- Department of Crop Genetics, John Innes Centre, Norwich Research Park, Norwich, England
Fusarium head blight (FHB) and Fusarium root rot (FRR) are important diseases of small-grain cereals caused by Fusarium species. While host response to FHB has been subject to extensive study, very little is known about response to FRR and the transcriptome responses of FHB and FRR have not been thoroughly compared. Brachypodium distachyon (Bd) is an effective model for investigating host responses to both FHB and FRR. In this study the transcriptome response of Bd to F. graminearum (Fg) infection of heads and roots was investigated. An RNA-seq analysis was performed on both Bd FHB and FRR during the early infection. Additionally, an RNA-seq analysis was performed on in vitro samples of Fg for comparison with Fg gene expression in planta. Differential gene expression and gene-list enrichment analyses were used to compare FHB and FRR transcriptome responses in both Bd and Fg. Differential expression of selected genes was confirmed using RT-qPCR. Most genes associated with receptor signalling, cell-wall modification, oxidative stress metabolism, and cytokinin and auxin biosynthesis and signalling genes were generally upregulated in FHB or were downregulated in FRR. In contrast, Bd genes involved in jasmonic acid and ethylene biosynthesis and signalling, and antimicrobial production were similarly differentially expressed in both tissues in response to infection. A transcriptome analysis of predicted Fg effectors with the same infected material revealed elevated expression of core tissue-independent genes including cell-wall degradation enzymes and the gene cluster for DON production but also several tissue-dependent genes including those for aurofusarin production and cutin degradation. This evidence suggests that Fg modulates its transcriptome to different tissues of the same host.
Introduction
Fusarium graminearum is a destructive plant pathogen that can cause the economically important disease Fusarium Head blight (FHB) in suitable Poaceae hosts like bread wheat (Triticum aestivum), barley (Hordeum vulgare), and rye (Secale cereale) (Parry et al., 1995). FHB disease is characterised by lesions in the floral bract and caryopsis caused by extensive cell death (Boddu et al., 2006; Lewandowski et al., 2006). Over time discoloration of the peduncle and bleaching of the inflorescence occur and infected florets will produce small and shrivelled kernels due to the destruction of starch and protein (Goswami and Kistler, 2004; Guenther and Trail, 2005). During FHB, F. graminearum behaves as a facultative hemibiotroph with a necrotrophic phase preceded by a biotrophic phase (Jansen et al., 2005; Brown et al., 2010). Most wheat plant tissues are susceptible to infection (Miedaner, 1997), and other Fusarium diseases include Fusarium Crown Rot (FCR), Fusarium Root Rot (FRR), and seedling blight. FRR causes necrosis of root tissue leading to reduced root, shoot length, biomass, and yield loss (Mergoum et al., 1998; Beccari et al., 2011; Wang et al., 2015). During FRR penetration, sporulation, and necrosis occur rapidly (Wang et al., 2015), and systemic migration of the pathogen via the vascular system can result in FCR and, in extreme cases, to FHB (Beccari et al., 2011; Wang et al., 2015). Due to the inherent difficulty in studying root diseases, much less is known about FRR than FHB.
Apart from affecting grain yield and quality of the cereal crop, F. graminearum can synthesise deoxynivalenol (DON), and nivalenol (NIV) mycotoxins that contaminate grain and pose a risk to human and animal consumers (Antonissen et al., 2014; Payros et al., 2016).
Fusarium graminearum is known to utilise a combination of cell wall-degrading enzymes (CWDEs) and DON to overcome host defences in small-grain cereals. Infection is also accompanied by an increase in secreted effectors, changes to pathogen molecular transport and signalling, and changes to secondary metabolite and nutrient metabolism (Kikot et al., 2009; Lysøe et al., 2011; Brown et al., 2017). Cereal hosts have been shown to respond to FHB through the deployment of numerous defences including deoxynivalenol (DON) detoxication, host metabolism changes, cell wall development changes, and synthesis of antimicrobial compounds and PATHOGENESIS-RELATED (PR) proteins (Boddu et al., 2006; Jia et al., 2009; Pasquet et al., 2014).
Transcriptomics is an effective tool to investigate molecular responses during pathogenesis. Several studies have investigated F. graminearum-induced changes in gene transcription during infection of wheat and barley (Hordeum vulgare) using methods such as microarray analysis and real-time quantitative PCR (RT-qPCR) (Boddu et al., 2006; Li and Yen, 2008; Jia et al., 2009). More recently, RNA-seq has been utilised to study wheat responses to FHB and FCR (Powell et al., 2017a; Pan et al., 2018; Wang L. et al., 2018). Likewise, several studies have investigated the transcriptome of F. graminearum during Fusarium head blight (FHB) disease of small grain cereals, in most cases through microarray analysis (Lysøe et al., 2011; Harris et al., 2016; Brown et al., 2017). RNA-seq technology has been used to-date, in terms of F. graminearum disease transcriptomics, for FRR (Ding et al., 2022) and FHB (Pan et al., 2018).
Of the canonical defence phytohormones, the consensus is that salicylic acid (SA) - regulated responses are associated with resistance to biotrophic pathogens whereas jasmonic acid (JA) and ethylene associated responses are linked to resistance to necrotrophic pathogens (Glazebrook, 2005; Bari and Jones, 2009; Pieterse et al., 2012). As a result of the hemibiotrophic lifestyle of F. graminearum, SA and JA/ethylene pathways have both been shown to be important for defence (Makandar et al., 2011; Ding et al., 2011; Wang L. et al., 2018). Many other phytohormones including abscisic acid (ABA), gibberellic acid (GA), auxin, and cytokinin have also been implicated in response to F. graminearum infection (Powell et al., 2017a; Pan et al., 2018; Wang L. et al., 2018). In a previous study we showed that exogenous application of many phytohormones induced significant effects on FHB and FRR resistance (Haidoulis and Nicholson, 2020). However the phytohormones SA, JA, and ethylene induced opposing effects on resistance to FHB and FRR whereas cytokinin and auxin induced similar effects in the two tissues.
Brachypodium distachyon (purple false brome) is a temperate monocotyledonous plant in the Pooideae sub-family. Similar to wheat, both B. distachyon roots and florets can be infected by F. graminearum infection which permits the investigation of both FHB and FRR in this host (Peraldi et al., 2011; Pasquet et al., 2014).
It is unclear whether F. graminearum differentially expresses genes and deploys tissue-specific effectors within the same host. The aim of the present study is to compare the transcriptomes of both host (B. distachyon) and pathogen (F. graminearum) during early FHB and FRR infection to identify similarities and differences related to the host tissue. We believe this to be the first study to examine both host and pathogen gene expression within different tissues in the same host. The data reveals that gene expression of B. distachyon defence and F. graminearum virulence occur in both a tissue-specific manner and non-specific manner suggesting that there are core processes alongside bespoke tissue-related responses in both host and pathogen.
Materials and methods
Plant and fungal material and growth conditions
The B. distachyon accession Bd3-1 was obtained from the John Innes Centre, Norwich, UK. B. distachyon seed preparation and growth conditions were the same as in (Haidoulis and Nicholson, 2020) except for a three day seed stratification for root assay.
The F. graminearum isolate PH1 was obtained from the John Innes Centre, Norwich, UK and used for all experiments. For the FRR assay mycelium inoculum and FHB conidial suspension, F. graminearum was prepared as in (Haidoulis and Nicholson, 2020). The F. graminearum in vitro control samples (1 x 104 conidia/ml) were grown in sterile Czapek-Dox liquid medium with 1 unit/ml penicillin-streptomycin for four days and incubated in a shaker at 25°C and 200 rpm.
Sample inoculation and preparation
For FRR assays, ten stratified seeds were placed on 9 cm2 filter paper square on a 50 ml 0.8% agar (Fischer Science) in square Petri-dishes. All plates were angled at 70° from the horizontal in a plant propagator tray and incubated for three days at 22°C (16h/8h - light/dark photoperiod) before inoculation. Mycelial slurry was prepared from blended seven-day old mycelium from a PDA Petri-dish with 1 ml sterile deionised water. After three days, Bd3-1 roots were inoculated at three positions per root with a mycelial PDA slurry or PDA slurry using a 10 ml syringe. After one day (1 dpi), the inoculum slurry was removed, and roots were rinsed with water (Supplementary Figure S1). For each biological replicate, ten roots were cut and frozen in liquid nitrogen. Three biological replicates were obtained for each treatment.
For FHB assays, Bd3-1 spikelets were sprayed at mid-anthesis with conidial inoculum (1 x 106 conidia/ml) in sterile distilled water before the dark photoperiod. Both inoculum suspension and water control were amended with 0.05% Tween 20. Plants were incubated at 22°C for 3 d at high humidity. After three days (3 dpi), for each biological replicate, three infected and mock inoculated spikelets from randomly selected plants were cut and immediately frozen in liquid nitrogen (Supplementary Figure S1). Three biological replicates were obtained for each treatment.
RNA was extracted from both spike and root tissues using QIAGEN RNAeasy kit as per the manufacturers protocol. RNA was then cleaned using Turbo DNA-free kits as per standard protocol with two rounds of Turbo DNAse treatment. RNA samples were quantified and quality checked using a Qubit and TapeStation (performed by Genewiz).
Library preparation RNA-seq and analysis
Library preparation was performed at Genewiz and sequenced using Illumina HiSeq, PE 2x150bp sequencing configuration with a single index per lane. RNA-seq Illumina reads FASTA data, obtained from Genewiz, were analysed on the Galaxy platform (Afgan et al., 2016). FastQC (G.V.0.72) was used for sample FASTA reads as a quality check. Using Trimmomatic (G.V.0.36.5), paired-end FASTA reads were trimmed with the settings: ‘Sliding window’ (4 bases), ‘leading’ and ‘trailing’ ends (3 bases each), and TrueSeq3 Illumina clip was used to remove Illumina adaptor sequences. Trimmed FASTA reads were quality checked again with FASTQC (G.V.0.72). Trimmed FASTA reads were aligned to the most recent version of the Bd assembly (Bd21 JGI v3.0)(Phytozome JGI V12.1.5, (Initiative, 2010; Goodstein et al., 2012)) using HISAT (G.V.2.10). Gene annotations were assigned using Stringtie v3.1 (G.V.1.3.4) with annotations (Phytozome JGI, B. distachyon v3.1 (Initiative, 2010; Goodstein et al., 2012)). Stringtie gene counts for FHB and FRR were differentially compared to respective control samples with DEseq2 (G.V.2.11.40.2).
The predicted functions for B. distachyon genes were obtained from Ensembl Genomes (Howe et al., 2020), UniProt (Consortium, 2018), BrachyPan (Goodstein et al., 2012), (Gordon et al., 2017), B. distachyon v3.1 from Phytozome JGI (V12.1.5) (Initiative, 2010; Goodstein et al., 2012), and (Kakei et al., 2015; Kouzai et al., 2016). The predicted gene functions within the heatmaps with a prefix and percentage homology denotes the percentage of B. distachyon sequence that matches the orthologous sequence obtained from Ensembl Genomes (Howe et al., 2020), were derived from the Arabidopsis Information Resource (TAIR10) database (Berardini et al., 2015), or from previous studies (Yazaki et al., 2004; Jain et al., 2005; Jain et al., 2006b; Jain et al., 2006a; Tsai et al., 2012). Then, if necessary, the B. distachyon homologue/s were identified within the Ensembl Genomes database (Howe et al., 2020) (At: A. thaliana TAIR10, Os: O. sativa RGSP-1.0, Hv: H. vulgare IBSC_v2, or Zm: Z. mays B73_RefGen_v4) and were then searched for within the RNA-seq dataset.
The same pipeline described above was used for F. graminearum reads with the same samples but aligned to the F. graminearum PH1 genome assembly and gene annotation (European Nucleotide Archive; GCA_900044135.1, study PRJEB5475 (King et al., 2015)). FHB and FRR sample gene counts were separately compared against the same F. graminearum in vitro control samples using DEseq2. The average of normalised gene counts per biological replicate from FHB and FRR DEseq2 is presented. Differentially expressed F. graminearum genes in FHB and FRR were filtered for potential effectors using the PH1 v5.0 secretome prediction script (Brown et al., 2012). The online databases Ensembl Genomes (Howe et al., 2020), UniProt (Consortium, 2018), and protein sequence BLAST (Sayers et al., 2020) were used to predict F. graminearum gene functions.
Time-course RT-qPCR
FHB and FRR samples were prepared as described before. Samples from different plants were harvested at 3 dpi, 5 dpi, and 7 dpi for FHB and 1 dpi, 3 dpi, and 5 dpi for FRR, with at least three biological replicates per treatment and time-point. For F. graminearum genes, only one time point was used for the in vitro control which was subsequently compared to all three infected FHB and FRR time-point samples. After sample RNA extraction using QIAGEN RNAeasy kit as per the manufacturers protocol and DNase treatment as described before, first strand synthesis of RNA was performed with Invitrogen SuperScript III Reverse Transcriptase (Invitrogen) as per the manufacturers protocol. Reverse transcriptase qPCR was performed with 2 µl cDNA, 5 µl of 2x SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich), 0.6 µl of 10 µM for each primer (Supplementary Tables S1, S9), in a final volume of 10 µl. Two to three technical replicates were analysed for each gene, biological replicate, and time-point combination. RT-qPCR reactions were prepared in a Framestar-480/384 well plate with BioRAD microseal B adhesive film. Thermocycling was carried out on a Roche LightCycler LC480 on SYBR green 1 scan mode with the following parameters: 300 s 95°C, 45 x (94°C 10 s, 58°C (or 60°C for Bradi1g57590) 10 s, 72°C 10 s, 75°C 2 s (single acquisition)) and subsequently analysis of dissociation by ramping 95°C 5s, 60°C 60 s, 97°C (continuous) followed by 40°C for 30 s. LC480 raw data was converted with LC480 conversion software and analysed for primer efficiency and Cq values with the LinRegPCR tool (Ruijter et al., 2009). Log fold changes were calculated from Cq values using the following equations: Δ Cq = (Gene of interest Cq – housekeeping gene Cq), ΔΔ Cq = (Infected treatment Δ Cq – mock treatment Δ Cq), Log2 fold change = Log2 (Primer efficiencyΔΔ Cq) (Pfaffl, 2001). ΔΔ Cq values with respective SE bars for each gene and treatment are listed in Supplementary Table S2 and Table S10.
All primers (Sigma Aldrich UK. Primers) for gene targets (Supplementary Table S1 and Table S9), unless otherwise stated, were designed using Primer 3 (Koressaar and Remm, 2007; Untergasser et al., 2012; Kõressaar et al., 2018) on a single CDS exonic region and avoiding untranslated (UTR) regions. The best housekeeping gene GAPDH (Supplementary Table S1) for these samples was experimentally determined and analysed on NormFinder in GenEx V6 using cDNA obtained from both control and infected root and spike material.
Statistics and software
The RNA-seq p-value and p-adj values were outputs from Galaxy DEseq2. A standard Student’s t-test on Microsoft Excel was used for time-course RT-qPCR Cq data. Heatmaps were prepared on R studio using ‘pheatmap’ and ‘rcolorbrewer’ package. The normalised transcript counts (reads) were transformed (Log2 (x + 1)) and then scaled per gene (row). Hierarchical clustering of genes (rows) used Euclidean distance metric with complete-linkage clustering. Volcano plots were prepared on R studio using the packages ‘ggplot2’, ‘ggrepel1’, and ‘EnhancedVolcano’ (Blighe et al., 2019). Venny V2.1 was used for Venn Diagrams and sorting treatment groups (Oliveros, 2018). Graphs for time-course experiment were prepared on GraphPad Prism V5. Gene ontology enrichment for B. distachyon was achieved using The Gene Ontology Resource (Database released 2021-08-18) and PANTHER Overrepresentation Test (Released 20210224) (Ashburner et al., 2000; Mi and Thomas, 2009; Mi et al., 2019; Mi et al., 2021; The Gene Ontology Consortium, 2021; Thomas et al., 2022). Gene Ontology enrichment plots were produced as described in (Supek et al., 2011; Bonnot et al., 2019). Fisher’s exact test type with FDR correction was used for GO enrichment and default REVIGO settings were used. Gene-list enrichment for Fusarium graminearum was performed using KOBAS 3.0 (Wu et al., 2006; Xie et al., 2011; Ai and Kong, 2018).
Results
Fusarium head blight and fusarium root rot display distinct global transcriptome responses to infection
The time points 3 days-post inoculation (dpi) for FHB and 1 dpi for FRR represent the earliest stage at which symptoms were visible for the two diseases. Differential gene expression analysis was performed on total gene counts of diseased B. distachyon floral and root tissues in comparison to respective mock-inoculated treatments. Coverage to the B. distachyon (accession Bd21) assembly was between 80-95% coverage. With no log-fold change threshold, there were 6,158 genes significantly differentially expressed in response to FHB (p-adj < 0.05) (Figure 1A), whereas 8,568 genes were significantly differentially expressed in response to FRR (p-adj < 0.05) (Figure 1B). Approximately 17% of the genes significantly differentially expressed in response to FRR exceeded the 2 Log-fold change threshold, whereas 29% of the genes significantly differentially expressed in response to FHB exceeded the 2 Log-fold change threshold.
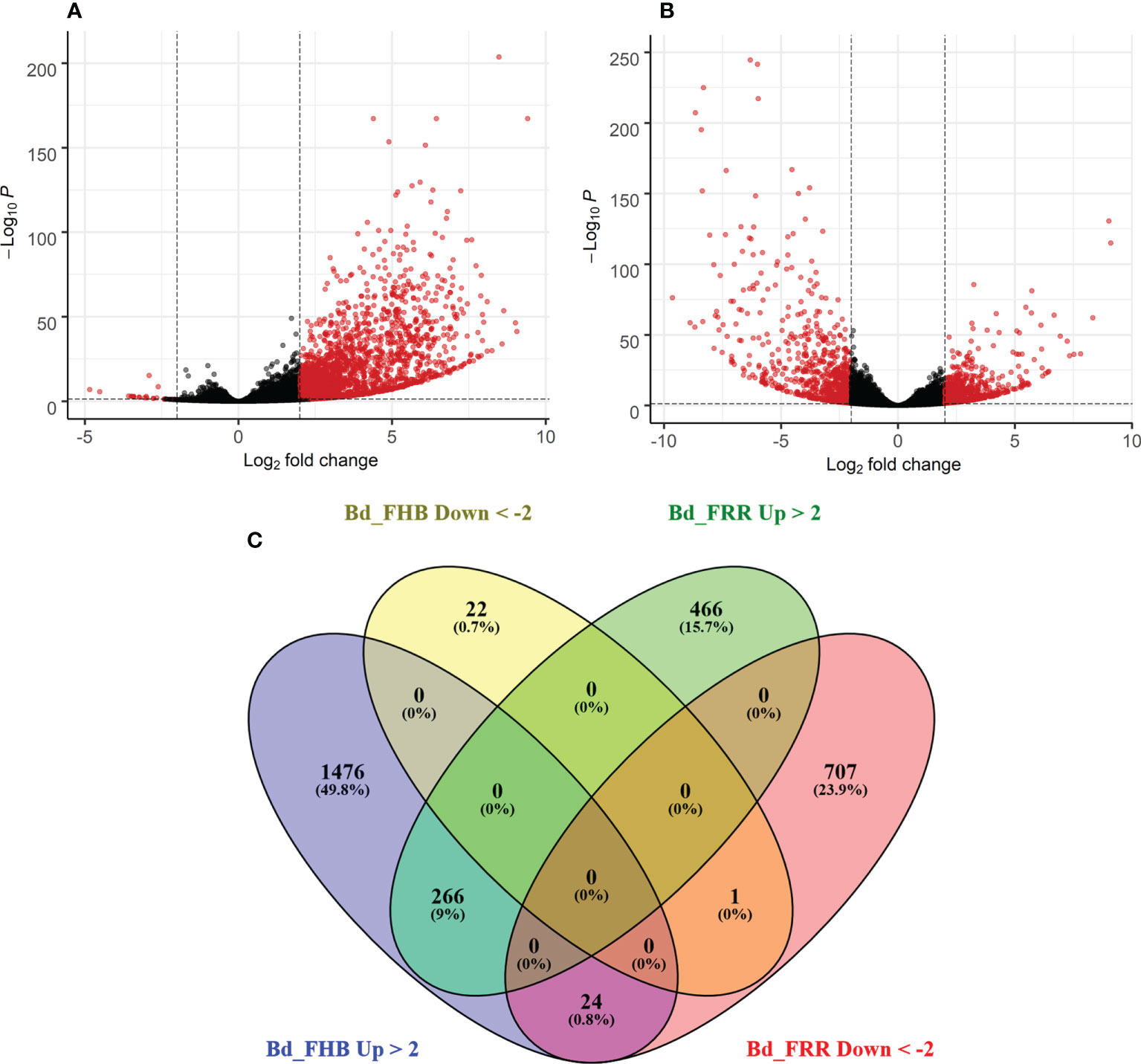
Figure 1 Summary of expression of B. distachyon genes in response to FHB and FRR. (A) Differential gene expression in response to FHB. (B) Differential gene expression in response to FRR. (A, B) Each dot represents a gene (Supplementary Table S3), excluding those without a p-adj value. The y-axis is the -Log10 of the p-adj value (p-adj < 0.05) denoted by a dotted line. The x-axis is the Log2 fold change with a cut-off of 2 denoted by two dotted lines. Thus a red dot denotes that a gene was statistically significantly differentially expressed. (C) A comparison of genes between FHB and FRR datasets. The threshold of -2 ≤ x ≥ 2 Log-fold change and p-adj < 0.05 was applied to all genes. Abbreviations: Bd (B. distachyon), Up (Upregulated), Down (Downregulated), FRR (Fusarium Root Rot), FHB (Fusarium Head Blight).
The transcriptome response between FHB and FRR was compared (Figure 1C). Following FHB infection the number of upregulated genes (1766) was much greater than the number of downregulated genes (23) (Figure 1A). In contrast, similar numbers of genes were upregulated and downregulated in response to FRR (Figure 1B). There were relatively few genes that were upregulated (226) or downregulated (1) in response to both FHB and FRR. There were more genes exclusively upregulated (466 genes) and downregulated (707 genes) in response to FRR but the most pronounced difference was observed for FHB where 1,476 genes were exclusively upregulated. In contrast, only 22 genes were exclusively and significantly downregulated in response to FHB. A small group of 24 genes were upregulated in response to FHB and downregulated in response to FRR (Figure 1C). Of these 24 (Supplementary Table S4), notable genes include a xyloglucan endotransglucosylase (Bradi3g31767), a pathogenesis-related protein 1 (Bradi3g53681), a disease resistance protein RPP13-related (Bradi1g29381), a peroxidase (Bradi5g27150), an endoglucanase (Bradi3g36210), a RING-type E3 ubiquitin transferase (Bradi3g52120), and an expansin (Bradi3g09960).
Gene ontology enrichment analysis reveals differentially expressed pathways in B. distachyon
Gene Ontology (GO) enrichment analysis was performed on the different treatment groups: FHB upregulated, FRR upregulated, and FRR downregulated (Figure 2). A total of 67 GO biological processes were significantly overrepresented in one or more of the treatment groups (Figure 2). Most GO terms were overrepresented in a tissue-specific manner. For overrepresented FHB-specific processes, notable GO terms were receptor signalling (GO:0007178, GO:0007167, GO:0007166), defence/immune response and response to microorganisms (GO:0009611, GO:0031347, GO:0002376, GO:0042742, GO:0002239, GO:0002229, GO:0009607, GO:0044419, GO:0006950), Glutathione metabolic process (GO:000674), indole compound biosynthesis and metabolism (GO:0042435, GO:0042430), jasmonic acid signalling (GO:2000022), oxylipin biosynthesis and metabolism (GO:0031408, GO:0031407), phenylpropanoid metabolism (GO:0009698), and phosphorylation-associated processes (GO:0000160, GO:0006468, GO:0016310, GO:0006793) (Figure 2). The pathways for response to stimulus and stress (GO:0050896, GO:0006950), and phosphorylation-associated processes (GO:0006468, GO:0016310, GO:0006793) showed the greatest number of significantly expressed genes expected in the pathway for the GO analysis (Figure 2). There were insufficient genes and no significant biological processes for the FHB-downregulated gene list. The GO pathways: cellular response to chemical stimulus (GO:0070887) and tryptophan biosynthesis (GO:0000162), were enriched exclusively in FRR upregulated genes (Figure 2). Only three GO processes were similarly overrepresented within the two tissues from upregulated gene: response to ethylene (GO:0009723), the ethylene-activated signalling pathway (GO:0009873) and aromatic acid family metabolic process (GO:0009073) (Figure 2). There were far more GO processes for downregulated FRR differentially expressed genes than upregulated genes, notably an overrepresentation in cellular oxidant detoxification (GO:0098869), cell wall modification (GO:0071669, GO:0046274, GO:0071554, GO:0006073, GO:0045229), fluid and water transport (GO:0042044, GO:0006833), response to toxic substance (GO:0009636), and carbohydrate metabolism (GO:0006073, GO:0044262, GO:0005976, GO:0005975) (Figure 2). The pathway biological process (GO:0008150) showed the greatest number of significantly expressed genes expected in the pathway for the GO analysis (Figure 2). Three GO processes for response to oxidative stress (GO:0006979), ROS metabolism (GO:0072593), and hydrogen peroxide detoxification (GO:0042744) were overrepresented in both upregulated and downregulated FRR-differentially expressed genes. These were among the most significant and greatest fold enrichment for downregulated processes (Figure 2).
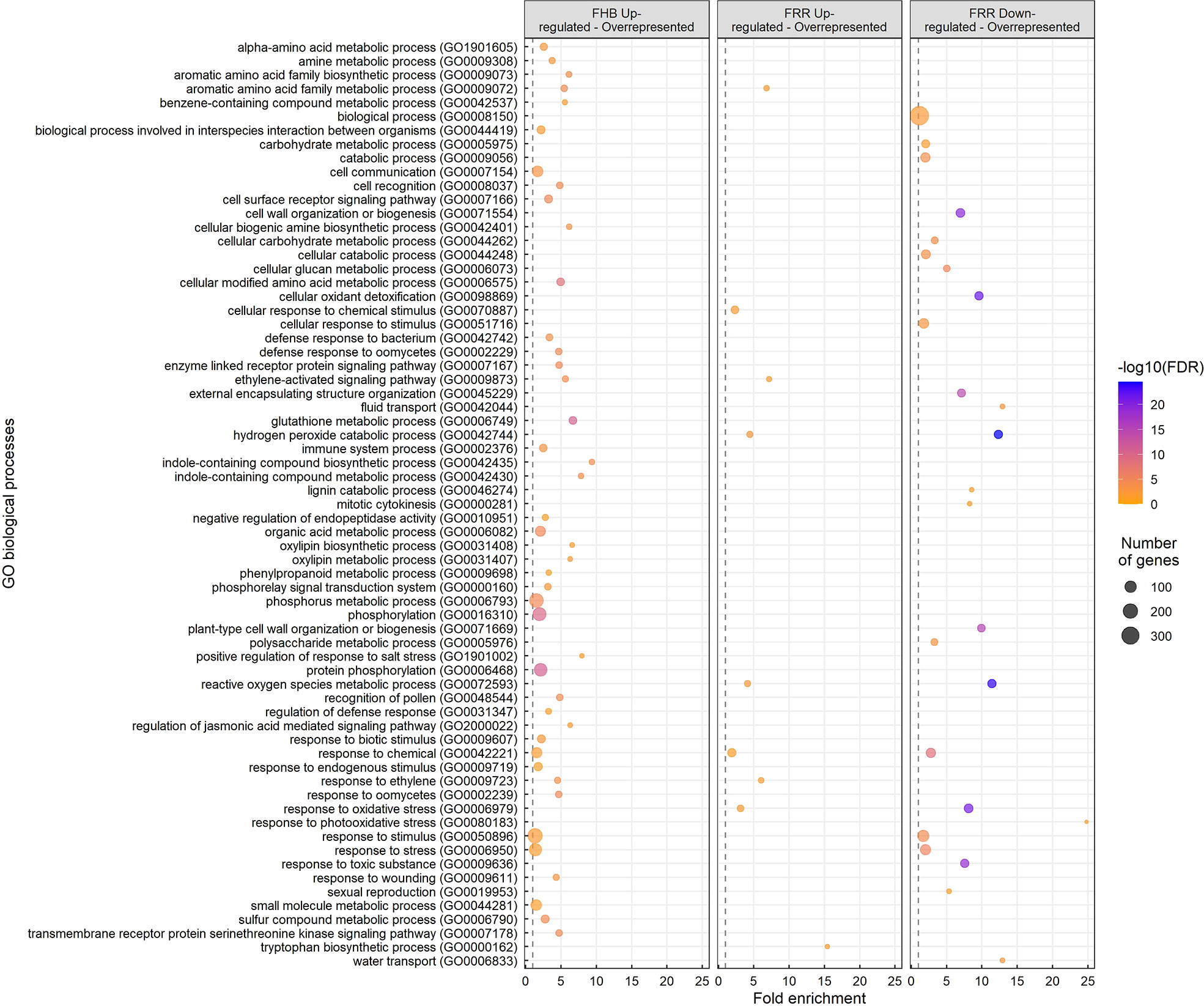
Figure 2 Overrepresented biological processes from significantly upregulated and downregulated genes in FHB and FRR. The dot size represents the number of genes within the process and the dot colour represents the enrichment significance (-log10 (False Discovery Rate (FDR) - corrected p-value)). The vertical dotted lines denote a fold enrichment threshold of 1 signifying that the biological processes are overrepresented. Data in Supplementary Table S5.
Several gene groups were differentially expressed in response to FHB and FRR
Significantly upregulated or downregulated B. distachyon genes (p-adj < 0.05) were grouped based on predicted function and compared between FHB and FRR. Differential expression was defined as having a log-fold threshold more than 2-fold or less than -2-fold change, and a p-adjusted value of less than 5% significance level (p < 0.05). Several genes were found to encode similar products and function within the same role. These were grouped and described in the following sub-sections, focusing on pathways that have roles in plant-pathogen interactions and defence.
Phytohormone-related genes
Phytohormones play important roles in Fusarium resistance. Exogenous application of JA, ethylene, auxin, and cytokinin was shown to significantly affect resistance to both FHB and FRR in B. distachyon (Haidoulis and Nicholson, 2020). Here we add to these results and show that phytohormone-related changes in response to infection also occur at the transcriptional level. In a similar fashion, based on predicted-function of genes, the phytohormones JA, ethylene, auxin, cytokinins, and to a lesser extent (ABA and SA) were the main phytohormones significantly altered in transcription to FHB and FRR in the RNA-seq data (Supplementary Table S6). The transcription of genes related to phytohormones SA, auxin, cytokinin, and ABA were mostly differentially expressed in FHB and FRR. Most SA and ABA – related genes were upregulated in FHB. Most auxin and cytokinin – related genes were downregulated in FRR and were either upregulated or not expressed in FHB. In contrast, ethylene, and to a lesser extent JA-related, transcription tended to be similar in FHB and FRR.
There were 20 jasmonic acid-related genes differentially expressed between FHB and FRR (Figure 3A). Only the JA-repressor JAZ genes (Bradi3g23190, Bradi4g31240), and the JA biosynthesis lipoxygenase LOX2 gene Bradi3g39980 were upregulated in both FHB and FRR (Figure 3A) while the remaining five JAZ genes increased in expression only in FHB (Figure 3A). Furthermore, all the biosynthetic OPR genes (Bradi1g05870, and Bradi1g05860), lipoxygenase genes, and 4CL were exclusively upregulated in FHB (Figure 3A). In contrast, the lipoxygenase genes Bradi1g09260 and Bradi1g11680, and Jasmonate O-methyltransferase (Bradi1g43080), were downregulated in response to FRR (Figure 3A). Overall, JA-biosynthetic and signalling genes appeared primarily FHB-responsive. Interestingly there were differences in basal expression of several JA-related genes in the two tissues. The genes Bradi3g43920, Brad1g72610, and Bradi4g20220 had much higher transcript counts in non-inoculated spikes as opposed to roots while all OPR genes, including Bradi1g09260, Bradi5g08650, Bradi5g11590, and Bradi3g37300 had much higher transcript counts in non-inoculated roots as opposed to spikes (Figure 3A).
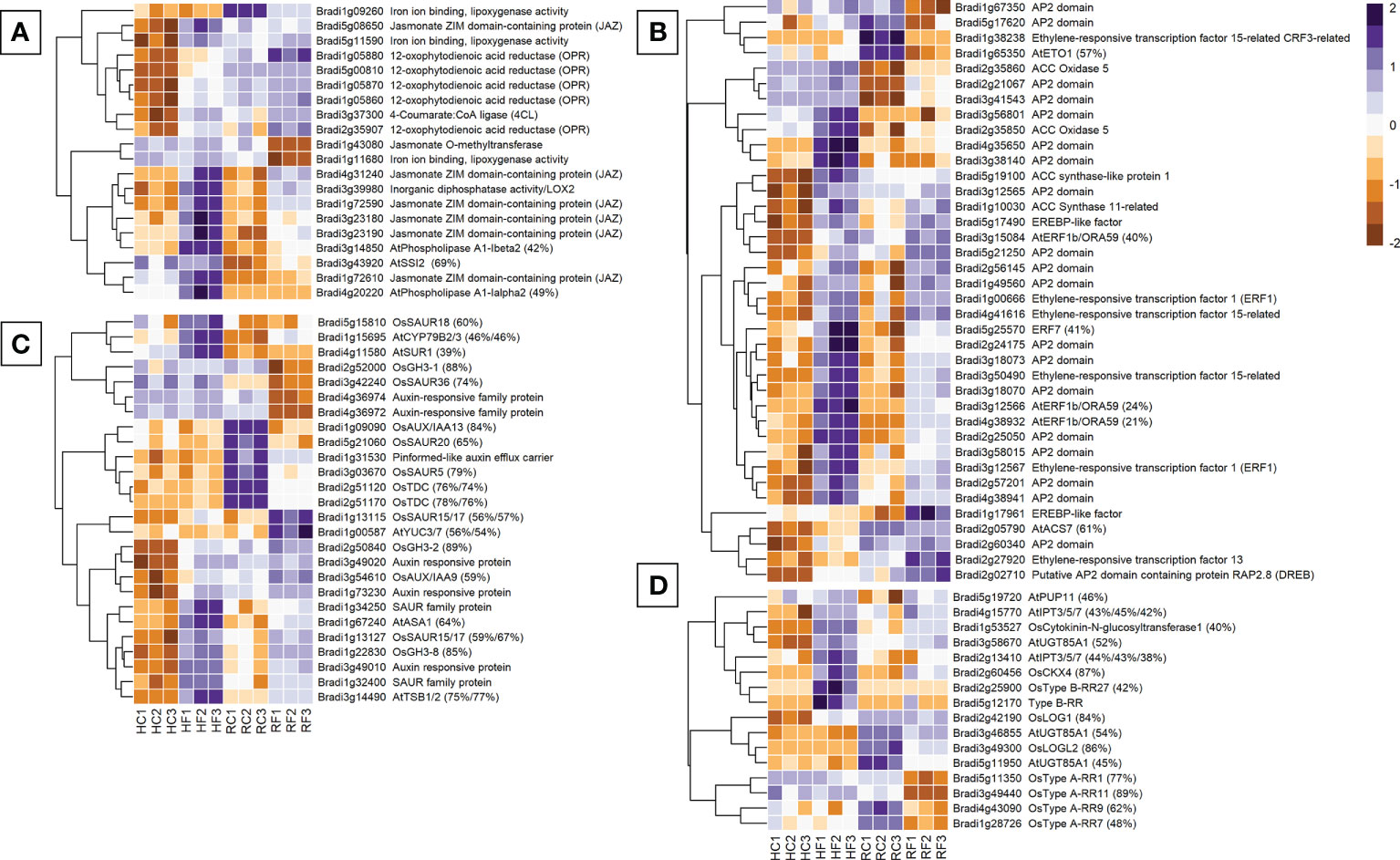
Figure 3 The most expressed or repressed phytohormone-associated B. distachyon genes. JA (A), ethylene (B), auxin (C), and cytokinin (D) related genes are displayed. These genes showed a log-fold change of -2 ≤ Log2 ≥ 2 with a p-adj < 0.05 in response to either FHB, FRR, or both. The scale bar on the right is the Z-score for all the heatmaps. Some gene functions have a prefix denoting that they were derived from A. thaliana (At) or O. sativa (Os) and they include the percentage of B. distachyon sequence that matches the homologous sequence. Three biological replicates for each of the four treatments are displayed as columns abbreviated as HC (Head-FHB control), HF (Head-FHB fungus), RC (Root-FRR Control), RF (Root-FRR fungus). The control samples (HC1-HC3, RC1-RC3) are normalised transcript counts from mock inoculated head (water with Tween 20) and root (PDA slurry) B. distachyon tissues and were separately analysed through the RNA-seq pipeline with the respective inoculated sample tissues.
Ethylene-associated genes were the largest phytohormone-related group expressed for FHB and FRR (38 genes) (Figure 3B). As for JA, most of the ethylene biosynthesis genes encoding ACS and ACO genes were upregulated in response to FHB (Bradi5g19100, Bradi2g35850, Bradi1g10030, Bradi2g05790). The biosynthesis regulator Bradi1g65350 (orthologues to AtETO1) was exclusively downregulated in response to FRR. Conversely, ACO5 (Bradi2g35860) was upregulated in response to FRR. Differential expression of many ethylene-associated genes was just below the threshold (Log-fold change > 2) for FRR (Supplementary Table S6). Most ethylene-associated genes encoded well known downstream transcription factors such as ERF1/ORA59 or other less characterised genes encoding proteins with AP2/EREBP domains (Broekaert et al., 2006). The majority showed similar expression patterns in response to FHB and FRR including ERF1 (Bradi3g12567) and ERF1b/ORA59 (Bradi4g38932). The genes ACS7 (Bradi2g05790), ERF15 (Bradi1g38238), and ETO1 (Bradi1g65350) had substantially higher basal expression in non-inoculated root tissues compared to non-inoculated spike tissues.
Expression of many of the 26 auxin-associated genes identified, increased in FHB but the response in FRR was mixed (Figure 3C). Auxin biosynthetic orthologues were mainly upregulated in FHB (AtSUR1 Bradi4g11580, AtTSB1/2 Bradi3g14490). However, while some were also upregulated (AtASA1 Bradi1g67240, AtYUC3/7 Bradi1g00587, AtCYP79B2/3 Bradi1g15695) others were downregulated in FRR (OsTDC, Bradi2g51120 Bradi2g51170). The biggest differences in response between FHB and FRR were for genes downstream of auxin biosynthesis. GH3 orthologues with known roles in resistance (Ding et al., 2008; Domingo et al., 2009; Fu et al., 2011) were differentially expressed with OsGH3-8 (Bradi1g22830) upregulated in both FHB and FRR, OsGH3-2 (Bradi2g50840) expressed in FHB only, and OsGH3-1 (Bradi2g52000) downregulated only in FRR. The auxin-response factor (ARF) genes Bradi3g49010, Bradi3g49020, and Bradi1g73230 were upregulated in FHB whereas Bradi4g36974 and Bradi4g36972 were downregulated in FRR. The response of SAUR genes was also highly variable. Bradi1g13127 was upregulated in both FHB and FRR, Bradi5g15810 was upregulated only in FHB, while Bradi5g21060 and Bradi3g42240 were downregulated only in FRR. Lastly, only one auxin transport gene was repressed (Pin-efflux carrier, Bradi1g31530) in FRR.
Rice orthologues were also used to identify cytokinin associated genes in B. distachyon (Tsai et al., 2012). From the total of 16 cytokinin-associated genes identified, almost all biosynthetic genes were upregulated in FHB (e.g. AtUGT85A1 Bradi3g58670, AtIPT3/5/7 Bradi2g13410, AtIPT3/5/7 Bradi4g15770, and OsLOG1 Bradi2g42190). In contrast many biosynthetic genes were downregulated in response to FRR (OsLOGL2 Bradi3g49300, AtUGT85A1 Bradi3g46855, AtUGT85A1 Bradi5g11950). The same was true for downstream signalling Response Regulator (RR) genes. All OsType B RR homologues were upregulated in FHB (Bradi2g25900, Bradi5g12170), whereas all OsType A RR homologues were significantly downregulated in FRR (Bradi4g43090, Bradi5g11350, Bradi3g49440, Bradi1g28726).
Salicylic acid (SA) is an important phytohormone involved in resistance to biotrophs and generally functions antagonistically to JA (Glazebrook, 2005; Bari and Jones, 2009; Pieterse et al., 2012). The Bd homologues of SA-responsive genes OsNPR4 (Bradi2g54340), NPR1 interacting (Bradi2g27670), AtCBP60g (Bradi4g05360), AtSARD1 (Bradi2g02310), AtGRX480 genes (Bradi2g08400 and Bradi2g46093) were exclusively upregulated in response to FHB (Supplementary Table S6). Likewise, several systemic acquired resistance (SAR)-associated genes MES1-encoding genes were upregulated in FHB (Bradi2g41070, Bradi3g44867, Bradi4g35382) whereas two were downregulated in FRR (Bradi2g52110 and Bradi4g09007). Bradi1g71530, orthologous to AtALD1, was also exclusively upregulated in FHB. Similarly, Bradi3g43920 (orthologous to AtSSI2 involved in SA and JA antagonism (Pieterse et al., 2012)) was upregulated exclusively in response to FRR (Figure 3A). Relatively few SA-related genes were significantly differentially expressed between FHB and FRR compared to the previous four phytohormones, but for the few that were, they were primarily upregulated in FHB as opposed to FRR.
Several ABA-associated genes were also identified as differentially expressed (Supplementary Table S6). The biosynthetic genes homologous to Arabidopsis genes AtNCED2/3/5/9 were generally upregulated in response to FHB (e.g. Bradi1g13760, Bradi1g51850, Bradi1g58580) whereas Bradi1g52740 (orthologous to AtAAO1/2/3/4) was downregulated in response to FRR. The homologue of the signalling gene AtAP2C1 was also upregulated in response to FHB. In contrast, downstream Bradi2g43056, Bradi2g60441, Bradi2g60490 (orthologous to homeostasis AtBG1 genes) were upregulated in response to both FHB and FRR. Overall, there was a substantial differential in expression of ABA biosynthesis and signalling genes between FHB and FRR. Very few GA and BR-associated genes were identified as being differentially expressed in response to either FHB or FRR (Supplementary Table S6).
Genes encoding pathogen sensing proteins
Receptor-related genes displayed one of the largest differences in expression between FHB and FRR. The majority were upregulated in FHB but either not expressed or downregulated in FRR. Those that were also upregulated in FRR did so at a much-reduced level relative to FHB (Figure 4A). Putative leucine rich receptor-like kinases (LRR-RLKs) (e.g. Bradi1g22650, Bradi4g11740, and Bradi2g19380), LRR-containing genes (e.g. Bradi3g55006, Bradi4g42828, and Bradi4g42839), and nucleotide binding leucine rich repeat (NBS-LRR) proteins (e.g. Bradi5g15560, Bradi1g56690, Bradi2g21360) were almost all exclusively expressed in FHB (Supplementary Table S7). Only the LRR-RLK Bradi1g32160 was significantly upregulated in FRR (Logfold > 2). The remaining LRR-RLKs were either not expressed or were downregulated in FRR (e.g. Bradi1g75430). LRR-RLKs can be grouped into specific classes (Rameneni et al., 2015; Liu et al., 2017). The LRR-RLK classes 8, 10b, 12, 14, and 15 were only expressed in FHB (except Bradi1g32160) (Figure 4A and Supplementary Table S7). On the other hand, LRR-RLK classes 7a, and 11 were exclusively downregulated in FRR. NBS-LRR-encoding genes were primarily upregulated in FHB and downregulated in response to FRR except for Bradi2g18840 which was highly expressed in both tissues (Figure 4A and Supplementary Table S7). Disease resistance rpp13-like proteins (Bradi1g29381, Bradi5g01167, Bradi4g21942 Bradi4g09247) showed different differential expression between FHB and FRR (Figure 4A and Supplementary Table S7). Many genes encoding wall-associated receptor kinase (WAK) galacturonan-binding genes (e.g. Bradi2g02440, Bradi2g02450, Bradi2g02537), WAK receptor-like proteins (e.g. Bradi2g17520, Bradi3g01170, Bradi3g39670), or WAK receptor-like protein kinases (e.g. Bradi3g39670, Bradi2g02470, Bradi2g17520) were exclusively upregulated in FHB (Supplementary Table S7). Increased expression of most of the WAK-related genes in FRR was only moderate [being below the Log Fold 2 threshold (Supplementary Table S7)].
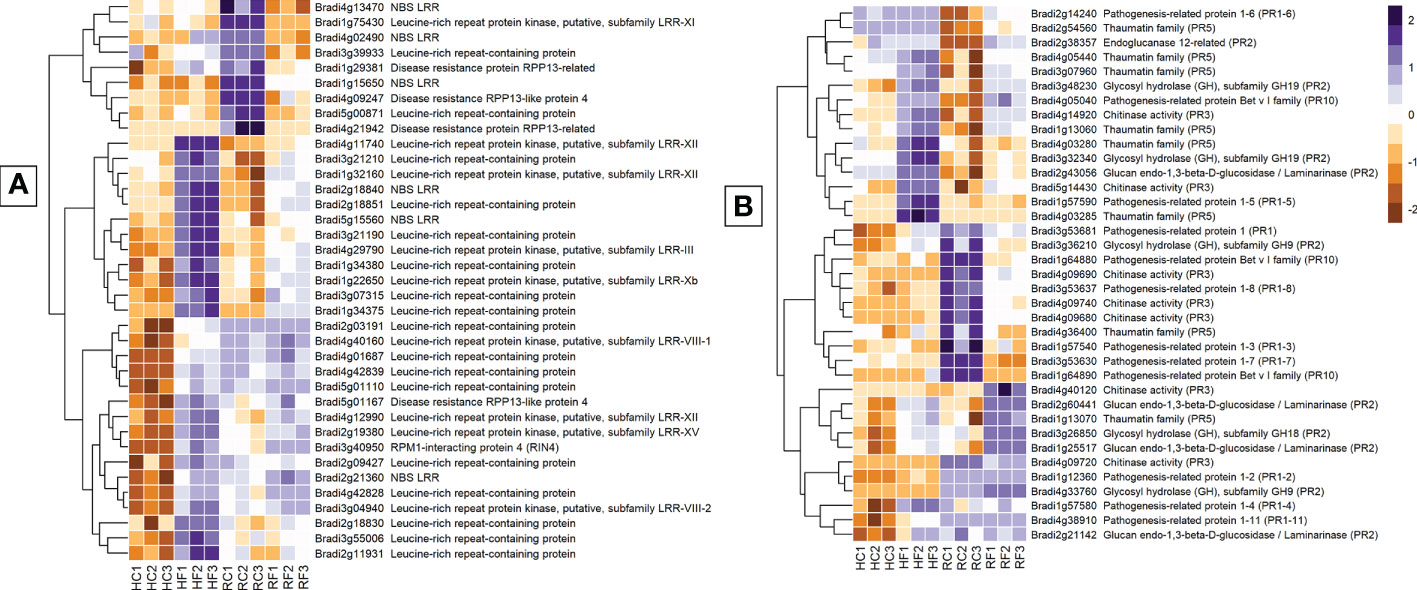
Figure 4 The most expressed or repressed receptor-related (A) and defence-associated (B) B. distachyon genes. These genes showed a log-fold change of -3 ≤ Log2 ≥ 3 (A) and -2 ≤ Log2 ≥ 2 (B) with a p-adj < 0.05 in response to either FHB, FRR, or both. A more restrictive Log-fold threshold is used for (A) due to a larger number of significantly expressed or repressed receptor-related genes. The scale bar on the right is the Z-score for all the heatmaps. Three biological replicates for each of the four treatments are displayed as columns abbreviated as HC (Head-FHB control), HF (Head-FHB fungus), RC (Root-FRR Control), RF (Root-FRR fungus). The control samples (HC1-HC3, RC1-RC3) are normalised transcript counts from mock inoculated head (Water with Tween 20) and root (PDA slurry) B. distachyon tissues and were separately analysed through the RNA-seq pipeline with the respective inoculated sample tissues.
Genes encoding PRs and other antimicrobial compounds
Pathogenesis-related (PR) genes are important constituents of resistance because of their antimicrobial properties (Stintzi et al., 1993). PR genes were either similarly or differently expressed between FHB and FRR depending on the PR class (Figure 4B). Most of the B. distachyon PR1 genes described were differentially expressed between FHB and FRR (Figure 4B). The PR gene Bradi3g53681 was upregulated in FHB but downregulated in FRR. Most PR genes identified (Figure 4B) were also categorised based on hormone responsiveness, with majority being responsive to JA (Kouzai et al., 2016). Glucanases (PR2) (e.g. Bradi2g60441) and chitinases (PR3) (e.g. Bradi5g14430) on the other hand were generally expressed in both tissues. However, there was a cluster of chitinase genes on chromosome 4 that were significantly downregulated in FRR only (Figure 4B, Supplementary Table S7). PR5 genes encode thaumatin and most PR5 genes were upregulated in FHB and/or FRR including Bradi1g13060 and Bradi1g13070. Bradi4g36400 was the only PR5 gene downregulated in FRR (Supplementary Table S7). PR10/Bet v 1 gene products are also involved in pathogen resistance (Agrawal and Agrawal, 2014). One PR10 gene (Bradi4g05040) was highly expressed in FHB and FRR while two PR10 genes (Bradi1g64890 and Bradi1g64880) were exclusively downregulated in response to FRR (Figure 4B).
Several other genes with predicted functions in secondary metabolite biosynthesis were identified (Supplementary Table S7). Those that were generally similarly differentially upregulated in both FHB and FRR include agmatine coumaroyltransferase (e.g. Bradi5g25166, Bradi3g23280, Bradi1g72220), phenylalanine ammonia lyase (e.g. Bradi3g47110, Bradi3g47120), tryptophan biosynthesis (Bradi1g55440, Bradi1g35600), and salt-stress/antifungal genes (Bradi1g25697, Bradi1g25552, Bradi5g03937) (Supplementary Table S7). There were, however, groups that showed different differential expression between FHB and FRR. These include flavonoid-related genes such as Bradi3g15700 and Bradi3g15690 that were highly downregulated in FRR. A cluster of six genes on chromosome 2 encoding Secologanin synthase-like genes were highly expressed in FHB but showed low or no expression in FRR (Supplementary Table S7). Lastly, one terpene related gene (Bradi3g35027) coding for alpha-humulene synthase was upregulated in FHB and downregulated in FRR (Supplementary Tables S4, S7).
Genes encoding proteins with structural and cell wall-modification functions
Genes involved in cell-wall modification were one of the most differentially expressed groups between FHB and FRR. Of all the structural genes, expansins showed the most substantial difference between FHB and FRR (Figure 5A). The majority of identifiable expansins were highly downregulated in FRR and were generally not expressed in FHB (Figure 5A, Supplementary Table S7). The most downregulated of these include Bradi5g04120 and Bradi3g27460. The downregulation of expansin genes in response to FRR may, in part reflect the very high read count of these genes in non-inoculated root samples (Figure 5A, Supplementary Table S7). Xyloglucan-related genes were also differentially expressed between FHB and FRR, for example, Bradi3g31767 was upregulated in FHB and highly downregulated in FRR (Figure 5A, Supplementary Table S7). Cellulose synthase related genes Bradi3g00491 and Bradi4g33090 were expressed in FHB whereas two others, Bradi3g34490 and Bradi1g25130, were downregulated in FRR (Figure 5A, Supplementary Table S7). Most pectin-associated genes involved in de-esterification of pectin (pectinesterase, pectinmethylesterase, pectinmethylesterase inhibitor) were also differentially expressed. Aside from the pectin methylesterase genes Bradi2g11850 Bradi3g24750, the remaining functionally related genes were not significantly differentially expressed in FHB. In contrast, all pectin-associated genes were either upregulated or downregulated in FRR. The lignin biosynthesis-related gene, cinnamoyl-CoA reductase (Bradi3g19670), was upregulated in both FHB and FRR (Figure 5A).
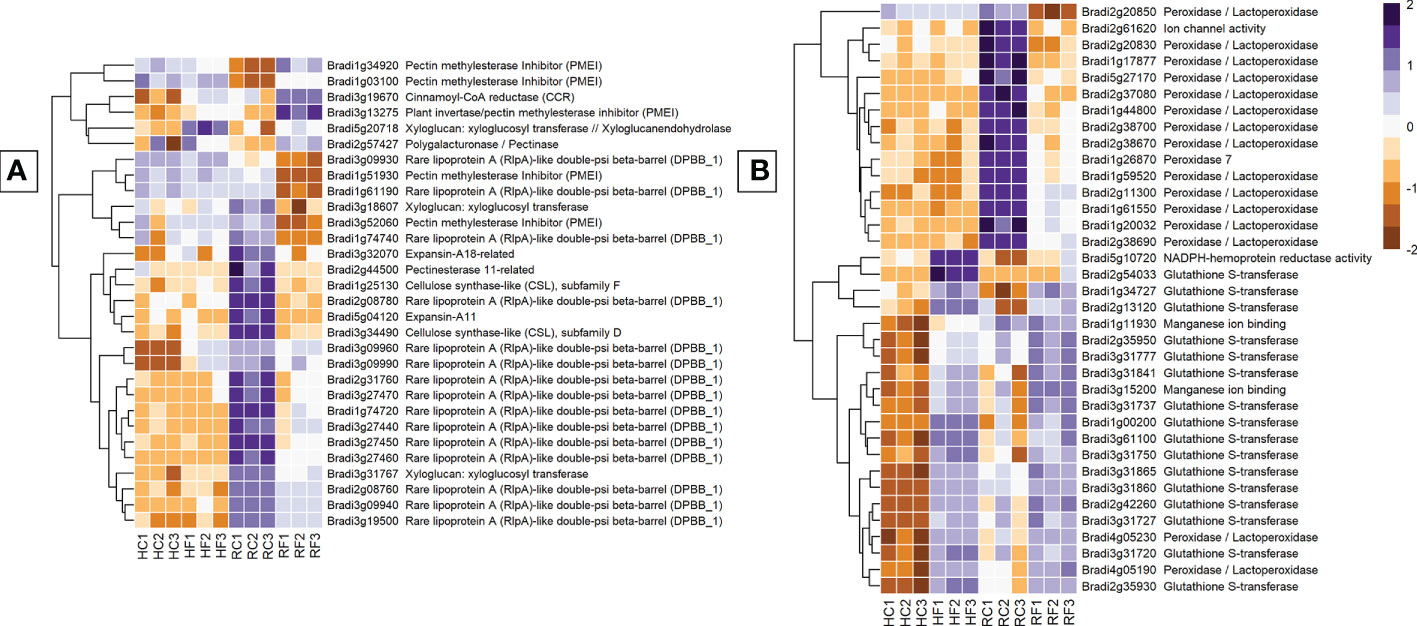
Figure 5 The most expressed or repressed cell-wall modification (A) and ROS-associated (B) B. distachyon genes. These genes showed a log-fold change of -3 ≤ Log2 ≥ 3 (A) and -5 ≤ Log2 ≥ 5 (B) with a p-adj < 0.05 in response to either FHB, FRR, or both. A more restrictive Log-fold threshold is used for (B) due to a larger number of significantly expressed or repressed ROS-associated genes. The scale bar on the right is the Z-score for all the heatmaps. Three biological replicates for each of the four treatments are displayed as columns abbreviated as HC (Head-FHB control), HF (Head-FHB fungus), RC (Root-FRR Control), RF (Root-FRR fungus). The control samples (HC1-HC3, RC1-RC3) are normalised transcript counts from mock inoculated head (Water with Tween 20) and root (PDA slurry) B. distachyon tissues and were separately analysed through the RNA-seq pipeline with the respective inoculated sample tissues.
Reactive oxygen species - associated genes
Genes associated with reactive oxygen species (ROS) and antioxidation were the most differentially expressed group of genes between FHB and FRR (Figure 5B). The number of peroxidase genes downregulated during early FRR was the most for any gene class. Out of 68 genes, 8 peroxidases (e.g. Bradi3g10460, Bradi1g26870, and Bradi2g09650) and 33 peroxidases/lactoperoxidases (e.g. Bradi1g17877, Bradi1g59520, and Bradi2g38690) were exclusively downregulated in FRR (Figure 5B, Supplementary Table S7). No peroxidase genes were downregulated in FHB (Log Fold > 2) and some were upregulated (Supplementary Table S7). The difference in response of expression of peroxidases between FHB and FRR may be partly explained by the very high read count of peroxidase genes in non-inoculated root samples (Figure 5B, Supplementary Table S7). Several glutathione-associated genes were upregulated in response to both FHB and FRR (Bradi1g34727, Bradi3g31841, Bradi2g35950, Supplementary Table S7). Most of the glutathione-associated genes, however, were exclusively upregulated in FHB being either downregulated or not significantly differentially expressed in FRR. Two classes of genes have been proposed as having roles in ROS metabolism of FCR (Powell et al., 2017b). These include germin-like proteins (Bradi1g04907, Bradi3g15200, Bradi3g15190) and oxalate oxidase genes (Bradi1g11930, Bradi1g11920). These genes were highly upregulated during FHB but were not differentially expressed in FRR (Figure 5B, Supplementary Table S7). Likewise, expression of four NADPH-hemoprotein reductase activity genes was upregulated in FHB (Supplementary Table S7). Finally, a few RBOHD genes were also upregulated exclusively in FHB except for Bradi4g05540 which was upregulated exclusively in FRR.
Transcription factors
Transcription factors showed different expression patterns between FHB and FRR tissues, depending on the class of transcription factor (Supplementary Table S7). The majority of WRKY, NAC, and MADS transcription factors were upregulated in FHB but not differentially expressed in FRR (Supplementary Table S7). The genes Bradi2g53760, Bradi2g00280, Bradi4g01950, and Bradi3g09810 encoding WRKY transcription factors were significantly expressed in FHB only. MYB, AP2 domain, EREBP, and ERF transcription factors were broadly upregulated in both FHB and FRR (e.g. Bradi2g38560, Bradi3g18070, Bradi5g17490, Bradi3g12567), however many were not upregulated in FRR (Supplementary Table S7). The bZIP (e.g. Bradi2g50220, Bradi3g09340, Bradi2g06790) and bHLH transcription factors (e.g. Bradi2g12490, Bradi1g70860, and Bradi5g23580) were generally downregulated in FRR but most showed no large change in expression in FHB (Supplementary Table S7).
Genes coding for ABC transporters and putative DON detoxification functional genes
DON detoxification is an important defence strategy to restrict Fusarium infection (Boutigny et al., 2008; Pasquet et al., 2016). The gene Bradi5g03300 encoding a UDP-glycosyltransferase can detoxify DON (Poppenberger et al., 2003; Pasquet et al., 2016). Several UDP glycosyltransferases, including Bradi5g03300, were highly expressed in FHB but showed either no significant expression or moderate upregulation (Log Fold > 1) in FRR (Supplementary Table S7).
ATP-binding cassette (ABC) transporters have important roles in defence and have been found to be involved in resistance to FHB (Walter et al., 2015). Several ABC transporters were identified in B. distachyon that increase in expression in response to FCR (Powell et al., 2017b). Seven out of the nine identified were upregulated in FHB but were not expressed in FRR (e.g. Bradi3g35390, Bradi2g04577, Bradi5g03460) (Supplementary Table S7).
Time-course analysis of differentially expressed Brachypodium genes
The FHB and FRR assays used to generate the RNA-seq data were repeated with the addition of two additional time-points and expression of a selection of genes were assessed using RT-qPCR to confirm the results from RNA-seq and to provide additional information on temporal aspects of expression. The selected genes were all differentially expressed between FHB and FRR in the RNA-seq except for WRKY45 (Bradi2g30695) which was not expressed in either FHB or FRR. These genes either have predicted roles with phytohormones or have roles in plant defence. In almost every instance, the expression differential at the first time point (Figure 6), was similar to that observed in the RNA-seq experiment (Supplementary Table S4). The results from the RT-qPCR confirmed those from the RNA-seq and showed that, for many of these genes, the expression differences between FHB and FRR were maintained over time. The SA-responsive WRKY45 (Bradi2g30695), SA-associated MES1 (Bradi2g52110), auxin responsive homolog OsAUX/IAA (Bradi1g09090), cytokinin-responsive homolog OsLOG1 (Bradi2g42190), cytokinin-responsive homolog OsA-RR9 (Bradi4g43090), and defence-associated ABC transporter (Bradi2g43120) showed differential expression patterns between FHB and FRR over the entire time course (Figure 6). In contrast, the genes JA and SA-responsive PR1-5 (Bradi1g57590), SA-associated NPR4 (Bradi2g54340), auxin-responsive homolog of OsGH3 (Bradi2g50840), the orthologue of TaCYP450 with a role in DON detoxification (Bradi2g44150), and defence-associated oxalate oxidase (Bradi1g11930) showed similar expression patterns at 3 dpi and 5 dpi for FHB and FRR (Figure 6) despite being differentially expressed at the earlier time points in the RNA-seq data (Supplementary Table S4). The JA-associated genes LOX2 (Bradi3g39980), and JAZ (Bradi4g31240) which were similarly expressed in FHB and FRR in the RNA-seq (Supplementary Table S6) were also similarly expressed at 3 dpi (Figure 6).
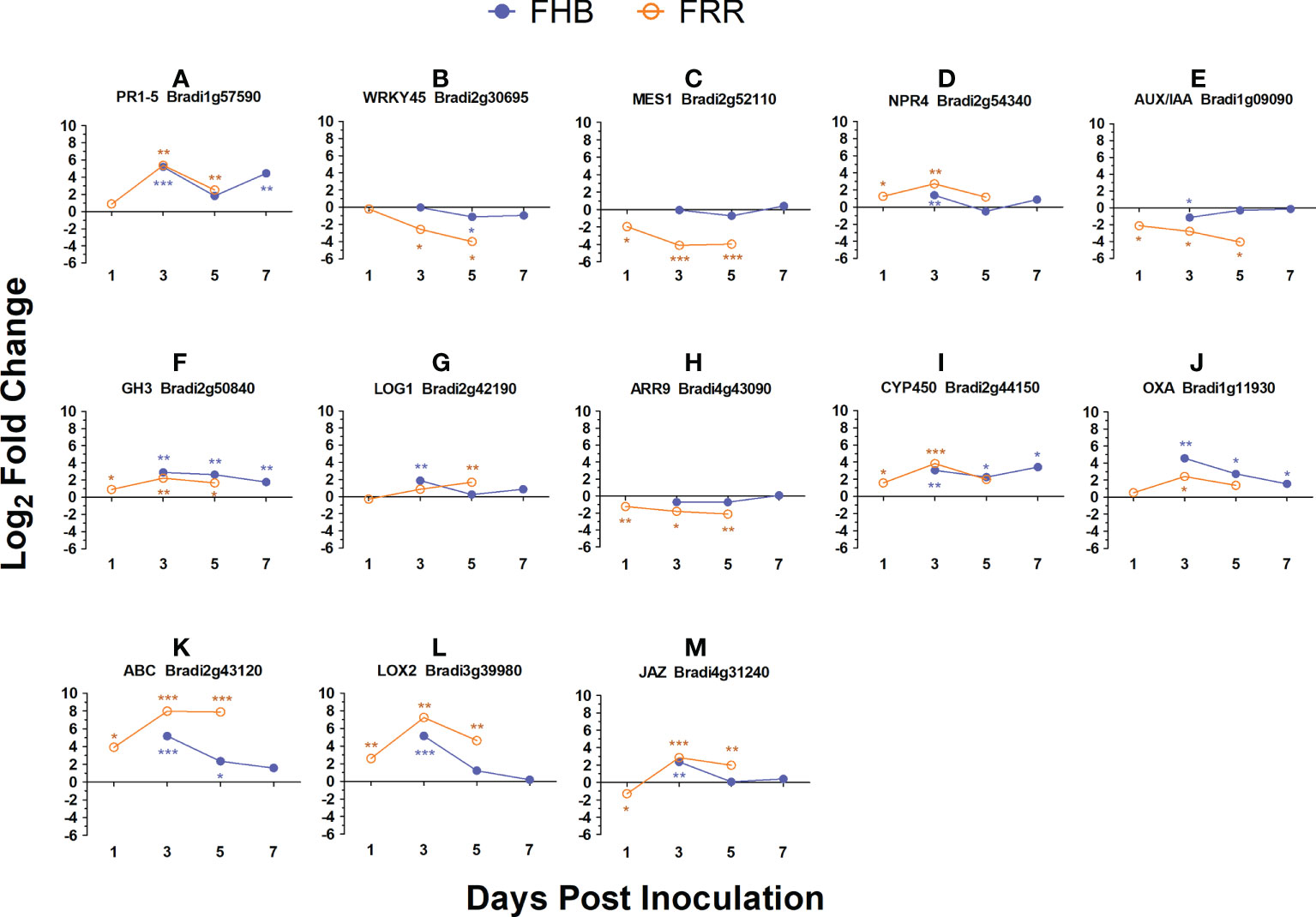
Figure 6 Expression time-course of differentially expressed B. distachyon genes in FHB and FRR. The blue lines denote FHB and orange lines denote FRR. The Log values presented are calculated by comparing infected tissue against mock-inoculated treatments. The reference housekeeping gene GAPDH was used. Each point is the average of three biological replicates and 2-3 technical replicates. Levels of significance relative to the mock-control, Cq t-test *p<0.05, **p<0.01, ***p < 0.001.
Differential expression of F. graminearum genes and predicted effectors
Gene transcript counts of F. graminearum in the FHB and FRR samples were compared against F. graminearum in vitro control samples to determine whether gene expression in the pathogen differed when infecting the two tissues (Supplementary Tables S11, S12). Differentially expressed genes were compared between FHB and FRR (Supplementary Table S13). Moderate proportions of the transcript reads mapped to the F. graminearum PH1 assembly, with 8% to 15% for FHB and FRR respectively. A total of 4,567 F. graminearum genes were significantly responsive in FHB or FRR, or in both (Figure 7A), however only 6% of these were functionally characterised on UniProt (Consortium, 2018). A total of 3,499 genes were significantly differentially expressed in spike infection (FHB), of which 1,919 (55%) were upregulated (Figure 7A), while 3,214 genes were differentially expressed during root infection (FRR), of which 1,815 (56%) were upregulated (Figure 7A). From the significantly differentially expressed F. graminearum genes, 1,167 (26%) were upregulated and 934 (21%) were downregulated in both root and spike tissues, respectively, relative to axenic culture medium (Figure 7A). Only 45 (1%) genes were upregulated in one tissue but downregulated in the other (Figure 7A). Gene-list enrichment was performed on all significantly upregulated and downregulated genes (Supplementary Table S15). Most pathways were similarly expressed between FHB and FRR (e.g. ‘metabolic pathways’, ‘pentose and glucuronate interconversion’, and ‘starch and sucrose metabolism’). The pathways ‘cyanoamino acid metabolism’ and ‘other glycan degradation’ were, however, exclusively upregulated in FHB whereas ‘arginine and proline metabolism’ was exclusively upregulated in FRR. No pathways were significantly downregulated in either FHB or FRR.
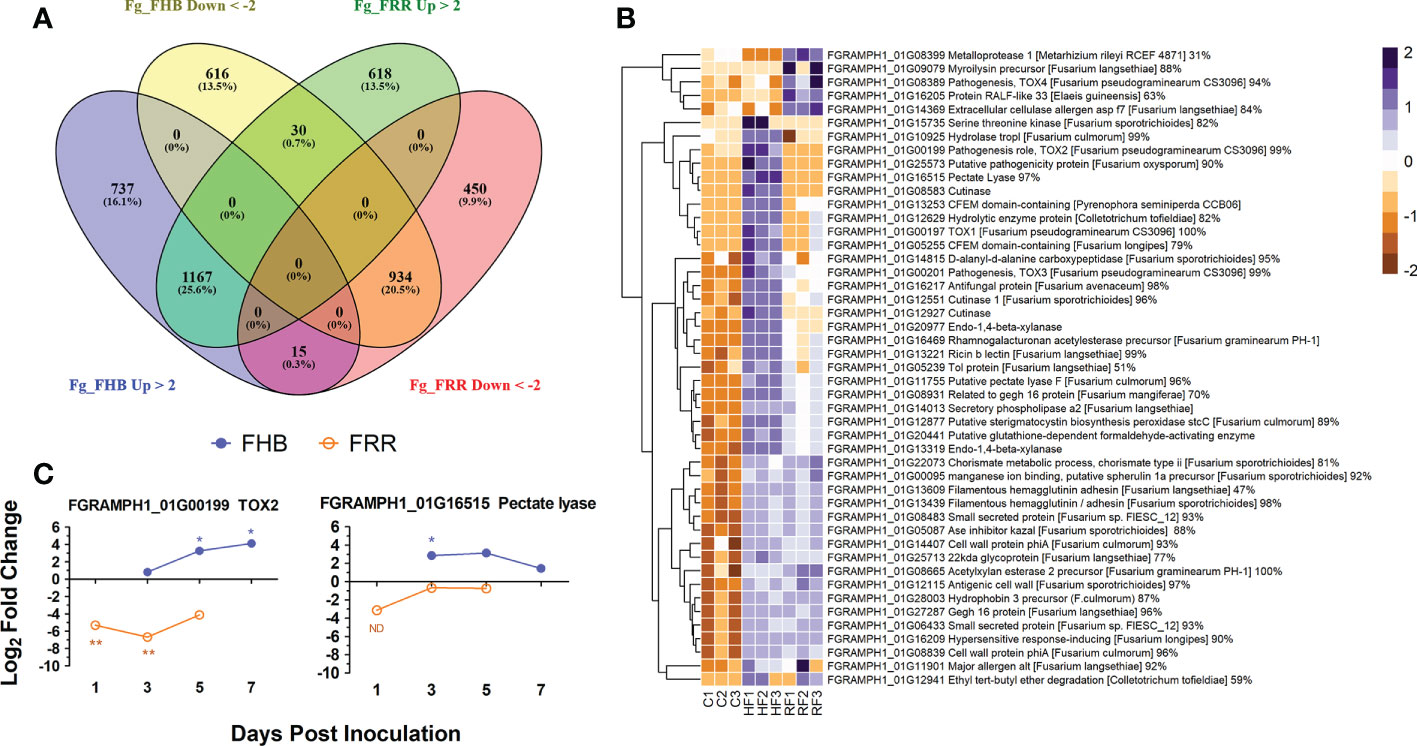
Figure 7 The differential expression of F. graminearum genes between FHB and FRR. (A) A summary of all significantly upregulated or downregulated F. graminearum genes in response to FHB and FRR. The threshold of -2 ≤ x ≥ 2 Log-fold change and p-adj < 0.05 was applied to all genes. (B) The most upregulated F. graminearum predicted-effector genes in FHB and FRR that have predicted functions. These genes have an expression of Log-fold increase ≥ 3 in FHB and/or FRR. Three biological replicates for each of the three treatments are displayed as columns. Control Treatment (C1-3) is the average normalised gene counts (two values per column/biological replicate) of in vitro treatments (four-day-old samples grown in Czapek-Dox Liquid media). The same in vitro sample was analysed separately (DESeq) with the FHB and FRR samples. (C) Expression time-course of two differentially expressed F. graminearum genes (FGRAMPH1_01G00199 (TOX2) and FGRAMPH1_01G16515 (pectate lyase)) in FHB and FRR identified in (B) Blue lines with solid circles denote FHB and orange lines with open circles denote FRR. Log values presented are calculated by comparing infected tissue against mock-inoculated treatments. The housekeeping gene GzUBH was used. Level of significance relative to the mock-control: Cq t-test *p < 0.05, **p < 0.01. “ND” (Not enough Data) denotes no statistical comparison due to insufficient data points in treatment. Bd (B. distachyon), Up (Upregulated), Down (Downregulated), FRR (Fusarium Root Rot), FHB (Fusarium Head Blight). C (in vitro control respective to FHB and FRR), HF (Head-FHB fungus), RF (Root-FRR fungus).
All the significantly expressed genes were filtered by association with the F. graminearum secretome database (Brown et al., 2012). Only 92 (3%) of FHB-responsive genes (Figure 7A) were classed as effectors and 37 (40%) of these were exclusively expressed or repressed in FHB. On the other hand, 70 (2%) of FRR-responsive genes (Figure 7A) were classed as effectors and 15 (21%) of these were exclusively expressed or repressed in root tissues (FRR) (Supplementary Table S14). Fifty-eight (54%) of all these effector-associated genes had a predicted function (From F. graminearum (UniProt (Consortium, 2018)) or through protein homology in different fungal species (BLAST (Sayers et al., 2020; Supplementary Table S14). The upregulated genes were then selected since they were predicted to be the effectors playing important roles in FHB and FRR virulence. A total of 80 F. graminearum genes were highly upregulated (Log2-fold ≥ 3) in FHB and/or FRR (Supplementary Table S14) and 47 of these with predicted functions are presented (Figure 7B). Many of the predicted functions were associated with cell-wall degradation and pathogenesis. The majority of F. graminearum predicted effectors were similarly expressed between FHB and FRR (Figure 7B). The predicted effectors FGRAMPH1_01G14013 (secretory phospholipase), FGRAMPH1_01G20977 (Endo-1,4-beta-xylanase), FGRAMPH1_01G16469 (rhamnogalacturonan acetylesterase precursor), FGRAMPH1_01G13253 (CFEM domain-containing), and FGRAMPH1_01G27287 (gEgh 16 protein) were among the most highly upregulated effectors in both FHB and FRR. A number of effectors, however, were upregulated in a tissue-specific manner. Nine predicted effectors (e.g. FGRAMPH1_01G00199 (TOX2), FGRAMPH1_01G16515 (Pectate Lyase), and FGRAMPH1_01G12927 (Cutinase)) were exclusively upregulated in FHB, whereas 5 (e.g. FGRAMPH1_01G08389 (TOX4), FGRAMPH1_01G08399 (Metalloprotease), and FGRAMPH1_01G09079 (myroilysin precursor)) were exclusively upregulated in FRR (Figure 7B).
The expression of two effector genes (FGRAMPH1_01G00199 (TOX2) and FGRAMPH1_01G16515 (Pectate Lyase)) that showed differential expression between FHB and FRR in the RNA-seq data set (Supplementary Table S14) were examined using RT-qPCR (Figure 7C). Although the absolute expression values differed between experiments for the same gene at 1 dpi for FRR and 3 dpi for FHB (Figures 6C, 7B), the differential expression between tissues was maintained over time (Figure 7C). TOX2 was downregulated in FRR but upregulated in FHB whereas pectate lyase was only significantly upregulated in FHB at 3 dpi (Figure 7C).
Trichothecene production is regulated by the Tri gene cluster (Kimura et al., 2003; Kimura et al., 2007). F. graminearum PH1 is a 15-acetylDON (ADON) producer (Kimura et al., 2007). The expression of the Tri5-gene cluster was investigated to identify any difference in transcription of DON associated genes between FHB and FRR. The essential DON biosynthetic triplet of genes (Tri4, Tri5, Tri11) were the most upregulated genes in both FHB and FRR (Table 1). The transcriptional regulators (Tri6, Tri10), and Tri12 transporter were also upregulated in both FHB and FRR and there were also high levels of expression of Tri3, Tri9 and Tri14 in both tissues (Table 1). Despite the low but significant expression in FHB, Tri8 was the only gene in the Tri5-gene cluster that was exclusively expressed in FHB (Table 1). Tri8 encodes a C-3 deacetylase that is involved in 15-ADON production (Alexander et al., 2011). The Tri7 and Tri13 genes in F. graminearum PH1 do not encode functional proteins as this isolate is a 15-AcDON chemotype and these genes are involved in the synthesis of nivalenol (Lee et al., 2002; Kimura et al., 2003). Neither of these genes exceeded the significance log-fold threshold in either FHB or FRR (Table 1).
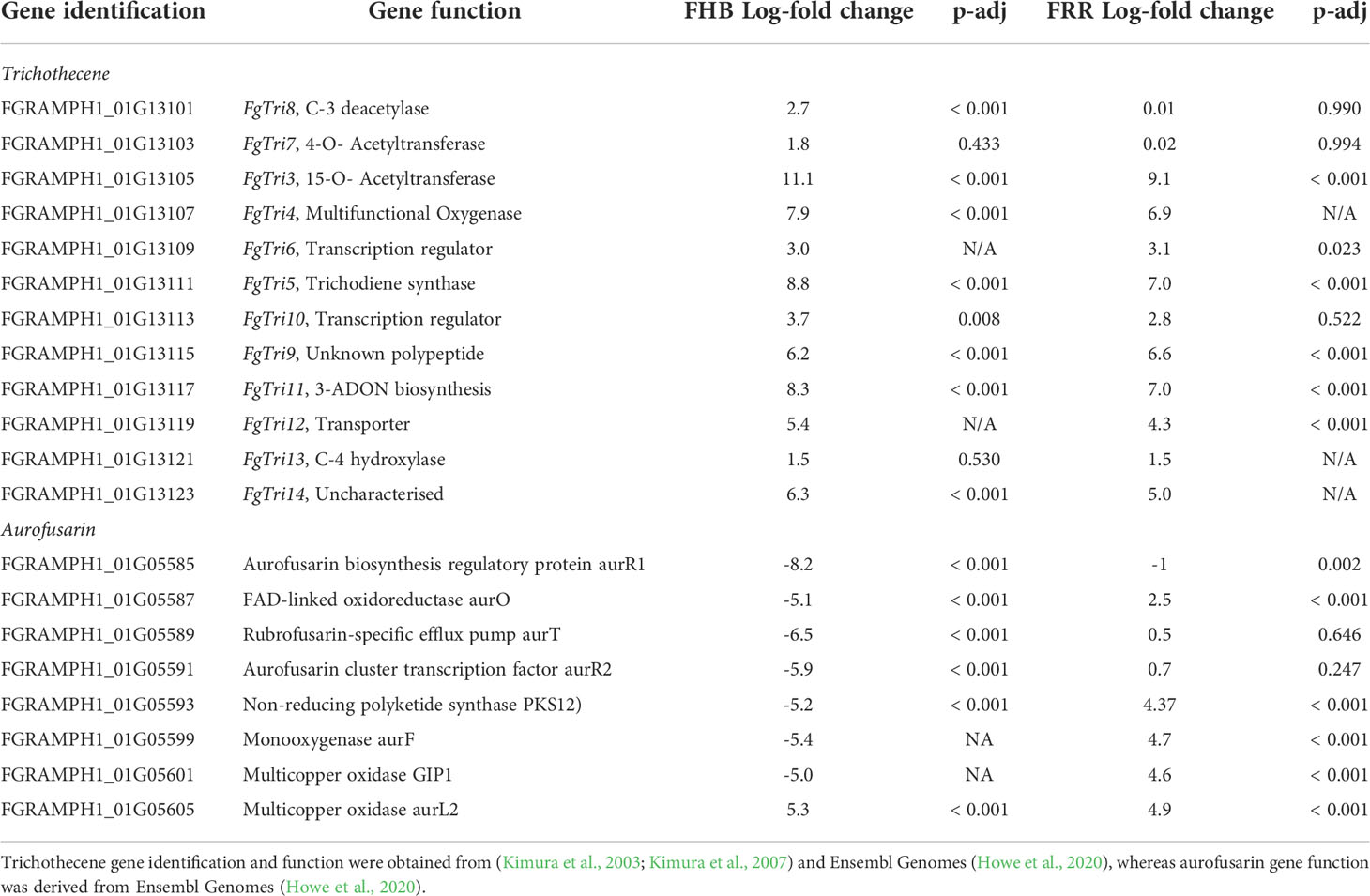
Table 1 Notable F. graminearum secondary metabolite clusters that were significantly expressed in FHB and FRR.
The cluster of genes involved in the synthesis of the red pigment aurofusarin were differentially expressed between FHB and FRR (Table 1). Most of the genes in the pathway were significantly downregulated in spike tissue (FHB) but upregulated in root tissue (FRR) (Table 1). Both FGRAMPH1_01G05593 (PKS12) and FGRAMPH1_01G05587 (aurO) (Table 1) were among the 30 genes that were highly upregulated in FRR and downregulated in FHB (Figure 7A). FGRAMPH1_01G05599 (aurF) and FGRAMPH1_01G05601 (GIP1) also displayed differentials in expression but the effects were not statistically significant for FHB (Table 1). The exceptions to this differential trend were FGRAMPH1_01G05605 (aurL2) which was significantly upregulated in both FHB and FRR, and FGRAMPH1_01G05589 (aurT) and FGRAMPH1_01G05591 (aurR2) which were downregulated in response to FHB but were not significantly upregulated in response to FRR (Table 1). Likewise, FGRAMPH1_01G05585 (aurR1) was significantly downregulated relative to in vitro conditions in both FHB and FRR, but to a lesser degree in FRR (Table 1).
Discussion
The transcriptome of B. distachyon exhibited many tissue-specific differences in response to infection of spike and root tissues by F. graminearum. The majority of differentially expressed genes were exclusively expressed or repressed in response to FHB or FRR (Figure 1C). Interestingly, while an equal number of genes were upregulated or downregulated in response to FRR (Figure 1A) the vast majority of the differentially expressed genes in FHB were upregulated (Figure 1A). The preponderance of upregulation over suppression of gene expression in response to FHB has also been reported with wheat (Buerstmayr et al., 2021). More genes were upregulated than downregulated in B. distachyon following infection of the base of seedlings (Fusarium Crown Rot (FCR)) by F. pseudograminearum (Powell et al., 2017a; Powell et al., 2017b) suggesting that the transcriptional response of FCR may be more like that to FHB than to FRR. Differences in the proportion of up and downregulated genes in different tissues has been observed in other host-pathogen interactions. Expression of more genes in A. thaliana roots was repressed in response to F. oxysporum infection than in response to leaf infection (Chen et al., 2014) and approximately half of the differentially expressed genes displayed root or leaf-specific expression following infection of A. thaliana by F. oxysporum (Lyons et al., 2015). Many pathogens exhibit organ specificity being able to infect some tissues but not others but it is unclear whether these effects are controlled by the host or pathogen (Strugala et al., 2015). The rice cultivar Tai-Nong is resistant to foliar infection by isolate 031 of Magnaporthe oryzae but is susceptible to root infection by the same isolate (Jansen et al., 2006). Additionally, root systems of the FHB-resistant wheat cv. Sumai 3 were found to be susceptible to FRR (Wang et al., 2015). In the present study we provide evidence of tissue-specific host responses to infection by F. graminearum as well as tissue-specific gene expression in the pathogen.
We previously showed that exogenous application of phytohormones induced tissue-specific effects on resistance to FHB and FRR in B. distachyon (Haidoulis and Nicholson, 2020) indicating that pathways controlled by phytohormones play different roles in resistance in spike and root tissues. More JA-responsive genes were significantly expressed and to a greater extent in FHB than in FRR (Figure 2, Figure 3A, Supplementary Table S6), although there were similar changes in expression of some biosynthesis and signalling genes (LOX and JAZ) in FHB and FRR (Supplementary Table S6, Figures 6L, M). For most biosynthesis genes (OPR and LOX), the basal level of expression was much higher in non-inoculated root tissues compared to non-inoculated spikes (Supplementary Table S6) which may account for the apparent reduced level of expression of JA-associated processes during infection in roots. Supporting these results, LOX and JAZ genes were also found to be upregulated in B. distachyon in response to FHB, FCR, or FRR (Pasquet et al., 2014; Powell et al., 2017b; Ding et al., 2021), in wheat to FHB (Sun et al., 2016; Pan et al., 2018; Wang L P. et al., 2018), and in wheat to FCR and FRR (Powell et al., 2017a; Wang Q. et al., 2018). Similar to JA, ethylene biosynthesis (AOC) and many signalling genes (ERF1 and AP2 domain containing genes) were upregulated in both FHB and FRR (Figure 3B, Supplementary Table S6). Genes with the same function were also upregulated in B. distachyon FCR and FRR (Powell et al., 2017b; Ding et al., 2021), B. distachyon FHB (Pasquet et al., 2014), and wheat FHB (Sun et al., 2016; Buhrow et al., 2021). Ethylene functions synergistically with JA signalling in plants by prioritizing JA signalling and fine-tuning resistance to necrotrophic pathogens (Bari and Jones, 2009; Pieterse et al., 2012) via ERF1/ORA59 transcription factors (Lorenzo et al., 2003; McGrath et al., 2005; Pieterse et al., 2012). Given the abundance of ethylene-related transcription factors upregulated in both FHB and FRR (Figure 3B) and the overrepresentation of ethylene-associated processes in FHB and FRR (Figure 2), the data suggests that the ethylene branch of JA signalling is activated in both tissues and that the JA/ethylene synergism is important in FHB and FRR. Supporting this, exogenous application of JA and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) had similar effects on resistance in both tissues (Haidoulis and Nicholson, 2020). In the present study, JA and ethylene - related gene expression was broadly similar in FHB and FRR (Figures 3A, B). There is a contrast in the effects of exogenous phytohormone application and gene expression in the different tissues for the same hormones (Supplementary Table S8). This may be caused by the different native states of expression of the phytohormone associated genes in spikes and roots which may dictate whether associated genes are significantly expressed or not on exposure to the pathogen. Alternatively, this may be due to a potentially contrasting lifestyle of F. graminearum in FHB and FRR. We previously suggested that F. graminearum may be behaving predominantly as a necrotroph in B. distachyon roots (Haidoulis and Nicholson, 2020). This may trigger a different host response in B. distachyon roots.
In contrast to JA and ethylene, there was minimal evidence for altered expression of genes involved in SA biosynthesis or signalling in FHB and FRR (Supplementary Table S6). The key SA-negative regulator NPR4 (Figure 6D) (Ding et al., 2018), and the SA-responsive WRKY transcription factor (Figure 6B) (Kakei et al., 2015; Kouzai et al., 2016) were upregulated and downregulated, respectively, in FHB and FRR. Together, these findings indicate that the JA, rather than the SA, pathway is predominantly expressed in both tissues at the onset of first symptoms. An absence or a very low amount of SA-related gene transcription was also identified in wheat, B. distachyon, and Arabidopsis in response to Fusarium infection (Lyons et al., 2015; Sun et al., 2016; Powell et al., 2017b). However, in contrast, SA signalling was suggested to be a significant factor in the B. distachyon response to FRR (Ding et al., 2021). The difference may be due to FRR sampling which was 4 dpi, later than in this study.
Unlike the canonical defence-associated phytohormones, expression of auxin- and cytokinin-associated genes generally differed between FHB and FRR. For auxin, two GO processes involved in indole-containing compound biosynthesis and metabolic processes were overrepresented in FHB but not in FRR (Figure 2). The signalling genes encoding AUX/IAA, ARF, and SAUR were generally upregulated in response to FHB but suppressed in FRR (Figure 3C, Figure 6E). Two of the same ARFs and a SAUR gene were also reported to be upregulated in FHB in B. distachyon (Pasquet et al., 2014), while two SAUR genes were similarly downregulated in FRR of B. distachyon (Ding et al., 2021). In contrast, auxin homeostasis-associated GH3 genes (Staswick et al., 2005) showed similar expression at 3 dpi and 5 dpi in FHB and FRR (Figure 6F). Rice GH3 homologues are associated with resistance responses (Domingo et al., 2009; Fu et al., 2011) and biosynthesis and auxin-related signalling can affect resistance to plant pathogens (Bari and Jones, 2009; Kazan and Manners, 2009). Lyons and colleagues (2015) concluded that auxins were important components of defence responses to F. oxysporum in both shoot and root tissues of Arabidopsis. We reported previously that exogenous application of auxin increased resistance to both FHB and FRR in B. distachyon (Haidoulis and Nicholson, 2020) and others have found similar effects on FHB in barley (Petti et al., 2012). In contrast, there is evidence that F. graminearum-induced FHB and leaf susceptibility increased following exogenous IAA application in wheat (Su et al., 2020). Furthermore, auxin content and signalling have been associated with FHB susceptibility (Wang L. et al., 2018; Brauer et al., 2019). Disruption of the auxin receptor gene TaTIR1 and attenuation of TaARF2 expression in wheat reduced susceptibility to FHB (Chen et al., 2016; Su et al., 2020). Auxin homeostasis may be important in resistance to FHB and FRR and exogenous application of auxin or disruption of signalling may both suppress the potential of the pathogen to manipulate levels of auxin in the host. This possibility is supported by the finding that F. graminearum can synthesise auxin (Luo et al., 2016).
Aside from a study by (Powell et al., 2017a), the role of cytokinins in F. graminearum infection has not been investigated beyond the exogenous application of cytokinins which enhanced FHB and FRR susceptibility (Haidoulis and Nicholson, 2020). Most biosynthesis genes (LOG1, IPT, UGT85A1) and signalling genes (Type A RRs and B RRs) were differentially expressed between the two tissues in response to F. graminearum (Figure 3D, Figures 6G, H), highlighting a difference in response to FHB and FRR in terms of cytokinin signalling. Both auxin and cytokinin-associated signalling genes tended to be upregulated in FHB but downregulated in FRR although exogenous application of these phytohormones had similar but contrasting effects on resistance in both tissues with auxin increasing resistance while cytokinin reduced resistance to both FHB and FRR (Supplementary Table S8). This may reflect differences in the infection strategy of F. graminearum between tissues, discussed in (Haidoulis and Nicholson, 2020), however further research is needed to understand the role of auxin and cytokinin in the defence response to F. graminearum.
Previous transcriptomic studies have shown substantial transcriptomic similarities between responses to FHB and the application of phytohormones on wheat (Qi et al., 2019; Buhrow et al., 2021). For example, expression in response to methyl JA and ethylene were very similar to those induced by FHB (Qi et al., 2019), supporting the JA and ethylene responsiveness observed in this study of FHB and FRR. The phytohormone ABA has been reported to induce many genes that are also expressed in response to FHB (Qi et al., 2019; Buhrow et al., 2021). In contrast in the present study, a relatively small number of ABA-responsive genes were expressed in FHB and FRR (Supplementary Table S6). It is unclear whether the ABA and FHB responses are linked or independent.
Several types of antimicrobial-associated genes were similarly expressed in FHB and FRR (Figure 4B). Five classes of PR genes were generally upregulated in both FHB and FRR (Figure 4B). Many of the same PR gene classes were similarly expressed in FHB of wheat (Pritsch et al., 2000; Pan et al., 2018; Wang L. et al., 2018; Buhrow et al., 2021), FCR of wheat (Powell et al., 2017b), FHB of barley (Boddu et al., 2006), and FHB, FCR, and FRR of B. distachyon (Pasquet et al., 2014; Powell et al., 2017b; Ding et al., 2021). Together the data suggests that PR gene expression is a common host response to most Fusarium diseases.
ROS-associated genes and pathways were the most differently expressed in the dataset. Most peroxidases, involved in the breakdown of hydrogen peroxide, were downregulated in FRR (Supplementary Table S7, Figure 5B). Several of these genes had previously been found to be highly downregulated as part of the response of B. distachyon to FRR (Ding et al., 2021). GO biological pathways for hydrogen peroxide catabolic process, ROS metabolic process, response to oxidative stress, and cellular oxidant detoxification were all highly downregulated in FRR (Figure 2). Together this suggests that in FRR there is predominantly a reduction in ROS metabolism, oxidative response, and oxidant detoxification. This observation may, in part, be due to the high basal expression of peroxidases in non-inoculated root samples compared to non-inoculated spikes (Figure 2). In contrast to FRR, most peroxidases were not significantly expressed in FHB (Supplementary Table S7, Figure 5B). However, genes involved in glutathione metabolism, mostly glutathione-S-transferases (GST) (Supplementary Tables S5, S7), were generally highly upregulated in FHB but only moderately upregulated in FRR (Figure 5B). Likewise, the GO process glutathione metabolic process was only overrepresented in FHB (Figure 2). Glutathione metabolism, was reported to be an important response to B. distachyon FCR (Powell et al., 2017b). Likewise several glutathione associated genes were upregulated in B. distachyon to FRR (Ding et al., 2021), and GST expression was upregulated in barley FHB (Boddu et al., 2006) and associated with resistance in wheat FHB (Pan et al., 2018). The high upregulation in FHB may be because of relatively low basal levels of GST in non-inoculated spikes compared to non-inoculated roots (Figure 2). There is, however, a notable difference in expression of these two different classes of enzymes with similar function between FHB and FRR (peroxidase and glutathione) which suggests a different ROS state between FHB and FRR.
It was possible to examine the transcriptomes of both F. graminearum and B. distachyon in the same material. This offered the opportunity to observe both the host and pathogen components of the interaction in the two tissues. Cell-wall degrading enzymes (CWDEs) are associated with both necrotrophic and hemibiotrophic pathogens (Zhao et al., 2014; Kabbage et al., 2015). F. graminearum is known to express an abundance of CWDE genes (Cuomo et al., 2007; Kikot et al., 2009) in symptomatic tissue (Brown et al., 2017). The role of CWDEs is likely as a means for nutrient acquisition and/or as effectors (Walton, 1994; Cuomo et al., 2007). Several CWDEs were among the putative F. graminearum effectors and increased in expression in both FHB and FRR (Figure 7B). Enhanced expression of CWDEs, including genes involved in pectin degradation (Figure 7B), have also been reported in several other studies with wheat, barley, and maize FHB (Lysøe et al., 2011; Harris et al., 2016; Brown et al., 2017; Pan et al., 2018) and B. distachyon FRR (Ding et al., 2022). An effective plant strategy to detect necrotrophic pathogens is the release of DAMPs like oligogalacturonides by wall-associated receptor kinases (WAKs) that bind galacturonan (He et al., 1998; Wang et al., 2014). WAKS serve as pectin debris receptors (Kohorn and Kohorn, 2012). Interestingly, expression of WAKs was generally only upregulated in response to FHB (Supplementary Table S7) despite similar expression of pectin-associated CDWEs by F. graminearum in both tissues (Figure 7B). Gadaleta and colleagues found a TaWAK2 gene associated with the FHB resistance QTL QFhb.mgb-2A in wheat (Gadaleta et al., 2019). Elevated expression of several WAK genes was reported in Bd roots at 5 dpi (Ding et al., 2021) suggesting that there may be a delayed response in root tissues.
The core Tri genes for DON production (Kimura et al., 2003) were upregulated in both FHB and FRR (Table 1). This cluster has been shown to be upregulated in FHB in several species (Lysøe et al., 2011; Harris et al., 2016; Brown et al., 2017; Pan et al., 2018) and in B. distachyon FRR (Ding et al., 2022). Wang and colleagues also reported the presence of DON in root tissues (Wang et al., 2015). Although F. graminearum has been shown to produce DON in both B. distachyon floral and root tissue during infection, there is evidence that it does not act as a virulence factor in roots (Ding et al., 2022). Furthermore DON does not act as a virulence factor in FHB of barley and maize (Maier et al., 2006). It is plausible that the influence of DON on virulence is tissue-specific despite its tissue-independent production. DON detoxification during early infection is an effective host strategy to increase resistance against FHB and FCR (Mandalà et al., 2019). Several UDP-glycosyltransferase (UGT) genes were found to be upregulated in FHB in the present study (Supplementary Table S7) and upregulation of many of these was also observed in other studies with FHB, FCR, and FRR (Pasquet et al., 2014; Powell et al., 2017b; Wang Q. et al., 2018). UDP-glucosyltransferases can glucosylate DON into the less toxic deoxynivalenol-3-O-glucose form (Pasquet et al., 2016). Interestingly, the gene encoding a UDP-glucosyltransferase that was highly expressed in FHB (Bradi5g03300) (Supplementary Table S7) has been shown to confer spike resistance to F. graminearum and root tolerance to DON in B. distachyon (Pasquet et al., 2016) and type 2 FHB resistance when expressed in wheat (Gatti et al., 2019). Bradi2g44150 (Figure 6I) is a homolog of TaCYP72A involved in resistance to DON (Gunupuru et al., 2018), and was significantly upregulated in FHB in the RNA-seq and at 3 dpi in both tissues (Figure 6I, Supplementary Table S7). If this homolog serves a similar DON resistance function in B. distachyon as in wheat (Gunupuru et al., 2018), then activation of DON-detoxification processes may be occurring similarly in both tissues.
F. graminearum can behave as a facultative hemibiotroph in spike tissues of wheat (Brown et al., 2010). Effectors were originally believed to be biotroph-specific however evidence is accumulating to suggest their importance for hemibiotrophic and necrotrophic pathogens (Amselem et al., 2011; Guyon et al., 2014; Kabbage et al., 2015). Plants often detect effectors using NBS-NLRs or LRR-RLK receptors. The receptor signalling GO processes were overrepresented only in FHB (Figure 2). Furthermore, the differential expression of many NBS-LRRs and LRR-RLKs was observed to be tissue-dependent in the present study (Figure 4A). For example, homologues of the Recognition of Peronospora 13 (RPP13) genes were differentially expressed between FHB and FRR (Figure 4A, Supplementary Table S7). RPP13 confers resistance to diseases such as downy mildew (Bittner-Eddy et al., 2000), and homologues of this gene were reported to be upregulated in response to B. distachyon FCR (Powell et al., 2017b). It is unclear why there is such a marked difference in the expression of genes involved in pathogen recognition in the two tissues, but it suggests that the host may be responding to different sets of effectors produced by F. graminearum in root and spike tissues.
A comparison of the different F. graminearum genes and effector-like genes in FHB and FRR revealed evidence for tissue-specific gene expression of components of the secretome. Of the F. graminearum predicted effector genes, 42% and 24% were exclusively expressed in FHB and FRR, respectively (Supplementary Table S14). This suggests that the expression of a large proportion of the F. graminearum secretome genes are controlled in a tissue-specific manner following infection of B. distachyon tissues. Genes encoding cutinases are present in relatively large numbers in hemibiotrophic and necrotrophic pathogens such as Gaeumannomyces graminis and Magnaporthe oryzae (Zhao et al., 2014). Two of the three expressed F. graminearum cutinases were exclusively upregulated in FHB (Supplementary Table S14). This finding is supported by reports that cutinases were expressed in wheat and barley FHB (Lysøe et al., 2011; Harris et al., 2016; Pan et al., 2018), but not in B. distachyon FRR (Ding et al., 2022). This difference between tissues is likely due to the cutin layer being present only on the epidermis of shoot tissues (Walton, 1994). The expression of one putative cutinase in roots (FGRAMPH1_01G12551) may also be associated with a different pathogenicity-associated role (Walton, 1994). Fusarium graminearum can produce the naphthoquinone aurofusarin, which is a red pigment and is synthesised by the aurofusarin gene cluster (Malz et al., 2005; Frandsen et al., 2006). The cluster of genes involved in aurofusarin biosynthesis (excluding FGRAMPH1_01G05605 (FGSG_02330)) were downregulated in FHB but the key genes were upregulated in FRR (Table 1). A similar result was observed for symptomatic wheat FHB with the same F. graminearum isolate (Brown et al., 2017). This suggests aurofusarin is being produced by F. graminearum PH1 during FRR pathogenesis of B. distachyon. A potential role of aurofusarin in FRR is not known although it was shown to not affect FHB virulence in wheat and barley (Kim et al., 2005; Malz et al., 2005). Four TOX genes are known in F. graminearum with TOX1, TOX2, and TOX3 being present in a gene cluster while TOX4 is located at the opposite end of the chromosome. The four genes showed tissue-specific differential expression with TOX1, TOX2, and TOX3 being upregulated in FHB while TOX4 was upregulated in FRR (Figure 7B). RT-qPCR demonstrated that the differential expression of TOX2 was maintained over time (Figure 7C). Expression of TOX1, TOX2, and TOX3 was reported to be upregulated in FHB of wheat and barley (Lysøe et al., 2011; Harris et al., 2016; Pan et al., 2018) whereas TOX4 was only expressed in wheat FHB (Lysøe et al., 2011; Harris et al., 2016; Pan et al., 2018). The role of TOX genes in F. graminearum virulence in FHB and FRR is unclear but the host/tissue-specific expression of the four genes is intriguing. The examples above hint at the possibility of a specialised secretome where F. graminearum has the capacity to recognise the type of host tissue it is infecting and deploy a bespoke array of effectors.
In this study, only one F. graminearum isolate was used for FHB and FRR assays. However, there are many F. graminearum isolates with different levels of virulence and mycotoxin chemotypes. It is not known whether the differences between infected tissues is isolate-dependent. Thus an analysis of the F. graminearum secretome and secondary metabolite clusters between tissues from different F. graminearum isolates merits further investigation. Likewise, these experiments were only performed on one susceptible B. distachyon accession (Peraldi et al., 2011) so it is still unclear whether the transcriptome response of the host are susceptibility or resistance-associated responses. Lastly, B. distachyon root and spike tissues are at different developmental stages during infection in this study. Plant age may play an important factor in the transcriptome responses of FRR. The symptoms of FHB and FRR has been shown to progress differently (Wang et al., 2015), thus it is unclear if there is an ideal time-point to use for FRR. Additional studies investigating adult root response to F. graminearum infection would be valuable to further compare FRR to FHB.
To summarise, the transcriptome response of B. distachyon and F. graminearum during early FHB and FRR was investigated and compared. There were similarities in B. distachyon spike and root tissues with increased expression of genes associated with antimicrobial compounds and JA and ethylene phytohormones-associated genes in both tissues. Significantly, however, there were several tissue-dependent responses including those involved with the phytohormones cytokinin and auxin, receptor signalling, cell-wall modification, and ROS metabolism and response. Likewise for F. graminearum, there were both core-genes that were expressed in a tissue-independent manner such as DON and CWDEs, while TOX genes, and those involved in cutin degradation, and aurofusarin biosynthesis were expressed in a tissue-specific manner. Overall, this study reveals both host defence responses and pathogen attack strategies in different tissues of the same host and highlights some potentially important tissue-specific aspects.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Contents of this study have been previously published in a thesis (Haidoulis, 2021) and as a pre-print on BioRxiv (Haidoulis and Nicholson, 2022a). The RNA-seq datasets can also be found at (Haidoulis and Nicholson, 2022b).
Author contributions
JH: Experimental hypotheses and design. Methods and experiments. Data analysis. Manuscript writing. Making figures and tables. PN: Supervision and guidance on experimental design and hypothesis. Providing equipment, resources, and laboratory space. Manuscript and figure proofreading, editing, and some writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the BBSRC (grant number: BB/M011216/1) and BASF SE at Limburgerhof in Germany as part of the PhD studentship of JH and is supported by the BBSRC Plant Health ISP (grant number: BBS/E/J/000PR9797) for PN.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1025161/full#supplementary-material.
References
Afgan, E., Baker, D., van den Beek, M., Blankenberg, D., Bouvier, D., Čech, M., et al. (2016). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, W3–W10. doi: 10.1093/nar/gkw343
Agarwal, P., Agarwal, P. K. (2014). Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Molecular Biology Reports. 41, 599–611. doi: 10.1007/s11033-013-2897-4
Ai, C., Kong, L. (2018). CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genomics 45, 489–504. doi: 10.1016/j.jgg.2018.08.002
Alexander, N. J., Mccormick, S. P., Waalwijk, C., van der Lee, T., Proctor, R. H. (2011). The genetic basis for 3-ADON and 15-ADON trichothecene chemotypes in fusarium. Fungal Genet. Biol. 48, 485–495. doi: 10.1016/j.fgb.2011.01.003
Amselem, J., Cuomo, C. A., Van Kan, J. A., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens sclerotinia sclerotiorum and botrytis cinerea. PloS Genet. 7, e1002230. doi: 10.1371/journal.pgen.1002230
Antonissen, G., Martel, A., Pasmans, F., Ducatelle, R., Verbrugghe, E., Vandenbroucke, V., et al. (2014). The impact of fusarium mycotoxins on human and animal host susceptibility to infectious diseases. Toxins 6, 430–452. doi: 10.3390/toxins6020430
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bari, R., Jones, J. D. G. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488. doi: 10.1007/s11103-008-9435-0
Beccari, G., Covarelli, L., Nicholson, P. (2011). Infection processes and soft wheat response to root rot and crown rot caused by fusarium culmorum. Plant Pathol. 60, 671–684.
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The arabidopsis information resource: making and mining the “gold standard” Annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Bittner-Eddy, P. D., Crute, I. R., Holub, E. B., Beynon, J. L. (2000). RPP13 is a simple locus in arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in peronospora parasitica. Plant J. 21, 177–188. doi: 10.1046/j.1365-313x.2000.00664.x
Blighe, K., Rana, S., Lewis, M. (2018). EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R Package Version 1. Available at:https://github.com/kevinblighe/EnhancedVolcano (Accessed June 2021).
Boddu, J., Cho, S., Kruger, W. M., Muehlbauer, G. J. (2006). Transcriptome analysis of the barley-fusarium graminearum interaction. Mol. Plant-Microbe Interact. 19, 407–417. doi: 10.1094/MPMI-19-0407
Bonnot, T., Gillard, M. B., Nagel, D. H. (2019). A simple protocol for informative visualization of enriched gene ontology terms. Bio-protocol 9, e3429. doi: 10.21769/BioProtoc.3429
Boutigny, A.-L., Richard-Forget, F., Barreau, C. (2008). Natural mechanisms for cereal resistance to the accumulation of fusarium trichothecenes. Eur. J. Plant Pathol. 121, 411–423. doi: 10.1007/s10658-007-9266-x
Brauer, E. K., Rocheleau, H., Balcerzak, M., Pan, Y., Fauteux, F., Liu, Z., et al (2019). Transcriptional and hormonal profiling of fusarium graminearum-infected wheat reveals an association between auxin and susceptibility. Physiol. Mol. Plant Pathol. 107, 33–39 doi: 10.1016/j.pmpp.2019.04.006
Broekaert, W. F., Delauré, S. L., de Bolle, M. F., Cammue, B. P. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416. doi: 10.1146/annurev.phyto.44.070505.143440
Brown, N. A., Antoniw, J., Hammond-Kosack, K. E. (2012). The predicted secretome of the plant pathogenic fungus fusarium graminearum: a refined comparative analysis. PloS One 7, e33731. doi: 10.1371/journal.pone.0033731
Brown, N. A., Evans, J., Mead, A., Hammond-Kosack, K. E. (2017). A spatial temporal analysis of the fusarium graminearum transcriptome during symptomless and symptomatic wheat infection. Mol. Plant Pathol. 18, 1295–1312 doi: 10.1111/mpp.12564
Brown, N. A., Urban, M., van de Meene, A. M. L., Hammond-Kosack, K. E. (2010). The infection biology of fusarium graminearum: Defining the pathways of spikelet to spikelet colonisation in wheat ears. Fungal Biol. 114, 555–571. doi: 10.1016/j.funbio.2010.04.006
Buerstmayr, M., Wagner, C., nosenko, T., Omony, J., Steiner, B., Nussbaumer, T., et al. (2021). Fusarium head blight resistance in European winter wheat: insights from genome-wide transcriptome analysis. BMC Genomics 22, 470. doi: 10.1186/s12864-021-07800-1
Buhrow, L. M., Liu, Z., Cram, D., Sharma, T., foroud, N. A., Pan, Y., et al. (2021). Wheat transcriptome profiling reveals abscisic and gibberellic acid treatments regulate early-stage phytohormone defense signaling, cell wall fortification, and metabolic switches following fusarium graminearum-challenge. BMC Genomics 22, 1–21. doi: 10.1186/s12864-021-08069-0
Chen, W., Kastner, C., Nowara, D., Oliveira-Garcia, E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. doi: 10.1093/jxb/erw263
Chen, Y. C., Kidd, B. N., Carvalhais, L. C., Schenk, P. M. (2014). Molecular defense responses in roots and the rhizosphere against fusarium oxysporum. Plant Signaling Behav. 9, e977710. doi: 10.4161/15592324.2014.977710
Consortium, T. U. (2018). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. doi: 10.1093/nar/gky1049
Cuomo, C. A., Güldener, U., Xu, J.-R., Trail, F., Turgeon, B. G., di Pietro, A., et al. (2007). The fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317, 1400–1402. doi: 10.1126/science.1143708
Ding, X., Cao, Y., Huang, L., Zhao, J., Xu, C., Li, X., et al. (2008). Activation of the indole-3-Acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20, 228–240. doi: 10.1105/tpc.107.055657
Ding, Y., Gardiner, D. M., Kazan, K. (2022). Transcriptome analysis reveals infection strategies employed by fusarium graminearum as a root pathogen. Microbiol. Res. 256, 126951. doi: 10.1016/j.micres.2021.126951
Ding, Y., Gardiner, D. M., Powell, J. J., Colgrave, M. L., Park, R. F., Kazan, K. (2021). Adaptive defence and sensing responses of host plant roots to fungal pathogen attack revealed by transcriptome and metabolome analyses. Plant Cell Environ. 44, 3756–3774 doi: 10.1111/pce.14195
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173, 1454–1467.e15. doi: 10.1016/j.cell.2018.03.044
Ding, L., Xu, H., Yi, H., Yang, L., Kong, Z., Zhang, L., et al. (2011). Resistance to hemi-biotrophic f. graminearum infection is associated with coordinated and ordered expression of diverse defense signaling pathways. PloS One 6, e19008–e19008. doi: 10.1371/journal.pone.0019008
Domingo, C., Andrés, F., Tharreau, D., Iglesias, D. J., Talón, M. (2009). Constitutive expression of OsGH3. 1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol. Plant-Microbe Interact. 22, 201–210. doi: 10.1094/MPMI-22-2-0201
Frandsen, R. J., Nielsen, N. J., Maolanon, N., Sørensen, J. C., Olsson, S., Nielsen, J., et al. (2006). The biosynthetic pathway for aurofusarin in fusarium graminearum reveals a close link between the naphthoquinones and naphthopyrones. Mol. Microbiol. 61, 1069–1080. doi: 10.1111/j.1365-2958.2006.05295.x
Fu, J., Liu, H., Li, Y., Yu, H., Li, X., Xiao, J., et al. (2011). Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 155, 589–602. doi: 10.1104/pp.110.163774
Gadaleta, A., Colasuonno, P., Giove, S. L., Blanco, A., Giancaspro, A. (2019). Map-based cloning of QFhb. mgb-2A identifies a WAK2 gene responsible for fusarium head blight resistance in wheat. Sci. Rep. 9, 6929. doi: 10.1038/s41598-019-43334-z
Gatti, M., Cambon, F., Tassy, C., Macadre, C., Guerard, F., Langin, T., et al. (2019). The brachypodium distachyon UGT Bradi5gUGT03300 confers type II fusarium head blight resistance in wheat. Plant Pathol. 68, 334–343. doi: 10.1111/ppa.12941
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gordon, S. P., Contreras-Moreira, B., Woods, D. P., Des Marais, D. L., Burgess, D., Shu, S., et al. (2017). Extensive gene content variation in the brachypodium distachyon pan-genome correlates with population structure. Nat. Commun. 8, 1–13. doi: 10.1038/s41467-017-02292-8
Goswami, R. S., Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/j.1364-3703.2004.00252.x
Guenther, J. C., Trail, F. (2005). The development and differentiation of gibberella zeae (anamorph: Fusarium graminearum) during colonization of wheat. Mycologia 97, 229–237. doi: 10.1080/15572536.2006.11832856
Gunupuru, L. R., Arunachalam, C., Malla, K. B., Kahla, A., Perochon, A., Jia, J., et al. (2018). A wheat cytochrome P450 enhances both resistance to deoxynivalenol and grain yield. PloS One 13, e0204992. doi: 10.1371/journal.pone.0204992
Guyon, K., Balagué, C., Roby, D., Raffaele, S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen sclerotinia sclerotiorum. BMC Genomics 15, 1–18. doi: 10.1186/1471-2164-15-336
Haidoulis, J. (2021). Investigating the role of phytohormones in fusarium head blight and fusarium root rot of brachypodium distachyon (University of East Anglia, Norwich).
Haidoulis, J. F., Nicholson, P. (2020). Different effects of phytohormones on fusarium head blight and fusarium root rot resistance in brachypodium distachyon. J. Plant Interact. 15, 335–344. doi: 10.1080/17429145.2020.1820592
Haidoulis, J., Nicholson, P. (2022a). Tissue-specific transcriptome responses to fusarium head blight and fusarium root rot. bioRxiv. doi: 10.1101/2022.04.07.487462
Haidoulis, J., Nicholson, P. (2022b). The transcriptome responses of fusarium head blight and fusarium root rot in B. distachyon (Dryad, Dataset). doi: 10.5061/dryad.cz8w9gj6k
Harris, L. J., Balcerzak, M., Johnston, A., Schneiderman, D., Ouellet, T. (2016). Host-preferential fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 120, 111–123. doi: 10.1016/j.funbio.2015.10.010
He, Z. H., He, D., Kohorn, B. D. (1998). Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14, 55–63. doi: 10.1046/j.1365-313X.1998.00092.x
Howe, K. L., Contreras-Moreira, B., de Silva, N., Maslen, G., Akanni, W., Allen, J., et al. (2020). Ensembl genomes 2020–enabling non-vertebrate genomic research. Nucleic Acids Res. 48, D689–D695. doi: 10.1093/nar/gkz890
Initiative, I. B. (2010). Genome sequencing and analysis of the model grass brachypodium distachyon. Nature 463, 763–768. doi: 10.1038/nature08747
Jain, M., Kaur, N., Garg, R., Thakur, J. K., Tyagi, A. K., Khurana, J. P. (2006a). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 47–59. doi: 10.1007/s10142-005-0005-0
Jain, M., Kaur, N., Tyagi, A. K., Khurana, J. P. (2006). The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics 6, 36–46. doi: 10.1007/s10142-005-0142-5
Jain, M., Tyagi, A. K., Khurana, J. P. (2006b). Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88, 360–371. doi: 10.1016/j.ygeno.2006.04.008
Jansen, M., Slusarenko, A. J., Schaffrath, U. (2006). Competence of roots for race-specific resistance and the induction of acquired resistance against magnaporthe oryzae. Mol. Plant Pathol. 7, 191–195. doi: 10.1111/j.1364-3703.2006.00331.x
Jansen, C., von Wettstein, D., Schäfer, W., Kogel, K.-H., Felk, A., Maier, F. J. (2005). Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted fusarium graminearum. Proc. Natl. Acad. Sci. U. States America 102, 16892–16897. doi: 10.1073/pnas.0508467102
Jia, H., Cho, S., Muehlbauer, G. J. (2009). Transcriptome analysis of a wheat near-isogenic line pair carrying fusarium head blight–resistant and–susceptible alleles. Mol. Plant-Microbe Interact. 22, 1366–1378. doi: 10.1094/MPMI-22-11-1366
Kabbage, M., Yarden, O., Dickman, M. B. (2015). Pathogenic attributes of sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. doi: 10.1016/j.plantsci.2014.12.018
Kakei, Y., Mochida, K., Sakurai, T., Yoshida, T., Shinozaki, K., Shimada, Y. (2015). Transcriptome analysis of hormone-induced gene expression in brachypodium distachyon. Sci. Rep. 5, 14476. doi: 10.1038/srep14476
Kazan, K., Manners, J. M. (2009). Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci. 14, 373–382. doi: 10.1016/j.tplants.2009.04.005
Kikot, G. E., Hours, R. A., Alconada, T. M. (2009). Contribution of cell wall degrading enzymes to pathogenesis of fusarium graminearum: a review. J. Basic Microbiol. 49, 231–241. doi: 10.1002/jobm.200800231
Kim, J.-E., Han, K.-H., Jin, J., Kim, H., Kim, J.-C., Yun, S.-H., et al. (2005). Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in gibberella zeae. Appl. Environ. Microbiol. 71, 1701–1708. doi: 10.1128/AEM.71.4.1701-1708.2005
Kimura, M., Tokai, T., O’Donnell, K., Ward, T. J., Fujimura, M., Hamamoto, H., et al. (2003). The trichothecene biosynthesis gene cluster of fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 539, 105–110. doi: 10.1016/S0014-5793(03)00208-4
Kimura, M., Tokai, T., Takahashi-Ando, N., Ohsato, S., Fujimura, M. (2007). Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol. Biochem. 71, 2105–2123. doi: 10.1271/bbb.70183
King, R., Urban, M., Hammond-Kosack, M. C. U., Hassani-Pak, K., Hammond-Kosack, K. E. (2015). The completed genome sequence of the pathogenic ascomycete fungus fusarium graminearum. BMC Genomics 16, 1–21. doi: 10.1186/s12864-015-1756-1
Kohorn, B. D., Kohorn, S. L. (2012). The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3, 88. doi: 10.3389/fpls.2012.00088
Kõressaar, T., Lepamets, M., Kaplinski, L., Raime, K., Andreson, R., Remm, M. (2018). Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics 34, 1937–1938. doi: 10.1093/bioinformatics/bty036
Koressaar, T., Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kouzai, Y., Kimura, M., Yamanaka, Y., Watanabe, M., Matsui, H., Yamamoto, M., et al. (2016). Expression profiling of marker genes responsive to the defence-associated phytohormones salicylic acid, jasmonic acid and ethylene in brachypodium distachyon. BMC Plant Biol. 16, 1–11. doi: 10.1186/s12870-016-0749-9
Lee, T., Han, Y.-K., Kim, K.-H., yun, S.-H., Lee, Y.-W. (2002). Tri13 and Tri7 determine deoxynivalenol-and nivalenol-producing chemotypes of gibberella zeae. Appl. Environ. Microbiol. 68, 2148–2154. doi: 10.1128/AEM.68.5.2148-2154.2002
Lewandowski, S. M., Bushnell, W. R., Evans, C. K. (2006). Distribution of mycelial colonies and lesions in field-grown barley inoculated with fusarium graminearum. Phytopathology 96, 567–581. doi: 10.1094/PHYTO-96-0567
Liu, P.-L., Du, L., Huang, Y., Gao, S.-M., Yu, M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17, 47. doi: 10.1186/s12862-017-0891-5
Li, G., Yen, Y. (2008). Jasmonate and ethylene signaling pathway may mediate fusarium head blight resistance in wheat. Crop Sci. 48, 1888–1896. doi: 10.2135/cropsci2008.02.0097
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Luo, K., Rocheleau, H., Qi, P. F., Zheng, Y. L., Zhao, H. Y., Ouellet, T. (2016). Indole-3-acetic acid in fusarium graminearum: Identification of biosynthetic pathways and characterization of physiological effects. Fungal Biol. 120, 1135–1145. doi: 10.1016/j.funbio.2016.06.002
Lyons, R., Stiller, J., Powell, J., Rusu, A., Manners, J. M., Kazan, K. (2015). Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in arabidopsis thaliana. PloS One 10, e0121902. doi: 10.1371/journal.pone.0121902
Lysøe, E., Seong, K.-Y., Kistler, H. C. (2011). The transcriptome of fusarium graminearum during the infection of wheat. Mol. Plant-Microbe Interact. 24, 995–1000. doi: 10.1094/MPMI-02-11-0038
Maier, F. J., Miedaner, T., Hadeler, B., Felk, A., Salomon, S., Lemmens, M., et al. (2006). Involvement of trichothecenes in fusarioses of wheat, barley and maize evaluated by gene disruption of the trichodiene synthase (Tri5) gene in three field isolates of different chemotype and virulence. Mol. Plant Pathol. 7, 449–461. doi: 10.1111/j.1364-3703.2006.00351.x
Makandar, R., Nalam, V. J., Lee, H., Trick, H. N., Dong, Y., Shah, J. (2011). Salicylic acid regulates basal resistance to fusarium head blight in wheat. Mol. Plant-Microbe Interact. 25, 431–439. doi: 10.1094/MPMI-09-11-0232
Malz, S., Grell, M. N., Thrane, C., Maier, F. J., Rosager, P., Felk, A., et al. (2005). Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the fusarium graminearum species complex. Fungal Genet. Biol. 42, 420–433. doi: 10.1016/j.fgb.2005.01.010
Mandalà, G., Tundo, S., Francesconi, S., Gevi, F., Zolla, L., Ceoloni, C., et al. (2019). Deoxynivalenol detoxification in transgenic wheat confers resistance to fusarium head blight and crown rot diseases. Mol. Plant-Microbe Interact. 32, 583–592. doi: 10.1094/MPMI-06-18-0155-R
McGrath, K. C., Dombrecht, B., Manners, J. M., Schenk, P. M., Edgar, C. I., Maclean, D. J., et al. (2005). Repressor-and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. doi: 10.1104/pp.105.068544
Mergoum, M., Hill, J., Quick, J. (1998). Evaluation of resistance of winter wheat to fusarium acuminatum by inoculation of seedling roots with single, germinated macroconidia. Plant Dis. 82, 300–302. doi: 10.1094/PDIS.1998.82.3.300
Mi, H., Ebert, D., Muruganujan, A., Mills, C., Albou, L.-P., Mushayamaha, T., et al. (2021). PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394–D403. doi: 10.1093/nar/gkaa1106
Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mills, C., Guo, T., et al. (2019). Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v. 14.0). Nature protocols 14:703–721. doi: 10.1038/s41596-019-0128-8
Mi, H., Thomas, P. (2009). PANTHER Pathway: An Ontology-Based Pathway Database Coupled with Data Analysis Tools. Protein Networks and Pathway Analysis. Methods in Molecular Biology. Nikolsky, Y., Bryant, J. Eds. Humana Press 563, 123–140. doi: 10.1007/978-1-60761-175-2_7
Miedaner, T. (1997). Breeding wheat and rye for resistance to fusarium diseases. Plant Breed. 116, 201–220. doi: 10.1111/j.1439-0523.1997.tb00985.x
Mi, H., Muruganujan, A., Ebert, D., Huang, X., Thomas, P. D. (2019). PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426. doi: 10.1093/nar/gky1038
Oliveros, J. (2018) An interactive tool for comparing lists with venn’s diagrams, (2007–2015). Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html (Accessed Accessed: 2018-2021).
Pan, Y. L., Liu, Z. Y., Rocheleau, H., Fauteux, F., Wang, Y. L., Mccartney, C., et al. (2018). Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genomics 19, 1–26. doi: 10.1186/s12864-018-5012-3
Parry, D. W., Jenkinson, P., Mcleod, L. (1995). Fusarium ear blight (scab) in small grain cereals–a review. Plant Pathol. 44, 207–238. doi: 10.1111/j.1365-3059.1995.tb02773.x
Pasquet, J. C., Changenet, V., Macadre, C., Boex-Fontvieille, E., Soulhat, C., Bouchabke-Coussa, O., et al. (2016). A brachypodium UDP-glycosyltransferase confers root tolerance to deoxynivalenol and resistance to fusarium infection. Plant Physiol. 172, 559–574. doi: 10.1104/pp.16.00371
Pasquet, J. C., Chaouch, S., Macadre, C., Balzergue, S., Huguet, S., Martin-Magniette, M. L., et al. (2014). Differential gene expression and metabolomic analyses of brachypodium distachyon infected by deoxynivalenol producing and non-producing strains of fusarium graminearum. BMC Genomics 15, 1–17. doi: 10.1186/1471-2164-15-629
Payros, D., Alassane-Kpembi, I., Pierron, A., Loiseau, N., Pinton, P., Oswald, I. P. (2016). Toxicology of deoxynivalenol and its acetylated and modified forms. Arch. Toxicol. 90, 2931–2957. doi: 10.1007/s00204-016-1826-4
Peraldi, A., Beccari, G., Steed, A., Nicholson, P. (2011). Brachypodium distachyon: a new pathosystem to study fusarium head blight and other fusarium diseases of wheat. BMC Plant Biol. 11, 1–14. doi: 10.1186/1471-2229-11-100
Petti, C., Reiber, K., Ali, S. S., Berney, M., Doohan, F. M. (2012). Auxin as a player in the biocontrol of fusarium head blight disease of barley and its potential as a disease control agent. BMC Plant Biol. 12, 1–9. doi: 10.1186/1471-2229-12-224
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45–e45. doi: 10.1093/nar/29.9.e45
Pieterse, C. M. J., Does, D. V. D., Zamioudis, C., Leon-Reyes, A., Wees, S. C. M. V. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Poppenberger, B., Berthiller, F., Lucyshyn, D., Sieberer, T., Schuhmacher, R., Krska, R., et al. (2003). Detoxification of the fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from arabidopsis thaliana. J. Biol. Chem. 278, 47905–47914. doi: 10.1074/jbc.M307552200
Powell, J. J., Carere, J., Fitzgerald, T. L., Stiller, J., Covarelli, L., Xu, Q., et al. (2017a). The fusarium crown rot pathogen fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum l.). Ann. Bot. 119, 853–867. doi: 10.1093/aob/mcw207
Powell, J. J., Carere, J., Sablok, G., Fitzgerald, T. L., Stiller, J., Colgrave, M. L., et al. (2017b). Transcriptome analysis of brachypodium during fungal pathogen infection reveals both shared and distinct defense responses with wheat. Sci. Rep. 7, 17212. doi: 10.1038/s41598-017-17454-3
Pritsch, C., Muehlbauer, G. J., Bushnell, W. R., Somers, D. A., Vance, C. P. (2000). Fungal development and induction of defense response genes during early infection of wheat spikes by fusarium graminearum. Mol. Plant-Microbe Interact. 13, 159–169. doi: 10.1094/MPMI.2000.13.2.159
Qi, P.-F., Jiang, Y.-F., Guo, Z.-R., Chen, Q., Ouellet, T., Zong, L.-J., et al. (2019). Transcriptional reference map of hormone responses in wheat spikes. BMC Genomics 20, 390. doi: 10.1186/s12864-019-5726-x
Rameneni, J. J., Lee, Y., Dhandapani, V., Yu, X., Choi, S. R., Oh, M.-H., et al. (2015). Genomic and post-translational modification analysis of leucine-rich-repeat receptor-like kinases in brassica rapa. PloS One 10, e0142255. doi: 10.1371/journal.pone.0142255
Ruijter, J., Ramakers, C., Hoogaars, W., Karlen, Y., Bakker, O., van den Hoff, M., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45–e45. doi: 10.1093/nar/gkp045
Sayers, E. W., Beck, J., Brister, J. R., Bolton, E. E., Canese, K., Comeau, D. C., et al. (2020). Database resources of the national center for biotechnology information. Nucleic Acids Res. 48, D9. doi: 10.1093/nar/gkz899
Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T., Maldonado, M. C., et al. (2005). Characterization of an arabidopsis enzyme family that conjugates amino acids to indole-3-Acetic acid. Plant Cell 17, 616–627. doi: 10.1105/tpc.104.026690
Stintzi, A., Heitz, T., Prasad, V., Wiedemann-Merdinoglu, S., Kauffmann, S., Geoffroy, P., et al. (1993). Plant ‘pathogenesis-related’proteins and their role in defense against pathogens. Biochimie 75, 687–706. doi: 10.1016/0300-9084(93)90100-7
Strugala, R., Delventhal, R., Schaffrath, U. (2015). An organ-specific view on non-host resistance. Front. Plant Sci. 6, 526. doi: 10.3389/fpls.2015.00526
Sun, Y., Xiao, J., Jia, X., Ke, P., He, L., Cao, A., et al. (2016). The role of wheat jasmonic acid and ethylene pathways in response to fusarium graminearum infection. Plant Growth Regul. 80, 69–77. doi: 10.1007/s10725-016-0147-1
Supek, F., Bošnjak, M., Škunca, N., Šmuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PloS One 6, e21800. doi: 10.1371/journal.pone.0021800
Su, P., Zhao, L., Li, W., Zhao, J., Yan, J., Ma, X., et al. (2020). Integrated metabolo-transcriptomics and functional characterization reveals that the wheat auxin receptor TIR1 negatively regulates defense against fusarium graminearum. J. Integr. Plant Biol. 63, 340–352. doi: 10.1111/jipb.12992
The Gene Ontology Consortium (2021). The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 49 (D1), D325–D334. doi: 10.1093/nar/gkaa1113
Thomas, P. D., Ebert, D., Muruganujan, A., Mushayahama, T., Albou, LP., Mi, H. P., et al. (2022). PANTHER: Making genome‐scale phylogenetics accessible to all. Protein Science. 31, 8–22. doi: 10.1002/pro.4218
Tsai, Y.-C., Weir, N. R., Hill, K., Zhang, W., Kim, H. J., Shiu, S.-H., et al. (2012). Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158, 1666–1684. doi: 10.1104/pp.111.192765
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115–e115. doi: 10.1093/nar/gks596
Walter, S., Kahla, A., Arunachalam, C., Perochon, A., Khan, M. R., Scofield, S. R. (2015). A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J Exp Bot. 66, 2583–2593. doi: 10.1093/jxb/erv048
Walton, J. D. (1994). Deconstructing the cell wall. Plant Physiol. 104, 1113–1118. doi: 10.1104/pp.104.4.1113
Wang, X., Jiang, N., Liu, J., Liu, W., Wang, G.-L. (2014). The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence 5, 722–732. doi: 10.4161/viru.29798
Wang, L., Li, Q., Liu, Z., Surendra, A., Pan, Y., Li, Y., et al. (2018). Integrated transcriptome and hormone profiling highlight the role of multiple phytohormone pathways in wheat resistance against fusarium head blight. PloS One 13, e0207036. doi: 10.1371/journal.pone.0207036
Wang, L. P., Li, Q., Liu, Z. Y., Surendra, A., Pan, Y. L., Li, Y. F., et al. (2018). Integrated transcriptome and hormone profiling highlight the role of multiple phytohormone pathways in wheat resistance against fusarium head blight. PloS One 13, 24. doi: 10.1371/journal.pone.0207036
Wang, Q., Shao, B., Shaikh, F. I., Friedt, W., Gottwald, S. (2018). Wheat resistances to fusarium root rot and head blight are both associated with deoxynivalenol- and jasmonate-related gene expression. Phytopathology 108, 602–616. doi: 10.1094/PHYTO-05-17-0172-R
Wang, Q., Vera Buxa, S., Furch, A., Friedt, W., Gottwald, S. (2015). Insights into triticum aestivum seedling root rot caused by fusarium graminearum. Mol. Plant-Microbe Interact. 28, 1288–1303. doi: 10.1094/MPMI-07-15-0144-R
Wu, J., Luo, J., Wei, L., Cai, T., Mao, X. (2006). KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, W720–W724. doi: 10.1093/nar/gkl167
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Yazaki, J., Shimatani, Z., Hashimoto, A., Nagata, Y., Fujii, F., Kojima, K., et al. (2004). Transcriptional profiling of genes responsive to abscisic acid and gibberellin in rice: phenotyping and comparative analysis between rice and arabidopsis. Physiol. Genomics 17, 87–100. doi: 10.1152/physiolgenomics.00201.2003
Keywords: fusarium root rot, fusarium head blight, Brachypodium distachyon, Fusarium graminearum, phytohormones, RNA-seq, RT-qPCR, transcriptome
Citation: Haidoulis JF and Nicholson P (2022) Tissue-specific transcriptome responses to Fusarium head blight and Fusarium root rot. Front. Plant Sci. 13:1025161. doi: 10.3389/fpls.2022.1025161
Received: 22 August 2022; Accepted: 29 September 2022;
Published: 24 October 2022.
Edited by:
Shaobin Zhong, North Dakota State University, United StatesReviewed by:
Marie Dufresne, Université Paris-Saclay, FranceMichele Christine Loewen, National Research Council Canada (NRC-CNRC), Canada
Copyright © 2022 Haidoulis and Nicholson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul Nicholson, Paul.Nicholson@jic.ac.uk
 John Francis Haidoulis
John Francis Haidoulis Paul Nicholson
Paul Nicholson