP2 were significantly downregulated in PrASIL11-overexpressing transgenic Arabidopsis. Therefore, lower activity of ACCase in the initial stage of the FA biosynthetic pathway might inhibit subsequent oil accumulation in transgenic seeds. Thereafter, malonyl-CoA and ACP are catalyzed by fatty acid synthase (FAS) complex into C16:0-ACP, in which ACP5, MCAMT, KASI, KAR, HAD, and MOD1 encode for the components (Li-Beisson et al., 2010). Our results showed that these genes involved in encoding FAS complex are down-regulated in PrASIL1- overexpressing transgenic Arabidopsis and tobacco, whereas the expression levels of PrMOD1 is increased in PrASIL1-silenced tree peony. The considerable downregulation of these FAS-encoding genes should decelerate FA biosynthesis in PrASIL11-overexpressed plants. Finally, 16:0-ACP is elongated by KASII to 18:0-ACP and then desaturated by SAD to generate 18:1-ACP (Lindqvist et al., 1996; Carlsson et al., 2002). The content of C16:0 in Arabidopsis KASII mutant (kas2) was remarkably increased (Gao et al., 2020), whereas the decreased activity of AAD5 resulted in a significant reduction in the level of C18:1 in mature seeds (Kazaz et al., 2020). Thus, the decrease of AtAAD5 expression led to the reduction of C18:1 in transgenic Arabidopsis, while the upregulation of PrSAD and PrKASII increased the C18:0 and C18:1 in PrASIL1-silenced tree peony. In the next step, Acyl-ACP thioesterases (FATs), such as FATA and FATB are required to catalyze the release of FA before they are transported from plastids to the endoplasmic reticulum for TAG synthesis. FATA generally releases ACP of the monounsaturated acyl chains with 18 carbons, while FATB acts on saturated acyl chains with 16 and 18 carbons (Jones et al., 1995; Dörmann et al., 2000; Salas and Ohlrogge, 2002). The reduction of FATA activity in Arabidopsis caused the decrease of oil content and change in FA composition of seeds (Moreno-Pérez et al., 2012). Consistently, the decline in C18:1 content of PrASIL1-overexpressed Arabidopsis seeds may be associated with the decrease in AtFATA expression, and the enhancement in C18:0 and C18:1 contents of PrASIL1-silenced plants may be due to the increased transcript levels of PrFATA and PrFATB. Hence, the suppression of several FA biosynthetic genes by PrASIL1 affects FA composition in transgenic plants.
The CoA esters synthesized in the plastid are transported to the ER that are modified by fatty acid desaturases. FAD2 is essential for polyunsaturated FAs biosynthesis from phospholipids (Okuley et al., 1994), while FAD3 possesses a crucial function in C18:3 biosynthesis (Shah et al., 1997). Therefore, the downregulation of endogenous FAD2 and FAD3 would individually inhibit the accumulation of C18:2 and C18:3 in transgenic Arabidopsis seeds and PrASIL11-overexpressed tobacco leaves. In contrast, increased levels of PrFAD3 expression supported C18:3 accumulation in PrASIL1-silenced tree peony (Figure 7F). The effect of PrASIL1 was more pronounced on FAD3 than FAD2 either in overexpression or silenced conditions. Further, our results showed that PrASIL1 largely repressed the expression of TAG biosynthesis genes and TAG storage gene, such as endogenous GPAT9, LPAAT1, DGAT1, PDAT2, and OLE3 (Figure 8). This could explain why over expression of PrASIL1 in Arabidopsis seeds and N. benthamiana leaves accumulated significantly less oil than wild-type plants. Therefore, PrASIL1 has an inhibitory function on TAG biosynthesis and storage-related genes, which ultimately decreases the oil accumulation.
Among the network of major TFs, ASIL1 was hypothesized as a negative regulator that indirectly down-regulates WRI1, FUS3, LEC1, and LEC2, which positively regulate seed maturation and oil accumulation in Arabidopsis (Weselake et al., 2009). Furthermore, LEC1 and LEC2 positively regulate FUS3 (Kroj et al., 2003; Kagaya et al., 2005), and all three of them are upstream positive regulators of WRI1 (Baud et al., 2007; Wang and Perry, 2013; Pelletier et al., 2017). As a master regulator of FA synthesis, WRI positively modulates the enzymatic reaction of glycolysis and FA biosynthesis by directly facilitating the expression of those genes (Focks and Benning, 1998; Cernac and Benning, 2004; Maeo et al., 2009). In transgenic Arabidopsis, the down-regulation of WRI1, FUS3, LEC1, and LEC2 were accompanied by low expression levels of various oil biosynthetic genes and significant reduction in seed oil content. Similarly, the oil quantity of PrASIL11-overexpressed tobacco leaves, together with the transcript levels of WRI1 and several oil biosynthetic genes, decreased (Figures 3E, 4). Since tree peony has no effective genetic transformation system, we used VIGS to silence PrASIL1 in leaf tissues to predict its role in seed oil accumulation of tree peony. In agreement with the effects of heterologous overexpression of PrASIL1, the silencing of PrASIL11 in tree peony leaves resulted in the increase of the expression level of oil synthetic genes along with the oil content.
Conclusion
The trihelix transcription factor PrASIL1 is highly expressed in the early and late seed maturation phases indicating its active regulatory role in suppressing rate of seed oil biosynthesis in those phases. Structurally, PrASIL1 shares conserved domains with its ortholog from Arabidopsis, which indicates that both of them might be acting as a negative regulator of seed oil biosynthesis and seed maturation. To this extent, our results from transient and stable transgene over-expression in tobacco and Arabidopsis showed that PrASIL1 reduced total fatty acids and altered fatty acid compositions, which in turn associated with the decrease in expression levels of several genes involved in oil metabolism. Especially the expression of master positive regulators, such as AtLEC1, AtLEC2, AtWRI1, and AtFUS3, were repressed in transgenic Arabidopsis. Hence PrASIL1 may act as a negative regulator of oil biosynthesis, functioning upstream of master regulatory genes. Additionally, silencing of PrASIL1 in tree peony leaves increased total fatty acids and affected fatty acid compositions are accompanied by increased expression of numerous genes in the oil biosynthesis pathway, which is in agreement with its negative role in oil metabolism. Taken together, PrASIL1 is likely to inhibit the expression of select oil biosynthesis genes by directly interacting with them or via the down-regulation of several master positive regulators. The mechanistic aspects of these interactions and target genes for PrASIL1 need further investigation. Nevertheless, it is clear that PrASIL1 acts as a negative regulator of oil accumulation at the transcription level and can be targeted for oil enhancement in other crops through gene manipulation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
WY and JH carried out the experiments. WY analyzed the data and wrote the manuscript. LN, QZ, and YaZ conceived and designed the experiments. AK, JB, and LX analyzed the data and revised the manuscript. YY, YuZ, and YX assisted with doing the experiments. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Natural Science Foundation of China (Grant Nos. 31972453, 31901357 and 31971690) and the Postdoctoral Fund of Shaanxi Province (Grant No. K3380220028).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Ziwei Xin for assistance with data analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.796181/full#supplementary-material
Footnotes
References
Adhikari, N. D., Bates, P. D., and Browse, J. (2016). WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol. 171, 179–191. doi: 10.1104/pp.15.01906
An, D., Kim, H., Ju, S., Go, Y. S., Kim, H. U., and Suh, M. C. (2017). Expression of camelina WRINKLED1 isoforms rescue the seed phenotype of the Arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 8:34. doi: 10.3389/fpls.2017.00034
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Baroja-Fernandez, E., Munoz, F. J., Li, J., Bahaji, A., Almagro, G., Montero, M., et al. (2012). Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc. Natl. Acad. Sci. U.S.A. 109, 321–326. doi: 10.1073/pnas.1117099109
Bates, P. D., Stymne, S., and Ohlrogge, J. (2013). Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 16, 358–364. doi: 10.1016/j.pbi.2013.02.015
Baud, S., Kelemen, Z., Thévenin, J., Boulard, C., Blanchet, S., To, A., et al. (2016). Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by L-AFL proteins in Arabidopsis seed. Plant Physiology 171, 1099–1112. doi: 10.1104/pp.16.00034
Baud, S., and Lepiniec, L. (2010). Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49, 235–249. doi: 10.1016/j.plipres.2010.01.001
Baud, S., Wuillème, S., Dubreucq, B., De Almeida, A., Vuagnat, C., Lepiniec, L., et al. (2007). Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana†. Plant J. 52, 405–419. doi: 10.1111/j.1365-313X.2007.03232.x
Baud, S. B., Wuill, Ã, Me, S., To, A., Rochat, C., and Lepiniec, L. C. (2009). Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 60, 933–947. doi: 10.1111/j.1365-313X.2009.04011.x
Behera, J. R., Rahman, M. M., Bhatia, S., Shockey, J., and Kilaru, A. (2021). Functional and predictive structural characterization of WRINKLED2, a unique oil biosynthesis regulator in avocado. Front. Plant Sci. 12:648494. doi: 10.3389/fpls.2021.648494
Browse, J., McConn, M., James, D. J., and Miquel, M. (1993). Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268, 16345–16351. doi: 10.1016/s0021-9258(19)85427-3
Cai, Y., Goodman, J. M., Pyc, M., Mullen, R. T., Dyer, J. M., and Chapman, K. D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27, 2616–2636. doi: 10.1105/tpc.15.00588
Carlsson, A. S., LaBrie, S. T., Kinney, A. J., Von Wettstein Knowles, P., and Browse, J. (2002). A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. Cell Mol. Biol. 29, 761–770. doi: 10.1046/j.1365-313X.2002.01253.x
Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Chen, M., Wang, Z., Zhu, Y., Li, Z., Hussain, N., Xuan, L., et al. (2012). The effect of transparent TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol. 160, 1023–1036. doi: 10.1104/pp.112.202945
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dörmann, P., Voelker, T. A., and Ohlrogge, J. B. (2000). Accumulation of palmitate in Arabidopsis mediated by the Acyl-Acyl carrier protein thioesterase FATB11. Plant Physiol. 123, 637–644. doi: 10.1104/pp.123.2.637
Duan, S., Jin, C., Li, D., Gao, C., Qi, S., Liu, K., et al. (2017). MYB76 inhibits seed fatty acid accumulation in Arabidopsis. Front. Plant Sci. 8:226. doi: 10.3389/fpls.2017.00226
Durrett, T. P., Benning, C., and Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Egert, S., Baxheinrich, A., Lee-Barkey, Y. H., Tschoepe, D., Wahrburg, U., and Stratmann, B. (2014). Effects of an energy-restricted diet rich in plant-derived α-linolenic acid on systemic inflammation and endothelial function in overweight-to-obese patients with metabolic syndrome traits. Br. J. Nutr. 112, 1315–1322. doi: 10.1017/S0007114514002001
Elahi, N., Duncan, R. W., and Stasolla, C. (2015). Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant Physiol. Biochem. 96, 222–230. doi: 10.1016/j.plaphy.2015.08.002
Elahi, N., Duncan, R. W., and Stasolla, C. (2016). Modification of oil and glucosinolate content in canola seeds with altered expression of Brassica napus LEAFY COTYLEDON1. Plant Physiol. Biochem. 100, 52–63. doi: 10.1016/j.plaphy.2015.12.022
Focks, N., and Benning, C. (1998). Wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism1. Plant Physiol. 118, 91–101. doi: 10.1104/pp.118.1.91
Gao, J., Lunn, D., Wallis, J. G., and Browse, J. (2020). Phosphatidylglycerol composition is central to chilling damage in the Arabidopsisfab1 mutant. Plant Physiol. 184, 1717–1730. doi: 10.1104/pp.20.01219
Gao, M., Lydiate, D. J., Li, X., Lui, H., Gjetvaj, B., Hegedus, D. D., et al. (2009). Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell 21, 54–71. doi: 10.1105/tpc.108.061309
Gilmartin, P. M., Memelink, J., Hiratsuka, K., Kay, S. A., and Chua, N. H. (1992). Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. Plant Cell 4, 839–849. doi: 10.1105/tpc.4.7.839
Graham, I. A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142. doi: 10.1146/annual.arplant.59.032607.092938
Grimberg, Å, Carlsson, A. S., Marttila, S., Bhalerao, R., and Hofvander, P. (2015). Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 15:192. doi: 10.1186/s12870-015-0579-1
Huang, B., and Liu, J. (2006). Cloning and functional analysis of the novel gene GhDBP3 encoding a DRE-binding transcription factor from Gossypium hirsutum. Biochim. Biophys. Acta Gene Struct. Express. 1759, 263–269. doi: 10.1016/j.bbaexp.2006.04.006
Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., and Yoshioka, H. (2011). Phosphorylation of the nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23, 1153–1170. doi: 10.1105/tpc.110.081794
Ji, X., Mao, X., Hao, Q., Liu, B., Xue, J., and Li, R. Z. (2018). Splice variants of the castor WRI1 gene upregulate fatty acid and oil biosynthesis when expressed in tobacco leaves. Int. J. Mol. Sci. 19:146. doi: 10.3390/ijms19010146
Johnston, M. L., Luethy, M. H., Miernyk, J. A., and Randall, D. D. (1997). Cloning and molecular analyses of the Arabidopsis thaliana plastid pyruvate dehydrogenase subunits. Biochim. Biophys. Acta 1321, 200–206. doi: 10.1016/s0005-2728(97)00059-5
Jones, A., Davies, H. M., and Voelker, T. A. (1995). Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 7, 359–371. doi: 10.1105/tpc.7.3.359
Kagaya, Y., Toyoshima, R., Okuda, R., Usui, H., Yamamoto, A., and Hattori, T. (2005). LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 46, 399–406. doi: 10.1093/pcp/pci048
Kaplan-Levy, R. N., Brewer, P. B., Quon, T., and Smyth, D. R. (2012). The trihelix family of transcription factors – light, stress and development. Trends Plant Sci. 17, 163–171. doi: 10.1016/j.tplants.2011.12.002
Kazaz, S., Barthole, G., Domergue, F., Ettaki, H., To, A., and Vasselon, D. (2020). Differential activation of partially redundant δ9 Stearoyl-ACP desaturase genes is critical for omega-9 monounsaturated fatty acid biosynthesis during seed development in Arabidopsis. Plant Cell 32, 3613–3637. doi: 10.1105/tpc.20.00554
Kim, H. U., Jung, S., Lee, K., Kim, E. H., Lee, S., Roh, K. H., et al. (2014). Ectopic overexpression of castor bean LEAFY COTYLEDON2 (LEC2) in Arabidopsis triggers the expression of genes that encode regulators of seed maturation and oil body proteins in vegetative tissues. FEBS Open Biol. 4, 25–32. doi: 10.1016/j.fob.2013.11.003
Kong, Q., and Ma, W. (2018). WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci. 272, 153–156. doi: 10.1016/j.plantsci.2018.04.013
Kong, Q., Singh, S. K., Mantyla, J. J., Pattanaik, S., Guo, L., Yuan, L., et al. (2020a). TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 interacts with WRINKLED1 to mediate seed oil biosynthesis. Plant Physiol. 184, 658–665. doi: 10.1104/pp.20.00547
Kong, Q., Yang, Y., Guo, L., Yuan, L., and Ma, W. (2020b). Molecular basis of plant oil biosynthesis: Insights gained from studying the WRINKLED1 transcription factor. Front. Plant Sci. 11:24. doi: 10.3389/fpls.2020.00024
Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130, 6065–6073. doi: 10.1242/dev.00814
Lee, H. G., Kim, H., Suh, M. C., Kim, H. U., and Seo, P. J. (2018). The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in Arabidopsis seeds. Plant Cell Physiol. 59, 1432–1442. doi: 10.1093/pcp/pcy073
Li, D., Jin, C., Duan, S., Zhu, Y., Qi, S., Liu, K., et al. (2017). MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 173, 1211–1225. doi: 10.1104/pp.16.01634
Li, Q., Shao, J., Tang, S., Shen, Q., Wang, T., Chen, W., et al. (2015). Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front. Plant Sci. 6:1015. doi: 10.3389/fpls.2015.01015
Li, R., Yu, K., Wu, Y., Tateno, M., Hatanaka, T., and Hildebrand, D. F. (2012). Vernonia DGATs can complement the disrupted oil and protein metabolism in epoxygenase-expressing soybean seeds. Metab. Eng. 14, 29–38. doi: 10.1016/j.ymben.2011.11.004
Li, Y., Beisson, F., Pollard, M., and Ohlrogge, J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67, 904–915. doi: 10.1016/j.phytochem.2006.02.015
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2010). Acyl-Lipid metabolism. Arabidopsis Book 8:e133. doi: 10.1199/tab.0133
Lindqvist, Y., Huang, W., Schneider, G., and Shanklin, J. (1996). Crystal structure of delta9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 15, 4081–4092. doi: 10.1002/j.1460-2075.1996.tb00783.x
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313x.2002.01394.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, L., Wei, W., Li, Q., Bian, X. A., and Zhang, J. (2021). A transcriptional regulatory module controls lipid accumulation in soybean. New Phytol. 231, 661–678. doi: 10.1111/nph.17401
Luerssen, H., Kirik, V., Herrmann, P., and Misera, S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764. doi: 10.1046/j.1365-313x.1998.00259.x
Ma, W., Kong, Q., Grix, M., Mantyla, J. J., Yang, Y., Benning, C., et al. (2015). Deletion of a C–terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J. 83, 864–874. doi: 10.1111/tpj.12933
Ma, W., Kong, Q., Mantyla, J. J., Yang, Y., Ohlrogge, J. B., and Benning, C. (2016). 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J. 88, 228–235. doi: 10.1111/tpj.13244
Maeo, K., Tokuda, T., Ayame, A., Mitsui, N., Kawai, T., Ishiguro, S., et al. (2009). An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 60, 476–487. doi: 10.1111/j.1365-313X.2009.03967.x
McGlew, K., Shaw, V., Zhang, M., Kim, R. J., and Yang, W. (2015). An annotated database of Arabidopsis mutants of acyl lipid metabolism. Plant Cell Rep. 34, 519–532. doi: 10.1007/s00299-014-1710-8
Moreno-Pérez, A. J., Venegas-Calerón, M., Vaistij, F. E., Salas, J. J., Larson, T. R., Garcés, R., et al. (2012). Reduced expression of FatA thioesterases in Arabidopsis affects the oil content and fatty acid composition of the seeds. Planta 235, 629–639. doi: 10.1007/s00425-011-1534-5
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148, 1042–1054. doi: 10.1104/pp.108.126342
Nookaraju, A., Pandey, S. K., Fujino, T., Kim, J. Y., Suh, M. C., and Joshi, C. P. (2014). Enhanced accumulation of fatty acids and triacylglycerols in transgenic tobacco stems for enhanced bioenergy production. Plant Cell Rep. 33, 1041–1052. doi: 10.1007/s00299-014-1582-y
Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 13839–13844. doi: 10.1073/pnas.96.24.13839
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., Browse, J., et al. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158. doi: 10.1105/tpc.6.1.147
Osorio, M. B., Bucker-Neto, L., Castilhos, G., Turchetto-Zolet, A. C., Wiebke-Strohm, B., Bodanese-Zanettini, M. H., et al. (2012). Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 35, 233–246. doi: 10.1590/S1415-47572012000200005
Pelletier, J. M., Kwong, R. W., Park, S., Le, B. H., Baden, R., Cagliari, A., et al. (2017). LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U.S.A. 114, E6710–E6719. doi: 10.1073/pnas.1707957114
Perisic, O., and Lam, E. (1992). A tobacco DNA binding protein that interacts with a light-responsive box II element. Plant Cell 4, 831–838. doi: 10.1105/tpc.4.7.831
Pidkowich, M. S., Nguyen, H. T., Heilmann, I., Ischebeck, T., and Shanklin, J. (2007). Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc. Natl. Acad. Sci. U.S.A. 104, 4742–4747. doi: 10.1073/pnas.0611141104
Reinders, A., Schulze, W., Kühn, C., Barker, L., Schulz, A., Ward, J. M., et al. (2002). Protein-protein interactions between sucrose transporters of different affinities colocalized in the same enucleate sieve element. Plant Cell 14, 1567–1577. doi: 10.1105/tpc.002428
Salas, J. J., and Ohlrogge, J. B. (2002). Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 403, 25–34. doi: 10.1016/S0003-9861(02)00017-6
Sanjaya, Durrett, T. P., Weise, S. E., and Benning, C. (2011). Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in transgenic Arabidopsis. Plant Biotechnol. J. 9, 874–883. doi: 10.1111/j.1467-7652.2011.00599.x
Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., Lepiniec, L., et al. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620. doi: 10.1111/j.1365-313X.2008.03461.x
Shah, S., Xin, Z. G., and Browse, J. (1997). Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol. 114, 1533–1539. doi: 10.1104/pp.114.4.1533
Shen, B., Allen, W. B., Zheng, P., Li, C., Glassman, K., Ranch, J., et al. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153, 980–987. doi: 10.1104/pp.110.157537
Shibata, M., Breuer, C., Kawamura, A., Clark, N. M., Rymen, B., Braidwood, L., et al. (2018). GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 145:v159707. doi: 10.1242/dev.159707
Snell, P., Grimberg, Å, Carlsson, A. S., and Hofvander, P. (2019). WRINKLED1 is subject to evolutionary conserved negative autoregulation. Front. Plant Sci. 10:387. doi: 10.3389/fpls.2019.00387
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Sun, L., Rodriguez, G. R., Clevenger, J. P., Illa-Berenguer, E., Lin, J., Blakeslee, J. J., et al. (2015). Candidate gene selection and detailed morphological evaluations offs8.1, a quantitative trait locus controlling tomato fruit shape. J. Exp. Bot. 66, 6471–6482. doi: 10.1093/jxb/erv361
To, A., Joubes, J., Thueux, J., Kazaz, S., Lepiniec, L., Baud, S., et al. (2020). AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 103, 660–676. doi: 10.1111/tpj.14759
To, A., Valon, C., Savino, G., Guilleminot, J., Devic, M., Giraudat, J., et al. (2006). A network of local and redundant gene regulation Governs Arabidopsis seed maturation. Plant Cell 18, 1642–1651. doi: 10.1105/tpc.105.039925
Troncoso Ponce, M. A., Kilaru, A., Cao, X., Durrett, T. P., Fan, J., Jensen, J. K., et al. (2011). Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68, 1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x
Tsukagoshi, H., Morikami, A., and Nakamura, K. (2007). Two b3 domain transcriptional repressors prevent Sugar-Inducible expression of seed maturation genes in Arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 104, 2543–2547. doi: 10.1073/pnas.0607940104
Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., et al. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220. doi: 10.1104/pp.104.045179
Vanhercke, T., El Tahchy, A., Liu, Q., Zhou, X., Shrestha, P., Divi, U. K., et al. (2014). Metabolic engineering of biomass for high energy density: Oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 12, 231–239. doi: 10.1111/pbi.12131
Wang, F., and Perry, S. E. (2013). Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264. doi: 10.1104/pp.112.212282
Weselake, R. J., Taylor, D. C., Rahman, M. H., Shah, S., Laroche, A., McVetty, P. B., et al. (2009). Increasing the flow of carbon into seed oil. Biotechnol. Adv. 27, 866–878. doi: 10.1016/j.biotechadv.2009.07.001
Willmann, M. R., Mehalick, A. J., Packer, R. L., and Jenik, P. D. (2011). MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 155, 1871–1884. doi: 10.1104/pp.110.171355
Wroblewski, T., Tomczak, A., and Michelmore, R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3, 259–273. doi: 10.1111/j.1467-7652.2005.00123.x
Xie, L., Zhang, Q., Sun, D., Yang, W., Hu, J., Niu, L., et al. (2019). Virus-induced gene silencing in the perennial woody Paeonia ostii. PeerJ 7:e7001. doi: 10.7717/peerj.7001
Yeap, W., Lee, F., Shabari Shan, D. K., Musa, H., Appleton, D. R., Kulaveerasingam, H., et al. (2017). WRI1-1, ABI5, NF-YA3 and NF-YC2 increase oil biosynthesis in coordination with hormonal signaling during fruit development in oil palm. Plant J. 91, 97–113. doi: 10.1111/tpj.13549
Yin, D.-D., Xu, W.-Z., Shu, Q.-Y., Li, S.-S., Wu, Q., Feng, C.-Y., et al. (2018). Fatty acid desaturase 3 (PsFAD3) from Paeonia suffruticosa reveals high α-linolenic acid accumulation. Plant Sci. 274, 212–222. doi: 10.1016/j.plantsci.2018.05.027
Zhang, H., and Ogas, J. (2009). An epigenetic perspective on developmental regulation of seed genes. Mol. Plant 2, 610–627. doi: 10.1093/mp/ssp027
Zhang, M., Cao, X., Jia, Q., and Ohlrogge, J. (2016). FUSCA3 activates triacylglycerol accumulation in Arabidopsis seedlings and tobacco BY2 cells. Plant J. 88, 95–107. doi: 10.1111/tpj.13233
Zhang, Q., Yu, R., Sun, D., Rahman, M., Xie, L., Hu, J., et al. (2019). Comparative transcriptome analysis reveals an efficient mechanism of α-Linolenic acid in tree peony seeds. Int. J. Mol. Sci. 20:65. doi: 10.3390/ijms20010065
Zhang, Q., Yu, R., Xie, L., Rahman, M. M., Kilaru, A., Niu, L. X., et al. (2018). Fatty acid and associated gene expression analyses of three tree peony species reveal key genes for α-Linolenic acid synthesis in seeds. Front. Plant Sci. 9:106. doi: 10.3389/fpls.2018.00106
Zhang, Y., Clemens, A., Maximova, S. N., and Guiltinan, M. J. (2014). The Theobroma cacao B3 domain transcription factor TcLEC2 plays a duel role in control of embryo development and maturation. BMC Plant Biol. 14:106. doi: 10.1186/1471-2229-14-106
Zhao, D., Li, T., Li, Z., Sun, J., and Tao, J. (2020). Characteristics of Paeonia ostii seed oil body and OLE17.5 determining oil body morphology. Food Chem. 319:126548. doi: 10.1016/j.foodchem.2020.126548
Keywords: PrASIL1, transcription factor, seed oil, tree peony, fatty acid biosynthesis
Citation: Yang W, Hu J, Behera JR, Kilaru A, Yuan Y, Zhai Y, Xu Y, Xie L, Zhang Y, Zhang Q and Niu L (2021) A Tree Peony Trihelix Transcription Factor PrASIL1 Represses Seed Oil Accumulation. Front. Plant Sci. 12:796181. doi: 10.3389/fpls.2021.796181
Received: 16 October 2021; Accepted: 22 November 2021;
Published: 10 December 2021.
Edited by:
Wei Ma, Nanyang Technological University, SingaporeReviewed by:
Jin-Song Zhang, State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (CAS), ChinaLiang Guo, Huazhong Agricultural University, China
Copyright © 2021 Yang, Hu, Behera, Kilaru, Yuan, Zhai, Xu, Xie, Zhang, Zhang and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanlong Zhang, zhangyanlong@nwafu.edu.cn; Qingyu Zhang, zhangqingyu@nwafu.edu.cn; Lixin Niu, niulixin@nwafu.edu.cn
†These authors have contributed equally to this work
 Weizong Yang1,2†
Weizong Yang1,2† Aruna Kilaru
Aruna Kilaru Yanlong Zhang
Yanlong Zhang Lixin Niu
Lixin Niu